Abstract
A disintegrin and metalloproteinase 10 (ADAM10) plays a major role in the ectodomain shedding of important surface molecules with physiological and pathological relevance including the amyloid precursor protein (APP), the cellular prion protein, and different cadherins. Despite its therapeutic potential, there is still a considerable lack of knowledge how this protease is regulated. We have previously identified tetraspanin15 (Tspan15) as a member of the TspanC8 family to specifically associate with ADAM10. Cell-based overexpression experiments revealed that this binding affected the maturation process and surface expression of the protease. Our current study shows that Tspan15 is abundantly expressed in mouse brain, where it specifically interacts with endogenous ADAM10. Tspan15 knockout mice did not reveal an overt phenotype but showed a pronounced decrease of the active and mature form of ADAM10, an effect which augmented with aging. The decreased expression of active ADAM10 correlated with an age-dependent reduced shedding of neuronal (N)-cadherin and the cellular prion protein. APP α-secretase cleavage and the expression of Notch-dependent genes were not affected by the loss of Tspan15, which is consistent with the hypothesis that different TspanC8s cause ADAM10 to preferentially cleave particular substrates. Analyzing spine morphology revealed no obvious differences between Tspan15 knockout and wild-type mice. However, Tspan15 expression was elevated in brains of an Alzheimer’s disease mouse model and of patients, suggesting that upregulation of Tspan15 expression reflects a cellular response in a disease state. In conclusion, our data show that Tspan15 and most likely also other members of the TspanC8 family are central modulators of ADAM10-mediated ectodomain shedding in vivo.
Electronic supplementary material
The online version of this article (10.1007/s00018-018-2791-2) contains supplementary material, which is available to authorized users.
Keywords: ADAM10, N-cadherin, Prion protein, Tetraspanin
Introduction
The metalloproteinase ADAM10, together with its close relative ADAM17, is one of the best-described members of the ADAM family of proteases [1]. ADAM10 is expressed in several mammalian cell lines and in a wide variety of organs, including the brain [2–4]. As a type I transmembrane protein, ADAM10 is synthesized in the endoplasmic reticulum (ER) as an inactive proform with a molecular weight of about 100 kDa. During transport to the plasma membrane, ADAM10 is N-glycosylated at four positions and undergoes maturation by removal of the prodomain mediated by the proprotein convertase furin and the proprotein convertase 7 (PC7) [5–7]. The resulting mature and active form of ADAM10 has a molecular weight of about 70 kDa and mediates substrate cleavage in late secretory compartments and at the plasma membrane [8]. The substrate spectrum of ADAM10 to date comprises more than 40 validated substrates, including adhesion molecules (e.g., cadherins), chemokines (e.g., CX3CL1), growth factors (e.g., EGF and TNFα), signalling receptors, and ligands (e.g., Notch receptor and Fas ligand) [9–11].
The regulation of ADAM10’s proteolytic activity and substrate specificity plays an important role in developmental and physiological processes and decides about its functions in health or disease. Tetraspanins were discovered to associate with ADAM10 and to regulate its activity and substrate specificity [12–19]. Tetraspanins are a group of evolutionary conserved transmembrane proteins with four membrane spanning domains forming a small and a large extracellular loop (LEL). In the case of the tetraspanin CD81, two pairs of transmembrane domains are separated and capped by the LEL, and within the membrane, an intramembrane pocket containing a cholesterol molecule was revealed [20]. A structural characteristic of tetraspanins is the formation of disulfide bonds in the LEL. This involves 4–8 cysteines with two of them being located in a highly conserved CCG motif. Through a wide-ranging network of interactions, including interactions with other tetraspanins and other integral membrane proteins, tetraspanins form tetraspanin-enriched microdomains (TEMs) within the membrane that contribute to the cell’s tetraspanin web [21, 22]. TEMs represent platforms, where tetraspanins organise their partner proteins in functional complexes. Increasing understanding of the tetraspanin biology in recent years revealed further important roles of tetraspanins in diverse processes such as immune response, fertilization, pathogen infection, and synapse formation [23].
Tspan15 is one of the first tetraspanins containing eight cysteine residues (TspanC8 group) which were identified as direct interaction partners of ADAM10 [12–14]. In cellulo experiments revealed Tspan15 as a potent regulator of ADAM10 proteolytic activity. By promoting ADAM10’s ER exit and stabilisation of its mature form, Tspan15 increases ADAM10 maturation and surface expression. This also enhances ADAM10-mediated ectodomain shedding of N-cadherin and APP in N2a, HEK293, and Cos7 cells [14, 24, 25]. In contrast, in the human osteosarcoma cell line U2OS-N1, Tspan15 was shown to be a negative regulator of Notch and APP processing, indicating that Tspan15 has cell-type-specific functions [24]. Moreover, Tspan15 expression modulates the composition of proteins which are associated with ADAM10 at the plasma membrane and influences their motility [24]. Despite the increased understanding of how Tspan15 regulates ADAM10-mediated shedding in vitro, its physiological functions and its role in ADAM10 regulation in vivo have not been addressed yet. To study the role of Tspan15 for ADAM10-mediated shedding events in vivo, Tspan15 expression was investigated and a Tspan15 knockout mouse model was generated and analyzed. Our data reveal that Tspan15 also has significant impact on the maturation and activity of ADAM10 in the central nervous system (CNS), which determines Tspan15 (and possibly the other members of the TspanC8 family) as essential chaperones modulating the function of this important cell surface protease in the brain.
Materials and methods
Animals
Tspan15 knockout mice were generated by transcription activator-like effector nuclease (TALEN)-mediated genome editing. TALEN were designed to target exon2 of the Tspan15 gene locus. The following TALEN-repeat domain sequences were used. Left TALEN: NN HD HD NG NG HD HD NG NN NN HD HD HD HD HD NN, right TALEN: NN NI NI HD NI NG NN NI HD HD NI HD HD HD HD HD NI NN NN NI. TALEN-RNA precursors were generated and microinjected into male nucleoli of zygotes isolated from C57BL/6N mice as described [26]. Founder mice were genotyped for TALEN-mediated frameshift mutations. Resulting from this, mice with a 104 bp deletion mutation inducing a translational stop codon within exon2 of the Tspan15 gene were used for further breeding and analysis. 5xFAD transgenic mice express three mutant forms of human APP695, APP KM670/671NL (Swedish), APP I716V (Florida), APP V717I (London), as well as two human presenilin-1 mutants M146L and L286V. 5xFAD mice were obtained from Jackson Laboratories and maintained on a C57BL/6N background.
Antibodies
The following antibodies were used: anti-Actin (IB, Sigma-Aldrich, Munich, Germany), anti-ADAM10 antibody EPR5622 recognizing the C-terminal part of ADAM10 (IB, Abcam, Cambridge, UK), self-made anti-ADAM10 antibody raised against the C-terminus of ADAM10 (IP, [14]), anti-APP-C-terminus B63.2 (IB, a kind gift of Wim Annaert, Leuven, Belgium), anti-GAPDH FL-335 (IB, Santa Cruz Biotechnology, Dallas, USA), anti-GFAP M0761 (IHC, DAKO, Hamburg, Germany), anti-Iba1 019-19741 (IHC, Wako, Neuss, Germany), anti-NeuN MAB377 (IHC, Millipore, Billerica, USA), anti-myc 9B11 (IB, Cell Signaling Technology, Danvers, USA), anti-N-cadherin (IB, BD Transduction Laboratories, Heidelberg, Germany), anti-LIMP2 L2T2 (IB, [27]), anti-PrPC POM1 and POM2 (IB, Prof. Dr. Aguzzi, Zürich, Switzerland), rabbit polyclonal anti-shed PrPC (IB, available from Prof. Dr. Glatzel/Dr. Altmeppen, Hamburg, Germany), anti-synaptophysin M7315 (IHC, DAKO, Hamburg, Germany), anti-beta-Tubulin E7 (IB, DSHB Iowa City, USA), anti-Tspan15 NBP1-92540 (IB, Novusbio, Littleton, USA), self-made anti-Tspan3 antibody raised against a synthetic peptide consisting of the last 19 amino acids of the Tspan3 C-terminus (IB, Pineda, Berlin, Germany), self-made anti-Tspan7 antibody raised against the 17 amino acids of the Tspan7 C-terminus (IB, Pineda, Berlin, Germany), self-made anti-Tspan15 T3CT (IP, antibody raised against a synthetic peptide corresponding to the last 15 amino acids of the C-terminus of murine Tspan15 (IB, Pineda, Berlin, Germany), and self-made anti-Tspan15 T2EL antibody produced against a synthetic peptide corresponding to amino acids 199–209 within the large extracellular region of murine Tspan15 (IB, Pineda, Berlin, Germany).
Cell culture, transfection, and plasmids
All cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM, Thermo Fisher Scientific, Waltham, USA) supplemented with 10% fetal calf serum (FCS) and 1% penicillin/streptomycin at 37 °C, 5% CO2 atmosphere, and 95% relative humidity. For transient transfections, respective amounts of DNA were mixed 1:3 (w/v) with polyethylenimine Max (PEI, Polysciences, Hirschberg, Germany) and added to the cells. For inhibitor treatment, cell culture media were supplemented with 3 µM of the selective ADAM10 inhibitor GI254023X (Sigma-Aldrich, Munich, Germany) or DMSO as control and cells incubated for 24 h at standard cell culture conditions. Mammalian expression vectors were pcDNA3.1Hygro(–) and pFrog3 derived from pcDNA3.1 (Thermo Fisher Scientific, Waltham, USA) [28]. The myc-epitope was inserted after the last amino acid of the Tspan15 coding sequence.
Protein extraction from cells and organs
Cells were washed three times with cold phosphate buffered saline (PBS), scraped off in PBS supplemented with 0.04% proteinase inhibitor cocktail (Complete™, Roche Diagnostics, Mannheim, Germany), and centrifuged for 20 min at 4000g. The cell pellet was resuspended in lysis buffer (1 mM EGTA, 5 mM Tris, 250 mM Saccharose, pH 7,4) containing 1% Triton X-100 and 0.04% proteinase inhibitor cocktail. Afterwards, cells were sonicated two times for 10 s at 60 Hz, incubated for 1 h on ice, and sonicated again. Cell lysates were centrifuged for 10 min at 14,000g at 4 °C to remove cellular debris and nuclei. For protein extraction from organs, tissues were isolated and homogenized with RIPA lysis buffer (20 mM Tris–HCl pH 7.5, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% NP40, 2.5 mM sodium deoxycholate, and 0.04% proteinase inhibitor cocktail) using the Precellys homogenization system (Bertin Corp., Rockville, USA). Thereafter, homogenates were sonicated two times for 10 s at 60 Hz, incubated on ice for 1 h, and centrifuged for 10 min at 14,000g at 4 °C. Protein concentration was measured using the Pierce™ BCA Protein Assay Kit (Thermo Fisher Scientific, Waltham, USA). Finally, lysates were prepared for SDS-PAGE by boiling with reducing SDS-PAGE loading buffer (625 mM Tris base, 10% SDS, 50% glycerol, and 250 mM DTT) for 5 min at 95 °C.
