Abstract
During ciliogenesis, the mother centriole transforms into a basal body competent to nucleate a cilium. The mother centriole and basal body possess sub-distal appendages (SDAs) and basal feet (BF), respectively. SDAs and BF are thought to be equivalent structures. In contrast to SDA assembly, little is known about the players involved in BF assembly and its assembly order. Furthermore, the contribution of BF to ciliogenesis is not understood. Here, we found that SDAs are distinguishable from BF and that the protein NPHP5 is a novel SDA and BF component. Remarkably, NPHP5 is specifically required for BF assembly in cells able to form basal bodies but is dispensable for SDA assembly. Determination of the hierarchical assembly reveals that NPHP5 cooperates with a subset of SDA/BF proteins to organize BF. The assembly pathway of BF is similar but not identical to that of SDA. Loss of NPHP5 or a BF protein simultaneously inhibits BF assembly and primary ciliogenesis, and these phenotypes could be rescued by manipulating the expression of certain components in the BF assembly pathway. These findings define a novel role for NPHP5 in specifically regulating BF assembly, a process which is tightly coupled to primary ciliogenesis.
Electronic supplementary material
The online version of this article (10.1007/s00018-019-03181-7) contains supplementary material, which is available to authorized users.
Keywords: Centrosome-centriole-distal, Appendage-subdistal, Appendage-microtubule-cell, Biology-superresolution
Introduction
The centrosome participates in the organization of the microtubule network in many eukaryotic cells and coordinates a number of microtubule-related processes such as cell division, cell polarity, cell motility, and cell signalling [1]. As a dynamic organelle, its structure and function are subjected to tight spatial and temporal regulation [2, 3]. A cell in G1 phase contains a single centrosome comprised of two centrioles, the mother and daughter centrioles, which are surrounded by the pericentriolar material (PCM) from which microtubules emanate and elongate. The mother centriole is structurally distinct from the daughter centriole in that the former possesses sub-distal appendages (SDAs) and distal appendages (DAs) [2, 4]. After centrosome duplication in S phase and maturation in G2 phase, two fully functional centrosomes with increased capacity to nucleate microtubules are formed. At the onset of mitosis, the two centrosomes separate, migrating to opposite poles and establishing the mitotic spindle. When a cell exits the cell cycle, the mother centriole transforms into the basal body, a structure essential for the nucleation of a cilium [2, 4]. At the molecular level, the mother centriole-to-basal body transformation is thought to entail targeting of TTBK2 to, followed by loss of CP110 from and recruitment of intraflagellar transport (IFT) proteins to, the mother centriole [5].
The basal body is accompanied by basal feet (BF) and transition fibers (TFs) [2, 4]. TFs and DAs are analogous structures that exist in a mutually exclusive manner. Nine DAs are present at the distal end of the mother centriole [6, 7], while nine TFs occupy a similar location at the basal body to dock vesicles/membranes during ciliogenesis [8, 9]. Likewise, BF and SDAs are believed to be equivalent but mutually exclusive. BF and SDAs project laterally from the sides of the basal body and mother centriole, respectively, near the distal end [7]. The number of SDAs and BF appears to vary depending on the cell type. Based on electron microscopy (EM) studies, SDA number can range from zero to more than nine. Furthermore, basal bodies that template motile cilia reportedly possess one basal foot which is significantly larger than an individual SDA [8, 9], whereas basal bodies that template primary cilia are alleged to contain one to several BF [10–12]. Thus, numerical and structural differences likely exist between BF and SDAs.
On the other hand, there seems to be considerable overlap between BF and SDAs in terms of function and molecular composition. Both BF and SDAs are able to nucleate and anchor microtubules [8, 13, 14]. The molecular composition of SDA is beginning to emerge and a handful of SDA proteins have been identified. ODF2, ninein, Cep170, centriolin, ε-tubulin, CCDC120, and CCDC68 are considered core SDA components based on immuno-EM and/or super-resolution microscopy studies [15–20], and a loss of any one of them compromises SDA assembly and/or function. Ablation of ODF2 also disrupts BF assembly [15], suggesting that this protein likely localizes to both SDAs and BF. Other proteins such as Kif3a, p150Glued, Sec15, CC2D2A, and Cep128 are found to be present at the sub-distal region of the mother centriole and play a critical role in SDA assembly and/or function [10, 21–23]. Kif3a and CC2D2A, in particular, are also involved in BF assembly [10, 21]. Another protein TCHP is enriched at the sub-distal to the medial region of centrioles, including the mother centriole, where it regulates SDA assembly in a positive manner [24]. Paradoxically, TCHP disappears from the basal body of quiescent cells and might play an inhibitory role in BF formation [25]. Identification of novel proteins that specifically localize to and/or participate in the assembly of SDAs or BF would greatly enhance our understanding of the similarities and differences between the two structures.
Proper assembly of SDAs entails the recruitment of various SDA components in a hierarchical manner [10, 22–24, 26]. While the assembly order of SDA components is not fully understood, it has been reported that (1) Kif3a is required for the localization of p150Glued and ninein to SDAs [10]; (2) ninein recruits Cep170 [26]; (3) ODF2 recruits TCHP which in turn recruits ninein to build SDAs [24]; and (4) ODF2 is required for the localization of Sec15 to SDAs [23]. By comparison, little is known about the players involved in BF assembly and their assembly hierarchy.
In contrast to TFs, the requirement of BF for ciliogenesis is controversial. The Tsukita group showed that basal bodies lacking BF can still template cilia, indicating that BF are not required for ciliogenesis [27]. Likewise, using CRISPR-mediated gene targeting to inactivate SDA/BF components, Mazo et al. found that SDAs/BF are not needed for cilia assembly [22]. Moreover, depletion of CCDC120 has no impact on ciliogenesis [18]. In contrast, several other studies showed that ablation of ODF2, ninein, Kif3a, or CC2D2A inhibits ciliogenesis [10, 15, 21, 26, 28–31], although it is not clear whether this is attributed to defects in cell cycle exit, mother centriole-to-basal body conversion, and/or BF formation. While the aforementioned proteins positively regulate ciliogenesis, TCHP is unique among SDA proteins in that it is a negative regulator of ciliogenesis, and a loss of this protein in cycling cells leads to aberrant formation of cilia [25].
Here, using three-dimensional structured illumination microscopy (3D-SIM), we found that SDAs are distinguishable from BF in normal diploid retinal pigmented epithelial cells (RPE-1), a well-established model for primary cilia assembly. We then identified the protein NPHP5 as a novel SDAs and BF component. NPHP5 specifically regulated BF assembly by coordinating with a subset of SDA/BF proteins in cell lines able to form basal bodies. In striking contrast, NPHP5 did not organize SDAs of mother centrioles. We determined the assembly pathway of BF and found it to be similar but not identical to the SDA assembly pathway, consistent with the notion that BF and SDAs are distinct entities. Finally, we observed a positive correlation between BF assembly and primary ciliogenesis, and demonstrated a tight coupling between these two processes.
Materials and methods
Cell culture and plasmids
Human RPE-1, ARPE-19, HK-2, HeLa, U2OS, PC-3, MCF-7, DU-145 and SAOS-2 cells were grown in DMEM (Wisent Inc, 319-005-CL) and supplemented with 10% FBS (Wisent Inc, 080150) at 37 °C in a humidified 5% CO2 atmosphere. The following proteins were expressed from plasmids in mammalian cells: pEGFP-C1, pEGFP-C1-NPHP5, pGL-FLKif3a (a gift from L. Wordeman; Addgene plasmid #13742), and pShuttle-CMV-GFP-ODF2 isoform 11 (a gift from K. Lee). ODF2 isoform 11 is also known as cenexin1.
Antibodies
Antibodies used in this study included rabbit anti-NPHP5 (IF: 1:100 unless otherwise indicated, Santa Cruz Biotechnology, sc-134804), mouse anti-NPHP5 (IF: 100, Abcam, ab69927), rabbit anti-Kif3a (IF and WB: 1:100, Proteintech, 13930-1-AP), mouse anti-ninein (IF and WB: 1:250, Santa Cruz Biotechnology, sc-376420), mouse anti-Cep170 (IF and WB: 1:100, Invitrogen, 41-3200), mouse anti-ODF2 (IF: 1:100, Santa Cruz Biotechnology, sc-393881), mouse anti-TCHP (IF and WB: 1:100, Santa Cruz Biotechnology, sc-515025) rabbit anti-TCHP (IF: 100, Proteintech, 25931-1-AP), rabbit anti-CC2D2A (IF: 1:100, Sigma-Aldrich, HPA044124), rabbit anti-Cep83 (IF: 1:100, Proteintech, 26013-1-AP), rabbit anti-Cep164 (IF: 1:500, a gift from E. Nigg), goat anti-Cep164 (1:100, Santa Cruz Biotechnology, sc-240226), mouse anti-α-tubulin (IF and WB: 1:1000, Sigma-Aldrich, T5168), rabbit anti-γ-tubulin (IF: 1:1000, Sigma-Aldrich, T3559), goat anti-γ-tubulin (1:100, Santa Cruz Biotechnology, sc-7396), rabbit anti-CP110 (IF: 1:500, Bethyl Laboratories, A301-344A), rabbit anti-CEP290 (IF: 1:500, Bethyl Laboratories, A301-659A), mouse anti-centrin (IF: 1:1000, Millipore, 04-1624), rabbit anti-POC5 (IF: 1:50, Bethyl Laboratories, A303-341A), rabbit anti-Sec15 (IF: 1:100, Sigma-Aldrich, SAB1104731), rabbit anti-GFP (IF: 1:500, Sigma-Aldrich, G1544), rabbit anti-IFT88 (IF: 1:100, Proteintech, 13967-1-AP), mouse anti-glutamylated tubulin (GT335) (IF: 1:1000, Adipogen life science, AG-20B-0020), rabbit anti-detyrosinated tubulin (DT) (IF: 1:1000, Millipore, AB3201), mouse anti-Ki67 (IF: 1:1000, Invitrogen, 7B11), and rabbit anti-Ki67 (IF: 1000, Cell Signalling, 12202).
