Abstract
Cysteine and aromatic residues are major structure-promoting residues. We assessed the abundance, structural coverage, and functional characteristics of the “non-smelly” proteins, i.e., proteins that do not contain cysteine residues (C-depleted) or cysteine and aromatic residues (CFYWH-depleted), across 817 proteomes from all domains of life. The analysis revealed that although these proteomes contained significant levels of the C-depleted proteins, with prokaryotes being significantly more enriched in such proteins than eukaryotes, the CFYWH-depleted proteins were relatively rare, accounting for about 0.05% of proteomes. Furthermore, CFYWH-depleted proteins were virtually never found in PDB. Depletion in cysteine and in aromatic residues was associated with the substantially increased intrinsic disorder levels across all domains of life. Archaeal and eukaryotic organisms with higher levels of the C-depleted proteins were shown to have higher levels of the intrinsic disorder and lower levels of structural coverage. We also showed that the “non-smelly” proteins typically did not independently fold into monomeric structures, and instead, they fold by interacting with nucleic acids as constituents of the ribosome and nucleosome complexes. They were shown to be involved in translation, transcription, nucleosome assembly, transmembrane transport, and protein folding functions, all of which are known to be associated with the intrinsic disorder. Our data suggested that, in general, structure of monomeric proteins is crucially dependent on the presence of cysteine and aromatic residues.
Electronic supplementary material
The online version of this article (10.1007/s00018-019-03292-1) contains supplementary material, which is available to authorized users.
Keywords: Intrinsically disordered proteins, Cysteine-depleted proteins, Nucleic acid-binding proteins, Proteins depleted in cysteine and aromatic residues, Protein structure, Protein function
Highlights
Cysteine-depleted proteins are abundant in all domains of life.
Prokaryotes are significantly enriched in cysteine-depleted proteins compared to eukaryotes.
Only about 0.05% of proteins are depleted in aromatic residues and cysteine.
Proteins depleted in aromatic residues and cysteine have high levels of intrinsic disorder.
Organisms with higher levels of cysteine-depleted proteins have higher levels of the intrinsic disorder.
“Non-smelly” proteins are involved in translation, transcription, nucleosome assembly, protein folding, and transmembrane transport functions.
Electronic supplementary material
The online version of this article (10.1007/s00018-019-03292-1) contains supplementary material, which is available to authorized users.
Introduction
It is accepted now that intrinsically disordered proteins (IDPs) and hybrid proteins containing ordered domains and functionally important intrinsically disordered proteins regions (IDPRs) occupy a significant part of any proteome across all kingdoms of life and viruses [1–6], being especially abundant in eukaryotes [2, 7]. Under physiological conditions, IDPs/IDPRs lack rigid 3D structure and, therefore, are typically not amenable to experimental structure determination by X-ray crystallography [8–10], which is by far the most commonly used technology to solve protein structures. As a result, they are considered as major constituents of the dark proteome [8, 11, 12]. While being disordered as a whole or in localized regions, these proteins have a number of important biological roles, especially in transcriptional and translational regulation, splicing, and signaling via cellular protein networks [13–15]. Furthermore, enhanced structural plasticity and exceptional spatiotemporal heterogeneity of IDPs/IDPRs define their mosaic structures, where different regions are disordered to different degrees. IDPs contain a multitude of potentially foldable, partially foldable, differently foldable or not foldable at all segments playing different roles in protein functionality [16, 17], and even containing ordered regions that need to undergo order-to-disorder transition to make protein active [16, 18, 19]. In cellular protein–protein interaction networks, IDPs/IDPRs often play a role of hubs [20–24] that are engaged in promiscuous interactions and regulate the structural and functional integrity of these networks [15, 25, 26]. Furthermore, because of this binding promiscuity [27] and the ability to gain very different structures at binding to different partners [28], IDPs/IDPRs can “rewire” protein–protein interaction networks in response to environmental changes [29].
Systematic comparative analyses of amino acid sequences of ordered proteins and IDPs revealed the presence of numerous important differences [7, 13, 30–33]. For examples, extended IDPs/IDPRs from different kingdoms of life were shown to be rich in polar and charged amino acids and deficient in hydrophobic residues [30, 33–35]. This also resulted in the elaboration of the concept of “order-promoting” (C, W, Y, I, F, V, L, H, T, and N) and “disorder-promoting” residues (A, G, D, M, K, R, S, Q, P, and E), i.e., residues more commonly found in ordered and disordered proteins/regions, respectively [36]. Because of the high relative enrichment of the amino acid sequences of ordered proteins and domains in cysteine, tryptophan, tyrosine, phenylalanine, and histidine, these residues are typically considered as strong order-promoting residues. Based on these observations, we hypothesized that structure and functionality of proteins can be noticeably dependent on the presence cysteine and aromatic residues in their amino acid sequences. One can argue that cysteine is important for protein structural stability only when another cysteine is present in the same chain, to enable disulfide bond formation. Observations below provide important evidence that this is not always correct. Intramolecular disulfide bonds are surely important stabilizing factors. For example, proteins and peptides containing cystine knot, which is a rotaxane-like structural motif containing three disulfide bridges, where a polypeptide region between two of those disulfides forms a loop, through which a third disulfide bond is threaded, are known to show a particularly high degree of structural stability [37, 38]. There are also numerous examples in the literature, where the importance of intramolecular disulfide bonds for protein thermal stability was demonstrated (as systemized in [39]). As a result, introduction of additional disulfide bonds is considered as an attractive protein engineering strategy for generating proteins (e.g., antibodies) with enhanced conformational stability [40]. Furthermore, dysregulated cellular redox conditions leading to the alterations in the formation of native disulfide bonds are directly linked to various human diseases [41]. However, even a single cysteine contributes to protein structure stability, as it can be engaged in the intermolecular disulfide bond, or can exist as a free thiol and serve as a part of a protein catalytic site, or as a site of various posttranslational modifications (e.g., S-hydroxylation (S-OH), disulfide bond formation, phosphorylation, S-acylation, S-prenylation, protein splicing, N-acetylation, N-ADP-ribosylation, amidation, S-archaeol cysteine, cysteine sulfinic acid (–SO2H) formation, methylation, N-myristoylation, nitrosylation, N-palmitoylation, S-palmitoylation, and S-glutathionylation) [42]. Furthermore, a single cysteine can be used for specific coordination of various ligands, e.g., metal ions. In fact, cysteine is known to show high affinity toward zinc ions (Zn2+), and the resulting cysteine-Zn2+ complexes are important for protein structure, catalysis, and regulation [43], as seen in the CH3-type zinc finger proteins [44, 45] and in redox switches [43]. On the other hand, CD3 motifs serve as an Mn2+ coordination group [46].
To check the validity of the hypothesis that the presence cysteine and aromatic residues is crucial for protein structure, we conducted here a comprehensive bioinformatics analysis of the “non-smelly” proteins, i.e., proteins depleted in cysteine and aromatic residues. Since cysteines are known to smell like rotten eggs [47], and since the side chains of W, Y, F, and H are aromatic (i.e., they contain aromatic ring systems, which are stable, cyclic, planar compounds with a ring of resonance bonds and which, unlike pure saturated hydrocarbons, might have specific odors/aroma), the proteins depleted in these residues are dubbed here as “non-smelly”. In this study, we assembled a data set of “non-smelly” proteins found in 817 complete proteomes, and also looked for such proteins in the Protein Data Bank (PDB) [48, 49].
Materials and methods
Data sets
We analyzed a data set of 817 complete proteomes, which are defined as collections of proteins encoded by the fully sequenced genome of a specific organism. We obtained these proteomes from the UniProt resource [50, 51]. They cover 276,733 proteins from 64 Archaean organisms, 5,077,609 proteins from 552 Bacterial organisms, and 4,208,817 proteins from 201 Eukaryotic organisms, for the total of 9,563,159 proteins. A complete list of the considered species is given in the Supplementary Materials. Table 1 provides a breakdown of these organisms into specific kingdoms/phyla.
Table 1.
Amount of cysteine-depleted (C-depleted) and cysteine and aromatic residues-depleted (CFYWH-depleted) proteins, disorder content and structural coverage for the considered 817 proteomes
| Domain of life | Kingdom/phylum of life | Number of Species | Median (per proteome) fraction of C-depleted proteins [%] | Median (per proteome) fraction of CFYWH-depleted proteins [%] | Median (per proteome) disorder content [%] | Median (per proteome) structural coverage [%] |
|---|---|---|---|---|---|---|
| Archaea | All | 64 | 28.48 | 0.06 | 5.88 | 53.10 |
| Crenarchaeota | 17 | 34.05 | 0.06 | 3.00 | 53.56 | |
| Other | 4 | 24.71 | 0.11 | 4.45 | 55.85 | |
| Euryarchaeota | 43 | 19.81 | 0.07 | 5.10 | 53.96 | |
| Bacteria | All | 552 | 18.65 | 0.04 | 5.45 | 55.85 |
| Firmicutes | 76 | 22.62 | 0.04 | 4.60 | 60.07 | |
| Actinobacteria | 70 | 21.97 | 0.07 | 10.30 | 59.45 | |
| Other | 108 | 19.20 | 0.03 | 4.70 | 57.70 | |
| Bacteroidetes | 44 | 18.67 | 0.02 | 3.45 | 54.02 | |
| Proteobacteria | 254 | 16.99 | 0.04 | 5.80 | 61.37 | |
| Eukaryota | All | 201 | 7.87 | 0.04 | 19.70 | 47.70 |
| Fungi | 84 | 9.64 | 0.03 | 21.55 | 46.33 | |
| Viridiplantae | 15 | 7.37 | 0.04 | 16.40 | 45.84 | |
| Other | 31 | 5.40 | 0.04 | 16.80 | 41.71 | |
| Metazoa | 71 | 4.64 | 0.08 | 19.50 | 62.78 |
We report median of these per-proteome measurements for the entire domains of life (in bold font) and several larger kingdoms/phyla. The domains of life and the phyla/kingdoms within each domain are arranged according to their overall fraction of C-depleted proteins
We also examined proteins with solved structures collected from PDB. We limited our analysis to the wild-type protein chains that have the expression tags removed and that exclude peptides (chain length > 30 residues), and which cover majority of the corresponding full protein chain from UniProt (> 60% coverage). We clustered the sequences of the considered PDB structures at 100% identity to remove duplicates. These steps ensure compatibility with the proteome-level analysis. We collected 99,461 chains that satisfy the aforementioned criteria (PDB data set), as well as two of its subsets that include 50,301 chains that are in complex with nucleic acids (PDB NA data set) and 7413 monomers, i.e., single-chain structures that do not interact with other proteins and nucleic acids (PDB monomer data set).
