Abstract
Transforming growth factor (TGF)-β signalling pathways are intensively investigated because of their diverse association with physiological and pathophysiological states. Smad transcription factors are the key mediators of TGF-β signalling. Smads can be directly phosphorylated in the carboxy terminal by the TGF-β receptor or in the linker region via multiple intermediate serine/threonine kinases. Growth factors in addition to hormones and TGF-β can activate many of the same kinases which can phosphorylate the Smad linker region. Historically, Smad linker region phosphorylation was shown to prevent nuclear translocation of Smads and inhibit TGF-β signalling pathways; however, it was subsequently shown that Smad linker region phosphorylation can be a driver of gene expression. This review will cover the signalling pathways of Smad linker region phosphorylation that drive the expression of genes involved in pathology and pathophysiology. The role of Smad signalling in cell biology is expanding rapidly beyond its role in TGF-β signalling and many signalling paradigms need to be re-evaluated in terms of Smad involvement.
Keywords: Nuclear translocation, G protein coupled receptors, Serine/threonine kinase receptors, Cancer, Alk 5, Cell signalling
Introduction
Transforming growth factor (TGF-β) is a pleiotropic growth factor with perhaps one of the widest range of physiological and pathophysiological roles of any biologically active polypeptide [1]. The general principles of TGF-β signalling were described in detail 20 years ago; however, because of its importance, TGF-β signalling has continued to be intensively investigated and it has even been used as a model system, for example, in the study of phenomena of whether cells respond to absolute or relative changes in signals [2].
Smad transcription factors, especially phosphorylated Smads, are key mediators of TGF-β signalling, but recent data have shown that the involvement of Smads in cellular signalling extends well beyond their role as mediators of TGF-β signalling. Smad transcription factors are divided into three distinct subgroups based on their role in signal transduction: the receptor-regulated Smads (R-Smads), common mediator Smads (Co-Smads) and the inhibitory Smads (I-Smads). The R-Smads comprise Smads 1–3,5, 8/9 and are phosphorylated in response to TGF-β and bone morphogenetic proteins (BMP) [3]. Smad4 is the only mammalian Co-Smad identified and it mediates signalling from both TGF-β and influences BMP signalling pathways [3]. I-Smads, Smad6 and Smad7 inhibit the activation of R-Smads via competitive binding with Smad4. TGF-β-regulated Smads, Smad2 and Smad3 are composed of N-terminal domain (MAD homology 1(MH1)), the linker region and a carboxy- terminal (MAD homology 2 (MH2)) domain that contains two serine residues directly phosphorylated by TGF-β type I receptor (TGFBR1) [3].
In Smad signalling pathways R-Smads, Smad2 and Smad3 are phosphorylated by TGF-β, and Smad1, Smad5 and Smad8 are phosphorylated in response to BMP. Canonically, TGF-β signalling involves the TGFBR1 also known as activin-like kinase (Alk) 5 that mediates the phosphorylation of R-Smads (Smad2 and Smad3) on the serine residues of the carboxyl terminal. Like other members of the TGF-β superfamily, BMPs elicit their effects through the activation of serine/threonine kinase receptors. Alk3 and Alk6 activate the R-Smads (Smad1, Smad5 and Smad8) in the carboxyl terminal. However, the Alk2 receptor only activates Smad1 and Smad5. The phosphorylation of the R-Smads in the carboxy terminal promotes their binding with Co-Smad (Smad4) to form a Smad complex that enters the nucleus for regulation of gene transcription. In addition to the canonical Smad signalling pathways, the TGF-β superfamily members can also signal via non-canonical Smad pathways. Mammalian Smad2 and Smad3 have a similar amino acid sequence with 66% and 96% identity in the MH1 and MH2 amino acid sequences, respectively. In contrast to the similarity in their amino acid sequences, there are two major differences between the overall structure of Smad2 and Smad3. Alignment of the MH1 domains of Smad2 and Smad3 reveals that there are two regions of amino acid sequences that are present in Smad2, but absent in Smad3. One of these is called GAG, a short N-terminal amino acid residue consisting of ten residues [4]. The other is a 30-residue sequence referred to as TID [4]. The removal of the GAG domain in Smad2 or the insertion in Smad3 had no effect on the transcriptional activity of the Smads [5]. However, the TID domain, rich in serine and threonine residues, prevents Smad2 from directly binding to DNA.
TGF-β-activated Smads are composed of the N-terminal domain, the central linker region and carboxy-terminal domain that contains three serine residues of which two are directly phosphorylated by TGFBR1. Non-canonical Smad signalling involves TGFBR1-mediated activation of multiple intermediate serine/threonine kinases which are part of cascades that phosphorylate serine and threonine residues in the linker region of R-Smads. In the context of non-canonical Smad signalling, many growth factors and hormones in addition to TGF-β can activate these same serine/threonine kinases that can directly or indirectly lead to the phosphorylation of the relevant target residues in the linker region of R-Smads [6, 7]; some of these kinases may be the same or different from those activated by TGF-β.
In the first descriptions of the role of Smad linker region phosphorylation, oncogenic RAS as well as epidermal growth factor (EGF) phosphorylated four serine/threonine sites in the linker region of Smad2 and Smad3. Notably, these stimuli do not phosphorylate the serine residues of the carboxyl terminal [8]. EGF-mediated Smad linker phosphorylation inhibits the translocation and accumulation of Smads in the nucleus, thus disrupting TGF-β signalling and functions [8]. Canonical signalling involving phosphorylation of the serine residues in the carboxyl terminal of R-Smads leads to the regulation of gene expression. The canonical signalling pathway can be modulated by TGF-β or other growth factors and hormones that activate cytosolic serine/threonine kinases which phosphorylate the Smad linker region residues. However, with new data emerging, it has become apparent that linker region-phosphorylated Smad2 can translocate to the nucleus and indeed stimulate gene expression [6, 9–13]. We showed the presence of linker region phosphorylated Smad2 in the nucleus of human vascular smooth muscle cells (VSMCs) treated with TGF-β [14].
