Abstract
Metamorphic transformation from larvae to adults along with the high fecundity is key to insect success. Insect metamorphosis and reproduction are governed by two critical endocrines, juvenile hormone (JH), and 20-hydroxyecdysone (20E). Recent studies have established a crucial role of microRNA (miRNA) in insect metamorphosis and oogenesis. While miRNAs target genes involved in JH and 20E-signaling pathways, these two hormones reciprocally regulate miRNA expression, forming regulatory loops of miRNA with JH and 20E-signaling cascades. Insect metamorphosis and oogenesis rely on the coordination of hormones, cognate genes, and miRNAs for precise regulation. In addition, the alternative splicing of genes in JH and 20E-signaling pathways has distinct functions in insect metamorphosis and oogenesis. We, therefore, focus in this review on recent advances in post-transcriptional regulation, with the emphasis on the regulatory role of miRNA and alternative splicing, in insect metamorphosis and oogenesis. We will highlight important new findings of miRNA interactions with hormonal signaling and alternative splicing of JH receptor heterodimer gene Taiman.
Keywords: Insect development, Reproduction, Juvenile hormone, Ecdysone, Non-coding RNA, Isoforms
Introduction
Insect metamorphosis is the fascinating biological process and highly successful strategy for environmental adaptation. Insects undergo metamorphosis for either gradual nymphal–adult transition in hemimetabolous orders or dramatic larval–pupal–adult transformation in holometabolous species. After metamorphosis, adult insects become sexually mature and capable of reproductive activity. The process of insect metamorphosis and reproduction is subject to regulation by internal factors such as endocrines and environmental changes such as anthropogenic stressors [1–3]. For a long time, research in insect metamorphosis and reproduction has focused on the endocrine regulation, with the emphasis on actions of two primary lipophilic hormones, the steroid hormone 20-hydroxyecdysone (20E, an active form of ecdysone), and the sesquiterpenoid juvenile hormone (JH). In juvenile stage, 20E initiates larval–pupal or nymphal–adult metamorphosis, while JH exerts its status quo function to keep the insect in its immature state and thus to ensure proper timing of metamorphosis by repressing the metamorphic action of 20E [4–8]. In adulthood, both 20E and JH can stimulate various aspects of female reproduction such as oocyte formation, previtellogenic development, vitellogenesis, and choriogenesis, depending on the great variances in reproductive strategies among different species [9–12]. As illustrated in Fig. 1, 20E acts through a heterodimeric receptor comprised of Ecdysone Receptor (EcR) and Ultraspiracle (USP) [13], leading to transcriptional activation of 20E early inducible genes including ecdysone-induced proteins 74 (E74), 75 (E75), 93F (E93), Broad-Complex (Br-C) and Ftz-f1 [14, 15]. The regulatory cascade initiated by 20E is evolutionarily conserved during metamorphosis and oogenesis across insect orders [9, 16–19].
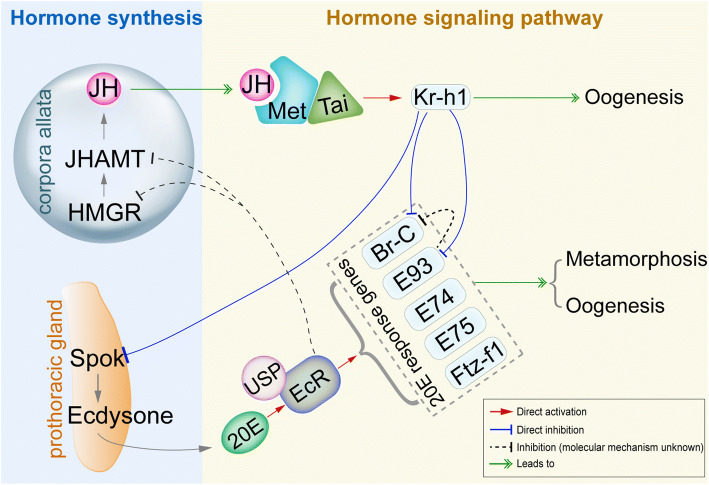
Fig. 1.
Schematic illustration of hormonal regulation in insect metamorphosis and oogenesis. 20E acts through EcR/USP to activate the transcription of 20E-early responsive genes E74, E75, Br-C, E93, and Ftz-f1 and initiate larval–pupal or nymphal–adult metamorphosis [14, 15, 19, 175, 176]. JH-Met/Tai receptor complex activates the transcription of primary JH-early responsive gene Kr-h1 [20, 21, 23, 28]. Kr-h1 prevents immature larvae from precocious larval–pupal metamorphosis by inhibiting Br-C expression in holometabolous insects [31, 32]. Kr-h1 also inhibits precocious adult metamorphosis by repressing E93 expression in both hemimetabolous and holometabolous insects [32–35]. In addition, Kr-h1 inhibits Spok expression in the prothoracic gland to inhibit ecdysone synthesis and prevent precocious metamorphosis [36, 37], while EcR suppresses JH biosynthesis in by inhibiting the expression of HMGR and JHAMT to ensure the onset of metamorphosis. In adults, JH exerts its vitellogenic role through Kr-h1 to promote insect vitellogenesis and oogenesis [10, 38, 39, 43]. 20E signaling cascade is evolutionarily conserved during metamorphosis and oogenesis across insect orders [9, 10, 16–18]
With respect to JH, it induces the heterodimerization of Methoprene-tolerant (Met) with Taiman (Tai) to form an active JH-receptor complex [20–22], consequently activating the transcription of JH-responsive genes [5, 23–27]. Krüppel homolog 1 (Kr-h1) is the primary JH-responsive gene that mediates anti-metamorphic action of JH [5, 28–30] (Fig. 1). Kr-h1 prevents immature larvae from precocious larval–pupal transformation by suppressing the expression of Br-C, known as the pupa-specifier gene in holometabolous insects [4, 31, 32]. Kr-h1 also inhibits precocious adult metamorphosis by repressing the transcription of E93, which is defined as an adult-specifier gene in both hemimetabolous and holometabolous insects [32–35]. Furthermore, Kr-h1 is shown to inhibit the synthesis of ecdysteroids by repressing the transcription of steroidogenic enzyme gene Spok in prothoracic glands to prevent precocious metamorphosis [36, 37], whereas EcR suppresses JH biosynthesis by inhibiting the expression of genes coding for JH acid methyltransferase (Jhamt) and HMG Coenzyme-A reductase (HMGR) in corpora allata to ensure the onset of metamorphosis [37]. The regulatory role of Kr-h1 in JH-mediated female reproduction has been reported in several insect species [10, 38–42] (Fig. 1). In the migratory locust Locusta migratoria, JH achieves its vitellogenic effect through Kr-h1 to promote vitellogenesis, ovarian development, and oocyte maturation [43, 44]. In the yellow fever mosquito Aedes aegypti, Kr-h1 transduces JH signaling as both activator and repressor to regulate the expression of JH-responsive genes involved in previtellogenic development as well as egg production after a blood meal [39, 45].
Besides the transcriptional regulation orchestrated by JH and 20E, microRNA (miRNA) has emerged as a critical regulator in insect metamorphosis and oogenesis [10, 46, 47]. In addition, RNA alternative splicing has been reported to control insect metamorphosis and oogenesis [44, 48, 49]. Furthermore, accumulating studies have been conducted to identify long non-coding RNA (lncRNA) [50–52] and circular RNA (circRNA) [53–55] potentially involved in insect development. However, to the best of our knowledge, the regulatory mechanisms of lncRNA in insect metamorphosis and oogenesis have been solely reported in Drosophila melanogaster, and lncRNA targets in JH or 20E signaling cascades have not been characterized [56]. Moreover, circRNA–target interactions in insect metamorphosis and oogenesis have not been experimentally determined. We, therefore, focus in this review on recent research progress in the regulatory role of miRNA and alternative splicing.
miRNA regulation of insect metamorphosis
miRNA, a class of approximately 22-nucleotide-long endogenous non-coding RNA, typically binds complementarily to the 3′-untranslated region (3′-UTR) or the coding sequence (CDS) of target mRNAs [57–61]. Insect miRNAs often act as post-transcriptional repressors through translation inhibition or mRNA degradation in a spatiotemporal way. However, literatures have reported that miRNAs can bind the 5′-UTR or CDS of mRNAs and upregulate the expression of target genes [62–65]. Pioneer analysis of phenotypes caused by loss-of-function mutants or RNAi-mediated knockdown of genes coding for Dicer1 and Argonaute1 (Ago1), two core enzymes for miRNA biogenesis and functioning, has revealed the essential role of miRNA in insect metamorphosis and reproduction [66, 67]. In the model organism D. melanogaster, null mutation of Dicer1 or Ago1 leads to marked defects in pupal formation and oocyte development [66, 68]. As well, in holometabolous beetle Tribolium castaneum and cotton bollworm Helicoverpa armigera, depletion of Dicer1 results in defective larval–pupal transition [69, 70]. Ago1 knockdown also causes impaired pupation in T. castaneum [69]. In hemimetabolous species like the cockroach Blattella germanica, silencing of Dicer1 in penultimate instar nymphs results in overall decline of miRNA expression along with abnormal metamorphosis [71]. Depletion of Dicer1 also causes retarded oogenesis in B. germanica [67]. Silencing of either Dicer1 or Ago1 in the migratory locust Locusta migratoria interferes with nymphal–adult transition in nymphs and vitellogenesis in adults [72, 73]. It has been recently reported that Regnase-1 RNase is essential for remodeling miRNA profiles during Drosophila larva-to-pupa transition [74]. Regnase-1 knockout flies failed to complete pupal-to-adult shift and died in puparium case [74]. Initially, miRNA identification and function study in insects have been mainly performed in D. melanogaster [75, 76]. In a recent review, Belles summarized miRNAs and the evolution of insect metamorphosis by comparing hemimetabolous and holometabolous insect species [47]. In the past few years, elucidation of miRNA function in mosquitoes and agriculturally important insects has attracted more attentions. This review of miRNAs is thereby to highlight recent advances in understanding the precise regulation of insect metamorphosis and oogenesis coordinated by miRNAs and hormones by focusing on physiological aspects.
Evolutionarily conserved miRNAs in insect metamorphosis
Evolutionarily conserved miRNAs are well known to tune the expression of analogous target genes in insect molting and metamorphosis. One example of such miRNAs is let-7, which is generally clustered with miR-100 and miR-125 in a primary miRNA transcript and polycistronically transcribed [77]. let-7 cluster (or let-7-Complex, let-7-C) was originally identified in the worm Caenorhabditis elegans as part of a pathway of heterochronic genes promoting stage-specific cell fate decisions [57]. As summarized in Fig. 2a, let-7 is essential to appropriate remodeling of abdominal neuromusculature during Drosophila metamorphosis [78, 79]. Both let-7 and miR-125 target Abrupt gene consequently regulating wing morphogenesis and proper timing of neuromuscular junction maturation during Drosophila larval-to-adult development [80]. let-7 and miR-125 also repress the expression of chronologically inappropriate morphogenesis (chinmo) gene to regulate sequential generation of mushroom body neurons, cell death, and eviction during Drosophila pupal morphogenesis [81, 82]. In addition, let-7 targets Abrupt gene, controlling neuron cell fate of mushroom body at prepupal–pupal stage [83]. In the silkworm Bombyx mori, knockdown of let-7 using a miRNA sponge with the binary GAL4/UAS system caused development arrest at larval–pupal transformation by targeting Ftz-f1 and E74 [84] (Fig. 2a). In the oriental fruit fly Bactrocera dorsalis, let-7 modulates metamorphosis by repressing E75 [85]. In hemimetabolous cockroach B. germanica, let-7/miR-100/miR-125 are expressed at high levels in the final nymphal instar, similar to the expression pattern of Br-C [86]. Depletion of let-7 or miR-100, but not miR-125 by specific antagomiR treatment brought about reduced wing size and malformed vein patterns [87] (Fig. 2a). In another hemimetabolous species, L. migratoria, let-7 together with miR-278 suppresses Kr-h1 expression, and application of let-7 or miR-278 agomiR in the penultimate instar nymph resulted in partially precocious metamorphosis [88] (Fig. 2a).
