Abstract
Quorum sensing (QS), a microbial cell-to-cell communication process, dynamically regulates a variety of metabolism and physiological activities. In this review, we provide an update on QS applications based on autoinducer molecules including acyl-homoserine lactones (AHLs), auto-inducing peptides (AIPs), autoinducer 2 (AI-2) and indole in population-level control of bacteria, and highlight the potential in developing novel clinical therapies. We summarize the development in the combination of various genetic circuits such as genetic oscillators, toggle switches and logic gates with AHL-based QS devices in Gram-negative bacteria. An overview is then offered to the state-of-the-art of much less researched applications of AIP-based QS devices with Gram-positive bacteria, followed by a review of the applications of AI-2 and indole based QS for interspecies communication among microbial communities. Building on these general-purpose QS applications, we highlight the disruptions and manipulations of QS devices as potential clinical therapies for diseases caused by biofilm formation, antibiotic resistance and the phage invasion. The last part of reviewed literature is dedicated to mathematical modelling for QS applications. Finally, the key challenges and future perspectives of QS applications in monoclonal synthetic biology and synthetic ecology are discussed.
Keywords: Cell–cell communication, Signaling molecule, Microbial community, Population control, Genetic circuit, Gut microbiota
Introduction
Quorum sensing (QS) is a cell–cell communication process, which is ubiquitous in fungi [1], bacteria and even viruses [2]. QS regulates a series of physiological and biochemical functions, such as biofilm formation, conjugation, competence, bacteriocin production and pathogenesis, achieved by microbes producing, secreting, sensing and responding to certain signal molecules which are called autoinducers (AIs) [3]. Generally, various AIs can be roughly divided into three types: (i) acylated homoserine lactones (AHLs) and the diffusible signaling factors (DSFs) utilized by Gram-negative bacteria; (ii) auto-inducing peptides (AIPs) utilized by Gram-positive bacteria; and (iii) autoinducer 2 (AI-2) and indole for interspecies communication of microbial communities [4]. Combining these AIs and their relevant QS devices with synthetic genetic circuits is of great importance to the dynamic control of bacterial populations and to the development of potential clinical therapies (Fig. 1).
Fig. 1.
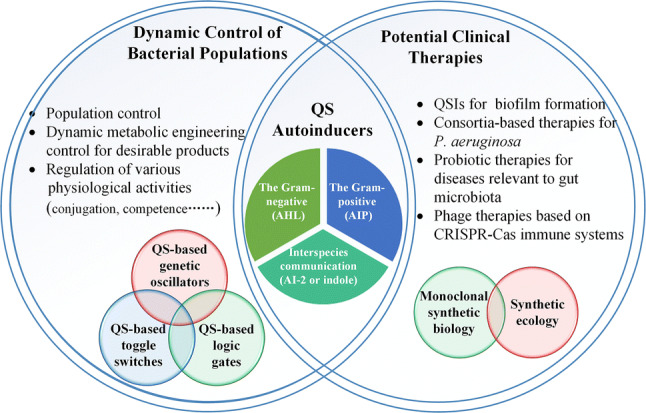
QS applications for dynamic control of bacteria populations and its potential clinical therapies for diseases. Dynamic control of bacterial populations includes three aspects, i.e., the population of bacteria control, dynamic metabolic engineering control and regulation of physiological activities. They mainly work on the combinations of various genetic circuits such as genetic oscillators, genetic toggle switches and genetic logic gates. Underpinned by the functioning of autoinducer molecules, i.e., AHLs, DSFs, AIPs, AI-2 and indole, the disruptions and manipulations of QS in the dynamic control of bacteria populations can be extended to be widely applied in monoclonal synthetic biology and synthetic ecology to develop potential clinical therapies
Dynamic control of bacterial populations usually includes population size control, dynamic metabolic engineering for desirable products and the regulation of various physiological activities [5] (Fig. 1). Metabolic control, a major issue in dynamic control of metabolic engineering, can be divided into static metabolic control and dynamic metabolic control [6]. Usually, static metabolic control involves natural or slightly modified control systems with knockout, weakening and overexpression of genes. Dynamic metabolic control utilizes genetic circuits such as toggle switches and sensor-regulator to achieve the dynamic adjustment of metabolic production of microbes [5]. According to the type of genetic circuits involved, either on–off or continuous dynamic metabolic control can be achieved [7]. A common strategy via introducing an on–off switch is to close the relevant competitive pathways when the bacteria population reaches a certain level. This type of genetic circuits has the disadvantages of requiring proper induction time, increasing production costs due to the addition of inducer and being incapable of sensing the changing environment continuously. To overcome these disadvantages, continuous dynamic metabolic control has been developed to up-regulate the desirable product using synthetic feedback loops, such as QS-based devices [7]. The implementation of dynamic metabolic control can be either pathway-specific or pathway-independent [8]. The pathway-specific implementations are achieved by detecting input and output changes of a relevant intermediate or byproduct [9], while the pathway-independent implementations are through nutrients in the medium or by QS [10, 11]. Compared to the pathway-specific implementations which are restricted to sense and dynamically control the metabolism of intracellular pathways, pathway-independent implementations allow microbes to respond to the changing extracellular environment and adjust accordingly their metabolism and physiological activities, with QS as an important enabler [8]. What’s more, significant advances have been made in synthetic biology which created synthetic pathways and circuits to control the expression levels of relevant genes, such as overexpressing the genes for producing glycosides [12] in engineered bacteria. Transcriptional toggle switches [13] and transcriptional oscillators [14] are involved in transcriptional regulation of genes, and genetic loops such as bistable positive feedback loops and RNA-based anti-switches are constructed into biological systems to control post-transcriptional regulation [15], metabolic flux distribution [16] and signaling proteins expression [17]. These QS-based genetic circuits not only make the synthetic systems more reliable and robust [18], but also provide new avenues to the dynamic control of bacterial populations [19].
With the increasingly recognized importance of pathogens and microbiota for human health, the QS-based monoclonal synthetic biology and synthetic ecology have enormous potential in promoting the development of potential clinical therapies for curing devastating diseases, tackling antimicrobial resistance [20] (Fig. 1). Many bacteria have been shown to have a tendency to organize in aggregates generally to adhere to surfaces to form biofilms, and biofilm formation is a principal virulence factor in many localized chronic infections [21, 22]. As shown by existing studies [23], QS and quorum quenching [4] can significantly affect biofilm formation. One of the most important factors causing the changes of microbiota is the use of antibiotics, which does not only alter the microbiota but also promote the emergence of antibiotics resistance [24]. It has recently been demonstrated that QS inhibition including quorum quenching and other QS-blocking approaches can decrease the production of virulence factor [25]. Therefore, coupling QS devices with microbial consortia in various environments such as the human gut has much therapeutic potential, for example, treating chronic infections [26] and relieving antimicrobial resistance [27]. Besides, as infections of phage increase at high cell density [28], QS devices have much potential to regulate the CRISPR-Cas (clustered regularly interspaced short palindromic repeats; CRISPR-associated) immune systems to monitor the development of diseases [29].
Recently, several QS-based reviews have been published which focus on aspects including QS signals transduction and architectures, dynamic control for metabolic engineering, applications of synthetic microbial consortia, socio-microbiology based on cell–cell communications, the applications of QS inhibitors in biofilm formation and the mathematical modelling for QS, as listed in Table 1. The purpose of this current review is to provide an updated summary of the more recent achievements in applying QS to the dynamic control of bacterial populations and in developing potential clinical therapies. We start by presenting the recent promising achievements, including bacteria population control, applications of the genetic toggle switches, synthetic genetic oscillators and genetic logic gates that apply QS devices which are based on AHLs signal transduction in Gram-negative bacteria. We then provide an overview of the AIP-based dynamic regulations of competence and virulence in Gram-positive bacteria, followed by that of AI-2-based and indole-based control of metabolism and physiological activities in microbial communities. Building on the review of these rather generic QS applications, we further highlight the important progress in the disruptions and manipulations of QS devices for therapeutic applications. The last part of the reviewed literature is on the mathematical modelling related to QS applications in the dynamic control of bacteria. Finally, we identify key challenges and suggest directions for future QS research.
Table 1.
Recent reviews focusing on QS
| Theme | Core content | References |
|---|---|---|
| QS signals transduction and architectures | Reviewed various signal–response systems and their applications in Gram-negative bacteria | [18] |
| Reviewed types of molecular mechanisms coupled with various QS devices in Gram-negative and Gram-positive bacteria, and some network architectures of QS circuits | [30] | |
| Reviewed various signal–response systems in Gram-positive bacteria | [31] | |
| Reviewed the mechanisms of intracellular pathway and extracellular pathway QS system, and the applications of regulating of conjugation, competence, bacteriocin production, and biofilm formation in Gram-positive bacteria | [32] | |
| Reviewed the function of indole in bacterial pathogenesis and eukaryotic immunity | [33] | |
| QS and its applications in marine microbes | [34] | |
| Reviewed the diversity, functions, biosynthetic pathways, and turnover systems for the diffusible signaling factors (DSF) family of QS signals | [35] | |
| Dynamic control for metabolic engineering | Reviewed various strategies such as QS system in the applications of dynamic metabolic engineering | [7] |
| Reviewed some dynamic control strategies coupling with QS to synchronize cellular activity | [36] | |
| Applications of synthetic microbial consortia | Reviewed some engineering cell–cell communication, mainly on QS, and synthetic microbial consortia for community composition, division of labor, and biofilm formation with QS system | [37] |
| Reviewed some typical synthetic microbial consortia by cell–cell communications, mainly on QS | [38] | |
| Discussed complex interactions and interplays in synthetic microbial ecology based on QS-based cell–cell communication | [39] | |
| Socio-microbiology based on cell-to-cell communications | Discussed the complex signal network and the cooperation with QS cheating phenotypes in bacterial. And reviewed various and feasible mechanisms that have been certified to stabilize QS-based cooperation in microbes | [40] |
| Reviewed the background and brief history of QS, and the applications of QS in socio-microbiology | [4] | |
| Applications of QS inhibitors in biofilm formation | Reviewed natural and synthetic quorum sensing inhibitors (QSIs) in various microbes | [25] |
| Reviewed applications of QS in biotechnology, especially for QSIs and some other biosensors | [41] | |
| Reviewed the mechanism for pathogenic biofilms formation, and discussed the current biofilm-targeting therapeutic strategies for disease which caused by microbial biofilms and drug tolerance | [42] | |
| Reviewed how bacteria deploy QS in realistic, complex and dynamically changing scenarios | [43] | |
| Mathematical modelling for QS | Reviewed the modeling approaches on a systemic level | [44] |
| Proposed the core principles of autoinducer systems in bacteria | [45] | |
| Reviewed various QS mathematical models | [46] |
QS applications in Gram-negative bacteria
QS for population control
QS-dependent activities are the result of density-dependent expression of both intra- and extracellular gene products [47]. It is essential for population-level dynamics and genetic-level regulation [19]. An et al. [48] and Goo et al. [49] certified that nutrients are typically limited and the environment is unfavorable for growth and metabolism of microbes in a crowded environment. Therefore, it is of much importance to control the cell density to optimize metabolic production.
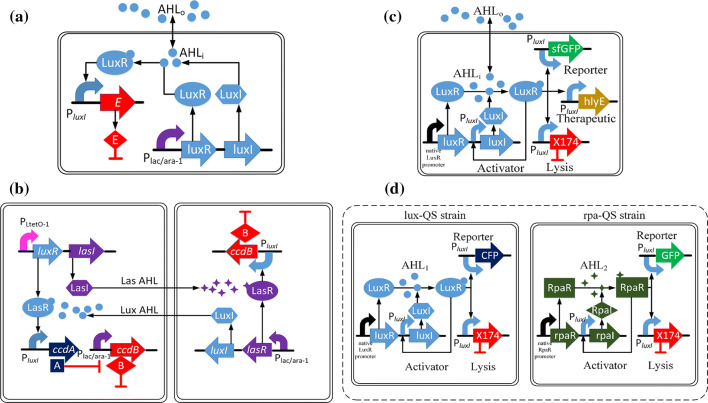
You et al. [50] proposed that cell–cell communication can be used to programme the dynamics of a population. Combining the cell survival and death genes to the LuxI/LuxR QS circuit, they built and characterized a ‘population control’ circuit that autonomously regulated the density of an Escherichia coli population (Fig. 2a). Balagadde et al. [51] applied two E. coli populations to construct a synthetic ecosystem (predator and prey) (Fig. 2b), which was based on two QS mechanisms (LuxI/LuxR and LasI/LasR). The predators will die following the expression of a suicide gene (ccdB) when the density of prey is low. As the prey density increases, AHL accumulates in the culture and eventually reaches a sufficiently high concentration, its combination with LuxR will then work to increase the expression of an antidote gene (ccdA) to rescue the predators. However, the predators will produce and accumulate Las AHL to a sufficient level to bind with LasR, which will, in turn, activate the expression of ccdB gene to kill the preys. The series of events leads to the oscillatory behavior of the two E. coli populations, which is typical for a two-strain ecosystem. More recently, AHL-based synthetic E. coli systems were used to test a general rule deduced for predicting coexistence and productivity of mutualistic communities [52]. Compared to the previous work by the same group [51], new features of these experimentally tested systems included the use of Isopropyl β-d-1-thiogalactopyranoside (IPTG) to induce the expression of CcdB (representing stress), and the application of anhydrotetracycline (aTc) to induce the QS module to impose the cooperation cost to both strains.
Fig. 2.
QS for the lysis and population control. a The signaling molecule 3OC6HSL (Lux AHL) is produced by the LuxI synthase. It will accumulate in the culture with the increased cell density of E. coli. When the concentration of the Lux AHL reaches a certain threshold, AHL will diffuse back into E. coli and be recognized by LuxR, a specific protein receptor to activate the transcriptional expression of the killer protein LacZa-ccdB to regulate the cell death of E. coli and consequently control the population density. b Based on the two QS mechanisms (LuxI/LuxR and LasI/LasR QS devices), two E. coli strains are engineered to construct a synthetic predator and prey ecosystem. c When the population reaches the critical threshold, the AHL will bind with LuxR to become AHL–LuxR complex. It will facilitate the expression of LuxI, gene ϕX174E for lysis, therapeutic gene for cytotoxic agents, and sfGFP for reporter. d Genetic circuits of a two-strain ecosystem including Lux-QS and Rpa-QS S. typhimurium strains
With the development of genetic circuits, the lysis genes can be coupled with new synthetic circuits such as genetic oscillators to realize various functions. Din et al. [53] integrated the lysis genes with a microbial drug delivery system [54] to form a synchronized lysis circuit (SLC) for controlling population levels and facilitating drug delivery using bacteria (Fig. 2c). The circuit includes a luxI promoter which promotes expression of LuxI, a therapeutic gene, a reporter gene and a lysis gene ϕX174 E. When AHL reaches a target threshold, the expression of the therapeutic gene and the reporter gene will be promoted, and bacteria will produce and release cytotoxic agents continually. At the same time, the number of bacterium will decrease due to the expression of the lysis gene. Then, a small number of remaining bacteria will begin to produce AHL again to restart this process in a cyclical fashion. Compared with existing drug delivery strategies, SLC can be used as a novel therapeutic technique to cure diseases through population control of bacteria.
In a separate study, two orthogonal QS devices and a population control mechanism were combined together to control the population densities of competitive microbes of Salmonella typhimurium strains [55] (Fig. 2d). lux and rpa QS devices can be integrated with two lysis genes to form SLCs in two bacterial strains to control bacteria population. When two competitive microbes, the Lux-QS strain and the Rpa-QS strain, are co-cultured, the latter has a significant growth advantage over the former. It was observed that from an initial population ratio of 100:1 between the Lux-QS strain and the Rpa-QS strain with the lysis gene, the population ratio became about 1:1 over 10 h. Without the lysis gene, the co-culture would be taken over by the Lux-QS strain. This demonstrates that the integration of two orthogonal QS devices to form an ‘ortholysis’ system is a potential strategy to stabilize competitive strains in co-cultures.
The above studies suggest that the combination of lysis genes, such as ccdB, and various QS devices (LuxI/LuxR, LasI/LasR, or RpaI/RpaR) can be applied to achieve the control of bacterial populations in either mon-culture or co-culture systems. Such control helps meet the essential requirement on population size needed for realizing various high-value metabolic production and microbial clinical treatments.
QS-based synthetic genetic oscillators
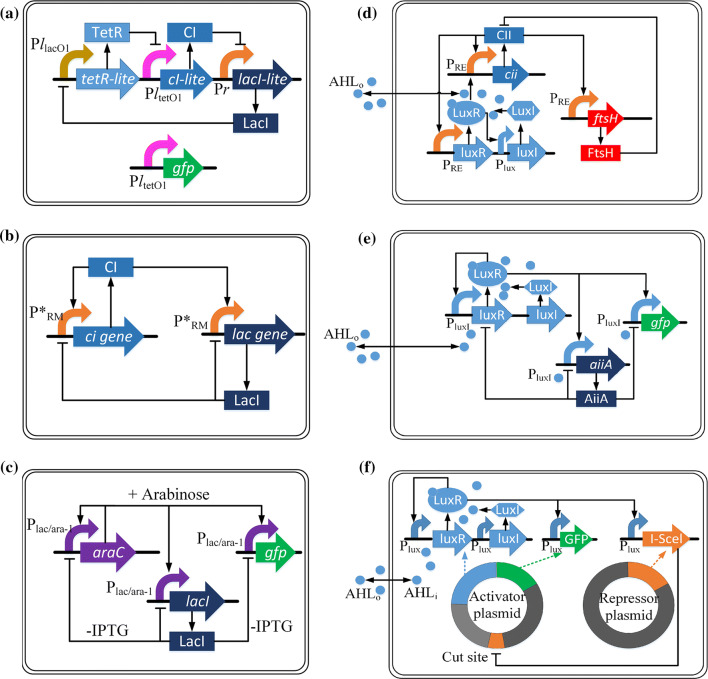
Using only a small amount of regulators in a large-scale genetic regulatory network, a relatively large number of genes and hence the complex cell behavior can be regulated [56, 57]. Many metabolic activities, such as population control, respiration, hormone secretion and circadian rhythms, are closely associated with synchronized oscillators [58–60]. Developing synthetic gene oscillators (SGOs) is one of the main research directions of the research on synthetic genetic regulatory networks [61]. The first SGO, the repressilator, was proposed in 2000 and is illustrated in Fig. 3a [14]. Unlike the repressilator where only repression exists, a relaxation oscillator has positive feedback loops which can promote the expression of negative feedback loops [62]. Hasty et al. [63] applied the common gene regulatory components (ci, lac and PRM) to construct a SGO model (Fig. 3b). Stricker et al. [64] developed a fast, robust and persistent engineered genetic oscillator in E. coli with the induction of IPTG and arabinose (Fig. 3c), and confirmed that its oscillatory period can be tuned by altering inducer levels, temperature and the media source.
