Abstract
The secondary and tertiary structures of a mRNA are known to effect hybridization efficiency and potency of antisense oligonucleotides in vitro. Additional factors including oligonucleotide stability and cellular uptake are also thought to contribute to antisense potency in vivo. Each of these factors can be affected by the sequence of the oligonucleotide. Although mRNA structure is presumed to be a critical determinant of antisense activity in cells, to date little direct experimental evidence has addressed the significance of structure. In order to determine the importance of mRNA structure on antisense activity, oligonucleotide target sites were cloned into a luciferase reporter gene along with adjoining sequence to form known structures. This allowed us to study the effect of target secondary structure on oligonucleotide binding in the cellular environment without changing the sequence of the oligonucleotide. Our results show that structure does play a significant role in determining oligonucleotide efficacy in vivo. We also show that potency of oligonucleotides can be improved by altering chemistry to increase affinity for the mRNA target even in a region that is highly structured.
INTRODUCTION
As the name implies, antisense oligonucleotides must hybridize to their target mRNA to specifically degrade the mRNA, usually via a RNase H-dependent mechanism (1). Thus, for an antisense oligonucleotide to be effective, the complementary target sequence must be available for hybridization. This is not always the case as the RNA target is not a single-stranded random coil but contains secondary and tertiary structures that have been shown to affect the affinity and rate of oligonucleotide hybridization (2–6). The antisense oligonucleotide may also need to compete with proteins that bind to the same site on the message (7).
Several factors are thought to influence antisense activity in cell culture and in vivo. These include chemical stability of the oligonucleotide (8), secondary structure of the oligonucleotide (9), oligonucleotide delivery and bioavailablity (10) and the proximity of the binding site to a functional site on the RNA such as the CAP or translational start site (11). The potency of an antisense oligonucleotide may also depend on the type of cell being targeted (12–14).
Identification of potent antisense sequences has often been based upon empirical approaches to oligonucleotide selection because the optimal target site on the mRNA cannot yet be predicted. Many investigators employ oligonucleotide ‘walks’, spacing oligonucleotides of a given length at intervals along the RNA and choosing the one with the most activity (15–21). It is generally assumed that active oligonucleotides are hybridizing to sequences that are available due to lack of secondary structure at the target site, however, to date little direct information has been gathered on the effect of RNA secondary structure on hybridization of antisense oligonucleotides in cells. While oligonucleotides have been targeted to known structures with varying degrees of success (7,22–24), in all of these cases activity was optimized by testing numerous oligonucleotides shifted either 5′ or 3′ of the initial site. These experiments change not only the target site on the mRNA, but also the sequence and base composition of oligonucleotides. Any or all of these changes might affect oligonucleotide potency.
In this study, we changed the structure surrounding particular oligonucleotide target sites so that the same oligonucleotide could be used to evaluate binding to target sites in the context of varying degrees of structure. This strategy abolished any effects that might be caused by changing the proximity of the oligonucleotide binding site to functional sites on the message. More importantly, any effects due to sequence and base composition of the oligonucleotide were eliminated, permitting direct evaluation of the contribution of RNA target structure to antisense potency. Results suggest that structure in the target mRNA does indeed have a significant and predictable effect on antisense activity.
MATERIALS AND METHODS
Oligonucleotide synthesis
Synthesis and purification of unmodified deoxyphosphorothioate or chimeric deoxyphosphorothioate/2′-O-methoxyethyl base oligonucleotides was performed using an Applied Biosystems 380B automated DNA synthesizer as previously described (25). Sequences of oligonucleotides and placement of 2′-O-methoxyethyl modifications are detailed in Table 1.
Table 1. Oligonucleotides used in the study.
P=S, unmodified deoxyphosphorothioate; P=S/MOE, chimeric deoxyphosphorothioate/2′-O-methoxyethyl base. 2′-O-methoxyethyl bases are bold in the oligonucleotide sequence. The amount of oligonucleotide bound to complementary target RNA at the concentrations listed as determined by gel shift assay is given in the last two columns.
Construction of luciferase expression clones
