Abstract
Ubiquitin ligases play an integral role in fine-tuning signaling cascades necessary for normal cell function. Aberrant regulation of ubiquitin ligases has been implicated in several neurodegenerative diseases, generally, due to mutations within the E3 ligase itself. Several proteomic-based methods have recently emerged to facilitate the rapid identification of ligase–substrate pairs—a previously challenging feat due to the transient nature of ligase–substrate interactions. These novel methods complement standard immunoprecipitations (IPs) and include proximity-dependent biotin identification (BioID), ubiquitin ligase–substrate trapping, tandem ubiquitin-binding entities (TUBEs), and a molecular trapping unit known as the NEDDylator. The implementation of these techniques is expected to facilitate the rapid identification of novel substrates of E3 ubiquitin ligases, a process that is likely to enhance our understanding of neurodegenerative diseases and highlight novel therapeutic targets for the treatment of neurodegenerative diseases.
Keywords: BioID, E3 ubiquitin ligases, Immunoprecipitation, Ligase trapping, NEDDylator, Proteomics, TUBE, Ubiquitylation
E3 ubiquitin ligases and neurodegeneration
Neurodegenerative diseases are characterised by the formation of insoluble protein inclusions within the brains of affected patients [1]. This includes Alzheimer’s disease (AD) [2], Parkinson’s disease (PD) [3, 4], Amyotrophic Lateral Sclerosis (ALS)/Frontotemporal Dementia (FTD) [5–7], and Huntington’s disease (HD) [8–11]. The accumulation of proteins, which may co-localise with protein degradation markers such as ubiquitin [5, 6, 9, 10, 12–14], implicates defective protein turnover systems as a common mechanism underlying neurodegenerative diseases.
The ubiquitin-proteasome system (UPS) is one of the major intracellular protein degradation systems [15–17], with defects frequently associated with neurodegenerative diseases [18]. This system is composed of ubiquitylation enzymes and the proteasome [19]. The process of ubiquitylation requires a cascade of enzymatic reactions involving ubiquitin-activating enzymes (E1s), ubiquitin-conjugating enzymes (E2s), and ubiquitin ligases (E3s) [20, 21]. To begin, an E1 generates a high-energy thioester bond between an internal active site cysteine residue and ubiquitin in an ATP-dependent manner [20]. This form of ubiquitin is transferred to a cysteine residue within an E2 (generating E2-ubiquitin), which then co-operates with an E3 to mediate the ubiquitylation of substrates [20]. To generate polyubiquitin chains on substrates, the E2 is recycled, dissociating from the E3 after ubiquitin transfer and reassociating once charged with a new ubiquitin molecule [22]. Through this process, E1, E2, and E3 ligases facilitate the mono-, multi-, and polyubiquitylation of substrates [21]. This modification of substrates leads to diverse ubiquitin chain topology on substrates [23], consequently, generating specific signals that influence a wide range of biological processes such as autophagic protein degradation [24], protein sub-cellular localisation [25], DNA repair [26], and cell signalling events [27].
Ubiquitin ligases or their components have been linked to several neurodegenerative diseases, typically by containing mutations in their encoding genes or by interacting with disease-associated proteins [28]. For example, mutations in the gene PARK2, which encodes for Parkin, is found in approximately 50% of patients with autosomal recessive juvenile parkinsonism [29, 30]. In addition, mutations in cyclin F and Ube3A have been linked to ALS/FTD [31, 32] and Angelman Syndrome [33], respectively. E3 ligases may also interact and mediate the ubiquitylation of proteins that are key factors in disease initiation and progression. For example, the E3 ligase, C-terminus of Hsc70-interacting protein (CHIP), recognises and ubiquitylates Tau [34], a protein that forms filamentous Tau inclusions in multiple neurodegenerative diseases [35].
Challenges associated with identifying ligase–substrate pairs
Given that defects in E3 ubiquitin ligases may be contributing factors in disease initiation and progression, it becomes important to identify substrates that are ubiquitylated by each E3 ligase. By doing so, it becomes possible to identify interactions that may influence the progression of disease. Identifying ligase and substrate pairs can be challenging, however, due to the nature of ligase–substrate interactions. Some challenges associated with identifying ligase–substrate pairs include: (1) transient interactions between the E3-ligase and substrate. Standard immunoprecipitations (IPs) are based on maintaining the interaction between protein-binding partners post-lysis [36]; however, ligase–substrate interactions are typically transient [37], making it more difficult to detect the presence of substrates following standard IPs. (2) Many proteins that are ubiquitylated by E3 ligases are rapidly degraded through the proteasome [15, 16]. As a result, the abundance of substrates may be relatively low, which necessitates the development of strategies that can stabilise protein substrates for subsequent proteomic identification. (3) Ubiquitylation does not always lead to protein degradation [23]. Thus, developing strategies that can be used to identify substrates targeted for proteasomal degradation may not necessarily be useful when targeting substrates that are tagged for other biological outcomes such as protein translocation. (4) Ubiquitylation is a reversible post-translational modification. Ubiquitylation of substrates is reversible due to the activity of deubiquitylases (DUBs), making it more challenging to identify ubiquitylated substrates [38]. (5) Many proteins can be ubiquitylated by more than one ligase [39–41]. This redundancy of protein ubiquitylation results in the need to validate specific ligase–substrate pairs.
Proteomic-based methods designed to identify ubiquitin ligase–substrate pairs
As a result of these difficulties, as well as the time-consuming nature of identifying and validating ligase–substrate pairs, it is estimated that only ~ 15% of ubiquitylated proteins have been matched with their corresponding ubiquitin ligase [42]. Over the years, several proteomic-based methods have emerged, which enable the identification of ligase–substrate pairs. IPs followed by mass spectrometry (IP-LC/MS) have been the primary route used to identify targets of a protein of interest [43] (see Sects. “Immunoprecipitations and ubiquitylation assays” and “Combining immunoprecipitations, bioinformatics, and structural data”). However, several new proteomics-based techniques have emerged, which enable the rapid identification of ligase–substrate pairs. These novel techniques include proximity-dependent biotin identification (BioID) [44, 45] (see Sect. “Proximity-dependent biotin identification (BioID)”), ubiquitin ligase–substrate trapping [46–48] (see Sect. “Ubiquitin ligase–substrate trapping”), use of tandem ubiquitin-binding entities (TUBEs) [49] (see Sect. “Tandem ubiquitin-binding entities”), and application of the NEDDylator [50] (see Sect. “NEDDylator”). These techniques followed by further biochemical studies greatly enhance the possibility to explore novel functions of E3 ligases. In the context of neurodegeneration, these methods can provide valuable insight into altered signalling pathways that can be a consequence of disease-causing mutations in E3 ligases. In addition, these methods enable the identification of ligase and substrate pairs that may assist in understanding the mechanisms by which ubiquitylated protein deposits are formed.
Immunoprecipitations and ubiquitylation assays
The initial route used to identify ligase–substrate pairs has been to detect the physical interaction between the two using IPs followed by mass spectrometry (IP-LC/MS) [43]. This method has previously been implemented to identify the interaction partners of F-box proteins—proteins that are the substrate recruitment units of multi-protein E3 ubiquitin ligases. For example, D’Angiolella et al. use IP-LC/MS workflows to identify interaction partners of the F-box protein and cyclin F, and have successfully identified and validated substrates such as RRM2 and CP110 [51, 52]. IP-LC/MS studies involve immunoprecipitating the E3 ligase or components of the E3 ligase from cell lysates and subsequently identifying co-IP partners using mass spectrometry. Notably, this method relies on quality antibodies of reliable specificity. Furthermore, identifying ligase–substrate pairs using this method relies on maintaining a stable interaction between the ligase and substrate. Thus, although this method has successfully been used to identify some substrates of E3 ligases, such as Skp1–Cul1–F-box(SCF)cyclin F, it is likely that many substrates cannot be identified due to their transient/weak interactions between the ligase–substrate pairs [37].
