Abstract
Etoposide-induced protein 2.4 (EI24), located on the endoplasmic reticulum (ER) membrane, has been proposed to be an essential autophagy protein. Specific ablation of EI24 in neuronal and liver tissues causes deficiency of autophagy flux. However, the molecular mechanism of the EI24-mediated autophagy process is still poorly understood. Like neurons and hepatic cells, pancreatic β cells are also secretory cells. Pancreatic β cells contain large amounts of ER and continuously synthesize and secrete insulin to maintain blood glucose homeostasis. Yet, the effect of EI24 on autophagy of pancreatic β cells has not been reported. Here, we show that the autophagy process is inhibited in EI24-deficient primary pancreatic β cells. Further mechanistic studies demonstrate that EI24 is enriched at the ER–mitochondria interface and that the C-terminal domain of EI24 is important for the integrity of the mitochondria-associated membrane (MAM) and autophagy flux. Overexpression of EI24, but not the EI24-ΔC mutant, can rescue MAM integrity and decrease the aggregation of p62 and LC3II in the EI24-deficient group. By mass spectrometry-based proteomics following immunoprecipitation, EI24 was found to interact with voltage-dependent anion channel 1 (VDAC1), inositol 1,4,5-trisphosphate receptor (IP3R), and the outer mitochondrial membrane chaperone GRP75. Knockout of EI24 impairs the interaction of IP3R with VDAC1, indicating that these proteins may form a quaternary complex to regulate MAM integrity and the autophagy process.
Electronic supplementary material
The online version of this article (10.1007/s00018-019-03236-9) contains supplementary material, which is available to authorized users.
Keywords: Phagophore, Autophagy initiation, Mitochondria-associated membrane, VDAC–IP3R complex
Introduction
Macroautophagy (hereafter referred to as autophagy) is an evolutionarily conserved and tightly regulated intracellular degradation and recycling process, and plays fundamental roles in maintaining cell growth, development, and homeostasis [1]. The process of autophagy has been well studied and dissected into several steps: phagophore initiation, elongation, autophagosome maturation, fusion with lysosomes to form autolysosomes, and degradation [2]. Dysregulation in any of these steps may lead to damage to the autophagy process, causing a variety of pathological diseases such as cancer, neurodegenerative disorders, and diabetes [3, 4].
Diabetes is a metabolic disease that mainly results from insufficient insulin secretion caused by dysfunction and/or reduction in the mass of pancreatic β cells [5]. In vivo and in vitro models containing a deletion of the autophagy-related gene 7 (ATG7), an essential molecule for the induction of autophagy, reveal progressive reduction of pancreatic β-cell mass and impaired glucose tolerance [6, 7]. Similar results are observed in mice with pancreas-specific disruption of autophagy-related gene 5 (ATG5), which develop reduced autophagy and an increased sign of cell necrosis [8]. Besides ATGs, our previous work found that etoposide-induced protein 2.4 (EI24) is essential for the survival and the mass of pancreatic β cells [9]. It has been reported that EI24 plays an important role in autophagy in neurons and the liver [10]. Neural-specific EI24 deficiency causes extensive neuronal loss in the brain and massive axon degeneration, while liver-specific depletion of EI24 leads to severe hepatomegaly with enlarged hepatocytes [10]. However, autophagy in a spleen-specific disruption of EI24 mice appears normal compared with neural and liver tissue-specific EI24 knockout (KO) mice [10], indicating that EI24 functions differently in different tissues. Pancreatic β cells are another kind of secretory cell that stores and releases insulin in response to increased blood glucose concentrations. However, the role of EI24 on the autophagy function of pancreatic β cells has not been reported.
Tian et al. reported that loss of function of EI24 in C. elegans causes severely retarded formation and closure of the phagophore membrane [11], suggesting that EI24 is involved in one of the earlier steps of autophagy. In the early events of autophagy, the initiation of autophagy begins with phagophore formation [12]. The source of the phagophore remains one of the most fundamental unresolved questions and is still hotly debated. In genetic screens in C. elegans, Tian et al. found that the earliest autophagic structures were surrounding the rough endoplasmic reticulum (ER) or the mitochondria, which revealed the two possible sources of the autophagosomal membrane [11]. Using electron tomography, a physical connection between the autophagosomes and the ER has been observed, suggesting that the phagophore initially associates with the ER [13]. It should be noted that about 30% of the autophagosome precursors were not part of the ER-isolated membrane complex by 3D tomographic assay [14], indicating that other membrane sources of the phagophore exist, such as the plasma membrane [15] and the Golgi [16]. More recently, the ER–mitochondria contact site has been proposed as the site of autophagosome formation [17]. Mounting evidence shows that proteins in the mitochondria-associated membrane (MAM) compartment are essential for proper autophagosome formation [18–20] and thus play a key role in autophagy. EI24 is an ER transmembrane protein [21] and whether it is involved in phagophore formation and the regulation of early events in autophagy has not been reported.
In this paper, we demonstrate that the autophagy process is inhibited in EI24 deficient primary pancreatic β cells, U-2 OS, and HEK293T cells. The C-terminal of EI24 is essential for the integrity of MAM and autophagy flux. We further show that EI24 interacts with IP3R, GRP57, and VDAC1. Deficiency of EI24 impairs interaction of IP3R with VDAC1, indicating that EI24 may form a quaternary complex with IP3R-GRP57-VDAC1 to facilitate the formation of ER–mitochondria associations that serve as phagophore initiation sites and promote autophagy flux.
