Abstract
Formins are a conserved family of proteins that primarily act to form linear polymers of actin. Despite their importance to the normal functioning of the cytoskeleton, for a long time, the only two formin genes known to be a genetic cause of human disorders were DIAPH1 and DIAPH3, whose mutation causes two distinct forms of hereditary deafness. In the last 10 years, however, the formin INF2 has emerged as an important target of mutations responsible for the appearance of focal segmental glomerulosclerosis, which are histological lesions associated with glomerulus degeneration that often leads to end-stage renal disease. In some rare cases, focal segmental glomerulosclerosis concurs with Charcot–Marie–Tooth disease, which is a degenerative neurological disorder affecting peripheral nerves. All known INF2 gene mutations causing disease map to the exons encoding the amino-terminal domain. In this review, we summarize the structure, biochemical features and functions of INF2, conduct a systematic and comprehensive analysis of the pathogenic INF2 mutations, including a detailed study exon-by-exon of patient cases and mutations, address the impact of the pathogenic mutations on the structure, regulation and known functions of INF2, draw a series of conclusions that could be useful for INF2-related disease diagnosis, and suggest lines of research for future work on the molecular mechanisms by which INF2 causes disease.
Electronic supplementary material
The online version of this article (10.1007/s00018-020-03550-7) contains supplementary material, which is available to authorized users.
Keywords: Focal segmental glomerulosclerosis, Charcot–Marie–Tooth disease, Chronic kidney disease, Peripheral neuropathy, Genetic disorder, Actin
The inverted formin 2 (INF2)
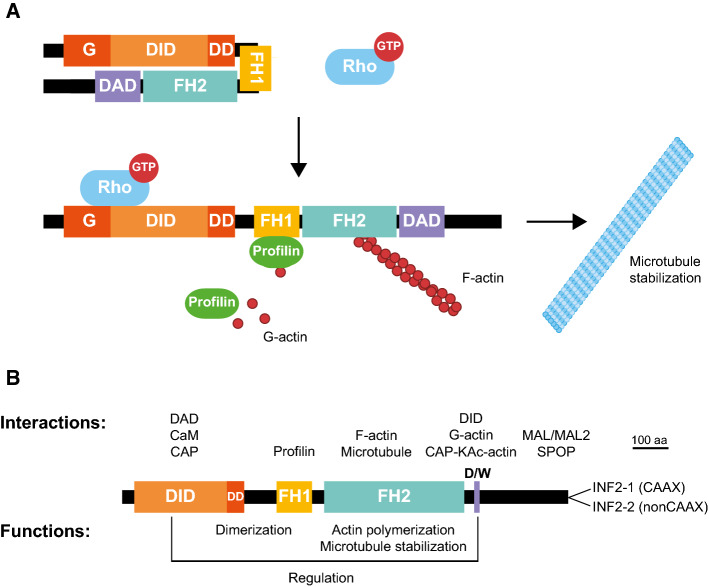
Formins are a family of proteins whose primary function is to polymerize monomeric globular actin (G-actin) into linear actin filaments [1, 2]. Humans express 15 different formins, which are classified into eight groups [3], the best characterized being that of diaphanous-related formins, which includes formins mDia 1–3 [4]. The defining feature of formins is the formin homology (FH) 2 domain, which catalyzes actin nucleation. Upstream of the FH2 domain is a proline-rich FH1 domain that binds profilin, which provisions G-actin to the FH2, while the diaphanous autoregulatory domain (DAD) is located downstream. The DAD interacts with a diaphanous inhibitory domain (DID) present at the amino-terminal region of the formin where it enables the molecule to be folded to yield an inactive state [5]. In diaphanous-related formins, as in most formins, the binding of a specific Rho-family GTPase in its GTP-loaded form to a region encompassing part of the DID and the most proximal part (called G domain) of an extension amino-terminal to the DID releases the DID–DAD interaction and unfolds the molecule to render it in an active form [1, 2] (Fig. 1a). In addition to their role in actin polymerization, formins bind to microtubules through their FH2 domain and, through mechanisms that are not well characterized, regulate microtubule stability and the alignment of microtubules with actin fibers [6–8].
Fig. 1.
Domains, regulation and function of the diaphanous-related formin INF2. a Structure and regulation of diaphanous-related formins. The inactive formin arises in a closed conformation by the interaction of the DID and the DAD. The binding of a specific Rho-family GTPase in its active GTP-loaded form to a region encompassing the G domain and part of the DID releases the autoinhibitory DID–DAD interaction and opens the molecule to exist in its active form. In the open conformation, the FH1 domain recruits profilin, which, in turn, feeds the FH2 domain with G-actin to form the actin filaments. The role of formins on microtubule stabilization is also indicated. b Organization of human INF2 with indication of the interactions and functions of the different domains. CAP cyclase-associated protein, CaM calmodulin, D/W, DAD/WH2, F-actin filamentous actin, G-actin globular actin, KAc-actin lysine-acetylated actin, SPOP Speckle-type POZ protein
In humans, the formin INF2 is expressed as two isoforms, INF2-1 and INF2-2, which differ in their carboxyl terminus (Fig. 1b). INF2-1, which has an 18-amino acid carboxyl-terminal sequence ending with a consensus CAAX box of prenylation, is farnesylated and localizes to the endoplasmic reticulum. INF2-2, which has at the carboxyl terminus an alternative 9-amino acid sequence containing basic residues, is cytosolic [9–11]. The overall domain organization of INF2 is similar to that of diaphanous-related formins with two differential features: the extension amino-terminal to the DID is shorter than in diaphanous-related formins, and the presence within its DAD of a Wiskott–Aldrich syndrome homology 2 (WH2) motif with high affinity for G-actin (Fig. 1b). The presence of this motif confers two specific features on INF2. First, in addition to catalyzing the formation of actin filaments, as do all other formins, INF2 is able to sever and depolymerize actin filaments [12, 13]. Second, the in vitro binding of G-actin to the DAD/WH2 competes with the binding to the DID and regulates the folding of the molecule and, subsequently, the activity of INF2 [14]. This property has led to INF2 being proposed as a sensor of subtle oscillations in the levels of G-actin that can fine-tune its actin polymerization activity [15]. In this way, INF2 could control actin homeostasis and, subsequently, the transcription mediated by the myocardin-related/serum response factor transcription factor complex of a large number of genes encoding products related to the cytoarchitecture [16–18].
The N-terminal region of INF2 interacts with calmodulin and mediates rapid and transient actin cytoskeleton remodeling in response to increased intracellular Ca2+ levels [19, 20]. The observation that INF2 is constitutively active in vitro but is inhibited in cells by the DID–DAD interaction enabled the identification of a complex of the cyclase-associated protein (CAP) and lysine-acetylated actin (KAc-actin) as a cellular inhibitor of INF2 [21]. Since CAP shows significant affinity for the N-terminal region of INF2 in vitro while CAP–KAc-actin binds the DAD, it has been proposed that CAP–KAc-actin inhibits INF2 by bridging the DID and the DAD [22]. Speckle-type POZ protein, which is an adaptor protein of the CUL3-RBX1 E3 ubiquitin ligase complex, interacts with a short peptide motif present at the C-terminal region of INF2 and promotes INF2 ubiquitination, but not degradation, and induces INF2 disassociation from the endoplasmic reticulum [23]. In addition, the C-terminal region of INF2 interacts with the MAL and MAL2 proteins [11, 24], which are components of the machinery for apical transport in polarized epithelial cells [11, 24–26].
INF2 binds to and stabilizes microtubules [27, 28] as do other formins [6], and regulates the acquisition of specific posttranslational modifications by microtubules [18, 28]. INF2 has been implicated in specialized pathways of vesicular transport in epithelial cells and T lymphocytes [11, 24], mitochondrial fission [29–33], podosome formation [34], extensive cytoplasmic and nuclear actin filament remodeling in response to increased levels of cytosolic Ca2+ [19, 35, 36], and Ca2+-promoted apical extrusion of apoptotic or transformed cells [37]. In addition, INF2 has shown to play a role in trophoblast invasion of the uterus for placentation [38], Ca2+-induced inhibition of the Hippo pathway in glioblastoma cells [39], cerebral ischemia–reperfusion injury [40, 41], and breast and prostate cancer migration and invasion [23, 42].
INF2-related disorders
Focal segmental glomeruloesclerosis (FSGS)
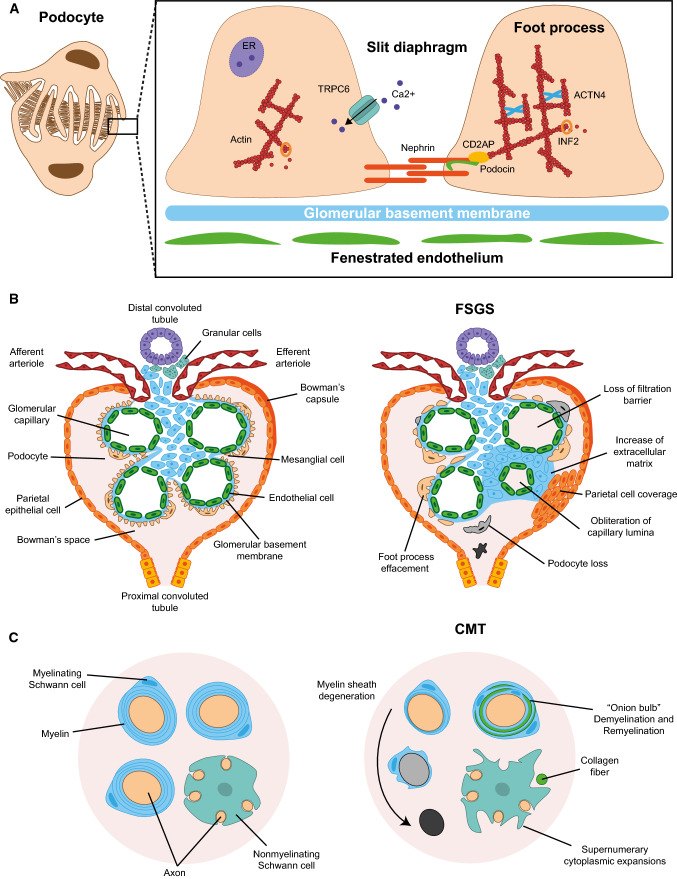
The glomerular filtration barrier allows the kidney to produce urine with a chemical composition that is distinct from that of the blood from which it is derived. The barrier consists of the glomerular basement membrane, capillary endothelial cells and podocytes [43]. Podocytes are terminally differentiated epithelial cells with a unique morphology containing elaborate projections, known as foot processes or pedicels, that closely wrap around the exterior of glomerular capillaries [44, 45]. The cell–cell contact between adjacent foot processes forms the slit diaphragm, which is the structure where blood is filtered (Fig. 2a). Podocytes have a very limited capacity to regenerate and proliferate [46]. Podocyte injury with the consequent alteration of the slit diaphragm is central to the development of kidney disease [47, 48].
Fig. 2.
Phenotypes caused by pathogenic mutations. a Schematic of podocyte structure (left panel) and magnification of the slit diaphragm formed between adjacent pedicels, indicating some of the proteins whose mutation causes FSGS (right panel). b Structure of a glomerulus in control (left panel) and in patients with FSGS (right panel), indicating the various glomerular structures and the typical alterations found in FSGS. c Myelinating and nonmyelinating cells in control (left panel) and in patients with FSGS + CMT caused by INF2 mutation (right panel). In the latter, the myelin sheath formed by Schwann cells degenerates progressively, leading to the appearance of “onion bulbs” and unmyelinated axons, and the nonmyelinating Schwann cells change their morphology and display supernumerary extensions
FSGS refers to a histological pattern of kidney injury characterized by scarring in localized regions of some, but not all, glomeruli (Fig. 2b). Patients with FSGS exhibit a progressive loss of podocytes, which causes glomerular dysfunction, and is initially manifested as proteinuria with or without other signs of nephrotic syndrome (Fig. 3), and that, in a more advanced stage, might result in end-stage renal disease (ESRD). FSGS has different etiologies, including those produced by circulating factors, metabolic misbalance, viral infection, medication, and genetic causes [49, 50]. The latter includes gene variants (e.g., APOL1) that, depending on the genetic background of the individual, influence the susceptibility to the development of FSGS, and usually include extra-renal manifestations (syndromic FSGS). There are also gene mutations whose effects are limited to the kidney and that present a Mendelian pattern of inheritance and have high penetrance (non-syndromic FSGS). Some of these genes encode protein components of the slit diaphragm (Fig. 2a) whose mutation is autosomal recessive (e.g., NPHS1, NPHS2 and CD2AP) and that, as a rule, produces onset of the disease during childhood [50, 51]. The NPHS2 gene, which encodes the raft-associated protein podocin, is the most frequently involved, accounting for up to 30% of the cases with autosomal recessive inheritance. Mutations of other genes encoding slit diaphragm-associated proteins are autosomal dominant and produce the onset and slow progression during adulthood. These genes are exemplified by ACTN4 and TRPC6, which encode the actin filament cross-linking protein α-actinin-4 and the transient receptor potential cation channel subfamily C member 6, respectively.
Fig. 3.
