Abstract
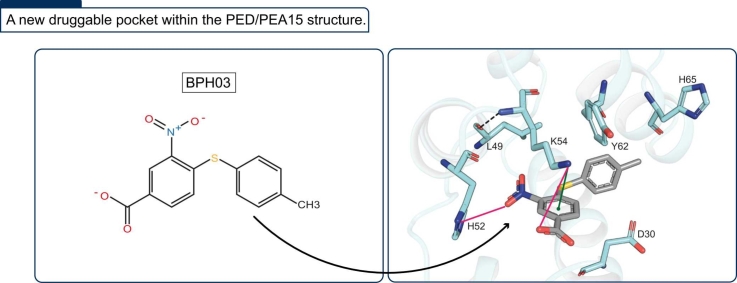
In a recent study, we have identified BPH03 as a promising scaffold for the development of compounds aimed at modulating the interaction between PED/PEA15 (Phosphoprotein Enriched in Diabetes/Phosphoprotein Enriched in Astrocytes 15) and PLD1 (phospholipase D1), with potential applications in type II diabetes therapy. PED/PEA15 is known to be overexpressed in certain forms of diabetes, where it binds to PLD1, thereby reducing insulin-stimulated glucose transport. The inhibition of this interaction reestablishes basal glucose transport, indicating PED as a potential target of ligands capable to recover glucose tolerance and insulin sensitivity. In this study, we employ computational methods to provide a detailed description of BPH03 interaction with PED, evidencing the presence of a hidden druggable pocket within its PLD1 binding surface. We also elucidate the conformational changes that occur during PED interaction with BPH03. Moreover, we report new NMR data supporting the in-silico findings and indicating that BPH03 disrupts the PED/PLD1 interface displacing PLD1 from its interaction with PED. Our study represents a significant advancement toward the development of potential therapeutics for the treatment of type II diabetes.
Keywords: Phosphoprotein Enriched in Diabetes/Phosphoprotein Enriched in Astrocytes 15, Type II diabetes, Molecular Dynamics, Cavity mapping, Nuclear Magnetic Resonance
Graphical abstract
1. Introduction
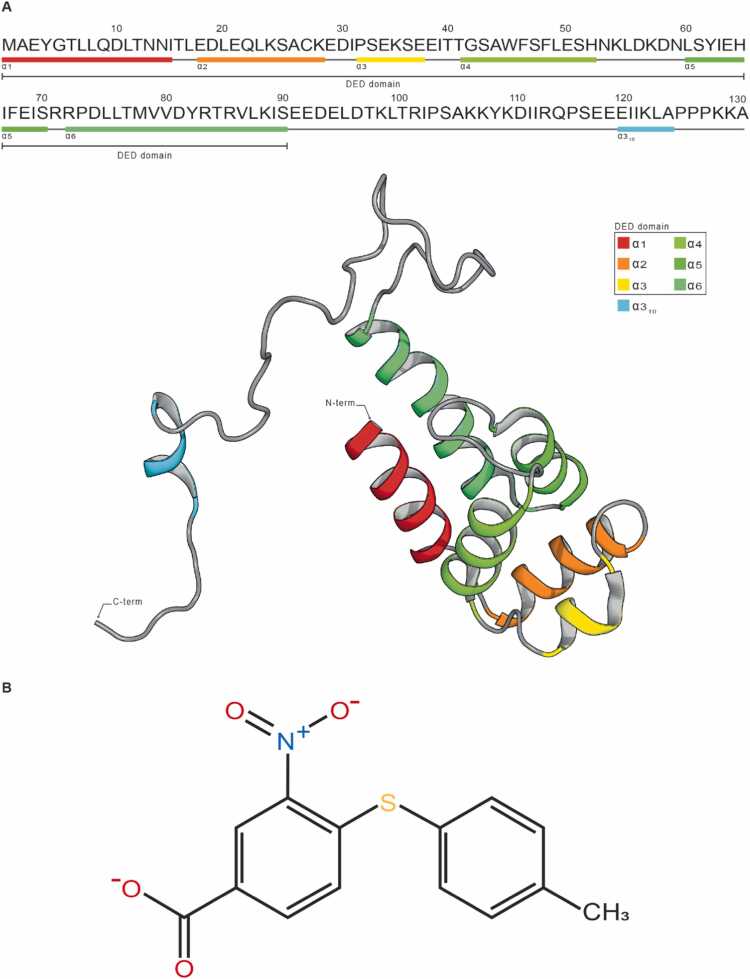
PED/PEA15 (hereinafter referred to as PED) is the Phosphoprotein Enriched in Diabetes/Phosphoprotein Enriched in Astrocytes 15 protein. It is a ∼ 15 kDa cytosolic protein expressed in human cells that is largely conserved in mammals [1], [2], [3], [4]. It is involved in numerous protein-protein interactions controlling the function of several effectors of key cellular functions, including glucose metabolism, proliferation, and apoptosis [1], [5], [6], [2], [7], [4], [8], [9], [10], [11], [12], [13], [14]. The NMR structure of PED reveals a canonic N-terminal Death Effector Domain (DED) composed of six amphipathic α-helices and a 40 residues C-terminal tail partially disordered [15], [11], [16], [17] (Fig. 1A). PED is commonly overexpressed in type 2 diabetes (T2D) as well as in several T2D-associated comorbidities, including cancer and some neurodegenerative disorders [4], [19], [20], [18]. Consequently, targeting PED presents potential therapeutic opportunities for T2D. PED is known to interact with PLD1 and PLD2, the phospholipase D isoforms.
Fig. 1.
PED and BPH03 structures. (A) Schematic representation of PED primary and secondary structure (Upper) and PED NMR structure (PDB ID: 1N3K (1N3K[15])) (Lower). The 3D structure of the protein consists of a six-helices bundle N-terminal of 90 residues, a long poorly structured C-terminal tail (40 residues) hosting a small 310 helix; the N-terminal globular region constitutes the DED domain (Death Effector Domain) that includes the 6 α-helices (α1–α6). (B) 2D structure of BPH03.
In cultured muscle, adipose cells and peripheral tissues of transgenic mice, high levels of PED increase the classical protein kinase C (PKC) isoform PKC-α [21] and influence negatively insulin-stimulated GLUT4 translocation and glucose transport, suggesting that overexpression of PED may contribute to insulin resistance in T2D [22]. Therefore, inhibiting PED and modulating its interaction with PLD1 can restore proper glucose transport identifying PED/PLD1 complex as a target for the development of new antidiabetics drugs [23], [24], [25], [26]. The interaction between PED and PLD1 involves the Death Effector Domain of PED as well as specific segments of the PED C-terminal tail [23], [24], [20], [25], [27]. On the PLD1 side, a C-terminal segment from residue 712 to 1074 (referred to as D4) has been demonstrated to interact with PED [23]. Inhibition of the PED/D4 interaction restores basal glucose transport in skeletal muscle cells with elevated levels of PED [25], [28], [27]. Hence, the PED/PLD1 interaction appears to be predominantly regulated by a limited number of interactions within the N-terminus of D4 [24], [25], [28], [27]. Indeed, D4α, a segment of D4 comprising residues 712–818, has been shown to have equivalent affinity for PED and to function similarly to D4 in cells overexpressing PED [23]. D4α has been also exploited to unveil the molecular determinants of PED/D4 interaction by solution NMR [24]. The binding interface between PED and D4α in phosphate buffer, as well as the conformational alterations in PED resulting from its interaction with D4, were delineated and subsequently validated in cellular lysates from HEK 293 cells expressing PLD1. These structural insights have facilitated the screening of a library of small molecules, leading to the identification of the 4-[(4-Methylphenyl)thio]-3-nitrobenzoic acid, a compound named BPH03 (Fig. 1B), as a promising candidate for the development of modulators capable of directly interfering with the binding interface between PED and PLD1 [29]. As the interference of protein-protein interactions with a small molecule is expected to be more successful in presence of compactly clustered interaction hot spots [30] we here report the detailed description of BPH03 interaction with PED evidencing, within its D4 binding surface, the presence of a hidden druggable pocket as well as the conformational changes taking place during PED/BPH03 interaction. These results were achieved using a combined experimental and in-silico approach. Starting from the recently conducted experimental screening of a molecular fragments library by NMR [29], molecular modeling techniques (cavity mapping, molecular docking, and molecular dynamics) were employed in continuous interplay with NMR experiments conducted to support the in-silico results. Given the overexpression of PED in various tissues of individuals with T2D [19], [27] all the obtained results are discussed in the perspective of being used as a starting point for rationally designing and developing the first inhibitors of the PED/PLD1 interaction for the treatment of T2D.