Fig. 3.
The applications of QS-based synthetic genetic oscillators. a When the promoter PllacO1 is successfully promoted, the TetR protein will inhibit the downstream PltetO1 promoter, the CI protein will not be expressed, the inhibition of the Pr promoter will be released, LacI protein will be expressed, and the state of the PllacO1 promoter will be changed from “turn on” to “turn off”. Then the expression of TetR protein will be inhibited, and the inhibition of the PltetO1 promoter will be released, CI protein will be expressed, Pr promoter will be inhibited, LacI protein expression will be inhibited, and promoter PllacO1 will resume its “turn on” state. This is the mechanism of the first repressilator. b A relaxation oscillator with several positive and negative feedback loops. CI protein promotes the expression of itself and of LacI protein, while LacI protein inhibits the expression of itself and of CI protein. c A relaxation oscillator with the induction of IPTG and arabinose. Arabinose promotes the expression of AraC, GFP and LacI protein, while LacI protein inhibits the expression of itself, GFP and CI protein without IPTG. d Network architecture of the proposed gene network. The relaxation oscillator includes CII protein, FtsH protein LuxI/LuxR type QS system. e The network architecture of a synchronized oscillation design with LuxI/LuxR type QS system. f The circuit diagrams of two-plasmid circuit which includes the activator and repressor plasmid. The activator plasmid keeps activating luxI promoter to activate the expression of I-SceI in the repressor plasmid. I-SceI can be used to negatively regulate the expression of some cut site genes on the activator plasmid, which will reduces its copy number
Due to the complexity of cellular interaction and variability, it is important to investigate population-level dynamics of microbes, such as synchronization [65, 66] and programmed population interactions [39, 67, 68]. To avoid the random phase drift and remove the effects of noise, it is desirable to introduce measures to coordinate and synchronize each cellular oscillator [69], and QS has been found to offer an important means for this task [70].
Mcmillen et al. [71] firstly combined a genetic relaxation oscillator, which is composed of promotor PRE, gene X (cii) and gene Y (ftsh), with the lux QS mechanism (Fig. 3d). Protein CII can be degraded by protein FtsH, while the complex of AHL and LuxR (LuxR-AHL) can activate the transcription of CII. When the concentration of AHL reaches its threshold, it will bind with another LuxR in other cells to regulate their CII level.
Danino et al. [17] applied a QS-based approach to synchronized oscillations at the colony level (Fig. 3e). In this circuit, luxI, aiiA and yemGFP genes were controlled by the luxI promoter. The LuxR–AHL complex activated the luxI promoter [72]. AHL was degraded by homoserine lactonase (AiiA).
As the DNA copy number changes with environmental pressures, it is a challenge to combine gene circuits without predictable dynamic control of gene expression [73]. Treating the DNA copy number as a circuit control element, Baumgart et al. [74] reported that the expression of some cut site genes on a plasmid can be repressed by a targeted nuclease (I-SceI) to reduce the copy number. They combined the negative feedback component with the positive feedback component of lux QS system to form a synthetic gene oscillator of the plasmid copy number (Fig. 3f).
Most SGOs, such as those introduced above, have been constructed to operate within single, isogenic cellular populations. Representing a step further, Prindle et al. [75] integrated a genetic relaxation oscillator, lux QS system and redox signaling (arsenite) to form coupled genetic ‘biopixels’ among different colonies (Fig. 4a). This genetic circuit consists of arsenite-responsive promoter, ArsR, lux QS system and arsenite. When there is no arsenite, ArsR will repress the expression of luxR, thus no fluorescence or oscillation is generated. The repression will be removed when there is a sufficient amount of arsenite, thereby the LuxR–AHL complex will promote oscillations and the expression of the fluorescence gene.
Fig. 4.
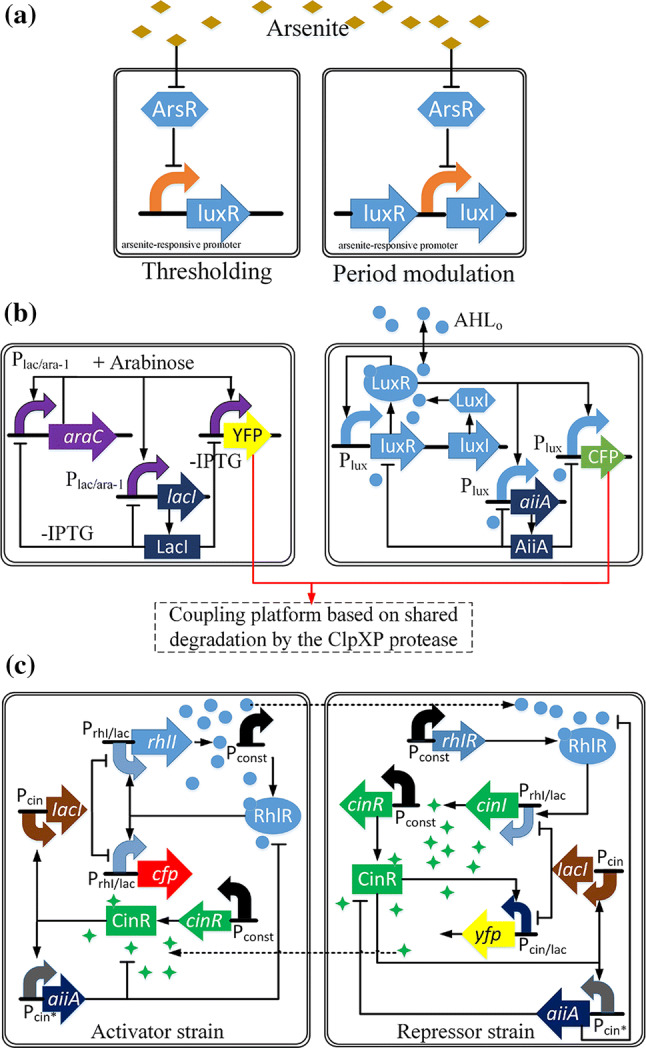
QS for synchronization of genetic oscillators. a A genetic relaxation oscillator consists of arsenite-responsive promoter, ArsR, lux QS system, and arsenite. b Coupling (i) genetic oscillators which includes promoter Plac/ara-1, lacI gene, arabinose and IPTG inducers and (ii) LuxI/LuxR type QS system to realize the synchronization by the ClpXP protease platform. c Genetic circuit diagrams of the activator with Rhl QS system and repressor strains with Cin QS system
Compared to the synchronization of genetic oscillators by means of coupling with standard transcription factor-based methods such as QS devices, work on the delay times of synchronization by competitive protein degradation is much less [76]. Prindle et al. developed a post-translational coupling platform which worked through shared degradation by the ClpXP protease [77] to couple various synthetic genetic modules rapidly and efficiently. This platform was used to integrate intracellular genetic oscillators (including Plac/ara-1, lacI and inducers) and the LuxI/LuxR-type QS system to realize synchronization (Fig. 4b).
As an example of synchronization with two QS devices, Chen et al. [78] constructed a SGO with an “activator” strain and a “repressor” strain to realize the emergent, population-level oscillations of two genetically distinct E. coli (Fig. 4c). The activator produces Rhl AHL, which promotes the transcription of target genes for both strains, while the repressor produces Cin AHL, which inhibits the transcription for both strains mediated by the LacI protein. Besides, there is another negative feedback loop in which the AiiA protein can degrade these two AHLs. These feedback loops were divided into four types of topologies to investigate the population-level dynamics of these two strains.
These studies have demonstrated that incorporating QS devices can indeed enrich the design and implementation of SGOs. Such systems hold much potential in regulating the synchronization of synthetic microbial consortia to benefit real applications in metabolic engineering, particularly the development and optimization of production pathways for high-value metabolites, as well as some medical applications, such as the drug release process for some probiotic therapies. On the other hand, these potential applications may require further optimization of the QS-based SGOs to achieve more accurate controls, beyond the feasibility already demonstrated by the proof-of-concept studies.
QS-based genetic toggle switches
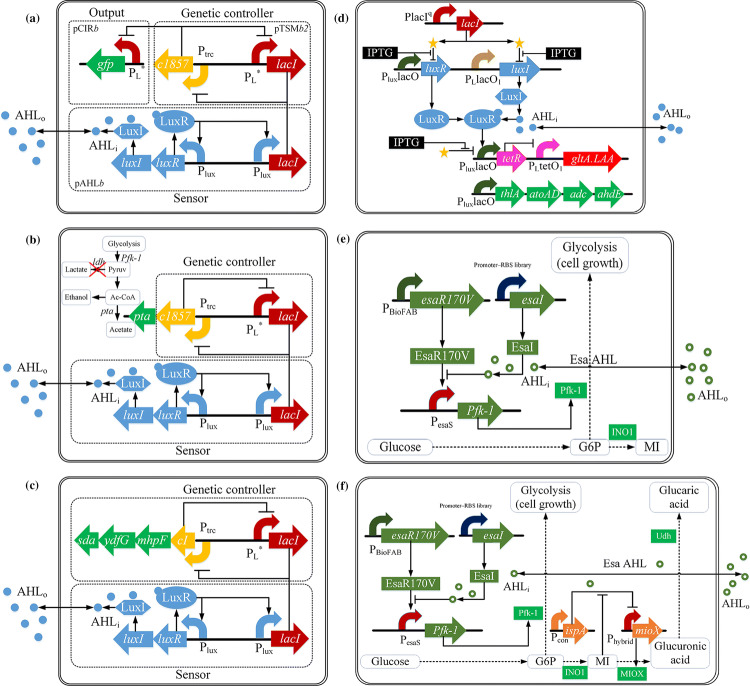
The main objective of metabolic engineering is to increase the yield and productivity of the desirable production through genetic engineering [79]. As a synthetic genetic circuit, a metabolic toggle switch (MTS) is often used to meet this goal [13, 80]. Demonstrating a modular design strategy, Kobayashi et al. [81] created four E. coli strains that contained a genetic toggle switch (Fig. 5a). Half of the strains were interfaced with a transgenic QS signaling pathway from Vibrio fischeri that detects AHLs. The genetic circuit combined the QS mechanism and the artificial ON/OFF genetic toggle switch. The QS circuit was composed of the luxI, luxR and lacI genes, and would work as follows: Initially, the concentration of the AHL is too low to function, the expression of the target gene, such as gfp, is maintained at the ‘OFF’ state; when AHL reaches a critical concentration, the gene will switch to the ‘ON’ state.
Fig. 5.
QS for the dynamic metabolic control in Gram-negative bacteria. a Genetic circuit diagram of a genetic toggle switch and QS signaling pathway. b Genetic circuit diagram of the integration of LuxI/LuxR type QS system, central carbon metabolism and a genetic toggle switch in E. coli. The genetic toggle switch consists of LacI and λCI proteins. Their expressions are inhibited mutually. The pta gene, relevant to ethanol production, is at the downstream of λcI gene. At low AHL concentration, λcI and pta genes express normally, while lacI gene is inhibited. When the concentration of the Lux AHLs reach a certain threshold, the inhibition of lacI gene will be released. Later on, the expression of λcI and pta genes will be repressed. c Genetic circuit diagram consists of the genetic controller for serine production and the QS sensor. d Design of synthetic genetic circuit coupling QS system and the genetic toggle switch. With the help of IPTG inducer, this genetic circuit can realize the switch flexibly between two pathways: isopropanol synthesis and the TCA cycle. e Schematic of dynamic control of cell growth and myo-inositol production. f Schematic of two-layer dynamic control of cell growth and d-glucaric acid production
Anesiadis et al. [82] proposed an integrated computational model and showed that a genetic toggle switch can be effectively employed in dynamic metabolic engineering to increase bioprocess productivity and yield (Fig. 5b). Anesiadis et al. [83] also applied the LuxI/LuxR-type QS system to reconstruct E. coli to improve the productivity of serine with an ON/OFF genetic toggle switch (Fig. 5c). The highest productivity of the final strain to produce serine was 29.6% higher than that of the previous mutant strain. To further the design, they integrated population response, dynamic metabolic regulation, and the DFBA metabolic modeling method to construct a mathematical model to analyze the global sensitivity.
Although the aforementioned on–off two-stage control strategies have taken into consideration the appropriate cell density or sufficiently high concentrations of AHLs into consideration to achieve the intended purposes, the desirable gene expression often requires a more accurate cell density to synchronize microbial growth and cellular activity. To further improve the desirable production, novel engineering strategies to manage the trade-off between cell growth and desirable production are needed. Soma et al. [10] constructed a synthetic lux system to achieve a dynamic switch of the metabolic flux between the TCA cycle and the isopropanol synthesis pathway (Fig. 5d), resulting in the yield and the conversion rate improved by 3 and 2.3 times, respectively. The threshold cell density was controlled by IPTG in this system.
Gupta et al. [8] introduced the QS circuits to control the expression of pfk-1 gene (which determines the carbon flux to glycolysis and cell growth) to identify the optimal point to switch off gene expression in terms of the desired times and cell densities (Fig. 5e). When there is no AHL, the transcriptional regulator EsaRI70V will bind to the PesaS promoter. As cell density increases, the accumulation of AHL will reduce the activity of EsaRI70V and turn off the expression of pfk-1 gene. As a result, most of the glucose will be switched to the target pathway to increase the titers of myo-inositol (MI).
Doong et al. [11] combined pathway-independent and pathway-specific strategies to form a control mechanism that involved two orthogonal and tunable dynamic regulation strategies, for the purpose of improving the production of d-glucaric acid (Fig. 5f). The pathway-independent strategy was based on the QS system to turn glucose utilization from the glycolysis to the production of d-glucaric acid at the threshold of AHL concentration. The pathway-specific strategy used myo-inositol as the intermediate metabolite sensor to achieve dynamic and autonomous control to improve the production of d-glucaric acid.
More recently, Honjo et al. [84] constructed an engineered microbial community composed of an E. coli strain producing beta-glucosidase (BGL) and the other E. coli strain producing isopropanol (IPA) using QS-dependent cell lysis circuits. Specifically, the BGL-producing strain will produce BGL to convert cellobiose to glucose as the carbon source when the desired cell density is reached by lysing itself. The IPA-producing strain grows with the help of the BGL release and can detect AHL produced by the BGL-producing strain to induce the expression of IPA. With a three-species consortium (Gluconobacter oxydans–Ketogulonicigenium vulgare–Bacillus megaterium) for vitamin C fermentation, Wang et al. [85] applied the LuxI/LuxR QS system to control the lysis of G. oxydans as the population level QS-based metabolic toggle switches for l-sorbose production and for the relieve of l-sorbose competition with K. vulgare, thus realizing one-step fermentation for vitamin C.
The above studies commonly feature the combination of synthetic QS systems and metabolic toggle switches, allowing the synthetic genetic circuit to be activated at the targeted cell density, and hence resulting in self-induced metabolic states switching. With the gradual maturation of the application of metabolic division of labor in metabolic engineering [86], the combined applications of engineered microbial communities and QS-induced metabolic toggle switches appear to offer a potentially promising direction for metabolic engineering and synthetic biology.
QS-based logic gates
Positive and negative feedback control loops are prevalent in physical systems. Analogously, various positive and negative feedback regulatory structures have been found in biological systems [87]. The amazing similarities and sophisticated connections between the two types of systems have attracted many researchers to develop biomolecular computing systems [88] which mainly include the design and simulation of various genetic circuits such as logic gates [89, 90]. Logic gates, such as Boolean logic gates, are the basic content and computing units of digital electronic circuits. Boolean logic gates for genetic circuits that have been involved in various applications mainly include AND, OR, NOR, NAND and XOR [91]. As early as 2002, an AND logic gate based on exogenously added signals has been established, activating the expression of GFP as the output based on the addition of two inputs, IPTG and aTc [92]. Step by step, various logic gates were constructed in microbes, such as AND logic gates in Pseudomonas aeruginosa [93] and Shewanella oneidensis [94]. Due to the ability of coordinating cell behavior at the population level, the QS devices such as lux, las and rhl have been combined with other genetic circuits to form various QS-based logic gates.
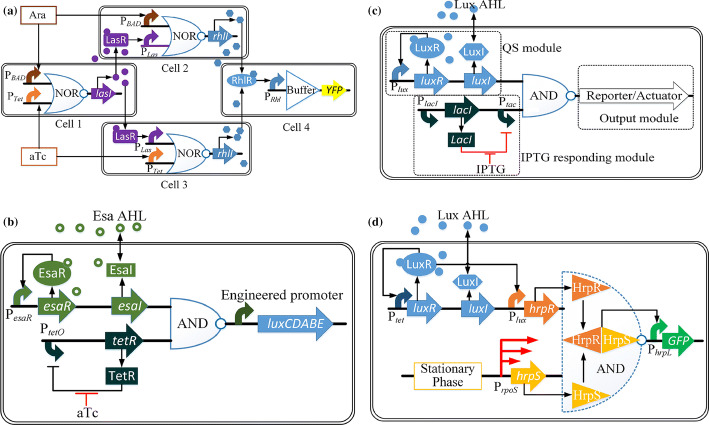
QS devices can be used as “wires” to combine genetic circuits to produce more complex computations in space, as shown by Tamsir et al. [95]. Firstly, based on previous work [96], they constructed the simplest NOR gates from NOT gates with the addition of a repressor. The inputs and the outputs of the NOR gates were designed to act as promoters to form multiple gates. Secondly, three NOR gates and a buffer gate in four separate E. coli cells were wired together to form an XOR gate via Las AHL and Rhl AHL from P. aeruginosa PAO1 (Fig. 6a).
