Abstract
Throughout the human life, the gut microbiota interacts with us in a number of different ways, thereby influencing our health status. The acquisition of such an interactive gut microbiota commences at birth. Medical and environmental factors including diet, antibiotic exposure and mode of delivery are major factors that shape the composition of the microbial communities in the infant gut. Among the most abundant members of the infant microbiota are species belonging to the Bifidobacterium genus, which are believed to confer beneficial effects upon their host. Bifidobacteria may be acquired directly from the mother by vertical transmission and their persistence in the infant gut is associated with their saccharolytic activity toward glycans that are abundant in the infant gut. Here, we discuss the establishment of the infant gut microbiota and the contribution of bifidobacteria to this early life microbial consortium.
Keywords: Microbiome, Microbiota, Bifidobacteria, Genomics, Metagenomics
Development of the human gut microbiota
Most of the microbial community found in the human body is located in the large intestine, where bacteria outnumber eukaryotic cells by about tenfold [69, 100]. It is well established that the gut microbiota performs a number of important activities that affect overall host metabolism and physiology. However, before it reaches its adult configuration, the gut microbiota undergoes a compositional progression that starts at birth and is aimed at establishing a mutually beneficial cohabitation with the host. In this context, the current data available on the progression of infant gut microbiota development has benefited from the latest technologies in DNA sequencing. It is generally accepted that, in healthy subjects, the gut microbiota is highly stable during adult life, slightly fluctuating on both sides of an equilibrium, also known as climax, whose composition appears to be host-specific [120, 144].
Although the intra-uterine environment is considered to be sterile, there is some evidence to support the occurrence of bacteria on or in the foetus [31, 59, 94]. It has been suggested that such microorganisms originate from prenatal colonization of the meconium [59, 71], where bacteria, such as Escherichia coli, Enterococcus faecium as well as Bifidobacterium spp. and Staphylococcus epidermidis, are present, which in turn may have ended up there as a result of translocation from the maternal gut [58]. Further evidence that corroborates the notion of bacterial colonization of the intra-uterine habitat is provided by PCR-based identification of Bifidobacterium and Lactobacillus species in the placenta of vaginally and caesarean section-delivered infants [116]. Despite these reports, there is still controversy regarding the presence of bacteria in the intra-uterine environment, and whether this can be taken as a general phenomenon, as a technical anomaly or as an atypical finding. Thus, we neither exclude that colonization of the infant gut in fact happens during prenatal growth, nor do we know if this is of any relevance or consequence to the colonization events that will happen immediately following birth.
External factors, such as drugs, illness, stress and heavy metal/toxin exposure, are considered to, or known to, influence the development of the infant gut microbiota. In this context, one particular animal trial involving infant monkeys, where mothers were exposed to severe stress, has shown that bifidobacterial populations are markedly reduced compared to control monkey infants born from non-stressed mothers [11]. Unfortunately, the low resolution of the methodology used in this study for the compositional characterization of the non-human primate gut microbiota did not allow species identification of these bifidobacterial taxa. Other studies have shown that the use of antibiotics during pregnancy does not significantly affect the corresponding infant gut microbiota [64, 94]. Thus, it is considered that external factors may substantially influence the infant gut microbiota, although such effects may nonetheless be subtle, yet impactful [73].
Key features of the infant gut microbiota
It has been shown that the microbiota residing in the infant gut is very different to that identified in the adult gut [73]. The exact age at which a stable adult-type microbiota is established has not yet been defined, although it has been suggested that this occurs between 1 and 3 years of age [62, 158]. Nevertheless, this process is believed to take several years and events, such as hormonal changes during puberty or modifications in eating habits, may affect the microbiota structure [2, 54].
During gut microbiota establishment, the first microbial actors that render the gastro intestinal environment fully anaerobic are facultative anaerobes, which include several members of the Enterobacteriaceae family [58, 73]. After the removal of oxygen, the infant gut undergoes extensive colonization driven by strictly anaerobic bacteria taxa, such as those belonging to the genera Bifidobacterium, Clostridium, Bacteroides and Ruminococcus [59, 73].
This initial colonization process of the infant gut is influenced by environmental factors including the duration of the gestation (being either full-term or pre-term), the delivery mode (i.e. vaginal or caesarian), the type of feeding (i.e. breast milk or formula milk) and the living environment [1, 73]. It has recently been demonstrated that there is a large inter-individual variability in the rate of microbiota progression [32]. Despite this, it has been possible to categorize the gut microbiota arrangements into three main and distinct types: cluster 1, which is rich in Firmicutes and in particular of Streptococcus; cluster 2, displaying high levels of Enterobacteriaceae family members, especially in Klebsiella; and cluster 3, which is characterized by high numbers of members of the genera Bifidobacterium and Collinsella, and a low abundance of Streptococcus and Klebsiella [32]. Notably, the composition and developmental progression of the early microbiota appears to have long-lasting implications on host health. Disruption of the early gut microbiota through antibiotic therapy [3, 85], or having followed a differential trajectory in gut microbiota acquisition following caesarian delivery [15, 55] are two events that are linked to increased adiposity in later life. In this context, the infant gut microbiota that displays a cluster 3 profile is associated with a lower risk of adiposity in later life [32].
Environmental influences on gut microbiota assembly of infants
An important external factor that influences the development of the infant gut microbiota is the mother herself. In fact, there is a strong link between the gut microbiota of the infant and that of the corresponding mother. In this context, evidence has been put forward that supports the notion of vertical transmission of specific members (e.g., bifidobacterial strains) of the gut microbiota of the mother to the neonate [78]. Furthermore, at 1 month of age, the gut microbiota of an infant is both functionally and phylogenetically very related to that of the mother [142]. Nevertheless, at 1 year of age significant phylogenetic differences are found, although substantial similarities between predicted microbiota functions between mother and child are still present [142].
The gestation time is another important factor in shaping the infant gut microbiota. In this context, comparison of the faecal microbiota of full-term versus pre-term infants revealed marked differences. Members of the family Enterobacteriaceae are significantly enriched in the gut microbiota of pre-term infants, whereas that of full-term infants exhibit a higher diversity with abundant occurrence of various genera, in particular Bifidobacterium, Lactobacillus and Streptococcus [7, 8].
The mode of delivery represents another key factor that impacts on the assembly of the microbial communities of the infant gut. Metagenomic analyses of the meconium revealed a strong link between the first microbial gut communities of the newborn, and those of the mother vaginal mucosa or that of the maternal skin in case of vaginal delivery or caesarian section, respectively [33]. This suggests that the intestinal environment of the neonate becomes populated by the first microorganisms it encounters. Generally, the numbers of bifidobacteria are particularly high in vaginally delivered babies [56]. However, in pre-term infants the delivery mode does not appear to influence the gut microbiota composition of the newborn [7, 112].
In addition, delayed gut colonization in caesarian-delivered infants seems to occur for certain bacterial groups, including members of the genera Bifidobacterium and Bacteroides [88].
Another key factor that has a marked impact on shaping the neonatal gut microbiota is the type of feeding (for a review see [73]). Several studies based on culture-dependent approaches have previously demonstrated that the gut microbiota of breast-fed infants has significantly higher counts of bifidobacteria and lactobacilli, and lower counts of Bacteroides, Clostridium coccoides group, Staphylococcus and members of the Enterobacteriaceae family, compared to that of formula-fed neonates [42, 103]. Recent metagenomic analyses of the infant gut microbiota indeed reinforced these original findings [74, 78, 138, 157]. The identification of specific oligosaccharides present in human milk, also known as human milk oligosaccharides (HMOs), that appear to act as powerful prebiotics for specific members of the gut microbiota, in particular bifidobacteria, represents a milestone in the area of human gut microbiota development. Notably, milk oligosaccharides are produced by all mammalian taxa, although oligosaccharide profiles are highly variable among species [128]. This suggests that oligosaccharides in mammalian milk have been subjected to natural selection [52, 53]. In human milk, HMOs, after lactose by ranked abundance, are one of the most abundant components of milk [52, 53]. Interestingly, human milk contains a greater diversity and higher abundance of oligosaccharides than any other mammal [52]. Furthermore, individual mothers produce HMO-containing milk of particular composition, which varies by the presence, abundance and proportion of particular oligosaccharide isomers [123].
HMOs are not digested by the mammalian enzymatic arsenal, and they therefore remain intact during the passage through the gastrointestinal tract to arrive in the colon where they undergo degradation performed by specific components of the gut microbiota [28]. In this context, it has been shown that bifidobacterial genomes belonging to the Bifidobacterium longum subsp. infantis, Bifidobacterium bifidum and Bifidobacterium breve taxa contain specific gene clusters encoding enzymes that are capable to hydrolyze (certain) HMOs [57, 66, 122, 141]. Notably, this HMO-metabolizing ability corresponds to the high abundance of these species in the gut of breast-fed infants [68, 74, 138]. Altogether, these data imply that mothers are not only feeding their child, but also nurture specific members of the gut microbiota, in particular specific bifidobacterial strains/species, which may represent a key development with long-lasting effects on host health.
The familial and social environment represents another source of bacteria that may affect early gut colonization. In this context, infants with older siblings display a lower occurrence of bacteria in their gut, yet contain a comparatively higher proportion of bifidobacteria [94]. Similarly, comparisons have been made between the faecal microbiota of children from different geographical regions, such as Amerindians vs Malawians and North Americans, which revealed that the microbiota very much depends on the immediate environment as well as the diet [158]. These analyses further confirm the dominance of bifidobacteria during the first stages of human life even in populations from very distinct geographical regions [158]. Notably, people living in proximity, i.e. in the same household, share a more similar microbiome composition, even though the extent of horizontal transmission, especially in the context of bifidobacterial communities, has not yet been determined.
Currently, there is no clear evidence suggesting that gut microbiota dysbiosis or gut microbiota-linked diseases (e.g., ulcerative colitis, and inflammatory bowel disease) can be transmitted to newborns by other members of the household. As has been shown for adults, drug-based therapies are known to modify the composition of the infant gut microbiota [30]. This can occur directly or indirectly through antibiotic treatment of the (breastfeeding) mother, although there is conflicting data in literature regarding this issue (see above in this review), thus requiring further study. Antibiotic treatment in infants is correlated with higher occurrence of enterobacteria and enterococci, and lower abundance of bifidobacteria [127], and such changes are persistent for up to 1 month after the cessation of the treatment. During the initial colonization phase of the neonate, antibiotics cause a marked reduction in complexity of the gut microbiota composition [154]. Antibiotic treatment of mothers in the prenatal or breastfeeding phases is associated with a less diverse infant gut microbiota characterized by a low abundance of members of Bacteroides and Atopobium genera [42]. Nevertheless, as described above, there are contradictory published data about this topic that will require additional analyses.
