Abstract
Endonuclease genes encoded in invasive introns are themselves supposed to be mobile elements which, during evolution, have colonized pre-existing introns converting them into invasive elements. This hypothesis is supported by numerous data concerning the LAGLI-DADG subclass of intronic endonucleases. Less is known about the GIY-YIG ORFs which constitute another family of endonucleases. In this paper we describe the presence of one optional GIY-YIG ORF in the second intron of the mitochondrial cytochrome b gene in the fungus Podospora curvicolla. We show that this GIY-YIG ORF is efficiently transferred from an ORF-containing intron to an ORF-less allele. We also show that the products of both the GIY-YIG ORF and the non-canonical LAGLI-DADG-GIY-YIG ORF, which is generated by its integration, have endonuclease activities which recognize and cut the insertion site of the optional sequence. This constitutes the first direct evidence for potential mobility of an intronic GIY-YIG endonuclease. We discuss the role that such a mobile sequence could have played during evolution.
INTRODUCTION
Movements of mobile elements are now considered as powerful drivers of evolution. Indeed, if they cause a deleterious effect on the cell, they may also increase the phenotypic capacities of the organism and allow adaptation to changing environmental conditions. For example, the transfer of integrons and mobile cassettes containing resistance genes are known to contribute to the spread of multi-antibiotic resistance in populations of pathogenic Gram-negative bacteria (1). In eukaryotes, there are also several documented cases in which the ancient insertion of transposable elements appears to contribute to the functional diversification of genes by supplying, for example, cis regulatory domains that alter expression patterns (2).
Endonuclease genes encoded in some mobile group I and group II introns are among the different elements which seem to play a role in the evolution of genomes. These endonucleases, which were first described in intronic mitochondrial sequences (reviewed in 3), promote the transfer (homing) (4) of their host introns from intron-containing alleles to cognate intron-less alleles by recognizing the exon–exon junction of the intron-less allele and creating a double-strand break in its vicinity (5–7). Repair of this double-strand break by a mechanism of gene conversion, which uses the intron-containing gene as matrix, leads to acquisition of the entire intronic sequence in the DNA (8).
The homing endonucleases are classified into four different groups, depending on the presence of conserved amino acid sequence blocks. The largest family is constituted of proteins which harbor two, often degenerate, copies (P1 and P2) of the conserved amino acids sequence LAGLI-DADG. Another family is constituted of endonucleases harboring GIY and YIG motifs that are separated by 10 or 11 amino acids. The two last groups are constituted of proteins containing H-N-H and H-Cys motifs (3,9,10).
Numerous lines of evidence suggest that these endonucleases are mobile elements, which have invaded pre-existing self-splicing introns during evolution. This provided the introns with their invasive properties. For example, it was shown that the same intron (or related introns) could contain endonucleases that belong to different subclasses, suggesting that the ORFs, on the one hand, and the core intronic sequences, on the other, are two distinct entities that evolved independently (11–13). Such a hypothesis is also supported by the finding that the proteins harboring LAGLI-DADG, GIY-YIG or H-N-H motifs are not exclusively encoded in group I intronic sequences but are also located in other genetic contexts (3,14). Indeed, such sequences were also identified as free-standing ORFs located between genes (15), as part of archeal introns (16) or as part of inteins in all three biological kingdoms (17,18).
The study of inteins gave further evidence for the intrinsic mobility of the LAGLI-DADG endonucleases. It was shown that although unrelated to the group I introns, some of these intein genes are also able to move and that they use for their mobility a mechanism quite similar to that of the intronic homing process (19,20). Furthermore, numerous structural, genetic and computational data indicate that the protein splicing domain and the LAGLI-DADG endonuclease domain harbored by the inteins are two separate entities, which have evolved independently (20–27). This suggests that, like group I mobile introns, the mobile inteins are composite genes that result from invasion of endonuclease ORFs into proteins having splicing activity. This was recently confirmed by the report of intein sequences lacking an endonuclease domain (22,28). Direct evidence for the intrinsic mobility of one intronic LAGLI-DADG ORF was also demonstrated in the mitochondrial genome of Podospora anserina (29).
The GIY-YIG endonucleases were initially found in the mitochondrial genome of fungi and in bacteriophage DNA, mostly as intronic but also as free-standing ORFs (12,30–32). GIY-YIG ORFs were also described in the mitochondrial genomes of Chlorogonium elongatum and Chlamydomonas eugametos (33,34) and in the chloroplast genome of Chlamydomonas reinhardtii (35). Recently GIY-YIG motifs were identified in bacterial gene products including several unidentified proteins and the UvrC gene encoding subunit c of the abc excinuclease complex (36,37; our unpublished results). A study of the best known GIY-YIG endonuclease, I-TevI, encoded in the intron of the thymidylate synthase gene of bacteriophage T4, showed that it is constituted of two separate domains that behave as distinct physical entities. The C-domain contains the DNA-binding activity, whereas the N-terminal domain, which possess the two GIY and YIG peptides, constitutes the catalytic domain (38). Although this GIY-YIG catalytic domain is conserved in different GIY-YIG proteins, the C-terminal binding domain is variable, suggesting that the GIY-YIG domain is a catalytic cassette that can be combined with different DNA-binding proteins to evolve as nucleases with various specificities (38).
In this paper we describe the presence in the mitochondrial DNA (mtDNA) of the fungus Podospora curvicolla of one optional ORF encoding a GIY-YIG protein. The presence of this sequence, which occurs inside an intron that already contains a typical LAGLI-DADG protein, generates a new sequence. The latter encodes two non-canonical intronic ORFs, the first one harboring a single P1 motif associated with a GIY-YIG motif and the second one harboring only one P2 motif. We show that during heteroplasmon formation experiments, the sequence encoding the optional GIY-YIG ORF is very efficiently transferred from the ORF-containing intron to the ORF-less allele. We also show that the products of both the non-canonical P1-GIY-YIG ORF and the GIY-YIG ORF have endonuclease activities (called I-PcI and I-PcII, respectively) which recognize and cut the insertion site of the optional sequence. Together these results strongly support the hypothesis that the GIY-YIG ORF is transferred through a homing process rather than by regular recombination and constitutes the first evidence for mobility of an intronic GIY-YIG endonuclease ORF.
MATERIALS AND METHODS
Strains and cultures conditions
Podospora curvicolla is a homothallic filamentous fungus. Strain A was kindly provided by Dr U. Kück while strains V and L were recovered in France by Prof. L. Belcour. The culture conditions were as for P.anserina (39). Hygromycin was directly added into agar medium before cooling whereas the phleomycin (30 µl at 10 mg/ml) was laid over the solid medium after cooling. The hygromycin- and phleomycin-resistant P.curvicolla strains used in protoplast fusion or vegetative fusion experiments were progenies obtained after self-fertilization of resistant strains recovered after transformation of the natural strains by the integrative pBC hygro and pBC phleo vectors (40).
