Abstract
SOX proteins form a large family of transcription factors related by a DNA-binding domain known as the HMG box. Some 30 Sox genes have been identified in mammals and orthologues have been found in a wide range of other metazoans. Sox genes are highly conserved and are known to play important roles in embryonic development, including roles in gonadal, central nervous system, neural crest and skeletal development. Several SOX genes have been implicated in human congenital diseases. We report here the isolation of Sox8 and its characterisation in mice and humans. This gene has a remarkably similar primary structure and genomic organisation to the campomelic dysplasia gene SOX9 and the Waardenburg–Shah syndrome gene SOX10. SOX8 protein is able to bind to canonical SOX target DNA sequences and activate transcription in vitro through two separate trans-activation regions. Further, Sox8 is expressed in the central nervous system, limbs, kidneys, gonads and craniofacial structures during mouse embryo development. Sox8 maps to the t complex on mouse chromosome 17 and to human chromosome 16p13.3, a region associated with the microphthalmia–cataract syndrome CATM and the α-thalassemia/mental retardation syndrome ATR-16.
INTRODUCTION
Within a decade of the discovery of the mammalian Y-linked sex-determining gene Sry, the Sry-related HMG box (Sox) family has emerged as one of the major developmental gene families, containing some 30 members to date (for a review see 1). SOX proteins are characterised by a highly conserved DNA-binding and bending domain, known as an HMG box due to its amino acid similarity to the DNA-binding domain of the high mobility group (HMG) proteins. Two main groups exist within the HMG superfamily, the HMG/UBF group and the TCF/MATA/SOX group. HMG/UBF proteins, including non-histone proteins such as HMG-1 and HMG-2, contain at least one HMG box, bind DNA non-specifically and are usually associated with organising chromatin structure. The TCF/MATA/SOX group on the other hand contains the transcription factors TCF1 and LEF1, the yeast mating type protein MATA1 and SOX proteins. Members of this group bind DNA sequence specifically and contain a single HMG box (2).
Sox genes are predominantly expressed during embryogenesis and have been implicated in many developmental processes and human congenital diseases. Sox9, for example, is widely expressed in the developing embryo and is involved in skeletogenesis, brain development and sex determination (3). Mutations in SOX9 are responsible for the human congenital disease campomelic dysplasia, characterised by a variety of skeletal dysgeneses and, in the majority of XY cases, partial or complete sex reversal (4,5). In contrast, Sox10 expression is associated with neural crest cells in the embryo and defects in Sox10 underlie the Dom (dominant megacolon) mouse mutant and the human neurocristopathy Waardenburg–Shah syndrome (6,7). The Sox gene family can be divided into eight subfamilies, A–H, on the basis of the HMG box amino acid sequence (8,9). These subfamilies appear to represent groups which are derived from common ancestors (for a review see 10). Sox9 and Sox10 are members of the same subfamily, group E, and share moderate sequence similarity and conserved functional motifs such as a C-terminal trans-activation domain.
The importance of Sox9 and Sox10 in both mouse and human development prompted us to investigate a previously uncharacterised member of group E, Sox8. A putative Sox8 gene, dubbed SoxP1, was identified in rainbow trout by Ito and co-workers (11), but very little is known about its function, expression or whether it is a genuine orthologue of mouse Sox8, hitherto known only as a PCR fragment of the HMG box region (8). Here we present the sequence, genomic structure and chromosomal localisation of both mouse and human Sox8 and note a striking similarity in sequence and genomic organisation to Sox9 and Sox10. We show that SOX8 protein is able to bind sequence specifically to DNA and is able to activate transcription, suggesting that SOX8 acts as a modular transcription factor. The expression pattern of Sox8 during mouse development, together with its location on human chromosome 16p13.3, implicate SOX8 in the aetiology of the α-thalassemia deletion variant ATR-16, characterised by haematological abnormalities, mental retardation and craniofacial defects.
MATERIALS AND METHODS
Isolation of genomic and cDNA clones
cDNA clones were obtained from an 11.5 days post coitum (d.p.c.) whole embryo cDNA library in λGT10 (Stratagene), using a partial HMG box probe of mouse Sox8 (nucleotides 1321–1496, GenBank accession no. AF191325) and a wash stringency of 0.1× SSC at 78°C. The same probe was used to obtain a single Sox8 clone from a genomic DNA library in λDashII, prepared from a partial MboI digest of mouse genomic DNA. The genomic clone was digested with EcoRI and subcloned. The 10 kb insert was analysed by Southern hybridisation, subcloning and sequencing.
DNA sequencing and analysis
All DNA sequencing was performed using the big-dye terminator automated sequencing kit (Perkin Elmer Applied Biosystems) and either M13 forward and reverse primers or Sox8-specific primers and PCR conditions: 96°C for 2 min, followed by 25 cycles of 96°C for 30 s, 50°C for 15 s, 60°C for 4 min, followed by a final 10 min extension at 72°C. Sequences were read using an ABI 377 sequencer at the Australian Genome Research Facility. A mouse expressed sequence tag (EST) (accession no. AA288697) was found in the EST database (db-est) by a Blast 2.0 search with nucleotides 842–1508 as the query. This clone was obtained from UK-HGMP and sequenced as above. Human SOX8 sequence (accession no. HS349H11) was obtained from a Blast 2.0 search of the NCBI high throughput genomic sequence (HTGS) database (http://www.ncbi.nlm.nih.gov/blast/ ), using mouse Sox8 sequence as the query.
Northern analysis
An adult multi-tissue northern blot (Clontech) was probed according to the manufacturer’s instructions using Sox8 cDNA (nucleotides 842–1288, accession no. AF191325) and the control actin cDNA as probes, with washing to 0.1× SSC at 65°C, and analysed by phosphorimaging.
