Abstract
Objectives
The aim of the study was to investigate, by quantitative PCR (qPCR), the presence of papillomavirus in feline viral plaques (VPs), Bowenoid in situ carcinoma (BISC), squamous cell carcinoma (SCC) and actinic keratosis (AK).
Methods
Twenty-nine cases with previously established diagnoses of feline VPs, BISC, invasive SCC and AK were selected from a dermatopathological database. A critical re-evaluation of diagnosis was performed by defining clear criteria toward carcinomatous vs non-carcinomatous, in situ vs invasive (if carcinomatous) and viral vs actinic. Cases were evaluated for p16 immunolocalisation. The presence of the target viral genes for Felis catus papillomavirus (FcaPV)-1, FcaPV-2, FcaPV-3 and FcaPV-4 was determined by qPCR. The data generated ΔΔCq values, which represent a normalised measure of DNA viral quantity. Samples with a positive ΔΔCq value were submitted to sequence analysis.
Results
Four VPs, 19 BISCs, four SCCs and one case of AK were included. By ΔΔCq analysis we found that all VPs were positive for FcaPV-1 or FcaPV-2; eight BISCs were positive for FcaPV-1, FcaPV-2 and FcaPV-4. FcaPV-2 was the most prevalent among the group of VPs and BISCs.
Conclusions and relevance
Using the ΔΔCq method we report the first evidence of FcaPV-1, FcaPV-2 and FcaPV-4 in Italy. FcaPV-2 was the most frequently detected; to a lesser extent, FcaPV-1 and FcaPV-4 were detected in the examined samples. FcaPV-3 was never associated with viral-induced lesions by ΔΔCq investigation. Compared with conventional PCR the ΔΔCq method has the advantage of establishing a possible role of the virus in the outcome of infection.
Introduction
Detection of Felis catus papillomaviruses (FcaPVs) from feline cutaneous lesions has been reported worldwide,1,2 and these viruses are considered to cause several feline skin conditions.1–5 These include a wide spectrum of proliferative skin lesions ranging from non-neoplastic viral plaques (VPs) to preneoplastic Bowenoid in situ carcinoma (BISC) and invasive squamous cell carcinoma (SSC). 6 A progression from non-neoplastic viral-induced lesions to overt neoplasia has also been documented to occur. 7 Four FcaPV types (FcaPV-1–FcaPV-4) have so far been described in cats, each having different anatomical distributions and presentations.1,8–11 The most frequent FcaPV, FcaPV-2 (belonging to the Dyothetapapillomavirus genus) was isolated from a cutaneous pigmented plaque and later related to cutaneous SCC.12–15 FcaPV-2 seems to play a role in a high proportion of nasal planum SCCs and animals with tumours associated with papillomaviral aetiology with p16 upregulation show increased survival compared with those attributable to ultraviolet radiation. 16 The relevance of FcaPV-2 in the pathogenesis of both premalignant and malignant lesions has been recently investigated by concurrently assessing the number of DNA viral copies and viral gene expression.5–7 However, the effective role of FcaPV-2 in the development of malignant lesions is still debated, especially in view of recent results indicating a high FcaPV-2 prevalence in healthy domestic cats.7,17 The role of other feline papillomavirus (PV) types, namely FcaPV-1, FcaPV-3 and FcaPV-4, remains obscure. FcaPV-1 belongs to the Lambdapapillomavirus genus and was identified by Tachezy et al in a hyperkeratotic cutaneous lesion from a Persian cat. 9 In the same year another FcaPV was also identified in a doemstic shorthair cat with papillomatosis. 11 More recently the virus was detected in multiple small, sessile, raised lesions on the ventral surface of the tongue in two 13-year-old domestic cats. 18 FcaPV-3 is a Taupapillomavirus, isolated for the first time from a cutaneous in situ carcinoma. 3 Recently, FcaPV-3 was detected in a BISC with a novel histological feature and a benign clinical behaviour. 19 The same histopathological changes were observed in a feline basal cell carcinoma, from which DNA sequences of a novel PV closely related to FcaPV-3 were detected by the authors. 4 FcaPV-4 belongs to genus Taupapillomavirus and has been detected in the oral cavity following a severe gingivitis. 10 The pathogenicity of FcaPV-1 and FcaPV-4 is still unclear and these viruses are rarely detected in oral inflammatory, as well as neoplastic, lesions. Therefore, an active role of FcaPV-1 and FcaPV-4 in carcinogenesis is still debated.