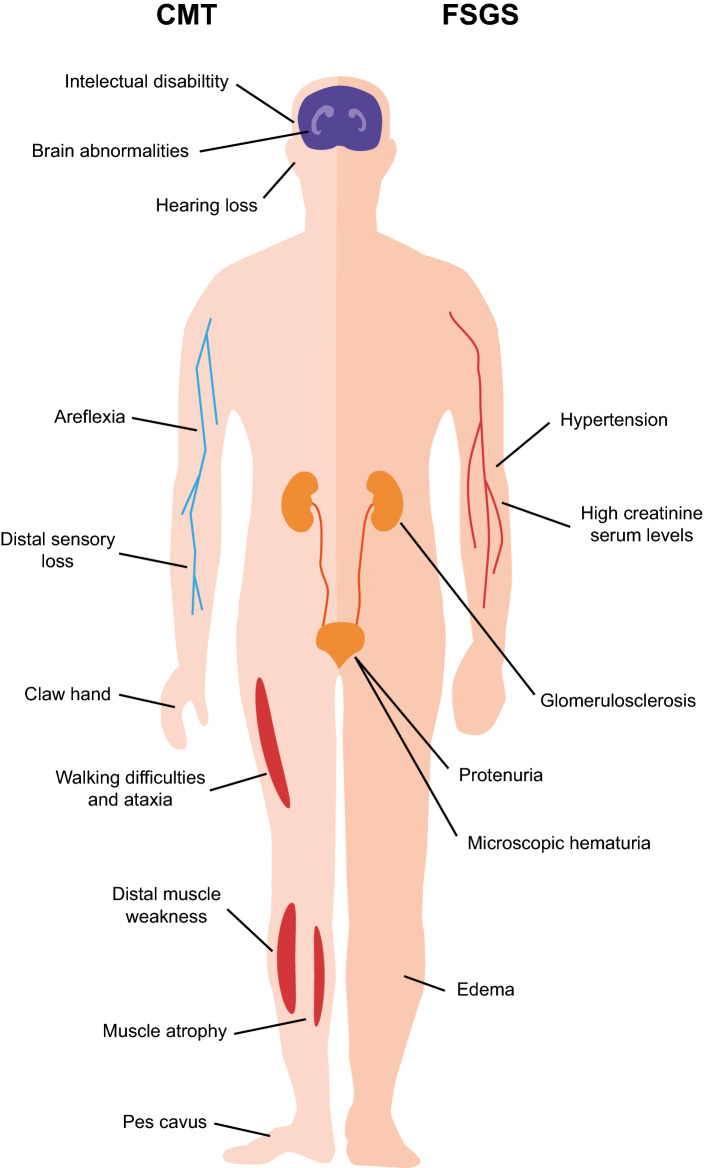
Symptoms of INF2-related diseases. Some of the main symptom characteristics of CMT and FSGS are indicated in the left and right parts of the schematic of the human body, respectively
Ten years have passed since the mutations in the INF2 gene were described as a cause of FSGS (FSGS5; OMIM 613,237) [52]. It has been estimated that INF2 mutations account for up to 17% of autosomal dominant familial FSGS cases and 1% of sporadic cases [53–55]. Onset of disease occurs in adolescence or adulthood, and proteinuria is typically moderate. How pathogenic INF2 mutations affect podocytes is not well understood, but it was observed that patients carrying these mutations present actin filament aggregations within the foot processes of the podocytes [52], indicating that changes in the regulation of the actin polymerization activity depend on the molecular basis of INF2-related FSGS.
Charcot–Marie–Tooth (CMT) disease combined with FSGS
CMT disease comprises a heterogeneous group of neurological disorders that affect the motor nerves and peripheral nervous system [56]. These neuropathies usually share clinical pattern of progressive distal muscle weakness, atrophy of distal extremities, loss of distal sensitivity, reduction or absence of tendon reflexes, and hand and foot deformities (Fig. 3). CMT is the most common inherited neurological disease, with a prevalence of 1 case in 2500 individuals [56, 57]. The onset of symptoms of the majority of patients with CMT occurs in the first or second decade, the severity varying significantly, from severe deficits in early childhood to only mild features in very late life. According to electrophysiological measurement of the motor nerve conduction velocity, CMT is classified as CMT1 or “demyelinating”, which is caused by abnormalities in the myelin sheath and exhibits slow velocity (< 38 m/s), CMT2 or “axonal”, in which the abnormalities are in the axon rather than in the myelin sheath and have normal velocity (45 m/s), and intermediate CMT, with a nerve conduction velocity between that of CMT1 and CMT2, and with demyelinating and axonal features. CMT1 and CMT2 account for approximately 30% and 20–40% of CMT cases, respectively [56].
More than 80 different genes are so far known to be associated with CMT [56, 57]. Most types of CMT are inherited in an autosomal dominant manner, but autosomal recessive and X-linked (CMTX) forms also occur. An analysis carried out by the Inherited Neuropathies Consortium (https://www.rarediseasesnetwork.org) of patients with a genetic diagnosis established that duplication of the PMP22 gene, which encodes a major integral component of the myelin sheath of peripheral nerves, and mutations in the MFN2 gene, which encodes the GTPase involved in mitochondria fission mitofusin 2, and in the GJB1 gene, which encodes the protein gap junction β-1/connexin 32. The mutations in these genes account for the majority of CMT1, CMT2 and CMTX cases, representing 61.6%, 7.0% and 10.7% of all CMT cases, respectively [58].
The association of CMT disease with FSGS (FSGS + CMT) was described for the first time more than 50 years ago [59], being the estimated prevalence of FSGS in CMT much higher than in the general population [60, 61]. Following the identification of mutations in the INF2 gene as a cause of FSGS, INF2 gene mutations were found in approximately 75% of the cases of FSGS + CMT [62]. INF2-related FSGS + CMT is formally called dominant intermediate CMT subtype E (CMTDIE; OMIM 614455) due to its dominant inheritance and intermediate motor nerve conduction velocity [63]. CMT symptoms in FSGS + CMT patients appear in childhood, and renal symptoms appear earlier than in patients with isolated FSGS [61].
Analysis of nerve biopsies from patients with CMTDIE showed pathological lesions suggestive of chronic demyelination and remyelination associated with progressive axonal loss (Fig. 2c). Electron microscopic analysis revealed a marked decrease of large myelinated fibers and fibers with an abnormally thin myelin sheath. In addition, there were many multilayered onion bulbs that consisted of flattened Schwann cell cytoplasm processes containing a few small axons, and many unusual whorl-like proliferations of nonmyelinating Schwann cell cytoplasm with supernumerary protrusions and an abnormal accumulation of β-actin in the cytoplasm [64]. It was proposed that INF2 dysfunction in the mutants causing CMT is the result of a defect of Schwann cell polarization leading to abnormal myelin formation and/or maintenance [64, 65]. Although, INF2 dysfunction also results in axonal loss, it is unclear whether this effect is secondary to the Schwann cell pathology or is a direct effect in the neurons.
The FSGS + CMT phenotype caused by mutations in INF2 may also encompass sensorineural hearing loss, with a several-fold greater prevalence than in overall CMT [62]. Mutation in the formin DIAPH1 [66] and DIAPH3 genes [67] produces deafness autosomal dominant 1 (OMIM 124900) and auditory neuropathy autosomal dominant 1 (OMIM 609129), respectively. The relationship between hearing loss and formin mutation can be explained as the function of hair cells in the inner ear is intimately related with the actin network [68]. Finally, a small number of patients with INF2-related FSGS + CMT present intellectual disability and central nervous system anomalies [62].
Description and classification of the reported cases with pathogenic mutations in the INF2 gene
A comprehensive bibliographic search for reported cases of FSGS and FSGS + CMT caused by mutation in the INF2 gene [52–55, 62, 69–102] is presented in Table S1, which shows data from the case collection, and Table S2, which summarizes specific information for each mutation, including whether it is registered in the Human Gene Mutation Database (HGMD®; https://www.hgmd.cf.ac.uk/ac) [103], which constitutes a comprehensive collection of published germline mutations in nuclear genes related to human disease, and in the Exome Variant Server (EVS) of the National Heart, Lung, and Blood Institute (NHLBI) GO Exome Sequencing Project, (ESP6500SI-V2; https://evs.gs.washington.edu/EVS), which allows to assess if they are rare non-pathogenic genetic variants. The cases represent single individuals or a group of members of the same family. As shown in Table 1, we classified the cases as: (a) familial, when more than one member of the family possessed a copy of the INF2 mutation or there was a familial record of related diseases but the INF2 gene was sequenced in only one member of the family; (b) sporadic, when INF2 from both progenitors were sequenced and did not present the mutation or one or both parents were not sequenced but they did not present any disease antecedents; (c) group with no familial study. INF2-related FSGS cases accounted for 70.3% of the total number, whereas FSGS + CMT cases made up 23.9% of the total. For each specific disorder, the percentage of familial cases of isolated FSGS (78.4%) was greater than that of FSGS + CMT cases (45.5%) (Table 1). This difference is probably a consequence of earlier ESRD progression in the latter, which makes it difficult to transmit the mutation to the next generation.
Table 1.
Summary of reported cases of INF2-related renal and neurological disease, classified by disease type and inheritance manner
| Disease | FSGS | FSGS + CMT | Other | Total |
|---|---|---|---|---|
| Number of cases | 97 (70.3) | 33 (23.9) | 8 (5.8) | 138 |
| Inheritance (%) | ||||
| Familial | 76 (78.4) | 15 (45.5) | 8 (100.0) | 99 (71.7) |
| Sporadic | 12 (12.4) | 18 (54.5) | 0 (0.0) | 30 (21.7) |
| Not determined | 9 (9.3) | 0 (0.0) | 0 (0.0) | 9 (6.5) |
The pedigrees with intrafamilial phenotypic variability reveal that INF2 mutations have incomplete penetrance. Thus, although INF2 mutations are autosomal dominant, one or more relatives of the families analyzed had the mutation but not FSGS, at least at the time of the study [52, 54, 55, 76, 80, 85, 104]. In some of the families with FSGS + CMT, at least one member with the mutant variant presents FSGS, but not the associated CMT [54, 76]. There are also some independent cases in which the same mutation manifests as FSGS or FSGS + CMT. An INF2 mutation, which involves the appearance of a cryptic splice site, is the only case so far reported to cause CMT with minimal or absent clinically relevant kidney dysfunction [74]. In conclusion, the development and progression of FSGS and FSGS + CMT caused by INF2 mutation could be influenced by the genetic background and environmental conditions.
Alport syndrome is a progressive hereditary glomerular disease, often accompanied by sensorineural hearing loss and ocular abnormalities, and characteristic focal thinning and lamellation of the glomerular basement membrane. It is caused by pathogenic variants in the COL4A3, COL4A4 and COL4A5 genes, which encode type IV collagen α3, α4, and α5 chains, respectively, that constitute the glomerular basement membrane [105, 106]. There are two cases in which patients with known FSGS-causing INF2 mutations present, in addition to FSGS, alterations in the glomerular basement membrane that mimic those of Alport syndrome [89, 95]. In one of these cases, a mutation in the COL4A5 gene that normally causes nonserious Alport syndrome co-occurs with a mutation in INF2, possibly with an additive effect on the clinical manifestations of nephropathy [92].
Tables S1 and S2 also include six INF2 mutations found in different disorders with renal dysfunction that were not classified as FSGS. One corresponds to a case that was diagnosed with minimal change nephropathy (MCN) [96], which presents effacement of the foot processes, but unlike FSGS, no podocyte loss [107]. Other mutations was found in a case categorized as an unclassified renal pathology, who was from a family with no signs of FSGS or extra-renal manifestations [81], and one case each of pathologies reported as focal glomerular obsolescent and minor glomerular abnormalities [101]. Atypical hemolytic uremic syndrome (aHUS; OMIM: 235400) is a disorder characterized by acute renal failure, microangiopathic hemolytic anemia, and thrombocytopenia [108]. Mutations in components or regulators of the complement cascade system increase the susceptibility to develop aHUS. Our analysis includes INF2 mutations found in two familial cases, both with pedigrees with an aHUS risk haplotype, which presented either aHUS alone or in combination with CMT [75].
