Abstract
Odorant-binding proteins (OBPs) are small soluble proteins that are thought to transport hydrophobic odorants across the aqueous sensillar lymph to olfactory receptors. A recent study revealed that OBP28a, one of the most abundant Drosophila OBPs, is not required for odorant transport, but acts in buffering rapid odour variation in the odorant environment. To further unravel and decipher its functional role, we expressed recombinant OBP28a and characterized its binding specificity. Using a fluorescent binding assay, we found that OBP28a binds a restricted number of floral-like chemicals, including ß-ionone, with an affinity in the micromolar range. We solved the X-ray crystal structure of OBP28a, which showed extensive conformation changes upon ligand binding. Mutant flies genetically deleted for the OBP28a gene showed altered responses to ß-ionone at a given concentration range, supporting its essential role in the detection of specific compounds present in the natural environment of the fly.
Electronic supplementary material
The online version of this article (10.1007/s00018-019-03300-4) contains supplementary material, which is available to authorized users.
Keywords: Drosophila melanogaster, Insect, Olfaction, Odorant, Pheromone, Odorant-protein-binding assay
Introduction
The perception of chemical cues allows insects to detect potential food sources, oviposition sites, and mating partners. Olfaction is mediated by olfactory-sensory neurons (OSNs) housed in sensilla borne by their antennae and maxillary palps. Olfactory sensilla are small hair-like structures organized into three main classes—trichoid, basiconic, and coeloconic—according to their size and morphology. Fruit odours [1] and amines/acids [2, 3] are detected by basiconic and coeloconic sensilla, respectively, whereas pheromones are generally detected by trichoid sensilla [4–6]. Under the protection of a permeable cuticular shell, each sensillum contains olfactory neuron dendrites bathing in an aqueous lymph. In Drosophila melanogaster, at least two classes of olfactory receptor proteins seem to be essential for the detection of volatile compounds. First, odorant receptors (Ors; ~ 60 genes), which are co-expressed with the OrCO co-receptor both in basiconic and trichoid sensilla. Ors are involved in the detection of the male sex pheromone (Z)-11-octadecenyl acetate (cis-vaccenyl acetate, cVA) and of general odorants [7, 8]. Second, ionotropic receptors (Irs; ~ 17 genes), which are expressed in coeloconic sensilla, can detect a variety of chemicals, such as alcohols, ammonia, organic acids, and amines [9, 10]. In addition to both families of transmembrane proteins, the Drosophila genome encodes a large and diverse family of 52 odorant-binding protein (OBP) genes [11–14]. Insect OBPs are small soluble proteins (approximately 14 kDa) abundantly secreted in the sensillar lymph by non-neuronal support cells; OBPs show binding activity towards odorants and pheromones [15, 16]. The binding properties of OBPs were investigated in many insect species: some of them show a broad selectivity, while others have a narrower specificity towards odorants [17, 18]. Surprisingly, several OBPs were found in the gustatory system [11, 19]. Despite a high sequence divergence between OBPs, structural studies revealed that all OBPs share a common overall structure made of a 6 α-helix core stabilized with three disulphide bridges shaping together an internal binding cavity [20–24]. Although OBPs have been deeply studied in many insect species, their physiological functions remain elusive. Their relatively low affinity for odorants taken together with their abundance in the sensilla lymph suggests that OBPs transport hydrophobic stimuli to the chemoreceptors across the aqueous sensilla lymph [17, 25, 26]. OBPs have also been proposed to be involved in the elimination of odorants after olfactory receptor binding [15].
In Drosophila melanogaster, the physiological role of OBP76a (also called LUSH) is necessary for the electrophysiological activation of chemosensory neurons and behavioural response to cVA [27], although the precise mechanism involved remains controversial [28]. OBP69a was shown to be a key player in the machinery modulating social responses in Drosophila [29]. Two other Drosophila OBPs (OBP57d and OBP57e) are necessary for the detection of hexanoic and octanoic acids, both compounds involved in the oviposition site preference diverging between D. melanogaster and D. sechellia species [30]. Recently, an unexpected role of OBP59a in humidity detection was revealed, supporting the idea of diverse roles for OBPs [31].
The recent characterization of the expression pattern of 10 abundant OBPs in Drosophila antennae showed that OBP28a is expressed in different subsets of basiconic sensilla (ab1–ab10). The electrophysiological responses of single ab8 sensilla to odorant molecules of different chemical classes, such as 1-octanol, butyric acid and ethyl acetate, tested at varied concentrations, showed a higher frequency of action potential in flies genetically deleted for the OBP28a gene compared to control flies [32]. This result indicates that OBP28a is not only required for the transport of the tested odorants, but can also buffer quantitative odour variations, depending on the odour concentration in the odorant environment.
To better understand the physiological role of OBP28a, we performed a multidisciplinary study based on structural, genetic, biochemical, behavioural, and electrophysiological methods in Drosophila melanogaster. First, to obtain enough OBP28a for further testing, we expressed this protein in the yeast Pichia pastoris. This recombinantly expressed protein allowed us to perform a wide screening array involving varied ligand compounds. We found that OBP28a binds a limited number of floral-like chemicals, including ß-ionone. The three-dimensional structure of purified OBP28a was solved in both ligand-free (apo) and bound states, revealing a large conformational reorganization induced by ligand binding after atypical remodelling of the binding cavity. The genetic deletion of the OBP28a gene also affected both behavioural and electrophysiological responses in mutant fruit flies at a specific range of ß-ionone concentrations.
Results
Heterologous expression and biophysical characterization of OBP28a
To elucidate the structure–function relationship of OBP28a, we heterologously secreted this protein using the methylotrophic yeast P. pastoris, which was previously used to express honeybee OBPs [33, 34]. The recombinant OBP28a was purified by ion-exchange chromatography followed by reverse-phase liquid chromatography (RPLC) (Supplemental Fig. 1). SDS-PAGE analysis of purified OBP28a revealed a > 99% purity of the recombinant protein with a molecular mass of around 13 kDa. This experimental molecular mass is in agreement with the theoretical molecular mass value of the mature protein as ascertained by mass spectrometry analysis (Supplemental Fig. 2). We used circular dichroism (CD) spectroscopy to confirm the folding of OBP28a. The OBP28a far-UV CD spectrum displayed a positive peak centred at 193 nm together with two negative peaks at 206 and 222 nm, which are characteristics of a protein containing a high number of helical secondary structures (Supplemental Fig. 2) and is consistent with the expected highly α-helical content of insect OBPs. Given that insect OBPs have been shown to form monomers or dimers [35], we tested the oligomeric state of OBP28a. Using size-exclusion chromatography (SEC) coupled with in-line multi-angle laser light scattering (MALS; SEC-MALS), we found that OBP28a elutes as a monodisperse peak with a measured mass of 15.7 kDa (Supplemental Fig. 2). This value is close to the monomer theoretical mass value, strongly suggesting that OBP28a acts as a monomer in solution at the tested concentration.
Overall structure of OBP28a
Our next goal was to determine the molecular features of OBP28a. We solved the crystal structure of OBP28a by molecular replacement and refined its structure to a 2.0 Å resolution with Rwork and Rfree of 19.0 and 23.6%, respectively (Table 1). We found two OBP28a protein molecules in the crystallographic asymmetric unit. More precisely, OBP28a forms an asymmetrical dimer with a surface of the crystal-packing interface between monomers of 515.8 Å2, representing ~ 8% of the total molecular surface. This low surface contact value suggests that the crystallographic OBP28a dimer has no biological significance, in agreement with a monomeric state of OBP28a, as demonstrated by SEC-MALS analysis (Supplemental Fig. 2). The two OBP28a configuration molecules (conformers) share the typical overall structure of OBPs with a 6 α-helix bundle connected with 3 disulphide bridges shaping a ligand-binding cavity (Fig. 1a). Analysis of the monomers revealed the presence of a residual electron density in the binding cavity of OBP28a, indicating the presence of a ligand. In the first monomer, the shape and size of the compound in the cavity are compatible with pentaethylene glycol, a Jeffamine derivative (1PE), used as a precipitating agent (Fig. 1b). In the second monomer (called apo-OBP28a), we identified a different and much smaller atom density, which can be attributed to one malonic acid molecule used as a sodium salt precipitant (Fig. 1c). Due to its very limited interaction with OBP28a, this molecule cannot be considered as a ligand. In addition, 27 amino acids belonging to all helices contributed to the inner surface of the cavity, which was delineated by 19 hydrophobic side chains, 3 polar non-charged side chains, and only 5 charged side chains (Supplemental Fig. 3).
