Abstract
HBO1 complexes are major acetyltransferase responsible for histone H4 acetylation in vivo, which belongs to the MYST family. As the core catalytic subunit, HBO1 consists of an N-terminal domain and a C-terminal MYST domain that are in charge of acetyl-CoA binding and acetylation reaction. HBO1 complexes are multimeric and normally consist of two native subunits MEAF6, ING4 or ING5 and two kinds of cofactors as chromatin reader: Jade-1/2/3 and BRPF1/2/3. The choices of subunits to form the HBO1 complexes provide a regulatory switch to potentiate its activity between histone H4 and H3 tails. Thus, HBO1 complexes present multiple functions in histone acetylation, gene transcription, DNA replication, protein ubiquitination, and immune regulation, etc. HBO1 is a co-activator for CDT1 to facilitate chromatin loading of MCM complexes and promotes DNA replication licensing. This process is regulated by mitotic kinases such as CDK1 and PLK1 by phosphorylating HBO1 and modulating its acetyltransferase activity, therefore, connecting histone acetylation to regulations of cell cycle and DNA replication. In addition, both gene amplification and protein overexpression of HBO1 confirmed its oncogenic role in cancers. In this paper, we review the recent advances and discuss our understanding of the multiple functions, activity regulation, and disease relationship of HBO1.
Keywords: BRD1, CDK11, Jade-1, KATs, MOF, ORC1, T cell, YAP1
Background
HBO1 (also known as KAT7, MYST2) is a canonical member of the MYST (MOZ, Ybf1/Sas3, Sas2 and Tip60) acetyltransferase family, which is responsible for the bulk acetylation of histone H4 and H3K14 [1–5]. HBO1 functions as the core catalytic subunit in multimeric complexes established by cofactors and accessory proteins. Thus, HBO1 affords multiple functions in various processes such as DNA replication, gene transcription, protein ubiquitination, immune regulation, stem cell pluripotent and self-renewal maintenance as well as embryonic development [3, 6].
Acetylation is a universal protein modification regulating various cellular events of cell cycle, gene transcription, signaling transduction, RNA splicing and cellular metabolism, etc. For histone, acetylation unfolds the chromatin to facilitate proteins accessing DNA that to be replicated or transcribed. Proteomics studies revealed that acetylation happens in every aspect of the cells and thousands of proteins were acetylated [7–9]. The addition or removal of the acetyl group at the lysine residue is realized by lysine acetyltransferase (KAT) and deacetyltransferase, respectively. Except for the non-canonical members, KATs mainly consists of three family: GCN5 (also known as KAT2A), CBP/p300 (also known as KAT3A, KAT3B), and MYST. Owing to their critical functions in transcriptional regulation and the abundant activities, CBP/p300 contributes to the acetylation of two-thirds of known sites [10]. In comparison, HBO1 is less represented and far from elucidated, although it is highly conserved and widely expressed from yeast to human. The mechanism regarding HBO1 acetyltransferase activation and activity regulation, subunits interactions, and substrate specificity is critical questions to be studied. In addition, the high expression of HBO1 in tissues such as testis and ovary and even cancers is recently focused and unraveled interesting discoveries.
In this review, we summarize the discovery of HBO1, analyze the structure and elucidate its important roles in histone acetylation, DNA replication, immunity and cancers. Interestingly, through the virtual simulation of the protein structure of HBO1, one possible way regarding of HBO1 activation and the regulatory role of N terminal-domain was proposed. The potential of HBO1 as a target for cancer therapy and strategies for inhibitor design were also discussed.
Structure and functions of HBO1
HBO1 is a member of the MYST acetyltransferase family
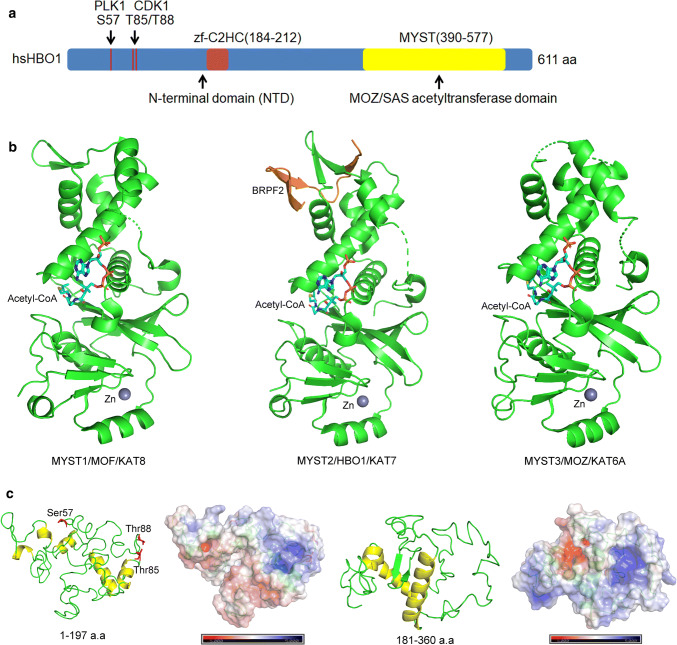
HBO1 was first identified and designated in a yeast two-hybrid analysis using DNA replication initiator subunit ORC1 as the bait protein from a HeLa cell cDNA library [11]. As the gene product of KAT7 (lysine acetyltransferase 7), human HBO1 is a 611 residue protein that mainly consists of two domains: an N-terminal domain (NTD) contains a short zf-C2HC DNA-binding motif that interacts with MCM2 and ORC1 [12], a C-terminal MYST acetyltransferase domain responsible for acetyl-CoA binding and acetylation reaction (Fig. 1a) [13]. MYST domain is a highly conserved acetyltransferase domain shared by the MYST family such as MYST1 (MOF/KAT8), MYST2 (HBO1/KAT7) and MYST3 (MOZ/KAT6A) (Fig. 1b). The MYST domain (from 340 to 607 a.a.) of HBO1 showed 61.567% identity with that of human KAT6A. Exceptionally, HBO1 comprises a cervical-loop structure proximity to the MYST domain that mediates the interaction with the N-terminal region (residues 31–80) of BRPF2 (also known as BRD1) [13], for BRPF2 is a cofactor directing HBO1 binding to the histone. In addition, the N-terminal region is highly conserved among BRPF1/2/3 and may provide a similar way for HBO1–BRPF interactions. In contrast, little is known about the NTD of HBO1, for no crystal structure is available. Apart from the short zf-C2HC motif, the sequence of NTD does not match any known motif. Nevertheless, through virtual simulation, we found that NTD exhibits abundant loop and a number of helix (Fig. 1c). In view of the evidence that in vitro-expressed full-length HBO1 exerts less acetylation activity compared to that of the separate MYST domain [14], NTD may provide a regulatory switch for HBO1 activity (discussed in “Acetylation and autoacetylation regulates HBO1 activity”). Thus, refined structure of the full-length protein and structure analysis will elucidate the mechanism of HBO1 activity regulation.
Fig. 1.
