Abstract
Mutations in the cystic fibrosis transmembrane conductance regulator (CFTR) gene cause the recessive genetic disease cystic fibrosis, where the chloride transport across the apical membrane of epithelial cells mediated by the CFTR protein is impaired. CFTR protein trafficking to the plasma membrane (PM) is the result of a complex interplay between the secretory and membrane recycling pathways that control the number of channels present at the membrane. In addition, the ion transport activity of CFTR at the PM is modulated through post-translational protein modifications. Previously we described that spleen tyrosine kinase (SYK) phosphorylates a specific tyrosine residue in the nucleotide-binding domain 1 domain and this modification can regulate the PM abundance of CFTR. Here we identified the underlying biochemical mechanism using peptide pull-down assays followed by mass spectrometry. We identified in bronchial epithelial cells that the adaptor protein SHC1 recognizes tyrosine-phosphorylated CFTR through its phosphotyrosine-binding domain and that the formation of a complex between SHC1 and CFTR is induced at the PM in the presence of activated SYK. The depletion of endogenous SHC1 expression was sufficient to promote an increase in CFTR at the PM of these cells. The results identify a SYK/SHC1 pathway that regulates the PM levels of CFTR channels, contributing to a better understanding of how CFTR-mediated chloride secretion is regulated.
Electronic supplementary material
The online version of this article (10.1007/s00018-020-03448-4) contains supplementary material, which is available to authorized users.
Keywords: CFTR, Chloride co-transport, Membrane traffic, Protein phosphorylation, SHC1, SYK
Introduction
Cystic fibrosis (CF) is the most common autosomal recessive disorder in the Caucasian population affecting more than 70,000 individuals worldwide, with an estimated incidence of one in 2500–4000 newborns [1]. CF is a multisystem disease affecting several organs and tissues caused by dysfunction of a single gene encoding the cystic fibrosis transmembrane conductance regulator (CFTR). This gene encodes the CFTR protein that functions as a chloride (Cl−) channel in the apical membrane of epithelial cells, for example, in lung, pancreas, and colon [2, 3]. The most severe disease phenotype occurs in the lungs because CFTR plays an important role in balancing Cl− secretion and sodium (Na+) absorption, which are essential for controlling airway hydration and the biophysical properties of the secreted mucus [4, 5]. Lack of CFTR function causes airway dehydration and increased mucus viscosity, compromising mucociliary clearance and promoting chronic infection and inflammation that eventually leads to respiratory failure.
CFTR is a member of the ATP-binding cassette (ABC) transporter family and is composed of five domains: two membrane-spanning domains (MSD1 and MSD2), each of them composed of six transmembrane segments (TM 1–6 and TM 7–12) that form the channel allowing Cl− (and also bicarbonate) to flow across the membrane; two cytosolic nucleotide-binding domains [nucleotide-binding domain 1 (NBD1) and NBD2], which regulate channel gating and where ATP is hydrolyzed; one regulatory domain (R) that mediates phosphorylation-dependent regulation of channel activity [6, 7].
More than 2000 mutations have been identified in the CFTR gene, which can affect various cellular processes required for the maturation and function of the CFTR protein, including protein folding, trafficking to the plasma membrane (PM) and channel activity [8]. A deletion of phenylalanine at position 508 in NBD1 is present in approximately 85% of CF patients in at least one allele. This F508del mutation causes protein misfolding and results in substantial endoplasmic reticulum (ER) retention and premature CFTR degradation, preventing most of the mutant protein from reaching the cell surface [2, 6, 9]. Compounds such as VX-809 have been recently identified that directly interact with F508del-CFTR and facilitate its folding leading to a partial rescue of its trafficking to the cell surface [10–12]. However, for improved clinical benefit, additional cellular processes need to be targeted, including those affecting the prevalence of CFTR once it reaches the PM [13–16]. To identify the suitability as therapeutic targets of such additional processes, more knowledge on basic CFTR biology is required. CFTR traffic, stability, and activation at the PM depend on numerous interactions involving the secretory and membrane recycling pathways, the cytoskeleton, and signaling mechanisms. The underlying network of proteins is very complex and far from fully characterized, but is known to include trafficking machinery components (Rab GTPases, SNAREs, PDZ-domain-containing proteins), other membrane proteins, as well as different types of molecular switches (such as kinases, phosphatases, and small GTPases) [17–21].
One additional mechanism found to modulate the PM prevalence of CFTR is its tyrosine phosphorylation by cytoplasmic non-receptor tyrosine kinases, which are frequently associated with transmembrane proteins at the inner plasma membrane surface. The CFTR protein contains 40 tyrosine (Tyr) residues and Tyr625 and 627 were found to contribute to CFTR activation by SRC or PYK2 [22, 23]. The spleen tyrosine kinase (SYK) was previously reported to specifically phosphorylate the NBD1 at Tyr512, the only tyrosine in the primary CFTR sequence that obeys the canonical SYK substrate motif (consensus Y-E/D-E/D-X) [24]. Phosphorylation by SYK led to decreased expression of CFTR at the PM and mutation of Tyr512 to the non-phosphorylatable residue Phe512 was sufficient to increase CFTR plasma membrane levels. However, the mechanism by which SYK phosphorylation regulates CFTR levels at the cell surface remained unknown. SYK has an important role in immune cell activation [25], including lung inflammatory processes, and its activation requires binding of its SH2 domains to phosphorylated immune receptor tyrosine-based activation motifs. In addition, SYK was also found to be expressed in airway epithelial cells [26, 27] and can also be activated by a distinct mechanism involving its recruitment to the cytoplasmic domain of clustered integrins with subsequent phosphorylation by SRC-family kinases [28]. Here we describe that experimental manipulation of SYK expression levels or SYK catalytic activity affects CFTR abundance at the PM and identifies the cytosolic adaptor protein SHC1 as a binding partner for SYK-phosphorylated CFTR.
Materials and methods
Cell culture and transfections
CFBE41o- cells stably expressing wt- (CFBE wt-CFTR) or F508del-CFTR (CFBE F508del-CFTR) [29] (gift from JP Clancy, University of Alabama, USA) were grown as standard monolayers in minimal essential medium (MEM) supplemented with l-glutamine, Earle’s salts (Gibco, Thermo Fisher Scientific, Waltham, USA), 10% (v/v) fetal bovine serum (FBS) and 2 μg/mL puromycin (Invitrogen, Thermo Fisher Scientific, Waltham, USA). CFBE41o- cells stably transduced with lentivirus encoding mCherry-Flag-tagged (CFBE mCherry-Flag-wt-CFTR cells) wt-CFTR under Tet-ON promotor were generated by ADV Bioscience LLC [30] and grown in MEM supplemented with l-glutamine, Earle’s salts (Gibco), 10% (v/v) FBS, 2 μg/mL puromycin and 10 mg/mL blasticidin (all from Invitrogen). Expression of CFTR constructs was induced by incubation for 24 h with 1 μg/mL doxycycline (dox) (Sigma-Aldrich, Madrid, Spain). HeLa (cervix carcinoma) cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) and Caco-2 cells in Roswell Park Memorial Institute (RPMI) 1640 Medium, both supplemented with 10% (v/v) FBS (both from Gibco). CFBE wt-CFTR and Caco-2 cells were further engineered to stably express the halide sensor YFP-F46L/H148Q/I152L (HS-YFP) [31], as described by AM Matos in [32]. Selected cell line clones were maintained in the same conditions as their respective parental cells, except for the addition of 400 μg/mL hygromycin (Invitrogen) as selection agent for HS-YFP expression. All cells were regularly checked for the absence of mycoplasm infection.
For transient cell transfection, 3 × 105 cells/cm2 CFBE monolayer cells were reverse-transfected using LipofectAMINE 2000 (Invitrogen), according to the manufacturer’s instructions. For ectopic expression of complementary DNA-encoding plasmids, a DNA/lipofectamine ratio of 1:3 (µg/µL) was used with 8 μg per 100-mm dish, 4 μg per 60-mm dish, or 2 μg per 35-mm dish. Constructs were supplemented with empty vector when required and cells were analyzed after 20 h for biochemical assays [33]. Transfection efficiencies were found to be around 90%, as determined microscopically using a GFP-encoding expression vector. In case of small interfering RNAs (siRNAs), 200 pmol were reverse transfected and cells analyzed 48 h after transfection. siRNAs used were sc-29501 and sc-29480 (Santa Cruz Biotechnology) against SYK and SHC, respectively, and the control luciferase siRNA (5′-CGU ACG CGG AAU ACU UCG ATT) (Eurofins Genomics, Ebersberg, Germany).
For drug treatments, cells were incubated for 1 h with the vehicle DMSO (Sigma-Aldrich) or with one of two different SYK inhibitors: BAY 61-3606 (2-(7-(3,4-dimethoxyphenyl)-imidazol[1,2-c]pyrimidin-5-ylamino) nicotinamide) (Calbiochem, San Diego, CA, USA) or PRT062607 (P505-15, BIIB057) (Selleck Chemicals, Munich, Germany) at a final concentration of 8 nM and 1 nM, respectively. The stock solutions were prepared at least 1000-fold in DMSO so that the DMSO concentration during cell treatment did not exceed 0.1% (v/v). Cells were treated for 24 h with 3 μM VX-809 (Selleck Chemicals) where indicated.
DNA plasmids and constructs
Human SHC1 cDNA was amplified from pcDNA3-SHC1 (gift from Enrica Migliaccio, Campos IFOM-IEO, Milan, Italy) by PCR with the primers EcoRI-Fw (5′ GAA TTC TTG AAC AAG CTG AGT GGA GG) and NotI-Rv (5′ CGG CCG TCA CAG TTT CCG CAC) and subcloned into the pcDNA3-Myc vector as an EcoRI/NotI fragment. SHC1 R401K, SHC1 R175K and SHC1 R401K/R175K were generated from pcDNA3-Myc-SHC1 by changing, respectively, codon 401 from CGG to AAG, codon 175 from AGA to AAA, or both codons, using the QuickChange Site-Directed Mutagenesis Kit (Stratagene La Jolla, CA, USA), according to the manufacturer’s instructions.
Previously published constructs used in this study were kinase-dead YFP-SYK-K402R (SYK kd) [24]. The Flag-tagged SYK phosphomimetic Y352D construct (SYK Y352D) [34] was a gift from Dimitar G. Efremov (ICGEB, Rome, Italy). All constructs were confirmed and verified by automated DNA sequencing.
Immunoprecipitation and western blot (WB) procedures
For co-immunoprecipitation experiments, CFBE mCherry-Flag-wt-CFTR cells were grown in 100-mm dishes, transfected as indicated above and incubated with 1 μg/mL of dox for 24 h. Then cells were placed on ice, washed three times with ice-cold phosphate-buffered saline–CM (PBS–CM: PBS, pH 8.0, containing 1 mM CaCl2 and 1 mM MgCl2) and left for 5 min in cold PBS–CM. Cells were incubated with agitation for 2 h with anti-Flag clone M2 (1:500, F3165, Sigma-Aldrich) and washed twice with ice-cold PBS–CM. Cells were again washed three times with PBS–CM and lysed in 500 μL lysis buffer (50 mM Tris–HCl, pH 7.5, 2 mM MgCl2, 100 mM NaCl, 10% glycerol, 1% NP-40, 0.01% SDS) supplemented with a protease inhibitor cocktail (composed of 1 mM PMSF, 1 mM 1,10-phenanthroline, 1 mM EGTA, 10 μM E64, and 10 μg/mL of each aprotinin, leupeptin, and pepstatin A (all from Sigma-Aldrich). Cells lysates were cleared at 3000×g for 5 min at 4 °C and about 450 μL lysate was added to 45 μL of streptavidin–agarose beads (Sigma-Aldrich) and rotated for 1 h at 4 °C to perform a pre-clearing of the lysates. After that, about 400 μL of the pre-cleared lysates was added to 15 μL protein G-conjugated magnetic beads (Invitrogen) and rotated for 1 h at 4 °C, centrifuged for 1 min at 3000×g and washed five times in cold lysis buffer. Proteins were solubilized from the beads in 20 μL 2 × SDS CFTR sample buffer [62.5 mM Tris–HCl, pH 6.8, 3% SDS, 10% glycerol, 0.02% bromophenol blue, 196.4 mM dithiothreitol (DTT)], with extra 100 mM DTT. Total protein was quantified by a modified micro-Lowry method, and 30 μg of total protein was loaded and separated in 10% SDS polyacrylamide gels in Protean III mini-gels (Bio-Rad, Hercules, CA, USA). Gels to assess CFTR expression contained 1% glycerol and were run at 4 °C [24, 33].
