Abstract
Natural reservoir hosts can sustain infection of pathogens without succumbing to overt disease. Multiple bat species host a plethora of viruses, pathogenic to other mammals, without clinical symptoms. Here, we detail infection of bat primary cells, immune cells, and cell lines with Dengue virus. While antibodies and viral RNA were previously detected in wild bats, their ability to sustain infection is not conclusive. Old-world fruitbat cells can be infected, producing high titres of virus with limited cellular responses. In addition, there is minimal interferon (IFN) response in cells infected with MOIs leading to dengue production. The ability to support in vitro replication/production raises the possibility of bats as a transient host in the life cycle of dengue or similar flaviviruses. New antibody serology evidence from Asia/Pacific highlights the previous exposure and raises awareness that bats may be involved in flavivirus dynamics and infection of other hosts.
Electronic supplementary material
The online version of this article (10.1007/s00018-019-03242-x) contains supplementary material, which is available to authorized users.
Keywords: Flavivirus, Immunity, Zoonosis, Pteropus, Disease, Bats
Introduction
In 1952, Reagen and Brueckner infected the Myotis lucifigus cave bat with Dengue virus (DENV)—Hawaii in vivo [1]. They successfully passaged the virus intra-cranially for six passages intra-bat. The bats displayed central nervous system defects and the virus produced was infectious and neutralised by DENV antibodies. While this was not a natural route of infection, it raised the possibility of natural infection/susceptibility of bats to DENV infection. In 1964, Shepherd and Williams infected the Angolan fruit bat (Rousettus angolensis), African straw-colored fruit bat (Eidolon helvum) and Egyptian fruit bat (Rousettus aegyptiacus) with Zika virus (ZIKV) [2]. Japanese encephalitis (JEV) virus infection has been established in Eptesicus fuscus, Myotis lucifugus and Pipistrellus subflavus bats, including validation of a sylvatic cycle with induced mosquito transmission [3]. Other flavivirus infection, such as St Louis encephalitis virus (SLEV), has been sustained in Eptesicus fuscus, even throughout hibernation [4]. Experimental infections of Yellow fever virus (YFV) and West Nile virus (WNV) (NY-99) show circulating virus in Eidolon Helvum, Rousettus sp, Eptesicus fuscus, and Tadarida brasiliensis. The same study also showed circulating Zika infection in Eidilon Helvum and Rousettus sp; however, there were low viremia and no clinical signs of disease to any virus [5, 6]. The absence of major symptoms in these subsequent infections is likely due to a more natural route of infection compared to intracranial passaging which artificially induces high titres across the blood–brain barrier. While species-specific factors may come into effect, there are also differences between flavivirus neurotropism that may play a role with regards to symptoms, [7–10]. Of note, to date, there have been 16 different flaviviruses isolated from bats in the wild [11].
There have been several studies detecting DENV-specific antibodies, neutralising antibodies, and even nucleic acids in bats [12–14]. There has been one experimental infection study of the great fruit-eating bats (Artibeus intermedius) with DENV2, and while there were signs of spleen, liver, and intestine damage, the virus was difficult to detect with limited sero-conversion [15]. The authors concluded that this species is not a suitable host for the virus. A close neighbour of this neotropical bat species (Artibeus jamaicensus) was evaluated at the cellular level along with two other related species and all displayed poor infection and replication [16]. The authors conclude that neotropical bat species may be incidental hosts during DENV transmission cycles and not capable of sustaining infection. However, the role of IndoMalayan and Australasian fruit bats in the course of DENV sylvatic infection has not been investigated.
Here, we report evidence for infection, active replication, and production of high titres from various cell lines of several bat species from IndoMalaya and Australasia. Different DENV strains readily infect and produce high viral titres/shedding. Intriguingly, DENV2 elicits a minimal immune response by qPCR, NanoString and proteomics in the Pteropus alecto bat cell line PakiT03 or primary immune cells. This fits with the role of a natural host of the pathogen [17–20]. In addition, we provide further serological evidence in bats of three species on the presence of DENV antibodies.
Materials and methods
Viruses and cells
Dengue virus stocks (DENV1 EDEN 2402, DENV2 New Guinea C, DENV2 Strain Thailand 16681, DENV3 EDEN 853, DENV4 SL2544) were propagated in C6/36 mosquito cells and clarified by centrifugation to remove cell debris. Nelson bay virus (p2) (NBV/PRV1NB) or Melaka virus (MelV/PRV3M) as a control was propagated in Vero E6 cells and clarified as previous. Viral titres were calculated upon infectivity in BHK and Vero E6 cells, respectively, by plaque assay. Pteropus alecto cell lines PakiT03 and PaLu have been published previously [21]. Eonycteris spelaea primary lung cell lines (EsLu) were generated by collagenase-type IV-digested lung tissue, grown out for selection of fibroblasts/epithelia and treated with SV40T antigen to attempt immortalisation (passages 2–10 were used so immortalisation status is not certain). Pteropus alecto primary lung was prepared similarly, but the initial p1 mixed cell suspension (including immune cells) was used. Pteropus alecto and Cynopterus brachyotis primary spleen cells were prepared by cutting and grinding the spleen through a 100 μm sieve and then resuspending in RPMI/10% FBS after RBC lysis (eBioScience, Thermo Scientific). The remaining adherent cells following growth from P. alecto were treated with SV40-T retrovirus for immortalisation of the PaSpT.01 spleen-derived fibroblast cell line. P. alecto bone-marrow-derived macrophage and dendritic cells were prepared as described previously [22].
Viral infection assays
Cells were plated onto 13 mm round glass coverslips #1.5 and allowed to adhere prior to infection. Infection was performed in 1% FBS in Fluorobrite-DMEM or phenol Red Free RPMI 1640 (Gibco, Thermo Scientific) for 2 h followed by rinsing with DPBS and fixing (viral entry) or incubating in 1% FBS/DMEM or RPMI for 24–72 h (replication) prior to either fixation or RNA extraction. Cells were fixed in 3.7% paraformaldehyde/1% glutaraldehyde (Sigma). RNA extraction was performed using the E.Z.N.A.® Total RNA Kit I (Omega Bio-Tek) and cDNA conversion/gDNA digestion with QuantiTect Reverse Transcription Kit (Qiagen) as per the manufacturer’s instructions. Samples for microscopy were permeabilised and labelled as previously described using anti 3H5/4G2 hybridoma TC supernatant (1:700), Alexa-647 Phalloidin (2 μl/200 ul), Hoechst 3342 (Thermo Scientific), anti β-actin (Sigma, #A2228), CellBrite membrane DiO (Biotium), or anti J2-dsRNA (1:1000) (SciCons) and labelled with anti-alexa 488/568/594/647 secondary antibodies (1:1000) or conjugated with Biotin/Alexa 488/568/647 Zenon labelling kits (Life Technologies). Tyramide signal amplification was performed with the TSA-594 or TSA-biotin labelling kits (Thermo Scientific) as per the manufacturer’s instruction followed by streptavidin-Northern Lights-637 (1:1000) (R&D systems). qPCR was performed using the SensiFAST SYBR no-ROX kit (BioLine) on a BioRad CFX96 machine. Primer sequences are available in the extended methods.
Innate immune stimulation
Virus infection was performed as previous at an MOI of 0.1 pfu/cell for 24 h in complete DMEM. Media were replaced with 1% FBS and cells were transfected with Fugene 6 (Promega): polyIC (InvivoGen) or 5′-ppp-dsRNA (InvivoGen) at a ratio of 3:1 (μl/μg) in OptiMEM (Gibco, ThermoFisher) to a final concentration on cells of 1 μg/ml for 6 h. RNA was collected as previous. The same transfection method was used for the stimulation of ligands in primary splenocytes (5 h treatment).
Viral plaque assays
Viral plaque assays were performed as previously described using a 2 h infection followed by rinsing and then adding a 1% methyl-cellulose (Sigma) overlay on BHK/PakiT03 cells in 2% FBS/DMEM for 4 days in triplicate in a 24-well plate with a tenfold dilution series.
Viral infection for label-free proteomics
Infections were performed as per previous in a T75 flask in biological triplicate and incubated for 48 h in 2% FBS/DMEM. Prior to harvesting cells were rinsed 3× in ice-cold Ca2+, Mg2+-free DPBS (Thermo Scientific) and harvested in 1.5 ml RIPA buffer (Pierce, Thermo Scientific). Details of the proteomics pipeline are in the extended methods section. The positive control sample was transfected with polyI:C (1 μg/ml, Roche) using Fugene 6 for 24 h and harvested at the same time.
Serology assays for DENV
Luciferase-based immunoprecipitation assays (LIPS) were performed as described previously for DENV2 NS1 protein fused to Renilla luciferase. The input was normalised to 1 million light units and 0.8 μl of neat bat sera was incubated with each antigen, prior to protein A/G capture and analysis using the single Renilla luciferase kit (Promega). Geometric mean of the samples ± 3 SD was used as the cutoff for baseline signal. DENV ELISA was performed using the cross-reactive DENV2 NS1 antigen as described previously [23]. 4 μl of 1:2 diluted bat sera was used for capture with the commercial bat IgG HRP (Novus Bio) antibody as detection and TMB substrate for visualisation (Thermo Scientific). VirScan was performed as previously described, see supplemental methods for further detail.