Fig. 6.
The applications of QS-based logic gates. a Genetic circuit of an XOR gate with three NOR gates and a buffer gate in four separate E. coli colonies. Arabinose (Ara) and anhydrotetracycline (aTc) are inputs and expression of LasI is the output for the first NOR gate in cell 1. Based on the first LasI input, Ara and aTc are regarded as the second inputs for the cell 2 and cell 3. The output of cell 2 and cell 3 is the expression of RhlI. The buffer gate responses to the RhlI input to express the reporter gene (YFP). b Schematic diagram of QS-dependent AND logic gate genetic circuits in P. stewartii. c Genetic circuits schematic diagram of the AND logic gate in S. oneidensis. The promoter Ptac is inhibited by LacI protein, and it can be relieved by IPTG addition in IPTG responding module. The AND logic gate functions by the combination of the IPTG responding module and QS regulation of LuxR–AHL complex (QS module). d Schematic diagram of the AND logic gate with the control of HrpR and HrpS in E. coli
Different from the logic gates which solely rely on two exogenously added signals such as IPTG and aTc [97], Shong et al. [98] developed a synthetic AND gate in response to the endogenous AHL signal and exogenously added IPTG or aTc (Fig. 6b). The esa QS system from Pantoea stewartii was applied to obtain the endogenous Esa AHL signal to avoid the disadvantages of the lux QS system. They showed that the downstream gene would not express without a second exogenous signal, hence demonstrating the function of this QS-dependent AND gate.
An AND logic gate combined with the lux QS system was constructed in Shewanella oneidensis to realize the application of a logic gate in microbial fuel cells (MFCs) [99]. They firstly integrated the IPTG responding module, a QS module and an output module (reporter or target gene mtrA) to form a synthetic AND gate (Fig. 6c) to control extracellular electron transfer in S. oneidensis. When the Lux AHL concentration reaches a critical threshold and binds with LuxR protein to facilitate the activation of promoter Plux, accompanied by the IPTG addition to release the inhibition of Ptac by LacI protein, this AND logic gate will be switched on to start extracellular electron transfer.
As stated earlier in “QS-based logic gates”, QS devices have been widely integrated with the synthetic genetic toggle switches in metabolic engineering to dynamically regulate and control the gene expression responsible for the desired production. As the cell’s physiological state affects metabolic regulation, the stationary phase sensing system and a QS system were combined by He et al. [100] to obtain an auto-induced AND gate for monitoring cell growth and polyhydroxybutyrate (PHB) production (Fig. 6d). PrpoS regulates gene expression in the stationary phase [101] and was chosen here to control the transcription of HrpS, forming one of the inputs of the AND gate. The other input was from a QS system that controlled the expression of HrpR. Promoter PhrpL, controlled by the complex of HrpR and HrpS, was the output.
As a broadly shared paradigm, the LuxI/LuxR-type QS system has been integrated with synthetic genetic oscillators, genetic toggle switches, and logic gates, revealing a wide range of possibilities and potential applications. It should be emphasized that, although some success has been achieved in applying QS-based engineering to specific microbes within the scope of monoclonal synthetic biology, complications arising from factors such as metabolic load and toxicity of metabolites could seriously limit what a single microbe can achieve. Therefore, we anticipate that, following some of the existing studies reviewed in this section, more explorations will be published on the engineering of microbial consortia with QS-based synthetic genetic circuits, as a practice of synthetic ecology, which realize the population-level synchronization in synthetic communities to overcome the limit of single species.
QS applications in Gram-positive bacteria
Gram-positive bacteria can also apply their own QS mechanism to regulate gene expression at the population level dynamically. Different from the AHLs adopted in the QS systems in Gram-negative bacteria, QS of Gram-positive bacteria is dependent on AIPs, also known as pheromones [3, 102]. The Gram-positive bacteria can thus sense AIPs to regulate their own metabolism in a changing environment [103]. Under certain conditions, AIPs of Gram-positive bacteria are produced in the cytoplasm and then secreted by the oligopeptide transport system to the extracellular medium. Thereafter, they are either detected at the bacterial surface by the extracellular pathway or re-internalized by the intracellular pathway [32].
As shown in “QS applications in Gram-negative bacteria”, the QS mechanisms in Gram-negative bacteria (including lux, las QS system and so on) are relatively well understood and have been shown to be applicable to the design and construction of genetic toggle switches, oscillators and logic gates. In contrast, QS in Gram-positive bacteria still has a number of unknown mechanisms [104], and there is much less research on the application of QS in Gram-positive bacteria than that of Gram-negative bacteria [41]. Recently, Marchand et al. [105] reported the first construction of a synthetic QS system to realize cell–cell communication among Gram-positive bacteria. They incorporated the agr QS system of Staphylococcus aureus into Bacillus megaterium to synthesize a genetic circuit for monitoring cell growth. In this circuit, the original P2 promoter for the expression of agrB, agrD, agrC, and agrA genes was replaced with the PxylA promoter. The AgrD protein was transported by ArgB and SipM protein to the culture medium to become mature AIPs, which would then be recognized by a two-component system (TCS). The TCS consisted of the AgrC receptor and the AgrA transcriptional activator which were used to regulate the P3 promoter and the expression of the target gene such as gfp reporter gene.
However, different from the aforementioned work [105], most of the other reported AIPs systems of Gram-positive bacteria are based on the up-regulation or down-regulation of their own native QS devices as opposed to “borrowing” one from a different species, as summarized in Table 2. These QS systems are disrupted and manipulated in their own specific bacteria. Compared with the AHLs, which can diffuse through the cell membrane freely, the secretion of AIPs requires the aid of the oligopeptides transport system [103]. Besides, it should be taken into consideration that the rates of diffusion of the larger oligopeptides are slower than the smaller AHLs, especially in a solid culture [31]. These differences between AHLs and AIPs are arguably among the reasons for fewer studies on the QS application with Gram-positive bacteria.
Table 2.
Recent QS applications in Gram-positive bacteria with native devices
| Pathways | AIPs | Bacteria species | Function controlled | References |
|---|---|---|---|---|
| Extracellular pathway | Agr type peptides | Clostridium botulinum | Neurotoxin production and sporulation | [106] |
| Clostridium acetobutylicum | Granulose formation, sporulation | [107] | ||
| Listeria monocytogenes | Population dynamics in soil | [108] | ||
| Clostridium perfringens | Virulence and toxin production | [109] | ||
| Staphylococcus epidermidis | Biofilms and infection | [110] | ||
| Clostridium perfringens | Toxin production and virulence | [111] | ||
| peptides that contain Gly–Gly motifs | Streptococcus thermophilus | Production of Blp Bacteriocins | [112] | |
| Streptococcus pneumoniae | Competence development | [113] | ||
| Streptococcus pneumoniae | Competence control | [114] | ||
| Streptococcus mutans | Competence control | [115] | ||
| Streptococcus pneumoniae | Competence control | [116] | ||
| Intracellular pathway | Rap/NprR/PlcR/PrgX (RNPP family) | Bacillus | Competence control (Rap) | [117] |
| Bacillus cereus group | protease production in sporulation (NprR) | [118] | ||
| Bacillus cereus | Necrotrophism (NprR) | [119] | ||
| Bacillus cereus | Virulence regulation (PlcR) | [120] | ||
| Bacillus cereus and Bacillus thuringiensis | Virulence and necrotrophic properties (NprR) | [121] | ||
| Enterococcus faecalis | Regulation of conjugation (PrgX) | [122] | ||
| Rgg-like family | Streptococci mutans | Competence control | [123] | |
| Streptococci genus | cross-talk between these different SHP/Rgg systems | [124] | ||
| Streptococcus genus | Competence control | [125] | ||
| Streptococcus thermophilus | Competence control | [126] | ||
| Streptococci genus | Competence control | [127] | ||
| Streptococcus mutans | Genetic competence | [128] |
QS for population-level control based on interspecies communication
QS signals can either be used by bacteria to form cooperation or exploited by individuals which do not secrete them; the latter is termed as cheating phenotypes [129]. QS cheating in microbes is one of the most important parts of QS research. As listed in Table 1, achievements on cheating phenotypes and AHL-based social interactions have been extensively reviewed by Whiteley et al. [4] and Asfahl et al. [40]. Different from the intra-species signal responses which are mainly dependent on AHLs and AIPs (as reviewed in “QS applications in Gram-negative bacteria” and “QS applications in Gram-positive bacteria”), the signals for interspecies communication are mainly autoinducer 2 (AI-2) [130–132] and indole [133, 134] that regulate cooperation and competition in microbial communities, which is the focus of this section.
AI-2-based communication
AI-2 is a product of the LuxS enzyme, which widely exists in Gram-positive and Gram-negative bacteria, and even in fungi [3]. LuxS enzymes synthesize 4,5-dihydroxy-2,3-pentanedione (DPD), which can be regarded as the precursor of AI-2 [135]. DPD is a by-product of the S-adenosylmethionine (SAM) metabolism, which is included in the activated methyl cycle (AMC). SAM can transfer methyl groups to methyl-transferases and substrates to produce S-adenosylhomocysteine (SAH). Catalyzed by a series of relevant protease, SAH can be converted to homocysteine and DPD [136]. DPD is a highly active molecule that can spontaneously cyclize into different DPD derivatives, which can be identified as a signal molecule, AI-2, by different bacteria [137]. Chen et al. [131] certified that AI-2 produced by Vibrio harveyi contains boron. In contrast, the AI-2 signals of S. typhimurium and E. coli [138] are non-borated cyclized. There are existing reviews [136, 137] which explain the mechanism of the AI-2 signaling systems at length.
Xavier et al. [139] testified that AI-2 can mediate two-way communication between E. coli and Vibrio harveyi in co-culture. The AI-2 produced by E. coli can be sensed by V. harveyi to induce bioluminescence, and reciprocally, the AI-2 produced by V. harveyi can be detected by E. coli to regulate its Lsr system. Armbruster et al. [140] found that Haemophilus influenzae and Moraxella catarrhalis, which are responsible for one of the common childhood infections named otitis media, have reciprocal effects on biofilm formation via the AI-2 QS signal. They pointed out that the former promotes the biofilm formation of the latter. This and the other studies show the potential of the exploration on AI-2-based control to further the understanding of interactions between microbes and hosts and to develop strategies to influence the physiological and biochemical functions of various pathogenic bacteria for curing the relevant diseases.
Indole-based communication
With l-tryptophan as the reactant, indole is synthetized by tryptophanase (TnaA) in many bacteria [141]. More recently, indole is regarded as a signaling molecule which is relevant to various bacterial physiology, such as biofilm formation [142], plasmid stability [143], population-based resistance [144], virulence [145], persister formation [146], spore formation [147], and cell division [148] in indole-producing bacteria. Yee et al. [149] had studied biofilm formation with indole-producing bacteria (E. coli) and non-indole-producing bacteria (P. fluorescens), where indole was converted to isoindigo by toluene o-monooxygenase (TOM), with the TOM gene introduced from the soil bacterium Burkholderia cepacia G4. Their results indicated that E. coli was present in a higher density when co-cultured with P. fluorescens which expressed TOM. Han et al. [150] proposed that indole oxidation by TOM will increase the electricity generation in an E. coli-catalyzed microbial fuel cell. Lee et al. [142] reported that indole increases the biofilm formation of P. aeruginosa that does not synthesize indole. Also, indole derivatives (Indole-3-acetaldehyde) from pathogen Rhodococcus sp. BFI 332 have been verified for inhibition to biofilm formation of E. coli O157:H7 [151]. Indole and its derivative 7-benzyloxyindole (7BOI) were investigated for their inhibition to the virulence of S. aureus [152]. Lee et al. [153] found that indole influences the growth, biofilm formation, antibiotic tolerance, and motility of Agrobacterium tumefaciens. Chu et al. [154] investigated the interaction and competitiveness between E. coli and P. aeruginosa in a mixed culture. They concluded that the major indole-based protection for the growth of E. coli in the mixed culture was due to direct inhibition of QS-based virulence factors, such as pyocyanin and elastase, in P. aeruginosa. Focusing on the host-microbe interactions of various bacteria and Caenorhabditis elegans, Lee et al. [155] concluded that indole and its derivatives will influence the egg-laying behavior, chemotaxis, and the survival of C. elegans. Further details and examples of indole-based QS can be found in several reviews, such as the one by Lee et al. [133] which covered at length indole-producing bacteria and the mechanisms and applications of the signaling systems based on indole and its derivatives. Recently, the perspectives on indole signaling systems have been expanded from interspecies to inter-kingdom. For example, Lee et al. [33] and Tomberlin et al. [156] provided overviews on how indole and its derivatives affect various physiological activities in fungi, insects, plants, and animals.
The functioning of AI-2 and Indole, as two main signaling molecules facilitating the inter-species communication, greatly expand the presence of QS in the microbial world. As the understanding of their mechanisms deepens, one would expect that engineering and medical innovations leveraging such knowledge will start to emerge, despite their current relative insignificance.
QS applications in potential clinical therapies
Biofilm formation and inhibition
Many bacteria have a tendency to be organized in aggregates, commonly adhering to surfaces to form biofilms [157]. Compared to the free-living counterparts, there are some unique properties in bacteria forming biofilms, such as antimicrobial tolerance [158]. In fact, biofilms formation has been widely regarded as one of the most important virulence factors for microbial toxicity and infections [159]. When there is a biofilm for bacteria within the human host, the infections will be hard to treat.
Many studies have reported that the QS of AI-2/luxS and DSF-based QS systems play an important role in biofilm development and disassembly [4, 42, 160, 161]. From the studies using flow-cell systems [160], QS and biofilms have been shown to be inextricably linked. Lebeer et al. [162] conducted the first investigation on the relationship between the LuxS and biofilm formation in Lactobacillus rhamnosus GG, which is one of the probiotics for human. They found that LuxS enzyme is crucial for the gastric stress resistance and the metabolism of L. rhamnosus GG. Sun et al. [163] found that the over-expression of luxS or AI-2 supplementation enhanced biofilm formation of Bifidobacterium longum NCC2705 by about 50%. Furthermore, exogenously addition of signaling molecule AI-2 was found to promote the biofilm formation of P. aeruginosa PAO1 [164], Helicobacter pylori [165], and Staphylococcus epidermidis [166]. Laganenka et al. [167] showed that self-produced AI-2 can mediate autoaggregation of E. coli to enhance bacterial stress resistance and promote biofilm formation. Papenfort et al. [168] discovered 3, 5-dimethylpyrazin-2-ol (DPO) in Vibrio cholerae which can be regarded as a new QS signal that regulates biofilm formation and virulence. Liu et al. [169] identified that d-Ribose can be applied to decrease the activity of AI-2 to inhibit biofilm formation of Lactobacillus paraplantarum L-ZS9. Besides, based on the DSF QS systems, Ryan et al. [170] proposed that DSF underpins the interspecies signaling between Stenotrophomonas maltophilia and P. aeruginosa, with the latter influencing the former’s biofilm formation. Dean et al. [171] demonstrated that the Burkholderia diffusible signal factor (BDSF) plays an important role in the inhibition and dispersion of biofilm formed by Francisella novicida. It has also been shown that DSF signaling regulates many functions that contribute to biofilm formation in Stenotrophomonas maltophilia [172] and Helicobacter pylori [173]. What’s more, DSF-based QS systems can be used to regulate antibiotic tolerance (e.g., [174]) and the production of virulence factors (e.g., [175]); this area has been reviewed at length [176].
Given the close connections between QS and biofilm formation, the development of novel antimicrobial therapies by QS inhibitors has attracted extensive attention from researchers [177]. QS inhibitors are functioned by the degradation of QS signals (quorum quenching) [178] or some other QS-blocking approaches. Shen et al. [179] synthesized a series of structural analogues of the substrate S-ribosylhomocysteine (SRH) and a 2-ketone intermediate to inhibit LuxS enzyme. Zhang et al. [180] proved that two small peptides, 5411 and 5906, could inhibit the AI-2 activity and influence biofilm formation and virulence of Edwardsiella tarda. Ni et al. [181] reviewed seven approaches to inhibiting QS pathways. Brackman et al. [182] certified that cinnamaldehyde and cinnamaldehyde derivatives can interact with the AI-2 QS signal by reducing the DNA-binding ability of LuxR. Brackman et al. [183] investigated the relationship between the susceptibility of biofilms to antibiotics and the antibiofilm effect of quorum sensing inhibitors (QSI) in vitro and in vivo model systems. Christensen et al. [184] discovered, by an high-throughput cell-free screen, three AHLs inhibitors which can be used as potential therapeutic agents for virulence and microbial infections. O’Loughlin et al. [185] reported that the meta-bromo-thiolactone (mBTL) can not only inhibit the production of pyocyanin and biofilm formation but also reduce the activity of two QS receptors, LasR and RhlR, in P. aeruginosa. Starkey et al. [186] identified several compounds as QSI (all with the structure of benzamide-benzimidazole) which can inhibit the expression of the mvfR QS system which is one of the key reasons for multidrug-resistant and antibiotic-tolerant infections. Ouyang et al. [187] also reported that quercetin can be applied to inhibit biofilm formation and virulence factors in P. aeruginosa.