Bifidobacteria and the gut microbiota of infants
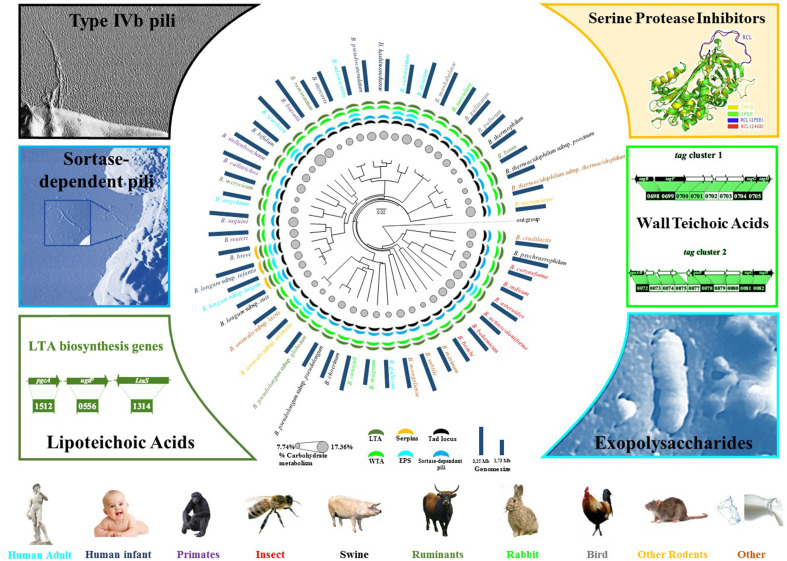
As outlined above, bifidobacteria are one of the most common inhabitants of the infant gut microbiota [75, 78, 87, 88, 104, 111, 138, 141, 150, 157]. Notably, the contribution of bifidobacteria to the GIT microbiota of human is higher in the large intestine, although they are found (at low numbers) in the oral cavity [151]. Bifidobacteria are Gram-positive microorganisms with a high G + C DNA content, that were first isolated from feces of a breast-fed infant by Tissier in 1899, and were then classified as a distinct genus belonging to the Actinobacteria phylum [67, 146]. Currently, the genus Bifidobacterium harbours 48 different taxa, 40 of which have been isolated from faecal samples of the gastro intestinal tract (GIT) contents of mammals, birds, or insects, while the remaining 8 originate from sewage and fermented milk [145, 148, 147, 149]. Bifidobacteria have been isolated from six different ecological niches including the GIT of humans, non-human mammals, birds, social insects, waste waters, human blood and oral cavity [141] (Fig. 1). Many of these ecological niches are not particularly related, suggestive of a rather broad ecological behaviour of bifidobacteria. However, it is possible to identify a common ecological overlap between all these habitats, represented by the fact that bifidobacterial hosts represent social animals whose offsprings enjoy parental care. Therefore, perhaps their ecological distribution is facilitated by direct transmission of bifidobacterial cells from mother/carer to offspring. This finding has recently been documented by the identification of bifidobacterial strains belonging to the Bifidobacterium breve, Bifidobacterium bifidum and Bifidobacterium longum subsp. longum that are shared between mothers and their respective children [78, 111]. Bifidobacteria encompass taxa, such as Bifidobacterium animalis subsp. lactis/animalis, Bifidobacterium adolescentis, Bifidobacterium dentium and Bifidobacterium catenulatum, which can be found in various animals, and are therefore also known as cosmopolitan bifidobacterial species [65]. In contrast, other species such as Bifidobacterium bifidum, Bifidobacterium breve and Bifidobacterium longum are known to be uniquely associated with human beings [131]. Among these, it is possible to distinguish bifidobacterial taxa that are typically found in adults, e.g. B. adolescentis and B. catenulatum, while others are much more commonly found in the gut of breast-fed infants, such as B. bifidum, B. breve and B. longum subsp. infantis [134, 138]. However, there is not a very strict infant vs adult division of bifidobacterial species. In fact, vertical transfer of bifidobacterial taxa, including adult-type members such as B. adolescentis [36] from mother to infant appears to be an important transmission route in microbiota establishment during the initial phase of life (see below).
Fig. 1.
Bifidobacterial phylogeny and proven or potential bifidobacterial interactions with its host. A supertree based on the alignment of 413 core COGs was constructed to obtain a robust phylogenetic reconstruction. The bifidobacterial (sub)species are typed with different colours consistent with their ecological origins. Pillars surrounding the tree represent approximate genome sizes. Circles in grey represent relative percentage of genes predicted to be involved in carbohydrate metabolism and transport. The presence of genes encoding various extracellular structures in the bifidobacterial genomes are represented by segments coloured according to the type of structures. Each external sector represents a different extracellular structure (potentially) involved in host interaction. For each extracellular structure, the corresponding image or genetic locus is indicated. The inset of the exopolysaccharide structure represents a cryo-SEM micrograph of a Bifidobacterium animalis subsp. lactis IPLA4549 cell modified as described elsewhere [105]
Bifidobacterial occurrence across the human lifespan
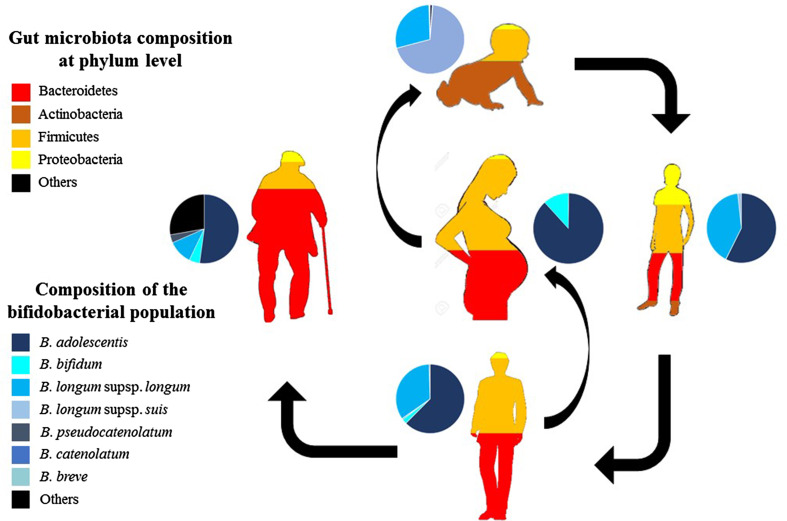
Bifidobacteria dominantly colonize the large intestine of (full-term, healthy and breast-fed) infants during the first weeks of life due to their competitive abilities with regards to breast milk (i.e. their ability to metabolize HMOs), as clearly demonstrated by metagenomic analyses [62, 104]. In breast-fed infants, the bifidobacterial population (on average across infants) has been shown to be dominated by B. breve species, followed by B. bifidum and B. longum subsp. infantis [74, 75, 88, 138] (Fig. 2). However, recent metagenomics-based investigations targeting the gut microbiota of breast-fed infants highlighted a high abundance of Operational Taxonomic Units belonging to B. longum subsp. longum species and B. pseudocatenulatum [74, 111, 157].
Fig. 2.
Evaluation of bifidobacterial distribution in the human gut as a function of age. Each cake diagram represents the relative abundance of various bifidobacterial (sub)species at the various stages of human life. The relative abundance of the various microbial phyla identified in the human gut at different ages are marked with different colours
The infant-to-adult gut microbiota transition coincides with a progressive reduction of breastfeeding and an increase of solid food intake, which represent dietary driving forces of microbial community structure [10]. During this weaning period, there is a progressive decrease of bifidobacteria even after 1 month of weaning; bifidobacteria are still dominant members of the gut microbiota [41, 42]. In contrast, bifidobacterial levels declined following weaning together with Clostridium perfringens and Clostridium difficile, while the abundance of other strictly anaerobic clostridia was shown to increase. Notably, 6 months after the start of weaning, the influence of feeding method and mode of delivery still persisted. In this context, a high abundance of bifidobacteria is linked with breastfeeding during the weaning period [41]. Overall, the gut microbiota of infants, due to dietary modifications, was shown to be more stable and homogenous after weaning, becoming more similar to the typical adult microbiota [62].
Notably, the switch from an infant to an adult gut microbiota, which happens, as mentioned above, between 1 and 2 years of age, is characterized by a high abundance of B. breve, although the particular reasons for this abundance are unknown [111].
The proportional abundance of bifidobacteria in the human colon declines as the host ages (Fig. 2), although ecological analyses based on FISH and metagenomic studies have pointed out that their presence in the adult colon is around 4.3 ± 4.4% of the overall microbiota [37, 83]. Notably, even if they are less numerous than other microorganisms in the adult gut, bifidobacteria appear to contribute a substantial proportion of the gut-associated enzymatic arsenal dedicated to the breakdown of dietary plant polysaccharides, such as starch and xylan [77, 79]. Thus, it has been postulated that bifidobacteria are important contributors to the gut glycobiome of human beings, by facilitating the metabolism of complex carbohydrates that are not digested by human hydrolytic enzymes or by the microbiota residing in the upper GIT compartments [77]. Furthermore, these bifidobacterial enzymatic activities either directly or indirectly, through modulation of the gut microbiota composition, lead to the production of short chain fatty acids (SCFA) [135]. In human adults, bifidobacterial communities are dominated by B. longum subsp. longum, B. adolescentis and B. catenulatum (Fig. 2). Culture-independent approaches based on the HITChip phylogenetic microarray (which encompasses over 4800 tiling oligonucleotides targeting the V1 or the V6 region of the 16S rRNA gene from 1132 microbial phylotypes belonging to the human gut microbiota) showed that faecal samples of adults contain a relatively stable bacterial population, both from a proportional and compositional perspective [102]. Extensive cataloguing of bifidobacterial populations present in human faecal samples of both adults and infants has provided a detailed delineation of the biodiversity of the bifidobacterial populations present in the human gut across a wide age range [133]. This work identified the most abundant bifidobacterial species, i.e., B. longum subsp. longum, present in the human gut, a finding that was confirmed in a subsequent metagenomic-based study [138]. Notably, apart from typical adult-type bifidobacterial species, the bifidobacterial population in the colon of adult human beings does contain, though at a low level, bifidobacterial species, such as B. bifidum and B. breve, that are typical of the infant gut [133, 138]. The distribution of bifidobacteria present in the large intestine of various human subjects of different ages underlines an inter-subject variability, yet high stability in the various colonic sites within the same individual [133]. These findings perfectly match the knowledge of the gut microbiota variability at intra- and inter-individual level [37].
Although faecal samples from infants are dominated by a relatively small number of bifidobacterial species and strains, they revealed a high inter-individual variation in taxonomic composition. In contrast, the bifidobacterial populations isolated from adults seem to be more complex, though they exhibit a remarkable conservation of bifidobacterial species and strains [133].
Metagenomic surveys of enteric microbial communities revealed the occurrence of many, as yet unclassified members of the human gut microbiota [101], which appear to be unculturable. Thus, it is highly plausible that such metagenomic sequences represent novel bifidobacterial taxa, which may contribute to the establishment and maintenance of human gut homeostasis. In this context, Turroni et al. evaluated the bifidobacterial composition of the human intestine by a microbiomic approach, through the analyses of five colonic mucosal samples from healthy adults [134]. This work identified various novel, i.e. unclassified, bifidobacterial 16S rRNA gene sequences, which are presumed to correspond to as yet undefined and thus novel bifidobacterial species.
Bifidobacterial contribution to the gut physiology
A key activity performed by bifidobacteria in the human gut is their contribution to the metabolism of glycans, which may be derived from the diet (e.g. complex plant carbohydrates) or the host (host-derived glycans). This important feature of bifidobacteria is in line with their saccharoclastic metabolism, whereby their energy and carbon demands are met by the fermentation of carbohydrates [77]. Bioinformatic analyses of gut commensals revealed that the genetic arsenal dedicated to the breakdown of complex, (host-)indigestible carbohydrates derived from the host, such as HMOs and mucin, or from the diet, are important contributors to the gut microbiome composition [72, 150] (Fig. 1).
As mentioned above, HMOs are selectively fermented by specific members of the infant gut microbiota, in particular by bifidobacterial species, such as B. longum subsp. infantis. The genome of B. longum subsp. infantis ATCC15697 encompasses a gene cluster predicted to encode for GHs and carbohydrate transporters necessary for importing and metabolizing HMOs, thus named as HMO cluster [122]. This HMO cluster encodes a variety of predicted or proven catabolic enzymes, such as fucosidases, sialidases, a β-hexosaminidase and β-galactosidases, as well as extracellular solute binding proteins and permeases, that are dedicated to HMO metabolism [121, 122, 159].
Interestingly, this enzymatic repertoire is not encoded by the chromosomes of the other two members of the B. longum phylogenetic group, i.e., B. longum subsp. longum and B. longum subsp. suis, which in contrast revealed a higher genomic capacity to utilize plant-derived glycans, including arabinoxylans [17, 21]. Furthermore, maternal genotypes that determine fucosylation patterns of HMOs have been shown to affect the assemblage of infants’ microbiota [14, 68]. Notably, as mentioned above, specific infant-associated bifidobacteria such as B. longum subsp. infantis can efficiently metabolize HMO components such as lacto-N-tetraose [122, 152], but they metabolize fucosyllactose (FL) at lower efficiency, this being the main component of HMOs [4, 19]. In this context, while B. longum subsp. infantis showed a homogeneous HMO-utilization pattern, characterized by the utilization of both FL as well as sialyllactose, more diverse and strain-specific patterns of HMO metabolism were observed for B. bifidum strains [50]. Only recently, it was shown that a key genetic factor encoding an ABC transporter for FL internalization is needed to utilize this HMO [74]. Such findings indicate that patterns of distribution of the FL transporter are responsible for changes in the gut microbiota composition in infants. It has been discovered that infants fed by non-secretor mothers, i.e., individuals with inactive alleles of the fucosyltransferase 2 gene, are delayed in their establishment of a bifidobacteria dominated-microbiota. It has been postulated that the reduction in HMO fucosylation in non-secretor mothers retards the infant gut colonization by particular bifidobacterial taxa specialized in the consumption of such fucosylated HMOs [68].
