Abstract
The genus Striga, also called “witchweed”, is a member of the family Orobanchaceae, which is a major family of root-parasitic plants. Striga can lead to the formation of seed stocks in the soil and to explosive expansion with enormous seed production and stability once the crops they parasitize are cultivated. Understanding the molecular mechanism underlying the communication between Striga and their host plants through natural seed germination stimulants, “strigolactones (SLs)”, is required to develop the technology for Striga control. This review outlines recent findings on the SL perception mechanism, which have been accumulated in Striga hermonthica by the similarity of the protein components that regulate SL signaling in nonparasitic model plants, including Arabidopsis and rice. HTL/KAI2 homologs were identified as SL receptors in the process of Striga seed germination. Recently, this molecular basis has further promoted the development of various types of SL agonists/antagonists as seed germination stimulants or inhibitors. Such chemical compounds are also useful to elucidate the dynamic behavior of SL receptors and the regulation of SL signaling.
Keywords: Hydrolase, Karrikin, Parasitic weed, Phytohormone, Shoot branching, Strigolactone analogs, Strigolactone mimics, Ubiquitin–proteasome system
Introduction
Orobanchaceae is a major family of root-parasitic plants and is divided into 90 genera (one genus is not parasitic) that are composed of more than 2000 species [1]. Each genus is widely distributed around the world with different host preferences and dependencies. Obligate parasites require parasitism to their host plants for survival, because they lack photosynthesis and/or other physiological functions. In contrast, facultative parasites can grow on their own, whereas they parasitize if host plants are located near them.
The genus Striga, also called “witchweed”, is a highly threatening obligate parasite of the family Orobanchaceae; its habitat has expanded over 40 countries in Africa, and some species have reached India and Pakistan beyond the Arabian Peninsula [2, 3]. Striga parasitizes major crops, including maize, millet, sorghum, and rice, as host plants, causing a 30–90% reduction in agricultural production [2, 4]. The crop damage caused by Striga is estimated to be 111–200 million dollars in rice alone [5] and adversely affect at least 100 million people in sub-Saharan Africa [4]. Striga produces over 50,000 very-tiny seeds (approx. 0.2‒0.5 mm) [6]. In addition, Striga seeds remain viable and dormant in the soil for over 14 years until they perceive germination signals from their host plants [7]. Because of these features, Striga can lead to the formation of seed stocks in the soil and to explosive expansion once their crop hosts are cultivated.
The molecular mechanism underlying the interaction between Striga and their host plants is required to develop the technology for Striga control. Three common strategies have been presented thus far: (1) utilizing host-acquired resistance; (2) suppressing the germination of Striga seeds; and (3) depleting the seed stocks in the soil. Based on the host preferences, various crops were screened and analyzed for resistance to Striga parasitism. For example, a rice cultivar (Nipponbare) exerts a potent post attachment resistance to Striga hermonthica [8]. The parasite penetrates the root cortex but cannot form a connection with the host xylem. Some other hosts induce a hypersensitive reaction at the site of infection to prevent Striga parasitism [9]. Regarding the ability to evade the Striga germination, some crops secrete modified germination signals that are not capable of inducing effective germination. LOW GERMINATION STIMULANT 1 (LGS1) was identified as a causative gene in sorghum [10]. After germination, the Striga radicle invades a host root in close proximity (2‒3 mm). In addition, the size of the endosperm is insufficiently large to maintain radicle growth for more than 7 days [6]. Based on the ecological features of Striga seeds, “suicidal germination stimulants” have been proposed to eliminate the Striga seeds [11]. In the suicidal germination stimulant method, synthetic germination stimulants are applied to the soil before sowing the host crops to induce the germination of Striga seeds. Because Striga are obligate parasites, they are not capable of surviving without hosts, and thus, the seed stocks could be depleted.
“Strigolactones (SLs)” and their molecular bases in Striga germination have been discovered in this past decade and lead to the chemical regulation of the obligate parasite, which are highlighted from various viewpoints in the recent review articles [12–18]. Here, we outline recent findings on the SL perception mechanism in Striga germination and the chemical compounds developed based on the action of SLs.
Strigolactones and their roles in the host recognition of root-parasitic plants
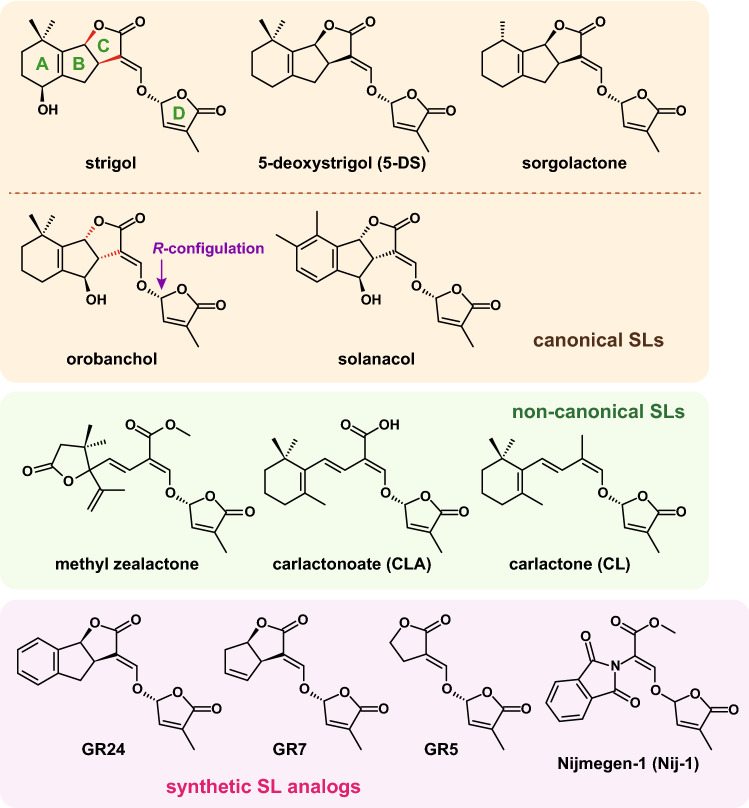
After invading a host root, the genus Striga initiates the formation of a structure called a haustorium for successful attachment and then grows underground for 4‒7 weeks, during which it causes severe damage to the host. Once emerged above ground, it rapidly develops flowers and seeds. Striga host sensing is achieved by the perception of SLs, which are exogenous compounds secreted by the host plants. In 1966, strigol was initially isolated as a natural SL from the root exudates of cotton [19] (Fig. 1). The structure of strigol was elucidated in 1972 [20], but its absolute configuration was not reported until 1985 [21]. Later, other germination stimulants were discovered with similar structures to strigol. For example, orobanchol, solanacol, and sorgolactone were identified from the root exudates of red clover [22], tobacco [23], and sorghum [24], respectively. Currently, 17 types of natural SLs have been proven to stimulate the germination of Striga and/or other Orobanche spp. [25].
Fig. 1.
Chemical structures of natural (canonical and non-canonical) SLs and synthetic SL analogs. The stereochemistry of the BC junction is different between the strigol and orobanchol classes, indicated by bonds colored in red
Canonical SLs are composed of a tricyclic lactone (ABC-ring) and a butenolide ring (D-ring) that are connected by an enol-ether linkage [11, 26] (Fig. 1). SLs are divided into two classes according to the stereochemistry of the BC junction on the ABC-ring. The SLs belonging to the “strigol class” have the same stereochemistry as strigol. In contrast, the “orobanchol class” is characterized by the opposite stereochemistry of the BC junction to the strigol class [27], which was initially observed in orobanchol [22]. The BC stereochemistry and other modifications of the ABC-ring affect the activity of SLs in Striga germination. Studies using different sorghum cultivars showed that the parasitism efficiency of S. hermonthica was positively correlated with the amount of 5-deoxystrigol (5-DS) production [28, 29] (Fig. 1). Another stereochemistry, the configuration of the D-ring at C-2′, is common to all of the natural SLs in both classes, which is limited to an R-configuration. The R-configuration is required for greater S. hermonthica germination activity compared to the germination promoted by the S-configuration.
