Abstract
For over a decade, multiple studies have disputed the notion of natural killer (NK) cells as purely innate lymphocytes by demonstrating that they are capable of putative antigen-specific immunological memory against multiple infectious agents including two critical global health priorities – human immunodeficiency virus (HIV) and influenza. However, the mechanisms underlying antigen specificity remain unknown. Herein, we demonstrate that antigen-specific human NK cell memory develops upon exposure to both HIV and influenza, unified by a conserved and epitope-specific targetable mechanism largely dependent on the activating CD94/NKG2C receptor and its ligand HLA-E, and confirm these findings by three rigorous assays. We validated the permanent acquisition of antigen-specificity by individual memory NK cells by single-cell cloning. We identified biomarkers of antigen-specific NK cell memory through complex immunophenotyping by 30-parameter flow cytometry showing elevated expression of KLRG1, α4β7, and NKG2C. Finally, we show individual HLA-E-restricted peptides that may constitute the dominant NK cell response in HIV-1- and influenza-infected persons in vivo. Our findings clarify the mechanisms behind antigen-specific memory NK cell responses, and suggest they could be targeted for future vaccines, cure strategies, or other therapeutic interventions.
INTRODUCTION
Natural killer (NK) cells are lymphocytes classically considered part of the innate immune system based on their ability to mediate very rapid and nonspecific responses to virally infected or neoplastic cells. A plethora of studies have provided compelling evidence for the significant contribution of NK cells to the immune control of human immunodeficiency virus (HIV) and influenza virus infections (1–3), two of the most critical global health priorities. NK cells represent a potent antiviral effector cell population that can promptly respond to HIV and influenza exposure without the need for prior antigen sensitization. The interest in harnessing NK cell functions for vaccine design and therapeutic interventions against these pathogens has dramatically increased in recent years, largely driven by a series of observations indicating that subpopulations of NK cells, called memory or adaptive NK cells, manifest multiple different forms of durable adaptive capabilities (4). This includes reports of true antigen-specific memory NK cells that can mediate recall responses against multiple infectious agents including HIV (5–7) and influenza (5, 8–10). Adaptive NK cell subsets have been associated with protective effects in people living with HIV (PLWH)(11–14) and exposure to influenza antigens induces protective influenza-specific memory NK cells in mouse models (5, 8). Thus, antigen-specific memory NK cells represent a third arm of the immune system that can be targeted by prophylactic or therapeutic interventions.
To date, only a few studies support the existence of true antigen-specific recall responses by NK cells in humans, which have been described against cytomegalovirus (CMV), varicella zoster virus (VZV), hepatitis A (HAV) and B (HBV) virus, and bacillus Calmette-Guerin (7, 15–19) and the opacity surrounding the mechanisms of memory NK cell formation represents a major obstacle to harnessing adaptive NK cell functions. Herein, we demonstrate that antigen-specific NK cell memory develops upon exposure to HIV and influenza in humans, as well as provide mechanistic evidence for memory NK cell recognition of target antigens at the single-cell and single-peptide level.
RESULTS
Evidence for human NK cells mediating HIV- and influenza antigen-specific responses.
Clear evidence of HIV- and influenza-specific recall responses by NK cells in humans is lacking. We first tested HIV antigen specificity using a modified intracellular cytokine staining (ICS) assay designed to uniquely quantify Gag-specific peripheral blood NK cells in viremic PLWH or healthy donors by CD107a upregulation and IFN-γ production against autologous B lymphoblastoid cell lines (BLCL) pulsed with a pool of HIV Gag peptides (Fig. 1A, Fig. S1A, Table S1). In PLWH, 0.5 to 6% of NK cells were reactive against HIV Gag , mirroring responses found in SIV-infected rhesus macaques (RM)(6), whereas in healthy donors, NK cell responses were generally undetectable above background. CD107a upregulation and IFN-γ production by HIV-specific NK cells were significantly correlated (Spearman r=0.8929; p=0.01), suggesting antigen-specific NK cell responses may not be exclusively cytotoxic. The capacity of NK cells isolated from PLWH to specifically react against HIV antigens was further confirmed using a flow cytometry-based cytotoxicity assay. To do so, we assessed the killing of autologous BLCL pulsed with a pool of HIV Gag peptides by untouched NK cells isolated either from PLWH or from healthy donors (Fig. 1B, Fig. S1B). Significant anti-HIV Gag activity by NK cells could only be detected in PLWH, while killing of BLCL pulsed with a pool of Epstein–Barr virus (EBV), CMV and influenza peptides was comparable between PLWH and healthy individuals. Potent lysis of major histocompatibility complex (MHC)-devoid K562 cells, a target cell line lacking MHC commonly used to evaluate NK cell cytotoxicity and that triggers non-antigen-specific stimulation, was similarly achieved by NK cells from PLWH and HIV-negative donors. PLWH included patients on combination antiretroviral therapy (cART) with undetectable viral loads, untreated viremic PLWH, as well as HIV elite controllers who achieve spontaneous control of viral replication in the absence of treatment (Table S1). NK cells from elite controllers with detectable HIV gag-specific cytotoxic activity displayed the most potent responses observed. These data showed that NK cells that can specifically respond to HIV antigens develop within PLWH and mediate potent anti-HIV activity, suggesting that HIV-specific NK cells might significantly contribute to the control of HIV. Using a similar ICS-based assay as described above, we next measured the ability of human NK cells to mediate antigen-specific responses to influenza, to which the vast majority, if not all, healthy adults likely have pre-existing immune memory (20–22). Enriched NK cells from donors were stimulated with peptide pools derived from the conserved matrix protein 1 (MP1) and nucleoprotein (NP) of the 2009 H1N1 pandemic strain (A/California/08/2009 and A/California/04/2009, respectively) (Fig. 1C, Fig. S1C). We detected positive NK cell responses against influenza in approximately 45% of HIV-negative donors who provided blood samples between 2017 and 2020. Altogether, these results suggested that peripheral blood NK cells can mediate antigen-specific recall responses in humans, a hallmark of memory.
Fig. 1. Human NK cells mediate antigen-specific responses against HIV and influenza.
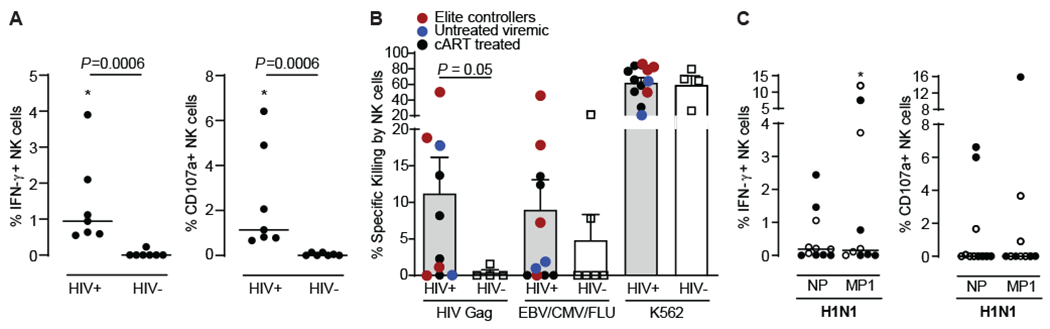
(A) Enriched NK cells from PLWH (N=7) or healthy donors (N=7) were co-cultured with autologous BLCL that had been pulsed with 2ug/mL peptide pools derived from HIV Gag (HIV-1 Consensus B; provided by the NIH AIDS Reagent Program) and NK cell responses assessed by ICS. Dead cells were excluded. Dot plots show proportions of IFN-γ+ and CD107a+ NK cells after subtracting background (unstimulated). (B) Autologous BLCL were pulsed with a pool of HIV Gag overlapping peptides or with the CEF (CMV, EBV and influenza) control peptide pool and were labeled with the CellTrace Violet dye. Mock-pulsed BLCL serving as intra-well controls were labeled with the green dye CFSE. Purified NK cells from PLWH (N=12) or healthy donors (N=6) were co-cultured with BLCL at 5:1 E:T ratios (equal mixture of pulsed target BLCL and unpulsed control BLCL) for 16 hours, and specific lysis of BLCL was determined by flow cytometry. Killing of HLA-deficient K562 cells was used as additional positive control. (C) Enriched NK cells from 11 HIV-negative healthy donors were incubated overnight with 2ug/mL peptide pools derived from influenza A/California/04/2009(H1N1) NP and A/California/08/2009(H1N1) MP1 and NK cell responses assessed by ICS. Dead cells were excluded. Dot plots show proportions of IFN-γ+ and CD107a+ NK cells after subtracting background (unstimulated). Full circle, positive for IgG antibodies against influenza A by ELISA. Empty circle, not tested for the presence of IgG antibodies against influenza A. Asterisks, significant differences compared to unstimulated controls. Data are represented as median and individual data points (A and C) or mean ± SEM and individual data points (B). Statistical significance was tested using Mann-Whitney U test for comparisons between PLWH (HIV+) and HIV-negative donors (HIV-) (A and B), or Wilcoxon signed-rank test for comparisons between unstimulated and stimulated NK cells (C). * p<0.05.
Single-NK cell cloning confirms antigen-specificity of memory NK cells.
