Abstract
Oxysterol-binding protein (OSBP) and OSBP-related proteins (ORPs) constitute a large eukaryotic gene family that transports and regulates the metabolism of sterols and phospholipids. The original classification of the family based on oxysterol-binding activity belies the complex dual lipid-binding specificity of the conserved OSBP homology domain (OHD). Additional protein- and membrane-interacting modules mediate the targeting of select OSBP/ORPs to membrane contact sites between organelles, thus positioning the OHD between opposing membranes for lipid transfer and metabolic regulation. This unique subcellular location, coupled with diverse ligand preferences and tissue distribution, has identified OSBP/ORPs as key arbiters of membrane composition and function. Here, we will review how molecular models of OSBP/ORP-mediated intracellular lipid transport and regulation at membrane contact sites relate to their emerging roles in cellular and organismal functions.
Keywords: Oxysterol-binding proteins, Membrane contact sites, Intracellular lipid transport, Cancer, Dyslipidemia, Metabolism, Viral replication
Introduction
Intracellular lipid transport creates and maintains the unique lipid composition of membranes that define organelle function and regulate communication with the external environment. While vesicular transport and membrane fusion are responsible for the bulk transfer of lipids and sterols, the specific and localized movement of individual lipid species between membranes is mediated by lipid transfer proteins (LTPs) [1]. While LTP gene families are numerous and diverse in structure, they share the ability to bind lipids within a hydrophobic fold for lipid transfer, substrate presentation or signaling [2]. Oxysterol-binding protein (OSBP) and OSBP-related proteins (ORPs) constitute a large family of eukaryotic LTPs that bind cholesterol, oxysterols and anionic phospholipids within a conserved hydrophobic binding domain. Additional membrane-targeting motifs allow OSBP/ORPs to simultaneously associate with a diverse set of protein and lipid partners in organelle membranes, thus positioning the lipid-binding domain in close apposition to donor and acceptor membranes to facilitate transfer or signaling functions. The potential for redundancy in this large gene family adds complexity to any analysis of individual OSBP/ORP function in normal and disease states. This review will focus on our current understanding of mammalian OSBP/ORP function at the cellular and systemic level and how these functions are exploited by pathogens and contribute to disease pathology.
Structural organization of OSBP/ORPs
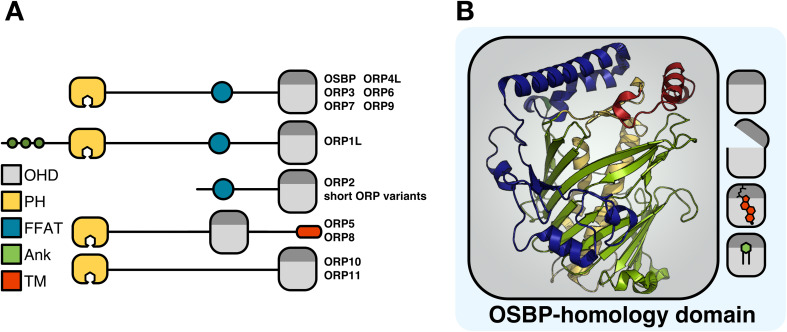
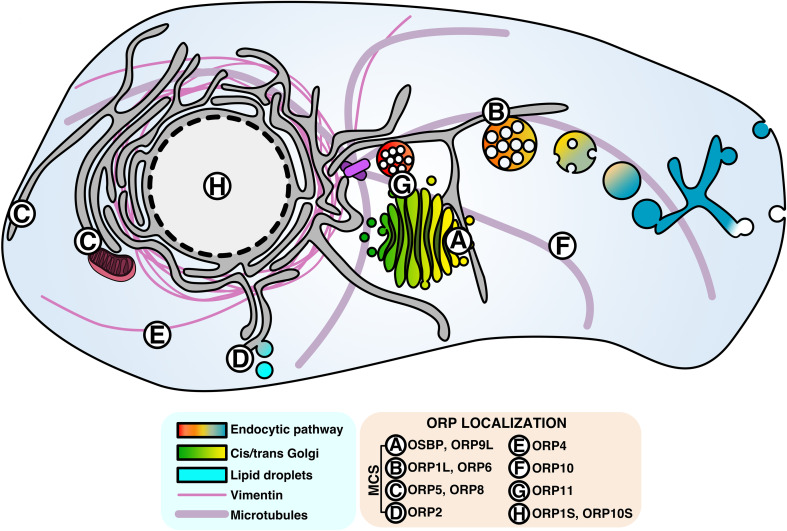
OSBP was identified in 1985 by Taylor and Kandutsch as a soluble receptor for oxysterols [3], which are oxidized cholesterol derivatives that promote cholesterol esterification and efflux and inhibit de novo cholesterol synthesis and uptake [4]. Cloned in 1989, OSBP was initially thought to be an oxysterol-regulated transcription factor due to a central leucine repeat reminiscent of some DNA binding proteins [5]. However, OSBP was later shown to undergo sterol-dependent localization to the Golgi apparatus [6]. In 1999, Ikonen and colleagues cloned the cDNAs for 6 human OSBP-related proteins (termed ORPs) [7], and the remaining members of the 12-gene family were cloned in 2001 [8]. Mammalian OSBP/ORPs contain a conserved C-terminal lipid-binding domain designated the OSBP homology domain (OHD) that harbors a highly conserved motif (EQVSHHPP) that is found in homologs across species (Fig. 1a). All but one of the OSBP genes encode full-length or long (L) forms that contain additional membrane-targeting domains (Fig. 1a). For example, all full-length OSBP/ORPs have an N-terminal pleckstrin homology (PH) domain that recognizes phosphatidylinositol phosphates (PIPs) with varying degrees of specificity and affinity. OSBP and ORPs 1, 2, 3, 4, 6, 7 and 9 also contain a two phenylalanines-in-an-acidic tract (FFAT) motif that interacts with the resident endoplasmic reticulum (ER) protein vesicle-associated membrane protein-associated protein (VAP). As a result, OSBP/ORPs that contain PH and FFAT motifs are peripherally anchored to the ER and to organelles enriched in PIPs, such as the Golgi apparatus, plasma membrane (PM) and endosomes. ORP5 and ORP8 provide a variation on this theme: both have C-terminal membrane-spanning domains that anchor them to the ER as well as PH domains that facilitate interaction with other organelles (Fig. 1a). Collectively, these dual membrane-binding properties could facilitate transient association–dissociation with individual membranes or simultaneous association at a membrane contact site (MCS) between organelles [2]. Based on experimental evidence, the localization of OSBP/ORPs at organelle MCS and with cytoskeletal networks is depicted in Fig. 2. How this unique subcellular localization is involved in lipid transfer and regulation is discussed below.
Fig. 1.
Structural organization of the OSBP/ORP family. a Domain structure of the OSBP/ORP family. OHD OSBP homology domsin, PH pleckstrin homology, FFAT two phenylalanines in an acidic tract, Ank ankyrin, TM transmembrane. b Shown is the structure of the S. cerevisiae Osh4p OHD composed of an incomplete β-barrel (green) flanked by two central helices (yellow), N-terminal region (blue) and α-helical lid (red). The OHD binds sterols and phospholipids competitively and in opposite orientations; the acyl chains of phospholipids are buried in the pocket, while the iso-octyl side chain of sterols interacts with the lid
Fig. 2.
Subcellular localization of the OSBP/ORP family. Aside from ORP5 and ORP8, all ORPs display partial cytosolic localization, but most also have discrete localization patterns, including membrane contact sites between organelles, on cytoskeletal elements and in the nucleus (see figure for details). For simplicity, only well-characterized localization patterns relevant to known or predicted protein functions have been included. ORP3 and ORP7 do not have well-defined localization patterns and have been omitted
Much of the structural and functional information we have for the OSBP/ORP family is derived from studies of the S. cerevisiae oxysterol-binding protein homolog (Osh) proteins (see “Structure and specificity of the OSBP homology domain”). Osh1p and Osh2p are long forms that contain N-terminal PH, FFAT and ankyrin motifs, Osh3p has PH, FFAT and Golgi dynamics (GOLD) motifs, while and Osh4p-Osh7p contain only the OHD. While the seven Osh proteins associate with different membranes and cellular processes [9], they are functionally redundant such that any one Osh protein can rescue the growth-arrest phenotype caused by deletion of all seven Osh proteins [10].
Studies in the cholesterol auxotrophs C. elegans and D. melanogaster, which each express four ORP homologs, revealed similar functional redundancy. Deletion of individual C. elegans obr1-to obr-4 homologs did not affect viability but deletion of all four caused embryonic lethality [11]. Deletion of individual Osbp homologs in D. melanogaster also yielded viable mutants [12]. However, Osbp deletion or overexpression in D. melanogaster caused aberrant cholesterol accumulation in the Golgi apparatus with resultant deleterious effects on spermatogenesis and wing expansion. These results also indicate that OSBP/ORPs are essential for lipid homeostasis in cells that lack de novo sterol biosynthesis [12, 13].
Structure and specificity of the OSBP homology domain
Osh4p was the first to have its structure solved by X-ray crystallography to reveal an OHD comprising an incomplete 19-strand beta-barrel (Fig. 1b, green) with an alpha-helical lid (Fig. 1b, red), which together form a hydrophobic binding cavity that accommodates cholesterol and oxysterols [14]. Cholesterol co-crystallized in the Osh4p OHD pocket is oriented ‘head-first’ (Fig. 1b); the 3-hydroxyl is coordinated at the bottom of the pocket while the iso-octyl side chain interacts with the lid in a closed position, making the pocket inaccessible to other ligands. Interestingly, protein–sterol interactions are mediated by water molecules that line the binding pocket, bridging basic residues at the base of the pocket with the 3-hydroxyl of the bound sterol. Indirect, water-mediated binding may explain the broad range of sterols that are bound by OSBP/ORP and Osh proteins.
Despite similarities in the sequence and structure of OHDs amongst the Osh and OSBP/ORP family, it is apparent that sterol binding is not a core activity [15]. Crystallization of the OHDs of Osh4p [16] and Osh3p [17] revealed that phosphatidylinositol 4-phosphate (PI(4)P) occupies the OHD ‘tail-first’ (Fig. 1b); the acyl chains extend into the binding pocket while the inositol 4-phosphate headgroup interacts with two histidine residues close to the entrance of the binding fold. Notably, the two histidine residues are part of the OSBP/ORP signature motif and not required for sterol binding [17], suggesting a primary function that involves the binding and transfer of anionic phospholipids. The “tail first” positioning of PI(4)P in the OHD suggests that the alpha-helical lid of the OHD is capable of inserting into membranes to excise embedded lipids for binding in the observed orientation [18]. This configuration for PI(4)P was confirmed for Osh1p, which also binds ergosterol [18]. While the structure of a mammalian OHD has not been solved, in vitro assays of lipid binding and extraction from liposomes have shown that mammalian OSBP/ORPs also competitively bind both sterols and phospholipids, including phosphatidylserine (PS) and PI(4)P (summarized in Table 1 and reviewed in [19]).
Table 1.
