Abstract
HMG box protein 1 (HBP1) is a transcription factor and a potent cell cycle inhibitor in normal and cancer cells. HBP1 activates or represses the expression of different cell cycle genes (such as CDKN2A, CDKN1A, and CCND1) through direct DNA binding, cofactor recruitment, chromatin remodeling, or neutralization of other transcription factors. Among these are LEF1, TCF4, and MYC in the WNT/beta-catenin pathway. HBP1 also contributes to oncogenic RAS-induced senescence and terminal cell differentiation. Collectively, these activities suggest a tumor suppressor function. However, HBP1 is not listed among frequently mutated cancer driver genes. Nevertheless, HBP1 expression is lower in several tumor types relative to matched normal tissues. Several micro-RNAs, such as miR-155, miR-17-92, and miR-29a, dampen HBP1 expression in cancer cells of various origins. The phosphatidylinositol-3 kinase (PI3K)/AKT pathway also inhibits HBP1 transcription by preventing FOXO binding to the HBP1 promoter. In addition, AKT directly phosphorylates HBP1, thereby inhibiting its transcriptional activity. Taken together, these findings place HBP1 at the center of a network of micro-RNAs and oncoproteins that control cell proliferation. In this review, we discuss our current understanding of HBP1 function in human physiology and diseases.
Keywords: Retinoblastoma protein RB, Ataxin, HDAC, Ubiquitin ligase, DNA methylation, Cell growth arrest
Introduction
The high-mobility group (HMG) box protein 1 (HBP1) cDNA was cloned serendipitously in 1994 in an effort to identify potassium channels by complementation of a mutant yeast strain [1]. HBP1 was devoid of transporter activity but had a predicted HMG box DNA-binding domain, suggesting a transcription factor function. Three years later, two independent groups identified mouse HBP1 in two-hybrid screens using pocket proteins of the retinoblastoma (RB) family as baits, suggesting for the first time a role for HBP1 in the cell cycle [2, 3]. Since then, a large number of studies have confirmed that HBP1 acts as a cell cycle inhibitor in cell culture and mouse models. Because it also promotes growth arrest in cancer cells, HBP1 has drawn much attention as a putative tumor suppressor. In this respect, HBP1 is part of an oncoprotein network that includes AKT, MYC, RAS, RB and β-catenin. HBP1 primarily functions as a transcriptional repressor, although activation of target genes has also been reported. This review will highlight recent progress in understanding the role of HBP1, as well as remaining questions regarding its mechanism of action and implication in human diseases.
HBP1 structure and transcriptional activity
The human HBP1 gene encodes a 514-aa protein, which is highly conserved in vertebrates [4]. HBP1 protein architecture is unique among human proteins, combining an ataxin homology (AXH) domain and the C-terminal HMG-box (Fig. 1). In addition, two nuclear localization signals (NLS) are present in the C-terminus.
Fig. 1.
HBP1 structure, posttranslational modifications and interactions. The HBP1 protein comprises three major domains: a transcriptional activation domain (in green), a transcriptional repression domain (in red), and an HMG-box DNA-binding domain (in blue). The activation and repression domains of HBP1 were defined by Lavender et al. [3] and Sampson et al. [26], respectively. Posttranslational modification sites of HBP1 are indicated below the HBP1 structure: Ac, acetylation (in orange); P, phosphorylation (in blue by PIM-1, in green by p38, in Bordeaux by AKT); Ub, ubiquitination (in gray). Proteins that directly interact with HBP1 are represented above HBP1 at their described binding site. See text for details
The HMG box is a basic DNA-binding domain of 75-aa residues, which are ±30% identical in members of the T-cell-specific transcription factor/lymphoid enhancer factor (TCF/LEF) and the SRY-related HMG box (SOX) families [5]. The HMG box of HBP1 is more distantly related to that of the HMGB1 family of alarmins. An analysis of HBP1-binding sites in several target gene promoters identified unrelated DNA sequences (Fig. 2). Several reports have suggested that HBP1 binds with a low affinity to the 5′-(A/T)(G/C)AATGGG-3′ sequence, which is related to classical HMG-box-binding sites, and with a higher affinity to 5′-TTCATTCATTCA-3′ [2, 6].
Fig. 2.

Mechanisms of transcriptional regulation by HBP1. HBP1 is mainly characterized as a transcriptional repressor, which sequesters other transcription factors, such as LEF, TCF4 or MYC (a), or directly represses gene expression by recruiting cofactors such as HDAC or Sin3 (b). HBP1 can also directly activate the transcription of genes, for instance CDKN2A, by recruiting CBP/p300 (c). HBP1 was reported to stabilize p53 by preventing its degradation by the MDM2 ligase, resulting in upregulation of the p53 target gene CDKN1A (D). Finally, by lowering the expression of DNMT1 and EZH2, HBP1 prevents the deposition of repressive epigenetic marks (methylation of DNA and histone H3K27, respectively), thereby increasing the expression of target genes (e). See text for details
In addition to its DNA-binding domain, HBP1 features a well-characterized transcriptional repressor region, which includes the AXH domain. This domain is also present in ataxin-1, a protein that interacts with chromatin modifiers and can aggregate, which plays a role in human neurodegenerative disorders [7]. de Chiara and colleagues [8] solved the structure of HBP1 AXH in solution and showed that it adopts an alternative fold compared with that of ataxin-1. As a result, HBP1 does not form aggregates but retains the ability to bind to RNA homopolymers. This intriguing capacity of HBP1 to bind to RNA has not been further investigated. This region also interacts with histone deacetylases (HDACs), such as HDAC4, which contributes to HBP1 repressor activity (Fig. 1) [9]. In addition to the AXH domain, several other motifs play roles in gene repression by HBP1. The corepressor Sin3 binds to a region located close to the AXH domain [10]. Sin3 functions as a molecular scaffold that recruits multiple chromatin-modifying enzymes, including HDACs. HBP1 also represses gene expression by neutralizing other transcription factors, such as LEF1, TCF4 or MYC (Figs. 1, 2).
In addition to this well-described repressor activity, HBP1 can also stimulate the expression of a few genes, such as CDKN2A, which encodes p16INK4A (Fig. 2) [9, 11]. HBP1 can recruit p300/CBP coactivators, which is consistent with its capacity to activate transcription [9]. In addition, Lavender et al. suggested that HBP1 includes a transactivator domain in its N-terminus. However, this domain is masked by two independent N- and C-terminal inhibitory regions, which must be deleted to identify HBP1 transcriptional activator potential in a reporter assay [3].
As mentioned above, HBP1 was cloned as a partner of the retinoblastoma protein RB and the related factor p130. Mutagenesis experiments identified two distinct RB-binding sites within HBP1: a high-affinity motif (LXCXE) in the N-terminal region and a low-affinity motif (IXCXE) in the repression domain (Fig. 1) [2, 3]. Mutation of the high-affinity site does not affect HBP1 function, while mutation of both sites prevents the regulation of several genes, including N-Myc and CDKN2A, suggesting that RB is required for several HBP1 functions [2, 12]. However, the AXH structure suggests that the second site is not accessible for direct binding by RB, questioning the interpretation of experiments performed with the double HBP1 mutant [8]. In addition, overexpression of RB blunts the ability of HBP1 to regulate transcription, suggesting that RB may also sequester HBP1 [3, 13]. The mechanisms whereby RB regulates HBP1 remain to be clarified.
