Abstract
Decidualization is a critical event for the blastocyst implantation, placental development and fetal growth and the normal term. In mice, the embryo implantation to the uterine epithelial would trigger the endometrial stromal cells to differentiate into decidual stromal cells. However, decidualization in women takes place from the secretory phase of each menstrual cycle and continues to early pregnancy if there is conceptus. Deficient decidualization is often associated with pregnancy specific complications and reproductive disorders. Dramatic changes occur in the gene expression profiles during decidualization, which is coordinately regulated by steroid hormones, growth factors, and molecular and epigenetic mechanisms. Recently, emerging evidences showed that epigenetic modifications, mainly including DNA methylation, histone modification, and non-coding RNAs, play an important role in the decidualization process via affecting the target genes’ expression. In this review, we will focus on the epigenetic modifications in decidualization and open novel avenues to predict and treat the pregnancy complications caused by abnormal decidualization.
Keywords: Decidualization, DNA methylation, Histone modification, Non-coding RNAs, Human endometrial stromal cells
Introduction
Decidualization represents the differentiation of endometrial fibroblast-like stromal cells into specialized secretory decidual cells that provide a nutritional and immunosuppressive matrix indispensable for embryo implantation and placental development. Human endometrial decidualization is triggered by the increase of the progesterone and the local cAMP production in the postovulatory phase in each cycle whether or not there is a conceptus, which is different from most mammals where decidualization takes place only after implantation [1]. The increase of progesterone and cAMP activates the transcription factor Forkhead box O1 (FOXO1) in endometrial stromal cells (ESCs), leading to cell cycle arrest and differentiation into decidual cells that will encapsulate and protect the feto/placental unit throughout pregnancy with intrinsic and specific endocrine and immunological functions [2]. It has been shown that decidual senescence, triggered by the activation of FOXO1, stimulates the transient pro-inflammatory response related to the endometrial receptivity, a critical step for successful pregnancy [3].
Following implantation, the endometrium undergoes a significant process of remodeling regulated by tissue remodeling enzymes known as Metalloproteinases (MMPs). Many of these MMPs are expressed by trophoblast cells to degrade the endometrial extracellular matrix (ECM) during the process of trophoblast invasion. In turn, decidual cells express tissue inhibitors of MMPs (TIMPs) [4] that will control the invasive process. This is an example of the close communication and mutual regulation between fetal cells and the maternal decidua. Alterations in this unique cell to cell communication might lead to abnormal placental development, deficient trophoblast invasion or excessive invasion that would bring out pregnancy complications such as recurrent pregnancy loss (RPL), preeclampsia (PE), preterm birth (PTB) or choriocarcinomas.
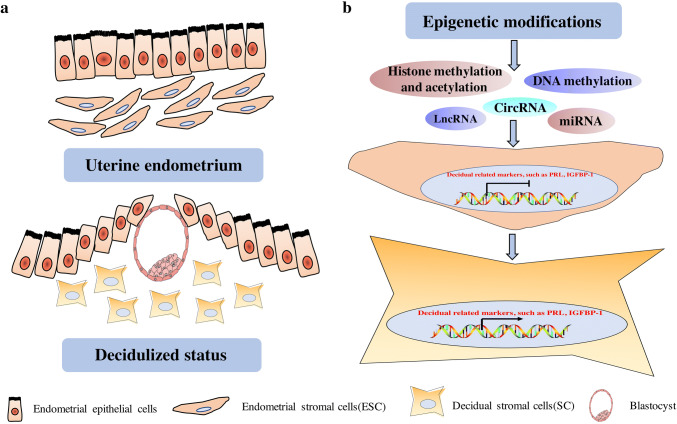
During decidualization, the endometrial stromal cells exhibit morphological and biochemical alterations to differentiate into secretory and immunological decidual stromal cells. In the histological changes, the decidualization accompanies the massive immune cells’ infiltration, including CD56+CD16− NK cells [1], CD163+ macrophages [5], T cells and dendritic cells [6, 7]. Decidual stromal cells have special cellular and ultrastructural characteristics with rounding nucleus, increasing number of nucleoli, expansion of the rough endoplasmic reticulum and Golgi apparatus and releasing of glycogen and lipid droplets in the dilating cytoplasm, which are contrary to the endometrial stromal cells [8] (Fig. 1a). In addition, decidual stromal cells also exhibit phagocytotic activity to engulf the ECM, which may contribute to the reprogramming of ECM [9]. These alterations prepare the receptive endometrium for the possible implantation embryos.
Fig. 1.
The morphology and epigenetic alterations during decidualization. a During decidualization, the ESC exhibit morphological and biochemical alterations to differentiate into secretory and immunological DSCs. DSCs have special cellular and ultrastructural characteristics with rounding nucleus, which is contrary to the ESCs. ESC endometrial stromal cells, DSCs decidual stromal cells. b Epigenetic modifications, including DNA methylation, histone methylation and acetylation, lnc RNAs, miRNAs and circRNAs, work on the endometrial stromal cells with inactivation of decidual-related markers to switch on the mode of transcripting the decidual-related markers, such as PRL and IGFBP-1 in decidual stromal cells. LncRNAs long non-coding RNAs, miRNAs micro RNAs, circRNAs circular RNAs
In 1978, it was found that amniotic prolactin (PRL) was specifically originated from the decidua and not from the pituitary [10, 11], amnion [12] and chorion [13]. Amniotic PRL is produced before implantation around day 22 of the regular menstrual cycle and is found in decidualizing stromal cells [14, 15]. PRL levels peaks between 18 and 26-week of gestation and it is 50–100-fold higher than the pituitary PRL [16, 17]. Decidual PRL has diverse functions associated with reproduction, metabolism, osmoregulation, immunoregulation and behavior [18]. In the nonpregnant endometrium, specifically during the late secretory phase, the PRL receptor (PRL-R) colocalizes with the transcription factor interferon regulatory factor 1(IRF1) in glandular epithelial compartment in late secretory phase of endometrium, which is regulated by PRL [19]. IRF1 is important for upregulated PD-L1 expression in tumor cells in vivo and in vitro [20], and as a target for PRL, it can be postulated that some of the modulatory functions of PRL are mediated through the IRF1 expression.
During pregnancy, PRL-Rs are expressed by multiple cell types at the maternal–fetal interface, such as the decidual stromal cells, cytotrophoblast, syncytiotrophoblast, amniotic epithelium cells and several immune cells present in the uterus [21, 22]. PRL can elicit the migration and invasion of the trophoblast cells [23], promote angiogenesis in the placenta and at the maternal spiral arteries [24]. In addition, PRL modulates dNK cell survival, regulate water transportation across the amnion toward the maternal side and contributes to the immune modulation necessary for the prevention of immune rejection of the semi-allogeneic fetus [12, 25]. Decidual PRL and PRL-R deficiency are associated with increased expression of the pro-inflammatory cytokine IL-6, which is potentially detrimental to pregnancy [26, 27].
IGF-binding protein-1 (IGFBP-1, formerly named as placental protein 12) is another highly produced decidual marker in amnion fluid [28]. IGFBP-1 modulates the bioavailability of phosphorylated IGF-1, which is also reported to stimulate trophoblast invasion [29, 30]. So, PRL and IGFBP-1 are now well-used markers as the putative decidualization of human endometrial stromal cells in vivo and in vitro [1]. Decidual cells transcriptome analysis of HESCs with decidualizing stimulus showed that the differentially expressed genes are supposed to be a broad spectrum of functions, such as cell cycle regulation, cytoskeletal remodeling, angiogenesis, immune modulation, oxidative stress defense, ion and water transport, responses to steroid hormones, deposition of extracellular matrix (ECM), modulation of transcription, epigenetic patterning, post-translational modifications, and growth factor, cytokine, and chemokine signaling [31]. Many of the regulatory functions described above are mediated by epigenetic modifications which control implantation, placentation, organ formation and fetal growth [32].