2. Materials and methods
2.1. Computational details
2.1.1. Docking studies
BPH03 was docked on the recently published X-ray structure of dually phosphorylated PED/PEA-15 in complex with ERK2 (resolution 1.93 Å – PDB code: 4IZA [11]). The retrieved .pdb file was prepared using the Protein Preparation Wizard tool, available from the Schrodinger Suite 2022-4 [31] for adding missing hydrogens atoms, reconstructing incomplete side chains, assigning favorable protonation states at physiological pH and performing a force field-based minimization of the 3D protein structures. The ligand was prepared using the LigPrep tool for generating all the possible ionization states and tautomers at a pH value of 7.0 ± 2.0 [32]. To this aim the OPLS4 force field was used [33]. The obtained file was employed for docking simulations performed by Grid-based ligand docking with energetics (GLIDE) [34], [35], [36], [37], [38]. In order to properly take into account putative conformational rearrangements of the protein binding site during molecular recognition, Induced Fit Docking (IFD) [39], [40] simulations were performed using the SP mode (all the used parameters are available in Table S1 - Supporting information) and all the default settings, building a cubic grid centered on the center of mass of H52 (predicted to be protonated during the protein preparation step) and H65 (predicted to be neutral and with the δ-N atom bound to an hydrogen atom), namely the residues mostly affected by the interaction with BPH03 as indicated by NMR data [29]. Furthermore, an inner box of 10 Å × 10 Å × 10 Å and an outer box of 30 Å × 30 Å × 30 Å were employed. 16 poses were generated and then subjected to molecular mechanics/generalized Born surface area calculations (MM-GBSA), one of the most widely used computational approaches for estimating the binding free energy of ligands to biological macromolecules [41]. Noteworthy, several studies showed that MM-GBSA scores exhibits a stronger correlation with experimental binding affinities compared to docking scores [42]. Furthermore, we used the OPLS4 force field, recently proved to provide, with respect to other force fields, more accurate predictions of binding affinities [43], [33]. Notice that during the IFD simulations flexibility was allowed for the residues D30, K54 and Y62. A thorough analysis of the crystal structure, in fact, revealed that their side chains protrude towards the hypothesized cavity, suggesting that they may potentially obstruct BPH03 binding. All generated poses, along with their respective docking and MM-GBSA scores (Table S2), are provided in the Supplementary material. It is worth noting that the substantial discrepancies between the two types of scores can be attributed to the fact that MM-GBSA score incorporates contributions from the implicit solvent Generalized Born model and solvent-accessible surface area, unlike the docking score. This integration potentially provides a more accurate representation of the physical processes involved in ligand binding.
2.1.2. Model system preparation
The PED-apo form and the PED-BPH03 complex returned by IFD simulation were subjected to Molecular Dynamics (MD) simulations to get more insights into BPH03 molecular recognition and to shed light into the protein conformational rearrangement taking place upon ligand binding. Using the system builder tool, available from the Schrodinger suite [44] the complexes were fully solvated into a minimized, orthorhombic TIP3P water-box. The simulation box was generated by using “Buffer” as method for calculating the box size. More specifically, we ensured that each protein atom maintained a distance larger than 10 Å from the box boundary, resulting in dimensions of 74.28 Å × 49.21 Å × 50.24 Å. Na+ and Cl- ions were added generating a 150 mM ionic concentration. In doing that, we obtained a neutral system including 17,545 atoms (PED-apo form) and 16,776 atoms (PED-BPH03). The default OLPS4 force field [43], [33] was used for both protein and ligand.
2.1.3. MD simulation protocol
MD simulations were performed on a GPU by using Desmond 7.2, implemented in the Schrodinger Suite 2022-4, as software [45], [44]. A non-bonded cut-off of 9 Å was used. All the prepared systems were minimized, equilibrated, and simulated using an isothermal isobaric ensemble (NPT, P = 1 atm, T = 300 K) with a Nosè–Hoover thermostat [46], [47] with a relaxation time of 1 ps and a Martyna-Tobias-Klein barostat [48] with a relaxion time of 2 ps. For each system, we performed 200 ns-long MD simulations replicas using a time step equal to 2 fs and storing the coordinates with a recording interval of 100 ps. In doing that, 2001 frames were generated and analyzed for each system.
