Abstract
Non-alcoholic fatty liver disease (NAFLD) is a metabolic liver disease that is thought to be reversible by changing the diet. To examine the impact of dietary changes on progression and cure of NAFLD, we fed mice a high-fat diet (HFD) or high-fructose diet (HFrD) for 9 weeks, followed by an additional 9 weeks, where mice were given normal chow diet. As predicted, the diet-induced NAFLD elicited changes in glucose tolerance, serum cholesterol, and triglyceride levels in both diet groups. Moreover, the diet-induced NAFLD phenotype was reversed, as measured by the recovery of glucose intolerance and high cholesterol levels when mice were given normal chow diet. However, surprisingly, the elevated serum triglyceride levels persisted. Metagenomic analysis revealed dietary-induced changes of microbiome composition, some of which remained altered even after reversing the diet to normal chow, as illustrated by species of the Odoribacter genus. Genome-wide DNA methylation analysis revealed a “priming effect” through changes in DNA methylation in key liver genes. For example, the lipid-regulating gene Apoa4 remained hypomethylated in both groups even after introduction to normal chow diet. Our results support that dietary change, in part, reverses the NAFLD phenotype. However, some diet-induced effects remain, such as changes in microbiome composition, elevated serum triglyceride levels, and hypomethylation of key liver genes. While the results are correlative in nature, it is tempting to speculate that the dietary-induced changes in microbiome composition may in part contribute to the persistent epigenetic modifications in the liver.
Electronic supplementary material
The online version of this article (10.1007/s00018-019-03114-4) contains supplementary material, which is available to authorized users.
Keywords: DNA methylation, Gut microbiome, NAFLD, Epigenetics, High-fat diet
Introduction
Non-alcoholic fatty liver disease (NAFLD) is a metabolic, inflammatory disease that affects the liver, in the absence of significant alcohol consumption. NAFLD is often associated with obesity. The global adoption of the ‘Western diet’, which is characterized by excessive fat and fructose consumption, has been suggested to contribute to NAFLD. NAFLD is currently a leading cause of chronic liver disease worldwide [1].
Lifestyle modifications are often effective when treating patients with NAFLD. Studies have shown that ≥ 10% reductions in body weight resulted in nearly a complete reduction in non-alcoholic steatohepatitis and improvements in liver fibrosis. Even modest weight loss (~ 5%) resulted in clinically beneficial effects on NAFLD symptoms [2]. Moreover, positive long-term effects (> 2 years) have been reported with a 6-month hypocaloric dietary intervention in patients with NAFLD [3]. However, while the majority of patients respond to dietary intervention therapies, there are a subset of patients that do not benefit from diet change and these patients have a higher mortality rate [4]. Thus, a better understanding of how lifestyle treatments affect the NAFLD syndrome at a systemic level is highly warranted.
In the twenty-first century, precision medicine must include the gut microbiome, because it is a major regulator of human health. Indeed, the microbiome was shown to be involved in lifestyle-induced diseases, such as NAFLD [5–7]. It has been observed that germ-free mice were resistant to certain forms of diet-induced obesity [8]. Moreover, significant differences in gut microbe composition have been reported for patients with NAFLD or NASH (non-alcoholic steatohepatitis) compared to healthy controls [9, 10]. Increased primary or secondary bile acid in NAFLD patient serum was highly correlated with gut microbiome composition changes [11]. Those findings suggested that there might be direct communication between the gut microbiome and liver metabolism. Interestingly and relevant to the current study, it has been found that changes in microbiome composition correlate with epigenetic modifications of the host genome. For example, changes in DNA methylation signatures in circulating lymphocytes were correlated with alterations in gut microbiome composition in pregnant women, which were relevant to lipid metabolism, inflammation, and obesity [12]. Other groups have reported that microbe-derived fatty acids may tune gene expression by changing the conformational three-dimensional structure of DNA [13].
DNA methylation is a type of epigenetic regulation that is closely linked to transcription factor binding and chromatin accessibility. In NAFLD, a close relationship has been reported between the insulin resistance phenotype and DNA methylation of insulin-linked genes [14]. Recently, a number of research groups have showed that a tight relationship between epigenetic modification and NAFLD development exists [15]. To this end, Murphy et al. reported that altered DNA methylation of genes can regulate the process of liver disease, and differences in DNA methylation can distinguish between advanced vs. mild NAFLD human patients [16]. Moreover, in mice, differences in their epigenetic profiles correlated with the susceptibility and development of hepatic steatosis of that mouse [17].
In the current study, we aimed to further understand the pathophysiology of NAFLD in genetically identical mice and to investigate the dynamics of persistent microbiome and DNA methylation patterns.
Methods
Mice and diets
Mice were maintained in air-conditioned isolated cages with 12-h light/dark cycles and ad libitum access to food and water. We followed the protocol approved by the Singhealth Institutional Animal Care and Use Committee, Singapore (2015/SHS/1040). The experimental plan is outlined in Figure S1 and the different diet compositions are detailed in Table 1. Briefly, 8-week-old male C57BL/6 J mice were given a high-fat diet (HFD) or high-fructose diet (HFrD) for 9 weeks to induce the NAFLD phenotype. Three-to-five mice were housed in one cage (NC n = 13, HFD n = 14, HFrD n = 11, R_NC n = 15, R_HFD n = 15, and R_HFrD n = 14). Thereafter, half of the cohorts from each treatment were killed, and tissue samples were collected for biological analysis. The remaining half of each treatment group was maintained for an additional 9 weeks, but they were fed normal chow (TD2918, Halan) to reverse the NAFLD phenotype. Throughout the experimental period, body weight and food intake were measured on a weekly basis.
Table 1.
Diet composition information
| % calories from fat | % calories from protein | % calories from carbohydrates | Source | |
|---|---|---|---|---|
| Normal chow diet (NC) | 18 | 19 | 59.9a | Specialty feeds |
| High fat diet (HFD) | 60.3 | 18.4 | 21.3 | TD06414 (Harlan) |
| High fructose diet (HFrD) | 13 | 20.2 | 66.8 | TD89247 (Harlan) |
| Reversal diet | 6.2 | 18.6 | 44.2b | TD2918 (Harlan) |
aCrude fibre 5.2%, acid detergent fibre 7.7%, neutral detergent fibre 15.5%)
bCrude fibre 3.5%, neutral detergent fibre 14.7%
Intraperitoneal glucose tolerance test (IPGTT)
Animals fasted overnight before undergoing each IPGTT. IPGTTs were conducted after 9 weeks and again after 18 weeks in all diet groups. Baseline blood glucose levels were measured prior to an intraperitoneal glucose injection (2 g/kg body weight) of 20% glucose. Blood glucose levels were then measured at the indicated timepoints.
Serum analysis
We collected whole blood samples and centrifuged them at 10,000 rpm for 10 min at 4ºC. The levels of trigylycerides (STA-396), Cholesterol (STA-384, Cell Biolabs) and acetate (MAK086, Sigma) in serum or liver tissue were measured with a colorimetric method as followed the manufacturer’s instructions.
Fecal DNA extraction and sequencing
Fecal samples were collected at 8 weeks and 19 weeks of feeding from four mice/group and stored at − 80 °C until analysis. Bacterial DNA was extracted with QIAamp Fast DNA Mini Kit (QIAGEN). Library preparation was performed according to Illumina’s TruSeq Nano DNA Sample Preparation protocol. The DNA samples were sheared on a Covaris E220 to ~ 450 bp, according to manufacturer recommendations, and each library was uniquely tagged with Illumina’s TruSeq HT DNA dual barcodes. Finished libraries were quantified with Invitrogen’s Picogreen assay, and the average library size was determined with a Bioanalyzer 2100 (Agilent). Library concentrations were normalized to 4 nM and validated by quantitative PCR (qPCR) on a ViiA-7 real-time thermocycler (Applied Biosystems), with the Kapa library quantification kit for Illumina platforms (Kapa Biosystems). The libraries were then pooled at equimolar concentrations and sequenced in two lanes on an Illumina HiSeq 2500 sequencer at a read length of paired-end 2 × 250 bp.
Sequencing assembly, alignment, and BLAST
The raw metagenomics Illumina paired-end reads were trimmed with cutadapt-1.8.1 software, with a quality cutoff of 20, and reads shorter than 30 base pairs were discarded. Next, the trimmed paired-end reads were locally assembled into a longer sequence, with a maximum mismatch density of 0.25, and maximum overlap of 10 base pairs, maximum mismatch density of 0.25, and maximum overlap of 251 base pairs. The merged metagenomics reads were then mapped to the mm10 mouse genome, as its sensitivity parameters. Any reads that could not be properly mapped to the mouse genome were separated and processed as microbiome reads. These microbiome reads were then aligned to sequences in the NCBI non-redundant protein database with the Diamond v. 0.85 tool. Based on these alignments, the microbial taxonomical classification was determined with the Lowest Common Ancestor (LCA) algorithm, implemented in MAGAN6, with the following classification parameters: maxmatches = 25, minscore = 120, and minsupport = 500.
