Abstract
Phosphatidylethanolamine-binding protein 1 (PEBP1), a small 21 kDa protein, is implicated in several key processes of the living cell. The deregulation of PEBP1, especially its downregulation, leads to major diseases such as cancer and Alzheimer’s disease. PEBP1 was found to interact with numerous proteins, especially kinases and GTPases, generally inhibiting their activity. To understand the basic functionality of this amazing small protein, we have considered several known processes that it modulates and we have discussed the role of each molecular target in these processes. Here, we propose that cortical actin organization, associated with membrane changes, is involved in the majority of the processes modulated by PEBP1. Furthermore, based on recent data, we summarize some key PEBP1-interacting proteins, and we report their respective functions and focus on their relationships with actin organization. We suggest that, depending on the cell status and environment, PEBP1 is an organizer of the actin-membrane composite material.
Keywords: Actin, Cytoskeleton, Membrane, Cell shape, Motility
Introduction
PhosphatidylEthanolamine-Binding Proteins (PEBPs) are a family of proteins expressed in a large number of living organisms such as bacteria, yeasts, parasites, plants, insects, and animals. Several PEBP isoforms were encountered in each specific species and appeared to be implicated in numerous cellular processes in response to the environment. In mammals, the main expressed isoform PEBP1 was first characterized in the brain where it is associated with membranes and phospholipids. Studies measuring the affinity of this isoform toward different phospholipids showed a preference for phosphatidylethanolamine (PE) [1, 2], and then designated PEBP1. The sequence, then the crystallographic structure of bovine [3], and human PEBP1 [4] were solved, revealing a small well-structured 21 kDa protein. A small accessible cavity has a high affinity for anions, such as phosphate, acetate, phosphorylethanolamine, or cacodilate molecules, and was identified as the binding site of PEBP1.
Subsequently, PEBP1 was described to be downregulated in numerous types of cancer, the disappearance of PEBP1 in tumor cells being concomitant with metastases development [5]. Thus, PEBP1 is considered a suppressor of metastases [6]. Since then, several studies have demonstrated the implication of PEBP1 in other diseases, such as Alzheimer’s disease, diabetes, nephropathies, and ciliopathies [7, 8]. Concerning the molecular role of PEBP1 in the cell, one major discovery was the inhibitory effect of PEBP1 toward Raf1, a kinase of the Raf/MEK/ERK signaling pathway [9], prompting the authors to redefine PEBP1 as a Raf Kinase Inhibitory Protein (RKIP). Afterwards, PEBP1 was described to regulate many other protein kinases implicated in various signaling pathways, particularly NF-kB [10] and PI3K/Akt/mTOR [11] which is strongly involved in cancer [12]. Surprisingly, in recent years, PEBP1 was described to influence a considerable number of cell physiological behaviors including cell growth and differentiation, proliferation, migration, motility, cell cycle, genomic stability, apoptosis, autophagy, drug resistance, spermatogenesis, mechanical and oxidative stress, adhesion, neuronal synapse, and immunity [7, 13].
To clarify the apparent discrepancy between the structural simplicity of PEBP1 and its multiple functions, we have inventoried and compared the various cellular processes in which it is implied. The comparison focused on both the functional and molecular aspects of the cell mechanisms. It became obvious that the primary common denominator for the vast majority of the processes and interacting proteins controlled by PEBP1 is cytoskeleton reorganization in close relationships with membrane remodeling [7].
In the present paper, based on recent data describing PEBP1 relationships with various molecular targets, we reinforce the idea that the likely principal functionality of PEBP1 is to regulate the transport of membrane proteins in coordination with cortical actin organization, particularly in cell motility. Moreover, the considered data suggest that the modulation of the actin skeleton, particularly cortical actin, is sufficient to explain the main PEBP1 features. Finally, we discuss the intertwined relationships between cortical actin and membrane, and we suggest the main role of PEBP1 in coordinating the actin cytoskeleton together with the cell shape.
Protein kinases of signaling pathways inhibited by PEBP1
An essential feature of PEBP1 is its ability to inhibit, by physical interactions, the kinases of the Raf/MEK/ERK cascade, and the inhibition of Raf1 and MEK interaction being the first to be described [9]. Since then, PEBP1 was described to regulate many other protein kinases implicated in various signaling pathways such as NF-kB, GPCRs, STAT3, GSK3 [8, 14], PI3K/Akt/mTOR [11, 15], Wnt [14], p38 [16], and Notch1 [17]. Thus, today, PEBP1 has been found to effect interplay with many, if not all, pivotal intracellular signaling cascades that control cellular growth, proliferation, division, differentiation, motility, apoptosis, genomic integrity, and therapeutic resistance [13]. In most cases, PEBP1 was demonstrated to regulate the phosphorylation cascades by inhibiting the kinases implied in each pathway. The inhibition is due to physical interaction of PEBP1 with the kinases, preventing them to interact with their cascade targets. However, one exception is GSK3β which is activated by PEBP1 [18]. The regulation of signaling pathways appeared to depend on the cell type [19], in agreement with the ability of PEBP1 to regulate the pathways that are actually activated in a given cell at a given time. Moreover, it is to note that PEBP1 may act on signaling pathways directly by inhibiting their kinases, but also indirectly, by binding proteins implicated in the regulation of diverse signaling pathways. Among them, the G protein-coupled receptor kinase 2 (GRK2) and the GTPase Rab8 are two main PEBP1 targets that regulate several signaling pathways.
Direct and indirect relationships between actin and PEBP1
Direct and indirect interactions between actin and PEBP1 have been suspected on several occasions. In this section, a first part is devoted to the data obtained by studying the PEBP1 interactome in gastric cancer cells [20], as well as the proteome of chondrocytes under mechanical stress [21], and the proteome of melanoma cells challenged by a conditioned medium of CENU-treated cells [22]. Then, in the second part of this section, we report the relationships between PEBP1 and various effectors of actin organization, such as Ras, RhoA, Rac1, CDC42, and Rab8.
Data from PEBP1 interactome and cellular proteomes
Monomeric actin is referred as globular or G-actin in contrast to filamentous or F-actin. In humans, six different isoforms of actin exist, encoded by six different genes. These include cytoplasmic beta and gamma-actins, skeletal and cardiac alpha-actins, as well as smooth muscle alpha- and gamma-actins (alpha-SMA and gamma-SMA, respectively). By mapping the interactome of overexpressed PEBP1 in a gastric cancer cell line, a total of 72 RKIP-interacting proteins were identified by MS/MS [20]. Among them, actin cytoplasmic 1 (beta-actin) was identified, as well as some actin-binding proteins such as filamin A and B, plectin-1, myosin-9, F-actin-capping protein subunit alpha (CAPZA1), and vinculin, suggesting the possible direct and indirect binding of PEBP1 to actin in gastric cancer.
Besides its possible direct and indirect interaction with actin, PEBP1 was also found to regulate the expression levels of actin isoforms or to be expressed together with actin isoforms. Thus, in hepatic fibrosis, in both liver and primary hepatic stellate cells, down-regulating PEBP1 expression resulted in liver fibrosis with increased level of alpha-SMA [23]. In another tissue, i.e., the neonatal mandibular condylar chondrocytes, 1 h of mechanical stress resulted first in the decrease of vimentin expression and changes in its phosphorylation, and then led to the subsequent decrease in expression of actin, PEBP1, and heat shock protein GRP75, revealing that mechanical stress produced inhibition effects on chondrocytes at the early stage [21]. In other series of experiment performed on melanoma tumors, bystander effects were induced by chloroethylnitrosourea (CENU) treatment and were associated with altered protein secretory activity of the CENU-treated tumor cells. Among de novo secreted proteins by CENU-treated cells, the most expressed were PEBP1, cardiovascular heat shock protein (cHsp), Rho-associated coiled-coil forming kinase 2 (ROCK), and fragments of actin, suggesting that these four proteins might be involved in cytoskeleton architecture disorders, decrease of cell proliferation, metabolite alterations, and in differentiation observed in treated tumor cells in vivo [22].
Relationships between PEBP1 and internal regulators of actin networks: Ras, Rho-GTPases, and their effectors
GTPases: Ras, RhoA, Rac1, and CDC42
Ras Recently, using AX3 cells of Dictyostelium submitted to a shallow gradient of chemoattractant, it was demonstrated that activation of Ras and F-actin is coupled through mutual positive feedback. Ras activation led to full excitation of Ras and subsequent F-actin at the side of the cell toward the chemoattractant, resulting in directed pseudopod extension and chemotaxis [24]. To our knowledge, the direct physical association of PEBP1 with Ras has not been described, but PEBP1 associates with signaling pathways regulated by Ras, such as Ras/Raf/MEK/ERK [9] and PI3K/Akt/mTOR [15].
Rho, Rac, CDC42 The Rho-GTPases subfamily is divided into Rho, Rac, and CDC42. These Rho-GTPases are major regulators of the actin cytoskeleton, with contractile actin-myosin stress fibers improved by RhoA, lamellipodia promoted by Rac1 and filopodia formation triggered by Cdc42 (Fig. 1) [25]. It is to note that no direct interaction between PEBP1 and the Rho-GTPases has been described. However, PEBP1 was found to interact with several Rho-GTPases effectors such as Raf1, GRK2, Rab8, the guanine exchange Factor (GEF) DOCK4, and the GTPase-activating proteins (GAPs) TBC1D5 and NF1. The main properties of all these effectors agree with their role played in actin organization.
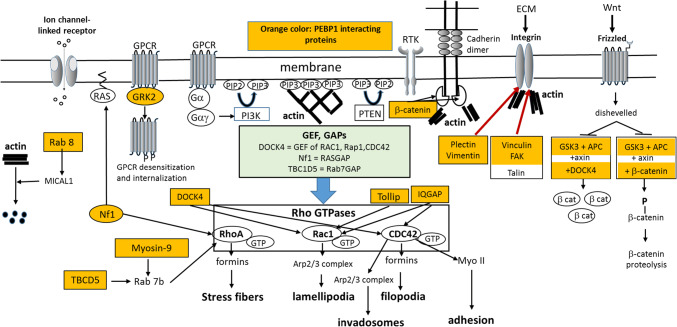
Fig. 1.
The role of PEBP1-interacting proteins in actin organization. Different types of receptors receive extracellular signals and trigger signaling that aside from phosphorylation cascades result in activation of small GTPases of the Rho family. Activated Rho-GTPases in turn lead to the formation of actin-based protrusions and adhesion and thus induce cell locomotion. The small GTPases are modulated by numerous regulatory proteins, such as guanine-nucleotide exchange factors (GEFs) and GTPase-activating proteins (GAPs). Note that for clarity of the figure, the phosphorylation cascades controlled by PEBP1 are not indicated. Note also that there is considerable cross-talk between the Rho-GTPases and several positive and negative feedback loops arising from actin remodeling (they are omitted in the figure for the sake of readability). The main known PEBP1-interacting proteins are highlighted in orange color. They appear to regulate several key points of actin organization from membrane receptors to Rho-GTPases
GEF and GAP effectors of GTPases: DOCK4, myosin-9, TBC1D5, and NF1
DOCK4 Recently, in models of triple-negative breast cancer, authors have identified the prometastatic gene Dedicator of Cytokinesis 4 (DOCK4) as a novel target of PEBP1 [26]. DOCK4 is a GEF which activates Rac, Rap1, and CDC42 (Fig. 1). DOCK4 regulates cellular β-catenin levels in response to the Wnt signal, and is required for Wnt-induced Rac activation, T-cell factor transcription, and cell migration [27]. In addition, DOCK4 promotes lateral filopodia formation [28], lung adenocarcinoma metastases [29], and the DOCK4-mediated Rac activation is required for breast cancer cell migration [30]. In the stereocilia of mice inner ear, it was shown that the splice variant DOCK4-Ex49 is an efficient Rac activator, suggesting that Rac and DOCK4 are members of a signaling pathway implicated in actin cytoskeleton organization [31]. In addition, in humans, DOCK4 mutant linked to autism and dyslexia leads to defective neuronal polarization and loss of neurite outgrowth, revealing that DOCK4 plays an important role in actin dynamics that drives neurite differentiation during early neuronal development [32].
