Abstract
The pre-initiation complex (pre-IC) has been proposed for two decades as an intermediate right before the maturation of the eukaryotic DNA replication fork. However, its existence and biochemical nature remain enigmatic. Here, through combining several enrichment strategies, we are able to isolate an endogenous dimeric CMG-containing complex (designated as d-CMG) distinct from traditional single CMG (s-CMG) and in vitro reconstituted dimeric CMG. D-CMG is assembled upon entry into the S phase and shortly matures into s-CMG/replisome, leading to the fact that only ~ 5% of the total CMG-containing complexes can be detected as d-CMG in vivo. Mass spectra reveal that RPA and DNA Pol α/primase co-purify with s-CMG, but not with d-CMG. Consistently, the former fraction is able to catalyze DNA unwinding and de novo synthesis, while the latter catalyzes neither. The two CMGs in d-CMG display flexibly orientated conformations under an electronic microscope. When DNA Pol α-primase is inactivated, d-CMG % rose up to 29%, indicating an incomplete pre-IC/fork transition. These findings reveal biochemical properties of the d-CMG/pre-IC and provide in vivo evidence to support the pre-IC/fork transition as a bona fide step in replication initiation.
Electronic supplementary material
The online version of this article (10.1007/s00018-019-03333-9) contains supplementary material, which is available to authorized users.
Keywords: DNA helicase, Intermediate, Chromatin, Supercomplex
Introduction
Eukaryotic cells exploit multilevel mechanisms to strictly control the initiation of DNA replication, to achieve proper transmission of their genomes during cell proliferation. As an engine of the replication machinery for all eukaryotes, Mcm2-7 (MCM) comprises the core of replicative helicase for unwinding the duplex genome [1, 2]. MCM is loaded onto the double-stranded origin DNA as a catalytically inactive, head-to-head double hexamer (DH) called pre-replication complex (pre-RC) in G1 phase [3–6]. Two co-activators, Cdc45 and the GINS heterotetramer (go ichi ni san, composed of Sld5, Psf1, Psf2 and Psf3), have been demonstrated to be essential to convert the DH into two single CMGs (s-CMG, Cdc45-MCM-GINS), which operate as an 11-subunit holo-helicase moving along the leading strand during S phase [7–14].
As cells proceed to S phase, the Dbf4-dependent Cdc7 protein kinase DDK phosphorylates the N-terminal tails of Mcm2/4/6 [15, 16], triggering their interaction with Sld3-Cdc45 and the assembly of the Cdc45-MCM-Sld3 (CMS) platform [17–20]. Then, Sld2 and Sld3 are phosphorylated by S-phase cyclin-dependent kinase (S-CDK), which promotes the formation and recruitment of the Sld2-Dpb11-Pol ε-GINS complex [18, 21]. It is conceivable that this step results in the replacement of Sld3 by GINS. Although these highly orchestrated events are believed to lead to CMG formation, a series of recent studies have demonstrated that the initial Cdc45 and GINS association is not sufficient for complete CMG maturation [22–25]. A so-called pre-initiation complex (pre-IC) has been proposed for over two decades to define the intermediate before replication initiation [26]. Using chromatin immunoprecipitation, Miyazawa-Onami et al. recently showed that pre-IC might exist in yeast cells [24], though it remains to be biochemically characterized.
In this study, through expanded tandem affinity purification approaches and glycerol sedimentation-velocity gradient centrifugation, we have isolated a dimeric CMG complex (d-CMG) that differs from the well-characterized s-CMG in the S phase yeast cells. Among all CMG-containing complexes, about 5% are detected to contain two copies of all Cdc45, MCM and GINS subunits (d-CMGs), while the rest are s-CMGs. Unexpectedly, a small fraction of Cdc45 and GINS exist as homodimers before being recruited onto the MCM DH, which might result in the assembly of d-CMG/pre-IC. With S phase progression, d-CMG segregates gradually and this, in turn, leads to the appearance of s-CMG/replisome. Revealed by mass spectrometry, the sequential changes of the components of these complexes are as follows: the d-CMG/pre-IC fractions do not contain RPA or DNA Pol α-primase, which co-purify in the s-CMG/replisome fractions exclusively; in contrast, both fractions have DNA Pol ε and Tof1/Mrc1/Csm3. Under the single-particle electron microscope (EM), our endogenous d-CMG fractions display a very different spectrum of conformations compared to the previously reported fly dimeric CMG complexes prepared by baculovirus-mediated co-expression of recombinant Cdc45, four GINS and six MCM subunits [22]. More importantly, the d-CMG/s-CMG transition is significantly affected in the absence of primase. These data suggest the pre-IC/fork transition as an essential step during eukaryotic replication firing.
Results
Identification of a new CMG-containing complex containing two copies of MCM
Pre-IC has been presumed to contain the same subunits as CMG. As a transient state between the MCM DH/pre-RC and CMG/replisome, we speculated whether pre-IC still contains two copies of MCM. Based on these hypotheses, we set out to test if there are any CMG-containing complexes with two MCMs in vivo.
To this end, an extra copy of MCM4 with a 5FLAG epitope under the control of its native promoter in a pRS317 plasmid was introduced into a yeast strain whose endogenous copy of MCM4 was tagged with a calmodulin-binding protein (CBP). This allowed isolation of a dimeric species of MCM through tandem affinity purification via calmodulin and anti-FLAG beads. The proteins eluted after each purification step were analyzed by western blotting. Little Cdc45 and Psf2 (a subunit of GINS) were detectable in the MCM DH complex, consistent with two previous independent reports (data not shown) [8, 24].
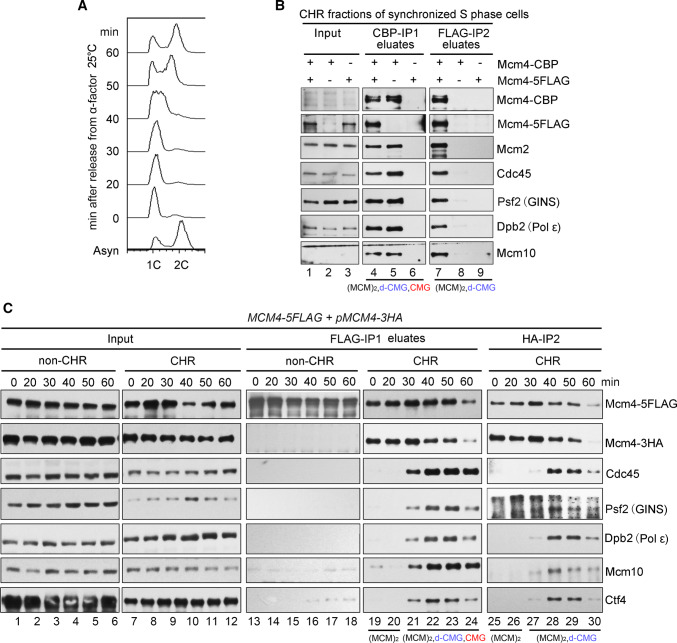
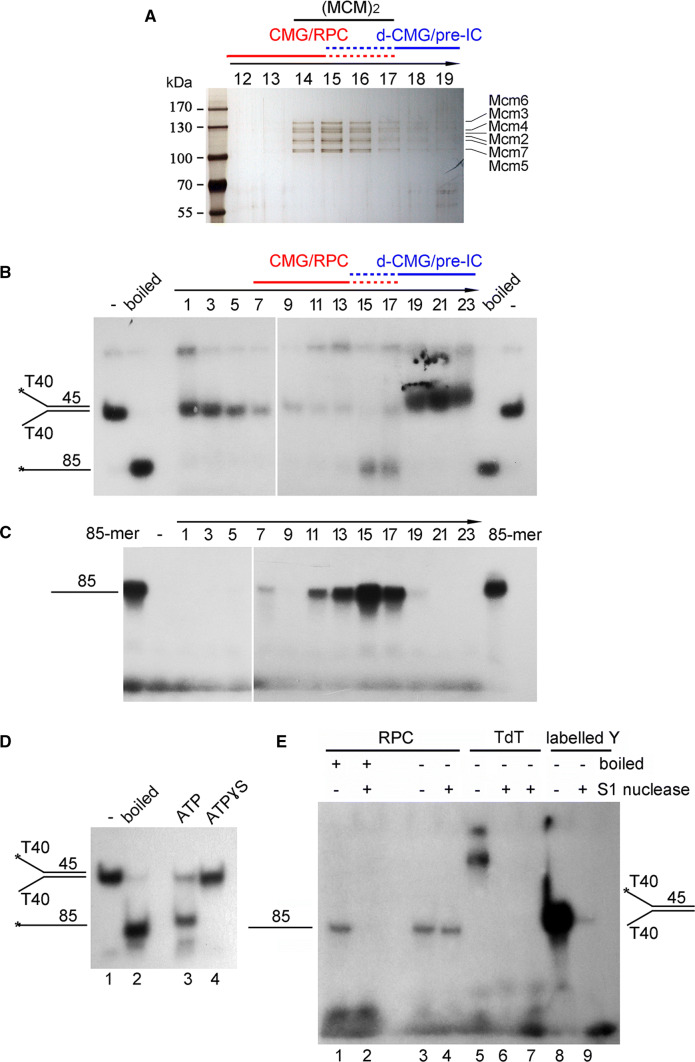
We speculated that the repeated failures to capture the pre-IC might be due to its intrinsic transient/unstable manner compared with the relatively stable MCM DH and CMG complexes. To overcome this difficulty, we made three major modifications to the purification procedure. First, we grew the yeast cells in dropout media at 25 °C to slow down the cell cycle progression. Second, the S phase cells were enriched through α-factor arrest and release for 40 min (Fig. 1a). Third, since pre-IC is assembled exclusively on chromatin, cellular proteins were fractionated into chromatin-bound (CHR) and non-chromatin-bound (non-CHR) ones as described previously [23]. The purification was usually carried out on CHR fractions unless otherwise specified. By combining all these strategies, after the first immunoprecipitation (Mcm4-CBP IP1) and ethylene glycol tetraacetic acid (EGTA) elution, we observed the co-purification of Mcm4-5FLAG, Cdc45 and Psf2 (GINS) with Mcm4-CBP (Fig. 1b, lane 4). This could be due to the co-existence of the MCM DH and CMG complexes. To further separate these complexes, we applied the CBP IP1 eluates (lanes 4–6) to the second IP (Mcm4-5FLAG-IP2), which allowed us to detect an endogenous dimeric MCM complex containing both Cdc45 and Psf2 (GINS) (Fig. 1b, lane 7). It was unlikely for the nonspecific association to occur under our experimental conditions since no protein is detected in the eluates of the controls harboring only one of the epitope tags on MCM4 (lanes 6, 8 and 9), e.g., excluding the putative contaminations like s-CMG (as in lanes 4 and 5). These results suggest the existence of a putative novel class of the CMG-containing complex in vivo which harbors two copies of MCM. We designated this complex as “dimeric CMG” (d-CMG) to distinguish it from CMG (single CMG, s-CMG) that has been well characterized to contain only one copy of each Cdc45, MCM and GINS subunit.