OSBP/ORP localization and function
| Name | Isoforms | Domains | Tissue expression | Localization | Ligands (kd) | Intracellular functions |
|---|---|---|---|---|---|---|
| OSBP (OSBP) | OHD, FFAT, PH | Ubiquitous [133] | Golgi, cytosol [6], ER [23] | 25OH (8 nM) [3, 5, 51], cholesterol (173 nM) [51], PI(4)P [25] | Catalyzes sterol/PI(4)P counter-current transport at ER/Golgi. Recruits CERT [24, 25, 27] and regulates SM synthesis [134] | |
| ORP1 (OSBPL1) | ORP1L | OHD, ANK, PH | Brain, lung [50] | LEL, ER [50, 52] | 25OH (83–97 nM) [40, 56], cholesterol (1.4 μM), PI(4)P [56] | Endosome positioning [52] and cholesterol efflux [56] via Rab7/RILP/p150GLUED assembly [53] |
| ORP1S | OHD | Heart, skeletal muscle, macrophages [50] | Cytosol [50], nucleus [51] | 25OH (8.4 nM) [50], PI(4)P [56] | LXR regulation [40] and sterol transport from PM to LD [125] | |
| ORP2 (OSBPL2) | OHD, VAP | Ubiquitous, enriched in CNS [62] | LD, ER, Golgi [62] | 22(R)OH (14 nM), 7-ketocholesterol (160 nM), cholesterol [58], 25OH (3.9 μM) [40] | Cholesterol efflux [62], endocytosis [63] and PM-to-ER sterol transport [125]. Regulates triglyceride hydrolysis [59] and actin dynamics [73]. Binds LXR and regulates cortisol biosynthesis [60] | |
| ORP3 (OSBPL3) | OHD, VAP, PH | Kidney, lymph nodes, thymus [135], macrophages [50] | Cytosol, ER, PM [135], nuclear envelope [136] | Photo-25OH (weak), photo-cholesterol [40] | Regulates actin dynamics via interaction with R-Ras [137], and PI levels [138] | |
| ORP4 (OSBP2) | ORP4L | OHD, FFAT, PH | Brain, heart, testis, skeletal muscle [111] | Cytosol, ER, vimentin [111] | 25OH (17–27 nM) [112, 113], cholesterol (68 nM) [113] | Regulates Ca2+ homeostasis [90, 115, 116]. Murine KO has oligo-astheno-teratozoospermia and sterility [117]. Silencing in cell culture causes apoptosis [113] |
| ORP4M | OHD, FFAT | N/A | Vimentin aggregates [113] | N/A | ||
| ORP4S | OHD | Brain, heart [111] | Vimentin aggregates [111] | 25OH (23 nM), cholesterol (60 nM) PI(4)P [113] | ||
| ORP5 (OSBPL5) | OHD, PH, TM | Heart, brain, lung, liver, kidney [139] | ER [39], PM [140], mitochondria [37] | Photo-25OH [40], All PIPs (0.8–26.8 μM) [36], PS [141] | Cholesterol efflux from endosomes. PS/PI(4)P counter-current transport [39] at ER/PM [140]. PS transport to mitochondria [37] | |
| ORP6 (OSBPL6) | OHD, FFAT, PH | Brain, skeletal muscle [135] | Nuclear envelope, cytosol, ER, PM [135], LEL [49] | Photo-25OH and photo-cholesterol (weak) [40] | Regulated by LXR. Involved in cholesterol efflux from early endosomal compartment [49] | |
| ORP7 (OSBPL7) | OHD, FFAT, PH | Gastrointestinal tract [135] | Cytosol, ER, PM [135] | Photo-25OH and photo-cholesterol (weak) [40] | Promotes proteosomal degradation of Golgi snare GS28 via 25OH-dependent interaction with GS28 partner protein GATE-16 [108] | |
| ORP8 (OSBPL8) | OHD, PH, TM | Macrophages, spleen, kidney, liver, brain [91] | ER, PM [140], mitochondria [37] | 25OH [91], All PIPs (1.7–6.0 μM) [36], PS [141] | Counter-current transport of PS/PI(4)P at ER/PM [140]. PS transport to mitochondria [37]. Involved in macrophage motility and hepatic lipid efflux [91, 93] | |
| ORP9 (OSBPL9) | ORP9L | OHD, FFAT, PH | Brain, heart, kidney, liver [142] | ER, Golgi [142] | Cholesterol [143], PI(4)P, cholestatrienol, dehydroergosterol [144] | Recruits ORP11 to the Golgi [137]. Regulates Golgi organization via cholesterol [143] or PI(4)P trafficking [144] |
| ORP9S | OHD, FFAT | Liver [142] | ER [142] | PI(4)P, cholestatrienol, dehydroergosterol [144] | Involved in Golgi organization, growth inhibition [143] and PI(4)P regulation [144] | |
| ORP10 (OSBPL10) | ORP10L | OHD, PH | N/A | Microtubules [85, 87], Golgi [87] | Photo-25OH [58], cholesterol [87], PS [141] | Dimerizes with ORP9L [87]. Negative regulator of hepatic lipid biosynthesis and apoB-100 secretion [85, 87] |
| ORP10S | OHD | N/A | Cytosol, nucleus [87] | N/A | N/A | |
| ORP11 (OSBPL11) | OHD, PH | Ovary, testis, brain, liver, kidney, stomach, adipose tissue [145] | Golgi, LEL [145] | N/A | Dimerizes with ORP9L [145]. involved in cellular triglyceride storage and induced during adipocyte differentiation [99] |
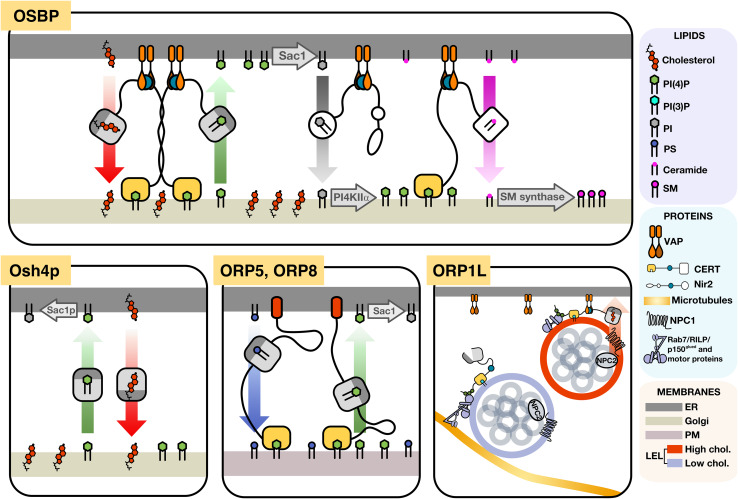
Dual-binding specificity for phospholipid and sterol ligands allows an Osh/OSBP/ORP to potentially exist in binary states; each ligand induces conformational changes in the OHD that direct protein and membrane association to potentially drive lipid exchange between organelles. This concept was first demonstrated for Osh4p, a prototypical OHD-only protein that does not physically bridge donor and acceptor membranes during a lipid transfer cycle. Liposome-based Osh4p lipid transfer assays showed that the rate of delivery of ergosterol from a donor membrane to an acceptor membrane increased when the acceptor membrane was enriched with anionic lipids [16]. However, Osh4p only dissociated from the acceptor membrane when bound to a molecule of ergosterol, therefore resulting in no net change in the ergosterol content of the acceptor membrane. However, when the acceptor membrane was enriched with PI(4)P, which is an anionic lipid and Osh4p ligand, ergosterol delivery to acceptor membranes was increased while the back-transfer of ergosterol was inhibited by competitive binding of PI(4)P, thereby driving directional transport of ergosterol to the acceptor membrane. Complementation studies in yeast suggest that Osh4p functions similarly in vivo (Fig. 3), transferring ER-derived cholesterol to the Golgi apparatus in exchange for Golgi-derived PI(4)P, which is then hydrolyzed in the ER by the PI(4)P phosphatase Sac1p [20]. In yeast, the phosphatidylinositol (PI) transfer protein Sec14p drives PI(4)P synthesis by providing substrate for the PI 4-kinase Pik1p. In the absence of Sec14p, unchecked Osh4p-mediated PI(4)P depletion causes cytotoxic failure of the secretory system [21].
Fig. 3.
Models of ORP function at membrane contact sites. OSBP localizes to closely apposed ER/Golgi membranes, where it exchanges PI(4)P and cholesterol. The resulting cholesterol enrichment on the Golgi activates PI4KIIα, which phosphorylates PI delivered by Nir2, into a PI(4)P pool that recruits the ceramide transfer protein CERT, resulting in ceramide delivery to sphingomyelin (SM) synthase and consequent SM synthesis. Consumption of PI(4)P in the endoplasmic reticulum (ER) by the Sac1 phosphatase maintains a concentration gradient that drives the PI(4)P/cholesterol counter-current transport. Yeast Osh4p functions similarly to OSBP but without tethering to the ER or Golgi apparatus. ORP5 and ORP8 are ER-tethered proteins that extend to the plasma membrane via PH domain interaction with PIPs, using the transport of PI(4)P to the ER to drive phosphatidylserine transport to the PM. ORP1L is localized to the late endosomes/lysosomes (LEL) via interaction between N-terminal ankyrin repeats and Rab7. When cholesterol is low in the limiting membrane of LEL, ORP1L promotes the assembly of a microtubule-tethering complex (Rab7/RILP/p150glued) that facilitates minus-end movement of the LEL. When cholesterol on the LEL limiting membrane is elevated, the complex dissembles and ORP1L tethers the LEL to the ER via VAP and facilitates cholesterol efflux to the ER
Lipid transport and metabolic regulation by OSBP at ER/Golgi contact sites
Oxysterol-binding protein is a prototypical dual membrane-interacting protein that transfers lipids and regulates lipid metabolism at MCS. Over a decade of research has described OSBP as a sterol-sensitive regulator of sphingomyelin (SM) and PI(4)P synthesis at the ER/Golgi interface (Fig. 3). OSBP is a cytosolic protein that associates transiently with the trans-Golgi and trans-Golgi network (TGN) via the PH domain [22], and the ER via interaction between its FFAT motif and the ER-resident protein VAP-A [23]. When cells are treated with exogenous oxysterols (a signal of excess cholesterol) or depleted of cholesterol to stimulate de novo synthesis, OSBP is associated with ER/Golgi MCS [6, 24]. There, OSBP transports ER-derived cholesterol to the Golgi and Golgi-derived PI(4)P to the ER, where it is degraded by the PI4P phosphatase Sac1 [25]. Cholesterol enrichment in the Golgi apparatus leads to recruitment of phosphatidylinositol-4-kinase (PI4K)IIα [26] and, consequently, a local increase in PI(4)P, which in turn leads to recruitment of ceramide transport protein (CERT) via PH and FFAT domain-dependent targeting to the MCS [27]. The transport of ceramide to the Golgi apparatus provides substrate for the synthesis of SM, thereby matching the increased transport of cholesterol to maintain the relative proportions of SM and cholesterol-rich microdomains, which are important for protein and lipid sorting in the secretory pathway [28]. For example, the biogenesis of carriers of the trans-Golgi network to the cell surface (CARTS) requires VAP localization to ER/Golgi MCS and lipid transfer mediated by OSBP [29]. Reduction in the levels of VAP, OSBP or CERT prevented the secretion of CARTS cargo, as did expression of OSBP or CERT with mutations in the FFAT motifs. This suggests a requirement for OSBP in the maintenance of cholesterol and sphingolipid-enriched membranes at TGN/ER contacts and in the resulting directional transport of CARTS cargoes.
Less than 1% of cellular cholesterol is present in the ER, where the sterol regulatory element binding protein (SREBP) pathway is subject to negative feedback inhibition by cholesterol and oxysterols [30]. Thus, subtle changes in sterol levels in the ER can initiate rapid changes in cholesterol biosynthetic capacity. While the ER is relatively cholesterol poor, there is a cholesterol gradient from the cis/medial to trans-Golgi to PM, which constitutes approximately 60–80% of cellular cholesterol. A model for the maintenance of heterogenous cholesterol distribution between the ER and TGN posits that OSBP couples the transport of PI(4)P down its concentration gradient from the TGN to ER to power the counter-transport of cholesterol against its concentration gradient from the ER to TGN. The hydrolysis of the 4-phosphate of PI(4)P by Sac1 in the ER maintains the PI(4)P gradient, effectively harnessing the energy of PI(4)P synthesis and hydrolysis to drive cholesterol transport. Inhibition of OSBP with OSW-1, a member of the antineoplastic saponins (termed ORPphilins) that compete for sterol binding to the OHD of ORP4 and OSBP [31], doubled the cellular mass of PIP, of which PI(4)P is the predominant species. This finding suggests that an OSBP-mediated ‘lipid pump’ mechanism could consume up to 50% of cellular PI(4)P mass to power this cholesterol transport process. Preventing the replenishment of PI(4)P by pharmacological inhibition of PI4KIIIβ caused oscillation of PI(4)P levels at the TGN. In this scenario, OSBP drives consumption of PI(4)P and, consequently, dissociates from the TGN until PI4KIIα activity eventually replenishes the TGN PI(4)P pool, at which point OSBP is recruited to MCS and restarts the cycle [32].
The phenotypes of OSBP silencing in cultured cells have indicated an important role in ER-to-Golgi cholesterol transport. These include reduction in TGN cholesterol content [26] leading to the mislocalization of Golgi tether proteins [33], and accumulation of PI(4)P in Golgi and endosomal membranes leading to disruption of retromer-mediated transport and increased actin nucleation [34]. Silencing of OSBP expression had no effect on the morphology of the cis/medial-Golgi or TGN; however, the perinuclear intra-Golgi v-SNARES GS28 and GS15 become dispersed in the cytosol. GS28 and GS15 traverse the Golgi in COP-I transport vesicles, the fusion of which requires cholesterol-dependent conformational changes in SNARE proteins. Treatment of cells with lovastatin, an inhibitor of cholesterol biosynthesis, replicated the effects of OSBP silencing on GS28 and GS15 localization, suggesting that mislocalization is the result of aberrant cholesterol distribution due to the absence of OSBP [34].
Early reports that OSBP overexpression increased cholesterol biosynthesis and inhibited cholesterol esterification suggested a role in cholesterol transport or regulation in the ER [22]. Recently, inhibition of OSBP by OSW-1 was shown to increase lipid droplet formation indicative of ER cholesterol accumulation and enhanced esterification. However, OSBP silencing in Chinese hamster ovary cells had no effect on cholesterol regulation or esterification in the ER [27]. Similarly, OSBP silencing in HeLa cells had no effect on regulation of cholesterol biosynthesis by oxysterols [35]. These data suggest that OSBP-mediated cholesterol export to the Golgi apparatus may involve a relatively small fraction of the ER cholesterol pool that has minimal impact on esterification and the SREBP regulatory machinery.
Lipid transfer by ORP5 and ORP8 at membrane contact sites
ORP5 and ORP8 associate with the ER via tail-anchored C-terminal transmembrane domains while the N-terminal PH domains interact with di- and tri-phosphorylated PIPs in the PM (Fig. 3) [36]. ORP5 and ORP8 are also localized at ER/mitochondria contact sites, but this activity is PH domain independent. Silencing of ORP5 and ORP8 caused defects in mitochondrial function and morphology without affecting the number of ER/PM or ER/mitochondrial MCS [37]. Together, these results suggest that ORP5 and ORP8 may also transfer PS from the ER to the mitochondria where it is decarboxylated to phosphatidylethanolamine required for mitochondrial structure and function [38].
The identification of ORP5 as a PS/PIP transfer protein contrasts an earlier study claiming that ORP5 was responsible for the transfer of low density lipoprotein (LDL)-cholesterol from the late endosomes/lysosomes (LEL) to the ER [39]. siRNA silencing of ORP5, but not ORP8, in HeLa cells caused accumulation of cholesterol in the limiting membrane of LEL and prevented cholesterol delivery to the ER, as measured by acyl-CoA: cholesterol acyltransferase activity [39]. Consistent with this activity, the ORP5 OHD displayed cholesterol binding and transfer activity in vitro [39, 40]. ORP5 interacted with the LEL cholesterol transports protein Niemann–Pick type C protein 1 (NPC1) [39] and, thus, could receive cholesterol from NPC1 at the LEL/ER membrane interface. Recently, the identification of an interaction between the mammalian target of rapamycin (mTOR) and the ORP5 OHD indicates that ORP5 could have a more complex role in nutrient sensing at LEL [41], perhaps related to the regulation of the mTOR by cholesterol [42]. Therefore, ORP5 appears to have lipid transfer activities at multiple MCS, but whether these are discrete or related activities is presently unclear.
Sterol transfer by ORP1L and ORP6 at endosome/ER contact sites
In addition to de novo cholesterol biosynthesis in the ER, cells also acquire exogenous cholesterol by the uptake and processing of lipoproteins, primarily LDL, in the endo-lysosomal pathway. After receptor-mediated endocytosis, the cholesterol esters in LDL are hydrolyzed in LEL [43]. The bulk of LDL-derived cholesterol is exported from the late endosomes before lysosomal maturation, as evidenced by the discrepancy in cholesterol content between the two compartments [44]. Cholesterol export from the LEL lumen to the external leaflet of the limiting membrane occurs by the concerted action of NPC protein 2 (NPC2), a soluble protein that binds cholesterol in the LEL lumen, and NPC1, a multi-pass transmembrane protein that receives cholesterol from NPC2 and transfers it to the limiting membrane of the LEL [45]. Once on the external leaflet of the limiting membrane, cholesterol is transferred to other organelles by poorly understood mechanisms. The bulk of LDL-derived cholesterol appears to flow to the ER [43], potentially at contact sites with LELs [46]. Indeed, quantitative imaging studies show that almost 100% of Rab7-positive late endosomes have frequent and sustained interactions with the ER [47]. The dual membrane-targeting ORP1L and ORP6 have been implicated in sterol transfer at these contacts.
Initial studies showed that ORP6 mRNA is increased in response to cholesterol loading in cultured cells [48]. Later studies confirmed that expression of ORP6 is regulated by the liver X receptor (LXR) and miR-33 and -27b, and increased under conditions of cholesterol loading in macrophages and animal models [49]. In ORP6-silenced and cholesterol-loaded macrophages, cholesterol accumulated in abnormally clustered early endosomes and cholesterol esterification in the ER was inhibited. Conversely, overexpression of ORP6 stimulated cholesterol esterification and cholesterol efflux from cells. Collectively this indicated that LXR activation of ORP6 is a mechanism to promote the efflux of sterols from early endosomes and suppress cholesterol biosynthesis.