DNA sequences encoding various oligonucleotide target sites and surrounding RNA structure were cloned into the luciferase expression vector pGL3-Control (Promega). For 5′-untranslated region (UTR) insertions, unique HindIII and NcoI sites in the vector were employed. Cleavage with the two enzymes releases a small portion of the 5′-UTR without affecting the promoter or the luciferase coding region. Inserts were prepared by annealing cDNA oligonucleotides containing the target sequences for known active antisense oligonucleotides and additional sequence necessary to form various RNA secondary structures. For cloning of 5132-S20 the following oligonucleotides were annealed at a concentration of 2 µg/µl by slow cooling from 95°C in 1× ligase buffer (30 mM Tris–HCl pH 7.8, 10 mM MgCl2): AGCTTGGCATTCCGGTGTAATGCATGTCACA- GGCGGGATTCGTCCCGCCTGTGACATGCATTTGTTG-GTAAAGAATTC and CATGGAATTCTTTACCAACAA-ATGCATGTCACAGGCGGGACGAATCCCGCCTGTGAC-ATGCATTACACCGGAATGCCA. This produces a double-stranded DNA fragment with a HindIII-compatible sticky 5′-end and an NcoI-compatible overhang on the 3′-end. An EcoRI site was also included near the 3′-end to allow analysis following cloning. The 5132 oligonucleotide target site is in bold. The insert was then diluted to 25 ng/µl and ligated into the pGL-3 vector prepared as described above. Transformants were checked for the proper insert by digesting with XbaI and EcoRI, which release a 1600 bp digestion product unique to plasmids with inserts. The following oligonucleotide pairs were used to construct the remaining clones. 5132-S0, AGCTTGGCATTCCGGTACAATGCATGTCACAGGCG- GGAGAATTC and CATGGAATTCTCCCGCCTGTGACA-TGAATTGTACCGGAATGCCA; 5132-S20-10-3, AGCTT-CGGAGGACATGCATTGAACGTCGATTTCGATCGACG-TTCAATGCATGTCACAGGCGGGACATGTTGGTAA-AGAATTC and CATGGAATTCTTTACCTTCATGTCCCG-CCTGTGACATGCATTGAACGTCGATCGAAATCGACG-TTCAATGCATGTCCTCCGA; 5132-S20-10-5, AGCTTGG-CATTCCGGTACAATGCATGTCACAGGCGGGAATCG-ACGTTCTTCGGAACGTCGATTCCCGCCTGTGAATTC and CATGGAATTCACAGGCGGGAATCGACGTTCCGAA-GAACGTCGATTCCCGCCTGTGACATGCATTGTACCGGAATGCCA; 5132-S10-14, AGCTTGGCATTCCGGT-ACAATGCATGTCACAGGCGGGATTCGGACATGCA-TTGTACCGTAAAGAATTC and CATGGAATTCTTTAC-GGTACAATGCATGTCCGAATCCCGCCTGTGACATGC-ATTGTACCGGAATGCCA; 5132-S10-4, AGCTTGGCAT-TCCGGTACAATGCATGTCACAGGCGGGATTCGTCC-CGCCTGTTGGTAAAGAATTC and CATGGAATTCTTT-ACCAACAGGCGGGACGAATCCCGCCTGTGACATGC-ATTGTACCGGAATGCCA; 2302-S20, AGCTTGAAAAG-TTCGTACTGACGGATGCCAGCTTGGGCTTCGGCCC-AAGCTGGCATCCGTCATGTTGGTAAAGAATTC and CATGGAATTCTTTACCAACATGACGGATGCCAGCTT-GGGCCGAAGCCCAAGCTGGCATCCGTCAGTACCGGA-ATGCA; 2302-S0, AGCTTGAAAAGTTCGTACTGACGG-ATGCCAGCTTGGGCTTCGCTAGACGGCGCTCTACA-CGCTGTTGGTAAAGAATTC and CATGGAATTCTTTA-CCAACAGCGTGTAGAGCGCCGTCTAGCGAAGCCCAAG-CTGGCATCCGTCAGTACGAACTTTTCA; 18119-S20, AGC-TTGGCATTCCGGTGTTGACACAAGATAGAGTTAAC-TTCGGTTAACTCTATCTTGTGTCATGTTGGTAAAGA-ATTC and CATGGAATTCTTTACCAACATGACACAAGATAGAGTTAACCGAAGTTAACTCTATCTTGTGTC-AACACCGGAATGCCA; 18119-S0, AGCTTGGCATTCC-GGTGTTGACACAAGATAGAGTTAACTTCGATCAAA-TCGATGTTATGCCATGTTGGTAAAGAATTC and CATG-GAATTCTTTACCAACATGGCATAACATCGATTTGATC-GAAGTTAACTCTATCTTGTGTCAACACCGGAATGCCA.
The XbaI site in the 3′-UTR of the pGL-3 plasmid was also used for certain constructs. In this case, the plasmid was cut to completion with the enzyme, then treated with alkaline phosphatase. Oligonucleotides were synthesized to include the target site and surrounding structure as above and, in addition, an EcoRI site was included near the 5′-end of the insert. When annealed, both ends of the insert have overhangs compatible with XbaI. Oligonucleotides were synthesized with 5′-terminal phosphates to allow ligation to the phosphatase-treated plasmid. Orientation of the insert was evaluated by digestion with EcoRI and HpaI, which cut 160 bp downstream of the XbaI site. 3′-5132-S20, CTAGAATCCCTTTCGGACAATG-CATGTCACAGGCGGGATTCGTCCCGCCTGTGACATGCATTTGCTAGTAATGAATTT and CTAGAAATTCAT-TACTAGCAAATGCATGTCACAGGCGGGACGAATCC-CGCCTGTGACATGCATTGTCCGAAACCAATT; 3′-5132-S0, CTAGAATTCCTTTCGGACAATGCATGTCACAGG-CGGGATTCGTTCTGACAGACTACTCAGGTTGCTAGT-AATGAATTT and CTAGAAATTCATTACTAGCAACCT-GAGTAGTCTGTCAGAACGAATCCCGCCTGTGACATG-CATTGTCCGAAACCAATT.
RNA folding and ΔG calculations
RNA structures were predicted for each insert described above using RNAStructure 2.52. (26–29). The entire luciferase RNA with modified 5′-UTR was also folded for each construct to confirm the absence of long range interactions that might affect local structure in the 5′-UTR. Overall stability of the oligonucleotide–RNA duplex formation was calculated using OligoWalk (30,31). The input RNA for this calculation was the fragment inserted between the HindIII and NcoI sites for each target. The overall ΔG37° computed is the sum of the unfavorable free energy required to open the RNA base pairs at the oligonucleotide binding site, the unfavorable free energy required to break up secondary structure in the oligonucleotide and the favorable free energy for pairing the antisense oligonucleotide to the target RNA. Thermodynamic parameters are not available for predicting secondary structure or hybrid duplex stabilities for modified oligonucleotides so DNA parameters were used for the unmodified deoxyphosphorothioate oligonucleotides and RNA parameters for the chimeric deoxyphosphorothioate/2′-O-methoxyethyl base oligonucleotides. Because the same oligonucleotide was used with each structure studied, any errors introduced by this approximation would contribute a constant free energy and would not affect relative values.
Luciferase assays
Plasmid (10 µg) was introduced into COS-7 cells at 70% confluency in a 10 cm dish using SuperFect Reagent (Qiagen). Following a 2 h treatment, cells were trypsinized and split into a 24-well plate. Cells were allowed to adhere for 1 h, then oligonucleotide was added in the presence of Lipofectin Reagent at 3 µg/ml/100 nM oligonucleotide. All oligonucleotide treatments were done in duplicate or triplicate. Following the 4 h oligonucleotide treatment, cells were washed and fresh DMEM + 10% FCS was added. The cells were incubated overnight at 37°C. The following morning cells were harvested in 150 µl of Passive Lysis Buffer (Promega). An aliquot of 60 µl of lysate was added to each well of a black 96-well plate then 50 µl Luciferase Assay Reagent (Promega) was added. Luminescence was measured using a Packard TopCount. Error bars represent the standard deviation from the mean of at least two independent oligonucleotide treatments.