Based on the literature, it can be seen that most cases of FcaPV infection have been documented in domestic felids from New Zealand, North and South America and, to a lesser extent from Europe. This might be reflective of the active research undertaken in these countries, without any epidemiological meaning. In Europe, PV infections have been reported in Switzerland in nine cases,12,20 in one case in Germany 21 and in one case in Italy. 22 Here we assess, by a combination of histology, immunohistochemistry and quantitative PCR (qPCR), the concurrent presence of FcaPV-1, FcaPV-2, FcaPV-3 and FcaPV-4 viral DNA and p16 immunostaining in 29 feline lesions collected in Italy, including non-neoplastic and preneoplastic, viral and non-viral-induced skin lesions, namely VPs, BISC, invasive SSC and actinic keratosis (AK). Two methods were applied to analyse the viral load data: absolute quantification (AQ) and relative quantification (RQ). The AQ was applied to determine the viral copy number per µg of DNA; however, by this method a wide range of values can be obtained without a cut-off value that would allow us to discriminate the presence of FcaPV as an innocent bystander or as responsible for viral lesions. To address this problem we approached the RQ by ΔΔCq calculation. 23 This method measures the presence of viral genome by relating the obtained values of each of the four FcaPVs in each lesion classified as VPs or BISC to those of SCC and AK.
Materials and methods
Biopsies
Feline skin biopsies have been identified from dermatopathological databases and 29 cases with a diagnosis of VPs, BISC, invasive SCC and AK were selected. The retrieved formalin-fixed cases were routinely processed for histopathology and 4 µm thick paraffin-embedded tissues sections were stained with haematoxylin and eosin. Before inclusion in the study, the selected samples were subjected to critical re-evaluation of diagnosis by defining clear distinctive criteria, such as carcinomatous vs non-carcinomatous, in situ vs invasive (if carcinomatous) and viral vs actinic. The established criteria were defined as described in the scientific literature and are summarised in Table 1.14,20,24 Data from signalment and lesion distribution, when available, were included.
Table 1.
Histological criteria for the diagnosis of viral plaque (VP), Bowenoid in situ carcinoma (BISC), squamous cell carcinoma (SCC) and actinic keratosis (AK)
| Type of lesion | Histopathological criteria |
|---|---|
| VP | Focal epidermal hyperplasia, koilocytes, clear cells, giant keratohyalin granules, cytoplasm enlarged by blue–greyish fibrillar material, hyperpigmentation |
| BISC | Epidermal and follicular dysplasia with upward in situ keratinocyte proliferation, loss of nuclear polarity, windblown nuclei, koilocytes, mitosis |
| SCC | Epidermal dysplasia and atypia with downward keratinocyte proliferation, keratinocyte invasion to the dermis through the basal lamina, mitosis |
| AK | Epidermal dysplasia and atypia with upward in situ keratinocyte proliferation, squamatisation, apoptosis |
P16 immunohistochemistry
Cases were evaluated for the detection of p16 by immunohistochemistry employing an ABC system (Vector Laboratories). Antigen unmasking was carried out at 120°C for 3 mins in a pressure cooker with a Tris-EDTA buffer (1.2 g/l Tris, 0.36 g/l EDTA, pH 9.0). Sections were pretreated for 10 mins with 1% H2O2 in 0.1 M phosphate-buffered saline (PBS; pH 7.4) to quench endogenous peroxidase activity, and blocked for 30 mins at room temperature (RT) in PBS with 2% normal horse serum (PK-7200; Vector Laboratories) and 0.05% TritonX-100. Sections were then incubated overnight at 4ºC with a 1:100 diluted monoclonal primary antibody p16-INK4a (mouse monoclonal IgG; BiorByt). After washing in PBS, an incubation with a secondary universal biotinylated anti-mouse/rabbit antibody (PK-7200; Vector Laboratories) was performed for 1 h at RT. Staining was visualised with a diaminobenzidine (SK-4105; Vector Laboratories) solution under a light microscope (Eclipse 80i; Nikon). The epidermis surrounding VP and BISC was used as an internal negative control; specificity of anti-human p16 for feline tissues has been previously documented.16,25
Sampling material for qPCR
Three 10 µm thick sections from the selected samples were cut with a microtome, placed onto slides and left unfixed. These slides were observed under an optic microscope. Tissue that was not relevant for the study was scraped off and the remaining tissue directly collected in a DNase-free 1.5 ml tube. To prevent carryover of contaminating DNA the microtome overlay was covered with a new piece of adhesive tape and a new blade was used for each sample.