SLs are instable in water and easily degrade into inactive ABC-rings and D-rings by breakdown of the enol-ether linkage [30], and it is difficult to isolate natural SLs, because only trace amounts are produced in plants [31]. On the other hand, the ABC-ring of natural SLs adopts a variable structure to some extent. This structural information led to the design of synthetic SL analog GR24 (Fig. 1), which has been widely used for research purposes because of its high Striga germination activity [30, 32]. Compounds lacking the A- or AB-ring of GR24, namely, GR7 or GR5, were also confirmed as retaining the ability to stimulate Striga germination. However, only the ABC-ring or D-ring do not show germination activity. These findings suggest that the CD part is bioactiphore of SLs as germination stimulants [11, 26, 30], which is used as a model for the dedicated design of active SL analogs [33]. Nijmegen-1 (Nij-1) is a first SL analog with the germination activity of root-parasitic plants although it has a carboxy methyl group as an open C-ring. Indeed, Nij-1 showed a considerable reduction of Striga seeds as suicidal germination stimulants in the field trials as well as GR24 and GR7, and also worked in tobacco fields infested by Orobanche ramose L. [11, 34]. Recently, new types of natural SLs lacking the A-, B-, or C-ring were isolated from the root exudates of several plants, such as maize and sunflower, as germination stimulants [13] (Fig. 1). These are called non-canonical SLs as which methyl zealactone, an SL biosynthetic precursor carlactone (CL), and its oxidized metabolite carlactonoate (CLA) are classified. Methyl zealactone has a carboxy methyl group connecting an enol-ether‒D-ring moiety similarly to Nij-1, and acts as a germination stimulant for root-parasitic plants [35].
Because the transformation method for Striga has not yet been established, a genetic approach is not available to identify the components related to SL-dependent germination using root-parasitic plants. Therefore, recent progress in this field has instead been achieved using chemical tools, including synthetic SL analogs/mimics and model plants that possess a similar pathway of SL signaling.
Similar molecular mechanisms of SL perception and signaling in parasitic and nonparasitic plants
Although SLs are secreted from host plants and used as gemination signals in root-parasitic plants, they also have important physiological roles, such as shoot branching suppression, root development, and stress responses, as endogenous phytohormones in the host plants [36–38]. The genetic components were initially identified by analyzing the phenotypes of mutants in model plants, including Arabidopsis and rice. The core regulatory components for SL signaling are believed to be three types of subcellular proteins: DWARF14 (D14) [39], D3/MORE AXILLARY GROWTH2 (MAX2) [40, 41], and D53/SUPPRESSOR OF MAX2 1 (SMAX1)-Like (SMXL) [42, 43].
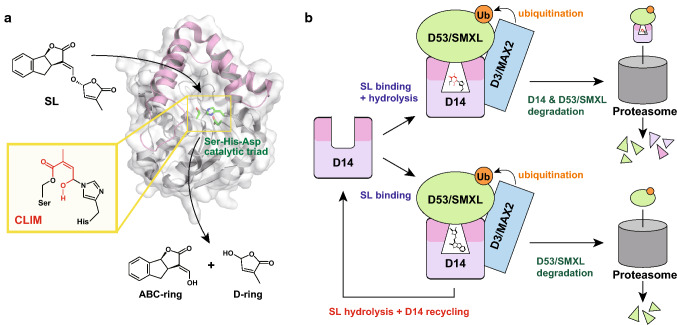
D14 is the most promising SL receptor and adopts an α/β hydrolase fold that is composed of a core domain with the conserved Ser-His-Asp catalytic triad and a helical cap domain [44–47] (Fig. 2a). Mutational analyses have shown that the catalytic triad is required for SL hydrolysis that breaks the bond between the ABC-ring and the D-ring [44, 47]. The remaining enzymatic activity is unusual in the ligand perception of phytohormone receptors [48]. SLs induce the interaction between D14 and D3/MAX2, which is a key molecular mechanism responsible for the activation of SL signaling (Fig. 2b). D3/MAX2 is a member of the leucine-rich repeat (LRR) family, composed of an N-terminal F-box domain and a C-terminal LRR domain with 19 LRRs in tandem [49, 50]. The F-box domain of MAX2 interacts with ARABIDOPSIS SKP-LIKE 1 (ASK1), which is a component of the SKP1-Cullin-F-box (SCF) ubiquitin ligase complex [51]. Therefore, D3/MAX2 is involved in SL signaling through a ubiquitin–proteasome system. According to this mechanistic insight, D53/SMXL and the DELLA protein SLENDER RICE1 (SLR1) were identified as SL-dependent interaction factors of D14 [42, 43, 46] (Fig. 2b). D53/SMXL is also degraded as a target substrate of D3/MAX2 in SL signaling, and the rice d53 mutant showed an exaggerated number of tillers compared to wild-type plants and was insensitive to exogenous treatment of 5-DS or GR24 [42, 43]. Although SMAX1 and seven SMXL proteins (SMXL2‒8) were identified in Arabidopsis, only SMXL6, 7, and 8 were responsible for the SL-dependent regulation of shoot branching [52, 53]. D53/SMXL belongs to the p-loop nucleoside triphosphate hydrolase superfamily, consisting of an N-terminal domain, a D1 ATPase domain, an M domain, and a D2 ATPase domain, similar to the chaperone protein HSP101. The D1 and D2 ATPase domains are responsible for SL-dependent interactions with D14 [43, 54]. In addition, the D2 ATPase domain contains the ethylene response element binding factor-associated amphiphilic repression (EAR) motifs for the interaction with TOPLESS corepressor, which regulates the transcription of many genes for plant development and phytohormone response [42, 55]. In fact, D53 interacts with rice TOPLESS-related proteins (TPRs), especially TPR2 [42]. These results propose that D53/SMXL acts as a repressor of SL signaling through the interaction of TOPLESS and TPRs. One of EAR motifs in D53 binds to the conserved TOPLESS domain (TPD) of TPR2 to mediate TPD tetramer–tetramer interaction, which promotes the assembly of D53–TPR2–nucleosome complex [55]. On the other hand, SLR1 is a repressor of gibberellin (GA) signaling in rice [56], and GA is a phytohormone that suppresses the shoot branching of plants. Therefore, the interaction between D14 and SLR1 suggests cross-talk between SL and GA signaling in a branching regulation [46].
Fig. 2.
Molecular function of D14 as an SL receptor. a SL hydrolysis in the catalytic pocket of D14. CLIM is an intermediate covalently bound to catalytic residues in the hydrolysis process. The helical cap domain is colored in pink. b Schematic diagrams of SL signaling mediated by D14, D3/MAX2, and D53/SMXL. In the upper pathway, SL hydrolysis is required to form ternary complexes of D14, D3/MAX2, and D53/SMXL. The lower pathway shows that SL is hydrolyzed for the deactivation and recycling of D14
Recent biochemical and structural analyses proposed the mechanistic basis for the SL-dependent formation of the D14‒D3/MAX2 complex that eventually degrades D53/SMXL through its ubiquitination [50, 57]. The interaction with MAX2/D3 in the presence of SLs induced a conformational change in the helical cap domain of Arabidopsis thaliana D14 (AtD14), and SL was broken down into the intermediate D-ring molecule (CLIM) at the active site [50] (Fig. 2b). CLIM was proposed to have structures that covalently link with the Ser and His residues of the catalytic triad (Fig. 2a). The covalent linkage of the His residue with the D-ring was also detected by mass spectrometry [57]. These findings concluded that SL hydrolysis was required for the complex formation of D14 with D3/MAX2. However, Seto et al. more recently reported that the AtD14 mutant at the catalytic Asp residue (D218A mutant) was still capable of interacting with MAX2 and SMXL7, complementing the atd14 mutant phenotype in an SL-dependent manner, although the D218A mutant lacked the hydrolytic activity of SL [58]. Based on these results, an intact SL molecule induces the formation of the D14‒D3/MAX2‒D53/SMXL complex, and D14 is deactivated after SL hydrolysis (Fig. 2b). Another possible mechanism of SL signaling deactivation involves AtD14 degradation from the MAX2-dependent proteasomal pathway after SL perception [59]. Thus, the accumulated evidence suggests the possibility of multiple SL perception mechanisms and interaction plasticity [46, 54]. According to these mechanisms, the bioactiphore of SLs is probably required for their hydrolysis to induce the formation of SL signaling complex and/or to deactivate the signaling.