Table 1.
Data collection and refinement of the OBP28a crystal structure
| PDB ID | 6QQ4 |
|---|---|
| Beamline | ID29 |
| Wavelength (Å) | 0.978 |
| Temperature (K) | − 173 °C |
| Detector | Pilatus3–6 M |
| Crystal–detector distance (mm) | 392.72 |
| Oscillation (°) | 0.1 |
| Total rotation range | 180 |
| Exposure time per image (s) | 0.020 |
| Space group | P21 |
| a, b, c (Å) | a = 34.5, b = 83.0, c = 45.5 |
| α, β, γ (°) | β = 98.9 |
| Mosaicity | 0.238 |
| Resolution range (Å) (last bin) | 2.00 (2.05–2.00) |
| No. of observed reflections (last bin) | 55,842 (4435) |
| No. of unique reflections (last bin) | 16,958 (1282) |
| Completeness (%) (last bin) | 98.1 (98.7) |
| Redundancy | 3.29 (3.46) |
| CC 1/2 (%) | 99.8 (75.4) |
| I/σ (I) (last bin) | 13.68 (1.99) |
| Rmeas (%) (last bin) | 6.6 (85.4) |
| Overall B factor from Wilson plot (Ų) | 34.4 |
| Refinement statistics | |
| Resolution (Å) | 2.0 |
| Rwork/Rfree (last bin) | 19.0/23.6 (27.3/32.8) |
| Number of atoms | |
| Protein | 1761 |
| Ligands/ions | 24 |
| Solvent | 162 |
| Average B factors (Ų) | |
| Protein atoms (chain A: chain B) | 41.0:40.4 |
| 1PE (chain A) | 52.5 |
| MLA (chain B) | 50.4 |
| Solvent | 45.0 |
| RMSD | |
| Bond lengths (Å) | 0.007 |
| Bond angles (°) | 1.08 |
| Ramachandran plot | |
| Ramachandran favoured (%) | 100 |
| Ramachandran outliers (%) | 0.0 |
Fig. 1.
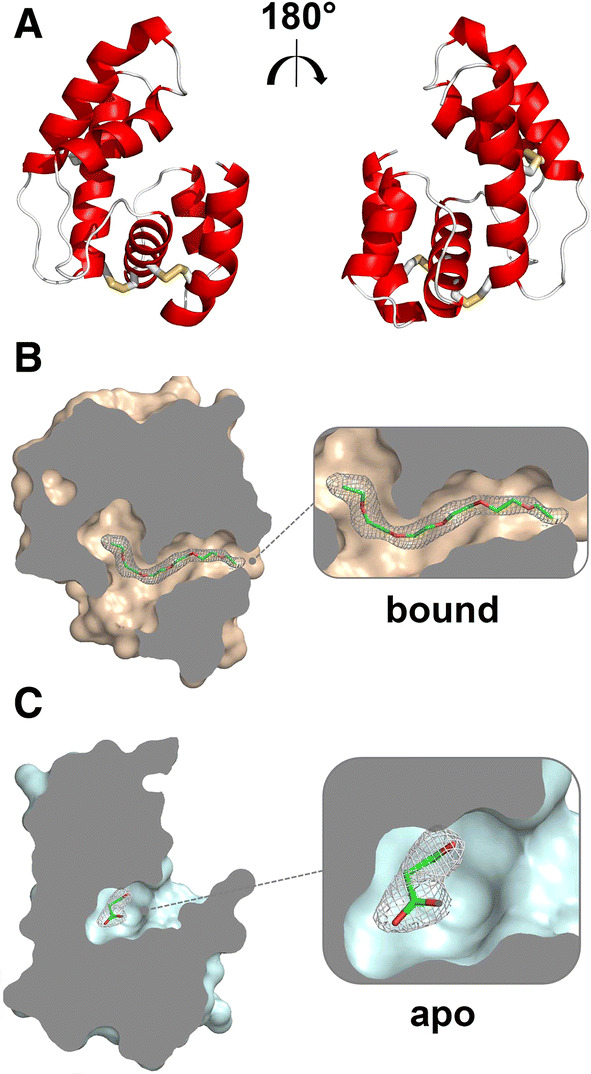
Crystal structure of OBP28a with a bound serendipitous ligand. a Rotated views (180°) of the tri-dimensional structure of OBP28a in cartoon representation. The three disulphide bridges are shown in yellow. b Slabbed view of OBP28a in complex with pentaethylene glycol. c Slabbed view of OBP28a in complex with malonic acid
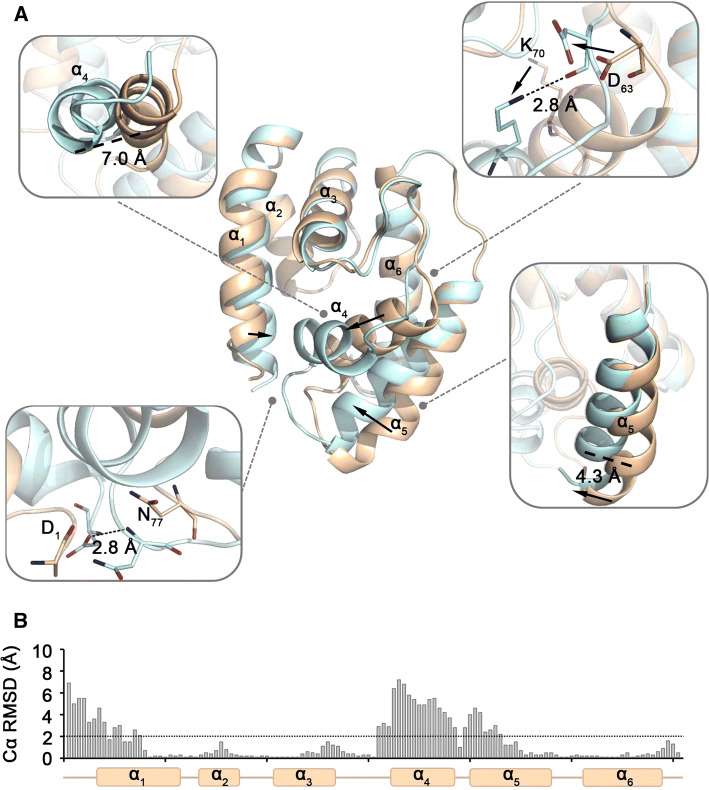
OBP28a crystals reveal two alternative conformations
OBP28a was crystallized in two alternative conformations in the same crystal and in the same conditions (e.g., pH, ionic force, and buffer) with the only dependence of ligand binding. These large conformational changes were only caused by the molecule binding into the cavity (here: pentaethylene glycol). The bound structure (Fig. 2a; Supplemental Movie) displayed a 120% increase in the solvent accessible volume compared to the apo structure (201 and 90 Å3, respectively). The structural reorganization of the α1, α4, and α5 helices can explain the extended conformation of the bound structure (Fig. 2b). In addition to the extended cavity conformation, the bound structure also showed the formation of a second entry in the cavity (Supplemental Fig. 4).
Fig. 2.
