Abstract
As the female gamete, meiotic oocytes provide not only half of the genome but also almost all stores for fertilization and early embryonic development. Because de novo mRNA transcription is absent in oocyte meiosis, protein-level regulations, especially the ubiquitin proteasome system, are more crucial. As the largest family of ubiquitin E3 ligases, Skp1–Cullin–F-box complexes recognize their substrates via F-box proteins with substrate-selected specificity. However, the variety of F-box proteins and their unknown substrates hinder our understanding of their functions. In this report, we find that Fbxo30, a new member of F-box proteins, is enriched in mouse oocytes, and its expression level declines substantially after the metaphase of the first meiosis (MI). Notably, depletion of Fbxo30 causes significant chromosome compaction accompanied by chromosome segregation failure and arrest at the MI stage, and this arrest is not caused by over-activation of spindle assembly checkpoint. Using immunoprecipitation and mass spectrometric analysis, we identify stem-loop-binding protein (SLBP) as a novel substrate of Fbxo30. SLBP overexpression caused by Fbxo30 depletion results in a remarkable overload of histone H3 on chromosomes that excessively condenses chromosomes and inhibits chromosome segregation. Our finding uncovers an unidentified pathway-controlling chromosome segregation and cell progress.
Electronic supplementary material
The online version of this article (10.1007/s00018-019-03038-z) contains supplementary material, which is available to authorized users.
Keywords: Cell cycle, Ubiquitination, F-box family, Chromosome condensation, SLBP
Introduction
Life begins with fertilization of an oocyte and a sperm. The former provides not only half the genome but also almost all stores for fertilization and early embryonic development [1, 2]. Thus, the quality of oocytes is essential for genomic delivery and life generation. In mammalian ovaries, fully grown oocytes in follicles are arrested at the dictyate stage of the first meiotic prophase, namely, the germinal vesicle (GV) stage. Upon stimulation with a surge in pituitary hormone, meiosis resumes with the signature GV breakdown (GVBD) and chromatin condensation [3]. During the prophase of the first meiosis (Pro-MI), the spindle starts its assembly accompanied by the capture of condensed chromosomes. When all chromosomes align at the equator of the spindle, the oocyte enters the metaphase of the first meiosis (MI), followed by chromosome segregation in anaphase I (AI) and polar body exclusion during the metaphase of the second meiosis (MII) [4–6]. Upon fertilization, the oocyte fuses with the sperm and begins early embryonic development. Errors in meiotic progression result in fertilization failure or embryonic development abnormalities, leading to early abortion and female infertility.
Successful oocyte meiosis requires a hierarchical network to ensure the cell cycle process and chromosome integrity [7, 8]. Because de novo mRNA transcription is absent in fully grown oocytes [9, 10], the regulation of proteins, especially the ubiquitin proteasome system, is critical. Accumulative evidence has shown that different sets of proteins are correlated with distinct characteristics of oocytes during different meiotic stages [11]. The progression of meiosis requires the timely degradation of specific proteins by fine regulatory mechanisms [12, 13]. The MI-to-AI transition is a typical process in which numerous proteins undergo programmed elimination, allowing chromosome segregation during meiotic division [14–17].
The specificity of the ubiquitin proteasome system is monitored by E3 ligases, including the major representative Skp1–Cullin–F-box (SCF) complex. The SCF complex is the largest family of ubiquitin ligases that ubiquitinate various substrate proteins [18, 19]. The complex consists of the invariant components S-phase kinase-associated protein 1 (SKP1), E3 ligase RBX1 and cullin1 and the variable F-box proteins that provide specific substrate selectivity [20, 21]. F-box proteins are defined by the presence of the F-box motif, which is a protein–protein interaction domain that recruits F-box proteins into the SCF complex [22, 23]. Although a broad range of F-box protein substrates exists in cells, many have not been identified. As a new member of F-box proteins, the functions of Fbxo30 and its substrates in oocytes are largely unknown.
In this study, we find that Fbxo30 is enriched in mouse oocytes and its expression level decreases after the MI stage. In cells, Fbxo30 is localized at the spindle during the Pro-MI and MI stages. Interestingly, Fbxo30 depletion causes significantly compacted chromosomes and chromosome segregation that is not resulted from spindle assembly checkpoint (SAC) activation. By immunoprecipitation and mass spectrometric analysis, we identify stem-loop-binding protein (SLBP), a stabilizer of histone mRNA by binding its 3′ untranslated region (UTR) stem-loop structure, as a novel substrate of Fbxo30. Upregulation of SLBP caused by Fbxo30 RNA interference (RNAi) results in a remarkable overload of histone H3 on chromosomes, leading to chromosome overcondensation and inhibiting chromosome segregation, and this defect could be rescued by SLBP knockdown (KD). Our study therefore reveals an unidentified function of Fbxo30 in the regulation of chromosome segregation and establishes a molecular link between Fbxo30 and oocyte maturation.
Materials and methods
Chemicals and antibodies
All chemicals and reagents were purchased from Sigma (St. Louis, MO, USA) except for those specifically mentioned as follows: anti-Fbxo30 antibody (Santa Cruz Biotechnology; Dallas, Texas, USA; Cat#: sc-138935; immunofluorescence (IF), 1:200; western blot (WB), 1:500), anti-SLBP antibody (Thermo Fisher Scientific; Waltham, Massachusetts, USA; Cat#: PA5-41819; IF, 1:100; WB, 1:1000), anti-histone H2A (Cell Signaling Technology; Danvers, Massachusetts, USA; Cat#: 12349; WB, 1:1000), anti-histone H2B (Cell Signaling Technology; Cat#: 12364; WB, 1:1000), anti-histone H3 antibody (Cell Signaling Technology; Cat#: 14269; IF, 1:400; WB, 1:1000), anti-histone H4 antibody (Cell Signaling Technology; Cat#: 13919; WB, 1:1000), and anti-Ubiquitin antibody (Cell Signaling Technology; Cat#: 3933; WB, 1:1000).
Oocyte collection and culture
ICR mice were kept and handled according to the policies published by the Ethics Committee of Peking University Health Science Center. After injections of 5 IU pregnant mare serum gonadotropin (PMSG) for 44–46 h, female ICR mice (4–6 weeks) were humanely euthanized. Fully grown oocytes arrested at the dictyate stage of the first meiotic prophase were collected from the ovaries of ICR mice [24]. Only those oocytes with clear GVs were cultured in M16 medium under mineral oil in a humidified atmosphere of 5% CO2 at 37 °C. Oocytes were harvested for subsequent analysis at the GV (0 h), GVBD (2 h), Pro-MI (6 h), MI (8 h), and MII (12 h) stages.
Microinjection and RNAi
We performed microinjection using a Diaphot ECLIPSE Ti-S inverted microscope (Nikon UK Ltd., Kingston upon Thames, Surrey, UK) equipped with MM-93B hydraulic three-dimensional micromanipulators (Narishige Inc., Sea Cliff, NY, USA). To knockdown Fbxo30 or SLBP, 25 μM non-targeting (control) or targeting small interfering RNA (siRNA) were microinjected into oocytes. During and after microinjection, oocytes were arrested at the GV stage with 2.5 μM milrinone for 16 h to allow siRNA interference followed by a thorough wash and cultured in fresh M16 medium. The Fbxo30 siRNA sequences were as follows: 5′-GCCACAAGAGACAUUGGCAUGUUAA-3′, 5′-GAGAUGUUGAUGAAGUGGCACAAUU-3′ and 5′-CCUUUGAACGAGUAGCUUGCUUAA-3′ (Invitrogen, USA). Slbp siRNA sequences: 5′-GAGAGAGAGGAAGUCAUCAUCGGGA-3′, 5′-GAGACGGAUGAAAGUGUCUUGAUGA-3′ and 5′-AAGAAGGAUGUGAUUUGCAAGAAAU-3′ (Invitrogen, USA).
