Abstract
Accurate delineation of Gross Tumor Volume (GTV) is crucial for radiotherapy. Deep learning-driven GTV segmentation technologies excel in rapidly and accurately delineating GTV, providing a basis for radiologists in formulating radiation plans. The existing 2D and 3D segmentation models of GTV based on deep learning are limited by the loss of spatial features and anisotropy respectively, and are both affected by the variability of tumor characteristics, blurred boundaries, and background interference. All these factors seriously affect the segmentation performance. To address the above issues, a Layer-Volume Parallel Attention (LVPA)-UNet model based on 2D-3D architecture has been proposed in this study, in which three strategies are introduced. Firstly, 2D and 3D workflows are introduced in the LVPA-UNet. They work in parallel and can guide each other. Both the fine features of each slice of 2D MRI and the 3D anatomical structure and spatial features of the tumor can be extracted by them. Secondly, parallel multi-branch depth-wise strip convolutions adapt the model to tumors of varying shapes and sizes within slices and volumetric spaces, and achieve refined processing of blurred boundaries. Lastly, a Layer-Channel Attention mechanism is proposed to adaptively adjust the weights of slices and channels according to their different tumor information, and then to highlight slices and channels with tumor. The experiments by LVPA-UNet on 1010 nasopharyngeal carcinoma (NPC) MRI datasets from three centers show a DSC of 0.7907, precision of 0.7929, recall of 0.8025, and HD95 of 1.8702 mm, outperforming eight typical models. Compared to the baseline model, it improves DSC by 2.14 %, precision by 2.96 %, and recall by 1.01 %, while reducing HD95 by 0.5434 mm. Consequently, while ensuring the efficiency of segmentation through deep learning, LVPA-UNet is able to provide superior GTV delineation results for radiotherapy and offer technical support for precision medicine.
Keywords: Nasopharyngeal carcinoma, Image segmentation, Deep learning, Convolutional network, Layer volume Attention-UNet
Highlights
-
•
Parallel 2D-3D strategy reduces anisotropy and spatial feature loss impacts.
-
•
Introduction of L(V)-MSCA addresses tumor variability and unclear edges.
-
•
Layer-Channel Attention elevates the network's focus on tumor-relevant structures.
-
•
LVPA-UNet has the potential to optimize radiation dose distribution planning.
1. Introduction
Intensity-modulated radiation Therapy (IMRT) employs high-energy rays or particle beams to target and eradicate cancer cells while avoiding the surrounding normal tissues to minimize damage [1]. Therefore, identifying and delineating Gross Tumor Volume (GTV) rapidly and accurately is a prerequisite for a good tumor radiotherapy plan formulation, effective dose delivery, and efficient and precise treatment. When the delineated GTV is smaller or larger than its actual size, under-treatment of tumor cells or damage of normal cells may occur by the corresponding biased radiation plan [2]. Currently, GTV delineation in MRIs is a predominantly manual, slice-by-slice process conducted by radiation oncologists, which is time-consuming and several hours may be needed for one complex case. It is difficult to achieve a consistent boundary judgment among radiation oncologists of varying expertise levels [3], which brings certain obstacles to the accurate formulation of the IMRT plan. In this situation, there is an urgent need for intelligent GTV segmentation methods, where machines replace humans to achieve precise and consistent delineation of GTV.
In recent years, deep learning technology has achieved significant breakthroughs and applications across various fields, including industry [4,5], agriculture [6], psychology [7,8], and medicine [9,10]. Especially in the medical field, deep learning technology provides intelligent and precise solutions for GTV segmentation. This technology is capable of learning the features of the image in depth, automatically identifying the boundary between tumor and normal tissue, and thus accurately locating and segmenting the GTV, which provides strong support for the formulation of the IMRT plan.
Existing deep learning-based GTV segmentation methods can be categorized into three types: The first approach deconstructs 3D images into a series of slices and employs 2D neural networks for precise segmentation by recognizing and differentiating minor structures and lesions. Subsequently, the segmented 2D slices are reassembled into cohesive 3D images [[11], [12], [13]]. However, due to the lack of spatial correlation between slices in the 2D network segmentation process, this might lead to an incoherent understanding of the 3D anatomical structure and loss of critical 3D spatial feature information, potentially causing inconsistencies in the predicted tumor areas across different slices. The second method employs 3D neural networks to segment 3D images directly [[14], [15], [16], [17], [18], [19], [20], [21], [22], [23], [24]], which is effective in maintaining continuity between slices and overall spatial correlation, providing a comprehensive perception of volumetric spatial features. However, this method might lean towards learning incorrect volumetric structural information due to the resolution difference between the x, y, and z axes in MRIs (image anisotropy), overlooking important details within slices and causing distortion in segmentation results [25,26]. The third approach utilizes hybrid 2D-3D networks, combining the capabilities of 2D networks to focus on fine details within slices with the advantages of 3D networks in understanding the overall spatial structures [[26], [27], [28]], effectively solving the issue of spatial feature loss encountered with the use of only the 2D networks, as well as the issue of the 3D networks in dealing with the anisotropy of the images [29].
Nonetheless, hybrid 2D-3D networks still share common issues with 2D and 3D networks, including: (a) Tumors exhibit significant variabilities in shapes and sizes, and the differences in gray level with surrounding tissues are not distinct, resulting in their boundaries appearing blurred in images. If the receptive fields of the networks are not wide enough, they may not be able to accurately capture the complete shape, structure, and boundary information of the tumors, and especially perform poorly when dealing with tumor scenes with slim edges or blurred boundaries. (b) In MRIs, tumors occupy a small spatial proportion, and the networks, while extracting large amounts of irrelevant background information, might overshadow critical semantic features of tumors, affecting the ability to differentiate details between tumors and normal tissues. In such cases, attention mechanisms that target specific regions become the key to addressing this issue [30,31].
To address the issues of the existing methods, this study proposes the Layer-Volume Parallel Attention (LVPA)-UNet network, which fully harnesses the strengths of hybrid 2D-3D networks. Built upon the classic UNet architecture, LVPA-UNet incorporates an LVPA module at each stage of the encoder, employing three strategies to accurately capture key tumor characteristics. Firstly, this study extends the multi-scale convolutional attention (MSCA) module [32] and proposes an integrated 2D-3D parallel workflow strategy that combines parallel Layer MSCA (L-MSCA) and Volume MSCA (V-MSCA). This parallel strategy facilitates collaboration between feature extraction processes across different dimensions, effectively extracting the anatomical structure information of tumors within slices and their volumetric spaces. It addresses issues of segmentation inconsistency between slices and morphological distortion, caused by the loss of spatial features and anisotropy, respectively. Secondly, for the modular design of L-MSCA and V-MSCA, 2D and 3D multi-branch depth-wise strip convolutions are respectively implemented. This design significantly expands the model's receptive field, enabling it to adapt to variations in tumor shape and size within slices and volumetric spaces, and enhances its ability to identify and process blurred boundaries. Thirdly, leveraging the concept of the dual attention mechanism [31,33], the Layer-Channel Attention module is proposed to focus on slices and channels that are closely related to tumors, while effectively suppressing background noise and interfering signals from lesion-free tissues, thus enhancing the detailed differentiation between tumors and normal tissues. The contributions of this study are as follows.
-
(a)
Based on the hybrid 2D-3D architecture, this study proposes an LVPA-UNet to realize GTV segmentation, which can assist radiation therapists in formulating more accurate radiation dose distribution plans.
-
(b)
For each stage of the encoder, this paper introduces three innovative strategies: 2D-3D parallel workflows, parallel multi-branch depth-wise strip convolutions, and Layer-Channel Attention—to address the issues that 2D, 3D, or hybrid 2D-3D networks face, including spatial feature loss, anisotropy, variable tumor characteristics, blurred boundaries, and background interference in MRIs. By integrating these three strategies, a significant improvement in the overall performance of the network is achieved.
-
(c)
In a performance comparison conducted on 1010 stage II NPC T1-weighted MRIs, LVPA-UNet outperforms eight typical models, showcasing its superior effectiveness in GTV segmentation.