RNA analysis
RNA levels were evaluated for each plasmid construct by northern analysis. Aliquots of 3 µg of each luciferase plasmid construct were co-transfected into cells along with 2 µg of pcmB7-2, a plasmid expressing murine B7-2 under control of the CMV promoter, using SuperFect Reagent (Qiagen). After 2 h the plasmid was removed and the cells incubated for an additional 4 h in complete medium. Total RNA was harvested using a ToTALLY RNA kit (Ambion) according to the manufacturer’s protocol. RNA was separated on a 1.2% agarose gel containing 1.1% formaldehyde, then transferred to nylon membranes. Blots were hybridized with [32P]dCTP random prime labeled cDNA probes specific for luciferase and murine B7-2 for 2 h in Rapid-hyb solution (Amersham). Blots were washed with 2× SSC containing 0.1% SDS at room temperature, followed by 0.1× SSC containing 0.1% SDS at 60°C. Quantitation of RNA expression was performed using a Molecular Dynamics PhosphorImager.
Synthesis of S20 RNAs for binding and RNase H experiments
Forty-four base RNAs were synthesized from oligonucleotide templates with T7 RNA polymerase. The bottom strand oligonucleotides were complementary to the 44 bases of each stem–loop followed by sequence complementary to the T7 promoter at the 3′-end. These were annealed with a 22 base oligonucleotide corresponding to the T7 promoter (AATTTA-ATACGACTCACTATAG) at a concentration of 100 µM each strand in 1× T7 buffer. The partially single strand template was used at 6 µM in a 20 µl reaction using a MaxiScript T7 polymerase kit (Ambion) and [α-32P]UTP. After 1 h incubation at 37°C, the RNAs were purified by gel electrophoresis. Unlabeled RNAs were also produced using a T7 MegaShortScript kit (Ambion) according to the manufacturer’s protocol. 5132 T7 template, AATGCATGTCACAGGCGGGACGAATCCCGC-CTGTGACATGCATTCCCTATAGTGAGTCGTATTAAATT; 2302 T7 template, TGACGGATGCCAGCTTGGGCCGAA-GCCCAAGCTGGCATCCGTCACCCTATAGTGAGTCGT-ATTAAATT.
For the structured RNAs, RNase H activity was determined by combining the cold structured target RNA at 5 µM and 25 000 c.p.m. of the labeled target RNA with 1 or 10 µM antisense oligonucleotide in 10 µl of 1× RNase H assay buffer (20 mM Tris–HCl pH 7.5, 10 mM MgCl2, 0.1 mM EDTA, 0.1 mM DTT). Reactions were incubated for 3 h at 37°C. Aliquots of 0.5 U of Escherichia coli RNase H (US Biochemical) were added and the reaction incubated for an additional 20 min at the same temperature. Reactions were then heated to 90°C for 2 min prior to separating products on a 10% polyacrylamide gel with 50% (w/v) urea.
Complementary 20 base RNAs were synthesized for each target at Genset (Paris). The RNAs were radioactively labeled with [γ-33P]ATP using polynucleotide kinase; labeled RNAs were purified on an acrylamide gel. Binding of oligonucleotide to complement was determined by gel mobility shift assay. 33P-end-labeled RNA (100 000 c.p.m.) was incubated with complementary oligonucleotide and 100 ng of tRNA carrier in 1× RNase H buffer for 1 h at room temperature. Bound was then separated from unbound by electrophoresis on a 12% native acrylamide gel run in 1× TBE at a constant power of 10 W at 4°C.
RESULTS
Effect of target structure
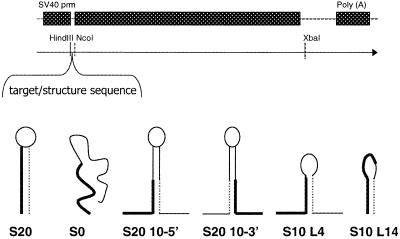
In order to evaluate the effect of structure in the mRNA target on activity of an antisense oligonucleotide in cells, the structural context of the target site was altered. The binding site for a previously identified antisense oligonucleotide (ISIS 5132) which inhibits the expression of human c-raf kinase (15) was cloned into the 5′-UTR of the luciferase reporter plasmid pGL3-Control as detailed in Materials and Methods. Sequence immediately adjacent to the target sequence was altered as outlined in Figure 1 to form various predicted RNA secondary structures that included the 5132 target sequence. These structures ranged from one in which the entire target site was sequestered in a 20 base stem closed by a UUCG tetraloop (S20) to one that had little predicted secondary structure likely to inhibit hybridization of 5132 to its target (S0). Like the S20 construct, S20-10-5 also had a 20 base stem with a tetraloop, however, only 10 bases of the target site were contained within the stem on the 5′-side. S20-10-3 was similar except that the target site was on the 3′-side of the stem; thus the opposite half of the target site was contained in the stem. S10-L4 has a 10 base stem complementary to the 5′-half of the oligonucleotide with a tetraloop, while S10-L14 had a 10 base stem complementary to the 3′-half of the oligonucleotide followed by a 14 base loop containing the remainder of the target sequence.
Figure 1.
(A) Cloning vector pGL3-Control. Modified 5′- and 3′-UTR sequences were cloned into either the HindIII and NcoI sites (5′-UTR) or the XbaI site as detailed in Materials and Methods. (B) Predicted structures of the resulting target sequences. Bold lines represent the oligonucleotide binding site, thin dashed lines represent sequence complementary to the oligonucleotide binding site.