Following electrophoresis, the separated proteins were transferred onto a PVDF membrane (BioRad) in a cooled Mini Trans-Blot cell (Bio-Rad) at 300 mA, followed by Coomassie Brilliant Blue staining to check for equal transfer. Membranes were blocked in TBS, 0.1% Triton X-100, 5% non-fat milk powder, probed using the indicated antibodies, and then incubated with a secondary peroxidase-conjugated antibody (BioRad) followed by chemiluminescence detection. Primary antibodies used for WB were mouse anti-GFP (11814560001) from Roche, mouse anti-GFP (ab1218), rabbit anti-SYK (ab40781) from Abcam, mouse anti-Flag clone M2 (F3165) and anti-Myc clone 9E10 (M5546) from Sigma-Aldrich, mouse anti-PCNA clone PC10 (NA03) from Calbiochem, mouse anti-CFTR clone 596 from Cystic Fibrosis Foundation Therapeutics; rabbit anti-Myc (sc-789), mouse anti-GRB2 (sc-8034), mouse anti-SHC (sc-967) and mouse anti-BSPRY (sc-377320) from Santa Cruz Biotechnology; mouse anti-NCK (610099) and mouse anti-DAB2 (610464) from BD Transduction Laboratories (San Jose, CA, USA). For densitometric quantification of band intensities, the luminescence film exposures from at least three independent experiments were digitalized and analyzed using the ImageJ software (NIH). The obtained values were normalized to the value of the loading control protein in the control sample.
Biotinylation of cell surface proteins
CFBE wt-CFTR and CFBE F508del-CFTR cells (either transfected or drug-treated as described above) were washed three times with warm culture medium to remove dead cells, and then placed on ice in a cold room. Cells were washed three times with ice-cold phosphate-buffered saline–CM (PBS–CM) (PBS, pH 8.0, containing 1 mM CaCl2 and 1 mM MgCl2) to ensure arrest of endocytic traffic. Then cells were incubated for 45 min with 0.5 mg/mL EZ-Link Sulfo-NHS-SS-Biotin (sc-212981, from Santa Cruz Biotechnology) in PBS–CM to label all cell surface proteins. Cells were rinsed twice and incubated for 15 min on ice in ice-cold Tris/glycine (100 mM Tris–HCl, pH 8.0, 150 mM NaCl, 1 mM MgCl2, 0.1 mM CaCl2, 10 mM glycine, 1% BSA) to quench the biotinylation reagent. Cells were again washed three times with cold PBS–CM and lysed in 250 μL pull-down buffer (50 mM Tris–HCl, pH 7.5, 100 mM NaCl, 10% glycerol, 1% NP-40) in the presence of the protease inhibitor cocktail (described above). The cell lysates were harvested at 9000×g at 4 °C for 5 min. An aliquot of 40 µL representing the total protein level was removed and added to 2 × SDS CFTR sample buffer (see above), while 200 µL lysate was added to 45 µL streptavidin–agarose beads (Sigma-Aldrich), previously incubated for 1 h in 1 mL cold pull-down buffer containing 2% non-fat milk powder, and washed three times in pull-down buffer. Lysates were incubated with the prepared beads for 1 h at 4 °C, the beads collected by centrifugation of 1 min at 3000×g, and washed four times in cold wash buffer (100 mM Tris–HCl, pH 7.5, 300 mM NaCl, 1% Triton X-100). Captured proteins were recovered in 20 µL of 2 × SDS CFTR sample buffer with extra 100 mM DTT and analyzed by WB with specific antibodies, as described above.
Peptide synthesis and peptide pull-down assays (PPD)
Biotinylated peptides were synthetized by Invatis (Cologne, Germany), and the identity and purity confirmed by mass spectrometry (MS) analysis. Peptides were designed as decamers bearing an N-terminal biotin on a tetrapeptide linker SGSG. The biotin–SGSG-cap enabled attachment to streptavidin beads in the pull-down assay. Peptides were synthesized in three variants: as “active” (tyrosine-phosphorylated—pY), “control” (non-phosphorylated—WT), and “mutant” [substitution of tyrosine (Y) by phenylalanine (F)—MUT]. Peptide sequences used were EGFR-pY-Biot-SSSGSGLPVPEY(p)INQSV; EGFR-WT-Biot-SSSGSGLPVPEYINQSV; EGFR-MUT-Biot-SSSGSGLPVPEFINQSV; CFTR-pY-Biot-SSSGSGIFGVSY(p)DEYRY; CFTR-WT-Biot-SSSGSGIFGVSYDEYRY; CFTR-MUT-Biot-SSSGSGIFGVSFDEYRY.
Immobilized streptavidin beads (120 µL) were coupled with 5 µg/µL of each of the biotinylated peptide and incubated for 2 h at 4 °C. CFBE wt-CFTR cells were grown in 100-mm dishes to capture endogenous proteins, and seeded in 60-mm dishes when exogenous proteins were used. Cells were lysed on ice in 750 μL lysis buffer (50 mM Tris–HCl, pH 7.5, 100 mM NaCl, 2 mM MgCl2, 1% NP-40, 10% glycerol) supplemented with a protease inhibitor cocktail (described above) and lysates were cleared by centrifugation at 9000×g at 4 °C for 5 min. An aliquot of 40 µL representing the total protein level was preserved and added to 2 × SDS CFTR sample buffer (see above), while the remaining 700 µL lysate was pre-cleared by adding to 100 µL streptavidin–agarose beads and incubating for 2 h at 4 °C. The pre-cleared lysates were collected by centrifugation of 1 min at 3000×g, added to the peptide-coupled beads and incubated for 2 h at 4 °C. The beads were collected by centrifugation of 1 min at 3000×g and washed nine times with wash buffer (50 mM Tris–HCl, pH 7.5, 150 mM NaCl, 2 mM MgCl2, 1% NP-40, 10% glycerol). Captured proteins were recovered in 60 µL of 2 × SDS sample buffer (100 mM Tris–HCl, pH 6.8, 4% SDS, 10% glycerol, 0.5 mg bromophenol blue, 130 mM DTT) and separated in a 10% gel by SDS-PAGE.
For proteomic identification, SDS-PAGE-separated proteins were silver stained, after gel fixation for 1 h in fixer solution (40% ethanol, 10% glacial acetic acid, 50% ddH2O) and washing overnight in ddH2O. Gels were sensitized in 0.02% sodium thiosulfate during 1 min, washed three times with ddH2O, then incubated for 20 min in cold 0.1% silver nitrate solution with 0.02% formaldehyde and washed twice with ddH2O. Gels were stained in 3% sodium carbonate solution with 0.05% formaldehyde until visible bands appeared, then washed with ddH2O and incubated for 5 min with 5% acetic acid and stored at 4 °C in 1% acetic acid until MS analysis. For identification of the complex mixture of proteins pulled down with different peptides, the whole samples were loaded on a SDS-PAGE gel, run for a distance of only 1 cm, and after silver staining the entire stained area was excised as one sample and sent to nano-LC–MS analysis using SCIEX TripleTOF 6000 system, outsourced to UniMS (ITQB Institute, Oeiras, Portugal).
Bioinformatic data analysis
Peptide pull-down (PPD) experiments were performed in triplicates, and for each replicate, the identification of each protein in MS spectra was characterized by a protein confidence score (PCS), detected with ProteinPilot™ Software (Sciex). For PCS > 1.3, the confidence in the identification of a particular protein in the sample is equal or higher than 95%. To conjugate this individual score with the detection of the same protein across replicates, we generated a combined confidence score (CCS). The CCS is characterized by five levels of confidence for protein detection: level 5 proteins are detected with PCS ≥ 1.3 in more than one replicate and not in the controls; level 4 proteins are detected in one replicate with PCS ≥ 1.3 and not in the controls; level 3 proteins are detected in more replicates than in controls, and with an average PCS higher than their respective the controls; level 2 proteins are detected in one replicate with PCS ≤ 1.3, and not in the controls; and level 1 proteins are detected in the same number of replicates and controls, with a replicate average PCS higher than the corresponding controls. All other cases are integrated in level 0, where proteins are not considered to be present in the set of replicates.
Human interactome networks between the detected proteins were built from two data sources: Agile Protein Interactomes DataServer (APID, https://apid.dep.usal.es/) and The Human Reference Protein Interactome Mapping Project (HuRI, https://interactome.baderlab.org/). APID collects physical protein interactions reported in the literature. We used all the interactions available in APID between human proteins. HuRI gives access to interactions detected experimentally through unbiased high-throughput pairwise protein interactions. Network analyses were conducted in R using functions from the iGraph package. Network visualizations were produced with Cytoscape.
Iodide influx assay
CFBE wt-CFTR or Caco-2 cells, expressing the HS-YFP halide sensor [31], were seeded as a monolayer in eight-well chamber slides (Nunc, Rochester, NY, USA). While Caco-2 cells were treated with SYK inhibitors as described above, CFBE cells were transfected with either siSYK (sc-29501), siSHC (sc-29480) or siLUC. After 48 h at 37 °C, cells were carefully washed twice with isomolar PBS (WPBS: 137 mM NaCl, 2.7 mM KCl, 0.7 mM CaCl2, 1.1 mM MgCl2, 1.5 mM KH2PO4, 8.1 mM Na2HPO4, pH 7.4), and incubated for 15 min in WPBS containing 1 μM indomethacin to reduce endogenous cAMP levels, and with or without 25 μM of CFTR inhibitor 172 (inh172). CFTR activity was then stimulated for 10 min by addition of 5 μM forskolin (Fsk). Cells were then transferred to a Leica TCS-SPE confocal microscope for time-lapse analysis. Each well was assayed individually for iodide influx by recording fluorescence continuously (500 ms per point), first for 10 s (baseline) and then for 60 s, after the rapid (≤ 1 s) addition of IPBS, in which NaCl was replaced by 137 mM NaI (final iodide concentration of 100 mM/plate well). Cells were kept at 37 °C up until being assayed at room temperature. After background subtraction, HS-YPF fluorescence recordings (F) were normalized to the initial average measured before addition of I− (F0). Quantification of fluorescence decay was performed on at least 24 individual cells per well, using ImageJ (NIH) as described [33]. The average fluorescence decay was fitted to an exponential decay function to derive the maximal slope that corresponds to initial influx of I− into the cells [31, 33]. All experiments were performed in triplicate.
Statistical analysis
Data were analyzed using one-way ANOVA followed by post hoc Tukey HSD tests for multiple comparisons or unpaired Student’s t tests when comparing two datasets. In both cases, statistical significance was accepted for P values < 0.05. Shown data reflect the means ± SEM from at least three independent experiments.
