Abstract
Circulating tumor cells (CTCs) are regarded as harbingers of metastases. Their ability to predict response to therapy, relapse, and resistance to treatment has proposed their value as putative diagnostic and prognostic indicators. CTCs represent one of the zeniths of cancer evolution in terms of cell survival; however, the triggers of CTC generation, the identification of potentially metastatic CTCs, and the mechanisms contributing to their heterogeneity and aggressiveness represent issues not yet fully deciphered. Thus, prior to enabling liquid biopsy applications to reach clinical prime time, understanding how the above mechanistic information can be applied to improve treatment decisions is a key challenge. Here, we provide our perspective on how CTCs can provide mechanistic insights into tumor pathogenesis, as well as on CTC clinical value. In doing so, we aim to (a) describe how CTCs disseminate from the primary tumor, and their link to epithelial–mesenchymal transition (EMT); (b) trace the route of CTCs through the circulation, focusing on tumor self-seeding and the possibility of tertiary metastasis; (c) describe possible mechanisms underlying the enhanced metastatic potential of CTCs; (d) discuss how CTC could provide further information on the tissue of origin, especially in cancer of unknown primary origin. We also provide a comprehensive review of meta-analyses assessing the prognostic significance of CTCs, to highlight the emerging role of CTCs in clinical oncology. We also explore how cell-free circulating tumor DNA (ctDNA) analysis, using a combination of genomic and phylogenetic analysis, can offer insights into CTC biology, including our understanding of CTC heterogeneity and tumor evolution. Last, we discuss emerging technologies, such as high-throughput quantitative imaging, radiogenomics, machine learning approaches, and the emerging breath biopsy. These technologies could compliment CTC and ctDNA analyses, and they collectively represent major future steps in cancer detection, monitoring, and management.
Keywords: Circulating tumor cell, Circulating tumor DNA, Disseminated tumor cell, Liquid biopsy, Epithelial–mesenchymal transition, Tumor self-seeding, Tumor evolution, Richter's syndrome
Introduction
A typical primary tumor is highly fragile. The metastatic spread of a primary tumor is triggered by tumor cells that break off and enter the peripheral bloodstream; these are termed circulating tumor cells (CTCs). First described by Thomas Ashworth in 1869, CTCs were then detected in the blood of a metastatic cancer patient. Ashworth noted similarities between the primary tumor and CTCs, and he proposed that CTCs may shed light on the origin of metastatic lesions within the same person [1]. CTCs are extraordinarily rare, occurring at a frequency of one CTC per billion normal blood cells [2], and they present with a comparatively short half-life [3]. They exist as either apoptotic or viable cells, with the latter comprising individual cells or clusters [4]. CTCs derived from different cancer types exhibit remarkable heterogeneity in terms of size, cell surface marker expression, and tumor-seeding potential [4, 5]. For example, based on surface marker expression, CTCs can be classified as (a) epithelial CTCs (ECTCs), (b) mesenchymal CTCs (MCTCs), and (c) epithelial–mesenchymal CTCs (EMCTCs) [6, 7]. Even though CTC isolation and evaluation techniques have tremendously improved in the last decade, challenges in isolating CTCs are still present. There is also still a lack of markers to differentiate clonally evolving CTCs that may possess enhanced metastatic potential. To this end, circulating tumor DNA (ctDNA) analysis or even exosomes (which represents another form of liquid biopsy biomarker not further discussed here-in) may be more informative in deducing the mechanisms of CTC dissemination and metastatic potential.
In this review, we aim to explore (a) what mechanistic insights CTCs can offer in better understanding the metastatic colonization process, and (b) how circulating tumor DNA (ctDNA) analysis can complement the approach of CTCs to add to the clinical relevance of liquid biopsy. (For our methodology, see, “Search strategy and selection criteria”).
Can circulating tumor cells overcome metastatic inefficiency?
CTCs obtain access to the circulation system very early during the metastatic process, even before a tumor is medically diagnosed [2, 3, 4]. They are subjected to numerous restrictive barriers, such as significant shear forces, loss of adhesion-dependent survival signals leading to anoikis (i.e., a type of programmed cell death occurring after detachment of the cell from extracellular matrix (ECM)), immune surveillance and clearance, apoptosis, and oxidative stress [8, 9]. Furthermore, CTCs are under constant evolutionary pressure not only to survive the journey through the systemic circulation, but also to acquire self-renewal, multipotency, and tumor-initiating capabilities [7]; thus, metastasis is a highly inefficient process. Prior to understanding of these barriers and pressures, we must first appreciate the value of CTCs for evaluation of the metastatic potential of a primary tumor.
Because of several important stressful factors (such as the size of blood and lymphatic vessels, the shear stress exercised, and the flow rates) (reviewed by Follain et al. [10]), most CTCs never survive the long journey to their target sites, with only a small proportion colonizing the distant host tissue. This is evident from the disproportionately large numbers of CTCs found in the circulation compared to the number of overt metastatic lesions that develop. Those that do eventually infiltrate the distant host tissue are termed disseminated tumor cells (DTCs) or isolated tumor cells (ITCs) [11], and they seem to represent the zenith of cancer evolution in terms of cell survival. Nevertheless, DTCs must continue to adapt to supportive niches and survive latency periods, eventually becoming established in a new target site. However, following the formation of metastases, the existing therapeutic approaches are most often unsuccessful to offer long-term benefits, suggesting that other types of approaches, e.g., via (neo)adjuvant and locoregional therapies, may represent a better option, with the constant notion that prevention, e.g., through early cancer detection, is always preferable than treatment of metastases [9].
A more thorough comprehension of the metastatic colonization process is urgently essential to help physicians choosing appropriate treatment options. As potential drivers and prognostic biomarkers of metastasis, understanding CTC biology and targeting the mechanisms that trigger their metastatic potential represent a viable approach [12, 13].
Epithelial–mesenchymal transition: the first step towards circulating tumor cell dissemination and metastatic self-sufficiency
Very few studies have explored the mechanism by which CTCs overcome the barriers to metastasis. Metastasis-initiating cells (MICs) represent a subpopulation of CTCs from primary human luminal breast cancer, and they are characterized by a unique surface marker signature that is distinct from the bulk of the CTCs found in the circulation. Based on studies in a xenograft assay of primary human luminal breast cancer, MICs were shown to lead to the formation of metastases to the bone, lung, and liver in mice; these findings were also confirmed in a small cohort of patients with breast cancer, in which MICs were linked to lower overall survival and higher number of metastatic sites [14]. A similar rare subset of CTCs harboring metastatic traits was identified in patients with late-stage, metastatic, castration-resistant prostate cancer [15]. However, as mentioned in [16], how much heterogeneous are the CTC; what are the cellular and molecular pathways that lead to both CTC intravasation and extravasation; what makes the metastatic CTC subpopulations vulnerable; what is the mechanism leading MIC generation; all these are research questions that should be fully elucidated.
CTC generation from a primary epithelial tumor is thought to be triggered by epithelial-to-mesenchymal transition (EMT). This complex process is hallmarked by an attenuation in epithelial properties, and an increase in mesenchymal or stem cell-like attributes, triggered by the accumulation of both epigenetic and genetic changes in the epithelial cells or due to metabolic cues from the surrounding stroma [17, 18]. It involves many cellular and molecular changes, including (a) loss of intercellular interactions and those between cells and extracellular matrices, and (b) increased migratory and invasive capabilities [19, 20]. It has also been perceived that a meta-stable state (partial EMT cell state) may demonstrate a higher potential for CTC generation (as further discussed by Agnoletto et al. [21]). To colonize a distant metastatic site, CTCs may acquire aggressive, tumor-initiating capacities, and stem cell properties following EMT [22, 23]. Indeed, transformed mammary epithelial cells from human patients which were subjected to EMT show stronger tumor-forming potential [22]. Thus, CTC subpopulations, such as the MICs that are positive for EMT markers, may represent prognostic markers of cancer progression.
Epithelial proteins, such as epithelial cellular adhesion molecule (EpCAM), E-cadherins and cytokeratins, are downregulated during EMT; whereas, mesenchymal markers such as vimentin, N-cadherin and fibronectin and EMT regulators, such as Twist 1, Snail 1, Slug, Zeb1/Zeb2, Akt, PI3K and FoxC 2, are upregulated [19, 20, 24]. These markers of EMT and stem cell markers, such as ALDH1, CD44, and CD133, are characterized by their overexpression in CTCs, regardless of cancer-type [21, 25]. Several reports provide evidence of associations (both positive and negative) with regards to the expression of markers of EMT and stemness on CTCs with tumor stage and metastasis, therapeutic response, and prognosis; however, to validate these associations, prospective clinical trials recruiting larger number of patients would be crucial [26–28].
To enable CTCs to metastasize, EMT and its regulatory networks—such as EMT-transcription factors (EMT-TFs), matrix extracellular molecules and the hypoxic tumor microenvironment—function together to induce morphological changes by reorganizing the cytoskeleton. These changes are accompanied by upregulated expression of matrix metallo-proteases (MMPs) that degrade the surrounding tissue and promote angiogenesis to increase tumor cell invasiveness and intravasation [19]. An example of cytoskeletal reorganization, single-cell polarity, is associated with changes in adhesion, migration, and metastatic capabilities [29, 30]. EMT also facilitates the avoidance of apoptosis, anoikis, and senescence to promote CTC survival in the blood circulation [20]. Core paracrine EMT-signaling pathways that determine mesenchymal/ stem cell fate in mammary epithelial cells also activate other EMT-TFs. The maintenance of this state is achieved by autocrine signaling from the same pathways, while their endogenous inhibitors are suppressed [31]. Such EMT events derive their origin in the primary tumor and are dependent on cues from the immediate microenvironment. However, this does not preclude the existence of a subpopulation of CTCs which undergo EMT after entering the peripheral circulation.
Clarification of this issue requires examination of two proposed metastatic models that involve EMT: the classic mesenchymal–epithelial transition (MET) model, and the collective migration model. The second model postulates the collaboration of mesenchymal-like with epithelial-like cells to formulate metastatic niches [19]. This model suggests an epithelial–mesenchymal plasticity, wherein CTCs adapt to different microenvironments with time- and spatially regulated dynamic interconversions between epithelial and mesenchymal states [21, 32, 33]. CTCs in the circulation undergo EMT as part of a cellular response to transforming growth factor-beta (TGF-β), following the release of the latter from activated platelets [34]. Similar spatial heterogeneity exists in CTCs of hepatocellular carcinoma, in which the epithelial phenotype was predominant at release; however, CTCs were then converted into a more mesenchymal phenotype during EMT in a Smad2- and β-catenin-dependent signaling [35]. Thus, it is likely that EMT events are initiated at the primary tumor site to induce dissemination of CTCs but continue within the circulation as a mechanism for CTC survival, which, in turn, is crucial for eventual metastasis.
Circulating tumor cell dissemination: individual cells, clusters, or both?
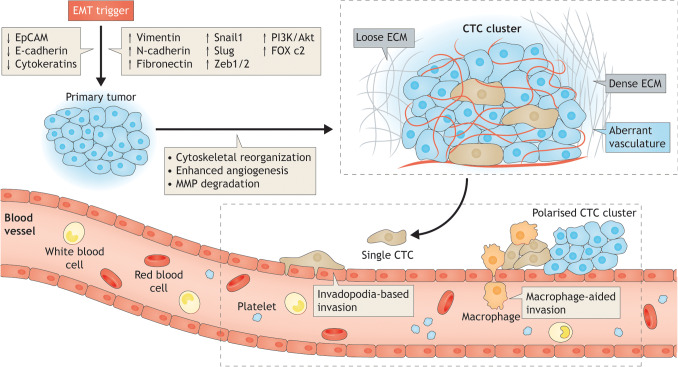
CTCs enter the circulation from a primary tumor either after an EMT event as individual cells or break off from the original cancer site as clusters of 2–50 cancer cells [3] (for a review on the mechanisms leading to the formation of CTCs clusters, please see Giuliano et al. [8]). They may also originate from primary or secondary metastatic sites in subsequent events. Very little is known about the exact triggers and the precise modes of CTC generation. One concept is cell jamming, wherein extracellular matrix (ECM) density correlates with the type of CTCs generated. Higher ECM density was associated with the generation of CTCs with a more mesenchymal phenotype and a collective migration mode; whereas, a low ECM density gave rise to single cells in an in vitro model [36]. In either case, access to the circulation could be via the porous vessels of the rapidly growing tumor cell mass [8]. Alternatively, CTC clusters might reach the circulation via invadopodia-based or macrophage-dependent mechanisms [37]. It has also been reported that, in principal, CTC clusters stem as oligoclonal precursors from the original cancerous site, and not through intravascular aggregation events; this could strengthen the evidence that single cells derived from the primary tumor do not aggregate within the circulation [3, 38]. A schematic diagram of the process of CTC dissemination from an established primary tumor is shown in Fig. 1.
Fig. 1.
How do CTCs disseminate from the primary tumor? Mechanisms behind CTC dissemination: the dissemination of CTCs from a primary tumor is triggered by a complex cellular process described as epithelial–mesenchymal transition (EMT). Cancer cells undergo EMT or adopt a stem cell-like fate by downregulating epithelial proteins and upregulating mesenchymal or stem cell markers. EMT markers reorganize the cytoskeleton, promote angiogenesis and express MMPs to facilitate degradation of surrounding tissue. The exact trigger for CTC dissemination is unknown. One possible trigger is termed cell jamming, where low extracellular matrix (ECM) density gives rise to single CTCs, while higher ECM density is associated with CTCs with a more mesenchymal phenotype, generating hybrid CTC clusters. Access to the circulation is likely through the aberrant vasculature of the primary tumors or sometimes via invadopodia-based or macrophage-dependent mechanisms. Both single cells and leading cells in hybrid CTC clusters exhibit a polarized phenotype, which is linked to their enhanced adhesive, migratory and metastatic capabilities. An understanding of the mechanisms behind CTC dissemination can aid in targeting specific steps in this cascade and preventing the development of metastases
Detection of individual CTCs has improved with advances in technology and the identification of specific cell surface markers [39–44]; however, this progress does not extend to the detection of CTC clusters. Technical limitations—such as the relatively low number of CTC clusters in circulation, and the possibility of dissociation and loss of cell viability during isolation, combined with limited knowledge about the various heterogenous CTC clusters—have made the detection and isolation of these aggregates challenging [8, 45]. Another possible barrier to efficient CTC cluster detection is the phenomenon of cloaking, wherein CTCs are covered by platelets, macrophages, or stromal cells [4, 46]. Although association with platelets within the circulation or stromal cells during collective migration helps CTC clusters survive the hazards of the immune system, it unfortunately hampers biomarker-based capture of CTCs. Recent advances in microfluidic devices have led to successful capture of CTC clusters, based mainly on size differences of the larger CTC clusters with single CTCs [46–49]. Additionally, the in vivo detection of CTC clusters using improved flow cytometry techniques has facilitated direct visualization of cancer progression in murine models [50]. Future improvements in the detection of CTCs in vivo, either as individual cells or clusters, will be invaluable for elucidation of their modes of generation and the development of strategies to target them at their source.