Agonists of G protein coupled receptor (GPCRs), protein tyrosine kinase receptors (PTKRs) and serine/threonine kinase receptors (S/TKR) can all mediate Smad2 linker region phosphorylation. Smad carboxy-terminal phosphorylation only arises from TGFBR1. The TGFBR1 is activated via direct agonist stimulation or via transactivation-dependent mechanisms in which a GPCR via intricate cell membrane mechanisms activates the TGFBR1 [7, 13, 15–17]. In these examples, it is difficult to interpret the separate roles for Smad carboxy and linker region phosphorylation. However, in the one specific example of thrombin signalling via protease-activated receptors (PAR)-1 in keratinocytes, thrombin had no effect on the carboxy-terminal phosphorylation of Smad2; this implies that the responses of increased plasminogen activator inhibitor (PAI)-1 expression and cell migration arose from thrombin-stimulated Smad linker region phosphorylation [6]. Furthermore, thrombin-stimulated PAI-1 expression was not blocked by the prototypical TGFBR1 inhibitor, SB431542, demonstrating that there is no role for TGFBR1 or Smad carboxy-terminal phosphorylation in this response [6]. These results provide evidence that it is now possible to identify the GPCR/PAR-1 serine/threonine kinase-mediated Smad linker region phosphorylation pathway as a signalling pathway in its own right and not simply a pathway for the regulation of the canonical TGF-β signalling pathway. In this review we examine, evaluate and present the evidence that the non-canonical Smad linker region phosphorylation pathway is a signalling pathway in its own right.
Historical perspective
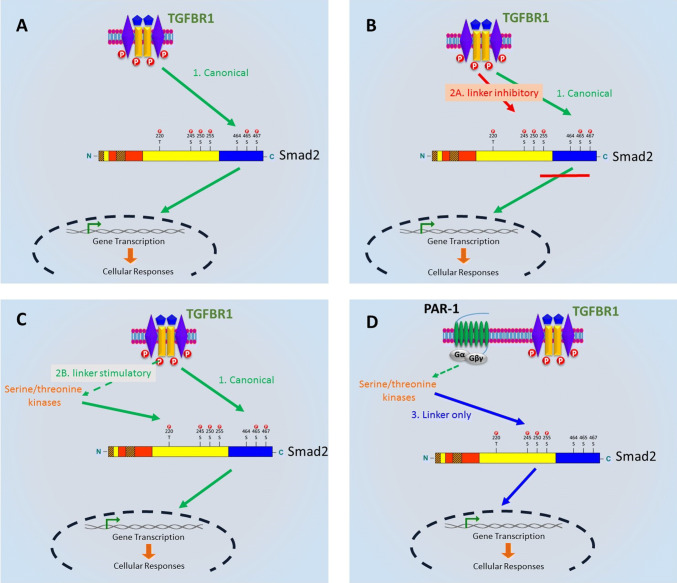
The first characterization of TGF-β and Smad signalling revealed carboxyl terminal phosphorylation of R-Smads, followed by nuclear translocation and regulation of gene transcription (Fig. 1a) [1]. Subsequently, it was shown that concomitant linker region phosphorylation of Smad linker and carboxyl terminal lead to nuclear exclusion and effectively inhibition of TGF-β signalling and cellular responses (Fig. 1b) [8, 18].
Fig. 1.
The evolution of Smad signalling. a Canonical TGF-β receptor (TGFBR1) signalling results in the direct phosphorylation of Smad in the carboxyl-terminal region. Phosphorylated Smad forms a complex with Smad4 and enters the nucleus to regulate transcription. b Phosphorylation of the Smad2 linker region inhibits nuclear translocation and TGF-β signalling [8]. c Activation of the TGFBR1 leads to the phosphorylation of the Smad2 linker region via activation of serine/threonine kinases, and phosphorylated Smad2 in vascular smooth muscle cells stimulates the expression of genes associated with atherosclerosis [10]. d G protein-coupled receptor agonist phosphorylated the Smad2 in the linker region in the absence of Smad carboxyl phosphorylation to regulate gene expression and migration [6]
However, in recent reports, it has become apparent that TGF-β can activate kinases which lead to the phosphorylation of Smads in the linker region and this dually carboxyl terminal and linker region phosphorylated entity can translocate to the nucleus and stimulate gene expression. In data from our laboratory, TGF-β treatment of human VSMCs leads to upregulation of the expression of the genes for two of the enzymes which are rate limiting in the polymerization and elongation of the glycosaminoglycan (GAG) chains on the proteoglycan, biglycan (Fig. 1c) [10, 14]. Inhibitors of Erk1/2 and p38 (but not Jnk) MAP kinases [14] block TGF-β-mediated increase in the length of GAG chains and expression of the GAG elongation enzymes; and the kinase-specific inhibition of GAG elongation correlated with the effects on Smad linker region phosphorylation in these cells [10, 14]. It was this observation that transformed the prevailing view of the role of non-canonical Smad signalling [11, 15, 19]. In VSMCs, GAG hyperelongation by TGF-β stimulation is dependent on both transcription and translation [20]. Our recent work using a chromatin immunoprecipitation assay shows that Smads bind to the promoter region of GAG-synthesizing genes [11]. This work also shows that for TGF-β-regulated GAG gene expression, the phosphorylation of Smad2 linker region facilitates gene expression. In VSMCs, thrombin via transactivation of the TGFBR1 and EGFR regulated GAG chain hyperelongation [21] and the expression of the genes for GAG chain elongation [7, 13, 19]. Mechanistic studies show that thrombin activates the PAR-1 leading to transactivation of the EGFR via matrix metalloproteinase to phosphorylate the Smad2 linker region [7]. This is an additional example of phosphorylation of the Smad linker region stimulating rather than inhibiting gene expression.
In VSMCs, thrombin transactivates both TGFBR1 (leading to phospho-Smad2C) and EGFR (leading to phospho-Erk1/2), so it is difficult to distinguish or separate the involvement of TGFBR1 action arising from thrombin-meditated transactivation of TGF-β (via TGFBR1 to stimulate carboxy-terminal phosphorylation of Smads). We have recently demonstrated in human keratinocytes that PAR-1 activation by thrombin only transactivates EGFR (leading to Smad linker region phosphorylation) and thrombin activation of PAR-1 does not transactivate TGFBR1 as we have observed in VSMCs [6]. PAR-1 transactivation of the EGFR sequentially stimulates Smad linker region phosphorylation, the expression of PAI-1 mRNA and keratinocyte migration [6]. So, it is Smad2 linker phosphorylation arising from EGFR transactivation that mediates the responses and there is no role for canonical carboxy-terminal Smad phosphorylation in this process because the response is not blocked by the prototypical TGFBR1 inhibitor, SB431542.