Escherichia coli strains SURE and DE3 (Stratagene) were used as hosts for cloning and expression of the I-PcI and I-PcII endonucleases, respectively.
Transformation of P.curvicolla
Protoplasts were prepared as described previously (41). Aliquots of 10 mg of each plasmid (pBC hygro or pBC phleo) containing the genes for resistance to hygromycin and phleomycin were added to 100 µl of protoplasts (108 protoplasts/ml) and incubated at room temperature for 15–20 min in 1 ml of 60% PEG 4000, 10 mM Tris–HCl (pH 7.5), 10 mM CaCl2. After incubation, the protoplasts were rinsed twice with 10 ml of 10 mM Tris–HCl, 10 mM CaCl2, 0.6 M saccharose, centrifuged at 3000 r.p.m. and plated on regeneration medium containing hygromycin or phleomycin. After regeneration, the transformants were transferred to hygromycin or phleomycin MR medium.
Mitochondrial DNA transfer
The transfer of mtDNA sequences between incompatible strains was carried out by protoplast fusion. The donor and recipient strains used in each transfer experiment were distinguishable by the presence of a hygromycin or phleomycin resistance gene introduced by transformation. Aliquots of 107 protoplasts of each strain were mixed and incubated at room temperature for 15–20 min in 2 ml of 60% PEG 4000, 10 mM Tris–HCl (pH 7.5), 10 mM CaCl2. After incubation, the protoplasts were rinsed twice with 20 ml of 10 mM Tris–HCl, 10 mM CaCl2, 0.6 M saccharose, centrifuged at 3000 r.p.m. and plated on regeneration medium containing both hygromycin and phleomycin. The few unstable heterokaryons able to grow were recovered and transferred to MR medium containing hygromycin or phleomycin depending on the nuclear genotype of the recipient strain to be selected. The recipient nuclear background of the growing cultures was confirmed by incompatibility tests with the two parental strains. The presence/absence of the cytb-i2l or cytb-i2s intron in these cultures was detected by digestion of their mtDNAs with HaeIII.
The transfer of mtDNA sequences between compatible P.curvicolla strains was carried out by vegetative fusion experiments analogous to those previously described for P.anserina (42,43). In each fusion, the recipient and the donor strains were distinguished by the presence of different nuclear marker resistance genes (hygromycin or phleomycin resistance gene). These fusions were carried out on MR solid medium containing hygromycin or phleomycin depending on the nuclear genotype of the recipient strain.
Mitochondrial DNA analysis
Small scale mtDNA extraction was by the method of Lecellier and Silar (44), large scale as previously described (45). The digestions with restriction enzymes, agarose gel electrophoresis and transfer to Hybond N membranes (Amersham) were carried out using standard conditions. Hybridization was carried out using as probe a 5′-32P-end-labeled oligonucleotide (oligonucleotide 1579, 5′-AGCAGCAAACATAGCTATAA- CACC-3′) corresponding to a sequence located in the downstream exon of the cytb-i2 intron.
A 5 kb BglII fragment containing the entire cytb-i2l intron of strain V and a 7 kb PstI fragment containing the exon 2–exon 3 junction of the cytochrome b gene of strain L were cloned by independent shotgun experiments into the pUC19 vector and sequenced with the Sequenase sequencing kit (US Biochemical) using specific synthetic oligonucleotides as primers. The cytb-i2s intron of strain A was amplified using primers located in flanking exonic sequences and cloned using a TA cloning kit (pMOS blue T vector kit; Amersham). Two independent clones obtained from two independent PCR amplifications were sequenced in order to detect errors introduced during the amplification by Taq polymerase.
Plasmid constructions
Construction of the two expression plasmids pI-PcI and pI-PcII. Two 822 and 1020 bp fragments corresponding, respectively, to the sequences of the I-PcI and I-PcII endonuclease genes with NdeI and BamHI linkers were prepared using PCR. The mtDNA of strain V was used as template. Aliquots of 50 pM of each primer (5′-CCCCCCCCCATATGCAAGTAAAAAGATTTA- TTGATGATGT-3′ and 5′-TTTTTTTGGATCCTTATTACTTAGATACTTTTATTATAAG-3′ for amplification of the I-PcI endonuclease gene and 5′-TTTTTTTTCATATGTATTTATCCCCTATATTATTCTTT-3′ and 5′-TTTTTTTG-GATCCTTATTACTTAGATACTTTTATTATAAG-3′ for amplification of the I-PcII endonuclease gene) and the high fidelity Pfu polymerase (Stratagene) were used in each experiment. PCR was carried out for 30 rounds in 50 µl with buffer supplied by the manufacturer in a Hybaid cycler (30 s at 95°C, 45 s at 58°C, 1 min at 72°C and 30 s at 95°C, 45 s at 48°C, 1 min at 72°C). The amplified fragments were purified from agarose gels by electroelution, desalted, concentrated by centrifugation on microcon 30 (Amicon) and cloned into the EcoRV site of the pBluescript SK+ vector (Stratagene). The unique TGA codon present in each sequence (which specifies tryptophan in the mitochondrial code but stop in the universal code) was modified to TGG by oligonucleotide site-directed mutagenesis. After confirmation by sequencing (Sequenase v.2.0; US Biochemical) each plasmid was digested with NdeI and BamHI and the inserts were subcloned independently into the pET11a expression vector (Stratagene).
Construction of plasmids pBC-a and pBC-b containing the target site for the I-PcI and I-PcII endonucleases. A 503 bp fragment surrounding the insertion site of the mobile sequence was prepared by PCR with the mtDNA from strain A as template, 50 mM of each primer (5′-AAGAGATAGTCCGACCATG-3′ and 5′-CCACGGGGAGTTAACAACTCTC-3′) and Pfu polymerase. After agarose gel electrophoresis, the amplified fragment was purified by electroelution and cloned into the EcoRV site of the pBC SK+ cloning vector (Stratagene). The orientation of the insert was determined by restriction analysis and its nucleotidic sequence was confirmed.
In vivo DNA endonuclease assay
Overnight cultures of E.coli DE3 strains transformed with two plasmids (one containing the sequence of the I-PcI or I-PcII endonuclease gene and the other containing the target site, pBC-a or pBC-b) were diluted 10-fold in fresh LB medium containing 50 µg/ml ampicillin and 34 µg/ml chloramphenicol. After 2 h incubation at 37°C, IPTG was added to a final concentration of 1 mM. After an additional 1 h incubation, the cells were collected by centrifugation and the total DNA prepared using a procedure previously described (5). The DNAs were digested with EcoRI, analyzed on agarose gels and transferred to Hybond N membrane (Amersham). The membrane was hybridized with a probe corresponding to the purified 503 bp amplified fragment surrounding the insertion site labeled with [α-32P]dATP by nick translation (Amersham).