Results
SYK activity modulates the cell surface expression of CFTR in human airway epithelial cells
Previously, we described in CFTR-transfected BHK-21 (baby hamster kidney) cells that SYK-mediated phosphorylation at the specific Tyr512 affected the abundance of CFTR in the PM [24]. To demonstrate that inhibition of SYK activity also interfered with CFTR PM abundance in a physiologically relevant cell line, bronchial epithelial CFBE41o- cells stably expressing human wild-type (wt)-CFTR [29] were used to modulate either the activity or the expression of SYK.
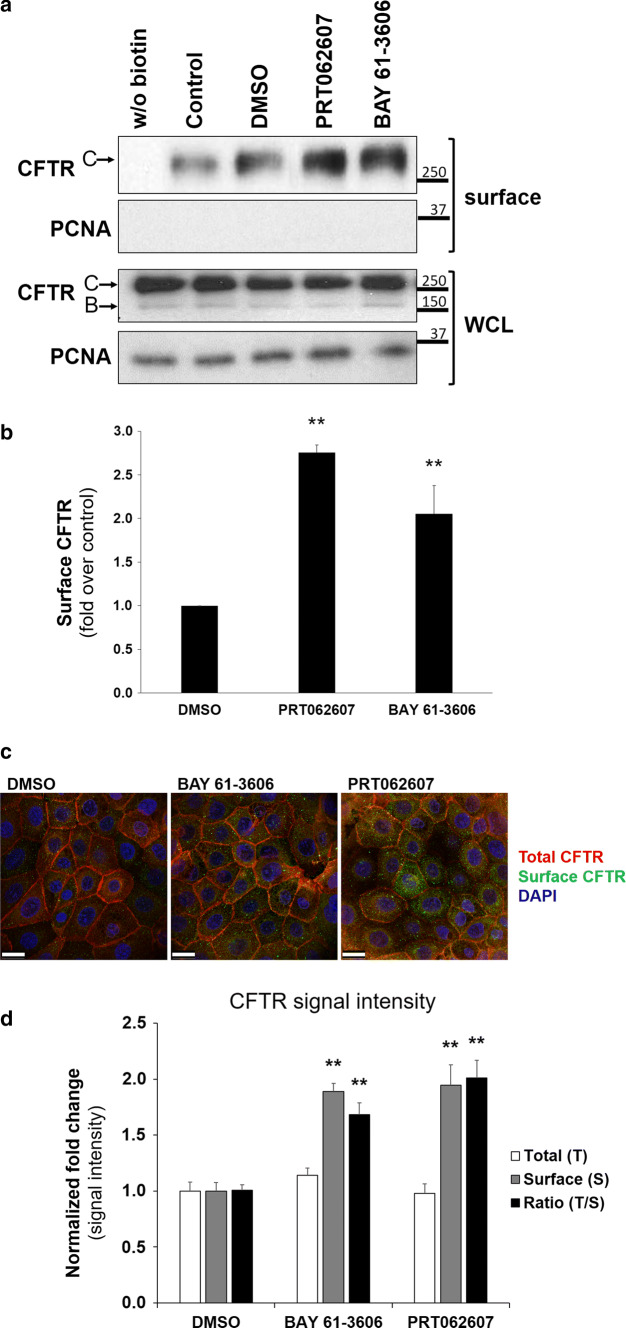
First, cells were treated for 1 h with either DMSO or with one of two specific SYK inhibitors (BAY 61-3606 and PRT062607). Then the expression of CFTR at the PM was analyzed by biotinylation of cell surface proteins. Data were compared to cells incubated with DMSO, because this was the solubilizing vehicle for the inhibitors and it can, by itself, act as a chemical chaperone that improves protein folding and may thus increase steady-state delivery of plasma membrane proteins. Under these conditions, treatment with both SYK inhibitors promoted a significant, two- to threefold increase in CFTR levels at the PM (Fig. 1a, b). In parallel, we analyzed the effect of both SYK inhibitors by confocal fluorescence microscopy. For this, we used CFBE cells that stably express a CFTR wt construct carrying a N-terminal mCherry tag and a Flag-epitope located at the fourth extracellular loop [30] so that anti-Flag antibodies on intact cells will exclusively bind to CFTR proteins at the cell surface (as described in Refs. [32, 34]). As shown in Fig. 1c, d, treatment of these cells with either SYK inhibitor promoted a significant increase in anti-Flag signal at the cell surface, consistent with increased abundance of CFTR at the PM.
Fig. 1.
Effect of pharmacological SYK inhibition on the levels of CFTR at the PM of CFBE cells. CFBE wt-CFTR cells were treated for 1 h with control solvent DMSO, or with one of the SYK inhibitors BAY 61-3606 (8 nM) or PRT062607 (1 nM). After the treatment, cell surface proteins were biotinylated, cells lysed, and proteins resolved by SDS-PAGE and detected by WB. a Detection of the indicated proteins in whole-cell lysates (WCL) or in the biotinylated protein fraction (surface). For CFTR, the mature, fully glycosylated and the immature, core-glycosylated protein are marked as bands C and B, respectively. The PCNA protein served as a loading (WCL) and contamination (surface) control, respectively. b Corresponding quantification of CFTR detection in the biotinylated cell surface fractions, obtained from at least three independent experiments. Densitometric analyses of the band intensities in a were expressed relative to DMSO-treated cells. All shown data represent means ± SEM; **P < 0.01. c After inhibitor treatment, mCherry-Flag-wt-CFTR expressing CFBE cells were fixed and analyzed by confocal fluorescence microscopy. Confocal images captured the cell surface plane of non-permeabilized cells and show an overlay from triple-labeled cells, where mCherry fluorescence reflects the total amount of CFTR, Alexa Fluor 488 fluorescence stains the amount of Flag-CFTR present at the cell surface and DAPI stains nuclei. The cell surface plane was selected after acquisition of a 0.2-μm Z-stack of 1-Airy confocal images. Note that inhibitor treatment increased the green signal representing Flag-tagged CFTR at the cell surface. White bars correspond to 25 μm. d Corresponding quantification of total and cell-surface located CFTR signal intensities in the images. ImageJ software (NIH) was used to delimit each single cell as an individual region-of-interest to allow measurement of mCherry and Alexa488 fluorescence signal intensities (3 independent experiments with ~ 120 cells analyzed from 5 representative microscopic fields per condition). Note the specific increase in cell surface-located CFTR after inhibitor treatment. Data shown in b and d represent means ± SEM (n ≥ 3).Significance was determined using one-way ANOVA (both P < 0.001), followed by post hoc Tukey’s tests (**P < 0.01)
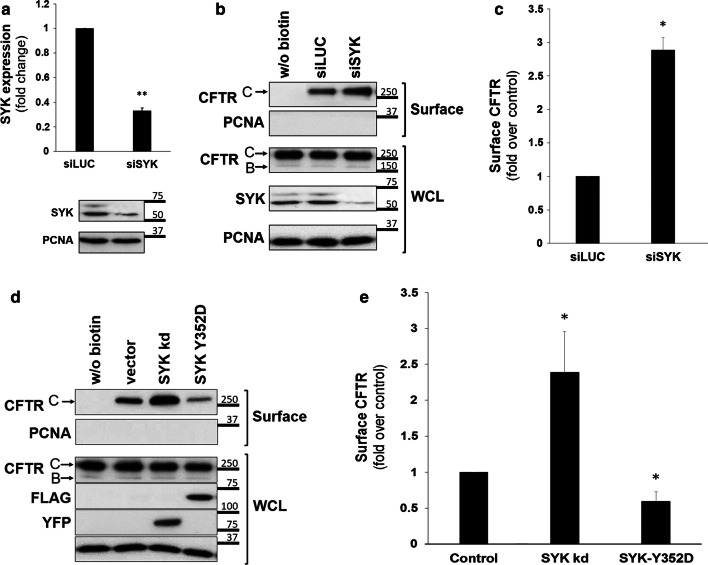
Second, specific siRNAs were used to deplete endogenous SYK expression in CFBE wt-CFTR cells, which decreased to approximately 30% of its normal levels (Fig. 2a). Under these conditions, we found a 2.9-fold increase in the amount of CFTR in the cell surface fraction (Fig. 2b, c), compatible with the result obtained by pharmacological inhibition of SYK activity. No additive effect was observed when SYK-depleted cells were treated for 1 h with BAY 61-3606 or PRT062607.
Fig. 2.
Effect of experimental manipulation of SYK expression levels and its catalytic activity on the levels of CFTR at the PM of CFBE wt-CFTR cells. a Assessment of the efficiency of siRNA-mediated knockdown of SYK. CFBE wt-CFTR cells were transfected as described under “Cell culture and transfections”and depletion of endogenous SYK protein levels analyzed by Western blot. Data quantification by Student’s t test (n = 6) is graphically displayed and shown as fold-change relative to siLUC (control). Note the successful downregulation of SYK expression by approximately 70%. b, c Effect of depleting endogenous SYK expression in CFBE wt-CFTR cells transfected with one of the indicated siRNAs. d, e Effect of overexpression of SYK mutants in cells transfected with empty-vector, inactive mutant YFP-SYK kd, or constitutively active mutant Flag-SYK Y352D. After the treatments described above, cell surface proteins were biotinylated, cells were lysed, and proteins resolved by SDS-PAGE and detected by WB. b, d Detection of the indicated proteins in whole-cell lysates (WCL) or in the biotinylated protein fraction (surface). For CFTR, the mature, fully glycosylated and the immature, core-glycosylated protein are marked as bands C and B, respectively. The PCNA protein served as a loading (WCL) and contamination (surface) control, respectively. c, e Corresponding quantification of CFTR detection in the biotinylated cell surface fractions, obtained from at least three independent experiments. a, c, e Densitometric analyses of the band intensities were expressed relative to the control sample (n ≥ 3) and analyzed either by unpaired Student’s T tests (a, c) or one-way ANOVA (P < 0.001), followed by post hoc Tukey’s tests (e). All shown data represent means ± SEM. *P < 0.05; **P < 0.01
Finally, SYK activity was modulated by transfecting cells with SYK mutants, either a kinase-dead (kd) or a constitutively active SYK mutant (SYK Y352D). In agreement with the data described above, we observed an over twofold increase in CFTR levels at the cell surface upon expression of SYK kd (Fig. 2d, e), while a proportional decrease was detected when cells were transfected with constitutively active SYK.
Identification of the biochemical mechanism underlying the effect of SYK phosphorylation on CFTR at the PM
The molecular details underlying the effect of SYK phosphorylation on CFTR at the PM remained unknown. We hypothesized that tyrosine phosphorylation would create binding sites for proteins containing phosphotyrosine-binding (PTB) domains, such as the SRC-homology 2 (SH2) or PTB domains. These binding proteins can act as adaptors for the cellular response to the tyrosine phosphorylation, an effect well studied in the assembly and activation of signaling events triggered upon growth factor stimulation of receptor tyrosine kinases (RTK). We thus set out to identify adaptor proteins in the proteome of bronchial epithelial cells using a PPD assay previously employed for the identification of GBR2 binding to the epidermal growth factor receptor (EGFR) [35]. GRB2 recognizes the activated tyrosine-phosphorylated receptor and recruits the guanine exchange factor SOS to activate the GTPase RAS at the PM [36].