Circulating tumor cells or disseminated tumor cells: Where do they go once in the circulation?
The terms DTC and ITC [11] have been used interchangeably to encompass cells that have broken off from their parent tissue and are no longer subject to the controls of their original microenvironment. CTCs frequently home to bone marrow via the hematogenous route or to the lymph nodes via the lymphatic system, and they may remain dormant for years without forming metastases, followed by an active aggressive phase [51, 52]. The presence of DTCs is linked to increased risk of relapse and to poor prognosis, in particular following resection of the primary tumor [53]. A combination of EMT/MET transition, acquisition of cancer stem cell (CSC)-like traits and the angiogenic switch cause DTCs to emerge from their quiescent state [54, 55]. It is possible that DTCs might re-enter circulation through the aberrant vasculature in the secondary metastatic site [11]. However, the destination of DTCs following entry into the circulation and whether they should be considered to represent CTCs again are unclear. Likewise, it is still unknown if CTCs could act as attractants of other CTCs, once establishing themselves into metastatic niches; this notion, so far relevant to inflammation, could be mediated by complex cytokine or other molecular networks.
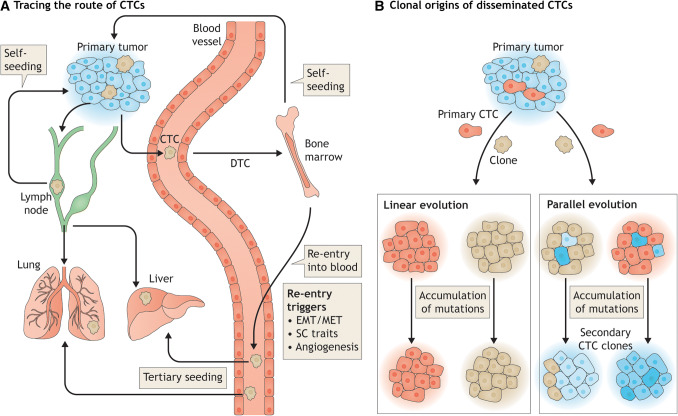
There are two possible routes for DTCs/CTCs once they re-enter circulation: onwards to a tertiary metastatic site or back to the primary tumor during the so-called process of tumor self-seeding (see “Route B: back to the primary”, below) (Fig. 2a). The destination of a secondary DTC/CTC is difficult to predict, as it could be associated with several parameters, such as the location of re-entry, the direction of blood flow, and the presence or induction of a favorable niche in which to establish metastasis. We can, however, work backward to try and reconstruct the path of already disseminated secondary DTCs/CTCs.
Fig. 2.
Where do CTCs go after entry into the circulation? a Tracing the route of CTCs: CTCs originating from a primary cancer site, enter the circulation, and migrate to a permissive secondary site in a distant location; these CTCs in a secondary target organ are then designated as disseminated tumor cells or DTCs. Most CTCs migrate to the bone marrow through the blood or to the lymph nodes via the lymphatic system, where they may remain dormant for prolonged periods without forming metastases, followed by an active aggressive phase. This phase can be triggered by a combination of EMT/MET transition, acquisition of stem cell traits and the angiogenic switch, which together lead to the development of overt metastases or the re-entry of CTCs/DTCs into the circulation. Following entry into the circulation, CTCs either migrate to a tertiary metastatic site (such as the lung or the liver) or return to the primary tumor, in a phenomenon termed tumor self-seeding. b Clonal origins of disseminated CTCs: Two models of clonal evolution are proposed prior to CTC dissemination. In the first model, CTCs undergo linear evolution, sharing genetic signatures with the parent tumor. In the second model, CTCs undergo parallel evolution, with early dissemination and the acquisition of newer mutations during the process, thus generating genetic signatures that differ vastly from that of the parent tumor. Analysis of disseminated CTCs can provide details of their origins, thus aiding our understanding the prognosis, as well as, the development of tailored therapies
Route A: to a ‘tertiary’ metastatic site?
CTCs and DTCs exhibit unique differences in their signatures due to adaptation of DTCs to the soil environment via clonal evolution. In the ‘seed and soil hypothesis’, soil means a subsequent secondary target organ to which the CTCs metastasize. Two models of evolution have been proposed, with one suggesting that DTCs disseminate early and undergo parallel/divergent evolution [56, 57]. Tracing the origin of such divergent DTCs might prove to be more difficult. Reiter et al. analyzed inter-metastatic genetic heterogeneity and mapped cancer phylogenies in treatment-naïve metastatic patients. Through mathematical modeling, it was shown that there was very little genetic heterogeneity in tumors with high dissemination rates because metastases are established before subclones in the primary tumor have had a chance to expand. The study, however, had several limitations, as it focused only on single-nucleotide substitutions or small indels (insertions/deletions), but not on copy number aberrations or modifications in the epigenome. In addition, they were unable to analyze undetectable micro-metastases [58]. Minimally invasive biopsy of DTCs and their genetic analysis from the bone marrow or lymph nodes might fill this gap.
In contrast, in another model, it has been proposed that DTCs evolve in a linear fashion, retaining much of the genetic signature of the primary tumor [59]. Breast cancer DTCs isolated 3 years post-diagnosis were shown to arise from lymph node metastasis, and they were identifiable via a combined analysis of copy number aberrations through single-cell sequencing and phylogenetic reconstruction. The original cancerous site and the metastatic lesion in the lymph node were distinct but still evolutionarily related, allowing the DTC origins to be traced [59]. These results highlight the value of DTCs for tracing their origins and, hence, developing appropriate therapeutic regimens. Interestingly, it is tempting to theorize that based on the above results, the possibility of tertiary metastases could exist. This term intends to describe the possibility of a metastasis from a secondary organ to another tertiary organ, considering the origin of tumor as primary source (Fig. 2b). Nonetheless, extended mathematical modeling would be required to affirm this hypothesis.
Route B: back to the primary tumor?
In a seminal paper in 2009, Kim et al. introduced the concept of tumor self-seeding, suggesting that dissemination of CTCs was, in fact, bi-directional. Given the aberrant vasculature of tumors and the fact that CTCs might not need to adapt to an unfamiliar microenvironment, the authors tested the hypothesis that CTCs might re-infiltrate an established tumor [60]. In contra-lateral seeding experiments in mice, CTCs shed into circulation seeded an established tumor mass with high efficiency. The authors also demonstrated that the metastatic cell progeny (which is known to mediate organ-specific metastasis with distinct gene sets) preferentially seeded the established tumor, irrespective of their organ tropisms [60]. This surprising discovery indicated that not all the genes in the organ-specific metastatic sub-populations are crucial for tumor self-seeding. It could also suggest that the recipient tumor mass may imitate a multiorgan environment having the ability to select all metastatic variants on the same time [61]. Additionally, these aggressive cells seeded only established tumors and not intact mammary glands. The latter results indicated that tumor-derived signals may be necessary to attract CTCs. Indeed, Kim et al. showed that IL-6 and IL-8 function as chemo-attractants for self-seeding CTCs; in turn, tumor infiltration by these CTCs is mediated by MMP-1 and fascin-1. In addition, invading CTCs were also shown to promote tumor growth rate by enhancing angiogenesis and recruiting favorable stromal components [60]. Others have also observed the self-seeding phenomenon, and they evaluated approaches, such as suppressing tumor-derived IL-6 or enhancing the endothelial barrier, which could block the return of the CTCs to their primary tumor [62, 63].
It is conceivable that this self-seeding model also applies to metastatic tumors, as well as, cross-infiltration of other tumor types following primary tumor resection. Kim et al. confirmed this phenomenon in extremely rare cases, in which a second tumor was present [60]. Under experimental conditions, it is difficult to determine whether a CTC has returned to the primary tumor site without first seeding a metastatic site (primary seeding), or instead, it has returned as a case of secondary seeding from metastatic tumors. Using mathematical models, Scott et al. showed that the possibility for secondary seeding is many folds higher than that for primary seeding [64]. CTCs must be responsive to cytokine signals from the primary tumor [64]. Strong reduction in CTC numbers due to filtration at capillary beds and the limited diffusional transport of cytokine signals make it impossible for upstream CTCs to sense these signals, suggesting that primary seeding is highly unlikely [63]. In contrast, secondary seeding avoids the filtration step, and, furthermore, CTCs might gain additional aggressive mutations during clonal expansion, thereby increasing the self-seeding potential [64].
The tumor self-seeding phenomenon could explain many of the mysteries of cancer metastasis [65]. Chief among them is how an apparently non-invasive benign lesion gives rise to an invasive lesion in a distal part of the same organ. This scenario has been observed in pancreatic intraepithelial neoplasia (PanIN), which need not be a spatially localized lesion, but whose spreading occurs throughout the organ’s ductal system [66]. It can be speculated that the PanIN lesions shed CTCs into the fluid of the pancreatic ductal system, resulting in the eventual development of pancreatic ductal adenocarcinoma (PDAC). Mutational and phylogenetic analysis revealed that PDAC and PanIN lesions derived from the same patient shared common ancestry and accumulated driver mutations resulting in cancer progression [66]. Alternatively, it is possible that CTCs use an entirely different mechanism, such as venous invasion (which is relatively common in pancreatic cancer), to replace the endothelial cells and grow along the inner walls of the vessels [67]. Isolating CTCs from pancreatic fluid or from pancreatic veins to confirm this phenomenon might at present be challenging, but not an impossibility. Under all circumstances, these observations may be unique to pancreatic cancer and, thus, not generalizable to other cancer types.
Similar localized metastatic expansion into the leptomeninges has been observed in medulloblastoma, a malignant embryonal tumor of the developing cerebellum. CTCs were thought to be shed from the original site of neoplasia into the cerebrospinal fluid (CSF) and to expand locally in the leptomeninges. However, recent evidence from mouse models revealed that CTCs from primary medulloblastoma were shed into the blood circulation and homed back to the leptomeninges [68]. Interestingly, CTCs or ctDNA isolated from metastatic medulloblastoma patients harbored genetic signatures that were either specific to the metastatic compartment alone or shared between both the primary and metastatic compartments [68]. Collectively, these results provide further evidence in support of a self-seeding phenomenon.
Finally, integrated RNA and DNA sequencing of tumor tissues in patients with breast cancer showed a diversity in the patterns of spreading and seeding; these observations supported both the linear and parallel evolution models described earlier [69]. That being said, higher sequencing depth would have been necessary to derive conclusions on more complex self-seeding processes that could have taken place [69]. Single-cell DNA sequencing techniques in combination with bulk exome and targeted deep-sequencing were more successful in distinguishing self-seeding versus early dissemination models of metastasis in two colorectal cancer patients, although the authors found no empirical evidence of self-seeding [70]. Observation of this phenomenon in humans is likely hampered by (a) the rarity of the self-seeding phenomenon, (b) the lack of biomarkers that differentiate self-seeding CTCs from other CTC types, and (c) the challenges in deep-sequencing of single CTCs.
Are circulating tumor cell clusters one step ahead in the metastasis race?
Many early preclinical studies have demonstrated a greater metastatic potential for CTC clusters compared to individual CTCs [71]. Suo et al. reported a dramatically increased number of CTC clusters compared to single CTCs during the later stages of disease in mouse models of prostate and liver cancer [50]. CTCs employ different mechanisms to enable survival within the blood circulation, as well as, acquire tumor-initiating capabilities, which contribute to the enhanced metastatic potential of a small subset of CTCs. Differences in physical characteristics, EMT status, and genetic profiles of CTC clusters compared with single CTCs are some examples of potential mechanisms [3, 33, 72] (Table 1).
Table 1.
Mechanisms contributing to enhanced metastatic potential of circulating tumor cells
| Mechanism | Description | References |
|---|---|---|
| Association with non-tumor cells | ||
| Tumor stromal cells (mainly activated fibroblasts) | Increase viability of circulating cancer cells, survival in blood circulation and at secondary metastatic site, and enhance metastatic potential | [73] |
| Endothelial cells | Enhance neo-angiogenesis, tumor growth, and metastatic capacity of primary tumors originating from tumor-endothelial spheroids | [74] |
| Platelets | Protect cancerous cells from shear stress-induced blood damage and attacks from the immune system | [71] |
| Leukocytes | Enhance metastasis | [71] |
| Neutrophils | Upregulate pro-tumorigenic genes of the cell cycle and DNA replication programs, and enhance metastatic potential | [75] |
| Collective migration | CTC clusters migrate as a collection of cells with epithelial and mesenchymal signatures, undergoing dynamic interconversion and enhancing metastasis | [3, 6, 33] |
| Upregulation of pro-metastatic genes and proteins | ||
| Adherens junction proteins | Plakoglobin and E-cadherin facilitate CTC cluster formation and contribute to enhanced metastasis | [3] |
| Keratin-14 positive CTCs engage in collective polyclonal invasion, upregulate desmosome and hemidesmosome complex genes, and enhance metastasis by modulating multiple metastasis effectors | [76] | |
| Carbohydrate antigens | CTCs aggregate at the endothelial surface via attachment to β-galactosidase-binding protein galectin-3, enhancing survival and metastatic growth | [38] |
| Pro-inflammatory cytokines | IL-6 and TNF-α lead to CTC aggregation and recruitment to inflamed endothelium, possibly contributing to enhanced metastatic potential, in breast cancer | [77] |
| Epigenetic control via DNA methylation | ||
| Tumor suppressors | CST6, BRMS1, and SOX17 encode proteins with potential tumor suppressor functions are methylated in CTCs and associated with tumor progression | [78] |
| Methylation of CST6, ITIH5, or RASSF1 is linked to poor survival outcomes in patients with metastatic breast cancer | [79] | |
| Apoptosis, angiogenesis, and VEGF signaling pathway proteins | Invasive CTCs from metastatic, castration-resistant prostate cancer display hypermethylation signals consistent with primary tumors in pathways implicated in tumor progression | [78] |
| Growth factors | Reduced methylation at the promoters of HGF and its receptor proto-oncogene c-MET were observed in cell lines created on the basis of CTCs isolated from a syngeneic murine hepatocellular carcinoma (HCC) mouse model | [78] |
| Stemness- and proliferation-associated transcription factors | Hypomethylation of OCT4, SOX2, NANOG and SIN3A binding sites in CTC clusters associated with worse prognosis in cancer of the breast | [80] |
CTC circulating tumor cell, IL-6 interleukin-6, TNF-α tumor necrosis factor-alpha, CST6 Cystatin M6, BRMS1 breast cancer metastasis suppressor 1, SOX17 SRY(sex-determining region Y)-box 17, ITIH5 inter-alpha-trypsin inhibitor heavy chain 5, RASSF1 Ras association domain family member 1, OCT4 octamer-binding transcription factor 4, SOX2 SRY (sex-determining region Y)-box 2, NANOG Nanog homeobox, SIN3A SIN3 transcription regulator family member A
In addition to active recruitment of supportive cells and accumulating genetic changes in favor of metastasis, the physical characteristics of the two CTC types could favor enhanced metastasis. Single CTCs have a relatively longer half-life (25–30 min) compared to CTC clusters in the circulation (6–10 min). This faster clearance rate of CTC clusters is associated with their size difference, entrapment within the microvasculature of distant organs, and subsequent initiation of secondary metastasis [3]. What exactly happens after CTC clusters are trapped in small capillaries has not been deciphered. According to current consensus, dissociation of CTC clusters is followed by extravasation across the vascular wall to a favorable niche in the secondary metastatic site. This involves loss of the advantages of multicellularity and cooperative interactions, suggesting that collective migration may be necessary only to traverse the circulation without any value in the equally hazardous soil environment (see description of seed and soil hypothesis above). A newly identified mechanism for extravasation, known as angiopellosis, which involves active remodeling of endothelial cells to expel circulating non-leukocytic cells, could help explain this conundrum [81]. Taking a cue from this exit strategy, the same group showed that CTC clusters maintain multicellularity during extravasation, which contributes to their enhanced metastatic potential in an in vivo zebrafish model [82].