Peptide N-glycosidase F (PNGase F) digestion
Cell lysates and mouse brain homogenates were prepared as described above. Equal protein amounts were incubated with 5× denaturing buffer (250 mM Na2PO4, pH 8.0, 1% SDS, 4% β-mercaptoethanol) for 5 min at 95 °C. Following, 5× Triton solution (7.5% Triton X-100, 50 mM EDTA) and proteinase inhibitor cocktail were added and samples were incubated with PNGase F (Roche, Diagnostics, Mannheim, Germany) or with ddH2O for 2 h at 37 °C. Finally, samples were denatured with SDS-PAGE loading buffer and further analyzed by SDS-PAGE and immunoblotting.
Co-immunoprecipitation experiments
Tissue lysates were prepared with immunoprecipitation buffer (120 mM NaCl, 50 mM Tris–HCl, 0.04% complete proteinase inhibitor cocktail, and pH 7.4) containing 0.5% NP-40 (Sigma-Aldrich, Munich, Germany) as described above. 60 µl of the total lysates were kept to control for protein expression. The residual lysate was used for immunoprecipitation. Therefore, equal protein amounts were incubated with primary antibodies over night at 4 °C under continuous rotation. For each sample, 70 µl of magnetic Protein G-coupled Dynabeads (Thermo Fisher Scientific, Waltham, USA) were washed with lysis buffer and incubated with SEA-blocking buffer (Thermo Fisher Scientific, Waltham, 1:1 diluted with lysis buffer) over night at 4 °C to reduce unspecific binding of proteins. Next day, Dynabeads were incubated with the lysate–antibody complexes for 30 min at room temperature and under continuous rotation. Beads were washed three times with lysis buffer and incubated with 50 µl reducing SDS-PAGE loading buffer for 20 min at 65 °C to elute precipitated proteins. Input controls and immunoprecipitates were separated on 10% SDS gels and further analyzed by immunoblotting.
SDS-PAGE and immunoblotting
Equal amounts of extracted proteins were loaded on 10% SDS-PAGE or 4–12% gradient BIS/Tris NuPAGE™ gels (Thermo Fisher Scientific, Waltham, USA) and separated for 1.5 h. Thereafter, proteins were transferred to nitrocellulose membranes (Whatman, GE Healthcare, Little Chalfont, UK) using the tank blotting system for 2 h at 0.8 A and 4 °C. For detection of APP-C-terminal fragments, blotting was performed at 0.25 A for 2 h at 4° C. Membranes were blocked with 5% milk powder in Tris-buffered saline (TBS) plus 0.1% Tween (TBS-T) to prevent unspecific antibody binding. Primary antibodies were diluted in 5% milk powder solution in TBS-T and incubated with membranes over night at 4 °C under continuous rotation. The next day, membranes were washed three times with TBS-T and incubated with peroxidase-coupled secondary antibodies diluted in 5% milk powder/TBS-T for 1 h at room temperature. Again, membranes were washed three times with TBS-T and subjected to detection using the Lumigen ECL Ultradetection solution (Lumigen Inc., Southfield, USA) in combination with the chemiluminescence detection system LAS4000 (GE Healthcare, Little Chalfont, UK). Signal intensities were quantified using the quantification software ImageJ.
Quantitative RT-PCR (qRT-PCR)
RNA was extracted from 30 mg tissue using the NucleoSpin® RNA Kit (Macherey-Nagel, Düren, Germany). According to the manufacturer’s instructions, 1 µg of extracted RNA was reverse transcripted using the RevertAid™ First Strand cDNA Synthesis Kit (Thermo Fisher Scientific, Waltham, USA) and Random Hexamer primer. Gene transcription was determined by real-time PCR analysis of 0.5 µl cDNA on a LightCycler® 480 Real-time PCR System (Roche Diagnostics, Mannheim, Germany) in 10 µl reaction volume as triplicates. Relative gene transcription was calculated using ΔCt values normalized to the transcription level of house-keeping genes tubulin and actin. The PCR efficiency of each assay was determined by serial dilution of standards and these values were used for calculation. t tests were performed on the mean ΔCt values of the technical triplicate measurements.
Histological and immunohistochemical analysis
Freshly extracted mouse brain hemispheres were immediately fixed in buffered 4% paraformaldehyde and embedded in paraffin blocks. 4 µm-thick sagittal sections were prepared. Hematoxylin/eosin (H&E) and immunohistochemical (IHC) stainings were performed according to routine laboratory procedures. To ensure comparability and reduce experimental error, for each staining, all samples were run in parallel using the automated Ventana Benchmark XT system (Ventana, Tucson, USA). Antigen retrieval was achieved by boiling sections for 30 min (“mild”; for GFAP and Iba1) or 1 h (“standard”, for NeuN and Synaptophysin) in citrate buffer (Cell Conditioning Solution, CC1, Ventana). Sections were incubated for 1 h with the primary antibodies diluted in antibody diluent solution (Zytomed, Berlin, Germany) containing 5% goat serum in 45% TBS pH 7.6, 0.1% Triton X-100 (Dianova, Hamburg, Germany): Secondary anti-mouse and anti-rabbit antibodies were purchased from Ventana. Staining was performed using the ultraView Universal DAB detection kit (Ventana) according to routine conditions of the machine; and samples were finally mounted and sealed with a coverslip. Samples were analyzed and representative pictures were taken with the digital DMD108 microscopic device (Leica, Wetzlar, Germany).
Ultrastructural analysis of hippocampal sections
Mice were perfused and sections prepared as described previously [29].
Localization of Tspan15 in primary hippocampal neurons
Primary hippocampal rat cultures were prepared as described previously [30]. At day in vitro (DIV) 13–15 cells were transfected with Tspan15-YPET with or without the Golgi satellite marker pGolt-mCherry [31] using lipofectamine 2000 according to the manufacturer’s instructions. 24 h after transfection neurons were imaged on a VisiScope TIRF/FRAP imaging system (Visitron Systems) based on Nikon Ti-E equipped with a perfect focus system (Nikon), Nikon CFI Apo TIRF 100×, 1.49 N.A. oil objective, a back focal TIRF scanner for suppression of interference fringes (Ilas-2, Roper Scientific France/PICT-IBiSA, Institut Curie) and controlled with VisiView software. Samples were illuminated and fluorophores activated by 488 and 561 nm laser lines.
Fluorescence was collected through an ET 405/488/561/640 Laser Quad Band filter with a The ORCA-Flash 4.0 LT sCMOS camera. Pictures were taken with 100 ms stream acquisition. For run length and velocity analysis, kymographs of 5 px (= 325 nm) wide lines along dendrites were created using the KymographClear plugin [32] in Fiji [33] and analyzed manually.
To stain mitochondria MitoTracker Red580 (Invitrogen, M22425) was applied to the cells in 1:10,000 dilution and single images taken 15–45 min afterwards. Quantification of co-localization of Tspan15 with pGolt or MitoTracker was performed manually. Tspan15 puncta were counted and considered as co-localizing with pGolt or MitoTracker if they show enrichment in fluorescence intensity.
Liquid chromatography–mass spectrometry (LC–MS) analysis
SDS-PAGE bands corresponding to full-length and cleaved Tspan15 were excised, cut into smaller cubes (2 mm3), destained, reduced (dithiothreitol), and alkylated (iodoacetamide) using standard protocols. Here, triethylammonium bicarbonate (TEAB) replaced traditionally used ammonium bicarbonate (ABC) as it is volatile and compatible with downstream reductive dimethylation labeling. Samples were labeled using 25 mM formaldehyde in 25 mM NaCNBH3 overnight in 100 mM HEPES buffer (pH 7.0) at 25 °C. The reaction was quenched using 1 M ethanolamine for 30 min. Afterwards, samples were washed with ammonium bicarbonate (ABC), dehydrated with acetonitrile (ACN), and dried down and deglycosylated overnight with the addition of PNGaseF (New England Biolabs, Ispwich, USA) at 37 °C in ABC buffer. Gel bands were washed extensively to remove released N-glycans and thereafter digested overnight with either 100 ng of chymotrypsin (0.1 M TEAB in 2 mM CaCl2) or 100 ng of trypsin (0.1 M TEAB). Peptides were extracted, dried via vacuum centrifugation and resuspended in 3% ACN and 0.1% trifluoroacetic acid, and separated across a Dionex UltiMate 3000 RSLCnano system (Thermo, Bremen, Germany) equipped with an Acclaim PepMap RSLC nanocolumn (C18, 100 Å, 2 μm, 75 μm × 500 mm) (Thermo) which was coupled online to a Q Exative Plus mass spectrometer (Thermo). The eluents used were: eluent A: 0.05% formic acid (FA), eluent B: 80% ACN + 0.04% FA; flow rate of 300 nl/min. The separation occurred over a programmed 90 min run. Chromatographic conditions were: 5% B for 2 min followed by a linear gradient from 5 to 50% B over 60 min; then, 50–95% B over 5 min, before a 10 min wash at 95% B, followed by a 13 min equilibration of the column at 5% B.
The MS analysis was performed in positive ion mode utilizing HCD as the fragmentation method. Full-scan MS acquisition was performed (resolution 70,000, AGC target 3e6, inject time 80 ms, scan range 300–2000 m/z in profile mode). Subsequent MS/MS via HCD with a normalized collision energy (NCE) of 27 (resolution 17,500, AGC target 1e5, max inject time 50 ms, isolation window 3.0 m/z) of the top 10 most intense ions was performed. Charge exclusion was enabled for unassigned, + 1 and > + 7 ions, and dynamic exclusion (15 s duration) and lock mass (445.12003 m/z) were enabled.
Database searches were performed on Proteome Discoverer software (v2.2.0.388) using the SequestHT search algorithm. The data were searched against the reviewed and canonical human database (07.04.2017) with contaminants (cRAP) and murine Tspan15 with a C-terminal myc tag appended to the database (20,319 sequences). Data were searched with semi-enzyme specificity [five miss-cleavages for chymotrypsin (FQYLM) and two miss-cleavages for trypsin (Arg-C specificity)] with fixed modification at lysine (dimethylation) and cysteine (carbamidomethylation) residues. Modification at the peptide N-term (dimethylation), methionine (oxidation) and asparagine (deamidation) residues was set as a variable modification. A precursor mass tolerance of 10 ppm and a fragment mass tolerance of 0.02 Da were used.
Statistical analysis
Statistical significances were analyzed with the statistical analysis software GraphPad Prism using the two-tailed unpaired Student’s t tests. Values are shown as mean ± standard deviation (SD) or standard error of the mean (SEM). Statistical significance was considered with p values as followed: *p < 0.05, **p < 0.01.