Computation of structural characteristics
We used computational methods to quantify content of putative intrinsic disorder (the fraction of residues that are predicted to be intrinsically disordered) and the current structural coverage (the fraction of proteins for which structure is available) on the whole-proteome scale. We evaluated the quality of the disorder content predictions using a large benchmark data set that was recently used in [52, 53] and which was originally published in [54]. We quantify the predictive quality by computing the mean absolute error (MAE) and Pearson correlation coefficient (PCC) between the disorder content predicted with the consensus and the native disorder content. The resulting MAE = 5.5% and PCC = 0.43, which suggests that the content predictions are relatively accurate and correlated with the native disorder content. Our consensus secures similar results for the subset of the benchmark proteins that have cysteine (MAE = 4.9% and PCC = 0.46) and that have above average cysteine content (MAE = 5.0% and PCC = 0.42).
Recent studies demonstrate that intrinsic disorder can be accurately predicted from protein sequences [54–58]. Furthermore, consensus-based approaches that combine outputs of several disorder predictors were shown to provide more accurate predictions when compared to single predictors [59–61]. For instance, consensus predictors more precisely quantify the disorder content (fraction of the disordered residues in a given protein sequence), reducing error by about 4% when compared to single predictors [60]. We applied a consensus of five complementary predictions produced by two popular tools, IUPred [62] and ESpritz [63]. They include results produced with two versions of the IUPred method, which were designed to predict long (≥ 30 consecutive residues) and short disordered regions, and three versions of ESpritz that focus on the three types of annotations of disorder using: DisProt database [64, 65], crystal structures from PDB, and NMR structures from PDB. These tools are characterized by competitive levels of predictive quality [54, 56] and short runtime, which is critical to facilitate processing of over 9.5 million protein sequences. The consensus prediction of disorder requires that at least 3 out of 5 predictions indicate intrinsic disorder. The same consensus was applied in several related studies [2, 8, 66–71]. Our methodology is also similar to the consensus-derived putative disorder in MobiDB [72, 73] and D2P2 [16] databases. We calculated the disorder content of a given data set of proteins (e.g., proteome) which is defined as a fraction of residues predicted as disordered among all residues in that data set.
We estimated the current structural coverage based on a computationally tractable approach proposed in [74] and recently used in [2, 8, 75]. For each protein, we ran three rounds of PSI-BLAST [76] searches against the sequences of protein structures from PDB. A given proteins sequence that has > 50 residues in length is annotated as having structure if it registers a hit in PDB with the E value < 0.001. In other words, structurally solved proteins are assumed to have at least one long segment of residues (representing at least one domain) that is sufficiently similar to a sequence of an already solved structure. The structural coverage of a proteome is defined as the fraction of the structurally solved sequences among all sequences in this proteome. Research shows that such PSI-BLAST-based estimates provide relatively accurate results. For instance, a similar PSI-BLAST-based approach failed to find templates (similar sequences that are structured) for only 3 out of 120 target proteins in CASP9 [77]. We recognize that there are more precise approaches to estimate structural coverage that are capable of finding remote homologs, such as I-TASSER [78, 79], HHpred [80, 81], and MODELLER [82, 83]. However, these tools could not be scaled to the size of our data set. To the best of our knowledge, the largest such attempt is the MODBASE resource that covers only 76 organisms [84]. We note that our estimates of the structural coverage are slightly underestimated by inadequately considering remote homologs. Nevertheless, this bias should be equally distributed across different proteomes, allowing us to perform comparative analyses between the corresponding organisms and domains of life.
Functional annotations using GO terms
We annotated protein functions and cellular locations that are associated with the proteins depleted in major order-promoting residues, such as cysteine, phenylalanine, tyrosine, tryptophan, and histidine. Such proteins were split into two groups, depleted in cysteine (C-depleted) and depleted in cysteine and aromatic residues: phenylalanine, tyrosine, tryptophan, and histidine (CFYWH-depleted). The corresponding functions/locations are significantly enriched in these proteins sets when compared with the proteins from the same domain of life. This analysis relies on the GO terms [85] collected from the UniProt resource. We excluded annotations with “potential”, “probable”, and “by similarity” qualifiers that are generated using computer predictions or indirect experimental evidence. We evaluated magnitude and statistical significance of the differences in the rates of occurrence of GO terms between the C-depleted (or CFYWH-depleted) proteins and a generic set of proteins in the same domain of life by following protocols defined in earlier related analyses [2, 3, 70]. This analysis was performed for each of the three types of GO terms: cellular components, biological processes, and molecular functions. We randomly selected half of the GO-annotated chains for a given C-/CFYWH-depleted protein set and compared them with the same number of chains/residues drawn at random from the same taxonomic domain. We ensured that proteins drawn from the same domain of life have the same chain length (with ± 10% tolerance), since the amount of intrinsic disorder, which indirectly affects protein function and location, is dependent on the chain length [86]. We repeated this 10 times and evaluated the significance of the differences in the 10 sets of counts for the corresponding GO terms. If these measurements are normal, based on the Anderson–Darling test at 0.05 significance, then we applied the paired t test (proteins sets are paired to match chain lengths) to evaluate the statistical significance of differences; otherwise, we utilized the non-parametric paired Wilcoxon rank sum test. We considered only the differences with p value < 0.001 which also have large magnitude, i.e., the average enrichment in the C-/CFYWH-depleted protein set must be larger than 30%. We analyzed the enrichment of GO terms for the entire set of C/CFYWH-depleted proteins as well as for the subsets of fully disordered C-/CFYWH-depleted proteins.
Results and discussion
Abundance of C-depleted and CFYWH-depleted proteins
We measured fraction of the C-depleted and the CFYWH-depleted proteins across the 817 proteomes and among the proteins in the three PDB-derived data sets. Table 1 summarizes these values across each domain of life and several larger kingdoms and phyla. About 28% proteins in Archaea, 19% in bacteria and 8% in eukaryota do not have cysteines. While we observe substantial differences in the abundance of the C-depleted proteins across the three domains of life, while these values are consistent across the kingdoms and phyla within each domain of life (Table 1). This observation suggests that this trend is broadly associated with domains of life. Only about 0.06% proteins in archaea and 0.04% in bacteria and eukaryota do not have cysteine and aromatic residues. Table 1 shows that the abundance of the CFYWH-depleted proteins is similar across Bacteria and Eukaryota, with Archaea having somehow elevated levels of such proteins.
Figure 1 summarizes the distribution of the per-proteome abundance of the C-/CFYWH-depleted proteins for the three domains of life. The numbers of the C-depleted proteins vary significantly between Archaea, Bacteria and Eukaryota (p values < 0.0001; Fig. 1a), while the numbers of the CFYWH-depleted proteins are not significantly different (p values ≥ 0.01; Fig. 1b). Our analysis has revealed that prokaryotes harbor significantly larger numbers of the C-depleted proteins compared to eukaryotes. Furthermore, we compared these rates with the corresponding rates for the proteins with known structures collected from PDB. About 35% of proteins in the PDB data set are depleted in cysteine (Fig. 1a), while only two proteins (0.002%) are depleted in cysteine and aromatic residues (Fig. 1b). The relatively high rate of the C-depleted proteins in PDB can be explained by two observations: about 2/3 of the PDB data set is composed of the prokaryotic proteins, because 51% of the proteins in this data set were solved in complex with nucleic acids (PDB NA data set). The effect of the latter factor is supported by our empirical finding that about 50% of the proteins in the PDB NA data set are depleted in cysteine, which is a substantial enrichment particularly when compared to the PDB monomer data set that has only about 19% of the C-depleted proteins (Fig. 1a). We also emphasize the lack of the CFYWH-depleted proteins in PDB (Fig. 1b). We found only two of them overall, with none in the PDB NA data set and only one among the monomers. Importantly, the levels of the presence of the CFYWH-depleted proteins in PDB are substantially lower when compared to the rates of the CFYWH-depleted proteins in whole proteomes, i.e., 0.002% in PDB vs. 0.04% in Eukaryotic and Bacterial proteomes (20-fold decrease) and 0.06% in Archaean proteomes (30-fold decrease).
Fig. 1.
Abundance of C-depleted (panel a) and CFYWH-depleted proteins (panel b) in the three domains of life and among the structurally solved proteins from PDB. The blue, red, and green bars show the median per-proteome fraction of C-/CFYWH-depleted proteins among the 64 Archaean, 552 Bacterial, and 201 eukaryotic organisms, respectively. The whiskers denote the first and third quartiles of these per-proteome fractions. Statistical significance of the differences for the per-proteome values between domains of life was assessed with the Wilcoxon test for unpaired data; distributions of the measured values are not normal. The black, dark gray and light gray bars show the fraction of the C-/CFYWH-depleted proteins among wild-type proteins chains from PDB (PDB data set), wild-type PDB proteins that interact with nucleic acids (PDB NA data set) and wild-type PDB monomers (PDB monomers data set), respectively
Altogether, these results demonstrate that the C-depleted proteins are significantly enriched in prokaryotes compared to eukaryotes and that they are often involved in protein–nucleic acids interactions and relatively rarely fold into monomer structures. On the other hand, the CFYWH-depleted proteins are equally abundant across the three domains of life and virtually never found in PDB. The latter suggest that they are hard to solve structurally.
CFYWH- and FYWH-depleted proteins in the PDB data set and their intrinsic disorder status
Our search for the CFYWH-depleted proteins in the PDB data set produced only two hits, a deletion mutant of the transcarboxylase biotin carrier subunit (also known as biotin carboxyl carrier protein, BCCP) from Propionibacterium freudenreichii subsp. shermanii (PDB ID: 1O78) and a molybdenum–pterin-binding protein 2 (molbindin-2 or MopII) from Clostridium pasteurianum (PDB ID: 1GUT). Functionally, BCCP serves as a carrier subunit of the transcarboxylase, which is a biotin-dependent 1200 kDa multi-subunit enzyme composed of 30 separable polypeptides [87]. Here, BCCP functions as a carboxyl group carrier to which biotin is covalently attached at Lys89. BCCP also binds the other two subunits of transcarboxylase to assist in the overall assembly of the enzyme [88]. NMR solution structure analysis revealed that the BCCP C-terminal domain (residues 51–123) is characterized by a compact β-sandwich structure, whereas the N-terminal region of the protein (residues 1–50) is disordered and does not have detectable structure [89]. Figure 2a represents the NMR solution structure of a CFYWH-depleted 10–48 deletion mutant (residues 1–9/49–123) of BCCP and shows that this protein contains six anti-parallel β-strands forming β-sandwich and a rather disordered N-terminal region. Furthermore, high flexibility was also detected at the C-terminal ‘β-finger’ segment of this deletion mutant that contains the Lys89 biotinylation site [90]. The second CFYWH-depleted protein in PDB, molbindin-2, is a bacterial protein that serves as an intracellular storage facility for molybdate. Figure 2b shows that this protein exists as a hexamer assembled as a trimer of dimers and binds up to eight molybdate ions with high affinity [91]. A protomer of this protein has a twisted anti-parallel β-sheet structure formed by five β-strands [91] (see Fig. 2c).