Results
EI24 deficiency impairs autophagy
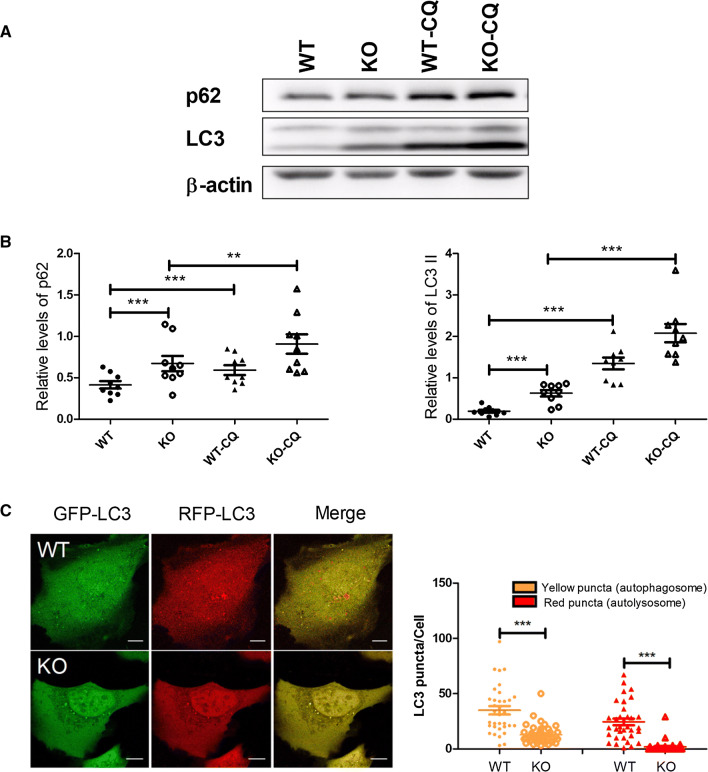
To investigate the role of EI24 on autophagy of pancreatic β cells, we generated β-cell-specific Ei24-deficient mice by crossing Ei24-conditional KO mice (Ei24flox/flox) with transgenic mice expressing Cre recombinase under the control of the β-cell-specific rat insulin promoter (Ins II) [9]. Histological analyses were performed by immunohistochemical staining of sections from the pancreas of wild-type (WT) and β-cell-specific EI24 knockout (KO) mice at 1 month of age. The results showed that p62, a hallmark of autophagy, was dramatically aggregated in the KO group compared to the WT group (Fig. 1a). p62 mRNA level was downregulated in EI24 KO pancreatic β cells (Fig. 1b), indicating that the accumulation of p62 protein is not due to the increased generation process. Western blot analysis of the levels of p62 and LC3II, a lipidated form of LC3I that associates with phagophore membranes, confirmed that autophagy was impaired severely (Fig. 1c, d). To explore the mechanism of how EI24 mediates the autophagy process, we knocked out EI24 using the CRISPR-Cas9 genomic-editing approach to generate HEK293T and U-2 OS cell lines (Fig. 1e, f), and found that levels of p62 and LC3II in EI24-deficiency groups were significantly higher (Fig. 1g, h), suggesting that the inhibition of autophagy by knocking out EI24 is a conserved phenomenon. With the treatment of chloroquine (CQ), which mainly inhibits autophagy by impairing autophagosome fusion with lysosome [22], the turnover of p62 and LC3II was further blocked (Fig. 2a, b), suggesting the impairment of the autophagy flux. Meanwhile, the KO group showed higher LC3 II levels vs WT, suggesting that the autophagosome formation was also impaired when the autophagic flux was blocked pharmacologically (Fig. 2a, b). To further monitor the function of EI24 in autophagy, the tandem mRFP–GFP–LC3 reporter system was used in live-cell imaging experiment based on different pH sensitivity of mRFP and GFP fluorescent proteins. We found a decrease in the number of yellow and red puncta in EI24 KO group which indicated that the autophagosome formation was reduced (Fig. 2c). However, the number of yellow puncta decreased by 63% after EI24 knockout, while the red puncta decreased by 91% (Fig. 2c). The result suggests that the autophagic flux is also impaired, because if the flux is not affected, but only the autophagosome formation is affected, the reduction rates of the number of yellow and red puncta before and after EI24 knockout should be the same. Moreover, to exclude the possibility that the accumulation of p62 is due to the dysfunction of lysosome in EI24 KO cells, the size and function of lysosome were analyzed. The results showed that the size of lysosome, the cathepsin enzyme activity, as well as the pH value were comparable between WT and KO groups, indicating that EI24 knockout does not affect lysosome function and the accumulation of p62 is because of the impaired autophagy (Supplementary Fig. 1a–c). Taken together, these data suggest that EI24 functions in both autophagosome formation and the autophagic flux.
Fig. 1.
EI24 deficiency impairs autophagy flux. a Co-staining of nucleus (blue), p62 (green), and insulin (red) in pancreas from 4-week-old mice. Bar = 50 μm. b Total RNA was prepared from the pancreas of WT and EI24 KO mice. Transcription levels of p62 mRNA are normalized to β-actin. c Islets were extracted and purified from WT and EI24 KO mice at 1 month of age. Proteins from islets were extracted and separated by SDS-PAGE and analyzed by immunoblotting with anti-p62 and anti-LC3 antibodies. d Quantification analysis of p62 and LC3II, normalized to β-actin. e Using the CRISPR-Cas9 genomic-editing approach, EI24 is effectively depleted in U-2 OS and f HEK293T cells. Sequence of guide RNA (sgRNA) is shown in pink. g, h Total protein from U-2 OS cells and HEK293T cells was extracted and separated by SDS-PAGE and immunoblotted with anti-p62 and anti-LC3 antibodies. Results shown are from three independent experiments. *p < 0.05, **p < 0.01
Fig. 2.
EI24 deficiency impairs the autophagy flux. a U-2 OS cells were treated with or without 100 μM chloroquine (CQ) for 6 h. Total protein was extracted and separated by SDS-PAGE and immunoblotted with anti-p62 and anti-LC3 antibodies. b Quantification analysis of p62 and LC3II, normalized to β-actin. c U-2 OS cells were transfected with the mRFP–GFP–LC3 plasmid, and the left panel represents fluorescent images of U-2 OS cells acquired by confocal microscopy. The right panel shows the quantification of the number of autophagosome (yellow) and autolysosome (red) puncta in WT and EI24 KO U-2 OS cells. WT (n = 34), KO (n = 40). Bar = 10 μm, **p < 0.01, ***p < 0.001
EI24 mediates MAM integrity of VDAC1–GRP75–IP3R complex formation
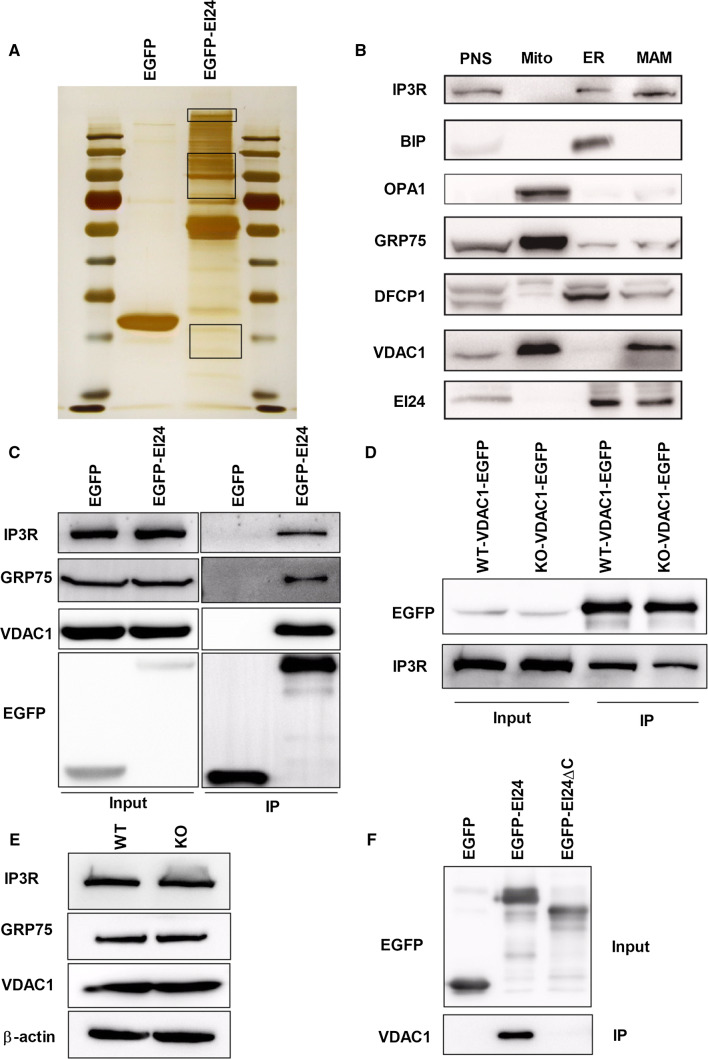
To study molecular mechanisms of EI24-mediated autophagy, we first used immunoprecipitation followed by mass spectrometry analysis to identify the binding partners for EI24 (Fig. 3a). To our surprise, among the interacting proteins, about 25% of the proteins belong to mitochondrial proteins (Table 1). Since EI24 is an ER-localized transmembrane protein (Supplementary Fig. 2a), we hypothesized that EI24 may mediate MAM formation. According to the data shown in Table 1, we noticed that GRP75, a mitochondria chaperone protein, was identified in the EI24 immunoprecipitation lane with 26 unique peptides and a higher score, but was absent in the negative control lanes (Table 1). It has been reported that GRP75 strengthens the interaction between the ER and mitochondria by forming a ternary bridging complex with VDAC1 and IP3R [23]. To confirm that the VDAC1–IP3R complex associates with EI24, we first analyzed the subcellular distribution of EI24 in U-2 OS cells and INS-I cells using Percoll density-gradient centrifugation. EI24, VDAC1, IP3R, and GRP75 co-existed in the same MAM fractions (Fig. 3b and Supplementary Fig. 2b). Immunoprecipitation experiments followed by immunoblot analysis were then performed. Results showed strong interactions between EI24 with IP3R, GRP75, or VDAC1 (Fig. 3c). Moreover, EI24 knockout compromised the interaction between VDAC1 and IP3R (Fig. 3d), and this phenomenon was not due to the changed levels of IP3R, GRP75, and VDAC1 in the WT and KO group (Fig. 3e), indicating that EI24 may form a quaternary complex with VDAC1–GRP75–IP3R to facilitate the ER–mitochondria association. To gain insight into the domain of EI24 involved in binding VDAC1, we generated a series of truncation mutants of EI24 by the dichotomy method and tested their ability to bind to VDAC1 by immunoprecipitation assays. Results showed almost no interactions of EI24 with VDAC1 after truncation of the C-terminal of EI24 (Fig. 3f), suggesting that the C-terminal of EI24 plays a very important role in the interaction of EI24 with VDAC1.
