Abstract
Mitotic kinesin-5 bipolar motor proteins perform essential functions in mitotic spindle dynamics by crosslinking and sliding antiparallel microtubules (MTs) apart within the mitotic spindle. Two recent studies have indicated that single molecules of Cin8, the Saccharomyces cerevisiae kinesin-5 homolog, are minus end-directed when moving on single MTs, yet switch directionality under certain experimental conditions (Gerson-Gurwitz et al., EMBO J 30:4942–4954, 2011; Roostalu et al., Science 332:94–99, 2011). This finding was unexpected since the Cin8 catalytic motor domain is located at the N-terminus of the protein, and such kinesins have been previously thought to be exclusively plus end-directed. In addition, the essential intracellular functions of kinesin-5 motors in separating spindle poles during mitosis can only be accomplished by plus end-directed motility during antiparallel sliding of the spindle MTs. Thus, the mechanism and possible physiological role of the minus end-directed motility of kinesin-5 motors remain unclear. Experimental and theoretical studies from several laboratories in recent years have identified additional kinesin-5 motors that are bidirectional, revealed structural determinants that regulate directionality, examined the possible mechanisms involved and have proposed physiological roles for the minus end-directed motility of kinesin-5 motors. Here, we summarize our current understanding of the remarkable ability of certain kinesin-5 motors to switch directionality when moving along MTs.
Keywords: Kinesin-5, Microtubules, In-vitro motility assays, Mitosis, Bidirectionality, Control of kinesin motility
Introduction
The precise segregation of chromosomes during mitosis is essential for maintaining genetic integrity and preventing defects that can lead to chromosome instability and cancer. Lower eukaryotes such as fungi, divide by closed mitosis in which the nuclear envelope remains intact, while higher eukaryotes, such as metazoans, undergo open mitosis in which the nuclear envelope breaks down (reviewed in [1–4]). Chromosomes are segregated by the mitotic spindle, an intracellular microtubule (MT)-based bipolar structure. MTs are comprised of αβ-tubulin heterodimers which are assembled into thirteen parallel protofilaments and exhibit dynamic instability manifested in alternating growth and shrinkage phases occurring primarily at the MT plus end, terminating with a β-tubulin subunit [5, 6]. Within mitotic spindles, MTs are arranged in an antiparallel bipolar array, emanating from the centrosomes or spindle pole bodies (SPBs) in yeast, with their minus ends concentrated at the SPBs.
Three types of MTs are found in the eukaryote spindles, with each exhibiting different architecture and function (Fig. 1), as reviewed previously [7, 8]. Kinetochore MTs (kMTs) capture kinetochores (protein structures where MTs attach to chromosomes) at their plus ends and control chromosome movement within the spindle [9, 10]. Astral or cytoplasmic MTs (cMTs) are captured at specific sites on the cell cortex and facilitate spindle positioning [11–13]. Interpolar MTs (iMTs) which span the two spindle poles overlap in an antiparallel array in the middle region of the spindle, termed the midzone [14–16]. iMTs are tightly focused in parallel arrays near the poles, as recently reviewed [8].
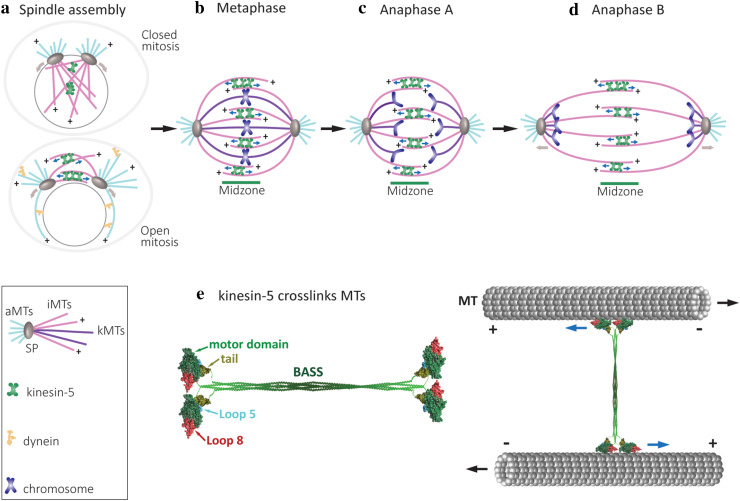
Fig. 1.
Schematic representation of the major roles of kinesin-5 motors in mitotic spindle dynamics. a Spindle-pole (SP) separation during spindle assembly in open and closed mitosis [150]. Kinesin-5 motors provide the spindle pole-separating forces by crosslinking and sliding antiparallel spindle MTs apart. The minus end-directed motor dynein (yellow shape) contributes to spindle-pole separation in cells dividing via open mitosis only [151, 152]. The direction of movement of the kinesin-5 motors and the spindle poles are indicated by the blue and brown arrows, respectively. b Metaphase, bipolar mitotic spindles with chromosomes congressed at the middle of the spindle. Three types of MTs, kinetochore MTs (kMTs, purple), astral or cytoplasmic MTs (cMTs, pink) and interpolar MTs (iMTs, light blue), are indicated by different colors. At metaphase, the chromosomes appear as pairs of sister chromatids that align in the middle of the spindle and are attached to kMTs by their kinetochores. Kinesin-5 motors crosslink antiparallel iMTs at the midzone and stabilize the spindle. In metazoans, kinesin-5 motors drive poleward flux and maintain spindle bipolarity [39, 84, 153]. c Anaphase A. Loss of cohesion between the sister chromatids marks the onset of anaphase, with sister chromatids being pulled to the opposite spindle poles. Anaphase A starts with poleward flux-based depolymerization of kMTs. d Anaphase B spindle elongation is marked by separation of the two opposing spindle poles, pulling along disjoined sister chromatids, leading to final chromosome segregation. Anaphase B results in a two- to five-fold increase in the distance between the spindle poles in both budding and fission yeast [154, 155]. Spindle elongation is mediated by cortical force generators, such as dynein motors attached to the cortex which translocate along cMTs, and kinesin-5-mediated forces produced by sliding antiparallel iMTs apart at the midzone. e Schematic presentation of a full-length kinesin-5 tetramer and its arrangement in crosslinking spindle MTs. Left: the model depicts the motor and tail domains at the end of the bipolar structure connected through the central stalk. The central stalk which mainly consists of a coiled-coil structure formed from the four helixes joining each motor domain to the corresponding tail domain, includes the bipolar-assembly (BASS) domain in the middle region, which is important for the organization of bipolar homo-tetrameric kinesin-5 complex [57, 110]. The models of Cin8 motor (aa 73–522) and tail (aa 970–1038) domains were construct by homology modelling using the Swiss Model server [152] and depicted using UCSF Chimera [155]. The motor domain is super-imposed on the cryo-electron microscopy structure of a S. pombe Cut7 motor domain-decorated MT in the AMP-PNP-bound state (PDB: 5M5I) [128]. The motor domains are represented in green, with the large loop 8 in motor domain of Cin8 (see below) in red and loop 5 in cyan; the tail domain is shown in olive green. Right: the pair of motor domains at each end interacts with the two antiparallel MTs and hence, crosslinks and slides them apart. Blue arrows indicate the direction of kinesin-5 head movement on the MTs; black arrows represent the directions of MT movement during antiparallel sliding
To properly segregate the genetic material, the mitotic spindle undergoes extensive morphological changes that are regulated and coordinated temporally and spatially in each mitotic cycle. The bipolar spindle is assembled through the separation of the two centrosomes or SPBs from their initial position close to one another to their final location, where one lies opposite the other and with interdigitating iMTs linking the two (Fig. 1a) [2, 17, 18]. This separation is primarily achieved by pushing forces applied from within the spindle, on the iMTs [19–23]. In higher eukaryotes that divide by open mitosis, additional pulling forces applied by cortex- or nuclear envelope-bound dynein motor proteins on cMTs have also been demonstrated to contribute to centrosome separation during spindle assembly (Fig. 1a) [24–29]. On the other hand, in yeast cells, the role of dynein in the initial SPB separation during spindle assembly had not been demonstrated (Fig. 1a). Following mitotic spindle assembly, the chromosomes are attached via their kinetochores to the plus end of MTs emanating from the SPBs. Properly attached chromosomes are congressed to the middle of the spindles by balanced forces applied by MTs emanating from each pole. At this stage (metaphase; Fig. 1b), the spindle assembly checkpoint is activated to prevent sister chromatid separation prior to proper attachment of all chromosomes, as reviewed elsewhere [30–33]. When all of the chromosomes are attached to kMTs from opposite poles and the spindle assembly checkpoint is inactivated, sister chromatids are separated by degradation of the cohesion complex [34–36] and move towards opposing poles via depolymerization of the kMTs at their plus ends. This process is termed as anaphase A (Fig. 1c) [37, 38]. Following anaphase A, the spindle elongates between two to five times its original length, depending on cell type, so as to further separate the two groups of chromatids. This process, termed anaphase B, is achieved primarily by pushing forces from within the spindle and by elongation/polymerization of iMTs at their plus ends (Fig. 1d) (reviewed in [39, 40]).
The mitotic kinesin-5 motors
One of the factors that govern the morphological changes mediated by the mitotic spindle is molecular motors that use ATP hydrolysis to move along the MT tracks, for review, see [41–47]. Among the mitotic motor proteins, members of the conserved kinesin-5 family, previously termed BimC proteins, have been shown to perform essential roles in the mitotic spindle dynamics of eukaryotic cells [20, 23, 48–54]. Kinesin-5 motors are homo-tetrameric proteins with two pairs of catalytic motor domains located at opposite sides of a central minifilament in the active motor complex [55–57]. This unique architecture enables kinesin-5 motors to crosslink and slide antiparallel spindle MTs apart by their plus end-directed motility on both of the MTs they crosslink (Fig. 1e) [58, 59], thus allowing them to perform their conserved functions in spindle assembly in fungi and higher eukaryotes [20, 21, 60–62], and maintenance of the bipolar spindle structure [54]. In some organisms, such as Saccharomyces cerevisiae, and mouse and Drosophila melanogaster embryos, kinesin-5 motors were shown to play a role in facilitating spindle elongation during anaphase B, likely by sliding apart the antiparallel MTs at the midzone [63–68]. However, in other cell types, such as in Caenorhabditis elegans and porcine kidney epithelial cells, kinesin-5 motors were shown to act as a “brake” that restricts the rate of spindle elongation during anaphase B [69, 70]. It is not clear while acting as a brake if kinesin-5 is immobile and only crosslink spindle MTs or whether they perform slow antiparallel MT sliding. Examination of spindle elongation rates in cells expressing kinesin-5 mutants impaired in their ATPase activity [71] should shed light on this question.