DNA extraction
DNA extraction was performed using a DNeasy Blood and Tissue kit (Qiagen) following the manufacturer’s instruction and applying preliminary removal of paraffin by extraction with xylene. DNA was eluted in 30 µl and each sample concentration was quantified using a Qubit spectrophotometer (ThermoFisher Scientific). DNA samples were stored at −20°C until analysis.
qPCR
The extracted DNAs were amplified using four specific sets of primers amplifying a portion of the L1 gene of the four types of FcaPV so far identified in the feline species (Table 2). To normalise the amount of DNA used for each sample to achieve a correct quantification of viral copy number, qPCR under the same conditions as for FcaPV but with a specific set of primers was run in parallel for the reference gene albumin (ALB). 26 The number of copies of the target viral gene, measured as Cq value, generates a ΔCq value when compared with the corresponding Cq value of the reference gene. Moreover, a ΔΔCq value was calculated comparing the ΔCq value of the sample of interest to ΔCq values obtained from the SCC and AK group, considered as the negative control group. The ΔΔCq value represents a normalised measure of DNA viral quantity and was calculated using REST software. 27 In this work, RQ using the 2−ΔΔCq method was adapted to estimate in each sample the fold change of FcaPV-1, FcaPV-2, FcaPV-3 and FcaPV-4 target viral gene copies relative to the albumin reference gene. 23
Table 2.
Primers nucleotide sequences
| Oligonucleotide name | Nucleotide sequence (5′-3′) | 5′ Nucleotide position | Reference sequence accession number |
|---|---|---|---|
| FcaPV1-F | AGGATGGTGACATGGTGGAT | 7143 | AF480454 |
| FcaPV1-R | TTTGCACTGTGTGTCTGCAA | 7246 | |
| FcaPV2-F | TACACGCGGTACCAATTTCA | 7191 | EU796884 |
| FcaPV2-R | AGAGTGACCACGCACACTTG | 7331 | |
| FcaPV3-F | AAGATTGGTATGGCGTTTGC | 5960 | JX972168 |
| FcaPV3-R | TTTGCCTTTCATCTGCTGTG | 6105 | |
| FcaPV4-F | ATGCAAATGGCCAGACTTTC | 6356 | KF147892 |
| FcaPV4-R | AAAAATGGCGGCAGTACAAC | 6453 | |
| Fel Alb F | GATGGCTGATTGCTGTGAGA | 3706 | NC_018726.2 |
| Fel Alb-R | CCCAGGAACCTCTGTTCATT | 3855 | GPC_000001738 |
FcaPV = Felis catus papillomavirus; Fel Alb = feline albumin; F = forward; R = reverse
Melting-curve analysis was performed in conjunction with each four specific FcaPV amplification protocols to determine if non-specific products were amplified during reaction. The specificity of the melting curve was compared with melting curves values obtained from a plasmid (pFcaPV) containing an amplicon of the four FcaPVs spanning the real-time products.
The plasmid was generated by inserting the four FcaPV gene segments into a pMA-T vector using GeneArt technology (ThermoFisher Scientific). Serial dilutions of pFcaPV plasmid ranging from 106–102 copies/5 µl were used to calculate the efficiency of qPCR for each set of FcaPV primers and compared with the efficiency calculated on five points of a two-fold serial dilution of a quantified DNA template for ALB. The efficiency of each qPCR assay was similar, ranging from 91–98% (average 94.6%). The lesions identified by histological classification as SCC/AK were presumed to serve as the negative control group and each sample belonging to this group was also individually tested by ΔΔCq analysis for each FcaPV type. Samples that had eventually scored a positive 2−ΔΔCq value were excluded. In addition, the viral copy number/µl of input DNA for each FcaPV type in each sample tested was calculated (AQ). The assays were performed in Rotorgene thermocycler (Corbett Research) using SSCO SYBR Green master mix (Bio-Rad) and 5 µl extracted DNA. All samples were tested in duplicate and the results were calculated using the mean Cq values. Samples positive for only one replica were considered as negative. All samples with a positive 2−ΔΔCq value were submitted to sequence analysis (BMR Genomics).
Results
Histologically, all VPs were recognised as areas of focal epidermal dysplasia with koilocytes and keratinocytes with cytoplasm enlarged by blue–greyish material. Koilocytes were recognised for their swollen cytoplasms and shrunken nuclei. 20 Regarding BISC diagnosis, the presence of koilocytes or koilocyte-like cells pointed to a viral origin in 17/20 cases, whereas in the remaining three cases diagnosis was formulated by other morphological details (Table 1). SCC and AK showed the morphological alterations listed in Table 1. Among the 20 BISCs cases, five VPs were also detected in the adjacent skin by histology and in the other five sites penetration of keratinocytes through the basement membrane into the dermis were observed, indicative of progression towards SCC.