OSBPL1 encodes two variants due to alternate promoter start sites: full-length ORP1L, which has N-terminal ankyrin repeats and a PH domain, and the truncated variant ORP1S that contains only the OHD [50]. ORP1S is a soluble sterol receptor that localizes to the nucleus in response to ligand binding and enhances the activity of LXR [51]. Initial studies of ORP1L described interactions with Rab7 in the LEL via the ankyrin motif and with VAP in the ER via the FFAT motif [52]. The functionality of ORP1L at ER/LEL contacts appears to depend on the level of cholesterol in the limiting membranes of each compartment. In instances where LDL is absent and LEL cholesterol is low, ORP1L, Rab7 and VAP are implicated in the transfer of cholesterol at contacts between the ER and a subpopulation of multivesicular endosomes involved in epidermal growth factor receptor sorting into intraluminal vesicles [46]. These contacts are formed by interaction between annexin A1 and S100A11 and are calcium dependent. In C. elegans, deletion of all four obr genes also resulted in abnormal multivesicular bodies with reduced cholesterol content, supporting a role for ORP1L in LEL maturation [11].
In a related sterol-sensing function of ORP1L, elevated cholesterol in the LEL (as in the case of NPC disease) initiates the formation of an ORP1L, Rab7 and Rab7-interacting lysosomal protein (RILP) complex that recruits the dynactin subunit p150Glued and βIII-spectrin to initiate minus-end transport on microtubules to the perinuclear region [53, 54]. Reduction of LEL cholesterol promotes a conformational change in ORP1L that increases VAP interactions and displaces p150Glued, leading to plus-end transfer of LEL. This sterol-sensing function of ORP1L thus regulates the positioning of LEL based on cholesterol content as well as endosome maturation via interaction with the homotypic fusion and vacuolar sorting (HOPS) complex [55].
ORP1L is also involved in the transfer of LDL-cholesterol from LEL-to-ER. ORP1L knockout cells display sequestration of perinuclear endosomes, defective cholesterol delivery to the ER and increased de novo cholesterol biosynthesis [56]. Contrary to a sensing function, ORP1L interacts with VAP on the ER when cholesterol is present on the limiting membrane of the LEL to establish a peripheral distribution of LELs and mediate cholesterol shuttling to the ER (Fig. 3). When the limiting membrane is depleted of cholesterol, ORP1L dissociates from VAP and promotes assembly of motor complexes that drive minus-end microtubule transport of LEL toward the perinuclear region [53, 55]. Thus, ORP1L has multiple transfer and regulatory activities that could result from its association with numerous, functionally distinct LEL-ER contact sites. However, as a cholesterol transfer protein, ORP1L appears to facilitate the bi-directional transfer of cholesterol between the LEL and ER depending on the sterol gradient.
ORP2 at ER/lipid droplet contact sites
The ER contains acyltransferases that synthesize cholesterol esters and triacylglycerol that coalesce within the bilayer of ER membranes, eventually budding to form specialized storage organelles called lipid droplets (LD) [57]. The lipids stored in LDs are a source of fatty acids for oxidation or can be packaged into lipoproteins for secretion and transport to other cells. ORP2 is composed of an OHD, which binds cholesterol and oxysterols, and an FFAT motif [58]. ORP2 is present at ER/LD contact sites [59] and dissociates from LDs upon ligand binding [60]. The function of ORP2 at ER/LD contacts is not well understood, but silencing of ORP2 leads to increased cellular cholesterol levels [60] and aberrant metabolism of triglycerides [58, 59] and fatty acids [61]. Overexpression of ORP2 increased cellular cholesterol efflux [62, 63] and LD dispersion [60]. Together, these studies suggest that ORP2 regulates LD positioning by establishing ER contacts in a sterol-dependent manner, which facilitates lipid exchange between the organelles.
Involvement of OSBP/ORPs in human disease
OSBP/ORPs display both ubiquitous and tissue-specific expression patterns (summarized in Table 1). Despite the size of the family and the potential for redundant function within specific tissues, high-throughput genetic and targeted functional studies have implicated OSBP/ORPs in a range of rare and common human disorders (summarized in Table 2). Genetic screens of cancerous tissues and cell lines have detected elevated expression of ORP1 [64], ORP2 [64], ORP4 [65, 66], ORP5 [67], ORP7 [64] and ORP10 [68, 69] and revealed that ORP3 is frequently mutated in metastatic versus nonmetastatic breast cancers [70]. In some cases, targeted research on individual ORPs has led to a limited mechanistic understanding of their contribution to the disease phenotype. The following section and Table 2 outline our current understanding of how OSBP/ORPs contribute to a range of human disorders.
Table 2.
OSBP/ORPs in human pathologies
| Protein | Pathology | Notes | Study type | References |
|---|---|---|---|---|
| Cholangiocarcinoma | Increased expression | qRT-PCR and immunohistochemistry | [64] | |
| OSBP | Amyotrophic lateral sclerosis | Overexpression rescues mutant VAP-B phenotype | Drosophila study | [81] |
| ORP1 | Dyslipidemia | Nonfunctional mutant is associated with HDL cholesterol < 1st percentile | Small cohort study (n = 80), genome sequencing and blood analysis | [88] |
| ORP2 | Autosomal dominant non-syndromic hearing loss | Truncation/deletion due to frameshift mutation causes hereditary hearing loss | Whole-exon sequencing of affected Chinese and German families | [71, 72] |
| Dyslipidemia | Expression inversely correlated with levels of hepatic steatosis marker Hsa-miR-855-5p and HDL cholesterol | microRNA profiling | [84] | |
| Amyotrophic lateral sclerosis | Overexpression rescues mutant VAP-B ER stress phenotype | Cell culture study | [80, 81] | |
| ORP3 | Glioblastoma | Expression associated with positive treatment response | RNA-seq | [146] |
| Breast cancer | More frequently mutated in metastatic breast cancer than early breast cancer | Next-gen seq | [70] | |
| Dyslipidemia | Gain-of-function mutation in LRH-1 induces NAFLD in mice via ORP3 expression | Mouse study | [83] | |
| Pancreatic ductal carcinoma | High expression associated with poor prognosis | Microarray database analysis, immunohistochemistry | [147] | |
| Metastasis | Highly expressed in disseminated tumor cells | Differential display PCR | [66, 110] | |
| ORP4 | T-cell acute lymphoblastic leukemia | High ORP4 expression provides energetic advantage via calcium signaling | Cell culture study, including patient samples | [116] |
| Chronic myeloid leukemia | ORP4 elevated in 80% of CLM patients studied | RT-PCR of patient samples | [65, 109] | |
| Spermatogenesis | ORP4 KO causes male infertility by oligo-astheno-teratozoospermia | Mouse KO study | [117] | |
| Cholangiocarcinoma | Increased expression | qRT-PCR and immunohistochemistry | [64] | |
| Pancreatic cancer | Expression positively correlated with cancer cell invasiveness and poor prognosis | Cell culture studies based on patient gene expression profiles, includes patient samples | [67, 102] | |
| Failures in meiosis II leading to recurrent triploidy | Rare OSBPL5 mutation may contribute to recurrent IVF failure by triploidy | Genotyping, whole-exome seq, methylation analysis | [147] | |
| ORP5 | Metastatic lung cancer | Increased expression in metastatic lung vs nonmetastatic lung tumor | 2D-DIGE/antibody screen, tissue microarrays, cell culture studies | [148] |
| Alcohol dependence | SNPs in OSBPL5 are associated with alcoholism | GWAS | [149] | |
| Dyslipidemia | Expression level in leukocytes is positively correlated with plasma lipid levels | Microarray expression profiling | [150] | |
| ORP6 | Dyslipidemia | Expression level is positively correlated with HDL cholesterol levels | Cell culture studies and patient screen | [49] |
| Alzheimer’s | SNPs in OSBPL6 are associated with familial Alzheimer’s | GWAS | [151] | |
| ORP7 | Cholangiocarcinoma | Increased expression | qRT-PCR and immunohistochemistry | [64] |
| Dyslipidemia | SNPs in OSBPL7 are associated with total cholesterol and LDL levels | GWAS and genotyping | [152, 153] | |
| Gastric cancer | Reduced expression | Cell culture study based on patient screen | [154] | |
| Cholangiocarcinoma | Increased expression | qRT-PCR and immunohistochemistry | [64] | |
| Dyslipidemia | SNPs in OSBPL8 associated with HDL cholesterol levels | GWAS | [155] | |
| Obesity-related miRNA-143 reduced insulin-induced PKB activation in liver cells via reduction of ORP8 levels | Mouse study | [94] | ||
| ORP8 | Activin A from epicardial adipose tissue reduced ORP8 levels via production of miRNA-143 | Cell culture study with primary rat cells | [95, 96] | |
| ORP8 KD reduced arterial lesion size by 20% in HFD-fed LDLR-KO mice | Mouse study | [92] | ||
| Lenz-Majewski disease mutations in PS synthesis alter counter-current transport by ORP8 | Cell culture study | [156] | ||
| Expression level in leukocytes is negatively correlated with plasma lipid levels | GWAS | |||
| Gastric cancers and hepatocellular carcinoma | ORP8 expression is low in gastric and hepatic cancers; overexpression decreases tumor growth and increases apoptosis by mitochondrial and ER stress | Cell culture and mouse studies based on patient expression profiles | [107, 154] | |
| ORP9 | Colorectal cancer | Elevated expression is positively correlated with survival | Whole genome expression profiling | [157] |
| Dyslipidemia | SNPs in OSBPL9 may be associated with increased risk of cerebral infarction | PCR–RFLP screen of Chinese cohort | [158] | |
| Dyslipidemia | SNPs in OSBPL10 associated with extreme high triglyceride levels (> 95th percentile), silencing increases lipogenesis and apoB-100 secretion in hepatic cell line | Cell culture experiments based on genotyping in Finnish dyslipidemic families | [85, 87] | |
| SNPs in OSBPL10 associated with hypercholesterolemia | Japanese cohort study | [159] | ||
| ORP10 | RNA expression in leukocytes is positively correlated with plasma lipid levels | GWAS | [150] | |
| SNP in OSBPL10 intron strongly associated with peripheral arterial disease | GWAS | [86] | ||
| Prostate cancer | Potential cancer marker | Microarray analysis | [68, 69] | |
| Hypertension | SNPs in OSBPL10 may contribute to hypertension | Next-generation linkage association | [160] | |
| Dyslipidemia | SNPs in OSBPL11 associated with metabolic syndrome | Genotyping of obese individuals | [97] | |
| ORP11 | Granular cholesterol staining in dermal tissue of individuals with homozygous OSBPL11 missense mutation | Case study of family with rare neurodegenerative disease | [98] |
ORP2 and hearing loss
ORP2 is ubiquitously expressed but enriched in cells of the central nervous system as well as cochlear inner and outer hair cells [71]. Whole-exome sequencing identified frameshift mutations in OSBPL2 that have been linked to familial autosomal dominant non-syndromic hearing loss [71, 72]. These mutations were predicted to introduce truncations and deletions in ORP2 that are deleterious to activity, but this was not confirmed experimentally and a functional link to the auditory system was not established [72]. However, ORP2 was recently identified as a regulator of cell morphology, attachment, motility and proliferation via actin remodeling through the RhoA signaling pathway [73]. CRISPR-Cas9-mediated ORP2 knockout in HUH7 hepatocarcinoma cells affected the expression of several RhoA effectors, including the Rho GTPase-activating protein ARHGAP12 and diaphanous-1, with which ORP2 interacts [73, 74]. Interestingly, ARHGAP12 is expressed at inner ear hair cell–cell junctions [75], while mutations in diaphanous-1 are causative in autosomal dominant, non-syndromic hearing loss in an extended Costa Rican family [76]. These studies suggest that interaction between ORP2 and diaphanous-1 could be essential to the function and morphology of cochlear hair cells. The connection between this disease phenotype and the purported role of ORP2 in LD function (see “ORP2 at ER/lipid droplet contact sites”) remains to be resolved.
ORP3 and OSBP in heritable amyotrophic lateral sclerosis
Amyotrophic lateral sclerosis (ALS) is a poorly understood sporadic neurodegenerative disorder of motor neurons in which 10% of cases are attributed to heritable genetic components [77]. One of these is ALS8, a subtype caused by a loss-of-function point mutation in the major sperm protein domain of VAP-B [78, 79], which interacts with the FFAT motif of partner proteins such as OSBP/ORPs. Mutant VAP-B forms aggregates with itself and wild-type VAP-A and VAP-B in detergent-insoluble vacuoles that sequester ER proteins, leading to mislocalization of Sac1 and defects in retrograde transport due to perinuclear and PM accumulation of PI(4)P [80]. Interestingly, overexpression of ORP3 in HeLa cells prevented mutant VAP-B aggregation and rescued both PI(4)P accumulation and secretory phenotypes [80]. Similar results were obtained by co-expressing mutant VAP-B and the D. melanogaster homolog of OSBP [81]. It is unclear whether the lipid-transport functions of ORP3 and OSBP contributed to the rescue of mutant VAP-B phenotypes, or if their overexpression titrates the mutant VAP-B, freeing wild-type VAP-A and B to interact with partner proteins.
ORPs and dyslipidemia
Dyslipidemia is defined as elevated levels of one or more lipoprotein species in the blood, which predisposes individuals to cardiovascular events, hepatic disease and metabolic syndrome. Dyslipidemias have genetic and lifestyle-related contributions, and can result from increased lipid production, decreased lipid clearance or increased lipid absorption from the diet. In cases of hypercholesterolemia, elevated low-density lipoprotein (LDL) imposes cardiovascular risk while elevated high density lipoprotein (HDL) is considered beneficial [82]. HDL and LDL levels are dependent on factors that regulate their metabolism in the circulation as well as by cellular cholesterol synthesis, uptake, processing and secretion. Genetic screens of dyslipidemic patients and functional studies on mouse and cell culture models have confirmed a role for the OSBP/ORP family in all steps affecting lipoprotein metabolism.
Studies have implicated OSBP/ORPs in lipid dysregulation without identifying specific mechanisms. ORP3 was recently identified as a liver receptor homolog-1 (LRH-1) target gene responsible for the increased de novo cholesterol synthesis and hepatic steatosis observed in mice expressing a gain-of-function LRH-1 mutant [83]. In a genetic screen of 871 Finnish adults, increased levels of the hepatic steatosis marker hsa-miR-885-5p were found to be inversely correlated with ORP2 expression and HDL cholesterol levels [84]. Studies of hyperlipidemic Finnish [85] and Japanese [86] families identified single nucleotide polymorphisms (SNPs) in the OSBPL10 gene that were linked to elevated plasma triglyceride and LDL levels. A correlation between SNPs in the OSBPL10 gene and incidence of peripheral arterial disease was also noted [86]. These findings prompted cell culture studies in which ORP10 was found to associate with microtubules and the Golgi apparatus to promote lipoprotein secretion [85, 87]. However, the connection between the OSBPL10 SNPs and plasma lipid levels remains obscure.
