Abstract
In recent years, a large number of circRNAs have been identified in mammalian cells with high-throughput sequencing technologies and bioinformatics. The aberrant expression of circRNAs has been reported in many human diseases including gastric cancer (GC). The number of GC-related circRNAs with validated biological functions and mechanisms of action is growing. CircRNAs are critically involved in GC cell proliferation, apoptosis, migration, and invasion. CircRNAs have been shown to function as regulators of parental gene transcription and alternative splicing and miRNA sponges. Moreover, circRNAs have been suggested to interact with proteins to regulate their expression level and activities. Several circRNAs have been identified to encode functional proteins. Due to their great abundance, high stability, tissue- and developmental-stage-specific expression patterns, and wide distribution in various body fluids and exosomes, circRNAs exhibit a great potential to be utilized as biomarkers for GC. Herein, we briefly summarize their biogenesis, properties and biological functions and discuss about the current research progress of circRNAs in GC with a focus on the potential application for GC diagnosis and therapy.
Keywords: CircRNA, Gastric cancer, miRNA sponge, Biomarker, Therapeutic target
Introduction
Gastric cancer (GC) is one of the most common malignant tumors with high morbidity and mortality worldwide. In 2018, there are estimated 1,033,701 new cases of GC and 782,685 cases of GC-related death, which ranks the fifth and third in cancer incidence rates and mortality rates, respectively [1]. Gastric tumorigenesis is a multifactorial, multicellular and multistep process [2]. Environmental factors (diet, exogenous chemicals), intragastric formation of carcinogens and Helicobacter pylori can induce the occurrence of GC [3]. The communication of GC cells and cancer microenvironment cells determines the development and progression of GC [4–6]. The detailed pathological mechanisms for GC remain not well understood. Currently, the diagnosis of GC is mainly dependent on imaging examination, serum tumor marker examination, endoscopy, and tissue biopsy [7]. However, as gold standard method, tissue biopsy can induce obvious surgical trauma and hysteresis. Besides, due to the fact that early GC lacks specific symptoms, most GC patients are diagnosed in the intermediate or terminal stage with poor prognosis [8]. Moreover, the main available treatments like radiotherapy and chemotherapy have strong side effects on GC patients and sometimes, the curative effect of chemotherapy is obvious but could not last for a long period due to drug resistance. Therefore, it is urgent to explore more specific and effective method for GC diagnosis and treatment.
CircRNAs, a novel type of RNAs with covalently closed loop structure, are becoming a new hotspot in the field of ncRNAs lately. Since they were first discovered in viruses as early as the 1970s [9], a handful of such RNAs were found and considered to be the rare errors in canonical and non-canonical RNA splicing. However, with recent advances in RNA deep sequencing technology and bioinformatics, a large amount of circRNAs have been revealed to be abundant in mammalian cells. Salzman et al. have confirmed that circRNAs are predominant transcripts of diverse human cell types [10]. Jeck et al. have identified over 25,000 circRNAs in human fibroblasts and considered them as stable, conserved and nonrandom products of RNA splicing which could be associated with complementary ALU repeats in bordering introns [11]. Afterwards, Memczak et al. and Hansen et al. both made breakthrough in circRNA biological functions researches and first proposed that circRNAs could act as miRNA sponge in gene expression and regulation and could function in posttranscriptional process [12, 13]. Since then, circRNAs have begun to attract wide attention of the public and relevant studies on their biogenesis, characteristics, functional mechanisms as well as potential application in clinical diagnosis and treatment have been carried out. Nowadays, an increasing number of circRNAs have been found to regulate gene expression at transcriptional, posttranscriptional and translational levels. They participate in many pathological processes such as Alzheimer’s disease, diabetes, atherosclerosis and glioma through regulating alternative splicing, sponging miRNAs, sequestering functional proteins or even encoding proteins [14–17]. Particularly, circRNAs play an important role in cancer growth, metastasis, recurrence and therapy resistance [18]. A variety of circRNAs have been reported to be aberrantly expressed in GC tissues or cell lines. Biological experiments proved that many of them harbor miRNA-binding sites and could act as miRNA sponges to suppress or promote GC progression. Given that circRNAs are of high abundance and stability, evolutionary species conservation as well as tissue specificity, and are widespread in various body fluids, exploring GC-related circRNAs as biomarkers or targets would create new possibilities for GC early diagnosis, prognosis and effective treatment.
In this review, we summarize the biogenesis, characteristics, biological functions and mechanisms of circRNAs. Then, we review GC-related circRNAs, and finally discuss their potential applications as biomarkers, therapeutic targets or tools.
Biogenesis, classification and properties of circRNAs
As a novel type of RNAs, circRNAs have circular structure which is generated from unique back-splicing biogenesis process with different molecular mechanisms. This circular structure and biogenesis mechanisms also confer many unique characteristics to them such as high abundance and stability, evolutionary species conservation, and tissues-specific expression patterns.
Biogenesis and classification
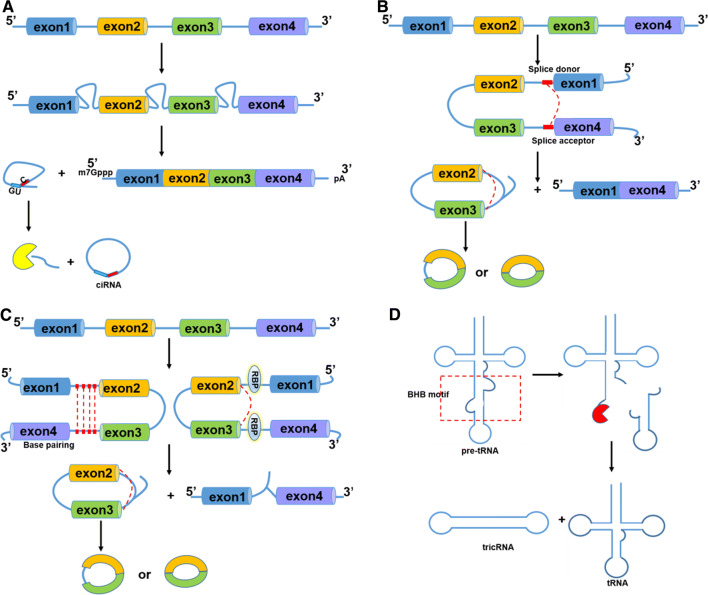
Alternative splicing of RNA is a basic gene expression event in eukaryotic cells. Pre-mRNA is catalyzed by spliceosomal machinery to remove introns and then exons with protein-encoding information are sequentially connected to form mature mRNA (Fig. 1a). With alternative splicing such as exon skipping, a gene generates several different mRNAs and proteins, which increases the diversity of cell transcriptome. Unlike canonical splicing of mRNA, circRNAs are generated by back-splicing process that ligates a downstream 5′ splice donor site with an upstream 3′ splice acceptor site to form a single-strand covalently closed loop. Then, the spliceosome removes all or part of introns and the rest of sequences are connected. Afterwards, three types of circRNAs are generated, including exonic circRNAs (ecircRNAs) which contain exon sequence only [10], intronic circRNAs (ciRNAs) which synthesized from introns, and exon–intron circular RNAs (EIciRNAs) which contain both exon and intron sequences [19, 20]. The detailed mechanisms for circRNA biogenesis have not been completely elucidated. Jeck et al. proposed two models of ecircRNA formation [11]. One is termed “lariat-driven circularization” and the other one is “intron-pairing-driven circularization” (Fig. 1b, c). In addition, Zhang et al. suggested that exon circularization is dependent on flanking intronic complementary sequences [21]. Liang et al. demonstrated that the miniature introns containing splice sites and short (30–40 nt) inverted repeats are sufficient for circRNA formation [22]. Both studies provide evidence to the models proposed above. It is generally believed that introns removed by spliceosome are unstable and can be degraded by exonucleases. While, ciRNAs generated from introns that form lariat structure by joining with both ends are resistant to exonuclease degradation (Fig. 1a) [19]. In addition, tRNA intronic circRNAs (tricRNAs) are another type of circRNAs which are generated during the splicing of pre-tRNA (Fig. 1d) [23].
Fig. 1.
Biogenesis of circRNA. a Canonical splicing. Pre-mRNA is spliced by spliceosome to remove intervening introns and leave only exons sequentially connected to form mature mRNA. The rest lariat intron can escape debranching and degradation with 7 nt GU-rich sequences near the 5′ splice site and 11 nt C-rich sequence near branch site and instead the 3′ “tail” downstream sequence is trimmed to form stable ciRNA. b Lariat-driven circularization. A downstream 5′ splice site of exon 4 is joined to an upstream 3′ splice site of exon 1 forming lariat structure. And then, the lariat containing skipped exons undergoes internal splicing of one intron to form EIciRNA or splicing of two introns to form ecircRNA. c Intron-pairing-driven circularization. Intronic motifs flanking circularized exon 2 and 3 have considerably complementary sequences which bring splicing sites close to form EIciRNA or eciRNA. RNA binding proteins (RBPs) driven circularization. RBPs with binding sites on flanking introns of exons 2 and 3 could bring two splicing sites close together to facilitate circularization. d The tRNA splicing endonuclease complex (TSEN) recognizes bulge–helix–bulge (BHB) motif and carries out intron excision of pre-tRNA and then release and ligate the end to form tRNA and tricRNA
The recent studies have indicated that some proteins are involved in circRNA biogenesis (Fig. 1c). MBL (muscleblind) is shown to have binding sites on flanking introns of its pre-mRNA, which bring two splicing sites close together to facilitate circularization [24]. Besides, ADAR1 can inhibit circRNA expression by Adenosine-to-Inosine (A-to-I) editing to diminish RNA pairing structure of flanking introns and decrease back-splicing efficiency [25]. RNA-binding protein QKI (quaking) is also demonstrated as a regulator of circRNA biogenesis during human epithelial–mesenchymal transition (EMT) where it binds to sites flanking circRNA-forming exons to induce exon circularization [26].
Properties
CircRNAs are usually widespread and many of them exist with high abundance. More than 25,000 different circRNAs have been identified in human fibroblasts and 14.4% of actively transcribed genes can generate ecircRNAs [11]. Most of them contain less than 5 exons and their size can range from under 100 nt to over 4000 nt [27]. Because of alternative circularization resulting from competition between different flanking complementary introns (ALU sequence), a gene can give rise to a single or many circRNA isoforms, which increases the number of circRNAs [21]. Moreover, the amount of some circRNAs is more abundant than its linear RNAs. In some cases, the abundance of circRNAs is more than tenfold of that of linear mRNAs [11]. Also, circRNAs have been discovered in several species including human, mouse, plants, Archaea, C. elegans and so on [11, 28, 29]. Additionally, circRNAs are located in different subcellular portions. EcircRNAs are mainly located in the cytoplasm while both ciRNAs and EIciRNAs are present in the nucleus of eukaryotic cells [10, 12, 19, 23]. However, the detailed mechanisms for the import/export of circRNAs are not clear. Huang et al. recently reported that the key modulators UAP56 or URH49 could control the nuclear export efficiency of long and short circRNAs, respectively [30].
CircRNAs are expressed across the tree of eukaryotic life, and thus presented to be evolutionarily conserved among distinct species [31]. Up to date, 69 circRNAs containing precisely conserved backsplice junctions have been identified between human and mouse [11]. Studies focusing on the sequence conservation within 3 subtypes of circRNAs suggested that ecircRNAs composed of coding sequence are overall conservative, while a few ciRNAs or intergenic circRNAs show a mild but abundant conserved nucleotides [12]. The conservation of splicing regulatory elements might be responsible for this phenomenon. Besides, complementary flanking introns such as ALU sequences facilitating circRNAs biogenesis are relatively conserved in animals or plants. Also, the miRNA target sites of circRNA sponges display few polymorphisms [32], which also ensure the conservation of circRNAs.
Typically, circRNAs have high stability and are more stable than the linear transcripts. They are covalently closed with neither 5′–3′ polarity nor polyadenylated tail; thus, they could resist degradation by RNase R, debranching enzyme or RNA exonuclease and are more stable than the linear transcripts [33, 34]. Their average half-life in cells is over 48 h, which is much longer than that of mRNA (< 10 h) [35]. Although it is still unclear how circRNAs are degraded and cleared, circRNAs are able to be targeted by siRNA [11]. Furthermore, circRNAs are also targets of microRNAs. For instance, CDR1as is targeted by miR-7 and subjected to the silencing activity of Ago2 and cleaved in a miR-671-dependent manner [36].