Results
Tspan15 is a glycosylated protein with a broad expression pattern which is proteolytically processed
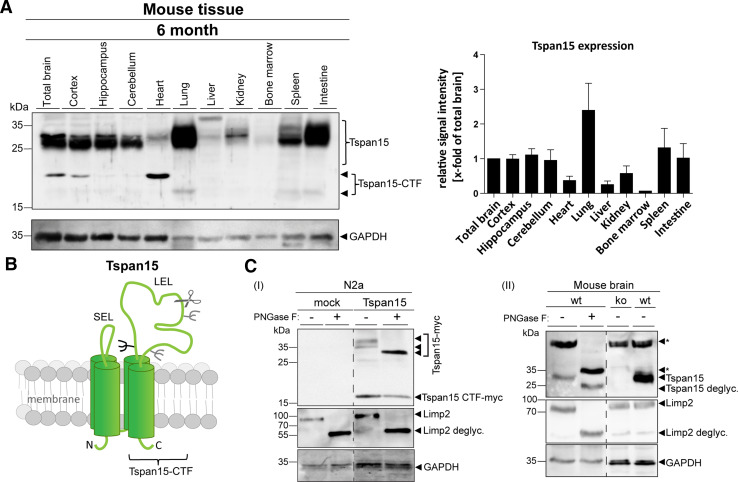
We have previously identified Tspan15 as an ADAM10 interacting protein using a split-ubiquitin yeast screening assay [14]. However, there is not much known about the in vivo role of this tetraspanin and how it affects the protease. To address potential functions of Tspan15, we first evaluated the expression in different mouse tissues and subregions of a 6-month-old mouse brain (Fig. 1a). Immunoblot analysis revealed that Tspan15 is strongly expressed in lung, spleen, intestine, and brain, whereas in heart, bone marrow, liver, and kidney, its expression is much lower. Within the brain, it is strongly expressed in all regions analyzed (i.e. cortex, hippocampus and cerebellum). Interestingly, in brain, lung, spleen, intestine, and even more pronounced in heart, we observed additional immunoreactive bands of around 16 and 20 kDa (heart and brain) which likely represent proteolytic fragments originating from a cut in the large extracellular loop of Tspan15 (Fig. 1b; Suppl. Fig. S1). A similar fragment was also observed after overexpression of a murine myc-tagged Tspan15 construct in N2a cells (Fig. 1c, I; Suppl. Fig. S2). Treatment of these cells with the selective ADAM10 inhibitor GI254023X did not reduce, but increased the occurrence of the Tspan15 CTF (Suppl. Fig. S2A), possibly excluding a role of ADAM10 in this cleavage event. The fuzzy nature of the full-length Tspan15 protein of around 30 kDa and its different mobility depending on the tissue suggest that it is glycosylated. This assumption was confirmed after PNGase F digestion of a myc-tagged version of Tspan15 expressed in N2a cells and endogenous Tspan15 in brain homogenates (Fig. 1c I, II). The molecular weight of the full-length Tspan15 protein, but not the carboxyterminal fragment (CTF) of Tspan15 (Fig. 1c, I), was reduced after deglycosylation with PNGase F, suggesting that Tspan15 is glycosylated at the single site (N118) at the beginning of the large extracellular domain (Fig. 1b).
Fig. 1.
Tspan15 is a glycosylated protein with a prominent expression in different murine tissues. a Indicated organs were isolated from a 6-month-old wild-type mouse; homogenates were prepared and subjected to immunoblot analysis. Using a C-terminal anti-Tspan15 antibody (NBP1-92540), Tspan15 expression was detected in all tissues. It was most abundantly expressed in lung, spleen, intestine, total brain, and all of the analyzed subregions (cortex, hippocampus, and cerebellum) of the brain. Less Tspan15 was detected in heart, liver, kidney, and bone marrow (representative immunoblot of n = 3 independent experiments, n = 1 for bone marrow). In addition, a Tspan15-C-terminal fragment (CTF) was visible in total brain, cortex, and heart. Protein loading was controlled by anti-GAPDH staining. For quantification, Tspan15 signal intensities were measured and normalized to total brain (set to one). b Schematic drawing of the full-length Tspan15 structure, showing its four transmembrane domains, the small (SEL), the large extracellular loop (LEL), the potential (black), and additionally predicted (grey) glycosylation sites. A possible cleavage site within the LEL, which generates a Tspan15 C-terminal fragment, is indicated with a scissor symbol. c Deglycosylation of Tspan15 using peptide N-glycosidase F (PNGase F) digestion. (I) N2a cells were transfected with a murine myc-tagged Tspan15 construct or an empty vector (mock). Cells were lysed and incubated with PNGase F (+) or left untreated (−). Using an anti-myc antibody, Tspan15-myc was detected at a molecular weight of 37 kDa in the Tspan15-myc-transfected untreated (Tspan15-myc, −), but not in the mock transfected samples. PNGase F treatment (Tspan15-myc, +) reduced the Tspan15-myc molecular weight to 32 kDa. In addition, a C-terminal fragment of Tspan15-myc appeared at around 17 kDa in both the PNGase F treated (+) and the untreated (−) Tspan15-myc-transfected sample, which indicates that no glycans exist in this stretch. (II) Brain homogenate of a wild-type (wt) mouse was prepared and subjected to PNGase F digestion. Removal of N-linked glycans decreased the molecular weight of endogenous Tspan15 from 30 kDa (wt, −) to 25 kDa (wt, +). Specificity of the anti-Tspan15 antibody (Tspan15 T2EL) was confirmed by the absence of the Tspan15 signal in a Tspan15 knockout (ko) control brain sample. Efficiency of the PNGase F digestion (I + II) was confirmed by the decreased molecular weight of the lysosomal integral membrane protein, LIMP-2, which is a highly glycosylated lysosomal protein. Immunoblots are representative for n = 3 independent experiments. Probing the immunoblots (I + II) for GAPDH confirmed equal protein loading. Asterisks (*) mark unspecific signals
Performing liquid chromatography–mass spectrometric analysis of a chymotryptic digest of the reductively dimethylated immunoprecipitated Tspan15 CTF (Suppl. Fig. S2B) revealed a prominent peptide with an N-terminally (reductively) dimethylated Isoleucine187. This provides a hint that a cleavage occurs between C186 and I187. Interestingly, the identified cleavage site is located C-terminal to a double cysteine and N-terminally of a glycosylated Asn189 which could indicate that this residue is carrying only a short carbohydrate chain. The peptide identified (Suppl. Fig. S2B) contains aspartate D189; this derives from a deamidation of the genetically encoded Asn189 during deglycosylation (PNGase F) which was performed prior to chymotryptic digestion and clearly leads to the conclusion that the CTF has been N-glycosylated at this residue. However, the data generated cannot rule out the presence of further cleavage sites.
Tspan15 expression increases with age and Tspan15 is transported in dendrites and axons of primary rat neurons
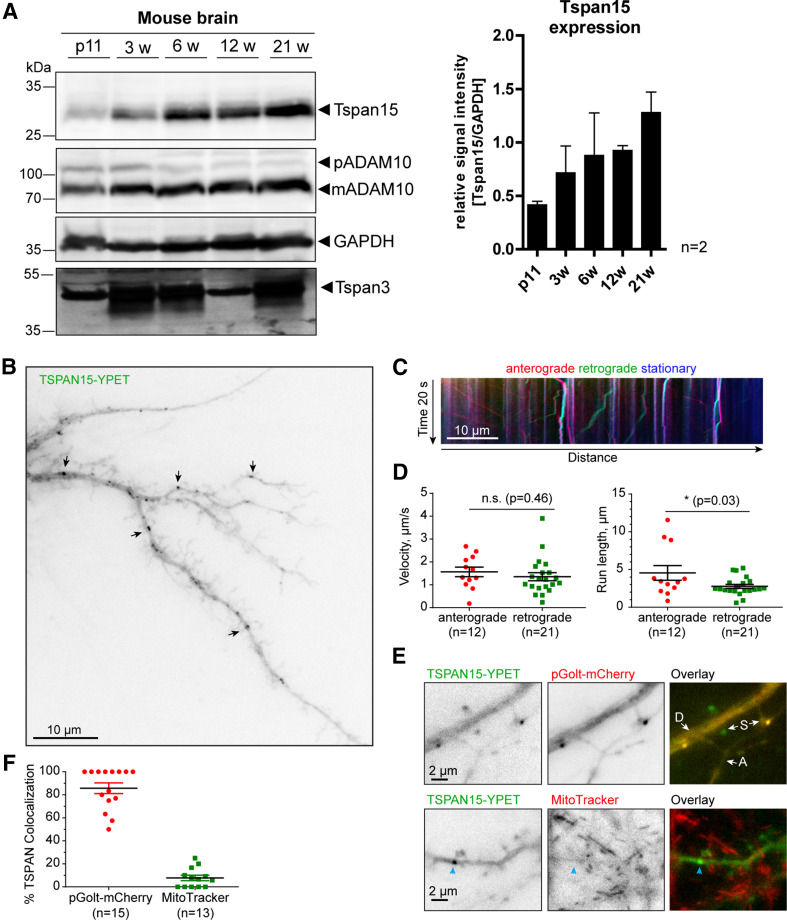
To correlate the expression of Tspan15 and a possible regulation of ADAM10, we analyzed the expression of both proteins at different ages. Interestingly, we found that Tspan15 expression increased from 11-day-old (11d) to 21-week-old (21w) mice. In addition, ADAM10 expression in the brain was low at 11 days and became more prominent after 3 weeks of age. In contrast to Tspan15 and ADAM10, the expression of the TspanC6 tetraspanin Tspan3 shows a higher expression in 3- (3w), 6- (6w), and 21-week-old and a low expression in 11d and 12w mice (Fig. 2a). Since the anti-Tspan15 antibody did not work in immunohistochemistry under different conditions (data not shown), we decided to express a yellow fluorescent protein optimized for FRET (YPET) fused to Tspan15 in primary hippocampal rat neurons. We observed distinct Tspan15 punctae in dendrites (Fig. 2b). Live cell imaging allowed to follow up the trafficking of Tspan15; and this analysis revealed a similar anterograde and retrograde velocity, but a longer anterograde path length as compared to the retrograde movement (Fig. 2c, d, also see: Supplementary Video S1). We found a partial co-localization with the Golgi marker pGolt-mCherry [31], but not with the mitochondrial marker MitoTracker, suggesting that Tspan15 passes the Golgi microsatellite system in dendrites (Fig. 2e, f). Tspan15 was also found near synaptic spines (Fig. 2b, e) where ADAM10 has been reported to act [29].
Fig. 2.