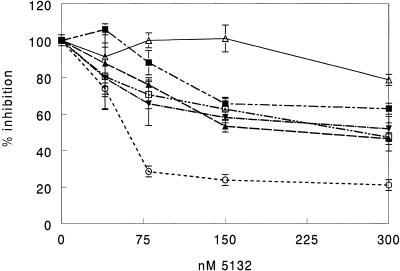
Each construct was transfected into COS-7 cells as detailed in Materials and Methods. Transfected cells were seeded in 24-well plates and treated with ISIS 5132 using cationic lipid at doses ranging from 40 to 300 nM. All oligonucleotide treatments were performed in duplicate or triplicate. Lysates from the treated cells were assayed for luciferase produced. Reduction in luciferase activity is correlated with degradation of mRNA mediated by antisense oligonucleotides. The results are shown in Figure 2. As expected, 5132 showed most activity against the construct with the least amount of secondary structure (S0). Against the S0 construct 5132 had an IC50 of ~60 nM. In contrast, 5132 showed very little activity against the S20 construct even at the highest concentration tested. The remaining constructs showed intermediate activity with all IC50 values in the 300–400 nM range. Differences in activity could not be reliably determined among the intermediate constructs. The predicted overall ΔG37° for invasion and binding of the target was –22.1 kcal mol–1 for the S0 construct and +6.1 kcal mol–1 for the S20 construct. Overall free energies for formation and binding of the intermediate constructs were similar to one another with ΔG37° values in the range –13.4 to –10.1 kcal mol–1. This correlates well with their intermediate levels of activity. An exception was the S20-10-5 construct, with a predicted overall ΔG37° of –1.4 kcal mol–1. One would expect antisense oligonucleotides to be less effective against this target than the other three with intermediate antisense susceptibility based upon the predicted free energies, however, this was not the case. Thus, oligonucleotide efficacy was qualitatively, but not quantitatively, correlated with predicted RNA structure and oligonucleotide binding thermodynamics.
Figure 2.
Inhibition of alternate structure clones by ISIS 5132. Cells were transfected with the luciferase reporter plasmids diagrammed in Figure 1, then treated in duplicate with oligonucleotide at doses ranging from 40 to 300 nM. Luciferase expression was measured the following day. Results are percent luciferase expression compared to the no oligonucleotide control. Open triangle, S20; open circle, S0; inverted closed triangle, S20-10-5; closed square, S20-10-3; closed triangle, S10-4; open square, S10-14.
To ensure that the observed effects were not the result of variations in the transcription efficiencies or mRNA stabilities of the constructs, RNA levels were evaluated for each construct by northern analysis as detailed in Materials and Methods. Cells were co-transfected with a second plasmid construct expressing a cDNA for murine B7-2 to account for variation in transfection efficiency. Luciferase RNA expression normalized to levels of the mB7-2 RNA varied by <30% from the control expression vector pGL-3 (data not shown). The variation was not correlated with the amount of structure in the construct.
Effect of sequence context
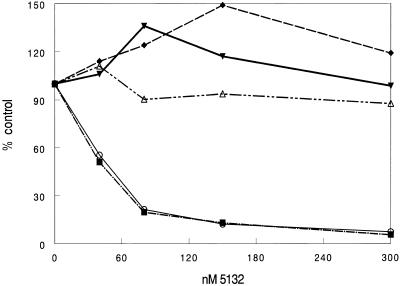
In order to determine if sequence context has an effect on oligonucleotide efficacy, the 5132 S0 and S20 sites were cloned into the 3′-UTR of pGL-3 using the unique XbaI site as detailed in Materials and Methods. The ability of 5132 to inhibit luciferase expression from these constructs was evaluated and compared with the original 5132 S0 and S20 constructs (Fig. 3). Placement of the target within the message had little effect on oligonucleotide potency. Luciferase production from the S20 constructs was not inhibited even at the highest dose of 5132 tested. This was comparable to the construct without a target site at all. On the other hand, inhibition of the S0 targets at either site in the message was almost identical at all doses. It seems to make little difference where the target is located within the RNA as long as RNA structure around the target site does not inhibit binding of the complementary oligonucleotide.
Figure 3.
Effect of sequence context on oligonucleotide efficacy. The 5132 S20 and S0 targets were inserted in the 3′ UTR of pGL3-Control as detailed in Materials and Methods. Unmodified plasmid as well as plasmid containing the 5132 target site and structure were transfected into Cos-7 cells. Following the transfection cells were treated with 5132 in the presence of cationic lipid for 4 h at doses ranging from 40 to 300 nM. Luciferase activity was measured the following day. Results are given as percent of no oligonucleotide control for each plasmid. Open triangle, S20; open circle, S0; closed inverted triangle, pGL3-Control; closed diamond, S20-3′; closed square, S0-3′.
Effect of oligonucleotide chemistry
To date the most well characterized class of antisense oligonucleotides are phosphorothioate oligodeoxynucleotides (32) that exert their activity primarily through a RNase H-mediated mechanism (33–35). More recently other types of nucleotide modifications have been designed with the intent of improving the metabolic stability of the oligonucleotide as well as increasing affinity for the target RNA. Usually the same properties that enhance the affinity of these analogs for RNA result in loss of RNase H activity. For example, 2′-O-methyl- and other 2′-modified oligonucleotides have been shown to exhibit greater metabolic stability and affinity for their RNA targets, but these compounds do not support RNase H activity (36–38). Recent antisense strategies have focused on chimeric oligonucleotides that contain both unmodified 2′-deoxy- and 2′-modified nucleotides within the oligomer to achieve the desired increases in metabolic stability and binding affinity yet still support RNase H activity (25,39–47). Usually, the analogs contain a region of modified nucleotides at one or both ends designed to prevent nuclease degradation and enhance affinity and an unmodified ‘gap’ region that retains the ability to direct RNase H cleavage.