Another host-produced glycan is represented by mucins, which make up much of the mucus layer that covers the intestinal mucosa. The main monosaccharide components in mucin-derived glycoproteins are N-acetylglucosamine, N-acetylgalactosamine, fucose and galactose, and these glycoproteins are frequently decorated with sialic acid and/or sulphate groups [126].
The repertoire of GH needed to cleave the mucin oligosaccharide chain into its constituent monosaccharides encompasses: (1) endo-α-N-acetylgalactosaminidase, catalyzing the breakdown of O-glycosidic α-linkages between galactosyl β-1-3N-acetylgalactosamine and serine or threonine residues typically found in mucin-type glycoproteins [47]; (2) fucosidases, releasing the α-linked L-fucose from the oligosaccharide core of the mucin structure [9]. Additional GHs involved in the metabolism of mucin include N-acetyl-β-hexosaminidases, β-galactosidases and sialidases [86]. Furthermore, the core oligosaccharide structure of mucin is composed of N-tetraose that is degraded by certain enteric bacteria including particular bifidobacteria into N-biose [152] and then transported in the cytoplasm through a specific ABC-type transporter, phosphorylated by a galacto-N-biose phosphorylase and finally metabolized in the glycolytic and amino sugar metabolic pathways [86].
Notably, only certain enteric bacteria have been shown to utilize mucins and in this context, within the genus Bifidobacterium only members of the B. bifidum species have been reported to efficiently metabolize mucin [106, 108, 130, 136]. In addition, the prediction of the pan-genome of the B. bifidum species together with functional genomic approaches have identified a gene set involved in mucin metabolism that is uniquely present in the genomes of members of the B. bifidum species, thus being part of the core genome of this species [35]. These findings provide an interesting example of co-evolution of a human gut commensal to the human intestine, where the glycans produced by the host serve as a carbon and energy source for this bifidobacterial species [130].
Another source of glycoproteins is represented by milk. Interestingly, milk glycoproteins have been shown to act as specific growth substrates for infant-type bifidobacteria. Recently, a cell wall-associated endo-β-N-acetylglucosaminidase (EndoBI-1) found in various infant-type bifidobacteria was identified as being able to remove a range of intact N-linked glycans, which serve as a sole source for the selective growth of bifidobacteria [60].
Enteric bacteria are also active in the metabolism of various complex, dietary-derived glycans, such as resistant starch, maltodextrin, maltotriose and amylopectin, which have escaped digestion by the relatively limited mammalian enzymatic repertoire. Starch consists of amylose and amylopectin moieties, with the former being a linear α-(1, 4) glucose chain with a plant-specific degree of polymerization of 200–6000, while the latter representing short linear α-(1, 4) glucose-linked chains with α-(1, 6)-linked side chains [143]. Other derivatives of starch are amylopectin, maltodextrin, maltotriose and maltose chains [143]. Hydrolysis of these polysaccharides is carried out by human gut commensals due to the combined action of amylases (EC 3.2.1.1, EC 3.2.1.2 and EC 3.2.1.3) and amylopullulanases (APU; EC 3.2.1.41). Starch degradation can be performed by various gut microbiota members, such as Ruminococcus bromii [161], Bacteroides thetaiotaomicron [26] and Roseburia inulinivorans [119], where all produce amylases and/or amylopullulanases. Although bifidobacteria are considered to represent relatively minor players in the adult gut microbiota, their important biological roles in the metabolism of dietary and host-derived glycans have only recently become appreciated [77].
In bifidobacteria, comparative and functional genomic analyses have highlighted the widespread occurrence of the above-mentioned starch-degrading enzymes, especially in the genomes of the adult-type Bifidobacterium adolescentis [36]. Glycobiome predictions of B. adolescentis have shown that this species, when compared to other human gut bifidobacteria, encodes the largest set of GH13 family enzymes, which is predicted to include amylase, pullulanase and cyclomaltodextrinase activities, thus suggesting excellent growth performance of this species on particular plant-derived carbohydrates [76, 77, 79]. These findings were corroborated by the investigation of fermentation profiles of members of the B. adolescentis species, revealing a preference for the utilization of different esose-containing sugars, such as galactose, mannose and glucose, as well as plant-derived glycans, such as starch, which are commonly present in the human diet [77].
In contrast to what has been described above, there are also bifidobacterial species such as B. breve displaying carbohydrate breakdown capabilities toward both dietary as well as host-derived glycans [89, 96–98, 110]. The prediction of the pan-genome of the B. breve taxon revealed variability for the presence of genes previously characterized as being involved in the utilization of the carbohydrates ribose, sucrose and raffinose, as well as the plant-derived polysaccharides starch, galactan and cellodextrin [17]. This diversity and flexibility in carbohydrate utilization is believed to enable the persistence of particular bifidobacterial species among the gut microbiota irrespective of host age or diet. In this context, the glycobiome of B. breve species is particularly rich in representatives of the GH13 family, which are typically enzymes responsible for the breakdown of substrates with alpha-glucopyranose units, such as pullulan, glycogen, maltodextrin, starch, and amylopectin, and their presence has been marked as a genetic feature that is typical of the B. breve taxon [61, 110]. Furthermore, (many) B. breve strains can efficiently utilize sialic acid as an energy source and can cross-feed on HMO degradation products allowing members of this bifidobacterial species to establish themselves as part of the breast-fed infant microbiota [39, 40].
Notably, the core genome of B. breve includes genes that are predicted to encode enzymes involved in the uptake and utilization of host-derived mono/oligosaccharides, in particular mucin and HMOs [17]. These encompass gene clusters (predicted to be) involved in the metabolism of sialic acid, lacto-N-biose, fucose and N-linked glycans. While B. breve is not known to be able to grow to any appreciable density on mucin or HMOs, host-derived mono/oligosaccharides may become available through hydrolytic activities of other bifidobacteria present in the gut (e.g. B. bifidum, thereby allowing B. breve strains to utilize such liberated carbohydrates through cross-feeding activities, see also below) [38–40, 137].
As described above, the occurrence and abundance of bifidobacteria in the human gut is modified during the human life span. In this context, factors that shape the development of the microbiota are still poorly understood, and the mechanisms by which these factors affect gut metabolites have not been extensively investigated. Even less information is available on how bifidobacteria affect metabolic profiles of the human host. It has been shown that HMO-utilizing bifidobacteria cause an increase in gut acetate levels, which is claimed to elicit various beneficial effects on the host, including protection against enteropathogens by improving gut barrier function [48]. Furthermore, a recent publication has highlighted how bifidobacterial dominance during the first stages of life is positively associated with linoleate metabolism, with a high level of trans-10, cis-12-conjugated linoleic acid (t10c12-CLA) detected in blood samples of infants [125].
Bile exerts a crucial role in the physiology of enteric bacteria and in influencing their activities, thus conditioning their functionality and influencing their survival [109]. In this context, gut microorganisms have evolved various genetic strategies to counteract the anti-microbial activity of bile salts [109], including the activity of bile-salt hydrolases (BSHs). BSHs act to de-conjugated glycine and taurine from their respective bile salts, and the corresponding de-conjugated acids can be further metabolized by other gut bacteria [27]. In bifidobacteria, an extensive genetic survey highlighted the occurrence of BSH-encoding genes in bifidobacterial species, in particular those isolated from primates [76, 77, 114, 115], corroborating the importance of this genetic trait for survival. Notably, BSH activity is also considered to play an important beneficial role because of its cholesterol-removing capacity [109]. In this context, the functional role of the bsh gene of B. bifidum PRL2010 in reducing the level of serum cholesterol has recently been studied in a murine model [160].
Cross-feeding activities operated by bifidobacteria
Bifidobacterial communities are considered to establish and maintain themselves in their ecological niche by means of trophic interactions with each other as well as with other members of the gut microbiota, leading to competition for or co-operative sharing of nutrients.
In general, microbe–microbe interactions can either positively or negatively influence the fitness of the affected organisms [93]. Large parts of these interactions depend on either the active or passive release of molecules (e.g. enzymes and metabolites) into the environment [81, 95]. Thanks to cross-feeding strategies, gut bacteria promote an expansion of their carbohydrate acquisition abilities, thereby positively influencing the ecological fitness of the overall gut microbiota. Cross-feeding activities in the gut are generally carried out by primary degraders such as bifidobacteria, which facilitated by partial extracellular hydrolysis of specific complex carbohydrates provide glycans (monosaccharides and/or oligosaccharides) to other microbial gut inhabitants [29]. The successive fermentative metabolism of these carbohydrates generates end-metabolites, such as acetate and lactate, which may act as substrates for secondary degraders like the butyrate-producing colonic bacteria [12, 34, 43, 82, 84].
Bifidobacterial interspecies cross-feeding activities have been observed between two infant-type bifidobacteria, Bifidobacterium bifidum PRL2010 and Bifidobacterium breve UCC2003, when cultivated on sialyl lactose as the unique carbon source [39, 40]. Previous studies have shown metabolic cross-feeding between B. adolescentis and lactate-utilizing, butyrate-producing Firmicutes, in particular Eubacterium hallii and Anaerostipes caccae [13]. These cross-feeding activities are relevant to host health as butyrate is widely regarded as a beneficial SCFA produced by components of the microbiota. In addition, bacterial cross-feeding opportunities as facilitated by members of the colonic microbiota are considered to be pivotal for carbohydrate turn-over in this ecological niche [29].
Recently, the cross-feeding activities of the bifidobacterial strains B. bifidum PRL2010, B. breve 12L, B. adolescentis 22L and B. thermophilum JCM7017 have been evaluated when they were cultivated in various combinations on plant-derived glycans, such as starch and xylan [137]. Co-cultivation assays coupled with transcriptomic and metabolomic analyses revealed that co-occurrence of the above-mentioned bifidobacterial strains causes enhanced metabolic activity of B. bifidum PRL2010, thus indicating that PRL2010 cells benefit from the presence of other bifidobacterial strains.
Cross-feeding phenotypes between bifidobacteria have been further confirmed in in vivo trials involving conventional mice, which were colonized with (combinations of) different infant-type bifidobacterial strains (B. bifidum PRL2010, B. longum subsp. infantis ATCC15697, B. adolescentis 22L and B. breve 12L) [135]. Interestingly, in this study transcriptomic experiments coupled with metagenomic analyses of single, dual or multiple-associations of bifidobacterial strains highlighted cross-feeding behaviour consisting of an expansion of the murine gut glycobiome towards its enzymatic potential related to the breakdown of the plant-derived carbohydrates xylo-oligosaccharides, arabinoxylan and starch, in addition to host-glycan substrates. Furthermore, these analyses revealed distinct ecological responses by the different bifidobacterial strains towards glycans, revealing ‘selfish’ behaviour of B. longum subsp. infantis ATCC15697 when considering its competitive harvesting of carbohydrates. This is in contrast to what was noticed for B. bifidum PRL2010, which actively participates in the extracellular breakdown of host-glycans, thereby releasing simple sugars in its environment that can be utilized by other members of the bifidobacterial community [135].
Thus, bifidobacteria play a key ecological role, acting directly or through cross-feeding strategies, in shaping the gut mammalian microbiome to an enrichment of saccharolytic microbiota. Co-colonization of germ-free mice with Eubacterium rectale, B. longum subsp. longum together with Bacteroides theiotaomicron, caused an alteration of glycan metabolism of Bact. theiotaomicron toward polysaccharides containing mannose [113, 124]. Furthermore, the transcriptomic changes observed upon the co-colonization of B. longum subsp. longum and Bact. thetaiotaomicron in polysaccharide-utilization genes imply a symbiosis between these microorganisms, where each species possesses a complementary set of GH activities, which when combined allow both to participate in a synergistic harvest of xylose and mannose-containing glycans [113, 124].
