Abstract
The increased production of the 42 aminoacids long beta-amyloid (Aβ42) peptide has been established as a causal mechanism of the familial early onset Alzheimer’s disease (AD). In contrast, the causal mechanisms of the late-onset AD (LOAD), that affects most AD patients, remain to be established. Indeed, Aβ42 accumulation has been detected more than 30 years before diagnosis. Thus, the mechanisms that control Aβ accumulation in LOAD likely go awry long before pathogenesis becomes detectable. Early on, APOE4 was identified as the biggest genetic risk factor for LOAD. However, since APOE4 is not present in all LOAD patients, genome-wide association studies of thousands of LOAD patients were undertaken to identify other genetic variants that could explain the development of LOAD. PICALM, BIN1, CD2AP, SORL1, and PLD3 are now with APOE4 among the identified genes at highest risk in LOAD that have been implicated in Aβ42 production. Recent evidence indicates that the regulation of the endocytic trafficking of the amyloid precursor protein (APP) and/or its secretases to and from sorting endosomes is determinant for Aβ42 production. Thus, here, we will review the described mechanisms, whereby these genetic risk factors can contribute to the enhanced endocytic production of Aβ42. Dissecting causal LOAD mechanisms of Aβ42 accumulation, underlying the contribution of each genetic risk factor, will be required to identify therapeutic targets for novel personalized preventive strategies.
Keywords: Late-onset Alzheimer’s disease, Trafficking, Endocytosis, APOE4, PICALM, BIN1, CD2AP, SORL1, PLD3
Introduction
Alzheimer’s disease (AD) is a progressive neurodegenerative disease that impairs memory, behavior and the ability to be independent. It is an overwhelming disease not only for patients but also for their caregivers and families. AD can be familial and rare with an early onset (eFAD) starting in the thirties; or very common affecting 1 in 10 elderlies with more than 65 years old, a late-onset AD (LOAD). The lack of an effective treatment and the increasing aging of the population has transformed LOAD into a health and socioeconomic problem.
Pathologically, AD is characterized by progressive accumulation of amyloid plaques and tau neurofibrillary tangles. However, it is the progressive synapse loss which better predicts cognitive decline with aging [1].
eFAD is caused by inheritance of familial mutations in amyloid precursor protein (APP) or presenilins 1 and 2 (PSEN1, PSEN2; γ-cleavage of APP), that lead to the excessive neuronal production of the longest form of beta-amyloid (Aβ42) or an increased ratio of Aβ42 over Aβ40. Aβ42 is more prone to oligomerize and the oligomers have been established as the most toxic species in AD [2]. Mice carrying eFAD mutations recapitulate cognitive memory deficits and develop amyloid plaques and tau neurofibrillary tangles, modeling essential AD features. In eFAD, synapses progressively become dysfunctional, lost and eventually, neurons degenerate due to progressive accumulation and aggregation of Aβ42 with aging [3–5]. At synapses, Aβ42 accumulates both extra- and intracellularly [6–13]. Aβ42 is generated by intracellular processing of APP in endosomes [14–18]. Upon production, Aβ is either secreted or retained in endosomes. Aβ42 accumulates intracellularly in multivesicular endosomes, altering the sorting and lysosomal degradation of endocytosed membrane receptors [10, 19]. Indeed, intracellular Aβ accumulation precedes extracellular Aβ deposits and abnormal tau phosphorylation and aggregation [9].
AD silent cellular mechanisms that lead to Aβ42 accumulation and synaptic dysfunction are predicted to begin more than 30 years before diagnosis. LOAD is a multifactorial disease, caused by a combination of genetic and lifestyle risk factors. The most important genetic risk factor is APOE4, identified in 1993 [20–23]. However, APOE4 is not present in all cases of LOAD and this prompted geneticists to search for other genetic risk factors. Genome-wide association studies (GWAS) of thousands of LOAD patients were undertaken to identify genetic variants (or single nucleotide polymorphisms, SNP) that could explain the development of LOAD [24]. Among the first identified genes at the highest risk in AD, were PICALM, BIN1, and CD2AP [25–28]. SORL1 was later found by meta-analysis of the large LOAD patients’ consortiums [28]. Genome sequencing of smaller cohorts identified rare variants in PLD3 associated with AD [29], although this association has yet to be confirmed in larger cohorts; for further details on the genetic associations, see a recent review by R. Guerreiro [30]. All together with APOE4 have been implicated in Aβ production and linked to endosomal trafficking. Here, we will review their impact on Aβ production at endosomes. Dissecting causal LOAD mechanisms of Aβ42 accumulation and synaptic dysfunction will be required to identify therapeutic targets and novel personalized preventive and curative strategies.
Endocytic production of Aβ
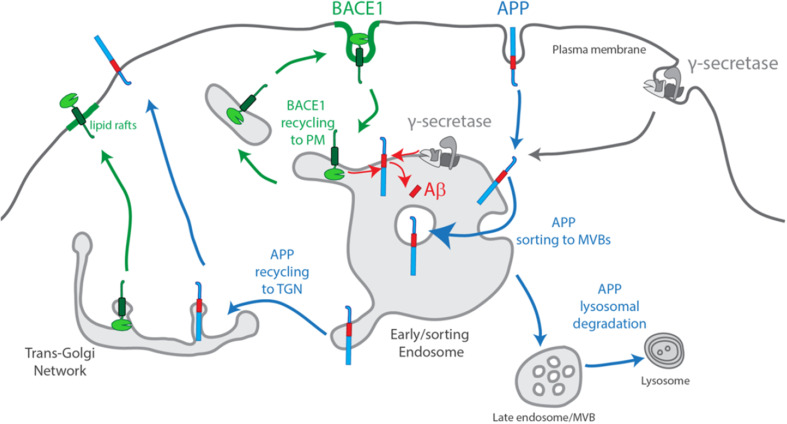
Normal Aβ production only occurs to a small extent, because the neuronal trafficking pathways of APP and BACE1 are largely segregated (Fig. 1). APP and BACE1, both transmembrane proteins, initiate their secretory pathway to the plasma membrane with their exit from the trans-Golgi network (TGN) in distinct post-Golgi carriers [31]. At the plasma membrane, evidence supports a segregation of APP from BACE1, with BACE1 being more present in membrane microdomains rich in cholesterol and flotillin (lipid rafts) than APP [31–33]. BACE1 and APP undergo endocytosis through different internalization mechanisms.
Fig. 1.
Scheme of normal endocytic production of Aβ. APP and BACE1 exit the Trans-Golgi Network (TGN) to the plasma membrane in separate post-Golgi secretory vesicles. At the plasma membrane, BACE1 prefers lipid rafts, and is endocytosed independently of APP. Less clear is γ-secretase complex assembly and endocytic trafficking. Upon endocytosis, APP, BACE1 and γ-secretase reach early/sorting endosomes. BACE1 recycles fast out of sorting endosomes to the plasma membrane, while APP is sorted into inner luminal vesicles during MVB biogenesis. Aβ production occurs upon acidification of sorting endosomes which favors BACE1 activity and APP processing at the endosomal limiting membrane. APP degradation occurs upon fusion with the lysosome
APP endocytosis is mostly clathrin-mediated [34]. The YENPTY motif in APP C-terminus is the sorting signal for endocytosis [35], and it is involved in the interaction of APP with auxiliary proteins [36]. There is evidence that a cholesterol/flotillin-dependent clustering of APP may stimulate the internalization via clathrin-dependent endocytosis to promote Aβ production [37]. Indeed, altering the lipid membrane composition in cholesterol, flotillin and caveolin-1 levels influenced the rate of APP processing and Aβ production [37–39]. Lipids can potentially alter APP endocytosis. APP endocytosis may also be regulated by protein interaction, such as with ApoE receptors [40–42].
BACE1 endocytosis occurs by a less defined mechanism, independently of clathrin, regulated by Arf6 [43] or by clathrin-mediated endocytosis [44, 45]. A dileucine acidic motif in BACE1 C-terminus is the sorting signal for endocytosis and endosomal trafficking [44, 46, 47].
APP and BACE1 endocytic vesicles are delivered to a common early endosome. Indeed, endocytosis has been shown to be required for the sequential processing of APP by BACE1 and by γ-secretase specifically in neurons, producing mainly Aβ40 and Aβ42 [48–51]. In non-neuronal cells, evidence indicates that the TGN is a preferential site for APP processing upon APP endocytosis [52]. Normally, APP processing is likely avoided by BACE1 sorting into endosomal tubules for the recycling pathway, whereas APP is sorted into intraluminal vesicles for the degradative pathway in a process termed multivesicular endosome (MVB) biogenesis [45, 53, 54]. Since endosomal acidification is required for optimal BACE1 activity [31], and γ-secretase is active at late-endosomes [55], it is likely that APP processing occurs during early endosome maturation [31, 43]. Indeed, APP processing and Aβ production increase by blocking APP sorting to MVBs intraluminal vesicles [54, 56] and by blocking BACE1 recycling [57]. Aβ can be secreted or retained within neurons in MVBs [7, 58].
In the past few years, the mechanisms whereby several LOAD genetic risk factors contribute to Aβ accumulation have started to be uncovered. As such, their impact deregulating the neuronal endosomal trafficking of APP and its secretases will be reviewed in the next section.
Regulators of endosomal trafficking identified as risk factors for AD
Apolipoprotein E4
APOE4 was identified associated with AD in 1993, and it remains the strongest genetic risk factor for LOAD [20–23]. APOE4 is one of the three polymorphic alleles of the APOE gene. The other alleles are APOE2 and APOE3. Apolipoprotein E (ApoE) is highly expressed in the brain, mainly by astrocytes [59]. Upon secretion, ApoE binds cholesterol and other lipids enabling their endocytosis, via ApoE receptors [60]. The three different protein isoforms, ApoE2, ApoE3, and ApoE4, have a different effect on AD pathogenesis. ApoE4 is pathological, while ApoE2 and ApoE3 are neuroprotective or neutral, respectively [61].