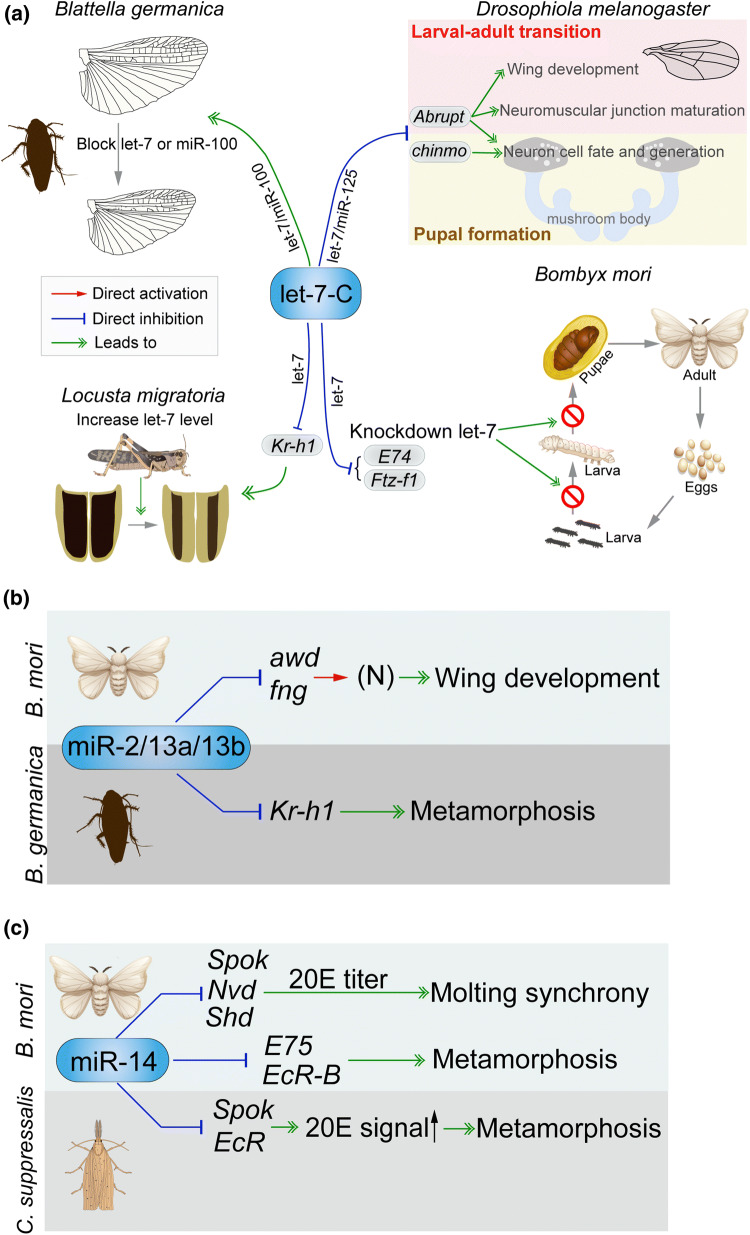
Fig. 2.
Regulatory role of selective miRNAs in insect metamorphosis. a let-7-Complex. In hemimetabolous Blattella germanica, let-7 and miR-100 antagomiR treatment resulted in defective wing formation during metamorphosis [86, 87]. In another hemimetabolous species Locusta migratoria, let-7 suppresses Kr-h1 expression, and let-7 agomiR treatment on penultimate instar nymphs caused partially precocious metamorphosis [88]. In holometabolous Drosophila melanogaster, let-7 and miR-125 target Abrupt and chinmo genes to regulate the morphogenesis of wing and nervous system [78–83]. In holometabolous Bombyx mori, let-7 downregulates E74 and Ftz-f1 genes to control larval–pupal transformation. b miR-2 family. In B. mori, miR-2/13a/13b modulate wing morphogenesis via suppressing the expression of Awd and Fng genes [90]. In B. germanica, miR-2/miR-13a/miR-13b eliminate residual transcripts of Kr-h1 at the final instar nymph to ensure the entry of metamorphosis [91]. c Canonical invertebrate miR-14 regulates molting synchrony and pre-pupation of B. mori by downregulating Spo, Nvd, Shd, EcR and E75 expression [95, 96]. In Chilo suppressalis, miR-14 targets Spo and EcR to control metamorphic development [97]
The miR-2 cluster comprising miR-2 family members (miR-2, miR-13a and miR-13b) and miR-71 is invertebrate specific, but conserved across insect orders [89]. In B. mori, miR-2, miR-13a, and miR-13b modulate wing morphogenesis via suppressing genes coding for abnormal wing disc (Awd) and fringe (Fng), two crucial factors in determining wing vein cells and margin patterning [90] (Fig. 2b). Overexpression of miR-2 cluster using GAL4/UAS system gave rise to deformed adult wings during metamorphosis [90]. In hemimetabolous B. germanica, the miR-2 family members are upregulated in the final instar nymph and coordinately suppress Kr-h1 expression by binding the 3′-UTR of Kr-h1 mRNA [91]. Importantly, along with the reduction of Kr-h1 transcription, miR-2/miR-13a/miR-13b eliminates residual transcripts of Kr-h1 at the final instar nymph, thus crucially contributing to the onset of metamorphosis [91] (Fig. 2b).
Another example of conserved miRNAs is miR-8. This miRNA is expressed at low levels in the prepupal/pupal stage of Drosophila and downregulates U-shaped (Ush), a PI3 kinase inhibitor in insulin pathway [92, 93]. Overexpression of miR-8 in UAS-miR-8 flies made pupae bigger [93], whereas loss of miR-8 function in miR-8-null alleles reduced metamorphic transition under heat stress [94]. For the canonical invertebrate miR-14, its expression reaches a peak at the prepupal stage of B. mori, coinciding with the increased titer of 20E that initiates metamorphosis. Overexpression of miR-14 using GAL4/UAS system caused prolonged larval development, while disruption of miR-14 using the CRISPR/Cas9 system resulted in precocious pre-pupation [95]. Another interesting aspect is that miR-14 regulates molting synchrony of B. mori by targeting genes in 20E synthesis pathway, including Spook (Spo), Neverland (Nvd) and Shade (Shd) [96] (Fig. 2c). In the rice stem borer Chilo suppressalis, miR-14 represses the expression of Spo and EcR as revealed by dual luciferase assays, and agomiR-14 treatment at the final larval instar caused apparent defects in metamorphic development [97] (Fig. 2c). During pupariation, the old chitin, a primary component of epidermis, needs to be degraded and replaced by newly synthesized chitin. In the brown planthopper Nilaparvata lugens, miR-8-5p and miR-2a-3p repress chitin biosynthesis pathway genes tmembranebound trehalase (Tre-2) and phosphoacetylglucosamine mutase (PAGM) to tune nymphal ecdysis and nymphal–adult shift [98]. In L. migratoria, two conserved miRNAs, miR-71 and miR-263, jointly regulate their target genes Chitin synthase 1 (Chs1) and Chitinase 10 (CHT10), respectively, to control chitin metabolism and molting process [99].
Lineage- and species-specific miRNAs in insect metamorphosis
During these years, the fast-growing progress has been made on action of lineage- and species-specific miRNAs in diverse insect species. Lineage-specific miR-2942 in mosquitos and miR-2768 in lepidopteran insects are found to modulate larval–pupal transition [100, 101]. miR-173 is identified as a novel miRNA targeting Ftz-f1 in hemimetabolous N. lugens. Administration of agomiR-173 caused nymph mortality, wing defects, and failure of nymphal–adult transition [102]. Several studies have reported the participation of non-conserved miRNAs in the molting and metamorphosis process by regulating chitin metabolism. In N. lugens, non-conserved miR-2703 inhibits the expression of Chs1a for metamorphic switch [103]. Another novel insect miRNA, miR-4924 regulates larval molting by repressing Chitinase 1 in the beet armyworm Spodoptera exigua [104]. Intriguingly, in the endoparasitic wasp Cotesia vestalis, both polydnavirus (PDV)-derived novel miR-22 and teratocyte-derived miR-281 downregulate EcR expression and influences metamorphosis of its host, the diamondback moth Plutella xylostella [105]. This finding suggests that miRNAs from both teratocytes and PDVs of parasite wasp share EcR as their common target in the host and control host insect metamorphosis. Thus, evolutionarily conserved and newly evolved miRNAs both target genes for insect molting and metamorphosis, indicating that these two categories of miRNAs use this pathway to achieve common and lineage/species-specific functions.
Interaction of miRNA and hormonal pathways
Interplay of miRNA with 20E signaling cascade
There is accumulating evidence of a link between hormone and miRNA in insect metamorphosis. Several miRNAs elicit their metamorphic or anti-metamorphic functions via targeting 20E pathway genes, including EcR, E74, E75, and Ftz-f1. Nevertheless, an increasing body of evidence supports the importance of 20E-responsive miRNAs and their critical roles in insect metamorphosis. In Drosophila, 20E modulates the upregulation of let-7 cluster and the downregulation of miR-34, which is indispensable for remodeling abdominal neuromusculature during larval–pupal–adult transition [78–80]. The expression of let-7 or let-7 cluster is activated by 20E, through EcR/USP binding to the upstream ecdysone response elements (EcREs) of let-7-C locus [82, 83, 106]. Moreover, let-7 represses the expression of Ftz-f1 and E74, which in turn influences larval ecdysis and pupal transition in B. mori [84]. In another dipteran species, Bactrocera dorsalis, let-7 is upregulated by 20E. Injection of let-7 antagomiR in the final instar larvae reduced E75 expression levels, accompanied by abnormal pupation and eclosion [85]. The canonical invertebrate miR-14 appears to be a general regulator in maintaining ecdysone homeostasis for normal development and metamorphosis. In Drosophila, miR-14 represses EcR expression for developmental timing, and reciprocally, 20E via EcR downregulates the expression of miR-14, thereby forming a positive autoregulatory loop to amplify the response [107] (Fig. 3a). This positive autoregulatory loop acts like a switch to boost its own activity through inhibiting the expression of miRNA and increasing the levels of its receptor, which is likely sensitive to subtle change of 20E titers. Interestingly, miR-14 suppresses EcR-B and E75 expression to modulate metamorphosis in B. mori, and its expression is enhanced by 20E but reduced by JH [95]. 20E is also demonstrated to alleviate miR-965-mediated repression of EcR, String and Wingless in D. melanogaster. At the onset of metamorphosis, 20E reduces the production of miR-965, which in turn enhances the levels of EcR to trigger histoblast proliferation during pupal development (Fig. 3a). Thus, similar to miR-14, miR-965 also buffers a positive regulatory loop in a mutual repression circuit [108]. A mutual repression between miR-281 and 20E signaling was observed in B. mori. miR-281 suppresses the expression of EcR-B but not EcR-A in Malpighian tubules, and its expression is further repressed by 20E, constituting a regulatory loop in the regulation of B. mori larval–pupal shift [109] (Fig. 3a).
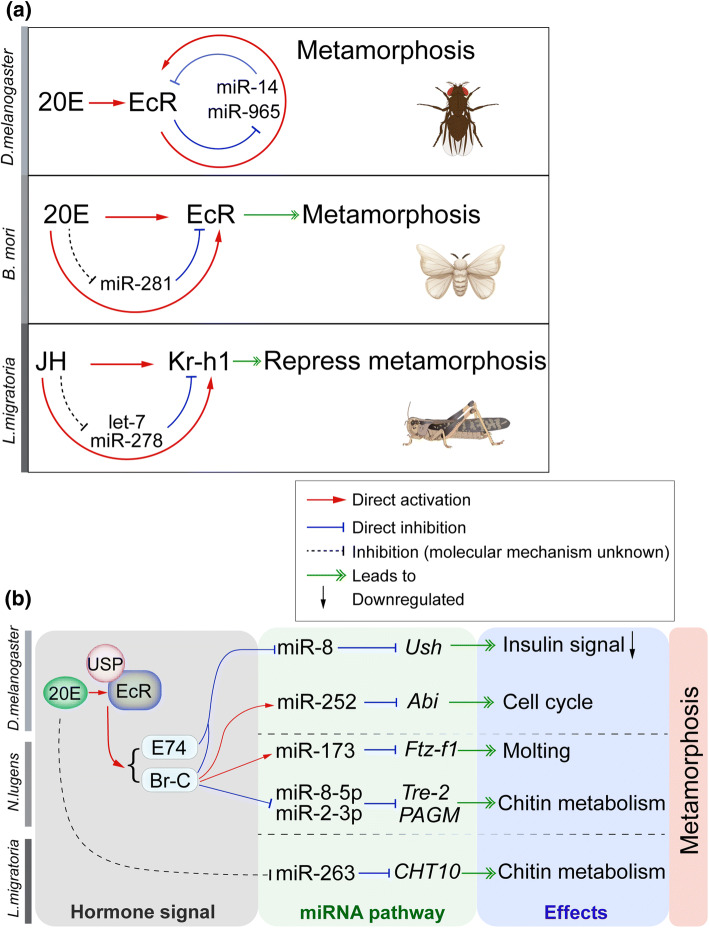
Fig. 3.
Interaction of miRNA and hormone pathways in insect metamorphosis. a Regulatory loops of miRNA and hormonal signaling cascades. In Drosophila, miR-14 downregulates EcR, and reciprocally 20E via EcR represses miR-14 expression, which forms a positive autoregulatory loop to amplify 20E signaling [107]. 20E, EcR and miR-965 also constitute a positive regulatory loop in a mutual repression circuit [108]. In addition, mutual repression between miR-281 and 20E-EcR was observed in Bombyx mori [109]. In Locusta migratoria, let-7 and miR-278 inhibit Kr-h1 expression, whereas let-7 and miR-278 are downregulated by JH, which work together to maintain an appropriate level of Kr-h1 in nymphs essential for preventing precocious metamorphosis [88]. b Experimentally confirmed miRNAs that are involved in 20E signaling pathway and regulate insect metamorphosis, represented by holometabolous D. melanogaster as well as hemimetabolous Nilaparvata lugens and L. migratoria
As illustrated in Fig. 3b, miR-8 is transcriptionally repressed by 20E-signaling cascade, including EcR, Br-C, and E74, which could determine the pupa size during Drosophila metamorphosis [93]. Moreover, such control of 20E-induced body size can be eliminated by overexpression or deletion of miR-8 [93]. In D. melanogaster, the expression of miR-252 is activated by 20E and Br-C, which in turn represses the expression of Abelson interacting protein (Abi). The subsequent reduction in Abi protein levels leads to a concomitant decrease of cyclins A and B, which is required for the successful completion of M-phase in the cell cycle during larva-to-adult metamorphosis [110]. In hemimetabolous N. lugens, miR-173, promoted by Br-C in response to 20E, regulates nymphal molting by targeting Ftz-f1 [102]. 20E via Br-C also negatively regulates miR-8-5p and miR-2a-3p to control the expression levels of Tre-2 and PAGM for chitin metabolism and nymphal ecdysis in N. lugens [98]. In L. migratoria, 20E treatment inhibits miR-263 expression and in turn stimulates the expression of its target gene Chitinase 10 to promote molting process [99].