2. Methodology
The purpose of this study is to develop and evaluate a novel model for predicting GTV in MRIs. A dataset comprising 1010 cases of stage II NPC from three research institutions has been gathered to train, validate, and test the proposed model LVPA-UNet, which is an end-to-end deep learning network designed specifically for GTV segmentation. The LVPA-UNet's performance improvement is confirmed by comparing it with typical models and through ablation experiments.
2.1. The overall architecture of LVPA-UNet
Fig. 1 shows the network architecture of the proposed LVPA-UNet. The input to LVPA-UNet is a 3D T1-weighted MRI, denoted as , where comprises slices of 2D spatial resolution . Given that the input modality is solely T1-weighted, the channel number is 1. Based on the UNet architecture, LVPA-UNet primarily comprises an encoder and a decoder. The encoder comprises multiple stages, each leveraging the LVPA module to generate high-level information through the integration of three advanced mechanisms. Specifically, these encoder enhancements include parallel 2D and 3D workflows to improve both 2D slice details and 3D spatial information, L-MSCA and V-MSCA [32] composed of parallel multi-branch depth-wise strip convolutions to expand the receptive field with a heightened focus on boundary processing and variable tumors, and Layer-Channel Attention module to increase attention on tumor-related slices and channels. Within the decoder module, each stage employs a skip connection to fuse information from corresponding stages in the encoder and decoder, progressively generating the segmentation map. The final high-resolution segmentation result is formed by combining maps from adjacent stages.
Fig. 1.
The overall architecture of the proposed LVPA-UNet.
Abbreviation: CNN = convolution neural network; L-M = L-MSCA module; V-M = V-MSCA module; LVPA = Layer-Volume Parallel Attention.
2.2. Network encoder
The encoder of LVPA-UNet consists of four stages, each encompassing an LVPA module that is designed to integrate information from 2D slices with 3D volumetric space and adaptively perceive highly correlated slices and channels with tumors. In constructing the encoder, specific considerations are given to the feature dimensions at each stage: (a) The Stem block [32,34] is incorporated with the intent to preserve detailed features at the original resolution as much as possible. Instead of simply halving , , and , they are tailored to be consistent with the size of the input MRI. (b) Considering that the dataset in this study suffers from anisotropy, with averaging of only 33.01 layers, it's imperative to minimize the loss of depth-related feature information due to a substantial reduction in . Hence, in stage I, is maintained to be consistent with the input image. (c) From stage II to stage IV, before the extraction of features by the LVPA module in each stage, overlapping patch embedding operations are configured to reduce resolution while extracting more enriched semantic features.
2.3. The architecture of the LVPA module
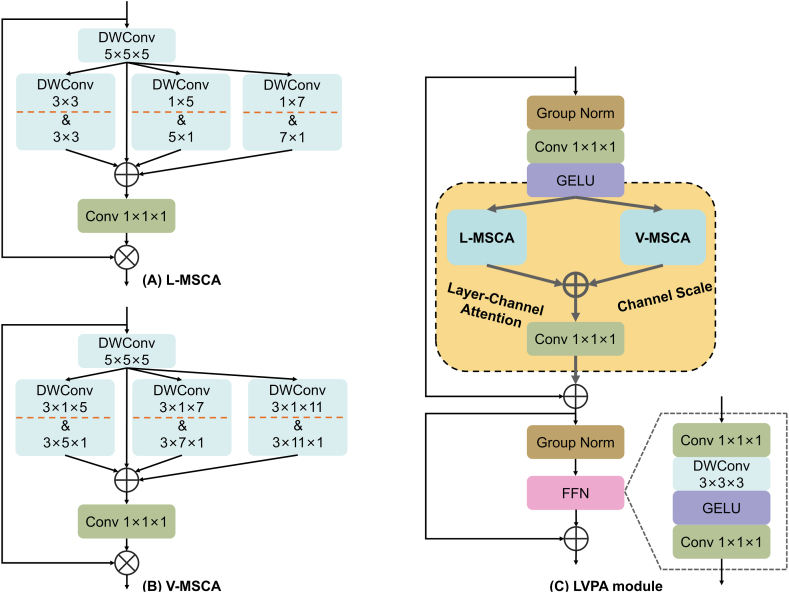
As depicted in Fig. 2, the LVPA module is a composite built from a stack of parallel L-MSCA, V-MSCA, Layer-Channel Attention, and auxiliary modules.
Fig. 2.
The architecture of the proposed LVPA module in detail. (A) denotes the L-MSCA, (B) denotes the V-MSCA, and (C) denotes the L-MSCA and V-MSCA are parallel in the yellow region of the LVPA module and then subsequently fused by Conv 1 × 1 × 1.
Abbreviation: Conv = convolution; DWConv = depth-wise convolution; FFN = feedforward neural network; GELU = gaussian error linear unit. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
2.3.1. The overview of L (V)-MSCA module
At the core of the LVPA module, inspired by Ref. [32], an innovative approach is taken to handle 3D images. This marked the inception of Layer MSCA (L-MSCA) and Volume MSCA (V-MSCA). As illustrated in Fig. 2(A and B), both L-MSCA and V-MSCA adopt similar architectural designs, encompassing four key components: (a) a depth-wise convolution serving as the conduit for local information aggregation; (b) multi-branch depth-wise strip convolutions engineered to capture semantic features of tumor at multiple scales and different shapes, thus expanding the receptive field; (c) a convolution to fuse information across different channel; and (d) a subsequent application of the output from the aforementioned stages as attention weights, through multiplication with the L (V)-MSCA module's input, effectuating a re-weighting of the input features for both L-MSCA and V-MSCA.
As shown in Fig. 2(C), the introduction of L-MSCA and V-MSCA integrates two key strategies of this study. The parallel approach fosters cooperation between 2D and 3D feature extraction processes, effectively extracting anatomical information within slices and their volumetric spaces, overcoming the loss of spatial features and the distortion caused by image anisotropy; the design of parallel multi-branch depth-wise strip convolutions expands the model's receptive field, adapting to the variability of tumor characteristics and enhancing the capability to handle ambiguous boundaries. Detailed descriptions of the L(V)-MSCA design are provided in sections 2.3.2, 2.3.3.
2.3.2. L-MSCA module
In the pursuit of robust segmentation from MRI data, one must account for the inherent anisotropy within the data. For instance, it is not uncommon to observe a glaring presence of a tumor in one slice and its complete absence in an adjacent one. This discontinuity poses a conundrum: an excessive reliance on 3D modules in segmentation networks could inadvertently sway the model toward volumetric information, at the peril of overshadowing crucial intra-slice features. Thus, there is a palpable necessity to devise a 2D-centric module that deftly captures the nuances within slices.
The L-MSCA module is improved based on [32], both modules for processing 2D images. However, what demands attention in the context of the NPC dataset is that NPC GTV generally occupies a minuscule fraction of the image and exhibits gray levels not significantly divergent from surrounding tissues, compounded by blurred boundaries. Thus, when constructing the L-MSCA module, it is necessary to give it a finer processing capability and slightly different corresponding parameters.
Fig. 2(A) delineates the structure of the L-MSCA module. For an input feature map , the module first applies a depth-wise convolution (DWConv) to assimilate local information. This operation can be succinctly expressed as:
| (1) |
where symbolizes the DWConv operation.
Subsequently, the feature maps are processed through an ensemble of three parallel depth-wise strip convolution branches coupled with an additional identity branch. Within these three branches, pairs of depth-wise strip convolutions are deployed, with convolutional pairs specifically configured as and , and , as well as and . The rationale behind utilizing pairs with larger kernels in strip convolutions is to optimize for flat or elongated targets, such as the heterogeneously shaped NPC GTV. This design also enables the inclusion of more information about the surrounding areas of tumors with variable shapes, thereby optimizing the handling of their ambiguous boundaries. Furthermore, employing and convolution sizes emulates the performance of convolutions, thereby achieving a broader receptive field with minimal computational overhead. Lastly, to counterbalance any deficiencies that depth-wise strip convolutions may exhibit in block object scenarios, a complementary branch with a pair of DWConvs is integrated. Taken together, this architecture, boasting four parallel branches, is exquisitely tailored to the context like NPC GTV, ensuring the rich excavation of multiscale and morphologically diverse information within the slices. The process is succinctly encapsulated in the following equation:
| (2) |
where represents a pair of DWConvs, with strip convolution kernel sizes set at and .