Magnetic resonance imaging (MRI) and fat quantification
MRI scan was performed at 8 week and 17 weeks of diet feeding. MRI images were acquired with a 1 T micro-MRI system (Mediso NanoScan PET/MRI, Mediso, Hungary). Mice were anesthetized by inhalation of a mixture of isoflurane (1–2%) and medical air. Physiological parameters were monitored for each animal throughout the period of anesthesia. The two-point Dixon MRI method was performed to generate in-phase (water + fat) and out-of-phase (water–fat) images, which allows isolated measurements of fat and water [18].
The MRI images were acquired with a three-dimensional spoiled gradient echo sequence with the following parameters: TR = 17.1 ms, TE (in-phase/out-phase) = 6.4/3.2 ms, flip angle (FA) = 10°, matrix size = 256 × 256 mm, field of view (FOV) = 60 × 60 mm, slice thickness = 1 mm, number of averages = 3, number of slices = 30. Imaging data were post-processed with the script provided in MATLAB (MathWorks, Natick, MA). The fat-only (F) and water-only (W) measurements were evaluated from the in-phase (IP) and out-of-phase (OP) images, respectively, as follows:
A hepatic fat fraction (FF) map was then generated, as follows:
To derive the mean FF from each subject, we performed a region-of-interest (ROI) analysis on the FF map, in which ROIs were identified, according to mouse liver anatomy [19]. Briefly, the mouse liver was divided into four functionally independent segments, each with its own vascular inflow, outflow, and biliary drainage. One ROI was placed in each segment, with reference to the anatomy visualized on T2-weighted images. Care was taken to avoid large blood vessels when selecting the ROIs.
Staining and steatosis scoring
Liver tissue was fixed with 4% formaldehyde and incubated overnight at 4 °C. Then, tissues were processed, paraffin embedded, and sectioned at a thickness of 4 µm. Hematoxylin–eosin (H&E) staining was performed on each section. Stained tissue sections were assessed using Kleiner and Brunt scoring system for steatosis (grades 0–3) and fibrosis (grades 0–3) analysis [20].
Sample preparation for genomic DNA and RNA extractions
Livers were dissected and immediately snap-frozen in liquid nitrogen before storage at − 80 °C. The next day, frozen liver samples were crushed in liquid nitrogen and homogenized. Next, DNA and RNA extractions were performed with the AllPrep DNA/RNA mini kit (Qiagen), as per the manufacturer’s instructions. The purity of DNA/RNA was confirmed with a Nanodrop 2000 (Thermo Fisher Scientific), and quality was determined with gel electrophoresis on a Bioanalyzer (Agilent). Prior to reduced representation bisulphite sequencing (RRBS-seq), genomic DNA was quantified with a Qubit HS Assay (Thermo Fisher Scientific). We used 500 ng of total RNA to synthesize cDNA with iscript II (Biorad). We used 5 ng of the resulting cDNA per reaction for quantitative real-time PCR (qRT-PCR) analyses.
Reduced representation bisulphite sequencing
Genome-wide DNA methylation was profiled with RRBS-seq, as previously described [21]. Briefly, 100 ng of liver genomic DNA was digested with 1 unit of Msp1. The fragmented DNA was end-repaired and A-tailed with the NEBNext Ultra I DNA library prep kit for the Illumina instrument (New England Biolabs). Lambda DNA (Thermo Fisher Scientific) was spiked into the DNA sample to serve as an internal control; this served to calculate the bisulfite conversion efficiency. The adapter ligated DNA was subjected to bisulfite conversion with the EpiTect fast bisulfite conversion kit (Qiagen, 59824) with the following cycling conditions: 2 cycles of (95 °C for 5 min, 60 °C for 10 min, 95 °C for 5 min, and 60 °C for 10 min), and then, the temperature was held at 20 °C. Bisulfite converted DNA was PCR-amplified in 14–16 cycles with 2.5 U of Pfu Turbo Cx Hotstart DNA polymerase (Agilent Technologies, 600410). The products were size selected for fragments between 200 bp and 500 bp, with Ampure Xp magnetic beads (Agencourt, A63880). Purified DNA was subjected to single-end sequencing with the Illumina Hiseq 2500 at a 1 × 101 bp read length.
Differential methylation processing and analysis
RRBS-seq reads were aligned to the mouse reference genome, mm9, with a Bismark16 and the default parameters. To ensure high-quality data, CpGs with q < 30 and read depths less than 5 × were filtered out before calculating the percentage methylation (PM). The PM was calculated for each covered cytosine residue by taking the ratio of reads identified as methylated cytosines, divided by the total number of methylated and unmethylated reads. To achieve an accurate estimate of the methylation level, a high read cutoff was applied to eliminate PCR effects. CpGs that had coverages higher than the 99.9 percentile of reading counts were removed with the filterByCoverage function in the methylKit17 package. Because methylation occurs almost exclusively in the CpG context, we focused only on cytosines in CpG dinucleotides. A differentially methylated CpG (dmCpG) was defined as a CpG with a q value < 0.01 and a methylation difference ≥ 10% between samples.
Western blot analysis
Protein samples (40 ug) were separated with 10% SDS-PAGE and transferred to a PVDF membrane. Western blot analysis was performed using APOA4 (Cell Signaling Technology) and β-ACTIN (Santa Cruz) overnight at 4 °C. The next day, blots were washed and incubated with HRP-labelled secondary antibody. Enhanced chemiluminescence was used for signal detection (Santa Cruz). The western blot experiment was repeated in triplicate.
Statistical analysis
Results are presented as mean ± SD and were assessed by two-way analysis of variance (ANOVA) using diet and weeks as fixed factors, or one-way ANOVA using diet as the fixed factor. P values of < 0.05 were considered significant.
Results
Reversibility of liver remodeling
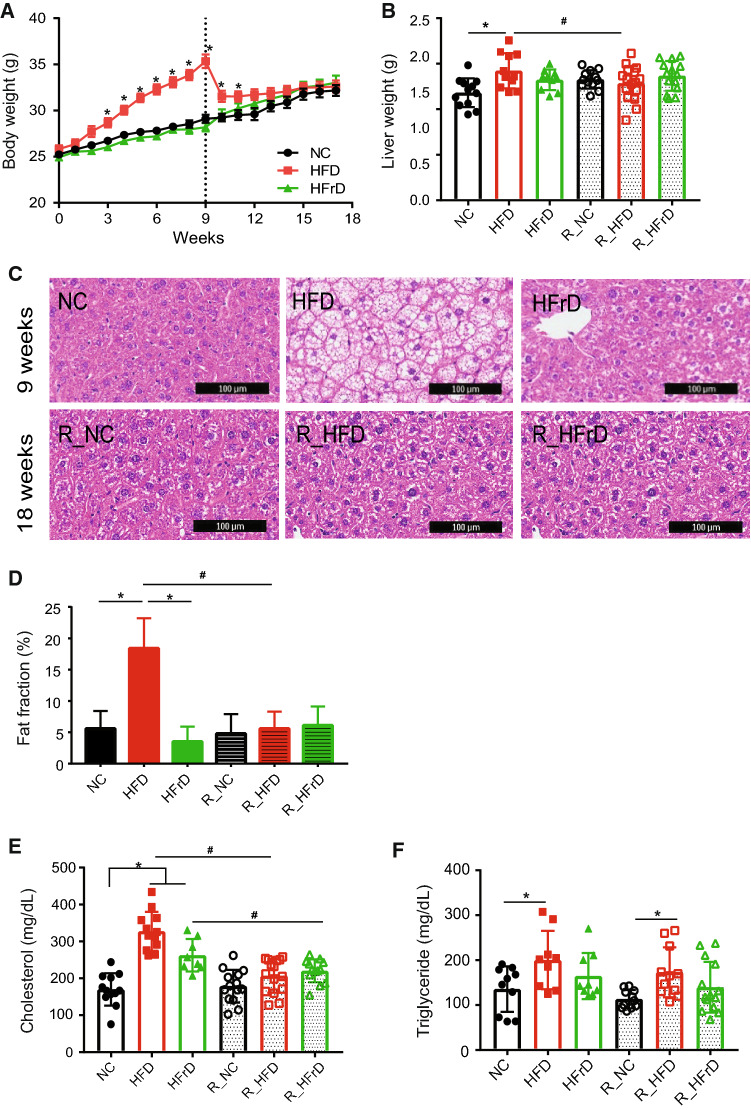
Following exposure to the dietary changes, the HFD group showed significant body weight gain compared to controls (NC group; Fig. 1a). In contrast, the HFrD group did not show any excess weight gain compared to controls, consistent with the previous reports [22]. Following the switch back to an NC diet, the reversal HFD group (R_HFD) showed a reduction in body weight within 2 weeks; in fact, they reached a body weight similar to that of the control group (R_NC; Fig. 1a). Food intake amounts in both the HFD and HFrD (Figure S2A) groups were similar than that of controls; however, caloric intake in the HFD group was significantly higher than in the HFrD or NC groups (Figure S2B).