Transmission EM
For ultra-structural characterization, cells were fixed with 2% glutaraldehyde (Electron Microscopy Sciences) and dehydrated through a series of graded ethanol dilutions. Samples were embedded in epoxy resin (Electron Microscopy Sciences). Ultrathin sections cut with a diamond knife (Diatome) (Electron Microscopy Sciences) on a Leica Microsystems UCT ultramicrotome were placed on formvar-coated nickel grids (Electron Microscopy Sciences) and stained with uranyl 2% (w/v) acetate and lead citrate (Electron Microscopy Sciences). Samples were observed with a FEI Tecnai 12 TEM (FEI) at an accelerating voltage of 120 kV and imaged with an AMT XR80C CCD camera (Advanced Microscopy Techniques Corp).
Immunoblotting and immunofluorescence
Immunoblotting and immunofluorescence were performed as described previously [32]. Cells were lysed in a lysis buffer (50 mM HEPES/pH 7.4, 250 mM NaCl, 5 mM EDTA/pH 8, 0.1% NP-40, 1 mM DTT, 0.5 mM PMSF, 2 μg/ml leupeptin, 2 μg aprotinin, 10 mM NaF, 50 mM β-glycerophosphate and 10% glycerol) at 4 °C for 30 min. Extracted proteins were recovered in the supernatant after centrifugation at 16,000g for 5 min. For immunoblotting, 100 μg of extract was used and proteins were analyzed by SDS-PAGE and immunoblotted with primary antibodies and horseradish peroxidase-conjugated secondary antibodies (Rockland Inc, 610-703-002 and 611-7302). For immunofluorescence staining, cells were fixed with cold methanol or 4% paraformaldehyde and permeabilized with 1% Triton X-100/PBS. Slides were blocked with 3% BSA in 0.1% Triton X-100/PBS and subsequently incubated with primary antibodies and secondary antibodies. Secondary antibodies used were Cy3- (Jackson Immunolabs, 711-165-151 and 715-165-152) or Alexa488- (Thermo Fisher Scientific, A11008, A11055, and A11001) conjugated donkey anti-mouse, anti-goat or anti-rabbit IgG). DAPI (Molecular Probes, D3571) stained for DNA and slides were mounted, observed, and photographed using a Leitz DMRB (Leica) microscope (100×, NA 1.3) equipped with a Retiga EXi cooled camera. Super-resolution 3D imaging was performed using an ELYRA PS.1 microscope (Carl Zeiss Microscopy) equipped with an alpha plan-apochromat 100×/1.46 oil DIC M27 immersion objective and 488 nm or 561 nm lasers. Image stacks of 2 µm in height with a z-distance of 0.116 µm were acquired with an Andor iXon 885 EMCCD camera. Each Z section was recorded with 5 grating rotation and 5 phase changes. Inner ring diameter was measured using Imaris 8.2 (Bitplane).
Quantitation of fluorescence intensity
A region of interest was drawn around a fluorescent spot in the vicinity of the centrosome. The area of the region of interest was used to determine the fluorescence intensity using Volocity6 (PerkinElmer). Image conditions were identical in all cases and none were saturated as confirmed by the pixel intensity range.
Microtubule re-growth assay
Cells were treated with 10 µM nocodazole (Sigma-Aldrich, M1404) for 1 h at 4 °C. After washing the cells several times with cold medium, they were placed in a pre-warmed medium at 37 °C. Cells were fixed at various time points (0, 1, 5, 20 and 60 min) after 37 °C and processed for immunofluorescence.
In situ PLA
Duolink in situ PLA kit (Sigma, DUO92101-1KT) was used per manufacturer’s instructions. In brief, cells grown on a glass coverslip were fixed, permeabilized, and incubated with blocking reagent for 1 h at room temperature. Thereafter, cells were incubated with primary antibody for 1 h at room temperature, washed with Duolink Wash Buffer A twice, incubated with Plus and Minus PLA probes in a preheated humidity chamber for 1 h at 37 °C, washed with Duolink Wash Buffer A twice, incubated with the ligation solution for 30 min at 37 °C, washed with Duolink Wash Buffer A twice, incubated with Duolink amplification solution for 100 min at 37 °C, washed with Duolink Wash Buffer B twice, incubated anti-γ-tubulin-FITC for 45 min, and washed with Duolink Wash Buffer B once. Slides were mounted with the Duolink mounting medium containing DAPI.
RNA interference and expression of recombinant proteins
Synthetic siRNA oligonucleotides were purchased from Dharmacon and the sequences were:
NS (non-specific): 5′-AATTCTCCGAACGTGTCACGT-3′; NPHP5 oligo2: 5′-ACCCAAGGATCTTATCTAT-3′ (used for knocking down NPHP5); NPHP5 oligo5: 5′-CCCTAAGAATTGACACAAA-3′ (targeted against 3′-UTR and used for knocking down NPHP5 in rescue experiments only); Cep290: 5′-AAATTAAGATGCTCACCGATT-3′; Kif3a: 5′-CAGATTGTCCTATGTTGCGCTGT-3′ (targeted against 3′-UTR); ODF2 oligo2: 5′-GGTCAAGATGCAAAAAGGT-3′ (used for knocking down ODF2); ODF2 oligo3′UTR: 5′-GGTCTTGTCCTTAGCTACTAG-3′ (targeted against 3′-UTR and used for knocking down ODF2 in rescue experiments only); TCHP: 5′-CAGGGCATTGTTCCATGGTTA-3′; Ninein: 5′-GCGGAGCTCTCTGAAGTTAAA-3′; and Cep170: 5′-GAAGGAATCCTCCAAGTCA-3′. siRNA transfection was performed using siIMPORTER (Millipore, 64-101) according to per manufacturer’s instructions. For RNA interference, cells were transfected with siRNA and harvested 72 h after transfection. For experiments involving RNA interference and recombinant protein expression, cells were transfected with siRNA at 0 h, transfected with an expression vector at 24 h, and harvested at 72 h time point.
Induction of primary cilia
Cells were induced to form primary cilia by serum withdrawal for at least 48 h. Under this condition, the majority of RPE-1 and ARPE-19 cells, along with a significant percentage of HK-2 and HeLa cells, entered quiescence and formed primary cilia. In contrast, very few U2OS, PC-3, and MCF-7 cells entered quiescence and formed primary cilia. A certain percentage of DU-145 and SAOS-2 cells entered quiescence yet they did not form primary cilia. Primary cilia were detected by staining cells with antibodies against IFT88, glutamylated tubulin (GT335), or detyrosinated tubulin (DT).
Results
SDA and BF are distinguishable
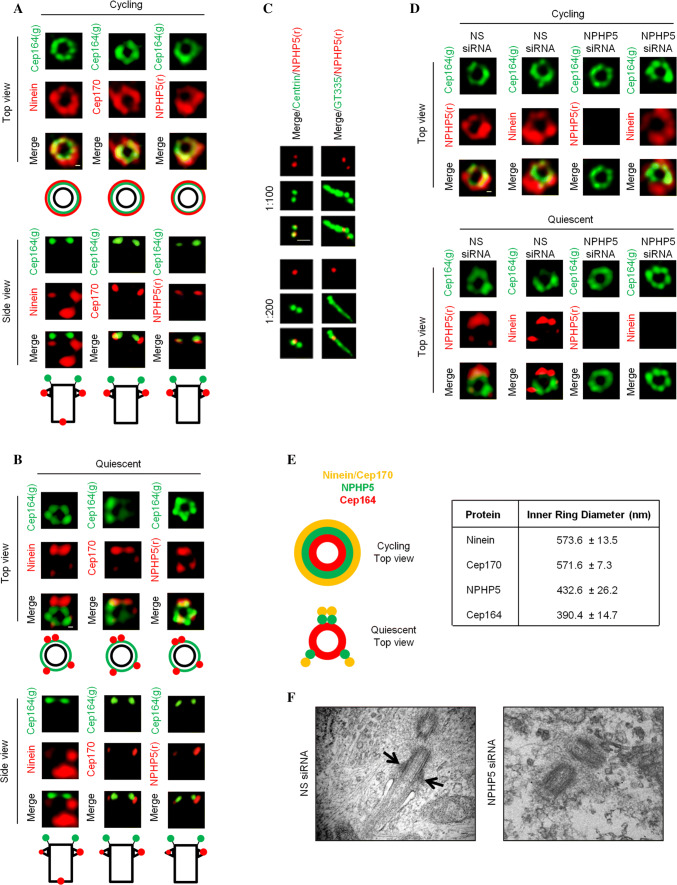
To explore the notion that SDAs of mother centrioles and BF of basal bodies might not be identical at the structural level, we used 3D-SIM to examine the localization pattern of two SDA/BF proteins, ninein and Cep170, in cycling versus quiescent RPE-1 cells. Cells positive and negative for Ki67 were deemed to be cycling and quiescent, respectively. Cells with one CP110 dot were deemed to possess basal bodies, whereas those with two/four CP110 dots possessed mother centrioles. When grown in the presence of serum, the majority of RPE-1 cells were cycling (73% of cells were Ki67 positive) and contained mother centrioles (78% of cells had two/four centriolar CP110 dots) not competent to template cilia (81% of cells lacked cilia) (Fig. S1). Under this condition, antibodies against ninein, Cep170, or a DA/TF protein Cep164 stained a ring-like structure, indicative of nine appendages, when viewed from the top (Fig. 1a, d, e). The ring diameter of Cep164 was substantially smaller than that of ninein or Cep170 (Fig. 1a, d, e), in agreement with published data [33, 34]. In the side view, two ninein or Cep170 dots corresponding to SDAs were located slightly proximal to Cep164 (Fig. 1a). An extra ninein dot representing the proximal end of the mother centriole could also be seen (Fig. 1a). In contrast, most RPE-1 cells grown in serum-free medium were quiescent (78% of cells were Ki67 negative) and possessed basal bodies (77% of cells with one CP110 dot) competent to template cilia (73% of cell had cilia) (Fig. S1). Under this condition, the ring-like structure of Cep164 remained (Fig. 1b, d, e), which is consistent with a previous report [35]. Remarkably, we observed four ninein or Cep170 dots instead of a ring from the top (Fig. 1b, d, e). Two of the four dots were very close to each other. These two dots, along with the remaining two dots, appeared to form three equidistant points on a circle (Fig. 1b, d, e). The side view picture showed one bright and one weak dots of ninein or Cep170 located proximal to Cep164 (Fig. 1b). Another ninein dot corresponding to the proximal end of the mother centriole was also observed (Fig. 1b). Together, these results indicate that SDAs and BF are morphologically different.