Myosin-9 Mapping the interactome of overexpressed PEBP1 in gastric cancer cell line revealed that 35 proteins have existing interactions with PEBP1 as first and second level neighbors [20]. Previously, we have discussed the main properties of each of these 35 PEBP1-related proteins [7]. Among them, Myosin-9 (MYH9) is of particular interest due to its well-documented role in actin organization [20]. MYH9 is expressed in most cells and tissues where it participates in a variety of processes requiring contractile force, such as cytokinesis, cell migration, polarization and adhesion, maintenance of cell shape, and signal transduction [33]. In humans, a recent study of de novo mutations found in multiple neurodevelopmental disorders revealed that MYH9 is one of only three affected genes shared by autism, schizophrenia, and intellectual disability [33]. MYH9 encodes the two heavy chains of the non-muscle myosin II NMIIA which is a fast motor able to cope with rapid actin polymerization, contributing to focal adhesion assembly near the leading edge. During cell spreading, NMIIA plays an important role in extracellular matrix (ECM) remodeling [34], cytoskeleton reorganization, focal contact formation, and lamellipodial retraction [35].
TBC1D5 Recently, in prostate cancer, using gene co-expression and co-localization methods [36], PEBP1 showed potentially interactions with TBC1 Domain Family Member 5 (TBC1D5). TBC1D5 is known to be implicated in autophagy, intracellular protein transport, positive regulation of receptor internalization, and regulation of cilium assembly. It was shown that TBC1D5 localizes to Rab7b-positive vesicles and displays GAP activity towards Rab7b in vitro. Rab7b is known to play a role in the transport and degradation of proteins in endosomes and lysosomes in mammalian cells [37] and to regulate actin remodeling, therefore acting on cell polarization, adhesion, and migration [38] (Fig. 1).
NF1 PEBP1 was also shown to potentially interact with the RasGAP neurofibromin NF1 [39] which is responsible for the genetic disease Neurofibromatosis type I. The evidence was provided that Nf1 mediates a cross-talk between Ras and Rho signaling pathways [39], (Fig. 1). Nf1 was also shown to increase cell motility by regulating actin filament dynamics through the inhibition of the Rho/ROCK/LIMK2/cofilin pathway and then connecting neurofibromatosis type I to actin cytoskeleton remodeling [39]. A two-hybrid screening led to the identification of LIMK2 as a binding protein of Nf1. Nf1/LIMK2 interaction partially inhibits the kinase activity of LIMK2 on cofilin. By interacting with NF1, PEBP1 may potentially act on LIMK2 activity and subsequent actin remodeling.
Other effectors of GTPases: Tollip and IQGAP1
Tollip The ways by which PEBP1 interacts with key gene products in epithelial-to-mesenchymal transition (EMT) and autophagy were investigated during the progression of prostate cancer [36]. The gene expression data set from three groups of prostate tissues, namely benign prostate hyperplasia, and primary and metastatic cancer, showed that PEBP1 potentially interacts with seven autophagy gene. Among them was Toll-interacting protein (Tollip) which is an inhibitory adaptor protein within Toll-like receptors (TLR). The TLR pathway is a part of the innate immune system that recognizes structurally conserved molecular patterns of microbial pathogens, leading to an inflammatory immune response [36]. During E. coli infection, functional studies demonstrated that Tollip, Tom1, and clathrin associate with Rac1 and localize at the site of bacterial entry. Tollip is an effector of Rac1 (Fig. 1) hijacked by pathogenic bacteria for efficient cell invasion, suggesting that Rac1 may modulate actin polymerization at the level of membrane-bound Tollip [40].
Ras GTPase-activating-like protein IQGAP1 In a gastric cancer cell line overexpressing PEBP1, seven proteins were identified as first-level neighbors of PEBP1 [20]. Among them, IQGAP1 drew particularly our attention as it is known to be involved in cancer biology, cell motility, and actin remodeling [41]. Despite its name, IQGAP1 is not a GAP, but stabilizes Rac1 and Cdc42 in their active GTP-bound forms and consequently modulates the cytoskeleton indirectly (Fig. 1). In addition, IQGAP1 enhances actin polymerization and colocalizes with actin in lamellipodia, wherein it stimulates actin assembly by forming complexes with FGFR1 (fibroblast growth factor receptor), N-WASP (neuronal Wiskott Aldrich Syndrome protein), and Arp2/3 (Actin-related protein 2/3) that promote nucleation of branched actin network [42].
To conclude, it appears that PEBP1 interacts with several effectors of Rho-GTPases, influencing their ability to drive the formation of lamellipodia, filopodia, stress fibers, and focal adhesion. All these actin structures are major actors in cell motility. Consequently, it seems likely that PEBP1 may participate in the formation of structures governing cell migration, giving an explanation for its key role in cell adhesion and motility.
Role of PEBP1 in internalization of receptors: the interplay with PKC, GRK2, and GPCRs
PEBP1 was demonstrated to inhibit Raf1 by direct interaction, preventing the kinase to target its downstream substrates [9]. It was demonstrated that the Protein Kinase C (PKC) isoforms – α, − βI, − βII, −γ, and atypical PKCζ phosphorylate PEBP1 at Ser153 [43]. The phosphorylated RKIP releases from Raf1 and then binds to GRK2 inhibiting its activity. The inhibition of GRK2 by the phosphorylated form of PEBP1 results in blocking the G protein-coupled receptors (GPCRs) internalization [44]. Thus, PEBP1 appeared to govern the fate of GPCRs in the cell.
PKC
It was demonstrated that various isoforms of PKC can phosphorylate PEBP1 at Ser153 [43]. Depending on isozyme and cell type, PKCs are either pro-mitogenic or inhibit cell cycle progression. For instance, PKCα and PKC∆ promote anti-mitotic responses in tumor cells, whereas PKCε is required for cancer cell survival. Moreover, during formation of lamellipodia, PKCα has been shown to interact with F-actin-binding proteins ERM which support cell shape extension [45].
GRK2
GRK2 phosphorylates activated GPCRs which causes them to dissociate from G proteins and leads to GPCR internalization (Fig. 1), a process that results in the transfer of receptors from the plasma membrane to membrane of the endosomal compartment. The inhibition of GRK2 by the phosphorylated form of PEBP1 blocks the GPCRs’ internalization [44]. Such a blocking have impacts on signal desensitization and also on prolonged receptor signaling which is required for biological effects of hormones like TSH, LH, and adrenaline.
However, GRK2 is not just implicated in GPCRs fate and phosphorylates diverse non-GPCRs substrates displaying a complex network of functional interactions with proteins involved in many cell processes and in signal transduction [46]. Among them are PI3K, MEK1, Akt, p38, calmodulin, HSP90 and tubulin, all of which interacting also with PEBP1. GRK2 interactome is involved in the modulation of cell cycle progression and in cell migration [47], since data have revealed GRK2 interactions with a variety of proteins related to cell cycle and cell motility, respectively. Moreover, evidence was provided that GRK2 regulates membrane protrusion and collective migration of a cell sheet during wound closure in MDCK cell monolayers [48]. By dynamically interacting with PI3K and HDAC6, GRK2 modulates cell polarity membrane and by phosphorylating the proteins ezrin and radixin (two ERM proteins), GRK2 contributes to local F-actin polymerization and membrane protrusion. Such an effect is due to ERMs that bridge the plasma membrane and actin filaments at the leading edge in a phosphorylation-dependent manner. Finally, it appears that GRK2 may modulate several aspects of cell migration in a stimuli and cellular context-dependent manner [47].
Interestingly, GRK2 has been described to bind various PEBP1-interacting proteins, among them are PI3K, Akt, MEK, ezrin (an ERM protein), adenomatous polyposis coli (APC) [46], and HDAC6 [49]. Since PEBP1 controls the activity of GRK2 and binds several of its targets, the involvement of PEBP1 in many cellular processes could be due, at least in part, to the subsequent important functions of GRK2 in cells. As an example, a recent paper showed that PEBP1 is constitutively phosphorylated and highly expressed in chronic lymphocytic leukemia (CLL) cells [50]. The disruption of PEBP1 by locostatin induced apoptosis in CLL cells, suggesting that the disruption of phosphorylated PEBP1 leads to GRK2 activation and consequently to MEK1 and Akt inhibition [50].
GPCRs
GPCRs constitute a large family of membrane receptors involved in various signaling cascades. The nearly 1000 GPCRs encoded by the human genome regulate virtually all physiological functions and induce signals at the cell surface [51]. In chemotaxis, chemoattractants are detected by GPCRs that link to heterotrimeric G proteins (Fig. 1); then, the signaling components direct reorganization of the actin cytoskeleton that, in turn, drives cell movement [52]. In D. discoideum and in mammalian cancer cells, various chemokines activate Rac through several GPCR signaling pathways, leading to the formation of the complex ELMO, Gβγ, and DOCK and subsequent transduction of GPCR signaling to control the reorganization of actin cytoskeleton [53].
In conclusion, PEBP1 may be phosphorylated by various isoforms of PKC, leading to GRK2 inhibition and subsequent blocking of GPCRs internalization. Furthermore, GRK2 regulates and phosphorylates numerous substrates that govern a large number of functions in the cell. Thus, by inhibiting GRK2 activity, PEBP1 may affect many cellular processes. In particular, GRK2 contributes to microtubule networks by targeting HDAC6 and to local F-actin polymerization-dependent membrane protrusion by targeting ERM proteins. By inhibiting GRK2 activity, phosphorylated PEBP1 may subsequently influence microtubule networks and F-actin polymerization.
PEBP1 in ciliogenesis: a linked role with Cep290 and Rab8
Photoreceptors are dynamic and highly compartimentalized sensory neurons of retina that degenerate in retinitis pigmentosa, leading to blindness. In retinal degeneration, the ciliary protein Cep290 appeared to be frequently mutated; thus, the rd16 mice, displaying a deletion of 299 amino acids in Cep290 was studied and revealed that photoreceptor degeneration is associated with aberrant accumulation of PEBP1 [54]. It was shown that PEBP1 interacts with Cep290 at the connecting cilium (transition zone) of photoreceptors and that the domain deleted in the rd16 mouse is more specifically implicated in the interaction with PEBP1. Further experiments in zebrafish and cultured cells demonstrated that accumulation of PEBP1 prevents cilia formation by interacting with Rab8A, a critical component of the protein trafficking machinery of photoreceptors. PEBP1 interacted preferably with the GDP-bound form of Rab8A, suggesting that the PEBP1/Rab8A(GDP) complex may need to dissociate for the release of Rab8A(GDP) and its further conversion to Rab8A(GTP) allowing appropriate ciliary transport [54].