In addition to the well-established functions of kinesin-5 motors in mitotic spindle dynamics that rely on their ability to slide antiparallel iMTs apart (Fig. 1), other intracellular functions have been described for kinesin-5 motors in different organisms. For instance, kinesin-5 is expressed in fully differentiated neurons [72]. Inhibition of kinesin-5 function in neuronal cells facilitates the growth of axons and dendrites [73–77], suggesting that kinesin-5 motor activity in these cells antagonizes the forces of other motors and provides a brake on the MT bundle mobility required for axon and dendrite growth [74]. Kinesin-5 motors have also been demonstrated to affect MT dynamics. Reports regarding this effect are, however, contradictory. On the one hand, kinesin-5 has been shown to stabilize MTs in vitro [78] and in vivo [79], likely due to the motors pausing at the MT plus ends and enhancing polymerization by stabilizing longitudinal tubulin–tubulin interactions [78]. On the other hand, in vivo evidence has demonstrated that the S. cerevisiae kinesin-5 Cin8 is a MT destabilizer [80, 81]. The mechanism of this effect is, however, not clear. In higher eukaryotic cells, kinesin-5 have been shown to affect the poleward turnover of tubulin (poleward flux) in kMTs and iMTs [82], thus contributing to the maintenance of spindle length, chromosome congression, and separation [67, 83, 84]. Poleward flux is not observed in fungal S. cerevisiae and Schizosaccharomyces pombe cells [85, 86], however, kinesin-5 motors have been shown to bind to kinetochores, focus kinetochore clusters and limit the length of the kMTs in S. cerevisiae [81, 87–89]. Finally, the S. cerevisiae kinesin-5 Kip1 is required for the segregation of the 2-μm plasmid [90–92], the function which can be related to the minus end-directed motility of Kip1 [79] (discussed below).
Kinesin motor proteins are defined by their conserved catalytic motor domains that contain ATP- and MT-binding sites (reviewed in [47, 93–97]. The motor domain is followed by a flexible 14–18 residue-long neck linker that contains family-specific features and is believed to interact directly with the catalytic domain and influence processivity and directionality [53, 98–103]. The subsequent stalk and tail domains are important for interactions with other subunits of the holoenzyme or with cargo molecules [104, 105]. The majority of the kinesin motors carry their catalytic domain at the N-terminus and have been shown to move towards the plus end of the MTs. Until recently [106, 107], the only exception to this rule were members of the kinesin-14 sub-family that carry their catalytic motor domains at the C-terminus and are minus end-directed, as reviewed elsewhere [108, 109].
Kinesin-5 motors are structurally adapted to mediate antiparallel MT sliding to perform their unique mitotic functions. These motors are unique in that they function as homotetramers, with pairs of catalytic motor domains located on opposite sides of a 60-nm-long rod-like minifilament structure, forming an elongated bipolar structure in the dumbbell-shaped molecule [55–57, 110] (Fig. 1e). This structure is believed to be essential for crosslinking and the sliding apart of two antiparallel MTs during mitosis [58, 59], as the monomeric and dimeric forms are non-functional in vivo [55, 111]. In addition, it had previously been demonstrated that tetrameric kinesin-5 chimeras containing the catalytic domains of kinesin-1 or chromokinesin exhibited MT crosslinking activity but were non-functional in spindle dynamics [112], indicating that the tetrameric structure is essential but not sufficient for kinesin-5 mitotic functions in spindle dynamics. The kinesin-5 stalk contains four regions of heptad repeat sequences that form an α-helical coiled-coil. The stalk region contains structural elements that are responsible for tetramerization of the kinesin-5 motors. Deletion studies on the S. cerevisiae kinesin-5 Cin8 and phylogenetic comparisons with other kinesin-5 proteins suggest that the coiled-coil region of 100–200 amino acids located immediately after the neck region is essential for self-interaction and sufficient for dimer formation [111]. After this coil, all kinesin-5 homologues exhibit an extended region of high and moderate coiled-coil probability. Besides coil 1, most kinesin-5 homologues present a 30–60 residue-long stretch at the end of the stalks that is essential for tetramerization. In two separate studies, Hildebrandt et al. and Tao et al. found that the central bipolar-assembly (BASS) domain that spans some 200 residues in the central part of the stalk is essential for kinesin-5 activity and cell viability [111, 113], indicating the importance of this domain in the organization of the bipolar homotetrameric kinesin-5 complex. In recent studies, this BASS domain was shown to form a bipolar minifilament, with deletion of this bipolar minifilament resulting in monomeric kinesin-5 proteins [57, 110]. Scholey et al. further elucidated the crystal structure of the D. melanogaster kinesin-5 Klp61F BASS domain and found that it consists of two antiparallel coiled-coils at its center, stabilized by alternating hydrophobic and ionic four-helical interfaces. The helixes emerge from the central part towards the N-terminal, where they bend, swap partners and form parallel coiled-coils offset by 90°. Based on this structure, these authors proposed that the central bass domain plays a role in transmitting forces between motors situated at the opposite ends of the molecule [57].
Bidirectional motility of kinesin-5 motors
The sliding apart of antiparallel MTs at the spindle midzone, and hence the separating of spindle poles by kinesin-5 motors, can only be accomplished by the plus end-directed motility of the catalytic motor domains in the homotetrameric kinesin-5 motor complex. Indeed, in vitro plus end-directed motility is a well-established characteristic of a number of kinesin-5 motors [58, 114–117]. Surprisingly, two independent studies have reported that the S. cerevisiae kinesin-5 Cin8 was minus end-directed, when moving as a single molecule on a single MT in high ionic strength conditions, and changed directionality in multi-motor MT gliding and antiparallel MT sliding assays (Fig. 2) [106, 107], as well as in low ionic strength conditions [106]. These reports broke the accepted dogma regarding kinesin directionality on two fronts. First, Cin8 was the first kinesin motor with an N-terminal motor domain that was reported to move processively towards the MT minus end. Second, kinesin motors were believed to be exclusively either plus or minus end-directed [108, 109], whereas Cin8 was reported to change directionality depending on the experimental conditions (Fig. 2) [106, 107, 118]. Following the first reports on Cin8 bidirectionality, the second S. cerevisiae kinesin-5 Kip1 and S. pombe Cut7 kinesin-5 homologs were also reported to be bidirectional and exhibit switchable directionality under certain conditions [79, 119]. Interestingly, S. pombe Cut7 was reported to be minus end-directed in both single molecule and multi-motor MT gliding assays [119]. The fact that bidirectional motility is a characteristic of several kinesin-5 motors suggests that such behavior is important for their in vivo functions.
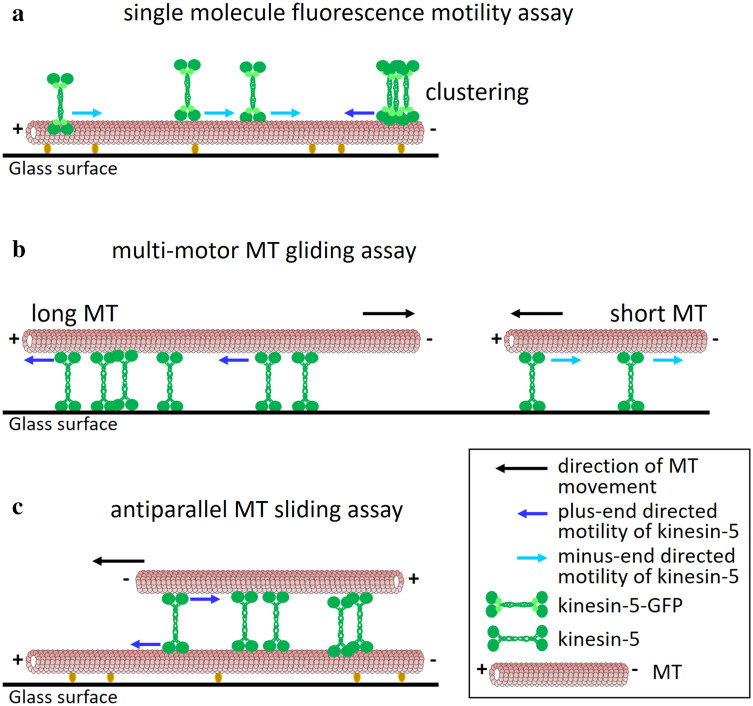
Fig. 2.
Different directionalities of kinesin-5 motors in the various types of motility assays. a Single molecule fluorescence motility assay. Fluorescently labeled MTs are immobilized on a glass surface and fluorescently labeled kinesin-5 motors [usually with green fluorescent protein (GFP)] move on the immobilized MTs, likely by interactions of one of the two pairs of catalytic domains with the immobilized MT. In such an assay, Cin8, Kip1 (in high ionic strength conditions) and Cut7 were shown to be minus end-directed [79, 106, 107, 119]. Accumulation of Cin8 in clusters on a single MT was shown to reverse directionality to plus end-directed motility [120]. b Multi-motor MT gliding assay. Motor proteins are immobilized to a glass surface and fluorescently labeled MTs undergo kinesin-5-driven motility. The directionality of the MTs is opposite to that of the immobilized kinesin-5 motors. When using long MTs in such an assay, Cin8 and Kip1 were shown to be plus end-directed [80, 106, 107, 115], while Cut7 was shown to be minus end-directed [119]. With shorter MTs, Cin8 exhibited minus end-directed motility [107]. c Antiparallel MT sliding assay. One set of MTs is immobilized to the surface while the other set, sometimes differently labeled, is free to float in solution. The free MTs undergo antiparallel sliding on top of the immobilized MTs, mediated by kinesin-5 motors that crosslink and walk on both MTs [58]. In such an assay, Cin8 was shown to undergo plus end-directed motility [106, 107, 120], similarly to kinesin-5 motors in higher eukaryotes [58, 116]
One of the conditions under which the S. cerevisiae kinesin-5 Cin8 was shown to switch from fast minus end- to slow plus end-directed motility is when the motor transitions from a state of being attached to a single MT to a state where it crosslinks two antiparallel MTs during Cin8-mediated antiparallel MT sliding (Fig. 2c) [106, 107, 120]. Because of the polarity of MTs in the spindle apparatus (Fig. 1), such a switch is necessary to produce the outward-directed force that separates the spindle poles during mitosis, one of the major functions of kinesin-5 motors [96, 97]. This switch, occurring upon the binding of the two pairs of motor domains to antiparallel MTs, is likely to be transduced by the stalk and BASS domain between the two pairs [57], and resembles the activation of Xenopus laevis kinesin-5 Eg5, which under high ionic strength conditions switches from diffusive to processive plus end-directed motility upon binding to the two MTs the protein crosslinks [116]. Interestingly, Aspergillus nidulans KlpA, a member of the minus end-directed kinesin-14 sub-family, was recently shown to be plus end-directed on the single molecule level but switched directionality in multi-motor MT gliding and antiparallel MT sliding assays [121], similar to S. cerevisiae Cin8 and Kip1 [79, 106, 107]. This indicates that context-dependent directionality switching is not unique to kinesin-5 motors and may reflect an adaptation of different kinesin sub-families to their physiological functions.
