Abstract
High-carbohydrate diets (HCD) can induce the occurrence of nonalcoholic fatty liver disease (NAFLD), characterized by dramatic accumulation of hepatic lipid droplets (LDs). However, the potential molecular mechanisms are still largely unknown. In this study, we investigated the role of autophagy in the process of HCD-induced changes of hepatic lipid metabolism, and to examine the process of underlying mechanisms during these molecular contexts. We found that HCD significantly increased hepatic lipid accumulation and activated autophagy. Using primary hepatocytes, we found that HG increased lipid accumulation and stimulated the release of NEFA by autophagy-mediated lipophagy, and that lipophagy significantly alleviated high glucose (HG)-induced lipid accumulation. Oxidative and endoplasmic reticulum (ER) stress pathways played crucial regulatory roles in HG-induced lipophagy activation and HG-induced changes of lipid metabolism. Further investigation found that HG-activated lipophagy and HG-induced changes of lipid metabolism were via enhancing carbohydrate response element-binding protein (ChREBP) DNA binding capacity at PPARγ promoter region, which in turn induced transcriptional activation of the key genes related to lipogenesis and autophagy. The present study, for the first time, revealed the novel mechanism for lipophagy mediating HCD-induced changes of lipid metabolism by oxidative stress and ER stress, and ChREBP/PPARγ pathways. Our study provided innovative evidence for the direct relationship between carbohydrate and lipid metabolism via ChREBP/PPARγ pathway.
Electronic supplementary material
The online version of this article (10.1007/s00018-019-03263-6) contains supplementary material, which is available to authorized users.
Keywords: Dietary carbohydrate, Lipid deposition, Lipid metabolism, Regulatory pathways, Lipophagy
Introduction
Non-alcoholic fatty liver disease (NAFLD), characterized by the excessive triglyceride (TG) accumulation in the livers, has been increasing in recent years [1]. The spectrum of NAFLD ranges from simple fatty liver to nonalcoholic steatohepatitis (NASH), which may result in liver fibrosis and eventually the development of hepatocellular carcinoma [2]. At present, treatment approaches are limited because of the unclear pathological mechanism of NAFLD. It is well known that the diet rich in carbohydrate along with other factors can induce hepatic NAFLD and NASH [3, 4]. However, the additional mechanisms of dietary carbohydrate leading to the NAFLD are not fully elucidated.
Autophagy is a highly conserved self-renewal process in eukaryotic cells, characterized by the engulfment of cytoplasmic materials into double-membrane vesicles (autophagosomes) for subsequent degradation in lysosomes [5]. Studies suggested that autophagy regulates lipid metabolism by eliminating TG and its activation plays a key inhibitory role in the development of NAFLD [6]. Moreover, lipophagy, one kind of autophagy, regulates lipid metabolism by breaking lipid droplets down and enhancing the rate of mitochondrial β-oxidation [6–8]. Impaired lipophagy will increase cellular lipid storage and result in the occurrence of NAFLD [9, 10].
Several signaling pathways were reported to regulate the autophagic process, such as oxidative stress, endoplasmic reticulum (ER) stress and PPARγ pathways. Oxidative stress, which results from an imbalance between free radical production and scavenging, induces the formation of autophagy [11]. Oxidative stress also accelerates lipid accumulation by disrupting mitochondrial functions and reducing oxidation of fatty acids [12–14]. The ER is the intracellular important organelle for the synthesis and folding of proteins. ER stress will occur if ER homeostasis is disrupted and unfolded/unprocessed proteins are accumulated within the ER. Increasing evidences demonstrated that ER stress might be associated with autophagy [15, 16], and involved in the development of NAFLD [17, 18]. The PPARγ is the important transcriptional factor which regulates autophagic process and lipid metabolism [19, 20].
Thus, in view of the important role of autophagy in regulating lipid metabolism, and the relevant signaling pathways can activate autophagy, we hypothesis that these molecular events mediated high-carbohydrate diets-induced changes of lipid accumulation in the liver. If our hypothesis can be confirmed, our results will provide insight into key pathological mechanisms contributing towards fatty liver occurrence.
Fish are the largest group of vertebrates in the world. During the evolution, fish were considered to experience the fish-specific genome duplication event (FSGD) [21]. By analyzing whole-genome sequence information, Gong et al. [22] found the FSGD in yellow catfish Pelteobagrus fulvidraco, an important freshwater omnivorous fish in China and other countries [23]. Some duplicated genes evolve new functions that in turn result in novel regulatory mechanism [21]. Therefore, using yellow catfish as a model, we hope to find some novel regulatory mechanism of metabolism. Moreover, previous study in our laboratory pointed that high dietary carbohydrate increased liver lipid deposition and developed fatty liver symptom in juvenile yellow catfish [23]. Accordingly, the present study investigated the mechanism of dietary carbohydrate influencing lipid metabolism and dissected the roles of lipophagy mediating carbohydrate-induced changes of lipid metabolism via oxidative stress, ER stress and ChREBP/PPARγ pathways.
Materials and methods
Animals feeding and sampling
The protocols of all animal and cells experiments were approved by the ethical guidelines of Huazhong Agricultural University for the care and use of laboratory animals. The experimental protocols were similar to those in our recent study [24], as described in Yang et al. [25]. Briefly, 270 uniformly sized yellow catfish (4.1 ± 0.01 g, mean ± SEM) were randomly stocked in 9 circular fiberglass tanks, 30 fish for each tank. They were fed with three experimental diets with dietary carbohydrate levels at 17.2% (low-carbohydrate diet, LCD), 22.8% (intermediate carbohydrate diet, ICD) and 30.2% (high-carbohydrate diet, HCD), respectively, and corn starch was used as the carbohydrate source (Supplementary Table 1). Each diet was assigned to three tanks in a completely randomized manner and fed to apparent satiation twice daily at 8:00 am and 4:00 pm for 10 weeks. During the experiment, water temperature ranged from 25.7 to 28.6 °C; dissolved oxygen and NH4-N were 6.07 ± 0.01 mg/L and 0.11 ± 0.01 mg/L, respectively.
At the end of the 10-week feeding trial, all fish were fasted for 24 h before sampling. Fish were anesthetized with MS-222 (100 mg/L water), and then, counted and weighed to determine survival, WG (weight gain) and SGR (specific growth rate). After obtaining the final total weight of fish in each tank, ten fish per tank were randomly collected, weighed and dissected on ice to obtain the liver and viscera samples for the calculation of VSI (viscerosomatic index), VAI (visceral adipose index), and HSI (hepatosomatic index). Another six fish were randomly selected from per tank, and the liver and blood were immediately collected. The livers were quickly frozen in liquid nitrogen and stored at − 80 °C for RNA and protein isolation. The blood was centrifuged at 3500 g min−1 for 10 min, and then serum was separated to determine glucose content. Another three fish per tank were randomly selected and the liver were collected, and then fixed in 10% neutral buffered formalin and 2.5% paraformaldehyde for histological, histochemical and ultrastructural observation, respectively. For analyzing hepatic enzyme activity and TG content, another six fish per tank were randomly selected and the liver were collected, and then were immediately frozen in liquid nitrogen and stored at − 80 °C for subsequent analysis.
Cell culture and treatments
Primary hepatocytes were isolated from P. fulvidraco liver and cultured as described previously [24]. To determine the mechanism of autophagy influencing the glucose-induced lipid deposition, we used the pharmacological autophagy–lysosomal pathway inhibitor 3-MA (S2767; Selleck Chemicals, Houston, TX, USA) or CQ (C6628; Millipore Sigma, Burlington, MA, USA), respectively, to incubate the hepatocytes. To illuminate the mechanism by which glucose induced autophagy and lipid accumulation, the corresponding inhibitors of the signaling pathways were used, including 4-PBA (ER stress inhibitor, SML0309; MilliporeSigma), NAC (ROS scavenger, S1623; Selleck Chemicals) and T0070907 (PPARγ inhibitor, S2871; Selleck Chemicals). The concentrations and incubation time for all experiments are reported in the Figure legends.
MTT assay for cell viability
The protocols for MTT assay were based on these described in our recent publication [26].
Histological, histochemical analysis, transmission electron microscopy (TEM) observation and Bodipy 493/503 staining
Hematoxylin–eosin (H&E staining), histochemical (Oil Red O staining) and Bodipy 493/503 (D3922; Thermo Fisher Scientific Waltham, MA, USA) staining for livers and hepatocytes were conducted according to the methods as previously described [14, 24]. For statistics of relative areas for hepatic vacuoles in H&E and lipid droplets in Oil red O staining, we randomly examined ten fields and each sample was quantified with Image J. The protocols for TEM observation have been described in Wei et al. [24]. For the Bodipy 493/503 staining, hepatocytes were observed with a laser scanning confocal microscope (Leica Microsystems, Wetzlar, Germany) to visualize the intensity of fluorescence. In those microscopes, scale bar was 10 μm with ×40 objective and ×3 zoom, and scale bar was 5 μm with ×40 objective and ×6 zoom, respectively. The green dots were defined as lipid droplet, which were quantified by Image-Pro Plus 6.0 (Media Cybernetics, Silver Spring, MD, USA).
Determination of the contents of TG, nonesterified fatty acid (NEFA) and serum glucose
TG, NEFA and serum glucose concentrations were determined with commercial kits (Nanjing Jiancheng Bioengineering Institute, Nanjing, China), according to the manufacturer’s instructions. Proteins concentrations were determined with the Bradford Protein Assay Kit of Nanjing Jiancheng Bioengineering Institute (Nanjing, China).