Results
Infection of PaKiT03 with minimal immune responses
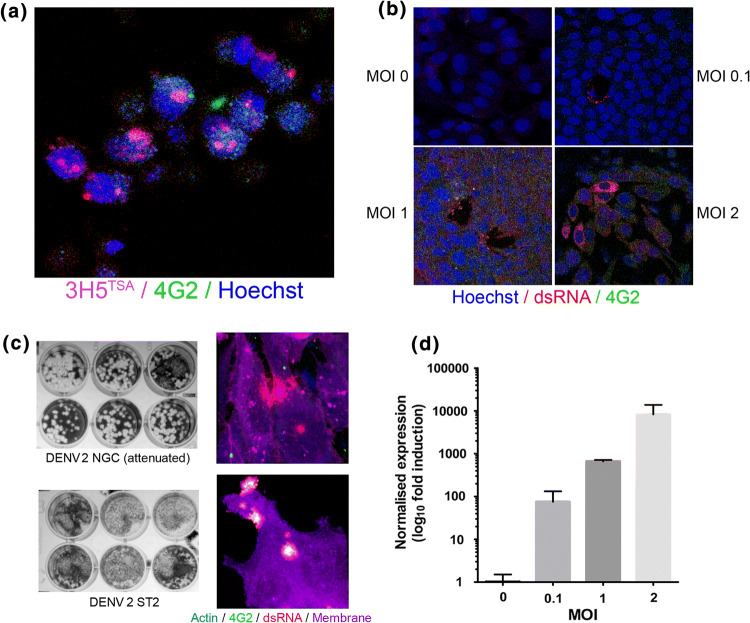
To examine the infectivity of Pteropus alecto cells for DENV, we studied viral entry (2 h) with DENV2 (NGC) in PakiT03 cells using a high MOI of 10 (Fig. 1a, S1a). Virus particles were detected by DENV antibodies (3H5, anti-PreM-E) with TSA signal amplification (~ 20×). Confocal microscopy was used to eliminate any surface-bound virus particles and example Z-slices are shown to indicate entry. To establish if active viral replication was underway, cells were labelled for cellular dsRNA (mab J2, anti-dsRNA) and virus (4G2) from viral replication (24 h) at several MOIs (Fig. 1b). The distinct localisation of dsRNA after infection correlates with previously published literature on replicating bodies [24–26]. As a measure of the complete replication cycle and production of infectious virus, supernatant from cells infected at an MOI of 1 (72 h) with two different DENV2 strains was plaqued onto BHK cells for 5 days (Fig. 1c, S1b). Matched confocal images of the infected cells are presented to show active replication (dsRNA staining). Confirmation of viral replication was performed by qPCR for DENV2 (NGC) NS2a across MOIs of 0.1, 1, 2 (Fig. 1d). Intriguingly, prolonged replication was observed (96 h) in PakiT03 cells with an MOI of 0.1, an MOI commonly used in the flavivirus field [27–31], showing active cytoplasmic dsRNA (J2), 3H5 staining, 4G2 staining (anti-E, intact), and yet, there was no observable CPE (DiC-imaging) at this dosage (Fig. S1c). Taken together, these data indicate that DENV2 can infect, replicate, produce infectious virus, and yet have minimal impact on the health of PakiT03 cells.
Fig. 1.
Productive DENV infection in bat cells. a Single confocal z-plane images of Dengue virus (NGC) entry into PakiT03 cells after 2 h at a MOI of 10 (washed, then fixed). Cells were stained with biotinylated 3H5 antibody (PreM-E) followed by TSA-Alexa594 amplification, Alexa-488 conjugated 4G2 antibody (Env) (green) and Hoechst3342 (blue). Membrane stain (Phalloidin-647) was visualised to ensure internalisation of virus particles but not included in the image. b J2 (dsRNA) replication after 24 h was visualised at various MOI’s (as indicated) with the J2-dsRNA antibody-alexa 594 conjugated (red), Alexa-488 conjugated 4G2 antibody (Env) (green) and counterstained with Hoechst 3342 (blue), as previously. c Productive infection was measured by titrating viral supernatant from PakiT03 cells after 48 h of infection with an MOI of 1 for either DENV2 (NGC) or DENV2 (ST2), infected cells were labelled with 4G2 (green), anti-βactin (blue), J2 (dsRNA) (red) or membrane DiO label (magenta)—right panel, as previously. Supernatant was titrated onto BHK cells for 5 days in 1% methyl-cellulose overlays. Example images are shown for MOI of 0.1, 1 and n = 3, d qPCR validation of viral replication at various MOI’s after 48 h of infection in PakiT03 cells using primers for NS2a, normalised to housekeeping gene expression (SNRPD3). One representative experiment is shown for n = 3
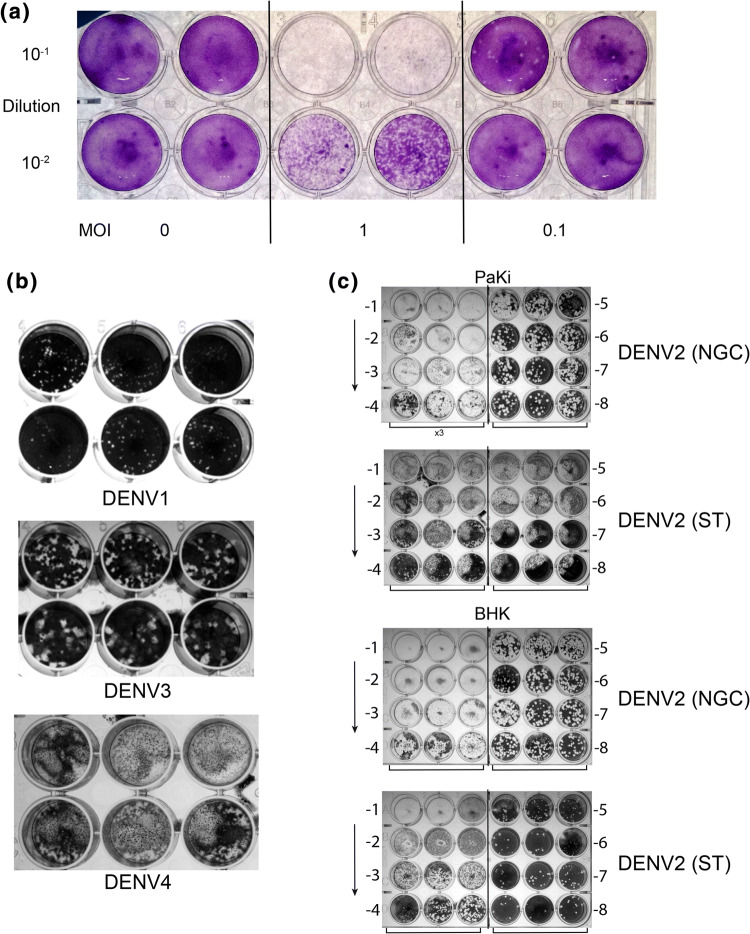
To more closely examine bat cells’ ability to produce infectious viral particles, we examined supernatant from cells infected for 24 h with an MOI of either 0.1 or 1 and titrated onto BHK cells (Fig. 2a). The short time frame was used to indicate a rapid production of virus particles. To calculate whether virus infection led to “productive” infection, supernatant collected after 72 h of infection in PakiT03 cells with an MOI of 0.1 was titrated onto BHK cells yielding a titre of 3.5 × 108 pfu/ml (Fig. S1d). The initial infection was approximately 4.98 × 105 pfu/ml which is approximately 3 × log lower than the output, indicating active production of virus particles to a level (in vitro) easily high enough for mosquito transmission [32, 33]. Active virus production was also observed for DENV 3 (EDEN 853), DENV1 (EDEN 2402) and DENV4 (SL2544) (Fig. 2b), indicating their potential to be amplifying hosts for multiple Dengue serotypes, although a slightly lower titre (106 pfu/ml) was observed compared to DENV2 with DENV4 failing to produce defined plaques in PakiT03 cells. The overall infectivity of PakiT03 cells compared to BHK-21, standard cells for Dengue titration, was titred side-by-side with DENV2 NGC and ST (Fig. 2c). The results indicate a similar infection capacity for these bat cells compared to BHK-21 cells, showing a high permissiveness for infection, in conjunction to production of high viral yields. Interestingly, strain ST yields “small” plaques in PakiT03 cells, though all other DENV strains tested produce much larger plaques.
Fig. 2.
