Abstract
ZNFX1 anti-sense RNA 1 (ZFAS1) has been indicated in the tumorigenesis of various human cancers. However, the role of ZFAS1 in prostate cancer (PCa) progression and the underlying mechanisms remain incompletely understood. In the present study, we discovered that ZFAS1 is upregulated in PCa and that ZFAS1 overexpression predicted poor clinical outcomes. ZFAS1 overexpression notably promoted the proliferation, invasion, and epithelial–mesenchymal transition of PCa cells. Furthermore, we not only discovered that miR-27a/15a/16 are targeted by ZFAS1, which binds to their miRNA-response elements, but also revealed their tumor suppressor roles in PCa. We also identified that the Hippo pathway transducer YAP1, as well as its cooperator, TEAD1, are common downstream targets of miR-27a/15a/16. In addition, H3K9 demethylase KDM3A was found to be another target gene of miR-27a. Importantly, YAP1, TEAD1, and KDM3A all act as strong c-Myc inducers in an androgen-independent manner. Taken together, we suggest a regulatory network in which ZFAS1 is capable of enhancing c-Myc expression by inducing the expression of YAP1, TEAD1, and KDM3A through crosstalk with their upstream miRNAs, thereby globally promoting prostate cancer tumorigenesis.
Electronic supplementary material
The online version of this article (10.1007/s00018-019-03226-x) contains supplementary material, which is available to authorized users.
Keywords: ZNFX1 anti-sense RNA 1, Prostate cancer, Competing endogenous RNAs, c-Myc
Introduction
Prostate cancer (PCa) ranks as the second most frequent cancer and is the fifth leading cause of cancer death in men around the world [1]. In the United States, prostate cancer is the most diagnosed cancer among men [1, 2]. Although diagnostic screening and improved treatments have been applied for the detection and management of PCa at an earlier stage, which results in the postponement of death for patients with metastatic PCa, it still remains an important health problem [3]. Studies have shown that androgen receptor (AR) plays a key role in the tumorigenesis and progression of prostate cancer, and aberrant AR activation causes resistance to androgen-deprivation therapy (ADT), which remains the main line of treatment for advanced prostate cancer [4, 5]. Therefore, identifying the molecular mechanism underlying AR activity and the progression of prostate cancer could greatly improve the treatment for castration-resistant prostate cancer (CRPC) [5].
Long noncoding RNAs (lncRNAs) are a class of non-protein-coding RNAs with lengths of more than 200 nucleotides [6]. Accumulating evidence has shown that lncRNAs participate in multiple biological processes, such as cell differentiation [7], autophagy [8], chemotherapy resistance [9], and chromatin modification [10]. Moreover, lncRNA expression is frequently dysregulated in various types of cancers, and specific lncRNAs can function as oncogenes or tumor suppressors in different cancers, including breast cancer [9], bladder cancer [11], hepatocellular carcinoma [12], and gastric cancer [10]. LncRNAs exert their functions through a variety of mechanisms, and recent studies have revealed that they serve as competing endogenous RNAs (ceRNAs) and crosstalk with mRNAs by sponging their common microRNAs (miRNAs), therefore regulating the expression of oncogene or tumor suppressor genes at the posttranscriptional level [13–15]. For example, in gastric cancer, KRTAP5-1/KRTAP5-2 Anti-sense RNA 1 (KRTAP5-AS1) and Tubulin Beta 2A Class IIa (TUBB2A) regulate Claudin 4 (CLDN4) gene expression through ceRNA-mediated miR-596 and miR-3620-3p degradation [16]; HOX Transcript Anti-sense RNA (HOTAIR) promotes renal carcinoma progression via the regulation of miR-138/200c/204/217 as a ceRNA [17]. Some lncRNAs have been highlighted in prostate cancer in terms of their functions as highly androgen-specific. Nuclear-enriched abundant transcript 1 (NEAT1) is regulated by estrogen receptor alpha (ERα) signaling and drives the oncogenic growth of PCa cells by altering the epigenetic landscape of target gene promoters to favor transcription [18]. HOTAIR binds to the AR protein, blocks its interaction with the E3 ubiquitin ligase MDM2 proto-oncogene (MDM2), and prevents AR ubiquitination and degradation [19]. Nonetheless, further work is necessary to identify the functional and prognostic significance of lncRNAs in prostate cancer.
ZNFX1 anti-sense RNA 1 (ZFAS1), a novel lncRNA transcribed from the anti-sense orientation of zinc-finger NFX-type containing 1 (ZNFX1), is located on chromosome 20q13.13 [20]. Recent studies have revealed that the aberrant expression of ZFAS1 contributes to the development, recurrence, and chemotherapeutic sensitivity of various human cancers, including gastric cancer [21], hepatocellular carcinoma [22], colorectal cancer [23], and ovarian cancer [24]. The mechanisms through which ZFAS1 is involved in tumorigenesis are reported as the participation in p53 signaling [23], cell cycle arrest [21], and epithelial–mesenchymal transition (EMT) [22, 23]. The proto-oncogene c-Myc is known to be widely involved in many cancers, in which its expression is estimated to be either elevated or deregulated in up to 70% of human cancers [25, 26]. It has been established that c-Myc mRNA and protein levels are upregulated in human prostate cancer tissues, and its expression is associated with the progression and recurrence of prostate cancer [27–29]. Multiple mechanisms are proposed to regulate c-Myc expression in cancers. Among them, lncRNAs, such as FILNC1 and CCAT1-L, crosstalk with c-Myc and regulate c-Myc expression at the transcriptional or posttranscriptional level [30, 31]. In addition, a recent study revealed that KDM3A, a histone demethylase, promotes prostate cancer cell proliferation and survival [32] by controlling c-Myc expression though recruiting AR to the c-Myc gene promoter and inducing H3K9 demethylation.
In the present study, we discovered that lncRNA ZFAS1 is essential for the proliferation and invasion of PCa cells. Mechanistically, it promotes c-Myc expression by serving as a ceRNA and releasing three key c-Myc regulators, KDM3A, YAP1, and TEAD1, from being suppressed by their common upstream microRNAs. Here, we propose a regulatory network in prostate cancer cells. c-Myc is regulated by ZFAS1 through ceRNA-mediated miRNA evasion. Our study unravels an undisclosed role of ZFAS1 in regulating c-Myc and promoting the tumorigenesis of prostate cancer.
Materials and methods
Tissues
The benign prostate tissues were freshly collected from 61 patients who were diagnosed with benign prostate hyperplasia (BPH) and underwent transurethral resection of prostate. A total of 52 prostate cancer tissues and matched non-tumorous adjacent prostate tissues were obtained from patients who were pathologically diagnosed with prostate cancer and received surgical resection. All the patients were hospitalized and received surgical treatment at the Urology Department at the First Hospital of China Medical University (Shenyang, China). The study was conducted according to an institutional review board-approved protocol (2012–33) by Medical Ethics Committee of the First Affiliated Hospital of China Medical University, and written informed consent was obtained from each patient for surgery and research purposes. The clinical pathological sections of prostate cancer with Gleason score 3–4 were provided by Department of Pathology at The First Hospital of China Medical University, and the pathological diagnosis and analysis of immunochemistry staining result in this study were made in collaboration with Department of Pathology.
Cell culture
LNCaP, PC-3, and RV-1 cells were purchased from the cell bank of Chinese Academy of Sciences (Shanghai, China). These cells are maintained in RPMI 1640 medium supplemented with 10% FBS and antibiotics, and regularly tested to ensure that they are mycoplasma-free.
RNA isolation and real-time quantitative PCR
Total RNA was extracted from cultured cell lines using TRIzol reagent (Invitrogen) and reverse transcribed with random primers using PrimeScript™ RT Master Mix (Perfect Real Time; Takara Biotechnology Co. Ltd., Dalian, China) according to the manufacturer’s instructions. For microRNA detection, cDNA synthesis and quantitative real-time PCR were performed using a mercury LNA™ Universal RT microRNA PCR kit (Exiqon, Skelstedet, Vedbaek, Denmark). qRT-PCR was performed using SYBR® Premix Ex Taq™ (Tli RNaseH Plus; Takara Biotechnology Co. Ltd., Dalian, China) and LightCyclerTM 480 II system (Roche, Basel, Switzerland). β-actin and U6 snRNA were employed as endogenous controls for mRNA/lncRNA and miRNA, respectively. The primers used to amplify the target genes are listed in Supplementary Table S3. The relative levels of gene expression were quantified and analyzed using LightCyclerTM 480 software 1.5.1.6.2 (Roche, Basel, Switzerland). The real-time value for each sample was averaged and compared using the 2-ΔΔCt method. Three independent experiments were performed to analyze the relative gene expression.
Luciferase reporter assay
Luciferase reporters were generated based on the psiCHECK2 vector (Promega) and a Dual Luciferase Reporter Assay Kit (Promega) according to the manufacturer’s protocol. To construct psiCHECK-ZFAS1, full-length sequences of ZFAS1 (1075nt, UCSC accession no. uc061xqc.1) were PCR amplified and subcloned into the psiCHECK2 vector. The luciferase reporter construct was co-transfected with agomir of miR-181a-5p, miR-181c-5p, miR-27a-3p, miR-15a-5p, miR-497-5p, and miR-16-5p into the PC-3 and RV-1 cells by Lipofectamine 3000 (Invitrogen) according to the manufacturer’s guidelines. To construct the psiCHECK-ZFAS1 mutant, sequence of ZFAS1 with the predicted MREs of either mR-27a/15a/16 mutated was subcloned into the psiCHECK2 vector, and the luciferase report was relatively co-transfected with negative control or agomir of miR-27a-3p, miR-15a-5p or miR-16-5p as indicated. To test the interaction between ZFAS1 and miR-27a/15a/16, wild MRE sequence of ZFAS1 gene and relative mutant type were cloned into the psiCHECK2 vector. The sequence was listed in Supplementary Fig. 3e. The relative luciferase activity was measured by Synergy HTX multi-mode microplate reader (BioTek).
Western blotting analysis
Antibodies to KDM3A (ab106456), c-Myc (ab32072), YAP1 (ab52771), TEAD1 (ab197589), and GAPDH (ab181602) (all from abcam) were used according to the manufacturer’s recommendations. Cells were harvested in RIPA lysis buffer (Beyotime, Beijing, China) and boiled for 10 min at 90 °C. 50 μg of protein extract from cultured cells were separated by 10% SDS–polyacrylamide gel electrophoresis (SDS-PAGE), and the gels were subsequently electrotransferred onto polyvinylidene difluoride (PVDF) membranes (Millipore), followed by incubating with the indicated primary antibodies in 5% nonfat milk in TBS-T overnight at 4 °C. On the next day, the membranes were washed for 15 min each and immediately incubated with anti-rabbit or anti-mouse horseradish peroxidase-conjugated secondary antibodies for 1 h at 37 °C. The immunobands were visualized using ECL reagents (Transgen Biotechnology, Beijing, China) on a MicroChemi Chemiluminescent Imaging System (DNR Bio-Imaging Systems, Mahale HaHamisha, Jerusalem, Israel). The densitometric values were calculated by Image J 1.46r software (Wayne Rasband, National Institutes of Health, Bethesda, MA, USA), and the ratios of target protein to GAPDH were used to conduct the statistical analysis.
