Neurodegeneration and ciliary degeneration are caused by mutation of the deglutamylase CCP1/CCPP-1 in humans and C. elegans. The conserved NIMA-related kinase NEKL-4/NEK10 can suppress or promote degeneration in an activity-dependent manner that involves cilia–mitochondria communication and that is independent of glutamylation.
Abstract
Ciliopathies are often caused by defects in the ciliary microtubule core. Glutamylation is abundant in cilia, and its dysregulation may contribute to ciliopathies and neurodegeneration. Mutation of the deglutamylase CCP1 causes infantile-onset neurodegeneration. In C. elegans, ccpp-1 loss causes age-related ciliary degradation that is suppressed by a mutation in the conserved NEK10 homolog nekl-4. NEKL-4 is absent from cilia, yet it negatively regulates ciliary stability via an unknown, glutamylation-independent mechanism. We show that NEKL-4 was mitochondria-associated. Additionally, nekl-4 mutants had longer mitochondria, a higher baseline mitochondrial oxidation state, and suppressed ccpp-1∆ mutant lifespan extension in response to oxidative stress. A kinase-dead nekl-4(KD) mutant ectopically localized to ccpp-1∆ cilia and rescued degenerating microtubule doublet B-tubules. A nondegradable nekl-4(PEST∆) mutant resembled the ccpp-1∆ mutant with dye-filling defects and B-tubule breaks. The nekl-4(PEST∆) Dyf phenotype was suppressed by mutation in the depolymerizing kinesin-8 KLP-13/KIF19A. We conclude that NEKL-4 influences ciliary stability by activating ciliary kinesins and promoting mitochondrial homeostasis.
Introduction
Cilia are microtubule-based organelles that project from most cell types in the body, including those in the nervous system (Lee and Gleeson, 2011). In the brain, motile cilia generate cerebrospinal flow (Spassky and Meunier, 2017) and neuronal primary cilia form synapses (Sheu et al., 2022). Ciliopathies, disorders of cilia, comprise a diverse group of diseases ranging from single-organ dysfunction (e.g., polycystic kidney disease and retinitis pigmentosa) to congenital malformations and developmental defects that affect multiple organ systems including the brain (e.g., Joubert syndrome and Bardet–Biedl syndrome) (Mill et al., 2023). The causes of ciliopathies vary widely, but many involve ultrastructural abnormalities in the microtubule-based structural core of the cilium called the axoneme. The axoneme is composed of a ring of nine outer A-B doublet microtubules surrounding a pair of central singlets or no central pair—the “9 + 2” or “9 + 0” formation in motile or primary/sensory cilia, respectively (Mill et al., 2023).
Ciliary microtubule structure and microtubule-based transport are regulated by the “Tubulin Code” via tubulin posttranslational modifications (PTMs) that are added and removed by enzymatic “writers” and “erasers” (McKenna et al., 2023). Glutamylation is an evolutionarily conserved PTM that occurs in cilia from protozoa to mammals. Dysregulated glutamylation (hypo- or hyperglutamylation) leads to neurodegeneration and ciliopathies such as idiopathic scoliosis (Mathieu et al., 2021), retinitis pigmentosa (Cehajic-Kapetanovic et al., 2022), and Joubert syndrome (He et al., 2018). Tubulin PTMs regulate microtubule-based transport and are interpreted by “readers” that include kinesin motors. The localization of proteins and other molecular cargoes along microtubules is of vital importance in the long processes of neurons, and dysregulation of this process may contribute to neurodegeneration (Guo et al., 2020). Mitochondria are transported along microtubules, making microtubule-based transport essential since mitochondrial ATP is required at synapses that may be at a great distance from the neuronal cell body (Panchal and Tiwari, 2019; Cheng and Sheng, 2021). Hyperglutamylation caused by mutation of the deglutamylase CCP1 results in early-onset neurodegeneration and defects in microtubule-based mitochondrial transport (Shashi et al., 2018; Magiera et al., 2018; Gilmore-Hall et al., 2019; Li et al., 2020).
The C. elegans CCP1 homolog CCPP-1 regulates microtubule stability in sensory cilia (O’Hagan et al., 2011). In the cilia of ccpp-1∆ mutants, B-tubules degenerate in amphid chemosensory neurons in an age-dependent and progressive manner. Glutamylation occurs preferentially on the B-tubule (Lechtreck and Geimer, 2000; Kubo et al., 2010; Kubo and Oda, 2017; Orbach and Howard, 2019; Alvarez Viar et al., 2023, Preprint), consistent with ccpp-1∆ hyperglutamylation-induced B-tubule defects. Previously, we identified the NIMA-related kinase NEK10/NEKL-4 as a genetic interactor of the CCPP-1 deglutamylase (Power et al., 2020). Loss-of-function mutations of nekl-4 suppress ccpp-1∆ progressive ciliary degeneration, as measured by fluorescent lipophilic dye-filling defects (Dyf) of the ciliated neurons. Conversely, overexpression (OE) of NEKL-4 results in a Dyf phenotype (Power et al., 2020; Li et al., 2021a). Endogenous NEKL-4 protein does not localize to the cilium or influence ciliary tubulin glutamylation, and the NEKL-4 mechanism of action remained unclear (Power et al., 2020). The mammalian NEKL-4 ortholog NEK10 has potential roles in cilia motility in airway epithelial cilia (Chivukula et al., 2020; Al Mutairi et al., 2020) and in regulating mitochondria localization and homeostasis in mammalian cell culture (Peres de Oliveira et al., 2020), but how these processes are connected remains elusive.
To understand NEKL-4/NEK10 function and to determine where and how NEKL-4 regulates ciliary stability, we used CRISPR/Cas9 genomic editing to generate NEKL-4 endogenously tagged fluorescent reporters and targeted mutations. We engineered individual mutations in the active site or the PEST domain of the NEKL-4 endogenously tagged fluorescent reporter strain to generate predicted kinase dead NEKL-4(KD) and non-degradable NEKL-4(PEST∆) mutants. We discovered that NEKL-4 was mitochondria-associated and that nekl-4 mutants displayed longer mitochondria, a higher baseline mitochondrial oxidation state, and suppression of the ccpp-1∆ mutant lifespan extension in response to oxidative stress. NEKL-4(KD) also localized to mitochondria, but ectopically localized to cilia of ccpp-1∆ deglutamylase and ttll-11∆ glutamylase mutants. Additionally, nekl-4(KD) suppressed the B-tubule degradation of ccpp-1∆ cilia and prevented ciliary degeneration. Conversely, the nekl-4(PEST∆) mutant resembled the ccpp-1∆ mutant with Dyf and ultrastructural defects, including B-tubule breaks. The nekl-4(PEST∆) Dyf defect was suppressed by mutation of the depolymerizing kinesin-8 KLP-13/KIF19A, suggesting that NEKL-4 may regulate ciliary stability by regulating this “reader” of the Tubulin Code. Combined, our data indicate that NEKL-4 negatively influences ciliary stability by two distinct but potentially related mechanisms: activation of ciliary kinesins and reduction of mitochondrial stress.
Results
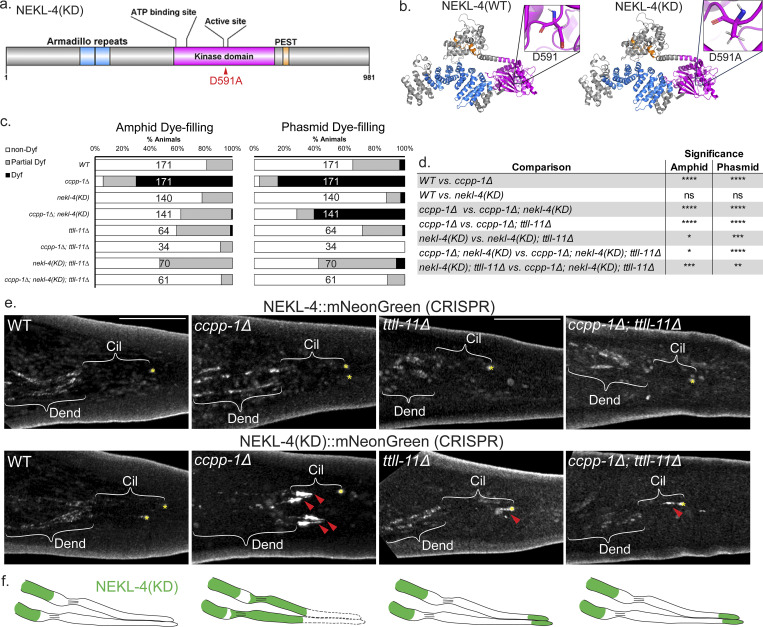
Kinase-dead NEKL-4(KD) suppresses the ccpp-1∆ dye-filling phenotype and ectopically localizes to cilia in ccpp-1∆ and ttll-11∆ mutants
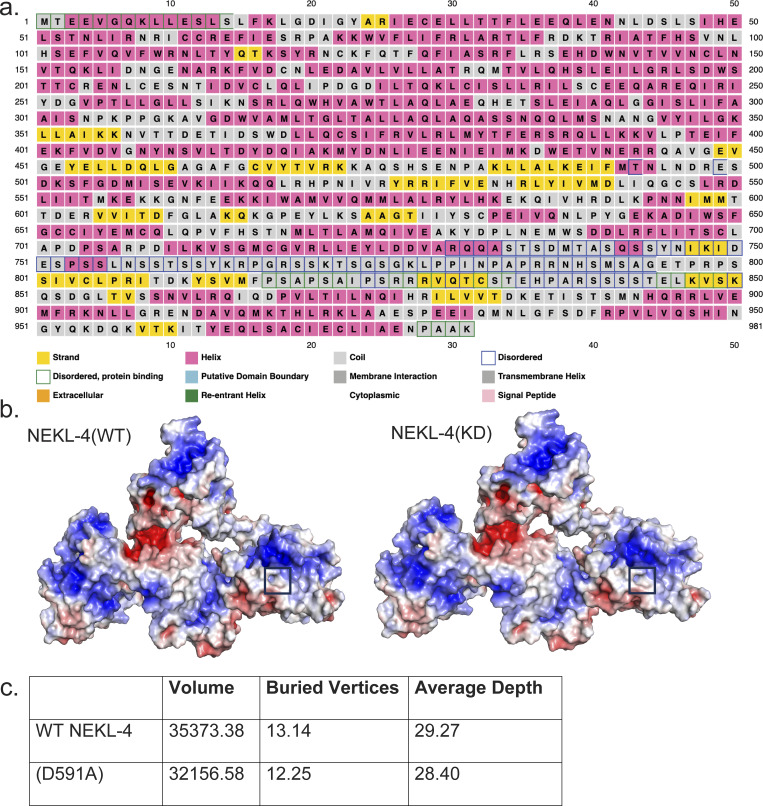
Our previous study was performed with null nekl-4 alleles that were either an early stop codon (my31) or a partial deletion that eliminates the ATP binding site in the kinase domain (tm4910, referred to as nekl-4∆) (Power et al., 2020). We wanted to determine if loss of NEKL-4 kinase activity was required or sufficient for the suppression of the ccpp-1∆ Dyf phenotype. To accomplish this, we used CRISPR/Cas9 to engineer a kinase-dead nekl-4 allele by mutating the active site predicted by InterPro from D591 to A591 (Fig. 1 a). Asp to Ala substitutions have also been used in other studies to create kinase-dead proteins (Lew et al., 2009; Muñoz et al., 2018). This active site-disrupted allele will be referred to as nekl-4(KD), and it was also endogenously tagged with mNeonGreen. This allele was used for most experiments other than those in which the fluorescent tag would interfere. In silico analysis based on the mammalian homolog NEK10 indicated that the D591A substitution does not alter the overall structure of NEKL-4 outside of the active site (Fig. 1 b and Fig S1). We performed a dye-filling assay to assess if nekl-4(KD) suppressed the ccpp-1∆ dye-filling phenotype. We found that nekl-4(KD) suppressed ccpp-1∆ Dyf to a similar extent as nekl-4(my31) and nekl-4∆ (Fig. 1, c and d [Power et al., 2020]). Additionally, Dyf suppression was maintained into later adulthood in ccpp-1∆; nekl-4(KD) animals, even as the Dyf phenotype worsened for ccpp-1∆ mutants, suggesting that loss of nekl-4 function protects neuronal cilia from age-related degeneration (Fig. S2 a). We observed some differences in Dyf suppression between the amphid and phasmid cilia. However, with respect to other phenotypes, amphid and phasmid cilia were similar. The amphid sensory organ houses 12 sensory neurons, which cannot easily be distinguished using live confocal imaging. The phasmid cilia are structurally comparable with amphids (Tran et al., 2023) and are easily distinguished as they are the only ciliated neurons in the posterior area of the worm other than the oxygen-sensing PQR neuron. Thus, for further experiments requiring live imaging, the phasmid neurons were used.
Figure 1.
Loss of NEKL-4 kinase activity suppresses ccpp-1∆ dye-filling defects (Dyf), and NEKL-4(KD) localizes to cilia in ccpp-1∆, ttll-11∆, and ccpp-1∆; ttll-11∆ mutant animals. (a) Schematic diagram of NEKL-4 domains showing the location of the D591A active site mutation. (b) Full-length NEKL-4 and NEKL-4(KD) predicted protein structures based on the amino-acid sequence and homolog structure information. Magenta = kinase domain, blue = armadillo repeat domain, orange = PEST sequence. The positions of the active site residue D591 and the mutant D591A are highlighted. (c) Dye-filling assays of Day 2 adult animals. (d) Significance of relevant comparisons in c. * indicates P ≤ 0.05, ** indicates P ≤ 0.01, *** indicates P ≤ 0.001, **** indicates P ≤ 0.0001 by Kruskal–Wallis one-way ANOVA with post hoc Dunn’s correction for multiple comparisons. (e) Confocal images of endogenously-tagged NEKL-4::mNeonGreen in the phasmid neurons. Scale bar = 10 µm. Cil = cilia, Dend = dendrites, red arrowhead = NEKL-4 ciliary localization, * = phasmid pore. (f) Schematics of NEKL-4(KD) localization in WT, ccpp-1∆, ttll-11∆, and ccpp-1∆; ttll-11∆ mutants. Dashed lines in the ccpp-1∆ panel represent ciliary degeneration.