Similarly, in vivo trials in axenic mice revealed that Bact. thetaiotaomicron and B. longum subsp. infantis induce the same genes during HMO consumption as those that are expressed to harvest host mucin [72]. Notably, Bact. thetaiotaomicron is able to degrade mucin, thereby providing a selective advantage to B. longum subsp. infantis (which on its own is not able to utilize mucins) when bi-associated with Bact. thetaiotaomicron [72].
Despite the ecological importance of the above-mentioned bifidobacterial cross-feeding activities, the current molecular knowledge of what drives such phenomena is still limited to in vitro models based on only a very small number of different taxa. In contrast, when placed in an in vivo context, such activities may underscore the concurrent and interacting metabolism of a substantial number of distinct bacterial species.
Extracellular structures encoded by bifidobacteria as effector molecules for host and microbiota dialogue
To date, only a small number of extracellular molecules produced by bifidobacterial genomes have been considered important for host interaction [107]. Such molecules include capsular polysaccharides as well as fimbriae/pili, which are covering or protruding from the bacterial cell surface, and which are therefore exposed to the external environment (for review see [141]) (Fig. 1). Notably, a large body of information is currently available from pathogenic bacteria or other human gut commensals such as Bacteroides [20, 63], where such external molecules are considered to play pivotal roles in survival, host interaction and gut colonization. Recently, it was shown that members of the B. bifidum species produce appendages known as sortase-dependent pili that modulate the interaction of B. bifidum cells with the human host as well as with other members of the gut microbiota [46, 139, 140]. Notably, publicly available B. bifidum chromosomal sequences have been assessed for the presence of sortase-dependent pili, which allowed the identification of three such pili-specifying loci. Notably, of these three identified loci, only two, named pil2 and pil3, were demonstrated to be genetically intact, whereas the third was shown to be non-functional as a consequence of a frameshift within the coding region of the gene encoding the predicted major pilus subunit. Transcriptome analyses carried out on B. bifidum PRL2010 cells upon colonization of conventional mice, i.e. mice with their intact gut microbiota, as well as upon contact with human cell lines, showed a clear transcriptional up-regulation of the pil2 and pil3 gene clusters. Furthermore, heterologous expression of the pil2- or pil3-associated genes in the non-piliated, Gram-positive host Lactococcus lactis demonstrated that both pilus types stimulate adhesion to human enterocytes through extracellular matrix (ECM) proteins and bacterial aggregation [139, 140]. In addition, competitive carbohydrate binding experiments showed that mannose and fucose act as potential receptors for Pil2 of B. bifidum PRL2010 in a fashion that is reminiscent of that previously described for other enteric bacteria [45], whereas the putative binding partners for Pil3 appear to encompass a larger set of carbohydrates.
Recently, co-aggregation between B. bifidum PRL2010 cells and other human gut microbiota members, i.e., bifidobacteria and lactobacilli, have been observed to be influenced by the production of pili [140]. Such findings suggest an important ecological role of these extracellular structures in the development of intestinal biofilms as well as in promoting cell–cell proximity, perhaps promoting the exchange of genetic material by conjugation [16, 17].
The pan-genome of an infant-type bifidobacterial species such as B. breve has been recently characterized by the sequencing of 13 strains, which revealed a closed structure [17]. Among the genetic repertoire that is shared by all B. breve genomes, is a member of the so-called type IVb or tight-adherence (Tad) pilus family [91, 141, 150]. Similar to the sortase-dependent pili (that are also present in the genomes of B. breve taxon), it has been shown that Tad pili are extracellular structures that are composed of assembled pilin subunits, which are believed to be linked by non-covalent interactions and attached to the membrane lipid bilayer. The latter is in contrast to sortase-dependent pili, which are covalently anchored to the cell wall and their subunit assembly involves the establishment of covalent bonds catalyzed by a specific sortase [118].
A functional characterization of the tad locus has been performed for the B. breve UCC2003 strain, demonstrating that this cluster is crucial for in vivo murine gut colonization [91].
Another extracellular structure that is known to modulate microbe-host interactions in the gut is the capsular exopolysaccharide (EPS) [20, 63]. The involvement of a bifidobacterial EPS in colonization and host interaction has been characterized for B. breve UCC2003 [44]. The chromosome of this particular gut commensal encompasses a gene cluster predicted to specify the production of two different cell surface-associated EPS structures [91]. Notably, site-directed mutagenesis facilitated the generation of an acapsular mutant B. breve UCC2003, which shows a significantly enhanced adaptive immune response compared to the EPS-producing wild type. Furthermore, it was shown that colonization of (wild type) B. breve UCC2003, in contrast to the acapsular mutant, reduced infection levels of the murine pathogen Citrobacter rodentium. These data implicate bifidobacterial EPS in modulating microbe-host cross talk, resulting in host-mediated immune tolerance of the commensal, while it also in an as yet unknown manner provides protection against a murine pathogen [44]. Recently, other studies have confirmed specific immune activity exerted by bifidobacterial EPS [5, 117].
Bifidobacteria have furthermore been shown to produce an interspecies signalling molecule known as the quorum sensing molecule AI-2 [18, 22]. The production of this quorum sensing molecule is dependent on LuxS, an enzyme that forms an essential part of the activated methyl cycle, involved in recycling S-adenosylhomocysteine [155]. Although the LuxS-encoding gene seems to be part of the core genome of the genus Bifidobacterium [76], it has only been characterized in B. breve UCC2003 [22]. Insertional inactivation and complementation experiments indeed demonstrated that a functional luxS gene is necessary for bifidobacterial AI-2 synthesis. Furthermore, these analyses showed that the UCC2003-luxS mutant, compared to the UCC2003 wild type strain, is less effective in iron sequestering and unable to colonize the murine gastrointestinal tract, while this mutant was also shown to confer less protection against Salmonella infection [22].
Other bifidobacterial proteins that are reported to be involved in microbe-host interactions include eukaryotic-type serine–protease inhibitors, also known as serpins, which are produced by various members of the human gut microbiota, including certain bifidobacteria (Fig. 1). Members of the serpin family regulate a wide range of signalling pathways in eukaryotes and some are recognized for their ability to suppress inflammatory responses by inhibiting elastase activity [99]. Genomic surveys involving bifidobacterial chromosome sequences have pointed out the occurrence of genes encoding serpin-like proteins among bifidobacteria, including B. breve, B. longum subsp. longum and B. longum subsp. infantis [132]. Furthermore, functional analyses involving transcriptomics as well as targeted gene inactivation performed in B. breve strains, highlighted that transcription of the serpin-encoding gene is greatly induced when B. breve cultures are exposed to various host-derived proteases, such as pancreatic elastase, human neutrophil elastase, thrombin, papain, kallikrein, trypsin, α-antitrypsin, chymotrypsin and plasmin [6, 132]. This finding is highly relevant since many of these proteases are normally found within the human gut, and thus the presence of a protease inhibitor may provide an ecological advantage to bifidobacteria since serpin activity may protect them against these host proteases.
Other extracellular structures that are considered important for microbe-host interaction are teichoic acids that are positioned on the cell surface either as lipoteichoic acid (LTA), when anchored to the cytoplasmic membrane with a glycolipid anchor, and/or as wall teichoic acid (WTA), when covalently linked to peptidoglycan [156] (Fig. 1). A large body of information currently exists for LTA produced by particular human gut commensals, such as lactobacilli, where their influence is mediated through immune-modulatory properties [51, 80]. In contrast, very little information is available for TA or LTA in bifidobacteria. Recently, in silico analyses involving the genomes of all 48 (sub)species that currently represent the genus Bifidobacterium, revealed the presence of genes responsible for TA biosynthesis, indicating that bifidobacteria contain both WTA as well as LTA [24]. Furthermore, transcriptome analyses of the infant gut commensal B. bifidum PRL2010 showed that transcription of the TA loci is modulated in response to environmental conditions reflecting those of the human gut [24].
Conclusions
During human infant years, the gut microbiota is subjected to rapid changes that ultimately lead to the establishment of a so-called climax [158]. It has been pointed out that the human gut microbiota influences several aspects of host metabolism. Although most of this causality is not yet fully understood at the molecular level, a strong relationship appears to exist between dysbiosis, deviations in gut microbiota composition and susceptibility to disease [92]. Preliminary information suggests this predictive importance of the gut microbiota with regards to gut-related diseases, and particularly how early microbiota composition is linked to long-lasting effects on health and disease [32]. Perturbations or dysbiosis of the early microbiota have been shown to elicit persistent effects, such as altered metabolic processes, brain function and immune-system development [23, 25, 49]. Unfortunately, it is unclear how interventions using probiotics, prebiotics or targeted antibiotics can be directed to manipulate the early microbiota to reduce or abolish risks factors and to increase overall infant health and consequently of adults. In this context, a new player, represented by (bacterio)phages, that is believed to influence microbiota homeostasis has come into the spotlight of scientific attention. Preliminary virome studies that have interrogated infant metagenomic datasets have revealed that phages may be exploited in preventing/treating gut dysbiosis by reducing ‘blooming’ of particular bacterial taxa [70]. Other interventions based on prebiotic compounds such as galactooligosaccharides, which simulate HMOs, produced variable results on bifidobacterial growth among the various taxa of this genus [90, 153]. Additionally, in our efforts to understand infant microbiota functionality, we should not merely restrict ourselves to cataloguing the various microbial groups found in the infant gut, but we should also attempt to isolate these bacteria, including the currently “unculturable” microorganisms. Such efforts may facilitate the development of novel therapeutic approaches based on targeted modification of the microbiota in infants in order to suppress the risks of disease in adults.
Notably, although bifidobacteria are present at low relative abundance in the adult gut microbiota, they may still dramatically influence the overall microbial community through their metabolic and physiological activities. In this context, it has previously been shown that bifidobacteria contribute to the human intestinal adult glycobiome [77]. Despite the observed and investigated functional contributions of bifidobacteria to the gut microbiota, we still know surprisingly little about these gut commensals [129]. In this context, it is worth mentioning that their real contribution to the biodiversity of the human gut microbiota may be underestimated by the limitations of the metagenomics approaches currently used to assess the detailed composition of complex gut communities. In fact, recent findings have highlighted that erroneous conclusions about the presence/absence as well as relative proportion of bifidobacteria are due to bias in the experimental procedures applied (e.g., sample storage, DNA extraction and PCR steps) [75, 138]. It is therefore crucial that optimized protocols, both facilitating metagenomics analyses as well as culturomics approaches, will be developed to address the true contributions of bifidobacteria in health and disease of its human host.
Acknowledgements
This work was funded by the EU Joint Programming Initiative—a Healthy Diet for a Healthy Life (JPI HDHL, http://www.healthydietforhealthylife.eu/) and the MIUR to MV. We thank GenProbio srl for financial support of the Laboratory of Probiogenomics. LM is supported by Fondazione Cariparma, Parma, Italy. SD is supported by Fondazione Caritro, Trento, Italy. DvS is a member of The APC Microbiome Institute funded by Science Foundation Ireland (SFI), through the Irish Government’s National Development Plan (Grant number SFI/12/RC/2273).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no competing interests.