Regulation of the bidirectionality of kinesin-5 motors
A recent study demonstrated that the forces produced by Cin8 in the plus-end and minus-end directions are similar [122], suggesting a similar mode of motility in the two directions. However, the molecular mechanism and regulation of bidirectionality of Cin8, Kip1 and Cut7 remain largely unknown. Several structural elements, discussed below, have been considered in terms of regulating the directionality of these motors (Fig. 3).
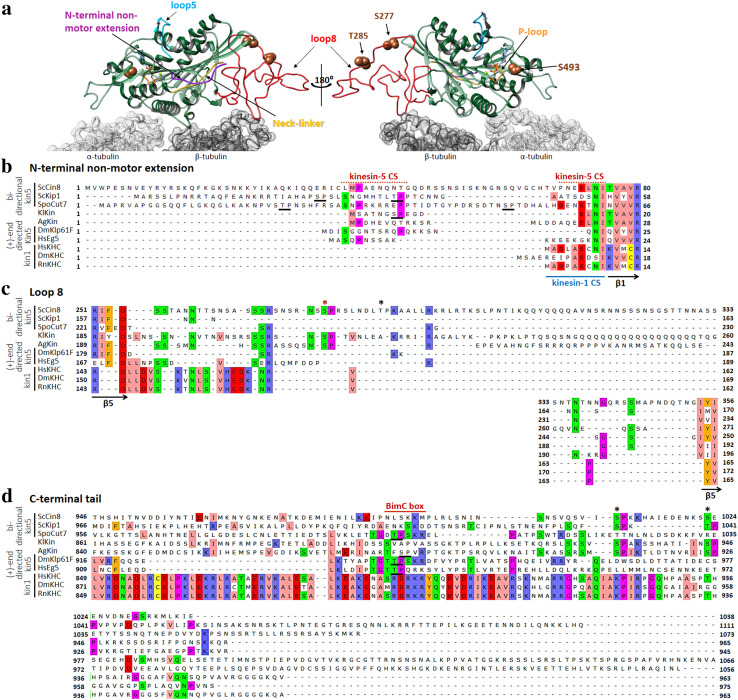
Fig. 3.
Structural features that affect the directionality of kinesin-5 motors. a A Cin8 motor bound to αβ-tubulin dimer. The model of Cin8 motor domain is super-imposed on the cryo-electron microscopy structure of a S. pombe Cut7 motor domain-decorated MT in the AMP-PNP-bound state (PDB: 5M5I) [128]. Except for the non-motor N-terminal extension, the structure of Cut7 was omitted from the overlay for clarity. α- and β-tubulin subunits are indicated in light and dark gray, respectively. The structural elements are highlighted in different colors, with the Cut7N-terminal extension in purple, the ATP-binding p-loop in orange, loop 5 in cyan, Cin8-specific loop 8 in red, the neck linker in yellow and the three Cdk1 phosphorylation site S277, T285 and S493 as brown spheres. The Cin8 model was created using the homology modelling Swiss-Model server [152], while the superimposition and molecular graphics were performed with UCSF Chimera [155]. b–d Amino acid sequence alignments of seven kinesin-5 homologs (top seven sequences) and three kinesin-1 homologs (bottom three sequences). Organisms are indicated on the left: Sc: Saccharomyces cerevisiae, Spo: Schizosaccharomyces pombe, Kl: Kluyveromyces lactis, Ag: Ashbya gossypii, Dm: Drosophila melanogaster, Hs: Homo sapiens, Rn: Rattus norvegicus. Numbers represent amino acid positions in the sequence of each homolog. The sequences were aligned using Unipro UGENE software and adjusted to align the consensus secondary structure elements depicted at the bottom of each alignment. The threshold for consensus residue highlighting was fixed at 30%. The directionalities of kinesin-5 proteins are represented on the left, and although the directionalities of KlKin and AgKin are unknown, they are included here as they contain the loop 8 insertion, similar to ScCin8. The UniProt ID of the sequences are: ScCin8–P27895, ScKip1–P28742, SpoCut7–P24339, KlKin–Q6CSH2, AgKin–Q8J1G7, DmKlp61F–P46863, HsEg5–P52732, HsKHC–P33176, DmKHC–P17210, RnKHC–Q2PQA9. b N-terminal non-motor extension. The first β-strand of the catalytic domain (β1) is indicated on the bottom. The nine residues that can form a β-sheet while interacting with the docked neck linker and form a neck cover strand (CS) in kinesin-1 motors [124] are indicated as kinesin-1 CS. The kinesin-5-specific N-terminal region that was shown to function as a CS [96, 99] is indicated above as kinesin-5 CS. The isoelectric pH values for the N-terminal extensions were calculated as follows: ScCin8—9.47, ScKip1–11.42, SpoCut7—9.39, KlKin—3.57, AgKin—4.58, DmKlp61F—9.99, HsEg5—9.82, HsKHC—3.67, DmKHC—4.00, RnKHC—3.67. The unique Cdk1 sites present in ScKip1, SpoCut7 and KlKin are underlined. The roles of these sites are unknown. The Cin8 sequence starts from methionine at position − 39, relative to the first methionine indicated in electronic databases, since previous publications reported that Cin8 is expressed from – 39M [106, 107, 111, 133], indicated here as the first methionine, M1. c Loop 8, with red and black asterisks indicating the conserved and non-conserved Cdk1 phosphorylation sites, respectively. The bordering β-strands are indicated at the bottom. d The C-terminal tail domain. The BimC boxes containing the Cdk1 phosphorylation site are underlined. Other Cdk1 sites are indicated by asterisks
N-terminal non-motor extension
Previous studies have indicated that sequences at the N-terminal non-motor region affect kinesin-1 and kinesin-5 motor function [99, 123–126]. Molecular dynamics simulations demonstrated a nine residue-long N-terminal region in kinesin-1 motors (termed the neck linker cover strand (CS), or β0) responsible for conformational change of the neck linker that is essential for force generation. Upon ATP binding, this region contributes to the formation of a β-sheet with the neck linker, the structure that was thought to be involved in stabilization of the motor domain-docked confirmation of the neck linker, important for the plus end-directed motility [123–126]. The N-terminal non-motor region of kinesin-5 motors contains longer extensions, compared to kinesin-1 (Fig. 3b). However, cryo-electron microscopy and kinetic experiments have indicated that the longer N-terminal region of the plus-end-directed kinesin-5 Eg5 is docked onto the motor domain with the neck linker under several nucleotide-based conditions [99], suggesting that although their lengths are different, the neck linker CS performs similar functions in stabilizing docked conformations of the neck linker of kinesin-1 and kinesin-5 motors. Moreover, a recent study had demonstrated that the neck linker of the plus-end-directed kinesin-5 Eg5 assumes different conformations, compared to kinesin-1, in some nucleotide-bound states [53]. Stabilization of these conformations in the Eg5 motor may occur through sequences in the neck linker CS, in a manner specific to plus-end-directed kinesin-5 motors (Fig. 3b). Sequence alignment reveals that the bidirectional kinesin-5 motors contain longer and divergent non-motor N-terminal extensions, compared to kinesin-1 and the plus-end-directed kinesin-5 motors (Fig. 3b). These extensions are present within and upstream to the neck linker CSs of kinesin-1 and plus-end-directed kinesin-5s (Fig. 3b). It is tempting to speculate that these additional sequences stabilize the conformation(s) of the neck linker (or other structural elements within the motor domain) compatible with bidirectional motility. Mutagenesis studies of the N-terminal region of bidirectional kinesin-5s will shed light on the function of this region in the bidirectional motility of kinesin-5 motors.
To date, the function of the non-motor N-terminal region has been studied in the bidirectional kinesin-5 Cut7 only. It was shown that partial or complete deletion of the N-terminal non-motor extension of Cut7 reduced its MT binding [127], however, these deletions had minor effects on minus end-directed motility in single molecule and multi-motor MT gliding motility assays [119, 127]. Interestingly, partial but not complete deletion of this region resulted in a non-functional kinesin-5 motor in vivo [127]. In support of this notion, another recent report indicated that the N-terminal extension is not involved in the directional stepping of Cut7 but is directly involved in binding MTs and increases drag, thereby slowing velocity [128]. However, in a cryo-EM reconstruction of the MT-bound Cut7 motor domain, the N-terminal extension was found to be only partially proximal to the neck linker [128]. Therefore, it remains unclear if the N-terminal extension of bidirectional kinesin-5 motors interacts with the neck linker in a similar manner as was suggested for other kinesin motors and whether it affects directionality.
Loop 8 and phosphorylation in the catalytic domain
Loop 8 of the kinesin motor catalytic motor domain [129] is considered to be part of the MT-binding domain as it faces the MT lattice in the MT-bound state of the kinesin motor domain [130, 131]. In the dimeric structure of kinesin-1, loop 8 of one motor domain was shown to interact via a salt-bridge with loop 10 of the other motor domain. This interaction was suggested to serve as an inter-subunit switch sensitive to the bound nucleotide state during the catalytic cycle of kinesin motors [131]. The amino acid sequence of S. cerevisiae Cin8 contains a large 99 amino acid insert in loop 8 (Fig. 3a), which is the largest insert among kinesin motors and is not essential for Cin8 function in vivo [20]. However, loop 8 of Cin8 was found to be important for the regulation of directionality, since replacement of this non-conserved large insert with the short loop 8 of the homologous Kip1 induced bias to the minus end-directionality of Cin8 in vitro [106]. Moreover, it has been recently demonstrated that the motor domain of Cin8 exhibits noncanonical binding to MTs with about four motor domains bound per αβ-tubulin dimer [132]. Deletion of the large loop 8 of Cin8 abolished this effect, reverting Cin8 to canonical binding of one motor domain to an αβ-tubulin dimer [132], indicating the importance of the large loop 8 of Cin8 in regulating its activity and binding to MTs.
Loop 8 of Cin8 contains two (S277 and T285) of the three Cdk1 phosphorylation sites in the motor domain of Cin8 that were implicated in regulating the in vivo functions of the protein [133–135]. Interestingly, the S277 site is conserved among fungal kinesin-5 homologs that contain inserts in loop 8, whereas the T285 site is unique to Cin8 (Fig. 3c). The third site, S493, is highly conserved among kinesin-5 motors and is located near the kinesin motor ATP-binding P-loop (Fig. 3a) [136]. It has been demonstrated that phospho-mimic mutations at these three sites promote minus end-directed motility of Cin8 in vitro [136], indicative of the directionality of kinesin-5 motors being, at least in part, regulated by phosphorylation of the motor domain. The fact that a Cin8 mutant lacking loop 8 behaves similarly to the phospho-mimic mutant indicates that loop 8 contains a molecular switch, possibly in the form of phosphorylation, which regulates the directionality of Cin8. High ionic strength conditions that induce minus end-directed motility of Cin8 [106, 118] may mimic the effects of phosphorylation by affecting the binding of Cin8 to MTs or other proteins that activate the molecular switch regulating the directionality.