Fig. 2.
Structural characterization of the CFYWH- and FYWH-depleted proteins found in PDB: a NMR solution structure of a CFYWH-depleted 10–48 deletion mutant (residues 1–9/49–123) of the transcarboxylase biotin carrier subunit (also known as biotin carboxyl carrier protein, BCCP) from Propionibacterium freudenreichii subsp. shermanii (PDB ID: 1O78); b crystal structure of the homohexameric molybdenum–pterin-binding protein 2 (molbindin-2 or MopII) from Clostridium pasteurianum (PDB ID: 1GUT); c aligned structures of the molbindin-2 protomers. Structures were aligned using a MultiProt server [127]. Plot was created using VMD platform [128]; d high-resolution cryo-electron microscopy structure of a 40S ribosomal protein S33 from Trypanosoma brucei brucei (strain 927/4 GUTat10.1). Structure of this protein was extracted from of the cryo-EM structure of bacterial ribosome (PDB ID: 4V8M-AZ). Plot was created using VMD platform [128]; e solution NMR structure of a 30S bacterial ribosomal protein S28E from Methanobacterium thermoautotrophicum (PDB ID: 1NE3); f aligned structures of a eukaryotic 40S ribosomal protein rpS28e from Tetrahymena thermophila (PDB ID: 4V5O-A1/B1 and PDB ID: 4BTS-A1/B1/C1/D1). Corresponding structures were extracted from the crystal structures of the eukaryotic 40S ribosomal subunit in complex with initiation factor 1 (PDB ID: 4V5O-A1/B1) and the crystal structure of the eukaryotic 40S ribosomal subunit in complex with eIF1 and eIF1A (PDB ID: 4BTS-A1/B1/C1/D1). Structures were aligned using a MultiProt server [127]. Plot was created using VMD platform [128]; g solution NMR structure of the pulmonary surfactant-associated polypeptide C (SP-C) solved in apolar solvent [a mixed solvent of C2H3Cl/C2H3OH/1 M HCl 32:64:5 (v/v)] (PDB ID: 1SPF); and h crystal structure of the metal-bound dimer rat metallothionein-2 (PDB ID: 4MT2)
Our analysis showed that the number of proteins in the PDB data set that are depleted in the aromatic residues (FYWH-depleted) is also very low. In fact, we found only 7 such proteins, which, in addition to the aforementioned BCCP and molbindin-2 were a ribosomal protein S33 from Trypanosoma brucei brucei (strain 927/4 GUTat10.1) (which is a part of the bacterial ribosome, high-resolution structure of which was solved by cryo-electron microscopy, PDB ID: 4V8M-AZ, see Fig. 2d), a bacterial ribosomal protein S28E from Methanobacterium thermoautotrophicum (PDB ID: 1NE3, see Fig. 2e), an eukaryotic 40S ribosomal protein rpS28e from Tetrahymena thermophila (PDB ID: 4V5O-A1/B1 and PDB ID: 4BTS-A1/B1/C1/D1, see Fig. 2f), the pulmonary surfactant-associated polypeptide C (SP-C, PDB ID: 1SPF, see Fig. 2g), and rat metallothionein-2 (PDB ID: 4MT2, see Fig. 2h). Three of these FYWH-depleted proteins are ribosomal proteins, with the solution NMR structure of one of which (S28E) being solved at pH 4.5, and with two others (S33 and rpS28e) being a part of the ribosomal subunit). One of them (metallothionein-2) is a metal-binding protein that does not have any regular secondary structure elements and whose 3D structure is stabilized by homodimerization and coordination of five cadmium ions, two zinc ions, and one sodium ion [92]. The last one is a membrane-embedded protein (SP-C), whose structure in apolar solvent (a mixed solvent of C2H3Cl/C2H3OH/1 M HCl 32:64:5 (v/v)) was solved by NMR [93].
Since all CFYWH- and FYWH-depleted proteins in the PDB data set are rather small and are characterized by strong amino acid biases [for example, metallothionein-2 possesses extremely high content of cysteine residues (32.8%)], next, we analyzed their intrinsic disorder predispositions using a set of commonly used per-residue disorder predictors, such as PONDR® VLXT [7], PONDR® VL3 [94], PONDR® VSL2 [95], IUPred_short [96] (yellow curve), IUPred_long [96], and PONDR® FIT [97]. Figure 3 indicates that many of these proteins are predicted to have high levels of intrinsic disorder. In fact, according to their mean disorder predisposition, they can be ranged as follows: S33 (0.60 ± 0.16) > rpS28e (0.52 ± 0.17) > BCCP (0.47 ± 0.14) = metallothionein-2 (0.47 ± 0.44) > S28E (0.46 ± 0.15) > molbindin (0.33 ± 0.17) > SP-C (0.16 ± 0.15). Low level of intrinsic disorder in SP-C was expected, since this is a transmembrane protein characterized by the high content of hydrophobic, order-promoting residues. Figure 3 also shows that although, generally, the outputs of the predictors used in this study agree with each other, the disorder profile generated for metallothionein-2 reflects noticeable “confusion”, where PONDR® VL3, PONDR® VSL2, and PONDR® FIT predicted this protein to be completely disordered, whereas IUPred_short and IUPred_long suggested that the metallothionein-2 is absolutely ordered. This discrepancy is defined by the highly biased amino acid sequence of this protein, which does not have aromatic residues, being instead heavily enriched in cysteine residues (20 of its 61 residues (32.8%) are cysteines).
Fig. 3.
Multiparametric analysis of the intrinsic disorder predisposition of the CFYWH- and FYWH-depleted proteins found in PDB by several common predictors of intrinsic disorder: PONDR® VLXT [7] (black curves), PONDR® VL3 [94] (red curves), PONDR® VSL2 [96] (green curves), IUPred_short [96] (yellow curves), IUPred_long [96] (blue curves), and PONDR® FIT [97] (pink curves). Dark cyan dashed line shows the mean disorder propensity calculated by averaging disorder profiles of individual predictors. Light pink shadow around the PONDR® FIT shows error distribution for this predictor, whereas light cyan shadow around the mean disorder curve shows error distribution for evaluation of mean disorder. In these analyses, the predicted intrinsic disorder scores above 0.5 are considered to correspond to the disordered residues/regions, whereas regions with the disorder scores between 0.2 and 0.5 are considered flexible. Analyzed proteins were a BCCP (residues 1–9/49–123 of UniProt ID: 02904); b molbindin-2 (UniProt ID: P08854); c 40S ribosomal protein S33 from Trypanosoma brucei (UniProt ID: Q57U30); d 30S ribosomal protein S28E from Methanobacterium thermoautotrophicum (UniProt ID: O26356); e 40S ribosomal protein rpS28e from Tetrahymena thermophila (UniProt ID: Q234G5); f pulmonary surfactant-associated polypeptide C (UniProt ID: P15785); and g metallothionein-2 (UniProt ID: P04355)
One can argue that CWYFH-depleted proteins could contain a higher number of other (non-WYFH) hydrophobic amino acids and still be folded. Unfortunately, the amount of currently available data related to such proteins is not sufficient for conducting reliable statistical analysis to check this hypothesis. In fact, almost complete lack of the non-smelly proteins in PDB, which has only two CWYFH-depleted and seven WYFH-depleted proteins, serves as an important indication that unique (foldable) protein structure requires cysteines and aromatic residues. Composition profiler-based [33] comparison of the amino acid compositions of two CWYFH-depleted proteins, BCCP, and molbindin-2, with the amino acid compositions of globular proteins in PDB revealed that these non-smelly proteins are significantly enriched in valines (p value < 0.05). Extending this analysis to all seven WYFH-depleted proteins showed that they are significantly enriched in valines and methionines. However, the levels of other hydrophobic residues (leucines and isoleucines) were not significantly increased. These data are insufficient for making unambiguous conclusion on the presence of the compensatory increase in the number of non-CWYFH hydrophobic amino acids in the non-smelly proteins. In general, since hydrophobic residues are order-promoting [36], one would expect that if such compensation would take place, then the resulting WYFH-depleted proteins with the increased content of non-WYFH hydrophobic residues would still be mostly ordered. However, we are showing here that proteins without CWYFH are more disordered than proteins with CWYFH (see below). This indicates that the proposed compensation is not globally observed. Of course, there could be some exceptions from the rule, but there is no such compensation, in general. Furthermore, there is a logical limit on how many hydrophobic residues one can put into a sequence that can fold into a soluble structure (there is the surface to volume ratio limiting the number of hydrophobic groups that can be protected from water by a surface layer of hydrophilic residues upon formation of a globular structure).
In summary, the number of the CFYWH- and FYWH-depleted proteins in PDB is vanishingly small. Despite being structurally characterized, and these proteins typically (with the noticeable exception of the pulmonary surfactant-associated polypeptide C, which is a highly hydrophobic, membrane binding protein) have rather content of intrinsic disorder. None of these proteins are enzymes. They are either oligomeric metal-binding proteins or ribosomal proteins engaged in interaction with ribosomal RNA and other ribosomal proteins, or parts of protein complexes, or transmembrane proteins. In other words, none of these seven proteins exist as a non-interacting monomer, suggesting that their structure is stabilized by interaction with binding partners. Therefore, it is safe to conclude that stable monomeric protein structure requires the inclusion of cysteine and aromatic residues.