Determination of enzymatic activities and indices of oxidative stress
Activities of several enzymes related to lipid metabolism, such as G6PD (glucose 6-phosphate dehydrogenase), 6PGD (6-phosphogluconate dehydrogenase), ME (malic enzyme), ICDH (isocitrate dehydrogenase), FAS (fatty acid synthase), and CPT I (carnitine palmitoyltransferase 1), were measured as previously described [24, 26]. One unit of enzyme activity, defined as the amount of enzyme that converts 1 μM of substrate to product per minute at 30 °C, was expressed as units per milligram of soluble protein. Protein concentrations were determined with the Bradford Protein Assay Kit (Nanjing, China).
We also measured several parameters involved in oxidative stress, such as SOD activity and the levels of MDA, ROS, GSH and GSSG. SOD activity was measured based on aerobic reduction of NBT at 535 nm by superoxide radicals following the method described in Pan et al. [14]. MDA level was measured using malondialdehyde (MDA) assay kit from Nanjing Jiancheng Bioengineering Institute (Nanjing, China) base on measuring thiobarbituric acid-malondialdehyde (TBA-MDA) complex absorbance at 532 nm. Intracellular ROS level was measured using oxidation-sensitive fluorescent probe DCFH-DA (287810; MilliporeSigma) as described previously [26]. Liver and intracellular GSH and GSSG levels were determined according to the studies by Giustarini et al. [27]. All these analyses were conducted in three replicates.
Detection of autophagic vesicles and free Ca2+ in the hepatocytes
Detection of autophagic vesicles and free Ca2+ in the hepatocytes was performed according to recent studies [24, 28, 29]. Fluo-4 AM (F14201; Thermo Fisher Scientific) was used to detect free Ca2+. AO (318337; MilliporeSigm), MDC (30432; MilliporeSigma) were used to detect the autophagic vesicles and Lyso-Tracker Red DND-99 (L7528; Thermo Fisher Scientific) was used to detect lysosomal activity based on the published protocols [28]. Briefly, the treated cells were incubated with the corresponding reagents for 30 min, followed by three PBS washes, and then observed with the laser scanning confocal microscope (Leica) to visualize the intensity of fluorescence (×40 objective and ×2 zoom, scale bar: 25 μm). The fluorescences of the stained cells were quantified on a CytoFlex Flow Cytometer (Beckman Coulter) and data analysis was performed with FlowJo v.10 software. The relative fluorescence intensity of Ca2+, and co-localization of lipid droplets and autolysosomes were quantified using Image J analysis software.
RNA isolation and quantitative real-time PCR (qRT-PCR)
Total RNA was isolated with Trizol reagent (Thermo Fisher Scientific) and transcribed into the cDNA with a Reverse Transcription Kit (Thermo Fisher Scientific). qPCR assays were performed based on the methods described in our recent publications [26]. The gene-specific primers are given in Supplementary Table 2. Ten housekeeping genes (18S rRNA, β-actin, HPRT, B2M, UBCE, TUBA, GAPDH, RPL7, TBP and ELFA) were selected to test their transcription stability. The relative expression of genes was calculated using the 2−ΔΔCt method when normalizing to the geometric mean of the best combination of two genes as analyzed by geNorm (https://genorm.cmgg.be/).
Western blotting
Based on the protocol described in our recent publication [24], we used western blot analysis to test protein expression levels, such as LC-3B, Beclin-1, SQSTM1/p62, BiP/GRP78 and PPARγ. Briefly, liver and cell lysates were prepared with RIPA buffer (Thermo Fisher Scientific). Proteins (40 μg from each sample) were then separated on 10 or 15% SDS–polyacrylamide gel (depending on the molecular size of proteins to be analyzed), transferred to PVDF membranes (Thermo Fisher Scientific), and then blocked with 8% (w/v) skimmed milk in TBST buffer (20 mM Tris–HCl, pH 7.5, 150 mM sodium chloride, 0.1% Tween 20) for 1 h and then washed thrice with TBST buffer for 10 min each, followed by incubation with specific primary antibodies including rabbit anti-LC3B (1:1000, ab51520; Abcam, Cambridge, MA, USA), anti-Beclin-1 (1:1000, #3738; Cell Signaling Technology, Danvers, MA, USA), anti-SQSTM1/p62 (1:500, #5114; Cell Signaling Technology), anti-BiP/GRP78 (1:500, #3183; Cell Signaling Technology), anti-PPARγ (1:500, ab59256; Abcam), anti-GAPDH (1:4000, #2118; Cell Signaling Technology), and anti-β-actin(1:2000, #4970; Cell Signaling Technology) for overnight at 4 °C, followed by incubating with goat anti-rabbit IRDye 800CW secondary antibody (1:20,000, 926-32211; Li-Cor Biosciences, Lincoln, NE, USA) or HRP-conjugated anti-rabbit IgG antibody (1:2000, #7074; Cell Signaling Technology). Immunoreactive bands were visualized via Odyssey Infrared Fluorescent Western Blots Imaging System (Li-Cor Bioscience) or enhanced chemiluminescence (Cell Signaling Technology) and quantified via densitometry using Image J (version 1.42, National Institutes of Health).
Immunofluorescence
We used immunofluorescence to analyze the distribution of LC3B in hepatocytes, based on the protocol of Cai et al. [30]. In brief, after the indicated treatments, cells were washed in PBS and fixed in 4% paraformaldehyde at room temperature for 15 min. Then, the cells were blocked for 1 h in 5% BSA, and followed by incubation with specific primary antibodies rabbit anti-LC3B (1:500, ab51520; Abcam) overnight at 4 °C. The cells were washed thrice with PBS for 5 min each time, followed by incubation with a Goat Anti-Rabbit IgG H&L (Alexa Fluor® 647, 1:500, ab150079; Abcam) secondary antibody for 60 min at room temperature in the dark. DAPI was used to stain the nucleus of hepatocytes. The images were captured with the laser scanning confocal microscope (Leica) (×40 objective and ×5 zoom, scale bar: 7.5 μm), and fluorescence intensity was quantified by software Image J.
Dual-luciferase reporter assay for detecting ChoRE at the PPARγ promoter region
Based on the PPARγ promoter characterized in our recent study [31], we constructed PPARγ promoter into the pGL3 basic vector with ClonExpress™ II One Step Cloning Kit (Vazyme, Piscataway, NJ, USA). The carbohydrate response element-binding protein (ChREBP) binding sites [carbohydrate response element (ChoRE), 5′-GGCACGTGTG-3′] in the PPARγ promoter from yellow catfish were predicted by JASPAR database (http://jaspar.genereg.net/). The site mutation of the ChoRE was performed with the Quick-Change Site-Directed Mutagenesis Kit (Vazyme). These mutated constructs were named mutation 1 (5′-ATGCGCGCAT -3′) and mutation 2 (5′-TAGGGCGCGA-3′). HEK 293T cells were transfected with different plasmids by using Lipofectamine 2000 Transfection Reagent (Thermo Fisher Scientific) in 24-well plates. Cells were collected to assay luciferase activity by the Dual-Luciferase Reporter Assay System (Promega, Minneapolis, MN, USA) according to the manufacturer’s instructions.
EMSA (electrophoretic mobility shift assay) analysis for detecting the direct binding of ChoRE at the PPARγ promoter
EMSA analysis for detecting the direct binding of ChoRE at the PPARγ promoter followed the protocols described in our recent publications [31, 32]. Nuclear proteins were prepared and determined by the bicinchoninic acid method. Each oligonucleotide duplex of ChoRE (5′-AGTGTGGCACGTGTGTAATA-3′) was incubated with 10-μg nuclear extracts according to the manufacturer’s instructions. The reaction mixture was incubated at room temperature for 30 min, and then detected by electrophoresis on 6% native polyacrylamide gels and ultimately transferred to a nylon membrane. The transferred DNA was cross-linked to the membrane and detected by chemiluminescence reaction. Competition analyses were performed using 100-fold of unlabeled oligonucleotide duplex, with or without the mutation. These mutated constructs were named mutation 1 (5′-AGTGTATGCGCGCATTAATA-3′) and mutation 2 (5′-AGTGTTAGGGCGCGATAATA-3′).
Statistical analysis
Results were expressed as mean ± S.E.M. Firstly, the normality of the different treatments was evaluated with the Shapiro–Wilk test. Then, all data were evaluated by one-way analysis of variance and further analyzed by post hoc Duncan’s multiple range testing to determine statistical significance. For the comparison between two groups, Student’s t tests were used (unpaired, two-tailed). The analysis was performed with SPSS 19.0 (IBM, Armonk, NY, USA). P < 0.05 was considered significant.
Results
Growth performance, morphological parameters and serum glucose
In the present study, the survival was 100% among the three treatments (Supplementary Table 3). WG, SGR and FI (feed intake) increased with dietary carbohydrate levels. However, the FCR (feed conversion rate) declined with the increase of dietary carbohydrate supplementation (P < 0.05). HSI tended to increase with dietary carbohydrate levels but the differences were not statistically significant among three treatments. VSI and VAI showed no significant differences among three treatments.
HCD increases liver lipid accumulation
The amount of cytoplasmic vacuolation in the liver was highest in HCD group than those in LCD and ICD groups (Supplementary Fig. 1A–C). HCD increased the amount of hepatic lipid droplets (Supplementary Fig. 1D–F) and hepatic TG content (Supplementary Fig. 1G). These observations were further confirmed by the areas quantified for lipid droplets in the H&E and Oil Red O staining (Supplementary Fig. 1H and I).
