Abstract
Zygosis is the generation of new biological individuals by the sexual fusion of gamete cells. Our current understanding of eukaryotic phylogeny indicates that sex is ancestral to all extant eukaryotes. Although sexual development is extremely diverse, common molecular elements have been retained. HAP2-GCS1, a protein that promotes the fusion of gamete cell membranes that is related in structure to certain viral fusogens, is conserved in many eukaryotic lineages, even though gametes vary considerably in form and behaviour between species. Similarly, although zygotes have dramatically different forms and fates in different organisms, diverse eukaryotes share a common developmental programme in which homeodomain-containing transcription factors play a central role. These common mechanistic elements suggest possible common evolutionary histories that, if correct, would have profound implications for our understanding of eukaryogenesis.
Keywords: Reproduction, Syngamy, Evolution, Homeoproteins, Mitochondria, Archaea
Introduction
Sex is a cyclical process that produces new eukaryotic individuals in two ways. In one phase of sex, zygosis (also known as syngamy and amphimixis [1]), two individuals are combined: two cells fuse, and then their nuclei fuse, forming a new individual with a doubled genetic content. In the other phase, meiosis, cells of this higher genetic content are divided to produce cells that have reverted to the lower ploidy level. At its simplest, this sexual cycle is restricted to single-celled individuals, but it always involves another, asexual replicative mode of individuation, in which cellular ploidy is changed only insofar as a new genome copy is produced before each division, so that each daughter cell reproduces its parent. However, even the simplest sexual cycles necessitate an elaborate sequence of events that must be carefully coordinated. Sex was ancestral to all extant eukaryotes (as far as is known), and understanding its conserved molecular foundations can help to shed light on some of the central structural and regulatory requirements that shaped the earliest eukaryotic cells, which must have been decisive in shaping the tremendous diversity of eukaryotic organisms that have evolved as their descendants [2–5].
Zygosis involves the pairing and fusion of gametes (Fig. 1, left), which are normally monoploid (possessing one genome copy), forming a prozygote cell [6]. The two nuclei then fuse to form the zygote (Fig. 1, right). The zygote then progresses to meiosis without dividing in haplontic organisms, or enters the mitotic cell cycle in diplontic and haplo-diplontic eukaryotes. Variations on this basic pattern of behaviour are seen in ciliates and certain fungi, in which cell fusions are transient and nuclei are exchanged without mixture of cytoplasms [7, 8]; and in social amoebae, in which prozygotes have a transient syncytial stage, where many gametes fuse simultaneously, mixing their cytoplasms thoroughly then dividing gradually to uni- and binucleate cells before nuclear fusion occurs [9–11]. Despite these elaborations of the basic pattern and the diversity of sexual cycles in general, recent research has illustrated common mechanisms that are widely conserved in very different eukaryotes and that, therefore, very likely reflect ancestral mechanisms governing zygosis. This review will highlight certain of these recent advances, mostly in unicellular eukaryotes, and discuss how they might deepen, and even transform our understanding of fundamental aspects of eukaryotic cell biology and evolution.
Fig. 1.
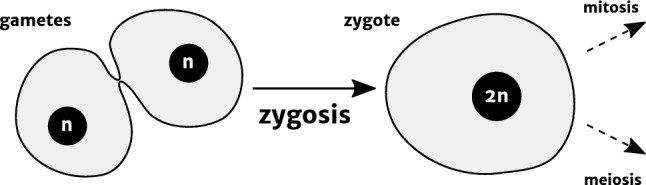
The rudiments of zygosis. Zygosis involves the fusion of two gametes (shown here immediately after the initial fusion of their membranes), followed by nuclear fusion of their nuclei to form a zygote, a new individual with twice the original genetic content (2n). Zygotes can either enter the mitotic cell cycle (in diplontic and haplo-diplontic organisms) or progress immediately to meiosis to regenerate haploid progeny (in haplontic organisms). Typically, but not always, gametes are monoploid and zygotes diploid
Conserved mechanisms of gamete fusion
Sex is advantageous and involves multiple cells, and so is a competitive process. Gametes might be slow or otherwise inefficient in pairing and fusing with compatible partners, and many will fail to fuse at all. Genetic variants that affect gamete fusion can, therefore, be expected to be under strong selection. Relatedly, we might also expect that gamete recognition genes might be relatively fast-evolving: positive selection on these genes could lead to frequent fixation of effective new variants. Variants that introduce new proteins into the recognition and fusion processes to increase the success rate of gamete fusion can also be selected for, which might promote the loss of ancestral genes if those can be completely replaced. Accordingly, gamete fusion and recognition mechanisms vary considerably between different organisms, even otherwise closely related species [12].
The most highly conserved gamete fusion gene, as far as is known, encodes the fusogen protein HAP2-GCS1. This gene is present in at least some species in almost all major eukaryotic lineages, though it appears to have been lost multiple times, notably being absent in the entire vertebrates and fungal lineages [5, 13]. HAP2-GCS1 is structurally related to viral class II fusion proteins [14–16]; these proteins, together classed as fusexins [14, 17], undergo multimerisation and substantial conformational changes to bring lipid bilayers together, and promote membrane fusion [18]. In the case of the viral fusogens, one of the two membranes is the envelope bilayer surrounding the infective virion, in which the fusogen protein is embedded, and the other is the endolysosomal membrane within target cells. Fusion of the membranes in this case allows the virion and its contents to enter the host cytoplasm. The viral fusexins are often activated by acidification of the endosome lumen after endocytosis [19, 20]. In the case of HAP2-GCS1, the two membranes to be fused are the plasma membranes of two gametes. In this fusexin, a hydrophobic or amphipathic polypeptide region comprising one or more loops at one extremity of the fusion structure inserts into the target membrane during the initial contact, allowing the subsequent merger of both membranes [21]. Substantial variation in the sequence and structure of the HAP2-GCS1 fusion loops exists between lineages [22, 23], presumably reflecting differences in target membrane composition combined with the long-term effects of sexual competition. Variable regions away from the fusion interface likely reflect different protein–protein interactions undergone by HAP2-GCS1 during its activation cycle [23].