References
- 1.Adlerberth I, Wold AE. Establishment of the gut microbiota in Western infants. Acta Paediatr. 2009;98:229–238. doi: 10.1111/j.1651-2227.2008.01060.x. [DOI] [PubMed] [Google Scholar]
- 2.Agans R, Rigsbee L, Kenche H, Michail S, Khamis HJ, Paliy O. Distal gut microbiota of adolescent children is different from that of adults. FEMS Microbiol Ecol. 2011;77:404–412. doi: 10.1111/j.1574-6941.2011.01120.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ajslev TA, Andersen CS, Gamborg M, Sorensen TI, Jess T. Childhood overweight after establishment of the gut microbiota: the role of delivery mode, pre-pregnancy weight and early administration of antibiotics. Int J Obes. 2011;35:522–529. doi: 10.1038/ijo.2011.27. [DOI] [PubMed] [Google Scholar]
- 4.Albrecht S, Schols HA, van den Heuvel EG, Voragen AG, Gruppen H. Occurrence of oligosaccharides in feces of breast-fed babies in their first six months of life and the corresponding breast milk. Carbohydr Res. 2011;346:2540–2550. doi: 10.1016/j.carres.2011.08.009. [DOI] [PubMed] [Google Scholar]
- 5.Altmann F, Kosma P, O’Callaghan A, Leahy S, Bottacini F, Molloy E, Plattner S, Schiavi E, Gleinser M, Groeger D, et al. Genome analysis and characterisation of the exopolysaccharide produced by Bifidobacterium longum subsp. longum 35624. PLoS One. 2016;11:e0162983. doi: 10.1371/journal.pone.0162983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alvarez-Martin P, O’Connell Motherway M, Turroni F, Foroni E, Ventura M, van Sinderen D. A two-component regulatory system controls autoregulated serpin expression in Bifidobacterium breve UCC2003. Appl Environ Microbiol. 2012;78:7032–7041. doi: 10.1128/AEM.01776-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arboleya S, Ang L, Margolles A, Yiyuan L, Dongya Z, Liang X, Solis G, Fernandez N, de Los Reyes-Gavilan CG, Gueimonde M. Deep 16S rRNA metagenomics and quantitative PCR analyses of the premature infant fecal microbiota. Anaerobe. 2012;18:378–380. doi: 10.1016/j.anaerobe.2012.04.013. [DOI] [PubMed] [Google Scholar]
- 8.Arboleya S, Sanchez B, Milani C, Duranti S, Solis G, Fernandez N, de los Reyes-Gavilan CG, Ventura M, Margolles A, Gueimonde M. Intestinal microbiota development in preterm neonates and effect of perinatal antibiotics. J Pediatr. 2015;166:538–544. doi: 10.1016/j.jpeds.2014.09.041. [DOI] [PubMed] [Google Scholar]
- 9.Ashida H, Miyake A, Kiyohara M, Wada J, Yoshida E, Kumagai H, Katayama T, Yamamoto K. Two distinct alpha-l-fucosidases from Bifidobacterium bifidum are essential for the utilization of fucosylated milk oligosaccharides and glycoconjugates. Glycobiology. 2009;19:1010–1017. doi: 10.1093/glycob/cwp082. [DOI] [PubMed] [Google Scholar]
- 10.Backhed F, Roswall J, Peng Y, Feng Q, Jia H, Kovatcheva-Datchary P, Li Y, Xia Y, Xie H, Zhong H, et al. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe. 2015;17:852. doi: 10.1016/j.chom.2015.05.012. [DOI] [PubMed] [Google Scholar]
- 11.Bailey MT, Lubach GR, Coe CL. Prenatal stress alters bacterial colonization of the gut in infant monkeys. J Pediatr Gastroenterol Nutr. 2004;38:414–421. doi: 10.1097/00005176-200404000-00009. [DOI] [PubMed] [Google Scholar]
- 12.Barcenilla A, Pryde SE, Martin JC, Duncan SH, Stewart CS, Henderson C, Flint HJ. Phylogenetic relationships of butyrate-producing bacteria from the human gut. Appl Environ Microbiol. 2000;66:1654–1661. doi: 10.1128/AEM.66.4.1654-1661.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Belenguer A, Duncan SH, Calder AG, Holtrop G, Louis P, Lobley GE, Flint HJ. Two routes of metabolic cross-feeding between Bifidobacterium adolescentis and butyrate-producing anaerobes from the human gut. Appl Environ Microbiol. 2006;72:3593–3599. doi: 10.1128/AEM.72.5.3593-3599.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bienenstock J, Buck RH, Linke H, Forsythe P, Stanisz AM, Kunze WA. Fucosylated but not sialylated milk oligosaccharides diminish colon motor contractions. PLoS One. 2013;8:e76236. doi: 10.1371/journal.pone.0076236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blustein J, Attina T, Liu M, Ryan AM, Cox LM, Blaser MJ, Trasande L. Association of caesarean delivery with child adiposity from age 6 weeks to 15 years. Int J Obes. 2013;37:900–906. doi: 10.1038/ijo.2013.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bottacini F, O’Connell Motherway M, Casey E, McDonnell B, Mahony J, Ventura M, van Sinderen D. Discovery of a conjugative megaplasmid in Bifidobacterium breve . Appl Environ Microbiol. 2015;81:166–176. doi: 10.1128/AEM.02871-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bottacini F, O’Connell Motherway M, Kuczynski J, O’Connell KJ, Serafini F, Duranti S, Milani C, Turroni F, Lugli GA, Zomer A, et al. Comparative genomics of the Bifidobacterium breve taxon. BMC Genom. 2014;15:170. doi: 10.1186/1471-2164-15-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bottacini F, Ventura M, van Sinderen D, O’Connell Motherway M. Diversity, ecology and intestinal function of bifidobacteria. Microb Cell Fact. 2014;13(Suppl 1):S4. doi: 10.1186/1475-2859-13-S1-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Castanys-Munoz E, Martin MJ, Prieto PA. 2′-Fucosyllactose: an abundant, genetically determined soluble glycan present in human milk. Nutr Rev. 2013;71:773–789. doi: 10.1111/nure.12079. [DOI] [PubMed] [Google Scholar]
- 20.Cerdeno-Tarraga AM, Patrick S, Crossman LC, Blakely G, Abratt V, Lennard N, Poxton I, Duerden B, Harris B, Quail MA, et al. Extensive DNA inversions in the B. fragilis genome control variable gene expression. Science. 2005;307:1463–1465. doi: 10.1126/science.1107008. [DOI] [PubMed] [Google Scholar]
- 21.Chaplin AV, Efimov BA, Smeianov VV, Kafarskaia LI, Pikina AP, Shkoporov AN. Intraspecies genomic diversity and long-term persistence of Bifidobacterium longum . PLoS One. 2015;10:e0135658. doi: 10.1371/journal.pone.0135658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Christiaen SE, O’Connell Motherway M, Bottacini F, Lanigan N, Casey PG, Huys G, Nelis HJ, van Sinderen D, Coenye T. Autoinducer-2 plays a crucial role in gut colonization and probiotic functionality of Bifidobacterium breve UCC2003. PLoS One. 2014;9:e98111. doi: 10.1371/journal.pone.0098111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clarke G, O’Mahony SM, Dinan TG, Cryan JF. Priming for health: gut microbiota acquired in early life regulates physiology, brain and behaviour. Acta Paediatr. 2014;103:812–819. doi: 10.1111/apa.12674. [DOI] [PubMed] [Google Scholar]
- 24.Colagiorgi A, Turroni F, Mancabelli L, Serafini F, Secchi A, van Sinderen D, Ventura M. Insights into teichoic acid biosynthesis by Bifidobacterium bifidum PRL2010. FEMS Microbiol Lett. 2015;362:fnv141. doi: 10.1093/femsle/fnv141. [DOI] [PubMed] [Google Scholar]
- 25.Cox LM, Yamanishi S, Sohn J, Alekseyenko AV, Leung JM, Cho I, Kim SG, Li H, Gao Z, Mahana D, et al. Altering the intestinal microbiota during a critical developmental window has lasting metabolic consequences. Cell. 2014;158:705–721. doi: 10.1016/j.cell.2014.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.D’Elia JN, Salyers AA. Effect of regulatory protein levels on utilization of starch by Bacteroides thetaiotaomicron . J Bacteriol. 1996;178:7180–7186. doi: 10.1128/jb.178.24.7180-7186.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.De Boever P, Wouters R, Verschaeve L, Berckmans P, Schoeters G, Verstraete W. Protective effect of the bile salt hydrolase-active Lactobacillus reuteri against bile salt cytotoxicity. Appl Microbiol Biotechnol. 2000;53:709–714. doi: 10.1007/s002530000330. [DOI] [PubMed] [Google Scholar]
- 28.De Leoz ML, Kalanetra KM, Bokulich NA, Strum JS, Underwood MA, German JB, Mills DA, Lebrilla CB. Human milk glycomics and gut microbial genomics in infant feces show a correlation between human milk oligosaccharides and gut microbiota: a proof-of-concept study. J Proteome Res. 2015;14:491–502. doi: 10.1021/pr500759e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.De Vuyst L, Leroy F. Cross-feeding between bifidobacteria and butyrate-producing colon bacteria explains bifidobacterial competitiveness, butyrate production, and gas production. Int J Food Microbiol. 2011;149:73–80. doi: 10.1016/j.ijfoodmicro.2011.03.003. [DOI] [PubMed] [Google Scholar]
- 30.Dethlefsen L, Huse S, Sogin ML, Relman DA. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 2008;6:e280. doi: 10.1371/journal.pbio.0060280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.DiGiulio DB, Romero R, Amogan HP, Kusanovic JP, Bik EM, Gotsch F, Kim CJ, Erez O, Edwin S, Relman DA. Microbial prevalence, diversity and abundance in amniotic fluid during preterm labor: a molecular and culture-based investigation. PLoS One. 2008;3:e3056. doi: 10.1371/journal.pone.0003056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dogra S, Sakwinska O, Soh SE, Ngom-Bru C, Bruck WM, Berger B, Brussow H, Lee YS, Yap F, Chong YS, et al. Dynamics of infant gut microbiota are influenced by delivery mode and gestational duration and are associated with subsequent adiposity. mBio. 2015;6:1–9. doi: 10.1128/mBio.02419-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N, Knight R. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci USA. 2010;107:11971–11975. doi: 10.1073/pnas.1002601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Duncan SH, Flint HJ. Proposal of a neotype strain (A1-86) for Eubacterium rectale. Request for an opinion. Int J Syst Evol Microbiol. 2008;58:1735–1736. doi: 10.1099/ijs.0.2008/004580-0. [DOI] [PubMed] [Google Scholar]
- 35.Duranti S, Milani C, Lugli GA, Turroni F, Mancabelli L, Sanchez B, Ferrario C, Viappiani A, Mangifesta M, Mancino W, et al. Insights from genomes of representatives of the human gut commensal Bifidobacterium bifidum . Environ Microbiol. 2015;17(7):2515–2531. doi: 10.1111/1462-2920.12743. [DOI] [PubMed] [Google Scholar]
- 36.Duranti S, Turroni F, Lugli GA, Milani C, Viappiani A, Mangifesta M, Gioiosa L, Palanza P, van Sinderen D, Ventura M. Genomic characterization and transcriptional studies of the starch-utilizing strain Bifidobacterium adolescentis 22L. Appl Environ Microbiol. 2014;80:6080–6090. doi: 10.1128/AEM.01993-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Egan M, Jiang H, O’Connell Motherway M, Oscarson S, van Sinderen D. Glycosulfatase-encoding gene cluster in Bifidobacterium breve UCC2003. Appl Environ Microbiol. 2016;82:6611–6623. doi: 10.1128/AEM.02022-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Egan M, Motherway MO, Kilcoyne M, Kane M, Joshi L, Ventura M, van Sinderen D. Cross-feeding by Bifidobacterium breve UCC2003 during co-cultivation with Bifidobacterium bifidum PRL2010 in a mucin-based medium. BMC Microbiol. 2014;14:282. doi: 10.1186/s12866-014-0282-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Egan M, O’Connell Motherway M, Ventura M, van Sinderen D. Metabolism of sialic acid by Bifidobacterium breve UCC2003. Appl Environ Microbiol. 2014;80:4414–4426. doi: 10.1128/AEM.01114-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fallani M, Amarri S, Uusijarvi A, Adam R, Khanna S, Aguilera M, Gil A, Vieites JM, Norin E, Young D, et al. Determinants of the human infant intestinal microbiota after the introduction of first complementary foods in infant samples from five European centres. Microbiology. 2011;157:1385–1392. doi: 10.1099/mic.0.042143-0. [DOI] [PubMed] [Google Scholar]