PCR and RT–PCR
All oligonucleotides were purchased desalted and RP-HPLC purified from Pacific Oligos (Lismore, Australia). Total RNA was isolated from embryonic tissue by the acid guanidinimum thiocyanate/phenol/chloroform method (12). cDNA was synthesised using AMV reverse transcriptase (Boehringer Mannheim) and oligo(dT) primers. PCR on embryonic cDNA was performed using primers RT-for (5′-TGGAGTCTGGTGCCTATGCCTGT-3′) and RT-rev (5′-GCCGAGCACTGCATCAGCTTTGT-3′) and a cycling program of 96°C for 2 min followed by 35 cycles of 96°C for 30 s, 65°C for 30 s, 72°C for 1 min, with a final extension of 5 min at 72°C. PCR products were analysed on 0.8–2% agarose gels depending on product size.
In situ hybridisation
Antisense and sense Sox8 digoxigenin-labelled RNA probes were prepared from the 3′-UTR and coding sequence (nucleotides 1546–2106). Whole mount hybridisations were carried out essentially as described (13).
Electrophoretic mobility shift assay (EMSA)
Murine SOX8 was produced by in vitro transcription/translation of full-length Sox8 cDNA in pcDNA3.1, using the TNT T7 quick-coupled transcription/translation system (Promega). Transcription/translation product of the same clone in reverse orientation was expressed as a control. Samples labelled with 35S were analysed by SDS–PAGE and autoradiography. EMSA was performed on unlabelled protein produced in parallel as described (14). The double-stranded oligonucleotides (SoCM, 5′-GATCAGACTGAGAACAAAGCGCTCTCACACGATC-3′, and SryC, 5′-GATCCGGACTAATAAACAATAAAGTC GACGGATC-3′) were labelled with 32P as described (14).
GAL4 trans-activation domain analysis
PCR was used to generate several fragments of mouse Sox8 for in-frame subcloning into the modified vector pGAL0 (15). The conditions used were: 96°C for 2 min followed by 35 cycles of 96°C for 1 min, 68°C for 1 min, 72°C for 90 s, followed by a final 5 min extension at 72°C. Primers used included:
Gal-A (5′-CGGAATTCGCCACCATGCTGGACATGAGT-3′); Gal-B (5′-GTTCTAGAGCCTTGAGCGTGCCGCCACC-3′); Gal-C (5′-GGAGAATTCTGAAGACTGGCCGGAGCGAC-3′); Gal-D (5′-CATCTAGATCAGGGTCGGGTCAGGG-3′); Gal-E (5′-CCGAATTCCCACAGATCAAGACGGAGC-3′); Gal-F (5′-TCGAATTCCTCTCTATGCCACCCGCAC-3′); Gal-G (5′-GGTCTAGACTGCTCCGTCTTGATCTGTG-3′).
All constructs were sequenced to ensure structural integrity. COS1 cells were transfected with these constructs using FuGENE 6 (Boehringer Mannheim) lipofectamine reagent, adding 2 µg of both the GAL0 fusion construct and the reporter plasmid (per well, 12-well plates) and incubating for 48 h. Cells were freeze/thawed and lysed by the addition of 100 µl lysis solution (50 mM HEPES pH 7.4, 1 mM CaCl2, 1 mM MgCl2, 2% Triton N-101) and incubation at 25°C for 5 min with shaking. An aliquot of 100 µl of luciferase reporter gene assay reagent (Boehringer Mannheim) was then added and the fluorescence of 180 µl detected on a Trilux MicroBeta 1450 luminometer. To confirm that all constructs were expressed, western analysis was performed as described previously (16) using rabbit anti-GAL4 (DBD) antibody (sc-577; Santa Cruz) at a concentration of 0.4 µg/ml and incubating overnight at 4°C. ECL detection was performed using a SuperSignal West Pico kit (Pierce) according to the manufacturer’s instructions
Chromosomal localisation
Mouse Sox8 was mapped using an interspecific mouse backcross panel from UK-HGMP. Mice were genotyped by PCR utilising an 18 bp deletion/insertion difference between Mus musculus musculus and Mus spretus. PCR was performed with primers momap-for (5′-AGATTCTCAGGGTGTGTGTGTGT-3′) and momap-rev (5′-TCCCGAAGTCCCCCTTTCCAGCA-3′), using the following conditions: 96°C for 2 min followed by 30 cycles of 96°C for 1 min, 65°C for 30 s, 72°C for 30 s. Products were analysed on a 3% agarose gel. Data were analysed by UK-HGMP and manual haplotype analysis. Mouse Sox8 was mapped on two YACs that spanned the linkage region using the same PCR procedure outlined above. Human SOX8 was mapped using the Genebridge4 radiation hybrid panel (UK-HGMP), using the primers humap-for (5′-CGAGGGACCTGCTTAGCCAC-3′) and humap-rev (5′-TGGGGGGAAGAAGCCGACAG-3′) and PCR program: 96°C for 2 min followed by 40 cycles of 96°C for 1 min, 65°C for 1 min, 72°C for 1 min, with a final extension of 5 min at 72°C. Products were electrophoresed on 2% agarose gels. Eighty-nine DNAs were typed and the results analysed by UK-HGMP (http://www.menu.hgmp.mrc.ac.uk/ ).