RESULTS
A chimeric P1-GIY-YIG protein translated from the cytb-i2l intron of mtDNA of P.curvicolla
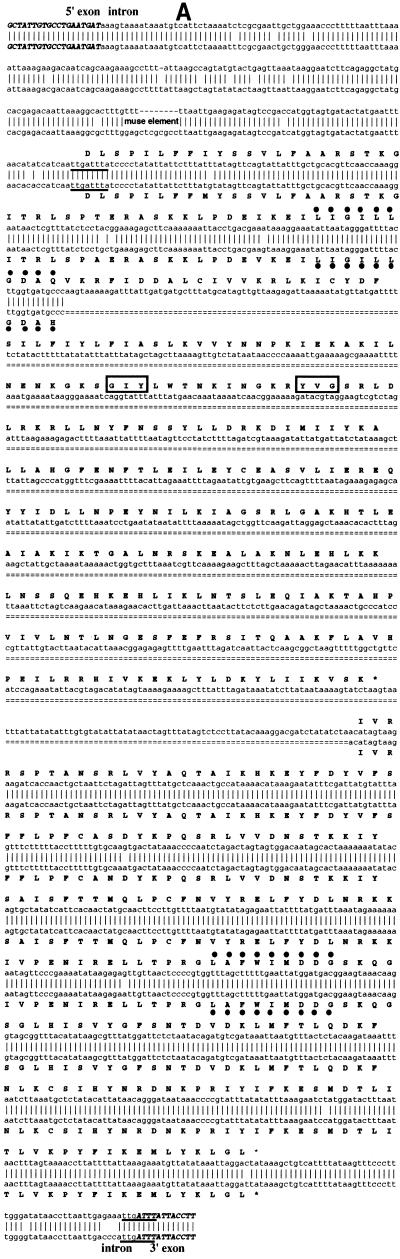
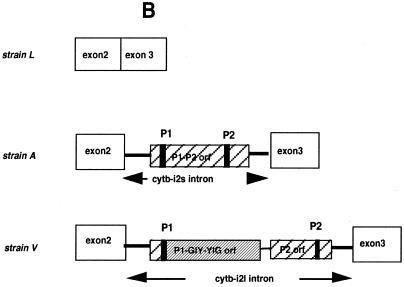
Our analysis of the mtDNA of three independent strains of the filamentous fungus P.curvicolla revealed that the cytochrome b (cytb) gene of this fungus may contain an optional sequence homologous to the 990 bp cytb-i3 intron of P.anserina (Pa cytb-i3) (46), which is mobile (Belcour and Rossignol, unpublished results). This sequence, which corresponds to the second intron of this gene in P.curvicolla (Pc cytb-i2) was present in the mtDNA of strain A but was absent in the mtDNA of strain L.
Interestingly, the mtDNA of another strain, V, contained a longer intronic sequence (~2000 bp) suggesting that one additional sequence could be inserted in this intron. To establish the nature of this insertion, the mtDNA fragments containing the short form, cytb-i2s, and the long form, cytb-i2l, of the intron were cloned and sequenced. The nucleotide sequence of Pc cytb-i2s exhibited a high degree of similarity (95%) to Pa cytb-i3, its P.anserina counterpart. It was shorter by 9 bp and this difference was essentially due to the lack of one mitochondrial ultra-short element (MUSE) which is hypothesized to be mobile (47). Like its P.anserina counterpart, the Pc cytb-i2s intron contains a free-standing ORF encoding a protein related to the LAGLI-DADG family (46). An alternative splice site allowing its expression was conserved in the upstream sequence, as previously identified in P.anserina (46).
As expected the Pc cytb-i2l intron shared a common sequence with its short counterpart Pc cytb-i2s. These two common sequences are 100% identical. An additional DNA fragment of 858 bp was inserted in this sequence. This could not be folded into an RNA secondary structure typical of group I or group II introns (48; Lisacek and d’Aubenton-Carafa, personal communication). However, this insert encoded a 261 amino acid polypeptide which was shown by Blast analysis (49) to share 39% identity and 53% similarity with the GIY-YIG polypeptide encoded by the second intron of the P.anserina cytochrome b gene. This GIY-YIG ORF was inserted in-phase with the LAGLI-DADG ORF just after the last codon of the first P1 motif (Fig. 1A). Consequently the Pc cytb-i2l intron potentially encoded two separate free-standing polypeptides, the first one harboring both one LAGLI-DADG (motif P1) and one GIY-YIG motif and the second one only one LAGLI-DADG (motif P2). The alternative splice site was conserved in the upstream sequence encoding the chimeric P1-GIY-YIG polypeptide, suggesting that it could be effectively expressed (Fig. 1A). In contrast, no typical alternative splice site nor ATG allowing its expression seems to be present in the upstream P2 ORF.
Figure 1.
(A) Comparison of the nucleotidic sequences of the cytb-i2l intron of P.curvicolla and of the cytb-i3 intron of P.anserina. (Top) Sequence of P.curvicolla. (Bottom) Sequence of P.anserina. The flanking exonic sequences are in bold italic. The constitutive 3′ splice site and the 3′ alternative splice site allowing expression of the P1-GIY-YIG ORF are underlined. Derived amino acids of each sequence are also compared. The GIY and YIG peptides are boxed whereas the P1 and P2 motifs are denoted by dots. (B) Organization of a part of the cytochrome b gene in the different natural isolates L, A and V of P.curvicolla. The ORFs identified in the cytb-i2s and cytb-i2l introns are represented by cross-hatched blocks. Motifs P1 and P2 are indicated by black bars.
Experiments were undertaken in order to determine if the cytb-i2s and cytb-i2l introns as well as the optional GIY-YIG ORF alone were mobile. In P.anserina, the transfer of mobile DNA sequences from one mitochondrial genome to another was shown to occur during the formation of a heteroplasmon between compatible strains. This process occurred without recombinational events between mtDNAs or nucleus migration (50). The natural isolates of P.curvicolla strains contained numerous incompatibility genes preventing such anastomosis formation and thus impaired the eventual transfer of mtDNA sequences. However, by protoplast fusion experiments analogous to that described in Neurospora crassa (51) we were able to construct several compatible strains of P.curvicolla which differed by the presence/absence of the cytb-i2s or cytb-i2l intron in their mtDNA. They were used as donor or recipient strains in vegetative transfer experiments analogous to that described in P.anserina.