Plasmids and transfection
The shRNAs were constructed using pLKO.1 vector according to Addgene TRC Cloning Protocol. The micro RNA agomir and antagomir were synergized by and purchased from GenePharma (Shanghai, China). Transfections were performed using the Lipofectamine 3000 Reagent (Invitrogen) following the manufacturer’s protocol. Final concentrations for miRNA agomir or plasmids were 50 nM and 0.75 μg/ml. Cells were cultured in a 6-well plate with 2 ml culture medium. The concentration for lentivirus transduction was 5 × 106 transducing units of lentivirus. The stable cell lines were constructed using puromycin (200 μg ml−1).
Engineered prostate cancer cells
RV-1 cells and PC-3 cells were used as parallel cell lines of prostate cancer in this study. ZFAS1 gene was subcloned into pLVX-IRES-zsGreen1 vector, and the empty vector was used as the control. Lentiviral vector-encoding ZFAS1 was packaged in 293 T cells by the calcium phosphate transfection. Prostate cancer cells were transduced with the supernatant of lentiviral particles. In this study, ZFAS1 transduced prostate cancer cells were named as pLVX-NC RV-1 or PC-3 cells, pLVX-ZFAS1 RV-1 or PC-3 cells. Similarly, miRNAs (miR-27a, miR-15a or miR-16) were overexpressed or knocked down in prostate cancer cells by transfecting the agomir or antagomir or the relative scramble control (miR-NC or Ant-miR) into the cells.
Proliferation assay
The capacity for cellular proliferation was measured using a Cell Counting Kit-8 (CCK-8) (Dojindo, Tokyo, Japan) and a cell colony formation assay, according to the manufacturer’s protocol. The absorbance value was measured at 450 nm to determine cell viability using a 96-well plate reader. For cell colony formation assay, the cells were plated in 24-well plates (300 cells per well) and incubated for 14 days in complete medium. Colonies were fixed with 10% formaldehyde for 10 min and stained with 1.0% crystal violet for 5 min. The number of colonies, defined as > 50 cells/colony, was counted.
Transwell invasion assay
The transwell invasion assay was performed using the transwell (Corning) and matrigel (BD Biosciences) according to the manufacturer’s instructions. Cells were re-suspended in RPMI 1640 containing 1% FBS, and 0.2 ml cell suspension (1 × 104/ml) was seeded into the top chamber, whereas 0.6 ml of RPMI 1640 containing 10% FBS was filled in the lower chamber, used as a chemoattractant. After 24 h of incubation at 37 °C with 5% CO2, the number of cells invading through the matrigel was counted in ten randomly selected visual fields, and the images were captured by a Leica DM3000 microscope (Leica).
Immunohistochemistry
Clinical pathological sections of prostate cancer tissue specimens from 30 PCa patients were provided by Department of Pathology at The First Hospital of China Medical University. The expression of KDM3A, YAP1, TEAD1, and c-Myc in tissue specimens was detected using an UltraSensitive™ SP (Mouse/Rabbit) IHC kit (Maxin-Bio, Fuzhou, Fujian, China) according to the manufacturer’s instructions. Briefly, sections were first dewaxed in xylene and ethanol, and antigen retrieval was performed using a microwave for 10 min at 100 °C. The sections were then incubated with antibodies for 1 h, followed by biotinylated anti-IgG antibody and streptavidin-biotinylated-complex/horseradish peroxidase. DAB and hematoxylin were used for nuclear staining. The images were then captured by upright metallurgical microscope (Olympus, Tokyo, Japan) under an original magnification of 200×. Scoring was done by two pathologists who counted the intensity of positive cells in a defined area. The staining intensity of target factors were assessed by two ways: To quantify ZFAS1, KDM3A, YAP1, TEAD1, or c-Myc staining, staining intensity was classified as: 0 (no staining), 1 (low staining), 2 (medium staining), and 3 (high staining). 0 and 1 were defined as low expression, and 2 and 3 were defined as high expression; to quantify staining of ZFAS1 and/or c-Myc, the cell-positive rate was counted in three random visual fields.
In situ hybridization
In situ hybridization (ISH) was performed using the ISH Kit (GenePharma, Shanghai, China) according to the manufacturer’s instructions. Briefly, formalin-fixed paraffin-embedded tissue slides were deparaffinized and deproteinated, and prehybridized in prehybridization solution for 2 h at 42 °C. Then, the sections were incubated in ZFAS1-probe solution over night at 42 °C. The slides were then exposed to a streptavidin–peroxidase reaction system and stained with DAB and hematoxylin. The images were captured by upright metallurgical microscope (Olympus, Tokyo, Japan) under an original magnification of 200×.
RNA immunoprecipitation
RV-1 cells were lysed by trizol reagent (Invitrogen), and total RNAs were extracted and prepared. 1 μg of each RNA sample was incubated without probe or with 5 μg of biotin-labeled MREs mutated ZFAS1 anti-sense probe or biotin-labeled ZFAS1 anti-sense probe overnight at 4 °C, and the complexes were isolated with streptavidin agarose beads (Invitrogen). The miRNAs present in the pull-down products were measured by real-time PCR using designed primers.
Animal experiments
BALB/c nude mice (4–6 weeks old, 14–16 g) were purchased from Beijing Vital River Experimental Animal Technology Co. Ltd., and housed in Department of laboratory animal science of China Medical University. The study was approved by Medical Laboratory Animal Welfare and Ethics Committee of China Medical University and the methods were carried out in accordance with the approved guidelines. The pLVX-NC-RV-1 and pLVX-ZFAS1-RV-1 cells were separately injected into the flanks of athymic nude mice to establish a xenograft tumor. When tumor sizes reached approximately 20 mm3 (approximately 1 week after inoculation), the mice were randomly divided into five groups (one was established by pLVX-NC-RV-1 cells and the other four by pLVX-ZFAS1-RV-1 cells; 5 mice/group), and the agomir of miR-27a-3p/miR-15a-5p/miR-16-5p (diluted in PBS at 2 μM) or 100 μL of negative control was injected intratumorally twice per week in either group for 5 weeks. Tumors were examined twice weekly; the length, width, and thickness were measured with calipers, and tumor volumes were calculated using the equation (Length × Width2)/2. 45 days after injection, the animals were euthanized, and the tumors were excised, weighed, paraffin-embedded, and subjected to staining assays.
Statistical analysis
Data are shown as mean ± SEM from at least three independent experiments. Statistical analyses involved Student’s t test, one-way ANOVA, and Kaplan–Meier analysis with SPSS 22 (IBM Corp., Armonk, NY) or GraphPad Prism 8.0.1 (GraphPad Software, Inc., La Jolla, CA). P < 0.05 was considered statistically significant.
Results
ZFAS1 expression correlates with PCa progression and is associated with poor clinical outcomes
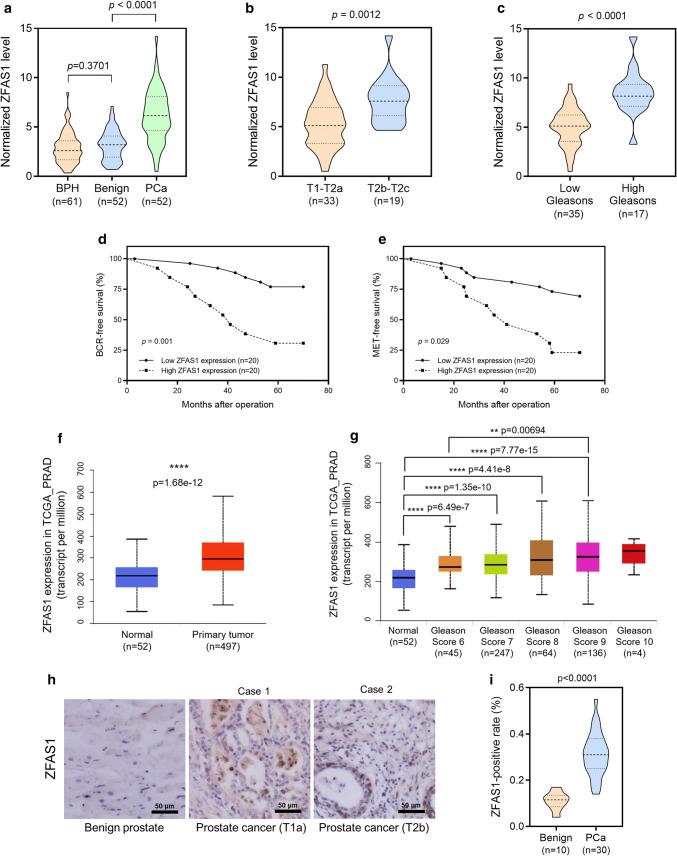
It has been reported that ZFAS1 expression is upregulated in many cancers. We first measured ZFAS1 expression levels using a cohort of clinical tissue specimens, including 61 prostate tissues from 61 patients diagnosed with benign prostatic hyperplasia (BPH; BPH is a noncancerous increase in size of the prostate, and the enlarged prostate may cause frequent urination, trouble starting to urinate, weak stream, inability to urinate, or loss of bladder control) and 52 pairs of PCa tissues with relative nontumor prostate tissues from 52 patients diagnosed with prostate cancer (Table 1). We found that ZFAS1 is significantly upregulated in PCa tissues compared to either prostate tissues from BPH or paired benign tissues (Fig. 1a). Next, we stratified the PCa tissues into different groups based on the pathological tumor stage or Gleason score. Unsurprisingly, the expression of ZFAS1 is much higher in tumors with an advanced T stage (T stage > T2a) and a high Gleason score (> 7) compared to those with a low T stage and Gleason score (Fig. 1b, c). Then, we analyzed whether ZFAS1 expression is associated with the clinical outcome of prostate cancer patients using Kaplan–Meier analysis. We divided the patients into two groups based on ZFAS1 expression and found that high ZFAS1 expression is significantly associated with unfavorable clinical outcomes, biochemical recurrence, and metastases in PCa patients. These data analyzed from our clinical database revealed that ZFAS1 expression is associated with PCa progression and poor clinical outcomes (Fig. 1d, e). To further verify this finding, we analyzed ZFAS1 expression based on the TCGA database using UALCAN software [33]. The results were consistent with our finding that ZFAS1 is significantly upregulated in primary prostate tumors (n = 497) compared with normal prostate tissues (n = 52), and is correlated with the clinical progression and poor outcomes of PCa (Fig. 1f, g). Furthermore, ZFAS1 expression was also measured by an in situ hybridization (ISH) staining assay in 30 human prostate cancer specimens. The representative images of ISH staining and analysis of the ZFAS1-positive rate showed that ZFAS1 is overexpressed in prostate cancer tissues compared with benign prostate tissues (Fig. 1h, i).