HAP2-GCS1 function has been characterised in several organisms. In plants, where the gene encoding the protein was first discovered, HAP2-GCS1 is expressed in male gametes and is necessary for fusion with the female gamete [24, 25]. This male-specific requirement for HAP2-GCS1 has also been demonstrated in the apicomplexan Plasmodium [26, 27], and seems likely to be conserved in metazoa, as well [28, 29]. In the isogamous green alga Chlamydomonas, the HAP2-GCS1 orthologue is again only required to function in the minus gamete, not the plus gamete [26]. In contrast, in the ciliate Tetrahymena, which has several mating types, HAP2-GCS1 is required in both of the paired gametes [30], in a deviation from the “virus-like” unidirectional function that occurs in plants, green algae, and others. The social amoeba Dictyostelium discoideum appears, puzzlingly, to be an intermediate case: this species has three mating types, and two of them express HAP2-GCS1 and require its function during fusion, but, in the third mating type, the protein is not necessary for fusion [31]. Although biochemical data are required to confirm it, this finding suggests that, in two of the three D. discoideum mating configurations, HAP2-GCS1 function is ‘conventionally’ unidirectional, while, in the third, it is bidirectional as in Tetrahymena.
The trigger of the fusogen activity of HAP2-GCS1 is not known and might vary between different organisms. In the flowering plant, Arabidopsis membrane localisation of the protein is a regulated step: HAP2-GCS1 is only delivered to the plasma membrane of male gametes after stimulation by proteins secreted by the female gamete [32]. In contrast, in the green alga, Chlamydomonas HAP2-GCS1 is constitutively present in a small region of the differentiated minus gamete plasma membrane [33]. The cytoplasmic C-terminal domain is important for the function of the protein: a cluster of cysteine residues therein, often found but not always in corresponding positions in different HAP2-GCS1 orthologues, is important for fusion in Chlamydomonas, and another mutation affecting the C-terminal domain interferes with targeting of the protein to the fusion site [33]. In Arabidopsis, positively charged residues in the C-terminal domain are required for efficient fusion [34] (but see also [35]).
As noted above, several eukaryotic lineages appear to have lost the HAP2-GCS1 gene, very likely after its function in gamete function was made redundant after the emergence of novel fusion-promoting mechanisms. In fungi, the membrane proteins Prm1 and Fig1 have been implicated as important regulators of fusion [36–40], but the precise mechanism of membrane fusion in this lineage remains elusive. In vertebrates, mechanisms of gamete recognition, and perhaps fusion, appear to be diverse, and perhaps fast-evolving [41–43]. It seems likely that further diversity will be discovered when other sexual lineages that lack HAP2-GCS1 homologues are examined.
Prevention of fusion—how do gametes fuse in twos, not threes, and more?
Sex is a cycle of doubling then halving the ploidy level, and so must normally involve fusions of pairs of gametes to form zygotes: uncontrolled ploidy increases through fusion of multiple cells are not commonly found (most likely because of costs to polyploidy [44]). If gametes are rare, this pairwise fusion will occur almost automatically, but since sex is competitive (as mentioned above), in many organisms, there is scope for multiple fusions to occur. Consequently, mechanisms have evolved to promote biparental sex, and triploid zygotes are rare, aberrant occurrences [45–47]. Remarkably, in Chlamydomonas, the HAP2-GCS1 protein is rapidly degraded after gamete fusion, along with another membrane protein called FUS1 [45]. The destruction of HAP2-GCS1 prevents fusion of the prozygote with plus gametes, and destruction of FUS1, which is expressed in plus gametes and important for gamete recognition [48], prevents fusion with minus gametes [45]. A recent study in the ascomycete yeast Schizosaccharomyces pombe demonstrated a rapid post-fusion transcriptional response, involving a homeodomain transcription factor in limiting the potential for polyploidy [49]; a pair of homeoproteins has also been implicated in preventing supernumerary fusion in the basidiomycete fungus Cryptococcus [50], and more generally, the frequent involvement of related transcription factors during zygosis (see below) suggests that this might be a common function in evolution.
In animals, the fusion of multiple sperm cells with a single egg occurs frequently in some species, but not others [51, 52]; the physical properties of the egg can alter after the initial fusion, limiting further entry, or degradation of supernumerary sperm-derived nuclei or pronuclei can occur in the egg cytoplasm, depending on the species [51]. Zygosis in flowering plants occurs through an orchestrated series of events including the repulsion of supernumerary pollen tubes from each ovule, as well as blocks to polyspermy; mechanisms underlying the former ‘polytubey’ are known [53, 54], but those limiting polyploidy remain unclear [55]. Finally, as noted above, social amoebae are unusual in forming frequent syncytia before gamete nuclei fuse [9–11]. Puzzlingly, haploid nuclei appear to coexist in these syncytia for up to 8 h as the syncytia gradually break apart to form binucleate cells, in which nuclear fusion occurs [10, 56]. How nuclei of different mating types might recognise each other in these cells, and how fusion between them might be controlled, are not known. Nuclear fusion during zygosis is another complex process, involving fusion of outer and inner nuclear membranes, along with their associated endoplasmic reticulum. These fusion events involve the KAR5-GEX1 protein in budding yeast, Chlamydomonas, and zebrafish, and its wide conservation across eukaryotes suggests that, like HAP2-GCS1, KAR5-GEX1 is ancestral to all eukaryotes [40, 57–59]. Complex regulation of nuclear fusion is important in certain fungi as well (apparently) in social amoebae [8, 11, 40]; it is conceivable that nuclear fusion was more complex ancestrally than in most extant organisms if, as some have hypothesised [60], early eukaryotes were multinucleate.
Zygote differentiation—a conserved ancestral function for homeoproteins
Across eukaryotes, zygotes have diverse features and fates: as mentioned earlier, they can immediately commit to meiosis or can enter a prolonged diploid (or polyploid) phase of the organism’s lifecycle, depending on the species. Whether this involves the entry into the diploid (or polyploid) mitotic cell cycle, or an immediate commitment to meiotic division, zygote-specific genes must be induced and then further changes in gene expression must occur after fusion of the gamete nuclei. Despite this diversity of zygotic fates in different lineages, it has become clear that a common element in the initial transition from the haploid to the diploid phase is present in several eukaryotic supergroups: an involvement of homeodomain-containing transcription factors [61].
