Abstract
Ubiquitin modification plays significant roles in protein fate determination, signaling transduction, and cellular processes. Over the past 2 decades, the number of studies on ubiquitination has demonstrated explosive growth. E3 ubiquitin ligases are the key enzymes that determine the substrate specificity and are involved in cancer. Several recent studies shed light on the functions and mechanisms of HECTD3 E3 ubiquitin ligase. This review describes the progress in the recent studies of HECTD3 in cancer and other diseases. We propose that HECTD3 is a potential biomarker and a therapeutic target, and discuss the future directions for HECTD3 investigations.
Keywords: Ubiquitination, HECTD3, Cancer, Inhibitors
Protein ubiquitination
Ubiquitination is a kind of protein posttranslational modification. Sequential reactions catalyzed by ubiquitin-activating enzyme (E1), ubiquitin-conjugating enzyme (E2), and ubiquitin ligase (E3) result in covalent conjugation of ubiquitin (Ub) molecules to a target protein to serve as a signal label that regulates the fate of a target protein. Therefore, ubiquitination participates in the regulation of numerous fundamental biological processes and is essential for maintenance of normal physiological activities of the organisms [1, 2]. A ubiquitin molecule forms an isopeptide bond to the side chain of a Lys (K) residue of a target protein through its C-terminal Gly residue [3]. Additional ubiquitin molecules can be attached to the previous ones through K residues (6, 11, 27, 29, 33, 48, and 63) or N-terminal Met residue to form a polymeric Ub chain [4, 5]. K48- and K63-linked polyubiquitin chains are the most common [6]. Ubiquitin modification can be removed by the deubiquitinating enzymes (DUBs).
Ubiquitination regulates protein stability, cellular localization, protein–protein interactions, trafficking, and activity depending on the site, number, linkage, and length of the modifications [3]. For example, degradation-independent functions are associated with monoubiquitination, linear ubiquitination, or K63-linked polyubiquitination. Ubiquitination plays a pivotal role in the regulation of cellular functions, such as cell cycle, apoptosis, DNA damage repair, transcription, endocytosis, and signaling [7–13]. Dysfunction of E3 and DUBs is an important factor contributing to the development and pathogenesis of multiple human diseases and E3 and DUBs are potential therapeutic targets [14–16]. Accumulating evidence indicates importance of ubiquitination in initiation, metastasis, and drug resistance in cancer [17–21].
E3 ubiquitin ligases
There are over 600 E3 ligases in the human body [22]. E3 ligases directly interact with the target proteins and are responsible for specificity of ubiquitination. Several E3 ligases, such as murine double minute 2 (MDM2) [23], SKP1-CUL1-F-box protein (SCF) complex (SCFSKP2 [24] and SCFFBW7 [25]), pVHL-elongin C-elongin B-cullin 2-RBX1 (VCB-CR) complex [26], and breast cancer type 1 susceptibility protein (BRCA1) [27], are known to play important roles in cancer. Targeting E3 ligases has become a novel avenue for drug development and cancer treatment. Currently, several MDM2 inhibitors have been tested for antitumor activity in clinical trials [28–30].
E3 ligases can be classified into RING (really interesting new genes), HECT (homologous to E6AP C terminus), and RBR (RING- IBR-RINGs) type E3s [3] according to their protein structure and functional mechanisms. RING E3 ligases contain a RING finger domain, but do not possess catalytic activity. They function as adaptor proteins that promote the ubiquitin transfer from E2 to a substrate protein. In contrast, HECT and RBR E3 ligases form an intermediate with ubiquitin through their own catalytic cysteine (Cys) and then transfer the ubiquitin moiety to a substrate protein [31, 32]. RBR family is comprised of a central in-between-RINGs (IBR) domain and two RING finger domains located at both termini [33].
HECT E3 family contains 28 members with common C-terminal HECT domain. Furthermore, HECT E3 ligases can be divided into three subfamilies: Nedd4 subfamily, HERC subfamily, and other HECT E3s [6, 31]. Nedd4 subfamily members contain a C2 domain and 2–4 WW domains. HERC subfamily members contain one or more RCC1 like domain (RLD) domains. Members of the other HECT subfamily are composed of diverse N-terminal domains. Conservative catalytic Cys located in the C-lobe of the HECT domain can form a thioester bond with ubiquitin, while the N-lobe is responsible for E2 enzyme binding [6, 34–36].
HECT-type E3 ligases promote ubiquitination of a large number of substrate proteins. In addition, they are regulated by intricate signals and widely implicated in cancer. E6-associated protein (E6-AP), the first identified HECT-type E3, targets tumor protein p53 for ubiquitin-dependent degradation after interaction with the E6 protein of HPV [37, 38]. Moreover, E6-AP promotes ubiquitination and degradation of tumor suppressor promyelocytic leukemia protein (PML) in B-cell lymphoma and prostate cancer [39–41]. In contrast, Mansour et al. found that E6-AP targets epithelial cell transforming 2 (ECT2) for proteasomal degradation and inhibits breast cancer metastasis [42]. Gene amplification and protein overexpression of WW domain containing E3 ubiquitin protein ligase 1 (WWP1) frequently occur in human prostate cancer and breast cancer [43, 44]. Substrates of WWP1 include tumor protein p63, Kruppel like factor 5 (KLF5), transforming growth factor beta receptor 1 (TGFβR1), and erb-b2 receptor tyrosine kinase 4 (ERBB4) [45–48]. Itchy E3 ubiquitin protein ligase (ITCH) mediates the ubiquitin–proteasome degradation of large tumor suppressor kinase 1 (LATS1) and regulates the Hippo pathway [49]. ITCH also targets H1.2 linker histone for polyubiquitination, which regulates DNA damage response in triple negative breast cancer (TNBC) [50]. SMAD-specific E3 ubiquitin protein ligase 2 (SMURF2) negatively regulates the TGFβ signaling pathway by promoting the degradation of transforming growth factor beta (TGFβ) receptor and SMAD family member 2 (SMAD2) [51, 52]. In addition, SMURF2 regulates monoubiquitination of histones through the ubiquitin-dependent degradation of ring finger protein 20 (RNF20) and is thus involved in maintenance of genomic stability and tumor suppression [53]. NEDD4 E3 ubiquitin protein ligase (NEDD4-1) and WW domain containing E3 ubiquitin protein ligase 2 (WWP2) are suggested to be potential oncoproteins by facilitating the degradation of phosphatase and tensin homolog (PTEN), a distinguished tumor suppressor [54, 55].
HECTD3
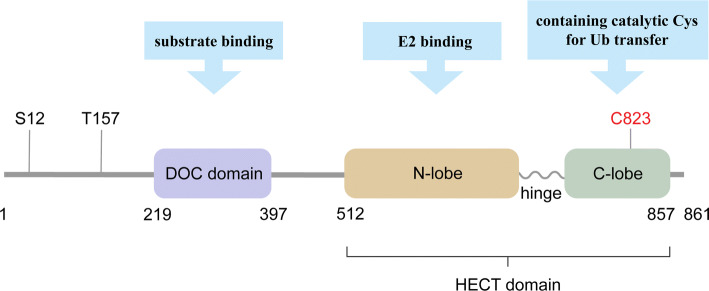
Homologous to the E6-associated protein carboxyl terminus domain containing 3 (HECTD3) is classified as the third subfamily of HECT E3s. HECTD3 is comprised of 861 amino acid residues, and contains an N-terminal DOC domain (219-397) and a C-terminal HECT domain (512-857) (Fig. 1). Like other HECT E3s, in its HECT domain, a flexible hinge links the N-lobe which contains E2 binding motif and the C-lobe where catalytic Cys (C832) locates in. The N-terminal DOC domain is responsible for substrate binding. In addition, there are two known phosphorylation sites (S12 and T157) at the N-terminus of HECTD3 (Fig. 1).
Fig. 1.
The diagram of HECTD3 protein structure. HECTD3 belongs to the other HECT E3s subfamily. It is comprised of a DOC domain and an HECT domain. In the HECT domain, a flexible hinge links the N-lobe, which contains the E2 binding site, and the C-lobe, which contains the catalytic Cys for Ub transfer. The DOC domain is responsible for substrate binding. Catalytic Cys and phosphorylation sites are indicated [60, 61, 111]
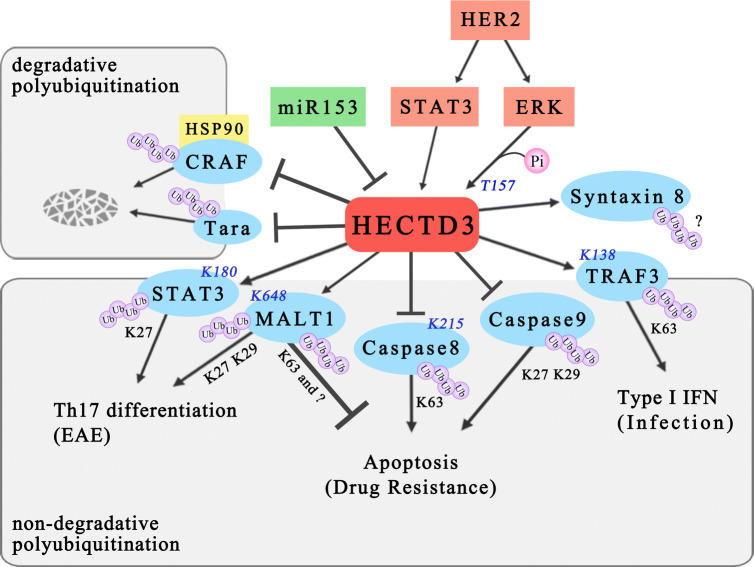
In recent years, the function of HECTD3 has received considerable attention. HECTD3 has been reported to modify a variety of substrate proteins, to be regulated by diverse factors, and to play crucial roles in cellular processes, such as apoptosis, drug resistance, and immunoreaction (Fig. 2, Table 1). HECTD3 may become a therapeutic target in cancer and other diseases.