Analysis of pathogenic cases and the corresponding INF2 mutation exon by exon
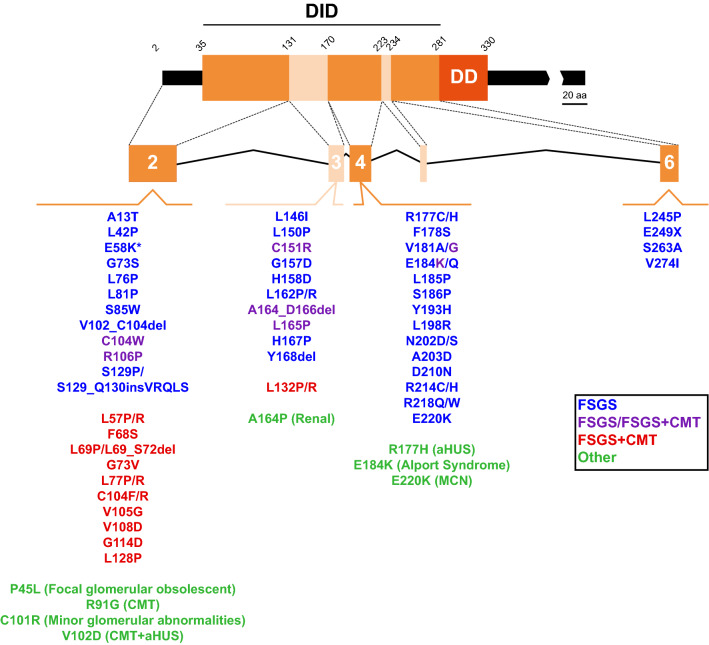
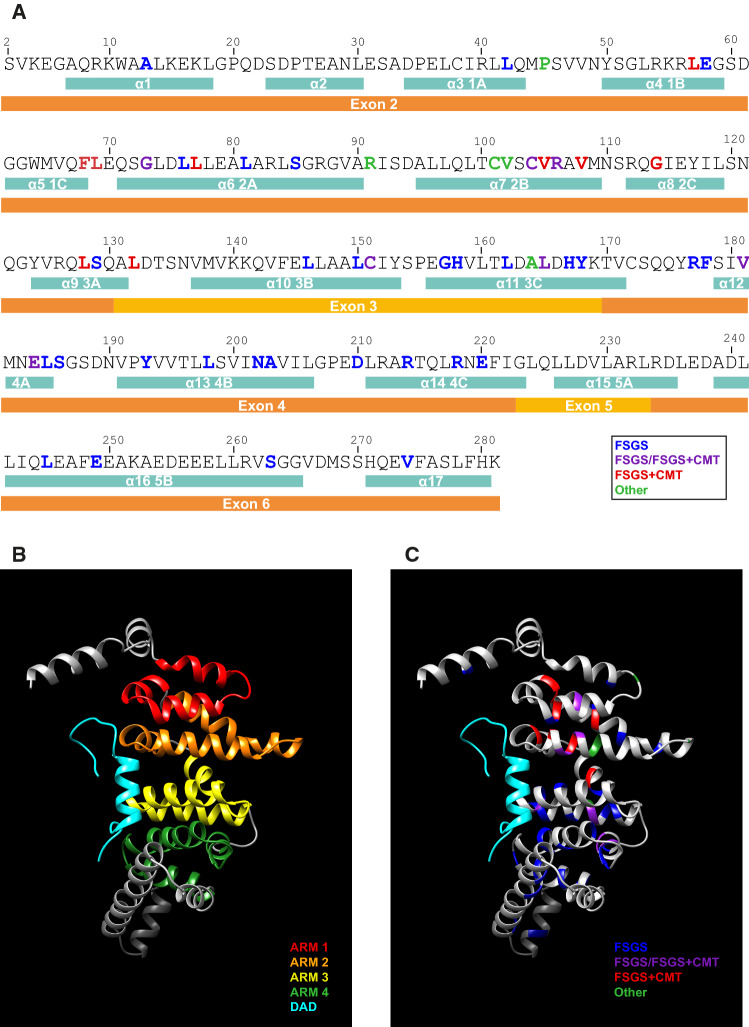
The INF2 gene spans approximately 33 kbp of the q32.33 region of chromosome 14. It consists of 23 exons, of which exon 1 is the only non-coding exon. Exon 22, which encodes the carboxyl terminus of INF2-1, is spliced out in INF2-2 mRNA to allow the sequence at the beginning of exon 23 to encode the specific carboxyl terminus of INF2-2 [11]. More than 60 pathogenic mutations in the INF2 gene are known, all of which map to a genomic region of 4,800 bp containing exons 2–6 that encode the amino-terminal extension and the DID (Fig. 4; Table 2). Mutations are most common in exon 2 (30 of 68, or 44.1%), followed by exons 4 (20 of 68, or 29.4%) and 3 (14 of 68, or 20.6%). It is of note that all the mutations capable of producing FSGS + CMT, except two (V181G and E184K), are concentrated in exons 2 and 3, whereas those causing only FSGS are distributed in exons 2, 3, 4 and 6. No mutations have so far been identified in exon 5, which is the smallest of the DID-coding exons.
Fig. 4.
Pathogenic mutations of the INF2 DID-encoding exons. The exons encoding different segments of the human INF2 DID are indicated. The INF2 mutations associated with isolated FSGS (blue), FSGS in some patients or FSGS + CMT in others (purple), FSGS + CMT (red), and other diseases (green) are listed. *The E58K mutation has been found only in association with the L42P mutation that causes FSGS and, therefore, may not be pathogenic. The exons are drawn to scale
Table 2.
INF2-related disease cases and mutations grouped by exon
| Exon (%) | 2 | 3 | 4 | 6 | Total | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Cases | Mutations | Cases | Mutations | Cases | Mutations | Cases | Mutations | Cases | Mutations | |
| 40 (29.0) | 30 (44.1) | 19 (13.8) | 14 (20.6) | 74 (53.6) | 20 (29.4) | 5 (3.6) | 4 (5.9) | 138 | 68 | |
| Disease | ||||||||||
| FSGS | 15 | 12 | 10 | 9 | 67 | 20 | 5 | 4 | 97 | 45 |
| FSGS + CMT | 21 | 16a | 8 | 5a | 4 | 2a | 0 | 0 | 33 | 23 |
| Other | 4 | 4 | 1 | 1 | 3 | 3b | 0 | 0 | 8 | 8 |
| Inheritance | ||||||||||
| Familial | 18 | 15 | 16 | 12 | 62 | 18 | 3 | 3 | 99 | 48 |
| Sporadic | 20 | 15c | 3 | 3c | 7 | 4c | 0 | 0 | 30 | 22 |
| Not determined | 2 | 3d | 0 | 0 | 5 | 4d | 2 | 2d | 9 | 9 |
aIncludes mutations (C104W, R106P, C151R; V181G, E184K) that can cause isolated FSGS and not only FSGS + CMT or FSGS/FSGS + CMT
bIncludes mutations (R177H, E184K, E220K) that can cause FSGS and not only “others”
cIncludes mutations (G114D, L132R, E184K, R218Q, E220K) that can cause familial disease and not only sporadic cases
dIncludes mutations (L42P, R106P, R177H, R218Q, E220K, S263A) that can cause sporadic or familial disease and not only “not determined” cases
Considering the percentage of total pathogenic cases corresponding to each exon (Table 2), rather than on the number of different mutations, we observed that 74 of 138 cases (53.6%) had mutations in exon 4. In contrast, exon 2, which is the exon with the highest number of mutations accounted for only 29.0% of cases (40 of 138 cases). Comparison of case frequency revealed that the majority of FSGS + CMT cases (21 of 33, or 63.6%) were caused by mutations in exon 2, whereas exon 4 was the most frequently involved in FSGS alone (67 of 97 cases, or 69.1%). It is also to be noted that mutations in exon 2 accounted for most of the total number of sporadic cases (20 of 30, or 66.7%), while mutations in exon 3 and exon 4 were associated with the highest percentage of familial cases (78 of 99, or 78.8%).
Recurrent mutations are most frequent in exon 4, the three most recurrent being R218Q, E220K and E184K, which, together, account for almost one quarter of all cases (Table 3). The differences in nationalities and ethnicities of these patients rule out a possible founder effect that could explain the recurrence of these mutations.
Table 3.
Recurrence of pathogenic INF2 DID mutations
| Mutation | Number of cases | % |
|---|---|---|
| R218Q | 14 | 10.1 |
| E220K | 11 | 8.0 |
| E184K | 8 | 5.8 |
| R214H | 6 | 4.4 |
| R177H | 5 | 3.6 |
| R214C | 5 | 3.6 |
| R218W | 5 | 3.6 |
| C151R | 4 | 2.9 |
| R177C | 4 | 2.9 |
| L42P | 3 | 2.2 |
| R106P | 3 | 2.2 |
| G114D | 3 | 2.2 |
| L128P | 3 | 2.2 |
| V181G | 3 | 2.2 |
| G73S | 2 | 1.5 |
| L77R | 2 | 1.5 |
| C104W | 2 | 1.5 |
| L132R | 2 | 1.5 |
| A164_D166del | 2 | 1.5 |
| S186P | 2 | 1.5 |
| L198R | 2 | 1.5 |
| S263A | 2 | 1.5 |
| All other mutations | 1 | 0.7 |
The great majority of the 68 reported pathogenic mutations in the exons coding the DID are missense (61 mutations, or 89.7%), the others being deletions (4 mutations, or 5.9%), and single cases each of a short in-frame insertion, a nonsense mutation and a mutation in a cryptic splicing site (Tables S1, S2). Between one quarter and one fifth of these mutations were changes of Leu to Pro, and nearly one tenth of Leu to Arg. All other mutations were miscellaneous (Table S3). Our findings are consistent with those a previous analysis [109] showing that Leu to Pro is one of the most frequent and harmful of amino acid replacements.
In silico analysis of the INF2 DID structure and effect of pathogenic mutations
An armadillo repeat (ARM) is an approximately 40 amino acid-long sequence that is tandemly repeated 3–20 times in more than 70 proteins involved in fundamental cellular processes, including cell–cell adhesion, cytoskeletal organization, nuclear import, and molecular signaling [110]. Although ARMs do not necessarily have high sequence identity, they share a related structure. They consist of three α-helices, the second and third being arranged in an antiparallel manner, roughly perpendicular to the first helix. Neighboring ARMs stack together to form an elongated super-helix in which the second helix of every ARM forms a highly conserved, concave surface involved in ligand binding [111]. The DID of most formins is organized into a variable number of ARMs [112]. Solving its crystal structure reveals that the DID of mDia1 is composed of four complete ARMs [112–114]. In mDia1, the DAD makes extensive hydrophobic contacts with the second helices of ARMs 2–4, interacting with a patch of exposed residues with very high conservation between different diaphanous-related formins [112–114].
On the basis of the crystal structure of mDia1 DID, the 280 amino-terminal amino acid sequence of INF2 is modeled with Phyre2 software [115] into 17 α-helices, of which α-helices 3–14 are organized into four complete ARMs and form the DID (Figs. 5a, b, S1A, B). Exon 2 encodes two predicted α-helices (α-helices 1 and 2) upstream of ARM1, the entire ARM1 and ARM2, and the first α-helix of ARM3, while exon 3 accounts for the second and third α-helices of ARM3, and exon 4 for the entire ARM4 (Fig. 5b). Nearly all the residues mutated in INF2-related disease are present in the α-helices of the DID ARMs, with only a few in inter-helix sequences (Fig. 5c). The mutation A13T is present in the first helix of the extension amino-terminal to the INF2 DID. Since it has been also found in controls in the NHLBI EVS, this variant has suggested being just a polymorphism without pathological implications [53].
Fig. 5.
In silico prediction of the INF2 DID structure, and localization of pathogenic mutations. a Primary structure of the INF2 DID and upstream amino-terminal sequence, indicating the predicted α-helices and the exons that encode them. The INF2 residues whose mutation causes FSGS alone (blue), FSGS in some cases or FSGS + CMT in others (purple), FSGS + CMT (red), and other diseases (green) are indicated. b The structure of the DID and DAD (cyan) of INF2 as modeled by Phyre2 (Fig. S1a, b). The graphic models were obtained with UCSF Chimera v1.13.1 [142]. The four ARMs are indicated by different colors: ARM1, red; ARM2, orange; ARM3, yellow; ARM4, green. c Position of the residues mutated in pathogenic INF2 in the DID structure. The residues mutated in isolated FSGS alone (blue), FSGS in some cases or FSGS + CMT in others (purple), FSGS + CMT (red), and other diseases (green) are indicated
Similar to what is known for mDia1 [112–114], our predicted structure of the INF2 DID suggests that there should be extensive hydrophobic contacts between the INF2 DID and DAD (Fig. S1C, D). Since Leu is overrepresented in ARMs relative to other amino acids [116], the abundance of Leu substitutions—around one third of the total number—among the pathogenic mutations found in the DID of INF2 is not surprising (Table S3).
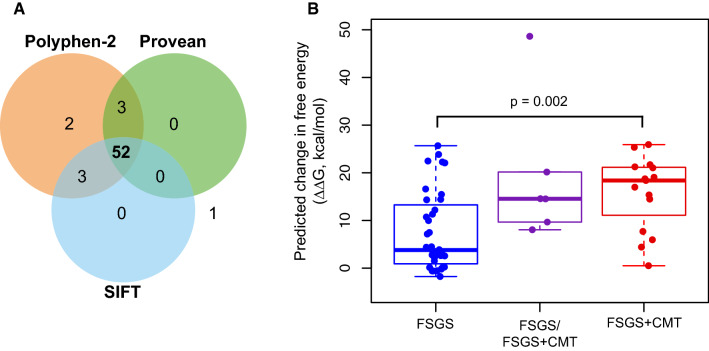
Several bioinformatics tools predict the possible impact of the amino acid substitutions on protein function. SIFT (v6.2.1; https://sift.bii.a-star.edu.sg) [117] is based on sequence homology and the physico-chemical similarity between the alternative amino acid. PROVEAN (v1.1.3; https://provean.jcvi.org) [118] evaluates clusters of sequences homologous with the protein of interest collected from the National Center for Biotechnology Information Non-Redundant Protein Database (https://www.ncbi.nlm.nih.gov/protein). PolyPhen-2 (v2; https://genetics.bwh.harvard.edu/pph2) [119], in addition to relying on sequence homologies and protein families annotated in the Pfam database (https://pfam.xfam.org), uses 3D structures from the Protein Data Bank (https://www.rcsb.org) when they are available. The majority of missense pathogenic mutations in the DID of INF2 (52 of 61, or 85.2%) reported so far are predicted to be “damaging” by these three algorithms (Fig. 6a; Table S2), suggesting that their substitution has an important impact on their function. The only mutation classified as “benign” by the tree programs (S263A) is present in two FSGS cases in which, at least in one of them, the mutation co-occurs with a mutation in the LAMB2 gene, which encodes lamin B2, a nuclear lamina component.
Fig. 6.