Clinical reports for each of the 29 cases fulfilling the established histological criteria were recorded. Breed, age, sex and anatomical distribution are summarised in Table 3. Twenty-five subjects were shorthair domestic cats, two were Persian and two were Maine Coons. Mean age at presentation was 6–20 years (median 11.6 years); 14/29 were female and 15 were male. A total of four VPs, 20 BISCs, four SCCs and one AK were detected.
Table 3.
Signalment, lesion distribution, p16 immunolocalisation and Felis catus papillomavirus (FcaPV) type assessed by ΔΔCq analysis in cats with diagnoses of viral plaque (VP), Bowenoid in situ carcinoma (BISC), squamous cell carcinoma (SCC) and actinic keratosis (AK)
| Case number |
Diagnosis | Breed | Age (years) | Sex | Lesion distribution |
p16 | FcaPV type |
2−ΔΔCq* |
|---|---|---|---|---|---|---|---|---|
| 1 | VP | DSH | 10 | M | Flank | + | 2 | 119 |
| 2 | VP | Persian | 15 | F | Groin | + | 2 | 53 |
| 3 | VP | DSH | 9 | M | Nose | + | 1 | 72 |
| 4 | VP | Maine Coon | 10 | F | Nose | + | 2 | 271 |
| 5 | BISC | DSH | 10 | F | Flank | + | − | |
| 6 | BISC | Maine Coon | 6 | M | Shoulder | + | 4 | 5836 |
| 7 | BISC | DSH | 13 | M | Nose | + | − | |
| 8 | BISC | DSH | 13 | F | Nose | + | 1 | 49 |
| 9 | BISC | DSH | 9 | F | Temporal | + | − | |
| 10 | BISC | DSH | 20 | F | Dorsal | + | ND | |
| 11 | BISC | DSH | 15 | M | Temporal | + | 2–4 | 224–331 |
| 12 | BISC | DSH | 15 | M | Nose | + | − | |
| 13 | BISC | DSH | 16 | F | Shoulder | + | − | |
| 14 | BISC | DSH | 12 | M | Ear | + | − | |
| 15 | BISC | DSH | 12 | M | Dorsum | + | 2 | 172 |
| 16 | BISC | DSH | 8 | F | Temporal | + | 1–2 | 58–167 |
| 17 | BISC | DSH | 10 | F | Temporal | + | − | |
| 18 | BISC | Persian | 15 | M | Eyelid | + | 2 | 211 |
| 19 | BISC | DSH | 10 | F | Ear | + | − | |
| 20 | BISC | DSH | 7 | F | Flank | + | − | |
| 21 | BISC | DSH | 10 | M | Nose | + | 2 | 159 |
| 22 | BISC | DSH | 15 | M | Shoulder | + | − | |
| 23 | BISC | DSH | 12 | M | Nose | + | 1–2 | 71–326 |
| 24 | BISC | DSH | 10 | M | Flank | + | − | |
| 25 | SCC | DSH | 9 | F | Ear | +/− | − | |
| 26 | SCC | DSH | 8 | M | Nose | +/− | − | |
| 27 | SCC | DSH | 15 | F | Ear | +/− | − | |
| 28 | SCC | DSH | 10 | M | Eyelid | +/− | − | |
| 29 | AK | DSH | 11 | F | Ear | +/− | − |
Indicates the fold change of FcaPV viral genomes normalised to albumin (ALB) compared with the negative control group (SCC and AK)
DSH = domestic shorthair; M = male; F = female; ND = not determined
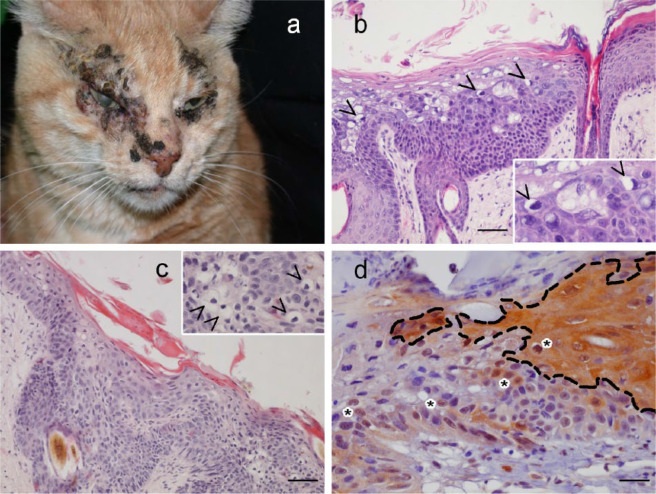
Lesions were observed in densely haired skin regions in nine cats (dorsum, flank, and shoulder), in areas not exposed to sunlight in one cat (groin) and in hypotrichotic and solar-exposed skin in 18 cats (ears, nose, eyelids, temporal region). All cases of SCC were observed in sun-exposed areas. Clinically, VPs were a few millimetres wide, alone or grouped, slightly raised, pigmented and non-pruritic lesions; BISCs were clinically larger than VPs, usually more than 1 cm in diameter, multifocally coalescing, raised and often verrucous, crusted and hyperpigmented. In SCC, erosive and ulcerative crusted lesions were clinically detected. In the only case in which AK was diagnosed, erythema, scales and crust were seen (Figure 1a–c).