HBO1 is a MYST lysine acetyltransferase. a HBO1 consists of the N-terminal domain (NTD) and MYST domain. b HBO1 comprises a conserved MYST domain in the C-terminus. Cartoon models indicate the highly conserved MYST domain of protein family members MYST1 (also known as MOF or KAT8) (177–449 a.a, PDB: 5WCI), HBO1 (also known as MYST2, or KAT7) (336–606 a.a, PDB: 5GK9), and MYST3 (also known as MOZ, or KAT6A) (497–780 a.a, PDB: 2OZU). BRPF2 (also called BRD1) binds to HBO1 in a cervical-loop structure proximity to the MYST domain to facilitate histone binding. c NTD of HBO1 consists of a number of loops and a small part of the helix. The structure is highly flexible, which may provide abundant conformation changes for HBO1 activity regulation or protein binding
HBO1 regulates gene transcription
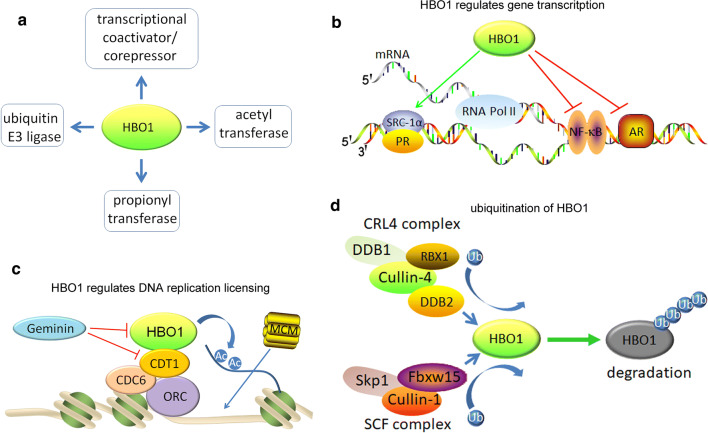
HBO1 is a multifunctional protein not only participates in protein acetylation but also regulates gene transcription [15], protein acylation (propionylation) [16], and protein ubiquitination [17] (Fig. 2a). Sas2 (MYST1 in humans) and Sas3 (MYST3 in humans) are MYST family homologs in yeast, which contains a conserve MYST domain and function as transcriptional silencers. Consistently, human HBO1 is identified as a partner of androgen receptor (AR), in which its serine-rich N-terminus is responsible for the association with AR. Besides, HBO1-AR interaction is ligand (dihydrotestosterone, etc.) promoted and the N-terminus of HBO1 acts as a suppressor in AR-mediated transcription [15] (Fig. 2b). Similarly, the transcriptional activity of NF-κB complexes is sequestrated by HBO1 through its N-terminal domain. However, this inhibition does not guaranty the direct association of HBO1 with NF-κB, and instead, HBO1 sequestrated an essential cofactor of NF-κB [18]. In contrast to AR and NF-κB, both progesterone receptor (PR) and steroid receptor co-activator (SRC1) interacts with the MYST domain of HBO1 through their N-terminal transactivation domains. HBO1 enhances coactivation of PR-mediated transcription through HBO1–PR–SRC1 complex-mediated chromatin remodeling [19, 20]. Recently, HBO1 is reported to participate in transcriptional regulation in alternative complexes such as HBO1-SIX1 and HBO1-Niam [21, 22]. Moreover, HBO1 encourages tissue-specific gene expression, for it was newly identified participating in intragenic histone acetylation and mediated Pol II binding in regulating the expression of endothelial VEGFR-2 [23]. However, the exact role and mechanism regarding of HBO1 acetyltransferase complexes in transcriptional activation or suppression remains elucidated, especially the function of HBO1 acetyltransferase activity in gene transcription regulation. Normally, HBO1-mediated histone acetylation enables the accession of transcriptional factors to the chromatin and regulates the initiation of transcription [24]. Alternatively, HBO1 complexes occupied the coding region to afford a direct role in transcriptional elongation [25]. Besides, HBO1 might acetylate the transcriptional factors and change their protein–protein interactions. Additional evidence is needed to clarify the role of HBO1 and its activity in regulating gene transcription.
Fig. 2.
HBO1 is a multifunctional acetyltransferase. a HBO1 displays the activities of acetyltransferase, propionyltransferase, transcriptional activation or repression and ubiquitin E3 ligase. b HBO1 affords transcriptional activation activity toward SRC-1α/PR, whereas suppresses the activity of NF-κB and AR. c In the assembly of prereplicative complexes in late mitosis, HBO1 binds to CDT1, recognizes and acetylates N-terminal tail of histone H4, and facilitates the loading of MCM complexes to the chromatin. Binding of Geminin to CDT1 or HBO1 inhibits both the licensing activity of CDT1 and acetyltransferase activity of HBO1, which might provide a strategy to inhibit DNA rereplication. d HBO1 is targeted for ubiquitin-mediated degradation by two E3 ubiquitin ligases CRL4 and SCF complexes
HBO1 facilitates chromatin loading of MCM complexes and promotes DNA replication licensing
Eukaryotic DNA replication licensing is a successive process involving the recognition of the replication origins, prereplicative complexes loading to the origins and initiation of replication forks. These tightly regulated steps require key factors such as ORCs, CDC6, CDT1 and MCM (minichromosome maintenance complexes) [26, 27]. Recent evidence employs the indispensable roles of HBO1 in chromosome remodeling and DNA replication [12, 28–30] (Fig. 2c). Loading of MCM complexes to chromatin is the final step of the prereplicative complexes assembly. CDT1 and CDC6 are two well-characterized factors in this process. Abrogation of HBO1 activity caused by either RNA interference or dominant negative mutation (S57A, etc.) did not affect the recruitment of ORC, CDC6 and CDT1 to replication origins, but remarkably impaired the loading of MCMs to the origins and subsequently delayed DNA replication licensing [28, 30, 31]. However, the mechanism regarding how HBO1 facilitate MCM loading and the involved protein–protein interactions remains elucidated. HBO1 is proposed to be the mediator of CDT1 and MCMs. Since HBO1 directly interacts with CDT1 and co-expression of HBO1 with CDT1 remarkably promotes replication licensing and causes rereplication; overexpression of HBO1 alone does not [28, 32]. In stress response, phosphorylation on CDT1 inhibited its interaction with HBO1 and prevented DNA replication [33]. Thus, HBO1 is a co-activator for CDT1 in DNA replication licensing.
Interestingly, CDT1–HBO1 interactions do not require the endogenous CDT1 inhibitor Geminin and in turn Geminin inhibits both the activity of CDT1 and HBO1 in DNA replication licensing. Similarly, in response to genotoxic stress, activated p53 physically interacted with HBO1 and inhibited its acetyltransferase activity to prevent DNA rereplication [14]. However, the function of HBO1 in DNA replication was first implicated from the interaction of ORC1 and HBO1 [11], for their physical interactions are confirmed both by yeast two-hybrid analysis and protein–protein immunoprecipitations. Thus, how CDT1, HBO1, and ORC1 act in concert with other replication factors to corporately establish the prereplicative complexes in the spatial–temporal order remains elucidated. Moreover, it is proposed that HBO1 performs its functions based on its cellular locations, since only the origin associated but not the other chromatin-associated HBO1 enables to promote DNA replication licensing [28]. In other words, HBO1 should distinguish different types of chromatin; there are cofactors potentiate the specificity of its activity. An interesting hypothesis is that HBO1 acts as a guide to switch on DNA replication origins through its acetyltransferase activity on histone tails [34]. However, the low absolute binding affinity of HBO1 with chromatin suggests that there are potential cofactors responsible for replication origin recognition. In fact, it was already reported that HBO1 regulates histone turnover and heterochromatin organization. Interaction with M18BP1 (Mis18-binding protein 1) bridges the acetylation activity of HBO1 to centromere chromatin and enhanced centromeric CENP-A assembly, thus preventing the spread of heterochromatin to centromere [35]. From the evidence aforementioned, dissecting location-specific activity of HBO1 on high-order chromatin structure will deepen our knowledge on the DNA replication initiation and chromosome organization.
HBO1 can be either ubiquitinated or act as an ubiquitin ligase
Two ubiquitin complexes are identified to destabilize HBO1, the CRL4 and SCF complexes (Fig. 2d). In response to DNA damage, HBO1 is degraded by DDB2-mediated CRL4 (Cullin-4/DDB1/RBX1) complexes that results in cell proliferation inhibition [36, 37], whereas in response to endotoxin, SCF (SKP1/Cullin-1/Fbxw15) complex-mediated HBO1 ubiquitination at Lys338 and degradation to regulate cell proliferation [38]. Phosphorylation is required in both the CRL4 and SCF complex-mediated HBO1 degradation pathways, although different kinases ATM/ATR or Mek1 are involved. On the contrast, USP25-mediated deubiquitination stabilizes HBO1 in response to lipopolysaccharide-induced inflammatory reaction, thus enhances HBO1-mediated inflammatory gene transcription [39]. In addition to be ubiquitinated, HBO1 performs as an E3 ubiquitin ligase toward ERα to promote its proteasomal degradation in breast cancers [17, 40]. Besides, the cofactors of HBO1 complexes are found to act as E3 ligase, for example Jade-1 ubiquitylated β-catenin and mediated inhibition of the Wnt pathway [41]. However, it is necessary to authenticate which domain of HBO1 actually affords the activity of ubiquitylation. And it is interesting to decipher the integrity of HBO1 complexes in the process of protein ubiquitination to distinguish whether it is HBO1 or its components independently exert E3 ubiquitin activity or relay on the complete HBO1 complexes.
HBO1 is required for T cell development and immune regulation
HBO1 associated histone acetylation participates in immune system regulation, especially for T cells. Depletion of HBO1 was associated with obvious abrogation of peripheral mature T cells, for HBO1 deficiency leads to decrease of the global H3K14 acetylation and impairs T cell survival. Thus, HBO1-mediated global H3K14 acetylation is critical to the normal development of the immune system [42]. Consistently in T cell development, depletion of BRPF2 in haematopoietic progenitors results in variegated CD8 expression due to the inefficient activation of Cd8 expression. Since as the subunit of HBO1 complexes, BRPF2 localized at the known enhancers of Cd8 gene and augmented the activity of HBO1 complexes for acetylation on H3K14 resulting in enhanced chromatin relaxation for subsequent transcriptional factors recruitments for full activation of Cd8 locus [43]. Moreover, in immune-related disease, HBO1 is upregulated in synovial fibroblasts, which are the key pathogenic factors contributing to the development and progression of rheumatoid arthritis (RA) [44]. Modulation of the histone acetylation could be an approach adopted by a human T cell leukemia virus mediated by its protein HBZ (HTLV-1 basic zipper factor) during pathogenesis. HBZ interacts with HBO1 and inhibits its acetylation activity to reduce p53-mediated transcription activation of p21/CDKN1A and Gadd45a, and subsequently delays G2-cell cycle arrest [45]. These results aforementioned indicated the immune regulatory functions of HBO1. Further research is needed to identify the factors connecting HBO1 to the immune-regulatory signaling pathways and unravel the functions of immune system-specific HBO1 activity.