Predicted effect of pathogenic missense INF2 mutations on DID stability. a Venn diagram of the analysis of DID-localized pathogenic missense mutations in INF2 by SIFT, PROVEAN and Polyphen-2 programs. The numbers inside the circles indicate the mutations scored as “damaging” by each of the algorithms. Only one mutation (S263A) is predicted as being “benign” by the three algorithms. b Predicted change in free energy (ΔΔG) caused by pathogenic missense mutations of the INF2 DID, calculated using Rosetta software for each of the mutations causing FSGS, FSGS in some cases or FSGS + CMT in others, and FSGS + CMT. Data were analyzed using unpaired Student’s t test. Given the small number of FSGS/FSGS + CMT cases, no statistical comparison with this group was attempted
Protein structure stability depends on the free energy (ΔG) of the folded state. Instead of the primary structure, as used by other algorithms, the Rosetta algorithm (Cyrus Bench®, https://cad.cyrusbio.com) relies on the tertiary structure of the protein of interest to predict the impact on protein stability and structure resulting from single amino acid substitutions by calculating the variation of free energy (ΔΔG) between the folded mutant and the structure of the wild-type protein [120]. The greater the difference, the more destabilizing the mutation is predicted to be. Consistent with the results of a previous analysis of a smaller number of mutations [72], the analysis with Rosetta software [120] of the 54 missense INF2 mutations indicates that the mutations causing FSGS + CMT are generally predicted to have a more destabilizing effect than those producing only FSGS. It is of note that the mutations that cause FSGS or FSGS + CMT, depending on the particular case, had an intermediate destabilizing effect (Fig. 6b; Table S2). These observations are consistent with the fact that FSGS + CMT mutations are more harmful than those producing isolated FSGS, because they produced earlier ESRD. Since the vast majority of highly destabilizing pathogenic mutations of INF2, which are precisely those that cause FSGS + CMT, are concentrated in ARM2 (Fig. 5b, c; Table S2), this ARM might be less tolerant to mutation than the other ARMs, or its integrity may be more essential for normal INF2 function.
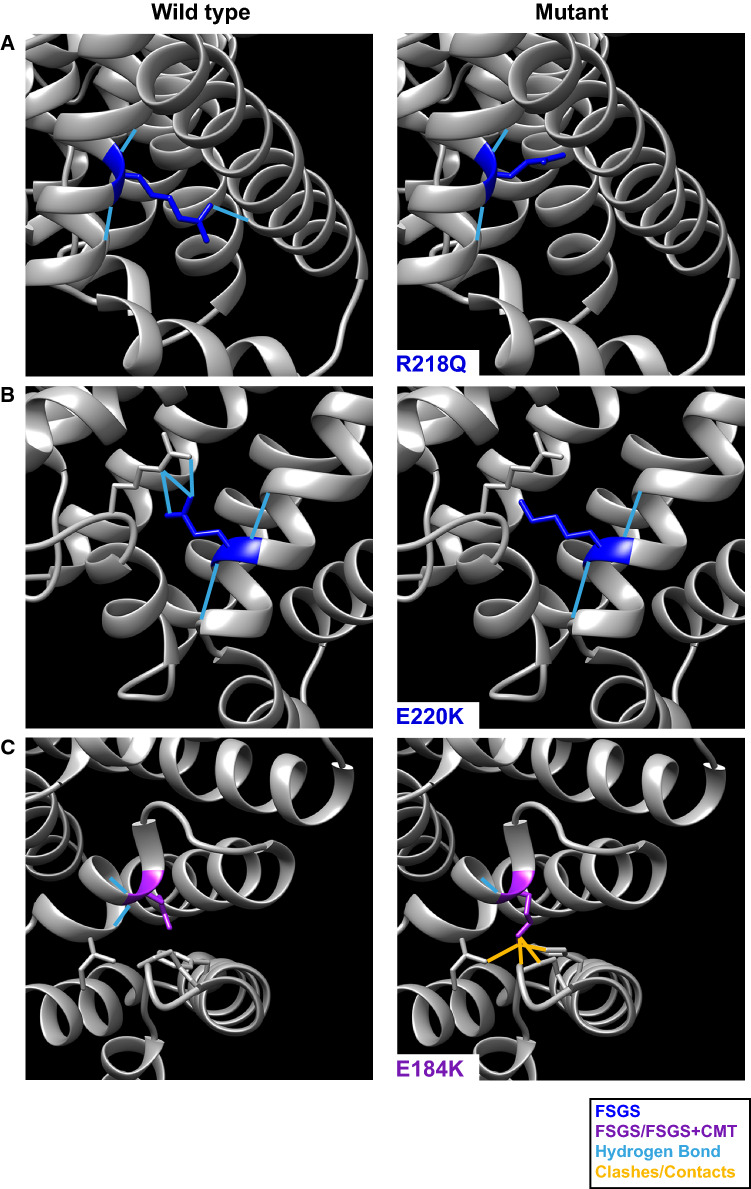
The pathogenic mutations of INF2 could break the corresponding α-helix and alter the DID structure, compromising its interaction with the DAD. As suggested by the comparison between the predicted structure of the DID of wild-type INF2 and that of the three most recurrent mutants (Fig. 7a–c), the pathogenic mutations of INF2 probably affect DID stability to different extent—more in the case of those causing FSGS + CMT than in those producing only FSGS—probably altering DAD binding. In summary, an alteration of the DID structure, with the consequent dysregulation of the INF2 activity, appears to be the cause of INF2-related disease.
Fig. 7.
Predicted structural changes caused by pathogenic INF2 mutations in the DID. a–c Close-up views (right panels) of the effect of the three most recurrent mutations (R218Q, E220K and E184K) on the structure of the INF2 DID, obtained using the Rotamers tool [143]. The close-up views of the corresponding residues in the wild-type protein are shown (left panels). Hydrogen bonds are represented in blue, possible clashes and contacts are indicated in yellow
INF2 variants outside the DID-coding exons
We excluded from our analysis the cases describing mutations found outside the genomic region encoding the INF2 DID (Table S4). These mutations are of three types: in intronic putative splice sites, missense variants, and a gene duplication (Table S4). Those in putative splice sites have been found in two cases: c.1736-6C>T, which relates to global glomerulosclerosis [96], and c.1735 + 2 T>G, which is associated with FSGS [94]. However, these mutations have not actually been shown to produce pathogenic splicing variants. There are also a large number of INF2 gene missense variants outside the DID-coding exons that have been compiled in open access databases such as the NHLBI EVS or The Human Variome Project (https://www.humanvariomeproject.org). Most of these variants are probably just genetic polymorphisms that are not associated with disease, but there is a small group of reported mutations outside the DID that have been associated with FSGS/chronic kidney disease (S483F, V557G, E593K, I685V, R689W, R877Q and P1096S) [96, 97, 121, 122] or CMT (R1045Q) [123]. Among this group, the replacements occur in residues present in the FH2 domain, with the exception of S483F (which maps to the FH1 domain) and R1045Q and P1096S (which map to the C-terminal region). Only the R689W variant has been determined to be damaging by the three predictors, SIFT, PROVEAN and Polyphen-2 (Table S5). Some of the cases associated with the variants outside the DID concur with amino acid substitutions in other proteins related to genetic FSGS or present more than one of this type of replacement in INF2 [97, 122] (Table S4). These observations suggest that the variants outside the DID may require a second hit in INF2 or in another FSGS-associated gene to produce renal disease. This could be also the case of the S263A mutation in the DID. Finally, it is of particular note that there was a single case of FSGS + CMT whose entire INF2 gene was duplicated [86], which suggests that increased levels of INF2 could mimic the effect of the pathogenic mutations.
Effect of the pathogenic mutations on the interactions and function of INF2
Most pathogenic INF2 mutations localized to the DID, where they could potentially alter its binding to the DAD or affect the association of INF2 with other proteins. Tables 4 and S6 summarize the in vitro effect of the pathogenic mutation on INF2 functions and the animal models, respectively, that have been used in research into INF2-related disease. These effects and models are discussed in detail below.
Table 4.
Summary of the main in vitro effects of INF2 pathogenic mutations
| Effect of mutations | References | |
|---|---|---|
| INF2 regulation | Impaired DID–DAD interaction | [125] |
| Loss of inhibition by CAP–KAc-actin | [21] | |
| Altered distribution of the N-terminal proteolytic fragment | [130] | |
| INF2 interactions | Reduced binding of calmodulin | [20] |
| Enhanced interaction with constitutively active Cdc24 | [62] | |
| Reduced interaction of the INF2 DID with the DAD of mDia in some, but not all, of the INF2 mutants assayed | [124] | |
| Increased interaction with G-actin, profilin and CapZ α-1 | [125] | |
| Protein distribution | Altered distribution of MAL, IQGAP1, podocin, and nephrin | [62, 126] |
| Actin cytoskeleton | Reduced cortical actin and stress fiber staining, and diffuse actin distribution | [62] |
| Increased perinuclear actin filament content | [20] | |
| Microtubule cytoskeleton | Diffuse microtubule staining, and reduced capacity to induce microtubule detyrosination | [27, 62] |
| Gene transcription | Reduced impairment of serum response factor-mediated transcription activation by mDia2 | [124] |
The DID of INF2 associates with the Rho-family GTPases Cdc42 and Rac1 in pull-down assays [11, 24, 124]. The association of INF2 with Cdc42 was enhanced in the five pathogenic INF2 mutants assayed [62]. However, as the interaction with Cdc42 seems not to be direct [14], it is unclear how the mutations strengthen the association. The association of INF2 with the scaffold protein IQGAP1 was not affected by the mutations [62], which is consistent with the observation that the interaction with IQGAP1 takes place through the carboxyl-terminal region of INF2 [27]. The analysis of three pathogenic INF2 mutants (E184K, S186P and R218Q) indicates that they weaken the autoinhibitory DID–DAD interaction [125], increasing the interaction of INF2 with G-actin, and also with profilin and the actin capping protein CapZ α-1. These findings suggest aberrant regulation of actin polymerization activity in pathogenic mutants of INF2 [125].
In addition to the intramolecular interaction of the DID with the DAD responsible for INF2 autoregulation, the DID of INF2 mediates an intermolecular association with the DAD of the mDia1-3 formins [124]. Immunostaining of a renal biopsy of a patient with the INF2 R218Q mutation revealed abnormal distribution of podocin and nephrin in glomeruli [126]. In vitro experiments in a podocyte-derived cell line suggest that INF2 transports nephrin to the cell surface by counteracting Rho/mDia, which is consistent with the inability of INF2 R218Q to modulate Rho/mDia1 [126]. Consistent with this finding, the expression of normal INF2, but not of E184K and R218Q, in a zebrafish in vivo model was found to rescue the defects in nephrogenesis and Dia activity caused by INF2 knockdown [127]. Unlike transgenic mice expressing active Rho [128], studies in knock-in mice expressing INF2 R218Q, which is the most recurrent pathogenic INF2 mutation (Table 3), did not reveal any renal pathology or proteinuria at baseline, and most podocytes appeared normal with foot processes and slit diaphragm structure intact. This result was taken to indicate that normal INF2 function is not required for glomerular development, although it was shown to be required for recovery from kidney injury [129]. However, since the INF2 R218Q mutation has low penetrance in humans (Table S1) [52, 54, 76], it is also possible that the mutation cannot alter INF2 function sufficiently in mice. It is of note that whereas the interaction of the INF2 DID with the mDia DAD is impaired in the INF2 E184K and R218Q mutants, the pathogenic S186 mutant maintains the capacity to interact with the mDia DAD [124]. Therefore, although the regulation of Rho/mDia by INF2 appears to be important in some of the INF2 mutants, it is not clear whether it is generally involved in INF2-related disease.
The effect of CAP–KAc-actin complex, an inhibitor of INF2 actin polymerization activity, has been studied in the presence of two disease-associated INF2 mutants (L77R and R218Q) [21, 22]. These INF2 mutants do not bind the CAP–KAc-actin inhibitor efficiently and, consequently, their expression result in a considerable increase in the content of actin filaments. Therefore, pathogenic mutations in the DID of INF2 appear to make INF2 refractory to inhibition by CAP–KAc-actin, placing them in a constitutively active state that could be the cause of the increased content of actin filaments in cells expressing the mutants [20].
Another aspect of INF2 regulation affected by disease-causing mutations was highlighted in a recent report [130] showing that in mouse and human podocytes INF2 undergoes cathepsin-mediated cleavage between Gly547 and Ser548, resulting in an N-terminal fragment that includes the DID and FH1 domains, and a C-terminal fragment containing the FH2 and DAD domains. These fragments displayed differential accumulation in podocytes: both fragments accumulated in the cell body, but only the DID-containing N-terminal fragment accumulated at foot processes. Although comparable levels of INF2 cleavage were observed in cells expressing pathogenic mutants (R218Q, E220K and S186P), there was a loss of the N-terminal fragment from foot processes in kidney tissue from an individual with FSGS due to R218Q mutation, whereas the distribution of the C-terminal fragment was unaltered. Therefore, it is possible the loss of the N-terminal fragment from the foot processes in the INF2 mutants may alter podocyte structure and function.
Ca2+ is a highly versatile second messenger of signal transduction that influences nearly every aspect of cellular life [131, 132], including cell functions such as motility, cytoskeleton remodeling, gene transcription, cell-to-cell communication and cell division, to mention but a few [133]. Given the importance of Ca2+, inappropriate responses to Ca2+ are often associated with disease [134]. Ca2+-sensing proteins, such as calmodulin, mediate Ca2+ responses [135]. INF2, through its interaction with calmodulin, mediates cytoplasmic and nuclear actin remodeling in response to the variation in intracellular Ca2+ levels [19, 20, 36]. Depending on the specific INF2 DID mutation, the association of pathogenic INF2 with calmodulin decreases to different extent and leads to deregulated activation and impaired response to increase cytosolic Ca2+ levels [20]. This implies that defective regulation of INF2 mutants by the concentration of Ca2+ might contribute to the pathological process.