The C-terminal tail domain
The tail domain of kinesin motors has been shown to modulate kinesin activity. For example, the tail domain of kinesin-1 has been reported to inhibit its motility [137]. The mechanism of this inhibition likely involves crosslinking of the two catalytic domains by the tail in the active kinesin dimer [138]. Direct evidence for interaction between the motor and tail domains in kinesin-5 motors has yet to be demonstrated. However, the tail domain of kinesin-5 motors from X. laevis and D. melanogaster has been shown to be involved in MT crosslinking [59, 139], suggesting direct interaction with MTs. Likewise, interaction of the tail domain with MTs has not been demonstrated for fungal kinesin-5 motors, however, the motor domain of S. cerevisiae Cin8 was shown to be essential for its intracellular functions [111], indicating that the tail domain alone is insufficient to produce MT crosslinking activity as part of the intracellular functions of Cin8. Kinesin-5 homologs contain a Cdk1 (p34/Cdc2) kinase phosphorylation site in the tail domain (Fig. 3d). In kinesin-5 motors of higher eukaryotes, the Cdk1 site is located within a conserved “BimC box” reportedly phosphorylated during mitosis [61]. Phospho-deficient mutants of human, D. melanogaster and X. laevis kinesin-5 homologs at this site did not localize to the spindle apparatus [61, 140, 141]. The role of Cdk1 phosphorylation in the tail is less clear in fungi as mutations at this site in S. cerevisiae and S. pombe kinesin-5 homologs produced no obvious phenotype [133, 142].
The influence of the tail domain of kinesin-5 motors on directionality has been addressed in several studies. In a study focused on the tail domain of S. cerevisiae Cin8 and involving a chimera of the kinesin-5 Cin8 and kinesin-1 from D. melanogaster, Duselder et al. found that although the bidirectionality of Cin8 was the inherent characteristic of the motor domain, control over bidirectionality was lost in Cin8 lacking the tail domain (Cin8Δtail) [143]. The motility of single Cin8Δtail molecules in high ionic strength conditions was slow, processive and bidirectional, in stark contrast to the behavior of wild-type Cin8. Moreover, Cin8Δtail was not found to properly cross-link MTs in vitro. In vivo, Cin8Δtail was unable to support viability of the cell when present as the sole kinesin-5 motor [143]. On the other hand, deletion of the tail domain of S. pombe Cut7 did not seem to affect minus end-directionality in a multi-motor MT gliding assay, although single molecule motility of the tailless Cut7 variant was not examined [119]. Since in the tetrameric complex, the C-terminal tail domains of kinesin-5 motors are likely to be found in close proximity to their N-terminal motor domains [57, 110], regulation of kinesin-5 motor motility through direct interactions between motor and tail domains had been suggested in a number of studies [19, 57, 110]. It is, therefore, tempting to speculate that specific interactions between the tails and motor domains regulate the directionality of bidirectional kinesin-5 motors. In support of this notion, it had been demonstrated that a dimeric version of Cin8 is bidirectional [143], while a truncated monomeric versions of Cut7 that only contain the motor domain and neck linker exclusively produce plus end-directed motility [128]. These reports indicate that the tetrameric kinesin-5 complex, which likely includes motor-tail interactions, is essential for the processive minus end-directed motility.
Inter-molecular interactions affect the directionality of bidirectional kinesin-5 motors
Several lines of evidence indicate that inter-molecular interactions between bidirectional kinesin-5 motors or between these motors and non-motor proteins affect their directionality. First, it has been suggested that in multi-motor MT gliding and antiparallel MT-sliding assays, the switch from fast minus end- to slow plus end-directed motility is induced by a coupling of S. cerevisiae Cin8 motors through the MT with which they interact [107]. This model was mainly based on the finding that in the multi-motor gliding assay, the switch in directionality was dependent on MT length, such that the longer the MT, the more frequent was the occurrence of plus end-directed motility, indicating that the number of motors interacting with the same MT determines directionality (Figs. 2b, 4a) [107]. This model was recently supported by a theoretical study [144]. The generality of this mechanism, however, remains unclear since S. pombe Cut7 was reported to be minus end-directed in both single molecule and multi-motor MT gliding assays [119].
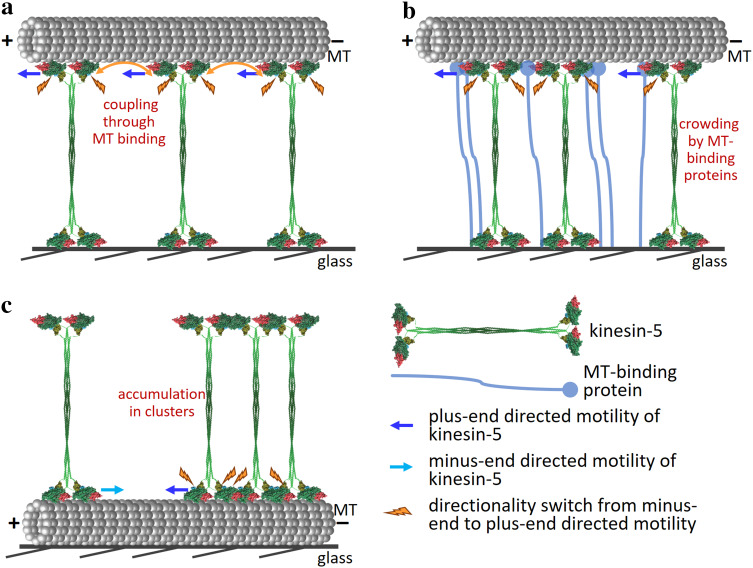
Fig. 4.
Suggested mechanism of directionality switch in yeast kinesin-5 motors. a Coupling through the MT. Directionality is altered as a result of coupling between motors through the MT with which they interact. The number of mechanically coupled motors interacting with the same MT determines motor directionality, such that the larger is the number, the higher is the probability for the motors to move in the plus-end direction [107, 122]. For a constant number of motors, the directionality switch is dependent on MT length. Orange arrows indicate coupling between motor domains through the MT, promoting plus end directional stepping. b Crowding by MT-binding (motor and non-motor) proteins. Bidirectionality depends on the extent of motor crowding on the microtubule lattice and is mediated through steric blocking or a proximity-sensing mechanism. The crowding of kinesin-5 motors by MT-binding proteins reverse the stepping direction towards the plus-end of the MTs [128]. This mechanism is independent of MT length. c Accumulation in clusters. Accumulation of Cin8 in clusters induces directionality switch from fast minus-end-directed to slow plus-end-directed motility [120]. The switch occurs due to specific inter-molecular Interactions between Cin8 tetramers in a cluster [120]. Alternatively, accumulation of Cin8 in clusters can produce crowding in the vicinity of the motor domains and induce a directionality switch similarly to the mechanism depicted in b. This mechanism is expected to be independent of MT length
Second, it has been recently proposed that the minus end-directed stepping action of Cut7 is selectively inhibited by collisions with neighboring proteins under crowded conditions, whereas its plus end-directed motility, being less “space-hungry”, is not [128]. Using in vitro MT gliding assays and total internal reflection fluorescence (TIRF) microscopy, [128] proposed that the proximity sensing mechanism regulates directional switching in Cut-7, which depends on the local density of motors. They demonstrated that crowding of Cut7 by motor and non-motor proteins, such as the dynein MT-binding domain and the kinesin-14 Klp2, can drive directional switching from minus end- to plus end-directed motility and suggested that such crowding acts in a steric blocking model (Fig. 4b). This model of directionality switch significantly differs from motor-coupling-based models [107, 144] since it depends on the interaction of kinesin-5 motors with motor and non-motor proteins bound to the MT and is independent of MT length. Finally, it has been recently demonstrated using a single molecule TIRF-based motility assay that in high ionic strength conditions, accumulation of Cin8 into clusters on MTs slowed motility in the minus end direction and induced a switch from minus end- to plus end-directed motility (Figs. 2a, 4c) [120]. While this mechanism of directionality switch should be independent of MT length, the point should be tested experimentally. This study further proposed that since the ability to move in both directions is an intrinsic property of Cin8 tetramers [106, 118, 143], specific inter-molecular interactions between Cin8 tetramers in a cluster control its directionality [120]. The recent report indicating that the Cin8 catalytic domain binds MTs in a super-stoichiometric ratio of approximately four motors per αβ-tubulin dimer within the MT lattice [132] supports the notion that specific interactions between kinesin-5 motors in a cluster can affect their motor activity. Such interactions could be mediated by the large loop 8 of Cin8 which was shown to induce non-canonical binding to MTs [132] or by the tail domain, deletion of which was shown to abolish the minus-end directionality preference of Cin8 under high ionic strength conditions [143].
The above summarized evidence demonstrates that the directionality of bidirectional kinesin-5 motors is sensitive to the environment. Although the mechanism of such collective control of directionality remains unclear, two alternative models can be proposed to explain this phenomenon (Fig. 4). One possibility is that directionality is controlled by mechanical communication/coupling between motors through the MT [107, 122, 144] (Fig. 4a), while the other possibility is that directionality regulation occurs through direct interaction of the catalytic domain of the kinesin-5 motor with other motor and non-motor proteins bound to the same MT [120, 128] (Fig. 4b, c). Interestingly, a recent study demonstrated that when found in high motor density on MTs, Cin8-induced MT gliding was plus-end-directed. In contrast, MTs moved in both directions when found at low density [122]. This finding is consistent with both models since higher motor density could potentially increase the coupling forces between motors through the MT and/or increase the chance of motor–motor interactions, thereby inducing a crowding/clustering effect. One of the predictions of the coupling-through-MT model is that application of an external load would affect the directionality of kinesin-5-driven MT motility. However, this recent study [122] had also demonstrated that under low motor density conditions, when Cin8-driven MT motility occurred in both the plus- and minus-end directions, application of force onto the moving MTs using an optical trap did not change MT directionality, suggesting that such directionality is load-independent. Work in coming years will likely help to distinguish between these two models and shed light on this remarkable environment-sensitive directionality switch.
A possible physiological role for the switchable directionality of kinesin-5 motors
The fact that the three bidirectional kinesin-5 motors reported thus far are expressed in fungal cells raises the possibility that bidirectional motility is required for the physiological function of kinesin-5 motors in these cells. A recent report indicated that before spindle assembly in S. cerevisiae cells, Cin8 accumulates near the spindle poles, at the minus end of nuclear MTs, while in assembled spindles, Cin8 is also distributed between the poles, on overlapping spindle MTs [120]. Similar localization patterns were reported for Cut7 in S. pombe [50]. Based on these findings, and the report that accumulation of Cin8 in clusters promotes antiparallel MT capture and sliding [120], it has been recently proposed that prior to spindle assembly, fungal kinesin-5 accumulates in clusters near the SPBs via their minus end-directed motility on single MTs. These kinesin-5 clusters capture MTs emanating from neighboring SPBs and promote their plus end-directed antiparallel sliding (Fig. 5) [120]. Alternatively, accumulation of kinesin-5 in clusters near SPBs can capture MTs from neighboring SPBs and function as cross-linkers, with the polymerizing MTs providing the force necessary for SPB separation [22]. This model implies that accumulation of kinesin-5 motors near SPBs at this initial stage is required to maximize the crosslinking of nuclear MTs emanating from the neighboring SPBs, which is the key step for kinesin-5-mediated SPB separation and spindle formation. The directionality switch and formation of clusters at the minus ends of nuclear MTs, near the SPB [120], can be also caused by motor crowding on the MT lattice and the proximity-sensing mechanism [128]. Therefore, it is possible that during this stage, other proteins near SPBs are also involved in inducing the directionality switching of kinesin-5 motors.
