Abstract
Cancer cells exhibit altered metabolic pathways, prominently featuring enhanced glycolytic activity to sustain their rapid growth and proliferation. Dysregulation of glycolysis is a well-established hallmark of cancer and contributes to tumor progression and resistance to therapy. Increased glycolysis supplies the energy necessary for increased proliferation and creates an acidic milieu, which in turn encourages tumor cells' infiltration, metastasis, and chemoresistance. Circular RNAs (circRNAs) have emerged as pivotal players in diverse biological processes, including cancer development and metabolic reprogramming. The interplay between circRNAs and glycolysis is explored, illuminating how circRNAs regulate key glycolysis-associated genes and enzymes, thereby influencing tumor metabolic profiles. In this overview, we highlight the mechanisms by which circRNAs regulate glycolytic enzymes and modulate glycolysis. In addition, we discuss the clinical implications of dysregulated circRNAs in cancer glycolysis, including their potential use as diagnostic and prognostic biomarkers. All in all, in this overview, we provide the most recent findings on how circRNAs operate at the molecular level to control glycolysis in various types of cancer, including hepatocellular carcinoma (HCC), prostate cancer (PCa), colorectal cancer (CRC), cervical cancer (CC), glioma, non-small cell lung cancer (NSCLC), breast cancer, and gastric cancer (GC). In conclusion, this review provides a comprehensive overview of the significance of circRNAs in cancer glycolysis, shedding light on their intricate roles in tumor development and presenting innovative therapeutic avenues.
Keywords: circRNAs, Glycolysis, Cancer, Metastasis, Chemoresistance
1. Introduction
Cancer has emerged as one of the significant causes of death in the modern era and has become a critical issue in public health all over the world [[1], [2], [3], [4]]. Modifications in both genetic and epigenetic characteristics contribute to the complexity of this disease [[5], [6], [7]]. Identifying molecular alterations in cancer genes and associated signaling pathways may provide more accurate diagnostic tools and therapeutic strategies [8,9]. This understanding can potentially contribute to advancements in cancer detection and therapy.
A recently identified family of ncRNAs with high levels of stability and covalent close is known as circRNAs [10]. These circRNAs have a fundamental role in scaffolding protein complexes, allowing them to control gene expression, including that of their parental genes. Additionally, they may operate as molecular sponges to sequester miRNAs in a sequence-specific method, which can impact alternative splicing processes and modify the interplay between RNAs and proteins [11]. There is growing evidence suggesting circRNAs play a role in several critical processes in the regulation of many different types of cancer, including breast cancer, HCC, osteosarcoma, gastric cancer, thyroid cancer, and pancreatic cancer, where they may act in an oncogenic manner and hence provide value as prospective clinical biomarkers and/or potential strategies for therapeutic action [[12], [13], [14], [15], [16], [17], [18], [19], [20]]. However, there are several significant limitations to their use in the clinic. Their circular form restricts their annotation, since it lacks comprehensive functional explanation [20]. This complicates their detection and biomedical application, presenting ongoing challenges.
Human malignancies often exhibit metabolic changes throughout their development [21]. For example, cancer cells often exhibit higher levels of glycolysis, a metabolic process that converts glucose into lactate. The Warburg effect, or aerobic glycolysis, is a mechanism that typically occurs in tumor cells. Anaerobic glycolysis is the most common type of glycolysis. This enables the generation of higher amounts of energy and materials necessary for increasing biosynthesis levels. Metabolic disorders and cancer may be caused by inherited genetic mutations or abnormal expression of several metabolic enzymes [22]. Many tumor cells depend on glycolysis as the main energy source and materials to maintain their functional conditions. Explosive growth, chemoresistance, and immune evasion are all aggressive characteristics linked to this metabolic reprogramming [[23], [24], [25]]. Based on these findings, scientists have begun exploring the potential of inhibiting glycolysis as a cancer treatment. It has been shown that some circRNAs, which are critical regulators in the development of cancer, may also influence glycolysis to modify gastric cancer malignancy [23,26,27].
The primary factor contributing to the failure of cancer treatments is the absence of efficient interventions and precise biological targets. Consequently, the discovery of new molecularly targeted therapies for cancer is an endeavor of great importance. In this summary, we discussed some of the most recent findings about the underlying mechanisms of the regulatory functions of circRNAs in regulating glycolysis in cancer cells.
2. CircRNAs biogenesis, and biological function and its role in health and disease
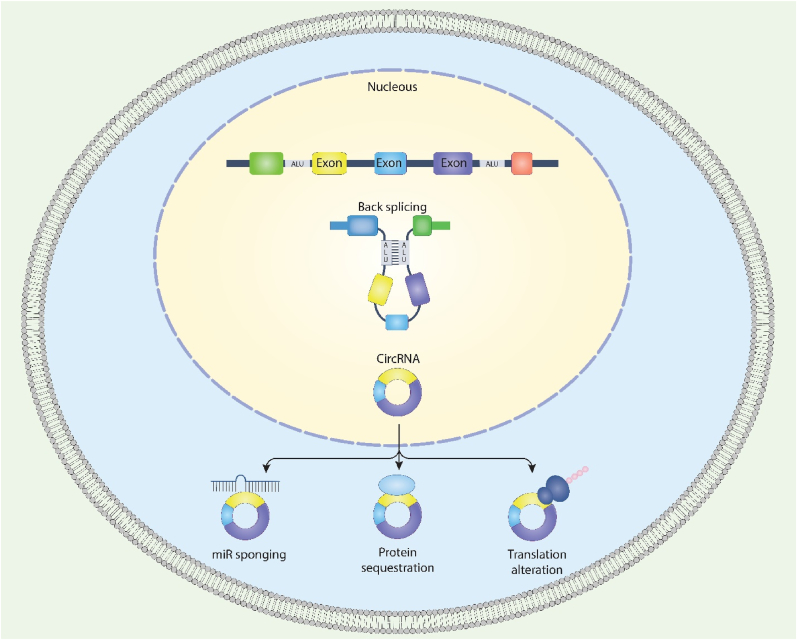
CircRNAs are produced by back-splicing, which involves the molecular linking of the spliced RNA's 5′ and 3′ ends [28]. Depending on their structural characteristics, circRNAs are now thought to fall into four classes: intergenic circRNAs, intronic circRNAs, exon-intron circRNAs (EIciRNAs), and exonic circRNAs (ecircRNAs). Indeed, circRNAs account for over 80 % of all circRNAs discovered to date, and the vast majority are found exclusively in the cytoplasm [29,30]. On the other hand, competing endogenous RNAs (ceRNAs) are found mainly in the nucleus [31], pointing to the possibility that they control gene expression. Read-through circRNAs, a new class of circular transcripts produced by tandem exons from genes' flanking regions, have recently been discovered [32]. Lariat-driven circularization, RBP-mediated circularization, and intron pairing-driven circularization are three putative hypotheses for circRNA biogenesis processes that have been accepted so far. Because RBPs play a crucial role in driving circRNA biosynthesis by controlling neighboring splice sites, the first model is dubbed RBP-mediated circularization (Fig. 1). For example, the splicing factors muscleblind (MBL) [33], quaking [34], and adenosine deaminase acting on RNA [35] are implicated in circRNA biogenesis. The second model of circRNA biosynthesis is a model in which intron splicing causes RNA rotation [36]. Alternate circularization, facilitated by the complementary pairing of both flanks of introns, may produce diverse circRNAs, such as ecircRNAs and EIciRNAs, since pre-mRNA flanking introns include inverted complementary sequences. Additionally, the flanking sequences of circRNAs often include longer introns [30], and the production of circRNAs is aided by the presence of rcs in longer introns [37]. The third form of circRNA biosynthesis is lariat-driven circularization, which similarly promotes the production of circRNAs [38]. For more information, please refer to the study conducted by Geng et al. [39].
Fig. 1.
Schematic illustration of circRNAs biogenesis and function.
CircRNA is implicated in controlling gene expression and plays a significant role in biological evolution, including forming miRNA sponges, endogenous RNAs, and biomarkers. CircRNAs additionally serve as a critical element in disease diagnosis function. Investigations have shown that circRNAs are associated with the growth of embryos, preeclampsia, sperm, endometrial cancer, and ovarian epithelial tumors. These findings imply that circRNAs can become pharmacological intervention targets or biomarkers for pathological conditions [40]. CircRNAs can act as a sponge for miRNA, bind to RNAs, and be translated into proteins. An increasing number of scientists have focused their attention on the interaction between circRNAs and miRNAs. Sry is a gene on the mouse Y chromosome responsible for determining sex. Capel and colleagues [41] discovered that Sry might be translated into circRNAs. Through the post-transcriptional regulation of RNA, RNA-binding proteins, also known as RBP, are implicated in many cellular activities, including growth, diversification, transformation, apoptosis, aging, and the reaction to oxidative stress [42]. For instance, a circRNA known as RasGEF1B may enhance the durability of intercellular cell adhesion molecule-1 in murine macrophages [43]. After initiation of translation, it has been shown, both in-vitro and in-vivo, that the 40S subunit of the eukaryotic ribosome can bind to engineered circRNAs that contain an IRES [44,45]. Transcripts produced from circRNAs in certain eukaryotic cells are promising, suggesting that these circRNAs may have the power to translate proteins [40]. For example, Legnini et al. [46] described that IRES-carrying circ-ZNF609 could translate proteins. For more details, refer to Geng et al. [39] and Liu et al. [40].
Many cellular functions, including embryogenesis, cell cycle regulation, cellular senescence, signaling pathways, and cellular responses to stress, are regulated by circRNAs [47]. CircRNAs have an important function in proper hemostasis by regulating embryogenesis, cellular stress, cell cycle progression, and metabolism [47]. For instance, circRNAs are abnormally abundant in the brain during embryogenesis, suggesting they play an essential part in forming the embryonic brain. At the embryonic stage, E115, CiRS-7 is significantly expressed in the cerebellum, but at E60, its expression is downregulated in the cerebral cortex of swine embryos [48]. Regarding metabolism, it has been suggested that increased expression of CDR1as through modulation of miR-7 in pancreatic β-cells alters β-cell function. Genes responsible for cell signaling (protein kinase C beta), cytoskeletal architecture (profilin 2, phosphatase, and actin regulator 1), and transcription factor (paired box 6) expression are all targeted by miR-7. Increased insulin production in mouse islets has been linked to overexpression of murine Pax6 expression via the sponging effect of CDR1as on miR-7, which might have far-reaching implications for beta cell function [49]. To survive, living organisms have a vital need for cellular balance. Both positive and negative effects of circRNAs on cellular homeostasis, including the regulation of proliferation, apoptosis, immunological response, and drug resistance, have been described. Potential functions for CDR1as include the regulation of p21-activated kinase 1, promotion of DNA repair, and suppression of apoptosis [47]. In addition, sustained nucleolar stress is induced in cultured human cells when a circRNAs is overexpressed from the locus encoding the lncRNA ANRIL [47].
Due to their fundamental function in gene regulation, reports of circRNA expression abnormalities in the context of disease are emerging. Numerous disorders, such as cancer, cardiovascular disease, neurological diseases, diabetes, and atherosclerosis, have been linked to modified circRNA expression, even though the underlying processes by which these diseases manifest are still poorly understood [47]. CircRNAs have been linked to senescence, malignancies, cardiovascular disease, osteoarthritis, preeclampsia, neurological diseases, and type 2 diabetes [47].
Investigations have demonstrated that circRNAs can perform a crucial function in the formation of tumors, enhanced cellular motility, and propagation [[50], [51], [52], [53], [54]]. Han et al. [55] reported that circINPP4B could function as a sponge for miR-30a to control Th17 cell development throughout the advancement of experimental autoimmune encephalomyelitis, which is a murine model of multiple sclerosis. Much research has been done on how circRNAs contribute to cardiovascular disease development [56,57]. It has been suggested that circRNAs may function in the body's defense against viral infection. Transfection of purified circRNAs produced in-vitro into mammalian cells resulted in strong stimulation of innate immune genes and protection against viral infection, as shown by Chen et al. [58]. These studies showed that circRNAs perform a crucial function in the pathophysiology of many diseases.
3. Cancer and glycolysis: the role of circRNAs
One of the fundamental characteristics found in cancer cells is reprogramming their metabolism. Although oxygen is present, tumor cells preferentially employ glycolysis, a less efficient metabolic route, to generate energy. The Warburg effect describes how dependence on aerobic glycolysis stimulates carcinogenesis and malignant development. Tumors undergo a metabolic switch to glycolysis for unknown reasons. It is becoming evident that numerous signal molecules are implicated in this process, such as oncogenes and tumor suppressors. However, the mechanism by which oncogenic signals decrease mitochondrial function and encourage the transition to glycolysis is still opaque [59].
There are many hypothesized processes by which tumor cells might sustain high levels of glycolytic flux [60]. Phosphofructokinase-1 (PFK1) is an essential enzyme for maintaining glycolytic flow. PFK2 is overexpressed in malignant cells and stimulates the synthesis of fructose-2,6-bisphosphate, which is a potent allosteric inducer of PFK1 and counteracts the inhibitory effects of high ATP concentrations on the enzyme. Second, lactate dehydrogenase (LDH) plays a crucial function in maintaining glycolysis by regenerating NAD+ and producing lactate. Furthermore, tumor cells exhibit elevated pyruvate kinase M2 (PKM2) levels. Delivery of glycolytic byproducts upstream of the pyruvate biosynthesis pathway is inhibited by allosteric and covalent repression of PKM2 [60]. Besides signal molecules and transcription factors, mTOR, Akt, c-Myc, and HIF-1α are the key regulators studied in depth [60].
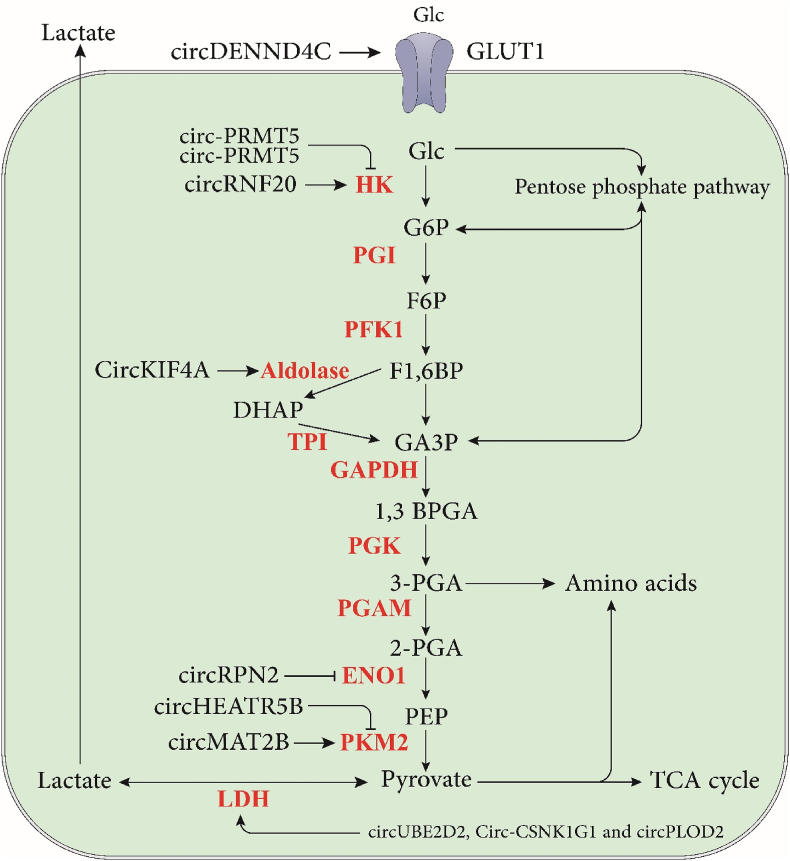
Recent data imply that circRNAs increase in tumor tissues, promoting tumor glycolysis, development, and metastases by altering cellular metabolism (Fig. 2) [59]. For example, in HCC, the circUBE2D2 [50], circMAT2B [61], circ_0008450 [62], and circ-PRMT5 [63] promote glycolysis, but some circRNAs, such as circRPN2 [64] inhibit glycolysis in HCC. Also, in CRC, the circPCLE1 [65], circDENND4C [53], and circ_0136666 [66] promote glycolysis; on the other hand, some circRNA such as CircTADA2A [67] inhibits glycolysis in CRC.
Fig. 2.
The schematic illustration of the immunomodulatory effects of circRNAs on the glycolysis pathway.
Exploring interactions between circRNA and glycolysis encounters technical challenges. The precise isolation and characterization of circRNAs are intricate due to their diverse structures and expression patterns. Additionally, understanding the complex mechanisms through which circRNAs regulate glycolysis requires sophisticated experimental techniques, such as advanced sequencing methods, functional assays, and bioinformatics tools. The regulatory influence of circRNAs on glycolysis may vary based on cancer type, stage, and microenvironmental factors. Hence, comprehending the context-specific effects of circRNAs on glycolytic pathways is crucial for understanding their roles in tumor progression and therapeutic responses. Future investigations should focus on uncovering the contextual factors influencing circRNA-glycolysis interactions to devise tailored therapeutic approaches.
Although circRNAs show promise as potential biomarkers for cancer diagnosis, prognosis, and predicting therapeutic responses, challenges persist in identifying robust circRNA signatures with clinical relevance. Validating circRNA biomarkers in large patient cohorts and standardizing detection assays are crucial steps toward their clinical application. Furthermore, integrating circRNA expression profiles with other omics data could enhance the predictive accuracy of circRNA-based biomarkers in cancer management. Utilizing circRNAs as therapeutic targets to modulate glycolysis in cancer requires the development of effective and specific targeting strategies. Designing circRNA-targeting agents that selectively inhibit oncogenic circRNAs or restore the function of tumor-suppressive circRNAs poses technical and logistical challenges. Future research should prioritize refining therapeutic delivery systems, optimizing drug formulations, and evaluating the safety and efficacy of circRNA-targeting therapies in both preclinical and clinical settings. Translating discoveries from the laboratory to clinical practice demands rigorous validation of circRNA-glycolysis interactions in clinical settings. Large-scale clinical studies are essential to assess the prognostic and predictive value of circRNA biomarkers and therapeutic targets across diverse patient populations.
Addressing these limitations and embracing future perspectives will deepen our understanding of the intricate interplay between circRNAs and glycolysis in cancer and pave the way for the development of innovative diagnostic tools and targeted therapies for combating cancer progression and improving patient outcomes. A deeper insight into the underpinning molecular processes in cancer pathogenesis may facilitate the development of novel medications for cancer treatment. In the next section, we present an in-depth discussion of the molecular involvement of circRNAs in the control of glycolysis in a wide range of cancer types, including HCC, GC, NSCLC, CRC, PCa, BC, CC, and glioma.
4. HCC
Among all kinds of liver cancer, HCC is the most common clinical tissue subtype, accounting for between 75 and 85 percent of all cases [68,69]. The primary risk factors for HCC incidence were thought to include alcoholism, aflatoxin exposure, obesity, hepatitis B/C virus infection, and smoking [70]. Remarkably, the clinical outcome and overall long-term survival of HCC patients have not remained ideal despite advances made in technical methods [71]. Furthermore, the fact that HCC was an ongoing and complex process involving many variables and developmental stages made it challenging to discover new treatments for HCC [72].
circRNAs have recently emerged as viable and promising biomarkers for detecting and managing cancer [73]. The fundamental processes that underlie the action of circRNA in the establishment and tumorigenesis are not yet fully understood [74]. In one study, 12 miRNAs anticipated to have anti-cancer impacts in HCC were examined, and it was shown that miR-338-3p could bind to circMAT2B. Additionally, 20 anticipated oncogenes were identified in HCC, and it was demonstrated that circMAT2B sponge inhibits endogenous miR-338-3p function by sequestering it and increasing the production of miR-338-3p, which in turn targets the oncogene PKM2. PKM2 expression has recently been shown to be significantly greater in HCC tissues. Also, a favorable association was detected between the patterns of PKM2 expression and circMAT2B expression [61]. For instance, activating PI3K/AKT/mTOR further upregulated PKM2 expression [61].
In conclusion, the oncogenic circMAT2B gene was characterized, and its function and mechanism were evaluated in the rewiring of glucose metabolism in HCC as well as other malignancies. The results show that circMAT2B is significantly elevated in HCC tissues and is associated with a poor outcome in individuals who had hepatectomy-treated HCC. In addition, circMAT2B enhances glycolysis and tumorigenesis in HCC cells, both in-vitro and in-vivo, when the cells are exposed to hypoxia. The mechanism by which circMAT2B boosts HCC glycolysis and tumorigenesis is mediated by regulating the miR-338-3p/PKM2 axis. This axis can potentially be a targeted therapy for HCC [61].
In another work, He et al. [75] investigated the role of circ_0000517 in HCC. According to the findings of this investigation, circ0000517 knockdown prevented epithelial-mesenchymal transition (EMT), which in turn prevented the invasion and migration of cancer cells. Besides, glycolysis is a significant feature of cancer and has been shown to contribute to the emergence of HCC. It was observed that inhibiting circ0000517 reduced glycolysis in HCC by identifying glycolysis biomarkers and particular enzymes (LDHA and HK2) [[76], [77], [78]]. The inhibition of circ0000517 had an anti-tumor effect on HCC in vitro, consistent with findings from an earlier investigation [79]. The most important way circRNA affects tumor progression is ceRNA interaction with circRNA/miRNA/mRNA [80]. A study showed that miR-326 was sponged through circ0000517, and its expression level was low in HCC. miR-326 has been discovered to be a potential target of circ0000517. In addition, findings supported the idea that miR-326 has a tumor-suppressing function in HCC, aligning with earlier research findings [81,82]. Furthermore, the regulating function of circ0000517 silencing on the advancement of HCC may be reversed by the reduced expression of miR-326. This suggests that circ0000517 could control the progression of HCC through sponging miR-326 [75]. Dysregulation of IGF1R remained a key focus in the research and management of HCC [83]. It has been reported that miR-326 is responsible for targeting IGF1R. Furthermore, studies have shown that IGF1R has a role in promoting the growth, movement, and infiltration of cancer cells, therefore contributing to the oncogenic properties of HCC, which was also in line with previous research findings [[84], [85], [86]].
In addition, overexpression of IGF1R causes a faster increase in glycolysis in HCC, such as in glioma and breast cancer [87,88]. Studies have shown that IGF1R may counteract the impact of miR-326, indicating that miR-326 specifically targets IGF1R and has a regulatory function in the progression of HCC. In addition, it has been discovered that circ_0000517 can modulate IGF1R levels by competitively interacting with miR-326 with miR-326 and supports the ceRNA network consisting of circ0000517/miR-326/IGF1R. Furthermore, the knockdown of circ_0000517 resulted in tumor growth inhibition, which was discovered by employing an in-vivo xenograft model [75]. Taken together, interference with circ_0000517 suppressed HCC development, perhaps through the miR-326/IGF1R axis, putting forward an innovative approach for the function of circ 0000517 in the advancement of HCC. Based on the findings of this investigation, circ_0000517 shows promise as an exciting new target for the treatment of HCC [75].
In hepatocarcinoma cells, Ding et al. [63] examined the impact of Circ-PRMT5 on the course of proliferation, motility, and glycolysis of HCC. This investigation showed that circ-PRMT5 was upregulated more in HCC tissues and cells than in the controls. Moreover, a negative correlation was found between circ-PRMT5 and the expression of miR-188-5p. The expression of circ-PRMT5 was inhibited in HCC cells, which resulted in the suppression of cell proliferation, motility, and glycolysis. It has been discovered that the overexpression of circ-PRMT5 may reduce the miR-188-5p-induced impact on HCC cells, giving fresh evidence for the crucial regulatory role that the circ-PRMT5/miR-188-5p regulatory axis plays in the growth, progression, emigration, and spread of cancer [63]. The elevation of miR-188-5p resulted in an apparent restriction of glucose uptake and the formation of lactate and ATP. This indicates that miR-188-5p may prevent the growth of HCC by inhibiting glycolysis.
Furthermore, suppressing the spreading and proliferation of HCC cells resulted in the increased expression of miR-188-5p [63]. Both Ma and colleagues [89] and Cheng colleagues [90] concluded that miR-188-5p is responsible for HCC antitumor activities. Similarly, functional studies found that miR-188-5p is a potent inhibitor of HK2 expression. miR-188-5p plays an essential part in glycolysis [91]; furthermore, it is involved in the abnormal regulation of HK2 expression in the metabolism of tumor cells, implying that HK2 might be a promising target for therapeutic intervention in the management of malignant tumors [92,93]. Overall, circ-PRMT5 has increased significantly in HCC tissues and cells compared to matched controls. Suppression of circ-PRMT5 may also efficiently impede the growth, movement, and glucose metabolism in HCC cells. The study demonstrated an interaction between circ-PRMT5, miR-188-5p, and HK2 in HCC. The circ-PRMT5/miR-188-5p/HK2 axis might potentially be used as a targeted treatment for HCC [63].
Recent research conducted by Du et al. [62] focused on the role that circ_0008450 plays in the pathogenesis of HCC. The rapidly expanding number of HCC cells often surpasses the formation of effective blood arteries, resulting in inadequate oxygen delivery to HCC tumors. This is generally a crucial issue because HCC tumors are malignant [94]. On the other hand, tumor cells respond to hypoxia by switching from oxidative glucose metabolism to glycolytic glucose metabolism to have enough energy and chemicals for their anabolic metabolism [95]. The present research found that HCC tissues and cells treated with hypoxia showed increased expression of the gene circ0008450. A correlation was found between high circ0008450 expression and the size of tumors, the tumor_node_metastasis (TNM) stage, the presence of lymphatic metastasis, and the existence of distant metastasis in patients with HCC. Additionally, the diagnostic value of the circulating circ_0008450 was found to be relatively high. According to these findings, has_circ0084927 has the potential to become an innovative biomarker for the detection of HCC [62].
In addition, suppression of circ_0008450 reduced tumorigenesis in vivo, reversed its effect on survival and glycolysis and increased the death rate in hypoxia-exposed HCC cells cultured in-vitro [62]. Based on these results, circ_0008450 might potentially serve as a diagnostic indicator for HCC, and it also seems that hypoxia-induced circ_0008450 plays a role in promoting the development of HCC. Evidence shows that circ_0008450 controls tumor development by functioning as a miRNA sponge [96,97]. This study's findings suggest that hypoxia-treated HCC cells, which were identified as circ_0008450 gene targets, have reduced miR-431 expression. Also, hypoxia-treated HCC cells could undo their effects of survival, apoptosis, and glycolysis by inhibiting miR-431. According to these findings, hypoxia-induced circ_0008450 was responsible for modulating the aggressive activity of HCC cells via miR-431 when the cells were exposed to hypoxic conditions [62]. It has been discovered that the scaffold protein known as AKAP1 plays a significant part in regulating mitochondrial activity [98]. According to research from the past, AKAP1 helped promote the mTOR pathway and contributed to the expansion of cancer cells [99]. The present study found that circ_0008450 might control AKAP1 via the use of miR-431. AKAP1 overexpression may counteract the impact of miR-431 on the survival, cell death, and glycolysis of HCC cells induced by hypoxia treatment. As a result, circ_0008450 was responsible for advancing HCC because it upregulated AKAP1 and sponged off of miR-431 when the conditions were hypoxic. In this study, however, only molecular biology research was carried out, and no clinical examination was performed on the materials. This is something to look into in further study. In sum, recent studies have shown that hypoxia-induced circ_0008450 promotes the aggressive behavior of HCC cells under hypoxic conditions via the miR-431/AKAP1 pathway. This suggests that circ_0008450 might potentially be used as a targeted treatment for HCC [62].
