Abstract
The maternal-to-zygotic transition (MZT) is essential for the developmental control handed from maternal products to newly synthesized zygotic genome in the earliest stages of embryogenesis, including maternal component (mRNAs and proteins) degradation and zygotic genome activation (ZGA). Various protein post-translational modifications have been identified during the MZT, such as phosphorylation, methylation and ubiquitination. Precise post-translational regulation mechanisms are essential for the timely transition of early embryonic development. In this review, we summarize recent progress regarding the molecular mechanisms underlying post-translational regulation of maternal component degradation and ZGA during the MZT and discuss some important issues in the field.
Keywords: MZT, ZGA, Phosphorylation, Ubiquitination, Histone modification
Introduction
Embryogenesis begins with the fertilization of a haploid egg by a haploid sperm. Then, a highly coordinated cascade of events is initiated in the earliest stages of embryogenesis. Maternal mRNAs and proteins accumulated in the egg during oogenesis control almost all aspects of initial embryonic development, but transcription of the zygotic genome is quiescent. After several rapid cell divisions, maternal mRNAs and proteins are eliminated, the zygotic genome is activated, and developmental control is transferred from maternal components to the gene products synthesized from the zygotic genome, defined as maternal-to-zygotic transition (MZT) [1–5]. The MZT involves two major processes, including the clearance of maternal mRNA and proteins, which are necessary for oocyte maturation and the earliest stages of embryogenesis but become unnecessary or possibly harmful in embryonic development, and zygotic genome activation (ZGA), which establishes new zygotic instructions for embryogenesis [1, 2, 4].
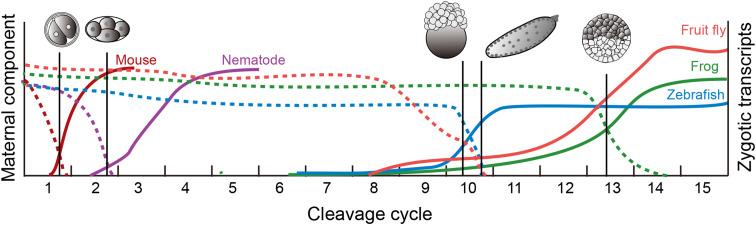
During the MZT, there are extensive variations in the timing and duration of these events among different species, which occur between 2 h (Drosophila) and 2 days (mouse) after fertilization (Fig. 1) [2]. In Caenorhabditis elegans, about 30% maternal mRNAs appear to decay before the four-cell stage, and the first zygotic transcription can be detected in the four-cell stage, a little over 2 h after fertilization [6, 7]. In Drosophila melanogaster, the maternally deposited mRNAs are destabilized upon egg activation and the zygotic transcription first initiates at the cleavage cycles 8 and increases rapidly until the cleavage cycles 14 [2, 8]. In zebrafish, the zygotic genome activation occurs at around ten cell cycles after fertilization, together with maternal mRNA clearance [9]. In Xenopus laevis, the MZT begins at fertilization, and degradation of maternal messages begins immediately after fertilization, and ends during gastrulation, the major ZGA occurs 6 h later [10]. In mouse, a large fraction of the maternally supplied mRNAs is degraded by the two-cell stage, and the first wave of transcription commencing occurs at the one-cell stage [2, 5]. In human, 10% of the expressed maternal mRNAs and 2% of the lncRNAs are eliminated between the four- and eight-cell stage, and the ZGA occurs at four- to eight-cell stage, 2 days after fertilization [5, 11].
Fig. 1.
Overview of the maternal-to-zygotic transition in several model organisms. Dashed curves represent the degradation profiles of destabilized maternal component in each species. The solid line curves illustrate cumulatively increase in zygotic gene expression
The MZT is one of the most complex and crucial developmental processes in the life of an organism and multiple mechanisms at different levels regulate the clearance of the maternal components and the activation of the zygotic genome, including transcriptional, post-transcriptional, translational, and post-translational regulation [12–19]. Because the zygotic genome is inactive in the early stages of embryogenesis, post-translational regulation is particularly important in the MZT. Various protein post-translational modifications have been identified during this stage, which are essential for the timely transition of embryonic development. The focus of this review is on recent advances in our understanding of the molecular mechanisms underlying the post-translational regulation of maternal mRNA-protein degradation and zygotic genome activation during the MZT.
Post-translational regulation of maternal mRNAs clearance
At the earliest stages of embryogenesis, the transcription of the zygotic genome is quiescent and the developmental processes in the embryo rely exclusively on maternal mRNAs, which are produced by immature oocytes or nurse cells and are regulated under the control of mRNA stability, translation, and localization [3, 18, 20, 21]. During the MZT, approximately 60% of maternal mRNA levels are considerably reduced in Drosophila [3, 4, 22] and up to 90% of maternal mRNA is eliminated in the two-cell stage of the mouse [23, 24]. The stability of maternal mRNAs is dependent on three main features: the mRNA sequence, the 7-methylguanylate (m7G) cap, and the length of the 3′ untranslated region (UTR) [4]. Multiple mechanisms for maternal mRNA clearance have been identified, including (1) RNA-binding proteins (RBPs) that direct the maternal degradation machinery to its target maternal mRNAs, (2) small RNA-induced silencing, (3) some signaling pathways in the early embryos, (4) spatial control of transcript clearance, and (5) m6A-dependent RNA decay, which was recently uncovered during zebrafish MZT [3, 4, 20, 25, 26]. Post-translational modifications have recently been reported to participate in the clearance of maternal mRNAs via the activation of maternal mRNA decay machinery, and the regulation of maternal mRNA clearance-related RNA-binding proteins and small RNA biogenesis.
Activation of maternal mRNA decay machinery
Little is known about the initiation of maternal mRNA decay. Maternal transcripts are destabilized in activated eggs, indicating that egg activation is necessary and sufficient to trigger maternal mRNA destabilization, while the exact link between them remains unclear [27]. Nonetheless, the post-translational cascade triggered upon egg activation has been identified to function in maternal transcript destabilization in many organisms, such as the Pan gu (PNG) Ser/Thr kinase complex, which regulates the translation of Smaug in Drosophila and the extracellular signal-regulated kinases 1 and 2 (ERK1/2) mediated phosphorylation is essential for the activation of BTG4 in mouse.