Fig. 1.
HFD significant increases in body weight and liver fat, which returned to normal after diet reversal to NC. a Body weight changes. *P < 0.05 for NC vs. HFD, two-way ANOVA. (n = 13–16 mice per group) b Liver weight changes. *P < 0.05 for NC vs. HFD, #P < 0.05 for HFD vs. R_HFD, one-way ANOVA (n = 13–16 mice per group). c Representative H&E stains show fat content in liver tissue. (Top row) after test diet; (bottom row) after reversal to NC diet. The scale bar is 100 µm. d Quantification of liver fat fraction based on MRI image analysis. *P < 0.05 NC vs. HFD and HFD vs. HFrD, #P < 0.05 HFD vs. R_HFD, one-way ANOVA. (n = 3–5 mice per group). e Total serum cholesterol level. *P < 0.05 NC vs. HFD or HFrD, #P < 0.05 HFD vs. R_HFD, HFrD vs. R_HFrD t test (n = 13–16 mice per group). f Serum triglyceride levels. *P < 0.05 for NC vs. HFD and R_NC vs. R_HFD one-way ANOVA. (n = 9–16 mice per group)
Analogous to the changes in weight gain in the HFD group, we observed an increase in liver weight, which reverted after the mice returned to the NC diet (Fig. 1b). The increased liver weight in the HFD group was confirmed by normalizing to the tibia length (Figure S2C).
H&E staining showed significant fat accumulation in liver tissue within the HFD group, which reverted to normal after exposure to the NC diet (Fig. 1c). Correlating the grading of hepatic steatosis by liver US and the NAS score, grade 1 (61.54%) and grade 3 (38.46%) steatosis was found in HFD fed mice, whereas only grade 1 (54.55%) was found in HFrD fed mice. After reversal diet, most of the mice showed as Grade 0 steatosis. A mixture of micro-vesicular and macro-vesicular steatosis were shown in both HFD and HFrD diet group as well as R_HFD and R_HFrD group. In addition, HFD and HFrD both group showed partial fibrosis (38.46% in HFD, 27.27% in HFrD) and became grade 0 after reversal diet (Table 2). Hence, changing the diet in an animal model was sufficient to reverse the histological features and induce weight reduction.
Table 2.
Grades of steatosis and fibrosis in liver tissue by the Brunt scoring
| NC (n = 10) | HFD (n = 13) | HFrD (n = 11) | R_NC (n = 15) | R_HFD (n = 16) | R_HFrD (n = 14) | |
|---|---|---|---|---|---|---|
| Steatosis | ||||||
| Grade 0 | 8 (80%) | 5 (45.45%) | 14 (93.33%) | 12 (75%) | 4 (28.57%) | |
| Grade 1 | 2 (20%) | 8 (61.54%)* | 6 (54.55%) | 1 (6.67%) | 4 (14%) | 10 (71.43%) |
| Grade 2 | ||||||
| Grade 3 | 5 (38.46%)* | |||||
| Micro-vesicular | 12 (92.31%) | 5 (45.45%) | 1 (6.67%) | 4 (25%) | 10 (71.43%) | |
| Macro-vesicular | 9 (69.23%) | 3 (36.36%) | 1 (6.25%) | |||
| Fibrosis | ||||||
| Grade 0 | 10(100%) | 8 (61.54%) | 8 (72.73%) | 14 (93.33%) | 15 (93.75%) | 14 (100%) |
| Grade 1 | 5 (38.46%) | 3 (27.27%) | 1 (6.67%) | 1 (6.25%) | ||
Steatosis: grade 0: < 5% hepatocytes involved, grade 1: 5–33% hepatocytes involved, grade 2: 33–66% hepatocytes involved, grade 3: > 66% hepatocytes involved
Fibrosis: grade 0: no fibrosis, grade 1: zone3 perivenular, perisinusoidal, or pericellular fibrosis; focal and extensive
*P significant NC vs HFD, one way ANOVA in steatosis < 0.05
MRI quantifications of the liver fat fraction (FF) also showed an increase in the HFD group, which returned to levels similar to those found in the R_NC group after the reversal diet (Fig. 1d). Interestingly, the HFrD group did not gain excess weight or show major signs of lipid accumulation in the liver during the HFrD feeding period (Fig. 1c). We also found that the HFD group had impaired glucose metabolism, which paralleled the lipid accumulations; again, glucose metabolism returned to control levels after the reversal diet (Figure S3A, B). We also performed mRNA-level analysis to confirm inflammatory marker genes. We found toll-like receptor 4 (TLR4) and Interleukin 18 (IL-18), well-known pro-inflammatory markers, to be significantly upregulated in both HFD and HFrD groups (Figure S3 C and D). These elevated inflammatory markers were significantly decreased after reversal diet.
Both the HFD and HFrD group displayed significantly elevated levels of serum cholesterol (Fig. 1e); however, HFD only induced elevated triglyceride levels in the serum (Fig. 1f). Interestingly, although the cholesterol levels returned to normal levels in the R_HFD group, the elevated triglyceride levels persisted, despite the diet reversal (Fig. 1f). These findings indicate that the dietary intervention only partially rescues the NAFLD phenotype, irrespective of whether NAFLD was induced by an HFD or HFrD.
NAFLD-induced persistent changes in the gut microbiome composition
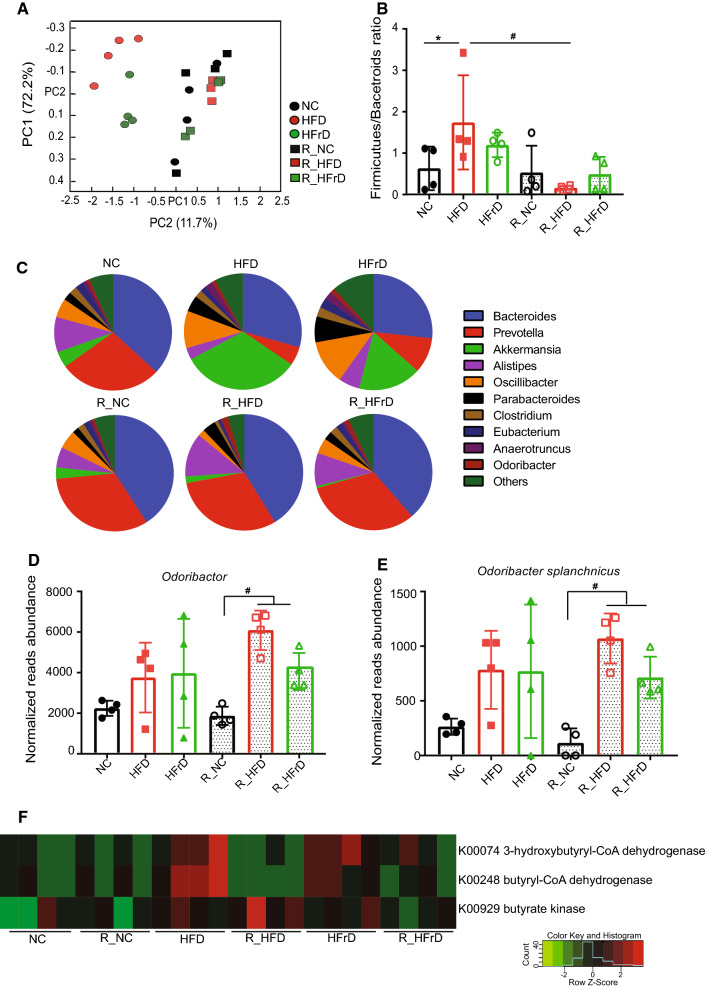
Research over the last two decades has demonstrated that alterations in microbiome composition were strongly correlated with changes in diet [23]. Therefore, it was of great interest to assess changes in the microbiome composition in different diet groups, before and after the dietary changes. Principle coordinate analysis (PCoA) plots showed that the microbiome composition changed after 8 weeks of HFD or HFrD; the HFD and HFrD clusters were clearly separate from the NC clusters (Fig. 2a). Interestingly, when we reversed the diet to NC, the HFD and HFrD clusters were similar to those in the R_NC group (Fig. 2a). We observed a significant increase in the ratio of Firmicutes/Bacteroidetes in the HFD group (Fig. 2b), consistent with the previous reports [24]. We also noticed a somewhat smaller increase in the Firmicutes/Bacteroidetes ratio in the HFrD group, possibly due to a smaller reduction in the amount of Bacteroidetes in this diet. A more detailed analysis revealed that the Prevotella and Alistipes genuses were considerably reduced in the HFD and HFrD groups, but they reverted with the change back to the NC diet (Fig. 2c and Figure S4A, B). These results were consistent with those in a previous report, which showed that Prevotella was reduced in children with NAFLD [25]. Another study showed that Alistipes was negatively correlated with liver cirrhosis [26]. Consequently, the persistent abundance of Alistipes in the reversal treatment groups warrants further investigation. Moreover, both the HFD and HFrD groups showed elevations in Oscillibacter, Parabacteroides, Clostridium, Anaerotruncus, Odoribacter, Desulfovibrio, and Akkermansia, and these species also recovered after the reversal diets (Fig. 2c and Figure S4A). Akkermansia showed the most significant increase among all the tested bacteria in both the HFD and HFrD diets (Figure S4C). This bacterium was previously reported to be positively correlated with metabolic disease. In that metagenomics study, Akkermansia was increased in rats fed a HFD and in patients with type 2 diabetes [27]. Nevertheless, a closer look showed that distinct trajectories in the diet groups appeared after reverting back to the NC. This finding indicated that diet-induced changes in the gut microbiome might have had a “priming effect”, which led to persistent metabolic effects.
