Abstract
The centromere is a specialized region on the chromosome that directs equal chromosome segregation. Centromeres are usually not defined by DNA sequences alone. How centromere formation and function are determined by epigenetics is still not fully understood. Active centromeres are often marked by the presence of centromeric-specific histone H3 variant, centromere protein A (CENP-A). How CENP-A is assembled into the centromeric chromatin during the cell cycle and propagated to the next cell cycle or the next generation to maintain the centromere function has been intensively investigated. In this review, we summarize current understanding of how post-translational modifications of CENP-A and other centromere proteins, centromeric and pericentric histone modifications, non-coding transcription and transcripts contribute to centromere function, and discuss their intricate relationships and potential feedback mechanisms.
Keywords: Centromere, Post-translational modifications, Histone modifications, Non-coding transcription, Centromeric transcript, Pericentric heterochromatin
Introduction
The centromere is a specialized region on the chromosome, where kinetochore proteins assemble into a complex that mediates chromosome attachment to microtubules. Centromeres that appear as one primary constriction on the mitotic chromosomes are called monocentromeres, which can be sub-divided into point and regional centromeres. Point centromeres, which are unique for budding yeast species, are short and occupy only one nucleosome [1]. Regional centromeres occupy a larger domain, but have widely variable sizes among humans, mice, chickens, frogs, fruit flies and fission yeast. Some algae, plants, insects and nematodes contain polycentromeres that have multiple primary constrictions or even holocentromeres that occupy the entire poleward faces of the chromosomes. In this review, most studies described were carried out in regional centromeres; while, a few examples from other centromere organizations were used for comparisons.
Centromeric DNA is the DNA sequence where canonical, endogenous centromeres form on. Centromeric DNA sequences are drastically different among species, despite having certain common characteristics (See [2] for an overview of the centromeric sequence diversity). Centromeric DNA of regional centromeres is flanked by highly condensed chromatin domains, which are called pericentric heterochromatin, that facilitate sister chromatid cohesion [3–7]. Both centromeric and heterochromatic DNA in regional centromeres consist of repetitive sequences. For example, in humans, arrays of α-satellite sequences constitute both the centromeric (mainly High Order Repeat (HOR) arrays) and pericentric regions (mainly monomeric α-satellites) (See [8] for a comprehensive discussion on human centromere sequences). In mice, the minor satellite DNA and the major satellite DNA form the centromere and pericentric heterochromatin, respectively.
In budding yeast species, point centromeres are originally thought to be mainly DNA sequence dependent. In organisms with regional centromeres, forming a centromere does not always require the exact endogenous centromeric DNA sequence, and having the centromeric DNA sequences does not always guarantee the formation of centromeres. Occasionally centromeres are found to reposition to regions outside of the canonical centromeric DNA, and the resulting functional, ectopic centromeres are referred to as neocentromeres. Cases of neocentromeres have been reported in human patients, who have lost the original centromeric sequences, or who have undergone centromere inactivation with their α-satellite DNA remaining intact on their chromosomes [9, 10] (See [11] for more clinical cases of human neocentromeres). On the other hand, in human patients with a dicentric chromosome, which can arise from fusion of chromosome fragments, either one of the two centromeres can become inactivated and the other can function as a monocentric chromosomes [12]. Neocentromeres have also been observed or induced experimentally in different organisms, e.g., by selecting for surviving cells after removing the original centromere [13] (See [14] for a comprehensive review of neocentromeres). The above examples suggest that centromere activity can be established or abolished epigenetically.
Most functional centromeres are marked by a centromeric-specific histone H3 variant centromere protein A (CENP-A) [12]. CENP-A plays a critical role in centromere specification, centromere maintenance and kinetochore assembly. CENP-A and the other constitutive centromere-associated network (CCAN) components, including CENP-C and CENP-T, are required for recruitment of the other kinetochore proteins, including the KMN network (the KNL-1, MIS-12 and NDC-80 complexes) [15]. Studies in human and fly regional centromeres showed that CENP-A is interspaced with H3. Like other canonical histones, CENP-A is diluted in half during DNA replication. However, unlike other canonical histones, which are often replenished in S phase, CENP-A replenishment is independent of DNA replication, though it also occurs only once every cell cycle at various times among organisms [16]. On the other hand, in budding yeast with point monocentromeres and in the nematode Caenorhabditis elegans with holocentromeres, all or most CENP-A on chromatin is turned over during each cell cycle [17, 18].
The molecular mechanism of recruiting new CENP-A to the correct position on the chromosome, and hence maintaining the centromere through cell cycles and generations has become clearer in the last decade [19]. In general, replenishment of new CENP-A at centromere involves licensing, deposition and stabilization. First, the original centromere position is primed by licensing factors, e.g., Mis18 complex (Mis18α, Mis18β, and Mis18BP1/KNL-2) in human cells [20], before CENP-A loading. Second, pre-nucleosomal CENP-A is deposited onto the primed centromeric chromatin, assisted by the CENP-A chaperone, which is HJURP in humans [21, 22], Scm3 in budding yeast and fission yeast [23, 24], CAL1 in Drosophila [25], and potentially RbAp46/48p55/LIN−53 in Drosophila and C. elegans [26, 27]. The CENP-A assembly by HJURP induced the removal of Mis18BP1 from human centromeres [28]. S. pombe RbAp46/48Mis16 binds to CENP-ACnp−1-H4 tetramer and Scm3, and then forms a complex with Mis18 [29]. Third, CENP-A-containing chromatin is stabilized by a maturation process, e.g., CENP-A monoubiquitination in Drosophila [30], or else CENP-A would be removed from the chromatin [31–33].
Besides the centromeric epigenetic marker CENP-A, there are multiple epigenetic mechanisms to regulate centromere activity. In this review, we summarize the current knowledge of epigenetic regulations on centromere function, focusing on post-translational modifications on CENP-A, histones, other centromeric and kinetochore proteins, the transcription process and the corresponding transcripts at centromeric and pericentric regions. We discuss the relationships and interdependencies of these epigenetic regulations, their feedback regulatory mechanisms and evolutionary origins.
Post-translational modifications (PTMs) of CENP-A
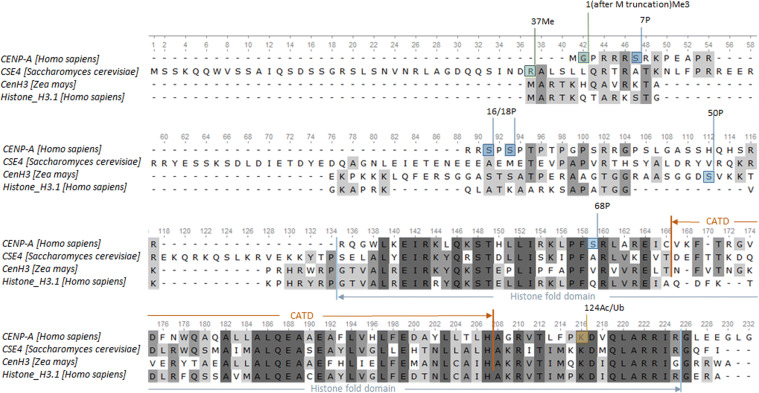
In the past decade, various post-translational modifications of CENP-A have been identified by mass spectrometry and characterized (Fig. 1 and Table 1). These modifications include methylation, acetylation, ubiquitination, and sumoylation. Modifications on CENP-A are dynamic during the cell cycle and contribute to CENP-A’s localization and function.
Fig. 1.
Alignment of CENP-A proteins in humans, budding yeast, and maize with histone H3 in humans to show the position of CENP-A post-translational modifications listed in Table 1. The multiple protein sequences are aligned in unipro UGENE (ClustalW). The amino acid sequences are extracted from NCBI: CENP-A isoform a [Homo sapiens]|NP_001800.1; CSE-4 [Saccharomyces cerevisiae]|NP_012875.2; centromeric histone H3 (CenH3) [Zea mays]|NP_001105520.1; histone H3.1 [Homo sapiens]|NP_003522.1. The region of CENP-A targeting domain (CATD) was adopted from [163], which is the loop-1 and alpha helix 2 of the CENP-A histone-fold domain. The region of histone-fold domain of CSE-4 was adopted from [164]. The histone modifications are labeled with the position number followed by the type of modification (green box: methylation; blue box: phosphorylation; beige box: acetylation or ubiquitination)
Table 1.
Post-translational modifications of centromere proteins and their functions
| Protein | Modification | Responsible enzyme | Species | Function | Reference |
|---|---|---|---|---|---|
| CENP-A | Phosphorylations | ||||
| Serine 7 phosphorylation | Aurora A/B | Humans | Is required for cytokinesis completion and correct Aurora B, 14–3-3, PP1γ1, and CENP-C localization | [54, 56–58] | |
| Non-phosphorylable mutant can bind centromere, does not affect CENP-C localization, nor long-term survival | [59] | ||||
| Serine 16/18 phosphorylation | ? | Humans | Compacts CENP-A nucleosomal array | [51] | |
| Serine 18 phosphorylation | CDK2 | Humans | Inhibits CENP-A localization to centromere | [35] | |
| Serine 68 phosphorylation | CDK1 | Humans | Inhibits HJURP association and CENP-A premature loading | [36, 38] | |
| Expression of non-phosphorylable mutant can rescue CENP-A null cells | [39] | ||||
| Serine 50 phosphorylation | ? | Maize | ? | [60] | |
| Methylations | |||||
| Glycine 1 tri-methylation | RCC1? | Humans | Interacts with DNA | [51, 61] | |
| Lysine 124 monomethylation | ? | Humans | ? | [50, 60] | |
| Arginine 37 methylation | ? | Budding yeast | Recruits inner and linker kinetochore proteins | [42, 61] | |
| Ubiquitinations | |||||
| Lysine 124 ubiquitination | CUL4A-RBX1-COPS8 | Humans | Localizes to centromere | [30, 47] | |
| Polyubiquitination | Psh1 or Slx5 | Budding yeast | Targets CENP-A for proteolysis | [42, 43, 51] | |
| Monoubiquitination | CUL3/ RDX | Fruit flies | Stabilizes CENP-A | [30, 51] | |
| Others | |||||
| Lysine 124 acetylation | HAT p300? | Humans | Increases nucleosome accessibility to chromatin remodelers and disrupts C-terminal binding to CENP-C | [49, 50] | |
| Sumoylation | Siz1/Siz2 | Budding Yeast | Primes CENP-A for polyubiquitination-related degradation | [42, 50] | |
| HJURP | Serine 412, 448 and 473 phosphorylation | CDK1 | Humans | Weakens interaction with M18β | [62, 63] |
| M18BP1 | Serine/threonine 24 phosphorylation | CDK1/2 | Humans | Prevents premature CENP-A loading | [64] |
| Threonine 40 & serine 110 phosphorylation | CDK1 | Humans | Prevents M18α:Mis18β binding | [65] | |
| Threonine 653 phosphorylation | CDK1/2 | Humans | Inhibits Mis18BP1 centromere localization | [28] | |
| Phosphorylation | ? (Not CDK1/2) | Xenopus | Inhibits Mis18BP1 interaction with CENP-A in metaphase | [68] | |
| Mis18 complex | Phosphorylation | Plk1 | Humans | Localizes Mis18 complex and loads CENP-A | [67] |
Proper timing of CENP-A deposition
CENP-A is loaded in late mitosis to G1 phase in vertebrates [34]. The cell cycle-dependent phosphorylation state of CENP-A is proposed to be critical for timely CENP-A deposition. Human CENP-A serine 18 (S18) is phosphorylated by Cyclin E1/CDK2 from G1 to S phase [35] and serine 68 is phosphorylated by CDK1 in early mitosis [36]. Both phosphorylations at CENP-A S18 and S68 prevent CENP-A’s premature recognition by HJURP and CENP-A loading onto the centromere [35, 36]. Overexpressed phosphomimicking S68E CENP-A cannot associate with HJURP; whereas, overexpressed non-phosphorylatable S68A CENP-A can associate with HJURP, but it is occasionally mislocalized to non-centromeric regions [36–38]. On the other hand, LacI-fused non-phosphorylatable S68Q CENP-A has reduced HJURP recruitment to the lacO array at an ectopic site outside the context of the centromere, as compared to wild-type CENP-A [39]. Non-phosphorylatable S68Q CENP-A mutant can localize to centromeres, and expression of this mutant by retrovirus at endogenous level can rescue cell lethality of CENP-A-null cells, suggesting that S68 phosphorylation is dispensable for centromere maintenance [39]. During mitotic exit, CENP-A S68 is dephosphorylated by Protein Phosphatase 1 alpha (PPla), so that CENP-A can be loaded onto the centromere [36]. Drosophila and Xenopus also load CENP-A at late mitosis to G1 phase, using similar temporal regulation by phosphorylation [40, 41]. Further studies are needed to determine whether phosphorylations also control the CENP-A loading time outside of late mitosis to G1 phase in other species (e.g., budding yeast CENP-ACse-4 loading occurs in S phase [17]), and in species that have complete CENP-A turnover every cell cycle.