Fig. 2.
Functions, mechanisms, and regulation of HECTD3. HECTD3 targets several apoptosis-related proteins (MALT1, caspase-8, and caspase-9) for nondegradative polyubiquitination and confers drug resistance to cancer cell. It also regulates the immune response through nondegradative polyubiquitination of STAT3, MALT1, and TRAF3. In addition to the nondegradative pathway, HECTD3 mediates ubiquitin–proteasome degradation of Tara and CRAF. HER2/ERK and miR-153 regulate the HECTD3 activity and expression. The linkage types of polyubiquitin chain and the modification sites in the substrate proteins are indicated
Table 1.
HECTD3-mediated protein ubiquitination
| Substrate | Ub linkage | Molecular function | Cellular function | References |
|---|---|---|---|---|
| Tara | – | proteasomal degradation | Cell cycle and formation of multipolar spindle | [56] |
| Syntaxin 8 | – | – | Neurodegenerative diseases | [57] |
| MALT1 | Non K48 | Increase in MALT1 protein stability | Antiapoptotic chemoresistance | [58] |
| K27, K29 | Activation of NF-κB signaling | Promotion of the differentiation of Th17 cells | [59] | |
| Caspase-8 | K63 | Inhibition of caspase-8 activity | Inhibition of extrinsic apoptosis | [60] |
| Caspase-9 | K27, K29 | Inhibition of caspase-9 activity | Inhibition of intrinsic apoptosis | [61] |
| CRAF | – | Proteasomal degradation | Restriction of MAPK pathway | [62] |
| TRAF3 | K63 | Activation of TBK1 | Promotion of type I interferon production and bacterial infection | [63] |
| Stat3 | K27 | Promotion of Stat3 activation | Induction of Th17 cell differentiation | [59] |
HECTD3-modified proteins
In 2008, Yu et al. reported HECTD3 as an E3 ligase of trio-associated repeat on actin (Tara) that promotes Tara ubiquitination and degradation [56]. In 2009, Zhang et al. found that syntaxin 8 is another HECTD3 substrate protein and HECTD3 may influence neurodegenerative diseases [57].
Li et al. demonstrated that MALT1 paracaspase (MALT1) is a bona fide HECTD3 substrate protein [58]. The HECTD3 DOC domain interacts with the MALT1 DD domain; therefore, HECTD3 increases MALT1 polyubiquitination and protein stability. This study suggested that the MALT1 ubiquitin chain linkage mediated by HECTD3 is not K48-linked, but is K63-linked and other K-linked. It has been speculated that the stabilizing effect of HECTD3 on MALT1 may antagonize the degradation mediated by other E3s. It is intriguing that an HECTD3 mutant without catalytic activity still promotes MALT1 ubiquitination. A possible explanation for this effect is that HECTD3 promotes ubiquitination of MALT1 by other E3 ligases, such as TNF receptor-associated factor 6 (TRAF6). It has been reported that TRAF6 promotes MALT1 ubiquitination [64]. However, this possibility needs additional supporting evidence.
In a recent study, Hectd3 was shown to modify Malt1 with K27- and K29-linked polyubiquitin chains while K648 is the ubiquitination site of Malt1, which is related to nuclear translocation of p65 and activation of NF-κB signaling pathway [59]. In addition, Hectd3 binds to the signal transducer and activator of transcription 3 (Stat3) linker region through the DOC domain and modifies Stat3 at K180 with K27-linked polyubiquitin chains; this modification is associated with phosphorylation of Stat3 at Y705 and expression of Retineic-acid-receptor-related orphan nuclear receptor gamma (RORγt) in T helper 17 (Th17) cells [59].
Our previous study suggested that HECTD3 specifically catalyzes K63-linked polyubiquitination of caspase-8 [60]. Ubiquitination prevents the recruitment of caspase-8 into TRAIL (TNF-related apoptosis-inducing ligand)-induced DISC (death-inducing signaling complex) for activation [60]. This study identified HECTD3 C823 as the catalytic Cys and caspase-8 K215 as the ubiquitination site [60]. Later, cullin 7 (CUL7), the scaffold protein of cullin-RING E3 ubiquitin ligase 7 (CRL7), was shown to ubiquitinate caspase-8 at the same site and to have similar antiapoptotic function [65]. In addition to caspase-8, HECTD3 ubiquitinates caspase-9 with K27- and K29-linked polyubiquitin chains and suppresses its activation [61].
HECTD3 was demonstrated to mediate heat shock protein 90 (HSP90) inhibitor-induced degradation of RAF proto-oncogene serine/threonine-protein kinase (CRAF), an HSP90 client kinase. HECTD3 interacts with HSP90 and CRAF and subsequently targets CRAF for degradation [62]. It is well established that chaperones are essential for correct folding of proteins and the ubiquitin–proteasome system is responsible for degradation of incorrectly folded proteins. It is unknown whether HECTD3 targets other chaperon clients.
Most recently, Li et al. reported that HECTD3 catalyzes K63-linked polyubiquitination of TNF receptor-associated factor 3 (TRAF3) at K138 [63]. HECTD3-mediated TRAF3 ubiquitination promotes the interaction between TRAF3 and TANK-binding kinase 1 (TBK1) and subsequent phosphorylation of TBK1 and interferon regulatory factor 3 (IRF3). Hectd3-deficient macrophages failed to produce type I interferon (IFN) in response to intracellular bacterial infection [63].
The regulation of HECTD3
Shu T et al. showed that erb-b2 receptor tyrosine kinase 2 (HER2) increased HECTD3 expression through activation of STAT3 [66]. STAT3 binds to the HECTD3 gene promoter to induce its transcription. It is well known that STAT3 can be activated by epidermal growth factor receptors (EGFRs) and is usually constitutively activated in cancer [67].
Li Y et al. reported that HECTD3 can be phosphorylated and activated by mitogen-activated protein kinase 1 (ERK, also known as MAPK) [61]. A PKC activator, phorbol-12-myristate-13-acetate (PMA), activates the Raf–MEK–ERK pathway and facilitates the phosphorylation of HECTD3 at Thr157 via ERK. The phosphorylation of HECTD3 increases the binding of HECTD3 and caspase-9 [61].
Wu X et al. demonstrated that microRNA-153 (miR-153) targets HECTD3 [68]. MiR-153 inhibits HECTD3 mRNA expression and enhances the sensitivity of TNBC cells to cisplatin. MiR153 is a well-known tumor suppressor in breast cancer. We reported that miR-153 targets several oncogenes, including KLF5, hypoxia inducible factor 1 subunit alpha (HIF1α), angiopoietin 1 (ANGPT1), and myeloid cell leukemia 1 (MCL1), thus boosting stemness, cell growth, and angiogenesis of breast cancer [69–71]. Consistently, the expression level of miR153 is positively associated with the 5-year survival rate and prognosis of breast cancer patients. Interestingly, miR-153 itself is regulated by endoplasmic reticulum (ER) stress. Hypoxia and drug treatment can directly induce miR-153 transcription through inositol requiring enzyme 1 alpha (IRE1α)/X-box-binding protein 1 (XBP1) [69, 70]. A possibility that ER stress inhibits HECTD3 expression through miR153 has not been tested.
The role of HECTD3 in cancer
Accumulated evidence suggests that HECTD3 has a prosurvival role in several types of cancer. HECTD3 has become a potential biomarker for cancer diagnosis and prognosis and a therapeutic target. In breast cancer, gene amplification leads to HECTD3 overexpression. The overexpression of HECTD3 was linked to cisplatin resistance through ubiquitination and stabilization of MALT1 [58]. This was the first study to reveal the prosurvival function of HECTD3 and MALT1. Recently, Ekambaram et al. demonstrated that in angiotensin II receptor-positive breast cancer, the activation of the CARMA3–Bcl10–MALT1 pathway promotes cancer cell proliferation and invasion [72]. Moreover, Lin et al. reported that the lack of MALT1 protease activity in Treg cells leads to inhibition of lymphoma growth [73]. Kawadler et al. showed that MALT1 can control the activation of caspase-8 and facilitate lymphocyte proliferation and survival [74]. In addition, HECTD3 promotes the survival of human breast cancer cells under extrinsic apoptotic stimuli via direct ubiquitination of caspase-8 [60]. In esophageal squamous cell carcinoma (ESCC) KYSE30 cells, HECTD3 overexpression results in cisplatin resistance through blockade of activation of caspase-9 [61]. In ovarian cancer cell lines and xenograft mouse models, downregulation of HECTD3 significantly facilitated carboplatin-induced apoptosis [66]. Given that HECTD3 confers apoptosis resistance and chemoresistance, HECTD3 inhibitors in combination with chemotherapeutic drugs may alleviate drug chemoresistance.
