Abstract
In eukaryotic cells, the site-specific 2′-O-ribose methy-lation of ribosomal RNAs (rRNAs) and the U6 spliceosomal small nuclear RNA (snRNA) is directed by small nucleolar RNAs (snoRNAs). The C and D box-containing 2′-O-methylation guide snoRNAs select the correct substrate nucleotide through formation of a long 10–21 bp interaction with the target rRNA and U6 snRNA sequences. Here, we report on the characterisation of two novel mammalian C/D box snoRNAs, called U83 and U84, that contain all the elements that are essential for accumulation and function of 2′-O-methylation guide snoRNAs. However, in contrast to all of the known 2′-O-methylation guide RNAs, the human, mouse and pig U83 and U84 snoRNAs feature no antisense elements complementary to rRNA or U6 snRNA sequences. The human U83 and U84 snoRNAs are not associated with maturing nucleolar pre-ribosomal particles, suggesting that they do not function in rRNA biogenesis. Since artificial substrate RNAs complementary to the evolutionarily conserved putative substrate recognition motifs of the U83 and U84 snoRNAs were correctly 2′-O-methy-lated in the nucleolus of mouse cells, we suggest that the new snoRNAs act as 2′-O-methylation guides for cellular RNAs other then rRNAs and the U6 snRNA.
INTRODUCTION
The eukaryotic nucleolus is a specialised subnuclear organelle devoted to the biogenesis of cytoplasmic ribosomes (1). This complex process includes the RNA polymerase I-directed synthesis of the long precursor ribosomal RNA (pre-rRNA), its nucleolytic processing into mature sized 18S, 5.8S and 25/28S rRNAs and packaging of the mature rRNAs with more than 80 ribosomal proteins. Formation of functional 18S, 5.8S and 25/28S rRNAs is assisted by many small nucleolar RNAs (snoRNAs) that occur in the form of ribonucleoprotein particles (snoRNPs) (reviewed in 2–5). A few snoRNAs are required for the nucleolytic processing of pre-rRNA, but the majority of them function as guide RNAs in the post-transcriptional modification of rRNAs (for reviews see 4,6). Many snoRNAs that contain the conserved H box and ACA sequence elements direct the site-specific pseudouridylation of rRNAs (7–9). Another large group of snoRNAs which share the C and D box motifs guide the synthesis of 2′-O-methylated nucleotides (10–13).
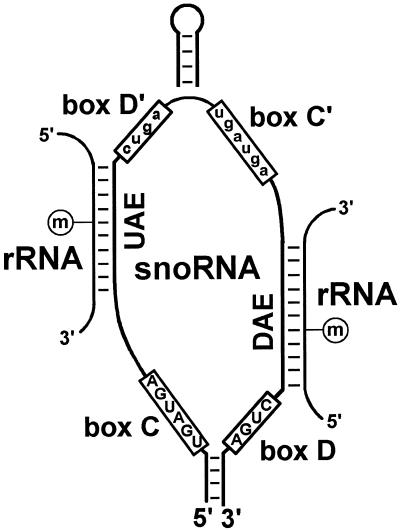
Both classes of rRNA modification guide snoRNAs select the substrate nucleotides through formation of direct base pairing interactions with rRNA sequences. The C and D box-containing 2′-O-methylation guide snoRNAs carry one or, less frequently, two 10–21 nt long rRNA recognition motifs, also known as antisense elements (14). The antisense elements are located either close to the 3′-end of the snoRNA (downstream antisense element) or on the 5′-half of the snoRNA (upstream antisense element) (Fig. 1). The downstream antisense element is followed by the D box and the upstream antisense element is flanked by the D′ box motif, which usually represents a slightly altered version of the D box (10,15). A Watson–Crick helix formed by the antisense element of the snoRNA and the complementary rRNA sequences places the D or D′ box of the snoRNA exactly 5 bp from the 2′-O-methylated nucleotide in the rRNA (Fig. 1). A putative methyltransferase enzyme, most likely the fibrillarin snoRNP protein (16,17), which binds directly or indirectly to the D and D′ box of the snoRNA selects the target nucleotide to be methylated. The C′ box and probably also the C box play a crucial role in the methytransfer reaction directed by the upstream or downstream antisense element, respectively (13).
Figure 1.
Selection of 2′-O-methylated nucleotides by box C/D snoRNAs. The consensus sequences of the conserved C, C′, D and D′ boxes are indicated. The upstream (UAE) and/or downstream (DAE) antisense element of the snoRNA forms a double helix with complementary rRNA sequences. A nucleotide in the rRNA that base pairs with the fifth nucleotide 5′ to the D or D′ box of the snoRNA is 2′-O-methylated. The position of an internal hairpin that frequently folds together the D′ and C′ boxes is indicated (13).