Figure S1.
In silico modeling of NEKL-4 and NEKL-4(KD). (a) Secondary structure prediction and prediction of intrinsically disordered regions of NEKL-4. (b) Surface Electrostatic Potential of NEKL-4 and NEKL-4(KD). Blue = Positive, Red = Negative (−5 to +5 kT/e). Boxed area is residue D591 or D591A. (c) Cleft analysis of NEKL-4 and NEKL-4(KD).
Figure S2.
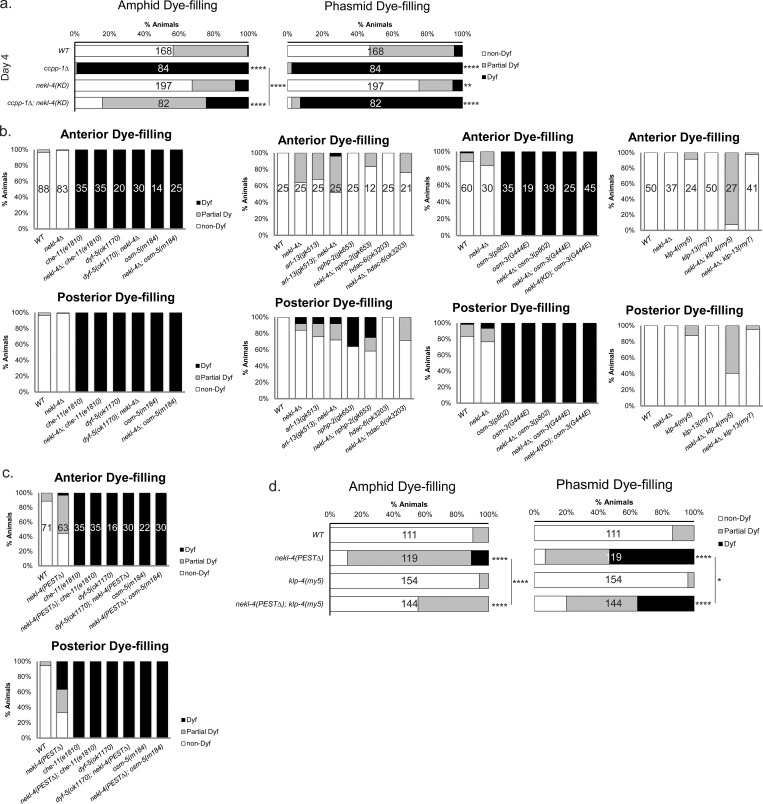
Additional dye-filling data for nekl-4(KD), nekl-4∆, and nekl-4(PEST∆) mutants. (a) nekl-4(KD) mutation continues to suppress the ccpp-1∆ dye-filling defect into later adulthood (Day 4). (b) Negative dye-filling interactions for nekl-4∆ and nekl-4(KD). (c) Negative dye-filling interactions for nekl-4(PEST∆). (d) Mutation of kinesin-3 KLP-4/KIF13 suppresses the nekl-4(PEST∆) Dyf phenotype. For all panels, * indicates P ≤ 0.05, ** indicates P ≤ 0.01, **** indicates P ≤ 0.0001 by Kruskal–Wallis one-way ANOVA with post hoc Dunn’s correction for multiple comparisons.
Next, we examined wild-type NEKL-4 and NEKL-4(KD) localization in ccpp-1(+) and ccpp-1∆ mutant backgrounds (Fig. 1 e). NEKL-4::mNG was expressed in most or all ciliated neurons and localized to dendrites, cell bodies, and axons, but was undetectable in cilia, consistent with previous results (Power et al., 2020). A similar distribution of NEKL-4::mNG was also observed in a ccpp-1∆ mutant background (Fig. 1 e). In the ccpp-1(+) background, NEKL-4(KD) resembled the NEKL-4 localization pattern, though the fluorescence was slightly dimmer. Intriguingly, in a ccpp-1∆ mutant background, NEKL-4(KD) exhibited a novel phenotype-localization to cilia in addition to the other parts of the neuron (Fig. 1, e and f). In hyperglutamylated ccpp-1∆ mutants, NEKL-4(KD) localization appeared restricted to the proximal region of the cilium. Notably, NEKL-4(KD) ectopic ciliary localization occurred specifically in this ccpp-1∆ Dyf-suppressed mutant, whereas the wild-type NEKL-4 protein did not show such ectopic localization. These observations suggest that the microtubule glutamylation state affects NEKL-4 localization.
To test whether loss of glutamylation affected NEKL-4 distribution, we examined NEKL-4 and NEKL-4(KD) in a ttll-11∆ glutamylase mutant background. In both ttll-11∆ and ccpp-1∆; ttll-11∆ mutants, NEKL-4 localization was similar to the WT ccpp-1(+); ttll-11(+) background (Fig. 1 e). In contrast, NEKL-4(KD) ectopically localized to the distal region of cilia of ttll-11∆ single and ccpp-1∆; ttll-11∆ double mutants. These results are consistent with perturbed microtubule glutamylation states affecting the ciliary localization of NEKL-4(KD). Additionally, the region of the cilium where NEKL-4(KD) ectopically located varied based on hyperglutamylation (proximal region in ccpp-1∆ single mutant) or hypoglutamylation (distal region in ttll-11∆ single and ccpp-1∆; ttll-11∆ double mutants).
Since nekl-4(KD) suppressed the ccpp-1∆ Dyf, we hypothesized that nekl-4 may genetically interact with other components of the glutamylation/deglutamylation machinery. We performed dye-filling assays on double and triple mutants of nekl-4(KD), ccpp-1∆, and glutamylase mutant ttll-11∆, which lack detectable ciliary glutamylation (Power et al., 2020). As published previously, ttll-11∆ was not Dyf in comparison with WT, and it suppressed the Dyf phenotype of ccpp-1∆ (Fig. 1, c and d). We found that the nekl-4(KD); ttll-11∆ double mutant dye-filling phenotype resembled the ttll-11∆ single mutant and that ccpp-1∆; nekl-4(KD); ttll-11∆ triple mutants were similar to ccpp-1∆; ttll-11∆ double mutants. However, the nekl-4(KD); ttll-11∆ mutants differed from the nekl-4(KD) mutant and ccpp-1∆; nekl-4(KD); ttll-11∆ triple mutants differed from both ccpp-1∆; nekl-4(KD) and nekl-4(KD); ttll-11∆ double mutants. Hence, the nekl-4(KD) mutation interacts with ttll-11∆ with respect to dye filling.
The nekl-4(KD) mutation does not directly affect ciliary glutamylation state
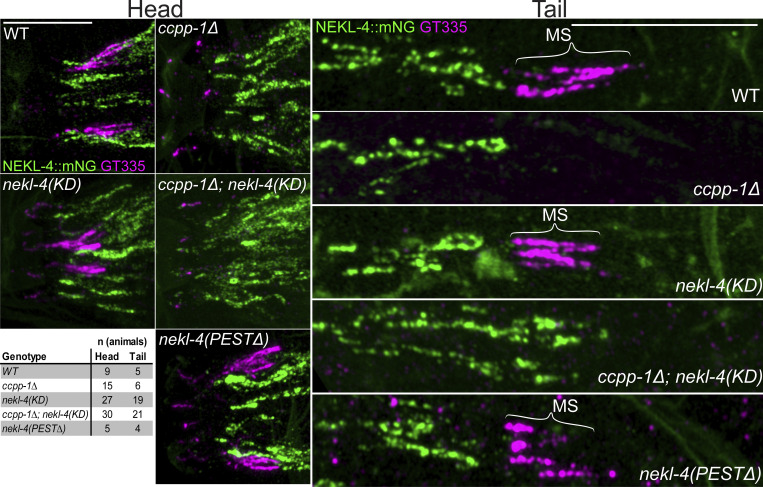
To determine if our nekl-4(KD) allele affected microtubule glutamylation, we visualized glutamylation in the phasmid cilia. We used GT335, a monoclonal antibody that detects branch-point glutamylation and labels the doublet microtubule region of C. elegans cilia (Wolff et al., 1992; Kimura et al., 2010; Wloga et al., 2017; O’Hagan et al., 2017). We previously showed that ccpp-1∆ deglutamylase mutants, contrary to what was expected, do not stain with GT335 in the amphid cilia, likely due to the loss of doublet microtubules and ciliary degeneration (O’Hagan et al., 2017; Power et al., 2020). We found that both nekl-4(my31) and nekl-4∆ stain with GT335, similarly to WT, indicating that loss of nekl-4 function did not affect glutamylation and did not suppress the glutamylation defects in ccpp-1∆ amphid cilia (Power et al., 2020). Similarly, nekl-4(KD) did not affect glutamylation in the amphid and phasmid cilia and did not suppress the loss of GT335 staining in ccpp-1∆ mutants (Fig. S3). nekl-4(PEST∆) mutant cilia also stained with GT335, but in a discontinuous pattern that appeared shorter than WT cilia, consistent with the dye-filling phenotype. This result is consistent with nekl-4(KD) suppression of ccpp-1∆ Dyf by a glutamylation-independent mechanism.
Figure S3.
Loss of NEKL-4 kinase activity does not suppress the loss of GT335 staining in ccpp-1∆ mutant amphid and phasmid cilia. Images of endogenously-tagged NEKL-4::mNeonGreen and GT335, a monoclonal antibody that detects branch point glutamylation, in the amphid, labial, and cephalic cilia. In most images, labial and cephalic cilia are stained but not visible since the images are adjusted to best show the amphids, which stain much brighter. Scale bars = 10 µm. MS = middle segment.
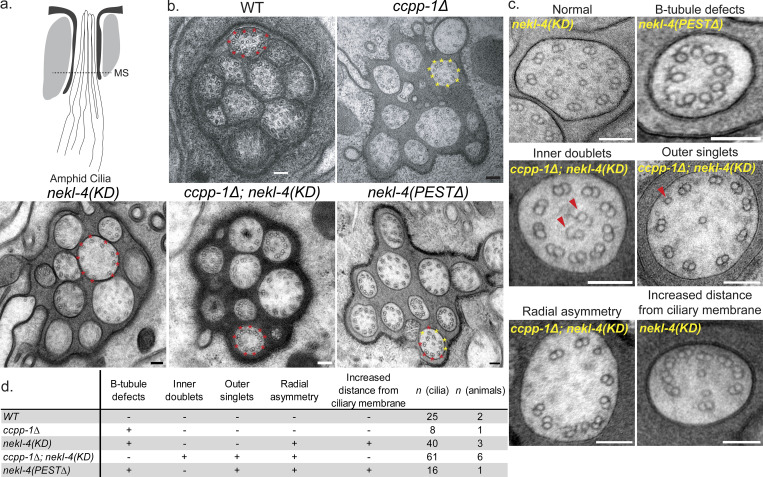
The nekl-4(KD) mutation suppresses B-tubule instability in ccpp-1∆ cilia and causes other axonemal microtubule abnormalities
NEKL-4(KD) did not impact ciliary glutamylation, yet ectopically localized to abnormally glutamylated cilia and suppressed the ccpp-1∆ Dyf phenotype. Therefore, we examined the microtubule ultrastructure of nekl-4(KD) and ccpp-1∆; nekl-4(KD) amphid channel cilia (Fig. 2, a and b). The WT amphid channel cilium contains nine microtubule doublets composed of complete A-tubules and incomplete B-tubules and a variable amount of microtubule singlets in the center. In adult ccpp-1∆ mutant amphid cilia, microtubule doublets typically have open, hook-shaped B-tubules, indicating that B-tubule stability is reduced due to hyperglutamylation (O’Hagan et al., 2011). We rarely observed abnormal B-tubules in nekl-4(KD) mutants, consistent with normal localization of doublet region-exclusive kinesin-2, IFT protein CHE-13/IFT57, and kinesin OSM-3/KIF17 in nekl-4 mutants (Fig. S4). In ccpp-1∆; nekl-4(KD) double mutants, the number of open B-tubule doublets was reduced in comparison with ccpp-1∆ (Fig. 2 b). In ccpp-1∆; nekl-4(KD), we observed ectopic microtubule singlets in the doublet region and ectopic doublets in the singlet region of some cilia (Fig. 2, c and d), similar to mutants of Tubulin Code readers MAPK mapk-15 and homodimeric kinesin-2 osm-3 (Xie et al., 2020; Dobbelaere et al., 2023). We also observed additional ciliary defects in nekl-4(KD), ccpp-1∆; nekl-4(KD), and nekl-4(PEST∆) mutants, including loss of radial symmetry and increased, irregular distances between the doublets and the ciliary membrane (Fig. 2 c). In general, these defects were not as severe as those in ccpp-1∆ mutants and did not cause a dye-filling phenotype, with the exception of the nekl-4(PEST∆) mutant described below.