S. hermonthica has the orthologous genes D14 (ShD14) and D3/MAX2 (ShMAX2), and hence evolutionally conserves a similar SL perception system to host plants. However, no obvious defects in seed germination were observed in the loss-of-function atd14 mutant. In Arabidopsis, HYPOSENSITIVE TO LIGHT/KARRIKIN INSENSITIVE2 (HTL/KAI2) was identified as a seed germination regulator that belongs to the closely related α/β-hydrolase of D14, and its loss-of-function mutant showed a hyperdormant phenotype [60, 61]. These genetic studies elucidated that HTL/KAI2 and MAX2 were necessary for the responses to exogenous SLs and other germination stimulants, including karrikins (KARs) [62–64], which are derived from the smoke of burning vegetation and induce seed germination after forest fires [65–67]. Although HTL/KAI2 is SL-responsive as well as D14, it recognizes different stereoisomers of SL [46, 63, 68]. D14 responses specific to naturally occurring SLs with R-configuration at C-2′ of the D-ring and HTL/KAI2 exhibit selectivity to non-natural S-configuration.
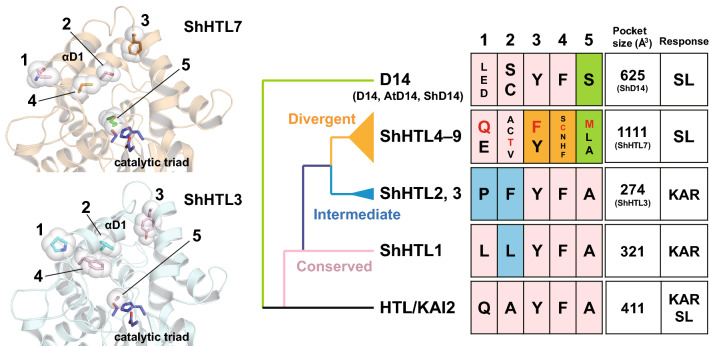
HTL/KAI2 paralogous genes are present in higher copy numbers in seed-parasitic plants than nonparasitic plants. In S. hermonthica, 11 HTL/KAI2 paralogs were identified and categorized into three phylogenetic clades: the conserved clade (ShHTL1/ShKAI2c), intermediate clade (ShHTL2 and ShHTL3/ShKAI2i), and divergent clade (ShHTL4/ShKAI2d3 to ShHTL11/ShKAI2d9) [69–71] (Fig. 3). All of them possess the conserved Ser-His-Asp catalytic triad and share high sequence similarity. However, ShHTLs belonging to different clades show different responses to SLs and/or KARs in cross-species complementation assays in which ShHTL genes were transformed into Arabidopsis htl/kai2 mutants [69–71]. ShHTL4–9 belong to a parasite-specific clade and recovered seed germination in the loss-of-function mutant by responding to SLs but not KARs. Therefore, ShHTL4–9 from divergent clades are proposed to be SL receptors in root-parasitic plants and are optimized to receive natural exogenous SLs. The SL/KAR responsiveness can be explained by the biochemical properties of ShHTLs among the three phylogenetic clades [72, 73]. The divergent clade (ShHTL4 and ShHTL7) showed hydrolytic activity toward GR24. In contrast, the conserved (ShHTL1) and intermediate (ShHTL3) clades possess KAR-binding properties without hydrolytic activity toward SLs (Fig. 3). According to the isothermal titration calorimetry (ITC) experiments, the binding affinity of ShHTL1 and ShHTL3 toward KAR1 was slightly higher than that of HTL/KAI2. These SL/KAR selectivities are correlated to the size of catalytic pockets (Fig. 3), which were evaluated by a comparative analysis with the crystal structures of ShHTLs belonging to the three clades [71, 73]. The pocket size is affected by the arrangement of helix αD1 on the helical cap domain and the types of residues located in the pocket [73] (Fig. 3). Among the ShHTLs of the divergent clade, ShHTL7, which has the largest pocket size compared to the remaining ShHTLs, has evolved with unbulky residues. These structural bases may help to develop the chemical compounds that can be used for separately controlling the different clades of ShHTLs to their function in the seed germination of root-parasitic plants.
Fig. 3.
Structural basis of the molecular evolution of ShHTLs. The catalytic pocket is narrowed by the residues in the blue box (positions 1 and 2 of the intermediate clade and position 1 of the conserved clade) to prevent SL binding. The residue at position 1 affects the arrangement of helix αD1 on the helical cap domain. The residues in the orange box (positions 3 and 4 of divergent clade) contribute to expanding the catalytic pocket to accept various types of SLs. D14 and ShHTLs of the divergent clade are not capable of binding KAR due to steric hindrance with the residues in the green box (position 5). The residues of ShHTL3 and ShHTL7 (colored in red) are shown on their structure models
ShHTL7 and ShHTL4 are capable of binding to ShMAX2 (the D3/MAX2 protein is conserved in S. hermonthica) in an SL-dependent manner, whereas this interaction with ShMAX2 is not detected in other clades of ShHTLs (ShHTL1 and ShHTL3) [73, 74]. These observations suggest that the SL-induced seed germination process of S. hermonthica shares a similar SL perception mechanism to nonparasitic plants using the divergent clades of ShHTLs instead of D14. However, the role of SL hydrolysis and downstream signaling components are not yet fully understood in seed germination stimulation through the divergent clades of ShHTLs, and the target molecules for ShMAX2-dependent proteolysis are likely to be different from shoot branching inhibition through D14.
Development of SL agonists and antagonists
To address the characterization of the parasitic SL receptors with a chemical biology approach because of unavailable genetic approaches, the fluorogenic SL mimic, Yoshimulactone Green (YLG) (Fig. 4), was designed based on the SL hydrolytic activity of the ShHTLs [70]. Competitive binding assays using YLG revealed that ShHTL6 and ShHTL7 exerted high sensitivities to GR24 and strong responses to various SLs [70, 71]. These characteristics are relative to larger pockets than the other ShHTLs, because there is less chance for steric hindrance to occur upon SL binding [73]. A variant of YLG with two D-rings (YLGW) was improved to eliminate background fluorescence and was applied to visualize the dynamic behavior of SL signaling in S. hermonthica by utilizing its activity as an SL agonist [70] (Fig. 4). Root elongation occurred after three phases with different fluorescent patterns (initial perception, wake-up wave, and pregermination pause). YLGW fluorescence was strongly detected at the root tip and then gradually spread toward the cotyledon during root elongation. Such a dynamic manner of YLGW fluorescence suggests that the induction of root germination is intricately regulated through SL receptor activation and degradation, amplification from ethylene production, and cell-to-cell signal spread [16].