Eliminating mislocalized CENP-A
Ubiquitination of CENP-A helps to further ensure that CENP-A’s distribution is exclusively on the existing centromeres in budding yeast and Drosophila. In budding yeast, ectopically localized CENP-ACse4 is first subjected to sumoylation by E3 ligases Siz1 and Siz2, followed by polyubiquitination mediated by E3 ubiquitin ligase Slx5, and it is then targeted for proteolysis [42]. When CENP-ACse4 is overexpressed, another budding yeast E3 ubiquitin ligase, Psh1, polyubiquitinates mislocalized CENP-ACse4 [43]. The presence of CENP-ACse4 chaperone Scm3 reduces CENP-ACse4 ubiquitination by Psh1 in vitro, suggesting that Scm3 protects CENP-ACse4 from degradation [44]. In Drosophila, E3 ubiquitin ligase SCFPpa destabilizes CENP-ACID and regulates CENP-ACID chromatin localization [45]. A recent study in human cells revealed that CENP-A can localize to chromosome arm regions that are transcriptionally active before S phase [46]. CENP-A not bound by CENP-C is stripped off from the nucleosome when the DNA is replicated [46]. Whether CENP-A ubiquitination and the DNA replication machinery coordinate to evict CENP-A is not clear.
Effects on CENP-A nucleosomal structure
While polyubiquitination removes ectopically loaded CENP-A, monoubiquitination is found to stabilize CENP-A on the centromere in Drosophila and human cells [30, 47]. In Drosophila, CENP-ACID is stabilized by E3 ligase CUL3/RDX-mediated monoubiquitination in a CAL1-dependent manner, potentially when CAL-1 forms a pre-nucleosomal complex with CENP-ACID during mitosis [30]. In humans, CENP-A monoubiquitination on the highly conserved lysine 124 (K124) residue by E3 ligase CUL4A-RBX1/ROC1 promotes CENP-A dimerization, increases CENP-A’s affinity to HJURP, and is required both for the maintenance of old CENP-A and the recruitment of newly synthesized CENP-A [47]. However, in another study, the CENP-A K124R mutant is still competent for centromere maintenance [39, 47, 48]. Ubiquitination on additional residues on CENP-A might also be involved in maintaining CENP-A stability.
Interestingly, different modifications can be found on human CENP-A lysine 124. CENP-A K124 acetylation, which tightens the histone core, was suggested to mediate nucleosome sliding and increase centromere accessibility before DNA replication [49, 50]. In early S phase, CENP-A K124 was found to be monomethylated, which is proposed to avoid over-replication of centromeric DNA [50]. Therefore, even the same residue, e.g., K124, on CENP-A can undergo modification transitions to tailor for specific functions at different cell cycle stages, adding complexity to epigenetic regulations by PTMs.
Several other post-translational modifications on human CENP-A were also implicated in directly affecting CENP-A nucleosomal structure. For instance, CENP-A glycine 1 tri-methylation was suggested to aid the interaction of CENP-A with the underlying α-satellite centromeric DNA, and serine 16 and 18 phosphorylations are proposed to form a local secondary salt bridge structure that compacts the CENP-A nucleosomal array [51].
Localization of other kinetochore proteins and mitotic progression
One of the main functions of CENP-A is recruiting kinetochore proteins to the centromere. The kinetochore recruitment requires both CENP-A targeting domain (CATD) and either the carboxyl- or amino-terminal tail of the CENP-A protein in humans. H3 chimeras containing the CENP-A CATD domain, with either CENP-A amino-terminal tail (residues 1–29) or carboxyl-terminal tail (residues 135–140), were sufficient to recruit and maintain kinetochore proteins on human endogenous centromeres or on neocentromeres [52, 53]. This result suggested that kinetochore proteins are redundantly recruited and maintained by the tails of CENP-A together with the CENP-A CATD domain. For the short CENP-A carboxyl-terminal tail outside the conserved histone-fold domain, no specific post-translational modification has been reported to date. CENP-A amino-terminal tail deletion causes serious mitotic defect, but this can be rescued when CENP-A amino-terminal tail is substituted with that of H3, which is also phosphorylatable [54]. At the amino-terminal tail, phosphorylation of human CENP-A serine 7 by Aurora A and Aurora B appears in prophase, peaks in metaphase, and levels off in anaphase [55, 56]. Overexpression of serine 7 non-phosphorylatable CENP-A mutant has been shown to cause chromosome misalignment at metaphase [56], sister chromatid cohesion defects [57], reduced localization of CENP-C [54] and a phospho-binding protein, 14–3–3, from the centromere [54], dispersed Aurora B localization away from the inner centromere to chromosome arms in prometaphase [56, 57], and dispersed PP1γ1 from the midbody to the anaphase chromatids as well [58]. Moreover, overexpression of serine 7 phosphomimicking CENP-A mutant has abolished PP1γ1 localization at the midbody in anaphase [58]. CENP-A phosphorylated at serine 7 has been shown to bind 14–3–3, which is proposed to bridge CENP-A to CENP-C [54]. However, the importance of S7p remains elusive as a recent study using auxin-induced degradation (AID) and gene editing system revealed the opposite result, in which cells with only S7A CENP-A mutant do not display any abnormalities in centromere function, including CENP-C recruitment, nor long-term survival defects [59]. Barra et al. attributed the previous reported S7A defects to having a suboptimal level of CENP-A [59]. In maize, serine 50 phosphorylation of CENP-A has a similar kinetics as CENP-A S7 phosphorylation in humans, with a sharp reduction at metaphase–anaphase transition, but the function of S50 in maize is not well understood [60]. Moreover, budding yeast CENP-ACse4 methylation at arginine 37 may be involved kinetochore recruitment and centromere function, as the absence of this methylation (Cse4-R37A mutant) results in slightly reduced levels of kinetochore proteins (Mtw1 and Ame1) on the centromere in cbf1 deletion background [61]. Cse4-R37A mutant also results in an increased plasmid loss rate in cells with plasmid lacking CDEI element, and synthetic lethality or growth defect when combined with COMA (Ctf19-Okp1-Mcm21-Ame1) mutants or cbf1 mutant [61].
Post-translational modifications on other proteins important for CENP-A centromere targeting
Many proteins involved in the CENP-A chromatin assembly process are also post-translationally modified (Table 1). CENP-A chaperone HJURP serine 412, 448 and 473 phosphorylations by CDK1 weaken the interaction between HJURP and Mis18β [62], and a non-phosphorylatable HJURP mutant precociously deposits CENP-A on to the centromere [63]. CDK1 and CDK2 also phosphorylate the centromere licensing factor Mis18BP1 to suppress its centromere localization during S, G2, and M phase [64]. Phosphorylation of Mis18BP1 at T653 prevents premature centromere targeting at G2 [28]. Phosphorylation of Mis18BP1 at T40 and S110 reduces its affinity to Mis18α and Mis18β, and a phosphomimicking mutant prevents new CENP-A recruitment [65, 66]. The inhibition of HJURP and Mis18BP1 localization to centromere in turns restricts the centromere licensing time to late mitosis or early G1, and prevents premature CENP-A chromatin assembly. In contrast, Polo-like Kinase 1 (PLK1) phosphorylates human Mis18 complex to promote its centromere localization in early G1, contributing to CENP-A deposition [67]. Taking these results together, CENP-A deposition, at least in humans, is likely to be controlled by integrated phosphorylation and dephosphorylation signals from CDK1, CDK2 and PLK1. In Xenopus, phosphorylation of Mis18BP1 also inhibit its association with CENP-A nucleosomes during metaphase, but this phosphorylation is independent of CDK [68]. It is worth to note that many of the above findings are yielded from organisms like humans and Drosophila, which have their CENP-A loaded in late G1 or anaphase [34, 40]. As the loading time of CENP-A varies among species, it will be interesting to further investigate whether the CENP-A loading mechanisms, as well as their regulations, are conserved in different species.
Histone modifications important for centromere function
Centromeric and pericentromeric histones H3 and H4 acetylation status
Many centromeric histone modifications have been identified as pertinent to centromere functions (Table 2). Histone acetylation, as commonly found at actively transcribed regions in euchromatin, neutralizes the positive charge in histones, weakening histone–DNA interactions, thereby opening up the chromatin [69]. One of the earliest studies in barley has shown that centromeric histones H3/H4 are hypoacetylated in mitosis [70]. In fission yeast, centromeric histones H3/H4 are maintained in hypoacetylated states by RbAp46/48Mis16 and Mis18 [20]. In budding yeast, H4 in the CENP-ACse4 nucleosome is hypoacetylated at K16 by Sir2, and this hypoacetylation is required to maintain kinetochore integrity and accurate chromosome segregation [71]. In humans and Drosophila, the repetitive centromeric chromatin also contains hypoacetylated H3/H4 [3]. The general hypoacetylated state at the centromere is consistent with the cytological observation that the centromeres are the more condensed part of the chromosomes.
Table 2.
Histone modifications of centromeric core chromatin and pericentric heterochromatin
| Location | Modifications | Function | Species | Reference |
|---|---|---|---|---|
| Centromeric and Pericentromeric | H3/H4 hypoacetylation | Promotes condensation of centromeric chromatin in barley; maintains kinetochore integrity and accurate chromosome segregation in budding yeast | Barley, fission yeast, budding yeast, humans, Drosophila | [3, 20, 70, 71] |
| Centromeric | H3 lysine 4 di-methylation | Targets HJURP to the centromere and maintains kinetochore | Humans, Drosophila, Arabidopsis, fission yeast, chicken, rice | [6, 73, 78, 79, 83] |
| H3 lysine 9 di- or tri-methylation | ? | Rice, chicken | [6, 87] | |
| H3 lysine 27 tri-methylation | ? | C. elegans | [18] | |
| H3 threonine 3 phosphorylation | Recruits CPC via Survivin binding and important for Aurora B function | Fission Yeast | [90, 92, 93] | |
| H2A serine 121/ threonine 120 or 133 phosphorylation | Localizes CPC component Shugoshin to centromere in fission yeast | Fission yeast, maize, wheat, rice, barley, L. elegans, humans | [91, 94–97] | |
| H4 acetylation | Primes the centromere prior to CENP-A deposition in humans | Rice, humans | [73, 74] | |
| H4 lysine 5 and 12 acetylations | Are required for CENP-A deposition | Chicken | [72] | |
| H4 lysine 20 monomethylation | Is required for CENP-T binding | Humans, chicken | [88] | |
| Pericentric | H3 lysine 9 di or tri-methylation | Is crucial for mitotic progression, sister chromatids cohesion, and kinetochore–microtubules interaction; Is required for heterochromatin formation (see Table 4) | Humans, Drosophila, Arabidopsis, fission yeast, maize | [3, 4, 79–82, 84, 85, 149, 164, 166] |
On the other hand, H4 lysine 5 and lysine 12 acetylations, which were dependent on RbAp48-HAT1 complex but not Mis18, were found to be enriched in non-repetitive centromeres in chicken DT40 cells [72]. Consistently, H4 acetylations (at lysine 5, 8, 12, and 16) were found in rice centromeres where active genes are present [73]. H4 acetylation mediated by the human Mis18 complex was suggested to prime or license the centromere prior to CENP-A deposition [74]. A study in barley has shown that centromeric H4 lysine 5 acetylation is dynamic across the cell cycle, in which H4 lysine 5 deacetylation in metaphase is proposed to promote condensation of centromeric chromatin [75] (See [76, 77] for reviews focusing on the dynamics of histone modifications).