To further investigate the existence of antigen-specific memory NK cells that can efficiently respond to HIV and influenza, and study these likely rare cells in more detail, we clonally expanded individual peripheral blood NK cells from 19 PLWH (4 cART-treated and 15 viremic untreated) and 10 healthy adults (Fig. S2). Using a cytotoxicity assay, we assessed the ability of single NK cell clones (NKCL) to lyse MHC-devoid K562 cells (to confirm normal NK cell function) or autologous BLCL pulsed with a pool of self-peptides (to exclude autoreactive NKCL), CMV pp65-, HIV Gag-, HIV Envelope (Env)-, and A/California/04/2009 H1N1 NP-derived overlapping peptides (Fig. 2, Fig. S3). As expected, NKCL could potently lyse K562 cells. A fourth of NKCL from PLWH and half of NKCL from healthy donors had little to no reactivity to unpulsed BLCL and/or BLCL pulsed with a self-peptide control and were responding to viral peptide pools (Fig. S4). Among these, approximately 25% of NKCL from PLWH (Fig. S4a–b) and 50% of NKCL from healthy adults (Fig. S4C–D) displayed over 10% antigen-specific killing against HIV Gag/Env or influenza NP, respectively. Strikingly, we were able to isolate NKCL with very robust anti-HIV (up to 87% specific lysis against Env) or anti-influenza H1N1 NP (up to 57% specific) cytotoxic activity. While HIV-specific responses could be detected in both untreated and treated PLWH, the magnitude of antigen-specific killing was overall higher in viremic untreated compared to treated participants (Fig. S5A–C). We could not detect HIV antigen-specific killing above background mediated by NKCL generated from HIV-negative healthy donors (Fig. S5D). Interestingly, among the 16 NKCL that displayed the highest antigen-specific lysis against HIV, 5 NKCL (31%) had over 15% specific killing against both HIV Env and HIV Gag or HIV Env and CMV pp65, while NKCL displaying over 50% antigen-specific killing were able to respond to either Gag or Env but never both (Fig. 2E). Together, these data demonstrate the existence of HIV- and influenza-specific NK cells in humans and show that NK cells displaying true and robust antigen-specific killing are found at the single cell level.
Fig. 2. Single-cell cloning of human antigen-specific NK cells.
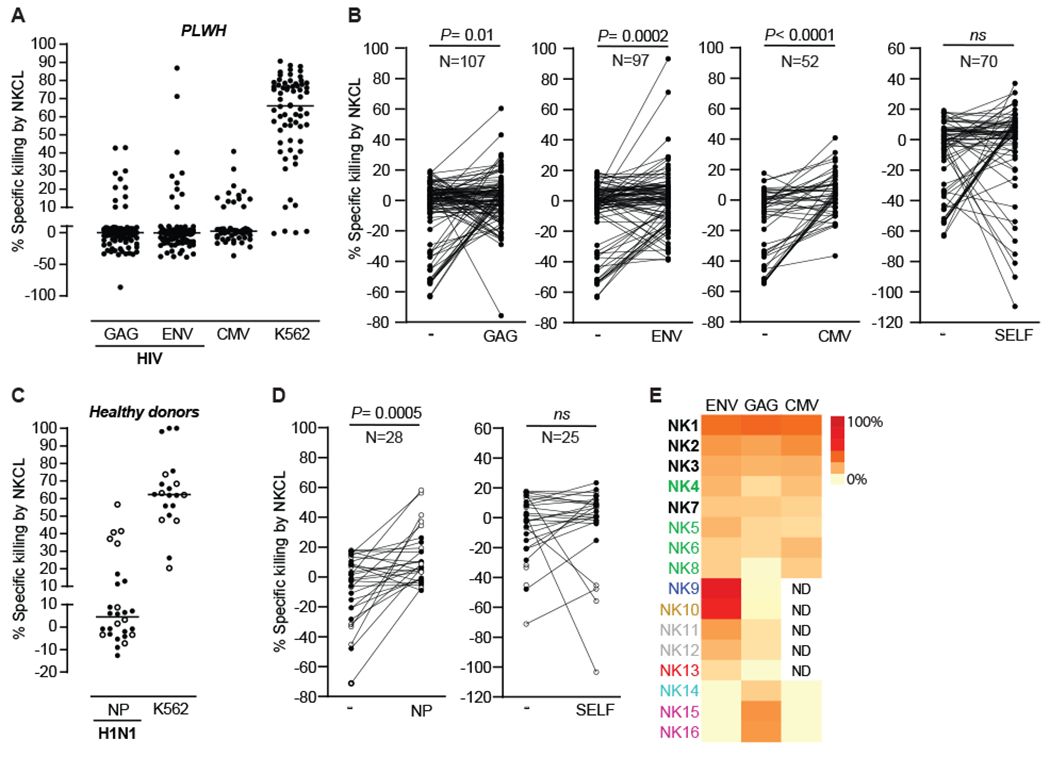
Percentages of antigen-specific lysis by 133 NKCL generated from 19 PLWH against HIV Env, HIV Gag and CMV pp65 (A) after subtracting background (mock-pulsed BLCL) and (B) compared to non-specific killing of mock-pulsed BLCL for each NKCL. Percentages of antigen-specific lysis by 28 NKCL generated from 10 healthy donors against influenza H1N1 NP (C) after subtracting background (mock-pulsed BLCL) and (D) compared to non-specific killing of mock-pulsed BLCL for each NKCL. Full circle, positive for IgG antibodies against influenza A. Empty circle, not tested for the presence of IgG antibodies against influenza A. CAM cytotoxicity assays were used to evaluate lysis after co-culture of NKCL with autologous BLCL pulsed with indicated peptide pools. Non-specific lysis was assessed by measuring killing of mock-pulsed autologous BLCL, self-peptides-pulsed BLCL (negative control) and HLA-E-deficient K562 cells (positive control). (E) Heatmap of antigen-specific lysis after subtracting background for 16 NKCL from 8 PLWH with the highest specific killing against HIV Gag and/or ENV. NKCL generated from the same PLWH are displayed in a unique color. NKCL displaying over 15% specific killing against both HIV Env and HIV Gag or HIV Env and CMV pp65 are indicated in bold font. Statistical significance was tested using Wilcoxon signed-rank test (B and D). ns, not significant.
Mechanisms of memory NK cell target recognition and killing are largely dependent on the activating NKG2C receptor.
To define the subset of peripheral blood NK cells that drives antigen-specific memory responses, we compared expression levels of 21 cell surface receptors between NKCL with anti-HIV Env/Gag activity and NKCL that did not react to those HIV antigens using advanced multiparameter flow cytometry (Table S2). Phenotypic analysis revealed that most HIV-specific NKCL express activating (i.e., NKp46, NKG2D, NKG2C) and inhibitory (NKG2A, CD85j) receptors typically found on NK cells, trafficking markers (i.e., α4β7, CXCR6, CCR5, CCR7), co-stimulatory molecules (i.e., 2B4, CD2, CD8), and various levels of maturation/activation markers (CD57, KLRG1, PD1, Tim-3, HLA-DR) or killer-cell immunoglobulin-like receptors (KIRs) (Fig. S6–S7). Compared to NKCL that did not react to HIV, HIV-specific NKCL had enhanced expression of KLRG1 and α4β7 and lower expression of CCR5 (Fig. S7). Furthermore, analysis using a non-linear dimensionality reduction algorithm (t-SNE) revealed distinct clustering for HIV reactive and non-HIV-reactive NKCL, with HIV-reactive NKCL being partly associated with cell clusters expressing high levels of the inhibitory receptor NKG2A or high levels of activating NKG2C and/or inhibitory KIR3DL1 (Fig. 3A and Fig. S8).
Fig. 3. Human antigen-specific NK cell responses are associated with NKG2C expression.
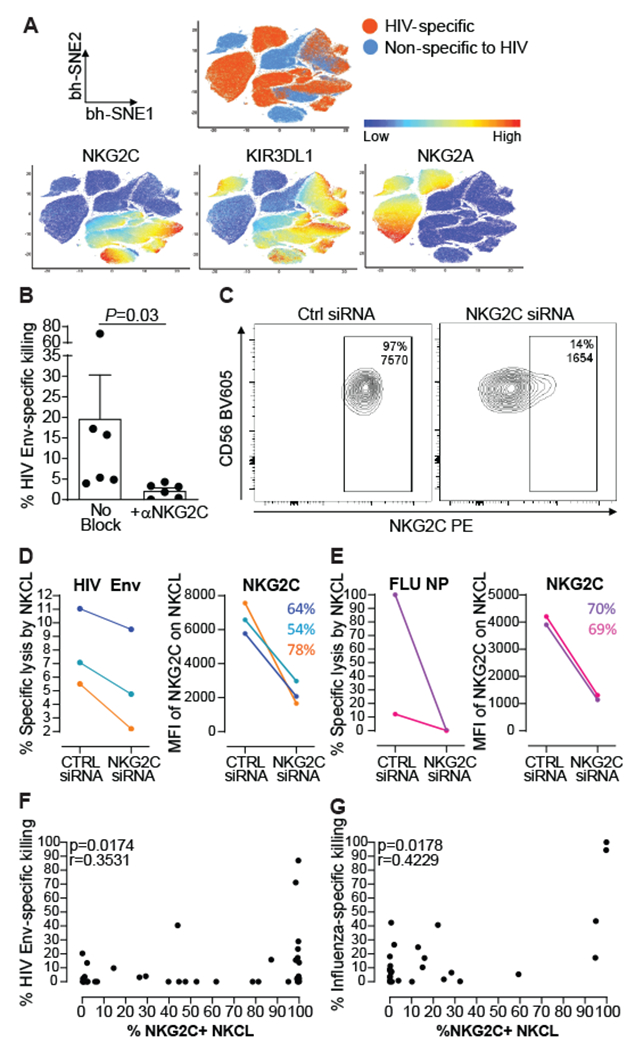
(A) Non-linear t-SNE (bh-SNE) plots of 14 NKCL from one untreated viremic PLWH, showing distinct clustering for 7 HIV reactive and 7 non-HIV-reactive NKCL, with HIV-reactive NKCL being partly associated with cell clusters expressing high levels of the inhibitory NKG2A or high levels of activating NKG2C and/or inhibitory KIR3DL1. (B) Antigen-specific killing of HIV Env-pulsed BLCL by 6 NKCL in the presence of isotype control or NKG2C blocking antibodies. Bars represent mean+SEM. Statistical significance was tested using Wilcoxon signed-rank test. (C) Representative flow cytometry plots showing NKG2C expression on one NKCL 48 hours after nucleofection with a scramble siRNA control (Ctrl siRNA) or with siRNA targeting NKG2C. (D) Percentages of lysis by 3 NKG2C+ NKCL reacting against HIV Env (left panel) and MFI of NKG2C on the respective NKCL (right panel) from 2 PLWH 48 hours following nucleofection with siRNA control or siRNA targeting NKG2C. (E) Percentages of lysis by 2 NKG2C+ NKCL reacting against influenza NP (A/AnnArbor/6/1960(H2N2)) (left panel) and MFI of NKG2C on the respective NKCL (right panel) from 1 healthy donor 48 hours following nucleofection with siRNA control or siRNA targeting NKG2C. Percentages of NKG2C down-modulation are indicated for each NKCL. Only NKCL displaying responses over 5% specific killing and at least 1.5-fold above the background (unpulsed) after nucleofection with Ctrl siRNA were included in the analysis. (F), (G) Spearman correlation analysis between frequencies of NKG2C+ NKCL and specific killing of BLCL pulsed with HIV Env (F) or influenza NP, MP1, or HA (G). Frequencies of NKG2C+ NKCL were determined as the proportion of NKCL beyond an arbitrary cut-off line set by at the right-hand side border of the NKG2C histogram in FMO controls (See Fig. S9).