Table 1.
Associations between ZFAS1 and clinicopathological characteristics of 52 PCa patients
| Characteristics | N | ZFAS1 expression | P | |
|---|---|---|---|---|
| Low | High | |||
| Total cases | 52 | 26 | 26 | |
| Age (year) | ||||
| ≤ 70 | 32 | 17 | 15 | 0.5686 |
| > 70 | 20 | 9 | 11 | |
| Gleason score | ||||
| ≤ 7 | 35 | 23 | 12 | 0.0011** |
| > 7 | 17 | 3 | 14 | |
| Serum PSA (ng/μl) | ||||
| ≤ 20 | 19 | 11 | 8 | 0.3876 |
| > 20 | 33 | 15 | 18 | |
| Tumor stage | ||||
| I–IIa | 33 | 20 | 12 | 0.0226* |
| IIb/IIc | 19 | 6 | 14 | |
| Bone metastasis (24 month) | ||||
| Yes | 29 | 10 | 19 | 0.012* |
| No | 23 | 16 | 7 | |
| Biochemical recurrence (24 month) | ||||
| Yes | 19 | 5 | 14 | 0.0095** |
| No | 33 | 21 | 12 | |
*P < 0.05, **P < 0.01, by Chi-square test
Fig. 1.
High expression level of ZFAS1 was associated with advanced tumor stages and poor prognosis in RCC patients. a Expression of ZFAS1 was detected in 61 benign prostate tissues (from 61 BPH patients) and 52 pairs of PCa tissues and the adjacent non-tumorous tissues (from 52 PCa patients) by real-time PCR analysis. b, c Expression of ZFAS1 in PCa tissues was compared between groups divided with tumor stages (low: T1–T2a, n = 33; high T2b–T2c, n = 19) and Gleason scores (high: > 7, n = 17). The data statistical significance is assessed by Student’s t test. d, e Kaplan–Meier analysis for the correlation between ZFAS1 expression level and BCR-free survival/MET-free survival. Survival data were collected from 40 patients with PCa staged from T2a to T2c. The patients were divided into low ZFAS1 expression group and high ZFAS1 expression group. The median expression level of ZFAS1 was used as the cut-off value. The data statistical significance is assessed by log-rank test. f ZFAS1 expression in normal prostate tissues and prostate cancer tumor tissues was compared based on TCGA_PRAD data set. g ZFAS1 expression levels were compared between normal prostate tissues and prostate cancer tissues with different Gleason scores, based on TCGA_PRAD data set. (ANOVA) ZFAS1 expression levels were also detected in 30 prostate cancer tissues and 10 non-tumorous prostate tissues by ISH assay. Representative images for ZFAS1 expressed in benign prostate tissue or PCa tissue were shown in h. The cell-positive rate was quantified and shown in i. Original magnification ×200. Scale bars = 50 μm. **p < 0.01, ****p < 0.0001
The biological function of ZFAS1 in vitro
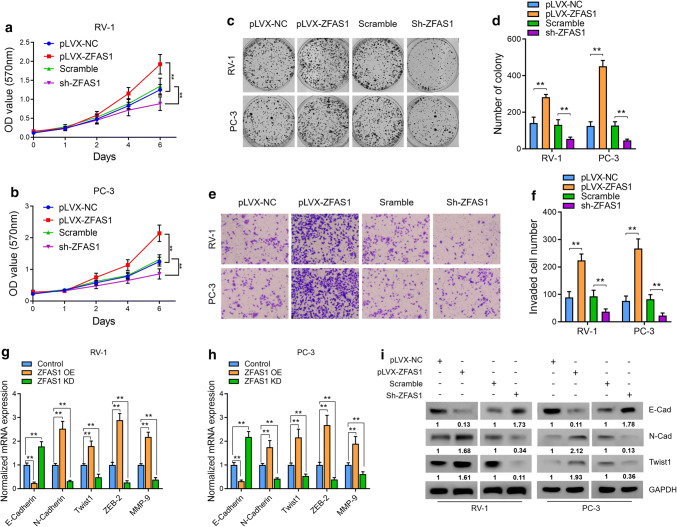
To test the biological function of ZFAS1 and further verify its role in PCa progression, we first explored whether its expression could be regulated by AR signaling. Two AR-positive PCa cell lines, RV-1 and LNCaP, were treated with 1 nM R1881, a commonly used AR signaling activator, or control vehicle for 24 h, followed by qRT-PCR analysis of ZFAS1 prostate-specific antigen (PSA). R1881 significantly upregulated PSA expression; however, no notable upregulation of ZFAS1 was observed in the presence of R1881 (Supplementary Fig. 1a). To further test the functional role of AR on ZFAS1, we next knocked down ZFAS1 in RV-1 and LNCaP cells via lentivirus transduction, and R1881 or the control vehicle was added to the cells. The results suggested that sh-ZFAS1 dramatically decreased the expression of ZFAS1 in the presence or absence of R1881, and ZFAS1 expression could not be rescued by treatment with synthetic androgen R1881 (Supplementary Fig. 1b). Therefore, we confirmed that ZFAS1 is not a target of AR signaling. We then stably overexpressed ZFAS1 (Supplementary Fig. 1c) or knocked down ZFAS1 via lentivirus transduction (Supplementary Fig. 1d) in two PCa cell lines, RV-1 and PC-3. By performing a cell proliferation assay using the Cell Counting Kit-8 (CCK-8), we found that ZFAS1 overexpression could significantly increase the proliferative capacity of PCa cells compared with parallel stable cell lines containing the empty vector pLVX-NC. In contrast, knockdown of ZFAS1 remarkably decreased the proliferation of PCa cells (Fig. 2a–d). To examine the effect of ZFAS1 on the cell invasion ability, we performed Transwell experiments. The results suggested that ZFAS1 overexpression significantly increased the invasion ability of PCa cells compared with pLVX-NC-transfected cells, and knockdown of ZFAS1 decreased the cell invasion ability compared with parallel stable cell lines transfected with scrambled shRNA (Fig. 2e, f).
Fig. 2.
ZFAS1 induced cell proliferative and invasive capacities as well as EMT of PCa cells in vitro. a, b Cell proliferation was assessed for 6 days using the Cell Counting Kit-8 assay in ZFAS1 overexpressed or knocked down PCa cells. c, d Cell colony formation assay was also performed to determine the proliferation of PC-3 and RV-1 cells. The number of colonies, defined as > 50 cells/colony, was counted for statistical analysis. e, f Transwell assay was used to evaluate the cell invasive capacity in ZFAS1 overexpressed or knocked down PCa cells, cells were counted in three random visual fields under the microscope, original magnification ×400. g, h mRNA expression levels of EMT-related markers were measured by real-time PCR analysis in ZFAS1 overexpressed or knocked down PCa cells. β-actin was used as the internal control, the Ct values for each group were compared using the 2-ΔΔCt method. OE: overexpression, KD: knockdown. i The translational levels of EMT markers were assessed by western blot analysis; GAPDH was used as the internal control. The gels were run under the same experimental conditions. The band intensities were calculated by Image J 1.46r software, and the ratio of target gene against GAPDH was used to conduct the statistical analysis. *P < 0.05 and **P < 0.01, as determined by Student’s t test
Since ZFAS1 could increase cell invasion and since ZFAS1 has been reported to induce EMT in gastric cancer [21] and colorectal cancer [23], we further studied whether ZFAS1 induces EMT in PCa cells. First, we performed qRT-PCR to measure the mRNA levels of several EMT-related markers, namely, E-cadherin, N-cadherin, Twist 1, zinc-finger E-box-binding homeobox (ZEB) 2, and matrix metallopeptidase (MMP) 9, in ZFAS1-overexpressing PCa cells. The results revealed that ZFAS1 overexpression increased the expression levels of N-cadherin, Twist 1, ZEB 2, and MMP 9, and decreased the expression level of E-cadherin compared with those in pLVX-NC-transfected cells. On the other hand, knockdown of ZFAS1 decreased the expression levels of N-cadherin, Twist 1, ZEB 2, and MMP 9, and increased the expression level of E-cadherin (Fig. 2g, h). We then performed western blot analysis to detect the protein levels of E-cadherin, N-cadherin, and Twist 1, and the results were consistent with the results from qRT-PCR (Fig. 2i). These data suggest that the overexpression of ZFAS1 induced EMT in PCa cells, thus promoting cell invasion. Given the tumorigenic role of ZFAS1 in both AR-positive and AR-negative cells, we concluded that ZFAS1 drives the progression of prostate cancer in an AR-independent manner, suggesting that ZFAS1 could act as a crucial regulator in CRPC.
Bioinformatic analysis of the involvement of ZFAS1 in promoting PCa tumorigenesis
As lncRNAs have been known to function as ceRNAs to sponge endogenous miRNAs, we then focused on examining whether ZFAS1 exerts its oncogenic function by interacting with miRNAs. Using the bioinformatic prediction software LncBase v.2 [34] from the DIANA tool, we predicted 152 miRNAs that may be targeted by ZFAS1 (Supplementary Table 1 and Supplementary Fig. 2a). Next, we performed pathway analysis using miRPath v.3 software [35] based on the predicted 152 miRNAs. In the top ten listed KEGG pathways in which these miRNAs might participate, we noticed that the prostate cancer pathway ranked fourth, and the Hippo signaling pathway ranked eighth. The Hippo signaling pathway is a conserved regulator of organ size, and this kinase cascade starts from the tumor suppressor Hippo [36, 37]. The oncoprotein YAP/TAZ is the downstream factor and major target of the Hippo kinase cascade. When activated, YAP/TAZ shuttles from the cytoplasm to the nucleus, interacts with transcriptional factors, primarily TEA domain (TEAD) family members, and triggers the activation of target genes, including the proto-oncogene c-Myc [36–38]. Therefore, YAP/TAZ is essential for cancer initiation and the growth of most solid tumors.