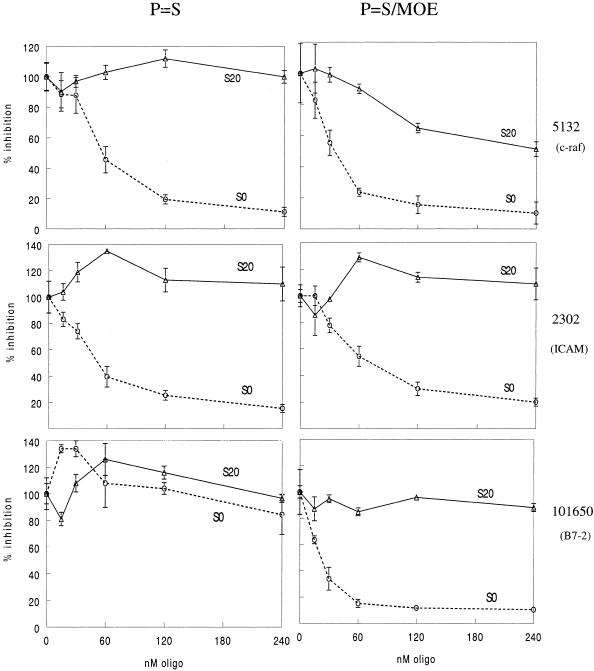
A chimeric antisense analog of 5132, 13650, was also evaluated in the luciferase reporter system. ISIS 13650 is a full phosphorothioate analog of 5132 with 2′-O-methoxyethyl substitutions at positions 1–6 and 15–20. Residues 7–14 are unmodified 2′-deoxy so they can serve as substrates for RNase H. The first panel of Figure 4 shows results for this oligonucleotide targeted to the 5132 site in the S0 and S20 constructs. When cells containing the S0 construct were assayed, 13650 was a slightly more potent inhibitor of luciferase production than was the full deoxyphosphorothioate oligonucleotide, 5132. However, when targeting the same site in the S20 construct, 13650 was clearly a more effective inhibitor than 5132. 13650 had an IC50 in the 300 nM range while no inhibition of luciferase activity was observed with 5132, even at the highest dose. Thus, while there was little advantage of the modified chemistry against an unstructured target, the increased binding affinity of the modified oligonucleotide led to modest inhibition of the highly structured S20 target site.
Figure 4.
S20 and S0 constructs for three oligonucleotide targets were transfected, then cells were treated in duplicate or triplicate with either unmodified oligonucleotide or chimeric deoxyphosphorothioate/2′-O-methoxyethyl base (P=S/MOE) oligonucleotide gap-mers at doses ranging from 25 to 240 nM. Luciferase expression was assayed 18–24 h after oligonucleotide treatment. Activity is given as percent of the no oligonucleotide control. Triangle, S20; circle, S0.
Effect of oligonucleotide/target sequence
In order to determine if this observation was sequence specific, constructs were made with the target site sequestered within a 20 base stem (S20) or with no predicted structure (S0) using two additional target sequences and corresponding oligonucleotides. Constructs were made to include sequence for oligonucleotides originally designed to target human ICAM-1 (ISIS 2302) and B7-2 (ISIS 101650) mRNAs. In cells where wild-type message is targeted, ISIS 2302, a deoxyphosphorothioate oligonucleotide, is a potent inhibitor of ICAM-1 (18), however the deoxyphosphorothioate ISIS 101650, which targets human B7-2, did not effectively inhibit B7-2 expression (data not shown). Chimeric deoxyphosphorothioate/2′-O-methoxyethyl base versions of both oligonucleotides effectively inhibited expression of their targeted wild-type genes in cells.
Each construct was transfected into COS-7 cells which were then treated with unmodified deoxyphosphorothioate or chimeric deoxyphosphorothioate/2′-O-methoxyethyl base oligonucleotides complementary to the target sequences at doses ranging from 30 to 300 nM. Luciferase activity was measured the following day. The results are shown in Figure 4. In all cases the chimeric oligonucleotides inhibited expression of the S0 constructs with similar IC50 values, in the 20–60 nM range. Activities of the unmodified deoxyphosphorothioate oligonucleotides against the S0 constructs were similar to the activities of the chimeras. The exception was the unmodified B7-2 oligonucleotide, 101650, which showed no activity even at the highest oligonucleotide dose. As with the 5132 experiments, the unmodified oligonucleotides showed no activity against the S20 targets. Unlike the c-raf chimeric oligonucleotide 13650, neither of the chimeric oligonucleotides targeting the ICAM-1 or B7-2 sites inhibited expression of the respective S20 constructs.
Evaluation of hybridization in vitro
Hybridization of each oligonucleotide to length-matched complementary RNAs was evaluated by gel shift assay. Each chemically synthesized RNA was end-labeled then incubated with oligonucleotide at 10 and 100 nM. Bound target was separated from free by electrophoresis on a native acrylamide gel. The results, summarized in Table 1, are given as the percent of target RNA bound at each of the two oligonucleotide concentrations. The data indicate that the B7-2 full deoxyphosphorothioate oligonucleotide, 101650, bound its target sequence with less affinity than the other unmodified oligonucleotides. This may account for the lack of activity observed with 101650 in the luciferase assay. All other oligonucleotides had similar affinities for their respective targets.
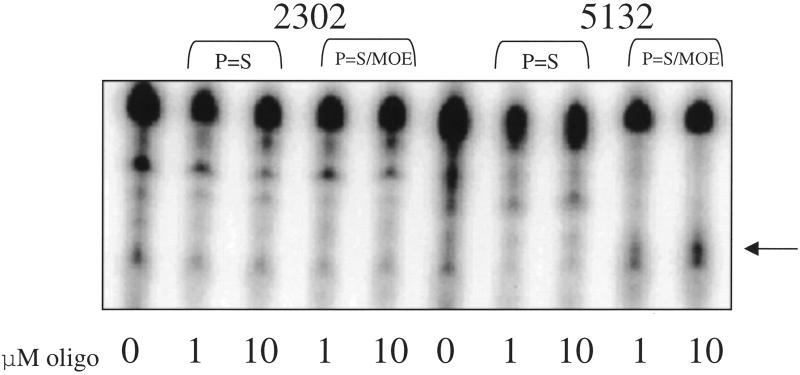
Oligonucleotide affinity for short, but structured, target RNAs was also determined. RNAs 44 nt in length were produced using T7 RNA polymerase from oligonucleotide templates corresponding to the S20 target regions for 5132 and 2302. Oligonucleotides were incubated with the structured target at 10 or 1 µM. RNase H was added to allow cleavage of the DNA–RNA hybrids formed. The results are shown in Figure 5. The unmodified oligonucleotides did not direct RNase H cleavage of either of the structured targets at the concentrations tested. A cleavage product of the 5132 S20 target was observed in the presence of the chimeric oligonucleotide 13650, however, no cleavage product was observed for the 2302 S20 target in the presence of its corresponding chimeric oligonucleotide 15839. These results are consistent with those previously observed with plasmid constructs in the luciferase assay.