Interestingly, Balakrishnan et al. showed that the poor patient survival and therapy response is predicted from the cluster phenotype, and not the presence of CTC clusters per se [83]. Three cluster phenotypes were identified based on cell density (very tight, tight, and loose) using a short-term culture method [83]. Within 12 weeks, tight clusters were associated with shorter overall survival and increased resistance to therapy [83]. Treatment response could also be monitored by dynamic changes in cluster phenotype from tight to loose clusters when therapy was favorable [83]. In another report, the creation of CTC clusters was inversely associated with higher levels of drug concentration and response to treatment, thus providing proof-of-concept for a robust anti-cancer drug screening system [84]. Such approaches could become tools for early prognosis and cancer treatment initiation in addition to improved monitoring of patient response and resistance to therapy.
Failure of targeted systemic therapies is a common phenomenon in breast cancer metastasis, possibly due to receptor conversion of progesterone receptor (PR), human epidermal growth factor receptor 2 (HER2), and estrogen receptor alpha (ERa) in the distant metastatic tissue compared to the primary tumor [85]. Such conversion could be the result of evolutionary pressure from local downregulation of receptor ligands induced by systemic hormone therapies [86]. Routine biopsy of distant metastases is impractical and challenging. CTCs may help predict receptor status after therapy and guide therapy decisions. The evidence for CTC receptor status before and after therapy is conflicting, either showing perfect concordance with the primary tumor or conversion [87–89]. Jordan and colleagues demonstrated the existence of discrete subpopulations of HER2-positive and HER2-negative cells in patients undergoing HER2-targeted therapy. These sub-populations had evolved to activate multiple signaling pathways and to undergo dynamic interconversion, thus acquiring drug resistance, and, in turn, the ability to establish metastasis more rapidly. This mixed population of cells was eliminated by simultaneous treatments targeting multiple pathways combined with systemic chemotherapy [88]. Similarly, CTC genotyping has been used as a pharmacodynamic monitoring tool to provide mechanistic insights into resistance to endocrine therapy [90]. If advances in CTC enumeration enable routine monitoring of CTC receptor status, real-time biopsy could become a reality for querying dynamic changes within metastatic CTCs to help inform decisions rather than resort to mere watchful waiting.
Unraveling the mechanisms that underlie the potential of CTC clusters to initiate secondary metastasis may help us developing approaches that could (a) reverse EMT in CTCs, (b) prevent stromal environment remodeling, (c) target specific proteins involved in CTC cluster formation, or (d) modulate DNA methylation patterns in CTCs; all of these are vital for arresting metastasis. In addition, global gene expression profiling of individual CTCs or clusters (alone or with other stromal and/or blood components) would also be essential to identify multiple key pathways leading to overt metastasis, and ultimately to develop targeted therapies.
In light of the translational medicine nature of the field, describing the mechanistic insights provided by CTC should be jointly discussed with the increasing value of CTC in clinical practice. Indeed, CTC research in the past decade has delineated the value of liquid biopsy in clinical oncology. Therefore, in the subsequent sections, we present the current evidence on the clinical relevance of CTCs. We also discuss how ctDNA analysis may be more informative in deducing the mechanisms of CTC dissemination and metastatic potential, especially once complemented by traditional imaging modalities and emerging computational, notably machine learning-based, approaches.
Current evidence for the clinical relevance of liquid biopsy
Liquid biopsies, defined as the analysis of blood to detect circulating tumor cells (CTCs) or circulating tumor DNA (ctDNA), can be helpful for screening, diagnosis, and prognosis, as well as, predicting early relapse, treatment response, and resistance of many solid cancers [91–98]; however, the precise frequency and time points for CTC examination during the follow-up of patients with cancer should be the objective of future clinical studies. For example, the value of liquid biopsy in the therapeutic approaches of small-cell lung cancer (SCLC) cannot be overstated. In this cancer type, obtaining tumor biopsies, in particular during relapse and after chemotherapy, is a major obstacle; hence, having more immediate access to CTCs and ctDNA can yield valuable information to guide SCLC treatment [99]. In the same context, the clinical relevance of CTCs was underscored by the United States Food and Drug Administration (US FDA) approval in 2008 of the CellSearch® CTC detection platform; this platform referred to monitoring disease and prognosis of patients with colon, breast, and prostate cancer. This technology can be used to predict survival probability and monitor treatment with a threshold of three CTCs/7.5 ml of blood for metastatic colorectal cancer, and five CTCs/7.5 ml blood for patients with breast cancer and metastatic prostate cancer [100–102]. Other studies have used the same CTC cut-offs to predict risk of relapse and therapy response [103–106]. Moreover, CTC clusters are linked to poor prognosis and therapeutic resistance in many different cancer types [107–109]. Detection of CTCs is linked to aggressive disease progression, reduced overall, as well as, disease-free survival, and increased mortality; it can also predict likelihood of relapse and treatment response. Particularly regarding prostate cancer, detection of the androgen-receptor isoform encoded by splice variant 7 (AR-V7) in CTC has been associated with resistance to enzalutamide and abiraterone. According to experts, this biomarker might have prognostic and plausible predictive role; however, its clinical utility has not been proven for routine use in clinical practice [110, 111]. Moreover, CTC numeration with the CellSearch® platform was embedded in phase III trials with the aim to reduce CTC count from detectable to 0, or from above 5 to below 5, as a surrogate biomarker for overall survival in metastatic prostate cancer, with promising results so far [112, 113]. Table 2 describes the accumulating evidence for the prognostic value of CTCs in various types of cancers.
Table 2.
Prognostic significance of CTCs in various cancer types based on published data from meta-analysis of CTC prognostic studies between 2010 and 2019
| Type of cancer | No. of studies (patients included) | Detection method | Prognostic parameter | HR/RR/OR (95% CI) | References |
|---|---|---|---|---|---|
| Colorectal cancer | 20 (3687) | RT-PCR, CellSearch, Epispot, CMx platform | Aggressive disease progression | 2.57 (1.64–4.02) | [118] |
| Reduced survival | 2.41 (1.66–3.51) | ||||
| 15 (3129) | RT-PCR, CellSearch, flow cytometry, membrane array | Mortality | 2.36 (1.87–2.97) | [119] | |
| Aggressive disease progression | 1.83 (1.42–2.36) | ||||
| 12 (2363) | RT-PCR | Overall survival | 3.07 (2.05–4.62) | [120] | |
| Disease-free survival | 2.58 (2.00–3.32) | ||||
| 11 (1847) | CellSearch | Overall survival | 2.00 (1.49–2.69) | [121] | |
| Disease-free survival | 1.80 (1.52–2.13) | ||||
| Breast cancer | 21 (2030) | CellSearch | Overall survival | 0.19 (0.10–0.31) | [122] |
| Distant disease-free survival | 0.25 (0.16–0.38) | ||||
| Loco-regional relapse-free interval | 0.49 (0.29–0.80) | ||||
| 4 (334) | HER2 + CTC detected by CellSearch, Veridex, RT-PCR | Overall survival in patients without metastasis | 2.27 (1.34–3.85) | [96] | |
| Progression-free survival in patients without metastasis | 2.87 (1.30–6.34) | ||||
| 49 (6825) | CellSearch, RT-PCR, other ICC | Disease-free survival (early stage) | 2.86 (2.19–3.75) | [123] | |
| Overall survival (early stage) | 2.78 (2.22–3.48) | ||||
| Progression-free survival (breast cancer with metastasis) | 1.78 (1.52–2.09) | ||||
| Overall survival (breast cancer with metastasis) | 1.78 (1.52–2.09) | ||||
| Lung cancer | 27 (2615) | RT-PCR, ISET, CellSearch | Overall survival (pre-treatment) | 2.61 (1.82–3.74) | [124] |
| Overall survival (post-treatment) | 4.19 (2.92–6.00) | ||||
| Progression-free survival (pre-treatment) | 2.01 (1.24–3.25) | ||||
| Progression-free survival (post-treatment) | 4.97 (3.05–8.11) | ||||
| 8 (453) | OBP-401 assay, quantitative RT-PCR, FISH, CellSearch, Cytelligen | Disease control rate (pre-treatment) | 2.56 (1.36–4.82) | [125] | |
| Disease control rate (during chemotherapy) | 9.08 (3.44–23.98) | ||||
| Objective response rate (during chemotherapy) | 1.72 (1.27–2.32) | ||||
| Pancreatic cancer | 13 (855) | Immunodetection, RT-PCR, CellSearch | Overall survival | 1.84 (1.37–2.45) | [126] |
| Disease-free survival | 1.93 (1.19–3.11) | ||||
| Prostate cancer | 33 (4170) | Immunohistochemistry, RT-PCR, CellSearch | Overall survival | 2.43 (2.07–2.86) | [127] |
| Disease-free survival | 2.15 (1.69–2.73) | ||||
| 10 (1206) | Immunodetection | Overall survival | 2.76 (2.28–3.34) | [128] | |
| 4 (486) | Immunomagnetic detection | Overall survival | 2.51 (1.96–3.21) | [103] | |
| Gastric cancer | 16 (1110) | CellSearch, ICC, FACS-ICC, MetaCell ICS, flow cytometry | Overall survival | 2.23 (1.86–2.66) | [129] |
| Progression-free survival | 2.02 (1.36–2.99) | ||||
| Esophageal cancer | 18 (1719) | RT-PCR, CellSearch, Immunofluorescence, flow cytometry, immunohistochemistry, immunomagnetic enrichment | Progression-free survival | 2.61 (2.08–3.28) | [130] |
| Overall survival | 2.50 (2.12–2.94) | ||||
| Relapse | 2.84 (1.81–4.44) | ||||
| Chemotherapy response | 0.64 (0.43–0.96) | ||||
| Head-and-neck cancer | 6 (429) | RT-PCR, CellSearch, ICC, flow cytometry | Recurrence-free survival | 4.88 (1.93–12.35) | [131] |
CTC circulating tumor cell, HR hazard ratio, OR odds ratio, RR risk ratio, RT-PCR real-time polymerase chain reaction, ICC immunocytochemistry, ISET isolation by size of tumor cells, FISH fluorescence in situ hybridization, FACS florescence activated cell sorting
Nonetheless, and despite the efficacy of molecular oncology approaches (including CTCs), it is disappointing that their availability for patient care is far from being optimal [114]. At this point, it is important to make a technical note, i.e., that CTCs which have undergone EMT may not be included in total CTC counts based on cell surface epithelial marker detection, resulting in underestimation of the risk of relapse. As noted in [115], combining total CTC count and the proportion of MCTCs detected seems to be a better predictor of therapeutic resistance and prognosis in metastatic breast cancer; this indicates that using multiple detection/isolation technologies simultaneously to enrich the CTC population provides enhanced prognostic value [116]. The advances in CTC detection and/or enrichment technologies have been comprehensively reviewed [39, 40, 42, 117]; however, it remains to be seen which of these technologies complement each other to provide more complete actionable knowledge.
If not circulating tumor cells, can the parallel universe of cell-free circulating tumor DNA inform us better?
In humans, circulating cell-free DNA (cfDNA) can be found principally in plasma and urine, with a miniscule proportion represented by circulating tumor DNA (ctDNA), which arises from the death of circulating tumor cells [92]. The ctDNA has not been shown to induce antibodies, as, in general, DNA per se is not known to be immunogenic; whereas, its origin has been attributed to (a) lysis of CTCs, (b) DNA spontaneously released from either primary or/and metastatic cancer into the circulation, and, (c) tumor necrosis or apoptosis leading to cell-derived leakage of DNA [132]. ctDNA analysis for non-invasive genomic profiling has been applied in several cancer types [133–138]. Advances in CTC detection and enumeration have been accompanied by an increasing interest in ctDNAs and the plasma cell-free DNA methylome (for a discussion on cancer detection technologies focusing on DNA methylation, which exceeds the scope of this review, see Sina et al. [139]); however, improvements in the clinical validity of different detection methods should be made to reach clinical prime time [40, 140]. To our best knowledge, most studies assessing the degree of concordance between CTC and ctDNA have reported, in general, a positive association between these two methods; for instance, they present a joint effect (< 17-fold) in patients with metastatic breast cancer [141–142].
These advances have been also characterized by efforts to standardize ctDNAs in terms of their analytical validity [143], as well as, the creation of several international collaborative projects and training programmes (such as the European Liquid Biopsy Academy) to further advance the transition of the liquid biopsy applications to clinical practice. The ability to enrich ctDNAs from the wider pool of cfDNA based on fragment length has facilitated their analysis [144].
On the one hand, key advances, such as the CancerSEEK platform, allow detection of somatic mutations and protein biomarkers in liquid biopsies with exquisite specificity to localize the organ of origin in early stage cancers [145, 146]. Other similar approaches, such as targeted error correction sequencing (TEC-Seq), PapSEEK, and UROSEEK, are also used to screen early stage cancers. Somatic mutation detection, however, is plagued by low sensitivity, especially in early stage cancers [147–149]. Moreover, such results should be validated in prospective studies (and not merely case–control studies), and among population groups with different prevalence of cancer; doing so is necessary because overestimations in sensitivity, specificity, and positive or negative predictive values may be otherwise produced. Furthermore, the identification of cancer-associated mutations in normal (notably aging) tissues has posed questions on the axiom that these mutations are specific to neoplastic tissue, and it could represent a major challenge for the ctDNA-based assays because of potential false-positive results [150]. Last, the position of these assays in the biomarker armamentarium should be re-considered in light of the so-called tissue-agnostic therapies (such as pembrolizumab, larotrectinib, and entrectinib). These therapies focus on the drug target and not the tissue of origin; for example, entrectinib is a tyrosine kinase inhibitor targeting ROS1, ALK receptor, and the three Trk proteins and is FDA-approved against solid tumors, harboring NTRK gene fusions, regardless of tumor origin [151–153].
On the other hand, techniques for investigating tissue- and cancer-type-specific epigenetic alterations based on plasma-free DNA methylomes may be more robust for cancer detection and classification [43]. Other tumor-derived circulating components, such as exosomes, CTC-related microRNAs, piRNAs, surrogates of immunologic cell death, as well as, circulating hybrid cells that can be detected in liquid biopsies, seem to also have clinical relevance [154–158].