Immunofluorescence microscopy and image analysis
Oocytes were fixed with 4% paraformaldehyde in PBS (pH 7.4) for 30 min and were permeabilized by 0.5% Triton X-100 in PBS for 25 min at room temperature. Then, oocytes were blocked in 1% BSA-supplemented PBS for 1 h at room temperature followed by incubation with primary antibodies overnight at 4 °C. After oocytes were washed three times in PBS containing 0.1% Tween 20 and 0.01% Triton X-100, they were labelled with the appropriate fluorescent secondary antibodies for 1 h at room temperature. After three washes, oocytes were counterstained with Hoechst 33342 (10 μg/ml) for 15 min or with the fluorescent α-tubulin antibody (1:200) for 1 h at room temperature. Immunofluorescence signals were observed using a confocal laser scanning microscope (Carl Zeiss 710, Jena, Germany). Instrument settings were kept constant for each replicate.
Chromosome spreading
Chromosome spreading was performed according to the previous report [25]. In brief, the zona pellucidae of oocytes was removed by brief exposure to acidic M2 medium (pH 2.0). After recovery in M2 medium for 5 min, oocytes were fixed in a drop of hypotonic fixation solution (1% paraformaldehyde in distilled H2O, pH 9.2, 0.15% Triton X-100 and 3 mM dithiothreitol) on a glass slide for 30 min. In the hypotonic buffer, oocytes burst and slowly melt onto the slide. Spreading of chromosomes was achieved when oocytes were positioned evenly across the slide and allowed to air dry at room temperature. The immunofluorescence and image analysis of histone H3 were performed as described in “Immunofluorescence microscopy and image analysis” above.
Three-dimensional modeling
Chromosome spreading of meiotic oocytes was imaged as a z-series at 0.5-µm intervals to capture the entire chromosome structure via three-dimensional (3D) confocal z-stacks. CZI files were imported, and chromosome status and H3 staining were analysed using the Imaris program (Bitplane, Belfast, United Kingdom) [26, 27]. Isosurface renderings of the chromosomes were created and rotated in 3D space to examine their alignment and morphology. 3D computerized reconstruction of H3 staining was modeled using the Surface Creation Wizard after manual threshold optimizations for location analyses.
Immunoprecipitation and western blot
Immunoprecipitation was performed with the indicated antibodies according to the standard protocol of the ProFound Mammalian Co-Immunoprecipitation kit (Pierce, USA). The immunoprecipitates were used for the following mass spectrometry analysis or western blot. For western blot, oocytes were lysed in 4× LDS sample buffer (Thermo Fisher Scientific, Waltham, Massachusetts, USA) containing protease inhibitor. After the proteins were boiled for 5 min, they were separated on SDS-PAGE and electrically transferred onto PVDF membranes (Millipore, Darmstadt, Germany). The membranes were blocked in TBST containing 5% skimmed milk for 1 h at room temperature and then incubated with the indicated primary antibodies overnight at 4 °C. After three washes in TBST, the membranes were incubated with the appropriate horse radish peroxidase (HRP)-conjugated secondary antibody for 1 h at room temperature. Finally, the membranes were detected by the enhanced chemiluminescence detection system (Amersham Biosciences, Piscataway, NJ, USA).
Isobaric tags for relative and absolute quantitation (iTRAQ)-based proteomic analysis
Oocytes (3000/sample) were digested with trypsin and subsequently processed using the 8-plex iTRAQ reagent (Applied Biosystems, Carlsbad, CA, USA) according to the manufacturer’s protocol. Peptides of each subject were labelled with different iTRAQ tags and incubated at room temperature for 2 h. Labelled peptides were pooled and separated into 12 fractions by nanoAcquity UPLC (Waters Corporation, 34 Maple Street Milford, MA, 01757 USA) using a Phenomenex AQ column (100 μm × 150 mm, 3 μm, C18) coupled with tandem mass spectrometry (LC–MS/MS) in a Q-Exactive high-resolution mass spectrometer (Thermo Fisher Scientific, Waltham, MA, USA). The mass spectrometer (MS) was operated in a data-dependent manner. Full MS scans were performed in the range of 300–2000 Da at a 70,000 resolution. For tandem MS (MS/MS) scans, the 10 most abundant ions with multiple charge states were selected. The isolation window was set as 2.0 Da, and the dynamic exclusion was 10 s. The MS/MS spectra were obtained using Mascot Distiller 2.4 (Matrix Science, London, UK), and proteins were identified in the UniProt human database by the Mascot search engine version 2.4 (Matrix Science, London, UK). All proteomic and bioinformatic analyses were performed by Junchen Company (Beijing, China) and Zhengdakangjian Company (Beijing, China). Proteins with at least two unique peptides were validated and selected for further quantitative analysis with Scaffold Q+ software. Proteins with at least two unique peptides whose fold change was greater than or equal to 1.5 with a p value less than 0.05 were considered to be differentially expressed.
Ubiquitination assay
For the construction of expression vectors encoding Myc-His-Fbxo30 and Flag-EGFP-SLBP, mouse Fbxo30 and SLBP cDNA were subcloned into pcDNA3.1/myc-His(−)A and pEGFP-N1-FLAG, respectively. Ubiquitin cDNA with a HA tag was cloned into pEF1a-Luc-IRES-Neo. After transfection with various mixtures of plasmids as indicated, 293T cells were treated with MG132 (4 μΜ) for 6 h to inhibit proteasome activity. Cells were lysed in lysis buffer containing 50 mM Tris–Cl (pH 7.5), 150 mM NaCl, 10% glycerol, 0.5% NP-40, 1 mM EDTA, and 0.5% SDS in the presence of protease inhibitors. After centrifugation, the supernatant was immunoprecipitated by an anti-Flag M2 affinity gel and eluted using a buffer with high ionic concentration (50 mM Tris–Cl, pH 7.5, 500 mM NaCl, 10% glycerol, 0.5% NP-40, and 1 mM EDTA). Ubiquitination analysis was carried out by immunoblotting with anti-ubiquitin, SLBP, and Fbxo30 antibodies. Whole-cell lysate (WCL) was used as input, and β-actin was used as a loading control.
Quantitative PCR
Total RNA was isolated from oocytes (200/sample) using the RNeasy Micro kit (QIAGEN, Germany). The RNA was then reverse-transcribed using a RevertAid Fit Strand cDNA Synthesis Kit (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s protocols to generate cDNA. Quantitative PCR analysis was performed using SYBR Green PCR master mix with the QuantStudio 3 Real-Time PCR system (Applied Biosystems, Singapore). The primer sequences were as follows: H2A, 5′-CGACGAGGAGCTCAACAAG-3′ (sense), 5′-ACTTGCCCTTCGCCTTATG-3′ (anti-sense); H2B, 5′-CGAAGAAGGGCTCCAAGAA-3′ (sense), 5′-CTTGTACACGTACACCGAGTAG-3′ (anti-sense); H3, 5′-GATCGCGCAGGACTTCAA-3′ (sense), 5′-GTCCTCAAACAGACCCACAA-3′ (anti-sense); H4, 5′-GGTTCTTCGCGATAACATCCA-3′ (sense), 5′-TCGTAGATGAGACCGGAGATG-3′ (anti-sense). The relative expression levels of targeted genes were calculated using the 2−ΔΔCT method.
Statistical analyses
All experiments were performed in triplicate unless otherwise indicated. Mean values and standard deviations were plotted. Student’s t test was used for statistical analyses.