Conformational changes of OBP28a upon ligand binding. a Conformational differences between apo- (in wheat) and bound-OBP28a (in light blue) were identified and indicated in each insert. b Cα RMSD along the sequence of OBP28a. The line indicates the mean RMSD Cα between both conformations
OBP28a displays a restrained binding profile
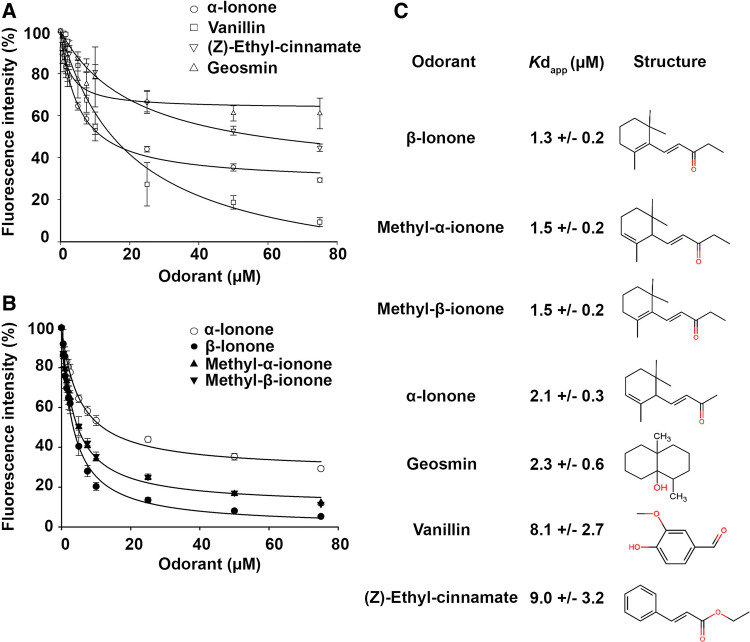
To measure the binding properties of OBP28a, we first tested its ability to bind the fluorescent probe N-phenyl-1-naphthylamine (NPN). The titration of OBP28a with NPN was saturable, leading to a dissociation constant of 1.31 ± 0.06 µM (Supplemental Fig. 5). A microplate-adapted competitive binding assay was performed for rapid screening of ligands for OBP28a. We measured the displacement of the NPN reporter probe to investigate the binding capacity of 103 odorant compounds (from diverse families of odorants, including pheromones) (Supplemental Fig. 6). The fluorescence intensity of the OBP28a-NPN complex was drastically reduced in the presence of the following 3 odorant molecules: (Z)-ethyl cinnamate, α-ionone, and vanillin. A reduction of a lesser amplitude was also detected with cinnamic acid, citronellyl tiglate, ethyl undecanoate, 1-octanol, and limonol, while all other tested compounds induced no significant effect. We found that OBP28a had a weak, or no binding capacity for dihydro-ß-ionone derivatives as observed with compounds structurally related to vanillin and (Z)-ethyl cinnamate (benzaldehyde, anisaldehyde, isovanillin, cinnamaldehyde, ethyl caproate, and ethyl butanoate; Data not shown). We further assessed the dose–response of (Z)-ethyl cinnamate, α-ionone, and vanillin using a cuvette mode fluorescence assay. Their apparent dissociation constants (Kdapp), deduced from the half-maximal values (IC50), were 9.0 ± 3.2 µM, 2.1 ± 0.3 µM, and 8.1 ± 2.7 µM, respectively (Fig. 3a). Other compounds structurally related to these odorants were also tested for their ability to bind OBP28a (Supplemental Fig. 7). Our binding curves revealed that OBP28a had a strong affinity to compounds closely related to ionone derivative odorants such as methyl-α-ionone and methyl-ß-ionone, (Kdapp values of 1.5 ± 0.2 µM and 1.5 ± 0.2 µM, respectively) with ß-ionone being the strongest ligand (Kdapp value of 1.3 ± 0.2 µM; Fig. 3b, c). We also tested the microbial compound, geosmin (trans-1,10-dimethyl-trans-9-decalol), which is detected by Drosophila with a remarkable sensitivity and selectivity [36]. Our binding curve revealed that OBP28a binds this compound with a Kdapp value of 2.3 ± 0.6 µM (Fig. 3A and C). In addition, we assayed the ability of pentaethylene glycol (co-crystallized ligand of OBP28a) to compete with the fluorescent probe NPN. We observed only a weak decrease of fluorescence indicating that pentaethylene glycol is a poor ligand for OBP28a (data not shown). Note that the values of fluorescent displacement are different between both cuvette and microplate modes. The fact that these two procedures rely on distinct parameters (i.e., different excitation and emission wavelengths, use of filters rather than monochromators) can explain the observed differences.
Fig. 3.
OBP28a displays a restrained ligand-binding profile. a Competitive binding curves of the three best ligands identified in the screening assay using cuvette mode assay. b Competitive binding curves of the ionone derivatives using cuvette mode assay. Note that methyl-ß-ionone and methyl-α-ionone curves overlap. c Affinities of OBP28a ligands with their respective structure
Predictive molecular interactions between OBP28a and ligands
To further understand the binding mode of OBP28a, we simulated the binding of the fluorescent probe NPN and the three odorants, ß-ionone, vanillin, and (Z)-ethyl cinnamate, using in silico blind molecular docking. The best conformation for the lowest free energy (Supplemental Table 1) for these ligands revealed that all ligands bind adjacent binding sites formed by residues belonging to the α1, α2, α3, α4, and α6 helices (Fig. 4). Our docking experiment also suggests the relevance of some amino-acid residues in ligand binding, such as arginine 113 (R113), which may be implicated in a hydrogen bond (N–H) between the R113 main chain and ß-ionone ligand (Fig. 4a). The R113 residue may also be involved in vanillin binding, given that a putative hydrogen bond can be made between the nitrogen of the guanidine of arginine and the (C=O) of the aldehyde moiety of vanillin (Fig. 4b). The binding of (Z)-ethyl cinnamate should differ, since this compound was docked in the binding cavity, wherein a hydrogen bond might be made between the (N–H) of glycine 89 (G89) and the (C=O) of the aldehyde moiety (Fig. 4c). In all docking cases, including NPN (Fig. 4d), it is worth noting that tyrosine 74 (Y74) and phenylalanine 112 (F112) participate in the stabilization of the aromatic cycle. This explains their systematic presence in the best OBP28a ligands.
Fig. 4.
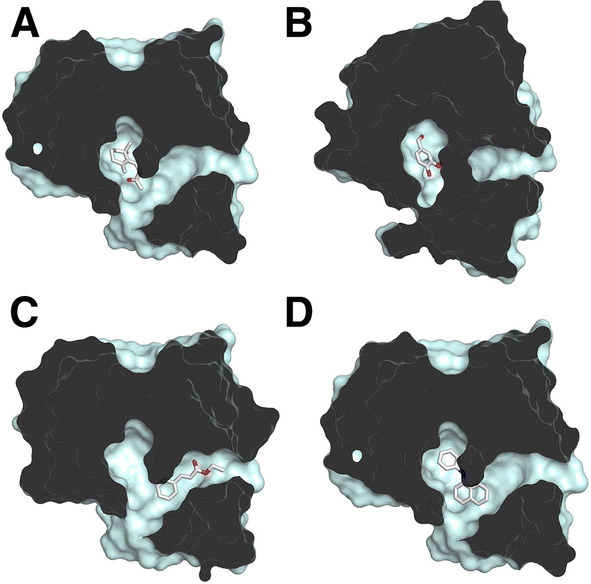
Molecular docking of OBP28a with identified ligands. Slabbed view of OBP28a bound with a ß-ionone, b vanillin, c (Z)-ethyl cinnamate, and d NPN
Behavioural responses in the absence of OBP28a
To investigate the behavioural role of OBP28a, we compared the olfactory preference to three ß-ionone concentrations (0.01, 0.05, and 1 mM) in mutant flies deleted for the OBP28a gene (OBP28a−) and in control flies (wCS). Two linked aspects of the olfactory preference were examined: (1) the motion of the fly to the choice point and (2) the preference either to ß-ionone or its solvent [in either olfactometer arm (Supplemental Fig. 8A)]. After 2 h, control flies showed a significant motion to the choice point at the three chosen ß-ionone concentrations, whereas OBP28a mutant flies showed no significant motion at the lower odorant concentrations (0.01 and 0.05 mM; Supplemental Fig. 8B). Moreover, while control flies significantly preferred 0.01 and 0.05 mM—but not 1 mM—ß-ionone, OBP28a− mutant flies did not show preference at any concentrations (Supplemental Fig. 8B).
ORN responses to ß-ionone depend on OBP28a expression
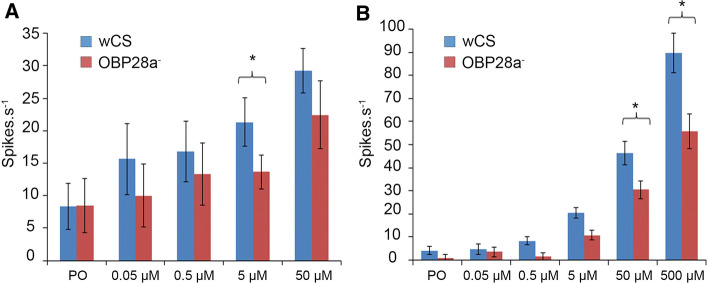
To further determine whether the altered olfactory response to ß-ionone of the OBP28a mutant flies depends on peripheral detection or central perception, we performed single-sensillum recordings (SSRs) in the sensilla known to be the most responsive to ß-ionone, i.e., ab4 basiconic sensilla on the antenna and pb2 sensilla on the maxillary palps [32, 37]. Physiological doses of ß-ionone (0.05–500 µM) were tested during brief exposure periods (0.5 s) to evoke natural stimulations (Fig. 5). When stimulated by ß-ionone, ab4B responses were significantly lower in the OBP28a mutant at the 5 µM dose, while they only showed a (non-significant) tendency to decrease at the 50 µM concentration (Fig. 5a). Moreover, pb2 sensilla also exhibited a significantly decreased response to the two higher ß-ionone stimulations (50 and 500 µM) (Fig. 5b).