The primary cilium is a solitary organelle that protrudes from the cell surface of most mammalian cell types during growth arrest. Interestingly, the centrosome can transform to a primary cilium when the cell exits the cell cycle and enters a quiescent state [55]. This is in agreement with the implication of PEBP1, Cep290, and Rab8 in primary cilium formation and in centrosome function, suggesting that specific molecular assemblies regulate the coordination between the cell cycle and primary cilium. Increasing evidence suggests that primary cilium is a key coordinator of signaling pathways during development and in tissue homeostasis. The defection of primary cilium is a major cause of human diseases and developmental disorders designated ciliopathies. Furthermore, it is to note that most of cancer cells are defective in primary cilium [55].
The primary cilium is a cell antenna which transduces signals, affecting multiple cellular processes. A large quantity of GPCRs, ion channels, and downstream effector molecules are sequestered and confined to its membrane and lumen [56]. Numerous signaling pathways have been linked to the cilium, including Hedgehog, Wnt, Notch, Hippo, mTOR, and TGFbeta. Therefore, there is no doubt that the vast majority of signaling pathways in vertebrates function through the primary cilium [57]. Ciliary membranes are partitioned into highly fluid membrane nanodomains that are delimited by filamentous actin and that form corrals, suggesting that the primary cilium is divided into functional domains, probably to segregate signaling cascades. Indeed, the confinement of the GPCRs within corrals may increase the likelihood of a given GPCR to encounter other proteins, such as a G protein, within the same corral. Thus, the corrals may help to generate local signaling nanodomains and could play regulatory roles in ciliary signaling by limiting the number of interactions between cascade components [58].
Primary cilium is also involved in mechanosensing. For example, the primary cilium is able to sense mechanical and chemical cues provided by the cellular environment [59]. Proteomic analyses were performed on collecting duct cell lines with or without cilia that were kept stationary or rotated to stimulate cilia bending. The expression of PEBP1 was significantly elevated in rotated cilia ( +) cells and elevated PEBP1 levels were associated with reduced cell proliferation, suggesting that ciliary movement may help to control the expression of PEBP1 and thus maintain cell differentiation. In polycystic kidney disease, loss of cilia and, therefore, sensitivity to flow may lead to reduced PEBP1 levels and contribute to the formation of cysts [60].
Primary cilium plays a role in autophagy and nutrient deprivation is a stimulus shared by both autophagy and formation of primary cilium. It was shown that part of the molecular machinery involved in ciliogenesis also participates in the early steps of the autophagic process. Abrogation of ciliogenesis partially inhibits autophagy, while blockage of autophagy enhances primary cilia growth during normal nutritional conditions, suggesting that basal autophagy regulates ciliary growth through the degradation of proteins required for intraflagellar transport [61].
Furthermore, primary cilium plays an important role in cell cycle as primary cilium disassembly is associated with cycle progression. Indeed, during G0 and G1 phases, primary cilia are assembled and function as important signaling hubs. Later, when the cell enters the S phase, ciliary disassembly begins; then, in M phase, the mother and daughter centrioles are duplicated and move to the spindle poles where they become centrosomes and the mitotic spindle [62]. Interestingly, PEBP1 controls the cell cycle in the two interphases G1/S and G2/M, when the primary cilium is disassembled [63].
In conclusion, PEBP1 prevents primary cilium formation by interacting with Rab8 at the transition zone of primary cilium, inhibiting the transport of proteins toward the axoneme and ciliary membrane. PEBP1 seems also to control the primary cilium disassembly during the two interphases G1/S and G2/M of cell cycle. Conversely, the cilia movements of renal collecting duct cell lines enhance expression of PEBP1 and subsequently maintain cell differentiation. Thus, the implication of PEBP1 in several cellular processes, such as signal transduction, mechanosensing, ciliary autophagy, and cell cycle, may be explained by its location and its activity at the primary cilium. On another hand, ciliary membranes appeared to be partitioned into membrane nanodomains delimited by F-actin, limiting the number of interactions between cascade components, and suggesting close and intertwined relationships between actin and primary cilium membrane.
PEBP1 in autophagy: LC3 and Rab8
LC3
In HeLa and H1299 cells, PEBP1 was demonstrated to negatively regulate starvation-induced autophagy by direct interaction with microtubule-associated protein 1 light chain 3β (LC3) [64]. In the membranes, LC3 is conjugated with phosphatidylethanolamine (PE) and is then designated LC3-II. It was demonstrated that PEBP1 interacts with LC3 through the LIR motif 55WDGL58 that is situated in an exposed external loop of PEBP1. Mutation of the LIR motif in PEBP1 disrupted its interaction with LC3 [64]. PEBP1 bound specifically to the unconjugated form of LC3. Conversely, PEBP1 phosphorylation at Ser153 caused dissociation of LC3 from the PEBP1-LC3 complex suggesting that, upon activation of autophagy, PEBP1 may be phosphorylated by kinases (such as PKC isoforms) to release LC3, allowing its conjugation with PE and subsequent autophagosome formation. PEBP1 lacking the LIR motif highly stimulated starvation-induced autophagy through the AKT-MTORC1-dependent pathway, indicating that PEBP1 prevents autophagy by regulating not only LC3 lipidation but also the Akt-mTOR-ULK pathway [64].
The role of LC3 was also observed at focal adhesions, where autophagy promotes cell migration. Indeed, the interaction between LC3 and the cargo receptor Neighbor of BRCA1 gene 1 protein (NBR1) induces the targeting of autophagosomes to focal adhesions. The subsequent sequestration of focal adhesion proteins results in focal adhesion disassembly and promotes cell migration [65]. It was shown that cilia-mediated autophagy may induce autophagosome formation from the ciliary pocket [61]. During autophagy, actin and LC3 co-localize at the isolation membrane of autophagosome [66] and LC3 recruits Junction-Mediating and Regulatory Protein (JMY) to the phagophore, promoting its actin nucleation activity [67, 68].
In sum, LC3 is a key protein in autophagosomes formation. It governs autophagy at focal adhesion, regulating subsequent cell migration, and it plays a role in primary cilium growth by directing autophagy at the ciliary pocket.
Rab8
In the preceding section, we have discussed the role of Rab8 as a regulator of primary cilium formation [54]. Additionally, as numerous Rab proteins, Rab8 is also implied in autophagy. Notably, Rab8B plays a key role in autophagosome maturation, while the recruitment of Rab8A by activated optineurin allows initiation of autophagy at the precise site of the material intended for degradation (protein aggregate, dysfunctional organelles, or intracellular pathogen) [69].
In conclusion, LC3 and Rab8 govern vesicle trafficking from the membrane and to the membrane, respectively. Since, they are able to interact directly with PEBP1, they strongly suggest the participation of PEBP1 in membrane remodeling.
Cell motility: focal adhesions, formation of protrusions and implication of FAK, soluble MMPs, MT1-MMP, and Rab8
Accumulative studies have shown that loss or downregulation of PEBP1 enhances motility and metastasis of tumor cells [70, 71], suggesting that PEBP1 is a general upstream regulator of cell motility [72] whose role is fundamental to its metastasis suppressive function in cancer [63]. Usually, cell migration is divided into several main steps: (1) protrusion based on actin polymerization force, (2) adhesion at the front, (3) actin–myosin-mediated contraction of the cytoplasm, (4) release of adhesions at the rear, and (5) forward translocation of the cell body and recycling of the motility machinery [73]. Though PEBP1 is probably involved in several of these steps, it was particularly described to play a key role in the formation of focal adhesions and protrusions to facilitate cell progression through the ECM.
Focal adhesions and FAK
Two major studies have suggested the role of PEBP1 in cell adhesion. First, overexpression of PEBP1 in Madin–Darby canine kidney (MDCK) epithelial cells increases adhesion to the substratum, while decreasing adhesion of the cells to one another [74]. In another series of experiments, Melanoma Differentiation Associated gene-9 (MDA-9), also known as syntenin, functions as a positive regulator of melanoma progression and metastasis. It was found that MDA-9 transcriptionally downregulated PEBP1 and that MDA-9 and PEBP1 physically interact, leading to a suppression of focal adhesion kinase (FAK) and c-Src phosphorylation, crucial steps to initiate signaling cascades leading to metastatic phenotype [75].
Formation of protrusions, soluble MMPs, MT1-MMP, and Rab8
In cancer, the motility of the cells is essential to enhance metastasis. The study of invadopodia, the hallmark of cancer, suggests a potential role of PEBP1 in cell motility. Invadopodia are formed by cancer cells during migration; they are F-actin protrusions located on the internal surface of cell membrane, near the ECM [76]. Invadopodia penetrate into ECM, and enzymatically degrade it, enhancing matrix remodeling and cell invasion across tissues. In cancer cells, invadopodia are considered to possess three functional domains: (1) the proteolytic domain, which ensures the metalloproteinase-based degradation of the ECM, (2) the invasive domain, which is the cytoskeleton-based mechanical penetration into the matrix, and (3) the adhesive domain, which is the integrin adhesome-based adhesion to the ECM. As described below, the first domain is of particular interest as it is based on the activity of several proteins whose expression levels are modulated by PEBP1 (such as MMPs and MT1-MMP) or that are known to bind PEBP1 (such as Rab8).
MMPs and MT1-MMP
The matrix degradation capacity of invadopodia is attributed to a variety of proteases, belonging to the metalloproteinase (MMPs), ADAM, and serine protease families. Among them, the soluble MMPs and the membrane-bound MT1-MMP are the main proteases associated with invadopodia activity [77]. Particularly, soluble MMP2 and MMP9 are secreted by the migrating cell and degrade ECM around invadopodia, while the membrane-bound MT1-MMP displays intrinsic collagenolytic activity, and can also cleave and activate some of the soluble MMPs. In breast cancer, polarized exocytosis and transport of vesicles of MT1-MMP to sites of invadopodia are regulated by Rab8.
The overall “mission” of invadopodia is the promotion of cell migration across connective tissue barriers. Focal degradation of the matrix by MT1-MMP and broader degradation by the soluble proteases reduce matrix barriers under and around invadopodia, conditioning these areas for the penetration step. Next, actin polymerization in the core of the invasive domain pushes against the plasma membrane, driving the penetration into the ECM [77].
Rab8
Independently of its role in polarized exocytosis and transport of vesicles of MT1-MMP in invadopodia, Rab8 drives also cell motility. To determine the directionality of cell migration, Rab8 regulates the establishment of cell polarity, turnover of focal adhesions, and actin cytoskeleton rearrangements [78]. Beside invadopodia, Rab8 is also implied in the formation of other protrusions including lamellipodia, filopodia, and podosomes. Lamellipodia are large and flat protrusions which are located at the leading edge of migratory cells. They sense the surrounding environment, drive cell locomotion, and are considered to be the initial protrusions in cancer cells. In melanoma cells A375, recent data showed that lipid rafts recruit β1 and β3 integrin to the leading edge, in a manner dependent on the actin cytoskeleton, thereby participating in lamellipodia formation. The migration of cancer cells is inhibited when the integrity of lipid rafts is disrupted, revealing the association between lipid rafts and lamellipodia formation and indicating that lamellipodia require the assembly of the actin cytoskeleton and the motility of membranes [79].