Results
The expression and localization of Fbxo30 during oocyte meiosis
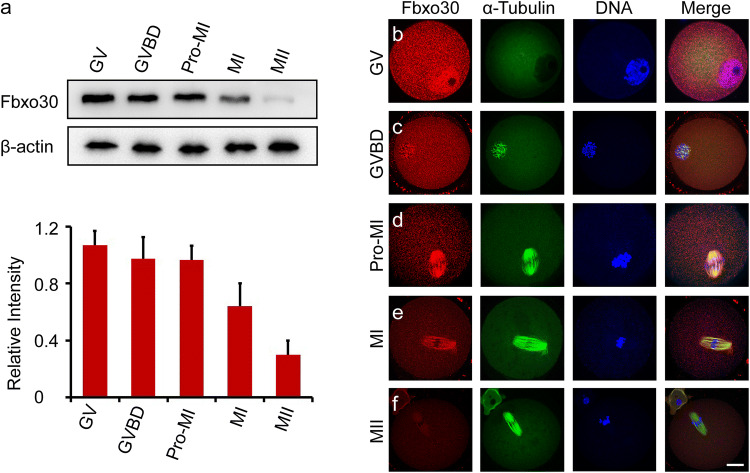
To investigate the role of Fbxo30 during meiosis, we first examined the expression level of Fbxo30. Western blot of Fbxo30 was performed on oocytes harvested at different stages. As shown in Fig. 1a, Fbxo30 showed relatively constant expression from the GV to Pro-MI stages. When oocytes entered MI, the expression level began to decrease. During the MII stage before fertilization, oocytes expressed the lowest level of this protein. Furthermore, we examined the subcellular localization of Fbxo30 in oocytes. Fbxo30 was mainly localized to the nucleus in GV-stage oocytes and was then distributed in the cytoplasm with heavier staining around chromosomes during GVBD (Fig. 1b, c, Fig. S1a). As the oocyte entered Pro-MI, Fbxo30 was obviously localized at the spindle (Fig. 1d, Fig. S1a). During MI, when the chromosomes aligned at the equatorial plate, spindle-localized Fbxo30 expression decreased (Fig. 1e). At the MII stage, when homologous chromosomes had already separated, little Fbxo30 was detected in the oocyte (Fig. 1f, Fig. S1a). The change in Fbxo30 expression levels before and after the MI stage implies a potential function of this protein during oocyte meiosis.
Fig. 1.
Expression and subcellular localization of Fbxo30 during oocyte meiosis. a Protein level of Fbxo30 during meiotic stages. Two hundred oocytes at each stage were harvested and lysed for western blot of Fbxo30. β-Actin was used as the loading control. The relative intensity of immunoreactive bands was quantified in the histogram below. The error bars represent the standard deviation. b–f Subcellular localization of Fbxo30 in meiotic stages. Oocytes at each meiotic stage were harvested for immunofluorescent staining of Fbxo30, microtubules, and DNA. Scale bar, 20 μm
Depletion of Fbxo30 blocks chromosome segregation and meiotic progress
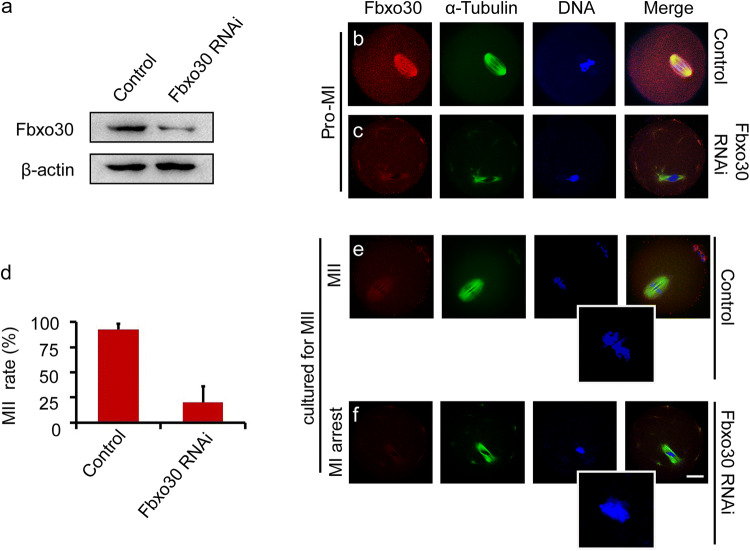
To gain insight into the role of Fbxo30 in meiosis, a loss-of-function experiment using gene-targeting siRNA microinjection was performed to deplete Fbxo30 in oocytes. For control oocytes, the same amount of non-targeting siRNA was microinjected. Western blot of Fbxo30 from Pro-MI oocytes confirmed the effective depletion of the protein (Fig. 2a). Consistently, immunofluorescent staining of Fbxo30 was almost undetectable in Fbxo30 RNAi oocytes before the MI stage (Fig. 2b, c). Fbxo30 RNAi did not affect the rate of GVBD during oocyte maturation (Fig. S1b). When cultured to the MII stage, nearly all control oocytes (92.3%) excluded the polar body and reached MII. In contrast, most Fbxo30 RNAi oocytes (80.6%) were not able to exclude the polar body. Focussing on the spindle and chromosomes revealed that homologous chromosomes did not separate and that polar body exclusion did not occur in these Fbxo30 knockdown (Fbxo30-KD) oocytes (Fig. 2d–f), demonstrating that Fbxo30 depletion caused meiotic arrest with chromosome segregation defects. In addition, a considerable portion of Fbxo30 RNAi oocytes (56%) showed abnormal spindle assembly beginning at the Pro-MI stage and especially in MI (Figs. 2b–f, 5b, Fig. S2). This abnormal assembly may have been caused by the overcondensation of chromosomes resulting in an inability of the chromosomes to be pulled and aligned by the spindle in a normal manner, thereby unbalancing the force between chromosomes and microtubules during spindle assembly. Furthermore, chromosome overcondensation may affect spindle assembly directly as an increasing number of studies have demonstrated that chromosomes contribute to spindle assembly in acentrosomal oocytes [4, 28, 29].
Fig. 2.
Effects of Fbxo30 depletion on meiotic progression. a Expression level of Fbxo30 in control and Fbxo30-KD oocytes. Two hundred oocytes in each group were harvested and lysed for western blot of Fbxo30. β-Actin was used as the loading control. b, c Immunofluorescent staining of Fbxo30 in control and Fbxo30-KD oocytes. Little Fbxo30 signal was detected in Fbxo30-KD meiotic oocytes. d MII rates of control and Fbxo30-KD oocytes. Each group of one repeat contained at least 100 oocytes. Statistical data were summarized from three independent repeats. The error bars represent the standard deviation. e, f Fbxo30 knockdown resulted in chromosome segregation failure and meiotic arrest. Oocytes in the control and Fbxo30-KD groups were cultured for 12 h followed by staining with Fbxo30, α-tubulin and Hoechst. Most of the Fbxo30-KD oocytes were arrested at the MI stage with chromosome segregation failure. The boxed regions denote the magnifications. Scale bar, 20 μm
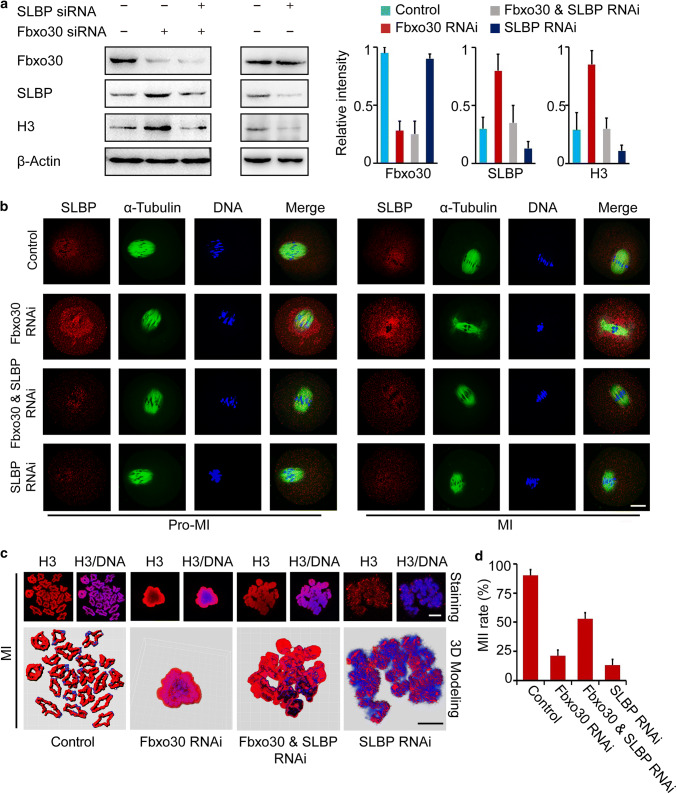
Fig. 5.
