Abstract
Hypoxia inducible factor-1α (HIF-1α) is a central molecule involved in mediating cellular processes. Alterations of HIF-1α and hypoxically regulated microRNAs (miRNAs) are correlated with patients’ outcome in various cancers, indicating their crucial roles on cancer development. Recently, an increasing number of studies have revealed the intricate regulations between miRNAs and HIF-1α in modulating a wide variety of processes, including proliferation, metastasis, apoptosis, and drug resistance, etc. miRNAs are a class of small noncoding RNAs which function as negative regulators by directly targeting mRNAs. Evidence shows that miRNAs can be regulated by HIF-1α at transcriptional level. In turn, HIF-1α itself can be modulated by many miRNAs whose alterations have been implicated in tumorigenesis, thus forming a reciprocal regulation network. These findings add a new layer of complexity to our understanding of HIF-1α regulatory networks. Here, we will provide a comprehensive overview of the current advances about the bidirectional interactions between HIF-1α and miRNAs in human cancers. Besides, the review will summarize the roles of miRNAs/HIF-1α crosstalk according to various cellular processes. Finally, the potential values of miRNAs/HIF-1α loops in clinical applications are discussed.
Electronic supplementary material
The online version of this article (10.1007/s00018-018-2941-6) contains supplementary material, which is available to authorized users.
Keywords: miRNAs, HIF-1α, Cancer, Proliferation, Apoptosis, Metastasis
Introduction
Hypoxia, a common feature of solid tumors in the middle-late stages, plays a significant role in tumorigenesis by promoting the formation of a neoplastic environment [1]. Tumor cells adapt to hypoxia partially through a transcriptional program orchestrated by the HIF family of transcription factors [2]. HIFs are heterodimers containing two subunits: an oxygen-modulated α-subunit (HIF-α) and a constitutively expressed β-subunit (HIF-β) [3]. Among three isoforms of HIFα (HIF-1α, -2α and -3α), HIF-1α is ubiquitously expressed in various cells and regarded as the major regulator of oxygen homeostasis [4]. The activity of HIF-1α can be regulated by different oxygen conditions [5, 6]. Under normal oxygen conditions, HIF-1α is hydroxylated by a family of prolyl-hydroxylases (PHDs) on two specific proline residues (Pro402 and Pro564) [7, 8]. Then, hydroxylated HIF-1α is recognized and targeted for rapid proteasomal degradation by the substrate recognition component of an E3-ubiquitin ligase, the von Hippel-Lindau (VHL) tumor suppressor [9, 10]. While under hypoxia, HIF-1α can escape proteasomal degradation due to the decreased activity of PHDs and inhibited hydroxylation [5, 7]. The level of HIF-1α is also regulated by other O2-dependent ways. For example, hydroxylation by inhibiting HIF-1 (FIH-1) impairs HIF-1α transcriptional activity because this modification inhibits the interaction between HIF-1α and coactivators p300/CBP [11]. Moreover, HIF-1α upregulation can be the result of increased reactive oxygen species (ROS) [12].
The increased HIF-1α dimerizes with HIF-1β can specifically bind to the promoters of many hypoxically regulated genes [13]. HIF-1α, therefore, modulates a wide variety of cellular processes, including metabolism, redox homeostasis, inflammation, and angiogenesis [7, 9]. Moreover, dysregulation of HIF-1α is also correlated to tumorigenesis and HIF-1α participates in different signaling pathways in human cancers [9, 14]. HIF-1α is widely involved in PI3K/Akt pathway, probable pinoresinol–lariciresinol reductase 3 (PLR-3)/NF-κB pathway, and Wnt signaling pathway [15–17].
HIF-1α is an attractive candidate for prognosis analysis. Cells with an increased level of HIF-1α may be less sensitive to apoptosis and show a more aggressive phenotype [18]. Elevated HIF-1α has been widely reported to be linked with human cancer progression [19]. Immunohistochemical analyses indicated that HIF-1α protein is commonly overexpressed in primary and metastatic tumor tissues compared to its absence in normal controls [20]. Moreover, overexpressed intracellular HIF-1α is associated with patient mortality and worse prognosis [19]. Significant correlations between HIF-1α upregulation and prognosis have been identified in cancers of the esophagus [21], breast [22], ovary [23] and head and neck [24], etc. In some types of cancer, HIF-1a protein is even considered as an independent prognostic indicator [25].
miRNAs are a class of single-stranded, endogenous, small noncoding RNAs (containing about 19–24 nucleotides) that regulate target mRNA through inhibiting its translation or promoting degradation [26, 27]. miRNAs play significant roles in a broad range of biological and pathological processes, including proliferation [28], metabolism [29], apoptosis [30] as well as drug resistance [26, 31]. miRNAs can simultaneously regulate several target mRNAs, and act as either tumor suppressors or oncogenes in human cancers [32]. Some studies have indicated that miRNAs might be the ideal biomarkers for early detection and prognosis prediction of cancers [33, 34].
To date, the significance of long non-coding RNAs (lncRNAs) is gradually being recognized [35]. Compared to miRNAs, lncRNAs are transcripts longer than 200 nucleotides with little or no protein-coding capacity [36]. lncRNAs are widely participated in the gene regulations at different levels, including epigenetic regulations [37], transcription and posttranscriptional processing [38]. Studies have demonstrated that lncRNAs potentially contribute to tumor progression [39]. The roles of lncRNAs as tumor suppressors or oncogenes have been confirmed in various cancer [40, 41]. Notably, pivotal roles of hypoxia-responsive lncRNAs (HRLs) in cancer progression have been found, and some lncRNAs even participate in the crosstalks of miRNAs and HIF-1α [35, 42]. Moreover, unique profiles of lncRNAs can also reflect cancer progression and may serve as indicators for patient’s outcome [39, 43].
miRNA/HIF-1α interactive regulations
Many studies have elucidated the novel roles of miRNAs/HIF-1α crosstalk in cancers. As a transcriptional factor, the HIF-1α/HIF-1β dimmer can bind to the promoters of miRNAs and regulate various cellular processes. In addition, tumor-related miRNAs can also modulate the activity of HIF-1α, finally exerting pro- or anti-tumor effects on tumor cells.
HIF-1α related miRNAs
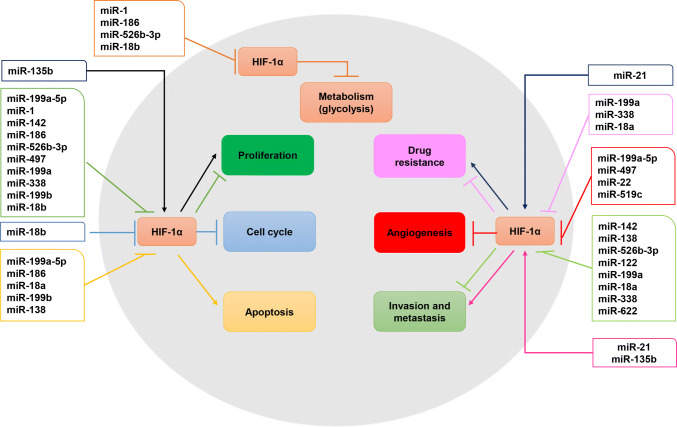
During the past few years, multiple studies have demonstrated that miRNAs can influence the tumorigenic phenotypes of cancer cells (such as invasion, metastasis, angiogenesis, and drug resistance) (Fig. 1; Tables 1, 2). More importantly, HIF-1α is confirmed as a common target gene of certain miRNAs (Fig. 2; Table S1).
Fig. 1.
The molecular regulatory networks about miRNAs regulate HIF-1α in human cancers. The miRNA/HIF-1α interactive relation play an intricate role in tumorigenicity including cell proliferation, cell cycle, apoptosis, metabolism, invasion and metastasis, angiogenesis, and drug resistance (different round colors showed). HIF-1α, hypoxia inducible factor-1α
Table 1.
Summary of reported miRNAs directly targeting HIF-1α in human cancers
| miRNAs | Cancer or cell types | Validation ways | Functions | References |
|---|---|---|---|---|
| Downregulated miRNAs | ||||
| miR-1 | Colorectal cancer | Targets 3′-UTR of HIF-1α (luciferase assay) | Inhibits proliferation; suppresses glycolysis | Xu et al. [28] |
| miR-199a-5p | HCC | Targets 3′-UTR of HIF-1α (luciferase assay) | Inhibits the Warburg effect; suppresses cell growth | Li et al. [29] |
| Prostate adenocarcinoma | Targets 3′-UTR of HIF-1α (luciferase assay) | Inhibits proliferation, motility, and angiogenesis; iromotes apoptosis | Zhong et al. [44] | |
| Malignant melanoma | Targets 3′-UTR of HIF-1α (luciferase assay) | Inhibits proliferation and growth; arrests cells in G1 phase | Yang et al. [116] | |
| miR-199a | Osteosarcoma | Targets 3′-UTR of HIF-1α (luciferase assay) | Reverses drug resistance | Keremu et al. [26] |
| Colorectal cancer | Western blot | Inhibits proliferation, migration and invasion | Ye et al. [46] | |
| EOC | Targets 3′-UTR of HIF-1α (luciferase assay) | Inhibits migration; inhibits ECM remodeling; inhibits cell growth | Joshi et al. [47] | |
| miR-199b | Prostate adenocarcinoma | Targets 3′-UTR of HIF-1α (luciferase assay) | Inhibits proliferation; increases cell death | Shang et al. [45] |
| miR-138 | Ovarian cancer | Targets 3′-UTR of HIF-1α (luciferase assay) | Suppresses invasion, migration, and metastasis | Yeh et al. [51] |
| GBC | Targets 3′-UTR of HIF-1α (luciferase assay) | Inhibits metastasis and EMT | Cai et al. [52] | |
| CCRCC | Targets 3′-UTR of HIF-1α (luciferase assay) | Increases apoptosis; reduces migration | Song et al. [53] | |
| miR-18b | Malignant melanoma | Targets 3′-UTR of HIF-1α (luciferase assay) | Decreases proliferation; induces cell cycle arrest; inhibits glycolysis | Chen et al. [54] |
| miR-18a | Breast cancer | Western blot and qRT-PCR | Reduces drug resistance | Li et al. [55] |
| Gastric cancer | Targets 3′-UTR of HIF-1α (luciferase assay) | Inhibits apoptosis and invasion | Wu et al. [56] | |
| miR-142 | Pancreatic cancer | Targets 3′-UTR of HIF-1α (luciferase assay) | Inhibits proliferation and invasion | Lu et al. [58] |
| miR-186 | Gastric cancer | Targets 3′-UTR of HIF-1α (luciferase assay) | Inhibits proliferation; promotes apoptosis; inhibits glycolysis | Liu et al. [59] |
| miR-526b-3p | Colon cancer | Targets 3′-UTR of HIF-1α (luciferase assay) | Suppresses proliferation, metastasis and glycolysis | Zhang et al. [60] |
| miR-22 | Colon cancer | Targets 3′-UTR of HIF-1α (luciferase assay) | Inhibits angiogenesis | Yamakuchi et al. [61] |
| miR-497 | Breast cancer | Western blot | Inhibits proliferation, growth, and angiogenesis | Wu et al. [62] |
| miR-622 | Lung cancer | Targets 3′-UTR of HIF-1α (luciferase assay) | Inhibits invasion and migration, | Cheng et al. [63] |
| miR-519c | Lung cancer | Targets 3′-UTR of HIF-1α (luciferase assay) | Reduces angiogenesis | Cha et al. [64] |
| miR-122 | HCC | Targets 3′-UTR of HIF-1α (luciferase assay) | Inhibits EMT | Ambade et al. [65] |
| miR-145 | HCC | Western blot and qRT-PCR | – | Takahashi et al. [66] |
| miR-338-3p | NPC | Targets 3′-UTR of HIF-1α (luciferase assay) | Inhibits proliferation, migration, and EMT; overcomes drug resistance | Shan et al. [117] |
| Upregulated miRNAs | ||||
| miR-135b | HNSCC | Western blot and qRT-PCR | Increases proliferation, migration, and colony formation | Zhang et al. [70] |
| miR-21 | Ovarian cancer | Western blot and qRT-PCR | Induces drug resistance | Xie et al. [71] |
| Breast cancer | Western blot and qRT-PCR | Promotes EMT | Han et al. [72] | |
CCRCC, clear cell renal cell carcinoma; EOC, epithelial ovarian cancer; GBC, gallbladder cancer cell; HCC, hepatocellular carcinoma; HNSCC, head and neck squamous cell carcinoma; NPC, nasopharyngeal carcinoma; HIF-1α, hypoxia inducible factor-1α; EMT, epithelial–mesenchymal transition; ECM, extracellular matrix; qRT-PCR, quantitative real-time polymerase chain reaction
Table 2.