The homing process is often accompanied by co-conversion of the flanking sequences (9). So, when nucleotide variations around the mobile sequences were observed between the donor and the recipient, transfer from one genome to another could be easily monitored by sequencing. Unfortunately, such a polymorphism does not exist in the vicinity of the cytb-i2 intron in P.curvicolla. Thus, only restriction and hybridization experiments could be exploited to monitor the transfer of this intron. In order to unambiguously distinguish the strains whose mtDNA received the mobile sequence from that whose mtDNA gave the mobile sequence, we used strains that differed by two nuclear selectable markers (hygromycin and phleomycin resistance genes). These markers were introduced into the strains by nuclear transformation. Furthermore, in order to minimize transfer of the complete mitochondrial genome from the donor to the recipient, fusion was carried out on medium allowing growth only of the recipient cells.
Mobility of the Pc cytb-i2l intron
A hygromycin-resistant strain of P.curvicolla containing the cytb-i2l intron (strain L1) was used as donor in vegetative fusion experiments with a phleomycin-resistant strain whose mtDNA is devoid of this intron (strain L2) (see Table 1). Lack of the cytb-i2 intron in the mtDNA of the L2 recipient strain was confirmed by the presence of a 5 kb HaeIII fragment which was replaced by a 7 kb fragment in strain L1 (Fig. 2B). Both fragments hybridized with a specific oligonucleotide probe (1579 probe) located in the exonic part of the cytochrome b gene. The mtDNAs of strains L1 and L2 were also distinguishable by another polymorphic marker, a 4.2 kb fragment which was specific to the donor strain and which could correspond to a fragment bearing an optional inserted sequence. Its appearance correlated with the disappearance of one of two 3.7 kb fragments of the L2 mtDNA, which co-migrated on the gels. (This disappearance, which appeared as a reduction in intensity of the band, was only visible on restriction analysis of mtDNA purified on a CsCl gradient: data not shown.)
Table 1. A summary of the nuclear and mitochondrial genotypes of the different compatible strains used in the vegetative fusion experiments.
| Strain | Nuclear | Mitochondrial |
|---|---|---|
| L1 | Hygromycin-resistant | cytb-i2(l) |
| L2 | Phleomycin-resistant | cytb-i2(–) |
| L3 | Phleomycin-resistant | cytb-i2(s) |
| L4 | Hygromycin-resistant | cytb-i2(–) |
These strains were constructed by nuclear transformation of the L isolate with plasmid pBC phleo or pBC hygro, which confer resistance to phleomycin and hygromycin, respectively. After transformation, protoplast fusions with incompatible strains of P.curvicolla A and L allowed us to introduce the different forms of the cytb-i2 intron into their mtDNA (see Materials and Methods). Cytb-i2(l) and cytb-i2(s) indicate that the mitochondrial genome contains the long or short form of the intron, respectively, and cytb-i2(–) that it lacks it.
Figure 2.
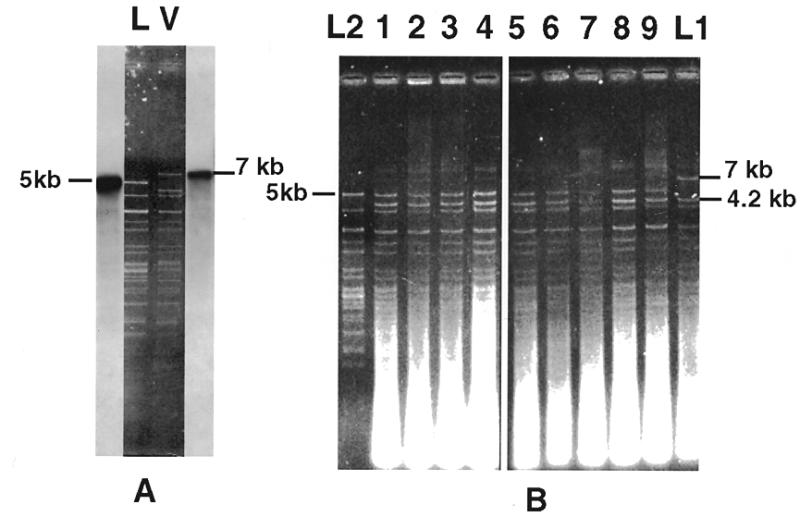
(A) HaeIII RFLP of the mtDNA of the two P.curvicolla natural isolates L and V. The mtDNA of the V strain, which contains the cytb-i2l intron, and the mtDNA of the L strain, which lacks it, were digested with HaeIII and hybridized with an oligonucleotide probe (1579). This probe corresponds to a sequence of the cytochrome b gene located downstream from the intron. A 7 kb fragment is labeled by this probe in the V strain whereas a 5 kb fragment is labeled in the L strain. (B) Ethidium bromide stained agarose gel of mtDNAs extracted from nine independent phleomycin-resistant cultures (lanes 1–9) that resulted from the vegetative fusion of L1 and L2. The 7 and 5 kb fragments which correspond to the sequences which contain or which lack this intron are indicated as well as the additional 4.2 kb fragment characteristic of the donor strain.
The vegetative fusion experiments between L1 and L2 were carried out on MR medium containing phleomycin using a protocol which, in the related species P.anserina, allowed transfer of mobile DNA without recombinational events between mtDNAs or nuclear migration (50). However, we could not exclude that, in P.curvicolla, such recombination could occur. In this case, we expected to recover four classes of mitochondrial genomes after fusion: two parental and two recombined genomes, one recombined genome containing only the 4.2 kb fragment and the other only the 7 kb fragment.
The mtDNA of 50 phleomycin-resistant cultures resulting from 50 independent fusions was extracted, digested with HaeIII and compared to the mtDNA of the two parental strains. Only one class of mitochondrial genome constituted of molecules containing the 4.2 kb fragment but lacking the 7 kb fragment were recovered after these fusions (Fig. 2B). A systematic loss of the cytb-i2l intron in the donor strain was unlikely to explain this result. Indeed, we never observed such a loss during vegetative growth of this strain or during fusions between this L1 strain and strain L3 (see below). The recovered mtDNA thus corresponded to the mtDNA of the recipient cells into which the 4.2 kb fragment was integrated. This attested to anastomosis formation between the two parental strains. Absence of the two parental mtDNAs and of one of the expected recombined molecules strongly suggested that, as previously shown in P.anserina (50), recombination did not occur or occurred at a low level during these anastomoses. Complete absence of the donor genome also indicated that the donor mtDNA did not survive during these fusions. Even if background recombination occurred during these fusions, the high rate of transmission of the 4.2 kb fragment (100%) strongly supported the hypothesis that it contained an invasive element which was transmitted through a homing process analogous to that described for mobile group I introns (5). The nature of this mobile element was not determined. In contrast, the complete absence (0/50) of the 7 kb fragment in the mtDNA of the recipient cells allowed us to conclude that the cytb-i2l intron was not mobile. This low rate also indicated that, even if recombination occurred, the mitochondrial region surrounding this intron was poorly transmitted under the experimental conditions used.