HCD increased lipogenesis and reduced lipolysis and fatty acid β-oxidation
Activities of lipogenic enzymes (G6PD, 6PGD and FAS) increased with dietary carbohydrate levels, but ICDH and ME activities showed no marked differences among the three treatments (Supplementary Fig. 2A). Glucose transporter 2 (GLUT2) is a high capacity transporter expressed in the livers [33]. The hepatic GLUT2 mRNA abundances for fish fed the HCD (Supplementary Fig. 2B) were higher than those in the LCD and ICD groups. The mRNA abundances of lipogenic genes (G6PD, FAS and ACCa), and the transcriptional factors related to lipogenesis (PPARγ, SREBP-1, LXRα and ChREBP) for fish fed the HCD (Supplementary Fig. 2C) were higher than those in the LCD and ICD groups. 6PGD mRNA levels tended to increase with dietary carbohydrate levels. mRNA abundances of the lipolytic genes CPT1A and HSL for fish fed the HCD were lower than those in the LCD and ICD groups. The mRNA abundances of mitochondria (ACADL, ACAD8) and peroxisomal β-oxidation (ACAA2) for fish fed the HCD were lower than those in the LCD group, but the mRNA level of ACADVL, ACADSB, HADHAB, ECSH1, ACOX1 and ACAA1 showed no marked differences among the three treatments (Supplementary Fig. 2D).
HCD triggers hepatic ER stress, UPR and oxidative stress
To determine whether dietary carbohydrate cause ER stress, UPR and oxidative stress, thereby mediating the change of lipid metabolism, we first examined the ultrastructural changes of the liver. HCD caused the swelling of ER (Fig. 1a), indicating the occurrence of ER stress. Then, we examined if HCD influenced the mRNA expression of marker genes of ER stress and UPR (Fig. 1b). The mRNA expression of liver GRP78/BiP was significantly increased in HCD group, further confirming HCD-induced occurrence of ER stress. The mRNA levels of hepatic PERK, eIF2α and ATF6 were significantly higher in the HCD group than those in LCD and ICD groups, indicating the HCD-induced activation of PERK–eIF2α and ATF6 pathways. Western blotting analysis indicated that the protein levels of ER stress markers (GRP78) were significantly higher in HCD group than those in ICD groups (Fig. 1c, d). SOD activities and the ratio of GSH/GSSG decreased with dietary carbohydrate levels (Fig. 1e, g), and that the MDA levels increased with dietary carbohydrate addition (Fig. 1f), indicating the activation of oxidative stress.
Fig. 1.
Dietary carbohydrate supplementation triggers ER stress, UPR and oxidative stress in the liver of yellow catfish. a Liver ultrastructure (TEM, original magnification ×10,000, bars, 1 μm). m mitochondria, nu hepatocyte nucleus, er endoplasmic reticulum, sm swelling and vesiculation of mitochondria, ser swelling of endoplasmic reticulum. b The relative mRNA expression of genes involved in ER stress and UPR. c Western blot analysis of ER stress marker GRP78. d Relative quantification of protein levels of GRP78 was normalized to GAPDH. e Hepatic SOD activity. f Hepatic MDA content. g The ratio of GSH/GSSG in the liver. Values are mean ± S.E.M. (n = 3 replicate tanks and was used as three biological replicates. At least three fish were sampled for each tank and used as technical replicates). mRNA expression values were normalized to housekeeping genes (β-actin and HPRT) expressed as a ratio of the LCD. P value was calculated by one-way ANOVA and further post hoc Duncan’s multiple range testing. Values without the same letter indicate significant difference among three treatments (P < 0.05)
HCD increased hepatic autophagosome formation
Given the evidence that autophagy regulates lipid metabolism [6], we investigated the effect of HCD on autophagy. TEM observations demonstrated that HCD increased the number of LDs, which was accompanied by increased autophagosome formation (Fig. 2a–c). Some LDs were combined with the autolysosome, indicating a direct interaction between autophagy and lipid metabolism (autophagy-mediated lipophagy). The mRNA levels of autophagy-related genes (ATG1A, ATG6, ATG4A, ATG5, ATG7, ATG8A, ATG8B, ATG9A, ATG9B and p62) were significantly higher in HCD group than those in LCD group (Fig. 2d). Moreover, the protein levels of autophagy markers (LC3B-II and Beclin-1) were significantly higher in HCD group than those in LCD group (Fig. 2e, f). These results confirmed that HCD induced autophagy.
Fig. 2.
Dietary carbohydrate supplementation increases hepatic autophagosome formation of yellow catfish. a–c Representative image of liver ultrastructure (TEM, Scale bars, 2 μm). Black arrows represent the autophagosome. d Relative mRNA levels of key hepatic autophagy-related genes. e Western blot analysis of LC3B and Beclin1. f Relative quantification of protein levels of LCB were normalized to β-actin and Beclin1 were normalized to GAPDH. Relative mRNA expression values were normalized to housekeeping genes (β-actin and HPRT) expressed as a ratio of the low-carbohydrate diet LCD. All data are expressed as mean ± S.E.M. (n = 3 replicate tanks and was used as three biological replicates. At least three fish were sampled for each tank and used as technical replicates). P value was calculated by one-way ANOVA and further post hoc Duncan’s multiple range testing. Values without the same letter indicate significant difference among three treatments (P < 0.05)
Lipophagy mediates high glucose (HG)-induced changes of lipid metabolism
To gain insight into the mechanisms of lipophagy mediating the high glucose-induced change in lipid metabolism, we isolated the primary hepatocytes from yellow catfish and conducted several in vitro experiments. The MTT assay showed that glucose concentrations of 5–25 mM had no significant influence on the viability of hepatocytes (Supplementary Fig. 3A). Thus, we choose the dose of 10-mM glucose as HG (high glucose) group in the in vitro experiments and the concentration showed optimum response in TG accumulation and also is also a physiologically relevant dose in the plasma of yellow catfish fed the HCD. HG incubation increased GLUT2 mRNA expression and TG content (Supplementary Fig. 3B and C). HG also up-regulated mRNA expression of Plin2 and Plin3 (the LD-specific coat protein) (Supplementary Fig. 3D). Bodipy 493/503 staining further confirmed that HG incubation led to elevated LD accumulation (Supplementary Fig. 3E and F).
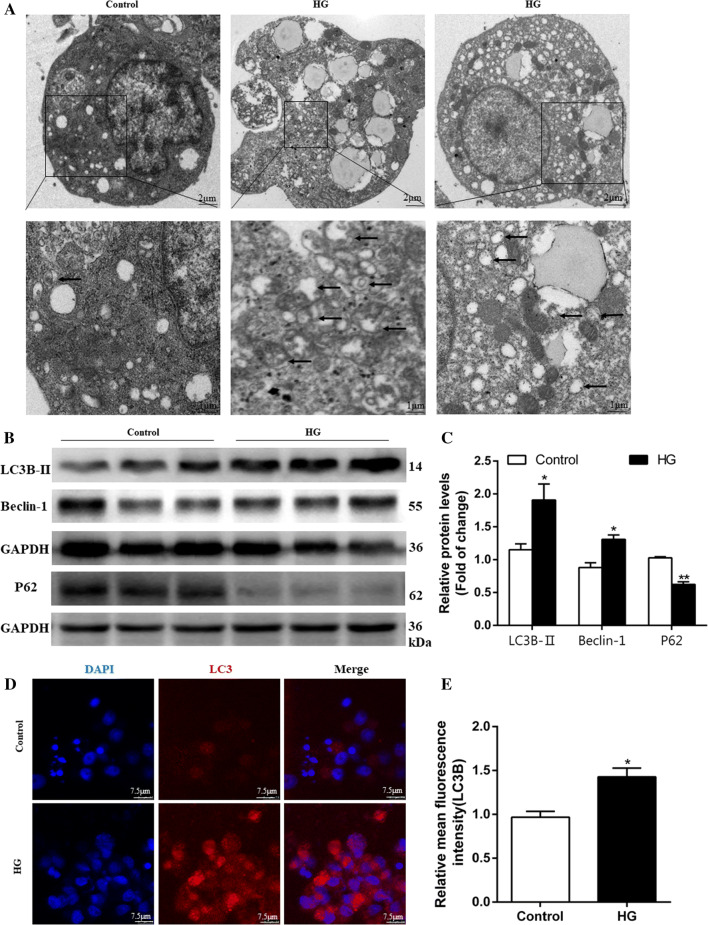
Then, we used seven methods to confirm the HG-induced autophagosome formation. MDC is widely used as a probe to label autophagic vacuoles; whereas, Lyso-Tracker Red is used for labeling lysosomes specifically. Depending on the acidity of AO, autophagic lysosomes presented as orange/red fluorescent vesicles, whereas nuclei appeared green [28]. AO, MDC, and Lyso-Tracker Red staining demonstrated that HG incubation increases intracellular acidic compartments (Supplementary Fig. 4A–G). Co-staining with MDC and Lyso-Tracker Red demonstrated that HG increased autophagosomes (blue dots), autolysosomal (red dots) formation and autophagic flux (purple dots), indicating HG-induced increase of autophagic flux (Supplementary Fig. 4G). Meanwhile, TEM observation confirmed that the intracellular acidic vesicles correlated with the formation of autophagosomes and that HG incubation increased autophagosomes (Fig. 3a). Immunoblot analysis of Beclin-1 and LC3B-II (autophagosome marker), and p62/SQSTM1 (autophagic flux protein) revealed that HG up-regulated the protein expression of Beclin-1 and LC3B-II, and down-regulated P62 protein expression (Fig. 3b, c), indicating that HG increased autophagosome formation and autophagic flux. Besides, we used immunofluorescence to assess autophagosome marker LC3, and found HG-induced increase of fluorescence intensity (Fig. 3d, e). Overall, these data confirmed that HG increased autophagosome formation.