HIF-1α regulates miRNAs in human cancers
| miRNAs | Cancer or cell types | Validation ways | Functions | miRNA targets | Group |
|---|---|---|---|---|---|
| Upregulated miRNAs | |||||
| miR-210 | Schwannoma | qRT-PCR | Inhibits apoptosis and autophagy; promotes angiogenesis and proliferation | EFNA3 | Wang et al. [75] |
| Ovarian cancer | – | Promotes invasion and metastasis | VMP1 | Liu et al. [76] | |
| Ovarian cancer | qRT-PCR | Promotes proliferation; inhibits apoptosis | PTPN1 | Li et al. [77] | |
| Gastrointestinal cancer | ChIP assay | Promotes migration | SPRED2 | Chen et al. [78] | |
| OPSCC | qRT-PCR | Influences metabolic reprogramming | ISCU | Saenz-de-Santa-Maria et al. [79] | |
| Paragangliomas | qRT-PCR | Influences metabolic reprogramming | – | Merlo et al. [80] | |
| CCRCC | qRT-PCR | Induces G2/M accumulation; Induces aneuploidy and multipolar spindle formation | E2F3 | Nakada et al. [81] | |
| HCC | Luciferase assay | Promotes invasion and migration | HIF-3α | Kai et al. [82] | |
| Colon cancer | qRT-PCR and Western blot | Enhances autophagy; reduces radiosensitivity | Bcl-2 | Sun et al. [83] | |
| Gastric cancer | qRT-PCR | Promotes EMT | HOXA9 | Yu et al. [84] | |
| Pancreatic cancer | qRT-PCR | – | E2F3; EFNA3; GIT2; MNT; ZNF462; EGR3 | Chen et al. [85] | |
| ESCC | qRT-PCR | Inhibits proliferation; induces G2/M phase cell cycle arrest | PLK1 | Li et al. [86] | |
| miR-21 | Cervical cancer | qRT-PCR and Western blot | Increases radioresistance | PTEN | Song et al. [87] |
| OSCC | ChIP assay | Promotes cell growth, migration and invasion | PTEN; Pdcd4; E-cadherin; Vimentin | Li et al. [88] | |
| Pancreatic cancer | ChIP assay | Increases proliferation; overrides cell cycle arrest; Enhances cell survival | – | Mace et al. [89] | |
| miR-191 | Lung cancer | qRT-PCR | Promotes proliferation and migration | NFIA | Zhao et al. [90] |
| miR-224 | Gastric cancer | Luciferase and ChIP assays | Promotes cell growth, migration and invasion | RASSF8 | He et al. [91] |
| miR-421 | Gastric cancer | Luciferase assay | Promotes metastasis; inhibits apoptosis; induces drug resistance | E-cadherin; Caspase-3 | Ge et al. [92] |
| miR-27a | Gastric cancer | ChIP and luciferase assays | Induces multidrug resistance | MDR1/P-gp; LRP; Bcl-2 | Zhao et al. [93] |
| miR-107 | Gastric cancer | qRT-PCR | – | – | Ayremlou et al. [94] |
| miR-382 | Gastric cancer | Luciferase assay | Promotes angiogenesis | PTEN | Seok et al. [95] |
| miR-183 | HCC | qRT-PCR and Western blot | Induces multidrug resistance | IDH2; SOCS6 | Wang et al. [96] |
| miR-494 | Lung cancer | ChIP assay | Promotes angiogenesis | PTEN; Akt/eNOS pathway | Mao et al. [97] |
| miR-145 | Bladder cancer | ChIP assay | Induces apoptosis | – | Blick et al. [98] |
| miR-184 | Glioma | qRT-PCR and Western blot | Suppresses apoptosis; facilitates invasion and migration. | FIH-1 | Yuan et al. [99] |
| miR-485-5p; miR-210-3p | STS | RT-PCR and microarrays | – | HIF-3α | Gits et al. [100] |
| miR-675-5p | Glioma | qRT-PCR and Western blot | Promotes angiogenesis | HuR/VHL? | Lo Dico et al. [107] |
| Downregulated miRNAs | |||||
| miR-199a-5p | HCC | qRT-PCR | Inhibits Warburg effect and cell growth | HIF-1α | Li et al. [29]. |
| miR-548an | Pancreatic cancer | ChIP assay | Inhibits proliferation and invasion | Vimentin | Zhu et al. [101] |
| miR-17; miR-20a | AML | qRT-PCR and Western blot | Inhibits cell differentiation; promotes proliferation | p21;STAT3 | He et al. [102] |
| miR-33a | HCC | Western blot | Inhibits invasion and migration | Twist1 | Guo et al. [103] |
| miR-101 | Pancreatic cancer | qRT-PCR and Western blot | Inhibits invasion and migration | EZH2 | Cao et al. [104] |
| miR-34a | Colorectal cancer | ChIP assay | Inhibits the EMT and metastasis | PPP1R11 | Li et al. [105] |
CCRCC, clear cell renal cell carcinoma; HCC, hepatocellular carcinoma; OPSCC, oropharyngeal squamous cell carcinomas; OSCC, oral squamous cell carcinoma; STS, soft tissue sarcomas; AML, acute myeloid leukemia; ESCC, esophageal squamous cell carcinoma cell; NFIA, nuclear factor 1α; EFNA3, Ephrin A3; ISCU; Iron–sulfur cluster assembly enzyme; HIF-1α, hypoxia inducible factor-1α; RASSF8, Ras association domain family member 8; IDH2, isocitrate dehydrogenase (NADP (+)) 2, mitochondrial; SOCS6, suppressor of cytokine signaling 6; STAT3, signal transducer and activator of transcription 3; PTEN, phosphatase and tensin homolog; Pdcd4, programmed cell death 4; HIF-3α, hypoxia inducible factor-3α; Twist1, twist family bHLH transcription factor 1; MDR1/P-gp, MDR1: multi-drug resistance gene1/P-glycoprotein; LRP, leucine-responsive global transcriptional regulator; Bcl-2, B cell leukemia/lymphoma 2; HOXA9, homeobox A9; SPRED2, sprouty related EVH1 domain containing 2; FIH-1, factor inhibiting hypoxia-inducible factor 1 alpha; VMP1, vacuole membrane protein 1; PTPN1, protein tyrosine phosphatase, non-receptor type 1; PPP1R11, protein phosphatase 1 regulatory inhibitor subunit 11; VHL, von Hippel-Lindau; E2F3, E2F transcription factor 3; GIT2, GIT ArfGAP 2; MNT, MAX network transcriptional repressor; ZNF462, zinc finger protein 462; EGR3, early growth response 3; PLK1, polo like kinase 1; EZH2, enhancer of zeste 2 polycomb repressive complex 2 subunit; EMT, epithelial–mesenchymal transition; ChIP, chromatin immunoprecipitation; qRT-PCR, quantitative real time polymerase chain reaction
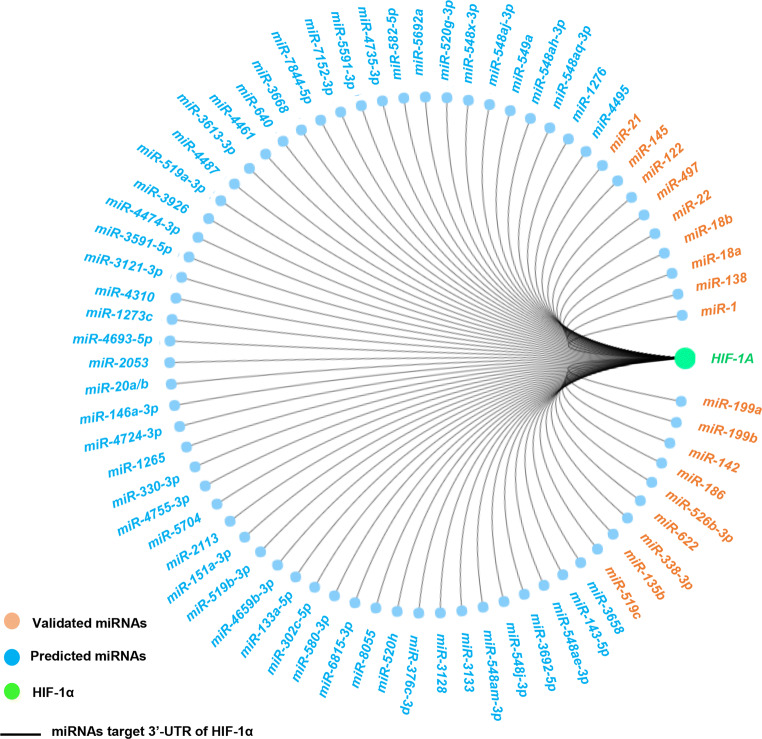
Fig. 2.
miRNAs target HIF-1α in human cancers (including the validated miRNAs and predicted miRNAs). Data publicly available from the miRDB (http://www.mirdb.org/). The components are colored as the key at the lower left. HIF-1α, hypoxia inducible factor-1α
miR-199
One of the most focused miRNAs is miR-199. Increased level of either miR-199a-5p or miR-199b can significantly inhibit the luciferase activity of HIF-1α in prostate adenocarcinoma [44, 45]. miR-199a-5p is reduced in hepatocellular carcinoma (HCC) tissues and cell lines, and HIF-1α is identified as the target of miR-199a-5p [29]. In colorectal cancer, overexpressed miR-199a suppresses tumor progression through the HIF-1α/vascular endothelial growth factor A (VEGF-A) pathway [46]. Moreover, HIF-1α is a direct target of miR-199a in osteosarcoma and ovarian cancer [26, 47].
miR-138
miR-138 is highly conserved miRNA among various species [48, 49]. Two miR-138 genes (miR-138-1 and miR-138-2) are mapped to chromosome 3p21.33 and 16q13, respectively [48]. It is believed that only miR-138-2 is functionally transcribed in human cells [50]. miR-138 plays a crucial role in human cancers [51–53]. In gallbladder cancer (GBC), the level of miR-138 is regulated by the long non-coding RNA LINC00152, and HIF-1α is confirmed as the target gene of miR-138 [52]. In ovarian cancer cells, miR-138 was reported to exert its anti-metastasis effect by targeting the 3′-UTR of HIF-1α [51]. miR-138 is markedly reduced in clear cell renal cell carcinoma (CCRCC), and HIF-1α is the target gene of miR-138 [53].
miR-18
The two members of miR-18 family (miR-18a miR-18b) have been found to function as tumor suppressors in different cancers [54–56]. miR-18a is downregulated in gastric cancer, and it can target HIF-1α [56]. miR-18a/HIF-1α feedback loop is closely related to tamoxifen sensitivity in estrogen receptor (ER)-positive cells. And this feedback loop can be modulated by a lncRNA, urothelial carcinoma-associated 1 (UCA1), which plays an oncogenic role in breast cancer [55]. Besides, miR-18b is downregulated in malignant melanoma tissues and cell lines, and HIF-1α is a direct target of miR-18b [54].
Other miRNAs
The 3′-UTRs of HIF-1α also contains the binding sites for other miRNAs. For instance, the base-pair between HIF-1α and miRNA seed region is evident with miR-1 [28, 57], miR-142 [58], and miR-186 [59]. Transfection of miR-526b-3p or miR-22 can decrease HIF-1α protein levels in colon cancer [60, 61]. In breast cancer cells, HIF-1α is identified as a representative target of miR-497 [62]. miR-622 and miR-519c are involved in lung carcinogenesis by targeting HIF-1α [63, 64]. In parallel to these, miR-122, miR-145, and miR-138 are also abnormally expressed in cancers and HIF-1α is identified as their direct target [51, 65, 66].
It is worth noting that most of miRNAs shows adverse expression levels with HIF-1α since miRNAs can function as negative gene regulators via the inhibition of RNAs translation or degradation of target mRNAs [67, 68]. However, some miRNAs present consistent levels with HIF-1α, which might due to the integrated effects of genes regulatory networks [69]. For example, upregulated miR-135b is positively correlated with HIF-1α expression and acts as a tumor promoter by increasing the level of HIF-1α [70]. In ovarian cancer and breast cancer cells, both HIF-1a and miR-21 are upregulated, and they can promote cancer progression [71, 72]. These findings indicate that there are complex regulatory networks between miRNAs and HIF-1α, and in-depth studies are needed.
HIF-1α regulates miRNAs
HIF-1α is one of the important transcriptional activators, and dysregulation of HIF-1α usually causes the alterations of its downstream genes, including miRNAs. Some hypoxamiRs carry a hypoxia response element (HRE) in their promoters which can bind to HIF-1α [69]. Just as mentioned above, some miRNAs are apt to inhibit HIF-1α and some of them are also regulated by HIF-1α (Table 2; Fig. 2).
miR-210
miR-210 is a key regulator of hypoxia and generally recognized as a robust HIF target [73]. Elevated miR-210 is detected in various cancers and acts on a number of targets [74], including B cell lymphoma-2 (Bcl-2), vacuole membrane protein 1 (VMP1) and iron–sulfur cluster assembly enzyme (ISCU). Hypoxia-induced miR-210 is involved in the development of schwannoma by targeting ephrin-A3 (EFNA3) [75]. In ovarian cancer, HIF-1α/miR-210 axis is an important contributor for tumor progression through acting on VMP1 and non-receptor type 1 (PTPN1) [76, 77]. Besides, both the HIF-1α and miR-210 are upregulated in the cells of gastrointestinal cancer, and the chromatin immunoprecipitation (ChIP) assay confirms that HIF-1α can enhance miR-210 transcript levels via directly binding to the HRE in promoter region of miR-210 [78]. In some cancers (oropharyngeal squamous cell carcinomas (OPSCC) and paragangliomas), HIF-1α induced miR-210 may influence the metabolic reprogramming [79, 80]. Also, miR-210 is found to be upregulated in CCRCC cells as a result of HIF-1α accumulation [81].