Recently, snoRNAs have also been implicated in site-specific modification of the U6 spliceosomal small nuclear RNA (snRNA). Ribose methylation of the C77 residue in an in vitro synthesised human U6 snRNA, after injection into Xenopus oocytes, is dependent on the presence of a C/D box snoRNA, termed mgU6-77 (18). The antisense elements of two other C/D box snoRNAs, mgU6-47 and mgU6-53, can position the mA47 and mA53 residues of the U6 snRNA for 2′-O-methylation (18,19). Moreover, the nucleolus contains all the factors that are required for the site-specific synthesis of the remaining five ribose-methylated nucleotides of mammalian U6 snRNA (19). The correct recognition of these methylation sites depends on short snRNA sequences located around the target nucleotide. These findings strongly support the idea that 2′-O-ribose methylation of the U6 snRNA is mediated exclusively by C/D box snoRNAs.
In this study, identification and characterisation of two novel C and D box-containing snoRNAs, termed U83 and U84, suggest that C/D box snoRNAs may also function in 2′-O-methylation of cellular RNAs other than the 18S, 5.8S and 25/28S rRNAs or the U6 spliceosomal RNA.
MATERIALS AND METHODS
Oligodeoxynucleotides
Oligonucleotides used for PCR amplification, DNA sequencing, primer extension or cloning were synthesised by the standard phosphoramidite method and purified on a 20% polyacrylamide–8 M urea gel. The following oligonucleotides were used in this study: 1, CTAGTCAAGGGTGATAGA; 2, TCGATCTATCACCCTTGA; 3, CTAGTCATGGGTGATAGA; 4, TCGATCTATCACCCATGA; 5, GCGCAAAGCGCTCAC-CTTTCG; 6, GCACTGAGGTGCTCCTGTTT; 7, GGAGCA-CCTCATGTGCA; 8, GCGCTTTGCGCAGTGAT; 9, NNNN-TATCACCCATG.
Expression constructs for transfection of mouse cells
Construction of the pW(Xb/Xh) mouse ribosomal minigene expression vector has been described (20,21). To obtain pW-U84t, oligodeoxynucleotides 1 and 2 were annealed and inserted into the XbaI and XhoI sites of pW(Xb/Xh). The same approach was used to generate pW-U83t (oligos 3 and 4). Transfection of plated mouse L929 cells (ATCC CCL1) was performed by the DEAE–dextran method (22).
RNA analyses
RNAs from human HeLa and mouse L929 cells or from the nuclear, nucleolar, nucleoplasmic and cytoplasmic fractions of HeLa cells were extracted by the guanidine thiocyanate/phenol-chloroform extraction method (23). Primer extension analysis of the 5′-termini of human U83 and U84 snoRNAs was performed using 5′-end-labelled oligonucleotides 5 and 6 as primers, respectively, and the samples were electrophoresed on denaturing 8% polyacrylamide gels. The 3′-terminal sequences of U83 and U84 were determined by the T4 RNA ligase–PCR approach as described (24), except that oligos 7 and 8 were used as U83- and U84-specific primers for the PCR amplification reaction.
RNase A/T1 mappings were performed as described (23). Antisense RNA probes for the human U3 snoRNA and the U4 snRNA were synthesised in vitro (24). To generate sequence-specific probes for the human U83 and U84 snoRNAs, the pU83 and pU84 recombinant plasmids carrying the full-length cDNAs of U83 and U84 were linearised with EcoRI and used as templates for in vitro transcription with T3 RNA polymerase. The probes were purified on a 6% sequencing gel.
Ribose-methylated nucleotides were mapped by primer extension analyses (25). To monitor 2′-O-methylation of the mouse ribosomal minigene transcripts, the 5′-end-labelled 3-oCAT2 oligonucleotide (20), complementary to the pW transcript downstream from the inserted target sequences, was used as primer. To detect a potential target RNA for the U83 2′-O-methylation guide snoRNA, 2 pmol of terminally labelled oligo 9 was annealed to 10 µg of HeLa total cellular or nuclear RNA and extended by AMV reverse transcriptase. The extended DNA products were analysed on a 6% sequencing gel.
Cell fractionation and glycerol gradient analyses
Human HeLa cells were fractionated into nuclear, nucleolar, nucleoplasmic and cytoplasmic fractions as described (26). Preparation and fractionation of HeLa cell extracts on 10–30% glycerol gradients were performed as described (26,27).