Figure 1.
(a) Multiple symmetrical, raised, hyperpigmented and crusted plaques on the face of a cat with Bowenoid in situ carcinoma (BISC). (b) Histopathology from a viral plaque in a cat, showing focal epidermal hyperplasia, absence of follicular wall involvement, and evident viral cytopathic effects (koilocytes are indicated by arrowheads and better viewed in the inset) (bar = 50 µm). (c) Histopathology from a BISC in a cat, showing epidermal and follicular dysplasia with upward in situ keratinocyte proliferation and a few koilocytes in the bottom right of the image (better viewed and indicated by arrowheads in the inset) (bar = 50 µm). (d) p16 immunohistochemistry; groups of keratinocytes (encircled by a dotted line) show cytoplasmic positivity, whereas positive nuclei are indicated by asterisks (ABC system, bar = 100 µm)
P16 was immunolocalised in the epidermis of all VPs and in the epidermis and follicular wall of the BISCs. No signal or only faint staining was detected in the SCC and AK lesions. Immunoreactivity was either nuclear and cytoplasmic; in VPs and BISCs the signal was strong, whereas in the SCCs immunoreactivity was faint and therefore considered as non-significant (Figure 1d).
By qPCR we were able to detect the reference gene ALB in 28/29 DNA samples. Only one sample classified as BISC was not suitable for molecular analysis and therefore it was excluded from further analysis.
By AQ, the presence of FcaPV was detected in low copy numbers in almost all cases (Table 4). By RQ (ΔΔCq method) a reduced number of samples was linked to viral lesions. In details, positive FcaPV 2−ΔΔCq values were detected in all VPs (4/4) and in 8/19 BISCs (Table 3). VPs were positive for FcaPV-2 in three samples and for FcaPV-1 in one sample. BISCs were positive for FcaPV-2 in three samples, and for FcaPV-4 and FcaPV-1 in one sample each. Interestingly, as not reported previously, in BISC lesions we found three cases of double infection. In two of these cases, FcaPV-1 and FcaPV-2 were detected simultaneously and in one case FcaPV-2 was present along with FcaPV-4 (Table 3).
Table 4.
Absolute viral load quantification expressed as viral copy number/µg of input DNA
| Case number | Type of lesion | FcaPV-1 | FcaPV-2 | FcaPV-3 | FcaPV-4 |
|---|---|---|---|---|---|
| 1 | VP | 10 | 153* | 10 | 20 |
| 2 | VP | 100* | 6 | ||
| 3 | VP | 167* | 33 | 23 | 28 |
| 4 | VP | 1130* | |||
| 5 | BISC | 30 | 48 | ||
| 6 | BISC | 18 | 55 | 26 | 3700* |
| 7 | BISC | 13 | 2 | 4 | |
| 8 | BISC | 201* | |||
| 9 | BISC | 42 | 12 | ||
| 10 | BISC | 15 | |||
| 11 | BISC | 87 | 1150* | 59 | 391* |
| 12 | BISC | 10 | |||
| 13 | BISC | 33 | 55 | 28 | 120 |
| 14 | BISC | ||||
| 15 | BISC | 14 | 436* | ||
| 16 | BISC | 176* | 210* | 13 | |
| 17 | BISC | 18 | |||
| 18 | BISC | 1120* | 79 | 45 | |
| 19 | BISC | 3 | |||
| 20 | BISC | 33 | 10 | ||
| 21 | BISC | 6 | 193* | ||
| 22 | BISC | ||||
| 23 | BISC | 310* | 1140* | 18 | 26 |
| 24 | BISC | ||||
| 25 | SSC | 6 | 2 | ||
| 26 | SSC | 12 | 10 | 17 | |
| 27 | SSC | 20 | 15 | 7 | 23 |
| 28 | SSC | 10 | 5 | 14 | 14 |
| 29 | AK | 12 | 18 | ||
| Mean viral load | 63 | 247.7 | 2.4 | 366.2 | |
| Mean ± SD viral load of ΔΔCq-positive samples | 213.5 ± 65.9 | 625.8 ± 491.7 | 2045.5 ± 2339.8 | ||
Positive ΔΔCq samples
FcaPV = Felis catus papillomavirus; VP = viral plaque; BISC = Bowenoid in situ carcinoma; SCC = squamous cell carcinoma; AK = actinic keratosis
In all cases with a positive 2−ΔΔCq value, the viral load was always >102 copies/µg DNA (FcaVP-1: range 167–310, SE 33.0; FcaPV-2: range 100–1150, SE 163.9; FcaPV-4: range 391–3700, SE 1654.5) (Table 4). Sequence analysis confirmed the FcaPV types as indicated by qPCR ΔΔCq analysis.