Figure 2.
nekl-4(KD) mutation suppresses B-tubule instability in ccpp-1∆ amphid cilia. (a) Diagram of the amphid cilia indicating the position of the cross-sections examined. MS = middle segment. (b) Examples of microtubule doublet structure in WT, ccpp-1∆, nekl-4(KD), ccpp-1∆; nekl-4(KD), and nekl-4(PEST∆) mutant amphid channels. The WT and ccpp-1∆ panels are reproduced from O’Hagan et al. (2011) and show a male amphid. All other genotypes show a hermaphrodite amphid. Red * = normal microtubule doublet, yellow * = microtubule doublet with broken or missing B-tubule in a representative cilium. Scale bars = 100 nm. (c) Examples of phenotypes seen in nekl-4(KD), ccpp-1∆; nekl-4(KD), and nekl-4(PEST∆) mutant cilia. Scale bars = 100 nm. (d) Table of phenotypes observed in each genotype and numbers of cilia and animals analyzed. The WT and ccpp-1∆ data are reproduced from O’Hagan et al. (2011).
Figure S4.
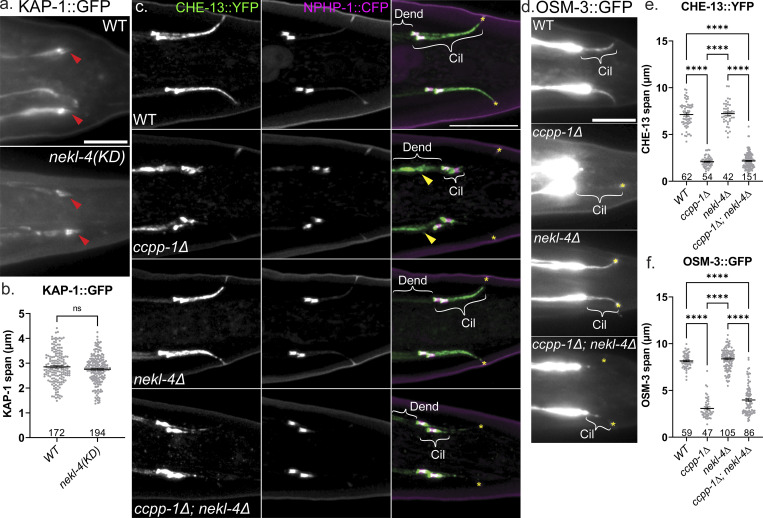
KAP-1, CHE-13, and OSM-3 spans are not altered in nekl-4 mutants. (a) Examples of KAP-1::GFP localization. Red arrowhead = phasmid cilia. Scale bar = 10 µm. (b) Quantification of KAP-1 span in the phasmid cilia. Not significant by Mann–Whitney test. (c) Confocal images of CHE-13 and NPHP-1, a transition zone protein. (d) Widefield images of OSM-3 in the phasmid cilia. (e) Quantification of the span of CHE-13 in the phasmid cilia. (f) Quantification of the span of OSM-3 in the phasmid cilia. For c and d, Cil = cilia, Dend = dendrites, yellow arrowhead = dendritic CHE-13 accumulation, * = phasmid pore. Scale bar = 10 µm. For e and f, mean ± SEM; **** indicates P ≤ 0.0001 by Kruskal–Wallis one-way ANOVA with post hoc Dunn’s correction for multiple comparisons. Note that in c, the dendritic accumulation of CHE-13 in ccpp-∆ mutants is suppressed by nekl-4∆.
The nekl-4(PEST∆) mutation causes progressive ciliary degeneration
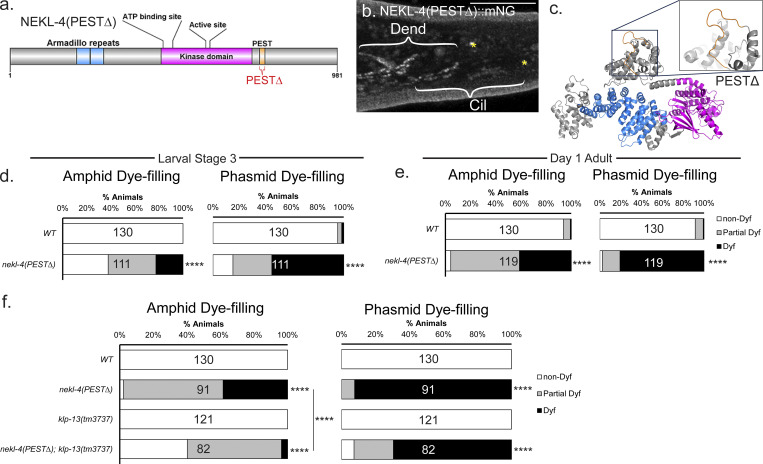
NEKL-4 influences ciliary stability, though the mechanism by which this function is performed remains unclear. In Tetrahymena, OE of NIMA-related kinases causes ciliary shortening (Wloga et al., 2006). In C. elegans, NEKL-4 OE causes a Dyf phenotype in a kinase activity-dependent manner, and nekl-4 is subject to regulation via RNA editing (Power et al., 2020; Li et al., 2021a). NEKL-4 contains a PEST sequence, which is usually found in proteins whose expression levels are rapidly and tightly controlled by ubiquitin-mediated degradation (Rechsteiner and Rogers, 1996; Power et al., 2020). NEK10 also contains a PEST sequence and is regulated by ubiquitination (Porpora et al., 2018). Based on these findings, we hypothesized that NEKL-4 may be negatively regulated by the PEST motif and that PEST deletion may result in NEKL-4 overactivity and ciliary microtubule instability. To test this hypothesis, we generated a CRISPR/Cas9 mutant with a deletion of the predicted PEST domain, which we refer to as nekl-4(PEST∆) (Fig. 3 a).
Figure 3.
NEKL-4(PEST∆) causes a progressive Dyf phenotype that is suppressed by mutation of kinesin-8 KLP-13/KIF19. (a) Schematic diagram of NEKL-4 showing the location of the PESTΔ mutation. (b) Fluorescence microscope image showing expression of NEKL-4(PEST∆)::mNG. Scale = 10 µm. Cil = cilia, Dend = dendrites, * = phasmid pore. (c) NEKL-4(PEST∆) modeled and predicted protein structure. Magenta = kinase domain, blue = armadillo repeat domain, orange = residues surrounding the removed PEST sequence. The removal of the residues within the PEST domain is highlighted. (d) Dye-filling assays of nekl-4(PEST∆) L3 larvae. (e) Dye-filling assays of nekl-4(PEST∆) Day 1 adults. For d and e, **** indicates P ≤ 0.0001 by Mann–Whitney test. (f) Dye-filling assays of nekl-4(PEST∆); klp-13 Day 1 adults. **** indicates P ≤ 0.0001 by Kruskal–Wallis one-way ANOVA with post hoc Dunn’s correction for multiple comparisons.
NEKL-4(PEST∆)::mNG was still expressed based on imaging of the endogenous mNeonGreen tag in the strain background and localized in a pattern similar to WT NEKL-4::mNG (Fig. 3 b). In silico analysis indicated that this mutation did not alter the overall structure of the protein outside of the removal of the PEST sequence (Fig. 3 c).
To determine the effect of the PEST sequence on NEKL-4 function, we performed dye-filling assays on nekl-4(PEST∆) stage 3 larvae (L3) and day one adults. We hypothesized that, similar to ccpp-1∆, nekl-4(PEST∆) might cause age-dependent ciliary degeneration. Wild-type L3-stage animals have fully developed amphid and phasmid neurons that are able to dye-fill (O’Hagan et al., 2011). At the L3 larval stage, nekl-4(PEST∆) mutants were Dyf, and the severity of this defect increased as animals reached adulthood (Fig. 3, d and e). nekl-4(PEST∆) phenocopied nekl-4(OE), consistent with dysregulated NEKL-4 causing ciliary instability. Supporting this hypothesis, we also found that nekl-4(PEST∆) amphid cilia displayed more broken or defective microtubule doublets compared with WT (Fig. 2, b–d).
Our data indicated that NEKL-4 negatively regulates ciliary stability through an unknown mechanism. To explore mechanisms by which NEKL-4 may influence ciliary stability, we considered potential NEKL-4 substrates identified by phosphoproteomic analysis of NEK10-depleted cultured cells (Chivukula et al., 2020). We focused on proteins identified in cultured cells that have both a C. elegans ortholog and a predicted or known ciliary function in mammals or C. elegans: OSM-5/IFT88, DYF-5/MAK, CHE-11/IFT140, KLP-4/KIF13, and KLP-13/KIF19 (Table S1). OSM-5, DYF-5, and CHE-11 have been extensively characterized as regulators of C. elegans ciliogenesis and cilium assembly (Qin et al., 2001; Burghoorn et al., 2007; Mijalkovic et al., 2018). Kinesin-3 KLP-4 is involved in neurotransmitter trafficking; KLP-13 is a depolymerizing kinesin-8 that negatively regulates cilium length (Magaletta et al., 2019; Park et al., 2021). We therefore examined genetic interactions between nekl-4 and these candidates. We found that the null nekl-4∆ allele did not modify the dye-filling phenotypes of any of the mutants, with the exception of a mild synthetic Dyf interaction with klp-4 (Fig. S2 b). nekl-4∆ also did not modify the dye-filling phenotypes of other genes implicated in the microtubule double region and B-tubule stability, namely arl-13, nphp-2, and hdac-6 (Warburton-Pitt et al., 2014), or the Dyf of ciliary kinesin-2 loss-of-function mutation osm-3(p802) or gain-of-function mutation osm-3(sa125) ([Perkins et al., 1986; Imanishi et al., 2006]; Fig. S2 b).
Next, we hypothesized that, since nekl-4(PEST∆) mutants may be Dyf due to NEKL-4 overactivity, loss of a NEKL-4 substrate may suppress this defect. We found no genetic interaction between nekl-4(PEST∆) and osm-5, che-11, or dyf-5 (Fig. S2 c). However, mutation of either klp-4 or klp-13 suppressed the Dyf defect of nekl-4(PEST∆) (Fig. 3 f and Fig. S2 d). KLP-13 is a cilia-localized microtubule-depolymerizing kinesin, and loss of klp-13 function results in a longer ciliary axoneme (Park et al., 2021). Based on these observations, we propose that NEKL-4 negatively regulates ciliary stability via interaction with ciliary kinesins, possibly promoting KLP-13 depolymerizing activity.
NEKL-4 is mitochondria-associated and influences mitochondrial morphology
Our results thus far indicate that NEKL-4 influences ciliary stability despite not localizing to cilia or affecting ciliary glutamylation. To begin determining the likely site of NEKL-4 function, we investigated the localization and function of mammalian CCP1 and NEK10. In neurons, loss of CCP1 results in fragmented mitochondria morphology and impaired mitochondrial transport (Gilmore-Hall et al., 2019). NEK10 is localized to mitochondria and promotes mitochondrial DNA integrity, mitochondrial respiration, and mitochondria fusion (Peres de Oliveira et al., 2020). We then sought to determine if CCPP-1 or NEKL-4 operated by similar mechanisms in C. elegans.
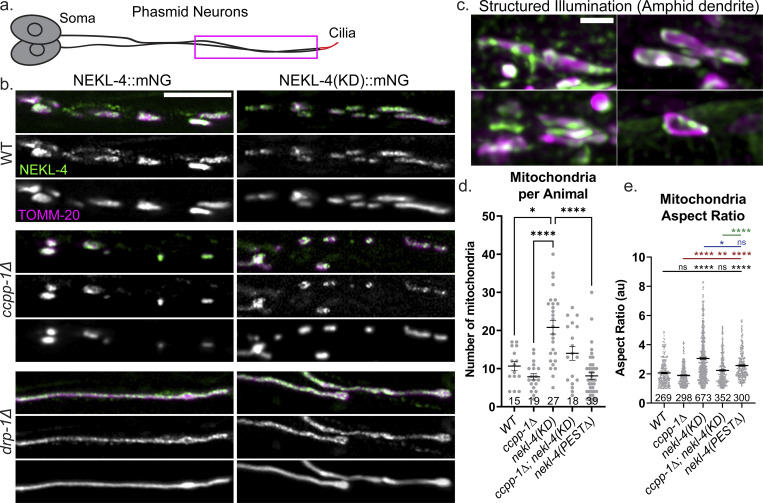
To determine if NEKL-4 is mitochondria-associated, we examined localization with a mitochondrial marker. We created an extrachromosomal transgene (osm-5p::TOMM-20::tagRFP) that expresses an outer mitochondrial membrane marker in the ciliated neurons and examined the colocalization of TOMM-20 with NEKL-4::mNG in the phasmid dendrites (Fig. 4 a). We found that the majority of NEKL-4 protein was associated with mitochondria (Fig. 4 b), though there was a small amount of NEKL-4 that was unassociated with mitochondria. NEKL-4::mNG was also cotransported with mitochondria in dendrites (Video 1). Mutation of the NEKL-4 active site nekl-4(KD) or ccpp-1∆ did not affect mitochondrial association (Fig. 4 b). To confirm the specificity of mitochondrial association, we imaged both markers in a dynamin-related protein drp-1Δ mutant background, which is defective in mitochondrial fission and in which mitochondria are hyperfused (Labrousse et al., 1999). In the drp-1Δ mutant, NEKL-4 and NEKL-4(KD) remained associated with mitochondria despite the drastic change in morphology, confirming the specificity of the association (Fig. 4 b). We predict that NEKL-4 is associated with the outer mitochondrial membrane, but is not embedded within it, based on structured illumination microscopy data (Fig. 4 c) and the lack of a predicted mitochondria localization signal or transmembrane domain within the protein. NEKL-4 mitochondrial localization in C. elegans ciliated sensory neurons is consistent with mammalian NEK10, which is mitochondria-associated in cultured mammalian cells (Peres de Oliveira et al., 2020).
Figure 4.