Fig. 1.
NPHP5 is a novel component of two distinguishable structures, SDAs and BF. a Cycling or b quiescent RPE-1 cells were stained with the indicated antibodies and images were acquired with 3D-SIM. Black circle and rectangle represent top-view and side-view of a mother centriole a or basal body b. Scale bar, 0.1 µm. c Quiescent RPE-1 cells were stained with the indicated antibodies. Two different dilutions of antibodies against NPHP5, 1:100 and 1:200, were used. Scale bar, 1 µm. d Cycling or quiescent RPE-1 cells transfected with NS (non-specific) or NPHP5 siRNAs were stained with the indicated antibodies and images were acquired with 3D-SIM. Scale bar, 0.1 µm. e (Left) 3D-SIM staining patterns of several proteins in cycling and quiescent RPE-1 cells are presented. (Right) Average inner ring diameter from the top view of cycling RPE-1 cells n = 5. f Quiescent RPE-1 cells transfected with NS or NPHP5 siRNAs were analyzed by transmission electron microscopy. Scale bar, 0.1 µm. Arrows point to BF
NPHP5 is a novel SDA and BF component
We and others previously reported NPHP5 as a centrosomal protein which localizes to the distal region of centrioles [36–38]. In particular, two NPHP5 dots could be seen in quiescent and cycling RPE-1 cells in G1 phase [36, 37]. Upon closer examination, one dot appeared to be brighter than the other, and the dot associated with the cilium-nucleating basal body was always more intense than the dot associated with the daughter centriole (Fig. 1c). Use of limiting amounts of antibody resulted in the disappearance of the weaker dot (Fig. 1c), suggesting that NPHP5 might be enriched at the mother centriole and basal body. We thus examined NPHP5 localization in greater detail using 3D-SIM. When viewed from the top, NPHP5 exhibited a ring-like structure reminiscent of ninein and Cep170 under cycling conditions (Fig. 1a, d, e). The NPHP5 ring had a smaller diameter than the ninein and Cep170 ring (Fig. 1e), suggesting that this protein is located closer to the outer surface of the centriole barrel. In the side view, two dots of NPHP5 were located slightly proximal to Cep164 (Fig. 1a). Under quiescent conditions, the top view picture revealed three NPHP5 dots (Fig. 1b, d, e) forming equidistant points on a circle. One dot was always brighter than the other two dots and might represent two smaller dots that could not be resolved by 3D-SIM. The side view pictures showed one bright and one weak dots of NPHP5 proximal to Cep164 (Fig. 1b). These patterns were similar to those of ninein and Cep170 but distinct from Cep164 (Fig. 1b, e). Thus, our results argue that NPHP5 is preferentially enriched at SDAs of mother centrioles and BF of basal bodies.
NPHP5 is required for BF assembly
Next, we explored whether NPHP5 might contribute to the assembly of SDAs and BF. Although depletion of NPHP5 with siRNA did not affect the ninein ring in cycling RPE-1 cells as revealed by 3D-SIM (Fig. 1d), it led to the disappearance of ninein dots in quiescent cells (Fig. 1d), suggesting that BF formation is compromised. To confirm this finding, we examined centrosomes/cilia in control and NPHP5-depleted quiescent cells by ultrathin section EM. Control cells had primary cilia emanating from BF-containing basal bodies (Fig. 1f). On the contrary, very few cells depleted of NPHP5 possessed cilia or BF (Fig. 1f). When EM images were analyzed, 50% of centrioles scored at random were expected to be basal bodies containing BF and 50% were daughter centrioles lacking BF. We found that 24 out of 51 (47%) control centrioles possessed BF, in contrast to 14 out of 56 (25%) centrioles from NPHP5-depleted cells. Furthermore, unlike Cep164 depletion, depletion of NPHP5 in quiescent cells did not compromise basal body formation since CP110 disappeared from one of the two centrioles (Fig. 2) and IFT88 was properly recruited to the centrosome (Fig. S2). Considering that the knockdown efficiency of NPHP5 was actually better in cycling cells than quiescent cells (Fig. S3), our data suggest that in addition to cilia formation, NPHP5 is specifically required for, and/or plays a prominent role in, BF assembly.
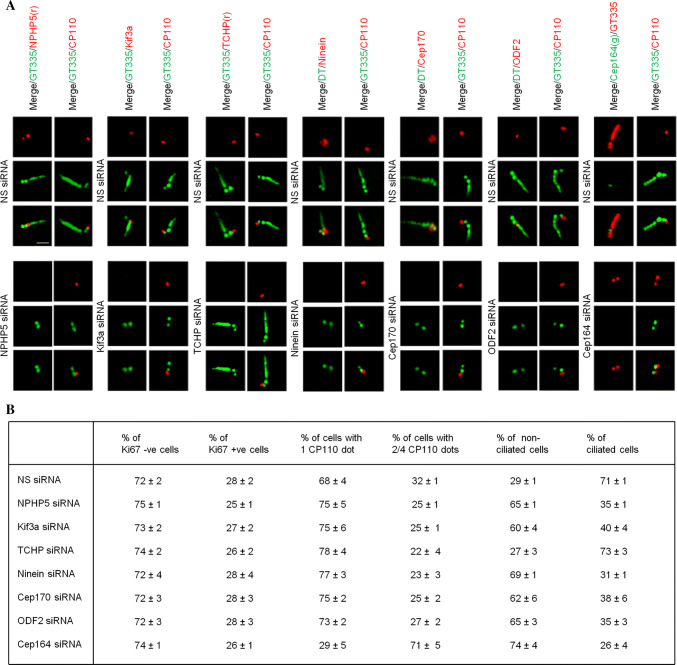
Fig. 2.
Ablation of BF components inhibits ciliogenesis without affecting entry into quiescence or basal body formation. a Quiescent RPE-1 cells transfected with NS (non-specific) or the indicated siRNAs targeting SDA/BF components (NPHP5, Kif3a, TCHP, ninein, Cep170, ODF2) or a DA/TF component (Cep164) were stained with the indicated antibodies. Scale bar, 1 µm. b Quiescent RPE-1 cells transfected with NS or the indicated siRNAs were stained with antibodies against CP110, Ki67 or glutamylated tubulin (GT335). The percentage of Ki67 negative versus positive cells, the percentage of cells with 1 CP110 dot versus 2/4 CP110 dots, and the percentage of non-ciliated versus ciliated cells are presented. At least 100 cells for each condition were scored, and the mean and standard error of three independent experiments are presented
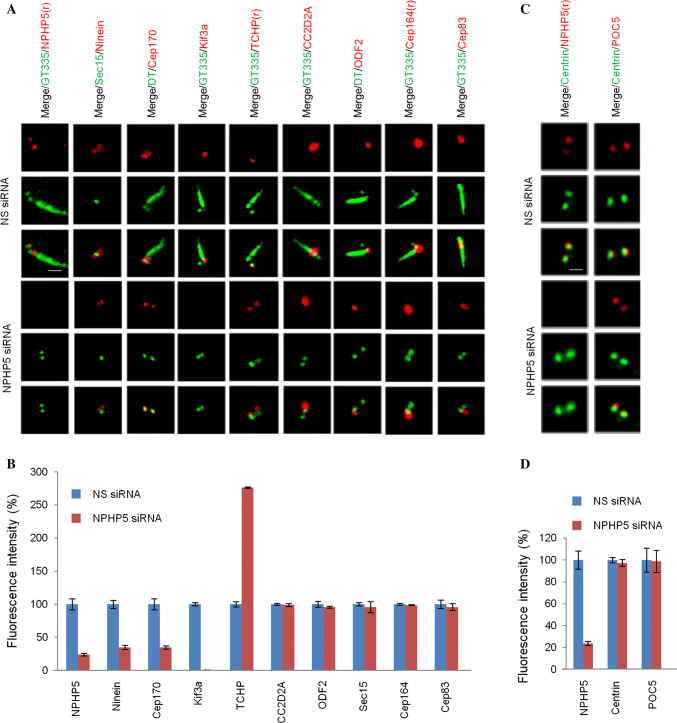
To confirm a critical role of NPHP5 in BF assembly, we studied the effects of depleting this protein on a panel of known SDA/BF markers in quiescent RPE-1 cells using epifluorescence microscopy. Ninein and Cep170 signals appeared as four dots, two correspondings to SDA/BF and the other two corresponding to the proximal end of centrioles (Fig. 3a and [15, 22, 24]). Upon NPHP5 depletion, ninein and Cep170 staining intensities were substantially reduced and two of the four dots were lost (Fig. 3a, b). The two remaining ninein dots did not overlap with Sec15 (Fig. 3a), a SDA/BF marker, suggesting that ninein remains at the proximal end of centrioles but is de-localized from BF. These results are consistent with our earlier 3D-SIM and EM studies (Fig. 1d, f) that BF assembly is compromised in NPHP5-depleted quiescent cells. For Kif3a, one single dot on BF was observed in control cells, and this signal completely disappeared in NPHP5-depleted cells (Fig. 3a, b). Of note, although ablation of NPHP5 affected the localization of ninein, Cep170, and Kif3a, it did not affect their protein levels (Fig. S4A and C). TCHP was present on the daughter centriole only in control cells (Fig. 3a). In cells depleted of NPHP5, TCHP was found on the daughter centriole and basal body (Fig. 3a), and its centrosomal staining (Fig. 3b) and protein level dramatically increased (Fig. S4A and C). We also found that depletion of NPHP5 in quiescent cells has no effect on the localization of three other SDA/BF proteins CC2D2A, ODF2, and Sec15 (Fig. 3a, b). Moreover, NPHP5 loss did not affect the localization of proteins residing at the TFs (Cep164 and Cep83; Fig. 3a, b) or within the distal lumen of centrioles (centrin and POC5; Fig. 3c, d). Thus, NPHP5 organizes BF by modulating the abundance of TCHP and the localization of a subset of SDA/BF proteins including Kif3a, ninein, Cep170, and TCHP.