Fig. 5.
ab4 sensillum a and pb2 sensillum b responses to ß-ionone measured by single-sensillum recordings. Action potentials recorded from control (wCS) and OBP28a mutant (OBP28a−) sensilla stimulated for 0.5 s with ß-ionone in paraffin oil solvent. Histograms represent the mean ± SEM. Letters indicate the differences for each genotype (ANOVA, Kruskal–Wallis, Dunn’s post hoc test), (*p < 0.05). N ≥ 9 for each data point. PO paraffin oil
Discussion
For long, OBPs have been thought to be involved in several functions pertaining to the transport and clearance of the ligand molecules present in the sensillar lymph [38–41]. It is now generally accepted that insect OBPs mediate the solubilization and transport of hydrophobic chemical cues to olfactory receptors and contribute to the sensitivity of the insect olfactory system. However, the complete physiological function of OBPs remains a matter of debate [16, 42]. Here, we have deepened the investigation of OBP functions by elucidating both the structure and function of OBP28a, one of the most abundant soluble proteins found in Drosophila melanogaster antennal sensilla [14, 32]. The yeast P. pastoris system was used to produce OBP28a, which is a well-conformed protein present as a monomer, similar to several other insect OBPs [17]. Insect OBPs can be secreted as homodimers [43, 44] or heterodimers [35, 45]. Using X-ray crystallography, we also deciphered a high-quality structure of OBP28a, both when devoid of a ligand (apo-OBP28a) and when bound to pentaethylene glycol (bound-OBP28a). The structure of OBP28a was similar to that of most known insect OBPs, with a folding consisting of a bundle of six helices, creating a hydrophobic-binding cavity stabilized by the three highly conserved disulphide bridges. The closest structurally related transporter molecules are the OBPs of the cockroach Leucophaea maderae (lmadPBP) and the LUSH protein of D. melanogaster (dmelOBP76a), which show only 22% and 20% amino-acid identity to OBP28a, respectively (Supplemental Fig. 4). The RMSD value allowed us to compare the structural proximity between these two structures. Despite a well-conserved overall structure, the structural comparison between apo-OBP28a, lmadPBP and dmelOBP76a (LUSH OBP) revealed significant conformational differences of OBP28a from lmadPBP and dmelOBP76a with an RMSD of 1.8 Å for both OBPs. Structural differences were mostly shown by the α1, α4, and α5 helices, which exhibited a clear shift that explains the more compact conformation of the OBP28a structure compared to the two other insect OBPs. Notably, the apo-OBP28a conformation showed a small ligand-binding cavity (i.e., ~ 90 Å3), similar to the apo-conformations shown by other OBPs with available 3D structures (Supplemental Fig. 4). The size of this small cavity was similar to that found in Apis mellifera amelASP1 and amelOBP14 and in L. maderae lmadPBP [22, 46, 47]. Moreover, OBP28a displayed a classical OBP binding cavity mostly composed of hydrophobic or semi-polar residues, thus allowing the fixation of lipophilic ligands (Fig. 1). This 201 Å3 binding cavity was larger than in bound-dmelOBP76a (~ 150 Å3), suggesting that OBP28a is capable of binding odorant molecules larger than small-chain alcohols [21].
We solved the OBP28a crystal structure in two alternative conformations from the same crystal, which consequently offers the possibility to better understand the relationship between the structural features of the OBP28a and its binding properties. In particular, the comparison of the apo- and bound-OBP28a forms revealed the structural reorganization of the α1, α4, and α5 helices depending on the ligand nature. This structural reorganization was visible when bound to pentaethylene glycol, a ligand with a poor in vitro affinity with OBP28a. However, the binding pocket of the bound-OBP28a resembled those of agamOBP1 and cquiOBP1 bound with their respective ligands [48–50]. Several studies have reported that structural OBP reorganizations can occur upon the nature of the ligand bound. For instance, dmelOBP76a, amelASP1, agamOBP1, and lmadPBP share a mechanism during which the N- and C-termini form a wall in the closed state with an opening involving the residues from α1, α3, and α4 helices [21–23, 46]. In addition, the Bombyx mori PBP (bmorPBP) shows a pH-dependent conformational change, where the C-terminus creates a helix filling its binding pocket, thus preventing binding of the bombykol pheromone [51]. Note also that OBP28a lacks the C-terminus extension present in bmorPBP, which makes a seventh helix, suggesting an alternative mechanism of ligand binding. Herein, upon ligand binding, α4 and α5 helices underwent a shift with one turn unwinding of α4, leading to the formation of a hydrogen bond between the N-terminus and ℓ4→5. This structural reorganization led to the formation of a double-ended “tunnel-shape” cavity, which is similar in agamOBP and aaegOBP that have a similar size to OBP28a [23, 49, 52]. The conformational change in OBP28a due to ligand binding was also observed in agamOBP20, wherein α1, α4, and α5 helices were subjected to a similar, but moderate movement [53].
Our fluorescence competitive binding assay revealed that OBP28a is a narrowly tuned OBP able to bind a restricted number of odorant molecules, including ß-ionone, geosmin, vanillin, or ethyl-(Z)-cinnamate. The measured dissociation constants were included in the micromolar range, similar to other insect OBPs. These floral-like odorants are attractive for Drosophila and this supports the expression pattern of OBP28a in olfactory organs and especially in basiconic sensilla known to detect fruit-like odours [1]. The study of a mutant deletion that encodes the region of the OBP28a gene revealed that this carrier protein affects the detection of varied odorant compounds, such as 1-octanol, γ-hexalactone, 2-pentanone, 2-pentanol, and 3-methylthio-1-propanol, ethyl acetate, and butyric acid, in given concentration ranges [32]. However, our fluorescence competitive binding assay did not allow us to detect a significant interaction between OBP28a and these compounds (data not shown), except for 1-octanol, which showed a relatively low affinity for OBP28a (Supplemental Fig. 6).
Nevertheless, our behavioural assays support the later report [32], indicating that the responses of the OBP28a− mutant flies are only altered at certain ligand concentrations such as for ß-ionone in our study. In our Y-maze assays, the mutant flies showed decreased sensitivity to the 0.01 and 0.05 mM ß-ionone concentrations. More precisely, with the tested concentrations, control flies frequently walked to the choice point, while mutant flies did not except with the 1 mM concentration. Moreover, unlike mutant flies, control flies showed a preference for the arm containing 0.01 and 0.05 mM ß-ionone. This indicates that the OBP28a deletion could induce an increased threshold relative to the ß-ionone detection. The lack of preference at higher concentrations may reflect the possibility that the detection system of flies is unable to distinguish the arm containing ß-ionone.
The data provided by the electrophysiological response of the ab4 and pb2 sensilla support this interpretation. Larter and collaborators have observed an increase in the magnitude of the electrophysiological responses in the OBP28a− mutant for a number of odorants across a wide range of concentrations [32]. In contrast, we found that the OBP28a− mutant showed a decreased response compared to the control, only at the highest ß-ionone tested concentrations. Our results are supported by a recent study, revealing that Bombyx mori PBP1 plays a role in enhancing the sensitivity—but not the selectivity—of sex pheromone detection [54].
Conclusion
In summary, our study provides novel insights into the deorphanization of OBP28a as well as the ligand-binding mechanism. Our structural and biochemical data taken with our behavioural and electrophysiological measurements indicate that OBP28a possibly changes the detection threshold of ß-ionone at a range compatible with biological concentrations. This supports the “classical” transporter function of OBP28a in olfaction, but also suggests its modulating effect in the detection of floral-like compounds at natural concentrations. Even if OBP28a is one of the best-characterized OBPs, some functional aspects need to be further investigated. All these elements will pave the way of further studies to decipher the multiple physiological roles of OBPs.