INF2 is necessary for the correct functioning of specialized routes of membrane trafficking mediated by the MAL and MAL2 proteins [11, 24], whose expression coincides in specific types of polarized epithelia (e.g., renal distal tubules), but diverges in other cell types (e.g., human T cells and hepatocytes). MAL is expressed in human Schwann cells but not in podocytes [136], whereas the opposite is true for MAL2 [137]. Immunohistochemical analysis confirmed the colocalization of INF2 and MAL2 in human podocytes, and of INF2 and MAL in normal human peripheral nerve serial sections and mouse Schwann cells [62]. In contrast to the localization of intact INF2, INF2 mutants in patients with FSGS + CMT localized diffusely throughout the cytoplasm and force MAL to acquire a diffused distribution as well [62]. Given that the trafficking of the nephrin and podocin complex to the slit diaphragm membrane is a raft-mediated process, that this trafficking is blocked in INF2 knockdown cells [126], and that MAL and MAL2 are involved in raft-mediated trafficking [25, 26], it is possible that the pathogenic mutations of INF2 could also impair transport mediated by MAL and MAL2 and thereby contribute to the alterations of podocyte and Schwann cell architecture as those found in INF2-linked diseases.
INF2 acts downstream of Rho/mDia to regulate microtubule stability in a process facilitated by the association of INF2 with IQGAP1 [27]. It is of note that, unlike intact INF2, neither E184K nor R218Q INF2 mutants were able to replace endogenous INF2 in microtubule detyrosination, which is a post-translational modification of stable microtubules [27]. This suggests that these pathogenic mutations of INF2 hamper the ability of INF2 to detyrosinate microtubules. Therefore, although the analysis of biological specimens from FSGS and FSGS + CMT patients suggests an alteration of the actin cytoskeleton is the main cause of disease, microtubules could also be affected.
Recent work has shown that INF2 functions, and the effects of disease-causing mutations, are evolutionarily conserved. Studies of the C. elegans “excretory canal”, a unicellular tube required for osmoregulation, showed that EXC-6, a worm INF2 homolog, regulates microtubule dynamics in vivo, and that human INF2, when activated by deletion of the DID and DAD domains or by introduction of disease-causing mutations, rescues exc-6 mutants [138]. Moreover, a second worm INF2 homolog, called INFT-2, was shown to regulate F-actin levels, and itself to be regulated by CYK-1, the worm mDia homolog [139]. Drosophila nephrocytes provide another in vivo genetic model for studying INF2 [20]. Nephrocytes are part of the fruit-fly excretory system, and have remarkable anatomical, molecular and functional similarity with vertebrate podocytes. In particular, the fly nephrocyte forms slit diaphragms similar to those of podocytes, which, unlike those of podocytes, they are formed across deep membrane invaginations of the plasma membrane [140, 141]. It is of note that the expression in Drosophila nephrocytes of pathogenic INF2 mutants leads to reorganization of the actin cytoskeleton and alters nephrin distribution in vivo, the extent of which is associated with the severity of the mutations in humans [20]. This effect on nephrin distribution in nephrocytes is reminiscent of that reported in a podocyte-derived cell line [126], suggesting defects in nephrin traffic. These studies suggest that future work using invertebrate models could help identify new INF2 regulators and functions relevant to FSGS.
Concluding remarks, diagnostic implications and future directions
It has been 10 years since INF2 mutations were reported to be a genetic cause of FSGS. Since then, the numbers of independent cases and reported pathogenic mutations in the DID have increased considerably. More than 60 different pathogenic missense mutations have so far been described, most of which map to exon 2, and to a lesser extent to exons 3, exon 4 and 6. Most of the mutations predicted to be the most destabilizing are located on exons 2 and 3, where they coincide with those causing FSGS + CMT. Those producing isolated FSGS are less destabilizing and are found throughout exons 2, 3, 4 and 6, whereas the mutations that, depending on each specific case, cause either FSGS or FSGS + CMT are predicted to have an intermediate destabilizing potential. Most of the mutations causing FSGS + CMT occur in the residues of the second ARM. Since ARMs 2–4 are responsible for the intramolecular autoinhibitory interaction of the DID with the DAD, an alteration of actin dynamics appears to be the main cause of INF2-related disease.
Despite the persistently low number of cases, the limited ethnic diversity of the patients studied (approximately 70% European or European descendants), and the probable underdiagnosis of CMT in some FSGS patients [70], our analysis allows us to make some relevant conclusions about the molecular diagnosis of INF2-related disease. FSGS + CMT cases are mostly associated with mutations in exon 2. Exon 3 mutations are mainly associated with familial cases and cause similar percentages of cases of FSGS and FSGS + CMT. Exon 4 mutations mainly correspond to familial FSGS. Our analysis also shows that the percentage of sporadic cases of FSGS + CMT is higher than that of FSGS alone. It would therefore be useful to screen for INF2 mutations in cases of FSGS + CMT, regardless of whether there is a familial history of the disease. Conversely, most INF2-related FSGS cases are actually familial, so molecular diagnosis could be restricted to patients with a family history of the disease. This analysis would also enable us to exclude as kidney donors those relatives at risk of developing FSGS [80].
The variable penetrance of some of the INF2 mutations provides a good example of the importance of the environmental and genetic background in the development of INF2-related pathologies. It seems plausible that a second mutation either in INF2 or in a gene conferring susceptibility to FSGS, or a specific risk haplotype related to FSGS, is necessary for the disease to appear in these cases. More detailed genetic and life style studies of families with this type of mutation could identify the factors that determine the severity of the disease and provide persuasive evidence to encourage patients to adopt lifestyle habits that would delay the development of ESRD [80].
The genetic origin of FSGS nephropathy caused by INF2 mutation means that drug-based therapies, such as treatment with steroids or cyclosporine A, applied to FSGS patients with other etiologies, is not effective for treating INF2-related disease. Kidney transplantation, with its consequent drawbacks, is the only option currently available for the complete correction of the renal defect in FSGS caused by pathogenic INF2.
Despite the enormous advances in recent years in our understanding of INF2 function under normal and pathological conditions, further work is needed to fully understand the molecular mechanism leading to the pathological conditions caused by INF2 mutation. The involvement of INF2 DID mutations in disease has been well established by a large number of genetic studies. It will be interesting to see whether INF2 variants found outside the DID are also involved in disease. INF2 and other formins have a dual role in the actin and microtubule cytoskeletons [7, 8]. Therefore, in addition to altering actin dynamics, the pathogenic mutations in the INF2 DID can influence microtubule dynamics. As a consequence of the involvement of INF2 in the cytoskeleton, processes such as membrane trafficking, cell polarity and the cell response to increased intracellular Ca2+ levels might be affected in INF2-related disease. These deserve further exploration. We hope that this effort, as well as helping to clarify the mechanism by which INF2 mutations cause cell dysfunction, will enable us to develop novel strategies for treating INF2-related disease.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We thank Dr Phil Mason for revising the English language of the manuscript. We thank Cyrus Biotechnology Inc. for a free trial of the Cyrys Bench® software. Research in the laboratory of MAA is supported by a Grant (PGC2018-095643-B-I00) from the Ministerio de Ciencia e Innovación, Fondo Europeo de Desarrollo Regional y Agencia Estatal de Investigación. A contract (FPU16/00935) from the Ministerio de Ciencia e Innovación to LL-d-H is also acknowledged.
Abbreviations
- aHUS
Atypical hemolytic uremic syndrome
- ARM
Armadillo
- CAP
Cyclase-associated protein
- CMT
Charcot–Marie–Tooth
- DAD
Diaphanous autoinhibitory domain
- DID
Diaphanous inhibitory domain
- ESRD
End-stage renal disease
- FH
Formin homology
- FSGS
Focal segmental glomerulosclerosis
- INF2
Inverted formin 2
- KAc-actin
Lysine-acetylated actin
- MCN
Minimal change nephropathy
- WH2
Wiskott–Aldrich syndrome homology 2
Compliance with ethical standards
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Goode BL, Eck MJ. Mechanism and function of formins in the control of actin assembly. Annu Rev Biochem. 2007;76:593–627. doi: 10.1146/annurev.biochem.75.103004.142647. [DOI] [PubMed] [Google Scholar]
- 2.Chesarone MA, DuPage AG, Goode BL. Unleashing formins to remodel the actin and microtubule cytoskeletons. Nat Rev Mol Cell Biol. 2010;11:62–74. doi: 10.1038/nrm2816. [DOI] [PubMed] [Google Scholar]
- 3.Schönichen A, Geyer M. Fifteen formins for an actin filament: a molecular view on the regulation of human formins. Biochim Biophys Acta Mol Cell Res. 2010;1803:152–163. doi: 10.1016/j.bbamcr.2010.01.014. [DOI] [PubMed] [Google Scholar]
- 4.Pruyne D. Revisiting the phylogeny of the animal formins: two new subtypes, relationships with multiple Wing Hairs proteins, and a lost human formin. PLoS ONE. 2016;11:e0164067. doi: 10.1371/journal.pone.0164067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alberts AS. Identification of a carboxyl-terminal diaphanous-related formin homology protein autoregulatory domain. J Biol Chem. 2001;276:2824–2830. doi: 10.1074/jbc.M006205200. [DOI] [PubMed] [Google Scholar]
- 6.Bartolini F, Gundersen GG. Formins and microtubules. Biochim Biophys Acta (Mol Cell Res) 2010;1803:164–173. doi: 10.1016/j.bbamcr.2009.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ishizaki T, Morishima Y, Okamoto M, Furuyashiki T, Kato T, Narumiya S. Coordination of microtubules and the actin cytoskeleton by the Rho effector mDia1. Nat Cell Biol. 2001;3:8–14. doi: 10.1038/35050598. [DOI] [PubMed] [Google Scholar]
- 8.Palazzo AF, Cook TA, Alberts AS, Gundersen GG. mDia mediates Rho-regulated formation and orientation of stable microtubules. Nat Cell Biol. 2001;3:723–729. doi: 10.1038/35087035. [DOI] [PubMed] [Google Scholar]
- 9.Chhabra ES, Ramabhadran V, Gerber SA, Higgs HN. INF2 is an endoplasmic reticulum-associated formin protein. J Cell Sci. 2009;122:1430–1440. doi: 10.1242/jcs.040691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ramabhadran V, Korobova F, Rahme GJ, Higgs HN. Splice variant-specific cellular function of the formin INF2 in maintenance of Golgi architecture. Mol Biol Cell. 2011;22:4822–4833. doi: 10.1091/mbc.E11-05-0457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Madrid R, Aranda JF, Rodríguez-Fraticelli AE, Ventimiglia L, Andres-Delgado L, Shehata M, Fanayan S, Shahheydari H, Gomez S, Jimenez A, Martin-Belmonte F, Byrne JA, Alonso MA. The formin INF2 regulates basolateral-to-apical transcytosis and lumen formation in association with Cdc42 and MAL2. Dev Cell. 2010;18:814–827. doi: 10.1016/j.devcel.2010.04.001. [DOI] [PubMed] [Google Scholar]
- 12.Chhabra ES, Higgs HN. INF2 is a WASP homology 2 motif-containing formin that severs actin filaments and accelerates both polymerization and depolymerization. J Biol Chem. 2006;281:26754–26767. doi: 10.1074/jbc.M604666200. [DOI] [PubMed] [Google Scholar]