Most circRNAs present tissue/developmental-stage-specific expression pattern. Their expression levels are significantly diverse in various cell types and are dynamically changed across the development of embryonic tissues. For example, ciRS-7 is highly expressed in brain tissues but lowly expressed or absent in non-neuronal tissues [12]. The repertoire of genes expressing circRNAs, the relative levels between circRNAs and linear transcripts, and even pattern of splice isoforms of circRNAs from each gene are cell-type specific [37]. Moreover, a recent study showed that circRNAs are highly abundant and dynamically expressed in a spatio-temporal manner in porcine fetal brain [38]. During human fetal development, fetal brain, such as frontal cortex with marked enrichment for genes, is the region where circRNA isoforms are predominant among various tissues [39]. All the researches above suggest that the distribution of circRNAs is regulated by an uncharacterized pathway which needs to be further explored.
Functions and mechanisms of circRNAs
Apart from its unique properties, the manners by which circRNAs participate in the regulation of biological processes further broaden our understanding of ncRNAs. So far, circRNAs have been demonstrated to serve as miRNA sponges, regulate alternative splicing, bind to RNA binding proteins (RBPs) as well as encode proteins.
MiRNA sponge
MiRNAs are able to directly bind to target mRNA in a base-pairing manner and trigger cleavage of mRNAs or repress mRNA translation [40]. CircRNAs located in cytoplasm also contain complementary miRNA binding sites, and thus serve as competitive inhibitors for miRNAs (Fig. 2d). CiRS-7 was first demonstrated to harbor 63 conserved binding sites for miR-7 and suppress its biological activity and functions [13]. Later, increasing studies have shown the existence and importance of ciRS-7 serving as miR-7 sponges in many pathophysiology process such as insulin secretion, myocardial infarction, hepatocellular carcinoma (HCC) and GC progression [15, 41–43]. In addition, several new cancer-related circRNA sponges have been discovered. CircITCH sponges miR-7, miR-17, and miR-214 in esophageal squamous cell carcinoma (ESCC) and exerts suppressive effects [42]. Hsa_circ_001569 targets miR-145 and promotes the proliferation and invasion of colorectal cancer [44]. CircMOT1 binds with miR-9 to inhibit HCC progression, while circTCF25 interacts with miR-103a and miR-107 to enhance the proliferation and migration of bladder carcinoma cells [45, 46]. These results collectively indicate that miRNA sponges may be a common function for circRNAs.
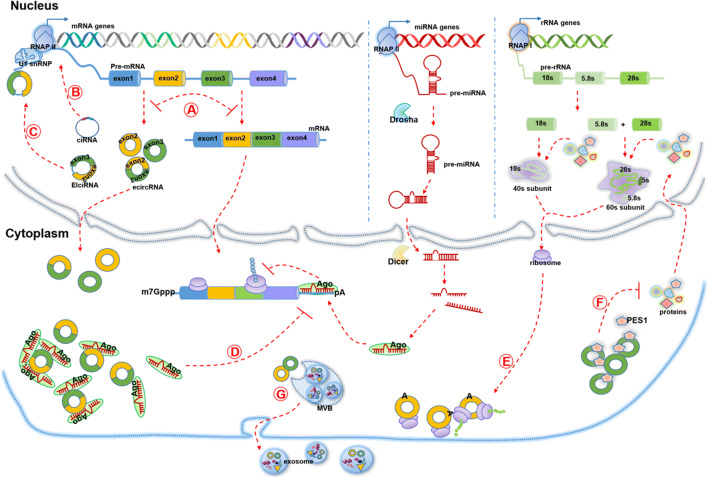
Fig. 2.
Functions and mechanisms of circRNA. a circRNAs’ backsplicing process forms three types of circRNAs and competes with linear RNAs’ canonical splicing to maintain the transcripts dynamic balance. Mature mRNA could be translated into proteins or inhibited by miRNAs which bound with Argonauto 2 (AGO2) protein. b CiRNAs accumulate to transcription sites and promote transcription by regulating elongation Pol II machinery. c EIciRNAs located in nucleus bind to U1 snRNA and form EIciRNA–U1 snRNP complexes to enhance gene transcription by interacting with Pol II transcription complex at the promoters of parental genes. d EcircRNAs exported into cytoplasm share some miRNA binding sites and sponge miRNAs to inhibit their effects on mRNA. e CircRNAs containing extensive m6A modification are sufficient to drive translation of circRNAs in a cap-independent fashion. f CircRNA (circANRIL) binds to PES1 which is essential to 36S and 32S pre-rRNA processing to mature 5.8S and 28S rRNA and thus impair exonuclease-mediated pre-rRNA processing and ribosome biogenesis. g CircRNAs could be packaged into exosomes and exported to extracellular space or circulating system
Although some researches still doubt the sponge function of circRNAs as most of circRNAs contain less than 10 miRNA binding sites [11], the other studies are in support of this notion. Many circRNAs harbor binding sites for multiple miRNAs and regulate them simultaneously and these evolutionarily conserved binding sites ensure the efficiency of targets [32]. Moreover, circRNAs are able to accumulate in cells and maintain its role for a relatively longer time due to high stability [47].
Regulation of alternative splicing
The backsplicing of circRNAs could compete with linear splicing of pre-mRNA for splicing sites (Fig. 2a). For example, circMbl, generated by second exon of MBL, has MBL binding sites in flanking introns [24]. MBL levels thus significantly influence circMbl biogenesis. The increase of canonical splicing efficiency results in the decrease of relevant circRNAs expression level, which indicates that circRNAs are involved in alternative splicing regulation. In addition, it is proposed that circRNAs also act as an “mRNA trap”. During the back-splicing process, circRNAs might sequester the translation start site and prevent the existence of certain normal linear transcripts that could be translated, which thereby reduce the expression level of certain proteins. For example, ecircRNAs produced from Fmn gene and DMD gene result in inactive mRNA transcripts with certain deletion mutations and lower levels of normal functional proteins [48, 49].
Regulation of parental gene transcription
EIciRNAs and ciRNAs harbor few binding sites for miRNAs and have distinct functions. EIciRNAs hold U1 snRNP through interaction with U1 snRNA, and then the EIciRNA–U1 snRNP complexes further interact with Pol II transcription complex at the promoters of parental genes to enhance gene transcription and expression (Fig. 2c) [20]. CiRNAs also positively regulate Pol II transcription. For example, ci-ankrd52, generated from gene ANKRD52, is capable of accumulating to its transcription sites and regulates elongation Pol II machinery acting as a positive regulator for transcription (Fig. 2b) [19].
Protein production
CircRNAs were initially recognized as non-coding RNAs [10, 11]. Only the engineered circRNAs inserted an internal ribosome entry sites (IRES) in upstream of the start codons of a protein could be translated in vitro and in vivo [50, 51]. While, extensive N6-methyladenosine (m6A) modification is sufficient to drive translation of circRNAs in a cap-independent fashion along with m6A reader YTHDF3 and translation initiation factors eIF4G2 and eIF3A (Fig. 2e) [52]. Further analyses showed that translatable circRNAs might be common in human transcriptome and a subset of circRNAs is found to be translated in vivo by performing ribosome footprinting and mass spectrometry [53]. Furthermore, circ-SHPRH was revealed to encode a new protein SHPRH-146aa which protects full-length SHPRH from degradation by the ubiquitin proteasome and suppresses glioma tumorigenesis [54].
However, the translational ability of circRNA remains controversial. Compared to linear transcripts, a circular structure may help ribosome recycling once translation is initiated and facilitate protein synthesis. While, some studies argue that this translation may be inefficient because a circular structure makes the beginning and the end of same ORF close to each other and thus, initiation and termination of translational process occur in the same place simultaneously [55]. Thus, new approaches should be developed to further study the coding potential of circRNAs.
Protein decoy
Several circRNAs were reported to bound and sequester proteins to suppress their function. For example, circMbl harbors several MBL binding sites and can sponge out the excessive MBL proteins to maintain its balance [24]. CircANRIL is able to bind to pescadillo homologue 1 (PES1), an essential 60S-preribosomal assembly factor, and then impair exonuclease-mediated pre-rRNA processing and ribosome biogenesis (Fig. 2f) [16]. Circ-Foxo3 binds to cyclin-dependent kinase 2(CDK2) and cyclin-dependent kinase inhibitor 1(p21) to form a ternary complex, which arrests the function of CDK2 and blocks cell cycle progression [56]. Circ-Foxo3 also has high binding affinity to anti-senescent ID-1, transcription factor E2F1, as well as anti-stress proteins FAK and HIF1a, and makes them retain in the cytoplasm and leads to increased cellular senescence [56]. Similarly, nuclear circRNA cia-cGAS is able to bound DNA sensor cGAS to block its enzymatic activity and prevent cGAS from recognizing self-DNA to maintain homeostasis [57]. CircPABPN1 binds to HuR and, thus, prevents HuR from binding to PABPN1 mRNA to lower PABPN1 translation [58]. However, not all of circRNAs interacting with proteins inhibit proteins functions. Ectopic circ-Amotl1 interacts with and stabilizes nuclear oncogene c-myc to upregulate c-myc targets and promote tumorgenesis [59].
Public database for circRNAs studies
Nowadays, many circRNAs-associated public databases have been developed. Many of them provide prediction on circRNAs’ possible biological functions or regulatory network in human diseases. For example, Circ2Traits provides information about human diseases-associated circRNAs [60]. StarBasev2.0 and circInteractome predict circRNA–miRNA–mRNA network and interaction between circRNA and RBPs [61, 62]. The protein-encoding ability could be predicted in CircRNADb and CSCD [63, 64]. Additionally, TRCirc starts to provide the regulatory information of circRNAs transcription [65]. Other databases and their available services are listed (Table 1), which are beneficial to data sharing and further study of circRNAs’ biological functions and applications. Despite this, the lack of standardized nomenclature still makes it difficult to search the same circRNA in all different databases and compare different studies. Also, curated database is still limited.
Table 1.
Public database for circRNAs
| Name | URL | Number of human circRNAs | Associated diseases | Resources available | Refs. |
|---|---|---|---|---|---|
| Circ2Traits | http://gyanxet-beta.com/circdb/ | 1953 | 105 diseases | Predicted disease-associated circRNAs and miRNA–circRNA–mRNA–lncRNA interactions | [60] |
| CircBase | http://circrna.org/ | Over 90000 | – | CircRNAs annotation | [66] |
| StarBaseV2.0 | http://starbase.sysu.edu.cn/ | Over 120000 | – | Predicted lncRNAs, circRNAs, miRNAs, mRNAs interaction networks | [61] |
| CircNet | http://circnet.mbc.nctu.edu.tw/; http://syslab5.nchu.edu.tw/CircNet/ | 212950 | – | CircRNAs annotation and predicted circRNA–miRNA–mRNA interaction networks | [67] |
| DeepBase v2.0 | http://rna.sysu.edu.cn/deepBase/ | 14867 | – | Small ncRNAs, lncRNAs and circRNAs annotation | [68] |
| CircInteractome | http://circinteractome.nia.nih.gov/ | Over 120000 | – | Predicted circRNA–miRNA interactions and RNA-binding proteins | [62] |
| CircRNADb | http://reprod.njmu.edu.cn/circrnadb | 32914 | – | CircRNAs annotation and protein-encoding ability | [63] |
| CSCD | http://gb.whu.edu.cn/CSCD | 1394023 | 19 cancers | Cancer-specific circRNAs annotation, cellular localization, protein-encoding ability, predicted circRNA–miRNA interactions and RNA-binding proteins | [64] |
| ExoRBase | http://www.exoRBase.org | 58330 | 5 diseases | Human blood exosome-derived circRNAs, lncRNAs and mRNAs annotation | [57] |
| CircRNADisease | http://cgga.org.cn:9091/circRNADisease/ | 330 | 48 diseases | Published disease-associated circRNAs annotation | [69] |
| CIRCpedia v2 | http://www.picb.ac.cn/rnomics/circpedia | 183943 | – | CircRNAs annotation including conservation and enrichment analysis | [70] |
| TRCirc | http://www.licpathway.net/TRCirc | 92375 | – | Regulatory information of circRNAs transcription | [65] |
CircRNAs profiles in gastric cancer
Through high-throughput sequencing technologies and bioinformatic analyses, lots of novel circRNAs have been identified in GC cells. Some of them are differentially expressed between GC tissues and adjacent normal tissues, which implies the important roles of circRNAs in GC development and progression.