2.1.4. MD simulation analysis
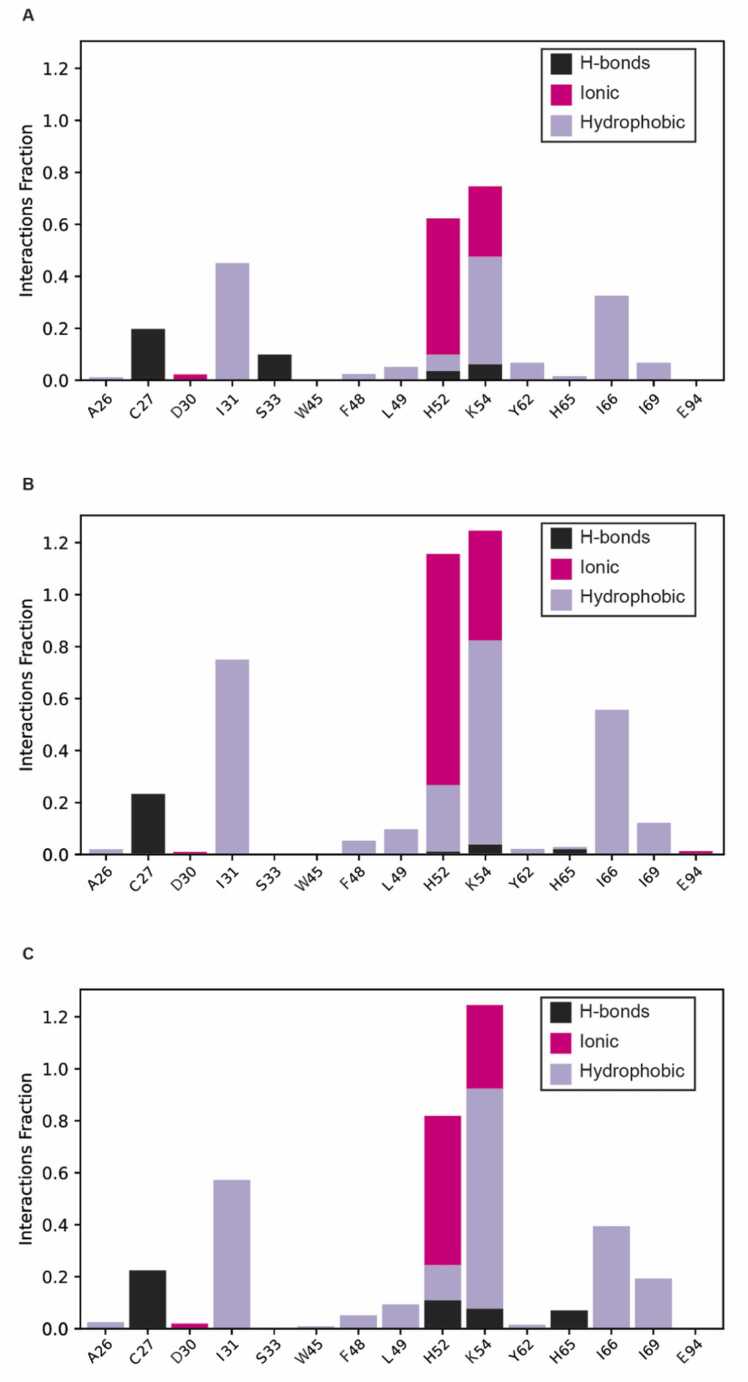
The obtained trajectories were analyzed using the trajectory player available in the Schrodinger Suite 2022-4. More specifically, the Root Mean Square Deviation (RMSD) of the alpha carbon atoms of the protein were computed using the ‘Compute Properties Over Trajectory’ tool. To better compare and measure similarity between the trajectories, the probability distribution was estimated for the computed RMSD. All the simulations were subjected to a trajectory clustering analysis, performed using the Desmond Trajectory Clustering tool [45], [49], available in the Schrodinger suite 2022-4, and a sampling structure frequency value of 10 (corresponding to 1 ns). Notice that this tool employs the 'Affinity Propagation Clustering' method reported by Brendan et al. [49]. 14 clusters of varying sizes were identified by the algorithm but only the five most populated ones were outputted, each containing the most representative frame of the respective clusters. More specifically, through an RMSD matrix of the sampled frames, the tool generates clusters with varying numbers of elements (sampled frames) that are sampled for their more likeness based on the RMSD valued among them. Subsequently all the sampled frames within the same cluster are averaged to obtain a unique frame, assumed to be the most representative frame of all the cluster. Finally, the representative structure of the cluster with the highest abundance of sampled frames was considered the most representative frame of the entire MD run simulation. Protein-ligand interactions for the PED-BPH03 complex were plotted in bar-plots using data from the Simulation Interaction Analysis (SID) tool, available from the Schrodinger Suite 2022-4. Interactions plotted refers to H-bonds, ionic, and hydrophobic interactions. Notice that the following criteria were adopted:
Hydrogen Bonds: a maximum distance of 2.5 Å between the donor and acceptor atoms (D—H···A); a donor angle of ≥ 120° between the donor-hydrogen-acceptor atoms (D—H···A); and an acceptor angle of ≥ 90° between the hydrogen-acceptor-bonded atoms (H···A—X).
Hydrophobic contacts: i) π-Cation - Aromatic and charged groups within 4.5 Å; ii) π-π - Two aromatic groups stacked face-to-face or face-to-edge; iii) other - a non-specific hydrophobic sidechain within 3.6 Å of a ligand's aromatic or aliphatic carbons.
Ionic/Salt bridge interactions: Distance between two oppositely charged atoms within 3.7 Å.
To better assess ligand complementary in the putative cavity binding site and validate the cavity analysed in all the performed simulations, the SiteMap [50], [51] tool, available from the Schrodinger Suite 2022-4, was used. This tool allowed us to highlight the regions within the binding site suitable for occupancy by hydrophobic groups or by ligand hydrogen-bond donors, acceptors, or metal-binding functionality. SiteMap returns two scores for each predicted cavity: (i) SiteScore, based on the cavity size, degree of enclosure, and hydrophobicity, and (ii) DScore, for assessing cavity druggability. Notice that the SiteScore has been constructed and calibrated based on 157 investigated submicromolar sites. In particular, a SiteScore greater than 0.80 has been found to reliably identify drug-binding sites compared to non-drug-binding one [50], [51].
2.2. NMR studies
15N-Labeled PED was expressed in Escherichia coli and purified as previously reported [28]. The In-cell NMR analysis of the protein-small molecules interactions [53], [52] were conducted on cell lysates prepared as reported in Farina et al. [29].
2.2.1. NMR samples
250 μM stock solutions of 15N-labeled PED were prepared in 20 mM sodium phosphate buffer (pH = 7.4). NMR samples (pH 7.4) contained HEK 293 cells lysates not expressing PLD1, 15N-labeled PED at 60 μM concentration, 0.02 % sodium azide. As previously reported [29] BPH03, dissolved in DMSO-d6 at 100 mM final concentration (stock solution), was added in small aliquots to the NMR samples attaining a final DMSO-d6 concentration of 5 %.
2.2.2. NMR spectroscopy
The 2D [1H, 15N] HSQC experiments were all acquired on a Bruker AVANCE III HD 600 MHz spectrometer equipped with a triple resonance Prodigy N2 cryoprobe having a z-axis pulse field gradient. Temperature was set at 298 K. Spectra (Fig. S1) were processed by means of TopSpin 4.1.0 software (Bruker) and analyzed with CARA [54] and SPARKY [55] software. PyMOL 2.5 [56] was used to visualize the protein structures. PED structural perturbations upon BPH03 addition were estimated by combining 1H (∆δH) and 15N (∆δN) Chemical Shift Perturbations (CSPs) as follows:
| ∆δHN/N = ((∆δHWH)2 + ((∆δNWN)2)1/2 |
in which WH and WN are the weighing factors for 1H and 15N shifts defined as WH = |γH/γH| = 1; and WN = |γN/γH| = 0.101. ∆δH and ∆δN are the chemical shift differences in ppm for 1H and 15N, respectively; γH and γN are the gyromagnetic ratios of the different nuclei [57].