Huang et al. [50] studied the effect of the circUBE2D2 gene in their research on HCC glycolysis and sorafenib resistance. Sorafenib, a molecularly targeted medication, is used to treat advanced types of HCC. It may prevent the formation of tumors and cancer relapse by preventing the propagation of malignant cells. Even if the therapeutic effect of this multi-targeted therapy was not as great as expected, it has the potential to prolong the life of HCC patients. Resistance to sorafenib is the main cause of the problem. Thus, removing resistance at the molecular level is the answer [50]. It has been suggested that circRNA circUBE2D2 may play a role in the resistance of HCC patients to sorafenib. Sorafenib tolerance was shown to be associated with aerobic glycolysis [100,101]; thus, the novel hypoxia-associated circUBE2D2 may be involved in sorafenib tolerance. In cellular analysis, gain and loss of function tests were performed to determine IC50 resistance to sorafenib and glycolytic ability. Based on the findings, increasing the expression level of circUBE2D2 increased glucose consumption, lactate production volume, and ECAR amount. Besides, knockdown circUBE2D2 decreased glucose intake, lactate output level, and ECAR amount. Therefore, their results indicated that circUBE2D2 is responsible for the increased rate of aerobic glycolysis and resistance to sorafenib [50].
There are many speculations about the role of ncRNA in sorafenib resistance of HCC. It has been found that HCC cells resistant to sorafenib have a specific circRNA expression pattern, and circRNAs with variable expression play a key role in this resistance [102]. It appears that ncRNA plays a crucial role in the sorafenib tolerance of HCC [50]. Researchers have identified many circRNAs as having a role in the pathophysiological process of HCC development. For example, circRNA circSLC3A2 is primarily found in the cytoplasm and has been shown to exert an oncogenic function by suppressing miR-490-3p and increasing PPM1F protein levels. This indicates that there is a significant association between circRNA circSLC3A2 and HCC patients having a shorter overall survival time [103]. These findings, collectively, provide evidence that circRNAs play an essential role as critical oncogenic components in HCC.
Chen et al. [104] studied the mechanism by which circ-CFH has a role in the pathophysiology of HCC. This research investigated the possible molecular mechanism of circ-CFH in the progression of HCC. The findings demonstrated an upsurge in circ-CFH expression in HCC cells and tissues. The circ-CFH molecule facilitated the malignant progression of HCC by acting as a sponge for miR-377-3p, leading to increased production of RNF38 in HCC cells [104]. The study's results indicate that circ-CFH exhibited considerable expression in both tissues and cells of HCC. Furthermore, this upregulation was shown to be closely associated with the TNM stage and the spread of malignancy. Remarkably, cancer cells acquire energy and large molecular building blocks via glycolysis, even when there is sufficient oxygen. This is strongly associated with resistance to currently used therapy and a poor prognosis in patients [105,106]. As a result, the processes of glycolysis, cell growth, motility, invasion, and apoptosis were evaluated in HCC cells. In the current study, the knockdown of circ-CFH in HCC cells in vitro increased apoptosis while decreasing cell survival, migration, and invasion. However, it did not affect glycolysis. Tumor growth in living organisms was reduced when the expression of circ-CFH was suppressed utilizing gene silencing technology. Studies conducted in a laboratory setting indicated that circ-CFH may function as a miR-1250-3p sponge to regulate the survival, growth, and invasion of hepatoblastoma cells [107].
Liu et al. [108] Liu et al. [104] observed that targeting miR-7 to inhibit circ-CFH lowered cell survival, proliferation, and invasiveness in their investigation of HCC cells. According to the findings of this study, it was hypothesized that circ-target CFH in HCC would be miR-377-3p. As a result, it was indicated that circ-CFH plays an essential role in HCC in conjunction with miR-377-3p. Upregulation of miR-377 stops HCC cells from proliferating and invading, according to Chen et al. [109]. According to their findings, miR-377-3p exhibited a decreased expression level in both HCC cells and tissue. Additionally, it had an inverse relationship with the amount of circ-CFH expression seen in HCC tissues. Furthermore, a reduction in miR-377-3p levels has been demonstrated to partly counteract the inhibitory effect of circ-CFH knockdown on the progression of HCC. Ultimately, circ-CFH functioned as a molecular sponge for miR-377-3p, hence regulating the development of HCC [104]. circ-CFH is an essential component in the carcinogenesis process carried out by HCC. The progression of HCC is impeded by the suppression of circ-CFH since this mechanism inhibits the processes of cell growth, cell death, motility, invasion, and glycolysis in HCC cells. The biological functions of Circ-CFH in HCC were mediated by a pathway controlled by miR-377-3p/RNF38. This finding provides a theoretical basis for developing alternative treatments for HCC.
Cai et al. [110] studied the function of CircRHBDD1 in metabolic reprogramming and found that it inhibits the effectiveness of immunotherapy in HCC through modifying m6A. This glycolytic transition is often regulated by circRNAs, which play a crucial role in this process. Tumor cells commonly take over glucose metabolism to ensure their continued survival [111]. It would be exciting to discover previously unknown circRNAs that have a role in HCC glycolysis. In a study carried out by Cai and colleagues, they evaluated the influence of circRHBDD1 on ECAR, OCR, GLUT1, HK2, and several metabolites associated with glycolysis. This was carried out by examinations that quantified the effects of both reduced and increased circRHBDD1 activity. The findings of the in-vitro research demonstrated that circRHBDD1 had a tight relationship with the increased glycolysis in HCC [110].
Furthermore, researchers developed PDX mouse models and proved the tumor-promoting properties of circRHBDD1 in vivo. PDX models are powerful tools that may be used to evaluate the effect of innovative therapeutic techniques in vivo, as they accurately recreate the original morphological and genetic features of the original tumors [112]. The PI3K/AKT signaling pathway is essential for regulating glucose metabolism and several metabolic events for cancer development. PIK3R1, a regulatory component of PI3K, enhances PI3K/AKT signaling to expedite the process of glycolysis [113]. CircularRHBDD1 was shown to activate PI3K/AKT signaling via the PIK3R1 receptor. The overexpression of circRHBDD1 increased the amount of PIK3R1 protein [110].
Neither overexpression nor knockdown of circRHBDD1 altered PIK3R1 mRNA levels statistically significantly. The inability of therapies with MG132 or CQ to restore the lower levels of PIK3R1 protein in circRHBDD1-silenced HCC cells suggests that circRHBDD1 may not alter the decomposition of PIK3R1 protein. Polysomic profiling analysis led to the discovery of the role of circRHBDD1 in promoting PIK3R1 translation. An RNA pull-down experiment was conducted, and then mass spectrometry was used to determine whether YTHDF1 interacted with circRHBDD1. This was performed to assess how circRHBDD1 regulated PIK3R1 translation [110]. m6A is involved in almost every stage of the life cycle of RNA, such as translation, splicing, and maintenance of mRNA [114]. YTHDF1, an m6A reader protein, can detect m6A sites and enhance the translation of targeted mRNA [115].
The pathogenesis of several cancer forms involves using YTHDF1 in a m6A-dependent manner [[116], [117], [118]]. The study demonstrated that YTHDF1 enhanced the translation of PIK3R1 mRNA in a way that depended on m6A modification. The use of several bioinformatics tools and meRIP assays reached this conclusion. The capacity of YTHDF1 to bind m6A is abrogated by functional ablation of the YTH domain. The study revealed that YTHDF1-Mut hindered the interaction between PIK3R1 mRNA and YTHDF1, hence abolishing the heightened levels of PIK3R1 protein production seen in cancerous cells. The pull-down experiment revealed that YTHDF1 is the protein that interacts with circRHBDD1 [110]. Nevertheless, other proteins might potentially bind with circRHBDD1 and participate in circRHBDD1-inducing metabolic wiring in an unknown mechanism [110].
Additional investigation is required to explore the involvement of m6A writers and erasers in the communication between YTHDF1 and PIK3R1 [110]. A subgroup of HCC patients with anti-PD-1 treatment exhibited significant enhancement in their clinical condition [119]. However, a considerable obstacle still exists in the form of restricted responsiveness to anti-PD-1 therapy in most patients [120]. Reprogramming the cancer cells' metabolism can modify the TME's characteristics [121]. The glycolytic action of cancerous cells generates lactate and acidifies the cancer environment, both of which are detrimental to the anticancer immune responses of T cells and NK cells [122]. According to several reports, the presence of aerobic glycolytic function in human malignancies is inversely related to the overall success of immunotherapy [123]. Evidence showed that the concurrent suppression of circRHBDD1 and anti-PD-1 therapy resulted in enhanced anti-tumor efficacy. The findings indicate that circRHBDD1 can potentially be a viable treatment option for HCC, either alone or in conjunction with immune checkpoint inhibition [110]. Ultimately, this study demonstrated a significant upregulation of circRHBDD1 expression in HCC, which enhanced glycolytic activity in HCC cell lines. The circRHBDD1 connected with YTHDF1 at the molecular level, resulting in an improved translation of PIK3R1 that is reliant on m6A. Suppression of circRHBDD1 may also enhance the effects of treatment with anti-PD-1 antibodies. Ultimately, focusing on the circRHBDD1/YTHDF1/PIK3R1 axis can serve as a therapeutic strategy for treating head and neck cancer [110].
Li et al. [64] examined the role of CircRPN2 in HCC growth and aerobic glycolysis in their most recent study. The researchers used circRNA-seq to detect circRNAs that exhibit differential expression in primary HCCs from patients with and without metastases. They discovered that circASAP1 plays a critical role in HCC dissemination by modulating the miR-326/miR-532-5p-MAPK1/CSF-1 signaling pathway [124]. The researchers investigated the function of circRPN2 in this work. According to the circRNA-seq data, circRPN2 expression was shown to be reduced in HCC metastasis. Notably, it has been demonstrated that circRPN2 is significantly reduced in HCC tissues, particularly in cases of HCC with metastases or relapses [64].
Furthermore, for the first time, it was shown that circRPN2 might limit the development and dissemination of HCC by promoting ENO1 degradation and modulating the miR-183-5p/FOXO1 axis. This is an important finding because it suggests that circRPN2 may be able to prevent the spread of HCC cells [64]. It has been observed that circRPN2-mediated knockdown of ENO1 inhibits HCC growth and metastasis, increases respiration, and strongly decreases glycolysis in HCC cells. Significantly, the effect of circRPN2 on glucose metabolism, progression, and release is reduced when ENOblock pharmacologically inhibits ENO1. Therefore, the results of their study suggest that circRPN2 may play an important role in HCC proliferation and glycolytic rewiring by interacting with ENO1 and directing the degradation of this protein [64].
Cancer cells may enhance their glucose uptake by using glycolysis, a metabolic pathway that supplies carbon for cell proliferation. This metabolic adaptation enables cancer cells to participate in autocrine communication and local metabolite-based paracrine communication, both facilitating tumor growth [125]. Consequently, inhibition of the glycolysis pathway may be an effective method for cancer treatment. New research has shown that the glycolytic phase catalyzed by ENO1 may be reversed [126]. In addition, whereas enolase consists of three isoforms, only ENO1 is somewhat stable. Consequently, there was a decrease in harmful effects and an enhancement in the selectiveness of ENO1-targeting substances, indicating that ENO1 might be a viable contender for suppressing glycolysis in cancer treatment. Enolase is an enzyme that catalyzes glycolysis, which is one of the processes that causes cancer to grow. Li and colleagues suppressed ENO1 by modulating circRPN2 levels and found effects on HCC metastasis and glycolysis that were comparable to those described in previous research [64].
Ultimately, these findings support the notion that ENO1 has great potential as a viable treatment choice for cancer. Significantly, they found that inhibiting ENO substantially decreased the development and spread of HCC in laboratory experiments and in vivo, indicating that it might potentially benefit patients with circRPN2-associated HCC [64]. Notably, the decrease in metabolic activity caused by ENO inhibition in HCC is similar to previous research findings [127]. Also, the particular mode of action might be complicated and have to be investigated in depth [128]. The AKT/mTOR signaling system impacts cellular biosynthetic and aerobic glycolytic processes in malignant tumors [129,130]. Metabolic changes are influenced by AKT downstream targets, including mTOR [131]. In line with these findings, cirRPN2 has been shown to suppress the AKT/mTOR signaling pathway in HCC cells through the hydrolysis of ENO1 to enhance the inhibitory effect on glycolysis. This was performed to inhibit HCC cell glycolysis. This results in a reduction in lactate generation and amelioration of cancer cell malignant characteristics. Pena et al. reported circRPN2-mediated suppression of the AKT/mTOR pathway using the AKT antagonist MK2206. Based on their theory, they found that MK2206 treatment decreased glycolytic rewiring and increased levels of pAKT and p-mTOR in circRPN2 knockdown cells. Circ_RPN2 inhibitor cells also had significant amounts of pAKT and p-mTOR. These results indicate that circRPN2 influences the rewiring of glycolysis and the progression of HCC via activating the AKT/mTOR pathway through ENO1 [64]. Yan and colleagues observed that circRPN2 can bind with miR-183-5p, a molecule that promotes tumor growth in several types of malignancies by specifically targeting FOXO1. The researchers evaluated the potential of circRPN2 to function as a miRNA sponge in HCC and found that circRPN2 can interact with miR-183-5p [132]. Prior studies have demonstrated that FOXO1 is crucial in regulating glucose metabolism and functions as an inhibitor of tumor growth. FOXO1 maintains glucose balance, promotes tumor growth, and regulates cell death. It is a crucial component of the STAT3 signaling pathway [133]. Using the FOXO1 antagonist (AS1842856), Li et al. showed that the circRPN2/miR-183-5p/FOXO1 axis facilitates circRPN2's impacts on glycolysis and the progression of HCC. This discovery aligns with the previously reported role of FOXO1, indicating that this pathway is responsible for facilitating the impact of circRPN2 on glycolysis and the development of HCC [64]. Ultimately, the collective evidence from the researchers suggests that circRPN2 is reduced in HCC patients who have tumor development or relapse. In addition, circRPN2 has a crucial role in modulating aerobic glycolysis, metastases expansion, and dissemination of HCC by promoting the degradation of ENO1 and controlling the miR-183-5p/FOXO1 signaling pathway [64]. Thus, it was postulated that circRPN2, either alone or in combination with FOXO1 and ENO1, might serve as a noteworthy and distinctive prognostic indicator for HCC.
5. GC
GC is a prevalent kind of cancer that is responsible for a significant number of tumor-related deaths globally. It is a major public health issue owing to its grim prognosis and unsatisfactory treatment results [134,135]. GC poses significant patient challenges and has a poor 5-year survival rate due to increased mortality and morbidity [136,137]. The prognosis of GC is still generally unfavorable despite the widespread use of numerous treatments, such as surgery, chemotherapy, and radiation [138]. Growing information points to a complex etiology for GC carcinogenesis [139]. Hence, investigating the biological traits and molecular mechanisms might be advantageous in developing therapies for GC.
According to increasing data volume, CircRNAs are critical in the Warburg effect occurring in GC. The findings of current microarray research showed several dysregulated circRNAs in the GC tissue [140]. Pu et al. [140] investigated the underlying molecular mechanism in this study of the control of glycolysis in GC. They found that GC tissue had higher levels of circCUL3, a new circRNA, than the adjacent normal specimens. This research demonstrated a correlation between the increased expression of circCUL3 and a worse prognosis in patients with GC. These findings indicate that circCUL3 may serve as a possible risk indicator in the GC population. The results of the gain- and loss-of-function experiments on cellular activities demonstrated that circCUL3 enhanced the proliferation of GC cells. Consequently, their research indicated that circCUL3 may function as a cancer-causing element in gastric cancer GC [140]. The data suggest that circCUL3 may act as a competitive endogenous RNA (ceRNA) for miR-515-5p. miR-515-5p targets STAT3, which in turn binds to the promoter region of HK2. The researchers determined that the circCUL3/miR-515-5p/STAT3/HK2 axis is involved in developing aerobic glycolysis in gastric cancer. HK2 plays a crucial function in both the Warburg effect and the progression of aerobic glycolysis. In the context of GC, miR-181b specifically acts on HK2 to inhibit its expression, reducing glucose uptake and lactate production [140]. In conclusion, the data demonstrate that circCUL3 uniquely alters the metabolic route of GC and offers a potential therapy for GC.
Yang et al. [51] explored the involvement of circUBE2Q2 in the control of glycolysis and autophagy in another study. It was confirmed that circUBE2Q2 may function as a sponge for miR-370-3p to regulate the malignant progression of GC. Yang et al. [51] demonstrate that STAT3 functions as a ceRNA for circUBE2Q2 and that the circUBE2Q2/miR-370-3p axis might contribute to the aggressive progression of GC. Subsequently, they verified that the circUBE2Q2/STAT3 axis can impede autophagy and stimulate glycolysis, thereby fostering the growth of GC. While their research has indicated that the circUBE2Q2/miR-370-3p/STAT3 axis interferes with autophagy and higher glycolysis rates may promote the development of GC, additional studies are necessary to determine if there is a consensus regulation between the autophagy process and glycolysis, as well as whether regulation is necessary. The connection between self-regulatory impacts mutually reinforces the favorable effects on GC [51]. Exosomes have been linked to the advancement of GC using autocrine mechanisms [141]. Exosomes containing increased circSHKBP1 were also shown to stimulate the growth of GC cells by Xie et al. [142]. In addition, they devised two co-cultivation techniques and confirmed the existence of circUBE2Q2 in exosomes generated by GC cells. The exosomal circUBE2Q2 was shown to promote metastases in live organisms and to have regulatory implications on the STAT3 signaling pathway and EMT in laboratory conditions. Therefore, their findings pointed out that the overexpression of circUE2Q2 in GC cells might lead to increased transportation of circUE2Q2 to exosomes, hence influencing GC biological processes and triggering downstream cascades via autocrine or paracrine mechanisms [51].
Nevertheless, the specific mechanism by which exosomal circUBE2Q2 stimulates the malignant proliferation of GC remains unexplained and requires more investigation [51]. Subsequent in-vivo studies showed that combining circUBE2Q2 knockdown with STAT3 antagonists resulted in a more pronounced reduction in tumor growth than circUBE2Q2 knockdown alone [51]. The circUBE2Q2/STAT3 axis has the potential to provide novel ideas for targeted treatment in GC. The observations indicate that circUBE2Q2 can potentially accelerate tumor growth and might serve as a new target for therapeutic interventions in GC [51].
The function of circ_0006089 in the pathophysiology of GC was investigated by Zhou et al. [143]. They observed that circ_0006089 intervention reduced tumor growth by inhibiting GC cell proliferation, proliferation, glycolysis, and revascularization. These findings supported previous findings that circ_0006089 had an oncogenic function in GC [144]. Circ_0006089 has been shown to sponge miR3613p. Research shows circ_0011385 downregulated miR3613p to promote HCC growth [145]. MiR3613p was discovered as a tumor suppressor in cervical cancer, suppressing cancer cell survival and proliferation [143], GC cell proliferation and metastasis, and angiogenesis [146,147]. Zhou et al. [143] discovered that miR3613p mimic significantly inhibited GC cell growth, glycolysis, dissemination, and revascularization, and its antagonist similarly inhibited the activity of circ0006089#2 in GC cells. The results validated the anticancer role of miR3613p in GC and demonstrated that circ_0006089 promoted the malignant characteristics of GC by directly interacting with miR3613p. In conclusion, Zhou et al. [143] found that circ_0006089 promoted GC cell proliferation, metastases, glycolysis, and revascularization by modulating the miR3613p/TGFB1 pathway. Collectively, their study demonstrated that circ0006089 functions as an oncogene and contributes to the development of GC's malignant phenotype. These findings enhance our comprehension of the origin of GC and support the idea that circ0006089 might serve as a promising therapeutic target for GC [143].
Li et al. [148] evaluated the involvement of circDNMT1 in the pathogenesis of GC in another investigation. circDNMT1 is a new circRNA linked to the advancement of breast cancer. In breast cancer tissues, circDNMT1 was shown to be highly expressed. CircDNMT1 functions as a sponge for miRNA, hence sustaining the highly propagating state of breast cancer via the regulation of the miR-485-3p/ZEB1 pathway [149]. Li et al. [148] revealed that circDNMT1 was overexpressed in GC. The expression profile of circDNMT1 suggests disease severity and poor survival in GC patients with clinical and pathological features and 3-year overall survival. CircDNMT1 was associated with a lower survival rate in GC patients.
Further research revealed that circDNMT1 knockdown inhibited GC development. On the other hand, its overexpression may encourage these aggressive activities. CircDNMT1 could potentially be a valuable target in the clinical setting of GC [148]. It has been hypothesized that circDNMT1 might aid tumorigenesis by changing glucose metabolisms. The expression of circDNMT1 was reduced, which decreased the glycolytic markers. The ECAR quantifies the total levels of glycolysis, while the OCR quantifies oxidative phosphorylation. Suppression of CircDNMT1 resulted in a reduction in ECAR and an increase in OCR. These findings suggest that GC cells depend on oxidative phosphorylation to provide additional energy when aerobic glycolysis is restricted. The results indicated that circDNMT1 has a crucial role in regulating GC glycolysis, both in laboratory settings and in vivo, potentially contributing to the development of tumors [148].
HIF-1 enhances the genesis and growth of GC by modulating the miR-17-5p/PDCD4 pathway [150]. It enhances revascularization by boosting VEGF-A expression and polarizing macrophages [151]. HIF-1α controls glycolytic function as a cellular sensor of the environment to maintain tumor growth and development [152,153]. Additionally, Increased HIF-1α accelerates glycolysis in cancer [154]. Li et al. [148] hypothesized that circDNMT1 accelerates GC development through sponging the miR-576-3p/HIF-1α axis. An assortment of interaction experiments showed that the 3′ untranslated region (UTR) of HIF-1α mRNA and miR-576-3p possess notable binding capabilities. HIF-1α upregulation diminished its inhibitory impact on GC malignant tendencies. Furthermore, after suppressing HIF-1α, the functionality of circDNMT1/miR-576-3p was no longer present. The data indicate that HIF-1α is the main focus of the circDNMT1/miR-576-3p axis [148].
The study performed by Li et al. [148] had certain limitations. First, glycolysis is a key mediator in the progression of cancer. Their investigation only confirmed that circDNMT1 increased GC aggressiveness and glycolysis simultaneously. Further evaluation is necessary to determine its contributions to the progression of glycolysis. Second, although HIF-1 may be a crucial downstream target of circDNMT1, it is not the sole target.
Further studies are needed to explore circRNA-miRNA pathways in GC. Third, whether this pathway regulates aggressiveness in other types of cancer remains to be determined. This research showed that circDNMT1 has an oncogenic function in GC [148]. CircDNMT1 was elevated in GC and linked to clinicopathological characteristics and poor outcomes in GC patients. In vitro and in vivo, targeting the 576-3p/HIF-1 axis boosted GC release, motility, invasiveness, and glycolysis. CircDNMT1 has the potential to serve as both a predictive biomarker and a treatment alternative in clinical trials for GC [148].
Zheng et al. focused on the role of a circular RNA, circRPS19, in GC and provided insights into its involvement in aerobic glycolysis and cell viability [155]. Loss-of-function assays demonstrate that silencing circRPS19 inhibits GC cells [155]. Also, this study results, specifically, circRPS19 knockdown leads to the suppression of cell proliferation and aerobic glycolysis [155]. Zheng et al. found that silencing of circRPS19 is associated with increased apoptosis of GC cells [155]. This suggests an anti-apoptotic role of circRPS19 in GC. Their study demonstrated that circRPS19 upregulates the expression of ubiquitin-specific processing protease 7 (USP7) [155]. USP7, in turn, stabilizes HK2 protein through deubiquitination. In vivo experiments confirm that circRPS19 promotes GC progression [155]. Zheng et al. observed that promotion includes an enhancement of aerobic glycolysis, which aligns with the in vitro findings [155]. Mechanistically, circRPS19 influences the miR-125a-5p/USP7 axis, suggesting a regulatory pathway [155]. This axis is implicated in the stabilization of HK2 and the promotion of aerobic glycolysis in GC. In summary, circRPS19 is identified as an upregulated circRNA in GC, and its silencing hinders cell proliferation, induces apoptosis, and suppresses aerobic glycolysis. The mechanism involves the regulation of the miR-125a-5p/USP7 axis, contributing to the stabilization of HK2. These findings position circRPS19 as a potential therapeutic target for GC, suggesting its critical role in the progression of this refractory disease.
6. Lung cancer
Lung cancer is the primary contributor to cancer-related mortality globally [156]. The majority of cases of lung cancer are identified as non-small-cell lung cancer (NSCLC) [156]. Despite the recent progress in the effectiveness of treatments for NSCLC, including chemotherapy, individualized therapy, and immunotherapy, the prognosis for NSCLC patients remains very poor, with a 5-year overall survival rate of about 17 % [68]. A thorough understanding of the mechanisms that affect carcinogenesis and development is required to enhance NSCLC diagnosis and therapy. In recent years, molecular targeted therapy has emerged as an essential method for improving the treatments and outcomes for patients with NSCLC [157,158]. A study conducted by Xu et al. (5) evaluated the influence of circ_0001421 in the pathogenesis of NSCLC. The researchers found that the expression of circ_0001421 and TMEM14A was up, whereas the expression of miR-409-3p was reduced in NSCLC. Circ_0001421 may boost NSCLC progression by continuing to act as a miR-409-3p sponge to enhance TMEM14A expression, implying that it has the possibility of becoming a molecular target in NSCLC therapies [159]. The accumulating data emphasized the function of circRNAs in the etiology of NSCLC [[160], [161], [162]]. The researchers identified and confirmed the miRNA target, miR-409-3p, of circ_0001421 in NSCLC, shedding light on the physiological mechanism of circ_0001421. An investigation has demonstrated that miR-409-3p has antineoplastic properties in numerous kinds of cancer [159]. Expression of circ_0001421 was shown to suppress glycolysis [159]. Furthermore, in-vivo studies have demonstrated that circ_0001421 promotes cancer development by activating the miR-409-3p/TMEM14A pathway [159]. The study discovered that circ_0001421 enhanced the formation of cell colonies, migration, invasion, and glycolysis in NSCLC via regulating the miR-409-3p/TMEM14A pathway. These findings indicate that circ_0001421 might be a potential and effective molecular treatment approach for NSCLC [159].