Fig. 5.
Possible roles for the bidirectional motility of some kinesin-5 motors in spindle assembly. Adapted from Shapira et al. [120]. During the initial stages of spindle assembly, the two SPBs are closely located and there is a small overlap of MTs emanating from the different poles. a Movement of kinesin-5 motors towards the SPBs due to their minus end-directed motility induces their accumulation in clusters near the SPBs at the minus ends of the MTs. b Clustering of kinesin-5 motors near the SPBs mediates the capture of MTs emanating from the adjoining SPB, thus maximizing the crosslinking of these MTs at the beginning of spindle assembly. c The capturing of MTs from neighboring poles and their antiparallel sliding creates the initial separation of the two SPBs. This separation, in turn, increases the overlap between interpolar MTs from neighboring poles, creating additional sites for Cin8 to bind, crosslink and slide the antiparallel MTs apart. d This provides additional SPB-separating force until the spindle is formed
The minus end-directed motility of kinesin-5 motors in yeast may represent an evolutionary adaptation allowing these proteins to perform their essential functions in spindle assembly. In higher eukaryotes, nuclear envelope breakdown occurs during open mitosis, with cytoplasmic dynein having been shown to be involved in the initial spindle-pole separation and, in cooperation with kinesin-5, in spindle assembly [145, 146]. In contrast, in S. cerevisiae and S. pombe cells which divide via closed mitosis without nuclear envelope disassembly, dynein does not play a role in mitotic spindle assembly [66, 147, 148]. Therefore, in yeast cells, with no external pulling force provided by dynein, the minus end-directed motility of kinesin-5 motors is required for the initial SPB separation needed for spindle assembly. Indeed, a recently proposed computational model suggests that the bidirectionality of kinesin-5 motors is essential for the generation and stability of spindle bipolarity, as well as for proper localization of the spindle [149], supporting the notion that the bidirectionality of fungal kinesin-5 motors is essential for their intracellular function in spindle assembly.
While the molecular mechanisms of the remarkable bidirectionality of certain kinesin-5 motors remain unclear, work conducted in recent years indicates that this bidirectionality is more common that was initially appreciated. It is also evident that the environment in the vicinity of the motors, such as being part of a cluster, other motor proteins coupled through the same MT or non-motor proteins, affects the directionality of these motors. Work in coming years should provide further insight into the physiological roles of this behavior in different organisms and shed light on the mechanism and regulation of the bidirectional motility of individual motors and their activity in ensembles.
Acknowledgements
This work was supported in part by the Israel Science Foundation (ISF) (Grant 165/13 to L.G.), the National Science Foundation (NSF-1615991), United States—Israel Binational Science Foundation (BSF Grant BSF-2015851 to L.G. and J.A.-B.) and the National Institutes of Health (Grant NIH-R01-GM11283 to J.A.-B.). This manuscript has been deposited in PMC for release after 12 months.
References
- 1.Boettcher B, Barral Y. The cell biology of open and closed mitosis. Nucleus. 2013;4:160–165. doi: 10.4161/nucl.24676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lim HH, Zhang T, Surana U. Regulation of centrosome separation in yeast and vertebrates: common threads. Trends Cell Biol. 2009;19:325–333. doi: 10.1016/j.tcb.2009.03.008. [DOI] [PubMed] [Google Scholar]
- 3.Sazer S, Lynch M, Needleman D. Deciphering the evolutionary history of open and closed mitosis. Curr Biol. 2014;24:R1099–R1103. doi: 10.1016/j.cub.2014.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zheng Y. A membranous spindle matrix orchestrates cell division. Nat Rev Mol Cell Biol. 2010;11:529–535. doi: 10.1038/nrm2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mitchison T, Kirschner M. Dynamic instability of microtubule growth. Nature. 1984;312:237–242. doi: 10.1038/312237a0. [DOI] [PubMed] [Google Scholar]
- 6.Walker RA, O’Brien ET, Pryer NK, Soboeiro MF, Voter WA, Erickson HP, Salmon ED. Dynamic instability of individual microtubules analyzed by video light microscopy: rate constants and transition frequencies. J Cell Biol. 1988;107:1437–1448. doi: 10.1083/jcb.107.4.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dumont S, Mitchison TJ. Force and length in the mitotic spindle. Curr Biol. 2009;19:R749–R761. doi: 10.1016/j.cub.2009.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Prosser SL, Pelletier L. Mitotic spindle assembly in animal cells: a fine balancing act. Nat Rev Mol Cell Biol. 2017;18:187–201. doi: 10.1038/nrm.2016.162. [DOI] [PubMed] [Google Scholar]
- 9.Maddox P, Straight A, Coughlin P, Mitchison TJ, Salmon ED. Direct observation of microtubule dynamics at kinetochores in Xenopus extract spindles: implications for spindle mechanics. J Cell Biol. 2003;162:377–382. doi: 10.1083/jcb.200301088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McDonald KL, O’Toole ET, Mastronarde DN, McIntosh JR. Kinetochore microtubules in PTK cells. J Cell Biol. 1992;118:369–383. doi: 10.1083/jcb.118.2.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grill SW, Howard J, Schaffer E, Stelzer EH, Hyman AA. The distribution of active force generators controls mitotic spindle position. Science. 2003;301:518–521. doi: 10.1126/science.1086560. [DOI] [PubMed] [Google Scholar]
- 12.O’Connell CB, Wang YL. Mammalian spindle orientation and position respond to changes in cell shape in a dynein-dependent fashion. Mol Biol Cell. 2000;11:1765–1774. doi: 10.1091/mbc.11.5.1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sheeman B, Carvalho P, Sagot I, Geiser J, Kho D, Hoyt MA, Pellman D. Determinants of S. cerevisiae dynein localization and activation: implications for the mechanism of spindle positioning. Curr Biol. 2003;13:364–372. doi: 10.1016/S0960-9822(03)00013-7. [DOI] [PubMed] [Google Scholar]
- 14.Mastronarde DN, McDonald KL, Ding R, McIntosh JR. Interpolar spindle microtubules in PTK cells. J Cell Biol. 1993;123:1475–1489. doi: 10.1083/jcb.123.6.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nazarova E, O’Toole E, Kaitna S, Francois P, Winey M, Vogel J. Distinct roles for antiparallel microtubule pairing and overlap during early spindle assembly. Mol Biol Cell. 2013;24:3238–3250. doi: 10.1091/mbc.e13-05-0232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Winey M, Mamay CL, O’Toole ET, Mastronarde DN, Giddings TH, Jr, McDonald KL, McIntosh JR. Three-dimensional ultrastructural analysis of the Saccharomyces cerevisiae mitotic spindle. J Cell Biol. 1995;129:1601–1615. doi: 10.1083/jcb.129.6.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kapoor TM. Metaphase spindle assembly. Biology (Basel) 2017;6:E8. doi: 10.3390/biology6010008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tanenbaum ME, Medema RH. Mechanisms of centrosome separation and bipolar spindle assembly. Dev Cell. 2010;19:797–806. doi: 10.1016/j.devcel.2010.11.011. [DOI] [PubMed] [Google Scholar]
- 19.Blangy A, Arnaud L, Nigg EA. Phosphorylation by p34cdc2 protein kinase regulates binding of the kinesin-related motor HsEg5 to the dynactin subunit p150. J Biol Chem. 1997;272:19418–19424. doi: 10.1074/jbc.272.31.19418. [DOI] [PubMed] [Google Scholar]
- 20.Hoyt MA, He L, Loo KK, Saunders WS. Two Saccharomyces cerevisiae kinesin-related gene products required for mitotic spindle assembly. J Cell Biol. 1992;118:109–120. doi: 10.1083/jcb.118.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mayer TU, Kapoor TM, Haggarty SJ, King RW, Schreiber SL, Mitchison TJ. Small molecule inhibitor of mitotic spindle bipolarity identified in a phenotype-based screen. Science. 1999;286:971–974. doi: 10.1126/science.286.5441.971. [DOI] [PubMed] [Google Scholar]
- 22.Rincon SA, Lamson A, Blackwell R, Syrovatkina V, Fraisier V, Paoletti A, Betterton MD, Tran PT. Kinesin-5-independent mitotic spindle assembly requires the antiparallel microtubule crosslinker Ase1 in fission yeast. Nat Commun. 2017;8:15286. doi: 10.1038/ncomms15286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roof DM, Meluh PB, Rose MD. Kinesin-related proteins required for assembly of the mitotic spindle. J Cell Biol. 1992;118:95–108. doi: 10.1083/jcb.118.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cytrynbaum EN, Scholey JM, Mogilner A. A force balance model of early spindle pole separation in Drosophila embryos. Biophys J. 2003;84:757–769. doi: 10.1016/S0006-3495(03)74895-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gönczy P, Pichler S, Kirkham M, Hyman AA. Cytoplasmic dynein is required for distinct aspects of MTOC positioning, including centrosome separation, in the one cell stage Caenorhabditis elegans embryo. J Cell Biol. 1999;147:135–150. doi: 10.1083/jcb.147.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Robinson JT, Wojcik EJ, Sanders MA, McGrail M, Hays TS. Cytoplasmic dynein is required for the nuclear attachment and migration of centrosomes during mitosis in Drosophila. J Cell Biol. 1999;146:597–608. doi: 10.1083/jcb.146.3.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Salina D, Bodoor K, Eckley DM, Schroer TA, Rattner JB, Burke B. Cytoplasmic dynein as a facilitator of nuclear envelope breakdown. Cell. 2002;108:97–107. doi: 10.1016/S0092-8674(01)00628-6. [DOI] [PubMed] [Google Scholar]
- 28.Sharp DJ, Rogers GC, Scholey JM. Cytoplasmic dynein is required for poleward chromosome movement during mitosis in Drosophila embryos. Nat Cell Biol. 2000;2:922–930. doi: 10.1038/35046574. [DOI] [PubMed] [Google Scholar]