- 13.Gurel PS, Ge P, Grintsevich EE, Shu R, Blanchoin L, Zhou ZH, Reisler E, Higgs HN. INF2-mediated severing through actin filament encirclement and disruption. Curr Biol. 2014;24:156–164. doi: 10.1016/j.cub.2013.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ramabhadran V, Hatch AL, Higgs HN. Actin monomers activate inverted formin 2 by competing with its autoinhibitory interaction. J Biol Chem. 2013;288:26847–26855. doi: 10.1074/jbc.M113.472415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fernandez-Barrera J, Alonso MA. Coordination of microtubule acetylation and the actin cytoskeleton by formins. Cell Mol Life Sci. 2018;75:3181–3191. doi: 10.1007/s00018-018-2855-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Olson EN, Nordheim A. Linking actin dynamics and gene transcription to drive cellular motile functions. Nat Rev Mol Cell Biol. 2010;11:353–365. doi: 10.1038/nrm2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Posern G, Treisman R. Actin' together: serum response factor, its cofactors and the link to signal transduction. Trends Cell Biol. 2006;16:588–596. doi: 10.1016/j.tcb.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 18.Fernandez-Barrera J, Bernabe-Rubio M, Casares-Arias J, Rangel L, Fernandez-Martin L, Correas I, Alonso MA. The actin-MRTF-SRF transcriptional circuit controls tubulin acetylation via α-TAT1 gene expression. J Cell Biol. 2018;217:929–944. doi: 10.1083/jcb.201702157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wales P, Schuberth CE, Aufschnaiter R, Fels J, Garcia-Aguilar I, Janning A, Dlugos CP, Schäfer-Herte M, Klingner C, Wälte M, Kuhlmann J, Menis E, Hockaday Kang L, Maier KC, Hou W, Russo A, Higgs HN, Pavenstädt H, Vogl T, Roth J, Qualmann B, Kessels MM, Martin DE, Mulder B, Wedlich-Söldner R. Calcium-mediated actin reset (CaAR) mediates acute cell adaptations. eLife. 2016;5:e19850. doi: 10.7554/eLife.19850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bayraktar S, Nehrig J, Menis E, Karli K, Janning A, Struk T, Halbritter J, Michgehl U, Krahn MP, Schuberth CE, Pavenstädt HP, Wedlich-Söldner RA. A deregulated stress response underlies distinct INF2 associated disease profiles. bioRxiv. 2019 doi: 10.1101/838086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mu A, Fung TS, Kettenbach AN, Chakrabarti R, Higgs HN. A complex containing lysine-acetylated actin inhibits the formin INF2. Nat Cell Biol. 2019;21:592–602. doi: 10.1038/s41556-019-0307-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mu A, Fung TS, Francomacaro LM, Huynh T, Kotila T, Svindrych Z, Higgs HN. Regulation of INF2-mediated actin polymerization through site-specific lysine acetylation of actin itself. Proc Natl Acad Sci USA. 2020;117:439–447. doi: 10.1073/pnas.1914072117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jin X, Wang J, Gao K, Zhang P, Yao L, Tang Y, Tang L, Ma J, Xiao J, Zhang E, Zhu J, Zhang B, Zhao S-m, Li Y, Ren S, Huang H, Yu L, Wang C. Dysregulation of INF2-mediated mitochondrial fission in SPOP-mutated prostate cancer. PLoS Genet. 2017;13:e1006748. doi: 10.1371/journal.pgen.1006748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Andres-Delgado L, Anton OM, Madrid R, Byrne JA, Alonso MA. Formin INF2 regulates MAL-mediated transport of Lck to the plasma membrane of human T lymphocytes. Blood. 2010;116:5919–5929. doi: 10.1182/blood-2010-08-300665. [DOI] [PubMed] [Google Scholar]
- 25.Puertollano R, Martin-Belmonte F, Millan J, de Marco MC, Albar JP, Kremer L, Alonso MA. The MAL proteolipid is necessary for normal apical transport and accurate sorting of the influenza virus hemagglutinin in Madin-Darby canine kidney cells. J Cell Biol. 1999;145:141–151. doi: 10.1083/jcb.145.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Marco MC, Martin-Belmonte F, Kremer L, Albar JP, Correas I, Vaerman JP, Marazuela M, Byrne JA, Alonso MA. MAL2, a novel raft protein of the MAL family, is an essential component of the machinery for transcytosis in hepatoma HepG2 cells. J Cell Biol. 2002;159:37–44. doi: 10.1083/jcb.200206033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bartolini F, Andres-Delgado L, Qu X, Nik S, Ramalingam N, Kremer L, Alonso MA, Gundersen GG. An mDia1-INF2 formin activation cascade facilitated by IQGAP1 regulates stable microtubules in migrating cells. Mol Biol Cell. 2016;27:1797–1808. doi: 10.1091/mbc.E15-07-0489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Andres-Delgado L, Anton OM, Bartolini F, Ruiz-Saenz A, Correas I, Gundersen GG, Alonso MA. INF2 promotes the formation of detyrosinated microtubules necessary for centrosome reorientation in T cells. J Cell Biol. 2012;198:1025–1037. doi: 10.1083/jcb.201202137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chakrabarti R, Ji W-K, Stan RV, de Juan SJ, Ryan TA, Higgs HN. INF2-mediated actin polymerization at the ER stimulates mitochondrial calcium uptake, inner membrane constriction, and division. J Cell Biol. 2018;217:251–268. doi: 10.1083/jcb.201709111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Korobova F, Ramabhadran V, Higgs HN. An actin-dependent step in mitochondrial fission mediated by the ER-associated formin INF2. Science. 2013;339:464–467. doi: 10.1126/science.1228360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Manor U, Bartholomew S, Golani G, Christenson E, Kozlov M, Higgs H, Spudich J, Lippincott-Schwartz J. A mitochondria-anchored isoform of the actin-nucleating spire protein regulates mitochondrial division. eLife. 2015;4:e08828. doi: 10.7554/eLife.08828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hatch AL, Gurel PS, Higgs HN. Novel roles for actin in mitochondrial fission. J Cell Sci. 2014;127:4549–4560. doi: 10.1242/jcs.153791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Korobova F, Gauvin TJ, Higgs HN. A role for myosin II in mammalian mtochondrial fission. Curr Biol. 2014;24:409–414. doi: 10.1016/j.cub.2013.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Panzer L, Trübe L, Klose M, Joosten B, Slotman J, Cambi A, Linder S. The formins FHOD1 and INF2 regulate inter- and intra-structural contractility of podosomes. J Cell Sci. 2016;129:298–313. doi: 10.1242/jcs.177691. [DOI] [PubMed] [Google Scholar]
- 35.Shao X, Li Q, Mogilner A, Bershadsky AD, Shivashankar GV. Mechanical stimulation induces formin-dependent assembly of a perinuclear actin rim. Proc Natl Acad Sci USA. 2015;112:E2595–E2601. doi: 10.1073/pnas.1504837112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang Y, Sherrard A, Zhao B, Melak M, Trautwein J, Kleinschnitz E-M, Tsopoulidis N, Fackler OT, Schwan C, Grosse R. GPCR-induced calcium transients trigger nuclear actin assembly for chromatin dynamics. Nat Commun. 2019;10:5271. doi: 10.1038/s41467-019-13322-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Takeuchi Y, Narumi R, Akiyama R, Vitiello E, Shirai T, Tanimura N, Kuromiya K, Ishikawa S, Kajita M, Tada M, Haraoka Y, Akieda Y, Ishitani T, Fujioka Y, Ohba Y, Yamada S, Hosokawa Y, Toyama Y, Matsui T, Fujita Y. Calcium wave promotes cell extrusion. Curr Biol. 2020;30:670–681.e676. doi: 10.1016/j.cub.2019.11.089. [DOI] [PubMed] [Google Scholar]
- 38.Lamm KYB, Johnson ML, Baker Phillips J, Muntifering MB, James JM, Jones HN, Redline RW, Rokas A, Muglia LJ. Inverted formin 2 regulates intracellular trafficking, placentation, and pregnancy outcome. eLife. 2018;7:e31150. doi: 10.7554/eLife.31150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu Z, Wei Y, Zhang L, Yee PP, Johnson M, Zhang X, Gulley M, Atkinson JM, Trebak M, Wang H-G, Li W. Induction of store-operated calcium entry (SOCE) suppresses glioblastoma growth by inhibiting the Hippo pathway transcriptional coactivators YAP/TAZ. Oncogene. 2019;38:120–139. doi: 10.1038/s41388-018-0425-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang Z, Yu J. Nurr1 exacerbates cerebral ischemia–reperfusion injury via modulating YAP-INF2-mitochondrial fission pathways. Int J Biochem Cell Biol. 2018;104:149–160. doi: 10.1016/j.biocel.2018.09.014. [DOI] [PubMed] [Google Scholar]
- 41.Zhao H, Pan W, Chen L, Luo Y, Xu R. Nur77 promotes cerebral ischemia, Äìreperfusion injury via activating INF2-mediated mitochondrial fragmentation. J Mol Histol. 2018;49:599–613. doi: 10.1007/s10735-018-9798-8. [DOI] [PubMed] [Google Scholar]
- 42.Heuser VD, Mansuri N, Mogg J, Kurki S, Repo H, Kronqvist P, Carpen O, Gardberg M. Formin proteins FHOD1 and INF2 in triple-negative breast cancer: association with basal markers and functional activities. Breast Cancer (Auckl) 2018;12:1178223418792247. doi: 10.1177/1178223418792247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Scott RP, Quaggin SE. The cell biology of renal filtration. J Cell Biol. 2015;209:199–210. doi: 10.1083/jcb.201410017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Welsh GI, Saleem MA. The podocyte cytoskeleton-key to a functioning glomerulus in health and disease. Nat Rev Nephrol. 2012;8:14. doi: 10.1038/nrneph.2011.151. [DOI] [PubMed] [Google Scholar]
- 45.Schlondorff J. How many Achilles' heels does a podocyte have? An update on podocyte biology. Nephrol Dial Transpl. 2015;30:1091–1097. doi: 10.1093/ndt/gfu214. [DOI] [PubMed] [Google Scholar]
- 46.Hermann P, Wilhelm K, Matthias K. Cell biology of the glomerular podocyte. Physiol Rev. 2003;83:253–307. doi: 10.1152/physrev.00020.2002. [DOI] [PubMed] [Google Scholar]
- 47.Assady S, Wanner N, Skorecki KL, Huber TB. New insights into podocyte biology in glomerular health and disease. J Am SocNephrol. 2017;28:1707. doi: 10.1681/asn.2017010027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Neal CR. Podocytes … What's under yours? (Podocytes and foot processes and how they change in nephropathy) Front Endocrinol. 2015 doi: 10.3389/fendo.2015.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rosenberg AZ, Kopp JB. Focal segmental glomerulosclerosis. Clin J Am Soc Nephrol. 2017;12:502–517. doi: 10.2215/cjn.05960616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fogo AB. Causes and pathogenesis of focal segmental glomerulosclerosis. Nat Rev Nephrol. 2015;11:76. doi: 10.3389/fendo.2015.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lepori N, Zand L, Sethi S, Fernandez-Juarez G, Fervenza FC. Clinical and pathological phenotype of genetic causes of focal segmental glomerulosclerosis in adults. Clin Kidney J. 2018;11:179–190. doi: 10.1093/ckj/sfx143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brown EJ, Schlondorff JS, Becker DJ, Tsukaguchi H, Tonna SJ, Uscinski AL, Higgs HN, Henderson JM, Pollak MR. Mutations in the formin gene INF2 cause focal segmental glomerulosclerosis. Nat Genet. 2010;42:72–76. doi: 10.1038/ng.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Barua M, Brown EJ, Charoonratana VT, Genovese G, Sun H, Pollak MR. Mutations in the INF2 gene account for a significant proportion of familial but not sporadic focal and segmental glomerulosclerosis. Kidney Int. 2013;83:316–322. doi: 10.1038/ki.2012.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Boyer O, Benoit G, Gribouval O, Nevo F, Tete M-J, Dantal J, Gilbert-Dussardier B, Touchard G, Karras A, Presne C, Grunfeld J-P, Legendre C, Joly D, Rieu P, Mohsin N, Hannedouche T, Moal V, Gubler M-C, Broutin I, Mollet G, Antignac C. Mutations in INF2 are a major cause of autosomal dominant focal segmental glomerulosclerosis. J Am Soc Nephrol. 2011;22:239–245. doi: 10.1681/asn.2010050518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gbadegesin RA, Lavin PJ, Hall G, Bartkowiak B, Homstad A, Jiang R, Wu G, Byrd A, Lynn K, Wolfish N, Ottati C, Stevens P, Howell D, Conlon P, Winn MP. Inverted formin 2 mutations with variable expression in patients with sporadic and hereditary focal and segmental glomerulosclerosis. Kidney Int. 2012;81:94–99. doi: 10.1038/ki.2011.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rossor AM, Polke JM, Houlden H, Reilly MM. Clinical implications of genetic advances in Charcot–Marie–Tooth disease. Nat Rev Neurol. 2013;9:562–571. doi: 10.1038/nrneurol.2013.179. [DOI] [PubMed] [Google Scholar]
- 57.Barreto LCLS, Oliveira FS, Nunes PS, de França Costa IMP, Garcez CA, Goes GM, Neves ELA, de Souza Siqueira Quintans J, de Souza Araújo AA. Epidemiologic study of Charcot–Marie–Tooth disease: a systematic review. Neuroepidemiology. 2016;46:157–165. doi: 10.1159/000443706. [DOI] [PubMed] [Google Scholar]
- 58.Hoyle JC, Isfort MC, Roggenbuck J, Arnold WD. The genetics of Charcot–Marie–Tooth disease: current trends and future implications for diagnosis and management. Appl Clin Genet. 2015;8:235–243. doi: 10.2147/tacg.s69969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lemieux G, Neemeh JA. Charcot–Marie–Tooth disease and nephritis. Can Med Assoc J. 1967;97:1193–1198. [PMC free article] [PubMed] [Google Scholar]