Mobility of the Pc cytb-i2s intron
By protoplast fusions between the incompatible A and L strains we were able to construct a phleomycin-resistant strain whose mtDNA harbored the short form of the Pc cytb-i2 intron (strain L3) (see Table 1). This strain was used as donor in independent vegetative fusion experiments with a compatible hygromycin-resistant recipient strain (L4) whose mtDNA was devoid of the intron. The presence or absence of the intron could be distinguished by hybridization experiments on the mtDNA digested with HaeIII using the radioactive 1579 probe, which revealed a 6 kb restriction fragment when the intron was present and a 5 kb fragment when it was absent.
The mtDNAs of hygromycin-resistant cultures resulting from these fusions were extracted, digested with HaeIII and hybridized with the 1579 probe. As seen in Figure 3, which presents part of these results, all (15 of 15) the mtDNA contained the 6 kb HaeIII fragment characteristic of presence of the intron. If either regular recombination or homing could both explain this result, the high frequency of transfer of this sequence from one genome to another (100%) leads us to favor the second hypothesis. This hypothesis is also supported by our previous results which showed that, even if a low level of background recombination occurred during fusion, it did not allow preferential transfer of the sequences surrounding the cytb-i2 intron.
Figure 3.
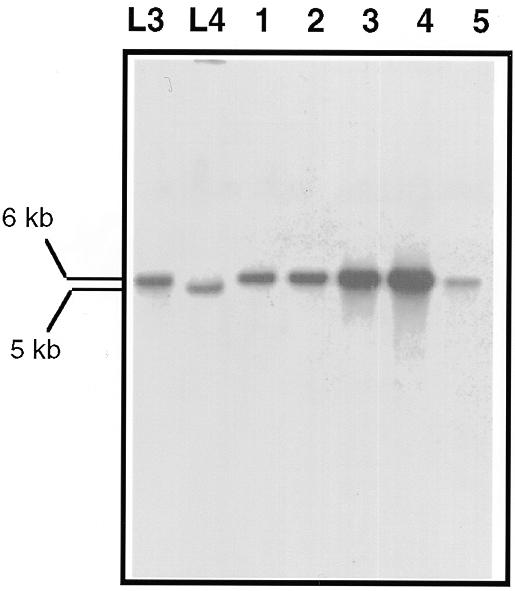
Southern analysis of mtDNAs extracted from hygromycin-resistant cultures resulting from independent vegetative fusions between L3 and L4. The mtDNAs were digested with HaeIII and hybridization was carried out with the 1579 probe.
Mobility of the GIY-YIG ORF
The presence/absence of the optional 858 bp sequence of the GIY-YIG ORF in cytb-i2s of the mtDNA of a P.curvicolla strain could be distinguished by the occurrence of a 7 or a 6 kb HaeIII restriction fragment which hybridized with the 1579 oligonucleotide probe. In order to test whether this ORF is a mobile entity, vegetative fusion experiments were performed using as donor the L1 hygromycin-resistant strain (see above) which contains the long cytb-i2l intron and as recipient a phleomycin-resistant L3 strain which contains the short cytb-i2s intron.
As seen in Figure 4, the mtDNAs extracted (14/14) from the phleomycin-resistant cultures resulting from these fusions contained the 7 kb HaeIII fragment characteristic of presence of the GIY-YIG ORF. An eventual transfer of the entire cytb-i2l intron is unlikely since this intron is not efficiently transmitted under our experimental conditions (see above). As to transfer of the cytb-i2s intron, the quantitative data are more consistent with the sequence of the GIY-YIG ORF being transferred by a homing process and thus alone being mobile.
Figure 4.
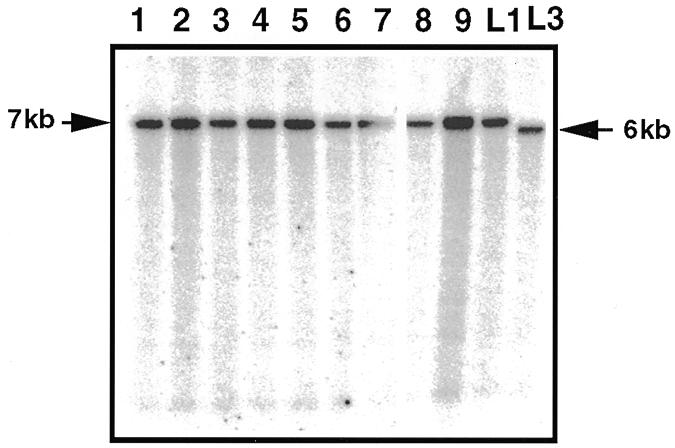
Southern hybridization of mtDNAs extracted from phleomycin-resistant cultures resulting from vegetative fusions between L1 and L3. The mtDNAs were digested with HaeIII and hybridized with the 1579 probe.
Both the P1-GIY-YIG and GIY-YIG proteins have endonuclease activity
The lack of polymorphism in the cytb-i2 intron and in the surrounding sequences does not allow us to present sequencing data supporting a homing process of the GIY-YIG ORF. However, if such a mechanism is really responsible of its highly efficient transfer, the protein encoded by this sequence would possess an endonucleolytic activity able to recognize and cut near its insertion site.
In order to determine whether the GIY-YIG or P1-GIY-YIG protein possesses endonucleolytic activity, which could promote mobility of the GIY-YIG element, we used a two plasmids system. The ORFs encoding the chimeric P1-GIY-YIG (I-PcI) and GIY-YIG (I-PcII) proteins were both amplified by PCR and cloned downstream from the T7 promoter of the pET11a E.coli expression vector (plasmids pI-PcI and pI-PcII, respectively). A 503 bp fragment surrounding the insertion site of the GIY-YIG mobile sequence was amplified by PCR and cloned in two different orientations into the pBC vector (plasmids pBC-a and pBC-b) (Fig. 5A). After induction by IPTG, total DNA was isolated from E.coli strains transformed with two different plasmids: one plasmid allowing expression of one ORF (pI-PcI or pI-PcII) and one plasmid containing the target sequence (pBC-a or pBC-b). These DNAs were digested with EcoRI, which cleaves at a single site in the pBC-a and pBC-b plasmids. After agarose gel electrophoresis and Southern blotting, the products were hybridized with a probe corresponding to the entire 503 bp target sequence. As seen in Figure 5B, whatever the ORF expressed (I-PcI or I-PcII), an extra band of ~300 bp was observed in the strains containing plasmid pBC-a. The size of this band decreased when the strains contained plasmid pBC-b. These results are consistent with the presence of a cleavage site located at or near the insertion site of the GIY-YIG mobile sequence, this cleavage being created by the hybrid P1-GIY-YIG protein as well as by the GIY-YIG protein.