RESULTS
Mammalian U83 and U84 C/D box snoRNAs lack sequences complementary to rRNAs
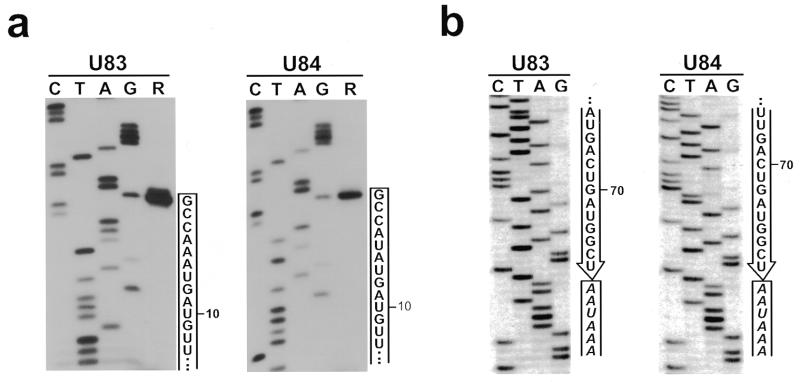
Partial sequences of two novel putative snoRNAs were obtained during characterisation of a cDNA library of human snoRNAs (10). Northern assays of human HeLa and mouse cellular RNAs revealed that the new RNAs contain about 75–80 nt, whose sequences are conserved in mammalian cells (data not shown). The 3′-terminal sequences of the two human RNAs were determined by the oligoribonucleotide ligation–PCR amplification procedure (24; Fig. 2b) and the correct 5′-ends were defined by primer extension analyses (Fig. 2a). The novel human RNAs, hereafter named U83 and U84, consist of 76 and 78 nt, respectively (Fig. 3). Database searches revealed that sequences 80–85% homologous to the human U83 and U84 RNAs are present in the fifth and first introns, respectively, of the pig and mouse BAT1 gene that encodes for a putative RNA helicase (GenBank accession nos Z34846 and AC007080). Moreover, a perfect copy of the U83 RNA was found in the fifth intron of the human BAT1 gene (GenBank accession no. AF029062). Unfortunately, the 5′-terminal part of the human BAT1 gene that may encompass the human U84 RNA gene has not yet been characterised. Nevertheless, we concluded that the U83 and U84 RNAs represent novel intron-encoded RNAs that are processed from the fifth and first introns of the mammalian BAT1 gene.
Figure 2.
Characterisation of human U83 and U84 snoRNAs. (a) Primer extension analyses of the 5′-termini of U83 and U84 RNAs. Terminally labelled oligonucleotides specific for U83 and U84 were annealed to human nuclear RNAs and extended by AMV reverse transcriptase (lanes R). Lanes C, T, A and G represent dideoxy sequencing reactions using the same oligonucleotides as primers and recombinant plasmids carrying the full-length cDNAs of U83 and U84 as templates. The extended products were separated on an 8% sequencing gel. The 5′-terminal sequences of the U83 and U84 RNAs are shown. (b) Determination of the 3′-terminal sequences of human U83 and U84 snoRNAs by the T4 RNA ligase/PCR procedure. The 3′-terminal sequences of the two snoRNAs and the 5′-terminal sequence of the oligoribonucleotide ligated to the U83 and U84 RNAs are shown.
Figure 3.
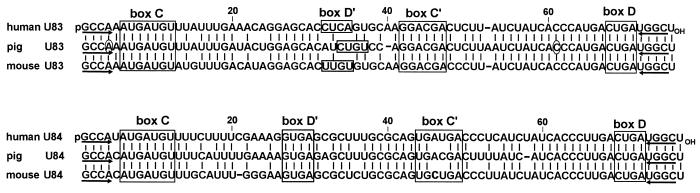
Alignments of human, pig and mouse U83 and U84 snoRNAs. The C and D boxes and the potential C′ and D′ box motifs are indicated. The vertical lines highlight nucleotides conserved in the aligned snoRNAs and dashes stand for gaps. Inverted arrows indicate sequences capable of forming base pairing interactions. The presence of an additional A and C residue in the pig U83 RNA at positions 4 and 61 (circled), as compared to the published sequence (GenBank accession no. Z34846), has been verified by sequence analysis of the appropriate region of the pig BAT1 locus (data not shown). Sequences of the human U83 and U84 snoRNAs have been deposited in the EMBL database (accession nos AJ243200 and AJ243199, respectively).
Alignments of the human, pig and mouse U83 and U84 RNAs are shown in Figure 3. These RNAs feature all of the elements, the C, C′, D and D′ box sequence motifs and a short 5′-, 3′-terminal helix structure, that are essential for nucleolar accumulation and function of intron-encoded 2′-O-methylation guide snoRNAs directing the modification of rRNAs and U6 snRNA (18,19,28–30). However, neither U83 nor U84 RNA possesses sequences that could form a long base pairing interaction with rRNA or U6 snRNA sequences. The best interaction, potentially formed by U83 (positions 58–66) or U84 (positions 60–68) snoRNA with the 18S rRNA sequence between positions 404 and 413, would consist of eight canonical base pairs interrupted by one non-canonical C:A or C:U base pair, respectively. Based on a recent functional analysis of the guide RNA and rRNA duplex (31), such a poor interaction would be insufficient to support the 2′-O-methylation reaction. Consistent with this conclusion, no 2′-O-methylated nucleotide was reported in this region of the human 18S rRNA (32).