Depletion of SLBP rescues meiotic arrest in Fbxo30-KD oocytes. a Protein levels of SLBP, H3, and Fbxo30 in control, Fbxo30-KD, Fbxo30/SLBP double-KD, and SLBP-KD oocytes. One hundred oocytes were harvested for each western blot sample. The relative intensity of the bands is summarized in the histogram. b Immunofluorescence of SLBP in control, Fbxo30-KD, Fbxo30/SLBP double-KD and SLBP-KD oocytes. Scale bar, 20 μm. c Chromosome spreading and 3D modeling of chromosomes from control, Fbxo30-KD, Fbxo30/SLBP double-KD, and SLBP-KD oocytes. Chromosome spreading was performed with MI-stage oocytes followed by immunostaining of H3. 3D models were constructed using the Imaris program. Isosurface renderings of the chromosomes were created and rotated in 3D space to assess chromosome morphology. 3D painting of H3 on chromosomes was created using the Surface Creation Wizard after manual threshold optimizations for location analyses. Scale bar, 5 μm. d MII rates of control, Fbxo30-KD, Fbxo30/SLBP double-KD and SLBP-KD oocytes. Each group of one repeat contained at least 100 oocytes. Statistical data were summarized from three independent repeats. Error bars represent the standard deviation
SLBP is a substrate of Fbxo30 in oocyte meiosis
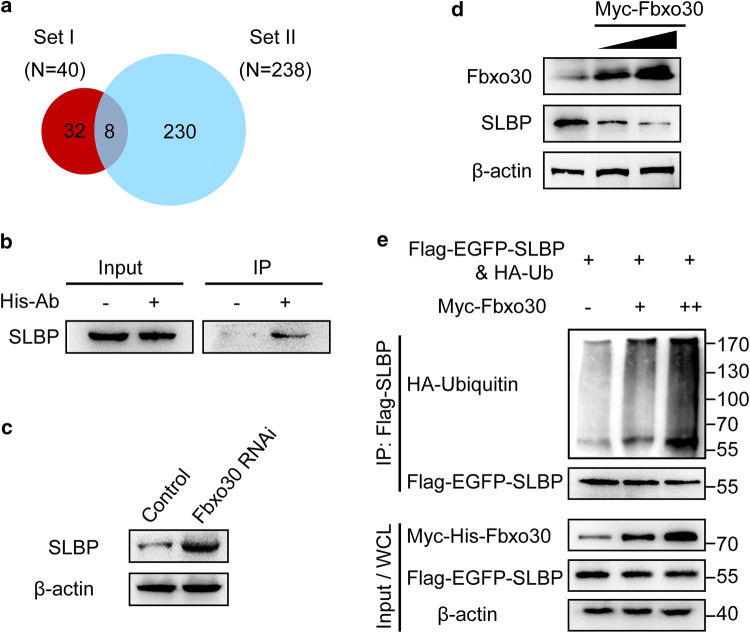
Because F-box proteins function for substrate selection in the ubiquitin proteasome system [20, 21], we investigated the potential substrate of Fbxo30 and its molecular mechanism in oocyte meiosis. The substrate selected by Fbxo30 undergoes degradation; therefore, Fbxo30 depletion would give rise to an increase in the protein level of this substrate. We therefore compared the proteome between control and Fbxo30 RNAi oocytes. In each group, 3000 MI-stage oocytes were harvested for iTRAQ-based quantitative proteomics analysis. After digestion with trypsin, the peptides were labelled with iTRAQ reagents. Labelled peptides were then pooled and separated into 12 fractions by nanoAcquity UPLC, followed by MS/MS analysis with a Q-Exactive high-resolution mass spectrometer. A total of 2306 proteins (FDR < 1%) were identified from the labelled samples in control and Fbxo30 RNAi oocytes (Table S1). To identify the potential substrate, we defined upregulated proteins in Fbxo30 RNAi oocytes as follows: fold change ≥ 1.5 and p value less than 0.05. Proteins were identified and relatively quantified in terms of at least two unique peptides. Based on the screening criteria above, 40 proteins (referred to as Set I) were identified to be more highly expressed in Fbxo30 RNAi oocytes than in control oocytes (Table S2).
To specifically target the substrate, we further performed an Fbxo30 immunoprecipitation experiment. Purified His-Fbxo30 was incubated with the lysate of Fbxo30 RNAi oocytes in the presence of the His-tag antibody and Protein A/G Sepharose beads. The associated proteins were pulled down and analysed by LC–MS/MS. Two hundred and thirty-eight Fbxo30-associated proteins (refer to Set II) were identified (Table S3). Among these proteins, eight candidates were found in Set I (Table S4; Fig. 3a). Of note, 13 unique peptides from SLBP were detected. To confirm the interaction between Fbxo30 and SLBP, we immunoblotted the rest of the immunoprecipitates pulled down by His-Fbxo30 described above using the SLBP antibody. As shown in Fig. 3b, His-Fbxo30 immunoprecipitated SLBP in the oocyte lysate. On the other hand, Fbxo30 knockdown resulted in an increase in SLBP expression in oocytes (Fig. 3c).
Fig. 3.
SLBP is a substrate of the Fbxo30-mediated ubiquitin proteasome system. a Screening of the substrate of Fbxo30 by mass spectrometry analysis. Set I includes the candidates significantly upregulated in Fbxo30-KD oocytes analysed by iTRAQ technique. Set II includes the candidates identified by LC–MS/MS that potentially interact with Fbxo30. The Venn diagram represents the overlap proteins from Set I and Set II. b SLBP interacted with Fbxo30. Purified His-Fbxo30 was incubated with the lysate of Fbxo30-KD oocytes in the presence of Protein A/G beads and the His antibody. The immunoprecipitate was blotted with an SLBP antibody. Ten percent of the WCL was loaded as the input. c Protein levels of SLBP in control and Fbxo30-KD oocytes were examined by western blot. d Overexpression of Fbxo30 led to a decrease in endogenous SLBP. 293T cells were transfected with Myc-Fbxo30 followed by western blot of SLBP. β-Actin was used as the loading control. e The ubiquitination level of SLBP was increased with Fbxo30 overexpression. 293T cells expressing Flag-EGFP-SLBP and HA-Ub were co-transfected with or without the Myc-Fbxo30 expression plasmid. Cellular lysate was immunoprecipitated by anti-FLAG M2 affinity gel (to bind Flag-SLBP) followed by western blot with the ubiquitin and SLBP antibody, respectively. Ten percent of the WCL was used as the input
To validate the relationship of the E3 ligase substrate between Fbxo30 and SLBP, we expressed Myc-Fbxo30 in 293T cells and tested the level of endogenous SLBP. As expected, increasing expression of Myc-Fbxo30 resulted in a decrease in the expression of SLBP (Fig. 3d). In parallel, 293T cells expressing Flag-SLBP and HA-Ub were co-transfected with Myc-Fbxo30 or empty vector, followed by proteasome inhibitor treatment. In the immunoprecipitates pulled down by anti-Flag beads, the ubiquitination level of Flag-SLBP increased in a dose-dependent manner with an increasing amount of Myc-Fbxo30 (Fig. 3e). These data indicated that SLBP interacted with Fbxo30 and was degraded by Fbxo30-mediated ubiquitination.