Figure 5.
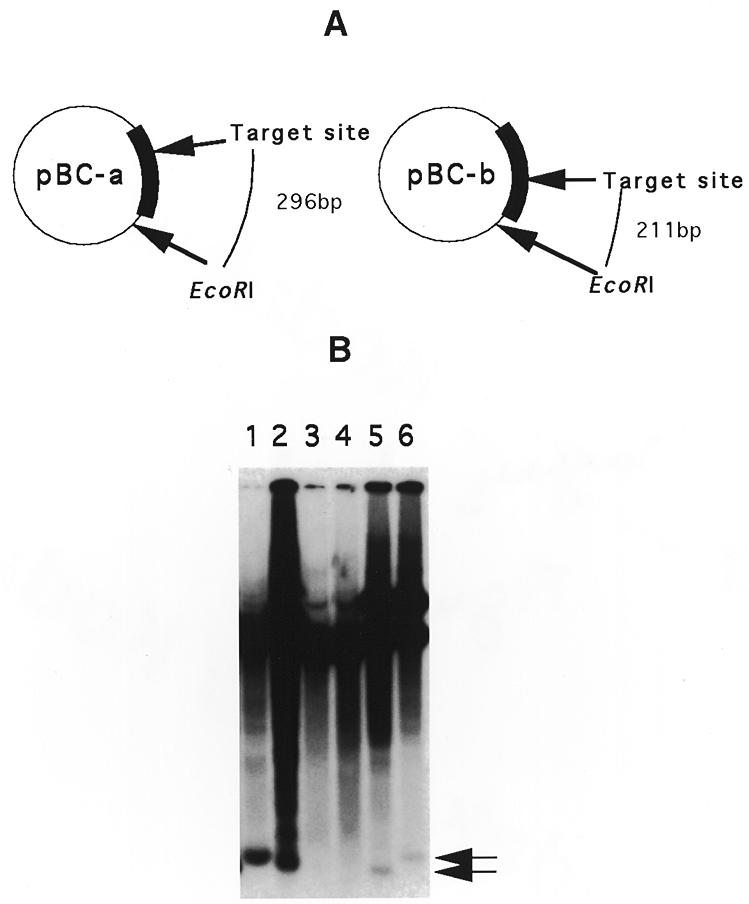
In vivo DNA endonuclease assay of endonucleases I-PcI and I-PcII. (A) Structure of the two plasmids containing the insertion site of the GIY-YIG ORF. A 503 bp sequence containing this sequence was inserted in two different orientations into the EcoRV site of the pBC vector. The localization of the unique EcoRI site as well as the target insertion site are indicated. The sizes of the restriction fragments generated after digestion by EcoRI if a double-strand break is generated at this target site are indicated. (B) Southern analysis of total DNAs extracted from E.coli strains harboring two different plasmids: one plasmid allowing the expression of one ORF (pI-PcI or pI-PcII) and one plasmid containing the target sequence (pBC-a or pBC-b). The total DNA was extracted after induction by IPTG, digested with EcoRI and hybridized with a radioactive probe that was made from the 503 bp fragment. Lanes 1 and 2, pI-PcII with vectors pBC-a and pBC-b, respectively; lanes 3 and 4, control vector pET11a lacking the endonuclease gene with pBC-a and pBC-b, respectively; lanes 5 and 6, pI-PcI with pBC-b and pBC-a, respectively. The arrows indicate the positions of additional fragments generated by the double-strand break at the target site.
DISCUSSION
In this paper we show that the second intron of the cytochrome b gene (cytb-i2) located in the mtDNA of the fungus P.curvicolla is an optional sequence which can exist in a short (cytb-i2s) or a long form (cytb-i2l). We show that the cytb-i2s intron encoding a LAGLI-DADG protein is efficiently transferred from one genome to another consistent with this intron being a group I mobile intron. In contrast, the cytb-i2l intron, which consists of the cytb-i2s intron into which an optional 858 bp sequence of a GIY-YIG ORF is inserted, appears not to be mobile. Insertion of the optional sequence occurs at the level of the first conserved P1 motif of the LAGLI-DADG protein and generates a non-canonical intronic ORF harboring both one LAGLI-DADG motif (P1 motif) and one GIY-YIG motif. Here we show that the sequence of this optional GIY-YIG ORF is efficiently transferred from one ORF-containing intron to an ORF-less intron. The hypothesis of mobility of this sequence is supported by the fact that both the hybrid P1-GIY-YIG ORF (I-PcI) and the GIY-YIG ORF (I-PcII) encode proteins that possess endonucleolytic activities able to recognize and cut in the vicinity of the insertion site of this mobile sequence.
Study of the well-known I-TevI endonuclease of phage T4, which belongs to the GIY-YIG subclass, has shown that this enzyme consists of two separate domains that behave as discrete physical entities (38). Whereas the N-terminal domain constitutes the catalytic domain, the C-terminal domain has the DNA-binding activity. Sharing of an identical C-terminal domain by the I-PcI and I-PcII endonucleases, which differ only by addition of 53 amino acids in their N-terminal part, could thus explain why the two enzymes recognize and bind the same DNA sequence. Each conserved motif, LAGLI-DADG or GIY-YIG, was shown to be involved in the catalytic activity of the enzymes which possess it (19,52–53). Some differences were observed between these activities (14). Whereas the LAGLI-DADG endonucleases generate a 4 nt 3′-OH overhang that is generally located immediately adjacent to their target site, the GIY-YIG endonucleases leave a 2 nt 3′-OH overhang at a site more distantly (15–30 nt) located from the recognized sequence (54). However, the simultaneous presence of one LAGLI-DADG and one GIY-YIG motif in the N-terminal part of the non-canonical I-PcII endonuclease raises the question of which amino acids are effectively involved in its endonucleolytic activity. Our data do not answer this question. It will thus be interesting to characterize the exact cleavage sites of the I-PcI and I-PcII enzymes.
Whatever the motif involved in the catalytic activity of the I-PcI endonuclease, we have shown that this enzyme, as well as the I-PcII endonuclease, cuts near the insertion site of the mobile GIY-YIG ORF. Both enzymes are thus able to promote the transfer of this sequence. However, whereas an alternative splice site can be identified in the cytb-i2l intron upstream of the sequence encoding the chimeric P1-GIY-YIG ORF, no alternative splice site nor ATG codon is present immediately upstream of the GIY-YIG ORF. This suggests that only the chimeric I-PcI endonuclease is expressed from the intron to promote mobility of the GIY-YIG endonuclease gene. Nevertheless, the observed endonucleolytic activity of the product of the GIY-YIG ORF strongly supports the hypothesis that, during evolution, this enzyme initiated the first integration of its own coding sequence into the pre-existing cytb-i2s intron.