U83 and U84 RNAs are nucleolar but are not associated with precursor ribosomal particles
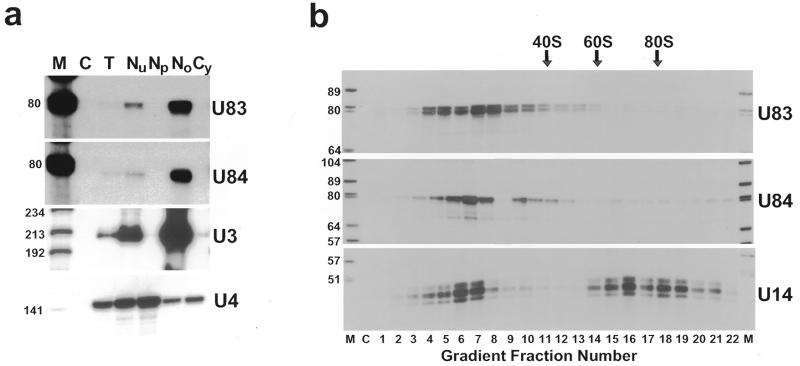
We assayed the intracellular distribution of the U83 and U84 RNAs, whose C and D box elements predict a nucleolar localisation. Human HeLa cells were fractionated into nuclear, nucleoplasmic, nucleolar and cytoplasmic fractions. RNAs obtained from each fraction as well as from whole HeLa cells were mapped by RNase A/T1 protection using antisense RNA probes specific for the U83 and U84 RNAs (Fig. 4a). As controls, we also measured the U3 snoRNA and the U4 spliceosomal RNA content of each fraction. The U83 and U84 RNAs, like the U3 snoRNA, were highly enriched in the nucleolar fraction of HeLa cells and were hardly detectable in the nucleoplasmic fraction where U4 snRNA accumulated, demonstrating that the U83 and U84 RNAs represent authentic snoRNAs.
Figure 4.
Human U83 and U84 RNAs are not associated with higher order nucleolar structures. (a) Intracellular localisation of human U83 and U84 snoRNAs. RNAs extracted either from human HeLa cells (T) or from nuclear (N), nucleoplasmic (Np), nucleolar (No) and cytoplasmic (Cy) fractions of HeLa cells were mapped by RNase A/T1 protection using sequence-specific antisense RNA probes as indicated. Lane C, control mapping with Escherichia coli tRNA. Lane M, size markers (a mixture of HaeIII- and TaqI-digested pBR322). (b) Sedimentation analyses of U83, U84 and U14 snoRNP particles. HeLa cell extract was fractionated on a 10–30% glycerol gradient. RNAs were isolated from each fraction and subjected to RNase A/T1 mapping with antisense RNA probes specific for the U83, U84 or U14 snoRNA. Positions of HeLa ribosome markers are indicated. Lanes C and M represent control mappings and size markers, respectively.
The nucleolar localisation of the U83 and U84 snoRNAs suggests that they may function in the nucleolar maturation of rRNAs. snoRNAs involved in the nucleolytic processing or nucleotide modification of rRNAs are associated with higher order structures that likely represent ribosomal particles undergoing maturation in the nucleolus (11,21,26,27,33,34). To assess whether the U83 and U84 snoRNAs are associated with large nucleolar structures, a HeLa cell extract was fractionated in a non-denaturing glycerol gradient (Fig. 4b). Distribution of the U83 and U84 snoRNAs as well as U14 snoRNA, which functions both in the nucleolytic processing and 2′-O-methylation of 18S rRNA (35,36), was investigated by RNase mapping. About half of the cellular U14 snoRNA sedimented together with large particles that showed sedimentation properties similar to the 60S and 80S cytoplasmic ribosomal particles. Very similar results were obtained with another snoRNA, U3 snoRNA (data not shown), which functions in the early processing of pre-rRNA (37–40). In contrast to the U14 and U3 snoRNAs, the U83 and U84 snoRNAs sedimented exclusively in 10–20S structures that likely represent snoRNP monoparticles. These findings indicate that the U83 and U84 snoRNPs are not associated with pre-ribosomal particles in the nucleolus.
U83 and U84 snoRNAs direct 2′-O-methylation of artificial substrate RNAs
A few C/D box snoRNPs, such as the U3 (37–40), U8 (41,42), U22 (43) and U14 (35,44) RNPs, function in the nucleolytic processing of rRNAs. These snoRNPs, similarly to the authentic rRNA 2′-O-methylation particles, also contain the fibrillarin protein that likely catalyses the 2′-O-methyltransfer reaction (16,17). This is contrary to the fact that the above mentioned snoRNPs, with the exception of U14, apparently do not function in rRNA methylation. We therefore examined whether U83 and U84 snoRNP particles possess the potential to direct RNA ribose methylation.
Sequences immediately preceding the D box motifs of the U83 and U84 snoRNAs are perfectly conserved in the human, mouse and pig RNAs (Fig. 2). We examined whether the mouse intracellular U83 and U84 snoRNAs are capable of directing site-specific 2′-O-methylation of properly designed artificial substrate RNAs in the nucleolus. To this end, two short DNA fragments (U83t and U84t) were inserted into a mouse ribosomal minigene (W) that possesses the RNA polymerase I promoter and terminator regions but lacks rRNA sequences (20; Fig. 5a). The RNA transcripts generated from the W-U83t and W-U84t constructs carry 15 nt long sequences that are perfectly complementary to the putative downstream antisense elements of the U83 or U84 snoRNAs. Since the RNA polymerase I-directed transcription of the ribosomal minigenes occurs in the nucleolus, the W-U83t and W-U84t RNAs are expected to accumulate in the nucleolus (19–21).
Figure 5.