NEKL-4 is mitochondria-associated and influences mitochondrial morphology. (a) Cartoon of one set of C. elegans phasmid neurons. Imaged area is boxed in magenta. (b) Confocal images of mutants showing NEKL-4::mNG and TOMM-20::tagRFP labeling the outer mitochondrial membrane. Scale bar = 5 µm. (c) Representative SIM images of NEKL-4::mNG and TOMM-20 colocalization on individual mitochondria in the amphid dendrites. Scale bar = 1 µm. (d and e) Quantification of the number of mitochondria in the phasmid dendrites per animal and the aspect ratio of each mitochondrion (length/width). Mean ± SEM; * indicates P ≤ 0.05, ** indicates P ≤ 0.01, **** indicates P ≤ 0.0001 by Kruskal–Wallis one-way ANOVA with post hoc Dunn’s correction for multiple comparisons.
Video 1.
NEKL-4 and mitochondria are co-transported in dendrites. Retrograde movement is shown beginning at the amphid distal dendrite (left, green arrow). Green is NEKL-4::mNG, magenta is TOMM-20::tagRFP. Scale bar = 10 µm. Frames acquired every 2.5 s, played back at 10 fps.
To examine mitochondrial morphological differences in nekl-4(KD) and ccpp-1∆ animals, we quantified the number and shape of mitochondria in the phasmid dendrites. We found that ccpp-1∆ mutant mitochondria appeared slightly fewer in number than WT and that nekl-4(KD) mutant mitochondria were significantly more numerous (Fig. 4 d). The number of mitochondria in the ccpp-1∆; nekl-4(KD) double mutant was similar to WT. In the nekl-4(PEST∆) mutant, mitochondria were similar in length to the ccpp-1∆ mutant. Additionally, we analyzed mitochondria morphology and measured their individual aspect ratios, which is the measurement of length divided by width. ccpp-1∆ mutant mitochondria appeared slightly more rounded than WT, although there was no significant difference. nekl-4(KD) mutant mitochondria were significantly longer than WT, ccpp-1∆, and ccpp-1∆; nekl-4(KD) mutants (Fig. 4 e). ccpp-1∆; nekl-4(KD) double mutant mitochondria were similar to WT, and nekl-4(PEST∆) mutant mitochondria were longer than WT but shorter than nekl-4(KD) mutants. Based on these findings, we conclude that nekl-4 and ccpp-1 have opposing effects on mitochondrial morphology and number.
NEKL-4 reduces oxidative stress in the ciliated neurons
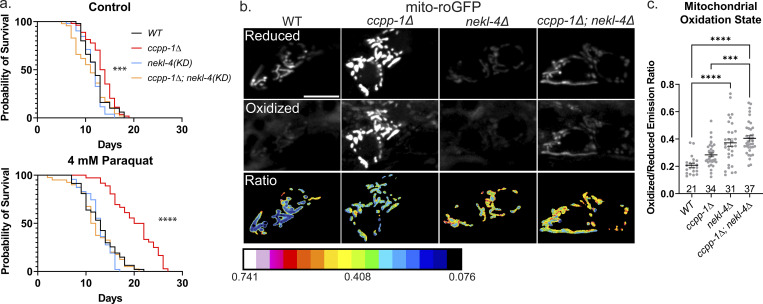
Once we established that nekl-4 and ccpp-1 mutations affect mitochondria morphology and localization, we then sought to determine if CCPP-1 and/or NEKL-4 were required for mitochondria function. Mitochondria are subject to oxidative stress, which affects ciliogenesis through multiple signaling pathways (Han et al., 2021; Bae et al., 2023; Ignatenko et al., 2023). Additionally, C. elegans ciliated chemosensory neurons may influence longevity through oxidative stress-induced signaling, and cilia mutants were resistant to mitochondrial stress and have an increased lifespan (Apfeld and Kenyon, 1999; Fujii et al., 2004). To determine if nekl-4(KD) and/or ccpp-1∆ mutants were sensitive to oxidative stress, we performed an adult chronic paraquat (PQ) assay. PQ is a compound that induces oxidative stress through the generation of reactive oxygen species (ROS). We plated Day 1 adult hermaphrodites on plates containing 4 mM PQ and measured the lifespan of individuals. On control plates, the lifespans of ccpp-1∆ and ccpp-1∆; nekl-4(KD) mutants were slightly longer and shorter than WT, respectively (Fig. 5 a). On PQ, the lifespan of all mutants was extended, consistent with previous studies showing an increase in longevity when exposed to mitochondrial stress (Maglioni et al., 2014; Schaar et al., 2015). On PQ, survival curves for nekl-4(KD) and ccpp-1∆; nekl-4(KD) mutants were comparable to WT, while the lifespan of ccpp-1∆ mutants was significantly extended (Fig. 5 a).
Figure 5.
ccpp-1∆ and nekl-4 mutations affect both sensitivity to oxidative stress and baseline oxidative stress in C. elegans ciliated sensory neurons. (a) Survival curves of control (top) and paraquat-exposed (bottom) worms, where Day 0 represents when Day 1 adult hermaphrodites were placed on plates. Control n = 50, 54, 52, 47; PQ n = 47, 36, 47, 40; *** indicates P ≤ 0.001, **** indicates P ≤ 0.0001 by Log-rank Mantel-Cox test. (b) Uniformly adjusted images of phasmid soma in the “reduced” channel (top) and “oxidized” channel (middle), as well as the ratio of oxidized/reduced (bottom). Scale bar = 5 µm. (c) Quantification of oxidized/reduced emission ratios, where one point represents one set of phasmid soma. Mean ± SEM; *** indicates P ≤ 0.001, **** indicates P ≤ 0.0001 by Kruskal–Wallis one-way ANOVA with post hoc Dunn’s correction for multiple comparisons.
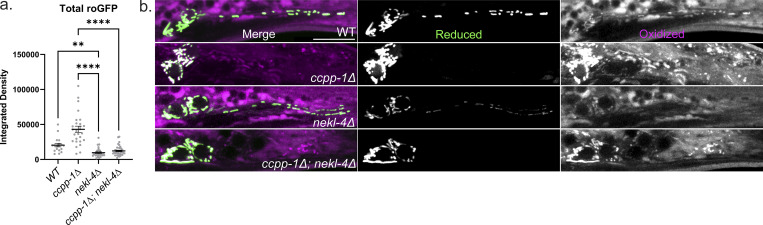
To further assess neuronal mitochondria stress in nekl-4 and ccpp-1∆ mutants, we visualized the oxidation state of mitochondria in the phasmid neurons using roGFP, a redox-sensitive GFP biosensor (Hanson et al., 2004). For this assay, we used the nekl-4∆ null allele to avoid interference from the mNeonGreen-tagged nekl-4(KD). We found that ccpp-1∆, nekl-4∆, and ccpp-1∆; nekl-4∆ mutant mitochondria all had an elevated oxidation state compared with WT, indicating that under baseline conditions, mitochondria in nekl-4∆, ccpp-1∆, and double mutants were under elevated oxidative stress (Fig. 5 b). Interestingly, though the lifespan extension phenotype of ccpp-1∆ under oxidative stress was suppressed by nekl-4∆ mutation, the same interaction was not observed when examining baseline oxidative stress. In fact, the oxidation state was higher in nekl-4∆ mutants than ccpp-1∆ mutants, and double mutants were similar to nekl-4∆ single mutants (Fig. 5 c). Of note, total levels of roGFP were slightly elevated in ccpp-1∆ mutants and significantly decreased in nekl-4∆ and ccpp-1∆; nekl-4∆ mutants, though the reason for this phenomenon is unclear (Fig. S5 a). Additionally, we did not observe dendritic mitochondria in ccpp-1∆ or ccpp-1∆; nekl-4∆ mutants, which may differ from our results in Fig. 4 due to differences in expression between the markers (Fig. S5 b).
Figure S5.
Differences in roGFP levels and dendritic localization in ccpp-1∆ and nekl-4∆ mutants. (a) Total roGFP signal measured in phasmid soma, calculated by adding the integrated densities of the 488 and 405 nm channels. Mean ± SEM; ** indicates P ≤ 0.01, **** indicates P ≤ 0.0001 by Kruskal–Wallis one-way ANOVA with post hoc Dunn’s correction for multiple comparisons. (b) Dendritic mitochondrial localization is defective in ccpp-1∆ and ccpp-1∆; nekl-4∆ mutants. Uncropped images of roGFP in the phasmid neurons. Scale bar = 10 µm.
Overall, our results present a fascinating finding: NEKL-4 does not localize to cilia, yet has a cilia-destabilizing function as well as a mitochondrial function. Additionally, CCPP-1 is a regulator of mitochondria morphology and transport, in addition to its role in ciliary microtubule stability. NEKL-4 and CCPP-1 appear to mainly perform opposing functions in both cilia and mitochondria, and the question of how these two phenomena are related poses an exciting new direction for future studies.
Discussion
Our previous genetic analyses indicated that NEKL-4 regulates ciliary stability in a manner downstream or independent of the glutamylation/deglutamylation machinery. In this study, we explored the NEKL-4 mechanism of action and discovered an unexpected role in mitochondrial homeostasis in the ciliated neurons. Recent studies have revealed extensive connections between cilia stability, neurodegeneration, and oxidative stress, and our study provides evidence of NEKL-4 involvement in the intersection of these processes. In this work, we were not able to directly connect the ciliary and mitochondrial phenotypes caused by mutation of ccpp-1 and nekl-4, and further research is necessary for a fully mechanistic understanding. We propose that NEKL-4 regulates multiple processes within the ciliated neurons via phosphorylation of multiple proteins that play diverse roles in ciliary stability and mitochondrial function.
NEKL-4 and the tubulin code
NEK kinases are regulators of microtubule dynamics, particularly during the cell cycle and ciliogenesis (Li et al., 2021b). One mechanism by which NEK kinases may act is likely to involve the tubulin code. Tubulin PTMs such as polyglutamylation influence microtubule stability and intracellular transport (O’Hagan et al., 2022; Zocchi et al., 2023). TTLL glutamylases and CCP deglutamylases act as writers and erasers, while motors and microtubule-associated proteins act as readers of the Tubulin Code (Verhey and Gaertig, 2007). NEKs may regulate writers, erasers, and/or readers of the tubulin code. For example, NEK5 negatively regulates TTLL4 through phosphorylation, and possibly other TTLLs (Melo-Hanchuk and Kobarg, 2021). In the trypanosomatid Crithidia fasciculata, the NIMA-related kinase CfNek performs polyglutamylase activity, directly modifying the Tubulin Code (Westermann and Weber, 2002). NEK kinases may phosphorylate tubulin. For example, NEK1 interacts with and phosphorylates α-tubulin, with loss of function variants being a major genetic cause of amyotrophic lateral sclerosis (Mann et al., 2023). NEK kinases may also regulate microtubule-associated proteins and motors, leading to changes in activity or stability. nekl-4 mutations do not affect glutamylation and do not modify the GT335 staining defect of ccpp-1∆ ([Power et al., 2020], Fig. S3). While NEKL-4 does not directly modify the glutamylation state of ciliary microtubules, we propose that NEKL-4 regulates other components of the tubulin code such as kinesins and microtubule-associated proteins. For example, the open B tubules in the ccpp-1∆ and nekl-4(PEST∆) mutants might result from the lack of delivery or incorporation of inner junction proteins such as FAP20 into the cilia (Chen et al., 2023). Determining the substrates of NEKL-4 is an important next step.
NEKL-4(KD) ectopic ciliary localization in glutamylation mutants and NEKL-4(PEST∆) genetic interaction with KLP-13
Wild-type NEKL-4::mNG and NEKL-4(KD)::mNG are notably absent from cilia, raising the question of how this NEK kinase regulates ciliary stability. One clue comes from the localization of NEKL-4(KD) in glutamylation-defective mutants. In ccpp-1∆ and ttll-11∆ single mutants and ccpp-1∆; ttll-11∆ double mutants, NEKL-4(KD) ectopically localizes to cilia. Though protein modeling did not reveal a significant difference in protein folding or surface electrostatic potential between NEKL-4(WT) and NEKL-4(KD) (Fig. S1), there is a predicted reduction in cleft volume and depth in NEKL-4(KD). We hypothesize that this change in the protein-binding cleft may influence NEKL-4 interactions with binding partners as well as substrates. Kinase-dead proteins are predicted to be unable to release their substrates (He et al., 2009; Hewezi et al., 2015). If this is the case for NEKL-4(KD), then NEKL-4(KD) may be carried into cilia in ccpp-1∆ and ttll-11∆ mutants via a “stuck” substrate, such as kinesin-8 KIF19/KLP-13. KIF19 is a predicted NEK10 substrate (Chivukula et al., 2020). In C. elegans phasmid neurons, KLP-13 localizes to the tips of cilia, where it likely performs its microtubule depolymerizing function (Park et al., 2021). In mammalian cells, KIF19A localizes to ciliary tips and performs a dual function as both a microtubule plus-end-directed motor and a depolymerizing kinesin to regulate ciliary length (Niwa et al., 2012). Since klp-13 mutation suppresses the ciliary degeneration of nekl-4(PEST∆) mutants, we hypothesize that NEKL-4 positively regulates KLP-13 depolymerizing activity to negatively regulate microtubule stability (see model in Fig. 6).