Fig. 2.
Dietary changes induced significant changes in gut microbiome. (n = 4 mice per group). a PCA plot shows principle gut components after dietary changes and return to NC. bFirmicutes/Bacteroides ratios in dietary groups. *P < 0.05 for NC vs. HFD, #P < 0.05 for HFD vs. R_HFD, one-way ANOVA. c Pie chart shows microbiome composition and relative abundances of each species for each diet group. Normalized abundances of dOdoribacter; #P < 0.05 for R_NC vs. R_HFD or R_HFrD, one-way ANOVA and eOdoribacter splanchnicus; #P < 0.05 for R_NC vs. R_HFD or R_HFrD, one-way ANOVA. f Heatmap shows differential expression of butyrate synthesis-related enzymes, identified with KEGG pathways analysis
Indeed, we observed persistent microbiome changes that occurred after the diet reversals. Although most of the microbiome composition changes with the diets reversed with the NC diet, we found that Parabacteroides goldsteinii remained elevated in the R_HFD group (Figure S4D). This bacterium was previously shown to be protective, by exerting anti-inflammatory actions [28]. In addition, Odoribacter remained significantly elevated in both reversal diet groups (Fig. 2d). More specifically, we found that the Odoribacter splanchnicus species was persistently elevated after the reversal diet (Fig. 2e). Odoribacter is a known butyrate producer [29]; therefore, we checked the levels of key butyrate synthesis enzymes, such as butyrate kinase, 3-hydroxybutyryl-CoA dehydrogenase, and butyryl-CoA dehydrogenase, which were identified based on a KEGG pathway analysis. These enzymes were upregulated in the HFD and HFrD groups (Fig. 2f). Moreover, a persistent increase in butyrate kinase was observed in the R_HFD group (Figure S4E). This increase could be due to the lasting presence of Odoribacter, which would have a long-term effect on butyrate production.
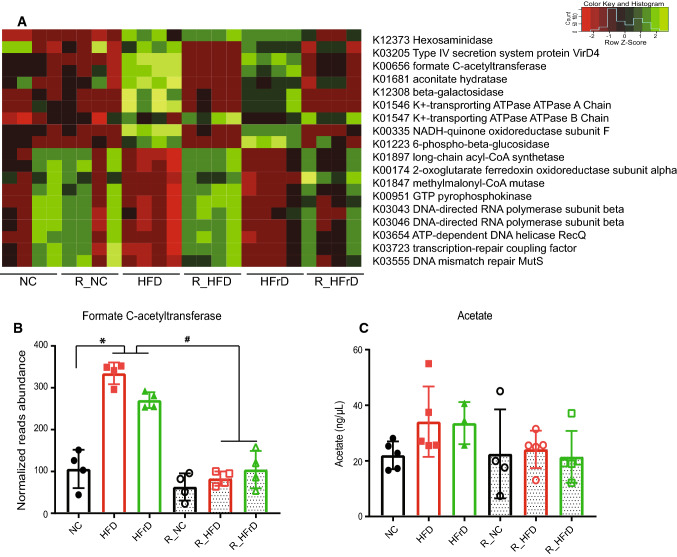
Based on another KEGG analysis, we observed that a set of genes related to the tricarboxylic acid (TCA) cycle and ATP generation were differentially expressed among the diet groups (Fig. 3a). Interestingly, formate C-acetyltransferase, which produces acetyl-CoA, was one of the most significantly enriched genes in the HFD and HFrD microbiomes (Fig. 3b). Acetyl-CoA is a TCA cycle precursor and a precursor of acetate. Acetate is involved in the biosynthesis of cholesterol and long chain fatty acids [30]. We observed an increase in acetate in the HFD and HFrD groups, although the change was not statistically significant (Fig. 3c).
Fig. 3.
Dietary changes induced changes in expression of genes related to metabolism. (n = 4 mice per group). a Heatmap shows differential expression of genes related to metabolism, identified in KEGG pathways analyses. b Normalized reads show abundance of formate C-acetyltransferase. *P < 0.05 for NC vs. HFD or HFrD, #P < 0.05 for HFD vs. R_HFD and HFD vs. R_HFrD, one-way ANOVA. c Serum acetate changes with diet
Persistent changes in liver methylation patterns after the HFD or HFrD
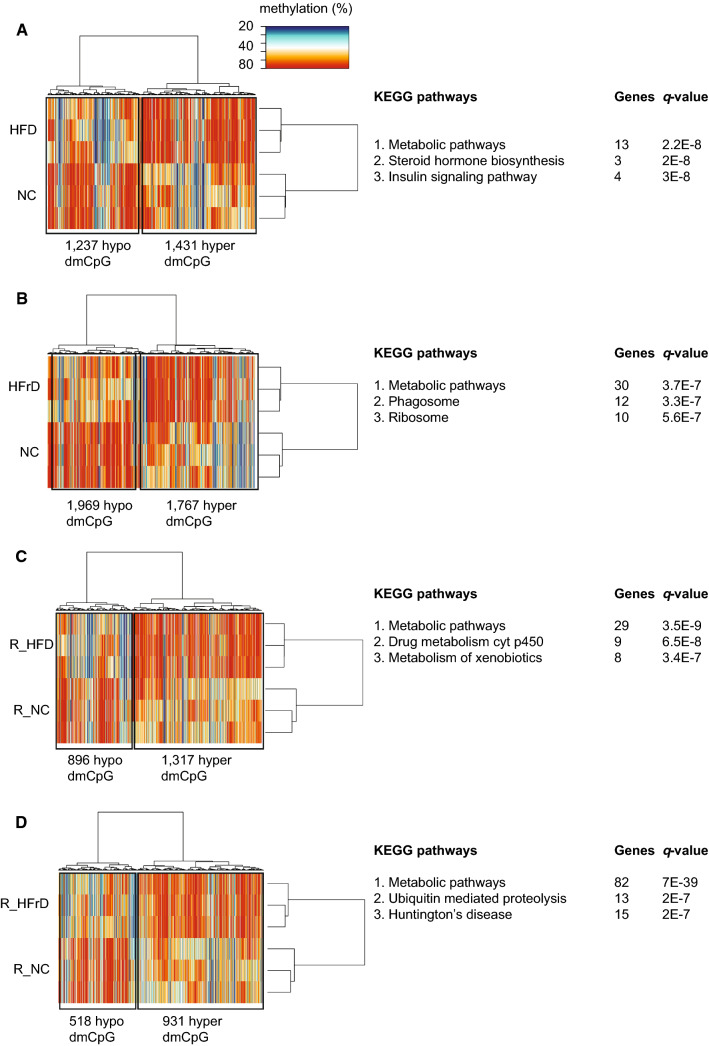
We next assessed whether dietary changes modified the epigenetic landscape in liver DNA. We performed genome-wide methylation study with RRBS-seq. Based on a cutoff value that indicated a 10% difference in methylation and an adjusted P value of ≤ 0.01, the test diets clustered with each other (Fig. 4a–d). These findings were consistent with the previous findings that global DNA methylation changes were induced in the liver by changes in the diet [31, 32]. A detailed analysis revealed that differentially methylated CpGs (dmCpGs) were found in intergenic or intronic regions, and promoter regions harbored 2–4% of dmCpGs (Figure S5A). When we plotted the average methylation levels of different genomic regions, we found that methylation was reduced around the transcription start site (TSS) in all samples (Figure S5B).
