Abstract
Recent genetic and technological advances have determined a role for chromatin structure in neurodevelopment. In particular, compounding evidence has established roles for CTCF and cohesin, two elements that are central in the establishment of chromatin structure, in proper neurodevelopment and in regulation of behavior. Genetic aberrations in CTCF, and in subunits of the cohesin complex, have been associated with neurodevelopmental disorders in human genetic studies, and subsequent animal studies have established definitive, although sometime opposing roles, for these factors in neurodevelopment and behavior. Considering the centrality of these factors in cellular processes in general, the mechanisms through which dysregulation of CTCF and cohesin leads specifically to neurological phenotypes is intriguing, although poorly understood. The connection between CTCF, cohesin, chromatin structure, and behavior is likely to be one of the next frontiers in our understanding of the development of behavior in general, and neurodevelopmental disorders in particular.
Keywords: CTCF, Cohesin, Condensin, Neurodevelopment, Chromatin structure, Behavior, Intellectual disability, SMC
Chromosome organization in general and in neurons
Chromatin organization in the nucleus of the eukaryotic cell is highly complex, and is tightly associated with nuclear functions including cell proliferation and differentiation. Many studies have described local genomic structure at the level of nucleosomes and histone modifications, and their impact on genome stability, dynamics, and expression. However, the importance of interactions between distant genomic regions and the assembly of chromatin domains to these properties is just beginning to be clarified, with the help of new technological advances. Several proteins have been identified as central players in the regulation chromatin organization and chromatin domain organization. These factors include the CCCTC-binding factor (CTCF) and members of the Structural Maintenance of Chromosome (SMC) complexes cohesin and condensin. Understanding the interplay between these factors and chromatin organization is an ongoing effort.
The chromosomes are not randomly organized in the nuclei, rather each chromosome occupies a specific territory in the nucleus. Examination of the interphase nucleus by microscopy reveals areas of highly condensed chromatin, referred to as heterochromatin, and less condensed chromatin, referred to as euchromatin. Modern molecular methods allow for more detailed characterization of the chromatin structure, including the existence of lamina-associated domains (LADs), which are associated with transcriptional inactivity, and topologically associated domains (TADs), which are associated with transcriptional activity [1]. TADS are formed by intra-chromosome interaction of remote genomic regions that divide the chromatin into distinct structural units and are located in the internal nuclear space. Promoter–enhancer interactions typically occur within the same TAD, and enhancers typically will not bind promoters found in a separate TAD. Therefore, the construction of a TAD allows for concerted gene regulation within a defined genomic region, and insulates that region from the influence of factors outside of that TAD. Chromosome structure dramatically changes when cells enter mitosis. Chromosome condensation and formation of distinct morphological bodies is accompanied with the disappearance of TADs and gene expression shutdown [2, 3].
Recent evidence suggests that neuronal differentiation induces changes in genomic organization that may be relevant to neuronal function and disease states. On a very general level, it has been found that differentiation from embryonic stem cells to post-mitotic neurons is accompanied by migration of the euchromatin from the inner regions of the nucleus to the periphery, and the opposite migration of heterochromatin to the inner regions of the nucleus [4, 5]. Another study used Hi-C technology to uncover genomic structure in the cortex of human fetuses, and discovered unique brain-specific TADs, and enhancer–promoter binding sites [6]. In addition, they found that schizophrenia-associated SNPs exist in several of the brain-specific enhancer–promoter interactions. A recent study has also added mechanistic insight, and determined that the methyltransferase SETDB1 regulates the formation of neuron-specific TADs [7]. Therefore, formation of specific TADS and genomic interactions may be involved in proper neurodevelopment and brain function.
Apart from the emerging studies suggesting neuron-specific chromatin structure, there is growing evidence that the proteins responsible for genomic organization, including CTCF and complexes of the Structural maintenance of chromosome (SMC), play important and specific roles in neurodevelopment and behavior. Both genetic studies of human neurodevelopmental conditions and animal models studies have uncovered roles for CTCF and SMC complexes in neurodevelopment and behavior. This review is focused on reviewing the recent developments in this field.
Factors involved in chromatin organization
The CCCTC-binding factor (CTCF) is a sequence-specific DNA binding protein with over 30,000 putative binding sites in the human genome [8]. The center of the protein contains eleven C2H2-type zinc fingers [9]. CTCF is involved in diverse roles in gene regulation, including gene activation, repression, genomic imprinting and V(D)J recombination [10–12].
SMC is an evolutionary conserved family of ATPases that regulates chromatin structure. The SMC complex cohesin is composed of four core subunits [SMC1, SMC3, RAD21 (SA1 or SA2)] and about 20 auxiliary proteins. These factors include the NIPBL/MAU2 cohesin loader, the PDS5 unloader, the ESCO acetyltransferase, HDAC8 deacetylase and the chromatin remodeler DDX11/Ch1R that are involved in cohesion establishment during S phase of the cell cycle. Cohesin is best known for its role in mitotic sister chromatid cohesion that allows proper segregation of the chromatids. However, cohesin plays central roles in other mitotic and non-mitotic processes that include DNA repair, recombination, chromosome condensation and regulation of gene expression [10, 13].
Another member of the SMC family is condensin which is involved in chromosome condensation. Mammalian cells contain two related condensins, called condensin I and II. These complexes share the core subunits SMC2 and SMC4 but differ in their regulatory subunits: NCAPH, NCAPG and NCAPD2, and NCAPH2, NCAPG2 and NCAPD3, respectively. Condensin I and II have distinct roles in the organization of mitotic chromosomes. New evidence suggests that condensin has a second role in transcription regulation during interphase [14, 15].
CTCF and Cohesin are intricately involved in TAD formation. The TAD stem is defined by both factors, although the exact mechanism through which TADs are formed is still under debate. One recent theory of how loops are formed in the DNA, called the “loop extrusion model”, states that a DNA loop moves through the doughnut-like hole in the cohesin complex, and only stops when it bumps into a previously bound CTCF. This will form a stable loop of DNA, where cohesin and CTCF are interacting at the base of the loop [16]. Other studies have recognized that cohesin and CTCF may each have different, more specific, roles in the formation of genomic structure. For example, downregulation of cohesin leads to reduced inter-TAD interactions but does not affect the structure of TADs. In contrast, depletion of CTCF blurs the boundaries of the TAD, and results in increased inter-domain interactions [17]. Similarly, condensin II is located at TAD boundaries, with the RNA polymerase III regulatory factor TFIIC. However, its role in TAD formation is less understood [18]. In addition, the effect of CTCF and cohesin depletion on transcription is distinct. While cohesin regulates expression by mediating interactions between genes and remote enhancers, CTCF controls expression by direct binding to promoters [17]. Knockdown of the condensin II subunit NCAPH2 in mouse inhibited the interaction between the two histone gene clusters, and was associated with reduced expression of histone genes [18]. Other studies implied that both CTCF and SMC complexes affect protein levels through direct interaction with components of the transcription and translation machineries. These factors include RNA polymerase II and III and transcription factors, such as RUNX1, TFIDD, YY1, TFCII [18–21]. Future studies are required to determine the mechanisms by which these, and similar interactions contribute to chromatin structure and transcription in the context of nerve cells growth and development.