One method that can be used in addition to a standard IP-LC/MS workflow to differentiate substrates from binding partners has been exemplified by Peschiaroli et al., who have identified putative substrates of β-TrCP1 and 2 (herein referred to as β-TrCP) using a dual-tagging strategy [53]. Notably, β-TrCP are well-studied F-box proteins that, similar to cyclin F, are capable of forming a multi-protein SCF E3 ubiquitin ligase (SCFβ-TrCP), which mediates ubiquitylation of substrates and thereby influences a range of downstream biological processes [54]. β-TrCP1 and 2 are examples of well-characterised E3 ligases, as over a decade of research has been devoted to identifying their substrates [55]. Over 50 substrates of β-TrCP have been characterised to date and new substrates are still being uncovered [56].
The process of the dual-tagging strategy begins with a standard IP of the ligase of interest, in this case β-TrCP. The immunoprecipitated ligase along with co-immunoprecipitated proteins are subsequently used in an in vitro ubiquitylation assay whereby all components required for ubiquitylation are incorporated in vitro. Notably, in this case, ubiquitin is tagged with a Flag-tag. As the in vitro ubiquitylation proceeds, associated substrates are ubiquitylated, after which they can be pulled down using Flag-tagged ubiquitin and identified using MS-based workflows [53, 57, 58]. Using this method, novel ubiquitylated substrates of β-TrCP were identified including Claspin [53], PCDC4 [58], and REST [57]. Importantly, validation of these substrates and further investigation into the biological implications of this ubiquitylation linked the E3 ligase to novel biological processes including termination of the DNA replication checkpoint response [53], proper activation of the spindle assembly checkpoint [57], and promotion of protein translation and cell growth [58].
Limitations and challenges
While this strategy has been useful in identifying novel substrates of SCFβ-TrCP, difficulties of this process can arise due to the two-step process involved. In the first step, the ubiquitylation process proceeds with low efficiency, and in the second step, there is low recovery of ubiquitylated proteins. Thus, although this process enables the discrimination between interaction partners and substrates, the number of substrates identified is likely to be limited.
Combining immunoprecipitations, bioinformatics, and structural data
Low et al. have also identified substrates of β-TrCP2 using IPs followed by proteomics, which they refer to as affinity–purification mass spectrometry (AP–MS). In this case, Low et al. identified high-confidence substrates of β-TrCP2 by applying a combination of structural information, bioinformatics platforms, and IPs. Three forms of β-TrCP2 were used in the approach, wild-type β-TrCP2, β-TrCP2 (R447A), and mutant β-TrCP2, with a deleted F-box. The latter are strategically placed mutations as the R447A mutation is situated in the substrate-binding area of β-TrCP2 which prevents binding and ubiquitylation of substrates [59, 60]. The F-box deletion in β-TrCP2 prevents the formation of a functional SCFβ-TrCP2 E3 ligase; however, it may still bind substrates [61]. This provides two controls which are necessary for comparison in this study and assisted Low et al., in eliminating binding partners from the list of candidate substrates.
To minimise non-specific binding, Low et al. used constructs that encode β-TrCP2 in frame with 2xFlag and 2xHA tags. These tags were used to sequentially IP β-TrCP2, using one tag and then the other. The idea is that the sequential IPs using antibodies recognising the HA-tag and then Flag-tag reduce the presence of non-specific proteins that are associated with each immunocomplex. The final immunocomplex is then subjected to standard proteomic analyses.
Identifying substrates
Using this method, Low et al. reported 1626 proteins that interacted with β-TrCP2 through the WD40 domain, or substrate-binding region. However, given the large reported number of substrates, it is likely that many of these proteins were non-specific interaction partners of secondary binding proteins. As a result, additional filtering processes and the use of bioinformatics platforms were required. These filtering processes involved using the Contaminant Repository for Affinity Purification (CRAPome) as well as Perseus software suites, which enabled the removal of common contaminating proteins as well as false-positives results. In addition, the resulting interaction partners were scanned for the presence of a conserved phosphodegron sequence, which is necessary for their recognition by β-TrCP. The phosphodegron sequence recognised by β-TrCP is well defined, which assisted in the identification of substrates. Using this information, the number of putative ligase substrates was reduced to 221 for further validation [61].
Limitations and challenges
Low et al. have established a workflow which effectively incorporates AP–MS, structural information, and degron mining to identify high-confidence substrates of β-TrCP2. The resulting list of putative substrates includes a series of known β-TrCP substrates. One consideration for the application of this method, however, is that it involves stringent wash steps which may come at a loss of weaker interaction partners. Furthermore, β-TrCP is a well-studied F-box protein with a solved crystal structure [59] and a well-defined substrate-binding region [61]. Thus, using this approach for poorly studied F-box proteins with fewer known substrates and little structural information may be more challenging.
Proximity-dependent biotin identification (BioID)
Proximity-dependent biotin identification (BioID) can be used to identify weak/transient protein interactions in live cells [45, 62]. This approach makes use of biotin ligase (BirA), a ~ 35 kDa DNA-binding protein ligase derived from Escherichia coli [63]. The process of biotinylation by this biotin ligase occurs through a two-step process. First, the biotin ligase generates a highly reactive biotinoyl-AMP intermediate in an ATP-dependent manner [64]. In its wild-type form, activated biotin is held within the active site of BirA until it is released to label a specific substrate [64]. BirA has since been engineered to generate a promiscuous variant of the biotin ligase (denoted as BirA*), which does not specifically biotinylate any particular substrate. BirA* contains an R118G mutation within its active site [45], which results in a reduced association with activated biotin [65]. As a result, the intermediate diffuses away from the active site of BirA* leading to biotinylation of any lysine residues in close proximity to BirA*. In this way, BirA* can biotinylate proteins in close proximity to BirA* in a promiscuous fashion. These biotinylated proteins can be effectively isolated with avidin/streptavidin conjugated to beads, before identification using proteomics.
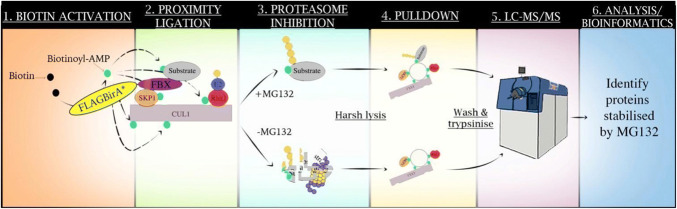
For the application of BioID, the protein of interest is fused in frame with BirA* before the fusion protein is expressed in live cells [45]. In recent years, Coyaud et al. have applied BioID as a complementary method to IPs to identify substrates of β-TrCP [44]. As shown in Fig. 1, Coyaud first expressed the β-TrCP–BirA* fusion protein in live cells. Once expressed, β-TrCP was still able to form a functional, multi-protein E3 ligase to ubiquitylate substrates. When biotin was introduced, it became activated by mutant BirA*, and due to the reduced affinity for activated biotin, lysine residues of surrounding proteins were biotinylated. This included lysine residues on binding partners, substrates, and proteins within the labelling radius.
Fig. 1.