- 42.Fallani M, Young D, Scott J, Norin E, Amarri S, Adam R, Aguilera M, Khanna S, Gil A, Edwards CA, et al. Intestinal microbiota of 6-week-old infants across Europe: geographic influence beyond delivery mode, breast-feeding, and antibiotics. J Pediatr Gastroenterol Nutr. 2010;51:77–84. doi: 10.1097/MPG.0b013e3181d1b11e. [DOI] [PubMed] [Google Scholar]
- 43.Falony G, Calmeyn T, Leroy F, De Vuyst L. Coculture fermentations of Bifidobacterium species and Bacteroides thetaiotaomicron reveal a mechanistic insight into the prebiotic effect of inulin-type fructans. Appl Environ Microbiol. 2009;75:2312–2319. doi: 10.1128/AEM.02649-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fanning S, Hall LJ, Cronin M, Zomer A, MacSharry J, Goulding D, Motherway MO, Shanahan F, Nally K, Dougan G, et al. Bifidobacterial surface-exopolysaccharide facilitates commensal-host interaction through immune modulation and pathogen protection. Proc Natl Acad Sci USA. 2012;109:2108–2113. doi: 10.1073/pnas.1115621109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Farfan MJ, Cantero L, Vidal R, Botkin DJ, Torres AG. Long polar fimbriae of enterohemorrhagic Escherichia coli O157:H7 bind to extracellular matrix proteins. Infect Immun. 2011;79:3744–3750. doi: 10.1128/IAI.05317-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Foroni E, Serafini F, Amidani D, Turroni F, He F, Bottacini F, O’Connell Motherway M, Viappiani A, Zhang Z, Rivetti C, et al. Genetic analysis and morphological identification of pilus-like structures in members of the genus Bifidobacterium. Microb Cell Fact. 2011;10(Suppl 1):S16. doi: 10.1186/1475-2859-10-S1-S16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fujita K, Oura F, Nagamine N, Katayama T, Hiratake J, Sakata K, Kumagai H, Yamamoto K. Identification and molecular cloning of a novel glycoside hydrolase family of core 1 type O-glycan-specific endo-alpha-N-acetylgalactosaminidase from Bifidobacterium longum . J Biol Chem. 2005;280:37415–37422. doi: 10.1074/jbc.M506874200. [DOI] [PubMed] [Google Scholar]
- 48.Fukuda S, Toh H, Hase K, Oshima K, Nakanishi Y, Yoshimura K, Tobe T, Clarke JM, Topping DL, Suzuki T, et al. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature. 2011;469:543–547. doi: 10.1038/nature09646. [DOI] [PubMed] [Google Scholar]
- 49.Funkhouser LJ, Bordenstein SR. Mom knows best: the universality of maternal microbial transmission. PLoS Biol. 2013;11:e1001631. doi: 10.1371/journal.pbio.1001631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Garrido D, Ruiz-Moyano S, Lemay DG, Sela DA, German JB, Mills DA. Comparative transcriptomics reveals key differences in the response to milk oligosaccharides of infant gut-associated bifidobacteria. Sci Rep. 2015;5:13517. doi: 10.1038/srep13517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Grangette C, Nutten S, Palumbo E, Morath S, Hermann C, Dewulf J, Pot B, Hartung T, Hols P, Mercenier A. Enhanced antiinflammatory capacity of a Lactobacillus plantarum mutant synthesizing modified teichoic acids. Proc Natl Acad Sci USA. 2005;102:10321–10326. doi: 10.1073/pnas.0504084102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hinde K, German JB. Food in an evolutionary context: insights from mother’s milk. J Sci Food Agric. 2012;92:2219–2223. doi: 10.1002/jsfa.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hinde K, Milligan LA. Primate milk: proximate mechanisms and ultimate perspectives. Evol Anthropol. 2011;20:9–23. doi: 10.1002/evan.20289. [DOI] [PubMed] [Google Scholar]
- 54.Hollister EB, Riehle K, Luna RA, Weidler EM, Rubio-Gonzales M, Mistretta TA, Raza S, Doddapaneni HV, Metcalf GA, Muzny DM, et al. Structure and function of the healthy pre-adolescent pediatric gut microbiome. Microbiome. 2015;3:36. doi: 10.1186/s40168-015-0101-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Huh SY, Rifas-Shiman SL, Zera CA, Edwards JW, Oken E, Weiss ST, Gillman MW. Delivery by caesarean section and risk of obesity in preschool age children: a prospective cohort study. Arch Dis Child. 2012;97:610–616. doi: 10.1136/archdischild-2011-301141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Huurre A, Kalliomaki M, Rautava S, Rinne M, Salminen S, Isolauri E. Mode of delivery—effects on gut microbiota and humoral immunity. Neonatology. 2008;93:236–240. doi: 10.1159/000111102. [DOI] [PubMed] [Google Scholar]
- 57.James K, Motherway MO, Bottacini F, van Sinderen D. Bifidobacterium breve UCC2003 metabolises the human milk oligosaccharides lacto-N-tetraose and lacto-N-neo-tetraose through overlapping, yet distinct pathways. Sci Rep. 2016;6:38560. doi: 10.1038/srep38560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jimenez E, Delgado S, Fernandez L, Garcia N, Albujar M, Gomez A, Rodriguez JM. Assessment of the bacterial diversity of human colostrum and screening of staphylococcal and enterococcal populations for potential virulence factors. Res Microbiol. 2008;159:595–601. doi: 10.1016/j.resmic.2008.09.001. [DOI] [PubMed] [Google Scholar]
- 59.Jimenez E, Marin ML, Martin R, Odriozola JM, Olivares M, Xaus J, Fernandez L, Rodriguez JM. Is meconium from healthy newborns actually sterile? Res Microbiol. 2008;159:187–193. doi: 10.1016/j.resmic.2007.12.007. [DOI] [PubMed] [Google Scholar]
- 60.Karav S, Parc AL, de Moura Bell JM, Rouquie C, Mills DA, Barile D, Block DE. Kinetic characterization of a novel endo-beta-N-acetylglucosaminidase on concentrated bovine colostrum whey to release bioactive glycans. Enzyme Microb Technol. 2015;77:46–53. doi: 10.1016/j.enzmictec.2015.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kelly ED, Bottacini F, O’Callaghan J, Motherway MO, O’Connell KJ, Stanton C, van Sinderen D. Glycoside hydrolase family 13 alpha-glucosidases encoded by Bifidobacterium breve UCC2003; A comparative analysis of function, structure and phylogeny. Int J Food Microbiol. 2016;224:55–65. doi: 10.1016/j.ijfoodmicro.2016.02.014. [DOI] [PubMed] [Google Scholar]
- 62.Koenig JE, Spor A, Scalfone N, Fricker AD, Stombaugh J, Knight R, Angenent LT, Ley RE. Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci USA. 2011;108(Suppl 1):4578–4585. doi: 10.1073/pnas.1000081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kuwahara T, Yamashita A, Hirakawa H, Nakayama H, Toh H, Okada N, Kuhara S, Hattori M, Hayashi T, Ohnishi Y. Genomic analysis of Bacteroides fragilis reveals extensive DNA inversions regulating cell surface adaptation. Proc Natl Acad Sci USA. 2004;101:14919–14924. doi: 10.1073/pnas.0404172101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lahtinen SJ, Boyle RJ, Kivivuori S, Oppedisano F, Smith KR, Robins-Browne R, Salminen SJ, Tang ML. Prenatal probiotic administration can influence Bifidobacterium microbiota development in infants at high risk of allergy. J Allergy Clin Immunol. 2009;123:499–501. doi: 10.1016/j.jaci.2008.11.034. [DOI] [PubMed] [Google Scholar]
- 65.Lamendella R, Santo Domingo JW, Kelty C, Oerther DB. Bifidobacteria in feces and environmental waters. Appl Environ Microbiol. 2008;74:575–584. doi: 10.1128/AEM.01221-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lee DW, Selamoglu N, Lanciano P, Cooley JW, Forquer I, Kramer DM, Daldal F. Loss of a conserved tyrosine residue of cytochrome b induces reactive oxygen species production by cytochrome bc1. J Biol Chem. 2011;286:18139–18148. doi: 10.1074/jbc.M110.214460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lee JH, O’Sullivan DJ. Genomic insights into bifidobacteria. Microbiol Mol Biol Rev. 2010;74:378–416. doi: 10.1128/MMBR.00004-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lewis ZT, Totten SM, Smilowitz JT, Popovic M, Parker E, Lemay DG, Van Tassell ML, Miller MJ, Jin YS, German JB, et al. Maternal fucosyltransferase 2 status affects the gut bifidobacterial communities of breastfed infants. Microbiome. 2015;3:13. doi: 10.1186/s40168-015-0071-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444:1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 70.Lugli GA, Milani C, Turroni F, Tremblay D, Ferrario C, Mancabelli L, Duranti S, Ward DV, Ossiprandi MC, Moineau S, et al. Prophages of the genus Bifidobacterium as modulating agents of the infant gut microbiota. Environ Microbiol. 2015;18(7):2196–2213. doi: 10.1111/1462-2920.13154. [DOI] [PubMed] [Google Scholar]
- 71.Madan JC, Salari RC, Saxena D, Davidson L, O’Toole GA, Moore JH, Sogin ML, Foster JA, Edwards WH, Palumbo P, et al. Gut microbial colonisation in premature neonates predicts neonatal sepsis. Arch Dis Child Fetal Neonatal Ed. 2012;97:F456–F462. doi: 10.1136/fetalneonatal-2011-301373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Marcobal A, Barboza M, Sonnenburg ED, Pudlo N, Martens EC, Desai P, Lebrilla CB, Weimer BC, Mills DA, German JB, et al. Bacteroides in the infant gut consume milk oligosaccharides via mucus-utilization pathways. Cell Host Microbe. 2011;10:507–514. doi: 10.1016/j.chom.2011.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Matamoros S, Gras-Leguen C, Le Vacon F, Potel G, de La Cochetiere MF. Development of intestinal microbiota in infants and its impact on health. Trends Microbiol. 2013;21:167–173. doi: 10.1016/j.tim.2012.12.001. [DOI] [PubMed] [Google Scholar]
- 74.Matsuki T, Yahagi K, Mori H, Matsumoto H, Hara T, Tajima S, Ogawa E, Kodama H, Yamamoto K, Yamada T, et al. A key genetic factor for fucosyllactose utilization affects infant gut microbiota development. Nat Commun. 2016;7:11939. doi: 10.1038/ncomms11939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Milani C, Hevia A, Foroni E, Duranti S, Turroni F, Lugli GA, Sanchez B, Martin R, Gueimonde M, van Sinderen D, et al. Assessing the fecal microbiota: an optimized ion torrent 16S rRNA gene-based analysis protocol. PLoS One. 2013;8:e68739. doi: 10.1371/journal.pone.0068739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Milani C, Lugli GA, Duranti S, Turroni F, Bottacini F, Mangifesta M, Sanchez B, Viappiani A, Mancabelli L, Taminiau B, et al. Genomic encyclopedia of type strains of the genus Bifidobacterium. Appl Environ Microbiol. 2014;80:6290–6302. doi: 10.1128/AEM.02308-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Milani C, Lugli GA, Duranti S, Turroni F, Mancabelli L, Ferrario C, Mangifesta M, Hevia A, Viappiani A, Scholz M, et al. Bifidobacteria exhibit social behavior through carbohydrate resource sharing in the gut. Sci Rep. 2015;5:15782. doi: 10.1038/srep15782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Milani C, Mancabelli L, Lugli GA, Duranti S, Turroni F, Ferrario C, Mangifesta M, Viappiani A, Ferretti P, Gorfer V, et al. Exploring vertical transmission of bifidobacteria from mother to child. Appl Environ Microbiol. 2015;81(20):7078–7087. doi: 10.1128/AEM.02037-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Milani C, Turroni F, Duranti S, Lugli GA, Mancabelli L, Ferrario C, van Sinderen D, Ventura M. Genomics of the genus bifidobacterium reveals species-specific adaptation to the glycan-rich gut environment. Appl Environ Microbiol. 2016;82:980–991. doi: 10.1128/AEM.03500-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mohamadzadeh M, Pfeiler EA, Brown JB, Zadeh M, Gramarossa M, Managlia E, Bere P, Sarraj B, Khan MW, Pakanati KC, et al. Regulation of induced colonic inflammation by Lactobacillus acidophilus deficient in lipoteichoic acid. Proc Natl Acad Sci USA. 2011;108(Suppl 1):4623–4630. doi: 10.1073/pnas.1005066107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Morris BE, Henneberger R, Huber H, Moissl-Eichinger C. Microbial syntrophy: interaction for the common good. FEMS Microbiol Rev. 2013;37:384–406. doi: 10.1111/1574-6976.12019. [DOI] [PubMed] [Google Scholar]