- 29.Vaisberg EA, Koonce MP, McIntosh JR. Cytoplasmic dynein plays a role in mammalian mitotic spindle formation. J Cell Biol. 1993;123:849–858. doi: 10.1083/jcb.123.4.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cleveland DW, Mao Y, Sullivan KF. Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell. 2003;112:407–421. doi: 10.1016/S0092-8674(03)00115-6. [DOI] [PubMed] [Google Scholar]
- 31.Diogo V, Teixeira J, Silva PM, Bousbaa H. Spindle assembly checkpoint as a potential target in colorectal cancer: current status and future perspectives. Clin Colorectal Cancer. 2016;23:30080–30089. doi: 10.1016/j.clcc.2016.06.006. [DOI] [PubMed] [Google Scholar]
- 32.Musacchio A. The molecular biology of spindle assembly checkpoint signaling dynamics. Curr Biol. 2015;25:R1002–R1018. doi: 10.1016/j.cub.2015.08.051. [DOI] [PubMed] [Google Scholar]
- 33.Tan AL, Rida PC, Surana U. Essential tension and constructive destruction: the spindle checkpoint and its regulatory links with mitotic exit. Biochem J. 2005;386:1–13. doi: 10.1042/BJ20041415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alexandru G, Uhlmann F, Mechtler K, Poupart MA, Nasmyth K. Phosphorylation of the cohesin subunit Scc1 by Polo/Cdc5 kinase regulates sister chromatid separation in yeast. Cell. 2001;105:459–472. doi: 10.1016/S0092-8674(01)00362-2. [DOI] [PubMed] [Google Scholar]
- 35.D’Amours D, Amon A. At the interface between signaling and executing anaphase—Cdc14 and the FEAR network. Genes Dev. 2004;18:2581–2595. doi: 10.1101/gad.1247304. [DOI] [PubMed] [Google Scholar]
- 36.Surana U, Amon A, Dowzer C, McGrew J, Byers B, Nasmyth K. Destruction of the CDC28/CLB mitotic kinase is not required for the metaphase to anaphase transition in budding yeast. EMBO J. 1993;12:1969–1978. doi: 10.1002/j.1460-2075.1993.tb05846.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Asbury CL. Anaphase A: disassembling microtubules move chromosomes toward spindle poles. Biology (Basel) 2017;6:E15. doi: 10.3390/biology6010015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meadows JC, Millar JB. Sharpening the anaphase switch. Biochem Soc Trans. 2015;43:19–22. doi: 10.1042/BST20140250. [DOI] [PubMed] [Google Scholar]
- 39.Scholey JM, Civelekoglu-Scholey G, Brust-Mascher I. Anaphase B. Biology (Basel) 2016;5:E51. doi: 10.3390/biology5040051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Winey M, Bloom K. Mitotic spindle form and function. Genetics. 2012;190:1197–1224. doi: 10.1534/genetics.111.128710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barlan K, Gelfand VI. Microtubule-based transport and the distribution, tethering, and organization of organelles. Cold Spring Harb Perspect Biol. 2017;9:a025817. doi: 10.1101/cshperspect.a025817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barton NR, Goldstein LS. Going mobile: microtubule motors and chromosome segregation. Proc Natl Acad Sci USA. 1996;93:1735–1742. doi: 10.1073/pnas.93.5.1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bloom GS, Endow SA. Motor proteins 1: kinesins. Protein Profile. 1995;2:1105–1171. [PubMed] [Google Scholar]
- 44.Hirokawa N, Noda Y, Okada Y. Kinesin and dynein superfamily proteins in organelle transport and cell division. Curr Opin Cell Biol. 1998;10:60–73. doi: 10.1016/S0955-0674(98)80087-2. [DOI] [PubMed] [Google Scholar]
- 45.Lu W, Gelfand VI. Moonlighting motors: kinesin, dynein, and cell polarity. Trends Cell Biol. 2017;27:505–514. doi: 10.1016/j.tcb.2017.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Milisav I. Dynein and dynein-related genes. Cell Motil Cytoskelet. 1998;39:261–272. doi: 10.1002/(SICI)1097-0169(1998)39:4<261::AID-CM2>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 47.Morfini G, Schmidt N, Weissmann C, Pigino G, Kins S. Conventional kinesin: biochemical heterogeneity and functional implications in health and disease. Brain Res Bull. 2016;126:347–353. doi: 10.1016/j.brainresbull.2016.06.009. [DOI] [PubMed] [Google Scholar]
- 48.Cole DG, Saxton WM, Sheehan KB, Scholey JM. A “slow” homotetrameric kinesin-related motor protein purified from Drosophila embryos. J Biol Chem. 1994;269:22913–22916. [PMC free article] [PubMed] [Google Scholar]
- 49.Enos AP, Morris NR. Mutation of a gene that encodes a kinesin-like protein blocks nuclear division in A. nidulans. Cell. 1990;60:1019–1027. doi: 10.1016/0092-8674(90)90350-N. [DOI] [PubMed] [Google Scholar]
- 50.Hagan I, Yanagida M. Kinesin-related cut7 protein associates with mitotic and meiotic spindles in fission yeast. Nature. 1992;356:74–76. doi: 10.1038/356074a0. [DOI] [PubMed] [Google Scholar]
- 51.Heck MM, Pereira A, Pesavento P, Yannoni Y, Spradling AC, Goldstein LS. The kinesin-like protein KLP61F is essential for mitosis in Drosophila. J Cell Biol. 1993;123:665–679. doi: 10.1083/jcb.123.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Muretta JM, Behnke-Parks WM, Major J, Petersen KJ, Goulet A, Moores CA, Thomas DD, Rosenfeld SS. Loop L5 assumes three distinct orientations during the ATPase cycle of the mitotic kinesin Eg5: a transient and time-resolved fluorescence study. J Biol Chem. 2013;288:34839–34849. doi: 10.1074/jbc.M113.518845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Muretta JM, Jun Y, Gross SP, Major J, Thomas DD, Rosenfeld SS. The structural kinetics of switch-1 and the neck linker explain the functions of kinesin-1 and Eg5. Proc Natl Acad Sci USA. 2015;112:E6606–E6613. doi: 10.1073/pnas.1512305112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Saunders WS, Hoyt MA. Kinesin-related proteins required for structural integrity of the mitotic spindle. Cell. 1992;70:451–458. doi: 10.1016/0092-8674(92)90169-D. [DOI] [PubMed] [Google Scholar]
- 55.Gordon DM, Roof DM. The kinesin-related protein Kip1p of Saccharomyces cerevisiae is bipolar. J Biol Chem. 1999;274:28779–28786. doi: 10.1074/jbc.274.40.28779. [DOI] [PubMed] [Google Scholar]
- 56.Kashina AS, Baskin RJ, Cole DG, Wedaman KP, Saxton WM, Scholey JM. A bipolar kinesin. Nature. 1996;379:270–272. doi: 10.1038/379270a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Scholey JE, Nithianantham S, Scholey JM, Al-Bassam J. Structural basis for the assembly of the mitotic motor Kinesin-5 into bipolar tetramers. Elife. 2014;3:e02217. doi: 10.7554/eLife.02217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kapitein LC, Peterman EJ, Kwok BH, Kim JH, Kapoor TM, Schmidt CF. The bipolar mitotic kinesin Eg5 moves on both microtubules that it crosslinks. Nature. 2005;435:114–118. doi: 10.1038/nature03503. [DOI] [PubMed] [Google Scholar]
- 59.van den Wildenberg SM, Tao L, Kapitein LC, Schmidt CF, Scholey JM, Peterman EJ. The homotetrameric kinesin-5 KLP61F preferentially crosslinks microtubules into antiparallel orientations. Curr Biol. 2008;18:1860–1864. doi: 10.1016/j.cub.2008.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Asraf H, Avunie-Masala R, Hershfinkel M, Gheber L. Mitotic slippage and expression of survivin are linked to differential sensitivity of human cancer cell-lines to the kinesin-5 inhibitor monastrol. PLoS One. 2015;10:e0129255. doi: 10.1371/journal.pone.0129255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Blangy A, Lane HA, d’Herin P, Harper M, Kress M, Nigg EA. Phosphorylation by p34cdc2 regulates spindle association of human Eg5, a kinesin-related motor essential for bipolar spindle formation in vivo. Cell. 1995;83:1159–1169. doi: 10.1016/0092-8674(95)90142-6. [DOI] [PubMed] [Google Scholar]
- 62.Leizerman I, Avunie-Masala R, Elkabets M, Fich A, Gheber L. Differential effects of monastrol in two human cell lines. Cell Mol Life Sci. 2004;61:2060–2070. doi: 10.1007/s00018-004-4074-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.FitzHarris G. Anaphase B precedes anaphase A in the mouse egg. Curr Biol. 2012;22:437–444. doi: 10.1016/j.cub.2012.01.041. [DOI] [PubMed] [Google Scholar]
- 64.Gerson-Gurwitz A, Movshovich N, Avunie R, Fridman V, Moyal K, Katz B, Hoyt MA, Gheber L. Mid-anaphase arrest in S. cerevisiae cells eliminated for the function of Cin8 and dynein. Cell Mol Life Sci. 2009;66:301–313. doi: 10.1007/s00018-008-8479-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Movshovich N, Fridman V, Gerson-Gurwitz A, Shumacher I, Gertsberg I, Fich A, Hoyt MA, Katz B, Gheber L. Slk19-dependent mid-anaphase pause in kinesin-5-mutated cells. J Cell Sci. 2008;121:2529–2539. doi: 10.1242/jcs.022996. [DOI] [PubMed] [Google Scholar]
- 66.Saunders WS, Koshland D, Eshel D, Gibbons IR, Hoyt MA. Saccharomyces cerevisiae kinesin- and dynein-related proteins required for anaphase chromosome segregation. J Cell Biol. 1995;128:617–624. doi: 10.1083/jcb.128.4.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sharp DJ, McDonald KL, Brown HM, Matthies HJ, Walczak C, Vale RD, Mitchison TJ, Scholey JM. The bipolar kinesin, KLP61F, cross-links microtubules within interpolar microtubule bundles of Drosophila embryonic mitotic spindles. J Cell Biol. 1999;144:125–138. doi: 10.1083/jcb.144.1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Straight AF, Sedat JW, Murray AW. Time-lapse microscopy reveals unique roles for kinesins during anaphase in budding yeast. J Cell Biol. 1998;143:687–694. doi: 10.1083/jcb.143.3.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Collins E, Mann BJ, Wadsworth P. Eg5 restricts anaphase B spindle elongation in mammalian cells. Cytoskeleton (Hoboken) 2014;71:136–144. doi: 10.1002/cm.21158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Saunders AM, Powers J, Strome S, Saxton WM. Kinesin-5 acts as a brake in anaphase spindle elongation. Curr Biol. 2007;17:R453–R454. doi: 10.1016/j.cub.2007.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kwok BH, Yang JG, Kapoor TM. The rate of bipolar spindle assembly depends on the microtubule-gliding velocity of the mitotic kinesin Eg5. Curr Biol. 2004;14:1783–1788. doi: 10.1016/j.cub.2004.09.052. [DOI] [PubMed] [Google Scholar]