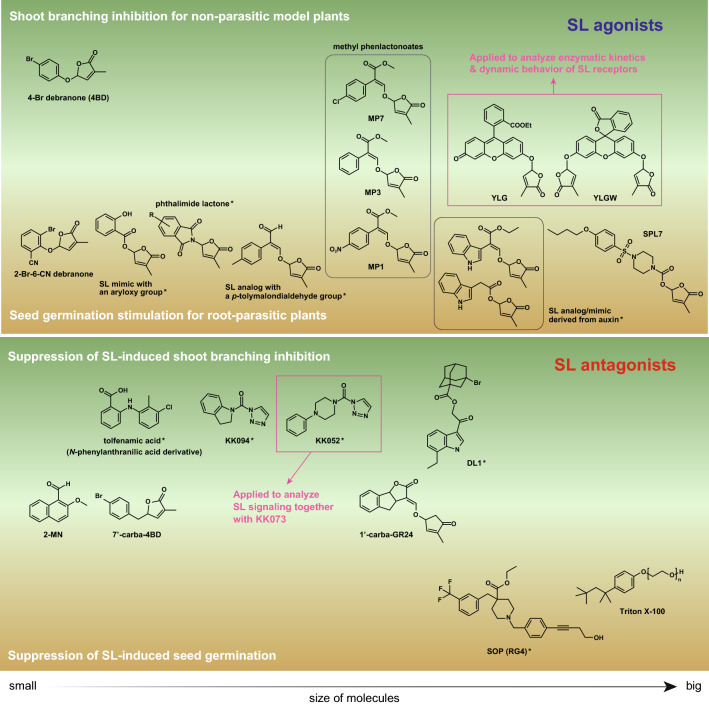
Fig. 4.
Synthetic SL agonists and antagonists categorized based on their effects on shoot branching (green field) and seed germination (ocher field). The compounds exhibiting both effects are located in the middle field. Only one effect was evaluated in the compounds with an asterisk, and the compounds in pink boxes were used to analyze the molecular mechanisms of the SL receptors
SLs play an essential role in the seed germination mechanisms of root-parasitic plants. However, the practical use of natural SLs is limited because of their uneconomical and complicated syntheses and instability; hence, artificial SL analogs, including GR24 and Nij-1, have been developed so far. In addition, synthetic compounds with defective bioactiphore of SLs have been explored as SL mimics that can exert a part of SL functions. Debranones (phenoxyfuranone compounds) are the simplest SL mimics and function as shoot branching inhibitors similar to GR24 and modestly induce the seed germination of Striga [75, 76] (Fig. 4). Chemical modification improved the stimulation activity of debranones for Striga seed germination [77]. The 2-bromo-6-nitrophenyl derivative was the most effective at inducing Striga seed germination among all tested debranones. In other synthetic SL analogs/mimics, SL analog with p-tolylmalondialdehyde group, and SL mimics with aryloxy or phthalimide groups effectively function as germination stimulants for Orobanche-type parasitic plants [78, 79]. Methyl phenlactonoates (MPs) are SL analogs that are designed by mimicking methyl CLA [80] (Fig. 4). Chemical modification affected the activity of the MPs in Striga seed germination and the shoot branching of the nonparasitic model plants. Among the tested MPs, MP1 showed the most efficient activity for Striga seed germination but lower activity than MP3 and MP7 for the inhibition of rice tiller outgrowths. Recently, hybrid-type SL analogs/mimics were newly designed by coupling the D-ring with another phytohormone auxin [81, 82] (Fig. 4) and stimulated the seed germination of three types of the parasitic plants, S. hermonthica, Phelipanche ramose, and Orobanche minor. The SL analog derived from auxin, especially, is more effective toward O. minor than GR24. The higher activity was observed in the analogs/mimics with a natural monomethylated D-ring than in the derivatives with the di- or trimethylated D-ring.
These SL analogs/mimics can be referred to as SL agonists that are targeted to the D14/HTL-type SL receptors in both parasitic and nonparasitic plants because of their common structural features. Uraguchi et al. designed an SL mimic, sphynolactone-7 (SPL7), to have a high affinity for ShHTL7 [83] (Fig. 4). In the competitive binding assays using YLG, the IC50 value for ShHTL7 was 0.31 μM, but almost all other ShHTLs and AtD14 showed an IC50 value of > 10 μM. SPL7 also showed a Striga-selective SL agonistic activity in the femtomolar range. These results support that ShHTL7 can play a crucial role in the SL-dependent seed germination of S. hermonthica, and is a major target for developing suicidal germination stimulants. Although there is no structural information of SPL7 bound to ShHTL7, it is speculated that sulfonyl piperazine moiety at the ABC-ring portion of SLs contributes to the selectivity to ShHTL7 and the femtomolar-rage potency [83]. Practical SL agonists as suicidal germination stimulants need to fulfill all requirements for Striga control in the field. Successful field trial for suicidal germination activity was performed in three SL analogs, Nij-1, MP1, and MP3 [84]. These formulated compounds reduced the number of emerged Striga plants to 39‒65% in farmer’s fields of pearl millet. Formulated MP1 also showed 42% reduction of Striga emergence in the sorghum field.
Many types of SL antagonists have also been discovered with in silico and/or chemical library screening to control the second tiller elongation of the host plants and the seed germination of S. hermonthica (Fig. 4). 2-Methoxy-1-naphthaldehyde (2-MN) was identified with in silico screening based on the structure of D14 complexed with the D-ring [46, 85]. 2-MN inhibits the D14‒D53 and D14‒SLR1 interactions to restore the second tiller elongation and suppresses the SL-dependent germination of Striga seeds [85]. DL1 and N-phenylanthranilic acid derivatives exert the inhibitory activity of the SL receptor and increase the number of shoot branches [86, 87]. These compounds were identified by high-throughput screening using a YLG competition assay or differential scanning fluorimetry (DSF). On the other hand, soporidine (SOP) and Triton X-100 act as SL antagonists that inhibit the germination of S. hermonthica in the presence of SLs [88, 89]. Triton X-100 specifically sealed the entrance of the catalytic pocket of ShHTL7.
Carba-SLs are the most simply designed SL antagonists, but their actions are suggestive of the mechanism of SL signaling activation [90]. In these compounds, the ether oxygen of the D-ring or the phenol ether oxygen of the synthetic SL agonists (GR24 and 4-bromodebranone) was replaced with a methylene group (Fig. 4). All carba-SLs are not cleaved by D14 hydrolytic activity; hence, they inhibit the interaction between D14 and D53 and suppress the SL-induced inhibition of rice tiller outgrowths. SL-induced seed germination of S. hermonthica is also suppressed by carba-SLs, which are capable of binding to the catalytic pocket of ShHTL7. Recently, Nakamura et al. reported SL agonists/antagonists of triazole urea derivatives (KK compounds) (Fig. 4), which are able to form a covalent bond with the catalytic Ser residue of D14 [91]. Among them, KK094 showed the most potent suppressing effect on the SL-dependent inhibition of second tiller elongation. KK052 also acts as an inhibitor of D14, whereas KK073, with a trifluoromethyl group on the benzene ring of KK052, is capable of inducing the complex formation of D14 with D53 or SLR1. Therefore, the antagonistic and agonistic effects on the SL receptor are switched by the chemical modification of KK compounds, suggesting that these discovered compounds could also be useful to further elucidate the molecular mechanisms of SL signaling.
Summary and future work
In addition to the development of SL analogs based on the bioactiphore of SLs, the understanding of D14 and ShHTLs as SL receptors has helped the design of several types of synthetic compounds that exert activity as SL agonists or antagonists. Some of these compounds have the potential to suppress the germination of Striga seeds or deplete the seed stocks in the soil as suicidal germination stimulants, because they show better stability compared with the natural SLs and/or selectivity toward root-parasitic plants, including S. hermonthica. On the other hand, further mechanistic findings may lead to a new approach to suppress the growth and expansion of root-parasitic plants. Based on the unraveled mechanism of SL-dependent seed germination in S. hermonthica, the SL perception system shares similar components between parasitic plants (ShHTLs and ShMAX2) and nonparasitic model plants (D14 and D3/MAX2). However, there are several remaining questions that are yet to be described: for example, the molecular mechanism from SL perception to signal transduction (including the target molecule of the SL receptor complex formed by ShHTLs and ShMAX2), the physiological meaning of the molecular diversity of ShHTLs, and the dynamic behavior of SL signaling in seed germination, which may be characteristic of parasitic plants.