Posttranslational modifications of HBP1
Multiple posttranslational modifications, including ubiquitination, phosphorylation and acetylation, regulate HBP1 protein abundance and transcriptional activity (Fig. 1). HBP1 is polyubiquitinated by ubiquitin ligases, which target it for degradation by proteasomes. This is mediated at least in part by the cullin–RING ligases (CRL) 3 and 4, which were reported to interact with HBP1 and ubiquitinate Lys16 and Lys398 [14, 15]. Consequently, proteasome inhibitors, such as MG-132, and the CRL inhibitor pevonedistat (MLN-4924) stabilize HBP1 in multiple cell types (reference [14] and our unpublished observations). Recently, a large E3 ligase complex named human glucose-induced degradation deficient (GID) or C-terminal to LisH (CTLH) was also shown to target HBP1 for degradation [16]. Taken together, these observations indicate that HBP1 protein levels are tightly controlled by the ubiquitin–proteasome pathway.
HBP1 phosphorylation on different sites also controls its degradation. The p38 mitogen-activated protein kinase (MAPK) phosphorylates HBP1 at Ser402, inducing its stabilization and, subsequently, a cell cycle arrest at the G1 phase. Phosphorylation at Ser402 protects HBP1 from proteasomal degradation but has no effect on its intrinsic transcriptional activity. A specific binding site for p38 MAPK was identified in the N-terminal region of HBP1 (amino acids 135-152) [17]. Another kinase that may prevent HBP1 degradation is PIM-1. Under oxidative stress, PIM-1 was shown to bind to HBP1 and phosphorylate Ser372 and Ser380, promoting its accumulation and transcriptional activation [18]. By contrast, phosphorylation by AKT on three sites, Ser380, Thr484 and Ser509, does not impact HBP1 expression but reduces its transcriptional activity [11]. The opposite effects of AKT and PIM-1 on HBP1 function are surprising because these two oncogenic kinases share a number of other substrates and regulate them in a similar manner [19].
In addition to ubiquitination and phosphorylation, acetylation also modulates HBP1 function. The acetyltransferase p300/CBP acetylates HBP1 on five lysine residues (Lys171, Lys297, Lys305, Lys307 and Lys419), increasing its transcriptional activity [9]. Acetylation of Lys419 appears to be essential for HBP1 transactivation, enhancing HBP1-mediated premature senescence. HDAC4 was shown to reverse this process by deacetylating HBP1 [9].
HBP1 target genes and cellular functions
HBP1 can regulate gene transcription by multiple mechanisms summarized in Fig. 2. We already mentioned direct binding to gene promoters and interaction with other transcription factors, such as MYC or TCF4. HBP1 also regulates chromatin remodeling, which may amplify its effects on the transcription of other target genes. For instance, HBP1 represses the expression of DNA methyltransferase 1 (DNMT1), leading to global DNA hypomethylation in some cell lines [20]. HBP1 also downregulates EZH2 expression and histone methylation during senescence [20, 21]. Furthermore, HBP1 promotes the expression of histone H10, which is a specific chromatin-associated protein [22]. The full list of reported HBP1 target genes is presented in Fig. 3.
Fig. 3.
HBP1 target genes. HBP1 target genes are classified according to their function (left). Reported direct HBP1-binding sites are indicated on the right side. Some target genes are regulated in an indirect manner through interactions with transcription factors or cofactors
Cell cycle and WNT signaling
HBP1 inhibits the progression from G1 to S phase of the cell cycle by regulating several key genes involved in cell proliferation (Fig. 3). HBP1 was shown to bind to the promoter of N-Myc and repress its expression (Fig. 2) [2]. HBP1 prevents c-Myc binding to the promoter of its target genes, such as E2F2 (Fig. 2) [23]. HBP1 also represses the promoter of the NCF1 gene, which encodes the NADPH oxidase p47phox, regulating reactive oxygen species (ROS) levels and response to growth factors [24]. Finally, HBP1 induces the expression of two cyclin-dependent kinase inhibitors, CDKN2A (p16INK4A) and CDKN1A (p21CIP1) [11, 21, 25]. The coordinated regulation of these genes by HBP1 produces cell growth arrest in many cell types (Fig. 4).
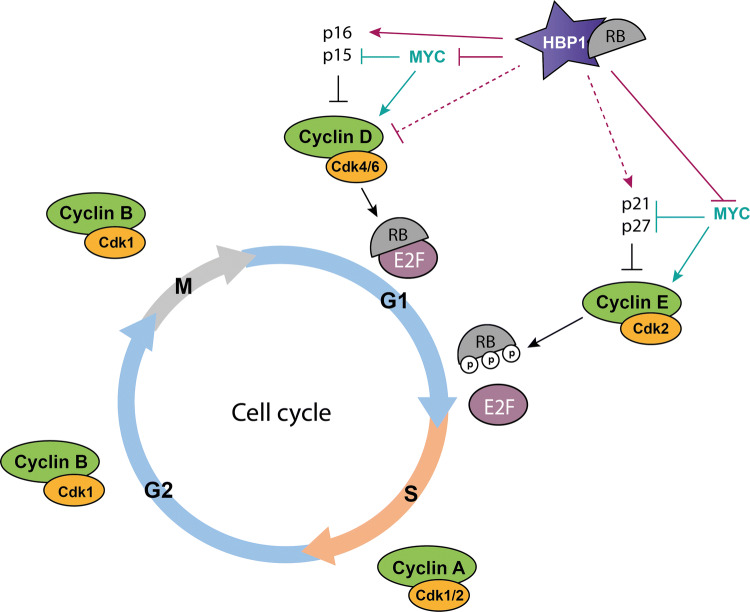
Fig. 4.
Cell cycle regulation by HBP1. HBP1 inhibits G1- to S-phase progression in many cell types. HBP1 upregulates the transcription of the cyclin-dependent kinase inhibitors p21 and p16, and represses the expression of cyclin D1. In addition, HBP1 inhibits the transcriptonal activity of MYC. HBP1 was shown to interact with the retinoblastoma protein RB, another key cell cycle regulator. Activation (arrows) and inhibition (crosshead arrows) are indicated. Dashed lines represent indirect regulation. P phosphorylation
In addition to direct gene regulation, HBP1 also inhibits the WNT/β-catenin pathway, which plays a particularly important role in the cell cycle (Fig. 5) [26]. This signaling pathway leads to the nuclear accumulation of β-catenin, which binds to TCF/LEF factors and subsequently activates genes such as c-Myc and cyclin D1. HBP1 binds to TCF4 and prevents the regulation of WNT target genes [27]. Multiple studies have confirmed that WNT pathway inhibition by HBP1 results in c-Myc downregulation. In addition, HBP1 can directly interact with the c-Myc and N-Myc proteins and inhibit their transcriptional activity (Fig. 2) [23, 28].