Fig. 3.
EI24 is a major interactor protein for the IP3R–GRP75–VDAC1 complex. a HEK293T cells transfected with EGFP and EGFP–EI24 were subjected to immunoprecipitation assays followed by a silver staining assay. The bands indicated with a rectangle were performed by mass spectrum analysis. b Subcellular fractions from U-2 OS cells were subjected to western blot analysis with the indicated antibodies. PNS post-nuclear supernatant, Mito mitochondria, ER endoplasmic reticulum, MAM mitochondria-associated membrane, OPA1 mitochondrial dynamin-like GTPase which is used as a marker of mitochondria, BIP binding immunoglobulin protein, the major chaperone of the ER lumen which is used as a marker of ER, DFCP1 double FYVE-containing protein 1, which is considered to be the site for autophagosome formation. c HEK293T cells transfected with EGFP or EGFP–EI24 were subjected to immunoprecipitation assays and western blot analysis with the indicated antibodies. d Interaction of VDAC1 with IP3R. WT and EI24 KO HEK293T cells were transfected with VDAC1–EGFP, subjected to immunoprecipitation assays and western blot analysis with the indicated antibodies. e Protein levels of IP3R, GRP75, and VDAC1 in WT and EI24 KO cells. f Interactions of VDAC1 with EI24 and EI24 mutations
Table 1.
Identification of proteins commonly associated with mitochondria found from EGFP to EI24 affinity purification in HEK293T cells
| Protein ID | Genes | Description | Peptide no. |
|---|---|---|---|
| P38646 | HSPA9 | GRP75_HUMAN Stress-70 protein, mitochondrial | 26 |
| P21796 | VDAC1 | VDAC1_HUMAN Voltage-dependent anion-selective channel protein 1 | 1 |
| P05141 | SLC25A5 | ADT2_HUMAN ADP/ATP translocase 2 | 4 |
| P40939 | HADHA | ECHA_HUMAN Trifunctional enzyme subunit alpha, mitochondrial | 27 |
| P12236 | SLC25A6 | ADT3_HUMAN ADP/ATP translocase 3 | 2 |
| Q00325;F8VVM2 | SLC25A3 | MPCP_HUMAN Phosphate carrier protein, mitochondrial | 2 |
| Q8TC07 | TBC1D15 | TBC15_HUMAN TBC1 domain family member 15 | 13 |
| P27348 | YWHAQ | 1433T_HUMAN 14-3-3 protein theta | 3 |
| P11021 | HSPA5 | GRP78_HUMAN 78 kDa glucose-regulated protein | 9 |
| P61981 | YWHAG | 1433G_HUMAN 14-3-3 protein gamma | 7 |
| P0DMV9;P0DMV8;A0A0G2JIW1 | HSPA1B;HSPA1A;HSPA1B | A0A0G2JIW1_HUMAN Heat shock 70 kDa protein 1B | 11 |
| Q9Y277 | VDAC3 | VDAC3_HUMAN Voltage-dependent anion-selective channel protein 3 | 4 |
| Q04917 | YWHAH | 1433F_HUMAN 14-3-3 protein eta | 3 |
| P31946 | YWHAB | 1433B_HUMAN 14-3-3 protein beta/alpha | 2 |
| P53007 | SLC25A1 | TXTP_HUMAN Tricarboxylate transport protein, mitochondrial | 4 |
| Q02978 | SLC25A11 | M2OM_HUMAN Mitochondrial 2-oxoglutarate/malate carrier protein | 2 |
| P35232 | PHB | PHB_HUMAN Prohibitin | 16 |
| Q9BQT8 | SLC25A21 | ODC_HUMAN Mitochondrial 2-oxodicarboxylate carrier | 2 |
| H3BPZ1;H3BS72;Q9P035 | HACD3 | H3BS72_HUMAN Very-long-chain (3R)-3-hydroxyacyl-CoA dehydratase | 6 |
| Q9UBX3;F6RGN5 | SLC25A10 | F6RGN5_HUMAN Mitochondrial dicarboxylate carrier | 3 |
| B0QYW5;F6RTR7;O43808 | SLC25A17 | PM34_HUMAN Peroxisomal membrane protein PMP34 | 2 |
| P38606 | ATP6V1A | VATA_HUMAN V-type proton ATPase catalytic subunit A | 4 |
| O43819 | SCO2 | SCO2_HUMAN Protein SCO2 homolog, mitochondrial | 3 |
| Q9H3P7 | ACBD3 | GCP60_HUMAN Golgi resident protein GCP60 | 2 |
| P45880 | VDAC2 | VDAC2_HUMAN Voltage-dependent anion-selective channel protein 2 | 4 |
| J3QTB2;Q9BRQ6;J3QTA6 | CHCHD6 | J3QTA6_HUMAN MICOS complex subunit | 1 |
| P08238 | HSP90AB1 | HS90B_HUMAN Heat shock protein HSP 90-beta | 3 |
| Q15046 | KARS | SYK_HUMAN Lysine–tRNA ligase | 3 |
| Q9UJS0 | SLC25A13 | CMC2_HUMAN Calcium-binding mitochondrial carrier protein Aralar2 | 4 |
| P21964 | COMT | COMT_HUMAN Catechol O-methyltransferase | 1 |
| P28331;B4DJ81 | NDUFS1 | NDUS1_HUMAN NADH–ubiquinone oxidoreductase 75 kDa subunit, mitochondrial | 4 |
| O95573 | ACSL3 | ACSL3_HUMAN Long-chain-fatty-acid–CoA ligase 3 | 4 |
| A0A0D9SF53;A0A0D9SFB3;A0A0D9SG12;O00571 | DDX3X | A0A0D9SF53_HUMAN ATP-dependent RNA helicase DDX3X | 4 |
| A8MX94;P09211 | GSTP1 | GSTP1_HUMAN Glutathione S-transferase P | 2 |
| P10809 | HSPD1 | CH60_HUMAN 60 kDa heat shock protein, mitochondrial | 4 |
| P49327 | FASN | FAS_HUMAN Fatty-acid synthase | 21 |
| O75746 | SLC25A12 | CMC1_HUMAN Calcium-binding mitochondrial carrier protein Aralar1 | 3 |
| P12277 | CKB | KCRB_HUMAN Creatine kinase B-type | 1 |
| Q7L8L6 | FASTKD5 | FAKD5_HUMAN FAST kinase domain-containing protein 5, mitochondrial | 4 |
| Q7Z7H8 | MRPL10 | RM10_HUMAN 39S ribosomal protein L10, mitochondrial | 1 |
| P25705 | ATP5A1 | ATPA_HUMAN ATP synthase subunit alpha, mitochondrial | 2 |
| P63104 | YWHAZ | 1433Z_HUMAN 14-3-3 protein zeta/delta | 5 |
| C9IY40;Q5T653 | MRPL2 | RM02_HUMAN 39S ribosomal protein L2, mitochondrial | 1 |
| P42704 | LRPPRC | LPPRC_HUMAN Leucine-rich PPR motif-containing protein, mitochondrial | 8 |
| Q5T3Q7;Q9H583 | HEATR1 | HEAT1_HUMAN HEAT repeat-containing protein 1 | 13 |
| P42345 | MTOR | MTOR_HUMAN Serine/threonine–protein kinase mTOR | 12 |