Discussion
In this retrospective study, we detected the concurrent presence of different types of PV, namely FcaPV-1, FcaPV-2 and FcaPV-4 in feline VPs and BISCs by the ΔΔCq method using specific primers. To the best of our knowledge only the concurrent presence of FcaPV-2 and FcaPV-3 has been reported from one BISC and one VP.3,22 However, differently from previous reports, we report here the presence of FcaPV-1 and FcaPV-4, alone or in association with FcaPV-2.
A limitation when performing PCR-based studies on PV infection is that the viral DNA is often detected and cannot be directly linked to an effective role of virus in pathogenesis and therefore it is not possible to fully establish whether the presence of a viral genome is uneventful (subclinical lesion). In particular, FcaPV-2 has recently been detected in a number of skin swabs from healthy cats, making it difficult to discern whether PVs cause cancer or are merely ‘innocent bystanders’. 7 The hypothesis that the presence of high viral loads likely represents an infectious state of PV is supported by Thomson et al, 7 who recently showed that the finding of high copy numbers of FcaPV-2 DNA within a lesion suggests that the detected virus may be responsible for the lesion. In fact, while high viral copy numbers were associated with E6/E7 gene expression, no gene expression was detected in association with low copy numbers, an indication of an incidental finding.
For the above-mentioned reasons, in this study we used a qPCR protocol applying a ΔΔCq method to investigate if and which FcaPVs induced lesions in cats. In this sense the results obtained are partially in line with the scientific literature, which so far indicates that FcaPV-2 is the major PV type implicated in skin preneoplastic and neoplastic lesions in cats. 7 As the genome organisation and the role of viral proteins within the replication cycle are considered similar even between different genera belonging to the Papillomaviridae family we assumed that a similar link is maintained for feline PVs other than FcaPV-2. Importantly, we reported positive 2−ΔΔCq values for FcaPV-1 and FcaPV-4 – two FcaPV types rarely reported.9–11,18
A comparison between our data and those obtained from other studies is difficult for two reasons: (1) the type of PCR primers used (specific vs consensus); (2) type of PCR used (conventional vs quantitative). A general prevalence ranging from 24–100% of FcaPV-2 in BISCs and VPs has been reported. 7,8,28,29 In our study, by using specific primers in a qPCR analysis applying the RQ method, we obtained a prevalence for FcaPV-2 in BISCs of 31.6% (n = 6/19) and for all FcaPV types of 42.1% (n = 8/19).
Failure to demonstrate PV DNA in every BISC has been previously explained via a carcinogenesis model in which PVs cause transformation; however, they are only present within the lesion for a short period. 30 Surprisingly, cases in which koilocytes were detected did not show any positive 2−ΔΔCq value; this result is likely due to: (1) the detection of koilocytes in very focal lesions that may not have been sampled when additional serial paraffin sections were prepared for DNA extraction and PCR analysis; (2) the presence of a PV variant undetectable by PCR.
Low viral copies for all PV types have been detected in almost all cases; however, the ΔΔCq analysis was necessary to identify the FcaPVs as the biological agent potentially causing the lesions. Positive 2−ΔΔCq values for the different PV types were detected in all VPs and in 42% of BISCs. It is noteworthy that the mean 2−ΔΔCq of positive samples differed among PV types: 62.5 for FcaPV-1, 189.1 FcaPV-2 and 3083.5 for FcaPV-4. Studies considering a larger panel of cases might be necessary to establish a further ΔΔCq cut-off value.
Here we document the presence of FcaPV-2 with a high prevalence in both VPs and BISCs; FcaPV-1 and FcaPV-4 were rescued less frequently. These last two PVs are rarely described as playing an active role in skin and oral mucosa lesions in cats.9,10,18,31 None of the lesions was associated with FcaPV-3 when adopting the ΔΔCq method, confirming the rare occurrence of this FcaPV type in Italy.