Finally, the feature maps from the four branches are amalgamated, serving as attention weights that undergo a Hadamard product with to reweigh the input features. This series of operations can be articulated as:
| (3) |
where denotes the output of the L-MSCA module, represents a convolution, and symbolizes the Hadamard product.
2.3.3. V-MSCA module
As shown in Fig. 2(B), V-MSCA's network structure is akin to the L-MSCA module; however, it distinguishes itself by employing 3D depth-wise strip convolutions in its three branches. To further augment the expression of multi-scale features, it exhibits a unique set of convolution kernel sizes distinct from L-MSCA. The V-MSCA module's calculations are captured in the following:
| (4) |
| (5) |
| (6) |
where and represent intermediate outputs, signifies the output of the V-MSCA module, and denotes a pair of DWConvs with strip convolution kernel sizes set at and .
2.3.4. Layer-Channel Attention
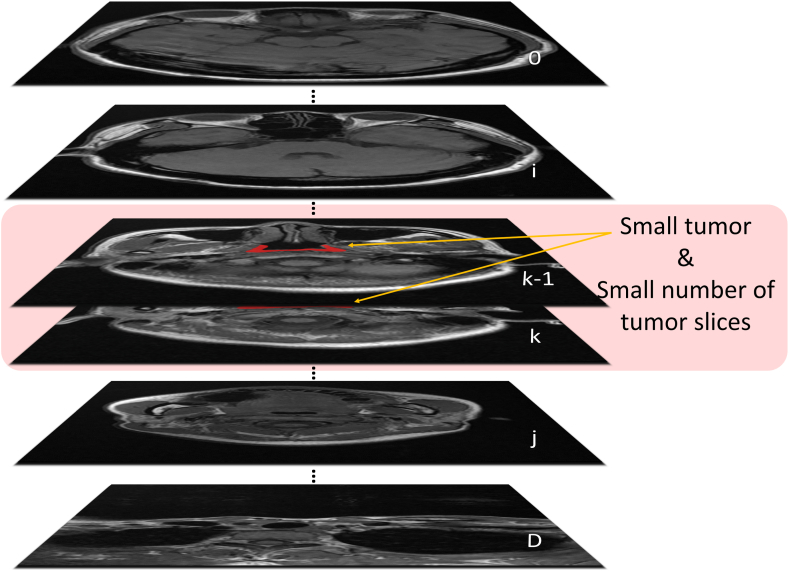
As depicted in Fig. 3, the slices that contain the NPC tumor constitute only a small fraction of the total slice count. Moreover, the small size of the tumor within these slices, combined with its indistinct boundaries when compared to normal tissue, presents a significant challenge. Indiscriminately applying sliding convolutional kernels across feature maps results in the extraction of information that is mixed with the complex background, negatively impacting the precision of GTV segmentation. Given these observations, a Layer-Channel Attention module is introduced, specifically tailored to the output of the L-MSCA module, as an effective countermeasure.
Fig. 3.
Visualization of tumor characteristics in MRI imaging. represents image depth, represents the slice number containing the NPC tumor, and the red markings indicate areas with the tumor. Note that the tumor is present only in a small portion of the total number of slices, and it appears small in size within these slices. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
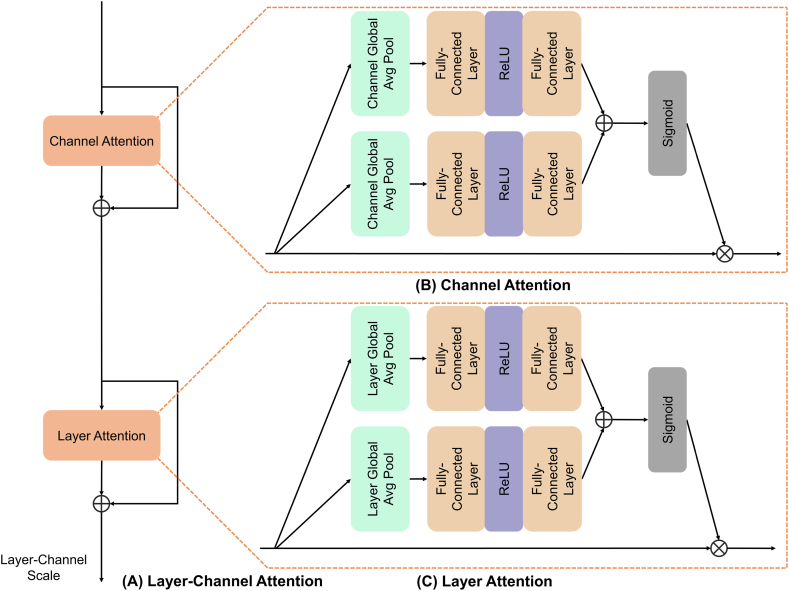
As shown in Fig. 4, inspired by the dual attention mechanism presented in Refs. [31,33], the conventional channel attention has been extended into a more sophisticated Layer-Channel Attention. Specifically, the input feature maps, denoted as , are sequentially passed through Channel Attention and Layer Attention, processing the features along the channel and layer dimensions. This endows the network with the ability to allocate varying weights to each channel and slice, thereby accentuating features that exhibit a strong correlation with the tumor and attenuating contributions from less relevant regions. Finally, a Layer-Channel Scale matrix of size is deployed, which further amplifies the importance of the strongly correlated channels and slices of the tumor to the overall segmentation. The specific formulation of the formula is as follows:
| (7) |
| (8) |
| (9) |
where represents Channel Attention, denotes Layer Attention, and signifies the Layer-Channel Scale. and are intermediate outputs after the application of Channel Attention and Layer Attention, respectively, whereas represents the final output subsequent to processing by the entire Layer-Channel Attention module.
Fig. 4.
The architecture of Layer-Channel Attention. Layer-Channel Attention is depicted in (A). Channel Attention (B) and Layer Attention (C) are similar in structure, but the scope of expression of their attention is different. This architecture automatically selects key features for multiple channels and layers.
Abbreviation: ReLU = rectified linear unit.
2.4. Network decoder
LVPA-UNet embraces the architecture of the decoder design delineated in Ref. [24] which is organized in a four-stage cascade that works together to gradually improve resolution and define the segmentation outlines. Each stage comprises three Conv3D-GroupNorm-ReLU (CGR) operations, where Group Normalization is chosen over Instance Normalization. Between adjacent CGR operations, upsampling methods based on trilinear interpolation and skip connections are implemented. These skip connections efficiently combine the high-level feature maps from the encoder with the corresponding stage of the decoder, effectively integrating multi-scale features. Notably, to safeguard the richness of depth information, The decoder treats in the same way as the encoder, with stages I and II preserving depth congruent to the input features.
Furthermore, the segmentation results of the previous layer are often instructive. As shown in the green flow in Fig. 1, the decoder part starts from stage III, downscaling the features extracted by the CNN block, realizing cross-channel information interaction and integration, and generating a single-channel rough segmentation result; the rough segmentation result is scaled up to the size of the result of the next layer, and the output of the CNN block of the next layer is fused into a higher resolution and more spatially detailed segmentation result to achieve the effect that the segmentation result of the former layer guides the latter layer. Repeat the operation until the final prediction map is generated.