Abbreviations
- 2-MN
2-Methoxy-1-naphthaldehyde
- 5-DS
5-Deoxystrigol
- ASK1
ARABIDOPSIS SKP-LIKE 1
- ATPase
Adenosine triphosphatase
- CL
Carlactone
- CLA
Carlactonoate
- CLIM
Covalently linked intermediate molecule
- D14
DWARF14
- DSF
Differential scanning fluorimetry
- EAR
Element binding factor-associated amphiphilic repression
- GA
Gibberellin
- HTL
HYPOSENSITIVE TO LIGHT
- ITC
Isothermal titration calorimetry
- KAI2
KARRIKIN INSENSITIVE2
- KAR
Karrikin
- LGS1
LOW GERMINATION STIMULANT 1
- LRR
Leucine-rich repeat
- MAX2
MORE AXILLARY GROWTH2
- MP
Methyl phenlactonoate
- Nij-1
Nijmegen-1
- SCF
SKP1-Cullin-F-box
- SL
Strigolactone
- SLR1
SLENDER RICE1
- SMAX1
SUPPRESSOR OF MAX2 1
- SMXL
SMAX1-Like
- SOP
Soporidine
- SPL7
Sphynolactone-7
- TPR
TOPLESS-related protein
- YLG
Yoshimulactone Green
- YLGW
YLG double
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Westwood JH, Yoder JI, Timko MP, dePamphilis CW. The evolution of parasitism in plants. Trends Plant Sci. 2010;15(4):227–235. doi: 10.1016/j.tplants.2010.01.004. [DOI] [PubMed] [Google Scholar]
- 2.Ejeta G. The Striga scourge in Africa: A growing pandemic. In: Ejeta G, Gressel J, editors. Integrating new technologies for Striga control: towards ending the witch-hunt. 1. Singapore: World Scientific; 2007. pp. 3–16. [Google Scholar]
- 3.Parker C. Parasitic weeds: a world challenge. Weed Sci. 2012;60(2):269–276. [Google Scholar]
- 4.Scholes JD, Press MC. Striga infestation of cereal crops—an unsolved problem in resource limited agriculture. Curr Opin Plant Biol. 2008;11(2):180–186. doi: 10.1016/j.pbi.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 5.Rodenburg J, Demont M, Zwart SJ, Bastiaans L. Parasitic weed incidence and related economic losses in rice in Africa. Agric Ecosyst Environ. 2016;235:306–317. [Google Scholar]
- 6.Berner DK, Kling JG, Singh BB. Striga research and control: a perspective from Africa. Plant Dis. 1995;79(7):652–660. [Google Scholar]
- 7.Atera E, Itoh K. Evaluation of ecologies and severity of Striga weed on rice in sub-Saharan Africa. Agric Biol J N Am. 2011;2(5):752–760. [Google Scholar]
- 8.Gurney AL, Slate J, Press MC, Scholes JD. A novel form of resistance in rice to the angiosperm parasite Striga hermonthica. New Phytol. 2006;169(1):199–208. doi: 10.1111/j.1469-8137.2005.01560.x. [DOI] [PubMed] [Google Scholar]
- 9.Li J, Timko MP. Gene-for-gene resistance in Striga-cowpea associations. Science. 2009;325(5944):1094. doi: 10.1126/science.1174754. [DOI] [PubMed] [Google Scholar]
- 10.Gobena D, Shimels M, Rich PJ, Ruyter-spira C, Bouwmeester H, Kanuganti S. Mutation in sorghum LOW GERMINATION STIMULANT 1 alters strigolactones and causes Striga resistance. Proc Natl Acad Sci USA. 2017;114(17):4471–4476. doi: 10.1073/pnas.1618965114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zwanenburg B, Mwakaboko AS, Reizelman A, Anilkumar G, Sethumadhavan D. Structure and function of natural and synthetic signalling molecules in parasitic weed germination. Pest Manag Sci. 2009;65(5):478–491. doi: 10.1002/ps.1706. [DOI] [PubMed] [Google Scholar]
- 12.Zwanenburg B, Blanco-Ania D. Strigolactones: new plant hormones in the spotlight. J Exp Bot. 2018;69(9):2205–2218. doi: 10.1093/jxb/erx487. [DOI] [PubMed] [Google Scholar]
- 13.Yoneyama K, Xie X, Yoneyama K, Kisugi T, Nomura T, Nakatani Y, Akiyama K, McErlean CSP. Which are the major players, canonical or non-canonical strigolactones? J Exp Bot. 2018;69(9):2231–2239. doi: 10.1093/jxb/ery090. [DOI] [PubMed] [Google Scholar]
- 14.Takahashi I, Asami T. Target-based selectivity of strigolactone agonists and antagonists in plants and their potential use in agriculture. J Exp Bot. 2018;69(9):2241–2254. doi: 10.1093/jxb/ery126. [DOI] [PubMed] [Google Scholar]
- 15.Brun G, Braem L, Thoiron S, Gevaert K, Goormachtig S, Delavault P. Seed germination in parasitic plants: what insights can we expect from strigolactone research? J Exp Bot. 2018;69(9):2265–2280. doi: 10.1093/jxb/erx472. [DOI] [PubMed] [Google Scholar]
- 16.Tsuchiya Y, Yoshimura M, Hagihara S. The dynamics of strigolactone perception in Striga hermonthica: a working hypothesis. J Exp Bot. 2018;69(9):2281–2290. doi: 10.1093/jxb/ery061. [DOI] [PubMed] [Google Scholar]
- 17.Lumba S, Holbrook-Smith D, McCourt P. The perception of strigolactones in vascular plants. Nat Chem Biol. 2017;13(6):599–606. doi: 10.1038/nchembio.2340. [DOI] [PubMed] [Google Scholar]
- 18.Waters MT, Gutjahr C, Bennett T, Nelson DC. Strigolactone signaling and evolution. Annu Rev Plant Biol. 2017;68:291–322. doi: 10.1146/annurev-arplant-042916-040925. [DOI] [PubMed] [Google Scholar]
- 19.Cook CE, Whichard LP, Turner B, Wall ME, Egley GH. Germination of Witchweed (Striga lutea Lour.): isolation and properties of a potent stimulant. Science. 1966;154(3753):1189–1190. doi: 10.1126/science.154.3753.1189. [DOI] [PubMed] [Google Scholar]
- 20.Cook CE, Whichard LP, Wall ME, Egley GH, Coggon P, Luhan PA, McPhail AT. Germination stimulants. II. The structure of strigol—a potent seed germination stimulant for witchweed (Striga lutea Lour.) J Am Chem Soc. 1972;94(17):6198–6199. [Google Scholar]
- 21.Brooks DW, Bevinakatti HS, Powell DR. The absolute structure of (+)-strigol. J Org Chem. 1985;50(20):3779–3781. [Google Scholar]
- 22.Yokota T, Sakai H, Okuno K, Yoneyama K, Takeuchi Y. Alectrol and orobanchol, germination stimulants for Orobanche minor, from its host red clover. Phytochemistry. 1998;49(7):1967–1973. [Google Scholar]
- 23.Xie X, Kusumoto D, Takeuchi Y, Yoneyama K, Yamada Y, Yoneyama K. 2′-epi-orobanchol and solanacol, two unique strigolactones, germination stimulants for root parasitic weeds, produced by tobacco. J Agric Food Chem. 2007;55(20):8067–8072. doi: 10.1021/jf0715121. [DOI] [PubMed] [Google Scholar]
- 24.Hauck C, Muller S, Schildknecht H. A germination stimulant for parasitic flowering plants from Sorghum bicolor, a genuine host plant. J Plant Physiol. 1992;139(4):474–478. [Google Scholar]
- 25.Xie X, Yoneyama K, Yoneyama K. The strigolactone story. Ann Rev Phytopathol. 2010;48:93–117. doi: 10.1146/annurev-phyto-073009-114453. [DOI] [PubMed] [Google Scholar]