The GIY-YIG subclass of endonucleases were first described as intronic or free-standing ORFs encoded by fungal mtDNA and in some bacteriophage genomes. Lateral transmission has thus been invoked to explain the presence of such related ORFs in these unrelated organisms (55). However, the finding of a GIY-YIG ORF in an intron located in the plastid DNA of C.reinhardtii suggested an ancient origin of these proteins which could be present in the common ancestor of cyanobacteria and proteobacteria before the endosymbiosis events which gave rise to plastids and mitochondria, respectively (35). Consistent with this hypothesis, which anticipated their discovery, several unidentified ORFs harboring a GIY-YIG motif were recently identified in the genomes of several divergent eubacteria (E.coli, Synechocystis spp. and Mycobacterium spp.), as well as in the N-terminal part of all UvrC proteins identified so far in the eubacterial genome (36,37; our unpublished results).
UvrC is one of the three subunits of the abc excision nuclease complex, which catalyzes the excision of UV-damaged nucleotide segments, producing oligomers having the modified base (56). The presence of a common motif between UvrC and a mobile endonuclease could reflect divergent adaptations of a common ancestral GIY-YIG protein. However, the results presented here, which indicate that an ancient insertion of a GIY-YIG mobile sequence could have generated a protein which is nowadays functional, suggest an alternative explanation. Indeed, it is tempting to speculate that a GIY-YIG ORF analogous to those identified in the genomes of the contemporary bacteria E.coli and Synechocystis spp. was already present in the common ancestor of all eubacteria before endosymbiosis and that it was already mobile. It thus could have invaded a sequence which could interact with DNA, creating a new protein which evolved as UvrC. In this hypothesis, direct transmission of mobile GIY-YIG ORFs and their subsequent loss, depending on the organism, could explain their distribution in unrelated organisms. If this hypothesis supports the idea of an ancient origin of the GIY-YIG mobile endonuclease it does not question the hypothesis that additional horizontal transfer of mobile GIY-YIG elements between species could also have occurred during evolution.
Through their ability to make nicks and breaks in the DNA, all endonuclease genes are able to give rise to mobile elements. This was confirmed by the construction of an artificial mobile DNA element from the EcoRI restriction endonuclease gene that was capable of specific insertion at a rate near that of mobile introns (57). One can also suppose that the UvrC gene could be the direct ancestor of mobile endonucleases. Cutting induced by UvrC could thus lead to duplication of its gene, one copy evolving as a mobile endonuclease. In this hypothesis the possibility that several independent duplication events could have occurred during evolution, before or after endosymbiosis, that could explain the presence of GIY-YIG ORFs in unrelated organisms cannot be excluded.
Whatever the evolutionary relationship between UvrC and the GIY-YIG endonucleases, the results presented here constitute the first experimental evidence for the mobility of an intronic GIY-YIG ORF. This is in agreement with previous results which indicate that the intergenic GIY-YIG endonuclease segE of T4 is preferentially inherited in the progeny of mixed infections between segE-containing and segE-lacking phage (58). It thus provides an additional strong argument for the theory, supported by a large amount of data concerning the LAGLI-DADG proteins, which supposes that endonuclease genes have invaded self-splicing introns and inteins converting them into invasive genetic elements. This has subsequently led to the spread of intronic sequences in genomes. However, we also show here that a second insertion event of an endonuclease gene into a mobile intron could, on the contrary, lead to its immobilization. This phenomenon may constitute a mechanism able to regulate intronic propagation that could help to explain why genomes are not saturated by intronic sequences. In conclusion, this work illustrates, once again, the important role that mobile elements play in the dynamics of the genome.
Acknowledgments
ACKNOWLEDGEMENTS
We are particularly indebted to Y. d’Aubenton-Carafa and Dr J. Cohen for their constant friendly support during this work. We also thank Drs P. Silar, L. Sperling and J. Lazowska for their critical reading of the manuscript and Dr U. Kück for the gift of the Podospora curvicolla A strain.
DDBJ/EMBL/GenBank accession nos AJ249984, AJ249985
REFERENCES
- 1.Recchia G.D. and Hall,R.M. (1997) Trends Microbiol., 5, 389–394. [DOI] [PubMed] [Google Scholar]
- 2.Kidwell M.G. and Lisch,D. (1997) Proc. Natl Acad. Sci. USA, 94, 7704–7711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lambowitz A.M. and Belfort,M. (1993) Annu. Rev. Biochem., 62, 587–622. [DOI] [PubMed] [Google Scholar]
- 4.Dujon B. (1989) Gene, 82, 91–114. [DOI] [PubMed] [Google Scholar]
- 5.Colleaux L., d’Auriol,L., Betermier,M., Cottarel,G., Jacquier,A., Galibert,F. and Dujon,B. (1986) Cell, 44, 521–533. [DOI] [PubMed] [Google Scholar]
- 6.Colleaux L., d’Auriol,L., Galibert,F. and Dujon,B. (1988) Proc. Natl Acad. Sci. USA, 85, 6022–6026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wenzlau J.M., Saldanha,R.J., Butow,R.A. and Perlman,P.S. (1989) Cell, 56, 421–430. [DOI] [PubMed] [Google Scholar]
- 8.Szostak J.W., Orr,W.T., Rothstein,R.J. and Stahl,F.W. (1983) Cell, 33, 25–35. [DOI] [PubMed] [Google Scholar]
- 9.Belfort M., Reaban,M.E., Coetzee,T. and Dalgaard,J.Z. (1995) J. Bacteriol., 177, 3897–3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Belfort M. and Perlman,P.S. (1995) J. Biol. Chem., 270, 30237–30240. [DOI] [PubMed] [Google Scholar]