A heterozygous loss-of-function mutation in ORP1L, which delivers lipoprotein-derived cholesterol from the LEL to the ER for storage and efflux (see “Sterol transfer by ORP1L and ORP6 at endosome/ER contact sites”), was identified in patients with extremely low levels of HDL cholesterol [88]. Individuals with the loss-of-function ORP1L mutation exhibited decreased plasma levels of apoA-1 as well as decreased cholesterol efflux to available apoA-1. Therefore, it remains unclear whether the mutation is associated with impaired cholesterol efflux by ATP binding cassette (ABC)A1 to apoA-1 or impaired secretion of apoA-1 from the liver or other tissues. The former mechanism is supported by ORP1L silencing experiments in cultured macrophages that showed significantly reduced efflux of [3H]cholesterol to apoA-1in the media [89]. Overall, these findings are consistent with a model in which ORP1L-mediated export of cholesterol from the LEL is coupled to efflux from cells by an ABCA1-dependent pathway. ORP6 may have a related function since its expression was positively correlated with HDL cholesterol levels in healthy human subjects and decreased in the atherosclerotic plaques of LDL receptor knockout mice [49].
Homozygous knockout of ORP4 in mice was also shown to reduce atherosclerotic lesion size by a mechanism independent of cholesterol efflux. Rather, ORP4 promotes macrophage survival in atherosclerotic plaques, attenuating oxysterol-induced cytotoxicity by a pro-survival calcium signaling pathway (“ORP4 in leukemia and metastasis”) [90]. ORP8 expression is increased in human atherosclerotic plaques [91] and the macrophage-specific knockout of ORP8 in LDLR-knockout mice attenuated arterial lesion size and inflammation [92]. A potential link between ORP8 and lesion formation was strengthened by the finding that ORP8 negatively regulated cholesterol efflux by cultured macrophages via transcriptional suppression of ABCA1 [91]. Consistent with this, chow-fed male OSBPL8 knockout mice had significantly elevated HDL levels relative to female mice on the same diet and both genders displayed significantly elevated HDL when fed a high-fat diet [89]. The regulatory activity of ORP8 in cultured hepatocytes could be mediated by interaction with the nucleoporin Nup62 [93], or via obesity-associated miR-142 and attenuation of insulin-stimulated protein kinase B/AKT activity in liver and smooth muscles cells [94–96]. Like ORP8, SNPs in OSBPL11 were associated with altered metabolism of lipids and glucose in obese populations [97], and siblings homozygous for a loss-of-function OSBPL11 missense mutation display abnormal dermal cholesterol deposits [98]. Interestingly, ORP8 and ORP11 normally follow opposing expression patterns during adipocyte differentiation (reduced and increased, respectively), and in cell culture studies the overexpression of ORP8 or silencing of ORP11 impedes triglyceride storage [99]. In general, these studies show that select ORPs are involved, either directly or indirectly, in pathways that remove or prevent accumulation of cholesterol in cells of the cardiovascular system, leading to reduced risk of atherosclerosis.
OSBP/ORPs in cancers
While genetic screens have associated the expression and/or mutation of individual ORPs with various cancers (summarized in Table 2), only ORP5, ORP8, ORP4 and OSBP were investigated to confirm a functional role in malignancy. Here, we review the current understanding of how these ORPs contribute to cancer cell invasion, evasion of apoptosis and proliferation.
ORP5 and invasiveness in pancreatic ductal carcinoma
Chemical induction of pancreatic ductal carcinoma in hamsters yielded two cell lines, PC1 and the more invasive PC1.0, that are used as models of the human disease [100, 101]. Analysis of mRNA profiles of the two cell lines revealed that OSBPL5 was differentially expressed in PC1.0 cells. Silencing of ORP5 in PC1.0 cells significantly decreased invasiveness while the converse result was obtained by ORP5 overexpression in PC1 cells [67]. Similar results were obtained in human pancreatic cancer cell lines [102]. In pancreatic cancer patients, increased OSBPL5 expression was correlated with a 50% reduction in 1-year survival, and a nearly 70% reduction in median survival time in patients with stage I, II and III cancers [67].
The mechanism by which ORP5 increases cancer cell invasiveness could involve upregulation of oncogene expression through effects on the cholesterol biosynthetic pathway. Under cholesterol-replete conditions, SREBP2 is sequestered in ER membranes, whereas lowering ER cholesterol promotes SREBP translocation to the Golgi apparatus for proteolytic processing to release the soluble transcription factor, which translocates to the nucleus and binds sterol response elements in gene promoters [103]. The promoter region of the histone deacetylase 5 gene HDAC5 has a sterol response element, placing oncogenic HDAC5 expression under sterol control [104]. Treatment with the HDAC inhibitor trichostatin-A significantly reduced cell growth in ORP5-positive pancreatic cancer cell lines [102]. However, the mechanistic link between ORP5 and SREBP2 is uncertain; ORP5 is implicated in the delivery of LDL-derived cholesterol to the ER (Sect. 2.4) [39, 40], which would potentially restrict SREBP2 activation and subsequent HDAC5 expression. Also, there is a synergistic relationship between the trichostatin-A and the cholesterol synthesis inhibitor simvastatin [102], which would have opposing functions if HDAC expression was being induced by the SREBP2 [104]. Future studies to evaluate how ORP5 affects SREBP2 processing in the ER, and the link to expression of oncogenic genes, are warranted.
ORP8 promotes apoptosis by the Fas receptor pathway
Fas receptor binding of autocrine or paracrine-secreted Fas ligand leads to the initial activation of caspase 8 and a resulting cascade of caspase autoproteolysis that ultimately leads to cell death [105]. Immunohistochemical analysis of liver tissue from patients with hepatocellular carcinoma (HCC) revealed that malignant cells contained less cell surface and more internal Fas receptor than non-malignant counterparts, implying that HCC cells may avoid apoptosis due to insufficient Fas receptor presentation [106]. ORP8 was identified as a necessary factor for the PM presentation of Fas in HCC cell lines and patient tissue samples in which ORP8 expression are reduced by miR-143 [107]. Expression of ORP8 in HCC cells induced Fas-mediated apoptosis by shifting cytosolic Fas to the PM and reduced the size of mouse tumor xenografts. However, the apoptotic effect of ORP8 may be specific to HCC; ORP8 expression was increased in a hamster model of cholangiocarcinoma [64], and ORP8 expression in HepG2 cells mediated cell-cycle inhibition by 25-hydroxycholesterol but did not induce apoptosis [108].
ORP4 in leukemia and metastasis
Elevated ORP4 transcripts in disseminated tumor cells and the leukocytes of patients with chronic myeloid leukemia first pointed to this ORP as a potential bio-marker or regulator of metastasis [65, 66, 109, 110]. Alternate promoter start sites in OSBPL2 produce three ORP4 variants; full-length ORP4L, and ORP4M and ORP4S that have truncated and deleted N-terminal PH domains, respectively [113]. Like OSBP, the ORP4 OHD binds cholesterol, oxysterols and PI(4)P [40, 111, 112], and ORP4L has a PH domain that recognizes Golgi-associated PI(4)P [113]. However, ORP4 is the only member of the ORP family required for the proliferation of cultured cells [113]. Silencing of ORP4L induced growth arrest and cell death in immortalized and cancerous cell lines, which was exacerbated by knockdown of all three variants. Moreover, steroidal antineoplastic saponins called the ORPphillins were identified as ORP4 antagonists that compete for sterol binding to the OHD [31].
The correlation between ORP4 expression and tumor dissemination may involve the epithelial-to-mesenchymal transition (EMT), the process by which epithelial cells develop mesenchymal characteristics such as increased motility and decreased cell–cell contact. EMT is accompanied by increased vimentin intermediate filament expression, which leads to increased motility and invasiveness that is oncogenic outside the context of development [114]. All three ORP4 variants interact with the vimentin intermediate filament network but ORP4M and ORP4S collapse the network, suggesting negative regulation of this interaction by the ORP4L PH domain [111–113]. Considering the regulatory role of vimentin in cell adhesion and motility, further study of a functional relationship with ORP4 in the context of metastasis is needed.
Yan and colleagues identified a potential mechanism for ORP4L-mediated proliferation in T cell acute lymphoblastic leukemia, in which ORP4L scaffolds a unique PM-associated phospholipase C (PLC)3β activation complex that produces inositol triphosphate required for ER calcium release and mitochondrial ATP production [115, 116]. In the absence of ORP4L, inositol triphosphate signaling is attenuated, leading to mitochondrial dysfunction and cell death. Importantly, ORP4L is virtually absent from normal T-cells, where inositol triphosphate production occurs via a G-protein-independent pathway involving PLCγ1. Thus, malignant T-cells and other cancerous cells that express abundant ORP4L appear to have acquired a novel signaling pathway that provides increased energy production for survival and proliferation. ORP4 knockout mice develop normally except for male sterility due to defective sperm elongation [117], which is consistent with high levels of ORP4 expression in testis. However, it is unclear why other tissues that express ORP4, such as the brain and retina, are unaffected in the knockout animals.
To summarize, many studies implicating ORPs in cancers are large-scale genomic analyses that have associated ORP gene SNPs with either cancer progression or survival (summarized in Table 2). However, the relation of these SNPs to gene transcription and protein expression is unknown. The limited number of analyses available suggests that those ORPs implicated in cancer initiation or progress do so downstream of their primary lipid binding/transfer functions. However, the well-defined role for ORP4L in the survival of lymphoblastic T-leukemia cells indicates that other scaffold/signaling mechanisms may apply.
OSBP/ORPs in viral infection, replication and release
Single-stranded RNA viruses replicate in the cytosol of host cells and employ the Golgi apparatus, ER and/or endosomal membranes as replication platforms, altering the morphology of the organelles using a combination of viral and host factors. The morphology of the hijacked host membranes, termed replication organelles, depends on the infecting virus. For example, dengue virus forms double-membrane vesicles of ER origin, while the single-membrane vesicles formed by poliovirus are ER-, Golgi- and lysosome-derived and arranged in rosettes [118]. As outlined below, OSBP, ORP1L, ORP4 and ORP10 contribute to viral egress or replication processes.
Currently, the most well-studied example of viral dependence on ORP function is the requirement for OSBP and ORP4 in the development the hepatitis C virus (HCV) replication organelle. HCV replication involves the translation of polyproteins, cleavage into active peptides and the establishment of a replication organelle, called the membranous web, from host membranes. A screen for host proteins associated with non-structural HCV proteins revealed that the ER-anchored non-structural viral protein NS5A interacts with both OSBP and VAP-A, and established that OSBP is required for the replication and egress of HCV particles [119]. Tai and colleagues determined that OSBP supplies the membranous web with cholesterol in a PI(4)P-dependent manner, similar to the exchange between the Golgi apparatus and ER (Sect. 2.1) [120]. The requirement for OSBP to supply cholesterol extends to the development of replication organelles by poliovirus and dengue [120], with concurrent studies showing similar results in picornavirus [121] and encephalomyocarditis virus infections [122]. These mechanistic findings conceptualized OSBP as a target for known and novel antiviral compounds; OSBP is an identified target of the anti-enteroviral compounds itraconazole and TTP-8307 [123, 124] and the antiviral properties of the ORPphilin OSW-1 are dependent on OSBP and ORP4 [125]. Interestingly, despite the dependence of OSW-1 antiviral activity on ORP4, ectopic expression of ORP4L or ORP4S inhibited the replication of HCV and produced infection-related lipid droplets on the membranous web [126], suggesting that ORP4 may contribute to cellular lipid metabolism by a different mechanism than OSBP. In addition to viral replication and egress, OSBP has also been implicated in viral entry. Treatment of cells with interferon-inducible transmembrane protein 3 inhibited the interaction between OSBP with VAP-A and cholesterol egress from the ER, leading to aberrant accumulation of cholesterol in the endocytic system that prevented cytosolic release of endocytosed vesicular stomatitis virus [127].
A study of dengue susceptibility in African, Asian and European populations revealed that SNPs which affect the expression of OSBPL10 are protective in African populations [128]. Functional studies using THP-1 monocytic leukemia cells demonstrated that ORP10 silencing significantly reduced viral replication and secretion [128].
Expression of an OHD deletion mutant of ORP1L reduced the infectivity of vesicular stomatitis virus presenting the Ebola glycoprotein by 50%, suggesting a requirement for ORP1L-mediated endosome motility in Ebola infection [55]. ORP1L was also identified as an interacting partner of a 2′–5′ oligonucleotide synthase viral host defence pathway against flavivirus infection, but the mechanism of action is unknown [129]. ORP1L was implicated in another innate immune response to adenovirus infection [130]. In this case, the viral receptor internalization and degradation complex protein RIDα, a GTP-Rab7 mimic, interacts with ORP1L to direct the transfer of LDL-cholesterol from LEL-to-ER independently of NPC1 [131]. The RIDα-ORP1L pathway for cholesterol transfer did not affect cholesterol biosynthesis in the ER, but directed cholesterol to lipid droplets and attenuated lipid raft-dependent pro-inflammatory signaling by Toll-like receptor 4 [130].
Concluding remarks
Changes in ORP expression and function are implicated in common and rare human disorders, and infections by common pathogenic viruses. An emerging recognition that OSBP/ORPs mediate lipid transport and regulate metabolism at MCS suggests that these pathologies may arise due to deleterious changes in the intracellular distribution of cholesterol, phospholipids and/or sphingolipids. Alternatively, considering that many ORPs exhibit nanomolar affinity for oxysterols, ORPs may act as high-affinity receptors that regulate signaling pathways via protein–protein or protein–membrane interactions. While both are attractive possibilities as primary functions of the ORP family, their multivalent roles suggest that these could be dichotomous functions. For example, sterol-binding by ORP1L [55] and ORP4L [90] alters interactions with their respective protein partners, making them candidate sterol-regulated scaffolding proteins for functional complexes at membranes. However, both have also been shown to bind and transport lipids in vitro [56, 113]. ER-to-Golgi cholesterol transfer by OSBP is required for synergy between sterol and sphingolipid metabolism, but OSBP also acts as a sterol-regulated scaffold for the dephosphorylation of phosphorylated extracellular signal-regulated kinase [132].