So far, several circRNA sequencing data have shown that circRNAs are abundant in GC tissues and many of them have aberrant expression levels which could be GC-related circRNAs. According to the circRNAs microarray data in 8 GC tissues and adjacent tissue samples, Sui et al. identified 467 differentially expressed circRNAs between GC tissues and adjacent normal tissues, of which 214 were significantly upregulated and 253 were downregulated in GC tissues [71]. Huang et al. found over 2000 circRNAs expressed in 3 paired GC and adjacent normal tissues, of which 100 circRNAs had over twofold change and were predicted to function in circRNA–miRNA–mRNA/gene network [72]. Fang et al. identified 306 differently expressed circRNAs (141 upregulated and 165 downregulated) between advanced GC and adjacent normal tissues, among which 273 were predicted to regulate target miRNAs [73]. Other circRNA sequencing data are listed (Table 2). Many factors affect the final results of sequencing data including clinical reasons (clinical sample resources), pre-analytical reasons (sample processing and preparing), analytical reasons (detection methods), and postanalytical reasons (methods or tools for circRNA validation). Although these data may have discrepancies due to factors above, most of them indicate that the down-regulated circRNAs are more than up-regulated ones in GC tissues. Moreover, function prediction results suggest that most of the circRNAs are able to act as miRNAs regulators in GC, which needs to be further confirmed in future studies.
Table 2.
Overview of circRNAs identified by RNA sequencing and microarrays in GC
| Sample | Special treatment | Detection method | Number of circRNAs | Number of circRNA differently expressed(≥ twofold change) | Refs. |
|---|---|---|---|---|---|
| 3 paired GC and GCN tissues | RNase R | CircRNA microarray | About 2000 | 100(16 upregulated, 84 downregulated) | [72] |
| 8 paired GC and GCN tissues | RNase R | CircRNA microarray | 4458 | 467(214 upregulated, 253 downregulated) | [71] |
| 3 paired GC and GCN tissues | – | CircRNA microarray | 5396 | 308(107 upregulated, 201 downregulated) | [74] |
| 3 paired III stage GC and GCN tissues | – | CircRNA microarray | – | 306(141 upregulated, 165 downregulated) | [73] |
| 3 paired GC and GCN tissues | Ribosome-depleted | RNA-seq | About 3000 | 180(82 upregulated, 98 downregulated) | [75] |
| 3 groups of gastric tissues(gastric tissue without cancer, GC tissue, GCN tissue) | Ribosome-depleted | RNA-seq | 736 | – | [76] |
| 5 paired GC and GCN tissues | RNase R | Microarray chip assay | 62998 | 713(191 upregulated, 522 downregulated) | [77] |
| 3 paired plasma of patients and healthy persons | – | CircRNA microarray | 5396 | 343(171 upregulated, 172 downregulated) | [78] |
| 6 paired GC and GCN tissues | – | CircRNA microarray | 2070 | 400(176 upregulated, 264 downregulated) | [79] |
CircRNAs in GC and molecular mechanisms
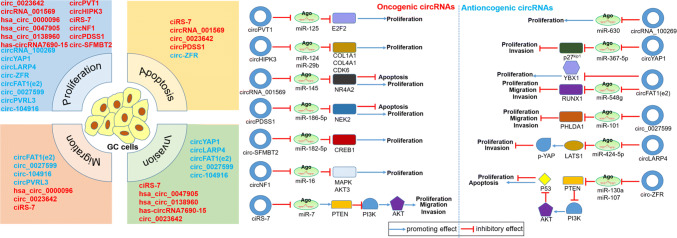
Recently, several oncogenic and antioncogenic circRNAs have been discovered to regulate the proliferation, migration, invasion and apoptosis of GC cells (Fig. 3). Most of them could regulate GC-related signaling pathways through miRNA sponges. Also, the circRNA interacting with RBPs in GC starts to be indentified.
Fig. 3.
The role and regulatory pathway of GC-related circRNAs. The schematic diagram depicts the known role of circRNAs in GC progression and the way that circRNAs involved in miRNA-associated gene regulatory pathway
Oncogenic circRNAs in GC
CiRS-7, one of the most studied circRNAs is miR-7 sponge. While, ciRS-7 was recently found to act as oncogenic circRNA by antagonizing miR-7-mediated PTEN/PI3K/AKT pathway in GC [80]. Overexpression of ciRS-7 could neutralize the suppressive effects of miR-7 on GC cells migration ability and reduce apoptotic cells induced by miR-7. Xenograft mice model experiment also showed that ciRS-7 is able to abrogate miR-7-mediated suppressive effects on tumor growth. Further studies suggested that ciRS-7 weakens miR-7 effects on upregulating expression of PTEN and downregulating the levels of PI3K and AKT phosphorylation of PTEN/PI3K/AKT pathway in vitro. As PTEN/PI3K/AKT pathway is involved in multiple tumors such as lung cancer, breast cancer and colorectal cancer [81–83]. PTEN acting as antioncogene negatively regulates PI3K/AKT signal pathway to affect cell proliferation, apoptosis, migration of tumor. Here, ciRS-7-miR-7-PTEN/PI3K/AKT regulatory axis in GC progression is firstly found.
CircPVT1, derived from a cancer susceptibility locus PVT1, was initially confirmed to be frequently upregulated in GC tissues partly due to DNA amplification [75]. Further studies indicated that circPVT1 might serve as a proliferative factor through sponging miR-125 in GC. Silencing circPVT1 selectively impairs proliferation of GC cells and decreases the expression of miR-125b target E2F2. Co-expression of circPVT1 and miR-125b attenuates the anti-proliferative effects of miR-125b. In addition, circPVT1 expression level is related to T4 stage tumor numbers and neural invasion. CircPVT1 level, along with TNM stage and tumor size, was verified as independent prognostic factors for GC patients.
CircHIPK3 might be involved in GC progression through circHIPK3-miR-124/miR-29b axis [84]. As known sponge for miR-124 and miR-29b, circHIPK3 are upregulated in GC tissues and related to T stage and Ming’s classification, and negatively correlated with miR-124/miR-29b in GC tissues and cell lines. Moreover, circHIPK3-miR-124/miR-29b axes were predicted and most of the target genes are closely related to cell proliferation. Gain and loss of function studies also verified that circHIPK3 promotes GC cell proliferation. Meanwhile, the axes target genes COL1A1, COL4A1 and CDK6, upregulated in GC tissues, could be potential biomarker for diagnosis and prognosis.
CircRNA_001569 was revealed as miR-145 sponge and could regulate cell proliferation and apoptosis through oncogene NR4A2 in GC [85]. CircRNA_001569 were found to be upregulated in GC tissues and cells and positively correlated with NR4A2, while negatively correlated with miR-145. Overexpression and knockdown experiment suggested that circRNA_001569 significantly promotes GC cell proliferation and suppresses cell apoptosis through decreasing miR-145 expression and increasing expression of miR-145 target NR4A2. Although circRNA_001569-miR-145 regulatory axis was previously reported in colorectal cancer [86], here, its promotive effects were also verified in GC progression.
CircPDSS1 promotes GC progression as miR-186-5p sponge [87]. CircPDSS1 is significantly upregulated in GC tissues and cells, and related with poor prognosis in GC patients. Gain and loss of function experiments indicated that circPDSS1 promotes GC cell proliferation and suppresses cell apoptosis. Moreover, bioinformatic prediction showed that circPDSS1 binds with miR-186-5p, while NEK2 could be miR-186-5p target. Functional assays demonstrated that circPDSS1 acts as oncogene through sponging miR-186-5p and increasing NEK2 expression level. As NEK2 is abnormally expressed in a wide variety of cancer and involved in tumorigenesis and metastasis [88], circPDSS1-miR-186-5p-NEK2 axis seems to be important in GC progression.
Circ-SFMBT2, derived from exons 5–8 of gene SFMBT2, was found to be upregulated in GC tissues, cell lines and plasma from GC patients, and positively associated with TNM stage [89]. Silencing circ-SFMBT2 in the cytoplasm markedly inhibits GC cell proliferation. Furthermore, circ-SFMBT2 was predicted and validated to target miR-182-5p and competitively share it with oncogene CREB1. Overexpression of miR-182-5p decreases CREB1 expression and suppresses cell proliferation. While, circ-SFMBT2 knockdown simultaneously enhances these effects as more functional miR-182-5p are released. Here, circ-SFMBT2 was revealed to regulate CREB1 expression through competing for miR-182-5p in GC.
CircNF1 (hsa_circ_0042881), derived from exons 2–8 of tumor-suppressive gene NF1, serves as a novel oncogenic circRNA through functioning as miR-16 sponge in GC [90]. CircNF1 was found to be markedly upregulated in GC tissues and cell lines and promote cell proliferation. Luciferase reporter assays showed that circNF1 binds to miR-16 and then derepresses its downstream target mRNA, MAP7 and AKT3. Additionally, the expressions of circNF1 and linear NF1 are not significantly correlated, suggesting that they might have independent roles in GC development. Here, novel circNF1 was demonstrated to promote GC progression through absorbing miR-16.
Antioncogenic circRNAs in GC
CircRNA_100269, transcribed from gene LPHN2, may serve as tumor suppressor in GC through sponge for miR-630. CircRNA_100269 was found to be significantly down-regulated in recurrent cancer tumors compared with non-recurrent cancer tissues and could be an independent predictor of early recurrence of stage III GC [91]. Later, further studies showed that miR-630 might be a direct target of circRNA_100269 [92]. Overexpression of circRNA_100269 suppresses GC cell growth, while co-expression of miR-630 and circRNA_100269 evidently increases the growth rates of GC cells as miR-630 could bind to circRNA_100269 and block its suppressive effects on GC cells. Although accumulating evidence indicated that miR-630 is overexpressed in various tumors and related to cell proliferation and invasion [93–95], here, a novel circRNA_100269-miR-630 regulatory axis were found to be involved in GC cell growth.
CircYAP1 might serve as potential suppressive factor for GC [96]. CircYAP1 (hsa_circ_0002320), selected from CircNet database, was found to be decreased in GC tissues. Patients with lower circYAP1 expression might have shorter survival time, suggesting that circYAP1 could serve as prognostic factors. Overexpression and knockdown experiments indicated that circYAP1 is able to inhibit GC proliferation and invasion in vitro and in vivo. Moreover, circYAP1-miR-367-5p-p27kip1 regulatory axes were predicted and validated in GC cells. CircYAP1 overexpression or silence could upregulate or downregulate target gene p27kip1, respectively, which could be reversed by miR-367-5p mimics or inhibitor. Although circYAP1 is derived from oncogene YAP1 [97], it could act as an anti-oncogene in GC.
CircLARP4 may act as a novel suppressive factor and a potential GC biomarker [98]. CircLARP4 derived from exon 9 and 10 regions of LARP4 is preferentially located in the cytoplasm and presents high stability (half-life exceeds 24 h). Moreover, ectopic upregulated miR-424-5p could promote GC cell growth and invasion through targeting LATS1 gene, while circLARP4 might serve as miR-424-5p sponge. Overexpression of circLARP4 would inhibit the ability of proliferation and invasion through counteracting miR-424-5p effects and revive expression of LATS1 and its downstream p-YAP in vitro. Hippo-Yap pathway is known to play a pivotal role in the regulation of tumor proliferation and metastasis [99]. By phosphorylating YAP, LATS1 could prevent YAP from transporting to nucleus to suppress oncogene transcription which has been confirmed in GC [100]. While, circLARP4 is a newly discovered molecular involved in modulating Hippo-Yap pathway as miR-424-5p sponge.
Circ-ZFR are markedly down-regulated in GC tissues and cells and acts as antioncogene [101]. Functional experiments showed that circ-ZFR promotes cell apoptosis and suppresses cell proliferation by arresting cell cycle and cell propagation. Meanwhile, circ-ZFR-miR-130a/miR-107-PTEN network was predicted and confirmed to regulate GC. Overexpression of circ-ZFR would weaken miR-130a/miR-107 effects on GC cell cycle, cell propagation and apoptosis. Transfection of miR-130a/miR-107 mimics promotes GC cell propagation and impedes apoptosis by targeting PTEN. Moreover, in vivo experiment approved that circ-ZFR is able to curb GC tumor growth and increase expression of PTEN and p53 through sponging miR-130a/miR-107. Here, circ-ZFR was verified as novel antioncogenic circRNA regulating GC proliferation and apoptosis through miR-130a/miR-107-PTEN regulatory network.