Currently, ctDNA is analyzed to identify the mutational landscape (and, in turn, predict treatment response) or to allow early detection of relapse; doing so is assisted with the use of sensitive PCR-based methods or second-generation sequencing [159]. Mutational analysis of ctDNA, however, provides only weak clues about their tissue origins [160]. Typically, ctDNAs occur at sizes of approximately 160 to < 200 bp, and they are fragmented or associated with nucleosomes. Snyder et al. attempted to elucidate the in vivo nucleosome footprint of ctDNAs by mapping nucleosome occupancy through deep-sequencing of cfDNA. They matched the epigenetic signature of ctDNA fragmentation patterns with gene expression and regulatory site profiles from reference datasets of diverse cancer types. Thus, they were able to non-invasively identify the anatomical origins of a patient’s cancer [160]. If matched primary and/or metastatic tumor genotypes are available, this method may help to obtain a better understanding of the molecular profile during the metastatic journey of their cancer. Recently, assays have been developed that focus on circulating cell-free nucleosomes, which are more stable than ctDNAs. The scientific basis behind these assays lies on detecting the differences in certain histone (e.g., H3.1) variants and the associated acetylation patterns between patients and healthy controls [161].
DNA evaluation of fragments for early interception (DELFI) is another approach that allows differentiation between healthy and tumor-derived ctDNA based on genome-wide analysis of fragmentation patterns [162]. The authors showed that these patterns could not only be used as biomarkers for cancer detection, but also to monitor patients during therapy. Coupled with machine learning, the DELFI method was also used to identify the tissue of origin of ctDNA with high sensitivity (75%). This approach was also successfully combined with traditional mutational analysis to enhance the sensitivity of cancer detection [162].
A recent comprehensive, integrative, and molecular analysis of over 10,000 tumors representing 33 types of cancer from The Cancer Genome Atlas (TCGA) demonstrated strong cluster patterns of anatomic origin, tissue type and histology. This study suggested that the patterns describing cell of origin are key deterministic patterns of cancer progression [163]. These data indicated that the molecular analysis of CTCs and ctDNAs should not only encompass mutation detection, e.g., using recently developed methods such as nucleosome mapping; rather, this analysis should entail other complementary approaches allowing the clustering of molecular data based, e.g., on DNA methylation levels, structural variations at the chromosomal level, miRNA expression levels, and so on, in order to make more well-informed decisions on effective cancer treatment. In addition, a recent thorough investigation of the mutational landscape in metastatic tumors revealed that (a) at least in more than half of lesions examined, whole-genome duplication events were present, (b) four out of five tumor-suppressor genes were inactivated, and, (c) almost all (> 95%) driver mutations were clonal [163].
The concept of a tissue of origin for CTCs and ctDNAs does not apply only to solid tumors [164]; for instance, what about lymphoproliferative cancers such as chronic lymphocytic leukemia (CLL)? Since CLL cells are themselves CTCs, their identification and monitoring of their clonal evolution represent a challenge. In around 2–10% of CLL patients, CLL is known to become transformed to a rather aggressive form of diffuse large B-cell lymphoma (DLBCL), which is named Richter’s transformation or Richter’s syndrome (RS). RS can be triggered by germline, clinical, or certain biological characteristics of CLL and sometimes by CLL therapy [165]. Currently, there is a lack of markers that are sufficiently sensitive and specific to differentiate between circulating CLL and DLBCL. Yeh et al. analyzed ctDNA using a combination of whole-genome sequencing and whole-exome sequencing of several clinical samples at baseline and after transformation to RS. Copy number alterations and single-nucleotide variants that are representative of the transformation event under the selective pressure of therapy were identifiable; these features might be useful for early identification of treatment failure in CLL [166]. In light of these findings, it might be worth speculating on the mechanistic clues that CTCs as exemplar could provide regarding RS. Could RS be a cellular dysregulation harboring modifications of immune cell repertoires, which could be captured by prospective analysis of an adequate number of samples from CLL patients at single-cell immuno-sequencing level? Or, could RS be a phenomenon implicating mechanobiological alterations, where CLL cells undergo changes, e.g., into their cytoskeleton, to resemble those in lymphoma, which, in turn, affect gene expression (via mechanobiological pathways)? In the latter cases, detecting the morphological changes of CLL-transforming cells would be crucial.
As far as brain tumors are concerned, detection of ctDNA in the blood of primary brain tumor patients remains challenging due to morbidity and cost constraints. However, tracing glioma evolution through analysis of ctDNA derived from the CSF of primary brain tumor patients is possible, and it can provide a comprehensive and genetically faithful representation of the tumor at the time of CSF collection [167]. Moreover, a recent method using the so-called circulating tumor-specific fluorescent extracellular vesicles was developed; these extracellular vesicles can be detected in the blood of brain cancer patients after consuming the imaging agent 5-aminolevulinic acid (5-ALA) [168]. Newer techniques, such as photoacoustic flow cytometry, may allow for in vivo diagnosis of circulating CSF markers to better judge leptomeningeal and brain metastasis [169]. Last, future studies should focus on how CTCs could interact with the recently described glymphatic (i.e., glial-lymphatic) and meningeal lymphatic systems, in particular, how the latter contribute to the dissemination of brain tumor CTCs in cervical lymph nodes [170, 171].
Support for circulating tumor cells analysis: complimentary approaches
Imaging methods including computed tomography (CT), magnetic resonance imaging (MRI), positron emission tomography (PET), and more recently, [18F]fluorodeoxyglucose PET/CT (FDG-PET/CT), and FDG-PET/MRI are increasingly used in clinical practice for cancer detection and prognosis, as well as, prediction of treatment responses [172].
Deciphering patient’s intratumoral and intertumoral heterogeneity during the initial stages of cancer formation is important for prognosis and choice of appropriate treatment [173–175]. To this end, the approach of single-cell genomics of CTCs could be helpful [176]. Furthermore, radiogenomics, which combines radio-imaging features with specific gene expression patterns, might prove to be a cost-effective complement to CTC analysis [177, 178]. Recognition of imaging traits that accurately predict primary tumor behaviour could improve prognosis, together with associating these traits with CTCs released very early in the metastatic process. Such associations between many types of imaging techniques and CTC detection using both non-EpCAM and EpCAM-based methods have already confirmed this as a viable complimentary approach [172, 179, 180]. The potential of emerging PET/MRI technology, which offers more soft tissue contact and less ionizing radiation exposure, as a similar complementary approach remains to be explored [181, 182].
Cytopathologic characterization of tumor biopsy specimens is routine in clinical practice. A principal example of point-of-care cytopathological diagnostics is the Papanikolaou (Pap) smear for cervical cancer diagnosis, which is still valid 90 years after its invention due to its simplicity [183]. However, not all cancer types are easily diagnosed with non-invasive techniques. With improvements in CTC detection methodologies, the so-called enrichment-free immunofluorescent staining with fiber-optic array scanning technology (FAST) has facilitated detailed cytomorphologic analysis [184, 185]. Pleiomorphic CTCs in the circulation of patients with metastatic breast and colorectal cancer were identified using this approach. These included (a) CTCs with both high and low nuclear–cytoplasmic ratio (N/C ratios), (b) varying degrees of apoptotic CTCs, and (c) CTC clusters consisting of mixed populations. In all cases, the sum of morphological types was revealed by comparing CTCs from primary and metastatic tumor samples, thus disabusing the notion of morphologically distinct clonal sub-populations [184, 185]. Recently, deep-learning algorithms that consider abnormal cell segmentation in Pap smears (and the frequency with which these clumps overlap with free-lying cells) have been developed to support correct diagnosis of cervical cancer, through reliable quantitative evaluation against a database of cervical cytology images [186, 187]. It is possible that similar approaches could be combined with the FAST-based cytomorphologic analysis of CTCs for objective classification of CTC origins and dissemination patterns, thus guiding prognosis and treatment decisions.
Last, in the pursuit for early detection of cancer, liquid biopsy may be complemented, in the future, with the so-called breath biopsy; the latter will analyze the patterns of volatile organic compounds stemming from pathological cellular processes, after detecting these compounds in breath and urine using advanced analytical methods [188, 189].
Conclusions and future trends
Research on CTCs is a promising field for advancing early cancer detection (also known as secondary prevention). Clinical trials as the gold-standard method to test and validate biomarkers and to assess their clinical utility are expected to play a critical role. Exponential progress in technology has yielded major improvements in the detection, enrichment, analysis and application of CTCs that will provide a more comprehensive understanding of cancer and its diagnosis, prevention, and treatment. Coupled with the use of microfluidic technology for improved detection and enrichment, genomic analysis of CTCs and ctDNAs is expected to reach clinical prime time in the future. In parallel, mammalian acoustic reporter genes for ultrasound imaging may allow non-invasive visualization of CTC location and function [190]. Bioinformatic approaches, such as machine learning methods to aid cancer prognosis and prediction, are also gaining traction [191]. Tremendous insights have been gained, using high-throughput genomic approaches, into the mechanisms underlying early metastatic seeding and the potential for CTC generation and dissemination [192] (in principal, through epithelial–mesenchymal transition [193]); for example, the ground-breaking concept of tumor self-seeding. Parkins et al. have very recently exploited this concept to engineer self-homing CTCs as a so-called theranostics tool to primary as well as, metastatic lesions [194]. Thus, CTCs offer tremendous opportunities to understand, prevent, and treat cancer. Moreover, while the great majority of studies have so far focused on CTCs, recent interest has emerged for the so-called Circulating Ensembles of Tumor Associated Cells (C-ETACs). These include immune cells (whose role in immunotherapy has been proven pivotal), fibroblasts, and tumor emboli, and they are implicated in high metastatic potential [195]. Future studies should focus on how CTCs and C-ETACs could be considered together in terms of their clinical (i.e., diagnostic and prognostic) importance. In addition, single-cell sequencing approaches coupled with sophisticated phylogenetic approaches (e.g., Bayesian methods) and the recent genetic lineage tracing approaches could help elucidating the evolution of primary tumors to CTCs and, in turn, to metastatic cells; while, the emerging application of shallow tumor RNA-sequencing could be assessed in terms of its clinical utility and cost-effectiveness before reaching clinical prime time [196, 197, 198, 199].
Search strategy and selection criteria
Our search strategy included papers between 2010 and 2019, except for seminal papers describing key phenomena or major discoveries. The contents presented in Tables 1 and 2 were identified using the following search strings: {meta-analysis} AND {type of cancer} AND {prognosis} OR {prognostic significance} and {mechanisms} AND {metastatic potential} AND {CTCs}, respectively.
Acknowledgements
This study was not supported by any funding or grant scheme. AFAM is highly indebted to Anna S. Gkika for her continuous support during this study. AFAM would like to dedicate this study to the memory of late Prof. George Vlastos, M.D., Ph.D., who served as Head of the Senology Unit, University Hospitals of Geneva, Switzerland. Prof. Vlastos provided the initial inspiration to AFAM to work on this topic.
Author contributions
AFAM, PG, ED, NAR, and AGP conceived the study. AFAM, PG, and ED performed literature search. NAR provided critical clinical input to this study. AGP supervised this study. AFAM, PG, and ED prepared first draft. NAR and AGP revised the manuscript for important intellectual content. AFAM, PG, ED, NAR, and AGP approved the final version of the manuscript.
Compliance with ethical standards
Conflict of interest
AFAM, ED, NAR, and AGP report no financial or other conflicts of interest. PDG has provided unrelated consulting/advisory role for the following entities in the last 2 years: Merck, Bristol-Myers Squibb, AstraZeneca, Clovis Oncology, EMD Serono, Seattle Genetics, Foundation Medicine, Driver, Pfizer, QED Therapeutics, Heron Therapeutics, Janssen, Bayer, Genzyme, Mirati Therapeutics, Exelixis, Roche, GlaxoSmithKline. Also, he has received Research Funding at his Institution from the following entities: Pfizer, Clovis Oncology, Bavarian Nordic, Immunomedics, Bristol-Myers Squibb, and Debiopharm Group.