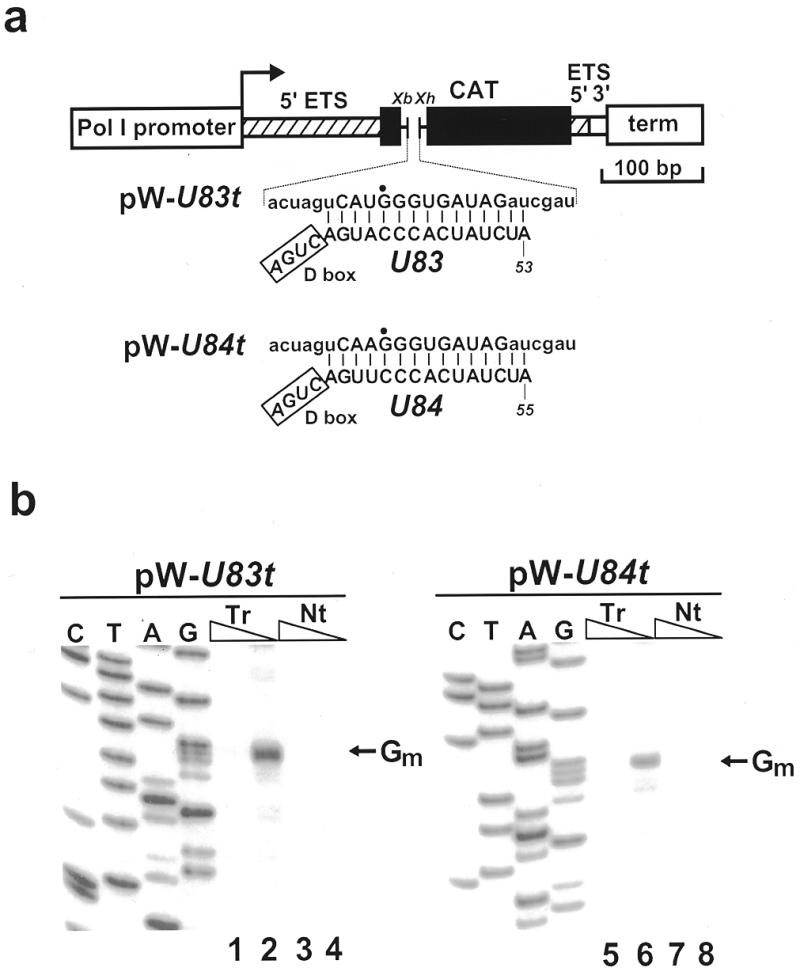
Ribose methylation of artificial substrate RNAs in mouse cells. (a) Schematic structure of the pW-U83t and pW-U84t expression constructs used for transfection of mouse cells. The RNA polymerase I promoter and terminator, the terminal regions of the 5′ (hatched box) and 3′ (open box) external transcribed sequences (ETS) and a fragment of the chloramphenicol acetyltransferase (CAT) gene are indicated. To generate pW-U83t and pW-U84t, appropriate synthetic DNA fragments were inserted into the XbaI (Xb) and XhoI (Xh) sites of pW(Xb/Xh). Nucleotides facilitating the cloning are in lower case letters. The potential base pairing interactions formed between the expressed artificial substrate RNAs and the putative antisense elements of the U83 and U84 snoRNA are shown. Nucleotides predicted to be 2′-O-methylated are indicated by closed circles. (b) Primer extension mapping of 2′-O-methylated nucleotides. A 5′-end-labelled deoxyoligonucleotide was annealed with RNAs extracted from mouse cells non-transfected (Nt) or transfected (Tr) with the indicated expression constructs and extended with AMV reverse transcriptase in the presence of 1 or 0.004 mM dNTPs (as indicated above the lanes). Lanes C, T, A and G are dideoxy sequencing reactions performed on the pW-U83t or pW-U84t expression constructs.
The pW-U83t and pW-U84t constructs were transfected into mouse cells and the state of ribose methylation of the expressed minigene transcripts was monitored by primer extension (Fig. 5b). In the presence of low concentration of dNTPs, ribose-methylated nucleotides interfere with the passage of reverse transcriptase and result in stops 1 nt before or at the modified nucleotide (25). When RNAs obtained from cells transfected with the pW-U83t or pW-U84t expression construct were analysed in the presence of 0.004 mM dNTPs, strong stop signals were obtained 1 nt before the G residues that are predicted to be 2′-O-methylated by the endogenous U83 and U84 snoRNPs (Fig. 5, lanes 2 and 6). In contrast, no reverse transcriptase stops were observed in the presence of 1 mM dNTPs (lanes 1 and 5) or during mapping of control RNAs derived from non-transfected mouse cells (lanes 3, 4, 7 and 8). These results show that the U83 and U84 snoRNAs are capable of directing site-specific 2′-O-methylation of artificial substrate RNAs and suggest that these snoRNAs likely function in the nucleolus as 2′-O-methylation guide RNAs.
In an attempt to detect a potential target RNA for the U83 methylation guide RNA, a terminally labelled oligodeoxynucleotide, identical to the U83 snoRNA sequences from position 56 to 66 and extended by four randomly synthesised 5′-terminal nucleotides, was annealed to human HeLa total cellular or nuclear RNAs. After incubation with AMV reverse transcriptase, an extended DNA product of ~82–83 nt was reproducibly detected in total cellular RNAs (Fig. 6, lane 2) and, more efficiently, in RNAs extracted from the nuclear fraction of HeLa cells (lane 1). To identify this RNA molecule and to investigate whether it really represents the natural substrate of the U83 2′-O-methylation guide snoRNP remains a challenge for the future.