CircFAT1(e2), derived from exon 2 of gene FAT1, inhibits GC progression through sponging miR-548g in the cytoplasm and binding to protein YBX1 in nucleus [102]. CircFAT1(e2) was confirmed to be downregulated in GC tissues and cell lines and correlated with clinical stage. Overexpression of circFAT1(e2) significantly suppresses GC cell proliferation, invasion and migration in vitro and tumor growth in vivo. Subcellular location results showed circFAT1(e2) is localized in both cytoplasm and nucleus of GC cells. In cytoplasm, circFAT1(e2) was predicted and validated to directly bind to miR-548g and negatively regulate miR-548g expression which promotes GC progression through RUNX1. In nucleus, circFAT1(e2) interacts with protein YBX1 and then downregulates its target genes, EGFP, c-Met, CDC25A. As oncoprotein YBX1 belongs to DNA- and RNA-binding family of transcription factors and is involved in many cellular processes including transcription, translation and DNA damage repair [103], here, circFAT1(e2) could bind to YXB1 and block its activities in GC.
Circ_0027599 suppresses GC cell proliferation and metastasis through circ_0027599-miR-101-PHLDA1 axis [104]. PHLDA1, as tumor suppressor, is downregulated in GC cell lines and inhibits cell proliferation, migration and invasion. While, circ_0027599 was found to reduce miR-101 expression and increase its target PHLDA1 levels. Overexpression circ_0027599 could suppress GC cell proliferation and metastasis through regulating miR-101 and derepressing its target tumor-suppressive gene PHLDA1.
Other circRNAs with unknown mechanisms
Hsa_circ_0000096, derived from gene HIAT1, presents oncogenic activity [105]. It is downregulated in GC tissues and cells and associated with gender, tumor invasion and TNM stage. Knockdown of hsa_circ_0000096 arrests cells at G0/G1 phase, and inhibits cell proliferation and migration. Xenograft nude mouse model experiment also confirmed these effects. Here, downregulated hsa_circ_0000096 acting as an oncogene is interesting as it is not consistent with that low expression of non-coding RNA in cancers generally acts as a suppressor gene.
Circ-104916, derived from gene NEK6, seems to be a novel tumor-suppressive circRNA. It is downregulated in GC tissues and associated with tumor stage, lymphatic metastasis, nerve invasion and invasion depth [93]. Overexpression of circ-104916 inhibits cell proliferation, migration and invasion capability. Meanwhile, circ-104916 might suppress EMT process to affect GC cell migration and invasion as the level of E-cadherin was upregulated, while N-cadherin, Vimentin and Slug were downregulated.
circRNA_0023642 (hsa_circ_0023642), one of the top 5 upregulated circRNAs in previous sequencing data, seems to have tumor-promotive function in GC [72, 106]. Knockdown of it inhibits cell proliferation, migration, invasion and induces cell apoptosis. Also, circRNA_0023642 might affect EMT signaling pathway in GC as downregulation of circRNA_0023642 could decrease the EMT-related gene.
CircPVRL3, derived from exons 4, 5, 6 and 7 of gene PVRL3, are involved in proliferation and migration of GC [107]. It is downregulated in GC tissues and cell lines and associated with TNM stage and prognosis. Knockdown of circPVRL3 would promote proliferation and migration. Moreover, bioinformatics prediction revealed its several potential functions. CircPVRL3 has a high degree of AGO occupancy and might serve as miRNA sponge. Meanwhile, circPVRL3 might function through RBPs complexes with FUS, LIN28A, PTB, and EIF4A3. Additionally, CircPVRL3 contains internal ribosome entry sites, ORF and m6A modification structure and might encode functional proteins. However, such potential functions have not been verified.
Through circRNA and mRNA co-expression network, hsa_circ_0047905, hsa_circ_0138960 and has-circRNA7690-15 were focused as these circRNAs interacting with more mRNAs might play more significant roles in GC [108]. They are upregulated in GC tissues and stable in the cytoplasm. Depletion of them downregulate their parent gene expression and suppress GC cells proliferation and invasion. In addition, these three circRNAs present good diagnostic value.
CircRNAs as diagnostic and prognostic biomarkers for GC
The high incidence and mortality of GC make new requirements for early diagnosis and the prognosis. CircRNAs show great potential to be used as cancer biomarkers. Firstly, they can be easily detected due to their high stability, abundance and conservation in human cells. Secondly, they often display tissue- and developmental-stage-specific expression. Thirdly, circRNAs are also present in human serum, plasma and other body fluids [109, 110], especially in microvesicles and exosomes [111], which can be easily detected by non-invasive method. Also, qRT-PCR with high sensitivity could facilitate detection of circRNAs in drops of fluids.
Furthermore, the clinical value of circRNAs as biomarkers has been explored in many studies. According to large clinical sample test including tissues and serum from GC patients and healthy controls, followed with clinicopathologic factors correlation analysis as well as prognostic and survival analysis, a group of potential circRNAs biomarkers are emerged for GC early diagnosis and prediction of recurrence and metastasis. Hsa_circ_0000096 and hsa_circ_0047905 present good diagnostic value as their area under the ROC curve (AUC) in GC tissues is up to 0.82 and 0.85, respectively [105, 108]. Plasma hsa_circ_0000520 presents a better diagnostic value than that in tissues as its AUC, sensitivity and specificity in plasma are 0.897, 82.4% and 84.4%, respectively, which are higher than that in tissues (0.613, 53.6%, 85.7%) [112]. While, hsa_circ_0000190 is opposite. Its expression is downregulated in GC tissues and plasma and associated with tumor diameter, lymphatic and distal metastasis, TNM stage as well as CA19-9 level [113]. The AUC of has_circ_0000190 in tissues is 0.75 much higher than 0.60 in plasma, suggesting a modest diagnostic value. Besides, the combination of has_circ_0001017 and has_circ_0061276 in tissues and plasma exhibits the best diagnostic value and its AUC reaches to 0.996, with 95.5% sensitivity and 95.7% specificity. Additionally, hsa_circ_0058246 was upregulated in GC patients with metastasis and recurrence [73], while patients with lower levels of circPVT1 had a significantly shorter OS and DFS (disease-free survival) [75]. Both of them could be used as a prognostic biomarker. Other circRNAs with potential diagnostic and prognostic value are listed (Table 3).
Table 3.
CircRNAs as diagnostic and prognostic biomarkers for GC
| CircRNA | CircBase ID | Sample | Dysregulation in GC | Clinicopathological association | Potential function | AUC | Sensitivity and specificity | Refs. |
|---|---|---|---|---|---|---|---|---|
| Hsa_circ_0003159 | Hsa_circ_0003159 | Paired GC and GCN tissues (n = 108) | ↓ | Gender, stage, distal metastasis, tumor node metastasis | Diagnosis | 0.75 | 85.2%, 56.5% | [121] |
| CircFAT1(e2) | Hsa_circ_0001461 | Paired GC and GCN tissues (n = 38) GC cell lines | ↓ | Metastasis, TNM stage | – | – | – | [102] |
| Circ-104916 | Hsa_circ_0008383 | Paired GC and GCN tissues (n = 70) GC cell lines | ↓ | Invasion depth, tumor stage, lymphatic metastasis | – | – | – | [93] |
| Hsa_circ_0000096 | Hsa_circ_0000096 | Paired GC and GCN tissues (n = 101) GC cell lines | ↓ | Gender, invasion, TNM stage | Diagnosis | 0.82 | – | [105] |
| CircRNA_100269 | Hsa_circ_0013048 | Paired GC and GCN tissues (n = 112) GC cell lines | ↓ | Histological subtype, node invasion number | Prognosis | – | – | [91] |
| CircLARP4 | Hsa_circ_0003222 | GC and GCN tissues (n = 80) GC cell lines | ↓ | Tumor size, lymphatic metastasis | Prognosis | – | – | [117] |
| CircYAP1 | Hsa_circ_0002320 | Paired GC and GCN tissues (n = 97) | ↓ | Tumor size, TNM stage | Prognosis | – | – | [96] |
| CircPVRL3 | Hsa_circ_0066779 | Paired GC and GCN tissues (n = 62) GC cell lines | ↓ | TNM stage | Diagnosis | 0.763 | 90.3%, 56.4% | [107] |
| Hsa_circ_0074362 | Hsa_circ_007436 | GC and GCN tissues (n = 127) Gastritis tissues (n = 83) GC cell lines | ↓ | CA19–9, lymphatic metastasis | Diagnosis | 0.63 | 36.2%, 84.3% | [122] |
| Hsa_circ_0001895 | Hsa_circ_0001895 | GC and GCN (n = 96) Gastric mucosa tissues (n = 35) Gastric dysplasia tissues (n = 30) GC cell lines | ↓ | Cell differentiation, Borrmann type, CEA | Diagnosis | 0.792 | 67.8%, 85.7% | [123] |
| Hsa_circ_0014717 | Hsa_circ_0014717 | Paired GC and GCN tissues (n = 96) Gastric juice (n = 38) Gastric ulcer (n = 30) Chronic atrophic gastritis (n = 15) GC tissues (n = 39) | ↓ | Tumor stage, distal metastasis, tissue CEA and CA19-9 | Diagnosis | 0.696 | 59.4%, 81.3% | [74] |
| Hsa_circ_0000705 | Hsa_circ_0000705 | Paired GC and GCN tissues (n = 96) Gastric mucosa tissues (n = 35) Gastric ulcer tissues (n = 16) Erosive gastritis tissues (n = 18) Gastric dysplasia tissues (n = 25) | ↓ | Tumor location, stage, Borrmann type, pathologic diagnosis, CA19-9 | Diagnosis | 0.719 | 64.6%, 69.8% | [124] |
| CircPDSS1 | Hsa_circ_0093398 | Paired GC and GCN tissues (n = 20) GC cell lines | ↑ | – | Prognosis | – | – | [87] |
| Hsa_circ_0058246 | Hsa_circ_0058246 | Paired GC and GCN tissues (n = 43) | ↑ | Lymph node invasion(>=16), recurrence | Prognosis | – | – | [73] |
| Hsa_circ_0047905, Hsa_circ_0138960, Has-circRNA7690-15 | Hsa_circ_0047905, Hsa_circ_0138960, - | GC and GCN tissues (n = 31) GC cell lines | ↑ | – | Diagnosis | 0.85, 0.647, 0.681 | – | [108] |
| CircPVT1 | Hsa_circ_0001821 | Paired GC and GCN tissues (n = 187) | ↑ | T4 stage tumors, neural invasion | Prognosis | – | – | [75] |
| CiRS-7 | Hsa_circ_0001946 | Paired GC and GCN tissues (n = 102) | ↑ | TNM stage, lymph node involvement, distant metastasis | Prognosis | – | – | [80] |
| CircHIPK3 | Hsa_circ_0000284 | Paired GC and GCN tissues (n = 63) | ↑ | T stage, Ming’s classification | – | – | – | [84] |
| CircRNA_001569 | Hsa_circ_001569 | Paired GC and GCN tissues (n = 30) GC cell lines | ↑ | Tumor size, clinical stage, invasion | – | – | – | [85] |
| Circ-SFMBT2 | Hsa_circ_0017639 | Paired GC and GCN tissues (n = 36) Plasma samples from 26 GC patients and 18 healthy controls GC cell lines | ↑(GC tissues) ↑(GC plasma) | Tissues: TNM stage | Diagnosis | 0.759 (Tissues) | 80.6%, 63.9% | [89] |
| Hsa_circ_0006633 | Hsa_circ_0006633 | Paired GC and GCN tissues (n = 96) Healthy gastric mucosa (n = 35) Gastritis mucosa (n = 51) Gastric dysplasia tissues (n = 20) Paired plasma samples from GC patients and healthy controls (n = 20) GC cell lines | ↓(GC, gastritis and dysplasia tissues and GC cells) ↑(GC plasma) | Tissues: Distal metastasis, CEA | Diagnosis | 0.741 (Tissues) | 60%, 81% | [125] |
| Hsa_circ_002059 | Hsa_circ_002059 | Paired GC and GCN tissues (n = 101) Paired pre- and post-operative plasma sample (n = 36) | ↓(GC tissues) ↑(preoperative plasma) | Tissues: TNM stage, distal metastasis, gender, age | Diagnosis | 0.73 | 81%, 62% | [126] |
| Hsa_circ_0001649 | Hsa_circ_0001649 | Paired GC and GCN tissues (n = 76) Paired preoperative and postoperative blood samples (n = 20) | ↓(GC tissues) ↓(preoperative plasma) | Tissues: Pathological differentiation | Diagnosis | 0.834 (Tissues) | 71.1%, 81.6% | [127] |
| Circ_0027599 | Hsa_circ_0027599 | Paired GC and GCN tissues (n = 78) Blood samples from GC patients and healthy controls | ↓(GC tissues) ↓(GC blood) | Tissues: Tumor size, invasion, lymph node distant metastasis, distant metastasis, TNM stage | – | – | – | [104] |
| Circ-KIAA1244 | Hsa_circ_0130810 | Paired GC and GCN tissues (n = 28) Plasma samples from 62 GC patients and matched 25 healthy controls GC cell lines | ↓(GC tissues and cell lines) ↓(GC serum) | Plasma: TNM stage, lymphatic metastasis | Diagnosis Prognosis | 0.748 (Plasma) | 77.4%, 68.0% | [128] |