Figure 5.
RNase cleavage of S20 targets. Forty-four base S20 RNA targets were synthesized and labeled as described in Materials and Methods. Targets were hybridized with unmodified deoxyphosphorothioate (P=S) or chimeric deoxyphosphorothioate/2′-O-methoxyethyl base (P=S/MOE) oligonucleotides complementary to the target sequences in 1× RNase H buffer at either 1 or 10 µM. Cleavage was initiated by the addition of 0.5 U RNase H. Cleaved target RNA was visualized by electrophoresis on a denaturing acrylamide gel. The arrow at the right of the figure indicates the position of the expected cleavage product.
DISCUSSION
The purpose of this study was to determine the effect of target secondary structure on antisense activity in the absence of changes to oligonucleotide sequence and structure. Evidence for the role of target structure in antisense activity has been provided by earlier studies that ‘walked’ antisense oligonucleotides along a target mRNA in an attempt to correlate antisense activity with predicted structure at the binding sites (5,48). Other studies have relied upon ‘walking’ oligonucleotides around a known structure in order to determine the binding site with the highest in vitro affinity for the target (7,49–51). Yet another strategy used by Milner et al. employed an oligonucleotide array to measure the potential for hybridization of each oligonucleotide to a structured target (52). While these methods often do identify oligonucleotides with the highest affinity for the target, they do not directly address the question of the role of structure in the binding of a particular oligonucleotide. Another limitation of these types of studies is that the sequence of the oligonucleotide changes as the target is moved around the structure. These sequence changes can affect chemical stability and secondary structure, which may in turn affect the in vivo potency of the oligonucleotide. In addition, the position of the target site relative to functional sites on the mRNA may also affect oligonucleotide potency. The current study differs from these earlier studies in that only target structure was varied, not oligonucleotide sequence or target position. In addition, by modifying the sequence of the mRNA near the target, we were able to create structures that likely represent extremes of stability possible with native mRNA. Furthermore, we were able to evaluate antisense inhibition of these structured targets in cell-based assays, which may be a more accurate predictor of in vivo activity than biochemical assays.
ISIS 5132 has previously been shown to be a potent inhibitor of the expression of human c-raf kinase (15). We cloned the target sequence for 5132 into the 5′-UTR of a luciferase reporter gene. Additional sequence was added adjacent to the target site sequence in order to build a stem–loop containing the entire binding site (S20) or to avoid as much structure as possible around the binding site (S0). Four intermediate structures in which 10 of the 20 target nucleotides were predicted to form a stem were also constructed. One critical assumption of this work is that the predicted target structures and calculated free energies actually correspond to the structures encountered by the antisense oligonucleotide in the cell. Although RNA structure predictions are not yet able to consistently predict all details of known structures, the success rate is improving (28,53). The S20 structure, a 20 nt stem closed by a UUCG tetraloop, is known to be extremely stable so we were comfortable in assuming that it was indeed the target structure formed in the cell. For the S0 structure, several structures with suboptimal free energy were examined and no structures with more than four contiguous paired nucleotides in the 20 nt target site were observed, supporting our classification of these targets as ‘unstructured’. Although exact structures for the S20-10-5′, S20-10-3′, S10-L4 and S10-L14 sites are not certain, examination of suboptimal structures confirmed the presence of the 10 and 20 nt stems with few base pairs in the unstructured part of the target site. Thus, although the free energies calculated for oligonucleotide binding to these sites are not quantitatively correct, the predicted rank order probably is.
We found that both antisense oligonucleotide 5132 and the 5132 chimera 13650 were more potent against the unstructured target (S0) than the structured target (S20) (Fig. 4). The targets partially contained within structures exhibited activity intermediate between that of the S20 and S0 targets (Fig. 2 for 5132, data not shown for 13650). When the 5132 targets and corresponding structures were moved from the 5′- to the 3′-UTR of the message, the results were not affected. This suggests that the structures formed as predicted even when the sequence outside the folded area was changed significantly. As long as there was little local structure that included the oligonucleotide target site, the position of the target site on the message had little effect on activity. However, strong local structure at the oligonucleotide binding site effectively inhibited antisense activity regardless of adjacent sequence context.
S0 and S20 constructs were also made for two additional target sites, 2302 (human ICAM-1) and 101650 (human B7-2). Antisense activity was evaluated for these constructs as well as the 5132 S20 and S0 constructs with unmodified deoxyphosphorothioate oligonucleotides (Fig. 4). In the case of the 5132 and 2302 constructs we found that antisense activity was higher for the unstructured target (S0) than for the structured target (S20). However, oligonucleotide 101650 was inactive against both the structured and unstructured targets. The inactivity of this compound even against the unstructured target is consistent with the lack of activity observed for this oligonucleotide against B7-2 mRNA in a cellular assay and the weak binding observed to a length-matched RNA complement (Table 1). Increased affinity of the oligonucleotide for its target sequence was accomplished by changing the chemistry. The chimeric deoxyphosphorothioate/2′-O-methoxyethyl base version of 101650, 18119, showed substantially increased binding to a length-matched complement, which in turn led to activity in the cellular luciferase reporter assay. However, the chimera 18119 was not active against the S20 target. It is likely that the increased affinity of 18119, as compared to 101650, was not enough to overcome the S20 structure, but it was able to make a weak binder active against the unstructured target.