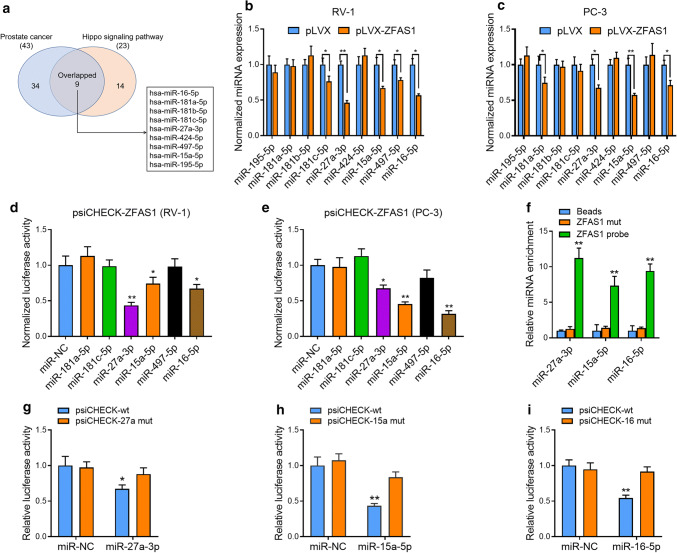
We then performed pathway intersection analysis based on miRNA participation in the prostate cancer signaling pathway (23 miRNAs) and Hippo signaling pathway (43 miRNAs). The results revealed that nine miRNAs overlapped in both pathways (Supplementary Table 2, Supplementary Figs. 2b, 3a). Next, we investigated whether ZFAS1 mRNA could physically interact with the nine candidate miRNAs. We overexpressed ZFAS1 in RV-1 and PC-3 cells and then tested the expression of all nine candidates. Among them, in response to ZFAS1 overexpression, miR-181c-5p, miR-27a-3p, miR-15a-5p, miR-497-5p, and miR-16-5p were significantly downregulated in RV-1 cells (Fig. 3b), whereas miR-181a-5p, miR-27a-5p, miR-15a-5p, and miR-16-5p were dramatically downregulated in PC-3 cells (Fig. 3c). Then, we cloned the full length of the ZFAS1 gene into the luciferase reporter construct psiCHECK-2 (psiCHECK-ZFAS1) and co-transfected PCa cells with psiCHECK-ZFAS1 and the agomir of each of the above ZFAS1-downregulated miRNAs or the common scramble control. The results suggested that miR-27a-3p, miR-15a-5p, and miR-16-5p could decrease the expression of ZFAS1 after the cells were transfected with the luciferase reporter gene (Fig. 3d, e). To further verify the interactions between the ZFAS1 gene and these miRNAs, we performed an RNA pull-down assay using a biotin-labeled ZFAS1 anti-sense probe. The results showed that miR-27a-3p/15a-5p/16-5p was enriched in the probe pull-down product (Fig. 3f), suggesting that ZFAS1 may be able to bind to these miRNAs via its putative miRNA-response elements (MREs).
Fig. 3.
ZFAS1 interacts with and regulates multiple tumor suppressor miRNAs in PCa cells. a Pathway analysis was performed to detect the biological function of ZFAS1-targeted miRNAs. Nine candidate miRNAs were discovered to overlap in “Prostate cancer” and “Hippo signaling pathway”. b, c Expression levels of candidate miRNAs were measured in ZFAS1 overexpressed PCa cells by real-time PCR analysis. U6 snRNA was used as the control. d, e Luciferase activity in PCa cells co-transfected with luciferase reporter-containing ZFAS1 and agomir of indicated miRNAs. Data are presented as the relative ratio of renilla luciferase activity and firefly luciferase activity. f RNA pull-down assay was performed to assess the interaction between ZFAS1 and indicated miRNAs. The miR-27a/15a/16 was enriched in biotinylated ZFAS1 anti-sense oligo pull-down product. g, h, i Luciferase activity in RV-1 cells co-transfected with agomir of miR-27a/15a/16 and luciferase reporter-containing ZFAS1 with relatively mutated MREs. The data statistical significance is assessed by Student’s t test. *P < 0.05, **P < 0.01
ZFAS1 possessed two predicted miR-27a-3p MREs and two predicted MREs for either miR-15a-5p or miR-16-5p (Supplementary Fig. 3a). It is notable that miR-15a and miR-16 are located close to one another on chromosome 13q14 and constitute clustered miRNAs [39]. In addition, they belong to the same miRNA family and function through the same seed sequence. We cloned the ZFAS1 gene into the luciferase reporter construct (Supplementary Fig. 3b–d) and mutated these MREs based on the predicted positions (Supplementary Fig. 3e). In RV-1 cells, the relative luciferase activity was decreased after the overexpression of miR-27a/15a/16, and the miRNA-mediated luciferase reduction was diminished after mutating the putative MREs (Fig. 3g–i). Next, we measured ZFAS1 expression in miR-27a/15a/16-overexpressing or knockdown PCa cells by qRT-PCR. The results suggested that the overexpression of either of the miRNAs could decrease the expression level of ZFAS1 (Supplementary Fig. 3f), and knockdown of either of the miRNAs could increase its expression (Supplementary Fig. 3g). Taken together, these data suggested that the ZFAS1 gene directly interacted with the miR-27a/15a/16 gene via binding to the putative MREs.
ZFAS1 enhances c-Myc expression by upregulating YAP1, TEAD1, and KDM3A gene expression via a regulatory network of ceRNAs
Given the function of ceRNAs in the prostate cancer cell line, we then aimed to determine how ZFAS1 promoted the oncogenesis of prostate cancer. To this end, we performed bioinformatic analysis to determine which downstream targets, especially the key oncogenes that may promote the progression of PCa, could be activated by ZFAS1-mediated miRNA evasion. We analyzed the possible common downstream target genes of miR-27a/15a/16 under the categories of the “prostate cancer pathway” and “Hippo signaling pathway” using the miRPath v.3 and TarBase v.8 bioinformatic software from the DIANA tool, and the results were confirmed by TargetScan software [40] (Supplementary Table 2). There are 42, 28, and 27 target genes for miR-27a-3p, miR-15-5p, and miR-16-5p, respectively, under the “Hippo signaling pathway”, and 29, 22, and 22 target genes for miR-27a-3p, miR-15-5p, and miR-16-5p, respectively, under the “prostate cancer pathway”. Among the predicted targets, we noted that Yes-associated protein 1 (YAP1) and TEAD domain transcription factor 1 (TEAD1) were common targets of all three miRNAs. YAP1 is a key effector and downstream target of the Hippo pathway. Moreover, YAP1 has been shown to increase the transcription of c-Myc and promote proliferation and the stem-cell-like properties of cancer cells [38]. In addition, as a transcription coactivator, YAP1 cooperates with other transcription factors to promote the transcription of target genes [37]. The TEAD family is one of the partner transcription factors of YAP1 [41]. The TEAD family members contain a TEA domain for DNA binding and a transactivation domain for the interaction with transcription coactivators [37, 42]. TEAD1 cooperates with YAP1 and enhances its transcriptional activation for c-Myc [41, 43]. Therefore, TEAD1 and YAP1 were proposed to function as cooperators to promote the tumorigenesis of multiple cancers, including gastric cancer [44], breast cancer [45], and colorectal cancer [46].
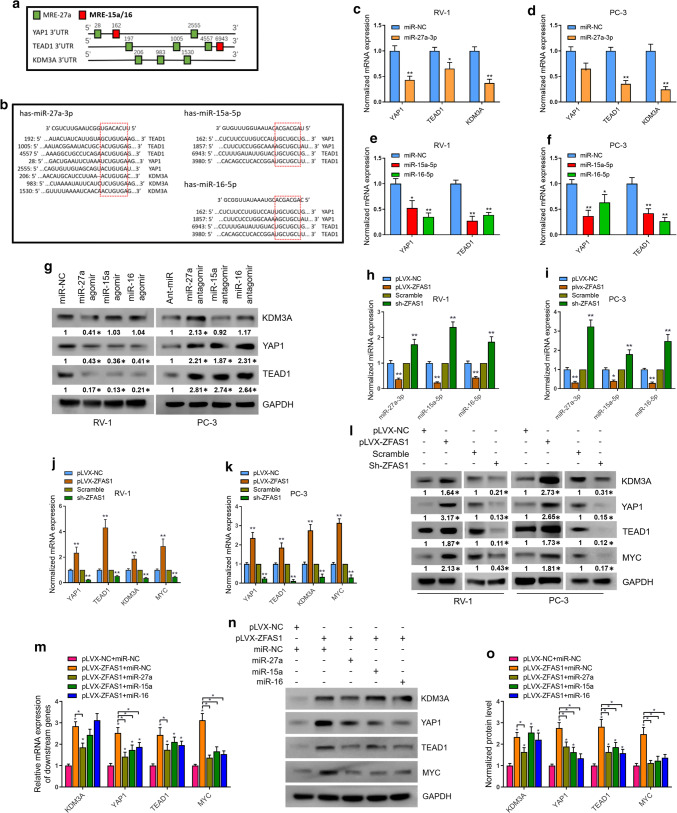
Another target gene that drew our attention was lysine demethylase 3A (KDM3A), also known as Jumonji domain-containing 1A (JMJD1A). KDM3A is a histone demethylase that removes the repressive H3K9 methylation marks (H3K9me1 or H3K9me2) to regulate gene expression [47]. KDM3A has been reported to play a key role in the proliferation and survival of prostate cancer cells [32]. Mechanistically, it interacts with AR to increase c-Myc expression levels. Knockdown of KDM3A leads to the downregulation of c-Myc. Taken together, we identified YAP1, TEAD1, and KDM3A as possible downstream oncogenes that can be activated by ZFAS1-mediated miRNA evasion. The predicted MREs in the 3′ UTR of these target genes are shown in Fig. 4a, b.
Fig. 4.