| Hsa_circ_0000190 | Hsa_circ_0000190 | Paired GC and GCN tissues (n = 104) Paired plasma samples from GC patients and healthy controls (n = 104) | ↓(GC tissues) ↓(GC plasma) | Tissues: Tumor diameter, lymphatic metastasis, distal metastasis, TNM stage, CA19-9 Plasma: CEA | Diagnosis | 0.75 (Tissues) 0.60 (Plasma) | 72.1%, 68.3% (Tissues) 41.4%, 87.5% (Plasma) | [113] |
| Hsa_circ_0000745 | Hsa_circ_0000745 | GC and GCN tissues (n = 80) Paired plasma samples from GC patients and healthy controls (n = 60) | ↓(GC tissues) ↓(GC plasma) | Tissues: Tumor differentiation Plasma: Tumor node metastasis, stage | Diagnosis | 0.683 (Plasma) | 85.5%, 45% | [114] |
| Hsa_circ_0000181 | Hsa_circ_0000181 | Paired GC and GCN tissues (n = 115) Paired plasma samples from GC patients and healthy controls (n = 102) | ↓(GC tissues) ↓(GC plasma) | Tissues: Tumor diameter, lymphatic metastasis, distal metastasis, CA19-9 Plasma: Differentiation, CA19-9 | Dignosis | 0.756 (Tissues) 0.582 (Plasma) | 53.9%, 85.2% (Tissues) 99.0%, 20.6% (Plasma) | [129] |
| Hsa_circ_0001017, Hsa_circ_0061276 | Hsa_circ_0001017, Hsa_circ_0061276 | Paired GC and GCN tissues (n = 121) Paired plasma samples from GC patients and healthy controls (n = 121) | ↓(GC tissues) ↓(GC plasma) | Tissues: Age, tumor size, TNM stage, distal metastasis, CEA Plasma: Gender, tumor size, differentiation, poor prognosis | Prognosis Diagnosis | 0.966 (Combinative AUC) | 95.5%, 95.7% | [78] |
| Hsa_circ_0000520 | Hsa_circ_0000520 | Paired GC and GCN tissues (n = 56) Plasma samples from 45 GC patients and matched 17 healthy controls | ↓(GC tissues) ↓(GC plasma) | Tissues: TNM stage Plasma: CEA | Diagnosis | 0.613 (Tissues) 0.897 (Plasma) | 53.6%, 85.7% (Tissues) 82.4%, 84.4% (Plasma) | [112] |
In addition, the combination of several circRNAs or a single circRNA with traditional diagnostic indicators could increase the sensitivity, specificity and accuracy of GC diagnosis and prognosis. The AUC of downregulated hsa_circ_0000096 and hsa_circ_002059 in GC tissues is 0.82 and 0.73, respectively, while increased to 0.91 when combined [105], indicating that the diagnostic value of a combination of circRNAs is higher than that of individual circRNA. Besides, Zhang et al. constructed a four-circRNA-based classifier and a new formula combined circRNA classifier with TNM stages to evaluate the risk of early recurrence of stage III GC after radical surgery and distinguish patients with a high risk from those with a low risk. The AUC of two classifiers was 0.763 and 0.711 as well as 0.866 and 0.818, respectively, in the two cohorts [91], suggesting that the prognostic value of circRNAs combined with traditional factors is much higher. Similarly, the AUC of downregulated hsa_circ_0000745 in plasma is 0.683, while increased to 0.775 in combination with CEA [114]. Although the potential of circRNAs to be promising GC biomarkers is emerging, their real value still needs to be further tested in a larger cohort of clinical samples. Furthermore, due to the lack of adequate biomarkers with high specificity and sensibility simultaneously, the novel diagnostic pattern combining circRNAs with other traditional biomarkers like CEA and CA19-9 might be one of the solutions. Additionally, circRNAs are enriched and stable in exosomes [111]. The presence of circRNAs in exosomes provides a new mechanism for the communication between cancer cells and the other cells [115]. Therefore, exosomes-derived circRNAs (ex-circRNAs) may be associated with cancer progression and could be a novel diagnostic and prognostic marker.
CircRNAs as potential therapeutic targets for GC
As their regulatory roles in cancer are gradually being unveiled, circRNAs might be developed as effective therapeutic targets. Several strategies based on the functions of circRNAs have been proposed to treat GC. Firstly, exogenous upregulation or downregulation of relevant circRNAs to regulate miRNA molecules might be a useful method. Several methods have been applied. For example, siRNA or shRNA targeting the specific backspliced sequence of circRNA is used to inhibit its expression [42]. CRISPR/Cas9 system could achieve knockout of circRNAs [116]. While, plasmid and lentiviral vector are used to increase circRNAs levels [75, 86, 117]. Liang et al. introduced how to construct effective overexpression vectors of circRNAs with short intronic repeat sequences [22]. However, how to control the process of endogenous circularization remains unknown. Additionally, synthetic circRNAs sponges might be a simple, effective and convenient strategy. Recently, synthetic circular RNA containing miR-21 binding sites was proved to achieve targeted loss of miRNA function and suppress GC cell proliferation in vitro, suggesting that synthetic circRNAs sponges could be potential therapeutic application in human patients [118]. Also, artificially controlled endogenous circularization may be another choice. Using an “mRNA trap” to sequester translation start site of dysfunctional mRNAs, the biogenesis of diseases-related proteins might be reduced [48]. Furthermore, applying circRNAs functioning as protein decoys to regulate disease-related proteins release and biological activity, or targeting encoding circRNAs involving in tumorigenesis or progression like circ-SHPRH might be potential treatment methods. Although such circRNAs have not been found and disclosed in GC recently, it will be promising application for future tumor treatment with further studies.
Using circRNAs as novel therapeutic targets broadens the universe of potential “druggable” targets. Nevertheless, the real clinical application of circRNAs as drugs or targets needs more detailed and complete experimental data such as safety and efficacy. Additionally, how to deliver engineered circRNAs in vivo safely is another question. Using modified exosomes which contain engineered circRNAs or siRNAs targeting circRNA might be an effective approach because a large amount of researches have proved that exosome-mediated delivery of RNA could be used for therapy purposes [119, 120]. The lipid bilayer structure of exosomes prevents RNAs from degradation and ensures the effective concentration of circRNAs. Moreover, the small size of exosome and its membrane structure are helpful for their absorbtion and fusion by cancer cells. Therefore, researches on circRNAs as therapeutic targets or tools will be one of the most noticeable areas in the field of circRNAs.
Challenges and perspectives
Nowadays, circRNAs are becoming new research frontier in tumor biology and therapy. Up to date, at least hundreds of circRNAs have been discovered to be aberrantly expressed in GC tissues. Many of them have been proved to modulate GC cell proliferation, migration, invasion and apoptosis and could be potential biomarker or effective therapeutic targets for GC. Although some new perspectives on the clinical application of circRNAs are generated rapidly, the experimental and clinical researches in GC still lag behind. Firstly, circRNAs mechanisms of circularization, degradation and cellular localization remain poorly understood. Besides, the number of circRNAs in GC with clear biological functions and mechanisms is limited. Novel functions or mechanisms except miRNA sponge need to be disclosed. Moreover, circRNAs effects on GC microenvironmental cells such as cancer-associated fibroblasts (CAFs) and macrophages are still not very clear as tumor microenvironment could affect tumor progression [130, 131]. Since exosomes have been verified to contain stable and enriched circRNAs, whether ex-circRNAs are critical to GC progression and can be developed as a novel biomarker, therapeutic targets for GC need further exploration. In addition, current studies on exploring circRNAs as GC biomarkers and therapeutic tools or targets are simple and limited. Standardized methodology for clinical circRNAs detecting is lacked. Systematical assessment including costs, accuracy, reproductivity, specificity and sensitivity of circRNAs biomarkers in larger samples and comparison with traditional tumor markers are insufficient. Synthetic circular RNA molecules seem to be effective as gene therapy methods, while the efficacy, safety and potential side effects are unknown. Moreover, clinical trials on circRNAs as biomarkers and therapeutic targets or tools remain to be carried out.
Exosome might extremely extend circRNAs’ studies and applications. Exosome, as nano-sized vesicles released by many cells especially tumor cells, could transfer signaling molecules including circRNAs to recipient cells and regulate cancer progression. For example, exosomal ciRS-133 derived from GC cells could be delivered into pre-adipocytes and promote white adipose browning through targeting miR-133/PRDM16 pathway [132]. Some researchers proposed that the concentration of circulating non-coding RNAs is too low to induce biological responses [133]. Nonetheless, several non-coding RNAs including circRNAs could regulate the same gene and multiple genes in same signaling pathway. Thus, exosomal circRNAs would be potential therapeutic targets. Also, membrane of exosome could protect inner signaling contents from nuclease and other environmental factors and make it easier to be detected in many biological fluids like blood, urine, saliva and breast milk [134]. Recent RNA sequencing data revealed that many circRNAs are stable and enriched in exosome and could be a promising biomarker for cancer diagnosis (Fig. 2g) [111]. Exosomal circ-PDE8A could be detected in blood circulation and are associated with progression and prognosis in pancreatic ductal adenocarcinoma patients [135]. Moreover, as natural intracellular communication mediators, exosomes have been proved to be good targeted drug delivery tool. Exosomes could even act as natural and effective carriers for small interfering RNAs (siRNAs) delivery of gene therapy [120]. Therefore, detection and modification of exosome contents provide a new direction to disease diagnosis and treatment. Also, exosomal circRNAs are expected to become remarkable biomarkers and therapy tools for GC.
Conclusion
With biological methods and informatics technologies along with further studies, more and more GC-related circRNAs along with the physiological and pathological functions have been identified. Moreover, circRNAs present great potential to be biomarker for GC diagnosis and prognosis as well as novel therapeutic targets or tools. Although the knowledge of precise mechanisms of circularization, degradation, cellular localization and functions in GC is still limited, the mystery of circRNAs will finally be solved, and novel diagnostic and therapeutic strategies based on circRNAs will serve clinical practice effectively in future.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grant 81572075, Grant 81972310), Zhenjiang Key Laboratory of High Technology Research on Exosomes Foundation and Transformation Application (Grant SS2018003), and Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
Abbreviations
- GC
Gastric cancer
- EcircRNAs
Exonic circRNAs
- CiRNAs
Intronic circRNAs
- EIciRNAs
Exon–intron circular RNAs
- TricRNAs
TRNA intronic circRNAs
- MBL
Muscleblind
- A-to-I
Adenosine-to-Inosine
- QKI
Quaking
- EMT
Epithelial-mesenchymal transition
- RBPs
RNA-binding proteins
- TSEN
TRNA splicing endonuclease complex
- BHB
Bulge–helix–bulge
- HCC
Hepatocellular carcinoma
- ESCC
Esophageal squamous cell carcinoma
- IRES
Internal ribosome entry sites
- CDK2
Cyclin-dependent kinase 2
- M6A
N6-methyladenosine
- PES1
Pescadillo homologue 1
- AGO2
Argonauto 2
- AUC
Area under the ROC curve
- Ex-circRNAs
Exosomes-derived circRNAs
- CAFs
Cancer-associated fibroblasts
- SiRNAs
Small interfering RNAs
Author contributions
RL and JJ collected literature and draft the manuscript. HS, XZ, HQ and WX reviewed and made significant revisions to the manuscript. All authors have read and approved the final manuscript.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rong Li and Jiajia Jiang are equal contributors.
Contributor Information
Xu Zhang, Email: xuzhang@ujs.edu.cn.