Shangguan et al. [163] analyzed the function of circSLC25A16 in glycolysis control in NSCLC. They discovered that the newly discovered circRNA, circSLC25A16, was highly elevated in NSCLC cells and tissue. Moreover, the significant increase in circSLC25A16 expression was linked to an unfavorable outcome in individuals with NSCLC. They demonstrated that the stage III-IV NSCLC group exhibited increased circSLC25A16 expression in the research trial. This discovery indicated that circSLC25A16 might be related to tumor grade. Based on the functional test, it was shown that CircSLC25A16 enhanced the glycolysis pattern of NSCLC cells, particularly by increasing glucose absorption, lactate generation, ATP levels, and ECAR production capacity. Concerning the cancer phenotype, mechanistic experiments demonstrated that circSLC25A16 enhanced the proliferation of NSCLC cells. circSLC25A16 has been discovered to function as an oncogene in developing NSCLC by promoting glycolysis and cellular proliferation [163].
According to Shangguan et al. [163] have shown that circSLC25A16 functions as a sponge for miR-488-3p and miR-488-3p. It specifically targets the 3′-UTR of HIF-1 mRNA, therefore creating the circSLC25A16/miR-488-3p/HIF-1α axis. Given that HIF-1α is the primary mediator responsible for the effects of circSLC25A16, it makes sense that circSLC25A16 modulates glycolysis via influencing HIF-1α. In addition, HIF-α is a hypoxia-related transcriptional regulator, and they discovered that HIF-1α might stimulate LDHA mRNA transcription. Furthermore, the HIF-1α/LDHA axis was recently identified in human malignancies [164]. The current study shows that circSLC25A16 is associated with a poor prognosis in NSCLC patients and has been increased in NSCLC tissue/cells. CircSLC25A16 increases LDHA transcription by interacting with miR-488-3p/HIF-1 [163]. The key circRNA and glycolysis molecular pathways that drive NSCLC progression and development are critical for successful treatment.
Yang et al. [165] identified circ_0006677 as the underlying molecular mechanism in the etiology of NSCLC. It was discovered that circ_0006677 functions as a crucial tumor suppressor by impeding NSCLC development and metabolic changes via the control of the miR-578/SOCS2 axis. Finally, NSCLC therapy and diagnostics might benefit from targeting circ_0006677/miR-578/SOCS2 signaling [165]. Yang et al. [165] applied the GEO dataset to show a decrease in circ_0006677 expression in NSCLC tissues. The lower expression of circ_0006677 was confirmed in NSCLC tissues and cell lines. Low levels of circ 0006677 were linked to tumor size, tumor severity, spread to other parts of the body, and unfavorable outlook in NSCLC patients, indicating that circRNAs might serve as a reliable diagnostic and prognostic marker for NSCLC [165]. Their study revealed a significant inverse correlation between the levels of circ_0006677 and miR-578 in NSCLC tissues [165]. Yang et al. [165] discovered that miR-578 hindered circ_0006677's ability to enhance the invasive characteristics and glucose metabolism of NSCLC cells. Their research results together corroborate the idea that 0006677 inhibits NSCLC advancement by acting as a miR-578 sponge [165].
Downregulation of SOCS2 has been seen in several cancers, and it impairs cytokine-induced signaling transmission [[166], [167], [168]]. Yang et al. [165] demonstrated that SOCS2 hinders the progression of NSCLC and is a key focus of miR-578. In NSCLC cells, the circ_0006677 inducer elevated SOCS2 protein levels, but miR-578 reduced it. Circ_0006677 could inhibit miR-578 to promote SOCS2 expression, suggesting a potential therapeutic target for NSCLC therapy [165]. Furthermore, their study demonstrated that circ_0006677 suppressed the proliferation of NSCLC by reducing glucose uptake and lactate production via the modulation of SOCS2. The findings support the significance of SOCS2 for circ_0006677-induced tumor suppression in NSCLC [165].
Zhang et al. [169] analyzed the function of circEHD2 in the development of NSCLC. This study examined the role of circEHD2 in NSCLC and found that circEHD2 may promote cell growth and glycolysis while suppressing autophagy and programmed cell death in NSCLC. The above results were achieved by sponging circEHD2 to modulate FOXK1 expression via miR-3186-3p [169]. CircEHD2 was discovered to be a pro-oncogene in NSCLC that promotes growth and glycolysis while suppressing autophagy [169]. FOXK1, an oncogenic gene, is present in several malignancies [[170], [171], [172]]. This research proved the role of FOXK1 as a proto-oncogene [169]. This study showed that inhibiting FOXK1 may counteract the inhibitory impact of circEHD2 knockdown on glycolysis and promotion of autophagy in NSCLC. Elevated glycolysis and reduced autophagy allow uncontrolled tumor progression [169]. This study is restricted by the lack of clinical data on circEHD2 in NSCLC, making it uncertain if targeting circEHD2 influences tumor development in NSCLC patients. Future studies should investigate the potential impact of circEHD2 on resistance to therapy and metastatic disease in NSCLC.
In another recent investigation, Chen et al. [173] investigated the function of circSHKBP1 in the progression of NSCLC through the modulation of glycolysis. Their research demonstrated that circSHKBP1 functions as an oncogene in NSCLC. Higher circSHKBP1 expression was detected in the tissues and serum exosomes of NSCLC patients. NSCLC growth, motility, invasiveness, cell survival, glycolysis, and macrophage polarization and engagement were all affected by exosomal circSHKBP1. Additionally, lymph node metastasis progress, TNM staging, and a poor prognosis were linked to circSHKBP1 overexpression [173]. Serum exosomes have a high level of CircSHKBP1, which influences the miR-582-3p/HUR/VEGF pathway, prevents HSP90 decomposition, and promotes GC carcinogenesis, as previously reported [142]. Exosome-transferred circSATB2 increases NSCLC cell growth, motility, and invasion [174]. Circ_0008928, a serum exosome-based marker, has been linked to cisplatin response, tumorigenesis, and glucose metabolism in NSCLC [174]. Chen et al. [173] discovered that circSHKBP1 is present in high exosome levels, and exosomal circSHKBP1 influences the activity of NSCLC cells and the characteristics of macrophages. Under both in-vitro and in-vivo circumstances, reducing circSHKBP1 resulted in reduced growth, movement, invasion, cell viability, glycolysis, polarization, and macrophage activation. Additionally, circSHKBP1 may function as an oncogene in NSCLC [173]. PKM2 is a key enzyme that promotes the progression of glycolysis and enhances tumor formation [175]. PKM2 might influence the tumor microenvironment and promote malignancy via exosomes. Exosomal PKM2 modifies the TME, facilitating the development of HCC [176]. Chen et al. [173] discovered that PKM2 is a downstream target of exosomal circSHKBP1 and functions as an oncogene in NSCLC. Exosomal circSHKBP1 derived from NSCLC cells promotes NSCLC growth, movement, invasiveness, cell viability, and glycolysis. It accelerates the process of macrophage polarization and recruiting by blocking miR-1294 and boosting PKM2 levels. CircSHKBP1 may act as an oncogene in NSCLC and is associated with a worse prognosis. In conclusion, circSHKBP1 might act as a prognostic marker for NSCLC [173].
Huang et al. focus on circRARS and its role in NSCLC, particularly emphasizing its expression, clinical significance, and functional impact on tumor progression [177]. The research revealed that circRARS is elevated in NSCLC tissues, and its expression is associated with a history of smoking, lymph node metastases, and advanced tumor stages [177]. CircRARS promotes the growth, spread, and movement of NSCLC cells in vitro functional assays [177]. These findings suggest a tumor-promoting role for circRARS in NSCLC progression. RNA pull-down assays reveal that circRARS can bind with LDHA [177]. Huang et al. demonstrated that circRARS positively regulates both the activity and transcriptional expression of LDHA in NSCLC cells [177]. The research found that reducing circRARS reduced glucose intake and lactate generation in NSCLC cells [177]. CircRARS compromises aerobic glycolysis, suggesting a role in modulating cellular metabolism [177]. By controlling LDHA activity, rescue experiments show that circRARS stimulates the growth of NSCLC cells. Furthermore, circRARS enhances glycolysis and tumor advancement in NSCLC via controlling LDHA, indicating a possible involvement in altering metabolic pathways in cancer cells [177]. In conclusion, the study provides insights into the role of circRARS in NSCLC, highlighting its association with clinical features, prognostic value, and functional impact on tumor progression. The identified circRARS-LDHA axis suggests a potential avenue for therapeutic intervention targeting metabolic pathways in NSCLC.
The possible pathways by which the circular RNA circ_0003028 contributes to NSCLC were examined by Shi et al. [178]. Circ_0003028 is identified as being upregulated in NSCLC tissues, suggesting its potential relevance to the disease [178]. In patients with NSCLC, an elevated level of circ_0003028 is linked to a poor overall prognosis [178]. In this study, circ_0003028 demonstrates high diagnostic potential, indicating its potential utility as a biomarker for NSCLC [178]. Results from the study by Shi et al. demonstrated that NSCLC cells overexpress circ_0003028, which inhibits apoptosis but enhances proliferation and glycolytic capability [178]. Also, they explained that silencing circ_0003028 has the opposite effect, suggesting a direct influence on tumor cell behavior [178]. According to the research findings, SLC5A1 is a target gene regulated by either miR-1305 or the miR-1322/SLC5A1 axis [178]. It would suggest that circ_0003028 accelerates malignant tendencies and glycolytic capability in NSCLC cells from a mechanistic viewpoint [178]. One possible explanation is that the miR-1305/SLC5A1 axis or miR-1322 is involved in the control of this process. Ultimately, the research highlights the relevance of circ_0003028 in NSCLC, focusing on its clinical importance and functional influence on tumor cell activity. The regulatory pathway consisting of circ_0003028, miR-1305, miR-1322, and SLC5A1 offers valuable information on prospective therapeutic avenues for NSCLC treatment and diagnostics.
Li et al. explored the role of circP4HB in lung adenocarcinoma (LUAD), focusing on its expression, clinical significance, and functional impact on tumor progression [179]. The results show that CircP4HB is overexpressed in LUAD tissues and that its expression is positively associated with lymph node metastases and advanced TNM stages [179]. There is a correlation between an elevated expression of circP4HB in LUAD and an unfavorable prognosis [179]. Both in vivo and in vitro experiments demonstrated that circP4HB increases LUAD growth, suggesting that it may play a role in tumor formation [179]. Also, upregulated circP4HB increases glucose consumption and lactate production in LUAD cells [179]. CircP4HB accelerates aerobic glycolysis, suggesting a role in the metabolic reprogramming characteristics of cancer cells. From a mechanistic standpoint, they showed that circP4HB interacts with PKM2 and mainly concentrates in the cytoplasm of LUAD cells [179].
Moreover, Li et al. indicated that circP4HB promotes M2 macrophage phenotype shift, indicating its influence on the TME [179]. In conclusion, the study underscores the significance of circP4HB in promoting LUAD progression through its interaction with PKM2, influencing glucose metabolism, and modulating the tumor microenvironment. The findings suggest that circP4HB could be a potential therapeutic target for LUAD.
7. Colorectal cancer (CRC)
CRC is a common cancer that regularly occurs in 40 % of people after treatment, generally with local tissue metastasis to the liver or lung [180]. Even though 80 % of recurrences occur during the first three years following radical surgery, CRC cells may disseminate during this time and may cause a relapse even years later [181]. As a result, boosting the quality of efficient colorectal cancer screening may help reduce CRC incidence and mortality [182]. Patients may now choose better drugs because of advances in molecular science, and genetic mutation studies should be used as a diagnostic guide [183]. However, the course of treatment for relapse is unknown, and different patients require individualized treatment to achieve sustained remission [184].
Zhang et al. [53] studied the molecular role of circDENND4C in CRC proliferation, migration, and glycolysis in their investigation. According to the study carried out by Liang et al. [185], circDENND4C is an HIF1α-associated circRNA that may stimulate breast cancer cell growth in hypoxic conditions. The roles of circDENND4C in CRC were initially assessed by Zhang and colleagues [53], and their findings showed that circDENND4C was elevated in CRC tissues and cells. The inhibition of circDENND4C significantly reduced cell proliferation, motility, and glycolysis [53]. Meanwhile, the earlier study found that the expression level of GLUT1 was elevated in CRC tissues and cells [186]. GLUT1 has been linked to the glycolysis pathway, and silencing GLUT1 inhibited cell growth, motility, and glycolysis of CRC tissues [187]. Studies revealed that GLUT1 upregulation abolished the impacts of circDENND4C knockdown on cell growth, emigration, and glycolysis. However, the molecular mechanism by which circDENND4C and GLUT1 interact was unknown [53].
The online programs StarBase 3.0 and TargetScan effectively determined the putative binding sites for circDENND4C and miR-760, GLUT1-3′UTR, and miR-760 [53]. Through the miR-760 sponge, circDENND4C was shown to modulate the effects of GLUT1 on the growth, motility, and glycolysis of CRC cells [186]. CircRNA 100290's action as a ceRNA for miR-378a facilitated the growth of oral squamous cell carcinoma cells via GLUT1 and glycolysis [187]. Similarly, Zhang et al. [53] indicated that circDENND4C influences CRC cell migration and proliferation via modulating glycolysis. These findings provide a foundation for developing new diagnostic tools and therapeutic techniques for CRC cells and a foundation for a more profound knowledge of the pathogenic mechanisms of CRC cells. Li et al. [66] researched the significance that circ_0136666 serves in the development of CRC. The putative regulation mechanism responsible for the Warburg effect in tumor cells remains unknown. This is the first time circ_0136666 has been shown to participate in the glycolysis of CRC cells, as discovered by Li et al. [66].
An interactome analysis identified miR-383 as a target of circ_0136666, which was confirmed by a dual-luciferase reporter assay. miR-383 has been shown to act as a tumor suppressor in ovarian, HCC, and GC [[188], [189], [190]]. It has been shown that depleting miR-383 counteracts both the boosting impact on CRC cell death and the antagonistic effect of lowering circ_0136666 on CRC cell proliferation and glycolysis. This suggests that circ_0136666 promotes CRC advancement via miR-383 [66]. The downstream components of miR-383 were examined to elucidate the regulatory mechanism by which miR-383 hinders the progression of CRC [66]. The Starbase program proposed CREB1 as a potential target of miR-383. The double luciferase reporter technique verified the interaction between miR-383 and CREB1 in 293T cells. Yan et al. [191] noticed that CREB1 promoted the proliferation and dissemination of CRC cells. Li et al. [66] discovered that CREB1 functions as an oncogene in CRC, consistent with previous research. Increased CREB1 expression reduced the inhibitory impact of elevated miR-383 levels on CRC cell proliferation and glucose metabolism while enhancing programmed cell death. They postulated that miR-383 prevents CRC development by increasing the expression of CREB1. It is theorized that circ_0136666 enhances the proliferation and glycolysis of CRC cells and suppresses apoptosis via interacting with miR-383/CREB1 axis [66]. Li et al. [66] used western blot experiments to assess the levels of glycolysis-related proteins (HK2 and LDHA) to explore the importance of the circ_0136666/miR-383/CREB1 axis in CRC cell glycolysis. Increasing the quantity of glycolysis-related proteins in CRC cells via the miR-383/CREB1 axis was the mechanism by which Circ_0136666 induced an increase in glycolysis. The influence of circ_0136666 on the growth of colorectal cancer tumors was investigated using a mouse xenograft model. Circ_0136666 enhanced CRC tumor development by overexpressing CREB1 through miR-383. In sum, circ_0136666 boosted CRC formation in-vitro and in-vivo by increasing tumor growth and glycolysis and inhibiting apoptosis via the miR-383/CREB1 axis [66].
Another recent research [67] investigated the molecular significance of circTADA2A in the development of CRC. Several cancers, including osteosarcoma and breast cancer, have been linked to CircTADA2A. It was reported by Wu et al. that circTADA2A was an oncogene in osteosarcoma. Furthermore, they found that circTADA2A promoted osteosarcoma cell motility and propagation via the miR-203a-3p/CREB3 axis. Furthermore, Xu and colleagues [192] demonstrated that circTADA2A reduced the proliferation and spread of breast cancer cells by acting on the miR-203a-3p/SOCS3 axis. The heterogeneity of the TME may be responsible for the various actions of circTADA2A in osteosarcoma and breast cancer. The researchers used information from the GEO dataset and seventy matched pairs of CRC and normal tissue to discover that circTADA2A was significantly downregulated in CRC [67]. It was also shown that the level of circTADA2A was decreased in CRC cells compared to NCM460 cells [67]. To assess the activity of circTADA2A in vivo, a murine xenograft model was developed [67]. The upregulation of circTADA2A confirmed the anticancer impact of circTADA2A in CRC and inhibited the development of CRC tumors in vivo. Similarly, an increase in the expression of circTADA2A in vitro enhanced the risk of death while hindering glycolysis and the cell cycle in CRC cells [67].
Researchers have discovered that KLF14 acts as a tumor suppressor in many different types of cancer. An insufficient amount of KLF14 was shown to be associated with centrosome amplification and malignancy, as discovered by Fan and colleagues [193]. In their study, Wang and colleagues [194] discovered that the lncRNA DGCR5 reduced the progression of HCC by acting on the miR-346/KLF14 axis. In the instance of CRC, Zhou et al. [195] found that KLF14, which functions as a downstream gene of the HAND2-AS1/miR-1275 axis, inhibited the advancement of CRC. KLF14 was shown to be a target of miR-374a-3p in patients with colorectal cancer, according to Li et al. [67]. Previous research [195,196] discovered that rescue tests demonstrated that KLF14 functions as a target of miR-374a-3p, which contributes to reducing the aggressiveness of CRC cells. Additional research has shown that circTADA2A may increase KLF14 expression by interacting with miR-374a-3p. It was demonstrated that CircTADA2A suppresses tumor growth in CRC. CircTADA2A increased the expression of KLF14 and targeted miR-374a-3p to induce apoptosis in CRC cells. This resulted in the inhibition of cellular proliferation, glycolysis, and progression of tumors. Based on the findings, the circTADA2A/miR-374a-3p/KLF14 axis has the potential to serve as a feasible target and biomarker for the treatment of CRC [67].
The role of circNOX4 in the development of CRC was examined by Wang et al. [197]. The results of earlier studies were confirmed, with CRC tissues and cell lines having much greater levels of circNOX4 than non-tumor tissues and cells [198]. It was discovered by Wang et al. [197] that there is a connection between high levels of circNOX4 expression and worse outcomes for individuals who have been diagnosed with CRC. Zhang et al. identified that pim1 increases CRC cell proliferation in the presence of glucose deprivation by promoting glycolysis. In this study, we investigated whether or not circNOX4 can control glycolysis and other processes in colorectal cancer cells. CRC cells are susceptible to the oncogenic effects of CircumNOX4, which include an increase in cell growth, motility, invasion, and glycolysis [197]. It has been shown that miR-485-5p may act as a tumor suppressor in several different forms of cancer [199,200]. In CRC cells, MiR-485-5p was shown to be a direct downstream gene of circNOX4 [197]. Further functional research revealed that via sponging miR-485-5p, circNOX4 may accelerate the development of CRC.
The CKS1 family, of which CKS1B is a part, has been shown to control the cell cycle process [201,202]. The study used the StarBase database to anticipate the binding sequence of CKS1B and miR-485-5p. Subsequently, dual-luciferase reporter and RIP experiments were conducted to validate this connection. CKS1B expression has been elevated in CRC cells and tissues, aligning with previous investigations [203]. The research discovered that elevating CKS1B expression diminishes the inhibitory effect of miR-485-5p mimic on CRC cell growth, motility, invasion, and glycolysis. Additionally, suppressing circNOX4 hindered CRC development in living organisms by affecting the miR-485-5p/CKS1B pathway [197]. It was found that circNOX4 may accelerate CRC development by increasing CKS1B expression in vitro and in vivo through miR-485-5p sponging. Additional study is required to ascertain how CKS1B enhances cell growth, migration, and glycolysis in CRC cells.
Yi et al. [65] investigated the involvement of circPCLE1 in the pathogenesis of CRC in another recent study. This study established the biological significance of the newly discovered circRNA PCLE1 in CRC via promoting TAM M2 polarization, CRC EMT, and glycolysis through binding competition with miR-485-5p to regulate ACTG1 [65]. Consistent with previous findings, our research showed that circPLCE1 is raised in CRC tissues and cells and is associated with lymph node metastases in CRC patients [204]. This study's results are consistent with other research showing that circPCLE1 promotes the growth of CRC cells and suppresses cell death [204]. This reinforces the idea that circPCLE1 acts as a proto-oncogene in CRC. This work enhanced glycolysis in CRC by introducing a new circular RNA called PCLE1. Previous research has shown that circNOX4 [197], circ0136666 [66], circTADA2A [67], and other circRNAs play critical roles in CRC glycolysis. CircPCLE1 processes more glucose but fails to provide sufficient energy for the spread and growth of CRC. Previous studies indicate that ACTG1 promotes glycolysis in HCC [205]. The study found circPCLE1 influences CRC glycolysis by competitively recruiting miR-485-5p to regulate ACTG1. This highlights the significance of ACTG1 in cancer glycolysis and indicates it could be a potential target for inhibiting cancer glycolysis in the future.
Zhuang et al. focused on circ_0053277 and its progression in CRC [206]. Their results showed that circ_0053277 is upregulated in colorectal cancer tissues and cells compared to normal counterparts [206]. Also, the knockdown of circ_0053277 leads to a significant reduction in CRC cell growth, aggression, migration, and angiogenesis [206]. Zhuang et al. found that circ_0053277 knockdown inhibits glycolysis in CRC cells, as evidenced by reduced glucose uptake and lactate production [206]. The current study demonstrated that circ_0053277 acts as a sponge for miR-520 h, implicating a regulatory relationship between circ_0053277 and this microRNA [206]. miR-520h targets HK1, forming a possible ceRNA network. Their results revealed that inhibiting miR-520 h restores the inhibitory effects of circ_0053277 knockdown on CRC cell progression [206]. Furthermore, the upregulation of HK1 reverses the inhibitory effects of miR-520h on CRC cell proliferation, angiogenesis, and dissemination [206]. Knockdown of circ_0053277 reduces CRC tumor growth in an in vivo xenograft model [206]. In summary, circ_0053277 promotes CRC progression by influencing various cellular processes, including proliferation, angiogenesis, migration, invasion, apoptosis, and glycolysis. The identified ceRNA network involving circ_0053277, miR-520 h, and HK1 provides insights into the molecular mechanisms underlying its regulatory functions in CRC. The findings propose circ_0053277 as a potential therapeutic target for CRC.
Xiao et al. investigated the functional role and underlying mechanism of circ_0087862 in CRC progression [207]. Their findings demonstrated that circ_0087862 and phosphoglycerate kinase 1 (PGK1) are upregulated in CRC tissues and cells [207]. Also, miR-296-3p is downregulated in CRC, creating a potential regulatory axis. Silencing circ_0087862 results in the suppression of cell proliferation, invasion, and glycolysis, along with promoting apoptosis in CRC cells [207]. These findings suggest that circ_0087862 is pivotal in facilitating tumorigenesis and glycolytic processes in CRC. In the current study, circ_0087862 is identified as a sponge for miR-296-3p, indicating a direct interaction and regulation between circ_0087862 and miR-296-3p [207].
Moreover, inhibition of miR-296-3p reverses the inhibitory effects of circ_0087862 silencing on cell proliferation, invasion, and glycolysis [207]. This suggests that the functional impact of circ_0087862 in CRC is, at least in part, mediated by its interaction with miR-296-3p. PGK1 is identified as a direct target of miR-296-3p. Overexpression of PGK1 counteracts the tumor-suppressive effects mediated by miR-296-3p, suggesting PGK1 as a downstream effector of this regulatory axis [207]. Knockdown of circ_0087862 inhibits tumorigenesis in vivo, emphasizing the clinical relevance and potential therapeutic impact of targeting circ_0087862 in CRC [207]. circ_0087862 is proposed to promote CRC progression through the miR-296-3p/PGK1 axis. In summary, circ_0087862 is identified as a key regulator in CRC, promoting tumorigenesis, invasion, and glycolysis through its interaction with miR-296-3p and subsequent regulation of PGK1. This circRNA holds promise as a potential target for therapeutic interventions in CRC.
8. Glioma
Glioma, the most common primary intracranial malignancy, accounts for over 80 % of brain malignancies [208]. Glioma is a primary cancer that affects the central nervous system and most often manifests itself in the brain. Glioma is frequently regarded as one of the kinds of cancer that presents one of the most significant therapeutic challenges because of the disease's fast development and the limits imposed on surgical intervention [209]. Temozolomide is frequently employed as an adjuvant chemotherapy for individuals with glioma, demonstrating excellent efficacy in glioma therapy. This is performed to prolong the survival time of people with glioma [210,211]. On the other hand, resistance to temozolomide is seen in about half of patients with glioma, and all patients ultimately experience therapy failure [212,213]. Consequently, elucidating the basis of disease development and tolerability of temozolomide in glioma is still critical [214].
A growing body of research indicates that many circRNAs function as oncogenes to regulate the cellular phenotype of glioma cells via multiple mechanisms [[215], [216], [217]] or perform the function of tumor suppressors [[218], [219], [220]]. CircPITX1, an oncogene found in glioma, was the main focus of this study by Guan et al. [217]. In accordance with the strategy that Lv et al. used for their research, circPITX1 was shown to be elevated in glioma tissues and in glioma U251 and LN229 cells. This was in contrast to normal tissues and NHA. This was consistent with the findings of a previous study [221]. They performed additional experiments to investigate the stability of circPITX1. The results showed that circPITX1 is resistant to RNase R digestion. Subsequent functional experiments were conducted to investigate the biological role of circPITX1 in glioma development. The researchers discovered that circPITX1 deficiency inhibited glioma cell survival, glycolysis, radioresistance clonogenicity, and tumorigenesis in-vivo, indicating that circPITX1 is an essential mediator of the development of gliomas [222]. The ceRNA hypothesis postulates that circRNA might hinder the expression of certain miRNAs, therefore influencing mRNA expression and performance. For instance, CircPITX1 stimulated miR-1304 to alter ERBB4 expression, expediting gliomas' development [223]. Xiao et al. [224] confirmed that miR-329-3p might block E2F1 in a targeted manner, therefore acting as a tumor suppressor and reducing the rate at which the cell cycle progresses in glioma. According to their findings, the expression of miR-329-3p decreased in glioma samples and in glioma U251 and LN229 cells compared to the respective controls; this finding was consistent with previous publications [224,225].
Additionally, it has been demonstrated that there was an inverse relationship between circPITX1 and miR-329-3p expression in glioma specimens. circPITX1 negatively regulates miR-329-3p, and miR-329-3p inhibitors might reverse the decrease of glycolysis and radioresistance of glioma cells caused by circPITX1 knockdown. This suggests that circPITX1's carcinogenic action was achieved by sponging miR-329-3p [222]. As the researchers showed in their study, both mRNA and protein levels of the NEK2 gene were increased in glioma tissues and cells. A subsequent experiment showed that NEK2-mediated inhibition of miR-329-effector 3p in reducing radioresistance and glycolysis could be reversed [222].