3. Experiments and results
3.1. Experimental data
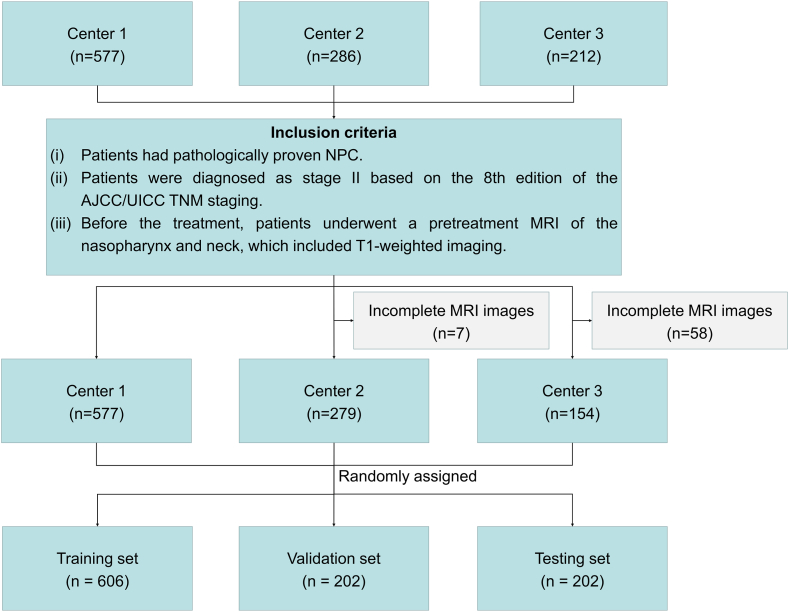
In this study, a retrospective review is carried out on 577 cases of stage II NPC from the Sun Yat-sen University Cancer Center (from May 2010 to July 2017), 286 cases from Fujian Province Cancer Hospital (from April 2008 to December 2016), and 212 cases from Jiangxi Province Cancer Hospital (from January 2010 to July 2017). As illustrated in Fig. 5, based on the inclusion and exclusion criteria, a total of 1010 NPC patients (median age, 45 years; 673 males, 337 females) with corresponding diagnostic MRIs are included in this study. Oncologists delineate the GTV on all MRI slices, serving as the ground truth for training and assessing the model's performance..
Fig. 5.
Flowchart of patient enrollment.
AbbreviationAJCC/UICC = American Joint Committee on Cancer/Union for International Cancer Control.
Experiments are conducted on T1-weighted MRIs of the head and neck region of 1010 NPC patients. The inter-voxel spacings in the MRIs are set to through linear interpolations, with average dimensions of . The data are randomly divided into three subsets: a training set (606 cases, 60 %), a validation set (202 cases, 20 %), and a testing set (202 cases, 20 %). Statistical information on the dataset is shown in Table 1.
Table 1.
Statistical information of the dataset.
| Characteristic | Training-Validation Set (n = 808) | Testing Set (n = 202) |
|---|---|---|
| Age (years), median (range) | 45 (19, 82) | 45 (15, 76) |
| Sex, No. (%) | ||
| Male | 533 (66.0) | 140 (69.3) |
| Female | 275 (34.0) | 62 (30.7) |
| Histopathology, No. (%) | ||
| WHO I | 50 (6.2) | 0 (0.0) |
| WHO II | 108 (13.4) | 13 (6.4) |
| WHO III | 650 (80.4) | 189 (93.6) |
| T, No. (%) | ||
| T1 | 399 (49.4) | 108 (53.5) |
| T2 | 409 (50.6) | 94 (46.5) |
Patients are categorized based on the 8th edition of the American Joint Committee on Cancer staging manual.
Abbreviations: WHO: World Health Organization.
3.2. Performance metrics
The segmentation performance is evaluated via four metrics, namely the Dice similarity coefficient (DSC) [35], 95 % Hausdorff distance (HD95) [36], precision, and recall.
The DSC is defined as follows:
| (10) |
where denotes the predicted output mask, and denotes the ground truth mask. A higher DSC indicates a more accurate segmentation. In binary segmentation tasks, the DSC can be derived from precision and recall and is computationally equivalent to the F1-score.
The HD95 assesses the spatial discrepancy between the predicted output and ground truth masks, with smaller values indicating better performance. Let and denote the boundaries of the predicted mask and the ground truth mask, respectively. The maximal HD is defined as:
| (11) |
where , . The value 95 % is employed to mitigate the influence of a very small subset of outliers.
In summary, among these four metrics, a higher DSC, precision, recall, and lower HD95 indicate superior segmentation performance.
3.3. Implementation details
In this study, for the sake of fair comparison, the settings for LVPA-UNet, as well as all other models used in comparative experiments, are consistent. Apart from differences in model architecture, all training and testing parameters are kept identical.
All experiments are conducted using PyTorch [37] and MONAI [38]. Training and testing are carried out on a Windows Server equipped with an Intel Xeon CPU E5-2650 v4 with 12 cores and an NVIDIA GeForce RTX 3090 with 24 GB memory. All data are subjected to min-max normalization to normalize pixel values to the range of 0–1 before use.
During the training process, in addition to normalization, random flipping and rotation are performed on the data to enhance the model's generalizability. Furthermore, random patches of are cropped from 3D image volumes, an approach devised to exploit as many voxels as possible within the constraints of GPU memory. The dimensions of essentially encompass all lesions, striving to optimize segmentation performance. All weights are initialized using the Kaiming method [39]. The loss function comprises a weighted sum of Dice loss and cross entropy loss [19]. An Adam optimizer and cosine scheduler are employed, and the learning rate is initially set to 0.0001. The training batch size is set to 1, with 300 training iterations. The model weights at the iteration with the highest Dice score on the validation set are selected as the final model weights.
For the testing process, the sliding window inference method from MONAI is used, with a window size of 32 × 256 × 256 and an overlap of 50 %. The final four metrics are then derived by averaging the evaluations across all test data.
3.4. Comparison with typical models
The proposed model is compared to eight typical segmentation models: 3D UNet [40], Lin et al. [23], Isensee et al. [24], the 2D implementation of Isensee et al. [24], TransBTS [20], VT-UNet [19], UNETR [18], and MPU-Net [41]. As shown in Table 2, LVPA-UNet outperforms all other models in every metric, reaching peak scores for DSC, HD95, precision, and recall at 0.7907, 1.8702 mm, 0.7929, and 0.8025, respectively. Compared to the top-performing model VT-UNet, LVPA-UNet demonstrates an increase in mean DSC (0.7907 vs 0.7758), precision (0.7929 vs 0.7690), and recall (0.8025 vs 0.7989), respectively. Compared to the previous advanced model by Lin et al. [23] from Sun Yat-sen University Cancer Center, LVPA-UNet showcases an increase in mean DSC (0.7907 vs 0.7754), precision (0.7695 vs 0.7633), and recall (0.8025 vs 0.7983), respectively. In terms of HD95, it achieves superior performance by recording a lower value compared to Isensee et al. [24] (1.8702 mm vs 2.4136 mm).
Table 2.
Comparison of evaluation metrics of various typical models.
| Models | DSC ↑ | HD95 (mm) ↓ | Precision ↑ | Recall ↑ |
|---|---|---|---|---|
| 3D UNet | 0.7172 ± 0.0955 | 5.7052 ± 26.80 | 0.7354 ± 0.1185 | 0.7223 ± 0.1317 |
| Isensee et al. [24] | 0.7693 ± 0.0893 | 2.4136 ± 3.5872 | 0.7633 ± 0.1143 | 0.7924 ± 0.1163 |
| 2D Isensee et al. [24] | 0.6520 ± 0.1463 | 7.9084 ± 10.2189 | 0.8081 ± 0.1134 | 0.5763 ± 0.1846 |
| Lin et al. [23] | 0.7754 ± 0.0954 | 2.6832 ± 4.7794 | 0.7695 ± 0.1177 | 0.7983 ± 0.1197 |
| TransBTS | 0.7744 ± 0.0929 | 8.6436 ± 87.3025 | 0.7583 ± 0.1107 | 0.8080 ± 0.1221 |
| UNETR | 0.7683 ± 0.0944 | 3.0822 ± 6.2588 | 0.7687 ± 0.1184 | 0.7850 ± 0.1199 |
| VT-UNet | 0.7758 ± 0.0906 | 2.5433 ± 5.1414 | 0.7690 ± 0.1073 | 0.7989 ± 0.1203 |
| MPU-Net | 0.7693 ± 0.0963 | 3.1479 ± 7.7695 | 0.7817 ± 0.1143 | 0.7747 ± 0.1279 |
| LVPA-UNet | 0.7907 ± 0.0937 | 1.8702 ± 2.8491 | 0.7929 ± 0.1088 | 0.8025 ± 0.1201 |
Fig. 6 depicts the visual comparison of segmentation outcomes between LVPA-UNet and other typical models. The first and second rows present two adjacent slices from the same MRI image. In the first row, the delineations of the thin tumor area by 3D UNet, Lin et al. [23], Isensee et al. [24], 2D implementation of Isensee et al. [24], and VT-UNet are visibly thinner than the ground truth, even fragmented. Only UNETR, MPU-Net, and LVPA-UNet manage to accurately depict this area, but UNETR shows a tendency toward overlapping boundaries. In the second row, 3D UNet's prediction noticeably deviates from the ground truth. While the predictions by Isensee et al. [24], Lin et al. [23], TransBTS, UNETR, MPU-Net, and VT-UNet are fairly similar and superior to 3D UNet, LVPA-UNet's prediction closely matches the ground truth, demonstrating optimal performance.