Fig. 1.
Identification of a CMG-containing complex harboring two MCMs. a A representative cell cycle profile of the samples used for protein complex purification. Cells (Strain LL45-1, Table S1) were grown in dropout media at 25 °C and synchronized by 7.5 μg/ml α factor. G1 arrested cells were released and continued growth for the indicated time. Cell cycle profiles were analyzed by flow cytometry. b The MCM4-CBP/pMCM4-5FLAG (Strain LL94-1, Table S1) cells were grown, synchronized in G1 by α-factor, and released into S phase at 25 °C for 40 min. The chromatin-bound protein fraction (CHR) was prepared and subjected to tandem affinity purification via calmodulin and anti-FLAG M2 resins. After three washes, the bound fractions were eluted from beads by 3 mM EGTA (labeled as CBP-IP1 eluates) and 2 mg/ml FLAG peptides (labeled as FLAG-IP2 eluates), respectively. The eluted samples were resolved on an SDS-polyacrylamide gel (PAGE) and detected via immunoblots using the indicated antibodies. Strains (LL94-2 and LL6-1, Table S1) harboring a single tag (either CBP or 5FLAG) on Mcm4 were applied as controls. The species of MCM-containing complexes obtained after each purification step are indicated for clarity. c The MCM4-5FLAG/pMCM4-3HA (LL45-1, Table S1) cells were grown, synchronized in G1 by α-factor (0 min) and released into S phase at 25 °C for the indicated time. Spheroplasts were fractionated into the non-chromatin-bound (non-CHR) and chromatin-bound (CHR) protein fractions. Mcm4-5FLAG and then Mcm4-3HA were precipitated consecutively in a similar procedure mentioned above. After three washes, the proteins specifically associated with beads were eluted by 2 mg/ml of FLAG peptide or boiled directly (for HA-IP) before western blotting. The species of MCM-containing complexes obtained after each purification step are indicated for clarity
Assembly and segregation of d-CMG during S phase
To test if d-CMG may be an intermediate between DH and s-CMG, we next examined the dynamic changes of these MCM-containing complexes throughout the cell cycle. Yeast cells were synchronized in G1 (0 min) by α-factor or released into S phase for 20, 30, 40, 50 or 60 min at 25 °C (Fig. 1c). We isolated the MCM-containing complexes from both CHR and non-CHR fractions of each sample. This time, the dimeric form of MCM was obtained by using the second set of affinity tags (FLAG and HA). MCM DHs, i.e., double-labeled FLAG/HA Mcm2-7 complexes, were detected exclusively in the chromatin fraction (Fig. 1c, lanes 19–24). No intermolecular MCM–MCM interaction and firing factors had ever been detected in the non-CHR fraction [23] (Fig. 1c, lanes 13–18), further corroborating the specificity of the tandem affinity purification used here. Meanwhile, the MCM DH already appeared in G1 phase, though no additional proteins were detected at this time point. After release into S phase for about 30 min, firing factors including Dpb2 (a subunit of DNA Pol ε), Cdc45 and Psf2 were first detected in the chromatin-associated MCM complexes. Next, to separate d-CMG from s-CMG, we then applied the FLAG peptide eluates (Fig. 1c, lanes 19–24) comprising all kinds of MCM-containing complexes (DH, d-CMG and s-CMG) to the second IP (Mcm4-3HA IP2). Before 30 min, MCM existed as the DH on chromatin (Fig. 1c, lanes 25–26). D-CMG appeared at 30 min and peaked at ~ 40 min, coincident with a decline in the MCM DH level (Fig. 1c, lanes 25–30). These results imply the remodeling of the MCM complexes during S phase.
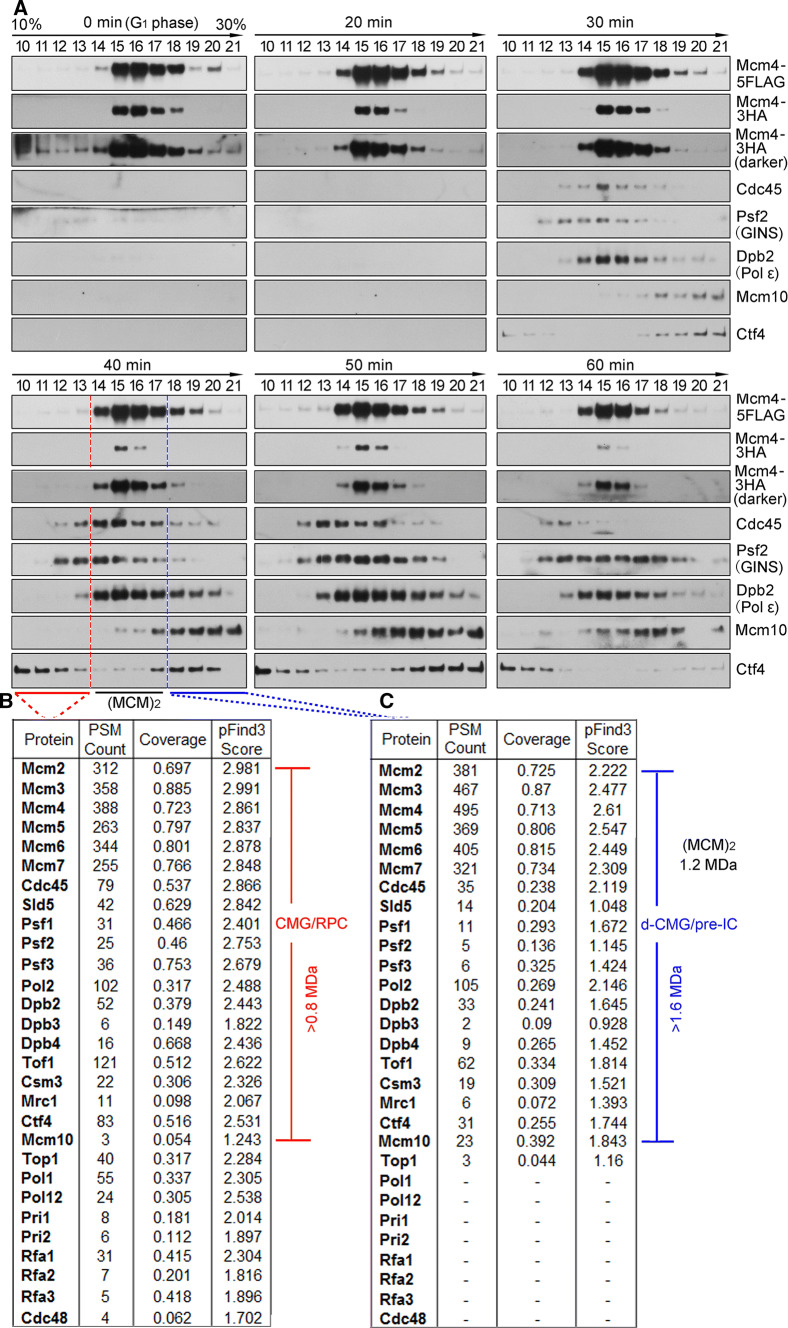
To further validate and characterize the different species of the MCM-containing complexes during the cell cycle, we next subjected the FLAG peptide eluates from the first IP of the CHR fractions (Fig. 1c, lanes 19–24) on a 10–30% glycerol sedimentation/velocity gradient. In G1 phase, only the MCM DH, peaking at fractions 15–17, was detected (Fig. 2a, 0 min, panel 1). These fractions sedimented more rapidly than a 669 kDa protein standard (fraction 13), identifying it as a double hexameric MCM (theoretically 1211 kDa), as shown previously [23]. 30 min after cells entered S phase; the MCM-containing complexes appeared to co-sediment with Cdc45 and GINS (Fig. 2a, panel 3). Although the separation was not complete, it appeared that there are at least two distinct populations of the CMG-containing complexes, one population migrating more slowly than the MCM DH (panel 4, fractions 10–13, red underline) and the other migrating faster (panel 4, fractions 18–21, blue underline). Moreover, Mcm10, an essential initiation factor known to bind the DH preferentially [23, 27], was primarily enriched in the MCM DH (fractions 14–17) and higher density gradients (fractions 18–21) as well. These results imply that the fast-sedimenting MCM complex may be the dimeric species of CMG (theoretically > 1.6 MDa).