Decidual transformation is a biphasic process, characterized initially by an acute-phase inflammatory signal, followed by a profound anti-inflammatory environment [1]. Recent studies have shown that steroid hormone-dependent transformation of the human endometrium is due to epigenetic modifiers and chromatin modifications [33, 34]. The epigenetic regulation mainly occurred in the transcriptional, post-transcriptional and post-translational processes, and included the DNA methylation, histone modification and non-coding RNA regulation. DNA methylation and histone modification are crucial for genome reprogramming during early pregnancy and type-specific gene expression [35]. The classical decidualization process is induced in vitro by the exposure of cyclic AMP and progesterone in HESCs with cells transiting from a proliferative to a differentiated HESC phenotype [36]. Epigenetics and histone modification profiles demonstrated that the epigenetic effectors expressed in the HESCs are significantly upregulated upon decidualization, which are encoded by histone modifiers and their cofactors and DNA methyltransferases (DMNTs) [37]. In addition, non-coding RNAs, including long non-coding RNA (lncRNAs), microRNA (miRNAs) and circular RNAs (cirRNAs), are also important epigenetic regulatory elements [38] in endometrial function [39] and may regulate the process of decidualization (Fig. 1b).
Recently, epigenetic regulation attracted more attention on its role in the decidualization-associated gene expression and the formation of receptive endometrium. In this review, we will summarize the insight of epigenetic modifications on the decidualization and endometrial receptivity during early pregnancy and also pregnancy-related complications, and therefore provide new predictive and therapeutic targets for the related disease (Table 1).
Table 1.
Epigenetic modifications for decidualization and endometrial receptivity
| Epigenetic factor | Function | References |
|---|---|---|
| DNMT | DNMTs were highly increased during pre-receptive phase of pregnancy but generally back to the baseline during receptive and post-implantation periods | [43, 47] |
| DNA demethylating agent 5-aza-CdR decreased DNMTs, and hypomethylated at Esr1, Pgr and Hoxa10, and further lead to the defected endometrial decidualization and stromal cells proliferation in mice | [44] | |
| DNMT inhibitor 5-AZA promote the non-receptive AN3-CA cells to differentiate into receptive decidual stromal cells by increasing E-cadgerin | [45] | |
| DNMT3B was downregulated in the DSC and attenuated the decidual-specific IGFBP-1 expression in HESC cells | [46] | |
| HESC treated with DNMT inhibitor 5-Aza-2′-deoxycytidine promoted the decidual-like morphology by upregulation of PRL and IGFBP-1 expression and inhibited the proliferation of ESC | [36] | |
| Inhibition of DNM abrogated the decidualization during peri- or post-implantation, and therefore lead to embryo loss | [36] | |
| DNMT3a and DNMT3b mRNA levels were significantly decreased in ESC by E + MPA treatment, and DNMTs decreased in the human endometrium during the secretory stage | [47] | |
| Histone methylation and acetylation | The dynamic changes of H3K27me3 indicated its essential role in the maintenance of uterine quiescence in early pregnancy and initiation of parturition in late pregnancy | [54] |
| EZH2 is gradually but markedly lost in differentiating human endometrium during the menstrual cycle, and results in the decrease of H3K27me3 at the TSS of PRL and IGFBP1 in response to 8-Br-cAMP and MPA, alone or combination | [51] | |
| The loss of H3K27me3 is companied by the enrichment of H3K27ac, and promote the decidual essential genes, such as WNT4, ZBTB16, PROK1 and GREB1 | [55] | |
| H3K27ac increases during decidualization of HESCs, and as an enhancer enriched in the promoter of IGFBP-1 | [56] | |
| cAMP regulates IGFBP-1 expression by recruitment of C/EBPβ, FOXO1, and p300 to the IGFBP-1 enhancer in ESC. Inhibition of C/EBPβ impaired the control of cAMP on the H3K27ac, chromatin opening, and p300 recruitment to the IGFBP-1 enhancer | [56, 57] | |
| HDAC-specific inhibitor TSA increased the expression of decidual markers such as PRL and IGFBP-1, and also TSA regulates TIMP-1 and TIMP-3 expression through TSA-induced acetylation at their promoters in HESCs | [4, 52] | |
| Long non-coding RNAs | The downregulated Lnc 473 inhibited in vitro decidualization, Lnc473 is strongly upregulated in in vitro decidualization of HESCs after decidual stimulus, which is regulated by the cAMP-PKA pathway through IL-11-mediated STAT3 phosphorylation | [67] |
| lncRNA H19, one of the first genes found to be transcribed into long non-coding RNAs, was downregulated in the endometrium at mid-luteal phase during the window of implantation in repeated implantation failure | [68] | |
| IFLMN analysis of endometrium of six RIFs and eight controls showed the differently expressed lncRNA–miRNA and miRNA–mRNA pairs | [69] | |
| Genome-wide RNA sequencing of the endometrium between RIF and control women identified the target genes of lncRNAs and identified 148 lnc RNAs corresponding to 147 cis-regulated target genes | [70] | |
| lncRNA PGK1P2 has a high sequence similarity to PGK1, PGK1P2 acted as a competing endogenous RNA (ceRNA) to inhibit PGK1 expression through miR-330-5p in human decidualization by regulating angiogenesis and glycolysis metabolism | [73] | |
| HK2P1 and its homologous gene HK2 are necessary for the endometrial decidualization. Inhibition of HK2P1 or HK2 was involved in the blocking the ESCs proliferation and differentiation by inhibiting glycolysis of ESCs and further impaired the decidualization due to competing for the endogeneous RNA miR-6887-3p | [74] | |
| microRNAs | miR-181, miR-183 and miR-200 are decreased in the decidualization process, which may target numerous decidualization markers, such as Prolactin, IGFBP-1, MPIF-1 and TIMP-3 secretion as well as HOXA10, COX2, SP1, C/EBPβ and FOXO1 expression in decidualized HESCs with decreased Dicer function | [87] |
| In human, loss of miR-542-3p promote the induction of major decidual marker genes, including IGFBP1, WNT4 and PRL | [88, 89] | |
| Overexpression of miR-542-3p inhibited the capacity of migration and invasion of endometriotic cells | [90] | |
| High levels of miRNA-136 suppressed cell proliferation and promoted apoptosis of MSCs targeting BCL2, and it also inhibited HUVEC capillary formation by suppressing VEGF | [91] | |
| miRNA-181b downregulated OPN and then affected the expression of decidualization- and angiogenesis- related genes in implantation group in IVF/ICSI-ET cycles | [92] | |
| Increased miRNA-181a promoted the hESC decidualization-related gene expression and morphological transformation by inhibiting KLF12 at the transcriptional and translational levels | [93] | |
| hsa-miR-222 participates in ESC differentiation by regulating the cell cycle of ESCs arrest in S phase | [94] | |
| In mice, overexpression of miR-290b-5p in mouse ESCs inhibited in vitro decidualization by targeting NDRG3 induced by estradiol and progesterone | [95] | |
| mmu-miR-96 is strongly expressed during decidualization in mouse uterine, and its overexpression may induce apoptosis of stromal cells via inhibiting Bcl2to impede decidualization | [96] | |
| Inhibition of miRNA-200 blocked the decidualization and prevented the mesenchymal–epithelial transition-like changes that accompanied decidual differentiation | [97] | |
| Circular RNAs | circRNA microarray identified that 856 circRNAs changes in the endometrium in RIF compared with normal controls | [103] |
| circRNAs profiles of the goat endometrium from pre-receptive to receptive phase found that critical regulatory relationships between circRNAs with miRNAs and corresponding mRNAs in the endometrial receptivity | [104] | |
| Modulation of circRNA-9119-miR-26a-PTGS2 expression in EECs may emerge as a potential target to regulate the development of endometrial receptivity | [105] | |
| circRNA–miRNA–mRNA negative correlation networks was also involved in the altered expression patterns of circular RNAs between implantation sites and interimplantation sites in early pregnant mice | [106] |