NKG2C is an activating NK cell receptor that forms heterodimers with CD94, is usually not co-expressed with its inhibitory counterpart NKG2A (23) (Fig. S14B), and interacts with HLA-E bound to a peptide (24–27). High cell surface expression of NKG2C/CD94 has been consistently linked to long-lived NK cells endowed with adaptive capabilities that develop upon human CMV infection (23, 28–31). Variants of CMV UL-40-derived peptides that mimic canonical HLA class-I (HLA-I)-derived leader peptides are presented by HLA-E and finely tune adaptive NKG2C+ NK cell functions (15, 18), suggesting HCMV-specific recognition via the NKG2C/HLA-E axis. Moreover, we previously showed that in non-human primates, HIV antigen-specific memory NK cell responses largely depend on NKG2A/C (6). As HIV-specific NKCL were associated with high NKG2C expression (Fig. 3A), we directly assessed the role of NKG2C in HIV antigen-specific memory NK cell responses. HIV Env-specific NKCL which were all positive for NKG2C, NKp30, and NKp44 were co-cultured with BLCL pulsed with HIV Env peptides in the presence of blocking antibodies against these activating receptors and specific lysis was measured (Fig. 3B, Fig. S9A). NKG2C blockade significantly decreased HIV Env-specific responses by NKCL, while blockade of natural cytotoxicity receptors either did not or marginally impacted HIV-specific NK cell responses, corroborating our previous findings (6). Furthermore, NKG2C knockdown in either HIV- or influenza-specific NKCL robustly decreased or completely abrogated killing of BLCL targets pulsed by HIV Env and influenza NP peptide pools, respectively (Fig. 3C–E). Accordingly, we found a positive correlation between HIV Env-specific killing and influenza-specific killing by NKCL and the proportions of NKG2C+ NKCL (Fig. 3F–G and Fig. S9B) and expression of NKG2C on NKCL (Fig. S9C–D), while these responses were not significantly associated with expression of NKG2A (Fig. S9E–F). Altogether, our results confirm that antigen-specific memory NK cell responses are largely dependent on NKG2C, likely indicative of an HLA-E-dependent recognition mechanism.
HIV- and influenza-derived peptides bind HLA-E, the ligand for NKG2C, and potently activate antigen-specific NK cells.
HLA-E presents largely conserved nonameric peptides derived from leader sequences of other HLA-I molecules. However, pathogen-derived peptides have been shown to bind HLA-E (32–35). To establish the relevance of an HLA-E-dependent recognition mechanism in antigen-specific memory NK cell responses against two unrelated viruses, we first screened for canonical peptides derived from HIV Env, Gag, and influenza NP that can stabilize HLA-E at the surface of K562 cells stably expressing HLA-E*0101 but lacking cell-intrinsic HLA-E-binding peptides (Fig. 4A, Fig. S10, Table S3). Peptide pulsing at various concentrations revealed that among all tested peptides (84 for HIV Env, 60 for HIV Gag, 18 for influenza NP), two HIV Env-derived and one influenza NP-derived peptide could stabilize HLA-E to the same extent as the positive control (CMV UL40 VMAPRTLFL), with additional peptides derived from HIV Env (n=3), HIV Gag (n=5), and influenza NP (n=5) conferring lower levels of HLA-E stabilization (Fig. 4A, Fig. S11A). Using competition-binding assays, we could demonstrate that peptides known to strongly bind HLA-E (VMAPRTLFL) could be displaced by a subset of HIV- and influenza-derived peptides with enhanced potential to stabilize HLA-E on K562 cells, further supporting the ability of these nonameric peptides to form stable complexes with HLA-E (Fig. S11B).
Fig. 4. Human antigen-specific memory NK cell responses are mediated via recognition of HLA-E-binding viral peptides.
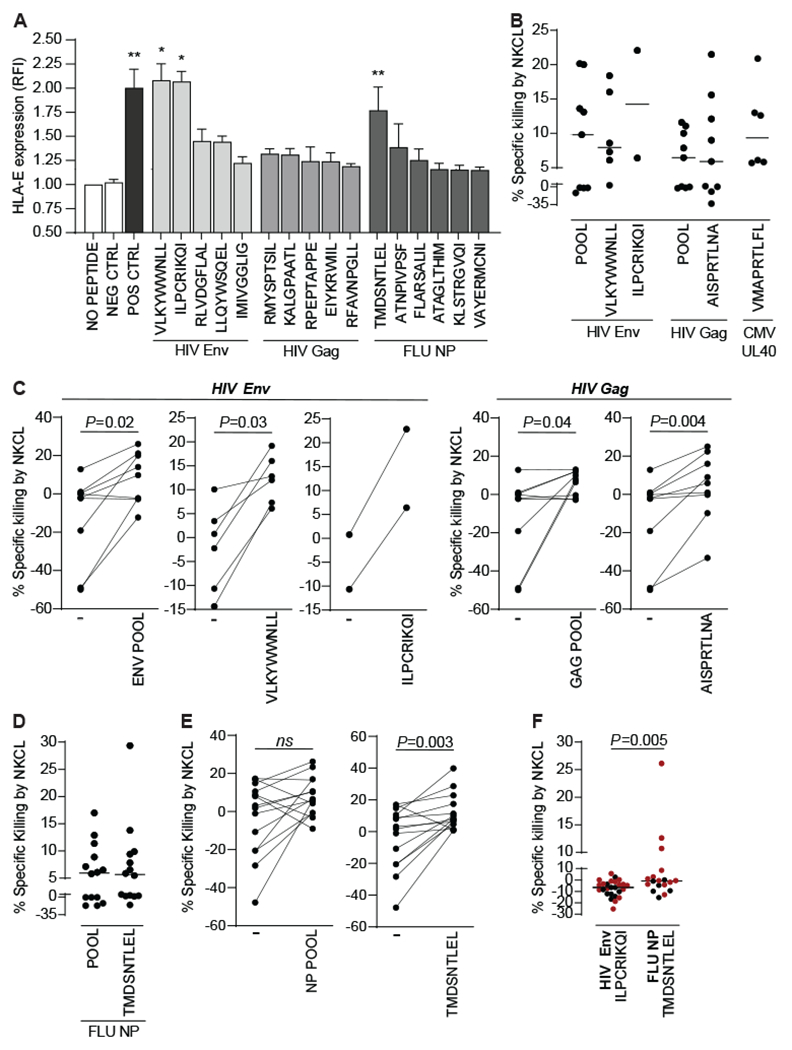
(A) K562 cells stably expressing HLA-E*0101 were either left mock pulsed or pulsed for 16h with 40nm of nonameric peptides derived from HIV consensus B Gag or Env or from A/California/04/2009(H1N1) NP. Controls included CMV pp65-derived NLVPMVATV that do not stabilize HLA-E (NEG), and VMAPRTLFL, a CMV/HLA-G leader sequence-derived peptide that stabilizes HLA-E (POS). HLA-E surface stabilization was assessed by flow cytometry. Bars represent the relative mean fluorescence intensity (RFI) + SEM of HLA-E on K562 HLA-E*01:01 cells pulsed with indicated individual peptides found to stabilize HLA-E and pooled from at least 2 distinct experiments as compared to HLA-E expression in the absence of peptide (No Peptide). The dotted line marks the cut off for increased HLA-E expression, defined as the mean value plus 2 standard deviations of non-stabilizing peptides. Asterisks, significant differences compared to K562 HLA-E*01:01 pulsed with non-binding peptides (NEG). *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001. Percentages of peptide-specific lysis by 17 NKCL from 5 untreated viremic PLWH (B) after subtracting non-specific lysis (killing of mock pulsed autologous BLCL) and (C), compared to non-specific killing of mock-pulsed BLCL for each NKCL. Percentages of peptide-specific lysis by 14 NKCL from 5 healthy donors (D) after subtracting non-specific lysis (killing of mock pulsed autologous BLCL) and (E), compared to non-specific killing of mock-pulsed BLCL for each NKCL. CAM cytotoxicity assays were used to evaluate lysis after co-culture of NKCL with autologous BLCL pulsed with peptide pools encompassing the whole HIV Env, HIV Gag or H1N1 NP sequence or with indicated single HLA-E-binding nonameric peptides derived from CMV UL40, HIV Gag, HIV Env, or H1N1 NP. (F) Percentages of peptide-specific lysis by 27 NKCL from 7 HIV-negative healthy donors after subtracting non-specific lysis (killing of mock pulsed autologous BLCL). CAM cytotoxicity assays were used to evaluate lysis after co-culture of NKCL with autologous BLCL pulsed with indicated single HLA-E-binding nonameric peptides derived from HIV Env or H1N1 NP. NKG2C+ NKCL are indicated in red. Statistical significance was tested using Kruskal Wallis test with Dunn’s multiple comparison test (A) or Wilcoxon signed-rank test (C, E and F).