Fbxo30 depletion causes an accumulation of SLBP and H3 in oocytes
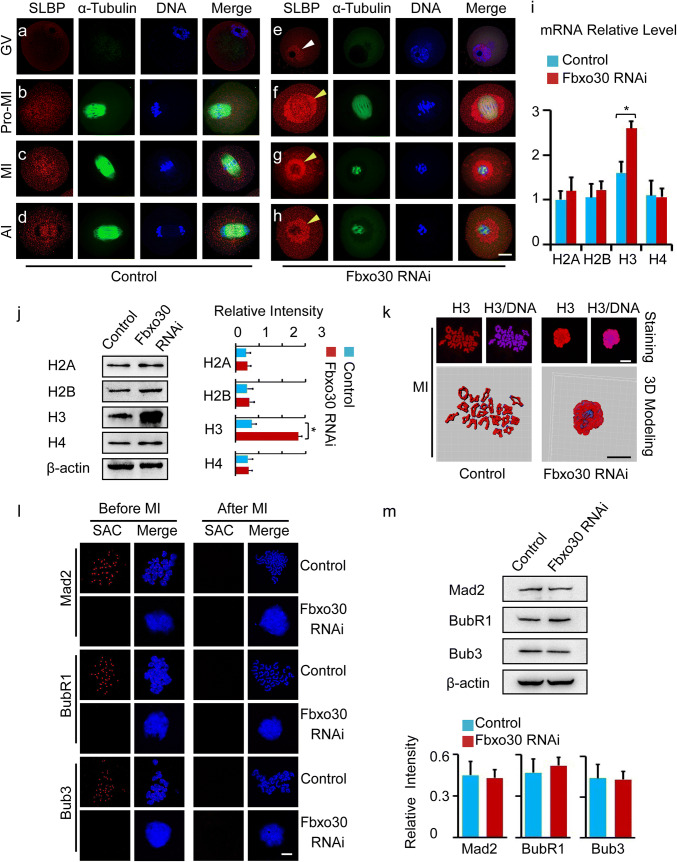
Because SLBP is a substrate of Fbxo30, we investigated the cellular behaviour of SLBP in oocytes. Before GVBD, SLBP was almost undetectable in whole oocytes (Fig. 4a). As oocytes reached Pro-MI and MI after GVBD, a mild SLBP signal was found around the spindle and chromosomes (Fig. 4b, c). During AI, when the chromosomes segregated, the expression of SLBP remained at a mild level (Fig. 4d). When Fbxo30 was depleted by siRNA, SLBP became detectable in the nucleus before GVBD (Fig. 4e). Notably, after GVBD, an abundance of SLBP was expressed and overloaded around the spindle and chromosomes in Fbxo30 RNAi oocytes. With further culture, the oocytes showed disorganized chromosomes surrounded by accumulated SLBP and were arrested at MI with chromosome segregation failure (Fig. 4f–h).
Fig. 4.
Knockdown of Fbxo30 results in the accumulation of SLBP and H3 in oocytes. a–h Subcellular distribution of SLBP in control and Fbxo30-KD oocytes at the GV (0 h), Pro-MI (6 h), MI (8 h), and AI (10 h) stages. Oocytes were stained with SLBP, microtubules, and DNA. The white arrowhead indicates the increased SLBP in the nuclei of Fbxo30-KD oocytes at the GV stage, while the yellow arrowheads indicate the accumulated SLBP around the spindle after GVBD. Scale bar, 20 μm. i mRNA expression level of core histones in control and Fbxo30-KD oocytes. Total RNA was extracted from 100 oocytes and then reverse-transcribed to cDNA. mRNA expression was examined by qPCR. j Protein expression of core histones in control and Fbxo30-KD oocytes. Two hundred oocytes were lysed, followed by western blotting of H2A, H2B, H3, and H4. β-Actin was used as a loading control. The relative intensity of the bands is summarized in the histogram. k Chromosome spreading and 3D modeling of chromosomes from control and Fbxo30-KD oocytes. Chromosome spreading was performed with MI-stage oocytes followed by immunostaining of H3. 3D models were constructed using the Imaris program. Isosurface renderings of the chromosomes were created and rotated in 3D space to assess chromosome morphology. 3D painting of H3 on chromosomes was created using the Surface Creation Wizard after manual threshold optimizations for location analyses. l The status of SAC before and after the MI stage from control and Fbxo30-KD oocytes. The spread of chromosomes from oocytes before or after MI (MII) stained for core SAC components Mad2, BubR1, and Bub3. Normally in control oocytes, the SAC was active (positive kinetochore staining) before MI and became inactive after chromosome segregation. SAC components failed to be loaded onto kinetochores before MI in Fbxo30-KD oocytes. Scale bar, 5 μm. m The expression of SAC proteins in control and Fbxo30-KD oocytes. Two hundred oocytes were harvested for each western blot sample of Mad2, BubR1, and Bub3. β-Actin was used as a loading control. The relative intensity of the bands is summarized in the histogram. No significant difference was observed between the control and Fbxo30-KD groups. Error bars represent the standard deviation. The asterisk indicates a significant difference
After nuclear envelope breakdown in eukaryotic cells, loose chromatins with long arms are transferred to compact chromosomes packaged into rod-shaped structures [30, 31]. This process ensures proper separation of chromosomes during cell division and is highly regulated by related factors, including histones [32–34]. By directly binding to the 3′ end of histone mRNA, SLBP participates in almost all aspects of histone mRNA metabolism and is essential for histone translation [35, 36]. We thus tested the expression of core histones in Fbxo30-KD oocytes in which SLBP protein was upregulated. The mRNA expression levels of histone H2A, H2B, and H4 were similar in control and Fbxo30 RNAi oocytes. However, the level of histone H3 in Fbxo30 RNAi oocytes was significantly higher than that in control oocytes (Fig. 4i). Moreover, western blot confirmed the overexpression of H3 at the protein level (Fig. 4j). Previous reports highlighted the importance of phosphorylation of serine 10 of H3 (pH3Ser10) [32, 33, 37]; thus, we examined this phosphorylation by western blot and chromosome spreading staining. As expected, pH3Ser10 in Fbxo30 RNAi oocytes was higher than that in the control (Fig. S3). As histone is required for chromosome packaging after nuclear envelope breakdown, we wondered the status of chromosomes when H3 was expressed abnormally in Fbxo30 RNAi oocytes. Chromosome spreading of meiotic oocytes was performed followed by analysis of chromosome status as well as H3 staining of chromosomes. In normal meiotic oocytes before AI, histone H3 was evenly expressed on whole individual chromosomes except for the region of the kinetochore. All 20 pairs of homologous chromosomes exhibited a strip-like shape, and no pairs combined with one another. In contrast, H3 expression in most Fbxo30 RNAi oocytes was not well organized and was found roughly surrounding the disorganized chromosomes. The chromosomes lost their strip-like shape and showed chaotic morphology. These chromosomes were condensed together, resulting in obvious combination or fusion with each other, which was confirmed by 3D modeling using the Imaris program (Fig. 4k). To clarify whether the “chromosome mess” phenotype could be due to MI arrest, we treated oocytes with nocodazole, a classic drug for arresting cells at MI stage. Although all the treated oocytes were arrested at MI (even for 12 h), none of them displayed hypercondensed chromsomes (Fig. S4a). In addition, to examining the progression of chromosome condensation, we detected chromosome status in meiotic oocytes by close time point staining. Oocytes were examined at early Pro-MI (2 h after GVBD), late Pro-MI (4 h after GVBD), and MI (6 h after GVBD). The bivalents were visualized by SMC3 and Hoechst 33342 co-staining. Specifically, Fbxo30 RNAi oocytes in early Pro-MI showed bivalents with similar morphology to control oocytes. Beginning in late Pro-MI, however, the bivalents in Fbxo30 RNAi oocytes seemed to be condensed with fuzzy localization of SMC3. When the oocytes processed to MI, chromosomes were further condensed, and nearly no bivalents could be detected (Fig. S4b).