BioID workflow used to identify substrates of β-TrCP. Flag-tagged BirA* is fused in frame with β-TrCP and the fusion protein is expressed in live cells, enabling the formation of a functional multi-protein E3 ligase (1). Biotin is activated by BirA*, leading to promiscuous biotinylation of surrounding proteins (2). Proteasome inhibition, using MG132, stabilises biotinylated substrates (3). Biotinylated proteins are isolated by streptavidin (4) and analysed using LC–MS/MS (5). Peptides identified are analysed using a suite of bioinformatics programs and proteins stabilised by MG132 are classified as potential substrates of β-TrCP (6)
(Image adapted from [44])
Enriching substrates
To distinguish substrates from binding partners or other biotinylated proteins, Coyaud et al. made use of proteasome inhibition by treating cells with MG132. β-TrCP mediates the ubiquitylation of substrates for proteasomal degradation [66]. Thus, in cases where cells are not treated with MG132, substrates are ubiquitylated and subsequently degraded by the proteasome. In contrast, when cells are treated with MG132, substrates accumulate allowing for their subsequent identification by mass spectrometry. When comparing these two data sets, biotinylated proteins that match a “substrate profile” were those which had accumulated in response to MG132 (when comparing the ratio of +MG132/untreated cells). These proteins represent likely substrates as they are within proximity to the E3 ligase and accumulate in response to proteasome inhibition.
Limitations and challenges
A limitation of this method is the promiscuity of the biotin ligase. The labelling kinetics of the BioID is slow, with the half-life of the intermediate in the minute range [67]. Thus, typical workflows using BioID require labelling timeframes for ~ 24 h [68] to generate decent amounts of sample for analysis. This results in a fairly large labelling radius. In addition, proteasome inhibition is used in this workflow, which leads to the biotinylation of proteins that may have accumulated during the 24 h labelling period. A challenge, therefore, exists in reducing the number of false-positive results.
Ubiquitin ligase–substrate trapping
Ubiquitin ligase–substrate trapping or “ligase trapping” is another proteomics-based strategy used to identify ligase–substrate pairs [46–48]. This method is similar to a standard IP; however, the ligase of interest is fused to a polyubiquitin-binding region which enables the enrichment of ubiquitylated substrates [47]. Specifically, the polyubiquitin-binding region is a ubiquitin-associated domain (UBA), a well-conserved domain of approximately 45 amino acids in length [69]. Proteins that contain UBA domains are frequently involved in facilitating proteasomal degradation by binding to polyubiquitylated proteins as well as subunits of the proteasome [70]. For example, a well-studied class of UBA containing proteins is the UbL–UBA family. Notably, the UBA domain within these proteins may enable stronger binding to polyubiquitin compared to monoubiquitin [71], a feature which makes them particularly useful for targeting polyubiquitylated proteins. For the application of substrate trapping, the small UBA domain derived from proteins, Rad23 or Dsk2, is encoded in frame with an E3 ligase of interest and expressed in live cells [46]. As substrates are ubiquitylated by the E3 ligase, the associated UBA binds to the polyubiquitin chain, thereby literally trapping the substrate with the ligase complex for further isolation and identification using mass spectrometry. Ligase trapping and similar applications have been applied in yeast and mammalian cells [46–48].
Ubiquitin ligase trapping in yeast
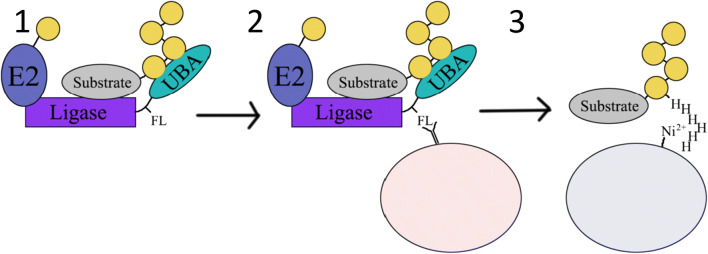
Ligase trapping was first established by Mark et al. [46] to identify substrates of 8 F-box proteins found in yeast. This ubiquitin trapping method involves expressing the ligase-UBA fusion protein in cells alongside ubiquitin modified with hexahistidine (His6)-tagged ubiquitin (Fig. 2). Once the fusion protein and tagged ubiquitin is expressed in yeast, substrates are ubiquitylated by the E3 ligase and become trapped by the UBA. A two-step purification process then follows to identify the trapped proteins. In the first step of purification, the E3 ligase and trapped substrate are isolated from cell lysates by immunoprecipitating the complex using antibodies recognising the Flag-tag. This co-immunoprecipitated complex is then subjected to a second purification step to minimise the number of contaminating proteins that may have been co-immunoprecipitated. To do this, the immunoprecipitated complex is exposed to buffers containing the chaotropic reagent, urea. By doing so, proteins are denatured and protein–protein interactions are disrupted. His6-tagged polyubiquitylated proteins can then be purified using a nickel–nitrilotriacetic acid (NiNTA) affinity column. This two-step purification procedure is followed by trypsin digestion before resulting peptides are analysed by mass spectrometry.
Fig. 2.
Ubiquitin ligase–substrate trapping. The E3 ligase is fused in frame to a UBA (green). When a substrate (grey) binds to the ligase and is ubiquitylated, the ubiquitin moieties (yellow) become associated with the UBA, thereby ‘trapping’ the substrate with the ligase and preventing it from being degraded by the proteasome (1). The complex can then be pulled down using antibodies recognising the Flag-tag (light pink) (2). The complex is subsequently denatured and the his6-ubiquitin-modified proteins are purified using Ni–NTA purification (light blue) (3). Upon trypsinisation, the substrates can be identified using proteomics
(Image adapted from [46])
Enriching for substrates in yeast
Given that this process physically traps ubiquitinated proteins in complex with the ligase of interest, the ubiquitylated proteins are highly likely to be substrates of the specific E3 ligase at hand. Furthermore, non-specific binding partners are diminished due to the two-step purification procedure. Due to the double enrichment strategy, it becomes easy to distinguish between substrates and non-specific binding partners using substrate trapping. Overall, Mark et al. were able to identify 17 known substrates and 18 novel substrates of the 8 F-box proteins. The four most abundant putative substrates of SCFSaf1 were vacuolar/lysosomal hydrolases. Previously, Saf1 was poorly characterised, with only one reported substrate. Using this method, Saf1 could be linked to novel processes involved in vacuolar/lysosomal hydrolase function [46].
Ubiquitin ligase trapping in mammalian cells
Substrate trapping has since been applied in a mammalian system by Loveless et al. [48] who have used the method to identify substrates of β-TrCP in HEK293 cells. Twelve previously known substrates were identified and 11 new substrates were uncovered.
Similar to the application of ligase trapping in yeast, the UBA domain of Rad23 was encoded in frame with β-TrCP and the fusion protein was expressed in modified HEK293 cells. The HEK293 Flp-In T-Rex cells used in this study stably express His6-tagged ubiquitin upon treatment with doxycycline or tetracycline. Thus, similar to the process used by Mark et al., trapped proteins can be identified using a two-step purification procedure—first by immunoprecipitating the E3 ligase using the attached Flag-tag, and subsequently pulling down his-tagged ubiquitylated proteins under denaturing conditions using an NiNTA affinity column or beads. Isolated proteins can then be identified using mass spectrometry workflows.
Enriching for substrates in mammalian cells
Similar to Coyaud et al., Loveless used proteasome inhibition prior to cell harvesting to increase the identification of polyubiquitylated proteins. One consideration, however, is that the protein of interest may bind many ubiquitin-conjugated proteins and this may lead to false-positive results. To demonstrate the specific functionality of the β-TrCP trap, the group expressed a known substrate of β-TrCP, ATF4, with an epitope tag in their cell line of interest. They then compare substrate trapping using β-TrCP traps as well as two unrelated F-box traps, in this case FBXO24 or Fbw7 traps. Notably, β-TrCP but not FBXO24 or Fbw7 traps could bind polyubiquitylated ATF4 demonstrating the specificity and functionality of this method [48].
Limitations and challenges
A drawback of this system is that the ligase is trapped in complex with the substrate. This potentially limits the number of substrates that can be identified. Another consideration when applying ligase trapping is that optimising the linker length and configuration between the F-box and UBA can be a time-consuming process. This is required, however, to ensure that the ligase does not interfere with substrate binding and polyubiquitin chain formation.