- 26.Zwanenburg B, Pospíšil T, Ćavar Zeljković S. Strigolactones: new plant hormones in action. Planta. 2016;243(6):1311–1326. doi: 10.1007/s00425-015-2455-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zwanenburg B, Nayak SK, Charnikhova TV, Bouwmeester HJ. New strigolactone mimics: structure–activity relationship and mode of action as germinating stimulants for parasitic weeds. Bioorg Med Chem Lett. 2013;23(18):5182–5186. doi: 10.1016/j.bmcl.2013.07.004. [DOI] [PubMed] [Google Scholar]
- 28.Yoneyama K, Arakawa R, Ishimoto K, Kim HI, Kisugi T, Xie X, Nomura T, Kanampiu F, Yokota T, Ezawa T, Yoneyama K. Difference in Striga-susceptibility is reflected in strigolactone secretion profile, but not in compatibility and host preference in arbuscular mycorrhizal symbiosis in two maize cultivars. New Phytol. 2015;206(3):983–989. doi: 10.1111/nph.13375. [DOI] [PubMed] [Google Scholar]
- 29.Mohemed N, Charnikhova T, Bakker EJ, van Ast A, Babiker AG, Bouwmeester HJ. Evaluation of field resistance to Striga hermonthica (Del.) Benth. in Sorghum bicolor (L.) Moench. The relationship with strigolactones. Pest Manag Sci. 2016;72(11):2082–2090. doi: 10.1002/ps.4426. [DOI] [PubMed] [Google Scholar]
- 30.Mangnus EM, Zwanenburg B. Tentative molecular mechanism for germination stimulation of Striga and Orobanche seeds by strigol and its synthetic analogs. J Agric Food Chem. 1992;40(6):1066–1070. [Google Scholar]
- 31.Yoneyama K, Xie X, Kim HI, Kisugi T, Nomura T, Sekimoto H, Yokota T, Yoneyama K. How do nitrogen and phosphorus deficiencies affect strigolactone production and exudation? Planta. 2012;235(6):1197–1207. doi: 10.1007/s00425-011-1568-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johnson AW, Gowda G, Hassanali A, Knox J, Monaco S, Razavi Z, Rosebery G. The preparation of synthetic analogues of strigol. J Chem Soc Perkin. 1981;1(6):1734–1743. [Google Scholar]
- 33.Nefkens GHL, Thuring JWJF, Beenakkers MFM, Zwanenburg B. Synthesis of a phthaloylglycine-derived strigol analogue and its germination stimulatory activity toward seeds of the parasitic weeds Striga hermonthica and Orobanche crenata. J Agric Food Chem. 1997;45(6):2273–2277. [Google Scholar]
- 34.Zwanenburg B, Mwakaboko AS, Kannan C. Suicidal germination for parasitic weed control. Pest Manag Sci. 2016;72(11):2016–2025. doi: 10.1002/ps.4222. [DOI] [PubMed] [Google Scholar]
- 35.Xie X, Kisugi T, Yoneyama K, Nomura T, Akiyama K, Uchida K, Yokota T, McErlean CSP, Yoneyama K. Methyl zealactonoate, a novel germination stimulant for root parasitic weeds produced by maize. Pestic Sci. 2017;42(2):58–61. doi: 10.1584/jpestics.D16-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gomez-Roldan V, Fermas S, Brewer PB, Puech-Pagès V, Dun EA, Pillot JP, Letisse F, Matusova R, Danoun S, Portais JC, Bouwmeester H, Bécard G, Beveridge CA, Rameau C, Rochange SF. Strigolactone inhibition of shoot branching. Nature. 2008;455(7210):189–194. doi: 10.1038/nature07271. [DOI] [PubMed] [Google Scholar]
- 37.Umehara M, Hanada A, Yoshida S, Akiyama K, Arite T, Takeda-Kamiya N, Magome H, Kamiya Y, Shirasu K, Yoneyama K, Kyozuka J, Yamaguchi S. Inhibition of shoot branching by new terpenoid plant hormones. Nature. 2008;455(7210):195–200. doi: 10.1038/nature07272. [DOI] [PubMed] [Google Scholar]
- 38.Brewer PB, Koltai H, Beveridge CA. Diverse roles of strigolactones in plant development. Mol Plant. 2013;6(1):18–28. doi: 10.1093/mp/sss130. [DOI] [PubMed] [Google Scholar]
- 39.Arite T, Umehara M, Ishikawa S, Hanada A, Maekawa M, Yamaguchi S, Kyozuka J. d14, a strigolactone-insensitive mutant of rice, shows an accelerated outgrowth of tillers. Plant Cell Physiol. 2009;50(8):1416–1424. doi: 10.1093/pcp/pcp091. [DOI] [PubMed] [Google Scholar]
- 40.Ishikawa S, Maekawa M, Arite T, Onishi K, Takamure I, Kyozuka J. Suppression of tiller bud activity in tillering dwarf mutants of rice. Plant Cell Physiol. 2005;46(1):79–86. doi: 10.1093/pcp/pci022. [DOI] [PubMed] [Google Scholar]
- 41.Stirnberg P, Furner IJ, Ottoline Leyser HM. MAX2 participates in an SCF complex which acts locally at the node to suppress shoot branching. Plant J. 2007;50(1):80–94. doi: 10.1111/j.1365-313X.2007.03032.x. [DOI] [PubMed] [Google Scholar]
- 42.Jiang L, Liu X, Xiong G, Liu H, Chen F, Wang L, Meng X, Liu G, Yu H, Yuan Y, Yi W, Zhao L, Ma H, He Y, Wu Z, Melcher K, Qian Q, Xu HE, Wang Y, Li J. DWARF 53 acts as a repressor of strigolactone signalling in rice. Nature. 2013;504(7480):401–405. doi: 10.1038/nature12870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou F, Lin Q, Zhu L, Ren Y, Zhou K, Shabek N, Wu F, Mao H, Dong W, Gan L, Ma W, Gao H, Chen J, Yang C, Wang D, Tan J, Zhang X, Guo X, Wang J, Jiang L, Liu X, Chen W, Chu J, Yan C, Ueno K, Ito S, Asami T, Cheng Z, Wang J, Lei C, Zhai H, Wu C, Wang H, Zheng N, Wan J. D14–SCFD3-dependent degradation of D53 regulates strigolactone signalling. Nature. 2013;504(7480):406–410. doi: 10.1038/nature12878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hamiaux C, Drummond RS, Janssen BJ, Ledger SE, Cooney JM, Newcomb RD, Snowden KC. DAD2 is an α/β hydrolase likely to be involved in the perception of the plant branching hormone, strigolactone. Curr Biol. 2012;22(21):2032–2036. doi: 10.1016/j.cub.2012.08.007. [DOI] [PubMed] [Google Scholar]
- 45.Kagiyama M, Hirano Y, Mori T, Kim SY, Kyozuka J, Seto Y, Yamaguchi S, Hakoshima T. Structures of D14 and D14L in the strigolactone and karrikin signaling pathways. Genes Cells. 2013;18(2):147–160. doi: 10.1111/gtc.12025. [DOI] [PubMed] [Google Scholar]
- 46.Nakamura H, Xue YL, Miyakawa T, Hou F, Qin HM, Fukui K, Shi X, Ito E, Ito S, Park SH, Miyauchi Y, Asano A, Totsuka N, Ueda T, Tanokura M, Asami T. Molecular mechanism of strigolactone perception by DWARF14. Nat Commun. 2013;4:2613. doi: 10.1038/ncomms3613. [DOI] [PubMed] [Google Scholar]
- 47.Zhao LH, Zhou XE, Wu ZS, Yi W, Xu Y, Li S, Xu TH, Liu Y, Chen RZ, Kovach A, Kang Y, Hou L, He Y, Xie C, Song W, Zhong D, Xu Y, Wang Y, Li J, Zhang C, Melcher K, Xu HE. Crystal structures of two phytohormone signal-transducing α/β hydrolases: karrikin-signaling KAI2 and strigolactone-signaling DWARF14. Cell Res. 2013;23(3):436–439. doi: 10.1038/cr.2013.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Miyakawa T, Tanokura M. Structural basis for the regulation of phytohormone receptors. Biosci Biotechnol Biochem. 2017;81(7):1261–1273. doi: 10.1080/09168451.2017.1313696. [DOI] [PubMed] [Google Scholar]