Consortia-based therapies for P. aeruginosa infection
Pseudomonas aeruginosa, a multidrug resistant pathogen, can cause disease in plants and animals, including humans [188]. Biofilms of P. aeruginosa can cause chronic opportunistic infections, especially for immunocompromised patients and the elderly. These biofilms also appear to protect the bacteria from traditional antibiotic therapies [189]. Therefore, research on the discovery of new treatments, such as consortia-based therapies against P. aeruginosa is much needed. In particular, engineering microbial consortia with genetic circuits based on QS devices to inhibit biofilm formation offers a potentially attractive therapeutic technique to deal with the infectious pathogens.
Taking the potential applications in bioremediation (among other applications) into consideration, Hong et al. [23] combined the LasI/LasR-type QS system from P. aeruginosa with biofilm dispersal genes to control biofilm displacement in a microfluidic device. The engineered microbial consortia consisted of disperser cells and initial colonizer cells. In a disperser cell, the LasI protein which is the precursor of the signaling molecule Las AHL is produced continuously, and the biofilm-dispersing Hha13D6 protein is encoded when the relevant gene is induced by IPTG. In an initial colonizer cell, the LasR protein is encoded, which couples with Las AHL to form a complex to promote the expression of another biofilm-dispersing protein, BdcAE50Q (Fig. 7a).
Fig. 7.
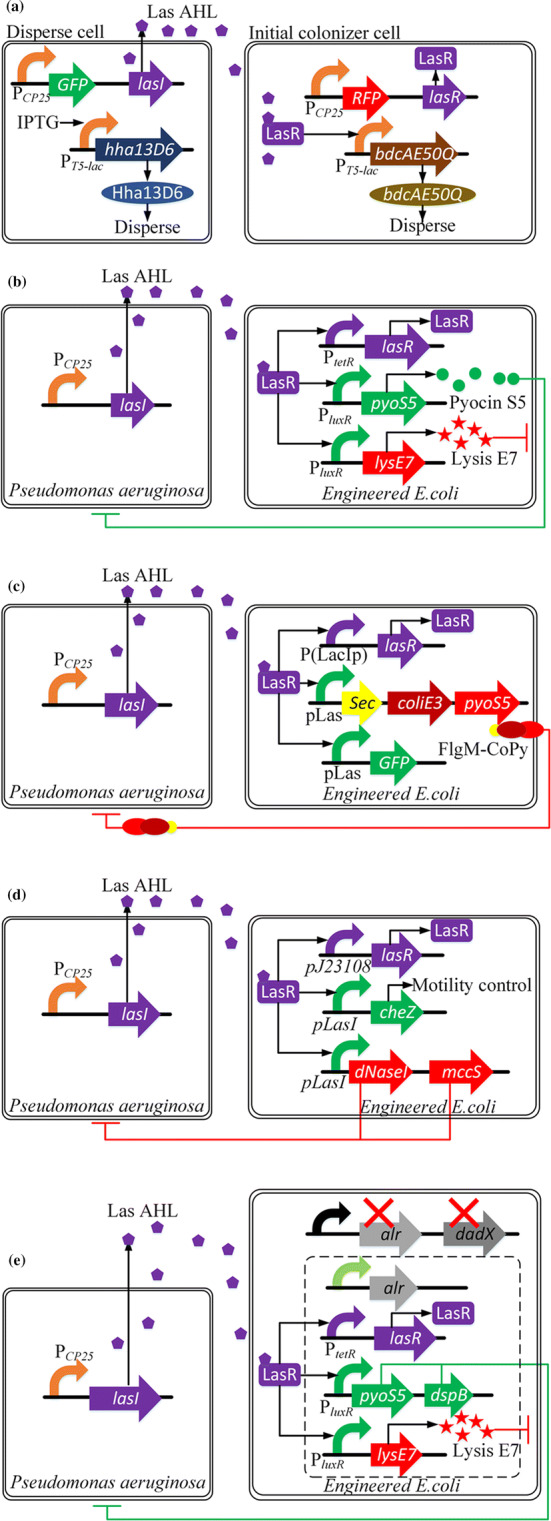
The applications of QS devices relevant to biofilm formation. a Diagram of genetic circuits of disperser cell and initial colonizer cell. b Schematic of genetic circuit coupling QS, killing, and lysing systems to kill P. aeruginosa. c Genetic circuit architectural of “sense-kill” system of P. aeruginosa with transport system of the bacteriocin CoPy. d Schematic of engineering E. coli to sense, migrate and kill P. aeruginosa. e Diagram of novel “sense-kill” genetic circuit coupling QS, dspB and pyoS5 gene (for killing), and lysing systems to kill P. aeruginosa
Saeidi et al. [190] integrated the LasI/LasR-type QS system, a lysing device for engineered E. coli, and pyocin for killing P. aeruginosa. The Las AHL is produced by P. aeruginosa and detected by the engineered E. coli. When the Las AHL concentration reaches a threshold, the Las LasR–AHL complex promotes the transcription of PluxR and then facilitates the expression of pyocin S5 and lysis E7 genes. E7 lysis protein will accumulate and then lyse E. coli cell to release S5 pyocin to inhibit biofilm formation and kill P. aeruginosa (Fig. 7b).
In a further study, Gupta et al. [191] integrated a secretion module to the genetic circuit “sense-kill” system for P. aeruginosa. They applied a novel pathogen-specific bacteriocin CoPy, the combination of Colicin E3 and Pyocin S3 to kill P. aeruginosa. What’s more, they utilized a secretion tag, FlgM, to transport the bacteriocin CoPy into the culture to kill P. aeruginosa (Fig. 7c).
Hwang et al. [192] combined the LasI/LasR-type QS system, motility control and a killing device to form a novel genetic circuit engineered into E. coli to kill P. aeruginosa. The Las LasR–AHL complex promotes the expression of gene lasI, gene cheZ (controlling the motility of E. coli toward P. aeruginosa), gene Dnasel (controlling biofilm degradation of P. aeruginosa), and gene mcsS (killing P. aeruginosa) (Fig. 7d).
Based on a previous work [190], Hwang et al. [193] introduced an auxotrophic marker in E. coli to avoid the horizontal gene transfer of the antibiotic resistance to other bacteria. The alr and dadX genes which help the interconversion of d-alanine and l-alanine were knocked out in the novel engineered E. coli. To construct a modified “sense-kill” system for P. aeruginosa, they first complemented the auxotrophic E. coli with an alr + plasmid (pEaak) to ensure the growth and other physiological activities and then added the dspB gene to the previous genetic circuits (Fig. 6b) to inhibit the biofilm formation more efficiently. The dspB gene encodes dispersin B (DspB), an anti-biofilm enzyme for degrading mature biofilms. It was proven that combining the dspB and pyoS5 genes together to disassemble biofilm formation is an efficient strategy to kill P. aeruginosa (Fig. 7e).
Probiotic therapies for diseases relevant to gut microbiota
Gut microbiota has been shown to clearly relate to a range of diseases and conditions of human, such as type 2 diabetes [194], cardiovascular disease [195], clostridium difficile infection (CDI) [196], epithelial tumors [197] and obesity [198]. With the large quantities of antibiotics widely used in the past decades, antibiotic resistance is currently ubiquitous and hard to deal with, especially in the human gut [20, 199, 200]. Much research has thus been focusing on finding alternative antimicrobial therapies. Taking S. typhimurium, enterohaemorrhagic E. coli (EHEC) and Clostridium difficile as representative microbes, Bäumler et al. [60] reviewed the interactions among the microbiota, the host and these three pathogenic bacteria when treated by antibiotics. Antibiotics treatment appears to increase free sialic acid (from the host) and succinate (from the microbiota) level. The elevated sialic acid and succinate in turn promote the expansion of the S. typhimurium and C. difficile, which will do harm to the intestinal epithelium cells (IECs). Besides, EHEC was found to use a fucose-sensing signaling-transduction QS system [201] to avoid the nutrient competition with commensal E. coli. To avoid the defect of antibiotics treatment leading to antibiotic resistance, multiple attempts have been made to develop probiotic therapies which utilize probiotic bacteria such as lactic acid bacteria [54] to serve as vectors for delivery of drug and signaling molecules [202]. As one of the most important ways for cell–cell communication, QS devices hold enormous potential in sensing the pathogens (stated in “Consortia-based therapies for P. aeruginosa infection”) for probiotic therapies.
Diarrhoeal diseases, caused by the invasion of Vibrio cholera, are nightmares for both children and adults [203]. Co-culturing the Ruminococcus obeum and Vibrio cholera in AKI medium, Hsiao et al. [204] found that the pathogenicity of V. cholerae was reduced by AI-2 of R. obeum. The results of experiments in the gnotobiotic mice model illustrated that the virulence of V. cholerae was regulated via a novel regulatory pathway in R. obeum. It relates to a new mechanism based on the VqmA virulence regulator rather than the known pathway based on HapR.
Considering that the gut microbiota mainly includes Bacteroidetes and Firmicutes, Thompson et al. [27] chose these two microbes to investigate the influence of AI-2 on the ratio of them. Firstly, they used streptomycin to induce gut dysbiosis, which had previously been applied to investigate the relationship between streptomycin treatment and colonization resistance in intestinal E. coli [205]. They subsequently engineered E. coli to manipulate the AI-2 level in the mouse intestine and investigated the influence on streptomycin-induced dysbiosis. By increasing the level of AI-2, the ratio of Firmicutes and Bacteroidetes was increased so as to relieve the strong effect of the antibiotic and restore the dysbiosis (Fig. 8a). Analogously, Xavier et al. also proposed that AI-2 can make some difference on the composition of gut microbiota in mouse [206].
Fig. 8.
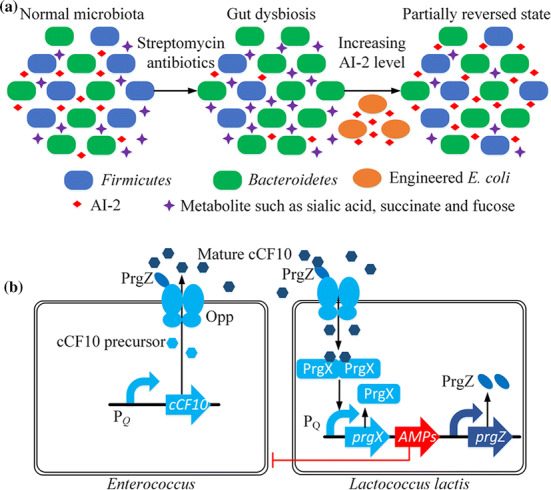
The applications of QS devices in the gut microbiota. a Balance of the gut microbiota. Once treated by antibiotics such as streptomycin, Firmicutes and AI-2 producing will reduce, while metabolite such as sialic acid, succinate and fucose will increase in the gut microbiota. Artificially increasing the levels of AI-2 produced by engineered E. coli reliefs the dysbiosis, increases the ratio of Firmicutes, and reverses state partially. b Diagram of genetic circuits of applying Lactococcus lactis to sense and kill Enterococcus
Lactococcus lactis and Enterococcus faecalis are ubiquitous in human gastrointestinal tract [207]. Borrero et al. [208] proposed using L. lactis, generally recognized as safe (GRAS) for human, to kill E. faecalis which is responsible for hospital-acquired infections such as enterococcal infections [209]. In this bi-directional system, L. lactis produces three antimicrobial peptides (AMPs), enterocin A, hiracin JM79 and enterocin P, to inhibit the growth of E. faecalis, including vancomycin-resistant enterococcus (VRE) strains. E. faecalis produces and secrets the sex pheromone cCF10 (as an AIP) to be detected by the engineered L. lactis (Fig. 8b). When the concentration of the sex pheromone cCF10 expressed by E. faecalis reaches the threshold, it will be imported by PrgZ protein and oligopeptide permease (Opp) system of the engineered L. lactis into its cytoplasm. cCF10 will then integrate with PrgX protein to form a PrgX–cCF10 complex which increases RNA polymerase access to PQ [104] and enhances the expression of the downstream antimicrobial peptides to kill E. faecalis.
As concluded by Coyte et al. [210], understanding the interactions between microbes, especially for the competition and cooperation among pathogens and probiotics, is key to revealing the mechanisms of gut microbiota-related diseases. To understand these complex processes, the studies reviewed above show that it is important to recognize the QS signaling molecules as the language or bonds that link together the members of the microbiome, and also as the bridge for between the bacteria and the host [211].
Therapies based on CRISPR-Cas immune systems
Bacteria often suffer from invasion by foreign mobile genetic elements such as bacteriophage infestation [212] and plasmids conjugation [213] which forms a basis for developing potential therapies to treat human diseases caused by pathogenic bacteria. Bacteria possess natural defense systems, named CRISPR-Cas immune systems [76], to reject foreign phages or plasmids (Fig. 9). The regulation of CRISPR-Cas immune systems can occur during different stages of an integrated network which involves pre-emptive warning, first contact, breaking the silence, detecting infection, and dedicated regulators [214]. Due to infections of phage increase at high cell density [28], QS devices have much potential to be applied to monitor the infections [29]. It is costly to continuously defend the whole CRISPR-Cas immune systems with an integrated network of various sensors [14]. Therefore, combining with QS devices, microbes can be “instructed” to regulate CRISPR-Cas immune systems only at high cell density, hence minimizing the cost. This is in line with the work of many researchers.
Fig. 9.
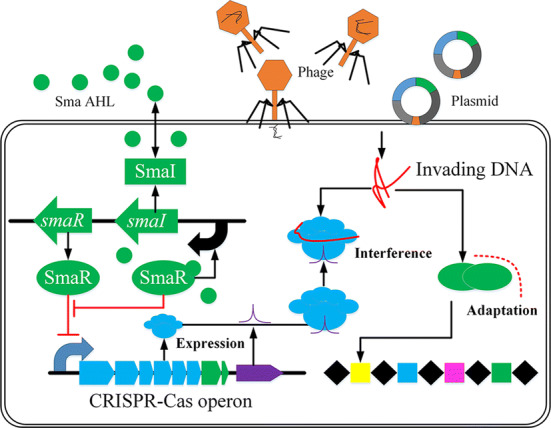
The schematic of QS regulation on CRISPR-Cas systems in Serratia. Mechanisms have been described in the text. The more specific introduction for the mechanisms of CRISPR-Cas immune systems can be found in the review paper (Patterson et al. [214])
In response to phage and plasmids invasion, this is in line with the work of Rossmann et al. [56] which proposed that AI‑2 regulates enterococcal pathogenicity and induces the horizontal gene transfer (HGT) of virulence genes by conjugation or transformation from phages to the commensal enterococci.
Høyland-Kroghsbo et al. [59] demonstrated that the CRISPR-Cas activity can be regulated by QS devices in the pathogen P. aeruginosa. The type I-F CRISPR-Cas system, LasI/LasR- and RhlI/RhlR-type QS devices were utilized to analyze the consequence of QS regulations on CRISPR-Cas activity. When lasI and rhlI genes were knocked out, the mutant exhibited obvious decreases in the expression of CRISPR-Cas relative to the wild type. Also, CRISPR-Cas activity restored to wild type levels when certain auto-inducers were artificially replenished. Therefore, it is possible to suppress the CRISPR-Cas immune system by QSIs as a cost-effective way to promote the killing of the pathogens by the phage.phage therapies.
Patterson et al. [215] combined the SmaI/smaR-type QS system and homologs of the LuxI/LuxR-type QS system with type I-E, I-F, and III-A CRISPR-Cas systems, respectively, to investigate the regulation effect of QS on HGT in Serratia sp. ATCC39006. The signaling molecule Sma AHL is produced by smaI gene and bonded with SmaR protein to form a SmaR–AHL complex. When the concentration of Sma AHL, Lux AHL homologs, is low at low cell density, the SmaR transcriptional regulator will act as a DNA-binding repressor for the CRISPR-Cas operon. As the cell density increases, Sma AHL accumulates in the culture and eventually reaches sufficiently high concentrations, and the SmaR–AHL complex will then inhibit the DNA binding activity of SmaR, which causes the expression of CRISPR-Cas to increase (Fig. 9).
The population-level resistance of bacteria upon invasion by foreign phages or pathogenic bacteria is commonly considered as important for maintaining the healthy state of the microbiome [214]. The development QS-based CRISPR-Cas technologies such as those reviewed above thus have the potential to bring useful additions to the toolbox for realizing population-level resistance.
Mathematical modelling for QS applications
Complementing experimental explorations and supported by the richness of biological information, a variety of synthetic genetic circuits and established databases [216], mathematical modelling has been widely applied in systems biology and synthetic biology to achieve systematic understandings of cellular behavior [217] and to optimize TYR for desirable products in engineering applications [218]. In particular, various modeling methods, such as flux balance analysis (FBA) [219], dynamic flux balance analysis (DFBA) [220], and sensitivity analysis have been combined with ordinary differential equations (ODE) [221–223] to construct mathematical models for QS. As listed in Table 1, there exist comprehensive reviews of deterministic and stochastic models and modelling approaches for QS devices, particularly of their molecular mechanisms. Not repeating these existing reviews, this section is intended to focus on the typical approaches and recent achievements of mathematical modelling of the applications of QS for bacterial population control as reviewed earlier in this work.
QS applications modelling in monoclonal synthetic biology
As a commonly shared paradigm for QS modelling, the LuxI/LuxR-type QS has been modelled by a number of researchers. To predict the function of the circuit illustrated earlier in Fig. 2a (and explained in “QS-based synthetic genetic oscillators”), You et al. built a mathematical model which includes cell growth, cell death, production and degradation of CcdB protein, and change of AHL concentration in this system [50]. The equations of the model are shown in Eqs. 1a–1c, which are derived following five assumptions:
Without IPTG induction, cell density changes will follow the logistic model;
When induced by IPTG, the rate of cell death will be proportional to the concentration of CcdB protein;
The generation rate of CcdB protein is proportional to the concentration of AHL, and the intracellular and extracellular AHL concentrations are equal;
The generation rate of AHL is proportional to cell density;
Degradation of CcdB protein and AHL follows a first-order kinetics.
| 1a |
| 1b |
| 1c |
A number of QS models similar to the above have subsequently been developed, making predictions based on the capture of the interactions between cell density, concentration of AIs, complex of AIs and their correspond receptors [44].