Fig. 6.
Comparison of segmentation results between various representative segmentation models and LVPA-UNet. Ground truths for GTV in the current slice annotated by oncologists are marked with blue lines, while model predictions are outlined in red. The numbers at the bottom of the image represent the DSC score for the GTV in the current slice. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Upon scrutinizing the adjacent slices in the first and second rows, it's evident that the size and shape of the tumor vary significantly, even in consecutive slices. The eight typical models exhibit varying degrees of error in outlining the tumor across these slices. Notably, the model proposed by Lin et al. [23] exhibits significant morphological distortion of the tumor between adjacent slices. The 2D implementation of Isensee et al. [24] exhibits inferior performance on the first slice and superior on the second compared to its 3D counterpart, indicating a loss of 3D features that leads to inconsistencies between adjacent slices. In stark contrast, LVPA-UNet's predictions align well with the ground truth in both slices. This illustrates the strength of LVPA-UNet's parallel and interactive 2D and 3D workflows, which integrate volumetric information into the fine edge details of 2D slices, adapting to the anisotropic image environment for enhanced segmentation performance.
As shown in the third row, the delineations of the thin tumor area by 3D UNet and the model proposed by Isensee et al. [24] are interrupted. TransBTS, UNETR, VT-UNet, and MPU-Net, despite connecting this thin tumor area, do not accurately represent its thickness, and they significantly miss the ambiguous region on the right. In contrast, LVPA-UNet proposed in this study adeptly handles both the thin tumor area and the ambiguous region on the right.
3.5. Ablation study on the effect of the LVPA module
To elucidate the effectiveness of the LVPA module in improving NPC GTV segmentation performance, different modules are integrated into the model in a stepwise manner. As Table 3 demonstrates, the work by Isensee et al. [24] denotes the baseline, ‘V’ indicates the exclusive application of V-MSCA in the encoder block, and ‘LV’ refers to the module featuring a parallel structure of L-MSCA and V-MSCA. ‘LVPA’ represents the LV module enhanced with the Layer-Channel Attention module. This progressive integration of components simultaneously improves segmentation performance. Notably, compared to the baseline model, LVPA-UNet achieves the best performance in terms of mean DSC (0.7907 vs 0.7693), mean HD95 (1.8702 mm vs 2.4136 mm), mean precision (0.7929 vs 0.7633) and mean recall (0.8025 vs 0.7924). These results manifest the superior performance of the LVPA module in NPC GTV segmentation.
Table 3.
Comparison of segmentation performance by stepwise integration of various modules into the model.
| Models | DSC ↑ | HD95 (mm) ↓ | Precision ↑ | Recall ↑ |
|---|---|---|---|---|
| Isensee et al. [24] | 0.7693 ± 0.0893 | 2.4136 ± 3.5872 | 0.7633 ± 0.1143 | 0.7924 ± 0.1163 |
| V | 0.7822 ± 0.0913 | 2.3478 ± 3.4176 | 0.7836 ± 0.1113 | 0.7952 ± 0.1168 |
| LV | 0.7865 ± 0.0925 | 2.3342 ± 6.0059 | 0.7827 ± 0.1106 | 0.8041 ± 0.1172 |
| LVPA | 0.7907 ± 0.0937 | 1.8702 ± 2.8491 | 0.7929 ± 0.1088 | 0.8025 ± 0.1201 |
Fig. 7 presents visual results that emerge from progressively integrating distinct modules into the model. As depicted in the first row, the baseline model delineates an area that deviates from the ground truth, interrupting the delineation in the thinned area of the tumor within the slice. The incorporation of V-MSCA brings the outlined area into closer alignment with the ground truth, and it bridges the delineation in the thinning target area within the slice, attributed to the strip design of MSCA's multi-branch depth-wise strip convolution that optimizes the segmentation of slender structures and their indistinct boundaries. Transitioning from V-MSCA to the parallel workflow of L-MSCA and V-MSCA enhances the detailed handling of the slices by leveraging the advantages of a 2D-3D parallel architecture. Ultimately, with the support of the Layer-Channel Attention module, LVPA adaptively promotes significant slices and channels while suppressing other non-important areas, resulting in segmentation outcomes that closely approximate the ground truth in the images.
Fig. 7.
Comparative visualization of segmentation results of different modules gradually integrated into the model. Ground truths for GTV in the current slice annotated by oncologists are marked with blue lines, while model predictions are outlined in red. The numbers at the bottom of the image represent the DSC score for the GTV in the current slice. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
In the second row, apart from the shrunken segmentation result of the baseline model, the proposed models closely align with the ground truth. While the V model marginally outperforms the LV model by 0.59 % in DSC, the fully integrated LVPA model achieves the best results. This underscores the necessity of integrating the complete set of modules in LVPA-UNet.
3.6. Ablation study on different designs of L-MSCA and V-MSCA
This study undertakes an ablation study on the distinct kernel designs within the multiple branches of the L-MSCA and V-MSCA in the LVPA module. Here, denotes a 3D depth-wise convolution and a 3D depth-wise convolution, while signifies a 2D depth-wise convolution and a depth-wise convolution. Table 4 presents the evaluation results for these different designs. The convolution parameters in the second row achieve the optimal scores in DSC, HD95, and precision. By selecting convolutional kernel geometries as exemplified in the second row, it is possible to capture both the comprehensive attributes and specific details of tumors across various dimensions and configurations, achieving a more appropriate receptive field that enhances the perception of indistinct boundaries. Consequently, this strategic choice can improve the overall GTV segmentation accuracy.
Table 4.
The effect of different L-MSCA and V-MSCA designs of LVPA modules.
| L-MSCA Design | V-MSCA Design | DSC ↑ | HD95 (mm) ↓ | Precision ↑ | Recall ↑ | ||||
|---|---|---|---|---|---|---|---|---|---|
| [3 × 1] | [5 × 1] | [7 × 1] | 3[3 × 1] | 3[5 × 1] | 3[7 × 1] | 0.7890 ± 0.0923 | 2.0439 ± 3.1675 | 0.7814 ± 0.1095 | 0.8116 ± 0.1187 |
| [3 × 1] | [5 × 1] | [7 × 1] | 3[5 × 1] | 3[7 × 1] | 3[11 × 1] | 0.7907 ± 0.0937 | 1.8702 ± 2.8491 | 0.7929 ± 0.1088 | 0.8025 ± 0.1201 |
| [5 × 1] | [7 × 1] | [11 × 1] | 3[3 × 1] | 3[5 × 1] | 3[7 × 1] | 0.7901 ± 0.0933 | 1.9259 ± 2.9413 | 0.7842 ± 0.1079 | 0.8103 ± 0.1202 |
| [7 × 1] | [11 × 1] | [21 × 1] | 3[3 × 1] | 3[5 × 1] | 3[7 × 1] | 0.7884 ± 0.0915 | 2.0829 ± 3.9428 | 0.7863 ± 0.1102 | 0.8049 ± 0.1172 |
4. Discussion
This study utilizes a dataset compiled from 1010 stage II NPC T1-weighted MRIs from three renowned medical centers, meticulously delineated by two experienced radiation therapy specialists. Utilizing deep learning technology, an LVPA-UNet segmentation network integrating three key strategic advantages is designed. Through training, an automated segmentation model targeting GTV is developed.