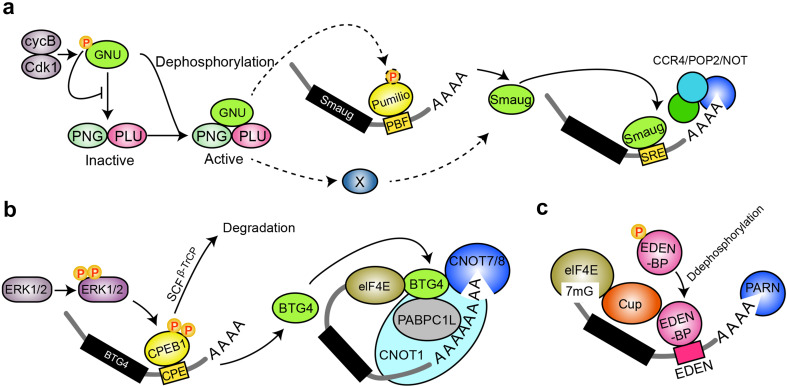
In Drosophila, the Pan gu (PNG) Ser/Thr kinase complex is a crucial regulator of the S phase to metaphase transition by ensuring adequate Cyclin B protein levels and consequently the activity of the cyclin-dependent kinase (CDK)/cyclin complex [28]. Following egg activation, the PNG kinase complex promotes the translation of a multifunctional and highly conserved RNA-binding protein (RBP), Smaug (SMG), to trigger maternal transcript clearance [27]. Smaug is responsible for both the translational repression and degradation of maternal mRNA by binding RNA stem loop structures, which are called Smaug recognition elements (SREs), through a sterile alpha motif (SAM) [29–31]. For maternal mRNA clearance, Smaug recruits the CCR4/POP2/NOT-deadenylase complex to initiate poly (A) tail shortening and subsequent mRNA elimination [32–34]. Smaug can also work together with piRNAs and their associated proteins to promote maternal mRNA decay [35]. Microarray analyses have shown that Smaug was required for the degradation of two-thirds of unstable maternal mRNAs [8]. PNG regulates the cytoplasmic polyadenylation of smg mRNA via the PUM (pumilio RNA-binding family member) [8], which is an RBP and has been implicated in the deadenylation and clearance of maternal mRNAs via binding with Pumilio-binding element (PBE) and the recruitment of Nanos and the deadenylation complex [3, 4, 36]. As in Xenopus, PUM1 can be phosphorylated during oocyte maturation and may induce a conformational change in the complex consisting of PUM1 and cytoplasmic polyadenylation element (CPE)-binding protein (CPEB) that targets CPEB for dissociation, which is required for the translational activation of cyclin B1 mRNA [37, 38]. Therefore, PNG may phosphorylate PUM to induce a conformational change and relieves the translational repression of Smaug. Because the restoration of polyadenylation in png mutants is not sufficient to rescue Smaug translation [8], one or more additional factors may act in parallel to modulate Smaug translation through smg mRNA’s 3′UTR (Fig. 2a). The activity of PNG could also be regulated by Cyclin B/CDK1-catalyzed phosphorylation on GNU, preventing binding to PNG–PLU and the activation of PNG kinase (Fig. 2a). Thus, meiotic completion promotes the dephosphorylation of GNU and PNG kinase activation to further regulate Smaug translation [39].
Fig. 2.
Mechanisms of post-translational regulation of maternal mRNA clearance. a PNG-mediated polyadenylation regulates smg mRNA translation. Cyclin B/CDK1-catalyzed phosphorylation on GNU inhibits the interaction between GNU and PNG–PLU, resulting in the inactivation of PNG kinase. Meiotic completion promotes GNU dephosphorylation and PNG kinase activation. The active PNG kinase phosphorylates PUM and one or more additional factors, which act in parallel through the smg mRNA’s 3′untranslated region (UTR) to promote the translation of Smaug. b ERK1/2 activates the translation of BTG4. Upon oocyte meiotic resumption, ERK1/2 is activated by upstream kinases and triggers CPEB1 phosphorylation and SCFβ-TrCP-dependent degradation. The phosphorylation and partial degradation of CPEB1 stimulates polyadenylation and translational activation of BTG4 to further promote maternal mRNA degradation by recruiting the RNA deadenylation complex CCR4-NOT. c Dephosphorylation of EDEN-BP regulates its deadenylation activity by deadenylase PARN and Cup. EDEN-BP is phosphorylated during oocyte maturation and calcium-dependently dephosphorylated following egg activation
A mitogen-activated protein kinase (MAPK) cascade and extracellular signal-regulated kinases 1 and 2 (ERK1/2) have also been found to activate the translation of BTG4 to promote maternal mRNA clearance in mouse [40]. Upon oocyte meiotic resumption, ERK1/2 is activated by upstream kinases and triggers CPEB1 phosphorylation and SCFβ-TrCP-dependent degradation. The phosphorylation and partial degradation of CPEB1 stimulates polyadenylation and translational activation of BTG4 (Fig. 2b), which belongs to the TOB/BTG family of proteins and promotes maternal mRNA degradation by recruiting the RNA deadenylation complex CCR4-NOT onto target mRNAs [41–43].
Regulation of RNA-binding proteins
RBPs regulate almost every step of RNA life, including RNA stability, translation, and localization, and are crucial for the temporal control of the maternal mRNA decay machinery [3, 20, 44, 45]. Several RBPs have been identified to bind to and direct the degradation of largely distinct subsets of maternal mRNAs [2–4], such as MEX-5/MEX-6 in C. elegans [46]; Smaug, Pumilio, AU-rich element-binding proteins (ARE-BPs) in Drosophila [8, 22, 31, 47]; embryonic deadenylation element-binding proteins (EDEN-BP) in Xenopus [48]; and Zinc Finger Protein C3H Type 36-Like 2 (ZFP36L2) and mouse-specific Y-box protein 2 (MSY2) in mouse [49, 50]. The post-translational modifications of RBPs have been found to function in maternal transcript destabilization in many organisms, including the phosphorylation of embryonic deadenylation element-binding protein (EDEN-BP) (Xenopus), cytosine–uridine–guanine-binding protein (CUG-BP) (human), and MSY2 (mouse).
In Xenopus, EDEN-BP recognizes embryonic deadenylation element (EDEN), which is rich in uridine/purine dinucleotides, to deadenylate the maternal transcripts upon fertilization [48, 51]. One hundred and fifty-eight maternal mRNAs as binding targets for EDEN-BP have been identified by microarrays analyses [51]. EDEN-dependent deadenylation is active in early Xenopus embryos, whereas the quantities of EDEN-BP remain constant from fertilization to the tadpole stage in Xenopus [48], suggesting that temporal and spatial regulation mechanisms are necessary for its precise activation. The dephosphorylation of EDEN-BP has been shown to regulate this sequence-specific deadenylation activity (Fig. 2c) [52]. EDEN-BP is phosphorylated during oocyte maturation and calcium-dependently dephosphorylated following egg activation. EDEN-dependent deadenylation is not influenced by M-phase promoting factor (MPF) reactivation, suggesting that this regulation does not depend directly on MPF activity [52].
CUG-BP, the homolog of EDEN-BP in human, interacts with PARN deadenylase to promote deadenylation, suggesting that EDEN-BP may also cooperate with PARN to promote deadenylation and further clearance of maternal transcripts [53]. The phosphorylation of CUG-BP catalyzed by myotonin-protein kinase (DMPK) in human somatic cells is involved in the regulation of its localization (nuclear versus cytoplasmic) [54], indicating that some kinases and phosphatases may also regulate the deadenylation activity of CUG-BP by modulating its localization.