To evaluate the effect of HLA-E-binding peptides on antigen-specific NK cell function, we compared killing by NKCL from PLWH of BLCL pulsed with peptide pools covering the whole HIV Gag and HIV Env with that of BLCL pulsed with single HLA-E-binding peptides. Tested single peptides included HIV Env-derived VLKYWWNLL and ILPCRIKQI or HIV Gag AISPRTLNA, a peptide that has been previously shown to significantly bind HLA-E and affect NK cell function (32, 33) (Fig 4B–C, Fig S11C–D, Fig. S12A–C). 4 out of 9 NKCL (44%) and 5 out of 6 NKCL (83%) reacted to single HLA-E-binding HIV Gag- and HIV Env-derived peptides (range of positive responses twice above the background: 6-21% Gag-specific and 6-22% Env-specific killing). Similarly, we evaluated killing of BLCL pulsed with either pools of 15-mer overlapping peptides encompassing the whole NP sequence or with TMDSNTLEL (all derived from A/California/7/2009 H1N1), by 14 NKCL from 5 healthy donors (Fig. 4D–E, Fig S11C–D, Fig. S12D–E). Nine NKCL (64%) reacted to TMDSNTLEL, among which 5 also displayed responses against the NP pool (range of positive responses twice above the background: 3-29% NP-specific killing). These results show that HIV Env-/Gag- and influenza NP-derived nonamers can bind HLA-E and potently activate patient-and healthy donor-derived memory NKCL. Importantly, NKG2C+ NKCL generated from HIV-negative donors were not able to react to HIV Env-derived HLA-E-binding ILPCRIKQI while a subset of them responded to influenza NP TMDSNTLEL (Fig. 4F). This suggests that peptide presentation through HLA-E is not sufficient to elicit killing by NKCL and that these responses likely depend on pre-exposure to a specific viral antigen, indicative of antigen-specific memory.
Next, we sought to confirm the in vivo relevance of the novel peptide responses we identified among single-cell clones. We showed stimulation with newly identified HIV Env- and influenza NP-derived HLA-E-binding peptides alone mostly promotes CD107a upregulation on a subset of primary NK cells in PLWH and healthy donors, respectively, with IFN-γ being produced to a lesser extent (Fig.5A–D, Fig. S13, Fig. S14). Degranulation responses to single HLA-E-binding peptides by NKG2C+ NK cells were significantly higher than those mediated by NKG2A+ NK cells or by NK cells negative for both receptors (Fig S14B–D). In healthy donors displaying enhanced CD107a upregulation on NK cells in response to TMDSNTLEL, NK cell degranulation was significantly impaired upon NKG2C blockade, further supporting a major role for the NKG2C/HLA-E axis in antigen-specific NK cell responses (Fig. 5E). CD107a upregulation in NK cells correlates significantly with NK cell-mediated cytotoxicity (36, 37) and in non-human primates, we also found a correlation between HIV-specific killing by memory NK cells and CD107a upregulation by memory NK cells (6). Thus, our results strongly indicate in vivo activation of cytotoxic antigen-specific adaptive NK cells by single HLA-E binding nonamers.
Fig. 5. HIV Env- and H1N1 NP-derived HLA-E-binding peptides elicit primary NK cell responses.
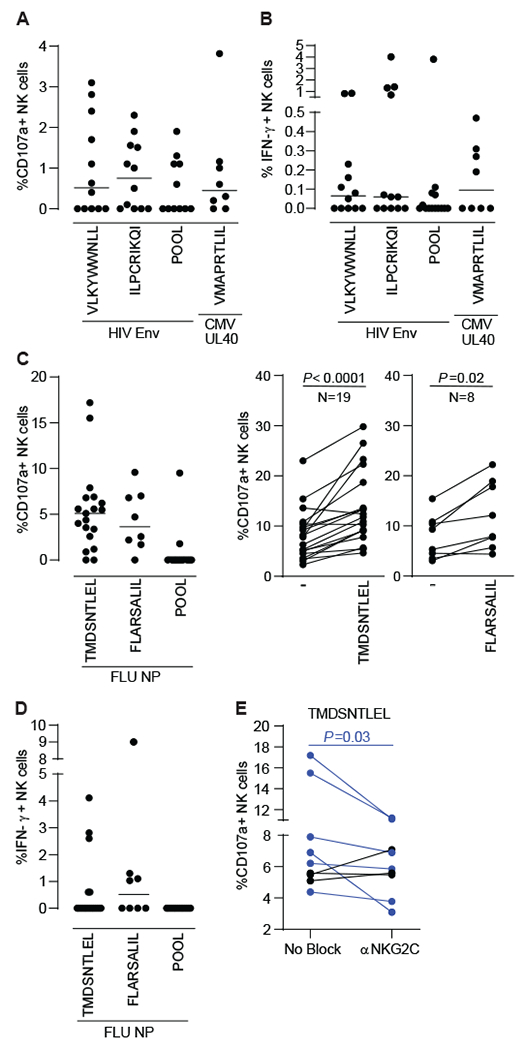
(A)(B) PBMCs samples from 12 PLWH (8 cART-treated and 4 untreated viremic) were incubated for 16h with 2μg/mL of 1 pool of 15 amino acid (aa) peptides overlapping by 11 aa and spanning HIV Env or single HIV Env- and CMV UL40-derived nonamers in the presence of CD107a antibodies. GolgiStop and GolgiPlug were added for the last 2 hours incubation prior to surface and intracellular cytokine staining to measure CD107a upregulation (A) and IFN-y production (B). Enriched NK cells from 19 healthy donors were incubated for 6 hours with 2μg/mL of 1 pool of 15 aa peptides overlapping by 11 aa and spanning H1N1 NP or 40uM of indicated single H1N1 NP-derived nonamers in the presence of CD107a antibodies, GolgiStop and GolgiPlug to evaluate CD107a upregulation (C) and IFN-γ production (D) by ICS. NK cell responses to H1N1 NP-derived TMDSNTLEL was evaluated in the presence of control or anti-NKG2C antibodies. Dot plots represent proportions of actively degranulating, cytotoxic NK cells, as judged by cell surface expression of CD107a (A, C left panel) or IFN-y producing NK cells (B, D) after subtracting proportions of CD107a+ and IFN-γ+ unstimulated NK cells, respectively. Bars represent the median. Paired plots compare CD107a expression between unstimulated and single-peptide-stimulated NK cells (C, right panels) or between NK cells stimulated with H1N1 NP-derived TMDSNTLEL in the presence of isotype control or anti-NKG2C antibodies after subtracting proportions of CD107a+ unstimulated NK cells (E). NK cells from all donors expressed NKG2C (PLWH: median 15.6%, range 2%-75%; healthy donors: median 10.3%, range 2%-48%). Statistical significance was tested using Wilcoxon signed-rank test (C and E).
Collectively, our work provides a dominant recognition mechanism underlying antigen-specific recall responses mediated by NK cells in humans, which have the potential to be harnessed for vaccine design or other therapeutic interventions.
DISCUSSION
Thus far, antigen-specific memory responses mediated by NK cells in humans have been described in VZV-experienced adults (7) as well as in HBV-infected or HAV/HBV-vaccinated individuals (17, 19). While the idea of primate NK cell memory has become well-accepted among immunologists, there have been no reported mechanisms describing how NK cells recognize and distinguish individual antigens, until now. Our study provides mechanistic evidence of human NK cell memory against pathogens of global public health importance that cannot currently be efficiently prevented by vaccination. We show that HLA-E possesses the previously unappreciated ability to bind HIV and influenza peptides and activate virus-specific NK cells. HLA-E has gained significant attention in the vaccine field as broad Mamu-E restricted CD8+ T cell responses have been implicated as immune correlates of protection in RM vaccinated with a CMV-vectored SIV vaccine (38, 39), and this overall function may be conserved between RM Mamu-E and human HLA-E (40). Vaccine strategies that mobilize HLA-E could induce HIV- or influenza-specific memory NK cells that recognize broad epitopes across viral antigens. One major issue for the development of an HIV vaccine or a universal influenza vaccine is the considerable variability of viral strains circulating among humans (41, 42). Antigen-specific NK cell responses relying on the NKG2/HLA-E axis is in accordance with the fact that in our studies, some antigen-specific NKCL were able to potently react to several antigens from the same as well as from different viruses. Further, we present primary HLA-E-mediated responses for both pathogens which may represent dominant memory NK cell responses in vivo that could be further targeted. This feature could be highly beneficial for vaccine design as it could allow a limited number of memory NK cells to target a broader range of HIV and influenza strains than memory T cells.