Involvement of CTCF and SMC complexes in the genetics of human neurodevelopmental disorders
Several genetic studies have begun to reveal the importance of CTCF in normal brain development and function. An initial genetic study identified de novo mutations in CTCF among four individuals who have intellectual disabilities [22]. Of these individuals, two carried different frameshift mutations, one carried a missense mutation, and one individual displayed a whole gene deletion. Although the genetic mutations were located on different position in the gene encoding CTCF (N-terminal region/C-terminal region/Zinc finger domain), the clinical features were similar. These individuals displayed intellectual disability, microcephaly, and growth retardation. In addition, two of the individuals displayed symptoms of autism. RNA-seq analysis of lymphocytes from these individuals revealed 698 downregulated genes, including genes involved in processes that are implicated in developmental and cognitive disorders, and 118 upregulated genes, including ribosomal genes. CTCF binding sites, as determined by Chip-seq, were more enriched in downregulated genes, compared to upregulated genes. Moreover, Chromatin Interaction Analysis by Paired-End Tag Sequencing (ChIA-PET) experimentation on the chronic myelogenous leukemia cell line K562 further determined that chromatin loops occupied by CTCF and RNA polymerase II are enriched for the genes that were downregulated in the cells of humans with mutations in CTCF. These molecular analyses indicate that mutations in CTCF may induce intellectual disabilities through destabilizing promoter–enhancer loops that are necessary for gene transcription. Recently, a second study has identified a genetic aberration in CTCF in an individual with intellectual disability, stunted growth, microcephaly, and developmental delay [23]. The patient displayed a de novo frameshift mutation that leads to a deletion in all 11 zinc-finger domains and the C-terminal region. A separate genetic study determined an association between Single Nucleotide Polymorphisms (SNPs) in the genomic vicinity of CTCF and schizophrenia [24]. The study detected strong association between SNPs in the genes CTCF and CACNB2 and schizophrenia, using a gene pathway-based approach. They found this association in multiple cohorts, including separate cohorts of 5040 and 5082 individuals of European ancestry. Therefore, multiple studies have implicated mutations in CTCF as causative factors in intellectual disabilities and there is some evidence to associate CTCF to schizophrenia.
Mutations in several cohesin-encoding subunits and auxiliary factors are associated with multi-systematic developmental disorders collectively called cohesinopathy. These disorders are characterized by overlapping phenotypes ranging from craniofacial abnormalities, cognitive impairment and growth retardation, and include Cornelia de Lange Syndrome (CdLS), Roberts syndrome (RBS), and Warsaw Breakage Syndrome (WBS). CdLS is a rare, genetically heterogeneous disorder. Clinical features include characteristic facial features and growth retardation. IQ mean is 53, ranging from below 30–102. Many CdLS patients demonstrate autistic and self-destructive tendencies. About 60% of all clinical cases are associated with a mutation in the cohesin loader NIPBL. Mutations in genes that encode the cohesin core subunits SMC1A, SMC3 and RAD21 are associated with 5%, 1–2% and < 1% of CdLS cases, respectively. Mutations in the gene encoding for cohesin auxiliary factor HDAC8 has been identified in about 4% of the clinical cases. The severity of the clinical phenotype ranges from severe in NIBPL-related cases to mild in SMC3-related cases [25]. The disorder is divided into five distinct subtypes based on the molecular genetics and clinical features. Most cases of CdLS are autosomal dominant while HDAC8 and SMC1A are X-linked. Most of the deleterious mutations are de novo, loss-of-function mutations that lead to haploinsufficiency. Cells from CdLS patients reveal global changes in gene expression [26].
Roberts syndrome (RBS) is an autosomal recessive disorder that is caused by a mutation in the ESCO2 acetyltransferase. Clinical features include severe pre- and postnatal growth retardation and development, microbrachycephaly and severe intellectual disability. In mammals, ESCO2 has a paralog called ESCO1. Cohesion establishment is the process in which ESCO2 and ESCO1 activates the chromatin tethering activity of cohesin through the acetylation of the SMC3 subunit of cohesin. Despite the mechanistic similarity of their action, ESCO1 and ESCO2 regulate distinct functions of cohesin. Mitotic cohesion establishment is mediated by ESCO2 while ESCO1 is involved in the formation of chromosome loops that are associated with transcriptional control [27]. Cells from RBS patients reveal premature separation of centromeres, increased sensitivity to DNA damage and reduced expression of rDNA genes, which in turn leads to a defect in protein translation [28]. WBS is a very rare disease with only a few cases reported. Similar to the other cohesinopathies, clinical features of WBS include developmental delays and severe intellectual disability. The molecular basis of the disorder is a mutation in the DNA helicase DDX11/Ch1R, which is involved in cohesin loading onto the chromosomes [29]. However, very little is known about this extremely rare syndrome, and its molecular basis [30, 31].
Only recently, a human disorder of extreme microcephaly and intellectual disability was identified in a cohort of patients, which is associated with mutations in the genes encoding for the condensin I subunits NCAPH and NCAPD2, and condensin II subunit NCAPD3 [32]. Mitotic chromosomes from patients’ fibroblasts revealed major defects in their structure, supporting the genotype and the phenotypic pathogenicity. Certain phenotypic features seem to be similar between individuals bearing mutations in CTCF or members of the cohesin complex. These features include growth retardation, microcephaly and cognitive impairment, although there are often differences in the severity of the phenotype. The molecular mechanism behind the neurological symptoms remains to be elucidated but could be due to changes in genome organization, de-compaction of chromatin and alteration in gene expression.