Fig. 3.
NPHP5 recruits Kif3a, ninein, and Cep170 to BF while preventing the recruitment of TCHP in quiescent RPE-1 cells. a, c Quiescent RPE-1 cells transfected with NS (non-specific) or NPHP5 siRNAs were stained with the indicated antibodies. Scale bar, 1 µm. b, d Fluorescence intensities of various proteins at the centrosome were quantitated and set to 100% in NS siRNA-transfected cells. For quantitation, at least 20 cells for each condition were analyzed, and the mean and standard error of three independent experiments are presented
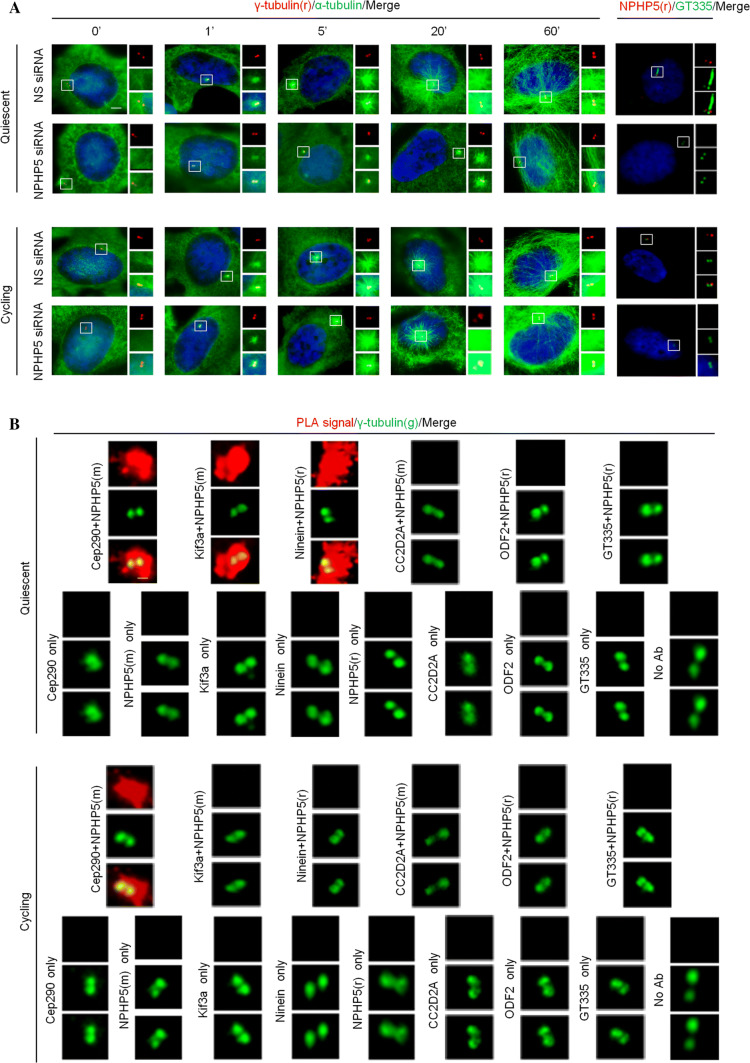
To validate the requirement of NPHP5 for BF as opposed to SDA assembly, four additional experiments were conducted. First, we confirmed that depletion of NPHP5 does not impinge on the localization of SDA proteins (ninein, Cep170, Kif3a, TCHP, CC2D2A, ODF2, and Sec15; Fig. S5A-B), DA proteins (Cep164 and Cep83; Fig. S5A-B), and distal centriolar lumen proteins (centrin and POC5; Fig. S5C-D), or the protein level of selected SDA proteins (Kif3a, ninein, Cep170, TCHP; Fig. S4A-B) in cycling RPE-1 cells. Second, as impaired SDA/BF assembly could affect microtubule nucleation and/or anchoring [13], we conducted a microtubule re-growth assay to assess SDA/BF function in control versus NPHP5-depleted RPE-1 cells. In control cycling and quiescent cells, an aster of microtubules radiating out from the centrosome, indicative of microtubule nucleation, was seen as early as 1′ after removal of the microtubule-depolymerizing drug nocodazole (Fig. 4a). 60′ after nocodazole washout, a large network of long microtubules centered at/near the centrosome, indicative of microtubule anchoring, was observed (Fig. 4a). Upon ablation of NPHP5, we found that the aster size is substantially smaller at early time points and microtubules are less focused around the centrosome at the 60′ time point in quiescent cells, but not in cycling cells (Fig. 4a). These results indicate that BF rather than SDA function is specifically impaired. Third, Cep290 is known to anchor NPHP5 to the centrosome [36, 38], and we further showed that a loss of the former disrupts the centrosomal localization of the latter in both cycling and quiescent RPE-1 cells (Fig. S6). Ablation of Cep290 reduced the number of ninein dots from four to two and the staining intensity of ninein in quiesent cells only (Fig. S6), phenotypes reminiscent of NPHP5 loss. Fourth, we reasoned that NPHP5 might cooperate and interact with SDA/BF proteins to assemble BF. In situ proximity ligation assays (PLA) were performed to assess the interaction between NPHP5 and Kif3a, ninein, CC2D2A, or ODF2 in cycling versus quiescent RPE-1 cells. Robust NPHP5:Kif3a and NPHP5:ninein PLA signals were detected in quiescent cells, but not in cycling cells (Fig. 4b), suggesting that NPHP5 binds and/or is close proximity to Kif3a and ninein when BF is assembled. In contrast, no NPHP5:CC2D2A or NPHP5:ODF2 signal was detected, while a strong NPHP5:Cep290 PLA signal observed under both cycling and quiescent conditions (Fig. 4b) was consistent with our previous results [39]. Taken together, these data further strengthen the role of NPHP5 in BF assembly.
Fig. 4.
NPHP5 is required for microtubule nucleation/anchoring and interacts with Kif3a and ninein at the centrosome in quiescent RPE-1 cells. a Quiescent or cycling RPE-1 cells transfected with NS (non-specific) or NPHP5 siRNAs and subjected to a microtubule re-growth assay were stained with the indicated antibodies and with DAPI (blue). Depletion of NPHP5 was monitored by staining cells with antibodies against NPHP5 (red) and glutamylated tubulin (GT335) (green). Scale bar, 2 µm. b In situ PLAs were performed on cycling or quiescent RPE-1 cells using the indicated combination of antibodies to reveal the location of close proximity/interaction (PLA signal, red) between two proteins. Cells were co-stained with γ-tubulin (green) to visualize the centrosome. No PLA signal was detected when NPHP5 and GT335 antibodies were used (negative control; no interaction between NPHP5 and glutamylated tubulin), one antibody was used, or two antibodies were missing (no Ab). As positive control, a robust PLA signal was detected using antibodies against NPHP5 and Cep290 in cycling and quiescent conditions. Scale bar, 1 µm
Cells able to form basal bodies require NPHP5 for BF assembly
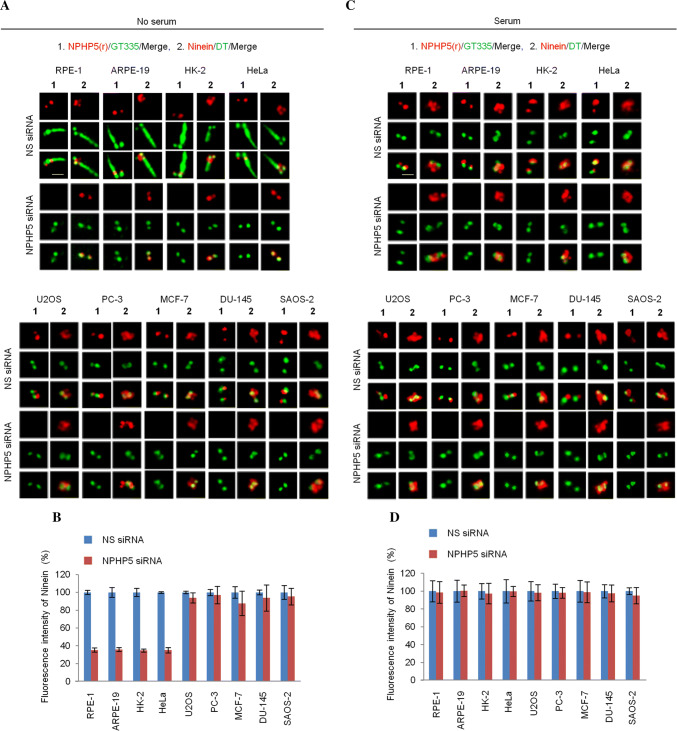
To assess whether the requirement of NPHP5 for BF assembly might extend beyond RPE-1 cells, we studied the consequences of depleting this protein in a number of different cell lines that are either able or unable to quiesce or form basal bodies. Similar to RPE-1, > 70% of ARPE-19 cells subjected to serum starvation were quiescent (Ki67 negative) and possessed basal bodies (one CP110 dot) that template cilia (Figs. 5a, S1 and S7). A significant percentage of serum-starved HK-2 (37–43%) or HeLa (31–36%) cells also exhibited the same properties (Figs. 5a, S1 and S7). Ablation of NPHP5 in quiescent RPE-1, ARPE-19, HK-2, and HeLa cells triggered a decrease in ninein dots (Fig. 5a) and ninein intensity (Fig. 5b), in addition to reduced ciliation (Fig. 5a), without affecting basal body formation or cell cycle exit (Fig. S7). In contrast, the same ablation in these four cell lines did not affect the number of ninein dots (Fig. 5c) or ninein intensity (Fig. 5d) under cycling conditions where cells were Ki67 positive and possessed mother centrioles (two CP110 dots) (Fig. S7) incompetent to template cilia. These results suggest that a loss of NPHP5 specifically disrupts BF assembly in quiescent cells that possess basal bodies. A second set of cell lines examined (U2OS, PC-3, MCF-7) did not readily undergo quiescence upon serum withdrawal (0–7% of cells were Ki67 negative) or form cilia (0–5% of cells had cilia) or basal bodies (4–10% of cells with one CP110 dot) (Figs. 5a, S1 and S7). In other words, these cells possessed mostly mother centrioles regardless of the absence or presence of serum (Fig. S7). Interestingly, the staining pattern and intensity of ninein in these cells remained unchanged upon NPHP5 depletion in serum or serum-free conditions (Fig. 5), reinforcing the idea that NPHP5 is not needed for the assembly of mother centriole-associated SDAs. Moreover, we studied two other cell lines (DU-145 and SAOS-2) that possessed unique properties: upon serum starvation, a significant percentage of cells (18–20%) entered quiescence, yet their mother centrioles could not be converted to basal bodies (96% of cells had two/four CP110 dots) and therefore lacked cilia (100% of cells lacked cilia) (Figs. 5a, S1 and S7). When focused on this particular quiescent cell population, we found that the number of ninein dots and ninein intensity are also unaffected by NPHP5 loss (Fig. 5a, b), indicating that this protein is dispensable in cells that lack basal bodies. Altogether, our data suggest that the ninein phenotype provoked by NPHP5 depletion can be attributed to a loss of BF in cell lines able to form basal bodies.