The genetic control of the haploid to diploid transition was first dissected in detail in the budding yeast Saccharomyces cerevisiae, in which proteins encoded at the mating-type locus were found to govern both haploid and diploid functions [62]. Two of these proteins, MATa1 and MATα2, contain homeodomains [63]. The genes encoding these proteins form part of different idiomorphs of the mating-type locus (that is, part of the different versions of the locus that determines mating type of haploid budding yeast cells), so they function independently in haploids. Upon gamete fusion, MATa1 and MATα2 bind each other to form a heterodimer that functions as a diploid-specific transcription factor [64]. As described above, a similar heterodimerisation involving a homeoprotein occurs at the same lifecycle stage in the distantly related ascomycete Schizosaccharomyces pombe; in this case, a homeoprotein named Pi binds a much shorter polypeptide, Mi [49, 65]; Mi is not recognisably homologous to homeodomain proteins, and its tertiary structure is not known, but it seems possible that the gene encoding it is a degenerate and truncated descendant of an ancestral homeodomain gene. In basidiomycete fungi, a similar process occurs to that founding in budding yeast: homeoproteins encoded by genes within different alleles of one mating-type locus (basidiomycetes typically have two such loci) are able to heterodimerise after cell fusion and activate dikaryon functions [66, 67].
Homeoproteins, which can be divided into two classes, TALE (three amino acid loop extension) and non-TALE [68, 69], also have important functions in establishing zygotic identity in plants and green algae. In Chlamydomonas, gametes fuse in a HAP2-GCS1-dependent process as described above, and then, zygotes differentiate into zygospores (Fig. 2, top). Zygotes do not divide by mitosis: sex in these algae is induced by nitrogen starvation, and zygospores can remain dormant for a period before undergoing meiotic divisions [70]. Like budding yeast and basidiomycetes, Chlamydomonas gametes of each mating type express a specific homeodomain transcription factor, one of the BELL and one of the KNOX class, although the genes encoding them are not located at the mating-type locus [71, 72]. Again, upon gamete fusion, the two homeoproteins, called GSM1 and GSP1, bind to each other to form a dimer that can only form in diploids heterozygous at the mating-type locus, and then activate zygotic gene expression [72]. Mutants lacking GSP1 fail to form zygospores after gamete fusion, instead remaining in the mitotic cell cycle (Fig. 2, bottom) [73]. The kinetics of degradation of HAP2-GCS1 and FUS1 in Chlamydomonas zygotes (mentioned above) are consistent with a process activated by fresh transcription after fusion, at least in part [45]. Remarkably, although land plants have dramatically elaborated lifecycles compared to their green algal ancestors, this ancestral function of homeoproteins in regulating zygote differentiation has been conserved in bryophytes. Physcomitrella mutants lacking two KNOX homeoproteins normally expressed in egg cells develop abnormally after zygosis, forming gametophyte-like (haploid-like) morphologies instead of diploid sporophytes [74]. Overexpressing a different, BELL class, homeoprotein in haploid cells of the same species produces sporophyte-like differentiation [75].
Fig. 2.
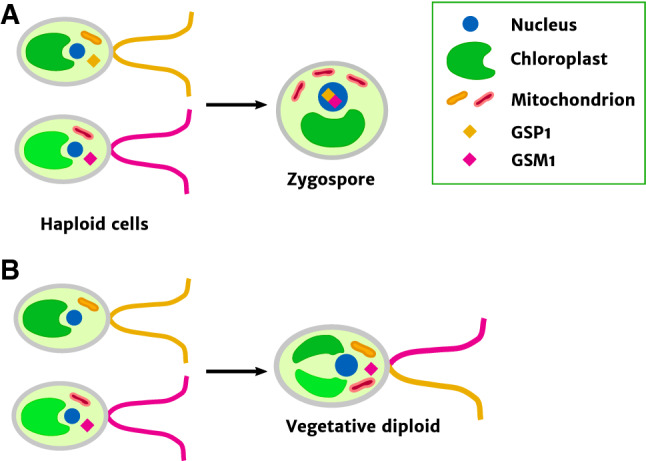
a Zygosis in Chlamydomonas. Two gametes, one of the plus mating type (upper) and one of the minus mating type (lower) fuse, ultimately forming a zygospore. bGSP1 mutants fail to complete zygosis. After fusion with a minus gamete, GSP1 mutant cells fail to limit mitochondrial and chloroplast inheritance in the normal fashion and do not differentiate into zygospores, resulting in vegetative diploid cells containing organelle genomes from both parents
The brown alga Ectocarpus, a member of the eukaryotic supergroup Heterokonta (also known as Stramenopiles), has a complex lifecycle involving multicellular haploid and diploid phases [76]. As in the very distantly related bryophytes and green algae, mutations affecting homeoproteins cause defects during zygosis: homozygous mutants in either the oro or sam genes cause diploid organisms to develop similarly to gametophytes (haploids) instead of sporophytes [77, 78]. The two homeoproteins heterodimerise, are TALE class like those with similar functions in plants and green algae, underscoring the conserved pattern of behaviours, although both proteins are expressed in both gamete classes unlike their fungal and green algal counterparts [78].
Finally, in the social amoeba Dictyostelium, like the fungi, genes located within the mating-type locus are essential for normal zygote development as well as mating [79, 80]. One pair of gametolgues in Dictyostelium discoideum is necessary for zygotic function in diploids formed from two of this species three mating types, and encodes proteins that have a homeodomain-like fold, but extremely divergent in sequence [80]. Their evolutionary history is not clear, nor their biochemical interactions, but they may also share ancestry with the homeoproteins with roles in zygosis in other eukaryotic lineages.