Recently, several examples [6, 22, 23] have emerged where it has been demonstrated that linker region phosphorylation of R-Smads can yield entities which are not carboxy terminal phosphorylated, but can modulate gene transcription. In the TH17 cell differentiation model, proximity ligation assays demonstrated that phosphorylated Smad2 linker region was in close proximity of STAT3 and histone acetyl-transferase p300, and in contrast C-terminal phosphorylated Smad2 was not [22]. Phosphorylation of Ser255 (in the Smad2 linker region) leads to formation of a complex with p300 and STAT3 to bind to the promotor regions of Rorc and Il17a genes [22]. This shows that the linker region of Smads induces cellular responses independent of carboxy-terminal phosphorylation and as such represents a signalling pathway in its own right.
GPCR transactivation-dependent signalling and Smad linker region phosphorylation
In the phenomenon of GPCR transactivation signalling, first discovered in the Ullrich laboratory [24] more than two decades ago and since studied in great detail, the paradigm is restricted to GPCR transactivation of tyrosine kinase cell surface receptors, most notably receptors for EGF, platelet-derived growth factor (PDGF), fibroblast growth factor and other tyrosine kinase agonists [25]. However, in the last decade we have discovered that GPCRs can transactivate serine threonine kinase cell surface receptors and specifically the TGFBR1 [26–28]. Although there were several earlier reports of GPCR agonists being associated with the generation of phospho-Smad2C, we suggested the model that this was transactivation signalling analogous to the original work of Ullrich and colleagues [24]. Our original studies were in VSMCs in which there is very strong and equivalent activation of both the tyrosine and the serine threonine kinase pathways—thrombin acting on PAR-1 leads to the activation of both EGFR and TGFBR1 to generate phospho-Erk1/2 and phospho-Smad2C, respectively, and both of these pathways drive the expression of genes for the polymerizing enzymes which are responsible for the synthesis of the chondroitin sulphate/dermatan sulphate GAG chains on the lipid-binding proteoglycan biglycan [7, 21, 29].
In the context of renal fibrosis, PAR-2 transactivation of EGFR and TGFBR1 was studied in human proximal tubular epithelial cells [30]. PAR-2 mediated phosphorylation of Smad2 was inhibited by a broad-spectrum matrix metalloproteinase (MMP) inhibitor, marmistat. However, in mouse proximal tubule cells, lysophosphatidic acid (LPA)-mediated Smad2C phosphorylation was unaffected by the broad-spectrum MMP inhibitor, GM6001 [31], and in VSMCs, PAR-1-mediated transactivation of the TGFBR1 was unaffected by GM6001 [21]. Re-examination of the role of MMPs provided the interpretation that in VSMCs, PAR-1-mediated phosphorylation of only the Smad2 in the linker region was dependent on MMPs [7] because it arose from classic EGFR transactivation. GPCR transactivation of PTKRs utilizes cell membrane mechanisms and MMPs. MMPs are involved in PAR-1 and PAR-2 (personal communication)-mediated phosphorylation of the Smad2 linker region [6, 7, 30].
In various cell types, GPCR agonists transactivate the TGFBR1 to phosphorylate Smad2 in the carboxy terminal [26, 27, 32]. However, we have screened numerous cell types treated with GPCR agonists and in some cells we could not detect Smad2 carboxy-terminal phosphorylation. Thus, the data available at present indicates that GPCR transactivation of TGFBR1 is an important, but not a universal cellular phenomenon. However, we have thus far universally observed the agonist-mediated phosphorylation of the linker region of Smad2/3. We also observed a Smad linker region phosphorylation mediated stimulation of gene expression, noting that this occurs without any activation of TGFBR1. From these and other studies, we tentatively developed the working hypothesis that the non-canonical Smad linker region phosphorylation pathway is a signalling pathway in its own right and not only a modulator of canonical TGF-β signalling. Below, we discuss GPCR-mediated Smad2 linker region phosphorylation in the absence of TGFBR1 and carboxy-terminal phosphorylation.
Signalling pathways leading to isolated Smad linker region phosphorylation
TGFBR1 not only phosphorylates the C-termini of R-Smads, but also activates multiple serine/threonine kinases, which then directly or indirectly phosphorylate various linker region residues of R-Smads [15, 33]. Smad-independent [34] or non-canonical cascades [35] refer to the signalling pathways resulting in the activation of serine/threonine kinases to phosphorylate the linker region of Smad transcription factors. The canonical TGF-β/BMP signalling pathway leading to Smad carboxy-terminal phosphorylation is dogmatically treated as the more important signalling pathway; however, recent observations suggest that the Smad linker region phosphorylation plays a more widespread role in cell biology than previously considered.
The Smad linker region can undergo regulatory phosphorylation by multiple kinases, including MAPKs, Erk, Jnk and p38, PI3 K, CDKs, ROCK and GSK3 [9, 10, 13, 36]. Among these kinases, MAPK and CDK are major groups of protein kinases showing preference for specific serine/threonine residues in the linker region [10, 11, 14]. Phosphorylation of the serine and threonine linker residues can act to both augment and antagonize R-Smads functioning in a context-dependent manner. For example, induction of the RAS protein or stimulation of Erk signalling prevents the nuclear accumulation of R-Smads and blocks gene transcription by Smads [37]. Phosphorylation of the same residue by p38, ROCK and Jnk enhances transcription by Smads [38]. In epithelial cells, TGFBR1 and RAS-associated kinases, including Erk, Jnk and CDK4, differentially phosphorylate Smad2 and Smad3 in the carboxy terminal, linker region or both [39]. The CDK-mediated phosphorylation events create variable Smad2/3 phospho-isoforms that can differentially interact with Smad4 and either translocate or be blocked from entering the nucleus to initiate the transcription of target genes.