In conclusion, PEBP1 appears to increase cell adhesion to the substratum while decreasing adhesion of the cells to one another, and to be an inhibitor of MDA-9/syntenin-mediated metastasis in melanoma. It is to note that the formation of cell protrusions requires the generation and turnover of actin filaments that arrange into specific, subcellular structures. The diverse protrusions share the transport of proteins to the membrane such as MT1-MMP in migratory cells and GPCRs in primary cilium. The traffic of vesicles which contain proteins is regulated by Rab8 activity. Then, by blocking Rab8 activity and also regulating MMPs expression, PEBP1 may likely play a crucial role in the formation and functionality of cellular protrusions. Considering that PEBP1 is strongly downregulated in cancer cells, it is tempting to suggest that in normal cells, PEBP1 regulates the expression of MMPs and is associated with various proteins, such as IQGAP and Rab8, to control cell migration across ECM. Consequently, normal cell migration under PEBP1 control may ensure development and differentiation of tissues and prevent invadopodia formation.
Coordination between actin filaments, microtubules, and intermediate filaments: roles of APC, Aurora B, and keratins 8/18
The following paragraphs focus on three proteins, interacting with PEBP1, which are recognized as cross-linkers between actin filaments and other constituents of the cell cytoskeleton such as microtubules and intermediate filaments.
APC
In models of triple-negative breast cancer (TNBC) [26], a network of stress kinases targeted by PEBP1 was identified and genes that correlate with PEBP1 expression in The Cancer Genome Atlas (TCGA) breast cancer were investigated. APC was identified as a novel PEBP1 target in in vitro and in vivo models of TNBC, and was demonstrated to be downstream of the PEBP1-stress network [26].
APC is a tumor suppressor that regulates cell protrusion and migration. In vivo APC localizes to actin-rich cortical protrusions and microtubule plus ends, and displays direct effects on the coordinated organization of actin and microtubule dynamics. It was shown that the C-terminal basic domain of APC stimulates actin assembly in cells and nucleates the formation of actin filaments in vitro. By changing only two residues in APC C-terminal basic domain, a mutant was created which abolishes actin nucleation activity without affecting microtubule interactions. As the wild-type APC, the APC-mutant localized to focal adhesions (FAs), but it was defective in promoting actin assembly and did not facilitate the FA turnover induced by microtubules. These data suggested a main role of APC in coordinating microtubules and actin nucleation at FAs to control cell migration [80, 81].
Another important molecular link between actin and microtubules comes from binding of the actin crosslinking IQGAP1 to APC. Indeed, IQGAP1 and APC each has independent connections to actin and microtubules, and both may act as scaffolds to coordinate actin–microtubule interactions during cell migration. Remarkably, IQGAP1 and APC also bind activated forms of Rac1 and Cdc42. Consistent with these protein interactions, IQGAP, APC, Rac1, and Cdc42 all accumulate at the leading edge of Vero cells (a monkey kidney epithelial line) during migration into a wound [82]. Thus, PEBP1, APC, and IQGAP are probably members of common molecular assemblies implied in cell migration.
Aurora B
In mammalian cells, PEBP1 was shown to regulate Aurora B kinase (AurB) and the spindle checkpoint [83]. PEBP1 was also found to be associated with centrosomes and with kinetochores which form the interface between microtubules of the mitotic spindle and chromosomes, regulating chromosomes movements during mitosis. In HeLa and H19-7 cells, PEBP1 depletion altered localization and kinase activity of AurB at kinetochores and led to a decrease in mitotic index, an acceleration in timing of the metaphase/anaphase transition and a defect in the spindle checkpoint. A direct interaction between PEBP1 and AurB was not proved, and thus, it was suggested that AurB kinase activity could be inhibited directly by ERK1/2 phosphorylation of AurB or indirectly by phosphorylation of other proteins required for recruitment of the kinase to the kinetochores [83].
In another note, by monitoring mitotic index and transit time from nuclear envelope breakdown to anaphase, it was demonstrated that PEBP1 depletion led to a defective spindle checkpoint and genomic instability, particularly in response to drugs that disrupt microtubule function [84]. Loss of PEBP1 during M phase leads to bypass of the spindle assembly checkpoint and the generation of chromosomal abnormalities. These results revealed a role for PEBP1 in ensuring the fidelity of chromosome segregation prior to cell division [85].
Keratin8/18
Keratin 8 and keratin 18 were identified as closely related to PEBP1 by mapping the interactome of overexpressed PEBP1 in a gastric cancer cell line. Then, the interaction complex PEBP1/Keratin 8 was confirmed by Western blot analysis and co-immunoprecipitation [20]. Keratins (Ks) are the intermediate filament (IF) proteins of epithelial cells, constituting the largest family of cytoskeletal proteins. Keratin IFs are heteropolymers that include at least two types of keratins which are coordinately expressed as pairs depending on the cell lineage and cell differentiation. In simple epithelial cells, K8/K18 IFs contribute to the cell-stiffness/ECM-rigidity mechanosensing interaction through actin cytoskeleton dynamics mediated by Rho. In hepatic cells, it was proposed that the interplay between K8/K18 IFs and Rho-mediated actin dynamics occurs at focal adhesions through a plectin–RACK1–Src connection [86]. The keratin network presents an exceptional plasticity which is obvious during the polarized keratin network assembly in the leading edge of migrating cells. Likewise, the plasticity of keratin network is proved in the dividing cells, wherein the rapid disassembly of the keratin cytoskeleton during prophase is followed by a subsequent re-assembly at the end of mitosis. Even in cells at equilibrium, a perpetual cycling of the keratin cytoskeleton is observed. Based on its binding capacity for actin filaments and keratins, it was suggested that plectin could link inward transport of keratins to actin retrograde flow in the cell periphery [87]. Interestingly, in addition to keratins 8 and 18, plectin was also identified as a PEBP1-interacting protein in a gastric cancer cell line [20] (Fig. 1).
In conclusion, the relationships of PEBP1 with APC, AuroraB, and keratins suggest a potential role of PEBP1 in the coordination of microtubules, intermediate filaments, and cortical actin in cellular processes, such as protrusions formation, cell migration, and mitosis.
Discussion
During the past 20 years, numerous papers have described the involvement of PEBP1 in various cellular processes, such as proliferation, differentiation, survival, and motility. PEBP1 was also implicated in several diseases, including cancer, Alzheimer disease, diabetes, and retinopathies. In cancer, PEBP1 was mainly associated with cell motility and metastases development. In addition, PEBP1 was shown to bind to various proteins, especially kinases of signaling pathways, GRK2, Rab8, LC3, effectors of Rho-GTPases, βcatenin, myosin 9, vimentin, and keratins. Altogether these findings gave to PEBP1, the image of a multifunctional protein which plays diverse roles in cells, thus raising the question of its basic features and modes of action [8, 13]. Previously, the goal of our work was to gather as much data from the literature as possible to clarify the PEBP1 key functions by deriving common features among the various cellular processes regulated by PEBP1 [7]. By discussing the molecular targets of PEBP1 that modulate multiple cellular functions, we have first gradually arrived to the conclusion that the main role of this small protein is to regulate important aspects of membrane remodeling and cytoskeleton organization. Here, based on recently identified targets of PEBP1, we focus on the implication of PEBP1 in vesicle trafficking and in actin organization. Furthermore, by examining the relationships between actin, membrane, and other cytoskeletal components (microtubules and intermediate filaments), we reinforce the concept that PEBP1 is a privileged regulator of cortical actin network in intimate connection with plasma membrane remodeling, throwing new light on its crucial role in cell motility. Finally, to complete the overview on the main features of PEBP1, we address thereafter some molecular aspects of the physical interactions between PEBP1 and its interacting targets, and at last, we summarize the main findings emerging from the discussed parallels between PEBP1 behavior and actin network organization.
Molecular grounds of PEBP1 interactions with its targets
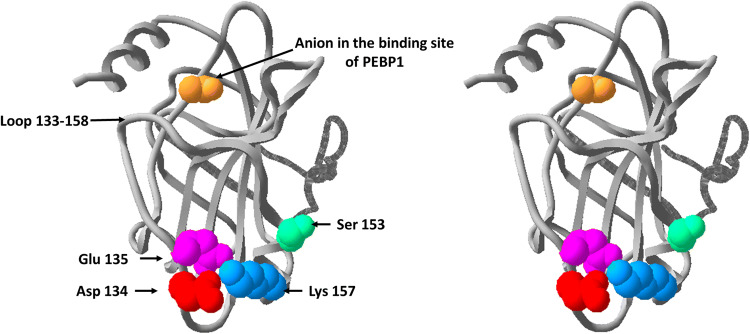
The interactions between PEBP1 and its protein targets were discovered essentially by studying cellular dysfunction and pathologies and also by performing methods such as two-hybrid screening, co-immunoprecipitation, co-localization, or enzyme inhibition. Existing data provide little information about the physical binding of PEBP1 with its interacting proteins. Crystallographic data [3, 4] and NMR studies [88] have demonstrated that several anionic ligands (acetate, phosphorylethanolamine, cacodylate, and nucleotides) are able to bind the small cavity of PEBP1 (Fig. 2). The binding of proteins proved to be more difficult to analyze, probably due to the dynamic and transient nature of the interactions between PEBP1 and its targets. Nevertheless, the interactions between PEBP1 and Raf1 have been studied, and the anion-binding site of PEBP1 was considered the main interacting region with the N-terminal of Raf1 [89]. Interestingly, PEBP1 binds to Raf1 or GRK2 according to its nonphosphorylated or phosphorylated state, respectively [44]. Recently, using NMR 1H-15 N HSQC technique, the spectra of PEBP1P74L (Pro74 mutated into Leu) and its Ser153 phosphorylated form were compared [90]. The PEBP1P74L mutant was used as it increases the phosphorylation rate of PEBP1. Peak movement, line broadering, and disappearance of peaks were observed for residues located near Ser153. The major conclusion was that, upon phosphorylation, the K157 side chain could rotate and form a salt bridge with pS153, while K157 forms a salt bridge with D134 and E135 in the crystal structure of the wild-type PEBP1 (Fig. 2). The disruption of the salt-bridge triad and the partial unfolding of the associated region could lead to the GRK2-binding competent state [90]. Given that the anion-binding site of PEBP1 was considered as the main interacting region with Raf1 N-terminal part, S153 may seem not situated at the direct interface between PEBP1 and Raf 1 [89]. Nevertheless, the segment 133–158 of PEBP1 that contains D134, E135, S153, and K157 is well exposed to solvent and, by protein–protein docking, it has been identified as a probable interaction zone with Raf1 [91]. Studies concerning numerous protein–protein interactions found that 87% of phosphorylation sites are either solvent accessible or interfacial. Among them, serine phosphorylation appears to be a major mechanism for regulating oligomeric protein interactions [92]. Thus, the interactions of PEBP1 with its targets are probably directed, at least in most part, by salt bridges which are themselves under the control of posttranslational modifications, especially phosphorylation. PEBP1-binding site, S153, and salt-bridge D134/E135-K157 are indicated on the 3D structure, as shown in Fig. 2.