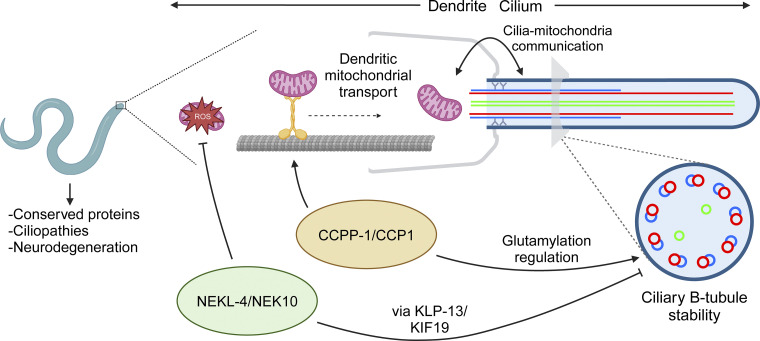
Figure 6.
Model for NEKL-4 function in relation to cilia, mitochondria, and CCPP-1. NEKL-4 functions in the ciliated neurons to reduce mitochondrial oxidative stress and destabilize ciliary B-tubules through the action of ciliary kinesins such as KLP-13. This ciliary function opposes the B-tubule stabilizing action of CCPP-1 through a pathway distinct from glutamylation regulation. CCPP-1 also promotes dendritic transport in the ciliated neurons, specifically the transport of mitochondria. Created with https://BioRender.com (license BE26DR2HV3).
A potential role of NEKL-4 in mediating cilia–mitochondria communication
NEKL-4(WT) did not detectably localize to cilia in any genetic backgrounds examined. Instead, NEKL-4 was mitochondria-associated, colocalized, and co-transported with the TOMM-20 outer mitochondrial membrane marker. In ciliated sensory neurons, NEKL-4 modulated both oxidative stress response and mitochondrial morphology. Similar to our findings for ccpp-1∆, CCP1 knockdown in cultured mammalian neurons results in mitochondria fragmentation and axonal transport defects (Magiera et al., 2018; Gilmore-Hall et al., 2019; Bodakuntla et al., 2020). Since we found a reduction in dendritic mitochondria in ccpp-1∆ mutants, we postulate that CCPP-1 plays a similar role in promoting mitochondrial transport in C. elegans’ ciliated neuron dendrites.
In different mammalian cell types, mitochondrial stress may promote either ciliogenesis or ciliary degeneration, but in general, increased ciliogenesis improves survival of cells when exposed to ROS (Han et al., 2021; Moruzzi et al., 2022; Ignatenko et al., 2023; Bear and Caspary, 2023; Bae et al., 2023). Cilia stability is influenced in opposing directions by mitochondrial dynamics: mitochondrial fragmentation, which increases ROS, increases ciliogenesis. Conversely, mitochondrial fusion decreases cilia length (Bae et al., 2019). In HeLa cells, NEK10 is mitochondria-localized and interacts with several mitochondrial proteins, the number of which increases in response to DNA damage-induced stress (Peres de Oliveira et al., 2020). The absence of NEK10 negatively impacts mitochondrial respiration and increases ROS, consistent with our findings that nekl-4 mutants experience higher baseline oxidative stress in the ciliated neurons. NEK10 loss-of-function mutation results in shorter mitochondria, in contrast to C. elegans nekl-4(KD) causing longer mitochondria, suggesting differences between NEKL-4 and its ortholog (Peres de Oliveira et al., 2020). Regardless, NEKL-4 and NEK10 appear to play evolutionarily conserved roles in interorganelle communication between cilia and mitochondria (Fig. 6).
Cilia communicate with other organelles to regulate diverse cellular processes. For example, ciliary–nuclear crosstalk is essential for organism development via signaling by Hedgehog (Gigante and Caspary, 2020) and other transcription factors (Nishimura et al., 2018). Cilia and the autophagy machinery share bidirectional communication to regulate ciliogenesis and proteolysis (Wiegering et al., 2019). In the nervous system, cilia are essential for modulating the creation of new synapses (Kumamoto et al., 2012), and, in the brain, cilia physically associate with axons to form synapses (Sheu et al., 2022). Cilia–mitochondria crosstalk also regulates cellular responses to mitochondrial stress (Silva and Cavadas, 2023), indicating that cilia are vital for signaling and sensing oxidative stress. NEKL-4 and CCPP-1 are expressed in the ciliated neurons (O’Hagan et al., 2011; Power et al., 2020) and appear to play a role in whole-organism oxidative stress response, leading us to hypothesize that these proteins act to mediate the communication between cilia and mitochondria, which then influences the signals sent to the rest of the animal by the ciliated neurons in response.
Our work offers insights into the multifunctional protein NEKL-4 and its roles in cilia and mitochondria. NEKL-4 both modulates ciliary stability and promotes mitochondrial homeostasis, similar to mammalian NEK10. Our work adds to the growing number of proteins that directly or indirectly regulate the tubulin code. Additionally, proteins that have ciliary functions may also influence processes in other subcellular locations, such as mitochondria. Further exploration of NEKL-4/NEK10 and other tubulin code-related components will provide insight into the communication between cilia and other organelles as well as the effects this communication may have on neurodegeneration and ciliopathies.
Limitations of study and future directions
Our study is limited to mainly genetic and imaging-based analysis. Future studies are required to understand the mechanistic relationship between mitochondrial defects and microtubules and cilia of ccpp-1∆ and nekl-4 mutants. It is no surprise that a NEKL kinase potentially phosphorylates numerous substrates and plays various roles in a living cell. On the other hand, the impairment of mitochondria will certainly affect the production of ATP, which will no doubt affect the motility of microtubule-based transport. An in vivo ATP biosensor would be useful in determining how mitochondrial function influences microtubule-based transport in dendrites and cilia in mitochondrial and cilia mutants. Additionally, it will be important to study age-related cilia degeneration/regeneration using extended time-lapse imaging studies of mitochondrial transport from the soma to the cilium and intraflagellar transport within cilia. Intraflagellar transport, sensory perception, and metabolism are decreased in aged C. elegans (Zhang et al., 2021). These age-related phenotypes can be countered by upregulation of the DAF-19/RFX transcription factor (Zhang et al., 2021), hinting at potential interventions for degenerating cilia function.
The nekl-4(PEST∆) and nekl-4(KD) mutants provide insight into the possible mechanisms of NEKL-4 function, which merits further study. We do not know the direct substrates of NEKL-4. NEK10 is a dual-specificity kinase that also phosphorylates tyrosine residues in addition to serine/threonine (van de Kooij et al., 2019). We do not know how similar the NEK10 motif is to the NEKL-4 motif, or if the kinesin KLP-13 is a direct substrate of NEKL-4. To identify potential substrates and interactors of NEKL-4, proximity labeling and/or phosphoproteomics could be employed utilizing wild-type NEKL-4 and the kinase-dead mutant. A powerful tool to identify physiological interactions is a forward genetic screen, similar to the genetic suppressor approach we used with the ccpp-1∆ mutant that originally identified NEKL-4 (Power et al., 2020). Since nekl-4(PEST∆) causes a significant Dyf phenotype, this mutant could be used in a dye-filling suppressor screen to identify genetic interactors. Discovery-based approaches will also be powerful to understand the relationship between ciliary signaling and mitochondrial homeostasis.
Materials and methods
C. elegans strains and maintenance
Nematodes were cultured on Nematode Growth Media (NGM) agar plates containing a lawn of OP50 E. coli and incubated at 20°C, as previously described (Brenner, 1974). The strains used are listed in Table S2. The strains used in supplemental material are listed in Table S3. Genetic sequence information was obtained from WormBase (Sternberg et al., 2024).
CRISPR constructs
For the creation of the nekl-4(KD) and nekl-4(PEST∆) CRISPR mutants (my66 and my120), we followed the Mello Lab protocol (Dokshin et al., 2018) and used ssOligos as donors (Table 1). The guide sequences were designed using CRISPOR (Haeussler et al., 2016), and silent mutation sites for PAM modification and diagnosis were located using WatCut silent mutation scanning. For nekl-4(KD), we modified the predicted active site (D591) to alanine, which is expected to eliminate NEKL-4 kinase activity without altering the gross structure of the protein. For nekl-4(PEST∆), we deleted the predicted PEST proteolytic degradation domain (17AA; 748–764, KIDESPSSLNSSTSSYK) using an ssOligo with the sequence encoding those residues removed. This mutant is recessive for the Dyf phenotype. Both mutants were generated in the nekl-4(my51[nekl-4::mNG]) background. CRISPR-generated strains were outcrossed at least 3X before use. For genotyping genetic crosses into strains not in the nekl-4(my51[nekl-4::mNG]) background, primers for NEKL-4::mNG were used for diagnosis of nekl-4(KD) and nekl-4(PEST∆).
Table 1.
Reagents used for generation of nekl-4(KD), nekl-4(PESTΔ), and pha-1 CRISPR mutations
| NEKL-4(KD) ssODN | 5′-CTCATAAGCTTTTATAATCTTTCAGATGATGTTAGCCCTTCGCTATTTGCACAAAGAAAAGCAAATTGTACATCGAGACTTAAAGCCAAATAACATAATGATGACGACAGACGAGCGAG-3′ |
| NEKL-4(KD) crRNA | 5′-UUCUUUGUGCAAAUAGCGAA-3′ |
| PESTΔ ssODN | 5′-TACTTCAGATATGACTGCTTCACAATCTAGTTATAATATCCGTCCTGGAAGGTCGTCAAAGACATCAGGGAGTGGAAAG-3′ |
| PESTΔ crRNA | 5′-GUGGAGGAAUUCAGGGAGGA-3′ |
| pha-1(e2123) ssODN | 5′-GTAATCAAAATACGAATCGAAGACTCAAAAAGAGTATACTGTATGATTACAGATGTTCATCAAGTTATTCATAAATCATTGATAGGTTCAG-3′ |
| pha-1(e2123) crRNA | 5′-AUGUUCAUCAAGUUAUUCAU-3′ |
We also followed the same protocol to recreate the pha-1(e2123) temperature-sensitive point mutation in a nekl-4∆ mutant background since the genes are the same linkage group. We validated that our version of the allele, pha-1(my82[pha-1(e2123)+SnaBI]), is temperature-sensitive before constructing strains or performing experiments with the mutant. Additionally, we inserted a silent mutation to create an additional SnaBI restriction enzyme cut site for diagnosis and differentiation between our my82 mutation and the original e2123 allele (Table 1).
Plasmid construction
For the creation of pKP06 (osm-5p::tomm-20::tagRFP), the tomm-20 coding sequence was fused to a 241 bp promoter from osm-5, which drives expression in all ciliated neurons. Both fragments were amplified from genomic DNA using primers with homology to pPD95.75 containing tagRFP or the other fragment. The stop codon for tomm-20 was removed using the reverse primer. pPD95.75 was amplified using primers with homology to osm-5p or tomm-20. Fragments were joined using Gibson assembly (Table 2).
Table 2.
Reagents used for construction of the osm-5p::TOMM-20::tagRFP marker
| Genomic osm-5p amplification (forward) | 5′-ATGGATACGCTAACAACTTGGGAAATTGAAAGACTCGAAC-3′ |
| Genomic osm-5p amplification (reverse) | 5′-GATTTGTTGAAACCAAGAATTGTGTCCGACATTTTGTATCATTAAGAAAAGTGTTC-3′ |
| Genomic tomm-20 amplification (forward) | 5′-GAACACTTTTCTTAATGATACAAAATGTCGGACACAATTCTTGGTTTCAACAAATC-3′ |
| Genomic tomm-20 amplification (reverse) | 5′-CTCCTTAATCAGCTCTTCGCCCTTAGACACCTCCAAGTCGTCGGTGTC-3′ |
| pPD95.75 amplification (forward) | 5′-GACACCGACGACTTGGAGGTGTCTAAGGGCGAAGAGCTGATTAAGGAG-3′ |
| pPD95.75 amplification (reverse) | 5′-GTTCGAGTCTTTCAATTTCCCAAGTTGTTAGCGTATCCAT-3′ |
For the creation of pKP07 (osm-5p::mito-roGFP), the previously described osm-5 promoter was amplified from genomic DNA using primers with homology to pOR823 containing dat-1p::mito-roGFP. pOR823 was amplified using primers with homology to osm-5p. The dat-1 promoter was not included in the amplification of the backbone. Fragments were joined using Gibson assembly. (Table 3).
Table 3.
Reagents used for construction of the osm-5p::mito-roGFP marker
| Genomic osm-5p amplification (forward) | 5′-CTGATCACGGTACCGCATGCCTGCAGGAAATTGAAAGACTCGAACCCCATAACT-3′ |
| Genomic osm-5p amplification (reverse) | 5′-GTACCAATACGACTCACTATAGAGCTCTTTGTATCATTAAGAAAAGTGTTC-3′ |
| pOR823 amplification (forward) | 5′-GAACACTTTTCTTAATGATACAAAGAGCTCTATAGTGAGTCGTATTGGTAC-3′ |
| pOR823 amplification (reverse) | 5′-AGTTATGGGGTTCGAGTCTTTCAATTTCCTGCAGGCATGCGGTACCGTGATCAG-3′ |
Dye-filling assays
We performed dye-filling assays using the following protocol described in Power et al. (2020). Prior to performing dye-filling assays, healthy, non-crowded plates containing many gravid hermaphrodite animals were washed and bleached to synchronize the worms. Assays were performed at least in triplicate and with genotypes blinded. Worms at desired ages were washed from the plate with 1 ml M9 buffer and transferred to 1.5 ml tubes. A stock solution of 2.5 mg/ml DiI (Cat. D282; Thermo Fisher Scientific) was added to a 1:250 dilution and worms were incubated with gentle shaking for 50 min at room temperature. Prior to scoring, worms were briefly spun at 2,000 RPM and plated on newly seeded NGM plates for ∼1 h to excrete excess dye. The presence and intensity of dye in the amphid and phasmid neurons were visually scored using a dissecting microscope. Worms scored as non-Dyf were similar in brightness to wild type, and Dyf worms had no staining. Partial Dyf worms had dye present, but were distinguishably less bright than wild type. Kruskal–Wallis one-way ANOVA analysis (where non-Dyf = 2, partial Dyf = 1. and Dyf = 0) and post-hoc Dunn’s multiple comparison test or Mann–Whitney test were performed in Prism (GraphPad Software).