The underlying mechanisms of action of ApoE4 in AD are still poorly understood [20, 22]. ApoE4 contribution to Aβ accumulation likely includes multiple mechanisms. Upregulation of Aβ production by ApoE4 is supported by several findings. Namely, exogenous ApoE4 increases Aβ accumulation by stimulation of APP endocytosis and processing, via ApoE receptor 2 (ApoER2) or lipoprotein receptor-related protein (LRP) but not low-density lipoprotein receptor (LDLR) [42, 62]. Importantly, ApoE3 and ApoE4 injection in hippocampus also increased APP processing [63]. In contrast, exogenous ApoE2, 3, and 4 were described to reduce extracellular Aβ, while APP processing increased [64]. Interestingly, the interaction between APP and γ-secretase complex can be up-regulated by expression of an ApoE interacting protein, TMC22, thus increasing Aβ production [65]. Remarkably, abnormal endosomes were described in the brain of AD patients with the APOE4 genotype [66] as well as in the aging brain of APOE4-humanized mice [67]. The mechanism underlying the increase of APP endocytosis in the presence of ApoE could be linked to alterations in the lipid membrane composition given ApoE function in lipid transport. However, experimental evidence suggests that the effect of ApoE4 on APP endocytosis/Aβ production is independent of its lipidation [42, 68].
Unexpectedly, a recent study from the Sudhof lab identified an ApoE4 mechanism independent of APP endocytosis. Instead, ApoE4 was found to boost APP transcription and thus Aβ production by activating a signal transduction pathway [69].
Aβ accumulation may also result from decreased clearance, since ApoE4 binds secreted Aβ less efficiently than ApoE3 and ApoE2, compromising Aβ uptake and lysosomal degradation [70–72]. Otherwise, ApoE4 may compete with Aβ for the same degradation pathways, without binding Aβ [73].
Alternative ApoE4 mechanisms, independent of Aβ, may exist as indicated by an ApoE4-dependent impairment of synaptic plasticity due to trapping of AMPA and NMDA receptors in intracellular compartments [74]. The uptake of cholesterol itself is compromised, since ApoE4 is lipidated less efficiently, which could, in turn, affect membrane trafficking [68].
Alternative ApoE4 mechanisms, independent of Aβ, may exist as indicated by an ApoE4-dependent impairment of synaptic plasticity due to trapping of AMPA and NMDA receptors in intracellular compartments [74]. The uptake of cholesterol itself is compromised, since ApoE4 is lipidated less efficiently, which could, in turn, affect membrane trafficking [68].
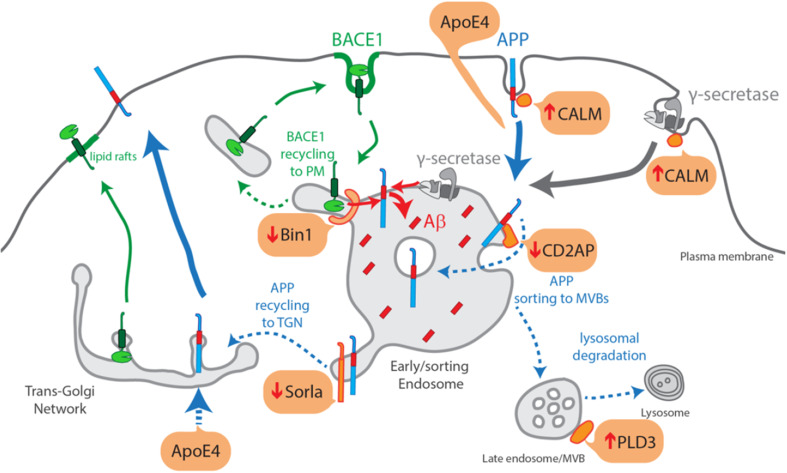
Taken together, as illustrated in Fig. 2, ApoE4 could mediate an increase in APP endocytosis via alterations in lipid membrane composition or via the increased APP in the secretory pathway due to ApoE4-dependent upregulation of APP transcription.
Fig. 2.
Scheme of the increased endocytic production of Aβ due to LOAD genetic risk factors. ApoE4-mediated increase in APP endocytosis and/or via increased APP in the secretory pathway due to increased APP transcription; Bin1 loss-of-function impedes BACE1 to recycle out of sorting endosomes; Sorla loss-of-function decreases APP recycling out of endosomes to the TGN; CD2AP loss-of-function decreases APP sorting into MVBs and lysosomal degradation; CALM loss-of-function increases APP and γ-secretase endocytosis and delivery to sorting endosomes; PLD3 loss-of-function affects lysosome morphology and perhaps APP processing at endosomes
PICALM
PICALM genetic variants were associated with LOAD by GWAS [25, 75–80]. A recent GWAS meta-analysis confirmed the association of one of the PICALM variants with higher risk of AD [81].
Correlative studies between PICALM variants and cognitive reserve, assessed based on brain volume and thickness, suggest that PICALM variants confer protection [82–84].
PICALM encodes for CALM (Clathrin Assembly Lymphoid Myeloid leukemia) [85], a cytosolic clathrin–endocytic adaptor [86]. CALM is ubiquitously expressed and is detected pre- and post-synaptically, while its neuron-specific homolog, AP180, is predominantly in presynaptic compartments [87, 88]. Despite their similarity, CALM and AP180 are not functionally redundant [86]. CALM interacts with clathrin and membrane lipids to promote the formation of endocytic vesicles [89, 90], while AP180 is more specifically implicated in synaptic vesicle retrieval [88]. CALM also functions in the retrieval of VAMP proteins, SNAREs that mediate fusion of exocytic vesicles from the plasma membrane [86, 90, 91].
Thus far, the data on the expression of CALM in AD are inconsistent, since it has been found decreased in the AD brain due to abnormal cleavage [92], but increased in the cortex of an eFAD mice model (Tg2576) [93]. Unpredictably, a modest increase in PICALM mRNA correlated with a protective genotype [94]. Moreover, PICALM depletion decreased amyloid plaques in the hippocampus of an eFAD mouse model (APP/PS1 mice) [95]. More research will be necessary to establish how CALM expression is altered in AD.
Mechanistically, evidence supports that CALM is required for clathrin-mediated endocytosis of APP and thus Aβ endocytic production [95–99]. Additional mechanisms include increased sorting of APP/APPCTFs for lysosomal degradation upon CALM overexpression [100]. Moreover, CALM may also be required for γ-secretase endocytosis, since CALM depletion increased nicastrin, a γ-secretase component, at the plasma membrane [98]. Alternatively, CALM deficiency decreases Aβ clearance across the murine blood–brain barrier (BBB) [101]. The decreased Aβ clearance could be due to a reduced endocytosis and recycling of Aβ bound to LRP1, impeding Aβ clearance by transcytosis across the microvessels epithelium [101]. CALM has also been shown to function at synapses, mediating the reclustering of synaptic vesicles proteins after exocytosis [102]. A role for CALM in cholesterol uptake has also been suggested, since CALM depletion alters LDL receptor endocytosis [103].
BIN1
BIN1 was first associated with LOAD by GWAS performed by Seshadri et al., 2010, which identified the most common SNP rs744373 in a locus within 30 kb of the gene BIN1 [104]. BIN1 association to LOAD was further confirmed in other large family-based GWAS [105], in candidate gene studies with independent cohorts [28, 80], and meta-analysis of multicenter datasets [28, 106, 107]. Subsequent analysis of GWAS patients found BIN1 associated with alterations in cortical thickness, lower scores on episodic memory and an earlier AD onset [79, 84, 108]. BIN1 sequencing identified rare coding variants associated with LOAD [109, 110].
BIN1 encodes for Bin1 (bridging integrator 1) first identified as an interactor of MYC, the oncoprotein [111]. BIN1 undergoes alternative splicing originating at least ten isoforms. All Bin1 isoforms are membrane-associated and share an N-terminal BAR domain, thus belonging to the BAR (Bin–Amphiphysin–Rvsp) family proteins. Through its BAR domain, Bin1 confers curvature to membranes, critical for its function in membrane tubulation and vesicle formation. The C-terminal SH3 domain mediates Bin1 interaction with proteins involved in endocytosis, such as dynamin [112, 113] and endophilin [114] that regulate membrane dynamics.
Importantly, in brain mainly the Bin1 neuronal-specific isoform (isoform 1) and at least one ubiquitous isoform (isoform 9) are expressed [115]. Neuronal BIN1 was almost simultaneously identified by different groups [113, 116–118]. Neuronal Bin1 was initially found enriched in brain synaptosomes and localizes to axon initial segments and nodes of Ranvier [116–118]. It is the longest isoform and contains a clathrin-associated protein-binding region (CLAP domain) [119]. Bin1 is very similar to amphiphysin, and dimerization with amphiphysin can enhance clathrin-mediated endocytosis [118]. Amphiphysin knockout mice present reduced levels of Bin1 and exhibit synaptic vesicle recycling defects [120]. BIN1 knockout mice die after birth, due to muscle defects, but embryonic primary neuronal cultures showed unaffected synapse morphology [121]. Ubiquitous BIN1 knockdown did not alter significantly endocytosis, instead increased defects in the recycling of transferrin receptor, in fibroblasts or HeLa cells [121, 122].
BIN1 mRNA transcripts were found increased in AD brains, maybe due to the augmented transcriptional activity of LOAD variants [123]. Higher BIN1 gene expression was found correlated with later onset and shorter disease duration [124]. Interestingly, the expression of BIN1 was highly correlated with that of CD2AP and PICALM [124]. Neuronal Bin1, but not ubiquitous Bin1, has been found decreased in LOAD [125–127].