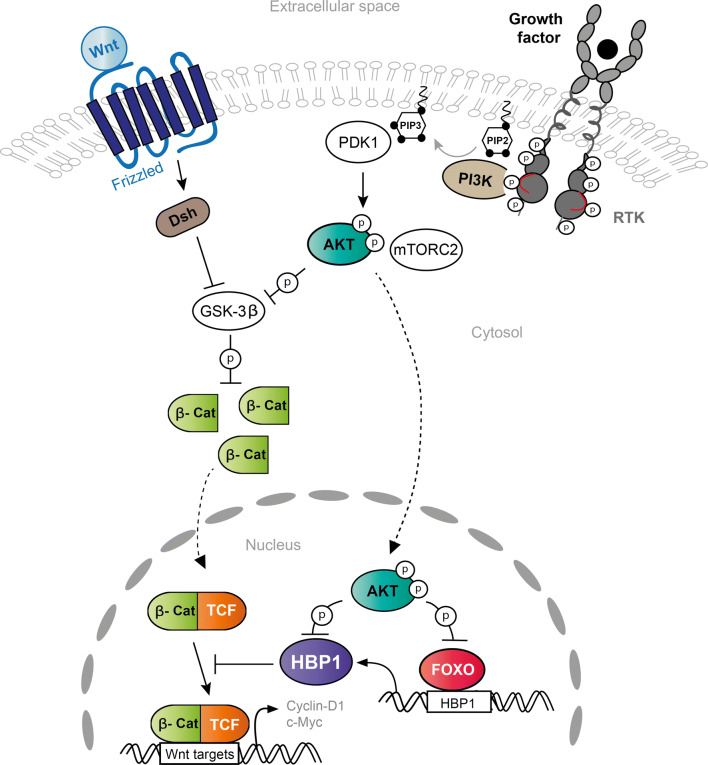
Fig. 5.
Interaction of HBP1 with the WNT/β-catenin and PI3K/AKT pathways. Activation of the WNT signaling pathway prevents β-catenin (β-cat) phosphorylation by GSK3β and degradation. In the nucleus, β-catenin binds to TCF/LEF factors and activates genes, including c-Myc and cyclin D1. HBP1 interacts with TCF and prevents TCF binding to WNT target genes. In addition, activation of the PI3K pathway by receptor tyrosine kinases (RTK) leads to AKT activation. AKT phosphorylates and inhibits GSK3β, FOXO transcription factors and HBP1. By doing so, AKT prevents FOXO binding to the HBP1 promoter, leading to a decrease in HBP1 expression, and blunts HBP1 activity by direct phosphorylation (P). Activation and inhibition are depicted by arrows and crosshead arrows, respectively, while cytosol/nuclear shuttling is represented by dashed lines
Oncogene-induced senescence
Several studies have suggested a role for HBP1 in RAS-induced premature senescence, which prevents early tumorigenic transition [12, 20, 25]. Oncogenic RAS activation triggers premature senescence in normal primary cells partly through the activation of p38 MAPK, which phosphorylates and stabilizes HBP1 [17]. This process depends on the association of HBP1 with RB [29] and allows HBP1 to directly regulate two target genes involved in premature senescence. First, HBP1 enhances the expression of CDKN2A (p16INK4A) [9, 25]. This process is facilitated by recruitment of HAT to the CDKN2A promoter and acetylation of HBP1 at Lys419 [9]. Second, as mentioned above, HBP1 downregulates DNMT1 expression, inducing global DNA hypomethylation, which is a critical process during cell senescence [20]. CDKN2A promoter demethylation was also suggested to facilitate HBP1 recruitment. In addition to the direct regulation of CDKN2A/p16INK4A and DNMT1, HBP1 contributes to cellular senescence by upregulating the expression of CDKN1A (p21CIP1) through indirect mechanisms (Figs. 2, 4): (1) HBP1 can stabilize p53, a direct activator of CDKN1A, by inhibiting its ubiquitination by MDM2; and (2) HBP1 reduces the methylation of the CDKN1A promoter by preventing TCF4-induced expression of the histone methyltransferase EZH2 and by directly repressing the expression of DNMT1 [20, 21]. In conclusion, HBP1 contributes to oncogene-induced senescence by multiple mechanisms, supporting a tumor suppressor function.
Cell differentiation
Lesage and colleagues had already noticed in 1994 that HBP1 expression increased during the differentiation of myogenic C2C12 cells and Ob1771 preadipocytes [1]. A number of additional reports have linked HBP1 to the differentiation of several cell lines (Fig. 3). As mentioned above, HBP1 positively regulates the transcription of histone H10, which is associated with terminal differentiation [22]. HBP1 also directly induces the expression of myeloperoxidase (MPO), a marker of myeloid differentiation, in the leukemic myeloid cell line K562 [30]. The same research group demonstrated that overexpression of HBP1 also increases the expression of megakaryocytic and erythroid markers, such as RUNX1 and GATA-1, respectively [31]. Similarly, HBP1 induces terminal adipocyte differentiation by regulating C/EBPα expression [32]. In contrast, overexpression of HBP1 prevents terminal differentiation of muscle C2C12 cells. In these cells, HBP1 inhibits the activity of the MyoD family of transcription factors [13]. Interestingly, this could be reverted by RB overexpression. Because these observations are mostly based on cell line studies, their physiological relevance remains to be established. More convincing is the report by Borrelli and colleagues, who showed that HBP1 contributes to the control of skin differentiation and stratification by p63, a transcription factor of the p53 family. HBP1 expression, which is repressed by p63, correlates with keratinocyte differentiation and cell cycle arrest in organotypic skin cultures [33]. This is also consistent with the higher expression of HBP1 in the upper layers of human skin.
Role of HBP1 in cancer
The role of HBP1 in cell cycle arrest and senescence supports the hypothesis that HBP1 acts as a tumor suppressor. However, the types of cancer gene alterations that are associated with bona fide tumor suppressors, such as TP53 and RB, have not been reported for the HBP1 gene. Paulson et al. described HBP1 mRNA variants that have lost part of the repression domain and the HMG-box in invasive breast cancer samples, but this observation has not been confirmed yet in large breast cancer genomics studies [34]. Nevertheless, the HBP1 chromosomal region (7q22-31) is frequently lost in breast cancer and myeloid leukemia [35–37]. Loss of heterozygosity at 7q31 has also been reported in ovarian, prostate and colon carcinomas [38–40]. However, one should bear in mind that other candidate tumor suppressor genes are present in this region. In addition to the deletion of the HBP1 locus, HBP1 mRNA expression is downregulated in human samples of different origins, including breast, prostate, colorectal and non-small cell lung cancers as well as myeloid leukemia, which is consistent with a tumor suppressor role [34, 41–43]. Lower HBP1 expression in human cancer tissues relative to normal ones may result from multiple processes that are described below.
Downregulation of HBP1 expression in cancer
Reduced expression of HBP1 was observed in invasive breast tumor samples and correlated with a poor prognosis [34]. In a separate study on breast cancer, Li and colleagues ascribed the decrease in HBP1 expression to miR-17-5p upregulation, promoting cell proliferation, invasion and migration [44]. We also observed a decrease in HBP1 expression in samples of breast cancers, as well as cervix cancers, relative to normal tissues, and we proposed an alternative mechanism involving FOXO transcription factor inhibition by the PI3K/AKT pathway [45]. A similar dual regulation of HBP1 by a microRNA (miR-17-92) and the PI3K pathway was reported in neuroblastoma [28]. In these tumors, low HBP1 level correlates with shorter patient survival. Additionally, reduced expression of HBP1 was reported in human prostate cancer samples relative to normal tissues. In these tumors, downregulation of HBP1 leads to higher levels of macrophage migration inhibitory factor (MIF), which may contribute to tumor progression [41]. A recent study showed that HBP1 enhances radiosensitivity in prostate cancer cell lines by inducing apoptosis, further supporting a role in this cancer type [46]. HBP1 expression was reduced in approximately 30% of non-small cell lung cancer (NSCLC) samples owing to hypermethylation of its promoter [42]. The study suggested that low HBP1 expression was one of the major determinants of NSCLC prognosis. In addition, low expression of HBP1 in NSCLC correlates with higher activity of β-catenin, which is associated with tumorigenesis [42]. Finally, lower expression of HBP1 mRNA was found in limited numbers of acute myeloid leukemia (AML) samples relative to hematopoietic stem cells and might be mediated by the overexpression of miR-29a [43]. HBP1 overexpression induces apoptosis and cell cycle arrest at the G1 phase in the K562 leukemic cell line [31]. In addition to miR-29a and miR-17-5p, other micro-RNAs target HBP1 expression in various models (Table 1).