High production of Dengue virus and infectivity with multiple strains. a Viral supernatant from PakiT03 cells post-infection (48 h) with DENV2 NGC as per Fig. 1c titrated onto BHK cells for 4 days with a methyl-cellulose overlay showing infective particles being produced from PakiT03 cells at 24 h with an MOI of 0.1 or 1. b Supernatant from DENV1,3,4 infected PakiT03 cells after 72 h with an MOI of 1, titrated onto BHK cells and left for 5 days in 1% methyl-cellulose as described previously. c DENV2 NGC and ST strains produced in PakiT03 cells for 96 h (Fig. 1c) after an initial MOI of 0.1 and then titrated onto both BHK cells and PakiT03 cells showing Paki cells are highly productive for DENV
Confirmation across cell types of multiple bat species
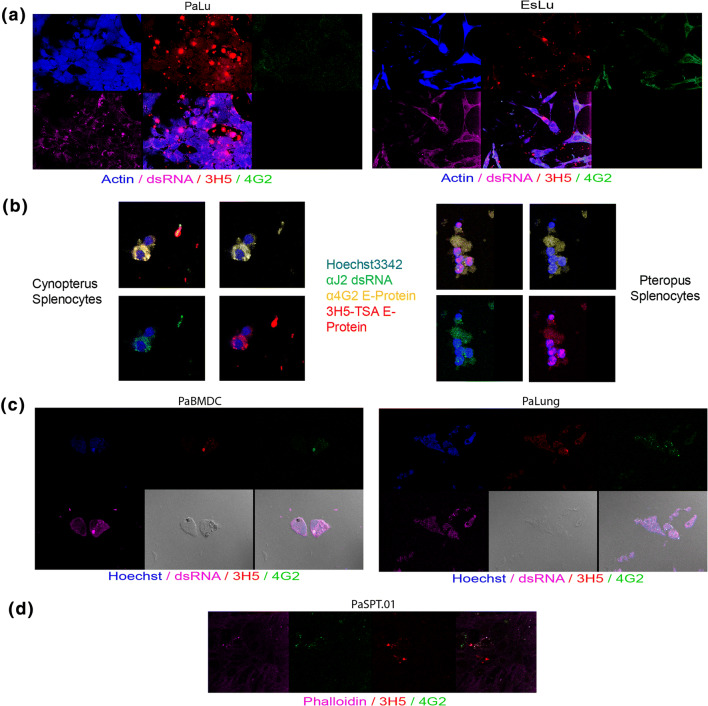
To ensure that infection was not cell line specific, we evaluated multiple cell lines from different species for their susceptibility to infection. Pteropus alecto lung cells (PaLu) and Eonycteris spelaea lung cells (EsLu) were infected as previously described for 48 h with an MOI of 1 (Fig. 3a, S2a, b). Active replication of DENV2 was observed as per PakiT03. Similarly, Cynopterus brachyotis and Pteropus alecto primary splenocytes were extracted and cultured overnight with DENV2 NGC at an MOI of 0.1 (Fig. 3b). Antibody labelling revealed active dsRNA replication along with DENV2 antibody staining in the majority of adherent splenocytes with 16 h of incubation. P. alecto FLT3-bone-marrow-derived dendritic cells showed active replication of DENV2 after 24 h of infection with an MOI of 0.1 (Fig. 3c). Distinct replicating centres can be observed with co-localisation of 3H5, 4G2, and J2 staining. Similarly, primary P. alecto lung cell suspensions indicated successful infection. Finally, P. alecto spleen-derived fibroblasts cells PaSpT.01 were infected for 48 h with an MOI of 0.1 (Fig. 3d) indicating a wide range of cell tropism that included fibroblast, epithelial, and stromal primary cells and cell lines, along with primary and in vitro-derived immune cells across three different species of bats.
Fig. 3.
Infectivity of multiple cell types, species and immune cells by Dengue virus. a Single-plane confocal images of PaLu (Pteropus alecto lung epithelial cells, left panel) and EsLu (Eonycteris spelaea lung epithelial cells) cells after 48 h infection with an MOI of 1 and labelled with Actin (blue), dsRNA (magenta), 3H5-594 (red) and 4G2 (green), as previously. b Primary splenocytes infected with DENV2 (NGC) at an MOI of 0.1 overnight for ~ 16 h, fixed and stained as previous (antibodies as indicated) for Cynopterus brachyotis (left panel) and Pteropus alecto (right panel). c Bone-marrow-derived dendritic cells from Pteropus alecto (FLT3-ligand, as previously published, left) infected with DENV2 (NGC) for 24 h with an MOI of 0.1 and labelled as per Fig. 2 (direct labelling without TSA amplification). Image includes DiC brightfield images and merged image as last picture. Similarly, for primary Pteropus alecto lung (mixed) cell suspension (right). d as per c for 48 h in PaSPT.01 (SV40T-immortalised spleen-derived fibroblast cells). Example images for one experiment are shown for n = 3 in a–d
Cellular responses to DENV infection
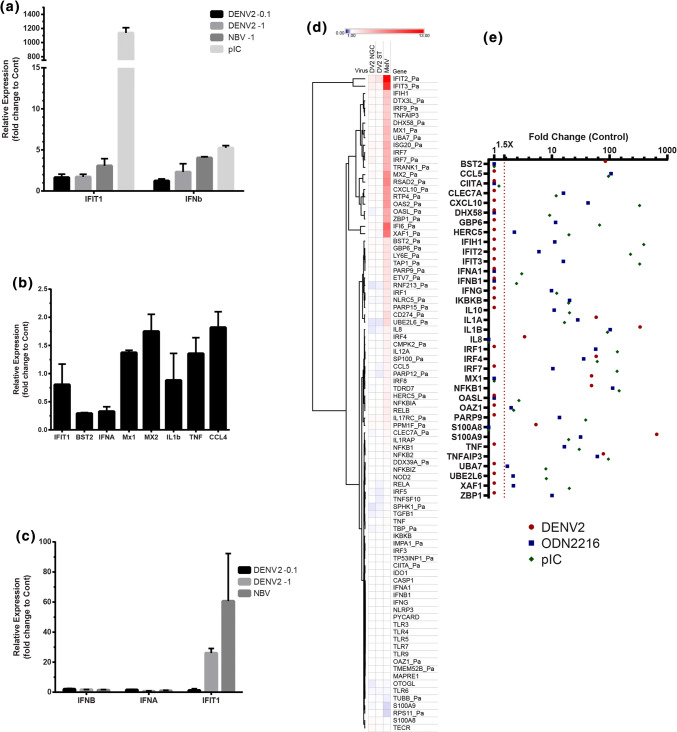
To evaluate potential cellular response to Dengue viruses, qPCR was performed on PakiT03 cells after 48 h with infection of DENV2 NGC at an MOI of 0.1 or 1, along with another RNA virus Nelson Bay Virus (NBV/Pteropus Orthoreovirus 1 NB/PRV1NB) or a positive control of dsRNA (transfected PolyI:C) (Fig. 4a). Levels of IFIT1 or IFNβ were quantified to examine the Interferon type I (IFN) response. Neither dose of DENV2 had a significant impact on gene expression compared to NBV or polyI:C. Matched RNA samples to Fig. 1b were examined for IFN-Regulated Gene (IRG) induction or cytokine gene induction, including IFIT1, BST2, IFNα, MX1, MX2, IL1β, TNF, and CCL4 (Fig. 4b) indicating, at most twofold gene induction, despite significant replication. This matched the lack of IFNα induction in BMDCs by Western Blot (Fig. S3a). This result seemed highly unusual as DENV was expected to trigger a strong immune response in PakiT03 cells, compared to a strong response triggered by other RNA stimulants or the response to DENV in in vitro cell culture of other mammals. The potential response of PakiT03 cells was confirmed at longer timepoints with IFNβ, IFNα, and IFIT1 expression at a DENV2 MOI of 0.1 or 1 compared to NBV infection with an MOI of 1 at 48 h (Fig. 4c). Only IFIT1 seemed to be induced by DENV2 at higher MOIs at this timepoint (Fig. S3b–f).
Fig. 4.
Limited immune response triggered by Dengue virus. a RT-qPCR of Paki cells infected with DENV2 NGC at an MOI of 0.1 or 1 (as indicated), NBV MOI of 1 or transfected with polyI:C (1 µg/ml) for 24 h. Relative expression is shown for IFIT1 and IFNβ relative to uninfected and normalised against SNRPD3 housekeeping gene. b RT-qPCR of several genes for DENV2 NGC infection at 24 h with an MOI of 0.1, as per 1D. Relative expression to control, normalised against SNRPD3 is shown for IFIT1, BST2, IFNα3, MX1, MX2, IL-1β, TNF and CCL4. c Relative expression as per a but at 48 h for IFNβ, IFNα3 and IFIT1 with DENV2 NGC at MOI of 0.1 or 1 and NBV MOI of 1. A representative experiment is shown for each, n = 3. d NanoString expression data from 200 ng RNA on a custom bat panel (expressed as average fold change relative to uninfected PakiT03 control cells) is shown as averages (n = 3) for a 24 h infection at an MOI of 1 for DENV2 (NGC), DENV2 (ST), or Melaka virus (MelV). Scale is from 0–10-fold change, as indicated. N = 4 each. e As for d from E. spelaea primary splenocytes infected with DENV2 NGC for 16 h or treated with ODN2216 (TLR9 agonist) or polyIC (TLR3/RIG-I) for 5 h. Data expressed as fold change (Log10) with 1.5× cutoff (red line)
To compare multiple genes and viruses, NanoString with a custom Bat Panel was performed on RNA from PakiT03 cells infected with an MOI of 1 for 24 h with either DENV2 (NGC), DENV2 (ST) and another dsRNA Orthoreovirus, MelV, as a positive control (Fig. 4d). While MelV is a potent inducer of the innate immune response, both flaviviruses barely induced an immune response, even with an MOI of 1. To examine the effects of DENV on innate immune stimulation, PakiT03 cells were infected for 24 h with an MOI of 0.1 (shown previously to exhibit replication at 24 h) prior to treatment with either polyIC or 5′ppp-dsRNA (RIG-I stimulant, PPP′) for 6 h to induce innate immune activation (Fig. S3g, h). The presence of DENV clearly decreases the gene expression in response to ligands, as well as basal expression of many IRGs and known anti-viral genes. Strong suppression of the transcription of BST2, OAS2, CXCL10, RSAD2, and IFIT3, all known to exhibit anti-viral activity against Dengue virus, was observed [34–38]. This also included downregulation of DHX58 (RIG-I) and IFIH1 (MDA5) themselves, known sensors of dsRNA from DENV [39–41]. This effect is partially alleviated by polyIC, suggesting that activation of multiple pattern recognition receptors (PRRs) by polyIC is better than solely activating RIG-I. There is minor activation of some NFκB-inducible genes, and this does not appear to be affected by the presence of DENV. These data correlate with the qPCR results (Fig. S3b–f) and suggest that this level of replication for DENV (in PakiT03) can produce virus particles yet successfully evade the bat innate immune response. As seen from the qPCR, an MOI of 2 has no visible suppression of MX-1, IFIT1, or IFNα, though there is still only limited gene induction. This suggests that there is a threshold of activation/suppression.