- 82.Morrison DJ, Mackay WG, Edwards CA, Preston T, Dodson B, Weaver LT. Butyrate production from oligofructose fermentation by the human faecal flora: what is the contribution of extracellular acetate and lactate? Br J Nutr. 2006;96:570–577. [PubMed] [Google Scholar]
- 83.Mueller S, Saunier K, Hanisch C, Norin E, Alm L, Midtvedt T, Cresci A, Silvi S, Orpianesi C, Verdenelli MC, et al. Differences in fecal microbiota in different European study populations in relation to age, gender, and country: a cross-sectional study. Appl Environ Microbiol. 2006;72:1027–1033. doi: 10.1128/AEM.72.2.1027-1033.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Munoz-Tamayo R, Laroche B, Walter E, Dore J, Duncan SH, Flint HJ, Leclerc M. Kinetic modelling of lactate utilization and butyrate production by key human colonic bacterial species. FEMS Microbiol Ecol. 2011;76:615–624. doi: 10.1111/j.1574-6941.2011.01085.x. [DOI] [PubMed] [Google Scholar]
- 85.Murphy R, Stewart AW, Braithwaite I, Beasley R, Hancox RJ, Mitchell EA, IPTS Group Antibiotic treatment during infancy and increased body mass index in boys: an international cross-sectional study. Int J Obes. 2014;38:1115–1119. doi: 10.1038/ijo.2013.218. [DOI] [PubMed] [Google Scholar]
- 86.Nishimoto M, Kitaoka M. Identification of N-acetylhexosamine 1-kinase in the complete lacto-N-biose I/galacto-N-biose metabolic pathway in Bifidobacterium longum . Appl Environ Microbiol. 2007;73:6444–6449. doi: 10.1128/AEM.01425-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Nuriel-Ohayon M, Neuman H, Koren O. Microbial changes during pregnancy, birth, and infancy. Front Microbiol. 2016;7:1031. doi: 10.3389/fmicb.2016.01031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.O’Callaghan A, van Sinderen D. Bifidobacteria and their role as members of the human gut microbiota. Front Microbiol. 2016;7:925. doi: 10.3389/fmicb.2016.00925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.O’Connell Motherway M, Fitzgerald GF, van Sinderen D. Metabolism of a plant derived galactose-containing polysaccharide by Bifidobacterium breve UCC2003. Microb Biotechnol. 2011;4:403–416. doi: 10.1111/j.1751-7915.2010.00218.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.O’Connell Motherway M, Kinsella M, Fitzgerald GF, van Sinderen D. Transcriptional and functional characterization of genetic elements involved in galacto-oligosaccharide utilization by Bifidobacterium breve UCC2003. Microb Biotechnol. 2013;6:67–79. doi: 10.1111/1751-7915.12011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.O’Connell Motherway M, Zomer A, Leahy SC, Reunanen J, Bottacini F, Claesson MJ, O’Brien F, Flynn K, Casey PG, Munoz JA, et al. Functional genome analysis of Bifidobacterium breve UCC2003 reveals type IVb tight adherence (Tad) pili as an essential and conserved host-colonization factor. Proc Natl Acad Sci USA. 2011;108:11217–11222. doi: 10.1073/pnas.1105380108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ottman N, Smidt H, de Vos WM, Belzer C. The function of our microbiota: who is out there and what do they do? Front Cell Infect Microbiol. 2012;2:104. doi: 10.3389/fcimb.2012.00104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Pande S, Shitut S, Freund L, Westermann M, Bertels F, Colesie C, Bischofs IB, Kost C. Metabolic cross-feeding via intercellular nanotubes among bacteria. Nat Commun. 2015;6:6238. doi: 10.1038/ncomms7238. [DOI] [PubMed] [Google Scholar]
- 94.Penders J, Thijs C, Vink C, Stelma FF, Snijders B, Kummeling I, van den Brandt PA, Stobberingh EE. Factors influencing the composition of the intestinal microbiota in early infancy. Pediatrics. 2006;118:511–521. doi: 10.1542/peds.2005-2824. [DOI] [PubMed] [Google Scholar]
- 95.Phelan VV, Liu WT, Pogliano K, Dorrestein PC. Microbial metabolic exchange—the chemotype-to-phenotype link. Nat Chem Biol. 2012;8:26–35. doi: 10.1038/nchembio.739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Pokusaeva K, Fitzgerald GF, van Sinderen D. Carbohydrate metabolism in Bifidobacteria. Genes Nutr. 2011;6:285–306. doi: 10.1007/s12263-010-0206-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pokusaeva K, Neves AR, Zomer A, O’Connell-Motherway M, MacSharry J, Curley P, Fitzgerald GF, van Sinderen D. Ribose utilization by the human commensal Bifidobacterium breve UCC2003. Microb Biotechnol. 2010;3:311–323. doi: 10.1111/j.1751-7915.2009.00152.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Pokusaeva K, O’Connell-Motherway M, Zomer A, Fitzgerald GF, van Sinderen D. Characterization of two novel alpha-glucosidases from Bifidobacterium breve UCC2003. Appl Environ Microbiol. 2009;75:1135–1143. doi: 10.1128/AEM.02391-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Potempa J, Korzus E, Travis J. The serpin superfamily of proteinase inhibitors: structure, function, and regulation. J Biol Chem. 1994;269:15957–15960. [PubMed] [Google Scholar]
- 100.Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Rajilic-Stojanovic M, de Vos WM. The first 1000 cultured species of the human gastrointestinal microbiota. FEMS Microbiol Rev. 2014;38:996–1047. doi: 10.1111/1574-6976.12075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Rajilic-Stojanovic M, Heilig HG, Molenaar D, Kajander K, Surakka A, Smidt H, de Vos WM. Development and application of the human intestinal tract chip, a phylogenetic microarray: analysis of universally conserved phylotypes in the abundant microbiota of young and elderly adults. Environ Microbiol. 2009;11:1736–1751. doi: 10.1111/j.1462-2920.2009.01900.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Rinne M, Kalliomaki M, Arvilommi H, Salminen S, Isolauri E. Effect of probiotics and breastfeeding on the bifidobacterium and lactobacillus/enterococcus microbiota and humoral immune responses. J Pediatr. 2005;147:186–191. doi: 10.1016/j.jpeds.2005.03.053. [DOI] [PubMed] [Google Scholar]
- 104.Roger LC, Costabile A, Holland DT, Hoyles L, McCartney AL. Examination of faecal Bifidobacterium populations in breast- and formula-fed infants during the first 18 months of life. Microbiology. 2010;156:3329–3341. doi: 10.1099/mic.0.043224-0. [DOI] [PubMed] [Google Scholar]
- 105.Ruas-Madiedo P, Gueimonde M, Arigoni F, de los Reyes-Gavilan CG, Margolles A. Bile affects the synthesis of exopolysaccharides by Bifidobacterium animalis. Appl Environ Microbiol. 2009;75:1204–1207. doi: 10.1128/AEM.00908-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ruas-Madiedo P, Gueimonde M, Fernandez-Garcia M, de los Reyes-Gavilan CG, Margolles A. Mucin degradation by Bifidobacterium strains isolated from the human intestinal microbiota. Appl Environ Microbiol. 2008;74:1936–1940. doi: 10.1128/AEM.02509-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ruiz L, Delgado S, Ruas-Madiedo P, Margolles A, Sanchez B. Proteinaceous molecules mediating bifidobacterium-host interactions. Front Microbiol. 2016;7:1193. doi: 10.3389/fmicb.2016.01193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ruiz L, Gueimonde M, Coute Y, Salminen S, Sanchez JC, de los Reyes-Gavilan CG, Margolles A. Evaluation of the ability of Bifidobacterium longum to metabolize human intestinal mucus. FEMS Microbiol Lett. 2011;314:125–130. doi: 10.1111/j.1574-6968.2010.02159.x. [DOI] [PubMed] [Google Scholar]
- 109.Ruiz L, Margolles A, Sanchez B. Bile resistance mechanisms in Lactobacillus and Bifidobacterium. Front Microbiol. 2013;4:396. doi: 10.3389/fmicb.2013.00396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ryan SM, Fitzgerald GF, van Sinderen D. Screening for and identification of starch-, amylopectin-, and pullulan-degrading activities in bifidobacterial strains. Appl Environ Microbiol. 2006;72:5289–5296. doi: 10.1128/AEM.00257-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Sabbioni A, Ferrario C, Milani C, Mancabelli L, Riccardi E, Di Ianni F, Beretti V, Superchi P, Ossiprandi MC. Modulation of the bifidobacterial communities of the dog microbiota by zeolite. Front Microbiol. 2016;7:1491. doi: 10.3389/fmicb.2016.01491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Salminen S, Gibson GR, McCartney AL, Isolauri E. Influence of mode of delivery on gut microbiota composition in seven year old children. Gut. 2004;53:1388–1389. doi: 10.1136/gut.2004.041640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Samuel BS, Gordon JI. A humanized gnotobiotic mouse model of host-archaeal-bacterial mutualism. Proc Natl Acad Sci USA. 2006;103:10011–10016. doi: 10.1073/pnas.0602187103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Sanchez B, Champomier-Verges MC, Anglade P, Baraige F, de Los Reyes-Gavilan CG, Margolles A, Zagorec M. Proteomic analysis of global changes in protein expression during bile salt exposure of Bifidobacterium longum NCIMB 8809. J Bacteriol. 2005;187:5799–5808. doi: 10.1128/JB.187.16.5799-5808.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Sanchez B, Champomier-Verges MC, Stuer-Lauridsen B, Ruas-Madiedo P, Anglade P, Baraige F, de los Reyes-Gavilan CG, Johansen E, Zagorec M, Margolles A. Adaptation and response of Bifidobacterium animalis subsp. lactis to bile: a proteomic and physiological approach. Appl Environ Microbiol. 2007;73:6757–6767. doi: 10.1128/AEM.00637-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Satokari R, Gronroos T, Laitinen K, Salminen S, Isolauri E. Bifidobacterium and Lactobacillus DNA in the human placenta. Lett Appl Microbiol. 2009;48:8–12. doi: 10.1111/j.1472-765X.2008.02475.x. [DOI] [PubMed] [Google Scholar]
- 117.Schiavi E, Gleinser M, Molloy E, Groeger D, Frei R, Ferstl R, Rodriguez-Perez N, Ziegler M, Grant R, Moriarty TF, et al. The surface-associated exopolysaccharide of Bifidobacterium longum 35624 plays an essential role in dampening host proinflammatory responses and repressing local TH17 responses. Appl Environ Microbiol. 2016;82:7185–7196. doi: 10.1128/AEM.02238-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Schreiner HC, Sinatra K, Kaplan JB, Furgang D, Kachlany SC, Planet PJ, Perez BA, Figurski DH, Fine DH. Tight-adherence genes of Actinobacillus actinomycetemcomitans are required for virulence in a rat model. Proc Natl Acad Sci USA. 2003;100:7295–7300. doi: 10.1073/pnas.1237223100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Scott KP, Martin JC, Chassard C, Clerget M, Potrykus J, Campbell G, Mayer CD, Young P, Rucklidge G, Ramsay AG, et al. Substrate-driven gene expression in Roseburia inulinivorans: importance of inducible enzymes in the utilization of inulin and starch. Proc Natl Acad Sci USA. 2011;108(Suppl 1):4672–4679. doi: 10.1073/pnas.1000091107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Seksik P, Lepage P, de la Cochetiere MF, Bourreille A, Sutren M, Galmiche JP, Dore J, Marteau P. Search for localized dysbiosis in Crohn’s disease ulcerations by temporal temperature gradient gel electrophoresis of 16S rRNA. J Clin Microbiol. 2005;43:4654–4658. doi: 10.1128/JCM.43.9.4654-4658.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Sela DA. Bifidobacterial utilization of human milk oligosaccharides. Int J Food Microbiol. 2011;149:58–64. doi: 10.1016/j.ijfoodmicro.2011.01.025. [DOI] [PubMed] [Google Scholar]