- 11.Mota E.M. and Collins,R.A. (1988) Nature, 332, 654–656. [DOI] [PubMed] [Google Scholar]
- 12.Shub D.A., Gott,J.M., Xu,M.Q., Lang,B.F., Michel,F., Tomaschewski,J., Pedersen,L.J. and Belfort,M. (1988) Proc. Natl Acad. Sci. USA, 85, 1151–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bell P.D., Quirk,S., Clyman,J. and Belfort,M. (1990) Nucleic Acids Res., 18, 3763–3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Belfort M. and Roberts,R.J. (1997) Nucleic Acids Res., 25, 3379–3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sharma M., Ellis,R.L. and Hinton,D.M. (1992) Proc. Natl Acad. Sci. USA, 89, 6658–6662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dalgaard J.Z., Garrett,R.A. and Belfort,M. (1993) Proc. Natl Acad. Sci. USA, 90, 5414–5417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hirata R., Ohsumk,Y., Nakano,A., Kawasaki,H., Suzuki,K. and Anraku,Y. (1990) J. Biol. Chem., 265, 6726–6733. [PubMed] [Google Scholar]
- 18.Kane P.M., Yamashiro,C.T., Wolczyk,D.F., Neff,N., Goebl,M. and Stevens,T.H. (1990) Science, 250, 651–657. [DOI] [PubMed] [Google Scholar]
- 19.Gimble F.S. and Stephens,B.W. (1995) J. Biol. Chem., 270, 5849–5856. [DOI] [PubMed] [Google Scholar]
- 20.Hodges R.A., Perler,F.B., Noren,C.J. and Jack,W.E. (1992) Nucleic Acids Res., 20, 6153–6157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Perler F.B., Xu,M.Q. and Paulus,H. (1997) Curr. Opin. Chem. Biol., 1, 292–299. [DOI] [PubMed] [Google Scholar]
- 22.Pietrokovski S. (1998) Protein Sci., 7, 64–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Duan X., Gimble,F.S. and Quiocho,F.A. (1997) Cell, 89, 555–564. [DOI] [PubMed] [Google Scholar]
- 24.Derbyshire V., Wood,D.W., Wu,W., Dansereau,J.T., Dalgaard,J.Z. and Belfort,M. (1997) Proc. Natl Acad. Sci. USA, 94, 11466–11471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hall T.M., Porter,J.A., Young,K.E., Koonin,E.V., Beachy,P.A. and Leahy,D.J. (1997) Cell, 91, 85–97. [DOI] [PubMed] [Google Scholar]
- 26.Dalgaard J.Z., Klar,A.J., Moser,M.J., Holley,W.R., Chatterjee,A. and Mian,I.S. (1997) Nucleic Acids Res., 25, 4626–4638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grindl W., Wende,W., Pingoud,V. and Pingoud,A. (1998) Nucleic Acids Res., 26, 1857–1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gorbalenya A.E. (1998) Nucleic Acids Res., 26, 1741–1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sellem C.H. and Belcour,L. (1997) Mol. Biol. Evol., 14, 518–526. [DOI] [PubMed] [Google Scholar]
- 30.Burger G. and Werner,S. (1985) J. Mol. Biol., 186, 231–242. [DOI] [PubMed] [Google Scholar]
- 31.Michel F. and Cummings,D.J. (1985) Curr. Genet., 10, 69–79. [DOI] [PubMed] [Google Scholar]
- 32.Paquin B., Laforest,M.J. and Lang,B.F. (1994) Proc. Natl Acad. Sci. USA, 91, 11807–11810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kroymann J. and Zetsche,K. (1997) Curr. Genet., 31, 414–418. [DOI] [PubMed] [Google Scholar]
- 34.Denovan W.E., Nedelcu,A.M. and Lee,R.W. (1998) Plant Mol. Biol., 36, 285–295. [DOI] [PubMed] [Google Scholar]
- 35.Paquin B., O’Kelly,C.J. and Lang,B.F. (1995) Curr. Genet., 28, 97–99. [DOI] [PubMed] [Google Scholar]
- 36.Aravind L., Walker,D.R. and Koonin,E.V. (1999) Nucleic Acids Res., 27, 1223–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kowalski J.C., Belfort,M., Stapleton,M.A., Holpert,M., Dansereau,J.T., Pietrokovski,S., Baxter,S.M. and Derbyshire,V. (1999) Nucleic Acids Res., 27, 2115–2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Derbyshire V., Kowalski,J.C., Dansereau,J.T., Hauer,C.R. and Belfort,M. (1997) J. Mol. Biol., 265, 494–506. [DOI] [PubMed] [Google Scholar]
- 39.Esser K. (1974) In King,R.C. (ed.), Handbook of Genetics. Plenum, New York, NY, pp. 531–551.
- 40.Silar P. (1995) Fungal Genet. Newsl., 42, 73. [Google Scholar]
- 41.Begueret J., Razanamparany,V., Perrot,M. and Barreau,C. (1984) Gene, 32, 487–492. [DOI] [PubMed] [Google Scholar]
- 42.Sainsard C.A., Begel,O. and Belcour,L. (1993) J. Mol. Biol., 234, 1–7. [DOI] [PubMed] [Google Scholar]
- 43.Jamet V.C., Boulay,J., Begel,O. and Silar,P. (1997) Curr. Genet., 31, 171–178. [DOI] [PubMed] [Google Scholar]
- 44.Lecellier G. and Silar,P. (1994) Curr. Genet., 25, 122–123. [DOI] [PubMed] [Google Scholar]
- 45.Koll F., Belcour,L. and Vierny,C. (1985) Plasmid, 14, 106–117. [DOI] [PubMed] [Google Scholar]
- 46.Cummings D.J., Michel,F. and McNally,K.L. (1989) Curr. Genet., 16, 407–418. [DOI] [PubMed] [Google Scholar]
- 47.Koll F., Boulay,J., Belcour,L. and d’Aubenton Carafa,Y. (1996) Nucleic Acids Res., 24, 1734–1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lisacek F., Diaz,Y. and Michel,F. (1994). J. Mol. Biol., 235, 1206–1217. [DOI] [PubMed] [Google Scholar]
- 49.Altschul S.F., Madden,T.L., Schaffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sellem C.H., d’Aubenton Carafa,Y., Rossignol,M. and Belcour,L. (1996) Genetics, 143, 777–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Collins R.A. and Saville,B.J. (1990) Nature, 345, 177–179. [DOI] [PubMed] [Google Scholar]
- 52.Henke R.M., Butow,R.A. and Perlman,P.S. (1995) EMBO J., 14, 5094–5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Turmel M., Otis,C., Cote,V. and Lemieux,C. (1997) Nucleic Acids Res., 25, 2610–2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chu F.K., Maley,G., Pedersen,L.J., Wang,A.M. and Maley,F. (1990) Proc. Natl Acad. Sci. USA, 87, 3574–3578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Michel F. and Dujon,B. (1986) Cell, 46, 323. [DOI] [PubMed] [Google Scholar]
- 56.Sancar A. (1996) Annu. Rev. Biochem., 65, 43–81. [DOI] [PubMed] [Google Scholar]
- 57.Eddy S.R. and Gold,L. (1992) Proc. Natl Acad. Sci. USA, 89, 1544–1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kadyrov F.A., Shlyapnikov,M.G. and Kryukov,V.M. (1997) FEBS Lett., 415, 75–80. [DOI] [PubMed] [Google Scholar]