Not only mutations in CTCF or the cohesin complex themselves affect brain development and function, rather modifications in their DNA binding site have also been implicated in neurological diseases or disorders. Friedreich ataxia (FRDA), in which there is progressive damage to the nervous system, is caused by expansion of triplet-repeats in the gene FXN [33, 34]. Several studies have shown that these changes in the FXN gene in humans are associated with a decrease in CTCF occupancy. A separate study found an association between a C/T SNP in a CTCF binding site in the gene HTR3A and schizophrenia in an Indian cohort [35]. Further analysis found that methylation on the C allele increases affinity for CTCF binding to the gene. Therefore, a combination of differential genetics and epigenetics at the CTCF binding site may influence the development of schizophrenia. A separate study also identified differential methylation in a CTCF binding site in the HTT gene, the causative gene of Huntington’s Disease (HD), in individuals with HD. Finally, it has been shown that CTCF directly binds to the promoter of FMR1, the causative gene in Fragile × syndrome, and can regulate its transcription [36–38]. Therefore, dysregulation of CTCF binding at genes responsible for neurodevelopmental phenotypes may also lead to human disorders.
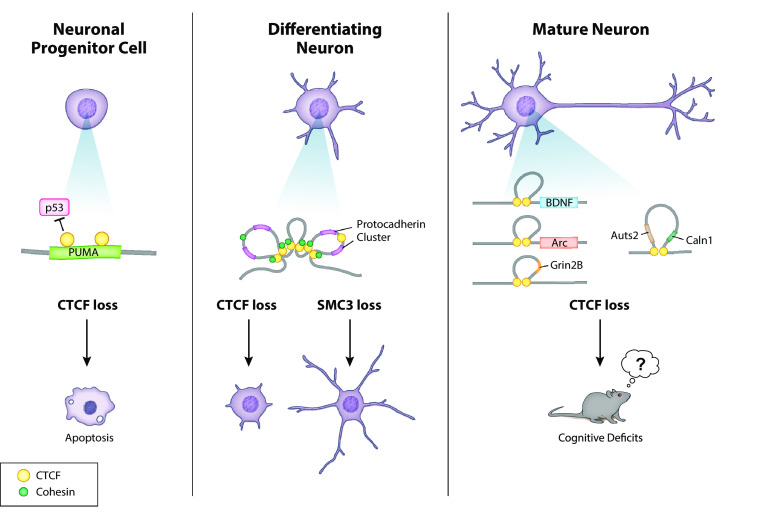
In addition, several proteins that complex with CTCF and/or cohesin are known to be involved in neurodevelopmental disorders. MeCP2, which binds specifically to methylated DNA, and can complex with both CTCF and cohesin in neurons, is the causative gene in Rett’s Disorder, a neurodevelopmental syndrome found mostly in girls [39]. MECP2 has been found to be a regulator of CTCF binding in neurons [40]. MECP2 and cohesin also form complex with ATRX, an ATP-dependent helicase [41]. Mutations in the gene ATRX cause X-linked mental retardation [42]. Another example is CHD8, an autism-associated gene [43] that encodes another ATP-dependent helicase. CHD8 binds CTCF and can influence CTCF binding to the DNA [44]. Knockdown of CHD8 protein was associated with CpG hypermethylation and histone hypoacetylation near CTCF binding sites which lead to changes in CTCF insulator and inhibitor activity [44]. These genetic studies point to the existence of a chromatin-binding complex, containing CTCF, cohesin, MECP2, among many others, that play a central role in proper neurodevelopment (Fig. 1).
Fig. 1. Roles of CTCF or cohesin in specific time points in neuronal development.
Independent studies have determined roles for CTCF and cohesin in genomic organization and cellular function at different time points in neuronal differentiation. While there is likely to be considerable overlap between these different functions, particularly at the level of gene organization and expression, during differentiation, several lines of evidence suggest specific functions for CTCF in proliferating, differentiating, and adult neuronal cells
In addition, dysregulation of condensin has also been implicated in neurodevelopmental disorders. Mutations in the gene MCPH1 are associated with autosomal recessive primary microcephaly. In addition, there is some evidence that MCPH2 has a role in the evolution of human brain size [45, 46]. Interestingly, a mutation in the gene leads to premature chromosome condensation in G2 phase and delayed decondensation in G1 phase of the cell cycle. This cellular outcome is mediated by untimely activation of condensin II, and provides an additional link between chromosome organization and neurodevelopment.
Implication of CTCF and cohesin in neurodevelopment and behavior from animal studies
While in vitro studies and human genetics have implicated that CTCF, cohesin and condensin play a role in neurodevelopment, animal studies are necessary to verify these roles and to understand the mechanism through which CTCF and cohesin specifically affect neurodevelopment. Since knockout of CTCF leads to lethality in the early stage of the embryo, several separate studies have developed conditional knockout (cKO) mice to determine the role of CTCF specifically in neurons and the brain [47–49]. One study used a Nestin-driven Cre line to delete CTCF specifically in neuronal precursor cells (NPCs) [50]. They determined that CTCF regulates the balance between NPC proliferation and differentiation, as well as is necessary for the survival of the NPCs. CTCF depletion in NPCs led to upregulation of the TP53 effector PUMA, resulting in apoptosis. As a result, the mice died approximately at the age of birth. A separate study used a Nex-Cre line to delete CTCF specifically in post-mitotic projection neurons [48]. Mice with a cKO of CTCF in post-mitotic projection neurons had defects in dendritic arborization and synapse formation which include a decrease in dendritic intersection, length and filopodia spine density. Moreover, they determined that CTCF regulates the stochastic expression of clustered protocadherins (Pcdhs) genes, which are essential for building functional neural networks in the brain (21–23). Furthermore, in the same study, mice displayed postnatal growth retardation and died approximately four weeks after the birth [48]. A recent study used a Camkiia-cre to delete CTCF specifically in post-mitotic excitatory forebrain neurons [51]. In contrast to the previous studies, the mice survived several months, until approximately 4 months of age. These mice displayed specific deficits in learning and memory, including spatial memory and fear memory. These findings were in correlation with impaired long-term potentiation and reduced spine density. RNA sequencing (RNA-seq) verified that protocadherin expression was also downregulated in the hippocampus of these mice. However, in addition, experience-dependent expression of key learning genes, BDNF, Arc, and Reln, was also attenuated in this mouse model. 4C chromosome capturing experiments determined dysregulation of chromatin looping in the BDNF and Arc genes in neurons lacking CTCF. Therefore, CTCF-dependent genomic structure may be mandatory for experience-dependant increases in learning-related genes. Therefore, several studies of CTCF knockout in neurons show an important role of CTCF in neurodevelopment and mature neuron function, which may be partly related to its regulation of the protocadherin genes, as well as regulation of cognition-related genes (Tables 1, 2).