Fig. 5.
NPHP5-mediated BF assembly is cell-type specific. RPE-1, ARPE-19, HK-2, HeLa, U2OS, PC-3, MCF-7, DU-145, and SAOS-2 cells transfected with NS (non-specific) or NPHP5 siRNAs and grown in the a absence or c presence of serum were stained with the indicated antibodies. Scale bar, 1 µm. b, d Fluorescence intensity of ninein at the centrosome was quantitated and set to 100% in NS siRNA-transfected cells. For quantitation, at least 20 cells for each condition were analyzed, and the mean and standard error of three independent experiments are presented
The BF and SDA assembly pathways are similar but not identical
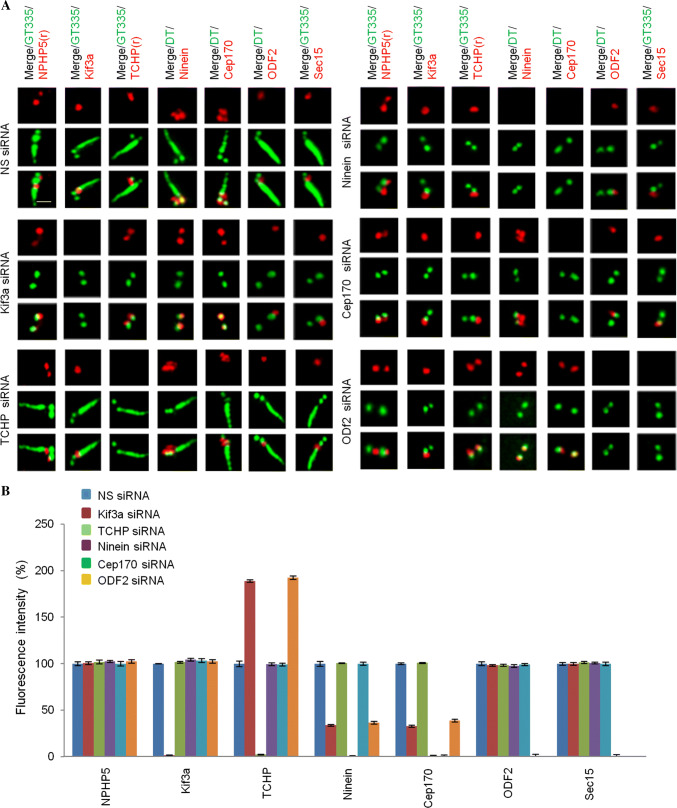
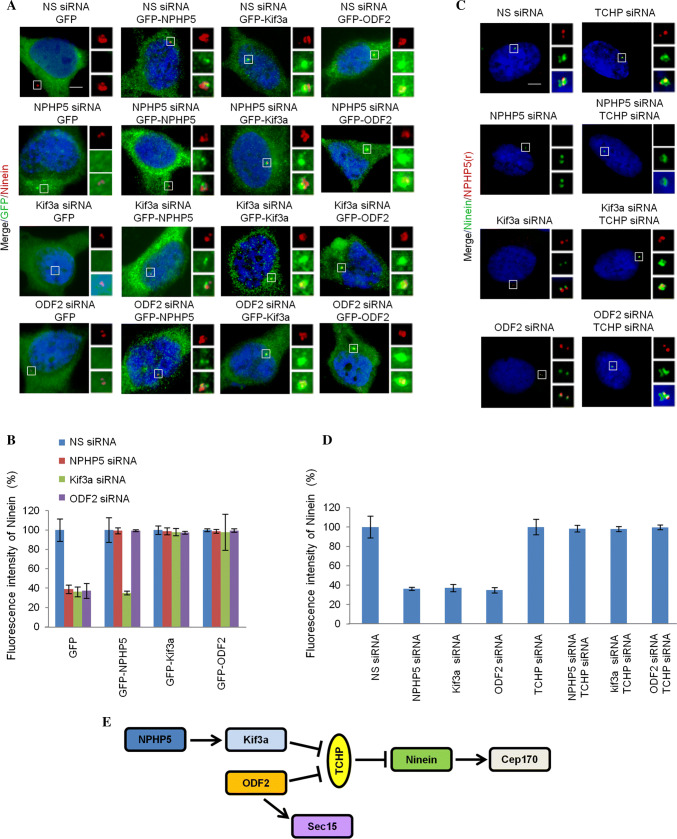
Having established the role of NPHP5 in BF assembly, we next investigated the hierarchical assembly of BF. Because of the similar protein makeup between BF and SDAs, we surmised that the assembly pathway of BF might resemble that of SDA. In terms of BF assembly, we showed that NPHP5 recruits Kif3a, ninein, and Cep170 to organize BF (Fig. 3a), prevents TCHP from being stabilized at, and recruited to, basal bodies (Figs. 3a, S4A and C), but has no effect on the recruitment of CC2D2A, ODF2, and Sec15 (Fig. 3a). We then proceeded to delineate the interrelationship between these proteins in BF assembly. First, we individually depleted Kif3a, TCHP, ninein, Cep170, or ODF2 in quiescent RPE-1 cells and showed that such depletion does not preclude entry into quiescence (Ki67 negative, Fig. 2b) or formation of basal bodies, as evidenced by the loss of one CP110 dot (Fig. 2a, b) and recruitment of IFT88 (Fig. S2). Second, we examined the effects of ablating one protein on the localization of other SDA/BF proteins. Depletion of Kif3a led to a loss of two ninein or Cep170 dots but had no effect on the localization of NPHP5, ODF2 or Sec15 (Fig. 6a, b). TCHP, on the other hand, persisted on both the daughter centriole and basal body (Fig. 6a, b). Depletion of TCHP does not impinge on the localization of NPHP5, Kif3a, ninein, Cep170, ODF2 or Sec15 (Fig. 6a, b). Depletion of ninein de-localized Cep170, but had negligible effects on the localization of NPHP5, Kif3a, TCHP, ODF2, or Sec15 (Fig. 6a, b). Depletion of Cep170 had no effect on the localization of NPHP5, Kif3a, TCHP, ninein, ODF2, or Sec15 (Fig. 6a, b). Depletion of ODF2 did not impinge on the localization of NPHP5 or Kif3a (Fig. 6a, b); rather, it reduced the number of ninein and Cep170 dots, induced the mislocalization of Sec15, and caused TCHP to remain on the daughter centriole and basal body (Fig. 6a, b). Next, a series of experiments were carried out in which we ablated one protein to disrupt BF formation and asked if over-expression or depletion of another SDA/BF protein could restore its formation. Depletion of NPHP5 in quiescent RPE-1 cells resulted in the reduction of ninein dots/intensity, and this phenotype could be rescued by over-expression of NPHP5 or Kif3a (Fig. 7a, b), co-depletion of TCHP (Fig. 7c, d), or interestingly, over-expression of ODF2 (Fig. 7a, b). Likewise, the rescue of the ninein phenotype provoked by ODF2 depletion was achieved by over-expression of ODF2 (Fig. 7a, b), co-depletion of TCHP (Fig. 7c, d), or over-expression of NPHP5 or Kif3a (Fig. 7a, b). Upon depletion of Kif3a in quiescent cells, only co-depletion of TCHP (Fig. 7c, d) or over-expression of Kif3a or ODF2 rescued the ninein phenotype (Fig. 7a, b), whereas over-expression of NPHP5 showed no rescue (Fig. 7a, b). These results suggest that NPHP5-Kif3a and ODF2 converge at the level of TCHP to regulate BF assembly. Taken together, we propose the following hierarchical pathway for BF assembly (Fig. 7e). NPHP5 recruits Kif3a, which prevents the recruitment of TCHP to basal bodies. ODF2 functions in parallel with NPHP5 and Kif3a to inhibit the recruitment of TCHP. Once TCHP is removed from basal bodies, ninein is brought in, followed by Cep170. Sec15 appears to be independently recruited to BF by ODF2, but we were unable to determine its precise relationship with other proteins in the BF assembly pathway due to sub-optimal knockdown efficiency.