For the 5132 constructs, use of chimeric oligonucleotide 13650 in the assay resulted in increased activity against both the S0 and S20 constructs. There was no measurable change in activity against the 2302 constructs tested with the corresponding chimeric oligonucleotide, 15839. This correlates with in vitro analysis of binding and RNase H cleavage. Affinity of 15839 for its length-matched RNA complement did not change significantly relative to the unmodified oligonucleotide 2302. In contrast, 13650 did show increased affinity for its length-matched RNA complement as measured by gel shift assay. This resulted in an increase in luciferase activity against both the S0 and S20 constructs. In addition, only 13650 was able to produce an RNase H cleavage product when incubated with its structured RNA target (Fig. 5).
The results of this study confirm that the structure of the mRNA target is an important factor in determining antisense efficacy in cells. Oligonucleotides were unable to invade a very stable stem–loop structure (S20), but generally showed good activity when impeded by little local structure (S0). For one oligonucleotide, 5132, the inhibitory effect of the S20 stem could be partially reduced by incorporation of high affinity chemistry. The structure of the mRNA clearly plays a large part in determining the efficacy of an antisense oligonucleotide in cells; thus discovery of active antisense oligonucleotides will require identification of unstructured sites in the cellular RNA. This will likely be accomplished using a combination of computational methods (27), cell-free mRNA assays (52,54,55) and simple oligonucleotide walks in cellular antisense assays.
REFERENCES
- 1.Crooke S.T. (1998) Antisense Nucleic Acid Drug Dev., 8, 133–134. [DOI] [PubMed] [Google Scholar]
- 2.Freier S.M. and Tinoco,I.J. (1975) Biochemistry, 14, 3310–3314. [DOI] [PubMed] [Google Scholar]
- 3.Fedor M.J. and Uhlenbeck,O.C. (1990) Proc. Natl Acad. Sci. USA, 87, 1668–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Herschlag D. and Cech,T.R. (1990) Biochemistry, 29, 10159–10171. [DOI] [PubMed] [Google Scholar]
- 5.Eckardt S., Romby,P. and Sczakiel,G. (1997) Biochemistry, 36, 12711–12721. [DOI] [PubMed] [Google Scholar]
- 6.Herschlag D. and Cech,T.R. (1990) Biochemistry, 29, 10172–10180. [DOI] [PubMed] [Google Scholar]
- 7.Ecker D.J., Vickers,T.A., Bruice,T.W., Freier,S.M., Jenison,R.D., Manoharan,M. and Zounes,M. (1992) Science, 257, 958–961. [DOI] [PubMed] [Google Scholar]
- 8.Taylor M.F., Paulauskis,J.D., Weller,D.D. and Kobzik,L. (1996) J. Biol. Chem., 271, 17445–17452. [DOI] [PubMed] [Google Scholar]
- 9.Shen L.X., Basilion,J.P. and Stanton,V.P.,Jr (1999) Proc. Natl Acad. Sci. USA, 96, 7871–7876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Manoharan M., Johnson,L.K., McGee,D.P.C., Guinosso,C.J., Ramasamy,K., Springer,R.H., Bennett,C.F., Ecker,D.J., Vickers,T., Cowsert,L. and Cook,P.D. (1992) Ann. N. Y. Acad. Sci., 660, 306–309. [DOI] [PubMed] [Google Scholar]
- 11.Baker B.F., Lot,S.S., Condon,T.P., Lesnik,E.A., Sasmor,H.M. and Bennett,C.F. (1997) J. Biol. Chem., 272, 11994–12000. [DOI] [PubMed] [Google Scholar]
- 12.Bennett C.F., Chiang,M.Y., Chan,H. and Grimm,S. (1993) J. Liposome Res., 3, 85–102. [Google Scholar]
- 13.Butler M., Stecker,K. and Bennett,C.F. (1997) Lab. Invest., 77, 379–388. [PubMed] [Google Scholar]
- 14.Graham M.J., Crooke,S.T., Monteith,D.K., Cooper,S.R., Lemonidis,K.M., Stecker,K.K., Martin,M.J. and Crooke,R.M. (1998) J. Pharmacol. Exp. Ther., 286, 447–458. [PubMed] [Google Scholar]
- 15.Monia B.P., Johnston,J.F., Geiger,T., Muller,M. and Fabbro,D. (1996) Nature Med., 2, 668–675. [DOI] [PubMed] [Google Scholar]
- 16.Tu G.-c., Qing-na,C., Zhou,F. and Israel,Y. (1998) J. Biol. Chem., 273, 25125–25131. [DOI] [PubMed] [Google Scholar]
- 17.Cotter F.E., Johnson,P., Hall,P., Pocock,C., Al Mahdi,N., Cowell,J.K. and Morgan,G. (1994) Oncogene, 9, 3049–3055. [PubMed] [Google Scholar]
- 18.Chiang M.Y., Chan,H., Zounes,M.A., Freier,S.M., Lima,W.F. and Bennett,C.F. (1991) J. Biol. Chem., 266, 18162–18171. [PubMed] [Google Scholar]
- 19.Johansson H.E., Belsham,G.J., Sproat,B.S. and Hentze,M.W. (1994) Nucleic Acids Res., 22, 4591–4598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bennett C.F., Condon,T.P., Grimm,S., Chan,H. and Chiang,M.-Y. (1994) J. Immunol., 152, 3530–3540. [PubMed] [Google Scholar]
- 21.Goodchild J., Agrawal,S., Civeira,M.P., Sarin,P.S., Sun,D. and Zamecnik,P.C. (1988) Proc. Natl Acad. Sci. USA, 85, 5507–5511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thierry A.R., Rahman,A. and Dritschilo,A. (1993) Biochem. Biophys. Res. Commun., 190, 952–960. [DOI] [PubMed] [Google Scholar]