Crosstalk of miRNA with JH signaling pathway
Although miRNA regulation in relation to 20E has been extensively studied in insects, few studies have explored the regulatory role of miRNA orchestrated with JH. Currently, only limited examples of miRNAs which interact with JH-signaling pathway have been reported. As previously introduced, the miR-2 family acts as the suppressor of Kr-h1, the master repressor of insect metamorphosis. In the final instar nymph of B. germanica, the declined titer of JH and elevated expression of miR-2 work together to remove Kr-h1 transcripts, ensuring the entry of metamorphosis [91]. In addition, in B. germanica, JH represses the expression of let-7, which contributes to wing development during metamorphosis [86, 87]. Our study has demonstrated that let-7 and miR-278 bind to the coding sequence of Kr-h1 and inhibit Kr-h1 expression in L. migratoria [88]. Intriguingly, the expression of these two miRNAs is repressed by JH. These observations together provide the evidence that while JH acts through Met/Tai to induce Kr-h1 transcription, JH also inhibits the expression of let-7 and miR-278 to maintain appropriate levels of Kr-h1 essential for preventing precocious metamorphosis, revealing a mechanism of precise regulation of miRNA and JH for insect metamorphosis. In B. germanica, 20E treatment on final instar nymphs reduces the expression levels of miR-252, but increases the expression of miR-1 and miR-100, whereas 20E plus JH treatment reduces the expression of miR-252, miR-100, miR-276, miR-190, miR-14, let-7, miR-125, and bantam [86]. In Drosophila S2 cells, the expression of let-7 and miR-125 is robustly induced by 20E, and such induction is repressed by subsequent application of JH [111]. By dual luciferase reporter assays, Qu et al. showed that let-7, miR-8, miR-14, miR-34, miR-278, and miR-304 bound to the 3′-UTR of Germ-cell expressed (Gce, the paralogue gene of Met) and downregulated its expression in D. melanogaster [112]. Interestingly, the expression of Met in the malaria mosquito Anopheles gambiae was shown to be repressed by miR-8, miR-14, miR-34, and miR-278, while only miR-29b was found to suppress Met expression in T. castaneum [112]. However, the functions of these miRNAs in metamorphosis are yet to be elucidated.
miRNA regulation of hormone synthesis
The role of JH and 20E in insect metamorphosis has been well studied, and several miRNAs have been found to be involved in regulation of some JH- or 20E-responsive genes. However, the interplay of miRNAs with JH and 20E synthesis pathways remains largely obscure. Recent studies have shown that miRNA can modulate the production of JH and 20E, which consequently regulates insect development and metamorphosis. MicroRNA bantam governs body size by connecting insulin signaling and ecdysone production in D. melanogaster [113]. Overexpression of bantam in ecdysone-producing cells induced pupal growth by inhibiting ecdysone production. On the contrary, bantam mutant flies displayed higher levels of ecdysone and reached metamorphosis with a delay [113]. In the screening of differentially expressed miRNAs during metamorphosis of C. suppressalis, He et al. demonstrated that two conserved miRNAs (bantam, and miR-9b), four lineage-specific miRNAs (miR-80, miR-89, miR-154, and miR-257), and one species-specific miRNA (miR-260) presumably modulated ecdysteroid biosynthesis by targeting three Halloween genes [114]. While miR-9b and miR-260 repressed the expression of Nvd and Disembodied (Dib), respectively, other five miRNAs (bantam, miR-154, miR-80, miR-89, and miR-25) jointly downregulated Spo expression [114]. In these agomiR-treated groups, 20E titers significantly declined and defective phenotypes of development, molting, and metamorphosis occurred [114]. The abundance of miR-14 elevates immediately after each ecdysis, efficiently suppressing 20E biosynthesis. Further experiments demonstrated that miR-14 repressed Spo expression to modulate 20E production and metamorphosis in C. suppressalis (Fig. 2c). Interestingly, in addition to Spo, miR-14 targets Nvd and Shd to regulate 20E synthesis and molting process in B. mori [96] (Fig. 2c). Thus, miRNA-14 acts as an efficient suppressor to switch off 20E production after ecdysis in these two lepidopteran insects.
Expression analyses of miRNAs in insects have indicated possible roles of some miRNAs in JH-synthesis pathway for metamorphosis. By dual luciferase reporter assays, Qu et al. identified a set of miRNAs that interact with Jhamt mRNAs in D. melanogaster, An. gambiae, and T. castaneum [112, 115]. In D. melanogaster, bantam, miR-252, and miR-304 were shown to downregulate Jhamt. Overexpression of bantam caused decreased levels of Jhamt transcript as well as reduced titers of JHB3 and JH III, accompanied by phenotypes of pupal lethality and malformed genital organs [112, 115]. In the mosquito An. gambiae, Jhamt is downregulated by miR-278. However, in T. castaneum, bantam, miR-252, and miR-304 target Jhamt [112]. These studies together imply an antagonistic role of miRNA in repressing JH biosynthesis, and the variation of miRNA-target interactions. A recent study described a comparative analysis of corpora allata and corpora cardiac-specific miRNAs with significant changes during metamorphosis of Ae. aegypti [116]. Bantam, miR-9a, miR-31, and miR-34 were predicted to downregulate genes coding for JH biosynthesis enzymes diphosphomevalonate decarboxylase (PP-MevD), arnesyl-pyrophosphate synthase (FPPS), HMGR, and aldehyde dehydrogenase (ALDH), respectively [116]. However, the regulatory mechanisms and function of these miRNAs in JH production and mosquito metamorphosis remain to be characterized.
Regulation of miRNA in insect oogenesis
Insects have two major types of ovarioles, panoistic ovarioles, and meroistic ovarioles [12, 117]. The panoistic ovaries have no nurse cells, whereas meroistic ovaries possess either nurse cells in the tropharioum (polytrophic ovarioles) or nutritive cords in a distance (teleotrophic ovarioles). Polytrophic ovarioles are present in advanced holometabolous insects such as Diptera (flies and mosquitos), and teleotrophic ovarioles are often seen in basal holometabolous orders such as Coleoptera (beetles) and Lepidoptera (moths and butterflies) as well as some hemimetabolous orders such as Hemiptera (bugs). Compared to that in insect metamorphosis, miRNA action in insect oogenesis has received less attention. Likewise, the function of miRNAs in insect oogenesis is described more in details on D. melanogaster [46, 75, 118]. Notably, miRNAs linked to insect oogenesis have been recently revealed in other important insects, represented by the panoistic L. migratoria and the meroistic Ae. aegypti.
miRNAs’ regulation in oogenesis of insects with panoistic ovarioles
L. migratoria, a representative of evolutionarily primitive insects with hemimetaboly and panoistic ovarioles, is the first insect that experimentally links miRNA regulation with JH signaling in oogenesis. Song et al. demonstrated that Dicer1 and Ago1 were expressed in response to JH. Depletion of Ago1 caused substantial reduction of Vg transcripts as well as severely impaired oocyte maturation and arrested ovarian growth [72]. Similar but mild phenotypes were also seen with Dicer1 knockdown [72]. These observations imply a crucial role of Dicer1/Ago1-dependent miRNAs in locust oogenesis. In the fat bodies of JH-deprived adult female locusts further treated with JH, the expression of two conserved miRNAs (miR-7, miR-8) and four specific miRNAs (miRNA-35, miR-57, miR-12, and miR-20) was significantly increased, while the expression levels of miR-2, miR-184, miR-13a as well as four specific miRNAs (miRNA-1, miRNA-6, miRNA-33, and miRNA-58) were significantly decreased [72]. In addition to transducing anti-metamorphic action of JH, Kr-h1 also plays an indispensable role in JH-stimulated insect reproduction [10, 43]. As reviewed in the previous section, let-7 and miR-278 act as the repressors of Kr-h1. Injection of let-7 and miR-278 agomiRs resulted in markedly reduced Vg protein levels, blocked oocyte maturation and impaired ovarian growth in L. migratoria [88] (Fig. 4a). Interestingly, let-7 and miR-278 were downregulated by JH and expressed at high levels after adult ecdysis. The increased JH titer and declined abundance of let-7 and miR-278 in both previtellogenic and vitellogenic phases thereby ensured high levels of Kr-h1 expression, consequently promoting oogenesis [88]. Notch is a crucial player in insect oogenesis [119–121]. Song et al. found that miR-2/13/71 cluster bound to the protein coding sequence of Notch mRNA and repressed its expression [122]. Moreover, miR-2/13/71 was downregulated by JH, and miR-2/13/71 expression significantly dropped after adult eclosion. When miR-2/13/71 agomiRs were applied, adult female locusts had significantly reduced Vg transcripts, inhibited oocyte maturation and blocked ovarian growth. Thus, the increase of JH and decline of miR-2/13/71 expression in vitellogenic adult female locusts ensure a high level of Notch, consequently contributing to successful oogenesis and egg production [122] (Fig. 4a). Collectively, the above two studies on L. migratoria have identified a regulatory mechanism by which JH stimulates insect oogenesis by repressing the expression of miRNAs that target genes promoting female reproduction. Another interesting finding is about miR-276, which promotes egg development rate and progeny hatching synchrony by upregulating a transcription coactivator gene, brahma in locusts [64] (Fig. 4a). miR-276 is expressed significantly higher in adult females of gregarious phase than solitarious phase. Injection of antagomiR-276 in gregarious adult females and treatment of agomiR-276 in solitarious females caused more heterochronic and synchronous hatching of progeny eggs, respectively [64]. This might partially explain why the egg-hatching time of gregarious locusts is more uniform compared with solitarious locusts. Dicer1 knockdown and expression analyses of miRNAs have indicated possible roles of miRNA in oogenesis of dictyopteran B. germanica. RNAi-mediated knockdown of Dicer1 in adult cockroaches inhibited epithelium development and ovarian growth. Moreover, depletion of Dicer1 caused significantly reduced expression of let-7, miR-100, miR-125, bantam, miR-184, miR-1, and miR-275, and let-7, some of which are predicted to play potential roles in cockroach oogenesis [67].
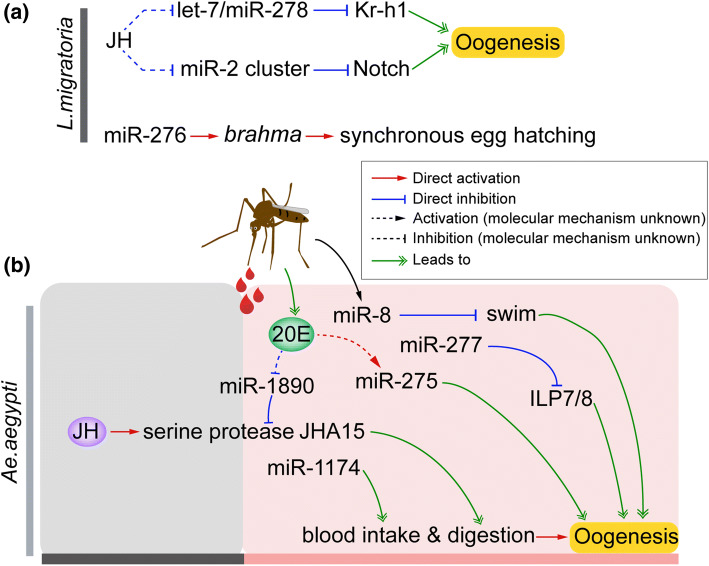
Fig. 4.