Furthermore, circPITX1 could upregulate the expression of NEK2 [217] by acting as a decoy for miR-329-3p. U251 and LN229 glioma cells were injected with a circPITX1 overexpression vector and 2-DG, a glycolysis blocker, to assess the level of radioresistance that developed by circPITX1. This was conducted to determine whether circPITX1 conferred radioresistance in glioblastoma. It seems that circPITX1 overexpression increases glycolysis and radioresistance, whereas 2-DG reverses the enhanced effect, indicating that circPITX1 knockdown reduces glycolysis to shield glioma cells from radiation treatment [222]. There are still significant gaps in the coverage provided by this study. Additional glycolysis-related genes that may signal the glycolytic pathway include monocarboxylate transporters 1 and 4 (MCT1, MCT4), glucose transporter isoform 1 (GLUT1), and HK1 [226,227]. Overall, circPITX1 was overexpressed in glioma and acted as an oncogene, as shown by suppression of circPITX1 in U251 and LN229 glioma cells, which resulted in reduced cell survival, radioresistance, glycolysis, cloning, and tumorigenesis. Glycolysis was suppressed, and radiosensitivity was enhanced in glioma cells thanks to the silencing of the circPITX1 gene that regulates the miR-329-3p/NEK2 axis [222].
The molecular function that circhHEATR5B plays in the control of glycolysis in glioma was the subject of a recent study by Song et al. [228]. They uncovered in this research that ZCRB1 upregulation enhanced newly emerging circHEATR5B expression without modifying its half-life. This indicates that ZCRB1 increased the expression circHEATR5B by boosting its establishment rather than impeding its degeneration [228]. They were curious whether HEATR5B-881aa might affect the development of GBM. Low expression of HEATR5B-881aa was observed in GBM and was shown to inhibit glycolysis and cell growth in GBM cells. They altered the start codon of circHEATR5B to show that these impacts were not caused by circHEATR5B itself. This mutation did not affect the expression or structure of circHEATR5B. All it did was prevent the protein from being appropriately translated. Rescue tests have shown that the inhibitory effect is due to the lack of the encoded HEATR5B-881aa rather than other potential functions of circHEATR5B. CircHEATR5B suppresses glycolysis and cell proliferation in GBM cells by producing HEATR5B-881aa [228]. They used Co-IP assays and mass spectrometry to investigate how HEATR5B-881aa modulated propagation and glycolysis in GBM cells. JMJD5 binds and stimulates PKM2 dimerization in breast cancer and controls glucose metabolism [229]. As a result, researchers checked PKM2 enzyme activity in GBM cells transfected with JMJD5. The results imply that JMJD5 might boost glycolysis and cell growth in GBM cells by promoting PKM2 dimerization. Dimerization of PKM2 reduces its enzymatic activity. Therefore, they detected PKM2 enzyme reactions in GBM cells transfected with JMJD5 [230,231].
HEATR5B-881aa was shown to decrease propagation and glycolysis in GBM cells by reducing the expression of JMJD5 at the protein level while not affecting JMJD5 mRNA expression. JMJD5 upregulation restored the suppression of glycolysis and propagation produced by HEATR5B-881aa. HEATR5B-881aa could reduce JMJD5 protein levels, although the underlying processes by which this occurred were not understood. Initially, JMJD5 and HEATR5B-881aa were found to interact directly with each other [228]. A higher level of S361 phosphorylation was associated with a stronger suppression of JMJD5 expression, further supporting the idea that HEATR5B-881aa inhibited JMJD5 employing S361 phosphorylation. Protein stability may be modified by phosphorylation [232]. This study provided the first evidence that low levels of the RNA-binding protein ZCRB1 inhibit circhHEATR5B synthesis, which in turn decreases the expression of the encoded HEATR5B-881aa and, finally, enhances JMJD5 stability. Excessive JMJD5 expression in GBM cells promotes aerobic glycolysis and cell proliferation via activating PKM2 dimerization. Findings from this study reveal a novel pathway via which ZCRB1/circHEATR5B/HEATR5B-881aa/JMJD5/PKM2 controls aerobic glycolysis and development in GBM cells, which may lead to new therapeutic approaches and targets for GBM treatment [228].
Luo et al. have investigated the role of CircKIF4A in accelerating glycolysis in glioma growth and temozolomide resistance [233]. Compared to normal tissues and cells, glioma tissues and cell lines showed significantly increased expression of circKIF4A. To find out how circKIF4A contributes to glioma formation, they utilized in-vitro and in-vivo loss-of-function and gain-of-function experiments. Decreased glioma cell growth and invasion potential were seen with inhibition of circKIF4A expression. Another finding was that glioma sensitivity to temozolomide was greatly enhanced by inhibiting circKIF4A. The rate of glycolysis was raised by upregulating CircKIF4A, which led to heightened glioma development and temozolomide resistance. Experiments with dual luciferase reporters and RNA immunoprecipitation were also performed to determine how circKIF4A functions in glioblastoma. ALDOA, an enzyme involved in the control of glycolysis, is regulated by circKIF4A in glioma cells through its interaction with miR-335-5p [234]. The endogenous RNA competition hypothesis postulates that mRNAs, pseudogenes, lncRNAs, and circRNAs may all control each other through a competitive sponge of common miRNAs [235]. Numerous malignant tumors have shown that miR-335-5p acts as a tumor suppressor. For example, we can mention the regulation of apoptosis and proliferation of breast cancer cells by miR-335-5p, which acts on multiple targets in the BRCA1 cascade [235]. ALDOA, a downstream target of miR-335-5p, accelerates glycolysis and stimulates human cancer growth and dissemination [236]. According to the findings of the study that was conducted by Luo and colleagues, CircKIF4A and ALDOA function as ceRNAs in glioma [233]. In summary, the results showed that overexpression of circKIF4A promotes glioma growth by binding miR-335-5p and increasing ALDOA expression. These findings have significant implications for the development of new therapeutic approaches.
Mu et al. investigated the role of circSOBP in glioma, focusing on its expression, functional impact, and underlying mechanisms [237]. The current study showed that circSOBP expression is significantly decreased in glioma cells and specimens, indicating its potential role in glioma pathogenesis [237]. Enhanced expression of circSOBP results in the mitigation of key glioma cell processes, including proliferation, invasion, migration, and glycolysis [237]. This suggests that circSOBP acts as a suppressor of glioma progression. Also, they found that circSOBP inhibits glycolysis in glioma cells, implicating its role in metabolic regulation [237]. In this study, circSOBP activates the MDA5-mediated IKKε/TBK1/IRF3 signaling pathway [237]. Mu et al. demonstrated that circSOBP binds to TKFC proteins, indicating a specific molecular interaction contributing to its regulatory functions [237]. This interaction likely plays a crucial role in modulating glioma metabolism and immune response. Their results showed that the MDA5-mediated signaling pathway activated by circSOBP leads to increased levels of interferon-I (IFN-I) [237]. Elevated IFN-I levels activate CD8+ T cells and NK cells in the immune response [237]. The results demonstrate that circSOBP inhibits glioma growth by increasing an MDA5-mediated immune system response and blocking glycolysis. Identifying circSOBP as a key player in glioma metabolism and immunological reprogramming suggests potential therapeutic strategies for tackling this challenging brain tumor.
Lei et al. examined the functional role and underlying molecular mechanisms of circYIPF6 in glioma [238]. The study findings showed that circYIPF6 is significantly upregulated in glioma tissues, suggesting its potential involvement in glioma progression [238]. They revealed that the knockdown of circYIPF6 suppresses glioma cell proliferation, indicating that circYIPF6 may promote cell growth [238]. CircYIPF6 knockdown also inhibits glycolysis, suggesting a potential role in altering metabolic processes in glioma cells [238]. CircYIPF6 acts as a sponge for miR-760, inhibiting its expression. The interaction between circYIPF6 and miR-760 suggests a regulatory axis in glioma [238]. Lei et al. results showed that the circYIPF6/miR-760 axis regulates the expression of the downstream target gene PTBP1 [238]. The suppression of PTBP1 by circYIPF6 suggests a mechanism through which circYIPF6 influences glioma cellular processes. The results of functional rescue examinations indicated that attenuating the regulatory impacts of circYIPF6 silencing in glioma cells may be accomplished by suppressing miR-760 and elevated levels of PTBP1 [238]. This lends credence to the idea that the circYIPF6/miR-760/PTBP1 axis plays a role in glioma cell growth, glycolysis, and apoptosis. Moreover, circYIPF6 knockdown effectively impedes glioma growth in a xenograft mouse model [238]. The in vivo findings validate the relevance of circYIPF6 in glioma progression. As a result, circYIPF6 modulates the miR-760/PTBP1 axis, which is responsible for glioma cell growth, glycolysis, and apoptosis. This research sheds light on circYIPF6's prospective function as a glioma therapy target.
9. Cervical cancer (CC)
CC is common and is mainly caused by human papillomavirus infection. Despite the widespread use of vaccination, the death rate from CC remains high [239]. Patients with advanced cancer have poor prognoses and treatment outcomes in CC treatment [240]. Statistics show that patients with metastases have a five-year survival rate of 16.5 %, while patients without metastases have a survival rate of 91.5 % [241]. For this reason, prevention of metastasis in people with CC increases clinical outcomes and chances of survival from this disease.
Wang et al. [54] addressed the role of circMYC in the development of CC. They discovered the oncogenic activities of circMYC in CC, including its involvement in increased cell proliferation, metastasis, and glycolysis. In addition, Wang et al. [54] studied the consequences of circMYC upregulation in CC tissues. They found a relationship between the upregulation and a poor prognosis and outcome. A newly discovered circRNA called circMYC has carcinogenic activity. Breast cancer cells have been shown to proliferate more in their presence [242]. circMYC has been shown to decrease radiosensitivity in nasopharyngeal cancer [243]. Metastasis and glycolysis were both shown to be increased by circMYC, as discovered by Wang et al. [54]. On the other hand, it was demonstrated that circMYC inhibited glycolysis in melanoma cells [244]. This variation may be attributable to the use of several cell lines derived from cancer.
Based on the findings, it was determined that the carcinogenic activity is mediated through the miR-577/MET axis [54]. A microRNA known as miR-577 can prevent the progression of several types of cancer by reducing cell proliferation and metastasis rates and stimulating apoptosis in cells [245,246]. The study also discovered that miR-577 expression was lower in CC cells and that inhibiting miR-577 had the impact of undoing the anticancer effects of circMYC silencing [54]. These data support the idea that restoring the expression of miR-577 could be a means to stop the spread of cancer. In this study, researchers investigated the relationship between circMYC and miR-577 related to CC. However, more miRNAs may be downstream targets of circMYC. Although only a few studies have focused on the relationship between circMYC and miRNA, it has been shown that circMYC may specifically target miR-1233 to control the glycolysis pathway in melanoma [244]. Therefore, further research is necessary to explore whether miR-577 is the exclusive target of circMYC in CC development. Overall, Wang et al. [54] have shown that circMYC/miR-577/MET modulates CC growth, metastasis, and glycolysis. In conclusion, they have proposed circMYC overexpression as a promising biomarker and target for cervical cancer therapy. Chen et al. [247] conducted another study in which they studied the underlying mechanisms of circCDK17 in the etiology of CC. The GSE102686 database was searched to discover circRNAs with varying expression degrees in CC tissues. Among the differentially expressed circRNA genes, the change in expression of circCDK17 was the most significant, according to the results of this study. Additionally, circCDK17 was chosen for further investigation. The subsequent analysis showed that reducing circCDK17 activity in CC cells decreased cell proliferation, migration, and invasion while inducing cell death [247].
Additionally, further investigation into the effects of circCDK17 inhibition on glycolysis was carried out [247]. The results showed that when circCDK17 is stopped, glycolysis slows down. Also, their results showed that most of the circCDK17 were in the nucleus. This suggests that circRNA plays a role in controlling the step after transcription. In addition, an experiment with a xenograft mouse model demonstrated that circCDK17 expression decreased, resulting in reduced tumor volume and mass [247]. The findings of this research indicated that the circCDK17 gene was connected to the miR-1294 gene [247]. According to specific pieces of evidence, miR-1294 served as a tumor suppressor [248,249]. Chen et al. [247] demonstrated a significant decrease in the expression of miR-1294 in CC tissues and cells. miR-1294 inhibitor decreased the inhibitory impacts of circCDK17 silencing on cell proliferation, motility, invasion, and glycolysis, as well as the boosting impact on cell death in CC. These data suggest that miR-1294 plays a negative regulatory function in the formation of CC, and circCDK17 influences CC advancement via interacting with miR-1294 [247]. The present results showed that YWHAZ plays an essential role in tumor growth [250,251]. During their research, it was discovered that YWHAZ could bind to miR-1294.
Experiments have also shown that YWHAZ rearrangement reverses miR-1294 mimetic effects on cell proliferation, motility, invasion, and death. This shows that YWHAZ had a role in the development of CC [247]. CircCDK17 and YWHAZ expression levels increased, but miR-1294 expression was reduced in CC cells or tissues. Blocking circCDK17 decreased cell growth, migration, invasion, and glucose metabolism, leading to cell death in CC via reducing YWHAZ expression through its interaction with miR-1294. Suppressing circCDK17 inhibited the formation of CC-associated tumors in vivo. These results provide a unique perspective on investigating circRNA-mediated therapies for treating CC [247].
Li et al. studied the potential mechanistic role of hsa circ _0018289 in the development of CC in their most recent work [252]. Extensive analytical experiments revealed that alteration of the miR-1294/ICMT axis is responsible for the suppressive effects of hsa _circ _0018289 silencing on CC cell proliferation, proliferation, glycolysis, and tumor growth [252]. They intended to use bioinformatics technologies to investigate the potential mechanism of hsa circ _0018289 in CC advancement. CircRNAs were used because they can function as microRNA decoys, modifying the regulation of certain genes. miR1294 stood out as very significant among these potential predictions. Both findings by Kan et al. [253] and the results presented by Li et al. [252] demonstrated the downregulation of miR-1294 in CC. Furthermore, its increased expression inhibited the proliferation and glycolysis of CC cells. Furthermore, blocking miR-1294 reduced the effectiveness of hsa_circ_0018289 knockdown in CC cells, suggesting that hsa_circ_0018289 promotes oncogenesis in CC cells by sequestering miR-1294 [252].
Significantly, miR1294 has ICMT as a target gene. Previous studies have shown that interfering with ICMT slows cell growth and promotes autophagy, indicating the protein's oncogenic importance in human malignancies [254]. There was a significant relationship between ICMT and the sensitivity of CC cells to chemotherapy [255]. Enhanced ICMT counteracted the effects of miR-1294, reducing its impact on the invasive activity of CC cells by creating the hsa_circ_0018289/miR-1294/ICMT axis [252]. Ultimately, the suppression of CC development, metastatic spread, glycolysis, and tumorigenesis by hsa_circ_0018289 knockdown suggests that hsa_circ_0018289 was a CC promoter, at least in part due to its downregulation of the miR-1294/ICMT axis. This was demonstrated by the fact that hsa_circ_0018289 knockdown suppresses tumor growth [252]. This research identified a possible therapeutic target for the prevention of CC.
Jia et al. studied the impact of circCCNB1 in CC, specifically examining its effects on cell growth, movement, invasion, glycolysis, and cell death [256]. The research discovered that circCCNB1 is significantly expressed in both squamous cell carcinoma and adenocarcinoma cells in CC tissues [256]. Downregulation of circCCNB1 suppresses cell growth, immigration, dissemination, and glycolysis in CC [256]. CircCCNB1 knockdown also induces cell apoptosis, suggesting a tumor-promoting role for circCCNB1. Jia et al. demonstrated that circCCNB1 acts as a decoy for miR-370-3p, inhibiting its production and activity in CC cells [256]. Also, circCCNB1 knockdown inhibits miR-370-3p suppression, leading to increased miR-370-3p levels. Elevated miR-370-3p levels result in decreased expression of SRY-box transcription factor 4 (SOX4) [256]. Their findings showed that blocking miR-370-3p may counteract the impacts of circCCNB1 reduction, leading to increased cell growth, movement, penetration, and glycolysis [256]. SOX4 overexpression negates the impact of miR-370-3p restoration, enhancing cell growth, movement, invasion, and glycolysis. Knocking down CircCCNB1 inhibits the development of CC in an in vivo xenograft model, validating its function in tumor suppression [256]. Jia et al. revealed that circCCNB1 knockdown blocks CC development by targeting the miR-370-3p/SOX4 pathway [256]. The regulatory axis involves circCCNB1 acting as a miR-370-3p sponge, leading to reduced suppression of SOX4 by miR-370-3p. In conclusion, the study highlights the significance of circCCNB1 in CC development, demonstrating its role as a miR-370-3p sponge and regulator of SOX4 expression. The identified circCCNB1/miR-370-3p/SOX4 pathway offers potential therapeutic avenues for CC intervention.
10. Breast cancer
Breast cancer has become the world's second-greatest cause of mortality for women, with a dramatic rise in recent years [257,258]. Despite remarkable progress in breast cancer therapy in recent decades, the disease continues to cause a high rate of death and disability [259,260]. The current body of literature demonstrates the complexity of the initiating elements leading to breast cancer, which include hormone imbalances, inheritance, and environment. In addition to the causes mentioned above, epigenetic alterations contribute significantly to the formation of breast cancer [261]. It is vital to unravel the molecular processes that are at play in order to get a better understanding of the genesis of breast cancer, particularly its proliferation and growth [262,263]. Moreover, there is an urgent need for clinically significant changes, such as improved coping strategies and, more specifically, focused therapies. New evidence lends credence to the fundamental roles that circRNA plays in breast cancer, which are discussed below.
Breast cancer tumor growth involves several pathogenic variables. In addition, circRNA may affect tumor characteristics, especially proliferation and aerobic glycolysis, the primary energy source for the breast cancer microenvironment [92]. There is growing evidence that circRNAs play important roles in breast cancer, particularly in apoptosis [264], inhibition of p53 mutational function [265], and dissemination [266]. Cao et al. [267] Cao et al. [258] found that the circRNA known as circRNF20 harbors the miRNA known as miR-487a, which enables it to perform the function of a miRNA sponge. In addition, miR-487a may target the 3′-UTR of HIF-1 to inhibit the production of HIF-1 protein. In addition, miR-487a might target the 3′-UTR of HIF-1α to decrease HIF-1α protein. The circRNF20/miR-487a/HIF-1α axis plays a key function in tumor energy metabolism since HIF-1α is essential for aerobic glycolysis [268,269]. Notably, HK2 transcription may be enhanced by targeting the HIF-1α promoter. Breast cancer cells are driven to glycolysis by the circRNF20/miR-487a/HIF-1α/HK2 axis. Overall, the results of this work clarify the importance of circRNF20 in breast cancer development and the Warburg effect. CircRNF20 acts as a miRNA sponge by sequestering miR-487a and guiding it to the 3′-UTR region of HIF-1α. Additionally, HIF-1 could bind to the HK2 promoter, which increased HK2 transcription. This research highlights the importance of the circRNF20/miR-487a/HIF-1/HK2 axis in the development of breast cancer. Zan et al. investigated how circ-CSNK1G1 contributes to the progression of TNBC [270]. The study examined the importance and operational process of circ-CSNK1G1 in TNBC, revealing a notable increase in circ-CSNK1G1 expression in both tumor tissues and cells. The circ-CSNK1G1-regulated miR-28-5p/LDHA axis has a crucial role in inhibiting TNBC development in functional experiments. Their study emphasized both the importance of circ-CSNK1G1 in TNBC and its possible mechanism of action [270]. The upregulation of circRNA CSNK1G1 was first identified in breast cancer tissues by a circRNA expression analysis [271].
Moreover, it was shown that CRC had elevated circ-CSNK1G1 expression [272]. Furthermore, their investigation validated the heightened expression of circ-CSNK1G1 in TNBC tissues and cells. In a research conducted by Zan and colleagues, circ-CSNK1G1 knockdown inhibited tumor cell proliferation, colony formation, motility, invasion, and glycolytic energy consumption. They also discovered that this inhibition also occurs in living organisms, halting tumor growth and progression. They determined that circ-CSNK1G1 is a major oncogenic component of TNBC [270].
As reported in previous research, the development of many malignancies, including HCC and CRC, was inhibited by miR-28-5p [273,274]. An exciting study has demonstrated the antitumor function of miR-28-5p in breast cancer. The findings indicate that the absence of miR-28-5p effectively reversed the proliferation, motility, and invasion of breast cancer cells that were suppressed by lnc-MCM3AP-AS1 [275]. Zan et al. [270] performed an experiment showing that the absence of miR-28-5p increased the inhibitory impact of circ-CSNK1G1 knockdown on cell growth, glycolysis, motility, invasion, and colony formation, whereas restoring miR-28-5p suppressed these malignant activities. Researchers have shown a strong correlation between LDHA expression and TNM grade and metastatic distance in breast cancer [276]. In addition, the role of LDHA in glucose metabolism is widely recognized. For example, breast tumor cells overexpressing LDHA have increased growth and glycolysis [277]. The ability of miR-30a-5p to lessen the induction of LDHA glycolysis in the body's energy metabolism prevented the development and proliferation of mammary tumors [278]. In a study performed by Zan et al. [263], similar results were obtained when LDHA was overexpressed; the impacts of miR-28a-5p recovery were nullified, and TNBC cell growth, motility, invading, and glycolysis were restored.
This study is the first investigation to establish the significance of circ-CSNK1G1 in TNBC. Zhan et al. have offered insights into the relationships and processes of circ-CSNK1G1 in TNBC [270], which elucidated the pathophysiology of TNBC from the point of view of circRNA deregulation. They only concentrate on a single miRNA/mRNA route that impacts circ-CSNK1G1. Additional study is required to clarify the involvement of various miRNAs and downstream mRNAs in circ-CSNK1G1-regulated complexes in TNBC [270].
Recent research by Li et al. [279] discovered the molecular role of Circ-RPPH1 in the development of breast cancer. Analysis of the GEO database showed that circ-RPPH1 was significantly higher in breast cancer tissues, indicating a possible link between circ-RPPH1 and the development of breast cancer. Analyzed studies observed an upregulation of circ-RPPH1 in breast cancer tissue in both clinical samples and cells. Additional proof that decreased circ-RPPH1 expression inhibits tumor development in vivo was discovered in a mouse xenograft model [279]. The research demonstrated that blocking circ-RPPH1 decreased glucose intake and lactate generation in cancer cells, indicating that reducing circ-RPPH1 levels inhibits glycolysis in breast cancer cells [279]. Recent studies indicate that glycolysis is a distinguishing feature of tumor cells. Cancer cells often rely on aerobic glycolysis for energy, even in the presence of oxygen, enabling them to outcompete normal cells for glucose and sustain their abnormal proliferation [280,281]. Reducing the amount of circ-RPPH1 successfully hindered the growth and spread of breast cancer in the study [279].
The possibility of using circ-RPPH1 as a miRNA sponge was also analyzed. Due to its involvement in the development of breast cancer, miR-328-3p was chosen for more investigation using bioinformatics [282,283]. Circ-RPPH1 functions as a ceRNA for miR-328-3p, regulating its expression in breast carcinoma cells. Re-expression of miR-328-3p attenuated cellular aggressive oncogenic properties and cellular glycolysis in breast cancer, despite miR-328-low 3p expression levels in breast cancer cells and tissues. Circ-RPPH1 inactivation in breast carcinoma cells was found to be reversed by inhibiting miR-328-3p, which restored its inhibitory action. HMGA2 was first identified as a target of miR-328-3p in breast tumor cells, followed by the investigation of other molecular targets of miR-328-3p [279]. Their research has shown that circ-RPPH1 enhances malignant cell traits and glycolysis in breast cancer cells by inhibiting miR-328-effect 3p on HMGA2, shedding light on breast cancer and proposing a novel treatment strategy for the disease.
11. Prostate cancer (PCa)
One of the most prevalent human urological tumors in recent years is aggressive PCa, which develops from the prostate's epithelial cells [284,285]. Most individuals with PCa have already progressed to a locally advanced stage or metastasized before diagnosis due to the lack of obvious early symptoms [286,287]. PCa often presents as urinary tract obstructive symptoms with bone discomfort and pathologic fractures following bone metastasis [288,289]. Understanding the molecular process of PCa has significant theoretical implications for PCa treatment. Certain studies have proven the importance of some dysregulated circRNAs in developing PCa [290]. However, the molecular basis by which most circRNAs contribute to PCa is currently not elucidated.
Many circRNAs have a significant role in the progression of PCa. CircFOXO3 was discovered to be increased in prostate cancer tissues, potentially enhancing cancer cell proliferation and inhibiting cell death via influencing the miR-29a-3p/SLC25A15 axis [291]. However, circUCK2 inhibited PCa growth and invasion by upregulating TET1 expression through the miR-767-5p sponge, suggesting that it may act as a tumor suppressor in PCa [292]. Ding et al. [293] selected circMID1, a circRNA predominantly expressed in H-PCa tissues, to investigate its role in PCa development. They confirmed the circular structure of circMID1 through an in vitro investigation. According to loss-of-function experiments, the knockdown of circMID1 inhibited the growth, glycolysis, motility, and invasion of PCa cells. Importantly, their results demonstrated that circMID1 downregulation decreased PCa carcinogenesis in vivo. These results provide further evidence that circMID1 participates in the growth of PCa tumors [293]. As shown in several studies, IGF1R has been identified as an oncogene that regulates the malignant development of cancer through the AKT signaling pathway [294,295]. CircMID1 has decreased the IGF1R and p-AKT/AKT expression, indicating that circMID1 could activate the IGF1R/AKT pathway. Prior studies have shown that YTHDC2 expression has a varying role in the malignant advancement of several cancers [296,297]. YTHDC2 expression was considerably greater in H-PCa tissues than in L-PCa tissues, and its regulation was influenced by circMID1 [293]. Thus, in line with other study findings, their results imply that YTHDC2 may favorably control the synthesis of IGF1R protein [298]. They employed bioinformatics to identify miRNAs that might be sequestered by circMID1 and regulate YTHDC2 and IGF1R. The researchers found that miR-330-3p might potentially communicate with circMID1, YTHDC2, and IGF1R. MiR-330-3p suppressed tumor development in several types of cancers [299,300]. Ding et al. found that blocking miR-330-3p partly reversed the inhibitory impact of circMID1 depletion on PCa development and IGF1R/AKT expression. Conversely, increasing pcDNA YTHDC2 reverted the influence of sh-circMID1 on PCa progression and IGF1R/AKT expression [293]. The results suggest that circMID1 regulates PCa progression via modulating the YTHDC2/IGF1R/AKT pathway by acting as a sponge for miR-330-3p. In conclusion, their study identified a new circular RNA that controls the progression of PCa. The research demonstrated that circMID1 enhances PCa cell growth, movement, invasion, and glycolysis to accelerate the development of tumors by influencing the miR-330-3p/YTHDC2/IGF1R/AKT pathway [293]. Overall, their results identified a novel prospective treatment option for PCa, indicating that targeting circMID1 suppression may be a viable way of managing PCa.