Fig. 2.
Dynamic changes of the MCM-containing complexes throughout the cell cycle. a The MCM4-5FLAG/pMCM4-3HA (LL45-1, Table S1) cells were synchronized and collected as described in Fig. 1c. The disparate forms of the MCM-containing complexes were isolated from CHR fractions via one-step purification (i.e., FLAG-IP and FLAG peptide elution) followed by 10–30% glycerol density gradient centrifugation. After centrifugation at 79,000g for 16 h, the total 4.8 ml sample was equally divided into 24 fractions (1–24, from top to bottom). 25 μl of each fraction (only fractions 10–21 are shown for simplicity) was analyzed by immunoblotting. The fraction number is indicated above each lane. Thyroglobulin (669 kDa) migrates at fraction 13 under this condition. Theoretical molecular weights of the complexes are calculated for the core components only: s-CMG (0.8 MDa), MCM DH (1.2 MDa) and d-CMG (1.6 MDa). The associating components such as DNA Pols, Mcm10, Mrc1/Tof1/Csm3 and Ctf4 are not included. b, c Mass spectra of the slow- and fast- sedimenting fractions. Fractions 10–13 (b) and 18–21 (c) were pooled before precipitating the proteins for LC–MS/MS analysis. The total number of identified peptides, coverage and score calculated by a newly developed unrestricted search engine pFind3 are summarized
To further test this interpretation, we then determined the composition of these two CMG-containing complexes by mass spectrometry. The slow-sedimenting fractions (e.g., 10–13) and fast-sedimenting fractions (e.g., 18–21) from the S-phase cells (40 min samples) were pooled separately prior to trypsin digestion. Apart from the essential initiation factors [28], other replication progression factors including the fork protection complex (Tof1-Mrc1-Csm3) required for efficient DNA replication were detected at 40 min, i.e., S phase, in both CMG-containing complexes. Strikingly, substantial RPA (Rfa1-Rfa2-Rfa3), DNA Pol α and primase (Pri1 and Pri2) were detected in the slow-sedimenting complex (Fig. 2b), but little in the fast-sedimenting one (Fig. 2c). The components identified in the slow-sedimenting complex were nearly the same as previous systematic mass spectra of the replication progression complex (RPC) and its associated factors [8]. Given that the loading of RPA and Pol α/primase requires unwound, single-stranded DNA, we infer from these results that the slow-sedimenting and fast-sedimenting species of the S-phase-specific CMG-containing complexes represent the active s-CMG/replisome and inactive d-CMG, respectively. Taken together, these data suggest that the MCM DH is initially assembled into a dimeric form of CMG before its transition into two monomeric active s-CMGs associated with additional fork progression proteins. Therefore, the d-CMG complex may represent the intermediate between the pre-RC and replisome, i.e., meet the definition of the pre-IC.
Pre-IC also contains two copies of Cdc45 and GINS
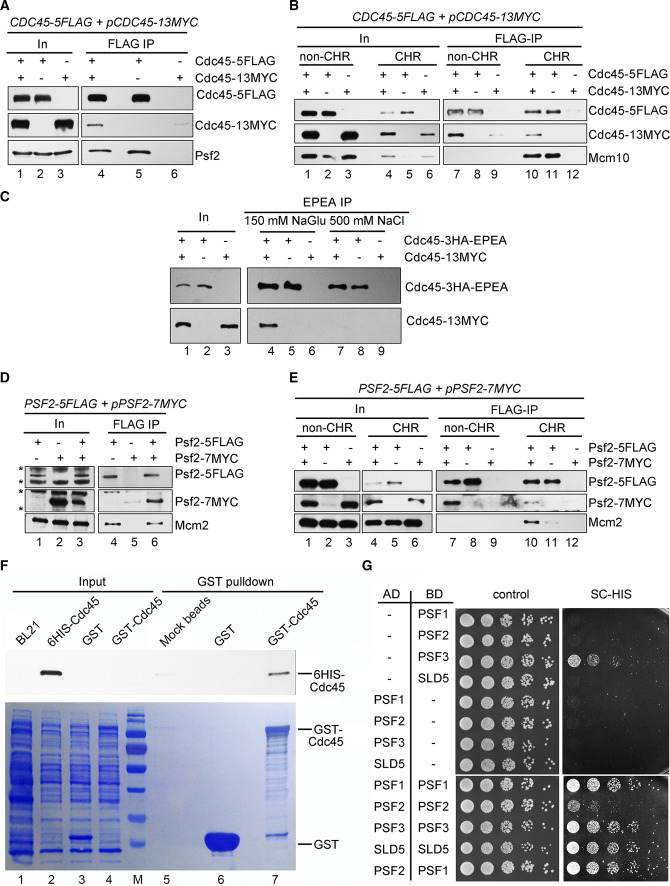
It is speculated that the pre-IC should harbor two molecules of Cdc45 and GINS as well. To test it, we applied the same dual tag strategy to Cdc45, i.e., Cdc45-5FLAG/Cdc45-13MYC. There was intermolecular interaction between Cdc45-5FLAG and Cdc45-13MYC in whole-cell extracts (WCE) (Fig. 3a), non-CHR and CHR fractions (Fig. 3b). This interaction is genuine because no contaminant bands were detectable in the mock IPs. Interestingly, it became sensitive to high salt wash, suggesting that the interaction in the context of either Cdc45 alone or d-CMG is not as tight as the MCM–MCM association within the DH (Fig. 3c). Moreover, the self-interaction of Cdc45 was direct, as shown by the pull-down assay using the purified recombinant proteins (Fig. 3f). Similarly, we were able to show that Psf2 (GINS) has self-interaction as well (Fig. 3d, e, g). These results indicate that Cdc45 and GINS may be present in a homodimer form by themselves.
Fig. 3.
Self-interactions of Cdc45 and GINS. a, b Cdc45-5FLAG was precipitated via M2 beads from WCE (a), non-CHR or CHR (b) fractions of the CDC45-5FLAG/pCDC45-13MYC cells (Strain LL85-1, Table S1). Co-precipitated proteins were detected via immunoblots against the indicated antibodies. Mcm10 was also probed to validate the fractionation of non-CHR and CHR-associated proteins given the fact that it binds to Cdc45 exclusively in the context of chromatin. In, input. c Cdc45-3HA-EPEA was precipitated via a C-tag affinity matrix from WCE of the CDC45-3HA-EPEA/pCDC45-13MYC cells (Strain LL111-1, Table S1). Co-precipitated proteins were detected as above. d, e Pfs2-5FLAG was precipitated via M2 beads from WCE (d), non-CHR or CHR (e) fractions of the PSF2-5FLAG/pPSF2-7MYC cells (LL67-1, Table S1). The precipitates were subjected to immunoblotting. Cross-reacting bands (d) are labeled by asterisks. Mcm2 was also probed to validate the fractionation of non-CHR and CHR-associated proteins. f GST pull-down analysis of Cdc45 self-interaction. Recombinant 6His-Cdc45 was incubated with GST-Cdc45 or GST alone in the presence of glutathione beads. After separation by SDS-PAGE, the bound proteins were analyzed via Coomassie Brilliant Blue staining and immunoblotting, respectively. g Yeast two-hybrid analysis of the self-interaction of the GINS subunits. The yeast cells expressing the indicated activation domain (AD)- and DNA binding domain (BD)- fusion proteins were grown at 30 °C on either SC-Trp-Leu or SC-Trp-Leu-His plates
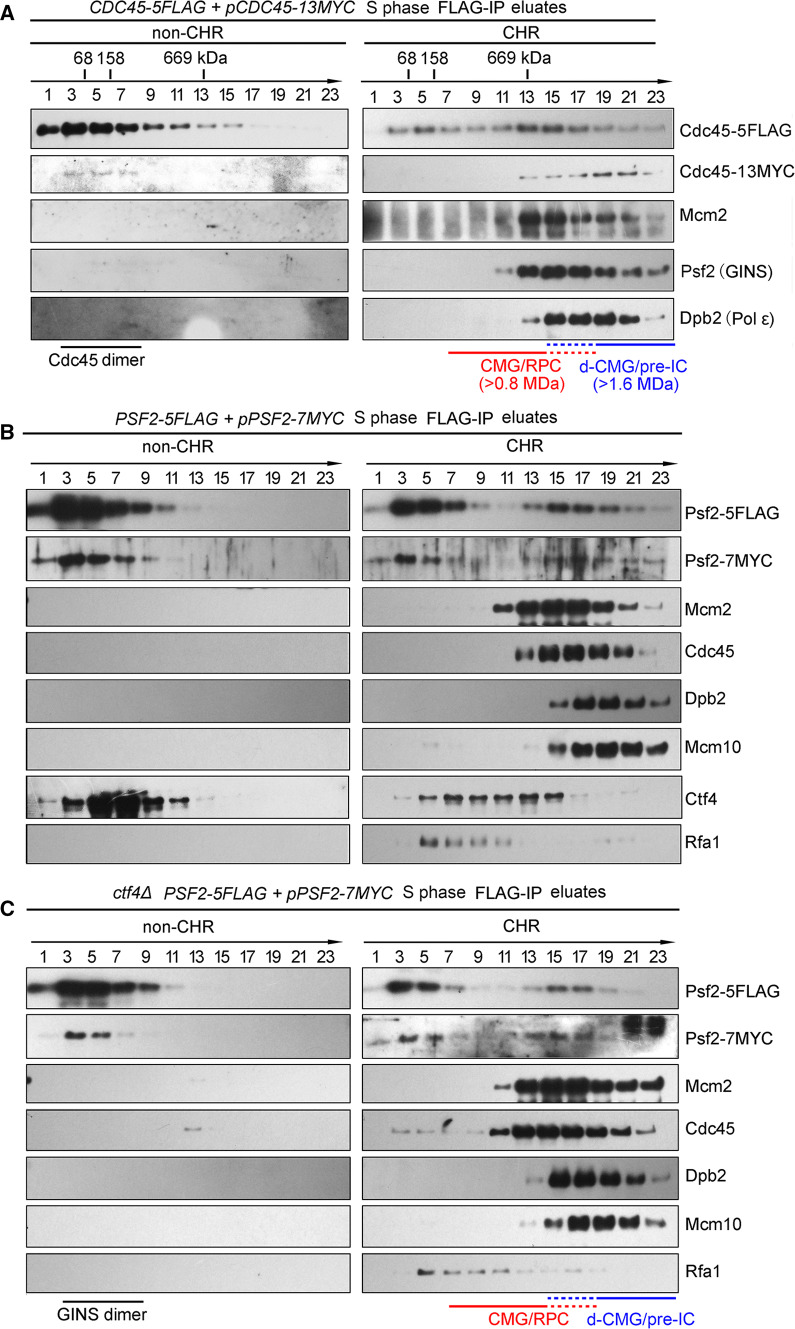
To further confirm this, we next purified the Cdc45-containing complexes from 20,000 OD600 units of the S-phase cells through FLAG-IP and peptide elution followed by glycerol gradient centrifugation. The interaction between Cdc45-5FLAG and Cdc45-13MYC is apparent in both non-CHR and CHR (Fig. 4a). In the non-CHR fraction, although most of the Cdc45 (74 kDa) sedimented as bull serum albumin (BSA, 68 kDa), a small fraction of them sedimented close to aldolase (158 kDa), equivalent to the predicted molecular weight of a Cdc45 homodimer (148 kDa). When we looked at the complexes purified from CHR fraction, Cdc45-5FLAG co-sedimented primarily with MCM, GINS and Dpb2, representing the CMG-containing complexes (Fig. 4a, right panel). Notably, there were also some fractions (15–23) containing the second copy of Cdc45 (Cdc45-13Myc), which sedimented relatively faster than the abundant s-CMGs (1.6 MDa vs. 0.8 MDa). These observations implicate that there are some dimerized CMG complexes, whereas most of them are s-CMGs in S phase.
Fig. 4.