Table 1.
Regulation of HBP1 expression by miRNAs
| Context | miRNA | Impact on HBP1 expression | References |
|---|---|---|---|
| Invasive breast cancer | ↗ miR-17-5p | ↓ (in cell lines) | [44] |
| Myc-induced lymphoma | ↗ miR-17-92 | ↓ (in mouse model) | [65] |
| Friend erythroleukemia | ↗ miR-17-92 | ↓ (in cell lines) | [66] |
| Neuroblastoma | ↗ miR-17-92 | ↓ (in cell lines) | [28] |
| Hepatocellular carcinoma | ↗ miR-21 | ↓ (in HepG2 cell line) | [67] |
| Acute myeloid leukemia | ↗ miR-29a | ↓ (in primary granulocytes) | [43] |
| Glioma | ↗ miR-96 | ↓ (in glioma specimens, primary astrocytes, and cell lines) | [68] |
| Colorectal carcinoma | ↗ miR-155 | ↓ (in cell lines) | [69] |
| Osteosarcoma | ↗ miR-155 | ↓ (in cell lines) | [70] |
| Glioma | ↗ miR-155 | ↓ (in cell lines) | [71] |
| Nasopharyngeal carcinoma | ↘ miR-29c | ↑ (in NPC specimens, mouse model, and cell lines) | [50] |
| T-cell acute lymphoblastic leukemia | ↘ miR-29a | ↑ (in mouse model) | [51] |
| ↘ miR-31 | |||
| ↘ miR-155 | |||
| Endothelial cells | miR-29a | ↓ (in HUVEC) | [72] |
| Atherosclerosis | ↗ miR-155 | ↓ (in RAW264 macrophages) | [73] |
| ↗ miR-19 | ↓ | [74] |
List of micro-RNAs that are up (↗) or down (↘) regulated in various model systems and target HBP1 transcripts
HBP1 as a target of the PI3K/AKT pathway
In addition to micro-RNAs, the phosphatidylinositol-3 kinase (PI3K) pathway can also downregulate HBP1 expression by inhibiting FOXO transcription factors. We showed that FOXO stimulates HBP1 expression [45]. In line with this result, we observed a significant correlation between the mRNA levels of FOXO1 and HBP1 in breast cancer samples. AKT, which phosphorylates and inactivates FOXO, decreases HBP1 expression [47]. A similar mechanism was described in invasive oral cancer and in neuroblastoma [28, 48]. In addition, we observed that AKT directly phosphorylates HBP1 on three conserved sites, namely Ser380, Thr484 and Ser509, inactivating HBP1 transcriptional activity (Fig. 1). We showed that this process favors the proliferation of glioblastoma cell lines [11]. Altogether, the PI3K/AKT pathway blocks HBP1 at the transcriptional and posttranslational levels. Since the PI3K pathway is one of the most commonly activated signaling cascades in human cancer, this mechanism may alleviate HBP1 cell cycle inhibitor activity in a broad spectrum of cancer types.
Is HBP1 a cancer driver gene?
Because systematic sequencing of human cancer genomes has not revealed a significant rate of HBP1 somatic gene alterations, HBP1 is not currently listed as a cancer driver gene [49]. Nevertheless, HBP1 may be subjected to a type of alteration that is missed by currently used methods (such as noncoding variants for instance). Even if HBP1 is not directly targeted by mutations, the results described above suggest that it plays an important role as a regulator of key oncogenic pathways, such as PI3K/AKT and WNT. However, most experiments have been performed in cancer cell lines, and evidence from mouse models is still lacking. The recent generation of various constitutive and conditional Hbp1-deficient mice will provide suitable models to support the HBP1 tumor suppressor hypothesis and open the quest for anti-HBP1 treatments.
Paradoxical role of HBP1 in some cancer models
Contrasting with the ample amount of data showing that HBP1 dampens tumor cell growth, a few studies recently reported that HBP1 may paradoxically promote tumor progression. For instance, HBP1 may facilitate nasopharyngeal carcinoma (NPC) growth by inducing cyclin D1 and D3 expression. High HBP1 expression correlates with the loss of miR-29c and poor overall survival rates of NPC patients [50]. Expression of HBP1 is also higher in patient samples of T-cell acute lymphoblastic leukemia (T-ALL) and may be related to the decreased expression of several miRNAs, including miR-29, miR-31 and miR-155 (Table 1). Consistently, overexpression of HBP1 in hematopoietic progenitor cells accelerates the rate of Notch1-driven T-ALL development in transplanted recipient mice, reducing survival [51]. These paradoxical results suggest a dual role of HBP1 in cancer development. Strikingly, this is reminiscent of the HBP1 target gene CDKN1A, which encodes p21, a potent cell cycle inhibitor and a mediator of p53 responses. P21 functions suggest a tumor suppressor role, but the gene is rarely mutated in human cancer and may paradoxically promote tumor development in the absence of p53 [52].
Physiological roles of HBP1 based on mouse models
To investigate the physiological role of HBP1 in vivo, different research groups have generated HBP1-deficient mice. Using a Hbp1-genetrap model, Spiller et al. demonstrated that C57BL/6 mice lacking HBP1 are viable and fertile [53]. We observed that homozygous Hbp1-deficient mice carrying another genetrap allele provided by the EUCOMM consortium on the CD-1 outbred background were also viable and fertile. However, the same homozygous genetrap allele led to perinatal lethality on the C57BL/6 background (Audrey de Rocca Serra, unpublished results). Future experiments will determine whether this discrepancy is due to differences in mouse strains or in gene targeting strategies. Perinatal lethality may be related to the phenotype described by Watanabe et al. Conditional deletion of HBP1 in neural stem cells of C57BL/6J/ICR mice leads to an impairment of cortical morphogenesis characterized by telencephalic vesicle dilatations and thinner cortical plates containing reduced numbers of neurons. In these mice, HBP1 was shown to control the duration of the neural stem cell cycle during cortical development [54]. These experiments support the data obtained in cell culture showing that HBP1 functions as a cell cycle inhibitor.
In accordance with its role in cell cycle progression in vivo, transgenic rat-HBP1 expression in the livers of C57BL/6 mice delayed the G1/S transition of hepatocytes during liver regeneration following partial hepatectomy [55]. Similarly, transgenic C57BL/10/CBA mice overexpressing HBP1 in thymocytes exhibit lower thymus cellularity despite an increase in the number of CD8+ T cells in the thymus [56]. However, this phenotype has not been confirmed in HBP1-deficient mice. Interestingly, in this study, HBP1 regulated the transcription of CD2 by binding to its T-cell-specific locus control region (LCR), which functions by recruiting chromatin-modifying and transcription complexes [6]. These results support the notion that HBP1 regulates chromatin remodeling.