Table 1.
CTCF, cohesin, and condensin complex genes that have been associated with neurodevelopmental disorders
| Gene name | Function | Case reports | Behavioral phenotype | References |
|---|---|---|---|---|
| CTCF | CTCF mediates chromosomal looping interactions and transcriptional gene regulation | Four cases identified in cohort of 400 individuals with intellectual disability; one case identified in United Arab Emirates | Intellectual disability, microcephaly, and growth retardation | [22, 23] |
| NIPBL | Cohesin loader into chromatin | 60% of CdLS, autosomal dominant disorder | Craniofacial abnormalities, cognitive impairment and growth retardation | [66] |
| SMC1A | Structural components of the cohesin ring | 5% of CdLS, X-linked disorder | ||
| SMC3 | Structural components of the cohesin ring | 1%-2% of CdLS, autosomal dominant disorder | ||
| RAD21 | Structural components of the cohesin ring | >1% of CdLS, autosomal dominant disorder | ||
| HDAC8 | Deacetylation of the SMC3 to facilitate renewal of the cohesin complex after its dissociation from chromatin | 4% of CdLS, X-linked disorder | ||
| ESCO2 | Acetyltransferase | RBS, rare disorder with approximately 150 affected individuals have been reported. autosomal recessive disorder | Growth retardation, limb and facial abnormalities, and severe intellectual disability | [67] |
| DDX11/Ch1R | DNA helicase, involved in cohesin loading onto the chromosomes | WBS, rare condition with at least four cases have been reported | Developmental delays, distinctive facial features and severe intellectual disability | [68, 69] |
| NCAPH and NCAPD2 | Condensin I subunits | Four cases identified | Extreme microcephaly and intellectual disability | [32] |
| NCAPH2 and NCAPD3 | Condensin II subunits |
Table 2.
Neurodevelopmental studies of CTCF or cohesin complex knockout in animal model system
| cKO mice model | Phenotype | Molecular finding | References |
|---|---|---|---|
| Nestin-driven Cre line- CTCF null specifically in neuronal precursor cells |
CTCF necessary for the survival of the NPCs Upregulation of the p53 effector PUMA, resulting in apoptosis Regulates the balance between NPC proliferation and differentiation |
[50] | |
| Nex-Cre line- CTCF null specifically in postmitotic projection neurons |
Postnatal growth retardation and abnormal behavior including changes in limb-clasping reflex Decrease in body weight and died approximately four weeks after the birth |
Defects in dendritic arborization and synapse formation Regulates the stochastic expression of clustered protocadherins (Pcdhs) genes |
[48] |
| Camkiia-cre line- CTCF null specifically in post-mitotic excitatory forebrain neurons |
Deficits in learning and memory, including spatial memory and fear memory Died approximately four months of age |
Defects in dendritic arborization and synapse formation Regulates the stochastic expression of clustered protocadherins (Pcdhs) genes Dysregulation of gene expression and chromatin looping in the BDNF and Arc |
[51] |
| Tau-cre line-SMC3 null specifically in neurons | More anxiety-related behavior |
Abnormal dendrite development and synaptic maturation Regulates neuronal development genes |
[58] |
| Full body heterozygous deletion (Nipbl ±) |
High mortality (75–80%) during the first weeks of life Small size, craniofacial anomalies, microbrachycephaly, heart defects, hearing abnormalities, delayed bone maturation and reduced body fat |
Regulates protocadherin beta (Pcdhb) gene expression | [70] |
| knockout of genes SMC1 and SA (piggyback-induced) in drosophila |
Reduced levels of the ecdysone receptor EcR-B1, a key regulator of axon pruning Disrupt axon pruning and causing neuroblast proliferation defects |
[55] |
Separate studies have recently highlighted how CTCF-dependent chromatin organization may be directly modulated by environmental events or pharmacological interventions. This opens up the interesting possibility that CTCF-dependent chromatin organization is not a static event, but rather may show plasticity, leading to downstream effects on behavior. In one such study, it was found that Grin2B, gene encoding a subunit of a glutamate receptor, contains a CTCF-dependent chromatin loop that can be disturbed by neuronal activation, therefore inducing gene expression [52]. In a separate study, it was found that cocaine consumption can disrupt a CTCF-dependent chromatin loop at the AUTS2 gene through inhibiting CTCF binding, and therefore inducing an increase in gene expression [53]. While previous studies have shown that genome structure is plastic during development, or by internal cues, such as circadian rhythms [54], these studies provide evidence that CTCF-dependent chromatin structure in the adult brain is a dynamic process that is regulated by external cues, and affects downstream behavioral phenotypes.
These various studies have shed light into the roles that CTCF plays at specific time points during neuronal development. Deletion of CTCF in neuronal precursor cells (NPCs) leads directly to apoptosis, similar to the effect of CTCF deletion in other dividing cell types [50]. In contrast, in one study, deletion of CTCF in post-mitotic neurons was tolerated for up to 14 weeks before apoptosis [51]. Therefore, CTCF is particularly necessary for the inhibition of apoptosis in NPCs, although it apparently has additive effects in post-mitotic neurons. An important note to consider is that all of these models have determined the role of complete CTCF knockout in specific cell types, while the human condition is caused by haploinsufficiency. Therefore, it is still difficult to pinpoint the exact mechanisms involved in the human condition, and is particularly not clear if haploinsufficiency would lead to any significant apoptotic processes.
RNA-seq of hippocampi from post-mitotic neuron-specific knockout in the Hirayama et al. study determined a strong dysregulation of protocadherin gene expression at P7 [48]. Interestingly, the same dysregulation of protocadherin gene expression was also determined in a separate model at 8 weeks of age, although the changes were not as robust [51]. Considering that protocadherins play an important role in the establishment of neuronal connectivity during development, it is notable that CTCF deletion has the strongest effects on protocadherin expression during the developmental time point. In contrast, gene ontology of RNA-seq results in the 8-week-old mice determined enrichment of cognition-related genes, which was not seen in the developing neurons. Together with the studies determining a role for CTCF in the expression of Grin2b and Auts2 in the adult brain, these findings further establish a temporal-specific role for CTCF in gene expression regulation.