Similar results were observed with primary E. spelaea splenocytes infected with DENV2 NGC (as per Fig. 3) when compared to common stimuli for monocyte/macrophage/DCs; either ODN2216 (TLR9) or polyIC (TLR3/RIG-I). While immune stimuli triggered potent induction of multiple genes, only limited gene induction was induced by DENV2 infection. BMDMs from P. alecto (as previously described) also exhibited limited gene induction to DENV2 NGC in culture, though these cells were previously shown to respond strongly to immune ligands (Fig. S4a) [22]. Unexpectedly, these cells also exhibited limited response to the same MOI of NBV, though NBV infection and its kinetics in macrophages are poorly characterised and this result may reflect only partial infectivity.
Quantitative proteomics post-infection with DENV
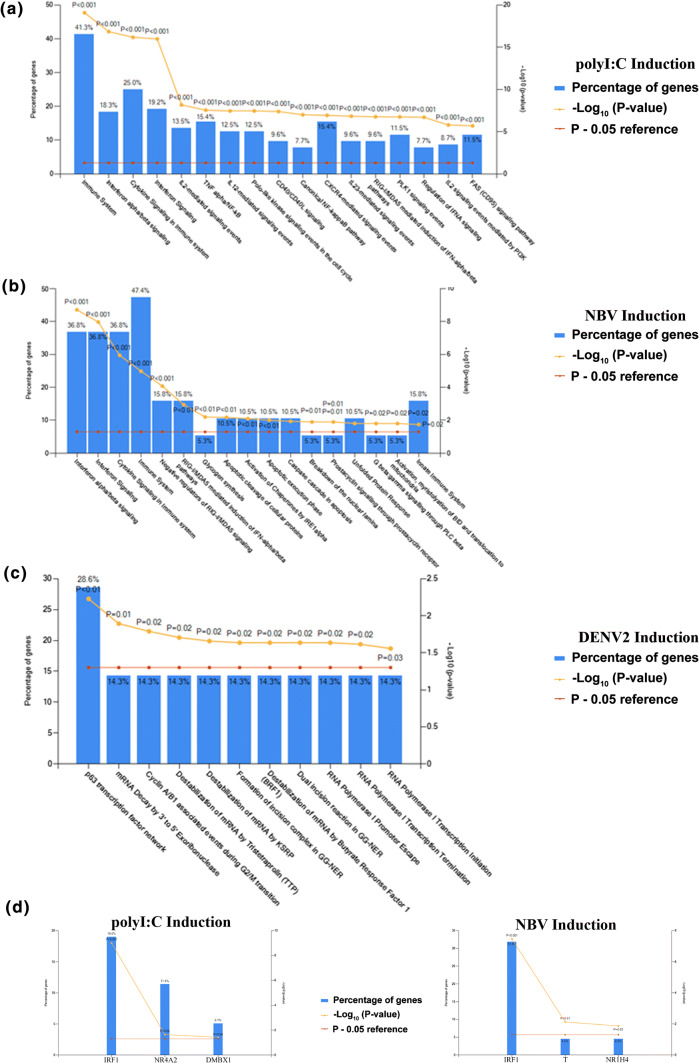
As antibodies to examine bat IFN responses at the protein level are lacking, we used quantitative whole-cell mass spectroscopy to examine en-bulk immune responses in PakiT03 cells. Table S1 contains the unique peptide count, MS count and normalised LFQ quantification for the 3721 proteins identified in 2 out of 3 replicates. DENV2 NGC samples were compared to either polyI:C transfected samples (24 h) or NBV-infected cells (matched MOI of 0.1 for 48 h). The significant pathways activated in PakiT03 cells by polyI:C include IFN activation, IFN signalling, immune signalling, cytokine signalling induction as expected, along with NFκB induction and RIG-I/MDA5 induction (Fig. 5a). NBV infection yielded similar results including negative regulation of RIG-I signalling, caspase/cell-death pathways (shown previously for Orthoreovirus infection [42]) and the unfolded protein response (Fig. 5b). Starkly, DENV2 infection did not yield any significant proteins, and the combination of mildly modified proteins indicated only one significant pathway, that of p63 transcription factor activation (Fig. 5c). The main transcription factors activated by polyIC treatment of PakiT03 cells included IRF1, NR4A2, and DMBX1 (Fig. 5d), while NBV activated IRF1, T, and NR1H4 (Fig. 5e), indicating minimal overlap with DENV2 infection but similarity between NBV and polyI:C. To confirm that infection was not impacted by DENV2-strain, the entire proteomics set was repeated for PakiT03 cells infected with DENV2 ST 16681 (a strain with a reliable protein reference in the database). Despite the entire DENV2 genome being covered by detected peptides for DENV2 (Table S2), there were only 6 significantly changed proteins with reliable coverage and unique peptide identities. This was not sufficient for pathway analysis, but indicated a possible involvement of endosomal re-assortment and Golgi recycling. This indicates a trend, whereby cells fully capable of an immune response were tolerant of DENV2 infection to such a degree that there was no visible cell death (at this MOI), no activation of an immune response or triggering of cell-death pathways and yet still producing large amounts of virus. Albeit at the cellular level, this result appears to be indicative of robust DENV production in P. alecto.
Fig. 5.
Quantitative proteomics of dengue infection reveals limited immune activation. a Pathway analysis (via reactome.org) of normalised LFQ protein counts from PakiT03 cells treated with polyI:C for 24 h (transfected) in biological triplicate. Graph overlay shows the percentage of genes from the pathway detected, the Z score p value as Log10 and the p = 0.05 threshold line (as indicated in the legend). b As per a for treatment with NBV at an MOI of 0.1 for 48 h. c As per a with DENV2 NGC at 48 h with an MOI of 0.1 d Transcription factor enrichment for pathways in a and e Transcription factors enriched for pathways in b
Serological investigation of DENV infection in different bats
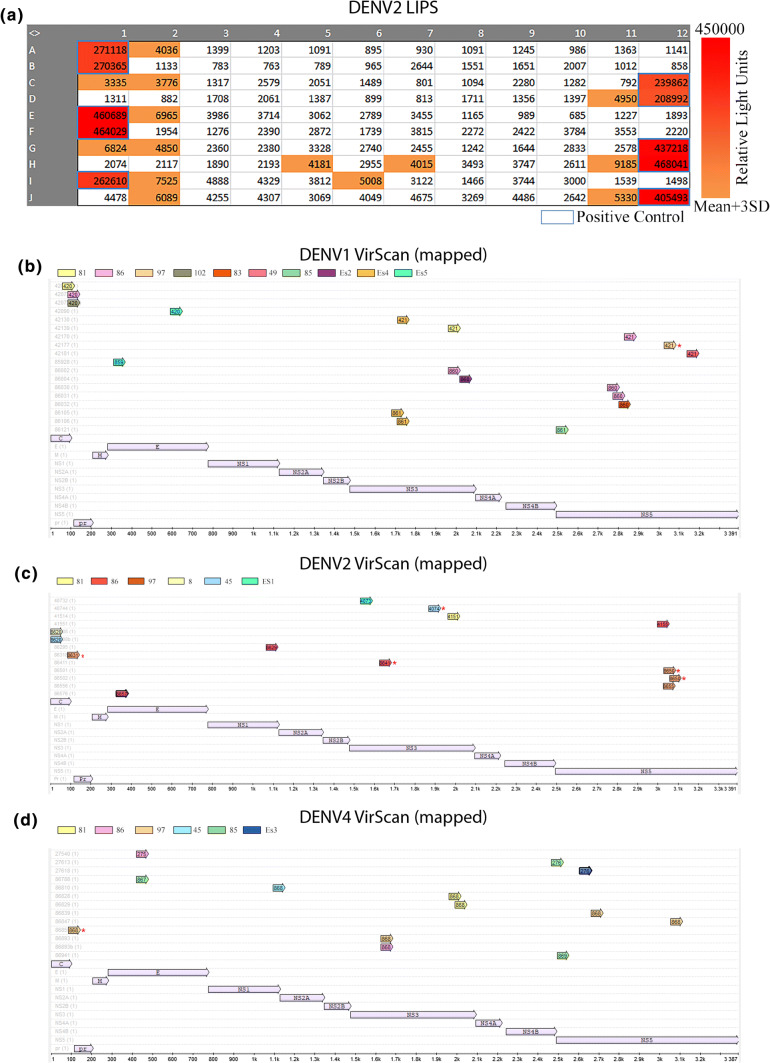
To evaluate any potential for DENV distribution/transmission by bats in the IndoMalaya region, heavily endemic for DENV, we performed luciferase immunoprecipitation system (LIPS)-serology for DENV2 NS1 antibodies in sera from E. spelaea and C. brachyotis bats in the region (Fig. 6a). The average body weight of bats used for this study was 45 g with all pregnant females excluded. A 53.9 g average was previously documented for the species [43], Blood collection from the wing yielded ~ 20–40 μl of sera. Due to limited serum availability, LIPS-based serology was used with only 2.4 μl of sera (for all replicates). 14/106 bats were sero-positive by LIPS. As this assay is mildly cross reactive with ZIKV, endemic to Singapore, samples were run concurrently with ZIKV NS1 (Fig. S5a). The relative ratio of DENV2 to ZIKV (normalised to positive control) signal was plotted accordingly (Fig. S5b). This indicated that most antibodies were against DENV2 with only two bats displaying stronger results for ZIKV NS1 over DENV. ZIKV NS1 in this assay produces a stronger signal (by RLU from equimolar protein) and has less signal to noise (lower values for mean + 4 SD), so further testing on these two samples would be required for confirmation. The data were, therefore, combined with the results for DENV2 NS1 IgG ELISA (mildly cross reactive with other Dengue Types but not other flaviviruses). The ELISA positive-IndoMalayan bats were additionally tested for all antibodies in their virome using VirScan phage-display peptide libraries covering linear epitopes of all viruses known to infect humans [44, 45] (combined bat/serology information in Supplement Table S3). Along with seven additional E. spelaea colony bat sera samples run on VirScan, these data for positive, unique, DENV-type specific peptides were mapped against the protein sequence to show coverage of antibodies detected against DENV1 (Fig. 6b), DENV2 (Fig. 6c), and DENV4 (Fig. 6d). Very few DENV3-specific peptides were detected. Conserved peptides covering multiple DENV-Types or flaviviruses were excluded. NS3 and NS5 were represented by the greatest peptide coverage with only a partially conserved region of NS5 detected with the same peptide in multiple bats (2/6). As ZIKV is not in this version of phage-display library, positive peptides were blasted against the ZIKV genome (Singapore SG-001) and while no peptides had 100% identity to ZIKV as expected, a few peptides with partial overlap (> 8 amino acid residues) were present and these are highlighted in Fig. 6. This mapping indicates a wide range of antibodies detected, covering multiple proteins, with different bats exhibiting antibodies against the same peptide region. Full VirScan coverage, including peptide counts for Z score > 10, and protein IDs are listed by bat for all flaviviruses in the library (DENV1-4, Kunjin, Kyasanur, Louping-ill, St Louis, Powassan, WNV, YFV, JEV) and available in Table S4.