Fig. 3.
High glucose concentration activates autophagy of hepatocytes from yellow catfish. The primary hepatocytes from P. fulvidraco were incubated in control (5 mM glucose) or HG (10 mM glucose) for 48 h in M199 medium. a Representative transmission electron microscope image of hepatocytes. Black arrows represent the autophagosome. b, c Western blot analysis of LC3B, Beclin1, and P62 protein levels (n = 3). d, e Representative confocal images and relative red fluorescence intensity showing LC3-II protein by immunofluorescence staining. Relative protein levels of LC3B, Beclin1, and P62 were normalized to GAPDH. All data are expressed as mean ± S.E.M. (n = 3 at least). P value was calculated by Student’s t tests. *P < 0.05, **P < 0.01, compared with control
To detect the role autophagy plays in regulating HCD-induced changes of lipid deposition, we treated cells with two autophagy inhibitors which block autophagy via distinct mechanisms: 3-methyladenine (3-MA, suppressing the activity of class III PI3K) and CQ (inhibition of lysosome function). 3-MA or CQ incubation showed no significant effect on cell viability (Supplementary Fig. 5A), and intracellular TG contents (Supplementary Fig. 5B). However, 3-MA and CQ pre-treatment tended to increase the HG-induced TG deposition although the differences did not reach statistical significance in the CQ pre-treated group. Bodipy 493/503 staining confirmed that 3-MA and CQ pre-incubation increased green mean fluorescence intensity and the number and size of LDs induced by HG (Supplementary Fig. 5C and K). Meanwhile, we quantified autophagy by flow cytometric analysis of the red/green fluorescence ratio using AO staining. 3-MA or CQ pretreatment alleviated the HG-induced increase in the red: green fluorescence ratio (Supplementary Fig. 5F and G). Confocal microscopic images of hepatocytes stained with AO indicated that 3-MA or CQ pretreatment alleviated the HG-induced autophagy (Supplementary Fig. 5J). MDC staining showed that 3-MA pretreatment alleviated but CQ pretreatment up-regulated the HG-induced increase in the fluorescence density (Supplementary Fig. 5D). This is understandable because 3-MA and CQ inhibited different processes of autophagy. 3-MA is an early-stage autophagy inhibitor and suppresses the activity of class III PI3K, and CQ is a late-stage autophagy inhibitor that blocks fusion of autophagosomes with lysosomes. Lyso-Tracker Red staining showed that 3-MA or CQ pretreatment alleviated the HG-induced increase in the fluorescence density (Supplementary Fig. 5E), and alleviated the HG-induced autophagy (Supplementary Fig. 5I). Taken together, all of these observations prove that autophagy mediated HG-induced changes of lipid deposition.
Lipophagy mediated HG-induced changes in lipid metabolism
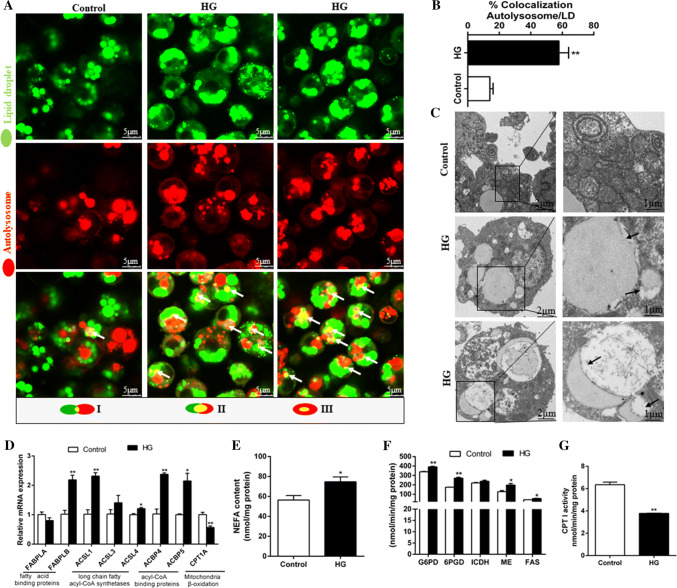
To determine whether LDs are associated with lipophagy, we performed the co-localization studies of autophagosomes, lysosomes and LDs. The colocalization of the autolysosomes and the LDs was observed in hepatocytes co-stained with Lyso-Tracker Red (red) and Bodipy 493/503 (green). HG induced the increase of LDs’ amounts and the colocalization of the autolysomes (red) and the LDs (green), which indicated the induction of lipophagy (yellow) (Fig. 4a, b). TEM analysis confirmed that LDs integrated with the autolysosomes in HG-Treated cells, indicating the occurrence of HG-induced lipophagy (Fig. 4c). To further confirm the role of lipophagy in HG-induced changes of lipid metabolism, we analyzed some parameters involved in lipid metabolism (Fig. 4d–g). HG induced the increases in intracellular NEFA content, indicating an increased flux in TG breakdown from LDs (Fig. 4e). HG incubation also up-regulated the mRNA levels of fatty acid-binding protein liver-B, long-chain fatty acyl-CoA synthetases (ACSL1 and -4), acyl-CoA binding proteins (ACBP-4 and -5), consistent with enhanced NEFA release (Fig. 4d). However, HG treatment decreased CPT I activity (lipolytic enzyme) and mRNA expression of CPT1A, and increased the activities of 6PGD, G6PD, ME, and FAS (lipogenic enzymes) (Fig. 4d, f, g). These indicated that HG activated lipogenesis and inhibited lipolysis which in turn increased TG synthesis, consistent with increased flux in TG breakdown from LDs, indicating that lipophagy plays a regulatory role in the dynamic balance of lipogenesis and lipolysis.
Fig. 4.
Lipophagy, which provided free fatty acids for the synthesis of TG but not for mitochondrial β-oxidation, alleviated HG-induced steatosis. The primary hepatocytes from P. fulvidraco were incubated in control (5-mM glucose) or HG (10-mM glucose) for 48 h in M199 medium. a The co-localization of the autolysosomes and the lipid droplets in hepatocytes co-stained with 50-nM Lyso-Tracker Red and 5 μg/ml BODIPY 493/503 (green) indicating the induction of lipophagy (yellow). b Schematic representation of the colocalization between autolysosome and LDs; c Representative TEM image of hepatocytes after control or HG incubation. Black arrows represent the lipophagy; d The mRNA levels of genes in the hepatocytes related to the fatty acid metabolism and mitochondria β-oxidation. e NEFA content in hepatocytes; f, g Activities of lipogenic (6PGD, G6PD, ICDH, ME and FAS) and lipolytic (CPTI) enzymes in the hepatocytes. Relative mRNA expression values were normalized to housekeeping genes (EIFA and RPL7) expressed as a ratio of the control. All data are expressed as mean ± S.E.M. (n = 3 at least). P value was calculated by Student’s t tests. *P < 0.05, **P < 0.01, compared with control
Oxidative stress mediated HG-induced lipophagy and lipid accumulation
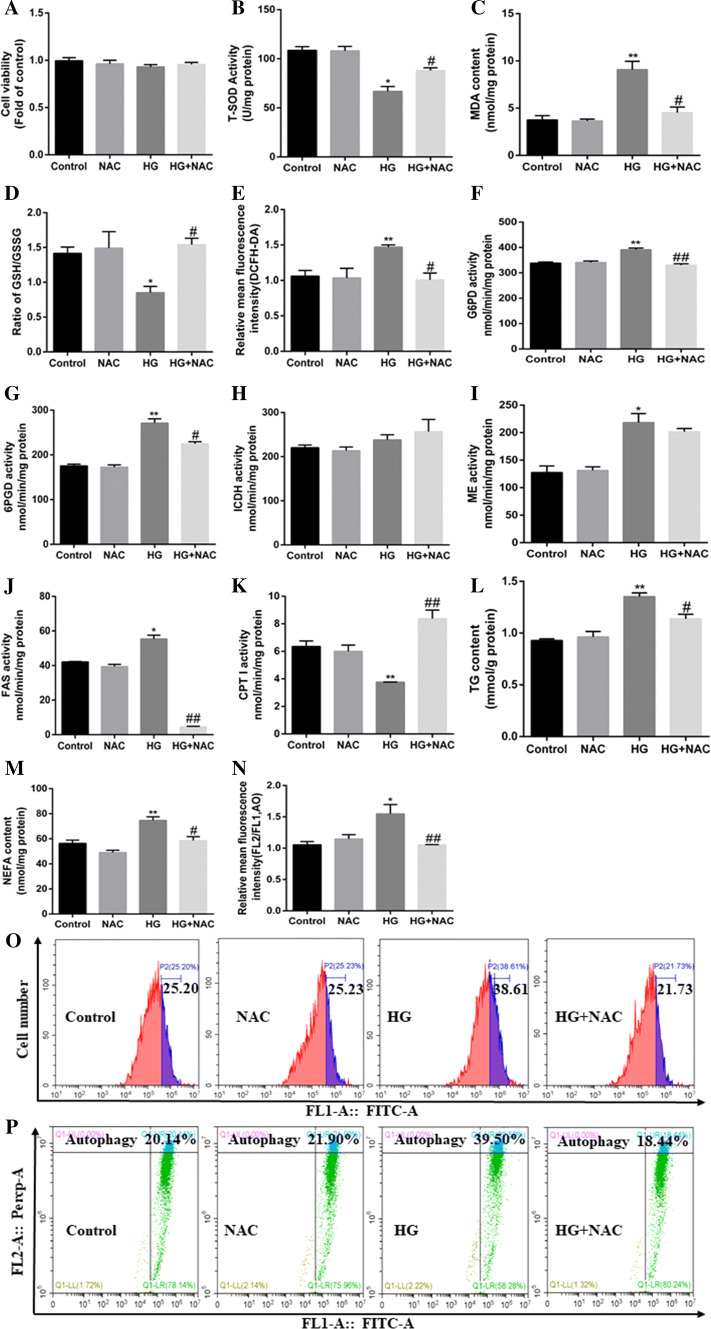
In an attempt to elucidate the mechanism of oxidative stress mediating HG-induced lipophagy and HG-induced changes of lipid metabolism, we used NAC (ROS scavenger) to block oxidative stress pathway. First, NAC did not significantly influence the viability (Fig. 5a) of hepatocytes. NAC pre-treatment abrogated the HG-induced reduction in SOD activity and the ratio of GSH/GSSG, and HG-induced increase in MDA and ROS contents (Fig. 5b–e, o). Also, NAC pretreatment alleviated the HG-induced increase in the contents of TG and NEFA, activities of lipogenic enzymes (G6PD, 6PGD, and FAS), and alleviated HG-induced reduction in CPT I activity (lipolytic enzyme) (Fig. 5f–m). Meantime, NAC pre-treatment alleviated the HG-induced increase in the mRNA expression of lipogenic genes and transcription factors (G6PD, 6PGD, ACCa, FAS, SREBP-1, PPARγ, and ChREBP), and alleviated the HG-induce reduction in mRNA expression of lipolytic genes (LPL, and CPT1A) (Supplementary Fig. 6A). Furthermore, NAC pretreatment alleviated the HG-induced increase in the red: green fluorescence ratio (Fig. 5n, p) and in relative mRNA expression of autophagy-related genes (Supplementary Fig. 6B). Overall, these data demonstrate that the oxidative stress played an important role in the HG-induced activation of lipophagy and HG-induced changes of lipid metabolism.