Wenrong Xu, Email: icls@ujs.edu.cn.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Karimi P, Islami F, Anandasabapathy S, Freedman ND, Kamangar F. Gastric cancer: descriptive epidemiology, risk factors, screening, and prevention. Cancer Epidemiol Biomark. 2014;23(5):700–713. doi: 10.1158/1055-9965.EPI-13-1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Triantafillidis JK, Cheracakis P. Gastric cancer: recent developments in its etiology and pathogenesis. Ann Gastroenterol. 2003;16(1):12–19. [Google Scholar]
- 4.Zhang X, Zhang W, Yuan X, Fu M, Qian H, Xu W. Neutrophils in cancer development and progression: roles, mechanisms, and implications (Review) Int J Oncol. 2016;49(3):857–867. doi: 10.3892/ijo.2016.3616. [DOI] [PubMed] [Google Scholar]
- 5.Wu L, Xu Z, Zhang B, Hui S, Xiao Y, Sun Y, Pan Z, Hui Q, Xu W. Exosomes derived from gastric cancer cells activate NF-κB pathway in macrophages to promote cancer progression. Tumor Biol. 2016;37(9):12169–12180. doi: 10.1007/s13277-016-5071-5. [DOI] [PubMed] [Google Scholar]
- 6.Xue Y, Jing H, Han Z, Ying W, Chong H, Wei L, Shi Y. One cell, multiple roles: contribution of mesenchymal stem cells to tumor development in tumor microenvironment. Cell Biosci. 2013;3(1):5. doi: 10.1186/2045-3701-3-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ajani JA, Bentrem DJ, Besh S, D’Amico TA, Das P, Denlinger C, Fakih MG, Fuchs CS, Gerdes H, Glasgow RE. Gastric cancer, version 2.2013: featured updates to the NCCN Guidelines. J Natl Compr Canc Netw. 2013;11(5):531–546. doi: 10.6004/jnccn.2013.0070. [DOI] [PubMed] [Google Scholar]
- 8.Chiurillo MA. Role of the Wnt/β-catenin pathway in gastric cancer: an in-depth literature review. World J Exp Med. 2015;5(2):84–102. doi: 10.5493/wjem.v5.i2.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sanger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci USA. 1976;73(11):3852–3856. doi: 10.1073/pnas.73.11.3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One. 2012;7(2):e30733. doi: 10.1371/journal.pone.0030733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, Marzluff WF, Sharpless NE. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19(2):141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, Loewer A, Ziebold U, Landthaler M, Kocks C, le Noble F, Rajewsky N. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 13.Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 14.Zhao Y, Alexandrov PN, Jaber V, Lukiw WJ. Deficiency in the ubiquitin conjugating enzyme UBE2A in Alzheimer’s disease (AD) is linked to deficits in a natural circular miRNA-7 sponge (circRNA; ciRS-7) Genes. 2016;7(12):116. doi: 10.3390/genes7120116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu H, Guo S, Li W, Yu P. The circular RNA Cdr1as, via miR-7 and its targets, regulates insulin transcription and secretion in islet cells. Sci Rep. 2015;5(1):12453. doi: 10.1038/srep12453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holdt LM, Stahringer A, Sass K, Pichler G, Kulak NA, Wilfert W, Kohlmaier A, Herbst A, Northoff BH, Nicolaou A. Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat Commun. 2016;7:12429. doi: 10.1038/ncomms12429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang M, Huang N, Yang X, Luo J, Yan S, Xiao F, Chen W, Gao X, Zhao K, Zhou H, Li Z, Ming L, Xie B, Zhang N. A novel protein encoded by the circular form of the SHPRH gene suppresses glioma tumorigenesis. Oncogene. 2018;37(13):1805–1814. doi: 10.1038/s41388-017-0019-9. [DOI] [PubMed] [Google Scholar]
- 18.Guarnerio J, Bezzi M, Jeong JC, Paffenholz SV, Berry K, Naldini MM, Lococo F, Tay Y, Beck AH, Pandolfi PP. Oncogenic role of fusion-circRNAs derived from cancer-associated chromosomal translocations. Cell. 2016;165(2):289–302. doi: 10.1016/j.cell.2016.03.020. [DOI] [PubMed] [Google Scholar]
- 19.Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH, Zhu S, Yang L, Chen LL. Circular intronic long noncoding RNAs. Mol Cell. 2013;51(6):792–806. doi: 10.1016/j.molcel.2013.08.017. [DOI] [PubMed] [Google Scholar]
- 20.Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, Zhong G, Yu B, Hu W, Dai L. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22(3):256–264. doi: 10.1038/nsmb.2959. [DOI] [PubMed] [Google Scholar]
- 21.Zhang XO, Wang HB, Zhang Y, Lu X, Chen LL, Yang L. Complementary sequence-mediated exon circularization. Cell. 2014;159(1):134–147. doi: 10.1016/j.cell.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 22.Liang D, Wilusz JE. Short intronic repeat sequences facilitate circular RNA production. Genes Dev. 2014;28(20):2233–2247. doi: 10.1101/gad.251926.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhipeng L, Filonov GS, Noto JJ, Schmidt CA, Hatkevich TL, Ying W, Jaffrey SR, Gregory AM. Metazoan tRNA introns generate stable circular RNAs in vivo. RNA. 2015;21(9):1554–1565. doi: 10.1261/rna.052944.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N, Kadener S. CircRNA biogenesis competes with pre-mRNA splicing. Mol Cell. 2014;56(1):55–66. doi: 10.1016/j.molcel.2014.08.019. [DOI] [PubMed] [Google Scholar]
- 25.Ivanov A, Memczak S, Wyler E, Torti F, Porath HT, Orejuela MR, Piechotta M, Levanon EY, Landthaler M, Dieterich C, Rajewsky N. Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep. 2015;10(2):170–177. doi: 10.1016/j.celrep.2014.12.019. [DOI] [PubMed] [Google Scholar]
- 26.Conn S, Pillman K, Toubia J, Conn V, Salmanidis M, Phillips C, Roslan S, Schreiber A, Gregory P, Goodall G. The RNA binding protein Quaking regulates formation of circRNAs. Cell. 2015;160(6):1125–1134. doi: 10.1016/j.cell.2015.02.014. [DOI] [PubMed] [Google Scholar]
- 27.Lasda E, Parker R. Circular RNAs: diversity of form and function. RNA. 2014;20(12):1829–1842. doi: 10.1261/rna.047126.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Danan M, Schwartz S, Edelheit S, Sorek R. Transcriptome-wide discovery of circular RNAs in Archaea. Nucleic Acids Res. 2012;40(7):3131–3142. doi: 10.1093/nar/gkr1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ye CY, Chen L, Liu C, Zhu QH, Fan L. Widespread noncoding circular RNAs in plants. New Phytol. 2015;208(1):88–95. doi: 10.1111/nph.13585. [DOI] [PubMed] [Google Scholar]
- 30.Huang C, Liang D, Tatomer DC, Wilusz JE. A length-dependent evolutionarily conserved pathway controls nuclear export of circular RNAs. Genes Dev. 2018;32(9–10):639–644. doi: 10.1101/gad.314856.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang PL, Bao Y, Yee MC, Barrett SP, Hogan GJ, Olsen MN, Dinneny JR, Brown PO, Salzman J. Circular RNA is expressed across the eukaryotic tree of life. PLoS One. 2014;9(6):e90859. doi: 10.1371/journal.pone.0090859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thomas LF. Circular RNAs are depleted of polymorphisms at microRNA binding sites. Bioinformatics. 2014;30(16):2243–2246. doi: 10.1093/bioinformatics/btu257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Suzuki H, Tsukahara T. A view of pre-mRNA splicing from RNase R resistant RNAs. Int J Mol Sci. 2014;15(6):9331–9342. doi: 10.3390/ijms15069331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vicens Quentin, Westhof Eric. Biogenesis of circular RNAs. Cell. 2014;159(1):13–14. doi: 10.1016/j.cell.2014.09.005. [DOI] [PubMed] [Google Scholar]
- 35.Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol. 2014;32(5):453–461. doi: 10.1038/nbt.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hansen TB, Wiklund ED, Bramsen JB, Villadsen SB, Statham AL, Clark SJ, Kjems J. MiRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. EMBO J. 2011;30(21):4414–4422. doi: 10.1038/emboj.2011.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO. Cell-type specific features of circular RNA expression. PLoS Genet. 2013;9(9):e1003777. doi: 10.1371/journal.pgen.1003777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Venø MT, Hansen TB, Venø ST, Clausen BH, Grebing M, Finsen B, Holm IE, Kjems J. Spatio-temporal regulation of circular RNA expression during porcine embryonic brain development. Genome Biol. 2015;16(1):245. doi: 10.1186/s13059-015-0801-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Szabo L, Morey R, Palpant NJ, Wang PL, Afari N, Jiang C, Parast MM, Murry CE, Laurent LC, Salzman J. Statistically based splicing detection reveals neural enrichment and tissue-specific induction of circular RNA during human fetal development. Genome Biol. 2015;16(1):126. doi: 10.1186/s13059-015-0690-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ambros V. The functions of animal microRNAs. Nature. 2004;431(7006):350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 41.Geng HH, Rui L, Su YM, Jie X, Min P, Cai XX, Ji XP. The circular RNA Cdr1as promotes myocardial infarction by mediating the regulation of miR-7a on its target genes expression. PLoS One. 2016;11(3):e0151753. doi: 10.1371/journal.pone.0151753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu L, Gong X, Sun L, Zhou Q, Lu B, Zhu L. The circular RNA Cdr1as act as an oncogene in hepatocellular carcinoma through targeting miR-7 expression. PLoS One. 2016;11(7):e0158347. doi: 10.1371/journal.pone.0158347. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 43.Pan H, Li T, Jiang Y, Pan C, Ding Y, Huang Z, Yu H, Kong D. Overexpression of circular RNA ciRS-7 abrogates the tumor suppressive effect of miR-7 on gastric cancer via PTEN/PI3K/AKT signaling pathway. J Cell Biochem. 2018;119(1):440–446. doi: 10.1002/jcb.26201. [DOI] [PubMed] [Google Scholar]
- 44.Xie H, Ren X, Xin S, Lan X, Lu G, Yuan L, Yang S, Zeng Z, Liao W, Ding YQ. Emerging roles of circRNA_001569 targeting miR-145 in the proliferation and invasion of colorectal cancer. Oncotarget. 2016;7(18):26680–26691. doi: 10.18632/oncotarget.8589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Han D, Li J, Wang H, Su X, Hou J, Gu Y, Qian C, Lin Y, Liu X, Huang M. Circular RNA MTO1 acts as the sponge of miR-9 to suppress hepatocellular carcinoma progression. Hepatology. 2017;66(4):1151–1164. doi: 10.1002/hep.29270. [DOI] [PubMed] [Google Scholar]
- 46.Zhong Z, Lv M, Chen J. Screening differential circular RNA expression profiles reveals the regulatory role of circTCF25-miR-103a-3p/miR-107-CDK6 pathway in bladder carcinoma. Sci Rep. 2016;6:30919. doi: 10.1038/srep30919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ebbesen KK, Kjems J, Hansen TB. Circular RNAs: identification, biogenesis and function. Biochim Biophys Acta 1859. 2016;1:163–168. doi: 10.1016/j.bbagrm.2015.07.007. [DOI] [PubMed] [Google Scholar]
- 48.Chao CW, Chan DC, Kuo A, Leder P. The mouse formin (Fmn) gene: abundant circular RNA transcripts and gene-targeted deletion analysis. Mol Med. 1998;4(9):614–628. [PMC free article] [PubMed] [Google Scholar]
- 49.Gualandi F, Trabanelli C, Rimessi P, Calzolari E, Toffolatti L, Patarnello T, Kunz G, Muntoni F, Ferlini A. Multiple exon skipping and RNA circularisation contribute to the severe phenotypic expression of exon 5 dystrophin deletion. J Med Genet. 2003;40(8):e100. doi: 10.1136/jmg.40.8.e100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang Y, Wang Z. Efficient backsplicing produces translatable circular mRNAs. RNA. 2015;21(2):172–179. doi: 10.1261/rna.048272.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen CY, Sarnow P. Initiation of protein synthesis by the eukaryotic translational apparatus on circular RNAs. Science. 1995;268(5209):415–417. doi: 10.1126/science.7536344. [DOI] [PubMed] [Google Scholar]