Guan et al. [301] studied the pathophysiological processes underlying CircTRRAP in PCa development. A recent study found that circTRRAP was increased in PCa specimens with spinal metastasis. Exosomal circTRRAP was shown to enhance cell EMT by interacting with miR-1 and upregulating MAP3K1 expression in PCa [302]. This study's findings verified the increased expression of circTRRAP in PCa samples in comparison to normal samples. Deleting circTRRAP in mice and xenograft models decreased PCa cell survival and hindered PCa cell proliferation, movement, invasiveness, and tumor growth. CircTRRAP enhances PCa progression and plays a role in glycolysis in PCa cells. It enhances the ability of PCa cells to invade and glycolysis via the miR-515-5p/HOXA1 pathway [301]. Silencing circTRRAP reduced glucose and lactate production uptake in PCa cells, suggesting circTRRAP's involvement in glycolysis in PCa [301]. Li and colleagues examined the potential connection between circTRRAP and miR-515-5p [301]. It has been discovered that MiR-515-5p acts as a suppressor in several different cancer types [140,303]. In PCa, overexpression of circ-0057553 enhanced tumor cell aerobic glycolysis, invasion, and motility through down-regulation of miR-515-5p [304]. Guan et al. [301] discovered that miR-515-5p was reduced in prostate cancer tissues and cells. Knocking down MiR-515-5p restored the inhibitory effects of circTRRAP silencing on carcinogenesis and glycolysis in PCa cells. The results confirmed that circTRRAP enhances the invasiveness and glycolysis of PCa cells by interacting with miR-515-5p [301].
Zhou et al. investigated the role of circROBO1 in PCa progression and resistance to enzalutamide, a common treatment for PCa [305]. The study seeks to fill the knowledge gap regarding the molecular pathways and roles of circRNAs in prostate cancer. It identified circROBO1 as notably upregulated in both prostate cancer cells and tissues. Inhibiting circROBO1 resulted in reduced proliferation of prostate cancer cells and enhanced sensitivity to enzalutamide treatment [305]. Significantly, the inhibition of circROBO1 notably decreased the glycolysis rate, which played a pivotal role in suppressing prostate cancer growth and overcoming resistance to enzalutamide [305]. The research findings shed light on the biological significance of the circROBO1-miR-556-5p-PGK1 axis in prostate cancer growth and resistance to enzalutamide [305]. This axis emerges as a promising therapeutic target for prostate cancer treatment [305]. Overall, Zhou et al. offer valuable insights into the involvement of circRNAs, particularly circROBO1, in the progression of prostate cancer and its resistance to enzalutamide, suggesting potential avenues for targeted therapy in addressing this cancer.
HOXA1 is part of the homeobox transcription factor family responsible for the coordinated regulation of pattern formation during embryogenesis, morphogenesis, and adipogenesis [306]. Growing data suggests that HOXA1 dysregulation has a role in cancer development [[307], [308], [309]]. An abundance of HOXA1 was found in PCa tissues and cells, according to Guan et al. [301], and higher amounts of HOXA1 decreased the aggressiveness of PCa cells and the miR-515-5p-mediated suppression of cellular glycolysis. Moreover, circTRRAP regulates HOXA1 expression by selectively sponging miR-515-5p, highlighting the network function of the circTRRAP/miR-515-5p/HOXA1 pathway in PCa cells [301]. As a whole, circTRRAP was found to be highly expressed in PCa cells and samples. Furthermore, the suppression of circTRRAP decreased the invasiveness and glycolysis of PCa cells by downregulating the expression of sponge-like HOXA1 and sequestering miR-515-5p. This research identified a novel route for PCa development.
12. CircRNAs regulate glycolysis and promote chemoresistance
The capacity of cancer cells to escape or adapt in the presence of treatments is known as chemoresistance and represents a significant hurdle that oncology is trying to understand and overcome. In response to conventional chemotherapy drugs, several molecular pathways have been found that allow cancer cells to maintain their longevity and escape apoptosis. These mechanisms allow cancer cells to resist the death process. These mechanisms consist of different signal transduction pathways and may enhance chemoresistance in response to various stimuli due to their capacity to initiate [310,311]. Consequently, elucidating these molecular processes, discovering therapeutic targets, screening the chemotherapy-sensitive population, and determining the effectiveness of chemical responses by cell culture and orthotopic models are critical for implementing targeted, personalized, and combination therapy [310].
Changes in glycolysis may alter the response of tumors to chemotherapy drugs, particularly increases in key enzymes and byproducts of this pathway. This leads to increased ATP production, which provides adequate energy for the cellular activities within the tumor cells [312,313]. This strategy increases the DNA damage repair process more effectively, increases the activity of enzymes related to phosphorylation, transport, and autophagy, and induces chemical resistance [314]. Additional clinical studies have been conducted to evaluate further the potential benefits of combining glycolysis-targeted drugs with clinical first- and second-line chemotherapy agents in treating malignancies [312].
Even though current advances in treatment, including combination drug administration, targeted therapies, and immunotherapies targeting malignant tumors, have helped reduce cancer-related mortality, cancer cells even now show the potential for drug resistance [315]. CircRNAs have been critical in the induction of chemoresistance in gastrointestinal cancers in recent years. A circRNA, circDDX17, was shown to sensitize CRC cells to 5-FU and induce apoptotic cell death via miR-31-5p/ankyrin-containing kidney axis 1 [316]. Evidence suggests that circRNAs may regulate glycolysis in tumor cells [317]. Wang et al. [317] recently analyzed the function of exosome-delivered circRNA in regulating glycolysis and chemoresistance in CRC. Recent advances that are targeting ABC transporters as therapeutic strategies, such as P-gp antagonists, RNA interference, and nanomedicine, are encouraging because they may provide a way to overcome chemoresistance in cancers. However, this field of study has not yet been performed [318], and the presentation of hybrid approaches [319] could not achieve its desired result [320]. In this research, the researchers investigated the consumption of ATP ABC drug efflux pumps instead of trying to disrupt the transporters themselves. Aerobic glycolysis is the primary energy source for most malignant solid tumors, especially CRC [321]. Evidence suggests that drug-resistant cells need more ATP to sustain their life-sustaining pathways when exposed to genotoxic stress, in contrast to drug-sensitive cells [322]. The drug-sensitive SW480 cell line showed increased glycolysis and ATP generation when PKM2 was elevated. The transporters may obtain the necessary power to remove oxaliplatin from cells, drastically decreasing their susceptibility to the medication [317]. The circRNA pattern showed differential expression in oxaliplatin-resistant CRC cells. ciRS-122, a circRNA, was anticipated to function as a miR-122 sponge. Drug-resistant cells release exosomes containing ciRS-122 to drug-sensitive cells, enhancing glycolysis and drug resistance via the upregulation of PKM2 expression and downregulation of miR-122 [317]. Wang et al. [310] employed si-ciRS-122 to block the ciRS-122/miR-122/PKM2 pathway to improve drug sensitivity. Isolation of exosomes from HEK293T cells to administer sicRS122 with improved bioactivity in vivo was a key step towards translational therapy. This study provided evidence that exosiRNA ciRS-122 may regulate the ciRS-122/miR-122/PKM2 pathway to inhibit glycolysis and overcome oxaliplatin resistance [317]. Overall, ciRS-122 was transported to oxaliplatin-sensitive cells through exosomes isolated from oxaliplatin-resistant CRC cells, where it enhanced PKM2 expression and improved glycolysis and treatment failure. Furthermore, in vivo targeting of ciRS122 with exosome-delivered siRNA increased treatment response, indicating a potentially novel method for reversing oxaliplatin resistance in CRC. Because chemoresistance causes cancer recurrence, tumor development, and death in cancer patients, it is critical to identify techniques to overcome inherent and acquired drug resistance. It is critical to understand the elements that lead to chemical resistance in order to successfully develop solutions to drug resistance. Researchers should also look towards developing medicines to limit circRNAs synthesis in tumor cells. They must also understand more about the role of circRNAs in the regulation of glycolysis in cancer chemoresistance.
13. Conclusion and future perpspective
Cancer is consistently ranked as one of the leading causes of death on a global scale, and during the past decade, a significant amount of experimental research has been initiated to discover new treatments or innovative methods for cancer diagnosis or prognosis. The outcomes of several types of cancer, including NSCLC, are still dismal, even with recent progress in targeted therapy, immunotherapy, and chemotherapy therapeutic interventions [68]. A greater understanding of the mechanisms that drive carcinogenesis and development is required to enhance NSCLC diagnosis and therapy. CircRNAs are a kind of lncRNA with a distinct structure and may be exploited as biomarkers in human cancers. Furthermore, the role of circRNA in carcinogenesis has been extensively established. Unsurprisingly, various circRNAs have distinct effects. Some circRNAs, for example, influence GC advancement as cancer-promoting agents, such as circ0002570 [323] and circC6orf132 [324], whereas others mediate GC growth as cancer inhibitory variables, such as circTNPO3 [325] and circGSK3B [326].
The importance of glycolysis as a source of energy production for tumor cells has been demonstrated by increasing research [327,328]. Many cancers rely on glycolysis as their primary mechanism for obtaining nutrients and energy. Some cancerous characteristics linked to this metabolic rewiring include accelerated growth, resistance to chemotherapy, and immune system evasion [[23], [24], [25]]. Since cancer cells depend on glycolysis to influence carcinogenesis, targeting glycolysis has a significant chance of being effective in treating malignancies [329]. This overview explained how circRNA could modulate cancer glycolysis by interacting with various molecular networks (for more details, see Table 1). Several circRNAs, such as circUBE2D2 [50], circ_0136666 [66], and circCDK17 [247], are examples of circRNAs that can promote glycolysis and the advancement of cancer. In addition, certain circRNAs, such as circRPN2 [64], circTADA2A [67], and circ_0006677 [165], can suppress glycolysis and the development of cancer. In general, the main reason for cancer treatment failure is the lack of effective therapies and precise molecular targets.
Table 1.
The role and action mechanisms of circRNAs in glycolysis in various types of cancer.
| Type of cancer | Circular RNA | Role in glycolysis | Mechanism of action | Ref |
|---|---|---|---|---|
| HCC | circBNC2 | Promotes glycolysis | The miR-217/HMGA2 axis is a mechanism by which circBNC2 promotes glycolysis and HCC stemness. | [330] |
| HCC | circRPN2 | Inhibits glycolysis | circRPN2 suppresses HCC glycolysis and spread by promoting the decomposition of ENO1 and controlling the miR-183–5p/FOXO1 pathway. | [64] |
| HCC | circUBE2D2 | Promotes glycolysis | circUBE2D2 had an oncogenic function by affecting the miR-889-3p/LDHA axis. | [50] |
| HCC | circMAT2B | Promotes Glycolysis | CircMAT2B advanced HCC development by increasing glycolysis via activating the circMAT2B/miR-338-3p/PKM2 pathway under hypoxic conditions. | [61] |
| HCC | circ_0008450 | Promotes glycolysis | By downregulating AKAP1 via miR-431, circ_0008450 suppression slowed the development of HCC in hypoxic environments. | [62] |
| HCC | circ-PRMT5 | Inhibits glycolysis | circ-PRMT5 may function as a miR-188-5p sponge to control HK2 expression. | [63] |
| HCC | circ-CFH | Promotes glycolysis | The circ-CFH/miR-377-3p/RNF38 axis mediated the development of HCC cells. | [104] |
| HCC | circRHBDD1 | Promotes Glycolysis | circRHBDD1 binds the m6A reader YTHDF1 to PIK3R1 mRNA, accelerating its translation in a m6A-dependent way. | [110] |
| HCC | circ_0000517 | Promotes Glycolysis | By regulating miR-326 and IGF1R, circ_0000517 knockdown inhibited the development of HCC in both in vitro and in vivo. | [75] |
| Liver cancer | circTUBGCP5 | Promotes Glycolysis | circ-TUBGCP5 significantly influences the aggressive characteristics and glycolysis of liver cancer by regulating the circThe TUBGCP5/miR-144-3p/ACSL4 axis | [331] |
| GC | circRPS19 | Promotes glycolysis | Through the miR-125a-5p/USP7 axis, circRPS19 stabilized the HK2 protein, causing aerobic glycolysis in GC cells and accelerating the growth of GC. | [155] |
| GC | circ_0067514 | Inhibits glycolysis | circ_0067514 inhibited the progression of GC and glycolysis by affecting the miR-654-3p/LATS2 pathway. | [332] |
| GC | circUBE2Q2 | Promotes Glycolysis | circUBE2Q2 controls GC development via the circUBE2Q2-miR-370-3p-STAT3 axis and facilitates tumor dissemination through exosomal communications. | [51] |
| GC | circ_0006089 | Promotes Glycolysis | GC development was facilitated via the circ_0006089/miR-361-3p/TGFB1 axis. | [143] |
| GC | circDNMT1 | Promotes Glycolysis | GC expansion, migration, invasion, and glycolysis were all reduced by circDNMT1 knockdown via the sponging miR-576-3p/HIF-1a axis. | [148] |
| GC | circCUL3 | Promotes Glycolysis | By controlling the STAT3/HK2 Axis, circCUL3 increases GC glycolysis development. | [140] |
| LUAD | circP4HB | Promotes Glycolysis | circP4HB may enhance the advancement of LUAD, suggesting circP4HB might be a promising treatment target for LUAD. | [179] |
| LUAD | Circ_0001777 | Inhibits glycolysis | Circ_0001777 inhibited the advancement of lung adenocarcinoma via interacting with miR-942-5p to promote PRICKLE2 expression. | [333] |
| NSCLC | circRARS | Promotes Glycolysis | circRARS can promote glycolysis and tumor progression in NSCLC by regulating LDHA. | [177] |
| NSCLC | Circ_0003028 | Promotes Glycolysis | Circ_0003028 may enhance the aggressive characteristics and glycolytic ability of NSCLC cells via a mechanism perhaps involving miR-1305 or the miR-1322/SLC5A1 axis. | [178] |
| NSCLC | circEHD2 | Promotes Glycolysis | Through the adsorption of miR-3186-3p, circEHD2 promotes FOXK1 and promotes the growth and glycolysis of NSCLC while inhibiting autophagy and apoptosis. | [169] |
| NSCLC | circSLC25A16 | Promotes Glycolysis | circSLC25A16 interacts with miR-488-3p/HIF-1α, which activates LDHA | [163] |
| NSCLC | circSHKBP1 | Promotes Glycolysis | Through the miR-1294/PKM2 axis, exosomal circSHKBP1 contributed to the advancement of NSCLC. | [173] |
| NSCLC | circ_0001421 | Promotes Glycolysis | Through modulating the miR-409-3p/TMEM14A axis, Circ 0001421 led to the development of NSCLC. | [159] |
| NSCLC | circMAGI3 | Promotes Glycolysis | circMAGI3 acted as a sponge for miR-515-5p, reducing HDGF expression and facilitating glycolysis in NSCLC. | [52] |
| NSCLC | circ_0006677 | Inhibits glycolysis | Through regulating the miR-578/SOSC2 axis, Circ 0006677 inhibits the development of NSCLC. | [165] |
| CRC | Circ_0053277 | Promotes Glycolysis | Circ_0053277 enhanced CRC cell proliferation, blood vessel formation, spread to other body parts, and glycolysis via the miR-520 h/HK1 pathway, suggesting that circ_0053277 might be a promising target for CRC therapy. | [334] |
| CRC | Circ_0087862 | Promotes Glycolysis | Circ_0087862 enhances cancer formation and glucose metabolism in CRC by acting as a decoy for miR-296-3p to control PGK1 levels. | [207] |
| CRC | circPCLE1 | Promotes Glycolysis | The miR-485-5p/ACTG1 axis is modulated by circPCLE1, which accelerates EMT, glycolysis in CRC, and M2 macrophage polarization. | [65] |
| CRC | circDENND4C | Promotes Glycolysis | circDENND4C promoted CRC cells' growth, movement, and glucose metabolism via controlling GLUT1 by acting as a sponge for miR-760. | [53] |
| CRC | circ_0136666 | Promotes Glycolysis | Circ_0136666 promoted the formation of colorectal cancer tumors using the circ_0136666/miR-383/CREB1 pathway in living organisms. | [66] |
| CRC | CircTADA2A | Inhibits glycolysis | CircTADA2A acted as a tumor suppressor in CRC by suppressing glycolysis and cell cycle progression and enhancing apoptosis of cancer cells via the miR-374a-3p/KLF14 pathway. | [67] |
| CRC | circNOX4 | Promotes Glycolysis | circNOX4 might serve as an oncogene in CRC by enhancing the growth, movement, infiltration, and glycolysis of CRC cells via the miR-485-5p/CKS1B pathway. | [197] |
| Colon cancer | circPLOD2 | Promotes Glycolysis | circPLOD2 facilitated the advancement of colon cancer by affecting the miR-513a-5p/SIX1/LDHA pathway. | [335] |
| Glioma | CircSOBP | Inhibits glycolysis | CircSOBP inhibits the advancement of glioma by interfering with glycolysis and enhancing the MDA5-mediated immune system reaction. | [237] |
| Glioma | circROBO1 | Promotes glycolysis | circROBO1 enhances glioma proliferation and resistance to enzalutamide by boosting glycolysis. | [336] |
| Glioma | CircYIPF6 | Promotes glycolysis | CircYIPF6 targets miR-760 to regulate the expression of PTBP1, influencing the proliferation, cell death, and glucose metabolism in glioma cells. | [238] |
| Glioma | CircKIF4A | Promotes glycolysis | circKIF4A promotes glioma advancement by binding to miR-335-5p and increasing ALDOA expression. | [233] |
| Glioma | circPITX1 | Promotes Glycolysis | Reducing glycolysis by CircPITX1 knockdown enhances radiation sensitivity in glioma by affecting the miR-329-3p/NEK2 axis. | [222] |
| Glioma | circHEATR5B | Inhibits glycolysis | Gymolysis and proliferation in gliomas are regulated via the ZCRB1/circHEATR5B/HEATR5B-881aa/JMJD5/PKM2 pathway. | [228] |
| CC | CircCCNB1 | Promotes Glycolysis | Downregulating CircCCNB1 inhibits the progression of CC via directing the miR-370-3p/SOX4 circuit. | [256] |
| CC | CircCDK17 | Promotes Glycolysis | In CC, reducing CircCDK17 expression increased cell death via the miR-1294/YWHAZ axis while downregulating cell growth, migration, invasion, and glycolysis. | [247] |
| CC | Hsa_circ_0018289 | Promotes Glycolysis | Hsa_circ_0018289 promotes cancer progression by controlling the miR-1294/ICMT pathway. | [252] |
| CC | circMYC | Promotes Glycolysis | circMYC enhances the development of CC via controlling the miR-577/MET axis. | [54] |
| BC | Circ-CSNK1G1 | Promotes Glycolysis | Through its regulation of the miR-28-5p/LDHA pathway, Circ-CSNK1G1 enhances cell movement, invasion, growth, and glycolysis metabolism throughout the progression of TNBC. | [270] |
| BC | circRNF20 | Promotes Glycolysis | circRNF20 sequesters miR-487a, functioning as a miRNA sponge. Subsequently, miR-487a targets the 3′-UTR of HIF-1α, which in turn binds to the promoter of HK2 and enhances its production. | [267] |
| BC | circ-RPPH1 | Promotes Glycolysis | Reducing Circ-RPPH1 inhibited aggressive cell characteristics and glycolysis via BC's miR-328-3p/HMGA2 pathway. | [279] |
| PCa | circROBO1 | Promotes Glycolysis | circROBO1 enhances PCa proliferation and resistance to enzalutamide by increasing glycolysis. | [305] |
| PCa | circLPAR3 | Promotes Glycolysis | By targeting JPT1 with microRNA-513b-5p, circular RNA LPAR3 promotes glycolytic activation while suppressing PCa radiosensitivity. | [337] |
| PCa | circTRRAP | Promotes Glycolysis | circTRRAP contributed to the advancement of PCa and glycolysis via regulating the miR-515-5p/HOXA1 pathway. | [301] |
| PCa | circMID1 | Promotes Glycolysis | circMID1 facilitated the advancement of PCa via controlling the miR-330-3p/YTHDC2/IGF1R/AKT pathway. | [293] |
| RCC | circCOL5A1 | Promotes Glycolysis | circCOL5A1 regulates the malignant behavior of RCC by modulating glycolysis. | [338] |
| RCC | Circ-CSPP1 | Promotes Glycolysis | Circular RNA CSPP1 promotes the development of RCC and glycolysis via interacting with RAC1 via microRNA-493-5p. | [339] |
Although the study of circRNAs in cancer glycolysis has yielded valuable insights into tumor progression and therapeutic approaches, several limitations persist that warrant attention in future research endeavors. Many studies investigating the role of circRNAs in cancer glycolysis have been hindered by small sample sizes and heterogeneous patient populations. Future investigations should strive to incorporate larger cohorts with well-defined clinical characteristics to bolster the reliability and applicability of results. Despite the identification of numerous circRNAs as regulators of glycolytic pathways in cancer, the functional roles of many circRNAs remain poorly understood. Future research efforts should concentrate on unraveling the specific mechanisms through which circRNAs modulate glycolysis, including their interactions with key regulatory proteins and signaling pathways. Moreover, the impact of circRNAs on cancer glycolysis may vary depending on the biological context and tumor microenvironment. Subsequent studies should explore circRNA expression patterns and functional effects across diverse cancer types, stages, and microenvironmental conditions to gain a deeper understanding of their context-specific roles in tumor metabolism. While circRNAs show promise as potential therapeutic targets for cancer treatment, strategies for therapeutically targeting circRNAs are still in their early stages of development. Future research should focus on developing innovative therapeutic approaches, such as circRNA-based inhibitors or delivery systems, and evaluating their efficacy in preclinical and clinical settings. Integrating circRNA expression profiles with other omics data, such as transcriptomics, proteomics, and metabolomics, can provide a more comprehensive understanding of the regulatory networks governing cancer glycolysis. Future studies should leverage multi-omics approaches to uncover novel circRNA-glycolysis interactions and identify potential biomarkers or therapeutic targets. Advances in bioinformatics and computational tools have facilitated the identification and analysis of circRNAs; however, there is still a need for improved algorithms and resources tailored specifically for studying circRNA-mediated regulation of glycolysis. Future research should focus on developing more sophisticated computational methods for predicting circRNA functions and interactions within the glycolytic network. Due to the inherent complexity of circRNA research, validation of experimental findings and reproducibility across independent studies are critical challenges. Future research should prioritize rigorous validation of circRNA-glycolysis interactions using complementary experimental techniques and independent patient cohorts to ensure the reliability and reproducibility of results. Therefore, discovering new molecular therapeutic targets for cancer is very important. As such, circRNAs may serve as a useful tool in the future for treatment, diagnosis, and prognosis. More research is needed to fully understand the role of circRNA in cancer development and how it contributes to its progression.
Ethics approval and consent to participate
Not applicable.
Consent for publication
All authors have read and agreed to the published version of the manuscript.
Funding
None.
Declaration of competing interest
The authors declare they have no conflict of interest.
Conflict of interest
None.
CRediT authorship contribution statement
Chou-Yi Hsu: Writing – review & editing, Writing – original draft, Investigation, Conceptualization. Ahmed Faisal: Writing – original draft, Methodology, Investigation. Sally Salih Jumaa: Methodology, Investigation. Nataliya Sergeevna Gilmanova: Methodology, Investigation. Mohammed Ubaid: Methodology, Investigation. Aya H. Athab: Methodology, Investigation. Rasoul Mirzaei: Writing – review & editing, Writing – original draft, Methodology, Investigation, Conceptualization. Sajad Karampoor: Writing – review & editing, Writing – original draft, Methodology, Investigation, Conceptualization.
Acknowledgment
None.
Abbreviations
- HCC
hepatocellular carcinoma
- ENO1
Enolase 1
- FOXO1
Forkhead box protein O1
- LDHA
Lactate dehydrogenase A
- PKM2
Pyruvate kinase M2
- AKAP1
A-kinase anchor protein 1
- HK2
Hexokinase 2
- PIK3R1
Phosphoinositide-3-Kinase Regulatory Subunit 1
- IGF1R
Insulin-like growth factor1 receptor
- GC
gastric cancer
- CC
cervical cancer
- BC
breast cancer
- Pca
prostate cancer
- CRC
colorectal cancer
- ALDOA
Aldolase A
- NEK2
NIMA-related kinase 2
- TNBC
triple-negative breast cancer
- HIF-1α
hypoxia-inducible factor-1α
- HK2
hexokinase II
- ACSL4
Acyl-CoA Synthetase Long Chain Family Member 4
- HMGA2
High Mobility Group AT-Hook 2
- EMT
epithelial mesenchymal transformation
- CRC
Colorectal cancer
- TAM
tumor-associated macrophage
- ACTG1
γ-Actin Gene
- SIX1
SIX Homeobox 1
- GLUT1
Glucose transporter 1
- LATS2
large tumor suppressor kinase 2
- TGFB1
transforming growth factor-β1
- STAT-3
signal transducer and activator of transcription 3
- HOXA1
homeobox A1
- NSCLC
Non-small cell lung cancer
- FOXK1
Forkhead Box K1
- PRICKLE2
prickle planar cell polarity protein 2
- BTG2
BTG anti-proliferation factor 2
- TMEM14A
transmembrane protein 14A, HDGF
- SOSC2
suppressor of cytokine signaling 2
- YWHAZ
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein zeta
Contributor Information
Rasoul Mirzaei, Email: rasul.micro92@gmail.com.
Sajad Karampoor, Email: sajadkarampour1987@gmail.com.