MSY2 is a DNA/RNA-binding protein that stabilizes mRNAs and inactivates the RNA degradation machinery in male mouse germ cells [55]. In mouse oocytes, MSY2 regulates the global stability of mRNA and the knockdown and disruption of Msy2 results in an approximate 20% decrease in the amount of total mRNA [56, 57]. Following fertilization, the amounts of msy2 mRNA and MSY2 protein decrease [58], suggesting that the decay of MYS2 is essential for maternal mRNA clearance. CDC2A-mediated phosphorylation of MSY2 has been proven to trigger the transition from maternal mRNA stability to instability [59]. Overexpression of a nonphosphorylatable form of MSY2 inhibits maternal mRNA clearance, whereas overexpression of a putative constitutively active form of MSY2 triggers maternal mRNA degradation in the absence of CDC2A. The phosphorylation of MSY2 may increase mRNA accessibility to the RNA degradation machinery as overexpressing the constitutively active form of MSY2 is far more sensitive to degradation by exogenous RNase. The decay of MYS2 upon fertilization may also be caused by a positive-feedback loop of CDK1-mediated phosphorylation of MSY2 that leads to the degradation of msy2 mRNA [59, 60]. Therefore, the CDC2A-mediated phosphorylation of MSY2 is essential for maternal mRNA clearance. CDC2A can also phosphorylate the mRNA-decapping complex, consisting of DCP1A and DCP2, which is essential for maternal mRNA degradation through removal of the 5′-monomethyl guanosine cap [61]. However, the significance of the maturation-associated phosphorylation of DCP1A and DCP2 requires further investigation.
Small RNA biogenesis
Small RNAs have emerged as widespread regulators of gene expression by interacting with mRNA and mediating translational repression, deadenylation, and mRNA destabilization [62]. Many small RNAs have been identified as mediators of maternal mRNA clearance, such as miR-35-42, miR-51-56, and the miR-58/80-82 family in C. elegans [63, 64]; the miR-309 cluster and piRNA in Drosophila [35, 65]; miR-430 in zebrafish [26, 66]; miR-18 and miR-427 in Xenopus; and miR-290 in mouse [67]. Dicer is crucial for the generation of functional miRNA or siRNA by cleaving the dsRNA or pre-miRNA into their final siRNA and miRNA forms, respectively [68, 69]. In C. elegans, the function of Dicer must be dynamically regulated during oocyte maturation and embryogenesis as the complete disruption of Dicer blocks oocyte meiotic maturation, whereas a reduction in Dicer function allows oogenesis to proceed but results in early embryonic death [70, 71]. In mouse, an expression analysis of Dicer ZP3-cKO oocytes indicated an overabundance of mRNA transcripts, which is consistent with a loss of miRNA inhibition of translation or mRNA degradation [67], suggesting a conserved functional role of Dicer in maternal mRNAs clearance.
The phosphorylation of Dicer, which is catalyzed by ERK/MPK-1, has been proven to be important for this dynamic regulation in C. elegans [71, 72]. During oogenesis, ERK/MPK-1 is active and phosphorylates Dicer on two conserved residues in its RNase IIIb and double-stranded RNA (dsRNA)-binding domains to trigger Dicer’s nuclear translocation and inhibit its function [71]. In oocyte maturation and fertilization, ERK/MPK-1 is inactivated, and Dicer is dephosphorylated and activated to generate small RNAs and further degrade maternal mRNAs [72]. The post-translational regulation of Dicer for maternal mRNAs clearance in other species still needs further exploration.
Post-translational regulation of maternal protein degradation
The maternal proteins stored in oocytes are essential for fertilization, the meiosis-to-mitosis transition, reprogramming, and the early stages of embryogenesis [2, 4, 5]. After fertilization, maternal proteins are quickly degraded, and a proteomic study reveals that zygotes show a reduction of approximately 900 proteins (~ 100,000 peptides) compared with oocytes in the second meiotic metaphase (MII) [73]. In eukaryotes, two major pathways for intracellular protein degradation exist, including the autophagy–lysosome pathway and the ubiquitin–proteasome pathway [74, 75]. Both of these proteolysis pathways are necessary for some maternal proteins degradation.
Autophagy regulates maternal protein degradation
Autophagy is a highly evolutionarily conserved membrane trafficking process in which cytoplasmic materials, such as long-lived proteins and organelles, are sequestered into a double-membrane autophagosome and then delivered to the lysosome for degradation [76, 77]. At least three types of autophagy have been defined, including macroautophagy, microautophagy, and chaperone-mediated autophagy [77–79]. The most extensively investigated form is macroautophagy, which is initiated from an isolated membrane followed by the formation of an autophagosome to engulf cytoplasmic cargos and then fuse with lysosomes for degradation [76, 77, 80]. More than 40 autophagy-related (ATG) proteins have been characterized, which consist of several functional units: the ULK1 kinase complex, PI3K complex, Atg9–Atg2–WIPI1/Atg18 complex, and two ubiquitin-like conjugation systems [81–83]. Recently, autophagy has been found to participate in both maternal protein and mRNA degradation.
In mouse, autophagy can be triggered by fertilization and transiently suppressed from the late one-cell to middle two-cell stages and then reactivated after the late two-cell stage. The disruption of autophagy in an oocyte fails to progress beyond the four- and eight-cell stages if the oocyte was fertilized by an Atg5-null sperm, indicating that autophagy is essential for preimplantation development [84]. Protein synthesis rates are decreased in autophagy-null embryos, which is likely due to amino acid insufficiency [84], suggesting that autophagy may promote maternal protein degradation for energy or amino acid recycling. During the establishment of anteroposterior (AP) polarity in the early embryogenesis of C. elegans, P-granules, large ribonucleoprotein complexes, localize in the germ line cytoplasm and are essential for cell fate determination [85]. In somatic cells, PGL-1, a marker of P-granules, is surrounded by autophagosomes, and a reduction of autophagy-related genes also blocks the degradation of GFP-PGL-1-positive granules in somatic cells, suggesting that autophagy may participate in the elimination of extra P-granule components in somatic blastomeres [86]. During this process, SEPA-1, which directly binds to the P-granule component PGL-3 and the autophagy protein LGG-1/Atg8, has been identified to function as a bridging molecule in the mediation of the specific recognition and degradation of P-granule components by autophagy [86]. Recently, autophagy has also been found to influence maternal mRNA degradation. In two-cell- and four-cell-stage of porcine parthenote embryos treated with 3-MA, an autophagy inhibitor, maternal mRNAs remain at high levels, but they were significantly reduced in embryos treated with rapamycin, which can activate autophagy [87]. However, the direct or indirect role of autophagy in maternal mRNA clearance is still unknown.
Ubiquitin–proteasome system regulates maternal protein degradation
The ubiquitin–proteasome system represents large machinery for protein degradation, consisting of protein ubiquitination components and a proteasome. Protein ubiquitination is an important post-translational modification in which ubiquitin, a 76-residue protein, is covalently attached to a lysine or a Ser/Thr residue in a target protein. Ubiquitination is achieved via three classes of enzymes working in sequence. E1 (ubiquitin-activating enzyme) forms a thioester bond between itself and the C-terminus of ubiquitin. Then, E2 (ubiquitin-conjugating enzyme) receives the activated ubiquitin from E1 by transthiolation. Finally, E3 (ubiquitin ligase) transfers ubiquitin from E2 to the target protein. Polyubiquitin chains conjugated on the target protein, especially Lys48-linked and Lys11-linked ubiquitin chains, can be recognized by the proteasome for further proteasomal degradation [88–91]. A proteomic analysis reveals that ubiquitination-related proteins are highly enriched in the mouse zygote [73], and many studies have identified some ubiquitin ligases with important roles in maternal protein degradation (Table 1) that are essential for meiosis-to-mitosis transition, spindle transition, and early embryonic development.
Table 1.