Tertiary structure prediction and model evaluation
We predicted the structure of NEKL-4 WT and its mutants using the I-TASSER server (Yang and Zhang, 2015). In I-TASSER, structural templates were first identified from the PDB database using multiple threading alignment approaches. The full-length structure models were then constructed by iterative fragment assembly simulations. The top five predicted models were rigorously evaluated by several model quality assessment programs including VERIFY3D (Lüthy et al., 1992), VoroMQA (Olechnovič and Venclovas, 2017), ProsaWEB (Wiederstein and Sippl, 2007), and ProQ3 (Uziela et al., 2016). These programs employ a variety of methods inclusive of deep learning approaches to evaluate the quality of the models. The final models were chosen by selecting the best evaluation profiles and their correlation with known functional features.
Surface electrostatic profiles of NEKL-4 and kinase dead mutant
PyMOL Version 2.0 (Schrödinger LLC) was used to visualize the protein models and its APBS plugin (Holst and Saied, 1993) to obtain the surface electrostatic profiles of both NEKL-4 and the Kinase Dead mutant.
Cleft analysis of NEKL-4 and kinase dead mutant
PDBsum (Laskowski, 2007) was utilized for visualization and analysis of both NEKL-4 and the Kinase Dead mutant’s clefts.
Antibody staining
We performed immunofluorescence microscopy using the following protocol described in Power et al. (2020). We synchronized animals by bleaching and fixing animals as Day 1 adults. Fixation was accomplished by washing animals from 3 NGM plates using M9 buffer, then washing animals with several changes of M9 over 1 h. Worms were chilled on ice before washing in ice-cold Ruvkun buffer (80 mM KCl, 20 mM NaCl, 10 mM EGTA, 5 mM spermidine-HCl, 15 mM PIPES, pH 7.4 and 25% methanol) plus 2% formaldehyde in 1.5 ml centrifuge tubes. The tubes were immersed in liquid nitrogen and melted under tap water to crack the worms’ cuticles. Worms were then washed twice with Tris-Triton buffer (100 mM Tris-HCl, pH 7.4, 1% Triton X-100, and 1 mM EDTA), suspended in Tris-Triton buffer + 1% β-mercaptoethanol, and incubated overnight at 37°C. The next day, worms were washed with 1X BO3 buffer (50 mM H3BO3, 25 mM NaOH) + 0.01% Triton and suspended in 1X BO3 + 0.01% Triton buffer + 10 mM DTT for 15 min with gentle agitation at room temperature. Worms were then washed with 1X BO3 buffer (50 mM H3BO3, 25 mM NaOH) + 0.01% Triton and suspended in 1X BO3 + 0.01% Triton buffer + 0.3% H2O2 for 15 min with gentle agitation at room temperature. After washing once with 1X BO3 + 0.01% Triton buffer, worms were washed for 15 min in Antibody buffer B (1X PBS, 0.1% BSA, 0.5% Triton X-100, 0.05% sodium azide, and 1 mM EDTA) with gentle agitation at room temperature. Fixed worms were stored in Antibody buffer A (1X PBS, 1% BSA, 0.5% Triton X-100, 0.05% sodium azide, 1 mM EDTA) at 4°C for up to 1 month before antibody staining. Animals were stained overnight at room temperature with a 1:600 final dilution (in Antibody Buffer A) of GT335 (Cat. AG-20B-0020-C100; Adipogen), a monoclonal antibody which binds the branch point of polyglutamylated substrates (Wolff et al., 1992). Stained worms were washed with several changes of Antibody B Buffer with gentle agitation at room temperature over several hours. After rinsing with Antibody Buffer A, Alexa-fluor 568-conjugated donkey anti-mouse secondary antibody (Cat. A10037; Invitrogen) was added at a final dilution of 1:2,500 and incubated for 2 h at room temperature with gentle agitation. Worms were then washed with several changes of Antibody Buffer B over several hours before mounting on 10% agarose pads for imaging.
Confocal imaging
Live animals were anesthetized with 10 mM levamisole and mounted on 10% agarose pads for imaging at room temperature. Confocal imaging was performed with a Zeiss LSM 880 inverted microscope with an Airyscan superresolution module using an LSM T-PMT detector and ZenBlack software (Carl Zeiss Microscopy). Laser intensity was adjusted to avoid saturated pixels. Images were acquired using a 63x/1.4 Oil Plan-Apochromat objective in Airyscan or Airyscan Fast mode and deconvolved using Airyscan processing. Image files were imported into Fiji/ImageJ with the BioFormats Importer plugin for linear adjustment of contrast and creation of maximum intensity projections.
Widefield imaging
Nematodes were mounted as above. Widefield images were acquired on a Zeiss Axio Observer with Colibri 7 LEDs and ZenBlue software (Carl Zeiss Microscopy) using a Photometrics Prime 95B sCMOS camera (Teledyne Photometrics). A 63x/1.4 Oil Plan-Apochromat objective was used for imaging.
Transmission electron microscopy
Three strains, PT3471 (nekl-4(KD)), PT3756 (nekl-4(PEST∆)), and PT3593 (ccpp-1∆; nekl-4(KD)), were grown for four generations on seeded OP50 agarose plates at 20°C, avoiding starvation at any time. Young adult worms were fixed by high-pressure freezing/freeze substitution (HPS/FS) (Hall et al., 2012) with a fixative of 2% osmium tetroxide, 0.1% uranyl acetate, and 2% H2O in acetone for a week at −90°C, then slowly warming stepwise to −60°C, −30°C, and 0°C. Samples were washed four times in 100% cold acetone, infiltrated in five changes into EMbed812 resin (Cat. 70020; Electron Microscopy Sciences) over 2 days, and cured at 60°C.
Twelve PT3471 (nekl-4(KD)), six PT3756 (nekl-4(PEST∆)), and 17 PT3593 (ccpp-1∆; nekl-4(KD)) young adult hermaphrodites were collected either at 70, 90, 150, or 200 nm transverse serial sections on Pioloform/Formvar carbon-coated wide slot grids from an RMC Ultramicrotome PowerTome PT XL microtome (Böckeler Instruments) starting nose tip to amphid base, covering about 25–40 µm of the amphid organ. Sections were poststained in 2% uranium acetate for 20–40 min and in 1:5 dilution aqueous Reynolds lead stain for 5 min. Most animals were imaged separately for the left and right amphids from the pore to the base of the amphid with Digital Micrograph software on a Hitachi H-7500 TEM with ATM CD camera, covering serial thin sections. To obtain electron tomograms, other animals were also viewed using SerialEM software on a JEM-1400Plus TEM (Jeol Ltd.) with Gatan Orius SC1000B bottom mount digital camera (Gatan Inc.) using serial thick sections. Serial amphid image stacks were aligned with TrakEM2, then processed and realigned with Etomo and 3dmod software (Mastronarde Group, University of Colorado, Boulder, CO) to produce tomograms.
Structured illumination microscopy (SIM)
Live Day 1 adult animals carrying osm-5p::tomm-20::tagRFP and endogenous NEKL-4::mNG were mounted as described above and imaged using a Zeiss Elyra 7 with Lattice SIM2 (Carl Zeiss Microscopy). We acknowledge Jessica Shivas and Nancy Kane for assistance with SIM imaging.
Automated mitochondrial morphology quantification
Images of animals carrying osm-5p::tomm-20::tagRFP were acquired using the Zeiss LSM 880 confocal microscope as described above, and maximum intensity projections were used for analysis. Mitochondria Analyzer, a Fiji/ImageJ macro, was used to quantify mitochondria morphology and number in phasmid dendrites (Chaudhry et al., 2020). Images were converted to 8-bit and cell bodies were trimmed. Settings used for batch 2D thresholding were as follows: Rolling 1.25 μm; max slope 1.8; gamma 0.8; block size 1.15 μm; and C-Value 5. Batch 2D analysis was then performed on a per-mito basis. After automatic analysis, data were cleaned by eliminating all mitochondria with >1 branch and/or ≠2 endpoints since mitochondria in the dendrites are unbranched and results indicating branches are likely an artifact of using Z-projected images for analysis. Mitochondria <0.3 µm were also eliminated as they are likely background noise or artifacts not eliminated by thresholding. Outliers were removed with the ROUT method (Q = 0.1%), and Kruskal-Wallis one-way ANOVA analysis and post-hoc Dunn’s multiple comparison test were performed in Prism (GraphPad Software).
Chronic PQ survival assay
The adult chronic PQ survival assay was performed as in Schaar et al. (2015). Worms were synchronized by bleaching, and Day 1 adults were plated on NGM plates containing 100 µM 5-Fluoro-2′-deoxyuridine (FuDR) (Cat. F0503; Sigma-Aldrich) with or without 4 mM methyl viologen dichloride hydrate (PQ) (Cat. 856177; Sigma-Aldrich). FuDR was used to inhibit the development of progeny since PQ causes an increased rate of internal hatching that may prematurely kill the worms. Control and PQ plates were seeded with 150 µl of 10X concentrated OP50 and allowed to dry before use. Adult worms were transferred to new plates every 7 days until there were no surviving animals. Worms were considered dead when unmoving and unresponsive to multiple touch stimuli. Worms that died during transfer or by crawling off the plate were censored. Data were visualized as Kaplan–Meier survival curves and analyzed using a log-rank Mantel-Cox test performed in Prism (GraphPad Software).
Mitochondrial redox state analysis
Nematodes carrying a mitochondria-targeted redox sensor expressed in the ciliated neurons (osm-5p::mito-roGFP) were mounted as above and imaged using the Zeiss LSM 880 confocal microscope. The mitochondria targeting signal for this biosensor was taken from aspartate aminotransferase. roGFP (reduction-oxidation-sensitive GFP) is a modified GFP that contains cysteines at certain residues that create two excitation peaks corresponding to different oxidation states (Hanson et al., 2004). By measuring the ratio of emissions at each wavelength, the relative oxidation state of mitochondria can be monitored. Phasmid cell bodies of Day 1 adult worms were imaged with 405 nm (oxidized) and 488 nm (reduced) excitation. Only mitochondria close to the coverslip were imaged to minimize differences in emission intensity caused by differences in the depth of the cells within the worm. For analysis, maximum intensity projections were created from three to five slices per worm. Channels were split and the 488 nm channel was duplicated and used to create ROIs including only mitochondria by thresholding. The mean fluorescence intensity of each channel within the ROI was measured, and the 405-nm intensity was divided by the 488 nm intensity to obtain the final emission ratio. To generate the ratio images, original images were converted to 32-bit and the 405 nm channel was divided by the 488 nm channel using the ImageJ image calculator and displayed using the “16 colors” LUT. Outliers were removed with the ROUT method (Q = 0.1%), and Kruskal-Wallis one-way ANOVA analysis and post-hoc Dunn’s multiple comparison tests were performed in Prism (GraphPad Software).
Online supplemental material
Fig. S1 shows the predicted secondary structure of NEKL-4, surface electrostatic potential of NEKL-4 and NEKL-4(KD), and cleft analysis of NEKL-4 and NEKL-4(KD). Fig. S2 shows additional dye-filling data and interactions for nekl-4(KD), nekl-4∆, and nekl-4(PEST∆) mutants. Fig. S3 shows that nekl-4(KD) does not suppress the loss of GT335 antibody staining in ccpp-1∆ mutant cilia. Fig. S4 shows that spans of IFT proteins are shortened in ccpp-1∆ mutant cilia and unaffected in nekl-4 mutants. Fig. S5 shows that roGFP expression level differs in ccpp-1∆ and nekl-4∆ mutants, and that ccpp-1∆ and ccpp-1∆; nekl-4∆ mutants lack dendritic mitochondria as visualized by roGFP. Video 1 shows the co-transport of NEKL-4 and mitochondria in dendrites. Table S1 lists candidate interactors of nekl-4 and their functions. Table S2 and Table S3 list strains used in the paper and supplemental material, respectively.
Supplementary Material
shows the candidate interactors of nekl-4 and their functions.
shows the strains used in this study.
shows the strains used in the supplemental material.
Acknowledgments
We thank the members of the Barr and Rongo laboratories for valuable input on this research, particularly Inna Nikonorova, Juan Wang, Jonathan Dietz, Joelle Smart, and Tatiana Popovitchenko. Additionally, KMP thanks his thesis committee: Monica Driscoll, Marc Gartenberg, and Bonnie Firestein. We thank Gloria Androwski (Barr laboratory) and Nanci Kane (Waksman Microscope Core Facilities) for technical assistance, as well as Christopher Nguyen for help with 3D visualization of our TEM data. Some strains were provided by the CGC, which was funded by the National Institutes of Health Office of Research Infrastructure Programs (P40 OD010440). We also thank WormBase (U41 HG002223) and WormAtlas (R24 OD010943) for valuable online resources.
This work was supported by National Institutes of Health (NIH) R01 DK059418, DK116606, and NS120745 (M.M. Barr), F31 NS122438 (K.M. Power), R01 GM101972 (C. Rongo), and R24 OD010943 (D.H. Hall).
Author contributions: K.M. Power: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Resources, Validation, Visualization, Writing—original draft, Writing—review and editing, K.C. Nguyen: Investigation, Resources, A. Silva Eichner: Validation, Writing—original draft, Writing—review and editing, S. Singh: Supervision, Validation, Writing—original draft, Writing—review and editing, D.H. Hall: Data curation, Funding acquisition, Resources, Supervision, Writing—review and editing, C. Rongo: Methodology, Supervision, Writing—review and editing, M.M. Barr: Conceptualization, Formal analysis, Funding acquisition, Project administration, Supervision, Validation, Visualization, Writing—original draft, Writing—review and editing.