ZFAS1-modulated ceRNA system regulates multiple oncogenes in PCa cells. a Schematic diagram demonstrating the position of putative MREs for miR-27a/15a/16 on 3′UTR of YAP1, TEAD1, and KDM3A. b Sequence of MREs for miR-27a/15/16 on 3′UTR of YAP1, TEAD1, and KDM3A. c-f Real-time PCR analysis was performed to assess the transcriptional levels of predicted target genes after overexpression of indicated miRNAs. g Translational levels of KDM3A, YAP1, and TEAD1 were measured in miRNAs overexpressed RV-1 cells and miRNAs knocked down PC-3 cells by western blot analysis. h–k Expression levels of miR-27a/15a/16 and predicted target genes were detected in ZFAS1 overexpressed or knocked down PCa cells by real-time PCR. l Protein expression levels of KDM3A, YAP1, TEAD1, and c-Myc were measured in ZFAS1 overexpressed or knocked down PCa cells. Transcriptional (m) and translational levels (n, o) of KDM3A, YAP1, TEAD1, and c-Myc were assessed in ZFAS1 overexpressed alone or ZFAS1 overexpressed with miRNAs restored RV-1 cells. The internal control genes were GAPDH for western blot analysis and β-actin/U6 snRNA for real-time PCR analysis. The gels were run under the same experimental conditions. The band intensities were calculated by Image J 1.46r software, and the ratio of target gene to GAPDH was used to conduct the statistical analysis. *P < 0.05 and **P < 0.01, as determined by Student’s t test
miR-27a, miR-15a, or miR-16 was overexpressed or knocked down by transfecting the tested cells with the agomirs or antagomirs of the respective miRNAs (Supplementary Fig. 4a–c). The results showed that the overexpression of miR-27a suppressed the mRNA levels of KDM3A, YAP1 and TEAD1 in RV-1 and PC-3 cell lines (Fig. 4c, d), and the overexpression of either miR-15a or miR-16 decreased the mRNA levels of YAP1 and TEAD1 in both cell lines (Fig. 4e, f). Western blot analysis further confirmed these results. The protein levels of YAP1 and TEAD1 were decreased in response to miR-27a/15a/16 overexpression, whereas the inhibition of miR-27a/15a/16 increased YAP1 and TEAD1 protein expression in PCa cells; KDM3A was translationally upregulated by miR-27a overexpression in RV-1 cells, and the inhibition of miR-27a increased KDM3A protein expression in PC-3 cells (Fig. 4g). Since we have identified that YAP1, TEAD1, and KDM3A are downstream targets of miR-27a/15a/16 and that ZFAS1 sponges with miR-27a/15a/16 and antagonizes their expression, we next sought to determine whether ZFAS1 could regulate the expression of YAP1, TEAD1, and KDM3A. We tested the levels of the above miRNAs in ZFAS1-overexpressing or ZFAS1-knockdown PCa cells. As expected, miR-27a/15a/16 was downregulated in response to ZFAS1 overexpression, and knockdown of ZFAS1 increased miR-27a/15a/16 expression levels (Fig. 4h, i). In addition, the mRNA levels of YAP1, TEAD1, and KDM3A were measured in ZFAS1-overexpressing or ZFAS1-knockdown cells, and the mRNA level of c-Myc was also examined. The results revealed that ZFAS1 overexpression significantly upregulated the mRNA levels of YAP1, TEAD1, KDM3A, and c-Myc, whereas ZFAS1 knockdown dramatically suppressed the expression of all four genes (Fig. 4j, k). YAP1, TEAD1, KDM3A, and c-Myc protein levels were assessed by western blot analysis. The results are consistent with the above mRNA level measurements (Fig. 4l). To verify that ZFAS1 regulated the expression of YAP1, TEAD1, KDM3A, and c-Myc via competitively binding to miR-27a/15a/16, we separately overexpressed miR-27a, miR-15a, and miR-16 in ZFAS1-overexpressing RV-1 cells and then observed the expression of YAP1, TEAD1, KDM3A, and c-Myc. The results showed that the overexpression of either miR-27a, miR-15a, or miR-16 partially abolished the upregulation of YAP1, TEAD1, KDM3A, and c-Myc, whose expression was induced by ZFAS1 overexpression (Fig. 4n–p). Taken together, we confirmed that ZFAS1 regulated the expression of YAP1, TEAD1, KDM3A, and c-Myc via a ceRNA regulatory network.
High levels of KDM3A, YAP1, and TEAD1 are correlated with c-Myc overexpression in human prostate cancer specimens
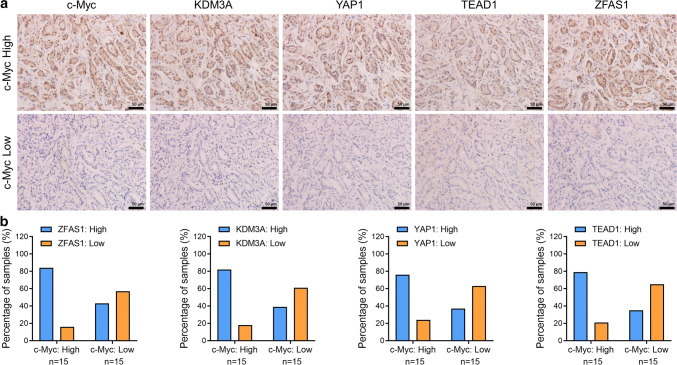
To examine the correlation between the expression levels of ZFAS1, KDM3A, YAP1, TEAD1, and c-Myc, ISH/IHC staining was performed in a subset of 30 human prostate cancer specimens with Gleason scores of 3–4 (Fig. 5a). Based on the respective staining intensities, we defined c-Myc samples as either c-Myc-low or -high. The staining intensities of ZFAS1, KDM3A, YAP1, and TEAD1 were likewise defined as either low or high. The analysis of ZFAS1, KDM3A, YAP1, TEAD1, and c-Myc staining levels (Fig. 5b) showed that c-Myc-high staining was positively correlated with a high expression of ZFAS1, KDM3A, YAP1, and TEAD1 in the PCa tumor samples (p < 0.05, Fisher’s exact test). Furthermore, data from the TCGA prostate cancer data set confirmed our results. The data suggested that the mRNA expression levels of ZFAS1, KDM3A, YAP1, and TEAD1 were positively correlated with c-Myc mRNA levels in 499 prostate cancer samples (Supplementary Fig. 5a–d). These data suggest a potential association of these factors in human prostate tumors.
Fig. 5.
High levels of ZFAS1, KDM3A, YAP1, and TEAD1 are associated with c-Myc overexpression in human prostate cancer specimens. a Representative image showing c-Myc, KDM3A, YAP1, TEAD1, and ZFAS1 immunohistochemistry (IHC)/in situ hybridization (ISH) staining of 30 human prostate cancer specimens. Staining was visualized by DAB with hematoxylin counterstaining. Scar bar = 50 μm. b Quantification of c-Myc, KDM3A, YAP1, TEAD1, and ZFAS1 staining in prostate cancer specimens. Staining intensity of each protein/gene was scored as 0 to3 (0: no staining, 1: weak staining, 2: medium staining, and 3: strong staining. 0 and 1 were classified as low expression, whereas 2 and 3 were defined as high expression. High c-Myc expression was correlated with high expression of either KDM3A, YAP1, TEAD1, or ZFAS1 (p < 0.05, Fisher’s exact test)
ZFAS1 drives the tumorigenesis of PCa cell lines through sponging miR-27a/15a/16 in vitro
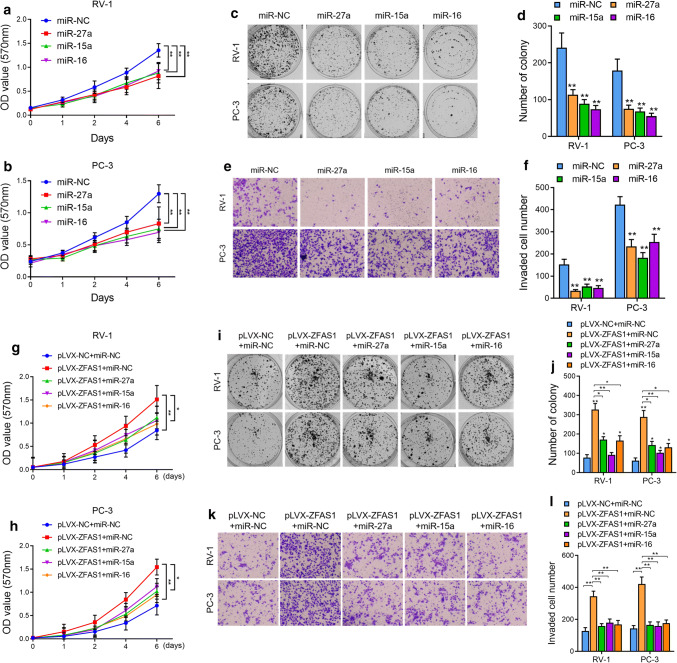
After identifying the miR-27a/15a/16 regulatory network through which ZFAS1 regulates c-Myc expression, we sought to determine whether ZFAS1 promoted the oncogenesis phenotypes of PCa cells via the above regulatory pathway. We first explored the roles of miR-27a/15a/16 in PCa cell proliferation. The colony formation assays showed that the overexpression of either miR-27a, miR-15a, or miR-16 significantly decreased the proliferative capacity of PCa cells compared with control cell lines (Fig. 6a–d). Moreover, knockdown of miR-27a, miR-15a, or miR-16 remarkably increased the proliferation of PCa cells (Supplementary Fig. 6a–d). The results of the transwell assays suggested that the overexpression of either miR-27a, miR-15a, or miR-16 significantly decreased the invasion ability of PCa cells (Fig. 6e, f), while the antagonization of either miR-27a, miR-15a, or miR-16 increased cell invasion (Fig. 6e, f). Next, we overexpressed miR-27a, miR-15a, or miR-16 in ZFAS1-overexpressing PCa cells and found that, compared with ZFAS1 overexpression alone, the restoration of either miR-27a, miR-15a, or miR-16 partially diminished the enhanced proliferation and invasion of PCa cells (Fig. 6g–i). The above data suggested that ZFAS1 overexpression promoted the proliferation and invasion capabilities of PCa cells by decreasing miR-27a/15a/16 levels in vitro.
Fig. 6.