Maternal proteins targeted for degradation in different organisms
| Substrate | E3 ligase | Function | References | |
|---|---|---|---|---|
| Mouse | Cyclin B | APC/C | Meiosis I and II | [99] |
| Cyclin B1 | RFPL4 | Meiosis I and II | [100] | |
| Emi2 | SCFβ-TrCP | Metaphase II exiting | [205] | |
| TAB1 | RNF114 | NF-κB pathway activation | [122] | |
| MATER | UCHL1 | Polyspermy block | [132] | |
| Xenopus | CyclinA, Cyclin B | APC/C(CORT) | Meiosis I and II | [102] |
| Mtrm | APC/C(CORT) | Polo kinase activation | [105] | |
| Cort | APC/C | Embryonic mitosis | [102] | |
| CPEB1 | SCFβ-TrCP | Maternal mRNAs polyadenylation | [129] | |
| C. elegans | MEI-1/katanin | CUL-3MEL-26 | Meiotic spindle degradation | [109] |
| EGG-3 | APC/C | MBK-2 activation | [117] | |
| OMA-1/2 | CRL1LIN-23, CRL1FBXB-3, CRL2ZYG-11 | ZGA | [175] |
Ubiquitin–proteasome system regulates the meiosis-to-mitosis transition
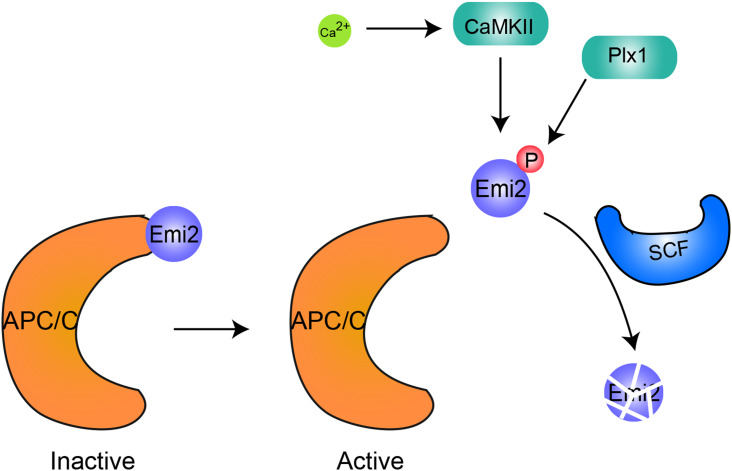
In most animals, mature oocytes are arrested at MII by Cyclin B, a maturation promoting factor (MPF), and after fertilization, the oocyte relieves the MII arrest and undergoes the transition from meiosis to mitosis. The degradation of MPF by the anaphase-promoting complex/cyclosome (APC/C), a multisubunit E3 ubiquitin ligase, is essential for this process [92]. In Xenopus and mouse, Emi2/XErp1, an essential cytostatic factor (CSF) component, has been identified to serve as a necessary protein for maintaining MII arrest before egg activation by inhibiting the activity of APC/C [93, 94]. Emi2/XErp1 contains three parts: an F-box domain, a C-terminal Zn2+-binding region (ZBR), and a D-box. The D-box and ZBR domain have been found to inhibit APC/C activity by binding the D-box receptor on the core of the APC/C and blocking the access of substrates to the APC/C [92]. In addition, phosphorylation of Emi2/XErp1 by the Mos/MAPK/p90Rsk pathway promotes the association of Emi2/XErp1 with the APC/C for further inactivation [92, 95]. Upon fertilization, Ca2+-induced calmodulin kinase II (CaMKII) phosphorylates Emi2/XErp1, and the phosphorylation of Emi2/XErp1 can be recognized by the polo-box domain (PBD) of polo-like kinase 1 (Plx1) [96, 97]. Plx1 further phosphorylates the DSGX3S motif of the Emi2/XErp1 protein, which is necessary for the recognition of the Skp1–Cullin–F-box (SCF) ubiquitin ligase complex [93]. SCF ubiquitin ligase catalyzes the ubiquitination of Emi2/XErp1 and promotes its degradation to reactivate the APC/C (Fig. 3) [93]. The active APC/C promotes the degradation of Cyclin B to exit MII [98, 99]. Some other E3 ligases have also been identified during this process, such as Ret Finger Protein-Like 4 (Rfpl4), a RING-type E3 ligase that accumulates in growing oocytes and quickly disappears during early embryonic cleavage. Rfpl4 could interact with Cyclin B1 and promote its degradation [100], indicating that Rfpl4 may regulate oocyte meiosis-to-mitosis transition. However, the precise mechanism still needs further investigation.
Fig. 3.
Emi2 regulates the activity of the APC/C to promote the metaphase II exit. Emi2 inhibits APC/C activity by binding the D-box receptor on the core of the APC/C and blocking the access of substrates to the APC/C. Upon fertilization, Ca2+-induced calmodulin kinase II (CaMKII) phosphorylates Emi2 and the phosphorylation of Emi2 can be recognized by the polo-box domain (PBD) of polo-like kinase 1 (Plx1) for further phosphorylation on the DSGX3S motif of Emi2. The phosphorylation of Emi2 is recognized by the Skp1–Cullin–F-box (SCF) ubiquitin ligase complex, which promotes its ubiquitination and further degradation to reactivate the APC/C
In Drosophila, the activation of the APC/C is regulated by a meiosis-specific adaptor protein Cortex, which acts as an activator of the APC/C and is required for cyclin degradation and the completion of meiosis during egg activation [101–104]. Cortex can interact with Polo kinase inhibitor matrimony and promote its degradation to activate Polo kinase, which is essential for chromosome segregation, centrosome dynamics, and cytokinesis [105]. During the meiosis-to-mitosis transition, Cortex need to be targeted for degradation by the APC/C following egg activation [102], which may allow mitotic Cdc20 (Fizzy) to regulate subsequent embryonic mitosis.
After the completion of meiosis triggered by fertilization, the spindle should transform from a small centrosomal meiotic spindle to a large mitotic spindle. In C. elegans, this spindle transition requires downregulation of the microtubule-severing katanin complex [106, 107]. The katanin complex is a p60–p80 heterodimeric complex consisting of a ‘catalytic’ AAA+ subunit (p60) and an ‘accessory’ subunit (p80), which are called MEI-1 and MEI-2, respectively, in C. elegans [107]. The degradation of katanin requires a cullin 3-based E3 ubiquitin ligase, a type of cullin-RING ligases (CRL), which are multisubunit enzymes composed of specific substrate-recognition modules dedicated to cullin-RING-based catalytic cores [108]. MEL-26 functions as a substrate-specific adaptor to recognize MEI-1 through its MATH (meprin and TRAF homology) domain and bind to the N terminus of CUL-3 through its Bric-a-brac, Tramtrack, and Broad (BTB) fold [109, 110]. Then, CUL-3 and MEL-26 can mediate the polyubiquitination of MEI-1 and promote its degradation [111, 112]. Some other types of post-translational modification have also been reported to modulate CUL-3MEL-26-mediated MEI-1 degradation. Nedd8 is a ubiquitin-like protein, and an enzymatic cascade similar to ubiquitination catalyzes neddylation [113]. As neddylation of cullin-RING ligases promotes the recruitment of the E2 into them, the neddylation and deneddylation of CUL-3 regulates the activity of cullin-RING ligases to further target MEI-1 for degradation [114]. The phosphorylation of MEI-1 catalyzed by MBK-2, a member of the dual-specificity tyrosine-related kinases (DYRKs), is also necessary for MEI-1 degradation [115, 116]. With high levels of MBK-2-dependent phosphorylation only after the completion of meiosis [117], MBK-2-mediated phosphorylation on MEI-1 may serve as a temporal regulator for the degradation of katanin after meiosis [112]. The activity of MBK-2 can also be regulated by ubiquitination. In oocytes, MBK-2 is sequestered in the cell cortex in an inactive state and depends on the pseudo-phosphatase EGG-3 [118]. EGG-3 recognizes EGG4/5 and uses their PTP domains to bind phosphorylated MBK-2 to prevent MBK-2 activation. Then, CDK-1-mediated phosphorylation activates MBK-2 during oocyte maturation [119, 120] and the APC/C activates the degradation of EGG-3 [117], which then releases active MBK-2 into the cytoplasm.