HBP1 is highly expressed in the embryonic mouse testis from approximately 12.5 days post-coitum, which corresponds to the embryonic cell cycle arrest of germ cells [53]. However, Hbp1-deficient germ cells proliferate correctly throughout development. In females, conditional ablation of Hbp1 in granulosa cells of C57BL/6 mice promotes follicle growth and oocyte production, suggesting that HBP1 prevents the early depletion of follicles and protects ovarian reserve in mice [57]. The opposite phenotype was observed in HBP1 transgenic mice. This effect was ascribed to the regulation of apoptosis and mitochondrial function by HBP1. In granulosa cells, HBP1 limits mitochondrial biogenesis and reduces oxygen consumption and respiration [57].
In conclusion, the available in vivo data confirm that HBP1 is a cell cycle inhibitor, but much more work is needed to understand the physiological function of this gene in detail.
Potential role of HBP1 in other human diseases
The human HBP1 gene is located within the locus 7q22-31, which is altered in rare cases of osteoarthritis, glaucoma, cardiovascular diseases and neurologic disorders. An analysis of osteoarthritis patients in particular revealed a significant reduction in HBP1 expression in cartilage and synovium associated with HBP1 allelic expression imbalance [58]. Multiple congenital anomalies, including severe glaucoma, were also reported to be associated with deletion encompassing the 7q21.3-q31.1 region, causing the codeletion of TAC1, HBP1 and a cytochrome P450 gene [59]. Recently, single-variant analysis of cardiovascular disease specimens associated genetic susceptibility of the 7q22 locus with atherosclerotic plaque characteristics. One variant (rs12539895), which correlates with a fat reduction in carotid plaques, alters COG5 and HBP1 expression in relevant cardiac tissues [60]. In addition, diverse alterations within the 7q21-q32 segment have often been associated with multiple clinical features, including developmental delay, mental retardation, microcephaly, palate defects, as well as ear and eye abnormalities [61–64]. Whether some of these features are related to HBP1 gene alteration remains to be established.
Conclusion
HBP1 mostly acts as a transcriptional repressor, although activation of transcription has also been reported. Experiments based on cell culture and mouse models have clearly established that HBP1 is a negative regulator of the cell cycle in multiple cell types. In this respect, the suggested roles of HBP1 in cell growth arrest associated with senescence and terminal cell differentiation make sense, even though the importance of HBP1 in this context has not yet been confirmed in vivo. HBP1 capacity to blunt the proliferation of cancer cell lines is also demonstrated by numerous reports and matches the lower HBP1 expression observed in cancer samples relative to normal tissues. HBP1 involvement in the WNT/β-catenin and PI3K pathways also supports a tumor suppressor function. However, recent results in specific cancer models suggest that HBP1 may play a dual role in tumorigenesis. Clearly, more work is needed to confirm whether alterations of the HBP1 gene drive cancer development in mice and patients. The recent development of mouse strains with conditional deletion of Hbp1 will provide better models to clarify the roles of this transcription factor in physiology and diseases.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflicts of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Lesage F, Hugnot JP, Amri EZ, Grimaldi P, Barhanin J, Lazdunski M. Expression cloning in K+ transport defective yeast and distribution of HBP1, a new putative HMG transcriptional regulator. Nucleic Acids Res. 1994;22:3685–3688. doi: 10.1093/nar/22.18.3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tevosian SG, Shih HH, Mendelson KG, Sheppard KA, Paulson KE, Yee AS. HBP1: a HMG box transcriptional repressor that is targeted by the retinoblastoma family. Genes Dev. 1997;11:383–396. doi: 10.1101/gad.11.3.383. [DOI] [PubMed] [Google Scholar]
- 3.Lavender P, Vandel L, Bannister AJ, Kouzarides T. The HMG-box transcription factor HBP1 is targeted by the pocket proteins and E1A. Oncogene. 1997;14:2721–2728. doi: 10.1038/sj.onc.1201243. [DOI] [PubMed] [Google Scholar]
- 4.Song X, Gao X, Lu J, Liang H, Su P, Li Q, Pang Y. High mobility group box transcription factor 1 (HBP1) from Lampetra japonica affects cell cycle regulation. Dev Growth Differ. 2018;60:146–157. doi: 10.1111/dgd.12426. [DOI] [PubMed] [Google Scholar]
- 5.Giese K, Amsterdam A, Grosschedl R. DNA-binding properties of the HMG domain of the lymphoid-specific transcriptional regulator LEF-1. Genes Dev. 1991;5:2567–2578. doi: 10.1101/gad.5.12b.2567. [DOI] [PubMed] [Google Scholar]
- 6.Zhuma T, Tyrrell R, Sekkali B, Skavdis G, Saveliev A, Tolaini M, Roderick K, Norton T, Smerdon S, Sedgwick S, Festenstein R, Kioussis D. Human HMG box transcription factor HBP1: a role in hCD2 LCR function. EMBO J. 1999;18:6396–6406. doi: 10.1093/emboj/18.22.6396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Chiara C, Giannini C, Adinolfi S, de Boer J, Guida S, Ramos A, Jodice C, Kioussis D, Pastore A. The AXH module: an independently folded domain common to ataxin-1 and HBP1. FEBS Lett. 2003;551:107–112. doi: 10.1016/S0014-5793(03)00818-4. [DOI] [PubMed] [Google Scholar]
- 8.de Chiara C, Menon RP, Adinolfi S, de Boer J, Ktistaki E, Kelly G, Calder L, Kioussis D, Pastore A. The AXH domain adopts alternative folds the solution structure of HBP1 AXH. Structure. 2005;13:743–753. doi: 10.1016/j.str.2005.02.016. [DOI] [PubMed] [Google Scholar]
- 9.Wang W, Pan K, Chen Y, Huang C, Zhang X. The acetylation of transcription factor HBP1 by p300/CBP enhances p16INK4A expression. Nucleic Acids Res. 2012;40:981–995. doi: 10.1093/nar/gkr818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Swanson KA, Knoepfler PS, Huang K, Kang RS, Cowley SM, Laherty CD, Eisenman RN, Radhakrishnan I. HBP1 and Mad1 repressors bind the Sin3 corepressor PAH2 domain with opposite helical orientations. Nat Struct Mol Biol. 2004;11:738–746. doi: 10.1038/nsmb798. [DOI] [PubMed] [Google Scholar]