Interestingly, the majority of the findings about the role of HIF-1α/miR-210 axis are found in gastrointestinal cancers, which may be due to the biological specificity. In HCC cells, HIF-1α/miR-210/HIF-3a regulatory circuit is closely related to the cancer metastasis [82]. Likewise, Sun Y and his colleagues have found that HIF-1α/miR-210/Bcl-2 pathway was responsible for the carcinogenesis of colon cancer [83]. miR-210 is regulated by HIF-1α in gastric cancer, and blocking this pathway might be conducive to inhibit tumor growth [84]. In pancreatic cancer cells, miR-210 expression is found to be induced by hypoxia through a HIF-1α-dependent pathway [85]. Besides, miR-210 is highly expressed under hypoxic conditions compared to normal oxygen conditions in esophageal cancer cells [86]. These facts highlight the importance of hypoxia/HIF-1α in regulating miR-210 expression during the carcinogenesis and provide critical clues for the treatment of gastrointestinal cancers [73].
miR-21
Apart from miR-210, miR-21 is another abundantly expressed hypoxamiR under the regulation of HIF-1α. Induction of miR-21 under hypoxia has been variably reported in cancer cells [87–89]. Song et al. have found that upregulated miR-21 in radioresistant cervical cancer was related to HIF-1α overexpression. Importantly, they proposed that there was a HIF-1α/miR-21 positive feedback loop through the PTEN/Akt/HIF-1α pathway in cervical cancer cells [87]. miR-21 shows a highest level among the differentially expressed miRNAs in hypoxic exosomes [88]. The expression level of miR-21 is dependent on both HIF-1α and HIF-2α in oral squamous cell carcinoma (OSCC) cells. In another study, Mace et al. have reported that miR-21 was significantly induced by hypoxia through HIF-1α upregulation in pancreatic cancer cells [89].
Others miRNAs
Using miRNA profiling techniques and bioinformatics analysis, more and more hypoxamiRs have been identified in recent years. For instance, miR-191 is found to be upregulated under chronic hypoxia and regulated by HIF-1α in non-small cell lung cancer (NSCLC) cells [90]. In gastric cancer cells, miR-224 is upregulated by HIF-1α at the transcriptional level [91]. Overexpressed miR-421, miR-27a, miR-107, and miR-382 are induced by the high level of HIF-1α in gastric cancer cells. The direct regulatory relationships of HIF-1α on these miRNAs are demonstrated by different approaches, including ChIP assay, luciferase assay, as well as qRT-PCR [92–95]. In HCC cells, the expression levels of miR-183 and HIF-1α are positively correlated, and miR-183 expression is affected by HIF-1α [96]. miR-494 is induced in lung cancer cells in response to hypoxia, likely via a HIF-1α-mediated mechanism [97]. In addition, HIF-1α can directly bind to the promoter region of miR-145 and HIF-1α promotes its expression in bladder cancer [98]. miR-184 is observed to be significantly upregulated in glioma under the regulation of HIF-1α [99]. In soft tissue sarcomas (STS), miR-485-5p and miR-210-3p are identified as the most increased miRNAs under hypoxia, and alterations of these two miRNAs can be influenced by HIF-1α [100].
There are also many downregulated miRNAs under hypoxia [69]. For instance, miR-548an, a tumor suppressor miRNA, is down-regulated by HIF-1α in pancreatic cancer cells, and it contributes to increasing vimentin level and facilitating the pancreatic tumorigenesis [101]. Hypoxia/HIF-1a can reduce the levels of miR-17 and miR-20a via downregulating c-Myc in acute myeloid leukemia (AML) cells [102]. miR-33a and miR-199a-5p are found to be reduced under the regulation of HIF-1α in HCC [29, 103]. Cao et al. have found that the deferoxamine mesylate (DFO)-induced HIF-1α and HIF-1β can decrease the level of miR-101 in pancreatic cancer cells [104]. Li et al. have demonstrated that downregulated miR-34a was necessary for the hypoxia-induced EMT, invasion and migration in colorectal cancer cells. HIF-1α can directly inhibit the expression of miR-34a in p53-defective colorectal cancer cells, whereas the level of miR-34a is increased in p53-proficient colorectal cancer cells under hypoxia [105] (Fig. 2). These findings suggest that the functions and correlations of miRNAs and HIF-1α in human cancer may be cell-type dependent.
The loops of miRNAs/HIF-1α
One of the prominent biological functions of miRNAs is to reinforce the inhibition of transcripts in cells. In turn, HIF-1α itself can be regulated by some miRNAs, thus forming the positive and negative feedback loops (Fig. 3). So far, these miRNAs include the miR-210 [73, 74, 82], miR-199a [29, 47, 106], miR-21 [87], miR-18a [55], miR-183 [96], and miR-138 [52]. Additional players in the loops of miRNAs/HIF-1α include miR-675-5p [107], miR-17-92 cluster [108], miR-20b [109, 110] as well as miR-519c [64].
Fig. 3.
Model of auto-regulatory feedback loop between several selected miRNAs and HIF-1α. The components of each feedback loop are colored according to genes type or genes functions (see the key at the middle left). DNM2, dynamin 2; GPD1L, glycerol-3-phosphate dehydrogenase 1 like; HIF-1α, hypoxia inducible factor-1α; HIF-3α, hypoxia inducible factor-3α; HUR, RNA-binding protein (RBP); IDH2, isocitrate dehydrogenase (NADP (+)) 2, mitochondrial; PPP1R11, protein phosphatase 1, regulatory subunit 11; PTEN, phosphatase and tensin homolog; SOCS6, suppressor of cytokine signaling 6; STAT3, signal transducer and activator of transcription 3; VHL, von Hippel-Lindau
Overexpressed miR-210 is a significant factor for HIF-1α stabilization during hypoxia, and increased activity of HIF-1α can trigger the accumulation of miR-210, thus forming a positive-feedback loop of HIF-1α amplification [73] (Fig. 3). Similarly, miR-675-5p, which is embedded in hypoxia-induced long non-coding RNA H19, is required to sustain the activity of HIF-1α [107]. In contrast to the positive-feedback loops mentioned above, there are also some miRNAs contribute to a negative-feedback loop which may affect the level of HIF-1α. For instance, miR-107 [111] and miR-155 [112] are reported to participate in the negative-feedback loop for the resolution of HIF-1α stabilization under hypoxia. P53-induced miR-107 can also repress HIF-1β expression and hypoxic signaling [111]. Other regulators which act on the negative-feedback loops refer to factor inhibiting HIF-1 (FIH-1) and cullin 2. Yuan et al. have demonstrated that FIH-1 (the asparagyl hydroxylase inhibitor of HIF-1α activity) was a target mRNA of miR-184 and upregulated FIH-1 might negatively control the protein level of HIF-1α [99]. Cullin 2 (CUL2) is a scaffolding protein which is critical to the assembly of the ubiquitin ligase system [113]. Hypoxia-induced miR-424 can target CUL2 to stabilize the HIF isoforms [114]. Given the increasing reports about the HIF-1α pathway in various cancers, it is highly anticipated that the loops of miRNAs/HIF-1α, directly or indirectly, provide new theoretical basis for clinical applications.
Molecular mechanisms of miRNAs/HIF-1α loops in cancers
As discussed earlier, miRNAs/HIF-1α loops are cancer type-dependent and play an intricate role in carcinogenesis. In this section, we will summarize the molecular mechanisms of these deregulated miRNAs/HIF-1α loops according to their various biological functions including cell proliferation and cell cycle, apoptosis, metastasis, and drug resistance (Fig. 1; Tables 1, 2).
Cell proliferation and cell cycle
Deregulation of the cell cycle underlies the uncontrolled cell proliferation is the marked feature of cancer cells [115]. In prostate adenocarcinoma, overexpressed miR-199a-5p inhibits tumor cell proliferation by targeting HIF-1α, and upregulated HIF-1α can partly rescue the proliferation inhibition caused by miR-199a-5p [44]. miR-199b can inhibit proliferation of prostate adenocarcinoma cells through affecting the level of HIF-1α [45]. In colorectal cancer cells and tissues, miR-199a acts as a tumor suppressor by directly targeting HIF-1α [46]. Likewise, upregulated miR-199a-5p is reported to inhibit proliferation of malignant melanoma cells, in part, by coordinately suppressing HIF-1α [116].
miR-18b is also reported to remarkably decrease proliferation of malignant melanoma cells by targeting the 3′-UTR of HIF-1α [54]. According to a recent report, ectopical expression of miR-1 exerts an anti-proliferative effect by targeting HIF-1α in colorectal cancer cell lines [28]. In pancreatic cancer cells, re-expression of miR-142 can significantly inhibit proliferation by downregulation of HIF-1α [58]. Meanwhile, the HIF-1α–histone deacetylase 1 (HDAC1) complex transcriptionally inhibits miR-548an expression under hypoxia, resulting in the high level of vimentin that facilitates the proliferation of pancreatic cancer cells [101]. Similarly, miR-17 and miR-20a are downregulated by HIF-1α, and they inhibit proliferation of AML cells [102]. Liu et al. showed that miR-186/HIF-1α axis is crucial to the proliferation of gastric cancer cells, upregulated miR-186 inhibits tumor cells proliferation through targeting HIF-1α [59]. miR-338-3p inhibits the cell growth and proliferation of nasopharyngeal carcinoma (NPC) cells via reduction of HIF-1α [117]. Additionally, upregulation of miR-526b-3p and miR-497 may inhibit proliferation of colon cancer cells and breast cancer cells by downregulating HIF-1α, respectively [60, 62].
However, miR-135b was found to increase proliferation of head and neck squamous cell carcinoma (HNSCC) cells by inhibiting the HIF-1α [70]. Similarly, some of hypoxamiRs commonly act as tumor promoters by targeting different mRNAs in cancer cells (Table 2). Upregulated miR-191 shows a promotive effect on NSCLC cell proliferation under the conditions of chronic hypoxia [90]. Overexpression of miR-21 is reported to remarkably promote proliferation of OSCC and pancreatic cancer cells [88, 89]. Upregulation of miR-210 promotes cell proliferation in schwannoma and ovarian cancer [75, 77]. Li et al. [86] as well as Tsuchiya et al. [118] have demonstrated that miR-210 can inhibit proliferation of ESCC cells by leading to cell cycle arrest in G1/G0 or G2/M phases, although it targets different mRNA [Polo-like kinase 1 (PLK1), Fibroblast Growth Factor Receptor-like 1 (FGFRL1)]. These results suggest that miRNAs/HIF-1α loops can either promote tumor proliferation or suppress tumor proliferation, depending on the cell types.
Apoptosis
Apoptosis is the programmed cell death that contributes to eliminating unnecessary cells and maintaining the healthy balance between cell survival and cell death in metazoan [119, 120]. Defect in apoptosis plays crucial roles in tumor progression, allowing tumor cells to survive over the intended lifespans [120]. Generally, the available evidence shows a predominantly anti-apoptosis role of these miRNAs in cancer cells [75, 77, 92, 99], whereas downregulation of them promoting apoptosis [44, 45, 53, 59] (Tables 1, 2). These studies have been performed in a series of cancer cells, including ovarian cancer and breast cancer. Although there are still significant gaps in our understanding of this process, a number of targets (including HIF-1α, FIH-1, caspase-3, and PTPN1.) have been identified to help explaining the role of hypoxamiRs in apoptosis. Moreover, despite the predominant evidence as mentioned above, recent studies also demonstrated some unusual cases. For instance, miR-18a is found to be downregulated in gastric cancer cells and inhibits apoptosis of tumor cells by targeting HIF-1α [56]. Hypoxia/HIF-1α-induced miR-145 can also induce cell apoptosis in bladder cancer [98].
Metabolism
Mitochondria, also named as the ‘‘energy factories’’, can generate majority of adenosine triphosphate (ATP) under normoxic conditions. Tumor cells shows a greater demand for energy and oxygen for their rapid proliferation and growth. Especially under hypoxia, tumor cells prefer to produce ATP through glycolysis pathway, which is less efficient compared with the oxidative phosphorylation pathway [121]. This phenomenon, also called “the Warburg effect”, reveals the relationships between metabolism and carcinogenesis. In this process, certain miRNAs and HIF-1α play a significant role either by decreasing the level of mitochondrial respiration and biogenesis related genes or increasing the activity of glycolytic enzymes [29, 69, 79].
For example, several studies have found that hypoxia-induced miR-210 may contribute to this metabolic shift through suppressing the level/functions of the genes or proteins in the mitochondrial electron transport chain (ETC). The targets of miR-210 in this metabolic shift include the ISCU [79], NADH dehydrogenase (ubiquinone) 1alpha subcomplex4 (NDUFA4) [122], succinate dehydrogenase complex, subunit D (SDHD) [122], cytochrome c oxidase assembly homolog 10 (COX10) [123], and glycerol-3-phosphate dehydrogenase 1-like (GPD1L) [69]. And these findings have been demonstrated in various cancer cells, including lung cancer, breast cancer, and colon cancer. Notably, some target genes like SDHD usually acts as a tumor suppressor, indicating a tumor promotion effect of miR-210. Evidence from a HIF-1α/miR-210/GPD1L circuit (Fig. 3) shows the close relationships of HIF-1α/miR-210 loop and metabolism in carcinogenesis. Besides, hypoxia-induced miR-210 is reported to influence metabolic reprogramming both in OPSCC [79] and paragangliomas cells [80], which is closely related to the Warburg effect.