3. Results and discussion
3.1. Induced Fit Docking (IFD) simulations suggest that protein conformational changes are required for PED-BPH03 binding
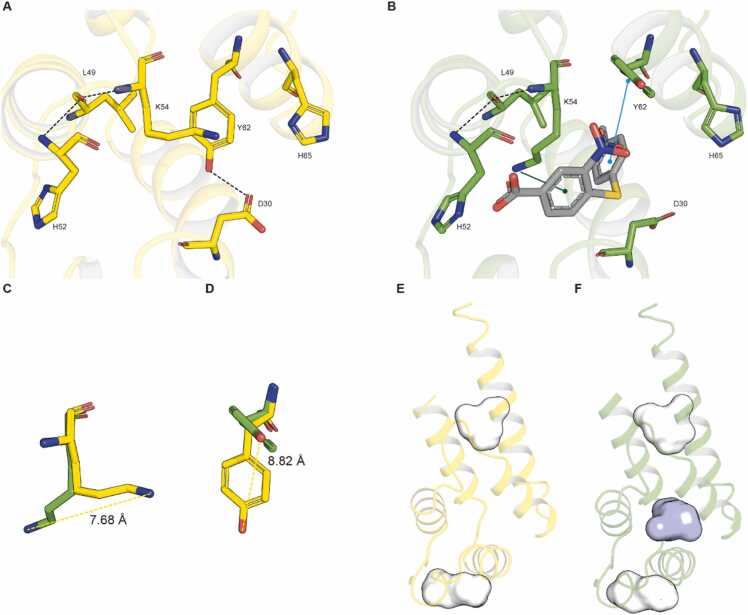
As a preliminary step, we conducted standard docking simulations of BPH03 within a cavity centered on H52 and H65. Consistently with the performed SiteMap predictions, indicating the absence of a druggable cavity near these residues, this simulation produced no poses. A closer examination of the crystal structure revealed that the conformation of D30, K54, and Y62 might obstruct BPH03 binding due to their side chains protruding towards the cavity (Fig. 1, Fig. 2). Based on these findings, we proceeded with an Induced Fit Docking calculation within the same cavity used for standard docking simulations, allowing full flexibility for the aforementioned three residues. 16 poses were generated, and the one with the best MM-GBSA score was selected. Notably, this protocol enabled the binding of the BPH03 ligand to the PED protein. A comparison between the crystal structure and the ligand-protein complex revealed a disrupted bond between D30 and Y62 (Fig. 2A and B), present in the crystal structure, allowing a conformational rearrangement of both K54 and Y62 sidechains (Fig. 2C and D). The top-scored pose yielded a MM-GBSA score of − 40.17 kcal/mol (docking score = − 5.753 kcal/mol) and underwent MD simulations. Remarkably, the conformational rearrangement induced by the IFD calculation led to the identification of a cavity in the proximity of H52 and H65 (absent in the starting crystal structure) (Fig. 2E and F). The significance of this cavity as a putative druggable pocket is supported by the promising SiteScore (0.749) and Dscore (0.746) returned [51].
Fig. 2.
Comparison between the PED-apo form and the top-scored docking pose of PED-BPH03 complex resulted from the applied Induced Fit Docking protocol. Hypothesized binding pocket for BPH03 in the PED-apo form (PDB ID: 4IZA[11]) and in the PED-BPH03 complex resulted from the performed Induced Fit Docking simulations are reported in yellow and green sticks, respectively (A, B). Notice that BPH03 is reported in grey sticks. Notice that the interactions are reported using this scheme: H-bonds in black-dotted line, π-stacking interactions in green line and π- π interactions in cyan line. The comparison between the two structures highlights the shift of two key residues to enable the ligand binding, K54 (C), and Y62 (D), respectively in the same above-mentioned colors scheme. SiteMap output returned by PED-apo form (PDB ID 4IZA[11]) (E) and PED-BPH03 complex returned by the performed IFD simulations (F). The pocket identified by NMR data and returned by PED-BPH03 complex only is shown as cyan surface.
3.2. Molecular dynamics simulations indicate that PED might be stabilized by BPH03
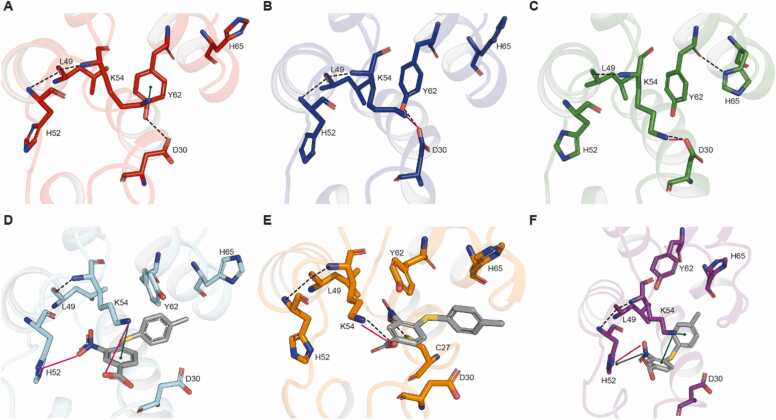
Encouraged by the obtained IFD data, we performed MD simulations of both PED-BPH03 complex and PED-apo form. The latter was used as reference for investigating the effect of BPH03 on the cavity conformation. Three 200-ns long trajectories were generated for each investigated system as replicates (hereinafter referred to as MD1-apo, MD2-apo, MD3-apo, MD1-com, MD2-com and MD3-com). Notice that the first 30 ns were removed from the analysis as necessary for model equilibration, in agreement with a previous study (see Fig. S2 in the Supporting information) [58]. More specifically, we performed a cluster analysis of the equilibrated MD trajectories (from 30 to 200 ns) to select the most representative frame for visual inspection and cavity mapping calculations. The visual inspection of the trajectories showed that BPH03 establishes salt-bridge interactions with H52 (found in 49.2 % of the frames belonging to MD1-com and 73.9% of those in MD3-com), and K54 (37.5 % of MD1-com, 32.3 % of MD2-com), hydrophobic and π-stacking interactions with H52 (17.5 % of MD3-com), and K54 (47.4 % of MD1-com, 60.1% of MD2-com), H-bond with C27 (17.6 % of MD2-com), and K54 (32.3 % of MD2-com) (Fig. 3). The interaction fractions calculated throughout the entire PED-BPH03 MD trajectories (Fig. 4) indicate that the predominant protein-ligand interactions are hydrophobic, with H52 (58.8 % of MD1-com, 88.3 % of MD2-com, 91.6 % of MD3-com), and K54 (94.2 % of MD1-com, 95.1 % of MD2-com, 96.5 % of MD3-com) being the most interacting residues. Notice that the interaction fraction indicates the duration for which a specific interaction persists throughout the trajectory snapshots. A fraction of 1.0 denotes that the interaction remains intact for the entire simulation duration, while fractions greater than 1.0 indicate residues capable of forming multiple contacts with the compound. The obtained data provide insights into a potential molecular recognition mechanism that aligns with the NMR solution experiments conducted by Farina et al. [29]. More intriguingly, the cavity mapping conducted on the representative frames of the three PED/PEA15-BPH03 MD runs yielded SiteScore and Dscore values always > 0.800 (0.964 and 0.988 for MD1-com; 0.802 and 0.804 for MD2-com, 0.820 and 0.802 for MD3-com, see Table S3 in the Supporting information), which are typical of pockets of interest for performing structure-based approaches (see Fig. S4 in the Supporting information) [51]. In contrast, SiteMap predicts, as mentioned, the considered cavity as non-druggable for the PED crystal structure taken as it is (PDB code: 4IZA [11]). It is noteworthy that the robustness and statistical significance of this data were reinforced by two additional 200 ns replicates for the PED-BPH03 complex (MD4-com and MD5-com), as indicated in the Supporting information (please refer to Table S3, Fig. S2 and Fig. S3 for details). Taken together, these findings strongly suggest the presence of a druggable cavity in the PED-PLD1 interaction region, warranting further characterization in subsequent studies.
Fig. 3.
Representative snapshots taken from the MD trajectories returned by PED-apo and PED-BPH03. Representative snapshots extracted from MD1-apo (A), MD2-apo (B), MD3-apo (C), MD1-com (D), MD2-com (E), MD3-com (F). Notice that BPH03 is reported always in grey stick representation. The interactions are reported using the current scheme: H-bonds in black-dotted line, ionic/salt-bridge interaction in purple line, π-stacking interactions in green line.
Fig. 4.