Regulation of HBO1
HBO1 acetyltransferase complexes and activity regulation
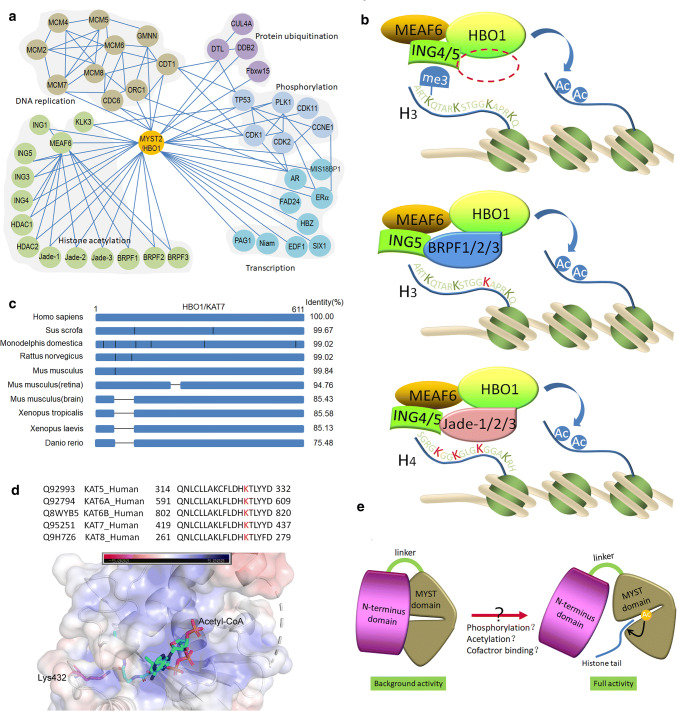
Multiple functions of HBO1 are realized by the formation of protein complexes with different cofactors or partner proteins. These proteins were manually selected and subjected to STRING analysis of functional protein–protein association (http://string.embl.de) [46]. The results showed that the above proteins mainly belong to the components of HBO1 acetylation complexes, DNA replication licensing factors, transcriptional factors, kinases and protein ubiquitination complexes (Fig. 3a). Cofactors of HBO1 such as MEAF6 (also known as hEAF6), ING1/3/4/5, Jade-1/2/3, and BRPF1/2/3 were introduced and connected to HBO1. The tumor suppressor p53, adipogenesis regulator FAD24 (factor for adipocyte differentiation 24, also called NOC3L) and cell cycle kinases CDK1, CDK2, CDK11 and PLK1 are linked to HBO1 [31, 47–50]. Moreover, cell growth inhibitor Niam and homeobox protein SIX1 that potentiates the Warburg effect by interaction with HBO1 are also presented [21, 22].
Fig. 3.
Regulation of HBO1 acetyltransferase activity. a References based protein–protein interaction networks of HBO1 with its cofactors and partner proteins. b HBO1 forms complexes with MEAF6, ING4, ING5 or BRPF1, BRPF2, BRPF3 or Jade-1, Jade-2, Jade-3, for binding to and acetylation histone H3 or H4. JADE1/2/3 directs acetylation toward the H4 tail (K5, K8 and K12), whereas BRPF1/2/3 targets H3 acetylation (K14). c Schematic diagram presented the sequence alignment of HBO1 among species. The vertical lines represent residue variants between the species. HBO1 is a conserved and widely expressed lysine acetyltransferase. However, there is a deletion (from 55 to 110 a.a.) in xenopus, zebrafish or partial tissues of mouse (brain/central nervous system or retina) in the N-terminus of HBO1. Interestingly, this sequence does not match any known motifs but consists of serine/threonine residues in rich abundance that may serve as phosphorylation sites for kinases such as CDKs and PLK1. d Lys432 is a conserved site in the MYST domain shared by MYST protein family located near the binding site of acetyl-CoA. It is autoacetylation and may regulate acetyltransferase activity and protein stability. e A proposed model suggests that NTD provides a regulatory switch for HBO1 activity. HBO1 includes two separate domains, the NTD (N-terminal domain) and MYST domain connecting by a hinge region. The close interaction of NTD with MYST domain rigidifies the full activation of HBO1 complexes, whereas conformation changes induced by modifications such as phosphorylation and acetylation, or binding of cofactor and substrate release MYST domain from the inhibition of NTD and subsequently switch the full activity of HBO1
From so far have been identified, HBO1 complexes mainly consist of accessory proteins MEAF6, ING4 or ING5, and two types of cofactors for chromatin binding: Jade-1/2/3 and BRPF1/2/3 (Fig. 3b). In fact, ING1, 3, 4, and 5 have been observed in HBO1 complexes [4, 51]. Both ING4 and ING5 are native subunits of HBO1 complexes belonging to the ING tumor suppressor family, which regulates cell cycle and apoptosis. ING4/5 forms homodimers or heterodimers through the N-terminal domain and recognizes histone H3 lysine 4 trimethylation (H3K4me3) through the C-terminal PHD domain to recruit HBO1 complexes to the histone tails [51–53]. The Jade family consists of Jade-1 (also called PHF17), Jade-2 (PHF15) and Jade-3 (PHF16) each containing two PHD domains. Jade-1 is the fundamental cofactor for HBO1 acetyltransferase complexes. Abrogation the expression of Jade-1 caused global decrease in acetylation level of histone H4, and in turn, co-expression of Jade-1 positively regulates HBO1 acetyltransferase activity and acts in concert with ING4/5 to promote HBO1-mediated histone acetylation [53–55]. Besides, Jade-1 dynamically binds to chromatin in a cell cycle-dependent manner and specifies HBO1 acetyltransferase activity on H4 [56–58]. In addition to PHD domain, BRPF1/2/3 comprises a bromodomain that binds to acetyllysine residues. BRPF proteins are well-established to potentiate HBO1 activity toward histone H3, especially Lys14 (H3K14) [59]. Depletion of the cofactors or disruption of their interactions with HBO1 will obviously impair its activity toward substrate in vitro and in vivo, for bacterial expressed HBO1 showed low activity without the binding of cofactors [13].
Phosphorylations regulate HBO1 activity and connect it to the cell cycle
The acetyltransferase activity of HBO1 is cell cycle oscillated that it is low in S phase, reaches to the maximum in G2/M, and maintains through G1, whereas the total protein level of HBO1 is slightly changed [30], indicating the post-transcriptional regulations on HBO1 activity. The N-terminal domain of HBO1 comprises abundant serine/threonine residues that can be phosphorylated. HBO1 is highly conserved and only a few residue variants exist between human, swine, canine and mouse (Fig. 3c). However, there is a sequence from 55 to 110 a.a in human HBO1 that does not match with any motifs but provides critical serine/threonine sites (Ser57, Thr85, Thr88, etc.), for kinases such as CDKs and PLK1 [31]. It is interesting to discover that in xenopus, zebrafish or mouse of special tissue (brain/central nervous system or retina) HBO1 coincidently lost these residues (Fig. 3c). Phosphorylation of Thr85/Thr88 by CDK1 provides a docking site for the subsequent phosphorylation of HBO1 on Ser57 by PLK1. The classical CDK1/PLK1-mediated sequential phosphorylation regulation is restricted to mitosis, when both CDK1 and PLK1 achieved their full activities. Inhibition of PLK1 or CDK1 with RNA interference decreases the acetylation levels of histone H4 and delays DNA replication, indicating the impairment of HBO1 activity [31]. In addition, CDK11 (also called CDC2L1) physically interacts and co-localizes with HBO1 in the nucleus. CDK11 phosphorylates HBO1 and strongly enhances its activity both in vitro and in vivo [49]. These results suggested that HBO1 is associated with the key events of the cell cycle, especially in mitosis through physical interaction with PLK1 and CDK1.