Fig. 6.
Hierarchical assembly of BF. a Quiescent RPE-1 cells transfected with NS (non-specific) or the indicated siRNAs targeting SDA/BF components (Kif3a, TCHP, ninein, Cep170, ODF2) were stained with the indicated antibodies. Scale bar, 1 µm. b Fluorescence intensities of various proteins at the centrosome were quantitated and set to 100% in NS siRNA-transfected cells. For quantitation, at least 20 cells for each condition were analyzed, and the mean and standard error of three independent experiments are presented
Fig. 7.
BF loss due to ablation of NPHP5, Kif3a or ODF2 can be rescued. a Quiescent RPE-1 cells transfected with NS (non-specific) or the indicated siRNAs and plasmid expressing the indicated protein were stained with the indicated antibodies and with DAPI (blue). Scale bar, 2 µm. b Fluorescence intensity of ninein at the centrosome was quantitated and set to 100% in NS siRNA-transfected cells. For quantitation, at least 20 cells for each condition were analyzed, and the mean and standard error of three independent experiments are presented. c Quiescent RPE-1 cells transfected with the indicated siRNAs were stained with the indicated antibodies and with DAPI (blue). Scale bar, 2 µm. d Fluorescence intensity of ninein at the centrosome was quantitated and set to 100% in NS siRNA-transfected cells. For quantitation, at least 20 cells for each condition were analyzed, and the mean and standard error of three independent experiments are presented. e Schematic model of BF assembly
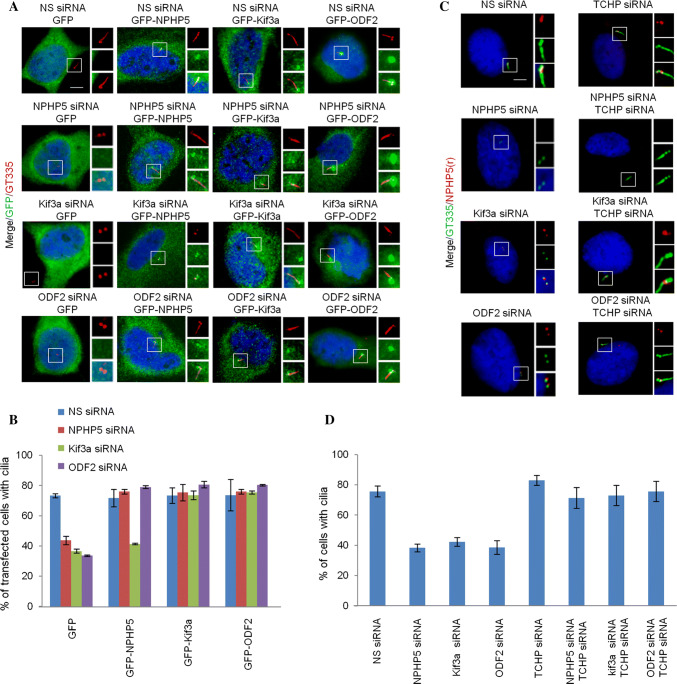
Our earlier data showed that NPHP5 is not required for SDA assembly (Fig. 1d and S5A-B). To study the hierarchical assembly of SDA, the effects of ablating one SDA protein on the localization of other SDA proteins were examined in cycling RPE-1 cells. Depletion of Kif3a had no effect on the localization any SDA protein examined (Fig. S8A-B). Depletion of TCHP is shown to trigger cell cycle arrest and cilia formation in a subpopulation of cells [25]. However, the remaining cells were not ciliated, and we observed loss of two ninein and Cep170 foci corresponding to SDAs (Fig. S8A-B). The localization of other SDA proteins NPHP5, Kif3a, ODF2 and Sec15 were not affected (Fig. S8A-B). Depletion of ninein de-localized Cep170 but had no impact on the localization of NPHP5, Kif3a, TCHP, ODF2, or Sec15 (Fig. S8A-B). Depletion of Cep170 had no effect on the localization of NPHP5, Kif3a, TCHP, ninein, ODF2, or Sec15 (Fig. S8A-B). Depletion of ODF2 did not impinge on the localization of NPHP5 or Kif3a; yet it de-localized TCHP, ninein, Cep170 and Sec15 from SDAs (Fig. S8A-B). These data suggest that the hierarchical pathway for SDA assembly does not involve NPHP5 and Kif3a. Rather, ODF2 independently recruits Sec15 and TCHP, which in turn recruits ninein and Cep170 in a sequential manner to build SDAs (Fig. S8C). Furthermore, our data support the notion that the BF assembly pathway is similar but not identical to the SDA assembly pathway.
BF assembly correlates with primary ciliogenesis
The relationship between BF assembly and ciliogenesis remains poorly defined. NPHP5 depletion in quiescent RPE-1 cells simultaneously prevented ciliogenesis and BF assembly (Figs. 1d, f, 2, 3a, b, 5a, b). Likewise, depletion of Kif3a, ninein, Cep170, or ODF2 in quiescent RPE-1 cells suppressed cilia formation in addition to BF assembly (Figs. 2, 6a, b). On the other hand, ablation of TCHP, a negative regulator of ciliogenesis, in quiescent RPE-1 cells neither enhanced ciliation nor BF assembly (Figs. 2, 6a, b). In light of a positive correlation between BF assembly and ciliogenesis, we investigated whether restoration of the former might reinstate the latter. Cilia loss associated with NPHP5 or ODF2 depletion in quiescent RPE-1 cells could be rescued by over-expression of NPHP5, Kif3a, or ODF2 (Fig. 8a, b), or co-depletion of TCHP (Fig. 8c, d). Interestingly, impaired ciliogenesis induced by Kif3a depletion was rescued by over-expression of Kif3a or ODF2 (Fig. 8a, b), or co-depletion of TCHP (Fig. 8c, d), but not by over-expression of NPHP5 (Fig. 8a, b). Our results suggest that BF formation is tightly coupled to cilia formation.
Fig. 8.
Cilia loss due to ablation of NPHP5, Kif3a or ODF2 can be rescued. a Quiescent RPE-1 cells transfected with NS (non-specific) or the indicated siRNAs and plasmid expressing the indicated protein were stained with the indicated antibodies and with DAPI (blue). Scale bar, 2 µm. b The percentage of GFP positive cells with cilia was scored. At least 100 cells for each condition were analyzed, and the mean and standard error of three independent experiments are presented. c Quiescent RPE-1 cells transfected with the indicated siRNAs were stained with the indicated antibodies and with DAPI (blue). Scale bar, 2 µm. d The percentage of cells with cilia was scored. At least 100 cells for each condition were analyzed, and the mean and standard error of three independent experiments are presented
Discussion
In this study, we demonstrated that SDAs and BF can be distinguished by their appearance. By using super-resolution microscopy, several SDA/BF components exhibit a ring-like structure in cells that possess mother centrioles. On the other hand, the same SDA/BF components exhibit three to four dots in cells that form basal bodies. These observations raise the intriguing possibility that the number of SDAs is greater than the number of BF. Future work will precisely determine the number of SDAs/BF and address how SDAs are modified into BFs.
SDAs are dynamic structures that can undergo extensive modification. During late G2/mitosis, SDAs are replaced by a halo [6, 40], and certain SDA proteins, including NPHP5, reportedly diffuse away or disappear from the centrosome/spindle poles [17, 26, 36, 37]. It is tempting to think that such modification is needed to prepare the cell for the next G0/G1 phase when the decision to assemble BF or reassemble SDAs has to be made.
Previous immuno-EM studies have shown that NPHP5 localizes to the outer segment and connecting cilium of photoreceptor cells [41]. The outer segment is a modified primary cilium, while the connecting cilium is equivalent to the transition zone of a primary cilium. In support of the transition zone location, NPHP5 directly interacts with a transition zone protein Cep290 and participates in trafficking the BBSome and its cargos to primary cilia [36, 38, 39, 42, 43]. In other studies, NPHP5 and Cep290 are reportedly localized to the distal region of the mother and daughter centrioles in non-ciliated cells [36–38]. These observations suggest that there could be at least three pools of Cep290/NPHP5, one at the transition zone, one at the daughter centriole, and one at the mother centriole. Our data revealed that the major pool of NPHP5 is localized to the mother centriole and basal body, and more precisely, to SDAs and BF. Future studies will determine whether Cep290 is present at SDAs/BF and whether it only serves to deliver NPHP5 to SDAs/BF.
One major finding from our study is that unlike other SDA/BF proteins known to date, NPHP5 is specifically required for BF assembly but not SDA assembly. We envision that although NPHP5 is targeted to SDAs of mother centrioles, this protein might be kept in an inactive state and barred from interacting with other SDA/BF proteins. During the conversion of mother centrioles to basal bodies, NPHP5 becomes activated, which allows it to interact with and recruit a subset of SDA/BF proteins for BF assembly. As NPHP5 is a relatively stable protein whose level does not fluctuate much in the cell cycle [37], it is plausible that its activation involves a post-translational mechanism. Further studies would be needed to decipher the mechanism by which NPHP5 transitions between inactive and active states.