We and Tomita’s lab found Bin1 depletion to increase Aβ production due to the accumulation of BACE1 in endosomes [128, 129]. A differential impact on the secretion of Aβ was observed. While in the Miyagawa study secreted Aβ42 and Aβ40 increased, we found a decrease in Aβ40, but not in Aβ42 secretion [128, 129]. More consensual was the increase in intracellular Aβ42 upon Bin1 depletion [128, 129]. Surprisingly, we found by subcellular analysis of Aβ42 accumulation that Aβ42 increased mainly in axons [129]. Both groups found BACE1 accumulating in early endosomes, suggesting that Bin1 controls Aβ production by regulating BACE1 trafficking [128, 129]. In the Miyagawa study, BACE1 levels increased in neurons depleted for Bin1 pointing to a function for Bin1 in controlling BACE1 degradation; however, the exact mechanism involved has yet to be investigated [128]. We found that Bin1 depletion led to an impaired BACE1 recycling, specifically in axons [129]. Mechanistically, Bin1 was found required for scission of BACE1 tubules from early endosomes enabling BACE1 recycling [129]. How this Bin1 role in BACE1 recycling affects BACE1 degradation needs to be investigated. Bin1 could also contribute to AD by playing a role in disease propagation, since Bin1 depletion increased tau propagation via an endosomal route [130].
CD2AP
CD2AP genetic variants were associated with LOAD by several GWAS [26, 79, 107]. CD2AP susceptibility loci correlate with AD progression [131]. Meta-analysis of GWAS studies confirmed CD2AP association and identified the non-coding variant, rs9346407, as the most frequent in LOAD patients [28, 132]. CD2AP sequencing identified rare coding variants in LOAD [110].
CD2AP encodes for CD2-associated protein (CD2AP), a membrane-associated scaffolding protein, first identified as a T cells adaptor protein [133]. CD2AP is an endocytic [134–137] and an actin cytoskeleton regulator [135, 138–140]. CD2AP may control endosome maturation and protein sorting for degradation via its actin regulation [135].
CD2AP is most expressed in kidney podocytes [141], where it anchors important adaptors of the slit diaphragm to the actin cytoskeleton [142]. Interestingly, podocytes, like neurons, have actin-rich protrusions and share actin regulators such as synaptopodin and drebrin [143, 144].
CD2AP is less expressed in the brain [141]; nevertheless, in situ hybridization clearly shows CD2AP mRNA expression in cortical and hippocampal neurons (Allen brain Atlas; ID 12488). CD2AP is detected in primary cortical neurons especially in dendrites, where it localizes to endosomes [129]. CD2AP expression in the LOAD brain has not been investigated, but there is evidence that it could be reduced as in the peripheral blood lymphocytes of a Chinese LOAD cohort [145].
The decreased CD2AP expression can increase intracellular exogenous Aβ40 and Aβ42 levels without increasing extracellular Aβ levels in neuroblastoma cells overexpressing APP [146]. We found that decreased CD2AP expression increased intracellular endogenous Aβ42 in wild-type neuroblastoma cells and particularly in dendrites of primary cortical neurons [129]. Importantly, the decreased function of CD2AP at dendritic endosomes was found responsible for an accumulation of APP at early endosomes limiting membrane [129]. The impaired sorting into multivesicular endosomes likely precluded an efficient degradation of APP by the lysosome and favored APP processing and Aβ production [129]. In young eAD transgenic mice (PS1/APP), CD2AP knockout did not alter Aβ accumulation nor amyloid plaques load [146]. Thus, the impact of CD2AP variants on the development of LOAD pathology needs to be assessed in a LOAD mouse model or in human neurons derived from fibroblasts of patients carrying CD2AP variants. Alternatively, CD2AP loss-of-function could have an impact on Aβ clearance, since it is detected in brain endothelial cells and CD2AP knockout mice have reduced blood–brain integrity [141, 147].
SORL1
SORL1 was initially associated with LOAD in candidate gene approaches and later in GWAS studies [28, 81, 148–153]. Subsequent sequencing studies identified rare missense variants in SORL1 both in eAD and LOAD [81, 152, 154–156].
SORL1 encodes for sortilin-related receptor with A-type repeats (Sorla), that belongs to the family of low-density lipoprotein receptors, as well as to the family of vacuolar protein sorting ten domain receptors (VPS10p) [156]. Sorla is a neuronal sorting receptor mainly found in sorting endosomes in the somatodendritic domain [157].
Sorla levels are decreased in AD [158, 159] and several underlying mechanisms have been identified: increased methylation of SORL1 in AD repressing gene expression [160]; the presence of shorter SORL1 splice variants in AD reducing full-length Sorla expression [161]; and the presence of SORL1 variants limiting the increase in Sorla expression upon brain-derived neurotrophic factor (BDNF) stimulation [154].
Sorla binds directly to APP, via an extracellular domain and via a motif in the cytosolic tail [162]. Sorla binding selects endocytosed APP to be retrogradely transported back to the TGN, reducing APP processing at endosomes and Aβ production [156, 157]. Evidence supports an important role for Sorla in removing APP from endosomes. Depletion of Sorla increases Aβ production and amyloid plaques load [163].
Human neurons carrying SORL1 AD variants showed decreased APP processing upon BDNF stimulation [154]. Some rare variants, such as p.Asn2174Ser, have been shown to decrease Sorla capacity to retrieve APP back to the TGN, increasing APP at endosomes and Aβ production [152]. The mechanism by which Sorla sorts APP back to the TGN has been shown to be dependent on the retromer. The retromer is a protein complex responsible for the formation of endosomal tubules enriched in APP and Sorla that upon scission will be transported back to the TGN [164, 165]. APP phosphorylation and dimerization have been shown to regulate APP trafficking dependent on Sorla [166, 167].
Alternatively, Sorla loss-of-function could decrease Aβ clearance, since Sorla binds to Aβ promoting its delivery to lysosome and degradation [158]. Interestingly, Sorla mediates the cellular uptake of cholesterol-loaded APOE, with a preference for APOE4 [168]. It is important to note that protective variants have also been identified, although their mechanism remains to be investigated [81].
Increasing Sorla could be a therapeutic approach, since it reduces Aβ concentration in mouse brain [158]. A promising study identified a Sorla activator, 6-shogoal, with therapeutic potential against AD [169].
PLD3
Rare variants in PLD3 were associated with increased LOAD risk [29, 170]. However, the association has not yet been replicated in AD [171] neither in eFAD [172]. PLD3 variants were weakly associated with cognitive decline and not with amyloid pathology [173, 174].
PLD3 encodes for phospholipase D3, a membrane-associated protein of the PLD family, which includes phospholipases D1 and D2, both involved in endocytic trafficking [175, 176]. Less studied, PLD3 does not have the PX and PH domains that localize PLD1 and 2 to membranes. While PLD1 and PLD2 produce phosphatidic acid, PLD3 has a conserved substitution in the lipase domain PLD3 that likely prevents its activity as a classical PLD [176]. PLD3 is a transmembrane glycoprotein associated with the endoplasmic reticulum, involved in its reorganization during myotube formation [177].
Importantly, PLD3 is highly expressed in hippocampus and cortex, regions more vulnerable to AD pathology [29, 178]. PLD3 mRNA and protein expression are decreased in LOAD patients brain [29, 179]. Notably, PLD3 accumulates in neuritic plaques [179]. Interestingly, depletion of PLD3 increased resistance to oxidative stress-dependent loss of cell viability [180].
PLD3 loss-of-function increased secretion of Aβ42 and Aβ40 [29]; however, recently, this result was not replicated in similar experimental conditions [181]. Instead, PLD3 was found enriched in lysosomes which became morphologically abnormal upon PLD3 loss-of-function [181]. Whether the lysosomal degradative activity is affected and whether it contributes to Aβ42 clearance instead of Aβ42 production will need to be further investigated.
Outlook
The studies of ApoE4, CALM, Bin1, CD2AP, Sorla, and PLD3 encoded by LOAD genetic risk factors reviewed here support that increased production of Aβ42 is a mechanism of LOAD. ApoE4 and loss-of-function of Bin1, CD2AP, CALM, Sorla, and PLD3 lead, by different mechanisms, to deregulation in intracellular trafficking of APP and/or of its secretases, to an increase in the retention of APP and/or its secretases in sorting endosomes, potentiating Aβ42 endocytic production (Fig. 2). However, this may not be the only causal mechanism of Aβ42 accumulation in LOAD, since at least two other mechanisms have been identified to be impaired by loss-of-function of the genetic risk factors: 1. defective clearance of Aβ42 through the BBB due to impaired endocytosis/transcytosis via sorting endosomes for ApoE4, CD2AP, CALM, and Sorla and 2. defective lysosomal clearance of Aβ42 for ApoE4, Sorla, PLD3 by neurons, and other brain cells. Additional mechanisms independent of Aβ may also occur in parallel, reflected by defects in glutamate receptors, cholesterol, and tau trafficking due to ApoE4, Sorla, and CALM. More research will be necessary to integrate the multiple ways by which the endocytic genetic risk factors contribute to AD development.
Most of the studies reviewed here used a knockdown or overexpression approach to study the role of the endocytic genetic risk factors in AD. The only variant associated with AD for which the impact on Aβ production has been determined is APOE4. It is critical in the future to identify functional variants for PICALM, BIN1, CD2AP, SORL1, and PLD3 to enable research aimed at validating or identifying the underlying mechanisms. Sequencing of such genetic risk factors has started identifying rare but predicted to be deleterious variants; however, the number of studies and patients sequenced is still very small. Moreover, given that AD is a human-specific disease, future research should consider using human neurons derived from patients or even genetically edited with patients’ mutations to dissect the causal mechanisms of LOAD.
Another aspect of major importance that should be addressed in the future is to determine whether the increase in Aβ42 triggered by the endocytic risk factors is sufficient to cause synaptic dysfunction, an earlier and functionally more relevant disease phenotype than amyloid plaques. Importantly, it is possible that aging together with the Aβ42 accumulation-triggered by genetic risk factors, will be sufficient to lead to the deposition of amyloid plaques, tangles formation and ultimately full-blown neurodegeneration. It is worthwhile mentioning the Model-AD initiative (https://model-ad.org/) which, by generating knock-in mice with the most promising genetic variants, may help to prove causality between endocytic deregulation and the development of LOAD.