- 49.Woo HR, Chung KM, Park JH, Oh SA, Ahn T, Hong SH, Jang SK, Nam HG. ORE9, an F-box protein that regulates leaf senescence in Arabidopsis. Plant Cell. 2001;13(8):1779–1790. doi: 10.1105/TPC.010061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yao R, Ming Z, Yan L, Li S, Wang F, Ma S, Yu C, Yang M, Chen L, Chen L, Li Y, Yan C, Miao D, Sun Z, Yan J, Sun Y, Wang L, Chu J, Fan S, He W, Deng H, Nan F, Li J, Rao Z, Lou Z, Xie D. DWARF14 is a non-canonical hormone receptor for strigolactone. Nature. 2016;536(7617):469–473. doi: 10.1038/nature19073. [DOI] [PubMed] [Google Scholar]
- 51.Zhao J, Wang T, Wang M, Liu Y, Yuan S, Gao Y, Yin L, Sun W, Peng L, Zhang W, Wan J, Li X. DWARF3 participates in an SCF complex and associates with DWARF14 to suppress rice shoot branching. Plant Cell Physiol. 2014;55(6):1096–1109. doi: 10.1093/pcp/pcu045. [DOI] [PubMed] [Google Scholar]
- 52.Wang L, Wang B, Jiang L, Liu X, Li X, Lu Z, Meng X, Wang Y, Smith SM, Li J. Strigolactone signaling in Arabidopsis regulates shoot development by targeting D53-like SMXL repressor proteins for ubiquitination and degradation. Plant Cell. 2015;27(11):3128–3142. doi: 10.1105/tpc.15.00605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Soundappan I, Bennett T, Morffy N, Liang Y, Stanga JP, Abbas A, Leyser O, Nelson DC. SMAX1-LIKE/D53 family members enable distinct MAX2-dependent responses to strigolactones and karrikins in Arabidopsis. Plant Cell. 2015;27(11):3143–3159. doi: 10.1105/tpc.15.00562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shabek N, Ticchiarelli F, Mao H, Hinds TR, Leyser O, Zheng N. Structural plasticity of D3–D14 ubiquitin ligase in strigolactone signalling. Nature. 2018;563(7733):652–656. doi: 10.1038/s41586-018-0743-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ma H, Duan J, Ke J, He Y, Gu X, Xu TH, Yu H, Wang Y, Brunzelle JS, Jiang Y, Rothbart SB, Xu HE, Li J, Melcher K. A D53 repression motif induces oligomerization of TOPLESS corepressors and promotes assembly of a corepressor–nucleosome complex. Sci Adv. 2017;3(6):e1601217. doi: 10.1126/sciadv.1601217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Itoh H, Ueguchi-Tanaka M, Sato Y, Ashikari M, Matsuoka M. The gibberellin signaling pathway is regulated by the appearance and disappearance of SLENDER RICE1 in nuclei. Plant Cell. 2002;14(1):57–70. doi: 10.1105/tpc.010319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.de Saint Germain A, Clavé G, Badet-Denisot MA, Pillot JP, Cornu D, Le Caer JP, Burger M, Pelissier F, Retailleau P, Turnbull C, Bonhomme S, Chory J, Rameau C, Boyer FD. An histidine covalent receptor and butenolide complex mediates strigolactone perception. Nat Chem Biol. 2016;12(10):787–794. doi: 10.1038/nchembio.2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Seto Y, Yasui R, Kameoka H, Tamiru M, Cao M, Terauchi R, Sakurada A, Hirano R, Kisugi T, Hanada A, Umehara M, Seo E, Akiyama K, Burke J, Takeda-Kamiya N, Li W, Hirano Y, Hakoshima T, Mashiguchi K, Noel JP, Kyozuka J, Yamaguchi S. Strigolactone perception and deactivation by a hydrolase receptor DWARF14. Nat Commun. 2019;10(1):191. doi: 10.1038/s41467-018-08124-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chevalier F, Nieminen K, Sanchez-Ferrero JC, Rodriguez ML, Chagoyen M, Hardtke CS, Cubas P. Strigolactone promotes degradation of DWARF14, an α/β hydrolase essential for strigolactone signaling in Arabidopsis. Plant Cell. 2014;26(3):1134–1150. doi: 10.1105/tpc.114.122903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sun XD, Ni M. HYPOSENSITIVE TO LIGHT, an alpha/beta fold protein, acts downstream of ELONGATED HYPOCOTYL 5 to regulate seedling de-etiolation. Mol Plant. 2011;4(1):116–126. doi: 10.1093/mp/ssq055. [DOI] [PubMed] [Google Scholar]
- 61.Waters MT, Nelson DC, Scaffidi A, Flematti GR, Sun YK, Dixon KW, Smith SM. Specialisation within the DWARF14 protein family confers distinct responses to karrikins and strigolactones in Arabidopsis. Development. 2012;139(7):1285–1295. doi: 10.1242/dev.074567. [DOI] [PubMed] [Google Scholar]
- 62.Nelson DC, Scaffidi A, Dun EA, Waters MT, Flematti GR, Dixon KW, Beveridge CA, Ghisalberti EL, Smith SM. F-box protein MAX2 has dual roles in karrikin and strigolactone signaling in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2011;108(21):8897–8902. doi: 10.1073/pnas.1100987108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Scaffidi A, Waters MT, Sun YK, Skelton BW, Dixon KW, Ghisalberti EL, Flematti GR, Smith SM. Strigolactone hormones and their stereoisomers signal through two related receptor proteins to induce different physiological responses in Arabidopsis. Plant Physiol. 2014;165(3):1221–1232. doi: 10.1104/pp.114.240036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Toh S, Holbrook-Smith D, Stokes ME, Tsuchiya Y, McCourt P. Detection of parasitic plant suicide germination compounds using a high-throughput Arabidopsis HTL/KAI2 strigolactone perception system. Chem Biol. 2014;21(8):988–998. doi: 10.1016/j.chembiol.2014.07.005. [DOI] [PubMed] [Google Scholar]
- 65.Flematti GR, Ghisalberti EL, Dixon KW, Trengove RD. A compound from smoke that promotes seed germination. Science. 2004;305(5686):977. doi: 10.1126/science.1099944. [DOI] [PubMed] [Google Scholar]
- 66.Flematti GR, Ghisalberti EL, Dixon KW, Trengove RD. Identification of alkyl substituted 2H-Furo[2,3-c]pyran-2-ones as germination stimulants present in smoke. J Agric Food Chem. 2009;57(20):9475–9480. doi: 10.1021/jf9028128. [DOI] [PubMed] [Google Scholar]
- 67.Nelson DC, Flematti GR, Ghisalberti EL, Dixon KW, Smith SM. Regulation of seed germination and seedling growth by chemical signals from burning vegetation. Annu Rev Plant Biol. 2012;63:107–130. doi: 10.1146/annurev-arplant-042811-105545. [DOI] [PubMed] [Google Scholar]
- 68.Flematti GR, Scaffidi A, Waters MT, Smith SM. Stereospecificity in strigolactone biosynthesis and perception. Planta. 2016;243(6):1361–1373. doi: 10.1007/s00425-016-2523-5. [DOI] [PubMed] [Google Scholar]
- 69.Conn CE, Bythell-Douglas R, Neumann D, Yoshida S, Whittington B, Westwood JH, Shirasu K, Bond CS, Dyer KA, Nelson DC. Convergent evolution of strigolactone perception enabled host detection in parasitic plants. Science. 2015;349(6247):540–543. doi: 10.1126/science.aab1140. [DOI] [PubMed] [Google Scholar]