Histone modifications at centromeric core versus pericentric heterochromatin
As mentioned in the introduction, CENP-A-containing centromeric core in regional centromeres is usually flanked by pericentric heterochromatin. Multiple approaches, including high-resolution imaging of stained chromatin fibers, pull-down of nucleosome using CENP-A antibody, and chromatin immunoprecipitation (chIP), have allowed us to distinguish the centromeric (CEN) chromatin from the pericentric heterochromatin by having distinct histone modification patterns. For example, in humans and Drosophila, while their CEN chromatin contains H3 lysine 4 dimethylation (H3K4me2), the pericentric heterochromatin consists of H3 lysine 9 di- or trimethylation (H3K9me2 or H3K9me3) [3, 78]. These modification patterns are consistently reported in other model organisms like the flowering plant Arabidopsis thaliana, fission yeast and maize [79–82]. A study in human artificial chromosomes (HACs) has demonstrated that H3K4me2 is required for targeting HJURP to the centromere and for kinetochore maintenance [83]. Mitotic-specific H3K9me3 at pericentric heterochromatin is crucial for mitotic progression and kinetochore–microtubule interaction in human cells [84, 85]. However, such distinct centromeric and pericentric histone modification pattern seems to be less obvious in chicken. In chicken DT40 cells, in which the centromeric chromatin are also mostly repetitive, H3K9me3 was robustly observed flanking the CENP-A subdomains on stretched chromatin fibers, but only low levels of H3K4me2 were detected in CEN chromatin by super-resolution microscopy [6]. Despite the difference in H3K9me3 pattern in human and chicken DT40 pericentric chromatin, one similarity among them is histone H4 lysine 20 monomethylation (H4K20me1) in CENP-A nucleosomes. H4K20me1 is usually associated with active transcription and is required for kinetochore assembly, probably through CENP-T binding [88, 89].
While H3K9me3 is enriched on chicken DT40 repetitive centromeres, it is not enriched on non-repetitive centromeres (Chromosome 5, 27 and Z) or neocentromeres [13]. In fact, the absence of H3K9me3 at non-repetitive neocentromeres has also been reported in a human cell line IMS13q [86]. On the other hand, on centromere 8 (Cen8) in rice, which contains a low abundance of highly repetitive satellite DNA, CENP-A centromeric chromatin is embedded in a larger H3K9me2 region [73, 87]. This whole H3K9me2 region, including CENP-A chromatin, is low in H3K4me2 [87], which is different from that in humans, flies and HACs.
Other than methylations, histone phosphorylations have also been reported. In maize, H3 serine 28 phosphorylation (H3S28P) occurs at pericentric chromatin from prometaphase to anaphase, and is proposed to function in sister chromatid cohesion [60]. In fission yeast pericentric chromatin, mitotic histone kinase Haspin phosphorylates H3 threonine 3 (H3T3P) [90], while spindle checkpoint kinase Bub1 phosphorylates H2A serine 121 (H2AS121P) [91]. These two phosphorylations are important for recruiting components in the chromosomal passenger complex (CPC), including Aurora B, and the cohesin protector, Shugoshin, to mediate sister chromatid bi-orientation and cohesion [92, 93]. In humans, the corresponding modification of H2AS121P is H2A threonine 120 phosphorylation (H2AT120P), which is also recognized by Shugoshin at the pericentric heterochromatin [94]. The corresponding phosphorylation in other plant species is either H2AT120P or H2AT133P [95–97]. In barley and Luzula elegans, H2AT120P is present on the centromeres, with a gap in between the sister chromatids, as shown by structured illumination microscopy [97]. Despite its different distribution and unclear function in cohesion in plants, this H2A modification represents a common epigenetic mark for centromeric chromatin in both monocentric (e.g., tomato, barley, rye, tobacco) and holocentric plant species (e.g., L. luzuloides and L. elegans)[96].
For the nematode C. elegans, which also have holocentromeres, CENP-AHCP−3 domains are positively correlated (r = 0.64) with heterochromatin mark H3K27me2/3, and negatively correlated (r = − 0.6) with actively transcribed mark H3K36me3, but the causal relationships between these histone modifications and CENP-AHCP-3 localization and their functions on the centromeres are not clear [18]. Taken together, some histone modification patterns at centromeric chromatin and pericentric heterochromatin are conserved among organisms, while some are unique. Their exact roles in specific species deserve further investigation.
Centromeric transcription
As a constricted region on the mitotic chromosome bound by kinetochores [98], the centromere was first conceived to have minimal transcriptional activity. Surprisingly, as opposed to this early speculation, transcription at the centromere is found to be compatible with centromere’s function in many species.
Centromeric transcripts as a structural and regulatory component for the centromere
Transcripts from the centromere have been detected, and are found to be required for centromere function in budding yeast [99, 100], fission yeast [101, 102], mammals [103, 104], insects [105, 106] and plants [73, 107]. The centromere-derived RNAs found in different organisms are heterogeneous in terms of their size, stability and the cell cycle timing in which they are transcribed (Table 3). RNAs originated from centromere can be as short as 21–23 nt for short interfering (si)RNAs in fission yeast, or over 5 kb for long non-coding (lnc) RNAs in humans [100, 105, 107, 108]. (See [109, 110] for reviews on the classifications of centromeric transcripts and their potential functions).
Table 3.
Organisms with known CENP-A containing centromeric transcription and its function
| Species | Transcription site | Enzyme | RNA Class | Centromeric function or characteristic of the RNA | Reference |
|---|---|---|---|---|---|
| Budding Yeast (Saccharomy-ces cerevisiae) | CEN DNA | RNAPII | > 200 nt non-coding RNA | Optimal expression level is required to maintain minichromosome stability and centromeric protein levels | [99, 100] |
| Fission Yeast (Schizosacch-aromyces pombe) | Central domain | RNAPII & Chd1 | ~ 0.5 kb non-coding RNA | Highly turnovered centromeric transcripts were detected in exosome mutants or centromeric protein mutants | [167] |
| Maize (Zea mays) | Centromeric CRM and CentC repeats | ? | 40–200 nt single stranded RNA | Associates tightly with the kinetochore | [107] |
| Promotes CENP-C localization to the kinetochore | [113] | ||||
| Beetles (Palorus ratzeburgii) | Centromeric PRAT repeats | RNAPII | 0.5–5 kb non-coding RNA | Is highly expressed and found in nucleus but function unknown | [105] |
| Fruit fly (Drosophila melanogaster) | Chromosome X centromeric Satellite III repeats | RNAPII & FACT | - | Is required for CENP-A loading | [106, 124] |
| 0.36–1.3 kb long non-coding RNA | Is required for CENP-A, CENP-C and kinetochore proteins localization, proper chromosome segregation and male-specific lethal complex localization | ||||
| Mouse (Mus musculus) | Minor satellites (Centromeric core) | ? | 200–400 nt non-coding RNA | Associates with CENP-A chromatin, Aurora B and Survivin in CPC and is required for Aurora B activity | [103, 114] |
| 120 nt non-coding RNA | Forced accumulation leads to chromosome missegregation and loss of sister-chromatid cohesion and mislocalizes Aurora B centromeric localization | [103, 114] | |||
| Tammar wallaby (Macropus eugenii) | Centromere-enriched retroelements (LINE, SINE, transposons) | ? | 35–42 nt Crasi RNA | Localizes to the centromere; ssRNAs and dsRNAs are required for CENP-B localization | [167] |
| Human (Homo sapiens) | Centromeric α-satellites | RNAPII | - | High level transcription inhibits kinetochore formation in HAC | [141] |
| 1.3 kb non-coding RNA | Interacts with CPC, CCAN components: INCENP and Aurora B | [104] | |||
| RNA knockdown increases Aurora B activity | [104] | ||||
| Is required for centromeric CENP-C, INCENP and Survivin loading and nucleolus CENP-C and INCENP targeting | [112] | ||||
| Frog (Xenopus laevis) | Frog Centromeric Repeat 1 | ? | 170 nt non-coding RNA | Associates with half of the centromeres and is required for normal Aurora-B localization to the inner centromere | [115] |
Centromeric ncRNAs may serve a structural or scaffolding role in the assembly of the kinetochore complex or in centromere targeting (Fig. 2d). The lncRNAs were found to associate with soluble HJURP and CENP-A, and the association is required for CENP-A centromere targeting [108]. A recent study also revealed that the α-satellite lncRNAs are cis-acting [111]. On the other hand, human centromeric RNAs have been found to bind and keep kinetochore proteins in the nucleolus [112]. The authors proposed that centromeric RNAs facilitate the preassembly of kinetochore proteins at the nucleolus, prior to their localization at the centromere. Centromeric RNAs’ structural role is likely conserved in other species. Maize centromeric ncRNAs co-precipitated with CENP-A chromatin [107]. CENP-C requires centromeric ncRNAs for its localization to maize, Drosophila and human centromeres [106, 112, 113].
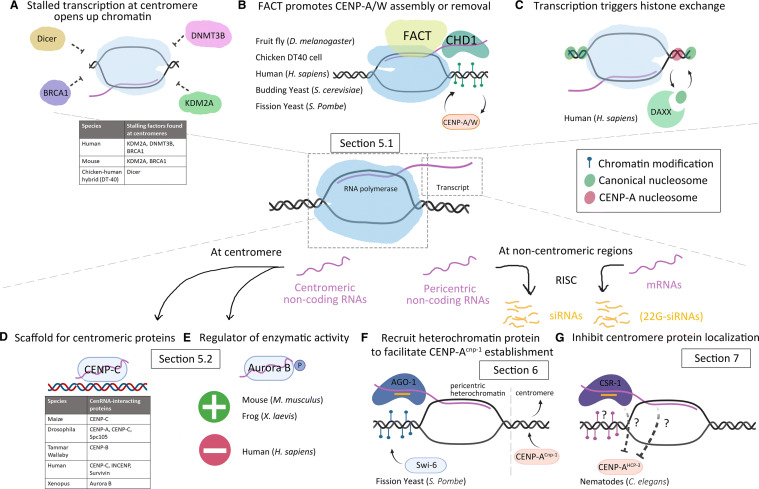
Fig. 2.
Proposed mechanisms of centromere regulation via transcription. (Top) Transcription process is proposed to introduce chromatin modifications by opening up chromatin, facilitating chromatin modifiers binding, and promoting histone exchange. a Centromeric RNAPII is associated with different stalling factors (see table), which may maintain an open chromatin state. b FACT is found on the centromere in human, chicken DT40 cells, and Drosophila [122–124]. In DT-40 cells, chromatin remodeling factor CHD1 is associated with FACT [123]. In Drosophila, FACT is required for new CENP-ACID loading [124]. In budding yeast, CENP-A degradation by E3 ubiquitin ligase requires FACT [126]. In fission yeast, FACT represses ectopic CENP-A localization [125]. c Histone exchange during transcription process is proposed to deposit new CENP-A or to remove ectopic CENP-A. Human transcription coupled H3.3 chaperone ATRX/DAXX is associated with ectopic centromere in human cancer cells [135]. (Bottom) Transcripts originated from centromeric or non-centromeric region have different forms and diverse effects in different organisms. d Long or short non-coding RNAs at the centromere are required for the targeting of some centromeric proteins (see table). e Centromere transcripts help to modulate Aurora B activity. The Aurora B kinase activity is promoted by cenRNAs in mouse and frog [114, 115], and is suppressed by cenRNAs in human cells [104]. f Non-coding RNAs are processed into short interfering RNAs (siRNAs) by the RNAi silencing complex (RISC), which can guide Argonaute binding onto the nascent transcript. In fission yeast, the AGO-1 RNAi machinery is involved in the recruitment of histone methyltransferase Clr4 for H3K9 methylation. H3K9me3 is then recognized by HP1 homolog, Swi6 [139, 149–152]. Swi6 is important in establishing pericentric heterochromatin and facilitating initial CENP-ACnp1 establishment in de novo centromeres. (G) In nematodes, the active transcribed regions are anti-correlated with the CENP-AHCP-3 localization [18]. CSR-1 RNAi pathway produces 22G-RNAs from germline mRNAs [160], which may facilitate chromatin modifications and inhibit HCP-3 binding. Related sections are highlighted for detailed descriptions and references
Centromeric non-coding RNAs may also serve a regulatory role in the activity of Aurora B kinase in CPC (Fig. 2e). Human α-satellite RNAs suppress Aurora B’s kinase activity [104]. In contrast, mouse minor satellite RNAs and Xenopus egg extracts’ centromeric non-coding transcripts fcr1 are required for Aurora B’s kinase activity [114, 115]. Despite the observed opposing effects on Aurora B function, knockdown of centromeric RNAs in humans, mice and Xenopus all leads to improper spindle attachments and chromosome missegregation [103, 104, 116]. Yet, further investigations are needed to reveal the exact role of centromeric RNAs in regulating centromeric protein activity and understand the mechanistic differences among organisms.