Regulatory role of selective miRNAs in insect oogenesis. a let-7, miR-278, and miR-2/13a/13b/71 play pivotal role in JH-dependent oogenesis of panoistic Locusta migratoria. JH inhibits the expression of let-7 and miR-278 that repress Kr-h1 expression. The elevated JH titer and declined abundance of let-7 and miR-278 in adult female locusts ensure high levels of Kr-h1 required for successful oogenesis and egg production [88]. Similar regulatory loop was also observed with JH, miR-2/13a/13b/71 and Notch [122]. Moreover, miR-276 promotes locust egg development and hatching synchrony by upregulating brahma gene [64]. b miR-8, miR-275, miR-1174, miR-1890, and miR-277 are dispensable for 20E-dependent blood intake, blood digestion, lipid metabolism and vitellogenin secretion in vitellogenic adult females of meositic Aedes aegypti. Dysfunction of these miRNAs caused severely defective oogenesis and egg production [130–134]
miRNAs’ regulation in oogenesis of insects with meroistic ovarioles
In D. melanogaster, null mutation of Dicer1 or Ago1 in Drosophila leads to obvious defects in oogenesis such as impaired oocyte maturation, abnormal proliferation of germline stem cells [66, 68, 123, 124], suggesting an important role of miRNAs in fruit fly oogenesis. Loss-of-function of let-7 or miR-124 in Drosophila influences development and remodeling of neuromuscular, causing impaired female fertility and attenuated egg production [78, 125]. The Drosophila Bithorax complex (BX-C) Hox cluster harbors two bidirectionally transcribed miRNAs, miR-iab-4, and miR-iab-48, which target Hox gene to control fruit fly fecundity [126]. miR-125 acts as a spatiotemporal coordinator between Notch and 20E signaling. 20E induces the expression of miR-125, which targets Tom, a negative regulator of Notch signaling to modulate Drosophila ovarian germline stem cell (GSC) niche formation [127]. miR-7 represses gene-encoding transcription factor, Tramtrack69 (Ttk69) to control developmental switch of follicle cells from endocycles to gene amplification during Drosophila oogenesis [128]. An ovary-enriched miRNA, miR-318 is upregulated by 20E and cooperates with Ttk69 to control differentiation of follicular epithelium during Drosophila oogenesis. Loss-of-function mutants of miR-318 show impaired choriogenins and female sterilization [129].
In polytrophic Ae. aegypti, recent studies have documented that miR-275, miR-1174, miR-1890, miR-277, and miR-8 play pivotal roles in 20E-dependent blood intake, blood digestion, lipid metabolism and Vg secretion in vitellogenic adult female mosquitoes, and dysfunction of these miRNAs causes severely defective oogenesis and egg production [130–134] (Fig. 4a). Disruption of 20E-regulated miRNA, miR-275 by its antagomiR treatment brought about severely impaired blood digestion and egg development [130] (Fig. 4b). Moreover, depletion of blood meal-triggered miR-8 resulted in reduced lipid accumulation, restrained follicle size and reduced egg number, by modulating Secreted wingless-interacting molecule (Swim) gene [134] (Fig. 4b). Prior to the blood meal, JH activates the expression of chymotrypsin-like serine protease (JHA15) in the midgut, which is essential for blood digestion after blood meal [135]. Lineage-specific miR-1890 is a negative regulator of JHA15 and downregulated by 20E. Increased titers of 20E after blood meal inhibits the expression of miR-1890 to maintain a high level of JHA15 required for blood digestion and consequent vitellogenesis and oocyte maturation [133] (Fig. 4b). Mosquito-specific miR-1174 is a midgut specifically expressed miRNA, which tunes Serine hydroxymethyltransferase (Shmt) gene for sugar absorption, blood intake and egg maturation [132] (Fig. 4b). miR-277 modulates insulin signal via targeting two insulin-like peptides, ILP7 and ILP8, which is essential for lipid metabolism and ovarian maturation of Ae. aegypti [131] (Fig. 4b). The miR-309/286/2944 cluster is upregulated after blood meals in mosquitos [136]. miR-309 represses the expression of gene coding for SIX homeobox 4 protein (SIX4), which is associated with maintenance of female GSCs. Genetic disruption of miR-309 by CRISPR/Cas9 system resulted in failure of primary follicle formation, suggesting that miR-309-targeted degradation of SIX4 mRNA in ovaries is required for successful switch from previtellogenic to postvitellogenic phases of ovarian development [137]. In the malaria mosquito Anopheles gambiae, injection of miR-309 antagomiR resulted in blocked oocyte development as well as declined egg number [136]. In the Asian tiger mosquito Ae. albopictus, disruption of miR-1891 and miR-286 reduces egg number and hatching rate, respectively [100]. In a comprehensive analysis to link miRNA identities with target genes during the gonadotrophic cycle of Ae. aegypti, miR-989 was identified as the most abundant miRNA in the ovary and shown to putatively target Vg, indicating the potential role of miR-989 in mosquito oogenesis [138].
The lepidoptera H. armigera bears polytrophic ovarioles. Supply of miR-2002b mimics led to reduced fecundity [139]. In N. Lugens, which has teleotrophic ovarioles, depletion of Dcr1 resulted in malformed follicle cells and undeveloped oocytes [140]. miR-4868b downregulates Glutamine synthase, and treatment of miR-4868b antagomiR led to reduced Vg accumulation, disrupted ovarian development, and declined fecundity [141]. Clearly, individual miRNAs in a variety of insect species act differentially to control different aspects of oogenesis.
Alternative splicing linked to insect metamorphosis and oogenesis
Alternative splicing is a process by which different combinations of exons produce multiple forms of mRNA from a single pre-mRNA [142, 143]. The well-known alternative splicing event related to insect metamorphosis and oogenesis is the generation of EcR, USP, E75, E74, and Br–C isoforms. While the isoforms of key players in 20E pathway have been well studied in the past two more decades, the isoforms of key players in JH pathway have been recently reported on Tai only. Here, we outline the alternative splicing of Tai. EcR and USP isoforms are highlighted in parallel. In D. melanogaster, EcR gene encodes splice variants EcR-A, EcR-B1, and EcR-B2 that share a common carboxy-terminal region including ligand-binding and DNA-binding sequences, but have varied amino-termini that influence receptor activation and repression properties [144]. EcR isoforms, especially EcR-A and EcR-B1, have been identified in a number of other insect species with distinct expression and function in a spatiotemporal-specific manner [145–150]. During larval stages, EcR-A is predominantly expressed in imaginal discs that develop into pupal and adult structures. EcR-B1 appears to be expressed in the tissues programmed for apoptosis, while EcR-B2 is mainly detected in Malpighian tubules. These isoforms mediate 20E signaling and coordinates insect metamorphosis [16, 146, 147, 151–153] and oogenesis [9, 16, 154]. Two USP isoforms, USP-1 and USP-2 (or USP-A and USP-B), have been found in diverse insect species [145, 146, 155–159]. Both EcR-A/USP-2 and EcR-B1/USP-1 heterodimers are present, but the effective combination of these isoforms upon 20E induction tends to vary in different tissues during molting, metamorphosis, and oogenesis [146, 158, 159].
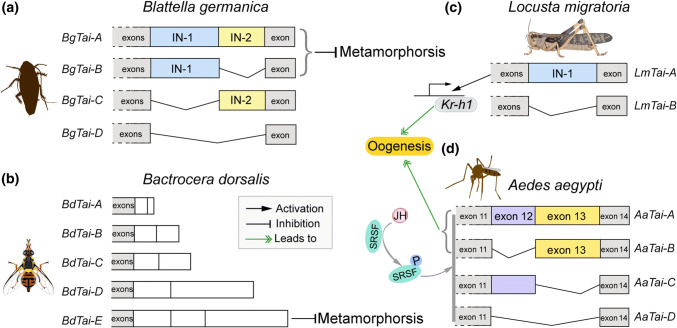
The alternative splicing of Tai [44, 48, 49] was a striking finding in the past years. Tai, orthologue to vertebrate steroid receptor coactivator (SRC), is a member of the basic–helix–loop–helix (bHLH)/Per-Arnt-Sim (PAS) family of transcription factors. JH induces the heterodimerization of Met and Tai to form a transcriptionally active complex to regulate the transcription of target genes in insect metamorphosis and reproduction [4, 5, 10]. No isoform of Met has been identified. However, two Met genes, Met1 and Met2, are present in the genomes of several lepidopteran insects [160–162]. D. melanogaster and its close relatives have two Met paralog genes, Met and Germ-cell expressed (Gce) [163, 164]. Vertebrate SRC has at least three isoforms with the splicing variants at the C-terminus region, which exhibit differential performances in hormone signaling [165–167]. The Tai isoforms were first identified in B. germanica, in which four isoforms are produced through the combination of two insertion domains (IN-1 and IN-2) near the C-terminus [49]. BgTai-A has both IN-1 and IN-2, whereas BgTai-B bears only IN-1 and BgTai-C harbors only IN-2. With respect to BgTai-D, neither IN-1 nor IN-2 is present (Fig. 5a). Depletion of all four isoforms in the nymph caused 100% mortality. However, specific knockdown of individual BgTai isoforms showed that along with Met, only BgTai-A and BgTai-B that have the IN-1-mediated anti-metamorphic signaling of JH. Lozano et al. further performed analysis of available Tai sequences and demonstrated that IN-1, but not IN-2 is present in Tai of D. melanogaster, T. castaneum, and A. mellifera, whereas neither IN-1 nor IN-2 is detected in B. mori Tai [49]. Five isoforms of Tai, derived from alternative splicing in the IN-1, were found in the dipteran Bactrocera dorsalis [168]. During the pre-final larval stage, simultaneous knockdown of all BdTai isoforms caused precocious metamorphosis. Moreover, specific depletion of BdTai-E that has the entire IN-1 resulted in phenotypes similar to that caused by knockdown of all Tai isoforms (Fig. 5b). However, no defective phenotype was observed with specific silencing of individual other four isoforms. The result suggests that BdTai isoform with the intact IN-1 is essential for the anti-metamorphic action of JH in the oriental fruit fly [169].
Fig. 5.
Tai isoforms and their role in metamorphosis and oogenesis. a In hemimetabolous Blattella germanica, four isoforms are produced through the combination of IN-1 and IN-2 domains. Depletion of all four isoforms in nymphs led to 100% mortality. Upon JH induction and Met binding, only BgTai-A and BgTai-B isoforms mediated the anti-metamorphic action of JH [49]. b Five splicing variants of Tai are found in holometabolous Bactrocera dorsalis. Specific knockdown of BdTai-E caused precocious metamorphosis, similar to that resulted from simultaneous depletion of all five isoforms [168]. c Two isoforms, LmTai-A and LmTai-B are identified in Locusta migratoria. Knockdown of both LmTai isoforms or LmTai-A alone but not LmTai-B resulted in defective phenotypes of oogenesis. Both isoforms dimerized with Met in the presence of JH and induced Kr-h1 transcription, but LmTai-A mediated a stronger transactivation than LmTai-B [44]. d In Aedes aegypti, the combinations of exon 12 (matching to IN-1) and exon 13 (matching to IN-2) produce 4 isoforms. JH acted via the RTK/PI3K/Akt and SRSF pathway to induce the production of AaTai-A and AaTai-B, which potentiated the 20E-EcR/USP transactivation and stimulated female reproduction [48]
Later, two isoforms, LmTai-A with IN-1 and LmTai-B without IN-1, were identified and experimentally elucidated for their function in the oogenesis of L. migratoria [44] (Fig. 5c). Interestingly, LmTai-A was expressed at levels about 50-fold higher than LmTai-B in the fat body of vitellogenic adult female locusts. Moreover, knockdown of both LmTai isoforms or LmTai-A alone but not LmTai-B resulted in the substantial reduction of Vg expression, arrested oocyte maturation, and blocked ovarian growth. Additional application of JH analogue on LmTai-A-depleted locusts was unable to restore the defective phenotypes to the normal levels, similar to that caused by knockdown of both two LmTai isoforms. Further studies demonstrated that, although both LmTai-A and LmTai-B dimerized with Met in the presence of JH and induced Kr-h1 transcription, LmTai-A mediated a stronger transactivation than Tai-B [44] (Fig. 5c). The results suggest that Tai-A with the IN-1 is more active than Tai-B without the IN-1 in transducing the vitellogenic JH signaling in L. migratoria. These data also suggest a central role of IN-1 domain in Tai function during locust oogenesis. The IN-1 domain encompasses a PRD-repeat motif rich in histidine and proline at the C-terminus, which has been reported to promote the dimerization of binding partners [170, 171].
Besides functioning as an obligatory component of functional JH–receptor complex, Tai serves as a transcriptional coactivator of the 20E-receptor EcR/USP [172, 173]. Ae. aegypti mosquito Tai has 14 exons, of which the combination of exon 12 (E12, matching to the IN-1 of Tai in B. germanica, D. melanogaster, T. castaneum, and L. migratoria) and exon 13 (E13, homologous to the IN-2 of Tai in B. germanica) yields four isoforms [48] (Fig. 5d). Accordingly, AaTai-A has both E12 and E13. AaTai-B carries only E13, whereas AaTai-C possesses only E12. Tai-D lacks both E12 and E13. Interestingly, JH, through the RTK/PI3 K/Akt-signaling pathway, stimulated the phosphorylation of serine/arginine-rich (pre-mRNA) splicing factor (SRSF) to induce AaTai alternative splicing for the production of the E13-containing isoforms AaTai-A and AaTai-B [48] (Fig. 5d). Consequently, AaTai-A and AaTai-B were abundant in the fat body of previtellogenic adult female mosquitoes. Depletion of AaTai-A or AaTai-B, either by inhibiting the alternative splicing or by isoform-specific RNAi, resulted in remarkably attenuated expression of 20E-responsive genes after blood feeding along with severely blocked oocyte development. Moreover, although four isoforms had similar capability to heterodimerize with Met in the presence of JH, AaTai-A and AaTai-B had a stronger capability for 20E-dependent binding to EcR/USP compared to other two isoforms [48]. Thus, AaTai-A/B but not AaTai-C/D potentiate the transcriptional activation by the 20E-receptor complex. These results indicate that JH-induced expression of AaTai-A/B in the fat body of adult females in the previtellogenic phase is essential for blood meal-induced and 20E-dependent egg production in mosquitoes (Fig. 5d). In light of above findings, Tai isoforms generated from alternative splicing play distinct roles in insect metamorphosis and oogenesis. While the alternative splicing motif, IN-1 is evolutionarily conserved, the presence of diverse Tai isoforms in different insects suggests the variation of alternative splicing of Tai across insect species.