However, Li et al. argued that HECTD3 may function as a tumor suppressor, because HECTD3 downregulates an HSP90 protein kinase client CRAF thus inhibiting the activation of MAPK [62]. Yu et al. reported that HECTD3 is the E3 ligase for Tara. Deletion of either Tara or HECTD3 results in the formation of multipolar spindle, indicating that HECTD3 may be important for maintenance of genomic stability [56]. It is possible that HECTD3 plays a context-dependent role.
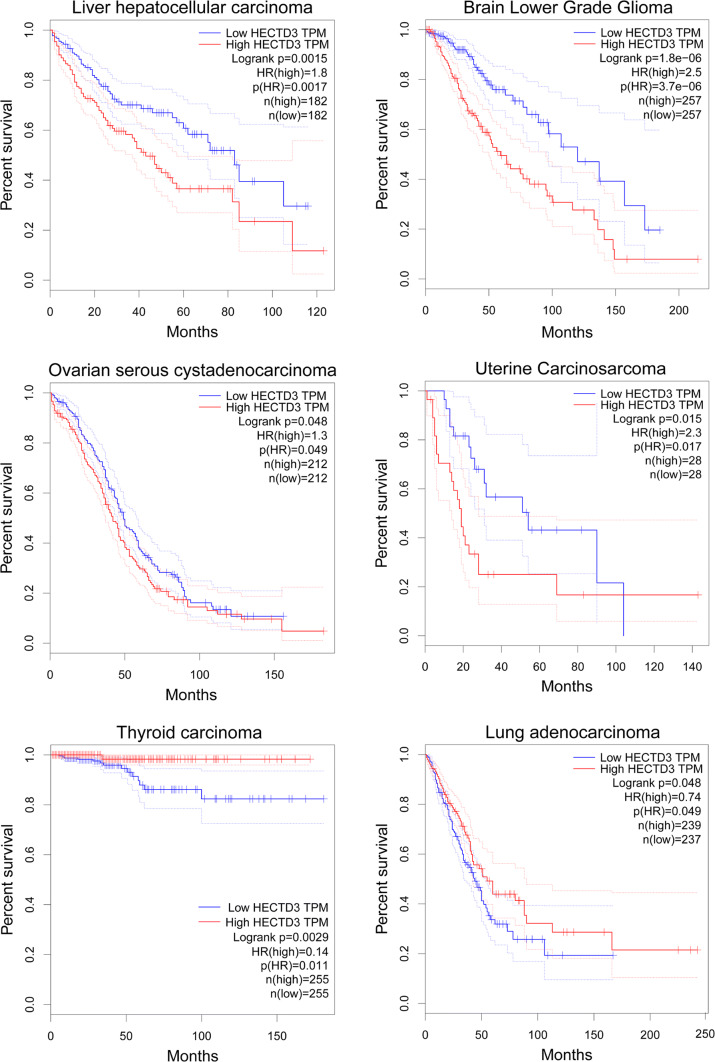
Moreover, we used GEPIA (Gene Expression Profiling Interactive Analysis, http://gepia.cancer-pku.cn/index.html) to analyze the prognosis values of HECTD3 mRNA expression levels in different cancer types. HECTD3 appears to act as both tumor-promoting and -suppressing factors in different types of cancer. As shown in Fig. 3, a high HECTD3 mRNA expression level is associated with poor prognosis in liver hepatocellular carcinoma (LIHC), brain lower grade glioma (LGG), ovarian serous cystadenocarcinoma (OV), and uterine carcinosarcoma (UCS). On the contrary, in thyroid carcinoma (THCA) and lung adenocarcinoma (LUAD), a high HECTD3 mRNA expression level is associated with favorable prognosis. The context-dependent roles of HECTD3 in different cancers should be investigated in the future.
Fig. 3.
Prognostic values of HECTD3 mRNA expression levels in different cancer types. A high HECTD3 mRNA expression level is associated with poor prognosis in liver hepatocellular carcinoma (LIHC), brain lower grade glioma (LGG), ovarian serous cystadenocarcinoma (OV) and uterine carcinosarcoma (UCS). On the contrary, it is associated with favorable outcomes in thyroid carcinoma (THCA) and lung adenocarcinoma (LUAD)
The role of HECTD3 in other diseases
In addition to cancer, HECTD3 may play significant roles in the immune system. HECTD3 is a potential target for multiple sclerosis and intracellular bacterial infection. First, HECTD3 positively regulates the production of type I IFN via ubiquitination of TRAF3 and activation of TBK1 in response to intracellular bacterial infection [63]. Hectd3 knockout limits the migration and dissemination of F. novicida-carrying macrophages and neutrophils, and promotes stronger defensive behavior in mice [63]. Second, Hectd3 promotes the activation of NF-κB and RORγt expression through ubiquitinating Malt1 and Stat3, respectively, and then facilitates Th17 cell differentiation [59]. Malt1 is an essential component of the Carma1–Bcl10–Malt1 (CBM) complex, which promotes NF-κB activation and Th17 pathogenicity in EAE [75]. Stat3 and RORγt are key transcription factors regulating Th17 cell differentiation. Therefore, Hectd3 global knockout mice have decreased symptoms of experimental autoimmune encephalomyelitis (EAE) [59]. This phenotype is similar to Malt1 knockout mice [75]. Finally, HECTD3 may be involved in neurodegenerative diseases by interacting with and ubiquitinating syntaxin 8 [57].
E3 ligase inhibitors
Following the approval of bortezomib, the first proteasome inhibitor, for the treatment of refractory hematologic malignancies [76, 77], great attention has been paid to the studies of the ubiquitin proteasome system. Oncogenic E3s are frequently overactivated in cancer by gene amplification and overexpression [78]. Several strategies can be used to target oncogenic E3s, such as inhibition of E3 expression, inhibition of E3 enzyme activity, disruption of the interaction between E3 and E2 or their substrates, or restraining the assembly of the E3 complex.
For example, a variety of small molecule inhibitors have been identified for MDM2. These inhibitors function through two mechanisms. One mechanism involves repression of the E3 ligase activity of MDM2. Another mechanism is based on interference in interaction between MDM2 and p53. For example, nutlins occupy the p53-binding site of MDM2, while RITA binds to the N-terminal region of p53 thus interfering with the interaction [79–81]. To date, several small molecule inhibitors of MDM2 are in clinical trials. In addition to RG7112 [82], multiple compounds, including MI-219, MI-319, MI-888, MI-77301, and APG-115, have been developed. MI-77301 and APG-115 have undergone phase I clinical trials [28, 29, 83–89]. RG7388 (idasanutlin) has entered a phase III clinical trial for the treatment of acute myeloid leukemia [30, 90].
In addition to traditional small molecule inhibitors of E3 ligases, emergence of several novel strategies provides more possibilities for cancer treatment. Ub variants (UbVs) can be developed as inhibitors or activators of E3s. Several studies screened specific UbVs against 20 HECT E3s and other E3s (Fig. 4). UbV inhibitors can hinder the E2-E3 binding or the assembly of the cullin1 (CUL1)-based E3 complexes [91–93]. These results indicate that UbVs may become a novel tool for inhibition of abnormal E3s in cancer. The delivery of UbVs in vivo, however, remains a challenge.
Fig. 4.
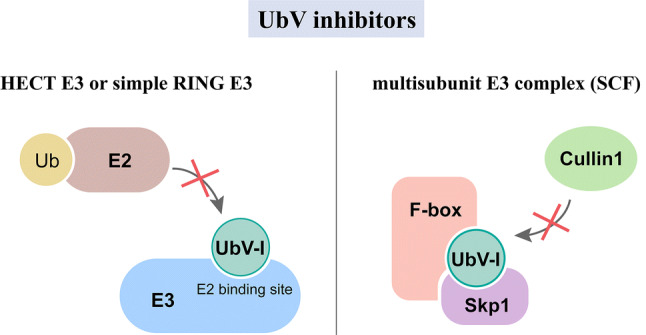
The functional mechanisms of UbV inhibitors. In the case of HECT E3s and simple RING E3s, UbV inhibitors interfere with the E2–E3 binding. In the case of the multisubunit E3 complex (i.e., SCF), UbV inhibitors suppress the assembly of the Cul1 subunit
Strategies of targeting HECTD3
As described above, HECTD3 inhibition may overcome chemoresistance in cancer, multiple sclerosis, and intracellular bacterial infection. Importantly, specific inhibition of HECTD3 should be safe, because Hectd3 whole-body knockout mice are normal and do not have detectable defects.
Various studies provide support and inspiration for HECTD3-targeted therapy. Several strategies can be considered for screening and development of HECTD3 inhibitors, including small molecule inhibitors, peptides, and proteins (Fig. 5).
Fig. 5.
Strategies to inhibit HECTD3-mediated ubiquitination process. ① Inhibition of the catalytic activity (e.g., heclin). ② Hijacking the E2-binding site (e.g., UbV inhibitor or bicyclic peptides). ③ Interference with the binding of specific substrates. ④ Obstruction of the ubiquitin transthiolation (e.g., clomipramine). ⑤ Interruption of polyubiquitin chain elongation (e.g., occupying the exosite). ⑥ Targeting the HECTD3 posttranslational modification or upstream factors. ⑦ Regulation of the autoinhibition mechanism
First, the catalytic activity of HECTD3 can be suppressed. It is well known that HECT E3 ligases possess intrinsic catalytic activity. Mund et al. identified two types of HECT E3 inhibitors that have different mechanisms of action and confirmed that targeting HECT domain for drug design is feasible [94]. One of the inhibitors, heclin, a small molecule compound with wider spectrum, suppresses multiple HECT E3 ligases via induction of spontaneous oxidation of a Cys residue of the E3 active site [94]. Possibility that heclin inhibits HECTD3 enzyme activity deserves investigation.