RESULTS
Sox8 sequence and genomic structure
To isolate mouse Sox8, cDNA and genomic libraries were screened with a probe corresponding to the central region of the mouse Sox8 HMG box. Primary screening revealed two identical cDNA clones that spanned nucleotides 843–1507 (GenBank accession no. AF191325). These clones overlapped with an EST clone spanning nucleotides 1–1171, the sequence of which concurred with those of the cDNA clones. The genomic library yielded a single clone of 14 kb from which the remainder of the open reading frame sequence was derived. Comparison of cDNA and genomic sequences, together with RT–PCR analysis, revealed the intron–exon boundaries, translation stop codon and putative translation initiation codon, which was preceded by an in-frame stop codon (nucleotide 1083). The deduced protein product of mouse Sox8 is 464 amino acids in length (Figs 1 and 2A and C).
Figure 1.
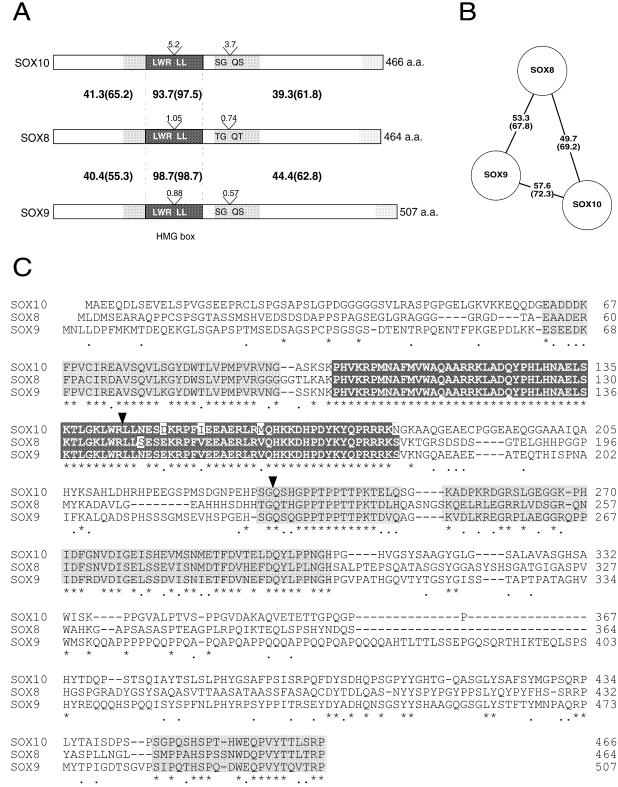
Structure and conservation of SOX8. Schematic representation of mouse SOX8 with the HMG box shaded light grey and the two trans-activation regions presented as hashed boxes (Fig. 3). Numbers refer to the first amino acid in each region. Amino acid identity between mouse (mo-SOX8), human (hu-SOX8) and trout (tr-SOXP1) SOX8 sequences are represented schematically below, with black denoting amino acid identity with the mouse sequence and white representing sequence divergence from the mouse sequence.
Figure 2.
Comparison of group E SOX proteins, SOX8, SOX9 and SOX10. (A) Schematic representation of mouse SOX8, SOX9 and SOX10 protein structures. Protein lengths, in amino acids (a.a.) are shown on the right. Intron positions are indicated (V), with the size in kb above and amino acid sequence flanking the splice sites below. The HMG boxes are shaded dark grey, while two conserved regions either side of the DNA-binding domain are shaded light grey. Amino acid identity (similarity in parentheses) between SOX8 and SOX9, and SOX8 and SOX10 is shown. (B) Schematic diagram of the relationship between SOX8, SOX9 and SOX10 proteins. Full-length amino acid sequence identity (percent) (similarity in parentheses) is indicated. (C) Amino acid sequence comparison of mouse SOX8, SOX9 and SOX10. The DNA-binding HMG box is boxed and shaded dark grey, with amino acid mismatches highlighted in white. Highly conserved regions, both N- and C-terminal of the HMG box, are shaded light grey. Amino acid identity between all three SOX proteins is indicated with an asterisk below the sequence, while conservative amino acid changes are indicated with a dot. Dashes (–) in the sequence represent gaps introduced to optimise sequence alignment. Amino acid numbering is on the right. Arrowheads show splice positions.
A search of the NCBI high throughput genomic sequence database yielded a sequence identified as human SOX8 on the basis of 100% amino acid sequence identity in the HMG box region. The amino acid sequence of human SOX8 shows 84.1% identity (91.4% similarity) overall with mouse SOX8 (Fig. 1). The open reading frame of human SOX8 is 1341 bp (producing a putative 447 amino acid protein), with intron–exon boundaries in the same positions as in mouse Sox8. The mouse protein also showed 81.6% similarity (72.4% identity) with the previously published trout sequence, SOXP1 (Fig. 1; 11), suggesting that tr-SoxP1 is orthologous to Sox8.
The genomic organisation of Sox8 was found to be remarkably similar to that of the other group E Sox genes Sox9 and Sox10. All are comprised of three exons of similar size and two introns in exactly cognate positions (Fig. 2A). Within this group, SOX9 and SOX10 are most similar, with 57.6% amino acid identity (>72% amino acid similarity), while SOX8 has a greater similarity to SOX9 (53.3% identity) than to SOX10 (49.7% identity) on the basis of amino acid sequence alone (Fig. 2B). Comparison of the three protein sequences revealed, in addition to the highly conserved HMG box, two highly conserved regions flanking the HMG box and a less well conserved region at the C-terminus (Fig. 2A and C).