Formation of d-CMG is independent on Ctf4. a Glycerol density gradient separation of the Cdc45-containing complexes. The CDC45-5FLAG/pCDC45-13MYC (LL85-1, Table S1) cells were cultured and released into S phase for 40 min at 25 °C after α-factor synchronization. Cells were then collected and fractionated. The Cdc45-containing complexes were purified by subjecting the Cdc45-5FLAG eluates of non-CHR and CHR fractions to centrifugation on a 10–30% glycerol density gradient as described in Fig. 2. The species of CMG-containing complexes obtained in different fractions are indicated. The dashed lines represent the overlapping fractions of d-CMG and CMG. The sedimenting positions of protein standards are labeled. b, c Glycerol density gradient separation of the GINS-containing complexes. WT (LL67-1, Table S1) (b) or ctf4Δ (LL149-1, Table S1) (c) cells in the PSF2-5FLAG/pPSF2-7MYC background were collected and fractionated basically as above. The GINS-containing complexes were purified and analyzed similarly
It is also worth noting that Ctf4 is co-purified with GINS on both non-CHR and CHR fractions, in agreement with the previous report that Ctf4 binds GINS directly (Fig. 4b) [29]. Given the fact that Ctf4 is a trimeric hub [30, 31], the small fraction of dimeric GINS or d-CMG may be mediated by Ctf4. To test this possibility, we examined their oligomeric status in the ctf4Δ cells. The sedimentation of the GINS dimer (210 kDa) in non-CHR and d-CMG in CHR was unchanged in the absence of Ctf4 (Fig. 4c, compare to b). This result indicates that both dimeric GINS and d-CMG are formed in a Ctf4-independent manner (either endogenously or artificially during the purification). Put together with the results in Figs. 1 and 2, these properties strongly support the existence of a dimerized form of the CMG-containing complex, which contains two copies of each of the Cdc45, MCM and GINS subunit.
D-CMG/pre-IC has neither helicase nor replication activities
The MCM-containing complexes purified in Figs. 2 and 4 from S-phase cells were also detected by silver staining. MCM subunits were relative clear in the purification of Mcm4-5FLAG (Fig. 5a), Cdc45-5FLAG (Figure S1A) or Psf2-5FLAG (Figure S1B), while the signals of Cdc45 and GINS were often weak. We next measured DNA helicase activity using 5′-32P-labeled partial duplex DNA as a substrate. The putative CMG/RPC fractions (e.g., 13–17) displayed clear unwinding activity on the Y-form substrate in the presence of ATP at 30 °C (Fig. 5b and S1C). The substrates became undetectable in fractions 7–11, suggesting degradation by nucleases, which are normal fork components (e.g., for Okazaki fragment maturation) as well. When ATP was replaced by ATPγS, the unwound products were barely detectable (Fig. 5d, lane 4). Since ATPγS is a slowly hydrolyzed analog of ATP, this result suggests that the single-stranded 85-mer is the product of DNA helicase activity in the purified CMG-containing complexes.
Fig. 5.
The fast-sedimenting fractions have little helicase or DNA synthesis activities. a Silver staining of the MCM-containing complexes purified from S phase (40 min) cells in Fig. 2a. The protein samples were resolved in an 8% polyacrylamide gel. The fraction number is indicated above each lane. The species of CMG-containing complexes obtained in different fractions are indicated. The dashed lines represent the overlapping fractions of d-CMG and CMG. b In vitro helicase assay. Each fraction from glycerol gradient centrifugation of the Psf2-5FLAG complexes in Fig. 4b was subjected to in vitro helicase assays as described in Experimental Procedures. A Y-shaped duplex DNA labeled at 5′-end with 32P was purified and used as a substrate. The products were analyzed by a native 8% polyacrylamide gel followed by autoradiography. Boiled substrates were loaded to indicate the migration of an 85-mer oligonucleotide. See Figure S1C for the results of independent biological repeats. c In vitro DNA synthesis assay. Each fraction was also applied to the same Y-form substrate without 32P-labeling for measuring DNA synthesis activity in the presence of all four kinds of NTPs and dNTPs including α–32P-dATP at 30 °C for 60 min. The reactions were quenched and resolved by a 20% polyacrylamide gel containing 8 M urea. The synthesized products were detected as 32P-dAMP incorporation by autoradiography. A 32P-labeled 85-mer was loaded as a size marker. See Figure S1D for the results of independent biological repeats. d In vitro helicase assay of fraction 15 was basically conducted as in (c) in the presence of either ATP or ATPɣS. e The 32P-dAMP incorporated products by RPC are resistant to S1 nuclease. In vitro DNA synthesis assays were performed as described above for both RPC fractions (11–17) and terminal deoxynucleotidyl transferase (TdT) enzymes. The final products were treated with S1 nuclease with or without boiling. The pre-labeled Y-DNA was digested by S1 nuclease as a control
Then, the unlabeled version of the same Y-form DNA substrate was used as a template to examine the in vitro DNA synthesis activity. After separation on a denaturing polyacrylamide gel, the products of replicated DNA were monitored by the incorporation of α-32P-dATP through autoradiography. As shown in Fig. 5c and S1D, in the presence of all four NTPs and dNTPs, substantial full-length (85-mer) DNA was produced by slow-sedimenting fractions (7–17), indicating an efficient de novo synthetic activity. Full-length products were hardly detected when NTPs or dNTPs were omitted (data not shown). Consistently, replication activities peaked around fraction 15 as well as helicase activities. Interestingly, fractions 7–11 showed clear synthesized products as well, probably due to the inhibition of nuclease activities in the presence of dNTPs.
Moreover, to exclude the possibility that α-32P-dAMP was incorporated by contaminating terminal deoxynucleotidyl transferase (TdT) activity, we incubated TdT with the unlabeled Y-shaped substrates in the presence of α-32P-dATP. The products much longer than 85-mer were detected (Fig. 5e, lane 5) and were very sensitive to single-stranded DNA-specific S1 nuclease (lanes 6 and 7). However, no products longer than 85-mer were observed for the putative CMG/RPC fractions (Fig. 5e, lanes 1 and 3). More importantly, 85-mer products could only be digested if they were boiled prior to S1 treatment (Fig. 5e, compare lanes 2 and 4). These results allow us to conclude that the products replicated by the CMG/RPC fractions (fractions 11–17, Fig. 5c) are bona fide duplex DNA. It is worth emphasizing that no primers were included in the reactions and that the RNA-dependent extension of DNA Pol α is usually limited to 10–12 nucleotides [32]. Therefore, the appearance of 85-mer products containing α-32P-dAMP should reflect at least the three kinds of essential activities in the DNA replication process, namely helicase, primase and polymerase, correlating well with the presence of CMG, Pri1/2, DNA Pol α and Pol ε in these putative s-CMG/RPC fractions as revealed by mass spectra (Fig. 2b).