Data availability
All data are present in the paper and/or the online supplemental material and will be made available upon request.
References
- Al Mutairi, F., Alkhalaf R., Alkhorayyef A., Alroqi F., Yusra A., Umair M., Nouf F., Khan A., Meshael A., Hamad A., et al. 2020. Homozygous truncating NEK10 mutation, associated with primary ciliary dyskinesia: A case report. BMC Pulm. Med. 20:141. 10.1186/s12890-020-1175-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez Viar, G., Klena N., Martino F., Nievergelt A., and Pigino G.. 2023. The tubulin nano-code: A protofilament-specific pattern of tubulin post-translational modifications regulates ciliary beating mechanics. bioRxiv. 10.1101/2023.06.28.546853 (Preprint posted June 28, 2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apfeld, J., and Kenyon C.. 1999. Regulation of lifespan by sensory perception in Caenorhabditis elegans. Nature. 402:804–809. 10.1038/45544 [DOI] [PubMed] [Google Scholar]
- Bae, J.-E., Jang S., Kim J.B., Hyung H., Park N.Y., Kim Y.H., Kim S.H., Kim S.H., Ha J.M., Oh G.S., et al. 2023. Enhanced primary ciliogenesis via mitochondrial oxidative stress activates AKT to prevent neurotoxicity in HSPA9/mortalin-depleted SH-SY5Y cells. Mol. Brain. 16:41. 10.1186/s13041-023-01029-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae, J.-E., Kang G.M., Min S.H., Jo D.S., Jung Y.-K., Kim K., Kim M.-S., and Cho D.-H.. 2019. Primary cilia mediate mitochondrial stress responses to promote dopamine neuron survival in a Parkinson’s disease model. Cell Death Dis. 10:952. 10.1038/s41419-019-2184-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bear, R.M., and Caspary T.. 2023. Cilia bent out of shape over dysfunctional astrocyte mitochondria. J. Cell Biol. 222:e202211123. 10.1083/jcb.202211123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodakuntla, S., Schnitzler A., Villablanca C., Gonzalez-Billault C., Bieche I., Janke C., and Magiera M.M.. 2020. Tubulin polyglutamylation is a general traffic-control mechanism in hippocampal neurons. J. Cell Sci. 133:jcs241802. 10.1242/jcs.241802 [DOI] [PubMed] [Google Scholar]
- Brenner, S. 1974. The genetics of Caenorhabditis elegans. Genetics. 77:71–94. 10.1093/genetics/77.1.71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burghoorn, J., Dekkers M.P.J., Rademakers S., de Jong T., Willemsen R., and Jansen G.. 2007. Mutation of the MAP kinase DYF-5 affects docking and undocking of kinesin-2 motors and reduces their speed in the cilia of Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA. 104:7157–7162. 10.1073/pnas.0606974104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cehajic-Kapetanovic, J., Martinez-Fernandez de la Camara C., Birtel J., Rehman S., McClements M.E., Charbel Issa P., Lotery A.J., and MacLaren R.E.. 2022. Impaired glutamylation of RPGRORF15 underlies the cone-dominated phenotype associated with truncating distal ORF15 variants. Proc. Natl. Acad. Sci. USA. 119:e2208707119. 10.1073/pnas.2208707119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhry, A., Shi R., and Luciani D.S.. 2020. A pipeline for multidimensional confocal analysis of mitochondrial morphology, function, and dynamics in pancreatic β-cells. Am. J. Physiol. Endocrinol. Metab. 318:E87–E101. 10.1152/ajpendo.00457.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, X.-T., and Sheng Z.-H.. 2021. Developmental regulation of microtubule-based trafficking and anchoring of axonal mitochondria in health and diseases. Dev. Neurobiol. 81:284–299. 10.1002/dneu.22748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Z., Li M., Zhu H., and Ou G.. 2023. Modulation of inner junction proteins contributes to axoneme differentiation. Proc. Natl. Acad. Sci. USA. 120:e2303955120. 10.1073/pnas.2303955120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chivukula, R.R., Montoro D.T., Leung H.M., Yang J., Shamseldin H.E., Taylor M.S., Dougherty G.W., Zariwala M.A., Carson J., Daniels M.L.A., et al. 2020. A human ciliopathy reveals essential functions for NEK10 in airway mucociliary clearance. Nat. Med. 26:244–251. 10.1038/s41591-019-0730-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobbelaere, J., Su T.Y., Erdi B., Schleiffer A., and Dammermann A.. 2023. A phylogenetic profiling approach identifies novel ciliogenesis genes in Drosophila and C. elegans. EMBO J. 42:e113616. 10.15252/embj.2023113616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dokshin, G.A., Ghanta K.S., Piscopo K.M., and Mello C.C.. 2018. Robust genome editing with short single-stranded and long, partially single-stranded DNA donors in Caenorhabditis elegans. Genetics. 210:781–787. 10.1534/genetics.118.301532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii, M., Matsumoto Y., Tanaka N., Miki K., Suzuki T., Ishii N., and Ayusawa D.. 2004. Mutations in chemosensory cilia cause resistance to paraquat in nematode Caenorhabditis elegans. J. Biol. Chem. 279:20277–20282. 10.1074/jbc.M313119200 [DOI] [PubMed] [Google Scholar]
- Gigante, E.D., and Caspary T.. 2020. Signaling in the primary cilium through the lens of the Hedgehog pathway. Wiley Interdiscip. Rev. Dev. Biol. 9:e377. 10.1002/wdev.377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmore-Hall, S., Kuo J., Ward J.M., Zahra R., Morrison R.S., Perkins G., and La Spada A.R.. 2019. CCP1 promotes mitochondrial fusion and motility to prevent Purkinje cell neuron loss in pcd mice. J. Cell Biol. 218:206–219. 10.1083/jcb.201709028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, W., Stoklund Dittlau K., and Van Den Bosch L.. 2020. Axonal transport defects and neurodegeneration: Molecular mechanisms and therapeutic implications. Semin. Cell Dev. Biol. 99:133–150. 10.1016/j.semcdb.2019.07.010 [DOI] [PubMed] [Google Scholar]
- Haeussler, M., Schönig K., Eckert H., Eschstruth A., Mianné J., Renaud J.-B., Schneider-Maunoury S., Shkumatava A., Teboul L., Kent J., et al. 2016. Evaluation of off-target and on-target scoring algorithms and integration into the guide RNA selection tool CRISPOR. Genome Biol. 17:148. 10.1186/s13059-016-1012-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall, D.H., Hartwieg E., and Nguyen K.C.Q.. 2012. Modern electron microscopy methods for C. elegans. Methods Cell Biol. 107:93–149. 10.1016/B978-0-12-394620-1.00004-7 [DOI] [PubMed] [Google Scholar]
- Hanson, G.T., Aggeler R., Oglesbee D., Cannon M., Capaldi R.A., Tsien R.Y., and Remington S.J.. 2004. Investigating mitochondrial redox potential with redox-sensitive green fluorescent protein indicators. J. Biol. Chem. 279:13044–13053. 10.1074/jbc.M312846200 [DOI] [PubMed] [Google Scholar]
- Han, Y.K., Kim J.S., Jang G., and Park K.M.. 2021. Cisplatin induces lung cell cilia disruption and lung damage via oxidative stress. Free Radic. Biol. Med. 177:270–277. 10.1016/j.freeradbiomed.2021.10.032 [DOI] [PubMed] [Google Scholar]
- He, K., Ma X., Xu T., Li Y., Hodge A., Zhang Q., Torline J., Huang Y., Zhao J., Ling K., and Hu J.. 2018. Axoneme polyglutamylation regulated by Joubert syndrome protein ARL13B controls ciliary targeting of signaling molecules. Nat. Commun. 9:3310. 10.1038/s41467-018-05867-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, S., Wang L., Miao L., Wang T., Du F., Zhao L., and Wang X.. 2009. Receptor interacting protein kinase-3 determines cellular necrotic response to TNF-alpha. Cell. 137:1100–1111. 10.1016/j.cell.2009.05.021 [DOI] [PubMed] [Google Scholar]
- Hewezi, T., Juvale P.S., Piya S., Maier T.R., Rambani A., Rice J.H., Mitchum M.G., Davis E.L., Hussey R.S., and Baum T.J.. 2015. The cyst nematode effector protein 10A07 targets and recruits host posttranslational machinery to mediate its nuclear trafficking and to promote parasitism in Arabidopsis. Plant Cell. 27:891–907. 10.1105/tpc.114.135327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holst, M., and Saied F.. 1993. Multigrid solution of the Poisson—Boltzmann equation. J. Comput. Chem. 14:105–113. 10.1002/jcc.540140114 [DOI] [Google Scholar]
- Ignatenko, O., Malinen S., Rybas S., Vihinen H., Nikkanen J., Kononov A., Jokitalo E.S., Ince-Dunn G., and Suomalainen A.. 2023. Mitochondrial dysfunction compromises ciliary homeostasis in astrocytes. J. Cell Biol. 222:e202203019. 10.1083/jcb.202203019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imanishi, M., Endres N.F., Gennerich A., and Vale R.D.. 2006. Autoinhibition regulates the motility of the C. elegans intraflagellar transport motor OSM-3. J. Cell Biol. 174:931–937. 10.1083/jcb.200605179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura, Y., Kurabe N., Ikegami K., Tsutsumi K., Konishi Y., Kaplan O.I., Kunitomo H., Iino Y., Blacque O.E., and Setou M.. 2010. Identification of tubulin deglutamylase among Caenorhabditis elegans and mammalian cytosolic carboxypeptidases (CCPs). J. Biol. Chem. 285:22936–22941. 10.1074/jbc.C110.128280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Kooij, B., Creixell P., van Vlimmeren A., Joughin B.A., Miller C.J., Haider N., Simpson C.D., Linding R., Stambolic V., Turk B.E., and Yaffe M.B.. 2019. Comprehensive substrate specificity profiling of the human Nek kinome reveals unexpected signaling outputs. Elife. 8:e44635. 10.7554/eLife.44635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo, T., and Oda T.. 2017. Electrostatic interaction between polyglutamylated tubulin and the nexin-dynein regulatory complex regulates flagellar motility. MBoC. 28:2260–2266. 10.1091/mbc.e17-05-0285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo, T., Yanagisawa H.-A., Yagi T., Hirono M., and Kamiya R.. 2010. Tubulin polyglutamylation regulates axonemal motility by modulating activities of inner-arm dyneins. Curr. Biol. 20:441–445. 10.1016/j.cub.2009.12.058 [DOI] [PubMed] [Google Scholar]
- Kumamoto, N., Gu Y., Wang J., Janoschka S., Takemaru K.-I., Levine J., and Ge S.. 2012. A role for primary cilia in glutamatergic synaptic integration of adult-born neurons. Nat. Neurosci. 15:399–405, S1. 10.1038/nn.3042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labrousse, A.M., Zappaterra M.D., Rube D.A., and van der Bliek A.M.. 1999. C. elegans dynamin-related protein DRP-1 controls severing of the mitochondrial outer membrane. Mol. Cell. 4:815–826. 10.1016/S1097-2765(00)80391-3 [DOI] [PubMed] [Google Scholar]
- Laskowski, R.A. 2007. Enhancing the functional annotation of PDB structures in PDBsum using key figures extracted from the literature. Bioinformatics. 23:1824–1827. 10.1093/bioinformatics/btm085 [DOI] [PubMed] [Google Scholar]
- Lechtreck, K.F., and Geimer S.. 2000. Distribution of polyglutamylated tubulin in the flagellar apparatus of green flagellates. Cell Motil. Cytoskeleton. 47:219–235. [DOI] [PubMed] [Google Scholar]
- Lee, J.E., and Gleeson J.G.. 2011. Cilia in the nervous system: Linking cilia function and neurodevelopmental disorders. Curr. Opin. Neurol. 24:98–105. 10.1097/WCO.0b013e3283444d05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lew, E.D., Furdui C.M., Anderson K.S., and Schlessinger J.. 2009. The precise sequence of FGF receptor autophosphorylation is kinetically driven and is disrupted by oncogenic mutations. Sci. Signal. 2:ra6. 10.1126/scisignal.2000021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, D., Liu Y., Yi P., Zhu Z., Li W., Zhang Q.C., Li J.B., and Ou G.. 2021a. RNA editing restricts hyperactive ciliary kinases. Science. 373:984–991. 10.1126/science.abd8971 [DOI] [PubMed] [Google Scholar]
- Li, J., Snyder E.Y., Tang F.H.F., Pasqualini R., Arap W., and Sidman R.L.. 2020. Nna1 gene deficiency triggers Purkinje neuron death by tubulin hyperglutamylation and ER dysfunction. JCI Insight. 5:e136078. 10.1172/jci.insight.136078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y.-Y., Guo L., and Han Z.-M.. 2021b. Roles of NEK family in cell cycle regulation. Yi Chuan. 43:642–653. 10.16288/j.yczz.20-421 [DOI] [PubMed] [Google Scholar]
- Lüthy, R., Bowie J.U., and Eisenberg D.. 1992. Assessment of protein models with three-dimensional profiles. Nature. 356:83–85. 10.1038/356083a0 [DOI] [PubMed] [Google Scholar]