C- and CFYWH-depleted proteins are enriched in intrinsic disorder
The empirical observation that C-/CFYWH-depleted proteins are relatively rare in PDB suggests that they could be intrinsically disordered [8, 10, 98]. We tested this hypothesis utilizing accurate putative annotations of disorder. Figure 4a compares the putative disorder content (% of disordered residues) in all complete proteomes with the putative disorder content in the C-depleted and the CFYWH-depleted proteins for each domains of life. We found that proteins depleted in cysteine have relatively high disorder content at 7.8% in Archaea, 11.0% in Bacteria, and 44.4% in Eukaryota. These are substantially higher amounts when compared to the corresponding complete proteomes. The increases relative to the complete proteomes range between (7.8–5.7)/5.7 = 37% in Achaea and (44.4–20.2)/20.2 = 120% in Eukaryota. The amounts of the putative intrinsic disorder are event higher among the CFYWH-depleted proteins, with 40.1% disorder content in Archaea, 60.9% in Bacteria, and over 80% in Eukaryota. When compared to the proteome-level disorder content, this corresponds to the relative increases by 604%, 867%, and 312%, respectively. Figure 4b compares fractions of the fully disordered proteins between the complete proteomes and the C-depleted and CFYWH-depleted protein data sets. The enrichment in the number of fully disordered protein is even more substantial than for the disorder content. About 0.2% of all proteins vs. 0.7% of the C-depleted proteins in Archaea are fully disordered (250% increase), 0.4% vs. 1.4% in Bacteria (250% increase), and 1.0% vs. 8.2% in Eukaryota (820% increase). The corresponding increases when comparing the whole-proteome-level amounts with the subset of the CFYWH-depleted proteins are approximately 8900% in Archaea and Bacteria and 6700% in Eukaryota. These results clearly demonstrate that the depletion in cysteine and in aromatic residues is associated with substantially elevated levels of intrinsic disorder across all domains of life.
Fig. 4.
Comparison of the disorder content (panel a) and fraction of fully disordered proteins (panel b) between complete proteomes, C-depleted proteins and CFYWH-depleted proteins in the three domains of life
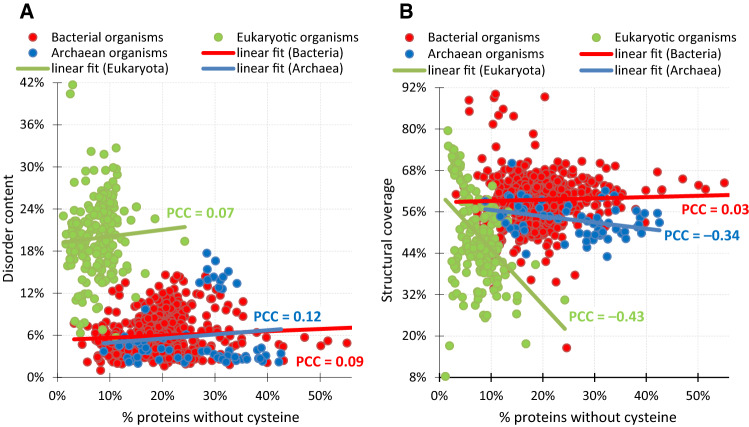
We further analyzed proteome-level relation between the disorder content and the abundance of the C-depleted proteins, see Fig. 5a. We did not pursue this analysis for the CFYWH-depleted proteins, since their numbers are small relative to the proteome sizes (Fig. 1a), and therefore, they do not make sufficient impact on the proteome-level measurements. Figure 5a reveals a slight increase in the disorder content for organisms with high levels of C-depleted proteins, i.e., the linear fit is sloped upwards and the Pearson correlation coefficients (PCCs) are positive consistently across the three domains of life. This is in agreement with the domain-level increase in the intrinsic disorder for the C-depleted proteins, as shown in Fig. 4a. We investigated whether this trends correlates with the current levels of structural coverage (% of proteins with at least partially known structures). Figure 5b shows relation between the current structural coverage and the amount of the C-depleted proteins for the three domains of life. We observe the modest correlations for Archaea (PCC = − 0.34) and Eukaryota (PCC = − 0.43), and no correlation for Bacteria (PCC = 0.03). This suggests that archaeal and eukaryotic organisms with higher levels of the C-depleted proteins are characterized by lower levels of structural coverage. Table 1 reveals that in case of the eukaryotes, this is driven by the high structural coverage and low fraction of the C-depleted proteins in metazoa. This, in turn, is related to a strong taxonomic bias in the PDB, where 44% of protein structures (61,323 out of the total of 138,194) are from metazoan organisms, in spite of the fact that only 10% of currently sequenced proteins (13,484,303 out of 134,315,728 in UniProt) are from this kingdom of life. One possible explanation for the lack of the correlation in Bacteria is that these proteins have high propensity for crystallization, particularly in contrast to the eukaryotic proteins [75]. This has substantial influence, since X-ray crystallography is the single biggest contributor to the protein structure determination efforts [99], i.e., 90.3% (124,770 out of 138,194) protein structures in PDB were solved using X-ray crystallography. The visible decline in the structural coverage for Archaean proteins, which also have high propensity for crystallization [75], is likely a result of the significantly higher amount of the C-depleted proteins (Fig. 1a) when compared to the Bacterial proteins. Overall, our empirical analysis reveals that archaeal and eukaryotic organisms with higher levels of the C-depleted proteins are characterized by higher levels of the intrinsic disorder and lower levels of the structural coverage.
Fig. 5.
Relation between proteome-level abundance of the C-depleted proteins and structural coverage (panel a) and disorder content (panel b). Each point represents a single proteome. Lines represent linear fit into the data for a specific domain of life, which is accompanied with the corresponding values of the Person correlation coefficient (PCC)
Functional analysis of C-depleted and CFYWH-depleted proteins
Figure 6 lists cellular location and functions that are enriched among the C-depleted proteins. The analysis is broken into three types of annotations: cellular components (at the top of the figure), molecular functions (in the middle of the figure), and biological processes (at the bottom of the figure) and performed separately for each domain of life. The C-depleted proteins in Archaea and Bacteria are primarily localized in membranes and ribosome, while in Eukaryota, they are also found in the nucleosome and nucleolus. These subcellular locations point to a high likelihood that C-depleted proteins are involved in the protein–RNA and protein–DNA interactions. Molecular functions listed in Fig. 6 reveal that indeed, they interact with the rRNAs and, in eukaryotes, with DNA, while also being involved in the transporter and motor functions. Since the C-depleted proteins are enriched in the intrinsic disorder, these observations are further supported by literature that suggests that IDPs and IDPRs play key roles in the protein–nucleic acids interactions [7, 30, 67, 68, 70, 100–106]. The biological processes associated with the C-depleted proteins are consistent with the aforementioned observations, and they cover translation, protein folding, nucleosome assembly, and protein transport. The subset of fully disordered C-depleted proteins (FD lines in Fig. 6) can be found in ribosome, nucleosome, and, in eukaryotes, in the chromatin. These proteins implement translation in Archaea and Bacteria, and they also carry out several other functions in Eukaryota, such as response to stress and spermatogenesis. Overall, our analysis reveals that protein–nucleic acids interactions underlie cellular functions and locations of the C-depleted proteins.
Fig. 6.
Cellular components (top of the figure), molecular functions (middle of the figure) and biological processes (bottom of the figure) that are significantly enriched in all C-depleted proteins (ALL lines) and among fully disordered C-depleted proteins (FD lines). The analysis is performed separately for eukaryotic species (green bars), bacterial species (red bars) archaeal species (blue bars). The y-axis lists five most frequent (the corresponding number of annotated proteins is shown inside the brackets) and significantly enriched functions/components (p value < 0.001 and enrichment > 30%). The x-axis shows the enrichment measured as relative increase in frequency when compared to the size and chain length matched set of randomly chosen proteins from the same domain of life. Details of the calculation are explained in the “Materials and methods” section. The functions/cellular components are sorted, within each group, by the number of annotated proteins
Figure 7 summarizes the major subcellular locations and functions that are enriched in the CFYWH-depleted proteins. These proteins are primarily found in ribosome in Archaea and Bacteria, while in Eukaryota, they are also located in the nucleus, particularly in the chromatin, by being part of the nucleosome complex. This is in agreement with the observations that these proteins are significantly enriched in disorder (Fig. 4) and that the nucleosome and ribosome complexes contain proteins enriched in disordered regions [67, 68, 70, 101]. The molecular functions and processes associated with the CFYWH-depleted proteins involve RNA and DNA binding in the context of translation, nucleosome assembly, and transcription. This is again consistent with earlier studies that revealed that the high levels of intrinsic disorder represent one of the important characteristics of the nucleic acid-binding proteins [7, 30, 67, 68, 70, 100–106]. Furthermore, the spatially and temporally coordinated action of many macromolecular complexes and proteins containing functionally significant IDPRs represents an important means for the control of transcription [107]. The major stages of transcription include chromatin remodeling that regulates the global accessibility of promoter DNA, action of regulatory transcription factors, co-activators/co-repressors, and the basal transcription machinery, and at each of these stages, intrinsically disordered proteins or proteins with IDPRs play very important regulatory roles [107]. Next, we discuss the role of disordered nucleic acid-binding proteins in each of these stages.
Fig. 7.
Cellular components (top of the figure), molecular functions (middle of the figure) and biological processes (bottom of the figure) that are significantly enriched in all CFYWH-depleted proteins (ALL lines) and among fully disordered CFYWH-depleted proteins (FD lines). The analysis is performed separately for eukaryotic species (green bars), bacterial species (red bars) archaeal species (blue bars). The y-axis lists five most frequent (the corresponding number of annotated proteins is shown inside the brackets) and significantly enriched functions/components (p value < 0.001 and enrichment > 30%). The x-axis shows the enrichment measured as relative increase in frequency when compared to the size and chain length matched set of randomly chosen proteins from the same domain of life. Details of the calculation are explained in “Materials and methods”. The functions/cellular components are sorted, within each group, by the number of annotated proteins
Formation of the nucleosomes, which are the basic structural units of chromatin, represents the primary step in the DNA condensation that is strongly protein intrinsic disorder-dependent. Nucleosomes are formed via association of small, highly basic nuclear proteins, core histones, with DNA in a specific stoichiometry. The formed nucleosomes are condensed together via action of the linker histones. Comprehensive bioinformatics analyses of 2007 histones from 746 species revealed that all the members of the histone family are highly disordered and utilize disorder for various functions, such as heterodimerization, formation of higher order oligomers, interaction with DNA and other proteins, and posttranslational modifications [101]. Among nuclear proteins that bind to nucleosomes, alter the structure of chromatin, and affect transcription are the members of a high mobility group N (HMGN) protein family of highly disordered chromatin modifying proteins [108]. In addition to HMGNs, many other IDPs and proteins containing functionally important IDPRs, including various chromatin modifying enzymes, are involved in the regulation of DNA accessibility [107].