Fig. 4.
Unsupervised hierarchical clustering of all differentially methylated CpGs (dmCpGs). We applied a cutoff of ± 10% change in methylation and an adjusted P value (q) ≤0.05. (Right panels) results of KEGG pathways analyses show the most significantly enriched pathways, in the following diet comparisons: a HFD vs. NC; b HFrD vs. NC; c R_HFD vs. R_NC; d R_HFrD vs. R_NC. (n = 3 mice per group)
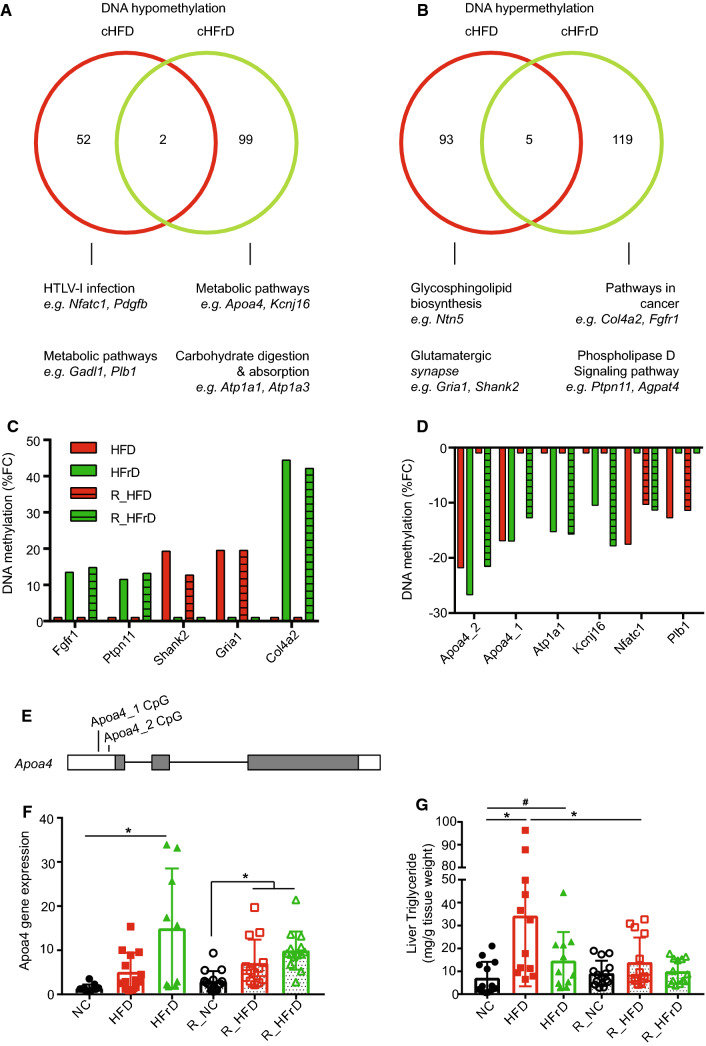
Interestingly, we noticed that a particular set of genes showed DNA methylation changes that persisted after reverting to the NC diet. With a principle coordinates analysis on common CpGs, we identified hypomethylated or hypermethylated genes in both the dietary and the reverse dietary groups. Among the most common hypomethylated promoters, we found 52 CpG loci in the HFD group (cHFD) and 99 CpG loci in the HFrD group (cHFrD). Only 2 CpG loci overlapped between the cHFD and cHFrD groups (Fig. 5a). Similarly, we found 93 hypermethylated promoters in the cHFD group and 119 hypermethylated promoters in the cHFrD group, with only 5 overlapping loci (Fig. 5b). A large fraction of the genes identified were linked to metabolic pathways, irrespective of the dietary regime. We analyzed the degree of methylation in these sets of hypermethylated (Fig. 5c) and hypomethylated genes (Fig. 5d), and connected them to metabolism-related KEGG pathways. This analysis revealed that the top persistently hypomethylated genes were Apoa4, Atp1a1, Kcnj16, Nfatc1, and Plb1, and the top persistently hypermethylated genes were Fgfr1, Ptpn11, Shank2, Gria1, and Col4a2.
Fig. 5.
Analysis of differential promoter methylation induced by diets. We applied a cutoff of ± 10% change in methylation and an adjusted P value of q ≤0.05. Methylation changes compared between HFD and R_HFD are termed cHFD; changes between HFrD and R_HFrD are termed cHFrD. a Venn diagram shows promoter CpG hypomethylation in cHFD (red) and cHFrD (green) and shared genes (n = 2); b Venn diagram shows promoter CpG hypermethylation on promoters for cHFD and cHFrD and shared genes (n = 5). (a, b, lower panels) Top KEGG pathways are shown for the promoters, where dmCpGs were found for cHFD and cHFrD. Fractional changes (FC) in the % methylation in a short list of c hypomethylated promoters and d hypermethylated promoters, selected based on relevance in KEGG pathways analyses. e Genomic diagram of the Apoa4 gene shows the locations of two hypomethylation sites; white bars indicate UTRs; grey bars indicate protein-coding exons. f Changes in Apoa4 gene expression with diets, measured with quantitative RT-PCR, and normalized to housekeeping gene expression levels. *P ≤ 0.05 for NC vs. HFrD, R_NC vs. R_HFD or R_HFrD, one-way ANOVA (n = 9–13 mice per group), g Triglyceride level in liver tissue. *P≤0.05 for NC vs. HFD, HFD vs. R_HFD, one-way ANOVA. #P≤0.05 NC vs. HFrD, t test (n = 12–16 mice per group)
Relevant to NAFLD and progression to liver cancer, we noticed that the Fgfr-1 gene was hypermethylated only in the HFrD and R_HFrD groups, but not in the HFD or R_HFD groups (Fig. 5c). The FGFR pathway is critical for HCC metastasis and survival [33, 34], and patients with HCC have elevated FGFR-1 levels [35]. HCC has been associated with high-fructose corn syrup [36]; this association might explain why it appeared in the HFrD/R_HFrD groups, but not the HFD/R_HFD groups. Prolonged changes in Fgfr-1 methylation associated with the HFrD might have influenced the upregulation of Fgfr-1 that was associated with HCC; this intriguing finding requires more mechanistic insight into the function of Fgfr-1 in NAFLD.
Among the hypomethylated genes, Apolipoprotein A4 (Apoa4), showed the highest fold change in both treatment groups (Fig. 5d). This hypomethylation was located at two CpG loci in the 5′-untranslated region (5′-UTR) of the Apoa4 gene (Fig. 5e). Promoter CpG hypomethylation is known to be involved in the upregulation of gene expression [37, 38]. Subsequently, we confirmed that Apoa4 gene expression was altered in these samples; the transcript was significantly upregulated in both the R_HFD and R_HFrD groups (Fig. 5f). We also found reduced TG levels in R_HFD group liver, as compared to R_NC group, indicating that there might be a functional level of Apoa4 in exporting TGs from the liver to the serum (Fig. 5f). When we investigated differences at protein levels of APOA4, we found an increase in APOA4 in HFD and HFrD groups (Figure S7); however, we saw no difference in R_HFD levels of APOA4 which may be due to the severity of the NAFLD at the point of reversal. In summary, we believe these epigenetic and expression changes to be initially driven by alterations in the microbiome caused by these diets, leading to long-term effects in liver epigenetics, and this relationship warrants further investigation.
Discussion
In this study, we report the dynamic responses of the gut microbiome and liver epigenetics following dietary-induced changes in the liver. We showed that after reversing from an HFD or HFrD to NC, elevations in serum triglyceride levels and gut Odoribacter levels persisted, and these elevations were associated with hypomethylation of the Apoa4 gene in liver DNA. These results suggest long-term dietary-induced changes in microbiome composition and liver DNA methylation, respectively. Importantly, we believe that this may have great relevance in how we view treatment of NAFLD in the clinic.