Fig. 2.
3D Structure of wild-type PEBP1 [3]. S153 is known to be the phosphorylation site of PEBP1 by PKC. The side chain of S153 is indicated in green color. Phosphorylated PEBP1 releases Raf1 and binds to GRK2. As seen here, in the wild-type structure, K157 interacts with D134 and E135. On the contrary, the phosphorylated S153 outcompetes D134 and E135 for interaction with K157 and allows the binding of PEBP1 to GRK2 [90]. D134, E135, and K157 are indicated in red, magenta, and blue colors, respectively. The binding site of PEBP1 toward anions is indicated by an acetate ion presented in gold color. The loop consisting of the segment 133–158 of PEBP1 that contains D134, E135, S153, and K157 is well exposed to solvent and, by protein–protein docking, has been identified as a possible interaction zone with Raf1 [91]
Relationships between cortical actin and membrane
The actin architecture beneath the membrane regulates the cell shape, but it also regulates the local and global composition of the membrane, and membrane models have demonstrated the influence of the underlying cortical actin on the diffusion of membrane components [93]. Membrane proteins and lipids display free diffusion in small local regions. Occasionally, they jump into another area, where they again display free diffusion. These areas of free diffusion coincide with the mesh size of the underlying actin network. This led to the conclusion that actin network, in connection with actin–membrane linker proteins, contribute to the plasma membrane compartmentalization. Furthermore, membrane tension and cortical actin organization influence each other, suggesting that local changes in membrane composition may act on membrane tension, while changes in tension may lead to modification in membrane composition. Thus, it was suggested that the plasma membrane and actin cortex have to be considered together, as a single composite that can change its composition locally and temporally [93].
Strikingly, there are a significant number of cytoskeleton structures described to be directly or indirectly modulated by PEBP1 that are related to actin cortex architectures; among them are invadopodia and cell adherens junctions. Particularly, invadopodia, the main features of migratory cells in cancer, consist of branched actin architecture. It is important to note that these actin cortex architectures influence shape and composition of the membrane [93]. By describing the role of PEBP1 in different cell processes, the preceding sections of this paper highlight the significance of the direct interaction between PEBP1 and Rab8, these two proteins being implied in major processes, such as ciliogenesis, autophagy, and cell motility. Particularly, activation of Rab8 was linked to the formation of the dynamic cell structures like filopodia, lamellipodia, ruffles, and primary cilia, suggesting that Rab8 has an important role in regulating cell migration by linking actin dynamics to membrane turnover [94]. Furthermore, depletion of Rab8 was also found to prevent maturation of cultured hippocampal neurons by inhibiting neurite outgrowth, due to a reduction of vesicles undergoing anterograde transported into neurites. Thus, membrane trafficking is likely to play an important role at different stages of neuron differentiation, particularly at neurite outgrowth, axon, and dendrite formation, and also at synapse formation [94]. The inhibition of Rab8 by PEBP1 could explain, at least in part, the important role of PEBP1 in brain function and development, such as synaptic plasticity of mammalian brain [95], neuronal differentiation [96], axonal regeneration after optic nerve crush [97], as well as enhanced neurite growth and neural differentiation in human neuroblastoma cells [98].
PEBP1 interacts with effectors regulating plasma membrane and cortical actin
The interplay of PEBP1 with proteins located from either side of plasma membrane or included in the membrane leaflet may explain its involvement in the response of the cell to its environment. Particularly, PEBP1 was shown to regulate the expression of several MMPs [7, 99], all enzymes that are capable of degrading all kinds of extracellular matrix proteins and that are thought to play a major role in cell proliferation, migration, differentiation, apoptosis, and host defense. PEBP1 participates also in sensing the plasma membrane status by acting on internalization of receptors and by controlling the formation of primary cilium that is a sensory organelle. Phosphorylated PEBP1 acts on GRK2 which is a main protein in cell life as it may modulate several facets of cell migration in a stimuli and cellular context-dependent manner [47]. PEBP1 was also found to associate with proteins that connect cortical actin to membrane, such as β-catenin and vinculin (Fig. 1). It should be remembered that the primary mechanical role of catenin isoforms is connecting cadherins to actin filaments, specifically in the adhesion junctions of epithelial cells [100]. Actin polymerization at the membrane is also governed by the regulation of PIP2/PIP3 levels. Proteins that link multiple actin filaments or mediate actin–membrane binding like alpha-actinin, vinculin, talin, or ezrin are activated by PIP2 [101, 102]. Among them, vinculin, identified as closely related to PEBP1 [20], is a cytoskeletal protein associated with cell–cell and cell–matrix junctions where it is involved in anchoring F-actin to the membrane. Frequently, the membrane appears to be bound to the actin architecture via linker proteins that associate with membrane proteins using protein–protein interaction motifs [93]; among them is another protein identified as closely related to PEBP1, namely filamin A [20].
PEBP1 spatio-temporally coordinates cortical actin and membrane organization
During cell migration, it was noted that actin is pushing as hard as it can, and that the leading edge advance is limited by the speed of the membrane to go forward [103]. Potentially, this may shed some light into the role of PEBP1 when inhibiting targets and particularly the kinases of signaling pathways. Indeed, it is likely that PEBP1 inhibits kinases and actin effectors the time necessary to enable the membrane achieving physical changes and then, ensuring the coordination between signaling cascades, actin organization, and cell shape. In the absence of any control, the actin skeleton may loosely push the membrane and activate cell motility, overriding the exterior constraints. In a way, it is what is observed in cancer, particularly during metastases development. Another similar case was reported that described the preferential interaction of PEBP1 with the GDP-bound form of Rab8A in the retina. Thus, the PEBP1/Rab8A(GDP) complex may need to dissociate for the release of Rab8A(GDP) and its subsequent conversion to Rab8A(GTP) that triggers the appropriate ciliary transport [54]. In this case, one might think that PEBP1 binds the inactive GDP-bound form of Rab8 the time required for the localization of the essential actors driving cilium formation. Similarly to Rab8, LC3 is also blocked in its inactive form by PEBP1, inhibiting autophagy at adherens junctions and subsequently cell motility [65]. Consequently, the depletion of PEBP1 in cancer may explain, at least in part, the increase of cancer cell motility and the development of metastases.
Actin seems to be capable of self-organization, particularly through Ras/actin and IQGAP/actin interactions. Consequently, the effectors of actin cytoskeleton appear the major drivers for directing actin networks toward specific functions. Repeatedly, PEBP1 was described as a scaffold protein that binds to kinases of various signaling pathways, generally inhibiting them. In addition, PEBP1 is a regulator of numerous effectors of actin organization such as GTPases, IQGAP, and GRK2. PEBP1 may also act as a switch between Raf1 and GRK2, two major kinases of cytoskeleton organization and cell motility. All these data are sufficient to explain the key role of PEBP1 in various cellular processes, notably cell motility, and strongly suggest that PEBP1 associates other proteins to participate in membrane remodeling and actin organization.
PEBP1 expression is dependent of actin status
Interestingly, actin organization determines the cell fate as survival, differentiation, and motility, especially via regulating expression of numerous translation effectors. PEBP1 was described to be a member of the loop NF-κB/Snail/YY1/PEBP1/PTEN identified to regulate EMT in cancer [104]. Interestingly, we have noticed that NF-kB, Snail, YY1, and PTEN are related to actin network.
Especially, it appears that each member of the loop known to control PEBP1 expression interplays with the status of actin. Therefore, the nuclear factor-kappa B (NF-kB) activation follows disruption of the actin cytoskeleton in human intestinal epithelial cells [105]. The zinc finger protein Snail, in IL-8-induced mesenchymal transition, stabilizes IL-8 via ELMO1 and a bipartite Rac GEF complex, activating Rac proteins and leading to actin polymerization and glioma cells migration and invasion [106]. The depolymerization of γ-Smooth Muscle actin filaments releases Yin Yang (YY1) to the nucleus to activate transcription [107]. Phosphatase and tensin homolog (PTEN) and PIP2 signaling appear to play a main role in myocyte hypertrophy by the regulation of actin filament dynamics [101]. An actin-binding protein linking synaptic dysfunction to cognitive deficits is Drebrin (DBN). The association of PTEN with DBN potentially governs essential processes during the reorganization of the actin cytoskeleton at the synapse [108]. Taken together with the identification of actin as closely related to PEBP1 [20], all these data strongly suggest that, directly or indirectly, PEBP1 expression may be controlled by the polymerization and depolymerization of actin.
Finally, it seems that the role of PEBP1 is dependent of cell type and cell state, according to the signaling pathways and the actin organization at work in the cell. This feature, which agrees with the control of PEBP1 expression by the polymerization and depolymerization of actin, is valuable to allow the adaption of the living cell to its environment. Another interesting impact of the modulation of PEBP1 expression by actin state is the competition observed between proliferation and differentiation. Because the global quantity of actin (both G and F forms) is constant in a given cell, the regulation of actin effectors is needed to balance the cell fate toward proliferation or differentiation. PEBP1 is probably one of these key effectors. As an example, it was observed that in adult hippocampal rat progenitor cells, overexpression of PEBP1 promoted neuronal and oligodendrocyte differentiation, whereas PEBP1 silencing promoted differentiation into astrocytes, suggesting that PEBP1 expression closely correlated with differentiation into neurons and oligodendrocytes, but not astrocytes [96].
Concluding remarks
Many of the cellular processes modulated by PEBP1 imply membrane changes (invagination, protrusion, engulfment, vesicle formation, or fusion) and all of them need fine-tuned cytoskeleton organization. These events impact all the cell life and are managed by numerous molecular complexes and signal exchanges between the cell and its environment. PEBP1 goes with actin from the cell outside up to the coordination of actin networks with microtubules and intermediate filaments, including membrane reorganization and modeling. It should be kept in mind that cell surface shape changes involve actin reorganization events. This is true for the plasma membrane and for large cell-spanning organelles like the endoplasmic reticulum, as well as for single vesicles and double membrane-encircled organelles like mitochondria [109]. By interacting with molecular assemblies, PEBP1 is one of the nodes where signaling pathways, actin cytoskeleton, and membrane meet. PEBP1 acts generally by direct interaction with protein targets, inhibiting their activity, this is the case for Rab8, LC3, and signaling kinases. This is true also for the phosphorylated form of PEBP1 which blocks GRK2 activity and the subsequent GPCRs internalization. We propose that the inhibitory action of PEBP1 leads to the coordination between the physical changes of the membrane and the actin organization. This is a crucial point during migration of cancer cells in which PEBP1 is downregulated. Indeed, the absence of coordination between membrane and cortical actin may explain, at least in part, the exceptional motility of the metastatic cells, because actin is pushing in the leading edge without considering the micro-environmental constraints. Thus, the apparent diverse functions of PEBP1 may be explained when taking into account the ability of this protein to accompany and direct the cortical actin reorganization in interweaving with membrane changes. In the future, this idea should be a main thread as it is likely that an in depth-understanding of PEBP1 function will emerge along with new knowledges on the dynamics of the actin-membrane composite material.