Among the most illustrative examples of IDPs related to the regulation of transcription (after the chromatin environment becomes accessible due to the actin of chromatin remodeling proteins) are transcription factors (TFs, which are also known as sequence-specific DNA-binding factors). TFs are multifunctional proteins which are crucial for the control of expression of specific genes and for the regulation of the gene activity in response to specific stimuli. They deliver their effects via binding to specific DNA sequences, recruiting the RNA polymerase to specific genes, controlling the transfer of genetic information from DNA to mRNA, and positively or negatively influencing the gene transcription either alone or in a complex with other proteins [109]. In general, the modular structure of TFs includes one or more DNA-binding domains (DBDs) for recognition and binding of the specific DNA sequences adjacent to the genes that they regulate, and one or more transactivation domains for recognition of the co-activators and/or other transcription factors. Computational analysis of several TF data sets revealed that between 82.6 and 94.1% of TFs possess long IDPRs, with the degree of disorder being significantly higher in eukaryotic FTs in comparison with their prokaryotic counterparts [110]. TFs also contain high levels of disorder-based protein interaction sites, molecular recognition features (MoRFs) [110]. Intrinsic disorder is not distributed evenly within the sequences of TFs. In fact, although in general, the DNA-binding domains are noticeably less disordered than the TF activation regions (or transactivator domains), the AT-hooks, and basic regions of DNA-binding domains of TFs are highly disordered [110]. In human TFs, almost 50% of the entire sequences are occupied by IDPRs [111]. Intrinsic disorder of transactivator domains is used in communication of TFs with other regulatory transcriptional proteins and has an important role in orchestrating the transcriptional assemblies [107]. Based on the high prevalence and versatility of intrinsic disorder in eukaryotic TFs, it has been concluded that these proteins can be used as important illustrations of various aspects of intrinsic disorder-based functionality [112].
At the next stage of transcription, co-activators and co-repressors define a cross-talk between chromatin, transcription factors, and the basal transcription machinery. Some of the co-activators can be considered as scaffolds containing multiple transcription factor-binding sites and thereby processing multiple transcriptional regulatory inputs. One of such co-activators, p300, is known to interact with over 50 proteins and possesses histone acetyltransferase activity. Another illustrative example of the importance of intrinsic disorder in the transcription regulation is the Mediator complex. This complex serves as an interface between gene-specific regulatory proteins and the general transcription machinery and it contains high levels of functional intrinsic disorder [113].
Similarly, many proteins related to translation (i.e., the process of ribosome-mediated biosynthesis of proteins from mRNA) are either intrinsically disordered or contain long IDPRs. For example, ribosomal proteins are considered as an important example of the exceptional functional versatility of the RNA-binding IDPs. Based on the comprehensive bioinformatics analyses of the 3411 ribosomal proteins from 32 species, it has been concluded that many ribosomal proteins are either intrinsically disordered as a whole or represent hybrids containing ordered and disordered domains and that intrinsic disorder is absolutely crucial for their various functions [68]. In agreement with these observations, our analysis showed that three of seven FYWH-depleted proteins, whose structure is present in PDB, are ribosomal proteins.
Taken together, our analysis revealed that although proteins that do not contain cysteines constitute rather large fraction of the analyzed proteomes (content of such C-depleted proteins is ranging from 8% proteins in Eukaryota to 19% in Bacteria and to 28% in Archaea), proteins that do not have cysteine and aromatic residues (CFYWH-depleted proteins) constitute only very minor fractions of 817 complete proteomes (about 0.06% proteins in Archaea and 0.04% proteins in Bacteria and Eukaryota). Archaeal and eukaryotic organisms with higher levels of the C-depleted proteins are predicted to have higher intrinsic disorder levels and lower structural coverage levels, whereas CFYWH-depleted proteins across all domains of life are characterized by the substantially increased levels of intrinsic disorder. Functional analysis revealed that the “non-smelly” proteins are often involved in protein–nucleic acids interactions. They are rarely present as independently folded monomeric structures and often serve as parts of the ribosome and nucleosome complexes. They are also found in cellular membranes. These C- and CFYWH-depleted “non-smelly” proteins are involved in translation, transcription, nucleosome assembly, transmembrane transport, and protein folding functions, all of which are known to be associated with the intrinsic disorder.
Finally, described in this article general inability of the “non-smelly” proteins to fold into self-organizing monomeric structures provides support to the hypothesis on the highly disordered nature of the primordial proteins, which is based on an intriguing correlation between the evolutions of genetic code and protein structure [114–116]. In fact, it was pointed out that the “prebiotic set” of amino acids (i.e., a set of amino acids that were generated by various abiotic processes) likely included 10 of 20 modern amino acids, such as A, D, E, G, I, L, P, S, T, and V [117, 118], many of which were disorder-promoting. Based on a combination of 40 different factors, Eduard Trifonov proposed the following temporal order of addition of the amino acids to the genetic code: G/A, V/D, P, S, E/L, T, R, N, K, Q, I, C, H, F, M, Y, and W [119]. This sorting underscores the correlation between the appearance of early amino acids (such as G, D, E, P, and S) in the primordial soup and their disorder-promoting tendencies in IDPs. In contrast, it seems that the major order-promoting residues, such as C, W, Y, F, and H, have been added to the genetic code at later evolutionary stages [114, 115]. In other words, primordial proteins were “non-smelly”. Similar inferences were also made by Brooks et al. in their study on the amino acid composition of last universal ancestral genomes [120]. In addition, it was pointed out that the emergence of the biosynthesis of aromatic amino acid enabled an early halophile-to-mesophile transition, emphasizing the potential role of aromatic residues in the adaptive spread of early life and suggesting a selective advantage for the incorporation of aromatic amino acids into the codon table [121]. Furthermore, the high prevalence of nucleic acid-binding-related functions among the modern “non-smelly” proteins can be considered as a kind of functional fossil, since nucleic acid binding and RNA chaperoning were proposed to be the first functions of primordial polypeptides [122, 123]. Such RNA chaperone activities of early proteins provided their carriers a significant selective advantage in the RNA world, where RNA, which is especially prone to misfolding [124, 125], was used for both information storage and catalysis [126].
Conclusions
We report the results of a comprehensive bioinformatics analysis of the prevalence and functionality of the “non-smelly” proteins (i.e., proteins that do not contain cysteine and aromatic residues, C- and CFYWH-depleted proteins) among 9,563,159 proteins from the 817 complete proteomes, and among the 99,461 PDB proteins with known 3D structures. This analysis revealed that prokaryotes are significantly enriched in the C-depleted proteins compared to eukaryotes. In fact, 28% proteins in Archaea and 19% in Bacteria vs. only 8% in Eukaryota do not have cysteines. In general, C-depleted proteins are often involved in protein–nucleic acids interactions and they relatively rarely fold into monomer structures. On the other hand, CFYWH-depleted proteins are rather rare, are equally distributed across the three domains of life, and are virtually never found in PDB. Only about 0.05% of proteins do not have cysteine and aromatic residues. Depletion in cysteine and in aromatic residues is associated with the substantially elevated levels of intrinsic disorder in proteins across all domains of life. Archaeal and eukaryotic organisms with higher levels of the C-depleted proteins have higher levels of the intrinsic disorder and lower levels of structural coverage. The C- and CFYWH-depleted proteins are part of the ribosome and nucleosome complexes and are also found in cellular membranes. They are involved in translation, transcription, nucleosome assembly, transmembrane transport and protein folding functions, all of which are known to be associated with the intrinsic disorder.
In line with highly disordered nature of the “non-smelly” proteins, is an important observation that such proteins are highly underrepresented in PDB. As a matter of fact, there are only two CFYWH-depleted proteins and five FYWH-depleted proteins among the hundred thousand proteins in the PDB data sets which were solved by X-ray crystallography, or NMR, or cryo-EM. Furthermore, only two of these proteins, deletion mutant of BCCP and ribosomal protein S28E, have structures that can be considered as a result of a spontaneous folding of a single polypeptide chain, whereas structures of the other “non-smelly” proteins are stabilized by binding of metal ions and self-oligomerization (metallothionein-2 and molbindin-2) or by inclusion into large ribonucleoprotein complexes (ribosomal proteins S33 and rpS28e), or by placing into the non-polar solvent (pulmonary surfactant-associated polypeptide C). These observations indicate that a self-foldable unique 3D-structure in a globular protein is crucially dependent on the presence of cysteine and aromatic residues in its amino acid sequence.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
This research was supported in part by the Qimonda Endowment and the National Science Foundation Grant 1617369 to Lukasz Kurgan.
Compliance with ethical standards
Conflict of interest
The authors have declared no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Lukasz Kurgan, Email: lkurgan@vcu.edu.