Despite being named for the founding member OSBP that binds oxysterols with high affinity, structural and binding analysis clearly indicates the Osh/OSBP/ORPs also bind phospholipids. OSBP/ORP function is contingent on the specificity of the OHD for lipid ligands, which is currently defined by in vitro binding of a limited set of available ligands that may not recapitulate in vivo specificity. Continued elucidation of the structure of OHDs and their associated ligands, particularly those from mammalian family members, will help define the repertoire of lipid ligands. Further studies are required to then reconcile in vitro analysis of lipid transport with in vivo studies of OSBP/ORP function to define their roles in normal and pathological conditions at the cellular and organismal level.
Abbreviations
- ALS
Amyotrophic lateral sclerosis
- ABC
ATP binding cassette
- CARTS
Carriers of the trans-Golgi network to the cell surface
- CERT
Ceramide transfer protein
- ER
Endoplasmic reticulum
- FFAT
Two phenylalanines in an acidic tract
- HDL
High density lipoprotein
- 25OH
25-Hydroxycholesterol
- HCC
Hepatocellular carcinoma
- HCV
Hepatitis C virus
- LDL
Low density lipoprotein
- LEL
Late endosomes–lysosomes
- LD
Lipid droplet
- LRH-1
Liver receptor homolog-1
- LXR
Liver X receptor
- LTP
Lipid transfer protein
- MCS
Membrane contact site
- mTOR
Mammalian target of rapamycin
- NPC
Niemann–Pick type C
- OSBP
Oxysterol-binding protein
- ORP
OSBP-related protein
- OHD
OSBP homology domain
- Osh
Oxysterol-binding protein homolog
- PM
Plasma membrane
- PI4K
Phosphatidylinositol-4-kinase
- PI
Phosphatidylinositol
- PI(4)P
Phosphatidylinositol 4-phosphate
- PIPs
Phosphatidylinositol phosphates
- PLC
Phospholipase C
- PS
Phosphatidylserine
- RILP
Rab7-interacting lysosomal protein
- SM
Sphingomyelin
- SNP
Single nucleotide polymorphisms
- SREBP
Sterol regulatory element binding protein
- TGN
Trans-Golgi network
- VAP
Vesicle-associated membrane protein-associated protein
References
- 1.Holthuis J, Menon AK. Lipid landscapes and pipelines in membrane homeostasis. Nature. 2014;510:48–57. doi: 10.1038/nature13474. [DOI] [PubMed] [Google Scholar]
- 2.Wong LH, Čopič A, Levine TP. Advances on the transfer of lipids by lipid transfer proteins. Trends Biochem Sci. 2017;42:516–530. doi: 10.1016/j.tibs.2017.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Taylor FR, Kandutsch AA. Oxysterol binding protein. Chem Phys Lipids. 1985;38:187–194. doi: 10.1016/0009-3084(85)90066-0. [DOI] [PubMed] [Google Scholar]
- 4.Brown MS, Dana SE, Goldstein JL. Cholesterol ester formation in cultured human fibroblasts. Stimulation by oxygenated sterols. J Biol Chem. 1975;250:4025–4027. [PubMed] [Google Scholar]
- 5.Dawson PA, Ridgway ND, Slaughter CA, Brown MS, Goldstein JL. cDNA cloning and expression of oxysterol-binding protein, an oligomer with a potential leucine zipper. J Biol Chem. 1989;264:16798–16803. [PubMed] [Google Scholar]
- 6.Ridgway ND, Dawson PA, Ho YK, Brown MS, Goldstein JL. Translocation of oxysterol binding protein to Golgi apparatus triggered by ligand binding. J Cell Biol. 1992;116:307–319. doi: 10.1083/jcb.116.2.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Laitinen S, Olkkonen VM, Ehnholm C, Ikonen E. Family of human oxysterol binding protein (OSBP) homologues. A novel member implicated in brain sterol metabolism. J Lipid Res. 1999;40:2204–2211. [PubMed] [Google Scholar]
- 8.Jaworski CJ, Moreira E, Li A, Lee R, Rodriguez IR. A family of 12 human genes containing oxysterol-binding domains. Genomics. 2001;78:185–196. doi: 10.1006/geno.2001.6663. [DOI] [PubMed] [Google Scholar]
- 9.Schulz TA, Prinz WA. Sterol transport in yeast and the oxysterol binding protein homologue (OSH) family. BBA Mol Cell Biol Lipids. 2007;1771:769–780. doi: 10.1016/j.bbalip.2007.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beh CT, Cool L, Phillips J, Rine J. Overlapping functions of the yeast oxysterol-binding protein homologues. Genetics. 2001;157:1117–1140. doi: 10.1093/genetics/157.3.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kobuna H, Inoue T, Shibata M, Gengyo-Ando K, Yamamoto A, Mitani S, Arai H. Multivesicular body formation requires OSBP-related proteins and cholesterol. PLoS Genet. 2010;6:e1001055. doi: 10.1371/journal.pgen.1001055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ma Z, Liu Z, Huang X. OSBP- and FAN-mediated sterol requirement for spermatogenesis in Drosophila. Development. 2010;137:3775–3784. doi: 10.1242/dev.049312. [DOI] [PubMed] [Google Scholar]
- 13.Ma Z, Liu Z, Huang X. Membrane phospholipid asymmetry counters the adverse effects of sterol overloading in the Golgi membrane of Drosophila. Genetics. 2012;190:1299–1308. doi: 10.1534/genetics.111.137687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Im Y, Raychaudhuri S, Prinz WA, Hurley JH. Structural mechanism for sterol sensing and transport by OSBP-related proteins. Nature. 2005;437:154–158. doi: 10.1038/nature03923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tong J, Manik MK, Yang H, Im YJ. Structural insights into nonvesicular lipid transport by the oxysterol binding protein homologue family. Biochim Biophys Acta. 2016;1861:928–939. doi: 10.1016/j.bbalip.2016.01.008. [DOI] [PubMed] [Google Scholar]
- 16.de Saint-Jean M, Delfosse V, Douguet D, Chicanne G, Payrastre B, Bourguet W, Antonny B, Drin G. Osh4p exchanges sterols for phosphatidylinositol 4-phosphate between lipid bilayers. J Cell Biol. 2011;195:965–978. doi: 10.1083/jcb.201104062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tong J, Yang H, Yang H, Eom SH, Im YJ. Structure of Osh3 reveals a conserved mode of phosphoinositide binding in oxysterol-binding proteins. Structure. 2013;21:1203–1213. doi: 10.1016/j.str.2013.05.007. [DOI] [PubMed] [Google Scholar]
- 18.Manik MK, Yang H, Tong J, Im YJ. Structure of yeast OSBP-related protein Osh1 reveals key determinants for lipid transport and protein targeting at the nucleus-vacuole junction. Structure. 2017;25:617–629000. doi: 10.1016/j.str.2017.02.010. [DOI] [PubMed] [Google Scholar]
- 19.Kentala H, Weber-Boyvat M, Olkkonen VM. OSBP-related protein family: mediators of lipid transport and signaling at membrane contact sites. Int Rev Cell Mol Biol. 2016;321:299–340. doi: 10.1016/bs.ircmb.2015.09.006. [DOI] [PubMed] [Google Scholar]
- 20.Guo S, Stolz LE, Lemrow SM, York JD. SAC1-like domains of yeast SAC1, INP52, and INP53 and of human synaptojanin encode polyphosphoinositide phosphatases. J Biol Chem. 1999;274:12990–12995. doi: 10.1074/jbc.274.19.12990. [DOI] [PubMed] [Google Scholar]
- 21.Fang M, Kearns BG, Gedvilaite A, Kagiwada S, Kearns M, Fung MK, Bankaitis VA. Kes1p shares homology with human oxysterol binding protein and participates in a novel regulatory pathway for yeast Golgi-derived transport vesicle biogenesis. EMBO J. 1996;15:6447–6459. doi: 10.1002/j.1460-2075.1996.tb01036.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lagace TA, Byers DM, Cook HW, Ridgway ND. Altered regulation of cholesterol and cholesteryl ester synthesis in Chinese-hamster ovary cells overexpressing the oxysterol-binding protein is dependent on the pleckstrin homology domain. Biochem J. 1997;326(Pt 1):205–213. doi: 10.1042/bj3260205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wyles JP, McMaster CR, Ridgway ND. Vesicle-associated membrane protein-associated protein-A (VAP-A) interacts with the oxysterol-binding protein to modify export from the endoplasmic reticulum. J Biol Chem. 2002;277:29908–29918. doi: 10.1074/jbc.M201191200. [DOI] [PubMed] [Google Scholar]
- 24.Peretti D, Dahan N, Shimoni E, Hirschberg K, Lev S. Coordinated lipid transfer between the endoplasmic reticulum and the Golgi complex requires the VAP proteins and is essential for Golgi-mediated transport. Mol Biol Cell. 2008;19:3871–3884. doi: 10.1091/mbc.e08-05-0498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mesmin B, Bigay J, Moser von Filseck J, Lacas-Gervais S, Drin G, Antonny B. A four-step cycle driven by PI(4)P hydrolysis directs sterol/PI(4)P exchange by the ER–Golgi Tether OSBP. Cell. 2013;155:830–843. doi: 10.1016/j.cell.2013.09.056. [DOI] [PubMed] [Google Scholar]
- 26.Banerji S, Ngo M, Lane CF, Robinson C-A, Minogue S, Ridgway ND. Oxysterol binding protein-dependent activation of sphingomyelin synthesis in the Golgi apparatus requires phosphatidylinositol 4-kinase IIα. Mol Biol Cell. 2010;21:4141–4150. doi: 10.1091/mbc.e10-05-0424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Perry RJ, Ridgway ND. Oxysterol-binding protein and vesicle-associated membrane protein–associated protein are required for sterol-dependent activation of the ceramide transport protein. Mol Biol Cell. 2006;17:2604–2616. doi: 10.1091/mbc.e06-01-0060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Slotte PJ. Biological functions of sphingomyelins. Prog Lipid Res. 2013;52:424–437. doi: 10.1016/j.plipres.2013.05.001. [DOI] [PubMed] [Google Scholar]
- 29.Wakana Y, Kotake R, Oyama N, Murate M, Kobayashi T, Arasaki K, Inoue H, Tagaya M. CARTS biogenesis requires VAP–lipid transfer protein complexes functioning at the endoplasmic reticulum–Golgi interface. Mol Biol Cell. 2015;26:4686–4699. doi: 10.1091/mbc.e15-08-0599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maxfield FR, Wüstner D. Intracellular cholesterol transport. J Clin Investig. 2002;110:891–898. doi: 10.1172/JCI0216500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Burgett AW, Poulsen TB, Wangkanont K, Anderson DR, Kikuchi C, Shimada K, Okubo S, Fortner KC, Mimaki Y, Kuroda M, Murphy JP, Schwalb DJ, Petrella EC, Cornella-Taracido I, Schirle M, Tallarico JA, Shair MD. Natural products reveal cancer cell dependence on oxysterol-binding proteins. Nat Chem Biol. 2011;7:639–647. doi: 10.1038/nchembio.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mesmin B, Bigay J, Polidori J, Jamecna D, Lacas-Gervais S, Antonny B. Sterol transfer, PI4P consumption, and control of membrane lipid order by endogenous OSBP. EMBO J. 2017;36:3156–3174. doi: 10.15252/embj.201796687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nishimura T, Uchida Y, Yachi R, Kudlyk T, Lupashin V, Inoue T, Taguchi T, Arai H. Oxysterol-binding protein (OSBP) is required for the perinuclear localization of intra-Golgi v-SNAREs. Mol Biol Cell. 2013;24:3534–3544. doi: 10.1091/mbc.e13-05-0250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dong R, Saheki Y, Swarup S, Lucast L, Harper JW, De Camilli P. Endosome-ER contacts control actin nucleation and retromer function through VAP-dependent regulation of PI4P. Cell. 2016;166:408–423. doi: 10.1016/j.cell.2016.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nishimura T, Inoue T, Shibata N, Sekine A, Takabe W, Noguchi N, Arai H. Inhibition of cholesterol biosynthesis by 25-hydroxycholesterol is independent of OSBP. Genes Cells. 2005;10:793–801. doi: 10.1111/j.1365-2443.2005.00879.x. [DOI] [PubMed] [Google Scholar]
- 36.Ghai R, Du X, Wang H, Dong J, Ferguson C, Brown AJ, Parton RG, Wu J-WW, Yang H. ORP5 and ORP8 bind phosphatidylinositol-4, 5-biphosphate (PtdIns(4,5)P 2) and regulate its level at the plasma membrane. Nat Commun. 2017;8:757. doi: 10.1038/s41467-017-00861-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Galmes R, Houcine A, Vliet AR, Agostinis P, Jackson CL, Giordano F. ORP5/ORP8 localize to endoplasmic reticulum–mitochondria contacts and are involved in mitochondrial function. EMBO Rep. 2016;17:800–810. doi: 10.15252/embr.201541108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Steenbergen R, Nanowski TS, Beigneux A, Kulinski A, Young SG, Vance JE. Disruption of the phosphatidylserine decarboxylase gene in mice causes embryonic lethality and mitochondrial defects. J Biol Chem. 2005;280:40032–40040. doi: 10.1074/jbc.M506510200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Du X, Kumar J, Ferguson C, Schulz TA, Ong YS, Hong W, Prinz WA, Parton RG, Brown AJ, Yang H. A role for oxysterol-binding protein-related protein 5 in endosomal cholesterol trafficking. J Cell Biol. 2011;192:121–135. doi: 10.1083/jcb.201004142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Suchanek M, Hynynen R, Wohlfahrt G, Lehto M, Johansson M, Saarinen H, Radzikowska A, Thiele C, Olkkonen VM. The mammalian oxysterol-binding protein-related proteins (ORPs) bind 25-hydroxycholesterol in an evolutionarily conserved pocket. Biochem J. 2007;405:473–480. doi: 10.1042/BJ20070176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Du X, Zadoorian A, Lukmantara IE, Qi Y, Brown AJ, Yang H. Oxysterol-binding protein-related protein 5 (ORP5) promotes cell proliferation by activation of mTORC1 signaling. J Biol Chem. 2018 doi: 10.1074/jbc.RA117.001558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu J, Dang Y, Ren YR, Liu JO. Cholesterol trafficking is required for mTOR activation in endothelial cells. Proc Natl Acad Sci USA. 2010;107:4764–4769. doi: 10.1073/pnas.0910872107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ikonen E. Cellular cholesterol trafficking and compartmentalization. Nat Rev Mol Cell Biol. 2008;9:125–138. doi: 10.1038/nrm2336. [DOI] [PubMed] [Google Scholar]