References
- 1.Siegel R.L., et al. Cancer death rates in US congressional districts. CA A Cancer J. Clin. 2015;65(5):339–344. doi: 10.3322/caac.21292. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R.L., et al. Colorectal cancer statistics, 2020. CA A Cancer J. Clin. 2020;70(3):145–164. doi: 10.3322/caac.21601. [DOI] [PubMed] [Google Scholar]
- 3.Joshi S.S., Badgwell B.D. Current treatment and recent progress in gastric cancer. CA A Cancer J. Clin. 2021;71(3):264–279. doi: 10.3322/caac.21657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ejam S.S., et al. Pathogenic role of 25-hydroxycholesterol in cancer development and progression. Future Oncol. 2022;18(39):4415–4442. doi: 10.2217/fon-2022-0819. [DOI] [PubMed] [Google Scholar]
- 5.Zhou S., Treloar A.E., Lupien M. Emergence of the noncoding cancer Genome: a target of genetic and epigenetic AlterationsThe noncoding cancer genome. Cancer Discov. 2016;6(11):1215–1229. doi: 10.1158/2159-8290.CD-16-0745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morel D., et al. Combining epigenetic drugs with other therapies for solid tumours—past lessons and future promise. Nature Reviews Clinical Oncology. 2020;17(2):91–107. doi: 10.1038/s41571-019-0267-4. [DOI] [PubMed] [Google Scholar]
- 7.Van Der Pol Y., Mouliere F. Toward the early detection of cancer by decoding the epigenetic and environmental fingerprints of cell-free DNA. Cancer Cell. 2019;36(4):350–368. doi: 10.1016/j.ccell.2019.09.003. [DOI] [PubMed] [Google Scholar]
- 8.Calses P.C., et al. Hippo pathway in cancer: aberrant regulation and therapeutic opportunities. Trends in cancer. 2019;5(5):297–307. doi: 10.1016/j.trecan.2019.04.001. [DOI] [PubMed] [Google Scholar]
- 9.Dhillon A.S., et al. MAP kinase signalling pathways in cancer. Oncogene. 2007;26(22):3279–3290. doi: 10.1038/sj.onc.1210421. [DOI] [PubMed] [Google Scholar]
- 10.Kristensen L.S., et al. The biogenesis, biology and characterization of circular RNAs. Nat. Rev. Genet. 2019;20(11):675–691. doi: 10.1038/s41576-019-0158-7. [DOI] [PubMed] [Google Scholar]
- 11.Panda A.C. Circular RNAs act as miRNA sponges. Circular RNAs. 2018:67–79. doi: 10.1007/978-981-13-1426-1_6. [DOI] [PubMed] [Google Scholar]
- 12.Qiu L., et al. Circular RNAs in hepatocellular carcinoma: biomarkers, functions and mechanisms. Life Sci. 2019;231:116660. doi: 10.1016/j.lfs.2019.116660. [DOI] [PubMed] [Google Scholar]
- 13.Misir S., et al. Circular RNAs serve as miRNA sponges in breast cancer. Breast Cancer. 2020;27(6):1048–1057. doi: 10.1007/s12282-020-01140-w. [DOI] [PubMed] [Google Scholar]
- 14.Li R., et al. CircRNA: a rising star in gastric cancer. Cell. Mol. Life Sci. 2020;77(9):1661–1680. doi: 10.1007/s00018-019-03345-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rong Z., et al. Circular RNA in pancreatic cancer: a novel avenue for the roles of diagnosis and treatment. Theranostics. 2021;11(6):2755. doi: 10.7150/thno.56174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li Z., et al. An update on the roles of circular RNAs in osteosarcoma. Cell Prolif. 2021;54(1) doi: 10.1111/cpr.12936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abdullah S.T., et al. Role of circular RNAs and gut microbiome in gastrointestinal cancers and therapeutic targets. Non-coding RNA research. 2023;9(1):236–252. doi: 10.1016/j.ncrna.2023.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang X., et al. Circular RNAs in breast cancer diagnosis, treatment and prognosis. Oncology Research. 2023;32(2):241. doi: 10.32604/or.2023.046582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yi Q., et al. Recent advances of exosomal circRNAs in cancer and their potential clinical applications. J. Transl. Med. 2023;21(1):516. doi: 10.1186/s12967-023-04348-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pisignano G., et al. Going circular: history, present, and future of circRNAs in cancer. Oncogene. 2023;42(38):2783–2800. doi: 10.1038/s41388-023-02780-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hakimi A.A., et al. An integrated metabolic atlas of clear cell renal cell carcinoma. Cancer Cell. 2016;29(1):104–116. doi: 10.1016/j.ccell.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abbaszadeh Z., Çeşmeli S. Ç.B. Avcı, Crucial players in glycolysis: cancer progress. Gene. 2020;726:144158. doi: 10.1016/j.gene.2019.144158. [DOI] [PubMed] [Google Scholar]
- 23.Liu J., et al. Circular RNA circ-MAT2B facilitates glycolysis and growth of gastric cancer through regulating the miR-515-5p/HIF-1α axis. Cancer Cell Int. 2020;20(1):1–12. doi: 10.1186/s12935-020-01256-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ganapathy-Kanniappan S. Linking tumor glycolysis and immune evasion in cancer: emerging concepts and therapeutic opportunities. Biochim. Biophys. Acta, Rev. Cancer. 2017;1868(1):212–220. doi: 10.1016/j.bbcan.2017.04.002. [DOI] [PubMed] [Google Scholar]
- 25.Chakraborty P.K., et al. MICU1 drives glycolysis and chemoresistance in ovarian cancer. Nat. Commun. 2017;8(1):1–16. doi: 10.1038/ncomms14634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao X., Tian Z., Liu L. circATP2B1 promotes aerobic glycolysis in gastric cancer cells through regulation of the miR-326 gene cluster. Front. Oncol. 2021;11:628624. doi: 10.3389/fonc.2021.628624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Y., et al. Knockdown of circBFAR inhibits proliferation and glycolysis in gastric cancer by sponging miR-513a-3p/hexokinase 2 axis. Biochem. Biophys. Res. Commun. 2021;560:80–86. doi: 10.1016/j.bbrc.2021.04.131. [DOI] [PubMed] [Google Scholar]
- 28.Braicu C., et al. Comprehensive analysis of circular RNAs in pathological states: biogenesis, cellular regulation, and therapeutic relevance. Cell. Mol. Life Sci. 2019;76(8):1559–1577. doi: 10.1007/s00018-019-03016-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Memczak S., et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 30.Jeck W.R., et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19(2):141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Y., et al. Circular intronic long noncoding RNAs. Mol. Cell. 2013;51(6):792–806. doi: 10.1016/j.molcel.2013.08.017. [DOI] [PubMed] [Google Scholar]
- 32.Vo J.N., et al. The landscape of circular RNA in cancer. Cell. 2019;176(4):869–881. doi: 10.1016/j.cell.2018.12.021. e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ashwal-Fluss R., et al. circRNA biogenesis competes with pre-mRNA splicing. Mol. Cell. 2014;56(1):55–66. doi: 10.1016/j.molcel.2014.08.019. [DOI] [PubMed] [Google Scholar]
- 34.Conn S.J., et al. The RNA binding protein quaking regulates formation of circRNAs. Cell. 2015;160(6):1125–1134. doi: 10.1016/j.cell.2015.02.014. [DOI] [PubMed] [Google Scholar]
- 35.Ivanov A., et al. Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep. 2015;10(2):170–177. doi: 10.1016/j.celrep.2014.12.019. [DOI] [PubMed] [Google Scholar]
- 36.Lee Y., Rio D.C. Mechanisms and regulation of alternative pre-mRNA splicing. Annu. Rev. Biochem. 2015;84:291. doi: 10.1146/annurev-biochem-060614-034316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang X.-O., et al. Complementary sequence-mediated exon circularization. Cell. 2014;159(1):134–147. doi: 10.1016/j.cell.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 38.Barrett S.P., Wang P.L., Salzman J. Circular RNA biogenesis can proceed through an exon-containing lariat precursor. Elife. 2015;4 doi: 10.7554/eLife.07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Geng X., et al. Circular RNA: biogenesis, degradation, functions and potential roles in mediating resistance to anticarcinogens. Epigenomics. 2020;12(3):267–283. doi: 10.2217/epi-2019-0295. [DOI] [PubMed] [Google Scholar]
- 40.Liu K.-S., et al. Biological functions of circular RNAs and their roles in occurrence of reproduction and gynecological diseases. Am. J. Tourism Res. 2019;11(1):1. [PMC free article] [PubMed] [Google Scholar]
- 41.Capel B., et al. Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell. 1993;73(5):1019–1030. doi: 10.1016/0092-8674(93)90279-y. [DOI] [PubMed] [Google Scholar]
- 42.Abdelmohsen K., et al. Posttranscriptional gene regulation by RNA-binding proteins during oxidative stress: implications for cellular senescence. Biol. Chem. 2008;389(3):243–255. doi: 10.1515/BC.2008.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ng W.L., et al. Inducible RasGEF1B circular RNA is a positive regulator of ICAM-1 in the TLR4/LPS pathway. RNA Biol. 2016;13(9):861–871. doi: 10.1080/15476286.2016.1207036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang Y., Wang Z. Efficient backsplicing produces translatable circular mRNAs. RNA. 2015;21(2):172–179. doi: 10.1261/rna.048272.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thomas L.F., Sætrom P. Circular RNAs are depleted of polymorphisms at microRNA binding sites. Bioinformatics. 2014;30(16):2243–2246. doi: 10.1093/bioinformatics/btu257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Legnini I., et al. Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol. Cell. 2017;66(1):22–37. doi: 10.1016/j.molcel.2017.02.017. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Haque S., Harries L.W. Circular RNAs (circRNAs) in health and disease. Genes. 2017;8(12):353. doi: 10.3390/genes8120353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Venø M.T., et al. Spatio-temporal regulation of circular RNA expression during porcine embryonic brain development. Genome Biol. 2015;16(1):1–17. doi: 10.1186/s13059-015-0801-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xu H., et al. The circular RNA Cdr1as, via miR-7 and its targets, regulates insulin transcription and secretion in islet cells. Sci. Rep. 2015;5(1):1–12. doi: 10.1038/srep12453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huang H., et al. Circular RNA circUBE2D2 functions as an oncogenic factor in hepatocellular carcinoma sorafenib resistance and glycolysis. Am. J. Tourism Res. 2021;13(6):6076. [PMC free article] [PubMed] [Google Scholar]
- 51.Yang J., et al. Circular RNA UBE2Q2 promotes malignant progression of gastric cancer by regulating signal transducer and activator of transcription 3-mediated autophagy and glycolysis. Cell Death Dis. 2021;12(10):1–16. doi: 10.1038/s41419-021-04216-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guo F., et al. Circular RNA circMAGI3 accelerates the glycolysis of non-small cell lung cancer through miR-515-5p/HDGF. Am. J. Tourism Res. 2020;12(7):3953. [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang Z., et al. Circular RNA circDENND4C facilitates proliferation, migration and glycolysis of colorectal cancer cells through miR-760/GLUT1 axis. Eur. Rev. Med. Pharmacol. Sci. 2020;24(5):2387–2400. doi: 10.26355/eurrev_202003_20506. [DOI] [PubMed] [Google Scholar]
- 54.Wang Z., et al. circMYC promotes cell proliferation, metastasis, and glycolysis in cervical cancer by up-regulating MET and sponging miR-577. Am. J. Tourism Res. 2021;13(6):6043. [PMC free article] [PubMed] [Google Scholar]
- 55.Han J., et al. The circular RNA circINPP4B acts as a sponge of miR-30a to regulate Th17 cell differentiation during progression of experimental autoimmune encephalomyelitis. Cell. Mol. Immunol. 2021;18(9):2177–2187. doi: 10.1038/s41423-021-00748-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Holdt L.M., Kohlmaier A., Teupser D. Circular RNAs as therapeutic agents and targets. Front. Physiol. 2018;9:1262. doi: 10.3389/fphys.2018.01262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhou M.-y., Yang J.-M., Xiong X.-d. The emerging landscape of circular RNA in cardiovascular diseases. J. Mol. Cell. Cardiol. 2018;122:134–139. doi: 10.1016/j.yjmcc.2018.08.012. [DOI] [PubMed] [Google Scholar]
- 58.Chen Y.G., et al. Sensing self and foreign circular RNAs by intron identity. Mol. Cell. 2017;67(2):228–238. doi: 10.1016/j.molcel.2017.05.022. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yu L., et al. The glycolytic switch in tumors: how many players are involved? J. Cancer. 2017;8(17):3430. doi: 10.7150/jca.21125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hamanaka R.B., Chandel N.S. Targeting glucose metabolism for cancer therapy. J. Exp. Med. 2012;209(2):211–215. doi: 10.1084/jem.20120162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li Q., et al. Circular RNA MAT2B promotes glycolysis and malignancy of hepatocellular carcinoma through the miR-338-3p/PKM2 axis under hypoxic stress. Hepatology. 2019;70(4):1298–1316. doi: 10.1002/hep.30671. [DOI] [PubMed] [Google Scholar]
- 62.Du Q., et al. Hypoxia-induced circular RNA hsa_circ_0008450 accelerates hepatocellular cancer progression via the miR-431/AKAP1 axis. Oncol. Lett. 2020;20(6):1. doi: 10.3892/ol.2020.12251. 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ding Z., et al. Circ-PRMT5 enhances the proliferation, migration and glycolysis of hepatoma cells by targeting miR-188-5p/HK2 axis. Ann. Hepatol. 2020;19(3):269–279. doi: 10.1016/j.aohep.2020.01.002. [DOI] [PubMed] [Google Scholar]
- 64.Li J., et al. CircRPN2 inhibits aerobic glycolysis and metastasis in hepatocellular carcinoma. Cancer Res. 2022;82(6):1055–1069. doi: 10.1158/0008-5472.CAN-21-1259. [DOI] [PubMed] [Google Scholar]
- 65.Yi B., et al. Circular RNA PLCE1 promotes epithelial mesenchymal transformation, glycolysis in colorectal cancer and M2 polarization of tumor-associated macrophages. Bioengineered. 2022;13(3):6243–6256. doi: 10.1080/21655979.2021.2003929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li Y., et al. circ_0136666 facilitates the progression of colorectal cancer via miR-383/CREB1 axis. Cancer Manag. Res. 2020;12:6795. doi: 10.2147/CMAR.S251952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li Z., et al. CircTADA2A suppresses the progression of colorectal cancer via miR-374a-3p/KLF14 axis. J. Exp. Clin. Cancer Res. 2020;39(1):1–14. doi: 10.1186/s13046-020-01642-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bray F., et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J. Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 69.Al-Awsi G.R.L., et al. The role of miRNA-128 in the development and progression of gastrointestinal and urogenital cancer. Future Oncol. 2023;18(38):4209–4231. doi: 10.2217/fon-2022-0574. [DOI] [PubMed] [Google Scholar]
- 70.Armstrong B.K. Cancer epidemiology and prevention. Int. J. Epidemiol. 2018;47(6):2097–2098. [Google Scholar]
- 71.Pan J., et al. Comparison of dexmedetomidine vs. remifentanil combined with sevoflurane during radiofrequency ablation of hepatocellular carcinoma: a randomized controlled trial. Trials. 2019;20(1):1–10. doi: 10.1186/s13063-018-3010-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang X.W., et al. Molecular pathogenesis of human hepatocellular carcinoma. Toxicology. 2002;181:43–47. doi: 10.1016/s0300-483x(02)00253-6. [DOI] [PubMed] [Google Scholar]
- 73.Yin Y., et al. Emerging roles of circRNA in formation and progression of cancer. J. Cancer. 2019;10(21):5015. doi: 10.7150/jca.30828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li X., Yang L., Chen L.-L. The biogenesis, functions, and challenges of circular RNAs. Mol. Cell. 2018;71(3):428–442. doi: 10.1016/j.molcel.2018.06.034. [DOI] [PubMed] [Google Scholar]
- 75.He S., et al. Circular RNA circ_0000517 regulates hepatocellular carcinoma development via miR-326/IGF1R axis. Cancer Cell Int. 2020;20(1):1–12. doi: 10.1186/s12935-020-01496-1. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 76.Fang G., et al. Inhibition of GSK-3β activity suppresses HCC malignant phenotype by inhibiting glycolysis via activating AMPK/mTOR signaling. Cancer Lett. 2019;463:11–26. doi: 10.1016/j.canlet.2019.08.003. [DOI] [PubMed] [Google Scholar]
- 77.Zhang Y., et al. The miR-873/NDFIP1 axis promotes hepatocellular carcinoma growth and metastasis through the AKT/mTOR-mediated Warburg effect. Am. J. Cancer Res. 2019;9(5):927. [PMC free article] [PubMed] [Google Scholar]
- 78.Sheng H., Tang W. Glycolysis inhibitors for anticancer therapy: a review of recent patents. Recent Pat. Anti-Cancer Drug Discov. 2016;11(3):297–308. doi: 10.2174/1574892811666160415160104. [DOI] [PubMed] [Google Scholar]
- 79.Zang H., et al. Circ_0000517 contributes to hepatocellular carcinoma progression by upregulating TXNDC5 via sponging miR-1296-5p. Cancer Manag. Res. 2020;12:3457. doi: 10.2147/CMAR.S244024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhong Y., et al. Circular RNAs function as ceRNAs to regulate and control human cancer progression. Mol. Cancer. 2018;17(1):1–11. doi: 10.1186/s12943-018-0827-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhao Q., et al. LncRNA SNHG3 promotes hepatocellular tumorigenesis by targeting miR-326. Tohoku J. Exp. Med. 2019;249(1):43–56. doi: 10.1620/tjem.249.43. [DOI] [PubMed] [Google Scholar]
- 82.Hu S., et al. MicroRNA-326 inhibits cell proliferation and invasion, activating apoptosis in hepatocellular carcinoma by directly targeting LIM and SH3 protein 1. Oncol. Rep. 2017;38(3):1569–1578. doi: 10.3892/or.2017.5810. [DOI] [PubMed] [Google Scholar]
- 83.Wang L., et al. Insulin-like growth factor I receptor: a novel target for hepatocellular carcinoma gene therapy. Mini Rev. Med. Chem. 2019;19(4):272–280. doi: 10.2174/1389557518666181025151608. [DOI] [PubMed] [Google Scholar]
- 84.Liu Q., et al. Long non-coding RNA NEAT1 promoted Hepatocellular Carcinoma cell proliferation and reduced apoptosis through the regulation of Let-7b-IGF-1R Axis. OncoTargets Ther. 2019;12:10401. doi: 10.2147/OTT.S217763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang X., et al. miR-29a-3p suppresses cell proliferation and migration by downregulating IGF1R in hepatocellular carcinoma. Oncotarget. 2017;8(49):86592. doi: 10.18632/oncotarget.21246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ye Y., et al. MicroRNA-495 suppresses cell proliferation and invasion of hepatocellular carcinoma by directly targeting insulin-like growth factor receptor-1. Exp. Ther. Med. 2018;15(1):1150–1158. doi: 10.3892/etm.2017.5467. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 87.Ter Braak B., et al. Insulin-like growth factor 1 receptor activation promotes mammary gland tumor development by increasing glycolysis and promoting biomass production. Breast Cancer Res. 2017;19(1):1–15. doi: 10.1186/s13058-017-0802-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wang B., et al. MicroRNA-7 directly targets insulin-like growth factor 1 receptor to inhibit cellular growth and glucose metabolism in gliomas. Diagn. Pathol. 2014;9(1):1–6. doi: 10.1186/s13000-014-0211-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ma J., et al. LncRNA PAPAS promotes hepatocellular carcinoma by interacting with miR-188-5p. J. Cell. Biochem. 2019;120(8):13494–13500. doi: 10.1002/jcb.28623. [DOI] [PubMed] [Google Scholar]
- 90.Cheng N., et al. LncRNA CASC11 promotes cancer cell proliferation in hepatocellular carcinoma by inhibiting miRNA-188-5p. Biosci. Rep. 2019;39(4) doi: 10.1042/BSR20190251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tan V.P., Miyamoto S. HK2/hexokinase-II integrates glycolysis and autophagy to confer cellular protection. Autophagy. 2015;11(6):963–964. doi: 10.1080/15548627.2015.1042195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Xu F., et al. miR-885-5p negatively regulates warburg effect by silencing hexokinase 2 in liver cancer. Mol. Ther. Nucleic Acids. 2019;18:308–319. doi: 10.1016/j.omtn.2019.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wang H., et al. Inhibition of glycolytic enzyme hexokinase II (HK2) suppresses lung tumor growth. Cancer Cell Int. 2016;16(1):1–11. doi: 10.1186/s12935-016-0280-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Rankin E., Nam J., Giaccia A. Hypoxia: signaling the metastatic cascade. Trends Cancer. 2016;2(6):295–304. doi: 10.1016/j.trecan.2016.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhang X., et al. Yes-associated protein (YAP) binds to HIF-1α and sustains HIF-1α protein stability to promote hepatocellular carcinoma cell glycolysis under hypoxic stress. J. Exp. Clin. Cancer Res. 2018;37(1):1–12. doi: 10.1186/s13046-018-0892-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wei H., et al. Circular RNA circ_0008450 upregulates CXCL9 expression by targeting miR-577 to regulate cell proliferation and invasion in nasopharyngeal carcinoma. Exp. Mol. Pathol. 2019;110:104288. doi: 10.1016/j.yexmp.2019.104288. [DOI] [PubMed] [Google Scholar]
- 97.Lin T., et al. Silencing of hsa_circ_0008450 represses hepatocellular carcinoma progression through regulation of microRNA-214-3p/EZH2 axis. Cancer Manag. Res. 2019;11:9133. doi: 10.2147/CMAR.S222716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Carlucci A., et al. Proteolysis of AKAP121 regulates mitochondrial activity during cellular hypoxia and brain ischaemia. EMBO J. 2008;27(7):1073–1084. doi: 10.1038/emboj.2008.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Rinaldi L., et al. Mitochondrial AKAP1 supports mTOR pathway and tumor growth. Cell Death Dis. 2017;8(6) doi: 10.1038/cddis.2017.241. p. e2842-e2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Feng J., et al. Emerging roles and the regulation of aerobic glycolysis in hepatocellular carcinoma. J. Exp. Clin. Cancer Res. 2020;39(1):1–19. doi: 10.1186/s13046-020-01629-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ma L., et al. Activator of thyroid and retinoid receptor increases sorafenib resistance in hepatocellular carcinoma by facilitating the Warburg effect. Cancer Sci. 2020;111(6):2028–2040. doi: 10.1111/cas.14412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wu M.-y., et al. Global transcriptomic study of circRNAs expression profile in sorafenib resistant hepatocellular carcinoma cells. J. Cancer. 2020;11(10):2993. doi: 10.7150/jca.39854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wang H., et al. CircSLC3A2 functions as an oncogenic factor in hepatocellular carcinoma by sponging miR-490-3p and regulating PPM1F expression. Mol. Cancer. 2018;17(1):1–13. doi: 10.1186/s12943-018-0909-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Chen Z., et al. circ-CFH promotes the development of HCC by regulating cell proliferation, apoptosis, migration, invasion, and glycolysis through the miR-377-3p/RNF38 axis. Open Life Sci. 2022;17(1):248–260. doi: 10.1515/biol-2022-0029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Vaitheesvaran B., et al. The Warburg effect: a balance of flux analysis. Metabolomics. 2015;11(4):787–796. doi: 10.1007/s11306-014-0760-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Yu M., et al. The prognostic value of GLUT1 in cancers: a systematic review and meta-analysis. Oncotarget. 2017;8(26):43356. doi: 10.18632/oncotarget.17445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Liu B.-H., et al. Expression profiling identifies circular RNA signature in hepatoblastoma. Cell. Physiol. Biochem. 2018;45(2):706–719. doi: 10.1159/000487163. [DOI] [PubMed] [Google Scholar]
- 108.Liu L., et al. Circ_0015756 promotes proliferation, invasion and migration by microRNA-7-dependent inhibition of FAK in hepatocellular carcinoma. Cell Cycle. 2019;18(21):2939–2953. doi: 10.1080/15384101.2019.1664223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Chen G., et al. MicroRNA-377 suppresses cell proliferation and invasion by inhibiting TIAM1 expression in hepatocellular carcinoma. PLoS One. 2015;10(3) doi: 10.1371/journal.pone.0117714. p. e0117714. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 110.Cai J., et al. CircRHBDD1 augments metabolic rewiring and restricts immunotherapy efficacy via m6A modification in hepatocellular carcinoma. Molecular Therapy-Oncolytics. 2022;24:755–771. doi: 10.1016/j.omto.2022.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Yu T., et al. CircRNAs in cancer metabolism: a review. J. Hematol. Oncol. 2019;12(1):1–10. doi: 10.1186/s13045-019-0776-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Invrea F., et al. Patient-derived xenografts (PDXs) as model systems for human cancer. Curr. Opin. Biotechnol. 2020;63:151–156. doi: 10.1016/j.copbio.2020.01.003. [DOI] [PubMed] [Google Scholar]
- 113.Lee J.-H., et al. EGFR-phosphorylated platelet isoform of phosphofructokinase 1 promotes PI3K activation. Mol. Cell. 2018;70(2):197–210. doi: 10.1016/j.molcel.2018.03.018. e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zaccara S., Ries R.J., Jaffrey S.R. Reading, writing and erasing mRNA methylation. Nat. Rev. Mol. Cell Biol. 2019;20(10):608–624. doi: 10.1038/s41580-019-0168-5. [DOI] [PubMed] [Google Scholar]
- 115.Lan Q., et al. The critical role of RNA m6A methylation in cancer. Cancer Res. 2019;79(7):1285–1292. doi: 10.1158/0008-5472.CAN-18-2965. [DOI] [PubMed] [Google Scholar]
- 116.Liu T., et al. The m6A reader YTHDF1 promotes ovarian cancer progression via augmenting EIF3C translation. Nucleic Acids Res. 2020;48(7):3816–3831. doi: 10.1093/nar/gkaa048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Hao L., et al. m6A-YTHDF1-mediated TRIM29 upregulation facilitates the stem cell-like phenotype of cisplatin-resistant ovarian cancer cells. Biochim. Biophys. Acta Mol. Cell Res. 2021;1868(1):118878. doi: 10.1016/j.bbamcr.2020.118878. [DOI] [PubMed] [Google Scholar]
- 118.Pi J., et al. YTHDF1 promotes gastric Carcinogenesis by controlling Translation of fzd7ythdf1/FZD7 Axis regulates gastric carcinogenesis. Cancer Res. 2021;81(10):2651–2665. doi: 10.1158/0008-5472.CAN-20-0066. [DOI] [PubMed] [Google Scholar]