In stark contrast, there were neither unwound (Fig. 5b and S1C) nor replicated DNA products (Fig. 5c and S1D) detectable in the fast-sedimenting fractions (19–23). The labeled substrates remained intact in fractions 19–23, indicating that the “missing” products are not due to nuclease degradation. It is also noteworthy that the fast-sedimenting fractions (19–23) consistently contain even more Cdc45, MCM and GINS than the slow-sedimenting fractions (7–13) as shown by immunoblots (Fig. 4). It is also worthy to note that other DNA helicases and polymerases have never been detected by MS in these purified complexes (e.g., Fig. 2b), although trace amounts of contaminants cannot be completely excluded. It is unlikely that such contaminants occur exclusively in the s-CMG complex. Thus, through comparing the CMG levels and replicated products in the fast- and slow- sedimenting fractions, we show the existence of a type of inactive huge CMG-containing complexes (d-CMG) in vivo.
D-CMGs display heterogeneous and flexibly oriented conformations
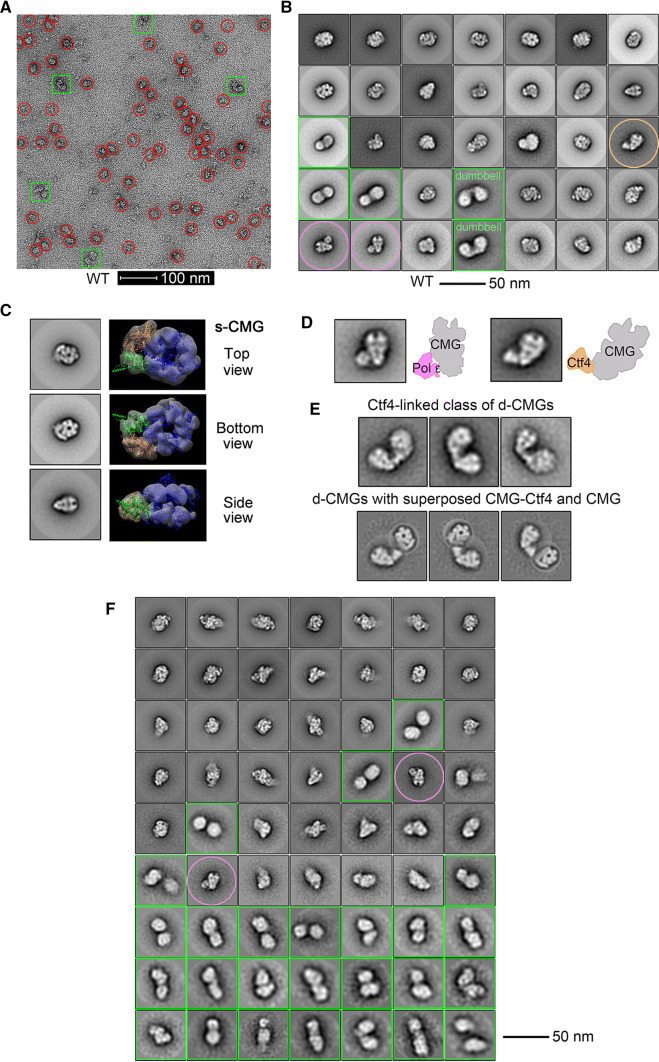
Next, we directly observed the above purified CMG-containing complexes using a transmission electron microscope after negative staining with uranyl acetate. The majority of the CMG-containing particles from the S-phase cells were homogeneous in size (20–23 nm) with a noticeable central channel from the top/bottom view (Fig. 6a–c), in good agreement with the high-resolution structure of s-CMG as reported recently [33, 34]. Interestingly, DNA Pol ε, Ctf4 and other components co-purified with s-CMG (Fig. 6b), representing relatively stable parts of RPCs. Consistent with recently resolved EM structures [35, 36], Pol ε associated with CMG through the C-terminal tier of the MCM complex and Ctf4 associated through GINS (Fig. 6d). These results corroborate that we have successfully purified the endogenous CMG-containing complexes. In addition to s-CMG, a proportion of particles appeared to have a markedly larger size (~ 35 nm), approximately twice the size of s-CMG (Fig. 6a, green squares vs. red circles). Unlike the MCM DH and s-CMG, the putative d-CMG displays markedly heterogeneous conformations, suggesting increased flexibility (Fig. 6b, green squares). It is in contrast to the d-CMG complex observed in the complex of co-expressed, recombinant fruit fly CMG constituent proteins, in which the MCMs associate stably with each other through the MCM N-termini, just as in its precursor MCM DH [22]. Moreover, the class averages of our representative d-CMG species showed that the two component CMGs are positioned in several different orientations (Fig. 6b, e). A sub-population of d-CMG, which we refer to as “dumbbell shaped”, revealed two MCM hexamers that appear to have detached from each other. Their association could be mediated by other components such as Ctf4 as computationally superposed in Fig. 6e. Given that Ctf4 is a trimeric hub directly associating with GINS, to exclude the artifactual formation of oligomeric CMGs during purification, we next monitored d-CMG species isolated in the ctf4Δ background. Indeed, CTF4 deletion abolished both CMG-Ctf4 and “dumbbell-shaped” d-CMG (Fig. 6f), indicating that this type of d-CMG is connected by Ctf4. However, as shown in Fig. 6f, in the absence of Ctf4, many other types of d-CMG persisted (green squares), consistent with the biochemical results shown in Fig. 4c. A preliminary 2D average nicely resolved densities for s-CMG, whereas the CMGs from the d-CMG particles were mostly smeared out (Fig. 6b, c, f). This implies that in our purified endogenous d-CMG, the tight association between the two single MCM hexamers [3–5] might have undergone conformational changes/rotations, resulting in partial disruption of the tightly associated MCM–MCM within the DH, in agreement with the very recent observation in vitro [25]. These data implicate that d-CMG likely undergoes multiple conformational changes before maturation into s-CMG helicases.
Fig. 6.
Single-particle EM analysis of the negatively stained CMG complexes. a A representative electron micrograph of the endogenous CMG-containing complexes (fractions 17–22) isolated from the S-phase cells of CDC45-5FLAG (LL85, Table S1) through the same purification procedure as described in Fig. 4. The putative single (s-CMG) and double (d-CMG) CMG particles are highlighted by red circles and green squares, respectively. b 2D class averages of all types of CMG-containing particles (38,787 in total). The putative CMG-DNA pol ε and CMG-Ctf4 particles are circled with pink and tawny, respectively. c S-CMG particles with top/bottom and side views. d S-CMG particles containing DNA Pol ε or Ctf4. e The dumbbell-shaped d-CMG particles (824 among total 6445 d-CMGs) with computationally superposed CMG-Ctf4 and CMG. f 2D class averages of the CMG particles (43,820 in total) purified endogenously from the S-phase ctf4Δ cells (LL-163, Table S1). The particles of putative d-CMG and CMG-DNA pol ε are labeled in green squares and pink circles, respectively
Only a small fraction of CMG-containing complexes are detected as d-CMGs
Till now we have isolated two distinct forms of the CMG-containing complexes, d-CMG and s-CMG. We then examined the relative percentages of them in yeast cells.
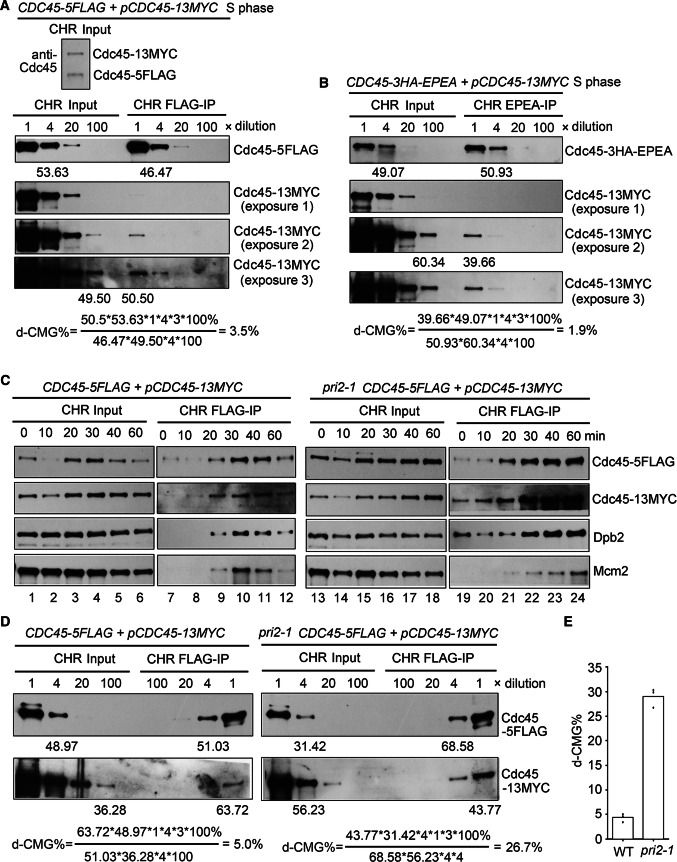
According to the results in Fig. 4a, most, if not all, Cdc45 on chromatin forms the CMG-containing complexes. Therefore, chromatin fractions of the S-phase cells (40 min) were precipitated by anti-FLAG beads, which can be considered as an enrichment of the CMG-containing complexes. First, the ratio of two different Cdc45 copies (Cdc45-5FLAG/Cdc45-13MYC) in the CHR fraction inputs was measured via anti-Cdc45 antibodies and was close to 1:1 (Fig. 7a, upper panel). Based on this, we postulated that there are equal amounts of three types of d-CMGs: Cdc45-5FLAG/Cdc45-13MYC (Type I), Cdc45-5FLAG/Cdc45-5FLAG (Type II) and Cdc45-13MYC/Cdc45-13MYC (Type III). Only Type I can be detected as d-CMGs in this procedure, which occupy approximately one-third of all d-CMGs. In the chromatin precipitates, the second copy of Cdc45 (Cdc45-13MYC) representing d-CMG was not apparent when the exposure was very low (Fig. 7a, exposure 1). This result is consistent with the previous work by Labib’s group [8], demonstrating that most of the CMG-containing complexes contain only one copy of Cdc45. However, d-CMG was visible with the darker exposures (Fig. 7a, exposures 2 and 3). To compare the relative levels of the differently tagged proteins, we quantified serial dilutions of the CHR fraction inputs and IPs. According to the band densities of Cdc45-5FLAG and Cdc45-13MYC in the inputs and IPs, we estimated the percentage of d-CMG based on the following formula:
(Formula 1)
Fig. 7.