The results of LVPA-UNet show a DSC of 0.7907, precision of 0.7929, recall of 0.8025, and a Hausdorff Distance HD95 of 1.8702 mm, surpassing the best existing model, with higher DSC by 1.49 %, greater precision by 2.39 %, improved recall by 0.36 %, and a reduced HD95 by 0.6731 mm. The model performance is enhanced continuously with the integration of modules during the ablation study: Firstly, a 1.29 % increase in DSC is obtained from the base model (Isensee et al. [24]) to the V model, which verifies that LVPA-UNet's strategy of multi-branch depth-wise strip convolutions improves the adaptability of the base model to the complex issues of elongated tumors, size variations, and ambiguous boundaries. Subsequently, a further 0.43 % increase in DSC is achieved when V is improved to LV, which thoroughly demonstrates the synergistic advantages of 2D-3D parallel workflows and parallel multi-branch depth-wise strip convolutions. Finally, an additional 0.42 % DSC increase is achieved with the segmentation algorithm from LV to LVPA by incorporating the Layer-Channel Attention module which can focus on slices with high tumor correlation. Overall, LVPA-UNet improves in DSC, precision, and recall by 2.14 %, 2.96 %, and 1.01 % respectively to the base model, while reducing HD95 by 0.5434 mm. The model successfully overcomes challenges commonly faced by normal 2D, 3D, or hybrid 2D-3D models, enables efficient and accurate segmentation for GTV automatically and consistently, and eliminates subjective judgment disparities.
From a medical perspective, the accurate delineation by experts from renowned medical institutions ensures the professionalism of the ground truth data, making it a solid foundation for the reliability of the segmentation model. Notably, while Lin et al. [23] have demonstrated their model's NPC GTV segmentation capabilities to be comparable with those of senior radiation oncologists, LVPA-UNet presented in this study outperforms it across all essential metrics. This advancement offers substantial technical support for the accurate formulation of IMRT radiation plans. Especially for remote areas without seasoned IMRT specialists, the reliable LVPA-UNet segmentation model can bring expert-level medical services to local patients.
From a perspective of universality, LVPA-UNet is endowed with natural generalizing genes for different tissues and organs by the inherent transferability of deep learning models. The solutions to address the issues of spatial feature loss and anisotropy, variability in tumor characteristics and blurred boundaries, as well as background interference, can be readily adapted to other similar 3D MRI or CT segmentation tasks, such as BraTS [42,43] and SPPIN [44]. This offers precise segmentation technology support for a wider range of clinical scenarios.
This study has several limitations.
-
(a)
The Diffusion Probabilistic Models (DPMs) [[45], [46], [47], [48]] have recently shown outstanding performance in the field of medical image segmentation. By simulating the diffusion process between image voxels, DPMs refine image features and capture contextual information, allowing them to implicitly recognize complex patterns and subtle changes within images. These models introduce unique optimization paths for GTV segmentation that differ from LVPA-UNet, demonstrating their distinct advantages. Consequently, by integrating the strengths of DPMs to enhance LVPA-UNet, there is potential for significant advancements in GTV segmentation performance.
-
(b)
The design of LVPA-UNet primarily focuses on optimizing the encoder stage, with limited depth optimization applied to skip connections and the decoder mechanism. There is a need to introduce attention mechanisms based on transformers or more complex fusion strategies in the future, to further enhance feature propagation [49,50]. Such advancements would not only allow for the full utilization of rich contextual information and long-distance dependencies during the decoding process but also more effectively restore spatial details in images.
-
(c)
The dataset utilized in this study is relatively limited in size. Given the importance of large datasets for enhancing the cross-domain adaptability and generalization capability of deep learning models, acquiring more extensive datasets is necessary. Moreover, LVPA-UNet currently relies solely on T1-weighted MRIs for training and evaluation. Incorporating additional MRI modalities such as T2-weighted and CET1-weighted would significantly enrich the information available to the model, providing more detailed analyses. However, the legal sensitivities associated with medical imaging data related to specific diseases like nasopharyngeal carcinoma impose restrictions and barriers on data collection and sharing, limiting the expansion of dataset size and modality diversity. In this context, exploring methods such as generative adversarial networks (GANs), DPMs, and text-to-image generation models [[51], [52], [53], [54]] to create synthetic images that closely resemble real data offers a viable solution to this challenge.
5. Conclusion
This study introduces LVPA-UNet, a network model for GTV segmentation. The model employs three strategies: parallel workflows, parallel multi-branch depth-wise strip convolutions, and Layer-Channel Attention mechanism, effectively addressing issues encountered in GTV segmentation, such as loss of spatial features and anisotropy, diversity in tumor size and shape, blurred boundaries, and background noise interference. By comparing the performance of the proposed model with eight typical models in NPC GTV segmentation tasks, the model's efficacy is successfully validated. From a medical application perspective, LVPA-UNet precisely delineates GTV, aiding in the creation of accurate radiation treatment plans, thereby improving patient treatment outcomes and quality of life. In terms of generalizability, LVPA-UNet applies to 3D segmentation tasks in similar images (e.g., MRI or CT). The synthetic images to expand multimodal information and increase data samples, investigate new skip connection and decoder network strategies will be the next work in the future, and the advantages of DPM will be also fully utilized to provide a more accurate and robust solution for the GTV segmentation task.
Ethics statement
This study receives ethics approval from the Ethics Committee of Sun Yat-Sen University Cancer Center, approval number B2021-229-01.
This study is a retrospective research project that analyzes medical records acquired from past diagnoses and treatments. The privacy and personal identity information of the subjects will be safeguarded. Consequently, this study is exempt from obtaining informed consent from patients.
Funding
This work is supported by Guangdong Basic and Applied Basic Research Foundation Grant (2023A1515011413).
Data availability statement
The source code of LVPA-UNet is available on https://github.com/XuHaoRran/LVPA-UNET. Data will be made available on request.
CRediT authorship contribution statement
Yu Zhang: Writing – review & editing, Writing – original draft, Validation, Project administration, Methodology, Investigation, Conceptualization. Hao-Ran Xu: Writing – review & editing, Writing – original draft, Visualization, Software, Methodology. Jun-Hao Wen: Investigation. Yu-Jun Hu: Investigation. Yin-Liang Diao: Writing – review & editing. Jun-Liang Chen: Visualization. Yun-Fei Xia: Writing – review & editing, Supervision, Project administration, Funding acquisition.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Contributor Information
Yu Zhang, Email: zhangyu@scau.edu.cn.
Yun-Fei Xia, Email: xiayf@sysucc.org.cn.