Inheritance of mitochondria and plastids during zygosis
In many cases, mitochondria and chloroplasts are inherited uniparentally during sex, usually through females in both animals and plants [81, 82]. Even in unicellular eukaryotes with gametes indistinguishable in size, more or less strict uniparental inheritance of these organelles is maintained. For instance, in Chlamydomonas, mitochondrial genomes are inherited only from the minus parent, while chloroplast genomes are inherited from the plus parent [83, 84], in a process depend on the aforementioned homeoprotein GSP1 (Fig. 2) [73]. Similarly, in the basidiomycete yeast Cryptococcus, two mating-type-specific homeodomain proteins are required for normal uniparental mitochondrial inheritance [85, 86]. In ascomycete yeasts, mitochondrial inheritance into the zygote is often biparental, but meiotic progeny revert quickly to homoplasmy (possessing a single mitotype) due to spatial segregation of mtDNA nucleoids. This segregation occurs immediately during meiosis in Schizosaccharomyces pombe and during the first mitotic divisions of meiotic progeny in Saccharomyces cerevisiae [87, 88]. In Dictyostelium, strict uniparental inheritance of mitochondria is not maintained, and because of the syncytial phase during gamete fusion mentioned earlier, mitochondrial genomes can be inherited laterally, so that meiotic progeny have three parents, their nuclear chromosomes recombined from two parents, and their mtDNA inherited from a third [11]. This unusual mode of inheritance may be related to the cannibalistic sexual development of zygotes in these amoeba [89] leading to selection for cytoplasmic genes that promote survival into progeny. It is possible that this feature of social amoebae, in which the mitochondrial genome seems to meet a strict definition of a selfish genetic element [90], is atavistic, resembling patterns of (proto-)mitochondrial inheritance early in eukaryotic evolution before strict controls on gamete fusion and organelle inheritance first emerged [11].
The evolution of zygosis and lifecycle regulators
Sexual lifecycles could have evolved originally in two ways: either meiosis (or another reductional mode of division) arose first as a mechanism for ploidy reduction in cells that had undergone serial reduplication of their genomes [91–93], or zygosis preceded it as a way to mask disadvantageous genetic variants [94]. One plausible hypothesis for the origin of zygosis suggests that cell fusion could have arisen as a by-product of a conjugative infectious process by which symbionts spread from cell to cell [11, 95, 96]. The realisation that HAP2-GCS1, the gamete fusogen very likely to be ancestral to all eukaryotes, is structurally related to viral fusion proteins provides a possible molecular basis for a parasitic origin of sex: co-option of a viral fusion protein [97]. It is, of course, also possible that this co-option could have occurred in the other direction; viruses frequently take possession of host genes [98]. Eukaryotes likely evolved from stem archaeal cells that acquired a bacterial symbiont that became the mitochondrion [99–101], though this is still a matter of contention (see [102–104]). This raises the possibility that elements specifically functioning in zygosis in eukaryotes, including HAP2-GCS1, could have archaeal (or archaeoviral) ancestry, as do components of meiosis like Spo11 [105, 106].
A related question concerns the origin of mating types: did unisexual, self-fertile individuals precede the evolution of genetically determined, distinct mating types [107]? The apparent ancestral role of HAP2-GCS1 does not settle this question, because it is not clear whether it originally functioned unilaterally or bilaterally in promoting fusion (both are found in extant eukaryotes, as described above). Mating types have been proposed to have evolved primarily as a way to cleanly trigger diploid-specific functions: [108] gamete fusion mixes cytoplasmic components from two differentiated haploid cell types, allowing fusion to be used as a logical AND gate. The roles of homeoproteins during zygosis in plants, green algae, brown algae, and fungi (and apparently divergent homeoproteins in social amoebae) might seem to support this idea, since the apparent conservation of this function again suggests that it could be ancestral to all eukaryotes. However, these homeoproteins are TALE class in plants, green algae, brown algae, and some fungal proteins (MATα2-like and Pi proteins in ascomycetes, HD1-type proteins in basidiomycetes), and non-TALE in other fungal proteins (MATa1-like proteins in ascomycetes and HD2-type proteins in basidiomycetes); the relationship of the social amoebae proteins is unclear. These differences make it difficult to exclude the possibility of convergent evolution. Again, if functions in lifecycle transitions could be ascribed to archaeal proteins related in structure to homeodomains [80], or if homeoproteins were found to be involved in zygosis in other eukaryotic lineages, and any widely conserved downstream targets of such proteins identified, the deep evolutionary picture could become clearer. The unclear evolutionary origins of fungal gamete fusion mechanisms, involving the apparent loss of ancestral HAP2-GCS1 function, and homeoprotein functions during zygosis might be elucidated by a broader examination including diverse fungal lineages, in the same way as a recent study demonstrating how a viral protein was co-opted to rewire ancestral mitotic cell cycle regulation [109].
Advances in our understanding of the molecular mechanisms of zygosis in diverse eukaryotes in recent years have transformed our understanding of the evolution of sex. It is to be hoped that further exploration of the biochemistry and cytology of the key components, ideally in even more eukaryotic lineages, along with the phylogenetic data that will result from ongoing genome and transcriptome sequencing efforts will allow us to address the important questions that so far remain unanswered and, perhaps, to identify further conserved ancestral genes.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Herranz G. Origin of the terms embryo, gamete and zygote. Zygote. 2012;20:313–320. doi: 10.1017/S0967199412000391. [DOI] [PubMed] [Google Scholar]