Acetylation and autoacetylation regulates HBO1 activity
HBO1 is multiple acetylated in which contains autoacetylation [9]. It is interesting to find out how HBO1 is acetylated and whether acetylation or autoacetylation regulates the activity of HBO1. Inhibition of histone deacetyltransferases (HDACs) by MS-275 or knockdown of SIRT1 resulted in two- or tenfold increase in the acetylation level of HBO1, respectively [9, 60], and this regulation mainly occurred on Lys199 (K199) within the residues “HLTGK (ac) HER” in humans (or Lys201 in mice). In contrast, acetylation at another conserve site Lys279 (or Lys277 in humans) in “RNSGLSK(ac)EQ” is downregulated in response to heat or X-ray treatments in mouse testis, indicating its functions in spermatogenesis [61]. The above two sites are specifically found in HBO1. In contrast, Lys432 is another conserve acetylation site but shared by the MYST protein family [9, 62, 63] (Fig. 3d). It is acetylated in high abundance and locates near the acetyl-CoA-binding site, suggesting its potential regulatory role in HBO1 activity or substrate/chromatin binding. However, studies in hMOF (also known as MYST1 or KAT8) showed that autoacetylation of Lys432 only slightly modulates enzyme activity [64], but regulates cellular protein stability [65]. In view of the highly conservation of this site among MYST proteins, a similar regulatory way may be used by HBO1. Hence, further evidence is needed to unravel the mechanism how acetylation regulates HBO1 activity and its interactions with cofactors or substrates. Apart from autoacetylation, it is remains unclear which KATs acetylate HBO1. Based on the database and sequence preference, KAT5 or KAT8 is predicted to acetylate Lys199 and Lys432 of HBO1 by software GPS-PAIL 2.0 [66]. Besides, HBO1 exerts significant acetyltransferase activity on proteins such as ORC2, MCM2, CDC6, and Geminin in in vitro assays, although non-histone substrates of HBO1 have not yet clearly identified, [30]. Thus, it is interesting to confirm the cellular acetyltransferase activity of HBO1 to the above factors and decipher the underlying regulatory mechanism involved.
Moreover, there is likely an intrinsic regulatory way of HBO1 acetylation activity. It is well established that separate MYST domain exerts higher activity than the full-length HBO1 containing an extra serine-rich N-terminal domain (NTD) [13, 14]. It is proposed that the close interaction of NTD with MYST domain serves as an impediment for the full acetyltransferase activity, whereas conformation changes by protein modifications such as phosphorylation (at S57, T85/T88, etc.), acetylation, and cofactor/substrate binding push away the NTD and switch on the full activity of the MYST domain (Fig. 3e). In other words, MYST domain is released from the inhibition of NTD and turned the rigidified status to the active form (Fig. 3e). Thus, dissecting the molecular structure of full-length HBO1 will greatly decipher the mechanism of HBO1 activity regulation and its cellular location-specific activity.
HBO1 and cancer
HBO1 is a potential anti-cancer target
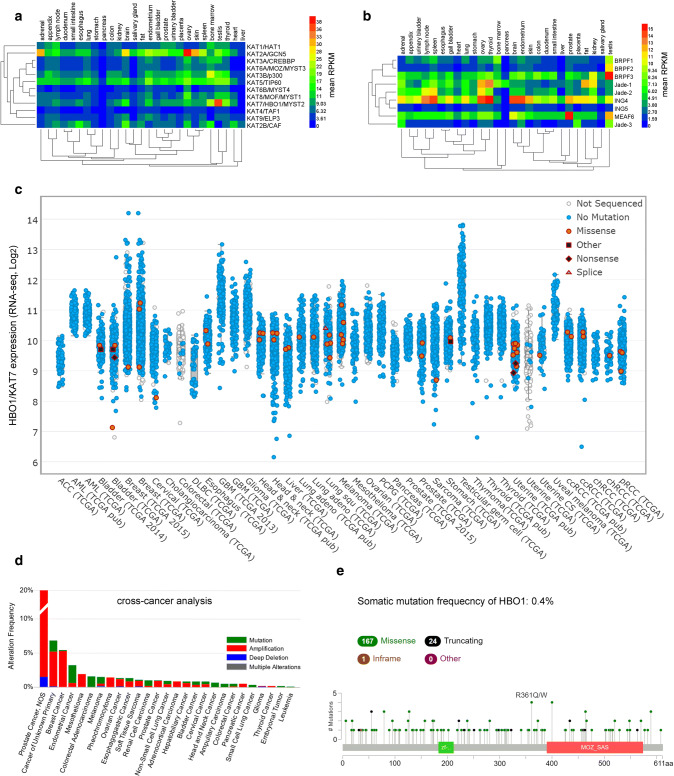
Using integrative RNA-seq data to classify gene expression across human tissues [67], we presented a heatmap to display the expressions of KATs. HBO1, KAT2A, KAT3B and KAT5 are the most abundant KATs expressed in human normal tissues, especially in the testis and ovary (Fig. 4a). Northern blotting confirmed the ubiquitous expression of HBO1 transcript, especially with high expression in the testis or ovary [15, 67]. In addition, the cofactors of HBO1 such as MEAF6, ING4, BRPF2, BRPF3, Jade-1, and Jade-2 all are coincidently expressed in testis, ovary, bone marrow and thyroid [67] (Fig. 4b). According to information from the integrative RNA-seq database (http://www.cbioportal.org) [68], HBO1 is overexpressed among cancers due to gene amplification (Fig. 4c-d). For instance, in prostate cancer, gene amplification occurs with a frequency of 20%. In contrast, HBO1 presents a somatic mutation frequency of 0.4%, in which 86.98% (167/192) is contributed by a missense mutation (Fig. 4e). It will be important to decipher the effects of these mutations on the activity of HBO1, as well as its binding affinity to the cofactors. Consistently, HBO1 is found to promote cell proliferation in bladder and breast cancer [69–71] or even contribute to gemcitabine resistance in pancreatic cancer [72]. The overexpression of HBO1 is connected to a poor prognosis in gastric cancer [73]. The components of HBO1 acetyltransferase complexes and related downstream pathways may also contribute to the activity of HBO1 in cell proliferation [21, 59, 74]. For example, Jade-2-mediated HBO1 acetylation activity enhanced the expression of mechano-transductor signaling factor YAP1 to modulate cell elasticity in ovarian cancer [75]. Besides, mutations in ING4 or ING5 destabilize the protein and contribute to tumorigenesis [76, 77]. However, HBO1 is essential for global acetylation of histone H3K4 and H4, thus the acetylation activity of HBO1 may also induce the expression of anti-cancer genes such as Brahma [78]. In acute myeloid leukemia, HBO1 expression is suppressed associated with the decease of global H4K5 acetylation [79]. Interestingly, a fusion of nucleoporin-98 (NUP98)-HBO1 was newly identified in a patient with chronic myelomonocytic leukemia (CMML). NUP98-HBO1 is sufficient to generate CMML pathogenesis through aberrant histone acetylation on the promoter of oncogene such as HOXA9 [80]. Therefore, it is necessary to decipher the functions of HBO1 in specific types of cancers, especially from different genetic backgrounds.
Fig. 4.
The expression of HBO1 and the KATs in normal tissue and cancers. a, b In normal tissue, HBO1 and its cofactors MEAF6, ING4, BRPF2, BRPF3, Jade-1, and Jade-2 are abundant and coincidently expressed in testis, ovary, bone marrow and thyroid. c HBO1 is highly expressed among cancers. d HBO1 gene is generally amplified in cancer genome. e Distribution of mutations in HBO1 from integrative sequencing data. Extended data can be accessed through http://www.cbioportal.org
Design of HBO1-targeting molecules and their applications
HBO1 is overexpressed in a number of cancers such as breast, prostate and gastric cancer. It is reasonable to interfere with the gene expression of HBO1 or block its acetyltransferase activity for the inhibition of cancer. Based on the structure, MYST domain of HBO1 contains two typical sites for molecule binding, the acetyl-CoA-binding site and histone tail binding site. However, the extraordinary similarity of MYST domain of HBO1 with the rest of MYST family members may act as a barrier for obtaining HBO1-specific molecules [81]. Therefore, innovative design of HBO1-targeting molecules should be useful. From virtual simulation, there are three possible sites for molecule design in the MYST domain (Fig. 5a), in which the BRPF2-binding site (1#, Fig. 5a) may be the optimal option. BRPF2 binds to HBO1 on the hinge connecting the NTD and MYST domain, thus it is reasonable to develop BRPF2-mimic peptides or molecules for disrupting HBO1–BRPF2 interaction and subsequently prevent the binding of HBO1 to chromatin. It should be an optimal strategy to overcome the difficulty of designing HBO1-specific inhibitor. In addition, the acetyl-CoA-binding site (2#) and the positive pocket site (3#) provide options (Fig. 5a). From aforementioned (“Acetylation and autoacetylation regulates HBO1 activity”), the interaction between the NTD and MYST domain may provide a regulatory switch for HBO1 activity. If this model works, it is possible to design molecules through which modulating the HBO1 activity by interfering the interactions of NTD-MYST domain. Besides, microRNA is another way to regulate HBO1 expression. For example, microRNA-548a-3p has been shown to target SIX1 to repress HBO1 acetyltransferase activity-mediated glycolytic function of SIX1 [21]. Interestingly, computational screen of miRNA targets indicates that Hsa-miR-548a-3p is also targeted to HBO1 3′ UTR with a 7mer-m8 site, suggesting the possible regulation of HBO1 by miRNA (Fig. 5b). In summary, further research in developing HBO1-targeting molecules will certainly contribute to understanding the mechanism of HBO1 regulation and have potential therapeutic applications.