The first study to determine a role for cohesin in neurodevelopment was performed in drosophila. They found that knockout of SMC1 induced a deficit of axonal pruning, which resulted in multiple axons in adult neurons [55]. In addition, SMC1 knockout induced deficits in dendritic targeting. Therefore, this initial study suggests that cohesin is necessary for proper formation of axons and dendrites. Similar to CTCF, knockout of most cohesin subunits leads to embryonic lethality in mice [49, 56]. The characterization of embryonic brain development in SA1 knockout mice revealed that cohesin regulates the expression of important genes in brain development including Myc and Protocadherins [57]. Alternatively, studies of conditional knockout mice in specific cell types can illuminate the role of cohesin in neurons. In a mouse study, deletion of SMC3 specifically in neurons induced greater dendritic complexity and a larger number of immature synapses in the cortex, which may be due to defects in synaptic pruning. The mice also displayed an increase in anxiety-like behavior. RNA-seq discovered a dysregulation of many genes involved in neuronal differentiation, morphogenesis, neurogenesis, and axon guidance [58]. Therefore, SMC3 has been identified as a modulator of synaptic maturation in multiple species.
Interestingly, deletion of the cohesin subunits in neurons displays many phenotypes which are the opposite of what is seen in deletion of CTCF in neurons. In particular, CTCF deletion leads to decreased dendritic complexity [48, 58], while SMC1 or SMC3 deletion leads to increased axons, more dendritic complexity, and deficits in pruning. It is not clear why such opposite phenotypes should be displayed. However, both CTCF and cohesin are involved in regulating the expression of protocadherins, proteins which are central in establishing the identity and complexity of neurons [48, 59, 60]. Of interest [59], Monahan et al. found that while there are many shared cohesin/CTCF binding sites within the Pcdh cluster, they also find binding sites that are unique to each of the two proteins, and conclude that cohesin and CTCF may have some different roles in Pcdh gene expression [59]. Therefore, this may be one mechanism as to how these two proteins have differential effects on neuronal differentiation.
Furthermore, the cohesin loader NIPBL, which is associated with most of the CdLS clinical cases, was recently found to interact with the neural transcription factor Zfp609 in brain development and to regulate cortical neuron migration. shRNA-mediated knockout in the prenatal cortical neurons induced a defect in neuronal migration [61]. In addition, another study found that mutated NDE1 induces DNA double-strand breaks in neuronal precursors, leading to the activation of P53-dependant apoptosis, and a decrease in the number of neurons in cortical layer II/III [62]. The severe phenotype of NIPBL knockdown and NDE1 mutation, which includes major deficits in the organization of the cortex, compared to the more minor phenotype of the SMC3 neuronal knockout, may help to explain why CdLS patients with NIPBL mutations have a more severe phenotype compared to those with SMC3 mutations. This further establishes that different members of the cohesin complex have specific roles in neuronal differentiation. Why these different subunits have such different roles is not yet clear.
Conclusions
Both human genetic evidence and animal model studies determine that CTCF and cohesin have critical roles in neurodevelopment and mammalian behavior. While it has already been well accepted that epigenetic modifications, including both DNA methylation and histone modifications, are crucial in driving neurodevelopment, these studies suggest that proper management of the chromatin structure is also critical. Considering the critical role for chromatin structure in development and genomic function in general, it is not yet clear why dysregulation of chromatin structure regulators would lead specifically to neurodevelopmental phenotypes. Therefore, future studies are needed to uncover specific roles for chromatin structure in neurons, or other brain cells, compared to other tissues. While it has historically been both difficult and expensive to map chromatin structure at the whole-genome level, there have been several recent technological advancements that will allow researchers to map genome structure in specific cell types, and possibly single neurons [63–65]. These studies should greatly advance our knowledge of the role of chromatin structure in neuronal function, and how dysregulation of CTCF and cohesin leads to specific behavioral phenotypes. In summary, connecting the dots between the molecular basis of chromatin structure, and neural system development and behavior will be a forthcoming challenge.
Acknowledgements
We would like to thank Anita Impagliazzo for providing professional graphical support in preparing the figure for this manuscript. This study has been supported by Israel Science Foundation Grant 1099/16 (I.O) and Israel Science Foundation Grant 898/17 (E.E).
References
- 1.Gonzalez-Sandoval A, Gasser SM. On tads and lads: spatial control over gene expression. Trends Genet. 2016;32:485–495. doi: 10.1016/j.tig.2016.05.004. [DOI] [PubMed] [Google Scholar]
- 2.Du Z, Zheng H, Huang B, Ma R, Wu J, Zhang X, He J, Xiang Y, Wang Q, Li Y, et al. Allelic reprogramming of 3D chromatin architecture during early mammalian development. Nature. 2017;547:232–235. doi: 10.1038/nature23263. [DOI] [PubMed] [Google Scholar]
- 3.Flyamer IM, Gassler J, Imakaev M, Brandão HB, Ulianov SV, Abdennur N, Razin SV, Mirny LA, Tachibana-Konwalski K. Single-nucleus Hi-C reveals unique chromatin reorganization at oocyte-to-zygote transition. Nature. 2017;544:110–114. doi: 10.1038/nature21711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aoto T, Saitoh N, Ichimura T, Niwa H, Nakao M. Nuclear and chromatin reorganization in the MHC-Oct3/4 locus at developmental phases of embryonic stem cell differentiation. Dev Biol. 2006;298:354–367. doi: 10.1016/j.ydbio.2006.04.450. [DOI] [PubMed] [Google Scholar]