Fig. 6.
Serological results for Dengue Virus in Singaporean bats. a DENV2 luciferase immunoprecipitation system (LIPS) results for DENV2 NS1 antigen on 106 bat sera from Singapore (Supplemental Table S3). Score is colour coded as indicated, colour is recorded for all hits above the background threshold on each run (mean + 3 SD). Positive control sera were human and macacque positive sera (protein A/G capture). b Peptide mapping results of peptides identified by phage-display library screening (VirScan) with a Z score > 10 from sera used in a displaying DENV1 unique peptide results mapped to DENV1 amino acid sequence. Colour indicates individual bat number. c as per b displaying DENV2 unique peptide results mapped to DENV2 amino acid sequence. d as per b displaying DENV4 unique peptide results mapped to DENV4 amino acid sequence. Each colour represents an individual bat (indicated in legend). Arrows represent mapped peptides. Asterisk represents peptides partially overlapping with ZIKV (not in Virscan 2.0). Protein name as indicated
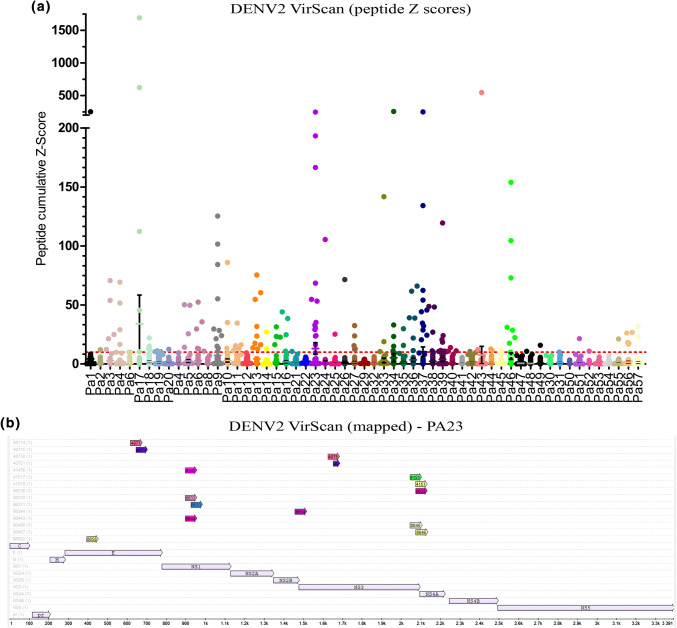
As we show reliable infection and productive infection in cells of P. alecto origin, we wanted to examine the potential exposure of Australasian bats to DENV, another region, where dengue is endemic. VirScan was performed on the sera of 57 seemingly healthy P. alecto individuals of similar age/size (mixed sex) collected in South-East Queensland. As there were too many peptides for mapping all bat samples to the DENV protein sequence the cumulative Z score of DENV2-unique peptides (for peptides, where at least one bat had a Z score > 10) are displayed (Fig. 7a). Several individuals show multiple DENV peptides with high Z scores. The peptide results for one individual bat (PA23) are mapped to the DENV2 ST 1688 protein sequence (Fig. 7b). While most flavivirus peptides aligned with DENV2, there were other DENV-Type specific hits and also other related flaviviruses detected (Table S5). These data indicate that these bats were exposed to DENV2 and developed an antibody response to multiple antigens, hinting at significant enough exposure to generate antibodies against multiple proteins.
Fig. 7.
Serology results of DENV2 positive bat sera from Australia. a Peptide Z scores for DENV2-unique peptides that have a Z scores > 10 in any of the Pteropus alecto bats. The cumulative Z scores of each peptide (individual Z scores for overlapping strain-specific peptides are added) is displayed per bat, bat number as indicated (different colour for each bat). Each dot represents one peptide. The Z scores cut-off line of 10 is displayed. Graph is in two segments as certain peptides have a very significant Z scores (multiple counts for the same peptide). Too many peptides were detected to map all. b Mapping of unique peptides (colored arrows) for one example individual bat (PA23) displaying all unique DE–NV2 peptides relevant to the amino acid sequence for DENV2
Discussion
DENV is the most important mosquito-transmitted virus across South-East Asia and Northern Australia [46]. The modelling of mosquito populations with human overlap can facilitate a public health effort to minimize the threat of this re-emerging pathogen through vector control [43, 47–49]. The sylvatic cycle of DENV, where replication can occur in other mammalian hosts such as monkeys and is driven by species other than Aedes aegypti, must be an important consideration when tracking the disease [50]. Natural sylvatic hosts and transient reservoirs of the virus are likely to tolerate infection and show only limited response to infection [51, 52], while at the same time requiring catholic vectors, that are not strictly anthropophilic, for transmission. A reservoir of infection includes all the hosts, either intermediate hosts or vectors, required to sustain a pathogen [52]. A reservoir host simply allows sustained infection and maintenance of the pathogen, although commonly this is associated with the absence of overt clinical symptoms or a limited immune response. Whether or not bats play a role as a sylvatic reservoir requires further investigation. The data presented here show multiple exposures of bats to DENV; however, there may yet be a role for bats as a transient host for the virus. Indeed, recent evidence shows that there are altered STING-induced responses to detecting DENV infection in both humans and primates [53]. We know from the previous work that STING-induced immune responses are also altered in the bat [54, 55]. Here, we provide evidence that several bat cell types of IndoMalayan and Australasian species can be infected with DENV, replicate, and produce infectious virus particles to a higher titre then the initial infection. In addition, P. alecto cell lines showed minimal immune response while tolerating virus infection and sustaining replication with minimal immunological effects. The lack of cellular immune responses suggests that there may be an important difference between these and Neotropical bats, which produce a robust response to DENV in vivo and rapid apoptosis in cell lines [15, 16]. While suppression of IRG activation and regulation of IFN induction are known to occur from various DENV proteins [56–60], this appears to be true in P. alecto at least, where DENV actively suppresses IFN activation. The active suppression of immune activation by DENV2 in PakiT03 cells suggest possible co-evolution of the virus with Australasian bats as a host or in the least, commonalities and conservation between these bats, the function of DENV proteins, and suppression of immunity in alternative hosts for DENV. While not directly showing reservoir-potential of this species, the data for productive in vitro infection suggest that further investigation is required to examine its role in transmission.