Transcription-coupled chromatin modifications
In line with the discovery of centromere transcripts, the process of transcription itself may have a role in centromere function. According to the characterization of the promoter sequence, the 5′ cap structure and the 3′ poly-A tail of the centromeric and pericentric RNAs, RNAPII is found to be responsible for their production in fission and budding yeast, beetles, and humans [99, 105, 117–120]. In humans, Bub1 recruits RNAPII to the centromeres, and the resulting transcription is important for the accumulation of Shugoshin [94]. Elongating RNAPII (phosphorylated at Ser2), RNAPII-specific transcription factor Carboxyl-terminal domain phosphatase 1 (CTDP1), and a FACT subunit Structure Specific Recognition Protein (SSRP1) are found to localize at the centromere on human mitotic chromosomes [119, 121]. RNAPII inhibition by α-amanitin reduces centromeric transcription and compromises kinetochore function in humans and beetles [105, 119]. Nonetheless, how RNAPII transcription functions at the centromere remains obscure. Potentially, RNAPII may serve as a platform to recruit chromatin modifiers, or stall to shape the chromatin environment (Fig. 2a). Alternatively, transcriptionally compatible chromatin may promote histone exchange by associating with the corresponding histone chaperones to facilitate CENP-A loading.
FACT associated with RNAPII was shown to facilitate centromere protein recruitment. FACT binds to the histone-fold domain of CENP-W directly, and stimulates CENP-T/W complex deposition at the centromere region in human cells [122] (Fig. 2b). In chicken DT40 cells, CENP-H recruits SSRP1 and its interacting chromatin remodeling factor CHD1 to the centromeres, which together facilitates the deposition of new CENP-A [123]. In Drosophila, FACT is recruited by the CENP-ACID chaperone, CAL-1, to promote RNAPII transcription, which destabilizes H3 nucleosomes and facilitates new CENP-ACID loading [124]. In fission yeast, FACT deletion induces the deposition of CENP-ACnp1 onto ectopic sites only, but does not affect CENP-ACnp1 on the endogenous centromeric regions, suggesting that FACT may act by promoting the correct localization of histone H3 in the genome [124, 125]. Consistently, in budding yeast, the degradation of CENP-ACse4 by E3 Ubiquitin ligase Psh1 requires FACT [126].
Evidences suggested that RNAPII may be stalled at centromeres. Depletion of RNAPII transcription restart factor Ubp3 or TFIIS leads to increased CENP-ACnp1 deposition onto fission yeast centromeres, suggesting that restart factors normally restricts CENP-ACnp1 deposition [127]. Potential transcription elongation repressors, including BRCA1, KDM2A, Dicer, and DNMT3B were found to localize to vertebrate centromeres [128–131] (Fig. 2a). The retention of RNAPII at the centromere may open up the centromeric DNA and facilitate the recruitment of CENP-A deposition factors or chromatin modifiers.
Transcription-coupled histone exchange is important for the turnover of some histone variants that are replication independent, such as CENP-A [132]. As DNA is being transcribed, histones are temporarily removed and reassembled, providing a chance to reload histones and incorporate specific variants with the help of chaperones that are associated with the RNA polymerase [133]. Centromeric chromatin is proposed to be pre-occupied by histone variant H3.3 after DNA replication and prior to new CENP-A loading [134]. This placeholder, H3.3, is loaded independent of replication, but is coupled to transcription [109, 134]. Besides, ectopic CENP-A domains in human cancer cells are found to be associated with transcription-coupled H3.3 chaperones ATRX/DAXX [135] (Fig. 2c). Whether transcription facilitates the replacement of H3.3 by CENP-A at a specific cell cycle stage remains to be determined.
Regulation of centromeric transcription
The transcription of centromeric RNAs has to be maintained but kept at a low level. At human centromeres, there is transcriptionally permissive mark H3K4me2, but not H3K4me3, which is a strong transcription histone mark [83]. Knocking down histone H3 lysine 4 demethylase KDM5A reduced α-satellite RNAs levels by half and resulted in genomic instability in humans [136]. Moreover, tethering a transcription suppressor to HAC α-satellites destabilizes the HAC, accompanied by a loss of H3K4me2 and a gain of H3K9me3 [83, 137]. This inactivation of HAC centromere can be rescued by p65-induced, H3K9ac-associated transcription [138]. Thus, the centromeric transcription level can be regulated by the landscape of histone modifications on the chromatin. In budding yeast, blocking centromeric transcription by lacI binding to lacOs flanking the centromere resulted in increased minichromosome loss [100]. These studies suggest that a low level of centromeric transcription is favorable for centromere function.
On the other hand, centromere function seems to be incompatible with strong centromeric transcription. Neocentromere formation repressed the expression of nearby genes in fission yeast [139, 140]. Conversely, induced expression of proximal genes repositioned the neocentromere in the yeast Candida albicans [141]. Therefore, the activity of the centromeres and nearby transcription may affect each other. Overexpression of the centromeric transcripts caused by adding a strong promoter [103, 142, 143], tethering a transcriptional activator to the centromere [137, 144], or removal of DNA methylation by DNMT3B depletion [83] also leads to chromosome missegregation. Even for budding yeast centromeres, which are originally thought to be genetically regulated, induced overexpression of centromeric RNAs reduces chromatin association of kinetochore proteins [145]. Overexpression of centromeric RNAs by deletion of centromeric transcription repressor, H2A.ZHtz1 or Cbf1, leads to reduced levels of centromeric CENP-ACse−4, CENP-CMif2 and AuroraBIpl[100]. It was proposed that the overexpressed centromeric RNAs may titrate centromeric proteins away and rendered their mislocalizations [103]. Knocking down centromeric RNAs in htz1 or cbf1 deletion mutant partially rescues minichromosome loss, suggesting a balanced level of centromeric RNAs is needed for centromere function [100]. In addition, centromeric transcript level was found to be up-regulated under stress conditions and in human cancer cells [131, 146]. The accumulation of centromeric transcripts may result from defects in RNA processing by exosome, as some centromeric RNAs, such as those in fission yeast, are known to have a high turnover rate [147, 148].
Transcription at pericentric heterochromatin
Non-coding transcription at the pericentric region remodels the chromatin to form heterochromatin and facilitate gene silencing. In fission yeast, Argonaute-bound, pericentromere-derived siRNAs recruit histone methyltransferase Clr4 to methylate H3 at lysine 9 [139, 149–152]. H3K9me in turn is recognized by the heterochromatin protein 1 (HP1) homolog, Swi6, which sets the boundary for pericentric and centromeric regions [81]. The pericentric heterochromatin is required for the initial establishment of CENP-ACnp1 domain, but not for its maintenance in fission yeast [81, 153] (Fig. 2f).
The links between the non-coding transcripts, the RNAi pathway, and heterochromatin formation may be conserved in other organisms. In mouse and chicken–human hybrid DT40 cells, knockdown of an RNAi component has resulted in an accumulation of long transcripts, destabilization of HP1 and reduction of H3K9 methylation at the pericentromere [128, 154, 155]. Pericentric heterochromatin formation is required for CENP-A loading in mice and Drosophila, but is dispensable in chicken–human hybrid cells, C. albicans, and C. elegans, suggesting that heterochromatin is not a universal prerequisite for centromere function [128, 141, 156–158, 160]. However, other siRNAs that originate from centromeric or pericentric chromatin may be involved in regulating centromere function (Table 4).
Table 4.
Relationship between transcription, RNAi, heterochromatin assembly and centromere function
| Species | Transcription site | RNAi pathway | Heterochromatin function of the transcription/ RNAi or phenotypes in mutant/ overexpression | Reference |
|---|---|---|---|---|
| Fission Yeast (Schizosaccharo-myces pombe) | Pericentric outer repeats dg region | Ago1 | RISC associates with Stc1 to recruit histone methyltransferase Clr4 for H3K9 methylation, H3K9me3 is then recognized by HP1 homolog Swi6, whereas H3K9me2 is important to maintain the siRNAs accumulation. Defective RNAi leads to chromosome missegregation | [101, 102, 150, 152, 153, 166, 169] |
| Transfer RNA alanine gene at the centromeric and pericentric boundary by RNA Pol III | - | Prevent spreading of heterochromatin | [170] | |
| Fruit fly (Drosophila melanogaster) | ? | Piwi | Piwi RNAi pathway is required for heterochromatin silencing and HP1 localization | [158, 171] |
| Chicken DT40-human hybrid cells | α-satellites | ? | Dicer deficient cells mislocalize cohesin Rad21 and Bub1, but do not affect CENP-A and CENP-C | [128] |
| Arabidopsis thaliana | 180 bp centromeric satellite repeats | ? | Dicer mutant abolishes 23–24 nt centromeric small RNAs, but do not affect H3K9me2 nor chromosome segregation | [79] |
| Tammar wallaby (Macropus eugenii) | Centromere-specific repetitive satellites | ? | Crasi RNAs are required for recruiting H3K9me3 at pericentric heterochromatin | [168] |
| Mouse (Mus musculus) | Minor satellites (Centromeric core) | ? | Dicer-null cells inhibit cell proliferation but not histone modification at the pericentromere | [154, 155] |
| Dicer knockdown reduces viability and differentiation ability | [155, 156] | |||
| Accumulation of 120nt RNAs affect distribution of heterochromatin marks H3K9me and HP1 | [103, 155] | |||
| Major satellites (Pericentromere) | ? | Histone methyltransferase Suv39h promotes H3K9me at pericentromere, and is required for viability and chromosome stability | [172] | |
| Association of HP1 to pericentromere is RNA-dependent | [154] | |||
| Human (Homo sapiens) | α-satellites | AGO2 | The AGO2 slicer activity is required for proper chromosome segregation | [173, 174] |
| Association of HP1a to pericentromere is RNA-dependent | [174] | |||
| Nematode (Caenorhabditis elegans) | Germline genes 22G-siRNA | CSR-1 | Is required for proper chromosome segregation, kinetochore organization and P-granule structure maintenance | [160] |
Relationship of the centromere with other RNA interference pathways
In holocentric C. elegans, CENP-AHCP−3 domains are inversely correlated with the transcribed regions in the germline, which are loci targeted by antisense 22G-RNAs generated by the Argonaute CSR-1 RNAi pathway [18, 160]. CSR-1 protects its targets from being silenced by other RNAi pathways and optimizes the target transcript levels [161, 164]. Disruption of CSR-1 RNAi pathway leads to chromosome missegregation, which is proposed to be caused by its target expression level changes [160]. However, CSR-1 and 22G-RNAs may potentially also affect CENP-AHCP−3 organization at the chromatin level (Fig. 2g).
Conclusion
Since the identification of CENP-A as a fundamental mark at the centromere, efforts have been made to unravel the mechanism that replenishes CENP-A after DNA replication, and hence maintains the centromere identity in every cell cycle. While many protein factors directly interacting with CENP-A have been identified, the post-translational modifications, chromatin environment, and transcriptional status that regulate precise temporal and spatial centromere assembly and function are just beginning to be understood.
Post-translational modifications of centromeric proteins, centromeric and pericentric histones are involved in governing the centromere function. These modifications can be highly dynamic with respect to the cell cycle phases. Phosphorylation on CENP-A and centromeric proteins often controls their physical interactions and deposition timing. Ubiquitination on CENP-A is crucial for its stability and restricting its localization. Methylations, acetylations and phosphorylations on specific centromeric histone proteins can influence the localization of other centromeric proteins.
While the functions of many histone modifications on the centromere are not completely clear yet, they can regulate centromeric transcription. Non-coding centromeric transcription event has been shown to be essential in several organisms. Most of the centromeric RNAs are transcribed by RNAPII at a tightly controlled, relatively low level. The transcription activity may function to recruit chromatin modifiers for regulating the centromeric chromatin environment, or the centromeric RNAs may function to facilitate kinetochore function as a structural or regulatory component. At the pericentric heterochromatin flanking the centromeric core, transcription, RNAi pathway and specific histone modifications may cooperate to facilitate sister chromatid cohesion, CENP-A loading or centromere establishment. Besides CENP-A, centromeric and pericentric histone modifications and transcription may function as conserved epigenetic mechanisms to create a favorable chromatin environment for centromere specification.
Following the discoveries of post-translational modifications of centromeric proteins, centromeric chromatin and transcriptional environment, future studies are anticipated to reveal more specific molecular functions of these epigenetic factors on centromere function, and the relationships among these different epigenetic pathways. How the centromeric marks are first established and then maintained? An active, functional centromere is likely to be crucial for the inheritance of these epigenetic marks, possibly constructing a positive feedback mechanism. It will be interesting to study how a functional centromere signals and regulates histone modifications and centromeric transcription, and it will be fascinating to understand how chromosomes are accurately transmitted from the beginning of eukaryotic life, via the propagation of epigenetic marks that define the centromere.