- Magaletta, M.E., Perkins K.J., Deuchler C.P., and Pieczynski J.N.. 2019. The Kinesin-3 motor, KLP-4, mediates axonal organization and cholinergic signaling in Caenorhabditis elegans. FASEB Bioadv. 1:450–460. 10.1096/fba.2019-00019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magiera, M.M., Bodakuntla S., Žiak J., Lacomme S., Marques Sousa P., Leboucher S., Hausrat T.J., Bosc C., Andrieux A., Kneussel M., et al. 2018. Excessive tubulin polyglutamylation causes neurodegeneration and perturbs neuronal transport. EMBO J. 37:e100440. 10.15252/embj.2018100440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maglioni, S., Schiavi A., Runci A., Shaik A., and Ventura N.. 2014. Mitochondrial stress extends lifespan in C. elegans through neuronal hormesis. Exp. Gerontol. 56:89–98. 10.1016/j.exger.2014.03.026 [DOI] [PubMed] [Google Scholar]
- Mann, J.R., McKenna E.D., Mawrie D., Papakis V., Alessandrini F., Anderson E.N., Mayers R., Ball H.E., Kaspi E., Lubinski K., et al. 2023. Loss of function of the ALS-associated NEK1 kinase disrupts microtubule homeostasis and nuclear import. Sci. Adv. 9:eadi5548. 10.1126/sciadv.adi5548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu, H., Patten S.A., Aragon-Martin J.A., Ocaka L., Simpson M., Child A., and Moldovan F.. 2021. Genetic variant of TTLL11 gene and subsequent ciliary defects are associated with idiopathic scoliosis in a 5-generation UK family. Sci. Rep. 11:11026. 10.1038/s41598-021-90155-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna, E.D., Sarbanes S.L., Cummings S.W., and Roll-Mecak A.. 2023. The tubulin code, from molecules to health and disease. Annu. Rev. Cell Dev. Biol. 39:331–361. 10.1146/annurev-cellbio-030123-032748 [DOI] [PubMed] [Google Scholar]
- Melo-Hanchuk, T.D., and Kobarg J.. 2021. Polyglutamylase activity of tubulin tyrosine ligase-like 4 is negatively regulated by the never in mitosis gene A family kinase never in mitosis gene A -related kinase 5. World J. Biol. Chem. 12:38–51. 10.4331/wjbc.v12.i3.38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mijalkovic, J., van Krugten J., Oswald F., Acar S., and Peterman E.J.G.. 2018. Single-molecule turnarounds of intraflagellar transport at the C. elegans ciliary tip. Cell Rep. 25:1701–1707.e2. 10.1016/j.celrep.2018.10.050 [DOI] [PubMed] [Google Scholar]
- Mill, P., Christensen S.T., and Pedersen L.B.. 2023. Primary cilia as dynamic and diverse signalling hubs in development and disease. Nat. Rev. Genet. 24:421–441. 10.1038/s41576-023-00587-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moruzzi, N., Valladolid-Acebes I., Kannabiran S.A., Bulgaro S., Burtscher I., Leibiger B., Leibiger I.B., Berggren P.-O., and Brismar K.. 2022. Mitochondrial impairment and intracellular reactive oxygen species alter primary cilia morphology. Life Sci. Alliance. 5:e202201505. 10.26508/lsa.202201505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz, I.M., Morgan M.E., Peltier J., Weiland F., Gregorczyk M., Brown F.C., Macartney T., Toth R., Trost M., and Rouse J.. 2018. Phosphoproteomic screening identifies physiological substrates of the CDKL5 kinase. EMBO J. 37:e99559. 10.15252/embj.201899559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura, Y., Kasahara K., Shiromizu T., Watanabe M., and Inagaki M.. 2018. Primary cilia as signaling hubs in health and disease. Adv. Sci. 6:1801138. 10.1002/advs.201801138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa, S., Nakajima K., Miki H., Minato Y., Wang D., and Hirokawa N.. 2012. KIF19A is a microtubule-depolymerizing kinesin for ciliary length control. Dev. Cell. 23:1167–1175. 10.1016/j.devcel.2012.10.016 [DOI] [PubMed] [Google Scholar]
- O’Hagan, R., Avrutis A., and Ramicevic E.. 2022. Functions of the tubulin code in the C. elegans nervous system. Mol. Cell. Neurosci. 123:103790. 10.1016/j.mcn.2022.103790 [DOI] [PubMed] [Google Scholar]
- O’Hagan, R., Piasecki B.P., Silva M., Phirke P., Nguyen K.C.Q., Hall D.H., Swoboda P., and Barr M.M.. 2011. The tubulin deglutamylase CCPP-1 regulates the function and stability of sensory cilia in C. elegans. Curr. Biol. 21:1685–1694. 10.1016/j.cub.2011.08.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Hagan, R., Silva M., Nguyen K.C.Q., Zhang W., Bellotti S., Ramadan Y.H., Hall D.H., and Barr M.M.. 2017. Glutamylation regulates transport, specializes function, and sculpts the structure of cilia. Curr. Biol. 27:3430–3441.e6. 10.1016/j.cub.2017.09.066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olechnovič, K., and Venclovas Č.. 2017. VoroMQA: Assessment of protein structure quality using interatomic contact areas. Proteins. 85:1131–1145. 10.1002/prot.25278 [DOI] [PubMed] [Google Scholar]
- Orbach, R., and Howard J.. 2019. The dynamic and structural properties of axonemal tubulins support the high length stability of cilia. Nat. Commun. 10:1838. 10.1038/s41467-019-09779-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panchal, K., and Tiwari A.K.. 2019. Mitochondrial dynamics, a key executioner in neurodegenerative diseases. Mitochondrion. 47:151–173. 10.1016/j.mito.2018.11.002 [DOI] [PubMed] [Google Scholar]
- Park, K., Li C., Tsiropoulou S., Gonçalves J., Kondratev C., Pelletier L., Blacque O.E., and Leroux M.R.. 2021. CDKL kinase regulates the length of the ciliary proximal segment. Curr. Biol. 31:2359–2373.e7. 10.1016/j.cub.2021.03.068 [DOI] [PubMed] [Google Scholar]
- Peres de Oliveira, A., Basei F.L., Slepicka P.F., de Castro Ferezin C., Melo-Hanchuk T.D., de Souza E.E., Lima T.I., Dos Santos V.T., Mendes D., Silveira L.R., et al. 2020. NEK10 interactome and depletion reveal new roles in mitochondria. Proteome Sci. 18:4. 10.1186/s12953-020-00160-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins, L.A., Hedgecock E.M., Thomson J.N., and Culotti J.G.. 1986. Mutant sensory cilia in the nematode Caenorhabditis elegans. Dev. Biol. 117:456–487. 10.1016/0012-1606(86)90314-3 [DOI] [PubMed] [Google Scholar]
- Porpora, M., Sauchella S., Rinaldi L., Delle Donne R., Sepe M., Torres-Quesada O., Intartaglia D., Garbi C., Insabato L., Santoriello M., et al. 2018. Counterregulation of cAMP-directed kinase activities controls ciliogenesis. Nat. Commun. 9:1224. 10.1038/s41467-018-03643-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power, K.M., Akella J.S., Gu A., Walsh J.D., Bellotti S., Morash M., Zhang W., Ramadan Y.H., Ross N., Golden A., et al. 2020. Mutation of NEKL-4/NEK10 and TTLL genes suppress neuronal ciliary degeneration caused by loss of CCPP-1 deglutamylase function. PLoS Genet. 16:e1009052. 10.1371/journal.pgen.1009052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin, H., Rosenbaum J.L., and Barr M.M.. 2001. An autosomal recessive polycystic kidney disease gene homolog is involved in intraflagellar transport in C. elegans ciliated sensory neurons. Curr. Biol. 11:457–461. 10.1016/S0960-9822(01)00122-1 [DOI] [PubMed] [Google Scholar]
- Rechsteiner, M., and Rogers S.W.. 1996. PEST sequences and regulation by proteolysis. Trends Biochem. Sci. 21:267–271. 10.1016/S0968-0004(96)10031-1 [DOI] [PubMed] [Google Scholar]
- Schaar, C.E., Dues D.J., Spielbauer K.K., Machiela E., Cooper J.F., Senchuk M., Hekimi S., and Van Raamsdonk J.M.. 2015. Mitochondrial and cytoplasmic ROS have opposing effects on lifespan. PLoS Genet. 11:e1004972. 10.1371/journal.pgen.1004972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shashi, V., Magiera M.M., Klein D., Zaki M., Schoch K., Rudnik-Schöneborn S., Norman A., Lopes Abath Neto O., Dusl M., Yuan X., et al. 2018. Loss of tubulin deglutamylase CCP1 causes infantile-onset neurodegeneration. EMBO J. 37:e100540. 10.15252/embj.2018100540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheu, S.-H., Upadhyayula S., Dupuy V., Pang S., Deng F., Wan J., Walpita D., Pasolli H.A., Houser J., Sanchez-Martinez S., et al. 2022. A serotonergic axon-cilium synapse drives nuclear signaling to alter chromatin accessibility. Cell. 185:3390–3407.e18. 10.1016/j.cell.2022.07.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva, D.F., and Cavadas C.. 2023. Primary cilia shape hallmarks of health and aging. Trends Mol. Med. 29:567–579. 10.1016/j.molmed.2023.04.001 [DOI] [PubMed] [Google Scholar]
- Spassky, N., and Meunier A.. 2017. The development and functions of multiciliated epithelia. Nat. Rev. Mol. Cell Biol. 18:423–436. 10.1038/nrm.2017.21 [DOI] [PubMed] [Google Scholar]
- Sternberg, P.W., Van Auken K., Wang Q., Wright A., Yook K., Zarowiecki M., Arnaboldi V., Becerra A., Brown S., Cain S., et al. 2024. WormBase 2024: Status and transitioning to alliance infrastructure. Genetics. 227:iyae050. 10.1093/genetics/iyae050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran, M.V., Khuntsariya D., Fetter R.D., Ferguson J.W., Wang J.T., Long A.F., Cote L.E., Wellard S.R., Vázquez-Martínez N., Sallee M.D., et al. 2023. MAP9/MAPH-9 supports axonemal microtubule doublets and modulates motor movement. Dev. Cell. 59:199–210.e11. 10.1016/j.devcel.2023.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uziela, K., Shu N., Wallner B., and Elofsson A.. 2016. ProQ3: Improved model quality assessments using Rosetta energy terms. Sci. Rep. 6:33509. 10.1038/srep33509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhey, K.J., and Gaertig J.. 2007. The tubulin code. Cell Cycle. 6:2152–2160. 10.4161/cc.6.17.4633 [DOI] [PubMed] [Google Scholar]
- Warburton-Pitt, S.R.F., Silva M., Nguyen K.C.Q., Hall D.H., and Barr M.M.. 2014. The nphp-2 and arl-13 genetic modules interact to regulate ciliogenesis and ciliary microtubule patterning in C. elegans. PLoS Genet. 10:e1004866. 10.1371/journal.pgen.1004866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermann, S., and Weber K.. 2002. Identification of CfNek, a novel member of the NIMA family of cell cycle regulators, as a polypeptide copurifying with tubulin polyglutamylation activity in Crithidia. J. Cell Sci. 115:5003–5012. 10.1242/jcs.00170 [DOI] [PubMed] [Google Scholar]
- Wiederstein, M., and Sippl M.J.. 2007. ProSA-web: Interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 35:W407–W410. 10.1093/nar/gkm290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiegering, A., Rüther U., and Gerhardt C.. 2019. The role of primary cilia in the crosstalk between the Ubiquitin⁻Proteasome system and autophagy. Cells. 8:241. 10.3390/cells8030241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wloga, D., Camba A., Rogowski K., Manning G., Jerka-Dziadosz M., and Gaertig J.. 2006. Members of the NIMA-related kinase family promote disassembly of cilia by multiple mechanisms. Mol. Biol. Cell. 17:2799–2810. 10.1091/mbc.e05-05-0450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wloga, D., Joachimiak E., Louka P., and Gaertig J.. 2017. Posttranslational modifications of tubulin and cilia. Cold Spring Harb. Perspect. Biol. 9:a028159. 10.1101/cshperspect.a028159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff, A., de Néchaud B., Chillet D., Mazarguil H., Desbruyères E., Audebert S., Eddé B., Gros F., and Denoulet P.. 1992. Distribution of glutamylated alpha and beta-tubulin in mouse tissues using a specific monoclonal antibody, GT335. Eur. J. Cell Biol. 59:425–432. [PubMed] [Google Scholar]
- Xie, C., Li L., Li M., Shao W., Zuo Q., Huang X., Chen R., Li W., Brunnbauer M., Ökten Z., et al. 2020. Optimal sidestepping of intraflagellar transport kinesins regulates structure and function of sensory cilia. EMBO J. 39:e103955. 10.15252/embj.2019103955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, J., and Zhang Y.. 2015. Protein structure and function prediction using I-TASSER. Curr. Protoc. Bioinformatics. 52:5.8.1–5.8.15. 10.1002/0471250953.bi0508s52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y., Zhang X., Dai Y., Song M., Zhou Y., Zhou J., Yan X., and Shen Y.. 2021. The decrease of intraflagellar transport impairs sensory perception and metabolism in ageing. Nat. Commun. 12:1789. 10.1038/s41467-021-22065-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zocchi, R., Compagnucci C., Bertini E., and Sferra A.. 2023. Deciphering the tubulin language: Molecular determinants and readout mechanisms of the tubulin code in neurons. Int. J. Mol. Sci. 24:2781. 10.3390/ijms24032781 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
shows the candidate interactors of nekl-4 and their functions.
shows the strains used in this study.
shows the strains used in the supplemental material.
Data Availability Statement
All data are present in the paper and/or the online supplemental material and will be made available upon request.