This study supports the exposure of Old-World fruit bats to flaviviruses and raises the possibility that they are a secondary or transient host, though not necessarily a sustainable sylvatic host. It was recently shown that canines can be infected with DENV and may possibly contribute to transmission in Thailand [61]. While there is minimal risk of a direct bat-to-human transmission, several species of mosquitoes (Culex and Aedes sp.) feed on bats [6, 14, 15, 50, 62]. Indeed, it appears as though bats are playing a role in arbovirus maintenance [63]. The volant nature of bats could provide a ready dispersal mechanism for these viruses when foraging or migrating. The Pteropus alecto flying fox is known to travel up to 150 km in a single day, rapidly outpacing typical DENV transmission dynamics via mosquitoes alone [64]. With respect to closely related flaviviruses, Culex annulirostris can transmit JEV to and from Pteropus alecto in experimental settings [65], and although there are limited data on the primary vector of DENV across the region, Aedes aegypti, it has been shown to feed on bats and transmit arboviruses [6]. There are several other Aedes (Stegomyia) vectors in the region that display a proclivity to feed on different mammals and are laboratory competent to transmit dengue [66] potentially maintaining sylvatic cycles. The titres produced from cell lines are sufficient for this notion, though whole-animal studies are needed. The first serology data on DENV (and ZIKV) in bats in the region provide evidence that bats across SE Asia and Australia have been exposed to DENV. Previous studies have focused largely on other flaviviruses such as JEV.
Taken together, these data highlight the need for greater surveillance and profiling of wildlife hosts and the vectoral capacity of mosquito populations, including host preference and vector competence. There is increasing evidence for bats being exposed to DENV around the world, and while Neotropical bats from South America may not be suitable hosts, it appears that there are large differences to the three bat species from SE Asia and Australia that were tested here. Neotropical bat cell lines showed very poor infectivity and generated a strong apoptotic response. One in vivo infection experiment suggests that they are not a suitable host; however, the authors also highlighted a clear subtype-dependency for DENV infection [15, 16]. The differences observed in IndoMalayan/Australasian bats may be due to co-evolution of the host and virus, as opposed to the relatively recent introduction of Dengue into the Neotropics [67–69]. Dengue-like illness is reported in a Chinese medical encyclopedia and added somewhere between 265 and 992 AD. The virus is believed to have come out of either Africa or Asia prior to that [70]. Neotropical bats diverge significantly from the old-world fruit bat ancestor of IndoMalayan and Australasian fruitbats [71–75]. There is, however, some limited movement in between populations that may explain transmission of pathogens [76]. Transmission studies in the appropriate bat species and more thorough identification of blood meals in mosquitoes will elucidate the full capacity of bats in transmission of DENV and DENV-like flaviviruses and incriminate possible vectors. If bats are indeed playing a role in flavivirus maintenance, we must consider how their dispersal impacts the introduction of arboviruses into new locales, and the subsequent risk of foci to human infections.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
This work was funded by the Singapore National Research Foundation Competitive Research Programme Grant (NRF2012NRF-CRP001-056 to LFW), National Medical Research Council of Singapore New Investigator’s Grant (NMRC/BNIG/2040/2015 to ATI, NMRC/BNIG/2005/2013 to IHM) and a National Medical Research Council Research Grant (ZRRF16006 to LFW and ATI). Many thanks to the following in helping with bat sample processing: NEA/NParks, Crameri Research Consulting, Prof. Joanne Meers of UQ, the Queensland Animal Science Precinct (QASP) team led by Hume Field, and Duke-NUS team members from LEZV/LOVE labs for collection of bat samples. We acknowledge the help from the SingHealth Advanced Bioimaging facility.
Abbreviations
- IFN
Interferon
- DENV
Dengue virus
- ZIKV
Zika virus
- JEV
Japanese encephalitis virus
- YFV
Yellow fever virus
- SLEV
St Louis encephalitis virus
- WNV
West Nile virus
- IRG
Interferon regulated gene
- PRR
Pattern recognition receptor
- BMDM
Bone-marrow-derived macrophage
- BMDC
Bone-marrow-derived dendritic cells
- LIPS
Luciferase immune-precipitation system
- SE
South east
Author contributions
ATI designed the study, performed experiments, analysed the data, and wrote the manuscript under supervision from LFW and with input from all authors. PR, KPS, KL, WNC, and SM performed experiments and analysed data. JLG and MLH performed proteomics studies. BPYHL, JHJN, IHM, and GJDS contributed with wild animal samples in Singapore, processing of bats, and generation of cell lines. BL and SJE designed the ViRScan/PhIP-Seq pipelines and contributed to the manuscript.
Compliance with ethical standards
Conflict of interest
We declare that there is no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Aaron T. Irving, Email: aaron.irving@duke-nus.edu.sg
Lin-Fa Wang, Email: Linfa.wang@duke-nus.edu.sg.
References
- 1.Reagan R, Brueckner AL. Studies of dengue fever virus in the cave bat (Myotus lucifugus) J Infect Dis. 1952;91(2):145–146. doi: 10.1093/infdis/91.2.145. [DOI] [PubMed] [Google Scholar]
- 2.Shepherd RC, Williams MC. Studies on viruses in East African bats (Chiroptera): 1. Haemagglutination inhibition and circulation of arboviruses. Zoonoses Res. 1964;3(3):125–139. [PubMed] [Google Scholar]
- 3.La Motte LC., Jr Japanese B encephalitis in bats during simulated hibernation. Am J Hyg. 1958;67(1):101–108. doi: 10.1093/oxfordjournals.aje.a119912. [DOI] [PubMed] [Google Scholar]
- 4.Herbold JR, Heuschele WP, Berry RL, Parsons MA. Reservoir of St. Louis encephalitis virus in Ohio bats. Am J Vet Res. 1983;44(10):1889–1893. [PubMed] [Google Scholar]
- 5.Davis A, Bunning M, Gordy P, Panella N, Blitvich B, Bowen R Experimental and natural infection of North American bats with West Nile virus. Am J Trop Med Hyg. 2005;73(2):467–469. doi: 10.4269/ajtmh.2005.73.467. [DOI] [PubMed] [Google Scholar]
- 6.Simpson DI, O’Sullivan JP. Studies on arboviruses and bats (Chiroptera) in East Africa: I Experimental infection of bats and virus transssion attempts in Aedes (Stegomyia) aegypti (Linnaeus) Ann Trop Med Parasitol. 1968;62(4):422–431. doi: 10.1080/00034983.1968.11686579. [DOI] [PubMed] [Google Scholar]
- 7.Hsieh JT, Rathore APS, Soundarajan G, St John AL. Japanese encephalitis virus neuropenetrance is driven by mast cell chymase. Nat Commun. 2019;10(1):706. doi: 10.1038/s41467-019-08641-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Puccioni-Sohler M, Rosadas C. Advances and new insights in the neuropathogenesis of dengue infection. Arq Neuropsiquiatr. 2015;73(8):698–703. doi: 10.1590/0004-282X20150074. [DOI] [PubMed] [Google Scholar]
- 9.Maximova OA, Pletnev AG. Flaviviruses and the central nervous system: revisiting neuropathological concepts. Annu Rev Virol. 2018;5(1):255–272. doi: 10.1146/annurev-virology-092917-043439. [DOI] [PubMed] [Google Scholar]
- 10.Mustafa YM, Meuren LM, Coelho SVA, de Arruda LB. Pathways exploited by flaviviruses to counteract the blood-brain barrier and invade the central nervous system. Front Microbiol. 2019;10:525. doi: 10.3389/fmicb.2019.00525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kading RC, Schountz T. Flavivirus infections of bats: potential role in zika virus ecology. Am J Trop Med Hyg. 2016;95(5):993–996. doi: 10.4269/ajtmh.16-0625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Thoisy B, Lacoste V, Germain A, Munoz-Jordan J, Colon C, Mauffrey JF, Delaval M, Catzeflis F, Kazanji M, Matheus S, Dussart P, Morvan J, Setien AA, Deparis X, Lavergne A. Dengue infection in neotropical forest mammals. Vector Borne Zoonotic Dis. 2009;9(2):157–170. doi: 10.1089/vbz.2007.0280. [DOI] [PubMed] [Google Scholar]