In general, MI arrest in oocytes requires SAC activation [15, 38, 39]. Therefore, we then examined SAC status. Core SAC proteins, including Mad2, BubR1, and Bub3, were obviously localized at the kinetochores of chromosomes before MI and disappeared after MI in control oocytes. Notably, none of the SAC proteins were detected at kinetochores before MI in Fbxo30 RNAi oocytes in either the chromosome spreading experiment (Fig. 4l) or whole-oocyte staining experiment (Fig. S5). However, the protein levels between control and Fbxo30 RNAi oocytes were similar (Fig. 4m), suggesting that Fbxo30 depletion affected the recruitment of SAC proteins at kinetochores rather than their expression in oocytes. These data indicated that H3 overexpression induced by Fbxo30 knockdown resulted in abnormal chromosome condensation blocking chromosome segregation and that the overcondensed chromosomes affected SAC localization at kinetochores.
Knockdown of SLBP restores meiotic progress in Fbxo30 RNAi oocytes
Because the depletion of Fbxo30 led to SLBP overexpression, which inhibited chromosome separation in oocyte meiosis, we examined whether a decrease in excess SLBP would restore meiosis. Thus, SLBP was knocked down in these Fbxo30 RNAi oocytes. As shown in Fig. 5a, SLBP knockdown not only decreased the overexpression of SLBP per se but also downregulated the expression of H3 in Fbxo30 RNAi oocytes. Consistently, the overloaded SLBP around the spindle disappeared in Fbxo30/SLBP double-KD oocytes (Fig. 5b). Due to the downregulation of H3, the chromosomes of these oocytes escaped from the compact disorganization (Fig. 5c), which allowed SAC proteins to be re-positioned on the kinetochores (Fig. S6). When cultured for MII, 53% oocytes with Fbxo30/SLBP double knockdown underwent chromosome segregation and reached MII (Fig. 5d). These results validated that Fbxo30-SLBP cascade in oocyte meiosis regulates histone H3 and chromosome segregation.
In oocytes with SLBP single knockdown, we observed decreased protein levels of SLBP per se and H3 (Fig. 5a). These oocytes showed similar GVBD rates to control oocytes (Fig. S1b). However, H3 was not sufficiently expressed to fully cover the chromosomes, and almost none of the chromosomes were condensed into strips as in the control condition (Fig. 5c). Due to chromosome alignment failure, most SLBP RNAi oocytes were arrested at MI and were unable to reach MII (Fig. 5d). These results suggested that either an excess or shortage of H3 disturbs oocyte maturation in different ways. Thus, SLBP single knockdown oocytes had defects different from Fbxo30/SLBP double knockdown oocytes. SLBP knockdown restored meiotic progress in Fbxo30 RNAi oocytes.
Discussion
Mammalian oocytes contain full components of maternal proteins required for subsequent fertilization, genomic reprogramming, and embryonic gene activation [11, 40]. Due to the absence of transcription during oocyte maturation, the ubiquitin proteasome system is critically important for the regulation of protein dynamics and degradation. Because substrate proteins are spatiotemporally targeted by the ubiquitin proteasome system, the substrate-selected specificity controlled by E3 ligases is significantly important [41]. As the best characterized E3 ligase, SCF is a highly diverse complex and fulfils specificity via variable adaptor F-box proteins that bind substrates [18]. At present, at least 38 F-box proteins have been identified in humans [23, 42]. However, many substrates have not been identified. In this study, we demonstrate a new degradation pathway mediated by Fbxo30 that is essential for oocyte maturation. Depletion of Fbxo30 causes an obvious failure of chromosome segregation and meiotic arrest. Based on the large family of F-box proteins, the SCF-mediated ubiquitin proteasome system may make critical contributions to the depletion of maternal materials in oocytes.
Histones are responsible for packaging and ordering DNA into nucleosomes and are the major protein components of chromatin. In proliferative cells, histones must be translated rapidly during the beginning of S phase when DNA is replicated and degraded at the end of S phase or when errors occur in DNA replication [43]. However, in oocytes and early embryos, histone mRNAs accumulate and are translated independent of DNA replication. These histone proteins are used to replace the protamine of sperm DNA and for the assembly of newly synthesized DNA in the early embryo [38, 44]. In this study, we found that SLBP is a substrate of SCF ubiquitin ligases targeted by Fbxo30. Depletion of Fbxo30 leads to an ineffective degradation of SLBP and, in turn, H3 overload in oocytes. The excess H3 protein causes abnormal chromosome condensation, arresting the meiotic cell cycle. Our findings imply that the potential for H3 translation from H3 mRNA is much higher than the actual translation observed in oocytes. Indeed, histone mRNAs are constantly transcribed throughout mammalian oogenesis [45, 46]. The abundant supply of histone mRNA is required for subsequent embryonic development but is not necessary for maturing oocytes per se. Thus, oocytes must restrict the protein expression of histone during meiosis because a higher expression of histone proteins puts pressure on chromosomes and consumes metabolic energy. The ubiquitin proteasome pathway mediated by Foxb30 thus functions as a guard to monitor the protein expression of H3 in oocytes. The triple-layer SCF-SLBP-H3 cascade not only maintains the enriched pool of H3 mRNA but also avoids robust expression of H3 protein before fertilization.
In recent decades, infertility has been one of the most harmful diseases for women and their families. A reported 8–10% of reproductive-aged couples suffer from infertility worldwide [47, 48]. However, due to the absence of molecular information, most infertile women cannot be treated precisely in the clinic. Errors in meiosis in women represent the leading cause of abortion and embryo defects [49]. Of note, most of the errors occur during the first meiotic division [50, 51], which largely depends on chromosome segregation. Previous studies have highlighted the role of SAC proteins in sensing the attachment between spindle microtubules and kinetochores of chromosomes that in turn inhibits chromosome segregation [52–55]. Our findings instead provide a new direction of segregation failure that is driven by overcondensed chromosomes. Functional mutations in SCF complexes, such as F-box proteins and numerous substrates of SCF, and histones required for chromosome packaging may cause defects in chromosome segregation and oocyte meiosis. Thus, our study may provide potential targets for the diagnosis and therapy of female infertility.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary material 1 (DOCX 42320 kb)
Acknowledgements
We are grateful to Dr. Hengyu Fan for the insightful comments and suggestions on the manuscript. Mass spectrometry experiments were carried out by Junchen Company (Beijing, China) and Zhengdakangjian Company (Beijing, China). This work was supported by the National Natural Science Foundation of China (NSFC) (81622035, 81871160), Key Special Fund of Ministry of Science and Technology of China (2017YFC1001501, 2016YFC1000604, 2018YFC1003800), and the Foundation for Innovative Research Groups of NSFC (81521002). ML is a member of the Thousand Young Talents Plan of China.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflicts of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Rong Li, Phone: +86 10 8226 6760, Email: roseli001@sina.com.
Mo Li, Phone: +86 10 8226 6760, Email: limo@hsc.pku.edu.cn.