- 52.Yang Y, Fan X, Mao M, Song X, Wu P, Zhang Y, Jin Y, Yang Y, Chen L, Wang Y. Extensive translation of circular RNAs driven by N6-methyladenosine. Cell Res. 2017;27(5):626–641. doi: 10.1038/cr.2017.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pamudurti NR, Bartok O, Jens M, Ashwalfluss R, Stottmeister C, Ruhe L, Hanan M, Wyler E, Perezhernandez D, Ramberger E. Translation of circRNAs. Mol Cell. 2017;66(1):9–21. doi: 10.1016/j.molcel.2017.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang M, Huang N, Yang X, Luo J, Yan S, Xiao F, Chen W, Gao X, Zhao K, Zhou H. A novel protein encoded by the circular form of the SHPRH gene suppresses glioma tumorigenesis. Oncogene. 2018;37(13):1805–1814. doi: 10.1038/s41388-017-0019-9. [DOI] [PubMed] [Google Scholar]
- 55.Granados-Riveron JT, Aquino-Jarquin G. The complexity of the translation ability of circRNAs. Biochim Biophys Acta 1859. 2016;10:1245–1251. doi: 10.1016/j.bbagrm.2016.07.009. [DOI] [PubMed] [Google Scholar]
- 56.Du WW, Yang W, Chen Y, Wu ZK, Foster FS, Yang Z, Li X, Yang BB. Foxo3 circular RNA promotes cardiac senescence by modulating multiple factors associated with stress and senescence responses. Eur Heart J. 2016;38(18):1402–1412. doi: 10.1093/eurheartj/ehw001. [DOI] [PubMed] [Google Scholar]
- 57.Xia P, Wang S, Ye B, Du Y, Li C, Xiong Z, Qu Y, Fan Z. A circular RNA protects dormant hematopoietic stem cells from DNA sensor cGAS-mediated exhaustion. Immunity. 2018;48(4):688–701. doi: 10.1016/j.immuni.2018.03.016. [DOI] [PubMed] [Google Scholar]
- 58.Abdelmohsen K, Panda AC, Munk R, Grammatikakis I, Dudekula DB, De S, Kim J, Noh JH, Kim KM, Martindale JL. Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation by CircPABPN1. RNA Biol. 2017;14(3):361–369. doi: 10.1080/15476286.2017.1279788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yang Q, Du WW, Wu N, Yang W, Awan FM, Fang L, Ma J, Li X, Zeng Y, Yang Z, Dong J, Khorshidi A, Yang BB. A circular RNA promotes tumorigenesis by inducing c-myc nuclear translocation. Cell Death Differ. 2017;24(9):1609–1620. doi: 10.1038/cdd.2017.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ghosal S, Das S, Sen R, Basak P, Chakrabarti J. Circ2Traits: a comprehensive database for circular RNA potentially associated with disease and traits. Front Genet. 2013;4:283. doi: 10.3389/fgene.2013.00283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li JH, Liu S, Zhou H, Qu LH, Yang JH. StarBase v20: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42:92–97. doi: 10.1093/nar/gkt1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dudekula DB, Panda AC, Grammatikakis I, De S, Abdelmohsen K, Gorospe M. CircInteractome: a web tool for exploring circular RNAs and their interacting proteins and microRNAs. RNA Biol. 2016;13(1):34–42. doi: 10.1080/15476286.2015.1128065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen X, Ping H, Tao Z, Guo X, Song X, Yan L. CircRNADb: a comprehensive database for human circular RNAs with protein-coding annotations. Sci Rep. 2016;6:34985. doi: 10.1038/srep34985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Xia S, Feng J, Chen K, Ma Y, Gong J, Cai F, Jin Y, Gao Y, Xia L, Chang H. CSCD: a database for cancer-specific circular RNAs. Nucleic Acids Res. 2018;46(D1):D925–D929. doi: 10.1093/nar/gkx863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tang Z, Li X, Zhao J, Qian F, Feng C, Li Y, Zhang J, Jiang Y, Yang Y, Wang Q, Li C. TRCirc: a resource for transcriptional regulation information of circRNAs. Brief Bioinform. 2018 doi: 10.1093/bib/bby083. [DOI] [PubMed] [Google Scholar]
- 66.Glažar P, Papavasileiou P, Rajewsky N. CircBase: a database for circular RNAs. RNA. 2014;20(11):1666–1670. doi: 10.1261/rna.043687.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liu YC, Li JR, Sun CH, Erik A, Chao RF, Lin FM, Weng SL, Sheng-Da H, Huang CC, Chao C. CircNet: a database of circular RNAs derived from transcriptome sequencing data. Nucleic Acids Res. 2016;44(D1):D209–D215. doi: 10.1093/nar/gkv940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zheng LL, Li JH, Jie W, Sun WJ, Liu S, Wang ZL, Hui Z, Yang JH, Qu LH. DeepBase v2.0: identification, expression, evolution and function of small RNAs, LncRNAs and circular RNAs from deep-sequencing data. Nucleic Acids Res. 2016;44(D1):D196–202. doi: 10.1093/nar/gkv1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhao Z, Wang K, Wu F, Wang W, Zhang K, Hu H, Liu Y, Jiang T. CircRNA disease: a manually curated database of experimentally supported circRNA-disease associations. Cell Death Dis. 2018;9(5):475. doi: 10.1038/s41419-018-0503-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dong R, Ma XK, Li GW, Yang L. CIRCpedia v2: an updated database for comprehensive circular RNA annotation and expression comparison. Genomics Proteomics Bioinform. 2018;16(4):226–233. doi: 10.1016/j.gpb.2018.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sui W, Shi Z, Xue W, Ou M, Zhu Y, Chen J, Lin H, Liu F, Dai Y. Circular RNA and gene expression profiles in gastric cancer based on microarray chip technology. Oncol Rep. 2017;37(3):1804–1814. doi: 10.3892/or.2017.5415. [DOI] [PubMed] [Google Scholar]
- 72.Huang YS, Jie N, Zou KJ, Weng Y. Expression profile of circular RNAs in human gastric cancer tissues. Mol Med Rep. 2017;16(3):2469–2476. doi: 10.3892/mmr.2017.6916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fang Y, Ma M, Wang J, Liu X, Wang Y. Circular RNAs play an important role in late-stage gastric cancer: circular RNA expression profiles and bioinformatics analyses. Tumour Biol. 2017;39(6):1010428317705850. doi: 10.1177/1010428317705850. [DOI] [PubMed] [Google Scholar]
- 74.Shao Y, Li J, Lu R, Li T, Yang Y, Xiao B, Guo J. Global circular RNA expression profile of human gastric cancer and its clinical significance. Cancer Med. 2017;6(6):1173–1180. doi: 10.1002/cam4.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chen J, Li Y, Zheng Q, Bao C, He J, Chen B, Lyu D, Zheng B, Xu Y, Long Z, Zhou Y, Zhu H, Wang Y, He X, Shi Y, Huang S. Circular RNA profile identifies circPVT1 as a proliferative factor and prognostic marker in gastric cancer. Cancer Lett. 2017;388:208–219. doi: 10.1016/j.canlet.2016.12.006. [DOI] [PubMed] [Google Scholar]
- 76.Vidal AF, Ribeirodossantos AM, Vinascosandoval T, Magalhães L, Pinto P, Anaissi AKM, Demachki S, Assumpção PPD, Santos SEBD, RibeirodosSantos Â. The comprehensive expression analysis of circular RNAs in gastric cancer and its association with field cancerization. Sci Rep. 2017;7(1):14551. doi: 10.1038/s41598-017-15061-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dang Y, Ouyang X, Zhang F, Wang K, Lin Y, Sun B, Wang Y, Wang L, Huang Q. Circular RNAs expression profiles in human gastric cancer. Sci Rep. 2017;7(1):9060. doi: 10.1038/s41598-017-09076-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li T, Shao Y, Fu L, Yi X, Zhu L, Sun W, Rui Y, Xiao B, Guo J. Plasma circular RNA profiling of patients with gastric cancer and their droplet digital RT-PCR detection. J Mol Med. 2018;96(1):85–96. doi: 10.1007/s00109-017-1600-y. [DOI] [PubMed] [Google Scholar]
- 79.Gu W, Sun Y, Zheng X, Ma J, Hu XY, Gao T, Hu MJ. Identification of gastric cancer-related circular RNA through microarray analysis and bioinformatics analysis. Biomed Res Int. 2018 doi: 10.1155/2018/2381680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pan H, Li T, Jiang Y, Pan C, Ding Y, Huang Z, Yu H, Kong D. Overexpression of circular RNA ciRS-7 abrogates the tumor suppressive effect of miR-7 on Gastric cancer via PTEN/PI3 K/AKT signaling pathway. J Cell Biochem. 2017;119(1):440–446. doi: 10.1002/jcb.26201. [DOI] [PubMed] [Google Scholar]
- 81.Pérezramírez C, Cañadasgarre M, Molina MÁ, Fausdáder MJ, Callejahernández MÁ. PTEN and PI3 K/AKT in non-small-cell lung cancer. Pharmacogenomics. 2015;16(16):1843–1862. doi: 10.2217/pgs.15.122. [DOI] [PubMed] [Google Scholar]
- 82.Wang F, Li L, Chen Z, Zhu M, Gu Y. MicroRNA-214 acts as a potential oncogene in breast cancer by targeting the PTEN-PI3K/Akt signaling pathway. Int J Mol Med. 2016;37(5):1421–1428. doi: 10.3892/ijmm.2016.2518. [DOI] [PubMed] [Google Scholar]
- 83.Zheng L, Zhang Y, Liu Y, Zhou M, Lu Y, Yuan L, Zhang C, Hong M, Wang S, Li X. MiR-106b induces cell radioresistance via the PTEN/PI3K/AKT pathways and p21 in colorectal cancer. J Transl Med. 2015;13:252. doi: 10.1186/s12967-015-0592-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cheng J, Zhuo H, Xu M, Wang L, Xu H, Peng J, Hou J, Lin L, Cai J. Regulatory network of circRNA–miRNA–mRNA contributes to the histological classification and disease progression in gastric cancer. J Transl Med. 2018;16(1):216. doi: 10.1186/s12967-018-1582-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Shen F, Liu P, Li N, Yi Z, Tie X, Zhang Y, Gao L, Xu Z. CircRNA_001569 promotes cell proliferation through absorbing miR-145 in gastric cancer. J Biochem. 2018;165(1):27–36. doi: 10.1093/jb/mvy079. [DOI] [PubMed] [Google Scholar]
- 86.Xie H, Ren X, Xin S, Lan X, Lu G, Lin Y, Yang S, Zeng Z, Liao W, Ding YQ. Emerging roles of circRNA_001569 targeting miR-145 in the proliferation and invasion of colorectal cancer. Oncotarget. 2016;7(18):26680–26691. doi: 10.18632/oncotarget.8589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ouyang Y, Li Y, Huang Y, Li X, Zhu Y, Long Y, Wang Y, Guo X, Gong K. CircRNA circPDSS1 promotes the gastric cancer progression by sponging miR-186-5p and modulating NEK2. J Cell Physiol. 2019;234(7):10458–10469. doi: 10.1002/jcp.27714. [DOI] [PubMed] [Google Scholar]
- 88.Hayward DG, Fry AM. Nek2 kinase in chromosome instability and cancer. Cancer Lett. 2006;237(2):155–166. doi: 10.1016/j.canlet.2005.06.017. [DOI] [PubMed] [Google Scholar]
- 89.Sun H, Xi P, Sun Z, Wang Q, Zhu B, Zhou J, Jin H, Zheng W, Tang W, Cao H, Cao X. Circ-SFMBT2 promotes the proliferation of gastric cancer cells through sponging miR-182-5p to enhance CREB1 expression. Cancer Manag Res. 2018;10:5725–5734. doi: 10.2147/CMAR.S172592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wang Z, Ma K, Pitts S, Cheng Y, Liu X, Ke X, Kovaka S, Ashktorab H, Smoot DT, Schatz M, Wang Z, Meltzer S. Novel circular RNA NF1 acts as a molecular sponge, promoting gastric cancer by absorbing miR-16. Endocr Relat Cancer. 2018;26(3):265–277. doi: 10.1530/ERC-18-0478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhang Y, Liu H, Li W, Yu J, Li J, Shen Z, Ye G, Qi X, Li G. CircRNA_100269 is downregulated in gastric cancer and suppresses tumor cell growth by targeting miR-630. Aging (Albany NY) 2017;9(6):1585–1594. doi: 10.18632/aging.101254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhang S, Zhang JY, Lu LJ, Wang CH, Wang LH. MiR-630 promotes epithelial ovarian cancer proliferation and invasion via targeting KLF6. Eur Rev Med Pharmacol Sci. 2017;21(20):4542–4547. [PubMed] [Google Scholar]