Figure 6.
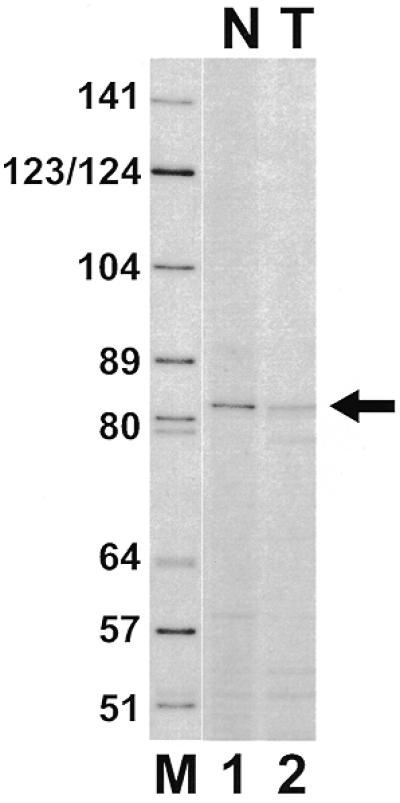
Primer extension analysis. RNAs obtained from human HeLa cells (T) or from a nuclear fraction of HeLa cells (N) were annealed with 5′-end-labelled oligo 9 and extended with AMV reverse transcriptase. The extended DNA products were size-fractionated on a 6% sequencing gel. Lane M, size marker.
DISCUSSION
We have identified two novel mammalian snoRNAs that represent new members of the family of C and D box-containing intron-encoded snoRNAs (Fig. 2). The mouse, pig and, most likely, human U83 and U84 snoRNAs are encoded in the fifth and first introns of the BAT1 putative RNA helicase gene, respectively.
More than 50 C/D box snoRNAs have been identified in vertebrates and their final number is predicted to exceed 100 (3–4). The vast majority of C/D box snoRNAs function as guide RNAs in the site-specific 2′-O-methylation of rRNAs. Selection of the ribosomal 2′-O-methylation sites is mediated by transient base pairing interactions formed between the target ribosomal sequences and the antisense elements of the guide snoRNAs (Fig. 1). The efficiency of the methyltransfer reaction greatly depends on the length and regularity of the snoRNA–rRNA duplex formed (31). The natural rRNA methylation guide snoRNAs possess at least 10 nt long rRNA antisense elements. A snoRNA–rRNA interaction composed of eight canonical and one non-canonical base pairs—the best interaction that could be drawn between the U83 or U84 snoRNA and mammalian 18S rRNA (see Results)—would not be expected to support rRNA methylation (31). In accordance with this conclusion, no 2′-O-methyl group was encountered in this particular region of the mammalian 18S rRNA (32). Hence, we concluded that neither the U83 nor the U84 snoRNA can direct ribose methylation of rRNAs in mammalian cells. A few C/D box snoRNAs, such as U3, U8, U14 and U22, function in the nucleolytic processing of pre-rRNAs (reviewed in 3,4). At the moment, we cannot unambiguously exclude the formal possibility that the U83 and U84 snoRNAs may function in the nucleolytic processing of rRNAs. However, in marked contrast to snoRNAs involved in rRNA modification or processing, the U83 and U84 snoRNAs are not associated with precursor ribosomal particles. This strongly argues against a role for these snoRNAs in rRNA processing.
A comparison of mammalian U83 and U84 snoRNAs revealed that, beside the C and D box motifs, the 3′-terminal regions preceding the D boxes show the highest degree of conservation (Fig. 2). In fact, these sequences are perfectly conserved in human, mouse and pig RNAs, underlining their functional importance. The demonstration that artificial substrate RNAs complementary to the conserved putative downstream antisense regions of the U83 and U84 snoRNAs are accurately 2′-O-methylated in mouse cells strongly supports the idea that the U83 and U84 snoRNAs function as 2′-O-ribose methylation guide RNAs in the cell (Fig. 5).
The notion that the U83 and U84 snoRNAs function as methylation guide RNAs raises questions about the nature of their target RNAs. The potential target recognition sequences of U83 and U84 show a striking similarity to each other (Fig. 2), indicating that these snoRNAs might direct the 2′-O-methylation of two sequence variants of the same cellular RNA. Primer extension analyses of human cellular RNAs revealed an RNA molecule that carries a sequence motif complementary to the putative RNA recognition motif of the U83 snoRNA (Fig. 6). Interestingly, the potential target RNA for the U83 guide snoRNA was detected mainly in the nuclear fraction of human cellular RNAs.