Second, it is possible to hijack the E2 binding site of HECTD3 or block the interaction between HECTD3 and specific substrate proteins. For instance, bicyclic peptides, another kind of inhibitors described in Mund et al.’s study, have high specificity against individual HECT E3 ligases by targeting the E2-binding sites [94].
Third, the ubiquitin transthiolation can be obstructed to halt the ubiquitination process. Rossi et al. demonstrated that clomipramine, an antidepressant, impedes the transthiolation of ubiquitin from E2 to HECT E3. Clomipramine irreversibly inhibited the Itch-mediated ubiquitination of p73 and showed anticancer activities in combination with chemotherapy drugs [95].
Fourth, extension of the ubiquitin chain can be blocked. Kathman et al. identified the first covalent inhibitor of Nedd4-1. The compound prevents Nedd4-1 from binding to ubiquitin by reacting with noncatalytic Cys627, which represses E3 progressivity and extension of the ubiquitin chains by occupying the exosite (a processivity site) [96]. This work provides a novel strategy for the development of HECT E3 inhibitors.
Fifth, the upstream positive regulators of HECTD3 can be inhibited. As mentioned above, ERK is one of the upstream factors of HECTD3 and can become a possible target for modulation of HECTD3 activity in cancer [61]. STAT3 may become another possible target for inhibition of HECTD3 expression. Recently, an STAT3 inhibitor, TTI-101, has entered phase I trial (NCT03195699). In addition, it is worthwhile to consider the use of gene therapy based on miRNA to control the transcriptional expression of HECTD3, e.g., by miR153 mimics. In addition to miR153, other miRNAs and regulators should be identified for efficient control of the HECTD3 expression level and activity.
Finally, we can inhibit the HECTD3 activity by regulating the autoinhibition mechanism and targeting its upstream factors. HECT E3 ligases need a “braking system” to appropriately switch between the active and inactive states to ensure proper functioning of E3 ligases and to elaborate regulation of cell signals. For example, HECT, UBA, and WWE domain containing E3 ubiquitin protein ligase 1 (HUWE1) activity is regulated by a conformational switch which causes suppression of the activity through self-dimerization and intramolecular interactions [97]. Several Nedd4 subfamily members lock themselves into a ground state T-shape through the WW2–WW3 linker and this braking effect can be relaxed by tyrosine phosphorylation [98]. For example, Abelson murine leukemia viral homolog 1 (c-Abl) can reduce the E3 ligase activation through phosphorylation of E6-AP at Y636 to alleviate the p53 degradation [99]. Hence, further characterization of HECTD3 protein structure and regulatory mechanism of HECTD3 is required.
Regardless of selected strategy, high-throughput screening technologies (HTS) will dramatically advance the development of HECTD3 inhibitors. The technologies include fragment-based drug discovery (FBDD), phage display technology, alpha screen technology, UbFluor, and fluorescence polarization assay for high-throughput screening (FP-HTS) [100–105]. Natural components from plants and microorganisms and FDA-approved drugs provide rich resources for drug discovery. DNA-encoded compound libraries (DECL) provide unprecedented space for the construction of compound libraries and drug screening [105, 106]. In silico and cell-based assays are important compound screening strategies [107–110]. We have developed an HTS method based on an in vitro HECTD3 self-ubiquitination assay and identified several natural compounds as potential HECTD3 inhibitors. Further experimental investigations are required to validate these HECTD3 inhibitors.
Summary and prospects
The ubiquitin system is crucial for maintenance of normal cellular biological progress. The system can not only target proteins for degradation but also mediate various proteasome-independent functions through the diversity of polyubiquitin chain linkages. E3 ligases have come to the central stage because of their numbers and substrate specificity. Several E3s are attractive therapeutic targets for cancer.
HECTD3 is an under-investigated HECT-type E3 with tremendous research value. Previous studies suggested that HECTD3 has a prosurvival function in cancer. One of the most important features of cancer is resistance to apoptosis and consequent drug resistance in clinic. Further studies are required to determine whether HECTD3 inhibitors have a synergistic effect in combination with chemotherapies in various types of cancer. It is worth investigating whether the expression of HECTD3 in patient specimens can be used as a prognosis biomarker to predict drug sensitivities. Of course, these inhibitors should be evaluated for treatment of multiple sclerosis and intracellular bacterial infection.
In addition to apoptosis and immune regulation, HECTD3 may have other functions depending on the context. A systematic understanding of the roles of HECTD3 remains incomplete. Hectd3 gene knockout mouse models will play important roles. It has been shown that Hectd3 gene whole-body knockout mice are resistant to intracellular bacterial infection and EAE [59]. Tissue-specific knockout and knock-in mouse models will be very useful to address the functions and mechanisms of Hectd3 in various organs and diseases.
Moreover, additional substrate proteins and potential upstream regulators need to be identified. Following that, identification of HECTD3 inhibitors with high specificity and strong efficacy is a major direction for future HECTD3 studies.
Acknowledgements
This study was supported in part by grants from the National Key R&D Program of China (2018YFC2000400) and the National Nature Science Foundation of China (81830087, U1602221, and 31771516 to Chen, C and 81773149 to Kong Y) and the Shenzhen Municipal Government of China (KQTD20170810160226082).
Abbreviations
- ACC
Adrenocortical carcinoma
- ANGPT1
Angiopoietin 1
- BRCA
Breast invasive carcinoma
- BRCA1
Breast cancer type 1 susceptibility protein
- c-Abl
Abelson murine leukemia viral homolog 1
- CHOL
Cholangiocarcinoma
- CRAF
RAF proto-oncogene serine/threonine-protein kinase
- CRL7
Cullin-RING E3 ubiquitin ligase 7
- CUL1
Cullin1
- CUL7
Cullin 7
- DECL
DNA-encoded compound libraries
- DISC
Death-inducing signaling complex
- DLBC
Lymphoid neoplasm diffuse large B-cell lymphoma
- DUB
Deubiquitinating enzyme
- E1
Ubiquitin-activating enzyme
- E2
Ubiquitin-conjugating enzyme
- E3
Ubiquitin ligase
- E6-AP
E6-associated protein
- EAE
Experimental autoimmune encephalomyelitis
- ECT2
Epithelial cell transforming 2
- EGFR
Epidermal growth factor receptor
- ER
Endoplasmic reticulum
- ERBB4
Erb-b2 receptor tyrosine kinase 4
- ERK
Mitogen-activated protein kinase 1
- ESCC
Esophageal squamous cell carcinoma
- FBDD
Fragment-based drug discovery
- FBW7
F-box and WD repeat domain containing 7
- FP-HTS
Fluorescence polarization assay for high-throughput screening
- HECT
Homologous to E6AP C terminus
- HECTD3
Homologous to the E6-associated protein carboxyl terminus domain containing 3
- HER2
Erb-b2 receptor tyrosine kinase 2
- HIF1α
Hypoxia inducible factor 1 subunit alpha
- HSP90
Heat shock protein 90
- HTS
High-throughput screening technologies
- HUWE1
HECT, UBA, and WWE domain containing E3 ubiquitin protein ligase 1
- IFN
Interferon
- IRE1α
Inositol requiring enzyme 1 alpha
- IRF3
Interferon regulatory factor 3
- ITCH
Itchy E3 ubiquitin protein ligase
- KLF5
Kruppel like factor 5
- LATS1
Large tumor suppressor kinase 1
- LIHC
Liver hepatocellular carcinoma
- LGG
Brain lower grade glioma
- LUAD
Lung adenocarcinoma
- MALT1
MALT1 paracaspase
- MCL1
Myeloid cell leukemia 1
- MDM2
Murine double minute 2
- miR-153
MicroRNA-153
- NEDD4-1
NEDD4 E3 ubiquitin protein ligase
- OV
Ovarian serous cystadenocarcinoma