- 72.Nadar VC, Ketschek A, Myers KA, Gallo G, Baas PW. Kinesin-5 is essential for growth-cone turning. Curr Biol. 2008;18:1972–1977. doi: 10.1016/j.cub.2008.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Baas PW, Matamoros AJ. Inhibition of kinesin-5 improves regeneration of injured axons by a novel microtubule-based mechanism. Neural Regen Res. 2015;10:845–849. doi: 10.4103/1673-5374.158351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jakobs M, Franze K, Zemel A. Force generation by molecular-motor-powered microtubule bundles; implications for neuronal polarization and growth. Front Cell Neurosci. 2015;9:441. doi: 10.3389/fncel.2015.00441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kahn OI, Sharma V, Gonzalez-Billault C, Baas PW. Effects of kinesin-5 inhibition on dendritic architecture and microtubule organization. Mol Biol Cell. 2015;26:66–77. doi: 10.1091/mbc.e14-08-1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Myers KA, Baas PW. Kinesin-5 regulates the growth of the axon by acting as a brake on its microtubule array. J Cell Biol. 2007;178:1081–1091. doi: 10.1083/jcb.200702074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nadar VC, Lin S, Baas PW. Microtubule redistribution in growth cones elicited by focal inactivation of kinesin-5. J Neurosci. 2012;32:5783–5794. doi: 10.1523/JNEUROSCI.0144-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chen Y, Hancock WO. Kinesin-5 is a microtubule polymerase. Nat Commun. 2015;6:8160. doi: 10.1038/ncomms9160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fridman V, Gerson-Gurwitz A, Shapira O, Movshovich N, Lakamper S, Schmidt CF, Gheber L. Kinesin-5 Kip1 is a bi-directional motor that stabilizes microtubules and tracks their plus-ends in vivo. J Cell Sci. 2013;126:4147–4159. doi: 10.1242/jcs.125153. [DOI] [PubMed] [Google Scholar]
- 80.Fridman V, Gerson-Gurwitz A, Movshovich N, Kupiec M, Gheber L. Midzone organization restricts interpolar microtubule plus-end dynamics during spindle elongation. EMBO Rep. 2009;10:387–393. doi: 10.1038/embor.2009.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gardner MK, Bouck DC, Paliulis LV, Meehl JB, O’Toole ET, Haase J, Soubry A, Joglekar AP, Winey M, Salmon ED, Bloom K, Odde DJ. Chromosome congression by Kinesin-5 motor-mediated disassembly of longer kinetochore microtubules. Cell. 2008;135:894–906. doi: 10.1016/j.cell.2008.09.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rogers GC, Rogers SL, Sharp DJ. Spindle microtubules in flux. J Cell Sci. 2005;118:1105–1116. doi: 10.1242/jcs.02284. [DOI] [PubMed] [Google Scholar]
- 83.Brust-Mascher I, Civelekoglu-Scholey G, Kwon M, Mogilner A, Scholey JM. Model for anaphase B: role of three mitotic motors in a switch from poleward flux to spindle elongation. Proc Natl Acad Sci USA. 2004;101:15938–15943. doi: 10.1073/pnas.0407044101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Brust-Mascher I, Sommi P, Cheerambathur DK, Scholey JM. Kinesin-5-dependent poleward flux and spindle length control in Drosophila embryo mitosis. Mol Biol Cell. 2009;20:1749–1762. doi: 10.1091/mbc.e08-10-1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Maddox PS, Bloom KS, Salmon ED. The polarity and dynamics of microtubule assembly in the budding yeast Saccharomyces cerevisiae. Nat Cell Biol. 2000;2:36–41. doi: 10.1038/71357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mallavarapu A, Sawin K, Mitchison T. A switch in microtubule dynamics at the onset of anaphase B in the mitotic spindle of Schizosaccharomyces pombe. Curr Biol. 1999;9:1423–1426. doi: 10.1016/S0960-9822(00)80090-1. [DOI] [PubMed] [Google Scholar]
- 87.De Wulf P, McAinsh AD, Sorger PK. Hierarchical assembly of the budding yeast kinetochore from multiple subcomplexes. Genes Dev. 2003;17:2902–2921. doi: 10.1101/gad.1144403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tytell JD, Sorger PK. Analysis of kinesin motor function at budding yeast kinetochores. JCB. 2006;172:861–874. doi: 10.1083/jcb.200509101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wargacki MM, Tay JC, Muller EG, Asbury CL, Davis TN. Kip3, the yeast kinesin-8, is required for clustering of kinetochores at metaphase. Cell Cycle. 2010;9:2581–2588. doi: 10.4161/cc.9.13.12076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cui H, Ghosh SK, Jayaram M. The selfish yeast plasmid uses the nuclear motor Kip1p but not Cin8p for its localization and equal segregation. J Cell Biol. 2009;185:251–264. doi: 10.1083/jcb.200810130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Prajapati HK, Rizvi SM, Rathore I, Ghosh SK. Microtubule-associated proteins, Bik1 and Bim1, are required for faithful partitioning of the endogenous 2 micron plasmids in budding yeast. Mol Microbiol. 2017;103:1046–1064. doi: 10.1111/mmi.13608. [DOI] [PubMed] [Google Scholar]
- 92.Rizvi SMA, Prajapati HK, Ghosh SK. The 2 micron plasmid: a selfish genetic element with an optimized survival strategy within Saccharomyces cerevisiae. Curr Genet. 2017;8:017–0719. doi: 10.1007/s00294-017-0719-2. [DOI] [PubMed] [Google Scholar]
- 93.Cochran JC. Kinesin motor enzymology: chemistry, structure, and physics of nanoscale molecular machines. Biophys Rev. 2015;7:269–299. doi: 10.1007/s12551-014-0150-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Cross RA. Review: mechanochemistry of the kinesin-1 ATPase. Biopolymers. 2016;105:476–482. doi: 10.1002/bip.22862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Friel CT, Howard J. Coupling of kinesin ATP turnover to translocation and microtubule regulation: one engine, many machines. J Muscle Res Cell Motil. 2012;33:377–383. doi: 10.1007/s10974-012-9289-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Goulet A, Moores C. New insights into the mechanism of force generation by kinesin-5 molecular motors. Int Rev Cell Mol Biol. 2013;304:419–466. doi: 10.1016/B978-0-12-407696-9.00008-7. [DOI] [PubMed] [Google Scholar]
- 97.Waitzman JS, Rice SE. Mechanism and regulation of kinesin-5, an essential motor for the mitotic spindle. Biol Cell. 2014;106:1–12. doi: 10.1111/boc.201300054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Endow SA, Waligora KW. Determinants of kinesin motor polarity. Science. 1998;281:1200–1202. doi: 10.1126/science.281.5380.1200. [DOI] [PubMed] [Google Scholar]
- 99.Goulet A, Behnke-Parks WM, Sindelar CV, Major J, Rosenfeld SS, Moores CA. The structural basis of force generation by the mitotic motor kinesin-5. J Biol Chem. 2012;287:44654–44666. doi: 10.1074/jbc.M112.404228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Henningsen U, Schliwa M. Reversal in the direction of movement of a molecular motor. Nature. 1997;389:93–96. doi: 10.1038/38022. [DOI] [PubMed] [Google Scholar]
- 101.Higuchi H, Endow SA. Directionality and processivity of molecular motors. Curr Opin Cell Biol. 2002;14:50–57. doi: 10.1016/S0955-0674(01)00293-9. [DOI] [PubMed] [Google Scholar]
- 102.Schief WR, Howard J. Conformational changes during kinesin motility. Curr Opin Cell Biol. 2001;13:19–28. doi: 10.1016/S0955-0674(00)00169-1. [DOI] [PubMed] [Google Scholar]
- 103.Shastry S, Hancock WO. Interhead tension determines processivity across diverse N-terminal kinesins. Proc Natl Acad Sci USA. 2011;108:16253–16258. doi: 10.1073/pnas.1102628108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Diefenbach RJ, Mackay JP, Armati PJ, Cunningham AL. The C-terminal region of the stalk domain of ubiquitous human kinesin heavy chain contains the binding site for kinesin light chain. Biochemistry. 1998;37:16663–16670. doi: 10.1021/bi981163r. [DOI] [PubMed] [Google Scholar]
- 105.Kanai Y, Dohmae N, Hirokawa N. Kinesin transports RNA: isolation and characterization of an RNA-transporting granule. Neuron. 2004;43:513–525. doi: 10.1016/j.neuron.2004.07.022. [DOI] [PubMed] [Google Scholar]
- 106.Gerson-Gurwitz A, Thiede C, Movshovich N, Fridman V, Podolskaya M, Danieli T, Lakamper S, Klopfenstein DR, Schmidt CF, Gheber L. Directionality of individual kinesin-5 Cin8 motors is modulated by loop 8, ionic strength and microtubule geometry. EMBO J. 2011;30:4942–4954. doi: 10.1038/emboj.2011.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Roostalu J, Hentrich C, Bieling P, Telley IA, Schiebel E, Surrey T. Directional switching of the Kinesin cin8 through motor coupling. Science. 2011;332:94–99. doi: 10.1126/science.1199945. [DOI] [PubMed] [Google Scholar]
- 108.Kasprzak AA, Hajdo L. Directionality of kinesin motors. Acta Biochim Pol. 2002;49:813–821. [PubMed] [Google Scholar]
- 109.Kull FJ. Motor proteins of the kinesin superfamily: structure and mechanism. Essays Biochem. 2000;35:61–73. doi: 10.1042/bse0350061. [DOI] [PubMed] [Google Scholar]
- 110.Acar S, Carlson DB, Budamagunta MS, Yarov-Yarovoy V, Correia JJ, Ninonuevo MR, Jia W, Tao L, Leary JA, Voss JC, Evans JE, Scholey JM. The bipolar assembly domain of the mitotic motor kinesin-5. Nat Commun. 2013;4:1343. doi: 10.1038/ncomms2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hildebrandt ER, Gheber L, Kingsbury T, Hoyt MA. Homotetrameric form of Cin8p, a Saccharomyces cerevisiae kinesin-5 motor, is essential for its in vivo function. J Biol Chem. 2006;281:26004–26013. doi: 10.1074/jbc.M604817200. [DOI] [PubMed] [Google Scholar]
- 112.Cahu J, Surrey T. Motile microtubule crosslinkers require distinct dynamic properties for correct functioning during spindle organization in Xenopus egg extract. J Cell Sci. 2009;122:1295–1300. doi: 10.1242/jcs.044248. [DOI] [PubMed] [Google Scholar]