Consistent with a role of NPHP5 in BF assembly, our microtubule re-growth assay revealed that microtubule nucleation/anchoring is impaired in NPHP5-depleted quiescent cells, which lack BF, but not in NPHP5-depleted cycling cells, which possess SDAs. SDAs/BF are thought to mediate microtubule nucleation/anchoring [13, 21, 24]; nevertheless, some studies have reported that SDAs/BF defects do not compromise nucleation/anchoring [15, 22, 27]. We speculate that these discrepancies could be due to differences in cell lines, kinetics of microtubule depolymerization/re-growth, and how ablation of a given SDA/BF protein affects the localization, stability, and/or activity of various nucleating (γ-tubulin) and anchoring factors (ninein, p150Glued, EB1, EB3) at the centrosome.
We further showed in this study that a loss of any BF component inhibits BF assembly without compromising basal body formation. We determined the assembly order of BF components and constructed, to our knowledge, the first-ever BF assembly pathway (Fig. 7e). There are noticeable differences between this pathway and the SDA assembly pathway (Figs. 7e and S8C). First, the relationship of TCHP with its neighbouring proteins differs between SDA and BF assembly. ODF2 recruits TCHP to SDAs but prevents TCHP from being recruited to BF. Kif3a also prevents TCHP recruitment to BF but is not required for TCHP localization to SDAs or SDA assembly, which is in contrast to a previous study conducted in MEF cells [10]. TCHP recruits ninein to SDAs but inhibits its recruitment to BF. Second, NPHP5 and Kif3a are specifically required to build BF, and NPHP5 has an additional role in destabilizing TCHP. The protein level of TCHP is known to be controlled by the ubiquitin–proteasome system. In cycling cells, polyubiquitination of TCHP by the ubiquitin ligase CRL3KCTD17 is inhibited by a novel SDA protein Ndel1 and counteracted by a deubiquitinase USP8 [44–47]. Upon quiescence, USP8 is inactivated and Ndel1 is degraded, thus allowing CRL3KCTD17 to polyubiquitinate TCHP. In light of these observations, it would be interesting to determine whether and how NPHP5, Kif3a or ODF2 modulates the activity/steady-state level of CRL3KCTD17, USP8 and/or Ndel1 to remove TCHP from BF in quiescent cells. Additional studies will also evaluate if the differential behaviour of TCHP towards SDA-associated ninein and BF-associated ninein might depend on its interaction with different forms of intermediate filaments.
In addition, we found a correlation between BF assembly and primary ciliogenesis by demonstrating that rescue of BF loss is sufficient to rescue cilia loss. How might BF assembly be linked to ciliogenesis? It is possible that BF defects prevent ciliogenesis by disrupting the microtubule network required for the transport of vesicles carrying ciliary building blocks to the basal body. Another possibility is that SDA/BF proteins we studied here are multifunctional and contribute to ciliogenesis through more than one mechanism. Besides organizing BF, Kif3a functions as an anterograde motor for IFT during ciliogenesis [30, 31]. Likewise, NPHP5 is thought to regulate ciliary trafficking at the transition zone [39], and its presence at the daughter centriole might implicate a role for daughter centriole-mediated ciliogenesis [48]. Moreover, TCHP activates Aurora A kinase which in turn might modulate the activity of HDAC6, leading to deacetylation and destabilization of axonemal microtubules [25, 49]. Further experiments will be needed to distinguish these possibilities.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We thank all members of the Tsang laboratory for constructive advice, and L. Wordeman, K. Lee, and E. Nigg for providing antibodies and plasmids. We are indebted to A. Das and J. Qian for their assistance on this project, H. Vali, K. Sears and J. Mui for their help with EM data acquisition and analysis, and D. Filion, E. Wee and M. Fu for their guidance with super-resolution microscopy. WYT was a Canadian Institutes of Health Research New Investigator and a Fonds de recherche Santé Junior 2 Research Scholar. This work was supported by the Canadian Institutes of Health Research and the Natural Sciences and Engineering Research Council of Canada to WYT.
Abbreviations
- 3D-SIM
Three dimensional-structured illumination microscopy
- BF
Basal feet
- DAs
Distal appendages
- DT
Anti-detyrosinated tubulin
- EM
Electron microscopy
- GT335
Anti-glutamylated tubulin
- IFT
Intraflagellar transport
- NS
Non-specific
- PCM
Pericentriolar material
- PLA
Proximity ligation assay
- RPE-1
Retinal pigmented epithelial cells
- SDAs
Sub-distal appendages
- TFs
Transition fibers
Author contributions
WYT designed all experiments with input from DH and MB. DH and MB performed the experiments and analyzed the results. WYT and DH wrote the paper, and all authors reviewed the paper.
Compliance with ethical standards
Conflict of interest
The authors declare no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Bornens M. The centrosome in cells and organisms. Science. 2012;335(6067):422–426. doi: 10.1126/science.1209037. [DOI] [PubMed] [Google Scholar]
- 2.Kobayashi T, Dynlacht BD. Regulating the transition from centriole to basal body. J Cell Biol. 2011;193(3):435–444. doi: 10.1083/jcb.201101005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nigg EA, Stearns T. The centrosome cycle: centriole biogenesis, duplication and inherent asymmetries. Nat Cell Biol. 2011;13(10):1154–1160. doi: 10.1038/ncb2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Seeley ES, Nachury MV. The perennial organelle: assembly and disassembly of the primary cilium. J Cell Sci. 2010;123(Pt 4):511–518. doi: 10.1242/jcs.061093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goetz SC, Liem KF, Jr, Anderson KV. The spinocerebellar ataxia-associated gene Tau tubulin kinase 2 controls the initiation of ciliogenesis. Cell. 2012;151(4):847–858. doi: 10.1016/j.cell.2012.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vorobjev IA, Chentsov Y. Centrioles in the cell cycle. I. Epithelial cells. J Cell Biol. 1982;93(3):938–949. doi: 10.1083/jcb.93.3.938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ibrahim R, Messaoudi C, Chichon FJ, Celati C, Marco S. Electron tomography study of isolated human centrioles. Microsc Res Tech. 2009;72(1):42–48. doi: 10.1002/jemt.20637. [DOI] [PubMed] [Google Scholar]
- 8.Anderson RG. The three-dimensional structure of the basal body from the rhesus monkey oviduct. J Cell Biol. 1972;54(2):246–265. doi: 10.1083/jcb.54.2.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kunimoto K, Yamazaki Y, Nishida T, Shinohara K, Ishikawa H, Hasegawa T, Okanoue T, Hamada H, Noda T, Tamura A, Tsukita S. Coordinated ciliary beating requires Odf2-mediated polarization of basal bodies via basal feet. Cell. 2012;148(1–2):189–200. doi: 10.1016/j.cell.2011.10.052. [DOI] [PubMed] [Google Scholar]
- 10.Kodani A, Salome Sirerol-Piquer M, Seol A, Garcia-Verdugo JM, Reiter JF. Kif3a interacts with Dynactin subunit p150 Glued to organize centriole subdistal appendages. EMBO J. 2013;32(4):597–607. doi: 10.1038/emboj.2013.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Odor DL, Blandau RJ. Observations on the solitary cilium of rabbit oviductal epithelium: its motility and ultrastructure. Am J Anat. 1985;174(4):437–453. doi: 10.1002/aja.1001740407. [DOI] [PubMed] [Google Scholar]
- 12.Hagiwara H, Ohwada N, Aoki T, Suzuki T, Takata K. The primary cilia of secretory cells in the human oviduct mucosa. Med Mol Morphol. 2008;41(4):193–198. doi: 10.1007/s00795-008-0421-z. [DOI] [PubMed] [Google Scholar]
- 13.Delgehyr N, Sillibourne J, Bornens M. Microtubule nucleation and anchoring at the centrosome are independent processes linked by ninein function. J Cell Sci. 2005;118(Pt 8):1565–1575. doi: 10.1242/jcs.02302. [DOI] [PubMed] [Google Scholar]
- 14.Gordon RE. Three-dimensional organization of microtubules and microfilaments of the basal body apparatus of ciliated respiratory epithelium. Cell Motil. 1982;2(4):385–391. doi: 10.1002/cm.970020407. [DOI] [PubMed] [Google Scholar]
- 15.Ishikawa H, Kubo A, Tsukita S. Odf2-deficient mother centrioles lack distal/subdistal appendages and the ability to generate primary cilia. Nat Cell Biol. 2005;7(5):517–524. doi: 10.1038/ncb1251. [DOI] [PubMed] [Google Scholar]
- 16.Mogensen MM, Malik A, Piel M, Bouckson-Castaing V, Bornens M. Microtubule minus-end anchorage at centrosomal and non-centrosomal sites: the role of ninein. J Cell Sci. 2000;113(Pt 17):3013–3023. doi: 10.1242/jcs.113.17.3013. [DOI] [PubMed] [Google Scholar]