Fig. 5.
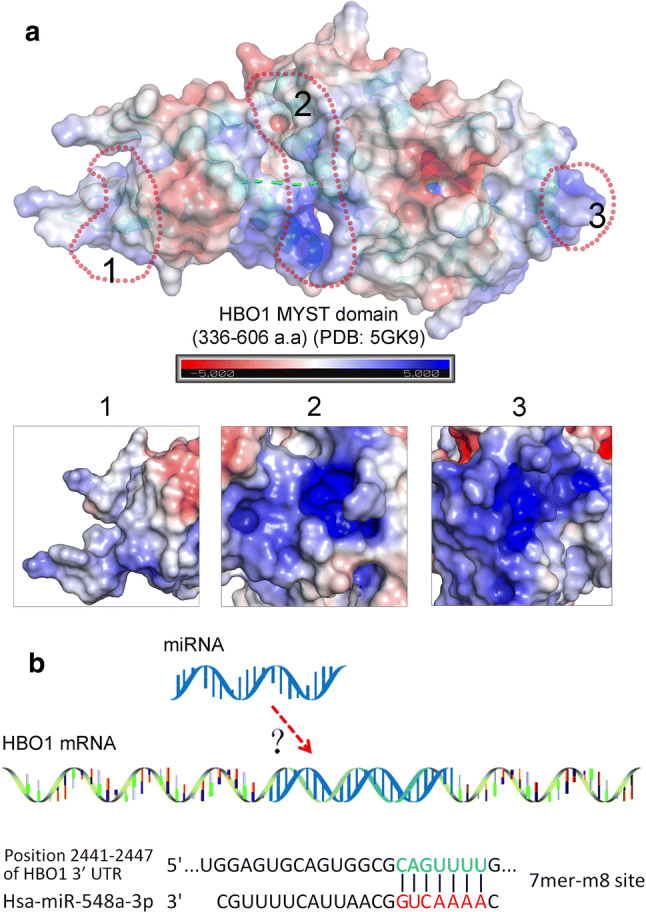
Design of targeting molecules and potential strategies for HBO1 inhibition. a MYST domain of HBO1 contains abundant sites for molecule binding. BRPF2-binding site (#1), acetyl-CoA-binding site (#2) and positive charge pocket (#3) provide potential sites for inhibitor binding. b miRNA is another approach for the regulation of HBO1 expression. For example, Hsa-miR-548a-3p is predicted to target on a 7mer-m8 site of 3′ UTR to inhibit HBO1 gene expression
Conclusion and perspectives
Compelling evidence increasingly deciphers the multiple functions of HBO1 in DNA replication, gene transcription, immune regulation, and cancers, etc. However, in-depth elucidation of the mechanism regarding HBO1 acetyltransferase activation and its substrate specificity, especially the role of NTD in HBO1 activity regulation is needed. Full-length structure of HBO1 available in future is critical to decipher the mechanism of HBO1 activation, the interaction between the NTD and MYST domain, the binding to cofactors and is required for designing HBO1-targeting molecule. Apart from histone H3 and H4, non-histone substrates of HBO1 remained to be characterized. High abundant acetylation was found in HBO1 and its cofactors such as ING4, ING5, Jade-1, BRPF2, and MEAF6. However, it is remains unclear whether the HBO1 activity that contributes to acetylation of these cofactors need to be characterized. Although MEAF6 is the native component of HBO1 complexes, how MEAF6 interact with HBO1 and its functions in HBO1 activation, activity regulation and substrate specificity, e.g., are far from well elucidated. In addition, tissue specific acetyltransferase activity of HBO1 and its relationship to disease requires special focus since HBO1 is highly expressed in testis or ovary. Although integrative RNA-seq data have indicated the gene amplification and overexpression of HBO1 in a number of cancers, its clear functions in cancer cell proliferation and invasive migration remain elucidated. Bona fide in vitro and in vivo evidence are needed to evaluate whether HBO1 is a promising target. Virtual screening and molecular design may be useful to obtain preliminary compounds that target HBO1. Collectively, in-depth elucidation of the properties of HBO1 and its protein complexes will help present a full story of HBO1 functions and pave a new avenue for applications such as disease therapy.
Abbreviations
- AR
Androgen receptor
- BRPF1
Bromodomain and PHD finger containing 1
- CDK1
Cyclin-dependent kinase 1
- ERα
Estrogen receptor α
- HAT
Histone acetyltransferase
- HBO1
Histone acetyltransferase binding to ORC1
- HBZ
HTLV-1 basic zipper factor
- ING4
Inhibitor of growth family member 4
- Jade-1
Jade family PHD finger 1
- KAT
Lysine acetyltransferase
- MCM
Minichromosome maintenance
- MEAF6
MYST/Esa1-associated factor 6
- MYST domain
(MOZ, Ybf1/Sas3, Sas2 and Tip60) domain
- M18BP1
Mis18-binding protein 1
- NF-κB
Nuclear factor κB
- NTD
N-terminal domain
- ORC1
Origin recognition complex 1
- PHD
Plant homology zinc finger domain
- PLK1
Polo-like kinase 1
- PR
Progesterone receptor
- SRC1
Steroid receptor co-activator 1
- USP25
Ubiquitin carboxyl-terminal hydrolase 25
Author contributions
RFL wrote and approved the manuscript. QQW performed the literature search and revised the manuscript.
Funding
This work was supported by Grants from the National Natural Science Foundation of China (21778038), and Shenzhen science and technology innovation committee program (JCYJ20180305163349116).
Compliance with ethical standards
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Availability of data and materials
Please contact corresponding author for data requests.