To validate the PPD assay, we ordered biotinylated synthetic EGFR peptides containing a biotin modification for coupling to streptavidin-coated beads. Three peptide versions were used: “active” (phosphorylated—pY), “control” (non-phosphorylated—WT) and “mutant” (substitution of tyrosine by phenylalanine—MUT) form. During initial optimization experiments, streptavidin beads were loaded with two different quantities (1 and 5 μg/μL) of biotinylated peptide prior to incubation with HeLa cell lysates to analyze the best condition for specific protein binding. To control for the non-specific binding of proteins, we incubated empty beads with the respective lysate. These assays demonstrated that GRB2 bound specifically to the EGFR phosphopeptide and not to the non-phosphorylated or mutant peptides. Moreover, the degree of depletion of GRB2 observed in the wash supernatants from the PPD fractions indicated that 5 μg/μL of peptide guaranteed an excess sufficient to capture most of the target protein.
Next, we used the active, control and mutant peptides containing the identified phosphotyrosine in the context of the CFTR primary sequence (see Fig. 3b) and performed the PPD using the physiologically relevant lysate of CFBE wt-CFTR cells. To identify which proteins bound specifically to the phosphorylated CFTR peptide, we sent the PPD fraction for MS analysis.
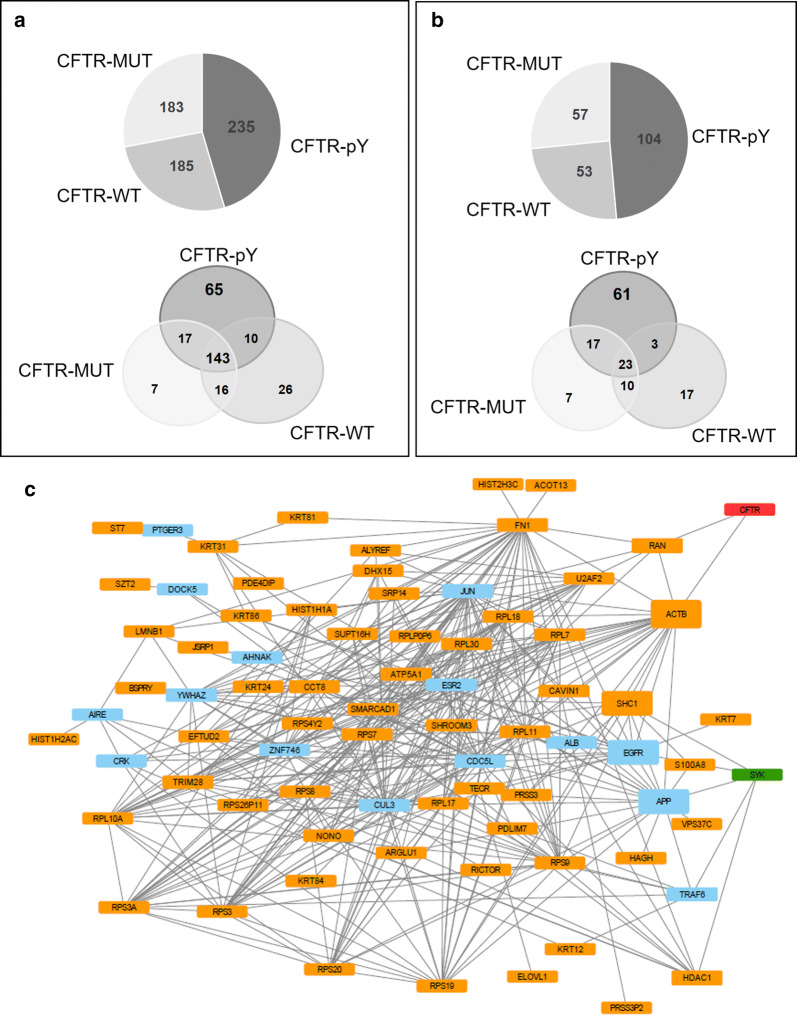
Fig. 3.
Graphic display of the number and network structure of proteins identified by MS in the CFTR interactome. Qualitative nanoLC MS/MS analysis was converted into Venn diagrams and indicate a the number of proteins in common between each synthetic peptide, as well as b interactors selected for a high-confidence score (CCS of 4 or 5). pY—tyrosine-phosphorylated peptide; WT—non-phosphorylated peptide; MUT—peptide with substitution of tyrosine by phenylalanine. c Subnetwork of proteins selected for direct interaction with SYK kinase and CFTR. Schematic representation of the 61 proteins connected through direct interactions with CFTR (in red) and SYK (in green). In blue are represented proteins that were required to allow the formation of a connected subnetwork. Node box height is proportional to the fractions of short paths that contain the node and link SYK and CFTR. Paths with four or less interactions of length were considered as short paths
Bioinformatic analysis of MS data identifies candidate proteins involved in the regulation of CFTR by SYK kinase
The qualitative nano-LC MS/MS analysis of the above-described PPD samples generated data sets in which each protein was characterized by a protein confidence score (PCS) attributed by the SCIEX proprietary ProteinPilot™ Software. For a PCS > 1.3, the confidence in the identification of that particular peptide is greater or equal than 95%. Altogether, this data analysis found 603 putative proteins associated with CFTR (Fig. 3a; Table S1), 235 of which identified from the phosphorylated peptide samples. Of these 235 proteins, 143 were also detected in the precipitates of the other two control peptides. This restricted the candidate list to 65 proteins that were specifically isolated with the “active” (pY) peptide.
Since experiments were performed in triplicate, we next created an algorithm to generate a CCS across the different replicates that produced five additional integrated confidence levels (for details see “Bioinformatic data analysis”). All proteins with a CCS of 4 or 5 (at least one replicate with PCS ≥ 1.3 and not present in control samples) were considered as a sublist of high-confidence putative CFTR-associated proteins.
Using this confidence filter, four proteins were excluded from the candidate list, and the 61 high-confidence score (CCS of 4 or 5) CFTR-associated proteins (Fig. 3b; Table S2) were then used to construct an integrated protein network by extracting known physical protein interactions from the human proteome databases. Most proteins in this sublist had known interactions between each other and where at a close distance from both SYK and CFTR. Using a minimum set of 15 extra proteins, it was possible to join the 61 high-confidence interactors, SYK kinase and CFTR in a densely connected subnetwork (Fig. 3c). Among the 61 high-confidence interactors, beta-actin (ACTB) and SRC-homology 2 domain containing transforming protein 1 (SHC1) were identified as strong mediators of the interaction between SYK and CFTR, as they were part of a greater number of short paths (with four or less interactions) linking these two proteins. To further refine our candidate list, we restricted this network to proteins that have a role in membrane trafficking, protein transport and regulation of ion channels or subcellular localization to plasma membrane, cytoplasm or ER (Table 1). This selected 12 proteins as the short list of candidates involved in mediating the effect of SYK-mediated phosphorylation on CFTR Tyr512.
Table 1.
Short list of high-confidence score proteins isolated with the CFTR phosphopeptide
| Peptide | Accession number | Protein name |
|---|---|---|
| CFTR-pY | Q5W0U4 | B box and SPRY domain-containing protein |
| Q9NR12 | PDZ and LIM domain protein 7 | |
| Q9H4L7 | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A containing DEAD/H box 1 | |
| Q96MG2 | Junctional sarcoplasmic reticulum protein 1 | |
| P29353 | SHC-transforming protein 1 | |
| Q9NWB6 | Arginine and glutamate-rich protein 1 | |
| P37108 | Signal recognition particle 14 kDa protein | |
| Q9NRC1 | Suppressor of tumorigenicity 7 protein | |
| Q9NPJ3 | Acyl-coenzyme A thioesterase 13 | |
| Q9BW60 | Elongation of very long chain fatty acids protein 1 | |
| Q8TF72 | Protein Shroom3 | |
| P98082 | Disabled homolog 2 | |
| A5D8V6 | Vacuolar protein sorting-associated protein 37C |
SHC1 binds specifically to the CFTR phosphopeptide
In cells, pTyr is specifically recognized by adaptor proteins containing one of two protein domains, SH2 or PTB. Among the list of candidate proteins shown in Table 1, only SHC1 is a known phosphotyrosine binding adaptor protein. SHC1 contains both an SH2 and a PTB domain and binds directly to tyrosine-phosphorylated RTKs [37–39]. SHC1 also was reported to interact directly with SYK [40] and was a highly relevant node in our network connecting SYK and CFTR (Fig. 3c).
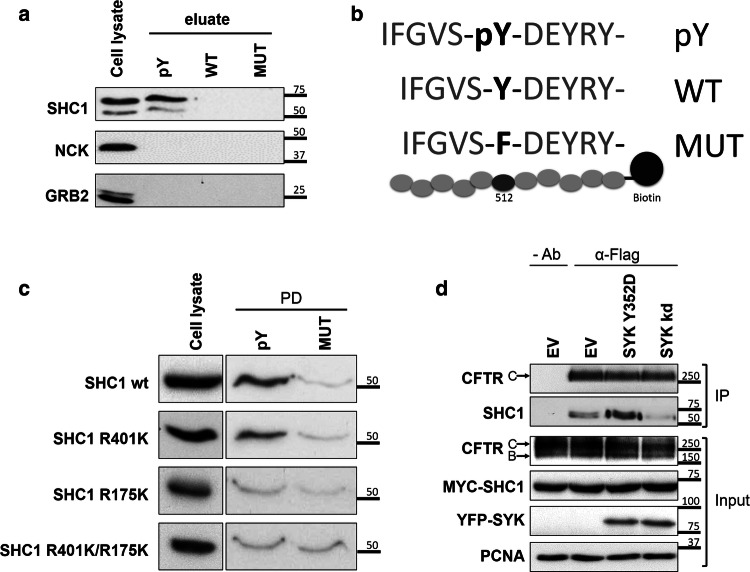
Having identified SHC1 as the putative candidate protein that binds to CFTR pTyr512, the other 11 proteins shown in Table 1 are likely part of an associated protein complex. To further validate the interaction of SHC1 with the CFTR phosphopeptide, we proceeded with the PPDs as above using CFBE wt-CFTR cell lysates, then separated the samples by SDS-PAGE and detected the respective endogenous SHC1 protein by WB with specific antibodies. As negative controls, we used NCK and GRB2, two well-known SH2 domain adaptor proteins, which were absent from the list of proteins pulled down with the CFTR peptides. The results confirmed that endogenous SHC1 binds specifically to the CFTR phosphopeptide (Fig. 4a, b), in contrast to the two negative controls, NCK and GRB2. The same binding specificity was observed when experiments were performed with lysates from HEK293 cells that express endogenous SHC1 but not CFTR protein (data not shown). We also confirmed that loading the streptavidin beads with 5 μg/μL of biotinylated peptide guaranteed an excess sufficient to capture most of the SHC1 protein.
Fig. 4.