- 44.Möbius W, van Donselaar E, Ohno-Iwashita Y, Shimada Y, Heijnen HFG, Slot JW, Geuze HJ. Recycling compartments and the internal vesicles of multivesicular bodies harbor most of the cholesterol found in the endocytic pathway. Traffic. 2003;4:222–231. doi: 10.1034/j.1600-0854.2003.00072.x. [DOI] [PubMed] [Google Scholar]
- 45.Vance JE, Karten B. Niemann–Pick C disease and mobilization of lysosomal cholesterol by cyclodextrin. J Lipid Res. 2014;55:1609–1621. doi: 10.1194/jlr.R047837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Eden ER, Sanchez-Heras E, Tsapara A, Sobota A, Levine TP, Futter CE. Annexin A1 tethers membrane contact sites that mediate er to endosome cholesterol transport. Dev Cell. 2016;37:473–483. doi: 10.1016/j.devcel.2016.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Friedman JR, DiBenedetto JR, West M, Rowland AA, Voeltz GK. Endoplasmic reticulum–endosome contact increases as endosomes traffic and mature. Mol Biol Cell. 2013;24:1030–1040. doi: 10.1091/mbc.e12-10-0733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lehto M, Laitinen S, Chinetti G, Johansson M, Ehnholm C, Staels B, Ikonen E, Olkkonen VM. The OSBP-related protein family in humans. J Lipid Res. 2001;42:1203–1213. [PubMed] [Google Scholar]
- 49.Ouimet M, Hennessy EJ, van Solingen C, Koelwyn GJ, Hussein MA, Ramkhelawon B, Rayner KJ, Temel RE, Perisic L, Hedin U, Maegdefessel L, Garabedian MJ, Holdt LM, Teupser D, Moore KJ. miRNA targeting of oxysterol-binding protein-like 6 regulates cholesterol trafficking and efflux. Arterioscler Thromb Vasc Biol. 2016;36:942–951. doi: 10.1161/ATVBAHA.116.307282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Johansson M, Bocher V, Lehto M, Chinetti G, Kuismanen E, Ehnholm C, Staels B, Olkkonen VM. The two variants of oxysterol binding protein-related protein-1 display different tissue expression patterns, have different intracellular localization, and are functionally distinct. Mol Biol Cell. 2003;14:903–915. doi: 10.1091/mbc.e02-08-0459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee S, Wang P-Y, Jeong Y, Mangelsdorf DJ, Anderson R, Michaely P. Sterol-dependent nuclear import of ORP1S promotes LXR regulated trans-activation of apoE. Exp Cell Res. 2012;318:2128–2142. doi: 10.1016/j.yexcr.2012.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Johansson M, Olkkonen VM. Assays for interaction between Rab7 and Oxysterol Binding Protein Related Protein 1L (ORP1L) Methods Enzymol. 2005;403:743–758. doi: 10.1016/S0076-6879(05)03065-X. [DOI] [PubMed] [Google Scholar]
- 53.Rocha N, Kuijl C, van der Kant R, Janssen L, Houben D, Janssen H, Zwart W, Neefjes J. Cholesterol sensor ORP1L contacts the ER protein VAP to control Rab7–RILP–p150Glued and late endosome positioning. J Cell Biol. 2009;185:1209–1225. doi: 10.1083/jcb.200811005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Johansson M, Rocha N, Zwart W, Jordens I, Janssen L, Kuijl C, Olkkonen VM, Neefjes J. Activation of endosomal dynein motors by stepwise assembly of Rab7-RILP-p150Glued, ORP1L, and the receptor betalll spectrin. J Cell Biol. 2007;176:459–471. doi: 10.1083/jcb.200606077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.van der Kant R, Fish A, Janssen L, Janssen H, Krom S, Ho N, Brummelkamp T, Carette J, Rocha N, Neefjes J. Late endosomal transport and tethering are coupled processes controlled by RILP and the cholesterol sensor ORP1L. J Cell Sci. 2013;126:3462–3474. doi: 10.1242/jcs.129270. [DOI] [PubMed] [Google Scholar]
- 56.Zhao K, Ridgway ND. Oxysterol-binding protein-related protein 1L regulates cholesterol egress from the endo-lysosomal system. Cell Rep. 2017;19:1807–1818. doi: 10.1016/j.celrep.2017.05.028. [DOI] [PubMed] [Google Scholar]
- 57.Walther TC, Chung J, Jr RV. Lipid droplet biogenesis. Annu Rev Cell Dev Biol. 2016;33:1–20. doi: 10.1146/annurev-cellbio-100616-060608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hynynen R, Suchanek M, Spandl J, Bäck N, Thiele C, Olkkonen VM. OSBP-related protein 2 is a sterol receptor on lipid droplets that regulates the metabolism of neutral lipids. J Lipid Res. 2009;50:1305–1315. doi: 10.1194/jlr.M800661-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Weber-Boyvat M, Kentala H, Peränen J, Olkkonen VM. Ligand-dependent localization and function of ORP-VAP complexes at membrane contact sites. Cell Mol Life Sci. 2015;72:1967–1987. doi: 10.1007/s00018-014-1786-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Escajadillo T, Wang H, Li L, Li D, Sewer MB. Oxysterol-related-binding-protein related Protein-2 (ORP2) regulates cortisol biosynthesis and cholesterol homeostasis. Mol Cell Endocrinol. 2016;427:73–85. doi: 10.1016/j.mce.2016.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Käkelä R, Tanhuanpää K, Laitinen S, Somerharju P, Olkkonen VM. Overexpression of OSBP-related protein 2 (ORP2) in CHO cells induces alterations of phospholipid species composition. Biochem Cell Biol. 2005;83:677–683. doi: 10.1139/o05-056. [DOI] [PubMed] [Google Scholar]
- 62.Laitinen S, Lehto M, Lehtonen S, Hyvärinen K, Heino S, Lehtonen E, Ehnholm C, Ikonen E, Olkkonen VM. ORP2, a homolog of oxysterol binding protein, regulates cellular cholesterol metabolism. J Lipid Res. 2002;43:245–255. [PubMed] [Google Scholar]
- 63.Hynynen R, Laitinen S, Käkelä R, Tanhuanpää K, Lusa S, Ehnholm C, Somerharju P, Ikonen E, Olkkonen VM. Overexpression of OSBP-related protein 2 (ORP2) induces changes in cellular cholesterol metabolism and enhances endocytosis. Biochem J. 2005;390:273–283. doi: 10.1042/BJ20042082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Loilome W, Wechagama P, Namwat N, Jusakul A, Sripa B, Miwa M, Kuver R, Yongvanit P. Expression of oxysterol binding protein isoforms in opisthorchiasis-associated cholangiocarcinoma: a potential molecular marker for tumor metastasis. Parasitol Int. 2012;61:136–139. doi: 10.1016/j.parint.2011.07.003. [DOI] [PubMed] [Google Scholar]
- 65.Silva N, Fournier M, Pimenta G, Pulcheri W, Spector N, da Carvalho M. HLM/OSBP2 is expressed in chronic myeloid leukemia. Int J Mol Med. 2003;12:663–666. [PubMed] [Google Scholar]
- 66.Fournier MV, da Costa GF, Paschoal ME, Ronco LV, Carvalho MG, Pardee AB, Giumaraes FC. Identification of a gene encoding a human oxysterol-binding protein-homologue: a potential general molecular marker for blood dissemination of solid tumors. Cancer Res. 1999;59:3748–3753. [PubMed] [Google Scholar]
- 67.Koga Y, Ishikawa S, Nakamura T, Masuda T, Nagai Y, Takamori H, Hirota M, Kanemitsu K, Baba Y, Baba H. Oxysterol binding protein-related protein-5 is related to invasion and poor prognosis in pancreatic cancer. Cancer Sci. 2008;99:2387–2394. doi: 10.1111/j.1349-7006.2008.00987.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dmitriev AA, Rosenberg EE, Krasnov GS, Gerashchenko GV, Gordiyuk VV, Pavlova TV, Kudryavtseva AV, Beniaminov AD, Belova AA, Bondarenko YN, Danilets RO, Glukhov AI, Kondratov AG, Alexeyenko A, Alekseev BY, Klein G, Senchenko VN, Kashuba VI. Identification of novel epigenetic markers of prostate cancer by NotI-microarray analysis. Dis Markers. 2015;2015:241301. doi: 10.1155/2015/241301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vozianov SO, Kashuba VI, Grygorenko VM, Gordiyuk VV, Danylets RO, Bondarenko YM, Vikarchuk MV. Identification of a new diagnostic marker of prostatic cancer, using NOTI-microchips. Klin Khir. 2016;8:54–57. [PubMed] [Google Scholar]
- 70.Lefebvre C, Bachelot T, Filleron T, Pedrero M, Campone M, Soria J-C, Massard C, Lévy C, Arnedos M, Lacroix-Triki M, Garrabey J, Boursin Y, Deloger M, Fu Y, Commo F, Scott V, Lacroix L, Dieci M, Kamal M, Diéras V, Gonçalves A, Ferrerro J-M, Romieu G, Vanlemmens L, Reynier M-A, Théry J-C, Du F, Guiu S, Dalenc F, Clapisson G, Bonnefoi H, Jimenez M, Tourneau C, André F. Mutational profile of metastatic breast cancers: a retrospective analysis. PLoS Med. 2016;13:e1002201. doi: 10.1371/journal.pmed.1002201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Thoenes M, Zimmermann U, Ebermann I, Ptok M, Lewis MA, Thiele H, Morlot S, Hess MM, Gal A, Eisenberger T, Bergmann C, Nürnberg G, Nürnberg P, Steel KP, Knipper M, Bolz H. OSBPL2 encodes a protein of inner and outer hair cell stereocilia and is mutated in autosomal dominant hearing loss (DFNA67) Orphanet J Rare Dis. 2015;10:15. doi: 10.1186/s13023-015-0238-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Xing G, Yao J, Wu B, Liu T, Wei Q, Liu C, Lu Y, Chen Z, Zheng H, Yang X, Cao X. Identification of OSBPL2 as a novel candidate gene for progressive nonsyndromic hearing loss by whole-exome sequencing. Genet Med. 2014;17:210–218. doi: 10.1038/gim.2014.90. [DOI] [PubMed] [Google Scholar]
- 73.Kentala H, Koponen A, Kivelä AM, Andrews R, Li C, Zhou Y, Olkkonen VM. Analysis of ORP2 knockout hepatocytes uncovers a novel function in actin cytoskeletal regulation. FASEB J. 2017 doi: 10.1096/fj.201700604R. [DOI] [PubMed] [Google Scholar]
- 74.Li D, Dammer EB, Lucki NC, Sewer MB. cAMP-stimulated phosphorylation of diaphanous 1 regulates protein stability and interaction with binding partners in adrenocortical cells. Mol Biol Cell. 2013;24:848–857. doi: 10.1091/mbc.e12-08-0597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rudnicki A, Isakov O, Ushakov K, Shivatzki S, Weiss I, Friedman LM, Shomron N, Avraham KB. Next-generation sequencing of small RNAs from inner ear sensory epithelium identifies microRNAs and defines regulatory pathways. BMC Genom. 2014;15:1–12. doi: 10.1186/1471-2164-15-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lynch ED, Lee MK, Morrow JE, Welcsh PL, León PE, King MC. Nonsyndromic deafness DFNA1 associated with mutation of a human homolog of the Drosophila gene diaphanous. Science. 1997;278:1315–1318. doi: 10.1126/science.278.5341.1315. [DOI] [PubMed] [Google Scholar]
- 77.Kiernan MC, Vucic S, Cheah BC, Turner MR, Eisen A, Hardiman O, Burrell JR, Zoing MC. Amyotrophic lateral sclerosis. Lancet. 2011;377:942–955. doi: 10.1016/S0140-6736(10)61156-7. [DOI] [PubMed] [Google Scholar]
- 78.Nishimura AL, Mitne-Neto M, Silva HCA, Richieri-Costa A, Middleton S, Cascio D, Kok F, Oliveira JRM, Gillingwater T, Webb J, Skehel P, Zatz M. A mutation in the vesicle-trafficking protein VAPB causes late-onset spinal muscular atrophy and amyotrophic lateral sclerosis. Am J Hum Genet. 2004;75:822–831. doi: 10.1086/425287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nishimura AL, Al-Chalabi A, Zatz M. A common founder for amyotrophic lateral sclerosis type 8 (ALS8) in the Brazilian population. Hum Genet. 2005;118:499–500. doi: 10.1007/s00439-005-0031-y. [DOI] [PubMed] [Google Scholar]
- 80.Darbyson A, Ngsee JK. Oxysterol-binding protein ORP3 rescues the amyotrophic lateral sclerosis-linked mutant VAPB phenotype. Exp Cell Res. 2016;341:18–31. doi: 10.1016/j.yexcr.2016.01.013. [DOI] [PubMed] [Google Scholar]
- 81.Moustaqim-Barrette A, Lin YQ, Pradhan S, Neely GG, Bellen HJ, Tsuda H. The amyotrophic lateral sclerosis 8 protein, VAP, is required for ER protein quality control. Hum Mol Genet. 2014;23:1975–1989. doi: 10.1093/hmg/ddt594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Huang L-H, Elvington A, Randolph GJ. The role of the lymphatic system in cholesterol transport. Front Pharmacol. 2015;6:182. doi: 10.3389/fphar.2015.00182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Stein S, Lemos V, Xu P, Demagny H, Wang X, Ryu D, Jimenez V, Bosch F, Lüscher TF, Oosterveer MH, Schoonjans K. Impaired SUMOylation of nuclear receptor LRH-1 promotes nonalcoholic fatty liver disease. J Clin Investig. 2017;127:583–592. doi: 10.1172/JCI85499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Raitoharju E, Seppälä I, Lyytikäinen L-P, Viikari J, Ala-Korpela M, Soininen P, Kangas AJ, Waldenberger M, Klopp N, Illig T, Leiviskä J, Loo B-M, Oksala N, Kähönen M, Hutri-Kähönen N, Laaksonen R, Raitakari O, Lehtimäki T. Blood hsa-miR-122-5p and hsa-miR-885-5p levels associate with fatty liver and related lipoprotein metabolism—the Young Finns Study. Sci Rep. 2016;6:38262. doi: 10.1038/srep38262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Perttilä J, Merikanto K, Naukkarinen J, Surakka I, Martin NW, Tanhuanpää K, Grimard V, Taskinen M-R, Thiele C, Salomaa V, Jula A, Perola M, Virtanen I, Peltonen L, Olkkonen VM. OSBPL10, a novel candidate gene for high triglyceride trait in dyslipidemic Finnish subjects, regulates cellular lipid metabolism. J Mol Med. 2009;87:825–835. doi: 10.1007/s00109-009-0490-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Koriyama H, Nakagami H, Katsuya T, Sugimoto K, Yamashita H, Takami Y, Maeda S, Kubo M, Takahashi A, Nakamura Y, Ogihara T, Rakugi H, Kaneda Y, Morishita R. Identification of evidence suggestive of an association with peripheral arterial disease at the OSBPL10 locus by genome-wide investigation in the Japanese population. J Atheroscler Thromb. 2010;17:1054–1062. doi: 10.5551/jat.4291. [DOI] [PubMed] [Google Scholar]