DNA methylation
DNA methylation (DNM) is one of the well-studied epigenetic mechanisms leading to stable inactivation of gene expression in mammals and is indispensable for normal development and successive progression of key biologic processes [35]. DNM is generally accompanied by the addition of 5-methylcytosine in the DNA strand via the enzymatic transfer of a methyl group to the 5′-cytosine of CpG catalyzed by DNA methyltransferases (DNMTs) (Dnmt1, Dnmt3a, and Dnmt3b) [40]. DNMT1 is an enzyme for the maintenance of methylation established and expressed constitutively during DNA replication [41], whereas Dnmt3a and Dnmt3b mainly contribute to the de novo methylation [42]. DNMTs were highly increased during prereceptive phase of pregnancy but generally back to the baseline during receptive and post-implantation periods. Moreover, lower Dnmt3a was detected in the stroma of implantation site (IS) when compared with inner-IS on embryo 5 in mice, whereas Dnmt1 and Dnmt3b were decreased in the luminal and glandular epithelia of IS. When detected in the folate-deficient diet (FDD) pseudo-pregnant mice, opposite expression levels of DNMTs were observed, which suggested the DNMTs may regulate the transcription of endometrial genes and decidualization during implantation in mice, especially in mice subjected to FDD [43]. Mice administrated with DNA demethylating agent 5-aza-2′-deoxycytidine (5-aza-CdR) lead to the decreased expression of DNMTs, and hypomethylated at the 5′ flanking regions of the estrogen receptor α (Esr1), progesterone receptor (Pgr), and homeobox A10 (Hoxa10), which are vital for control of endometrial changes, and further reduced the implantation sites in dose-dependent manner due to the defected endometrial decidualization and stromal cells proliferation [44]. Inhibition of DNA methylation by 5-AZA resulted in the non-receptive AN3-CA endometrial epithelial carcinoma cell lines differentiated into receptive decidual stromal cell(DSC)-like to BeWo cell spheroid attachment by enhancing the expression of E-cadherin, which suggested the endometrial receptivity in human pregnancy is epigenetically regulated [45]. Colorimetric and long interspersed nuclear element 1 methylation analysis showed there are no global changes in DNA methylation levels upon differentiation of HESCs [37]. However, the complex epigenetic regulation at the specific locus supports the acquisition of a decidual endometrial phenotype. In vitro study in HESCs has shown that downregulation of DNA methyltransferase DNMT3B in the DSC attenuates the decidual-specific IGFBP-1 expression [46]. Another study demonstrated that treatment of HESC with DNA methylation inhibitor 5-Aza-2′-deoxycytidine (AZA) promoted the differentiation of the fibroblast-like stromal cells into decidual-like morphology characterized by upregulation of PRL and IGFBP-1 expression, and inhibition the proliferation of endometrial stromal cells (ESC) [36]. In addition, inhibition of DNA methylation (DNM) abrogated the decidualization during the peri- or post-implantation and therefore led to embryo loss [36]. Besides, DNMT3a and DNMT3b mRNA levels were significantly decreased in ESC by estradiol and medroxyprogesterone acetate (E + MPA) treatment, and DNMTs decreased in the human endometrium during the secretory stage [47]. These findings suggested that decreased DNA methylation in secretory phase is necessary for the successful decidualization in secretory phase and embryo attachment during the early stages of pregnancy.
Histone methylation and acetylation
Histone methylation and acetylation are two major events related to histone modifications [48, 49]. The histone lysine methylation and acetylation are of great significance to the chromatin structures and genome organization and further regulate the gene expression. Transcription activation and silence depend on the lysine sites and methylation status [48]. In particular, di- and tri-methylation histone 3 lysine 27(H3K27me2/3) correlated with condensed and transcriptionally silence and is mediated by methyltransferase Enhancer of Zeste Homolog 2 (EZH2) [50–52]. By contrast, histone acetylation is related to transcription activation through open chromatin accessibility [48]. Histone acetylation is often catalyzed by histone acetyltransferases (HATs) and histone deacetylases (HDACs) [53]. Here we mainly focus on the histone 3 lysine 27 acetylation (H3K27ac).
The dynamic generation and erasure of repressive trimethylated lysine 27 of histone 3 (H3K27me3) indicated its essential role in the maintenance of uterine quiescence by transcriptionally silencing the parturition-related genes, type 1 immunity and promoting the wound-healing response in early pregnancy, and initiation of parturition in late pregnancy. Pharmacological inhibition of H3K27me3 prevented the term parturition and decreased pup viability in preterm model while delivery [54], which suggested the important role of H3K27me3 during pregnancy. Ezh2 is gradually but markedly lost in differentiating human endometrium during the menstrual cycle, which in turn results in the decrease H3K27me3 of at the transcriptional start site (TSS) of the widely used decidual-specific marker genes, PRL and IGFBP1 in response to 8-Bromo-cAMP (8-Br-cAMP) and the progestin medroxyprogesteroneacetate (MPA), alone or combination. Genome-wide analysis showed that the loss and gain of the inhibitory H3K27me3 caused by Ezh2 in the decidualizing cells at the specific loci are highly associated with the transcriptional regulators [51]. Moreover, the loss of H3K27me3 is reciprocally accompanied by the enrichment of acetylation at the same lysine site (H3K27ac), and promote the expression of the decidual essential genes, such as WNT4, ZBTB16, PROK1 and GREB1 [55], indicating active reprogramming converts from a suppressive to a transcriptionally permissive chromatin in decidualizing stromal cells.
H3K27ac increases during decidualization of HESCs, and there is an enrichment of H3K27ac in the distal upstream region (− 4701 to − 7501 bp) of the IGFBP-1 promoter controlling the expression of IGFBP-1. Deletion of the distal region of IGFBP-1 promoter by CRISPR/Cas-9 markedly reduced the IGFBP-1 expression, suggesting its role as an IGFBP-1 enhancer [56]. cAMP regulates IGFBP-1 expression and the mechanism is thought to be mediated by the recruitment of the transcriptional regulators CCAAT enhancer-binding protein β (C/EBPβ), FOXO1, and p300 to the IGFBP-1 enhancer in endometrial stroma cells. Inhibition of C/EBPβ impaired the control of cAMP on the H3K27ac, chromatin opening, and p300 recruitment to the IGFBP-1 enhancer, suggesting that the levels of H3K27ac in the distal upstream region and promoter of IGFBP-1 support its expression during decidualization [56, 57]. Trichostatin A (TSA), a specific HDAC inhibitor increases the expression of the most widely studied decidual markers such as PRL and IGFBP-1, suggesting that TSA is a potential enhancer of decidualization through the promotion of acetylation status of histone in the promoter of decidual marker genes [52]. Furthermore, it has been shown that TSA regulates TIMP-1 and TIMP-3 expression through TSA-induced acetylation at their promoters in HESCs, a mechanism that allows decidual cells to restrain trophoblast invasion [4].
Abnormal responsiveness of HESCs to decidualization is one of the characteristics observed in patients with recurrent pregnancy loss (RPL), suggesting that a possible underlying problem in these patients is a defect of the normal process of cell maturation/differentiation. Indeed, evaluation of samples obtained from patients with RPL revealed significantly decreased methylation in the defined CA-rich motifs in the genome [58]. Together, all these findings indicate that histone methylation and acetylation play critical roles in the transformation of endometrial stromal cells into decidual stromal cells [32] and histone modification enzymes are essential epigenetic modifiers for embryo implantation and a healthy pregnancy.