Re-introduction of tumor suppressor miRNAs rescued the ZFAS1-enhanced tumor genetic phenotypes of PCa cells. Cell proliferation was assessed in miR-27a/15a/16 overexpressed PCa cells by CCK-8 assay (a, b) and cell colony formation assay (c, d). e, f Cell invasive capacity was assessed by transwell assay in miR-27a/15a/16 overexpressed PCa cells. Cell proliferation was measured in ZFAS1 overexpressed alone or ZFAS1 overexpressed with miRNAs re-introduced PCa cells by CCK-8 (g, h) and cell colony formation assay (i, j). k, l Cell invasive ability was assessed in ZFAS1 overexpressed alone or ZFAS1 overexpressed with miRNAs re-introduced PCa cells by transwell assay. For cell colony formation essay, the number of colonies, defined as > 50 cells/colony, was counted for statistical analysis. For transwell assay, cells were counted in three random visual fields under the microscope, original magnification × 400. *P < 0.05 and **P < 0.01, as determined by Student’s t test
ZFAS1 promotes in vivo tumor growth in a xenograft mouse model
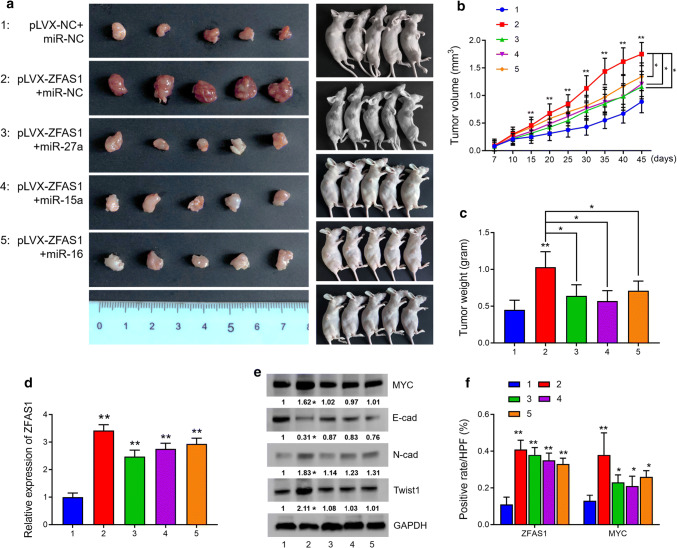
To evaluate the biological function of ZFAS1 in vivo, RV-1 cells were subcutaneously injected into nude mice. There were five groups in total: Group 1 (pLVX-NC + miR-NC) was injected with pLVX-NC-RV-1 cells followed by intratumoral injection of miR-NC; Group 2 (pLVX-ZFAS1 + miR-NC) was injected with pLVX-ZFAS1-RV-1 cells followed by intratumoral injection of miR-NC; Group 3 (pLVX-ZFAS1 + miR-27a) was injected with pLVX-ZFAS1-RV-1 cells followed by intratumoral injection of the agomir of miR-27a; Group 4 (pLVX-ZFAS1 + miR-15a) was injected with pLVX-ZFAS1-RV-1 cells followed by intratumoral injection of the agomir of miR-15a; and Group 5 (pLVX-ZFAS1 + miR-16) was injected with pLVX-ZFAS1-RV-1 cells followed by intratumoral injection of the agomir of miR-16. We found that the tumor lumps in the ZFAS1-overexpressing group were significantly larger and heavier than those in the control group, and that miR-27a, miR-15a, and miR-16 could partially reduce the tumor growth caused by ZFAS1 overexpression. Forty-five days after injection, the mice were sacrificed, and the volume and weight of the tumor lumps in each group were measured (Fig. 7a–c). The tumors were then subjected to RNA/protein extraction or staining assays. In each group, ZFAS1 expression was measured by qRT-PCR or ISH staining, and c-Myc expression was measured by western blot analysis or IHC staining. Expression of E-cadherin, N-cadherin, and Twist1 was measured by western blot analysis, as well. The results suggested that the expression of ZFAS1, c-Myc and the EMT-related proteins was significantly elevated in response to ZFAS1 overexpression. Moreover, the re-overexpression of either miR-27a, miR-15a, or miR-16 partially rescued expression of c-Myc and the EMT-related proteins in Groups 3–5 (Fig. 7d–f, representative images of IHC/ISH staining are shown in Supplementary Fig. 7). These results demonstrated that ZFAS1 overexpression promoted c-Myc expression as well as in vivo tumor growth and that the reintroduction of miR-27a, miR-15a or miR-16 partially rescued tumor growth and c-Myc expression.
Fig. 7.
ZFAS1 overexpression promoted xenograft tumor growth in vivo. a In vivo tumor lumps of xenograft mouse models composed of pLVX-NC-RV-1 cells or pLVX-ZFAS1-RV-1 cells, 1 week after inoculation, the negative control (miR-NC) or agomir of miR-27a/15a/16 was intratumorally injected twice per week. 45th day after injection, tested mice were sacrificed and each tumor lump was removed from the body, and images were captured (the tested mice were divided as: 1, injected with pLVX-NC-RV-1 cells followed with intratumor injection of miR-NC; 2, injected with pLVX-ZFAS1-RV-1 cells followed with intratumor injection of miR-NC; 3, injected with pLVX-ZFAS1-RV-1 cells followed with intratumor injection of miR-27a agomir; 4, injected with pLVX-ZFAS1-RV-1 cells followed with intratumor injection of miR-15a agomir; 5, injected with pLVX-ZFAS1-RV-1 cells followed with intratumor injection of miR-16 agomir). b The tumor growth curves for in vivo tumor volumes. Data are shown as mean ± SD of the tumor volumes, n = 5, *P < 0.05 and **P < 0.01 (ANOVA). c The mean tumor weight of each group. Data are shown as mean ± SD of the tumor weights, n = 5. *P < 0.05, **P < 0.01 (ANOVA). d Expression of ZFAS1 was measured in tumor extracts by qRT-PCR analysis. e Protein expression of c-Myc, E-cad, N-cad, and Twist1 was detected in tumor extracts by western blot analysis (n = 5). Tumor lumps were also subjected to staining assay, and expression of ZFAS1/c-Myc was detected in tumor samples by ISH/IHC staining. The cell-positive rates of ZFAS1 and c-Myc in different groups were quantified and demonstrated in f. *P < 0.05 and **P < 0.01 (Student’s t test)
To further examine the effect of ZFAS1 on tumor metastasis in vivo, we injected the control or ZFAS1-overexpressed RV-1 cells into the tested mice via the tail vein. The mice were sacrificed 50 days after injection, and lung metastatic lesions could be observed on the surface of the lungs. However, no significant difference was found in the number of lung metastatic nodules between the two groups (Supplementary Fig. 8). This result may suggest that ZFAS1 promotes PCa progression mainly by stimulating the tumor growth.
Discussion
The c-Myc oncogene contributes to the oncogenesis of many human cancers, including prostate cancer [25]. c-Myc mRNA and protein levels are upregulated in prostate cancer [48, 49], and c-Myc overexpression promotes the development and progression of prostate cancer [50–52]. For example, the prostate-specific overexpression of c-Myc alone promotes tumor development in mouse prostates [28] and combining c-Myc activation with Pten loss induces the development of genomic instability and the metastatic potential of PCa [29]. c-Myc is widely accepted as a central driver of tumorigenesis in PCa. Nonetheless, the mechanisms underlying the regulation of c-Myc by the ncRNA-mediated network remain elusive. Our study reveals that c-Myc is under the control of ZFAS1, a novel lncRNA-induced ceRNA network. ZFAS1 promotes c-Myc expression by antagonizing miR-27a, miR-15a, and miR-16, and diminishing their epigenetic silencing effects on the KDM3A and YAP1–TEAD1 complex. Furthermore, the assessment of ZFAS1 in a cohort of clinical samples supports that ZFAS1 expression is elevated in PCa tissues and is correlated with poor clinical outcomes and that high ZFAS1 expression is positively correlated with high c-Myc expression.
Long noncoding RNAs are emerging as important regulators of gene expression. They function by modulating transcriptional or posttranscriptional expression, such as influencing histone modifications and controlling mRNA stability and translation. It has been identified that lncRNA could act as a component of the transcriptional network of MYC. For example, a novel lncRNA, termed c-Myc-inhibitory factor (lncRNA-MIF, RP11-320M2.1), reportedly decreases c-Myc expression by sponging miR-586 and increasing Fbxw7 expression, thereby inhibiting cell proliferation and cell cycle progression [53]. Another novel lncRNA termed c-Myc upregulated lncRNA (MYU, LOC100128881) has been recently discovered to be a direct target of c-Myc and plays a critical role in the c-Myc-dependent proliferation of colon cancer cells [54]. ZFAS1 was recently identified as a vital oncogenic lncRNA in multiple cancer types. Inconsistent with its tumor-suppressing role in breast cancer, ZFAS1 expression is frequently upregulated in a wide variety of human cancers, such as colorectal, gastric, ovarian, and liver cancers. In addition, the upregulation of ZFAS1 is commonly associated with advanced TNM stages, lymph-node metastasis, and poor clinical outcomes [20–23]. Nonetheless, the biological functions of ZFAS1 in prostate cancer have not been characterized. By analyzing the prostate cancer data set from TCGA, Chen [55] reported that ZFAS1 is upregulated in prostate cancer, and a high level of ZFAS1 indicates a worse survival possibility of PCa patients. He also suggested that ZFAS1 is targeted by miR-940 via NAA10 and RPL28, and the co-expression network could drive the progression of PCa. Our study further confirmed that ZFAS1 is upregulated in PCa tissues, and the analysis based on ZFAS1 expression as well as the clinicopathological features of the patients from our clinical data suggest that ZFAS1 may serve as a biomarker for the prediction of the clinical outcomes of PCa patients. Androgen and androgen receptors are the central drivers for the initiation and progression of prostate cancer. To clarify whether ZFAS1 is a downstream factor of androgen signaling, we performed androgen stimulation in two AR-positive PCa cell lines, RV-1 and LNCaP. However, no positive results were observed. This suggests that ZFAS1 drives the carcinogenesis of prostate cancer in an androgen-independent manner.
Using bioinformatic analysis, we identified nine miRNAs that could potentially interact with ZFAS1 and facilitate PCa progression. Subsequent expression profiling confirmed that miR-27a-3p, miR-15a-5p, and miR-16-5p are putative targets of ZFAS1. MicroRNAs are well known to have tumor-suppressing functions and are dysregulated in cancers. In particular, miRNAs encoded by the miR-15a/16 cluster have been demonstrated to be tumor suppressors, and the expression of these genes inhibits cell proliferation promotes the apoptosis of cancer cells and suppresses tumorigenicity both in vitro and in vivo [39, 56]. miR-15a and miR-16 form an miRNA cluster at the chromosomal region 13q14, which is frequently deleted in cancer cells [39]. miR-15a and miR-16 function as putative tumor suppressors by targeting a variety of oncogenes, such as BCL2, CCND1, CCNE1, and CDK4-6 [39, 56, 57]. In prostate cancer, the loss of miR-15a and miR-16, together with increased miR-21 expression, activates TGF-β signaling and promotes the spread of prostate cancer and bone lesions [57]. This evidence supports our findings that ZFAS1 promotes the progression of PCa through sponging and decreasing the expression of tumor suppressor genes, miR-15a and miR-16. In addition, we revealed a novel mechanism through which miR-15a and miR-16 enhance the proliferation and invasion capacities of PCa cells and in vivo xenograft tumor growth. We proposed that the YAP1–TEAD1 complex serves as a downstream effector of the ZFAS1-miR-15a/miR-16 regulatory network, which regulates c-Myc activation in prostate cancer. Notably, it has been reported that the expression of miR-15a and miR-16 is repressed by c-Myc and is associated with tumorigenesis [58, 59]. Therefore, we proposed that ZFAS1-mediated miR-15a and miR-16 downregulation induces c-Myc activation, which would further enhance the c-Myc-induced repression of miR-15a and miR-16.
miR-27a is located on chromosome 19, and has been shown to play important roles in multiple tumor types, such as breast cancer, colorectal cancer, and cervical cancer [60–62]. However, whether miR-27a acts as a tumor suppressor or an oncogene in prostate cancer remains controversial. Fletcher et al. reported that the overexpression of miR-27a increases the expression of AR-targeted genes and promotes prostate cancer cell growth by directly regulating prohibitin (PHB) expression [63]. In contrast, another study revealed that miR-27a functions as a tumor suppressor by repressing MAP2K4, and inhibits the proliferation and migration of PCa cells [64]. Despite these contradictory findings, miR-27a has been identified as an AR target gene and participates in AR signaling [64, 65]. Our data suggested that the overexpression of miR-27a-3p globally inhibited the proliferative and invasive capacities of PCa cells. Furthermore, we discovered that KDM3A, YAP1, and TEAD1, which act as oncogenes by inducing the expression of c-Myc, are putative target genes of miR-27a-3p. In addition, we demonstrated that the overexpression of miR-27a could partially diminish the ZFAS1-enhanced tumorigenic phenotypes of PCa. Collectively, our results suggest that miR-27a is a tumor suppressor in PCa and acts as a key mediator in the network of ZFAS1-induced c-Myc activation.