- 11.Bollaert E, Johanns M, Herinckx G, de Rocca Serra A, Vandewalle VA, Havelange V, Rider MH, Vertommen D, Demoulin JB. HBP1 phosphorylation by AKT regulates its transcriptional activity and glioblastoma cell proliferation. Cell Signal. 2018;44:158–170. doi: 10.1016/j.cellsig.2018.01.014. [DOI] [PubMed] [Google Scholar]
- 12.Zhang X, Kim J, Ruthazer R, McDevitt MA, Wazer DE, Paulson KE, Yee AS. The HBP1 transcriptional repressor participates in RAS-induced premature senescence. Mol Cell Biol. 2006;26:8252–8266. doi: 10.1128/MCB.00604-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shih HH, Tevosian SG, Yee AS. Regulation of differentiation by HBP1, a target of the retinoblastoma protein. Mol Cell Biol. 1998;18:4732–4743. doi: 10.1128/MCB.18.8.4732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Emanuele MJ, Elia AE, Xu Q, Thoma CR, Izhar L, Leng Y, Guo A, Chen YN, Rush J, Hsu PW, Yen HC, Elledge SJ. Global identification of modular cullin-RING ligase substrates. Cell. 2011;147:459–474. doi: 10.1016/j.cell.2011.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wagner SA, Beli P, Weinert BT, Nielsen ML, Cox J, Mann M, Choudhary C. A proteome-wide, quantitative survey of in vivo ubiquitylation sites reveals widespread regulatory roles. Mol Cell Proteom. 2011;10(M111):013284. doi: 10.1074/mcp.M111.013284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lampert F, Stafa D, Goga A, Soste MV, Gilberto S, Olieric N, Picotti P, Stoffel M, Peter M. The multi-subunit GID/CTLH E3 ubiquitin ligase promotes cell proliferation and targets the transcription factor Hbp1 for degradation. Elife. 2018;7:e35528. doi: 10.7554/eLife.35528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xiu M, Kim J, Sampson E, Huang CY, Davis RJ, Paulson KE, Yee AS. The transcriptional repressor HBP1 is a target of the p38 mitogen-activated protein kinase pathway in cell cycle regulation. Mol Cell Biol. 2003;23:8890–8901. doi: 10.1128/MCB.23.23.8890-8901.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang S, Cao Z, Xue J, Li H, Jiang W, Cheng Y, Li G, Zhang X. A positive feedback loop between Pim-1 kinase and HBP1 transcription factor contributes to hydrogen peroxide-induced premature senescence and apoptosis. J Biol Chem. 2017;292:8207–8222. doi: 10.1074/jbc.M116.768101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Amaravadi R, Thompson CB. The survival kinases Akt and Pim as potential pharmacological targets. J Clin Invest. 2005;115:2618–2624. doi: 10.1172/JCI26273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pan K, Chen Y, Roth M, Wang W, Wang S, Yee AS, Zhang X. HBP1-mediated transcriptional regulation of DNA methyltransferase 1 and its impact on cell senescence. Mol Cell Biol. 2013;33:887–903. doi: 10.1128/MCB.00637-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen Y, Pan K, Wang P, Cao Z, Wang W, Wang S, Hu N, Xue J, Li H, Jiang W, Li G, Zhang X. HBP1-mediated regulation of p21 protein through the Mdm2/p53 and TCF4/EZH2 pathways and its impact on cell senescence and tumorigenesis. J Biol Chem. 2016;291:12688–12705. doi: 10.1074/jbc.M116.714147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lemercier C, Duncliffe K, Boibessot I, Zhang H, Verdel A, Angelov D, Khochbin S. Involvement of retinoblastoma protein and HBP1 in histone H1(0) gene expression. Mol Cell Biol. 2000;20:6627–6637. doi: 10.1128/MCB.20.18.6627-6637.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Escamilla-Powers JR, Daniel CJ, Farrell A, Taylor K, Zhang X, Byers S, Sears R. The tumor suppressor protein HBP1 is a novel c-myc-binding protein that negatively regulates c-myc transcriptional activity. J Biol Chem. 2010;285:4847–4858. doi: 10.1074/jbc.M109.074856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Berasi SP, Xiu M, Yee AS, Paulson KE. HBP1 repression of the p47phox gene: cell cycle regulation via the NADPH oxidase. Mol Cell Biol. 2004;24:3011–3024. doi: 10.1128/MCB.24.7.3011-3024.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li H, Wang W, Liu X, Paulson KE, Yee AS, Zhang X. Transcriptional factor HBP1 targets P16(INK4A), upregulating its expression and consequently is involved in Ras-induced premature senescence. Oncogene. 2010;29:5083–5094. doi: 10.1038/onc.2010.252. [DOI] [PubMed] [Google Scholar]
- 26.Sampson EM, Haque ZK, Ku MC, Tevosian SG, Albanese C, Pestell RG, Paulson KE, Yee AS. Negative regulation of the Wnt-beta-catenin pathway by the transcriptional repressor HBP1. EMBO J. 2001;20:4500–4511. doi: 10.1093/emboj/20.16.4500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Elfert S, Weise A, Bruser K, Biniossek ML, Jagle S, Senghaas N, Hecht A. Acetylation of human TCF4 (TCF7L2) proteins attenuates inhibition by the HBP1 repressor and induces a conformational change in the TCF4::DNA complex. PLoS One. 2013;8:e61867. doi: 10.1371/journal.pone.0061867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Claeys S, Denecker G, Durinck K, Decaesteker B, Mus LM, Loontiens S, Vanhauwaert S, Althoff K, Wigerup C, Bexell D, Dolman E, Henrich KO, Wehrmann L, Westerhout EM, Demoulin JB, Kumps C, Van Maerken T, Laureys G, Van Neste C, De Wilde B, De Wever O, Westermann F, Versteeg R, Molenaar JJ, Pahlman S, Schulte JH, De Preter K, Speleman F. ALK positively regulates MYCN activity through repression of HBP1 expression. Oncogene. 2018 doi: 10.1038/s41388-018-0595-3. [DOI] [PubMed] [Google Scholar]
- 29.Zhang Y, Gao Y, Zhao L, Han L, Lu Y, Hou P, Shi X, Liu X, Tian B, Wang X, Huang B, Lu J. Mitogen-activated protein kinase p38 and retinoblastoma protein signalling is required for DNA damage-mediated formation of senescence-associated heterochromatic foci in tumour cells. FEBS J. 2013;280:4625–4639. doi: 10.1111/febs.12435. [DOI] [PubMed] [Google Scholar]
- 30.Lin KM, Zhao WG, Bhatnagar J, Zhao WD, Lu JP, Simko S, Schueneman A, Austin GE. Cloning and expression of human HBP1, a high mobility group protein that enhances myeloperoxidase (MPO) promoter activity. Leukemia. 2001;15:601–612. doi: 10.1038/sj.leu.2402071. [DOI] [PubMed] [Google Scholar]
- 31.Yao CJ, Works K, Romagnoli PA, Austin GE. Effects of overexpression of HBP1 upon growth and differentiation of leukemic myeloid cells. Leukemia. 2005;19:1958–1968. doi: 10.1038/sj.leu.2403918. [DOI] [PubMed] [Google Scholar]