Acknowledgements
We thank Isabelle Callebaut (IMPMC) for helpful comments and fruitful remarks. We sincerely thank William Sacks (IMPMC) for critical reading and improvement of the manuscript. We thank also the members of the “Canceropôle du grand Ouest” for stimulating and constructive discussions throughout these last few years.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Bernier I, Tresca JP, Jollès P. Ligand-binding studies with a 23 kDa protein purified from bovine brain cytosol. Biochim Biophys Acta. 1986;871(1):19–23. doi: 10.1016/0167-4838(86)90128-7. [DOI] [PubMed] [Google Scholar]
- 2.Schoentgen F, Saccoccio F, Jollès J, Bernier I, Jollès P. Complete amino acid sequence of a basic 21-kDa protein from bovine brain cytosol. Eur J Biochem. 1987;166(2):333–338. doi: 10.1111/j.1432-1033.1987.tb13519.x. [DOI] [PubMed] [Google Scholar]
- 3.Serre L, Vallée B, Bureaud N, Schoentgen F, Zelwer C. Crystal structure of the Phosphatidylethanolamine-binding protein from bovine brain: a novel structural class of phospholipid-binding proteins. Structure. 1998;6(10):1255–1265. doi: 10.1016/s0969-2126(98)00126-9. [DOI] [PubMed] [Google Scholar]
- 4.Banfield MJ, Barker JJ, Perry AC, Brady RL. Function from structure? The crystal structure of human phosphatidylethanolamine-binding protein suggests a role in membrane signal transduction. Structure. 1998;6(10):1245–1254. doi: 10.1016/s0969-2126(98)00125-7. [DOI] [PubMed] [Google Scholar]
- 5.Lamiman K, Keller JM, Mizokami A, Zhang J, Keller ET. Survey of Raf kinase inhibitor protein (RKIP) in multiple cancer types. Crit Rev Oncog. 2014;19(6):455–468. doi: 10.1615/critrevoncog.2014011987. [DOI] [PubMed] [Google Scholar]
- 6.Keller ET, Fu Z, Yeung K, Brennan M. Raf kinase inhibitor protein: a prostate cancer metastasis suppressor gene. Cancer Lett. 2004;207(2):131–137. doi: 10.1016/j.canlet.2004.02.006. [DOI] [PubMed] [Google Scholar]
- 7.Schoentgen F, Jonic S (2018) PEBP1/RKIP: from multiple functions to a common role in cellular processes. arXiv:1802.02378 [q-bio.SC]
- 8.Rajkumar K, Nichita A, Anoor PK, Raju S, Singh SS, Burgula S. Understanding perspectives of signalling mechanisms regulating PEBP1 function. Cell Biochem Funct. 2016;34(6):394–403. doi: 10.1002/cbf.3198. [DOI] [PubMed] [Google Scholar]
- 9.Yeung K, Seitz T, Li S, Janosch P, McFerran B, Kaiser C, Fee F, Katsanakis KD, Rose DW, Mischak H, Sedivy JM, Kolch W. Suppression of Raf-1 kinase activity and MAP kinase signaling by RKIP. Nature. 1999;401(6749):173–177. doi: 10.1038/43686. [DOI] [PubMed] [Google Scholar]
- 10.Tang H, Park S, Sun SC, Trumbly R, Ren G, Tsung E, Yeung KC. RKIP inhibits NF-kappaB in cancer cells by regulating upstream signaling components of the IkappaB kinase complex. FEBS Lett. 2010;584(4):662–668. doi: 10.1016/j.febslet.2009.12.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maresch J, Birner P, Zakharinov M, Toumangelova-Uzeir K, Natchev S, Guentchev M. Additive effect on survival of Raf kinase inhibitor protein and signal transducer and activator of transcription 3 in high-grade glioma. Cancer. 2011;117(11):2499–2504. doi: 10.1002/cncr.25799. [DOI] [PubMed] [Google Scholar]
- 12.Wall AA, Luo L, Hung Y, Tong SJ, Condon ND, Blumenthal A, Sweet MJ, Stow JL. Small GTPase Rab8a-recruited phosphatidylinositol 3-kinase γ regulates signaling and cytokine outputs from endosomal Toll-like receptors. J Biol Chem. 2017;292(11):4411–4422. doi: 10.1074/jbc.M116.766337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Al-Mulla F, Bitar MS, Taqi Z, Yeung KC. RKIP: much more than Raf kinase inhibitory protein. J Cell Physiol. 2013;228(8):1688–1702. doi: 10.1002/jcp.24335. [DOI] [PubMed] [Google Scholar]
- 14.Shin SY, Rath O, Zebisch A, Choo SM, Kolch W, Cho KH. Functional roles of multiple feedback loops in extracellular signal-regulated kinase and Wnt signaling pathways that regulate epithelial-mesenchymal transition. Cancer Res. 2010;70(17):6715–6724. doi: 10.1158/0008-5472.CAN-10-1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin X, Bai F, Nie J, Lu S, Lu C, Zhu X, Wei J, Lu Z, Huang Q. Didymin alleviates hepatic fibrosis through inhibiting ERK and PI3K/Akt pathways via regulation of Raf kinase inhibitor protein. Cell Physiol Biochem. 2016;40(6):1422–1432. doi: 10.1159/000453194. [DOI] [PubMed] [Google Scholar]
- 16.Lai R, Gu M, Jiang W, Lin W, Xu P, Liu Z, Huang H, An H, Wang X. Raf kinase inhibitor protein preferentially promotes TLR3-triggered signaling and inflammation. J Immunol. 2017;198(10):4086–4095. doi: 10.4049/jimmunol.1601672. [DOI] [PubMed] [Google Scholar]
- 17.Noh HS, Hah YS, Ha JH, Kang MY, Zada S, Rha SY, Kang SS, Kim HJ, Park JY, Byun JH, Hahm JR, Shin JK, Jeong SH, Lee YJ, Kim DR. Regulation of the epithelial to mesenchymal transition and metastasis by Raf kinase inhibitory protein-dependent Notch1 activity. Oncotarget. 2016;7(4):4632–4646. doi: 10.18632/oncotarget.6728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Al-Mulla F, Bitar MS, Al-Maghrebi M, Behbehani AI, Al-Ali W, Rath O, Doyle B, Tan KY, Pitt A, Kolch W. Raf kinase inhibitor protein RKIP enhances signaling by glycogen synthase kinase-3β. Cancer Res. 2011;71(4):1334–1343. doi: 10.1158/0008-5472.CAN-10-3102. [DOI] [PubMed] [Google Scholar]
- 19.Gavilán E, Sánchez-Aguayo I, Daza P, Ruano D. GSK-3β signaling determines autophagy activation in the breast tumor cell line MCF7 and inclusion formation in the non-tumor cell line MCF10A in response to proteasome inhibition. Cell Death Dis. 2013;4:e572. doi: 10.1038/cddis.2013.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gu H, Zhan X, Zhang G, Yan L, Cho WC, Li M, Liu T, Chen Z. Mapping the interactome of overexpressed RAF kinase inhibitor protein in a gastric cancer cell line. BMC cancer. 2013 doi: 10.1186/1471-2407-13-536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li H, Yang HS, Wu TJ, Zhang XY, Jiang WH, Ma QL, Chen YX, Xu Y, Li S, Hua ZC. Proteomic analysis of early-response to mechanical stress in neonatal rat mandibular condylar chondrocytes. J Cell Physiol. 2010;223(3):610–622. doi: 10.1002/jcp.22052. [DOI] [PubMed] [Google Scholar]
- 22.Demidem A, Morvan D, Madelmont JC. Bystander effects are induced by CENU treatment and associated with altered protein secretory activity of treated tumor cells: a relay for chemotherapy? Int J Cancer. 2006;119(5):992–1004. doi: 10.1002/ijc.21761. [DOI] [PubMed] [Google Scholar]
- 23.Huang Q, Liang C, Wei L, Nie J, Lu S, Lu C, Zhuo L, Lu Z, Lin X. Raf kinase inhibitory protein down-expression exacerbates hepatic fibrosis in vivo and in vitro. Cell Physiol Biochem. 2016;40(1–2):49–61. doi: 10.1159/000452524. [DOI] [PubMed] [Google Scholar]
- 24.van Haastert PJ, Keizer-Gunnink I, Kortholt A. Coupled excitable Ras and F-actin activation mediates spontaneous pseudopod formation and directed cell movement. Mol Biol Cell. 2017;28(7):922–934. doi: 10.1091/mbc.E16-10-0733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nobes CD, Hall A. Rho, rac, and cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia, and filopodia. Cell. 1995;81(1):53–62. doi: 10.1016/0092-8674(95)90370-4. [DOI] [PubMed] [Google Scholar]
- 26.Yesilkanal AE, Rabe DC, Tiwari P, Frankenberger C, Johnson GL, Rosner M (2018) A novel approach for antimetastatic therapies against TNBC utilizing a physiologic suppressor [abstract]. In: Proceedings of the American Association for Cancer Research Annual Meeting 78(13 Suppl):Abstract nr 4179
- 27.Upadhyay G, Goessling W, North TE, Xavier R, Zon LI, Yajnik V. Molecular association between β-catenin degradation complex and Rac guanine exchange factor DOCK4 is essential for Wnt/β-catenin signaling. Oncogene. 2008;27(44):5845–5855. doi: 10.1038/onc.2008.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Abraham S, Scarcia M, Bagshaw RD, McMahon K, Grant G, Harvey T, Yeo M, Esteves FO, Thygesen HH, Jones PF, Speirs V, Hanby AM, Selby PJ, Lorger M, Dear TN, Pawson T, Marshall CJ, Mavria G. A Rac/Cdc42 exchange factor complex promotes formation of lateral filopodia and blood vessel lumen morphogenesis. Nat Commun. 2015 doi: 10.1038/ncomms8286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu JR, Tai Y, Jin Y, Hammell MC, Wilkinson JE, Roe JS, Vakoc CR, Van Aelst L. TGFb/ Smad signaling through DOCK4 facilitates lung adenocarcinoma metastasis. Genes Dev. 2015;29(3):250–261. doi: 10.1101/gad.248963.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hiramoto-Yamaki N, Takeuchi S, Ueda S, Harada K, Fujimoto S, Negishi M, Katoh H. Ephexin4 and EphA2 mediate cell migration through a RhoG-dependent mechanism. J Cell Biol. 2010;190(3):461–477. doi: 10.1083/jcb.201005141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yan D, Li F, Hall ML, Sage C, Hu WH, Giallourakis C, Upadhyay G, Ouyang XM, Du LL, Bethea JR, Chen ZY, Yajnik V, Liu XZ. An isoform of GTPase regulator DOCK4 localizes to the stereocilia in the inner ear and binds to harmonin (USH1C) J Mol Biol. 2006;357(3):755–764. doi: 10.1016/j.jmb.2006.01.017. [DOI] [PubMed] [Google Scholar]