In TH17 cells, phosphorylated Smad2 in the linker region serves as a STAT3 transcription factor co-activator, whereas unphosphorylated Smad3 in the carboxy terminal serves as a STAT3 co-repressor [22]. Treatment with PI3K inhibitor reduced phosphorylation of Thr179/220 residue of the Smad protein irrespective of Smad2/3 C-terminal phosphorylation in both human embryonic stem cells and tumor cell lines [23]. Utilizing both pharmacological and genetic approaches, mTORC2 was identified as a key mediator in regulating Smad2/3 linker region Thr220/179. Together, the Thr220/179 residue of the Smad2/3 linker region can be readily phosphorylated by PI3K/mTORC2 signalling pathways in the absence of Smad2/3 carboxy activation [23].
More recently, primary human mesenchymal stem cells stimulated with TGF-β, IL1β or a combination of the two agonists phosphorylated Smad2 [40]. IL1β induced the phosphorylation of both the Thr220 and the three serine residues of Smad2 in the linker region; however, these agonists had no effect on Smad2C phosphorylation within a 24 h time frame [40]. Palatal mesenchymal cells from TAK-1 knockout mice exhibit reduced Smad carboxy and linker region phosphorylation [41]; in primary mesenchymal stem cells TAK-1 plays a crucial role in mediating IL Iβ-induced TGF-β signalling via the Smad linker region [40]. Together, these results show that the linker region-phosphorylated Smad, in the absence of phosphorylation of the carboxy terminal, can translocate to the nucleus and regulate gene transcription.
In the context of extracellular matrix synthesis and proteoglycan modification, the stimulation of biglycan expression as well as the hyperelongation of GAG chains is dependent on Smad2 linker region phosphorylation [10, 13, 14, 19]. Phosphorylation of Smad has emerged as a critical step in the signalling pathways that control the synthesis of biglycan, both the core protein and GAG chains. More recently, we have used flavopiridol, a broad-spectrum CDK inhibitor, to study the role of linker region phosphorylation in TGF-β-stimulated synthesis of biglycan [11]. Flavopiridol blocked TGF-β-mediated linker region phosphorylation at both the serine and the threonine residues. TGF-β stimulated the binding of the Smad to the promoter region of the C4ST-1 and CHSY1 genes and this response was inhibited by flavopiridol. The increase in the binding of the Smad to the promoter regions of the two genes shows that in this context, the transcription factor can translocate into the nucleus to regulate gene transcription [11]. The GPCR agonist, thrombin, transactivates EGFR and TGFBR1 leading to the enhanced synthesis of proteoglycans and an increase in mRNA expression of these two GAG-synthesizing genes [7, 19]. The Smad2 linker region served as a point for integrating GPCR-mediated signalling via transactivation of the EGFR and TGFBR1, and phosphorylation of specific linker region residues led to downstream expression of individual genes associated with the initiation and elongation of proteoglycan GAG chains [13].
The Smad linker region is associated with a number of pathophysiological conditions, including cancer development [8], cardiovascular disease [10, 14], Alzheimer’s [42], renal fibrosis [43], esophagitis mucosae [44], endothelial dysfunction [9], wound healing [6] and inflammation-related diseases [22, 40]. Greater appreciation of the Smad2 linker region as a signalling pathway in its own right will allow for more in-depth studies of this transcription factor and may lead to the identification of therapeutic targets for intervention in human disease.
Non-TGF-β/TGFBR1-mediated Smad linker phosphorylation
Phosphorylation of the linker region of Smad transcription factors was originally described as a parallel opposing or modulating mechanism of canonical TGF-β/BMP effector signalling. However, Smad linker phosphorylation and downstream function entirely independent of TGF-β/BMP signals were reported in the earliest studies of Smad function. Kretzschmar et al. [45] were the first to demonstrate that specific serine residues in Smad1 linker could be phosphorylated by Erk1/2 independently of BMP signals. Simultaneous mutation of the linker region PXSP motifs and the BMP receptor carboxy target motif SSVS resulted in complete loss of Smad1 phosphorylation both in the presence and absence of stimulation by BMPs. Erk-mediated phosphorylation of Smad1 linker inhibited the accumulation and activity of Smad1 in the nucleus. In the following years, it was reported that PTKR-mediated Smad linker phosphorylation can generate activating signals; both hepatocyte growth factor (HGF) and EGF are able to trigger rapid phosphorylation, within 15 min, of Smad1 and Smad2 linker proteins by kinases downstream of MEK1 [46]. Blocking with TGF-β-neutralizing antibody had no effect on HGF-mediated Smad linker phosphorylation and downstream responses which confirmed that the signalling was independent and not due to the autocrine or paracrine action of TGF-β.
The linker phosphorylation sites differ between Smad proteins with the Smad1-type linkers containing a cluster of four PXSP sequences, which are classical MAPK sites, and the Smad2-type linkers containing one PXTP followed by three XXSP sequences [47] that can be phosphorylated by another serine threonine kinase Ca2+-calmodulin-dependent kinase II [48]. In 2004 Matsuura et al. [49] showed in mouse embryonic fibroblasts that cyclin-dependent kinases CDK4 and CDK2 phosphorylate Thr178 and Ser212 in Smad3 linker region and inhibit cell cycle progression from G1 to S phase [49]. In rat hepatic stellate cells, Yoshida et al. [50] demonstrated that PDGF-activated Jnk directly phosphorylates Smad2/3 linker and stimulates expression of PAI-1. The Smad3 linker region was subsequently shown to contain a discrete DNA-binding domain that is capable of mediating transcriptional activation [51]. In human aortic endothelial cells, Smad2 is preferentially phosphorylated by the serine/threonine kinase Akt in the linker region and then preferentially localized to the nucleus under shear stress raised from 2 to 10 dyn/cm2 [52]. These findings emphasize the potential importance of independent linker region phosphorylation in different cell signalling contexts and outcomes.