Long non-coding RNAs
So far, there is no official consensus to clarify the sequence and biological features to define long non-coding RNAs (lncRNAs). Nevertheless, however, it was generally considered that transcripts do not translate proteins and longer than 200 nucleotides are thought to be lncRNAs [59], which is longer than smaller non-coding RNAs [60, 61]. However, lncRNAs are usually shorter than protein-coding RNAs [62] and have fewer exons [59]. They regulate gene expression at different levels, including chromatin modification, protein localization and activity, alternative splicing and increasing mRNA stability by prevent the 3′ UTR of mRNAs from miRNA binding [63, 64]. Besides, they also function as scaffolds, signals, and antisense decoys [65]. Nowadays, emerging studies have documented the existence and the important roles of lncRNAs in human early embryo development [66], including decidualization. The downregulated LNC473 inhibited in vitro decidualization, which is the first time to report the relationship between lncRNA and human decidualizaition. Long intergenic non-coding LINC00473 (LINC473) is strongly upregulated in in vitro decidualization of HESCs after decidual stimulus, which is regulated by the cAMP-PKA pathway through IL-11-mediated STAT3 phosphorylation [67]. In addition, lncRNA H19, one of the first genes found to be transcribed into long non-coding RNAs, and integrin β3 protein was downregulated in the endometrium at mid-luteal phase during the window of implantation in repeated implantation failure [68]. Implantation failure-related lncRNA–mRNA network (IFLMN) analysis of the endometrium of six RIFs and eight controls, showed the differently expressed lncRNA–miRNA and miRNA–mRNA pairs and functional enrichment results achieved six key lncRNAs (NONHSAT083203.2, NONHSAT212577.1, NONHSAT035952.2, NONHSAT193031.1, NONHSAT053761.2, and NONHSAT025064.2) and their ceRNA sub-networks, which are involved in the vascular proliferation and apoptosis [69]. In line with this study, Genome-Wide RNA Sequencing of the endometrium between RIF and control women identified the target genes of lncRNAs and identified 148 lnc RNAs corresponding to 147 cis-regulated target genes. These cis-regulated target genes-lncRNAs are classified into several pathways, such as the tumor necrosis factor signaling pathway, the Toll-like receptor signaling pathway, and the NF-kappa B (NF-κB) signaling pathway [70]. These studies showed that the lncRNAs may be involved in the endometrial receptivity and further lead to RIF [71], but the specific molecular mechanism of these lncRNAs during uterine receptivity establishment needs further investigation. LncRNAs are not only related to the pathology of RIF, and also participate in other pregnancy complications, including PE. Glycolysis is considered as an important metabolism of endometrium epithelial cells during decidualization [72]. Pseudogene 2 (PGK1P2), a long non-coding RNA (lncRNA), has a high sequence similarity to Phosphoglycerate kinase 1(PGK1), which is an enzyme involved in the glycolytic pathway, and therefore acquires an ability for sequence-specific regulation. PGK1P2 acted as a competing endogenous RNA (ceRNA) to inhibit PGK1 expression through miR-330-5p in human decidualization by regulating angiogenesis and glycolysis metabolism and further resulted in the occurrence of PE [73]. In addition, long non-coding RNA HK2P1 (hexokinase 2 pseudogene 1) and its homologous gene HK2 (hexokinase 2) are necessary for the endometrial decidualization. Inhibition of HK2P1 or HK2 was involved in the blocking the ESCs proliferation and differentiation by inhibiting glycolysis of ESCs and further impaired the decidualization due to competing for the endogenous RNA miR-6887-3p [74]. These findings provide innovative ideas for the occurrence and a new regulating axis for the prediction of PE [75, 76]. Together, lncRNAs may develop into the predictive biomarkers of the endometrium receptivity and decidualization associated with pregnancy complications.
microRNAs
miRNAs are small non-coding RNAs with length ~ 22 nt, functioning as post-transcriptional regulator of gene expression by binding to the 3′ untranslated regions (UTRs) of the target mRNA [77], which could lead to gene either mRNA degradation or translational repression [78]. Almost all types of cells could secrete miRNAs and the density of extracellular miRNAs is related to the physiological and pathological situations [79]. miRNAs are expressed not only in the blood plasma and serum [80], but also detected in other body fluids [81]. Emerging studies showed that the extracellular miRNAs existed in stable forms and avoided being cleaved by endogenous RNases, which are feasible to be non-invasive biomarkers detecting and monitoring various physio-pathological conditions, including cancers and pregnancy complications [82]. Dicer and Drosha are critical RNase III proteins for mature miRNA processing. They were dynamically changed across the menstrual cycle and reduced levels of during receptive phase in the primary unexplained infertility [39], which suggested the dysregulated miRNA biogenesis machinery may lead to the failure of endometrial receptivity and further infertility. The alterations of miRNA expression have been described for several gynecological disorders, including recurrent pregnancy loss (RPL), preeclampsia (PE) [83] and endometriosis [84]. Studies on animal models and human have confirmed different miRNAs regulate multiple physiological processes by regulating the gene expression alterations during different phases of endometrial cycle, including decidualization which is associated with the endometrium receptivity and identified in endometrial biopsies as well as in endometrial fluids [85]. Decidualization is involved in massive proliferation, differentiation and apoptosis of the stromal cells [86]. miRNAs participate in the decidualization by targeting the decidual-related marker genes. The analysis of decidualized HESCs showed miR-181, miR-183 and miR-200 are decreased in the decidualization process, which may target numerous decidualization markers, such as Prolactin, IGFBP-1, MPIF-1 and TIMP-3 secretion as well as HOXA10, COX2, SP1, C/EBPβ and FOXO1 expression in decidualized HESCs with decreased Dicer function [87]. In human, loss of miR-542-3p promotes the induction of major decidual marker genes, including IGFBP1, WNT4 and PRL [88, 89]. In addition, overexpression of miR-542-3p inhibited the capacity of migration and invasion of endometriotic cells [90]. High levels of miRNA-136 suppressed cell proliferation and promoted apoptosis of Mesenchymal stem cells (MSCs) targeting B‑cell lymphoma 2(BCL2), and it also inhibited HUVEC capillary formation by suppressing VEGF to be a potential causal factor of PE [91]. miRNA-181b downregulated osteopontin (OPN) and then affected the expression of decidualization- and angiogenesis-related genes in implantation group in IVF/ICSI-ET cycles [92]. Moreover, increased miRNA-181a promoted the hESC decidualization-related gene expression and morphological transformation by inhibiting Krüppel-like factor 12 (KLF12) at the transcriptional and translational levels [93]. Microarray analysis of non-induced endometrial stromal cells (ESCs) and induced ESCs found that hsa-miR-222 participates in ESC differentiation by regulating the cell cycle of ESCs arrest in S phase [94]. In mice, overexpression of miR-290b-5p in mouse endometrial stromal cells (ESCs) inhibited in vitro decidualization by targeting the N-myc downstream-regulated gene 3 (NDRG3) induced by estradiol and progesterone [95]. mmu‑miR‑96 is strongly expressed during decidualization in mouse uterine, and its overexpression may induce apoptosis of stromal cells via inhibiting Bcl2, an anti‑apoptotic gene, to impede decidualization [96]. miRNA-200 family is significantly downregulated in mouse endometrial stromal cells before implantation. Inhibition of miRNA-200 blocked the decidualization and prevented the mesenchymal–epithelial transition-like changes that accompanied decidual differentiation [97]. These emerging studies showed miRNAs play important roles in decidualization and further promote the formation of receptivity of endometrium and pregnancy success.
Circular RNAs
The first circular RNAs (circRNAs) were to be identified in Viroids (plant pathogens) from 1976 [98]. circRNAs are covalently closed specific molecules of endogenous non-coding RNAs (ncRNAs). The most specificity, different from other ncRNAs, is lacking both a 5′ cap and a 3′ tail. Many circRNAs are expressed in a tissue-specific manner with high stability and at low levels [99] likely due to far less backsplicing than canonical splicing [100]. circRNAs are naturally resistant to degradation by exonucleases and therefore accumulate at stable status due to their long half-lives [101, 102]. The biogenesis of circRNAs is regulated by the combinatorial action of RNA-binding proteins, which allow circular RNAs to be expressed in unique patterns [101]. Many eukaryotic genes could generate circRNAs. CircRNAs are reported to regulate gene expression in mammals by sponge miRNAs at transcriptional and post-translational levels [102]. Liu et al. reported that circRNA microarray identified that 856 circRNAs changes in the endometrium in RIF compared with normal controls, 7 of which may exhibit important role in the development and progression of RIF [103]. Illumina Solexa technology analyzed the circRNAs profiles of the goat endometrium from pre-receptive to receptive phase found that critical regulatory relationships between circRNAs with miRNAs and corresponding mRNAs in the endometrial receptivity [104]. One specific study showed that circRNA-9119 decreased levels of miR-26a by acting as a microRNA sponge, and that miR-26a downregulated the expression of PTGS2 via the predicted target site in endometrial epithelial cells (EECs) of dairy goats in vitro, therefore, modulation of circRNA-9119-miR-26a-PTGS2 expression in EECs may emerge as a potential target to regulate the development of endometrial receptivity [105]. In addition, circRNA–miRNA–mRNA negative correlation networks was also involved in the altered expression patterns of circular RNAs between implantation sites and interimplantation sites in early pregnant mice [106]. These findings suggested that circRNAs might participate in endometrial receptivity and further promote the embryo attachment in mammals.
Conclusions
Recent years, emerging studies reported epigenetic modification factors and their expression and regulations at the peri-implantation phase of endometrial receptivity and decidualization (summarized in Table 1). However, the underlying mechanisms of their physiological functions are still a big challenge in this field [107]. The epigenetic control of endometrial receptivity and decidualization mainly target the expression of marker genes and the combination of in vivo and in vitro studies would be used to uncover the ministry of the genetic regulation network involved in the endometrial changes during the pre-implantation and early pregnancy. Moreover, miRNA and circRNAs can be stably detected in the serum and other body fluids and might act as biomarkers for the predication and potential treatment for the pregnancy complications caused by the deficiency of endometrial receptivity and decidualization.