Ubiquitin–proteasome system in other maternal protein degradation
To analyze the maternal proteome in depth and identify crucial ubiquitination-related proteins in it, we previously developed and performed one-dimensional sodium dodecyl sulfate polyacrylamide gel electrophoresis (1D SDS-PAGE) and reverse-phase liquid chromatography–tandem mass spectrometry (RP-LC–MS/MS) to analyze the mature oocyte proteome [121]. A set of 625 different mouse maternal proteins has been identified and RNF114 (Ring finger 114) protein was identified as one of the predominantly expressed proteins in the later stages of mouse oocyte maturation [121], indicating that RNF114 is important for maternal protein degradation. RNF114 is predominately expressed in oocytes and in early embryonic development. The knockdown of Rnf114 results in decreased early embryonic developmental competence. Through an unbiased screening of a protein microarray with more than 9000 proteins, 13 potential RNF114 substrates have been identified. Further ubiquitination and overexpression analyses have shown that TAB1 may be the major substrate of RNF114 during the MZT [122]. TAB1 is an activator of the MAP kinase kinase kinase (MAPKKK) MAP3K7/TAK1, which is an intracellular hub molecule that regulates both the nuclear factor-κB (NF-κB) and mitogen-activated protein kinase (MAPK) signaling pathways [123, 124]. The activation of the NF-κB pathway is crucial to the early development of the mouse embryo, and a previous study showed that TAB1 negatively regulated NF-κB pathway activity [125, 126]. We also found a decreased protein level of the inhibitory subunit IκBα accompanied by the upregulated phosphorylated form and the ratio of phosphorylated IκBα to total IκBα increased gradually from MII to the four-cell stage, indicating the activation of the NF-κB pathway during the MZT [122]. Therefore, RNF114-mediated ubiquitination and degradation of TAB1 may be essential to the activation of the NF-κB pathway during the MZT, directly linking maternal clearance to early embryonic development.
Some other maternal proteins have also been identified to be degraded via ubiquitin–proteasome system, such as CPEB and MATER. CPEB is a cytoplasmic polyadenylation element-binding protein that binds to the cytoplasmic polyadenylation element (CPE) and plays an important role in the translational control of maternal mRNAs in the early animal development [20]. During Xenopus oocyte maturation, two waves of phosphorylation of CPEB occur and play a role in differential mRNA translation or proper meiotic progression [20]. At an early stage of oocyte maturation, CPEB is phosphorylated by the kinase Aurora A on serine 174, which facilitates PARN expulsion from the RNP complex and Gld2-catalyzed polyadenylation, activating a class of maternal mRNAs [127, 128]. Then, CDC2 catalyzes six additional phosphorylation events on CPEB, which may cause conformational changes of CPEB and allow T125 phosphorylation. Polo-like kinase Plx1 binds CPEB at the phosphorylated Thr125 residue and facilitates TSG motif phosphorylation. β-TrCP, the F-box protein of SCFβ-TrCP, specifically recognizes the phosphorylated TSG motif, thereby targeting CPEB for degradation [129]. The polyubiquitination-dependent degradation of CPEB1 may cause a change in the CPEB/CPE ratio and result in activation of another class of mRNAs [129]. Recently, the peptidyl-prolyl cis–trans isomerase Pin1 has also been identified as an important factor in the promotion of CPEB destruction by interacting with CPEB and inducing its isomerization [130]. Maternal antigen that embryos require (MATER, Nlrp5) is one of the first characterized maternal effect proteins in mouse and combines with FLOPED, TLE6, and FILIA to create a subcortical maternal complex, which is essential for successful preimplantation development [5, 131]. In “gracile axonal dystrophy” (gad) female mice, which carry an intragenic deletion of Ubiquitin C-terminal hydrolase L1 (UCHL1), the MATER protein level increases significantly [132], suggesting that the ubiquitination of MATER is important for its stability. However, the precise mechanisms require further investigation.
Degradation of maternal plasma membrane (PM) proteins
In addition to the autophagy–lysosome pathway and the ubiquitin–proteasome pathway, endocytosis has also been found to participate in the degradation of a subset of maternal PM proteins after fertilization [133], including CAV-1 [134], RME-2 [135, 136], CHS-1 [137], and EGG-1 [138]. PM proteins destined for degradation are sorted into intraluminal vesicles (ILVs) at the endosomal membrane and form multivesicular bodies (MVBs). Then, MVBs fuse with lysosomes for degradation [139]. The sorting of maternal PM proteins to the lysosomal pathway is a selective process and maternal SNB-1 is targeted to the PM upon exocytosis but does not undergo lysosomal degradation [140]. Ubiquitination has been found to modulate the endocytosis, monoubiquitination, or K63-linked polyubiquitination of the cargo proteins required for sorting into the MVB pathway and internalization from the PM [141–143]. In C. elegans, K63-linked polyubiquitination is strongly induced on the endosomes of one-cell-stage embryos, which is regulated by UBC-13 and UEV-1 and induced by fertilization. In ubc-13 and uev-1 mutants, K63-linked polyubiquitylation is reduced and PM proteins are internalized from the PM, but they are inefficiently sorted to the MVB and targeted to lysosomes for degradation [144]. Therefore, UBC-13 and UEV-1-mediated K63-linked polyubiquitylation is required for efficient MVB sorting of maternal PM proteins and further lysosome degradation.
Post-translational regulation of ZGA
After the elimination of a subset of maternal mRNAs and proteins, zygotic genome transcription is initiated, commonly referred to as ZGA, and early embryonic developmental control passes to the zygotic genome [1, 145, 146]. ZGA is regulated in a precisely timed manner. Three broad classes of activation models have been proposed to explain ZGA timing: (1) the nucleocytoplasmic (N/C) ratio model in which a threshold ratio of nuclear components to cytoplasmic volume alleviates transcriptional repression [147], (2) the maternal clock model in which maternally deposited activating or derepressing transcription factors may determine the timing of gene expression [148], and (3) the de novo establishment of chromatin states permitting zygotic genome transcription [145, 146]. Post-translational regulation is essential for ZGA and recent findings have identified in N/C ratio-related regulation, transcriptional repression and activation, regulation of chromatin state remodeling, and histone modification [1, 145, 146].