Acknowledgements
We thank Inês Figueira for revising the manuscript. The Almeida lab has been supported by a Marie Curie Integration Grant (334366 TrafficInAD FP7-PEOPLE-2012-CIG; Marie Curie Actions, EC). iNOVA4Health—UID/Multi/04462/2013, a program financially supported by Fundação para a Ciência e Tecnologia (FCT)/Ministério da Educação e Ciência, through national funds and co-funded by FEDER under the PT2020 Partnership Agreement is acknowledged. CGA is funded by Investigator FCT (IF/00998/2012, FCT). FM has been the recipient of an FCT doctoral fellowship (PD/BD/128344/2017), CP has been the recipient of an FCT doctoral fellowship (SFRH/BD/128374/2017) and TB has been the recipient of an FCT doctoral fellowship (SFRH/BD/131513/2017).
Bibliography
- 1.Tampellini D, Gouras GK. Synapses, synaptic activity and intraneuronal abeta in Alzheimer’s disease. Front Aging Neurosci. 2010 doi: 10.3389/fnagi.2010.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benilova I, Karran E, De Strooper B. The toxic Aβ oligomer and Alzheimer’s disease: an emperor in need of clothes. Nat Neurosci. 2012;15:349–357. doi: 10.1038/nn.3028. [DOI] [PubMed] [Google Scholar]
- 3.Almeida CG, Tampellini D, Takahashi RH, et al. Beta-amyloid accumulation in APP mutant neurons reduces PSD-95 and GluR1 in synapses. Neurobiol Dis. 2005;20:187–198. doi: 10.1016/j.nbd.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 4.Snyder EM, Nong Y, Almeida CG, et al. Regulation of NMDA receptor trafficking by amyloid-beta. Nat Neurosci. 2005;8:1051–1058. doi: 10.1038/nn1503. [DOI] [PubMed] [Google Scholar]
- 5.Mucke L, Selkoe DJ. Neurotoxicity of amyloid β-protein: synaptic and network dysfunction. Cold Spring Harb Perspect Med. 2012;2:a006338. doi: 10.1101/cshperspect.a006338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Takahashi RH, Nagao T, Gouras GK. Plaque formation and the intraneuronal accumulation of β-amyloid in Alzheimer’s disease. Pathol Int. 2017;67:185–193. doi: 10.1111/pin.12520. [DOI] [PubMed] [Google Scholar]
- 7.Takahashi RH, Milner TA, Li F, et al. Intraneuronal Alzheimer abeta42 accumulates in multivesicular bodies and is associated with synaptic pathology. Am J Pathol. 2002;161:1869–1879. doi: 10.1016/S0002-9440(10)64463-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pensalfini A, Albay R, Rasool S, et al. Intracellular amyloid and the neuronal origin of Alzheimer neuritic plaques. Neurobiol Dis. 2014;71:53–61. doi: 10.1016/j.nbd.2014.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gouras GK, Almeida CG, Takahashi RH. Intraneuronal Abeta accumulation and origin of plaques in Alzheimer’s disease. Neurobiol Aging. 2005;26:1235–1244. doi: 10.1016/j.neurobiolaging.2005.05.022. [DOI] [PubMed] [Google Scholar]
- 10.Almeida CG, Takahashi RH, Gouras GK. Beta-amyloid accumulation impairs multivesicular body sorting by inhibiting the ubiquitin-proteasome system. J Neurosci. 2006;26:4277–4288. doi: 10.1523/JNEUROSCI.5078-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sahlin C, Lord A, Magnusson K, et al. The Arctic Alzheimer mutation favors intracellular amyloid-beta production by making amyloid precursor protein less available to alpha-secretase. J Neurochem. 2007;101:854–862. doi: 10.1111/j.1471-4159.2006.04443.x. [DOI] [PubMed] [Google Scholar]
- 12.Norvin D, Kim G, Baker-Nigh A, Geula C. Accumulation and age-related elevation of amyloid-β within basal forebrain cholinergic neurons in the rhesus monkey. Neuroscience. 2015;298:102–111. doi: 10.1016/j.neuroscience.2015.04.011. [DOI] [PubMed] [Google Scholar]
- 13.LaFerla FM, Green KN, Oddo S. Intracellular amyloid-beta in Alzheimer’s disease. Nat Rev Neurosci. 2007;8:499–509. doi: 10.1038/nrn2168. [DOI] [PubMed] [Google Scholar]
- 14.Mecozzi VJ, Berman DE, Simoes S, et al. Pharmacological chaperones stabilize retromer to limit APP processing. Nat Chem Biol. 2014;10:443–449. doi: 10.1038/nchembio.1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matsuda S, Matsuda Y, Snapp EL, D’Adamio L. Maturation of BRI2 generates a specific inhibitor that reduces APP processing at the plasma membrane and in endocytic vesicles. Neurobiol Aging. 2011;32:1400–1408. doi: 10.1016/j.neurobiolaging.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun M, Asghar SZ, Zhang H. The polarity protein Par3 regulates APP trafficking and processing through the endocytic adaptor protein Numb. Neurobiol Dis. 2016;93:1–11. doi: 10.1016/j.nbd.2016.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang X, Song W. The role of APP and BACE1 trafficking in APP processing and amyloid-β generation. Alzheimers Res Ther. 2013;5:46. doi: 10.1186/alzrt211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Takahashi K, Niidome T, Akaike A, et al. Phosphorylation of amyloid precursor protein (APP) at Tyr687 regulates APP processing by alpha- and gamma-secretase. Biochem Biophys Res Commun. 2008;377:544–549. doi: 10.1016/j.bbrc.2008.10.013. [DOI] [PubMed] [Google Scholar]
- 19.Tammineni P, Jeong YY, Feng T, et al. Impaired axonal retrograde trafficking of the retromer complex augments lysosomal deficits in Alzheimer’s disease neurons. Hum Mol Genet. 2017;26:4352–4366. doi: 10.1093/hmg/ddx321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saunders AM, Strittmatter WJ, Schmechel D, et al. Association of apolipoprotein E allele epsilon 4 with late-onset familial and sporadic Alzheimer’s disease. Neurology. 1993;43:1467–1472. doi: 10.1212/WNL.43.8.1467. [DOI] [PubMed] [Google Scholar]
- 21.Corder EH, Saunders AM, Strittmatter WJ, et al. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science. 1993;261:921–923. doi: 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- 22.Strittmatter WJ, Saunders AM, Schmechel D, et al. Apolipoprotein E: high-avidity binding to beta-amyloid and increased frequency of type 4 allele in late-onset familial Alzheimer disease. Proc Natl Acad Sci USA. 1993;90:1977–1981. doi: 10.1073/pnas.90.5.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rebeck GW, Reiter JS, Strickland DK, Hyman BT. Apolipoprotein E in sporadic Alzheimer’s disease: allelic variation and receptor interactions. Neuron. 1993;11:575–580. doi: 10.1016/0896-6273(93)90070-8. [DOI] [PubMed] [Google Scholar]
- 24.Chouraki V, Seshadri S, Theodore Friedmann JCD and SFG (2014) Chapter five—genetics of Alzheimer’s disease. Advances in Genetics. Academic Press, Cambridge, pp 245–294 [DOI] [PubMed]
- 25.Harold D, Abraham R, Hollingworth P, et al. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer’s disease. Nat Genet. 2009;41:1088–1093. doi: 10.1038/ng.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hollingworth P, Harold D, Sims R, et al. Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer’s disease. Nat Genet. 2011;43:429–435. doi: 10.1038/ng.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Naj AC, Jun G, Beecham GW, et al. Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer’s disease. Nat Genet. 2011;43:436–441. doi: 10.1038/ng.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lambert JC, Ibrahim-Verbaas CA, Harold D, et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat Genet. 2013;45:1452–1458. doi: 10.1038/ng.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cruchaga C, Karch CM, Jin SC, et al. Rare coding variants in the phospholipase D3 gene confer risk for Alzheimer’s disease. Nature. 2014;505:550–554. doi: 10.1038/nature12825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carmona S, Hardy J, Guerreiro R. The genetic landscape of Alzheimer disease. Handb Clin Neurol. 2018;148:395–408. doi: 10.1016/B978-0-444-64076-5.00026-0. [DOI] [PubMed] [Google Scholar]
- 31.Das U, Scott DA, Ganguly A, et al. Activity-induced convergence of APP and BACE-1 in acidic microdomains via an endocytosis-dependent pathway. Neuron. 2013;79:447–460. doi: 10.1016/j.neuron.2013.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ehehalt R, Keller P, Haass C, et al. Amyloidogenic processing of the Alzheimer beta-amyloid precursor protein depends on lipid rafts. J Cell Biol. 2003;160:113–123. doi: 10.1083/jcb.200207113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kalvodova L, Kahya N, Schwille P, et al. Lipids as modulators of proteolytic activity of BACE: involvement of cholesterol, glycosphingolipids, and anionic phospholipids in vitro. J Biol Chem. 2005;280:36815–36823. doi: 10.1074/jbc.M504484200. [DOI] [PubMed] [Google Scholar]
- 34.Koo EH, Squazzo SL. Evidence that production and release of amyloid beta-protein involves the endocytic pathway. J Biol Chem. 1994;269:17386–17389. [PubMed] [Google Scholar]
- 35.Lai A, Sisodia SS, Trowbridge IS. Characterization of sorting signals in the beta-amyloid precursor protein cytoplasmic domain. J Biol Chem. 1995;270:3565–3573. doi: 10.1074/jbc.270.8.3565. [DOI] [PubMed] [Google Scholar]
- 36.Van der Kant R, Goldstein LSB. Cellular functions of the amyloid precursor protein from development to dementia. Dev Cell. 2015;32:502–515. doi: 10.1016/j.devcel.2015.01.022. [DOI] [PubMed] [Google Scholar]