DNA binding and transcriptional trans-activation by SOX8 protein
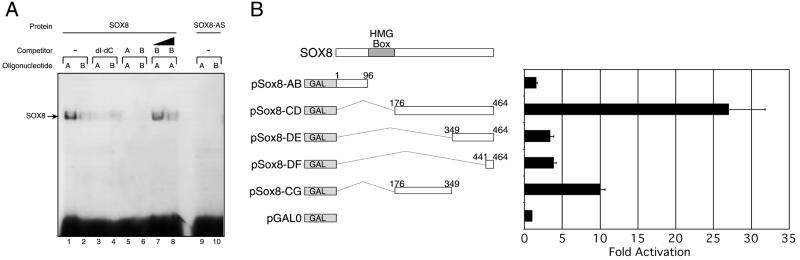
SOX proteins studied to date are characterised by the ability to bind to the core DNA sequence motifs AACAAT and AACAAAG or minor variants (9,17–21). We therefore wished to determine whether SOX8 is able to bind these sequences, which we have termed SryC and SoCM, respectively (21,22). In vitro transcription/translation followed by EMSA (Fig. 3A) demonstrated that full-length native mouse SOX8 is able to bind both oligonucleotides in the presence or absence of the non-specific competitor poly(dI·dC) (Fig. 3A, lanes 1–4). Furthermore, addition of the same unlabelled oligonucleotide abolished binding (Fig. 3A, lanes 5 and 6). A higher affinity of SOX8 for the SoCM oligonucleotide (AACAAAG) was confirmed by cross-competition experiments in which equimolar and twice molar amounts of unlabelled SryC were competed off by the addition of labelled SoCM (Fig. 3A, lanes 7 and 8).
Figure 3.
DNA binding and trans-activation by SOX8. (A) DNA binding. Lanes 1–8 contain equal amounts of full-length mouse SOX8, lanes 9 and 10 contain equal amounts of in vitro transcription/translation product of an antisense Sox8 construct (SOX8-AS). Lanes 1–8 all contain 2 ng of labelled oligonucleotide. Unlabelled non-specific competitor DNAs were included in lanes 3 and 4, as shown. Lanes 5 and 6 contain a 100-fold excess of unlabelled competitor oligonucleotide. Lanes 7 and 8 contain equimolar and twice molar amounts, respectively, of unlabelled oligonucleotide B. Oligonucleotide A contains the SoCM consensus sequence AACAAAG and B contains the SryC sequence AACAAT (21). The position of the SOX8 protein–DNA complex is indicated on the left. (B) GAL4 trans-activation assay. COS1 cells were transfected with the GAL4 fusion constructs shown on the left. The numbers represent mouse SOX8 amino acid intervals fused with GAL4. The right panel shows luciferase activity produced by each GAL4–SOX8 construct, relative to GAL0 = 1, with standard errors calculated from four replicate transfections. Similar results were observed in three independent experiments, each involving triplicate or quadruplicate determinations.
The two closest relatives of SOX8, namely SOX9 and SOX10, each contain transcriptional trans-activation domains at their C-termini (21,23). We tested the ability of SOX8 to activate transcription by co-transfecting GAL4–SOX8 fusion constructs and a Gal4UAS–luciferase reporter construct into COS1 cells and assaying the resultant luciferase reporter levels. All constructs lacked the DNA-binding HMG box as it is known to interfere with the activation assay (22). Western analysis confirmed that all GAL constructs were expressed equally (data not shown). Amino acids 176–464 of SOX8 were able to trans-activate transcription some 30-fold relative to GAL0 alone (vector containing no Sox8 fragment, Fig. 3B). Nested deletions of this region showed reduced trans-activation. A region comprising the 174 amino acids immediately C-terminal of the HMG box showed a 10-fold increase in trans-activation compared with GAL0. The C-terminus of SOX8 (amino acids 349–464 and 441–464, Fig. 3B) exhibited only limited trans-activation (~4-fold increase), in contrast to the findings for SOX9 and SOX10 (21,23,24).
Expression of mouse Sox8
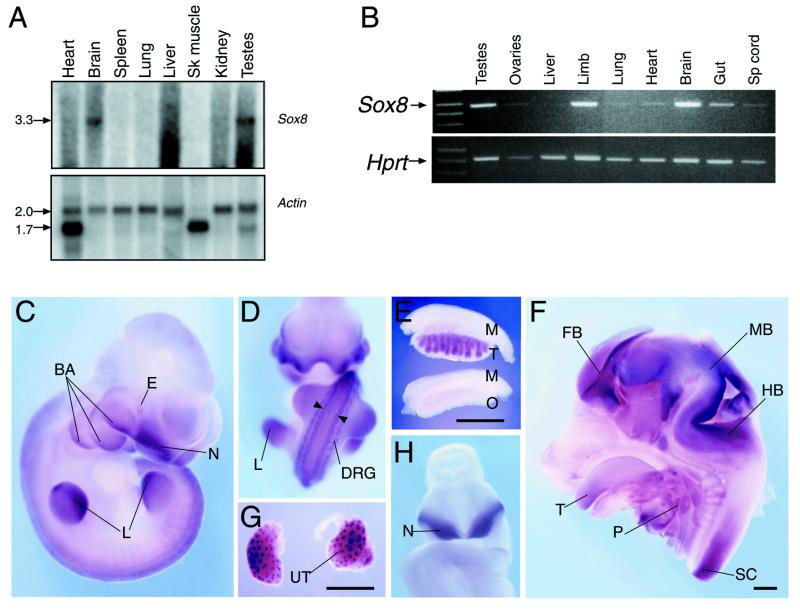
Sox8 expression in adult tissue was examined by probing an adult multi-tissue northern blot. A single 3.3 kb transcript was detected in brain and testes, with little or no detectable expression in other tissues (Fig. 4A).
Figure 4.