- 87.Nissilä E, Ohsaki Y, Weber-Boyvat M, Perttilä J, Ikonen E, Olkkonen VM. ORP10, a cholesterol binding protein associated with microtubules, regulates apolipoprotein B-100 secretion. BBA Mol Cell Biol Lipids. 2012;1821:1472–1484. doi: 10.1016/j.bbalip.2012.08.004. [DOI] [PubMed] [Google Scholar]
- 88.Motazacker MM, Pirhonen J, van Capelleveen JC, Weber-Boyvat M, Kuivenhoven J, Shah S, Hovingh KG, Metso J, Li S, Ikonen E, Jauhiainen M, Dallinga-Thie GM, Olkkonen VM. A loss-of-function variant in OSBPL1A predisposes to low plasma HDL cholesterol levels and impaired cholesterol efflux capacity. Atherosclerosis. 2016;249:140–147. doi: 10.1016/j.atherosclerosis.2016.04.005. [DOI] [PubMed] [Google Scholar]
- 89.Vihervaara T, Uronen R-L, Wohlfahrt G, Björkhem I, Ikonen E, Olkkonen VM. Sterol binding by OSBP-related protein 1L regulates late endosome motility and function. Cell Mol Life Sci. 2011;68:537–551. doi: 10.1007/s00018-010-0470-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhong W, Pan G, Wang L, Li S, Ou J, Xu M, Li J, Zhu B, Cao X, Ma H, Li C, Xu J, Olkkonen VM, Staels B, Yan D. ORP4L facilitates macrophage survival via G-protein-coupled signaling. Circ Res. 2016;119:1296–1312. doi: 10.1161/CIRCRESAHA.116.309603. [DOI] [PubMed] [Google Scholar]
- 91.Yan D, Mäyränpää MI, Wong J, Perttilä J, Lehto M, Jauhiainen M, Kovanen PT, Ehnholm C, Brown AJ, Olkkonen VM. OSBP-related protein 8 (ORP8) suppresses ABCA1 expression and cholesterol efflux from macrophages. J Biol Chem. 2008;283:332–340. doi: 10.1074/jbc.M705313200. [DOI] [PubMed] [Google Scholar]
- 92.van Kampen E, Beaslas O, Hildebrand RB, Lammers B, Berkel TJC, Olkkonen VM, Eck M. Orp8 deficiency in bone marrow-derived cells reduces atherosclerotic lesion progression in LDL receptor knockout mice. PLoS One. 2014;9:e109024. doi: 10.1371/journal.pone.0109024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhou T, Li S, Zhong W, Vihervaara T, Béaslas O, Perttilä J, Luo W, Jiang Y, Lehto M, Olkkonen VM, Yan D. OSBP-related Protein 8 (ORP8) regulates plasma and liver tissue lipid levels and interacts with the nucleoporin Nup62. PLoS One. 2011;6:e21078. doi: 10.1371/journal.pone.0021078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Jordan SD, Krüger M, Willmes DM, Redemann N, Wunderlich TF, Brönneke HS, Merkwirth C, Kashkar H, Olkkonen VM, Böttger T, Braun T, Seibler J, Brüning JC. Obesity-induced overexpression of miRNA-143 inhibits insulin-stimulated AKT activation and impairs glucose metabolism. Nat Cell Biol. 2011;13:434–446. doi: 10.1038/ncb2211. [DOI] [PubMed] [Google Scholar]
- 95.Blumensatt M, Greulich S, de Wiza D, Mueller H, Maxhera B, Rabelink MJ, Hoeben RC, Akhyari P, Al-Hasani H, Ruige JB, Ouwens MD. Activin A impairs insulin action in cardiomyocytes via up-regulation of miR-143. Cardiovasc Res. 2013;100:201–210. doi: 10.1093/cvr/cvt173. [DOI] [PubMed] [Google Scholar]
- 96.Blumensatt M, Wronkowitz N, Wiza C, Cramer A, Mueller H, Rabelink MJ, Hoeben RC, Eckel J, Sell H, Ouwens MD. Adipocyte-derived factors impair insulin signaling in differentiated human vascular smooth muscle cells via the upregulation of miR-143. BBA Mol Basis Dis. 2014;1842:275–283. doi: 10.1016/j.bbadis.2013.12.001. [DOI] [PubMed] [Google Scholar]
- 97.Bouchard L, Faucher G, Tchernof A, Deshaies Y, Marceau S, Lescelleur O, Biron S, Bouchard C, Pérusse L, Vohl MC. Association of OSBPL11 gene polymorphisms with cardiovascular disease risk factors in obesity. Obesity. 2009;17:1466–1472. doi: 10.1038/oby.2009.109. [DOI] [PubMed] [Google Scholar]
- 98.Kara B, Köroğlu Ç, Peltonen K, Steinberg RC, Genç H, Hölttä-Vuori M, Güven A, Kanerva K, Kotil T, Solakoğlu S, Zhou Y, Olkkonen VM, Ikonen E, Laiho M, Tolun A. Severe neurodegenerative disease in brothers with homozygous mutation in POLR1A. Eur J Hum Genet. 2017;25:315–323. doi: 10.1038/ejhg.2016.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhou Y, Robciuc MR, Wabitsch M, Juuti A, Leivonen M, Ehnholm C, Yki-Järvinen H, Olkkonen VM. OSBP-related proteins (ORPs) in human adipose depots and cultured adipocytes: evidence for impacts on the adipocyte phenotype. PLoS One. 2012;7:e45352. doi: 10.1371/journal.pone.0045352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Egami H, Takiyama Y, Cano M, Houser WH, Pour PM. Establishment of hamster pancreatic ductal carcinoma cell line (PC-1) producing blood group-related antigens. Carcinogenesis. 1989;10:861–869. doi: 10.1093/carcin/10.5.861. [DOI] [PubMed] [Google Scholar]
- 101.Pour PM, Egami H, Takiyama Y. Patterns of growth and metastases of induced pancreatic cancer in relation to the prognosis and its clinical implications. Gastroenterology. 1991;100:529–536. doi: 10.1016/0016-5085(91)90226-B. [DOI] [PubMed] [Google Scholar]
- 102.Ishikawa S, Nagai Y, Masuda T, Koga Y, Nakamura T, Imamura Y, Takamori H, Hirota M, Funakosi A, Fukushima M, Baba H. The role of oxysterol binding protein-related protein 5 in pancreatic cancer. Cancer Sci. 2010;101:898–905. doi: 10.1111/j.1349-7006.2009.01475.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Espenshade PJ. SREBPs: sterol-regulated transcription factors. J Cell Sci. 2006;119:973–976. doi: 10.1242/jcs02866. [DOI] [PubMed] [Google Scholar]
- 104.Eckschlager T, Plch J, Stiborova M, Hrabeta J. Histone deacetylase inhibitors as anticancer drugs. Int J Mol Sci. 2017;18:1414. doi: 10.3390/ijms18071414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wajant H. The Fas signaling pathway: more than a paradigm. Science. 2002;296:1635–1636. doi: 10.1126/science.1071553. [DOI] [PubMed] [Google Scholar]
- 106.Higaki K, Yano H, Kojiro M. Fas antigen expression and its relationship with apoptosis in human hepatocellular carcinoma and noncancerous tissues. Am J Pathol. 1996;149:429–437. [PMC free article] [PubMed] [Google Scholar]
- 107.Zhong W, Qin S, Zhu B, Pu M, Liu F, Wang L, Ye G, Yi Q, Yan D. Oxysterol-binding protein-related protein 8 (ORP8) increases sensitivity of hepatocellular carcinoma cells to Fas-mediated apoptosis. J Biol Chem. 2015;290:8876–8887. doi: 10.1074/jbc.M114.610188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zhong W, Zhou Y, Li J, Mysore R, Luo W, Li S, Chang M-S, Olkkonen VM, Yan D. OSBP-related protein 8 (ORP8) interacts with Homo sapiens sperm associated antigen 5 (SPAG5) and mediates oxysterol interference of HepG2 cell cycle. Exp Cell Res. 2014;322:227–235. doi: 10.1016/j.yexcr.2014.01.002. [DOI] [PubMed] [Google Scholar]
- 109.Silva N, Pimenta G, Pulcheri W, Fournier M, Spector N, da Carvalho CM. Detection of messenger RNA in leukocytes or plasma of patients with chronic myeloid leukemia. Oncol Rep. 2001;8:693–696. doi: 10.3892/or.8.3.693. [DOI] [PubMed] [Google Scholar]
- 110.Vlems FA, Diepstra JHS, Cornelissen I, Ligtenberg MJL, Wobbes T, Punt CJA, van Krieken J, Ruers TJM, van Muijen GNP. Investigations for a multi-marker RT-PCR to improve sensitivity of disseminated tumor cell detection. Anticancer Res. 2003;23:179–186. [PubMed] [Google Scholar]
- 111.Wang C, JeIley L, Ridgway ND. Oxysterol-binding-protein (OSBP)-related protein 4 binds 25-hydroxycholesterol and interacts with vimentin intermediate filaments. Biochem J. 2002;361:461–472. doi: 10.1042/bj3610461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wyles JP, Perry RJ, Ridgway ND. Characterization of the sterol-binding domain of oxysterol-binding protein (OSBP)-related protein 4 reveals a novel role in vimentin organization. Exp Cell Res. 2007;313:1426–1437. doi: 10.1016/j.yexcr.2007.01.018. [DOI] [PubMed] [Google Scholar]
- 113.Charman M, Colbourne TR, Pietrangelo A, Kreplak L, Ridgway ND. Oxysterol-binding protein (OSBP)-related protein 4 (ORP4) is essential for cell proliferation and survival. J Biol Chem. 2014;289:15705–15717. doi: 10.1074/jbc.M114.571216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lowery J, Kuczmarski ER, Herrmann H, Goldman RD. Intermediate filaments play a pivotal role in regulating cell architecture and function. J Biol Chem. 2015;290:17145–17153. doi: 10.1074/jbc.R115.640359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Li J-W, Xiao Y-L, Lai C-F, Lou N, Ma H-L, Zhu B-Y, Zhong W-B, Yan D-G. Oxysterol-binding protein-related protein 4L promotes cell proliferation by sustaining intracellular Ca2+ homeostasis in cervical carcinoma cell lines. Oncotarget. 2016;7:65849–65861. doi: 10.18632/oncotarget.11671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Zhong W, Yi Q, Xu B, Li S, Wang T, Liu F, Zhu B, Hoffmann PR, Ji G, Lei P, Li G, Li J, Li J, Olkkonen VM, Yan D. ORP4L is essential for T-cell acute lymphoblastic leukemia cell survival. Nat Commun. 2016;7:12702. doi: 10.1038/ncomms12702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Udagawa O, Ito C, Ogonuki N, Sato H, Lee S, Tripvanuntakul P, Ichi I, Uchida Y, Nishimura T, Murakami M, Ogura A, Inoue T, Toshimori K, Arai H. Oligo-astheno-teratozoospermia in mice lacking ORP4, a sterol-binding protein in the OSBP-related protein family. Genes Cells. 2014;19:13–27. doi: 10.1111/gtc.12105. [DOI] [PubMed] [Google Scholar]
- 118.Miller S, Krijnse-Locker J. Modification of intracellular membrane structures for virus replication. Nat Rev Microbiol. 2008;6:363–374. doi: 10.1038/nrmicro1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Amako Y, Sarkeshik A, Hotta H, Yates J, Siddiqui A. Role of oxysterol binding protein in hepatitis C virus infection. J Virol. 2009;83:9237–9246. doi: 10.1128/JVI.00958-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Wang H, Perry JW, Lauring AS, Neddermann P, Francesco R, Tai AW. Oxysterol-binding protein is a phosphatidylinositol 4-kinase effector required for HCV replication membrane integrity and cholesterol trafficking. Gastroenterology. 2014;146:1373–1385. doi: 10.1053/j.gastro.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Arita M. Phosphatidylinositol-4 kinase III beta and oxysterol-binding protein accumulate unesterified cholesterol on poliovirus-induced membrane structure. Microbiol Immunol. 2014;58:239–256. doi: 10.1111/1348-0421.12144. [DOI] [PubMed] [Google Scholar]
- 122.Dorobantu CM, Albulescu L, Harak C, Feng Q, van Kampen M, Strating JRPM, Gorbalenya AE, Lohmann V, van der Schaar HM, van Kuppeveld FJM. Modulation of the host lipid landscape to promote RNA virus replication: the picornavirus encephalomyocarditis virus converges on the pathway used by hepatitis C virus. PLoS Pathog. 2015;11:e1005185. doi: 10.1371/journal.ppat.1005185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Albulescu L, Bigay J, Biswas B, Weber-Boyvat M, Dorobantu CM, Delang L, van der Schaar HM, Jung Y-S, Neyts J, Olkkonen VM, van Kuppeveld F, Strating J. Uncovering oxysterol-binding protein (OSBP) as a target of the anti-enteroviral compound TTP-8307. Antivir Res. 2017;140:37–44. doi: 10.1016/j.antiviral.2017.01.008. [DOI] [PubMed] [Google Scholar]
- 124.Strating J, van der Linden L, Albulescu L, Bigay J, Arita M, Delang L, Leyssen P, van der Schaar HM, Lanke K, Thibaut H, Ulferts R, Drin G, Schlinck N, Wubbolts RW, Sever N, Head SA, Liu JO, Beachy PA, De Matteis MA, Shair MD, Olkkonen VM, Neyts J, van Kuppeveld F. Itraconazole inhibits enterovirus replication by targeting the oxysterol-binding protein. Cell Rep. 2015;10:600–615. doi: 10.1016/j.celrep.2014.12.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Albulescu L, Strating J, Thibaut H, van der Linden L, Shair MD, Neyts J, van Kuppeveld F. Broad-range inhibition of enterovirus replication by OSW-1, a natural compound targeting OSBP. Antivir Res. 2015;117:110–114. doi: 10.1016/j.antiviral.2015.02.013. [DOI] [PubMed] [Google Scholar]