- 113.Tao L, Mogilner A, Civelekoglu-Scholey G, Wollman R, Evans J, Stahlberg H, Scholey JM. A homotetrameric kinesin-5, KLP61F, bundles microtubules and antagonizes Ncd in motility assays. Curr Biol. 2006;16:2293–2302. doi: 10.1016/j.cub.2006.09.064. [DOI] [PubMed] [Google Scholar]
- 114.Düselder A, Thiede C, Schmidt CF, Lakämper S. Neck-linker length dependence of processive kinesin-5 motility. J Mol Biol. 2012;423:159–168. doi: 10.1016/j.jmb.2012.06.043. [DOI] [PubMed] [Google Scholar]
- 115.Gheber L, Kuo SC, Hoyt MA. Motile properties of the kinesin-related Cin8p spindle motor extracted from Saccharomyces cerevisiae cells. J Biol Chem. 1999;274:9564–9572. doi: 10.1074/jbc.274.14.9564. [DOI] [PubMed] [Google Scholar]
- 116.Kapitein LC, Kwok BH, Weinger JS, Schmidt CF, Kapoor TM, Peterman EJ. Microtubule cross-linking triggers the directional motility of kinesin-5. J Cell Biol. 2008;182:421–428. doi: 10.1083/jcb.200801145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lakämper S, Thiede C, Düselder A, Reiter S, Korneev MJ, Kapitein LC, Peterman EJ, Schmidt CF. The effect of monastrol on the processive motility of a dimeric kinesin-5 head/kinesin-1 stalk chimera. J Mol Biol. 2010;399:1–8. doi: 10.1016/j.jmb.2010.03.009. [DOI] [PubMed] [Google Scholar]
- 118.Thiede C, Fridman V, Gerson-Gurwitz A, Gheber L, Schmidt CF. Regulation of bi-directional movement of single kinesin-5 Cin8 molecules. Bioarchitecture. 2012;2:70–74. doi: 10.4161/bioa.20395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Edamatsu M. Bidirectional motility of the fission yeast kinesin-5, Cut7. Biochem Biophys Res Commun. 2014;446:231–234. doi: 10.1016/j.bbrc.2014.02.106. [DOI] [PubMed] [Google Scholar]
- 120.Shapira O, Goldstein A, Al-Bassam J, Gheber L. A potential physiological role for bi-directional motility and motor clustering of mitotic kinesin-5 Cin8 in yeast mitosis. J Cell Sci. 2017;130:725–734. doi: 10.1242/jcs.195040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Popchock AR, Tseng KF, Wang P, Karplus PA, Xiang X, Qiu W. The mitotic kinesin-14 KlpA contains a context-dependent directionality switch. Nat Commun. 2017;8:13999. doi: 10.1038/ncomms13999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Fallesen T, Roostalu J, Duellberg C, Pruessner G, Surrey T. Ensembles of bidirectional kinesin Cin8 produce additive forces in both directions of movement. Biophys J. 2017;113:2055–2067. doi: 10.1016/j.bpj.2017.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Geng Y-Z, Li T, Ji Q, Yan S. Simulation study of interactions between kinesin’s neck linker and motor domain. Cell Mol Bioeng. 2014;7:99–105. doi: 10.1007/s12195-014-0320-4. [DOI] [Google Scholar]
- 124.Hwang W, Lang MJ, Karplus M. Force generation in kinesin hinges on cover-neck bundle formation. Structure. 2008;16:62–71. doi: 10.1016/j.str.2007.11.008. [DOI] [PubMed] [Google Scholar]
- 125.Khalil AS, Appleyard DC, Labno AK, Georges A, Karplus M, Belcher AM, Hwang W, Lang MJ. Kinesin’s cover-neck bundle folds forward to generate force. Proc Natl Acad Sci. 2008;105:19247–19252. doi: 10.1073/pnas.0805147105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Yi-Zhao G, Qing J, Shu-Xia L, Shi-Wei Y. Initial conformation of kinesin’s neck linker. Chin Phys B. 2014;23:108701. doi: 10.1088/1674-1056/23/10/108701. [DOI] [Google Scholar]
- 127.Edamatsu M. Molecular properties of the N-terminal extension of the fission yeast kinesin-5, Cut7. Genet Mol Res. 2016;15:15017799. doi: 10.4238/gmr.15017799. [DOI] [PubMed] [Google Scholar]
- 128.Britto M, Goulet A, Rizvi S, von Loeffelholz O, Moores CA, Cross RA. Schizosaccharomyces pombe kinesin-5 switches direction using a steric blocking mechanism. Proc Natl Acad Sci USA. 2016;113:E7483–E7489. doi: 10.1073/pnas.1611581113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Sablin EP, Kull FJ, Cooke R, Vale RD, Fletterick RJ. Crystal structure of the motor domain of the kinesin-related motor ncd. Nature. 1996;380:555–559. doi: 10.1038/380555a0. [DOI] [PubMed] [Google Scholar]
- 130.Gigant B, Wang W, Dreier B, Jiang Q, Pecqueur L, Pluckthun A, Wang C, Knossow M. Structure of a kinesin-tubulin complex and implications for kinesin motility. Nat Struct Mol Biol. 2013;20:1001–1007. doi: 10.1038/nsmb.2624. [DOI] [PubMed] [Google Scholar]
- 131.Kozielski F, Sack S, Marx A, Thormahlen M, Schonbrunn E, Biou V, Thompson A, Mandelkow EM, Mandelkow E. The crystal structure of dimeric kinesin and implications for microtubule-dependent motility. Cell. 1997;91:985–994. doi: 10.1016/S0092-8674(00)80489-4. [DOI] [PubMed] [Google Scholar]
- 132.Bell KM, Cha HK, Sindelar CV, Cochran JC. The yeast kinesin-5 Cin8 interacts with the microtubule in a noncanonical manner. J Biol Chem. 2017;12:797662. doi: 10.1074/jbc.M117.797662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Avunie-Masala R, Movshovich N, Nissenkorn Y, Gerson-Gurwitz A, Fridman V, Koivomagi M, Loog M, Hoyt MA, Zaritsky A, Gheber L. Phospho-regulation of kinesin-5 during anaphase spindle elongation. J Cell Sci. 2011;124:873–878. doi: 10.1242/jcs.077396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Chee MK, Haase SB. B-cyclin/CDKs regulate mitotic spindle assembly by phosphorylating kinesins-5 in budding yeast. PLoS Genet. 2010;6:e1000935. doi: 10.1371/journal.pgen.1000935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Goldstein A, Siegler N, Goldman D, Judah H, Valk E, Koivomagi M, Loog M, Gheber L. Three Cdk1 sites in the kinesin-5 Cin8 catalytic domain coordinate motor. Cell Mol Life Sci. 2017;28:017–2523. doi: 10.1007/s00018-017-2523-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Shapira O, Gheber L. Motile properties of the bi-directional kinesin-5 Cin8 are affected by phosphorylation in its motor domain. Sci Rep. 2016;6:25597. doi: 10.1038/srep25597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Hackney DD. Highly processive microtubule-stimulated ATP hydrolysis by dimeric kinesin head domains. Nature. 1995;377:448–450. doi: 10.1038/377448a0. [DOI] [PubMed] [Google Scholar]
- 138.Kaan HY, Hackney DD, Kozielski F. The structure of the kinesin-1 motor-tail complex reveals the mechanism of autoinhibition. Science. 2011;333:883–885. doi: 10.1126/science.1204824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Weinger JS, Qiu M, Yang G, Kapoor TM. A nonmotor microtubule binding site in kinesin-5 is required for filament crosslinking and sliding. Curr Biol. 2011;21:154–160. doi: 10.1016/j.cub.2010.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Goshima G, Vale RD. Cell cycle-dependent dynamics and regulation of mitotic kinesins in Drosophila S2 cells. Mol Biol Cell. 2005;16:3896–3907. doi: 10.1091/mbc.e05-02-0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Sawin KE, Mitchison TJ. Mutations in the kinesin-like protein Eg5 disrupting localization to the mitotic spindle. Proc Natl Acad Sci USA. 1995;92:4289–4293. doi: 10.1073/pnas.92.10.4289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Drummond DR, Hagan IM. Mutations in the bimC box of Cut7 indicate divergence of regulation within the bimC family of kinesin related proteins. J Cell Sci. 1998;111(Pt 7):853–865. doi: 10.1242/jcs.111.7.853. [DOI] [PubMed] [Google Scholar]
- 143.Duselder A, Fridman V, Thiede C, Wiesbaum A, Goldstein A, Klopfenstein DR, Zaitseva O, Janson ME, Gheber L, Schmidt CF. Deletion of the tail domain of the kinesin-5 Cin8 affects its directionality. J Biol Chem. 2015;19:620799. doi: 10.1074/jbc.M114.620799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Saito N, Kaneko K. Embedding dual function into molecular motors through collective motion. Sci Rep. 2017;7:44288. doi: 10.1038/srep44288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Clift D, Schuh M. A three-step MTOC fragmentation mechanism facilitates bipolar spindle assembly in mouse oocytes. Nat Commun. 2015;6:7217. doi: 10.1038/ncomms8217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Sommi P, Cheerambathur D, Brust-Mascher I, Mogilner A. Actomyosin-dependent cortical dynamics contributes to the prophase force-balance in the early Drosophila embryo. PLoS One. 2011;6:e18366. doi: 10.1371/journal.pone.0018366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Courtheoux T, Gay G, Reyes C, Goldstone S, Gachet Y, Tournier S. Dynein participates in chromosome segregation in fission yeast. Biol Cell. 2007;99:627–637. doi: 10.1042/BC20070047. [DOI] [PubMed] [Google Scholar]
- 148.Li YY, Yeh E, Hays T, Bloom K. Disruption of mitotic spindle orientation in a yeast dynein mutant. Proc Natl Acad Sci USA. 1993;90:10096–10100. doi: 10.1073/pnas.90.21.10096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Blackwell R, Edelmaier C, Sweezy-Schindler O, Lamson A, Gergely ZR, O’Toole E, Crapo A, Hough LE, McIntosh JR, Glaser MA, Betterton MD. Physical determinants of bipolar mitotic spindle assembly and stability in fission yeast. Sci Adv. 2017;3:e1601603. doi: 10.1126/sciadv.1601603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Adams AE, Pringle JR. Relationship of actin and tubulin distribution to bud growth in wild-type and morphogenetic-mutant Saccharomyces cerevisiae. J Cell Biol. 1984;98:934–945. doi: 10.1083/jcb.98.3.934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Allan VJ. Cytoplasmic dynein. Biochem Soc Trans. 2011;39:1169–1178. doi: 10.1042/BST0391169. [DOI] [PubMed] [Google Scholar]
- 152.Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 153.Ferenz NP, Gable A, Wadsworth P. Mitotic functions of kinesin-5. Semin Cell Dev Biol. 2010;21:255–259. doi: 10.1016/j.semcdb.2010.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.McCully EK, Robinow CF. Mitosis in the fission yeast Schizosaccharomyces pombe: a comparative study with light and electron microscopy. J Cell Sci. 1971;9:475–507. doi: 10.1242/jcs.9.2.475. [DOI] [PubMed] [Google Scholar]
- 155.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]