- 70.Tsuchiya Y, Yoshimura M, Sato Y, Kuwata K, Toh S, Holbrook-Smith D, Zhang H, McCourt P, Itami K, Kinoshita T, Hagihara S. Probing strigolactone receptors in Striga hermonthica with fluorescence. Science. 2015;349(6250):864–868. doi: 10.1126/science.aab3831. [DOI] [PubMed] [Google Scholar]
- 71.Toh S, Holbrook-Smith D, Stogios PJ, Onopriyenko O, Lumba S, Tsuchiya Y, Savchenko A, McCourt P. Structure-function analysis identifies highly sensitive strigolactone receptors in Striga. Science. 2015;350(6257):203–207. doi: 10.1126/science.aac9476. [DOI] [PubMed] [Google Scholar]
- 72.Xu Y, Miyakawa T, Nakamura H, Nakamura A, Imamura Y, Asami T, Tanokura M. Structural basis of unique ligand specificity of KAI2-like protein from parasitic weed Striga hermonthica. Sci Rep. 2016;6:31386. doi: 10.1038/srep31386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Xu Y, Miyakawa T, Nosaki S, Nakamura A, Lyu Y, Nakamura H, Ohto U, Ishida H, Shimizu T, Asami T, Tanokura M. Structural analysis of HTL and D14 proteins reveals the basis for ligand selectivity in Striga. Nat Commun. 2018;9(1):3947. doi: 10.1038/s41467-018-06452-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yao R, Wang F, Ming Z, Du X, Chen L, Wang Y, Zhang W, Deng H, Xie D. ShHTL7 is a non-canonical receptor for strigolactones in root parasitic weeds. Cell Res. 2017;27(6):838–841. doi: 10.1038/cr.2017.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fukui K, Ito S, Ueno K, Yamaguchi S, Kyozuka J, Asami T. New branching inhibitors and their potential as strigolactone mimics in rice. Bioorg Med Chem Lett. 2011;21(16):4905–4908. doi: 10.1016/j.bmcl.2011.06.019. [DOI] [PubMed] [Google Scholar]
- 76.Fukui K, Ito S, Asami T. Selective mimics of strigolactone actions and their potential use for controlling damage caused by root parasitic weeds. Mol Plant. 2013;6(1):88–99. doi: 10.1093/mp/sss138. [DOI] [PubMed] [Google Scholar]
- 77.Fukui K, Yamagami D, Ito S, Asami T. A taylor-made design of phenoxyfuranone-type strigolactone mimic. Front Plant Sci. 2017;8:936. doi: 10.3389/fpls.2017.00936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zwanenburg B, Mwakaboko AS. Strigolactone analogues and mimics derived from phthalimide, saccharine, p-tolylmalondialdehyde, benzoic and salicylic acid as scaffolds. Bioorg Med Chem. 2011;19(24):7394–7400. doi: 10.1016/j.bmc.2011.10.057. [DOI] [PubMed] [Google Scholar]
- 79.Cala A, Ghooray K, Fernandez-Aparicio M, Molinillo JM, Galindo JC, Rubiales D, Macias FA. Phthalimide-derived strigolactone mimics as germinating agents for seeds of parasitic weeds. Pest Manag Sci. 2016;72(11):2069–2081. doi: 10.1002/ps.4323. [DOI] [PubMed] [Google Scholar]
- 80.Jamil M, Kountche BA, Haider I, Guo X, Ntui VO, Jia KP, Ali S, Hameed US, Nakamura H, Lyu Y, Jiang K, Hirabayashi K, Tanokura M, Arold ST, Asami T, Al-Babili S. Methyl phenlactonoates are efficient strigolactone analogs with simple structure. J Exp Bot. 2018;69(9):2319–2331. doi: 10.1093/jxb/erx438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Blanco-Ania D, Mateman JJ, Hýlová A, Spíchal L, Debie LM, Zwanenburg B. Hybrid-type strigolactone analogues derived from auxins. Pest Manag Sci. 2019 doi: 10.1002/ps.5553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hýlová A, Pospíšil T, Spíchal L, Mateman JJ, Blanco-Ania D, Zwanenburg B. New hybrid type strigolactone mimics derived from plant growth regulator auxin. N Biotechnol. 2019;48:76–82. doi: 10.1016/j.nbt.2018.08.001. [DOI] [PubMed] [Google Scholar]
- 83.Uraguchi D, Kuwata K, Hijikata Y, Yamaguchi R, Imaizumi H, Am S, Rakers C, Mori N, Akiyama K, Irle S, McCourt P, Kinoshita T, Ooi T, Tsuchiya Y. A femtomolar-range suicide germination stimulant for the parasitic plant Striga hermonthica. Science. 2018;362(6420):1301–1305. doi: 10.1126/science.aau5445. [DOI] [PubMed] [Google Scholar]
- 84.Kountche BA, Jamil M, Yonli D, Nikiema MP, Blanco-Ania D, Asami T, Zwanenburg B, Al-Babili S. Suicidal germination as a control strategy for Striga hermonthica (Benth.) in smallholder farms of sub‐Saharan Africa. Plants People Planet. 2019;1:107–118. [Google Scholar]
- 85.Mashita O, Koishihara H, Fukui K, Nakamura H, Asami T. Discovery and identification of 2-methoxy-1-naphthaldehyde as a novel strigolactone-signaling inhibitor. J Pestic Sci. 2016;41(3):71–78. doi: 10.1584/jpestics.D16-028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yoshimura M, Sato A, Kuwata K, Inukai Y, Kinoshita T, Itami K, Tsuchiya Y, Hagihara S. Discovery of shoot branching regulator targeting strigolactone receptor DWARF14. ACS Cent Sci. 2018;4(2):230–234. doi: 10.1021/acscentsci.7b00554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hamiaux C, Drummond RSM, Luo Z, Lee HW, Sharma P, Janssen BJ, Perry NB, Denny WA, Snowden KC. Inhibition of strigolactone receptors by N-phenylanthranilic acid derivatives: structural and functional insights. J Biol Chem. 2018;293(17):6530–6543. doi: 10.1074/jbc.RA117.001154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Holbrook-Smith D, Toh S, Tsuchiya Y, McCourt P. Small-molecule antagonists of germination of the parasitic plant Striga hermonthica. Nat Chem Biol. 2016;12(9):724–729. doi: 10.1038/nchembio.2129. [DOI] [PubMed] [Google Scholar]
- 89.Hameed US, Haider I, Jamil M, Kountche BA, Guo X, Zarban RA, Kim D, Al-Babili S, Arold ST. Structural basis for specific inhibition of the highly sensitive ShHTL7 receptor. EMBO Rep. 2018;19(9):e45619. doi: 10.15252/embr.201745619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Takeuchi J, Jiang K, Hirabayashi K, Imamura Y, Wu Y, Xu Y, Miyakawa T, Nakamura H, Tanokura M, Asami T. Rationally designed strigolactone analogs as antagonists of the D14 receptor. Plant Cell Physiol. 2018;59(8):1545–1554. doi: 10.1093/pcp/pcy087. [DOI] [PubMed] [Google Scholar]
- 91.Nakamura H, Hirabayashi K, Miyakawa T, Kikuzato K, Hu W, Xu Y, Jiang K, Takahashi I, Niiyama R, Dohmae N, Tanokura M, Asami T. Triazole ureas covalently bind to strigolactone receptor and antagonize strigolactone responses. Mol Plant. 2019;12(1):44–58. doi: 10.1016/j.molp.2018.10.006. [DOI] [PubMed] [Google Scholar]