Prospects and future directions
Insects provide excellent organisms to decipher the molecular basis of gene regulation at the post-transcriptional level. The collective studies have established the importance of miRNA and alternative splicing in insect metamorphosis and reproduction. It is widely recognized that the relationship between miRNAs and their targets is multiple to multiple [174]. Different miRNAs work together to co-regulate a common target gene, enforcing the effects on a given phenotype [114]. The effects of a single miRNA perturbation might be masked by changing the expression of relevant target genes. Moreover, miRNAs appear to be integrated in negative or positive feedback loops. Thus, the involvement of miRNAs in JH and 20E-orchestrated metamorphosis and oogenesis confers the reduction of transcriptional noise and fine-tuning of gene expression profiles to achieve the precise control of these processes. Though hundreds of miRNAs in diverse insect species have been identified and their target genes have been predicted, only a limited number of miRNA–target interactions have been experimentally determined. This is also true for the interactions of miRNA with JH and 20E-signaling pathways. It remains difficulty to uncover in vivo functions of individual miRNAs in non-model insects. Though a variety of databases and algorithms are available to predict miRNA-target interactions, a large number of false positives are still yielded. The fact that a single gene is likely targeted by multiple miRNAs and vice versa, each miRNA potentially targets multiple genes adds to the complexity of revealing the miRNA/gene regulatory axis. Certainly, the regulatory network of miRNA, JH, and 20E signal cascades in insect metamorphosis and reproduction need to be further dissected. In addition, more research on linear- or species-specific miRNAs in non-model insects is required to construct the physiological aspects of regulatory networks governing insect metamorphosis and reproduction. The comparative studies are also expected to explore the evolution of coordination of hormonal and post-transcriptional regulations in metamorphosis and oogenesis across insect orders. Beside miRNAs, a number of lncRNAs and circRNAs have been identified in various orders of insects by large-scale sequencing and target prediction. It will be urgent to experimentally document the effects of lncRNA and circRNA on metamorphosis and oogenesis. With the advantage of next generation sequencing platforms, bioinformatic toolkits and genetic manipulations such as Gal4-UAS and CRISPR/Cas-9 systems, studies on non-model insects are becoming common, which will help researchers to unveil the myriad of processes regulated by miRNA, lncRNA, and circRNA.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (NSFC) Grants 31630070 and 31702063.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Debecker S, Dinh KV, Stoks R. Strong delayed interactive effects of metal exposure and warming: latitude-dependent synergisms persist across metamorphosis. Environ Sci Technol. 2017;51(4):2409–2417. doi: 10.1021/acs.est.6b04989. [DOI] [PubMed] [Google Scholar]
- 2.Heneberg P, Bogusch P, Astapenková A. The effects of contact exposure to azole fungicides on insect metamorphosis. Crop Protect. 2019;121:66–72. doi: 10.1016/j.cropro.2019.03.012. [DOI] [Google Scholar]
- 3.Wesner JS, Kraus JM, Schmidt TS, Walters DM, Clements WH. Metamorphosis enhances the effects of metal exposure on the mayfly, Centroptilum triangulifer. Environ Sci Technol. 2014;48(17):10415–10422. doi: 10.1021/es501914y. [DOI] [PubMed] [Google Scholar]
- 4.Jindra M, Belles X, Shinoda T. Molecular basis of juvenile hormone signaling. Curr Opin Insect Sci. 2015;11:39–46. doi: 10.1016/j.cois.2015.08.004. [DOI] [PubMed] [Google Scholar]
- 5.Jindra M, Palli SR, Riddiford LM. The juvenile hormone signaling pathway in insect development. Annu Rev Entomol. 2013;58:181–204. doi: 10.1146/annurev-ento-120811-153700. [DOI] [PubMed] [Google Scholar]
- 6.Riddiford LM. Cellular and molecular actions of juvenile-hormone I. General-considerations and premetamorphic actions. Adv Insect Physiol. 1994;24:213–274. doi: 10.1016/S0065-2806(08)60084-3. [DOI] [Google Scholar]
- 7.Zhou B, Hiruma K, Shinoda T, Riddiford LM. Juvenile hormone prevents ecdysteroid-induced expression of broad complex RNAs in the epidermis of the tobacco hornworm, Manduca sexta. Dev Biol. 1998;203(2):233–244. doi: 10.1006/dbio.1998.9059. [DOI] [PubMed] [Google Scholar]
- 8.Zhou X, Riddiford LM. Broad specifies pupal development and mediates the ‘status quo’ action of juvenile hormone on the pupal-adult transformation in Drosophila and Manduca. Development. 2002;129(9):2259–2269. doi: 10.1242/dev.129.9.2259. [DOI] [PubMed] [Google Scholar]
- 9.Raikhel AS, Brown MR, Belles X. Hormonal control of reproductive processes. In: Gilbert LI, editor. Comprehensive molecular insect science. Amsterdam: Elsevier; 2005. pp. 433–491. [Google Scholar]
- 10.Roy S, Saha TT, Zou Z, Raikhel AS. Regulatory pathways controlling female insect reproduction. Annu Rev Entomol. 2018;63:489–511. doi: 10.1146/annurev-ento-020117-043258. [DOI] [PubMed] [Google Scholar]
- 11.Belles X. Vitellogenesis directed by juvenile hormone. Reprod Biol Invertebr. 2005;12:157–197. [Google Scholar]
- 12.Wyatt GR, Davey KG. Cellular and molecular actions of juvenile hormone. II. Roles of juvenile hormone in adult insects. Adv Insect Physiol. 1996;26:1–155. doi: 10.1016/S0065-2806(08)60030-2. [DOI] [Google Scholar]
- 13.Yao TP, Forman BM, Jiang Z, Cherbas L, Chen JD, McKeown M, Cherbas P, Evans RM. Functional ecdysone receptor is the product of EcR and Ultraspiracle genes. Nature. 1993;366(6454):476–479. doi: 10.1038/366476a0. [DOI] [PubMed] [Google Scholar]
- 14.Buszczak M, Segraves WA. Insect metamorphosis: out with the old, in with the new. Curr Biol. 2000;10(22):R830–R833. doi: 10.1016/s0960-9822(00)00792-2. [DOI] [PubMed] [Google Scholar]
- 15.Yamanaka N, Rewitz KF, O’Connor MB. Ecdysone control of developmental transitions: lessons from Drosophila research. Annu Rev Entomol. 2013;58:497–516. doi: 10.1146/annurev-ento-120811-153608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schwedes CC, Carney GE. Ecdysone signaling in adult Drosophila melanogaster. J Insect Physiol. 2012;58(3):293–302. doi: 10.1016/j.jinsphys.2012.01.013. [DOI] [PubMed] [Google Scholar]
- 17.Swevers L. An update on ecdysone signaling during insect oogenesis. Curr Opin Insect Sci. 2019;31:8–13. doi: 10.1016/j.cois.2018.07.003. [DOI] [PubMed] [Google Scholar]
- 18.Belles X, Piulachs MD. Ecdysone signalling and ovarian development in insects: from stem cells to ovarian follicle formation. Biochim Biophys Acta. 2015;1849(2):181–186. doi: 10.1016/j.bbagrm.2014.05.025. [DOI] [PubMed] [Google Scholar]
- 19.Truman JW, Riddiford LM. Endocrine insights into the evolution of metamorphosis in insects. Annu Rev Entomol. 2002;47:467–500. doi: 10.1146/annurev.ento.47.091201.145230. [DOI] [PubMed] [Google Scholar]
- 20.Charles JP, Iwema T, Epa VC, Takaki K, Rynes J, Jindra M. Ligand-binding properties of a juvenile hormone receptor, Methoprene-tolerant. Proc Natl Acad Sci USA. 2011;108(52):21128–21133. doi: 10.1073/pnas.1116123109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li M, Mead EA, Zhu J. Heterodimer of two bHLH-PAS proteins mediates juvenile hormone-induced gene expression. Proc Natl Acad Sci USA. 2011;108(2):638–643. doi: 10.1073/pnas.1013914108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jindra M, Uhlirova M, Charles JP, Smykal V, Hill RJ. Genetic evidence for function of the bHLH-PAS protein Gce/Met As a Juvenile hormone receptor. PLoS Genet. 2015;11(7):e1005394. doi: 10.1371/journal.pgen.1005394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cui Y, Sui Y, Xu J, Zhu F, Palli SR. Juvenile hormone regulates Aedes aegypti Krüppel homolog 1 through a conserved E box motif. Insect Biochem Mol Biol. 2014;52:23–32. doi: 10.1016/j.ibmb.2014.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guo W, Wu Z, Song J, Jiang F, Wang Z, Deng S, Walker VK, Zhou S. Juvenile hormone-receptor complex acts on mcm4 and mcm7 to promote polyploidy and vitellogenesis in the migratory locust. PLoS Genet. 2014;10(10):e1004702. doi: 10.1371/journal.pgen.1004702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li K, Jia QQ, Li S. Juvenile hormone signaling—a mini review. Insect Sci. 2019;26(4):600–606. doi: 10.1111/1744-7917.12614. [DOI] [PubMed] [Google Scholar]
- 26.Wu Z, Guo W, Xie Y, Zhou S. Juvenile hormone activates the transcription of cell-division-cycle 6 (Cdc6) for polyploidy-dependent insect vitellogenesis and oogenesis. J Biol Chem. 2016;291(10):5418–5427. doi: 10.1074/jbc.M115.698936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu Z, Guo W, Yang L, He Q, Zhou S. Juvenile hormone promotes locust fat body cell polyploidization and vitellogenesis by activating the transcription of Cdk6 and E2f1. Insect Biochem Mol Biol. 2018;102:1–10. doi: 10.1016/j.ibmb.2018.09.002. [DOI] [PubMed] [Google Scholar]
- 28.Kayukawa T, Minakuchi C, Namiki T, Togawa T, Yoshiyama M, Kamimura M, Mita K, Imanishi S, Kiuchi M, Ishikawa Y, Shinoda T. Transcriptional regulation of juvenile hormone-mediated induction of Kruppel homolog 1, a repressor of insect metamorphosis. Proc Natl Acad Sci USA. 2012;109(29):11729–11734. doi: 10.1073/pnas.1204951109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lozano J, Belles X. Conserved repressive function of Kruppel homolog 1 on insect metamorphosis in hemimetabolous and holometabolous species. Sci Rep. 2011;1:163. doi: 10.1038/srep00163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Konopova B, Smykal V, Jindra M. Common and distinct roles of juvenile hormone signaling genes in metamorphosis of holometabolous and hemimetabolous insects. PLoS One. 2011;6(12):e28728. doi: 10.1371/journal.pone.0028728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kayukawa T, Nagamine K, Ito Y, Nishita Y, Ishikawa Y, Shinoda T. Kruppel homolog 1 inhibits insect metamorphosis via direct tanscriptional repression of Broad-complex, a pupal specifier gene. J Biol Chem. 2016;291(4):1751–1762. doi: 10.1074/jbc.M115.686121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Urena E, Chafino S, Manjon C, Franch-Marro X, Martin D. The occurrence of the holometabolous pupal stage requires the interaction between E93, Kruppel-homolog 1 and Broad-complex. PLoS Genet. 2016;12(5):e1006020. doi: 10.1371/journal.pgen.1006020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Belles X, Santos CG. The MEKRE93 (Methoprene tolerant-Kruppel homolog 1-E93) pathway in the regulation of insect metamorphosis, and the homology of the pupal stage. Insect Biochem Mol Biol. 2014;52:60–68. doi: 10.1016/j.ibmb.2014.06.009. [DOI] [PubMed] [Google Scholar]
- 34.Gujar H, Palli SR. Kruppel homolog 1 and E93 mediate Juvenile hormone regulation of metamorphosis in the common bed bug, Cimex lectularius. Sci Rep. 2016;6:26092. doi: 10.1038/srep26092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kayukawa T, Jouraku A, Ito Y, Shinoda T. Molecular mechanism underlying juvenile hormone-mediated repression of precocious larval-adult metamorphosis. Proc Natl Acad Sci USA. 2017;114(5):1057–1062. doi: 10.1073/pnas.1615423114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang T, Song W, Li Z, Qian W, Wei L, Yang Y, Wang W, Zhou X, Meng M, Peng J, Xia Q, Perrimon N, Cheng D. Kruppel homolog 1 represses insect ecdysone biosynthesis by directly inhibiting the transcription of steroidogenic enzymes. Proc Natl Acad Sci USA. 2018;115(15):3960–3965. doi: 10.1073/pnas.1800435115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu S, Li K, Gao Y, Liu X, Chen W, Ge W, Feng Q, Palli SR, Li S. Antagonistic actions of juvenile hormone and 20-hydroxyecdysone within the ring gland determine developmental transitions in Drosophila. Proc Natl Acad Sci USA. 2018;115(1):139–144. doi: 10.1073/pnas.1716897115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Santos CG, Humann FC, Hartfelder K. Juvenile hormone signaling in insect oogenesis. Curr Opin Insect Sci. 2019;31:43–48. doi: 10.1016/j.cois.2018.07.010. [DOI] [PubMed] [Google Scholar]