Our results are in support of the previous findings that HFD-induced NAFLD in mouse model, accompanied with increased body weight, fatty liver, and hyperglycemia [32, 39]. However, HFrD induced elevated serum cholesterol without significant fat accumulation in liver or body weight changes over the time period studied (Fig. 1). High intake of fructose is known to elicit changes in lipid metabolism; for example, Masateru Ushio et al. have reported that high-fructose diet-induced liver steatosis without body weight increase [40]. An additional study reported similar findings with no body weight increase, no change in triglyceride levels, and hyperglycemia [22]. In contrast, experimental exposure to high-fructose corn syrup (HFCS), which is commonly used to sweeten soft drinks, has reported significant phenotypic changes in obesity and increased triglyceride levels [41]. Our study shows a disruption in lipid metabolism due to an excess of the fructose component in HFrD group and is, therefore, consistent with earlier findings.
At the microbiome level, we noticed a persistent enrichment of Odoribacter, at the genus level, and of Odoribacter splanchnicus, at the species level, in both the R_HFD and R_HFrD groups (Fig. 2d, e). These results corroborate earlier findings of a persistent change in microbiome composition following changes in the dietary regime [42]. While the mechanisms, underlying the persistent changes in microbiome composition, remain to be identified, it is interesting to note that Odoribacter is well known for its ability to produce butyrate [29]. Given that butyrate is known to induce changes in DNA methylation by its ability to act as a histone deacetylate inhibitor [43], we found Odoribacter to be increased (Fig. 2d, e) and, in parallel, increased butyrate kinase genes with KEGG analysis (Fig. 2f) in both diet group. Highly speculatively, the increase in butyrate producing bacteria Odoribacter in the HFD group correlates with changes in DNA methylation, suggesting a possible link between microbes and changes in methylation patterns observed in this study.
The identification of persistent changes in liver DNA methylation in genes belonging to the DNA methyltransferases and the family of 10–11 translocation enzymes (TETs). These genes play key roles in de novo methylation and maintaining DNA methylation [43]. Our analysis show that all of these enzymes were elevated in the liver in both the R_HFD and R_HFrD groups with Tet2 and Tet3 enzymes being the most significantly increased in both groups (Figure S6). That is, the elevated expression of the Tet genes may one of several mechanisms to explain the de novo methylation activity observed in the R_HFD and R_HFrD groups.
The Apoa4 gene is predominantly expressed in intestine and liver, and is reported to be associated with active intestinal lipid absorption and chylomicron assembly [44]. Furthermore, Apoa4 is linked tightly with hepatic triglyceride export due to its ability to promote VLDL particle expansion within the secretory pathway [45]. Thus, the increased Apoa4 expression in the reversal group (Fig. 5f) may, therefore, execute a similar function in our study by exporting triglyceride from the liver to the serum. Indeed, the triglyceride level in R_HFD serum remains significantly increased compare with R_NC serum, but the liver triglyceride in R_HFD reverted to normal levels supporting the increased function of triglyceride export from the liver (Figs. 1f, 5g).
In conclusion, our results show persistent diet-induced changes in microbiome composition, dysregulation of the triglyceride regulation pathway and changes in liver DNA methylation, which have implications for future treatment regimes of NAFLD in humans.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary material 1 (PDF 355 kb) Figure S1. Illustration of the experimental schematic
Supplementary material 2 (PDF 393 kb) Figure S2. Daily food intakes for each diet. (A) Amount of daily food intake per mouse for 15 weeks. Food intake measured 2 times/week for 24 h and makes average for 1 week. (B) Daily caloric intake. *P < 0.05 for NC vs. HFD, two-way ANOVA (C) Liver weight, normalized to the tibia length (TA). *P < 0.05 for NC vs. HFD, one-way ANOVA
Supplementary material 3 (PDF 436 kb) Figure S3. IPGTT in mice show diet effects on glucose tolerance. Blood glucose levels were measured during IPGTTs, administered after (A) test diets and (B) reversal diets *P < 0.05 for NC vs. HFD, # < 0.05 for NC vs. HFrD, two-way ANOVA. IL18 (C) TLR4 (D) gene expression measured with quantitative RT-PCR, and normalized to housekeeping gene expression levels. * P≤0.05 for NC vs. HFD or HFrD, R_NC vs. R_HFD, one-way ANOVA
Supplementary material 4 (PDF 10945 kb) Figure S4. Changes in gut microbiome induced by diet changes. (A) Bubble chart shows top 30 bacterial genera from each diet group. Bubble sizes represent the relative size of gut bacteria populations. Bar graphs show changes in (B) Prevotella *P < 0.05 NC vs. HFD or HFrD; #P < 0.05 HFD vs. R_HFD; and HFrD vs. R_HFrD; one-way ANOVA; (C) Akkermansia *P < 0.05 NC vs. HFD or HFrD; #P < 0.05 HFD vs. R_HFD; and HFrD vs. R_HFrD, one-way ANOVA; and (D) Parabacteroides goldsteinii *P < 0.05 NC vs. HFrD; #P < 0.05 R_HFD vs. R_NC or R_HFrD. (E) Normalized abundance of butyrate kinase in gut microbiome; #P < 0.05 R-HFD vs. R_NC or R_HFrD
Supplementary material 5 (PDF 475 kb) Figure S5. Sites of differential methylation in DNA from mice fed HFD and HFrD diets. (A) Pie charts show the distribution of sites, where significant dmCpGs are found in DNA, during test diets and after returning to NC. UTR = untranslated region. (B) Traces show the average DNA methylation levels around the TSS and TES regions, where all significant dmCpGs are found, TSS = transcription start site, TES = transcription end site. dmCpG significance level was P≤0.05
Supplementary material 6 (PDF 386 kb) Figure S6. Heat map shows changes in gene expression levels for enzymes critical in DNA methylation, for each diet. Expression was measured with quantitative RT-PCR, normalized to housekeeping gene expression levels, and respective controls (NC and R_NC). Red indicates minimal change in gene expression; blue is a ~ twofold increase in gene expression
Supplementary material 7 (PDF 681 kb) Figure S7. (A) Representative western blot of APOA4 in the liver (n = 2) (B) Relative intensity of APOA4 in the liver (n = 8-10 mice per group). Levels were normalized with corresponding β-actin levels
Acknowledgements
We thank our colleagues, Alicia Kang, Llanto Elma Faylon, Kok Huan Teo, and Norhashimah Binte Sulaiman from Nanyang Technological University for their assistance in animal maintaining and data collections that greatly contributed the manuscript. We thank Li Yiqing from National University Health System for his assistance with animal experiments. We thank Saraf Sahil, Loh Jie Hua, and Sam Xin Xiu from Singapore General Hospital for their great help in steatosis scoring. We also thank Zenia Tiang from the Genome Institute of Singapore with her help in library submission for sequencing. This work was supported by the LKC School of Medicine Start Up Grant, MOE TIER 1 Grant, Grant from SCELSE and EU Grant TORNADO awarded to Sven Pettersson. This work also was supported a CSIRC award (National Medical Research Council Singapore) and a SPF Grant (Biomedical Research Council Singapore) awarded to Roger Foo.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Hyejin Kim and Oliver Worsley co-first authors in this manuscript.
Contributor Information
Roger SY Foo, Phone: +65-6601 1036, Email: foosyr@gis.a-star.edu.sg.
Pierce Kah Hoe Chow, Phone: +65 6576 2151, Email: pierce.chow.k.h@singheath.com.sg.
Sven Pettersson, Phone: +65 6904 7067, Email: Sven.Pettersson@ki.se.