Ethical guidelines
Not applicable.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Nieva JJ, Kuhn P. Fluid biopsy for solid tumors: a patients companion for lifelong characterization of their disease. Futur Oncol. 2012;8:989–998. doi: 10.2217/fon.12.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yu M, Stott S, Toner M, Maheswaran S, Haber DA. Circulating tumor cells: approaches to isolation and characterization. J Cell Biol. 2011;192:373–382. doi: 10.1083/jcb.201010021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aceto N, Bardia A, Miyamoto DT, Donaldson MC, Wittner BS, Spencer JA, Yu M, Pely A, Engstrom A, Zhu H, et al. Circulating tumor cell clusters are oligoclonal precursors of breast cancer metastasis. Cell. 2014;158:1110–1122. doi: 10.1016/j.cell.2014.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen L, Bode AM, Dong Z. Circulating tumor cells: moving biological insights into detection. Theranostics. 2017;7:2606–2619. doi: 10.7150/thno.18588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gkountela S, Szczerba B, Donato C, Aceto N. Recent advances in the biology of human circulating tumour cells and metastasis. ESMO Open. 2016;1:1–9. doi: 10.1136/esmoopen-2016-000078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu M, Bardia A, Wittner BS, Stott SL, Smas ME, Ting DT, Isakoff SJ, Ciciliano JC, Wells MN, Shah AM, et al. Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science. 2013;339:580–584. doi: 10.1126/science.1228522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barriere G, Fici P, Gallerani G, Fabbri F, Zoli W, Rigaud M. Circulating tumor cells and epithelial, mesenchymal and stemness markers: characterization of cell subpopulations. Ann Transl Med. 2014;2:1–8. doi: 10.3978/j.issn.2305-5839.2014.10.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Giuliano M, Shaikh A, Lo HC, Arpino G, De Placido S, Zhang XH, Cristofanilli M, Schiff R, Trivedi MV. Perspective on circulating tumor cell clusters: why it takes a village to metastasize. Cancer Res. 2018;78:845–852. doi: 10.1158/0008-5472.CAN-17-2748. [DOI] [PubMed] [Google Scholar]
- 9.Massagué J, Obenauf AC. Metastatic colonization by circulating tumour cells. Nature. 2016;529:298–306. doi: 10.1038/nature17038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Follain G, Herrmann D, Harlepp S, Hyenne V, Osmani N, Warren SC, Timpson P, Goetz JG (2019) Fluids and their mechanics in tumour transit: shaping metastasis. Nat Rev Cancer. https://www.ncbi.nlm.nih.gov/pubmed/31780785. [DOI] [PubMed]
- 11.Bork U, Grützmann R, Rahbari NN, Schölch S, Distler M, Reissfelder C, Koch M, Weitz J. Prognostic relevance of minimal residual disease in colorectal cancer. World J Gastroenterol. 2014;20:10296–10304. doi: 10.3748/wjg.v20.i30.10296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alix-Panabières C, Pantel K. Characterization of single circulating tumor cells. FEBS Lett. 2017;591:2241–2250. doi: 10.1002/1873-3468.12662. [DOI] [PubMed] [Google Scholar]
- 13.Cortés-Hernández LE, Eslami-S Z, Alix-Panabières C. Circulating tumor cell as the functional aspect of liquid biopsy to understand the metastatic cascade in solid cancer. Mol Asp Med. 2019 doi: 10.1016/j.mam.2019.07.008. [DOI] [PubMed] [Google Scholar]
- 14.Baccelli I, Schneeweiss A, Riethdorf S, Stenzinger A, Schillert A, Vogel V, Klein C, Saini M, Bäuerle T, Wallwiener M, et al. Identification of a population of blood circulating tumor cells from breast cancer patients that initiates metastasis in a xenograft assay. Nat Biotechnol. 2013;31:539–544. doi: 10.1038/nbt.2576. [DOI] [PubMed] [Google Scholar]
- 15.Yao X, Choudhury AD, Yamanaka YJ, Adalsteinsson VA, Gierahn TM, Williamson CA, Lamb CR, Taplin ME, Nakabayashi M, Chabot MS, et al. Functional analysis of single cells identifies a rare subset of circulating tumor cells with malignant traits. Integr Biol (United Kingdom) 2014;6:388–398. doi: 10.1039/c3ib40264a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Castro-Giner F, Scheidmann MC, Aceto N. Beyond enumeration: Functional and computational analysis of circulating tumor cells to investigate cancer metastasis. Front Med. 2018;5:1–7. doi: 10.3389/fmed.2018.00034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Investig. 2009;119:1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mentis AA, Kararizou E. Metabolism and cancer: an up-to-date review of a mutual connection. Asian Pac J Cancer Prev. 2010;11:1437–1444. [PubMed] [Google Scholar]
- 19.Jie XX, Zhang XY, Xu CJ. Epithelial-to-mesenchymal transition, circulating tumor cells and cancer metastasis: mechanisms and clinical applications. Oncotarget. 2017;8:81558–81571. doi: 10.18632/oncotarget.18277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tiwari N, Gheldof A, Tatari M, Christofori G. EMT as the ultimate survival mechanism of cancer cells. Semin Cancer Biol. 2012;22:194–207. doi: 10.1016/j.semcancer.2012.02.013. [DOI] [PubMed] [Google Scholar]
- 21.Agnoletto C, Corrà F, Minotti L, Baldassari F, Crudele F, Cook WJJ, Di Leva G, D’Adamo AP, Gasparini P, Volinia S. Heterogeneity in circulating tumor cells: the relevance of the stem-cell subset. Cancers (Basel) 2019;11:9–12. doi: 10.3390/cancers11040483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mani SA, Guo W, Liao M, Eaton EN, Zhou AY, Brooks M, Reinhard F, Zhang CC, Campbell LL, Polyak K, et al. EMT creates cells with the properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ye X, Tam WL, Shibue T, Kaygusuz Y, Reinhardt F, Ng Eaton E, Weinberg RA. Distinct EMT programs control normal mammary stem cells and tumour-initiating cells. Nature. 2015;525:256–260. doi: 10.1038/nature14897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Werner S, Stenzl A, Pantel K, Todenh T. Isolation and molecular characterization of circulating tumor cells. Adv Exp Med Biol. 2017;994:205–228. doi: 10.1007/978-3-319-55947-6_11. [DOI] [PubMed] [Google Scholar]
- 25.Theodoropoulos PA, Polioudaki H, Agelaki S, Kallergi G, Saridaki Z, Mavroudis D, Georgoulias V. Circulating tumor cells with a putative stem cell phenotype in peripheral blood of patients with breast cancer. Cancer Lett. 2010;288:99–106. doi: 10.1016/j.canlet.2009.06.027. [DOI] [PubMed] [Google Scholar]
- 26.Zhang X, Wei L, Li J, Zheng J, Zhang S, Zhou J. Epithelial-mesenchymal transition phenotype of circulating tumor cells is associated with distant metastasis in patients with NSCLC. Mol Med Rep. 2019;19:601–608. doi: 10.3892/mmr.2018.9684. [DOI] [PubMed] [Google Scholar]
- 27.Milano A, Mazzetta F, Valente S, Ranieri D, Leone L, Botticelli A, Onesti CE, Lauro S, Raffa S, Torrisi MR et al (2018) Molecular detection of EMT markers in circulating tumor cells from metastatic non-small cell lung cancer patients: potential role in clinical practice. Anal Cell Pathol. https://www.ncbi.nlm.nih.gov/pubmed/29682444 [DOI] [PMC free article] [PubMed]
- 28.Satelli A, Mitra A, Brownlee Z, Xia X, Bellister S, Overman MJ, Kopetz S, Ellis LM, Meng QH, Li S. Epithelial–mesenchymal transitioned circulating tumor cells capture for detecting tumor progression. Clin Cancer Res. 2015;21:899–906. doi: 10.1158/1078-0432.CCR-14-0894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lorentzen A, Becker PF, Kosla J, Saini M, Weidele K, Ronchi P, Klein C, Wolf MJ, Geist F, Seubert B, et al. Single cell polarity in liquid phase facilitates tumour metastasis. Nat Commun. 2018;9:887. doi: 10.1038/s41467-018-03139-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heikenwalder M, Lorentzen A. The role of polarisation of circulating tumour cells in cancer metastasis. Cell Mol Life Sci. 2019;76:3765–3781. doi: 10.1007/s00018-019-03169-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scheel C, Eaton EN, Li SHJ, Chaffer CL, Reinhardt F, Kah KJ, Bell G, Guo W, Rubin J, Richardson AL, et al. Paracrine and autocrine signals induce and maintain mesenchymal and stem cell states in the breast. Cell. 2011;145:926–940. doi: 10.1016/j.cell.2011.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Francart ME, Lambert J, Vanwynsberghe AM, Thompson EW, Bourcy M, Polette M, Gilles C. Epithelial–mesenchymal plasticity and circulating tumor cells: travel companions to metastases. Dev Dyn. 2018;247:432–450. doi: 10.1002/dvdy.24506. [DOI] [PubMed] [Google Scholar]
- 33.Jolly MK, Boareto M, Huang B, Jia D, Lu M, Onuchic JN, Levine H, Ben-Jacob E. Implications of the hybrid epithelial/mesenchymal phenotype in metastasis. Front Oncol. 2015;5:1–19. doi: 10.3389/fonc.2015.00155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Labelle M, Hynes RO. The initial hours of metastasis: the importance of cooperative host-tumor cell interactions during hematogenous dissemination. Cancer Discov. 2012;2:1091–1099. doi: 10.1158/2159-8290.CD-12-0329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sun YF, Guo W, Xu Y, Shi YH, Gong ZJ, Ji Y, Du M, Zhang X, Hu B, Huang A, et al. Circulating tumor cells from different vascular sites exhibit spatial heterogeneity in epithelial and mesenchymal composition and distinct clinical significance in hepatocellular carcinoma. Clin Cancer Res. 2017;24:547–559. doi: 10.1158/1078-0432.CCR-17-1063. [DOI] [PubMed] [Google Scholar]
- 36.Haeger A, Krause M, Wolf K, Friedl P. Cell jamming: Collective invasion of mesenchymal tumor cells imposed by tissue confinement. Biochim Biophys Acta. 2014;1840:2386–2395. doi: 10.1016/j.bbagen.2014.03.020. [DOI] [PubMed] [Google Scholar]
- 37.Eddy RJ, Weidmann MD, Sharma VP, Condeelis JS. Tumor cell invadopodia: invasive protrusions that orchestrate metastasis. Trends Cell Biol. 2017;27:595–607. doi: 10.1016/j.tcb.2017.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Glinsky VV, Glinsky GV, Glinskii OV, Huxley VH, Turk JR, Mossine VV, Deutscher SL, Pienta KJ, Quinn TP. Intravascular metastatic cancer cell homotypic aggregation at the sites of primary attachment to the endothelium. Cancer Res. 2003;63:3805–3811. [PubMed] [Google Scholar]
- 39.Shen Z, Wu A, Chen X. Current detection technologies for circulating tumor cells. Chem Soc Rev. 2017;46:2038–2056. doi: 10.1039/c6cs00803h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bankó P, Lee SY, Nagygyörgy V, Zrínyi M, Chae CH, Cho DH, Telekes A. Technologies for circulating tumor cell separation from whole blood. J Hematol Oncol. 2019;12:1–20. doi: 10.1186/s13045-019-0735-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bailey PC, Martin SS. Insights on CTC biology and clinical impact emerging from advances in capture technology. Cells. 2019;8:553. doi: 10.3390/cells8060553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang QQ, Chen XX, Jiang W, Jin SL, Wang XY, Liu W, Guo SS, Guo JC, Zhao XZ. Sensitive and specific detection of circulating tumor cells promotes precision medicine for cancer. J Cancer Metastasis Treat. 2019;5:34. [Google Scholar]
- 43.Shen SY, Singhania R, Fehringer G, Chakravarthy A, Roehrl MHA, Chadwick D, Zuzarte PC, Borgida A, Wang TT, Li T, et al. Sensitive tumour detection and classification using plasma cell-free DNA methylomes. Nature. 2018;563:579–583. doi: 10.1038/s41586-018-0703-0. [DOI] [PubMed] [Google Scholar]
- 44.Chen H (2019) Capturing and clinical applications of circulating tumor cells with wave microfluidic chip. Appl Biochem Biotechnol. https://www.ncbi.nlm.nih.gov/pubmed/31782091 [DOI] [PubMed]
- 45.Zou D, Cui D. Advances in isolation and detection of circulating tumor cells based on microfluidics. Cancer Biol Med. 2018;15:335–353. doi: 10.20892/j.issn.2095-3941.2018.0256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sarioglu AF, Aceto N, Kojic N, Donaldson MC, Hamza B, Engstrom A, Zhu H, Sundaresan TK, David T, Luo X, et al. A microfluidic device for label-free, physical capture of circulating tumor cell-clusters. Nat Methods. 2015;12:685–691. doi: 10.1038/nmeth.3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Au SH, Edd J, Stoddard AE, Wong KHK, Fachin F, Maheswaran S, Haber DA, Stott SL, Kapur R, Toner M. Microfluidic isolation of circulating tumor cell clusters by size and asymmetry. Sci Rep. 2017;7:1–10. doi: 10.1038/s41598-017-01150-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cheng SB, Xie M, Chen Y, Xiong J, Liu Y, Chen Z, Guo S, Shu Y, Wang M, Yuan BF, et al. Three-dimensional scaffold chip with thermosensitive coating for capture and reversible release of individual and cluster of circulating tumor cells. Anal Chem. 2017;89:7924–7932. doi: 10.1021/acs.analchem.7b00905. [DOI] [PubMed] [Google Scholar]
- 49.Chiu TK, Chao AC, Chou WP, Liao CJ, Wang HM, Chang JH, Chen PH, Wu MH. Optically-induced-dielectrophoresis (ODEP)-based cell manipulation in a microfluidic system for high-purity isolation of integral circulating tumor cell (CTC) clusters based on their size characteristics. Sens Actuators B Chem. 2018;258:1161–1173. [Google Scholar]
- 50.Suo Y, Xie C, Zhu X, Fan Z, Yang Z, He H, Wei X. Proportion of circulating tumor cell clusters increases during cancer metastasis. Cytom Part A. 2017;91:250–253. doi: 10.1002/cyto.a.23037. [DOI] [PubMed] [Google Scholar]