- PMA
Phorbol-12-myristate-13-acetate
- PML
Promyelocytic leukemia protein
- PTEN
Phosphatase and tensin homolog
- RBR
RING-IBR-RINGs
- RING
Really interesting new genes
- RLD
RCC1 like domain
- RNF20
Ring finger protein 20
- RORγt
Retineic-acid-receptor-related orphan nuclear receptor γ
- SCF
SKP1-CUL1-F-box protein
- SKP2
S-phase kinase-associated protein 2
- SMAD2
SMAD family member 2
- SMURF2
SMAD specific E3 ubiquitin protein ligase 2
- Stat3
Signal transducer and activator of transcription 3
- Tara
Trio-associated repeat on actin
- TBK1
TANK binding kinase 1
- TGFβ
Transforming growth factor β
- TGFβR1
Transforming growth factor β receptor 1
- Th17
T helper 17
- THCA
Thyroid carcinoma
- THYM
Thymoma
- TNBC
Triple negative breast cancer
- TRAF3
TNF receptor-associated factor 3
- TRAF6
TNF receptor-associated factor 6
- TRAIL
TNF-related apoptosis-inducing ligand
- Ub
Ubiquitin
- UCEC
Uterine Corpus Endometrial Carcinoma
- UCS
Uterine Carcinosarcoma
- UbV
Ub variant
- UCS
Uterine carcinosarcoma
- VCB-CR
pVHL-elongin C-elongin B-cullin 2-RBX1
- VHL
Von Hippel–Lindau disease tumor suppressor
- WWP1
WW domain containing E3 ubiquitin protein ligase 1
- WWP2
WW domain containing E3 ubiquitin protein ligase 2
- XBP1
X-box binding protein 1
Compliance with ethical standards
Conflict of interest
The authors have no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Pickart CM. Mechanisms underlying ubiquitination. Annu Rev Biochem. 2001;70:503–533. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- 2.Rape M. Ubiquitylation at the crossroads of development and disease. Nat Rev Mol Cell Biol. 2018;19(1):59–70. doi: 10.1038/nrm.2017.83. [DOI] [PubMed] [Google Scholar]
- 3.Komander D, Rape M. The ubiquitin code. Annu Rev Biochem. 2012;81:203–229. doi: 10.1146/annurev-biochem-060310-170328. [DOI] [PubMed] [Google Scholar]
- 4.Tokunaga F, Sakata S-i, Saeki Y, Satomi Y, Kirisako T, Kamei K, Nakagawa T, Kato M, Murata S, Yamaoka S, Yamamoto M, Akira S, Takao T, Tanaka K, Iwai K (2009) Involvement of linear polyubiquitylation of NEMO in NF-κB activation. Nat Cell Biol 11:123. 10.1038/ncb1821. https://www.nature.com/articles/ncb1821#supplementary-information [DOI] [PubMed]
- 5.Trempe JF. Reading the ubiquitin postal code. Curr Opin Struct Biol. 2011;21(6):792–801. doi: 10.1016/j.sbi.2011.09.009. [DOI] [PubMed] [Google Scholar]
- 6.Rotin D, Kumar S. Physiological functions of the HECT family of ubiquitin ligases. Nat Rev Mol Cell Biol. 2009;10(6):398–409. doi: 10.1038/nrm2690. [DOI] [PubMed] [Google Scholar]
- 7.Haglund K, Dikic I. Ubiquitylation and cell signaling. EMBO J. 2005;24(19):3353–3359. doi: 10.1038/sj.emboj.7600808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoeller D, Hecker CM, Dikic I. Ubiquitin and ubiquitin-like proteins in cancer pathogenesis. Nat Rev Cancer. 2006;6(10):776–788. doi: 10.1038/nrc1994. [DOI] [PubMed] [Google Scholar]
- 9.Mukhopadhyay D, Riezman H. Proteasome-independent functions of ubiquitin in endocytosis and signaling. Science (New York, NY) 2007;315(5809):201–205. doi: 10.1126/science.1127085. [DOI] [PubMed] [Google Scholar]
- 10.Huen MS, Sy SM, Chen J. BRCA1 and its toolbox for the maintenance of genome integrity. Nat Rev Mol Cell Biol. 2010;11(2):138–148. doi: 10.1038/nrm2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vucic D, Dixit VM, Wertz IE. Ubiquitylation in apoptosis: a post-translational modification at the edge of life and death. Nat Rev Mol Cell Biol. 2011;12(7):439–452. doi: 10.1038/nrm3143. [DOI] [PubMed] [Google Scholar]
- 12.Gilberto S, Peter M. Dynamic ubiquitin signaling in cell cycle regulation. J Cell Biol. 2017;216(8):2259–2271. doi: 10.1083/jcb.201703170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Senft D, Qi J, Ronai ZA. Ubiquitin ligases in oncogenic transformation and cancer therapy. Nat Rev Cancer. 2018;18(2):69–88. doi: 10.1038/nrc.2017.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Popovic D, Vucic D, Dikic I. Ubiquitination in disease pathogenesis and treatment. Nat Med. 2014;20(11):1242–1253. doi: 10.1038/nm.3739. [DOI] [PubMed] [Google Scholar]
- 15.He M, Zhou Z, Wu G, Chen Q, Wan Y. Emerging role of DUBs in tumor metastasis and apoptosis: therapeutic implication. Pharmacol Ther. 2017;177:96–107. doi: 10.1016/j.pharmthera.2017.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harrigan JA, Jacq X, Martin NM, Jackson SP. Deubiquitylating enzymes and drug discovery: emerging opportunities. Nat Rev Drug Discov. 2018;17(1):57–78. doi: 10.1038/nrd.2017.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chan CH, Li CF, Yang WL, Gao Y, Lee SW, Feng Z, Huang HY, Tsai KK, Flores LG, Shao Y, Hazle JD, Yu D, Wei W, Sarbassov D, Hung MC, Nakayama KI, Lin HK. The Skp2-SCF E3 ligase regulates Akt ubiquitination, glycolysis, herceptin sensitivity, and tumorigenesis. Cell. 2012;149(5):1098–1111. doi: 10.1016/j.cell.2012.02.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dubrez L, Rajalingam K. IAPs and cell migration. Semin Cell Dev Biol. 2015;39:124–131. doi: 10.1016/j.semcdb.2015.02.015. [DOI] [PubMed] [Google Scholar]
- 19.Kim H, Frederick DT, Levesque MP, Cooper ZA, Feng Y, Krepler C, Brill L, Samuels Y, Hayward NK, Perlina A, Piris A, Zhang T, Halaban R, Herlyn MM, Brown KM, Wargo JA, Dummer R, Flaherty KT, Ronai ZA. Downregulation of the ubiquitin ligase RNF125 underlies resistance of melanoma cells to BRAF inhibitors via JAK1 deregulation. Cell Rep. 2015;11(9):1458–1473. doi: 10.1016/j.celrep.2015.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Randle SJ, Laman H. F-box protein interactions with the hallmark pathways in cancer. Semin Cancer Biol. 2016;36:3–17. doi: 10.1016/j.semcancer.2015.09.013. [DOI] [PubMed] [Google Scholar]
- 21.Yang L, Chen J, Huang X, Zhang E, He J, Cai Z. Novel insights Into E3 ubiquitin ligase in cancer chemoresistance. Am J Med Sci. 2018;355(4):368–376. doi: 10.1016/j.amjms.2017.12.012. [DOI] [PubMed] [Google Scholar]
- 22.Buetow L, Huang DT. Structural insights into the catalysis and regulation of E3 ubiquitin ligases. Nat Rev Mol Cell Biol. 2016;17(10):626–642. doi: 10.1038/nrm.2016.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wade M, Li Y-C, Wahl GM. MDM2, MDMX and p53 in oncogenesis and cancer therapy. Nat Rev Cancer. 2013;13(2):83–96. doi: 10.1038/nrc3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frescas D, Pagano M. Deregulated proteolysis by the F-box proteins SKP2 and beta-TrCP: tipping the scales of cancer. Nat Rev Cancer. 2008;8(6):438–449. doi: 10.1038/nrc2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu W, Taranets L, Popov N. Regulating Fbw7 on the road to cancer. Semin Cancer Biol. 2016;36:62–70. doi: 10.1016/j.semcancer.2015.09.005. [DOI] [PubMed] [Google Scholar]
- 26.Gossage L, Eisen T, Maher ER. VHL, the story of a tumour suppressor gene. Nat Rev Cancer. 2015;15(1):55–64. doi: 10.1038/nrc3844. [DOI] [PubMed] [Google Scholar]
- 27.Li ML, Greenberg RA. Links between genome integrity and BRCA1 tumor suppression. Trends Biochem Sci. 2012;37(10):418–424. doi: 10.1016/j.tibs.2012.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang S, Sun W, Zhao Y, McEachern D, Meaux I, Barrière C, Stuckey JA, Meagher JL, Bai L, Liu L, Hoffman-Luca CG, Lu J, Shangary S, Yu S, Bernard D, Aguilar A, Dos-Santos O, Besret L, Guerif S, Pannier P, Gorge-Bernat D, Debussche L. SAR405838: an optimized inhibitor of MDM2-p53 interaction that induces complete and durable tumor regression. Can Res. 2014;74(20):5855–5865. doi: 10.1158/0008-5472.Can-14-0799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aguilar A, Lu J, Liu L, Du D, Bernard D, McEachern D, Przybranowski S, Li X, Luo R, Wen B, Sun D, Wang H, Wen J, Wang G, Zhai Y, Guo M, Yang D, Wang S (2017) Discovery of 4-((3′R,4′S,5′R)-6″-Chloro-4′-(3-chloro-2-fluorophenyl)-1′-ethyl-2″-oxodispiro[cyclohexane-1,2′-pyrrolidine-3′,3″-indoline]-5′-carboxamido)bicyclo[2.2.2]octane-1-carboxylic Acid (AA-115/APG-115): a potent and orally active murine double minute 2 (MDM2) inhibitor in clinical development. J Med Chem 60 (7):2819–2839. 10.1021/acs.jmedchem.6b01665 [DOI] [PMC free article] [PubMed]