PED-BPH03 protein-ligand interactions summary. Protein-ligand interactions (or contacts) returned by the performed MD simulations and reported as H-bonds (black bars), Ionic (purple bars), and Hydrophobic (violet bars) interactions: MD1-com (A), MD2-com (B), and MD3-com (C). The stacked bars charts are reported as interactions fraction in function of the protein residues observed to make stable or multiple contacts with the protein. The interaction fraction values are normalized over the course of the trajectory: for example, the K54 in the bar chart referred to MD1 run (A), with a value of 0.8, suggests that the mentioned residue has interactions with the protein for the 80% of the simulation time.
3.3. BPH03 interaction displaces PLD1
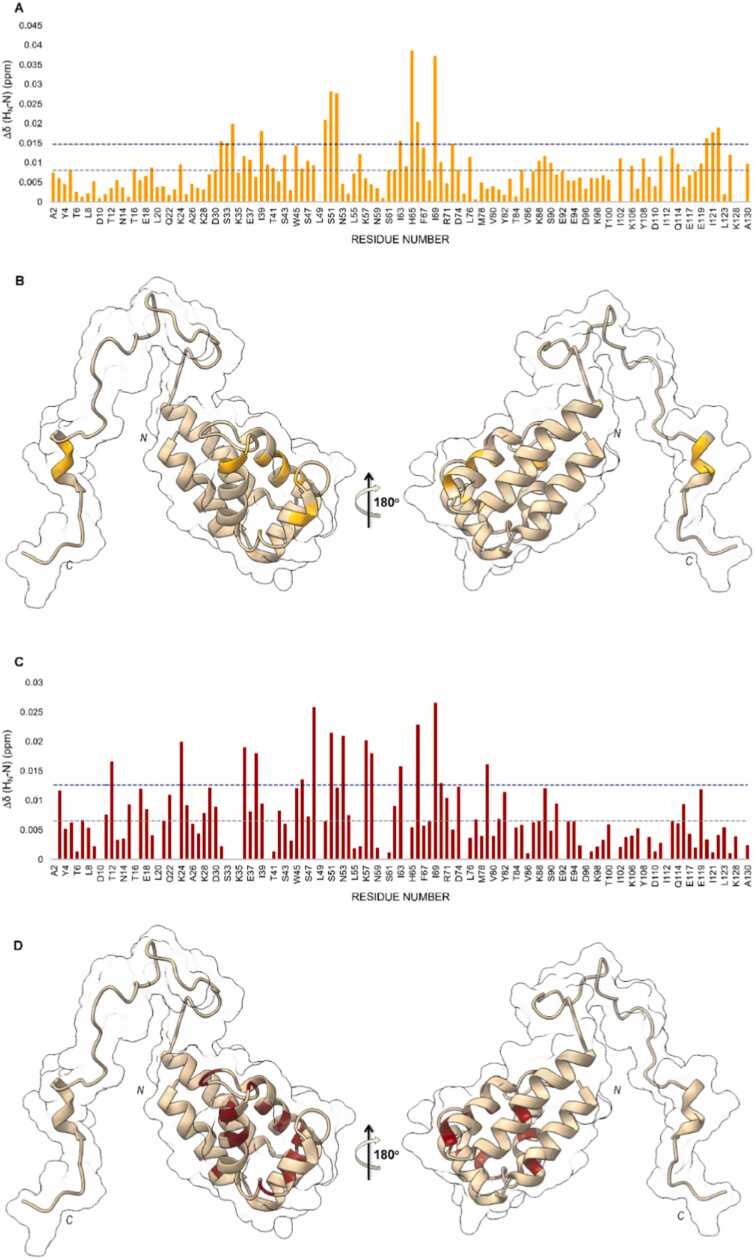
A previous NMR study [29] conducted in presence of two cellular lysates, a reference lysate attained from HEK 293 cells not expressing PLD1 and an interaction lysate obtained from the heterologous expression of PLD1 in the same cells, has led to the identification PED surface involved in the interaction with PLD1. This interaction site, previously identified in vitro by using D4α [23] is mainly constituted by α1, α3, α4 helices, the loops connecting α1 to α2 and α3 to α4 and the N-terminal part of the α4α5 loop. The involvement of the C-terminal poorly structured tail stabilizes the interaction. The same study has also allowed the identification, within a focused library of compounds, of BPH03 as the best scaffold for development of new compounds capable to interfere with PED/PLD1 interaction. BPH03 was found to be able to interact within the PED/PLD1 binding interface. Interestingly, experiments conducted in phosphate buffer outlined PED’s most perturbed residues upon BPH03 addition as exclusively belonging to the N-terminal DED domain of PED. In particular, residues K28 and I31 within the α2α3 loop, S36 within the α3-helix, E37, E38, and I39 in the α3α4 loop, E50 and S51 in the α4-helix, H52 in the α4α5 loop, and the segment spanning from I63 to I69 within the α5-helix were identified as perturbed by the presence of BPH03. The same experiments conducted in the presence of PLD1 (Figs. 5A and B), i.e., in the interaction lysate, while revealing the same interaction surface found within the DED domain in phosphate buffer, also demonstrated the perturbation of PED’s C-terminal tail. This tail is implicated in the interaction of PED with PLD1 in the interaction lysate but not involved in PED/BPH03 interaction in phosphate buffer. PED/BPH03 interaction in the reference lysate remained to be investigated. Thus, Figs. 5C and 5D report the results of these new experiments that indicate that, in these conditions, the C-terminal tail is not involved in BPH03 interaction with PED. Indeed, BPH03 interaction affects exclusively residues located on secondary structures within the DED domain: α1-helix (T12), α2-helix (K24), α3-helix (S36), α3α4loop (E38), α4-helix (F46, F48, S51), α4α5loop (N53, K57, D58), α5-helix (I63, I66, I69, S70), α6-helix (V79). In light of these findings, the chemical shifts perturbations of residues within the C-terminal tail observed in the interaction buffer when PED is in presence of BPH03 require a more accurate molecular explanation. The comparison of the two 1H 15N-HSQC spectra acquired in the reference and in the interaction lysates has allowed the identification of PLD1 binding interface within PED’s structure. This latter includes the DED domain and the C-terminal tail. The addition of BPH03 gives back the same binding surface identified in the phosphate buffer that partially overlaps PED/PLD1 interface. In no other conditions (i.e. phosphate buffer and reference lysate) BPH03 significantly perturbs the C-terminal tail. This strongly suggests that in the tested conditions BPH03, by intruding within PED/PLD1 interface, can displace PLD1 from PED’s interaction site. This displacement in turn leads to the observed chemical shifts perturbations in the interaction buffer involving the C-terminal tail.
Fig. 5.
PED-BPH03 interactions in cellular environment. Chemical Shift Perturbation (CSP) of PED in interacting lysate in presence of BPH03 (A) mapped on PED 3D structure (B) (1N3K[15]; CSP of PED in reference lysate upon addition of BPH03 (C) mapped on PED 3D structure (D). The blue and grey dashed lines indicate the average CSP (CSPavg) and CSPavg + SD (Standard Deviation), respectively.