- 51.Pantel K, Alix-Panabières C. Bone marrow as a reservoir for disseminated tumor cells: a special source for liquid biopsy in cancer patients. Bonekey Rep. 2014;3:1–6. doi: 10.1038/bonekey.2014.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sai B, Xiang J. Disseminated tumour cells in bone marrow are the source of cancer relapse after therapy. J Cell Mol Med. 2018;22:5776–5786. doi: 10.1111/jcmm.13867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dasgupta A, Lim AR, Ghajar CM. Circulating and disseminated tumor cells: harbingers or initiators of metastasis? Mol Oncol. 2017;11:40–61. doi: 10.1002/1878-0261.12022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Weidenfeld K, Barkan D. EMT and stemness in tumor dormancy and outgrowth: are they intertwined processes? Front Oncol. 2018;8:1–6. doi: 10.3389/fonc.2018.00381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ghajar CM, Peinado H, Mori H, Matei IR, Evason KJ, Brazier H, Almeida D, Koller A, Hajjar KA, Stainier DYR, et al. The perivascular niche regulates breast tumour dormancy. Nat Cell Biol. 2013;15:807–817. doi: 10.1038/ncb2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schmidt-kittler O, Ragg T, Daskalakis A, Granzow M, Ahr A, Blankenstein TJF, Bischoff J, Harich D, Kaufmann M, Diebold J, et al. From latent disseminated cells to overt metastasis: genetic analysis of systemic breast cancer progression. Proc Natl Acad Sci USA. 2003;100:7737–7742. doi: 10.1073/pnas.1331931100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Heyde A, Reiter JG, Naxerova K, Nowak MA. Consecutive seeding and transfer of genetic diversity in metastasis. Proc Natl Acad Sci USA. 2019;116:14129–14137. doi: 10.1073/pnas.1819408116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Reiter JG, Makohon-Moore AP, Gerold JM, Heyde A, Attiyeh MA, Kohutek ZA, Tokheim CJ, Brown A, DeBlasio RM, Niyazov J, et al. Minimal functional driver gene heterogeneity among untreated metastases. Science. 2018;361:1033–1037. doi: 10.1126/science.aat7171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Demeulemeester J, Kumar P, Møller EK, Nord S, Wedge DC, Peterson A, Mathiesen RR, Fjelldal R, Zamani Esteki M, Theunis K, et al. Tracing the origin of disseminated tumor cells in breast cancer using single-cell sequencing. Genome Biol. 2016;17:1–15. doi: 10.1186/s13059-016-1109-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kim M, Oskarsson T, Acharyya S, Nguyen DX, Xiang H, Norton L, Massagué J. Tumor self seeding by circulating cancer cells. Cell. 2009;139:1315–1326. doi: 10.1016/j.cell.2009.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Aguirre-Ghiso JA. On the theory of tumor self-seeding: Implications for metastasis progression in humans. Breast Cancer Res. 2010;12:1–2. doi: 10.1186/bcr2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dondossola E, Crippa L, Colombo B, Ferrero E, Corti A. Chromogranin A regulates tumor self-seeding and dissemination. Cancer Res. 2012;72:449–459. doi: 10.1158/0008-5472.CAN-11-2944. [DOI] [PubMed] [Google Scholar]
- 63.Zhang Y, Ma Q, Liu T, Guan G, Zhang K, Chen J, Jia N, Yan S, Chen G, Liu S, et al. Interleukin-6 suppression reduces tumour self-seeding by circulating tumour cells in a human osteosarcoma nude mouse model. Oncotarget. 2016;7:446–458. doi: 10.18632/oncotarget.6371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Scott JG, Basanta D, Anderson ARA, Gerlee P. A mathematical model of tumour selfseeding reveals secondary metastatic deposits as drivers of primary tumour growth. J R Soc Interface. 2013;10:1–9. doi: 10.1098/rsif.2013.0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Comen E, Norton L, Massagué J. Clinical implications of cancer self-seeding. Nat Rev Clin Oncol. 2011;8:369–377. doi: 10.1038/nrclinonc.2011.64. [DOI] [PubMed] [Google Scholar]
- 66.Makohon-Moore AP, Matsukuma K, Zhang M, Reiter JG, Gerold JM, Jiao Y, Sikkema L, Attiyeh MA, Yachida S, Sandone C, et al. Precancerous neoplastic cells can move through the pancreatic ductal system. Nature. 2018;561:201–205. doi: 10.1038/s41586-018-0481-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hruban RH, Gaida MM, Thompson E, Hong SM, Noë M, Brosens LAA, Jongepier M, Offerhaus GJA, Wood LD. Why is pancreatic cancer so deadly? The pathologist’s view. J Pathol. 2019;248:131–141. doi: 10.1002/path.5260. [DOI] [PubMed] [Google Scholar]
- 68.Garzia L, Kijima N, Morrissy AS, De Antonellis P, Guerreiro-Stucklin A, Holgado BL, Wu X, Wang X, Parsons M, Zayne K, et al. A hematogenous route for medulloblastoma leptomeningeal metastases. Cell. 2018;172:1050–1062. doi: 10.1016/j.cell.2018.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ullah I, Karthik GM, Alkodsi A, Kjällquist U, Stålhammar G, Lövrot J, Martinez NF, Lagergren J, Hautaniemi S, Hartman J, et al. Evolutionary history of metastatic breast cancer reveals minimal seeding from axillary lymph nodes. J Clin Investig. 2018;128:1355–1370. doi: 10.1172/JCI96149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Leung ML, Davis A, Gao R, Casasent A, Wang Y, Sei E, Vilar E, Maru D, Kopetz S, Navin NE. Single-cell DNA sequencing reveals a latedissemination model in metastatic colorectal cancer. Genome Res. 2017;27:1287–1299. doi: 10.1101/gr.209973.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hong Y, Fang F, Zhang Q. Circulating tumor cell clusters: what we know and what we expect (Review) Int J Oncol. 2016;49:2206–2216. doi: 10.3892/ijo.2016.3747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cho EH, Wendel M, Luttgen M, Yoshioka C, Marrinucci D, Lazar D, Schram E, Nieva J, Bazhenova L, Morgan A, et al. Characterization of circulating tumor cell aggregates identified in patients with epithelial tumors. Phys Biol. 2012;9:1–13. doi: 10.1088/1478-3975/9/1/016001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Duda DG, Duyverman AMMJ, Kohno M, Snuderl M, Steller EJA, Fukumura D, Jain RK. Malignant cells facilitate lung metastasis by bringing their own soil. Proc Natl Acad Sci USA. 2010;107:21677–21682. doi: 10.1073/pnas.1016234107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Upreti M, Jamshidi-Parsian A, Koonce NA, Webber JS, Sharma SK, Asea AAA, Mader MJ, Griffin RJ. Tumor-endothelial cell three-dimensional spheroids: new aspects to enhance radiation and drug therapeutics. Transl Oncol. 2011;4:365–376. doi: 10.1593/tlo.11187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Szczerba BM, Castro-Giner F, Vetter M, Krol I, Gkountela S, Landin J, Scheidmann MC, Donato C, Scherrer R, Singer J, et al. Neutrophils escort circulating tumour cells to enable cell cycle progression. Nature. 2019;566:553–557. doi: 10.1038/s41586-019-0915-y. [DOI] [PubMed] [Google Scholar]
- 76.Cheung KJ, Padmanaban V, Silvestri V, Schipper K, Cohen JD, Fairchild AN, Gorin MA, Verdone JE, Pienta KJ, Bader JS, et al. Polyclonal breast cancer metastases arise from collective dissemination of keratin 14-expressing tumor cell clusters. Proc Natl Acad Sci USA. 2016;113:E854–E863. doi: 10.1073/pnas.1508541113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Geng Y, Chandrasekaran S, Hsu JW, Gidwani M, Hughes AD, King MR. Phenotypic switch in blood: effects of pro-inflammatory cytokines on breast cancer cell aggregation and adhesion. PLoS One. 2013;8:1–10. doi: 10.1371/journal.pone.0054959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pixberg CF, Schulz WA, Stoecklein NH, Neves RPL. Characterization of DNA methylation in circulating tumor cells. Genes (Basel) 2015;6:1053–1075. doi: 10.3390/genes6041053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Benezeder T, Tiran V, Treitler AAN, Suppan C, Rossmann C, Stoeger H, Cote RJ, Datar RH, Balic M, Dandachi N. Multigene methylation analysis of enriched circulating tumor cells associates with poor progression-free survival in metastatic breast cancer patients. Oncotarget. 2017;8:92483–92496. doi: 10.18632/oncotarget.21426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gkountela S, Castro-Giner F, Szczerba BM, Vetter M, Landin J, Scherrer R, Krol I, Scheidmann MC, Beisel C, Stirnimann CU, et al. Circulating tumor cell clustering shapes DNA methylation to enable metastasis seeding. Cell. 2019;176:98–112. doi: 10.1016/j.cell.2018.11.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Allen TA, Gracieux D, Talib M, Tokarz DA, Hensley MT, Cores J, Vandergriff A, Tang J, de Andrade JBM, Dinh PU, et al. Angiopellosis as an alternative mechanism of cell extravasation. Stem Cells. 2017;35:170–180. doi: 10.1002/stem.2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Allen TA, Asad D, Amu E, Hensley MT, Cores J, Vandergriff A, Tang J, Dinh PU, Shen D, Qiao L, et al. Circulating tumor cells exit circulation while maintaining multicellularity, augmenting metastatic potential. J. Cell Sci. 2019;132:jcs231563. doi: 10.1242/jcs.231563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Balakrishnan A, Koppaka D, Anand A, Deb B, Grenci G, Viasnoff V, Thompson EW, Gowda H, Bhat R, Rangarajan A, et al. Circulating tumor cell cluster phenotype allows monitoring response to treatment and predicts survival. Sci Rep. 2019;9:7933. doi: 10.1038/s41598-019-44404-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Khoo BL, Grenci G, Jing T, Bena Lim Y, Lee SC, Thiery JP, Han J, Lim CT. Liquid biopsy and therapeutic response: circulating tumor cell cultures for evaluation of anticancer treatment. Sci Adv. 2016;2:1–15. doi: 10.1126/sciadv.1600274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hoefnagel LDC, van de Vijver MJ, van Slooten HJ, Wesseling P, Wesseling J, Westenend PJ, Bart J, Seldenrijk CA, Nagtegaal ID, Oudejans J, et al. Receptor conversion in distant breast cancer metastases. Breast Cancer Res. 2010;12:R75. doi: 10.1186/bcr2645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Timmer M, Werner JM, Röhn G, Ortmann M, Blau T, Cramer C, Stavrinou P, Krischek B, Mallman P, Goldbrunner R. Discordance and conversion rates of progesterone-, estrogen-, and HER2/neu-receptor status in primary breast cancer and brain metastasis mainly triggered by hormone therapy. Anticancer Res. 2017;37:4859–4865. doi: 10.21873/anticanres.11894. [DOI] [PubMed] [Google Scholar]
- 87.Munzone E, Nolé F, Goldhirsch A, Botteri E, Esposito A, Zorzino L, Curigliano G, Minchella I, Adamoli L, Cassatella MC, et al. Changes of HER2 status in circulating tumor cells compared with the primary tumor during treatment for advanced breast cancer. Clin Breast Cancer. 2010;10:392–397. doi: 10.3816/CBC.2010.n.052. [DOI] [PubMed] [Google Scholar]
- 88.Fehm T, Müller V, Aktas B, Janni W, Schneeweiss A, Stickeler E, Lattrich C, Löhberg CR, Solomayer E, Rack B, et al. HER2 status of circulating tumor cells in patients with metastatic breast cancer: a prospective, multicenter trial. Breast Cancer Res Treat. 2010;124:403–412. doi: 10.1007/s10549-010-1163-x. [DOI] [PubMed] [Google Scholar]
- 89.Jordan NV, Bardia A, Wittner BS, Benes C, Ligorio M, Zheng Y, Yu M, Sundaresan TK, Licausi JA, Desai R, et al. HER2 expression identifies dynamic functional states within circulating breast cancer cells. Nature. 2016;537:102–106. doi: 10.1038/nature19328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Paoletti C, Larios JM, Muñiz MC, Aung K, Cannell EM, Darga EP, Kidwell KM, Thomas DG, Tokudome N, Brown ME, et al. Heterogeneous estrogen receptor expression in circulating tumor cells suggests diverse mechanisms of fulvestrant resistance. Mol Oncol. 2016;10:1078–1085. doi: 10.1016/j.molonc.2016.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lianidou ES, Markou A, Strati A. The role of CTCs as tumor biomarkers. Adv Exp Med Biol. 2015;867:341–367. doi: 10.1007/978-94-017-7215-0_21. [DOI] [PubMed] [Google Scholar]
- 92.Stewart CM, Kothari PD, Mouliere F, Mair R, Somnay S, Benayed R, Zehir A, Weigelt B, Dawson SJ, Arcila ME, et al. The value of cell-free DNA for molecular pathology. J Pathol. 2018;244:616–627. doi: 10.1002/path.5048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Luchini C, Veronese N, Nottegar A, Cappelletti V, Daidone MG, Smith L, Parris C, Brosens LAA, Caruso MG, Cheng L, et al. Liquid biopsy as surrogate for tissue for molecular profiling in pancreatic cancer: a meta-analysis towards precision medicine. Cancers (Basel) 2019;11:1152. doi: 10.3390/cancers11081152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Pantel K, Alix-panabières C. Liquid biopsy and minimal residual disease—latest advances and implications for cure. Nat Rev Clin Oncol. 2019;16:409–424. doi: 10.1038/s41571-019-0187-3. [DOI] [PubMed] [Google Scholar]
- 95.Siravegna G, Mussolin B, Venesio T, Marsoni S, Seoane J, Dive C, Papadopoulos N, et al. How liquid biopsies can change clinical practice in oncology. Ann Oncol. 2019;30:1580–1590. doi: 10.1093/annonc/mdz227. [DOI] [PubMed] [Google Scholar]
- 96.Wang CH, Chang CJ, Yeh KY, Chang PH, Huang JS. The prognostic value of HER2-positive circulating tumor cells in breast cancer patients: a systematic review and meta-analysis. Clin Breast Cancer. 2017;17:341–349. doi: 10.1016/j.clbc.2017.02.002. [DOI] [PubMed] [Google Scholar]
- 97.Mattox AK, Bettegowda C, Zhou S, Papadopoulos N, Kinzler KW, Vogelstein B. Applications of liquid biopsies for cancer. Sci Transl Med. 2019;11:1–4. doi: 10.1126/scitranslmed.aay1984. [DOI] [PubMed] [Google Scholar]
- 98.Lyu M, Zhou J, Ning K, Ying B. The diagnostic value of circulating tumor cells and ctDNA for gene mutations in lung cancer. Onco Targets Ther. 2019;12:2539–2552. doi: 10.2147/OTT.S195342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Blackhall F, Frese KK, Simpson K, Kilgour E, Brady G, Dive C. Will liquid biopsies improve outcomes for patients with small-cell lung cancer? Lancet Oncol. 2018;19:e470–e481. doi: 10.1016/S1470-2045(18)30455-8. [DOI] [PubMed] [Google Scholar]
- 100.Danila DC, Heller G, Gignac GA, Gonzalez-Espinoza R, Anand A, Tanaka E, Lilja H, Schwartz L, Larson S, Fleisher M, et al. Circulating tumor cell number and prognosis in progressive castration-resistant prostate cancer. Clin Cancer Res. 2007;13:7053–7058. doi: 10.1158/1078-0432.CCR-07-1506. [DOI] [PubMed] [Google Scholar]
- 101.Cohen SJ, Punt CJA, Iannotti N, Saidman BH, Sabbath KD, Gabrail NY, Picus J, Morse M, Mitchell E, Miller MC, et al. Relationship of circulating tumor cells to tumor response, progression-free survival, and overall survival in patients with metastatic colorectal cancer. J Clin Oncol. 2008;26:3213–3221. doi: 10.1200/JCO.2007.15.8923. [DOI] [PubMed] [Google Scholar]
- 102.Riethdorf S, Fritsche H, Müller V, Rau T, Schindlbeck C, Rack B, Janni W, Coith C, Beck K, Jänicke F, et al. Detection of circulating tumor cells in peripheral blood of patients with metastatic breast cancer: a validation study of the cell search system. Clin Cancer Res. 2007;13:920–928. doi: 10.1158/1078-0432.CCR-06-1695. [DOI] [PubMed] [Google Scholar]
- 103.Wang F, Yang XQ, YangS WBC, Feng MH, Tu JC. A higher number of circulating tumor cells (CTC) in peripheral blood indicates poor prognosis in prostate cancer patients—a meta-analysis. Asian Pac J Cancer Prev. 2011;12:2629–2635. [PubMed] [Google Scholar]