- 17.Guarguaglini G, Duncan PI, Stierhof YD, Holmstrom T, Duensing S, Nigg EA. The forkhead-associated domain protein Cep170 interacts with Polo-like kinase 1 and serves as a marker for mature centrioles. Mol Biol Cell. 2005;16(3):1095–1107. doi: 10.1091/mbc.e04-10-0939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang N, Xia Y, Zhang D, Wang S, Bao Y, He R, Teng J, Chen J. Hierarchical assembly of centriole subdistal appendages via centrosome binding proteins CCDC120 and CCDC68. Nat Commun. 2017;8:15057. doi: 10.1038/ncomms15057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gromley A, Jurczyk A, Sillibourne J, Halilovic E, Mogensen M, Groisman I, Blomberg M, Doxsey S. A novel human protein of the maternal centriole is required for the final stages of cytokinesis and entry into S phase. J Cell Biol. 2003;161(3):535–545. doi: 10.1083/jcb.200301105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chang P, Giddings TH, Jr, Winey M, Stearns T. Epsilon-tubulin is required for centriole duplication and microtubule organization. Nat Cell Biol. 2003;5(1):71–76. doi: 10.1038/ncb900. [DOI] [PubMed] [Google Scholar]
- 21.Veleri S, Manjunath SH, Fariss RN, May-Simera H, Brooks M, Foskett TA, Gao C, Longo TA, Liu P, Nagashima K, Rachel RA, Li T, Dong L, Swaroop A. Ciliopathy-associated gene Cc2d2a promotes assembly of subdistal appendages on the mother centriole during cilia biogenesis. Nat Commun. 2014;5:4207. doi: 10.1038/ncomms5207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mazo G, Soplop N, Wang WJ, Uryu K, Tsou MF. Spatial control of primary ciliogenesis by subdistal appendages alters sensation-associated properties of cilia. Dev Cell. 2016;39(4):424–437. doi: 10.1016/j.devcel.2016.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hehnly H, Chen CT, Powers CM, Liu HL, Doxsey S. The centrosome regulates the Rab11- dependent recycling endosome pathway at appendages of the mother centriole. Curr Biol. 2012;22(20):1944–1950. doi: 10.1016/j.cub.2012.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ibi M, Zou P, Inoko A, Shiromizu T, Matsuyama M, Hayashi Y, Enomoto M, Mori D, Hirotsune S, Kiyono T, Tsukita S, Goto H, Inagaki M. Trichoplein controls microtubule anchoring at the centrosome by binding to Odf2 and ninein. J Cell Sci. 2011;124(Pt 6):857–864. doi: 10.1242/jcs.075705. [DOI] [PubMed] [Google Scholar]
- 25.Inoko A, Matsuyama M, Goto H, Ohmuro-Matsuyama Y, Hayashi Y, Enomoto M, Ibi M, Urano T, Yonemura S, Kiyono T, Izawa I, Inagaki M. Trichoplein and Aurora A block aberrant primary cilia assembly in proliferating cells. J Cell Biol. 2012;197(3):391–405. doi: 10.1083/jcb.201106101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Graser S, Stierhof YD, Lavoie SB, Gassner OS, Lamla S, Le Clech M, Nigg EA. Cep164, a novel centriole appendage protein required for primary cilium formation. J Cell Biol. 2007;179(2):321–330. doi: 10.1083/jcb.200707181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tateishi K, Yamazaki Y, Nishida T, Watanabe S, Kunimoto K, Ishikawa H, Tsukita S. Two appendages homologous between basal bodies and centrioles are formed using distinct Odf2 domains. J Cell Biol. 2013;203(3):417–425. doi: 10.1083/jcb.201303071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chang J, Seo SG, Lee KH, Nagashima K, Bang JK, Kim BY, Erikson RL, Lee KW, Lee HJ, Park JE, Lee KS. Essential role of Cenexin1, but not Odf2, in ciliogenesis. Cell Cycle. 2013;12(4):655–662. doi: 10.4161/cc.23585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hung HF, Hehnly H, Doxsey S. The mother centriole appendage protein cenexin modulates lumen formation through spindle orientation. Curr Biol. 2016;26(9):1248. doi: 10.1016/j.cub.2016.04.033. [DOI] [PubMed] [Google Scholar]
- 30.Marszalek JR, Ruiz-Lozano P, Roberts E, Chien KR, Goldstein LS. Situs inversus and embryonic ciliary morphogenesis defects in mouse mutants lacking the KIF3A subunit of kinesin-II. Proc Natl Acad Sci USA. 1999;96(9):5043–5048. doi: 10.1073/pnas.96.9.5043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takeda S, Yonekawa Y, Tanaka Y, Okada Y, Nonaka S, Hirokawa N. Left-right asymmetry and kinesin superfamily protein KIF3A: new insights in determination of laterality and mesoderm induction by kif3A−/− mice analysis. J Cell Biol. 1999;145(4):825–836. doi: 10.1083/jcb.145.4.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hossain D, Javadi Esfehani Y, Das A, Tsang WY. Cep78 controls centrosome homeostasis by inhibiting EDD-DYRK2-DDB1(Vpr)(BP) EMBO Rep. 2017;18(4):632–644. doi: 10.15252/embr.201642377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sonnen KF, Schermelleh L, Leonhardt H, Nigg EA. 3D-structured illumination microscopy provides novel insight into architecture of human centrosomes. Biol Open. 2012;1(10):965–976. doi: 10.1242/bio.20122337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mojarad BA, Gupta GD, Hasegan M, Goudiam O, Basto R, Gingras AC, Pelletier L. CEP19 cooperates with FOP and CEP350 to drive early steps in the ciliogenesis programme. Open Biol. 2017;7(6):170114. doi: 10.1098/rsob.170114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kobayashi T, Kim S, Lin YC, Inoue T, Dynlacht BD. The CP110-interacting proteins Talpid3 and Cep290 play overlapping and distinct roles in cilia assembly. J Cell Biol. 2014;204(2):215–229. doi: 10.1083/jcb.201304153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barbelanne M, Song J, Ahmadzai M, Tsang WY. Pathogenic NPHP5 mutations impair protein interaction with Cep290, a prerequisite for ciliogenesis. Hum Mol Genet. 2013;22(12):2482–2494. doi: 10.1093/hmg/ddt100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Das A, Qian J, Tsang WY. USP9X counteracts differential ubiquitination of NPHP5 by MARCH7 and BBS11 to regulate ciliogenesis. PLoS Genet. 2017;13(5):e1006791. doi: 10.1371/journal.pgen.1006791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sang L, Miller JJ, Corbit KC, Giles RH, Brauer MJ, Otto EA, Baye LM, Wen X, Scales SJ, Kwong M, Huntzicker EG, Sfakianos MK, Sandoval W, Bazan JF, Kulkarni P, Garcia-Gonzalo FR, Seol AD, O’Toole JF, Held S, Reutter HM, Lane WS, Rafiq MA, Noor A, Ansar M, Devi AR, Sheffield VC, Slusarski DC, Vincent JB, Doherty DA, Hildebrandt F, Reiter JF, Jackson PK. Mapping the NPHP-JBTS-MKS protein network reveals ciliopathy disease genes and pathways. Cell. 2011;145(4):513–528. doi: 10.1016/j.cell.2011.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barbelanne M, Hossain D, Chan DP, Peranen J, Tsang WY. Nephrocystin proteins NPHP5 and Cep290 regulate BBSome integrity, ciliary trafficking and cargo delivery. Hum Mol Genet. 2015;24(8):2185–2200. doi: 10.1093/hmg/ddu738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Uzbekov R, Prigent C. Clockwise or anticlockwise? Turning the centriole triplets in the right direction! FEBS Lett. 2007;581(7):1251–1254. doi: 10.1016/j.febslet.2007.02.069. [DOI] [PubMed] [Google Scholar]
- 41.Otto EA, Loeys B, Khanna H, Hellemans J, Sudbrak R, Fan S, Muerb U, O’Toole JF, Helou J, Attanasio M, Utsch B, Sayer JA, Lillo C, Jimeno D, Coucke P, De Paepe A, Reinhardt R, Klages S, Tsuda M, Kawakami I, Kusakabe T, Omran H, Imm A, Tippens M, Raymond PA, Hill J, Beales P, He S, Kispert A, Margolis B, Williams DS, Swaroop A, Hildebrandt F. Nephrocystin-5, a ciliary IQ domain protein, is mutated in Senior-Loken syndrome and interacts with RPGR and calmodulin. Nat Genet. 2005;37(3):282–288. doi: 10.1038/ng1520. [DOI] [PubMed] [Google Scholar]
- 42.Craige B, Tsao CC, Diener DR, Hou Y, Lechtreck KF, Rosenbaum JL, Witman GB. CEP290 tethers flagellar transition zone microtubules to the membrane and regulates flagellar protein content. J Cell Biol. 2010;190(5):927–940. doi: 10.1083/jcb.201006105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang TT, Su J, Wang WJ, Craige B, Witman GB, Tsou MF, Liao JC. Superresolution pattern recognition reveals the architectural map of the ciliary transition zone. Sci Rep. 2015;5:14096. doi: 10.1038/srep14096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kasahara K, Kawakami Y, Kiyono T, Yonemura S, Kawamura Y, Era S, Matsuzaki F, Goshima N, Inagaki M. Ubiquitin-proteasome system controls ciliogenesis at the initial step of axoneme extension. Nat Commun. 2014;5:5081. doi: 10.1038/ncomms6081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Inaba H, Goto H, Kasahara K, Kumamoto K, Yonemura S, Inoko A, Yamano S, Wanibuchi H, He D, Goshima N, Kiyono T, Hirotsune S, Inagaki M. Ndel1 suppresses ciliogenesis in proliferating cells by regulating the trichoplein-Aurora A pathway. J Cell Biol. 2016;212(4):409–423. doi: 10.1083/jcb.201507046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kasahara K, Aoki H, Kiyono T, Wang S, Kagiwada H, Yuge M, Tanaka T, Nishimura Y, Mizoguchi A, Goshima N, Inagaki M. EGF receptor kinase suppresses ciliogenesis through activation of USP8 deubiquitinase. Nat Commun. 2018;9(1):758. doi: 10.1038/s41467-018-03117-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nishimura Y, Kasahara K, Shiromizu T, Watanabe M, Inagaki M. Primary cilia as signaling hubs in health and disease. Adv Sci (Weinh) 2019;6(1):1801138. doi: 10.1002/advs.201801138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Loukil A, Tormanen K, Sutterlin C. The daughter centriole controls ciliogenesis by regulating Neurl-4 localization at the centrosome. J Cell Biol. 2017;216(5):1287–1300. doi: 10.1083/jcb.201608119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pugacheva EN, Jablonski SA, Hartman TR, Henske EP, Golemis EA. HEF1-dependent Aurora A activation induces disassembly of the primary cilium. Cell. 2007;129(7):1351–1363. doi: 10.1016/j.cell.2007.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.