Expression of murine Sox8. (A) Northern blot showing expression in adult mouse tissues. (Upper) Sox8 probe; (lower) β-actin. Band sizes, in kb, are represented on the left. Sk muscle refers to skeletal muscle. (B) RT–PCR analysis of 13.5 d.p.c. foetal tissues. (Upper) Sox8; (lower) Hprt. Sp cord refers to spinal cord. (C–H) Whole mount in situ hybridisation analysis of Sox8 expression. (C) 10.5 d.p.c. mouse embryo, side view. BA, branchial arches; E, eye; N, nasal invagination; L, limb. (D) 10.5 d.p.c. whole embryo, anterior view. L, limb; DRG, dorsal root ganglia. Arrowheads indicate two stripes of expression at the lateral margins of the dorsal spinal cord. (E) 13.5 d.p.c. testes and ovaries. M, mesonephros; T, testis; O, ovary. (F) 13.5 d.p.c. head, bisected, medial view. FB, forebrain; MB, midbrain; HB, hindbrain; T, tongue; P, pharyngeal region; SP, spinal cord. (G) 13.5 d.p.c. kidneys. UT, ureteric tree. (H) 9.5 d.p.c. whole embryo, anterior view. N, nasal invagination. Bars in (E), (F) and (G) represent 1 mm.
Sox genes described to date have important roles in embryonic development and are directly responsible for several human congenital diseases (4,7,25). In order to investigate the possible role(s) of Sox8 in development, we analysed its expression during mouse embryogenesis. RT–PCR analysis revealed expression at all stages tested (9.5–15.5 d.p.c.; data not shown). Assay of dissected organs at 13.5 d.p.c. revealed expression in the brain, gut, limb and testes, and barely detectable expression in the liver, ovaries, spinal cord, lung and heart (Fig. 4B).
Sox8 expression in mouse embryos was further analysed by whole mount in situ hybridisation. Strong expression was observed at 10.5 d.p.c. in the ventral nasal invaginations, branchial arches, limbs, eye, dorsal root ganglia and in two parallel stripes at the lateral margins of the dorsal spinal cord (Fig. 4C and D). At 13.5 d.p.c. male-specific expression in the gonads, associated with testis cords (Fig. 4E), and expression in the forebrain, midbrain, hindbrain and spinal cord, as well as the pharyngeal region and tongue, was observed (Fig. 4F). Expression in the kidneys at 13.5 d.p.c. (Fig. 4G) was observed at the tips of the ureteric trees. Strong Sox8 expression was also apparent in the ventral nasal region of 9.5 d.p.c. embryos (Fig. 4H). These in situ data confirmed and extended the expression profile determined by RT–PCR.
Chromosomal localisation of mouse and human SOX8
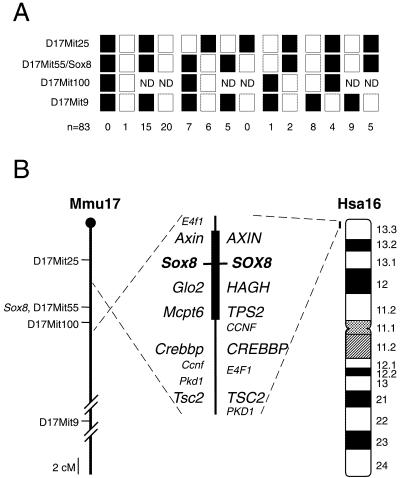
Sox8 was mapped in mice using the EUCIB interspecific backcross, exploiting an 18 bp difference in the first intron between M.musculus and M.spretus. Recombination and haplotype analysis of 83 mice (Fig. 5A) revealed a location on proximal mouse chromosome 17, 5.8 ± 3.2 cM from the anonymous marker D17Mit25 and 29.1 ± 6.3 cM from D17Mit9. Sox8 co-segregated with two other markers, D17Mit55 and D17Mit43, placing it within the t complex (26), a 20 cM region notable for several inversions and deletions and transmission ratio distortion (Fig. 5B). Further confirmation of the location of Sox8 was achieved by physically mapping Sox8 on two yeast artificial chromosomes, 156-B5 and 240-D4, known to span this region of the t complex (UK-HGMP, http://www.menu.hgmp.mrc.ac.uk/ ).
Figure 5.
Chromosomal location of human and mouse Sox8. (A) Recombination analysis of mouse interspecific backcrosses. Filled squares represent the M.musculus Sox8 allele, open squares represent the M.spretus allele, with numbers below representing the number of backcross progeny mice typed for each combination. Sox8 also co-segregated with the anonymous marker locus D17Mit43. ND is used when no data were recorded for D17Mit100. (B) Comparative maps of mouse chromosome 17 (Mmu17) and human chromosome 16 (Hsa16). The positions of conserved syntenic genes are represented along the central axis with Sox8 positioned between Mcpt6 (human TPS2) and Axin. The dashed line represents the orientation on the chromosome. Mouse anonymous marker loci are shown to the left of chromosome Mmu17. Ideogram band labels are positioned to the right of human chromosome 16.
Human SOX8 was mapped by typing 89 DNAs from the Genebridge4 radiation hybrid panel. This analysis revealed that SOX8 is located on chromosome 16, within 5 cM of the telomere of 16p, in band 16p13.3 between the marker loci D16S521 and D16S3024 (Fig. 5B). This is a region of conserved synteny with the t complex region on mouse chromosome 17 where Sox8 was localised, further confirming our mapping results.
DISCUSSION
Despite the number and diversity of mammalian Sox genes, certain structural themes have emerged within each of the eight subfamilies, groups A–H, described to date (1,9). For example, group D Sox genes (Sox5, Sox6, Sox13 and Sox23) all contain leucine zippers (27–30), while the members of group C (Sox4, Sox11, Sox22 and Sox24) encode highly conserved N- and C-terminal amino acid sequences and group G Sox genes (Sox15 and Sox20) encode proteins with a common proline/glutamine/serine-rich region at the C-terminus (1). In the case of group B, comprising Sox1, Sox2, Sox3, Sox14, Sox19 and Sox21, some functional similarity is also apparent since all are involved in neural development (1,31–36).