- 37.Schneider A, Rajendran L, Honsho M, et al. Flotillin-dependent clustering of the amyloid precursor protein regulates its endocytosis and amyloidogenic processing in neurons. J Neurosci. 2008;28:2874–2882. doi: 10.1523/JNEUROSCI.5345-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marquer C, Devauges V, Cossec J-C, et al. Local cholesterol increase triggers amyloid precursor protein-Bace1 clustering in lipid rafts and rapid endocytosis. FASEB J. 2011;25:1295–1305. doi: 10.1096/fj.10-168633. [DOI] [PubMed] [Google Scholar]
- 39.Kang MJ, Chung YH, Hwang CI, et al. Caveolin-1 upregulation in senescent neurons alters amyloid precursor protein processing. Exp Mol Med. 2006;38:126–133. doi: 10.1038/emm.2006.16. [DOI] [PubMed] [Google Scholar]
- 40.Zerbinatti CV, Bu G. LRP and Alzheimer’s disease. Rev Neurosci. 2005;16:123–135. doi: 10.1515/REVNEURO.2005.16.2.123. [DOI] [PubMed] [Google Scholar]
- 41.Pietrzik CU, Busse T, Merriam DE, et al. The cytoplasmic domain of the LDL receptor-related protein regulates multiple steps in APP processing. EMBO J. 2002;21:5691–5700. doi: 10.1093/emboj/cdf568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ye S, Huang Y, Müllendorff K, et al. Apolipoprotein (apo) E4 enhances amyloid beta peptide production in cultured neuronal cells: apoE structure as a potential therapeutic target. Proc Natl Acad Sci USA. 2005;102:18700–18705. doi: 10.1073/pnas.0508693102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sannerud R, Declerck I, Peric A, et al. ADP ribosylation factor 6 (ARF6) controls amyloid precursor protein (APP) processing by mediating the endosomal sorting of BACE1. Proc Natl Acad Sci USA. 2011;108:E559–E568. doi: 10.1073/pnas.1100745108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Prabhu Y, Burgos PV, Schindler C, et al. Adaptor protein 2-mediated endocytosis of the β-secretase BACE1 is dispensable for amyloid precursor protein processing. Mol Biol Cell. 2012;23:2339–2351. doi: 10.1091/mbc.E11-11-0944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chia PZC, Toh WH, Sharples R, et al. Intracellular itinerary of internalised β-secretase, BACE1, and its potential impact on β-amyloid peptide biogenesis. Traffic. 2013;14:997–1013. doi: 10.1111/tra.12088. [DOI] [PubMed] [Google Scholar]
- 46.Pastorino L, Ikin AF, Nairn AC, et al. The carboxyl-terminus of BACE contains a sorting signal that regulates BACE trafficking but not the formation of total A(beta) Mol Cell Neurosci. 2002;19:175–185. doi: 10.1006/mcne.2001.1065. [DOI] [PubMed] [Google Scholar]
- 47.He X, Zhu G, Koelsch G, et al. Biochemical and structural characterization of the interaction of memapsin 2 (beta-secretase) cytosolic domain with the VHS domain of GGA proteins. Biochemistry. 2003;42:12174–12180. doi: 10.1021/bi035199h. [DOI] [PubMed] [Google Scholar]
- 48.Cirrito JR, Kang J-E, Lee J, et al. Endocytosis is required for synaptic activity-dependent release of amyloid-beta in vivo. Neuron. 2008;58:42–51. doi: 10.1016/j.neuron.2008.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zou L, Wang Z, Shen L, et al. Receptor tyrosine kinases positively regulate BACE activity and Amyloid-beta production through enhancing BACE internalization. Cell Res. 2007;17:389–401. doi: 10.1038/cr.2007.5. [DOI] [PubMed] [Google Scholar]
- 50.Yan R, Vassar R. Targeting the β secretase BACE1 for Alzheimer’s disease therapy. Lancet Neurol. 2014;13:319–329. doi: 10.1016/S1474-4422(13)70276-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rajendran L, Schneider A, Schlechtingen G, et al. Efficient inhibition of the Alzheimer’s disease beta-secretase by membrane targeting. Science. 2008;320:520–523. doi: 10.1126/science.1156609. [DOI] [PubMed] [Google Scholar]
- 52.Choy RW-Y, Cheng Z, Schekman R. Amyloid precursor protein (APP) traffics from the cell surface via endosomes for amyloid β (Aβ) production in the trans-Golgi network. Proc Natl Acad Sci USA. 2012;109:E2077–E2082. doi: 10.1073/pnas.1208635109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Buggia-Prévot V, Fernandez CG, Udayar V, et al. A function for EHD family proteins in unidirectional retrograde dendritic transport of BACE1 and Alzheimer’s disease Aβ production. Cell Rep. 2013;5:1552–1563. doi: 10.1016/j.celrep.2013.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Morel E, Chamoun Z, Lasiecka ZM, et al. Phosphatidylinositol-3-phosphate regulates sorting and processing of amyloid precursor protein through the endosomal system. Nat Commun. 2013;4:2250. doi: 10.1038/ncomms3250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sannerud R, Esselens C, Ejsmont P, et al. Restricted Location of PSEN2/γ-Secretase Determines Substrate Specificity and Generates an Intracellular Aβ Pool. Cell. 2016;166:193–208. doi: 10.1016/j.cell.2016.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Edgar JR, Willén K, Gouras GK, Futter CE. ESCRTs regulate amyloid precursor protein sorting in multivesicular bodies and intracellular amyloid-β accumulation. J Cell Sci. 2015;128:2520–2528. doi: 10.1242/jcs.170233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Udayar V, Buggia-Prévot V, Guerreiro RL, et al. A paired RNAi and RabGAP overexpression screen identifies Rab11 as a regulator of β-amyloid production. Cell Rep. 2013;5:1536–1551. doi: 10.1016/j.celrep.2013.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Takahashi RH, Almeida CG, Kearney PF, et al. Oligomerization of Alzheimer’s beta-amyloid within processes and synapses of cultured neurons and brain. J Neurosci. 2004;24:3592–3599. doi: 10.1523/JNEUROSCI.5167-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Oropeza RL, Wekerle H, Werb Z. Expression of apolipoprotein E by mouse brain astrocytes and its modulation by interferon-gamma. Brain Res. 1987;410:45–51. doi: 10.1016/S0006-8993(87)80018-5. [DOI] [PubMed] [Google Scholar]
- 60.Bu G. Apolipoprotein E and its receptors in Alzheimer’s disease: pathways, pathogenesis and therapy. Nat Rev Neurosci. 2009;10:333–344. doi: 10.1038/nrn2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Conejero-Goldberg C, Gomar JJ, Bobes-Bascaran T, et al. APOE2 enhances neuroprotection against Alzheimer’s disease through multiple molecular mechanisms. Mol Psychiatry. 2014;19:1243–1250. doi: 10.1038/mp.2013.194. [DOI] [PubMed] [Google Scholar]
- 62.He X, Cooley K, Chung CHY, et al. Apolipoprotein receptor 2 and X11 alpha/beta mediate apolipoprotein E-induced endocytosis of amyloid-beta precursor protein and beta-secretase, leading to amyloid-beta production. J Neurosci. 2007;27:4052–4060. doi: 10.1523/JNEUROSCI.3993-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hoe H-S, Pocivavsek A, Dai H, et al. Effects of apoE on neuronal signaling and APP processing in rodent brain. Brain Res. 2006;1112:70–79. doi: 10.1016/j.brainres.2006.07.035. [DOI] [PubMed] [Google Scholar]
- 64.Irizarry MC, Deng A, Lleo A, et al. Apolipoprotein E modulates gamma-secretase cleavage of the amyloid precursor protein. J Neurochem. 2004;90:1132–1143. doi: 10.1111/j.1471-4159.2004.02581.x. [DOI] [PubMed] [Google Scholar]
- 65.Hopkins PCR, Sáinz-Fuertes R, Lovestone S. The impact of a novel apolipoprotein E and amyloid-β protein precursor-interacting protein on the production of amyloid-β. J Alzheimers Dis. 2011;26:239–253. doi: 10.3233/JAD-2011-102115. [DOI] [PubMed] [Google Scholar]
- 66.Cataldo AM, Peterhoff CM, Troncoso JC, et al. Endocytic pathway abnormalities precede amyloid β deposition in sporadic Alzheimer’s disease and Down syndrome: differential effects of APOE genotype and presenilin mutations. Am J Pathol. 2000;157:277–286. doi: 10.1016/S0002-9440(10)64538-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhao N, Liu C-C, Van Ingelgom AJ, et al. Apolipoprotein E4 impairs neuronal insulin signaling by trapping insulin receptor in the endosomes. Neuron. 2017;96(115–129):e5. doi: 10.1016/j.neuron.2017.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rapp A, Gmeiner B, Hüttinger M. Implication of apoE isoforms in cholesterol metabolism by primary rat hippocampal neurons and astrocytes. Biochimie. 2006;88:473–483. doi: 10.1016/j.biochi.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 69.Huang Y-WA, Zhou B, Wernig M, Südhof TC. Apoe2, apoe3, and apoe4 differentially stimulate APP transcription and aβ secretion. Cell. 2017;168(427–441):e21. doi: 10.1016/j.cell.2016.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Castellano JM, Kim J, Stewart FR, et al. Human apoE isoforms differentially regulate brain amyloid-β peptide clearance. Sci Transl Med. 2011;3:89ra57. doi: 10.1126/scitranslmed.3002156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fryer JD, Simmons K, Parsadanian M, et al. Human apolipoprotein E4 alters the amyloid-beta 40:42 ratio and promotes the formation of cerebral amyloid angiopathy in an amyloid precursor protein transgenic model. J Neurosci. 