- 13.Machain-Williams C, Lopez-Uribe M, Talavera-Aguilar L, Carrillo-Navarrete J, Vera-Escalante L, Puerto-Manzano F, Ulloa A, Farfan-Ale JA, Garcia-Rejon J, Blitvich BJ, Lorono-Pino MA. Serologic evidence of flavivirus infection in bats in the Yucatan Peninsula of Mexico. J Wildl Dis. 2013;49(3):684–689. doi: 10.7589/2012-12-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vicente-Santos A, Moreira-Soto A, Soto-Garita C, Chaverri LG, Chaves A, Drexler JF, Morales JA, Alfaro-Alarcon A, Rodriguez-Herrera B, Corrales-Aguilar E. Neotropical bats that co-habit with humans function as dead-end hosts for dengue virus. PLoS Negl Trop Dis. 2017;11(5):e0005537. doi: 10.1371/journal.pntd.0005537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Perea-Martinez L, Moreno-Sandoval HN, Moreno-Altamirano MM, Salas-Rojas M, Garcia-Flores MM, Arechiga-Ceballos N, Tordo N, Marianneau P, Aguilar-Setien A. Experimental infection of Artibeus intermedius bats with serotype-2 dengue virus. Comp Immunol Microbiol Infect Dis. 2013;36(2):193–198. doi: 10.1016/j.cimid.2012.12.002. [DOI] [PubMed] [Google Scholar]
- 16.Moreira-Soto A, Soto-Garita C, Corrales-Aguilar E. Neotropical primary bat cell lines show restricted dengue virus replication. Comp Immunol Microbiol Infect Dis. 2017;50:101–105. doi: 10.1016/j.cimid.2016.12.004. [DOI] [PubMed] [Google Scholar]
- 17.Bean AG, Baker ML, Stewart CR, Cowled C, Deffrasnes C, Wang LF, Lowenthal JW. Studying immunity to zoonotic diseases in the natural host—keeping it real. Nat Rev Immunol. 2013;13(12):851–861. doi: 10.1038/nri3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuzmin IV, Schwarz TM, Ilinykh PA, Jordan I, Ksiazek TG, Sachidanandam R, Basler CF, Bukreyev A. Innate immune responses of bat and human cells to filoviruses: commonalities and distinctions. J Virol. 2017 doi: 10.1128/jvi.02471-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mishra A, Vijayakumar P, Raut AA. Emerging avian influenza infections: current understanding of innate immune response and molecular pathogenesis. Int Rev Immunol. 2017;36(2):89–107. doi: 10.1080/08830185.2017.1291640. [DOI] [PubMed] [Google Scholar]
- 20.Ploquin MJ, Silvestri G, Muller-Trutwin M. Immune activation in HIV infection: what can the natural hosts of simian immunodeficiency virus teach us? Curr Opin HIV AIDS. 2016;11(2):201–208. doi: 10.1097/COH.0000000000000238. [DOI] [PubMed] [Google Scholar]
- 21.Crameri G, Todd S, Grimley S, McEachern JA, Marsh GA, Smith C, Tachedjian M, De Jong C, Virtue ER, Yu M, Bulach D, Liu JP, Michalski WP, Middleton D, Field HE, Wang LF. Establishment, immortalisation and characterisation of pteropid bat cell lines. PLoS One. 2009;4(12):e8266. doi: 10.1371/journal.pone.0008266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou P, Chionh YT, Irac SE, Ahn M, Jia Ng JH, Fossum E, Bogen B, Ginhoux F, Irving AT, Dutertre CA, Wang LF. Unlocking bat immunology: establishment of Pteropus alecto bone marrow-derived dendritic cells and macrophages. Sci Rep. 2016;6:38597. doi: 10.1038/srep38597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reller ME, Bodinayake C, Nagahawatte A, Devasiri V, Kodikara-Arachichi W, Strouse JJ, Broadwater A, Ostbye T, de Silva A, Woods CW. Unsuspected dengue and acute febrile illness in rural and semi-urban southern Sri Lanka. Emerg Infect Dis. 2012;18(2):256–263. doi: 10.3201/eid1802.110962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chatel-Chaix L, Cortese M, Romero-Brey I, Bender S, Neufeldt CJ, Fischl W, Scaturro P, Schieber N, Schwab Y, Fischer B, Ruggieri A, Bartenschlager R. Dengue virus perturbs mitochondrial morphodynamics to dampen innate immune responses. Cell Host Microbe. 2016;20(3):342–356. doi: 10.1016/j.chom.2016.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jain B, Chaturvedi UC, Jain A. Role of intracellular events in the pathogenesis of dengue; an overview. Microb Pathog. 2014;69–70:45–52. doi: 10.1016/j.micpath.2014.03.004. [DOI] [PubMed] [Google Scholar]
- 26.Balinsky CA, Schmeisser H, Wells AI, Ganesan S, Jin T, Singh K, Zoon KC. IRAV (FLJ11286), an interferon-stimulated gene with antiviral activity against dengue virus, interacts with MOV10. J Virol. 2017 doi: 10.1128/jvi.01606-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thiemmeca S, Tamdet C, Punyadee N, Prommool T, Songjaeng A, Noisakran S, Puttikhunt C, Atkinson JP, Diamond MS, Ponlawat A, Avirutnan P. Secreted NS1 protects dengue virus from mannose-binding lectin-mediated neutralization. J Immunol. 2016;197(10):4053–4065. doi: 10.4049/jimmunol.1600323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang WK, Sung TL, Tsai YC, Kao CL, Chang SM, King CC. Detection of dengue virus replication in peripheral blood mononuclear cells from dengue virus type 2-infected patients by a reverse transcription-real-time PCR assay. J Clin Microbiol. 2002;40(12):4472–4478. doi: 10.1128/jcm.40.12.4472-4478.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cheng MS, Lau SH, Chan KP, Toh CS, Chow VT. Impedimetric cell-based biosensor for real-time monitoring of cytopathic effects induced by dengue viruses. Biosens Bioelectron. 2015;70:74–80. doi: 10.1016/j.bios.2015.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pattanakitsakul SN, Rungrojcharoenkit K, Kanlaya R, Sinchaikul S, Noisakran S, Chen ST, Malasit P, Thongboonkerd V. Proteomic analysis of host responses in HepG2 cells during dengue virus infection. J Proteome Res. 2007;6(12):4592–4600. doi: 10.1021/pr070366b. [DOI] [PubMed] [Google Scholar]
- 31.Ludert JE, Mosso C, Ceballos-Olvera I, del Angel RM. Use of a commercial enzyme immunoassay to monitor dengue virus replication in cultured cells. Virol J. 2008;5:51. doi: 10.1186/1743-422X-5-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zompi S, Harris E. Animal models of dengue virus infection. Viruses. 2012;4(1):62–82. doi: 10.3390/v4010062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Romo H, Kenney JL, Blitvich BJ, Brault AC. Restriction of Zika virus infection and transmission in Aedes aegypti mediated by an insect-specific flavivirus. Emerg Microbes Infect. 2018;7(1):181. doi: 10.1038/s41426-018-0180-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hsu YL, Shi SF, Wu WL, Ho LJ, Lai JH. Protective roles of interferon-induced protein with tetratricopeptide repeats 3 (IFIT3) in dengue virus infection of human lung epithelial cells. PLoS One. 2013;8(11):e79518. doi: 10.1371/journal.pone.0079518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Helbig KJ, Carr JM, Calvert JK, Wati S, Clarke JN, Eyre NS, Narayana SK, Fiches GN, McCartney EM, Beard MR. Viperin is induced following dengue virus type-2 (DENV-2) infection and has anti-viral actions requiring the C-terminal end of viperin. PLoS Negl Trop Dis. 2013;7(4):e2178. doi: 10.1371/journal.pntd.0002178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fallahi P, Elia G. Interferon-gamma-induced protein 10 in dengue virus infection. Clin Ter. 2016;167(6):e186–e191. doi: 10.7417/CT.2016.1966. [DOI] [PubMed] [Google Scholar]
- 37.Lin RJ, Yu HP, Chang BL, Tang WC, Liao CL, Lin YL. Distinct antiviral roles for human 2′,5′-oligoadenylate synthetase family members against dengue virus infection. J Immunol. 2009;183(12):8035–8043. doi: 10.4049/jimmunol.0902728. [DOI] [PubMed] [Google Scholar]
- 38.Pan XB, Han JC, Cong X, Wei L. BST2/tetherin inhibits dengue virus release from human hepatoma cells. PLoS One. 2012;7(12):e51033. doi: 10.1371/journal.pone.0051033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sprokholt J, Helgers LC, Geijtenbeek TB. Innate immune receptors drive dengue virus immune activation and disease. Future Virol. 2017;13(4):287–305. doi: 10.2217/fvl-2017-0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Urcuqui-Inchima S, Cabrera J, Haenni AL. Interplay between dengue virus and toll-like receptors, RIG-I/MDA5 and microRNAs: implications for pathogenesis. Antivir Res. 2017;147:47–57. doi: 10.1016/j.antiviral.2017.09.017. [DOI] [PubMed] [Google Scholar]
- 41.Sprokholt JK, Kaptein TM, van Hamme JL, Overmars RJ, Gringhuis SI, Geijtenbeek TBH. RIG-I-like receptor activation by dengue virus drives follicular T helper cell formation and antibody production. PLoS Pathog. 2017;13(11):e1006738. doi: 10.1371/journal.ppat.1006738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mok L, Wynne JW, Tachedjian M, Shiell B, Ford K, Matthews DA, Bacic A, Michalski WP. Proteomics informed by transcriptomics for characterising differential cellular susceptibility to Nelson Bay orthoreovirus infection. BMC Genom. 2017;18(1):615. doi: 10.1186/s12864-017-3994-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hodgkison R, Balding ST, Zubaid A, Kunz TH. Temporal Variation in the relative abundance of fruit bats (Megachiroptera: Pteropodidae) in relation to the availability of food in a lowland Malaysian rain forest. Biotropica. 2004;36(4):522–533. [Google Scholar]