In principle, QS is not only dictated by reactions of various molecules but also affected by diffusion. Basu et al. [224] designed a synthetic multicellular system with QS devices, which consists of “sender” and “receiver” cells. Due to the diffusion of AHL, the concentration of AHL generated by the “sender” decreases gradually from the cell to the periphery. Consequently, the “receiver” cells in different regions respond to different concentrations of AHLs and express different colors of fluorescent protein, thus forming different colors and different ring-like patterns.
Building on the two models mentioned above, Anesiadis et al. [82] constructed a mechanistic model (Eqs. 2a–2e) for investigating the dynamics of the genetic circuit including a genetic toggle switch (introduced earlier in Fig. 5a). The same research group [83] further integrated the QS model (Eqs. 2a–2e) and a DFBA model to maximize serine production.
| 2a |
| 2b |
| 2c |
| 2d |
| 2e |
As shown earlier in Fig. 5e and explained in “QS-based genetic toggle switches”, a synthetic genetic toggle switch can be designed and applied for MI production (Eqs. 3a–3c). Gupta et al. [8] modified the population control equation Eq. 1a by removing the lysis term and adding a variable which denotes the fractional Pfk-1 activity to predict the circuit’s function.
| 3a |
| 3b |
| 3c |
QS applications modelling in synthetic ecology
Based on the aforementioned relatively simple models for monoclonal synthetic biology, several more complex models for combinatorial quorum sensing [70] in synthetic ecology have been reported. Building on the model of You et al. [50], Balagadde et al. [51] modelled the dynamics of a synthetic E. coli predator–prey system, which includes two QS devices (lux QS and las QS) (introduced earlier in Fig. 2b). Further, Song et al. [225], from the group of You, expanded their model to investigate the spatiotemporal dynamics of the predator–prey system by adding the factors of chemical diffusion, cellular motility, and nutrient consumption. To study the dynamics of emergent genetic oscillations in a synthetic microbial consortium (introduced earlier in Fig. 4c), Chen et al. [78] developed a model depicting the system with three compartments, namely the intracellular space of the activator strain, the intracellular space of the repressor strain, and the extracellular space, to reduce the difficulty of modelling. Scott et al. [55] combined agent-based modeling and deterministic modeling to describe the population-level dynamics of synchronized oscillations (introduced earlier in Fig. 2d). Based on the models derived from two-strain consortia, Kong et al. [226] developed models to investigate the dynamics of three- and four-strain ecosystems induced by nisin, a QS molecule of Gram-positive bacteria.
QS stochastic modelling
Stochastic models for QS are needed to account for the natural stochasticity in gene expression. In particular, when the rates of gene expression are relatively low and the amount of the reactants is relatively small, the effects of stochasticity on the system’s behavior cannot be ignored [227]. It thus becomes necessary to adopt a stochastic model to more faithfully capture the nature of genetic circuits such as synthetic genetic oscillators [56, 58]. Tian et al. [228] recoganized the importance of noise in the switching of bistable systems, such as the QS-based genetic toggle switch (Fig. 5a). They developed quantitative stochastic models for large-scale genetic regulatory networks by introducing Poisson random variables into deterministic models to analyze the influence of noise. The model developed by Baumgart et al. [74] considered the dynamics of the concentrations of LuxI, I-SceI and GFP, and was used for conducting robustness analysis of a stochastic process that involved the positive feedback of QS, negative feedback of plasmid copy number, and intracellular delay in feedback, which were integrated to describe the oscillator in DNA copy number control (mechanism shown in Fig. 3f).
The modelling studies reviewed in this section show that lumped-parameter (i.e., well-mixed), deterministic models, which are relatively simple, can already predict the effect of applications of QS-based genetic circuits in metabolic engineering and to offer useful insights on the dynamics of such systems. On the other hand, more advanced modelling schemes, such as those taking into account spatial heterogeneity and stochasticity, could offer more faithful and more detailed representation, which would become particularly important when such complexities play a decisive role in shaping the behavour and function of a QS-based system.
Summary and future perspectives
QS devices have been shown to be able to play a key role in engineered dynamic control of metabolism and various microbial physiological activities, such as the TYR (titer, yield and rate) increase of desirable metabolites, microbiota synchronization, and regulation of sporulation, virulence, competence and toxin production. These applications of QS have been realized by five main types of signaling molecules, namely AHLs, DSFs, AIPs, AI-2 and indole. Genetic oscillators, toggle switches and logic gates have been constructed by coupling with AHL-based QS devices in Gram-negative bacteria. With Gram-positive bacteria, both extracellular and intracellular pathway AIP-based QS devices have been developed for controlling physiological activities. AI-2 and indole, on the other hand, can be regarded as languages of communication in microbial communities. Engineered applications of QS in microbes can be either “constructive” (through introducing new/foreign or enhancing existing QS mechanisms) or “destructive” (through inhibiting existing QS mechanisms). In constructive cases, the QS devices could be either natural or synthetic. Furthermore, a synthetic QS device can be either constructed partly from naturally occurring QS modules or synthesized from scratch. Among other areas, both constructive and destructive QS applications have found increasing potential in developing clinical therapies for a range of diseases caused by biofilm formation, antibiotic resistance and phage invasion.
For QS to be more effective and more widely applicable, further work is needed to address several general challenges. First, the target QS modulators must recognize precisely the corresponding signals in an extremely miscellaneous pool. Second, compared to the long-range effective electrical signals [229, 230], quenching problems are common in chemical signals in QS devices; answering questions such as how to realize the spatiotemporal and tempo control of signaling molecules is essential, too. Third, there are couplings or crosstalk between different QS signals and receptors (e.g., AHL-based QS and indole-based QS affect each other); understanding how to decouple these pathways is necessary and important to achieve their applications with larger scale genetic oscillators in demanding tasks such as investigating the population-level dynamics of microbes. Fourth, the cheating behaviors aggravate the complexity of microbial communities; the questions of how to constrain cheaters when desirable and how to exploit cheating behaviors to inhibit QS are not only theoretically interesting for microbial ecology but also practically important for the development of QSIs. Finally, nonlinearities and stochasticity need to be considered and resolved appropriately in QS modelling.
With synthetic biology making great leaps forward, the future perspectives of QS applications are expected to keep pace with it. Currently, most of the engineered dynamic regulation mechanisms build on AHL-based QS devices of Gram-negative bacteria. In contrast, existing QS-based interventions in Gram-positive bacteria are mostly in the form of up-regulating or down-regulating naturally occurring processes, and there is very limited research on synthetic QS in this type of bacteria. On the other hand, Gram-positive bacteria, such as Lactococcus lactis have been viewed as important candidates for building engineered microbial cell factories and vaccine delivery systems, and they play an irreplaceable role in metabolic engineering and medical applications, such as producing dairy fermentations [231, 232], industrial products and various vaccines [233]. Given their significance, more research is needed to discover, study and apply the QS mechanisms in Gram-positive bacteria. In particular, one may expect that there is great potential to couple certain synthetic TCS-based or AIP-based QS devices with the original pathways in Gram-positive bacteria to improve the TYR of desirable products in Gram-positive cell factories and to develop future therapeutic systems.
As a broad perspective for future research, the exploration of QS for population-level control of bacteria and its potential in therapeutic applications may proceed in horizontal and vertical dimensions, respectively. Horizontally, QS applications can be centered on multi-circuit systems (the combination of various QS-based genetic circuits), multi-production (two or more products from one single cell), and microbial communities (engineering microbiota to produce one or more products). Vertically, QS applications in medical therapeutics have the potential to advance by addressing the interactions between microbes (virus, bacterium and fungi) and the host [212, 234–236]. The understanding of these interactions will accelerate the development of more reliable strategies for manipulating the microbiota against infectious diseases and antibiotic resistance, which is of great significance for human health. In the future horizontal and vertical developments of QS applications, mathematic models are expected to continue their supporting role to construct QS networks, by not only embracing nonlinearities and stochasticity, but also integrating across component, circuit, cellular and community levels and addressing the interactions between microbes and their hosts and other environments, to offer the power of making holistic predictions.
Acknowledgements
This study was supported by the National Key Research and Development Project of China (2017YFD0201400), the National Natural Science Foundation of China (31570089, 31170076), and the Funds for Creative Research Groups of China (21621004), Dr. Jianjun Qiao was supported by The New Century Outstanding Talent Support Program, Education Ministry of China.
List of symbols
- [A]
Intracellular AHL concentration (mM)
- [C]
Intracellular CI protein concentration (mM)
- [E]
Intracellular CcdB protein concentration (mM)
- [L]
Intracellular LacR concentration (mM)
- [LuxR]
Intracellular LuxR concentration (mM)
- [R]
Intracellular AHL/LuxR complex concentration (mM)
- N
The cell density (CFU ml−1)
- Nm
The maximum cell density (CFU ml−1)
- Fpfk
The fractional Pfk-1 activity (U/mg)
- Kd
The cumulative dissociation constant
- X
Biomass concentration (g L−1)
- n1, n2
Transcription factor cooperativity/multimerization
- αC
CI protein synthesis rate constant (μM min−1)
- αL1, αL2
LacR protein synthesis rate constants (μM min−1)
- βC
CI repression coefficient (mM)
- βL
LacR repression coefficient (mM)
- d
Cell death rate (nM−1 h−1)
- dA, dE
AHL and CcdB protein decay constant (min−1)
- dC
CI protein decay constant (min−1)
- dL, dR
LacR and LuxR–AHL complex decay constants (min−1)
- k
Growth rate (h−1)
- kE
CcdB protein production rate constant (h−1)
- vA
AHL production rate constant (nM mL h−1)
- θR
LuxR/AHL activation coefficient (mM)
- ρR
LuxR/AHL dimerization constant (μM−3 min−1)
Compliance with ethical standards
Conflict of interest
The authors declare no competing financial interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Aidong Yang, Email: aidong.yang@eng.ox.ac.uk.
Jianjun Qiao, Email: jianjunq@tju.edu.cn.
References
- 1.Homer CM, Summers DK, Goranov AI, Clarke SC, Wiesner DL, Diedrich JK, Moresco JJ, Toffaletti D, Upadhya R, Caradonna I, Petnic S, Pessino V, Cuomo CA, Lodge JK, Perfect J, Yates JR, 3rd, Nielsen K, Craik CS, Madhani HD. Intracellular action of a secreted peptide required for fungal virulence. Cell Host Microbe. 2016;19:849–864. doi: 10.1016/j.chom.2016.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Erez Z, Steinberger-Levy I, Shamir M, Doron S, Stokar-Avihail A, Peleg Y, Melamed S, Leavitt A, Savidor A, Albeck S. Communication between viruses guides lysis-lysogeny decisions. Nature. 2017;541:488–493. doi: 10.1038/nature21049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Melissa B, Miller BL. Quorum sensing in bacteria. Annu Rev Microbiol. 2001;55(1):165–199. doi: 10.1146/annurev.micro.55.1.165. [DOI] [PubMed] [Google Scholar]
- 4.Whiteley M, Diggle SP, Greenberg EP. Progress in and promise of bacterial quorum sensing research. Nature. 2017;551:313–320. doi: 10.1038/nature24624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chubukov V, Gerosa L, Kochanowski K, Sauer U. Coordination of microbial metabolism. Nat Rev Microbiol. 2014;12:327–340. doi: 10.1038/nrmicro3238. [DOI] [PubMed] [Google Scholar]
- 6.Holtz WJ, Keasling JD. Engineering static and dynamic control of synthetic pathways. Cell. 2010;140:19–23. doi: 10.1016/j.cell.2009.12.029. [DOI] [PubMed] [Google Scholar]
- 7.Venayak N, Anesiadis N, Cluett WR, Mahadevan R. Engineering metabolism through dynamic control. Curr Opin Biotechnol. 2015;34:142–152. doi: 10.1016/j.copbio.2014.12.022. [DOI] [PubMed] [Google Scholar]
- 8.Gupta A, Reizman IM, Reisch CR, Prather KL. Dynamic regulation of metabolic flux in engineered bacteria using a pathway-independent quorum-sensing circuit. Nat Biotechnol. 2017;35:273–279. doi: 10.1038/nbt.3796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu D, Evans T, Zhang F. Applications and advances of metabolite biosensors for metabolic engineering. Metab Eng. 2015;31:35–43. doi: 10.1016/j.ymben.2015.06.008. [DOI] [PubMed] [Google Scholar]
- 10.Soma Y, Hanai T. Self-induced metabolic state switching by a tunable cell density sensor for microbial isopropanol production. Metab Eng. 2015;30:7–15. doi: 10.1016/j.ymben.2015.04.005. [DOI] [PubMed] [Google Scholar]
- 11.Doong SJ, Gupta A, Prather KLJ. Layered dynamic regulation for improving metabolic pathway productivity in escherichia coli. Proc Natl Acad Sci USA. 2018;115(12):2964–2969. doi: 10.1073/pnas.1716920115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu X, Li XB, Jiang J, Liu ZN, Qiao B, Li FF, Cheng JS, Sun X, Yuan YJ, Qiao J. Convergent engineering of syntrophic Escherichia coli coculture for efficient production of glycosides. Metab Eng. 2018;47:243–253. doi: 10.1016/j.ymben.2018.03.016. [DOI] [PubMed] [Google Scholar]
- 13.Gardner TS, Cantor CR, Collins JJ. Construction of a genetic toggle switch in Escherichia coli. Nature. 2000;403(6767):339–342. doi: 10.1038/35002131. [DOI] [PubMed] [Google Scholar]
- 14.Elowitz MB, Leibler S. A synthetic oscillatory network of transcriptional regulators. Nature. 2000;403:335–338. doi: 10.1038/35002125. [DOI] [PubMed] [Google Scholar]
- 15.Win MN, Smolke CD. A modular and extensible rna-based gene-regulatory platform for engineering cellular function. Proc Natl Acad Sci USA. 2007;104:14283–14288. doi: 10.1073/pnas.0703961104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dueber JE, Wu GC, Malmirchegini GR, Moon TS, Petzold CJ, Ullal AV, Prather KL, Keasling JD. Synthetic protein scaffolds provide modular control over metabolic flux. Nat Biotechnol. 2009;27(8):753–759. doi: 10.1038/nbt.1557. [DOI] [PubMed] [Google Scholar]
- 17.Danino T, Mondragon-Palomino O, Tsimring L, Hasty J. A synchronized quorum of genetic clocks. Nature. 2010;463:326–330. doi: 10.1038/nature08753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Papenfort K, Bassler BL. Quorum sensing signal–response systems in gram-negative bacteria. Nat Rev Microbiol. 2016;14:576–588. doi: 10.1038/nrmicro.2016.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Diggle SP, Griffin AS, Campbell GS, West SA. Cooperation and conflict in quorum-sensing bacterial populations. Nature. 2007;450:411–414. doi: 10.1038/nature06279. [DOI] [PubMed] [Google Scholar]
- 20.Modi SR, Collins JJ, Relman DA. Antibiotics and the gut microbiota. J Clin Investig. 2014;124:4212–4218. doi: 10.1172/JCI72333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Flemming HC, Wingender J, Szewzyk U, Steinberg P, Rice SA, Kjelleberg S. Biofilms: an emergent form of bacterial life. Nat Rev Microbiol. 2016;14:563–575. doi: 10.1038/nrmicro.2016.94. [DOI] [PubMed] [Google Scholar]