4. Conclusions
The multifunctional scaffold PED is a small protein that interacts with other partner proteins regulating several cellular functions. In particular, this protein is known to interact in apoptotic processes with FADD and caspase 8 [6], [7] in the ERK/MAPK kinase cascade with ERK1/2 [15], [5], [31], [16] and in type II diabetes with the phospholipase D isoforms PLD1 and PLD2 [23], [24], [19], [25], [28], [27]. The interaction between PED and PLD1 has been extensively characterized through previous NMR studies conducted both in vitro, utilizing a segment of PLD1 named D4α, and in a cellular lysate environment employing cells overexpressing the entire PLD1. These investigations have revealed that the PED interacting surface is primarily defined by the helices α1, α3, and α4 of the DED domain, with the α5-helix being subject to allosteric effects. Notably, the interaction between PED and PLD1 is stabilized by a portion of the unstructured C-terminal tail. Identification of these "hot spots" crucial for mediating the PED/PLD1 binding facilitated the screening of a diverse library of small compounds, resulting in the discovery of BPH03 as a potential lead compound capable of directly interfering with the PED/PLD1 binding interface. However, while significant, this finding does not fully enable the application of rational design techniques, which rely on comprehensive structural insights into the interaction between PED and BPH03, including details related to the targeted cavity and ligand binding mode. To address this gap, we employed a combined experimental and theoretical approach, allowing us to: i) elucidate the molecular recognition mechanism of BPH03, shedding light on the conformational rearrangements of PED upon ligand binding, and ii) investigate whether the conformational changes induced by BPH03 binding affect the interaction with PLD1. Our in-depth examination of BPH03 interaction surface on PED revealed the absence of a distinct druggable pocket. Notably, the side chains of residues D30, K54, and Y62, projecting toward the potential cavity, appeared to obstruct BPH03 binding. Through in-silico studies, we showed that upon BPH03 binding, the bond between D30 and Y62 is disrupted, leading to conformational rearrangements of both K54 and Y62. These rearrangements, in turn, facilitated the identification of a druggable cavity near H52 and H65, which was not present in the initial PED structure, thus aligning well with the NMR data. Furthermore, the NMR data collected in cellular lysate presented here demonstrate that BPH03 binding in the presence of PLD1 perturbs PED C-terminus, which was shown to remain unaffected upon BPH03 binding in the absence of PLD1. Given that the interaction of the C-terminal tail is a crucial aspect of PED binding to PLD1, these findings collectively indicate that, under the conditions tested, BPH03 is capable of displacing PLD1 binding to PED. In summary, our study highlights a previously unrecognized druggable pocket within the PED structure, the specific conformation of which is crucial for ligand binding. Notably, we have provided the .pdb file resulting from the IFD calculations as Supporting information, ready for structured-based virtual screening procedures. Such a conformation can be utilized in the search for novel and more effective inhibitors of the PED/PLD1 interaction, with potential applications in the management of T2D.
CRediT authorship contribution statement
Annarita Del Gatto: Formal analysis. Luciano Pirone: Formal analysis. Gianluca D'Abrosca: Formal analysis. Domenico Alberga: Methodology, Writing – original draft, Writing – review & editing. Ivan Mercurio: Methodology, Writing – original draft. Laura Zaccaro: Formal analysis. Giuseppe Felice Mangiatordi: Conceptualization, Formal analysis, Supervision, Writing – original draft, Writing – review & editing. Gaetano Malgieri: Conceptualization, Writing – original draft, Writing – review & editing. Maria della Valle: Formal analysis. Michele Saviano: Conceptualization, Formal analysis, Supervision, Writing – original draft, Writing – review & editing. Carla Isernia: Formal analysis. Roberto Fattorusso: Conceptualization, Formal analysis. Sonia Di Gaetano: Formal analysis. Luigi Russo: Formal analysis. Emilia Maria Pedone: Formal analysis.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was performed within the project “Potentiating the Italian Capacity for Structural Biology Services in Instruct Eric (ITACA.SB)” (Project no. IR0000009), call MUR 3264/2021 PNRR M4/C2/L3.1.1, and the Mission 4, component 2 “From Research to Business”; Investment 3.3 “Introduction of innovative doctorates that respond to the innovation needs of businesses and promote the recruitment of researchers by the companies” PNRR funded by the European Union NextGenerationEU. This work was funded by grant PRIN 2022 PNRR num P2022AW2H9 to GM from Ministero dell'Università e della Ricerca.
Declaration of Competing Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Ethics approval and consent to participate
Not applicable.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.csbj.2024.04.063.
Contributor Information
Michele Saviano, Email: michele.saviano@cnr.it.
Giuseppe Felice Mangiatordi, Email: giuseppefelice.mangiatordi@cnr.it.
Appendix A. Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Data Availability
The.pdb files resulting from the IFD calculations are available as Supporting information.
PED_IFD_all_poses.zip:.pdb files of all the poses returned by the performed IFD calculation.
PED_APO-form.pdb:.pdb file of the PED-APO form upon preparation.
Moreover, the data that support the findings of this study are available from the authors upon request.
References
- 1.Araujo H., Danziger N., Cordier J., Glowinski J., Chneiweiss H. Characterization of PEA-15, a major substrate for protein kinase C in astrocytes. J Biol Chem. 1993;268:5911–5920. [PubMed] [Google Scholar]
- 2.Danziger N., Yokoyama M., Jay T., Cordier J., Glowinski J., Chneiweiss H. Cellular expression, developmental regulation, and phylogenic conservation of PEA-15, the astrocytic major phosphoprotein and protein kinase C substrate. J Neurochem. 1995 Mar;64(3):1016–1025. doi: 10.1046/j.1471-4159.1995.64031016.x. [DOI] [PubMed] [Google Scholar]
- 3.Estellé A., Yokoyama M., Nothias F., Vincent J.-D., Glowinski J., Vernier P., et al. The major astrocytic phosphoprotein PEA-15 is encoded by two mRNAs conserved on their full length in mouse and human*. J Biol Chem. 1996 doi: 10.1074/jbc.271.25.14800. [DOI] [PubMed] [Google Scholar]
- 4.Fiory F., Formisano P., Perruolo G., Beguinot F. Frontiers: PED/PEA-15, a multifunctional protein controlling cell survival and glucose metabolism. Am J Physiol Endocrinol Metab. 2009;297:592–601. doi: 10.1152/ajpendo.00228.2009. [DOI] [PubMed] [Google Scholar]
- 5.Callaway K., Abramczyk O., Martin L., Dalby K.N. The anti-apoptotic protein PEA-15 is a tight binding inhibitor of ERK1 and ERK2, which blocks docking interactions at the D-recruitment site. Biochemistry. 2007;46:9187–9198. doi: 10.1021/bi700206u. [DOI] [PubMed] [Google Scholar]
- 6.Condorelli G., Vigliotta G., Cafieri A., Trencia A., Andalò P., Oriente F., Miele C., Caruso M., Formisano P., Beguinot F. Oncogene. 1999 Aug;18(31):4409–4415. doi: 10.1038/sj.onc.1202831. [DOI] [PubMed] [Google Scholar]