Only a small fraction of the CMG-containing complexes can be detected as d-CMG. a The Cdc45-5FLAG CHR input and CHR IP of the S-phase cells (25 °C, 40 min) were obtained as in Fig. 4a. The relative protein levels in each sample were quantified via dilutions and immunoblots. b The experiments were basically performed as above, except in another strain (CDC45-3HA-EPEA/pCDC45-13MYC) (LL111-1, Table S1). c The experiments were basically performed as in (a) in both WT and pri2-1 background (LL226-1, Table S1) collected at the indicated time points after G1 release at 37 °C. d Dilution and calculation of d-CMG % in the S-phase cells of WT and pri2-1. e The quantification of d-CMG in WT and pri2-1 S-phase cells from three biological repeats
In wild-type (WT) S phase (25 °C, 40 min) cells, d-CMG occupied only less than 5%. Similar results were obtained in an independent EPEA-IP experiment regardless of a different pair of tags that was used (Fig. 7b). Consistent with the observations shown in Fig. 4, these data suggest that during S phase, a very small portion of the CMG-containing complexes can be detected as d-CMGs, whereas most of them become s-CMGs.
Accumulation of d-CMG/pre-IC in the absence of DNA Pol α-primase
Next, we asked whether the d-CMG/s-CMG transition can be blocked in the absence of DNA Pol α-primase. For this purpose, we introduced a temperature-sensitive mutant of DNA Pol α (pri2-1) into the strain used in Fig. 7a. As shown in Figure S2A, after releasing pri2-1 cells from G1-arrest to the non-permissive condition (37 °C), the S-phase progression was significantly inhibited. The cell cycle-specific redistribution of replication proteins on chromatin was monitored by western blotting. The levels of initiation proteins on chromatin rose at the same time during S phase in WT and pri2-1. In WT, the chromatin-associated levels of initiation factors, Cdc45, Mcm2 and Dpb2, exhibited a clear fluctuating pattern, peaking at about 30 min and then decreasing (Fig. 7c, lanes 1–6), correlating well with the S-phase/G2 progression (Figure S2A). All CMG components started to decline around 40 min, correlating well with the increasing G2 cells (Figure S2A), reflecting the replisome disassembly during replication termination [37–39]. On the contrary, when Pri2 was inactivated, none of these factors showed apparent cell cycle-dependent changes after loading onto chromatin; instead, the levels of replication proteins at various time points remained constant (Fig. 7c, lanes 13–18).
We then precipitated Cdc45-5FLAG from CHR fractions. As shown in the right panel of Fig. 7c, the amounts of Cdc45-13MYC in the CHR precipitates reached a similar peak level at 30 min in both WT (lanes 7–12) and pri2-1 (lanes 19–24) cells, in agreement with the function of primase after the d-CMG/pre-IC assembly. In WT, after 30 min, Cdc45-13MYC rapidly decreased in the Cdc45-FLAG-IP, as did other initiation factors (Fig. 7c, lanes 10–12). This result indicates that in WT, d-CMG shows an S-phase regulated assembly/segregation pattern. However, without primase, Cdc45-13MYC and other factors predominantly persisted throughout the cell cycle (lanes 22–24), indicating compromised segregation of d-CMG. Accordingly, the percentage of d-CMGs increased from about 5% in WT to 29% in pri2-1 during S phase (Fig. 7d,e and S2B). These results validate that there is a bona fide d-CMG/s-CMG transition process in vivo, which is significantly inhibited in the absence of key replisome components like primase.
Discussion
Here, we have identified a new class of the CMG-containing complex designated as d-CMG in vivo, which has at least four main properties distinct from the traditional CMG (s-CMG).
(i) Although both complexes share the same core subunits, d-CMG and s-CMG are distinct in subunit stoichiometry and composition. D-CMG contains two copies of each of the Cdc45, MCM and GINS subunits, whereas s-CMG bears only one copy of them. Moreover, RPA and Pol α/primase are present in s-CMG/replisome, but are not detectable in d-CMG/pre-IC.
(ii) D-CMG has neither DNA helicase nor replication activity, which is the key characteristic according to the original definition of the pre-IC [26].
(iii) About a small fraction of total cellular CMG-containing complexes can be detected as d-CMGs, whereas the rest are s-CMG/replisomes, in line with the transient and/or unstable intermediate nature of the pre-IC. Meanwhile, it is necessary to emphasize that d-CMG is not detectable until a combination of several enrichment strategies was used in this study. The tight head-to-head association within an MCM DH is disrupted upon Cdc45 and GINS association, as observed by Diffley’s group in vitro [25]. Therefore, the intermolecular interaction between the two CMGs in the d-CMG/pre-IC might have been dramatically loosened or interrupted (e.g., when they are indirectly connected by a short fragment of DNA). This may explain why different groups including the initial stage of our group have repeatedly failed to show its existence for years both in vivo [8, 24] and in vitro [25]. Alternatively, Cdc45 and GINS may not be able to separate the MCM DH in vivo as efficiently as they do in vitro, due to the different conditions such as protein stoichiometry/oligomerization/post-translational modifications, the activities of DDK and S-CDK and chromatin structure in vivo.
(iv) D-CMG is assembled and rapidly separated during S phase. Although s-CMG is also an S-phase-specific complex, it remains stable throughout the elongation stage of chromosome replication.
All these characters of the d-CMG supercomplex fit the definition of the pre-IC, i.e., the intermediate between the pre-RC and replication fork [26].
Although an excessive Benzonase (500 U/ml final concentration) was used to prepare the native chromatin fraction, it is possible that the two CMGs within the detected d-CMG might be connected by a short DNA fragment that is not accessible to the nuclease. In the absence of DNA Pol α-primase, the DNA fragment connecting two CMGs may become shorter, increasing the resistance to Benzonase digestion.
It is also worth pointing out that the endogenous d-CMG identified in this study exhibits heterogeneous and flexible conformations, which is distinct from the case of the d-CMG/DNA complexes prepared by the reconstitution of 11 CMG baculovirus expressed Drosophila melanogaster CMG subunits and DNA reported previously [22]. They could represent different stages of d-CMG assembly and development. Supporting this, only a small proportion of the CMG-containing particles exist as dimers in both studies. It is also not surprising that d-CMG has multiple flexible conformations given the starkly different structures of its precursor MCM DH and its product s-CMG observed to date. The two tilted and twisted MCM hexamers might have undergone rotation, thus resulting in detached and positioned in different orientations in most types of d-CMG. Technique improvements are necessary to solve its high-resolution structure(s) in the future.
Materials and methods
Strain and plasmid construction
Saccharomyces cerevisiae strains and plasmids/oligomers used in this study are listed in Tables S1 and S2, respectively.
Cell synchronization, flow cytometry, WCE preparation, immunoprecipitation (IP), GST pull-down and yeast two-hybrid assays were performed as previously described [23].
Native chromatin fractionation
Native chromatin fractionation was performed as previously described [23] with minor modifications. Yeast cells of 2000 OD600 units were spheroplasted by 75 U/ml lyticase. Crude extract was prepared by Triton X-100 treatment and fractionated via sucrose cushion in 5 ml of EBX-3 buffer {50 mM HEPES/KOH pH7.4, 150 mM NaGlu, 2.5 mM MgOAc, 0.1 mM ZnOAc, 5 mM NaF, 1 mM NaVO4, 10 mM β-Glycerophosphate, 3 mM ATP, 1 mM DTT, 1 mM PMSF, Protease inhibitor tablets (EDTA free, Roche)}. The supernatant contains non-chromatin-bound proteins (non-CHR), while the chromatin is in the pellet. Chromatin-bound proteins (CHR) were released by dissolving the pellet with EBX-3 buffer supplemented with 500 U/ml of Benzonase (Sigma) for 60 min at 4 °C.