Tspan15 expression increases with age and Tspan15 is transported in hippocampal dendrites. a Murine brain homogenates from 11-day-old (11d), 3-, 6-, 12- and 21-week-old (3, 6, 12, and 21w) wild-type mice were prepared; and the expression of Tspan15, ADAM10, and Tspan3 was analyzed by immunoblotting. Detection of Tspan15 using the Tspan15 T2EL antibody revealed a low Tspan15 expression in 11-day-old animals (11d), which increased with 3 weeks and was strongest in 21-week-old mice. Similarly, ADAM10 expression (detected with ADAM10 EPR5622) was low in 11-day-old mice and increased in older mice (n = 2). In contrast, Tspan3 showed a prominent expression at all age stages, but was strongest in 3-, 6-, and 21-week-old mice. Protein loading was controlled by staining with an anti-GAPDH antibody. b Hippocampal neuron (DIV15) transfected with Tspan15-YPET. Arrows indicate examples of Tspan15-positive dendritic punctae. c Kymograph showing dendritic trafficking of Tspan15. Position of particles along a dendritic stretch (x-axis) during 20 s of image acquisition (y-axis). Stationary particles shown as vertical lines (blue), anterograde (red), and retrograde (green) as diagonals. The slope correlates with transport speed. d Quantification of average velocity (on the left) and run length (on the right) for anterogradely and retrogradely moving Tspan15 vesicles (n is number of moving vesicles, data from 7 dendrites, 3 cells). Mean ± SEM. Two-tailed Student’s t test. e Tspan15 punctae are partially co-localizing with the satellite Golgi marker pGolt-mCherry, but not with mitochondria (MitoTracker). D dendrite, A axon, S dendritic spine. Blue arrow indicates Tspan15 cluster. Also see Supplementary Video S1. f Quantification of co-localization of Tspan15 puncta with pGolt-mCherry or MitoTracker. Mean ± SEM [n is number of analyzed neurons, data from three independent cultures (pGolt-mCherry), two coverslips (MitoTracker)]
Tspan15 interacts with endogenous ADAM10 in the brain
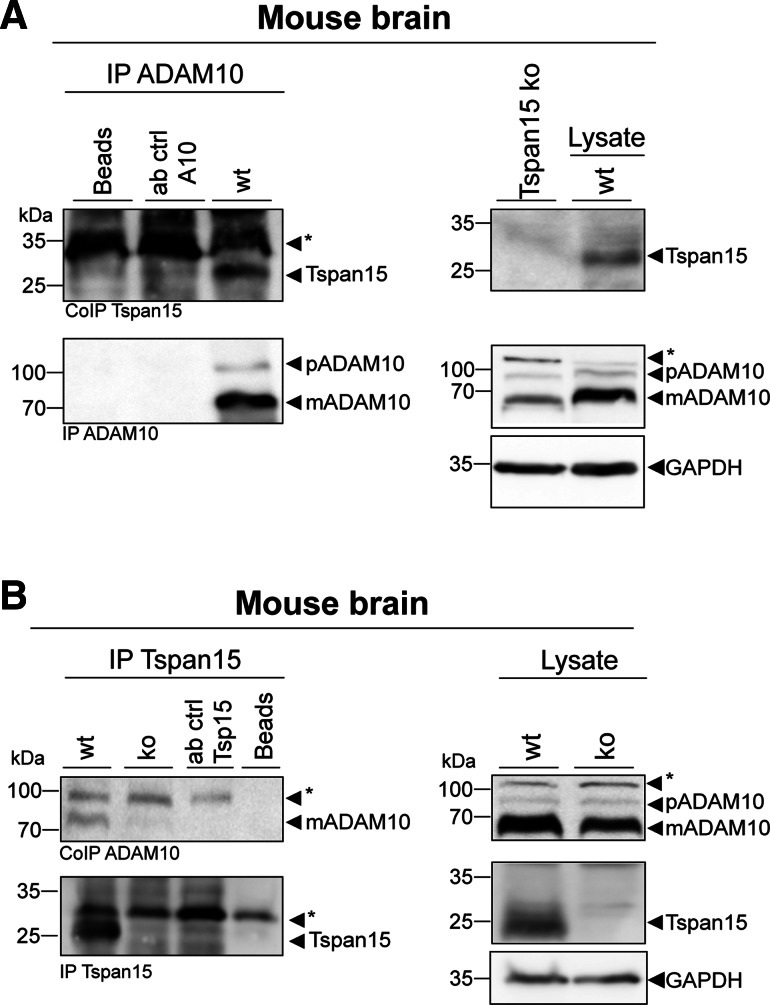
Experiments in overexpressing cells revealed the physical interaction of Tspan15 and ADAM10 [14]. To address the question of whether this association can also be demonstrated at the endogenous level in mouse brain, we precipitated ADAM10 and found the pro- and mature form of the protease (Fig. 3a, IP ADAM10). Tspan15, which was absent in Tspan15 knockout mouse brain lysates (see below), could also be found in this wild-type brain precipitate (Fig. 3a, CoIP Tspan15). Endogenous Tspan15 could also be precipitated from wild-type, but not from Tspan15 knockout brain lysates, confirming specificity of the Tspan15 precipitation (Fig. 3b, IP Tspan15). Likewise, co-precipitated mature ADAM10 was found in the wild-type, but not in the Tspan15 knockout precipitate (Fig. 3b, CoIP ADAM10). These findings demonstrate the specific interaction of both proteins also under endogenous conditions in the murine brain (Fig. 3a, b).
Fig. 3.
Tspan15 interacts with ADAM10 in murine brain. a Using a C-terminal specific anti-ADAM10 antibody (ADAM10 Pin1), ADAM10 was precipitated from total brain homogenate of a wild-type (wt) mouse. After immunoblotting, efficient immunoprecipitation was confirmed by detection of pro- (p) and mature (m) ADAM10 in the precipitate (IP ADAM10) using the ADAM10 EPR5622 antibody. In the same immunoprecipitate, Tspan15 was detected with an anti-Tspan15 antibody (CoIP Tspan15, Tspan15 T2EL), confirming Tspan15 as an interaction partner of ADAM10 (immunoblot is representative for two independent experiments). b Other way around immunoprecipitation of Tspan15 from wild-type (wt) and Tspan15 knockout (ko) brain homogenates was performed using a self-made C-terminal Tspan15 antibody (anti-Tspan15 T3CT). A Tspan15-specific signal was detected (anti-Tspan15 T2EL) in the wild-type, but not in the knockout precipitate (IP Tspan15). Likewise, co-immunoprecipitation of mature ADAM10 (CoIP ADAM10) was observed in the wild-type, but not in the Tspan15 knockout precipitate. A lysate-free sample containing lysis buffer and the anti-ADAM10 (a) or the anti-Tspan15 T3CT (b) antibody, respectively, served as antibody control (ab ctrl). Unspecific protein binding to the beads was controlled by incubation of beads with the homogenate and without antibody (a + b, beads). Expression of ADAM10 and Tspan15 was verified in the respective wild-type lysates (blots on the right). In addition, specificity of the anti-Tspan15 antibody (Tspan15 T2EL) was confirmed by the absence of a signal in a Tspan15 knockout (ko) brain sample. Asterisks (*) mark unspecific signals
Tspan15 deletion in mice
To further study the role of Tspan15 for ADAM10-mediated shedding events in vivo, a Tspan15 knockout mouse model was generated and analyzed. To achieve a gene-specific knockout of Tspan15, transcription activator-like effector nuclease (TALEN)-mediated genome editing was performed (Suppl. Fig. S3A). The TALEN-RNA was designed to target exon2 of the Tspan15 gene locus and microinjected into zygotes of a C57BL/6N founder mouse. Genomic DNA from offspring mice was isolated and analyzed for the presence of frameshift mutations. PCR and sequence analysis of Tspan15 exon2 revealed a 104 bp deletion mutation in the TALEN-targeted region, which was sufficient to introduce a premature translational stop codon (Suppl. Fig. S3B). To validate that the observed frameshift mutation was efficient to cause loss of the Tspan15 protein, the expression level of Tspan15 was analyzed in different organs isolated from a homozygous Tspan15Δ104 knockout mouse. Immunoblotting followed by detection of Tspan15 with an anti-Tspan15 antibody revealed prominent signals at around 30 kDa in the tested wild-type (wt) tissues, which correspond to endogenous Tspan15 (Suppl. Fig. S3C). No signals were detected in the respective Tspan15 knockout (ko) samples, confirming the successful deletion of the Tspan15 protein in various tissues.
During breeding of Tspan15 knockout mice, it was conspicuous that an increased number of animals died within the first weeks after birth. To calculate the postnatal mortality rate, the number of total born mice and mice, that were still alive 3 weeks after birth, was determined (Suppl. Fig. S3D). Calculation of the percentage of total alive to total born animals revealed a slightly increased mortality rate of 16% within the first 3 weeks after birth, which could indicate the occurrence of early developmental defects. However, genotype analysis of the surviving mice did not reveal apparent differences and corresponds to the expected Mendelian distribution with 24% wild-type (wt), 50% heterozygous (+/−), and 26% knockout (ko) animals.
Tspan15 deficiency impairs ADAM10 maturation and shedding activity in the brain
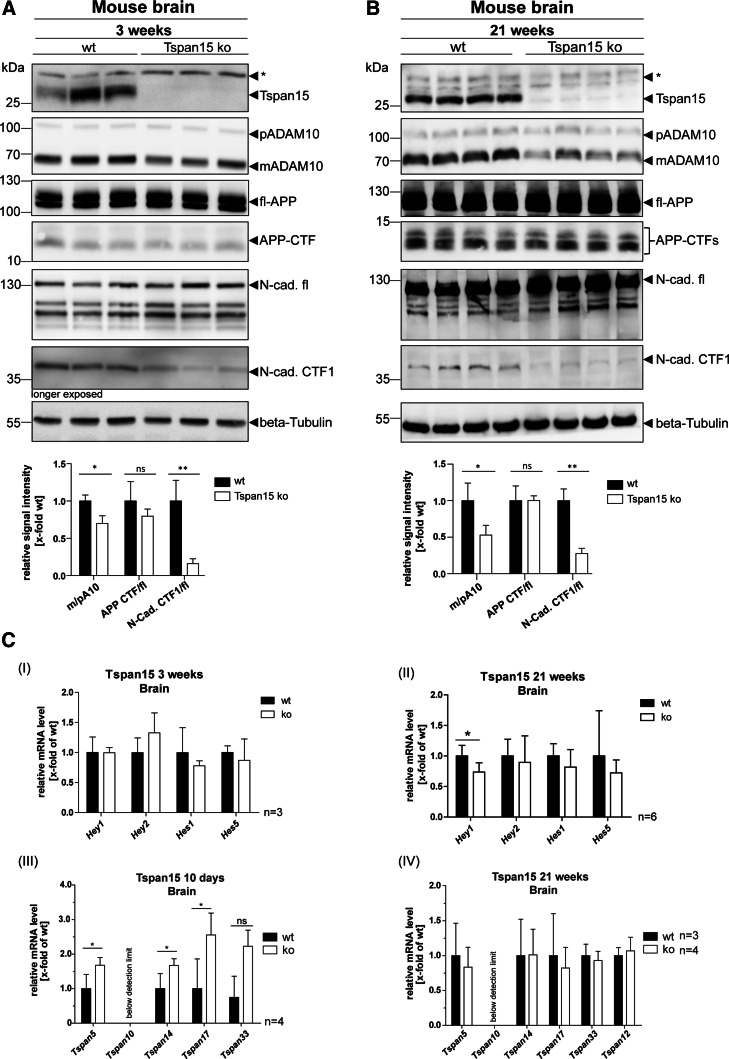
Since Tspan15 revealed a prominent expression in the brain (Fig. 1a), where the function of ADAM10 is well characterized (reviewed in Ref. [1]), the influence of Tspan15 deficiency in the brain was further analyzed. First, the impact of Tspan15 on ADAM10 maturation and proteolytic activity was analyzed in brain homogenates of 3-week-old wild-type and knockout mice. Therefore, tissue lysates were prepared and the expression of ADAM10 was analyzed by immunoblot analysis (Fig. 4a). Staining with an anti-Tspan15 antibody led to prominent signals at 30 kDa in the wild-type (wt) samples, whereas no signals were detected in the Tspan15 knockout (Tspan15 ko) samples, confirming the absence of Tspan15. In correlation with the loss of Tspan15, detection of ADAM10 in Tspan15 knockout samples revealed clearly decreased signal intensities of mature (m) ADAM10, compared to wild-type samples (Fig. 4a). In contrast, similar signal intensities of immature pro- (p) ADAM10 were observed in both wild-type and Tspan15 knockout samples. To evaluate whether the decreased appearance of mature ADAM10 has an influence on the shedding of ADAM10 substrates, proteolytic processing of APP and N-cadherin, two well-described ADAM10 substrates, was analyzed. Detection of APP with a C-terminal anti-APP antibody revealed no differences in the signal intensity of the full-length (fl-) APP and the APP-C-terminal fragment (CTF) in wild-type and Tspan15 knockout samples. In contrast to APP, detection of N-cadherin revealed an increased expression of full-length N-cadherin (N-cad.fl) in the brains of Tspan15 knockout mice compared to wild-type samples. Using the same antibody and a longer exposure time, a significant decrease of the ADAM10-generated N-cadherin cleavage product, CTF1 (N-cad. CTF1), was also observed in the Tspan15 knockout samples (Fig. 4a).