- 39.Ojani R, Fu X, Ahmed T, Liu P, Zhu J. Kruppel homologue 1 acts as a repressor and an activator in the transcriptional response to juvenile hormone in adult mosquitoes. Insect Mol Biol. 2018;27(2):268–278. doi: 10.1111/imb.12370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang WN, Ma L, Liu C, Chen L, Xiao HJ, Liang GM. Dissecting the role of Kruppel homolog 1 in the metamorphosis and female reproduction of the cotton bollworm, Helicoverpa armigera. Insect Mol Biol. 2018 doi: 10.1111/imb.12389. [DOI] [PubMed] [Google Scholar]
- 41.Yue Y, Yang RL, Wang WP, Zhou QH, Chen EH, Yuan GR, Wang JJ, Dou W. Involvement of Met and Kr-h1 in JH-Mediated Reproduction of Female Bactrocera dorsalis (Hendel) Front Physiol. 2018;9:482. doi: 10.3389/fphys.2018.00482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tang Y, He H, Qu X, Cai Y, Ding W, Qiu L, Li Y. RNA interference-mediated knockdown of the transcription factor Kruppel homolog 1 suppresses vitellogenesis in Chilo suppressalis. Insect Mol Biol. 2019 doi: 10.1111/imb.12617. [DOI] [PubMed] [Google Scholar]
- 43.Song J, Wu Z, Wang Z, Deng S, Zhou S. Kruppel-homolog 1 mediates juvenile hormone action to promote vitellogenesis and oocyte maturation in the migratory locust. Insect Biochem Mol Biol. 2014;52:94–101. doi: 10.1016/j.ibmb.2014.07.001. [DOI] [PubMed] [Google Scholar]
- 44.Wang Z, Yang L, Song J, Kang L, Zhou S. An isoform of Taiman that contains a PRD-repeat motif is indispensable for transducing the vitellogenic juvenile hormone signal in Locusta migratoria. Insect Biochem Mol Biol. 2017;82:31–40. doi: 10.1016/j.ibmb.2017.01.009. [DOI] [PubMed] [Google Scholar]
- 45.Shin SW, Zou Z, Saha TT, Raikhel AS. bHLH-PAS heterodimer of methoprene-tolerant and Cycle mediates circadian expression of juvenile hormone-induced mosquito genes. Proc Natl Acad Sci USA. 2012;109(41):16576–16581. doi: 10.1073/pnas.1214209109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lucas KJ, Zhao B, Liu S, Raikhel AS. Regulation of physiological processes by microRNAs in insects. Curr Opin Insect Sci. 2015;11:1–7. doi: 10.1016/j.cois.2015.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Belles X. MicroRNAs and the evolution of insect metamorphosis. Annu Rev Entomol. 2017;62:111–125. doi: 10.1146/annurev-ento-031616-034925. [DOI] [PubMed] [Google Scholar]
- 48.Liu P, Fu X, Zhu J. Juvenile hormone-regulated alternative splicing of the taiman gene primes the ecdysteroid response in adult mosquitoes. Proc Natl Acad Sci USA. 2018;115(33):E7738–E7747. doi: 10.1073/pnas.1808146115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lozano J, Kayukawa T, Shinoda T, Belles X. A role for Taiman in insect metamorphosis. PLoS Genet. 2014;10(10):e1004769. doi: 10.1371/journal.pgen.1004769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu Y, Cheng T, Liu C, Liu D, Zhang Q, Long R, Zhao P, Xia Q. Systematic identification and characterization of long non-coding RNAs in the Silkworm. Bombyx mori. PLoS One. 2016;11(1):e0147147. doi: 10.1371/journal.pone.0147147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xiao H, Yuan Z, Guo D, Hou B, Yin C, Zhang W, Li F. Genome-wide identification of long noncoding RNA genes and their potential association with fecundity and virulence in rice brown planthopper, Nilaparvata lugens. BMC Genom. 2015;16:749. doi: 10.1186/s12864-015-1953-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhu B, Xu M, Shi H, Gao X, Liang P. Genome-wide identification of lncRNAs associated with chlorantraniliprole resistance in diamondback moth Plutella xylostella (L.) BMC Genomics. 2017;18(1):380. doi: 10.1186/s12864-017-3748-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N, Kadener S. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell. 2014;56(1):55–66. doi: 10.1016/j.molcel.2014.08.019. [DOI] [PubMed] [Google Scholar]
- 54.Gan H, Feng T, Wu Y, Liu C, Xia Q, Cheng T. Identification of circular RNA in the Bombyx mori silk gland. Insect Biochem Mol Biol. 2017;89:97–106. doi: 10.1016/j.ibmb.2017.09.003. [DOI] [PubMed] [Google Scholar]
- 55.Tholken C, Thamm M, Erbacher C, Lechner M. Sequence and structural properties of circular RNAs in the brain of nurse and forager honeybees (Apis mellifera) BMC Genomics. 2019;20(1):88. doi: 10.1186/s12864-018-5402-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li K, Tian Y, Yuan Y, Fan X, Yang M, He Z, Yang D. Insights into the functions of LncRNAs in Drosophila. Int J Mol Sci. 2019 doi: 10.3390/ijms20184646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403(6772):901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 58.Vella MC, Choi EY, Lin SY, Reinert K, Slack FJ. The C. elegans microRNA let-7 binds to imperfect let-7 complementary sites from the lin-41 3′UTR. Genes Dev. 2004;18(2):132–137. doi: 10.1101/gad.1165404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chi SW, Zang JB, Mele A, Darnell RB. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature. 2009;460(7254):479–486. doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M, Jr, Jungkamp AC, Munschauer M, Ulrich A, Wardle GS, Dewell S, Zavolan M, Tuschl T. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141(1):129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tay Y, Zhang J, Thomson AM, Lim B, Rigoutsos I. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature. 2008;455(7216):1124–1128. doi: 10.1038/nature07299. [DOI] [PubMed] [Google Scholar]
- 62.Zhou X, Duan X, Qian J, Li F. Abundant conserved microRNA target sites in the 5′-untranslated region and coding sequence. Genetica. 2009;137(2):159–164. doi: 10.1007/s10709-009-9378-7. [DOI] [PubMed] [Google Scholar]
- 63.Yang M, Wei Y, Jiang F, Wang Y, Guo X, He J, Kang L. MicroRNA-133 inhibits behavioral aggregation by controlling dopamine synthesis in locusts. PLoS Genet. 2014;10(2):e1004206. doi: 10.1371/journal.pgen.1004206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.He J, Chen Q, Wei Y, Jiang F, Yang M, Hao S, Guo X, Chen D, Kang L. MicroRNA-276 promotes egg-hatching synchrony by up-regulating brm in locusts. Proc Natl Acad Sci USA. 2016;113(3):584–589. doi: 10.1073/pnas.1521098113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318(5858):1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- 66.Azzam G, Smibert P, Lai EC, Liu JL. Drosophila Argonaute 1 and its miRNA biogenesis partners are required for oocyte formation and germline cell division. Dev Biol. 2012;365(2):384–394. doi: 10.1016/j.ydbio.2012.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tanaka ED, Piulachs MD. Dicer-1 is a key enzyme in the regulation of oogenesis in panoistic ovaries. Biol Cell. 2012;104(8):452–461. doi: 10.1111/boc.201100044. [DOI] [PubMed] [Google Scholar]
- 68.Nakahara K, Kim K, Sciulli C, Dowd SR, Minden JS, Carthew RW. Targets of microRNA regulation in the Drosophila oocyte proteome. Proc Natl Acad Sci USA. 2005;102(34):12023–12028. doi: 10.1073/pnas.0500053102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wu W, Xiong W, Li C, Zhai M, Li Y, Ma F, Li B. MicroRNA-dependent regulation of metamorphosis and identification of microRNAs in the red flour beetle, Tribolium castaneum. Genomics. 2017;109(5–6):362–373. doi: 10.1016/j.ygeno.2017.06.001. [DOI] [PubMed] [Google Scholar]
- 70.Rahimpour H, Moharramipour S, Asgari S, Mehrabadi M. The microRNA pathway core genes are differentially expressed during the development of Helicoverpa armigera and contribute in the insect’s development. Insect Biochem Mol Biol. 2019;110:121–127. doi: 10.1016/j.ibmb.2019.05.005. [DOI] [PubMed] [Google Scholar]
- 71.Gomez-Orte E, Belles X. MicroRNA-dependent metamorphosis in hemimetabolan insects. Proc Natl Acad Sci USA. 2009;106(51):21678–21682. doi: 10.1073/pnas.0907391106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Song J, Guo W, Jiang F, Kang L, Zhou S. Argonaute 1 is indispensable for juvenile hormone mediated oogenesis in the migratory locust, Locusta migratoria. Insect Biochem Mol Biol. 2013;43(9):879–887. doi: 10.1016/j.ibmb.2013.06.004. [DOI] [PubMed] [Google Scholar]
- 73.Wang YL, Yang ML, Jiang F, Zhang JZ, Kang L. MicroRNA-dependent development revealed by RNA interference-mediated gene silencing of LmDicer1 in the migratory locust. Insect Sci. 2013;20(1):53–60. doi: 10.1111/j.1744-7917.2012.01542.x. [DOI] [PubMed] [Google Scholar]
- 74.Zhu L, Liao SE, Fukunaga R. Drosophila Regnase-1 RNase is required for mRNA and miRNA profile remodelling during larva-to-adult metamorphosis. RNA Biol. 2019;16(10):1386–1400. doi: 10.1080/15476286.2019.1630799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shcherbata HR. miRNA functions in stem cells and their niches: lessons from the Drosophila ovary. Curr Opin Insect Sci. 2019;31:29–36. doi: 10.1016/j.cois.2018.07.006. [DOI] [PubMed] [Google Scholar]
- 76.Chawla G, Sokol NS. MicroRNAs in Drosophila development. Int Rev Cell Mol Biol. 2011;286:1–65. doi: 10.1016/b978-0-12-385859-7.00001-x. [DOI] [PubMed] [Google Scholar]
- 77.Roush S, Slack FJ. The let-7 family of microRNAs. Trends Cell Biol. 2008;18(10):505–516. doi: 10.1016/j.tcb.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 78.Sokol NS, Xu P, Jan YN, Ambros V. Drosophila let-7 microRNA is required for remodeling of the neuromusculature during metamorphosis. Genes Dev. 2008;22(12):1591–1596. doi: 10.1101/gad.1671708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tennessen JM, Thummel CS. Developmental timing: let-7 function conserved through evolution. Curr Biol. 2008;18(16):R707–R708. doi: 10.1016/j.cub.2008.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Caygill EE, Johnston LA. Temporal regulation of metamorphic processes in Drosophila by the let-7 and miR-125 heterochronic microRNAs. Curr Biol. 2008;18(13):943–950. doi: 10.1016/j.cub.2008.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wu YC, Chen CH, Mercer A, Sokol NS. Let-7-complex microRNAs regulate the temporal identity of Drosophila mushroom body neurons via chinmo. Dev Cell. 2012;23(1):202–209. doi: 10.1016/j.devcel.2012.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jiang S. A regulator of metabolic reprogramming: microRNA let-7. Transl Oncol. 2019;12(7):1005–1013. doi: 10.1016/j.tranon.2019.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kucherenko MM, Barth J, Fiala A, Shcherbata HR. Steroid-induced microRNA let-7 acts as a spatio-temporal code for neuronal cell fate in the developing Drosophila brain. EMBO J. 2012;31(24):4511–4523. doi: 10.1038/emboj.2012.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ling L, Ge X, Li Z, Zeng B, Xu J, Aslam AF, Song Q, Shang P, Huang Y, Tan A. MicroRNA Let-7 regulates molting and metamorphosis in the silkworm, Bombyx mori. Insect Biochem Mol Biol. 2014;53:13–21. doi: 10.1016/j.ibmb.2014.06.011. [DOI] [PubMed] [Google Scholar]
- 85.Peng W, Zheng WW, Tariq K, Yu SN, Zhang HY. MicroRNA Let-7 targets the ecdysone signaling pathway E75 gene to control larval-pupal development in Bactrocera dorsalis. Insect Sci. 2019;26(2):229–239. doi: 10.1111/1744-7917.12542. [DOI] [PubMed] [Google Scholar]
- 86.Rubio M, de Horna A, Belles X. MicroRNAs in metamorphic and non-metamorphic transitions in hemimetabolan insect metamorphosis. BMC Genomics. 2012;13:386. doi: 10.1186/1471-2164-13-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rubio M, Belles X. Subtle roles of microRNAs let-7, miR-100 and miR-125 on wing morphogenesis in hemimetabolan metamorphosis. J Insect Physiol. 2013;59(11):1089–1094. doi: 10.1016/j.jinsphys.2013.09.003. [DOI] [PubMed] [Google Scholar]