| P04049 | RAF1 | RAF1_HUMAN RAF proto-oncogene serine/threonine–protein kinase | 2 |
| B4DNK4;P14618 | PKM | KPYM_HUMAN Pyruvate kinase PKM | 2 |
| Q7L4E1 | MIGA2 | MIGA2_HUMAN Mitoguardin 2 | 2 |
| Q92538 | GBF1 | GBF1_HUMAN Golgi-specific brefeldin A-resistance guanine nucleotide exchange factor 1 | 6 |
| P16435 | POR | NCPR_HUMAN NADPH–cytochrome P450 reductase | 2 |
| P33121;B7Z3Z9;D6RER0;E7EPM6 | ACSL1 | ACSL1_HUMAN Long-chain-fatty-acid–CoA ligase 1 | 1 |
| P43304 | GPD2 | GPDM_HUMAN Glycerol-3-phosphate dehydrogenase, mitochondrial | 1 |
| A0A140T9T7;Q03518 | TAP1 | A0A140T9T7_HUMAN Antigen peptide transporter 1 | 2 |
EI24-deficiency impairs calcium transfer from the ER to the mitochondria
It has been reported that MAM plays a pivotal role in different functions such as mitochondrial dynamics, lipid processing, calcium buffering, as well as mitophagy [24–26]. To investigate whether the compromised integrity of MAM caused by EI24 deficiency influences other functions besides autophagy, we first analyzed the dynamics of mitochondria. We found that there was no significant difference of the expression levels of mitochondrial fission and fusion markers between WT and KO groups (Supplementary Fig. 3a, b). Moreover, since the mitochondrial circularity and interconnectivity were usually used as indicators for mitochondrial morphological changes [27], the values of mitochondrial circularity (4π × area/perimeter2) and the interconnectivity (area/perimeter) were analyzed. The results showed that the circularity and interconnectivity of mitochondria were comparable between WT and KO groups, which further demonstrated that the dynamics of mitochondria were not influenced by EI24 (Supplementary Fig. 3c, d).
Mitochondria are partially autonomous organelles that depend on the import of lipid from ER to maintain membrane formation, although phosphatidylglycerols (PG), cardiolipins (CL), and phosphatidylethanolamines (PE) can be synthesized by mitochondrial enzymes [24]. The mitochondrial phospholipid composition in WT and KO groups was analyzed by liquid chromatography–mass spectrometry. There was no significant difference in almost all the phospholipid, including CL, PG, PE, phosphatidylcholines (PC), plasmalogen PC, phosphatidylinositols (PI), phosphatidylserines (PS), except for a slight decrease of phosphatidic acid (PA) level (Supplementary Fig. 4). Considering that PA accounts for a very small fraction (0.1%) of the total mitochondrial phospholipid, the function of slightly decreased PA in EI24 KO group is unknown and will be studied in the future.
To measure the mitochondrial calcium, we developed a ratiometric mitochondria calcium sensor by fusing CEPIA2mt [28] with an SNAP-tag to dynamically quantify local calcium of mitochondria in live cells [29]. We found that the mitochondria Ca2+ was increased obviously under the resting condition in EI24 KO group, while could be rescued back to the WT level after overexpressing EI24 in KO group (Fig. 4a). Next, we checked the effect of EI24 on the calcium transfer from the ER to the mitochondrial using histamine stimulation. We found that the level of mitochondrial Ca2+ was increased rapidly in both WT and KO group upon histamine stimulation, but the maximal [Ca2+]mito of KO group was extremely lower compared with WT group (Fig. 4b, c). The signal of mitochondrial Ca2+ showed a slower raise in EI24-overexpression group, but to a higher platform compared with WT group (Fig. 4b, c). These results indicated that although the increased autophagy flux in EI24 overexpression group does not be the results of a mitochondrial calcium overload, EI24 indeed affects the calcium transfer between the ER and the mitochondria. This may because EI24 mediates MAM formation through VDAC1–GRP75–IP3R complex.
Fig. 4.

EI24 regulates the calcium transfer from the ER to the mitochondria. a Relative level of mitochondria calcium at resting state measured by the ratio of fluorescence intensity of 488 nm/647 nm using flow cytometry. WT: 2 × 104 cells/time, four-time repetition; KO: 2 × 104 cells/time, eight-time repetition; KO–EGFP–EI24: 2 × 104 cells/time, eight-time repetition. b Representative curves of mitochondria calcium with 100 μM histamine treatment. c Relative quantification of CEPIA2mt-SNAP fluorescence peak values of ΔFmax/F0. WT (n = 35), KO (n = 45), KO–EGFP–EI24 (n = 37), *p < 0.05, ***p < 0.001
We further checked whether there was difference in mitophagy between WT and EI24 KO cells. Very recently Gelmetti et al. reported that PTEN-induced kinase 1 (PINK1), a mitochondrial serine/threonine–protein kinase, promoted the enhancement of MAM and the formation of omegasomes [26]. As shown in Supplementary Fig. 5a, b, the co-localization of LC3 with MitoTracker was comparable between WT and EI24 KO groups. In response to CCCP, PINK1 selectively accumulated on the surface of damaged mitochondria both in WT and KO groups, which were not be observed without CCCP treatment (Supplementary Fig. 5c, d). These results suggested that EI24-mediated MAM formation has no effect on mitophagy process.