- 32.Xiao Y, Peng Y, Wan J, Tang G, Chen Y, Tang J, Ye WC, Ip NY, Shi L. The atypical guanine nucleotide exchange factor Dock4 regulates neurite differentiation through modulation of Rac1 GTPase and actin dynamics. J Biol Chem. 2013;288(27):20034–20045. doi: 10.1074/jbc.M113.458612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Newell-Litwa KA, Horwitz R, Lamers ML. Non-muscle myosin II in disease: mechanisms and therapeutic opportunities. Dis Model Mech. 2015;8(12):1495–1515. doi: 10.1242/dmm.022103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shutova MS, Svitkina TM. Common and specific functions of non-muscle Myosin II paralogs in cells. Biochemistry (Mosc) 2018;83(12):1459–1468. doi: 10.1134/S0006297918120040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Betapudi V. Myosin II motor proteins with different functions determine the fate of lamellipodia extension during cell spreading. PLoS ONE. 2010;5(1):e8560. doi: 10.1371/journal.pone.0008560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ahmed M, Lai TH, Zada S, Hwang JS, Pham TM, Yun M, Kim DR. Functional linkage of RKIP to the epithelial to mesenchymal transition and autophagy during the development of prostate cancer. Cancers (Basel) 2018 doi: 10.3390/cancers10080273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Borg Distefano M, Hofstad Haugen L, Wang Y, Perdreau-Dahl H, Kjos I, Jia D, Morth JP, Neefjes J, Bakke O, Progida C. TBC1D5 controls the GTPase cycle of Rab7b. J Cell Sci. 2018 doi: 10.1242/jcs.216630. [DOI] [PubMed] [Google Scholar]
- 38.Borg M, Bakke O, Progida C. A novel interaction between Rab7b and actomyosin reveals a dual role in intracellular transport and cell migration. J Cell Sci. 2014 doi: 10.1242/jcs.155861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vallée B, Doudeau M, Godin F, Gombault A, Tchalikian A, de Tauzia ML, Bénédetti H. Nf1 RasGAP inhibition of LIMK2 mediates a new cross-talk between Ras and Rho pathways. PLoS ONE. 2012 doi: 10.1371/journal.pone.0047283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Visvikis O, Boyer L, Torrino S, Doye A, Lemonnier M, Lorès P, Rolando M, Flatau G, Mettouchi A, Bouvard D, Veiga E, Gacon G, Cossart P, Lemichez E. Escherichia coli producing CNF1 toxin hijacks Tollip to trigger Rac1-dependent cell invasion. Traffic. 2011;12(5):579–590. doi: 10.1111/j.1600-0854.2011.01174.x. [DOI] [PubMed] [Google Scholar]
- 41.Rotoli D, Morales M, Ávila J, Maeso MDC, García MDP, Mobasheri A, Martín-Vasallo P. Commitment of scaffold proteins in the onco-biology of human colorectal cancer and liver metastases after oxaliplatin-based chemotherapy Int J Mol Sci. 2017 doi: 10.3390/ijms18040891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Benseñor LB, Kan HM, Wang N, Wallrabe H, Davidson LA, Cai Y, Schafer DA, Bloom GS. IQGAP1 regulates cell motility by linking growth factor signaling to actin assembly. J Cell Sci. 2007;120(Pt 4):658–669. doi: 10.1242/jcs.03376. [DOI] [PubMed] [Google Scholar]
- 43.Corbit KC, Trakul N, Eves EM, Diaz B, Marshall M, Rosner MR. Activation of Raf-1 signaling by protein kinase C through a mechanism involving Raf kinase inhibitory protein. J Biol Chem. 2003;278:13061–13068. doi: 10.1074/jbc.M210015200. [DOI] [PubMed] [Google Scholar]
- 44.Lorenz K, Lohse MJ, Quitterer U. Protein kinase C switches the Raf kinase inhibitor from Raf-1 to GRK-2. Nature. 2003;426(6966):574–579. doi: 10.1038/nature02158. [DOI] [PubMed] [Google Scholar]
- 45.Foerster F, Braig S, Moser C, Kubisch R, Busse J, Wagner E, Schmoeckel E, Mayr D, Schmitt S, Huettel S, Zischka H, Mueller R, Vollmar AM. Targeting the actin cytoskeleton: selective antitumor action via trapping PKCε. Cell Death Dis. 2014 doi: 10.1038/cddis.2014.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Penela P, Murga C, Ribas C, Lafarga V. Mayor F Jr (2010) The complex G protein-coupled receptor kinase 2 (GRK2) interactome unveils new physiopathological targets. Br J Pharmacol. 2010;160(4):821–832. doi: 10.1111/j.1476-5381.2010.00727.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Penela P, Nogués L, Mayor F., Jr Role of G protein-coupled receptor kinases in cell migration. Curr Opin Cell Biol. 2014;27:10–17. doi: 10.1016/j.ceb.2013.10.005. [DOI] [PubMed] [Google Scholar]
- 48.Kahsai AW, Zhu S. Fenteany G (2010) G protein-coupled receptor kinase 2 activates radixin, regulating membrane protrusion and motility in epithelial cells. Biochim Biophys Acta. 1803;2:300–310. doi: 10.1016/j.bbamcr.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lafarga V, Mayor F, Jr, Penela P. The interplay between G protein-coupled receptor kinase 2 (GRK2) and histone deacetylase 6 (HDAC6) at the crossroads of epithelial cell motility. Cell Adh Migr. 2012;6(6):495–501. doi: 10.4161/cam.21585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Crassini K, Pyke T, Shen Y, Stevenson WS, Christopherson RI, Mulligan SP, Best OG. Inhibition of the Raf-1 kinase inhibitory protein (RKIP) by locostatin induces cell death and reduces the CXCR4-mediated migration of chronic lymphocytic leukemia cells. Leuk Lymphoma. 2018 doi: 10.1080/10428194.2018.1455974. [DOI] [PubMed] [Google Scholar]
- 51.Calebiro D, Godbole A. Internalization of G-protein-coupled receptors: implication in receptor function, physiology and diseases. Best Pract Res Clin Endocrinol Metab. 2018;32(2):83–91. doi: 10.1016/j.beem.2018.01.004. [DOI] [PubMed] [Google Scholar]
- 52.Jin T. Gradient sensing during chemotaxis. Curr Opin Cell Biol. 2013;25(5):532–537. doi: 10.1016/j.ceb.2013.06.007. [DOI] [PubMed] [Google Scholar]
- 53.Xu X, Jin T. ELMO proteins transduce G protein-coupled receptor signal to control reorganization of actin cytoskeleton in chemotaxis of eukaryotic cells. Small GTPases. 2017 doi: 10.1080/21541248.2017.1318816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Murga-Zamalloa CA, Ghosh AK, Patil SB, Reed NA, Chan LS, Davuluri S, Peränen J, Hurd TW, Rachel RA, Khanna H. Accumulation of the Raf-1 kinase inhibitory protein (RKIP) is associated with Cep290-mediated photoreceptor degeneration in ciliopathies. J Biol Chem. 2011;286(32):28276–28286. doi: 10.1074/jbc.M111.237560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Satir P, Pedersen LB, Christensen ST. The primary cilium at a glance. J Cell Sci. 2010;123(Pt 4):499–503. doi: 10.1242/jcs.050377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Singla V, Reiter JF. The primary cilium as the cell's antenna: signaling at a sensory organelle. Science. 2006;313(5787):629–633. doi: 10.1126/science.1124534. [DOI] [PubMed] [Google Scholar]
- 57.Wheway G, Nazlamova L, Hancock JT. signaling through the primary cilium. Front Cell Dev Biol. 2018;6:8. doi: 10.3389/fcell.2018.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lee S, Tan HY, Geneva II, Kruglov A, Calvert PD. Actin filaments partition primary cilia membranes into distinct fluid corrals. J Cell Biol. 2018;217(8):2831–2849. doi: 10.1083/jcb.201711104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Seeger-Nukpezah T, Golemis EA. The extracellular matrix and ciliary signaling. Curr Opin Cell Biol. 2012;24(5):652–661. doi: 10.1016/j.ceb.2012.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sas KM, Janech MG, Favre E, Arthur JM, Bell PD. Cilia movement regulates expression of the Raf-1 kinase inhibitor protein. Am J Physiol Renal Physiol. 2011;300(5):F1163–1170. doi: 10.1152/ajprenal.00602.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pampliega O, Orhon I, Patel B, Sridhar S, Díaz-Carretero A, Beau I, Codogno P, Satir BH, Satir P, Cuervo AM. functional interaction between autophagy and ciliogenesis. Nature. 2013;502(7470):194–200. doi: 10.1038/nature12639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Higgins M, Obaidi I, McMorrow T. Primary cilia and their role in cancer. Oncol Lett. 2019;17(3):3041–3047. doi: 10.3892/ol.2019.9942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Al-Mulla F, Bitar MS, Taqi Z, Rath O, Kolch W. Raf kinase inhibitory protein (RKIP) modulates cell cycle kinetics and motility. Mol Biosyst. 2011;7(3):928–941. doi: 10.1039/c0mb00208a. [DOI] [PubMed] [Google Scholar]
- 64.Noh HS, Hah YS, Zada S, Ha JH, Sim G, Hwang JS, Lai TH, Nguyen HQ, Park JY, Kim HJ, Byun JH, Hahm JR, Kang KR, Kim DR. PEBP1, a Raf kinase inhibitory protein, negatively regulates starvation-induced autophagy by direct interaction with LC3. Autophagy. 2016;12(11):2183–2196. doi: 10.1080/15548627.2016.1219013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kenific CM, Wittmann T, Debnath J. autophagy in adhesion and migration. J Cell Sci. 2016;129(20):3685–3693. doi: 10.1242/jcs.188490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mi N, Chen Y, Wang S, Chen M, Zhao M, Yang G, Ma M, Su Q, Luo S, Shi J, Xu J, Guo Q, Gao N, Sun Y, Chen Z, Yu L. CapZ regulates autophagosomal membrane shaping by promoting actin assembly inside the isolation membrane. Nat Cell Biol. 2015;17(9):1112–1123. doi: 10.1038/ncb3215. [DOI] [PubMed] [Google Scholar]
- 67.Hu X, Mullins RD. LC3 and STRAP regulate actin filament assembly by JMY during autophagosome formation. J Cell Biol. 2019;218(1):251–266. doi: 10.1083/jcb.201802157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liu X, Klionsky DJ. Regulation of JMY’s actin nucleation activity by TTC5/STRAP and LC3 during autophagy. Autophagy. 2019;15(3):373–374. doi: 10.1080/15548627.2018.1564417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Corbier C, Sellier C. C9ORF72 is a GDP/GTP exchange factor for Rab8 and Rab39 and regulates autophagy. Small GTPases. 2017;8(3):181–186. doi: 10.1080/21541248.2016.1212688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Keller ET. Metastasis suppressor genes: a role for Raf kinase inhibitor protein (RKIP) Anticancer Drugs. 2004;15(7):663–669. doi: 10.1097/01.cad.0000136877.89057.b9. [DOI] [PubMed] [Google Scholar]
- 71.He QY, Yi HM, Yi H, Xiao T, Qu JQ, Yuan L, Zhu JF, Li JY, Wang YY, Li LN, Feng J, Lu SS, Xiao ZQ. Reduction of RKIP expression promotes nasopharyngeal carcinoma invasion and metastasis by activating Stat3 signaling. Oncotarget. 2015;6(18):16422–16436. doi: 10.18632/oncotarget.3847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bement WM. A role for RKIP in cell motility. Chem Biol. 2005;12(9):953–954. doi: 10.1016/j.chembiol.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 73.Sheetz MP, Felsenfeld D, Galbraith CG, Choquet D. Cell migration as a five-step cycle. Biochem Soc Symp. 1999;65:233–243. [PubMed] [Google Scholar]