In oral keratinocytes, EGF promotes Erk1/2 activation and Smad2 linker phosphorylation that is blocked by the MEK1/2 inhibitor U0126. This pathway inhibits TGFβ1-mediated N-cadherin expression in normal oral keratinocytes, but does not block in malignant oral squamous carcinoma cells [53]. Melanoma cells exhibit constitutive Smad2 and Smad3 linker phosphorylation and this is attributed to an aberrant TGF-β-based autocrine loop that overrides TGF-β-mediated growth inhibition [18]. However, there is also evidence showing that human melanoma cells have TGF-β-independent Smad linker phosphorylation, which can be induced by the glutamate release inhibitor, riluzole, through GSK3 activation [54]. In fibro-carcinogenesis seen in chronic viral hepatitis of hepatocellular carcinoma, cytokine-driven Jnk activation triggers linker phosphorylation of Smad3 that promotes disease progression [55]. As a disease marker, low pSmad3C and high pSmad3L cell staining are predictive of human hepatocellular carcinoma development [55]. Taken together, these findings provide clear evidence that Smad linker phosphorylation can be a driving force in malignant cells.
We have recently reported on the mechanism of thrombin-stimulated PAI-1 expression and cell migration in keratinocytes [6]. Thrombin stimulated PAI-1 expression in HaCaT cells through EGFR transactivation and subsequent Erk1/2-dependent phosphorylation of Ser250 in the linker region of Smad2 [6]. Importantly, thrombin treatment does not result in an increase in carboxy-terminal phosphorylated Smad2, so thrombin does not transactivate TGFBR1 in these cells; furthermore, thrombin-stimulated expression of PAI-1 (tenfold) is not blocked by the TGFBR1 inhibitor, SB431542, under conditions in which TGF-β stimulation of PAI-1 (100-fold) is completely blocked by SB431542. SB431542 is a highly efficacious inhibitor of TGFBR1, so carboxy-terminal phosphorylation of Smad2 cannot be occurring, meaning that the signalling pathway for thrombin-stimulated PAI-1 expression involves Smad linker region phosphorylation, nuclear translocation and activation of PAI-1 gene expression in the absence of carboxy-terminal Smad phosphorylation. This data are the clearest demonstration that Smad linker region phosphorylation is a signalling pathway in its own right.
The importance of the specificity of Smad phospho-antibodies and signalling pathway interpretations
For cell biology and in particular cell signalling studies, there is a huge array of phospho-antibodies targeting an enormous range of epitopes, often multiple phospho-epitopes on the same protein. In many reports, activation is used synonymously with phosphorylation, but the latter is more specific and is preferred. There are many targets of kinases where different phosphorylation sites have little or no impact on the interpretation of the signalling pathways involved, because they indicate that the target is phosphorylated and the kinase activity is increased sometimes by several orders of magnitude. There are, however, some pathways in which the specific phospho-epitopes have profound implications for the integration and interpretation of the signalling pathway involved.
Smad has three domains: MH1, MH2 and the proline rich linker region. R-Smads have a characteristic Ser-Ser-X-Ser sequence at the C-terminus (MH2) which can be phosphorylated, resulting in the activation of downstream signalling pathways. Antibodies measuring the phosphorylation of carboxy-terminal residues are commercially available. The Smad linker region has at least four residues which are phosphorylated. In Smad2, a threonine in position 220 and three serine residues in positions 245, 250 and 255 can be phosphorylated. In Smad3 there is a threonine residue in position 179 and three serine residues at positions 204, 208 and 213 [56]. Early studies of the Smad linker region utilized an antibody which targeted the three serine residues; however, Sekimoto and colleagues [56] generated nine antibodies to detect various phosphorylation sites of the Smad2 and Smad3 linker region.
The availability of individual antibodies targeting distinct phospho-isoforms has greatly expanded the scope for studying TGF-β signalling. Linker region phosphorylation may allosterically regulate intramolecular interactions between the MH1 and MH2 domains, and intermolecular interactions between Smads and other molecules. Differential localization of protein kinases that can phosphorylate Smad2/3, whether in the cytoplasm or nucleus, also affects multiple signalling responses that determine the functional outcome [57]. In rat gastric epithelial cells, treatment with TGF-β and HGF resulted in nuclear translocation of phosphorylated Smad2 linker residues Thr220 and Ser255, but not Ser245 and Ser250, and with Smad3 linker region residues Thr179 and Ser208 and Ser213 were translocated to the nucleus but not Ser204 [56]. These results show the specificity of Smad2/3 linker signalling pathways. In bovine aortic endothelial cells, a specific signalling pathway was identified to regulate the mRNA expression of PAI-1 [9]. TGF-β via CDKs and p38 phosphorylates the Ser245 and Ser255 residue to regulate the mRNA expression of PAI-1 [9]. In VSMCs specific residues of the Smad2 linker region regulate different genes and functions in the context of cardiovascular disease [10]. Phosphorylation of the Thr220 Smad2 linker was associated with the regulation of the gene involved in the initiation of GAG chain synthesis [10]; however, the serine residues are involved in the elongation of the GAG chains [11]. More recently in human keratinocyte cells, the Smad2 linker region residue Ser250 was shown to regulate thrombin-mediated PAI-1 expression and cell migration [6].
In describing the pathway through which thrombin stimulates PAR-1 to mediate transactivation of the TGFBR1 leading to the formation of pSmad2C, we concluded that MMPs were not involved in this pathway; this was important because MMPs were well known to be involved in GPCR transactivation of EGFR so this was a major point of distinction of these two signalling pathways [58]. Specifically, we used a phospho antibody which targeted the Ser465/467 residues in the carboxy terminal of Smad2. In VSMCs, thrombin-stimulated increase in pSmad2C was not blocked by MMP inhibitor, GM6001, in the same cells in which GM6001 blocked thrombin-mediated increase in pErk1/2, a transactivation response which is known to involve MMPs [21]. Concomitantly to our work, Cheng et al. [30] found that the thrombin-stimulated increase in phosphoSmads was blocked by marmistat, an inhibitor of MMPs. This conflicting experimental data lead to a diametrically opposed conclusion as to the involvement of MMPs in PAR-1 transactivation of TGFBR1 [30]. To investigate this dilemma, we studied the effect of thrombin on both carboxy-terminal and linker region phosphorylation of Smad2 using phospho-antibodies to the appropriate regions [7]. We confirmed our earlier work by showing that the carboxy-terminal phosphorylation of Smad2 was not blocked by GM6001; however, we found that the linker region phosphorylation was indeed blocked by the MMP inhibitor, GM6001 [7]. Consistent with the hypothesis being brought forward in this paper, the data confirmed that linker region phosphorylation occurred as PAR-1 to EGFR transactivation (as described above) where activation of EGFR led to increased pErk1/2 and direct phosphorylation of Smad2 in the linker region [6]. The conclusion is that MMPs are involved in PAR-1-mediated transactivation of EGFR, but they are not involved in PAR-1-mediated transactivation of TGFBR1.