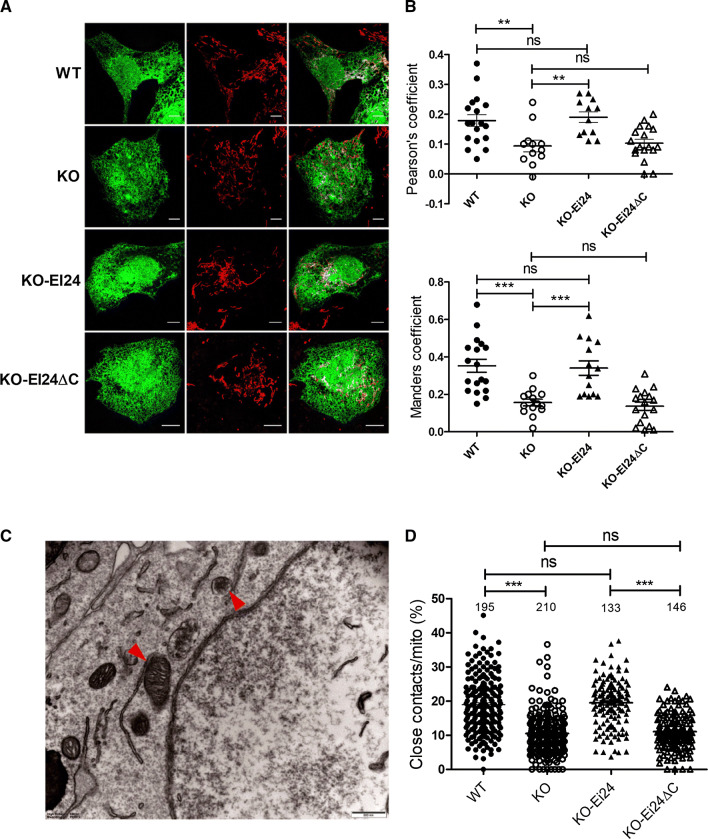
Fig. 5.
C-terminal of EI24 is essential for MAM formation. a Confocal microscopy analysis of U-2 OS cells transfected with EGFP–ER and labeled with MitoTracker (red). KO–EI24: EI24–KO cells were overexpressed with EI24, KO–EI24ΔC:EI24–KO cells were overexpressed with EI24ΔC. b Data analysis shown with Pearson’s coefficient and Manders coefficient. Mean ± SEM. c Representative EM images of U-2 OS cells. The red arrowheads indicate the ER–mitochondria contacts (closely apposed within 30 nm and a contact length ≥ 20 nm). Bar = 500 nm. d Quantification of the ratio (%) of ER–mitochondria contacts to the mitochondrial surface area. Numbers of cells are indicated on the figure. **p < 0.01, ***p < 0.001
C-terminal of EI24 is essential for VDAC1–IP3R complex-induced MAM formation
Since EI24 interacts with both VDAC1 and IP3R, we aimed to determine whether EI24 mediates the VDAC1–IP3R complex-induced MAM formation. The ER and mitochondria were labeled simultaneously by fluorescence proteins, and their co-localization, which indicates contact between the ER and mitochondria, was analyzed by Pearson’s coefficient and Manders coefficient. We found that KO EI24 significantly reduced the fraction of ER that colocalized with mitochondria (Fig. 5a, b). Overexpression of EI24, but not the C-terminal region of the EI24 deletion mutant (EI24-ΔC), could increase the MAM number to WT levels (Fig. 5a, b). To further confirm these results, we next monitored the number of ER–mitochondria contact sites (closely apposed within 30 nm and a contact length ≥ 20 nm) by high-resolution electron microscopy (Fig. 5c), and representative images at all of conditions were showed in Fig. 6a. Approximately 19% of the mitochondrial surface was closely associated with the ER in WT cells (Fig. 5d), while EI24 deficiency significantly reduced this value to 10% (Fig. 5d). Overexpression of EI24 in KO cells could rescue and significantly increase the co-localization of ER and mitochondria, while transfection of EI24-ΔC mutant had no effect on the integrity of ER–mitochondria associations (Fig. 5d), which further demonstrates that the C-terminal of EI24 is essential for the integrity of MAM. We next examined the length of MAM, as well as the distance between mitochondria and the nearest ER. It is very interesting that although there was no significant difference in the number of contacts per mitochondria and the averaged minimum ER–mitochondria distance between WT and EI24 overexpression groups (Figs. 5d, 6c), the length of MAM was elongated obviously in EI24 overexpression group (Fig. 6b). The results suggested that the total amount of MAM was increased when EI24 was overexpressed, which further proved the EI24 function in MAM formation.
Fig. 6.
EI24 regulates the ER–mitochondria contacts. a Representative EM images of WT, KO, KO with EI24 overexpression, and KO with EI24-ΔC group. The red arrowheads indicate the ER–mitochondria contacts (closely apposed within 30 nm and a contact length ≥ 20 nm). Bar = 2 μm. b Quantification of the length of MAM, and the distance between mitochondria and the nearest ER (c). *p < 0.05, **p < 0.01, ***p < 0.001
C-terminal of EI24 is essential for autophagy flux
Considering the possibility that phagophore formation begins at the MAM site, we studied the correlation of MAM number with autophagy flux. Results showed that the overexpression of EI24, but not EI24-ΔC, decreased the level of p62 and LC3II (Fig. 7a, b). The average number of p62 and LC3 punctate per cells by immunostaining was also significantly decreased in the EI24 overexpression group, but not in the EI24-ΔC group (Fig. 7c, d), further indicating that the C-terminal of EI24 is essential for autophagy flux.
Fig. 7.
C-terminal of EI24 is essential for autophagy flux. a U-2 OS cells, stable transfected with EGFP, EGFP–EI24, or EGFP–EI24ΔC, were applied using western blot analysis with the indicated antibodies. b Quantification analysis of p62 and LC3II, normalized to β-actin. c U-2 OS cells, stably transfected with EGFP, EGFP–EI24, or EGFP–EI24ΔC, were co-stained with nucleus (blue) and p62 (red) by immunostaining assay. d U-2 OS cells, stably transfected with mApple, mApple-EI24, or mApple-EI24ΔC, were co-stained with nucleus (blue) and LC3 (green) by immunostaining assay. Bar = 50 μm
Discussion
With the deepening understanding of autophagy flux in many contexts, some diseases, such as neurodegenerative disease, cancer, and ischemia–reperfusion injury, have come to the forefront in autophagy therapeutics research [30–32]. More recently, an emerging body of evidence demonstrates that dysfunction of autophagy is associated with obesity and diabetes [7, 33, 34]. Thus, autophagy has become a highly sought-after drug target for diabetic treatment. We previously demonstrated that EI24 protects pancreatic β cells from apoptosis and prevents the progression of diabetes [9]. However, whether EI24 mediates autophagy of pancreatic β cells and which step of autophagy flux EI24 is involved in is still unknown. Here, we showed that EI24, especially the C-terminus of EI24, plays an extremely important role in the autophagy of pancreatic β cells. Using biochemical and electron microscope methods, we also found that the C-terminus of EI24 regulates the integrity of IP3R–GRP75–VDAC1 complex; we also found a high degree of juxtaposition between the ER and mitochondria, which has been proposed as the site of autophagosome formation that mediates the initiation of autophagy flux.