- 74.Mc Henry KT, Montesano R, Zhu S, Beshir AB, Tang HH, Yeung KC, Fenteany G. Raf kinase inhibitor protein positively regulates cell-substratum adhesion while negatively regulating cell-cell adhesion. J Cell Biochem. 2008;103(3):972–985. doi: 10.1002/jcb.21470. [DOI] [PubMed] [Google Scholar]
- 75.Das SK, Bhutia SK, Sokhi UK, Azab B, Su ZZ, Boukerche H, Anwar T, Moen EL, Chatterjee D, Pellecchia M, Sarkar D, Fisher PB. Raf kinase inhibitor RKIP inhibits MDA-9/syntenin-mediated metastasis in melanoma. Cancer Res. 2012;72(23):6217–6226. doi: 10.1158/0008-5472.CAN-12-0402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Alblazi KM, Siar CH. Cellular protrusions - lamellipodia, filopodia, invadopodia and podosomes—and their roles in progression of orofacial tumours: current understanding. Asian Pac J Cancer Prev. 2015;16(6):2187–2191. doi: 10.7314/apjcp.2015.16.6.2187. [DOI] [PubMed] [Google Scholar]
- 77.Revach OY, Geiger B. The interplay between the proteolytic, invasive, and adhesive domains of invadopodia and their roles in cancer invasion. Cell Adh Migr. 2014;8(3):215–225. doi: 10.4161/cam.27842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bravo-Cordero JJ, Cordani M, Soriano SF, Díez B, Muñoz-Agudo C, Casanova-Acebes M, Boullosa C, Guadamillas MC, Ezkurdia I, González-Pisano D, Del Pozo MA, Montoya MC. A novel high-content analysis tool reveals Rab8-driven cytoskeletal reorganization through Rho GTPases, calpain and MT1-MMP. J Cell Sci. 2016;129(8):1734–1749. doi: 10.1242/jcs.174920. [DOI] [PubMed] [Google Scholar]
- 79.Bi J, Wang R, Zeng X. Lipid rafts regulate the lamellipodia formation of melanoma A375 cells via actin cytoskeleton-mediated recruitment of β1 and β3 integrin. Oncol Lett. 2018;16(5):6540–6546. doi: 10.3892/ol.2018.9466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Juanes MA, Bouguenina H, Eskin JA, Jaiswal R, Badache A, Goode BL. Adenomatous polyposis coli nucleates actin assembly to drive cell migration and microtubule-induced focal adhesion turnover. J Cell Biol. 2017;216(9):2859–2875. doi: 10.1083/jcb.201702007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Okada K, Bartolini F, Deaconescu AM, Moseley JB, Dogic Z, Grigorieff N, Gundersen GG, Goode BL. Adenomatous polyposis coli protein nucleates actin assembly and synergizes with the formin mDia1. J Cell Biol. 2010;189(7):1087–1096. doi: 10.1083/jcb.201001016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tirnauer JS. A new cytoskeletal connection for APC: linked to actin through IQGAP. Dev Cell. 2004;7(6):778–780. doi: 10.1016/j.devcel.2004.11.012. [DOI] [PubMed] [Google Scholar]
- 83.Eves EM, Shapiro P, Naik K, Klein UR, Trakul N, Rosner MR. Raf kinase inhibitory protein regulates aurora B kinase and the spindle checkpoint. Mol Cell. 2006;23(4):561–574. doi: 10.1016/j.molcel.2006.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Eves EM, Rosner MR. MAP kinase regulation of the mitotic spindle checkpoint. Methods Mol Biol. 2010;661:497–505. doi: 10.1007/978-1-60761-795-2_31. [DOI] [PubMed] [Google Scholar]
- 85.Rosner MR. MAP kinase meets mitosis: a role for Raf Kinase Inhibitory Protein in spindle checkpoint regulation. Cell Div. 2007;2:1. doi: 10.1186/1747-1028-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bordeleau F, Myrand Lapierre ME, Sheng Y, Marceau N. Keratin 8/18 regulation of cell stiffness-extracellular matrix interplay through modulation of Rho-mediated actin cytoskeleton dynamics. PLoS ONE. 2012 doi: 10.1371/journal.pone.0038780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Moch M, Windoffer R, Schwarz N, Pohl R, Omenzetter A, Schnakenberg U, Herb F, Chaisaowong K, Merhof D, Ramms L, Fabris G, Hoffmann B, Merkel R, Leube RE. Effects of plectin depletion on keratin network dynamics and organization. PLoS ONE. 2016 doi: 10.1371/journal.pone.0149106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tavel L, Jaquillard L, Karsisiotis AI, Saab F, Jouvensal L, Brans A, Delmas AF, Schoentgen F, Cadene M, Damblon C. Ligand binding study of human PEBP1/RKIP: interaction with nucleotides and Raf-1 peptides evidenced by NMR and mass spectrometry. PLoS ONE. 2012 doi: 10.1371/journal.pone.0036187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Park S, Rath O, Beach S, Xiang X, Kelly SM, Luo Z, Kolch W, Yeung KC. Regulation of RKIP binding to the N-region of the Raf-1 kinase. FEBS Lett. 2006;580(27):6405–6412. doi: 10.1016/j.febslet.2006.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Skinner JJ, Wang S, Lee J, Ong C, Sommese R, Sivaramakrishnan S, Koelmel W, Hirschbeck M, Schindelin H, Kisker C, Lorenz K, Sosnick TR, Rosner MR. Conserved salt-bridge competition triggered by phosphorylation regulates the protein interactome. Proc Natl Acad Sci USA. 2017;114(51):13453–13458. doi: 10.1073/pnas.1711543114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Martin J, Lavery R. Arbitrary protein-protein docking targets biologically relevant interfaces. BMC Biophys. 2012 doi: 10.1186/2046-1682-5-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Nishi H, Fong JH, Chang C, Teichmann SA, Panchenko AR. Regulation of protein-protein binding by coupling between phosphorylation and intrinsic disorder: analysis of human protein complexes. Mol Biosyst. 2013;9(7):1620–1626. doi: 10.1039/c3mb25514j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Köster DV, Mayor S. Cortical actin and the plasma membrane: inextricably intertwined. Curr Opin Cell Biol. 2016;38:81–89. doi: 10.1016/j.ceb.2016.02.021. [DOI] [PubMed] [Google Scholar]
- 94.Peränen J. Rab8 GTPase as a regulator of cell shape. Cytoskeleton (Hoboken) 2011;68(10):527–539. doi: 10.1002/cm.20529. [DOI] [PubMed] [Google Scholar]
- 95.Ling HH, Mendoza-Viveros L, Mehta N, Cheng HY. Raf kinase inhibitory protein (RKIP): functional pleiotropy in the mammalian brain. Crit Rev Oncog. 2014;19(6):505–516. doi: 10.1615/critrevoncog.2014011899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sagisaka T, Matsukawa N, Toyoda T, Uematsu N, Kanamori T, Wake H, Borlongan CV, Ojika K. Directed neural lineage differentiation of adult hippocampal progenitor cells via modulation of hippocampal cholinergic neurostimulating peptide precursor expression. Brain Res. 2010;1327:107–117. doi: 10.1016/j.brainres.2010.02.071. [DOI] [PubMed] [Google Scholar]
- 97.Wei J, Jiang H, Gao H, Wang G. Raf-1 Kinase Inhibitory Protein (RKIP) promotes retinal ganglion cell survival and axonal regeneration following optic nerve Crush. J Mol Neurosci. 2015;57(2):243–248. doi: 10.1007/s12031-015-0612-8. [DOI] [PubMed] [Google Scholar]
- 98.Hellmann J, Rommelspacher H, Mühlbauer E, Wernicke C. Raf kinase inhibitor protein enhances neuronal differentiation in human SH-SY5Y cells. Dev Neurosci. 2010;32(1):33–46. doi: 10.1159/000236595. [DOI] [PubMed] [Google Scholar]
- 99.Bildyug N. Matrix metalloproteinases: an emerging role in regulation of actin microfilament system. Biomol Concepts. 2016;7(5–6):321–329. doi: 10.1515/bmc-2016-0022. [DOI] [PubMed] [Google Scholar]
- 100.Buckley CD, Tan J, Anderson KL, Hanein D, Volkmann N, Weis WI, Nelson WJ, Dunn AR. The minimal cadherin–catenin complex binds to actin filaments under force. Science. 2014 doi: 10.1126/science.1254211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Li J, Tanhehco EJ, Russell B. Actin dynamics is rapidly regulated by the PTEN and PIP2 signaling pathways leading to myocyte hypertrophy. Am J Physiol Heart Circ Physiol. 2014;307(11):H1618–1625. doi: 10.1152/ajpheart.00393.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Papakonstanti EA, Stournaras C. Cell responses regulated by early reorganization of actin cytoskeleton. FEBS Lett. 2008;582(14):2120–2127. doi: 10.1016/j.febslet.2008.02.064. [DOI] [PubMed] [Google Scholar]
- 103.Graziano BR, Weiner OD. Self-organization of protrusions and polarity during eukaryotic chemotaxis. Curr Opin Cell Biol. 2014;30:60–67. doi: 10.1016/j.ceb.2014.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Shvartsur A, Givechian KB, Garban H, Bonavida B. Overexpression of RKIP and its cross-talk with several regulatory gene products in multiple myeloma. J Exp Clin Cancer Res. 2017 doi: 10.1186/s13046-017-0535-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Németh ZH, Deitch EA, Davidson MT, Szabó C, Vizi ES, Haskó G. Disruption of the actin cytoskeleton results in nuclear factor-kappaB activation and inflammatory mediator production in cultured human intestinal epithelial cells. J Cell Physiol. 2004;200(1):71–81. doi: 10.1002/jcp.10477. [DOI] [PubMed] [Google Scholar]
- 106.Zhang B, Shi L, Lu S, Sun X, Liu Y, Li H, Wang X, Zhao C, Zhang H, Wang Y. Autocrine IL-8 promotes F-actin polymerization and mediate mesenchymal transition via ELMO1-NF-kB-Snail signaling in glioma. Cancer Biol Ther. 2015;16(6):898–911. doi: 10.1080/15384047.2015.1028702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Favot Laure, Hall Susan M., Haworth Sheila G., Kemp Paul R. Cytoplasmic YY1 Is Associated with Increased Smooth Muscle-Specific Gene Expression. The American Journal of Pathology. 2005;167(6):1497–1509. doi: 10.1016/S0002-9440(10)61236-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kreis P, Hendricusdottir R, Kay L, Papageorgiou IE, van Diepen M, Mack T, Ryves J, Harwood A, Leslie NR, Kann O, Parsons M, Eickholt BJ. Phosphorylation of the actin binding protein Drebrin at S647 is regulated by neuronal activity and PTEN. PLoS ONE. 2013 doi: 10.1371/journal.pone.0071957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Steffen A, Stradal TE, Rottner K. Signalling pathways controlling cellular actin organization. Handb Exp Pharmacol. 2017;235:153–178. doi: 10.1007/164_2016_35. [DOI] [PubMed] [Google Scholar]