References
- 1.Fatemeh Hadizadeh EF, Adibi Peyman. Nonalcoholic fatty liver disease: diagnostic biomarkers. World J Gastrointest Pathophysiol. 2017;8:11–26. doi: 10.4291/wjgp.v8.i2.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Romero-Gomez MZ-S, Trenell SM. Treatment of NAFLD with diet, physical activity and exercise. J Hepatol. 2017;67:829–846. doi: 10.1016/j.jhep.2017.05.016. [DOI] [PubMed] [Google Scholar]
- 3.Haufe S, Hass V, Utz W, Birkenfeld AL, Jeran S, Bohnke J, Mahler A, Luft FC, Schulz-Menger J, Boschmann M, Jordan J, Engeli S. Long-lasting improvements in liver fat and metabolism despite body weight regain after dietary weight loss. Diabetes Care. 2013;36:3786–3792. doi: 10.2337/dc13-0102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pugh CJS, Jones VS, Richardson H, Shojaee-Moradie P, Umpleby F, Green AM, Cable DJ, Trenell NT, Kemp MI, Cuthbertson GJ. Exercise-induced improvements in liver fat and endothelial function are not sustained 12 months following cessation of exercise supervision in nonalcoholic fatty liver disease. Int J Obes (Lond) 2016;40:1927–1930. doi: 10.1038/ijo.2016.123. [DOI] [PubMed] [Google Scholar]
- 5.Nicholson JK, Holmes E, Kinross J, Burcelin R, Gibson G, Jia W, Pettersson S. Host-gut microbiota metabolic interactions. Science. 2012;336:1262–1267. doi: 10.1126/science.1223813. [DOI] [PubMed] [Google Scholar]
- 6.Kundu P, Blacher E, Elinav E, Pettersson S. Our gut microbiome: the evolving inner self. Cell. 2017;171:1481–1493. doi: 10.1016/j.cell.2017.11.024. [DOI] [PubMed] [Google Scholar]
- 7.Leung C, Rivera L, Furness JB, Angus PW. The role of the gut microbiota in NAFLD. Nat Rev Gastroenterol Hepatol. 2016;13:412–425. doi: 10.1038/nrgastro.2016.85. [DOI] [PubMed] [Google Scholar]
- 8.Backhed F, Manchester JK, Semenkovich CF, Gordon JI. Mechanisms underlying the resistance to diet-induced obesity in germ-free mice. PNAS. 2007;104:979–984. doi: 10.1073/pnas.0605374104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Raman M, Ahmed I, Gillevet PM, Probert CS, Ratcliffe NM, Smith S, Greenwood R, Sikaroodi M, Lam V, Crotty P, Bailey J, Myers RP, Rioux KP. Fecal microbiome and volatile organic compound metabolome in obese humans with nonalcoholic fatty liver disease. Clin Gastroenterol Hepatol. 2013;11(868–75):e1–e3. doi: 10.1016/j.cgh.2013.02.015. [DOI] [PubMed] [Google Scholar]
- 10.Zhu L, Baker SS, Gill C, Liu W, Alkhouri R, Baker RD, Gill SR. Characterization of gut microbiomes in nonalcoholic steatohepatitis (NASH) patients: a connection between endogenous alcohol and NASH. Hepatology. 2013;57:601–609. doi: 10.1002/hep.26093. [DOI] [PubMed] [Google Scholar]
- 11.Jiao N, Baker SS, Chapa-Rodriguez A, Liu W, Nugent CA, Tsompana M, Mastrandrea L, Buck MJ, Baker RD, Genco RJ, Zhu R, Zhu L. Suppressed hepatic bile acid signalling despite elevated production of primary and secondary bile acids in NAFLD. Gut. 2018;67:1881–1891. doi: 10.1136/gutjnl-2017-314307. [DOI] [PubMed] [Google Scholar]
- 12.Kumar H, Lund R, Laiho A, Lundelin K, Ley RE, Isolauri E, Salminen S. Gut microbiota as an epigenetic regulator: pilot study based on whole-genome methylation analysis. MBio. 2014;5:e02113–e02114. doi: 10.1128/mBio.02113-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krautkramer KA, Kreznar JH, Romano KA, Vivas EI, Barrett-Wilt GA, Rabaglia ME, Keller MP, Attie AD, Rey FE, Denu JM. Diet-microbiota interactions mediate global epigenetic programming in multiple host tissues. Mol Cell. 2016;64:982–992. doi: 10.1016/j.molcel.2016.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sookoian S, Rosselli MS, Gemma C, Burgueno AL, Fernandez Gianotti T, Castano GO, Pirola CJ. Epigenetic regulation of insulin resistance in nonalcoholic fatty liver disease: impact of liver methylation of the peroxisome proliferator-activated receptor gamma coactivator 1alpha promoter. Hepatology. 2010;52:1992–2000. doi: 10.1002/hep.23927. [DOI] [PubMed] [Google Scholar]
- 15.Del Campo JA, Gallego-Duran R, Gallego P, Grande L. Genetic and epigenetic regulation in nonalcoholic fatty liver disease (NAFLD) Int J Mol Sci. 2018;19:911–921. doi: 10.3390/ijms19030911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murphy SK, Yang H, Moylan CA, Pang H, Dellinger A, Abdelmalek MF, Garrett ME, Ashley-Koch A, Suzuki A, Tillmann HL, Hauser MA, Diehl AM. Relationship between methylome and transcriptome in patients with nonalcoholic fatty liver disease. Gastroenterology. 2013;145:1076–1087. doi: 10.1053/j.gastro.2013.07.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pogribny IP, Tryndyak VP, Bagnyukova TV, Melnyk S, Montgomery B, Ross SA, Latendresse JR, Rusyn I, Beland FA. Hepatic epigenetic phenotype predetermines individual susceptibility to hepatic steatosis in mice fed a lipogenic methyl-deficient diet. J Hepatol. 2009;51:176–186. doi: 10.1016/j.jhep.2009.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gg H. Multipoint dixon technique for water and fat proton and susceptibility imaging. J Magn Reson Imaging. 1991;1:521–530. doi: 10.1002/jmri.1880010504. [DOI] [PubMed] [Google Scholar]
- 19.Fiebig T, Boll H, Figueiredo G, Kerl HU, Nittka S, Groden C, Kramer M, Brockmann MA. Three-dimensional in vivo imaging of the murine liver: a micro-computed tomography-based anatomical study. PLoS One. 2012;7:e31179. doi: 10.1371/journal.pone.0031179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Andrzej Gabriel MK, Ziółkowski Adam. Histopathological features and current scoring systmes for semiquantitative assessment of nonalcoholic fatty liver disease. Exp Clin Hepatol. 2008;4:48–54. [Google Scholar]
- 21.Gu H, Bock C, Mikkelsen TS, Jager N, Smith ZD, Tomazou E, Gnirke A, Lander ES, Meissner A. Genome-scale DNA methylation mapping of clinical samples at single-nucleotide resolution. Nat Methods. 2010;7:133–136. doi: 10.1038/nmeth.1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tillman EJ, Morgan DA, Rahmouni K, Swoap SJ. Three months of high-fructose feeding fails to induce excessive weight gain or leptin resistance in mice. PLoS One. 2014;9:e107206. doi: 10.1371/journal.pone.0107206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clarke SF, Murphy EF, O’Sullivan O, Ross RP, O’Toole PW, Shanahan F, Cotter PD. Targeting the microbiota to address diet-induced obesity: a time dependent challenge. PLoS One. 2013;8:e65790. doi: 10.1371/journal.pone.0065790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fukui H. Gut microbiota and host reaction in liver diseases. Microorganisms. 2015;3:759–791. doi: 10.3390/microorganisms3040759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Michail S, Lin M, Frey MR, Fanter R, Paliy O, Hilbush B, Reo NV. Altered gut microbial energy and metabolism in children with non-alcoholic fatty liver disease. FEMS Microbiol Ecol. 2015;91:1–9. doi: 10.1093/femsec/fiu002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qin N, Yang F, Li A, Prifti E, Chen Y, Shao L, Guo J, Le Chatelier E, Yao J, Wu L, Zhou J, Ni S, Liu L, Pons N, Batto JM, Kennedy SP, Leonard P, Yuan C, Ding W, Chen Y, Hu X, Zheng B, Qian G, Xu W, Ehrlich SD, Zheng S, Li L. Alterations of the human gut microbiome in liver cirrhosis. Nature. 2014;513:59–64. doi: 10.1038/nature13568. [DOI] [PubMed] [Google Scholar]
- 27.Qin J, Li Y, Cai Z, Li S, Zhu J, Zhang F, Liang S, Zhang W, Guan Y, Shen D, Peng Y, Zhang D, Jie Z, Wu W, Qin Y, Xue W, Li J, Han L, Lu D, Wu P, Dai Y, Sun X, Li Z, Tang A, Zhong S, Li X, Chen W, Xu R, Wang M, Feng Q, Gong M, Yu J, Zhang Y, Zhang M, Hansen T, Sanchez G, Raes J, Falony G, Okuda S, Almeida M, LeChatelier E, Renault P, Pons N, Batto JM, Zhang Z, Chen H, Yang R, Zheng W, Li S, Yang H, Wang J, Ehrlich SD, Nielsen R, Pedersen O, Kristiansen K, Wang J. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. 2012;490:55–60. doi: 10.1038/nature11450. [DOI] [PubMed] [Google Scholar]