Some miRNAs may play a different role as compared with miR-210. Restoration of miR-1 level can inhibit glycolysis in colorectal cancer cells via targeting HIF-1α, suggesting an anti-tumorigenic role of miR-1 [28]. Similarly, other reports have found that miR-186 [59], miR-526b-3p [60], and miR-18b [54] might inhibit glycolysis by downregulating the level of HIF-1α in cancer cells. miR-199-5a/HIF-1α loop shows a suppressive effect on Warburg effect of HCC cells under hypoxia [29].
Metastasis and invasion
Cancer metastasis is commonly known as the spread of tumor cells to distant parts of the body from its original site [52, 124]. Metastasis and invasion are highly dependant on the tumor–stromal interaction, such as extracellular matrix (ECM) remodeling and epithelial–mesenchymal transition (EMT) [52, 82]. Recent studies on cancer metastasis highlighted the significant role of reciprocal regulations between miRNAs and HIF-1α. Study conducted by Ye et al. has revealed a link between miR-199a and HIF-1α in colorectal cancer, and ectopic expression of miR-199a could decrease migratory and invasive capacity of cancer cells by targeting HIF-1α [46]. Likewise, miR-199a can inhibit metastasis of epithelial ovarian cancer (EOC) cells via suppressing the expression of HIF-1α [47]. In pancreatic cancer, upregulation of miR-142 shows an anti-metastatic role by targeting HIF-1α [58], similar results are also found in studies referred to miR-548an [101] and miR-101 [104]. miR-138/HIF-1α axis is demonstrated to reduce the migration of CCRCC cells [53]. miR-138 can also suppress the invasion and metastasis of ovarian cancer cells by downregulating HIF-1α [51, 52]. Besides, miR-526b-3p [60], miR-18a [56], miR-338-3p [117], and miR-622 [63] may also inhibit the invasion and metastasis of cancer cells through targeting HIF-1α.
There are also some reports on the pro-metastasis role of HIF-1α-related miRNAs in human cancers. Hypoxia induced miR-210 promotes invasion and migration in HCC [82], gastrointestinal cancer [78], and ovarian cancer [76]. Moreover, hypoxia-induced miR-184 can facilitate the invasion and migration of glioma cells by targeting FIH-1 [99]. Ectopic expression of HIF-1α and miR-21 contributes to the migration and invasion of OSCC [88]. In gastric cancer, both hypoxia induced miR-224 [91] and miR-421 [92] can promote migration and invasion of cancer cells. Under hypoxia, upregulated miR-191 can promote migration of lung cancer cells by targeting nuclear factor 1α (NFIA) [90].
As for EMT and ECM, overexpression of miR-138 can inhibit EMT in GBC cells, at part, by targeting HIF-1α [52]. In HCC cells, restoration of miR-122 inhibits the EMT by targeting HIF-1α [65]. miR-338-3p can inhibit EMT in NPC cells via downregulating HIF-1α [117]. In colorectal cancer, low level of miR-34a is necessary for the hypoxia-induced EMT, and miR-34a can reduce the expression of protein phosphatase 1 regulatory inhibitor subunit 11(PPP1R11) to prevent activation of signal transducer and activator of transcription (STAT3) [105]. miR-199a can inhibit ECM remodeling in EOC cells through inhibition of HIF-1α [47]. Recently, a novel feedback circuit consisting of HIF-1α/miR-210/HIF-3α is found to promote ECM remodeling and metastasis in HCC cells [82]. However, HIF-1α induced miR-210 can promote EMT by directly targeting HOXA9 in gastric cancer cells [84]. Han et al. have demonstrated that the formation of cancer stem cells (CSC)-like cells biological features, especially EMT, are associated with overexpression of HIF-1α, and both of which are regulated by miR-21 [48].
Angiogenesis
Angiogenesis is a multistep process of blood vessels neoformation which provides oxygen and nutrients to cells, allowing discharge of metabolic waste [125]. During the neoplastic processes, increased demand for nutrients of tumor cells drives a disruption of the balance between proangiogenic and antiangiogenic factors, causing the maintenance of primary tumors and the promotion of metastasis at secondary sites [126]. Hypoxia is a well-known trigger for its role in promoting neo-vessel formation. Recent studies have extensively explored the mechanisms of angiogenesis in tumor cells, and miRNAs/HIF-1α loops are found to be involved in the various steps of the angiogenic response. miR-199a-5p may inhibit the angiogenesis of prostate adenocarcinoma cells by targeting HIF-1α [44]. Upregulation of miR-497 can significantly inhibit the angiogenesis of breast cancer cells via inhibiting HIF-1α [62]. Silencing miR-22 enhances the angiogenesis of colon cancer cells through upregulation of HIF-1α and subsequently facilitates the development of colon cancer [61]. Besides, miR-519c can inhibit the expression level of HIF-1α, followed by reduced angiogenesis in lung cancer cells [64].
Some miRNAs may promote the angiogenesis in human cancers. For instance, HIF-1α/miR-210/EFNA3 axis may promote the angiogenesis of schwannoma cells, and this pathway can be taken as effective targets for clinical auxiliary treatment of neurilemmoma [75]. HIF-1α induced miR-494 promotes the angiogenesis of lung cancer cells by suppressing PTEN and Akt/eNOS pathway [97]. Long non-coding RNA H19-derived miR-675-5p is required to induce hypoxia-mediated angiogenesis in glioma [107]. In gastric cancer cells, upregulated miR-382 promotes angiogenesis via targeting PTEN and reducing the secretion of vascular endothelial growth factors (VEGFs) [71]. Notably, researchers have confirmed a positive feedback between VEGF and miR-210 in non-tumor cells [127]. If confirmed in cancer cells, this finding may promote the development of anti-VEGF therapy (such as using miR-210 inhibitors).
Drug resistance and radioresistance
Multidrug resistance (MDR) and radioresistance remains the major obstacle to successful cancer therapy. Generally, anoxic cells present to be more chemoresistant and radioresistant than normoxic cells [128]. Activation of the HIF-1a related pathway may cause an increased chemoresistance [129, 130] and radioresistance [131] in tumor cells. For example, HIF-1a regulates the transcription of many genes involved in chemoresistance (ABC transporter genes) and radioresistance (p53, p21) in cancer cells [128]. Tumor cells resisting to chemotherapies and radioresistance is highly dependent on some mutational genes which might be the therapeutic targets. miR-21 can decrease the levels of the P-glycoprotein (P-gp) and HIF-1α proteins, thus increasing the sensitivity of the A2780/taxol cells to paclitaxel [71]. HIF-1α induced miR-21 can increase the radioresistance in cervical cancer cells [87]. In gastric cancer cells, both miR-421 and miR-27a are identified to induce drug resistance by targeting MDR-related genes, including MDR1/P-gp, and LRP. In HCC, two feedback loops, HIF-1α/miR-183/IDH2 and HIF-1α/miR-183/SOCS6/p-STAT3, are responsible for miR-183 upregulation, and upregulated miR-183 can induce MDR through suppressing SOCS6 [96].
Under some circumstances, some miRNAs/HIF-1α axis may play a positive role in reversing MDR. For instance, miR-199a can reverse the cisplatin (CDDP) resistance in OS cells, at least, by targeting HIF-1α [26]. In NPC, the cells pre-transfected with miR-338-3p can overcome hypoxia-mediated CDDP resistance [117]. miR-18a/HIF-1α axis negatively contributes to acquired tamoxifen resistance in the breast cancer cells [55]. Upregulation of miR-210 may reduce radioresistance of colon cancer cells by targeting Bcl-2 [83].
Although these findings show a great promise in reversing MDR and radioresistance, there are still obstacles to put certain knowledge into clinical practice, including lack of the effective delivery systems, inconsistent outcomes, and few clinical trials. In the future, great progresses of this field should be expected.
Clinical applications
Diagnostic value for cancers
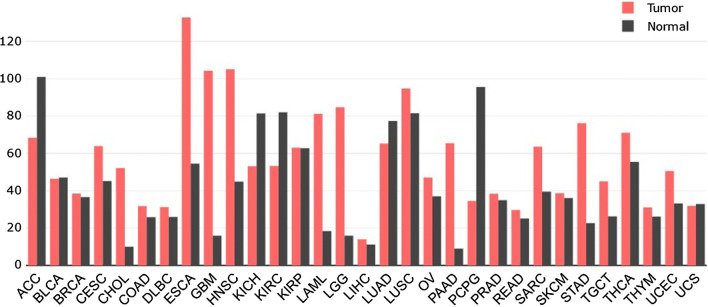
Increasing incidence rates of various cancers impose a heavy burden to most countries, better diagnostic tools, therefore, are needed. Many studies have showed strong evidence that miRNAs/HIF-1α loops were commonly dysregulated in cancers. Figure 4 shows the expression abundance of HIF-1α in various cancers tissues, which might be used to classify cancer subtypes.
Fig. 4.
Expression profile of HIF-1α across all tumor samples and paired normal tissues. Data publicly available from the Cancer Genome Atlas (TCGA) database (https://cancergenome.nih.gov/) were analyzed for the expression level of HIF-1α in carcinoma group and adjacent group of different types of cancers. The height of bar represents the median expression of certain tumor type or normal tissue. ACC, adrenocortical carcinoma; BLCA, bladder urothelial carcinoma; BRCA, breast invasive carcinoma; CESC, cervical squamous cell carcinoma and endocervical adenocarcinoma; CHOL, cholangiocarcinoma; COAD, colon adenocarcinoma; DLBC, lymphoid neoplasm diffuse large B-cell lymphoma; ESCA, esophageal carcinoma; GBM, glioblastoma multiforme; HNSC, head and neck squamous cell carcinoma; KICH, kidney chromophobe; KIRC, kidney renal clear cell carcinoma; KIRP, kidney renal papillary cell carcinoma; LAML, acute myeloid leukemia; LGG; brain lower grade glioma; LIHC, liver hepatocellular carcinoma; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; OV, ovarian serous cystadenocarcinoma; PAAD, pancreatic adenocarcinoma; PCPG, pheochromocytoma and paraganglioma; PRAD, prostate adenocarcinoma; READ, rectum adenocarcinoma; SARC, Sarcoma; SKCM, skin cutaneous melanoma; STAD, stomach adenocarcinoma; TGCT, testicular germ cell tumors; THCA, thyroid carcinoma; THYM, Thymoma; UCEC, uterine corpus endometrial carcinoma; UCS, uterine carcinosarcoma; HIF-1α, hypoxia inducible factor-1α
More importantly, circulating miRNAs are stable and easily detected, it may outperform other biomarkers when used to classify poorly differentiated tumors. Under hypoxic conditions, specific expression patterns of miRNAs may be the valuable biomarkers for cancer detection. miR-15a/16, miR-34a, miR-126 and miR-128 are significantly downregulated in A549 cells (lung cancer cell line) as comparted to control cells, while miR-21 and miR-210 are upregulated [132]. Distinct miRNAs signature (upregulated miR-210, miR-218, miR-224 and miR-452) is associated with melanoma subtypes and they are all regulated by HIF-1α [133]. Likewise, Blick et al. have identified a HIF-1α-related miRNAs signature in bladder cancer (upregulated miR-210, miR-193b, miR-145, miR-125-3p, miR-708 and miR-517a) which might be helpful to distinguish bladder cancer patients [98]. In gastric cancer, miR-107 is significantly higher in tumor tissues and serum as compared to matched normal controls. Its correlation with HIF-1α expression is also indicated in gastric cancer patients [94]. Upregulated miR-210 is even considered as a common risk factor for gastrointestinal cancer [78, 134].
Prognostic value for cancers
The expression of certain miRNAs has been reported to be associated with the clinical outcome in several cancers. Moreover, HIF-1α is differently expressed in various cancer tissues, and the role of HIF-1α on predicting overall survival (OS) may be variable. Therefore, the combination of miRNAs and HIF-1α seems to be more valuable.
One of the most extensive hypoxamiRs for predicting prognosis is miR-210. Overexpression of miR-210 is prevalent in HCC and breast cancer patients, and is significantly associated with worse survival with shorter disease-free (DFS) and OS [82, 135]. Serum level of miR-210 varies widely among mCRPC patients, and it correlates with the treatment response by evaluating the change in prostate specific antigen (PSA) [136]. Besides, coordinated activation of HIF-1α, CAIX, miR-210 and ISCU in OPSCC is associated with high recurrence rate and low OS of surgically treated patients [79].
Dysregulated miR-199a/HIF-1a regulatory loop (miR-199a low/HIF-1a high) is an independent prognostic factor for poor survival of patients with HCC and pancreatic cancer [29, 106]. Circulating exosomal miR-21 levels are correlated with the T stage, lymph node metastasis, and HIF-1a expression in OSCC patients [88]. The gastric cancer patients with low level of miR-421 have a significant longer OS [92]. miR-548an is significantly correlated with increased tumor size, advanced TNM stage, distant metastasis, and poor prognosis [101]. Lack of miR-526b-3p expression in colon cancer is positively related to the advanced stage and poor prognosis [60]. The expression of HIF-1α is positively correlated with the stage of pancreatic cancer, and miR-142 is negatively correlated with lymphatic metastasis [58]. miR-138 directly targets SRY-related high mobility group box 4 (SOX4) and HIF-1a, and the patients with miR-138 low/SOX4 high signature are predominant in late stage and tend to have malignant phenotypes [51]. In addition, increased HIF-N (HIF-1α in nucleus) can be a negative prognostic factor for both disease-specific survival (DSS) and progression-free survival (PFS) in prostate adenocarcinoma patients [44]. However, the role of miRNAs and HIF-1α on prognosis analysis still need in-depth studies, more pre-clinical and clinical researches with longer follow-up and larger cohorts should be carried out.