In this report we describe the third member of Sox group E, Sox8, in mice and humans. This gene shares a common genomic organisation with the two other group E genes Sox9 and Sox10, with the positions of the two introns absolutely conserved. Furthermore, the intron–exon boundaries differ from those in other intron-containing Sox groups (group D, Sox5, Sox6, Sox13 and Sox23; group F, Sox7, Sox17 and Sox18), providing further evidence for a common evolutionary ancestor for group E Sox genes and likely also for each of the Sox subfamilies. It will be of interest to determine whether a gene with similar sequence and genomic organisation exists in simpler organisms that may represent the postulated progenitor of group E Sox genes in vertebrates.
Group E Sox genes are also conserved at the sequence level, encoding proteins with several conserved regions that may be functionally important. Two regions are particularly highly conserved, a 35 amino acid hydrophobic region N-terminal to the HMG box and a 19 amino acid hydrophilic region C-terminal to the HMG box. These regions may provide additional sequence-specific DNA-binding or -bending properties, which in turn may allow specific interaction with other components of the transcriptional machinery. Alternatively, they may be directly involved in protein–protein binding with associated cofactors.
Several SOX proteins are modular transcription factors, with separable DNA-binding and transcriptional trans-activation domains (21–24,36). We have demonstrated that mouse SOX8 binds the SOX consensus motif (A/T)(A/T)CAA(A/T)G and is able to trans-activate transcription in cultured cells. While SOX9 and SOX10 have strong trans-activation domains at their C-termini (21,23,24), the C-terminus of SOX8 activated transcription only weakly. However, we identified a second, more potent trans-activation region in SOX8 (amino acids 176–349) that activates transcription 10-fold above GAL0. The two regions together appear to act synergistically to give a combined level of trans-activation some 30-fold above GAL0. It will be important to determine whether both regions are required in native SOX8 in vivo. McDowall et al. (37) have described a second trans-activation domain in SOX9 consisting entirely of proline, glutamine and alanine residues which contributes to the trans-activation ability of native SOX9. Curiously, this domain is poorly conserved between species and is completely absent in SOX8. Several SOX proteins, such as SOX9 (38), SOX10 (39) and SOX2 (34,40), have been shown to require additional cell-specific cofactors to enhance transcription. If different factors were to associate with the two trans-activation regions of SOX8, then tissue-specific expression of these factors during development would represent a new mechanism of transcriptional control by SOX proteins.
The expression profile of Sox8 during mouse development suggests roles in central nervous system, limb and facial development. Interestingly, expression in the developing gonads was specific to the testis cords, implicating Sox8 in male sex determination or differentiation or male germ cell development. To date two other Sox genes have been implicated in these processes, namely Sry, the mammalian Y-linked testis-determining gene (41–43), and Sox9 (4,5,44,45). During development, Sox8 is co-expressed in some regions with either Sox9 or Sox10, for example in the testis cords, kidneys and dorsal root ganglia. Whether this represents Sox gene complementation or some degree of functional redundancy, as suggested for Sox5 and Sox6 (27) and Sox1, Sox2 and Sox3 (33), is still unclear.
Mouse Sox8 was mapped to chromosome 17, specifically within the t complex. The t complex is a 20 cM region characterised by four non-overlapping inversions which are maintained on a wild-type background by transmission ratio distortion, whereby heterozygous males pass on a copy of the t complex to the next generation at a rate of greater than 90%. The mechanism by which this is achieved is still unclear, but is believed to involve specific sperm–egg donor–acceptor interactions (for a review see 26). Another characteristic of the t complex is the accumulation of several t complex-associated mutants, two of which, tw18 and th20, are located near Sox8 (46). The tw18 mutant has been shown to contain a deletion in the region of Sox8 and a duplication at the distal end of the t complex (26). Homozygous tw18 embryos die prior to organogenesis and develop abnormalities after formation of the primitive streak. Snow and Bennett observed abnormalities in mesoderm cell migration, an apparent block in mitosis and abnormalities in the plane of cell division in tw18 homozygotes (47). The th20 mutant is a recessive T-related deletion encompassing the tufted locus (tf) and the nearby Ki and Hba-ps4 loci (48), showing not only the abnormal hair phenotype of the tf mutant but also reduced fertility (48,49). It will be of interest to determine whether Sox8 is deleted in either of these mutants.
Human SOX8 was mapped near the telomere of chromosome 16p, a region of conserved synteny with mouse proximal chromosome 17. This region is rich in disease loci (Online Inheritance in Man, http://www.ncbi.nlm.nih.gov/Omim/ ), however, the genes responsible for all but two of these have been identified. One disease for which no gene has been identified is CATM, involving microphthalmia and congenital cataract. SOX8 is expressed weakly in the developing eye and remains a candidate gene for this disease. However, it is difficult to envisage mutations in this gene affecting only the eye, given the broad developmental profile of expression revealed by our study. The other disease for which there is no known gene candidate is haemoglobin H-related mental retardation, also known as ATR-16, which is a complex syndrome involving mild α-thalassemia associated with facial malformation and mental retardation (50). ATR-16 is caused by deletion of a portion of the distal short arm of chromosome 16, with four different breakpoints identified (50). The symptoms have been ascribed to the deletion of one to four of the α-globin genes, together with deletion of another unknown gene(s) within this region (50). The expression of Sox8 in the brain and spinal cord of developing mice, along with strong expression in the first and second branchial arches and nasal invaginations, is consistent with an involvement in this disease. However, since this syndrome is always associated with large deletions, it will not be possible to firmly identify SOX8 as the gene responsible for ATR-16 by analysis of patient DNA. Targeted inactivation of Sox8 in mice will be required to reveal the phenotypic consequences of Sox8 mutation.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Josephine Bowles, Jane Olsson, Murray Hargrave and Toshiya Yamada for help with interpretation of data and Jonathon Harris, Shen-Liang Chen and George Muscat for their input into the trans-activation domain analysis. This research was supported by a Special Research Centre grant from the Australian Research Council (ARC). G.S. is supported by an Australian Post-graduate Award. M.B. is supported by the Ministry of Education and Culture, Spain. P.K. is an ARC Australian Senior Research Fellow.