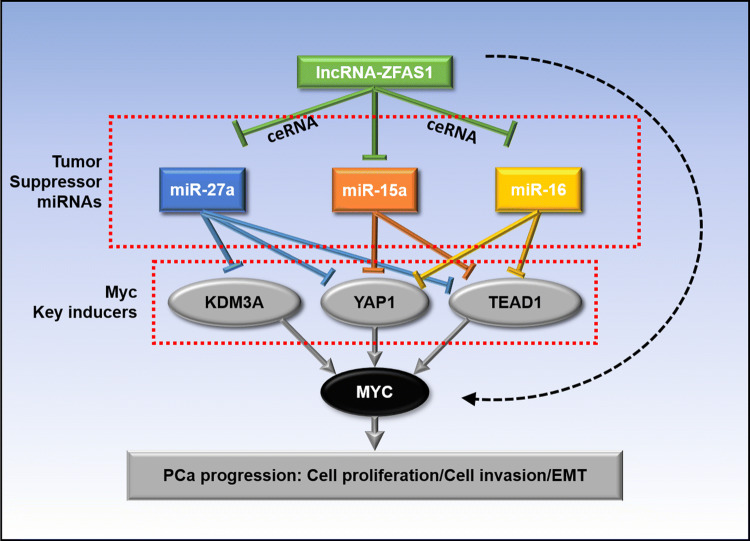
In summary, ZFAS1 overexpression in prostate cancer promoted cell proliferation and invasion as well as xenograft tumor growth, suggesting a critical tumorigenic role of ZFAS1 in PCa and even in CRPC. Mechanistically, ZFAS1 binds to miR-27a, miR-15a, and miR-16, and antagonizes their expression. miR-27a, miR-15a, and miR-16 serve as tumor suppressors in PCa, and their loss enhances tumor cell proliferation and invasion. More importantly, miR-27a, miR-15a, and miR-16 are key downstream effectors of ZFAS1. Their downregulation by ZFAS1 increases the expression of KDM3A, YAP1, and TEAD1, which are strong c-Myc inducers in PCa (Fig. 8). Therefore, we demonstrated an androgen-independent regulatory role of ZFAS1 for c-Myc expression in prostate cancer, indicating that ZFAS1 can be a potential biomarker and therapeutic target in CRPC.
Fig. 8.
Mechanistic diagram illustrates the function of lnc-ZFAS1 in PCa. ZFAS1 enhances c-Myc expression by upregulating KDM3A, YAP1, and TEAD1 expression via competitively sponging and inhibiting miR-27a/15a/16 expression, thereby promotes PCa progression
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
This work was financially supported by grant from the National Natural Science Foundation of China (Grant No. 81702505). The funding bodies played no role in the design or interpretation of the study.
Abbreviations
- ZFAS1
ZNFX1 anti-sense RNA 1
- YAP1
Yes-associated protein 1
- TEAD1
TEA domain transcription factor 1
- KDM3A
Lysine demethylase 3A
- PCa
Prostate cancer
- ceRNA
Competing endogenous RNA
- AR
Androgen receptor
- PSA
Prostate-specific antigen
Authors’ contributions
XC and CP conceived and designed the study. XC and CP performed the experiments. XC and CL analyzed the data and prepared the manuscript. ZZ and X Liu contributed the experimental reagents and materials. X Lin did the pathological diagnosis and provided the pathological sections. All authors have read and approved the final version of the manuscript.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Barry MJ, Simmons LH. Prevention of prostate cancer morbidity and mortality: primary prevention and early detection. Med Clin North Am. 2017;101(4):787–806. doi: 10.1016/j.mcna.2017.03.009. [DOI] [PubMed] [Google Scholar]
- 3.Robinson D, Van Allen EM, Wu YM, et al. Integrative clinical genomics of advanced prostate cancer. Cell. 2015;162(2):454. doi: 10.1016/j.cell.2015.06.053. [DOI] [PubMed] [Google Scholar]
- 4.Mitsiades N. A road map to comprehensive androgen receptor axis targeting for castration-resistant prostate cancer. Cancer Res. 2013;73(15):4599–4605. doi: 10.1158/0008-5472.CAN-12-4414. [DOI] [PubMed] [Google Scholar]
- 5.Sharma NL, Massie CE, Ramos-Montoya A, et al. The androgen receptor induces a distinct transcriptional program in castration-resistant prostate cancer in man. Cancer Cell. 2013;23(1):35–47. doi: 10.1016/j.ccr.2012.11.010. [DOI] [PubMed] [Google Scholar]
- 6.Tsai MC, Spitale RC, Chang HY. Long intergenic noncoding RNAs: new links in cancer progression. Cancer Res. 2011;71(1):3–7. doi: 10.1158/0008-5472.CAN-10-2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ulitsky I, Bartel DP. lincRNAs: genomics, evolution, and mechanisms. Cell. 2013;154(1):26–46. doi: 10.1016/j.cell.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang K, Liu CY, Zhou LY, et al. APF lncRNA regulates autophagy and myocardial infarction by targeting miR-188-3p. Nat Commun. 2015;6:6779. doi: 10.1038/ncomms7779. [DOI] [PubMed] [Google Scholar]
- 9.Xue X, Yang YA, Zhang A, et al. LncRNA HOTAIR enhances ER signaling and confers tamoxifen resistance in breast cancer. Oncogene. 2016;35(21):2746–2755. doi: 10.1038/onc.2015.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun M, Nie F, Wang Y, et al. LncRNA HOXA11-AS Promotes Proliferation and Invasion of Gastric Cancer by Scaffolding the Chromatin Modification Factors PRC2, LSD1, and DNMT1. Cancer Res. 2016;76(21):6299–6310. doi: 10.1158/0008-5472.CAN-16-0356. [DOI] [PubMed] [Google Scholar]
- 11.Xue M, Chen W, Xiang A, et al. Hypoxic exosomes facilitate bladder tumor growth and development through transferring long non-coding RNA-UCA1. Mol Cancer. 2017;16(1):143. doi: 10.1186/s12943-017-0714-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang H, Huo X, Yang XR, et al. STAT3-mediated upregulation of lncRNA HOXD-AS1 as a ceRNA facilitates liver cancer metastasis by regulating SOX4. Mol Cancer. 2017;16(1):136. doi: 10.1186/s12943-017-0680-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Du Z, Sun T, Hacisuleyman E, et al. Integrative analyses reveal a long noncoding RNA-mediated sponge regulatory network in prostate cancer. Nat Commun. 2016;7:10982. doi: 10.1038/ncomms10982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karreth FA, Reschke M, Ruocco A, et al. The BRAF pseudogene functions as a competitive endogenous RNA and induces lymphoma in vivo. Cell. 2015;161(2):319–332. doi: 10.1016/j.cell.2015.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tay Y, Kats L, Salmena L, et al. Coding-independent regulation of the tumor suppressor PTEN by competing endogenous mRNAs. Cell. 2011;147(2):344–357. doi: 10.1016/j.cell.2011.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Song YX, Sun JX, Zhao JH, et al. Non-coding RNAs participate in the regulatory network of CLDN4 via ceRNA mediated miRNA evasion. Nat Commun. 2017;8(1):289. doi: 10.1038/s41467-017-00304-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ding J, Yeh CR, Sun Y, et al. Estrogen receptor β promotes renal cell carcinoma progression via regulating LncRNA HOTAIR-miR-138/200c/204/217 associated CeRNA network. Oncogene. 2018;37(37):5037–5053. doi: 10.1038/s41388-018-0175-6. [DOI] [PubMed] [Google Scholar]
- 18.Chakravarty D, Sboner A, Nair SS, et al. The oestrogen receptor alpha-regulated lncRNA NEAT1 is a critical modulator of prostate cancer. Nat Commun. 2014;5:5383. doi: 10.1038/ncomms6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang A, Zhao JC, Kim J, et al. LncRNA HOTAIR enhances the androgen-receptor-mediated transcriptional program and drives castration-resistant prostate cancer. Cell Rep. 2015;13(1):209–221. doi: 10.1016/j.celrep.2015.08.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Askarian-Amiri ME, Crawford J, French JD, et al. SNORD-host RNA Zfas1 is a regulator of mammary development and a potential marker for breast cancer. RNA. 2011;17(5):878–891. doi: 10.1261/rna.2528811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou H, Wang F, Chen H, et al. Increased expression of long-noncoding RNA ZFAS1 is associated with epithelial-mesenchymal transition of gastric cancer. Aging (Albany NY) 2016;8(9):2023–2038. doi: 10.18632/aging.101048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li T, Xie J, Shen C, et al. Amplification of long noncoding RNA ZFAS1 promotes metastasis in hepatocellular carcinoma. Cancer Res. 2015;75(15):3181–3191. doi: 10.1158/0008-5472.CAN-14-3721. [DOI] [PubMed] [Google Scholar]
- 23.Wang W, Xing C. Upregulation of long noncoding RNA ZFAS1 predicts poor prognosis and prompts invasion and metastasis in colorectal cancer. Pathol Res Pract. 2016;212(8):690–695. doi: 10.1016/j.prp.2016.05.003. [DOI] [PubMed] [Google Scholar]
- 24.Liu R, Zeng Y, Zhou CF, et al. Long noncoding RNA expression signature to predict platinum-based chemotherapeutic sensitivity of ovarian cancer patients. Sci Rep. 2017;7(1):18. doi: 10.1038/s41598-017-00050-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dang CV. MYC on the path to cancer. Cell. 2012;149(1):22–35. doi: 10.1016/j.cell.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hsieh AL, Walton ZE, Altman BJ, Stine ZE, Dang CV. MYC and metabolism on the path to cancer. Semin Cell Dev Biol. 2015;43:11–21. doi: 10.1016/j.semcdb.2015.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gil J, Kerai P, Lleonart M, et al. Immortalization of primary human prostate epithelial cells by c-Myc. Cancer Res. 2005;65(6):2179–2185. doi: 10.1158/0008-5472.CAN-03-4030. [DOI] [PubMed] [Google Scholar]
- 28.Ellwood-Yen K, Graeber TG, Wongvipat J, et al. Myc-driven murine prostate cancer shares molecular features with human prostate tumors. Cancer Cell. 2003;4(3):223–238. doi: 10.1016/s1535-6108(03)00197-1. [DOI] [PubMed] [Google Scholar]