Targeted therapy for cancers
Single miRNA can regulate several target genes, while a mRNA can be modulated by multiple miRNAs simultaneously [27]. It is easy to understand that miRNAs can be directly or indirectly regulated by some genes, including their target mRNAs. To our knowledge, the regulatory feedback loops, such as miR-124/p63 [137], E2F transcription factor 3 b (E2F3b)/miR-200b [138], miR-139/Jun proto-oncogene (Jun) [139], signal transducer and activator of transcription 1(STAT1)/miR-155-5p [140], miR-203/Snail family transcriptional repressor 1(SNAI1) [141], play important roles in tumorigenesis. It seems that individually targeting a single miRNA or one target mRNA may be much weaker in cancer treatment, whereas a combination of multiple miRNAs regulating the same target genes may be a logical approach [27]. Therefore, a better understanding of the reciprocal regulation between miRNAs and HIF-1α is needed.
There is a positive association between the levels of miR-182 and ER, as well as progesterone receptor (PR) in breast cancer patients, especially in ER and PR positive cases [142]. Moreover, there is an improvement in 5-year OS in patients receiving radiotherapy (RT) and cotinamide (CON) as compared with RT alone in patients with high miR-210 expression [143]. These facts indicate that the level of HIF-1α-related miRNAs may influence the curative effects of cancer treatments. However, the major problem regarding miRNA/HIF-1α loop/axis is whether they can be efficiently targeted in cancer cells. It is conceivable that some miRNAs inhibitors or mimics in combination with the anticancer agents may be an available strategy for cancer treatment in the next years.
For example, transfection of miR-199a mimic into osteosarcoma cells can significantly sensitize cells to CDDP through inhibition of HIF-1α [26]. miR-186/HIF-1α axis seems to be a promising therapeutic target in an in vivo xenograft tumor experiment of gastric cancer [59]. In glioma, miR-675-5p might be a new therapeutic target for hypoxia-mediated angiogenesis since miR-675-5p inhibitor is able to counteract hypoxia process [107]. miR-135b mimics can act as a tumor promoter by targeting the HIF pathway in genetically defined mouse model of HNSCC [70]. AntagomiR-21 can effectively suppress tumor growth and angiogenesis in VEGFR2-luc mouse breast tumor model and bioluminescent imaging can be used as a tool for noninvasively and continuously monitoring tumor angiogenesis in vivo [144]. Yu et al. have proposed that miR-210 mimic was an ideal molecular to block the invasion and metastasis of gastric cancer [84]. Besides, miR-210 might open up a new therapeutic avenue to hypoxia GSCs [145].
Conclusion and perspective
Here, we attempt to briefly evaluate the available evidence on the bidirectional interactions between HIF-1α and miRNAs in human cancers, the role of miRNAs/HIF-1α crosstalk in cancer progression, and the clinical values of miRNAs and HIF-1α in cancer management. miRNAs can regulate the activity of HIF-1α, and HIF-1α can in turn modulate many miRNAs, thus the crosstalk between HIF-1α and miRNAs plays a crucial role in the tumor progression. To date, while many studies have confirmed miRNAs/HIF-1α loops/axis involved in carcinogenesis, some problems are still existed. For example, most of the studies only focus on the temporal alterations of miRNAs and HIF-1α under hypoxia, whereas the crosstalk between HIF-1α and miRNAs maybe a dynamic process. Furthermore, the majority of studies on miRNAs/HIF-1α loops/axis are performed in one or two types of cancer cell lines, it is difficult to confirm whether the findings are universal or cancer type-dependent. Besides, whether the difference of miRNA target sites can influence the miRNAs/HIF-1α interaction is largely unknown [146]. Finally, although we have made great efforts to explore the effective small molecules that target miRNAs/HIF-1α pathway, none of these molecules can effectively restore the levels of miRNAs and HIF-1α as their exclusive targets and the off-target effects.
Although many important questions refer to miRNAs/HIF-1α crosstalk still remain to be answered, the potential of miRNAs and HIF-1α being the clinical tools in future emphasizes the significance of the studies discussed here. From the perspective of researchers, the following points may be available. First, more basic and pre-clinical studies should be performed, and we should pay more attention to follow the dynamic alterations in studies. Second, new technologies and research tools are strongly needed. For example, bioinformatics analysis may greatly promote the development of cancer research studies in the next few years. New technologies like next-generation sequencing may be powerful tools for researches. Third, to minimize the errors in basic studies and clinical trials, strict criterions are also required, including standard operation procedures, standard platforms, and so on. Finally, data sharing and large sample analysis can improve the credibility of our studies.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
This study was supported in part by Grant from the National Natural Scientific Foundation of China (81171923), Grant from the State Key Laboratory of Cancer Biology (CBSKL2014Z13) and Grant from the National Clinical Research Center for Digestive Diseases(2015BAI13B07). It was not supported by any private or public company or organization.
Compliance with ethical standards
Conflict of interest
The authors declare no conflict of interests.
Footnotes
Wanli Yang, Jiaojiao Ma and Wei Zhou have contributed equally to this work.
References
- 1.Semenza GL. Targeting HIF-1 for cancer therapy. Nat Rev Cancer. 2003;3(10):721–732. doi: 10.1038/nrc1187. [DOI] [PubMed] [Google Scholar]
- 2.Semenza GL. Hypoxia-inducible factor 1 (HIF-1) pathway. Sci STKE. 2007;2007(407):cm8. doi: 10.1126/stke.4072007cm8. [DOI] [PubMed] [Google Scholar]
- 3.Jaakkola P, Mole DR, Tian YM, Wilson MI, Gielbert J, Gaskell SJ, von Kriegsheim A, Hebestreit HF, Mukherji M, Schofield CJ, Maxwell PH, Pugh CW, Ratcliffe PJ. Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science (New York, NY) 2001;292(5516):468–472. doi: 10.1126/science.1059796. [DOI] [PubMed] [Google Scholar]
- 4.Semenza GL. Hydroxylation of HIF-1: oxygen sensing at the molecular level. Physiology (Bethesda, Md) 2004;19:176–182. doi: 10.1152/physiol.00001.2004. [DOI] [PubMed] [Google Scholar]
- 5.Chan DA, Sutphin PD, Yen SE, Giaccia AJ. Coordinate regulation of the oxygen-dependent degradation domains of hypoxia-inducible factor 1 alpha. Mol Cell Biol. 2005;25(15):6415–6426. doi: 10.1128/mcb.25.15.6415-6426.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.D’Angelo G, Duplan E, Boyer N, Vigne P, Frelin C. Hypoxia up-regulates prolyl hydroxylase activity: a feedback mechanism that limits HIF-1 responses during reoxygenation. J Biol Chem. 2003;278(40):38183–38187. doi: 10.1074/jbc.M302244200. [DOI] [PubMed] [Google Scholar]
- 7.Majmundar AJ, Wong WJ, Simon MC. Hypoxia-inducible factors and the response to hypoxic stress. Mol Cell. 2010;40(2):294–309. doi: 10.1016/j.molcel.2010.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaelin WG, Jr, Ratcliffe PJ. Oxygen sensing by metazoans: the central role of the HIF hydroxylase pathway. Mol Cell. 2008;30(4):393–402. doi: 10.1016/j.molcel.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 9.Semenza GL. Defining the role of hypoxia-inducible factor 1 in cancer biology and therapeutics. Oncogene. 2010;29(5):625–634. doi: 10.1038/onc.2009.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Latif F, Tory K, Gnarra J, Yao M, Duh FM, Orcutt ML, Stackhouse T, Kuzmin I, Modi W, Geil L, et al. Identification of the von Hippel-Lindau disease tumor suppressor gene. Science (New York, NY) 1993;260(5112):1317–1320. doi: 10.1126/science.8493574. [DOI] [PubMed] [Google Scholar]
- 11.Webb JD, Coleman ML, Pugh CW. Hypoxia, hypoxia-inducible factors (HIF), HIF hydroxylases and oxygen sensing. Cell Mol Life Sci. 2009;66(22):3539–3554. doi: 10.1007/s00018-009-0147-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chandel NS, McClintock DS, Feliciano CE, Wood TM, Melendez JA, Rodriguez AM, Schumacker PT. Reactive oxygen species generated at mitochondrial complex III stabilize hypoxia-inducible factor-1alpha during hypoxia: a mechanism of O2 sensing. J Biol Chem. 2000;275(33):25130–25138. doi: 10.1074/jbc.M001914200. [DOI] [PubMed] [Google Scholar]
- 13.Lau KW, Tian YM, Raval RR, Ratcliffe PJ, Pugh CW. Target gene selectivity of hypoxia-inducible factor-alpha in renal cancer cells is conveyed by post-DNA-binding mechanisms. Br J Cancer. 2007;96(8):1284–1292. doi: 10.1038/sj.bjc.6603675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nagaraju GP, Bramhachari PV, Raghu G, El-Rayes BF. Hypoxia inducible factor-1alpha: its role in colorectal carcinogenesis and metastasis. Cancer Lett. 2015;366(1):11–18. doi: 10.1016/j.canlet.2015.06.005. [DOI] [PubMed] [Google Scholar]
- 15.Haque I, Banerjee S, Mehta S, De A, Majumder M, Mayo MS, Kambhampati S, Campbell DR, Banerjee SK. Cysteine-rich 61-connective tissue growth factor-nephroblastoma-overexpressed 5 (CCN5)/Wnt-1-induced signaling protein-2 (WISP-2) regulates microRNA-10b via hypoxia-inducible factor-1alpha-TWIST signaling networks in human breast cancer cells. J Biol Chem. 2011;286(50):43475–43485. doi: 10.1074/jbc.M111.284158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chai ZT, Kong J, Zhu XD, Zhang YY, Lu L, Zhou JM, Wang LR, Zhang KZ, Zhang QB, Ao JY, Wang M, Wu WZ, Wang L, Tang ZY, Sun HC. MicroRNA-26a inhibits angiogenesis by down-regulating VEGFA through the PIK3C2alpha/Akt/HIF-1alpha pathway in hepatocellular carcinoma. PLoS One. 2013;8(10):e77957. doi: 10.1371/journal.pone.0077957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang C, Tian W, Meng L, Qu L, Shou C. PRL-3 promotes gastric cancer migration and invasion through a NF-kappaB-HIF-1alpha-miR-210 axis. J Mol Med (Berlin, Germany) 2016;94(4):401–415. doi: 10.1007/s00109-015-1350-7. [DOI] [PubMed] [Google Scholar]
- 18.Brahimi-Horn MC, Ben-Hail D, Ilie M, Gounon P, Rouleau M, Hofman V, Doyen J, Mari B, Shoshan-Barmatz V, Hofman P, Pouyssegur J, Mazure NM. Expression of a truncated active form of VDAC1 in lung cancer associates with hypoxic cell survival and correlates with progression to chemotherapy resistance. Can Res. 2012;72(8):2140–2150. doi: 10.1158/0008-5472.can-11-3940. [DOI] [PubMed] [Google Scholar]
- 19.Zhong H, De Marzo AM, Laughner E, Lim M, Hilton DA, Zagzag D, Buechler P, Isaacs WB, Semenza GL, Simons JW. Overexpression of hypoxia-inducible factor 1alpha in common human cancers and their metastases. Can Res. 1999;59(22):5830–5835. [PubMed] [Google Scholar]
- 20.Shen XH, Qi P, Du X. Long non-coding RNAs in cancer invasion and metastasis. Mod Pathol. 2015;28(1):4–13. doi: 10.1038/modpathol.2014.75. [DOI] [PubMed] [Google Scholar]
- 21.Munipalle PC, Viswanath YK, Davis PA, Scoones D. Prognostic value of hypoxia inducible factor 1alpha in esophageal squamous cell carcinoma. Dis Esophagus. 2011;24(3):177–181. doi: 10.1111/j.1442-2050.2010.01122.x. [DOI] [PubMed] [Google Scholar]
- 22.Marton I, Knezevic F, Ramic S, Milosevic M, Tomas D. Immunohistochemical expression and prognostic significance of HIF-1alpha and VEGF-C in neuroendocrine breast cancer. Anticancer Res. 2012;32(12):5227–5232. [PubMed] [Google Scholar]