Validation of the interaction of SHC1 with tyrosine-phosphorylated CFTR. a WB analysis of the protein fraction isolated from CFBE wt-CFTR cell lysates using an affinity peptide pull-down (PPD) assay with the indicated synthetic peptides. Note the specific binding of both endogenous SHC1 isoforms to the tyrosine-phosphorylated CFTR peptide, whereas two other SH2 domain containing proteins (GRB2, NCK) did bind the CFTR peptides. pY—phosphorylated peptide; WT—non-phosphorylated peptide; MUT- peptide with substitution of tyrosine by phenylalanine. b Graphic display of the three CFTR peptide sequences (pY, WT and MUT) used for the PPD assay. c The PTB domain of SHC1 is required for association with CFTR phosphotyrosine domain. WB analysis following the PPD assay with lysates from CFBE wt-CFTR cells previously transfected with the indicated SHC1 mutants. Note that the point mutation R175K in the SHC1 PTB domain abolishes binding to the phosphorylated peptide. d SHC1 forms a protein complex with CFTR depending on SYK protein kinase activity. CFBE mCherry-Flag-wt-CFTR cells were co-transfected with Myc-SHC1 and either constitutively active SYK Y352D or SYK kd mutants. The fraction of PM-localized CFTR was immunoprecipitated with anti-Flag antibody (IP) and the amount of SHC1 co-precipitating with CFTR analyzed by WB. Note that the presence of active SYK strongly increased the co-precipitation of SHC1 with the mature, fully glycosylated CFTR-band C. Input- whole cell lysates; IP- immunoprecipitated fractions; Ab- antibody. All data shown are representative for at least three independent experiments
SHC1 binds to tyrosine-phosphorylated CFTR through its PTB domain
The adaptor protein SHC1 possesses two domains known to bind phosphorylated tyrosine residues, a C-terminal SH2 domain and an adjacent PTB domain. To determine which domain was involved in CFTR binding, p52SHC1 was subcloned and mutated to generate three distinct constructs: a PTB domain-mutant with an arginine to lysine substitution at residue 175 (SHC1 R175K [41]), an SH2 domain-mutant with an arginine to lysine substitution at residue 401 (SHC1 R401K), [42, 43] or a double mutant (SHC1 R175K/R401K). These SHC1 mutants were transfected into CFBE wt-CFTR cells and then tested in PPDs for their ability to bind the CFTR phosphopeptide. We observed that the mutation of arginine to lysine at residue 175 eliminated binding to the phosphorylated CFTR peptide. This allowed the conclusion that the phosphorylated CFTR tyrosine residue was specifically recognized by the SHC1 PTB domain (Fig. 4c).
SHC1 can associate with full-length CFTR
To provide evidence that SHC1 can not only bind to the CFTR peptide but also associate with full-length CFTR protein, we modulated SYK activity and then immunoprecipitated the PM-associated CFTR pool and probed for the presence of SHC1. For this, we used CFBE cells that stably express a CFTR wt construct carrying an N-terminal-fused mCherry tag and a Flag-epitope located at the fourth extracellular loop [30]. The extracellular tag allows to incubate live cells at 4 °C with anti-Flag antibody, which will exclusively bind to CFTR proteins at the PM. After thorough washing, cells can be lysed with a detergent-containing buffer and incubated with protein G-conjugated beads to selectively immunoprecipitate the antibody-labelled proteins at the membrane. This method allows for selective enrichment of PM-associated CFTR-containing protein complexes. These cells were first co-transfected with Myc-SHC1 together with either a control vector, SYK kd, or the active SYK Y352D mutant, then CFTR was immunoprecipitated from the PM as described above and analyzed by WB. Because both mutants alter the amount of CFTR at the PM (see Fig. 2), comparable amounts of immunoprecipitated CFTR were loaded on the gel, so that the presence of co-precipitating Myc-SHC1 could be compared more easily. We found that the presence of constitutively active SYK Y352D clearly promoted the co-immunoprecipitation of SHC1 with CFTR (Fig. 4d). In contrast, the amount of co-immunoprecipitated SHC1 observed in control-transfected cells was further reduced in the presence of SYK kd, indicating its dominant-negative effect on endogenous SYK. Although no formal proof for direct interaction, these experiments strongly support the idea that SHC1 forms a complex with CFTR that depends on the activity of kinase SYK.
Endogenous SHC1 modulates CFTR cell surface expression
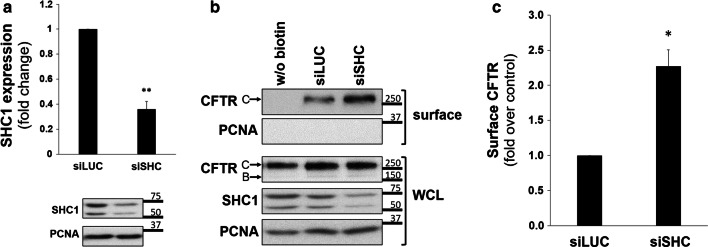
Next, we tested the physiological relevance of SHC1 expression for the PM levels of CFTR. CFBE wt-CFTR cells expressed endogenous SHC1, and their transfection with specific siRNA oligonucleotides successfully reduced SHC1 expression down to 36% (Fig. 5a). Under these conditions, cell surface proteins of CFBE wt-CFTR cells were biotinylated to determine the PM levels of CFTR. When the isolated fraction of biotinylated cell surface proteins was analyzed by WB we observed that depleting the expression of endogenous SHC1 led to a significant > twofold increase in CFTR PM levels (Fig. 5b, c).
Fig. 5.
SHC1 downregulation modulates CFTR levels at the cell surface. a Assessment of the efficiency of siRNA-mediated knockdown of SHC1. CFBE wt-CFTR cells were transfected as described under “Cell culture and transfections”and depletion of endogenous SHC1 protein levels analyzed by Western blot. Data quantification (n = 6), analyzed by Student’s t test, is graphically displayed and shown as fold-change relative to siLUC (control). Note the successful downregulation of SHC1 expression by approximately 61%. b Assessment of SHC1 siRNA-mediated knockdown efficiency in CFBE wt-CFTR cells after 48 h of transfection with either siLUC or siSHC. The indicated proteins were detected in whole-cell lysates (WCL) or in the biotinylated protein fraction (surface). For CFTR, the mature, fully glycosylated and the immature, core-glycosylated protein are marked as bands C and B, respectively. The PCNA protein served as a loading and contamination control, respectively. Note the successful downregulation of SHC1 expression. c Corresponding graph with quantification of CFTR detection in the biotinylated cell surface fraction. Densitometric values of band intensities of at least three independent experiments were expressed relative to the siLUC control and analyzed by Student’s t test. All shown data represent means ± SEM; *P < 0.05; **P < 0.01
SYK and SHC1 operate in the same pathway
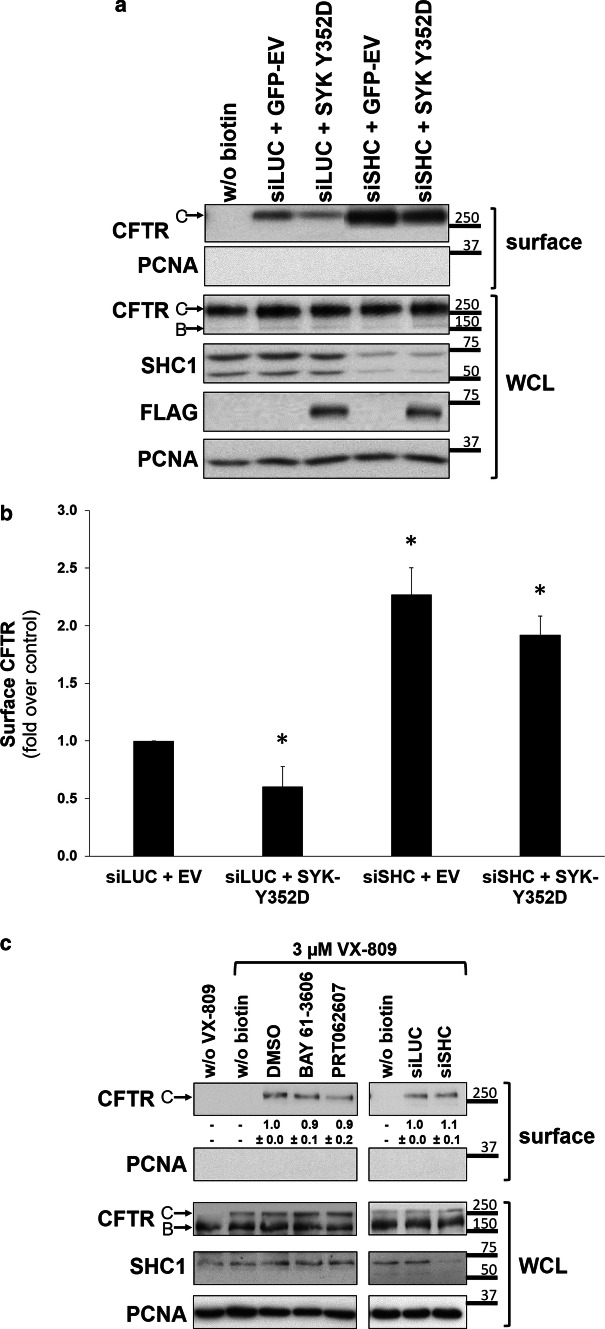
The data described above showed that protein kinase SYK as well as the adaptor SHC1 were able to modulate the amount of CFTR at the PM. To determine whether both events operate in the same pathway, we investigated if the inhibitory effect of the constitutively active SYK Y352D on CFTR PM abundance was dependent on the presence of the adaptor SHC1. For this, CFBE wt-CFTR cells were co-transfected with control siLUC or siSHC and the constitutively active mutant SYK Y352D. Then the expression levels of CFTR at the PM were determined as before. The expression of wt-SYK had no significant effect on CFTR levels at the PM (data not shown). As shown in Fig. 6 expression of constitutively active SYK alone decreased CFTR surface levels, whereas downregulation of SHC1 alone promoted a significant increase in CFTR levels at the cell surface. Importantly, the co-transfection of siSHC was able to revert the effect of constitutively active SYK, which was no longer able to decrease surface CFTR levels compared to control cells (Fig. 6a, b).
Fig. 6.
SYK and SHC1 operate in the same pathway of CFTR. a Effect of overexpression of a constitutively active SYK Y352D mutant alone or in combination with depletion of endogenous SHC1 expression. Cells were co-transfected with constitutively active mutant SYK Y352D or a control vector (YFP-EV) and either siLUC or siSHC. Note the successful downregulation of SHC1 expression. After 48 h, cell surface proteins were biotinylated, cells were lysed and proteins resolved by SDS-PAGE. WB was used to detect the indicated proteins in whole-cell lysates (WCL) or in the biotinylated protein fraction (surface). The PCNA protein served as a loading and contamination control, respectively. For CFTR, the mature, fully glycosylated and the immature, core-glycosylated protein are marked as bands C and B, respectively. b Corresponding quantification of CFTR detection in the biotinylated cell surface fraction that was obtained from at least five independent experiments. Statistical significance was calculated by one-way ANOVA (P < 0.001), followed by post hoc Tukey’s tests. Shown are mean values ± SEM. *P < 0.05. c Effect of experimental manipulation of the SYK/SHC1 pathway on the levels of F508del-CFTR at the PM. CFBE F508del-CFTR cells were first incubated at 37 °C for 24 h with 3 µM VX-809, a recently licensed therapeutic drug that rescues F508del-CFTR processing and increases its PM expression to detectable levels. Then cells were treated for 1 h with control solvent (DMSO) or SYK inhibitors BAY 61-3606 or PRT062607 (left-sided lanes). Alternatively, cells were transfected first with siSHC1 or control siLUC, then after 24 h incubated with 3 µM VX-809, and analyzed 24 h later (right-sided lanes). After the described treatments, cell surface proteins were biotinylated, cells lysed, and proteins resolved by SDS-PAGE and the indicated proteins detected by WB in whole-cell lysates (WCL) or in the biotinylated protein fraction (surface). The PCNA protein served as a loading and contamination control, respectively. Note the successful downregulation of SHC1 expression. The quantification of rescued F508del-CFTR bands in the biotinylated cell surface fraction is indicated beneath the top panels
Based on the above evidence that a SYK/SHC1 pathway modulates the levels of CFTR at the PM, we asked whether this would also affect F508del-CFTR, the most common mutation in CF patients. Because F508del causes the CFTR protein to misfold and become prematurely degraded, only a small portion of the mutant protein reaches the PM. For this reason, CFBE F508del-CFTR cells were incubated at 37 °C for 24 h with 3 μM VX-809, a recently licensed therapeutic drug that rescues F508del-CFTR processing and increases its PM expression to detectable levels by relaxing the ER quality control [9, 10, 44]. Cells were either previously transfected with siSHC1 or subsequently treated for 1 h with SYK inhibitors BAY 61-3606 or PRT062607; however, none of these treatments improved the amount of F508del-CFTR protein rescued to the PM (Fig. 6c).