References
- 1.Stitzel ML, Seydoux G. Regulation of the oocyte-to-zygote transition. Science. 2007;316(5823):407–408. doi: 10.1126/science.1138236. [DOI] [PubMed] [Google Scholar]
- 2.Matzuk MM, Burns KH, Viveiros MM, Eppig JJ. Intercellular communication in the mammalian ovary: oocytes carry the conversation. Science. 2002;296(5576):2178–2180. doi: 10.1126/science.1071965. [DOI] [PubMed] [Google Scholar]
- 3.Tripathi A, Kumar KV, Chaube SK. Meiotic cell cycle arrest in mammalian oocytes. J Cell Physiol. 2010;223(3):592–600. doi: 10.1002/jcp.22108. [DOI] [PubMed] [Google Scholar]
- 4.Dumont J, Desai A. Acentrosomal spindle assembly and chromosome segregation during oocyte meiosis. Trends Cell Biol. 2012;22(5):241–249. doi: 10.1016/j.tcb.2012.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schuh M, Ellenberg J. Self-organization of MTOCs replaces centrosome function during acentrosomal spindle assembly in live mouse oocytes. Cell. 2007;130(3):484–498. doi: 10.1016/j.cell.2007.06.025. [DOI] [PubMed] [Google Scholar]
- 6.Fan HY, Sun QY. Involvement of mitogen-activated protein kinase cascade during oocyte maturation and fertilization in mammals. Biol Reprod. 2004;70(3):535–547. doi: 10.1095/biolreprod.103.022830. [DOI] [PubMed] [Google Scholar]
- 7.Nebreda AR, Ferby I. Regulation of the meiotic cell cycle in oocytes. Curr Opin Cell Biol. 2000;12(6):666–675. doi: 10.1016/S0955-0674(00)00150-2. [DOI] [PubMed] [Google Scholar]
- 8.Handel MA, Schimenti JC. Genetics of mammalian meiosis: regulation, dynamics and impact on fertility. Nat Rev Genet. 2010;11(2):124–136. doi: 10.1038/nrg2723. [DOI] [PubMed] [Google Scholar]
- 9.De La Fuente R. Chromatin modifications in the germinal vesicle (GV) of mammalian oocytes. Dev Biol. 2006;292(1):1–12. doi: 10.1016/j.ydbio.2006.01.008. [DOI] [PubMed] [Google Scholar]
- 10.Chen J, Melton C, Suh N, Oh JS, Horner K, Xie F, Sette C, Blelloch R, Conti M. Genome-wide analysis of translation reveals a critical role for deleted in azoospermia-like (Dazl) at the oocyte-to-zygote transition. Genes Dev. 2011;25(7):755–766. doi: 10.1101/gad.2028911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang S, Kou Z, Jing Z, Zhang Y, Guo X, Dong M, Wilmut I, Gao S. Proteome of mouse oocytes at different developmental stages. Proc Natl Acad Sci USA. 2010;107(41):17639–17644. doi: 10.1073/pnas.1013185107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Suzumori N, Burns KH, Yan W, Matzuk MM. RFPL4 interacts with oocyte proteins of the ubiquitin-proteasome degradation pathway. Proc Natl Acad Sci USA. 2003;100(2):550–555. doi: 10.1073/pnas.0234474100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu C, Ji SY, Sha QQ, Sun QY, Fan HY. CRL4-DCAF1 ubiquitin E3 ligase directs protein phosphatase 2A degradation to control oocyte meiotic maturation. Nat Commun. 2015;6:8017. doi: 10.1038/ncomms9017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nabti I, Grimes R, Sarna H, Marangos P, Carroll J. Maternal age-dependent APC/C-mediated decrease in securin causes premature sister chromatid separation in meiosis II. Nat Commun. 2017;8:15346. doi: 10.1038/ncomms15346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McGuinness BE, Anger M, Kouznetsova A, Gil-Bernabe AM, Helmhart W, Kudo NR, Wuensche A, Taylor S, Hoog C, Novak B, Nasmyth K. Regulation of APC/C activity in oocytes by a Bub1-dependent spindle assembly checkpoint. Curr Biol. 2009;19(5):369–380. doi: 10.1016/j.cub.2009.01.064. [DOI] [PubMed] [Google Scholar]
- 16.Lane SI, Yun Y, Jones KT. Timing of anaphase-promoting complex activation in mouse oocytes is predicted by microtubule-kinetochore attachment but not by bivalent alignment or tension. Development. 2012;139(11):1947–1955. doi: 10.1242/dev.077040. [DOI] [PubMed] [Google Scholar]
- 17.Peters JM. The anaphase promoting complex/cyclosome: a machine designed to destroy. Nat Rev Mol Cell Biol. 2006;7(9):644–656. doi: 10.1038/nrm1988. [DOI] [PubMed] [Google Scholar]
- 18.Cardozo T, Pagano M. The SCF ubiquitin ligase: insights into a molecular machine. Nat Rev Mol Cell Biol. 2004;5(9):739–751. doi: 10.1038/nrm1471. [DOI] [PubMed] [Google Scholar]
- 19.Yen HC, Elledge SJ. Identification of SCF ubiquitin ligase substrates by global protein stability profiling. Science. 2008;322(5903):923–929. doi: 10.1126/science.1160462. [DOI] [PubMed] [Google Scholar]
- 20.Frescas D, Pagano M. Deregulated proteolysis by the F-box proteins SKP2 and beta-TrCP: tipping the scales of cancer. Nat Rev Cancer. 2008;8(6):438–449. doi: 10.1038/nrc2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zheng N, Schulman BA, Song L, Miller JJ, Jeffrey PD, Wang P, Chu C, Koepp DM, Elledge SJ, Pagano M, Conaway RC, Conaway JW, Harper JW, Pavletich NP. Structure of the Cul1–Rbx1–Skp1–F boxSkp2 SCF ubiquitin ligase complex. Nature. 2002;416(6882):703–709. doi: 10.1038/416703a. [DOI] [PubMed] [Google Scholar]
- 22.Skowyra D, Craig KL, Tyers M, Elledge SJ, Harper JW. F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell. 1997;91(2):209–219. doi: 10.1016/S0092-8674(00)80403-1. [DOI] [PubMed] [Google Scholar]
- 23.Kipreos ET, Pagano M. The F-box protein family. Genome Biol. 2000;1(5):REVIEWS3002. doi: 10.1186/gb-2000-1-5-reviews3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yin S, Ai J-S, Shi L-H, Wei L, Yuan J, Ouyang Y-C, Hou Y, Chen D-Y, Schatten H, Sun Q-Y. Shugoshin1 may play important roles in separation of homologous chromosomes and sister chromatids during mouse oocyte meiosis. PLoS One. 2008;3(10):e3516. doi: 10.1371/journal.pone.0003516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hodges CA, Hunt PA. Simultaneous analysis of chromosomes and chromosome-associated proteins in mammalian oocytes and embryos. Chromosoma. 2002;111(3):165–169. doi: 10.1007/s00412-002-0195-3. [DOI] [PubMed] [Google Scholar]
- 26.Li H, Guo F, Rubinstein B, Li R. Actin-driven chromosomal motility leads to symmetry breaking in mammalian meiotic oocytes. Nat Cell Biol. 2008;10(11):1301–1308. doi: 10.1038/ncb1788. [DOI] [PubMed] [Google Scholar]
- 27.Xie B, Zhang L, Zhao H, Bai Q, Fan Y, Zhu X, Yu Y, Li R, Liang X, Sun QY, Li M, Qiao J. Poly(ADP-ribose) mediates asymmetric division of mouse oocyte. Cell Res. 2018;28(4):462–475. doi: 10.1038/s41422-018-0009-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schneider I, Lenart P. Chromosome segregation: is the spindle all about microtubules? Curr Biol. 2017;27(21):R1168–R1170. doi: 10.1016/j.cub.2017.09.022. [DOI] [PubMed] [Google Scholar]