Author contributions
HL design, text and drawings. XH text and drawings. GM and AL design, text revision and final approval.
Funding
This work was supported by National Key Research & Developmental Program of China (2018YFC1003900; 2018YFC1003904), and National Natural Science Foundation of China (No. 81871186).
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Gellersen B, Brosens JJ. Cyclic decidualization of the human endometrium in reproductive health and failure. Endocr Rev. 2014;35(6):851–905. doi: 10.1210/er.2014-1045. [DOI] [PubMed] [Google Scholar]
- 2.Norwitz ER, Bonney EA, Snegovskikh VV, Williams MA, Phillippe M, Park JS, Abrahams VM. Molecular regulation of parturition: the role of the decidual clock. Cold Spring Harb Perspect Med. 2015;5(11):a023143. doi: 10.1101/cshperspect.a023143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Park Y, Nnamani MC, Maziarz J, Wagner GP. Cis-regulatory evolution of forkhead Box O1 (FOXO1), a terminal selector gene for decidual stromal cell identity. Mol Biol Evol. 2016;33(12):3161–3169. doi: 10.1093/molbev/msw193. [DOI] [PubMed] [Google Scholar]
- 4.Estella C, Herrer I, Atkinson SP, Quinonero A, Martinez S, Pellicer A, Simon C. Inhibition of histone deacetylase activity in human endometrial stromal cells promotes extracellular matrix remodelling and limits embryo invasion. PLoS One. 2012;7(1):e30508. doi: 10.1371/journal.pone.0030508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Russell P, Sacks G, Tremellen K, Gee A. The distribution of immune cells and macrophages in the endometrium of women with recurrent reproductive failure. III: further observations and reference ranges. Pathology. 2013;45(4):393–401. doi: 10.1097/PAT.0b013e328361429b. [DOI] [PubMed] [Google Scholar]
- 6.King A. Uterine leukocytes and decidualization. Hum Reprod Update. 2000;6(1):28–36. doi: 10.1093/humupd/6.1.28. [DOI] [PubMed] [Google Scholar]
- 7.Rieger L, Honig A, Sutterlin M, Kapp M, Dietl J, Ruck P, Kammerer U. Antigen-presenting cells in human endometrium during the menstrual cycle compared to early pregnancy. J Soc Gynecol Investig. 2004;11(7):488–493. doi: 10.1016/j.jsgi.2004.05.007. [DOI] [PubMed] [Google Scholar]
- 8.Kajihara T, Tanaka K, Oguro T, Tochigi H, Prechapanich J, Uchino S, Itakura A, Sucurovic S, Murakami K, Brosens JJ, Ishihara O. Androgens modulate the morphological characteristics of human endometrial stromal cells decidualized in vitro. Reprod Sci. 2014;21(3):372–380. doi: 10.1177/1933719113497280. [DOI] [PubMed] [Google Scholar]
- 9.Cornillie FJ, Lauweryns JM, Brosens IA. Normal human endometrium. An ultrastructural survey. Gynecol Obstet Invest. 1985;20(3):113–129. doi: 10.1159/000298983. [DOI] [PubMed] [Google Scholar]
- 10.Golander A, Hurley T, Barrett J, Hizi A, Handwerger S. Prolactin synthesis by human chorion-decidual tissue: a possible source of prolactin in the amniotic fluid. Science. 1978;202(4365):311–313. doi: 10.1126/science.694535. [DOI] [PubMed] [Google Scholar]
- 11.Riddick DH, Luciano AA, Kusmik WF, Maslar IA. De novo synthesis of prolactin by human decidua. Life Sci. 1978;23(19):1913–1921. doi: 10.1016/0024-3205(78)90557-X. [DOI] [PubMed] [Google Scholar]
- 12.Taii S, Ihara Y, Mori T. Identification of the mRNA coding for prolactin in the human decidua. Biochem Biophys Res Commun. 1984;124(2):530–537. doi: 10.1016/0006-291X(84)91586-9. [DOI] [PubMed] [Google Scholar]
- 13.Clements J, Whitfeld P, Cooke N, Healy D, Matheson B, Shine J, Funder J. Expression of the prolactin gene in human decidua-chorion. Endocrinology. 1983;112(3):1133–1134. doi: 10.1210/endo-112-3-1133. [DOI] [PubMed] [Google Scholar]
- 14.Maslar IA, Riddick DH. Prolactin production by human endometrium during the normal menstrual cycle. Am J Obstet Gynecol. 1979;135(6):751–754. doi: 10.1016/0002-9378(79)90386-7. [DOI] [PubMed] [Google Scholar]
- 15.Daly DC, Maslar IA, Riddick DH. Prolactin production during in vitro decidualization of proliferative endometrium. Am J Obstet Gynecol. 1983;145(6):672–678. doi: 10.1016/0002-9378(83)90572-0. [DOI] [PubMed] [Google Scholar]
- 16.Kletzky OA, Rossman F, Bertolli SI, Platt LD, Mishell DR., Jr Dynamics of human chorionic gonadotropin, prolactin, and growth hormone in serum and amniotic fluid throughout normal human pregnancy. Am J Obstet Gynecol. 1985;151(7):878–884. doi: 10.1016/0002-9378(85)90665-9. [DOI] [PubMed] [Google Scholar]
- 17.Ben-Jonathan N, Mershon JL, Allen DL, Steinmetz RW. Extrapituitary prolactin: distribution, regulation, functions, and clinical aspects. Endocr Rev. 1996;17(6):639–669. doi: 10.1210/edrv-17-6-639. [DOI] [PubMed] [Google Scholar]
- 18.Goffin V, Binart N, Touraine P, Kelly PA. Prolactin: the new biology of an old hormone. Annu Rev Physiol. 2002;64:47–67. doi: 10.1146/annurev.physiol.64.081501.131049. [DOI] [PubMed] [Google Scholar]
- 19.Jabbour HN, Critchley HO, Yu-Lee LY, Boddy SC. Localization of interferon regulatory factor-1 (IRF-1) in nonpregnant human endometrium: expression of IRF-1 is up-regulated by prolactin during the secretory phase of the menstrual cycle. J Clin Endocrinol Metab. 1999;84(11):4260–4265. doi: 10.1210/jcem.84.11.6142. [DOI] [PubMed] [Google Scholar]
- 20.Shao L, Hou W, Scharping NE, Vendetti FP, Srivastava R, Roy CN, Menk AV, Wang Y, Chauvin JM, Karukonda P, Thorne SH, Hornung V, Zarour HM, Bakkenist CJ, Delgoffe GM, Sarkar SN. IRF1 inhibits antitumor immunity through the upregulation of PD-L1 in the tumor cell. Cancer Immunol Res. 2019;7(8):1258–1266. doi: 10.1158/2326-6066.CIR-18-0711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu-Lee LY. Molecular actions of prolactin in the immune system. Proc Soc Exp Biol Med. 1997;215(1):35–52. doi: 10.3181/00379727-215-44111. [DOI] [PubMed] [Google Scholar]
- 22.Maaskant RA, Bogic LV, Gilger S, Kelly PA, Bryant-Greenwood GD. The human prolactin receptor in the fetal membranes, decidua, and placenta. J Clin Endocrinol Metab. 1996;81(1):396–405. doi: 10.1210/jcem.81.1.8550784. [DOI] [PubMed] [Google Scholar]
- 23.Stefanoska I, Jovanovic Krivokuca M, Vasilijic S, Cujic D, Vicovac L. Prolactin stimulates cell migration and invasion by human trophoblast in vitro. Placenta. 2013;34(9):775–783. doi: 10.1016/j.placenta.2013.06.305. [DOI] [PubMed] [Google Scholar]
- 24.Corbacho AM, Martinez De La Escalera G, Clapp C. Roles of prolactin and related members of the prolactin/growth hormone/placental lactogen family in angiogenesis. J Endocrinol. 2002;173(2):219–238. doi: 10.1677/joe.0.1730219. [DOI] [PubMed] [Google Scholar]
- 25.Jabbour HN, Critchley HO. Potential roles of decidual prolactin in early pregnancy. Reproduction. 2001;121(2):197–205. doi: 10.1530/rep.0.1210197. [DOI] [PubMed] [Google Scholar]
- 26.Reese J, Binart N, Brown N, Ma WG, Paria BC, Das SK, Kelly PA, Dey SK. Implantation and decidualization defects in prolactin receptor (PRLR)-deficient mice are mediated by ovarian but not uterine PRLR. Endocrinology. 2000;141(5):1872–1881. doi: 10.1210/endo.141.5.7464. [DOI] [PubMed] [Google Scholar]