Fig. 4.
Loss of Tspan15 reduces ADAM10 maturation and N-cadherin shedding. Brain homogenates of a 3-week-old and b 21-week-old wild-type (wt) and Tspan15 knockout (ko) mice were prepared and analyzed by immunoblot. Tspan15-deficiency in Tspan15 ko samples was confirmed by the absence of specific signals after staining with an anti-Tspan15 antibody (Tspan15 T2EL). Using an anti-ADAM10 antibody (ADAM10 EPR5622) expression of pro- (p) and mature (m) ADAM10 was detected in all samples. Compared to respective wild-type samples, expression of mADAM10 was clearly reduced in 3-week-old (a) and 21-week-old (b) Tspan15 knockout mice. Staining for APP with a C-terminal anti-APP antibody revealed no differences between Tspan15 knockout and wild-type mice (a + b) in the expression of full-length (fl-) APP and the APP-C-terminal fragment (CTF). Expression of full-length N-cadherin (N-cad.-fl) and the C-terminal fragment CTF1 generated by ADAM10 (N-cad. CTF1) was monitored with an anti-N-cadherin antibody. N-cadherin-CTF1 production was significantly reduced in 3-week-old (a) and 21-week-old (b) Tspan15 knockout mice compared to respective wild-type samples. Beta-Tubulin staining was used to control for equal protein loading. Asterisks (*) mark unspecific signals. Quantitative analysis was performed by calculation of relative signal intensities for ADAM10 (m/p), APP (CTF/fl), and N-cadherin (CTF1/fl). c (I–II) Tspan15-deficiency has only a minor effect on Notch downstream gene expression. To analyze the influence of the Tspan15 knockout on Notch shedding, the transcription level of Notch downstream genes Hey1, Hey2, Hes1, and Hes5 was analyzed in Tspan15 wild-type (wt) and knockout (ko) mice. mRNA was isolated from 3-week-old (a) and 21-week-old (b) animals. After reverse transcription into cDNA, relative mRNA levels were measured by qRT-PCR. ΔCt values were calculated and normalized to wild-type samples. (I) Loss of Tspan15 had no significant influence on the relative transcription levels of Hey1, Hey2, Hes1, and Hes5 in 3-week-old mice (n = 3). (II) In 21-week-old mice, Tspan15-deficiency only increased the relative mRNA level of Hey1 but had no effect on Hey2, Hes1, and Hes5 (n = 6). Statistical significance was tested using Student’s t test (*p < 0.05). (III–IV) Loss of Tspan15 in young mice increases transcription levels of other TspanC8 members. Relative transcription levels of TspanC8 tetraspanins 5, 10, 14, 17, and 33 were analyzed. mRNA was isolated from brains of 10-day-old (a) and 21-week-old (b) Tspan15 wild-type (wt) and knockout (ko) mice, reverse transcribed into cDNA, and TspanC8 transcription levels were measured by qRT-PCR. ΔCt values were calculated and normalized to wild-type samples. (III) The relative mRNA levels of Tspan5, 14, and 17, but not Tspan33, were significantly increased in 10-day-old Tspan15 ko mice compared to respective wt samples (n = 3). (IV) In 21-week-old mice, no significant differences in the transcription levels of the respective TspanC8 tetraspanins were observed between Tspan15 ko (n = 4) and wt (n = 3) mice. Tspan10 mRNA levels were below the detection limit (a + b). Values of wt samples were set to one and data are shown as mean values ± SD. Statistical significance was analyzed by Student’s t test (*p < 0.05, **p < 0.01)
To validate these findings in adult mice, the expression of ADAM10, APP, and N-cadherin was also assessed in brain homogenates of 21-week-old mice (Fig. 4b). Similar to the previous findings, the signal intensities of mature ADAM10 were significantly reduced in brains of Tspan15 knockout mice compared to wild-type samples. Interestingly, with a 50% reduction in the relative signal intensity of m/p ADAM10 (0.5 ± 0.06), this effect was even more pronounced in 21-week-old mice compared to 3-week-old mice (0.7 ± 0.06). The reduction of mature ADAM10 in 21-week-old animals was again accompanied by a clearly decreased appearance of the N-cadherin CTF1, but did not influence the expression of fl-APP and APP-CTFs in Tspan15 knockout samples (Fig. 4b).
In addition to N-cadherin and APP, the mRNA levels of Notch downstream target genes Hey1, Hey2, Hes1, and Hes5 were analyzed by qRT-PCR in 3- and 21-week-old mice (Fig. 4c, I + II). Loss of Tspan15 had no significant influence on the transcription levels of Notch downstream genes in 3-week-old mice and only reduced the mRNA level of Hey1 in 21-week-old Tspan15 knockout mice.
In combination with the observation that loss of Tspan15 had a more pronounced effect on the expression of mature ADAM10 and substrate cleavage in older mice, mRNA levels of TspanC8 tetraspanins 5, 14, 17, and 33 are significantly increased in 3-week-old Tspan15 knockout mice, but unchanged in 21-week-old mice (Fig. 4c, III + IV). This indicates that other TspanC8 members could partially compensate for the loss of Tspan15 during the early developmental stages, but not in aged mice.
In conclusion, the knockout of Tspan15 in mice led to a decreased expression of mature ADAM10 and significantly reduced shedding of N-cadherin. This effect was present in 3-week-old mice and more pronounced in 21-week-old Tspan15 knockout animals. Surprisingly, loss of Tspan15 and the concomitant reduction in mature ADAM10 did not influence the proteolytic processing of APP in young and adult mice and had only a minor effect on the transcription levels of Notch target genes in 21-week-old mice.
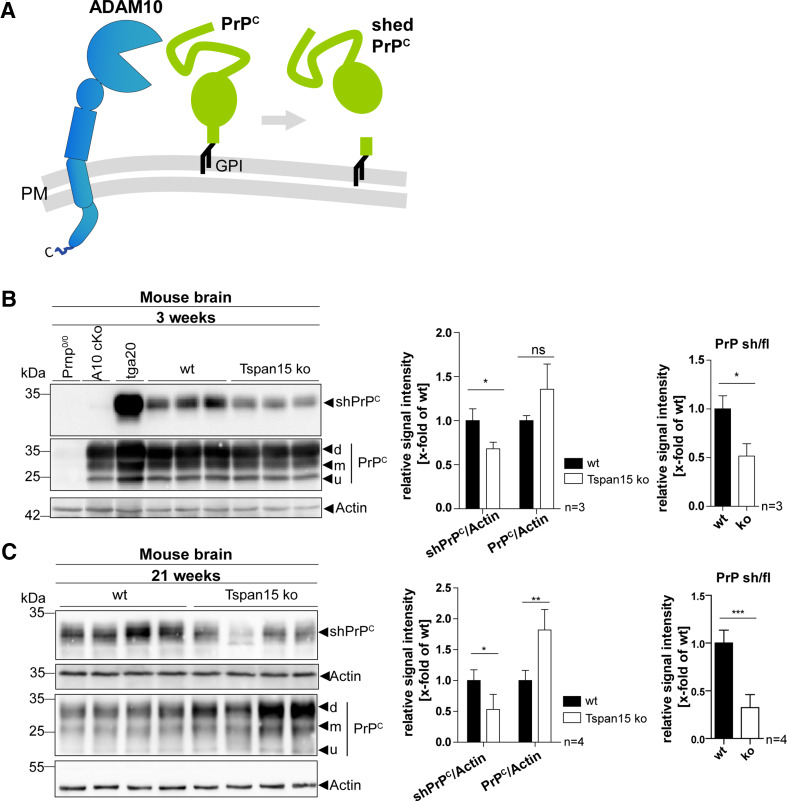
The shedding of the cellular prion protein is regulated by Tspan15
Due to the fact that a neuron-specific knockout of ADAM10 in mice prevented shedding of the cellular prion protein (PrPC) [34], it was further of interest, whether the decreased maturation of ADAM10 in Tspan15 knockout mice has a similar effect on PrPC shedding as observed for N-cadherin. The appearance of GPI-anchored full-length PrPC and its soluble shed form (shPrPC), which is released upon ADAM10 cleavage, was analyzed (Fig. 5a). Brain homogenates were prepared from 3-week-old wild-type and Tspan15 knockout mice (Fig. 5b). In addition, brain homogenates of PrPc-deficient mice (Prnp0/0), ADAM10 conditional knockout mice (A10 cKO), and PrPc overexpressing mice (tga20) were used as controls. Tissue lysates were subjected to immunoblotting, and expression of full-length PrPC and shPrPC was analyzed (Fig. 5b). Using an anti-shPrPC antibody, raised against the new carboxy-terminus upon ADAM10-mediated cleavage, shPrPC was detected in tga20 homogenates, but not in PrPc-deficient and A10 cKO samples, indicating specificity of the antibody (Fig. 5b). In comparison to wild-type (wt) homogenates, the shPrPC signal intensities were significantly reduced in the brains of Tspan15 knockout (ko) mice. Staining for full-length PrPC, using a C-terminal anti-PrPC antibody, revealed no significant differences of PrPC (di-, mono-, and unglycosylated) signal intensities in wild-type and Tspan15 knockout brains at this age (Fig. 5b).
Fig. 5.
Tspan15 deficiency reduces shedding of the cellular prion protein (PrPC). a Schematic drawing showing ADAM10 and the cellular prion protein (PrPC) with its GPI anchor at the plasma membrane (PM). ADAM10-mediated shedding of PrPC occurs in close proximity to the PM and releases nearly full-length PrPC as a shed, soluble form (shPrPC). b, c Immunoblot analysis of the proteolytic processing of PrPC in brain homogenates prepared from 3-week-old (b) and 21-week-old (c) Tspan15 wild-type (wt) and knockout (ko) mice. Shed PrPC (shPrPC) was detected with an antibody specifically directed against the novel C-terminus generated through cleavage by ADAM10 (shed PrPC). Membrane-bound di (d)-, mono- (m), and unglycosylated (u) full-length PrPC was detected with a C-terminal anti-PrPC antibody (Pom1,2). As controls, brain samples from PrP-deficient mice (Prnp0/0), conditional ADAM10 knockout mice (A10 cKO) with a postnatal deletion in forebrain neurons and PrPC overexpressing mice (tga20) were used (b). For quantitative analysis, the relative signal intensities of shPrPC/actin (b, c), and PrPC/actin (b + c) were calculated. Values of wt samples were set to one and mean values of n = 3 (a) and n = 4; (b) samples are shown ± SD. Statistical significance was analyzed by Student’s t test (*p < 0.05, **p < 0.01)
However, analysis of PrPC shedding in 21-week-old mice (Fig. 5c) not only revealed significantly reduced levels of shPrPC in Tspan15 knockout brain homogenates. At this age, reduced shedding of PrPC was also paralleled by a significant increase of full-length PrPC (di-, mono-, and unglycosylated).