- 23.Shen W, Li HL, Liu L, Cheng JX. Expression levels of PTEN, HIF-1alpha, and VEGF as prognostic factors in ovarian cancer. Eur Rev Med Pharmacol Sci. 2017;21(11):2596–2603. [PubMed] [Google Scholar]
- 24.Gong L, Zhang W, Zhou J, Lu J, Xiong H, Shi X, Chen J. Prognostic value of HIFs expression in head and neck cancer: a systematic review. PLoS One. 2013;8(9):e75094. doi: 10.1371/journal.pone.0075094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ben Lassoued A, Beaufils N, Dales JP, Gabert J. Hypoxia-inducible factor-1alpha as prognostic marker. Expert Opin Med Diagn. 2013;7(1):53–70. doi: 10.1517/17530059.2012.719022. [DOI] [PubMed] [Google Scholar]
- 26.Keremu A, Aini A, Maimaitirexiati Y, Liang Z, Aila P, Xierela P, Tusun A, Moming H, Yusufu A. Overcoming cisplatin resistance in osteosarcoma through the miR-199a-modulated inhibition of HIF-1alpha. Biosci Rep. 2017 doi: 10.1042/bsr20170080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gao Y, Feng B, Lu L, Han S, Chu X, Chen L, Wang R. MiRNAs and E2F3: a complex network of reciprocal regulations in human cancers. Oncotarget. 2017 doi: 10.18632/oncotarget.17364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu W, Zhang Z, Zou K, Cheng Y, Yang M, Chen H, Wang H, Zhao J, Chen P, He L, Chen X, Geng L, Gong S. MiR-1 suppresses tumor cell proliferation in colorectal cancer by inhibition of Smad3-mediated tumor glycolysis. Cell Death Dis. 2017;8(5):e2761. doi: 10.1038/cddis.2017.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li B, He L, Zuo D, He W, Wang Y, Zhang Y, Liu W, Yuan Y. Mutual regulation of MiR-199a-5p and HIF-1alpha modulates the warburg effect in hepatocellular carcinoma. J Cancer. 2017;8(6):940–949. doi: 10.7150/jca.17496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu N, Xia WY, Liu SS, Chen HY, Sun L, Liu MY, Li LF, Lu HM, Fu YJ, Wang P, Wu H, Gao JX. MicroRNA-101 targets von Hippel-Lindau tumor suppressor (VHL) to induce HIF1alpha mediated apoptosis and cell cycle arrest in normoxia condition. Sci Rep. 2016;6:20489. doi: 10.1038/srep20489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dang K, Myers KA. The role of hypoxia-induced miR-210 in cancer progression. Int J Mol Sci. 2015;16(3):6353–6372. doi: 10.3390/ijms16036353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang Q, Zhang RW, Sui PC, He HT, Ding L. Dysregulation of non-coding RNAs in gastric cancer. World J Gastroenterol. 2015;21(39):10956–10981. doi: 10.3748/wjg.v21.i39.10956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li M, Wang Y, Song Y, Bu R, Yin B, Fei X, Guo Q, Wu B. MicroRNAs in renal cell carcinoma: a systematic review of clinical implications (Review) Oncol Rep. 2015;33(4):1571–1578. doi: 10.3892/or.2015.3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ang C, O’Reilly EM, Abou-Alfa GK. MicroRNA, hypoxic stress and hepatocellular carcinoma: future directions. J Gastroenterol Hepatol. 2011;26(11):1586–1588. doi: 10.1111/j.1440-1746.2011.06903.x. [DOI] [PubMed] [Google Scholar]
- 35.Ling H, Fabbri M, Calin GA. MicroRNAs and other non-coding RNAs as targets for anticancer drug development. Nat Rev Drug Discov. 2013;12(11):847–865. doi: 10.1038/nrd4140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schmitz SU, Grote P, Herrmann BG. Mechanisms of long noncoding RNA function in development and disease. Cell Mol Life Sci. 2016;73(13):2491–2509. doi: 10.1007/s00018-016-2174-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Suzuki H, Maruyama R, Yamamoto E, Niinuma T, Kai M. Relationship between noncoding RNA dysregulation and epigenetic mechanisms in cancer. Adv Exp Med Biol. 2016;927:109–135. doi: 10.1007/978-981-10-1498-7_4. [DOI] [PubMed] [Google Scholar]
- 38.Yang Z, Guo X, Li G, Shi Y, Li L. Long noncoding RNAs as potential biomarkers in gastric cancer: opportunities and challenges. Cancer Lett. 2016;371(1):62–70. doi: 10.1016/j.canlet.2015.11.011. [DOI] [PubMed] [Google Scholar]
- 39.Dong L, Qi P, Xu MD, Ni SJ, Huang D, Xu QH, Weng WW, Tan C, Sheng WQ, Zhou XY, Du X. Circulating CUDR, LSINCT-5 and PTENP1 long noncoding RNAs in sera distinguish patients with gastric cancer from healthy controls. Int J Cancer. 2015;137(5):1128–1135. doi: 10.1002/ijc.29484. [DOI] [PubMed] [Google Scholar]
- 40.Huang J, Zhou N, Watabe K, Lu Z, Wu F, Xu M, Mo YY. Long non-coding RNA UCA1 promotes breast tumor growth by suppression of p27 (Kip1) Cell Death Dis. 2014;5:e1008. doi: 10.1038/cddis.2013.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li T, Yang XD, Ye CX, Shen ZL, Yang Y, Wang B, Guo P, Gao ZD, Ye YJ, Jiang KW, Wang S. Long noncoding RNA HIT000218960 promotes papillary thyroid cancer oncogenesis and tumor progression by upregulating the expression of high mobility group AT-hook 2 (HMGA2) gene. Cell Cycle (Georgetown, Tex) 2017;16(2):224–231. doi: 10.1080/15384101.2016.1261768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shih JW, Kung HJ. Long non-coding RNA and tumor hypoxia: new players ushered toward an old arena. J Biomed Sci. 2017;24(1):53. doi: 10.1186/s12929-017-0358-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang P, Ning S, Zhang Y, Li R, Ye J, Zhao Z, Zhi H, Wang T, Guo Z, Li X. Identification of lncRNA-associated competing triplets reveals global patterns and prognostic markers for cancer. Nucleic Acids Res. 2015;43(7):3478–3489. doi: 10.1093/nar/gkv233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhong J, Huang R, Su Z, Zhang M, Xu M, Gong J, Chen N, Zeng H, Chen X, Zhou Q. Downregulation of miR-199a-5p promotes prostate adeno-carcinoma progression through loss of its inhibition of HIF-1alpha. Oncotarget. 2017 doi: 10.18632/oncotarget.18315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shang W, Chen X, Nie L, Xu M, Chen N, Zeng H, Zhou Q. MiR199b suppresses expression of hypoxia-inducible factor 1alpha (HIF-1alpha) in prostate cancer cells. Int J Mol Sci. 2013;14(4):8422–8436. doi: 10.3390/ijms14048422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ye H, Pang L, Wu Q, Zhu Y, Guo C, Deng Y, Zheng X. A critical role of mir-199a in the cell biological behaviors of colorectal cancer. Diagn Pathol. 2015;10:65. doi: 10.1186/s13000-015-0260-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Joshi HP, Subramanian IV, Schnettler EK, Ghosh G, Rupaimoole R, Evans C, Saluja M, Jing Y, Cristina I, Roy S, Zeng Y, Shah VH, Sood AK, Ramakrishnan S. Dynamin 2 along with microRNA-199a reciprocally regulate hypoxia-inducible factors and ovarian cancer metastasis. Proc Natl Acad Sci USA. 2014;111(14):5331–5336. doi: 10.1073/pnas.1317242111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Obernosterer G, Leuschner PJ, Alenius M, Martinez J. Post-transcriptional regulation of microRNA expression. RNA (New York, NY) 2006;12(7):1161–1167. doi: 10.1261/rna.2322506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weber MJ. New human and mouse microRNA genes found by homology search. FEBS J. 2005;272(1):59–73. doi: 10.1111/j.1432-1033.2004.04389.x. [DOI] [PubMed] [Google Scholar]
- 50.Jin Y, Chen D, Cabay RJ, Wang A, Crowe DL, Zhou X. Role of microRNA-138 as a potential tumor suppressor in head and neck squamous cell carcinoma. Int Rev Cell Mol Biol. 2013;303:357–385. doi: 10.1016/b978-0-12-407697-6.00009-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yeh YM, Chuang CM, Chao KC, Wang LH. MicroRNA-138 suppresses ovarian cancer cell invasion and metastasis by targeting SOX4 and HIF-1alpha. Int J Cancer. 2013;133(4):867–878. doi: 10.1002/ijc.28086. [DOI] [PubMed] [Google Scholar]
- 52.Cai Q, Wang Z, Wang S, Weng M, Zhou D, Li C, Wang J, Chen E, Quan Z. Long non-coding RNA LINC00152 promotes gallbladder cancer metastasis and epithelial–mesenchymal transition by regulating HIF-1alpha via miR-138. Open Biol. 2017 doi: 10.1098/rsob.160247. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 53.Song T, Zhang X, Wang C, Wu Y, Cai W, Gao J, Hong B. MiR-138 suppresses expression of hypoxia-inducible factor 1alpha (HIF-1alpha) in clear cell renal cell carcinoma 786-O cells. Asian Pac J Cancer Prev. 2011;12(5):1307–1311. [PubMed] [Google Scholar]
- 54.Chen Y, Zhang Z, Luo C, Chen Z, Zhou J. MicroRNA-18b inhibits the growth of malignant melanoma via inhibition of HIF-1alpha-mediated glycolysis. Oncol Rep. 2016;36(1):471–479. doi: 10.3892/or.2016.4824. [DOI] [PubMed] [Google Scholar]
- 55.Li X, Wu Y, Liu A, Tang X. Long non-coding RNA UCA1 enhances tamoxifen resistance in breast cancer cells through a miR-18a-HIF1alpha feedback regulatory loop. Tumour Biol. 2016;37(11):14733–14743. doi: 10.1007/s13277-016-5348-8. [DOI] [PubMed] [Google Scholar]
- 56.Wu F, Huang W, Wang X. microRNA-18a regulates gastric carcinoma cell apoptosis and invasion by suppressing hypoxia-inducible factor-1alpha expression. Exp Ther Med. 2015;10(2):717–722. doi: 10.3892/etm.2015.2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Han C, Shen JK, Hornicek FJ, Kan Q. Duan Z (2017) Regulation of microRNA-1 (miR-1) expression in human cancer. Biochem Biophys Acta. 1860;2:227–232. doi: 10.1016/j.bbagrm.2016.12.004. [DOI] [PubMed] [Google Scholar]
- 58.Lu Y, Ji N, Wei W, Sun W, Gong X, Wang X. MiR-142 modulates human pancreatic cancer proliferation and invasion by targeting hypoxia-inducible factor 1 (HIF-1alpha) in the tumor microenvironments. Biol Open. 2017;6(2):252–259. doi: 10.1242/bio.021774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu L, Wang Y, Bai R, Yang K, Tian Z. MiR-186 inhibited aerobic glycolysis in gastric cancer via HIF-1alpha regulation. Oncogenesis. 2016;5:e224. doi: 10.1038/oncsis.2016.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang R, Zhao J, Xu J, Wang J, Jia J. miR-526b-3p functions as a tumor suppressor in colon cancer by regulating HIF-1alpha. Am J Transl Res. 2016;8(6):2783–2789. [PMC free article] [PubMed] [Google Scholar]
- 61.Yamakuchi M, Yagi S, Ito T, Lowenstein CJ. MicroRNA-22 regulates hypoxia signaling in colon cancer cells. PLoS One. 2011;6(5):e20291. doi: 10.1371/journal.pone.0020291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wu Z, Cai X, Huang C, Xu J, Liu A. miR-497 suppresses angiogenesis in breast carcinoma by targeting HIF-1alpha. Oncol Rep. 2016;35(3):1696–1702. doi: 10.3892/or.2015.4529. [DOI] [PubMed] [Google Scholar]
- 63.Cheng CW, Chen PM, Hsieh YH, Weng CC, Chang CW, Yao CC, Hu LY, Wu PE, Shen CY. Foxo3a-mediated overexpression of microRNA-622 suppresses tumor metastasis by repressing hypoxia-inducible factor-1alpha in ERK-responsive lung cancer. Oncotarget. 2015;6(42):44222–44238. doi: 10.18632/oncotarget.5826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cha ST, Chen PS, Johansson G, Chu CY, Wang MY, Jeng YM, Yu SL, Chen JS, Chang KJ, Jee SH, Tan CT, Lin MT, Kuo ML. MicroRNA-519c suppresses hypoxia-inducible factor-1alpha expression and tumor angiogenesis. Can Res. 2010;70(7):2675–2685. doi: 10.1158/0008-5472.can-09-2448. [DOI] [PubMed] [Google Scholar]
- 65.Ambade A, Satishchandran A, Szabo G. Alcoholic hepatitis accelerates early hepatobiliary cancer by increasing stemness and miR-122-mediated HIF-1alpha activation. Sci Rep. 2016;6:21340. doi: 10.1038/srep21340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Takahashi K, Yan IK, Haga H, Patel T. Modulation of hypoxia-signaling pathways by extracellular linc-RoR. J Cell Sci. 2014;127(Pt 7):1585–1594. doi: 10.1242/jcs.141069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kulshreshtha R, Ferracin M, Negrini M, Calin GA, Davuluri RV, Ivan M. Regulation of microRNA expression: the hypoxic component. Cell Cycle (Georgetown, Tex) 2007;6(12):1426–1431. doi: 10.4161/cc.6.12.4410. [DOI] [PubMed] [Google Scholar]