- 93.Li J, Zhen L, Zhang Y, Zhao L, Liu H, Cai D, Chen H, Yu J, Qi X, Li G. Circ-104916 is downregulated in gastric cancer and suppresses migration and invasion of gastric cancer cells. Onco Targets Ther. 2017;10:3521–3529. doi: 10.2147/OTT.S136347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhao JJ, Chen PJ, Duan RQ, Li KJ, Wang YZ, Li Y. miR-630 functions as a tumor oncogene in renal cell carcinoma. Arch of Med Sci. 2016;12(3):473–478. doi: 10.5114/aoms.2016.59918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhang JW, Li Y, Zeng XC, Zhang T, Fu BS, Yi HM, Zhang Q, Jiang N. MiR-630 overexpression in hepatocellular carcinoma tissues is positively correlated with alpha-fetoprotein. Med Sci Monit. 2015;21:667–673. doi: 10.12659/MSM.892515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Liu H, Liu Y, Bian Z, Zhang J, Zhang R, Chen X, Huang Y, Wang Y, Zhu J. Circular RNA YAP1 inhibits the proliferation and invasion of gastric cancer cells by regulating the miR-367-5p/p27 Kip1 axis. Mol Cancer. 2018;17(1):151. doi: 10.1186/s12943-018-0902-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kang W, Tong JH, Lung RW, Dong Y, Zhao J, Liang Q, Zhang L, Pan Y, Yang W, Pang JC. Targeting of YAP1 by microRNA-15a and microRNA-16-1 exerts tumor suppressor function in gastric adenocarcinoma. Mol Cancer. 2015;14(1):52. doi: 10.1186/s12943-015-0323-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhang J, Liu H, Hou L, Wang G, Zhang R, Huang Y, Chen X, Zhu J. Circular RNA_LARP4 inhibits cell proliferation and invasion of gastric cancer by sponging miR-424-5p and regulating LATS1 expression. Mol Cancer. 2017;16(1):151. doi: 10.1186/s12943-017-0719-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yimlamai D, Fowl BH, Camargo FD. Emerging evidence on the role of the Hippo/YAP pathway in liver physiology and cancer. J Hepatol. 2015;63(6):1491–1501. doi: 10.1016/j.jhep.2015.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhou GX, Li XY, Zhang Q, Zhao K, Zhang CP, Xue CH, Yang K, Tian ZB. Effects of the hippo signaling pathway in human gastric cancer. Asian Pac J Cancer Prev. 2013;14(9):5199–5205. doi: 10.7314/apjcp.2013.14.9.5199. [DOI] [PubMed] [Google Scholar]
- 101.Liu T, Liu S, Xu Y, Shu R, Wang F, Chen C, Zeng Y, Luo H. Circular RNA-ZFR inhibited cell proliferation and promoted apoptosis in gastric cancer by sponging miR-130a/miR-107 and modulating PTEN. Cancer Res Treat. 2018;50(4):1396–1417. doi: 10.4143/crt.2017.537. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 102.Fang J, Hong H, Xue X, Zhu X, Jiang L, Qin M, Liang H, Gao L. A novel circular RNA, circFAT1(e2), inhibits gastric cancer progression by targeting miR-548 g in the cytoplasm and interacting with YBX1 in the nucleus. Cancer Lett. 2019;442:222–232. doi: 10.1016/j.canlet.2018.10.040. [DOI] [PubMed] [Google Scholar]
- 103.Prabhu L, Hartley A-V, Martin M, Warsame F, Sun E, Lu T. Role of post-translational modification of the Y box binding protein 1 in human cancers. Genes Dis. 2015;2(3):240–246. doi: 10.1016/j.gendis.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wang L, Shen J, Jiang Y. Circ_0027599/PHDLA1 suppresses gastric cancer progression by sponging miR-101-3p.1. Cell Biosci. 2018;8(1):58. doi: 10.1186/s13578-018-0252-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Li P, Chen H, Chen S, Mo X, Li T, Xiao B, Rui Y, Guo J. Circular RNA 0000096 affects cell growth and migration in gastric cancer. Br J Cancer. 2017;116(5):626–633. doi: 10.1038/bjc.2016.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zhou LH, Yang YC, Zhang RY, Wang P, Pang MH, Liang LQ. CircRNA_0023642 promotes migration and invasion of gastric cancer cells by regulating EMT. Eur Rev Med Pharmacol Sci. 2018;22(8):2297–2303. doi: 10.26355/eurrev_201804_14818. [DOI] [PubMed] [Google Scholar]
- 107.Sun HD, Xu ZP, Sun ZQ, Zhu B, Wang Q, Zhou J, Jin H, Zhao A, Tang WW, Cao XF. Down-regulation of circPVRL3 promotes the proliferation and migration of gastric cancer cells. Sci Rep. 2018;8(1):10111. doi: 10.1038/s41598-018-27837-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lai Z, Yang Y, Yan Y, Li T, Li YS, Wang Z, Shen Z, Ye YJ, Jiang KW, Wang S. Analysis of co-expression networks for circular RNAs and mRNAs reveals that circular RNAs hsa_circ_0047905, hsa_circ_0138960 and has-circRNA7690-15 are candidate oncogenes in gastric cancer. Cell Cycle. 2017;16(23):2301–2311. doi: 10.1080/15384101.2017.1380135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Memczak S, Papavasileiou P, Peters O, Rajewsky N. Identification and characterization of circular RNAs as a new class of putative biomarkers in human blood. PLoS One. 2015;10(10):e0141214. doi: 10.1371/journal.pone.0141214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lin X, Lo HC, Wong DTW, Xiao X. Noncoding RNAs in human saliva as potential disease biomarkers. Front Genet. 2015;6:175. doi: 10.3389/fgene.2015.00175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J, Chen D, Gu J, He X, Huang S. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 2015;25(8):981–984. doi: 10.1038/cr.2015.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Sun H, Tang W, Rong D, Jin H, Fu K, Zhang W, Liu Z, Cao H, Cao X. Hsa_circ_0000520, a potential new circular RNA biomarker, is involved in gastric carcinoma. Cancer Biomark. 2018;21(2):299–306. doi: 10.3233/CBM-170379. [DOI] [PubMed] [Google Scholar]
- 113.Chen S, Li T, Zhao Q, Xiao B, Guo J. Using circular RNA hsa_circ_0000190 as a new biomarker in the diagnosis of gastric cancer. Clin Chim Acta. 2017;466:167–171. doi: 10.1016/j.cca.2017.01.025. [DOI] [PubMed] [Google Scholar]
- 114.Huang M, He YR, Liang LC, Huang Q, Zhu ZQ. Circular RNA hsa_circ_0000745 may serve as a diagnostic marker for gastric cancer. World J Gastroenterol. 2017;23(34):6330–6338. doi: 10.3748/wjg.v23.i34.6330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lasda E, Parker R. Circular RNAs co-precipitate with extracellular vesicles: a possible mechanism for circRNA clearance. PLoS One. 2016;11(2):e0148407. doi: 10.1371/journal.pone.0148407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Piwecka M, Glažar P, Hernandez-Miranda LR, Memczak S, Wolf SA, Rybak-Wolf A, Filipchyk A, Klironomos F, Cerda Jara CA, Fenske P. Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science. 2017 doi: 10.1126/science.aam8526. [DOI] [PubMed] [Google Scholar]
- 117.Yao Z, Luo J, Hu K, Lin J, Huang H, Wang Q, Zhang P, Xiong Z, He C, Huang Z. ZKSCAN1 gene and its related circular RNA (circZKSCAN1) both inhibit hepatocellular carcinoma cell growth, migration, and invasion but through different signaling pathways. Mol Oncol. 2017;11(4):422–437. doi: 10.1002/1878-0261.12045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Liu X, Abraham JM, Cheng Y, Wang Z, Wang Z, Zhang G, Ashktorab H, Smoot DT, Cole RN, Boronina TN, DeVine LR, Talbot CC, Jr, Liu Z, Meltzer SJ. Synthetic circular RNA functions as a miR-21 sponge to suppress gastric carcinoma cell proliferation. Mol Ther Nucleic Acids. 2018;13:312–321. doi: 10.1016/j.omtn.2018.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Qin J, Xu Q. Functions and applications of exosomes. Acta Pol Pharm. 2014;71(4):537–543. [PubMed] [Google Scholar]
- 120.Kumar L, Verma S, Vaidya B, Gupta V. Exosomes: natural carriers for siRNA delivery. Curr Pharm Des. 2015;21(31):4556–4565. doi: 10.2174/138161282131151013190112. [DOI] [PubMed] [Google Scholar]
- 121.Tian M, Chen R, Li T, Xiao B. Reduced expression of circRNA hsa_circ_0003159 in gastric cancer and its clinical significance. J Clin Lab Anal. 2018;32(3):e22281. doi: 10.1002/jcla.22281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Xie Y, Shao Y, Sun W, Ye G, Zhang X, Xiao B, Guo J. Downregulated expression of hsa_circ_0074362 in gastric cancer and its potential diagnostic values. Biomark Med. 2018;12(1):11–20. doi: 10.2217/bmm-2017-0114. [DOI] [PubMed] [Google Scholar]
- 123.Shao Y, Chen L, Lu R, Zhang X, Xiao B, Ye G, Guo J. Decreased expression of hsa_circ_0001895 in human gastric cancer and its clinical significances. Tumour Biol. 2017;39(4):1010428317699125. doi: 10.1177/1010428317699125. [DOI] [PubMed] [Google Scholar]
- 124.Shao Y, Yang Y, Lu R, Xiao B, Ye G, Guo J. Identification of tissue-specific circRNA hsa_circ_0000705 as an indicator for human gastric cancer. Int J Clin Exp Pathol. 2017;10(3):3151–3156. [Google Scholar]
- 125.Lu R, Shao Y, Ye G, Xiao B, Guo J. Low expression of hsa_circ_0006633 in human gastric cancer and its clinical significances. Tumour Biol. 2017;39(6):1010428317704175. doi: 10.1177/1010428317704175. [DOI] [PubMed] [Google Scholar]
- 126.Li P, Chen S, Chen H, Mo X, Li T, Shao Y, Xiao B, Guo J. Using circular RNA as a novel type of biomarker in the screening of gastric cancer. Clin Chim Acta. 2015;444:132–136. doi: 10.1016/j.cca.2015.02.018. [DOI] [PubMed] [Google Scholar]
- 127.Li WH, Song YC, Zhang H, Zhou ZJ, Xie X, Zeng QN, Guo K, Wang T, Xia P, Chang DM. Decreased expression of hsa_circ_00001649 in gastric cancer and its clinical significance. Dis Markers. 2017;2017:4587698. doi: 10.1155/2017/4587698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Tang W, Fu K, Sun H, Rong D, Wang H, Cao H. CircRNA microarray profiling identifies a novel circulating biomarker for detection of gastric cancer. Mol Cancer. 2018;17(1):137. doi: 10.1186/s12943-018-0888-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Zhao Q, Chen S, Li T, Xiao B, Zhang X. Clinical values of circular RNA 0000181 in the screening of gastric cancer. J Clin Lab Anal. 2018;32(4):e22333. doi: 10.1002/jcla.22333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lee K, Hwang H, Nam KT. Immune response and the tumor microenvironment: how they communicate to regulate gastric cancer. Gut Liver. 2014;8(2):131–139. doi: 10.5009/gnl.2014.8.2.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Wu X, Tao P, Zhou Q, Li J, Yu Z, Wang X, Li J, Li C, Yan M, Zhu Z, Liu B, Su L. IL-6 secreted by cancer-associated fibroblasts promotes epithelial-mesenchymal transition and metastasis of gastric cancer via JAK2/STAT3 signaling pathway. Oncotarget. 2017;8(13):20741–20750. doi: 10.18632/oncotarget.15119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Zhang H, Zhu L, Bai M, Liu Y, Zhan Y, Deng T, Yang H, Sun W, Wang X, Zhu K, Fan Q, Li J, Ying G, Ba Y. Exosomal circRNA derived from gastric tumor promotes white adipose browning by targeting the miR-133/PRDM16 pathway. Int J Cancer. 2019;144(10):2501–2515. doi: 10.1002/ijc.31977. [DOI] [PubMed] [Google Scholar]
- 133.Bär C, Thum T, Gonzalo-Calvo D. Circulating miRNAs as mediators in cell-to-cell communication. Epigenomics. 2019;11(2):111–113. doi: 10.2217/epi-2018-0183. [DOI] [PubMed] [Google Scholar]
- 134.Kim KM, Abdelmohsen K, Mustapic M, Kapogiannis D, Gorospe M. RNA in extracellular vesicles. Wiley Interdiscip Rev RNA. 2017;8(4):e1413. doi: 10.1002/wrna.1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Li Z, Yanfang W, Li J, Jiang P, Peng T, Chen K, Zhao X, Zhang Y, Zhen P, Zhu J, Li X. Tumor-released exosomal circular RNA PDE8A promotes invasive growth via the miR-338/MACC1/MET pathway in pancreatic cancer. Cancer Lett. 2018;432:237–250. doi: 10.1016/j.canlet.2018.04.035. [DOI] [PubMed] [Google Scholar]