In addition to the lack of mechanism previously described for NK cell memory, the field also continues to search for comprehensive biomarkers to distinguish adaptive subpopulations of NK cells from classical NK cells. CD49a, CXCR6, NKG2C, and suppressed gamma signaling chain have been implicated to partially delineate these populations, but particularly for true antigen-specific memory NK cells these phenotypes maybe be incomplete (43). Notably, previous studies have identified markers for antigen-specific memory NK cells associated with the liver, a putative site for antigen-specific NK cell maintenance (5, 7, 19), while our studies focused on peripheral blood NK cells. This major difference may explain why we did not observe any association between CXCR6 expression and antigen-specific responses by NK cells. However, we document multiple phenotypes that advance biomarker usage for NK cell memory. KLRG1 has been described as a marker of NK cell activation (44), which defines subsets of memory-like NK cells with protective functions against Mycobacterium tuberculosis (45) and subsets of NK cells that mediate antigen-specific memory responses against HBV antigens (17). Upregulation of KLRG1 on HIV-specific NKCL further suggests that KLRG1 might be a useful biomarker for identifying antigen-specific memory NK cells among bulk NK cells. Additionally, although it did not reach statistical significance, α4β7 expression trended to be upregulated on HIV-specific NKCL, and this corroborates previous observations that HIV and SIV infection specifically mobilize mucosal-homing NK cells (46, 47).
Not surprisingly, due to the mechanism described in these studies and others, NKG2C was also highly expressed on memory NK cells, and we acknowledge a possible overlap between antigen-specific memory NK cells described in this manuscript and the previously described CMV-driven adaptive NKG2C+ NK cell subsets. However, we want to emphasize that true antigen-specific memory NK cells are scarce in the peripheral blood, and therefore it is expected that only a minor portion of all NKG2C+ NK cells are endowed with antigen-specific memory features. Thus, these cells can also be found in CMV seronegative individuals with low proportions of NKG2C+ NK cells. A possible model to support the scarcity of antigen-specific memory NK cells observed in our studies is that upon viral infection, only a few viral peptides can be presented by HLA-E, and an even smaller proportion of these peptides can mediate a potent interaction with CD94/NKG2C and activate NK cells. This is supported by recent studies that identified CMV- and human-derived peptides that stabilize HLA-E and selectively activate CD94/NKG2C+ NK cells while mediating minimal inhibitory effects through CD94/NKG2A (48). Such rare events that rely on strong activating viral ligands may trigger a first set of epigenetic alterations, which affect the functionality of a small subset of CD94/NKG2C+ NK cells, similarly to what has been proposed for CMV-driven adaptive NK cells (49). These epigenetic signatures are further modified overtime as these NK cells are triggered by strong CD94/NKG2C ligands from other infections or viral reactivation that modify the HLA-E ligandome and generate, in our case, NK cells with enhanced responsiveness to viral peptides via CD94/NKG2C stimulation.
Moreover, antigen-specific memory NK cell responses likely do not solely depend on the NKG2C/HLA-E axis, and other pathways may be additive or alternative to this mechanism. Besides triggering NK cell activation through interactions between HLA-I ligands and activating NK cell receptors, influenza- or HIV-derived peptides may also disrupt inhibitory signals mediated through NKG2A (33, 50) or other inhibitory molecules (51) and HLA-E or their cognate HLA-I ligand, respectively, further promoting NK cell responses. This possible mechanism is consistent with HIV-reactive NKCL being partly associated with high expression levels of the inhibitory NKG2A and KIR3DL1 receptors (Fig. 3A and Fig. S8), and with the fact that in our studies, potent NK cell responses to HLA-E stabilizing peptides was occasionally observed in the absence of NKG2C expression. A potential role for NKG2A in human NK cell memory is further supported by studies demonstrating that in African green monkeys, the natural host of SIV, a subset of terminally differentiated adaptive NKG2A+ NK cells displaying enhanced MHC-E-restricted cytotoxicity is associated with control of SIVagm infection in secondary lymphoid tissues (52). Activating KIRs represent another particularly important family of receptors that may regulate memory NK cell function. Specific KIR genes expressed in conjunction with their HLA ligands have been clearly associated with improved control of viral infections (53), and while the interaction of inhibitory KIRs with HLA ligands has been studied in detail, ligands for most activating KIRs remain elusive. Future studies beyond the scope of this work will be required to determine if KIR-mediated recognition is complementary or independent to this highly dominant HLA-E-dependent mechanism.
Overall, our efforts have addressed two major deficits to the field of immunology: we demonstrated that a conserved and epitope-specific targetable mechanism predominantly dependent on NKG2C/HLA-E underlies true antigen-specific memory NK cell responses in humans, and, together with previous observations, our detailed phenotypic analyses suggest that complex phenotypes may be the most effective at tracking antigen-specific memory NK cells in vivo rather than individual markers.
MATERIALS AND METHODS
Study design
The aim of this study was to identify memory NK cells that develop against HIV-1 and influenza viruses in humans and to characterize the mechanisms underlying their antigen-specific responses. We used high-dimensional flow cytometry, single NK cell cloning and functional assays to interrogate protein expression and peptide-specific reactivity of memory NK cell subsets. The data collectively supported the existence of antigen-specific recall responses by human NK cells that mainly rely on the NKG2/HLA-E axis. For each experiment, the number of biological replicates and statistical significance are indicated in figure legends. Investigators were not blinded during experimental procedures due to the nature of the study, but individual experiments were blinded to assessment of outcomes. Numbers of participants for ex vivo experiments were determined on the basis of previous expertise.
Flow cytometry-based antigen-specific NK cell killing assay
EBV-transformed BLCL were generated as autologous targets for antigen-specific NK cell assays using standard protocols. BLCL functional capacity was confirmed by processing of DQ-Ovalbumin (Molecular Probes). K562 cells and autologous BLCL were labeled with 1uM CellTrace™ Violet Cell Proliferation Kit (Invitrogen) for 20 minutes according to manufacturers’ instructions. CellTrace Violet-labelled BLCL were then pulsed for 1.5 hours at 37°C with 2ug/mL HIV Gag (HIV-1 Consensus B; by the NIH AIDS Reagent Program) or CEF (CMV, EBV and influenza)-derived overlapping peptide pools in culture medium consisting in RPMI-1640 supplemented with 2mM L-glutamine, 100ug/ml streptomycin, 100U/ml penicillin and 5% human serum, washed with PBS and resuspended in culture medium. Unpulsed BLCL served as internal control and were labeled with 2μM CellTrace™ CFSE Cell Proliferation Kit (Invitrogen) for 15 minutes according to manufacturers’ instructions, washed, and resuspended in culture medium. NK cells were purified using AutoMACS NK cell enrichment kit (Miltenyi) and resuspended in culture medium with 0.1ng/mL recombinant human IL-15. 350,000-400,000 NK cells were co-cultured with BLCL at 10:1 E:T ratio containing an equal mixture of pulsed target cells and unpulsed control cells or K562 cells for 16 hours at 1M/mL, and subsequently stained using CD3-A700 (UCHT1), CD19-BV711 (HIB19), CD16-BV785 (3G8), and CD56-PEcy7 (B159). Specific lysis of BLCL was calculated as follows: (% sample lysis with NK effectors - % basal lysis without NK effectors) / (100 - % basal lysis without NK effectors) (Fig. S2).
Analysis of primary NK cell responses to viral peptides by intracellular cytokine staining
To measure primary antigen-specific NK cell responses, cryopreserved PBMC from healthy donors or PLWH were thawed and used either unfractionated or after CD3+ T cell depletion using the EasySep Human CD3 Positive Selection Kit II and the protocol for Using EasySep Positive Selection Kits for Cell Depletion provided by the manufacturer (STEMCELL Technologies). Autologous BLCL were pulsed for 1.5 to 2 hours at 37°C with 2μg/mL of peptide pools consisting of 15-mer sequences with an 11 aa overlap covering the complete sequence of HIV Gag (NIH AIDS Reagent Program) (Fig. 1A). After washing, peptide-loaded BLCL were co-cultured with enriched NK cells at a 1:1 E:T ratio at 37°C for 15 hours with CD107a-FITC (H4A3) (BD Biosciences). Unfractionated or CD3-depleted PBMC were stimulated directly with 2μg/mL of peptide pools consisting of 15-mer sequences with 11 aa overlap covering the complete sequence of the influenza virus A/California/04/09(H1N1) NP (PepTivator, Miltenyi), A/California/08/2009(H1N1) MP1 (PepMix, JPT Peptide Technologies), CMV pp65 (NIH AIDS Reagent Program), HIV Env and HIV Gag (both from NIH AIDS Reagent Program), or with 20-40μM of single nonameric synthetic peptides of ≥98% purity derived from influenza NP, HIV Env, HIV Gag, or CMV UL40 (Thermofisher) without the addition of BLCL (Fig. 1C, Fig. 5, Fig. S13). In some instances, 1ng/mL of recombinant human IL-15 (R&D) was also added (Fig 1C, Fig 5C–E). Unfractionated PBMC or CD3-depleted PBMCs were co-cultured with peptides at 37°C for 6 to 15 hours with CD107a-BV786 (BD Biosciences). 1μL/mL GolgiPlug (BD Biosciences) and 0.7μL/mL GolgiStop (BD Biosciences) were added for the whole 6 hours of incubation or, when incubation lasted over 6 hours, for the last 2 hours of incubation. For blockade experiments, CD3-depleted PBMC were incubated for 10 minutes at room temperature with 2.5μg of Human BD Fc Block followed by 20 minutes at 37°C in the presence of 10μg/mL of anti-NKG2C purified antibody (R&D, clone 134522) or isotype control antibodies prior to stimulation with peptides (Fig. 5E). At the end of the incubation, cells were stained first with the LIVE/DEAD® Fixable Blue Dead Cell Stain Kit (Invitrogen), then with BD Biosciences CD3-BV510 or CD3-A700 (UCHT1), CD14-BV421 (M5E2), CD19-BV421 (HIB19), CD16-APC-Cy7 (3G8), CD56-BV605 (NCAM16.2 or B159), Beckman Coulter NKG2A-PE-Cy7 (Z199), and Miltenyi NKG2C-PE (REA205) to gate on NK cell subsets, and finally fixed, permeabilized (Thermofisher Fix and Perm), and stained with BD Biosciences IFN-γ-FITC (B27) antibodies to detect intracellular cytokines. In all assays described above, incubation in the presence of 5μg/mL of phytohemagglutinin (PHA) or PMA/ionomycin were used as positive controls and unstimulated NK cells or, when applicable, unpulsed BLCL served as negative controls and for background subtraction. Positive responses were defined as %IFN-γ+ or %CD107a+ NK cells twice above the background (unstimulated/unpulsed) and over 1% after background subtraction. A Fluorescence Minus One (FMO) control and PHA-stimulated PBMC were used to set the gates for positive cytokine responses. Acquisition of data was performed on a BD LSRII instrument or a FACSymphony Analyzer (X-50) (BD Biosciences). Data was analyzed using Flow Jo v.10.8.1.