- 29.Hubbard GK, Mutton LN, Khalili M, et al. Combined MYC activation and Pten loss are sufficient to create genomic instability and lethal metastatic prostate cancer. Cancer Res. 2016;76(2):283–292. doi: 10.1158/0008-5472.CAN-14-3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xiang JF, Yin QF, Chen T, et al. Human colorectal cancer-specific CCAT1-L lncRNA regulates long-range chromatin interactions at the MYC locus. Cell Res. 2014;24(5):513–531. doi: 10.1038/cr.2014.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xiao ZD, Han L, Lee H, et al. Energy stress-induced lncRNA FILNC1 represses c-Myc-mediated energy metabolism and inhibits renal tumor development. Nat Commun. 2017;8(1):783. doi: 10.1038/s41467-017-00902-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fan L, Peng G, Sahgal N, et al. Regulation of c-Myc expression by the histone demethylase JMJD1A is essential for prostate cancer cell growth and survival. Oncogene. 2016;35(19):2441–2452. doi: 10.1038/onc.2015.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chandrashekar DS, Bashel B, Sah B, et al. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19(8):649–658. doi: 10.1016/j.neo.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paraskevopoulou MD, Vlachos IS, Karagkouni D, et al. DIANA-LncBase v2: indexing microRNA targets on non-coding transcripts. Nucleic Acids Res. 2016;44(D1):D231–D238. doi: 10.1093/nar/gkv1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vlachos IS, Zagganas K, Paraskevopoulou MD, et al. DIANA-miRPath v3.0: deciphering microRNA function with experimental support. Nucleic Acids Res. 2015;43(1):460–466. doi: 10.1093/nar/gkv403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pan D. The hippo signaling pathway in development and cancer. Dev Cell. 2010;19(4):491–505. doi: 10.1016/j.devcel.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zanconato F, Cordenonsi M, Piccolo S. YAP/TAZ at the roots of cancer. Cancer Cell. 2016;29(6):783–803. doi: 10.1016/j.ccell.2016.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moroishi T, Hansen CG, Guan KL. The emerging roles of YAP and TAZ in cancer. Nat Rev Cancer. 2015;15(2):73–79. doi: 10.1038/nrc3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bonci D, Coppola V, Musumeci M, et al. The miR-15a-miR-16-1 cluster controls prostate cancer by targeting multiple oncogenic activities. Nat Med. 2008;14(11):1271–1277. doi: 10.1038/nm.1880. [DOI] [PubMed] [Google Scholar]
- 40.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120(1):15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 41.Liu-Chittenden Y, Huang B, Shim JS, et al. Genetic and pharmacological disruption of the TEAD-YAP complex suppresses the oncogenic activity of YAP. Genes Dev. 2012;26(12):1300–1305. doi: 10.1101/gad.192856.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Koontz LM, Liu-Chittenden Y, Yin F, et al. The Hippo effector Yorkie controls normal tissue growth by antagonizing scalloped-mediated default repression. Dev Cell. 2013;25(4):388–401. doi: 10.1016/j.devcel.2013.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Santucci M, Vignudelli T, Ferrari S, et al. The hippo pathway and YAP/TAZ-TEAD protein–protein interaction as targets for regenerative medicine and cancer treatment. J Med Chem. 2015;58(12):4857–4873. doi: 10.1021/jm501615v. [DOI] [PubMed] [Google Scholar]
- 44.Jiao S, Wang H, Shi Z, et al. A peptide mimicking VGLL4 function acts as a YAP antagonist therapy against gastric cancer. Cancer Cell. 2014;25(2):166–180. doi: 10.1016/j.ccr.2014.01.010. [DOI] [PubMed] [Google Scholar]
- 45.Wang Z, Wu Y, Wang H, et al. Interplay of mevalonate and Hippo pathways regulates RHAMM transcription via YAP to modulate breast cancer cell motility. Proc Natl Acad Sci USA. 2014;111(1):E89–E98. doi: 10.1073/pnas.1319190110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang L, Shi S, Guo Z, et al. Overexpression of YAP and TAZ is an independent predictor of prognosis in colorectal cancer and related to the proliferation and metastasis of colon cancer cells. PLoS One. 2013;8(6):e65539. doi: 10.1371/journal.pone.0065539. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 47.Goda S, Isagawa T, Chikaoka Y, Kawamura T, Aburatani H. Control of histone H3 lysine 9 (H3K9) methylation state via cooperative two-step demethylation by Jumonji domain containing 1A (JMJD1A) homodimer. J Biol Chem. 2013;288(52):36948–36956. doi: 10.1074/jbc.M113.492595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gurel B, Iwata T, Koh CM, et al. Nuclear MYC protein overexpression is an early alteration in human prostate carcinogenesis. Mod Pathol. 2008;21(9):1156–1167. doi: 10.1038/modpathol.2008.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Koh CM, Bieberich CJ, Dang CV, Nelson WG, Yegnasubramanian S, De Marzo AM. MYC and prostate cancer. Genes Cancer. 2010;1(6):617–628. doi: 10.1177/1947601910379132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cho H, Herzka T, Zheng W, et al. RapidCaP, a novel GEM model for metastatic prostate cancer analysis and therapy, reveals myc as a driver of Pten-mutant metastasis. Cancer Discov. 2014;4(3):318–333. doi: 10.1158/2159-8290.CD-13-0346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang J, Kobayashi T, Floc’h N, et al. B-Raf activation cooperates with PTEN loss to drive c-Myc expression in advanced prostate cancer. Cancer Res. 2012;72(18):4765–4776. doi: 10.1158/0008-5472.CAN-12-0820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim J, Roh M, Doubinskaia I, Algarroba GN, Eltoum IE, Abdulkadir SA. A mouse model of heterogeneous, c-MYC-initiated prostate cancer with loss of Pten and p53. Oncogene. 2012;31(3):322–332. doi: 10.1038/onc.2011.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang P, Cao L, Fan P, Mei Y, Wu M. LncRNA-MIF, a c-Myc-activated long non-coding RNA, suppresses glycolysis by promoting Fbxw7-mediated c-Myc degradation. EMBO Rep. 2016;17(8):1204–1220. doi: 10.15252/embr.201642067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kawasaki Y, Komiya M, Matsumura K, et al. MYU, a target lncRNA for Wnt/c-Myc signaling, mediates induction of CDK6 to promote cell cycle progression. Cell Rep. 2016;16(10):2554–2564. doi: 10.1016/j.celrep.2016.08.015. [DOI] [PubMed] [Google Scholar]
- 55.Chen X, Yang C, Xie S, Cheung E. Long non-coding RNA GAS5 and ZFAS1 are prognostic markers involved in translation targeted by miR-940 in prostate cancer. Oncotarget. 2018;9:1048–1062. doi: 10.18632/oncotarget.23254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Aqeilan RI, Calin GA, Croce CM. miR-15a and miR-16-1 in cancer: discovery, function and future perspectives. Cell Death Differ. 2010;17(2):215–220. doi: 10.1038/cdd.2009.69. [DOI] [PubMed] [Google Scholar]
- 57.Bonci D, Coppola V, Patrizii M, et al. A microRNA code for prostate cancer metastasis. Oncogene. 2016;35(9):1180–1192. doi: 10.1038/onc.2015.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chang TC, Yu D, Lee YS, et al. Widespread microRNA repression by Myc contributes to tumorigenesis. Nat Genet. 2008;40(1):43–50. doi: 10.1038/ng.2007.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xue G, Yan HL, Zhang Y, et al. c-Myc-mediated repression of miR-15-16 in hypoxia is induced by increased HIF-2α and promotes tumor angiogenesis and metastasis by upregulating FGF2. Oncogene. 2015;34(11):1393–1406. doi: 10.1038/onc.2014.82. [DOI] [PubMed] [Google Scholar]
- 60.Tang W, Yu F, Yao H, et al. miR-27a regulates endothelial differentiation of breast cancer stem like cells. Oncogene. 2014;33(20):2629–2638. doi: 10.1038/onc.2013.214. [DOI] [PubMed] [Google Scholar]
- 61.Colangelo T, Polcaro G, Ziccardi P, et al. The miR-27a-calreticulin axis affects drug-induced immunogenic cell death in human colorectal cancer cells. Cell Death Dis. 2016;7:e2108. doi: 10.1038/cddis.2016.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sun Y, Yang X, Liu M, Tang H. B4GALT3 up-regulation by miR-27a contributes to the oncogenic activity in human cervical cancer cells. Cancer Lett. 2016;375(2):284–292. doi: 10.1016/j.canlet.2016.03.016. [DOI] [PubMed] [Google Scholar]
- 63.Fletcher CE, Dart DA, Sita-Lumsden A, Cheng H, Rennie PS, Bevan CL. Androgen-regulated processing of the oncomir miR-27a, which targets Prohibitin in prostate cancer. Hum Mol Genet. 2012;21(14):3112–3127. doi: 10.1093/hmg/dds139. [DOI] [PubMed] [Google Scholar]
- 64.Mo W, Zhang J, Li X, et al. Identification of novel AR-targeted microRNAs mediating androgen signalling through critical pathways to regulate cell viability in prostate cancer. PLoS One. 2013;8(2):e56592. doi: 10.1371/journal.pone.0056592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wan X, Huang W, Yang S, et al. Androgen-induced miR-27A acted as a tumor suppressor by targeting MAP2K4 and mediated prostate cancer progression. Int J Biochem Cell Biol. 2016;79:249–260. doi: 10.1016/j.biocel.2016.08.043. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.