- 2.Cavalier-Smith T. Cell cycles, diplokaryosis and the archezoan origin of sex. Arch Protistenk. 1995;145:189–207. [Google Scholar]
- 3.Kondrashov AS. Evolutionary genetics of life cycles. Annu Rev Ecol Syst. 1997;28:391–435. [Google Scholar]
- 4.Dacks J, Roger AJ. The first sexual lineage and the relevance of facultative sex. J Mol Evol. 1999;48:779–783. doi: 10.1007/pl00013156. [DOI] [PubMed] [Google Scholar]
- 5.Speijer D, Lukeš J, Eliáš M. Sex is a ubiquitous, ancient, and inherent attribute of eukaryotic life. PNAS. 2015;112:8827–8834. doi: 10.1073/pnas.1501725112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pixell-Goodrich HLM. Memoirs: on the life-history of the sporozoa of spatangoids, with observation on some allied forms. J Cell Sci. 1915;s2(61):81–104. [Google Scholar]
- 7.Dobell C. A commentary on the genetics of the ciliate protozoa. J Gen. 1914;4:131–190. [Google Scholar]
- 8.Raper JR. Sexual versatility and evolutionary processes in Fungi. Mycologia. 1959;51:107–124. [Google Scholar]
- 9.Saga Y, Okada H, Yanagisawa K. Macrocyst development in Dictyostelium discoideum. II. Mating-type-specific cell fusion and acquisition of fusion-competence. J Cell Sci. 1983;60:157–168. doi: 10.1242/jcs.60.1.157. [DOI] [PubMed] [Google Scholar]
- 10.Ishida K, Hata T, Urushihara H. Gamete fusion and cytokinesis preceding zygote establishment in the sexual process of Dictyostelium discoideum. Dev Growth Differ. 2005;47:25–35. doi: 10.1111/j.1440-169x.2004.00776.x. [DOI] [PubMed] [Google Scholar]
- 11.Bloomfield G, Paschke P, Okamoto M, et al. Triparental inheritance in Dictyostelium. Proc Natl Acad Sci USA. 2019;116:2187–2192. doi: 10.1073/pnas.1814425116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Swanson WJ, Vacquier VD. The rapid evolution of reproductive proteins. Nat Rev Genet. 2002;3:137–144. doi: 10.1038/nrg733. [DOI] [PubMed] [Google Scholar]
- 13.Wong JL, Johnson MA. Is HAP2-GCS1 an ancestral gamete fusogen? Trends Cell Biol. 2010;20:134–141. doi: 10.1016/j.tcb.2009.12.007. [DOI] [PubMed] [Google Scholar]
- 14.Valansi C, Moi D, Leikina E, et al. Arabidopsis HAP2/GCS1 is a gamete fusion protein homologous to somatic and viral fusogens. J Cell Biol. 2017;216:571–581. doi: 10.1083/jcb.201610093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pinello JF, Lai AL, Millet JK, et al. Structure-function studies link class II viral fusogens with the ancestral gamete fusion protein HAP2. Curr Biol. 2017;27:651–660. doi: 10.1016/j.cub.2017.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fédry J, Liu Y, Péhau-Arnaudet G, et al. The ancient gamete fusogen HAP2 is a eukaryotic class II fusion protein. Cell. 2017;168:904–915.e10. doi: 10.1016/j.cell.2017.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hernández JM, Podbilewicz B. The hallmarks of cell–cell fusion. Development. 2017;144:4481–4495. doi: 10.1242/dev.155523. [DOI] [PubMed] [Google Scholar]
- 18.Modis Y. Relating structure to evolution in class II viral membrane fusion proteins. Curr Opin Virol. 2014;5:34–41. doi: 10.1016/j.coviro.2014.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kielian M, Helenius A. pH-induced alterations in the fusogenic spike protein of Semliki Forest virus. J Cell Biol. 1985;101:2284–2291. doi: 10.1083/jcb.101.6.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lescar J, Roussel A, Wien MW, et al. The fusion glycoprotein shell of semliki forest virus: an icosahedral assembly primed for fusogenic activation at endosomal pH. Cell. 2001;105:137–148. doi: 10.1016/s0092-8674(01)00303-8. [DOI] [PubMed] [Google Scholar]
- 21.Feng J, Dong X, Pinello J, et al. Fusion surface structure, function, and dynamics of gamete fusogen HAP2. Elife. 2018;7:e39772. doi: 10.7554/eLife.39772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fedry J, Forcina J, Legrand P, et al. Evolutionary diversification of the HAP2 membrane insertion motifs to drive gamete fusion across eukaryotes. PLoS Biol. 2018;16:e2006357. doi: 10.1371/journal.pbio.2006357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baquero E, Fedry J, Legrand P, et al. Species-specific functional regions of the green alga gamete fusion protein HAP2 revealed by structural studies. Structure. 2019;27:113–124.e4. doi: 10.1016/j.str.2018.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mori T, Kuroiwa H, Higashiyama T, Kuroiwa T. GENERATIVE CELL SPECIFIC 1 is essential for angiosperm fertilization. Nat Cell Biol. 2006;8:64–71. doi: 10.1038/ncb1345. [DOI] [PubMed] [Google Scholar]
- 25.von Besser K, Frank AC, Johnson MA, Preuss D. Arabidopsis HAP2 (GCS1) is a sperm-specific gene required for pollen tube guidance and fertilization. Development. 2006;133:4761–4769. doi: 10.1242/dev.02683. [DOI] [PubMed] [Google Scholar]
- 26.Liu Y, Tewari R, Ning J, et al. The conserved plant sterility gene HAP2 functions after attachment of fusogenic membranes in Chlamydomonas and Plasmodium gametes. Genes Dev. 2008;22:1051–1068. doi: 10.1101/gad.1656508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hirai M, Arai M, Mori T, et al. Male fertility of malaria parasites is determined by GCS1, a plant-type reproduction factor. Curr Biol. 2008;18:607–613. doi: 10.1016/j.cub.2008.03.045. [DOI] [PubMed] [Google Scholar]
- 28.Steele RE, Dana CE. Evolutionary history of the HAP2/GCS1 gene and sexual reproduction in metazoans. PLoS One. 2009;4:e7680. doi: 10.1371/journal.pone.0007680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ebchuqin E, Yokota N, Yamada L, et al. Evidence for participation of GCS1 in fertilization of the starlet sea anemone Nematostella vectensis: implication of a common mechanism of sperm–egg fusion in plants and animals. Biochem Biophys Res Commun. 2014;451:522–528. doi: 10.1016/j.bbrc.2014.08.006. [DOI] [PubMed] [Google Scholar]