The causal relationship between autophagy and the diabetic process remains unclear. In both genetic diabetic mice (ob/ob and db/db) and a dietary model (fed with high-fat diet) of obesity, several autophagy indicators, such as ATG5, ATG7, and ATG6, are markedly decreased in the liver, and the level of p62 is elevated in ob/ob mice compared to lean controls [35]. This reflects the notion that persistent high concentrations of glucose and/or obesity lead to imbalances in nutrients and energy status, which may cause homeostasis disorders and defective autophagy. However, pancreatic β cell-specific Atg7 knockout mice (Atg7Δβ cell) develop hyperglycemia and hyperinsulinemia accompanied with accumulated p62, but not diabetes. The characteristics of diabetes become apparent after crossing Atg7Δβ cell mice with ob/ob mice [8], indicating that impaired autophagy in pancreatic β cells is a cause in the progression from obesity to diabetes. Our previous data showed that the content of insulin in β cells and the secreted insulin level in blood are significantly decreased in 12-week-old EI24 KO mice [9]. In this study, our results from the immunoreactivity of insulin in adolescents in mice (4 weeks of age) indicate that the content of insulin and the mass of pancreatic β cells is comparable in the EI24 KO group and WT group (Fig. 1a), while autophagy flux is significantly impaired as early as 4 weeks of age (Fig. 1c, d). Thus, we believe that impaired autophagy is a precipitating factor in the deterioration of diabetes.
Several different pairs of protein complexes have been proposed as ER–mitochondria tethers, such as the outer mitochondrial membrane VDAC1, which interacts with the IP3R on the ER through the molecular chaperone GRP75 [36], and mitofusin 2 (MFN2) on the ER, which forms homotypic and/or heterotypic complexes with MFN1 or MFN2 on the surface of mitochondria [37]. The ER-resident protein vesicle-associated membrane protein-associated protein-B (VAPB) has been reported to interact with the mitochondrial protein tyrosine phosphatase-interacting protein-51 (PTPIP51) to regulate ER–mitochondria associations [18]. However, complete loss of IP3R does not affect ER–mitochondria contacts in DT40 cells [38], and increased proximity between ER–mitochondria is observed when MFN2 levels are reduced [39]. These controversial reports indicate that the formation of MAM is dependent on the cell type and/or processing conditions. Recently, Zheng et al. showed that under DNA damage, EI24 interacts with VDAC2, but not VDAC1 and VDAC3 [40]. However, our findings reveal that upon physiological conditions, EI24 binds to VDAC1 and facilitates the formation of MAM through its C-terminal region, suggesting that the formation and composition of the MAM may be tightly regulated and highly relevant to physiological and pathological conditions. IP3R–GRP75–VDAC1 has been reported to regulate ER-to-mitochondrial Ca2+ shuttling and mitochondrial ATP production [23, 41]. EI24 depletion impairs the integrity of the IP3R–GRP75–VDAC1 complex, which may further disequilibrate calcium homeostasis and fundamentally change cellular function, including cell death, by influencing mitochondrial calcium uptake. It will be interesting to confirm whether EI24 plays important roles in other MAM pairs; these experiments will be performed in future studies.
In this paper, we demonstrated that the C-terminal of Ei24 positively impacts the formation of IP3R–GRP75–VDAC1 complex and the process of autophagy flux. Much remains to be understood, but through these results, we revealed that EI24 mediates autophagy flux through regulating the integrity of MAM. Such a mechanism may highlight a novel approach for pharmacological intervention that may be used for the treatment of diabetes.
Materials and methods
Cell culture and transfection
Primary pancreatic β cells were dispersed from islets and cultured in RPMI 1640 medium (Gibco) as previously reported [9]. HEK293T cells were maintained in DMEM (Invitrogen) containing 10% FBS in an incubator at 37 °C with 5% CO2. U-2 OS cells were cultured in McCoy’s 5A Medium Modified (MCMM) with 10% FBS at 37 °C and 5% CO2. INS-I cells were cultured in RPMI 1640 medium supplemented with 10% FBS, 1 mM sodium pyruvate, and 50 μM β mercaptoethanol at 37 °C and 5% CO2.
For the generation of CRISPR/Cas9 KO cell lines, gRNA-expressing plasmids were constructed using the p × 458 vector. Cells were transiently transfected with the appropriate plasmid by Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer’s instructions and screened in the presence of 2.5 μg/mL puromycin. Single knockout clones were verified by immunoblotting and sequencing of the PCR fragment. The following gRNA sequences were used for human EI24: GAGAGGAGCAGCGTCGAAGAA.
For transient transfections, plasmid transfection was carried out with Lipofectamine 2000 according to the manufacturer’s instructions. For stable cell lines, U-2 OS cells were transfected with the indicated plasmid and were cultured in a six-well plate for drug screening using 0.5 mg/mL G418 solution (Sigma). The culture medium was replaced every 2 days. Following 1 week of screening, 10% of cells with the highest fluorescence intensity were collected by fluorescence-activated cell sorting and cultured in MCMM with 10% FBS and 0.5 mg/mL G418 solution at 37 °C and 5% CO2.
Plasmid construction
The EGFP–EI24 plasmid was constructed as previously reported [9]. The sequence encoding the VDAC1 transcript was amplified from HEK293T cell cDNA and cloned into the AgeI/NotI sites of pEGFP-N1 (Addgene) to produce the VDAC1–EGFP vector. EI24 mutants were constructed by overlap PCR. To label the ER, the ER-targeting signal sequence and ER retention signal KDEL were constructed and in frame with EGFP as EGFP–ER.
Subcellular fractionation
Five plates (15-cm diameter) containing U-2 OS cells or INS-I cells were washed with ice-cold PBS, collected, and homogenized using a Dounce Tissue Grinder (Thermo Fisher) in ice-cold buffer A (250 mM sucrose, 20 mM HEPES, 1 mM EDTA, pH 7.4) containing protease inhibitor cocktail (Sigma). The homogenate was centrifuged twice at 600×g for 5 min at 4 °C, and 5% of the supernatant was collected and marked as the total fraction. The rest of the supernatant was centrifuged at 10,300×g for 10 min at 4 °C to pellet the crude mitochondrial extraction. The resultant supernatant was further centrifuged two times at 10,300×g for 10 min at 4 °C to collect the crude ER extraction (supernatant). For pure ER isolation, the crude ER extraction was centrifuged at 100,000×g for 1 h at 4 °C (Beckman SW40 rotor). The pellet was resuspended with buffer A and collected as the ER compartment.
For mitochondria and MAM isolation, the crude mitochondrial pellet was gently resuspended in 300 μL of ice-cold buffer B (0.25 M mannitol, 0.5 mM EGTA, and 5 mM HEPES, pH 7.4), layered on 11 mL of 30% Percoll solution (225 mM mannitol, 1 mM EGTA, 25 mM HEPES, pH 7.4), and centrifuged at 95,000×g for 1 h at 4 °C (Beckman SW40 rotor). The top band was collected, diluted five times with buffer B, and centrifuged at 6300×g for 10 min at 4 °C. The supernatant was further centrifuged at 100,000×g for 1 h at 4 °C (Beckman SW40 rotor). The pellet was resuspended with buffer B and collected as the MAM fractionation. The bottom band in the Percoll gradient was collected and diluted five times with buffer B and centrifuged at 6300×g for 10 min at 4 °C. The pellet was resuspended with buffer B and collected as the mitochondria fractionation.
MAM number collection
Cells were transfected with EGFP–ER using Lipofectamine 2000 in MCMM. At 24 h post-transfection, cells were stained by Mito Tracker (Thermo Fisher Scientific) according to the manufacturer’s instructions. Images were taken by FV1200 laser scanning confocal microscopy, and the interactions between mitochondria and ER were calculated using the Image J software. The co-localization data sets were compared using Pearson’s coefficient.