- 23.Laptev A.V., Lu,Z., Colige,A. and Prockop,D.J. (1994) Biochemistry, 33, 11033–11039. [DOI] [PubMed] [Google Scholar]
- 24.Lima W.F., Monia,B.P., Ecker,D.J. and Freier,S.M. (1992) Biochemistry, 31, 12055–12061. [DOI] [PubMed] [Google Scholar]
- 25.Monia B.P., Lesnik,E.A., Gonzalez,C., Lima,W.F., McGee,D., Guinosso,C.J., Kawasaki,A.M., Cook,P.D. and Freier,S.M. (1993) J. Biol. Chem., 268, 14514–14522. [PubMed] [Google Scholar]
- 26.Jaeger J.A., Turner,D.H. and Zuker,M. (1989) Proc. Natl Acad. Sci. USA, 86, 7706–7710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mathews D.H., Andre,T.C., Kim,J., Turner,D.H. and Zuker,M. (1998) ACS Symp. Ser., 682, 246–257. [Google Scholar]
- 28.Walter A.E., Turner,D.H., Kim,J., Lyttle,M.H., Mueller,P., Mathews,D.H. and Zuker,M. (1994) Proc. Natl Acad. Sci. USA, 91, 9218–9222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zuker M. (1989) Science, 244, 48–52. [DOI] [PubMed] [Google Scholar]
- 30.SantaLucia J., Allawi,H.T. and Senevirtatne,P.A. (1996) Biochemistry, 35, 3555–3562. [DOI] [PubMed] [Google Scholar]
- 31.Sugimoto N., Nakano,S.-i., Katoh,M., Matsumura,A., Nakamuta,H., Ohmichi,T., Yoneyama,M. and Sasaki,M. (1995) Biochemistry, 34, 11211–11216. [DOI] [PubMed] [Google Scholar]
- 32.Crooke S.T. (1995) In Cuello,A. and Collier,B. (eds), Pharmacological Science: Perspectives on Research and Therapy in the Late 1990s, Proceedings of the 12th International Congress on Pharmacology. Birkhauser, Basel, Switzerland, pp. 393–399.
- 33.Stein C.A., Subasinghe,C., Shinozuka,K. and Cohen,J.S. (1988) Nucleic Acids Res., 16, 3209–3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Toulme J.-J. and Helene,C. (1988) Gene, 72, 51–58. [DOI] [PubMed] [Google Scholar]
- 35.Monia B.P., Johnston,J.F., Ecker,D.J., Zounes,M.A., Lima,W.F. and Freier,S.M. (1992) J. Biol. Chem., 267, 19954–19962. [PubMed] [Google Scholar]
- 36.Cook P.D. (1998) Annu. Rep. Med. Chem., 33, 313–325. [Google Scholar]
- 37.Kogoma T., Subia,N.L. and von Meyenburg,K. (1985) Mol. Gen. Genet., 200, 103–109. [DOI] [PubMed] [Google Scholar]
- 38.Sproat B.S. and Lamond,A.I. (1993) In Crooke,S.T. and Lebleu,B. (eds), Antisense Research and Applications. CRC Press, Boca Raton, FL, pp. 351–362.
- 39.Inoue H., Hayase,Y., Iwai,S. and Ohtsuke,E. (1987) FEBS Lett., 215, 327–330. [DOI] [PubMed] [Google Scholar]
- 40.Furdon P.J., Dominski,Z. and Kole,R. (1989) Nucleic Acids Res., 17, 9193–9204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tidd D.M., Hawley,P., Warenius,H.M. and Gibson,I. (1988) Anticancer Drug Des., 3, 117–127. [PubMed] [Google Scholar]
- 42.Tidd D.M. and Warenius,H.M. (1989) Br. J. Cancer, 60, 343–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Quartin R.S., Brakel,C.L. and Wetmur,J.G. (1989) Nucleic Acids Res., 17, 7253–7262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Agrawal S., Mayrand,S.H., Zamecnik,P.C. and Pederson,T. (1990) Proc. Natl Acad. Sci. USA, 87, 1401–1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hayase Y., Inoue,H. and Ohtsuka,E. (1990) Biochemistry, 29, 8793–8797. [DOI] [PubMed] [Google Scholar]
- 46.Dagle J.M., Walder,J.A. and Weeks,D.L. (1990) Nucleic Acids Res., 18, 4751–4757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Baker C. (1990) Nucleic Acids Res., 18, 3537–3542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stull R.A., Taylor,L.A. and Szoka,F.C.,Jr (1992) Nucleic Acids Res., 20, 3501–3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hamilton S.P., Pitts,A.E., Katipally,R.R., Jia,X., Rutter,J.P., Davies,B.A., Shay,J.W., Wright,W.E. and Corey,D.R. (1997) Biochemistry, 36, 11873–11880. [DOI] [PubMed] [Google Scholar]
- 50.Petryshyn R.F., Ferrenz,A.G. and Li,J. (1997) Nucleic Acids Res., 25, 2672–2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lima W.F., Brown-Driver,V., Fox,M., Hanecak,R. and Bruice,T.W. (1997) J. Biol. Chem., 272, 626–638. [PubMed] [Google Scholar]
- 52.Milner N., Mir,K.U. and Southern,E.M. (1997) Nature Biotechnol., 15, 537–541. [DOI] [PubMed] [Google Scholar]
- 53.Mathews D.H., Sabina,J., Zuker,M. and Turner,D.H. (1999) J. Mol. Biol., 288, 911–940. [DOI] [PubMed] [Google Scholar]
- 54.Matveeva O., Felden,B., Audlin,S., Gesteland,R.F. and Atkins,J.F. (1997) Nucleic Acids Res., 25, 5010–5016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ho S.P., Britton,D.H.O., Stone,B.A., Behrens,D.L., Leffet,L.M., Hobbs,F.W., Miller,J.A. and Trainor,G.L. (1996) Nucleic Acids Res., 24, 1901–1907. [DOI] [PMC free article] [PubMed] [Google Scholar]