Methods
Drosophila stocks
The strains used in the behavioural and electrophysiological experiments (wCS control and OBP28a mutant) were generously provided by John Carlson [32]. All strains were raised on Drosophila standard medium under controlled conditions (24.5 ± 0.5 °C, 65 ± 5% relative humidity). Flies were tested in similar conditions.
Recombinant protein expression of OBP28a
The cDNA sequence encoding Drosophila melanogaster OBP28a (UniProt ID: P54195) was synthesized with codon usage optimized for Pichia pastoris by GeneArt® Gene Synthesis (Life Technologies). The synthetic cDNA of OBP28a was cloned into the XhoI and NotI sites of the pPIC9 plasmid, generating the construct pPIC9-OBP28a with the α-factor secretion signal fused to the mature OBP28a sequence without the Glu–Ala–Glu–Ala spacer peptide. The plasmid pPIC9-OBP28a was linearized with BglII and transferred into the Pichia pastoris GS115 strain by electroporation as previously described. Having identified the best OBP28a-producing transformant, large-scale OBP production was achieved as previously reported [34]. Briefly, the best clone was used to inoculate 20 mL of buffered minimal glycerol (BMGY) medium (1% w/v yeast extract, 2% w/v peptone, 1.34% w/v yeast nitrogen base with ammonium sulphate without amino acids (YNB), 4 mg/mL D-biotin, 100 mM potassium phosphate, pH 6.0, and 1% v/v glycerol) in sterile 50 mL Erlenmeyer flasks, which were then incubated at 28.9 °C, 280 rpm for 16 h. The cells were harvested by centrifugation and used to inoculate 4 L of BMGY medium equally distributed in eight 2 L-baffled glass flasks. The culture was performed under the same conditions as the pre-culture until the OD600 nm > 30. Cells were then pelleted at 7400 × g for 10 min at room temperature and resuspended in buffered minimal methanol (BMM) medium (1.34% w/v YNB, 4 mg/mL D-biotin, 100 mM potassium phosphate, and pH 6.0, 1% v/v methanol) before continuing the incubation for 2 days as previously described. During the induction period, methanol was fed twice a day to maintain a concentration of 0.5% v/v. The yeast supernatant containing secreted OBP28a was clarified by centrifugation at 21,600 × g for 20 min at 4 °C and by filtration (0.22 µm).
Purification of recombinant OBP28a
The yeast supernatant containing OBP28a was dialyzed in three successive steps against 8 L of 20 mM ammonium acetate, pH 4.0 at 4 °C for 2 days and then loaded onto a 5 mL sulfopropyl cation exchange column (GE Healthcare Biosciences, Uppsala, Sweden). The column was washed with 20 mM ammonium acetate, pH 4.0, and elution was performed using an increasing NaCl gradient (from 0 to 1 M). A second dialysis was performed against water adjusted to pH 4.0 using formic acid. OBP28a was purified using RPLC as previously described [55]. Fractions containing OBP28a were pooled, dialyzed extensively against HPLC-grade water, and stored at -20 °C before lyophilization. All buffers used for purification were prepared with HLPC-grade water. SDS-PAGE analysis was performed using the method of Schägger and von Jagow [56].
Biophysical analysis
Circular dichroism (CD) spectra were recorded using a JASCO spectrometer J-815 equipped with a Peltier temperature control system (JASCO, Lisses, France) in a 0.1 mm-thick quartz cell. Measurements were performed in 10 mM phosphate buffer (pH 7.4) with a protein concentration of 1.0 mg/mL. Spectra were recorded over a range of 180–260 nm at 20 °C with a data pitch of 0.5 nm and a scanning speed of 100 nm/min. Each protein spectrum was the result of 10 accumulations. Data were corrected for buffer contributions and converted into molar ellipticity. The secondary structure proportions were estimated using the deconvolution K2D algorithm on the DichroWeb program (http://dichroweb.cryst.bbk.ac.uk/html/home.shtml) [57]. The oligomeric state of OBP28a was analysed by size-exclusion chromatography—multi-angle laser light scattering (SEC-MALS) using a MinDAWN Treos laser (Wyatt Technology, Santa Barbara, CA USA) coupled with a UV/Vis detector (UV4070, JASCO, Lisses, France) and a refractive index detector (RI-4030, JASCO, Lisses, France). Prior to SEC-MALS analysis, the protein was injected in a KW-803 HPLC column (Shodex, Munich, Germany) at 0.5 mL/min in a buffer of 50 mM KH2PO4/K2HPO4 and 150 mM NaCl at pH 6.5 using an HPLC pump (PU 2080 plus, JASCO). Molecular weight determination was performed using ASTRA VI software (Wyatt Technology, Santa Barbara, CA USA). Mass spectrometry analysis was performed using infusion of OBP28a on an LTQ-Orbitrap Discovery mass spectrometer (Thermo Fisher, Waltham, MA USA).
Fluorescence-based binding assay
The ligand-binding properties of OBP28a were investigated using a competitive fluorescent binding assay using N-phenyl-1-naphthylamide (NPN) as a fluorescent probe. The binding affinity of OBP28a for NPN was determined by increasing the final concentration from 0 to 30 µM of 1 mM NPN stock solution (dissolved in 10% methanol) into a 2 µM protein sample in 50 mM phosphate buffer (pH 7.4). The excitation wavelength of NPN was 337 nm, and emission was recorded between 380 and 450 nm using a Cary Eclipse spectrofluorometer (Varian Instrument, Les Ulis, France), a 1-cm light path and a quartz cuvette. The excitation and emission slits were both 5 nm. The competitive binding of ligand was performed using 2 µM of NPN as a fluorescent probe, 2 µM of OBP28a with a final concentration of each ligand added to the sample ranging from 0 to 75 µM from a ligand stock solution of 1 mM (dissolved in 10% methanol) and 10 mM (dissolved in 100% methanol). The fluorescence intensities at the maximum fluorescence emission 410 nm were plotted against ligand concentrations. Binding of chemical compounds was estimated using the fluorescence displacement, assuming that 100% of the protein was active with a stoichiometry of 1:1 (protein:ligand) saturation. The apparent dissociation constant Kdapp was calculated from the corresponding IC50 values (the concentration of ligands halving the initial fluorescence value of NPN) using the equation Kdapp = [IC50]/(1 + [NPN]/KNPN), where [NPN] is the free concentration of NPN and KNPN is the dissociation constant of the complex OBP28a/NPN. Binding fluorescence data were collected from three independent experiments. To verify that fluorescence displacement is ligand-binding specific, each chemical was tested in the presence of 2 µM NPN (dissolved in 60% ethanol), and the resulting fluorescence, if present, was subtracted from the experimental spectra.
A microplate-adapted competitive binding assay was performed for rapid screening of ligands for OBP28a. This binding assay was performed using a Victor3 V microplate reader (PerkinElmer Life sciences, Courtaboeuf, France) at room temperature in a 96-well microplate format, using 25- and 5-nm slits for excitation and emission, respectively. The excitation wavelength was 340 nm, and the fluorescence emission intensity was recorded at 415 nm. Each well contained 200 µL of 4 µM OBP28a with 4 µM NPN dissolved in 50 mM phosphate buffer, pH 7.4. Five µL of each ligand was prepared as previously described, added to the microplate to a final concentration of 40 µM, and then mixed. Ligand-binding assays were performed in three independent experiments. Raw results were analysed using the Sigmaplot software.
Crystallization
OBP28a was concentrated at 20–25 mg/mL. Crystallization assays were performed using the hanging-drop vapor diffusion method setup in grease-sealed 24-well Linbro plates (Hampton research, Aliso Viejo, CA USA). PACT premier and JCSG + (Molecular Dimensions) commercial screen were used in a first approach with 3 ratios of protein:reservoir (1 µL:1 µL, 2 µL:1 µL, and 1 µL:2 µL). The Linbro wells were filled with 500 µL of screening solution. The plates were incubated at 20 °C and monitored weekly under a microscope, Nikon Tie (Nikon Instruments, Amsterdam, The Netherlands). The best hit consisting of needles was identified from the commercial screen JCSG + and contained 2.4 M sodium malonate at pH 7.0, 0.1 M HEPES, and 0.5% Jeffamine ED-2003. This condition was optimized for pH (from 7 to 8), sodium malonate concentration (from 2.5 to 3 M), and Jeffamine (from 0 to 1%) in a 24-well Linbro plate using the same protocol as described for the commercial screen. Two weeks later, larger needles (approximately 100 µm lengths) were obtained and manually separated to optimize their growth. Forty-eight hours after their separation, crystals were used for data collection.