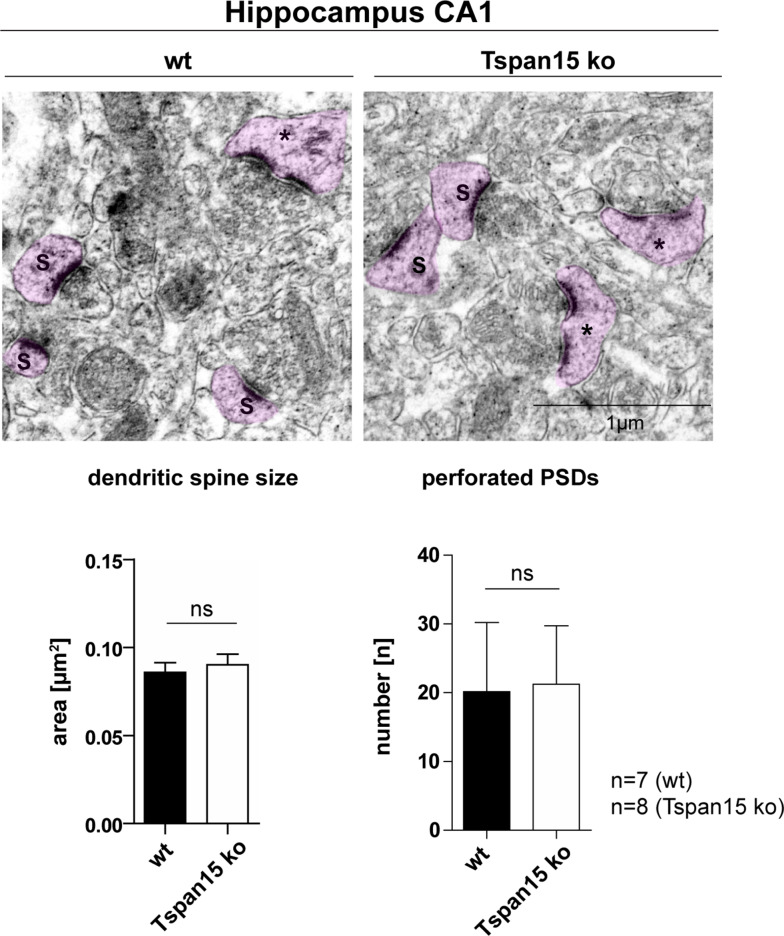
Morphology of synapses in Tspan15-deficient brains is unaltered
Previously, we showed that the postnatal neuronal depletion of ADAM10 in mice is associated with morphological changes of synaptic spine structure and leads to learning deficits and impairs synaptic plasticity [29]. In an ultrastructural analysis with transmission electron microscopy, we further revealed that ADAM10 deficiency in hippocampal neurons leads to an enlargement of dendritic spine heads with a shrunken and stubby shape [29]. To investigate whether the reduced presence of mature ADAM10 in Tspan15-deficient mice could have an effect on the morphology of dendritic spines, ultrastructural analysis of hippocampal neurons of Tspan15 knockout and wild-type mice was performed (Fig. 6). The electron micrographs revealed dendritic spine heads with a continuous postsynaptic density (S) and spines with a discontinuous or “perforated” (*) postsynaptic density (PSD), but showed no obvious difference in the overall morphology of dendritic spine heads. This was further supported after measuring dendritic spine head size and number of perforated PSDs, which were both unaltered by the loss of Tspan15 in comparison to wild-type samples (Fig. 6). In addition, histological analysis of the Tspan15 wild-type and knockout brains using H&E and DAB staining for the glia-specific protein GFAP, the microglia-specific protein Iba1, and synaptophysin did not reveal any apparent alterations in Tspan15-deficient mice (see Suppl. Fig. S4).
Fig. 6.
Loss of Tspan15 has no effect on the morphology of dendritic spines. Representative electron micrographs of the hippocampal CA1 region of wild-type (wt) and Tspan15-deficient mice (ko). Dendritic spines with a continuous (S) and perforated (*) postsynaptic density (PSD) are highlighted in pink. The size of dendritic spines (S) of n = 7 wild-type (wt) and n = 8 Tspan15 knockout (Tspan15 ko) animals was measured using ImageJ. Mean values ± SD of the measured spine area are shown in µm2. The number of perforated PSDs (*) of wild-type (wt) and Tspan15 knockout (ko) animals was counted. Data are shown as mean values ± SD. Statistical significance was analyzed by Student’s t test. No significant differences (ns) were observed
Although the loss of Tspan15 clearly decreased the expression of mature ADAM10 and the cleavage of important neuronal molecules, such as N-cadherin and PrPC, knockout of Tspan15 had no apparent influence on the overall hippocampal morphology and the ultrastructure of dendritic spines. It should be noted that in aged mice, no overt neurological symptoms were observed (Suppl. Fig. S5).
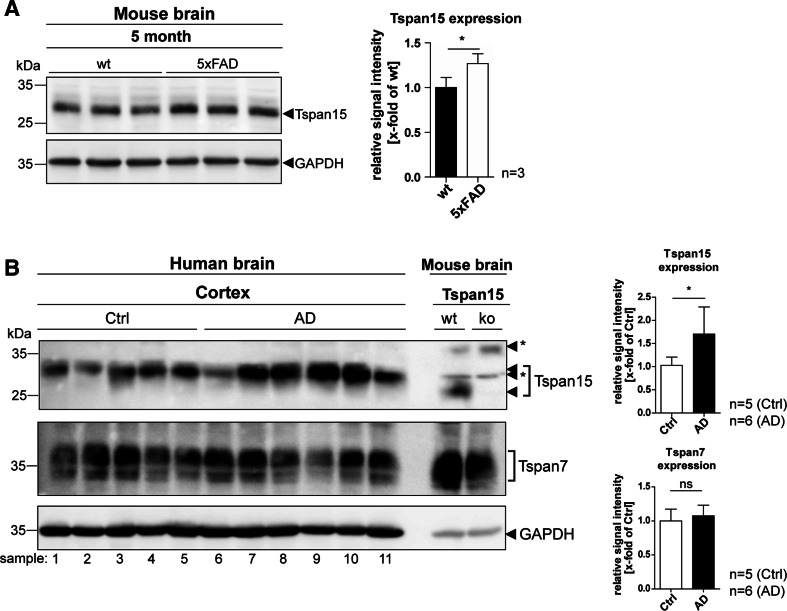
Tspan15 expression is increased in AD mouse models and in cortical AD patient material
The physiologically relevant α-secretase of APP, ADAM10, plays a central role in the context of Alzheimer’s disease [35, 36]. While reduced ADAM10 activity contributes to AD pathology, upregulation of ADAM10 activity is thought to be a promising therapeutic approach to treat AD [37, 38]. Although Tspan15-deficiency did not show an influence on the proteolytic processing of APP in our mouse model, the above shown experiments confirmed Tspan15 as an important regulator of ADAM10 maturation and activity in vivo. To test whether Tspan15 is associated with AD pathology, Tspan15 expression was analyzed in the brains of 5xFAD mice, a well-established model for AD. In comparison to age-matched wild-type samples, Tspan15 expression was significantly increased in the brains of 5xFAD mice (Fig. 7a). Importantly, in addition to murine AD samples, expression of Tspan15 expression was also significantly increased in prefrontal cortex samples of human AD patients compared to age-matched controls, whereas the expression level of a different tetraspanin, Tspan7, was similar in both groups (Fig. 7b, more detailed information about the patient material is provided in Suppl. Table S1).
Fig. 7.
Tspan15 expression is upregulated in Alzheimer’s disease model mice and patients’ brains. a Immunoblot analysis of brain homogenates of 5-month-old wild-type (wt) and 5xFAD mice. Detection of Tspan15 (Tspan15 T2EL) and subsequent quantification of signal intensities revealed an increased Tspan15 expression in 5xFAD compared to wild-type samples. b Tspan15 expression was analyzed in post-mortem prefrontal cortex samples of human Alzheimer’s disease (AD, n = 6) patients and patients without diagnosed neurodegeneration (Ctrl, n = 5). Tspan15 wild-type (wt) and knockout (ko) brain homogenates were used to confirm specificity of the anti-Tspan15 antibody (NBP1-92540). In addition, Tspan7 expression was analyzed with a self-made anti-Tspan7 antibody. Equal protein loading was verified by GAPDH staining. Values are shown as mean ± SD. Statistical significance was tested using Student’s t test (*p < 0.05). Numbers indicate patient samples as given in Suppl. Table S1. Asterisks (*) mark unspecific signals
Discussion
Tetraspanins have emerged as interactors of proteases and critical regulators of the processing of APP and other surface proteins [16]. Tspan15 is one of several tetraspanins controlling ADAM10 maturation, its trafficking to the cell surface, and substrate cleavage. Based on yeast and overexpression experiments, Tspan15 was found as an ADAM10 interaction partner [13, 14]. In particular, Tspan15 promoted N-cadherin cleavage after overexpression in several cell lines [14, 25]. Tspan15 was also identified as a negative regulator of Notch activity and, depending on the cell type, Tspan15 expression reduced or promoted APP proteolytic processing [14]. Despite an increasing appreciation of the functions of Tspan15 and other TspanC8 tetraspanins modulating ADAM10 activity and substrate cleavage, their general physiological relevance and their specific roles in modulation of ADAM10 are not well characterized.
Here, we demonstrate that Tspan15 is an important regulator of ADAM10 in the murine brain, where—as in lung and intestine—it is highly expressed and interacts with ADAM10. To better understand the role of Tspan15 for ADAM10-mediated shedding in vivo, we generated a Tspan15 knockout mouse model, which is after Tspan33 [13, 39], the second TspanC8 member depleted in mice. Tspan15-deficient mice did not display an overt phenotype, but it was conspicuous that a considerable fraction (16%) of the born animals died within the first 3 weeks, which likely results from a general enhanced postnatal lethality during the early developmental stages.
Although TspanC8 tetraspanins (except for Tspan10 and Tspan17) have redundant functions in terms of ADAM10 maturation and trafficking [12, 13], it was rather surprising that in Tspan15-deficient brains, a significant reduction of mature ADAM10 was revealed. This finding underscores the predominant role of Tspan15 for ADAM10 maturation in vivo. As previously demonstrated, Tspan15 binding to ADAM10 occurs already in the ER. This binding event triggers the ER export and surface delivery of the protease. An ER retention mutant of Tspan15 prevented the ER exit of ADAM10 [14]. Since the proform of ADAM10 is cleaved within the Golgi, a lack of Tspan15 causes a reduced transport of the protease from the ER and a decreased proteolytic maturation and generation of the active/cell surface expressed form of the protease. Our in vivo results analyzing the Tspan15 knockout brains revealed a reduced mature and active ADAM10 protease strongly confirming our cell-based experiments and supporting the model of Tspan15 (TspanC8) mediating the ER cell surface transport of ADAM10.
Analysis of the transcription levels of TspanC8 genes further revealed that loss of Tspan15 is likely partially compensated in younger animals, while no such compensation (i.e., increased mRNA expression of other TspanC8 family members) was found in older mice. In this regard, it should be noted that loss of Tspan15 did not completely abolish ADAM10 maturation, demonstrating that also other Tspan15-independent mechanisms control ADAM10 maturation. These mechanisms likely involve TspanC8 members Tspan5, 14, which also promote ADAM10 maturation and activity in cell-based assays and are highly expressed in different mouse brain cell types, including neurons, oligodendrocytes, and astrocytes as revealed by RNA sequencing [18].