Glycerol density gradient centrifugation
The native protein complexes in the peptide eluates after FLAG-IPs were concentrated and applied to the top of a 10–30% glycerol gradient in EBX-3 buffer without protease inhibitors. The gradients were centrifuged in a P55ST2 swinging bucket rotor (Hitachi CP100NX ultracentrifuge) at 79,000g for 16 h using slow deceleration. Following centrifugation, 24 fractions (200 μl each) were collected manually from top to bottom of the gradient. As molecular weight markers, a mixture of BSA (68 kDa), aldolase (158 kDa) and thyroglobulin (669 kDa) was centrifuged in a separate tube. Thyroglobulin (669 kDa) migrates at the fraction 13 under this condition. The fractions containing different species of the MCM complexes were pooled and processed for mass spectrometry, in vitro helicase and de novo DNA synthesis assays described below.
Helicase assays
The helicase activity was measured using a 5′-32P-labeled 85 bp Y-shaped DNA substrate (Table S2) bearing a single-stranded 3′-dT(40) tail with some modifications from [40]. Briefly, each reaction (37 μl) contains 0.5 nM 5′-32P-labeled Y-shaped DNA and 30 μl protein fraction collected from glycerol gradient centrifugation in a final helicase buffer (25 mM HEPES–KOH (pH 7.6); 150 mM potassium glutamate; 10 mM magnesium acetate; 0.1 mM EDTA; 2 mM DTT; 2 mM ATP). Reactions were conducted at 30 °C for 60 min before adding 4 μl quench buffer (200 mM EDTA, 1% sodium dodecyl sulfonate (SDS) and 0.1% bromophenol blue). Products were then separated on a native 8% polyacrylamide gel in 0.5 × TBE before autoradiography.
De novo DNA synthesis and S1 nuclease-resistant assays
The DNA synthesis activity of each fraction from glycerol gradient centrifugation was measured using an unlabeled version of the Y-shaped DNA used in the helicase assays. Synthesis reactions (40 μl) contain 0.5 nM unlabeled Y-form DNA and 33 μl of each fraction from glycerol gradient centrifugation in a final synthesis buffer (40 mM HEPES–KOH (pH 7.6); 150 mM potassium glutamate; 10 mM magnesium acetate; 2 mM DTT; 2 mM ATP) plus four NTPs (200 μM each), four dNTPs (40 μM dGTP/dCTP/dTTP and 4 μM dATP) and 40 nM α-32P-dATP. Reactions were conducted at 30 °C for 60 min.
For terminal deoxynucleotidyl transferase (TdT) assay, the reactions (30 μl each) contain 0.5 nM unlabeled Y-form DNA and 0.17 U/μl TdT (New England Biolabs) in a final buffer with 1 × TdT reaction buffer, 1 μM dATP and 55 nM α-32P-dATP. Reactions were conducted at 37 °C for 60 min before being inactivated at 75 °C for 20 min.
For S1 nuclease treatment, the synthesized products by the RPC fractions or TdT were subjected to S1 nuclease digestion before analysis. S1 nuclease (final concentration 1 U/μl) was incubated at 25 °C for 30 min with 40 μl synthesis reaction with or without prior boiling. The reactions were stopped by adding 6 μl quench buffer (200 mM EDTA and 0.1% bromophenol blue). All reaction products were separated on a 20% polyacrylamide gel containing 8 M urea in 1 × TBE before autoradiography.
MS sample preparation
Proteins were precipitated with 25% trichloroacetic acid (TCA) for at least 30 min on ice. The protein pellets were washed twice with 500 μl ice-cold acetone, air dried, and then resuspended in 8 M urea, 20 mM methylamine, and 100 mM Tris, pH 8.5. After reduction (5 mM TCEP, room temperature, 20 min) and alkylation (10 mM iodoacetamide, room temperature, 15 min in the dark), the samples were diluted to 2 M urea with 100 mM Tris, pH 8.5, and digested with trypsin at 1/50 (w/w) enzyme/substrate ratio at 37 °C for 16–18 h. The digestion was then stopped by adding formic acid to 5% (final concentration).
LC–MS/MS analysis
All protein samples were analyzed using an EASY-nLC 1000 system (Thermo Fisher Scientific, Waltham, MA) interfaced with a Q-Exactive mass spectrometer (Thermo Fisher Scientific). Peptides were loaded on a pre-column (75 μm ID, 4 cm long, packed with ODS-AQ 12 nm-10 μm beads) and separated on an analytical column (75 μm ID, 12 cm long, packed with Luna C18 1.9 μm 100 Å resin) with a 60 min linear gradient at a flow rate of 200 nl/min as follows: 0–5% B in 2 min, 5–30% B in 43 min, 30–80% B in 5 min, 80% B for 10 min (A = 0.1% FA, B = 100% ACN, 0.1% FA). Spectra were acquired in data-dependent mode: the top ten most intense precursor ions from each full scan (resolution 70,000) were isolated for HCD MS2 (resolution 17,500; NCE 27) with a dynamic exclusion time of 30 s. The AGC targets for the MS1 and MS2 scans were 3e6 and 1e5, respectively, and the maximum injection times for MS1 and MS2 were both 60 ms. Precursors with 1 + , more than 7 + or unassigned charge states were excluded.
The MS data were searched against a Uniprot S. cerevisiae protein database (downloaded from Uniprot on 2013-04-03) using an updated version of pFind [41] with the following parameters: instrument, HCD-FTMS; precursor mass tolerance, 20 ppm; fragment mass tolerance 20 ppm; open search mode; peptide length, minimum 6 amino acids and maximum 100 amino acids; peptide mass, minimum 600 and maximum 10,000 Da; enzyme, trypsin, with up to three missed cleavage sites. The results were filtered by requiring FDR < 1% at the spectral level and spectra count ≥ 2.
Electron microscopy
The CMG-containing complexes were isolated from 17 to 22 fractions from glycerol density gradient centrifugation and concentrated by ultrafiltration. Negative staining of the samples deposited on carbon-coated grids was conducted with 2% uranyl acetate. Grids were examined using an FEI Tecnai F20 microscope operated at 200 kV, and images were recorded at a nominal magnification of 50,000 × using a 4 k × 4 k charge-coupled device (CCD) camera (UltraScan 4000, Gatan), resulting in a 1.7 Å pixel size at the specimen level. EMAN2 was used for manual particle picking and micrograph screening [42]. The 2D classification, 3D classification and 3D refinement were performed using RELION1.4 [43]. Artificial CMG dimers were generated by relating the two CMG atomic models (PDB code: 3JC5) in UCSF Chimera [44], with the selected projection of the resulting dimer model matching the observed 2D class averages.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We thank Dr. Costa for sharing unpublished data and superposed CMG-Ctf4 structure shown in Fig. 6e; Dr. Karim Labib for insightful discussion; Drs. Karim Labib, Stephen Bell, and Li-Lin Du for reagents; Drs. Ning Gao, Hao Wu, Qun He, Yisui Xia, Ms. Yawen Bai, and members of the Lou lab for help and comments on the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (http://www.nsfc.gov.cn/) 31630005 and 31770084 to HL, 31771382 to QC, 31800066 to JZ; the China Postdoctoral Science Foundation (http://jj.chinapostdoctor.org.cn/) 2018M640201 to JZ; the National Basic Research Program (973 Program) of China (http://www.most.gov.cn/) 2014CB849801 to MQD; Program for Extramural Scientists of the State Key Laboratory of Agrobiotechnology (http://en.cau.edu.cn/colleges/biological-science/) (2018SKLAB6-5) to MQD; Opening Project of the State Key Laboratory of Microbial Resources (http://english.im.cas.cn/) to HL.
Compliance with ethical standards
Conflict of interest
The authors declare no competing financial interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Lu Liu, Yue Zhang, and Jingjing Zhang have contributed equally to this work.