Electron microscopy
U-2 OS cells were fixed in 2.5% glutaraldehyde in 0.1 M cacodylate buffer (pH 7.2) for 30 min at room temperature and then overnight at 4 °C. After being washed by ddH2O for three times, cells were post-fixed in 1% osmium tetroxide and 1.5% potassium ferrocyanide for 1 h at room temperature. The cells were then washed with distilled water and placed in 1% thiocarbohydrazide for 1 h at room temperature, followed by incubation with 1% osmium tetroxide for 1 h at room temperature. Staining was performed using 2% aqueous uranyl acetate for 2 h at room temperature. After washing with ddH2O, the cells were further dehydrated with a graded series of ethanol solutions and embedded in epoxy resin. Ultrathin sections were examined using a 120-kV electron microscopy (H-7650B, Hitachi) at 80 kV, and images were captured with an AMT CCD camera (XR-41) using the Digital Micrograph software.
Immunoprecipitation
HEK293T cells were transfected with the appropriate plasmids. After 24 h, cells were collected and lysed in lysis buffer (50 mM Tris–HCl, 150 mM NaCl, 1 mM EDTA, and 1% Triton X-100, pH 7.4) containing protease inhibitor cocktail (Sigma) on ice and centrifuged at 12,000 rpm for 5 min at 4 °C. The supernatants were incubated with GFP-Trap_A beads (ChromoTek) on a rotator for 2 h at 4 °C. Next, the precipitated samples were washed three times with washing buffer (50 mM Tris–HCl, 300 mM NaCl, 1 mM EDTA, 1% Triton X-100, pH 7.4) and eluted in 0.1 M glycine (pH 2.0). After neutralization by 1 M Tris base, the immunoprecipitated samples were analyzed by western blotting.
Protein extraction and western blotting
This procedure has previously been described in detail [42]. Briefly, the immunoprecipitated samples or cell lysate samples were subjected to SDS-PAGE electrophoresis and transferred onto polyvinylidene difluoride membranes (Millipore). The membranes were then blocked with skim milk, probed with primary and secondary antibodies and exposed using CLiNX chemiluminescence imager (Clinx Science Instruments). Antibodies against EI24 (Sigma), VDAC1 (Abcam), IP3R (Cell Signaling Technology), GRP75 (Abcam), p62 (Abcam), LC3 (Abcam), and β-actin (Cell-Signaling Technology) were used according to the manufacturer’s protocols.
Immunohistochemistry and immunostaining assays
Pancreatic tissue was fixed, dehydrated, and sectioned as previously reported [9]. After being incubated with indicated primary antibodies overnight at 4 °C and washed three times with PBS, the tissue sections were stained with fluorescently labeled secondary antibodies for 1 h at room temperature. For immunostaining assays, cells seeded on fibronectin-coated glass cover slips were fixed with 4% PFA for 15 min at 37 °C, and permeabilized in 0.1% Triton X-100 (Sigma) for 5 min at room temperature. After blocking with 5% goat serum for 1 h at room temperature, cells were incubated with indicated first antibodies, followed by washing with PBS and incubating with fluorescently labeled secondary antibodies. Images were acquired using an Olympus FV1200 Laser Scanning Confocal Microscope (Olympus) with a 60× (NA = 1.40) oil objective. The images were quantified and analyzed using the ImageJ software (National Institutes of Health).
Mass spectrometric analysis
Proteins immunoprecipitated by GFP-Trap_A beads were stained by silver stain according to the manufacturer’s protocols (Thermoscientific). The protein bands on the SDS-PAGE gel were then de-stained and in-gel digested with sequencing grade trypsin (10 ng/mL trypsin, 50 mM ammonium bicarbonate, pH 8.0) overnight at 37 °C. Peptides were extracted with 5% formic acid/60% acetonitrile and 0.1% formic acid/75% acetonitrile sequentially and then concentrated to 20 mL. The extracted peptides were separated on an analytical capillary column (diameter 75 μm, 15 cm) packed with 3 μm ReproSil-Pur C18-AQ (Dr. Maisch). A Waters nanoAcquity UPLC system (Waters) was used to generate the following HPLC gradient: 0–30% B in 75 min, 30–70% B in 10 min (A = 0.1% formic acid in water, B = 0.1% formic acid in 80% acetonitrile with 20% water). The eluted peptides were sprayed into an LTQ ORBITRAP Velos mass spectrometer (ThermoFisher Scientific) equipped with a nano-ESI ion source. Database searches were performed on Proteome Discoverer (version 1.4.0.288, Thermo Fischer Scientific) with Uniprot-Human-2017 database. The search parameters were as follows: 20 ppm mass tolerance for precursor ions 0.6 Da mass tolerances for product ions and two missed cleavage sites were allowed for trypsin digestion. Cysteine alkylation was set as an immobilization modification and methionine oxidation was set as a variable modification. The search results were filtered with both the peptide significance threshold and expectation value to be below 0.01.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
This project was supported by the National Key R&D Program of China (2017YFA0505300 and 2016YFA0501500), the National Natural Science Foundation of China (31421002, 21778069 and 31401174), Project of the Chinese Academy of Sciences (XDB08030203) and Project of Chinese Academy of Sciences-Peking University Leading Cooperation Team.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflicts of interest regarding the contents of this article.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Lin Yuan and Qi Liu contributed equally to this work.
Change history
11/21/2019
In the published article, few errors were noticed and this has been corrected with this erratum publication.