- 88.Song J, Li W, Zhao H, Gao L, Fan Y, Zhou S. The microRNAs let-7 and miR-278 regulate insect metamorphosis and oogenesis by targeting the juvenile hormone early-response gene Kruppel-homolog 1. Development. 2018 doi: 10.1242/dev.170670. [DOI] [PubMed] [Google Scholar]
- 89.Marco A, Hooks K, Griffiths-Jones S. Evolution and function of the extended miR-2 microRNA family. RNA Biol. 2012;9(3):242–248. doi: 10.4161/rna.19160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ling L, Ge X, Li Z, Zeng B, Xu J, Chen X, Shang P, James AA, Huang Y, Tan A. MiR-2 family targets awd and fng to regulate wing morphogenesis in Bombyx mori. RNA Biol. 2015;12(7):742–748. doi: 10.1080/15476286.2015.1048957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lozano J, Montanez R, Belles X. MiR-2 family regulates insect metamorphosis by controlling the juvenile hormone signaling pathway. Proc Natl Acad Sci USA. 2015;112(12):3740–3745. doi: 10.1073/pnas.1418522112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hyun S, Lee JH, Jin H, Nam J, Namkoong B, Lee G, Chung J, Kim VN. Conserved MicroRNA miR-8/miR-200 and its target USH/FOG2 control growth by regulating PI3 K. Cell. 2009;139(6):1096–1108. doi: 10.1016/j.cell.2009.11.020. [DOI] [PubMed] [Google Scholar]
- 93.Jin H, Kim VN, Hyun S. Conserved microRNA miR-8 controls body size in response to steroid signaling in Drosophila. Genes Dev. 2012;26(13):1427–1432. doi: 10.1101/gad.192872.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kennell JA, Cadigan KM, Shakhmantsir I, Waldron EJ. The microRNA miR-8 is a positive regulator of pigmentation and eclosion in Drosophila. Dev Dyn. 2012;241(1):161–168. doi: 10.1002/dvdy.23705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Liu Z, Ling L, Xu J, Zeng B, Huang Y, Shang P, Tan A. MicroRNA-14 regulates larval development time in Bombyx mori. Insect Biochem Mol Biol. 2018;93:57–65. doi: 10.1016/j.ibmb.2017.12.009. [DOI] [PubMed] [Google Scholar]
- 96.He K, Xiao H, Sun Y, Situ G, Xi Y, Li F. microRNA-14 as an efficient suppressor to switch off ecdysone production after ecdysis in insects. RNA Biol. 2019;16(9):1313–1325. doi: 10.1080/15476286.2019.1629768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.He K, Xiao H, Sun Y, Ding S, Situ G, Li F. Transgenic microRNA-14 rice shows high resistance to rice stem borer. Plant Biotechnol J. 2019;17(2):461–471. doi: 10.1111/pbi.12990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chen J, Liang Z, Liang Y, Pang R, Zhang W. Conserved microRNAs miR-8-5p and miR-2a-3p modulate chitin biosynthesis in response to 20-hydroxyecdysone signaling in the brown planthopper, Nilaparvata lugens. Insect Biochem Mol Biol. 2013;43(9):839–848. doi: 10.1016/j.ibmb.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 99.Yang M, Wang Y, Jiang F, Song T, Wang H, Liu Q, Zhang J, Zhang J, Kang L. miR-71 and miR-263 jointly regulate target genes Chitin synthase and Chitinase to control locust molting. PLoS Genet. 2016;12(8):e1006257. doi: 10.1371/journal.pgen.1006257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Puthiyakunnon S, Yao Y, Li Y, Gu J, Peng H, Chen X. Functional characterization of three MicroRNAs of the Asian tiger mosquito, Aedes albopictus. Parasites Vectors. 2013;6(1):230. doi: 10.1186/1756-3305-6-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhang X, Zheng Y, Jagadeeswaran G, Ren R, Sunkar R, Jiang H. Identification and developmental profiling of conserved and novel microRNAs in Manduca sexta. Insect Biochem Mol Biol. 2012;42(6):381–395. doi: 10.1016/j.ibmb.2012.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Chen J, Li TC, Pang R, Yue XZ, Hu J, Zhang WQ. Genome-wide screening and functional analysis reveal that the specific microRNA nlu-miR-173 regulates molting by targeting Ftz-F1 in Nilaparvata lugens. Front Physiol. 2018;9:1854. doi: 10.3389/fphys.2018.01854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Chen J, Li T, Pang R. miR-2703 regulates the chitin biosynthesis pathway by targeting chitin synthase 1a in Nilaparvata lugens. Insect Mol Biol. 2019 doi: 10.1111/imb.12606. [DOI] [PubMed] [Google Scholar]
- 104.Zhang YL, Huang QX, Yin GH, Lee S, Jia RZ, Liu ZX, Yu NT, Pennerman KK, Chen X, Guo AP. Identification of microRNAs by small RNA deep sequencing for synthetic microRNA mimics to control Spodoptera exigua. Gene. 2015;557(2):215–221. doi: 10.1016/j.gene.2014.12.038. [DOI] [PubMed] [Google Scholar]
- 105.Wang ZZ, Ye XQ, Shi M, Li F, Wang ZH, Zhou YN, Gu QJ, Wu XT, Yin CL, Guo DH, Hu RM, Hu NN, Chen T, Zheng BY, Zou JN, Zhan LQ, Wei SJ, Wang YP, Huang JH, Fang XD, Strand MR, Chen XX. Parasitic insect-derived miRNAs modulate host development. Nat Commun. 2018;9(1):2205. doi: 10.1038/s41467-018-04504-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Chawla G, Sokol NS. Hormonal activation of let-7-C microRNAs via EcR is required for adult Drosophila melanogaster morphology and function. Development. 2012;139(10):1788–1797. doi: 10.1242/dev.077743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Varghese J, Cohen SM. microRNA miR-14 acts to modulate a positive autoregulatory loop controlling steroid hormone signaling in Drosophila. Genes Dev. 2007;21(18):2277–2282. doi: 10.1101/gad.439807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Verma P, Cohen SM. miR-965 controls cell proliferation and migration during tissue morphogenesis in the Drosophila abdomen. eLife. 2015 doi: 10.7554/elife.07389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Jiang J, Ge X, Li Z, Wang Y, Song Q, Stanley DW, Tan A, Huang Y. MicroRNA-281 regulates the expression of ecdysone receptor (EcR) isoform B in the silkworm, Bombyx mori. Insect Biochem Mol Biol. 2013;43(8):692–700. doi: 10.1016/j.ibmb.2013.05.002. [DOI] [PubMed] [Google Scholar]
- 110.Lim DH, Lee S, Han JY, Choi MS, Hong JS, Seong Y, Kwon YS, Lee YS. Ecdysone-responsive microRNA-252-5p controls the cell cycle by targeting Abi in Drosophila. FASEB J. 2018;32(8):4519–4533. doi: 10.1096/fj.201701185RR. [DOI] [PubMed] [Google Scholar]
- 111.Garbuzov A, Tatar M. Hormonal regulation of Drosophila microRNA let-7 and miR-125 that target innate immunity. Fly. 2010;4(4):306–311. doi: 10.4161/fly.4.4.13008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Qu Z, Bendena WG, Nong W, Siggens KW, Noriega FG, Kai ZP, Zang YY, Koon AC, Chan HYE, Chan TF, Chu KH, Lam HM, Akam M, Tobe SS, Lam Hui JH. MicroRNAs regulate the sesquiterpenoid hormonal pathway in Drosophila and other arthropods. Proc Biol Sci. 2017 doi: 10.1098/rspb.2017.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Boulan L, Martin D, Milan M. bantam miRNA promotes systemic growth by connecting insulin signaling and ecdysone production. Curr Biol. 2013;23(6):473–478. doi: 10.1016/j.cub.2013.01.072. [DOI] [PubMed] [Google Scholar]
- 114.He K, Sun Y, Xiao H, Ge C, Li F, Han Z. Multiple miRNAs jointly regulate the biosynthesis of ecdysteroid in the holometabolous insects, Chilo suppressalis. RNA. 2017;23(12):1817–1833. doi: 10.1261/rna.061408.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Qu Z, Bendena WG, Tobe SS, Hui JHL. Juvenile hormone and sesquiterpenoids in arthropods: biosynthesis, signaling, and role of MicroRNA. J Steroid Biochem Mol Biol. 2018;184:69–76. doi: 10.1016/j.jsbmb.2018.01.013. [DOI] [PubMed] [Google Scholar]
- 116.Nouzova M, Etebari K, Noriega FG, Asgari S. A comparative analysis of corpora allata-corpora cardiaca microRNA repertoires revealed significant changes during mosquito metamorphosis. Insect Biochem Mol Biol. 2018;96:10–18. doi: 10.1016/j.ibmb.2018.03.007. [DOI] [PubMed] [Google Scholar]
- 117.Raikhel AS, Dhadialla TS. Accumulation of yolk proteins in insect oocytes. Annu Rev Entomol. 1992;37:217–251. doi: 10.1146/annurev.en.37.010192.001245. [DOI] [PubMed] [Google Scholar]
- 118.Asgari S. MicroRNA functions in insects. Insect Biochem Mol Biol. 2013;43(4):388–397. doi: 10.1016/j.ibmb.2012.10.005. [DOI] [PubMed] [Google Scholar]
- 119.Baumer D, Strohlein NM, Schoppmeier M. Opposing effects of Notch-signaling in maintaining the proliferative state of follicle cells in the telotrophic ovary of the beetle Tribolium. Front Zool. 2012;9(1):15. doi: 10.1186/1742-9994-9-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Irles P, Elshaer N, Piulachs MD. The Notch pathway regulates both the proliferation and differentiation of follicular cells in the panoistic ovary of Blattella germanica. Open Biol. 2016;6(1):150197. doi: 10.1098/rsob.150197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Xu J, Gridley T. Notch signaling during Oogenesis in Drosophila melanogaster. Genet Res Int. 2012;2012:648207. doi: 10.1155/2012/648207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Song J, Li W, Zhao H, Zhou S. Clustered miR-2, miR-13a, miR-13b and miR-71 coordinately target Notch gene to regulate oogenesis of the migratory locust Locusta migratoria. Insect Biochem Mol Biol. 2019;106:39–46. doi: 10.1016/j.ibmb.2018.11.004. [DOI] [PubMed] [Google Scholar]
- 123.Yang L, Chen D, Duan R, Xia L, Wang J, Qurashi A, Jin P, Chen D. Argonaute 1 regulates the fate of germline stem cells in Drosophila. Development. 2007;134(23):4265–4272. doi: 10.1242/dev.009159. [DOI] [PubMed] [Google Scholar]
- 124.Jin Z, Xie T. Dcr-1 maintains Drosophila ovarian stem cells. Curr Biol. 2007;17(6):539–544. doi: 10.1016/j.cub.2007.01.050. [DOI] [PubMed] [Google Scholar]
- 125.Wang C, Feng T, Wan Q, Kong Y, Yuan L. miR-124 controls Drosophila behavior and is required for neural development. Int J Dev Neurosci. 2014;38:105–112. doi: 10.1016/j.ijdevneu.2014.08.006. [DOI] [PubMed] [Google Scholar]
- 126.Garaulet DL, Castellanos MC, Bejarano F, Sanfilippo P, Tyler DM, Allan DW, Sanchez-Herrero E, Lai EC. Homeotic function of Drosophila Bithorax-complex miRNAs mediates fertility by restricting multiple Hox genes and TALE cofactors in the CNS. Dev Cell. 2014;29(6):635–648. doi: 10.1016/j.devcel.2014.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Yatsenko AS, Shcherbata HR. Stereotypical architecture of the stem cell niche is spatiotemporally established by miR-125-dependent coordination of Notch and steroid signaling. Development. 2018 doi: 10.1242/dev.159178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Huang YC, Smith L, Poulton J, Deng WM. The microRNA miR-7 regulates Tramtrack69 in a developmental switch in Drosophila follicle cells. Development. 2013;140(4):897–905. doi: 10.1242/dev.080192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Ge W, Deng Q, Guo T, Hong X, Kugler JM, Yang X, Cohen SM. Regulation of pattern formation and gene amplification during Drosophila oogenesis by the miR-318 microRNA. Genetics. 2015;200(1):255–265. doi: 10.1534/genetics.115.174748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Bryant B, Macdonald W, Raikhel AS. microRNA miR-275 is indispensable for blood digestion and egg development in the mosquito Aedes aegypti. Proc Natl Acad Sci USA. 2010;107(52):22391–22398. doi: 10.1073/pnas.1016230107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Ling L, Kokoza VA, Zhang C, Aksoy E, Raikhel AS. MicroRNA-277 targets insulin-like peptides 7 and 8 to control lipid metabolism and reproduction in Aedes aegypti mosquitoes. Proc Natl Acad Sci USA. 2017;114(38):E8017–E8024. doi: 10.1073/pnas.1710970114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Liu S, Lucas KJ, Roy S, Ha J, Raikhel AS. Mosquito-specific microRNA-1174 targets serine hydroxymethyltransferase to control key functions in the gut. Proc Natl Acad Sci USA. 2014;111(40):14460–14465. doi: 10.1073/pnas.1416278111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Lucas KJ, Zhao B, Roy S, Gervaise AL, Raikhel AS. Mosquito-specific microRNA-1890 targets the juvenile hormone-regulated serine protease JHA15 in the female mosquito gut. RNA Biol. 2015;12(12):1383–1390. doi: 10.1080/15476286.2015.1101525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Lucas KJ, Roy S, Ha J, Gervaise AL, Kokoza VA, Raikhel AS. MicroRNA-8 targets the Wingless signaling pathway in the female mosquito fat body to regulate reproductive processes. Proc Natl Acad Sci USA. 2015;112(5):1440–1445. doi: 10.1073/pnas.1424408112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Bian G, Raikhel AS, Zhu J. Characterization of a juvenile hormone-regulated chymotrypsin-like serine protease gene in Aedes aegypti mosquito. Insect Biochem Mol Biol. 2008;38(2):190–200. doi: 10.1016/j.ibmb.2007.