- 29.Heald R, Tournebize R, Blank T, Sandaltzopoulos R, Becker P, Hyman A, Karsenti E. Self-organization of microtubules into bipolar spindles around artificial chromosomes in Xenopus egg extracts. Nature. 1996;382(6590):420–425. doi: 10.1038/382420a0. [DOI] [PubMed] [Google Scholar]
- 30.Koshland D, Strunnikov A. Mitotic chromosome condensation. Annu Rev Cell Dev Biol. 1996;12:305–333. doi: 10.1146/annurev.cellbio.12.1.305. [DOI] [PubMed] [Google Scholar]
- 31.Belmont AS. Mitotic chromosome structure and condensation. Curr Opin Cell Biol. 2006;18(6):632–638. doi: 10.1016/j.ceb.2006.09.007. [DOI] [PubMed] [Google Scholar]
- 32.Wilkins BJ, Rall NA, Ostwal Y, Kruitwagen T, Hiragami-Hamada K, Winkler M, Barral Y, Fischle W, Neumann H. A cascade of histone modifications induces chromatin condensation in mitosis. Science. 2014;343(6166):77–80. doi: 10.1126/science.1244508. [DOI] [PubMed] [Google Scholar]
- 33.Swain JE, Ding J, Brautigan DL, Villa-Moruzzi E, Smith GD. Proper chromatin condensation and maintenance of histone H3 phosphorylation during mouse oocyte meiosis requires protein phosphatase activity. Biol Reprod. 2007;76(4):628–638. doi: 10.1095/biolreprod.106.055798. [DOI] [PubMed] [Google Scholar]
- 34.Tada K, Susumu H, Sakuno T, Watanabe Y. Condensin association with histone H2A shapes mitotic chromosomes. Nature. 2011;474(7352):477–483. doi: 10.1038/nature10179. [DOI] [PubMed] [Google Scholar]
- 35.Marzluff WF, Wagner EJ, Duronio RJ. Metabolism and regulation of canonical histone mRNAs: life without a poly(A) tail. Nat Rev Genet. 2008;9(11):843–854. doi: 10.1038/nrg2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marzluff WF, Duronio RJ. Histone mRNA expression: multiple levels of cell cycle regulation and important developmental consequences. Curr Opin Cell Biol. 2002;14(6):692–699. doi: 10.1016/S0955-0674(02)00387-3. [DOI] [PubMed] [Google Scholar]
- 37.Krapivinsky G, Krapivinsky L, Manasian Y, Clapham DE. The TRPM7 chanzyme is cleaved to release a chromatin-modifying kinase. Cell. 2014;157(5):1061–1072. doi: 10.1016/j.cell.2014.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tadros W, Lipshitz HD. The maternal-to-zygotic transition: a play in two acts. Development. 2009;136(18):3033–3042. doi: 10.1242/dev.033183. [DOI] [PubMed] [Google Scholar]
- 39.Wassmann K, Niault T, Maro B. Metaphase I arrest upon activation of the Mad2-dependent spindle checkpoint in mouse oocytes. Curr Biol. 2003;13(18):1596–1608. doi: 10.1016/j.cub.2003.08.052. [DOI] [PubMed] [Google Scholar]
- 40.Zhang P, Ni X, Guo Y, Guo X, Wang Y, Zhou Z, Huo R, Sha J. Proteomic-based identification of maternal proteins in mature mouse oocytes. BMC Genom. 2009;10:348. doi: 10.1186/1471-2164-10-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weissman AM. Themes and variations on ubiquitylation. Nat Rev Mol Cell Biol. 2001;2(3):169–178. doi: 10.1038/35056563. [DOI] [PubMed] [Google Scholar]
- 42.Cenciarelli C, Chiaur DS, Guardavaccaro D, Parks W, Vidal M, Pagano M. Identification of a family of human F-box proteins. Curr Biol. 1999;9(20):1177–1179. doi: 10.1016/S0960-9822(00)80020-2. [DOI] [PubMed] [Google Scholar]
- 43.Henikoff S, Ahmad K. Assembly of variant histones into chromatin. Annu Rev Cell Dev Biol. 2005;21:133–153. doi: 10.1146/annurev.cellbio.21.012704.133518. [DOI] [PubMed] [Google Scholar]
- 44.Allard P, Champigny MJ, Skoggard S, Erkmann JA, Whitfield ML, Marzluff WF, Clarke HJ. Stem-loop binding protein accumulates during oocyte maturation and is not cell-cycle-regulated in the early mouse embryo. J Cell Sci. 2002;115(23):4577–4586. doi: 10.1242/jcs.00132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Clarke HJ, Bustin M, Oblin C. Chromatin modifications during oogenesis in the mouse: removal of somatic subtypes of histone H1 from oocyte chromatin occurs post-natally through a post-transcriptional mechanism. J Cell Sci. 1997;110(Pt 4):477–487. doi: 10.1242/jcs.110.4.477. [DOI] [PubMed] [Google Scholar]
- 46.Allard P, Champigny MJ, Skoggard S, Erkmann JA, Whitfield ML, Marzluff WF, Clarke HJ. Stem-loop binding protein accumulates during oocyte maturation and is not cell-cycle-regulated in the early mouse embryo. J Cell Sci. 2002;115:4577–4586. doi: 10.1242/jcs.00132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mascarenhas MN, Flaxman SR, Boerma T, Vanderpoel S, Stevens GA. National, regional, and global trends in infertility prevalence since 1990: a systematic analysis of 277 health surveys. PLoS Med. 2012;9(12):e1001356. doi: 10.1371/journal.pmed.1001356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Inhorn MC, Patrizio P. Infertility around the globe: new thinking on gender, reproductive technologies and global movements in the 21st century. Hum Reprod Update. 2015;21(4):411–426. doi: 10.1093/humupd/dmv016. [DOI] [PubMed] [Google Scholar]
- 49.Hunt PA, Hassold TJ. Human female meiosis: what makes a good egg go bad? Trends Genet. 2008;24(2):86–93. doi: 10.1016/j.tig.2007.11.010. [DOI] [PubMed] [Google Scholar]
- 50.Nagaoka SI, Hodges CA, Albertini DF, Hunt PA. Oocyte-specific differences in cell-cycle control create an innate susceptibility to meiotic errors. Curr Biol. 2011;21(8):651–657. doi: 10.1016/j.cub.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kitajima TS, Ohsugi M, Ellenberg J. Complete kinetochore tracking reveals error-prone homologous chromosome biorientation in mammalian oocytes. Cell. 2011;146(4):568–581. doi: 10.1016/j.cell.2011.07.031. [DOI] [PubMed] [Google Scholar]
- 52.Qiao J, Wang ZB, Feng HL, Miao YL, Wang Q, Yu Y, Wei YC, Yan J, Wang WH, Shen W, Sun SC, Schatten H, Sun QY. The root of reduced fertility in aged women and possible therapentic options: current status and future perspects. Mol Asp Med. 2014;38:54–85. doi: 10.1016/j.mam.2013.06.001. [DOI] [PubMed] [Google Scholar]
- 53.Wang ZB, Schatten H, Sun QY. Why is chromosome segregation error in oocytes increased with maternal aging? Physiology (Bethesda) 2011;26(5):314–325. doi: 10.1152/physiol.00020.2011. [DOI] [PubMed] [Google Scholar]
- 54.Lu Y, Dai X, Zhang M, Miao Y, Zhou C, Cui Z, Xiong B. Cohesin acetyltransferase Esco2 regulates SAC and kinetochore functions via maintaining H4K16 acetylation during mouse oocyte meiosis. Nucleic Acids Res. 2017;45(16):9388–9397. doi: 10.1093/nar/gkx563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Touati SA, Buffin E, Cladiere D, Hached K, Rachez C, van Deursen JM, Wassmann K. Mouse oocytes depend on BubR1 for proper chromosome segregation but not for prophase I arrest. Nat Commun. 2015;6:6946. doi: 10.1038/ncomms7946. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material 1 (DOCX 42320 kb)