- 28.Chang CJ, Lin CS, Lu CC, Martel J, Ko YF, Ojcius DM, Tseng SF, Wu TR, Chen YY, Young JD, Lai HC. Ganoderma lucidum reduces obesity in mice by modulating the composition of the gut microbiota. Nat Commun. 2015;6:7489. doi: 10.1038/ncomms8489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gomez-Arango LF, Barrett HL, McIntyre HD, Callaway LK, Morrison M, Dekker Nitert M, Group ST. Increased systolic and diastolic blood pressure is associated with altered gut microbiota composition and butyrate production in early pregnancy. Hypertension. 2016;68:974–981. doi: 10.1161/HYPERTENSIONAHA.116.07910. [DOI] [PubMed] [Google Scholar]
- 30.Subramanian S, Goodspeed L, Wang S, Kim J, Zeng L, Ioannou GN, Haigh WG, Yeh MM, Kowdley KV, O’Brien KD, Pennathur S, Chait A. Dietary cholesterol exacerbates hepatic steatosis and inflammation in obese LDL receptor-deficient mice. J Lipid Res. 2011;52:1626–1635. doi: 10.1194/jlr.M016246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yoon A, Tammen SA, Park S, Han SN, Choi S-W. Genome-wide hepatic DNA methylation changes in high-fat diet-induced obese mice. Nutr Res Pract. 2017;11:105–113. doi: 10.4162/nrp.2017.11.2.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou D, Hlady RA, Schafer MJ, White TA, Liu C, Choi J-H, Miller JD, Roberts LR, LeBrasseur NK, Robertson KD. High fat diet and exercise lead to a disrupted and pathogenic DNA methylome in mouse liver. Epigenetics. 2017;12:55–69. doi: 10.1080/15592294.2016.1261239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jo J-C, Choi EK, Shin J-S, Moon J-H, Hong S-W, Lee H-R, Kim S-M, Jung S-A, Lee D-H, Jung SH, Lee S-H, Kim JE, K-p Kim, Hong YS, Suh Y-A, Jang SJ, Choi EK, Lee JS, Jin D-H, Kim TW. Targeting FGFR pathway in human hepatocellular carcinoma: expressing pFGFR and pMET for antitumor activity. Mol Cancer Ther. 2015;14:2613–2622. doi: 10.1158/1535-7163.MCT-14-0780. [DOI] [PubMed] [Google Scholar]
- 34.Cheng AL, Shen YC, Zhu AX. Targeting fibroblast growth factor receptor signaling in hepatocellular carcinoma. Oncology. 2011;81:372–380. doi: 10.1159/000335472. [DOI] [PubMed] [Google Scholar]
- 35.Wang J, Li J, Wang X, Zheng C, Ma W. Downregulation of microRNA-214 and overexpression of FGFR-1 contribute to hepatocellular carcinoma metastasis. Biochem Biophys Res Commun. 2013;439:47–53. doi: 10.1016/j.bbrc.2013.08.032. [DOI] [PubMed] [Google Scholar]
- 36.Laguna JC, Alegret M, Roglans N. Simple sugar intake and hepatocellular carcinoma: epidemiological and mechanistic insight. Nutrients. 2014;6:5933–5954. doi: 10.3390/nu6125933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 38.Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat Rev Genet. 2008;9:465–476. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- 39.Park EY, Choi H, Yoon JY, Lee IY, Seo Y, Moon HS, Hwang JH, Jun HS. Polyphenol-rich fraction of Ecklonia cava improves nonalcoholic fatty liver disease in high fat diet-fed mice. Mar Drugs. 2015;13:6866–6883. doi: 10.3390/md13116866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ushio M, Nishio Y, Sekine O, Nagai Y, Maeno Y, Ugi S, Yoshizaki T, Morino K, Kume S, Kashiwagi A, Maegawa H. Ezetimibe prevents hepatic steatosis induced by a high-fat but not a high-fructose diet. Am J Physiol Endocrinol Metab. 2013;305:E293–E304. doi: 10.1152/ajpendo.00442.2012. [DOI] [PubMed] [Google Scholar]
- 41.Bocarsly ME, Powell ES, Avena NM, Hoebel BG. High-fructose corn syrup causes characteristics of obesity in rats: increased body weight, body fat and triglyceride levels. Pharmacol Biochem Behav. 2010;97:101–106. doi: 10.1016/j.pbb.2010.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thaiss CA, Itav S, Rothschild D, Meijer M, Levy M, Moresi C, Dohnalova L, Braverman S, Rozin S, Malitsky S, Dori-Bachash M, Kuperman Y, Biton I, Gertler A, Harmelin A, Shapiro H, Halpern Z, Aharoni A, Segal E, Elinav E. Persistent microbiome alterations modulate the rate of post-dieting weight regain. Nature. 2016;540:544–551. doi: 10.1038/nature20796. [DOI] [PubMed] [Google Scholar]
- 43.Deaton AM, Bird A. CpG islands and the regulation of transcription. Genes Dev. 2011;25:1010–1022. doi: 10.1101/gad.2037511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang F, Kohan AB, Lo CM, Liu M, Howles P, Tso P. Apolipoprotein A-IV: a protein intimately involved in metabolism. J Lipid Res. 2015;56:1403–1418. doi: 10.1194/jlr.R052753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.VerHague MA, Cheng D, Weinberg RB, Shelness GS. Apolipoprotein A-IV expression in mouse liver enhances triglyceride secretion and reduces hepatic lipid content by promoting very low density lipoprotein particle expansion. Arterioscler Thromb Vasc Biol. 2013;33:2501–2508. doi: 10.1161/ATVBAHA.113.301948. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material 1 (PDF 355 kb) Figure S1. Illustration of the experimental schematic
Supplementary material 2 (PDF 393 kb) Figure S2. Daily food intakes for each diet. (A) Amount of daily food intake per mouse for 15 weeks. Food intake measured 2 times/week for 24 h and makes average for 1 week. (B) Daily caloric intake. *P < 0.05 for NC vs. HFD, two-way ANOVA (C) Liver weight, normalized to the tibia length (TA). *P < 0.05 for NC vs. HFD, one-way ANOVA
Supplementary material 3 (PDF 436 kb) Figure S3. IPGTT in mice show diet effects on glucose tolerance. Blood glucose levels were measured during IPGTTs, administered after (A) test diets and (B) reversal diets *P < 0.05 for NC vs. HFD, # < 0.05 for NC vs. HFrD, two-way ANOVA. IL18 (C) TLR4 (D) gene expression measured with quantitative RT-PCR, and normalized to housekeeping gene expression levels. * P≤0.05 for NC vs. HFD or HFrD, R_NC vs. R_HFD, one-way ANOVA
Supplementary material 4 (PDF 10945 kb) Figure S4. Changes in gut microbiome induced by diet changes. (A) Bubble chart shows top 30 bacterial genera from each diet group. Bubble sizes represent the relative size of gut bacteria populations. Bar graphs show changes in (B) Prevotella *P < 0.05 NC vs. HFD or HFrD; #P < 0.05 HFD vs. R_HFD; and HFrD vs. R_HFrD; one-way ANOVA; (C) Akkermansia *P < 0.05 NC vs. HFD or HFrD; #P < 0.05 HFD vs. R_HFD; and HFrD vs. R_HFrD, one-way ANOVA; and (D) Parabacteroides goldsteinii *P < 0.05 NC vs. HFrD; #P < 0.05 R_HFD vs. R_NC or R_HFrD. (E) Normalized abundance of butyrate kinase in gut microbiome; #P < 0.05 R-HFD vs. R_NC or R_HFrD
Supplementary material 5 (PDF 475 kb) Figure S5. Sites of differential methylation in DNA from mice fed HFD and HFrD diets. (A) Pie charts show the distribution of sites, where significant dmCpGs are found in DNA, during test diets and after returning to NC. UTR = untranslated region. (B) Traces show the average DNA methylation levels around the TSS and TES regions, where all significant dmCpGs are found, TSS = transcription start site, TES = transcription end site. dmCpG significance level was P≤0.05
Supplementary material 6 (PDF 386 kb) Figure S6. Heat map shows changes in gene expression levels for enzymes critical in DNA methylation, for each diet. Expression was measured with quantitative RT-PCR, normalized to housekeeping gene expression levels, and respective controls (NC and R_NC). Red indicates minimal change in gene expression; blue is a ~ twofold increase in gene expression
Supplementary material 7 (PDF 681 kb) Figure S7. (A) Representative western blot of APOA4 in the liver (n = 2) (B) Relative intensity of APOA4 in the liver (n = 8-10 mice per group). Levels were normalized with corresponding β-actin levels