- 5.Le Gros MA, Clowney EJ, Magklara A, Yen A, Markenscoff-Papadimitriou E, Colquitt B, Myllys M, Kellis M, Lomvardas S, Larabell CA. Soft X-ray tomography reveals gradual chromatin compaction and reorganization during neurogenesis in vivo. Cell Rep. 2016;17:2125–2136. doi: 10.1016/j.celrep.2016.10.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Won H, de la Torre-Ubieta L, Stein JL, Parikshak NN, Huang J, Opland CK, Gandal MJ, Sutton GJ, Hormozdiari F, Lu D, et al. Chromosome conformation elucidates regulatory relationships in developing human brain. Nature. 2016;538:523–527. doi: 10.1038/nature19847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jiang Y, Loh YE, Rajarajan P, Hirayama T, Liao W, Kassim BS, Javidfar B, Hartley BJ, Kleofas L, Park RB, et al. The methyltransferase SETDB1 regulates a large neuron-specific topological chromatin domain. Nat Genet. 2017;49:1239–1250. doi: 10.1038/ng.3906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bao L, Zhou M, Cui Y. CTCFBSDB: a CTCF-binding site database for characterization of vertebrate genomic insulators. Nucleic Acids Res. 2008;36:D83–D87. doi: 10.1093/nar/gkm875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Klenova EM, Nicolas RH, Paterson HF, Carne AF, Heath CM, Goodwin GH, Neiman PE, Lobanenkov VV. CTCF, a conserved nuclear factor required for optimal transcriptional activity of the chicken c-myc gene, is an 11-Zn-finger protein differentially expressed in multiple forms. Mol Cell Biol. 1993;13:7612–7624. doi: 10.1128/MCB.13.12.7612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nasmyth K, Haering CH. Cohesin: its roles and mechanisms. Annu Rev Genet. 2009;43:525–558. doi: 10.1146/annurev-genet-102108-134233. [DOI] [PubMed] [Google Scholar]
- 11.Wendt KS, Peters JM. How cohesin and CTCF cooperate in regulating gene expression. Chromosome Res. 2009;17:201–214. doi: 10.1007/s10577-008-9017-7. [DOI] [PubMed] [Google Scholar]
- 12.Lee BK, Iyer VR. Genome-wide studies of CCCTC-binding factor (CTCF) and cohesin provide insight into chromatin structure and regulation. J Biol Chem. 2012;287:30906–30913. doi: 10.1074/jbc.R111.324962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Onn I, Heidinger-Pauli JM, Guacci V, Unal E, Koshland DE. Sister chromatid cohesion: a simple concept with a complex reality. Annu Rev Cell Dev Biol. 2008;24:105–129. doi: 10.1146/annurev.cellbio.24.110707.175350. [DOI] [PubMed] [Google Scholar]
- 14.Rana V, Bosco G. Condensin regulation of genome architecture. J Cell Physiol. 2017;232:1617–1625. doi: 10.1002/jcp.25702. [DOI] [PubMed] [Google Scholar]
- 15.Kalitsis P, Zhang T, Marshall KM, Nielsen CF, Hudson DF. Condensin, master organizer of the genome. Chromosome Res. 2017;25:61–76. doi: 10.1007/s10577-017-9553-0. [DOI] [PubMed] [Google Scholar]
- 16.Dolgin E. DNA’s secret weapon against knots and tangles. Nature. 2017;544:284–286. doi: 10.1038/544284a. [DOI] [PubMed] [Google Scholar]
- 17.Zuin J, Dixon JR, van der Reijden MI, Ye Z, Kolovos P, Brouwer RW, van de Corput MP, van de Werken HJ, Knoch TA, van IJcken wf, et al. Cohesin and CTCF differentially affect chromatin architecture and gene expression in human cells. Proc Natl Acad Sci USA. 2014;111:996–1001. doi: 10.1073/pnas.1317788111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yuen KC, Slaughter BD, Gerton JL. Condensin II is anchored by TFIIIC and H3K4me3 in the mammalian genome and supports the expression of active dense gene clusters. Sci Adv. 2017;3:e1700191. doi: 10.1126/sciadv.1700191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Donohoe ME, Zhang LF, Xu N, Shi Y, Lee JT. Identification of a Ctcf cofactor, Yy1, for the X chromosome binary switch. Mol Cell. 2007;25:43–56. doi: 10.1016/j.molcel.2006.11.017. [DOI] [PubMed] [Google Scholar]
- 20.Liu Z, Scannell DR, Eisen MB, Tjian R. Control of embryonic stem cell lineage commitment by core promoter factor, TAF3. Cell. 2011;146:720–731. doi: 10.1016/j.cell.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marsman J, O’Neill AC, Kao BR, Rhodes JM, Meier M, Antony J, Mönnich M, Horsfield JA. Cohesin and CTCF differentially regulate spatiotemporal runx1 expression during zebrafish development. Biochim Biophys Acta. 2014;1839:50–61. doi: 10.1016/j.bbagrm.2013.11.007. [DOI] [PubMed] [Google Scholar]
- 22.Gregor A, Oti M, Kouwenhoven EN, Hoyer J, Sticht H, Ekici AB, Kjaergaard S, Rauch A, Stunnenberg HG, Uebe S, et al. De novo mutations in the genome organizer CTCF cause intellectual disability. Am J Hum Genet. 2013;93:124–131. doi: 10.1016/j.ajhg.2013.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bastaki F, Nair P, Mohamed M, Malik EM, Helmi M, Al-Ali MT, Hamzeh AR. Identification of a novel CTCF mutation responsible for syndromic intellectual disability—a case report. BMC Med Genet. 2017;18:68. doi: 10.1186/s12881-017-0429-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Juraeva D, Haenisch B, Zapatka M, Frank J, GROUP Investigators, PSYCH-GEMS SCZ Working Group, Witt SH, Mühleisen TW, Treutlein J, Strohmaier J et al (2014) Integrated pathway-based approach identifies association between genomic regions at CTCF and CACNB2 and schizophrenia. PLoS Genet 10:e1004345 [DOI] [PMC free article] [PubMed]
- 25.Mannini L, Cucco F, Quarantotti V, Krantz ID, Musio A. Mutation spectrum and genotype-phenotype correlation in Cornelia de Lange syndrome. Hum Mutat. 2013;34:1589–1596. doi: 10.1002/humu.22430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deardorff MA, Noon SE, Krantz ID (1993) Cornelia de lange syndrome. In: GeneReviews(®), Pagon RA, Adam MP, Ardinger HH, Wallace SE, Amemiya A, Bean LJH, Bird TD, Ledbetter N, Mefford HC, Smith RJH et al (eds). University of Washington, Seattle