- 27.Bao L, Tessier C, Prigent-Tessier A, Li F, Buzzio OL, Callegari EA, Horseman ND, Gibori G. Decidual prolactin silences the expression of genes detrimental to pregnancy. Endocrinology. 2007;148(5):2326–2334. doi: 10.1210/en.2006-1643. [DOI] [PubMed] [Google Scholar]
- 28.Rutanen EM, Koistinen R, Wahlstrom T, Bohn H, Ranta T, Seppala M. Synthesis of placental protein 12 by human decidua. Endocrinology. 1985;116(4):1304–1309. doi: 10.1210/endo-116-4-1304. [DOI] [PubMed] [Google Scholar]
- 29.Carter AM, Hills F, O’Gorman DB, Roberts CT, Sooranna SR, Watson CS, Westwood M. The insulin-like growth factor system in mammalian pregnancy—a workshop report. Placenta. 2004;25(Suppl A):S53–S56. doi: 10.1016/j.placenta.2004.01.019. [DOI] [PubMed] [Google Scholar]
- 30.Giudice LC. Maternal-fetal conflict—lessons from a transgene. J Clin Invest. 2002;110(3):307–309. doi: 10.1172/JCI0216389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Salker MS, Nautiyal J, Steel JH, Webster Z, Sucurovic S, Nicou M, Singh Y, Lucas ES, Murakami K, Chan YW, James S, Abdallah Y, Christian M, Croy BA, Mulac-Jericevic B, Quenby S, Brosens JJ. Disordered IL-33/ST2 activation in decidualizing stromal cells prolongs uterine receptivity in women with recurrent pregnancy loss. PLoS One. 2012;7(12):e52252. doi: 10.1371/journal.pone.0052252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Logan PC, Ponnampalam AP, Rahnama F, Lobie PE, Mitchell MD. The effect of DNA methylation inhibitor 5-Aza-2′-deoxycytidine on human endometrial stromal cells. Hum Reprod. 2010;25(11):2859–2869. doi: 10.1093/humrep/deq238. [DOI] [PubMed] [Google Scholar]
- 33.Zelenko Z, Aghajanova L, Irwin JC, Giudice LC. Nuclear receptor, coregulator signaling, and chromatin remodeling pathways suggest involvement of the epigenome in the steroid hormone response of endometrium and abnormalities in endometriosis. Reprod Sci. 2012;19(2):152–162. doi: 10.1177/1933719111415546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Munro SK, Farquhar CM, Mitchell MD, Ponnampalam AP. Epigenetic regulation of endometrium during the menstrual cycle. Mol Hum Reprod. 2010;16(5):297–310. doi: 10.1093/molehr/gaq010. [DOI] [PubMed] [Google Scholar]
- 35.Li E. Chromatin modification and epigenetic reprogramming in mammalian development. Nat Rev Genet. 2002;3(9):662–673. doi: 10.1038/nrg887. [DOI] [PubMed] [Google Scholar]
- 36.Gao F, Ma X, Rusie A, Hemingway J, Ostmann AB, Chung D, Das SK. Epigenetic changes through DNA methylation contribute to uterine stromal cell decidualization. Endocrinology. 2012;153(12):6078–6090. doi: 10.1210/en.2012-1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grimaldi G, Christian M, Quenby S, Brosens JJ. Expression of epigenetic effectors in decidualizing human endometrial stromal cells. Mol Hum Reprod. 2012;18(9):451–458. doi: 10.1093/molehr/gas020. [DOI] [PubMed] [Google Scholar]
- 38.Nelissen EC, van Montfoort AP, Dumoulin JC, Evers JL. Epigenetics and the placenta. Hum Reprod Update. 2011;17(3):397–417. doi: 10.1093/humupd/dmq052. [DOI] [PubMed] [Google Scholar]
- 39.Loke H, Rainczuk K, Dimitriadis E. MicroRNA biogenesis machinery is dysregulated in the endometrium of infertile women suggesting a role in receptivity and infertility. J Histochem Cytochem. 2019;67(8):589–599. doi: 10.1369/0022155419854064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet. 2010;11(3):204–220. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69(6):915–926. doi: 10.1016/0092-8674(92)90611-F. [DOI] [PubMed] [Google Scholar]
- 42.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99(3):247–257. doi: 10.1016/S0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 43.Ding YB, He JL, Liu XQ, Chen XM, Long CL, Wang YX. Expression of DNA methyltransferases in the mouse uterus during early pregnancy and susceptibility to dietary folate deficiency. Reproduction. 2012;144(1):91–100. doi: 10.1530/REP-12-0006. [DOI] [PubMed] [Google Scholar]
- 44.Ding YB, Long CL, Liu XQ, Chen XM, Guo LR, Xia YY, He JL, Wang YX. 5-Aza-2′-deoxycytidine leads to reduced embryo implantation and reduced expression of DNA methyltransferases and essential endometrial genes. PLoS One. 2012;7(9):e45364. doi: 10.1371/journal.pone.0045364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rahnama F, Thompson B, Steiner M, Shafiei F, Lobie PE, Mitchell MD. Epigenetic regulation of E-cadherin controls endometrial receptivity. Endocrinology. 2009;150(3):1466–1472. doi: 10.1210/en.2008-1142. [DOI] [PubMed] [Google Scholar]
- 46.Logan PC, Ponnampalam AP, Steiner M, Mitchell MD. Effect of cyclic AMP and estrogen/progesterone on the transcription of DNA methyltransferases during the decidualization of human endometrial stromal cells. Mol Hum Reprod. 2013;19(5):302–312. doi: 10.1093/molehr/gas062. [DOI] [PubMed] [Google Scholar]
- 47.Yamagata Y, Asada H, Tamura I, Lee L, Maekawa R, Taniguchi K, Taketani T, Matsuoka A, Tamura H, Sugino N. DNA methyltransferase expression in the human endometrium: down-regulation by progesterone and estrogen. Hum Reprod. 2009;24(5):1126–1132. doi: 10.1093/humrep/dep015. [DOI] [PubMed] [Google Scholar]
- 48.Baylin SB, Jones PA. A decade of exploring the cancer epigenome—biological and translational implications. Nat Rev Cancer. 2011;11(10):726–734. doi: 10.1038/nrc3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Alam H, Gu B, Lee MG. Histone methylation modifiers in cellular signaling pathways. Cell Mol Life Sci. 2015;72(23):4577–4592. doi: 10.1007/s00018-015-2023-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jin Y, Huo B, Fu X, Hao T, Zhang Y, Guo Y, Hu X. LSD1 collaborates with EZH2 to regulate expression of interferon-stimulated genes. Biomed Pharmacother. 2017;88:728–737. doi: 10.1016/j.biopha.2017.01.055. [DOI] [PubMed] [Google Scholar]
- 51.Grimaldi G, Christian M, Steel JH, Henriet P, Poutanen M, Brosens JJ. Down-regulation of the histone methyltransferase EZH2 contributes to the epigenetic programming of decidualizing human endometrial stromal cells. Mol Endocrinol. 2011;25(11):1892–1903. doi: 10.1210/me.2011-1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sakai N, Maruyama T, Sakurai R, Masuda H, Yamamoto Y, Shimizu A, Kishi I, Asada H, Yamagoe S, Yoshimura Y. Involvement of histone acetylation in ovarian steroid-induced decidualization of human endometrial stromal cells. J Biol Chem. 2003;278(19):16675–16682. doi: 10.1074/jbc.M211715200. [DOI] [PubMed] [Google Scholar]
- 53.Gonzalez-Ramirez I, Soto-Reyes E, Sanchez-Perez Y, Herrera LA, Garcia-Cuellar C. Histones and long non-coding RNAs: the new insights of epigenetic deregulation involved in oral cancer. Oral Oncol. 2014;50(8):691–695. doi: 10.1016/j.oraloncology.2014.04.006. [DOI] [PubMed] [Google Scholar]