SYK and SHC1 affect the cellular CFTR-dependent chloride transport capacity
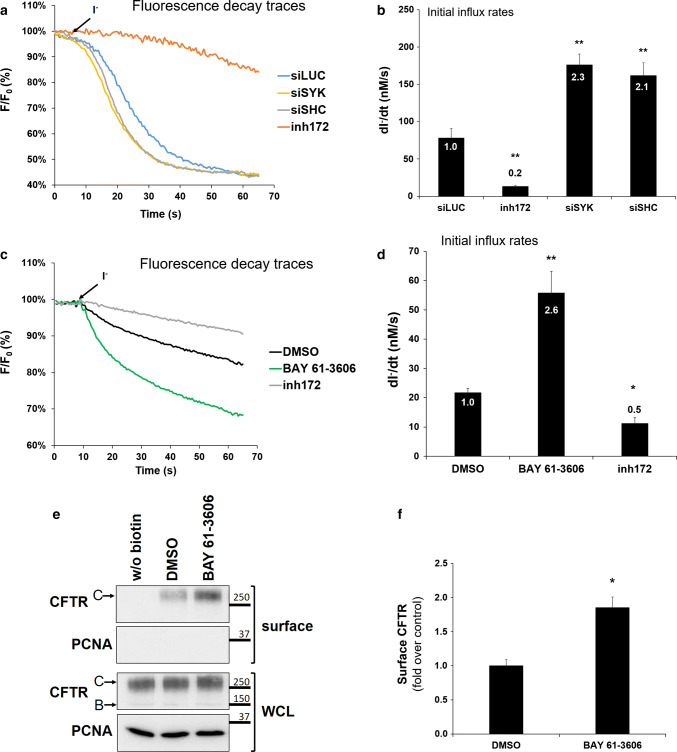
The observed changes in CFTR levels at the PM could correlate with the cellular ion transport capacity. To demonstrate this, we evaluated how SHC1 or SYK downregulation impacted the ion transport function of wt-CFTR and used an iodide influx assay based on a halide-sensitive yellow fluorescent protein (HS-YFP) version [31, 33]. CFBE wt-CFTR cells stably expressing the HS-YFP halide sensor were transfected first with either control siLUC, or siSHC1, or siSYK. Forty-eight hours later, CFTR activity in these cells was stimulated with 5 μM Fsk. Extracellular chloride was then replaced by iodide and the in vivo fluorescence decay resulting from iodide influx (proportional to the Fsk-induced CFTR-mediated chloride efflux [31, 32]) was recorded under a fluorescence microscope (Fig. 7a). The initial rates of iodide influx were calculated by fitting the fluorescence decay curves to the exponential decay function. We observed that the downregulation of SHC1 or SYK promoted a 2.1-fold or 2.3-fold increase in the CFTR-dependent iodide influx rates, respectively. The fluorescence decay was prevented in the presence of the CFTR inhibitor 172 (inh172) showing the specificity of the observed effect (Fig. 7b). This is consistent with the observed > twofold increase in CFTR abundance at the PM that we described in Figs. 2 and 5 although we cannot rule out that SYK-phosphorylation, or the interaction with SHC1, may also have an effect on CFTR channel gating.
Fig. 7.
SYK and SHC1 affect the cellular CFTR-dependent chloride transport capacity. Fluorescence recordings of iodide-induced fluorescence decay of HS-YFP a in CFBE wt-CFTR cells transfected with either siLUC, or siSYK, or siSHC, and b in intestinal Caco-2 cells treated with SYK inhibitor. Cells were stimulated for 10 min with Fsk, with or without previous incubation with 25 μM to inh172 for 15 min, followed by continuous fluorescence recording in the presence of 100 mM iodide. b, d Summary of iodide influx rates calculated by fitting the iodide assay results to exponential decay curves. Statistical significance was calculated by one-way ANOVA (P < 0.001), followed by post hoc Tukey’s tests. All shown data represent means ± SEM (n ≥ 3); *P < 0.05; **P < 0.01. e Caco-2 cells were treated for 1 h with control solvent DMSO or with SYK inhibitor BAY 61-3606 (8 nM). After the treatment, cell surface proteins were biotinylated, cells lysed, and proteins resolved by SDS-PAGE and detected by WB, as described in Fig. 1. f Quantification of CFTR bands detected in the biotinylated cell surface fraction in e. Data represent means ± SEM of at least three independent experiments, expressed relatively to DMSO control. Significance was analyzed by Student’s t test (*P < 0.05)
Because CFBE cells represent a model with high expression of the CFTR protein [29], we asked whether the effect of the SYK pathway could also be demonstrated in a different cell line and at endogenous CFTR expression levels. For this, we tested the intestinal cell line Caco-2, as a well-studied model for the gastrointestinal complications in CF patients [45]. As shown in Fig. 7c, d, the incubation with a SYK inhibitor of Caco-2 cells expressing the HS-YFP sensor promoted a 2.6-fold increase in the CFTR-dependent iodide influx rate. Under these experimental conditions, the amount of CFTR detected by protein biotinylation at the cell surface also increased roughly twofold (Fig. 7e, f). These results confirm in a different cell type and at endogenous CFTR expression level that the identified SYK pathway modulates CFTR abundance at the cell surface and the corresponding cellular CFTR-dependent chloride transport.
Discussion
The results presented in this work identified mechanistic details of SYK-mediated phosphorylation of Tyr512 in the CFTR chloride channel. Although abundant evidence has been reported that protein phosphorylation regulates CFTR gating activity, this mostly involves the phosphorylation at serine or threonine residues by protein kinase (PK) such as PKA, PKC, casein kinase 2, or AMP-dependent PK [19]. Phosphorylation on tyrosine residues is a less frequent modification. The proto-oncogene tyrosine-protein kinase SRC or proline-rich tyrosine kinase (PYK) 2 was reported to modulate CFTR gating [23] by phosphorylation of Tyr625 and Tyr627 of the NBD1 domain [22]. Here we described that SYK-mediated phosphorylation at Tyr512 regulates the PM abundance of CFTR in bronchial epithelial and intestinal cells.
First, our data demonstrate that experimental manipulation of SYK expression levels or SYK catalytic activity affects CFTR abundance. Second and as a major novelty, we identified the cytosolic adaptor protein SHC1 as a binding partner for SYK-phosphorylated CFTR. The identification of SHC1 was achieved through a proteome-wide biochemical approach using synthetic peptides of the primary sequence around the phosphorylated CFTR Tyr512 for a pull-down assay to selectively enrich interacting proteins. Although the number of proteins enriched in each replicate was variable, we found the same putative candidate proteins in different replicates. A replicate combining confidence score was then established that allowed to define 61 high-confidence candidate proteins (Table S2) and these could be connected with a subnetwork of proteins mediating the interaction between the hubs CFTR and SYK. The fact that most of the top-12 selected candidate proteins (Table 1) are adaptor proteins or proteins involved in cellular trafficking supports the relevance of our bait peptide-based interactome analysis.
Several hundred CFTR-associated proteins were previously identified in MS-based interatomic studies from transfected BHK [46] and bronchial epithelial cells [47]. These data included CFTR-binding chaperones and many previously unrecognized interactors. Among the 61 high-confidence score (CCS of 4 or 5) CFTR-associated proteins, 11 had also been identified in these previous studies (see Table S2), including SRP14, T-complex protein 1, ATP5A1, SZT2, S100-A8 and Trans-enoyl-CoA reductase. However, these CFTR interactomes reflected a whole-cell approach to find CFTR-associated proteins and thus many of the top-12 candidate proteins described in Table 1 were not found. Although this may be due to detection limits of MS, it most likely reflects the fact that our PPD approach was enriched in proteins that associate with CFTR after a particular regulatory event—Tyr512 phosphorylation. The pool of pTyr512-CFTR molecules is much smaller than the intracellular pool of immature, core-glycosylated CFTR protein and may thus be underrepresented in the interactomes identified by others [46, 47]. Consistently, SYK kinase that was previously described as a modulator of CFTR PM localization [24] and activity [48], was also not detected in that whole-cell CFTR interactomes.
Following the identification of SHC1 as a CFTR interactor, we validated its functional relevance for regulating the amount of CFTR at the cell surface. In particular, we found that RNA interference-mediated SHC1 depletion was sufficient to affect CFTR PM levels (Fig. 5). In addition, we were able to demonstrate the formation of a protein complex between SHC1 and the PM-localized pool of CFTR, under conditions of increased tyrosine phosphorylation by the expression of a constitutively active SYK mutant (Fig. 6). Furthermore, SHC1 was required downstream of SYK for the observed effect on CFTR (Fig. 6), providing strong evidence for a novel SYK/SHC1 pathway.
The SHC1 gene expresses various transcripts that encode three protein isoforms, differing only in the length of their N-terminus (p66SHC1, p52SHC1 and p46SHC1) [38, 49]. All three SHC1 isoforms share the PTB and SH2 domains that both can bind phosphorylated tyrosine residues in target proteins [50–53]. To clarify which of the two PTB domains bound to CFTR, we carried out PPD experiments using specific domain-inactivating mutants. Only the mutation of arginine 175 in the PTB domain resulted in loss of SHC1 binding to the CFTR phosphopeptide (Fig. 4c).
The SHC1 PTB domain is strictly pTyr dependent [54] and the motif NPXY has been considered as a canonical binding motif for PTB-domain-containing proteins [55, 56]. SHC1 recruitment is particularly well studied following the activation of the EGFR, where it leads to the activation of adaptor proteins GRB2 and SOS, which promotes the GTP-loading of RAS and subsequent stimulation of the MAPK kinase cascade [39, 57]. For this, SHC1 recognizes two phosphotyrosine motifs: pY1148 in the motif LDNPEY represents a typical NPXY motif [58, 59], but the pY1173 motif of EGFR (AENAEY) is less conserved and can be bound by either the PTB or SH2 domain of SHC1 [42]. Our results identified a different sequence motif in CFTR (IFGVSpYDE) that is recognized by the PTB domain of SHC1. This finding can be reconciled with the main structural information available on the SHC1 PTB domain: (1) a strong preference for phosphotyrosine due to ionic coordination with SHC1 Arg175 [60], (2) hydrophobic interactions at positions −2 and −3 with the PTB backbone, (3) an intramolecular hydrogen bond between residues −3 and −1 conferring a type I β-turn peptide conformation propensity, and (4) a large hydrophobic residue such as isoleucine at the −5 position [60, 61]. Considering the identified CFTR motif, (1) preferred binding to the phosphorylated tyrosine was observed, (2) residues −3 and −2 are glycine and valine, the latter compatible with hydrophobic interactions, (3) an intramolecular hydrogen bond could be formed by serine at position −1, and (4) the large hydrophobic residue at position −5 is indeed isoleucine. Interestingly, the ligand-free SHC PTB-domain structure revealed a highly disordered conformation, which was suggested to allow conformational flexibility of the binding pocket with a functional role for binding various different target peptides [62]. We dare to speculate that the type of motif found in CFTR is non-canonical and could mediate functions not related to RAS signaling, but rather the recruitment of proteins involved in endosome traffic or endocytosis.