- 7.Estellés A., Yokoyama M., Nothias F., Vincent J.D., Glowinski J., Vernier P., Chneiweiss H. The major astrocytic phosphoprotein PEA-15 is encoded by two mRNAs conserved on their full length in mouse and human. J Biol Chem. 1996;271(25):14800–14806. doi: 10.1074/jbc.271.25.14800. Jun 21. [DOI] [PubMed] [Google Scholar]
- 8.Formstecher E., Ramos J.W., Fauquet M., Calderwood D.A., Hsieh J.-C., Canton B., et al. PEA-15 mediates cytoplasmic sequestration of ERK MAP kinase biological outputs by phosphorylating nuclear and cyto-plasmic substrates. Nuclear substrates include tran-scription factors such as Elk-1, and cytoplasmic sub. Dev Cell. 2001 doi: 10.1016/s1534-5807(01)00035-1. [DOI] [PubMed] [Google Scholar]
- 9.Kitsberg D., Formstecher E., Fauquet M., Kubes M., Cordier J., Canton B., et al. Knock-out of the neural death effector domain protein PEA-15 demonstrates that its expression protects astrocytes from TNF-induced apoptosis. J Neurosci. 1999;19(19):8244–8251. doi: 10.1523/JNEUROSCI.19-19-08244.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kubes M., Cordier J., Glowinski J., Girault J.A., Chneiweiss H. Endothelin induces a calcium-dependent phosphorylation of PEA-15 in intact astrocytes: identification of Ser104 and Ser116 phosphorylated, respectively, by protein kinase C and calcium/calmodulin kinase II in vitro. J Neurochem. 1998;71:1307–1314. doi: 10.1046/j.1471-4159.1998.71031307.x. [DOI] [PubMed] [Google Scholar]
- 11.Mace P.D., Wallez Y., Egger M.F., Dobaczewska M.K., Robinson H., Pasquale E.B., et al. Structure of ERK2 bound to PEA-15 reveals a mechanism for rapid release of activated MAPK. Nat Commun. 2013;4 doi: 10.1038/ncomms2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ramos J.W., Hughes P.E., Renshaw M.W., Schwartz M.A., Formstecher E., Chneiweiss H., et al. Death effector domain protein PEA-15 potentiates ras activation of extracellular signal receptor-activated kinase by an adhesion-independent mechanism. Mol Biol Cell. 2000;11(9):2863–2872. doi: 10.1091/mbc.11.9.2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Trencia A., Perfetti A., Cassese A., Vigliotta G., Miele C., Oriente F., et al. Protein kinase B/Akt binds and phosphorylates PED/PEA-15, stabilizing its antiapoptotic action. Mol Cell Biol. 2003;23:4511–4521. doi: 10.1128/MCB.23.13.4511-4521.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xiao C., Yang B.F., Asadi N., Beguinot F., Hao C. Tumor necrosis factor-related apoptosis-inducing ligand-induced death-inducing signaling complex and its modulation by c-FLIP and PED/PEA-15 in glioma cells. J Biol Chem. 2002;277:25020–25025. doi: 10.1074/jbc.M202946200. [DOI] [PubMed] [Google Scholar]
- 15.Hill JM, Vaidyanathan H, Ramos JW, Ginsberg MH, Werner MH . Recognition of ERK MAP kinase by PEA-15 reveals a common docking site within the death domain and death effector domain. EMBO J. Vol. 2;21(no. 23). p. 6494–504. [DOI] [PMC free article] [PubMed]
- 16.Twomey E.C., Cordasco D.F., Wei Y. Profound conformational changes of PED/PEA-15 in ERK2 complex revealed by NMR backbone dynamics. Biochim Biophys Acta Proteins Proteom. 2012;1824:1382–1393. doi: 10.1016/j.bbapap.2012.07.001. [DOI] [PubMed] [Google Scholar]
- 17.Twomey E.C., Wei Y. High-definition NMR structure of PED/PEA-15 death effector domain reveals details of key polar side chain interactions. Biochem Biophys Res Commun. 2012;424:141–146. doi: 10.1016/j.bbrc.2012.06.091. [DOI] [PubMed] [Google Scholar]
- 19.Fiory F., Spinelli R., Raciti G.A., Parrillo L., D’esposito V., Formisano P., et al. Targetting PED/PEA-15 for diabetes treatment. Expert Opin Ther Targets. 2017 doi: 10.1080/14728222.2017.1317749. [DOI] [PubMed] [Google Scholar]
- 20.Valentino R., Lupoli G.A., Raciti G.A., Oriente F., Farinaro E., Della Valle E., et al. The PEA15 gene is overexpressed and related to insulin resistance in healthy first-degree relatives of patients with type 2 diabetes. Diabetologia. 2006;49:3058–3066. doi: 10.1007/s00125-006-0455-5. [DOI] [PubMed] [Google Scholar]
- 18.Condorelli G., Vigliotta G., Iavarone C., Caruso M., Tocchetti C.G., Andreozzi F., et al. PED/PEA-15 gene controls glucose transport and is overexpressed in type 2 diabetes mellitus. EMBO J. 1998;17:3858–3866. doi: 10.1093/emboj/17.14.3858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vigliotta G., Miele C., Santopietro S., Portella G., Perfetti A., Maitan M.A., et al. Overexpression of the ped/pea-15 Gene Causes Diabetes by Impairing Glucose-Stimulated Insulin Secretion in Addition to Insulin Action. Mol Cell Biol. 2004;24:5005–5015. doi: 10.1128/mcb.4.11.5005-5015.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miele C., Raciti G.A., Cassese A., Romano C., Giacco F., Oriente F., et al. PED/PEA-15 regulates glucose-induced insulin secretion by restraining potassium channel expression in pancreatic β-cells. Diabetes. 2007;56:622–633. doi: 10.2337/db06-1260. [DOI] [PubMed] [Google Scholar]
- 23.Doti N., Cassese A., Marasco D., Paturzo F., Sabatella M., Viparelli F., et al. Residues 762-801 of PLD1 mediate the interaction with PED/PEA15. Mol Biosyst. 2010;6:2039–2048. doi: 10.1039/c005272h. [DOI] [PubMed] [Google Scholar]
- 24.Farina B., Doti N., Pirone L., Malgieri G., Pedone E.M., Ruvo M., et al. Molecular basis of the PED/PEA15 interaction with the C-terminal fragment of phospholipase D1 revealed by NMR spectroscopy. Biochim Biophys Acta Proteins Proteom. 2013;1834 doi: 10.1016/j.bbapap.2013.04.012. [DOI] [PubMed] [Google Scholar]
- 25.Viparelli F., Cassese A., Doti N., Paturzo F., Marasco D., Dathan N.A., et al. Targeting of PED/PEA-15 molecular interaction with phospholipase D1 enhances insulin sensitivity in skeletal muscle cells. J Biol Chem. 2008;283:21769–21778. doi: 10.1074/jbc.M803771200. [DOI] [PubMed] [Google Scholar]
- 26.Zhang Y., Redina O., Altshuller Y.M., Yamazaki M., Ramos J., Chneiweiss H., et al. Regulation of expression of phospholipase D1 and D2 by PEA-15, a novel protein that interacts with them. J Biol Chem. 2000;275:35224–35232. doi: 10.1074/jbc.M003329200. [DOI] [PubMed] [Google Scholar]
- 27.Viparelli F., Doti N., Sandomenico A., Marasco D., Dathan N.A., Miele C., et al. Expression and purification of the D4 region of PLD1 and characterization of its interaction with PED-PEA15. Protein Expr Purif. 2008;59:302–308. doi: 10.1016/j.pep.2008.02.012. [DOI] [PubMed] [Google Scholar]
- 28.Viparelli F., Doti N., Sandomenico A., Marasco D., Dathan N.A., Miele C., et al. Expression and purification of the D4 region of PLD1 and characterization of its interaction with PED-PEA15. Protein Expr Purif. 2008;59:302–308. doi: 10.1016/j.pep.2008.02.012. [DOI] [PubMed] [Google Scholar]
- 29.Farina B., Pirone L., D’Abrosca G., Della Valle M., Russo L., Isernia C., et al. Screening a molecular fragment library to modulate the PED/PEA15-phospholipase D1 interaction in cellular lysate environments. ACS Chem Biol. 2021;16:2798–2807. doi: 10.1021/acschembio.1c00688. [DOI] [PubMed] [Google Scholar]
- 30.Arkin M.M.R., Wells J.A. Small-molecule inhibitors of protein-protein interactions: Progressing towards the dream. Nat Rev Drug Discov. 2004 doi: 10.1038/nrd1343. [DOI] [PubMed] [Google Scholar]
- 31.Madhavi Sastry G., Adzhigirey M., Day T., Annabhimoju R., Sherman W. Protein and ligand preparation: parameters, protocols, and influence on virtual screening enrichments. J Comput Aided Mol Des. 2013;27:221–234. doi: 10.1007/s10822-013-9644-8. [DOI] [PubMed] [Google Scholar]
- 32.Schrödinger Release 2022-4 LigPrep. Schrödinger, LLC, New York, NY; 2022.