- 30.Cole ES, Cassidy-Hanley D, Pinello JF, et al. Function of the male-gamete-specific fusion protein HAP2 in a seven-sexed ciliate. Curr Biol. 2014;24:2168–2173. doi: 10.1016/j.cub.2014.07.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Okamoto M, Yamada L, Fujisaki Y, et al. Two HAP2-GCS1 homologs responsible for gamete interactions in the cellular slime mold with multiple mating types: implication for common mechanisms of sexual reproduction shared by plants and protozoa and for male-female differentiation. Dev Biol. 2016;415:6–13. doi: 10.1016/j.ydbio.2016.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sprunck S, Rademacher S, Vogler F, et al. Egg cell–secreted EC1 triggers sperm cell activation during double fertilization. Science. 2012;338:1093–1097. doi: 10.1126/science.1223944. [DOI] [PubMed] [Google Scholar]
- 33.Liu Y, Pei J, Grishin N, Snell WJ. The cytoplasmic domain of the gamete membrane fusion protein HAP2 targets the protein to the fusion site in Chlamydomonas and regulates the fusion reaction. Development. 2015;142:962–971. doi: 10.1242/dev.118844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wong JL, Leydon AR, Johnson MA. HAP2(GCS1)-dependent gamete fusion requires a positively charged carboxy-terminal domain. PLoS Genet. 2010;6:e1000882. doi: 10.1371/journal.pgen.1000882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mori T, Hirai M, Kuroiwa T, Miyagishima S. The functional domain of GCS1-based gamete fusion resides in the amino terminus in plant and parasite species. PLoS One. 2010;5:e15957. doi: 10.1371/journal.pone.0015957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Heiman MG, Walter P. Prm1p, a pheromone-regulated multispanning membrane protein, facilitates plasma membrane fusion during yeast mating. J Cell Biol. 2000;151:719–730. doi: 10.1083/jcb.151.3.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jin H, Carlile C, Nolan S, Grote E. Prm1 prevents contact-dependent lysis of yeast mating pairs. Eukaryot Cell. 2004;3:1664–1673. doi: 10.1128/EC.3.6.1664-1673.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aguilar PS, Engel A, Walter P. The plasma membrane proteins Prm1 and Fig1 ascertain fidelity of membrane fusion during yeast mating. Mol Biol Cell. 2007;18:547–556. doi: 10.1091/mbc.E06-09-0776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aguilar PS, Baylies MK, Fleissner A, et al. Genetic basis of cell-cell fusion mechanisms. Trends Genet. 2013;29:427–437. doi: 10.1016/j.tig.2013.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fu C, Heitman J. PRM1 and KAR5 function in cell–cell fusion and karyogamy to drive distinct bisexual and unisexual cycles in the Cryptococcus pathogenic species complex. PLoS Genet. 2017;13:e1007113. doi: 10.1371/journal.pgen.1007113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Aydin H, Sultana A, Li S, et al. Molecular architecture of the human sperm IZUMO1 and egg JUNO fertilization complex. Nature. 2016;534:562–565. doi: 10.1038/nature18595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ohto U, Ishida H, Krayukhina E, et al. Structure of IZUMO1-JUNO reveals sperm-oocyte recognition during mammalian fertilization. Nature. 2016;534:566–569. doi: 10.1038/nature18596. [DOI] [PubMed] [Google Scholar]
- 43.Herberg S, Gert KR, Schleiffer A, Pauli A. The Ly6/uPAR protein Bouncer is necessary and sufficient for species-specific fertilization. Science. 2018;361:1029–1033. doi: 10.1126/science.aat7113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Comai L. The advantages and disadvantages of being polyploid. Nat Rev Genet. 2005;6:836–846. doi: 10.1038/nrg1711. [DOI] [PubMed] [Google Scholar]
- 45.Liu Y, Misamore MJ, Snell WJ. Membrane fusion triggers rapid degradation of two gamete-specific, fusion-essential proteins in a membrane block to polygamy in Chlamydomonas. Development. 2010;137(9):1473–1481. doi: 10.1242/dev.044743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nakel T, Tekleyohans DG, Mao Y, et al. Triparental plants provide direct evidence for polyspermy induced polyploidy. Nat Commun. 2017;8:1033. doi: 10.1038/s41467-017-01044-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Grossniklaus U. Polyspermy produces tri-parental seeds in maize. Curr Biol. 2017;27:R1300–R1302. doi: 10.1016/j.cub.2017.10.059. [DOI] [PubMed] [Google Scholar]
- 48.Ferris PJ, Woessner JP, Goodenough UW. A sex recognition glycoprotein is encoded by the plus mating-type gene fus1 of Chlamydomonas reinhardtii. MBoC. 1996;7:1235–1248. doi: 10.1091/mbc.7.8.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vještica A, Merlini L, Nkosi PJ, Martin SG. Gamete fusion triggers bipartite transcription factor assembly to block re-fertilization. Nature. 2018;560:397–400. doi: 10.1038/s41586-018-0407-5. [DOI] [PubMed] [Google Scholar]
- 50.Hull CM, Boily M-J, Heitman J. Sex-specific homeodomain proteins Sxi1alpha and Sxi2a coordinately regulate sexual development in Cryptococcus neoformans. Eukaryot Cell. 2005;4:526–535. doi: 10.1128/EC.4.3.526-535.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rothschild L. Polyspermy. Q Rev Biol. 1954;29:332–342. doi: 10.1086/400393. [DOI] [PubMed] [Google Scholar]
- 52.Bianchi E, Wright GJ. Sperm meets egg: the genetics of mammalian fertilization. Annu Rev Genet. 2016;50:93–111. doi: 10.1146/annurev-genet-121415-121834. [DOI] [PubMed] [Google Scholar]
- 53.Beale KM, Leydon AR, Johnson MA. Gamete fusion is required to block multiple pollen tubes from entering an Arabidopsis ovule. Curr Biol. 2012;22:1090–1094. doi: 10.1016/j.cub.2012.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Maruyama D, Völz R, Takeuchi H, et al. Rapid elimination of the persistent synergid through a cell fusion mechanism. Cell. 2015;161:907–918. doi: 10.1016/j.cell.2015.03.018. [DOI] [PubMed] [Google Scholar]