Recently, C/D box snoRNAs have been demonstrated to function in the 2′-O-methylation of U6 spliceosomal snRNA (18,19). Most likely, synthesis of the eight 2′-O-methylated nucleotides of mammalian U6 snRNA is directed exclusively by C/D box snoRNAs (19). Moreover, guide RNAs, most probably H/ACA box snoRNAs, have been implicated in the synthesis of the pseudouridine residues present in the U6 snRNA and the U3 snoRNA (19). These results show that snoRNAs can function in the post-transcriptional modification of various classes of cellular RNAs. Besides maturing rRNAs and authentic snoRNAs, several small RNAs are believed to appear, at least transiently, in the nucleolus (reviewed in 45). Some yeast precursor tRNAs cycle through the nucleolus to undergo nucleolytic processing (46). A fraction of mammalian telomerase (47), signal recognition particle (48) and RNase P (49) RNAs has been found in the nucleolus. Upon microinjection into the nucleoplasm, in vitro synthesised signal recognition particle (50), RNase MRP (51), RNase P (49) and U6 (A.Narayanan, R.Terns and M.Terns, personal communication) RNAs transiently localise to the nucleolus, suggesting that the nucleolus may function in the biogenesis of these RNAs. Unfortunately, computer-aided inspection of vertebrate small RNA sequences, including U snRNAs, snoRNAs, the telomerase, RNase P and MRP RNAs, as well as the cytoplasmic signal recognition particle RNA and the known tRNAs, failed to identify a potential substrate for the U83 or U84 2′-O-methylation guide snoRNAs. Of course, it is also possible that the U83 and U84 snoRNAs function in the 2′-O-methylation of some as yet unidentified snRNAs (52).
In summary, characterisation of the U83 and U84 2′-O-methylation guide snoRNAs that most likely function in the post-transcriptional modification of an as yet unidentified cellular RNA lends further support to the recently emerging idea that the nucleolus is a multifunctional subnuclear organelle that functions in the maturation of different classes of cellular RNAs.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Y. de Preval for synthesis of oligodeoxynucleotides. B. E. Jády was funded by the French Government and the Hungarian Research Foundation (OTKA, T 029042). This work was supported by the Centre National de la Recherche Scientifique and by grants from la Ligue Nationale Contre le Cancer, l’Association pour la Recherche sur le Cancer and the Hungarian Research Foundation (OTKA, T 029042).
DDBJ/EMBL/GenBank accession nos AJ243199, AJ243200
REFERENCES
- 1.Hadjiolov A.A. (1985) The Nucleolus and Ribosome Biogenesis. Springer-Verlag, Vienna, Austria.
- 2.Maxwell E.S. and Fournier,M.J. (1995) Annu. Rev. Biochem., 64, 897–934. [DOI] [PubMed] [Google Scholar]
- 3.Sollner-Webb B., Tycowski,K.T. and Steitz,J.A. (1996) In Zimmermann,R.A. and Dahlberg,A.E. (eds), Structure, Evolution, Processing and Function in Protein Biosynthesis. CRC Press, Boca Raton, FL, pp. 469–490.
- 4.Tollervey D. and Kiss,T. (1997) Curr. Opin. Cell Biol., 9, 337–342. [DOI] [PubMed] [Google Scholar]
- 5.Weinstein L.B. and Steitz,J.A. (1999) Curr. Opin. Cell Biol., 11, 378–384. [DOI] [PubMed] [Google Scholar]
- 6.Smith C.M. and Steitz,J.A. (1997) Cell, 89, 669–672. [DOI] [PubMed] [Google Scholar]
- 7.Ganot P., Bortolin,M.L. and Kiss,T. (1997) Cell, 89, 799–809. [DOI] [PubMed] [Google Scholar]
- 8.Ni J., Tien,A.L. and Fournier,M.J. (1997) Cell, 89, 565–573. [DOI] [PubMed] [Google Scholar]
- 9.Bortolin M.L., Ganot,P. and Kiss,T. (1999) EMBO J., 18, 457–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kiss-László Z., Henry,Y., Bachellerie,J.P., Caizergues-Ferrer,M. and Kiss,T. (1996) Cell, 85, 1077–1088. [DOI] [PubMed] [Google Scholar]
- 11.Cavaillé J., Nicoloso,M., Bachellerie,J.P. (1996) Nature, 383, 732–735. [DOI] [PubMed] [Google Scholar]
- 12.Tycowski K.T., Smith,C.M., Shu,M.D. and Steitz,J.A. (1996) Proc. Natl Acad. Sci. USA, 93, 14480–14485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kiss-László Z., Henry,Y. and Kiss,T. (1998) EMBO J., 17, 797–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bachellerie J.P., Michot,B., Nicoloso,M., Balakin,A., Ni,J. and Fournier,M.J. (1995) Trends Biochem. Sci., 20, 261–264. [DOI] [PubMed] [Google Scholar]
- 15.Tycowski K.T., Shu,M.D. and Steitz,J.A. (1996) Nature, 379, 464–466. [DOI] [PubMed] [Google Scholar]
- 16.Tollervey D., Lehtonen,H., Jansen,R., Kern,H. and Hurt,E.C. (1993) Cell, 72, 443–457. [DOI] [PubMed] [Google Scholar]
- 17.Niewmierzycka A. and Clarke,S. (1999) J. Biol. Chem., 274, 814–824. [DOI] [PubMed] [Google Scholar]
- 18.Tycowski K., You,Z.H., Graham,P.J. and Steitz,J.A. (1998) Mol. Cell, 2, 629–638. [DOI] [PubMed] [Google Scholar]
- 19.Ganot P., Jády,B.E., Bortolin,M.L., Darzacq,X. and Kiss,T. (1999) Mol. Cell. Biol., 19, 6906–6917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hadjiolova K.V., Normann,A., Cavaillé,J., Soupene,E., Mazan,S., Hadjiolov,A.A., Bachellerie,J.P. (1994) Mol. Cell. Biol., 14, 4044–4056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ganot P., Caizergues-Ferrer,M. and Kiss,T. (1997) Genes Dev., 11, 941–956. [DOI] [PubMed] [Google Scholar]
- 22.Selden R.F. (1992) In Ausubel,F.M., Brent,R., Kingston,R.E., Moore,D.D. and Seidman,J.G. (eds), Current Protocols in Molecular Biology. John Wiley & Sons, New York, NY, pp. 9.2.1–9.2.4.