- 32.Chan CY, Yu P, Chang FT, Chen ZH, Lee MF, Huang CY. Transcription factor HMG box-containing protein 1 (HBP1) modulates mitotic clonal expansion (MCE) during adipocyte differentiation. J Cell Physiol. 2018;233:4205–4215. doi: 10.1002/jcp.26237. [DOI] [PubMed] [Google Scholar]
- 33.Borrelli S, Candi E, Hu B, Dolfini D, Ravo M, Grober OM, Weisz A, Dotto GP, Melino G, Vigano MA, Mantovani R. The p63 target HBP1 is required for skin differentiation and stratification. Cell Death Differ. 2010;17:1896–1907. doi: 10.1038/cdd.2010.59. [DOI] [PubMed] [Google Scholar]
- 34.Paulson KE, Rieger-Christ K, McDevitt MA, Kuperwasser C, Kim J, Unanue VE, Zhang X, Hu M, Ruthazer R, Berasi SP, Huang CY, Giri D, Kaufman S, Dugan JM, Blum J, Netto G, Wazer DE, Summerhayes IC, Yee AS. Alterations of the HBP1 transcriptional repressor are associated with invasive breast cancer. Cancer Res. 2007;67:6136–6145. doi: 10.1158/0008-5472.CAN-07-0567. [DOI] [PubMed] [Google Scholar]
- 35.Liang H, Fairman J, Claxton DF, Nowell PC, Green ED, Nagarajan L. Molecular anatomy of chromosome 7q deletions in myeloid neoplasms: evidence for multiple critical loci. Proc Natl Acad Sci USA. 1998;95:3781–3785. doi: 10.1073/pnas.95.7.3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koike M, Tasaka T, Spira S, Tsuruoka N, Koeffler HP. Allelotyping of acute myelogenous leukemia: loss of heterozygosity at 7q31.1 (D7S486) and q33-34 (D7S498, D7S505) Leuk Res. 1999;23:307–310. doi: 10.1016/S0145-2126(98)00159-3. [DOI] [PubMed] [Google Scholar]
- 37.Driouch K, Briffod M, Bieche I, Champeme MH, Lidereau R. Location of several putative genes possibly involved in human breast cancer progression. Cancer Res. 1998;58:2081–2086. [PubMed] [Google Scholar]
- 38.Zenklusen JC, Thompson JC, Troncoso P, Kagan J, Conti CJ. Loss of heterozygosity in human primary prostate carcinomas: a possible tumor suppressor gene at 7q31.1. Cancer Res. 1994;54:6370–6373. [PubMed] [Google Scholar]
- 39.Zenklusen JC, Weitzel JN, Ball HG, Conti CJ. Allelic loss at 7q31.1 in human primary ovarian carcinomas suggests the existence of a tumor suppressor gene. Oncogene. 1995;11:359–363. [PubMed] [Google Scholar]
- 40.Zenklusen JC, Thompson JC, Klein-Szanto AJ, Conti CJ. Frequent loss of heterozygosity in human primary squamous cell and colon carcinomas at 7q31.1: evidence for a broad range tumor suppressor gene. Cancer Res. 1995;55:1347–1350. [PubMed] [Google Scholar]
- 41.Chen YC, Zhang XW, Niu XH, Xin DQ, Zhao WP, Na YQ, Mao ZB. Macrophage migration inhibitory factor is a direct target of HBP1-mediated transcriptional repression that is overexpressed in prostate cancer. Oncogene. 2010;29:3067–3078. doi: 10.1038/onc.2010.97. [DOI] [PubMed] [Google Scholar]
- 42.Tseng RC, Huang WR, Lin SF, Wu PC, Hsu HS, Wang YC. HBP1 promoter methylation augments the oncogenic beta-catenin to correlate with prognosis in NSCLC. J Cell Mol Med. 2014;18:1752–1761. doi: 10.1111/jcmm.12318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Han YC, Park CY, Bhagat G, Zhang J, Wang Y, Fan JB, Liu M, Zou Y, Weissman IL, Gu H. microRNA-29a induces aberrant self-renewal capacity in hematopoietic progenitors, biased myeloid development, and acute myeloid leukemia. J Exp Med. 2010;207:475–489. doi: 10.1084/jem.20090831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li H, Bian C, Liao L, Li J, Zhao RC. miR-17-5p promotes human breast cancer cell migration and invasion through suppression of HBP1. Breast Cancer Res Treat. 2011;126:565–575. doi: 10.1007/s10549-010-0954-4. [DOI] [PubMed] [Google Scholar]
- 45.Coomans de Brachene A, Bollaert E, Eijkelenboom A, de Rocca Serra A, van der Vos KE, Burgering BM, Coffer PJ, Essaghir A, Demoulin JB. The expression of the tumour suppressor HBP1 is down-regulated by growth factors via the PI3K/PKB/FOXO pathway. Biochem J. 2014;460:25–34. doi: 10.1042/BJ20131467. [DOI] [PubMed] [Google Scholar]
- 46.Chen Y, Wang Y, Yu Y, Xu L, Zhang Y, Yu S, Li G, Zhang Z. Transcription factor HBP1 enhances radiosensitivity by inducing apoptosis in prostate cancer cell lines. Anal Cell Pathol (Amst) 2016;2016:7015659. doi: 10.1155/2016/7015659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Coomans de Brachene A, Demoulin JB. FOXO transcription factors in cancer development and therapy. Cell Mol Life Sci. 2016;73:1159–1172. doi: 10.1007/s00018-015-2112-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chan CY, Huang SY, Sheu JJ, Roth MM, Chou IT, Lien CH, Lee MF, Huang CY. Transcription factor HBP1 is a direct anti-cancer target of transcription factor FOXO1 in invasive oral cancer. Oncotarget. 2017;8:14537–14548. doi: 10.18632/oncotarget.14653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bailey MH, Tokheim C, Porta-Pardo E, Sengupta S, Bertrand D, Weerasinghe A, Colaprico A, Wendl MC, Kim J, Reardon B, Ng PK, Jeong KJ, Cao S, Wang Z, Gao J, Gao Q, Wang F, Liu EM, Mularoni L, Rubio-Perez C, Nagarajan N, Cortes-Ciriano I, Zhou DC, Liang WW, Hess JM, Yellapantula VD, Tamborero D, Gonzalez-Perez A, Suphavilai C, Ko JY, Khurana E, Park PJ, Van Allen EM, Liang H, Group MCW, N Cancer Genome Atlas Research. Lawrence MS, Godzik A, Lopez-Bigas N, Stuart J, Wheeler D, Getz G, Chen K, Lazar AJ, Mills GB, Karchin R, Ding L. Comprehensive characterization of cancer driver genes and mutations. Cell. 2018;173:371–385 e18. doi: 10.1016/j.cell.2018.02.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.He S, Yang S, Niu M, Zhong Y, Dan G, Zhang Y, Ma H, Xiong W, Zhou M, Zhou Y, Xiang B, Li G, Shuai C, Peng S. HMG-box transcription factor 1: a positive regulator of the G1/S transition through the Cyclin-CDK-CDKI molecular network in nasopharyngeal carcinoma. Cell Death Dis. 2018;9:100. doi: 10.1038/s41419-017-0175-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sanghvi VR, Mavrakis KJ, Van der Meulen J, Boice M, Wolfe AL, Carty M, Mohan P, Rondou P, Socci ND, Benoit Y, Taghon T, Van Vlierberghe P, Leslie CS, Speleman F, Wendel HG. Characterization of a set of tumor suppressor microRNAs in T cell acute lymphoblastic leukemia. Sci Signal. 2014;7:ra111. doi: 10.1126/scisignal.2005500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Georgakilas AG, Martin OA, Bonner WM. p21: a two-faced genome guardian. Trends Mol Med. 2017;23:310–319. doi: 10.1016/j.molmed.2017.02.001. [DOI] [PubMed] [Google Scholar]