Generation of primary NK cell clones (NKCL)
Single primary human NK cells were cloned by limiting dilution or single-cell sorting using a protocol adapted from a previously reported method (54) (Fig. S2). Briefly, NK cells were isolated from fresh or cryopreserved PBMC via negative selection (EasySep Cell Separation; StemCell Technologies), added to a mix of irradiated feeders consisting of freshly isolated allogeneic PBMC combined with log-phase-growth RPMI 8866 cells (Sigma-Aldrich) at a 10:1 ratio in cloning medium. NKCL were stained for flow cytometric phenotyping using BD Biosciences CD3-A700 (UCHT1), CD16-APC-Cy7 (3G8), CD56-BV605 (NCAM16.2), Beckman Coulter NKG2A-PE-Cy7 (Z199), R&D NKG2C-PE (134591), or Miltenyi NKG2C-PE (REA205). Only NKCL that were CD3−CD56+ were used directly for subsequent assays. Importantly, NKCL generated using this procedure maintained their receptor profile and functional characteristics following expansion (Fig. S2B–C). For instance, NKCL generated from the same individual differ in their response to HLA-I-devoid K562 target cells, and, according to their KIR3DL1 expression, to 221 cells expressing a single HLA-B allele (Fig. S2C).
Human Subjects
De-identified and coded blood samples from the majority of HIV-negative healthy donors used in this study were collected under IRB-approved protocols and delivered to us by Research Blood Components, LLC or Zen-Bio, Inc. Additional healthy donors and all PLWH were recruited from outpatient clinics at MGH and affiliated Boston-area hospitals, at UAB University Hospital, and at Duke University Medical Center. The respective institutional review boards approved this study, and all subjects gave written informed consent. A total of 4 elite controllers, 27 untreated viremic chronic progressors, and 21 cART-treated chronically infected PLWH were studied (Table S1). Elite controllers were defined as having plasma HIV RNA levels of <50 copies/ml in the absence of antiretroviral therapy, on at least three determinations over at least a year. Untreated viremic chronic progressors were defined as subjects having untreated HIV infection for >1 year with plasma viral loads of >2,000 copies/ml for at least 1 year; cART-treated chronically infected had HIV RNA levels below the limit of detection for standard assays. PBMC were isolated from whole blood by Ficoll-Hypaque density gradient centrifugation. Presence of IgG against influenza A H1N1 (strain Beijing 265/95) and H3N2 (strain Sydney 5/97) antigens was assessed in healthy donors with available cryopreserved plasma using the GenWay influenza A IgG ELISA kit (Genway Biotech). Plasma samples from PLWH were sent to Quest Diagnostics for evaluation of IgG and IgM against CMV.
Calcein acetoxymethyl (AM)-based antigen-specific NK cell killing assay for NKCL
Autologous BLCL were stained with 10μm Calcein AM (Invitrogen) for 1 hour, then washed three times prior to be pulsed with overlapping peptide pools encompassing HIV-1 Consensus B Gag or Env (NIH AIDS Reagent Program), influenza A/California/04/2009(H1N1) NP (PepTivator, Miltenyi Biotec), influenza A/California/08/2009(H1N1) MP1 (PepMix™, JPT Peptide Technologies), CMV pp65 (NIH AIDS Reagent Program), and Myelin-oligodendrocyte glycoprotein (MOG) negative control (PepMix™, JPT Peptide Technologies) or with individual peptides. Peptide-pulsed BLCL or mock-pulsed controls were then incubated with or without NKCL at a 5:1 E:T ratio. For blockade experiments, 10μg/mL of the following blocking antibodies or isotype control antibodies were added to NKCL prior to co-culture with BLCL: anti-NKG2C purified antibody (R&D, clone 134522), anti-NKp44 purified antibody (BioLegend, clone P44-8), anti-NKp30 purified antibody (BioLegend, clone P30-15), purified mouse IgG1, κ Isotype control (MOPC-21) and purified Mouse IgG2b, κ Isotype control (MG2b-57). Supernatant were harvested after 4 hours of incubation at 37°C. Release of CAM into the supernatant was measured using the VICTOR3 multilabel plate reader or the VICTOR Nivo Multimode Plate Reader (PerkinElmer) (excitation 485nm, absorption 530nm). The percent-specific lysis was calculated as follows: (test release – spontaneous release)/(maximum release - spontaneous release) x 100.
Advanced Polychromatic flow cytometry for the phenotypic characterization of NKCL
Cryopreserved NKCL were thawed and stained with antibodies detailed in Table S2. Data were acquired on a FACSymphony A5 flow cytometer (BD Biosciences). Flow Cytometry Standard (FCS) files were exported as FCS3.0 from DiVa 8.0.1. FCS files were loaded and compensated in FlowJo by using single‐stained controls (Compbeads incubated with fluorescently conjugated antibodies). After compensation and gating, live NKCL were exported as FCS3.0 files for subsequent t-SNE analysis using CytoDRAV (https://github.com/ReevesLab/CytoDRAV).
HLA-E stabilization assay
A selection of nonameric peptides derived from subtype B HIV Gag (n=60) (GenBank accession number ABR15476.1) and Env (n=84) (UniProtKB accession number A0A160I6K4) as well as influenza A/California/04/2009(H1N1) NP (n=18) (UniProtKB accession number C3W5S2) predicted to bind HLA-E*01:01 by the NetMHCpan 4.0 and Immune Epitope Database servers were synthesized using the PEPotec Immuno Custom Peptide Libraries service provided by Thermofisher Scientific (Table S3). Analyses were conducted using influenza NP, HIV Gag, and HIV Env sequences matching those of the peptide pools used in our functional studies. Crude synthetic peptides were used for the initial screen (Fig. S10), while synthetic peptides of ≥98% purity were used to confirm HLA-E stabilization and for all functional assays (Fig. 4–5, Fig. S11–13). The ability of individual peptides to stabilize surface expression of HLA-E was assessed using K562-HLA-E*01:01 transfectants generated in the laboratory of Dr. Michaela Müller-Trutwin (52). These cells are engineered to express high levels of HLA-E while simultaneously lacking cell-intrinsic HLA-E-binding peptides, and therefore serve as a cellular model for HLA-E surface stabilization by exogenous peptides. K562-HLA-E*01:01 were pulsed with 40μM of individual synthetic peptides for 16 hours at 26°C in serum-free IMDM. Cell surface expression levels of HLA-E were assessed by flow cytometry staining using anti-HLA-E-BV421 antibody (3D12). HLA-E expression upon pulsing with different viral antigen-derived peptides was compared to that upon pulsing with positive control peptide VL9 (VMAPRTLIL), a canonical CMV UL40/HLA-Cw3 leader sequence-derived peptide that stabilizes HLA-E, and negative control peptide NLVPMVATV, a CMV pp65-derived nonamer that does not stabilize HLA-E. Data were acquired on an LSRII instrument (BD Biosciences) or a FACSymphony Analyzer (X-50) (BD Biosciences), and analyzed using FlowJo software v10.8.1 (Treestar).
For the peptide-binding competition assays, K562-E*01:01 cells were incubated overnight at 26°C with VMAPRTLFL-FITC (ProImmune Inc.) at 10μM and increasing concentrations (from 10μM to 100μM) of non-FITC competitor peptides of ≥98% purity (TMDSNTLEL, ILPCRIKQI, VLKYWWNLL, FLARSALIL, NLVPMVATV) were added to the culture binding medium (IMDM, 1% Penicillin-Streptomycin, without FBS). The expression of VMAPRTLFL-FITC bound to HLA-E was assessed by flow cytometry. Cells were acquired using a FACSymphony Analyzer (X-50) (BD Biosciences), and FlowJo software (version 10.8.1, Tree Star, Ashland, OR) was used for all analyses.
siRNA-mediated specific knockdown of NKG2C in NKCL
200,000 to 1,000,000 NKCL per condition were plated and nucleofected with 5μM SMARTpool human KLRC2 siRNA (Dharmacon Horizon Discovery) or non-targeting siRNA negative control (Dharmacon Horizon Discovery). Nucleofection was conducted using P3 Primary Cell 4D-Nucleofector X Kit (Lonza Bioscience) and the 4D-Nucleofector X Unit (Lonza Bioscience, program code CM-137 (55)) according to the manufacturer’s manual. After nucleofection, cells were cultured in pre-warmed recovery media (NK MACS media (Miltenyi) supplemented with 500U/mL recombinant human IL-2 (R&D Systems)). 48 hours after nucleofection, an aliquot of NKCL was stained using BD Biosciences CD3-A700 (clone UCHT1), CD16-APC-Cy-7 (clone 3G8), CD56-BV605 (clone NCAM16.2), Beckman Coulter NKG2A-PE-Cy7 (clone Z199), and Miltenyi NKG2C-PE (clone REA205) to assess NKG2C expression via flow cytometry. Only NKCL displaying at least 70% knock down of NKG2C were used in CAM cytotoxicity assays.