- 33.Lu C., Wu C., Ghoreishi D., Chen W., Wang L., Damm W., et al. OPLS4: improving force field accuracy on challenging regimes of chemical space. J Chem Theory Comput. 2021;17:4291–4300. doi: 10.1021/acs.jctc.1c00302. [DOI] [PubMed] [Google Scholar]
- 34.Friesner R.A., Banks J.L., Murphy R.B., Halgren T.A., Klicic J.J., Mainz D.T., et al. Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J Med Chem. 2004;47:1739–1749. doi: 10.1021/jm0306430. [DOI] [PubMed] [Google Scholar]
- 35.Friesner R.A., Murphy R.B., Repasky M.P., Frye L.L., Greenwood J.R., Halgren T.A., et al. Extra precision glide: docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. J Med Chem. 2006;49:6177–6196. doi: 10.1021/jm051256o. [DOI] [PubMed] [Google Scholar]
- 36.Halgren T.A., Murphy R.B., Friesner R.A., Beard H.S., Frye L.L., Pollard W.T., et al. Glide: a new approach for rapid, accurate docking and scoring. 2. Enrichment Factors in Database Screening. J Med Chem. 2004;47:1750–1759. doi: 10.1021/jm030644s. [DOI] [PubMed] [Google Scholar]
- 37.Schrödinger Release 2022-4 Glide QM-Polarized Ligand Docking protocol.
- 38.Yang Y., Yao K., Repasky M.P., Leswing K., Abel R., Shoichet B.K., Jerome S.V. Efficient Exploration of Chemical Space with Docking and Deep Learnin. J Chem Theory Comput. 2021;17(11):7106–7119. doi: 10.1021/acs.jctc.1c00810. Nov 9. [DOI] [PubMed] [Google Scholar]
- 39.Schrödinger Release 2022-4 Induced Fit Docking protocol.
- 40.Sherman W., Day T., Jacobson M.P., Friesner R.A., Farid R. Novel procedure for modeling ligand/receptor induced fit effects. J Med Chem. 2006;49:534–553. doi: 10.1021/jm050540c. [DOI] [PubMed] [Google Scholar]
- 41.Cho A.E., Guallar V., Berne B.J., Friesner R. Importance of accurate charges in molecular docking: Quantum Mechanical/Molecular Mechanical (QM/MM) approach. J Comput Chem. 2005;26:915–931. doi: 10.1002/jcc.20222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Genheden S., Ryde U. The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities. Expert Opin Drug Discov. 2015;10:449–461. doi: 10.1517/17460441.2015.1032936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kaminski G.A., Friesner R.A., Tirado-Rives J., Jorgensen W.L. Evaluation and reparametrization of the OPLS-AA force field for proteins via comparison with accurate quantum chemical calculations on peptides. J Phys Chem B. 2001;105:6474–6487. [Google Scholar]
- 44.Schrödinger Release 2022-4 Desmond Molecular Dynamics System.
- 45.Bowers K.J., Chow E., Xu H., Dror R.O., Eastwood M.P., Gregersen B.A., Klepeis J.L., Kolossvary I., Moraes M.A., Sacerdoti F.D., Salmon J.K., Shan Y., Shaw D.E. Scalable algorithms for molecular dynamics simulations on commodity clusters. Proceedings of the ACM/IEEE Conference on Supercomputing. 2006;((SC06),):11–17. November. [Google Scholar]
- 46.Hoover W.G. Canonical dynamics: equilibrium phase-space distributions. Phys Rev A Gen Phys. 1985;31(3):1695–1697. doi: 10.1103/physreva.31.1695. [DOI] [PubMed] [Google Scholar]
- 47.Nosé S. A unified formulation of the constant temperature molecular dynamics methods. J Chem Phys. 1984;81:511–519. [Google Scholar]
- 48.Martyna G.J., Tobias D.J., Klein M.L. Constant pressure molecular dynamics algorithms. J Chem Phys. 1994;101(1994):4177–4189. [Google Scholar]
- 49.Frey B.J., Dueck D. Clustering by passing messages between data points. Science. 2007;315(5814):972–976. doi: 10.1126/science.1136800. [DOI] [PubMed] [Google Scholar]
- 50.Halgren T. New method for fast and accurate binding-site identification and analysis. Chem Biol Drug Des. 2007;69:146–148. doi: 10.1111/j.1747-0285.2007.00483.x. [DOI] [PubMed] [Google Scholar]
- 51.Halgren T.A. Identifying and characterizing binding sites and assessing druggability. J Chem Inf Model. 2009;49:377–389. doi: 10.1021/ci800324m. [DOI] [PubMed] [Google Scholar]
- 53.Kumar A., Kuhn L.T., Balbach J. In-cell NMR: analysis of protein–small molecule interactions, metabolic processes, and protein phosphorylation. Int J Mol Sci. 2019;20(2):378. doi: 10.3390/ijms20020378. 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Barbieri L., Luchinat E., Banci L. Protein interaction patterns in different cellular environments are revealed by in-cell NMR. Sci Rep. 2015;5 doi: 10.1038/srep14456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Keller R. Cantina Verl; 2004. The computer aided resonance assignment tutorial. [1966-] [Google Scholar]
- 55.Lee W., Tonelli M., Markley J.L. NMRFAM-SPARKY: enhanced software for biomolecular NMR spectroscopy. Bioinformatics. 2015;31:1325–1327. doi: 10.1093/bioinformatics/btu830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schrodinger, LLC. The PyMOL molecular graphics system, version 2.5; 2010.
- 57.Williamson M.P. Using chemical shift perturbation to characterise ligand binding. Prog Nucl Magn Reson Spectrosc. 2013;73:1–16. doi: 10.1016/j.pnmrs.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 58.Ikedife J., He J., Wei Y. PEA-15 engages in allosteric interactions using a common scaffold in a phosphorylation-dependent manner. Sci Rep. 2022;12 doi: 10.1038/s41598-021-04099-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Data Availability Statement
The.pdb files resulting from the IFD calculations are available as Supporting information.
PED_IFD_all_poses.zip:.pdb files of all the poses returned by the performed IFD calculation.
PED_APO-form.pdb:.pdb file of the PED-APO form upon preparation.
Moreover, the data that support the findings of this study are available from the authors upon request.