Conclusions
The implications of these findings and interpretations are profound—there are a multitude of serine/threonine kinases which can phosphorylate multiple residues in the linker region of Smad2/3 and it is likely that these lead to very specific alterations in the expression of individual or sets of genes which may be involved in pathophysiological processes. The role of Smad linker region phosphorylation was discovered and described in terms of its role in modulating canonical TGFBR1/Smad signalling. There are, however, several layers of complexity that are imposed on this simple paradigm. Phosphorylation of the Smad2 linker region, however, can occur independently of TGFBR1. In the context of transactivation-dependent signalling, GPCRs can transactivate PTKRs to phosphorylate Smad2 linker region. Direct activation of PTKRs involved the activation of intracellular serine/threoning kinases to mediate Smad linker region phosphorylation. In the studies covered in this review, Smad linker region phosphorylation occurring independently of carboxy-terminal phosphorylation drives the expression of genes in functional signalling pathways. Thus, hitherto unimagined therapeutic targets, various serine/threonine kinases which phosphorylate Smad2/3 in the linker region, may arise from a reimagining of Smad transcription factor signalling as addressed in this review.
Abbreviations
- BMP
Bone morphogenetic proteins
- C4ST-1
Chondroitin 4-O-sulfotransferase 1
- CHSY1
Chondroitin synthase 1
- EGF
Epidermal growth factor
- EGFR
Epidermal growth factor receptor
- GAG
Glycosaminoglycan
- GPCR
G protein-coupled receptor
- MMP
Matrix metalloproteinase
- PAI-1
Plasminogen activator inhibitor-1
- PAR
Protease-activated receptor
- PDGF
Platelet-derived growth factor
- PTKR
Protein tyrosine kinase receptor
- S/TKR
Serine/threonine kinase receptor
- TGFBR1
Transforming growth factor-β receptor type 1
- TGF-β
Transforming growth factor-β
- VSMCs
Vascular smooth muscle cells
Funding
DK was supported by the NHMRC (APP1160925) and National Heart Foundation Fellowship (102129). Support was received from the University of Queensland through a personal support package to PJL and by the University of Queensland Early Career Grant (DK) (Grant no. 1832825).
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Massague J. TGF-beta signal transduction. Annu Rev Biochem. 1998;67:753–791. doi: 10.1146/annurev.biochem.67.1.753. [DOI] [PubMed] [Google Scholar]
- 2.Frick CL, et al. Sensing relative signal in the Tgf-beta/Smad pathway. Proc Natl Acad Sci USA. 2017;114(14):E2975–E2982. doi: 10.1073/pnas.1611428114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Massague J, Seoane J, Wotton D. Smad transcription factors. Genes Dev. 2005;19(23):2783–2810. doi: 10.1101/gad.1350705. [DOI] [PubMed] [Google Scholar]
- 4.Dennler S, Huet S, Gauthier JM. A short amino-acid sequence in MH1 domain is responsible for functional differences between Smad2 and Smad3. Oncogene. 1999;18(8):1643–1648. doi: 10.1038/sj.onc.1202729. [DOI] [PubMed] [Google Scholar]
- 5.Liu L, et al. Smad2 and Smad3 have differential sensitivity in relaying TGFbeta signaling and inversely regulate early lineage specification. Sci Rep. 2016;6:21602. doi: 10.1038/srep21602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Talati N, et al. Thrombin promotes PAI-1 expression and migration in keratinocytes via ERK dependent Smad linker region phosphorylation. Cell Signal. 2018;47:37–43. doi: 10.1016/j.cellsig.2018.03.009. [DOI] [PubMed] [Google Scholar]
- 7.Kamato D, et al. Protease activated receptor-1 mediated dual kinase receptor transactivation stimulates the expression of glycosaminoglycan synthesizing genes. Cell Signal. 2016;28(1):110–119. doi: 10.1016/j.cellsig.2015.11.003. [DOI] [PubMed] [Google Scholar]
- 8.Kretzschmar M, et al. A mechanism of repression of TGFbeta/Smad signaling by oncogenic Ras. Genes Dev. 1999;13(7):804–816. doi: 10.1101/gad.13.7.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kamato D, et al. Transforming growth factor beta-mediated site-specific Smad linker region phosphorylation in vascular endothelial cells. J Pharm Pharmacol. 2014;66(12):1722–1733. doi: 10.1111/jphp.12298. [DOI] [PubMed] [Google Scholar]
- 10.Rostam MA, et al. The role of specific Smad linker region phosphorylation in TGF-beta mediated expression of glycosaminoglycan synthesizing enzymes in vascular smooth muscle. Cell Signal. 2016;28(8):956–966. doi: 10.1016/j.cellsig.2016.05.002. [DOI] [PubMed] [Google Scholar]
- 11.Rostam MA, et al. Flavopiridol inhibits TGF-beta-stimulated biglycan synthesis by blocking linker region phosphorylation and nuclear translocation of Smad2. J Pharmacol Exp Ther. 2018;365(1):156–164. doi: 10.1124/jpet.117.244483. [DOI] [PubMed] [Google Scholar]
- 12.Kamato D, et al. (S)-[6]-Gingerol inhibits TGF-beta-stimulated biglycan synthesis but not glycosaminoglycan hyperelongation in human vascular smooth muscle cells. J Pharm Pharmacol. 2013;65(7):1026–1036. doi: 10.1111/jphp.12060. [DOI] [PubMed] [Google Scholar]
- 13.Kamato D, et al. Individual Smad2 linker region phosphorylation sites determine the expression of proteoglycan and glycosaminoglycan synthesizing genes. Cell Signal. 2019;53:365–373. doi: 10.1016/j.cellsig.2018.11.005. [DOI] [PubMed] [Google Scholar]