- 119.Zhang H., et al. Regulatory mechanisms of immune checkpoints PD-L1 and CTLA-4 in cancer. J. Exp. Clin. Cancer Res. 2021;40(1):1–22. doi: 10.1186/s13046-021-01987-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Waidmann O. Recent developments with immunotherapy for hepatocellular carcinoma. Expet Opin. Biol. Ther. 2018;18(8):905–910. doi: 10.1080/14712598.2018.1499722. [DOI] [PubMed] [Google Scholar]
- 121.Dey P., Kimmelman A.C., DePinho R.A. Metabolic codependencies in the tumor MicroenvironmentMetabolic codependencies in cancer. Cancer Discov. 2021;11(5):1067–1081. doi: 10.1158/2159-8290.CD-20-1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Renner K., et al. Restricting glycolysis preserves T cell effector functions and augments checkpoint therapy. Cell Rep. 2019;29(1):135–150. doi: 10.1016/j.celrep.2019.08.068. e9. [DOI] [PubMed] [Google Scholar]
- 123.Cascone T., et al. Increased tumor glycolysis characterizes immune resistance to adoptive T cell therapy. Cell Metabol. 2018;27(5):977–987. doi: 10.1016/j.cmet.2018.02.024. e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Hu Z.Q., et al. Circular RNA sequencing identifies CircASAP1 as a key regulator in hepatocellular carcinoma metastasis. Hepatology. 2020;72(3):906–922. doi: 10.1002/hep.31068. [DOI] [PubMed] [Google Scholar]
- 125.Hsu P.P., Sabatini D.M. Cancer cell metabolism: warburg and beyond. Cell. 2008;134(5):703–707. doi: 10.1016/j.cell.2008.08.021. [DOI] [PubMed] [Google Scholar]
- 126.Dai J., et al. Alpha-enolase regulates the malignant phenotype of pulmonary artery smooth muscle cells via the AMPK-Akt pathway. Nat. Commun. 2018;9(1):1–16. doi: 10.1038/s41467-018-06376-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Yang T., et al. Enolase 1 regulates stem cell-like properties in gastric cancer cells by stimulating glycolysis. Cell Death Dis. 2020;11(10):1–13. doi: 10.1038/s41419-020-03087-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Jung D.-W., et al. A unique small molecule inhibitor of enolase clarifies its role in fundamental biological processes. ACS Chem. Biol. 2013;8(6):1271–1282. doi: 10.1021/cb300687k. [DOI] [PubMed] [Google Scholar]
- 129.Ma Y., et al. Circular RNA PRKCI silencing represses esophageal cancer progression and elevates cell radiosensitivity through regulating the miR-186-5p/PARP9 axis. Life Sci. 2020;259:118168. doi: 10.1016/j.lfs.2020.118168. [DOI] [PubMed] [Google Scholar]
- 130.DeBerardinis R.J., et al. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metabol. 2008;7(1):11–20. doi: 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 131.Pena A., et al. Pharmacological inhibition of mTOR kinase reverses right ventricle remodeling and improves right ventricle structure and function in rats. Am. J. Respir. Cell Mol. Biol. 2017;57(5):615–625. doi: 10.1165/rcmb.2016-0364OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Yan R., et al. miR-183-5p promotes proliferation and migration in hepatocellular carcinoma by targeting IRS1 and its association with patient survival. Int. J. Biol. Markers. 2020;35(3):83–89. doi: 10.1177/1724600820951572. [DOI] [PubMed] [Google Scholar]
- 133.Zheng M., et al. STAT3 promotes invasion and aerobic glycolysis of human oral squamous cell carcinoma via inhibiting FoxO1. Front. Oncol. 2019;9:1175. doi: 10.3389/fonc.2019.01175. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 134.Bonelli P., et al. Precision medicine in gastric cancer. World J. Gastrointest. Oncol. 2019;11(10):804. doi: 10.4251/wjgo.v11.i10.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Gao Y., et al. Association between liquid biopsy and prognosis of gastric cancer patients: a systematic review and meta-analysis. Front. Oncol. 2019;9:1222. doi: 10.3389/fonc.2019.01222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Jones J.O., Smyth E.C. Gastroesophageal cancer: navigating the immune and genetic terrain to improve clinical outcomes. Cancer Treat Rev. 2020;84:101950. doi: 10.1016/j.ctrv.2019.101950. [DOI] [PubMed] [Google Scholar]
- 137.Zheng L.-N., et al. Prognostic significance of malignant ascites in gastric cancer patients with peritoneal metastasis: a systemic review and meta-analysis. World Journal of Clinical Cases. 2019;7(20):3247. doi: 10.12998/wjcc.v7.i20.3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Symeonidis D., et al. Current role of lymphadenectomy in gastric cancer surgery. J BUON. 2019;24(5):1761–1767. [PubMed] [Google Scholar]
- 139.Wang Q., Liu G., Hu C. Molecular classification of gastric adenocarcinoma. Gastroenterol. Res. 2019;12(6):275–282. doi: 10.14740/gr1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Pu Z., et al. Circular RNA circCUL3 accelerates the warburg effect progression of gastric cancer through regulating the STAT3/HK2 axis. Mol. Ther. Nucleic Acids. 2020;22:310–318. doi: 10.1016/j.omtn.2020.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Qu J.-L., et al. Gastric cancer exosomes promote tumour cell proliferation through PI3K/Akt and MAPK/ERK activation. Dig. Liver Dis. 2009;41(12):875–880. doi: 10.1016/j.dld.2009.04.006. [DOI] [PubMed] [Google Scholar]
- 142.Xie M., et al. Exosomal circSHKBP1 promotes gastric cancer progression via regulating the miR-582-3p/HUR/VEGF axis and suppressing HSP90 degradation. Mol. Cancer. 2020;19(1):1–22. doi: 10.1186/s12943-020-01208-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Zhou Y., et al. circ_0006089 promotes gastric cancer growth, metastasis, glycolysis, and angiogenesis by regulating miR-361-3p/TGFB1. Cancer Sci. 2022;113(6):2044. doi: 10.1111/cas.15351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Sun Y., et al. Circular RNA circ_ASAP2 regulates drug sensitivity and functional behaviors of cisplatin-resistant gastric cancer cells by the miR-330-3p/NT5E axis. Anti Cancer Drugs. 2021;32(9):950–961. doi: 10.1097/CAD.0000000000001087. [DOI] [PubMed] [Google Scholar]
- 145.Ni C., et al. Hsa_circ_0011385 knockdown represses cell proliferation in hepatocellular carcinoma. Cell Death Discovery. 2021;7(1):1–8. doi: 10.1038/s41420-021-00664-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Cui X., et al. Long noncoding RNA SNHG22 induces cell migration, invasion, and angiogenesis of gastric Cancer cells via microRNA-361-3p/HMGA1/Wnt/β-catenin Axis. Cancer Manag. Res. 2020;12:12867. doi: 10.2147/CMAR.S281578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Xin H., Yan Z., Cao J. Long non-coding RNA ABHD11-AS1 boosts gastric cancer development by regulating miR-361-3p/PDPK1 signalling. J. Biochem. 2020;168(5):465–476. doi: 10.1093/jb/mvaa065. [DOI] [PubMed] [Google Scholar]
- 148.Li H., et al. circDNMT1 promotes malignant progression of gastric cancer through targeting miR-576-3p/hypoxia inducible factor-1 alpha axis. Front. Oncol. 2022:12. doi: 10.3389/fonc.2022.817192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Xie C., et al. CircRNA DNA methyltransferase 1 silence inhibits breast cancer development by regulating micoRNA-485-3p/zinc finger E-box binding homeobox 1 axis. J. Obstet. Gynaecol. Res. 2021;47(3):1068–1081. doi: 10.1111/jog.14639. [DOI] [PubMed] [Google Scholar]
- 150.Zhao J., et al. The HIF-1A/miR-17-5p/PDCD4 axis contributes to the tumor growth and metastasis of gastric cancer. Signal Transduct. Targeted Ther. 2020;5(1):1–3. doi: 10.1038/s41392-020-0132-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Mu G., et al. Calmodulin 2 facilitates Angiogenesis and Metastasis of gastric Cancer via STAT3/HIF-1A/VEGF-A mediated macrophage polarization. Front. Oncol. 2021:3713. doi: 10.3389/fonc.2021.727306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Du Y., et al. Long noncoding RNA MIR210HG promotes the Warburg effect and tumor growth by enhancing HIF-1α translation in triple-negative breast cancer. Front. Oncol. 2020;10:580176. doi: 10.3389/fonc.2020.580176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Sun J., et al. Upregulation of LncRNA PVT1 facilitates pancreatic ductal adenocarcinoma cell progression and glycolysis by regulating MiR-519d-3p and HIF-1A. J. Cancer. 2020;11(9):2572. doi: 10.7150/jca.37959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Weng M.-l., et al. Fasting inhibits aerobic glycolysis and proliferation in colorectal cancer via the Fdft1-mediated AKT/mTOR/HIF1α pathway suppression. Nat. Commun. 2020;11(1):1–17. doi: 10.1038/s41467-020-15795-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Zheng X., et al. circRPS19 affects HK2-mediated aerobic glycolysis and cell viability via the miR-125a-5p/USP7 pathway in gastric cancer. Int. J. Oncol. 2023;63(2):1–14. doi: 10.3892/ijo.2023.5546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Sève P., Reiman T., Dumontet C. The role of βIII tubulin in predicting chemoresistance in non-small cell lung cancer. Lung Cancer. 2010;67(2):136–143. doi: 10.1016/j.lungcan.2009.09.007. [DOI] [PubMed] [Google Scholar]
- 157.Fujimoto J., Wistuba I.I. Seminars in Diagnostic Pathology. Elsevier; 2014. Current concepts on the molecular pathology of non-small cell lung carcinoma. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Prabhu V.V., Prabhu V. Epidermal growth factor receptor tyrosine kinase: a potential target in treatment of non-small-cell lung carcinoma. J. Environ. Pathol. Toxicol. Oncol. 2017;36(2) doi: 10.1615/JEnvironPatholToxicolOncol.2017018341. [DOI] [PubMed] [Google Scholar]
- 159.Xu Y., et al. Circular RNA circ_0001421 contributes to colony formation, migration, invasion and glycolysis of non-small cell lung cancer via the miR-409-3p/TMEM14A axis. Arch. Med. Sci. 2020 [Google Scholar]
- 160.Wang Y., et al. Circular RNA SMARCA5 inhibits the proliferation, migration, and invasion of non-small cell lung cancer by miR-19b-3p/HOXA9 axis. OncoTargets Ther. 2019;12:7055. doi: 10.2147/OTT.S216320. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 161.Huang M.-S., et al. Hsa_circ_0001946 inhibits lung cancer progression and mediates cisplatin sensitivity in non-small cell lung cancer via the nucleotide excision repair signaling pathway. Front. Oncol. 2019;9:508. doi: 10.3389/fonc.2019.00508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Gao P., et al. Circular RNA circ_0074027 indicates a poor prognosis for NSCLC patients and modulates cell proliferation, apoptosis, and invasion via miR-185-3p mediated BRD4/MADD activation. J. Cell. Biochem. 2020;121(3):2632–2642. doi: 10.1002/jcb.29484. [DOI] [PubMed] [Google Scholar]
- 163.Shangguan H., et al. Circular RNA circSLC25A16 contributes to the glycolysis of non-small-cell lung cancer through epigenetic modification. Cell Death Dis. 2020;11(6):1–10. doi: 10.1038/s41419-020-2635-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Jin J., et al. Cardamonin inhibits breast cancer growth by repressing HIF-1α-dependent metabolic reprogramming. J. Exp. Clin. Cancer Res. 2019;38(1):1–16. doi: 10.1186/s13046-019-1351-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Yang B., et al. CircRNA circ_0006677 Inhibits the Progression and Glycolysis in non–small-cell lung Cancer by Sponging miR-578 and regulating SOCS2 expression. Front. Pharmacol. 2021:839. doi: 10.3389/fphar.2021.657053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Rico-Bautista E., Flores-Morales A., Fernández-Pérez L. Suppressor of cytokine signaling (SOCS) 2, a protein with multiple functions. Cytokine Growth Factor Rev. 2006;17(6):431–439. doi: 10.1016/j.cytogfr.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 167.Das R., et al. MicroRNA-194 promotes prostate cancer Metastasis by inhibiting SOCS2miR-194 promotes prostate cancer metastasis. Cancer Res. 2017;77(4):1021–1034. doi: 10.1158/0008-5472.CAN-16-2529. [DOI] [PubMed] [Google Scholar]
- 168.Chhabra Y., et al. A growth hormone receptor SNP promotes lung cancer by impairment of SOCS2-mediated degradation. Oncogene. 2018;37(4):489–501. doi: 10.1038/onc.2017.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Zhang F., et al. Circular RNA Eps15-homology domain containing protein 2 motivates proliferation, glycolysis but refrains autophagy in non-small cell lung cancer via crosstalk with microRNA-3186-3p and forkhead box K1. Bioengineered. 2022;13(3):6464–6475. doi: 10.1080/21655979.2022.2031385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Liu Z., et al. Ubiquitylation of autophagy receptor Optineurin by HACE1 activates selective autophagy for tumor suppression. Cancer Cell. 2014;26(1):106–120. doi: 10.1016/j.ccr.2014.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Zheng S., et al. Long non-coding RNA HUMT hypomethylation promotes lymphangiogenesis and metastasis via activating FOXK1 transcription in triple-negative breast cancer. J. Hematol. Oncol. 2020;13(1):1–15. doi: 10.1186/s13045-020-00852-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Cui H., et al. Knockdown of FOXK1 suppresses liver cancer cell viability by inhibiting glycolysis. Life Sci. 2018;213:66–73. doi: 10.1016/j.lfs.2018.10.018. [DOI] [PubMed] [Google Scholar]
- 173.Chen W., et al. Exosomal circSHKBP1 participates in non-small cell lung cancer progression through PKM2-mediated glycolysis. Molecular Therapy-Oncolytics. 2022;24:470–485. doi: 10.1016/j.omto.2022.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Ge W.-L., et al. The YY1/miR-548t-5p/CXCL11 signaling axis regulates cell proliferation and metastasis in human pancreatic cancer. Cell Death Dis. 2020;11(4):1–18. doi: 10.1038/s41419-020-2475-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Wong N., et al. PKM2 contributes to cancer metabolism. Cancer Lett. 2015;356(2):184–191. doi: 10.1016/j.canlet.2014.01.031. [DOI] [PubMed] [Google Scholar]
- 176.Hou P.-p., et al. Ectosomal PKM2 promotes HCC by inducing macrophage differentiation and remodeling the tumor microenvironment. Mol. Cell. 2020;78(6):1192–1206. doi: 10.1016/j.molcel.2020.05.004. e10. [DOI] [PubMed] [Google Scholar]
- 177.Li H., et al. Circular RNA, circular RARS, promotes aerobic glycolysis of non-small-cell lung cancer by binding with LDHA. Thoracic Cancer. 2023;14(4):389–398. doi: 10.1111/1759-7714.14758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Shi L., et al. Circ_0003028 enhances the proliferation and glycolytic capacity and suppresses apoptosis in non-small cell lung cancer cells via the miR-1305/miR-1322-SLC5A1 axis. Ann. Transl. Med. 2023;11(5) doi: 10.21037/atm-23-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Li H., et al. Circular RNA P4HB promotes glycolysis and tumor progression by binding with PKM2 in lung adenocarcinoma. Respir. Res. 2023;24(1):252. doi: 10.1186/s12931-023-02563-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Augestad K., et al. Metastatic spread pattern after curative colorectal cancer surgery. A retrospective, longitudinal analysis. Cancer epidemiology. 2015;39(5):734–744. doi: 10.1016/j.canep.2015.07.009. [DOI] [PubMed] [Google Scholar]
- 181.Sosa M.S., Bragado P., Aguirre-Ghiso J.A. Mechanisms of disseminated cancer cell dormancy: an awakening field. Nat. Rev. Cancer. 2014;14(9):611–622. doi: 10.1038/nrc3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Kaminski M.F., et al. Optimizing the quality of colorectal cancer screening worldwide. Gastroenterology. 2020;158(2):404–417. doi: 10.1053/j.gastro.2019.11.026. [DOI] [PubMed] [Google Scholar]
- 183.Prabhakaran S., et al. Precision medicine in colorectal surgery. Surgical Oncology Clinics. 2020;29(1):23–34. doi: 10.1016/j.soc.2019.09.001. [DOI] [PubMed] [Google Scholar]
- 184.Augestad K.M., Merok M.A., Ignatovic D. Tailored treatment of colorectal cancer: surgical, molecular, and genetic considerations. Clin. Med. Insights Oncol. 2017;11 doi: 10.1177/1179554917690766. 1179554917690766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Liang G., et al. HIF1α-associated circDENND4C promotes proliferation of breast cancer cells in hypoxic environment. Anticancer Res. 2017;37(8):4337–4343. doi: 10.21873/anticanres.11827. [DOI] [PubMed] [Google Scholar]
- 186.Wei X., et al. DT-13 inhibited the proliferation of colorectal cancer via glycolytic metabolism and AMPK/mTOR signaling pathway. Phytomedicine. 2019;54:120–131. doi: 10.1016/j.phymed.2018.09.003. [DOI] [PubMed] [Google Scholar]
- 187.Chen X., et al. Circle RNA hsa_circRNA_100290 serves as a ceRNA for miR-378a to regulate oral squamous cell carcinoma cells growth via Glucose transporter-1 (GLUT1) and glycolysis. J. Cell. Physiol. 2019;234(11):19130–19140. doi: 10.1002/jcp.28692. [DOI] [PubMed] [Google Scholar]
- 188.Zhu C., Huang Q., Zhu H. miR-383 inhibited the cell cycle progression of gastric cancer cells via targeting cyclin E2. DNA Cell Biol. 2019;38(8):849–856. doi: 10.1089/dna.2019.4624. [DOI] [PubMed] [Google Scholar]
- 189.Chen L., et al. miR-383 inhibits hepatocellular carcinoma cell proliferation via targeting APRIL. Tumor Biol. 2016;37(2):2497–2507. doi: 10.1007/s13277-015-4071-1. [DOI] [PubMed] [Google Scholar]
- 190.Han R., et al. miR-383 inhibits ovarian cancer cell proliferation, invasion and aerobic glycolysis by targeting LDHA. Neoplasma. 2017;64(2):244–252. doi: 10.4149/neo_2017_211. [DOI] [PubMed] [Google Scholar]
- 191.Yan L., et al. A CREB1/miR-433 reciprocal feedback loop modulates proliferation and metastasis in colorectal cancer. Aging (Albany NY) 2018;10(12):3774. doi: 10.18632/aging.101671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Xu J.-Z., et al. circTADA2As suppress breast cancer progression and metastasis via targeting miR-203a-3p/SOCS3 axis. Cell Death Dis. 2019;10(3):1–16. doi: 10.1038/s41419-019-1382-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Fan G., et al. Loss of KLF14 triggers centrosome amplification and tumorigenesis. Nat. Commun. 2015;6(1):1–13. doi: 10.1038/ncomms9450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Wang Y.G., et al. LncRNA DGCR5 represses the development of hepatocellular carcinoma by targeting the miR-346/KLF14 axis. J. Cell. Physiol. 2019;234(1):572–580. doi: 10.1002/jcp.26779. [DOI] [PubMed] [Google Scholar]
- 195.Zhou J., et al. LncRNA HAND2-AS1 sponging miR-1275 suppresses colorectal cancer progression by upregulating KLF14. Biochem. Biophys. Res. Commun. 2018;503(3):1848–1853. doi: 10.1016/j.bbrc.2018.07.125. [DOI] [PubMed] [Google Scholar]
- 196.Wu G., et al. The KLF14 transcription factor regulates glycolysis by downregulating LDHB in colorectal cancer. Int. J. Biol. Sci. 2019;15(3):628. doi: 10.7150/ijbs.30652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Wang X., et al. Circular RNA NOX4 promotes the development of colorectal cancer via the microRNA-485-5p/CKS1B axis. Oncol. Rep. 2020;44(5):2009–2020. doi: 10.3892/or.2020.7758. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 198.Yuan W., et al. Identification and characterization of circRNAs as competing endogenous RNAs for miRNA-mRNA in colorectal cancer. PeerJ. 2019;7:e7602. doi: 10.7717/peerj.7602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Wang M., et al. miR-485-5p suppresses breast cancer progression and chemosensitivity by targeting survivin. Biochem. Biophys. Res. Commun. 2018;501(1):48–54. doi: 10.1016/j.bbrc.2018.04.129. [DOI] [PubMed] [Google Scholar]
- 200.Kang M., et al. miR-485-5p acts as a negative regulator in gastric cancer progression by targeting flotillin-1. Am. J. Tourism Res. 2015;7(11):2212. [PMC free article] [PubMed] [Google Scholar]
- 201.Hayles J., et al. The fission yeast cell cycle control gene cdc2: isolation of a sequence suc1 that suppresses cdc2 mutant function. Mol. Gen. Genet. MGG. 1986;202(2):291–293. doi: 10.1007/BF00331653. [DOI] [PubMed] [Google Scholar]
- 202.Lee E.-K., et al. Cell-cycle regulator Cks1 promotes hepatocellular Carcinoma by supporting NF-κB–Dependent Expression of interleukin-8skp2-independent Control of IL-8 by Cks1 in liver cancer. Cancer Res. 2011;71(21):6827–6835. doi: 10.1158/0008-5472.CAN-10-4356. [DOI] [PubMed] [Google Scholar]
- 203.Hwang J.-S., et al. MicroRNA-1258 inhibits the proliferation and migration of human colorectal cancer cells through suppressing CKS1B expression. Genes. 2019;10(11):912. doi: 10.3390/genes10110912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Chen Z., et al. CircPLCE1 facilitates the malignant progression of colorectal cancer by repressing the SRSF2-dependent PLCE1 pre-RNA splicing. J. Cell Mol. Med. 2021;25(15):7244–7256. doi: 10.1111/jcmm.16753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Yan Y., et al. RRAD suppresses the Warburg effect by downregulating ACTG1 in hepatocellular carcinoma. OncoTargets Ther. 2019;12:1691. doi: 10.2147/OTT.S197844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Zhuang J., et al. Circular RNA (circ) _0053277 Contributes to colorectal cancer cell growth, angiogenesis, Metastasis and glycolysis. Mol. Biotechnol. 2023:1–15. doi: 10.1007/s12033-023-00936-3. [DOI] [PubMed] [Google Scholar]
- 207.Xiao W., Li P. Circ_0087862 promotes tumorigenesis and glycolysis in colorectal cancer by sponging miR-296-3p to regulate PGK1 expression. Pathol. Res. Pract. 2023;248:154695. doi: 10.1016/j.prp.2023.154695. [DOI] [PubMed] [Google Scholar]
- 208.Ostrom Q.T., et al. The epidemiology of glioma in adults: a “state of the science” review. Neuro Oncol. 2014;16(7):896–913. doi: 10.1093/neuonc/nou087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Hervey-Jumper S.L., Berger M.S. Maximizing safe resection of low-and high-grade glioma. Journal of neuro-oncology. 2016;130(2):269–282. doi: 10.1007/s11060-016-2110-4. [DOI] [PubMed] [Google Scholar]
- 210.Karachi A., et al. Temozolomide for immunomodulation in the treatment of glioblastoma. Neuro Oncol. 2018;20(12):1566–1572. doi: 10.1093/neuonc/noy072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Bagherian A., et al. Combination therapy with nanomicellar-curcumin and temozolomide for in vitro therapy of glioblastoma multiforme via Wnt signaling pathways. J. Mol. Neurosci. 2020;70(10):1471–1483. doi: 10.1007/s12031-020-01639-z. [DOI] [PubMed] [Google Scholar]
- 212.Lim M., et al. Current state of immunotherapy for glioblastoma. Nat. Rev. Clin. Oncol. 2018;15(7):422–442. doi: 10.1038/s41571-018-0003-5. [DOI] [PubMed] [Google Scholar]
- 213.Meng X., et al. Dual functionalized brain-targeting nanoinhibitors restrain temozolomide-resistant glioma via attenuating EGFR and MET signaling pathways. Nat. Commun. 2020;11(1):1–15. doi: 10.1038/s41467-019-14036-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Khani P., et al. Genetic and epigenetic contribution to astrocytic gliomas pathogenesis. J. Neurochem. 2019;148(2):188–203. doi: 10.1111/jnc.14616. [DOI] [PubMed] [Google Scholar]
- 215.Jin P., et al. CircRNA circHIPK3 serves as a prognostic marker to promote glioma progression by regulating miR-654/IGF2BP3 signaling. Biochem. Biophys. Res. Commun. 2018;503(3):1570–1574. doi: 10.1016/j.bbrc.2018.07.081. [DOI] [PubMed] [Google Scholar]
- 216.Shi F., et al. CircRNA hsa-circ-0014359 promotes glioma progression by regulating miR-153/PI3K signaling. Biochem. Biophys. Res. Commun. 2019;510(4):614–620. doi: 10.1016/j.bbrc.2019.02.019. [DOI] [PubMed] [Google Scholar]
- 217.Peng H., et al. circCPA4 acts as a prognostic factor and regulates the proliferation and metastasis of glioma. J. Cell Mol. Med. 2019;23(10):6658–6665. doi: 10.1111/jcmm.14541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Lu H., et al. FBXW7 circular RNA regulates proliferation, migration and invasion of colorectal carcinoma through NEK2, mTOR, and PTEN signaling pathways in vitro and in vivo. BMC Cancer. 2019;19(1):1–8. doi: 10.1186/s12885-019-6028-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Begum S., et al. Novel tumour suppressive protein encoded by circular RNA, circ-SHPRH, in glioblastomas. Oncogene. 2018;37(30):4055–4057. doi: 10.1038/s41388-018-0230-3. [DOI] [PubMed] [Google Scholar]
- 220.Li X., Diao H. Circular RNA circ_0001946 acts as a competing endogenous RNA to inhibit glioblastoma progression by modulating miR-671-5p and CDR1. J. Cell. Physiol. 2019;234(8):13807–13819. doi: 10.1002/jcp.28061. [DOI] [PubMed] [Google Scholar]
- 221.Lv X., et al. Circular RNA circ-PITX1 promotes the progression of glioblastoma by acting as a competing endogenous RNA to regulate miR-379–5p/MAP3K2 axis. Eur. J. Pharmacol. 2019;863:172643. doi: 10.1016/j.ejphar.2019.172643. [DOI] [PubMed] [Google Scholar]
- 222.Guan Y., et al. Circular RNA circPITX1 knockdown inhibits glycolysis to enhance radiosensitivity of glioma cells by miR-329-3p/NEK2 axis. Cancer Cell Int. 2020;20(1):1–13. doi: 10.1186/s12935-020-01169-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Chen M., et al. Circular RNA circ_0074026 indicates unfavorable prognosis for patients with glioma and facilitates oncogenesis of tumor cells by targeting miR-1304 to modulate ERBB4 expression. J. Cell. Physiol. 2020;235(5):4688–4697. doi: 10.1002/jcp.29347. [DOI] [PubMed] [Google Scholar]
- 224.Xiao B., et al. MiRNA-329 targeting E2F1 inhibits cell proliferation in glioma cells. J. Transl. Med. 2013;11(1):1–10. doi: 10.1186/1479-5876-11-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Qiu S., et al. Interactions of miR-323/miR-326/miR-329 and miR-130a/miR-155/miR-210 as prognostic indicators for clinical outcome of glioblastoma patients. J. Transl. Med. 2013;11(1):1–11. doi: 10.1186/1479-5876-11-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Koch A., et al. Characterization of glycolysis-related gene expression in malignant melanoma. Pathol. Res. Pract. 2020;216(1):152752. doi: 10.1016/j.prp.2019.152752. [DOI] [PubMed] [Google Scholar]