- 60.Paul M, Fernandez D, Pryse-Phillips W, Gault M. Charcot–Marie–Tooth disease and nephropathy in a mother and daughter with a review of the literature. Nephron. 1990;54:80–85. doi: 10.1159/000185814. [DOI] [PubMed] [Google Scholar]
- 61.De Rechter S, De Waele L, Levtchenko E, Mekahli D. Charcot–Marie–Tooth: are you testing for proteinuria? Eur J Paed Neurol. 2015;19:1–5. doi: 10.1016/j.ejpn.2014.08.004. [DOI] [PubMed] [Google Scholar]
- 62.Boyer O, Nevo F, Plaisier E, Funalot B, Gribouval O, Benoit G, Cong EH, Arrondel C, Tête M-J, Montjean R, Richard L, Karras A, Pouteil-Noble C, Balafrej L, Bonnardeaux A, Canaud G, Charasse C, Dantal J, Deschenes G, Deteix P, Dubourg O, Petiot P, Pouthier D, Leguern E, Guiochon-Mantel A, Broutin I, Gubler M-C, Saunier S, Ronco P, Vallat J-M, Alonso MA, Antignac C, Mollet G. INF2 mutations in Charcot–Marie–Tooth disease with glomerulopathy. N Engl J Med. 2011;365:2377–2388. doi: 10.1056/NEJMoa1109122. [DOI] [PubMed] [Google Scholar]
- 63.Liu L, Zhang R. Intermediate Charcot–Marie–Tooth disease. Neurosci Bull. 2014;30:999–1009. doi: 10.1007/s12264-014-1475-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mathis S, Bi F, Boyer O, Lacroix C, Marcorelles P, Magy L, Richard L, Antignac C, Vallat J-M. Neuropathologic characterization of INF2-related Charcot–Marie–Tooth disease: evidence for a Schwann cell actinopathy. J Neuropathol Exp Neurol. 2014;73:223–233. doi: 10.1097/NEN.0000000000000047. [DOI] [PubMed] [Google Scholar]
- 65.Tricaud N. Myelinating Schwann cell polarity and mechanically-driven myelin sheath elongation. Front Cell Neurosci. 2018 doi: 10.3389/fncel.2017.00414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lynch ED, Lee MK, Morrow JE, Welcsh PL, Leon PE, King M-C. Nonsyndromic deafness DFNA1 associated with mutation of a human homolog of the Drosophila gene diaphanous. Science. 1997;278:1315–1318. doi: 10.1126/science.278.5341.1315. [DOI] [PubMed] [Google Scholar]
- 67.Schoen CJ, Emery SB, Thorne MC, Ammana HR, Sliwerska E, Arnett J, Hortsch M, Hannan F, Burmeister M, Lesperance MM. Increased activity of Diaphanous homolog 3 (DIAPH3)/diaphanous causes hearing defects in humans with auditory neuropathy and Drosophila. Proc Natl Acad Sci USA. 2010;107:13396–13401. doi: 10.1073/pnas.1003027107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Drummond MC, Belyantseva IA, Friderici KH, Friedman TB. Actin in hair cells and hearing loss. Hear Res. 2012;288:89–99. doi: 10.1016/j.heares.2011.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mademan I, Deconinck T, Dinopoulos A, Voit T, Schara U, Devriendt K, Meijers B, Lerut E, De Jonghe P, Baets J. De novo INF2 mutations expand the genetic spectrum of hereditary neuropathy with glomerulopathy. Neurology. 2013;81:1953–1958. doi: 10.1212/01.wnl.0000436615.58705.c9. [DOI] [PubMed] [Google Scholar]
- 70.Roos A, Weis J, Korinthenberg R, Fehrenbach H, Hausler M, Zuchner S, Mache C, Hubmann H, Auer-Grumbach M, Senderek J. Inverted formin 2-related Charcot–Marie–Tooth disease: extension of the mutational spectrum and pathological findings in Schwann cells and axons. J Peripher Nerv Syst. 2015;20:52–59. doi: 10.1111/jns.12106. [DOI] [PubMed] [Google Scholar]
- 71.Toyota K, Ogino D, Hayashi M, Taki M, Saito K, Abe A, Hashimoto T, Umetsu K, Tsukaguchi H, Hayasaka K. INF2 mutations in Charcot–Marie–Tooth disease complicated with focal segmental glomerulosclerosis. J Peripher Nerv Syst. 2013;18:97–98. doi: 10.1111/jns5.12014. [DOI] [PubMed] [Google Scholar]
- 72.Rodriguez PQ, Lohkamp B, Celsi G, Mache CJ, Auer-Grumbach M, Wernerson A, Hamajima N, Tryggvason K, Patrakka J. Novel INF2 mutation p. L77P in a family with glomerulopathy and Charcot–Marie–Tooth neuropathy. Ped Nephrol. 2013;28:339–343. doi: 10.1007/s00467-012-2299-1. [DOI] [PubMed] [Google Scholar]
- 73.Xie J, Hao X, Azeloglu EU, Ren H, Wang Z, Ma J, Liu J, Ma X, Wang W, Pan X, Zhang W, Zhong F, Li Y, Meng G, Kiryluk K, He JC, Gharavi AG, Chen N. Novel mutations in the inverted formin 2 gene of Chinese families contribute to focal segmental glomerulosclerosis. Kidney Int. 2015;88:593–604. doi: 10.1038/ki.2015.106. [DOI] [PubMed] [Google Scholar]
- 74.Echaniz-Laguna A, Latour P. A cryptic splicing mutation in the INF2 gene causing Charcot–Marie–Tooth disease with minimal glomerular dysfunction. J Peripher Nerv Sys. 2019;24:120–124. doi: 10.1111/jns.12308. [DOI] [PubMed] [Google Scholar]
- 75.Challis RC, Ring T, Xu Y, Wong EKS, Flossmann O, Roberts ISD, Ahmed S, Wetherall M, Salkus G, Brocklebank V, Fester J, Strain L, Wilson V, Wood KM, Marchbank KJ, Santibanez-Koref M, Goodship THJ, Kavanagh D. Thrombotic microangiopathy in inverted formin 2-mediated renal dsease. J Am Soc Nephrol. 2017;28:1084–1091. doi: 10.1681/asn.2015101189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Caridi G, Lugani F, Dagnino M, Gigante M, Iolascon A, Falco M, Graziano C, Benetti E, Dugo M, Del Prete D, Granata A, Borracelli D, Moggia E, Quaglia M, Rinaldi R, Gesualdo L, Ghiggeri GM. Novel INF2 mutations in an Italian cohort of patients with focal segmental glomerulosclerosis, renal failure and Charcot–Marie–Tooth neuropathy. Nephrol Dial Transplant. 2014;29(Suppl 4):iv80–iv86. doi: 10.1093/ndt/gfu071. [DOI] [PubMed] [Google Scholar]
- 77.Park HJ, Kim HJ, Hong YB, Nam SH, Chung KW, Choi BO. A novel INF2 mutation in a Korean family with autosomal dominant intermediate Charcot–Marie–Tooth disease and focal segmental glomerulosclerosis. J Peripher Nerv Syst. 2014;19:175–179. doi: 10.1111/jns5.12062. [DOI] [PubMed] [Google Scholar]
- 78.Laurin LP, Lu M, Mottl AK, Blyth ER, Poulton CJ, Weck KE. Podocyte-associated gene mutation screening in a heterogeneous cohort of patients with sporadic focal segmental glomerulosclerosis. Nephrol Dial Transplant. 2014;29:2062–2069. doi: 10.1093/ndt/gft532. [DOI] [PubMed] [Google Scholar]
- 79.Varner JD, Chryst-Stangl M, Esezobor CI, Solarin A, Wu G, Lane B, Hall G, Abeyagunawardena A, Matory A, Hunley TE, Lin JJ, Howell D, Gbadegesin R. Genetic testing for steroid-resistant-nephrotic syndrome in an outbred population. Front Pediatr. 2018;6:307. doi: 10.3389/fped.2018.00307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Munch J, Grohmann M, Lindner TH, Bergmann C, Halbritter J. Diagnosing FSGS without kidney biopsy—a novel INF2-mutation in a family with ESRD of unknown origin. BMC Med Genet. 2016;17:73. doi: 10.1186/s12881-016-0336-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Buscher AK, Celebi N, Hoyer PF, Klein HG, Weber S, Hoefele J. Mutations in INF2 may be associated with renal histology other than focal segmental glomerulosclerosis. Pediatr Nephrol. 2018;33:433–437. doi: 10.1007/s00467-017-3811-4. [DOI] [PubMed] [Google Scholar]
- 82.Gribouval O, Boyer O, Hummel A, Dantal J, Martinez F, Sberro-Soussan R, Etienne I, Chauveau D, Delahousse M, Lionet A, Allard J, Pouteil Noble C, Tete MJ, Heidet L, Antignac C, Servais A. Identification of genetic causes for sporadic steroid-resistant nephrotic syndrome in adults. Kidney Int. 2018;94:1013–1022. doi: 10.1016/j.kint.2018.07.024. [DOI] [PubMed] [Google Scholar]
- 83.Santin S, Bullich G, Tazon-Vega B, Garcia-Maset R, Gimenez I, Silva I, Ruiz P, Ballarin J, Torra R, Ars E. Clinical utility of genetic testing in children and adults with steroid-resistant nephrotic syndrome. Clin J Am Soc Nephrol. 2011;6:1139–1148. doi: 10.2215/CJN.05260610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sanchez-Ares M, Garcia-Vidal M, Antucho EE, Julio P, Eduardo VM, Lens XM, Garcia-Gonzalez MA. A novel mutation, outside of the candidate region for diagnosis, in the inverted formin 2 gene can cause focal segmental glomerulosclerosis. Kidney Int. 2013;83:153–159. doi: 10.1038/ki.2012.325. [DOI] [PubMed] [Google Scholar]
- 85.Weber S, Buscher AK, Hagmann H, Liebau MC, Heberle C, Ludwig M, Rath S, Alberer M, Beissert A, Zenker M, Hoyer PF, Konrad M, Klein HG, Hoefele J. Dealing with the incidental finding of secondary variants by the example of SRNS patients undergoing targeted next-generation sequencing. Pediatr Nephrol. 2016;31:73–81. doi: 10.1007/s00467-015-3167-6. [DOI] [PubMed] [Google Scholar]
- 86.Dohrn MF, Glockle N, Mulahasanovic L, Heller C, Mohr J, Bauer C, Riesch E, Becker A, Battke F, Hortnagel K, Hornemann T, Suriyanarayanan S, Blankenburg M, Schulz JB, Claeys KG, Gess B, Katona I, Ferbert A, Vittore D, Grimm A, Wolking S, Schols L, Lerche H, Korenke GC, Fischer D, Schrank B, Kotzaeridou U, Kurlemann G, Drager B, Schirmacher A, Young P, Schlotter-Weigel B, Biskup S. Frequent genes in rare diseases: panel-based next generation sequencing to disclose causal mutations in hereditary neuropathies. J Neurochem. 2017;143:507–522. doi: 10.1111/jnc.14217. [DOI] [PubMed] [Google Scholar]
- 87.Sadowski CE, Lovric S, Ashraf S, Pabst WL, Gee HY, Kohl S, Engelmann S, Vega-Warner V, Fang H, Halbritter J, Somers MJ, Tan W, Shril S, Is F, Lifton RP, Bockenhauer D, El-Desoky S, Kari JA, Zenker M, Kemper MJ, Mueller D, Fathy HM, Soliman NA, Group SS, Hildebrandt F. A single-gene cause in 29.5% of cases of steroid-resistant nephrotic syndrome. J Am Soc Nephrol. 2015;26:1279–1289. doi: 10.1681/asn.2014050489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Jin S, Wang W, Wang R, Lv H, Zhang W, Wang Z, Jiao J, Yuan Y. INF2 mutations associated with dominant inherited intermediate Charcot–Marie–Tooth neuropathy with focal segmental glomerulosclerosis in two Chinese patients. Clin Neuropathol. 2015;34:275–281. doi: 10.5414/NP300835. [DOI] [PubMed] [Google Scholar]
- 89.Rood IM, Bongers EM, Lugtenberg D, Klein IH, Steenbergen EJ, Wetzels JF, Deegens JK. Familial focal segmental glomerulosclerosis: mutation in inverted formin 2 mimicking Alport syndrome. Neth J Med. 2016;74:82–85. [PubMed] [Google Scholar]
- 90.Tan W, Lovric S, Ashraf S, Rao J, Schapiro D, Airik M, Shril S, Gee HY, Baum M, Daouk G, Ferguson MA, Rodig N, Somers MJG, Stein DR, Vivante A, Warejko JK, Widmeier E, Hildebrandt F. Analysis of 24 genes reveals a monogenic cause in 11. 1% of cases with steroid-resistant nephrotic syndrome at a single center. Pediatr Nephrol. 2018 doi: 10.1007/s00467-017-3801-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ogino D, Hashimoto T, Hattori M, Sugawara N, Akioka Y, Tamiya G, Makino S, Toyota K, Mitsui T, Hayasaka K. Analysis of the genes responsible for steroid-resistant nephrotic syndrome and/or focal segmental glomerulosclerosis in Japanese patients by whole-exome sequencing analysis. J Hum Genet. 2016;61:137–141. doi: 10.1038/jhg.2015.122. [DOI] [PubMed] [Google Scholar]
- 92.Shang S, Peng F, Wang T, Wu X, Li P, Li Q, Chen XM. Genotype-phenotype correlation and prognostic impact in chinese patients with Alport syndrome. Mol Genet Genom Med. 2019;7:e00741–e00741. doi: 10.1002/mgg3.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Safarikova M, Stekrova J, Honsova E, Horinova V, Tesar V, Reiterova J. Mutational screening of inverted formin 2 in adult-onset focal segmental glomerulosclerosis or minimal change patients from the Czech Republic. BMC Med Genet. 2018;19:147. doi: 10.1186/s12881-018-0667-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gast C, Pengelly RJ, Lyon M, Bunyan DJ, Seaby EG, Graham N, Venkat-Raman G, Ennis S. Collagen (COL4A) mutations are the most frequent mutations underlying adult focal segmental glomerulosclerosis. Nephrol Dial Transplant. 2016;31:961–970. doi: 10.1093/ndt/gfv325. [DOI] [PubMed] [Google Scholar]
- 95.Lee HK, Han KH, Jung YH, Kang HG, Moon KC, Ha IS, Choi Y, Cheong HI. Variable renal phenotype in a family with an INF2 mutation. Pediatr Nephrol. 2011;26:73–76. doi: 10.1007/s00467-010-1644-5. [DOI] [PubMed] [Google Scholar]
- 96.Lipska BS, Iatropoulos P, Maranta R, Caridi G, Ozaltin F, Anarat A, Balat A, Gellermann J, Trautmann A, Erdogan O, Saeed B, Emre S, Bogdanovic R, Azocar M, Balasz-Chmielewska I, Benetti E, Caliskan S, Mir S, Melk A, Ertan P, Baskin E, Jardim H, Davitaia T, Wasilewska A, Drozdz D, Szczepanska M, Jankauskiene A, Higuita LM, Ardissino G, Ozkaya O, Kuzma-Mroczkowska E, Soylemezoglu O, Ranchin B, Medynska A, Tkaczyk M, Peco-Antic A, Akil I, Jarmolinski T, Firszt-Adamczyk A, Dusek J, Simonetti GD, Gok F, Gheissari A, Emma F, Krmar RT, Fischbach M, Printza N, Simkova E, Mele C, Ghiggeri GM, Schaefer F. Genetic screening in adolescents with steroid-resistant nephrotic syndrome. Kidney Int. 2013;84:206–213. doi: 10.1038/ki.2013.93. [DOI] [PubMed] [Google Scholar]