Conflict of interest
The authors declare no conflicts of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Feng Y, Vlassis A, Roques C, Lalonde ME, Gonzalez-Aguilera C, Lambert JP, Lee SB, Zhao X, Alabert C, Johansen JV, Paquet E, Yang XJ, Gingras AC, Cote J, Groth A. BRPF3–HBO1 regulates replication origin activation and histone H3K14 acetylation. EMBO J. 2016;35(2):176–192. doi: 10.15252/embj.201591293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mishima Y, Miyagi S, Saraya A, Negishi M, Endoh M, Endo TA, Toyoda T, Shinga J, Katsumoto T, Chiba T, Yamaguchi N, Kitabayashi I, Koseki H, Iwama A. The Hbo1–Brd1/Brpf2 complex is responsible for global acetylation of H3K14 and required for fetal liver erythropoiesis. Blood. 2011;118(9):2443–2453. doi: 10.1182/blood-2011-01-331892. [DOI] [PubMed] [Google Scholar]
- 3.Kueh AJ, Dixon MP, Voss AK, Thomas T. HBO1 is required for H3K14 acetylation and normal transcriptional activity during embryonic development. Mol Cell Biol. 2011;31(4):845–860. doi: 10.1128/MCB.00159-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Doyon Y, Cayrou C, Ullah M, Landry AJ, Cote V, Selleck W, Lane WS, Tan S, Yang XJ, Cote J. ING tumor suppressor proteins are critical regulators of chromatin acetylation required for genome expression and perpetuation. Mol Cell. 2006;21(1):51–64. doi: 10.1016/j.molcel.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 5.Sapountzi V, Cote J. MYST-family histone acetyltransferases: beyond chromatin. Cell Mol Life Sci. 2011;68(7):1147–1156. doi: 10.1007/s00018-010-0599-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim MS, Cho HI, Park SH, Kim JH, Chai YG, Jang YK. The histone acetyltransferase Myst2 regulates Nanog expression, and is involved in maintaining pluripotency and self-renewal of embryonic stem cells. FEBS Lett. 2015;589(8):941–950. doi: 10.1016/j.febslet.2015.02.029. [DOI] [PubMed] [Google Scholar]
- 7.Wang Q, Zhang Y, Yang C, Xiong H, Lin Y, Yao J, Li H, Xie L, Zhao W, Yao Y, Ning ZB, Zeng R, Xiong Y, Guan KL, Zhao S, Zhao GP. Acetylation of metabolic enzymes coordinates carbon source utilization and metabolic flux. Science. 2010;327(5968):1004–1007. doi: 10.1126/science.1179687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao S, Xu W, Jiang W, Yu W, Lin Y, Zhang T, Yao J, Zhou L, Zeng Y, Li H, Li Y, Shi J, An W, Hancock SM, He F, Qin L, Chin J, Yang P, Chen X, Lei Q, Xiong Y, Guan KL. Regulation of cellular metabolism by protein lysine acetylation. Science. 2010;327(5968):1000–1004. doi: 10.1126/science.1179689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325(5942):834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 10.Narita T, Weinert BT, Choudhary C. Functions and mechanisms of non-histone protein acetylation. Nat Rev Mol Cell Biol. 2019;20(3):156–174. doi: 10.1038/s41580-018-0081-3. [DOI] [PubMed] [Google Scholar]
- 11.Iizuka M, Stillman B. Histone acetyltransferase HBO1 interacts with the ORC1 subunit of the human initiator protein. J Biol Chem. 1999;274(33):23027–23034. doi: 10.1074/jbc.274.33.23027. [DOI] [PubMed] [Google Scholar]
- 12.Burke TW, Cook JG, Asano M, Nevins JR. Replication factors MCM2 and ORC1 interact with the histone acetyltransferase HBO1. J Biol Chem. 2001;276(18):15397–15408. doi: 10.1074/jbc.m011556200m011556200. [DOI] [PubMed] [Google Scholar]
- 13.Tao Y, Zhong C, Zhu J, Xu S, Ding J. Structural and mechanistic insights into regulation of HBO1 histone acetyltransferase activity by BRPF2. Nucleic Acids Res. 2017;45(10):5707–5719. doi: 10.1093/nar/gkx142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iizuka M, Sarmento OF, Sekiya T, Scrable H, Allis CD, Smith MM. Hbo1 links p53-dependent stress signaling to DNA replication licensing. Mol Cell Biol. 2008;28(1):140–153. doi: 10.1128/mcb.00662-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sharma M, Zarnegar M, Li X, Lim B, Sun Z. Androgen receptor interacts with a novel MYST protein, HBO1. J Biol Chem. 2000;275(45):35200–35208. doi: 10.1074/jbc.M004838200. [DOI] [PubMed] [Google Scholar]
- 16.Han Z, Wu H, Kim S, Yang X, Li Q, Huang H, Cai H, Bartlett MG, Dong A, Zeng H, Brown PJ, Yang XJ, Arrowsmith CH, Zhao Y, Zheng YG. Revealing the protein propionylation activity of the histone acetyltransferase MOF (males absent on the first) J Biol Chem. 2018;293(9):3410–3420. doi: 10.1074/jbc.RA117.000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iizuka M, Susa T, Tamamori-Adachi M, Okinaga H, Okazaki T. Intrinsic ubiquitin E3 ligase activity of histone acetyltransferase Hbo1 for estrogen receptor alpha. Proc Jpn Acad Ser B Phys Biol Sci. 2017;93(7):498–510. doi: 10.2183/pjab.93.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Contzler R, Regamey A, Favre B, Roger T, Hohl D, Huber M. Histone acetyltransferase HBO1 inhibits NF-kappaB activity by coactivator sequestration. Biochem Biophys Res Commun. 2006;350(1):208–213. doi: 10.1016/j.bbrc.2006.09.030. [DOI] [PubMed] [Google Scholar]
- 19.Georgiakaki M, Chabbert-Buffet N, Dasen B, Meduri G, Wenk S, Rajhi L, Amazit L, Chauchereau A, Burger CW, Blok LJ, Milgrom E, Lombes M, Guiochon-Mantel A, Loosfelt H. Ligand-controlled interaction of histone acetyltransferase binding to ORC-1 (HBO1) with the N-terminal transactivating domain of progesterone receptor induces steroid receptor coactivator 1-dependent coactivation of transcription. Mol Endocrinol. 2006;20(9):2122–2140. doi: 10.1210/me.2005-0149. [DOI] [PubMed] [Google Scholar]
- 20.Vija L, Meduri G, Comperat E, Vasiliu V, Izard V, Ferlicot S, Boukari K, Camparo P, Viengchareun S, Constancis E, Dumitrache C, Lombes M, Young J. Expression and characterization of androgen receptor coregulators, SRC-2 and HBO1, during human testis ontogenesis and in androgen signaling deficient patients. Mol Cell Endocrinol. 2013;375(1–2):140–148. doi: 10.1016/j.mce.2013.05.004. [DOI] [PubMed] [Google Scholar]
- 21.Li L, Liang Y, Kang L, Liu Y, Gao S, Chen S, Li Y, You W, Dong Q, Hong T, Yan Z, Jin S, Wang T, Zhao W, Mai H, Huang J, Han X, Ji Q, Song Q, Yang C, Zhao S, Xu X, Ye Q. Transcriptional regulation of the Warburg effect in cancer by SIX1. Cancer Cell. 2018;33(3):368–385. doi: 10.1016/j.ccell.2018.01.010. [DOI] [PubMed] [Google Scholar]
- 22.Pardo M, Yu L, Shen S, Tate P, Bode D, Letney BL, Quelle DE, Skarnes W, Choudhary JS. Myst2/Kat7 histone acetyltransferase interaction proteomics reveals tumour-suppressor Niam as a novel binding partner in embryonic stem cells. Sci Rep. 2017;7(1):8157. doi: 10.1038/s41598-017-08456-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yan MS, Turgeon PJ, Man HJ, Dubinsky MK, Ho JJD, El-Rass S, Wang YD, Wen XY, Marsden PA. Histone acetyltransferase 7 (KAT7)-dependent intragenic histone acetylation regulates endothelial cell gene regulation. J Biol Chem. 2018;293(12):4381–4402. doi: 10.1074/jbc.RA117.001383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sterner DE, Berger SL. Acetylation of histones and transcription-related factors. Microbiol Mol Biol Rev. 2000;64(2):435–459. doi: 10.1128/MMBR.64.2.435-459.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saksouk N, Avvakumov N, Champagne KS, Hung T, Doyon Y, Cayrou C, Paquet E, Ullah M, Landry AJ, Cote V, Yang XJ, Gozani O, Kutateladze TG, Cote J. HBO1 HAT complexes target chromatin throughout gene coding regions via multiple PHD finger interactions with histone H3 tail. Mol Cell. 2009;33(2):257–265. doi: 10.1016/j.molcel.2009.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen X, Liu G, Leffak M. Activation of a human chromosomal replication origin by protein tethering. Nucleic Acids Res. 2013;41(13):6460–6474. doi: 10.1093/nar/gkt368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sugimoto N, Fujita M. Molecular mechanism for chromatin regulation during MCM loading in mammalian cells. Adv Exp Med Biol. 2017;1042:61–78. doi: 10.1007/978-981-10-6955-0_3. [DOI] [PubMed] [Google Scholar]