Fig. 5.
Mitochondrial oxidative stress pathway mediated HG-induced autophagy and lipid accumulation in the primary hepatocytes of yellow catfish. The primary hepatocytes from P. fulvidraco were incubated in control (5-mM glucose) or HG (10-mM glucose) for 48 h in M199 medium with or without 2-h pretreatment with a ROS scavenger (0.5-mM NAC). a Cell viability. b–d Activity of T-SOD, MDA content, and ratio of GSH/GSSG. e The intracellular ROS was quantified by calculating FL1 (green) mean fluorescence intensity (DCFH-DA fluorescent staining). f–k Activities of lipogenic (6PGD, G6PD, ICDH, ME and FAS) and lipolytic (CPTI) enzymes. l TG content. m NEFA content. n The autophagy was quantified by flow cytometric analysis of red/green (FL2/FL1) fluorescence ratio (acridine orange fluorescent staining, 1 μM). o The presence of DCFH-DA-stained intracellular ROS was determined by flow cytometry analysis of green fluorescence intensity. p The presence of acridine orange-stained intracellular autophagic vacuole was determined by flow cytometry analysis of red/green (FL2/FL1) fluorescence ratio. All data are expressed as mean ± S.E.M. (n = 3 at least). P value was calculated by Student’s t tests. *P < 0.05, **P < 0.01, compared with control; #P < 0.05, ##P < 0.01, compared with HG group
ER stress was involved in HG-induced lipophagy activation and lipid metabolism
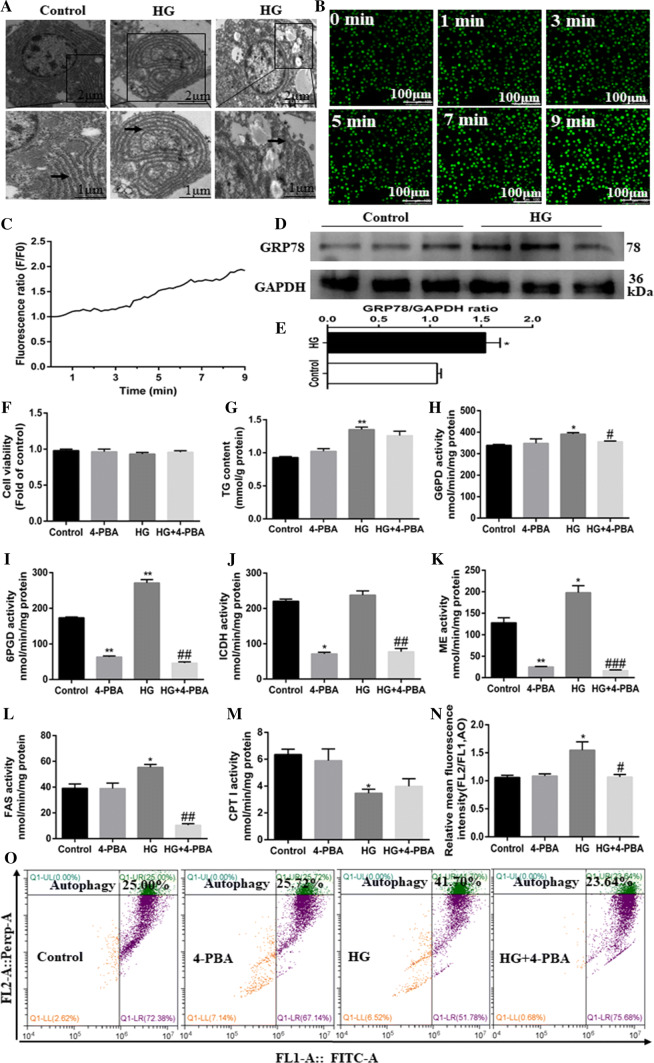
Next, we explored the mechanism of ER stress mediating HG-induced changes of lipid metabolism and lipophagy. Firstly, TEM observation revealed that HG treatment induced swelling and altered morphology of ER (Fig. 6a). ER is the main depot for intracellular free Ca2+ and Ca2+ will be released from ER to cytoplasm when ER stress occurs. We found that HG treatment led to the time-dependent increase in Ca2+ fluxes (Fig. 6b, c), and up-regulated the protein expression of GRP78/Bip (ER stress marker protein) (Fig. 6d, e). These results confirmed that HG induced ER stress. As a chemical chaperone, 4-PBA was reported to alleviate ER stress [30]. We found that 4-PBA had no marked effect on cell viability (Fig. 6f). However, 4-PBA pretreatment significantly suppressed the HG-induced increase in mRNA expression of GRP78, PERK, eIF2α, ATF4 and IRE1α (Supplementary Fig. 7A). 4-PBA pretreatment reduced HG-induced increase of TG content although the differences were not statistically significant (Fig. 6g). Furthermore, 4-PBA pretreatment markedly abolished the HG-induced increase in activities of lipogenic enzymes (G6PD and FAS) (Fig. 6h–l), but had no significant effect on HG-induced reduction in CPT I activity (lipolytic enzyme) (Fig. 6m). 4-PBA pre-treatment also alleviated the HG-induced increase in mRNA expression of lipogenic genes and transcription factors (6PGD, ACCa, FAS, SREBP-1, PPARγ, and ChREBP), and HG-induced reduction in mRNA expression of lipolytic PPARα (Supplementary Fig. 7B). In addition, 4-PBA pretreatment alleviated the HG-induced increase in the red: green fluorescence ratio (Fig. 6n, o) and mRNA expression of autophagy-related genes (Supplementary Fig. 7C). Taken together, all these results support that ER stress mediated HG-induced lipophagy activation and changes of lipid accumulation.
Fig. 6.