Nucleocytoplasmic ratio-related regulation
In most species, complete or nearly complete cytokinesis follows every cleavage [2]. The enormous cell volumes are decreased two fold with each division cycle, causing the nuclear-to-cytoplasmic ratio to double, and ZGA repressors in the cytoplasm may be titrated [147], such as histones [149], replication factors [150], DNA methyltransferase xDnmt1 [151], and Tramtrack (TTK) [152]. An increased N/C ratio is achieved by rapid early embryonic cell cycles in frogs and other aquatic organisms, which are simplified versions of the cell cycles of somatic cells [147, 153, 154]. These rapid cell cycles differ from the canonical cell cycle, including a lack of gap phases (G1 and G2) and cytoplasmic volume growth, which is entirely controlled by maternally provided mRNA and protein in an autonomous manner [154]. The phosphorylation-related kinase, CDK, has been identified to participate in this process.
CDK activity is essential for cell cycle regulation. Compared with somatic cell cycles, embryonic cell cycles only contain CDK2-Cyclins A/E, which mediate DNA replication and centrosome duplication, and CDK1-Cyclins A/B, which mediate mitotic progression [155–157]. In Xenopus, CDK1 and CDK2 are inhibited via Wee1 kinase-mediated phosphorylation at relatively low levels and Cdc25 phosphatases eliminate this inhibitory phosphorylation during the cleavage stages, maintaining CDK1 in a primed state for activation upon cyclin binding and mitotic entry [157–160]. The activity of CDK in early stage embryos is also predominately regulated by cyclin protein synthesis and degradation [157]. The translation of cyclin protein requires polyadenylation mediated by the phosphorylation of CPEB by Aurora [127, 128]. The degradation of cyclin protein depends on APC/C ligase [92]. CDK1 also regulates Cyclin B protein levels through a CDK1-APC/C negative feedback loop, decreasing the burden of Cyclin B degradation at anaphase and supporting the rapid cyclin oscillations in embryonic cell cycles [161].
Regulation of transcriptional activators
Transcriptional activators are required for transcription regulation, which should be central components of ZGA timing modulation [1, 145], including the TFIID complex [162] and TATA-binding protein (TBP) [163–167], which bind the promoter of zygotic genes to activate transcription. Among them, many pioneer transcriptional factors have been identified as essential in the activation of zygotic genomic transcription, including Zelda in D. melanogaster [168], Nanog, Pou5f1, and Sox19b in zebrafish [169, 170], DUX in mouse, and DUX4 in humans [171–173]. Similar to the functional roles of transcriptional factors in somatic cells, the regulation of ZGA-related transcriptional factors includes import into the nucleus, remodeling of nucleosomes, and recruitment and elongation of RNA polymerase (RNAP) [145]. Recently, phosphorylation and ubiquitination have been found to regulate the import of ZGA-related transcriptional factors into the nucleus.
In C. elegans, TATA-binding protein-associated factor 4 (TAF-4) is an essential factor for TFIID formation and function [174]. During the first two cleavages cycles, OMA-1 and OMA-2 phosphorylated by the kinase MBK-2 bind to TAF-4 and sequester it in the cytoplasm, subsequently preventing TFIID formation into a stable DNA-bound complex [162]. At the four-cell stage, phosphorylated OMA-1/2 are normally degraded and then TAF-4 returns to the nucleus to form TFIID and promotes the transcriptional activation of the zygotic genome [162]. CRL1LIN-23, CRL1FBXB-3, and CRL2ZYG-11 have been found to be related to the degradation of OMA-1 [175], but the precise mechanisms require further investigation. In mouse, Yap1, the Hippo pathway transcriptional regulator, has been reported to control zygotic genome transcription. Maternal deletion of Yap1 results in the downregulation of approximately 3000 ZGA transcripts [145]. The import of YAP into the nucleus is also regulated by its phosphorylation and dephosphorylation. YAP is phosphorylated at serine 112 by protein kinase A and excluded from the nucleus, whereas dephosphorylated YAP can be transported to the nucleus [176].
Chromatin remodeling allows ZGA
Chromatin consists of DNA wound around nucleosomes, which are octamers of the histones H2A, H2B, H3, and H4, and is joined together by histone linker protein H1. Chromatin conformation is necessary for transcriptional activity [177]. In Xenopus, although the zygotic genome is not activated after fertilization, plasmids microinjected into Xenopus fertilized eggs could be transcripted within 10 min [178], indicating that the chromatin state is essential for ZGA. A genome-wide map of accessible chromatin in mouse preimplantation embryos using ATAC-seq revealed that many gene loci show large open chromatin domains over entire transcribed units in late one-cell and early two-cell embryos. Along with gene expression during ZGA, sharp open chromatin peaks are gradually limited around promoter regions from the two-cell stage to the inner cell masses (ICM) blastocyst stage, indicating a unique spatiotemporal chromatin conformation during minor ZGA [179]. Recently, the three-dimensional structure of chromatin during the early mammalian development has been analyzed by an optimized Hi-C (genome-wide chromosome conformation capture) approach. Mature oocytes lack detectable topologically associating domains (TADs), which play important roles in regulating transcription and DNA replication [180]. After fertilization, the higher order structure of chromatin is significantly diminished, and then, chromatin organization is gradually re-established as slow TAD consolidation and chromatin compartment segregation. The establishment of the TAD structure requires DNA replication, but not zygotic genome activation. Therefore, during the early mammalian development, chromatin may exist in a markedly relaxed state after fertilization, followed by progressive maturation of higher order chromatin conformation (Fig. 4) [181, 182].
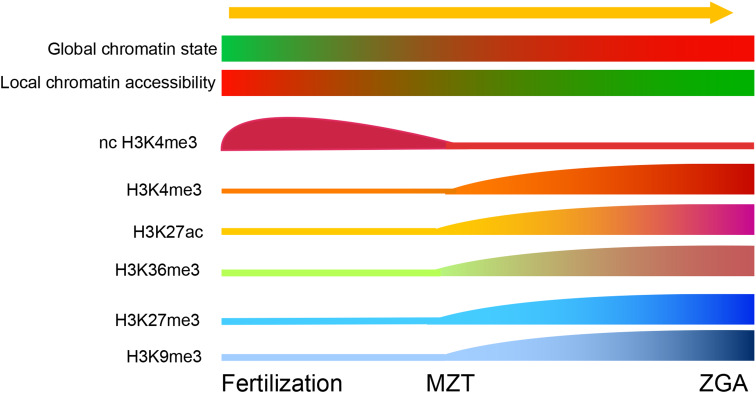
Fig. 4.
Chromatin states and histone modification during ZGA. The global chromatin is more accessible during the early stages of embryogenesis, gradually changing to a more compact conformation. Local chromatin accessibility and transcription-related histone modifications appear during ZGA
Chromatin conformation is highly regulated by histone variants and histone post-transcriptional modifications, such as methylation, acetylation, and ubiquitination [1, 183, 184]. The exchange of histone variants may stimulate promoter nucleosome changes to regulate the chromatin conformation [145].