- 28.Miotto B, Struhl K. HBO1 histone acetylase is a coactivator of the replication licensing factor Cdt1. Genes Dev. 2008;22(19):2633–2638. doi: 10.1101/gad.1674108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miotto B, Struhl K. HBO1 histone acetylase activity is essential for DNA replication licensing and inhibited by Geminin. Mol Cell. 2010;37(1):57–66. doi: 10.1016/j.molcel.2009.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iizuka M, Matsui T, Takisawa H, Smith MM. Regulation of replication licensing by acetyltransferase Hbo1. Mol Cell Biol. 2006;26(3):1098–1108. doi: 10.1128/mcb.26.3.1098-1108.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu ZQ, Liu X. Role for Plk1 phosphorylation of Hbo1 in regulation of replication licensing. Proc Natl Acad Sci U S A. 2008;105(6):1919–1924. doi: 10.1073/pnas.0712063105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wong PG, Glozak MA, Cao TV, Vaziri C, Seto E, Alexandrow M. Chromatin unfolding by Cdt1 regulates MCM loading via opposing functions of HBO1 and HDAC11–geminin. Cell Cycle. 2010;9(21):4351–4363. doi: 10.4161/cc.9.21.13596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miotto B, Struhl K. JNK1 phosphorylation of Cdt1 inhibits recruitment of HBO1 histone acetylase and blocks replication licensing in response to stress. Mol Cell. 2011;44(1):62–71. doi: 10.1016/j.molcel.2011.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chadha GS, Blow JJ. Histone acetylation by HBO1 tightens replication licensing. Mol Cell. 2010;37(1):5–6. doi: 10.1016/j.molcel.2009.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ohzeki J, Shono N, Otake K, Martins NM, Kugou K, Kimura H, Nagase T, Larionov V, Earnshaw WC, Masumoto H. KAT7/HBO1/MYST2 regulates CENP-A chromatin assembly by antagonizing SUV39h1-mediated centromere inactivation. Dev Cell. 2016;37(5):413–427. doi: 10.1016/j.devcel.2016.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Niida H, Matsunuma R, Horiguchi R, Uchida C, Nakazawa Y, Motegi A, Nishimoto K, Sakai S, Ohhata T, Kitagawa K, Moriwaki S, Nishitani H, Ui A, Ogi T, Kitagawa M. Phosphorylated HBO1 at UV irradiated sites is essential for nucleotide excision repair. Nat Commun. 2017;8:16102. doi: 10.1038/ncomms16102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Matsunuma R, Niida H, Ohhata T, Kitagawa K, Sakai S, Uchida C, Shiotani B, Matsumoto M, Nakayama KI, Ogura H, Shiiya N, Kitagawa M. UV damage-induced phosphorylation of HBO1 triggers CRL4DDB2-mediated degradation to regulate cell proliferation. Mol Cell Biol. 2015;36(3):394–406. doi: 10.1128/MCB.00809-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zou C, Chen Y, Smith RM, Snavely C, Li J, Coon TA, Chen BB, Zhao Y, Mallampalli RK. SCF(Fbxw15) mediates histone acetyltransferase binding to origin recognition complex (HBO1) ubiquitin-proteasomal degradation to regulate cell proliferation. J Biol Chem. 2013;288(9):6306–6316. doi: 10.1074/jbc.M112.426882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Long C, Lai Y, Li J, Huang J. Zou C (2018) LPS promotes HBO1 stability via USP25 to modulate inflammatory gene transcription in THP-1 cells. Biochim Biophys Acta Gene Regul Mech. 1861;9:773–782. doi: 10.1016/j.bbagrm.2018.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Iizuka M, Susa T, Takahashi Y, Tamamori-Adachi M, Kajitani T, Okinaga H, Fukusato T, Okazaki T. Histone acetyltransferase Hbo1 destabilizes estrogen receptor alpha by ubiquitination and modulates proliferation of breast cancers. Cancer Sci. 2013;104(12):1647–1655. doi: 10.1111/cas.12303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chitalia VC, Foy RL, Bachschmid MM, Zeng L, Panchenko MV, Zhou MI, Bharti A, Seldin DC, Lecker SH, Dominguez I, Cohen HT. Jade-1 inhibits Wnt signalling by ubiquitylating beta-catenin and mediates Wnt pathway inhibition by pVHL. Nat Cell Biol. 2008;10(10):1208–1216. doi: 10.1038/ncb1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Newman DM, Voss AK, Thomas T, Allan RS. Essential role for the histone acetyltransferase KAT7 in T cell development, fitness, and survival. J Leukoc Biol. 2017;101(4):887–892. doi: 10.1189/jlb.1MA0816-338R. [DOI] [PubMed] [Google Scholar]
- 43.Mishima Y, Wang C, Miyagi S, Saraya A, Hosokawa H, Mochizuki-Kashio M, Nakajima-Takagi Y, Koide S, Negishi M, Sashida G, Naito T, Ishikura T, Onodera A, Nakayama T, Tenen DG, Yamaguchi N, Koseki H, Taniuchi I, Iwama A. Histone acetylation mediated by Brd1 is crucial for Cd8 gene activation during early thymocyte development. Nat Commun. 2014;5:5872. doi: 10.1038/ncomms6872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gao S, Qi X, Li J, Sang L. Upregulated KAT7 in synovial fibroblasts promotes Th17 cell differentiation and infiltration in rheumatoid arthritis. Biochem Biophys Res Commun. 2017;489(2):235–241. doi: 10.1016/j.bbrc.2017.05.143. [DOI] [PubMed] [Google Scholar]
- 45.Wright DG, Marchal C, Hoang K, Ankney JA, Nguyen ST, Rushing AW, Polakowski N, Miotto B, Lemasson I. Human T-cell leukemia virus type-1-encoded protein HBZ represses p53 function by inhibiting the acetyltransferase activity of p300/CBP and HBO1. Oncotarget. 2016;7(2):1687–1706. doi: 10.18632/oncotarget.6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jensen LJ, Kuhn M, Stark M, Chaffron S, Creevey C, Muller J, Doerks T, Julien P, Roth A, Simonovic M, Bork P, von Mering C. STRING 8—a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 2009;37(Database issue):D412–D416. doi: 10.1093/nar/gkn760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ochiai N, Nishizuka M, Osada S, Imagawa M. Fad24, a positive regulator of adipogenesis, is required for S phase re-entry of C2C12 myoblasts arrested in G0 phase and involved in p27(Kip1) expression at the protein level. Biol Pharm Bull. 2016;39(5):807–814. doi: 10.1248/bpb.b15-00954. [DOI] [PubMed] [Google Scholar]
- 48.Duong MT, Akli S, Macalou S, Biernacka A, Debeb BG, Yi M, Hunt KK, Keyomarsi K. Hbo1 is a cyclin E/CDK2 substrate that enriches breast cancer stem-like cells. Cancer Res. 2013;73(17):5556–5568. doi: 10.1158/0008-5472.CAN-13-0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zong H, Li Z, Liu L, Hong Y, Yun X, Jiang J, Chi Y, Wang H, Shen X, Hu Y, Niu Z, Gu J. Cyclin-dependent kinase 11(p58) interacts with HBO1 and enhances its histone acetyltransferase activity. FEBS Lett. 2005;579(17):3579–3588. doi: 10.1016/j.febslet.2005.05.039. [DOI] [PubMed] [Google Scholar]
- 50.Johmura Y, Osada S, Nishizuka M, Imagawa M. FAD24 acts in concert with histone acetyltransferase HBO1 to promote adipogenesis by controlling DNA replication. J Biol Chem. 2008;283(4):2265–2274. doi: 10.1074/jbc.m707880200. [DOI] [PubMed] [Google Scholar]
- 51.Hung T, Binda O, Champagne KS, Kuo AJ, Johnson K, Chang HY, Simon MD, Kutateladze TG, Gozani O. ING4 mediates crosstalk between histone H3 K4 trimethylation and H3 acetylation to attenuate cellular transformation. Mol Cell. 2009;33(2):248–256. doi: 10.1016/j.molcel.2008.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ormaza G, Rodriguez JA, Ibanez de Opakua A, Merino N, Villate M, Gorrono I, Rabano M, Palmero I, Vilaseca M, Kypta R, Vivanco MDM, Rojas AL, Blanco FJ. The tumor suppressor ing5 is a dimeric, bivalent recognition molecule of the histone H3K4me3 mark. J Mol Biol. 2019;431(12):2298–2319. doi: 10.1016/j.jmb.2019.04.018. [DOI] [PubMed] [Google Scholar]
- 53.Palacios A, Moreno A, Oliveira BL, Rivera T, Prieto J, Garcia P, Fernandez-Fernandez MR, Bernado P, Palmero I, Blanco FJ. The dimeric structure and the bivalent recognition of H3K4me3 by the tumor suppressor ING4 suggests a mechanism for enhanced targeting of the HBO1 complex to chromatin. J Mol Biol. 2010;396(4):1117–1127. doi: 10.1016/j.jmb.2009.12.049. [DOI] [PubMed] [Google Scholar]