- 22.Yan J, Bassler BL. Surviving as a community: antibiotic tolerance and persistence in bacterial biofilms. Cell Host Microbe. 2019;26:15–21. doi: 10.1016/j.chom.2019.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hong SH, Hegde M, Kim J, Wang X, Jayaraman A, Wood TK. Synthetic quorum-sensing circuit to control consortial biofilm formation and dispersal in a microfluidic device. Nat Commun. 2012;3:613–620. doi: 10.1038/ncomms1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cho I, Yamanishi S, Cox L, Methé BA, Zavadil J, Li K, Gao Z, Mahana D, Raju K, Teitler I. Antibiotics in early life alter the murine colonic microbiome and adiposity. Nature. 2012;488:621–626. doi: 10.1038/nature11400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kalia VC. Quorum sensing inhibitors: an overview. Biotechnol Adv. 2013;31:224–245. doi: 10.1016/j.biotechadv.2012.10.004. [DOI] [PubMed] [Google Scholar]
- 26.Ma S, Zhou Z. Recent advances in the discovery of pqsd inhibitors as antimicrobial agents. ChemMedChem. 2017;12:420–425. doi: 10.1002/cmdc.201700015. [DOI] [PubMed] [Google Scholar]
- 27.Thompson JA, Oliveira RA, Djukovic A, Ubeda C, Xavier KB. Manipulation of the quorum sensing signal ai-2 affects the antibiotic-treated gut microbiota. Cell Rep. 2015;10:1861–1871. doi: 10.1016/j.celrep.2015.02.049. [DOI] [PubMed] [Google Scholar]
- 28.Abedon ST. Spatial vulnerability: bacterial arrangements, microcolonies, and biofilms as responses to low rather than high phage densities. Viruses. 2012;4:663–687. doi: 10.3390/v4050663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Semenova E, Severinov K. Come together: Crispr-cas immunity senses the quorum. Mol Cell. 2016;64:1013–1015. doi: 10.1016/j.molcel.2016.11.037. [DOI] [PubMed] [Google Scholar]
- 30.Hawver LA, Jung SA, Ng WL. Specificity and complexity in bacterial quorum-sensing systems. FEMS Microbiol Rev. 2016;40:738–752. doi: 10.1093/femsre/fuw014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Monnet V, Gardan R. Quorum-sensing regulators in gram-positive bacteria: ‘Cherchez le peptide’. Mol Microbiol. 2015;97:181–184. doi: 10.1111/mmi.13060. [DOI] [PubMed] [Google Scholar]
- 32.Monnet V, Juillard V, Gardan R. Peptide conversations in gram-positive bacteria. Crit Rev Microbiol. 2014;42:339–351. doi: 10.3109/1040841X.2014.948804. [DOI] [PubMed] [Google Scholar]
- 33.Lee JH, Wood TK, Lee J. Roles of indole as an interspecies and interkingdom signaling molecule. Trends Microbiol. 2015;23:707–718. doi: 10.1016/j.tim.2015.08.001. [DOI] [PubMed] [Google Scholar]
- 34.Hmelo LR. Quorum sensing in marine microbial environments. Ann Rev Mar Sci. 2017;9:257–281. doi: 10.1146/annurev-marine-010816-060656. [DOI] [PubMed] [Google Scholar]
- 35.Zhou L, Zhang LH, Camara M, He YW. The dsf family of quorum sensing signals: diversity, biosynthesis, and turnover. Trends Microbiol. 2017;25:293–303. doi: 10.1016/j.tim.2016.11.013. [DOI] [PubMed] [Google Scholar]
- 36.Xu P. Production of chemicals using dynamic control of metabolic fluxes. Curr Opin Biotechnol. 2017;53:12–19. doi: 10.1016/j.copbio.2017.10.009. [DOI] [PubMed] [Google Scholar]
- 37.Shong J, Diaz MRJ, Collins CH. Towards synthetic microbial consortia for bioprocessing. Curr Opin Biotechnol. 2012;23:798–802. doi: 10.1016/j.copbio.2012.02.001. [DOI] [PubMed] [Google Scholar]
- 38.Song H, Ding MZ, Jia XQ, Ma Q, Yuan YJ. Synthetic microbial consortia: from systematic analysis to construction and applications. Chem Soc Rev. 2014;43:6954–6981. doi: 10.1039/c4cs00114a. [DOI] [PubMed] [Google Scholar]
- 39.Dolinsek J, Goldschmidt F, Johnson DR. Synthetic microbial ecology and the dynamic interplay between microbial genotypes. FEMS Microbiol Rev. 2016;40:961–979. doi: 10.1093/femsre/fuw024. [DOI] [PubMed] [Google Scholar]
- 40.Asfahl KL, Schuster M, Gibbs K. Social interactions in bacterial cell–cell signaling. FEMS Microbiol Rev. 2017;41:92–107. doi: 10.1093/femsre/fuw038. [DOI] [PubMed] [Google Scholar]
- 41.Choudhary S, Schmidt-Dannert C. Applications of quorum sensing in biotechnology. Appl Microbiol Biotechnol. 2010;86:1267–1279. doi: 10.1007/s00253-010-2521-7. [DOI] [PubMed] [Google Scholar]
- 42.Koo H, Allan RN, Howlin RP, Stoodley P, Hall-Stoodley L. Targeting microbial biofilms: current and prospective therapeutic strategies. Nat Rev Microbiol. 2017;15:740–755. doi: 10.1038/nrmicro.2017.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mukherjee S, Bassler BL. Bacterial quorum sensing in complex and dynamically changing environments. Nat Rev Microbiol. 2019;17:371–382. doi: 10.1038/s41579-019-0186-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Goryachev AB. Understanding bacterial cell–cell communication with computational modeling. Chem Rev. 2011;111:238–250. doi: 10.1021/cr100286z. [DOI] [PubMed] [Google Scholar]
- 45.Hense BA, Schuster M. Core principles of bacterial autoinducer systems. Microbiol Mol Biol Rev. 2015;79:153–169. doi: 10.1128/MMBR.00024-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pérez-Velázquez J, Gölgeli M, García-Contreras R. Mathematical modelling of bacterial quorum sensing: a review. Bull Math Biol. 2016;78:1–55. doi: 10.1007/s11538-016-0160-6. [DOI] [PubMed] [Google Scholar]
- 47.Boyer M, Wisniewski-Dye F. Cell–cell signalling in bacteria: not simply a matter of quorum. FEMS Microbiol Ecol. 2009;70:1–19. doi: 10.1111/j.1574-6941.2009.00745.x. [DOI] [PubMed] [Google Scholar]
- 48.An JH, Goo E, Kim H, Seo YS, Hwang I. Bacterial quorum sensing and metabolic slowing in a cooperative population. Proc Natl Acad Sci USA. 2014;111:14912–14917. doi: 10.1073/pnas.1412431111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Goo E, Majerczyk CD, An JH, Chandler JR, Seo YS, Ham H, Lim JY, Kim H, Lee B, Jang MS, Greenberg EP, Hwang I. Bacterial quorum sensing, cooperativity, and anticipation of stationary-phase stress. Proc Natl Acad Sci USA. 2012;109:19775–19780. doi: 10.1073/pnas.1218092109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.You L, Cox RS, 3rd, Weiss R, Arnold FH. Programmed population control by cell–cell communication and regulated killing. Nature. 2004;428:868–871. doi: 10.1038/nature02491. [DOI] [PubMed] [Google Scholar]
- 51.Balagadde FK, Song H, Ozaki J, Collins CH, Barnet M, Arnold FH, Quake SR, You L. A synthetic escherichia coli predator–prey ecosystem. Mol Syst Biol. 2008;4:187–194. doi: 10.1038/msb.2008.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu F, Lopatkin AJ, Needs DA, Lee CT, Mukherjee S, You L. A unifying framework for interpreting and predicting mutualistic systems. Nat Commun. 2019;10:242–251. doi: 10.1038/s41467-018-08188-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Din MO, Danino T, Prindle A, Skalak M, Selimkhanov J, Allen K, Julio E, Atolia E, Tsimring LS, Bhatia SN. Synchronized cycles of bacterial lysis for in vivo delivery. Nature. 2016;536:81–85. doi: 10.1038/nature18930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mays ZJ, Nair NU. Synthetic biology in probiotic lactic acid bacteria: at the frontier of living therapeutics. Curr Opin Biotechnol. 2018;53:224–231. doi: 10.1016/j.copbio.2018.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Scott SR, Din MO, Bittihn P, Xiong L, Tsimring LS, Hasty J. A stabilized microbial ecosystem of self-limiting bacteria using synthetic quorum-regulated lysis. Nat Microbiol. 2017;2:17083–17091. doi: 10.1038/nmicrobiol.2017.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Woods ML, Leon M, Perezcarrasco R, Barnes CP. A statistical approach reveals designs for the most robust stochastic gene oscillators. ACS Synth Biol. 2016;5:459–470. doi: 10.1021/acssynbio.5b00179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jörg DJ, Morelli LG, Jülicher F. Chemical event chain model of coupled genetic oscillators. Phys Rev E. 2018;97(3–1):1–11. doi: 10.1103/PhysRevE.97.032409. [DOI] [PubMed] [Google Scholar]
- 58.Potvin-Trottier L, Lord ND, Vinnicombe G, Paulsson J. Synchronous long-term oscillations in a synthetic gene circuit. Nature. 2016;538:514–517. doi: 10.1038/nature19841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tu BP, Mcknight SL. Metabolic cycles as an underlying basis of biological oscillations. Nat Rev Mol Cell Biol. 2006;7:696–701. doi: 10.1038/nrm1980. [DOI] [PubMed] [Google Scholar]
- 60.Mondragon-Palomino O, Danino T, Selimkhanov J, Tsimring L, Hasty J. Entrainment of a population of synthetic genetic oscillators. Science. 2011;333:1315–1319. doi: 10.1126/science.1205369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Purcell O, Savery NJ, Grierson CS, Bernardo MD. A comparative analysis of synthetic genetic oscillators. J R Soc Interface. 2010;7(52):1503–1524. doi: 10.1098/rsif.2010.0183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Barkai N, Leibler S. Circadian clocks limited by noise. Nature. 2000;403(6767):267–268. doi: 10.1038/35002258. [DOI] [PubMed] [Google Scholar]
- 63.Hasty J, Dolnik M, Rottschäfer V, Collins JJ. Synthetic gene network for entraining and amplifying cellular oscillations. Phys Rev Lett. 2002;88(14):1–4. doi: 10.1103/PhysRevLett.88.148101. [DOI] [PubMed] [Google Scholar]
- 64.Stricker J, Cookson S, Bennett MR, Mather WH, Tsimring LS, Hasty J. A fast, robust and tunable synthetic gene oscillator. Nature. 2008;456:516–519. doi: 10.1038/nature07389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kellogg R, Tay S. Noise facilitates transcriptional control under dynamic inputs. Cell. 2015;160:381–392. doi: 10.1016/j.cell.2015.01.013. [DOI] [PubMed] [Google Scholar]
- 66.Tokuda IT, Okamoto A, Matsumura R, Takumi T, Akashi M. Potential contribution of tandem circadian enhancers to nonlinear oscillations in clock gene expression. Mol Biol Cell. 2017;28(17):2333–2342. doi: 10.1091/mbc.E17-02-0129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wintermute EH, Silver PA. Dynamics in the mixed microbial concourse. Genes Dev. 2010;24:2603–2614. doi: 10.1101/gad.1985210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xavier JB. Social interaction in synthetic and natural microbial communities. Mol Syst Biol. 2011;7:483–493. doi: 10.1038/msb.2011.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chuang JS. Engineering multicellular traits in synthetic microbial populations. Curr Opin Chem Biol. 2012;16:370–378. doi: 10.1016/j.cbpa.2012.04.002. [DOI] [PubMed] [Google Scholar]
- 70.Scott SR, Hasty J. Quorum sensing communication modules for microbial consortia. ACS Synth Biol. 2016;5:969–977. doi: 10.1021/acssynbio.5b00286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mcmillen D, Kopell N, Hasty J, Collins JJ. Synchronizing genetic relaxation oscillators by intercell signaling. Proc Natl Acad Sci USA. 2002;99:679–684. doi: 10.1073/pnas.022642299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Waters CM, Bassler BL. Quorum sensing: cell-to-cell communication in bacteria. Annu Rev Cell Dev Biol. 2005;21:319–346. doi: 10.1146/annurev.cellbio.21.012704.131001. [DOI] [PubMed] [Google Scholar]
- 73.Slager J, Kjos M, Attaiech L, Veening JW. Antibiotic-induced replication stress triggers bacterial competence by increasing gene dosage near the origin. Cell. 2014;157:395–406. doi: 10.1016/j.cell.2014.01.068. [DOI] [PubMed] [Google Scholar]
- 74.Baumgart L, Mather W, Hasty J. Synchronized DNA cycling across a bacterial population. Nat Genet. 2017;49(8):1282–1285. doi: 10.1038/ng.3915. [DOI] [PubMed] [Google Scholar]
- 75.Prindle A, Samayoa P, Razinkov I, Danino T, Tsimring LS, Hasty J. A sensing array of radically coupled genetic/’biopixels/’. Nature. 2012;481:39–44. doi: 10.1038/nature10722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Prindle A, Selimkhanov J, Li H, Razinkov I, Tsimring LS, Hasty J. Rapid and tunable post-translational coupling of genetic circuits. Nature. 2014;508:387–391. doi: 10.1038/nature13238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Levchenko I, Seidel M, Sauer RT, Baker TA. A specificity-enhancing factor for the clpxp degradation machine. Science. 2000;289:2354–2356. doi: 10.1126/science.289.5488.2354. [DOI] [PubMed] [Google Scholar]
- 78.Chen Y, Kim JK, Hirning AJ, Josić K, Bennett MR. Emergent genetic oscillations in a synthetic microbial consortium. Science. 2015;349(6251):986–989. doi: 10.1126/science.aaa3794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Feist AM, Palsson BO. The biomass objective function. Curr Opin Microbiol. 2010;13:344–349. doi: 10.1016/j.mib.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Soma Y, Tsuruno K, Wada M, Yokota A, Hanai T. Metabolic flux redirection from a central metabolic pathway toward a synthetic pathway using a metabolic toggle switch. Metab Eng. 2014;23:175–184. doi: 10.1016/j.ymben.2014.02.008. [DOI] [PubMed] [Google Scholar]
- 81.Kobayashi H, Kaern M, Araki M, Chung K, Gardner TS, Cantor CR, Collins JJ. Programmable cells: interfacing natural and engineered gene networks. Proc Natl Acad Sci USA. 2004;101:8414–8419. doi: 10.1073/pnas.0402940101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Anesiadis N, Cluett WR, Mahadevan R. Dynamic metabolic engineering for increasing bioprocess productivity. Metab Eng. 2008;10:255–266. doi: 10.1016/j.ymben.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 83.Anesiadis N, Kobayashi H, Cluett WR, Mahadevan R. Analysis and design of a genetic circuit for dynamic metabolic engineering. ACS Synth Biol. 2013;2:442–452. doi: 10.1021/sb300129j. [DOI] [PubMed] [Google Scholar]
- 84.Honjo H, Iwasaki K, Soma Y, Tsuruno K, Hamada H, Hanai T. Synthetic microbial consortium with specific roles designated by genetic circuits for cooperative chemical production. Metab Eng. 2019;55:268–275. doi: 10.1016/j.ymben.2019.08.007. [DOI] [PubMed] [Google Scholar]
- 85.Wang EX, Liu Y, Ma Q, Dong XT, Ding MZ, Yuan YJ. Synthetic cell–cell communication in a three-species consortium for one-step vitamin c fermentation. Biotechnol Lett. 2019;41:951–961. doi: 10.1007/s10529-019-02705-2. [DOI] [PubMed] [Google Scholar]
- 86.Tsoi R, Dai Z, You L. Emerging strategies for engineering microbial communities. Biotechnol Adv. 2019;37(6):1–9. doi: 10.1016/j.biotechadv.2019.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Nandagopal N, Elowitz MB. Synthetic biology: integrated gene circuits. Science. 2011;333:1244–1248. doi: 10.1126/science.1207084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Benenson Y. Biomolecular computing systems: principles, progress and potential. Nat Rev Genet. 2012;13(7):455–468. doi: 10.1038/nrg3197. [DOI] [PubMed] [Google Scholar]
- 89.Moon TS, Lou C, Tamsir A, Stanton BC, Voigt CA. Genetic programs constructed from layered logic gates in single cells. Nature. 2012;491:249–253. doi: 10.1038/nature11516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Baig H, Madsen J. Simulation approach for timing analysis of genetic logic circuits. ACS Synth Biol. 2017;6(7):1169–1179. doi: 10.1021/acssynbio.6b00296. [DOI] [PubMed] [Google Scholar]
- 91.Nielsen AA, Der BS, Shin J, Vaidyanathan P, Paralanov V, Strychalski EA, Ross D, Densmore D, Voigt CA. Genetic circuit design automation. Science. 2016;352(aac7341):1–11. doi: 10.1126/science.aac7341. [DOI] [PubMed] [Google Scholar]
- 92.Hasty J, McMillen D, Collins JJ. Engineered gene circuits. Nature. 2002;420:224–230. doi: 10.1038/nature01257. [DOI] [PubMed] [Google Scholar]
- 93.Li Z, Rosenbaum MA, Venkataraman A, Tam TK, Katz E, Angenent LT. Bacteria-based and logic gate: a decision-making and self-powered biosensor. Chem Commun. 2011;47:3060–3062. doi: 10.1039/c0cc05037g. [DOI] [PubMed] [Google Scholar]
- 94.Arugula MA, Shroff N, Katz E, He Z. Molecular and logic gate based on bacterial anaerobic respiration. Chem Commun. 2012;48:10174–10176. doi: 10.1039/c2cc35595g. [DOI] [PubMed] [Google Scholar]
- 95.Tamsir A, Tabor JJ, Voigt CA. Robust multicellular computing using genetically encoded nor gates and chemical /`wires/’. Nature. 2010;469:212–215. doi: 10.1038/nature09565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yokobayashi Y, Weiss R, Arnold F. Directed evolution of a genetic circuit. Proc Natl Acad Sci USA. 2002;99:16587–16591. doi: 10.1073/pnas.252535999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Miyamoto T, Razavi S, Derose R, Inoue T. Synthesizing biomolecule-based boolean logic gates. ACS Synth Biol. 2013;2:72–82. doi: 10.1021/sb3001112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Shong J, Collins CH. Quorum sensing-modulated and-gate promoters control gene expression in response to a combination of endogenous and exogenous signals. ACS Synth Biol. 2014;3:238–246. doi: 10.1021/sb4000965. [DOI] [PubMed] [Google Scholar]
- 99.Hu Y, Yang Y, Katz E, Song H. Programming the quorum sensing-based and gate in shewanella oneidensis for logic gated-microbial fuel cells. Chem Commun. 2015;51:4184–4187. doi: 10.1039/c5cc00026b. [DOI] [PubMed] [Google Scholar]
- 100.He X, Chen Y, Liang Q, Qi Q. An autoinduced and-gate controlling metabolic pathway dynamically in response to microbial communities and cell physiological state. ACS Synth Biol. 2017;6:463–470. doi: 10.1021/acssynbio.6b00177. [DOI] [PubMed] [Google Scholar]