Data collection and structure determination
A cryoprotectant solution consisting of the crystallization solution (i.e., 2.5 M sodium malonate) supplemented with 50% (v/v) PEG 6000 was added to the drop. The crystal was mounted onto a LithoLoop (Molecular Dimensions, Newmarket, Suffolk UK) and flash-frozen in liquid nitrogen. X-ray diffraction intensities were collected on the ID29 beamline at European Synchrotron Radiation Facility (ESRF, Grenoble, France) using a wavelength of 0.978 Å and a Pilatus3–6 M detector with 0.02 s exposures. Diffraction data were collected from 1800 images; individual frames consisted of 0.1° steps over a range of 180°. X-ray diffraction data were integrated and scaled using the XDS suite [58] (Table 1). OBP28a crystals belonged to the monoclinic space group P21, with unit-cell parameters of a = 34.6 Å, b = 83.7 Å, c = 46.5 Å, and β = 99.5°. OBP28a was approximately 13 kDa protein, and the calculated Matthews coefficient suggested 2 monomers per asymmetric unit (with 2.37 Å3·Da−1, corresponding to 48.10% solvent content). The molecular replacement was first tried with PHASER using the structure of the Leucophaea maderae Pheromone-Binding Protein (lmadPBP) as a model (22% sequence identity with OBP28a, PDB ID: 1ORG) [46] either with the full-length protein or with different deletion models (without loops, N-terminal helices, C-terminal helices, etc.). Unfortunately, high sequence divergence prevented us to use molecular replacement. None of the two expected molecules were correctly placed in the asymmetric unit. MrBUMP [59] was thus used to automatically generate different input models for molecular replacement. By searching the homologous structure, MrBUMP software created a set of suitable search models that were used for molecular replacement. The best solution was obtained for two molecules in the asymmetric unit (Rfree = 33.29%). Despite the good values for the observed R factors, the generated model was highly incomplete due to side chains, and more than 40% of the protein was missing. Manual model improvement and reconstruction were performed using Coot [60]. Refinement with REFMAC [61] and PHENIX software [62] was also performed. POLYGON from the PHENIX software was used to assess the quality of the final structure [63]. Restraints of the OBP28a ligand (i.e., the pentaethylene glycol issued from Jeffamine) were generated with the PRODRG server [64]. The ligand-binding cavity and volume were visualized with Castp [65]. The crystal-packing interface surface between OBP28a monomers was calculated with the PISA server from the EMBL server (http://www.ebi.ac.uk/pdbe/prot_int/pistart.html). All figures were created using PyMOL software, and the structure was deposited on the RCSB Protein Data Bank under the code 6QQ4.
Molecular docking
Prior to docking experiments, the protein was prepared using the dockprep plugin from USCF chimera software [66]. Ligands were taken from the ZINC database and prepared with the SWISSDOCK server or with PRODRG [64]. Docking experiments were performed with the best ligands of OBP28a versus the whole protein. Clusters with the best free energy (ΔG) displayed the ligand in the binding cavity of OBP28a. For docking experiments, both monomers were used and docked versus NPN. Docking experiments versus bound-OBP28a yielded the best binding energy, and the bound-OBP28a was used for all the docking experiments. Binding energies were used to choose the best conformation and to calculate the theoretical inhibition constant (Ki) with formula Ki = exp(∆G/RT), where ∆G is the Gibbs free enthalpy, R is the gas constant, and T is standard temperature. Docked ligands were visualized using PyMOL.
Olfactory preference behaviour
0–8 h After hatching, male flies were collected and kept individually in fresh food vials. When 5–7 days, and after 16 h of starvation (on a wet piece of filter paper), their preference for ß-ionone was tested in a Y-shaped olfactometer. After a brief anaesthesia on ice, individual flies were introduced into the straight arm of the device, which was divided into two arms either containing the ß-ionone (+ solvent) or the solvent alone (7% ethanol). Immediately before the test, a filter paper previously incubated at 25 °C for 30 min in standard medium and impregnated with 8 µL of each tested substance was deposited in each olfactometer arm. The test was performed for 2 h under far-red light (LED bulbs) with three ß-ionone concentrations ranging from 0.01 to 1 mM. After starting the test, and every 30 min, the position of the fly was noted relative to its position in the device in (1) the straight tube (between the starting point and the choice point) and (2) the preference tube (arm chosen). Flies were rarely seen retreating in our device. Control and OBP28a mutant flies as well as different concentrations of ß-ionone were simultaneously tested. The arms containing the ß-ionone and the control substance were always swapped between olfactometers, and assays performed on 1 day included both genotypes and different ß-ionone concentrations. All experiments were performed between 9 and 11 a.m. The significant differences were determined with an exact Fisher test.
Electrophysiology
Single-sensillum recording was performed with standard methods [67]. A female fly (3–6 days) was restrained in a 200 µL pipette tip with only the head protruding from the narrow end. The pipette tip was fixed with dental wax on a microscope glass slide (ventral side upward), and either one antenna or one maxillary palp was maintained on a piece of glass slide with a glass capillary. Afterwards, this preparation was placed under a light microscope (BX51WI, Olympus, Tokyo, Japan) equipped with a 100X magnification objective (LMPlanN 100X, Olympus) and × 15 eyepieces. To reduce the desiccation of the antenna (or palp), a 1.5 L·min−1 charcoal-filtered and humidified air flux was constantly delivered with a 7 mm diameter glass tube positioned 1.5 cm from the preparation. Stimulation devices were built by placing a 1 cm2 filter paper a Pasteur pipette and dropping 10 μL of ß-ionone (diluted in paraffin oil) onto the paper before closing the pipette with a 1 mL plastic pipette tip. Odorant stimulations were performed by inserting the tip of the Pasteur pipette into a hole in the permanent air flux glass tube and generating a 500 ms air pulse (0.6 L min−1). The reference electrode was inserted into the eye of the fly by a manually controlled micromanipulator. The recording electrode (tungsten) was inserted at the base of the ab4 or pb2 sensilla using a motor-controlled micromanipulator (Patchstar, Scientifica). The electrical signal was amplified using an EX-1 amplifier (Dagan, Minneapolis, MN, USA), digitized through a Digidata 1440A acquisition board (Molecular Devices, Sunnyvale, CA, USA) and then recorded and analysed using the pCLAMP 10 software (Molecular Devices). The net responses of ab4/pb2 neurons were calculated by subtracting the spontaneous firing rate (500 ms before stimulation) from the firing rate during the odorant stimulation (500 ms after the onset of the stimulation). We also subtracted from the experimental values, and the background responses measured to paraffin oil (data not shown).
Accession number
The atomic coordinates and structure factors have been deposited in the Protein Data Bank under accession number 6QQ4.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We thank Dr. J. R. Carlson for Drosophila lines; Dr. C. Everaerts for help with the statistics; and Dr. T. Tanimura for discussion and critical reading. The ESRF is acknowledged for access to beamlines via its in-house research program. Mass spectrometry experiments were performed by the Plateforme d’Analyse Protéomique de Paris Sud-Ouest (PAPPSO, Micalis Institute, INRA, AgroParisTech, Université Paris-Saclay, 78350 Jouy-en-Josas, France).
Abbreviations
- OBPs
Odorant-binding proteins
- OSNs
Olfactory-sensory neurons
- Ors
Odorant receptors
- cVA
Cis-vaccenyl acetate
- RPLC
Reverse-phase liquid chromatography
- CD
Circular dichroism
- SEC
Size-exclusion chromatography
- MALS
Multi-angle laser light scattering
- 1PE
Pentaethylene glycol
- NPN
N-phenyl-1-naphthylamine
- SSR
Single-sensillum recording
- lmadPBP
Pheromone-binding protein from Leucophaea maderae
- dmelOBP76a
Odorant-binding protein 76a from D. melanogaster
- amelASP1
Pheromone-binding protein 1 from Apis mellifera
- amelOBP14
Odorant-binding protein 14 from Apis mellifera
- bmorPBP
Pheromone-binding protein from Bombyx mori
- agamOBP1
Odorant-binding protein 1 from Anopheles gambiae
- cquiOBP1
Odorant-binding protein 1 from Culex pipiens quinquefasciatus
- BMGY
Buffered minimal glycerol
- YNB
Yeast nitrogen base
- BMM
Buffered minimal methanol
Author contributions
JFF and LB designed the research. DG, KR, FN, NP, SF, GG, and TC performed the research. DG, KR, FN, SF, GG, TC, and MM analysed the data. The manuscript was written by DG, KR, JFF, and LB. All authors read and approved the final manuscript.