- 55.Tekleyohans DG, Mao Y, Kägi C, et al. Polyspermy barriers: a plant perspective. Curr Opin Plant Biol. 2017;35:131–137. doi: 10.1016/j.pbi.2016.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Okada H, Hirota Y, Moriyama R, et al. Nuclear fusion in multinucleated giant cells during the sexual development of Dictyostelium discoideum. Dev Biol. 1986;118:95–102. [Google Scholar]
- 57.Abrams EW, Zhang H, Marlow FL, et al. Dynamic assembly of brambleberry mediates nuclear envelope fusion during early development. Cell. 2012;150:521–532. doi: 10.1016/j.cell.2012.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ning J, Otto TD, Pfander C, et al. Comparative genomics in Chlamydomonas and Plasmodium identifies an ancient nuclear envelope protein family essential for sexual reproduction in protists, fungi, plants, and vertebrates. Genes Dev. 2013;27:1198–1215. doi: 10.1101/gad.212746.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rogers JV, Rose MD. Kar5p is required for multiple functions in both inner and outer nuclear envelope fusion in Saccharomyces cerevisiae. G3 (Bethesda) 2014;5:111–121. doi: 10.1534/g3.114.015800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Garg SG, Martin WF. Mitochondria, the cell cycle, and the origin of sex via a syncytial eukaryote common ancestor. Genome Biol Evol. 2016;8:1950–1970. doi: 10.1093/gbe/evw136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bowman JL, Sakakibara K, Furumizu C, Dierschke T. Evolution in the cycles of life. Annu Rev Genet. 2016;50:133–154. doi: 10.1146/annurev-genet-120215-035227. [DOI] [PubMed] [Google Scholar]
- 62.Strathern J, Hicks J, Herskowitz I. Control of cell type in yeast by the mating type locus: the α1–α2 hypothesis. J Mol Biol. 1981;147:357–372. doi: 10.1016/0022-2836(81)90488-5. [DOI] [PubMed] [Google Scholar]
- 63.Shepherd JCW, McGinnis W, Carrasco AE, et al. Fly and frog homoeo domains show homologies with yeast mating type regulatory proteins. Nature. 1984;310:70. doi: 10.1038/310070a0. [DOI] [PubMed] [Google Scholar]
- 64.Dranginis AM. Binding of yeast al and α2 as a heterodimer to the operator DNA of a haploid-specific gene. Nature. 1990;347:682. doi: 10.1038/347682a0. [DOI] [PubMed] [Google Scholar]
- 65.Kelly M, Burke J, Smith M, et al. Four mating-type genes control sexual differentiation in the fission yeast. EMBO J. 1988;7:1537–1547. doi: 10.1002/j.1460-2075.1988.tb02973.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kämper J, Reichmann M, Romeis T, et al. Multiallelic recognition: nonself-dependent dimerization of the bE and bW homeodomain proteins in Ustilago maydis. Cell. 1995;81:73–83. doi: 10.1016/0092-8674(95)90372-0. [DOI] [PubMed] [Google Scholar]
- 67.Banham AH, Asante-Owusu RN, Gottgens B, et al. An N-terminal dimerization domain permits homeodomain proteins to choose compatible partners and initiate sexual development in the mushroom Coprinus cinereus. Plant Cell. 1995;7:773–783. doi: 10.1105/tpc.7.6.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Derelle R, Lopez P, Guyader HL, Manuel M. Homeodomain proteins belong to the ancestral molecular toolkit of eukaryotes. Evolut Dev. 2007;9:212–219. doi: 10.1111/j.1525-142X.2007.00153.x. [DOI] [PubMed] [Google Scholar]
- 69.Joo S, Wang MH, Lui G, et al. Common ancestry of heterodimerizing TALE homeobox transcription factors across Metazoa and Archaeplastida. BMC Biol. 2018;16:136. doi: 10.1186/s12915-018-0605-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Levine RP, Ebersold WT. The genetics and cytology of Chlamydomonas. Annu Rev Microbiol. 1960;14:197–216. doi: 10.1146/annurev.mi.14.100160.001213. [DOI] [PubMed] [Google Scholar]
- 71.Goodenough UW, Armbrust EV, Campbell AM, Ferris PJ. Molecular genetics of sexuality in Chlamydomonas. Annu Rev Plant Physiol Plant Mol Biol. 1995;46:21–44. [Google Scholar]
- 72.Lee J-H, Lin H, Joo S, Goodenough U. Early sexual origins of homeoprotein heterodimerization and evolution of the plant KNOX/BELL family. Cell. 2008;133:829–840. doi: 10.1016/j.cell.2008.04.028. [DOI] [PubMed] [Google Scholar]
- 73.Nishimura Y, Shikanai T, Nakamura S, et al. Gsp1 triggers the sexual developmental program including inheritance of chloroplast DNA and mitochondrial DNA in Chlamydomonas reinhardtii. Plant Cell. 2012;24:2401–2414. doi: 10.1105/tpc.112.097865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sakakibara K, Ando S, Yip HK, et al. KNOX2 genes regulate the haploid-to-diploid morphological transition in land plants. Science. 2013;339:1067–1070. doi: 10.1126/science.1230082. [DOI] [PubMed] [Google Scholar]
- 75.Horst NA, Katz A, Pereman I, et al. A single homeobox gene triggers phase transition, embryogenesis and asexual reproduction. Nat Plants. 2016;2:15209. doi: 10.1038/nplants.2015.209. [DOI] [PubMed] [Google Scholar]
- 76.Coelho SM, Peters AF, Charrier B, et al. Complex life cycles of multicellular eukaryotes: new approaches based on the use of model organisms. Gene. 2007;406:152–170. doi: 10.1016/j.gene.2007.07.025. [DOI] [PubMed] [Google Scholar]
- 77.Coelho SM, Godfroy O, Arun A, et al. OUROBOROS is a master regulator of the gametophyte to sporophyte life cycle transition in the brown alga Ectocarpus. PNAS. 2011;108:11518–11523. doi: 10.1073/pnas.1102274108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Arun A, Coelho SM, Peters AF, et al. Convergent recruitment of TALE homeodomain life cycle regulators to direct sporophyte development in land plants and brown algae. eLife. 2019;8:e43101. doi: 10.7554/eLife.43101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bloomfield G, Skelton J, Ivens A, et al. Sex determination in the social amoeba Dictyostelium discoideum. Science. 2010;330:1533–1536. doi: 10.1126/science.1197423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hedgethorne K, Eustermann S, Yang J-C, et al. Homeodomain-like DNA binding proteins control the haploid-to-diploid transition in Dictyostelium. Sci Adv. 2017;3:e1602937. doi: 10.1126/sciadv.1602937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Birky CW. The inheritance of genes in mitochondria and chloroplasts: laws, mechanisms, and models. Annu Rev Genet. 2001;35:125–148. doi: 10.1146/annurev.genet.35.102401.090231. [DOI] [PubMed] [Google Scholar]