- 68.Voellenkle C, Rooij J, Guffanti A, Brini E, Fasanaro P, Isaia E, Croft L, David M, Capogrossi MC, Moles A, Felsani A, Martelli F. Deep-sequencing of endothelial cells exposed to hypoxia reveals the complexity of known and novel microRNAs. RNA (New York, NY) 2012;18(3):472–484. doi: 10.1261/rna.027615.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gee HE, Ivan C, Calin GA, Ivan M. HypoxamiRs and cancer: from biology to targeted therapy. Antioxid Redox Signal. 2014;21(8):1220–1238. doi: 10.1089/ars.2013.5639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang L, Sun ZJ, Bian Y, Kulkarni AB. MicroRNA-135b acts as a tumor promoter by targeting the hypoxia-inducible factor pathway in genetically defined mouse model of head and neck squamous cell carcinoma. Cancer Lett. 2013;331(2):230–238. doi: 10.1016/j.canlet.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Xie Z, Cao L, Zhang J. miR-21 modulates paclitaxel sensitivity and hypoxia-inducible factor-1alpha expression in human ovarian cancer cells. Oncol Lett. 2013;6(3):795–800. doi: 10.3892/ol.2013.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Han M, Wang Y, Liu M, Bi X, Bao J, Zeng N, Zhu Z, Mo Z, Wu C, Chen X. MiR-21 regulates epithelial–mesenchymal transition phenotype and hypoxia-inducible factor-1alpha expression in third-sphere forming breast cancer stem cell-like cells. Cancer Sci. 2012;103(6):1058–1064. doi: 10.1111/j.1349-7006.2012.02281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yang W, Ma J, Zhou W, Zhou X, Cao B, Fan D, Hong L. Biological implications and clinical value of mir-210 in gastrointestinal cancer. Expert Rev Gastroenterol Hepatol. 2017;11(6):539–548. doi: 10.1080/17474124.2017.1309281. [DOI] [PubMed] [Google Scholar]
- 74.Ren CX, Leng RX, Fan YG, Pan HF, Wu CH, Ye DQ. MicroRNA-210 and its theranostic potential. Expert Opin Ther Targets. 2016;20(11):1325–1338. doi: 10.1080/14728222.2016.1206890. [DOI] [PubMed] [Google Scholar]
- 75.Wang Z, Deng M, Liu Z, Wu S. Hypoxia-induced miR-210 promoter demethylation enhances proliferation, autophagy and angiogenesis of schwannoma cells. Oncol Rep. 2017;37(5):3010–3018. doi: 10.3892/or.2017.5511. [DOI] [PubMed] [Google Scholar]
- 76.Liu T, Zhao L, Chen W, Li Z, Hou H, Ding L, Li X. Inactivation of von Hippel-Lindau increases ovarian cancer cell aggressiveness through the HIF1alpha/miR-210/VMP1 signaling pathway. Int J Mol Med. 2014;33(5):1236–1242. doi: 10.3892/ijmm.2014.1661. [DOI] [PubMed] [Google Scholar]
- 77.Li L, Huang K, You Y, Fu X, Hu L, Song L, Meng Y. Hypoxia-induced miR-210 in epithelial ovarian cancer enhances cancer cell viability via promoting proliferation and inhibiting apoptosis. Int J Oncol. 2014;44(6):2111–2120. doi: 10.3892/ijo.2014.2368. [DOI] [PubMed] [Google Scholar]
- 78.Chen KC, Liao YC, Wang JY, Lin YC, Chen CH, Juo SH. Oxidized low-density lipoprotein is a common risk factor for cardiovascular diseases and gastroenterological cancers via epigenomical regulation of microRNA-210. Oncotarget. 2015;6(27):24105–24118. doi: 10.18632/oncotarget.4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Saenz-de-Santa-Maria I, Bernardo-Castineira C, Secades P, Bernaldo-de-Quiros S, Rodrigo JP, Astudillo A, Chiara MD. Clinically relevant HIF-1alpha-dependent metabolic reprogramming in oropharyngeal squamous cell carcinomas includes coordinated activation of CAIX and the miR-210/ISCU signaling axis, but not MCT1 and MCT4 upregulation. Oncotarget. 2017;8(8):13730–13746. doi: 10.18632/oncotarget.14629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Merlo A, Bernardo-Castineira C, Saenz-de-Santa-Maria I, Pitiot AS, Balbin M, Astudillo A, Valdes N, Scola B, Del Toro R, Mendez-Ferrer S, Piruat JI, Suarez C, Chiara MD. Role of VHL, HIF1A and SDH on the expression of miR-210: implications for tumoral pseudo-hypoxic fate. Oncotarget. 2017;8(4):6700–6717. doi: 10.18632/oncotarget.14265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nakada C, Tsukamoto Y, Matsuura K, Nguyen TL, Hijiya N, Uchida T, Sato F, Mimata H, Seto M, Moriyama M. Overexpression of miR-210, a downstream target of HIF1alpha, causes centrosome amplification in renal carcinoma cells. J Pathol. 2011;224(2):280–288. doi: 10.1002/path.2860. [DOI] [PubMed] [Google Scholar]
- 82.Kai AK, Chan LK, Lo RC, Lee JM, Wong CC, Wong JC, Ng IO. Down-regulation of TIMP2 by HIF-1alpha/miR-210/HIF-3alpha regulatory feedback circuit enhances cancer metastasis in hepatocellular carcinoma. Hepatology (Baltimore, MD) 2016;64(2):473–487. doi: 10.1002/hep.28577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sun Y, Xing X, Liu Q, Wang Z, Xin Y, Zhang P, Hu C, Liu Y. Hypoxia-induced autophagy reduces radiosensitivity by the HIF-1alpha/miR-210/Bcl-2 pathway in colon cancer cells. Int J Oncol. 2015;46(2):750–756. doi: 10.3892/ijo.2014.2745. [DOI] [PubMed] [Google Scholar]
- 84.Yu P, Fan S, Huang L, Yang L, Du Y. MIR210 as a potential molecular target to block invasion and metastasis of gastric cancer. Med Hypotheses. 2015;84(3):209–212. doi: 10.1016/j.mehy.2014.12.024. [DOI] [PubMed] [Google Scholar]
- 85.Chen WY, Liu WJ, Zhao YP, Zhou L, Zhang TP, Chen G, Shu H. Induction, modulation and potential targets of miR-210 in pancreatic cancer cells. Hepatobil Pancreat Dis Int. 2012;11(3):319–324. doi: 10.1016/S1499-3872(12)60168-4. [DOI] [PubMed] [Google Scholar]
- 86.Li CL, Zhou XL, Wang YD, Jing SW, Yang CR, Sun GG, Liu Q, Cheng YJ, Wang L. miR-210 regulates esophageal cancer cell proliferation by inducing G(2)/M phase cell cycle arrest through targeting PLK1. Mol Med Rep. 2014;10(4):2099–2104. doi: 10.3892/mmr.2014.2416. [DOI] [PubMed] [Google Scholar]
- 87.Song L, Liu S, Zhang L, Yao H, Gao F, Xu D, Li Q. MiR-21 modulates radiosensitivity of cervical cancer through inhibiting autophagy via the PTEN/Akt/HIF-1alpha feedback loop and the Akt-mTOR signaling pathway. Tumour Biol. 2016;37(9):12161–12168. doi: 10.1007/s13277-016-5073-3. [DOI] [PubMed] [Google Scholar]
- 88.Li L, Li C, Wang S, Wang Z, Jiang J, Wang W, Li X, Chen J, Liu K, Li C, Zhu G. Exosomes derived from hypoxic oral squamous cell carcinoma cells deliver miR-21 to normoxic cells to elicit a prometastatic phenotype. Can Res. 2016;76(7):1770–1780. doi: 10.1158/0008-5472.can-15-1625. [DOI] [PubMed] [Google Scholar]
- 89.Mace TA, Collins AL, Wojcik SE, Croce CM, Lesinski GB, Bloomston M. Hypoxia induces the overexpression of microRNA-21 in pancreatic cancer cells. J Surg Res. 2013;184(2):855–860. doi: 10.1016/j.jss.2013.04.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhao J, Qiao CR, Ding Z, Sheng YL, Li XN, Yang Y, Zhu DY, Zhang CY, Liu DL, Wu K, Zhao S. A novel pathway in NSCLC cells: miR191, targeting NFIA, is induced by chronic hypoxia, and promotes cell proliferation and migration. Mol Med Rep. 2017;15(3):1319–1325. doi: 10.3892/mmr.2017.6100. [DOI] [PubMed] [Google Scholar]
- 91.He C, Wang L, Zhang J, Xu H. Hypoxia-inducible microRNA-224 promotes the cell growth, migration and invasion by directly targeting RASSF8 in gastric cancer. Mol Cancer. 2017;16(1):35. doi: 10.1186/s12943-017-0603-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ge X, Liu X, Lin F, Li P, Liu K, Geng R, Dai C, Lin Y, Tang W, Wu Z, Chang J, Lu J, Li J. MicroRNA-421 regulated by HIF-1alpha promotes metastasis, inhibits apoptosis, and induces cisplatin resistance by targeting E-cadherin and caspase-3 in gastric cancer. Oncotarget. 2016;7(17):24466–24482. doi: 10.18632/oncotarget.8228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhao Q, Li Y, Tan BB, Fan LQ, Yang PG, Tian Y. HIF-1alpha induces multidrug resistance in gastric cancer cells by inducing MiR-27a. PLoS One. 2015;10(8):e0132746. doi: 10.1371/journal.pone.0132746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ayremlou N, Mozdarani H, Mowla SJ, Delavari A. Increased levels of serum and tissue miR-107 in human gastric cancer: correlation with tumor hypoxia. Cancer Biomark Sect A Dis Mark. 2015;15(6):851–860. doi: 10.3233/cbm-150529. [DOI] [PubMed] [Google Scholar]
- 95.Seok JK, Lee SH, Kim MJ, Lee YM. MicroRNA-382 induced by HIF-1alpha is an angiogenic miR targeting the tumor suppressor phosphatase and tensin homolog. Nucleic Acids Res. 2014;42(12):8062–8072. doi: 10.1093/nar/gku515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wang XJ, Zhang DL, Fu C, Wei BZ, Li GJ. MiR-183 modulates multi-drug resistance in hepatocellular cancer (HCC) cells via miR-183-IDH2/SOCS6-HIF-1alpha feedback loop. Eur Rev Med Pharmacol Sci. 2016;20(10):2020–2027. [PubMed] [Google Scholar]
- 97.Mao G, Liu Y, Fang X, Liu Y, Fang L, Lin L, Liu X, Wang N. Tumor-derived microRNA-494 promotes angiogenesis in non-small cell lung cancer. Angiogenesis. 2015;18(3):373–382. doi: 10.1007/s10456-015-9474-5. [DOI] [PubMed] [Google Scholar]
- 98.Blick C, Ramachandran A, McCormick R, Wigfield S, Cranston D, Catto J, Harris AL. Identification of a hypoxia-regulated miRNA signature in bladder cancer and a role for miR-145 in hypoxia-dependent apoptosis. Br J Cancer. 2015;113(4):634–644. doi: 10.1038/bjc.2015.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yuan Q, Gao W, Liu B, Ye W. Upregulation of miR-184 enhances the malignant biological behavior of human glioma cell line A172 by targeting FIH-1. Cell Physiol Biochem. 2014;34(4):1125–1136. doi: 10.1159/000366326. [DOI] [PubMed] [Google Scholar]
- 100.Gits CM, van Kuijk PF, de Rijck JC, Muskens N, Jonkers MB, van Wilfred IF, Mathijssen RH, Verweij J, Sleijfer S, Wiemer EA. MicroRNA response to hypoxic stress in soft tissue sarcoma cells: microRNA mediated regulation of HIF3alpha. BMC Cancer. 2014;14:429. doi: 10.1186/1471-2407-14-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhu S, He C, Deng S, Li X, Cui S, Zeng Z, Liu M, Zhao S, Chen J, Jin Y, Chen H, Deng S, Liu Y, Wang C, Zhao G. MiR-548an, transcriptionally downregulated by HIF1alpha/HDAC1, suppresses tumorigenesis of pancreatic cancer by targeting vimentin expression. Mol Cancer Ther. 2016;15(9):2209–2219. doi: 10.1158/1535-7163.mct-15-0877. [DOI] [PubMed] [Google Scholar]
- 102.He M, Wang QY, Yin QQ, Tang J, Lu Y, Zhou CX, Duan CW, Hong DL, Tanaka T, Chen GQ, Zhao Q. HIF-1alpha downregulates miR-17/20a directly targeting p21 and STAT3: a role in myeloid leukemic cell differentiation. Cell Death Differ. 2013;20(3):408–418. doi: 10.1038/cdd.2012.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Guo XF, Wang AY, Liu J. HIFs-MiR-33a-Twsit1 axis can regulate invasiveness of hepatocellular cancer cells. Eur Rev Med Pharmacol Sci. 2016;20(14):3011–3016. [PubMed] [Google Scholar]
- 104.Cao P, Deng Z, Wan M, Huang W, Cramer SD, Xu J, Lei M, Sui G. MicroRNA-101 negatively regulates Ezh2 and its expression is modulated by androgen receptor and HIF-1alpha/HIF-1beta. Mol Cancer. 2010;9:108. doi: 10.1186/1476-4598-9-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Li H, Rokavec M, Jiang L, Horst D, Hermeking H. Antagonistic effects of p53 and HIF1A on microRNA-34a regulation of PPP1R11 and STAT3 and hypoxia-induced epithelial to mesenchymal transition in colorectal cancer cells. Gastroenterology. 2017;153(2):505–520. doi: 10.1053/j.gastro.2017.04.017. [DOI] [PubMed] [Google Scholar]