Funding
This work was partly supported by grants from the Institut National de la Recherche Agronomique, of the Centre National de la Recherche Scientifique, of the Université de Bourgogne-Franche Comté, the Bourgogne-Franche Comté Regional Council (PARI 2010–2011–2012, AGRALE1 Project), and a postdoctoral fellowship from the Bourgogne Regional Council (D.G.). Fellowship for PhD to K.R. (INRA + Bourgogne-Franche Comté Regional Council).
Compliance with ethical standards
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Daniel Gonzalez and Karen Rihani have contributed equally to this work.
References
- 1.de Bruyne M, Foster K, Carlson JR. Odor coding in the Drosophila antenna. Neuron. 2001;30:537–552. doi: 10.1016/S0896-6273(01)00289-6. [DOI] [PubMed] [Google Scholar]
- 2.Ai M, Min S, Grosjean Y, et al. Acid sensing by the Drosophila olfactory system. Nature. 2010;468:691–695. doi: 10.1038/nature09537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yao CA, Ignell R, Carlson JR. Chemosensory coding by neurons in the coeloconic sensilla of the Drosophila antenna. J Neurosci. 2005;25:8359–8367. doi: 10.1523/JNEUROSCI.2432-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clyne P, Grant A, O’Connell R, Carlson JR. Odorant response of individual sensilla on the Drosophila antenna. Invertebr Neurosci. 1997;3:127–135. doi: 10.1007/BF02480367. [DOI] [PubMed] [Google Scholar]
- 5.Dweck HKM, Ebrahim SAM, Thoma M, et al. Pheromones mediating copulation and attraction in Drosophila. Proc Natl Acad Sci. 2015;112:E2829–E2835. doi: 10.1073/pnas.1504527112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Younus F, Fraser NJ, Coppin CW, et al. Molecular basis for the behavioral effects of the odorant degrading enzyme Esterase 6 in Drosophila. Sci Rep. 2017;7:1–12. doi: 10.1038/srep46188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Larsson MC, Domingos AI, Jones WD, et al. Or83b encodes a broadly expressed odorant receptor essential for Drosophila olfaction. Neuron. 2004;43:703–714. doi: 10.1016/j.neuron.2004.08.019. [DOI] [PubMed] [Google Scholar]
- 8.Benton R, Sachse S, Michnick SW, Vosshall LB. Atypical membrane topology and heteromeric function of Drosophila odorant receptors in vivo. PLoS Biol. 2006;4:240–257. doi: 10.1109/ICARCV.2014.7064338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Benton R, Vannice KS, Gomez-Diaz C, Vosshall LB. Variant ionotropic glutamate receptors as chemosensory receptors in Drosophila. Cell. 2009;136:149–162. doi: 10.1016/j.cell.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hussain A, Zhang M, Üçpunar HK, et al. Ionotropic chemosensory receptors mediate the taste and smell of polyamines. PLoS Biol. 2016;14:e1002454. doi: 10.1371/journal.pbio.1002454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Galindo K, Smith DP. A large family of divergent Drosophila odorant-binding proteins expressed in gustatory and olfactory sensilla. Genetics. 2001;159:1059–1072. doi: 10.1093/genetics/159.3.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hekmat-Scafe DS, Scafe CR, McKinney AJ, Tanouye MA. Genome-wide analysis of the odorant-binding protein gene family in Drosophila melanogaster. Genome Res. 2002;12:1357–1369. doi: 10.1101/gr.239402.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vieira FG, Rozas J. Comparative genomics of the odorant-binding and chemosensory protein gene families across the arthropoda: origin and evolutionary history of the chemosensory system. Genome Biol Evol. 2011;3:476–490. doi: 10.1093/gbe/evr033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anholt RRH, Williams TI. The soluble proteome of the Drosophila antenna. Chem Senses. 2010;35:21–30. doi: 10.1093/chemse/bjp073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leal WS. Odorant reception in insects: roles of receptors, binding proteins, and degrading enzymes. Annu Rev Entomol. 2013;58:373–391. doi: 10.1146/annurev-ento-120811-153635. [DOI] [PubMed] [Google Scholar]
- 16.Pelosi P, Iovinella I, Zhu J, et al. Beyond chemoreception: diverse tasks of soluble olfactory proteins in insects. Biol Rev. 2018;93:184–200. doi: 10.1111/brv.12339. [DOI] [PubMed] [Google Scholar]
- 17.Pelosi P, Zhou JJ, Ban LP, Calvello M. Soluble proteins in insect chemical communication. Cell Mol Life Sci. 2006;63:1658–1676. doi: 10.1007/s00018-005-5607-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu Z, Lin J, Zhang H, Zeng X. BdorOBP83a-2 mediates responses of the oriental fruit fly to semiochemicals. Front Physiol. 2016;7:1–15. doi: 10.3389/fphys.2016.00452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jeong YT, Shim J, Oh SR, et al. An odorant-binding protein required for suppression of sweet taste by bitter chemicals. Neuron. 2013;79:725–737. doi: 10.1016/j.neuron.2013.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Horst R, Damberger F, Luginbühl P, et al. NMR structure reveals intramolecular regulation mechanism for pheromone binding and release. Proc Natl Acad Sci. 2001;98:14374–14379. doi: 10.1073/pnas.251532998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kruse SW, Zhao R, Smith DP, Jones DNM. Structure of a specific alcohol-binding site defined by the odorant binding protein LUSH from Drosophila melanogaster. Nat Struct Biol. 2003;10:694–700. doi: 10.1038/nsb960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lartigue A, Gruez A, Briand L, et al. Sulfur single-wavelength anomalous diffraction crystal structure of a pheromone-binding protein from the honeybee Apis mellifera L. J Biol Chem. 2004;279:4459–4464. doi: 10.1074/jbc.M311212200. [DOI] [PubMed] [Google Scholar]
- 23.Wogulis M, Morgan T, Ishida Y, et al. The crystal structure of an odorant binding protein from Anopheles gambiae: evidence for a common ligand release mechanism. Biochem Biophys Res Commun. 2006;339:157–164. doi: 10.1016/j.bbrc.2005.10.191. [DOI] [PubMed] [Google Scholar]
- 24.Lescop E, Briand L, Pernollet JC, Guittet E. Structural basis of the broad specificity of a general odorant-binding protein from honeybee. Biochemistry. 2009;48:2431–2441. doi: 10.1021/bi802300k. [DOI] [PubMed] [Google Scholar]
- 25.Fan J, Francis F, Liu Y, et al. An overview of odorant-binding protein functions in insect peripheral olfactory reception. Genet Mol Res. 2011;10:3056–3069. doi: 10.4238/2011.December.8.2. [DOI] [PubMed] [Google Scholar]
- 26.Swarup S, Williams TI, Anholt RRH. Functional dissection of Odorant binding protein genes in Drosophila melanogaster. Gene Brain Behav. 2011;10:648–657. doi: 10.1111/j.1601-183X.2011.00704.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laughlin JD, Ha TS, Jones DNM, Smith DP. Activation of pheromone-sensitive neurons is mediated by conformational activation of pheromone-binding protein. Cell. 2008;133:1255–1265. doi: 10.1016/j.cell.2008.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gomez-Diaz C, Reina JH, Cambillau C, Benton R. Ligands for pheromone-sensing neurons are not conformationally activated odorant binding proteins. PLoS Biol. 2013;11:e1001546. doi: 10.1371/journal.pbio.1001546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bentzur A, Shmueli A, Omesi L, et al. Odorant binding protein 69a connects social interaction to modulation of social responsiveness in Drosophila. PLoS Genet. 2018;14:e1007328. doi: 10.1371/journal.pgen.1007328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matsuo T, Sugaya S, Yasukawa J, et al. Odorant-binding proteins OBP57d and OBP57e affect taste perception and host-plant preference in Drosophila sechellia. PLoS Biol. 2007;5:0985–0996. doi: 10.1371/journal.pbio.0050118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun JS, Larter NK, Chahda S, et al. Humidity response depends on the small soluble protein Obp59a in Drosophila. eLife7. 2018;7:39249. doi: 10.7554/eLife.39249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Larter NK, Sun JS, Carlson JR. Organization and function of Drosophila odorant binding proteins. Elife. 2016;5:e20242. doi: 10.7554/eLife.20242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Briand L, Perez V, Huet JC, et al. Optimization of the production of a honeybee odorant-binding protein by Pichia pastoris. Protein Expr Purif. 1999;15:362–369. doi: 10.1006/prep.1998.1027. [DOI] [PubMed] [Google Scholar]