Acknowledgements
This work was supported by the General Research Grant [grant numbers 17126717, 17113418] and the Collaborative Research Fund [grant numbers C7058-18G]. We thank our lab members for critical reading and helpful suggestions on the manuscript.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Furuyama S, Biggins S. Centromere identity is specified by a single centromeric nucleosome in budding yeast. Proc Natl Acad Sci. 2007;104(37):14706–14711. doi: 10.1073/pnas.0706985104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Henikoff S, Ahmad K, Malik HS. The centromere paradox: stable inheritance with rapidly evolving DNA. Science. 2001;293(5532):1098–1102. doi: 10.1126/science.1062939. [DOI] [PubMed] [Google Scholar]
- 3.Sullivan BA, Karpen GH. Centromeric chromatin exhibits a histone modification pattern that is distinct from both euchromatin and heterochromatin. Nat Struct Mol Biol. 2004;11(11):1076–1083. doi: 10.1038/nsmb845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bernard P, Maure JF, Partridge JF, Genier S, Javerzat JP, Allshire RC. Requirement of heterochromatin for cohesion at centromeres. Science. 2001;294(5551):2539–2542. doi: 10.1126/science.1064027. [DOI] [PubMed] [Google Scholar]
- 5.Blower MD, Sullivan BA, Karpen GH. Conserved organization of centromeric chromatin in flies and humans. Dev Cell. 2002;2(3):319–330. doi: 10.1016/s1534-5807(02)00135-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ribeiro SA, Vagnarelli P, Dong Y, Hori T, McEwen BF, Fukagawa T, Flors C, Earnshaw WC. A super-resolution map of the vertebrate kinetochore. Proc Natl Acad Sci USA. 2010;107(23):10484–10489. doi: 10.1073/pnas.1002325107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jin W, Lamb JC, Zhang W, Kolano B, Birchler JA, Jiang J. Histone modifications associated with both A and B chromosomes of maize. Chromosome Res. 2008;16(8):1203–1214. doi: 10.1007/s10577-008-1269-8. [DOI] [PubMed] [Google Scholar]
- 8.Aldrup-MacDonald ME, Sullivan BA. The past, present, and future of human centromere genomics. Genes. 2014;5(1):33–50. doi: 10.3390/genes5010033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Voullaire LE, Slater HR, Petrovic V, Choo KH. A functional marker centromere with no detectable alpha-satellite, satellite III, or CENP-B protein: activation of a latent centromere? Am J Hum Genet. 1993;52(6):1153–1163. [PMC free article] [PubMed] [Google Scholar]
- 10.Amor DJ, Choo KH. Neocentromeres: role in human disease, evolution, and centromere study. Am J Hum Genet. 2002;71(4):695–714. doi: 10.1086/342730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marshall OJ, Chueh AC, Wong LH, Choo KHA. Neocentromeres: new insights into centromere structure, disease development, and karyotype evolution. Am J Hum Genet. 2008;82(2):261–282. doi: 10.1016/j.ajhg.2007.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Earnshaw WC, Rothfield N. Identification of a family of human centromere proteins using autoimmune sera from patients with scleroderma. Chromosoma. 1985;91(3–4):313–321. doi: 10.1007/BF00328227. [DOI] [PubMed] [Google Scholar]
- 13.Shang W-H, Hori T, Martins NM, Toyoda A, Misu S, Monma N, Hiratani I, Maeshima K, Ikeo K, Fujiyama A. Chromosome engineering allows the efficient isolation of vertebrate neocentromeres. Dev Cell. 2013;24(6):635–648. doi: 10.1016/j.devcel.2013.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fukagawa T, Earnshaw WC. Neocentromeres. Curr Biol. 2014;24(19):R946–R947. doi: 10.1016/j.cub.2014.08.032. [DOI] [PubMed] [Google Scholar]
- 15.Cheeseman IM, Desai A. Molecular architecture of the kinetochore–microtubule interface. Nat Rev Mol Cell Biol. 2008;9(1):33–46. doi: 10.1038/nrm2310. [DOI] [PubMed] [Google Scholar]
- 16.Shelby RD, Monier K, Sullivan KF. Chromatin assembly at kinetochores is uncoupled from DNA replication. J Cell Biol. 2000;151(5):1113–1118. doi: 10.1083/jcb.151.5.1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pearson CG, Yeh E, Gardner M, Odde D, Salmon ED, Bloom K. Stable kinetochore-microtubule attachment constrains centromere positioning in metaphase. Curr Biol. 2004;14(21):1962–1967. doi: 10.1016/j.cub.2004.09.086. [DOI] [PubMed] [Google Scholar]
- 18.Gassmann R, A Rechtsteiner, KW Yuen, A Muroyama, T Egelhofer, L Gaydos, F Barron, Maddox P, A. Essex, J. Monen, S. Ercan, JD Lieb, K Oegema, S Strome, Desai AP (2012) An inverse relationship to germline transcription defines centromeric chromatin in C. elegans. Nature, 484(7395), 534–7. [DOI] [PMC free article] [PubMed]
- 19.Nechemia-Arbely Y, Fachinetti D, Cleveland DW. Replicating centromeric chromatin: spatial and temporal control of CENP-A assembly. Exp Cell Res. 2012;318(12):1353–1360. doi: 10.1016/j.yexcr.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hayashi T, Fujita Y, Iwasaki O, Adachi Y, Takahashi K, Yanagida M. Mis16 and Mis18 are required for CENP-A loading and histone deacetylation at centromeres. Cell. 2004;118(6):715–729. doi: 10.1016/j.cell.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 21.Dunleavy EM, Roche D, Tagami H, Lacoste N, Ray-Gallet D, Nakamura Y, Daigo Y, Nakatani Y, Almouzni-Pettinotti G. HJURP is a cell-cycle-dependent maintenance and deposition factor of CENP-A at centromeres. Cell. 2009;137(3):485–497. doi: 10.1016/j.cell.2009.02.040. [DOI] [PubMed] [Google Scholar]
- 22.Foltz DR, Jansen LE, Bailey AO, Yates JR, 3rd, Bassett EA, Wood S, Black BE, Cleveland DW. Centromere-specific assembly of CENP-a nucleosomes is mediated by HJURP. Cell. 2009;137(3):472–484. doi: 10.1016/j.cell.2009.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Camahort R, Li B, Florens L, Swanson SK, Washburn MP, Gerton JL. Scm3 is essential to recruit the histone h3 variant cse4 to centromeres and to maintain a functional kinetochore. Mol Cell. 2007;26(6):853–865. doi: 10.1016/j.molcel.2007.05.013. [DOI] [PubMed] [Google Scholar]
- 24.Williams JS, Hayashi T, Yanagida M, Russell P. Fission yeast Scm3 mediates stable assembly of Cnp1/CENP-A into centromeric chromatin. Mol Cell. 2009;33(3):287–298. doi: 10.1016/j.molcel.2009.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen C-C, Dechassa ML, Bettini E, Ledoux MB, Belisario C, Heun P, Luger K, Mellone BG. CAL1 is the drosophila CENP-A assembly factor. J Cell Biol. 2014;204(3):313–329. doi: 10.1083/jcb.201305036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Furuyama T, Dalal Y, Henikoff S. Chaperone-mediated assembly of centromeric chromatin in vitro. Proc Natl Acad Sci. 2006;103(16):6172–6177. doi: 10.1073/pnas.0601686103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee BCH, Lin Z, Yuen KWY. RbAp46/48 LIN-53 is required for holocentromere assembly in caenorhabditis elegans. Cell Rep. 2016;14(8):1819–1828. doi: 10.1016/j.celrep.2016.01.065. [DOI] [PubMed] [Google Scholar]
- 28.Stankovic A, Guo LY, Mata JF, Bodor DL, Cao X-J, Bailey AO, Shabanowitz J, Hunt DF, Garcia BA, Black BE. A dual inhibitory mechanism sufficient to maintain cell-cycle-restricted CENP-A assembly. Mol Cell. 2017;65(2):231–246. doi: 10.1016/j.molcel.2016.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.An S, P Koldewey, J Chik, L Subramanian, U-S Cho, Mis16 switches function from a histone H4 chaperone to a CENP-ACnp1-specific assembly factor through Eic1 interaction. Structure, 2018. 26(7): 960e4–971e4. [DOI] [PMC free article] [PubMed]
- 30.Bade D, Pauleau A-L, Wendler A, Erhardt S. The E3 ligase CUL3/RDX controls centromere maintenance by ubiquitylating and stabilizing CENP-A in a CAL1-dependent manner. Dev Cell. 2014;28(5):508–519. doi: 10.1016/j.devcel.2014.01.031. [DOI] [PubMed] [Google Scholar]
- 31.Maddox PS, Hyndman F, Monen J, Oegema K, Desai A. Functional genomics identifies a Myb domain–containing protein family required for assembly of CENP-A chromatin. J Cell Biol. 2007;176(6):757–763. doi: 10.1083/jcb.200701065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lagana A, Dorn JF, De Rop V, Ladouceur AM, Maddox AS, Maddox PS. A small GTPase molecular switch regulates epigenetic centromere maintenance by stabilizing newly incorporated CENP-A. Nat Cell Biol. 2010;12(12):1186–1193. doi: 10.1038/ncb2129. [DOI] [PubMed] [Google Scholar]
- 33.Falk SJ, Guo LY, Sekulic N, Smoak EM, Mani T, Logsdon GA, Gupta K, Jansen LE, Van Duyne GD, Vinogradov SA, Lampson MA, Black BE. Chromosomes CENP-C reshapes and stabilizes CENP-A nucleosomes at the centromere. Science. 2015;348(6235):699–703. doi: 10.1126/science.1259308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jansen LE, Black BE, Foltz DR, Cleveland DW. Propagation of centromeric chromatin requires exit from mitosis. J Cell biol. 2007;176(6):795–805. doi: 10.1083/jcb.200701066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takada M, Zhang W, Suzuki A, Kuroda TS, Yu Z, Inuzuka H, Gao D, Wan L, Zhuang M, Hu L. FBW7 loss promotes chromosomal instability and tumorigenesis via cyclin E1/CDK2–mediated phosphorylation of CENP-A. Can Res. 2017;77(18):4881–4893. doi: 10.1158/0008-5472.CAN-17-1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu Z, Zhou X, Wang W, Deng W, Fang J, Hu H, Wang Z, Li S, Cui L, Shen J, Zhai L, Peng S, Wong J, Dong S, Yuan Z, Ou G, Zhang X, Xu P, Lou J, Yang N, Chen P, Xu RM, Li G. Dynamic phosphorylation of CENP-A at Ser68 orchestrates its cell-cycle-dependent deposition at centromeres. Dev Cell. 2015;32(1):68–81. doi: 10.1016/j.devcel.2014.11.030. [DOI] [PubMed] [Google Scholar]
- 37.Wang K, Yu Z, Liu Y, Li G. Ser68 phosphorylation ensures accurate cell-cycle-dependent CENP-A deposition at centromeres. Dev Cell. 2017;40:5–6. doi: 10.1016/j.devcel.2016.12.015. [DOI] [PubMed] [Google Scholar]
- 38.Zhao H, Winogradoff D, Bui M, Dalal Y, Papoian GA. Promiscuous histone mis-assembly is actively prevented by chaperones. J Am Chem Soc. 2016;138(40):13207–13218. doi: 10.1021/jacs.6b05355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fachinetti D, Logsdon GA, Abdullah A, Selzer EB, Cleveland DW, Black BE. CENP-A modifications on Ser68 and Lys124 are dispensable for establishment, maintenance, and long-term function of human centromeres. Dev Cell. 2017;40(1):104–113. doi: 10.1016/j.devcel.2016.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schuh M, Lehner CF, Heidmann S. Incorporation of drosophila CID/CENP-A and CENP-C into centromeres during early embryonic anaphase. Curr Biol. 2007;17(3):237–243. doi: 10.1016/j.cub.2006.11.051. [DOI] [PubMed] [Google Scholar]
- 41.Bernad R, Sánchez P, Rivera T, Rodríguez-Corsino M, Boyarchuk E, Vassias I, Ray-Gallet D, Arnaoutov A, Dasso M, Almouzni G, Losada A. Xenopus HJURP and condensin II are required for CENP-A assembly. J Cell Biol. 2011;192(4):569–582. doi: 10.1083/jcb.201005136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ohkuni K, Takahashi Y, Fulp A, Lawrimore J, Au W-C, Pasupala N, Levy-Myers R, Warren J, Strunnikov A, Baker RE. SUMO-targeted ubiquitin ligase (STUbL) Slx5 regulates proteolysis of centromeric histone H3 variant Cse4 and prevents its mislocalization to euchromatin. Mol Biol Cell. 2016;27(9):1500–1510. doi: 10.1091/mbc.E15-12-0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ranjitkar P, Press MO, Yi X, Baker R, MacCoss MJ, Biggins S. An E3 ubiquitin ligase prevents ectopic localization of the centromeric histone H3 variant via the centromere targeting domain. Mol Cell. 2010;40(3):455–464. doi: 10.1016/j.molcel.2010.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hewawasam G, Shivaraju M, Mattingly M, Venkatesh S, Martin-Brown S, Florens L, Workman JL, Gerton JL. Psh1 is an E3 ubiquitin ligase that targets the centromeric histone variant Cse4. Mol Cell. 2010;40(3):444–454. doi: 10.1016/j.molcel.2010.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moreno-Moreno O, Medina-Giró S, Torras-Llort M, Azorín F. The F box protein partner of paired regulates stability of drosophila centromeric histone H3, CenH3 CID. Curr Biol. 2011;21(17):1488–1493. doi: 10.1016/j.cub.2011.07.041. [DOI] [PubMed] [Google Scholar]