References
- 1.Parker MW, Botchan MR, Berger JM. Mechanisms and regulation of DNA replication initiation in eukaryotes. Crit Rev Biochem Mol Biol. 2017;52(2):107–144. doi: 10.1080/10409238.2016.1274717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bleichert F, Botchan MR, Berger JM. Mechanisms for initiating cellular DNA replication. Science. 2017 doi: 10.1126/science.aah6317. [DOI] [PubMed] [Google Scholar]
- 3.Evrin C, Clarke P, Zech J, Lurz R, Sun J, Uhle S, Li H, Stillman B, Speck C. A double-hexameric MCM2-7 complex is loaded onto origin DNA during licensing of eukaryotic DNA replication. Proc Natl Acad Sci. 2009;106(48):20240–20245. doi: 10.1073/pnas.0911500106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Remus D, Beuron F, Tolun G, Griffith JD, Morris EP, Diffley JF. Concerted loading of Mcm2-7 double hexamers around DNA during DNA replication origin licensing. Cell. 2009;139(4):719–730. doi: 10.1016/j.cell.2009.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li N, Zhai Y, Zhang Y, Li W, Yang M, Lei J, Tye B-K, Gao N. Structure of the eukaryotic MCM complex at 3.8 A. Nature. 2015;524(7564):186–191. doi: 10.1038/nature14685. [DOI] [PubMed] [Google Scholar]
- 6.Coster G, Diffley JFX. Bidirectional eukaryotic DNA replication is established by quasi-symmetrical helicase loading. Science. 2017;357(6348):314–318. doi: 10.1126/science.aan0063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moyer SE, Lewis PW, Botchan MR. Isolation of the Cdc45/Mcm2-7/GINS (CMG) complex, a candidate for the eukaryotic DNA replication fork helicase. Proc Natl Acad Sci USA. 2006;103(27):10236–10241. doi: 10.1073/pnas.0602400103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gambus A, Jones RC, Sanchez-Diaz A, Kanemaki M, van Deursen F, Edmondson RD, Labib K. GINS maintains association of Cdc45 with MCM in replisome progression complexes at eukaryotic DNA replication forks. Nat Cell Biol. 2006;8(4):358–366. doi: 10.1038/ncb1382. [DOI] [PubMed] [Google Scholar]
- 9.Pacek M, Tutter AV, Kubota Y, Takisawa H, Walter JC. Localization of MCM2-7, Cdc45, and GINS to the site of DNA unwinding during eukaryotic DNA replication. Mol Cell. 2006;21(4):581–587. doi: 10.1016/j.molcel.2006.01.030. [DOI] [PubMed] [Google Scholar]
- 10.Ilves I, Petojevic T, Pesavento JJ, Botchan MR. Activation of the MCM2-7 helicase by association with Cdc45 and GINS proteins. Mol Cell. 2010;37(2):247–258. doi: 10.1016/j.molcel.2009.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yardimci H, Loveland AB, Habuchi S, van Oijen AM, Walter JC. Uncoupling of sister replisomes during eukaryotic DNA replication. Mol Cell. 2010;40(5):834–840. doi: 10.1016/j.molcel.2010.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Costa A, Ilves I, Tamberg N, Petojevic T, Nogales E, Botchan MR, Berger JM. The structural basis for MCM2-7 helicase activation by GINS and Cdc45. Nat Struct Mol Biol. 2011;18(4):471–477. doi: 10.1038/nsmb.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Riera A, Barbon M, Noguchi Y, Reuter LM, Schneider S, Speck C. From structure to mechanism—understanding initiation of DNA replication. Genes Dev. 2017;31(11):1073–1088. doi: 10.1101/gad.298232.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.O’Donnell ME, Li H. The ring-shaped hexameric helicases that function at DNA replication forks. Nat Struct Mol Biol. 2018 doi: 10.1038/s41594-018-0024-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sheu YJ, Stillman B. Cdc7-Dbf4 phosphorylates MCM proteins via a docking site-mediated mechanism to promote S phase progression. Mol Cell. 2006;24(1):101–113. doi: 10.1016/j.molcel.2006.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sheu YJ, Stillman B. The Dbf4-Cdc7 kinase promotes S phase by alleviating an inhibitory activity in Mcm4. Nature. 2010;463(7277):113–117. doi: 10.1038/nature08647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heller RC, Kang S, Lam WM, Chen S, Chan CS, Bell SP. Eukaryotic origin-dependent DNA replication in vitro reveals sequential action of DDK and S-CDK kinases. Cell. 2011;146(1):80–91. doi: 10.1016/j.cell.2011.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tanaka S, Araki H. Helicase activation and establishment of replication forks at chromosomal origins of replication. Cold Spring Harb Perspect Bio. 2013;5(12):a010371. doi: 10.1101/cshperspect.a010371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deegan TD, Yeeles JT, Diffley JF. Phosphopeptide binding by Sld3 links Dbf4-dependent kinase to MCM replicative helicase activation. EMBO J. 2016 doi: 10.15252/embj.201593552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fang D, Cao Q, Lou H. Sld3-MCM interaction facilitated by Dbf4-dependent kinase defines an essential step in eukaryotic DNA replication initiation. Front Microbiol. 2016;7:885. doi: 10.3389/fmicb.2016.00885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Siddiqui K, On KF, Diffley JF. Regulating DNA replication in eukarya. Cold Spring Harb Perspect. 2013 doi: 10.1101/cshperspect.a012930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Costa A, Renault L, Swuec P, Petojevic T, Pesavento JJ, Ilves I, MacLellan-Gibson K, Fleck RA, Botchan MR, Berger JM. DNA binding polarity, dimerization, and ATPase ring remodeling in the CMG helicase of the eukaryotic replisome. Elife. 2014;3:e03273. doi: 10.7554/elife.03273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Quan Y, Xia Y, Liu L, Cui J, Li Z, Cao Q, Chen XS, Campbell JL, Lou H. Cell-cycle-regulated interaction between Mcm10 and double hexameric Mcm2-7 is required for helicase splitting and activation during S Phase. Cell Rep. 2015;13(11):2576–2586. doi: 10.1016/j.celrep.2015.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miyazawa-Onami M, Araki H, Tanaka S. Pre-initiation complex assembly functions as a molecular switch that splits the Mcm27 double hexamer. EMBO Rep. 2017;18(10):1752–1761. doi: 10.15252/embr.201744206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Douglas ME, Ali FA, Costa A, Diffley JFX. The mechanism of eukaryotic CMG helicase activation. Nature. 2018;555:265. doi: 10.1038/nature25787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zou L, Stillman B. Formation of a preinitiation complex by S-phase cyclin CDK-dependent loading of Cdc45p onto chromatin. Science. 1998;280(5363):593–596. doi: 10.1126/science.280.5363.593. [DOI] [PubMed] [Google Scholar]
- 27.Douglas ME, Diffley JFX. Recruitment of Mcm10 to sites of replication initiation requires direct binding to the minichromosome maintenance (MCM) complex. J Biol Chem. 2016;291(11):5879–6888. doi: 10.1074/jbc.M115.707802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yeeles JT, Deegan TD, Janska A, Early A, Diffley JF. Regulated eukaryotic DNA replication origin firing with purified proteins. Nature. 2015;519(7544):431–435. doi: 10.1038/nature14285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gambus A, van Deursen F, Polychronopoulos D, Foltman M, Jones RC, Edmondson RD, Calzada A, Labib K. A key role for Ctf4 in coupling the MCM2-7 helicase to DNA polymerase alpha within the eukaryotic replisome. EMBO J. 2009;28(19):2992–3004. doi: 10.1038/emboj.2009.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Simon AC, Zhou JC, Perera RL, van Deursen F, Evrin C, Ivanova ME, Kilkenny ML, Renault L, Kjaer S, Matak-Vinkovic D, Labib K, Costa A, Pellegrini L. A Ctf4 trimer couples the CMG helicase to DNA polymerase alpha in the eukaryotic replisome. Nature. 2014;510(7504):293–297. doi: 10.1038/nature13234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Villa F, Simon AC, Ortiz Bazan MA, Kilkenny ML, Wirthensohn D, Wightman M, Matak-Vinkovic D, Pellegrini L, Labib K. Ctf4 is a hub in the eukaryotic replisome that Links multiple CIP-box proteins to the CMG helicase. Mol Cell. 2016 doi: 10.1016/j.molcel.2016.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Perera RL, Torella R, Klinge S, Kilkenny ML, Maman JD, Pellegrini L. Mechanism for priming DNA synthesis by yeast DNA polymerase alpha. Elife. 2013;2:e00482. doi: 10.7554/eLife.00482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Georgescu R, Yuan Z, Bai L, de Luna Almeida Santos R, Sun J, Zhang D, Yurieva O, Li H, O’Donnell ME. Structure of eukaryotic CMG helicase at a replication fork and implications to replisome architecture and origin initiation. Proc Natl Acad Sci USA. 2017 doi: 10.1073/pnas.1620500114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yuan Z, Bai L, Sun J, Georgescu R, Liu J, O’Donnell ME, Li H. Structure of the eukaryotic replicative CMG helicase suggests a pumpjack motion for translocation. Nat Struct Mol Biol. 2016 doi: 10.1038/nsmb.3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sun J, Shi Y, Georgescu RE, Yuan Z, Chait BT, Li H, O’Donnell ME. The architecture of a eukaryotic replisome. Nat Struct Mol Biol. 2015;22(12):976–982. doi: 10.1038/nsmb.3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou JC, Janska A, Goswami P, Renault L, Abid Ali F, Kotecha A, Diffley JF, Costa A. CMG-Pol epsilon dynamics suggests a mechanism for the establishment of leading-strand synthesis in the eukaryotic replisome. Proc Natl Acad Sci USA. 2017;114(16):4141–4146. doi: 10.1073/pnas.1700530114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maric M, Maculins T, De Piccoli G, Labib K. Cdc48 and a ubiquitin ligase drive disassembly of the CMG helicase at the end of DNA replication. Science. 2014;346(6208):1253596. doi: 10.1126/science.1253596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Priego Moreno S, Bailey R, Campion N, Herron S, Gambus A. Polyubiquitylation drives replisome disassembly at the termination of DNA replication. Science. 2014;346(6208):477–481. doi: 10.1126/science.1253585. [DOI] [PubMed] [Google Scholar]
- 39.Dewar JM, Budzowska M, Walter JC. The mechanism of DNA replication termination in vertebrates. Nature. 2015;525:345. doi: 10.1038/nature14887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xia Y, Niu Y, Cui J, Fu Y, Chen X, Lou H, Cao Q. The helicase activity of hyperthermophilic archaeal MCM is enhanced at high temperatures by lysine methylation. Front Microbiol. 2015;6:1247. doi: 10.3389/fmicb.2015.01247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chi H, He K, Yang B, Chen Z, Sun R-X, Fan S-B, Zhang K, Liu C, Yuan Z-F, Wang Q-H, Liu S-Q, Dong M-Q, He S-M. pFind–Alioth: a novel unrestricted database search algorithm to improve the interpretation of high-resolution MS/MS data. J Proteomics. 2015;125:89–97. doi: 10.1016/j.jprot.2015.05.009. [DOI] [PubMed] [Google Scholar]
- 42.Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, Rees I, Ludtke SJ. EMAN2: an extensible image processing suite for electron microscopy. J Struct Biol. 2007;157(1):38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 43.Scheres SHW. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol. 2012;180(3):519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera—A visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.