- 14.Burch ML, et al. TGF-beta stimulates biglycan synthesis via p38 and ERK phosphorylation of the linker region of Smad2. Cell Mol Life Sci. 2010;67(12):2077–2090. doi: 10.1007/s00018-010-0315-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kamato D, et al. Transforming growth factor-beta signalling: role and consequences of Smad linker region phosphorylation. Cell Signal. 2013;25(10):2017–2024. doi: 10.1016/j.cellsig.2013.06.001. [DOI] [PubMed] [Google Scholar]
- 16.Dayati P, et al. G protein coupled receptors can transduce signals through carboxy terminal and linker region phosphorylation of Smad transcription factors. Life Sci. 2018;199:10–15. doi: 10.1016/j.lfs.2018.03.004. [DOI] [PubMed] [Google Scholar]
- 17.Kamato D, et al. RNA sequencing to determine the contribution of kinase receptor transactivation to G protein coupled receptor signalling in vascular smooth muscle cells. PLoS One. 2017;12(7):e0180842. doi: 10.1371/journal.pone.0180842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cohen-Solal KA, et al. Constitutive Smad linker phosphorylation in melanoma: a mechanism of resistance to transforming growth factor-beta-mediated growth inhibition. Pigment Cell Melanoma Res. 2011;24(3):512–524. doi: 10.1111/j.1755-148X.2011.00858.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kamato D, et al. Mechanisms of PAR-1 mediated kinase receptor transactivation: Smad linker region phosphorylation. J Cell Commun Signal. 2019 doi: 10.1007/s12079-019-00527-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang SN, et al. Growth factor-mediated hyper-elongation of glycosaminoglycan chains on biglycan requires transcription and translation. Arch Physiol Biochem. 2009;115(3):147–154. doi: 10.1080/13813450903110754. [DOI] [PubMed] [Google Scholar]
- 21.Burch ML, et al. Thrombin-mediated proteoglycan synthesis utilizes both protein-tyrosine kinase and serine/threonine kinase receptor transactivation in vascular smooth muscle cells. J Biol Chem. 2013;288(10):7410–7419. doi: 10.1074/jbc.M112.400259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoon JH, et al. Phosphorylation status determines the opposing functions of Smad2/Smad3 as STAT3 cofactors in TH17 differentiation. Nat Commun. 2015;6:7600. doi: 10.1038/ncomms8600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu JS, et al. PI3 K/mTORC2 regulates TGF-beta/Activin signalling by modulating Smad2/3 activity via linker phosphorylation. Nat Commun. 2015;6:7212. doi: 10.1038/ncomms8212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Daub H, et al. Role of transactivation of the EGF receptor in signalling by G-protein-coupled receptors. Nature. 1996;379(6565):557–560. doi: 10.1038/379557a0. [DOI] [PubMed] [Google Scholar]
- 25.Blume-Jensen P, Hunter T. Oncogenic kinase signalling. Nature. 2001;411(6835):355–365. doi: 10.1038/35077225. [DOI] [PubMed] [Google Scholar]
- 26.Little PJ, et al. Endothelin-1 stimulation of proteoglycan synthesis in vascular smooth muscle is mediated by endothelin receptor transactivation of the transforming growth factor-[beta] type I receptor. J Cardiovasc Pharmacol. 2010;56(4):360–368. doi: 10.1097/FJC.0b013e3181ee6811. [DOI] [PubMed] [Google Scholar]
- 27.Sharifat N, et al. Endothelin-1 (ET-1) stimulates carboxy terminal Smad2 phosphorylation in vascular endothelial cells by a mechanism dependent on ET receptors and de novo protein synthesis. J Pharm Pharmacol. 2017;69(1):66–72. doi: 10.1111/jphp.12654. [DOI] [PubMed] [Google Scholar]
- 28.Chaplin R, et al. Insights into cellular signalling by G protein coupled receptor transactivation of cell surface protein kinase receptors. J Cell Commun Signal. 2017;11(2):117–125. doi: 10.1007/s12079-017-0375-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Afroz R, et al. Signalling pathways regulating galactosaminoglycan synthesis and structure in vascular smooth muscle: Implications for lipoprotein binding and atherosclerosis. Pharmacol Ther. 2018;187:88–97. doi: 10.1016/j.pharmthera.2018.02.005. [DOI] [PubMed] [Google Scholar]
- 30.Chung H, et al. Proteinase-activated receptor-2 transactivation of epidermal growth factor receptor and transforming growth factor-beta receptor signaling pathways contributes to renal fibrosis. J Biol Chem. 2013;288(52):37319–37331. doi: 10.1074/jbc.M113.492793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Geng H, et al. Lysophosphatidic acid increases proximal tubule cell secretion of profibrotic cytokines PDGF-B and CTGF through LPA2- and Galphaq-mediated Rho and alphavbeta6 integrin-dependent activation of TGF-beta. Am J Pathol. 2012;181(4):1236–1249. doi: 10.1016/j.ajpath.2012.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Little PJ, et al. Integrating the GPCR transactivation-dependent and biased signalling paradigms in the context of PAR-1 signalling. Br J Pharmacol. 2015;173(20):2992–3000. doi: 10.1111/bph.13398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rezaei HB, et al. Cell biology of Smad2/3 linker region phosphorylation in vascular smooth muscle. Clin Exp Pharmacol Physiol. 2012;39(8):661–667. doi: 10.1111/j.1440-1681.2011.05592.x. [DOI] [PubMed] [Google Scholar]
- 34.Derynck R, Zhang YE. Smad-dependent and Smad-independent pathways in TGF-beta family signalling. Nature. 2003;425(6958):577–584. doi: 10.1038/nature02006. [DOI] [PubMed] [Google Scholar]