- 101.Takayanagi Y, Tanaka K, Takahashi H. Structure of the 5′ upstream region and the regulation of the rpos gene of Escherichia coli. Mol Genet Genomics. 1994;243:525–531. doi: 10.1007/BF00284200. [DOI] [PubMed] [Google Scholar]
- 102.Keller L, Surette MG. Communication in bacteria: an ecological and evolutionary perspective. Nat Rev Microbiol. 2006;4:249–258. doi: 10.1038/nrmicro1383. [DOI] [PubMed] [Google Scholar]
- 103.Papadimitriou K, Alegria A, Bron PA, de Angelis M, Gobbetti M, Kleerebezem M, Lemos JA, Linares DM, Ross P, Stanton C, Turroni F, van Sinderen D, Varmanen P, Ventura M, Zuniga M, Tsakalidou E, Kok J. Stress physiology of lactic acid bacteria. Microbiol Mol Biol Rev. 2016;80:837–890. doi: 10.1128/MMBR.00076-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Cook LC, Federle MJ. Peptide pheromone signaling in streptococcus and enterococcus. FEMS Microbiol Rev. 2014;38:473–492. doi: 10.1111/1574-6976.12046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Marchand N, Collins CH. Synthetic quorum sensing and cell–cell communication in gram-positive bacillus megaterium. ACS Synth Biol. 2016;5:597–606. doi: 10.1021/acssynbio.5b00099. [DOI] [PubMed] [Google Scholar]
- 106.Cooksley CM, Davis IJ, Winzer K, Chan WC, Peck MW, Minton PN. Regulation of neurotoxin production and sporulation by a putative agrbd signaling system in proteolytic clostridium botulinum. Appl Environ Microbiol. 2010;76:4448–4460. doi: 10.1128/AEM.03038-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Steiner E, Scott J, Minton NP, Winzer K. An agr quorum sensing system that regulates granulose formation and sporulation in clostridium acetobutylicum. Appl Environ Microbiol. 2012;78:1113–1122. doi: 10.1128/AEM.06376-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Vivant AL, Garmyn D, Gal L, Piveteau P. The agr communication system provides a benefit to the populations of listeria monocytogenes in soil. Front Cell Infect Microbiol. 2014;4(160):1–7. doi: 10.3389/fcimb.2014.00160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ma M, Li J, Mcclane BA. Structure-function analysis of peptide signaling in the clostridium perfringens agr-like quorum sensing system. J Bacteriol. 2015;197:1807–1818. doi: 10.1128/JB.02614-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Yang T, Talgan Y, Paharik AE, Horswill AR, Blackwell HE. Structure-function analyses of a staphylococcus epidermidis autoinducing peptide reveals motifs critical for agrc-type receptor modulation. ACS Chem Biol. 2016;11:1982–1991. doi: 10.1021/acschembio.6b00120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Yu Q, Lepp D, Mehdizadeh GI, Wu T, Zhou H, Yin X, Yu H, Prescott JF, Nie SP, Xie MY. The agr-like quorum sensing system is required for necrotic enteritis pathogenesis in poultry caused by clostridium perfringens. Infect Immun. 2017;85:e00975-16. doi: 10.1128/IAI.00975-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Fontaine L, Boutry C, Guédon E, Guillot A, Ibrahim M, Grossiord B, Hols P. Quorum-sensing regulation of the production of blp bacteriocins in Streptococcus thermophilus. J Bacteriol. 2007;189:7195–7205. doi: 10.1128/JB.00966-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Håvarstein LS. Increasing competence in the genus streptococcus. Mol Microbiol. 2010;78:541–544. doi: 10.1111/j.1365-2958.2010.07380.x. [DOI] [PubMed] [Google Scholar]
- 114.Mirouze N, Bergé MA, Soulet AL, Mortierbarrière I, Quentin Y, Fichant G, Granadel C, Noirotgros MF, Noirot P, Polard P. Direct involvement of dpra, the transformation-dedicated reca loader, in the shut-off of pneumococcal competence. Proc Natl Acad Sci USA. 2013;110:352–361. doi: 10.1073/pnas.1219868110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Reck M, Tomasch J, Wagner-Dobler I. The alternative sigma factor sigx controls bacteriocin synthesis and competence, the two quorum sensing regulated traits in streptococcus mutans. PLoS Genet. 2015;11:e1005353. doi: 10.1371/journal.pgen.1005353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Moreno-GãmS Sorg RA, Domenech A, Kjos M, Weissing FJ, van Doorn GS, Veening JW. Quorum sensing integrates environmental cues, cell density and cell history to control bacterial competence. Nat Commun. 2017;8:854–865. doi: 10.1038/s41467-017-00903-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Gallego del Sol F, Marina A. Structural basis of rap phosphatase inhibition by phr peptides. PLoS Biol. 2013;11:e1001511. doi: 10.1371/journal.pbio.1001511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Perchat S, Dubois T, Zouhir S, Gominet M, Poncet S, Lemy C, Aumont-Nicaise M, Deutscher J, Gohar M, Nessler S. A cell–cell communication system regulates protease production during sporulation in bacteria of the bacillus cereus group. Mol Microbiol. 2011;82:619–633. doi: 10.1111/j.1365-2958.2011.07839.x. [DOI] [PubMed] [Google Scholar]
- 119.Zouhir S, Perchat S, Nicaise M, Perez J, Guimaraes B, Lereclus D, Nessler S. Peptide-binding dependent conformational changes regulate the transcriptional activity of the quorum-sensor nprr. Nucleic Acids Res. 2013;41:7920–7933. doi: 10.1093/nar/gkt546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Grenha R, Slamti L, Nicaise M, Refes Y, Lereclus D, Nessler S. Structural basis for the activation mechanism of the plcr virulence regulator by the quorum-sensing signal peptide papr. Proc Natl Acad Sci USA. 2013;110:1047–1052. doi: 10.1073/pnas.1213770110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Slamti L, Perchat S, Huillet E, Lereclus D. Quorum sensing in bacillus thuringiensis is required for completion of a full infectious cycle in the insect. Toxins. 2014;6:2239–2255. doi: 10.3390/toxins6082239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Chen Y, Bandyopadhyay A, Kozlowicz BK, Haemig HAH, Tai A, Hu WS, Dunny GM. Mechanisms of peptide sex pheromone regulation of conjugation in enterococcus faecalis. Microbiologyopen. 2017;6(e492):1–13. doi: 10.1002/mbo3.492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Mashburn-Warren L, Morrison DA, Federle MJ. A novel double-tryptophan peptide pheromone controls competence in streptococcus spp. via an rgg regulator. Mol Microbiol. 2010;78:589–606. doi: 10.1111/j.1365-2958.2010.07361.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Fleuchot B, Guillot A, Mézange C, Besset C, Chambellon E, Monnet V, Gardan R. Rgg-associated shp signaling peptides mediate cross-talk in streptococci. PLoS One. 2013;8:e66042. doi: 10.1371/journal.pone.0066042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Parashar V, Aggarwal C, Federle MJ, Neiditch MB. Rgg protein structure-function and inhibition by cyclic peptide compounds. Proc Natl Acad Sci USA. 2015;112:5177–5182. doi: 10.1073/pnas.1500357112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Haustenne L, Bastin G, Hols P, Fontaine L. Modeling of the comrs signaling pathway reveals the limiting factors controlling competence in Streptococcus thermophilus. Front Microbiol. 2015;6(1413):1–20. doi: 10.3389/fmicb.2015.01413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Talagas A, Fontaine L, Ledesma-García L, Mignolet J, Li de la Sierra-Gallay I, Lazar N, Aumont-Nicaise M, Federle MJ, Prehna G, Hols P, Nessler S. Structural insights into streptococcal competence regulation by the cell-to-cell communication system comrs. PLoS Pathog. 2016;12:e1005980. doi: 10.1371/journal.ppat.1005980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Underhill SAM, Shields RC, Kaspar JR, Haider M, Burne RA, Hagen SJ. Intracellular signaling through the comrs system in streptococcus mutans genetic competence. mSphere. 2018;3(5):e00444-18. doi: 10.1128/mSphere.00444-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Garcíacontreras R, Nuñezlópez L, Jassochávez R, Kwan BW, Belmont JA, Rangelvega A, Maeda T, Wood TK. Quorum sensing enhancement of the stress response promotes resistance to quorum quenching and prevents social cheating. ISME J. 2014;9(1):115–125. doi: 10.1038/ismej.2014.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Bassler BL, Wright M, Showalter RE, Silverman MR. Intercellular signalling in vibrio harveyi: sequence and function of genes regulating expression of luminescence. Mol Microbiol. 1993;9:773–786. doi: 10.1111/j.1365-2958.1993.tb01737.x. [DOI] [PubMed] [Google Scholar]
- 131.Chen X. Structural identification of a bacterial quorum-sensing signal containing boron. Nature. 2002;415:545–549. doi: 10.1038/415545a. [DOI] [PubMed] [Google Scholar]
- 132.Sedlmayer F, Hell D, Muller M, Auslander D, Fussenegger M. Designer cells programming quorum-sensing interference with microbes. Nat Commun. 2018;9(1822):1–13. doi: 10.1038/s41467-018-04223-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Lee JH, Lee J. Indole as an intercellular signal in microbial communities. FEMS Microbiol Rev. 2010;34:426–444. doi: 10.1111/j.1574-6976.2009.00204.x. [DOI] [PubMed] [Google Scholar]
- 134.Wang D, Ding X, Rather PN. Indole can act as an extracellular signal in Escherichia coli. J Bacteriol. 2001;183:4210–4216. doi: 10.1128/JB.183.14.4210-4216.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Bassler BL, Losick R. Bacterially speaking. Cell. 2006;125:237–246. doi: 10.1016/j.cell.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 136.De Keersmaecker SC, Sonck K, Vanderleyden J. Let luxs speak up in ai-2 signaling. Trends Microbiol. 2006;14:114–119. doi: 10.1016/j.tim.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 137.Pereira CS, Thompson JA, Xavier KB. Ai-2-mediated signalling in bacteria. FEMS Microbiol Rev. 2013;37:156–181. doi: 10.1111/j.1574-6976.2012.00345.x. [DOI] [PubMed] [Google Scholar]
- 138.Xavier KB, Bassler BL. Regulation of uptake and processing of the quorum-sensing autoinducer ai-2 in Escherichia coli. J Bacteriol. 2005;187:238–248. doi: 10.1128/JB.187.1.238-248.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Xavier KB, Bassler BL. Interference with ai-2-mediated bacterial cell–cell communication. Nature. 2005;437:750–753. doi: 10.1038/nature03960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Armbruster CE, Hong W, Pang B, Weimer KED, Juneau RA, Turner J, Swords WE. Indirect pathogenicity of haemophilus influenzae and moraxella catarrhalis in polymicrobial otitis media occurs via interspecies quorum signaling. Mbio. 2010;1:119–121. doi: 10.1128/mBio.00102-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Newton WA, Snell EE. Formation and interrelationships of tryptophanase and tryptophan synthetases in Escherichia coli. J Bacteriol. 1965;89:355–364. doi: 10.1128/jb.89.2.355-364.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Lee J, Jayaraman A, Wood TK. Indole is an inter-species biofilm signal mediated by sdia. BMC Microbiol. 2007;7:1–15. doi: 10.1186/1471-2180-7-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Chant EL, Summers DK. Indole signalling contributes to the stable maintenance of Escherichia coli multicopy plasmids. Mol Microbiol. 2007;63:35–43. doi: 10.1111/j.1365-2958.2006.05481.x. [DOI] [PubMed] [Google Scholar]
- 144.Lee HH, Molla MN, Cantor CR, Collins JJ. Bacterial charity work leads to population-wide resistance. Nature. 2010;467:82–85. doi: 10.1038/nature09354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Jintae L, Can A, Cirillo SLG, Cirillo JD, Wood TK. Indole and 7-hydroxyindole diminish Pseudomonas aeruginosa virulence. Microb Biotechnol. 2010;2:75–90. doi: 10.1111/j.1751-7915.2008.00061.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Vega NM, Allison KR, Khalil AS, Collins JJ. Signaling-mediated bacterial persister formation. Nat Chem Biol. 2011;8:431. doi: 10.1038/nchembio.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Kim YG, Lee JH, Cho MH, Lee J. Indole and 3-indolylacetonitrile inhibit spore maturation in paenibacillus alvei. BMC Microbiol. 2011;11:119. doi: 10.1186/1471-2180-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Catalin C, Field CM, Silvia PF, Keyser UF, Summers DK. Indole prevents Escherichia coli cell division by modulating membrane potential. Biochim Biophys Acta Biomembr. 2012;1818:1590–1594. doi: 10.1016/j.bbamem.2012.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Yee DC, Maynard JA, Wood TK. Rhizoremediation of trichloroethylene by a recombinant, root-colonizing pseudomonas fluorescens strain expressing toluene ortho-monooxygenase constitutively. Appl Environ Microbiol. 1998;64:112–118. doi: 10.1128/aem.64.1.112-118.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Han TH, Cho MH, Lee J. Indole oxidation enhances electricity production in an E. coli-catalyzed microbial fuel cell. Biotechnol Bioprocess E. 2014;19:126–131. [Google Scholar]
- 151.Lee JH, Kim YG, Kim CJ, Lee JC, Cho MH, Lee J. Indole-3-acetaldehyde from Rhodococcus sp. Bfi 332 inhibits Escherichia coli o157:h7 biofilm formation. Appl Microbiol Biotechnol. 2012;96:1071–1078. doi: 10.1007/s00253-012-3881-y. [DOI] [PubMed] [Google Scholar]
- 152.Lee JH, Cho HS, Kim Y, Kim JA, Banskota S, Cho MH, Lee J. Indole and 7-benzyloxyindole attenuate the virulence of Staphylococcus aureus. Appl Microbiol Biotechnol. 2013;97:4543–4552. doi: 10.1007/s00253-012-4674-z. [DOI] [PubMed] [Google Scholar]
- 153.Lee JH, Kim YG, Baek KH, Cho MH, Lee J. The multifaceted roles of the interspecies signalling molecule indole in Agrobacterium tumefaciens. Environ Microbiol. 2015;17:1234–1244. doi: 10.1111/1462-2920.12560. [DOI] [PubMed] [Google Scholar]
- 154.Chu W, Zere TR, Weber MM, Wood TK, Whiteley M, Hidalgo-Romano B, Jr, Valenzuela E, Mclean RJ. Indole production promotes Escherichia coli mixed-culture growth with Pseudomonas aeruginosa by inhibiting quorum signaling. Appl Environ Microbiol. 2012;78:411–419. doi: 10.1128/AEM.06396-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Lee JH, Kim YG, Kim M, Kim E, Choi H, Kim Y, Lee J. Indole-associated predator–prey interactions between the nematode Caenorhabditis elegans and bacteria. Environ Microbiol. 2017;19:1776–1790. doi: 10.1111/1462-2920.13649. [DOI] [PubMed] [Google Scholar]
- 156.Tomberlin JK, Crippen TL, Wu G, Griffin AS, Wood TK, Kilner RM. Indole: an evolutionarily conserved influencer of behavior across kingdoms. BioEssays. 2017;39(1600203):1–12. doi: 10.1002/bies.201600203. [DOI] [PubMed] [Google Scholar]
- 157.Costerton JW, Cheng KJ, Geesey GG, Ladd TI, Nickel JC, Dasgupta M, Marrie TJ. Bacterial biofilms in nature and disease. Annu Rev Microbiol. 1987;41:435–464. doi: 10.1146/annurev.mi.41.100187.002251. [DOI] [PubMed] [Google Scholar]
- 158.Van AH, Van DP, Coenye T. Molecular mechanisms of antimicrobial tolerance and resistance in bacterial and fungal biofilms. Trends Microbiol. 2014;22:326–333. doi: 10.1016/j.tim.2014.02.001. [DOI] [PubMed] [Google Scholar]
- 159.Lebeaux D, Ghigo JM, Beloin C. Biofilm-related infections: bridging the gap between clinical management and fundamental aspects of recalcitrance toward antibiotics. Microbiol Mol Biol Rev. 2014;78:510–543. doi: 10.1128/MMBR.00013-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Parsek MR, Greenberg EP. Sociomicrobiology: the connections between quorum sensing and biofilms. Trends Microbiol. 2005;13:27–33. doi: 10.1016/j.tim.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 161.Deng YY, Wu JE, Tao F, Zhang LH. Listening to a new language: Dsf-based quorum sensing in gram-negative bacteria. Chem Rev. 2011;111:160–173. doi: 10.1021/cr100354f. [DOI] [PubMed] [Google Scholar]
- 162.Lebeer S, Claes IJ, Verhoeven TL, Shen C, Lambrichts I, Ceuppens JL, Vanderleyden J, De Keersmaecker SC. Impact of luxs and suppressor mutations on the gastrointestinal transit of Lactobacillus rhamnosus gg. Appl Environ Microbiol. 2008;74:4711–4718. doi: 10.1128/AEM.00133-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Sun Z, He X, Brancaccio VF, Yuan J, Riedel CU. Bifidobacteria exhibit luxs-dependent autoinducer 2 activity and biofilm formation. PLoS One. 2014;9:e88260. doi: 10.1371/journal.pone.0088260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Li H, Li X, Wang Z, Fu Y, Ai Q, Dong Y, Yu J. Autoinducer-2 regulates pseudomonas aeruginosa pao1 biofilm formation and virulence production in a dose-dependent manner. BMC Microbiol. 2015;15:1–8. doi: 10.1186/s12866-015-0529-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Anderson JK, Huang JY, Wreden C, Sweeney EG, Goers J, Remington SJ, Guillemin K. Chemorepulsion from the quorum signal autoinducer-2 promotes Helicobacter pylori biofilm dispersal. mBio. 2015;6:e00379. doi: 10.1128/mBio.00379-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Xue T, Ni J, Shang F, Chen X, Zhang M. Autoinducer-2 increases biofilm formation via an ica- and bhp-dependent manner in staphylococcus epidermidis rp62a. Microbes Infect. 2015;17:345–352. doi: 10.1016/j.micinf.2015.01.003. [DOI] [PubMed] [Google Scholar]