- 122.Sela DA, Chapman J, Adeuya A, Kim JH, Chen F, Whitehead TR, Lapidus A, Rokhsar DS, Lebrilla CB, German JB, et al. The genome sequence of Bifidobacterium longum subsp. infantis reveals adaptations for milk utilization within the infant microbiome. Proc Natl Acad Sci USA. 2008;105:18964–18969. doi: 10.1073/pnas.0809584105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Smilowitz JT, Lebrilla CB, Mills DA, German JB, Freeman SL. Breast milk oligosaccharides: structure-function relationships in the neonate. Annu Rev Nutr. 2014;34:143–169. doi: 10.1146/annurev-nutr-071813-105721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Sonnenburg JL, Chen CT, Gordon JI. Genomic and metabolic studies of the impact of probiotics on a model gut symbiont and host. PLoS Biol. 2006;4:e413. doi: 10.1371/journal.pbio.0040413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Stewart CJ, Embleton ND, Marrs EC, Smith DP, Nelson A, Abdulkadir B, Skeath T, Petrosino JF, Perry JD, Berrington JE, et al. Temporal bacterial and metabolic development of the preterm gut reveals specific signatures in health and disease. Microbiome. 2016;4:67. doi: 10.1186/s40168-016-0216-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Tailford LE, Crost EH, Kavanaugh D, Juge N. Mucin glycan foraging in the human gut microbiome. Front Genet. 2015;6:81. doi: 10.3389/fgene.2015.00081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Tanaka S, Kobayashi T, Songjinda P, Tateyama A, Tsubouchi M, Kiyohara C, Shirakawa T, Sonomoto K, Nakayama J. Influence of antibiotic exposure in the early postnatal period on the development of intestinal microbiota. FEMS Immunol Med Microbiol. 2009;56:80–87. doi: 10.1111/j.1574-695X.2009.00553.x. [DOI] [PubMed] [Google Scholar]
- 128.Tao N, Wu S, Kim J, An HJ, Hinde K, Power ML, Gagneux P, German JB, Lebrilla CB. Evolutionary glycomics: characterization of milk oligosaccharides in primates. J Proteome Res. 2011;10:1548–1557. doi: 10.1021/pr1009367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Turroni F, Berry D, Ventura M. Editorial: bifidobacteria and their role in the human gut microbiota. Front Microbiol. 2016;7:2148. doi: 10.3389/fmicb.2016.00865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Turroni F, Bottacini F, Foroni E, Mulder I, Kim JH, Zomer A, Sanchez B, Bidossi A, Ferrarini A, Giubellini V, et al. Genome analysis of Bifidobacterium bifidum PRL2010 reveals metabolic pathways for host-derived glycan foraging. Proc Natl Acad Sci USA. 2010;107:19514–19519. doi: 10.1073/pnas.1011100107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Turroni F, Duranti S, Bottacini F, Guglielmetti S, Van Sinderen D, Ventura M. Bifidobacterium bifidum as an example of a specialized human gut commensal. Front Microbiol. 2014;5:437. doi: 10.3389/fmicb.2014.00437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Turroni F, Foroni E, O’Connell Motherway M, Bottacini F, Giubellini V, Zomer A, Ferrarini A, Delledonne M, Zhang Z, van Sinderen D, et al. Characterization of the serpin-encoding gene of Bifidobacterium breve 210B. Appl Environ Microbiol. 2010;76:3206–3219. doi: 10.1128/AEM.02938-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Turroni F, Foroni E, Pizzetti P, Giubellini V, Ribbera A, Merusi P, Cagnasso P, Bizzarri B, de’Angelis GL, Shanahan F, et al. Exploring the diversity of the bifidobacterial population in the human intestinal tract. Appl Environ Microbiol. 2009;75:1534–1545. doi: 10.1128/AEM.02216-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Turroni F, Marchesi JR, Foroni E, Gueimonde M, Shanahan F, Margolles A, van Sinderen D, Ventura M. Microbiomic analysis of the bifidobacterial population in the human distal gut. ISME J. 2009;3:745–751. doi: 10.1038/ismej.2009.19. [DOI] [PubMed] [Google Scholar]
- 135.Turroni F, Milani C, Duranti S, Mancabelli L, Mangifesta M, Viappiani A, Lugli GA, Ferrario C, Gioiosa L, Ferrarini A, et al. Deciphering bifidobacterial-mediated metabolic interactions and their impact on gut microbiota by a multi-omics approach. Isme J. 2016;10(7):1656–1668. doi: 10.1038/ismej.2015.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Turroni F, Milani C, van Sinderen D, Ventura M. Genetic strategies for mucin metabolism in Bifidobacterium bifidum PRL2010: an example of possible human-microbe co-evolution. Gut Microbes. 2011;2:183–189. doi: 10.4161/gmic.2.3.16105. [DOI] [PubMed] [Google Scholar]
- 137.Turroni F, Ozcan E, Milani C, Mancabelli L, Viappiani A, van Sinderen D, Sela DA, Ventura M. Glycan cross-feeding activities between bifidobacteria under in vitro conditions. Front Microbiol. 2015;6:1030. doi: 10.3389/fmicb.2015.01030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Turroni F, Peano C, Pass DA, Foroni E, Severgnini M, Claesson MJ, Kerr C, Hourihane J, Murray D, Fuligni F, et al. Diversity of bifidobacteria within the infant gut microbiota. PLoS One. 2012;7:e36957. doi: 10.1371/journal.pone.0036957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Turroni F, Serafini F, Foroni E, Duranti S, O’Connell Motherway M, Taverniti V, Mangifesta M, Milani C, Viappiani A, Roversi T, et al. Role of sortase-dependent pili of Bifidobacterium bifidum PRL2010 in modulating bacterium-host interactions. Proc Natl Acad Sci USA. 2013;110(27):11151–11156. doi: 10.1073/pnas.1303897110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Turroni F, Serafini F, Mangifesta M, Arioli S, Mora D, van Sinderen D, Ventura M. Expression of sortase-dependent pili of Bifidobacterium bifidum PRL2010 in response to environmental gut conditions. FEMS Microbiol Lett. 2014;357:23–33. doi: 10.1111/1574-6968.12509. [DOI] [PubMed] [Google Scholar]
- 141.Turroni F, Ventura M, Butto LF, Duranti S, O’Toole PW, Motherway MO, van Sinderen D. Molecular dialogue between the human gut microbiota and the host: a Lactobacillus and Bifidobacterium perspective. Cell Mol Life Sci. 2014;71:183–203. doi: 10.1007/s00018-013-1318-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Vaishampayan PA, Kuehl JV, Froula JL, Morgan JL, Ochman H, Francino MP. Comparative metagenomics and population dynamics of the gut microbiota in mother and infant. Genome Biol Evol. 2010;2:53–66. doi: 10.1093/gbe/evp057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.van der Maarel MJ, van der Veen B, Uitdehaag JC, Leemhuis H, Dijkhuizen L. Properties and applications of starch-converting enzymes of the alpha-amylase family. J Biotechnol. 2002;94:137–155. doi: 10.1016/S0168-1656(01)00407-2. [DOI] [PubMed] [Google Scholar]
- 144.Vanhoutte T, Huys G, Brandt E, Swings J. Temporal stability analysis of the microbiota in human feces by denaturing gradient gel electrophoresis using universal and group-specific 16S rRNA gene primers. FEMS Microbiol Ecol. 2004;48:437–446. doi: 10.1016/j.femsec.2004.03.001. [DOI] [PubMed] [Google Scholar]
- 145.Ventura M, Canchaya C, Fitzgerald GF, Gupta RS, van Sinderen D. Genomics as a means to understand bacterial phylogeny and ecological adaptation: the case of bifidobacteria. Antonie Van Leeuwenhoek. 2007;91:351–372. doi: 10.1007/s10482-006-9122-6. [DOI] [PubMed] [Google Scholar]
- 146.Ventura M, Canchaya C, Tauch A, Chandra G, Fitzgerald GF, Chater KF, van Sinderen D. Genomics of Actinobacteria: tracing the evolutionary history of an ancient phylum. Microbiol Mol Biol Rev. 2007;71:495–548. doi: 10.1128/MMBR.00005-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Ventura M, Turroni F, Lugli GA, van Sinderen D. Bifidobacteria and humans: our special friends, from ecological to genomics perspectives. J Sci Food Agric. 2014;94(2):163–168. doi: 10.1002/jsfa.6356. [DOI] [PubMed] [Google Scholar]
- 148.Ventura M, Turroni F, Canchaya C, Vaughan EE, O’Toole PW, van Sinderen D. Microbial diversity in the human intestine and novel insights from metagenomics. Front Biosci. 2009;14:3214–3221. doi: 10.2741/3445. [DOI] [PubMed] [Google Scholar]
- 149.Ventura M, Turroni F, Lugli GA, van Sinderen D. Bifidobacteria and humans: our special friends, from ecological to genomics perspectives. J Sci Food Agric. 2014;94:163–168. doi: 10.1002/jsfa.6356. [DOI] [PubMed] [Google Scholar]
- 150.Ventura M, Turroni F, Motherway MO, MacSharry J, van Sinderen D. Host-microbe interactions that facilitate gut colonization by commensal bifidobacteria. Trends Microbiol. 2012;20:467–476. doi: 10.1016/j.tim.2012.07.002. [DOI] [PubMed] [Google Scholar]
- 151.Ventura M, Turroni F, Zomer A, Foroni E, Giubellini V, Bottacini F, Canchaya C, Claesson MJ, He F, Mantzourani M, et al. The Bifidobacterium dentium Bd1 genome sequence reflects its genetic adaptation to the human oral cavity. PLoS Genet. 2009;5:e1000785. doi: 10.1371/journal.pgen.1000785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Wada J, Ando T, Kiyohara M, Ashida H, Kitaoka M, Yamaguchi M, Kumagai H, Katayama T, Yamamoto K. Bifidobacterium bifidum lacto-N-biosidase, a critical enzyme for the degradation of human milk oligosaccharides with a type 1 structure. Appl Environ Microbiol. 2008;74:3996–4004. doi: 10.1128/AEM.00149-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Watson D, O’Connell Motherway M, Schoterman MH, van Neerven RJ, Nauta A, van Sinderen D. Selective carbohydrate utilization by lactobacilli and bifidobacteria. J Appl Microbiol. 2013;114:1132–1146. doi: 10.1111/jam.12105. [DOI] [PubMed] [Google Scholar]
- 154.Westerbeek EA, van den Berg A, Lafeber HN, Knol J, Fetter WP, van Elburg RM. The intestinal bacterial colonisation in preterm infants: a review of the literature. Clin Nutr. 2006;25:361–368. doi: 10.1016/j.clnu.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 155.Xavier KB, Bassler BL. LuxS quorum sensing: more than just a numbers game. Curr Opin Microbiol. 2003;6:191–197. doi: 10.1016/S1369-5274(03)00028-6. [DOI] [PubMed] [Google Scholar]
- 156.Xia G, Kohler T, Peschel A. The wall teichoic acid and lipoteichoic acid polymers of Staphylococcus aureus. Int J Med Microbiol IJMM. 2010;300:148–154. doi: 10.1016/j.ijmm.2009.10.001. [DOI] [PubMed] [Google Scholar]
- 157.Yassour M, Vatanen T, Siljander H, Hamalainen AM, Harkonen T, Ryhanen SJ, Franzosa EA, Vlamakis H, Huttenhower C, Gevers D, et al. Natural history of the infant gut microbiome and impact of antibiotic treatment on bacterial strain diversity and stability. Sci Transl Med. 2016;8:343ra381. doi: 10.1126/scitranslmed.aad0917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Baldassano RN, Anokhin AP, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Yoshida E, Sakurama H, Kiyohara M, Nakajima M, Kitaoka M, Ashida H, Hirose J, Katayama T, Yamamoto K, Kumagai H. Bifidobacterium longum subsp. infantis uses two different beta-galactosidases for selectively degrading type-1 and type-2 human milk oligosaccharides. Glycobiology. 2012;22:361–368. doi: 10.1093/glycob/cwr116. [DOI] [PubMed] [Google Scholar]
- 160.Zanotti I, Turroni F, Piemontese A, Mancabelli L, Milani C, Viappiani A, Prevedini G, Sanchez B, Margolles A, Elviri L, et al. Evidence for cholesterol-lowering activity by Bifidobacterium bifidum PRL2010 through gut microbiota modulation. Appl Microbiol Biotechnol. 2015;99:6813–6829. doi: 10.1007/s00253-015-6564-7. [DOI] [PubMed] [Google Scholar]
- 161.Ze X, Duncan SH, Louis P, Flint HJ. Ruminococcus bromii is a keystone species for the degradation of resistant starch in the human colon. ISME J. 2012;6:1535–1543. doi: 10.1038/ismej.2012.4. [DOI] [PMC free article] [PubMed] [Google Scholar]