In Xenopus, the H1 linker variant, H1M, persists in the chromatin until its somatic variants are synthesized at the MZT. The embryonic linker histone H1M is required for proper chromosome architecture and transcription initiation by potentially generating a less stable chromatin structure as H1M lacks multiple CDK1 phosphorylation sites and cannot bind to nuclear import receptors RanBP7 and importin β, which are necessary for H1 to condense chromatin during interphase [185]. The repressive H2A variant, macroH2A, is present in developing and mature mouse oocytes, but it is removed from the maternal genome after fertilization. The maternal macroH2A appears to contribute to its transcriptional silence and macroH2A is progressively lost as the embryo becomes transcriptionally active [186]. MacroH2A variants have a major function in maintaining nuclear organization and heterochromatin architecture [187].
In mammals, maternal and paternal genomes harbor distinct chromatin modifications and each parental genome contributes differently to the establishment of the chromatin conformation in the zygote, which is essential for zygotic gene expression [145, 188]. In maternal chromosomes, pericentric heterochromatin is marked by H3K9me3 and H4K20me3, which is established by the Suv39h and Suv4–20h histone methyltransferases (HMTs) in oocytes and by HP1 beta loaded onto chromatin upon gamete fusion [189]. Maternal chromosomes also provide polycomb repressive complex 1 (PRC1) components to paternal heterochromatin independent of the PRC2 complex, which mediates transcription repression by inhibiting chromatin remodeling or mediating chromatin compaction [190]. Nonetheless, it is accompanied by H3K27me3 of the paternal genome. This process is dependent on the RNF2 component of the PRC1 complex and is functionally critical for the regulation of transcription [189], indicating that H2Aub may also participate in this process. Therefore, the MZT is accompanied by nucleus-wide remodeling of chromatin of the paternal genome by maternally inherited components. In paternal chromosomes, protamines are replaced by maternally supplied histones after fertilization [191], but species-dependent amounts of histone and histone modification still exist in paternal chromosomes [145, 192] and paternal histone modification at particular genes may also influence zygotic gene expression [145]. For example, H3K27me3 and H3K4me3 in sperm may have roles in the establishment of a poised chromatin state in the embryo prior to ZGA in zebrafish [193]. In Drosophila, the oocytes also could transmit the repressive histone mark H3K27me3 to their offspring that promotes the aberrant accumulation of the active histone mark H3K27ac at regulatory regions and precocious activation of lineage-specific genes at zygotic genome activation, suggesting that maternally inherited H3K27me3 is essential for the activation of enhancers and lineage-specific genes during development [194].
Histone modification during ZGA
Histone modification is essential for the regulation of gene expression and many types of histone modification have been identified during ZGA (Fig. 4) [177, 195, 196]. Histones H3K4me3 and H3K27me3 are two important histone modifiers associated with transcription activation and repression, respectively [197]. H3K36me3 marks regions of transcriptional elongation [177]. In zebrafish, H3K4me3, H3K27me3, and H3K36me3 are not detected before the MZT [198]. After ZGA, H3K4me3, H3K27me3, and H3K36me3 emerge and approximately 80% of genes are marked by H3K4me3, including both zygotically expressed genes and inactive genes [198]. These gene loci exhibiting both H3K4me3 (transcription activation-associated marker) and H3K27me3 (transcription repression-associated marker) are similar to the “bivalent” chromatin domains in pluripotent mouse embryonic stem cells [199], which are presumed to poise genes for activation while keeping them repressed [198]. In addition, many inactive genes are associated with H3K4me3 but not H3K27me3, and H3K4me3 markers can form in the absence of both sequence-specific transcriptional activators and the stable association of RNA polymerase II, indicating that these monovalent domains may also poise genes for activation by creating a platform for the transcriptional machinery [198]. In mouse, some noncanonical H3K4me3 markers, which exist as broad peaks at promoters and in a large number of distal loci with low fold enrichment, are widely observed in full-grown and mature oocytes and are reduced in the two-cell stage. The re-establishment of canonical H3K4me3 occurs at the late two-cell stage. The removal of noncanonical H3K4me3 requires zygotic genome transcription, and the downregulation of noncanonical H3K4me3 by the overexpression of the H3K4me3 demethylases KDM5A and KDM5B is required for normal zygotic genome activation [199–202]. Two other transcription activation and repression histone modifiers, H3K27ac and H3K9me3, which are similar to H3K4me3 and H3K27me3, are also detectable in two-and eight-cell stage embryos [201].
Future perspectives on post-translational regulation of the MZT
The maternal-to-zygotic transition is a complex and highly coordinated development process that includes both material decay and generation. The temporal and spatial regulation mechanisms during the MZT are necessary for successful embryogenesis. Because there was no transcription, the post-translational regulation during MZT might play key roles to regulate both material decay and zygotic generation. While this mechanism has not been thoroughly investigated during MZT, less systematic work was performed to analyze the proteomics of phosphorylation, ubiquitination, acetylation, or other post-translational modifications during MZT. Except the above-mentioned modifications, the other post-translational modifications are even more poorly investigated during MZT, such as the SUMOylation, amidation, sulfation, N-myristoylation, and S-nitrosylation. Recently, the O-glycosylation of some mitochondrial TCA cycle enzymes has been reported to be essential for ZGA by regulating their localization, and protein palmitoylation has been found to induce the clearance of the maternal mRNA by activating miR-430 expression [203, 204]. However, the precise mechanisms are still largely unknown. Therefore, the identification of novel post-translational modifications and the investigation of their functional roles in MZT are necessary for further understanding the mechanisms underlying MZT. Although many researchers work on histone modifications during ZGA, most of their works are descriptive and correlative; detailed functional mechanism studies are required for further investigation. As many post-translational modifications could work with each other, the crosstalk between different types of post-translational modifications may also be essential for the temporal and spatial regulation of MZT. With the rapid development of proteomics, high-throughput sequencing, and gene editing tools, many open questions in the field may be addressed in the near future:
What initiates the MZT, degradation, or creation?
How is degradation linked with creation, particularly in terms of transcription?
How can an old network be renovated to build a new regulatory network?
What is the relationship between the autophagy–lysosome system and the ubiquitin–proteasome system during MZT?
What is the mechanism underlying substrate selection by the two degradation systems?
Are there new post-translational regulations during the MZT?
What are the key regulators of the epigenetic regulations during the MZT?
The answers to the above questions would certainly expand our knowledge regarding the MZT, deepen our understanding of the initiation of life, and help us to determine new strategies to treat infertility or other related diseases.
Acknowledgements
This work was supported by the Strategic Priority Research Program of the Chinese Academy of Sciences (Grant no. XDA16020700), the National Key R&D Program of China (Grant no. 2016YFA0500901) and the National Natural Science Foundation of China (Grant nos. 31471277, 31471107, 91649202, and 31771501).
Contributor Information
Ran Huo, Phone: +86-25-86862038, Email: huoran@njmu.edu.cn.
Wei Li, Phone: +86-10-64807529, Email: leways@ioz.ac.cn.