Vladimir N. Uversky, Email: vuversky@health.usf.edu
References
- 1.Xue B, Williams RW, Oldfield CJ, Dunker AK, Uversky VN. Archaic chaos: intrinsically disordered proteins in Archaea. BMC Syst Biol. 2010;4(Suppl 1):S1. doi: 10.1186/1752-0509-4-S1-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peng Z, Yan J, Fan X, Mizianty MJ, Xue B, Wang K, Hu G, Uversky VN, Kurgan L. Exceptionally abundant exceptions: comprehensive characterization of intrinsic disorder in all domains of life. Cell Mol Life Sci. 2015;72(1):137–151. doi: 10.1007/s00018-014-1661-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ward JJ, Sodhi JS, McGuffin LJ, Buxton BF, Jones DT. Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J Mol Biol. 2004;337(3):635–645. doi: 10.1016/j.jmb.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 4.Dunker AK, Obradovic Z, Romero P, Garner EC, Brown CJ. Intrinsic protein disorder in complete genomes. Genome Inform Ser Workshop Genome Inform. 2000;11:161–171. [PubMed] [Google Scholar]
- 5.Peng Z, Mizianty MJ, Kurgan L. Genome-scale prediction of proteins with long intrinsically disordered regions. Proteins. 2014;82(1):145–158. doi: 10.1002/prot.24348. [DOI] [PubMed] [Google Scholar]
- 6.Yan J, Mizianty MJ, Filipow PL, Uversky VN, Kurgan L. RAPID: fast and accurate sequence-based prediction of intrinsic disorder content on proteomic scale. Biochim Biophys Acta. 2013;1834(8):1671–1680. doi: 10.1016/j.bbapap.2013.05.022. [DOI] [PubMed] [Google Scholar]
- 7.Dunker AK, Lawson JD, Brown CJ, Williams RM, Romero P, Oh JS, Oldfield CJ, Campen AM, Ratliff CM, Hipps KW, Ausio J, Nissen MS, Reeves R, Kang C-H, Kissinger CR, Bailey RW, Griswold MD, Chiu W, Garner EC, Obradovic Z. Intrinsically disordered protein. J Mol Graph Model. 2001;19(1):26–59. doi: 10.1016/S1093-3263(00)00138-8. [DOI] [PubMed] [Google Scholar]
- 8.Hu G, Wang K, Song J, Uversky VN, Kurgan L. Taxonomic landscape of the dark proteomes: whole-proteome scale interplay between structural darkness, intrinsic disorder, and crystallization propensity. Proteomics. 2018 doi: 10.1002/pmic.201800243. [DOI] [PubMed] [Google Scholar]
- 9.Linding R, Jensen LJ, Diella F, Bork P, Gibson TJ, Russell RB. Protein disorder prediction: implications for structural proteomics. Structure. 2003;11(11):1453–1459. doi: 10.1016/j.str.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 10.Oldfield CJ, Xue B, Van YY, Ulrich EL, Markley JL, Dunker AK, Uversky VN. Utilization of protein intrinsic disorder knowledge in structural proteomics. Biochim Biophys Acta. 2013;1834(2):487–498. doi: 10.1016/j.bbapap.2012.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bhowmick A, Brookes DH, Yost SR, Dyson HJ, Forman-Kay JD, Gunter D, Head-Gordon M, Hura GL, Pande VS, Wemmer DE, Wright PE, Head-Gordon T. Finding our way in the dark proteome. J Am Chem Soc. 2016;138(31):9730–9742. doi: 10.1021/jacs.6b06543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kruger R. Illuminating the dark proteome. Cell. 2016;166(5):1074–1077. doi: 10.1016/j.cell.2016.08.012. [DOI] [PubMed] [Google Scholar]
- 13.Uversky VN, Dunker AK. Understanding protein non-folding. Biochim Biophys Acta. 2010;1804(6):1231–1264. doi: 10.1016/j.bbapap.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Uversky VN, Oldfield CJ, Dunker AK. Intrinsically disordered proteins in human diseases: introducing the D2 concept. Annu Rev Biophys. 2008;37:215–246. doi: 10.1146/annurev.biophys.37.032807.125924. [DOI] [PubMed] [Google Scholar]
- 15.Fuxreiter M, Toth-Petroczy A, Kraut DA, Matouschek A, Lim RY, Xue B, Kurgan L, Uversky VN. Disordered proteinaceous machines. Chem Rev. 2014;114(13):6806–6843. doi: 10.1021/cr4007329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oates ME, Romero P, Ishida T, Ghalwash M, Mizianty MJ, Xue B, Dosztanyi Z, Uversky VN, Obradovic Z, Kurgan L, Dunker AK, Gough J. D2P2: database of disordered protein predictions. Nucleic Acids Res. 2013;41:D508–D516. doi: 10.1093/nar/gks1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Romero PR, Zaidi S, Fang YY, Uversky VN, Radivojac P, Oldfield CJ, Cortese MS, Sickmeier M, LeGall T, Obradovic Z, Dunker AK. Alternative splicing in concert with protein intrinsic disorder enables increased functional diversity in multicellular organisms. Proc Natl Acad Sci USA. 2006;103(22):8390–8395. doi: 10.1073/pnas.0507916103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jakob U, Kriwacki R, Uversky VN. Conditionally and transiently disordered proteins: awakening cryptic disorder to regulate protein function. Chem Rev. 2014;114(13):6779–6805. doi: 10.1021/cr400459c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yan J, Dunker AK, Uversky VN, Kurgan L. Molecular recognition features (MoRFs) in three domains of life. Mol BioSyst. 2016;12(3):697–710. doi: 10.1039/c5mb00640f. [DOI] [PubMed] [Google Scholar]
- 20.Patil A, Kinoshita K, Nakamura H. Hub promiscuity in protein–protein interaction networks. Int J Mol Sci. 2010;11(4):1930–1943. doi: 10.3390/ijms11041930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gsponer J, Babu MM. The rules of disorder or why disorder rules. Prog Biophys Mol Biol. 2009;99(2–3):94–103. doi: 10.1016/j.pbiomolbio.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 22.Haynes C, Oldfield CJ, Ji F, Klitgord N, Cusick ME, Radivojac P, Uversky VN, Vidal M, Iakoucheva LM. Intrinsic disorder is a common feature of hub proteins from four eukaryotic interactomes. PLoS Comput Biol. 2006;2(8):e100. doi: 10.1371/journal.pcbi.0020100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu G, Wu Z, Uversky VN, Kurgan L. Functional analysis of human hub proteins and their interactors involved in the intrinsic disorder-enriched interactions. Int J Mol Sci. 2017 doi: 10.3390/ijms18122761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dunker AK, Cortese MS, Romero P, Iakoucheva LM, Uversky VN. Flexible nets. The roles of intrinsic disorder in protein interaction networks. FEBS J. 2005;272(20):5129–5148. doi: 10.1111/j.1742-4658.2005.04948.x. [DOI] [PubMed] [Google Scholar]
- 25.Barabasi AL, Oltvai ZN. Network biology: understanding the cell’s functional organization. Nat Rev Genet. 2004;5(2):101–113. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- 26.Barabasi AL, Albert R. Emergence of scaling in random networks. Science. 1999;286(5439):509–512. doi: 10.1126/science.286.5439.509. [DOI] [PubMed] [Google Scholar]
- 27.Vavouri T, Semple JI, Garcia-Verdugo R, Lehner B. Intrinsic protein disorder and interaction promiscuity are widely associated with dosage sensitivity. Cell. 2009;138(1):198–208. doi: 10.1016/j.cell.2009.04.029. [DOI] [PubMed] [Google Scholar]
- 28.Oldfield CJ, Meng J, Yang JY, Yang MQ, Uversky VN, Dunker AK. Flexible nets: disorder and induced fit in the associations of p53 and 14-3-3 with their partners. BMC Genom. 2008;9(Suppl 1):S1. doi: 10.1186/1471-2164-9-S1-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Buljan M, Chalancon G, Dunker AK, Bateman A, Balaji S, Fuxreiter M, Babu MM. Alternative splicing of intrinsically disordered regions and rewiring of protein interactions. Curr Opin Struct Biol. 2013;23(3):443–450. doi: 10.1016/j.sbi.2013.03.006. [DOI] [PubMed] [Google Scholar]
- 30.Uversky VN, Gillespie JR, Fink AL. Why are “natively unfolded” proteins unstructured under physiologic conditions? Proteins. 2000;41(3):415–427. doi: 10.1002/1097-0134(20001115)41:3<415::aid-prot130>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 31.Dunker AK, Garner E, Guilliot S, Romero P, Albrecht K, Hart J, Obradovic Z, Kissinger C, Villafranca JE. Protein disorder and the evolution of molecular recognition: theory, predictions and observations. Pac Symp Biocomput. 1998;3:473–484. [PubMed] [Google Scholar]
- 32.Radivojac P, Iakoucheva LM, Oldfield CJ, Obradovic Z, Uversky VN, Dunker AK. Intrinsic disorder and functional proteomics. Biophys J. 2007;92(5):1439–1456. doi: 10.1529/biophysj.106.094045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vacic V, Uversky VN, Dunker AK, Lonardi S. Composition profiler: a tool for discovery and visualization of amino acid composition differences. BMC Bioinform. 2007;8:211. doi: 10.1186/1471-2105-8-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Romero P, Obradovic Z, Li X, Garner EC, Brown CJ, Dunker AK. Sequence complexity of disordered protein. Proteins. 2001;42(1):38–48. doi: 10.1002/1097-0134(20010101)42:1<38::AID-PROT50>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 35.Campen A, Williams RM, Brown CJ, Meng J, Uversky VN, Dunker AK. TOP-IDP-scale: a new amino acid scale measuring propensity for intrinsic disorder. Protein Pept Lett. 2008;15(9):956–963. doi: 10.2174/092986608785849164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Williams RM, Obradovi Z, Mathura V, Braun W, Garner EC, Young J, Takayama S, Brown CJ, Dunker AK. The protein non-folding problem: amino acid determinants of intrinsic order and disorder. Pac Symp Biocomput. 2001;6:89–100. doi: 10.1142/9789814447362_0010. [DOI] [PubMed] [Google Scholar]
- 37.Daly NL, Craik DJ. Bioactive cystine knot proteins. Curr Opin Chem Biol. 2011;15(3):362–368. doi: 10.1016/j.cbpa.2011.02.008. [DOI] [PubMed] [Google Scholar]
- 38.Craik DJ, Daly NL, Waine C. The cystine knot motif in toxins and implications for drug design. Toxicon. 2001;39(1):43–60. doi: 10.1016/S0041-0101(00)00160-4. [DOI] [PubMed] [Google Scholar]
- 39.Trivedi MV, Laurence JS, Siahaan TJ. The role of thiols and disulfides on protein stability. Curr Protein Pept Sci. 2009;10(6):614–625. doi: 10.2174/138920309789630534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hagihara Y, Saerens D. Engineering disulfide bonds within an antibody. Biochim Biophys Acta. 2014;1844(11):2016–2023. doi: 10.1016/j.bbapap.2014.07.005. [DOI] [PubMed] [Google Scholar]