References
- 1.Yu L, Chen Y, Tooze SA. Autophagy pathway: cellular and molecular mechanisms. Autophagy. 2018;14(2):207–215. doi: 10.1080/15548627.2017.1378838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Farre JC, Subramani S. Mechanistic insights into selective autophagy pathways: lessons from yeast. Nat Rev Mol Cell Biol. 2016;17(9):537–552. doi: 10.1038/nrm.2016.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eskelinen EL, Saftig P. Autophagy: a lysosomal degradation pathway with a central role in health and disease. Biochim Biophys Acta. 2009;1793(4):664–673. doi: 10.1016/j.bbamcr.2008.07.014. [DOI] [PubMed] [Google Scholar]
- 4.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132(1):27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen CG, et al. Human beta cell mass and function in diabetes: recent advances in knowledge and technologies to understand disease pathogenesis. Mol Metab. 2017;6(9):943–957. doi: 10.1016/j.molmet.2017.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jung HS, et al. Loss of autophagy diminishes pancreatic beta cell mass and function with resultant hyperglycemia. Cell Metab. 2008;8(4):318–324. doi: 10.1016/j.cmet.2008.08.013. [DOI] [PubMed] [Google Scholar]
- 7.Sheng QF, et al. Autophagy protects pancreatic beta cell mass and function in the setting of a high-fat and high-glucose diet. Sci Rep. 2017;7:16348. doi: 10.1038/s41598-017-16485-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Diakopoulos KN, et al. Impaired autophagy induces chronic atrophic pancreatitis in mice via sex- and nutrition-dependent processes. Gastroenterology. 2015;148(3):626. doi: 10.1053/j.gastro.2014.12.003. [DOI] [PubMed] [Google Scholar]
- 9.Yuan L, et al. Etoposide-induced protein 2.4 functions as a regulator of the calcium ATPase and protects pancreatic beta-cell survival. J Biol Chem. 2018;293(26):10128–10140. doi: 10.1074/jbc.RA118.002399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao YG, et al. The p53-induced gene Ei24 is an essential component of the basal autophagy pathway. J Biol Chem. 2012;287(50):42053–42063. doi: 10.1074/jbc.M112.415968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tian Y, et al. C elegans screen identifies autophagy genes specific to multicellular organisms. Cell. 2010;141(6):1042–1055. doi: 10.1016/j.cell.2010.04.034. [DOI] [PubMed] [Google Scholar]
- 12.Xie ZP, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nat Cell Biol. 2007;9(10):1102–1109. doi: 10.1038/ncb1007-1102. [DOI] [PubMed] [Google Scholar]
- 13.Hayashi-Nishino M, et al. A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nat Cell Biol. 2009;11(12):1433–1437. doi: 10.1038/ncb1991. [DOI] [PubMed] [Google Scholar]
- 14.Yla-Anttila P, et al. 3D tomography reveals connections between the phagophore and endoplasmic reticulum. Autophagy. 2009;5(8):1180–1185. doi: 10.4161/auto.5.8.10274. [DOI] [PubMed] [Google Scholar]
- 15.Ravikumar B, et al. Plasma membrane contributes to the formation of pre-autophagosomal structures. Nat Cell Biol. 2010;12(8):747–757. doi: 10.1038/ncb2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bodemann BO, et al. RalB and the exocyst mediate the cellular starvation response by direct activation of autophagosome assembly. Cell. 2011;144(2):253–267. doi: 10.1016/j.cell.2010.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hamasaki M, et al. Autophagosomes form at ER–mitochondria contact sites. Nature. 2013;495(7441):389–393. doi: 10.1038/nature11910. [DOI] [PubMed] [Google Scholar]
- 18.Stoica R, et al. ER–mitochondria associations are regulated by the VAPB–PTPIP51 interaction and are disrupted by ALS/FTD-associated TDP-43. Nat Commun. 2014;5:3996. doi: 10.1038/ncomms4996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gomez-Suaga P, et al. The ER–mitochondria tethering complex VAPB–PTPIP51 regulates autophagy. Curr Biol. 2017;27(3):371–385. doi: 10.1016/j.cub.2016.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arasaki K, Tagaya M. Legionella blocks autophagy by cleaving STX17 (syntaxin 17) Autophagy. 2017;13(11):2008–2009. doi: 10.1080/15548627.2017.1371395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lieu K, et al. The p53-induced factor Ei24 inhibits nuclear import through an importin beta-binding-like domain. J Cell Biol. 2014;205(3):301–312. doi: 10.1083/jcb.201304055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mauthe M, et al. Chloroquine inhibits autophagic flux by decreasing autophagosome–lysosome fusion. Autophagy. 2018;14(8):1435–1455. doi: 10.1080/15548627.2018.1474314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Szabadkai G, et al. Chaperone-mediated coupling of endoplasmic reticulum and mitochondrial Ca2+ channels. J Cell Biol. 2006;175(6):901–911. doi: 10.1083/jcb.200608073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Flis VV, Daum G. Lipid transport between the endoplasmic reticulum and mitochondria. Cold Spring Harb Perspect Biol. 2013;5(6):a013235. doi: 10.1101/cshperspect.a013235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mejia EM, Hatch GM. Mitochondrial phospholipids: role in mitochondrial function. J Bioenerg Biomembr. 2016;48(2):99–112. doi: 10.1007/s10863-015-9601-4. [DOI] [PubMed] [Google Scholar]
- 26.Gelmetti V, et al. PINK1 and BECN1 relocalize at mitochondria-associated membranes during mitophagy and promote ER–mitochondria tethering and autophagosome formation. Autophagy. 2017;13(4):654–669. doi: 10.1080/15548627.2016.1277309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Y, et al. P32 regulates mitochondrial morphology and dynamics through parkin. Neuroscience. 2011;199:346–358. doi: 10.1016/j.neuroscience.2011.10.003. [DOI] [PubMed] [Google Scholar]
- 28.Suzuki J, et al. Imaging intraorganellar Ca2+ at subcellular resolution using CEPIA. Nat Commun. 2014;5:4153. doi: 10.1038/ncomms5153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luo C, et al. A genetically encoded ratiometric calcium sensor enables quantitative measurement of the local calcium microdomain in the endoplasmic reticulum. Biophys Rep. 2019;5(1):31–42. [Google Scholar]
- 30.Levy JMM, Thorburn A. Targeting autophagy during cancer therapy to improve clinical outcomes. Pharmacol Ther. 2011;131(1):130–141. doi: 10.1016/j.pharmthera.2011.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Papadakis M, et al. Tsc1 (hamartin) confers neuroprotection against ischemia by inducing autophagy. Nat Med. 2013;19(3):351–357. doi: 10.1038/nm.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chong CR, et al. A clinical drug library screen identifies astemizole as an antimalarial agent. Nat Chem Biol. 2006;2(8):415–416. doi: 10.1038/nchembio806. [DOI] [PubMed] [Google Scholar]
- 33.Sarparanta J, Garcia-Macia M, Singh R. Autophagy and mitochondria in obesity and type 2 diabetes. Curr Diabetes Rev. 2017;13(4):352–369. doi: 10.2174/1573399812666160217122530. [DOI] [PubMed] [Google Scholar]
- 34.Lim H, et al. A novel autophagy enhancer as a therapeutic agent against metabolic syndrome and diabetes. Nat Commun. 2018;9:1438. doi: 10.1038/s41467-018-03939-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang L, et al. Defective hepatic autophagy in obesity promotes ER stress and causes insulin resistance. Cell Metab. 2010;11(6):467–478. doi: 10.1016/j.cmet.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rowland AA, Voeltz GK. Endoplasmic reticulum-mitochondria contacts: function of the junction. Nat Rev Mol Cell Biol. 2012;13(10):607–625. doi: 10.1038/nrm3440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.de Brito OM, Scorrano L. Mitofusin 2 tethers endoplasmic reticulum to mitochondria. Nature. 2008;456(7222):605–610. doi: 10.1038/nature07534. [DOI] [PubMed] [Google Scholar]
- 38.Csordas G, et al. Structural and functional features and significance of the physical linkage between ER and mitochondria. J Cell Biol. 2006;174(7):915–921. doi: 10.1083/jcb.200604016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Filadi R, et al. Mitofusin 2 ablation increases endoplasmic reticulum–mitochondria coupling. Proc Natl Acad Sci USA. 2015;112(17):E2174–E2181. doi: 10.1073/pnas.1504880112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zheng P, et al. DNA damage triggers tubular endoplasmic reticulum extension to promote apoptosis by facilitating ER–mitochondria signaling. Cell Res. 2018;28(8):833–854. doi: 10.1038/s41422-018-0065-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Betz C, et al. mTOR complex 2-Akt signaling at mitochondria-associated endoplasmic reticulum membranes (MAM) regulates mitochondrial physiology. Proc Natl Acad Sci USA. 2013;110(31):12526–12534. doi: 10.1073/pnas.1302455110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yuan L, et al. Low-level shear stress induces human mesenchymal stem cell migration through the SDF-1/CXCR42 axis via MAPK signaling pathways. Stem Cells Dev. 2013;22(17):2384–2393. doi: 10.1089/scd.2012.0717. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.