- 54.Foy RL, Song IY, Chitalia VC, Cohen HT, Saksouk N, Cayrou C, Vaziri C, Cote J, Panchenko MV. Role of Jade-1 in the histone acetyltransferase (HAT) HBO1 complex. J Biol Chem. 2008;283(43):28817–28826. doi: 10.1074/jbc.m801407200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Han J, Lachance C, Ricketts MD, McCullough CE, Gerace M, Black BE, Cote J, Marmorstein R. The scaffolding protein JADE1 physically links the acetyltransferase subunit HBO1 with its histone H3–H4 substrate. J Biol Chem. 2018;293(12):4498–4509. doi: 10.1074/jbc.RA117.000677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Siriwardana NS, Meyer R, Havasi A, Dominguez I, Panchenko MV. Cell cycle-dependent chromatin shuttling of HBO1–JADE1 histone acetyl transferase (HAT) complex. Cell Cycle. 2014;13(12):1885–1901. doi: 10.4161/cc.28759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Havasi A, Haegele JA, Gall JM, Blackmon S, Ichimura T, Bonegio RG, Panchenko MV. Histone acetyl transferase (HAT) HBO1 and JADE1 in epithelial cell regeneration. Am J Pathol. 2013;182(1):152–162. doi: 10.1016/j.ajpath.2012.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Calvi BR. HBO1:JADE1 at the cell cycle chromatin crossroads. Cell Cycle. 2014;13(15):2322. doi: 10.4161/cc.29832. [DOI] [PubMed] [Google Scholar]
- 59.Lalonde ME, Avvakumov N, Glass KC, Joncas FH, Saksouk N, Holliday M, Paquet E, Yan K, Tong Q, Klein BJ, Tan S, Yang XJ, Kutateladze TG, Cote J. Exchange of associated factors directs a switch in HBO1 acetyltransferase histone tail specificity. Genes Dev. 2013;27(18):2009–2024. doi: 10.1101/gad.223396.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen Y, Zhao W, Yang JS, Cheng Z, Luo H, Lu Z, Tan M, Gu W, Zhao Y. Quantitative acetylome analysis reveals the roles of SIRT1 in regulating diverse substrates and cellular pathways. Mol Cell Proteom. 2012;11(10):1048–1062. doi: 10.1074/mcp.M112.019547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xie C, Shen H, Zhang H, Yan J, Liu Y, Yao F, Wang X, Cheng Z, Tang TS, Guo C. Quantitative proteomics analysis reveals alterations of lysine acetylation in mouse testis in response to heat shock and X-ray exposure. Biochim Biophys Acta. 2018;1866(3):464–472. doi: 10.1016/j.bbapap.2017.11.011. [DOI] [PubMed] [Google Scholar]
- 62.Sun B, Guo S, Tang Q, Li C, Zeng R, Xiong Z, Zhong C, Ding J. Regulation of the histone acetyltransferase activity of hMOF via autoacetylation of Lys274. Cell Res. 2011;21(8):1262–1266. doi: 10.1038/cr.2011.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yuan H, Rossetto D, Mellert H, Dang W, Srinivasan M, Johnson J, Hodawadekar S, Ding EC, Speicher K, Abshiru N, Perry R, Wu J, Yang C, Zheng YG, Speicher DW, Thibault P, Verreault A, Johnson FB, Berger SL, Sternglanz R, McMahon SB, Cote J, Marmorstein R. MYST protein acetyltransferase activity requires active site lysine autoacetylation. EMBO J. 2012;31(1):58–70. doi: 10.1038/emboj.2011.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yang C, Wu J, Sinha SH, Neveu JM, Zheng YG. Autoacetylation of the MYST lysine acetyltransferase MOF protein. J Biol Chem. 2012;287(42):34917–34926. doi: 10.1074/jbc.M112.359356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Peng L, Ling H, Yuan Z, Fang B, Bloom G, Fukasawa K, Koomen J, Chen J, Lane WS, Seto E. SIRT1 negatively regulates the activities, functions, and protein levels of hMOF and TIP60. Mol Cell Biol. 2012;32(14):2823–2836. doi: 10.1128/MCB.00496-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Deng W, Wang C, Zhang Y, Xu Y, Zhang S, Liu Z, Xue Y. GPS-PAIL: prediction of lysine acetyltransferase-specific modification sites from protein sequences. Sci Rep. 2016;6:39787. doi: 10.1038/srep39787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fagerberg L, Hallstrom BM, Oksvold P, Kampf C, Djureinovic D, Odeberg J, Habuka M, Tahmasebpoor S, Danielsson A, Edlund K, Asplund A, Sjostedt E, Lundberg E, Szigyarto CA, Skogs M, Takanen JO, Berling H, Tegel H, Mulder J, Nilsson P, Schwenk JM, Lindskog C, Danielsson F, Mardinoglu A, Sivertsson A, von Feilitzen K, Forsberg M, Zwahlen M, Olsson I, Navani S, Huss M, Nielsen J, Ponten F, Uhlen M. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol Cell Proteomics. 2014;13(2):397–406. doi: 10.1074/mcp.M113.035600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, Cerami E, Sander C, Schultz N. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chen Z, Zhou L, Wang L, Kazobinka G, Zhang X, Han X, Li B, Hou T. HBO1 promotes cell proliferation in bladder cancer via activation of Wnt/beta-catenin signaling. Mol Carcinog. 2018;57(1):12–21. doi: 10.1002/mc.22715. [DOI] [PubMed] [Google Scholar]
- 70.Iizuka M, Takahashi Y, Mizzen CA, Cook RG, Fujita M, Allis CD, Frierson HF, Jr, Fukusato T, Smith MM. Histone acetyltransferase Hbo1: catalytic activity, cellular abundance, and links to primary cancers. Gene. 2009;436(1–2):108–114. doi: 10.1016/j.gene.2009.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hu X, Stern HM, Ge L, O’Brien C, Haydu L, Honchell CD, Haverty PM, Peters BA, Wu TD, Amler LC, Chant J, Stokoe D, Lackner MR, Cavet G. Genetic alterations and oncogenic pathways associated with breast cancer subtypes. Mol Cancer Res. 2009;7(4):511–522. doi: 10.1158/1541-7786.MCR-08-0107. [DOI] [PubMed] [Google Scholar]
- 72.Song B, Liu XS, Rice SJ, Kuang S, Elzey BD, Konieczny SF, Ratliff TL, Hazbun T, Chiorean EG, Liu X. Plk1 phosphorylation of orc2 and hbo1 contributes to gemcitabine resistance in pancreatic cancer. Mol Cancer Ther. 2013;12(1):58–68. doi: 10.1158/1535-7163.MCT-12-0632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang Y, Chen S, Tian W, Zhang Q, Jiang C, Qian L, Liu Y. High-expression HBO1 predicts poor prognosis in gastric cancer. Am J Clin Pathol. 2019;152(4):517–526. doi: 10.1093/ajcp/aqz065. [DOI] [PubMed] [Google Scholar]
- 74.Yang XJ. MOZ and MORF acetyltransferases: molecular interaction, animal development and human disease. Biochim Biophys Acta. 2015;1853(8):1818–1826. doi: 10.1016/j.bbamcr.2015.04.014. [DOI] [PubMed] [Google Scholar]
- 75.Quintela M, Sieglaff DH, Gazze AS, Zhang A, Gonzalez D, Francis L, Webb P, Conlan RS. HBO1 directs histone H4 specific acetylation, potentiating mechano-transduction pathways and membrane elasticity in ovarian cancer cells. Nanomedicine. 2019;17:254–265. doi: 10.1016/j.nano.2019.01.017. [DOI] [PubMed] [Google Scholar]
- 76.Moreno A, Palacios A, Orgaz JL, Jimenez B, Blanco FJ, Palmero I. Functional impact of cancer-associated mutations in the tumor suppressor protein ING4. Carcinogenesis. 2010;31(11):1932–1938. doi: 10.1093/carcin/bgq171. [DOI] [PubMed] [Google Scholar]
- 77.Cengiz B, Gunduz E, Gunduz M, Beder LB, Tamamura R, Bagci C, Yamanaka N, Shimizu K, Nagatsuka H. Tumor-specific mutation and downregulation of ING5 detected in oral squamous cell carcinoma. Int J Cancer. 2010;127(9):2088–2094. doi: 10.1002/ijc.25224. [DOI] [PubMed] [Google Scholar]
- 78.Kahali B, Gramling SJ, Marquez SB, Thompson K, Lu L, Reisman D. Identifying targets for the restoration and reactivation of BRM. Oncogene. 2014;33(5):653–664. doi: 10.1038/onc.2012.613. [DOI] [PubMed] [Google Scholar]
- 79.Sauer T, Arteaga MF, Isken F, Rohde C, Hebestreit K, Mikesch JH, Stelljes M, Cui C, Zhou F, Gollner S, Baumer N, Kohler G, Krug U, Thiede C, Ehninger G, Edemir B, Schlenke P, Berdel WE, Dugas M, Muller-Tidow C. MYST2 acetyltransferase expression and Histone H4 Lysine acetylation are suppressed in AML. Exp Hematol. 2015;43(9):794–802. doi: 10.1016/j.exphem.2015.05.010. [DOI] [PubMed] [Google Scholar]
- 80.Hayashi Y, Harada Y, Kagiyama Y, Nishikawa S, Ding Y, Imagawa J, Shingai N, Kato N, Kitaura J, Hokaiwado S, Maemoto Y, Ito A, Matsui H, Kitabayashi I, Iwama A, Komatsu N, Kitamura T, Harada H. NUP98-HBO1-fusion generates phenotypically and genetically relevant chronic myelomonocytic leukemia pathogenesis. Blood Adv. 2019;3(7):1047–1060. doi: 10.1182/bloodadvances.2018025007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Guo LL, Yu SY, Li M. Functional analysis of HBO1 in tumor development and inhibitor screening. Int J Mol Med. 2016;38(1):300–304. doi: 10.3892/ijmm.2016.2617. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Please contact corresponding author for data requests.