- 167.Laganenka L, Colin R, Sourjik V. Corrigendum: chemotaxis towards autoinducer 2 mediates autoaggregation in Escherichia coli. Nat Commun. 2016;7:13979. doi: 10.1038/ncomms13979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Papenfort K, Silpe JE, Schramma KR, Cong JP, Seyedsayamdost MR, Bassler BL. A vibrio cholerae autoinducer-receptor pair that controls biofilm formation. Nat Chem Biol. 2017;13:551–557. doi: 10.1038/nchembio.2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Liu L, Wu R, Zhang J, Shang N, Li P. D-ribose interferes with quorum sensing to inhibit biofilm formation of Lactobacillus paraplantarum l-zs9. Front Microbiol. 2017;8:1860. doi: 10.3389/fmicb.2017.01860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Ryan RP, Fouhy Y, Garcia BF, Watt SA, Niehaus K, Yang L, Tolker-Nielsen T, Dow JM. Interspecies signalling via the Stenotrophomonas maltophilia diffusible signal factor influences biofilm formation and polymyxin tolerance in Pseudomonas aeruginosa. Mol Microbiol. 2008;68:75–86. doi: 10.1111/j.1365-2958.2008.06132.x. [DOI] [PubMed] [Google Scholar]
- 171.Dean SN, Chung MC, van Hoek ML. Burkholderia diffusible signal factor signals to Francisella novicida to disperse biofilm and increase siderophore production. Appl Environ Microbiol. 2015;81:7057–7066. doi: 10.1128/AEM.02165-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.An SQ, Tang JL. Diffusible signal factor signaling regulates multiple functions in the opportunistic pathogen Stenotrophomonas maltophilia. BMC Res Notes. 2018;11:569–575. doi: 10.1186/s13104-018-3690-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Krzyzek P, Gosciniak G. A proposed role for diffusible signal factors in the biofilm formation and morphological transformation of Helicobacter pylori. Turk J Gastroenterol. 2018;29:7–13. doi: 10.5152/tjg.2017.17349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Deng YY, Lim A, Lee J, Chen SH, An SW, Dong YH, Zhang LH. Diffusible signal factor (dsf) quorum sensing signal and structurally related molecules enhance the antimicrobial efficacy of antibiotics against some bacterial pathogens. BMC Microbiol. 2014;14:51–59. doi: 10.1186/1471-2180-14-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Barel V, Chalupowicz L, Barash I, Sharabani G, Reuven M, Dror O, Burdman S, Manulis-Sasson S. Virulence and in planta movement of Xanthomonas hortorum pv. Pelargonii are affected by the diffusible signal factor (dsf)-dependent quorum sensing system. Mol Plant Pathol. 2015;16:710–723. doi: 10.1111/mpp.12230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Dow JM. Diffusible signal factor-dependent quorum sensing in pathogenic bacteria and its exploitation for disease control. J Appl Microbiol. 2017;122:2–11. doi: 10.1111/jam.13307. [DOI] [PubMed] [Google Scholar]
- 177.Allen RC, Popat R, Diggle SP, Brown SP. Targeting virulence: can we make evolution-proof drugs? Nat Rev Microbiol. 2014;12:300–308. doi: 10.1038/nrmicro3232. [DOI] [PubMed] [Google Scholar]
- 178.Dong YH, Wang LH, Xu JL, Zhang HB, Zhang XF, Zhang LH. Quenching quorum-sensing-dependent bacterial infection by an n-acyl homoserine lactonase. Nature. 2001;411:813–817. doi: 10.1038/35081101. [DOI] [PubMed] [Google Scholar]
- 179.Shen G, Rajan R, Zhu J, Bell CE, Pei D. Design and synthesis of substrate and intermediate analogue inhibitors of s-ribosylhomocysteinase. J Med Chem. 2006;49:3003–3011. doi: 10.1021/jm060047g. [DOI] [PubMed] [Google Scholar]
- 180.Zhang M, Jiao X, Hu Y, Sun L. Attenuation of edwardsiella tarda virulence by small peptides that interfere with luxs/autoinducer type 2 quorum sensing. Appl Environ Microbiol. 2009;75:3882–3890. doi: 10.1128/AEM.02690-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Ni N, Li M, Wang J, Wang B. Inhibitors and antagonists of bacterial quorum sensing. Med Res Rev. 2009;29:65–124. doi: 10.1002/med.20145. [DOI] [PubMed] [Google Scholar]
- 182.Brackman G, Defoirdt T, Miyamoto C, Bossier P, Calenbergh SV, Nelis H, Coenye T. Cinnamaldehyde and cinnamaldehyde derivatives reduce virulence in vibriospp. By decreasing the DNA-binding activity of the quorum sensing response regulator luxr. BMC Microbiol. 2008;8:149. doi: 10.1186/1471-2180-8-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Brackman G, Cos P, Maes L, Nelis HJ, Coenye T. Quorum sensing inhibitors increase the susceptibility of bacterial biofilms to antibiotics in vitro and in vivo. Antimicrob Agents Chemother. 2011;55:2655–2661. doi: 10.1128/AAC.00045-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Christensen QH, Grove TL, Booker SJ, Greenberg EP. A high-throughput screen for quorum-sensing inhibitors that target acyl-homoserine lactone synthases. Proc Natl Acad Sci USA. 2013;110:13815–13820. doi: 10.1073/pnas.1313098110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.O’Loughlin CT, Miller LC, Siryaporn A, Drescher K, Semmelhack MF, Bassler BL. A quorum-sensing inhibitor blocks pseudomonas aeruginosa virulence and biofilm formation. Proc Natl Acad Sci USA. 2013;110:17981–17986. doi: 10.1073/pnas.1316981110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Starkey M, Lepine F, Maura D, Bandyopadhaya A, Lesic B, He J, Kitao T, Righi V, Milot S, Tzika A. Identification of anti-virulence compounds that disrupt quorum-sensing regulated acute and persistent pathogenicity. PLoS Pathog. 2014;10:e1004321. doi: 10.1371/journal.ppat.1004321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Ouyang J, Sun F, Feng W, Sun Y, Qiu X, Xiong L, Liu Y, Chen Y. Quercetin is an effective inhibitor of quorum sensing, biofilm formation and virulence factors in Pseudomonas aeruginosa. J Appl Microbiol. 2016;120:966–974. doi: 10.1111/jam.13073. [DOI] [PubMed] [Google Scholar]
- 188.Zerfaß C, Chen J, Soyer OS. Engineering microbial communities using thermodynamic principles and electrical interfaces. Curr Opin Biotechnol. 2018;50:121–127. doi: 10.1016/j.copbio.2017.12.004. [DOI] [PubMed] [Google Scholar]
- 189.McCardell RD, Huang S, Green LN, Murray RM. Control of bacterial population density with population feedback and molecular sequestration. 2017 doi: 10.1101/225045. [DOI] [Google Scholar]
- 190.Saeidi N, Wong CK, Lo TM, Nguyen HX, Ling H, Leong SS, Poh CL, Chang MW. Engineering microbes to sense and eradicate Pseudomonas aeruginosa, a human pathogen. Mol Syst Biol. 2011;7(521):1–11. doi: 10.1038/msb.2011.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Gupta S, Bram EE, Weiss R. Genetically programmable pathogen sense and destroy. ACS Synth Biol. 2013;2:715–723. doi: 10.1021/sb4000417. [DOI] [PubMed] [Google Scholar]
- 192.Hwang IY, Tan MH, Koh E, Ho CL, Poh CL, Chang MW. Reprogramming microbes to be pathogen-seeking killers. ACS Synth Biol. 2014;3:228–237. doi: 10.1021/sb400077j. [DOI] [PubMed] [Google Scholar]
- 193.Hwang IY, Koh E, Wong A, March JC, Bentley WE, Lee YS, Chang MW. Engineered probiotic Escherichia coli can eliminate and prevent Pseudomonas aeruginosa gut infection in animal models. Nat Commun. 2017;8:15028. doi: 10.1038/ncomms15028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Zhao L, Zhang F, Ding X, Wu G, Lam YY, Wang X, Fu H, Xue X, Lu C, Ma J. Gut bacteria selectively promoted by dietary fibers alleviate type 2 diabetes. Science. 2018;359:1151–1156. doi: 10.1126/science.aao5774. [DOI] [PubMed] [Google Scholar]
- 195.Wilck N, Matus MG, Kearney SM, Olesen SW, Forslund K, Bartolomaeus H, Haase S, Mähler A, Balogh A, Markó L. Salt-responsive gut commensal modulates th17 axis and disease. Nature. 2017;551:585–589. doi: 10.1038/nature24628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Crotty MP, Jackson PJ. Terminal room disinfection: how much betr can it get? Lancet. 2017;389:765–766. doi: 10.1016/S0140-6736(16)32412-6. [DOI] [PubMed] [Google Scholar]
- 197.Routy B, Le CE, Derosa L, Cpm D, Alou MT, DaillãRe R, Fluckiger A, Messaoudene M, Rauber C, Roberti MP. Gut microbiome influences efficacy of pd-1-based immunotherapy against epithelial tumors. Science. 2018;359:91–97. doi: 10.1126/science.aan3706. [DOI] [PubMed] [Google Scholar]
- 198.Zou J, Chassaing B, Singh V, Pellizzon M, Ricci M, Fythe MD, Kumar MV, Gewirtz AT. Fiber-mediated nourishment of gut microbiota protects against diet-induced obesity by restoring il-22-mediated colonic health. Cell Host Microbe. 2018;23:41–53. doi: 10.1016/j.chom.2017.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Sun Z, Grimm V, Riedel CU. Ai-2 to the rescue against antibiotic-induced intestinal dysbiosis? Trends Microbiol. 2015;23:327–328. doi: 10.1016/j.tim.2015.04.002. [DOI] [PubMed] [Google Scholar]
- 200.Kamada N, Kim YG, Sham HP, Vallance BA, Puente JL, Martens EC, Núñez G. Regulated virulence controls the ability of a pathogen to compete with the gut microbiota. Science. 2012;336:1325–1329. doi: 10.1126/science.1222195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Dandekar AA, Greenberg EP. Bacterial quorum sensing and metabolic incentives to cooperate. Science. 2012;338:264–266. doi: 10.1126/science.1227289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Bongaerts GP, Severijnen RS. A reassessment of the propatria study and its implications for probiotic therapy. Nat Biotechnol. 2016;34:55–63. doi: 10.1038/nbt.3436. [DOI] [PubMed] [Google Scholar]
- 203.Subramanian S, Huq S, Yatsunenko T, Haque R, Mahfuz M, Alam MA, Benezra A, Destefano J, Meier MF, Muegge BD. Persistent gut microbiota immaturity in malnourished bangladeshi children. Nature. 2014;510:417–421. doi: 10.1038/nature13421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Hsiao A, Ahmed AM, Subramanian S, Griffin NW, Drewry LL, Jr, Petri WA, Haque R, Ahmed T, Gordon JI. Members of the human gut microbiota involved in recovery from vibrio cholerae infection. Nature. 2014;515:423–426. doi: 10.1038/nature13738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Daniel R, Rubens JR, Sarpeshkar R, Lu TK. Synthetic analog computation in living cells. Nature. 2013;497:619–623. doi: 10.1038/nature12148. [DOI] [PubMed] [Google Scholar]
- 206.Bivar Xavier K. Bacterial interspecies quorum sensing in the mammalian gut microbiota. C R Biol. 2018;341:297–299. doi: 10.1016/j.crvi.2018.03.006. [DOI] [PubMed] [Google Scholar]
- 207.Gilmore MS, Lebreton F, Van SW. Genomic transition of enterococci from gut commensals to leading causes of multidrug-resistant hospital infection in the antibiotic era. Curr Opin Microbiol. 2013;16:10–16. doi: 10.1016/j.mib.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Borrero J, Chen Y, Dunny GM, Kaznessis YN. Modified lactic acid bacteria detect and inhibit multiresistant enterococci. ACS Synth Biol. 2015;4:299–306. doi: 10.1021/sb500090b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Arias CA, Murray BE. The rise of the enterococcus: beyond vancomycin resistance. Nat Rev Microbiol. 2012;10:266–278. doi: 10.1038/nrmicro2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Coyte KZ, Rakoff-Nahoum S. Understanding competition and cooperation within the mammalian gut microbiome. Curr Biol. 2019;29:538–544. doi: 10.1016/j.cub.2019.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Li Q, Ren Y, Fu X. Inter-kingdom signaling between gut microbiota and their host. Cell Mol Life Sci. 2019;76:2383–2389. doi: 10.1007/s00018-019-03076-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Mirzaei MK, Maurice CF. Menage a trois in the human gut: interactions between host, bacteria and phages. Nat Rev Microbiol. 2017;15:397–408. doi: 10.1038/nrmicro.2017.30. [DOI] [PubMed] [Google Scholar]
- 213.Bondydenomy J, Pawluk A, Maxwell KL, Davidson AR. Bacteriophage genes that inactivate the crispr/cas bacterial immune system. Nature. 2013;493:429–432. doi: 10.1038/nature11723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Patterson AG, Yevstigneyeva MS, Fineran PC. Regulation of crispr-cas adaptive immune systems. Curr Opin Microbiol. 2017;37:1–7. doi: 10.1016/j.mib.2017.02.004. [DOI] [PubMed] [Google Scholar]
- 215.Patterson A, Jackson S, Taylor C, Evans G, Salmond GC, Przybilski R, Staals RJ, Fineran P. Quorum sensing controls adaptive immunity through the regulation of multiple crispr-cas systems. Mol Cell. 2016;64:1102–1108. doi: 10.1016/j.molcel.2016.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Brophy JA, Voigt CA. Principles of genetic circuit design. Nat Methods. 2014;11:508–520. doi: 10.1038/nmeth.2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Buescher JM. Global network reorganization during dynamic adaptations of bacillus subtilis metabolism. Science. 2012;335:1099–1103. doi: 10.1126/science.1206871. [DOI] [PubMed] [Google Scholar]
- 218.Woolston BM, Edgar S, Stephanopoulos G. Metabolic engineering: past and future. Annu Rev Chem Biomol Eng. 2013;4:259–288. doi: 10.1146/annurev-chembioeng-061312-103312. [DOI] [PubMed] [Google Scholar]
- 219.Orth JD, Thiele I, Palsson BØ. What is flux balance analysis? Nat Biotechnol. 2010;28:245–248. doi: 10.1038/nbt.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Meadows AL, Karnik R, Lam H, Forestell S, Snedecor B. Application of dynamic flux balance analysis to an industrial escherichia coli fermentation. Metab Eng. 2010;12:150–160. doi: 10.1016/j.ymben.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 221.Hill A. The combinations of haemoglobin with oxygen and with carbon monoxide. I. Biochem J. 1913;7:577–586. doi: 10.1042/bj0150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Schmidt SK, Simkins S, Alexander M. Models for the kinetics of biodegradation of organic compounds not supporting growth. Appl Environ Microbiol. 1985;50:323–331. doi: 10.1128/aem.50.2.323-331.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.English BP, Min W, van Oijen AM, Lee KT, Luo G, Sun H, Cherayil BJ, Kou SC, Xie XS. Ever-fluctuating single enzyme molecules: Michaelis-menten equation revisited. Nat Chem Biol. 2006;2:87–94. doi: 10.1038/nchembio759. [DOI] [PubMed] [Google Scholar]
- 224.Basu Subhayu, Gerchman Yoram, Collins Cynthia H, Arnold FH, Weiss R. A synthetic multicellular system for programmed pattern formation. Nature. 2005;434:1130–1134. doi: 10.1038/nature03461. [DOI] [PubMed] [Google Scholar]
- 225.Song H, Stephen P, Meagan G, You L. Spatiotemporal modulation of biodiversity in a synthetic chemical-mediated ecosystem. Nat Chem Biol. 2009;5:929–935. doi: 10.1038/nchembio.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Kong W, Meldgin DR, Collins JJ, Lu T. Designing microbial consortia with defined social interactions. Nat Chem Biol. 2018;14:821–829. doi: 10.1038/s41589-018-0091-7. [DOI] [PubMed] [Google Scholar]
- 227.Mads K, Elston TC, Blake WJ, Collins JJ. Stochasticity in gene expression: from theories to phenotypes. Nat Rev Genet. 2005;6:451–464. doi: 10.1038/nrg1615. [DOI] [PubMed] [Google Scholar]
- 228.Tian T, Burrage K. Stochastic models for regulatory networks of the genetic toggle switch. Proc Natl Acad Sci USA. 2006;103:8372–8377. doi: 10.1073/pnas.0507818103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Prindle A, Liu J, Asally M, Ly S, Garciaojalvo J, Süel GM. Ion channels enable electrical communication within bacterial communities. Nature. 2015;527:59–63. doi: 10.1038/nature15709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Popkin G. Bacteria use brainlike bursts of electricity to communicate. New York: Quanta Magazine; 2017. [Google Scholar]
- 231.Liu J, Zhou J, Wang L, Ma Z, Zhao G, Ge Z, Zhu H, Qiao J. Improving nitrogen source utilization from defatted soybean meal for nisin production by enhancing proteolytic function of Lactococcus lactis f44. Sci Rep. 2017;7:6189. doi: 10.1038/s41598-017-06537-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Wu H, Song S, Tian K, Zhou D, Wang B, Liu J, Zhu H, Qiao J. A novel small rna s042 increases acid tolerance in Lactococcus lactis f44. Biochem Biophys Res Commun. 2018;500:544–549. doi: 10.1016/j.bbrc.2018.04.069. [DOI] [PubMed] [Google Scholar]
- 233.Song AA, In LLA, Lim SHE, Rahim RA. A review on Lactococcus lactis: from food to factory. Microb Cell Fact. 2017;16:55. doi: 10.1186/s12934-017-0669-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Guo CJ, Chang FY, Wyche TP, Backus KM, Acker TM, Funabashi M, Mao T, Donia MS, Nayfach S, Pollard KS. Discovery of reactive microbiota-derived metabolites that inhibit host proteases. Cell. 2017;168:517–526. doi: 10.1016/j.cell.2016.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Foster KR, Schluter J, Coyte KZ, Rakoff-Nahoum S. The evolution of the host microbiome as an ecosystem on a leash. Nature. 2017;548:43–51. doi: 10.1038/nature23292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Fernandez L, Rodriguez A, Garcia P. Phage or foe: an insight into the impact of viral predation on microbial communities. ISME J. 2018;12:1171–1179. doi: 10.1038/s41396-018-0049-5. [DOI] [PMC free article] [PubMed] [Google Scholar]