- 44.Larman HB, Zhao Z, Laserson U, Li MZ, Ciccia A, Gakidis MAM, Church GM, Kesari S, LeProust EM, Solimini NL, Elledge SJ. Autoantigen discovery with a synthetic human peptidome. Nat Biotechnol. 2011;29:535. doi: 10.1038/nbt.1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mohan D, Wansley DL, Sie BM, Noon MS, Baer AN, Laserson U, Larman HB. PhIP-Seq characterization of serum antibodies using oligonucleotide-encoded peptidomes. Nat Protoc. 2018;13(9):1958–1978. doi: 10.1038/s41596-018-0025-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shepard DS, Undurraga EA, Halasa YA. Economic and disease burden of dengue in southeast Asia. PLOS Negl Trop Dis. 2013;7(2):e2055. doi: 10.1371/journal.pntd.0002055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zheng TT, Nie LF. Modelling the transmission dynamics of two-strain dengue in the presence awareness and vector control. J Theor Biol. 2018;443:82–91. doi: 10.1016/j.jtbi.2018.01.017. [DOI] [PubMed] [Google Scholar]
- 48.Pepin KM, Leach CB, Marques-Toledo C, Laass KH, Paixao KS, Luis AD, Hayman DT, Johnson NG, Buhnerkempe MG, Carver S, Grear DA, Tsao K, Eiras AE, Webb CT. Utility of mosquito surveillance data for spatial prioritization of vector control against dengue viruses in three Brazilian cities. Parasit Vectors. 2015;8:98. doi: 10.1186/s13071-015-0659-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jing Y, Wang X, Tang S, Wu J. Data informed analysis of 2014 dengue fever outbreak in Guangzhou: impact of multiple environmental factors and vector control. J Theor Biol. 2017;416:161–179. doi: 10.1016/j.jtbi.2016.12.014. [DOI] [PubMed] [Google Scholar]
- 50.Vasilakis N, Cardosa J, Hanley KA, Holmes EC, Weaver SC. Fever from the forest: prospects for the continued emergence of sylvatic dengue virus and its impact on public health. Nat Rev Microbiol. 2011;9(7):532–541. doi: 10.1038/nrmicro2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mandl JN, Ahmed R, Barreiro LB, Daszak P, Epstein JH, Virgin HW, Feinberg MB. Reservoir host immune responses to emerging zoonotic viruses. Cell. 2015;160(1–2):20–35. doi: 10.1016/j.cell.2014.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ashford RW. When is a reservoir not a reservoir? Emerg Infect Dis. 2003;9(11):1495–1496. doi: 10.3201/eid0911.030088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stabell AC, Meyerson NR, Gullberg RC, Gilchrist AR, Webb KJ, Old WM, Perera R, Sawyer SL. Dengue viruses cleave STING in humans but not in nonhuman primates, their presumed natural reservoir. Elife. 2018 doi: 10.7554/elife.31919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mozzi A, Pontremoli C, Forni D, Clerici M, Pozzoli U, Bresolin N, Cagliani R, Sironi M. OASes and STING: adaptive evolution in concert. Genome Biol Evol. 2015;7(4):1016–1032. doi: 10.1093/gbe/evv046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xie J, Li Y, Shen X, Goh G, Zhu Y, Cui J, Wang LF, Shi ZL, Zhou P. Dampened STING-dependent interferon activation in bats. Cell Host Microbe. 2018;23(3):297–301.e294. doi: 10.1016/j.chom.2018.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Green AM, Beatty PR, Hadjilaou A, Harris E. Innate immunity to dengue virus infection and subversion of antiviral responses. J Mol Biol. 2014;426(6):1148–1160. doi: 10.1016/j.jmb.2013.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jones M, Davidson A, Hibbert L, Gruenwald P, Schlaak J, Ball S, Foster GR, Jacobs M. Dengue virus inhibits alpha interferon signaling by reducing STAT2 expression. J Virol. 2005;79(9):5414–5420. doi: 10.1128/JVI.79.9.5414-5420.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bui TT, Moi ML, Nabeshima T, Takemura T, Nguyen TT, Nguyen LN, Pham HTT, Nguyen TTT, Manh DH, Dumre SP, Mizukami S, Hirayama K, Tajima S, Le MTQ, Aoyagi K, Hasebe F, Morita K. A single amino acid substitution in the NS4B protein of dengue virus confers enhanced virus growth and fitness in human cells in vitro through IFN-dependent host response. J Gen Virol. 2018;99(8):1044–1057. doi: 10.1099/jgv.0.001092. [DOI] [PubMed] [Google Scholar]
- 59.Castillo Ramirez JA, Urcuqui-Inchima S. Dengue virus control of type I IFN responses: a history of manipulation and control. J Interferon Cytokine Res. 2015;35(6):421–430. doi: 10.1089/jir.2014.0129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Best SM. The many faces of the flavivirus NS5 protein in antagonism of type I interferon signaling. J Virol. 2017 doi: 10.1128/jvi.01970-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Thongyuan S, Kittayapong P. First evidence of dengue infection in domestic dogs living in different ecological settings in Thailand. PLoS One. 2017;12(8):e0180013. doi: 10.1371/journal.pone.0180013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ryan PA, Martin L, Mackenzie JS, Kay BH. Investigation of gray-headed flying foxes (Pteropus poliocephalus) (Megachiroptera: Pteropodidae) and mosquitoes in the ecology of Ross River virus in Australia. Am J Trop Med Hyg. 1997;57(4):476–482. doi: 10.4269/ajtmh.1997.57.476. [DOI] [PubMed] [Google Scholar]
- 63.Kading RC, Kityo RM, Mossel EC, Borland EM, Nakayiki T, Nyakarahuka L, Ledermann JP, Panella NA, Gilbert AT, Crabtree MB, Peterhans JK, Towner JS, Amman BR, Sealy TK, Nichol ST, Powers AM, Lutwama JJ, Miller BR. Neutralizing antibodies against flaviviruses, Babanki virus, and Rift Valley fever virus in Ugandan bats. Infect Ecol Epidemiol. 2018;8(1):1439215. doi: 10.1080/20008686.2018.1439215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Breed AC, Field HE, Smith CS, Edmonston J, Meers J. Bats without borders: long-distance movements and implications for disease risk management. EcoHealth. 2010;7(2):204–212. doi: 10.1007/s10393-010-0332-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.van den Hurk AF, Smith CS, Field HE, Smith IL, Northill JA, Taylor CT, Jansen CC, Smith GA, Mackenzie JS. Transmission of Japanese Encephalitis virus from the black flying fox, Pteropus alecto, to Culex annulirostris mosquitoes, despite the absence of detectable viremia. Am J Trop Med Hyg. 2009;81(3):457–462. doi: 10.4269/ajtmh.2009.81.457. [DOI] [PubMed] [Google Scholar]
- 66.Rosen L, Roseboom LE, Gubler DJ, Lien JC, Chaniotis BN. Comparative susceptibility of mosquito species and strains to oral and parenteral infection with dengue and Japanese encephalitis viruses. Am J Trop Med Hyg. 1985;34(3):603–615. doi: 10.4269/ajtmh.1985.34.603. [DOI] [PubMed] [Google Scholar]
- 67.Brathwaite Dick O, San Martin JL, Montoya RH, del Diego J, Zambrano B, Dayan GH. The history of dengue outbreaks in the Americas. Am J Trop Med Hyg. 2012;87(4):584–593. doi: 10.4269/ajtmh.2012.11-0770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gubler DJ. Dengue/dengue haemorrhagic fever: history and current status. Novartis Found Symp. 2006;277:3–16. doi: 10.1002/0470058005.ch2. [DOI] [PubMed] [Google Scholar]
- 69.Ooi EE, Gubler DJ. Dengue in Southeast Asia: epidemiological characteristics and strategic challenges in disease prevention. Cad Saude Publica. 2009;25(Suppl 1):S115–124. doi: 10.1590/S0102-311X2009001300011. [DOI] [PubMed] [Google Scholar]
- 70.Gubler DJ. Dengue and dengue hemorrhagic fever. Clin Microbiol Rev. 1998;11(3):480–496. doi: 10.1128/CMR.11.3.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lei M, Dong D. Phylogenomic analyses of bat subordinal relationships based on transcriptome data. Sci Rep. 2016;6:27726. doi: 10.1038/srep27726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shen B, Han X, Jones G, Rossiter SJ, Zhang S. Adaptive evolution of the myo6 gene in old world fruit bats (Family: Pteropodidae) PLoS One. 2013;8(4):e62307. doi: 10.1371/journal.pone.0062307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shen B, Han X, Zhang J, Rossiter SJ, Zhang S. Adaptive evolution in the glucose transporter 4 gene Slc2a4 in Old World fruit bats (Family: Pteropodidae) PLoS One. 2012;7(4):e33197. doi: 10.1371/journal.pone.0033197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gunnell GF, Smith R, Smith T. 33 million year old Myotis (Chiroptera, Vespertilionidae) and the rapid global radiation of modern bats. PLoS One. 2017;12(3):e0172621. doi: 10.1371/journal.pone.0172621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yin Q, Zhu L, Liu D, Irwin DM, Zhang S, Pan YH. Molecular evolution of the nuclear factor (erythroid-derived 2)-Like 2 gene Nrf2 in old world fruit bats (Chiroptera: Pteropodidae) PLoS One. 2016;11(1):e0146274. doi: 10.1371/journal.pone.0146274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hamilton PB, Cruickshank C, Stevens JR, Teixeira MM, Mathews F. Parasites reveal movement of bats between the new and old worlds. Mol Phylogenet Evol. 2012;63(2):521–526. doi: 10.1016/j.ympev.2012.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.