- 35.Zhang YE. Non-Smad pathways in TGF-beta signaling. Cell Res. 2009;19(1):128–139. doi: 10.1038/cr.2008.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mohamed R, et al. Transforming growth factor-beta1 mediated CHST11 and CHSY1 mRNA expression is ROS dependent in vascular smooth muscle cells. J Cell Commun Signal. 2018;13(2):225–233. doi: 10.1007/s12079-018-0495-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grimm OH, Gurdon JB. Nuclear exclusion of Smad2 is a mechanism leading to loss of competence. Nat Cell Biol. 2002;4(7):519–522. doi: 10.1038/ncb812. [DOI] [PubMed] [Google Scholar]
- 38.Kamaraju AK, Roberts AB. Role of Rho/ROCK and p38 MAP kinase pathways in transforming growth factor-beta-mediated Smad-dependent growth inhibition of human breast carcinoma cells in vivo. J Biol Chem. 2005;280(2):1024–1036. doi: 10.1074/jbc.M403960200. [DOI] [PubMed] [Google Scholar]
- 39.Horiguchi K, et al. Role of Ras signaling in the induction of snail by transforming growth factor-beta. J Biol Chem. 2009;284(1):245–253. doi: 10.1074/jbc.M804777200. [DOI] [PubMed] [Google Scholar]
- 40.van den Akker GG, et al. Interleukin 1 beta-induced SMAD2/3 linker modifications are TAK1 dependent and delay TGFbeta signaling in primary human mesenchymal stem cells. Cell Signal. 2017;40:190–199. doi: 10.1016/j.cellsig.2017.09.010. [DOI] [PubMed] [Google Scholar]
- 41.Yumoto K, et al. TGF-beta-activated kinase 1 (Tak1) mediates agonist-induced Smad activation and linker region phosphorylation in embryonic craniofacial neural crest-derived cells. J Biol Chem. 2013;288(19):13467–13480. doi: 10.1074/jbc.M112.431775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ueberham U, et al. Altered subcellular location of phosphorylated Smads in Alzheimer’s disease. Eur J Neurosci. 2006;24(8):2327–2334. doi: 10.1111/j.1460-9568.2006.05109.x. [DOI] [PubMed] [Google Scholar]
- 43.Sun YB, et al. Endothelial dysfunction exacerbates renal interstitial fibrosis through enhancing fibroblast Smad3 linker phosphorylation in the mouse obstructed kidney. PLoS One. 2013;8(12):e84063. doi: 10.1371/journal.pone.0084063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Takahashi Y, et al. Phosphorylation of Smad2/3 at the specific linker threonine residue indicates slow-cycling esophageal stem-like cells before re-entry to the cell cycle. Dis Esophagus. 2016;29(2):107–115. doi: 10.1111/dote.12277. [DOI] [PubMed] [Google Scholar]
- 45.Kretzschmar M, Doody J, Massague J. Opposing BMP and EGF signalling pathways converge on the TGF-beta family mediator Smad1. Nature. 1997;389(6651):618–622. doi: 10.1038/39348. [DOI] [PubMed] [Google Scholar]
- 46.de Caestecker MP, et al. Smad2 transduces common signals from receptor serine-threonine and tyrosine kinases. Genes Dev. 1998;12(11):1587–1592. doi: 10.1101/gad.12.11.1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Massague J. Integration of Smad and MAPK pathways: a link and a linker revisited. Genes Dev. 2003;17(24):2993–2997. doi: 10.1101/gad.1167003. [DOI] [PubMed] [Google Scholar]
- 48.Wicks SJ, et al. Inactivation of smad-transforming growth factor beta signaling by Ca(2 +)-calmodulin-dependent protein kinase II. Mol Cell Biol. 2000;20(21):8103–8111. doi: 10.1128/mcb.20.21.8103-8111.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Matsuura I, et al. Cyclin-dependent kinases regulate the antiproliferative function of Smads. Nature. 2004;430(6996):226–231. doi: 10.1038/nature02650. [DOI] [PubMed] [Google Scholar]
- 50.Yoshida K, et al. Transforming growth factor-beta and platelet-derived growth factor signal via c-Jun N-terminal kinase-dependent Smad2/3 phosphorylation in rat hepatic stellate cells after acute liver injury. Am J Pathol. 2005;166(4):1029–1039. doi: 10.1016/s0002-9440(10)62324-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Harikrishnan KN, et al. Brahma links the SWI/SNF chromatin-remodeling complex with MeCP2-dependent transcriptional silencing. Nat Genet. 2005;37(3):254–264. doi: 10.1038/ng1516. [DOI] [PubMed] [Google Scholar]
- 52.Shepherd RD, Kos SM, Rinker KD. Flow-dependent Smad2 phosphorylation and TGIF nuclear localization in human aortic endothelial cells. Am J Physiol Heart Circ Physiol. 2011;301(1):H98–H107. doi: 10.1152/ajpheart.00668.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Diamond ME, et al. Differential growth factor regulation of N-cadherin expression and motility in normal and malignant oral epithelium. J Cell Sci. 2008;121(Pt 13):2197–2207. doi: 10.1242/jcs.021782. [DOI] [PubMed] [Google Scholar]
- 54.Abushahba W, et al. Non-canonical Smads phosphorylation induced by the glutamate release inhibitor, riluzole, through GSK3 activation in melanoma. PLoS One. 2012;7(10):e47312. doi: 10.1371/journal.pone.0047312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Murata M, et al. Linker phosphorylation of Smad3 promotes fibro-carcinogenesis in chronic viral hepatitis of hepatocellular carcinoma. World J Gastroenterol. 2014;20(41):15018–15027. doi: 10.3748/wjg.v20.i41.15018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sekimoto G, et al. Reversible Smad-dependent signaling between tumor suppression and oncogenesis. Cancer Res. 2007;67(11):5090–5096. doi: 10.1158/0008-5472.CAN-06-4629. [DOI] [PubMed] [Google Scholar]
- 57.Matsuzaki K. Smad phospho-isoforms direct context-dependent TGF-β signaling. Cytokine Growth Factor Rev. 2013;24(4):385–399. doi: 10.1016/j.cytogfr.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 58.Kamato D, et al. The expansion of GPCR transactivation-dependent signalling to include serine/threonine kinase receptors represents a new cell signalling frontier. Cell Mol Life Sci. 2015;72(4):799–808. doi: 10.1007/s00018-014-1775-0. [DOI] [PMC free article] [PubMed] [Google Scholar]