2005;25:2803–2810. doi: 10.1523/JNEUROSCI.5170-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wildsmith KR, Holley M, Savage JC, et al. Evidence for impaired amyloid β clearance in Alzheimer’s disease. Alzheimers Res Ther. 2013;5:33. doi: 10.1186/alzrt187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Verghese PB, Castellano JM, Garai K, et al. ApoE influences amyloid-β (Aβ) clearance despite minimal apoE/Aβ association in physiological conditions. Proc Natl Acad Sci USA. 2013;110:E1807–E1816. doi: 10.1073/pnas.1220484110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chen Y, Durakoglugil MS, Xian X, Herz J. ApoE4 reduces glutamate receptor function and synaptic plasticity by selectively impairing ApoE receptor recycling. Proc Natl Acad Sci USA. 2010;107:12011–12016. doi: 10.1073/pnas.0914984107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jun G, Naj AC, Beecham GW, et al. Meta-analysis confirms CR1, CLU, and PICALM as alzheimer disease risk loci and reveals interactions with APOE genotypes. Arch Neurol. 2010;67:1473–1484. doi: 10.1001/archneurol.2010.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Carrasquillo MM, Belbin O, Hunter TA, et al. Replication of CLU, CR1, and PICALM associations with alzheimer disease. Arch Neurol. 2010;67:961–964. doi: 10.1001/archneurol.2010.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ferrari R, Moreno JH, Minhajuddin AT, et al. Implication of common and disease specific variants in CLU, CR1, and PICALM. Neurobiol Aging. 2012;33(1846):e7–e18. doi: 10.1016/j.neurobiolaging.2012.01.110. [DOI] [PubMed] [Google Scholar]
- 78.Moreno DJ, Ruiz S, Ríos Á, et al. Association of GWAS top genes with late-onset Alzheimer’s disease in Colombian population. Am J Alzheimers Dis Other Demen. 2017;32:27–35. doi: 10.1177/1533317516679303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Naj AC, Jun G, Reitz C, et al. Effects of multiple genetic loci on age at onset in late-onset Alzheimer disease: a genome-wide association study. JAMA Neurol. 2014;71:1394–1404. doi: 10.1001/jamaneurol.2014.1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lee JH, Cheng R, Barral S, et al. Identification of novel loci for Alzheimer disease and replication of CLU, PICALM, and BIN1 in Caribbean Hispanic individuals. Arch Neurol. 2011;68:320–328. doi: 10.1001/archneurol.2010.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wang Z, Lei H, Zheng M, et al. Meta-analysis of the Association between Alzheimer Disease and Variants in GAB2, PICALM, and SORL1. Mol Neurobiol. 2016;53:6501–6510. doi: 10.1007/s12035-015-9546-y. [DOI] [PubMed] [Google Scholar]
- 82.Xu W, Wang H-F, Tan L, et al. The impact of PICALM genetic variations on reserve capacity of posterior cingulate in AD continuum. Sci Rep. 2016;6:24480. doi: 10.1038/srep24480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mengel-From J, Christensen K, McGue M, Christiansen L. Genetic variations in the CLU and PICALM genes are associated with cognitive function in the oldest old. Neurobiol Aging. 2011;32(554):e7–e11. doi: 10.1016/j.neurobiolaging.2010.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Biffi A, Anderson CD, Desikan RS, et al. Genetic variation and neuroimaging measures in Alzheimer disease. Arch Neurol. 2010;67:677–685. doi: 10.1001/archneurol.2010.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Dreyling MH, Martinez-Climent JA, Zheng M, et al. The t(10;11)(p13;q14) in the U937 cell line results in the fusion of the AF10 gene and CALM, encoding a new member of the AP-3 clathrin assembly protein family. Proc Natl Acad Sci USA. 1996;93:4804–4809. doi: 10.1073/pnas.93.10.4804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Miller SE, Sahlender DA, Graham SC, et al. The molecular basis for the endocytosis of small R-SNAREs by the clathrin adaptor CALM. Cell. 2011;147:1118–1131. doi: 10.1016/j.cell.2011.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yao PJ, Petralia RS, Bushlin I, et al. Synaptic distribution of the endocytic accessory proteins AP180 and CALM. J Comp Neurol. 2005;481:58–69. doi: 10.1002/cne.20362. [DOI] [PubMed] [Google Scholar]
- 88.Vanlandingham PA, Barmchi MP, Royer S, et al. AP180 couples protein retrieval to clathrin-mediated endocytosis of synaptic vesicles. Traffic. 2014;15:433–450. doi: 10.1111/tra.12153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Meyerholz A, Hinrichsen L, Groos S, et al. Effect of clathrin assembly lymphoid myeloid leukemia protein depletion on clathrin coat formation. Traffic. 2005;6:1225–1234. doi: 10.1111/j.1600-0854.2005.00355.x. [DOI] [PubMed] [Google Scholar]
- 90.Sahlender DA, Kozik P, Miller SE, et al. Uncoupling the functions of CALM in VAMP sorting and clathrin-coated pit formation. PLoS One. 2013;8:e64514. doi: 10.1371/journal.pone.0064514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Koo SJ, Markovic S, Puchkov D, et al. SNARE motif-mediated sorting of synaptobrevin by the endocytic adaptors clathrin assembly lymphoid myeloid leukemia (CALM) and AP180 at synapses. Proc Natl Acad Sci USA. 2011;108:13540–13545. doi: 10.1073/pnas.1107067108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ando K, Brion J-P, Stygelbout V, et al. Clathrin adaptor CALM/PICALM is associated with neurofibrillary tangles and is cleaved in Alzheimer’s brains. Acta Neuropathol. 2013;125:861–878. doi: 10.1007/s00401-013-1111-z. [DOI] [PubMed] [Google Scholar]
- 93.Thomas RS, Lelos MJ, Good MA, Kidd EJ. Clathrin-mediated endocytic proteins are upregulated in the cortex of the Tg2576 mouse model of Alzheimer’s disease-like amyloid pathology. Biochem Biophys Res Commun. 2011;415:656–661. doi: 10.1016/j.bbrc.2011.10.131. [DOI] [PubMed] [Google Scholar]
- 94.Parikh I, Fardo DW, Estus S. Genetics of PICALM expression and Alzheimer’s disease. PLoS One. 2014;9:e91242. doi: 10.1371/journal.pone.0091242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Xiao Q, Gil S-C, Yan P, et al. Role of phosphatidylinositol clathrin assembly lymphoid-myeloid leukemia (PICALM) in intracellular amyloid precursor protein (APP) processing and amyloid plaque pathogenesis. J Biol Chem. 2012;287:21279–21289. doi: 10.1074/jbc.M111.338376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Boehm C, Kaden D, St. George-Hyslop P. Picalm but not bin1 alters the secretion of beta-amyloid peptide. Alzheimers Dement. 2012;8:P652. doi: 10.1016/j.jalz.2012.05.2175. [DOI] [Google Scholar]
- 97.Thomas RS, Henson A, Gerrish A, et al. Decreasing the expression of PICALM reduces endocytosis and the activity of β-secretase: implications for Alzheimer’s disease. BMC Neurosci. 2016;17:50. doi: 10.1186/s12868-016-0288-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kanatsu K, Morohashi Y, Suzuki M, et al. Decreased CALM expression reduces Aβ42 to total Aβ ratio through clathrin-mediated endocytosis of γ-secretase. Nat Commun. 2014;5:3386. doi: 10.1038/ncomms4386. [DOI] [PubMed] [Google Scholar]
- 99.Kanatsu K, Hori Y, Takatori S, et al. Partial loss of CALM function reduces Aβ42 production and amyloid deposition in vivo. Hum Mol Genet. 2016;25:3988–3997. doi: 10.1093/hmg/ddw239. [DOI] [PubMed] [Google Scholar]
- 100.Tian Y, Chang JC, Fan EY, et al. Adaptor complex AP2/PICALM, through interaction with LC3, targets Alzheimer’s APP-CTF for terminal degradation via autophagy. Proc Natl Acad Sci USA. 2013;110:17071–17076. doi: 10.1073/pnas.1315110110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhao Z, Sagare AP, Ma Q, et al. Central role for PICALM in amyloid-β blood-brain barrier transcytosis and clearance. Nat Neurosci. 2015;18:978–987. doi: 10.1038/nn.4025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Gimber N, Tadeus G, Maritzen T, et al. Diffusional spread and confinement of newly exocytosed synaptic vesicle proteins. Nat Commun. 2015;6:8392. doi: 10.1038/ncomms9392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Mercer JL, Argus JP, Crabtree DM, et al. Modulation of PICALM levels perturbs cellular cholesterol homeostasis. PLoS One. 2015;10:e0129776. doi: 10.1371/journal.pone.0129776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Seshadri S, Fitzpatrick AL, Ikram MA, et al. Genome-wide analysis of genetic loci associated with Alzheimer disease. JAMA. 2010;303:1832–1840. doi: 10.1001/jama.2010.574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wijsman EM, Pankratz ND, Choi Y, et al. Genome-wide association of familial late-onset Alzheimer’s disease replicates BIN1 and CLU and nominates CUGBP2 in interaction with APOE. PLoS Genet. 2011;7:e1001308. doi: 10.1371/journal.pgen.1001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hu X, Pickering E, Liu YC, et al. Meta-analysis for genome-wide association study identifies multiple variants at the BIN1 locus associated with late-onset Alzheimer’s disease. PLoS One. 2011;6:e16616. doi: 10.1371/journal.pone.0016616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kamboh MI, Demirci FY, Wang X, et al. Genome-wide association study of Alzheimer’s disease. Transl Psychiatry. 2012;2:e117. doi: 10.1038/tp.2012.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Barral S, Bird T, Goate A, et al. Genotype patterns at PICALM, CR1, BIN1, CLU, and APOE genes are associated with episodic memory. Neurology. 2012;78:1464–1471. doi: 10.1212/WNL.0b013e3182553c48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Tan M-S, Yu J-T, Jiang T, et al. Genetic variation in BIN1 gene and Alzheimer’s disease risk in Han Chinese individuals. Neurobiol Aging. 2014;35(1781):e1–e8. doi: 10.1016/j.neurobiolaging.2014.01.151. [DOI] [PubMed] [Google Scholar]