References
- 1.Wong K.C.W., Hui E.P., Lo K.-W., Lam W.K.J., Johnson D., Li L., Tao Q., Chan K.C.A., To K.-F., King A.D., Ma B.B.Y., Chan A.T.C. Nasopharyngeal carcinoma: an evolving paradigm. Nat. Rev. Clin. Oncol. 2021;18:679–695. doi: 10.1038/s41571-021-00524-x. [DOI] [PubMed] [Google Scholar]
- 2.Chen A.M., Chin R., Beron P., Yoshizaki T., Mikaeilian A.G., Cao M. Inadequate target volume delineation and local–regional recurrence after intensity-modulated radiotherapy for human papillomavirus-positive oropharynx cancer. Radiother. Oncol. 2017;123:412–418. doi: 10.1016/j.radonc.2017.04.015. [DOI] [PubMed] [Google Scholar]
- 3.Harrison K., Pullen H., Welsh C., Oktay O., Alvarez-Valle J., Jena R. Machine learning for auto-segmentation in radiotherapy planning. Clin. Oncol. 2022;34:74–88. doi: 10.1016/j.clon.2021.12.003. [DOI] [PubMed] [Google Scholar]
- 4.Chen Z., Yang J., Chen L., Feng Z., Jia L. Efficient railway track region segmentation algorithm based on lightweight neural network and cross-fusion decoder. Autom. ConStruct. 2023;155 doi: 10.1016/j.autcon.2023.105069. [DOI] [Google Scholar]
- 5.Feng Z., Yang J., Chen Z., Kang Z. LRseg: an efficient railway region extraction method based on lightweight encoder and self-correcting decoder. Expert Syst. Appl. 2024;238 doi: 10.1016/j.eswa.2023.122386. [DOI] [Google Scholar]
- 6.Zhang Y., Niu J., Huang Z., Pan C., Xue Y., Tan F. High-precision detection for sandalwood trees via improved YOLOv5s and StyleGAN. Agriculture. 2024;14 doi: 10.3390/agriculture14030452. [DOI] [Google Scholar]
- 7.Wang H., Li B., Wu S., Shen S., Liu F., Ding S., Zhou A. Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR) 2023. Rethinking the learning paradigm for dynamic facial expression recognition; pp. 17958–17968. [Google Scholar]
- 8.Liu F., Wang H.-Y., Shen S.-Y., Jia X., Hu J.-Y., Zhang J.-H., Wang X.-Y., Lei Y., Zhou A.-M., Qi J.-Y., Li Z.-B. OPO-FCM: a computational affection based OCC-PAD-OCEAN federation cognitive modeling approach. IEEE Transactions on Computational Social Systems. 2023;10:1813–1825. doi: 10.1109/TCSS.2022.3199119. [DOI] [Google Scholar]
- 9.Ren C.-X., Xu G.-X., Dai D.-Q., Lin L., Sun Y., Liu Q.-S. Cross-site prognosis prediction for nasopharyngeal carcinoma from incomplete multi-modal data. Med. Image Anal. 2024 doi: 10.1016/j.media.2024.103103. [DOI] [PubMed] [Google Scholar]
- 10.Hu Y.-J., Zhang L., Xiao Y.-P., Lu T.-Z., Guo Q.-J., Lin S.-J., Liu L., Chen Y.-B., Huang Z.-L., Liu Y., Su Y., Liu L.-Z., Gong X.-C., Pan J.-J., Li J.-G., Xia Y.-F. MRI-based deep learning model predicts distant metastasis and chemotherapy benefit in stage II nasopharyngeal carcinoma. iScience. 2023;26 doi: 10.1016/j.isci.2023.106932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cai J., Tang Y., Lu L., Harrison A.P., Yan K., Xiao J., Yang L., Summers R.M. Medical Image Computing and Computer Assisted Intervention–MICCAI 2018: 21st International Conference, Granada, Spain, September 16-20, 2018, Proceedings, Part IV 11. Springer; 2018. Accurate weakly-supervised deep lesion segmentation using large-scale clinical annotations: slice-propagated 3d mask generation from 2d recist; pp. 396–404. [Google Scholar]
- 12.Poudel R.P., Lamata P., Montana G. Revised Selected Papers 1. Springer; 2017. Recurrent fully convolutional neural networks for multi-slice MRI cardiac segmentation, in: reconstruction, segmentation, and analysis of medical images: first international workshops, RAMBO 2016 and HVSMR 2016, held in conjunction with MICCAI 2016, athens, Greece, october 17, 2016; pp. 83–94. [Google Scholar]
- 13.Wang W., Chen C., Wang J., Zha S., Zhang Y., Li J. Computer Vision–ECCV 2022: 17th European Conference, Tel Aviv, Israel, October 23–27, 2022, Proceedings, Part XXI. Springer; 2022. Med-DANet: dynamic architecture network for efficient medical volumetric segmentation; pp. 506–522. [Google Scholar]
- 14.Li J., Chen J., Tang Y., Wang C., Landman B.A., Zhou S.K. Transforming medical imaging with Transformers? A comparative review of key properties, current progresses, and future perspectives. Med. Image Anal. 2023 doi: 10.1016/j.media.2023.102762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bilic P., Christ P., Li H.B., Vorontsov E., Ben-Cohen A., Kaissis G., Szeskin A., Jacobs C., Mamani G.E.H., Chartrand G. others, the liver tumor segmentation benchmark (lits) Med. Image Anal. 2023;84 doi: 10.1016/j.media.2022.102680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen J., Lu Y., Yu Q., Luo X., Adeli E., Wang Y., Lu L., Yuille A.L., Zhou Y. 2021. Transunet: Transformers Make Strong Encoders for Medical Image Segmentation. arXiv Preprint arXiv:2102.04306. [Google Scholar]
- 17.Hatamizadeh A., Nath V., Tang Y., Yang D., Roth H.R., Xu D. Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries: 7th International Workshop, BrainLes 2021, Held in Conjunction with MICCAI 2021, Virtual Event, September 27, 2021, Revised Selected Papers, Part I. Springer; 2022. Swin unetr: swin transformers for semantic segmentation of brain tumors in mri images; pp. 272–284. [Google Scholar]
- 18.Hatamizadeh A., Tang Y., Nath V., Yang D., Myronenko A., Landman B., Roth H.R., Xu D. Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision. 2022. Unetr: transformers for 3d medical image segmentation; pp. 574–584. [Google Scholar]
- 19.Peiris H., Hayat M., Chen Z., Egan G., Harandi M. Medical Image Computing and Computer Assisted Intervention–MICCAI 2022: 25th International Conference, Singapore, September 18–22, 2022, Proceedings, Part V. Springer; 2022. A robust volumetric transformer for accurate 3d tumor segmentation; pp. 162–172. [Google Scholar]
- 20.Wang W., Chen C., Ding M., Yu H., Zha S., Li J. Medical Image Computing and Computer Assisted Intervention–MICCAI 2021: 24th International Conference, Strasbourg, France, September 27–October 1, 2021, Proceedings, Part I 24. Springer; 2021. Transbts: multimodal brain tumor segmentation using transformer; pp. 109–119. [Google Scholar]
- 21.Xie Y., Zhang J., Shen C., Xia Y. Medical Image Computing and Computer Assisted Intervention–MICCAI 2021: 24th International Conference, Strasbourg, France, September 27–October 1, 2021, Proceedings, Part III 24. Springer; 2021. Cotr: efficiently bridging cnn and transformer for 3d medical image segmentation; pp. 171–180. [Google Scholar]
- 22.Yu X., Yang Q., Zhou Y., Cai L.Y., Gao R., Lee H.H., Li T., Bao S., Xu Z., Lasko T.A., Abramson R.G., Zhang Z., Huo Y., Landman B.A., Tang Y. UNesT: local spatial representation learning with hierarchical transformer for efficient medical segmentation. Med. Image Anal. 2023 doi: 10.1016/j.media.2023.102939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin L., Dou Q., Jin Y.-M., Zhou G.-Q., Tang Y.-Q., Chen W.-L., Su B.-A., Liu F., Tao C.-J., Jiang N. others, Deep learning for automated contouring of primary tumor volumes by MRI for nasopharyngeal carcinoma. Radiology. 2019;291:677–686. doi: 10.1148/radiol.2019182012. [DOI] [PubMed] [Google Scholar]
- 24.Isensee F., Kickingereder P., Wick W., Bendszus M., Maier-Hein K.H. Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries: Third International Workshop, BrainLes 2017, Held in Conjunction with MICCAI 2017, Quebec City, QC, Canada, September 14, 2017, Revised Selected Papers 3. Springer; 2018. Brain tumor segmentation and radiomics survival prediction: contribution to the brats 2017 challenge; pp. 287–297. [Google Scholar]
- 25.Li X., Chen H., Qi X., Dou Q., Fu C.-W., Heng P.A. H-DenseUNet: hybrid densely connected UNet for liver and tumor segmentation from CT volumes. 2018. http://arxiv.org/abs/1709.07330 [DOI] [PubMed]