- 30.So WV, Ou Yang T-H, Yang X, Zhi J. Lack of UGT polymorphism association with idasanutlin pharmacokinetics in solid tumor patients. Cancer Chemother Pharmacol. 2019;83(1):209–213. doi: 10.1007/s00280-018-3741-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sluimer J, Distel B. Regulating the human HECT E3 ligases. Cell Mol Life Sci. 2018;75(17):3121–3141. doi: 10.1007/s00018-018-2848-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zheng N, Shabek N. Ubiquitin ligases: structure, function, and regulation. Annu Rev Biochem. 2017;86:129–157. doi: 10.1146/annurev-biochem-060815-014922. [DOI] [PubMed] [Google Scholar]
- 33.Wenzel DM, Lissounov A, Brzovic PS, Klevit RE. UBCH7 reactivity profile reveals parkin and HHARI to be RING/HECT hybrids. Nature. 2011;474(7349):105–108. doi: 10.1038/nature09966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lorenz S. Structural mechanisms of HECT-type ubiquitin ligases. Biol Chem. 2018;399(2):127–145. doi: 10.1515/hsz-2017-0184. [DOI] [PubMed] [Google Scholar]
- 35.Scheffner M, Nuber U, Huibregtse JM. Protein ubiquitination involving an E1–E2–E3 enzyme ubiquitin thioester cascade. Nature. 1995;373(6509):81–83. doi: 10.1038/373081a0. [DOI] [PubMed] [Google Scholar]
- 36.Huang L, Kinnucan E, Wang G, Beaudenon S, Howley PM, Huibregtse JM, Pavletich NP. Structure of an E6AP-UbcH7 complex: insights into ubiquitination by the E2-E3 enzyme cascade. Science (New York, NY) 1999;286(5443):1321–1326. doi: 10.1126/science.286.5443.1321. [DOI] [PubMed] [Google Scholar]
- 37.Huibregtse JM, Scheffner M, Beaudenon S, Howley PM. A family of proteins structurally and functionally related to the E6-AP ubiquitin-protein ligase. Proc Natl Acad Sci USA. 1995;92(11):5249. doi: 10.1073/pnas.92.11.5249-a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Scheffner M, Huibregtse JM, Vierstra RD, Howley PM. The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell. 1993;75(3):495–505. doi: 10.1016/0092-8674(93)90384-3. [DOI] [PubMed] [Google Scholar]
- 39.Louria-Hayon I, Alsheich-Bartok O, Levav-Cohen Y, Silberman I, Berger M, Grossman T, Matentzoglu K, Jiang YH, Muller S, Scheffner M, Haupt S, Haupt Y. E6AP promotes the degradation of the PML tumor suppressor. Cell Death Differ. 2009;16(8):1156–1166. doi: 10.1038/cdd.2009.31. [DOI] [PubMed] [Google Scholar]
- 40.Wolyniec K, Shortt J, de Stanchina E, Levav-Cohen Y, Alsheich-Bartok O, Louria-Hayon I, Corneille V, Kumar B, Woods SJ, Opat S, Johnstone RW, Scott CL, Segal D, Pandolfi PP, Fox S, Strasser A, Jiang YH, Lowe SW, Haupt S, Haupt Y. E6AP ubiquitin ligase regulates PML-induced senescence in Myc-driven lymphomagenesis. Blood. 2012;120(4):822–832. doi: 10.1182/blood-2011-10-387647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Paul PJ, Raghu D, Chan AL, Gulati T, Lambeth L, Takano E, Herold MJ, Hagekyriakou J, Vessella RL, Fedele C, Shackleton M, Williams ED, Fox S, Williams S, Haupt S, Gamell C, Haupt Y. Restoration of tumor suppression in prostate cancer by targeting the E3 ligase E6AP. Oncogene. 2016;35(48):6235–6245. doi: 10.1038/onc.2016.159. [DOI] [PubMed] [Google Scholar]
- 42.Mansour M, Haupt S, Chan AL, Godde N, Rizzitelli A, Loi S, Caramia F, Deb S, Takano EA, Bishton M, Johnstone C, Monahan B, Levav-Cohen Y, Jiang YH, Yap AS, Fox S, Bernard O, Anderson R, Haupt Y. The E3-ligase E6AP represses breast cancer metastasis via regulation of ECT2-Rho signaling. Can Res. 2016;76(14):4236–4248. doi: 10.1158/0008-5472.Can-15-1553. [DOI] [PubMed] [Google Scholar]
- 43.Chen C, Sun X, Guo P, Dong XY, Sethi P, Zhou W, Zhou Z, Petros J, Frierson HF, Vessella RL, Atfi A, Dong JT. Ubiquitin E3 ligase WWP1 as an oncogenic factor in human prostate cancer. Oncogene. 2007;26(16):2386–2394. doi: 10.1038/sj.onc.1210021. [DOI] [PubMed] [Google Scholar]
- 44.Chen C, Zhou Z, Ross JS, Zhou W, Dong JT. The amplified WWP1 gene is a potential molecular target in breast cancer. Int J Cancer. 2007;121(1):80–87. doi: 10.1002/ijc.22653. [DOI] [PubMed] [Google Scholar]
- 45.Li Y, Zhou Z, Chen C. WW domain-containing E3 ubiquitin protein ligase 1 targets p63 transcription factor for ubiquitin-mediated proteasomal degradation and regulates apoptosis. Cell Death Differ. 2008;15(12):1941–1951. doi: 10.1038/cdd.2008.134. [DOI] [PubMed] [Google Scholar]
- 46.Chen C, Sun X, Guo P, Dong XY, Sethi P, Cheng X, Zhou J, Ling J, Simons JW, Lingrel JB, Dong JT. Human Kruppel-like factor 5 is a target of the E3 ubiquitin ligase WWP1 for proteolysis in epithelial cells. J Biol Chem. 2005;280(50):41553–41561. doi: 10.1074/jbc.M506183200. [DOI] [PubMed] [Google Scholar]
- 47.Komuro A, Imamura T, Saitoh M, Yoshida Y, Yamori T, Miyazono K, Miyazawa K. Negative regulation of transforming growth factor-beta (TGF-beta) signaling by WW domain-containing protein 1 (WWP1) Oncogene. 2004;23(41):6914–6923. doi: 10.1038/sj.onc.1207885. [DOI] [PubMed] [Google Scholar]
- 48.Li Y, Zhou Z, Alimandi M, Chen C. WW domain containing E3 ubiquitin protein ligase 1 targets the full-length ErbB4 for ubiquitin-mediated degradation in breast cancer. Oncogene. 2009;28(33):2948–2958. doi: 10.1038/onc.2009.162. [DOI] [PubMed] [Google Scholar]
- 49.Salah Z, Melino G, Aqeilan RI. Negative regulation of the Hippo pathway by E3 ubiquitin ligase ITCH is sufficient to promote tumorigenicity. Can Res. 2011;71(5):2010–2020. doi: 10.1158/0008-5472.Can-10-3516. [DOI] [PubMed] [Google Scholar]
- 50.Chang L, Shen L, Zhou H, Gao J, Pan H, Zheng L, Armstrong B, Peng Y, Peng G, Zhou BP, Rosen ST, Shen B (2019) ITCH nuclear translocation and H1.2 polyubiquitination negatively regulate the DNA damage response. Nucleic Acids Res 47(2):824–842. 10.1093/nar/gky1199 [DOI] [PMC free article] [PubMed]
- 51.Lin X, Liang M, Feng XH. Smurf2 is a ubiquitin E3 ligase mediating proteasome-dependent degradation of Smad2 in transforming growth factor-beta signaling. J Biol Chem. 2000;275(47):36818–36822. doi: 10.1074/jbc.C000580200. [DOI] [PubMed] [Google Scholar]
- 52.Tang LY, Yamashita M, Coussens NP, Tang Y, Wang XC, Li CL, Deng CX, Cheng SY, Zhang YE. Ablation of Smurf2 reveals an inhibition in TGF-beta signalling through multiple mono-ubiquitination of Smad3. EMBO J. 2011;30(23):4777–4789. doi: 10.1038/emboj.2011.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Blank M, Tang Y, Yamashita M, Burkett SS, Cheng SY, Zhang YE. A tumor suppressor function of Smurf2 associated with controlling chromatin landscape and genome stability through RNF20. Nat Med. 2012;18(2):227–234. doi: 10.1038/nm.2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang X, Trotman LC, Koppie T, Alimonti A, Chen Z, Gao Z, Wang J, Erdjument-Bromage H, Tempst P, Cordon-Cardo C, Pandolfi PP, Jiang X. NEDD4-1 is a proto-oncogenic ubiquitin ligase for PTEN. Cell. 2007;128(1):129–139. doi: 10.1016/j.cell.2006.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Maddika S, Kavela S, Rani N, Palicharla VR, Pokorny JL, Sarkaria JN, Chen J. WWP2 is an E3 ubiquitin ligase for PTEN. Nat Cell Biol. 2011;13(6):728–733. doi: 10.1038/ncb2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yu J, Lan J, Zhu Y, Li X, Lai X, Xue Y, Jin C, Huang H. The E3 ubiquitin ligase HECTD3 regulates ubiquitination and degradation of Tara. Biochem Biophys Res Commun. 2008;367(4):805–812. doi: 10.1016/j.bbrc.2008.01.022. [DOI] [PubMed] [Google Scholar]
- 57.Zhang L, Kang L, Bond W, Zhang N. Interaction between syntaxin 8 and HECTd3, a HECT domain ligase. Cell Mol Neurobiol. 2009;29(1):115–121. doi: 10.1007/s10571-008-9303-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li Y, Chen X, Wang Z, Zhao D, Chen H, Chen W, Zhou Z, Zhang J, Zhang J, Li H, Chen C (2013) The HECTD3 E3 ubiquitin ligase suppresses cisplatin-induced apoptosis via stabilizing MALT1. Neoplasia 15(1):39-IN15. 10.1593/neo.121362 [DOI] [PMC free article] [PubMed]