DDBJ/EMBL/GenBank accession nos AF191325, HS349H11
REFERENCES
- 1.Wegner M. (1999) Nucleic Acids Res., 26, 1409–1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Laudet V., Stehelin,D. and Clevers,H. (1993) Nucleic Acids Res., 21, 2493–2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wright E., Hargrave,M.R., Christiansen,J., Cooper,L., Kun,J., Evans,T., Gangadharan,U., Greenfield,A. and Koopman,P. (1995) Nature Genet., 9, 15–20. [DOI] [PubMed] [Google Scholar]
- 4.Foster J.W., Dominguez-Steglich,M.A., Guioli,S., Kwok,C., Weller,P.A., Weissenbach,J., Mansour,S., Young,I.D., Goodfellow,P.N., Brook,J.D. and Schafer,A.J. (1994) Nature, 372, 525–530. [DOI] [PubMed] [Google Scholar]
- 5.Wagner T., Wirth,J., Meyer,J., Zabel,B., Held,M., Zimmer,J., Pasantes,J., Bricarelli,F.D., Keutel,J., Hustert,E., Wolf,U., Tommerup,N., Schempp,W. and Scherer,G. (1994) Cell, 79, 1111–1120. [DOI] [PubMed] [Google Scholar]
- 6.Pingault V., Bondurand,N., Kuhlbrodt,K., Goerich,D.E., Préhu,M.-O., Puliti,A., Herbarth,B., Hermans-Borgmeyer,I., Legius,E., Matthijs,G., Amiel,J., Lyonnet,S., Ceccherini,I., Romeo,G., Clayton Smith,J., Read,A.P., Wegner,M. and Goossens,M. (1998) Nature Genet., 18, 171–173. [DOI] [PubMed] [Google Scholar]
- 7.Southard-Smith E.M., Kos,L. and Pavan,W.J. (1998) Nature Genet., 18, 60–64. [DOI] [PubMed] [Google Scholar]
- 8.Wright E.M., Snopek,B. and Koopman,P. (1993) Nucleic Acids Res., 21, 744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Osaki E., Nishina,Y., Inazawa,J., Copeland,N.G., Gilbert,D., Jenkins,N., Ohsufi,M., Tezuka,T., Yoshida,M. and Semba,K. (1999) Nucleic Acids Res., 27, 2503–2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Soullier S., Jay,P., Poulat,F., Vanacker,J.M., Berta,P. and Laudet,V. (1999) J. Mol. Evol., 48, 517–527. [DOI] [PubMed] [Google Scholar]
- 11.Ito M., Ishikawa,M., Suzuki,S., Takamatsu,N. and Shiba,T. (1995) FEBS Lett., 377, 37–40. [DOI] [PubMed] [Google Scholar]
- 12.Chomczynski P. and Sacchi,N. (1987) Anal. Biochem., 162, 156–159. [DOI] [PubMed] [Google Scholar]
- 13.Hargrave M. and Koopman,P. (1999) In Darby,I. (ed.), Methods in Molecular Biology, Vol. 123, In Situ Hybridization Protocols, 2nd Edn. Humana Press, Totowa, NJ, pp. 279–289.
- 14.Schreiber E., Matthias,P., Muller,M.M. and Schaffner,W. (1989) Nucleic Acids Res., 17, 6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kato G.J., Barrett,J., Villa,G.M. and Dang,C.V. (1990) Mol. Cell. Biol., 10, 5914–5920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Downes M., Carozzi,A.J. and Muscat,G.E. (1995) Mol. Endocrinol., 9, 1666–1678. [DOI] [PubMed] [Google Scholar]
- 17.Denny P., Swift,S., Connor,F. and Ashworth,A. (1992) EMBO J., 11, 3705–3712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harley V.R., Lovell-Badge,R. and Goodfellow,P.N. (1994) Nucleic Acids Res., 22, 1500–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kanai Y., Kanai,A.M., Noce,T., Saido,T.C., Shiroishi,T., Hayashi,Y. and Yazaki,K. (1996) J. Cell Biol., 133, 667–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lefebvre V., Huang,W., Harley,V.R., Goodfellow,P.N. and De Crombrugghe,B. (1997) Mol. Cell. Biol., 17, 2336–2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ng L.-J., Wheatley,S., Muscat,G.E.O., Conway-Campbell,J., Bowles,J., Wright,E., Bell,D.M., Tam,P.P.L., Cheah,K.S.E. and Koopman,P. (1997) Dev. Biol., 183, 108–121. [DOI] [PubMed] [Google Scholar]
- 22.Hosking B.M., Muscat,G.E.O., Koopman,P.A., Dowhan,D.H. and Dunn,T.L. (1995) Nucleic Acids Res., 23, 2626–2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pusch C., Husten,E., Pfeifer,D., Sudbeck,P., Kist,R., Roe,B., Wang,Z., Balling,R., Blin,N. and Scherer,G. (1998) Hum. Genet., 103, 115–123. [DOI] [PubMed] [Google Scholar]
- 24.Südbeck P., Schmitz,M.L., Baeuerle,P.A. and Scherer,G. (1996) Nature Genet., 13, 230–232. [DOI] [PubMed] [Google Scholar]