Either cytoplasmic or nuclear p16 signals were detected in all cases where cytopathic effects were found (VPs, BISC), whereas p16 immunolocalisation was present with faint and mainly cytoplasmic signal in cases of SCCs. No p16-immunoreactivity was found in the case of AK. Despite the difficulties of interpretation when the signal was faint, these results are in line with what has already been documented. About half of the SCCs studied by Thomson et al 7 were negative for p16 immunostaining. A strong association between FcaPV-2 E6/E7 gene expression and p16 immunostaining has recently been found in feline SCC, with 18/20 (90%) E6/E7-positive SCCs being also positive for p16 vs 13/40 (33%) E6/E7-negative SCCs. 7 E6/7 gene expression investigation has not been carried out in our study, but our results might reflect this correlation.
Conclusions
ΔΔCq analysis has proved to be necessary to identify FcaPVs as biological agents potentially causing lesions. Based on this method the presence of FcaPV-2 is confirmed to be the most representative FcaPV in feline skin lesions referable for diagnosis of VPs and BISC in Italy. FcaPV-1 and FcaPV-4 were detected to a lesser extent in our examined samples.
Acknowledgments
The authors thank all colleagues who referred the cases.
Footnotes
Accepted: 15 August 2017
The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: This study was financially supported by Fondi di Ateneo University of Pisa.
References
- 1. Sundberg JP, Van Ranst M, Montali R, et al. Feline papillomas and papillomaviruses. Vet Pathol 2000; 37: 1–10. [DOI] [PubMed] [Google Scholar]
- 2. Gil da, Costa RM, Peleteiro MC, Pires MA, et al. An update on canine, feline and bovine papillomaviruses. Transbound Emerg Dis 2016; 64: 1371–1379. [DOI] [PubMed] [Google Scholar]
- 3. Munday JS, Dunowska M, Hills SF, et al. Genomic characterization of Felis catus papillomavirus-3: a novel papillomavirus detected in a feline Bowenoid in situ carcinoma. Vet Microbiol 2013; 165: 319–325. [DOI] [PubMed] [Google Scholar]
- 4. Munday JS, French A, Thomson N. Detection of DNA sequences from a novel papillomavirus in a feline basal cell carcinoma. Vet Dermatol 2017; 28: 236–e60. [DOI] [PubMed] [Google Scholar]
- 5. Munday JS, Gwyther S, Thomson NA, et al. Bilateral pre-auricular papillary squamous cell carcinomas associated with papillomavirus infection in a domestic cat. Vet Dermatol 2017; 28: 232–e58. [DOI] [PubMed] [Google Scholar]
- 6. Scott DW, Miller WH, Griffin CE, et al. Muller & Kirk’s small animal dermatology. St Louis, MO: Saunders Elsevier, 2016. [Google Scholar]
- 7. Thomson NA, Munday JS, Dittmer KE. Frequent detection of transcriptionally active Felis catus papillomavirus 2 in feline cutaneous squamous cell carcinomas. J Gen Virol 2016; 97: 1189–1197. [DOI] [PubMed] [Google Scholar]
- 8. Munday JS, Kiupel M, French AF, et al. Detection of papillomaviral sequences in feline Bowenoid in situ carcinoma using consensus primers. Vet Dermatol 2007; 18: 241–245. [DOI] [PubMed] [Google Scholar]
- 9. Tachezy R, Duson G, Rector A, et al. Cloning and genomic characterization of Felis domesticus papillomavirus type 1. Virology 2002; 301: 313–321. [DOI] [PubMed] [Google Scholar]
- 10. Dunowska M, Munday JS, Laurie RE, et al. Genomic characterisation of Felis catus papillomavirus 4, a novel papillomavirus detected in the oral cavity of a domestic cat. Virus Genes 2014; 48: 111–119. [DOI] [PubMed] [Google Scholar]
- 11. Terai M, Burk RD. Felis domesticus papillomavirus, isolated from a skin lesion, is related to canine oral papillomavirus and contains a 1.3 kb non-coding region between the E2 and L2 open reading frames. J Gen Virol 2002; 83: 2303–2307. [DOI] [PubMed] [Google Scholar]
- 12. Lange CE, Tobler K, Markau T, et al. Sequence and classification of FdPV2, a papillomavirus isolated from feline Bowenoid in situ carcinomas. Vet Microbiol 2009; 137: 60–65. [DOI] [PubMed] [Google Scholar]