The most frequent mutation in CF patients is the deletion of F508del, which leads to an increased proteolytic degradation of the misfolded mutant CFTR protein. Nevertheless, the mutant protein possesses some residual chloride channel activity and could significantly reduce disease severity if its retention at the PM could be improved [63]. Thus, it was interesting to understand if protein kinase SYK and the adaptor SHC1 would improve the PM retention of pharmacologically rescued F508del-CFTR. We recently described that either depletion of endogenous calpain 1 or its chemical inhibition dramatically improved the functional rescue of F508del-CFTR at the cell surface of airway cells [32]; however, depletion of endogenous SHC1 or pharmacological inhibition of SYK kinase activity under the same experimental conditions showed no effect on the surface levels of F508del-CFTR (Fig. 6c). This may be related to the fact that the SYK phosphorylation motif at Tyr512 is in close vicinity to the F508 deletion and may thus be affected by the misfolding. Although our results revealed no significant association between F508del-CFTR and SYK or SHC1, it will be interesting to evaluate the effect on other mutant CFTR proteins, as recent proteomic data revealed that protein interactions differ depending on which CFTR mutation is present [64]. Interestingly, a single-cell atlas based on single-cell transcriptional profiling of the airway epithelium was recently described [65]. These publicly available data allowed us to confirm that SYK and SHC1 transcripts are similarly expressed across all human bronchial epithelial cell types, including ionocytes with high levels of CFTR expression.
SYK inhibitors are currently used in clinical trials against B-cell malignancies [66] and to treat allergic rhinitis [67]. Since CF is characterized by chronic lung inflammation due to impaired clearance of the thick mucus that forms in the absence of CFTR-mediated chloride transport [68] and SYK also functions in the lung inflammatory response [26], SYK inhibition may be double beneficial for CF patients. However, it is essential to understand in more detail the underlying mechanism and find new interactors, like SHC1, that could regulate wt-CFTR but also F508del-CFTR, to reduce disease severity in the large number of patients that carry this mutation.
Several of the top hit proteins identified in Table 1 are involved in processes such as folding, trafficking, docking and degradation, such as SHROOM3 (Q8TF72), SRP14 (P37108), adaptor protein PDZ and LIM domain protein 7 (Q9NR12), and VPS37C (A5D8V6). By crossing all high-confidence-scoring proteins (Table S2) with the published whole CFTR interactome dataset, we further observed that four of our hits were also previously identified [47]. These candidates will be interesting to study in the near future since the data shown here focused on the validation of the adaptor protein SHC1.
In conclusion, we identified SHC1 as a specific binding protein for tyrosine-phosphorylated CFTR and showed that a SYK/SHC1 pathway exists to modulate the amount of CFTR at the PM. In addition, we identified further putative CFTR binding partners by building a protein interaction network. Our results contribute to the extensive work of the last decade on the complex mechanisms of CFTR biogenesis, processing, trafficking, and plasma membrane prevalence. They may also impact on therapeutic approaches to increase the PM retention time of CFTR, which is most relevant in case of mutant proteins that reach the PM but are rapidly endocytosed or contribute with residual ion transport activity [2].
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
This work was supported by Fundação para a Ciência e Tecnologia (FCT), Portugal, through Grants PTDC/BIA-CEL/28408/2017 to PJ and UID/MULTI/04046/2019 to the research unit BioISI, and fellowship SFRH/BD/52488/2014 from the BioSYS Ph.D. programme PD65-2012 to CAL and SFRH/BPD/94322/2013 to PB. The authors acknowledge the following colleagues for providing reagents used in this study: A. M. Matos, Lisbon; J. P. Clancy, University of Alabama, USA; Dimitar G. Efremov, ICGEB, Rome; Enrica Migliaccio, Campos IFOM-IEO, Milan, Italy.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Paulo Matos and Peter Jordan contributed equally.
Change history
8/1/2024
A Correction to this paper has been published: 10.1007/s00018-024-05360-7
References
- 1.Bobadilla JL, Macek M, Fine JP, Farrell PM (2002) Cystic fibrosis: a worldwide analysis of CFTR mutations—correlation with incidence data and application to screening. Hum Mutat 19:575–606. 10.1002/humu.10041 10.1002/humu.10041 [DOI] [PubMed] [Google Scholar]
- 2.Rowe SM, Miller S, Sorscher EJ (2005) Cystic fibrosis. N Engl J Med 352:1992–2001. 10.1056/NEJMra043184 10.1056/NEJMra043184 [DOI] [PubMed] [Google Scholar]
- 3.Saint-Criq V, Gray MA (2017) Role of CFTR in epithelial physiology. Cell Mol Life Sci 74:93–115. 10.1007/s00018-016-2391-y 10.1007/s00018-016-2391-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Donaldson SH, Boucher RC (2003) Update on pathogenesis of cystic fibrosis lung disease. Curr Opin Pulm Med 9:486–491 10.1097/00063198-200311000-00007 [DOI] [PubMed] [Google Scholar]
- 5.Ehre C, Ridley C, Thornton DJ (2014) Cystic fibrosis: an inherited disease affecting mucin-producing organs. Int J Biochem Cell Biol 52:136–145. 10.1016/j.biocel.2014.03.011 10.1016/j.biocel.2014.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Riordan JR (2008) CFTR Function and Prospects for Therapy. Annu Rev Biochem 77:701–726. 10.1146/annurev.biochem.75.103004.142532 10.1146/annurev.biochem.75.103004.142532 [DOI] [PubMed] [Google Scholar]
- 7.Liu F, Zhang Z, Csanády L et al (2017) Molecular structure of the human CFTR ion channel. Cell 169:85–95.e8. 10.1016/j.cell.2017.02.024 10.1016/j.cell.2017.02.024 [DOI] [PubMed] [Google Scholar]
- 8.Amaral MD, Farinha CM (2013) Rescuing mutant CFTR: a multi-task approach to a better outcome in treating cystic fibrosis. Curr Pharm Des 19:3497–3508. 10.2174/13816128113199990318 10.2174/13816128113199990318 [DOI] [PubMed] [Google Scholar]
- 9.Farinha CM, Matos P, Amaral MD (2013) Control of cystic fibrosis transmembrane conductance regulator membrane trafficking: not just from the endoplasmic reticulum to the golgi. FEBS J 280:4396–4406. 10.1111/febs.12392 10.1111/febs.12392 [DOI] [PubMed] [Google Scholar]
- 10.Bell SC, De Boeck K, Amaral MD (2015) New pharmacological approaches for cystic fibrosis: promises, progress, pitfalls. Pharmacol Ther 145:19–34. 10.1016/j.pharmthera.2014.06.005 10.1016/j.pharmthera.2014.06.005 [DOI] [PubMed] [Google Scholar]
- 11.Clancy JP, Rowe SM, Accurso FJ et al (2012) Results of a phase IIa study of VX-809, an investigational CFTR corrector compound, in subjects with cystic fibrosis homozygous for the F508del-CFTR mutation. Thorax 67:12–18. 10.1136/thoraxjnl-2011-200393 10.1136/thoraxjnl-2011-200393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sala MA, Jain M (2018) Tezacaftor for the treatment of cystic fibrosis. Expert Rev Respir Med 12:725–732. 10.1080/17476348.2018.1507741 10.1080/17476348.2018.1507741 [DOI] [PubMed] [Google Scholar]
- 13.Zhang W, Zhang X, Zhang YH et al (2016) Lumacaftor/ivacaftor combination for cystic fibrosis patients homozygous for Phe508del-CFTR. Drugs Today 52:229–237. 10.1358/dot.2016.52.4.2467205 10.1358/dot.2016.52.4.2467205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matos A, Matos P (2018) Combination therapy in Phe508del CFTR: how many will be enough? J Lung Health Dis 2:9–16 10.29245/2689-999X/2017/1.1113 [DOI] [Google Scholar]
- 15.Amaral MD (2015) Novel personalized therapies for cystic fibrosis: treating the basic defect in all patients. J Intern Med 277:155–166. 10.1111/joim.12314 10.1111/joim.12314 [DOI] [PubMed] [Google Scholar]
- 16.Farinha CM, Matos P (2016) Repairing the basic defect in cystic fibrosis—one approach is not enough. FEBS J 283:246–264. 10.1111/febs.13531 10.1111/febs.13531 [DOI] [PubMed] [Google Scholar]
- 17.Guggino WB, Stanton BA (2006) New insights into cystic fibrosis: molecular switches that regulate CFTR. Nat Rev Mol Cell Biol 7:426–436. 10.1038/nrm1949 10.1038/nrm1949 [DOI] [PubMed] [Google Scholar]
- 18.Moniz S, Sousa M, Moraes BJ et al (2013) HGF stimulation of Rac1 signaling enhances pharmacological correction of the most prevalent cystic fibrosis mutant F508del-CFTR. ACS Chem Biol 8:432–442. 10.1021/cb300484r 10.1021/cb300484r [DOI] [PubMed] [Google Scholar]
- 19.Farinha CM, Swiatecka-Urban A, Brautigan DL, Jordan P (2016) Regulatory crosstalk by protein kinases on CFTR trafficking and activity. Front Chem 4:1. 10.3389/fchem.2016.00001 10.3389/fchem.2016.00001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Farinha CM, Canato S (2017) From the endoplasmic reticulum to the plasma membrane: mechanisms of CFTR folding and trafficking. Cell Mol Life Sci 74:39–55. 10.1007/s00018-016-2387-7 10.1007/s00018-016-2387-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Farinha CM, Matos P (2018) Rab GTPases regulate the trafficking of channels and transporters—a focus on cystic fibrosis. Small GTPases 9:136–144. 10.1080/21541248.2017.1317700 10.1080/21541248.2017.1317700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Billet A, Jia Y, Jensen TJ et al (2016) Potential sites of CFTR activation by tyrosine kinases. Channels 10:247–251. 10.1080/19336950.2015.1126010 10.1080/19336950.2015.1126010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Billet A, Jia Y, Jensen T et al (2015) Regulation of the cystic fibrosis transmembrane conductance regulator anion channel by tyrosine phosphorylation. FASEB J 29:3945–3953. 10.1096/fj.15-273151 10.1096/fj.15-273151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mendes AI, Matos P, Moniz S et al (2011) Antagonistic regulation of cystic fibrosis transmembrane conductance regulator cell surface expression by protein kinases WNK4 and spleen tyrosine kinase. Mol Cell Biol 31:4076–4086. 10.1128/MCB.05152-11 10.1128/MCB.05152-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mócsai A, Ruland J, Tybulewicz VLJ (2010) The SYK tyrosine kinase: a crucial player in diverse biological functions. Nat Rev Immunol 10:387–402. 10.1038/nri2765 10.1038/nri2765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ulanova M, Puttagunta L, Marcet-Palacios M et al (2005) Syk tyrosine kinase participates in beta1-integrin signaling and inflammatory responses in airway epithelial cells. Am J Physiol Lung Cell Mol Physiol 288:L497–507. 10.1152/ajplung.00246.2004 10.1152/ajplung.00246.2004 [DOI] [PubMed] [Google Scholar]