Tandem ubiquitin-binding entities
Yoshida et al. used a similar ubiquitin-binding approach to identify substrates of FBXO21. Here, Yoshida et al. use tandem-repeated ubiquitin-binding entities (TUBEs) to enrich polyubiquitylated substrates [49]. TUBEs are based on UBA domains whereby four UBAs are bound to the protein of interest. In addition to having higher affinity to tetra-ubiquitin compared to single ubiquitin molecules, TUBEs protect polyubiquitylated proteins from being degraded by the proteasome or undergoing de-ubiquitylation by DUBs [72]. Once the ligase and TUBEs are co-expressed in mammalian cells, the TUBEs are able to capture polyubiquitylated proteins. The TUBEs (attached to polyubiquitylated proteins) are enriched by standard IP procedures using antibodies recognising the Flag-tag before the isolated proteins are trypsin digested and analysed using mass spectrometry. Notably, the use of trypsin-resistant TUBEs (TR-TUBEs) prevents the trypsin digestion of the highly abundant TUBE proteins, a process that maximises the identification of substrates by MS as they will not be masked by the presence of high abundant proteins belonging to TR-TUBEs.
Enriching for substrates
As a negative control, Yoshida et al. have mutated the F-box domain of FBXO21. This mutation prevents the formation of a functional E3 ligase, and thus, substrates could be defined as those that increased when comparing the wild-type control to the F-box mutant. Using this method, Yoshida et al. identified and validated TARS and EID1 as substrates of FBXO21. Both were strong candidates as they met the criteria for being potential substrates and could be reproducibly enriched and identified.
In addition to capturing ubiquitylated substrates, this assay can be used to obtain information about the ubiquitylated residues, further providing evidence that these substrates are ubiquitylated. To do so, Yoshida et al. used an antibody that recognises a ubiquitin remnant motif, K-Ɛ-GG, a lysine modified with two glycine residues. Using this method, Yoshida et al. found that > 95% of identified peptides captured contained the K-Ɛ-GG motif, further allowing them to map out the ubiquitin sites.
Limitations and challenges
This TUBE approach does not identify substrates in complex with the E3 ligase at hand; however, an advantage is that there is no need to characterise and optimise a fusion protein between the UBA domain and F-box of interest. Rather, investigators co-express TR-TUBE and variations of the F-box protein of interest. Nevertheless, overexpression of an F-box protein may lead to a cascade of ubiquitylation reactions, many of which may not be directly caused by the overexpressed ligase itself.
NEDDylator
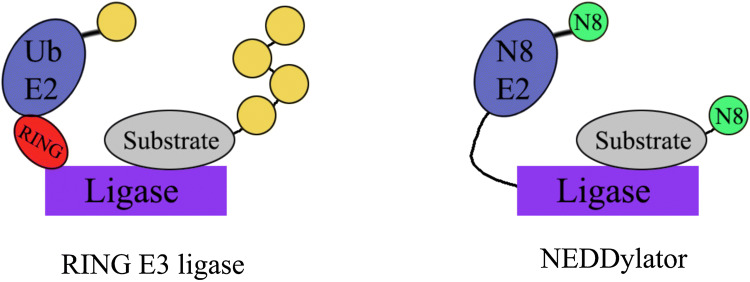
An alternative approach for the physical identification of substrates makes use of the NEDDylator [50]. This process works by fusing a modified E2 ligase to the F-box of interest. E2 ligases are integral to the cascade of events leading up to substrate ubiquitylation. Normally, the E2 ligase is responsible for binding the activated ubiquitin and positioning the activated ubiquitin close to the substrate by docking on the E3 ligase complex [73]. In the application by Zhuang et al., the E2 is modified, such that it is primed with a homolog of ubiquitin, NEDD8, instead of ubiquitin itself. Although this modification is similar to ubiquitin, NEDD8 modifications are rare and stable [50]. Thus, as the substrate interacts with the ligase, E2-NEDD8 facilitates the transfer of NEDD8 to the substrate, rather than ubiquitin (Fig. 3).
Fig. 3.
The concept of the NEDDylator (right) is based on the fusion of a ligase of interest with a modified E2-ligase. Under normal circumstances (left), E2 ligase brings activated ubiquitin (yellow) to the E3-ligase, such that ubiquitin is transferred on to substrate (grey). In comparison, the NEDDylator enables the transfer of His-biotin-tagged NEDD8 (green) to the substrate, enabling isolation of NEDDylated substrates by pull-downs
Substrate enrichment strategy
Although the functional E3 ligase loses the ability to ubiquitylate substrates, it is now able to transfer NEDD8 onto substrates. These NEDDylated proteins can be isolated using His-biotin-tagged NEDD8 (HB-NEDD8) under denaturing conditions, reducing the number of background interaction partners identified using LC–MS/MS. Furthermore, due to the low abundance of NEDD8 in mammalian cells, the enrichment of background-NEDDylated proteins is low. Finally, the transfer of a NEDD8 moiety rather than ubiquitin stabilises tagged substrates that are otherwise labelled for rapid proteasomal degradation.
Limitations and challenges
This workflow elegantly enables the precise NEDDylation of E3 ligase substrates; however, much work must go into engineering the fusion protein. Creating the NEDDylator for human XIAP required removal of the RING domain (435-497) from XIAP to prevent association with E2 primed with ubiquitin. Furthermore, the N-terminus of XIAP was fused via a flexible linker to the E2 carrying NEDD8, another aspect which must be optimised to ensure that NEDD8 can be transferred from the E2 to substrate. Although laborious, these are important considerations when constructing any NEDDylator chimeric proteins, as the engineered protein needs to be able to form an active ligase complex, capable of NEDDylating proteins brought to the complex by the F-box protein.
This workflow could become more challenging for the application of larger, multi-protein SCF complexes as they require additional protein interaction partners to generate the functional E3 ligase. In addition, in an SCF E3 ligase complex, the F-box protein can be switched with over 60 other F-box proteins in mammalian cells. Thus, the NEDDylated proteins may not be specific to any given F-box protein.
Comparison of techniques
A comparison of MS-based methods used to identify substrates of E3 ligases is summarised in Table 1. Although there are a variety of methods that are available to identify substrates, the use of β-TrCP in HEK293 cells enables some comparison between these methods. However, a major factor is the amount of starting material used and the stringency applied in classifying substrates. The best comparison of a standard IP to a novel method comes from Coyaud et al., as they implement both BioID and IP-MS to identify substrates of β-TrCP. Coyaud et al. demonstrated that a greater number of substrates were identified by the BioID approach compared to a standard IP approach. Furthermore, some putative substrates that could be identified by BioID were not observed by standard IPs, demonstrating the ability of BioID to identify transient protein interactions.
Table 1.
Proteomic techniques used to identify substrates of E3 ligases using MS
| Approach | Cell type | Substrate definition | Ligase | Validation approach/s | Substrates | References |
|---|---|---|---|---|---|---|
| IP-MS, in vitro ubiquitylation assay | HEK293 | Co-interacting partners subsequently ubiquitylated in vitro | β-TrCP | In vitro ubiquitylation assay, CHX chase assay and β-TrCP1 and 2 gene silencing using dsRNA | 4 known, 1 novel | [53] |
| IP-MS | HEK293 | Strategic mutations that affect known substrate binding. Substrates are also defined by presence of phosphodegron | β-TrCP | In vitro ubiquitylation study, degron mining | 27 known, 221 putative substrates | [61] |
| BioID | HEK293 Flp-in T-Rex | Biotinylated proteins that are stabilised by MG132 treatment | β-TrCP | CHX assays, in vivo ubiquitylation assay | 17 previously reported substrates or interaction partners and > 50 novel putative substrate | [44] |
| Ubiquitin trapping | Yeast | Polyubiquitylated proteins that are trapped | 8 F-box proteins | Cycloheximide (CHX) chase assay, in vivo ubiquitylation assays | 18 known, 17 new | [46] |
| Ubiquitin trapping | HEK293 Flp-in T-Rex | Proteins that are enriched after substrate trapping | β-TrCP | CHX chase assays | 12 known, 11 new | [48] |
| TR-TUBE | HEK293 | TUBE-binding proteins that decreased in abundance when FBXO21 was inactive | FBXO21 | In vivo ubiquitylation assay, siRNA knockdown of protein of FBXO21, CHX chase assays | 3 highly reproducible targets were selected from MS experiments | [49] |
| NEDDylator | Jurkat cells | Substrates are NEDD8 modified by the fusion protein. Lists are also filtered for substrates containing a known binding motif | Inhibitors of apoptosis proteins (IAPs) | In vitro ubiquitylation assay | Over 50 putative IAP substrates. | [50] |
Overall, methods such as BioID can identify the highest number of putative substrates; yet, it is worth considering that biotinylation of proximal proteins may lead to the inclusion of false-positive results at a higher rate than methods such as ubiquitin ligase trapping and the NEDDylator. The latter methods proceed through a more targeted approach and are followed by stringent multi-step purification procedures or purifications under denaturing conditions. This typically results in the identification of fewer but more confident putative substrates.