- 97.Bullich G, Trujillano D, Santin S, Ossowski S, Mendizabal S, Fraga G, Madrid A, Ariceta G, Ballarin J, Torra R, Estivill X, Ars E. Targeted next-generation sequencing in steroid-resistant nephrotic syndrome: mutations in multiple glomerular genes may influence disease severity. Eur J Human Genet. 2015;23:1192–1199. doi: 10.1038/ejhg.2014.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yao T, Udwan K, John R, Rana A, Haghighi A, Xu L, Hack S, Reich HN, Hladunewich MA, Cattran DC, Paterson AD, Pei Y, Barua M. Integration of genetic testing and pathology for the diagnosis of adults with FSGS. Clin J Am Soc Nephrol. 2019;14:213. doi: 10.2215/cjn.08750718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wang M, Chun J, Genovese G, Knob AU, Benjamin A, Wilkins MS, Friedman DJ, Appel GB, Lifton RP, Mane S, Pollak MR. Contributions of rare gene variants to familial and sporadic FSGS. J Am Soc Nephrol. 2019;30:1625. doi: 10.1681/asn.2019020152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Fu J, Ma M, Pang M, Yang L, Li G, Song J, Zhang J. Analysis of a pedigree with autosomal dominant intermediate Charcot–Marie–Tooth disease type E and nephropathy. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2019;36:918–921. doi: 10.3760/cma.j.issn.1003-9406.2019.09.016. [DOI] [PubMed] [Google Scholar]
- 101.Nagano C, Yamamura T, Horinouchi T, Aoto Y, Ishiko S, Sakakibara N, Shima Y, Nakanishi K, Nagase H, Iijima K, Nozu K. Comprehensive genetic diagnosis of Japanese patients with severe proteinuria. Sci Rep. 2020;10:270. doi: 10.1038/s41598-019-57149-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Larsen CP, Durfee T, Wilson JD, Beggs ML. A custom targeted next-generation sequencing gene panel for the diagnosis of genetic nephropathies. Am J Kidney Dis. 2016;67:992–993. doi: 10.1053/j.ajkd.2015.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Stenson PD, Mort M, Ball EV, Evans K, Hayden M, Heywood S, Hussain M, Phillips AD, Cooper DN. The human gene mutation database: towards a comprehensive repository of inherited mutation data for medical research, genetic diagnosis and next-generation sequencing studies. Hum Genet. 2017;136:665–677. doi: 10.1007/s00439-017-1779-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Snoek R, Nguyen TQ, van der Zwaag B, van Zuilen AD, Kruis HME, van Gils-Verrij LA, Goldschmeding R, Knoers NVAM, Rookmaaker MB, van Eerde AM. Importance of genetic diagnostics in adult-onset focal segmental glomerulosclerosis. Nephron. 2019;142:351–358. doi: 10.1159/000499937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Nozu K, Nakanishi K, Abe Y, Udagawa T, Okada S, Okamoto T, Kaito H, Kanemoto K, Kobayashi A, Tanaka E, Tanaka K, Hama T, Fujimaru R, Miwa S, Yamamura T, Yamamura N, Horinouchi T, Minamikawa S, Nagata M, Iijima K. A review of clinical characteristics and genetic backgrounds in Alport syndrome. Clin Exp Nephrol. 2019;23:158–168. doi: 10.1007/s10157-018-1629-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kashtan C. Alport syndrome: facts and opinions. F1000Research. 2017;6:50. doi: 10.12688/f1000research.9636.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Barisoni L, Schnaper HW, Kopp JB. A proposed taxonomy for the podocytopathies: a reassessment of the primary nephrotic diseases. Clin J Am Soc Nephrol. 2007;2:529–542. doi: 10.2215/cjn.04121206. [DOI] [PubMed] [Google Scholar]
- 108.Kavanagh D, Goodship TH, Richards A. Atypical hemolytic uremic syndrome. Sem Nephrol. 2013;33:508–530. doi: 10.1016/j.semnephrol.2013.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Petukh M, Kucukkal TG, Alexov E. On human disease-causing amino acid variants: statistical study of sequence and structural patterns. Hum Mutat. 2015;36:524–534. doi: 10.1002/humu.22770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Peifer M, Berg S, Reynolds AB. A repeating amino acid motif shared by proteins with diverse cellular roles. Cell. 1994;76:789–791. doi: 10.1016/0092-8674(94)90353-0. [DOI] [PubMed] [Google Scholar]
- 111.Tewari R, Bailes E, Bunting KA, Coates JC. Armadillo-repeat protein functions: questions for little creatures. Trends Cell Biol. 2010;20:470–481. doi: 10.1016/j.tcb.2010.05.003. [DOI] [PubMed] [Google Scholar]
- 112.Otomo T, Otomo C, Tomchick DR, Machius M, Rosen MK. Structural basis of Rho GTPase-mediated activation of the formin mDia1. Mol Cell. 2005;18:273–281. doi: 10.1016/j.molcel.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 113.Lammers M, Rose R, Scrima A, Wittinghofer A. The regulation of mDia1 by autoinhibition and its release by Rho-GTP. EMBO J. 2005;24:4176–4187. doi: 10.1038/sj.emboj.7600879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Rose R, Weyand M, Lammers M, Ishizaki T, Ahmadian MR, Wittinghofer A. Structural and mechanistic insights into the interaction between Rho and mammalian Dia. Nature. 2005;435:513–518. doi: 10.1038/nature03604. [DOI] [PubMed] [Google Scholar]
- 115.Kelley LA, Sternberg MJE. Protein structure prediction on the Web: a case study using the Phyre server. Nat Prot. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 116.Gul IS, Hulpiau P, Saeys Y, van Roy F. Metazoan evolution of the armadillo repeat superfamily. Cell Mol Life Sci. 2017;74:525–541. doi: 10.1007/s00018-016-2319-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Sim N-L, Kumar P, Hu J, Henikoff S, Schneider G, Ng PC. SIFT web server: predicting effects of amino acid substitutions on proteins. Nucl Acids Res. 2012;40:W452–W457. doi: 10.1093/nar/gks539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Choi Y, Chan AP. PROVEAN web server: a tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics. 2015;31:2745–2747. doi: 10.1093/bioinformatics/btv195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kellogg EH, Leaver-Fay A, Baker D. Role of conformational sampling in computing mutation-induced changes in protein structure and stability. Proteins. 2011;79:830–838. doi: 10.1002/prot.22921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Buscher AK, Beck BB, Melk A, Hoefele J, Kranz B, Bamborschke D, Baig S, Lange-Sperandio B, Jungraithmayr T, Weber LT, Kemper MJ, Tonshoff B, Hoyer PF, Konrad M, Weber S. Rapid response to cyclosporin A and favorable renal outcome in nongenetic versus genetic steroid-resistant nephrotic syndrome. Clin J Am Soc Nephrol. 2016;11:245–253. doi: 10.2215/CJN.07370715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.McCarthy HJ, Bierzynska A, Wherlock M, Ognjanovic M, Kerecuk L, Hegde S, Feather S, Gilbert RD, Krischock L, Jones C, Sinha MD, Webb NJ, Christian M, Williams MM, Marks S, Koziell A, Welsh GI, Saleem MA, Group RUSS. Simultaneous sequencing of 24 genes associated with steroid-resistant nephrotic syndrome. Clin J Am Soc Nephrol. 2013;8:637–648. doi: 10.2215/CJN.07200712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Pehlivan D, Beck CR, Okamoto Y, Harel T, Akdemir ZHC, Jhangiani SN, Withers MA, Goksungur MT, Carvalho CMB, Czesnik D, Gonzaga-Jauregui C, Wiszniewski W, Muzny DM, Gibbs RA, Rautenstrauss B, Sereda MW, Lupski JR. The role of combined SNV and CNV burden in patients with distal symmetric polyneuropathy. Genet Med. 2016;18:443–451. doi: 10.1038/gim.2015.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Sun H, Schlondorff JS, Brown EJ, Higgs HN, Pollak MR. Rho activation of mDia formins is modulated by an interaction with inverted formin 2 (INF2) Proc Natl Acad Sci USA. 2011;108:2933–2938. doi: 10.1073/pnas.1017010108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Rollason R, Wherlock M, Heath JA, Heesom KJ, Saleem MA, Welsh GI. Disease causing mutations in inverted formin 2 regulate its binding to G-actin, F-actin capping protein (CapZ alpha-1) and profilin 2. Biosci Rep. 2016;36:e00302–e00302. doi: 10.1042/bsr20150252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Sun H, Schlondorff J, Higgs HN, Pollak MR. Inverted formin 2 regulates actin dynamics by antagonizing Rho/diaphanous-related formin signaling. J Am Soc Nephrol. 2013;24:917–929. doi: 10.1681/asn.2012080834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Sun H, Al-Romaih K, MacRae CA, Pollak MR. Human kidney disease-causing INF2 mutations perturb Rho/Dia signaling in the glomerulus. EBioMedicine. 2014;1:107–115. doi: 10.1016/j.ebiom.2014.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Zhu L, Jiang R, Aoudjit L, Jones N, Takano T. Activation of RhoA in podocytes induces focal segmental glomerulosclerosis. J Am Soc Nephrol. 2011;22:1621–1630. doi: 10.1681/asn.2010111146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Subramanian B, Sun H, Yan P, Charoonratana VT, Higgs HN, Wang F, Lai K-MV, Valenzuela DM, Brown EJ, Schlöndorff JS, Pollak MR. Mice with mutant Inf2 show impaired podocyte and slit diaphragm integrity in response to protamine-induced kidney injury. Kidney Int. 2016;90:363–372. doi: 10.1016/j.kint.2016.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Subramanian B, Chun J, Perez-Gill C, Yan P, Stillman IE, Higgs HN, Alper SL, Schlöndorff JS, Pollak MR. FSGS-causing INF2 mutation impairs cleaved INF2 N-fragment functions in podocytes. J Am Soc Nephrol. 2020;31:374–391. doi: 10.1681/asn.2019050443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Berridge MJ, Bootman MD, Roderick HL. Calcium signalling: dynamics, homeostasis and remodelling. Nat Rev Mol Cell Biol. 2003;4:517–529. doi: 10.1038/nrm1155. [DOI] [PubMed] [Google Scholar]
- 132.Carafoli E, Krebs J. Why calcium? How calcium became the best communicator. J Biol Chem. 2016;291:20849–20857. doi: 10.1074/jbc.R116.735894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Berridge MJ. Unlocking the secrets of cell signaling. Annu Rev Physiol. 2005;67:1–21. doi: 10.1146/annurev.physiol.67.040103.152647. [DOI] [PubMed] [Google Scholar]
- 134.Berridge MJ. Calcium signalling remodelling and disease. Biochem Soc Trans. 2012;40:297–309. doi: 10.1042/BST20110766. [DOI] [PubMed] [Google Scholar]
- 135.Clapham DE. Calcium signaling. Cell. 2007;131:1047–1058. doi: 10.1016/j.cell.2007.11.028. [DOI] [PubMed] [Google Scholar]
- 136.Marazuela M, Acevedo A, Adrados M, Garcia-Lopez MA, Alonso MA. Expression of MAL, an integral protein component of the machinery for raft-mediated pical transport, in human epithelia. J Histochem Cytochem. 2003;51:665–673. doi: 10.1177/002215540305100512. [DOI] [PubMed] [Google Scholar]
- 137.Marazuela M, Acevedo A, Garcia-Lopez MA, Adrados M, de Marco MC, Alonso MA. Expression of MAL2, an integral protein component of the machinery for basolateral-to-apical transcytosis, in human epithelia. J Histochem Cytochem. 2004;52:243–252. doi: 10.1177/002215540405200212. [DOI] [PubMed] [Google Scholar]
- 138.Shaye DD, Greenwald I. The disease-associated formin INF2/EXC-6 organizes lumen and cell outgrowth during tubulogenesis by regulating F-actin and microtubule cytoskeletons. Dev Cell. 2015;32:743–755. doi: 10.1016/j.devcel.2015.01.009. [DOI] [PubMed] [Google Scholar]
- 139.Shaye DD, Greenwald I. A network of conserved formins, regulated by the guanine exchange factor EXC-5 and the GTPase CDC-42, modulates tubologenesis in vivo. Development. 2016;143:4173–4181. doi: 10.1242/dev.141861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Weavers H, Prieto-Sanchez S, Grawe F, Garcia-Lopez A, Artero R, Wilsch-Bräuninger M, Ruiz-Gomez M, Skaer H, Denholm B. The insect nephrocyte is a podocyte-like cell with a filtration slit diaphragm. Nature. 2009;457:322–326. doi: 10.1038/nature07526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Carrasco-Rando M, Prieto-Sanchez S, Culi J, Tutor AS, Ruiz-Gomez M. A specific isoform of Pyd/ZO-1 mediates junctional remodeling and formation of slit diaphragms. J Cell Biol. 2019;218:2294–2308. doi: 10.1083/jcb.201810171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera- visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 143.Shapovalov MV, Dunbrack RL., Jr A smoothed backbone-dependent rotamer library for proteins derived from adaptive kernel density estimates and regressions. Structure. 2011;19:844–858. doi: 10.1016/j.str.2011.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.