- 26.Tang H., Liu X., Han K., Xie X., Chen X., Qian H., Liu Y., Sun S., Bai N. Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision. 2021. Spatial context-aware self-attention model for multi-organ segmentation; pp. 939–949. [Google Scholar]
- 27.Xia F., Peng Y., Wang J., Chen X. A 2.5D multi-path fusion network framework with focusing on z-axis 3D joint for medical image segmentation. Biomed. Signal Process Control. 2024;91 doi: 10.1016/j.bspc.2024.106049. [DOI] [Google Scholar]
- 28.Zhao J., Xing Z., Chen Z., Wan L., Han T., Fu H., Zhu L. Uncertainty-Aware multi-dimensional mutual learning for brain and brain tumor segmentation. IEEE Journal of Biomedical and Health Informatics. 2023;27:4362–4372. doi: 10.1109/JBHI.2023.3274255. [DOI] [PubMed] [Google Scholar]
- 29.Zhang Y., Liao Q., Ding L., Zhang J. Bridging 2D and 3D segmentation networks for computation-efficient volumetric medical image segmentation: an empirical study of 2. 5 D solutions, Computerized Medical Imaging and Graphics. 2022;99 doi: 10.1016/j.compmedimag.2022.102088. [DOI] [PubMed] [Google Scholar]
- 30.Xie Y., Yang B., Guan Q., Zhang J., Wu Q., Xia Y. 2023. Attention Mechanisms in Medical Image Segmentation: A Survey. arXiv Preprint arXiv:2305.17937. [Google Scholar]
- 31.Liu Y., Zhang Z., Yue J., Guo W. SCANeXt: enhancing 3D medical image segmentation with dual attention network and depth-wise convolution. Heliyon. 2024;10 doi: 10.1016/j.heliyon.2024.e26775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guo M.-H., Lu C.-Z., Hou Q., Liu Z., Cheng M.-M., Hu S.-M. Segnext: rethinking convolutional attention design for semantic segmentation. arXiv Preprint arXiv:2209. 2022 [Google Scholar]
- 33.Tang P., Zu C., Hong M., Yan R., Peng X., Xiao J., Wu X., Zhou J., Zhou L., Wang Y. DA-DSUnet: dual attention-based dense SU-net for automatic head-and-neck tumor segmentation in MRI images. Neurocomputing. 2021;435:103–113. [Google Scholar]
- 34.Wang R.J., Li X., Ling C.X. Pelee: a real-time object detection system on mobile devices. Adv. Neural Inf. Process. Syst. 2018;31 [Google Scholar]
- 35.Ronneberger O., Fischer P., Brox T. Medical Image Computing and Computer-Assisted Intervention–MICCAI 2015: 18th International Conference, Munich, Germany, October 5-9, 2015, Proceedings, Part III 18. Springer; 2015. U-net: convolutional networks for biomedical image segmentation; pp. 234–241. [Google Scholar]
- 36.Huttenlocher D.P., Klanderman G.A., Rucklidge W.J. Comparing images using the Hausdorff distance. IEEE Trans. Pattern Anal. Mach. Intell. 1993;15:850–863. [Google Scholar]
- 37.Paszke A., Gross S., Chintala S., Chanan G., Yang E., DeVito Z., Lin Z., Desmaison A., Antiga L., Lerer A. 2017. Automatic Differentiation in PyTorch. [Google Scholar]
- 38.Cardoso M.J., Li W., Brown R., Ma N., Kerfoot E., Wang Y., Murrey B., Myronenko A., Zhao C., Yangothers D. MONAI: an open-source framework for deep learning in healthcare. arXiv Preprint arXiv:2211.02701. 2022 doi: 10.48550/arXiv.2211.02701. [DOI] [Google Scholar]
- 39.He K., Zhang X., Ren S., Sun J. Proceedings of the IEEE International Conference on Computer Vision. 2015. Delving deep into rectifiers: surpassing human-level performance on imagenet classification; pp. 1026–1034. [Google Scholar]
- 40.Çiçek Ö., Abdulkadir A., Lienkamp S.S., Brox T., Ronneberger O. Medical Image Computing and Computer-Assisted Intervention–MICCAI 2016: 19th International Conference, Athens, Greece, October 17-21, 2016, Proceedings, Part II 19. Springer; 2016. 3D U-Net: learning dense volumetric segmentation from sparse annotation; pp. 424–432. [Google Scholar]
- 41.Yu Z., Han S. 3D medical image segmentation based on multi-scale MPU-net. arXiv Preprint arXiv:2307.05799. 2023 doi: 10.48550/arXiv.2307.05799. [DOI] [Google Scholar]
- 42.Kazerooni A.F., Khalili N., Liu X., Haldar D., Jiang Z., Anwar S.M., Albrecht J., Adewole M., Anazodo U., Anderson H., Bagheri S., Baid U., Bergquist T., Borja A.J., Calabrese E., Chung V., Conte G.-M., Dako F., Eddy J., Ezhov I., Familiar A., Farahani K., Haldar S., Iglesias J.E., Janas A., Johansen E., Jones B.V., Kofler F., LaBella D., Lai H.A., Leemput K.V., Li H.B., Maleki N., McAllister A.S., Meier Z., Menze B., Moawad A.W., Nandolia K.K., Pavaine J., Piraud M., Poussaint T., Prabhu S.P., Reitman Z., Rodriguez A., Rudie J.D., Sanchez-Montano M., Shaikh I.S., Shah L.M., Sheth N., Shinohara R.T., Tu W., Viswanathan K., Wang C., Ware J.B., Wiestler B., Wiggins W., Zapaishchykova A., Aboian M., Bornhorst M., de Blank P., Deutsch M., Fouladi M., Hoffman L., Kann B., Lazow M., Mikael L., Nabavizadeh A., Packer R., Resnick A., Rood B., Vossough A., Bakas S., Linguraru M.G. The brain tumor segmentation (BraTS) challenge 2023. Focus on Pediatrics (CBTN-CONNECT-DIPGR-ASNR-MICCAI BraTS-PEDs) 2024 doi: 10.48550/arXiv.2305.17033. [DOI] [Google Scholar]
- 43.Baid U., Ghodasara S., Mohan S., Bilello M., Calabrese E., Colak E., Farahani K., Kalpathy-Cramer J., Kitamura F.C., Pati S., others The RSNA-ASNR-MICCAI BraTS 2021 benchmark on brain tumor segmentation and radiogenomic classification. arXiv Preprint arXiv:2107.02314. 2021 doi: 10.48550/arXiv.2107.02314. [DOI] [Google Scholar]
- 44.Buser M.A.D., van der Steeg A.F.W., Simons D.C., Wijnen M.H.W.A., Littooij A.S., ter Brugge A.H., Vos I.N., van der Velden B.H.M. 2023. Surgical Planning in Pediatric Neuroblastoma. [DOI] [Google Scholar]
- 45.Wu J., Fu R., Fang H., Zhang Y., Yang Y., Xiong H., Liu H., Xu Y. In: Medical Imaging with Deep Learning. Oguz I., Noble J., Li X., Styner M., Baumgartner C., Rusu M., Heinmann T., Kontos D., Landman B., Dawant B., editors. PMLR; 2024. MedSegDiff: medical image segmentation with diffusion probabilistic model; pp. 1623–1639.https://proceedings.mlr.press/v227/wu24a.html [Google Scholar]
- 46.Wu J., Ji W., Fu H., Xu M., Jin Y., Xu Y. 2023. Medsegdiff-v2: Diffusion Based Medical Image Segmentation with Transformer. arXiv Preprint arXiv:2301.11798. [Google Scholar]
- 47.Xing Z., Wan L., Fu H., Yang G., Zhu L. Diff-unet: a diffusion embedded network for volumetric segmentation. arXiv Preprint arXiv:2303.10326. 2023 doi: 10.48550/arXiv.2303.10326. [DOI] [Google Scholar]
- 48.Kazerouni A., Aghdam E.K., Heidari M., Azad R., Fayyaz M., Hacihaliloglu I., Merhof D. Diffusion models in medical imaging: a comprehensive survey. Med. Image Anal. 2023;88 doi: 10.1016/j.media.2023.102846. [DOI] [PubMed] [Google Scholar]
- 49.Aghdam E.K., Azad R., Zarvani M., Merhof D. 2023 IEEE 20th International Symposium on Biomedical Imaging (ISBI) 2023. Attention swin U-net: cross-contextual attention mechanism for skin lesion segmentation; pp. 1–5. [DOI] [Google Scholar]
- 50.Rahman M.M., Marculescu R. Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision (WACV) 2023. Medical image segmentation via cascaded attention decoding; pp. 6222–6231. [Google Scholar]
- 51.Makhlouf A., Maayah M., Abughanam N., Catal C. The use of generative adversarial networks in medical image augmentation. Neural Comput. Appl. 2023;35:24055–24068. [Google Scholar]
- 52.Chen Z., Yang J., Feng Z., Zhu H. RailFOD23: a dataset for foreign object detection on railroad transmission lines. Sci. Data. 2024;11:72. doi: 10.1038/s41597-024-02918-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim K., Na Y., Ye S.-J., Lee J., Ahn S.S., Park J.E., Kim H. Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision (WACV) 2024. Controllable text-to-image synthesis for multi-modality MR images; pp. 7936–7945. [Google Scholar]
- 54.Dayarathna S., Islam K.T., Uribe S., Yang G., Hayat M., Chen Z. Deep learning based synthesis of MRI, CT and PET: review and analysis. Med. Image Anal. 2024;92 doi: 10.1016/j.media.2023.103046. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The source code of LVPA-UNet is available on https://github.com/XuHaoRran/LVPA-UNET. Data will be made available on request.