- 59.Cho JJ, Xu Z, Parthasarathy U, Drashansky TT, Helm EY, Zuniga AN, Lorentsen KJ, Mansouri S, Cho JY, Edelmann MJ, Duong DM, Gehring T, Seeholzer T, Krappmann D, Uddin MN, Califano D, Wang RL, Jin L, Li H, Lv D, Zhou D, Zhou L, Avram D. Hectd3 promotes pathogenic Th17 lineage through Stat3 activation and Malt1 signaling in neuroinflammation. Nat Commun. 2019;10(1):701. doi: 10.1038/s41467-019-08605-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li Y, Kong Y, Zhou Z, Chen H, Wang Z, Hsieh YC, Zhao D, Zhi X, Huang J, Zhang J, Li H, Chen C. The HECTD3 E3 ubiquitin ligase facilitates cancer cell survival by promoting K63-linked polyubiquitination of caspase-8. Cell Death Dis. 2013;4:e935. doi: 10.1038/cddis.2013.464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li Y, Wu X, Li L, Liu Y, Xu C, Su D, Liu Z. The E3 ligase HECTD3 promotes esophageal squamous cell carcinoma (ESCC) growth and cell survival through targeting and inhibiting caspase-9 activation. Cancer Lett. 2017;404:44–52. doi: 10.1016/j.canlet.2017.07.004. [DOI] [PubMed] [Google Scholar]
- 62.Li Z, Zhou L, Prodromou C, Savic V, Pearl LH. HECTD3 mediates an HSP90-dependent degradation pathway for protein kinase clients. Cell Rep. 2017;19(12):2515–2528. doi: 10.1016/j.celrep.2017.05.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li F, Li Y, Liang H, Xu T, Kong Y, Huang M, Xiao J, Chen X, Xia H, Wu Y, Zhou Z, Guo X, Hu C, Yang C, Cheng X, Chen C, Qi X. HECTD3 mediates TRAF3 polyubiquitination and type I interferon induction during bacterial infection. J Clin Invest. 2018;128(9):4148–4162. doi: 10.1172/JCI120406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Oeckinghaus A, Wegener E, Welteke V, Ferch U, Arslan SC, Ruland J, Scheidereit C, Krappmann D. Malt1 ubiquitination triggers NF-kappaB signaling upon T-cell activation. EMBO J. 2007;26(22):4634–4645. doi: 10.1038/sj.emboj.7601897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kong Y, Wang Z, Huang M, Zhou Z, Li Y, Miao H, Wan X, Huang J, Mao X, Chen C. CUL7 promotes cancer cell survival through promoting Caspase-8 ubiquitination. Int J Cancer. 2019 doi: 10.1002/ijc.32239. [DOI] [PubMed] [Google Scholar]
- 66.Shu T, Li Y, Wu X, Li B, Liu Z. Down-regulation of HECTD3 by HER2 inhibition makes serous ovarian cancer cells sensitive to platinum treatment. Cancer Lett. 2017;411:65–73. doi: 10.1016/j.canlet.2017.09.048. [DOI] [PubMed] [Google Scholar]
- 67.Yu H, Jove R. The STATs of cancer—new molecular targets come of age. Nat Rev Cancer. 2004;4(2):97–105. doi: 10.1038/nrc1275. [DOI] [PubMed] [Google Scholar]
- 68.Wu X, Li L, Li Y, Liu Z. MiR-153 promotes breast cancer cell apoptosis by targeting HECTD3. Am J Cancer Res. 2016;6(7):1563–1571. [PMC free article] [PubMed] [Google Scholar]
- 69.Liu R, Shi P, Nie Z, Liang H, Zhou Z, Chen W, Chen H, Dong C, Yang R, Liu S, Chen C. Mifepristone suppresses basal triple-negative breast cancer stem cells by down-regulating KLF5 expression. Theranostics. 2016;6(4):533–544. doi: 10.7150/thno.14315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liang H, Xiao J, Zhou Z, Wu J, Ge F, Li Z, Zhang H, Sun J, Li F, Liu R, Chen C. Hypoxia induces miR-153 through the IRE1alpha-XBP1 pathway to fine tune the HIF1alpha/VEGFA axis in breast cancer angiogenesis. Oncogene. 2018;37(15):1961–1975. doi: 10.1038/s41388-017-0089-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liang H, Ge F, Xu Y, Xiao J, Zhou Z, Liu R, Chen C. miR-153 inhibits the migration and the tube formation of endothelial cells by blocking the paracrine of angiopoietin 1 in breast cancer cells. Angiogenesis. 2018;21(4):849–860. doi: 10.1007/s10456-018-9630-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ekambaram P, Lee JL, Hubel NE, Hu D, Yerneni S, Campbell PG, Pollock N, Klei LR, Concel VJ, Delekta PC, Chinnaiyan AM, Tomlins SA, Rhodes DR, Priedigkeit N, Lee AV, Oesterreich S, McAllister-Lucas LM, Lucas PC. The CARMA3-Bcl10-MALT1 signalosome drives NFκB activation and promotes aggressiveness in angiotensin II receptor-positive breast cancer. Can Res. 2018;78(5):1225–1240. doi: 10.1158/0008-5472.Can-17-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cheng L, Deng N, Yang N, Zhao X, Lin X (2019) Malt1 protease is critical in maintaining function of regulatory T cells and may be a therapeutic target for antitumor immunity. J Immunol (Baltimore, Md: 1950) 202(10):3008–3019. 10.4049/jimmunol.1801614 [DOI] [PubMed]
- 74.Kawadler H, Gantz MA, Riley JL, Yang X. The paracaspase MALT1 controls caspase-8 activation during lymphocyte proliferation. Mol Cell. 2008;31(3):415–421. doi: 10.1016/j.molcel.2008.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Brüstle A, Brenner D, Knobbe CB, Lang PA, Virtanen C, Hershenfield BM, Reardon C, Lacher SM, Ruland J, Ohashi PS, Mak TW. The NF-κB regulator MALT1 determines the encephalitogenic potential of Th17 cells. J Clin Invest. 2012;122(12):4698–4709. doi: 10.1172/jci63528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Paramore A, Frantz S. Bortezomib. Nat Rev Drug Discov. 2003;2(8):611–612. doi: 10.1038/nrd1159. [DOI] [PubMed] [Google Scholar]
- 77.Adams J. The development of proteasome inhibitors as anticancer drugs. Cancer Cell. 2004;5(5):417–421. doi: 10.1016/S1535-6108(04)00120-5. [DOI] [PubMed] [Google Scholar]
- 78.Qi J, Ronai ZA. Dysregulation of ubiquitin ligases in cancer. Drug Resist Updat. 2015;23:1–11. doi: 10.1016/j.drup.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Issaeva N, Bozko P, Enge M, Protopopova M, Verhoef LGGC, Masucci M, Pramanik A, Selivanova G. Small molecule RITA binds to p53, blocks p53–HDM-2 interaction and activates p53 function in tumors. Nat Med. 2004;10(12):1321–1328. doi: 10.1038/nm1146. [DOI] [PubMed] [Google Scholar]
- 80.Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z, Kong N, Kammlott U, Lukacs C, Klein C, Fotouhi N, Liu EA. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science (New York, NY) 2004;303(5659):844–848. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- 81.Yang Y, Ludwig RL, Jensen JP, Pierre SA, Medaglia MV, Davydov IV, Safiran YJ, Oberoi P, Kenten JH, Phillips AC, Weissman AM, Vousden KH. Small molecule inhibitors of HDM2 ubiquitin ligase activity stabilize and activate p53 in cells. Cancer Cell. 2005;7(6):547–559. doi: 10.1016/j.ccr.2005.04.029. [DOI] [PubMed] [Google Scholar]
- 82.Ray-Coquard I, Blay JY, Italiano A, Le Cesne A, Penel N, Zhi J, Heil F, Rueger R, Graves B, Ding M, Geho D, Middleton SA, Vassilev LT, Nichols GL, Bui BN. Effect of the MDM2 antagonist RG7112 on the P53 pathway in patients with MDM2-amplified, well-differentiated or dedifferentiated liposarcoma: an exploratory proof-of-mechanism study. Lancet Oncol. 2012;13(11):1133–1140. doi: 10.1016/s1470-2045(12)70474-6. [DOI] [PubMed] [Google Scholar]
- 83.Shangary S, Qin D, McEachern D, Liu M, Miller RS, Qiu S, Nikolovska-Coleska Z, Ding K, Wang G, Chen J, Bernard D, Zhang J, Lu Y, Gu Q, Shah RB, Pienta KJ, Ling X, Kang S, Guo M, Sun Y, Yang D, Wang S. Temporal activation of p53 by a specific MDM2 inhibitor is selectively toxic to tumors and leads to complete tumor growth inhibition. Proc Natl Acad Sci USA. 2008;105(10):3933–3938. doi: 10.1073/pnas.0708917105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Azmi AS, Aboukameel A, Banerjee S, Wang Z, Mohammad M, Wu J, Wang S, Yang D, Philip PA, Sarkar FH, Mohammad RM (2010) MDM2 inhibitor MI-319 in combination with cisplatin is an effective treatment for pancreatic cancer independent of p53 function. Eur J Cancer (Oxford, England: 1990) 46(6):1122–1131. 10.1016/j.ejca.2010.01.015 [DOI] [PMC free article] [PubMed]