- 25.Jäger R.J., Anvret,M., Hall,K. and Scherer,G. (1990) Nature, 348, 452–454. [DOI] [PubMed] [Google Scholar]
- 26.Silver L.M. (1985) Annu. Rev. Genet., 19, 179–208. [DOI] [PubMed] [Google Scholar]
- 27.Connor F., Wright,E., Denny,P., Koopman,P. and Ashworth,A. (1995) Nucleic Acids Res., 23, 3365–3372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takamatsu N., Kanda,H., Tsuchiya,I., Yamada,S., Ito,M., Kabeno,S., Shiba,T. and Yamashita,S. (1995) Mol. Cell. Biol., 15, 3759–3766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roose J., Korver,W., Oving,E., Wilson,A., Wagenaar,G., Markman,M., Lamers,W. and Clevers,H. (1998) Nucleic Acids Res., 26, 469–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wunderle V.M., Critcher,R., Ashworth,A. and Goodfellow,P.N. (1996) Genomics, 36, 354–358. [DOI] [PubMed] [Google Scholar]
- 31.Vriz S., Joly,C., Boulekbache,H. and Condamine,H. (1996) Mol. Brain Res., 40, 221–228. [DOI] [PubMed] [Google Scholar]
- 32.Rex M., Uwanogho,D.A., Orme,A., Scotting,P.J. and Sharpe,P.T. (1997) Mech. Dev., 66, 39–53. [DOI] [PubMed] [Google Scholar]
- 33.Collignon J., Sockanathan,S., Hacker,A., Cohen-Tannoudji,M., Norris,D., Rastan,S., Episkopou,V., Stevanovic,M., Goodfellow,P.N. and Lovell-Badge,R. (1996) Development, 122, 509–520. [DOI] [PubMed] [Google Scholar]
- 34.Kamachi Y., Sockanathan,S., Liu,Q., Breitman,M., Lovell-Badge,R. and Kondoh,H. (1995) EMBO J., 14, 3510–3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hargrave M., Karunaratne,A., Cox,L., Wood,S., Koopman,P. and Yamada,T. (2000) Dev. Biol., in press. [DOI] [PubMed] [Google Scholar]
- 36.Uchikawa M., Kamachi,Y. and Kondoh,H. (1999) Mech. Dev., 84, 103–108. [DOI] [PubMed] [Google Scholar]
- 37.McDowall S., Argentaro,A., Ranganathan,S., Weller,P., Mertin,S., Mansour,S., Tolmie,J. and Harley,V. (1999) J. Biol. Chem., 274, 24023–24030. [DOI] [PubMed] [Google Scholar]
- 38.Lefebvre V., Li,P. and de Crombrugghe,B. (1998) EMBO J., 17, 5718–5733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kuhlbrodt K., Herbarth,B., Sock,E., Hermans-Borgmeyer,I. and Wegner,M. (1998) J. Neurosci., 18, 237–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yuan H., Corbi,N., Basilico,C. and Dailey,L. (1995) Genes Dev., 9, 2635–2645. [DOI] [PubMed] [Google Scholar]
- 41.Sinclair A.H., Berta,P., Palmer,M.S., Hawkins,J.R., Griffiths,B.L., Smith,M.J., Foster,J.W., Frischauf,A.-M., Lovell-Badge,R. and Goodfellow,P.N. (1990) Nature, 346, 240–244. [DOI] [PubMed] [Google Scholar]
- 42.Koopman P., Gubbay,J., Vivian,N., Goodfellow,P. and Lovell-Badge,R. (1991) Nature, 351, 117–121. [DOI] [PubMed] [Google Scholar]
- 43.Gubbay J., Collignon,J., Koopman,P., Capel,B., Economou,A., Münsterberg,A., Vivian,N., Goodfellow,P. and Lovell-Badge,R. (1990) Nature, 346, 245–250. [DOI] [PubMed] [Google Scholar]
- 44.Kent J., Wheatley,S.C., Andrews,J.E., Sinclair,A.H. and Koopman,P. (1996) Development, 122, 2813–2822. [DOI] [PubMed] [Google Scholar]
- 45.Morais da Silva S., Hacker,A., Harley,V., Goodfellow,P., Swain,A. and Lovell-Badge,R. (1996) Nature Genet., 14, 62–68. [DOI] [PubMed] [Google Scholar]
- 46.Barkley J., King,T.F., Crossley,P.H., Barnard,R.C., Larin,Z., Lehrach,H. and Little,P.F.R. (1996) Genomics, 36, 39–46. [DOI] [PubMed] [Google Scholar]
- 47.Snow M.H.L. and Bennett,D. (1978) J. Embryol. Exp. Morphol., 47, 39–52. [PubMed] [Google Scholar]
- 48.Fox H.S., Silver,L.M. and Martin,G.R. (1984) Immunogenetics, 19, 125–130. [DOI] [PubMed] [Google Scholar]
- 49.Silver L.M., Cisek,L., Jackson,C. and Lukralle,D. (1985) Genet. Res., 45, 107–112. [DOI] [PubMed] [Google Scholar]
- 50.Wilkie A.O.M., Buckle,V.J., Harris,P.C., Lamb,J., Barton,N.J., Reeders,S.T., Lindenbaum,R.H., Nicholls,R.D., Barrow,M., Bethlenfalvay,N.C., Hutz,M.H., Tolmie,J.L., Weatherall,D.J. and Higgs,D.R. (1990) Am. J. Hum. Genet., 46, 1112–1126. [PMC free article] [PubMed] [Google Scholar]