In addition to the decreased level of mature ADAM10, the shedding of the ADAM10 substrates N-cadherin and the cellular prion protein (PrPC) were also reduced in Tspan15-deficient mice. ADAM10-mediated N-cadherin shedding is essential for cell adhesion and has important functions in the activation of β-catenin signalling [40]. As a neuronal adhesion molecule, N-cadherin is associated with synaptic adhesion, spine morphology, and plasticity [41–43]. A conditional knockout (cKO) of ADAM10 in the brain reduced N-cadherin shedding and caused severe developmental defects in the cortex, learning deficits, impaired network formation, and led to changes in dendritic spine morphology [29, 36]. Despite reduced N-cadherin shedding and localization near synaptic spines, our morphological analyses in the Tspan15 knockout mice did not reveal obvious synaptic changes of hippocampal CA1 neurons. It could be that the remaining ADAM10 activity—as revealed by the residual mature form of the protease seen in immunoblot—is sufficient to cleave enough N-cadherin and other synaptic cell surface molecule [11] to allow an apparently normal development of synapses, an assumption which is supported by the observation that ADAM10 heterozygote mice lack an obvious neurological phenotype. More detailed functional studies have to be performed to understand if Tspan15 deficiency may have some milder impact on synaptic transmission.
The physiological function of PrPC is not yet well understood. It is proposed to function in neurogenesis, synapse formation and differentiation of neuronal precursor cells, maintenance of myelin, and in the immune system [44–46]. Its role in neurodegenerative diseases is well documented (reviewed in Refs. [47, 48]). By conversion into the misfolded pathological form, PrP scrapie (PrPSc), the prion protein is the main causative agent of transmissible prion diseases in humans and other mammalian species [49]. ADAM10 is the main (if not only) sheddase of PrPC and releases a soluble GPI-anchorless form of PrPC [50]. Depletion of ADAM10 in neurons abolished PrPC shedding and resulted in an accumulation of full-length PrPC in the early secretory pathway and at the plasma membrane [34, 50]. After prion infection of mice deficient for neuronal ADAM10, an increased conversion of PrPC into PrPSc and shortened incubation times to prion disease, despite a simultaneously reduced spreading of pathology, were observed [34]. It is conceivable and supported by our observations of a decreased PrPC shedding that the knockout of Tspan15 could have a similar effect in prion disease. Detailed analyses of whether Tspan15, indeed, influences shedding and thus pathology in prion disease are of importance in this regard.
It should be noted that according to our immunoblot analyses, APP proteolytic processing was not affected by the loss of Tspan15 in the brain. This is in line with the previous cell-based findings, which revealed that Tspan15 has only a minor role in APP processing and that its ability to promote APP shedding rather seems to depend on the analyzed cell type or the rate of APP (over)expression [14].
It will be of interest to see how disease progression and amyloid pathology will develop in AD mouse models in a situation of Tspan15-deficiency. On the other hand, a direct or indirect role of Tspan15 is also suggested by our finding of its increased expression in the brains of 5xFAD mice and AD patients. However, such a finding could, as well, simply reflect an apparently non-successful compensatory response in patients’ brains to counterbalance the excessive neurotoxic production of amyloid beta peptides during the course of disease.
Taken together, our loss of function studies identified Tspan15 as an important regulator of ADAM10 maturation and shedding activity in vivo. The finding that loss of Tspan15 reduces N-cadherin and PrPC shedding, while it has no or only a minor effect on the proteolytic processing of APP and Notch, provides the first in vivo evidence for the hypothesis of a substrate specific role of TspanC8 tetraspanins, which supports the idea that binding of a TspanC8 constrains ADAM10 in a conformation, where it favours cleavage of specific substrates [17–19, 25]. The lack of an overt mouse phenotype implicates that Tspan15 is only one of many players to modulate ADAM10 activity (and possibly also other proteins expressed at the cell surface). Lack of Tspan15 in mice may, therefore, be efficiently compensated and only subtle alterations of the physiological function of ADAM10 and its substrate cleavage are found. How Tspan15 regulates ADAM10 activity in vivo and whether additional substrates are affected by the loss of Tspan15 will be interesting questions for future experiments. Moreover, it will be worthwhile to perform a more detailed analysis of its importance in pathological processes, such as prion and Alzheimer’s disease. Since ADAM10 is an interesting drug target, it may, as well, be wise to study if specific tetraspanins, including Tspan15, can be exploited to modulate ADAM10 activity in a more substrate- and tissue-specific manner to avoid severe side-effects.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
This work was supported by the Deutsche Forschungsgemeinschaft (DFG) Sonderforschungsbereich 877 [projects A3 (P.S.), Z2 (T.K., A.T.) and A12 (P.S., H.A., and M.G.)], the Creutzfeldt-Jakob Disease Foundation, Inc., and the Werner-Otto-Stiftung (to H.A.). R.S. was supported by CZ.1.05/2.1.00/19.0395 (MEYS) and Academy of Sciences of the Czech Republic (RVO 68378050). M. Mikhaylova is supported by grants from the Deutsche Forschungsgemeinschaft [DFG Emmy-Noether-Programm (Ml 1923/1-1) and FOR2419 (MI 1923/2-1)]. We gratefully thank Dr. Sebastian Wetzel and Dr. Dirk Schmidt-Arras for cloning the Tspan15-YPET construct. We also thank Kristin Hartmann (Mouse Pathology Core Unit, UKE, Hamburg) and Emanuela Szpotowicz and Chundamani Raithore (Prague) for valuable technical support.
Abbreviations
- AD
Alzheimer’s disease
- ADAM
A Disintegrin And Metalloproteinase
- APP
Amyloid precursor protein
- CA1
CA1 layer of the hippocampus
- CC
Corpus callosum
- CNS
Central nervous system
- cKO
Conditional knockout
- CTF
C-terminal fragment
- Cx
Cortex
- fl
Full-length
- GFAP
Glial fibrillary acidic protein
- GPI
Glycosylphosphatidylinositol
- IB
Immunoblot
- Iba1
Ionized calcium-binding adaptor molecule 1
- IP
Immunoprecipitation
- ko
Knockout
- LEL
Large extracellular loop
- LIMP-2
Lysosomal integral membrane protein-2
- NeuN
Neuronal neuclei
- PrPC
Cellular prion protein
- PrPSc
Scrapie PrP
- shPrPC
Shed PrPC
- PNGase F
Peptide N-glycosidase F
- PSD
Postsynaptic density
- SEL
Small extracellular loop
- Tspan
Tetraspanin
- TALEN
Transcription activator-like effector nuclease
- TEM
Tetraspanin-enriched microdomain
- wt
Wild-type
- 5xFAD
5 Familial AD mutations
Compliance with ethical standards
Ethical standards
All animal studies were performed according to the guidelines of the Federation of European Laboratory Animal Science Associations (FELASA) and were approved by the animal welfare committee of the Ministry of Energy, Agriculture, the Environment, Nature and Digitalization, Schleswig-Holstein, Germany (protocols V242.7224.121-3, V242-80867/2016). The use of specimens and basic clinical information were in agreement with the regulations and ethical standards at the contributing hospitals and written consent by patients or relatives was obtained where necessary.
Conflict of interest
The authors declare that they have no conflict of interest.
References
- 1.Saftig P, Lichtenthaler SF. The alpha secretase ADAM10: a metalloprotease with multiple functions in the brain. Prog Neurobiol. 2015;135:1–20. doi: 10.1016/j.pneurobio.2015.10.003. [DOI] [PubMed] [Google Scholar]
- 2.Howard L, Lu X, Mitchell S, Griffiths S, Glynn P. Molecular cloning of MADM: a catalytically active mammalian disintegrin-metalloprotease expressed in various cell types. Biochem J. 1996;317(Pt 1):45–50. doi: 10.1042/bj3170045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Karkkainen I, Rybnikova E, Pelto-Huikko M, Huovila AP. Metalloprotease-disintegrin (ADAM) genes are widely and differentially expressed in the adult CNS. Mol Cell Neurosci. 2000;15:547–560. doi: 10.1006/mcne.2000.0848. [DOI] [PubMed] [Google Scholar]
- 4.Marcinkiewicz M, Seidah NG. Coordinated expression of beta-amyloid precursor protein and the putative beta-secretase BACE and alpha-secretase ADAM10 in mouse and human brain. J Neurochem. 2000;75:2133–2143. doi: 10.1046/j.1471-4159.2000.0752133.x. [DOI] [PubMed] [Google Scholar]
- 5.Anders A, Gilbert S, Garten W, Postina R, Fahrenholz F. Regulation of the alpha-secretase ADAM10 by its prodomain and proprotein convertases. FASEB J. 2001;15:1837–1839. doi: 10.1096/fj.01-0007fje. [DOI] [PubMed] [Google Scholar]
- 6.Escrevente C, Morais VA, Keller S, Soares CM, Altevogt P, Costa J. Functional role of N-glycosylation from ADAM10 in processing, localization and activity of the enzyme. Biochim Biophys Acta. 2008;1780:905–913. doi: 10.1016/j.bbagen.2008.03.004. [DOI] [PubMed] [Google Scholar]
- 7.Lopez-Perez E, Seidah NG, Checler F. Proprotein convertase activity contributes to the processing of the Alzheimer’s beta-amyloid precursor protein in human cells: evidence for a role of the prohormone convertase PC7 in the constitutive alpha-secretase pathway. J Neurochem. 1999;73:2056–2062. [PubMed] [Google Scholar]
- 8.Lammich S, Kojro E, Postina R, Gilbert S, Pfeiffer R, Jasionowski M, Haass C, Fahrenholz F. Constitutive and regulated alpha-secretase cleavage of Alzheimer’s amyloid precursor protein by a disintegrin metalloprotease. Proc Natl Acad Sci USA. 1999;96:3922–3927. doi: 10.1073/pnas.96.7.3922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reiss K, Saftig P. The “a disintegrin and metalloprotease” (ADAM) family of sheddases: physiological and cellular functions. Semin Cell Dev Biol. 2009;20:126–137. doi: 10.1016/j.semcdb.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 10.Weber S, Saftig P. Ectodomain shedding and ADAMs in development. Development. 2012;139:3693–3709. doi: 10.1242/dev.076398. [DOI] [PubMed] [Google Scholar]
- 11.Kuhn PH, Colombo AV, Schusser B, Dreymueller D, Wetzel S, Schepers U, Herber J, Ludwig A, Kremmer E, Montag D et al (2016) Systematic substrate identification indicates a central role for the metalloprotease ADAM10 in axon targeting and synapse function. Elife 5 [DOI] [PMC free article] [PubMed]