Validation methods
Regardless of the method used, there is an important need to validate the interaction between ligases and potential substrates. There are several ways that potential targets of E3 ligases can be validated. In the examples from this review (see Table 1), approaches include in vitro or in vivo ubiquitylation assays, siRNA knockdown assays, cycloheximide (CHX) chase assays, and degron mining. In vitro ubiquitylation studies using recombinant proteins are considered to be the gold standard; however, these do not account for post-translational modifications and feedback regulation by DUBs, which would affect functionality of each component.
In addition to validating the interaction between the ligase–substrate pairs, further biological assays are also required to understand the impact of substrate ubiquitylation on biological processes. While identifying novel substrates is an important step in characterising an E3 ligase, the biological significance may be of greater value when teasing out the mechanisms that link defects in E3 ligases with disease.
Application of proteomic approaches for understanding molecular mechanisms underlying neurodegenerative diseases
Identifying ligase and substrate pairs may have important real-world outcomes when it comes to developing novel therapeutics for the treatment of neurodegenerative diseases. Not only does this process assist in identifying substrates that may be dysregulated due to enzyme dysfunction, the process may also assist in identifying ubiquitylation events that lead to the deposition of ubiquitylated inclusions that are characteristics of disease. For example, Parkin may bind and ubiquitylate TDP-43, consequently influencing the aggregation of TDP-43 [74]; a protein found in ubiquitylated inclusions in the post-mortem tissue of most ALS patients and more than half of FTD patients [75]. In addition, the E3-ligase CHIP may collaborate with chaperones to ubiquitylate and degrade Tau [34]. In mouse models of Tauopathies, CHIP removes aberrantly phosphorylated Tau species [76], whilst the silencing of CHIP leads to an increase in Tau levels [77]. In agreement with this, CHIP protein levels are inversely proportional to the accumulation of sarkosyl-insoluble Tau in AD patients [78]. Together the data suggest that protein quality-control mechanisms in neurons may be an important regulator of tauopathies. Accordingly, CHIP and associated chaperones have become attractive targets for the management of tauopathies [34].
Given that these ligases may directly impact the proteostasis of key pathological proteins, E3 ligases themselves have also become potential targets for the management of disease. For example, Parkin has been considered to be a target for the treatment of PD [79]. With the identification of the crystal structure of Parkin, several insights have been gained in terms of mechanisms that lead to ligase activation and the means by which mutations may impair structure/function and protein–protein interactions [80–82]. Accordingly, small molecule drugs may be designed, based on the crystal structure, to regulate Parkin activity [80] as well as ligase–substrate interactions. The enzymatic activity of SCFcyclin F is also affected by ALS-causing mutations. Specifically, the S621G mutation leads to increased ligase activity [83], a result contributing to the accumulation of K48-linked ubiquitylated proteins [84] and accumulation of proteins intended for proteasomal degradation [31]. The ligase activity and substrates of cyclin F may, therefore, also be seen as potential therapeutic targets for the treatment of ALS and FTD. Difficulties in developing therapeutic targets for cyclin F, however, lie in the fact that cyclin F forms part of a larger multi-protein ubiquitin ligase complex [85]. In addition, the crystal structure of cyclin F, which could enable rational drug design, has not yet been elucidated.
Conclusions and future prospects
In the last few years, the standard procedure that has been used to identify ligase–substrate pairs has evolved significantly, enabling the rapid identification of multiple ligase–substrate pairs. It is expected that as the field moves forward, more substrates are characterised, and clearer substrate profiles are obtained, the identification of ligase–substrate pairs may involve a series of additional steps, such as applying known information concerning protein expression profiles, degron sequences, and structural information. By doing so, it will become easier to identify high-confidence substrates using more effective but less stringent methods.
It is possible that, in the next 5 years, bioinformatics will become increasingly popular for the identification and validation of these pairs. Substrates of ligases may display typical characteristics, which include the presence of phosphodegrons, expression profiles, biological roles, interaction sites, and localisation—qualities that may be quickly cross-referenced against experimentally obtained results that are curated over many years. This process will be highly complementary to laboratory-based methods, and may be an efficient way to quickly identify if proteins are likely substrates of a given E3 ligase.
Given that neurodegenerative diseases are characterised by the deposition of insoluble protein inclusions tagged by protein degradation markers, a key direction in the field is to identify processes that may lead to the deposition of insoluble proteins and/or mediate the removal of such deposits. A benefit of many of the emerging methods is the ability to identify substrates of ligases that are insoluble in standard IP buffers. This feature makes them particularly promising, especially when challenged with the increasing insolubility of some substrates that occurs during disease. A consideration, however, is that each ligase may mediate the ubiquitylation of multiple substrates. Thus, identifying single ligase–substrate pairs may not reveal the breadth of functions that may be defective due to disease-causing mutations, and, as a result, a variety of interactions will require validation and further assessment to understand the complete impact of ligase dysfunction in neurodegenerative diseases.
Overall, the application of these novel methods enables the rapid identification of multiple putative substrates of E3 ligases and will assist not only in characterising novel biological roles of E3 ligases, but will also assist in determining processes that may be perturbed when E3 ligases contain disease-causing mutations. Ultimately, identifying processes that are managed by E3 ligases will help linking mutations in E3 ligases with disease pathogenesis and also provide novel targets for therapeutic intervention of neurodegenerative diseases.
Acknowledgements
SLR holds a Macquarie University Research Training Program Scholarship. This research was supported by Grant-in-Aid funding from the Motor Neurone Disease Research Institute of Australia (GIA1628, GIA1638, and GIA1715), National Health & Medical Research Council (APP1095215 and APP1107644), and donations made towards MND research at Macquarie University.
Abbreviations
- AD
Alzheimer’s disease
- ALS
Amyotrophic lateral sclerosis
- AP–MS
Affinity purification–mass spectrometry
- BioID
Proximity-dependent biotin identification
- CHX
Cycloheximide
- CRAPome
Contaminant repository for affinity purification
- DUBs
Deubiquitylases
- E1
Ubiquitin-activating enzyme
- E2
Ubiquitin-conjugating enzyme
- E3
Ubiquitin ligase
- FTD
Frontotemporal dementia
- His6
Hexahistidine
- HB-NEDD8
Histidine–biotin-tagged NEDD8
- HD
Huntington’s disease
- IP
Immunoprecipitation
- IP-LC/MS
Immunoprecipitation followed by mass spectrometry
- MS
Mass spectrometry
- NiNTA
Nickel–nitrilotriacetic acid
- PD
Parkinson’s disease
- TR-TUBE
Trypsin-resistant tandem ubiquitin-binding entities
- TUBE
Tandem ubiquitin-binding entity
- UBA
Ubiquitin-associated domain
- UPS
Ubiquitin-proteasome system
Author contributions
SLR, MPM, AL, and RC conceptualised the article content. SLR wrote the manuscript. MM, MPM, BS, MM, AL, and RC assisted in revising and editing the manuscript.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Roger Chung and Albert Lee are equal last authors.