- 54.Nancy P, Siewiera J, Rizzuto G, Tagliani E, Osokine I, Manandhar P, Dolgalev I, Clementi C, Tsirigos A, Erlebacher A. H3K27me3 dynamics dictate evolving uterine states in pregnancy and parturition. J Clin Invest. 2018;128(1):233–247. doi: 10.1172/JCI95937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Katoh N, Kuroda K, Tomikawa J, Ogata-Kawata H, Ozaki R, Ochiai A, Kitade M, Takeda S, Nakabayashi K, Hata K. Reciprocal changes of H3K27ac and H3K27me3 at the promoter regions of the critical genes for endometrial decidualization. Epigenomics. 2018;10(9):1243–1257. doi: 10.2217/epi-2018-0006. [DOI] [PubMed] [Google Scholar]
- 56.Tamura I, Jozaki K, Sato S, Shirafuta Y, Shinagawa M, Maekawa R, Taketani T, Asada H, Tamura H, Sugino N. The distal upstream region of insulin-like growth factor-binding protein-1 enhances its expression in endometrial stromal cells during decidualization. J Biol Chem. 2018;293(14):5270–5280. doi: 10.1074/jbc.RA117.000234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tamura I, Asada H, Maekawa R, Tanabe M, Lee L, Taketani T, Yamagata Y, Tamura H, Sugino N. Induction of IGFBP-1 expression by cAMP is associated with histone acetylation status of the promoter region in human endometrial stromal cells. Endocrinology. 2012;153(11):5612–5621. doi: 10.1210/en.2012-1420. [DOI] [PubMed] [Google Scholar]
- 58.Lucas ES, Dyer NP, Murakami K, Lee YH, Chan YW, Grimaldi G, Muter J, Brighton PJ, Moore JD, Patel G, Chan JK, Takeda S, Lam EW, Quenby S, Ott S, Brosens JJ. Loss of endometrial plasticity in recurrent pregnancy loss. Stem Cells. 2016;34(2):346–356. doi: 10.1002/stem.2222. [DOI] [PubMed] [Google Scholar]
- 59.Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, Lagarde J, Veeravalli L, Ruan X, Ruan Y, Lassmann T, Carninci P, Brown JB, Lipovich L, Gonzalez JM, Thomas M, Davis CA, Shiekhattar R, Gingeras TR, Hubbard TJ, Notredame C, Harrow J, Guigo R. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22(9):1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Housman G, Ulitsky I. Methods for distinguishing between protein-coding and long noncoding RNAs and the elusive biological purpose of translation of long noncoding RNAs. Biochim Biophys Acta 1859. 2016;1:31–40. doi: 10.1016/j.bbagrm.2015.07.017. [DOI] [PubMed] [Google Scholar]
- 61.Mattick JS, Rinn JL. Discovery and annotation of long noncoding RNAs. Nat Struct Mol Biol. 2015;22(1):5–7. doi: 10.1038/nsmb.2942. [DOI] [PubMed] [Google Scholar]
- 62.Pauli A, Valen E, Lin MF, Garber M, Vastenhouw NL, Levin JZ, Fan L, Sandelin A, Rinn JL, Regev A, Schier AF. Systematic identification of long noncoding RNAs expressed during zebrafish embryogenesis. Genome Res. 2012;22(3):577–591. doi: 10.1101/gr.133009.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Klinge CM. Non-coding RNAs: long non-coding RNAs and microRNAs in endocrine-related cancers. Endocr Relat Cancer. 2018;25(4):R259–R282. doi: 10.1530/ERC-17-0548. [DOI] [PubMed] [Google Scholar]
- 64.Chen YG, Satpathy AT, Chang HY. Gene regulation in the immune system by long noncoding RNAs. Nat Immunol. 2017;18(9):962–972. doi: 10.1038/ni.3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Geisler S, Coller J. RNA in unexpected places: long non-coding RNA functions in diverse cellular contexts. Nat Rev Mol Cell Biol. 2013;14(11):699–712. doi: 10.1038/nrm3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bouckenheimer J, Assou S, Riquier S, Hou C, Philippe N, Sansac C, Lavabre-Bertrand T, Commes T, Lemaitre JM, Boureux A, De Vos J. Long non-coding RNAs in human early embryonic development and their potential in ART. Hum Reprod Update. 2016;23(1):19–40. doi: 10.1093/humupd/dmw035. [DOI] [PubMed] [Google Scholar]
- 67.Liang XH, Deng WB, Liu YF, Liang YX, Fan ZM, Gu XW, Liu JL, Sha AG, Diao HL, Yang ZM. Non-coding RNA LINC00473 mediates decidualization of human endometrial stromal cells in response to cAMP signaling. Sci Rep. 2016;6:22744. doi: 10.1038/srep22744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zeng H, Fan X, Liu N. Expression of H19 imprinted gene in patients with repeated implantation failure during the window of implantation. Arch Gynecol Obstet. 2017;296(4):835–839. doi: 10.1007/s00404-017-4482-x. [DOI] [PubMed] [Google Scholar]
- 69.Feng C, Shen JM, Lv PP, Jin M, Wang LQ, Rao JP, Feng L. Construction of implantation failure related lncRNA-mRNA network and identification of lncRNA biomarkers for predicting endometrial receptivity. Int J Biol Sci. 2018;14(10):1361–1377. doi: 10.7150/ijbs.25081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen MY, Liao GD, Zhou B, Kang LN, He YM, Li SW. Genome-wide profiling of long noncoding RNA Expression patterns in women with repeated implantation failure by RNA sequencing. Reprod Sci. 2019;26(1):18–25. doi: 10.1177/1933719118756752. [DOI] [PubMed] [Google Scholar]
- 71.Fan LJ, Han HJ, Guan J, Zhang XW, Cui QH, Shen H, Shi C. Aberrantly expressed long noncoding RNAs in recurrent implantation failure: a microarray related study. Syst Biol Reprod Med. 2017;63(4):269–278. doi: 10.1080/19396368.2017.1310329. [DOI] [PubMed] [Google Scholar]
- 72.Tsai JH, Chi MM, Schulte MB, Moley KH. The fatty acid beta-oxidation pathway is important for decidualization of endometrial stromal cells in both humans and mice. Biol Reprod. 2014;90(2):34. doi: 10.1095/biolreprod.113.113217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tong J, Yang J, Lv H, Lv S, Zhang C, Chen ZJ. Dysfunction of pseudogene PGK1P2 is involved in preeclampsia by acting as a competing endogenous RNA of PGK1. Pregnancy Hypertens. 2018;13:37–45. doi: 10.1016/j.preghy.2018.05.003. [DOI] [PubMed] [Google Scholar]
- 74.Lv H, Tong J, Yang J, Lv S, Li WP, Zhang C, Chen ZJ. Dysregulated pseudogene HK2P1 may contribute to preeclampsia as a competing endogenous RNA for hexokinase 2 by impairing decidualization. Hypertension. 2018;71(4):648–658. doi: 10.1161/HYPERTENSIONAHA.117.10084. [DOI] [PubMed] [Google Scholar]
- 75.Yang X, Meng T. Long noncoding RNA in preeclampsia: transcriptional noise or innovative indicators? Biomed Res Int. 2019;2019:5437621. doi: 10.1155/2019/5437621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tong J, Zhao W, Lv H, Li WP, Chen ZJ, Zhang C. Transcriptomic profiling in human decidua of severe preeclampsia detected by RNA sequencing. J Cell Biochem. 2018;119(1):607–615. doi: 10.1002/jcb.26221. [DOI] [PubMed] [Google Scholar]
- 77.Treiber T, Treiber N, Meister G. Regulation of microRNA biogenesis and its crosstalk with other cellular pathways. Nat Rev Mol Cell Biol. 2019;20(1):5–20. doi: 10.1038/s41580-018-0059-1. [DOI] [PubMed] [Google Scholar]