- 12.Dornier E, Coumailleau F, Ottavi JF, Moretti J, Boucheix C, Mauduit P, Schweisguth F, Rubinstein E. TspanC8 tetraspanins regulate ADAM10/Kuzbanian trafficking and promote Notch activation in flies and mammals. J Cell Biol. 2012;199:481–496. doi: 10.1083/jcb.201201133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haining EJ, Yang J, Bailey RL, Khan K, Collier R, Tsai S, Watson SP, Frampton J, Garcia P, Tomlinson MG. The TspanC8 subgroup of tetraspanins interacts with A disintegrin and metalloprotease 10 (ADAM10) and regulates its maturation and cell surface expression. J Biol Chem. 2012;287:39753–39765. doi: 10.1074/jbc.M112.416503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Prox J, Willenbrock M, Weber S, Lehmann T, Schmidt-Arras D, Schwanbeck R, Saftig P, Schwake M. Tetraspanin15 regulates cellular trafficking and activity of the ectodomain sheddase ADAM10. Cell Mol Life Sci. 2012;69:2919–2932. doi: 10.1007/s00018-012-0960-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seipold L, Damme M, Prox J, Rabe B, Kasparek P, Sedlacek R, Altmeppen H, Willem M, Boland B, Glatzel M, Saftig P. Tetraspanin 3: a central endocytic membrane component regulating the expression of ADAM10, presenilin and the amyloid precursor protein. Biochim Biophys Acta. 2017;1864:217–230. doi: 10.1016/j.bbamcr.2016.11.003. [DOI] [PubMed] [Google Scholar]
- 16.Seipold L, Saftig P. The emerging role of tetraspanins in the proteolytic processing of the amyloid precursor protein. Front Mol Neurosci. 2016;9:149. doi: 10.3389/fnmol.2016.00149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matthews AL, Noy PJ, Reyat JS, Tomlinson MG. Regulation of A disintegrin and metalloproteinase (ADAM) family sheddases ADAM10 and ADAM17: the emerging role of tetraspanins and rhomboids. Platelets. 2017;28:333–341. doi: 10.1080/09537104.2016.1184751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matthews AL, Szyroka J, Collier R, Noy PJ, Tomlinson MG. Scissor sisters: regulation of ADAM10 by the TspanC8 tetraspanins. Biochem Soc Trans. 2017;45:719–730. doi: 10.1042/BST20160290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saint-Pol J, Eschenbrenner E, Dornier E, Boucheix C, Charrin S, Rubinstein E. Regulation of the trafficking and the function of the metalloprotease ADAM10 by tetraspanins. Biochem Soc Trans. 2017;45:937–944. doi: 10.1042/BST20160296. [DOI] [PubMed] [Google Scholar]
- 20.Zimmerman B, Kelly B, McMillan BJ, Seegar TCM, Dror RO, Kruse AC, Blacklow SC. Crystal structure of a full-length human tetraspanin reveals a cholesterol-binding pocket. Cell. 2016;167(1041–1051):e1011. doi: 10.1016/j.cell.2016.09.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zuidscherwoude M, Gottfert F, Dunlock VM, Figdor CG, van den Bogaart G, van Spriel AB. The tetraspanin web revisited by super-resolution microscopy. Sci Rep. 2015;5:12201. doi: 10.1038/srep12201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Charrin S, le Naour F, Silvie O, Milhiet PE, Boucheix C, Rubinstein E. Lateral organization of membrane proteins: tetraspanins spin their web. Biochem J. 2009;420:133–154. doi: 10.1042/BJ20082422. [DOI] [PubMed] [Google Scholar]
- 23.Hemler ME. Tetraspanin functions and associated microdomains. Nat Rev Mol Cell Biol. 2005;6:801–811. doi: 10.1038/nrm1736. [DOI] [PubMed] [Google Scholar]
- 24.Jouannet S, Saint-Pol J, Fernandez L, Nguyen V, Charrin S, Boucheix C, Brou C, Milhiet PE, Rubinstein E. TspanC8 tetraspanins differentially regulate the cleavage of ADAM10 substrates, Notch activation and ADAM10 membrane compartmentalization. Cell Mol Life Sci. 2016;73:1895–1915. doi: 10.1007/s00018-015-2111-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Noy PJ, Yang J, Reyat JS, Matthews AL, Charlton AE, Furmston J, Rogers DA, Rainger GE, Tomlinson MG. TspanC8 tetraspanins and A disintegrin and metalloprotease 10 (ADAM10) interact via their extracellular regions: evidence for distinct binding mechanisms for different TspanC8 proteins. J Biol Chem. 2016;291:3145–3157. doi: 10.1074/jbc.M115.703058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kasparek P, Krausova M, Haneckova R, Kriz V, Zbodakova O, Korinek V, Sedlacek R. Efficient gene targeting of the Rosa26 locus in mouse zygotes using TALE nucleases. FEBS Lett. 2014;588:3982–3988. doi: 10.1016/j.febslet.2014.09.014. [DOI] [PubMed] [Google Scholar]
- 27.Zunke F, Andresen L, Wesseler S, Groth J, Arnold P, Rothaug M, Mazzulli JR, Krainc D, Blanz J, Saftig P, Schwake M. Characterization of the complex formed by beta-glucocerebrosidase and the lysosomal integral membrane protein type-2. Proc Natl Acad Sci USA. 2016;113:3791–3796. doi: 10.1073/pnas.1514005113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gunther W, Luchow A, Cluzeaud F, Vandewalle A, Jentsch TJ. ClC-5, the chloride channel mutated in Dent’s disease, colocalizes with the proton pump in endocytotically active kidney cells. Proc Natl Acad Sci USA. 1998;95:8075–8080. doi: 10.1073/pnas.95.14.8075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Prox J, Bernreuther C, Altmeppen H, Grendel J, Glatzel M, D’Hooge R, Stroobants S, Ahmed T, Balschun D, Willem M, et al. Postnatal disruption of the disintegrin/metalloproteinase ADAM10 in brain causes epileptic seizures, learning deficits, altered spine morphology, and defective synaptic functions. J Neurosci. 2013;33(12915–12928):12928a. doi: 10.1523/JNEUROSCI.5910-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spilker C, Nullmeier S, Grochowska KM, Schumacher A, Butnaru I, Macharadze T, Gomes GM, Yuanxiang P, Bayraktar G, Rodenstein C, et al. A Jacob/Nsmf gene knockout results in hippocampal dysplasia and impaired BDNF signaling in dendritogenesis. PLoS Genet. 2016;12:e1005907. doi: 10.1371/journal.pgen.1005907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mikhaylova M, Bera S, Kobler O, Frischknecht R, Kreutz MR. A dendritic golgi satellite between ERGIC and retromer. Cell Rep. 2016;14:189–199. doi: 10.1016/j.celrep.2015.12.024. [DOI] [PubMed] [Google Scholar]
- 32.Mangeol P, Prevo B, Peterman EJ. KymographClear and KymographDirect: two tools for the automated quantitative analysis of molecular and cellular dynamics using kymographs. Mol Biol Cell. 2016;27:1948–1957. doi: 10.1091/mbc.e15-06-0404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Altmeppen HC, Prox J, Krasemann S, Puig B, Kruszewski K, Dohler F, Bernreuther C, Hoxha A, Linsenmeier L, Sikorska B et al (2015) The sheddase ADAM10 is a potent modulator of prion disease. Elife 4 [DOI] [PMC free article] [PubMed]
- 35.Kuhn PH, Wang H, Dislich B, Colombo A, Zeitschel U, Ellwart JW, Kremmer E, Rossner S, Lichtenthaler SF. ADAM10 is the physiologically relevant, constitutive alpha-secretase of the amyloid precursor protein in primary neurons. EMBO J. 2010;29:3020–3032. doi: 10.1038/emboj.2010.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jorissen E, Prox J, Bernreuther C, Weber S, Schwanbeck R, Serneels L, Snellinx A, Craessaerts K, Thathiah A, Tesseur I, et al. The disintegrin/metalloproteinase ADAM10 is essential for the establishment of the brain cortex. J Neurosci. 2010;30:4833–4844. doi: 10.1523/JNEUROSCI.5221-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Suh J, Choi SH, Romano DM, Gannon MA, Lesinski AN, Kim DY, Tanzi RE. ADAM10 missense mutations potentiate beta-amyloid accumulation by impairing prodomain chaperone function. Neuron. 2013;80:385–401. doi: 10.1016/j.neuron.2013.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Postina R, Schroeder A, Dewachter I, Bohl J, Schmitt U, Kojro E, Prinzen C, Endres K, Hiemke C, Blessing M, et al. A disintegrin-metalloproteinase prevents amyloid plaque formation and hippocampal defects in an Alzheimer disease mouse model. J Clin Investig. 2004;113:1456–1464. doi: 10.1172/JCI20864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Heikens MJ, Cao TM, Morita C, Dehart SL, Tsai S. Penumbra encodes a novel tetraspanin that is highly expressed in erythroid progenitors and promotes effective erythropoiesis. Blood. 2007;109:3244–3252. doi: 10.1182/blood-2006-09-046672. [DOI] [PubMed] [Google Scholar]
- 40.Reiss K, Maretzky T, Ludwig A, Tousseyn T, de Strooper B, Hartmann D, Saftig P. ADAM10 cleavage of N-cadherin and regulation of cell-cell adhesion and beta-catenin nuclear signalling. EMBO J. 2005;24:742–752. doi: 10.1038/sj.emboj.7600548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xie Z, Photowala H, Cahill ME, Srivastava DP, Woolfrey KM, Shum CY, Huganir RL, Penzes P. Coordination of synaptic adhesion with dendritic spine remodeling by AF-6 and kalirin-7. J Neurosci. 2008;28:6079–6091. doi: 10.1523/JNEUROSCI.1170-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Togashi H, Abe K, Mizoguchi A, Takaoka K, Chisaka O, Takeichi M. Cadherin regulates dendritic spine morphogenesis. Neuron. 2002;35:77–89. doi: 10.1016/S0896-6273(02)00748-1. [DOI] [PubMed] [Google Scholar]
- 43.Mendez P, De Roo M, Poglia L, Klauser P, Muller D. N-cadherin mediates plasticity-induced long-term spine stabilization. J Cell Biol. 2010;189:589–600. doi: 10.1083/jcb.201003007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Isaacs JD, Jackson GS, Altmann DM. The role of the cellular prion protein in the immune system. Clin Exp Immunol. 2006;146:1–8. doi: 10.1111/j.1365-2249.2006.03194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Steele AD, Emsley JG, Ozdinler PH, Lindquist S, Macklis JD. Prion protein (PrPc) positively regulates neural precursor proliferation during developmental and adult mammalian neurogenesis. Proc Natl Acad Sci USA. 2006;103:3416–3421. doi: 10.1073/pnas.0511290103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bremer J, Baumann F, Tiberi C, Wessig C, Fischer H, Schwarz P, Steele AD, Toyka KV, Nave KA, Weis J, Aguzzi A. Axonal prion protein is required for peripheral myelin maintenance. Nat Neurosci. 2010;13:310–318. doi: 10.1038/nn.2483. [DOI] [PubMed] [Google Scholar]
- 47.Aguzzi A, Calella AM. Prions: protein aggregation and infectious diseases. Physiol Rev. 2009;89:1105–1152. doi: 10.1152/physrev.00006.2009. [DOI] [PubMed] [Google Scholar]
- 48.Soto C, Satani N. The intricate mechanisms of neurodegeneration in prion diseases. Trends Mol Med. 2011;17:14–24. doi: 10.1016/j.molmed.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Collinge J. Prion diseases of humans and animals: their causes and molecular basis. Annu Rev Neurosci. 2001;24:519–550. doi: 10.1146/annurev.neuro.24.1.519. [DOI] [PubMed] [Google Scholar]
- 50.Altmeppen HC, Prox J, Puig B, Kluth MA, Bernreuther C, Thurm D, Jorissen E, Petrowitz B, Bartsch U, De Strooper B, et al. Lack of a-disintegrin-and-metalloproteinase ADAM10 leads to intracellular accumulation and loss of shedding of the cellular prion protein in vivo. Mol Neurodegener. 2011;6:36. doi: 10.1186/1750-1326-6-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.