ER stress pathway mediated HG-induced lipid deposition and autophagy of the primary hepatocytes from yellow catfish. Hepatocytes were incubated in control (5-mM glucose) or HG (10-mM glucose) for 48 h in M199 medium with or without 2-h pretreatment with an ER stress inhibitor (100-μM 4-PBA). a Representative TEM images. Black arrow pointing to endoplasmic reticulum. b Representative confocal microscopy image of hepatocytes stained with Ca2+ fluorescent probe (Fluo-4 AM, 4 μM), showing a time-dependent changes in green fluorescence levels of primary hepatocytes. c Schematic represent quantification of the Fluo-4 AM staining. d, e Western blot analysis of GRP78/Bip protein levels (n = 3). f Cell viability. g TG content. h–m Activities of lipogenic (6PGD, G6PD, ICDH, ME and FAS) and lipolytic (CPTI) enzymes. n The autophagy was quantified by flow cytometric analysis of red/green (FL2/FL1) fluorescence ratio (acridine orange fluorescent staining, 1 μM). o The presence of acridine orange-stained intracellular autophagic vacuole was determined by flow cytometry analysis of red/green (FL2/FL1) fluorescence ratio. All data are expressed as mean ± S.E.M. (n = 3 at least). P value was calculated by Student’s t tests. *P < 0.05, **P < 0.01, compared with control; #P < 0.05, ##P < 0.01, ###P < 0.001, compared with HG group
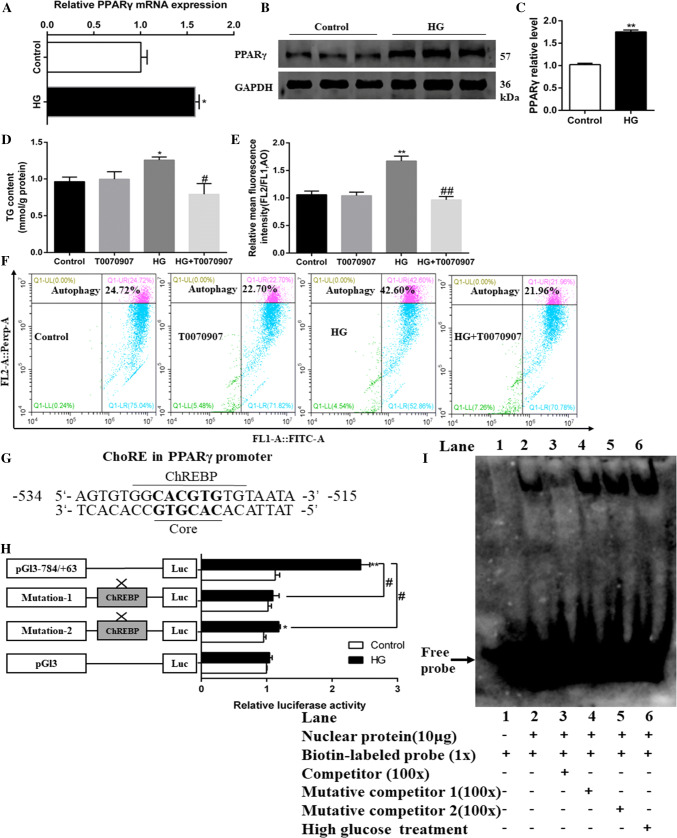
HG triggers the activation of lipophagy and changes of lipid metabolism through PPARγ pathway
To determine whether HG influenced mRNA expression of lipogenesis-related transcription factors, the mRNA levels of PPARγ, SREBP-1, PPARα, and ChREBP were measured (Supplementary Fig. 6A). PPARγ was the key integrator of lipid metabolism and autophagy [20, 34]. The present study indicated that HG significantly up-regulated the mRNA and protein levels of PPARγ (Fig. 7a–c). Next, we used T0070907 (PPARγ-specific inhibitor) to explore the effects of PPARγ signaling on lipid metabolism and autophagy. As expected, T0070907 pretreatment abolished the HG-induced TG accumulation (Fig. 7d) and alleviated HG-induced increase in the mRNA levels of lipogenic genes (G6PD, 6PGD, ACCa, FAS, SREBP-1 and ChREBP) (Supplementary Fig. 8A). T0070907 pretreatment also alleviated HG-induced increase in mRNA expression of genes involved in autophagosome membrane initiation (ATG1B and ATG6), autophagosome membrane expansion (ATG3, ATG4B and ATG5), vesicle recycling (ATG13 and ATG9A) and cargo recruitment (ATG8B and p62) (Supplementary Fig. 8B). Additionally, T0070907 pretreatment alleviated HG-induced increase of the red: green fluorescence ratio (Fig. 7e, f). All of these results indicated that PPARγ mediated HG-induced lipophagy activation and changes of lipid metabolism.
Fig. 7.
HG activates autophagy and lipid accumulation through PPARγ pathway in hepatocytes of yellow catfish. a The mRNA levels of PPARγ. b, c Western blot analysis of PPARγ protein levels. d TG content of the primary hepatocytes from P. fulvidraco were incubated in control (5-mM glucose) or HG (10-mM glucose) for 48 h in M199 medium with or without 2-h pretreatment with 1 μM T0070907 (PPARγ inhibitor). e The autophagy was quantified by flow cytometric analysis of red/green (FL2/FL1) fluorescence ratio using 1-μM acridine orange fluorescent staining. f The presence of acridine orange-stained intracellular autophagic vacuole was demonstrated by flow cytometry analysis of red–green (FL2/FL1) fluorescence ratio. g ChREBP binding sequence (ChoRE) located at − 534 bp to − 515 bp of PPARγ promoter region of yellow catfish. h Site-mutation analysis of ChREBP binding sites on pGl3-PPARγ − 784/+ 63 vectors. i EMSA of putative PPARγ binding sequences (ChoRE). The 5′-biotin labeled double-stranded oligomers were incubated with nuclear protein. A 100-fold excess of the competitor and mutative competitor oligomers was added to the competition and mutant competition assay, respectively. All data were expressed as mean ± S.E.M. (n = 3 at least). P value was calculated by Student’s t tests. *P < 0.05, **P < 0.01, compared with control; #P < 0.05, ##P < 0.01, compared with HG group. The comparison between other groups is shown in the figure
In an attempt to elucidate the mechanisms of HG activating lipophagy- and lipid metabolism-relevant genes at the transcriptional level, we further explored how HG modulated PPARγ signaling. ChREBP has emerged as a major mediator of intracellular glucose-sensory transcriptional activator and regulates gene expression by binding to the ChoRE motif of target genes [35]. By analyzing the promoter regions of PPARγ obtained in our laboratory [31], we found that ChoRE was located at − 534 to − 515 bp of PPARγ promoter region of yellow catfish, which consisted of the evolutionarily conserved core sequence CACGTG (Fig. 7g). Thus, we used luciferase reporter system to estimate whether HG could transcriptionally regulate the PPARγ promoter activity, and site-mutation analysis of ChREBP binding sites on pGl3-PPARγ-784/+63 vectors (mutations 1 and 2) was used to estimate the importance of the putative ChoRE sequences. Interestingly, our results revealed that HG incubation remarkably enhanced the luciferase activity of PPARγ promoter (Fig. 7h), and the mutation of ChoRE significantly reduced the luciferase activity of the PPARγ promoter. The mutation of site 1 (Mutation-1) did not significantly affect the HG-induced changes of the relative luciferase activity, but the mutation-2 at ChoRE binding site suppressed the HG-induced increase of luciferase activity, suggesting that ChREBP transactivated PPARγ by binding to the ChoRE motif in the PPARγ promoter region. Thus, the ChoRE motif was important for the HG-induced mRNA expression of PPARγ. Next, we used EMSA assay to determine whether ChREBP can directly bind with the promoter region of PPARγ. Our EMSA analysis indicated that the putative ChoRE sequences of the PPARγ promoter could directly bind with nuclear extract; the direct interaction can be disrupted by unlabeled wild-type and restored by the mutant probes (Fig. 7i). Moreover, HG incubation can significantly enhance the binding activity of ChREBP to ChoRE (Fig. 7i, lane 6), suggesting that the − 534 to − 515 bp region of PPARγ promoter could react with ChREBP. Together, these evidences confirmed that HG-activated lipophagy and lipid metabolism occurs via enhancing ChREBP DNA binding to the PPARγ promoter region.
Discussion
In agreement with many other studies [35, 36], our study indicated that high dietary carbohydrate supplementation induced the lipid accumulation. Furthermore, our result indicated that HCD increased hepatic TG accumulation via the upregulation of lipogenesis and the downregulation of lipolysis. Similarly, Postic et al. [35] reported that a diet rich in carbohydrate stimulates lipogenic pathways. Here, increasing dietary carbohydrate levels also up-regulate dietary energy content, which in turn will increase lipid deposition and influence lipid metabolism, as reported in other studies [23, 37].
Autophagy is important for regulating energy homeostasis and lipid content in hepatocytes [6]. The present study indicated that high carbohydrate induced autophagosome formation and that autophagy mediated HG-induced changes of lipid metabolism. Similarly, Gou et al. [38] showed that the expression of LC3B-II was markedly increased, and the autophagic vacuoles in cytoplasm were markedly accumulated when the HK2 cells were treated with high glucose. Emerging evidence indicates that LC3B-II, an autophagosome protein, is co-localized with lipid droplets; lipid droplet-specific autophagy has since been termed lipophagy [6]. The present study found that HG incubation activated lipophagy and increased intracellular NEFA content, indicating an increased flux in TG breakdown from LDs. Thus, our study indicated that lipophagy regulated HG-induced changes in lipid metabolism.
To explore the mechanism of high carbohydrate addition inducing autophagy, we determine the HG-induced changes of oxidative and ER stress. Our data indicated that ROS and ER stress mediated the HG-induced autophagy. Similarly, Wang et al. [39] pointed out that ER stress triggered autophagy. Other studies pointed out that ER stress was involved in lipid accumulation [29, 40], which was also observed in the present study. Oxidative stress and ROS production together with ER stress caused lipid peroxidation and resulted in autophagy through several distinct mechanisms involving autophagy-related genes and antioxidant enzymes [11]. The decreased GSH/GSSG ratio and SOD activity were hallmarks of oxidative stress and is involved in autophagy [41, 42]. Chen et al. [41] indicated that increasing oxidative stress upregulated autophagy. Zhao et al. [43] pointed out that oxidative stress mediated fructose-induced TG deposition in BRL-3A cells. In the present study, pretreatment with antioxidant NAC impaired the reduction in GSH/GSSG ratio and SOD activity, and significantly abrogated ROS production, and in turn, prevented autophagy activation and lipid accumulation, suggesting the important role of oxidative stress in mediating HG-induced lipophagy and lipid metabolism.