References
- 1.Ross CA, Poirier MA. Protein aggregation and neurodegenerative disease. Nat Med. 2004;10(Suppl):S10–S17. doi: 10.1038/nm1066. [DOI] [PubMed] [Google Scholar]
- 2.Iwatsubo T, Odaka A, Suzuki N, Mizusawa H, et al. Visualization of A beta 42(43) and A beta 40 in senile plaques with end-specific A beta monoclonals: evidence that an initially deposited species is A beta 42(43) Neuron. 1994;13:45–53. doi: 10.1016/0896-6273(94)90458-8. [DOI] [PubMed] [Google Scholar]
- 3.Spillantini MG, Crowther RA, Jakes R, Cairns NJ, et al. Filamentous alpha-synuclein inclusions link multiple system atrophy with Parkinson’s disease and dementia with Lewy bodies. Neurosci Lett. 1998;251:205–208. doi: 10.1016/S0304-3940(98)00504-7. [DOI] [PubMed] [Google Scholar]
- 4.Spillantini MG, Crowther RA, Jakes R, Hasegawa M, et al. alpha-Synuclein in filamentous inclusions of Lewy bodies from Parkinson’s disease and dementia with lewy bodies. Proc Natl Acad Sci USA. 1998;95:6469–6473. doi: 10.1073/pnas.95.11.6469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arai T, Hasegawa M, Akiyama H, Ikeda K, et al. TDP-43 is a component of ubiquitin-positive tau-negative inclusions in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Biochem Biophys Res Commun. 2006;351:602–611. doi: 10.1016/j.bbrc.2006.10.093. [DOI] [PubMed] [Google Scholar]
- 6.Neumann M, Sampathu DM, Kwong LK, Truax AC, et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314:130–133. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- 7.Behrouzi R, Liu X, Wu D, Robinson AC, et al. Pathological tau deposition in motor neurone disease and frontotemporal lobar degeneration associated with TDP-43 proteinopathy. Acta Neuropathol Commun. 2016;4:33. doi: 10.1186/s40478-016-0301-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davies SW, Turmaine M, Cozens BA, DiFiglia M, et al. Formation of neuronal intranuclear inclusions underlies the neurological dysfunction in mice transgenic for the HD mutation. Cell. 1997;90:537–548. doi: 10.1016/S0092-8674(00)80513-9. [DOI] [PubMed] [Google Scholar]
- 9.Becher MW, Kotzuk JA, Sharp AH, Davies SW, et al. Intranuclear neuronal inclusions in Huntington’s disease and dentatorubral and pallidoluysian atrophy: correlation between the density of inclusions and IT15 CAG triplet repeat length. Neurobiol Dis. 1998;4:387–397. doi: 10.1006/nbdi.1998.0168. [DOI] [PubMed] [Google Scholar]
- 10.DiFiglia M, Sapp E, Chase KO, Davies SW, et al. Aggregation of huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain. Science. 1997;277:1990–1993. doi: 10.1126/science.277.5334.1990. [DOI] [PubMed] [Google Scholar]
- 11.Gutekunst CA, Li SH, Yi H, Mulroy JS, et al. Nuclear and neuropil aggregates in Huntington’s disease: relationship to neuropathology. J Neurosci. 1999;19:2522–2534. doi: 10.1523/JNEUROSCI.19-07-02522.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hasegawa M, Fujiwara H, Nonaka T, Wakabayashi K, et al. Phosphorylated alpha-synuclein is ubiquitinated in alpha-synucleinopathy lesions. J Biol Chem. 2002;277:49071–49076. doi: 10.1074/jbc.M208046200. [DOI] [PubMed] [Google Scholar]
- 13.Lowe J, Lennox G, Jefferson D, Morrell K, et al. A filamentous inclusion body within anterior horn neurones in motor neurone disease defined by immunocytochemical localisation of ubiquitin. Neurosci Lett. 1988;94:203–210. doi: 10.1016/0304-3940(88)90296-0. [DOI] [PubMed] [Google Scholar]
- 14.Lowe J, Blanchard A, Morrell K, Lennox G, et al. Ubiquitin is a common factor in intermediate filament inclusion bodies of diverse type in man, including those of Parkinson’s disease, Pick’s disease, and Alzheimer’s disease, as well as Rosenthal fibres in cerebellar astrocytomas, cytoplasmic bodies in muscle, and mallory bodies in alcoholic liver disease. J Pathol. 1988;155:9–15. doi: 10.1002/path.1711550105. [DOI] [PubMed] [Google Scholar]
- 15.Lecker SH, Goldberg AL, Mitch WE. Protein degradation by the ubiquitin-proteasome pathway in normal and disease states. J Am Soc Nephrol JASN. 2006;17:1807–1819. doi: 10.1681/ASN.2006010083. [DOI] [PubMed] [Google Scholar]
- 16.Saeki Y. Ubiquitin recognition by the proteasome. J Biochem. 2017;161:113–124. doi: 10.1093/jb/mvw091. [DOI] [PubMed] [Google Scholar]
- 17.Cuervo AM, Wong ES, Martinez-Vicente M. Protein degradation, aggregation, and misfolding. Mov Disord. 2010;25(Suppl 1):S49–S54. doi: 10.1002/mds.22718. [DOI] [PubMed] [Google Scholar]
- 18.Ciechanover A, Kwon YT. Degradation of misfolded proteins in neurodegenerative diseases: therapeutic targets and strategies. Exp Mol Med. 2015;47:e147. doi: 10.1038/emm.2014.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tsakiri EN, Trougakos IP. The amazing ubiquitin-proteasome system: structural components and implication in aging. Int Rev Cell Mol Biol. 2015;314:171–237. doi: 10.1016/bs.ircmb.2014.09.002. [DOI] [PubMed] [Google Scholar]
- 20.Ye Y, Rape M. Building ubiquitin chains: E2 enzymes at work. Nat Rev Mol Cell Biol. 2009;10:755–764. doi: 10.1038/nrm2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sluimer J, Distel B. Regulating the human HECT E3 ligases. Cell Mol Life Sci CMLS. 2018;75:3121–3141. doi: 10.1007/s00018-018-2848-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eletr ZM, Huang DT, Duda DM, Schulman BA, et al. E2 conjugating enzymes must disengage from their E1 enzymes before E3-dependent ubiquitin and ubiquitin-like transfer. Nat Struct Mol Biol. 2005;12:933–934. doi: 10.1038/nsmb984. [DOI] [PubMed] [Google Scholar]
- 23.Akutsu M, Dikic I, Bremm A. Ubiquitin chain diversity at a glance. J Cell Sci. 2016;129:875–880. doi: 10.1242/jcs.183954. [DOI] [PubMed] [Google Scholar]
- 24.Tan JM, Wong ES, Kirkpatrick DS, Pletnikova O, et al. Lysine 63-linked ubiquitination promotes the formation and autophagic clearance of protein inclusions associated with neurodegenerative diseases. Hum Mol Genet. 2008;17:431–439. doi: 10.1093/hmg/ddm320. [DOI] [PubMed] [Google Scholar]
- 25.d’Azzo A, Bongiovanni A, Nastasi T. E3 ubiquitin ligases as regulators of membrane protein trafficking and degradation. Traffic. 2005;6:429–441. doi: 10.1111/j.1600-0854.2005.00294.x. [DOI] [PubMed] [Google Scholar]
- 26.Morris JR, Solomon E. BRCA1: BARD1 induces the formation of conjugated ubiquitin structures, dependent on K6 of ubiquitin, in cells during DNA replication and repair. Hum Mol Genet. 2004;13:807–817. doi: 10.1093/hmg/ddh095. [DOI] [PubMed] [Google Scholar]
- 27.McDowell GS, Philpott A. Non-canonical ubiquitylation: mechanisms and consequences. Int J Biochem Cell Biol. 2013;45:1833–1842. doi: 10.1016/j.biocel.2013.05.026. [DOI] [PubMed] [Google Scholar]
- 28.Ardley HC, Robinson PA. The role of ubiquitin-protein ligases in neurodegenerative disease. Neuro-degener Dis. 2004;1:71–87. doi: 10.1159/000080048. [DOI] [PubMed] [Google Scholar]