- 41.Bechtel TJ, Weerapana E. From structure to redox: the diverse functional roles of disulfides and implications in disease. Proteomics. 2017 doi: 10.1002/pmic.201600391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Darling AL, Uversky VN. Intrinsic disorder and posttranslational modifications: the darker side of the biological dark matter. Front Genet. 2018;9:158. doi: 10.3389/fgene.2018.00158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pace NJ, Weerapana E. Zinc-binding cysteines: diverse functions and structural motifs. Biomolecules. 2014;4(2):419–434. doi: 10.3390/biom4020419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Krishna SS, Majumdar I, Grishin NV. Structural classification of zinc fingers: survey and summary. Nucleic Acids Res. 2003;31(2):532–550. doi: 10.1093/nar/gkg161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Negi S, Itazu M, Imanishi M, Nomura A, Sugiura Y. Creation and characteristics of unnatural CysHis3-type zinc finger protein. Biochem Biophys Res Commun. 2004;325(2):421–425. doi: 10.1016/j.bbrc.2004.10.045. [DOI] [PubMed] [Google Scholar]
- 46.Harding MM. The architecture of metal coordination groups in proteins. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 5):849–859. doi: 10.1107/S0907444904004081. [DOI] [PubMed] [Google Scholar]
- 47.Laska M. Olfactory perception of 6 amino acids by human subjects. Chem Senses. 2010;35(4):279–287. doi: 10.1093/chemse/bjq017. [DOI] [PubMed] [Google Scholar]
- 48.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The protein data bank. Nucleic Acids Res. 2000;28(1):235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Burley SK, Berman HM, Kleywegt GJ, Markley JL, Nakamura H, Velankar S. Protein data bank (PDB): the single global macromolecular structure archive. Methods Mol Biol. 2017;1607:627–641. doi: 10.1007/978-1-4939-7000-1_26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.The UniProt C. UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2017;45(D1):D158–D169. doi: 10.1093/nar/gkw1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.UniProt C UniProt: a hub for protein information. Nucleic Acids Res. 2015;43(Database issue):D204–D212. doi: 10.1093/nar/gku989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hu G, Wu Z, Oldfield CJ, Wang C, Kurgan L. Quality assessment for the putative intrinsic disorder in proteins. Bioinformatics. 2019;35(10):1692–1700. doi: 10.1093/bioinformatics/bty881. [DOI] [PubMed] [Google Scholar]
- 53.Katuwawala A, Oldfield CJ, Kurgan L. Accuracy of protein-level disorder predictions. Brief Bioinform. 2019;46:48. doi: 10.1093/bib/bbz100. [DOI] [PubMed] [Google Scholar]
- 54.Walsh I, Giollo M, Di Domenico T, Ferrari C, Zimmermann O, Tosatto SC. Comprehensive large-scale assessment of intrinsic protein disorder. Bioinformatics. 2015;31(2):201–208. doi: 10.1093/bioinformatics/btu625. [DOI] [PubMed] [Google Scholar]
- 55.Monastyrskyy B, Kryshtafovych A, Moult J, Tramontano A, Fidelis K. Assessment of protein disorder region predictions in CASP10. Proteins. 2014;82(Suppl 2):127–137. doi: 10.1002/prot.24391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Peng ZL, Kurgan L. Comprehensive comparative assessment of in silico predictors of disordered regions. Curr Protein Pept Sci. 2012;13(1):6–18. doi: 10.2174/138920312799277938. [DOI] [PubMed] [Google Scholar]
- 57.Meng F, Uversky VN, Kurgan L. Comprehensive review of methods for prediction of intrinsic disorder and its molecular functions. Cell Mol Life Sci. 2017;74(17):3069–3090. doi: 10.1007/s00018-017-2555-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Meng F, Uversky V, Kurgan L. Computational prediction of intrinsic disorder in proteins. Curr Protoc Protein Sci. 2017;88:2–16. doi: 10.1002/cpps.28. [DOI] [PubMed] [Google Scholar]
- 59.Peng Z, Kurgan L. On the complementarity of the consensus-based disorder prediction. Pac Symp Biocomput. 2012;17:176–187. [PubMed] [Google Scholar]
- 60.Fan X, Kurgan L. Accurate prediction of disorder in protein chains with a comprehensive and empirically designed consensus. J Biomol Struct Dyn. 2014;32(3):448–464. doi: 10.1080/07391102.2013.775969. [DOI] [PubMed] [Google Scholar]
- 61.Necci M, Piovesan D, Dosztanyi Z, Tosatto SCE. MobiDB-lite: fast and highly specific consensus prediction of intrinsic disorder in proteins. Bioinformatics. 2017;33(9):1402–1404. doi: 10.1093/bioinformatics/btx015. [DOI] [PubMed] [Google Scholar]
- 62.Dosztanyi Z, Csizmok V, Tompa P, Simon I. The pairwise energy content estimated from amino acid composition discriminates between folded and intrinsically unstructured proteins. J Mol Biol. 2005;347(4):827–839. doi: 10.1016/j.jmb.2005.01.071. [DOI] [PubMed] [Google Scholar]
- 63.Walsh I, Martin AJ, Di Domenico T, Tosatto SC. ESpritz: accurate and fast prediction of protein disorder. Bioinformatics. 2012;28(4):503–509. doi: 10.1093/bioinformatics/btr682. [DOI] [PubMed] [Google Scholar]
- 64.Piovesan D, Tabaro F, Micetic I, Necci M, Quaglia F, Oldfield CJ, Aspromonte MC, Davey NE, Davidovic R, Dosztanyi Z, Elofsson A, Gasparini A, Hatos A, Kajava AV, Kalmar L, Leonardi E, Lazar T, Macedo-Ribeiro S, Macossay-Castillo M, Meszaros A, Minervini G, Murvai N, Pujols J, Roche DB, Salladini E, Schad E, Schramm A, Szabo B, Tantos A, Tonello F, Tsirigos KD, Veljkovic N, Ventura S, Vranken W, Warholm P, Uversky VN, Dunker AK, Longhi S, Tompa P, Tosatto SC. DisProt 7.0: a major update of the database of disordered proteins. Nucleic Acids Res. 2016;D1:D219–D227. doi: 10.1093/nar/gkw1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vucetic S, Obradovic Z, Vacic V, Radivojac P, Peng K, Iakoucheva LM, Cortese MS, Lawson JD, Brown CJ, Sikes JG, Newton CD, Dunker AK. DisProt: a database of protein disorder. Bioinformatics. 2005;21(1):137–140. doi: 10.1093/bioinformatics/bth476. [DOI] [PubMed] [Google Scholar]
- 66.Na I, Meng F, Kurgan L, Uversky VN. Autophagy-related intrinsically disordered proteins in intra-nuclear compartments. Mol BioSyst. 2016;12(9):2798–2817. doi: 10.1039/c6mb00069j. [DOI] [PubMed] [Google Scholar]
- 67.Meng F, Na I, Kurgan L, Uversky VN. Compartmentalization and functionality of nuclear disorder: intrinsic disorder and protein–protein interactions in intra-nuclear compartments. Int J Mol Sci. 2016 doi: 10.3390/ijms17010024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Peng Z, Oldfield CJ, Xue B, Mizianty MJ, Dunker AK, Kurgan L, Uversky VN. A creature with a hundred waggly tails: intrinsically disordered proteins in the ribosome. Cell Mol Life Sci. 2014;71(8):1477–1504. doi: 10.1007/s00018-013-1446-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hu G, Wu Z, Wang K, Uversky VN, Kurgan L. Untapped potential of disordered proteins in current druggable human proteome. Curr Drug Targets. 2016;17(10):1198–1205. doi: 10.2174/1389450116666150722141119. [DOI] [PubMed] [Google Scholar]
- 70.Wang C, Uversky VN, Kurgan L. Disordered nucleiome: abundance of intrinsic disorder in the DNA- and RNA-binding proteins in 1121 species from Eukaryota, Bacteria and Archaea. Proteomics. 2016;16(10):1486–1498. doi: 10.1002/pmic.201500177. [DOI] [PubMed] [Google Scholar]
- 71.Peng Z, Uversky VN, Kurgan L. Genes encoding intrinsic disorder in Eukaryota have high GC content. Intrinsically Disord Proteins. 2016;4(1):e1262225. doi: 10.1080/21690707.2016.1262225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Di Domenico T, Walsh I, Martin AJM, Tosatto SCE. MobiDB: a comprehensive database of intrinsic protein disorder annotations. Bioinformatics. 2012;28(15):2080–2081. doi: 10.1093/bioinformatics/bts327. [DOI] [PubMed] [Google Scholar]
- 73.Potenza E, Di Domenico T, Walsh I, Tosatto SC. MobiDB 2.0: an improved database of intrinsically disordered and mobile proteins. Nucleic Acids Res. 2015;43(Database issue):D315–D320. doi: 10.1093/nar/gku982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vitkup D, Melamud E, Moult J, Sander C. Completeness in structural genomics. Nat Struct Biol. 2001;8(6):559–566. doi: 10.1038/88640. [DOI] [PubMed] [Google Scholar]
- 75.Mizianty MJ, Fan X, Yan J, Chalmers E, Woloschuk C, Joachimiak A, Kurgan L. Covering complete proteomes with X-ray structures: a current snapshot. Acta Crystallogr D Biol Crystallogr. 2014;70(Pt 11):2781–2793. doi: 10.1107/S1399004714019427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mariani V, Kiefer F, Schmidt T, Haas J, Schwede T. Assessment of template based protein structure predictions in CASP9. Proteins. 2011;79(Suppl 10):37–58. doi: 10.1002/prot.23177. [DOI] [PubMed] [Google Scholar]
- 78.Yang J, Yan R, Roy A, Xu D, Poisson J, Zhang Y (2015) The I-TASSER Suite: protein structure and function prediction. Nat Methods 12(1):7–8. 10.1038/nmeth.3213. http://www.nature.com/nmeth/journal/v12/n1/abs/nmeth.3213.html#supplementary-information [DOI] [PMC free article] [PubMed]
- 79.Yang J, Zhang Y. I-TASSER server: new development for protein structure and function predictions. Nucleic Acids Res. 2015;43(W1):W174–W181. doi: 10.1093/nar/gkv342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Soding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33:W244–W248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hildebrand A, Remmert M, Biegert A, Soding J. Fast and accurate automatic structure prediction with HHpred. Proteins. 2009;77(Suppl 9):128–132. doi: 10.1002/prot.22499. [DOI] [PubMed] [Google Scholar]
- 82.Webb B, Sali A. Protein structure modeling with MODELLER. Methods Mol Biol. 2017;1654:39–54. doi: 10.1007/978-1-4939-7231-9_4. [DOI] [PubMed] [Google Scholar]
- 83.Sali A, Potterton L, Yuan F, van Vlijmen H, Karplus M. Evaluation of comparative protein modeling by MODELLER. Proteins. 1995;23(3):318–326. doi: 10.1002/prot.340230306. [DOI] [PubMed] [Google Scholar]