Next, we further explored the direct relationship between HG-induced lipophagy activation and the HG-induced changes of lipid metabolism. Studies pointed out that PPARγ was a key regulator of the transcriptional control of genes involved in lipid metabolism and autophagy [20, 34]. PPARγ activation can induce autophagy and lipid accumulation [20, 44]. Our present study found that HG upregulated the mRNA and protein expression of PPARγ; Moreover, HG-activated lipophagy and lipid accumulation occur via enhancing ChREBP DNA binding to the PPARγ promoter region, which in turn induced transcriptional activation of the key autophagy- and lipogenesis-related genes [44, 45]. ChREBP is considered to be a pivotal sensor protein of intracellular glucose, specifically binding to the ChoRE promoter region of its downstream target genes [35]. Postic et al. [35] showed that ChREBP was required for the carbohydrate-induced transcriptional activation of enzymes involved in TG synthesis. Therefore, ChREBP has emerged as a major mediator of glucose action on lipogenic gene expression and appears to act as a central “bridge” which connected the glucose and metabolic process. Here, we identified the putative ChoRE motif (5′-GGCACGTGTG-3′) in the PPARγ promoter region at − 534 and − 515 bp. Moreover, the dual-luciferase reporter assay and EMSA assay revealed a direct link between ChREBP and PPARγ, which indicated that endogenous ChREBP protein was recruited to the putative binding sites of PPARγ.
Conclusion
Combining our data, we proposed a model suggesting a novel mechanism of high-carbohydrate diets inducing lipid accumulation (Supplementary Fig. 9). HCD induced lipogenesis and suppressed lipolysis, and activated lipophagy, oxidative and ER stress; lipophagy mediated HCD-induced changes of lipid metabolism via oxidative and ER stress pathways. Meanwhile, HG enhanced the ChREBP DNA binding capability at the ChoRE of PPARγ promoter region, which in turn induced transcriptional activation of the key autophagy- and lipogenesis-related genes. It is noteworthy to point out that high dietary carbohydrate increased energy level, which potentially influences lipid metabolism and lipophagy. The mechanism of dietary energy levels influencing lipid metabolism and lipophagy remained to be investigated.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
This work was supported by the National Key R&D Program of China (2018YFD0900400) and Fundamental Research Funds for the Central Universities, China (Grant no. 2662018PY089).
Abbreviations
- 3-MA
3-Methyl adenine
- 4-PBA
4-Phenylbutyric acid
- 6PGD
6-Phosphogluconate dehydrogenase
- ACCa
Acetyl-CoA carboxylase a
- ACSL
Acyl-CoA synthetase long-chain
- AO
Acridine orange
- ATF
Activating transcription factor
- ATG
Autophagy-related gene
- BSA
Bovine serum albumin
- CF
Condition factor
- ChREBP
Carbohydrate response element-binding protein
- CPT-1
Carnitine palmitoyltransferase-1
- CQ
Chloroquine
- DCFH2-DA
2′,7′-Dichlorodihydrofluorescein diacetate
- eIF2α
Eukaryotic translation initiation factor 2α
- ERS
Endoplasmic reticulum stress
- FABPL
Fatty acid-binding protein liver
- FAS
Fatty acid synthase
- FBW
Final mean body weight
- FCR
Feed conversion rate
- FFA
Free fatty acids
- FI
Feed intake
- G6PD
Glucose 6-phosphate dehydrogenase
- GLUT
Glucose transporter
- GRP78
Glucose-regulated protein 78
- GSH
Glutathione
- GSSG
Glutathione disulfide
- HCD
High-carbohydrate diet
- H&E
Hematoxylin and eosin
- HG
High glucose
- HSI
Hepatosomatic index
- HSL
Hormone-sensitive lipase
- ICD
Intermediate carbohydrate diet
- ICDH
Isocitrate dehydrogenase
- IRE1α
Inositol requiring 1α
- LCD
Low-carbohydrate diet
- LD
Lipid droplet
- LXR a
Liver x receptor a
- MAP1LC3B
Microtubule-associated proteins 1A/1B light chain 3B
- IBW
Initial mean body weight
- M199
Medium-199
- MDA
Malondialdehyde
- MDC
Monodansylcadaverine
- ME
Malic enzyme
- NAC
N-acetyl-l-cysteine
- NAFLD
Nonalcoholic fatty liver disease
- NEFA
Nonesterified fatty acid
- OD
Optical density
- ORO
Oil red O
- PERK
Protein kinase R (PKR)-like ER kinase
- PPAR
Peroxisome proliferator-activated receptor
- PVDF
Polyvinylidene difluoride
- ROS
Reactive oxygen species
- S.E.M
Standard error of the mean
- SGR
Specific growth rate
- SOD
Superoxide dismutase
- SREBP-1
Sterol regulatory element-binding proteins-1
- TG
Triglyceride
- UPR
Unfolded protein response
- VAI
Visceral adipose index
- VSI
Viscerosomatic index
- WG
Weight gain
- XBP1
X-box binding protein 1
Authors’ contributions
ZL and TZ designed the experiments. TZ carried out animal and cell experiments and sample analysis with the help of KW, YHX, GHC, and CCW; TZ, ZL and CH analyzed data; TZ wrote the manuscript, and ZL and CH revised the manuscript. All the authors read and approved the manuscript.
Compliance with ethical standards
Conflict of interest
No potential conflict of interest were disclosed.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Fabbrini E, Sullivan S, Klein S. Obesity and nonalcoholic fatty liver disease: biochemical, metabolic, and clinical implications. Hepatology. 2010;51:679–689. doi: 10.1002/hep.23280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Farrell GC, Larter CZ. Non-alcoholic fatty liver: from steatosis to cirrhosis. Hepatology. 2006;43:S99–S112. doi: 10.1002/hep.20973. [DOI] [PubMed] [Google Scholar]
- 3.Abdelmalek MF, Suzuki A, Guy C, Unalp-Arida A, Colvin R, Johnson RJ, Diehl AM. Increased fructose consumption is associated with fibrosis severity in patients with nonalcoholic fatty liver disease. Hepatology. 2010;51:1961–1971. doi: 10.1002/hep.23535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Neuschwander-Tetri BA. Carbohydrate intake and nonalcoholic fatty liver disease. Curr Opin Clin Nutr. 2013;16:446–452. doi: 10.1097/MCO.0b013e328361c4d1. [DOI] [PubMed] [Google Scholar]
- 5.Levine B, Klionsky DJ. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev Cell. 2004;6:463–477. doi: 10.1016/s1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- 6.Singh R, Kaushik S, Wang YJ, Xiang YQ, Novak I, Komatsu M, Tanaka K, Cuervo AM, Czaja MJ. Autophagy regulates lipid metabolism. Nature. 2009;458:1131–1135. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bechmann LP, Hannivoort RA, Gerken G, Hotamisligil GS, Trauner M, Canbay A. The interaction of hepatic lipid and glucose metabolism in liver diseases. J Hepatol. 2012;56:952–964. doi: 10.1016/j.jhep.2011.08.025. [DOI] [PubMed] [Google Scholar]
- 8.Dong HQ, Czaja MJ. Regulation of lipid droplets by autophagy. Trends Endocrinol Metab. 2011;22:234–240. doi: 10.1016/j.tem.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Singh R, Cuervo AM. Lipophagy: connecting autophagy and lipid metabolism. Int J Cell Biol. 2012;2012:282041. doi: 10.1155/2012/282041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lavallard VJ, Gual P. Autophagy and non-alcoholic fatty liver disease. Biomed Res Int. 2014;2014:120179. doi: 10.1155/2014/120179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Azad MB, Chen YQ, Gibson SB. Regulation of autophagy by reactive oxygen species (ROS): implications for cancer progression and treatment. Antioxid Redox Signal. 2009;11:777–790. doi: 10.1089/ars.2008.2270. [DOI] [PubMed] [Google Scholar]
- 12.Kawai D, Takaki A, Nakatsuka A, Wada J, Tamaki N, Yasunaka T, Koike K, Tsuzaki R, Matsumoto K, Miyake Y, et al. Hydrogen-rich water prevents progression of nonalcoholic steatohepatitis and accompanying hepatocarcinogenesis in mice. Hepatology. 2012;56:912–921. doi: 10.1002/hep.25782. [DOI] [PubMed] [Google Scholar]
- 13.Panieri E, Santoro MM. ROS homeostasis and metabolism: a dangerous liason in cancer cells. Cell Death Dis. 2016;7:e2253. doi: 10.1038/cddis.2016.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pan YX, Luo Z, Zhuo MQ, Wei CC, Chen GH, Song YF. Oxidative stress and mitochondrial dysfunction mediated Cd-induced hepatic lipid accumulation in zebrafish Danio rerio. Aquat Toxicol. 2018;199:12–20. doi: 10.1016/j.aquatox.2018.03.017. [DOI] [PubMed] [Google Scholar]
- 15.Wang H, Sun RQ, Zeng XY, Zhou X, Li SP, Jo E, Molero JC, Ye JM. Restoration of autophagy alleviates hepatic ER stress and impaired insulin signalling transduction in high fructose-fed male mice. Endocrinology. 2015;156:169–181. doi: 10.1210/en.2014-1454. [DOI] [PubMed] [Google Scholar]