- 82.Breton S, Stewart DT. Atypical mitochondrial inheritance patterns in eukaryotes. Genome. 2015;58:423–431. doi: 10.1139/gen-2015-0090. [DOI] [PubMed] [Google Scholar]
- 83.Sager R, Lane D. Molecular basis of maternal inheritance. PNAS. 1972;69:2410–2413. doi: 10.1073/pnas.69.9.2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Boynton JE, Harris EH, Burkhart BD, et al. Transmission of mitochondrial and chloroplast genomes in crosses of Chlamydomonas. PNAS. 1987;84:2391–2395. doi: 10.1073/pnas.84.8.2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yan Z, Hull CM, Heitman J, et al. SXI1alpha controls uniparental mitochondrial inheritance in Cryptococcus neoformans. Curr Biol. 2004;14:R743–R744. doi: 10.1016/j.cub.2004.09.008. [DOI] [PubMed] [Google Scholar]
- 86.Yan Z, Hull CM, Sun S, et al. The mating type-specific homeodomain genes SXI1α and SXI2a coordinately control uniparental mitochondrial inheritance in Cryptococcus neoformans. Curr Genet. 2007;51:187–195. doi: 10.1007/s00294-006-0115-9. [DOI] [PubMed] [Google Scholar]
- 87.Nunnari J, Marshall WF, Straight A, et al. Mitochondrial transmission during mating in Saccharomyces cerevisiae is determined by mitochondrial fusion and fission and the intramitochondrial segregation of mitochondrial DNA. MBoC. 1997;8:1233–1242. doi: 10.1091/mbc.8.7.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mehta K, Ananthanarayanan V (2019) Cortical tethering of mitochondria by the dynein anchor Mcp5 enables uniparental mitochondrial inheritance during fission yeast meiosis. bioRxiv 525196 [DOI] [PMC free article] [PubMed]
- 89.Bloomfield G. Sex in dictyostelia. In: Romeralo M, Baldauf S, Escalante R, editors. Dictyostelids: evolution, genomics and cell biology. Berlin: Springer; 2013. pp. 129–148. [Google Scholar]
- 90.Hurst GD, Werren JH. The role of selfish genetic elements in eukaryotic evolution. Nat Rev Genet. 2001;2:597–606. doi: 10.1038/35084545. [DOI] [PubMed] [Google Scholar]
- 91.Cleveland LR. The origin and evolution of meiosis. Science. 1947;105:287–289. doi: 10.1126/science.105.2724.287. [DOI] [PubMed] [Google Scholar]
- 92.Hurst LD, Nurse P. A note on the evolution of meiosis. J Theor Biol. 1991;150:561–563. doi: 10.1016/s0022-5193(05)80447-3. [DOI] [PubMed] [Google Scholar]
- 93.Kondrashov AS. Gradual origin of amphimixis by natural selection. Lect Math Life Sci. 1994;25:27–51. [Google Scholar]
- 94.Crow JF, Kimura M. Evolution in sexual and asexual populations. Am Nat. 1965;99:439–450. [Google Scholar]
- 95.Hickey DA. Selfish DNA: a sexually-transmitted nuclear parasite. Genetics. 1982;101:519–531. doi: 10.1093/genetics/101.3-4.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hickey DA. Molecular symbionts and the evolution of sex. J Hered. 1993;84:410–414. doi: 10.1093/oxfordjournals.jhered.a111363. [DOI] [PubMed] [Google Scholar]
- 97.Clark T. HAP2/GCS1: mounting evidence of our true biological EVE? PLoS Biol. 2018;16:e3000007. doi: 10.1371/journal.pbio.3000007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Koonin EV, Dolja VV, Krupovic M. Origins and evolution of viruses of eukaryotes: the ultimate modularity. Virology. 2015;479–480:2–25. doi: 10.1016/j.virol.2015.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Williams TA, Foster PG, Cox CJ, Embley TM. An archaeal origin of eukaryotes supports only two primary domains of life. Nature. 2013;504:231–236. doi: 10.1038/nature12779. [DOI] [PubMed] [Google Scholar]
- 100.Raymann K, Brochier-Armanet C, Gribaldo S. The two-domain tree of life is linked to a new root for the Archaea. PNAS. 2015;112:6670–6675. doi: 10.1073/pnas.1420858112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zaremba-Niedzwiedzka K, Caceres EF, Saw JH, et al. Asgard archaea illuminate the origin of eukaryotic cellular complexity. Nature. 2017;541:353–358. doi: 10.1038/nature21031. [DOI] [PubMed] [Google Scholar]
- 102.Cunha VD, Gaia M, Gadelle D, et al. Lokiarchaea are close relatives of Euryarchaeota, not bridging the gap between prokaryotes and eukaryotes. PLoS Genet. 2017;13:e1006810. doi: 10.1371/journal.pgen.1006810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Spang A, Eme L, Saw JH, et al. Asgard archaea are the closest prokaryotic relatives of eukaryotes. PLoS Genet. 2018;14:e1007080. doi: 10.1371/journal.pgen.1007080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Cunha VD, Gaia M, Nasir A, Forterre P. Asgard archaea do not close the debate about the universal tree of life topology. PLoS Genet. 2018;14:e1007215. doi: 10.1371/journal.pgen.1007215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Bergerat A, de Massy B, Gadelle D, et al. An atypical topoisomerase II from archaea with implications for meiotic recombination. Nature. 1997;386:414. doi: 10.1038/386414a0. [DOI] [PubMed] [Google Scholar]
- 106.Goodenough U, Heitman J. Origins of eukaryotic sexual reproduction. Cold Spring Harb Perspect Biol. 2014;6:a016154. doi: 10.1101/cshperspect.a016154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Heitman J. Evolution of sexual reproduction: a view from the fungal kingdom supports an evolutionary epoch with sex before sexes. Fungal Biol Rev. 2015;29:108–117. doi: 10.1016/j.fbr.2015.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Perrin N. What uses are mating types? The “Developmental Switch” model. Evolution. 2012;66:947–956. doi: 10.1111/j.1558-5646.2011.01562.x. [DOI] [PubMed] [Google Scholar]
- 109.Medina EM, Turner JJ, Gordân R, et al. Punctuated evolution and transitional hybrid network in an ancestral cell cycle of fungi. Elife. 2016;5:e09492. doi: 10.7554/eLife.09492. [DOI] [PMC free article] [PubMed] [Google Scholar]