Statistics
Statistical tests used in this study include Wilcoxon signed-rank test, Mann-Whitney U test, Spearman nonparametric correlation, and Kruskal Wallis test, as specified in figure legends. All reported p-values, except for Fig. 4A, are raw unadjusted p-values. Fig. 4A p-values were adjusted using Dunn’s Correction. No confidence intervals were reported in this study. Significance was determined using an unadjusted or adjusted p-value threshold of 0.05. All statistical tests were performed in GraphPad Prism software as two-tailed tests. Nonparametric tests were utilized due to data not being normally distributed. Bar graphs are reported with mean plus standard error of the mean (SEM). Scatter dot plot bars represent the median value of the data.
Supplementary Material
Acknowledgements
We thank all the patients who contributed to this study making this work possible. We are grateful to Dr. Guido Ferrari for kindly providing patient samples. The following reagents were obtained through the NIH HIV Reagent Program, Division of AIDS, NIAID, NIH: Peptide Pool, Human Immunodeficiency Virus Type 1 Subtype B (Consensus) Env Region, ARP-12540, and Gag Region, ARP-12425; Human Cytomegalovirus (HCMV) pp65, ARP-11549; Cytomegalovirus, Epstein-Barr Virus and Influenza Virus (CEF) Control, ARP-9808, contributed by DAIDS, NIAID.
Funding
This research was supported by National Institutes of Health (NIH) grants: R01AI116363, R21AI137835, R01AI158516 (to S.J.) and R01AI120828, R01AI143457, R01AI161010, P01AI162242 (to R.K.R.) as well as by the NIH-funded BEAT-HIV Martin Delaney Collaboratory to cure HIV-1 infection by Combination Immunotherapy UM1 AI164570. We also acknowledge support from the CVVR Flow Cytometry and Harvard University Center for AIDS Research Advanced Laboratory Technologies Core (P30 AI060354) as well as from the Duke Human Vaccine Institute Research Flow Cytometry Shared Resource Facility (Durham, NC) directed by Dr. Derek W. Cain.
Footnotes
Competing Interests
The authors declare no competing financial interests.
REFERENCES
- 1.Florez-Alvarez L, Hernandez JC, Zapata W, NK Cells in HIV-1 Infection: From Basic Science to Vaccine Strategies. Front Immunol 9, 2290 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Scully E, Alter G, NK Cells in HIV Disease. Current HIV/AIDS reports 13, 85–94 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jost S, Altfeld M, Control of Human Viral Infections by Natural Killer Cells. Annual review of immunology, (2013). [DOI] [PubMed] [Google Scholar]
- 4.Mujal AM, Delconte RB, Sun JC, Natural Killer Cells: From Innate to Adaptive Features. Annu Rev Immunol 39, 417–447 (2021). [DOI] [PubMed] [Google Scholar]
- 5.Paust S, Gill HS, Wang BZ, Flynn MP, Moseman EA, Senman B, Szczepanik M, Telenti A, Askenase PW, Compans RW, von Andrian UH, Critical role for the chemokine receptor CXCR6 in NK cell-mediated antigen-specific memory of haptens and viruses. Nat Immunol 11, 1127–1135 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reeves RK, Li H, Jost S, Blass E, Schafer JL, Varner V, Manickam C, Eslamizar L, Altfeld M, von Andrian UH, Barouch DH, Antigen-specific NK cell memory in rhesus macaques. Nature immunology 16, 927–932 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nikzad R, Angelo LS, Aviles-Padilla K, Le DT, Singh VK, Bimler L, Vukmanovic-Stejic M, Vendrame E, Ranganath T, Simpson L, Haigwood NL, Blish CA, Akbar AN, Paust S, Human natural killer cells mediate adaptive immunity to viral antigens. Sci Immunol 4, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li T, Wang J, Wang Y, Chen Y, Wei H, Sun R, Tian Z, Respiratory Influenza Virus Infection Induces Memory-like Liver NK Cells in Mice. Journal of immunology 198, 1242–1252 (2017). [DOI] [PubMed] [Google Scholar]
- 9.Dou Y, Fu B, Sun R, Li W, Hu W, Tian Z, Wei H, Influenza vaccine induces intracellular immune memory of human NK cells. PloS one 10, e0121258 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zheng J, Wen L, Yen HL, Liu M, Liu Y, Teng O, Wu WF, Ni K, Lam KT, Huang C, Yang J, Lau YL, Perlman S, Peiris M, Tu W, Phenotypic and Functional Characteristics of a Novel Influenza Virus Hemagglutinin-Specific Memory NK Cell. J Virol 95, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ma M, Wang Z, Chen X, Tao A, He L, Fu S, Zhang Z, Fu Y, Guo C, Liu J, Han X, Xu J, Chu Z, Ding H, Shang H, Jiang Y, NKG2C(+)NKG2A(-) Natural Killer Cells are Associated with a Lower Viral Set Point and may Predict Disease Progression in Individuals with Primary HIV Infection. Front Immunol 8, 1176 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gondois-Rey F, Cheret A, Granjeaud S, Mallet F, Bidaut G, Lecuroux C, Ploquin M, Muller-Trutwin M, Rouzioux C, Avettand-Fenoel V, Moretta A, Pialoux G, Goujard C, Meyer L, Olive D, NKG2C(+) memory-like NK cells contribute to the control of HIV viremia during primary infection: Optiprim-ANRS 147. Clin Transl Immunology 6, e150 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thomas R, Low HZ, Kniesch K, Jacobs R, Schmidt RE, Witte T, NKG2C deletion is a risk factor of HIV infection. AIDS Res Hum Retroviruses 28, 844–851 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peppa D, Pedroza-Pacheco I, Pellegrino P, Williams I, Maini MK, Borrow P, Adaptive Reconfiguration of Natural Killer Cells in HIV-1 Infection. Front Immunol 9, 474 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hammer Q, Ruckert T, Borst EM, Dunst J, Haubner A, Durek P, Heinrich F, Gasparoni G, Babic M, Tomic A, Pietra G, Nienen M, Blau IW, Hofmann J, Na IK, Prinz I, Koenecke C, Hemmati P, Babel N, Arnold R, Walter J, Thurley K, Mashreghi MF, Messerle M, Romagnani C, Peptide-specific recognition of human cytomegalovirus strains controls adaptive natural killer cells. Nat Immunol 19, 453–463 (2018). [DOI] [PubMed] [Google Scholar]
- 16.Suliman S, Geldenhuys H, Johnson JL, Hughes JE, Smit E, Murphy M, Toefy A, Lerumo L, Hopley C, Pienaar B, Chheng P, Nemes E, Hoft DF, Hanekom WA, Boom WH, Hatherill M, Scriba TJ, Bacillus Calmette-Guerin (BCG) Revaccination of Adults with Latent Mycobacterium tuberculosis Infection Induces Long-Lived BCG-Reactive NK Cell Responses. Journal of immunology 197, 1100–1110 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wijaya RS, Read SA, Truong NR, Han S, Chen D, Shahidipour H, Fewings NL, Schibeci S, Azardaryany MK, Parnell GP, Booth D, van der Poorten D, Lin R, George J, Douglas MW, Ahlenstiel G, HBV vaccination and HBV infection induces HBV-specific natural killer cell memory. Gut, (2020). [DOI] [PubMed] [Google Scholar]
- 18.Rolle A, Meyer M, Calderazzo S, Jager D, Momburg F, Distinct HLA-E Peptide Complexes Modify Antibody-Driven Effector Functions of Adaptive NK Cells. Cell Rep 24, 1967–1976 e1964 (2018). [DOI] [PubMed] [Google Scholar]
- 19.Stary V, Pandey RV, Strobl J, Kleissl L, Starlinger P, Pereyra D, Weninger W, Fischer GF, Bock C, Farlik M, Stary G, A discrete subset of epigenetically primed human NK cells mediates antigen-specific immune responses. Sci Immunol 5, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park JK, Han A, Czajkowski L, Reed S, Athota R, Bristol T, Rosas LA, Cervantes-Medina A, Taubenberger JK, Memoli MJ, Evaluation of Preexisting Anti-Hemagglutinin Stalk Antibody as a Correlate of Protection in a Healthy Volunteer Challenge with Influenza A/H1N1pdm Virus. mBio 9, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dugan HL, Guthmiller JJ, Arevalo P, Huang M, Chen YQ, Neu KE, Henry C, Zheng NY, Lan LY, Tepora ME, Stovicek O, Bitar D, Palm AE, Stamper CT, Changrob S, Utset HA, Coughlan L, Krammer F, Cobey S, Wilson PC, Preexisting immunity shapes distinct antibody landscapes after influenza virus infection and vaccination in humans. Sci Transl Med 12, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bodewes R, de Mutsert G, van der Klis FR, Ventresca M, Wilks S, Smith DJ, Koopmans M, Fouchier RA, Osterhaus AD, Rimmelzwaan GF, Prevalence of antibodies against seasonal influenza A and B viruses in children in Netherlands. Clin Vaccine Immunol 18, 469–476 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guma M, Angulo A, Vilches C, Gomez-Lozano N, Malats N, Lopez-Botet M, Imprint of human cytomegalovirus infection on the NK cell receptor repertoire. Blood 104, 3664–3671 (2004). [DOI] [PubMed] [Google Scholar]
- 24.Sullivan LC, Clements CS, Beddoe T, Johnson D, Hoare HL, Lin J, Huyton T, Hopkins EJ, Reid HH, Wilce MC, Kabat J, Borrego F, Coligan JE, Rossjohn J, Brooks AG, The heterodimeric assembly of the CD94-NKG2 receptor family and implications for human leukocyte antigen-E recognition. Immunity 27, 900–911 (2007). [DOI] [PubMed] [Google Scholar]