- 16.Madaro L, Marrocco V, Carnio S, Sandri M, Bouché M. Intracellular signaling in ER stress-induced autophagy in skeletal muscle cells. FASEB J. 2013;27:1990–2000. doi: 10.1096/fj.12-215475. [DOI] [PubMed] [Google Scholar]
- 17.Rutkowski DT, Wu J, Back SH, Callaghan MU, Ferris SP, Iqbal J, Clark R, Miao H, Hassler JR, Fornek J, et al. UPR pathways combine to prevent hepatic steatosis caused by ER stress-mediated suppression of transcriptional master regulators. Dev Cell. 2008;15:829–840. doi: 10.1016/j.devcel.2008.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Werstuck GH, Lentz SR, Dayal S, Hossain GS, Sood SK, Shi YY, Zhou J, Maeda N, Krisans SK, Malinow MR, et al. Homocysteine-induced endoplasmic reticulum stress causes dysregulation of the cholesterol and triglyceride biosynthetic pathways. J Clin Invest. 2001;107:1263–1273. doi: 10.1172/JCI11596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Patel L, Pass I, Coxon P, Downes CP, Smith SA, Macphee CH. Tumor suppressor and anti-inflammatory actions of PPARγ agonists are mediated via upregulation of PTEN. Curr Biol. 2001;11:764–768. doi: 10.1016/s0960-9822(01)00225-1. [DOI] [PubMed] [Google Scholar]
- 20.Zheng JL, Zhuo MQ, Luo Z, Pan YX, Song YF, Huang C, Zhu QL, Hu W, Chen QL. Peroxisome proliferator-activated receptor gamma (PPARγ) in yellow catfish Pelteobagrus fulvidraco: molecular characterization, mRNA expression and transcriptional regulation by insulin in vivo and in vitro. Gen Comp Endocr. 2015;212:51–62. doi: 10.1016/j.ygcen.2014.12.020. [DOI] [PubMed] [Google Scholar]
- 21.Meyer A, Van de Peer Y. From 2R to 3R: evidence for a fish-specific genome duplication (FSGD) BioEssays. 2005;27:937–945. doi: 10.1002/bies.20293. [DOI] [PubMed] [Google Scholar]
- 22.Gong G, Dan C, Xiao S, Guo W, Huang P, Xiong Y, Wu J, He Y, Zhang J, Li X, et al. Chromosomal-level assembly of yellow catfish genome using third-generation DNA sequencing and Hi-C analysis. GigaScience. 2018;7:giy120. doi: 10.1093/gigascience/giy120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ye WJ, Tan XY, Chen YD, Luo Z. Effects of dietary protein to carbohydrate ratios on growth and body composition of juvenile yellow catfish, Pelteobagrus fulvidraco (Siluriformes, Bagridae, Pelteobagrus) Aquac Res. 2009;40:1410–1418. [Google Scholar]
- 24.Wei CC, Luo Z, Hogstrand C, Xu YH, Wu LX, Chen GH, Pan YX, Song YF. Zinc reduces hepatic lipid deposition and activates lipophagy via Zn2+/MTF-1/PPARα and Ca2+/CaMKKβ/AMPK pathways. FASEB J. 2018;32:6666–6680. doi: 10.1096/fj.201800463. [DOI] [PubMed] [Google Scholar]
- 25.Yang SB, Tan XY, Zhang DG, Cheng J, Luo Z. Identification of ten SUMOylation-related genes from yellow catfish Pelteobagrus fulvidraco, and their transcriptional responses to carbohydrate addition in vivo and in vitro. Front Physiol. 2018;9:1544. doi: 10.3389/fphys.2018.01544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu K, Luo Z, Hogstrand C, Chen GH, Wei CC, Li DD. Zn stimulates the phospholipids biosynthesis via the pathways of oxidative and endoplasmic reticulum stress in the intestine of freshwater teleost yellow catfish. Environ Sci Technol. 2018;52:9206–9214. doi: 10.1021/acs.est.8b02967. [DOI] [PubMed] [Google Scholar]
- 27.Giustarini D, Dalle-donne I, Milzani A, Fanti P, Rossi R. Analysis of GSH and GSSG after derivatization with n-ethylmaleimide. Nat Protoc. 2013;8:1660–1669. doi: 10.1038/nprot.2013.095. [DOI] [PubMed] [Google Scholar]
- 28.Klionsky DJ, Abdelmohsen K, Abe A, Abedin MJ, Abeliovich H, Arozena AA, Adachi H, Adams CM, Adams PD, Adeli K, et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition) Autophagy. 2016;12:1–222. doi: 10.1080/15548627.2015.1100356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Song YF, Luo Z, Zhang LH, Hogstrand C, Pan YX. Endoplasmic reticulum stress and disturbed calcium homeostasis are involved in copper-induced alteration in hepatic lipid metabolism in yellow catfish Pelteobagrus fulvidraco. Chemosphere. 2016;144:2443–2453. doi: 10.1016/j.chemosphere.2015.11.031. [DOI] [PubMed] [Google Scholar]
- 30.Cai XY, Liu YL, Hu YQ, Liu XZ, Jiang HY, Yang SH, Shao Z, Xia Y, Xiong L. Ros-mediated lysosomal membrane permeabilization is involved in bupivacaine-induced death of rabbit intervertebral disc cells. Redox Biol. 2018;18:65–76. doi: 10.1016/j.redox.2018.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu K, Tan XY, Xu YH, Chen GH, Zhuo MQ. Functional analysis of promoters of genes in lipid metabolism and their transcriptional response to STAT3 under leptin signals. Genes. 2018;9:334. doi: 10.3390/genes9070334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu YH, Luo Z, Wu K, Fan YF, You WJ, Zhang LH. Structure and functional analysis of promoters from two liver isoforms of CPT I in grass carp Ctenopharyngodon idella. Int J Mol Sci. 2017;18:E2405. doi: 10.3390/ijms18112405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thorens B. Glucose transporters in the regulation of intestinal, renal, and liver glucose fluxes. Am J Physiol. 1996;270:G541–G553. doi: 10.1152/ajpgi.1996.270.4.G541. [DOI] [PubMed] [Google Scholar]
- 34.Assumpção JAF, Magalhães KG, Corrêa JR. The role of pparγ and autophagy in ros production, lipid droplets biogenesis and its involvement with colorectal cancer cells modulation. Cancer Cell Int. 2017;17:82. doi: 10.1186/s12935-017-0451-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Postic C, Dentin R, Denechaud PD, Girard J. ChREBP, a transcriptional regulator of glucose and lipid metabolism. Annu Rev Nutr. 2007;27:179–192. doi: 10.1146/annurev.nutr.27.061406.093618. [DOI] [PubMed] [Google Scholar]
- 36.Chen B, Zheng YM, Zhang JP. Comparative study of different diets-induced NAFLD models of zebrafish. Front Endocrinol. 2018;9:366. doi: 10.3389/fendo.2018.00366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dias J, Alvarez MJ, Diez A, Arzel J, Corraze G, Bautista JM, Kaushik SJ. Regulation of hepatic lipogenesis by dietary protein/energy in juvenile European sea bass (Dicentrarchus labrax) Aquaculture. 1998;161:169–186. [Google Scholar]
- 38.Gou R, Chen JT, Sheng SF, Wang RQ, Fang YD, Yang ZJ, Wang L, Tang L. Kim-1 mediates high glucose-induced autophagy and apoptosis in renal tubular epithelial cells. Cell Physiol Biochem. 2016;38:2479–2488. doi: 10.1159/000445598. [DOI] [PubMed] [Google Scholar]
- 39.Wang H, Sun RQ, Camera D, Zeng XY, Jo E, Chan SM, Herbert TP, Molero JC, Ye JM. Endoplasmic reticulum stress up-regulates Nedd4-2 to induce autophagy. FASEB J. 2016;30:2549–2556. doi: 10.1096/fj.201500119. [DOI] [PubMed] [Google Scholar]
- 40.Lebeaupin C, Vallée D, Hazari Y, Hetz C, Chevet E, Bailly-Maitre B. Endoplasmic reticulum stress signaling and the pathogenesis of non-alcoholic fatty liver disease. J Hepatol. 2018;69:927–947. doi: 10.1016/j.jhep.2018.06.008. [DOI] [PubMed] [Google Scholar]
- 41.Chen Y, Azad MB, Gibson SB. Superoxide is the major reactive oxygen species regulating autophagy. Cell Death Differ. 2009;16:1040–1052. doi: 10.1038/cdd.2009.49. [DOI] [PubMed] [Google Scholar]
- 42.Zhang Z, Guo M, Zhao S, Shao J, Zheng S. ROS-JNK1/2-dependent activation of autophagy is required for the induction of anti-inflammatory effect of dihydroartemisinin in liver fibrosis. Free Radic Biol Med. 2016;101:272–283. doi: 10.1016/j.freeradbiomed.2016.10.498. [DOI] [PubMed] [Google Scholar]
- 43.Zhao XJ, Yu HW, Yang YZ, Wu WY, Chen TY, Jia KK, Kang LL, Jiao RQ, Kong LD. Polydatin prevents fructose-induced liver inflammation and lipid deposition through increasing miR-200a to regulate Keap1/Nrf2 pathway. Redox Biol. 2018;18:124–137. doi: 10.1016/j.redox.2018.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rovito D, Giordano C, Vizza D, Plastina P, Barone I, Casaburi I, Lanzino M, De AF, Sisci D, Mauro L, et al. Omega-3 PUFA ethanolamides DHEA and EPA induce autophagy through PPARγ activation in MCF-7 breast cancer cells. J Cell Physiol. 2013;228:1314–1322. doi: 10.1002/jcp.24288. [DOI] [PubMed] [Google Scholar]
- 45.Zhou J, Zhang W, Liang B, Casimiro MC, Whitaker-Menezes D, Wang M, Lisanti MP, Lanza-Jacoby S, Pestell RG, Wang C. PPARγ activation induces autophagy in breast cancer cells. Int J Biochem Cell Biol. 2009;41:2334–2342. doi: 10.1016/j.biocel.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.