- 85.Zhao Y, Liu L, Sun W, Lu J, McEachern D, Li X, Yu S, Bernard D, Ochsenbein P, Ferey V, Carry JC, Deschamps JR, Sun D, Wang S. Diastereomeric spirooxindoles as highly potent and efficacious MDM2 inhibitors. J Am Chem Soc. 2013;135(19):7223–7234. doi: 10.1021/ja3125417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhao Y, Yu S, Sun W, Liu L, Lu J, McEachern D, Shargary S, Bernard D, Li X, Zhao T, Zou P, Sun D, Wang S. A potent small-molecule inhibitor of the MDM2-p53 interaction (MI-888) achieved complete and durable tumor regression in mice. J Med Chem. 2013;56(13):5553–5561. doi: 10.1021/jm4005708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lu J, McEachern D, Li S, Ellis MJ, Wang S. Reactivation of p53 by MDM2 Inhibitor MI-77301 for the treatment of endocrine-resistant breast cancer. Mol Cancer Ther. 2016;15(12):2887–2893. doi: 10.1158/1535-7163.MCT-16-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Nör F, Warner KA, Zhang Z, Acasigua GA, Pearson AT, Kerk SA, Helman JI, Sant’Ana Filho M, Wang S, Nör JE. Therapeutic Inhibition of the MDM2-p53 Interaction Prevents Recurrence of Adenoid Cystic Carcinomas. Clin Cancer Res. 2017;23(4):1036–1048. doi: 10.1158/1078-0432.Ccr-16-1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Andrews A, Warner K, Rodriguez-Ramirez C, Pearson AT, Nör F, Zhang Z, Kerk S, Kulkarni A, Helman JI, Brenner JC, Wicha MS, Wang S, Nör JE. Ablation of cancer stem cells by therapeutic inhibition of the MDM2-p53 interaction in mucoepidermoid carcinoma. Clin Cancer Res. 2019;25(5):1588–1600. doi: 10.1158/1078-0432.Ccr-17-2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ding Q, Zhang Z, Liu JJ, Jiang N, Zhang J, Ross TM, Chu XJ, Bartkovitz D, Podlaski F, Janson C, Tovar C, Filipovic ZM, Higgins B, Glenn K, Packman K, Vassilev LT, Graves B. Discovery of RG7388, a potent and selective p53-MDM2 inhibitor in clinical development. J Med Chem. 2013;56(14):5979–5983. doi: 10.1021/jm400487c. [DOI] [PubMed] [Google Scholar]
- 91.Zhang W, Wu KP, Sartori MA, Kamadurai HB, Ordureau A, Jiang C, Mercredi PY, Murchie R, Hu J, Persaud A, Mukherjee M, Li N, Doye A, Walker JR, Sheng Y, Hao Z, Li Y, Brown KR, Lemichez E, Chen J, Tong Y, Harper JW, Moffat J, Rotin D, Schulman BA, Sidhu SS. System-wide modulation of HECT E3 ligases with selective ubiquitin variant probes. Mol Cell. 2016;62(1):121–136. doi: 10.1016/j.molcel.2016.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gorelik M, Orlicky S, Sartori MA, Tang X, Marcon E, Kurinov I, Greenblatt JF, Tyers M, Moffat J, Sicheri F, Sidhu SS. Inhibition of SCF ubiquitin ligases by engineered ubiquitin variants that target the Cul1 binding site on the Skp1–F-box interface. Proc Natl Acad Sci. 2016;113(13):3527. doi: 10.1073/pnas.1519389113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gabrielsen M, Buetow L, Nakasone MA, Ahmed SF, Sibbet GJ, Smith BO, Zhang W, Sidhu SS, Huang DT. A general strategy for discovery of inhibitors and activators of RING and U-box E3 ligases with ubiquitin variants. Mol Cell. 2017;68(2):456.e410–470.e410. doi: 10.1016/j.molcel.2017.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Mund T, Lewis MJ, Maslen S, Pelham HR. Peptide and small molecule inhibitors of HECT-type ubiquitin ligases. Proc Natl Acad Sci USA. 2014;111(47):16736–16741. doi: 10.1073/pnas.1412152111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rossi M, Rotblat B, Ansell K, Amelio I, Caraglia M, Misso G, Bernassola F, Cavasotto CN, Knight RA, Ciechanover A, Melino G. High throughput screening for inhibitors of the HECT ubiquitin E3 ligase ITCH identifies antidepressant drugs as regulators of autophagy. Cell Death Dis. 2014;5(5):e1203. doi: 10.1038/cddis.2014.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kathman SG, Span I, Smith AT, Xu Z, Zhan J, Rosenzweig AC, Statsyuk AV. A small molecule that switches a ubiquitin ligase from a processive to a distributive enzymatic mechanism. J Am Chem Soc. 2015;137(39):12442–12445. doi: 10.1021/jacs.5b06839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sander B, Xu W, Eilers M, Popov N, Lorenz S (2017) A conformational switch regulates the ubiquitin ligase HUWE1. eLife 6. 10.7554/elife.21036 [DOI] [PMC free article] [PubMed]
- 98.Chen Z, Jiang H, Xu W, Li X, Dempsey DR, Zhang X, Devreotes P, Wolberger C, Amzel LM, Gabelli SB, Cole PA. A tunable brake for HECT ubiquitin ligases. Mol Cell. 2017;66(3):345.e346–357.e346. doi: 10.1016/j.molcel.2017.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chan AL, Grossman T, Zuckerman V, Campigli Di Giammartino D, Moshel O, Scheffner M, Monahan B, Pilling P, Jiang YH, Haupt S, Schueler-Furman O, Haupt Y. c-Abl phosphorylates E6AP and regulates its E3 ubiquitin ligase activity. Biochemistry. 2013;52(18):3119–3129. doi: 10.1021/bi301710c. [DOI] [PubMed] [Google Scholar]
- 100.Hamzeh-Mivehroud M, Alizadeh AA, Morris MB, Church WB, Dastmalchi S. Phage display as a technology delivering on the promise of peptide drug discovery. Drug Discov Today. 2013;18(23–24):1144–1157. doi: 10.1016/j.drudis.2013.09.001. [DOI] [PubMed] [Google Scholar]
- 101.Ungermannova D, Lee J, Zhang G, Dallmann HG, McHenry CS, Liu X. High-throughput screening alphascreen assay for identification of small-molecule inhibitors of ubiquitin E3 Ligase SCFSkp2-Cks1. J Biomol Screen. 2013;18(8):910–920. doi: 10.1177/1087057113485789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Erlanson DA, Fesik SW, Hubbard RE, Jahnke W, Jhoti H. Twenty years on: the impact of fragments on drug discovery. Nat Rev Drug Discov. 2016;15(9):605–619. doi: 10.1038/nrd.2016.109. [DOI] [PubMed] [Google Scholar]
- 103.Gu L, Zhang H, Liu T, Zhou S, Du Y, Xiong J, Yi S, Qu CK, Fu H, Zhou M. Discovery of dual inhibitors of MDM2 and XIAP for cancer treatment. Cancer Cell. 2016;30(4):623–636. doi: 10.1016/j.ccell.2016.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Krist DT, Park S, Boneh GH, Rice SE, Statsyuk AV. UbFluor: a mechanism-based probe for HECT E3 ligases. Chem Sci. 2016;7(8):5587–5595. doi: 10.1039/C6SC01167E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Veggiani G, Gerpe MCR, Sidhu SS, Zhang W (2019) Emerging drug development technologies targeting ubiquitination for cancer therapeutics. Pharmacol Ther [DOI] [PMC free article] [PubMed]
- 106.Franzini RM, Neri D, Scheuermann J. DNA-encoded chemical libraries: advancing beyond conventional small-molecule libraries. Acc Chem Res. 2014;47(4):1247–1255. doi: 10.1021/ar400284t. [DOI] [PubMed] [Google Scholar]
- 107.Rognan D. The impact of in silico screening in the discovery of novel and safer drug candidates. Pharmacol Ther. 2017;175:47–66. doi: 10.1016/j.pharmthera.2017.02.034. [DOI] [PubMed] [Google Scholar]
- 108.Li Y, Xie P, Lu L, Wang J, Diao L, Liu Z, Guo F, He Y, Liu Y, Huang Q, Liang H, Li D, He F. An integrated bioinformatics platform for investigating the human E3 ubiquitin ligase-substrate interaction network. Nat Commun. 2017;8(1):347. doi: 10.1038/s41467-017-00299-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Herman AG, Hayano M, Poyurovsky MV, Shimada K, Skouta R, Prives C, Stockwell BR. Discovery of Mdm2-MdmX E3 ligase inhibitors using a cell-based ubiquitination assay. Cancer Discov. 2011;1(4):312–325. doi: 10.1158/2159-8290.Cd-11-0104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Tian M, Zeng T, Liu M, Han S, Lin H, Lin Q, Li L, Jiang T, Li G, Lin H, Zhang T, Kang Q, Deng X, Wang H-R. A cell-based high-throughput screening method based on a ubiquitin-reference technique for identifying modulators of E3 ligases. J Biol Chem. 2019;294(8):2880–2891. doi: 10.1074/jbc.ra118.003822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zhou H, Di Palma S, Preisinger C, Peng M, Polat AN, Heck AJ, Mohammed S. Toward a comprehensive characterization of a human cancer cell phosphoproteome. J Proteome Res. 2013;12(1):260–271. doi: 10.1021/pr300630k. [DOI] [PubMed] [Google Scholar]