- 27.Alomer RM, da Silva EML, Chen J, Piekarz KM, McDonald K, Sansam CG, Sansam CL, Rankin S. Esco1 and Esco2 regulate distinct cohesin functions during cell cycle progression. Proc Natl Acad Sci USA. 2017;114:9906–9911. doi: 10.1073/pnas.1708291114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gordillo M, Vega H, Jabs EW (1993) Roberts Syndrome. In GeneReviews(®), Pagon RA, Adam MP, Ardinger HH, Wallace SE, Amemiya A, Bean LJH, Bird TD, Ledbetter N, Mefford HC, Smith RJH et al (eds).University of Washington, Seattle
- 29.Rudra S, Skibbens RV. Chl1 DNA helicase regulates Scc2 deposition specifically during DNA-replication in Saccharomyces cerevisiae. PLoS One. 2013;8:e75435. doi: 10.1371/journal.pone.0075435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Banerji R, Skibbens RV, Iovine MK. How many roads lead to cohesinopathies? Dev Dyn. 2017;246(11):881–888. doi: 10.1002/dvdy.24510. [DOI] [PubMed] [Google Scholar]
- 31.Cucco F, Musio A. Genome stability: what we have learned from cohesinopathies. Am J Med Genet C Semin Med Genet. 2016;172:171–178. doi: 10.1002/ajmg.c.31492. [DOI] [PubMed] [Google Scholar]
- 32.Martin CA, Murray JE, Carroll P, Leitch A, Mackenzie KJ, Halachev M, Fetit AE, Keith C, Bicknell LS, Fluteau A, et al. Mutations in genes encoding condensin complex proteins cause microcephaly through decatenation failure at mitosis. Genes Dev. 2016;30:2158–2172. doi: 10.1101/gad.286351.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.De Biase I, Chutake YK, Rindler PM, Bidichandani SI. Epigenetic silencing in Friedreich ataxia is associated with depletion of CTCF (CCCTC-binding factor) and antisense transcription. PLoS One. 2009;4:e7914. doi: 10.1371/journal.pone.0007914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Al-Mahdawi S, Sandi C, Mouro Pinto R, Pook MA. Friedreich ataxia patient tissues exhibit increased 5-hydroxymethylcytosine modification and decreased CTCF binding at the FXN locus. PLoS One. 2013;8:e74956. doi: 10.1371/journal.pone.0074956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jajodia A, Singh KD, Singhal A, Vig S, Datta M, Singh Y, Karthikeyan M, Kukreti R. Methylation of a HTR3A promoter variant alters the binding of transcription factor CTCF. RSC Adv. 2015;5:45710–45717. doi: 10.1039/C5RA04455C. [DOI] [Google Scholar]
- 36.De Souza RA, Islam SA, McEwen LM, Mathelier A, Hill A, Mah SM, Wasserman WW, Kobor MS, Leavitt BR. DNA methylation profiling in human Huntington’s disease brain. Hum Mol Genet. 2016;25:2013–2030. doi: 10.1093/hmg/ddw076. [DOI] [PubMed] [Google Scholar]
- 37.Naumann A, Kraus C, Hoogeveen A, Ramirez CM, Doerfler W. Stable DNA methylation boundaries and expanded trinucleotide repeats: role of DNA insertions. J Mol Biol. 2014;426:2554–2566. doi: 10.1016/j.jmb.2014.04.025. [DOI] [PubMed] [Google Scholar]
- 38.Lanni S, Goracci M, Borrelli L, Mancano G, Chiurazzi P, Moscato U, Ferrè F, Helmer-Citterich M, Tabolacci E, Neri G. Role of CTCF protein in regulating FMR1 locus transcription. PLoS Genet. 2013;9:e1003601. doi: 10.1371/journal.pgen.1003601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet. 1999;23:185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- 40.Kernohan KD, Vernimmen D, Gloor GB, Bérubé NG. Analysis of neonatal brain lacking ATRX or MeCP2 reveals changes in nucleosome density, CTCF binding and chromatin looping. Nucleic Acids Res. 2014;42:8356–8368. doi: 10.1093/nar/gku564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kernohan KD, Jiang Y, Tremblay DC, Bonvissuto AC, Eubanks JH, Mann MR, Bérubé NG. ATRX partners with cohesin and MeCP2 and contributes to developmental silencing of imprinted genes in the brain. Dev Cell. 2010;18:191–202. doi: 10.1016/j.devcel.2009.12.017. [DOI] [PubMed] [Google Scholar]
- 42.Gibbons RJ, Picketts DJ, Villard L, Higgs DR. Mutations in a putative global transcriptional regulator cause X-linked mental retardation with alpha-thalassemia (ATR-X syndrome) Cell. 1995;80:837–845. doi: 10.1016/0092-8674(95)90287-2. [DOI] [PubMed] [Google Scholar]
- 43.Bernier R, Golzio C, Xiong B, Stessman HA, Coe BP, Penn O, Witherspoon K, Gerdts J, Baker C, Vulto-van Silfhout AT, et al. Disruptive CHD8 mutations define a subtype of autism early in development. Cell. 2014;158:263–276. doi: 10.1016/j.cell.2014.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ishihara K, Oshimura M, Nakao M. CTCF-dependent chromatin insulator is linked to epigenetic remodeling. Mol Cell. 2006;23:733–742. doi: 10.1016/j.molcel.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 45.Shi L, Li M, Lin Q, Qi X, Su B. Functional divergence of the brain-size regulating gene MCPH1 during primate evolution and the origin of humans. BMC Biol. 2013;11:62. doi: 10.1186/1741-7007-11-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Trimborn M, Schindler D, Neitzel H, Hirano T. Misregulated chromosome condensation in MCPH1 primary microcephaly is mediated by condensin II. Cell Cycle. 2006;5:322–326. doi: 10.4161/cc.5.3.2412. [DOI] [PubMed] [Google Scholar]
- 47.Heath H, Ribeiro de Almeida C, Sleutels F, Dingjan G, van de Nobelen S, Jonkers I, Ling KW, Gribnau J, Renkawitz R, Grosveld F, et al. CTCF regulates cell cycle progression of alphabeta T cells in the thymus. EMBO J. 2008;27:2839–2850. doi: 10.1038/emboj.2008.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hirayama T, Tarusawa E, Yoshimura Y, Galjart N, Yagi T. CTCF is required for neural development and stochastic expression of clustered Pcdh genes in neurons. Cell Rep. 2012;2:345–357. doi: 10.1016/j.celrep.2012.06.014. [DOI] [PubMed] [Google Scholar]