- 104.Hall CS, Karhade M, Laubacher BA, Kuerer HM, Krishnamurthy S, DeSnyder S, Anderson AE, Valero V, Ueno NT, Li Y, et al. Circulating tumor cells and recurrence after primary systemic therapy in stage III inflammatory breast cancer. J Natl Cancer Inst. 2015;107:11–14. doi: 10.1093/jnci/djv250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Li J, Fu W, Zhang W, Li P. High number of circulating tumor cells predicts poor survival of cutaneous melanoma patients in China. Med Sci Monit. 2018;24:324–331. doi: 10.12659/MSM.904770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lozar T, Gersak K, Cemazar M, Kuhar CG, Jesenko T. The biology and clinical potential of circulating tumor cells. Radiol Oncol. 2019;53:131–147. doi: 10.2478/raon-2019-0024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Rostami P, Kashaninejad N, Moshksayan K, Saidi MS, Firoozabadi B, Nguyen NT. Novel approaches in cancer management with circulating tumor cell clusters. J Sci Adv Mater Devices. 2019;4:1–18. [Google Scholar]
- 108.Murlidhar V, Reddy RM, Fouladdel S, Zhao L, Ishikawa MK, Grabauskiene S, Zhang Z, Lin J, Chang AC, Carrott P, et al. Poor prognosis indicated by venous circulating tumor cell clusters in early-stage lung cancers. Cancer Res. 2017;77:5194–5206. doi: 10.1158/0008-5472.CAN-16-2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Larsson AM, Jansson S, Bendahl PO, Levin Tykjaer Jörgensen C, Loman N, Graffman C, Lundgren L, Aaltonen K, Rydén L. Longitudinal enumeration and cluster evaluation of circulating tumor cells improve prognostication for patients with newly diagnosed metastatic breast cancer in a prospective observational trial. Breast Cancer Res. 2018;20:1–14. doi: 10.1186/s13058-018-0976-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Antonarakis ES, Lu C, Wang H, Luber B, Nakazawa M, Roeser JC, Chen Y, Mohammad TA, Chen Y, Fedor HL, Lotan TL, Zheng Q, De Marzo AM, Isaacs JT, Isaacs WB, Nadal R, Paller CJ, Denmeade SR, Carducci MA, Eisenberger MA, Luo J. AR-V7 and resistance to enzalutamide and abiraterone in prostate cancer. N Engl J Med. 2014;371(11):1028–1038. doi: 10.1056/NEJMoa1315815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bastos DA, Antonarakis ES. CTC-derived AR-V7 detection as a prognostic and predictive biomarker in advanced prostate cancer. Expert Rev Mol Diagn. 2018;18:155–163. doi: 10.1080/14737159.2018.1427068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Scher HI, Heller G, Molina A, Attard G, Danila DC, Jia X, Peng W, Sandhu SK, Olmos D, Riisnaes R, McCormack R, Burzykowski T, Kheoh T, Fleisher M, Buyse M, de Bono JS. Circulating tumor cell biomarker panel as an individual-level surrogate for survival in metastatic castration-resistant prostate cancer. J Clin Oncol Off J Am Soc Clin Oncol. 2015;33:1348–1355. doi: 10.1200/JCO.2014.55.3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Heller G, McCormack R, Kheoh T, Molina A, Smith MR, Dreicer R, Saad F, de Wit R, Aftab DT, Hirmand M, Limon A, Fizazi K, Fleisher M, de Bono JS, Scher HI. Circulating tumor cell number as a response measure of prolonged survival for metastatic castration-resistant prostate cancer: a comparison with prostate-specific antigen across five randomized phase III clinical trials. J Clin Oncol Off J Am Soc Clin Oncol. 2018;36:572–580. doi: 10.1200/JCO.2017.75.2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Oxnard GR, West HJ, King JC (2019) Association of patient characteristics and tumor genomics with clinical outcomes among patients with non-small cell lung cancer using a clinicogenomic database. JAMA Oncol. https://www.ncbi.nlm.nih.gov/pubmed/31647499 [DOI] [PMC free article] [PubMed]
- 115.Guan X, Ma F, Li C, Wu S, Hu S, Huang J, Sun X, Wang J, Luo Y, Cai R, et al. The prognostic and therapeutic implications of circulating tumor cell phenotype detection based on epithelial-mesenchymal transition markers in the first-line chemotherapy of HER2-negative metastatic breast cancer. Cancer Commun. 2019;39:1–10. doi: 10.1186/s40880-018-0346-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Gorges TM, Stein A, Quidde J, Hauch S, Röck K, Riethdorf S, Joosse SA, Pantel K. Improved detection of circulating tumor cells in metastatic colorectal cancer by the combination of the cell Search® system and the AdnaTest®. PLoS One. 2016;11:1–13. doi: 10.1371/journal.pone.0155126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ferreira MM, Ramani VC, Jeffrey SS. Circulating tumor cell technologies. Mol Oncol. 2016;10:374–394. doi: 10.1016/j.molonc.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lu YJ, Wang P, Peng J, Wang X, Zhu YW, Shen N. Meta-analysis reveals the prognostic value of circulating tumour cells detected in the peripheral blood in patients with non-metastatic colorectal cancer. Sci Rep. 2017;7:1–9. doi: 10.1038/s41598-017-01066-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Tan Y, Wu H. The significant prognostic value of circulating tumor cells in colorectal cancer: a systematic review and meta-analysis. Curr Probl Cancer. 2018;42:95–106. doi: 10.1016/j.currproblcancer.2017.11.002. [DOI] [PubMed] [Google Scholar]
- 120.Yang C, Zou K, Zheng L, Xiong B. Prognostic and clinicopathological significance of circulating tumor cells detected by rt-pcr in non-metastatic colorectal cancer: a meta-analysis and systematic review. BMC Cancer. 2017;17:725. doi: 10.1186/s12885-017-3704-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Huang X, Gao P, Song Y, Sun J, Chen X, Zhao J, Xu H, Wang Z. Meta-analysis of the prognostic value of circulating tumor cells detected with the cell search system in colorectal cancer. BMC Cancer. 2015;15:202. doi: 10.1186/s12885-015-1218-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Bidard FC, Michiels S, Riethdorf S, Mueller V, Esserman LJ, Lucci A, Naume B, Horiguchi J, Gisbert-Criado R, Sleijfer S, et al. Circulating tumor cells in breast cancer patients treated by neoadjuvant chemotherapy: a meta-analysis. J Natl Cancer Inst. 2018;110:560–567. doi: 10.1093/jnci/djy018. [DOI] [PubMed] [Google Scholar]
- 123.Zhang L, Riethdorf S, Wu G, Wang T, Yang K, Peng G, Liu J, Pantel K. Meta-analysis of the prognostic value of circulating tumor cells in breast cancer. Clin Cancer Res. 2012;18:5701–5710. doi: 10.1158/1078-0432.CCR-12-1587. [DOI] [PubMed] [Google Scholar]
- 124.Ma XL, Xiao ZL, Liu L, Liu XX, Nie W, Li P, Chen NY, Wei YQ. Meta-analysis of circulating tumor cells as a prognostic marker in lung cancer. Asian Pac J Cancer Prev. 2012;13:1137–1144. doi: 10.7314/apjcp.2012.13.4.1137. [DOI] [PubMed] [Google Scholar]
- 125.Wu ZX, Liu Z, Jiang HL, Pan HM, Han WD. Circulating tumor cells predict survival benefit from chemotherapy in patients with lung cancer. Oncotarget. 2016;7:67586–67596. doi: 10.18632/oncotarget.11707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Stephenson D, Nahm C, Chua T, Gill A, Mittal A, de Reuver P, Samra J. Circulating and disseminated tumor cells in pancreatic cancer and their role in patient prognosis: a systematic review and meta-analysis. Oncotarget. 2017;8:107223–107236. doi: 10.18632/oncotarget.19928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Ma XL, Xiao ZL, Li X, Wang F, Zhang J, Zhou R, Wang J, Liu L. The prognostic role of circulating tumor cells and disseminated tumor cells in patients with prostate cancer: a systematic review and meta-analysis. Tumor Biol. 2014;35:5551–5560. doi: 10.1007/s13277-014-1731-5. [DOI] [PubMed] [Google Scholar]
- 128.Zheng Y, Zhang C, Wu J, Cheng G, Yang H, Hua L, Wang Z. Prognostic value of circulating tumor cells in castration resistant prostate cancer: a meta-analysis. Urol J. 2016;13:2881–2888. [PubMed] [Google Scholar]
- 129.Zou K, Yang S, Zheng L, Wang S, Xiong B (2016) Prognostic role of the circulating tumor cells detected by cytological methods in gastric cancer: a meta-analysis. Biomed Res Int. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5098053/ [DOI] [PMC free article] [PubMed]
- 130.Hou J, Zou K, Yang C, Leng X, Xu Y. Clinicopathological and prognostic significance of circulating tumor cells in patients with esophageal cancer: a meta-analysis. Onco Targets Ther. 2018;11:8053–8061. doi: 10.2147/OTT.S175855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Cho J, Lee GJ, Kim H, Moon UY, Kim M, Kim S, Baek K, Jeong H. Differential impact of circulating tumor cells on disease recurrence and survivals in patients with head and neck squamous cell carcinomas: an updated meta-analysis. PLoS One. 2018;13:1–11. doi: 10.1371/journal.pone.0203758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Stroun M, Maurice P, Vasioukhin V, et al. The origin and mechanism of circulating DNA. Ann N Y Acad Sci. 2000;906:161–168. doi: 10.1111/j.1749-6632.2000.tb06608.x. [DOI] [PubMed] [Google Scholar]
- 133.Grivas P, Lalani AKA, Pond GR, Nagy RJ, Faltas B, Agarwal N, Gupta SV, Drakaki A, Vaishampayan UN, Wang J, et al. Circulating tumor DNA alterations in advanced urothelial carcinoma and association with clinical outcomes: a pilot study. Eur Urol Oncol. 2019 doi: 10.1016/j.euo.2019.02.004. [DOI] [PubMed] [Google Scholar]
- 134.Giannopoulou L, Mastoraki S, Buderath P, Strati A, Pavlakis K, Kasimir-Bauer S, Lianidou ES. ESR1 methylation in primary tumors and paired circulating tumor DNA of patients with high-grade serous ovarian cancer. Gynecol Oncol. 2018;150:355–360. doi: 10.1016/j.ygyno.2018.05.026. [DOI] [PubMed] [Google Scholar]
- 135.Sonpavde G, Agarwal N, Pond GR, Nagy RJ, Nussenzveig RH, Hahn AW, Sartor O, Gourdin TS, Nandagopal L, Ledet EM, et al. Circulating tumor DNA alterations in patients with metastatic castration-resistant prostate cancer. Cancer. 2019;125:1459–1469. doi: 10.1002/cncr.31959. [DOI] [PubMed] [Google Scholar]
- 136.Tzanikou E, Markou A, Politaki E, Koutsopoulos A, Psyrri A, Mavroudis D, Georgoulias V, Lianidou E. PIK3CA hotspot mutations in circulating tumor cells and paired circulating tumor DNA in breast cancer: a direct comparison study. Mol Oncol. 2019;13:2515–2530. doi: 10.1002/1878-0261.12540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Barata PC, Koshkin VS, Funchain P, Sohal D, Pritchard A, Klek S, Adamowicz T, Gopalakrishnan D, Garcia J, Rini B, Grivas P. Next-generation sequencing (NGS) of cell-free circulating tumor DNA and tumor tissue in patients with advanced urothelial cancer: a pilot assessment of concordance. Ann Oncol. 2017;28:2458–2463. doi: 10.1093/annonc/mdx405. [DOI] [PubMed] [Google Scholar]
- 138.Agarwal N, Pal SK, Hahn AW, Nussenzveig RH, Pond GR, Gupta SV, Wang J, Bilen MA, Naik G, Ghatalia P, Hoimes CJ, Gopalakrishnan D, Barata PC, Drakaki A, Faltas BM, Kiedrowski LA, Lanman RB, Nagy RJ, Vogelzang NJ, Boucher KM, Vaishampayan UN, Sonpavde G, Grivas P. Characterization of metastatic urothelial carcinoma via comprehensive genomic profiling of circulating tumor DNA. Cancer. 2018;124:2115–2124. doi: 10.1002/cncr.31314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Sina AAI, Carrascosa LG, Trau M. DNA methylation-based point-of-care cancer detection: challenges and possibilities. Trends Mol Med. 2019;25:955–966. doi: 10.1016/j.molmed.2019.05.014. [DOI] [PubMed] [Google Scholar]
- 140.Mamdouhi T, Twomey JD, McSweeney KM, Zhang B. Fugitives on the run: circulating tumor cells (CTCs) in metastatic diseases. Cancer Metastasis Rev. 2019;38:297–305. doi: 10.1007/s10555-019-09795-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Ye Z, Wang C, Wan S, Mu Z, Zhang Z, Abu-Khalaf MM, Fellin FM, Silver DP, Neupane M, Jaslow RJ, Bhattacharya S, Tsangaris TN, Chervoneva I, Berger A, Austin L, Palazzo JP, Myers RE, Pancholy N, Toorkey D, Yao K, Krall M, Li X, Chen X, Fu X, Xing J, Hou L, Wei Q, Li B, Cristofanilli M, Yang H. Association of clinical outcomes in metastatic breast cancer patients with circulating tumour cell and circulating cell-free DNA. Eur J Cancer. 2019;106:133–143. doi: 10.1016/j.ejca.2018.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Punnoose EA, Atwal S, Liu W, Raja R, Fine BM, Hughes BG, Hicks RJ, Hampton GM, Amler LC, Pirzkall A, Lackner MR. Evaluation of circulating tumor cells and circulating tumor DNA in non-small cell lung cancer: association with clinical endpoints in a phase II clinical trial of pertuzumab and erlotinib. Clin Cancer Res. 2012;18:2391–2401. doi: 10.1158/1078-0432.CCR-11-3148. [DOI] [PubMed] [Google Scholar]
- 143.Lampignano R, Neumann MHD, Weber S, Kloten V, Herdean A, Voss T, Groelz D, Babayan A, Tibbesma M, Schlumpberger M, et al. Multicenter evaluation of circulating cell-free DNA extraction and downstream analyses for the development of standardized (Pre)analytical work flows. Clin Chem. 2019;66:1–12. doi: 10.1373/clinchem.2019.306837. [DOI] [PubMed] [Google Scholar]
- 144.Mouliere F, Chandrananda D, Piskorz AM, Elizabeth K, Morris J, Ahlborn LB, Mair R, Marass F, Heider K, Wan JCM, et al. Europe PMC funders group enhanced detection of circulating tumor DNA by fragment size analysis. Sci Transl Med. 2018;10:1–28. doi: 10.1126/scitranslmed.aat4921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Cohen JD, Li L, Wang Y, Thoburn C, Danilova L, Douville C, Javed AA, Wong F, Mattox A, Hruban RH, et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science. 2018;359:926–930. doi: 10.1126/science.aar3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Groot VP, Mosier S, Javed AA, Teinor JA, Gemenetzis G, Ding D, Haley LM, Yu J, Burkhart RA, Hasanain A, et al. Circulating tumor DNA as a clinical test in resected pancreatic cancer. Clin Cancer Res. 2019;25:4973–4984. doi: 10.1158/1078-0432.CCR-19-0197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Phallen J, Sausen M, Adleff V, Leal A, Hruban C, Anagnostou V, Fiksel J, Cristiano S, Papp E, Speir S, et al. Direct detection of early-stage cancers using circulating tumor DNA. Sci Transl Med. 2019;9:1–25. doi: 10.1126/scitranslmed.aan2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Wang Y, Li L, Douville C, Cohen JD, Yen TT, Kinde I, Sundfelt K, Kjær SK, Hruban RH, Shih IM, et al. Evaluation of liquid from the papanicolaou test and other liquid biopsies for the detection of endometrial and ovarian cancers. Obstet Gynecol Surv. 2018;73:463–464. doi: 10.1126/scitranslmed.aap8793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Rodriguez Pena MDC, Springer SU, Taheri D, Li L, Tregnago AC, Eich ML, Eltoum IEA, VandenBussche CJ, Papadopoulos N, Kinzler KW, et al. Performance of novel non-invasive urine assay UroSEEK in cohorts of equivocal urine cytology. Virchows Arch. 2019 doi: 10.1007/s00428-019-02654-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Kennedy SR, Zhang Y, Risques RA. Cancer-associated mutations but no cancer: insights into the early steps of carcinogenesis and implications for early cancer detection. Trends Cancer. 2019;5:531–540. doi: 10.1016/j.trecan.2019.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Luoh S, Flaherty KT. When tissue is no longer the issue: tissue-agnostic cancer therapy comes of age. Ann Intern Med. 2018;169:233–239. doi: 10.7326/M17-2832. [DOI] [PubMed] [Google Scholar]