- 27.Wang X, Lau C, Wiehler S et al (2006) Syk is downstream of intercellular adhesion molecule-1 and mediates human rhinovirus activation of p38 MAPK in airway epithelial cells. J Immunol 177:6859–6870. 10.4049/jimmunol.177.10.6859 10.4049/jimmunol.177.10.6859 [DOI] [PubMed] [Google Scholar]
- 28.Woodside DG, Obergfell A, Leng L et al (2001) Activation of Syk protein tyrosine kinase through interaction with integrin β cytoplasmic domains. Curr Biol 11:1799–1804. 10.1016/S0960-9822(01)00565-6 10.1016/S0960-9822(01)00565-6 [DOI] [PubMed] [Google Scholar]
- 29.Illek B, Maurisse R, Wahler L et al (2008) Cl transport in complemented CF bronchial epithelial cells correlates with CFTR mRNA expression levels. Cell Physiol Biochem 22:57–68. 10.1159/000149783 10.1159/000149783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Botelho HM, Uliyakina I, Awatade NT et al (2015) Protein traffic disorders: an effective high-throughput fluorescence microscopy pipeline for drug discovery. Sci Rep 5:9038. 10.1038/srep09038 10.1038/srep09038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Galietta LJV, Haggie PM, Verkman AS (2001) Green fluorescent protein-based halide indicators with improved chloride and iodide affinities. FEBS Lett 499:220–224. 10.1016/S0014-5793(01)02561-3 10.1016/S0014-5793(01)02561-3 [DOI] [PubMed] [Google Scholar]
- 32.Matos AM, Pinto FR, Barros P et al (2019) Inhibition of calpain 1 restores plasma membrane stability to pharmacologically rescued Phe508del-CFTR variant. J Biol Chem. 10.1074/jbc.RA119.008738 10.1074/jbc.RA119.008738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Loureiro CA, Matos AM, Dias-Alves  et al (2015) A molecular switch in the scaffold NHERF1 enables misfolded CFTR to evade the peripheral quality control checkpoint. Sci Signal 8:ra48. 10.1126/scisignal.aaa1580 10.1126/scisignal.aaa1580 [DOI] [PubMed] [Google Scholar]
- 34.Carsetti L, Laurenti L, Gobessi S et al (2009) Phosphorylation of the activation loop tyrosines is required for sustained Syk signaling and growth factor-independent B-cell proliferation. Cell Signal 21:1187–1194. 10.1016/j.cellsig.2009.03.007 10.1016/j.cellsig.2009.03.007 [DOI] [PubMed] [Google Scholar]
- 35.Schulze WX, Mann M (2004) A novel proteomic screen for peptide–protein interactions. J Biol Chem 279:10756–10764. 10.1074/jbc.M309909200 10.1074/jbc.M309909200 [DOI] [PubMed] [Google Scholar]
- 36.Rozakis-Adcock M, Fernley R, Wade J et al (1993) The SH2 and SH3 domains of mammalian Grb2 couple the EGF receptor to the Ras activator mSos1. Nature 363:83–85. 10.1038/363083a0 10.1038/363083a0 [DOI] [PubMed] [Google Scholar]
- 37.Yaffe MB (2002) Phosphotyrosine-binding domains in signal transduction. Nat Rev Mol Cell Biol 3:177–186. 10.1038/nrm759 10.1038/nrm759 [DOI] [PubMed] [Google Scholar]
- 38.Rozakis-Adcock M, McGlade J, Mbamalu G et al (1992) Association of the Shc and Grb2/Sem5 SH2-containing proteins is implicated in activation of the Ras pathway by tyrosine kinases. Nature 360:689–692. 10.1038/360689a0 10.1038/360689a0 [DOI] [PubMed] [Google Scholar]
- 39.Schlessinger J, Lemmon MA (2003) SH2 and PTB domains in tyrosine kinase signaling. Sci STKE 2003:RE12. 10.1126/stke.2003.191.re12 10.1126/stke.2003.191.re12 [DOI] [PubMed] [Google Scholar]
- 40.Jabril-Cuenod B, Zhang C, Scharenberg AM et al (1996) Syk-dependent phosphorylation of Shc: a potential link between FcϵRI and the Ras/mitogen-activated protein kinase signaling pathway through SOS and Grb2. J Biol Chem 271:16268–16272. 10.1074/jbc.271.27.16268 10.1074/jbc.271.27.16268 [DOI] [PubMed] [Google Scholar]
- 41.van der Geer P, Wiley S, Gish GD et al (1996) Identification of residues that control specific binding of the Shc phosphotyrosine-binding domain to phosphotyrosine sites. Proc Natl Acad Sci USA 93:963–968. 10.1073/pnas.93.3.963 10.1073/pnas.93.3.963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sakaguchi K, Okabayashi Y, Kido Y et al (1998) Shc phosphotyrosine-binding domain dominantly interacts with epidermal growth factor receptors and mediates Ras activation in intact cells. Mol Endocrinol 12:536–543. 10.1210/mend.12.4.0094 10.1210/mend.12.4.0094 [DOI] [PubMed] [Google Scholar]
- 43.Vanderlaan RD, Hardy WR, Kabir MG et al (2011) The ShcA phosphotyrosine docking protein uses distinct mechanisms to regulate myocyte and global heart function. Circ Res 108:184–193. 10.1161/CIRCRESAHA.110.233924 10.1161/CIRCRESAHA.110.233924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Denning GM, Anderson MP, Amara JF et al (1992) Processing of mutant cystic fibrosis transmembrane conductance regulator is temperature-sensitive. Nature 358:761–764. 10.1038/358761a0 10.1038/358761a0 [DOI] [PubMed] [Google Scholar]
- 45.Kunzelmann K, Mall M (2002) Electrolyte transport in the mammalian colon: mechanisms and implications for disease. Physiol Rev 82:245–289. 10.1152/physrev.00026.2001 10.1152/physrev.00026.2001 [DOI] [PubMed] [Google Scholar]
- 46.Wang X, Venable J, LaPointe P et al (2006) Hsp90 cochaperone Aha1 downregulation rescues misfolding of CFTR in cystic fibrosis. Cell 127:803–815. 10.1016/j.cell.2006.09.043 10.1016/j.cell.2006.09.043 [DOI] [PubMed] [Google Scholar]
- 47.Pankow S, Bamberger C, Calzolari D et al (2015) ∆F508 CFTR interactome remodelling promotes rescue of cystic fibrosis. Nature 528:510–516. 10.1038/nature15729 10.1038/nature15729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Luz S, Kongsuphol P, Mendes AI et al (2011) Contribution of casein kinase 2 and spleen tyrosine kinase to CFTR trafficking and protein kinase A-induced activity. Mol Cell Biol 31:4392–4404. 10.1128/MCB.05517-11 10.1128/MCB.05517-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pelicci G, Lanfrancone L, Grignani F et al (1992) A novel transforming protein (SHC) with an SH2 domain is implicated in mitogenic signal transduction. Cell 70:93–104. 10.1016/0092-8674(92)90536-L 10.1016/0092-8674(92)90536-L [DOI] [PubMed] [Google Scholar]
- 50.Bonfini L, Migliaccio E, Pelicci G et al (1996) Not all Shc’s roads lead to Ras. Trends Biochem Sci 21:257–261. 10.1016/S0968-0004(96)10033-5 10.1016/S0968-0004(96)10033-5 [DOI] [PubMed] [Google Scholar]
- 51.Migliaccio E, Mele S, Salcini AE et al (1997) Opposite effects of the p52shc/p46shc and p66shc splicing isoforms on the EGF receptor-MAP kinase-fos signalling pathway. EMBO J 16:706–716. 10.1093/emboj/16.4.706 10.1093/emboj/16.4.706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Migliaccio E, Giorgio M, Mele S et al (1999) The p66shc adaptor protein controls oxidative stress response and life span in mammals. Nature 402:309–313. 10.1038/46311 10.1038/46311 [DOI] [PubMed] [Google Scholar]
- 53.Luzi L, Confalonieri S, Di Fiore PP, Pelicci PG (2000) Evolution of Shc functions from nematode to human. Curr Opin Genet Dev 10:668–674. 10.1016/S0959-437X(00)00146-5 10.1016/S0959-437X(00)00146-5 [DOI] [PubMed] [Google Scholar]
- 54.Uhlik MT, Temple B, Bencharit S et al (2005) Structural and evolutionary division of phosphotyrosine binding (PTB) domains. J Mol Biol 345:1–20. 10.1016/j.jmb.2004.10.038 10.1016/j.jmb.2004.10.038 [DOI] [PubMed] [Google Scholar]
- 55.Shoelson SE (1997) SH2 and PTB domain interactions in tyrosine kinase signal transduction. Curr Opin Chem Biol 1:227–234. 10.1016/S1367-5931(97)80014-2 10.1016/S1367-5931(97)80014-2 [DOI] [PubMed] [Google Scholar]
- 56.Wagner MJ, Stacey MM, Liu BA, Pawson T (2013) Molecular mechanisms of SH2- and PTB-domain-containing proteins in receptor tyrosine kinase signaling. Cold Spring Harb Perspect Biol 5:a008987. 10.1101/cshperspect.a008987 10.1101/cshperspect.a008987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pawson T, Gish GD, Nash P (2001) SH2 domains, interaction modules and cellular wiring. Trends Cell Biol 11:504–511. 10.1016/S0962-8924(01)02154-7 10.1016/S0962-8924(01)02154-7 [DOI] [PubMed] [Google Scholar]
- 58.Batzer AG, Rotin D, Ureña JM et al (1994) Hierarchy of binding sites for Grb2 and Shc on the epidermal growth factor receptor. Mol Cell Biol 14:5192–5201. 10.1128/mcb.14.8.5192 10.1128/mcb.14.8.5192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mandiyan V, O’Brien R, Zhou M et al (1996) Thermodynamic studies of SHC phosphotyrosine interaction domain recognition of the NPXpY motif. J Biol Chem 271:4770–4775. 10.1074/jbc.271.9.4770 10.1074/jbc.271.9.4770 [DOI] [PubMed] [Google Scholar]
- 60.Zhou M-M, Ravichandran KS, Olejniczak ET et al (1995) Structure and ligand recognition of the phosphotyrosine binding domain of Shc. Nature 378:584–592. 10.1038/378584a0 10.1038/378584a0 [DOI] [PubMed] [Google Scholar]
- 61.Farooq A, Zhou M-M (2004) PTB or not to be: promiscuous, tolerant and Bizarro domains come of age. IUBMB Life 56:547–557. 10.1080/15216540400013895 10.1080/15216540400013895 [DOI] [PubMed] [Google Scholar]
- 62.Farooq A, Zeng L, Yan KS et al (2003) Coupling of folding and binding in the PTB domain of the signaling protein Shc. Structure 11:905–913. 10.1016/S0969-2126(03)00134-5 10.1016/S0969-2126(03)00134-5 [DOI] [PubMed] [Google Scholar]
- 63.Amaral MD (2005) Processing of CFTR: traversing the cellular maze—how much CFTR needs to go through to avoid cystic fibrosis? Pediatr Pulmonol 39:479–491. 10.1002/ppul.20168 10.1002/ppul.20168 [DOI] [PubMed] [Google Scholar]
- 64.Hutt DM, Loguercio S, Campos AR, Balch WE (2018) A proteomic variant approach (ProVarA) for personalized medicine of inherited and somatic disease. J Mol Biol 430:2951–2973. 10.1016/j.jmb.2018.06.017 10.1016/j.jmb.2018.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Plasschaert LW, Žilionis R, Choo-Wing R et al (2018) A single-cell atlas of the airway epithelium reveals the CFTR-rich pulmonary ionocyte. Nature 560:377–381. 10.1038/s41586-018-0394-6 10.1038/s41586-018-0394-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liu D, Mamorska-Dyga A (2017) Syk inhibitors in clinical development for hematological malignancies. J Hematol Oncol 10:145. 10.1186/s13045-017-0512-1 10.1186/s13045-017-0512-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Masuda ES, Schmitz J (2008) Syk inhibitors as treatment for allergic rhinitis. Pulm Pharmacol Ther 21:461–467. 10.1016/j.pupt.2007.06.002 10.1016/j.pupt.2007.06.002 [DOI] [PubMed] [Google Scholar]
- 68.Belcher CN, Vij N (2010) Protein processing and inflammatory signaling in cystic fibrosis: challenges and therapeutic strategies. Curr Mol Med 10:82–94 10.2174/156652410791065408 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.