- 23.Goodall G.J., Wiebauer,K. and Filipowicz,W. (1990) Methods Enzymol., 181, 148–161. [DOI] [PubMed] [Google Scholar]
- 24.Kiss T. and Filipowicz,W. (1993) EMBO J., 12, 2913–2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maden B.E., Corbett,M.E., Heeney,P.A., Pugh,K. and Ajuh,P.4M. (1995) Biochimie, 77, 22–29. [DOI] [PubMed] [Google Scholar]
- 26.Tyc K. and Steitz,J.A. (1989) EMBO J., 8, 3113–3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kiss T., Marshallsay,C. and Filipowicz,W. (1992) EMBO J., 11, 3737–3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Caffarelli E., Fatica,A., Prislei,S., De Gregorio,E., Fragapane,P. and Bozzoni,I. (1996) EMBO J., 15, 1121–1131. [PMC free article] [PubMed] [Google Scholar]
- 29.Watkins N.J., Leverette,R.D., Xia,L., Andrews,M.T. and Maxwell,E.S. (1996) RNA, 2, 118–133. [PMC free article] [PubMed] [Google Scholar]
- 30.Cavaillé J. and Bachellerie,J.P. (1996) Biochimie, 78, 443–456. [DOI] [PubMed] [Google Scholar]
- 31.Cavaillé J. and Bachellerie,J.P. (1998) Nucleic Acids Res., 26, 1576–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maden B.E. (1990) Prog. Nucleic Acid Res. Mol. Biol., 39, 241–301. [DOI] [PubMed] [Google Scholar]
- 33.Epstein P., Reddy,R. and Busch,H. (1984) Biochemistry, 23, 5421–5425. [DOI] [PubMed] [Google Scholar]
- 34.Kiss T., Bortolin,M.L. and Filipowicz,W. (1996) Mol. Cell. Biol., 16, 1391–1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li H.D., Zagorski,J. and Fournier,M.J. (1990) Mol. Cell. Biol., 10, 1145–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dunbar D.A. and Baserga,S.J. (1998) RNA, 4, 195–204. [PMC free article] [PubMed] [Google Scholar]
- 37.Kass S., Tyc,K., Steitz,J.A. and Sollner-Webb,B. (1990) Cell, 60, 897–908. [DOI] [PubMed] [Google Scholar]
- 38.Hughes J.M.X. and Ares,M.,Jr (1991) EMBO J., 10, 4231–4239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Savino R. and Gerbi,S.A. (1990) EMBO J., 9, 2299–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mougey E.B., Pape,L.K. and Sollner-Webb,B. (1993) Mol. Cell. Biol., 13, 5990–5998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peculis B.A. and Steitz,J.A. (1993) Cell, 73, 1233–1245. [DOI] [PubMed] [Google Scholar]
- 42.Peculis B.A. (1997) Mol. Cell. Biol., 17, 3702–3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tycowski K.T., Shu,M.-D. and Steitz,J.A. (1994) Science, 266, 1558–1561. [DOI] [PubMed] [Google Scholar]
- 44.Liang W.-Q. and Fournier,M.J. (1995) Genes Dev., 9, 2433–2443. [DOI] [PubMed] [Google Scholar]
- 45.Pederson T. (1998) Nucleic Acids Res., 26, 3871–3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bertrand E., Houser-Scott,F., Kendall,A., Singer,R.H. and Engelke,D.R. (1998) Genes Dev., 12, 2463–2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mitchell J.R., Cheng,J. and Collins,K. (1999) Mol. Cell. Biol., 19, 567–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reddy R., Li,W.Y., Henning,D., Choi,Y.C., Nohga,K. and Busch,H. (1981) J. Biol. Chem., 256, 8452–8457. [PubMed] [Google Scholar]
- 49.Jacobson M.R., Cao,L.G., Taneja,K., Singer,R.H., Wang,Y.L. and Pederson,T. (1997) J. Cell Sci., 110, 829–837. [DOI] [PubMed] [Google Scholar]
- 50.Jacobson M.R. and Pederson,T. (1998) Proc. Natl Acad. Sci. USA, 95, 7981–7986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jacobson M.R., Cao,L.-G., Wang,Y.-L. and Pederson,T. (1995) J. Cell Biol., 131, 1649–1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yu Y.-T., Tarn,W.-Y. Yario,T.A. and Steitz,J.A. (1996) Exp. Cell Biol., 229, 276–281. [Google Scholar]