- 152.Hierro C, Matos I, Martin-Liberal J, Ochoa de Olza M, Garralda E. Agnostic-histology approval of new drugs in oncology: are we already there? Clin Cancer Res. 2019;25:3210–3219. doi: 10.1158/1078-0432.CCR-18-3694. [DOI] [PubMed] [Google Scholar]
- 153.Mullard A. FDA notches up third tissue-agnostic cancer approval. Nat Rev Drug Discov. 2019;18:737. doi: 10.1038/d41573-019-00155-z. [DOI] [PubMed] [Google Scholar]
- 154.Hoshino A, Costa-Silva B, Shen TL, Rodrigues G, Hashimoto A, Tesic Mark M, Molina H, Kohsaka S, Di Giannatale A, Ceder S, et al. Tumour exosome integrins determine organotropic metastasis. Nature. 2015;527:329–335. doi: 10.1038/nature15756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Markou A, Zavridou M, Sourvinou I, Yousef G, Kounelis S, Malamos N. Direct comparison of metastasis-related miRNAs expression levels in circulating tumor cells, corresponding plasma, and primary tumors of breast cancer patients. Clin Chem. 2016;62:1002–1011. doi: 10.1373/clinchem.2015.253716. [DOI] [PubMed] [Google Scholar]
- 156.Economopoulou P, Koutsodontis G, Strati A, Kirodimos E, Giotakis E, Maragoudakis P, Prikas C, Papadimitriou N, Perisanidis C, Gagari E, et al. Surrogates of immunologic cell death (ICD) and chemoradiotherapy outcomes in head and neck squamous cell carcinoma (HNSCC) Oral Oncol. 2019;94:93–100. doi: 10.1016/j.oraloncology.2019.05.019. [DOI] [PubMed] [Google Scholar]
- 157.Sutton TL, Walker BS, Wong MH. Circulating hybrid cells join the fray of circulating cellular biomarkers. Cell Mol Gastroenterol Hepatol. 2019;8:595–607. doi: 10.1016/j.jcmgh.2019.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Mentis AFA, Dardiotis E, Romas NA, Papavassiliou AG. PIWI family proteins as prognostic markers in cancer: a systematic review and meta-analysis. Cell Mol Life Sci. 2019 doi: 10.1007/s00018-019-03403-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Neumann MHD, Bender S, Krahn T, Schlange T. ctDNA and CTCs in liquid biopsy—current status and where we need to progress. Comput Struct Biotechnol J. 2018;16:190–195. doi: 10.1016/j.csbj.2018.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Snyder MW, Kircher M, Hill AJ, Daza RM, Shendure J. Cell-free DNA comprises an in vivo nucleosome footprint that informs its tissues-of-origin. Cell. 2016;164:57–68. doi: 10.1016/j.cell.2015.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Zhang H, Han J, Kang B, Burgess R, Zhang Z. Human histone acetyltransferase 1 protein preferentially acetylates H4 histone molecules in H3.1-H4 over H3.3-H4. J Biol Chem. 2012;287:6573–6581. doi: 10.1074/jbc.M111.312637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Cristiano S, Leal A, Phallen J, Fiksel J, Adleff V, Bruhm DC, Jensen SØ, Medina JE, Hruban C, White JR, et al. Genome-wide cell-free DNA fragmentation in patients with cancer. Nature. 2019;570:385–389. doi: 10.1038/s41586-019-1272-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Hoadley KA, Yau C, Hinoue T, Wolf DM, Lazar AJ, Drill E, Shen R, Taylor AM, Andrew D, Akbani R, et al. Cell-of-origin patterns dominate the molecular classification of 10,000 tumors from 33 types of cancer. Cell. 2018;173:291–304. doi: 10.1016/j.cell.2018.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Priestley P, Baber J, Lolkema MP, Steeghs N, De Bruijn E, Shale C, Duyvesteyn K, et al. Pan-cancer whole-genome analyses of metastatic solid tumours. Nature. 2019;575:210–216. doi: 10.1038/s41586-019-1689-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Parikh SA, Kay NE, Shanafelt TD. How we treat Richter syndrome. Blood. 2014;123:1647–1657. doi: 10.1182/blood-2013-11-516229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Yeh P, Hunter T, Sinha D, Ftouni S, Wallach E, Jiang D, Chan YC, Wong SQ, Silva MJ, Vedururu R, et al. Circulating tumour DNA reflects treatment response and clonal evolution in chronic lymphocytic leukaemia. Nat Commun. 2017;8:1–7. doi: 10.1038/ncomms14756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Miller AM, Shah RH, Pentsova EI, Pourmaleki M, Briggs S, Distefano N, Zheng Y, Skakodub A, Mehta SA, Campos C, et al. Tracking tumour evolution in glioma through liquid biopsies of cerebrospinal fluid. Nature. 2019;565:654–658. doi: 10.1038/s41586-019-0882-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Jones PS, Yekula A, Lansbury E, Small JL, Ayinon C, Mordecai S, Hochberg FH, Tigges J, Delcuze B, Charest A, et al. Characterization of plasma-derived protoporphyrin-IX-positive extracellular vesicles following 5-ALA use in patients with malignant glioma. EBioMedicine. 2019;48:23–35. doi: 10.1016/j.ebiom.2019.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Sindeeva OA, Verkhovskii RA, Sarimollaoglu M, Afanaseva GA, Fedonnikov AS, Osintsev EY, Kurochkina EN, Gorin DA, Deyev SM, Zharov VP, et al. New frontiers in diagnosis and therapy of circulating tumor markers in cerebrospinal fluid in vitro and in vivo. Cells. 2019;8:1195. doi: 10.3390/cells8101195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Louveau A, Plog BA, Antila S, Alitalo K, Nedergaard M, Kipnis J. Understanding the functions and relationships of the glymphatic system and meningeal lymphatics. J Clin Investig. 2017;127:3210–3219. doi: 10.1172/JCI90603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Harrison IF, Siow B, Akilo AB, Evans PG, Ismail O, Ohene Y, Nahavandi P, Thomas DL, Lythgoe MF, Wells JA. Non-invasive imaging of CSF-mediated brain clearance pathways via assessment of perivascular fluid movement with diffusion tensor MRI. Elife. 2018;7:1–14. doi: 10.7554/eLife.34028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Alunni-Fabbroni M, Müller V, Fehm T, Janni W, Rack B. Monitoring in metastatic breast cancer: is imaging outdated in the era of circulating tumor cells? Breast Care. 2014;9:16–21. doi: 10.1159/000360438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Liu J, Dang H, Wang XW. The significance of intertumor and intratumor heterogeneity in liver cancer. Exp Mol Med. 2018;50:e416. doi: 10.1038/emm.2017.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Mentis AFA, Boziki M, Grigoriadis N, Papavassiliou AG. Helicobacter pylori infection and gastric cancer biology: tempering a double-edged sword. Cell Mol Life Sci. 2019;76:2477–2486. doi: 10.1007/s00018-019-03044-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.El-deiry WS, Taylor B, Neal JW. Tumor evolution, heterogeneity, and therapy for our patients with advanced cancer: how far have we come? Am Soc Clin Oncol Educ Book. 2017;37:e8–e15. doi: 10.1200/EDBK_175524. [DOI] [PubMed] [Google Scholar]
- 176.Rossi E, Zamarchi R. Single-cell analysis of circulating tumor cells: how far have we come in the omics era? Front Genet. 2019;10:1–12. doi: 10.3389/fgene.2019.00958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Rutman AM, Kuo MD. Radiogenomics: creating a link between molecular diagnostics and diagnostic imaging. Eur J Radiol. 2009;70:232–241. doi: 10.1016/j.ejrad.2009.01.050. [DOI] [PubMed] [Google Scholar]
- 178.Hesketh RL, Zhu AX, Oklu R. Hepatocellular carcinoma: can circulating tumor cells and radiogenomics deliver personalized care? Am J Clin Oncol Cancer Clin Trials. 2015;38:431–436. doi: 10.1097/COC.0000000000000123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Nair VS, Keu KV, Luttgen MS, Kolatkar A, Vasanawala M, Kuschner W, Bethel K, Iagaru AH, Hoh C, Shrager JB, et al. An observational study of circulating tumor cells and 18F-FDG PET uptake in patients with treatment-naive non-small cell lung cancer. PLoS One. 2013;8:e67733. doi: 10.1371/journal.pone.0067733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Yu JQ, Cristofanilli M. Circulating tumor cells and PET. J Nucl Med. 2011;52:1501–1504. doi: 10.2967/jnumed.111.097683. [DOI] [PubMed] [Google Scholar]
- 181.Tabouret-Viaud C, Botsikas D, Delattre BMA, Mainta I, Amzalag G, Rager O, Vinh-Hung V, Miralbell R, Ratib O. PET/MR in breast cancer. Semin Nucl Med. 2015;45:304–321. doi: 10.1053/j.semnuclmed.2015.03.003. [DOI] [PubMed] [Google Scholar]
- 182.Ehman EC, Johnson GB, Villanueva-meyer JE, Cha S, Leynes AP, Eric P, Larson HTA. Mechanism of oxidative conversion of Amplex Red to resorufin: pulse radiolysis and enzymatic studies. Free Radic Biol Med. 2017;46:1247–1262. [Google Scholar]
- 183.Bettigole C. The thousand-dollar Pap smear. N Engl J Med. 2013;369:1486–1487. doi: 10.1056/NEJMp1307295. [DOI] [PubMed] [Google Scholar]
- 184.Marrinucci D, Bethel K, Bruce RH, Curry DN, Hsieh B, Humphrey M, Krivacic RT, Kroener J, Kroener L, Ladanyi A, et al. Case study of the morphologic variation of circulating tumor cells. Hum Pathol. 2007;38:514–519. doi: 10.1016/j.humpath.2006.08.027. [DOI] [PubMed] [Google Scholar]
- 185.Marrinucci D, Bethel K, Lazar D, Fisher J, Huynh E, Clark P, Bruce R, Nieva J, Kuhn P. Cytomorphology of circulating colorectal tumor cells: a small case series. J Oncol. 2010;2010:1–7. doi: 10.1155/2010/861341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Araújo FHD, Silva RRV, Ushizima DM, Rezende MT, Carneiro CM, Campos Bianchi AG, Medeiros FNS. Deep learning for cell image segmentation and ranking. Comput Med Imaging Graph. 2019;72:13–21. doi: 10.1016/j.compmedimag.2019.01.003. [DOI] [PubMed] [Google Scholar]
- 187.Lu Z, Carneiro G, Bradley AP, Ushizima D, Nosrati MS, Bianchi AGC, Carneiro CM, Hamarneh G. Evaluation of three algorithms for the segmentation of overlapping cervical cells. IEEE J Biomed Heal Inform. 2017;21:441–450. doi: 10.1109/JBHI.2016.2519686. [DOI] [PubMed] [Google Scholar]
- 188.Mozdiak E, Wicaksono AN, Covington JA, Arasaradnam RP. Colorectal cancer and adenoma screening using urinary volatile organic compound (VOC) detection: early results from a single-centre bowel screening population (UK BCSP) Tech Coloproctol. 2019;23:343–351. doi: 10.1007/s10151-019-01963-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Abderrahman B. Exhaled breath biopsy: a new cancer detection paradigm. Future Oncol. 2019;15:1679–1682. doi: 10.2217/fon-2019-0091. [DOI] [PubMed] [Google Scholar]
- 190.Farhadi A, Ho GH, Sawyer DP, Bourdeau RW, Shapiro MG. Ultrasound imaging of gene expression in mammalian cells. Science. 2019;365:1469–1475. doi: 10.1126/science.aax4804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Kourou K, Exarchos TP, Exarchos KP, Karamouzis MV, Fotiadis DI. Machine learning applications in cancer prognosis and prediction. Comput Struct Biotechnol J. 2015;13:8–17. doi: 10.1016/j.csbj.2014.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Hu Z, Ding J, Ma Z, Sun R, Seoane JA, Scott Shaffer J, Suarez CJ, Berghoff AS, Cremolini C, Falcone A, Loupakis F, Birner P, Preusser M, Lenz HJ, Curtis C. Quantitative evidence for early metastatic seeding in colorectal cancer. Nat Genet. 2019;51:1113–1122. doi: 10.1038/s41588-019-0423-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Mentis AFA, Kararizou E. Epithelial-mesenchymal transition and cancerogenesis. Iatriki. 2010;8:379–385. [Google Scholar]
- 194.Parkins KM, Dubois VP, Kelly JJ, Chen Y, Foster PJ, Ronald JA. Engineering “self-homing” circulating tumour cells as novel cancer theranostics. bioRxiv. 2019 doi: 10.1101/746685v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Akolkar D, Patil D, Crook T, Limaye S, Page R, Datta V, Patil R, Sims C, Ranade A, Fulmali P et al (2019). Circulating ensembles of tumor associated cells: a redoubtable new systemic hallmark of cancer. Int J Cancer. https://www.ncbi.nlm.nih.gov/pubmed/31785151 [DOI] [PMC free article] [PubMed]
- 196.Baron CS, van Oudenaarden A. Unravelling cellular relationships during development and regeneration using genetic lineage tracing. Nat Rev Mol Cell Biol. 2019;20:753–765. doi: 10.1038/s41580-019-0186-3. [DOI] [PubMed] [Google Scholar]
- 197.Gerstung M, Jolly C, Leshchiner I, Dentro SC, Gonzalez S, Rosebrock D, Mitchell TJ, Rubanova Y, Anur P, Yu K, Tarabichi M, Deshwar A, Wintersinger J, Kleinheinz K, Vázquez-García I, Haase K, Jerman L, Sengupta S, Macintyre G, Malikic S, Donmez N, Livitz DG, Cmero M, Demeulemeester J, Schumacher S, Fan Y, Yao X, Lee J, Schlesner M, Boutros PC, Bowtell DD, Zhu H, Getz G, Imielinski M, Beroukhim R, Sahinalp SC, Ji Y, Peifer M, Markowetz F, Mustonen V, Yuan K, Wang W, Morris QD, PCAWG Evolution & Heterogeneity Working Group. Spellman PT, Wedge DC, Van Loo P, PCAWG Consortium The evolutionary history of 2658 cancers. Nature. 2020;578:122–128. doi: 10.1038/s41586-019-1907-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Jiao W, Atwal G, Polak P, Karlic R, Cuppen E, PCAWG Tumor Subtypes and Clinical Translation Working Group. Danyi A, de Ridder J, van Herpen C, Lolkema MP, Steeghs N, Getz G, Morris Q, Stein LD, PCAWG Consortium A deep learning system accurately classifies primary and metastatic cancers using passenger mutation patterns. Nat Commun. 2020;11:728. doi: 10.1038/s41467-019-13825-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Milanez-Almeida P, Martins AJ, Germain RN, Tsang JS. Cancer prognosis with shallow tumor RNA sequencing. Nat Med. 2020;26:188–192. doi: 10.1038/s41591-019-0729-3. [DOI] [PubMed] [Google Scholar]