- 13. Ravens PA, Vogelnest LJ, Tong LJ, et al. Papillomavirus-associated multicentric squamous cell carcinoma in situ in a cat: an unusually extensive and progressive case with subsequent metastasis. Vet Dermatol 2013; 24: 642–e162. [DOI] [PubMed] [Google Scholar]
- 14. Munday JS. Papillomaviruses in felids. Vet J 2014; 199: 340–347. [DOI] [PubMed] [Google Scholar]
- 15. Altamura G, Corteggio A, Pacini L, et al. Transforming properties of Felis catus papillomavirus type 2 E6 and E7 putative oncogenes in vitro and their transcriptional activity in feline squamous cell carcinoma in vivo. Virology 2016; 496: 1–8. [DOI] [PubMed] [Google Scholar]
- 16. Munday JS, Gibson I, French AF. Papillomaviral DNA and increased p16CDKN2A protein are frequently present within feline cutaneous squamous cell carcinomas in ultraviolet-protected skin. Vet Dermatol 2011; 22: 360–366. [DOI] [PubMed] [Google Scholar]
- 17. Geisseler M, Lange CE, Favrot C, et al. Geno- and seroprevalence of Felis domesticus papillomavirus type 2 (FdPV2) in dermatologically healthy cats. BMC Vet Res 2016; 12: 147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Munday JS, Fairley RA, Mills H, et al. Oral papillomas associated with Felis catus papillomavirus type 1 in 2 domestic cats. Vet Pathol 2015; 52: 1187–1190. [DOI] [PubMed] [Google Scholar]
- 19. Munday JS, Fairley R, Atkinson K. The detection of Felis catus papillomavirus 3 DNA in a feline Bowenoid in situ carcinoma with novel histologic features and benign clinical behavior. J Vet Diagn Investig 2016; 28: 612–615. [DOI] [PubMed] [Google Scholar]
- 20. Wilhelm S, Degorce-Rubiales F, Godson D, et al. Clinical, histological and immunohistochemical study of feline viral plaques and Bowenoid in situ carcinomas. Vet Dermatol 2006; 17: 424–431. [DOI] [PubMed] [Google Scholar]
- 21. Egberink HF, Berrocal A, Bax HA, et al. Papillomavirus associated skin lesions in a cat seropositive for feline immunodeficiency virus. Vet Microbiol 1992; 31: 117–125. [DOI] [PubMed] [Google Scholar]
- 22. Alberti A, Tore G, Scagliarini A, et al. Regressing multiple viral plaques and skin fragility syndrome in a cat coinfected with Fcapv2 and FcaPV3. Case Reports Vet Med 2015; 2015: 1–5. [Google Scholar]
- 23. Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative CT method. Nat Protoc 2008; 3: 1101–1108. [DOI] [PubMed] [Google Scholar]
- 24. Levison DA, Parratt D. Skin diseases of the dog and cat. Oxford: Blackwell Science, 2005. [Google Scholar]
- 25. Carter RT, Giudice C, Dubielzig RR, et al. Telomerase activity with concurrent loss of cell cycle regulation in feline post-traumatic ocular sarcomas. J Comp Pathol 2005; 133: 235–245. [DOI] [PubMed] [Google Scholar]
- 26. Helfer-Hungerbuehler AK, Widmer S, Hofmann-Lehmann R. GAPDH pseudogenes and the quantification of feline genomic DNA equivalents. Mol Biol Int 2013; 2013: 587680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pfaffl MW, Horgan GW, Dempfle L. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res 2002; 30: e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nespeca G, Grest P, Rosenkrantz WS, et al. Detection of novel papillomavirus-like sequences in paraffin-embedded specimens of invasive and in situ squamous cell carcinomas from cats. Am J Vet Res 2006; 67: 2036–2041. [DOI] [PubMed] [Google Scholar]
- 29. Munday JS, Peters-Kennedy J. Consistent detection of Felis domesticus papillomavirus 2 DNA sequences within feline viral plaques. J Vet Diagn Invest 2010; 22: 946–949. [DOI] [PubMed] [Google Scholar]
- 30. Smith KT, Campq MS. ‘Hit and run’ transformation of mouse C127 cells by bovine papillomavirus type 4: the viral DNA is required for the initiation but not for maintenance of the transformed phenotype. Virology 1988; 164: 39–47. [DOI] [PubMed] [Google Scholar]
- 31. Munday JS, French AF, Gibson IR, et al. The presence of p16 CDKN2A protein immunostaining within feline nasal planum squamous cell carcinomas is associated with an increased survival time and the presence of papillomaviral DNA. Vet Pathol 2013; 50: 269–273. [DOI] [PubMed] [Google Scholar]