- 46.Nechemia-Arbely Y, Miga KH, Shoshani O, Aslanian A, McMahon MA, Lee AY, Fachinetti D, Yates JR, Ren B, Cleveland DW. DNA replication acts as an error correction mechanism to maintain centromere identity by restricting CENP-A to centromeres. Nat Cell Biol. 2019;21(6):743. doi: 10.1038/s41556-019-0331-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Niikura Y, Kitagawa R, Ogi H, Abdulle R, Pagala V, Kitagawa K. CENP-A K124 ubiquitylation is required for CENP-A deposition at the centromere. Dev Cell. 2015;32(5):589–603. doi: 10.1016/j.devcel.2015.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Niikura Y, Kitagawa R, Kitagawa K. CENP-A ubiquitylation is required for CENP-A deposition at the centromere. Dev Cell. 2017;40(1):7–8. doi: 10.1016/j.devcel.2016.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bui M, Dimitriadis EK, Hoischen C, An E, Quenet D, Giebe S, Nita-Lazar A, Diekmann S, Dalal Y. Cell-cycle-dependent structural transitions in the human CENP-A nucleosome in vivo. Cell. 2012;150(2):317–326. doi: 10.1016/j.cell.2012.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bui M, Pitman M, Nuccio A, Roque S, Donlin-Asp PG, Nita-Lazar A, Papoian GA, Dalal Y. Internal modifications in the CENP-A nucleosome modulate centromeric dynamics. Epigenetics Chromatin. 2017;10:17. doi: 10.1186/s13072-017-0124-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bailey AO, Panchenko T, Sathyan KM, Petkowski JJ, Pai PJ, Bai DL, Russell DH, Macara IG, Shabanowitz J, Hunt DF, Black BE, Foltz DR. Posttranslational modification of CENP-A influences the conformation of centromeric chromatin. Proc Natl Acad Sci USA. 2013;110(29):11827–11832. doi: 10.1073/pnas.1300325110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fachinetti D, Folco HD, Nechemia-Arbely Y, Valente LP, Nguyen K, Wong AJ, Zhu Q, Holland AJ, Desai A, Jansen LE. A two-step mechanism for epigenetic specification of centromere identity and function. Nat Cell Biol. 2013;15(9):1056. doi: 10.1038/ncb2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Logsdon GA, Barrey EJ, Bassett EA, DeNizio JE, Guo LY, Panchenko T, Dawicki-McKenna JM, Heun P, Black BE (2015) Both tails and the centromere targeting domain of CENP-A are required for centromere establishment. J Cell Biol, jcb. 201412011. [DOI] [PMC free article] [PubMed]
- 54.Goutte-Gattat D, Shuaib M, Ouararhni K, Gautier T, Skoufias DA, Hamiche A, Dimitrov S. Phosphorylation of the CENP-A amino-terminus in mitotic centromeric chromatin is required for kinetochore function. Proc Natl Acad Sci USA. 2013;110(21):8579–8584. doi: 10.1073/pnas.1302955110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li D, Liu R, Song L, Zhou H, Chen J, Huang X. The special location of p-H3 and p-CENP-A on heterochromatin during mitosis in MCF-7. Mol Biol Rep. 2008;35(4):657–662. doi: 10.1007/s11033-007-9136-9. [DOI] [PubMed] [Google Scholar]
- 56.Kunitoku N, Sasayama T, Marumoto T, Zhang D, Honda S, Kobayashi O, Hatakeyama K, Ushio Y, Saya H, Hirota T. CENP-A phosphorylation by Aurora-A in prophase is required for enrichment of Aurora-B at inner centromeres and for kinetochore function. Dev Cell. 2003;5(6):853–864. doi: 10.1016/s1534-5807(03)00364-2. [DOI] [PubMed] [Google Scholar]
- 57.Eot-Houllier G, Magnaghi-Jaulin L, Fulcrand G, Moyroud F-X, Monier S, Jaulin C. Aurora A-dependent CENP-A phosphorylation at inner centromeres protects bioriented chromosomes against cohesion fatigue. Nature communications. 2018;9(1):1888. doi: 10.1038/s41467-018-04089-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zeitlin SG, Shelby RD, Sullivan KF. CENP-A is phosphorylated by Aurora B kinase and plays an unexpected role in completion of cytokinesis. J Cell Biol. 2001;155(7):1147–1157. doi: 10.1083/jcb.200108125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Barra V, Logsdon GA, Scelfo A, Hoffmann S, Hervé S, Aslanian A, Nechemia-Arbely Y, Cleveland DW, Black BE, Fachinetti D. Phosphorylation of CENP-A on serine 7 does not control centromere function. Nature communications. 2019;10(1):175. doi: 10.1038/s41467-018-08073-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang X, Li X, Marshall JB, Zhong CX, Dawe RK. Phosphoserines on maize CENTROMERIC HISTONE H3 and histone H3 demarcate the centromere and pericentromere during chromosome segregation. Plant Cell. 2005;17(2):572–583. doi: 10.1105/tpc.104.028522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Samel A, Cuomo A, Bonaldi T, Ehrenhofer-Murray AE. Methylation of CenH3 arginine 37 regulates kinetochore integrity and chromosome segregation. Proc Natl Acad Sci USA. 2012;109(23):9029–9034. doi: 10.1073/pnas.1120968109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang J, Liu X, Dou Z, Chen L, Jiang H, Fu C, Fu G, Liu D, Zhang J, Zhu T, Fang J, Zang J, Cheng J, Teng M, Ding X, Yao X. Mitotic regulator Mis18beta interacts with and specifies the centromeric assembly of molecular chaperone holliday junction recognition protein (HJURP) The Journal of biological chemistry. 2014;289(12):8326–8336. doi: 10.1074/jbc.M113.529958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Muller S, Montes de Oca R, Lacoste N, Dingli F, Loew D, Almouzni G. Phosphorylation and DNA binding of HJURP determine its centromeric recruitment and function in CenH3(CENP-A) loading. Cell Rep. 2014;8(1):190–203. doi: 10.1016/j.celrep.2014.06.002. [DOI] [PubMed] [Google Scholar]
- 64.Silva MC, Bodor DL, Stellfox ME, Martins NM, Hochegger H, Foltz DR, Jansen LE. Cdk activity couples epigenetic centromere inheritance to cell cycle progression. Dev Cell. 2012;22(1):52–63. doi: 10.1016/j.devcel.2011.10.014. [DOI] [PubMed] [Google Scholar]
- 65.Pan D, Klare K, Petrovic A, Take A, Walstein K, Singh P, Rondelet A, Bird AW, Musacchio A. CDK-regulated dimerization of M18BP1 on a Mis18 hexamer is necessary for CENP-A loading. Elife. 2017;6:e23352. doi: 10.7554/eLife.23352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Spiller F, Medina-Pritchard B, Abad MA, Wear MA, Molina O, Earnshaw WC, Jeyaprakash AA. Molecular basis for Cdk1-regulated timing of Mis18 complex assembly and CENP-A deposition. EMBO Rep. 2017;18(6):894–905. doi: 10.15252/embr.201643564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.McKinley KL, Cheeseman IM. Polo-like kinase 1 licenses CENP-A deposition at centromeres. Cell. 2014;158(2):397–411. doi: 10.1016/j.cell.2014.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.French BT, Westhorpe FG, Limouse C, Straight AF. Xenopus laevis M18BP1 directly binds existing CENP-A nucleosomes to promote centromeric chromatin assembly. Dev Cell. 2017;42(2):190–199.e10. doi: 10.1016/j.devcel.2017.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shahbazian MD, Grunstein M. Functions of site-specific histone acetylation and deacetylation. Annu Rev Biochem. 2007;76:75–100. doi: 10.1146/annurev.biochem.76.052705.162114. [DOI] [PubMed] [Google Scholar]
- 70.Jasencakova Z, Meister A, Schubert I. Chromatin organization and its relation to replication and histone acetylation during the cell cycle in barley. Chromosoma. 2001;110(2):83–92. doi: 10.1007/s004120100132. [DOI] [PubMed] [Google Scholar]
- 71.Choy JS, Acuna R, Au WC, Basrai MA. A role for histone H4K16 hypoacetylation in Saccharomyces cerevisiae kinetochore function. Genetics. 2011;189(1):11–21. doi: 10.1534/genetics.111.130781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shang W-H, Hori T, Westhorpe FG, Godek KM, Toyoda A, Misu S, Monma N, Ikeo K, Carroll CW, Takami Y (2016) Acetylation of histone H4 lysine 5 and 12 is required for CENP-A deposition into centromeres. Nature communications. 7 [DOI] [PMC free article] [PubMed]
- 73.Yan H, Jin W, Nagaki K, Tian S, Ouyang S, Buell CR, Talbert PB, Henikoff S, Jiang J. Transcription and histone modifications in the recombination-free region spanning a rice centromere. Plant Cell. 2005;17(12):3227–3238. doi: 10.1105/tpc.105.037945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fujita Y, Hayashi T, Kiyomitsu T, Toyoda Y, Kokubu A, Obuse C, Yanagida M. Priming of centromere for CENP-A recruitment by human hMis18alpha, hMis18beta, and M18BP1. Dev Cell. 2007;12(1):17–30. doi: 10.1016/j.devcel.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 75.Wako T, Fukuda M, Furushima-Shimogawara R, Belyaev ND, Fukui K. Cell cycle-dependent and lysine residue-specific dynamic changes of histone H4 acetylation in barley. Plant Mol Biol. 2002;49(6):645–653. doi: 10.1023/a:1015554124675. [DOI] [PubMed] [Google Scholar]
- 76.Müller S, Almouzni G. Chromatin dynamics during the cell cycle at centromeres. Nat Rev Genet. 2017;18(3):192–208. doi: 10.1038/nrg.2016.157. [DOI] [PubMed] [Google Scholar]
- 77.Fukagawa T, William CE. The centromere: chromatin foundation for the kinetochore machinery. Dev Cell. 2014;30(5):496–508. doi: 10.1016/j.devcel.2014.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mravinac B, Sullivan LL, Reeves JW, Yan CM, Kopf KS, Farr CJ, Schueler MG, Sullivan BA. Histone modifications within the human X centromere region. PLoS ONE. 2009;4(8):e6602. doi: 10.1371/journal.pone.0006602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.May BP, Lippman ZB, Fang Y, Spector DL, Martienssen RA. Differential regulation of strand-specific transcripts from Arabidopsis centromeric satellite repeats. PLoS Genet. 2005;1(6):e79. doi: 10.1371/journal.pgen.0010079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cam HP, Sugiyama T, Chen ES, Chen X, FitzGerald PC, Grewal SI. Comprehensive analysis of heterochromatin- and RNAi-mediated epigenetic control of the fission yeast genome. Nat Genet. 2005;37(8):809–819. doi: 10.1038/ng1602. [DOI] [PubMed] [Google Scholar]
- 81.Folco HD, Pidoux AL, Urano T, Allshire RC. Heterochromatin and RNAi are required to establish CENP-A chromatin at centromeres. Science. 2008;319(5859):94–97. doi: 10.1126/science.1150944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Shi J, Dawe RK. Partitioning of the maize epigenome by the number of methyl groups on histone H3 lysines 9 and 27. Genetics. 2006;173(3):1571–1583. doi: 10.1534/genetics.106.056853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bergmann JH, Rodriguez MG, Martins NM, Kimura H, Kelly DA, Masumoto H, Larionov V, Jansen LE, Earnshaw WC. Epigenetic engineering shows H3K4me2 is required for HJURP targeting and CENP-A assembly on a synthetic human kinetochore. EMBO J. 2011;30(2):328–340. doi: 10.1038/emboj.2010.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chu L, Zhu T, Liu X, Yu R, Bacanamwo M, Dou Z, Chu Y, Zou H, Gibbons GH, Wang D, Ding X, Yao X. SUV39H1 orchestrates temporal dynamics of centromeric methylation essential for faithful chromosome segregation in mitosis. J Mol Cell Biol. 2012;4(5):331–340. doi: 10.1093/jmcb/mjs023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.McManus KJ, Biron VL, Heit R, Underhill DA, Hendzel MJ. Dynamic changes in histone H3 lysine 9 methylations identification of a mitosis-specific function for dynamic methylation in chromosome congression and segregation. J Biol Chem. 2006;281(13):8888–8897. doi: 10.1074/jbc.M505323200. [DOI] [PubMed] [Google Scholar]