- 34.Briand L, Swasdipan N, Nespoulous C, et al. Characterization of a chemosensory protein (ASP3c) from honeybee (Apis mellifera L.) as a brood pheromone carrier. Eur J Biochem. 2002;269:4586–4596. doi: 10.1046/j.1432-1033.2002.03156.x. [DOI] [PubMed] [Google Scholar]
- 35.Qiao H, He X, Schymura D, et al. Cooperative interactions between odorant-binding proteins of Anopheles gambiae. Cell Mol Life Sci. 2011;68:1799–1813. doi: 10.1007/s00018-010-0539-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stensmyr MC, Dweck HKM, Farhan A, et al. A Conserved dedicated olfactory circuit for detecting harmful microbes in Drosophila. Cell. 2012;151:1345–1357. doi: 10.1016/j.cell.2012.09.046. [DOI] [PubMed] [Google Scholar]
- 37.Dweck HKM, Ebrahim SAM, Khallaf MA, et al. Olfactory channels associated with the Drosophila maxillary palp mediate short- and long-range attraction. Elife. 2016;5:e14925. doi: 10.7554/eLife.14925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vogt RG, Rybczynski R, Lerner MR. Molecular cloning and sequencing of general odorant-binding proteins GOBP1 and GOBP2 from the tobacco hawk moth Manduca sexta: comparisons with other insect OBPs and their signal peptides. J Neurosci. 1991;11:2972–2984. doi: 10.1523/JNEUROSCI.11-10-02972.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pelosi P, Maida R. Odorant-binding proteins in insects. Comp Biochem Physiol B Biochem Mol Biol. 1995;111:503–514. doi: 10.1016/0305-0491(95)00019-5. [DOI] [PubMed] [Google Scholar]
- 40.Steinbrecht RA, Laue M, Ziegelberger G. Immunolocalization of insect odorant-binding proteins—a comparative-study. Chem Senses. 1995;20:109–110. [Google Scholar]
- 41.Krieger J, von Nickisch-Rosenegk E, Mameli M, et al. Binding proteins from the antennae of Bombyx mori. Insect Biochem Mol Biol. 1996;26:297–307. doi: 10.1016/0965-1748(95)00096-8. [DOI] [PubMed] [Google Scholar]
- 42.Sun JS, Xiao S, Carlson JR. The diverse small proteins called odorant-binding proteins. R Soc Open Biol. 2018;8:180–208. doi: 10.1098/rsob.180208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ban L, Scaloni A, D’Ambrosio C, et al. Biochemical characterization and bacterial expression of an odorant-binding protein from Locusta migratoria. Cell Mol Life Sci. 2003;60:390–400. doi: 10.1007/s000180300032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Honson N, Johnson MA, Oliver JE, et al. Structure-activity studies with pheromone-binding proteins of the gypsy moth, Lymantria dispar. Chem Senses. 2003;28:479–489. doi: 10.1093/chemse/28.6.479. [DOI] [PubMed] [Google Scholar]
- 45.Andronopoulou E, Labropoulou V, Douris V, et al. Specific interactions among odorant-binding proteins of the African malaria vector Anopheles gambiae. Insect Mol Biol. 2006;15:797–811. doi: 10.1111/j.1365-2583.2006.00685.x. [DOI] [PubMed] [Google Scholar]
- 46.Lartigue A, Gruez A, Spinelli S, et al. The Crystal structure of a cockroach pheromone-binding protein suggests a new ligand binding and release mechanism. J Biol Chem. 2003;278:30213–30218. doi: 10.1074/jbc.m304688200. [DOI] [PubMed] [Google Scholar]
- 47.Spinelli S, Lagarde A, Iovinella I, et al. Crystal structure of Apis mellifera OBP14, a C-minus odorant-binding protein, and its complexes with odorant molecules. Insect Biochem Mol Biol. 2012;42:41–50. doi: 10.1016/j.ibmb.2011.10.005. [DOI] [PubMed] [Google Scholar]
- 48.Tsitsanou KE, Thireou T, Drakou CE, et al. Anopheles gambiae odorant binding protein crystal complex with the synthetic repellent DEET: implications for structure-based design of novel mosquito repellents. Cell Mol Life Sci. 2012;69:283–297. doi: 10.1007/s00018-011-0745-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Drakou CE, Tsitsanou KE, Potamitis C, et al. The crystal structure of the AgamOBP1•Icaridin complex reveals alternative binding modes and stereo-selective repellent recognition. Cell Mol Life Sci. 2016;74:319–338. doi: 10.1007/s00018-016-2335-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mao Y, Xu X, Xu W, et al. Crystal and solution structures of an odorant-binding protein from the southern house mosquito complexed with an oviposition pheromone. Proc Natl Acad Sci. 2010;107:19102–19107. doi: 10.1073/pnas.1012274107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Damberger FF, Michel E, Ishida Y, et al. Pheromone discrimination by a pH-tuned polymorphism of the Bombyx mori pheromone-binding protein. Proc Natl Acad Sci. 2013;110:18680–18685. doi: 10.1073/pnas.1317706110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Leite NR, Krogh R, Xu W, et al. Structure of an odorant-binding protein from the mosquito Aedes aegypti suggests a binding pocket covered by a pH-sensitive “Lid”. PLoS One. 2009;4:e8006. doi: 10.1371/journal.pone.0008006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ziemba BP, Murphy EJ, Edlin HT, Jones DNM. A novel mechanism of ligand binding and release in the odorant binding protein 20 from the malaria mosquito Anopheles gambiae. Protein Sci. 2013;22:11–21. doi: 10.1002/pro.2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shiota Y, Sakurai T, Daimon T, et al. In vivo functional characterisation of pheromone binding protein-1 in the silkmoth, Bombyx mori. Sci Rep. 2018;8:13529. doi: 10.1038/s41598-018-31978-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Danty E, Briand L, Michard-Vanhée C, et al. Cloning and expression of a queen pheromone-binding protein in the honeybee: an olfactory-specific, developmentally regulated protein. J Neurosci. 1999;19:7468–7475. doi: 10.1523/JNEUROSCI.19-17-07468.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schägger H. Tricine–SDS-PAGE. Nat Protoc. 2006;1:16–22. doi: 10.1038/nprot.2006.4. [DOI] [PubMed] [Google Scholar]
- 57.Whitmore L, Wallace BA. DICHROWEB, an online server for protein secondary structure analyses from circular dichroism spectroscopic data. Nucl Acids Res. 2004;32:W668–W673. doi: 10.1093/nar/gkh371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kabsch W. XDS. Acta Crystallogr Sect D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Keegan RM, Winn MD. MrBUMP: an automated pipeline for molecular replacement. Acta Crystallogr Sect D: Biol Crystallogr. 2007;64:119–124. doi: 10.1107/S0907444907037195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr Sect D: Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 61.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr Sect D: Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 62.Adams PD, Grosse-Kunstleve RW, Hung LW, et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr Sect D: Biol Crystallogr. 2002;58:1948–1954. doi: 10.1107/S0907444902016657. [DOI] [PubMed] [Google Scholar]
- 63.Urzhumtseva L, Afonine PV, Adams PD, Urzhumtsev A. Crystallographic model quality at a glance. Acta Crystallogr Sect D: Biol Crystallogr. 2009;65:297–300. doi: 10.1107/S0907444908044296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.van Aalten DMF, Bywater R, Findlay JB, et al. PRODRG, a program for generating molecular topologies and unique molecular descriptors from coordinates of small molecules. J Comput Aided Mol Des. 1996;10:255–262. doi: 10.1007/BF00355047. [DOI] [PubMed] [Google Scholar]
- 65.Liang J, Edelsbrunner H, Woodward C. Anatomy of protein pockets and cavities: measurement of binding site geometry and implications for ligand design. Protein Sci. 1998;7:1884–1897. doi: 10.1002/pro.5560070905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pettersen EF, Goddard TD, Huang CC, et al. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 67.Stensmyr MC, Dekker T, Hansson BS. Evolution of the olfactory code in the Drosophila melanogaster subgroup. Proc R Soc B Biol Sci. 2003;270:2333–2340. doi: 10.1098/rspb.2003.2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.