- 126.Park I-W, Ndjomou J, Wen Y, Liu Z, Ridgway ND, Kao CC, He JJ. Inhibition of HCV replication by oxysterol-binding protein-related protein 4 (ORP4) through interaction with HCV NS5B and alteration of lipid droplet formation. PLoS One. 2013;8:e75648. doi: 10.1371/journal.pone.0075648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Amini-Bavil-Olyaee S, Choi Y, Lee J, Shi M, Huang IC, Farzan M, Jung JU. The antiviral effector IFITM3 disrupts intracellular cholesterol homeostasis to block viral entry. Cell Host Microbe. 2013;13:452–464. doi: 10.1016/j.chom.2013.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Sierra B, Triska P, Soares P, Garcia G, Perez AB, Aguirre E, Oliveira M, Cavadas B, Regnault B, Alvarez M, Ruiz D, Samuels DC, Sakuntabhai A, Pereira L, Guzman MG. OSBPL10, RXRA and lipid metabolism confer African-ancestry protection against dengue haemorrhagic fever in admixed Cubans. PLoS Pathog. 2017;13:e1006220. doi: 10.1371/journal.ppat.1006220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Courtney SC, Di H, Stockman BM, Liu H, Scherbik SV, Brinton MA. Identification of novel host cell binding partners of Oas1b, the protein conferring resistance to flavivirus-induced disease in mice. J Virol. 2012;86:7953–7963. doi: 10.1128/JVI.00333-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Cianciola NL, Chung S, Manor D, Carlin CR. Adenovirus modulates toll-like receptor 4 signaling by reprogramming ORP1L-VAP protein contacts for cholesterol transport from endosomes to the endoplasmic reticulum. J Virol. 2017;91:e01904. doi: 10.1128/JVI.01904-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Cianciola NL, Greene DJ, Morton RE, Carlin CR. Adenovirus RIDα uncovers a novel pathway requiring ORP1L for lipid droplet formation independent of NPC1. Mol Biol Cell. 2013;24:3309–3325. doi: 10.1091/mbc.e12-10-0760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Wang P-Y, Weng J, Lee S, Anderson RGW. The N terminus controls sterol binding while the C terminus regulates the scaffolding function of OSBP. J Biol Chem. 2008;283:8034–8045. doi: 10.1074/jbc.M707631200. [DOI] [PubMed] [Google Scholar]
- 133.Moreira EF, Jaworski C, Li A, Rodriguez IR. Molecular and biochemical characterization of a novel oxysterol-binding protein (OSBP2) highly expressed in retina. J Biol Chem. 2001;276:18570–18578. doi: 10.1074/jbc.M011259200. [DOI] [PubMed] [Google Scholar]
- 134.Storey MK, Byers DM, Cook HW, Ridgway ND. Cholesterol regulates oxysterol binding protein (OSBP) phosphorylation and Golgi localization in Chinese hamster ovary cells: correlation with stimulation of sphingomyelin synthesis by 25-hydroxycholesterol. Biochem J. 1998;336:247–256. doi: 10.1042/bj3360247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Lehto M, Tienari J, Lehtonen S, Lehtonen E, Olkkonen VM. Subfamily III of mammalian oxysterol-binding protein (OSBP) homologues: the expression and intracellular localization of ORP3, ORP6, and ORP7. Cell Tissue Res. 2004;315:39–57. doi: 10.1007/s00441-003-0817-y. [DOI] [PubMed] [Google Scholar]
- 136.Lehto M, Hynynen R, Karjalainen K, Kuismanen E, Hyvärinen K, Olkkonen VM. Targeting of OSBP-related protein 3 (ORP3) to endoplasmic reticulum and plasma membrane is controlled by multiple determinants. Exp Cell Res. 2005;310:445–462. doi: 10.1016/j.yexcr.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 137.Lehto M, Mäyränpää MI, Pellinen T, Ihalmo P, Lehtonen S, Kovanen PT, Groop P-H, Ivaska J, Olkkonen VM. The R-Ras interaction partner ORP3 regulates cell adhesion. J Cell Sci. 2008;121:695–705. doi: 10.1242/jcs.016964. [DOI] [PubMed] [Google Scholar]
- 138.Vihervaara T, Käkelä R, Liebisch G, Tarasov K, Schmitz G, Olkkonen VM. Modification of the lipidome in RAW264.7 macrophage subjected to stable silencing of oxysterol-binding proteins. Biochimie. 2013;95:538–547. doi: 10.1016/j.biochi.2012.05.004. [DOI] [PubMed] [Google Scholar]
- 139.Higashimoto K, Soejima H, Yatsuki H, Joh K, Uchiyama M, Obata Y, Ono R, Wang Y, Xin Z, Zhu X, Masuko S, Ishino F, Hatada I, Jinno Y, Iwasaka T, Katsuki T, Mukai T. Characterization and imprinting status of OBPH1/Obph1 gene: implications for an extended imprinting domain in human and mouse. Genomics. 2002;80:575–584. doi: 10.1006/geno.2002.7006. [DOI] [PubMed] [Google Scholar]
- 140.Chung J, Torta F, Masai K, Lucast L, Czapla H, Tanner LB, Narayanaswamy P, Wenk MR, Nakatsu F, Camilli P. PI4P/phosphatidylserine countertransport at ORP5- and ORP8-mediated ER–plasma membrane contacts. Science. 2015;349:428–432. doi: 10.1126/science.aab1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Maeda K, Anand K, Chiapparino A, Kumar A, Poletto M, Kaksonen M, Gavin A-CC. Interactome map uncovers phosphatidylserine transport by oxysterol-binding proteins. Nature. 2013;501:257–261. doi: 10.1038/nature12430. [DOI] [PubMed] [Google Scholar]
- 142.Wyles JP, Ridgway ND. VAMP-associated protein-A regulates partitioning of oxysterol-binding protein-related protein-9 between the endoplasmic reticulum and Golgi apparatus. Exp Cell Res. 2004;297:533–547. doi: 10.1016/j.yexcr.2004.03.052. [DOI] [PubMed] [Google Scholar]
- 143.Ngo M, Ridgway ND. Oxysterol binding protein–related protein 9 (ORP9) is a cholesterol transfer protein that regulates golgi structure and function. Mol Biol Cell. 2009;20:1388–1399. doi: 10.1091/mbc.e08-09-0905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Liu X, Ridgway ND. Characterization of the sterol and phosphatidylinositol 4-phosphate binding properties of Golgi-associated OSBP-related protein 9 (ORP9) PLoS One. 2014;9:e108368. doi: 10.1371/journal.pone.0108368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Zhou Y, Li S, Mäyränpää MI, Zhong W, Bäck N, Yan D, Olkkonen VM. OSBP-related protein 11 (ORP11) dimerizes with ORP9 and localizes at the Golgi-late endosome interface. Exp Cell Res. 2010;316:3304–3316. doi: 10.1016/j.yexcr.2010.06.008. [DOI] [PubMed] [Google Scholar]
- 146.Erdem-Eraslan L, van den Bent MJ, Hoogstrate Y, Naz-Khan H, Stubbs A, van der Spek P, Böttcher R, Gao Y, de Wit M, Taal W, Oosterkamp HM, Walenkamp A, Beerepoot LV, Hanse M, Buter J, Honkoop AH, van der Holt B, Vernhout RM, Smitt P, Kros JM, French PJ. Identification of patients with recurrent glioblastoma who may benefit from combined bevacizumab and CCNU therapy: a report from the BELOB Trial. Cancer Res. 2016;76:525–534. doi: 10.1158/0008-5472.CAN-15-0776. [DOI] [PubMed] [Google Scholar]
- 147.Li H, Wang X, Fang Y, Huo Z, Lu X, Zhan X, Deng X, Peng C, Shen B (2014) Integrated expression profiles analysis reveals novel predictive biomarker in pancreatic ductal adenocarcinoma. Oncotarget 5 [DOI] [PMC free article] [PubMed]
- 148.Nagano K, Imai S, Zhao X, Yamashita T, Yoshioka Y, Abe Y, Mukai Y, Kamada H, Nakagawa S, Tsutsumi Y, Tsunoda S-I. Identification and evaluation of metastasis-related proteins, oxysterol binding protein-like 5 and calumenin, in lung tumors. Int J Oncol. 2015;47:195–203. doi: 10.3892/ijo.2015.3000. [DOI] [PubMed] [Google Scholar]
- 149.Edenberg HJ, Koller DL, Xuei X, Wetherill L, McClintick JN, Almasy L, Bierut LJ, Bucholz KK, Goate A, Aliev F, Dick D, Hesselbrock V, Hinrichs A, Kramer J, Kuperman S, Nurnberger JI, Rice JP, Schuckit MA, Taylor R, Webb TB, Tischfield JA, Porjesz B, Foroud T. Genome-wide association study of alcohol dependence implicates a region on chromosome 11. Alcohol Clin Exp Res. 2010;34:840–852. doi: 10.1111/j.1530-0277.2010.01156.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Ma J, Dempsey AA, Stamatiou D, Marshall KW, Liew C-C. Identifying leukocyte gene expression patterns associated with plasma lipid levels in human subjects. Atherosclerosis. 2007;191:63–72. doi: 10.1016/j.atherosclerosis.2006.05.032. [DOI] [PubMed] [Google Scholar]
- 151.Herold C, Hooli BV, Mullin K, Liu T, Roehr JT, Mattheisen M, Parrado AR, Bertram L, Lange C, Tanzi RE. Family-based association analyses of imputed genotypes reveal genome-wide significant association of Alzheimer’s disease with OSBPL6, PTPRG, and PDCL3. Mol Psychiatry. 2016;21:1608–1612. doi: 10.1038/mp.2015.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Teslovich TM, Musunuru K, Smith AV, Edmondson AC, Stylianou IM, Koseki M, Pirruccello JP, Ripatti S, Chasman DI, Willer CJ, Johansen CT, Fouchier SW, Isaacs A, Peloso GM, Barbalic M, Ricketts SL, Bis JC, Aulchenko YS, Thorleifsson G, Feitosa MF, Chambers J, Orho-Melander M, Melander O, Johnson T, Li X, Guo X, Li M, Shin Cho Y, Jin Go M, Jin Kim Y, Lee JY, Park T, Kim K, Sim X, Twee-Hee Ong R, Croteau-Chonka DC, Lange LA, Smith JD, Song K, Hua Zhao J, Yuan X, Luan J, Lamina C, Ziegler A, Zhang W, Zee RY, Wright AF, Witteman JC, Wilson JF, Willemsen G, Wichmann HE, Whitfield JB, Waterworth DM, Wareham NJ, Waeber G, Vollenweider P, Voight BF, Vitart V, Uitterlinden AG, Uda M, Tuomilehto J, Thompson JR, Tanaka T, Surakka I, Stringham HM, Spector TD, Soranzo N, Smit JH, Sinisalo J, Silander K, Sijbrands EJ, Scuteri A, Scott J, Schlessinger D, Sanna S, Salomaa V, Saharinen J, Sabatti C, Ruokonen A, Rudan I, Rose LM, Roberts R, Rieder M, Psaty BM, Pramstaller PP, Pichler I, Perola M, Penninx BW, Pedersen NL, Pattaro C, Parker AN, Pare G, Oostra BA, O’Donnell CJ, Nieminen MS, Nickerson DA, Montgomery GW, Meitinger T, McPherson R, McCarthy MI, et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–713. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Willer G, Schmidt CJ, Sengupta EM, Peloso S, Gustafsson GM, Kanoni S, Ganna S, Chen A, Buchkovich J, Mora ML, Beckmann S, Bragg-Gresham JS, Chang JL, Demirkan H-Y, Hertog A, Do HM, Donnelly R, Ehret LA, Esko GB, Feitosa T, Ferreira MF, Fischer T, Fontanillas K, Fraser P, Freitag RM, Gurdasani DF, Heikkilä D, Hyppönen K, Isaacs E, Jackson A, Johansson AU, Johnson A, Kaakinen T, Kettunen M, Kleber J, Li ME, Luan X, Lyytikäinen JA, Magnusson L-P, Mangino PKE, Mihailov M, Montasser E, Müller-Nurasyid ME, Nolte M, O’Connell IM, Palmer JR, Perola CD, Petersen M, Sanna A-K, Saxena S, Service R, Shah SK, Shungin S, Sidore D, Song C, Strawbridge C, Surakka RJ, Tanaka I, Teslovich T, Thorleifsson TM, den Herik G, Voight EG, Volcik BF, Waite KA, Wong LL, Wu A, Zhang Y, Absher W, Asiki D, Barroso G, Been I, Bolton LF, Bonnycastle JL, Brambilla LL, Burnett P, Cesana MS, Dimitriou G, Doney M, Döring ASF, Elliott A, Epstein P, Eyjolfsson SE, Gigante G, Goodarzi B, Grallert MO, Gravito H, Groves ML, Hallmans CJ, Hartikainen G, Hayward A-L, Hernandez C, Hicks D, Holm AA, Hung H, Illig Y-J, Jones T, Kaleebu MR, Kastelein P, Khaw JJP et al (2013) Discovery and refinement of loci associated with lipid levels. Nat Genet 45:1274–1283 [DOI] [PMC free article] [PubMed]
- 154.Guo X, Zhang L, Zhang Y, Zhang D, Qin L, Dong S, Li G. Oxysterol binding protein-related protein 8 inhibits gastric cancer growth through induction of ER stress, inhibition of Wnt signaling and activation of apoptosis. Oncol Res. 2016;25:799–808. doi: 10.3727/096504016X14783691306605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Ma L, Yang J, Runesha BH, Tanaka T, Ferrucci L, Bandinelli S, Da Y. Genome-wide association analysis of total cholesterol and high-density lipoprotein cholesterol levels using the Framingham Heart Study data. BMC Med Genet. 2010;11:1–11. doi: 10.1186/1471-2350-11-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Sohn M, Ivanova P, Brown AH, Toth DJ, Varnai P, Kim Y, Balla T. Lenz-Majewski mutations in PTDSS1 affect phosphatidylinositol 4-phosphate metabolism at ER-PM and ER-Golgi junctions. Proc Natl Acad Sci USA. 2016;113:4314–4319. doi: 10.1073/pnas.1525719113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Aziz N, Mokhtar NM, Harun R, Mollah M, Rose I, Sagap I, Tamil A, Ngah W, Jamal R. A 19-Gene expression signature as a predictor of survival in colorectal cancer. BMC Med Genet. 2016;9:58. doi: 10.1186/s12920-016-0218-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Li L, Qu G, Wang M, Huang Q, Liu Y. Association of oxysterol binding protein-related protein 9 polymorphism with cerebral infarction in Hunan Han population. Ir J Med Sci. 2014;183:439–448. doi: 10.1007/s11845-013-1035-6. [DOI] [PubMed] [Google Scholar]
- 159.Koriyama H, Nakagami H, Katsuya T, Akasaka H, Saitoh S, Shimamoto K, Ogihara T, Kaneda Y, Morishita R, Rakugi H. Variation in OSBPL10 is associated with dyslipidemia. Hypertens Res. 2010;33:511–514. doi: 10.1038/hr.2010.28. [DOI] [PubMed] [Google Scholar]
- 160.Stewart WCL, Huang Y, Greenberg DA, Vieland VJ. Next-generation linkage and association methods applied to hypertension: a multifaceted approach to the analysis of sequence data. BMC Proc. 2014;8:1–4. doi: 10.1186/1753-6561-8-S1-S111. [DOI] [PMC free article] [PubMed] [Google Scholar]