- 84.Pieper U, Webb BM, Dong GQ, Schneidman-Duhovny D, Fan H, Kim SJ, Khuri N, Spill YG, Weinkam P, Hammel M, Tainer JA, Nilges M, Sali A. ModBase, a database of annotated comparative protein structure models and associated resources. Nucleic Acids Res. 2014;42:D336–D346. doi: 10.1093/nar/gkt1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G, The Gene Ontology Consortium Gene ontology: tool for the unification of biology. Nat Genet. 2000;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Howell M, Green R, Killeen A, Wedderburn L, Picascio V, Rabionet A, Peng ZL, Larina M, Xue B, Kurgan L, Uversky VN. Not that rigid midgets and not so flexible giants: on the abundance and roles of intrinsic disorder in short and long proteins. J Biol Syst. 2012;20(4):471–511. doi: 10.1142/S0218339012400086. [DOI] [Google Scholar]
- 87.Hennessey JP, Jr, Johnson WC, Jr, Bahler C, Wood HG. Subunit interactions of transcarboxylase as studied by circular dichroism. Biochemistry. 1982;21(4):642–646. doi: 10.1021/bi00533a007. [DOI] [PubMed] [Google Scholar]
- 88.Shenoy BC, Wood HG. Purification and properties of the synthetase catalyzing the biotination of the aposubunit of transcarboxylase from Propionibacterium shermanii. FASEB J. 1988;2(8):2396–2401. doi: 10.1096/fasebj.2.8.3360240. [DOI] [PubMed] [Google Scholar]
- 89.Reddy DV, Shenoy BC, Carey PR, Sonnichsen FD. High resolution solution structure of the 1.3S subunit of transcarboxylase from Propionibacterium shermanii. Biochemistry. 2000;39(10):2509–2516. doi: 10.1021/bi9925367. [DOI] [PubMed] [Google Scholar]
- 90.Jank MM, Sadowsky JD, Peikert C, Berger S. NMR studies on the solution structure of a deletion mutant of the transcarboxylase biotin carrier subunit. Int J Biol Macromol. 2002;30(5):233–242. doi: 10.1016/S0141-8130(02)00033-8. [DOI] [PubMed] [Google Scholar]
- 91.Schuttelkopf AW, Harrison JA, Boxer DH, Hunter WN. Passive acquisition of ligand by the MopII molbindin from Clostridium pasteurianum: structures of apo and oxyanion-bound forms. J Biol Chem. 2002;277(17):15013–15020. doi: 10.1074/jbc.M201005200. [DOI] [PubMed] [Google Scholar]
- 92.Braun W, Vasak M, Robbins AH, Stout CD, Wagner G, Kagi JH, Wuthrich K. Comparison of the NMR solution structure and the X-ray crystal structure of rat metallothionein-2. Proc Natl Acad Sci USA. 1992;89(21):10124–10128. doi: 10.1073/pnas.89.21.10124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Johansson J, Szyperski T, Curstedt T, Wuthrich K. The NMR structure of the pulmonary surfactant-associated polypeptide SP-C in an apolar solvent contains a valyl-rich alpha-helix. Biochemistry. 1994;33(19):6015–6023. doi: 10.1021/bi00185a042. [DOI] [PubMed] [Google Scholar]
- 94.Peng K, Radivojac P, Vucetic S, Dunker AK, Obradovic Z. Length-dependent prediction of protein intrinsic disorder. BMC Bioinform. 2006;7:208. doi: 10.1186/1471-2105-7-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Peng K, Vucetic S, Radivojac P, Brown CJ, Dunker AK, Obradovic Z. Optimizing long intrinsic disorder predictors with protein evolutionary information. J Bioinform Comput Biol. 2005;3(1):35–60. doi: 10.1142/s0219720005000886. [DOI] [PubMed] [Google Scholar]
- 96.Dosztanyi Z, Csizmok V, Tompa P, Simon I. IUPred: web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics. 2005;21(16):3433–3434. doi: 10.1093/bioinformatics/bti541. [DOI] [PubMed] [Google Scholar]
- 97.Xue B, Dunbrack RL, Williams RW, Dunker AK, Uversky VN. PONDR-FIT: a meta-predictor of intrinsically disordered amino acids. Biochim Biophys Acta. 2010;1804(4):996–1010. doi: 10.1016/j.bbapap.2010.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Oldfield CJ, Ulrich EL, Cheng Y, Dunker AK, Markley JL. Addressing the intrinsic disorder bottleneck in structural proteomics. Proteins. 2005;59(3):444–453. doi: 10.1002/prot.20446. [DOI] [PubMed] [Google Scholar]
- 99.Grabowski M, Niedzialkowska E, Zimmerman MD, Minor W. The impact of structural genomics: the first quindecennial. J Struct Funct Genom. 2016;17(1):1–16. doi: 10.1007/s10969-016-9201-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Basu S, Bahadur RP. A structural perspective of RNA recognition by intrinsically disordered proteins. Cell Mol Life Sci. 2016;73(21):4075–4084. doi: 10.1007/s00018-016-2283-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Peng Z, Mizianty MJ, Xue B, Kurgan L, Uversky VN. More than just tails: intrinsic disorder in histone proteins. Mol BioSyst. 2012;8(7):1886–1901. doi: 10.1039/c2mb25102g. [DOI] [PubMed] [Google Scholar]
- 102.Varadi M, Zsolyomi F, Guharoy M, Tompa P. Functional advantages of conserved intrinsic disorder in RNA-binding proteins. PLoS One. 2015;10(10):e0139731. doi: 10.1371/journal.pone.0139731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Tompa P. Intrinsically unstructured proteins. Trends Biochem Sci. 2002;27(10):527–533. doi: 10.1016/S0968-0004(02)02169-2. [DOI] [PubMed] [Google Scholar]
- 104.Dunker AK, Brown CJ, Lawson JD, Iakoucheva LM, Obradovic Z. Intrinsic disorder and protein function. Biochemistry. 2002;41(21):6573–6582. doi: 10.1021/bi012159+. [DOI] [PubMed] [Google Scholar]
- 105.Chowdhury S, Zhang J, Kurgan L. In silico prediction and validation of novel RNA binding proteins and residues in the human proteome. Proteomics. 2018 doi: 10.1002/pmic.201800064. [DOI] [PubMed] [Google Scholar]
- 106.Wu Z, Hu G, Yang J, Peng Z, Uversky VN, Kurgan L. In various protein complexes, disordered protomers have large per-residue surface areas and area of protein-, DNA- and RNA-binding interfaces. FEBS Lett. 2015;589(19 Pt A):2561–2569. doi: 10.1016/j.febslet.2015.08.014. [DOI] [PubMed] [Google Scholar]
- 107.Fuxreiter M, Tompa P, Simon I, Uversky VN, Hansen JC, Asturias FJ. Malleable machines take shape in eukaryotic transcriptional regulation. Nat Chem Biol. 2008;4(12):728–737. doi: 10.1038/nchembio.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Rochman M, Taher L, Kurahashi T, Cherukuri S, Uversky VN, Landsman D, Ovcharenko I, Bustin M. Effects of HMGN variants on the cellular transcription profile. Nucleic Acids Res. 2011;39(10):4076–4087. doi: 10.1093/nar/gkq1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Latchman DS. Transcription factors: an overview. Int J Biochem Cell Biol. 1997;29(12):1305–1312. doi: 10.1016/S1357-2725(97)00085-X. [DOI] [PubMed] [Google Scholar]
- 110.Liu J, Perumal NB, Oldfield CJ, Su EW, Uversky VN, Dunker AK. Intrinsic disorder in transcription factors. Biochemistry. 2006;45(22):6873–6888. doi: 10.1021/bi0602718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Minezaki Y, Homma K, Kinjo AR, Nishikawa K. Human transcription factors contain a high fraction of intrinsically disordered regions essential for transcriptional regulation. J Mol Biol. 2006;359(4):1137–1149. doi: 10.1016/j.jmb.2006.04.016. [DOI] [PubMed] [Google Scholar]
- 112.Staby L, O’Shea C, Willemoes M, Theisen F, Kragelund BB, Skriver K. Eukaryotic transcription factors: paradigms of protein intrinsic disorder. Biochem J. 2017;474(15):2509–2532. doi: 10.1042/bcj20160631. [DOI] [PubMed] [Google Scholar]
- 113.Toth-Petroczy A, Oldfield CJ, Simon I, Takagi Y, Dunker AK, Uversky VN, Fuxreiter M. Malleable machines in transcription regulation: the mediator complex. PLoS Comput Biol. 2008;4(12):e1000243. doi: 10.1371/journal.pcbi.1000243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Di Mauro E, Dunker AK, Trifonov EN. Disorder to order, non-life to life: in the beginning there was a mistake. In: Seckbach J, editor. Genesis—In the beginning. Precursors of life, chemical models and early biological evolution. Dordrecht: Springer; 2012. [Google Scholar]
- 115.Uversky VN. A decade and a half of protein intrinsic disorder: biology still waits for physics. Protein Sci. 2013;22(6):693–724. doi: 10.1002/pro.2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kulkarni P, Uversky VN. Intrinsically disordered proteins: the dark horse of the dark proteome. Proteomics. 2018;18(21–22):e1800061. doi: 10.1002/pmic.201800061. [DOI] [PubMed] [Google Scholar]
- 117.Longo LM, Blaber M. Protein design at the interface of the pre-biotic and biotic worlds. Arch Biochem Biophys. 2012;526(1):16–21. doi: 10.1016/j.abb.2012.06.009. [DOI] [PubMed] [Google Scholar]
- 118.Longo LM, Blaber M. Prebiotic protein design supports a halophile origin of foldable proteins. Front Microbiol. 2014 doi: 10.3389/fmicb.2013.00418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Trifonov EN. Consensus temporal order of amino acids and evolution of the triplet code. Gene. 2000;261(1):139–151. doi: 10.1016/S0378-1119(00)00476-5. [DOI] [PubMed] [Google Scholar]
- 120.Brooks DJ, Fresco JR, Lesk AM, Singh M. Evolution of amino acid frequencies in proteins over deep time: inferred order of introduction of amino acids into the genetic code. Mol Biol Evol. 2002;19(10):1645–1655. doi: 10.1093/oxfordjournals.molbev.a003988. [DOI] [PubMed] [Google Scholar]
- 121.Longo LM, Tenorio CA, Kumru OS, Middaugh CR, Blaber M. A single aromatic core mutation converts a designed “primitive” protein from halophile to mesophile folding. Protein Sci. 2015;24(1):27–37. doi: 10.1002/pro.2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Poole AM, Jeffares DC, Penny D. The path from the RNA world. J Mol Evol. 1998;46(1):1–17. doi: 10.1007/PL00006275. [DOI] [PubMed] [Google Scholar]
- 123.Tompa P, Csermely P. The role of structural disorder in the function of RNA and protein chaperones. FASEB J. 2004;18(11):1169–1175. doi: 10.1096/fj.04-1584rev. [DOI] [PubMed] [Google Scholar]
- 124.Treiber DK, Williamson JR. Beyond kinetic traps in RNA folding. Curr Opin Struct Biol. 2001;11(3):309–314. doi: 10.1016/S0959-440X(00)00206-2. [DOI] [PubMed] [Google Scholar]
- 125.Cristofari G, Darlix JL. The ubiquitous nature of RNA chaperone proteins. Prog Nucleic Acid Res Mol Biol. 2002;72:223–268. doi: 10.1016/S0079-6603(02)72071-0. [DOI] [PubMed] [Google Scholar]
- 126.Gilbert W. Origin of life—the RNA world. Nature. 1986;319(6055):618. doi: 10.1038/319618a0. [DOI] [Google Scholar]
- 127.Shatsky M, Nussinov R, Wolfson HJ. A method for simultaneous alignment of multiple protein structures. Proteins. 2004;56(1):143–156. doi: 10.1002/prot.10628. [DOI] [PubMed] [Google Scholar]
- 128.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14(1):33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.