- 78.Carthew RW, Sontheimer EJ. Origins and mechanisms of miRNAs and siRNAs. Cell. 2009;136(4):642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kosaka N, Iguchi H, Ochiya T. Circulating microRNA in body fluid: a new potential biomarker for cancer diagnosis and prognosis. Cancer Sci. 2010;101(10):2087–2092. doi: 10.1111/j.1349-7006.2010.01650.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O’Briant KC, Allen A, Lin DW, Urban N, Drescher CW, Knudsen BS, Stirewalt DL, Gentleman R, Vessella RL, Nelson PS, Martin DB, Tewari M. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA. 2008;105(30):10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Weber JA, Baxter DH, Zhang S, Huang DY, Huang KH, Lee MJ, Galas DJ, Wang K. The microRNA spectrum in 12 body fluids. Clin Chem. 2010;56(11):1733–1741. doi: 10.1373/clinchem.2010.147405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Liang J, Wang S, Wang Z. Role of microRNAs in embryo implantation. Reprod Biol Endocrinol. 2017;15(1):90. doi: 10.1186/s12958-017-0309-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhu XM, Han T, Sargent IL, Yin GW, Yao YQ. Differential expression profile of microRNAs in human placentas from preeclamptic pregnancies vs normal pregnancies. Am J Obstet Gynecol. 2009;200(6):661e661–661e667. doi: 10.1016/j.ajog.2008.12.045. [DOI] [PubMed] [Google Scholar]
- 84.Pan Q, Luo X, Toloubeydokhti T, Chegini N. The expression profile of micro-RNA in endometrium and endometriosis and the influence of ovarian steroids on their expression. Mol Hum Reprod. 2007;13(11):797–806. doi: 10.1093/molehr/gam063. [DOI] [PubMed] [Google Scholar]
- 85.Ferlita A, Battaglia R, Andronico F, Caruso S, Cianci A, Purrello M, Pietro CD. Non-coding RNAs in endometrial physiopathology. Int J Mol Sci. 2018;19(7):2120. doi: 10.3390/ijms19072120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bazer FW, Spencer TE, Johnson GA, Burghardt RC. Uterine receptivity to implantation of blastocysts in mammals. Front Biosci (Schol Ed) 2011;3:745–767. doi: 10.2741/s184. [DOI] [PubMed] [Google Scholar]
- 87.Estella C, Herrer I, Moreno-Moya JM, Quinonero A, Martinez S, Pellicer A, Simon C. miRNA signature and Dicer requirement during human endometrial stromal decidualization in vitro. PLoS One. 2012;7(7):e41080. doi: 10.1371/journal.pone.0041080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tochigi H, Kajihara T, Mizuno Y, Mizuno Y, Tamaru S, Kamei Y, Okazaki Y, Brosens JJ, Ishihara O. Loss of miR-542-3p enhances IGFBP-1 expression in decidualizing human endometrial stromal cells. Sci Rep. 2017;7:40001. doi: 10.1038/srep40001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kimura M, Kajihara T, Mizuno Y, Sato T, Ishihara O. Loss of high-mobility group N5 contributes to the promotion of human endometrial stromal cell decidualization. Reprod Med Biol. 2018;17(4):493–499. doi: 10.1002/rmb2.12226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sultana S, Kajihara T, Mizuno Y, Sato T, Oguro T, Kimura M, Akita M, Ishihara O. Overexpression of microRNA-542-3p attenuates the differentiating capacity of endometriotic stromal cells. Reprod Med Biol. 2017;16(2):170–178. doi: 10.1002/rmb2.12028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ji L, Zhang L, Li Y, Guo L, Cao N, Bai Z, Song Y, Xu Z, Zhang J, Liu C, Ma X. MiR-136 contributes to pre-eclampsia through its effects on apoptosis and angiogenesis of mesenchymal stem cells. Placenta. 2017;50:102–109. doi: 10.1016/j.placenta.2017.01.102. [DOI] [PubMed] [Google Scholar]
- 92.Wang XB, Qi QR, Wu KL, Xie QZ. Role of osteopontin in decidualization and pregnancy success. Reproduction. 2018;155(5):423–432. doi: 10.1530/REP-17-0782. [DOI] [PubMed] [Google Scholar]
- 93.Zhang Q, Zhang H, Jiang Y, Xue B, Diao Z, Ding L, Zhen X, Sun H, Yan G, Hu Y. MicroRNA-181a is involved in the regulation of human endometrial stromal cell decidualization by inhibiting Kruppel-like factor 12. Reprod Biol Endocrinol. 2015;13:23. doi: 10.1186/s12958-015-0019-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Qian K, Hu L, Chen H, Li H, Liu N, Li Y, Ai J, Zhu G, Tang Z, Zhang H. Hsa-miR-222 is involved in differentiation of endometrial stromal cells in vitro. Endocrinology. 2009;150(10):4734–4743. doi: 10.1210/en.2008-1629. [DOI] [PubMed] [Google Scholar]
- 95.Yang Q, Zhang X, Shi Y, He YP, Sun ZG, Shi HJ, Wang J. Increased expression of NDRG3 in mouse uterus during embryo implantation and in mouse endometrial stromal cells during in vitro decidualization. Reprod Sci. 2018;25(8):1197–1207. doi: 10.1177/1933719117737843. [DOI] [PubMed] [Google Scholar]
- 96.Yang Y, Xie Y, Wu M, Geng Y, Li R, Xu L, Liu X, Pan Y. Expression of mmu-miR-96 in the endometrium during early pregnancy and its regulatory effects on stromal cell apoptosis via Bcl2. Mol Med Rep. 2017;15(4):1547–1554. doi: 10.3892/mmr.2017.6212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Jimenez PT, Mainigi MA, Word RA, Kraus WL, Mendelson CR. miR-200 regulates endometrial development during early pregnancy. Mol Endocrinol. 2016;30(9):977–987. doi: 10.1210/me.2016-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sanger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci USA. 1976;73(11):3852–3856. doi: 10.1073/pnas.73.11.3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Westholm JO, Miura P, Olson S, Shenker S, Joseph B, Sanfilippo P, Celniker SE, Graveley BR, Lai EC. Genome-wide analysis of drosophila circular RNAs reveals their structural and sequence properties and age-dependent neural accumulation. Cell Rep. 2014;9(5):1966–1980. doi: 10.1016/j.celrep.2014.10.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhang Y, Xue W, Li X, Zhang J, Chen S, Zhang JL, Yang L, Chen LL. The biogenesis of nascent circular RNAs. Cell Rep. 2016;15(3):611–624. doi: 10.1016/j.celrep.2016.03.058. [DOI] [PubMed] [Google Scholar]
- 101.Wilusz JE. Circular RNAs: unexpected outputs of many protein-coding genes. RNA Biol. 2017;14(8):1007–1017. doi: 10.1080/15476286.2016.1227905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 103.Liu L, Li L, Ma X, Yue F, Wang Y, Wang L, Jin P, Zhang X. Altered circular RNA expression in patients with repeated implantation failure. Cell Physiol Biochem. 2017;44(1):303–313. doi: 10.1159/000484887. [DOI] [PubMed] [Google Scholar]
- 104.Song Y, Zhang L, Liu X, Niu M, Cui J, Che S, Liu Y, An X, Cao B. Analyses of circRNA profiling during the development from pre-receptive to receptive phases in the goat endometrium. J Anim Sci Biotechnol. 2019;10:34. doi: 10.1186/s40104-019-0339-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zhang L, Liu X, Che S, Cui J, Liu Y, An X, Cao B, Song Y. CircRNA-9119 regulates the expression of prostaglandin-endoperoxide synthase 2 (PTGS2) by sponging miR-26a in the endometrial epithelial cells of dairy goat. Reprod Fertil Dev. 2018;30(12):1759–1769. doi: 10.1071/RD18074. [DOI] [PubMed] [Google Scholar]
- 106.Zhang S, Ding Y, He J, Zhang J, Liu X, Chen X, Su Y, Wang Y, Gao R. Altered expression patterns of circular RNAs between implantation sites and interimplantation sites in early pregnant mice. J Cell Physiol. 2019;234(6):9862–9872. doi: 10.1002/jcp.27675. [DOI] [PubMed] [Google Scholar]
- 107.Kong S, Zhou C, Bao H, Ni Z, Liu M, He B, Huang L, Sun Y, Wang H, Lu J. Epigenetic control of embryo-uterine crosstalk at peri-implantation. Cell Mol Life Sci. 2019;76(24):4813–4828. doi: 10.1007/s00018-019-03245-8. [DOI] [PMC free article] [PubMed] [Google Scholar]