- 110.Vardarajan BN, Ghani M, Kahn A, et al. Rare coding mutations identified by sequencing of Alzheimer disease genome-wide association studies loci. Ann Neurol. 2015;78:487–498. doi: 10.1002/ana.24466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Sakamuro D, Elliott KJ, Wechsler-Reya R, Prendergast GC. BIN1 is a novel MYC-interacting protein with features of a tumour suppressor. Nat Genet. 1996;14:69–77. doi: 10.1038/ng0996-69. [DOI] [PubMed] [Google Scholar]
- 112.David C, McPherson PS, Mundigl O, de Camilli P. A role of amphiphysin in synaptic vesicle endocytosis suggested by its binding to dynamin in nerve terminals. Proc Natl Acad Sci USA. 1996;93:331–335. doi: 10.1073/pnas.93.1.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Leprince C, Romero F, Cussac D, et al. A new member of the amphiphysin family connecting endocytosis and signal transduction pathways. J Biol Chem. 1997;272:15101–15105. doi: 10.1074/jbc.272.24.15101. [DOI] [PubMed] [Google Scholar]
- 114.Micheva KD, Kay BK, McPherson PS. Synaptojanin forms two separate complexes in the nerve terminal. Interactions with endophilin and amphiphysin. J Biol Chem. 1997;272:27239–27245. doi: 10.1074/jbc.272.43.27239. [DOI] [PubMed] [Google Scholar]
- 115.Prokic I, Cowling BS, Laporte J. Amphiphysin 2 (BIN1) in physiology and diseases. J Mol Med. 2014;92:453–463. doi: 10.1007/s00109-014-1138-1. [DOI] [PubMed] [Google Scholar]
- 116.Ramjaun AR, Micheva KD, Bouchelet I, McPherson PS. Identification and characterization of a nerve terminal-enriched amphiphysin isoform. J Biol Chem. 1997;272:16700–16706. doi: 10.1074/jbc.272.26.16700. [DOI] [PubMed] [Google Scholar]
- 117.Butler MH, David C, Ochoa GC, et al. Amphiphysin II (SH3P9; BIN1), a member of the amphiphysin/Rvs family, is concentrated in the cortical cytomatrix of axon initial segments and nodes of ranvier in brain and around T tubules in skeletal muscle. J Cell Biol. 1997;137:1355–1367. doi: 10.1083/jcb.137.6.1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wigge P, Köhler K, Vallis Y, et al. Amphiphysin heterodimers: potential role in clathrin-mediated endocytosis. Mol Biol Cell. 1997;8:2003–2015. doi: 10.1091/mbc.8.10.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ramjaun AR, McPherson PS. Multiple amphiphysin II splice variants display differential clathrin binding: identification of two distinct clathrin-binding sites. J Neurochem. 1998;70:2369–2376. doi: 10.1046/j.1471-4159.1998.70062369.x. [DOI] [PubMed] [Google Scholar]
- 120.Di Paolo G, Sankaranarayanan S, Wenk MR, et al. Decreased synaptic vesicle recycling efficiency and cognitive deficits in amphiphysin 1 knockout mice. Neuron. 2002;33:789–804. doi: 10.1016/S0896-6273(02)00601-3. [DOI] [PubMed] [Google Scholar]
- 121.Muller AJ, Baker JF, DuHadaway JB, et al. Targeted disruption of the murine Bin1/Amphiphysin II gene does not disable endocytosis but results in embryonic cardiomyopathy with aberrant myofibril formation. Mol Cell Biol. 2003;23:4295–4306. doi: 10.1128/MCB.23.12.4295-4306.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Pant S, Sharma M, Patel K, et al. AMPH-1/Amphiphysin/Bin1 functions with RME-1/Ehd1 in endocytic recycling. Nat Cell Biol. 2009;11:1399–1410. doi: 10.1038/ncb1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Chapuis J, Hansmannel F, Gistelinck M, et al. Increased expression of BIN1 mediates Alzheimer genetic risk by modulating tau pathology. Mol Psychiatry. 2013;18:1225–1234. doi: 10.1038/mp.2013.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Karch CM, Jeng AT, Nowotny P, et al. Expression of novel Alzheimer’s disease risk genes in control and Alzheimer’s disease brains. PLoS One. 2012;7:e50976. doi: 10.1371/journal.pone.0050976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Glennon EBC, Whitehouse IJ, Miners JS, et al. BIN1 is decreased in sporadic but not familial Alzheimer’s disease or in aging. PLoS One. 2013;8:e78806. doi: 10.1371/journal.pone.0078806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Holler CJ, Davis PR, Beckett TL, et al. Bridging integrator 1 (BIN1) protein expression increases in the Alzheimer’s disease brain and correlates with neurofibrillary tangle pathology. J Alzheimers Dis. 2014;42:1221–1227. doi: 10.3233/JAD-132450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.De Rossi P, Buggia-Prévot V, Clayton BLL, et al. Predominant expression of Alzheimer’s disease-associated BIN1 in mature oligodendrocytes and localization to white matter tracts. Mol Neurodegener. 2016;11:59. doi: 10.1186/s13024-016-0124-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Miyagawa T, Ebinuma I, Morohashi Y, et al. BIN1 regulates BACE1 intracellular trafficking and amyloid-β production. Hum Mol Genet. 2016;25:2948–2958. doi: 10.1093/hmg/ddw146. [DOI] [PubMed] [Google Scholar]
- 129.Ubelmann F, Burrinha T, Salavessa L, et al. Bin1 and CD2AP polarise the endocytic generation of beta-amyloid. EMBO Rep. 2017;18:102–122. doi: 10.15252/embr.201642738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Calafate S, Flavin W, Verstreken P, Moechars D. Loss of bin1 promotes the propagation of tau pathology. Cell Rep. 2016;17:931–940. doi: 10.1016/j.celrep.2016.09.063. [DOI] [PubMed] [Google Scholar]
- 131.Shulman JM, Chen K, Keenan BT, et al. Genetic susceptibility for Alzheimer disease neuritic plaque pathology. JAMA Neurol. 2013;70:1150–1157. doi: 10.1001/jamaneurol.2013.2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Chen H, Wu G, Jiang Y, et al. Analyzing 54,936 samples supports the association between CD2AP rs9349407 polymorphism and Alzheimer’s disease susceptibility. Mol Neurobiol. 2015;52:1–7. doi: 10.1007/s12035-014-8834-2. [DOI] [PubMed] [Google Scholar]
- 133.Dustin ML, Olszowy MW, Holdorf AD, et al. A novel adaptor protein orchestrates receptor patterning and cytoskeletal polarity in T-cell contacts. Cell. 1998;94:667–677. doi: 10.1016/S0092-8674(00)81608-6. [DOI] [PubMed] [Google Scholar]
- 134.Cormont M, Metón I, Mari M, et al. CD2AP/CMS regulates endosome morphology and traffic to the degradative pathway through its interaction with Rab4 and c-Cbl. Traffic. 2003;4:97–112. doi: 10.1034/j.1600-0854.2003.40205.x. [DOI] [PubMed] [Google Scholar]
- 135.Gauthier NC, Monzo P, Gonzalez T, et al. Early endosomes associated with dynamic F-actin structures are required for late trafficking of H. pylori VacA toxin. J Cell Biol. 2007;177:343–354. doi: 10.1083/jcb.200609061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Monzo P, Mari M, Kaddai V, et al. (2005) CD2AP, Rabip4, and Rabip4′: Analysis of Interaction with Rab4a and Regulation of Endosomes Morphology. Meth Enzymol. Academic Press, Cambridge, pp 107–118 [DOI] [PubMed]
- 137.Kobayashi S, Sawano A, Nojima Y, et al. The c-Cbl/CD2AP complex regulates VEGF-induced endocytosis and degradation of Flt-1 (VEGFR-1) FASEB J. 2004;18:929–931. doi: 10.1096/fj.03-0767fje. [DOI] [PubMed] [Google Scholar]
- 138.Lynch DK, Winata SC, Lyons RJ, et al. A Cortactin-CD2-associated protein (CD2AP) complex provides a novel link between epidermal growth factor receptor endocytosis and the actin cytoskeleton. J Biol Chem. 2003;278:21805–21813. doi: 10.1074/jbc.M211407200. [DOI] [PubMed] [Google Scholar]
- 139.Tang VW, Brieher WM. FSGS3/CD2AP is a barbed-end capping protein that stabilizes actin and strengthens adherens junctions. J Cell Biol. 2013;203:815–833. doi: 10.1083/jcb.201304143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Zhao J, Bruck S, Cemerski S, et al. CD2AP links cortactin and capping protein at the cell periphery to facilitate formation of lamellipodia. Mol Cell Biol. 2013;33:38–47. doi: 10.1128/MCB.00734-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Li C, Ruotsalainen V, Tryggvason K, et al. CD2AP is expressed with nephrin in developing podocytes and is found widely in mature kidney and elsewhere. Am J Physiol Renal Physiol. 2000;279:F785–F792. doi: 10.1152/ajprenal.2000.279.4.F785. [DOI] [PubMed] [Google Scholar]
- 142.Wolf G, Stahl RAK. CD2-associated protein and glomerular disease. The Lancet. 2003;362:1746–1748. doi: 10.1016/S0140-6736(03)14856-8. [DOI] [PubMed] [Google Scholar]
- 143.Peitsch WK, Hofmann I, Endlich N, et al. Cell biological and biochemical characterization of drebrin complexes in mesangial cells and podocytes of renal glomeruli. J Am Soc Nephrol. 2003;14:1452–1463. doi: 10.1097/01.ASN.0000069222.63700.DE. [DOI] [PubMed] [Google Scholar]
- 144.Kobayashi N. Mechanism of the process formation; podocytes vs. neurons. Microsc Res Tech. 2002;57:217–223. doi: 10.1002/jemt.10077. [DOI] [PubMed] [Google Scholar]