- 106.Wang X, Ren H, Zhao T, Ma W, Dong J, Zhang S, Xin W, Yang S, Jia L, Hao J. Single nucleotide polymorphism in the microRNA-199a binding site of HIF1A gene is associated with pancreatic ductal adenocarcinoma risk and worse clinical outcomes. Oncotarget. 2016;7(12):13717–13729. doi: 10.18632/oncotarget.7263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lo Dico A, Costa V, Martelli C, Diceglie C, Rajata F, Rizzo A, Mancone C, Tripodi M, Ottobrini L, Alessandro R, Conigliaro A. MiR675-5p acts on HIF-1alpha to sustain hypoxic responses: a new therapeutic strategy for glioma. Theranostics. 2016;6(8):1105–1118. doi: 10.7150/thno.14700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Taguchi A, Yanagisawa K, Tanaka M, Cao K, Matsuyama Y, Goto H, Takahashi T. Identification of hypoxia-inducible factor-1 alpha as a novel target for miR-17-92 microRNA cluster. Can Res. 2008;68(14):5540–5545. doi: 10.1158/0008-5472.can-07-6460. [DOI] [PubMed] [Google Scholar]
- 109.Lei Z, Li B, Yang Z, Fang H, Zhang GM, Feng ZH, Huang B. Regulation of HIF-1alpha and VEGF by miR-20b tunes tumor cells to adapt to the alteration of oxygen concentration. PLoS One. 2009;4(10):e7629. doi: 10.1371/journal.pone.0007629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Cascio S, D’Andrea A, Ferla R, Surmacz E, Gulotta E, Amodeo V, Bazan V, Gebbia N, Russo A. miR-20b modulates VEGF expression by targeting HIF-1 alpha and STAT3 in MCF-7 breast cancer cells. J Cell Physiol. 2010;224(1):242–249. doi: 10.1002/jcp.22126. [DOI] [PubMed] [Google Scholar]
- 111.Yamakuchi M, Lotterman CD, Bao C, Hruban RH, Karim B, Mendell JT, Huso D, Lowenstein CJ. P53-induced microRNA-107 inhibits HIF-1 and tumor angiogenesis. Proc Natl Acad Sci USA. 2010;107(14):6334–6339. doi: 10.1073/pnas.0911082107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Bruning U, Cerone L, Neufeld Z, Fitzpatrick SF, Cheong A, Scholz CC, Simpson DA, Leonard MO, Tambuwala MM, Cummins EP, Taylor CT. MicroRNA-155 promotes resolution of hypoxia-inducible factor 1alpha activity during prolonged hypoxia. Mol Cell Biol. 2011;31(19):4087–4096. doi: 10.1128/mcb.01276-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ghosh G, Subramanian IV, Adhikari N, Zhang X, Joshi HP, Basi D, Chandrashekhar YS, Hall JL, Roy S, Zeng Y. Hypoxia-induced microRNA-424 expression in human endothelial cells regulates HIF-α isoforms and promotes angiogenesis. J Clin Investig. 2010;120(11):4141–4154. doi: 10.1172/JCI42980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ghosh G, Subramanian IV, Adhikari N, Zhang X, Joshi HP, Basi D, Chandrashekhar YS, Hall JL, Roy S, Zeng Y, Ramakrishnan S. Hypoxia-induced microRNA-424 expression in human endothelial cells regulates HIF-alpha isoforms and promotes angiogenesis. J Clin Invest. 2010;120(11):4141–4154. doi: 10.1172/jci42980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Williams GH, Stoeber K. The cell cycle and cancer. J Pathol. 2012;226(2):352–364. doi: 10.1002/path.3022. [DOI] [PubMed] [Google Scholar]
- 116.Yang X, Lei S, Long J, Liu X, Wu Q. MicroRNA-199a-5p inhibits tumor proliferation in melanoma by mediating HIF-1alpha. Mol Med Rep. 2016;13(6):5241–5247. doi: 10.3892/mmr.2016.5202. [DOI] [PubMed] [Google Scholar]
- 117.Shan Y, Li X, You B, Shi S, Zhang Q, You Y. MicroRNA-338 inhibits migration and proliferation by targeting hypoxia-induced factor 1alpha in nasopharyngeal carcinoma. Oncol Rep. 2015;34(4):1943–1952. doi: 10.3892/or.2015.4195. [DOI] [PubMed] [Google Scholar]
- 118.Tsuchiya S, Fujiwara T, Sato F, Shimada Y, Tanaka E, Sakai Y, Shimizu K, Tsujimoto G. MicroRNA-210 regulates cancer cell proliferation through targeting fibroblast growth factor receptor-like 1 (FGFRL1) J Biol Chem. 2011;286(1):420–428. doi: 10.1074/jbc.M110.170852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Elmore S. Apoptosis: a review of programmed cell death. Toxicol Pathol. 2007;35(4):495–516. doi: 10.1080/01926230701320337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Hassan M, Watari H, AbuAlmaaty A, Ohba Y, Sakuragi N. Apoptosis and molecular targeting therapy in cancer. Biomed Res Int. 2014;2014:150845. doi: 10.1155/2014/150845. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 121.Xu XD, Shao SX, Jiang HP, Cao YW, Wang YH, Yang XC, Wang YL, Wang XS, Niu HT. Warburg effect or reverse Warburg effect? A review of cancer metabolism. Oncol Res Treat. 2015;38(3):117–122. doi: 10.1159/000375435. [DOI] [PubMed] [Google Scholar]
- 122.Puissegur MP, Mazure NM, Bertero T, Pradelli L, Grosso S, Robbe-Sermesant K, Maurin T, Lebrigand K, Cardinaud B, Hofman V, Fourre S, Magnone V, Ricci JE, Pouyssegur J, Gounon P, Hofman P, Barbry P, Mari B. miR-210 is overexpressed in late stages of lung cancer and mediates mitochondrial alterations associated with modulation of HIF-1 activity. Cell Death Differ. 2011;18(3):465–478. doi: 10.1038/cdd.2010.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Chen Z, Li Y, Zhang H, Huang P, Luthra R. Hypoxia-regulated microRNA-210 modulates mitochondrial function and decreases ISCU and COX10 expression. Oncogene. 2010;29(30):4362–4368. doi: 10.1038/onc.2010.193. [DOI] [PubMed] [Google Scholar]
- 124.Li JY, Zhang Y, Zhang WH, Jia S, Kang Y, Zhu XY. Differential distribution of miR-20a and miR-20b may underly metastatic heterogeneity of breast cancers. Asian Pac J Cancer Prev. 2012;13(5):1901–1906. doi: 10.7314/APJCP.2012.13.5.1901. [DOI] [PubMed] [Google Scholar]
- 125.Giordano G, Febbraro A, Venditti M, Campidoglio S, Olivieri N, Raieta K, Parcesepe P, Imbriani GC, Remo A, Pancione M. Targeting angiogenesis and tumor microenvironment in metastatic colorectal cancer: role of aflibercept. Gastroenterol Res Pract. 2014;2014:526178. doi: 10.1155/2014/526178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Carmeliet P. Angiogenesis in life, disease and medicine. Nature. 2005;438(7070):932–936. doi: 10.1038/nature04478. [DOI] [PubMed] [Google Scholar]
- 127.Alaiti MA, Ishikawa M, Masuda H, Simon DI, Jain MK, Asahara T, Costa MA. Up-regulation of miR-210 by vascular endothelial growth factor in ex vivo expanded CD34 + cells enhances cell-mediated angiogenesis. J Cell Mol Med. 2012;16(10):2413–2421. doi: 10.1111/j.1582-4934.2012.01557.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Yasuda H. Solid tumor physiology and hypoxia-induced chemo/radio-resistance: novel strategy for cancer therapy: nitric oxide donor as a therapeutic enhancer. Nitric Oxide. 2008;19(2):205–216. doi: 10.1016/j.niox.2008.04.026. [DOI] [PubMed] [Google Scholar]
- 129.Comerford KM, Wallace TJ, Karhausen J, Louis NA, Montalto MC, Colgan SP. Hypoxia-inducible factor-1-dependent regulation of the multidrug resistance (MDR1) gene. Can Res. 2002;62(12):3387–3394. [PubMed] [Google Scholar]
- 130.Comerford KM, Cummins EP, Taylor CT. c-Jun NH2-terminal kinase activation contributes to hypoxia-inducible factor 1alpha-dependent P-glycoprotein expression in hypoxia. Can Res. 2004;64(24):9057–9061. doi: 10.1158/0008-5472.can-04-1919. [DOI] [PubMed] [Google Scholar]
- 131.Moeller BJ, Dewhirst MW. HIF-1 and tumour radiosensitivity. Br J Cancer. 2006;95(1):1–5. doi: 10.1038/sj.bjc.6603201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Tafsiri E, Darbouy M, Shadmehr MB, Cho WC, Karimipoor M. Abberant expression of oncogenic and tumor-suppressive microRNAs and their target genes in human adenocarcinoma alveolar basal epithelial cells. J Cancer Res Ther. 2016;12(1):395–400. doi: 10.4103/0973-1482.148673. [DOI] [PubMed] [Google Scholar]
- 133.Hwang HW, Baxter LL, Loftus SK, Cronin JC, Trivedi NS, Borate B, Pavan WJ. Distinct microRNA expression signatures are associated with melanoma subtypes and are regulated by HIF1A. Pigment Cell Melanoma Res. 2014;27(5):777–787. doi: 10.1111/pcmr.12255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Ke HL, Li WM, Lin HH, Hsu WC, Hsu YL, Chang LL, Huang CN, Li CC, Chang HP, Yeh HC, Li CF, Wu WJ. Hypoxia-regulated MicroRNA-210 overexpression is associated with tumor development and progression in upper tract urothelial carcinoma. Int J Med Sci. 2017;14(6):578–584. doi: 10.7150/ijms.15699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Camps C, Buffa FM, Colella S, Moore J, Sotiriou C, Sheldon H, Harris AL, Gleadle JM, Ragoussis J. hsa-miR-210 Is induced by hypoxia and is an independent prognostic factor in breast cancer. Clin Cancer Res. 2008;14(5):1340–1348. doi: 10.1158/1078-0432.ccr-07-1755. [DOI] [PubMed] [Google Scholar]
- 136.Cheng HH, Mitchell PS, Kroh EM, Dowell AE, Chery L, Siddiqui J, Nelson PS, Vessella RL, Knudsen BS, Chinnaiyan AM, Pienta KJ, Morrissey C, Tewari M. Circulating microRNA profiling identifies a subset of metastatic prostate cancer patients with evidence of cancer-associated hypoxia. PLoS One. 2013;8(7):e69239. doi: 10.1371/journal.pone.0069239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Liu K, Yao H, Lei S, Xiong L, Qi H, Qian K, Liu J, Wang P, Zhao H. The miR-124-p63 feedback loop modulates colorectal cancer growth. Oncotarget. 2017;8(17):29101–29115. doi: 10.18632/oncotarget.16248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Gao Y, Chen L, Song H, Chen Y, Wang R, Feng B. A double-negative feedback loop between E2F3b and miR-200b regulates docetaxel chemosensitivity of human lung adenocarcinoma cells. Oncotarget. 2016;7(19):27613–27626. doi: 10.18632/oncotarget.8376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Zhang Y, Shen WL, Shi ML, Zhang LZ, Zhang Z, Li P, Xing LY, Luo FY, Sun Q, Zheng XF, Yang X, Zhao ZH. Involvement of aberrant miR-139/Jun feedback loop in human gastric cancer. Biochem Biophys Acta 1853. 2015;2:481–488. doi: 10.1016/j.bbamcr.2014.12.002. [DOI] [PubMed] [Google Scholar]
- 140.Lin CC, Jiang W, Mitra R, Cheng F, Yu H, Zhao Z. Regulation rewiring analysis reveals mutual regulation between STAT1 and miR-155-5p in tumor immunosurveillance in seven major cancers. Sci Rep. 2015;5:12063. doi: 10.1038/srep12063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Moes M, Le Bechec A, Crespo I, Laurini C, Halavatyi A, Vetter G, Del Sol A, Friederich E. A novel network integrating a miRNA-203/SNAI1 feedback loop which regulates epithelial to mesenchymal transition. PLoS One. 2012;7(4):e35440. doi: 10.1371/journal.pone.0035440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Chiang CH, Chu PY, Hou MF, Hung WC. MiR-182 promotes proliferation and invasion and elevates the HIF-1alpha-VEGF-A axis in breast cancer cells by targeting FBXW7. Am J Cancer Res. 2016;6(8):1785–1798. [PMC free article] [PubMed] [Google Scholar]
- 143.Irlam-Jones JJ, Eustace A, Denley H, Choudhury A, Harris AL, Hoskin PJ, West CM. Expression of miR-210 in relation to other measures of hypoxia and prediction of benefit from hypoxia modification in patients with bladder cancer. Br J Cancer. 2016;115(5):571–578. doi: 10.1038/bjc.2016.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Zhao D, Tu Y, Wan L, Bu L, Huang T, Sun X, Wang K, Shen B. In vivo monitoring of angiogenesis inhibition via down-regulation of mir-21 in a VEGFR2-luc murine breast cancer model using bioluminescent imaging. PLoS One. 2013;8(8):e71472. doi: 10.1371/journal.pone.0071472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Yang W, Wei J, Guo T, Shen Y, Liu F. Knockdown of miR-210 decreases hypoxic glioma stem cells stemness and radioresistance. Exp Cell Res. 2014;326(1):22–35. doi: 10.1016/j.yexcr.2014.05.022. [DOI] [PubMed] [Google Scholar]
- 146.Lytle JR, Yario TA, Steitz JA. Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5′ UTR as in the 3′ UTR. Proc Natl Acad Sci USA. 2007;104(23):9667–9672. doi: 10.1073/pnas.0703820104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.