- 49.Moore JM, Rabaia NA, Smith LE, Fagerlie S, Gurley K, Loukinov D, Disteche CM, Collins SJ, Kemp CJ, Lobanenkov VV, et al. Loss of maternal CTCF is associated with peri-implantation lethality of Ctcf null embryos. PLoS One. 2012;7:e34915. doi: 10.1371/journal.pone.0034915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Watson LA, Wang X, Elbert A, Kernohan KD, Galjart N, Bérubé NG. Dual effect of CTCF loss on neuroprogenitor differentiation and survival. J Neurosci. 2014;34:2860–2870. doi: 10.1523/JNEUROSCI.3769-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sams DS, Nardone S, Getselter D, Raz D, Tal M, Rayi PR, Kaphzan H, Hakim O, Elliott E. Neuronal CTCF is necessary for basal and experience-dependent gene regulation, memory formation, and genomic structure of BDNF and arc. Cell Rep. 2016;17:2418–2430. doi: 10.1016/j.celrep.2016.11.004. [DOI] [PubMed] [Google Scholar]
- 52.Bharadwaj R, Peter CJ, Jiang Y, Roussos P, Vogel-Ciernia A, Shen EY, Mitchell AC, Mao W, Whittle C, Dincer A, et al. Conserved higher-order chromatin regulates NMDA receptor gene expression and cognition. Neuron. 2014;84:997–1008. doi: 10.1016/j.neuron.2014.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Engmann O, Labonté B, Mitchell A, Bashtrykov P, Calipari ES, Rosenbluh C, Loh YE, Walker DM, Burek D, Hamilton PJ, et al. Cocaine-induced chromatin modifications associate with increased expression and three-dimensional looping of Auts2. Biol Psychiatry. 2017;82(11):794–805. doi: 10.1016/j.biopsych.2017.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Aguilar-Arnal L, Hakim O, Patel VR, Baldi P, Hager GL, Sassone-Corsi P. Cycles in spatial and temporal chromosomal organization driven by the circadian clock. Nat Struct Mol Biol. 2013;20:1206–1213. doi: 10.1038/nsmb.2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schuldiner O, Berdnik D, Levy JM, Wu JS, Luginbuhl D, Gontang AC, Luo L. piggyBac-based mosaic screen identifies a postmitotic function for cohesin in regulating developmental axon pruning. Dev Cell. 2008;14:227–238. doi: 10.1016/j.devcel.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Singh VP, Gerton JL. Cohesin and human disease: lessons from mouse models. Curr Opin Cell Biol. 2015;37:9–17. doi: 10.1016/j.ceb.2015.08.003. [DOI] [PubMed] [Google Scholar]
- 57.Remeseiro S, Cuadrado A, Gómez-López G, Pisano DG, Losada A. A unique role of cohesin-SA1 in gene regulation and development. EMBO J. 2012;31:2090–2102. doi: 10.1038/emboj.2012.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fujita Y, Masuda K, Bando M, Nakato R, Katou Y, Tanaka T, Nakayama M, Takao K, Miyakawa T, Tanaka T, et al. Decreased cohesin in the brain leads to defective synapse development and anxiety-related behavior. J Exp Med. 2017;214:1431–1452. doi: 10.1084/jem.20161517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Monahan K, Rudnick ND, Kehayova PD, Pauli F, Newberry KM, Myers RM, Maniatis T. Role of CCCTC binding factor (CTCF) and cohesin in the generation of single-cell diversity of protocadherin-α gene expression. Proc Natl Acad Sci USA. 2012;109:9125–9130. doi: 10.1073/pnas.1205074109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hirabayashi T, Yagi T. Protocadherins in neurological diseases. Adv Neurobiol. 2014;8:293–314. doi: 10.1007/978-1-4614-8090-7_13. [DOI] [PubMed] [Google Scholar]
- 61.Van den Berg DL, Azzarelli R, Oishi K, Martynoga B, Urbán N, Dekkers DH, Demmers JA, Guillemot F. Nipbl Interacts with Zfp609 and the integrator complex to regulate cortical neuron migration. Neuron. 2017;93:348–361. doi: 10.1016/j.neuron.2016.11.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Houlihan SL, Feng Y. The scaffold protein Nde1 safeguards the brain genome during S phase of early neural progenitor differentiation. elife. 2014;3:e03297. doi: 10.7554/eLife.03297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ou HD, Phan S, Deerinck TJ, Thor A, Ellisman MH, O’Shea CC. ChromEMT: visualizing 3D chromatin structure and compaction in interphase and mitotic cells. Science. 2017;357:eaag0025. doi: 10.1126/science.aag0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Beagrie RA, Scialdone A, Schueler M, Kraemer DC, Chotalia M, Xie SQ, Barbieri M, de Santiago I, Lavitas LM, Branco MR, et al. Complex multi-enhancer contacts captured by genome architecture mapping. Nature. 2017;543:519–524. doi: 10.1038/nature21411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stevens TJ, Lando D, Basu S, Atkinson LP, Cao Y, Lee SF, Leeb M, Wohlfahrt KJ, Boucher W, O’Shaughnessy-Kirwan A, et al. 3D structures of individual mammalian genomes studied by single-cell Hi-C. Nature. 2017;544:59–64. doi: 10.1038/nature21429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Boyle MI, Jespersgaard C, Brøndum-Nielsen K, Bisgaard AM, Tümer Z. Cornelia de Lange syndrome. Clin Genet. 2015;88:1–12. doi: 10.1111/cge.12499. [DOI] [PubMed] [Google Scholar]
- 67.Vega H, Waisfisz Q, Gordillo M, Sakai N, Yanagihara I, Yamada M, van Gosliga D, Kayserili H, Xu C, Ozono K, et al. Roberts syndrome is caused by mutations in ESCO2, a human homolog of yeast ECO1 that is essential for the establishment of sister chromatid cohesion. Nat Genet. 2005;37(5):468–470. doi: 10.1038/ng1548. [DOI] [PubMed] [Google Scholar]
- 68.Capo-Chichi JM, Bharti SK, Sommers JA, Yammine T, Chouery E, Patry L, Rouleau GA, Samuels ME, Hamdan FF, Michaud JL, et al. Identification and biochemical characterization of a novel mutation in DDX11 causing Warsaw breakage syndrome. Hum Mutat. 2013;34:103–107. doi: 10.1002/humu.22226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Van der Lelij P, Chrzanowska KH, Godthelp BC, Rooimans MA, Oostra AB, Stumm M, Zdzienicka MZ, Joenje H, de Winter JP. Warsaw breakage syndrome, a cohesinopathy associated with mutations in the XPD helicase family member DDX11/ChlR1. Am J Hum Genet. 2010;86:262–266. doi: 10.1016/j.ajhg.2010.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kawauchi S, Calof AL, Santos R, Lopez-Burks ME, Young CM, Hoang MP, Chua A, Lao T, Lechner MS, Daniel JA, et al. Multiple organ system defects and transcriptional dysregulation in the Nipbl(±) mouse, a model of Cornelia de Lange Syndrome. PLoS Genet. 2009;5:e1000650. doi: 10.1371/journal.pgen.1000650. [DOI] [PMC free article] [PubMed] [Google Scholar]