- 25.Lee N, Goodlett DR, Ishitani A, Marquardt H, Geraghty DE, HLA-E surface expression depends on binding of TAP-dependent peptides derived from certain HLA class I signal sequences. J Immunol 160, 4951–4960 (1998). [PubMed] [Google Scholar]
- 26.Llano M, Lee N, Navarro F, Garcia P, Albar JP, Geraghty DE, Lopez-Botet M, HLA-E-bound peptides influence recognition by inhibitory and triggering CD94/NKG2 receptors: preferential response to an HLA-G-derived nonamer. Eur J Immunol 28, 2854–2863 (1998). [DOI] [PubMed] [Google Scholar]
- 27.Braud VM, Allan DS, O’Callaghan CA, Soderstrom K, D’Andrea A, Ogg GS, Lazetic S, Young NT, Bell JI, Phillips JH, Lanier LL, McMichael AJ, HLA-E binds to natural killer cell receptors CD94/NKG2A, B and C. Nature 391, 795–799 (1998). [DOI] [PubMed] [Google Scholar]
- 28.Hendricks DW, Balfour HH Jr., Dunmire SK, Schmeling DO, Hogquist KA, Lanier LL, Cutting edge: NKG2C(hi)CD57+ NK cells respond specifically to acute infection with cytomegalovirus and not Epstein-Barr virus. Journal of immunology 192, 4492–4496 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lopez-Verges S, Milush JM, Schwartz BS, Pando MJ, Jarjoura J, York VA, Houchins JP, Miller S, Kang SM, Norris PJ, Nixon DF, Lanier LL, Expansion of a unique CD57(+)NKG2Chi natural killer cell subset during acute human cytomegalovirus infection. Proceedings of the National Academy of Sciences of the United States of America 108, 14725–14732 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee J, Zhang T, Hwang I, Kim A, Nitschke L, Kim M, Scott JM, Kamimura Y, Lanier LL, Kim S, Epigenetic modification and antibody-dependent expansion of memory-like NK cells in human cytomegalovirus-infected individuals. Immunity 42, 431–442 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schlums H, Cichocki F, Tesi B, Theorell J, Beziat V, Holmes TD, Han H, Chiang SC, Foley B, Mattsson K, Larsson S, Schaffer M, Malmberg KJ, Ljunggren HG, Miller JS, Bryceson YT, Cytomegalovirus infection drives adaptive epigenetic diversification of NK cells with altered signaling and effector function. Immunity 42, 443–456 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nattermann J, Nischalke HD, Hofmeister V, Kupfer B, Ahlenstiel G, Feldmann G, Rockstroh J, Weiss EH, Sauerbruch T, Spengler U, HIV-1 infection leads to increased HLA-E expression resulting in impaired function of natural killer cells. Antivir Ther 10, 95–107 (2005). [DOI] [PubMed] [Google Scholar]
- 33.Davis ZB, Cogswell A, Scott H, Mertsching A, Boucau J, Wambua D, Le Gall S, Planelles V, Campbell KS, Barker E, A Conserved HIV-1-Derived Peptide Presented by HLA-E Renders Infected T-cells Highly Susceptible to Attack by NKG2A/CD94-Bearing Natural Killer Cells. PLoS pathogens 12, e1005421 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hannoun Z, Lin Z, Brackenridge S, Kuse N, Akahoshi T, Borthwick N, McMichael A, Murakoshi H, Takiguchi M, Hanke T, Identification of novel HIV-1-derived HLA-E-binding peptides. Immunol Lett 202, 65–72 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grant EJ, Nguyen AT, Lobos CA, Szeto C, Chatzileontiadou DSM, Gras S, The unconventional role of HLA-E: The road less traveled. Mol Immunol 120, 101–112 (2020). [DOI] [PubMed] [Google Scholar]
- 36.Aktas E, Kucuksezer UC, Bilgic S, Erten G, Deniz G, Relationship between CD107a expression and cytotoxic activity. Cell Immunol 254, 149–154 (2009). [DOI] [PubMed] [Google Scholar]
- 37.Alter G, Malenfant JM, Altfeld M, CD107a as a functional marker for the identification of natural killer cell activity. J Immunol Methods 294, 15–22 (2004). [DOI] [PubMed] [Google Scholar]
- 38.Hansen SG, Ford JC, Lewis MS, Ventura AB, Hughes CM, Coyne-Johnson L, Whizin N, Oswald K, Shoemaker R, Swanson T, Legasse AW, Chiuchiolo MJ, Parks CL, Axthelm MK, Nelson JA, Jarvis MA, Piatak M Jr., Lifson JD, Picker LJ, Profound early control of highly pathogenic SIV by an effector memory T-cell vaccine. Nature 473, 523–527 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hansen SG, Marshall EE, Malouli D, Ventura AB, Hughes CM, Ainslie E, Ford JC, Morrow D, Gilbride RM, Bae JY, Legasse AW, Oswald K, Shoemaker R, Berkemeier B, Bosche WJ, Hull M, Womack J, Shao J, Edlefsen PT, Reed JS, Burwitz BJ, Sacha JB, Axthelm MK, Fruh K, Lifson JD, Picker LJ, A live-attenuated RhCMV/SIV vaccine shows long-term efficacy against heterologous SIV challenge. Sci Transl Med 11, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu HL, Wiseman RW, Hughes CM, Webb GM, Abdulhaqq SA, Bimber BN, Hammond KB, Reed JS, Gao L, Burwitz BJ, Greene JM, Ferrer F, Legasse AW, Axthelm MK, Park BS, Brackenridge S, Maness NJ, McMichael AJ, Picker LJ, O’Connor DH, Hansen SG, Sacha JB, The Role of MHC-E in T Cell Immunity Is Conserved among Humans, Rhesus Macaques, and Cynomolgus Macaques. J Immunol 200, 49–60 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stephenson KE, Wagh K, Korber B, Barouch DH, Vaccines and Broadly Neutralizing Antibodies for HIV-1 Prevention. Annu Rev Immunol 38, 673–703 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Estrada LD, Schultz-Cherry S, Development of a Universal Influenza Vaccine. J Immunol 202, 392–398 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Paust S, Blish CA, Reeves RK, Redefining Memory: Building the Case for Adaptive NK Cells. J Virol 91, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fogel LA, Sun MM, Geurs TL, Carayannopoulos LN, French AR, Markers of nonselective and specific NK cell activation. J Immunol 190, 6269–6276 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Venkatasubramanian S, Cheekatla S, Paidipally P, Tripathi D, Welch E, Tvinnereim AR, Nurieva R, Vankayalapati R, IL-21-dependent expansion of memory-like NK cells enhances protective immune responses against Mycobacterium tuberculosis. Mucosal Immunol 10, 1031–1042 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reeves RK, Evans TI, Gillis J, Johnson RP, Simian immunodeficiency virus infection induces expansion of alpha4beta7+ and cytotoxic CD56+ NK cells. Journal of virology 84, 8959–8963 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sips M, Sciaranghella G, Diefenbach T, Dugast AS, Berger CT, Liu Q, Kwon D, Ghebremichael M, Estes JD, Carrington M, Martin JN, Deeks SG, Hunt PW, Alter G, Altered distribution of mucosal NK cells during HIV infection. Mucosal immunology 5, 30–40 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huisman BD, Guan N, Ruckert T, Garner L, Singh NK, McMichael AJ, Gillespie GM, Romagnani C, Birnbaum ME, High-throughput characterization of HLA-E-presented CD94/NKG2x ligands reveals peptides which modulate NK cell activation. Nat Commun 14, 4809 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rolle A, Jager D, Momburg F, HLA-E Peptide Repertoire and Dimorphism-Centerpieces in the Adaptive NK Cell Puzzle? Front Immunol 9, 2410 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hammer Q, Dunst J, Christ W, Picarazzi F, Wendorff M, Momayyezi P, Huhn O, Netskar HK, Maleki KT, Garcia M, Sekine T, Sohlberg E, Azzimato V, Aouadi M, Karolinska C-SG, Severe C-GG, Degenhardt F, Franke A, Spallotta F, Mori M, Michaelsson J, Bjorkstrom NK, Ruckert T, Romagnani C, Horowitz A, Klingstrom J, Ljunggren HG, Malmberg KJ, SARS-CoV-2 Nsp13 encodes for an HLA-E-stabilizing peptide that abrogates inhibition of NKG2A-expressing NK cells. Cell Rep 38, 110503 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cassidy SA, Cheent KS, Khakoo SI, Effects of Peptide on NK cell-mediated MHC I recognition. Front Immunol 5, 133 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huot N, Rascle P, Petitdemange C, Contreras V, Sturzel CM, Baquero E, Harper JL, Passaes C, Legendre R, Varet H, Madec Y, Sauermann U, Stahl-Hennig C, Nattermann J, Saez-Cirion A, Le Grand R, Keith Reeves R, Paiardini M, Kirchhoff F, Jacquelin B, Muller-Trutwin M, SIV-induced terminally differentiated adaptive NK cells in lymph nodes associated with enhanced MHC-E restricted activity. Nat Commun 12, 1282 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Savoy SKA, Boudreau JE, The Evolutionary Arms Race between Virus and NK Cells: Diversity Enables Population-Level Virus Control. Viruses 11, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cella M, Colonna M, Cloning human natural killer cells. Methods in molecular biology 121, 1–4 (2000). [DOI] [PubMed] [Google Scholar]
- 55.Rautela J, Surgenor E, Huntington ND, Drug target validation in primary human natural killer cells using CRISPR RNP. J Leukoc Biol 108, 1397–1408 (2020). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