- 53.Spiller CM, Wilhelm D, Jans DA, Bowles J, Koopman P. Mice lacking Hbp1 function are viable and fertile. PLoS One. 2017;12:e0170576. doi: 10.1371/journal.pone.0170576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Watanabe N, Kageyama R, Ohtsuka T. Hbp1 regulates the timing of neuronal differentiation during cortical development by controlling cell cycle progression. Development. 2015;142:2278–2290. doi: 10.1242/dev.120477. [DOI] [PubMed] [Google Scholar]
- 55.Shih HH, Xiu M, Berasi SP, Sampson EM, Leiter A, Paulson KE, Yee AS. HMG box transcriptional repressor HBP1 maintains a proliferation barrier in differentiated liver tissue. Mol Cell Biol. 2001;21:5723–5732. doi: 10.1128/MCB.21.17.5723-5732.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sekkali B, Szabat E, Ktistaki E, Tolaini M, Roderick K, Harker N, Patel A, Williams K, Norton T, Kioussis D. Human high mobility group box transcription factor 1 affects thymocyte development and transgene variegation. J Immunol. 2005;175:5203–5212. doi: 10.4049/jimmunol.175.8.5203. [DOI] [PubMed] [Google Scholar]
- 57.Dong Z, Huang M, Liu Z, Xie P, Dong Y, Wu X, Qu Z, Shen B, Huang X, Zhang T, Li J, Liu J, Yanase T, Zhou C, Xu Y. Focused screening of mitochondrial metabolism reveals a crucial role for a tumor suppressor Hbp1 in ovarian reserve. Cell Death Differ. 2016;23:1602–1614. doi: 10.1038/cdd.2016.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Raine EV, Wreglesworth N, Dodd AW, Reynard LN, Loughlin J. Gene expression analysis reveals HBP1 as a key target for the osteoarthritis susceptibility locus that maps to chromosome 7q22. Ann Rheum Dis. 2012;71:2020–2027. doi: 10.1136/annrheumdis-2012-201304. [DOI] [PubMed] [Google Scholar]
- 59.Martinez-Jacobo L, Cordova-Fletes C, Ortiz-Lopez R, Rivas F, Saucedo-Carrasco C, Rojas-Martinez A. Delineation of a de novo 7q21.3q31.1 deletion by CGH-SNP arrays in a girl with multiple congenital anomalies including severe glaucoma. Mol Syndromol. 2013;4:285–291. doi: 10.1159/000353510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.van der Laan SW, Siemelink MA, Haitjema S, Foroughi Asl H, Perisic L, Mokry M, van Setten J, Malik R, Dichgans M, Worrall BB, MCotISG Consortium. Samani NJ, Schunkert H, Erdmann J, Hedin U, Paulsson-Berne G, Bjorkegrenn JLM, de Borst GJ, Asselbergs FW, den Ruijter FW, de Bakker PIW, Pasterkamp G. Genetic susceptibility loci for cardiovascular disease and their impact on atherosclerotic plaques. Circ Genom Precis Med. 2018;11:e002115. doi: 10.1161/CIRCGEN.118.002115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Young RS, Weaver DD, Kukolich MK, Heerema NA, Palmer CG, Kawira EL, Bender HA. Terminal and interstitial deletions of the long arm of chromosome 7: a review with five new cases. Am J Med Genet. 1984;17:437–450. doi: 10.1002/ajmg.1320170207. [DOI] [PubMed] [Google Scholar]
- 62.Tzschach A, Menzel C, Erdogan F, Schubert M, Hoeltzenbein M, Barbi G, Petzenhauser C, Ropers HH, Ullmann R, Kalscheuer V. Characterization of a 16 Mb interstitial chromosome 7q21 deletion by tiling path array CGH. Am J Med Genet A. 2007;143:333–337. doi: 10.1002/ajmg.a.31601. [DOI] [PubMed] [Google Scholar]
- 63.Fagan K, Gill A, Henry R, Wilkinson I, Carey B. A summary of 7q interstitial deletions and exclusion mapping of the gene for beta-glucuronidase. J Med Genet. 1989;26:619–625. doi: 10.1136/jmg.26.10.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Al-Hassnan ZN, Al-Bakheet A, Abu-Dheim N, Al-Younes B, Colak D, Kaya N. A novel interstitial microdeletion of 7q22.1-7q22.3 detected by array comparative genomic hybridization. Am J Med Genet A. 2011;155A:3128–3131. doi: 10.1002/ajmg.a.34298. [DOI] [PubMed] [Google Scholar]
- 65.Li Y, Choi PS, Casey SC, Dill DL, Felsher DW. MYC through miR-17-92 suppresses specific target genes to maintain survival, autonomous proliferation, and a neoplastic state. Cancer Cell. 2014;26:262–272. doi: 10.1016/j.ccr.2014.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kayali S, Giraud G, Morle F, Guyot B. Spi-1, Fli-1 and Fli-3 (miR-17-92) oncogenes contribute to a single oncogenic network controlling cell proliferation in friend erythroleukemia. PLoS One. 2012;7:e46799. doi: 10.1371/journal.pone.0046799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wu H, Ng R, Chen X, Steer CJ, Song G. MicroRNA-21 is a potential link between non-alcoholic fatty liver disease and hepatocellular carcinoma via modulation of the HBP1-p53-Srebp1c pathway. Gut. 2016;65:1850–1860. doi: 10.1136/gutjnl-2014-308430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yan Z, Wang J, Wang C, Jiao Y, Qi W, Che S. miR-96/HBP1/Wnt/beta-catenin regulatory circuitry promotes glioma growth. FEBS Lett. 2014;588:3038–3046. doi: 10.1016/j.febslet.2014.06.017. [DOI] [PubMed] [Google Scholar]
- 69.Wan YC, Li T, Han YD, Zhang HY, Lin H, Zhang B. MicroRNA-155 enhances the activation of Wnt/beta-catenin signaling in colorectal carcinoma by suppressing HMG-box transcription factor 1. Mol Med Rep. 2016;13:2221–2228. doi: 10.3892/mmr.2016.4788. [DOI] [PubMed] [Google Scholar]
- 70.Sun X, Geng X, Zhang J, Zhao H, Liu Y. miR-155 promotes the growth of osteosarcoma in a HBP1-dependent mechanism. Mol Cell Biochem. 2015;403:139–147. doi: 10.1007/s11010-015-2344-z. [DOI] [PubMed] [Google Scholar]
- 71.Yan Z, Che S, Wang J, Jiao Y, Wang C, Meng Q. miR-155 contributes to the progression of glioma by enhancing Wnt/beta-catenin pathway. Tumour Biol. 2015;36:5323–5331. doi: 10.1007/s13277-015-3193-9. [DOI] [PubMed] [Google Scholar]
- 72.Yang Z, Wu L, Zhu X, Xu J, Jin R, Li G, Wu F. MiR-29a modulates the angiogenic properties of human endothelial cells. Biochem Biophys Res Commun. 2013;434:143–149. doi: 10.1016/j.bbrc.2013.03.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tian FJ, An LN, Wang GK, Zhu JQ, Li Q, Zhang YY, Zeng A, Zou J, Zhu RF, Han XS, Shen N, Yang HT, Zhao XX, Huang S, Qin YW, Jing Q. Elevated microRNA-155 promotes foam cell formation by targeting HBP1 in atherogenesis. Cardiovasc Res. 2014;103:100–110. doi: 10.1093/cvr/cvu070. [DOI] [PubMed] [Google Scholar]
- 74.Chen H, Li X, Liu S, Gu L, Zhou X. MircroRNA-19a promotes vascular inflammation and foam cell formation by targeting HBP-1 in atherogenesis. Sci Rep. 2017;7:12089. doi: 10.1038/s41598-017-12167-z. [DOI] [PMC free article] [PubMed] [Google Scholar]