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Fu X, Dimopoulos G, Zhu J. Association of microRNAs with Argonaute proteins in the malaria mosquito Anopheles gambiae after blood ingestion. Sci Rep. 2017;7(1):6493. doi: 10.1038/s41598-017-07013-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Zhang Y, Zhao B, Roy S, Saha TT, Kokoza VA, Li M, Raikhel AS. microRNA-309 targets the Homeobox gene SIX4 and controls ovarian development in the mosquito Aedes aegypti. Proc Natl Acad Sci USA. 2016;113(33):E4828–E4836. doi: 10.1073/pnas.1609792113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Zhang X, Aksoy E, Girke T, Raikhel AS, Karginov FV. Transcriptome-wide microRNA and target dynamics in the fat body during the gonadotrophic cycle of Aedes aegypti. Proc Natl Acad Sci USA. 2017;114(10):E1895–E1903. doi: 10.1073/pnas.1701474114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Jayachandran B, Hussain M, Asgari S. An insect trypsin-like serine protease as a target of microRNA: utilization of microRNA mimics and inhibitors by oral feeding. Insect Biochem Mol Biol. 2013;43(4):398–406. doi: 10.1016/j.ibmb.2012.10.004. [DOI] [PubMed] [Google Scholar]
- 140.Zhang X, Lu K, Zhou Q. Dicer1 is crucial for the oocyte maturation of telotrophic ovary in Nilaparvata lugens (STAL) (Hemiptera: Geometroidea) Arch Insect Biochem Physiol. 2013;84(4):194–208. doi: 10.1002/arch.21136. [DOI] [PubMed] [Google Scholar]
- 141.Fu X, Li T, Chen J, Dong Y, Qiu J, Kang K, Zhang W. Functional screen for microRNAs of Nilaparvata lugens reveals that targeting of glutamine synthase by miR-4868b regulates fecundity. J Insect Physiol. 2015;83:22–29. doi: 10.1016/j.jinsphys.2015.11.003. [DOI] [PubMed] [Google Scholar]
- 142.Blencowe BJ. Alternative splicing: new insights from global analyses. Cell. 2006;126(1):37–47. doi: 10.1016/j.cell.2006.06.023. [DOI] [PubMed] [Google Scholar]
- 143.Wang Z, Burge CB. Splicing regulation: from a parts list of regulatory elements to an integrated splicing code. RNA. 2008;14(5):802–813. doi: 10.1261/rna.876308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Talbot WS, Swyryd EA, Hogness DS. Drosophila tissues with different metamorphic responses to ecdysone express different ecdysone receptor isoforms. Cell. 1993;73(7):1323–1337. doi: 10.1016/0092-8674(93)90359-x. [DOI] [PubMed] [Google Scholar]
- 145.Wang SF, Li C, Zhu J, Miura K, Miksicek RJ, Raikhel AS. Differential expression and regulation by 20-hydroxyecdysone of mosquito ultraspiracle isoforms. Dev Biol. 2000;218(1):99–113. doi: 10.1006/dbio.1999.9575. [DOI] [PubMed] [Google Scholar]
- 146.Huang LX, Gong YJ, Gu J, Zeng BJ, Huang LH, Feng QL. Expression, subcellular localization and protein-protein interaction of four isoforms of EcR/USP in the common cutworm. Insect Sci. 2015;22(1):95–105. doi: 10.1111/1744-7917.12101. [DOI] [PubMed] [Google Scholar]
- 147.Xu QY, Deng P, Zhang Q, Li A, Fu KY, Guo WC, Li GQ. Ecdysone receptor isoforms play distinct roles in larval-pupal-adult transition in Leptinotarsa decemlineata. Insect Sci. 2019 doi: 10.1111/1744-7917.12662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Tan A, Palli SR. Edysone receptor isoforms play distinct roles in controlling molting and metamorphosis in the red flour beetle, Tribolium castaneum. Mol Cell Endocrinol. 2008;291(1):42–49. doi: 10.1016/j.mce.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Watanabe T, Takeuchi H, Kubo T. Structural diversity and evolution of the N-terminal isoform-specific region of ecdysone receptor-A and-B1 isoforms in insects. BMC Evol Biol. 2010;10:17. doi: 10.1186/1471-2148-10-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Tan YA, Xiao LB, Zhao J, Xiao YF, Sun Y, Bai LX. Ecdysone receptor isoform-B mediates soluble trehalase expression to regulate growth and development in the mirid bug, Apolygus lucorum (Meyer-Dur) Insect Mol Biol. 2015;24(6):611–623. doi: 10.1111/imb.12185. [DOI] [PubMed] [Google Scholar]
- 151.Chen CH, Pan J, Di YQ, Liu W, Hou L, Wang JX, Zhao XF. Protein kinase C delta phosphorylates ecdysone receptor B1 to promote gene expression and apoptosis under 20-hydroxyecdysone regulation. Proc Natl Acad Sci USA. 2017;114(34):E7121–E7130. doi: 10.1073/pnas.1704999114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Lenaerts C, Van Wielendaele P, Peeters P, Vanden Broeck J, Marchal E. Ecdysteroid signalling components in metamorphosis and development of the desert locust, Schistocerca gregaria. Insect Biochem Mol Biol. 2016;75:10–23. doi: 10.1016/j.ibmb.2016.05.003. [DOI] [PubMed] [Google Scholar]
- 153.Tan A, Palli SR. Ecdysone [corrected] receptor isoforms play distinct roles in controlling molting and metamorphosis in the red flour beetle, Tribolium castaneum. Mol Cell Endocrinol. 2008;291(1–2):42–49. doi: 10.1016/j.mce.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Hult EF, Huang J, Marchal E, Lam J, Tobe SS. RXR/USP and EcR are critical for the regulation of reproduction and the control of JH biosynthesis in Diploptera punctata. J Insect Physiol. 2015;80:48–60. doi: 10.1016/j.jinsphys.2015.04.006. [DOI] [PubMed] [Google Scholar]
- 155.Maestro O, Cruz J, Pascual N, Martin D, Belles X. Differential expression of two RXR/ultraspiracle isoforms during the life cycle of the hemimetabolous insect Blattella germanica (Dictyoptera, Blattellidae) Mol Cell Endocrinol. 2005;238(1–2):27–37. doi: 10.1016/j.mce.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 156.Jindra M, Huang JY, Malone F, Asahina M, Riddiford LM. Identification and mRNA developmental profiles of two ultraspiracle isoforms in the epidermis and wings of Manduca sexta. Insect Mol Biol. 1997;6(1):41–53. doi: 10.1046/j.1365-2583.1997.00153.x. [DOI] [PubMed] [Google Scholar]
- 157.Kapitskaya M, Wang S, Cress DE, Dhadialla TS, Raikhel AS. The mosquito ultraspiracle homologue, a partner of ecdysteroid receptor heterodimer: cloning and characterization of isoforms expressed during vitellogenesis. Mol Cell Endocrinol. 1996;121(2):119–132. doi: 10.1016/0303-7207(96)03847-6. [DOI] [PubMed] [Google Scholar]
- 158.Lan Q, Hiruma K, Hu X, Jindra M, Riddiford LM. Activation of a delayed-early gene encoding MHR3 by the ecdysone receptor heterodimer EcR-B1-USP-1 but not by EcR-B1-USP-2. Mol Cell Biol. 1999;19(7):4897–4906. doi: 10.1128/MCB.19.7.4897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Parthasarathy R, Palli SR. Stage- and cell-specific expression of ecdysone receptors and ecdysone-induced transcription factors during midgut remodeling in the yellow fever mosquito, Aedes aegypti. J Insect Physiol. 2007;53(3):216–229. doi: 10.1016/j.jinsphys.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 160.Cheng D, Meng M, Peng J, Qian W, Kang L, Xia Q. Genome-wide comparison of genes involved in the biosynthesis, metabolism, and signaling of juvenile hormone between silkworm and other insects. Genet Mol Biol. 2014;37(2):444–459. doi: 10.1590/S1415-47572014005000006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Kayukawa T, Shinoda T. Functional characterization of two paralogous JH receptors, methoprene-tolerant 1 and 2, in the silkworm, Bombyx mori (Lepidoptera: Bombycidae) Appl Entomol Zool. 2015 doi: 10.1007/s13355-015-0345-8. [DOI] [Google Scholar]
- 162.Jouraku A, Yamamoto K, Kuwazaki S, Urio M, Suetsugu Y, Narukawa J, Miyamoto K, Kurita K, Kanamori H, Katayose Y, Matsumoto T, Noda H. KONAGAbase: a genomic and transcriptomic database for the diamondback moth, Plutella xylostella. BMC Genomics. 2013;14:464. doi: 10.1186/1471-2164-14-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Baumann A, Fujiwara Y, Wilson TG. Evolutionary divergence of the paralogs Methoprene tolerant (Met) and germ cell expressed (gce) within the genus Drosophila. J Insect Physiol. 2010;56(10):1445–1455. doi: 10.1016/j.jinsphys.2010.05.001. [DOI] [PubMed] [Google Scholar]
- 164.Abdou MA, He Q, Wen D, Zyaan O, Wang J, Xu J, Baumann AA, Joseph J, Wilson TG, Li S, Wang J. Drosophila Met and Gce are partially redundant in transducing juvenile hormone action. Insect Biochem Mol Biol. 2011;41(12):938–945. doi: 10.1016/j.ibmb.2011.09.003. [DOI] [PubMed] [Google Scholar]
- 165.Dasgupta S, O’Malley BW. Transcriptional coregulators: emerging roles of SRC family of coactivators in disease pathology. J Mol Endocrinol. 2014;53(2):R47–R59. doi: 10.1530/JME-14-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Xu J, Wu RC, O’Malley BW. Normal and cancer-related functions of the p160 steroid receptor co-activator (SRC) family. Nat Rev Cancer. 2009;9(9):615–630. doi: 10.1038/nrc2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.York B, O’Malley BW. Steroid receptor coactivator (SRC) family: masters of systems biology. J Biol Chem. 2010;285(50):38743–38750. doi: 10.1074/jbc.R110.193367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Liu H, Li HM, Yue Y, Song ZH, Wang JJ, Dou W. The alternative splicing of BdTai and its involvement in the development of Bactrocera dorsalis (Hendel) J Insect Physiol. 2017;101:132–141. doi: 10.1016/j.jinsphys.2017.07.012. [DOI] [PubMed] [Google Scholar]
- 169.Li Y, Zheng F, Xiao X, Xie F, Tao D, Huang C, Liu D, Wang M, Wang L, Zeng F, Jiang G. CircHIPK3 sponges miR-558 to suppress heparanase expression in bladder cancer cells. EMBO Rep. 2017;18(9):1646–1659. doi: 10.15252/embr.201643581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Cao Y, Wang C. The COOH-terminal transactivation domain plays a key role in regulating the in vitro and in vivo function of Pax3 homeodomain. J Biol Chem. 2000;275(13):9854–9862. doi: 10.1074/jbc.275.13.9854. [DOI] [PubMed] [Google Scholar]
- 171.Sonnenfeld MJ, Delvecchio C, Sun X. Analysis of the transcriptional activation domain of the Drosophila tango bHLH-PAS transcription factor. Dev Genes Evol. 2005;215(5):221–229. doi: 10.1007/s00427-004-0462-9. [DOI] [PubMed] [Google Scholar]
- 172.Bai J, Uehara Y, Montell DJ. Regulation of invasive cell behavior by taiman, a Drosophila protein related to AIB1, a steroid receptor coactivator amplified in breast cancer. Cell. 2000;103(7):1047–1058. doi: 10.1016/s0092-8674(00)00208-7. [DOI] [PubMed] [Google Scholar]
- 173.Zhu J, Chen L, Sun G, Raikhel AS. The competence factor beta Ftz-F1 potentiates ecdysone receptor activity via recruiting a p160/SRC coactivator. Mol Cell Biol. 2006;26(24):9402–9412. doi: 10.1128/MCB.01318-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Hashimoto Y, Akiyama Y, Yuasa Y. Multiple-to-multiple relationships between microRNAs and target genes in gastric cancer. PLoS One. 2013;8(5):e62589. doi: 10.1371/journal.pone.0062589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Gilbert LI, Rybczynski R, Warren JT. Control and biochemical nature of the ecdysteroidogenic pathway. Annu Rev Entomol. 2002;47:883–916. doi: 10.1146/annurev.ento.47.091201.145302. [DOI] [PubMed] [Google Scholar]
- 176.Spindler KD, Honl C, Tremmel C, Braun S, Ruff H, Spindler-Barth M. Ecdysteroid hormone action. Cell Mol Life Sci. 2009;66(24):3837–3850. doi: 10.1007/s00018-009-0112-5. [DOI] [PMC free article] [PubMed] [Google Scholar]