- 227.Zhang Z., et al. MFN1-dependent alteration of mitochondrial dynamics drives hepatocellular carcinoma metastasis by glucose metabolic reprogramming. Br. J. Cancer. 2020;122(2):209–220. doi: 10.1038/s41416-019-0658-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Song J., et al. A novel protein encoded by ZCRB1-induced circHEATR5B suppresses aerobic glycolysis of GBM through phosphorylation of JMJD5. J. Exp. Clin. Cancer Res. 2022;41(1):1–20. doi: 10.1186/s13046-022-02374-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Wang H.-J., et al. JMJD5 regulates PKM2 nuclear translocation and reprograms HIF-1α–mediated glucose metabolism. Proc. Natl. Acad. Sci. USA. 2014;111(1):279–284. doi: 10.1073/pnas.1311249111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Gao M., et al. Trametinib inhibits the growth and aerobic glycolysis of Glioma cells by targeting the PKM2/c-Myc Axis. Front. Pharmacol. 2021:2947. doi: 10.3389/fphar.2021.760055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Hua Q., et al. Hypoxia-induced lncRNA-AC020978 promotes proliferation and glycolytic metabolism of non-small cell lung cancer by regulating PKM2/HIF-1α axis. Theranostics. 2020;10(11):4762. doi: 10.7150/thno.43839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Wu C., et al. Global and site-specific effect of phosphorylation on protein turnover. Dev. Cell. 2021;56(1):111–124. doi: 10.1016/j.devcel.2020.10.025. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Luo K., et al. CircKIF4A promotes glioma growth and temozolomide resistance by accelerating glycolysis. Cell Death Dis. 2022;13(8):1–11. doi: 10.1038/s41419-022-05175-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Tay Y., Rinn J., Pandolfi P.P. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505(7483):344–352. doi: 10.1038/nature12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Heyn H., et al. MicroRNA miR-335 is crucial for the BRCA1 regulatory cascade in breast cancer development. Int. J. Cancer. 2011;129(12):2797–2806. doi: 10.1002/ijc.25962. [DOI] [PubMed] [Google Scholar]
- 236.Chang Y.-C., et al. Roles of aldolase family genes in human cancers and diseases. Trends Endocrinol. Metabol. 2018;29(8):549–559. doi: 10.1016/j.tem.2018.05.003. [DOI] [PubMed] [Google Scholar]
- 237.Mu M., et al. CircSOBP suppresses the progression of glioma by disrupting glycolysis and promoting the MDA5-mediated immune response. iScience. 2023;26(10) doi: 10.1016/j.isci.2023.107897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Lei D., Xiao W., Zhang B. CircYIPF6 regulates glioma cell proliferation, apoptosis, and glycolysis through targeting miR-760 to modulate PTBP1 expression. Transl. Neurosci. 2023;14(1) doi: 10.1515/tnsci-2022-0271. 20220271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Hu Z., Ma D. The precision prevention and therapy of HPV-related cervical cancer: new concepts and clinical implications. Cancer Med. 2018;7(10):5217–5236. doi: 10.1002/cam4.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Wang J.-y., Chen L.-j. The role of miRNAs in the invasion and metastasis of cervical cancer. Biosci. Rep. 2019;39(3) doi: 10.1042/BSR20181377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Li H., Wu X., Cheng X. Advances in diagnosis and treatment of metastatic cervical cancer. Journal of gynecologic oncology. 2016;27(4) doi: 10.3802/jgo.2016.27.e43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Ruan H., et al. Comprehensive characterization of circular RNAs iñ 1000 human cancer cell lines. Genome Med. 2019;11(1):1–14. doi: 10.1186/s13073-019-0663-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Luo Y., et al. Diagnostic value of exosomal circMYC in radioresistant nasopharyngeal carcinoma. Head Neck. 2020;42(12):3702–3711. doi: 10.1002/hed.26441. [DOI] [PubMed] [Google Scholar]
- 244.Jin C., et al. CircMYC regulates glycolysis and cell proliferation in melanoma. Cell Biochem. Biophys. 2020;78(1):77–88. doi: 10.1007/s12013-019-00895-0. [DOI] [PubMed] [Google Scholar]
- 245.Xue K., et al. MiR-577 inhibits papillary thyroid carcinoma cell proliferation, migration and invasion by targeting SphK2. Eur. Rev. Med. Pharmacol. Sci. 2017;21(17):3794–3800. [PubMed] [Google Scholar]
- 246.Dong X., et al. Long intergenic non-protein coding RNA 1094 promotes initiation and progression of glioblastoma by promoting microRNA-577-regulated stabilization of brain-derived neurotrophic factor. Cancer Manag. Res. 2020;12:5619. doi: 10.2147/CMAR.S256147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Chen R., et al. CircCDK17 knockdown inhibits tumor progression and cell glycolysis by downregulaing YWHAZ expression through sponging miR-1294 in cervical cancer. J. Ovarian Res. 2022;15(1):1–13. doi: 10.1186/s13048-022-00952-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Zhang Z., et al. Hsa_circ_0004370 promotes esophageal cancer progression through miR-1294/LASP1 pathway. Biosci. Rep. 2019;39(5) doi: 10.1042/BSR20182377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Zhang Z., et al. MicroRNA-1294 targets HOXA9 and has a tumor suppressive role in osteosarcoma. Eur. Rev. Med. Pharmacol. Sci. 2018;22(24):8582–8588. doi: 10.26355/eurrev_201812_16621. [DOI] [PubMed] [Google Scholar]
- 250.Gan Y., Ye F., He X.-X. The role of YWHAZ in cancer: a maze of opportunities and challenges. J. Cancer. 2020;11(8):2252. doi: 10.7150/jca.41316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Neal C.L., et al. 14-3-3ζ overexpression defines high risk for breast cancer recurrence and promotes cancer cell survival. Cancer Res. 2009;69(8):3425–3432. doi: 10.1158/0008-5472.CAN-08-2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Li Y., et al. CircRNA hsa_circ_0018289 exerts an oncogenic role in cervical cancer progression through miR-1294/ICMT axis. J. Clin. Lab. Anal. 2022 doi: 10.1002/jcla.24348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.He J., Lv X., Zeng Z. A potential disease monitoring and prognostic biomarker in cervical cancer patients: The clinical application of circular RNA_0018289. J. Clin. Lab. Anal. 2020;34(8) doi: 10.1002/jcla.23340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Barrett S.P., Salzman J. Circular RNAs: analysis, expression and potential functions. Development. 2016;143(11):1838–1847. doi: 10.1242/dev.128074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Wang M., et al. Inhibition of isoprenylcysteine carboxylmethyltransferase induces autophagic-dependent apoptosis and impairs tumor growth. Oncogene. 2010;29(35):4959–4970. doi: 10.1038/onc.2010.247. [DOI] [PubMed] [Google Scholar]
- 256.Jia C., et al. CircCCNB1 knockdown Blocks the Progression of cervical Cancer by Acting as competing endogenous RNA in the miR-370-3p/SOX4 pathway. Ann. Clin. Lab. Sci. 2023;53(1):94–105. [PubMed] [Google Scholar]
- 257.Thompson B., et al. Breast cancer disparities among women in underserved communities in the USA. Current breast cancer reports. 2018;10(3):131–141. doi: 10.1007/s12609-018-0277-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Ashoor A., Monti S., Pezzella M. Fibromatosis, a benign breast disease mimicking carcinoma. A case report. International journal of surgery case reports. 2017;41:392–397. doi: 10.1016/j.ijscr.2017.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Galluzzi L., et al. Molecular mechanisms of cell death: recommendations of the nomenclature committee on cell death 2018. Cell Death Differ. 2018;25(3):486–541. doi: 10.1038/s41418-017-0012-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Marinko T. Pericardial disease after breast cancer radiotherapy. Radiol. Oncol. 2019;53(1):1–5. doi: 10.2478/raon-2018-0035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Gurrapu S., et al. Sema4C/PlexinB2 signaling controls breast cancer cell growth, hormonal dependence and tumorigenic potential. Cell Death Differ. 2018;25(7):1259–1275. doi: 10.1038/s41418-018-0097-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Xie Z., et al. Gemcitabine-based chemotherapy as a viable option for treatment of advanced breast cancer patients: a meta-analysis and literature review. Oncotarget. 2018;9(6):7148. doi: 10.18632/oncotarget.23426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Kowalczyk L., et al. Adverse events of trastuzumab emtansine (T-DM1) in the treatment of HER2-positive breast cancer patients. Breast Care. 2017;12(6):401–408. doi: 10.1159/000480492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Liang H.-F., et al. Circular RNA circ-ABCB10 promotes breast cancer proliferation and progression through sponging miR-1271. Am. J. Cancer Res. 2017;7(7):1566. [PMC free article] [PubMed] [Google Scholar]
- 265.Fang L., et al. Enhanced breast cancer progression by mutant p53 is inhibited by the circular RNA circ-Ccnb1. Cell Death Differ. 2018;25(12):2195–2208. doi: 10.1038/s41418-018-0115-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Wu J., et al. CircIRAK3 sponges miR-3607 to facilitate breast cancer metastasis. Cancer Lett. 2018;430:179–192. doi: 10.1016/j.canlet.2018.05.033. [DOI] [PubMed] [Google Scholar]
- 267.Cao L., et al. Circular RNA circRNF20 promotes breast cancer tumorigenesis and Warburg effect through miR-487a/HIF-1α/HK2. Cell Death Dis. 2020;11(2):1–10. doi: 10.1038/s41419-020-2336-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Wang Y., et al. SOX2 promotes hypoxia-induced breast cancer cell migration by inducing NEDD9 expression and subsequent activation of Rac1/HIF-1α signaling. Cell. Mol. Biol. Lett. 2019;24(1):1–12. doi: 10.1186/s11658-019-0180-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.De Francesco E.M., Maggiolini M., Musti A.M. Crosstalk between Notch, HIF-1α and GPER in breast cancer EMT. Int. J. Mol. Sci. 2018;19(7):2011. doi: 10.3390/ijms19072011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Zan X., et al. Circ-CSNK1G1 promotes cell proliferation, migration, invasion and glycolysis metabolism during triple-negative breast cancer progression by modulating the miR-28-5p/LDHA pathway. Reprod. Biol. Endocrinol. 2022;20(1):1–14. doi: 10.1186/s12958-022-00998-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Matoulkova E., et al. The role of the 3'untranslated region in post-transcriptional regulation of protein expression in mammalian cells. RNA Biol. 2012;9(5):563–576. doi: 10.4161/rna.20231. [DOI] [PubMed] [Google Scholar]
- 272.Ding B., et al. Whole-transcriptome analysis reveals a potential hsa_circ_0001955/hsa_circ_0000977-mediated miRNA-mRNA regulatory sub-network in colorectal cancer. Aging (Albany NY) 2020;12(6):5259. doi: 10.18632/aging.102945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Zhou S.L., et al. miR-28-5p-IL-34-macrophage feedback loop modulates hepatocellular carcinoma metastasis. Hepatology. 2016;63(5):1560–1575. doi: 10.1002/hep.28445. [DOI] [PubMed] [Google Scholar]
- 274.Almeida M.I., et al. Strand-specific miR-28-5p and miR-28-3p have distinct effects in colorectal cancer cells. Gastroenterology. 2012;142(4):886–896. doi: 10.1053/j.gastro.2011.12.047. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Chen Q., et al. LncRNA MCM3AP-AS1 promotes breast cancer progression via modulating miR-28-5p/CENPF axis. Biomed. Pharmacother. 2020;128:110289. doi: 10.1016/j.biopha.2020.110289. [DOI] [PubMed] [Google Scholar]
- 276.Huang X., et al. High expressions of LDHA and AMPK as prognostic biomarkers for breast cancer. Breast. 2016;30:39–46. doi: 10.1016/j.breast.2016.08.014. [DOI] [PubMed] [Google Scholar]
- 277.Xiao X., et al. The miR-34a-LDHA axis regulates glucose metabolism and tumor growth in breast cancer. Sci. Rep. 2016;6(1):1–9. doi: 10.1038/srep21735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Li L., et al. miR-30a-5p suppresses breast tumor growth and metastasis through inhibition of LDHA-mediated Warburg effect. Cancer Lett. 2017;400:89–98. doi: 10.1016/j.canlet.2017.04.034. [DOI] [PubMed] [Google Scholar]
- 279.Li J., Li Y., Cheng H. Circ-RPPH1 knockdown retards breast cancer progression via miR-328-3p-mediated suppression of HMGA2. Clin. Breast Cancer. 2022;22(3):e286–e295. doi: 10.1016/j.clbc.2021.08.009. [DOI] [PubMed] [Google Scholar]
- 280.Xu X.D., et al. Warburg effect or reverse Warburg effect? A review of cancer metabolism. Oncol. Res. Treat. 2015;38(3):117–122. doi: 10.1159/000375435. [DOI] [PubMed] [Google Scholar]
- 281.Kroemer G., Pouyssegur J. Tumor cell metabolism: cancer's Achilles' heel. Cancer Cell. 2008;13(6):472–482. doi: 10.1016/j.ccr.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 282.Xiao B., et al. Glutamate metabotropic receptor 4 (GRM4) inhibits cell proliferation, migration and invasion in breast cancer and is regulated by miR-328-3p and miR-370-3p. BMC Cancer. 2019;19(1):1–10. doi: 10.1186/s12885-019-6068-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Liu T., et al. Circular RNA hsa_circRNA_002178 silencing retards breast cancer progression via microRNA-328-3p-mediated inhibition of COL1A1. J. Cell Mol. Med. 2020;24(3):2189–2201. doi: 10.1111/jcmm.14875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Grozescu T., Popa F. Prostate cancer between prognosis and adequate/proper therapy. Journal of medicine and life. 2017;10(1):5. [PMC free article] [PubMed] [Google Scholar]
- 285.Rebbeck T.R. Seminars in Radiation Oncology. Elsevier; 2017. Prostate cancer genetics: variation by race, ethnicity, and geography. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Chang A.J., et al. High-risk prostate cancer—classification and therapy. Nat. Rev. Clin. Oncol. 2014;11(6):308–323. doi: 10.1038/nrclinonc.2014.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Wang G., et al. Genetics and biology of prostate cancer. Gene Dev. 2018;32(17–18):1105–1140. doi: 10.1101/gad.315739.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Komura K., et al. Current treatment strategies for advanced prostate cancer. Int. J. Urol. 2018;25(3):220–231. doi: 10.1111/iju.13512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Holm H.V., et al. Modern treatment of metastatic prostate cancer. Tidsskrift for Den norske legeforening. 2017;137(11):803–805. doi: 10.4045/tidsskr.16.0265. [DOI] [PubMed] [Google Scholar]
- 290.Greene J., et al. Differential circRNA expression signatures may serve as potential novel biomarkers in prostate cancer. Front. Cell Dev. Biol. 2021;9:605686. doi: 10.3389/fcell.2021.605686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Kong Z., et al. Circular RNA circFOXO3 promotes prostate cancer progression through sponging miR-29a-3p. J. Cell Mol. Med. 2020;24(1):799–813. doi: 10.1111/jcmm.14791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Xiang Z., et al. CircRNA-UCK2 increased TET1 inhibits proliferation and invasion of prostate cancer cells via sponge MiRNA-767-5p. Open Med. 2019;14(1):833–842. doi: 10.1515/med-2019-0097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Ding Y., Wang M., Yang J. Circular RNA midline-1 (circMID1) promotes proliferation, migration, invasion and glycolysis in prostate cancer. Bioengineered. 2022;13(3):6293–6308. doi: 10.1080/21655979.2022.2037367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Du P., et al. Linc00210 enhances the malignancy of thyroid cancer cells by modulating miR-195-5p/IGF1R/Akt axis. J. Cell. Physiol. 2020;235(2):1001–1012. doi: 10.1002/jcp.29016. [DOI] [PubMed] [Google Scholar]
- 295.Li X.-X., et al. Klotho suppresses growth and invasion of colon cancer cells through inhibition of IGF1R-mediated PI3K/AKT pathway. Int. J. Oncol. 2014;45(2):611–618. doi: 10.3892/ijo.2014.2430. [DOI] [PubMed] [Google Scholar]
- 296.Li Y., et al. The m6A reader protein YTHDC2 is a potential biomarker and associated with immune infiltration in head and neck squamous cell carcinoma. PeerJ. 2020;8 doi: 10.7717/peerj.10385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Zhuang J., Lin C., Ye J. m6A RNA methylation regulators contribute to malignant progression in rectal cancer. J. Cell. Physiol. 2020;235(9):6300–6306. doi: 10.1002/jcp.29626. [DOI] [PubMed] [Google Scholar]
- 298.He J.-J., et al. m6A reader YTHDC2 promotes radiotherapy resistance of nasopharyngeal carcinoma via activating IGF1R/AKT/S6 signaling axis. Front. Oncol. 2020;10:1166. doi: 10.3389/fonc.2020.01166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Jin Z., et al. miR-330-3p suppresses liver cancer cell migration by targeting MAP2K1. Oncol. Lett. 2019;18(1):314–320. doi: 10.3892/ol.2019.10280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Huang Y., et al. HLA-F-AS1/miR-330-3p/PFN1 axis promotes colorectal cancer progression. Life Sci. 2020;254:117180. doi: 10.1016/j.lfs.2019.117180. [DOI] [PubMed] [Google Scholar]
- 301.Li Z., et al. CircTRRAP (hsa_circ_0081234) participates in prostate cancer progression and glycolysis by HOXA1 via functioning as a miR-515-5p sponge. Applied Biological Chemistry. 2022;65(1):1–12. [Google Scholar]
- 302.Zhang G., et al. Inhibition of circ_0081234 reduces prostate cancer tumor growth and metastasis via the miR-1/MAP 3 K1 axis. J. Gene Med. 2022;24(8):e3376. doi: 10.1002/jgm.3376. [DOI] [PubMed] [Google Scholar]
- 303.Qiao K., et al. LINC00673 is activated by YY1 and promotes the proliferation of breast cancer cells via the miR-515-5p/MARK4/Hippo signaling pathway. J. Exp. Clin. Cancer Res. 2019;38(1):1–15. doi: 10.1186/s13046-019-1421-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Zhang Y., et al. Circ_0057553/miR-515-5p regulates prostate cancer cell proliferation, apoptosis, migration, invasion and aerobic glycolysis by targeting YES1. OncoTargets Ther. 2020;13:11289. doi: 10.2147/OTT.S272294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Zhou Z., et al. circROBO1 promotes prostate cancer growth and enzalutamide resistance via accelerating glycolysis. J. Cancer. 2023;14(13):2574. doi: 10.7150/jca.86940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Tischfield M.A., et al. Homozygous HOXA1 mutations disrupt human brainstem, inner ear, cardiovascular and cognitive development. Nat. Genet. 2005;37(10):1035–1037. doi: 10.1038/ng1636. [DOI] [PubMed] [Google Scholar]
- 307.Yuan C., et al. Elevated HOXA1 expression correlates with accelerated tumor cell proliferation and poor prognosis in gastric cancer partly via cyclin D1. J. Exp. Clin. Cancer Res. 2016;35(1):1–16. doi: 10.1186/s13046-016-0294-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Li Q., et al. Over-expressed lncRNA HOTAIRM1 promotes tumor growth and invasion through up-regulating HOXA1 and sequestering G9a/EZH2/Dnmts away from the HOXA1 gene in glioblastoma multiforme. J. Exp. Clin. Cancer Res. 2018;37(1):1–15. doi: 10.1186/s13046-018-0941-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Wardwell-Ozgo J., et al. HOXA1 drives melanoma tumor growth and metastasis and elicits an invasion gene expression signature that prognosticates clinical outcome. Oncogene. 2014;33(8):1017–1026. doi: 10.1038/onc.2013.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Zheng H.-C. The molecular mechanisms of chemoresistance in cancers. Oncotarget. 2017;8(35):59950. doi: 10.18632/oncotarget.19048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Senthebane D.A., et al. The role of tumor microenvironment in chemoresistance: to survive, keep your enemies closer. Int. J. Mol. Sci. 2017;18(7):1586. doi: 10.3390/ijms18071586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Hamadneh L., et al. Upregulation of PI3K/AKT/PTEN pathway is correlated with glucose and glutamine metabolic dysfunction during tamoxifen resistance development in MCF-7 cells. Sci. Rep. 2020;10(1):1–7. doi: 10.1038/s41598-020-78833-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Liu X., et al. Elevated hexokinase II expression confers acquired resistance to 4-hydroxytamoxifen in breast cancer cells. Mol. Cell. Proteomics. 2019;18(11):2273–2284. doi: 10.1074/mcp.RA119.001576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Bray F., et al. The ever-increasing importance of cancer as a leading cause of premature death worldwide. Cancer. 2021;127(16):3029–3030. doi: 10.1002/cncr.33587. [DOI] [PubMed] [Google Scholar]
- 315.Vasan N., Baselga J., Hyman D.M. A view on drug resistance in cancer. Nature. 2019;575(7782):299–309. doi: 10.1038/s41586-019-1730-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Ren T., et al. CircDDX17 reduces 5-fluorouracil resistance and hinders tumorigenesis in colorectal cancer by regulating miR-31-5p/KANK1 axis. Eur. Rev. Med. Pharmacol. Sci. 2020;24(4):1743–1754. doi: 10.26355/eurrev_202002_20351. [DOI] [PubMed] [Google Scholar]
- 317.Wang X., et al. Exosome-delivered circRNA promotes glycolysis to induce chemoresistance through the miR-122-PKM2 axis in colorectal cancer. Mol. Oncol. 2020;14(3):539–555. doi: 10.1002/1878-0261.12629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Bar-Zeev M., Livney Y.D., Assaraf Y.G. Targeted nanomedicine for cancer therapeutics: towards precision medicine overcoming drug resistance. Drug Resist. Updates. 2017;31:15–30. doi: 10.1016/j.drup.2017.05.002. [DOI] [PubMed] [Google Scholar]
- 319.Li W., et al. Overcoming ABC transporter-mediated multidrug resistance: Molecular mechanisms and novel therapeutic drug strategies. Drug Resist. Updates. 2016;27:14–29. doi: 10.1016/j.drup.2016.05.001. [DOI] [PubMed] [Google Scholar]
- 320.Robey R.W., et al. Revisiting the role of ABC transporters in multidrug-resistant cancer. Nat. Rev. Cancer. 2018;18(7):452–464. doi: 10.1038/s41568-018-0005-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Schell J.C., et al. A role for the mitochondrial pyruvate carrier as a repressor of the Warburg effect and colon cancer cell growth. Mol. Cell. 2014;56(3):400–413. doi: 10.1016/j.molcel.2014.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Zhou Y., et al. Intracellular ATP levels Are a pivotal Determinant of Chemoresistance in colon cancer CellsIntracellular ATP in cancer cell chemoresistance. Cancer Res. 2012;72(1):304–314. doi: 10.1158/0008-5472.CAN-11-1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Yang L., et al. Circular RNA circ-0002570 accelerates cancer Progression by regulating VCAN via MiR-587 in gastric cancer. Front. Oncol. 2021:11. doi: 10.3389/fonc.2021.733745. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 324.Chen W., Ji Y. CircC6orf132 facilitates proliferation, migration, invasion, and Glycolysis of gastric cancer cells under Hypoxia by Acting on the miR-873-5p/PRKAA1 Axis. Front. Genet. 2021:12. doi: 10.3389/fgene.2021.636392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Yu T., et al. Circular RNA circ-TNPO3 suppresses metastasis of GC by acting as a protein decoy for IGF2BP3 to regulate the expression of MYC and SNAIL. Mol. Ther. Nucleic Acids. 2021;26:649–664. doi: 10.1016/j.omtn.2021.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Ma X., et al. CircGSK3B promotes RORA expression and suppresses gastric cancer progression through the prevention of EZH2 trans-inhibition. J. Exp. Clin. Cancer Res. 2021;40(1):1–16. doi: 10.1186/s13046-021-02136-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Ganapathy-Kanniappan S., Geschwind J.-F.H. Tumor glycolysis as a target for cancer therapy: progress and prospects. Mol. Cancer. 2013;12(1):1–11. doi: 10.1186/1476-4598-12-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Moreno-Sánchez R., et al. Energy metabolism in tumor cells. FEBS J. 2007;274(6):1393–1418. doi: 10.1111/j.1742-4658.2007.05686.x. [DOI] [PubMed] [Google Scholar]
- 329.Pelicano H., et al. Glycolysis inhibition for anticancer treatment. Oncogene. 2006;25(34):4633–4646. doi: 10.1038/sj.onc.1209597. [DOI] [PubMed] [Google Scholar]
- 330.Feng Y., et al. Circular RNA circBNC2 facilitates glycolysis and stemness of hepatocellular carcinoma through the miR-217/high mobility group AT-hook 2 (HMGA2) axis. Heliyon. 2023;9(6) doi: 10.1016/j.heliyon.2023.e17120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Dai W., et al. circTUBGCP5 promotes liver cancer progression and glycolysis by up-regulating the expression of ACSL4. Applied Biological Chemistry. 2022;65(1):1–13. [Google Scholar]
- 332.Jiang S.-F., Li R.-R. hsa_circ_0067514 suppresses gastric cancer progression and glycolysis via miR-654-3p/LATS2 axis. Neoplasma. 2022:220225. doi: 10.4149/neo_2022_220225N209. N209-220225N209. [DOI] [PubMed] [Google Scholar]
- 333.Zhu L., et al. Circular RNA Circ_0001777 suppresses lung adenocarcinoma progression in Vitro and in vivo. Biochem. Genet. 2022:1–21. doi: 10.1007/s10528-022-10284-7. [DOI] [PubMed] [Google Scholar]
- 334.Zhuang J., et al. Circular RNA (circ)_0053277 Contributes to colorectal cancer cell growth, angiogenesis, Metastasis and glycolysis. Mol. Biotechnol. 2023 doi: 10.1007/s12033-023-00936-3. [DOI] [PubMed] [Google Scholar]
- 335.Li Y., Ma H. circRNA PLOD2 promotes tumorigenesis and Warburg effect in colon cancer by the miR-513a-5p/SIX1/LDHA axis. Cell Cycle. 2022:1–15. doi: 10.1080/15384101.2022.2103339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Zhou Z., et al. circROBO1 promotes prostate cancer growth and enzalutamide resistance via accelerating glycolysis. J. Cancer. 2023;14(13):2574–2584. doi: 10.7150/jca.86940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Chen Y.Y., Luo L.P., Deng K.C. Circular RNA LPAR3 targets JPT1 via microRNA-513b-5p to facilitate glycolytic activation but repress prostate cancer radiosensitivity. Acta Biochim. Pol. 2023;70(1):153–162. doi: 10.18388/abp.2020_6379. [DOI] [PubMed] [Google Scholar]
- 338.Xie Q., et al. hsa_circ_0003596, as a novel oncogene, regulates the malignant behavior of renal cell carcinoma by modulating glycolysis. Eur. J. Med. Res. 2023;28(1):315. doi: 10.1186/s40001-023-01288-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Zhang D., et al. Circular RNA CSPP1 motivates renal cell carcinoma carcinogenesis and the Warburg effect by targeting RAC1 through microRNA-493-5p. Acta Biochim. Pol. 2023;70(3):693–701. doi: 10.18388/abp.2020_6299. [DOI] [PubMed] [Google Scholar]