- 86.Alonso A, Hasson D, Cheung F, Warburton PE. A paucity of heterochromatin at functional human neocentromeres. Epigenetics Chromatin. 2010;3(1):6. doi: 10.1186/1756-8935-3-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Nagaki K, Cheng Z, Ouyang S, Talbert PB, Kim M, Jones KM, Henikoff S, Buell CR, Jiang J. Sequencing of a rice centromere uncovers active genes. Nat Genet. 2004;36(2):138–145. doi: 10.1038/ng1289. [DOI] [PubMed] [Google Scholar]
- 88.Hori T, Shang WH, Toyoda A, Misu S, Monma N, Ikeo K, Molina O, Vargiu G, Fujiyama A, Kimura H, Earnshaw WC, Fukagawa T. Histone H4 Lys 20 monomethylation of the CENP-A nucleosome is essential for kinetochore assembly. Dev Cell. 2014;29(6):740–749. doi: 10.1016/j.devcel.2014.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bailey AO, Panchenko T, Shabanowitz J, Lehman SM, Bai DL, Hunt DF, Black BE, Foltz DR. Identification of the post-translational modifications present in centromeric chromatin. Mol Cell Proteomics. 2016;15(3):918–931. doi: 10.1074/mcp.M115.053710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wang F, Dai J, Daum JR, Niedzialkowska E, Banerjee B, Stukenberg PT, Gorbsky GJ, Higgins JM. Histone H3 Thr-3 phosphorylation by Haspin positions Aurora B at centromeres in mitosis. Science. 2010;330(6001):231–235. doi: 10.1126/science.1189435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kawashima SA, Yamagishi Y, Honda T, Ishiguro K, Watanabe Y. Phosphorylation of H2A by Bub1 prevents chromosomal instability through localizing shugoshin. Science. 2010;327(5962):172–177. doi: 10.1126/science.1180189. [DOI] [PubMed] [Google Scholar]
- 92.Yamagishi Y, Honda T, Tanno Y, Watanabe Y. Two histone marks establish the inner centromere and chromosome bi-orientation. Science. 2010;330(6001):239–243. doi: 10.1126/science.1194498. [DOI] [PubMed] [Google Scholar]
- 93.Kelly AE, Ghenoiu C, Xue JZ, Zierhut C, Kimura H, Funabiki H. Survivin reads phosphorylated histone H3 threonine 3 to activate the mitotic kinase Aurora B. Science. 2010;330(6001):235–239. doi: 10.1126/science.1189505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Liu H, Qu Q, Warrington R, Rice A, Cheng N, Yu H. Mitotic transcription installs Sgo1 at centromeres to coordinate chromosome segregation. Mol Cell. 2015;59(3):426–436. doi: 10.1016/j.molcel.2015.06.018. [DOI] [PubMed] [Google Scholar]
- 95.Dong Q, Han F. Phosphorylation of histone H2A is associated with centromere function and maintenance in meiosis. Plant J. 2012;71(5):800–809. doi: 10.1111/j.1365-313X.2012.05029.x. [DOI] [PubMed] [Google Scholar]
- 96.Demidov D, Schubert V, Kumke K, Weiss O, Karimi-Ashtiyani R, Buttlar J, Heckmann S, Wanner G, Dong Q, Han F, Houben A. Anti-phosphorylated histone H2AThr120: a universal microscopic marker for centromeric chromatin of mono- and holocentric plant species. Cytogenet Genome Res. 2014;143(1–3):150–156. doi: 10.1159/000360018. [DOI] [PubMed] [Google Scholar]
- 97.Wanner G, Schroeder-Reiter E, Ma W, Houben A, Schubert V (2015) The ultrastructure of mono- and holocentric plant centromeres: an immunological investigation by structured illumination microscopy and scanning electron microscopy. Chromosoma. [DOI] [PubMed]
- 98.Karp G, Pruitt NL. Cell and molecular biology: concepts and experiments. New York: Wiley; 1996. [Google Scholar]
- 99.Ohkuni K, Kitagawa K. Endogenous transcription at the centromere facilitates centromere activity in budding yeast. Curr Biol. 2011;21(20):1695–1703. doi: 10.1016/j.cub.2011.08.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ling YH, Yuen KWY. Point centromere activity requires an optimal level of centromeric noncoding RNA. Proc Natl Acad Sci. 2019;116(13):6270–6279. doi: 10.1073/pnas.1821384116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Volpe T, Schramke V, Hamilton GL, White SA, Teng G, Martienssen RA, Allshire RC. RNA interference is required for normal centromere function in fission yeast. Chromosome Res. 2003;11(2):137–146. doi: 10.1023/a:1022815931524. [DOI] [PubMed] [Google Scholar]
- 102.Djupedal I, Kos-Braun IC, Mosher RA, Söderholm N, Simmer F, Hardcastle TJ, Fender A, Heidrich N, Kagansky A, Bayne E. Analysis of small RNA in fission yeast; centromeric siRNAs are potentially generated through a structured RNA. EMBO J. 2009;28(24):3832–3844. doi: 10.1038/emboj.2009.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bouzinba-Segard H, Guais A, Francastel C. Accumulation of small murine minor satellite transcripts leads to impaired centromeric architecture and function. Proc Natl Acad Sci USA. 2006;103(23):8709–8714. doi: 10.1073/pnas.0508006103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ideue T, Cho Y, Nishimura K, Tani T. Involvement of satellite I noncoding RNA in regulation of chromosome segregation. Genes Cells. 2014;19(6):528–538. doi: 10.1111/gtc.12149. [DOI] [PubMed] [Google Scholar]
- 105.Pezer Z, Ugarkovic D. RNA Pol II promotes transcription of centromeric satellite DNA in beetles. PLoS ONE. 2008;3(2):e1594. doi: 10.1371/journal.pone.0001594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Rošić S, Köhler F, Erhardt S. Repetitive centromeric satellite RNA is essential for kinetochore formation and cell division. J Cell Biol. 2014;207(3):335–349. doi: 10.1083/jcb.201404097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Topp CN, Zhong CX, Dawe RK. Centromere-encoded RNAs are integral components of the maize kinetochore. Proc Natl Acad Sci USA. 2004;101(45):15986–15991. doi: 10.1073/pnas.0407154101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Quénet D, Dalal Y. A long non-coding RNA is required for targeting centromeric protein A to the human centromere. Elife. 2014;3:e03254. doi: 10.7554/eLife.03254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Gent JI, Dawe RK. RNA as a structural and regulatory component of the centromere. Annu Rev Genet. 2012;46:443–453. doi: 10.1146/annurev-genet-110711-155419. [DOI] [PubMed] [Google Scholar]
- 110.Chan FL, Wong LH. Transcription in the maintenance of centromere chromatin identity. Nucleic Acids Res. 2012;40(22):11178–11188. doi: 10.1093/nar/gks921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.McNulty SM, Sullivan LL, Sullivan BA. Human centromeres produce chromosome-specific and array-specific alpha satellite transcripts that are complexed with CENP-A and CENP-C. Dev Cell. 2017;42(3):226–240.e6. doi: 10.1016/j.devcel.2017.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wong LH, Brettingham-Moore KH, Chan L, Quach JM, Anderson MA, Northrop EL, Hannan R, Saffery R, Shaw ML, Williams E, Choo KH. Centromere RNA is a key component for the assembly of nucleoproteins at the nucleolus and centromere. Genome Res. 2007;17(8):1146–1160. doi: 10.1101/gr.6022807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Du Y, Topp CN, Dawe RK. DNA binding of centromere protein C (CENPC) is stabilized by single-stranded RNA. PLoS Genet. 2010;6(2):e1000835. doi: 10.1371/journal.pgen.1000835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ferri F, Bouzinba-Segard H, Velasco G, Hube F, Francastel C. Non-coding murine centromeric transcripts associate with and potentiate Aurora B kinase. Nucleic Acids Res. 2009;37(15):5071–5080. doi: 10.1093/nar/gkp529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Blower MD. Centromeric transcription regulates Aurora-B localization and activation. Cell Rep. 2016;15(8):1624–1633. doi: 10.1016/j.celrep.2016.04.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Grenfell AW, Heald R, Strzelecka M (2016) Mitotic noncoding RNA processing promotes kinetochore and spindle assembly in Xenopus. J Cell Biol. [DOI] [PMC free article] [PubMed]
- 117.Kato H, Goto DB, Martienssen RA, Urano T, Furukawa K, Murakami Y. RNA polymerase II is required for RNAi-dependent heterochromatin assembly. Science. 2005;309(5733):467–469. doi: 10.1126/science.1114955. [DOI] [PubMed] [Google Scholar]
- 118.Djupedal I, Portoso M, Spahr H, Bonilla C, Gustafsson CM, Allshire RC, Ekwall K. RNA Pol II subunit Rpb7 promotes centromeric transcription and RNAi-directed chromatin silencing. Genes Dev. 2005;19(19):2301–2306. doi: 10.1101/gad.344205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Chan FL, Marshall OJ, Saffery R, Kim BW, Earle E, Choo KH, Wong LH. Active transcription and essential role of RNA polymerase II at the centromere during mitosis. Proc Natl Acad Sci USA. 2012;109(6):1979–1984. doi: 10.1073/pnas.1108705109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Ling YH, Yuen KWY (2020) Point centromere activity requires an optimal level of centromeric non-coding RNA. Proc Natl Acad Sci (In press). [DOI] [PMC free article] [PubMed]
- 121.Foltz DR, Jansen LE, Black BE, Bailey AO, Yates JR, 3rd, Cleveland DW. The human CENP-A centromeric nucleosome-associated complex. Nat Cell Biol. 2006;8(5):458–469. doi: 10.1038/ncb1397. [DOI] [PubMed] [Google Scholar]
- 122.Prendergast L, Müller S, Liu Y, Huang H, Dingli F, Loew D, Vassias I, Patel DJ, Sullivan KF, Almouzni G. The CENP-T/-W complex is a binding partner of the histone chaperone FACT. Genes Dev. 2016;30(11):1313–1326. doi: 10.1101/gad.275073.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Okada M, Okawa K, Isobe T, Fukagawa T. CENP-H-containing complex facilitates centromere deposition of CENP-A in cooperation with FACT and CHD1. Mol Biol Cell. 2009;20(18):3986–3995. doi: 10.1091/mbc.E09-01-0065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Chen CC, Bowers S, Lipinszki Z, Palladino J, Trusiak S, Bettini E, Rosin L, Przewloka MR, Glover DM, O'Neill RJ, Mellone BG. Establishment of centromeric chromatin by the CENP-A assembly factor CAL1 requires FACT-mediated transcription. Dev Cell. 2015;34(1):73–84. doi: 10.1016/j.devcel.2015.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Choi ES, Strålfors A, Catania S, Castillo AG, Svensson JP, Pidoux AL, Ekwall K, Allshire RC. Factors that promote H3 chromatin integrity during transcription prevent promiscuous deposition of CENP-A Cnp1 in fission yeast. PLoS Genet. 2012;8(9):e1002985. doi: 10.1371/journal.pgen.1002985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Deyter GM, Biggins S. The FACT complex interacts with the E3 ubiquitin ligase Psh1 to prevent ectopic localization of CENP-A. Genes Dev. 2014;28(16):1815–1826. doi: 10.1101/gad.243113.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Catania S, Pidoux AL, Allshire RC. Sequence features and transcriptional stalling within centromere DNA promote establishment of CENP-A chromatin. PLoS Genet. 2015;11(3):e1004986. doi: 10.1371/journal.pgen.1004986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Fukagawa T, Nogami M, Yoshikawa M, Ikeno M, Okazaki T, Takami Y, Nakayama T, Oshimura M. Dicer is essential for formation of the heterochromatin structure in vertebrate cells. Nat Cell Biol. 2004;6(8):784–791. doi: 10.1038/ncb1155. [DOI] [PubMed] [Google Scholar]