- 29.Klein C, Westenberger A. Genetics of Parkinson’s disease. Cold Spring Harbor Perspect Med. 2012;2:a008888. doi: 10.1101/cshperspect.a008888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kitada T, Asakawa S, Hattori N, Matsumine H, et al. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature. 1998;392:605–608. doi: 10.1038/33416. [DOI] [PubMed] [Google Scholar]
- 31.Williams KL, Topp S, Yang S, Smith B, et al. CCNF mutations in amyotrophic lateral sclerosis and frontotemporal dementia. Nature Commun. 2016;7:11253. doi: 10.1038/ncomms11253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsai PC, Liao YC, Chen PL, Guo YC, et al. Investigating CCNF mutations in a Taiwanese cohort with amyotrophic lateral sclerosis. Neurobiol Aging. 2018;62:243 e241–243 e246. doi: 10.1016/j.neurobiolaging.2017.09.031. [DOI] [PubMed] [Google Scholar]
- 33.Greer PL, Hanayama R, Bloodgood BL, Mardinly AR, et al. The Angelman syndrome protein Ube3A regulates synapse development by ubiquitinating arc. Cell. 2010;140:704–716. doi: 10.1016/j.cell.2010.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Petrucelli L, Dickson D, Kehoe K, Taylor J, et al. CHIP and Hsp70 regulate tau ubiquitination, degradation and aggregation. Hum Mol Genet. 2004;13:703–714. doi: 10.1093/hmg/ddh083. [DOI] [PubMed] [Google Scholar]
- 35.Lee VM, Trojanowski JQ. Neurodegenerative tauopathies: human disease and transgenic mouse models. Neuron. 1999;24:507–510. doi: 10.1016/S0896-6273(00)81106-X. [DOI] [PubMed] [Google Scholar]
- 36.Lin JS, Lai EM. Protein-protein interactions: co-immunoprecipitation. Methods Mol Biol. 2017;1615:211–219. doi: 10.1007/978-1-4939-7033-9_17. [DOI] [PubMed] [Google Scholar]
- 37.Pierce NW, Kleiger G, Shan SO, Deshaies RJ. Detection of sequential polyubiquitylation on a millisecond timescale. Nature. 2009;462:615–619. doi: 10.1038/nature08595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Clague MJ, Barsukov I, Coulson JM, Liu H, et al. Deubiquitylases from genes to organism. Physiol Rev. 2013;93:1289–1315. doi: 10.1152/physrev.00002.2013. [DOI] [PubMed] [Google Scholar]
- 39.Abbas T, Sivaprasad U, Terai K, Amador V, et al. PCNA-dependent regulation of p21 ubiquitylation and degradation via the CRL4Cdt2 ubiquitin ligase complex. Genes Dev. 2008;22:2496–2506. doi: 10.1101/gad.1676108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morishima Y, Wang AM, Yu Z, Pratt WB, et al. CHIP deletion reveals functional redundancy of E3 ligases in promoting degradation of both signaling proteins and expanded glutamine proteins. Hum Mol Genet. 2008;17:3942–3952. doi: 10.1093/hmg/ddn296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smith A, Simanski S, Fallahi M, Ayad NG. Redundant ubiquitin ligase activities regulate wee1 degradation and mitotic entry. Cell Cycle. 2007;6:2795–2799. doi: 10.4161/cc.6.22.4919. [DOI] [PubMed] [Google Scholar]
- 42.Li Y, Xie P, Lu L, Wang J, et al. An integrated bioinformatics platform for investigating the human E3 ubiquitin ligase–substrate interaction network. Nature Commun. 2017;8:347. doi: 10.1038/s41467-017-00299-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Harper JW, Tan MK. Understanding cullin-RING E3 biology through proteomics-based substrate identification. Mol Cell Proteom MCP. 2012;11:1541–1550. doi: 10.1074/mcp.R112.021154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Coyaud E, Mis M, Laurent EM, Dunham WH, et al. BioID-based identification of Skp cullin F-box (SCF)beta-TrCP1/2 E3 ligase substrates. Mol Cell Proteom MCP. 2015;14:1781–1795. doi: 10.1074/mcp.M114.045658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Roux KJ, Kim DI, Raida M, Burke B. A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J Cell Biol. 2012;196:801–810. doi: 10.1083/jcb.201112098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mark KG, Simonetta M, Maiolica A, Seller CA, et al. Ubiquitin ligase trapping identifies an SCF(Saf1) pathway targeting unprocessed vacuolar/lysosomal proteins. Mol Cell. 2014;53:148–161. doi: 10.1016/j.molcel.2013.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mark KG, Loveless TB, Toczyski DP. Isolation of ubiquitinated substrates by tandem affinity purification of E3 ligase-polyubiquitin-binding domain fusions (ligase traps) Nat Protoc. 2016;11:291–301. doi: 10.1038/nprot.2016.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Loveless TB, Topacio BR, Vashisht AA, Galaang S, et al. DNA damage regulates translation through beta-TRCP targeting of CReP. PLoS Genet. 2015;11:e1005292. doi: 10.1371/journal.pgen.1005292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yoshida Y, Saeki Y, Murakami A, Kawawaki J, et al. A comprehensive method for detecting ubiquitinated substrates using TR-TUBE. Proc Natl Acad Sci USA. 2015;112:4630–4635. doi: 10.1073/pnas.1422313112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhuang M, Guan S, Wang H, Burlingame AL, et al. Substrates of IAP ubiquitin ligases identified with a designed orthogonal E3 ligase, the NEDDylator. Mol Cell. 2013;49:273–282. doi: 10.1016/j.molcel.2012.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.D’Angiolella V, Donato V, Forrester FM, Jeong YT, et al. Cyclin F-mediated degradation of ribonucleotide reductase M2 controls genome integrity and DNA repair. Cell. 2012;149:1023–1034. doi: 10.1016/j.cell.2012.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.D’Angiolella V, Donato V, Vijayakumar S, Saraf A, et al. SCF(Cyclin F) controls centrosome homeostasis and mitotic fidelity through CP110 degradation. Nature. 2010;466:138–142. doi: 10.1038/nature09140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Peschiaroli A, Dorrello NV, Guardavaccaro D, Venere M, et al. SCFbetaTrCP-mediated degradation of Claspin regulates recovery from the DNA replication checkpoint response. Mol Cell. 2006;23:319–329. doi: 10.1016/j.molcel.2006.06.013. [DOI] [PubMed] [Google Scholar]
- 54.Fuchs SY, Spiegelman VS, Kumar KG. The many faces of beta-TrCP E3 ubiquitin ligases: reflections in the magic mirror of cancer. Oncogene. 2004;23:2028–2036. doi: 10.1038/sj.onc.1207389. [DOI] [PubMed] [Google Scholar]
- 55.Kim TY, Siesser PF, Rossman KL, Goldfarb D, et al. Substrate trapping proteomics reveals targets of the betaTrCP2/FBXW11 ubiquitin ligase. Mol Cell Biol. 2015;35:167–181. doi: 10.1128/MCB.00857-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mavrommati I, Faedda R, Galasso G, Li J, et al. beta-TrCP- and casein kinase II-mediated degradation of cyclin F controls timely mitotic progression. Cell reports. 2018;24:3404–3412. doi: 10.1016/j.celrep.2018.08.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Guardavaccaro D, Frescas D, Dorrello NV, Peschiaroli A, et al. Control of chromosome stability by the beta-TrCP-REST-Mad2 axis. Nature. 2008;452:365–369. doi: 10.1038/nature06641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dorrello NV, Peschiaroli A, Guardavaccaro D, Colburn NH, et al. S6K1- and betaTRCP-mediated degradation of PDCD4 promotes protein translation and cell growth. Science. 2006;314:467–471. doi: 10.1126/science.1130276. [DOI] [PubMed] [Google Scholar]