- 145.Tao Q-Q, Liu Z-J, Sun Y-M, et al. Decreased gene expression of CD2AP in Chinese patients with sporadic Alzheimer’s disease. Neurobiol Aging. 2017 doi: 10.1016/j.neurobiolaging.2017.03.013. [DOI] [PubMed] [Google Scholar]
- 146.Liao F, Jiang H, Srivatsan S, et al. Effects of CD2-associated protein deficiency on amyloid-β in neuroblastoma cells and in an APP transgenic mouse model. Mol Neurodegener. 2015;10:12. doi: 10.1186/s13024-015-0006-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Cochran JN, Rush T, Buckingham SC, Roberson ED. The Alzheimer’s disease risk factor CD2AP maintains blood-brain barrier integrity. Hum Mol Genet. 2015;24:6667–6674. doi: 10.1093/hmg/ddv371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Rogaeva E, Meng Y, Lee JH, et al. The neuronal sortilin-related receptor SORL1 is genetically associated with Alzheimer disease. Nat Genet. 2007;39:168–177. doi: 10.1038/ng1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Lee JH, Cheng R, Schupf N, et al. The association between genetic variants in SORL1 and Alzheimer disease in an urban, multiethnic, community-based cohort. Arch Neurol. 2007;64:501–506. doi: 10.1001/archneur.64.4.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Feng X, Hou D, Deng Y, et al. SORL1 gene polymorphism association with late-onset Alzheimer’s disease. Neurosci Lett. 2015;584:382–389. doi: 10.1016/j.neulet.2014.10.055. [DOI] [PubMed] [Google Scholar]
- 151.Piscopo P, Tosto G, Belli C, et al. SORL1 gene is associated with the conversion from mild cognitive impairment to Alzheimer’s disease. J Alzheimers Dis. 2015;46:771–776. doi: 10.3233/JAD-141551. [DOI] [PubMed] [Google Scholar]
- 152.Verheijen J, Van den Bossche T, van der Zee J, et al. A comprehensive study of the genetic impact of rare variants in SORL1 in European early-onset Alzheimer’s disease. Acta Neuropathol. 2016;132:213–224. doi: 10.1007/s00401-016-1566-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Miyashita A, Koike A, Jun G, et al. SORL1 is genetically associated with late-onset Alzheimer’s disease in Japanese, Koreans and Caucasians. PLoS One. 2013;8:e58618. doi: 10.1371/journal.pone.0058618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Young JE, Boulanger-Weill J, Williams DA, et al. Elucidating molecular phenotypes caused by the SORL1 Alzheimer’s disease genetic risk factor using human induced pluripotent stem cells. Cell Stem Cell. 2015;16:373–385. doi: 10.1016/j.stem.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Reitz C, Tokuhiro S, Clark LN, et al. SORCS1 alters amyloid precursor protein processing and variants may increase Alzheimer’s disease risk. Ann Neurol. 2011;69:47–64. doi: 10.1002/ana.22308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Schmidt V, Subkhangulova A, Willnow TE. Sorting receptor SORLA: cellular mechanisms and implications for disease. Cell Mol Life Sci. 2017;74:1475–1483. doi: 10.1007/s00018-016-2410-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Klinger SC, Højland A, Jain S, et al. Polarized trafficking of the sorting receptor SorLA in neurons and MDCK cells. FEBS J. 2016;283:2476–2493. doi: 10.1111/febs.13758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Caglayan S, Takagi-Niidome S, Liao F, et al. Lysosomal sorting of amyloid-β by the SORLA receptor is impaired by a familial Alzheimer’s disease mutation. Sci Transl Med. 2014;6:223ra20. doi: 10.1126/scitranslmed.3007747. [DOI] [PubMed] [Google Scholar]
- 159.Andersen OM, Rudolph I-M, Willnow TE. Risk factor SORL1: from genetic association to functional validation in Alzheimer’s disease. Acta Neuropathol. 2016;132:653–665. doi: 10.1007/s00401-016-1615-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Yu L, Chibnik LB, Srivastava GP, et al. Association of Brain DNA methylation in SORL1, ABCA7, HLA-DRB5, SLC24A4, and BIN1 with pathological diagnosis of Alzheimer disease. JAMA Neurol. 2015;72:15–24. doi: 10.1001/jamaneurol.2014.3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Grear KE, Ling I-F, Simpson JF, et al. Expression of SORL1 and a novel SORL1 splice variant in normal and Alzheimers disease brain. Mol Neurodegener. 2009;4:46. doi: 10.1186/1750-1326-4-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Fjorback AW, Seaman M, Gustafsen C, et al. Retromer binds the FANSHY sorting motif in SorLA to regulate amyloid precursor protein sorting and processing. J Neurosci. 2012;32:1467–1480. doi: 10.1523/JNEUROSCI.2272-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Dodson SE, Andersen OM, Karmali V, et al. Loss of LR11/SORLA enhances early pathology in a mouse model of amyloidosis: evidence for a proximal role in Alzheimer’s disease. J Neurosci. 2008;28:12877–12886. doi: 10.1523/JNEUROSCI.4582-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Bhalla A, Vetanovetz CP, Morel E, et al. The location and trafficking routes of the neuronal retromer and its role in amyloid precursor protein transport. Neurobiol Dis. 2012;47:126–134. doi: 10.1016/j.nbd.2012.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Bonifacino JS, Hurley JH. Retromer. Curr Opin Cell Biol. 2008;20:427–436. doi: 10.1016/j.ceb.2008.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Eggert S, Gonzalez AC, Thomas C, et al. Dimerization leads to changes in APP (amyloid precursor protein) trafficking mediated by LRP1 and SorLA. Cell Mol Life Sci. 2017;75:1–22. doi: 10.1007/s00018-017-2625-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Vieira SI, Rebelo S, Esselmann H, et al. Retrieval of the Alzheimer’s amyloid precursor protein from the endosome to the TGN is S655 phosphorylation state-dependent and retromer-mediated. Mol Neurodegener. 2010;5:40. doi: 10.1186/1750-1326-5-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Yajima R, Tokutake T, Koyama A, et al. ApoE-isoform-dependent cellular uptake of amyloid-β is mediated by lipoprotein receptor LR11/SorLA. Biochem Biophys Res Commun. 2015;456:482–488. doi: 10.1016/j.bbrc.2014.11.111. [DOI] [PubMed] [Google Scholar]
- 169.Na J-Y, Song K, Lee J-W, et al. Sortilin-related receptor 1 interacts with amyloid precursor protein and is activated by 6-shogaol, leading to inhibition of the amyloidogenic pathway. Biochem Biophys Res Commun. 2017;484:890–895. doi: 10.1016/j.bbrc.2017.02.029. [DOI] [PubMed] [Google Scholar]
- 170.Schulte EC, Kurz A, Alexopoulos P, et al. Excess of rare coding variants in PLD3 in late- but not early-onset Alzheimer’s disease. Hum Genome Var. 2015;2:14028. doi: 10.1038/hgv.2014.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Van der Lee SJ, Holstege H, Wong TH, et al. PLD3 variants in population studies. Nature. 2015;520:E2–E3. doi: 10.1038/nature14038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Cacace R, Van den Bossche T, Engelborghs S, et al. Rare variants in PLD3 do not affect risk for early-onset Alzheimer disease in a European consortium cohort. Hum Mutat. 2015;36:1226–1235. doi: 10.1002/humu.22908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Wang C, Wang H-F, Tan M-S, et al. Impact of common variations in PLD3 on neuroimaging phenotypes in non-demented elders. Mol Neurobiol. 2016;53:4343–4351. doi: 10.1007/s12035-015-9370-4. [DOI] [PubMed] [Google Scholar]
- 174.Lin E, Tsai S-J, Kuo P-H, et al. Association and interaction effects of Alzheimer’s disease-associated genes and lifestyle on cognitive aging in older adults in a Taiwanese population. Oncotarget. 2017;8:24077–24087. doi: 10.18632/oncotarget.15269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Donaldson JG. Phospholipase D in endocytosis and endosomal recycling pathways. Biochim Biophys Acta. 2009;1791:845–849. doi: 10.1016/j.bbalip.2009.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Jenkins GM, Frohman MA. Phospholipase D: a lipid centric review. Cell Mol Life Sci. 2005;62:2305–2316. doi: 10.1007/s00018-005-5195-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Osisami M, Ali W, Frohman MA. A role for phospholipase D3 in myotube formation. PLoS One. 2012;7:e33341. doi: 10.1371/journal.pone.0033341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Pedersen KM, Finsen B, Celis JE, Jensen NA. Expression of a novel murine phospholipase D homolog coincides with late neuronal development in the forebrain. J Biol Chem. 1998;273:31494–31504. doi: 10.1074/jbc.273.47.31494. [DOI] [PubMed] [Google Scholar]
- 179.Satoh J-I, Kino Y, Yamamoto Y, et al. PLD3 is accumulated on neuritic plaques in Alzheimer’s disease brains. Alzheimers Res Ther. 2014;6:70. doi: 10.1186/s13195-014-0070-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Nagaoka-Yasuda R, Matsuo N, Perkins B, et al. An RNAi-based genetic screen for oxidative stress resistance reveals retinol saturase as a mediator of stress resistance. Free Radic Biol Med. 2007;43:781–788. doi: 10.1016/j.freeradbiomed.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 181.Fazzari P, Horre K, Arranz AM, et al. PLD3 gene and processing of APP. Nature. 2017;541:E1–E2. doi: 10.1038/nature21030. [DOI] [PubMed] [Google Scholar]