- 129.Frescas D, Guardavaccaro D, Kuchay SM, Kato H, Poleshko A, Basrur V, Elenitoba-Johnson KS, Katz RA, Pagano M. KDM2A represses transcription of centromeric satellite repeats and maintains the heterochromatic state. Cell Cycle. 2008;7(22):3539–3547. doi: 10.4161/cc.7.22.7062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Gopalakrishnan S, Sullivan BA, Trazzi S, Della Valle G, Robertson KD. DNMT3B interacts with constitutive centromere protein CENP-C to modulate DNA methylation and the histone code at centromeric regions. Hum Mol Genet. 2009;18(17):3178–3193. doi: 10.1093/hmg/ddp256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Zhu Q, Pao GM, Huynh AM, Suh H, Tonnu N, Nederlof PM, Gage FH, Verma IM. BRCA1 tumour suppression occurs via heterochromatin-mediated silencing. Nature. 2011;477(7363):179–184. doi: 10.1038/nature10371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Schwartz BE, Ahmad K. Transcriptional activation triggers deposition and removal of the histone variant H33. Genes Dev. 2005;19(7):804–814. doi: 10.1101/gad.1259805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Ahmad K, Henikoff S. Epigenetic consequences of nucleosome dynamics. Cell. 2002;111(3):281–284. doi: 10.1016/s0092-8674(02)01081-4. [DOI] [PubMed] [Google Scholar]
- 134.Dunleavy EM, Almouzni G, Karpen GH (2011) H3.3 is deposited at centromeres in S phase as a placeholder for newly assembled CENP-A in G(1) phase. Nucleus 2(2):146–157 [DOI] [PMC free article] [PubMed]
- 135.Athwal RK, Walkiewicz MP, Baek S, Fu S, Bui M, Camps J, Ried T, Sung MH, Dalal Y. CENP-A nucleosomes localize to transcription factor hotspots and subtelomeric sites in human cancer cells. Epigenet Chromatin. 2015;8:2. doi: 10.1186/1756-8935-8-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Huang C, Cheng J, Bawa-Khalfe T, Yao X, Chin YE, Yeh ET. SUMOylated ORC2 recruits a histone demethylase to regulate centromeric histone modification and genomic stability. Cell Rep. 2016;15(1):147–157. doi: 10.1016/j.celrep.2016.02.091. [DOI] [PubMed] [Google Scholar]
- 137.Nakano M, Cardinale S, Noskov VN, Gassmann R, Vagnarelli P, Kandels-Lewis S, Larionov V, Earnshaw WC, Masumoto H. Inactivation of a human kinetochore by specific targeting of chromatin modifiers. Dev Cell. 2008;14(4):507–522. doi: 10.1016/j.devcel.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Molina O, Vargiu G, Abad MA, Zhiteneva A, Jeyaprakash AA, Masumoto H, Kouprina N, Larionov V, Earnshaw WC (2016) Epigenetic engineering reveals a balance between histone modifications and transcription in kinetochore maintenance. Nat Commun 7. [DOI] [PMC free article] [PubMed]
- 139.Allshire RC. RNA interference, heterochromatin, and centromere function. Cold Spring Harb Symp Quant Biol. 2004;69:389–395. doi: 10.1101/sqb.2004.69.389. [DOI] [PubMed] [Google Scholar]
- 140.Allshire RC, Javerzat J-P, Redhead NJ, Cranston G. Position effect variegation at fission yeast centromeres. Cell. 1994;76(1):157–169. doi: 10.1016/0092-8674(94)90180-5. [DOI] [PubMed] [Google Scholar]
- 141.Thakur J, Sanyal K. Efficient neocentromere formation is suppressed by gene conversion to maintain centromere function at native physical chromosomal loci in Candida albicans. Genome Res. 2013;23(4):638–652. doi: 10.1101/gr.141614.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Bergmann JH, Jakubsche JN, Martins NM, Kagansky A, Nakano M, Kimura H, Kelly DA, Turner BM, Masumoto H, Larionov V, Earnshaw WC. Epigenetic engineering: histone H3K9 acetylation is compatible with kinetochore structure and function. J Cell Sci. 2012;125(Pt 2):411–421. doi: 10.1242/jcs.090639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Hill A, Bloom K. Genetic manipulation of centromere function. Mol Cell Biol. 1987;7(7):2397–2405. doi: 10.1128/mcb.7.7.2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Cardinale S, Bergmann JH, Kelly D, Nakano M, Valdivia MM, Kimura H, Masumoto H, Larionov V, Earnshaw WC. Hierarchical inactivation of a synthetic human kinetochore by a chromatin modifier. Mol Biol Cell. 2009;20(19):4194–4204. doi: 10.1091/mbc.E09-06-0489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Collins KA, Castillo AR, Tatsutani SY, Biggins S. De novo kinetochore assembly requires the centromeric histone H3 variant. Mol Biol Cell. 2005;16(12):5649–5660. doi: 10.1091/mbc.E05-08-0771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Vourc’h C, Biamonti G (2011) Transcription of satellite DNAs in mammals, in long non-coding RNAs, Springer, Berlin, pp 95–118. [DOI] [PubMed]
- 147.Choi IR, Ostrovsky M, Zhang G, White KA. Regulatory activity of distal and core RNA elements in Tombusvirus subgenomic mRNA2 transcription. J Biol Chem. 2001;276(45):41761–41768. doi: 10.1074/jbc.M106727200. [DOI] [PubMed] [Google Scholar]
- 148.Ling YH, Yuen KWY (2019) Centromeric non-coding RNA as a hidden epigenetic factor of the point centromere. Curr Genet 1–7. [DOI] [PubMed]
- 149.Lachner M, O'Sullivan RJ, Jenuwein T. An epigenetic road map for histone lysine methylation. J Cell Sci. 2003;116(Pt 11):2117–2124. doi: 10.1242/jcs.00493. [DOI] [PubMed] [Google Scholar]
- 150.Hall IM, Shankaranarayana GD, Noma K, Ayoub N, Cohen A, Grewal SI. Establishment and maintenance of a heterochromatin domain. Science. 2002;297(5590):2232–2237. doi: 10.1126/science.1076466. [DOI] [PubMed] [Google Scholar]
- 151.Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410(6824):120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- 152.Nakayama J, Rice JC, Strahl BD, Allis CD, Grewal SI. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science. 2001;292(5514):110–113. doi: 10.1126/science.1060118. [DOI] [PubMed] [Google Scholar]
- 153.Kagansky A, Folco HD, Almeida R, Pidoux AL, Boukaba A, Simmer F, Urano T, Hamilton GL, Allshire RC. Synthetic heterochromatin bypasses RNAi and centromeric repeats to establish functional centromeres. Science. 2009;324(5935):1716–1719. doi: 10.1126/science.1172026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Maison C, Bailly D, Peters AH, Quivy JP, Roche D, Taddei A, Lachner M, Jenuwein T, Almouzni G. Higher-order structure in pericentric heterochromatin involves a distinct pattern of histone modification and an RNA component. Nat Genet. 2002;30(3):329–334. doi: 10.1038/ng843. [DOI] [PubMed] [Google Scholar]
- 155.Murchison EP, Partridge JF, Tam OH, Cheloufi S, Hannon GJ. Characterization of Dicer-deficient murine embryonic stem cells. Proc Natl Acad Sci USA. 2005;102(34):12135–12140. doi: 10.1073/pnas.0505479102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Kanellopoulou C, Muljo SA, Kung AL, Ganesan S, Drapkin R, Jenuwein T, Livingston DM, Rajewsky K. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev. 2005;19(4):489–501. doi: 10.1101/gad.1248505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Brower-Toland B, Findley SD, Jiang L, Liu L, Yin H, Dus M, Zhou P, Elgin SC, Lin H. Drosophila PIWI associates with chromatin and interacts directly with HP1a. Genes Dev. 2007;21(18):2300–2311. doi: 10.1101/gad.1564307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Gu T, Elgin SC. Maternal depletion of Piwi, a component of the RNAi system, impacts heterochromatin formation in Drosophila. PLoS Genet. 2013;9(9):e1003780. doi: 10.1371/journal.pgen.1003780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Yuen KW, Nabeshima K, Oegema K, Desai A. Rapid de novo centromere formation occurs independently of heterochromatin protein 1 in C elegans embryos. Curr Biol. 2011;21(21):1800–1807. doi: 10.1016/j.cub.2011.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Claycomb JM, Batista PJ, Pang KM, Gu W, Vasale JJ, van Wolfswinkel JC, Chaves DA, Shirayama M, Mitani S, Ketting RF, Conte D, Mello CC. The Argonaute CSR-1 and its 22G-RNA cofactors are required for holocentric chromosome segregation. Cell. 2009;139(1):123–134. doi: 10.1016/j.cell.2009.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Gerson-Gurwitz A, Wang S, Sathe S, Green R, Yeo GW, Oegema K, Desai A. A small RNA-catalytic Argonaute pathway tunes germline transcript levels to ensure embryonic divisions. Cell. 2016;165(2):396–409. doi: 10.1016/j.cell.2016.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Wedeles CJ, Wu MZ, Claycomb JM. Protection of germline gene expression by the C Elegans Argonaute CSR-1. Dev Cell. 2013;27(6):664–671. doi: 10.1016/j.devcel.2013.11.016. [DOI] [PubMed] [Google Scholar]
- 163.Black BE, Foltz DR, Chakravarthy S, Luger K, Woods VL, Cleveland DW. Structural determinants for generating centromeric chromatin. Nature. 2004;430(6999):578–582. doi: 10.1038/nature02766. [DOI] [PubMed] [Google Scholar]
- 164.Keith KC, Baker RE, Chen Y, Harris K, Stoler S, Fitzgerald-Hayes M. Analysis of primary structural determinants that distinguish the centromere-specific function of histone variant Cse4p from histone H3. Mol Cell Biol. 1999;19(9):6130–6139. doi: 10.1128/mcb.19.9.6130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Bernatavichute YV, Zhang X, Cokus S, Pellegrini M, Jacobsen SE. Genome-wide association of histone H3 lysine nine methylation with CHG DNA methylation in Arabidopsis thaliana. PLoS ONE. 2008;3(9):e3156. doi: 10.1371/journal.pone.0003156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Jih G, Iglesias N, Currie MA, Bhanu NV, Paulo JA, Gygi SP, Garcia BA, Moazed D. Unique roles for histone H3K9me states in RNAi and heritable silencing of transcription. Nature. 2017;547(7664):463–467. doi: 10.1038/nature23267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Choi ES, Strålfors A, Castillo AG, Durand-Dubief M, Ekwall K, Allshire RC. Identification of noncoding transcripts from within CENP-A chromatin at fission yeast centromeres. J Biol Chem. 2011;286(26):23600–23607. doi: 10.1074/jbc.M111.228510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Carone DM, Longo MS, Ferreri GC, Hall L, Harris M, Shook N, Bulazel KV, Carone BR, Obergfell C, O'Neill MJ, O'Neill RJ. A new class of retroviral and satellite encoded small RNAs emanates from mammalian centromeres. Chromosoma. 2009;118(1):113–125. doi: 10.1007/s00412-008-0181-5. [DOI] [PubMed] [Google Scholar]
- 169.Bayne EH, White SA, Kagansky A, Bijos DA, Sanchez-Pulido L, Hoe K-L, Kim D-U, Park H-O, Ponting CP, Rappsilber J. Stc1: a critical link between RNAi and chromatin modification required for heterochromatin integrity. Cell. 2010;140(5):666–677. doi: 10.1016/j.cell.2010.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Scott KC, White CV, Willard HF. An RNA polymerase III-dependent heterochromatin barrier at fission yeast centromere 1. PLoS ONE. 2007;2(10):e1099. doi: 10.1371/journal.pone.0001099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Pal-Bhadra M, Leibovitch BA, Gandhi SG, Rao M, Bhadra U, Birchler JA, Elgin SC. Heterochromatic silencing and HP1 localization in Drosophila are dependent on the RNAi machinery. Science. 2004;303(5658):669–672. doi: 10.1126/science.1092653. [DOI] [PubMed] [Google Scholar]
- 172.Peters AH, O'Carroll D, Scherthan H, Mechtler K, Sauer S, Schofer C, Weipoltshammer K, Pagani M, Lachner M, Kohlmaier A, Opravil S, Doyle M, Sibilia M, Jenuwein T. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell. 2001;107(3):323–337. doi: 10.1016/s0092-8674(01)00542-6. [DOI] [PubMed] [Google Scholar]
- 173.Huang C, Wang X, Liu X, Cao S, Shan G. RNAi pathway participates in chromosome segregation in mammalian cells. Cell Discovery. 2015;1:15029. doi: 10.1038/celldisc.2015.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Muchardt C, Guillemé M, Seeler JS, Trouche D, Dejean A, Yaniv M. Coordinated methyl and RNA binding is required for heterochromatin localization of mammalian HP1α. EMBO Rep. 2002;3(10):975–981. doi: 10.1093/embo-reports/kvf194. [DOI] [PMC free article] [PubMed] [Google Scholar]