- 59.Wu G, Xu G, Schulman BA, Jeffrey PD, et al. Structure of a beta-TrCP1-Skp1-beta-catenin complex: destruction motif binding and lysine specificity of the SCF(beta-TrCP1) ubiquitin ligase. Mol Cell. 2003;11:1445–1456. doi: 10.1016/S1097-2765(03)00234-X. [DOI] [PubMed] [Google Scholar]
- 60.Kruiswijk F, Yuniati L, Magliozzi R, Low TY, et al. Coupled activation and degradation of eEF2K regulates protein synthesis in response to genotoxic stress. Sci Signal. 2012;5:ra40. doi: 10.1126/scisignal.2002718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Low TY, Peng M, Magliozzi R, Mohammed S, et al. A systems-wide screen identifies substrates of the SCFbetaTrCP ubiquitin ligase. Sci Signal. 2014;7:rs8. doi: 10.1126/scisignal.2005882. [DOI] [PubMed] [Google Scholar]
- 62.Roux KJ, Kim DI, Burke B. BioID: a screen for protein-protein interactions. Curr Protocols Protein Sci. 2013;74:19–23. doi: 10.1002/0471140864.ps1923s74. [DOI] [PubMed] [Google Scholar]
- 63.Li Y, Sousa R. Expression and purification of E. coli BirA biotin ligase for in vitro biotinylation. Protein Expr Purif. 2012;82:162–167. doi: 10.1016/j.pep.2011.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cronan JE., Jr Biotination of proteins in vivo. A post-translational modification to label, purify, and study proteins. J Biol Chem. 1990;265:10327–10333. [PubMed] [Google Scholar]
- 65.Kwon K, Streaker ED, Ruparelia S, Beckett D. Multiple disordered loops function in corepressor-induced dimerization of the biotin repressor. J Mol Biol. 2000;304:821–833. doi: 10.1006/jmbi.2000.4249. [DOI] [PubMed] [Google Scholar]
- 66.Frescas D, Pagano M. Deregulated proteolysis by the F-box proteins SKP2 and beta-TrCP: tipping the scales of cancer. Nat Rev Cancer. 2008;8:438–449. doi: 10.1038/nrc2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rhee HW, Zou P, Udeshi ND, Martell JD, et al. Proteomic mapping of mitochondria in living cells via spatially restricted enzymatic tagging. Science. 2013;339:1328–1331. doi: 10.1126/science.1230593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Varnaite R, MacNeill SA. Meet the neighbors: mapping local protein interactomes by proximity-dependent labeling with BioID. Proteomics. 2016;16:2503–2518. doi: 10.1002/pmic.201600123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hofmann K, Bucher P. The UBA domain: a sequence motif present in multiple enzyme classes of the ubiquitination pathway. Trends Biochem Sci. 1996;21:172–173. doi: 10.1016/S0968-0004(96)30015-7. [DOI] [PubMed] [Google Scholar]
- 70.Su V, Lau AF. Ubiquitin-like and ubiquitin-associated domain proteins: significance in proteasomal degradation. Cell Mol Life Sci CMLS. 2009;66:2819–2833. doi: 10.1007/s00018-009-0048-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wilkinson CR, Seeger M, Hartmann-Petersen R, Stone M, et al. Proteins containing the UBA domain are able to bind to multi-ubiquitin chains. Nat Cell Biol. 2001;3:939–943. doi: 10.1038/ncb1001-939. [DOI] [PubMed] [Google Scholar]
- 72.Hjerpe R, Aillet F, Lopitz-Otsoa F, Lang V, et al. Efficient protection and isolation of ubiquitylated proteins using tandem ubiquitin-binding entities. EMBO Rep. 2009;10:1250–1258. doi: 10.1038/embor.2009.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Heride C, Urbe S, Clague MJ. Ubiquitin code assembly and disassembly. Curr Biol CB. 2014;24:R215–R220. doi: 10.1016/j.cub.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 74.Hebron ML, Lonskaya I, Sharpe K, Weerasinghe PP, et al. Parkin ubiquitinates Tar-DNA binding protein-43 (TDP-43) and promotes its cytosolic accumulation via interaction with histone deacetylase 6 (HDAC6) J Biol Chem. 2013;288:4103–4115. doi: 10.1074/jbc.M112.419945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ling SC, Polymenidou M, Cleveland DW. Converging mechanisms in ALS and FTD: disrupted RNA and protein homeostasis. Neuron. 2013;79:416–438. doi: 10.1016/j.neuron.2013.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dickey CA, Patterson C, Dickson D, Petrucelli L. Brain CHIP: removing the culprits in neurodegenerative disease. Trends Mol Med. 2007;13:32–38. doi: 10.1016/j.molmed.2006.11.003. [DOI] [PubMed] [Google Scholar]
- 77.Dickey CA, Koren J, Zhang YJ, Xu YF, et al. Akt and CHIP coregulate tau degradation through coordinated interactions. Proc Natl Acad Sci USA. 2008;105:3622–3627. doi: 10.1073/pnas.0709180105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sahara N, Murayama M, Mizoroki T, Urushitani M, et al. In vivo evidence of CHIP up-regulation attenuating tau aggregation. J Neurochem. 2005;94:1254–1263. doi: 10.1111/j.1471-4159.2005.03272.x. [DOI] [PubMed] [Google Scholar]
- 79.Kubo S, Hatano T, Takanashi M, Hattori N. Can parkin be a target for future treatment of Parkinson’s disease? Expert Opin Therapeutic Targets. 2013;17:1133–1144. doi: 10.1517/14728222.2013.827173. [DOI] [PubMed] [Google Scholar]
- 80.Wauer T, Simicek M, Schubert A, Komander D. Mechanism of phospho-ubiquitin-induced PARKIN activation. Nature. 2015;524:370–374. doi: 10.1038/nature14879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Trempe JF, Sauve V, Grenier K, Seirafi M, et al. Structure of parkin reveals mechanisms for ubiquitin ligase activation. Science. 2013;340:1451–1455. doi: 10.1126/science.1237908. [DOI] [PubMed] [Google Scholar]
- 82.Gladkova C, Maslen SL, Skehel JM, Komander D. Mechanism of parkin activation by PINK1. Nature. 2018;559:410–414. doi: 10.1038/s41586-018-0224-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lee A, Rayner SL, DeLuca A, Gwee SSL, et al. Casein kinase II phosphorylation of cyclin F at serine 621 regulates the Lys48-ubiquitylation E3 ligase activity of the SCF((cyclin F)) complex. Open Biol. 2017;7:170058. doi: 10.1098/rsob.170058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lee A, Rayner SL, Gwee SSL, De Luca A, et al. Pathogenic mutation in the ALS/FTD gene, CCNF, causes elevated Lys48-linked ubiquitylation and defective autophagy. Cell Mol life Sci CMLS. 2018;75:335–354. doi: 10.1007/s00018-017-2632-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Galper J, Rayner SL, Hogan AL, Fifita JA, et al. Cyclin F: a component of an E3 ubiquitin ligase complex with roles in neurodegeneration and cancer. Int J Biochem Cell Biol. 2017;89:216–220. doi: 10.1016/j.biocel.2017.06.011. [DOI] [PubMed] [Google Scholar]