References
- 1.Lee MT, et al. Zygotic genome activation during the maternal-to-zygotic transition. Annu Rev Cell Dev Biol. 2014;30:581–613. doi: 10.1146/annurev-cellbio-100913-013027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tadros W, Lipshitz HD. The maternal-to-zygotic transition: a play in two acts. Development. 2009;136:3033–3042. doi: 10.1242/dev.033183. [DOI] [PubMed] [Google Scholar]
- 3.Walser CB, Lipshitz HD. Transcript clearance during the maternal-to-zygotic transition. Curr Opin Genet Dev. 2011;21:431–443. doi: 10.1016/j.gde.2011.03.003. [DOI] [PubMed] [Google Scholar]
- 4.Yartseva V, Giraldez AJ. The maternal-to-zygotic transition during vertebrate development: a model for reprogramming. Curr Top Dev Biol. 2015;113:191–232. doi: 10.1016/bs.ctdb.2015.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li L, et al. The maternal to zygotic transition in mammals. Mol Aspects Med. 2013;34:919–938. doi: 10.1016/j.mam.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baugh LR, et al. Composition and dynamics of the Caenorhabditis elegans early embryonic transcriptome. Development. 2003;130:889–900. doi: 10.1242/dev.00302. [DOI] [PubMed] [Google Scholar]
- 7.Robertson S, Lin R. The maternal-to-zygotic transition in C. elegans. Curr Top Dev Biol. 2015;113:1–42. doi: 10.1016/bs.ctdb.2015.06.001. [DOI] [PubMed] [Google Scholar]
- 8.Tadros W, et al. SMAUG is a major regulator of maternal mRNA destabilization in Drosophila and its translation is activated by the PAN GU kinase. Dev Cell. 2007;12:143–155. doi: 10.1016/j.devcel.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 9.Wragg J, Muller F. Transcriptional regulation during zygotic genome activation in zebrafish and other anamniote embryos. Adv Genet. 2016;95:161–194. doi: 10.1016/bs.adgen.2016.05.001. [DOI] [PubMed] [Google Scholar]
- 10.Yang J, et al. The xenopus maternal-to-zygotic transition from the perspective of the germline. Curr Top Dev Biol. 2015;113:271–303. doi: 10.1016/bs.ctdb.2015.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yan LY, et al. Single-cell RNA-seq profiling of human preimplantation embryos and embryonic stem cells. Nat Struct Mol Biol. 2013;20:1131. doi: 10.1038/nsmb.2660. [DOI] [PubMed] [Google Scholar]
- 12.Sheets MD. Building the future: post-transcriptional regulation of cell fate decisions prior to the xenopus midblastula transition. Curr Top Dev Biol. 2015;113:233–270. doi: 10.1016/bs.ctdb.2015.06.008. [DOI] [PubMed] [Google Scholar]
- 13.Nothias JY, et al. Regulation of gene-expression at the beginning of mammalian development. J Biol Chem. 1995;270:22077–22080. doi: 10.1074/jbc.270.38.22077. [DOI] [PubMed] [Google Scholar]
- 14.Majumder S, Depamphilis ML. A unique role for enhancers is revealed during early mouse development. BioEssays. 1995;17:879–889. doi: 10.1002/bies.950171010. [DOI] [PubMed] [Google Scholar]
- 15.Schultz RM. The molecular foundations of the maternal to zygotic transition in the preimplantation embryo. Hum Reprod Update. 2002;8:323–331. doi: 10.1093/humupd/8.4.323. [DOI] [PubMed] [Google Scholar]
- 16.Pauli A, et al. Non-coding RNAs as regulators of embryogenesis. Nat Rev Genet. 2011;12:136–149. doi: 10.1038/nrg2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Svoboda P, Flemr M. The role of miRNAs and endogenous siRNAs in maternal-to-zygotic reprogramming and the establishment of pluripotency. EMBO Rep. 2010;11:590–597. doi: 10.1038/embor.2010.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Laver JD, et al. Regulation and function of maternal gene products during the maternal-to-zygotic transition in Drosophila. Curr Top Dev Biol. 2015;113:43–84. doi: 10.1016/bs.ctdb.2015.06.007. [DOI] [PubMed] [Google Scholar]
- 19.Stebbins-Boaz B, Richter JD. Translational control during early development. Crit Rev Eukaryot Gene Expr. 1997;7:73–94. doi: 10.1615/CritRevEukarGeneExpr.v7.i1-2.50. [DOI] [PubMed] [Google Scholar]
- 20.Barckmann B, Simonelig M. Control of maternal mRNA stability in germ cells and early embryos. Bba Gene Regul Mech. 2013;1829:714–724. doi: 10.1016/j.bbagrm.2012.12.011. [DOI] [PubMed] [Google Scholar]
- 21.Marlow FL (2010) Maternal control of development in vertebrates: my mother made me do it! Morgan & Claypool Life Sciences, San Rafael [PubMed]
- 22.Thomsen S, et al. Genome-wide analysis of mRNA decay patterns during early Drosophila development. Genome Biol. 2010;11:R93. doi: 10.1186/gb-2010-11-9-r93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hamatani T, et al. Dynamics of global gene expression changes during mouse preimplantation development. Dev Cell. 2004;6:117–131. doi: 10.1016/S1534-5807(03)00373-3. [DOI] [PubMed] [Google Scholar]
- 24.Schier AF. The maternal-zygotic transition: death and birth of RNAs. Science. 2007;316:406–407. doi: 10.1126/science.1140693. [DOI] [PubMed] [Google Scholar]
- 25.Zhao BS, et al. m6A-dependent maternal mRNA clearance facilitates zebrafish maternal-to-zygotic transition. Nature. 2017;542:475–478. doi: 10.1038/nature21355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Giraldez AJ, et al. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science. 2006;312:75–79. doi: 10.1126/science.1122689. [DOI] [PubMed] [Google Scholar]
- 27.Tadros W, et al. Regulation of maternal transcript destabilization during egg activation in Drosophila. Genetics. 2003;164:989–1001. doi: 10.1093/genetics/164.3.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee LA, et al. A genetic screen for suppressors and enhancers of the Drosophila PAN GU cell cycle kinase identifies cyclin B as a target. Genetics. 2001;158:1545–1556. doi: 10.1093/genetics/158.4.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smibert CA, et al. Smaug protein represses translation of unlocalized nanos mRNA in the Drosophila embryo. Genes Dev. 1996;10:2600–2609. doi: 10.1101/gad.10.20.2600. [DOI] [PubMed] [Google Scholar]
- 30.Aviv T, et al. The RNA-binding SAM domain of Smaug defines a new family of post-transcriptional regulators. Nat Struct Biol. 2003;10:614–621. doi: 10.1038/nsb956. [DOI] [PubMed] [Google Scholar]
- 31.Gotze M, Wahle E. Smaug destroys a huge treasure. Genome Biol. 2014;15:R4. doi: 10.1186/gb4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Semotok JL, et al. Smaug recruits the CCR4/POP2/NOT deadenylase complex to trigger maternal transcript localization in the early drosophila embryo. Curr Biol. 2005;15:284–294. doi: 10.1016/j.cub.2005.01.048. [DOI] [PubMed] [Google Scholar]
- 33.Zaessinger S, et al. Oskar allows nanos mRNA translation in Drosophila embryos by preventing its deadenylation by Smaug/CCR4. Development. 2006;133:4573–4583. doi: 10.1242/dev.02649. [DOI] [PubMed] [Google Scholar]
- 34.Semotok JL, et al. Drosophila maternal Hsp83 mRNA destabilization is directed by multiple SMAUG recognition elements in the open reading frame. Mol Cell Biol. 2008;28:6757–6772. doi: 10.1128/MCB.00037-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rouget C, et al. Maternal mRNA deadenylation and decay by the piRNA pathway in the early Drosophila embryo. Nature. 2010;467:U1128–U1144. doi: 10.1038/nature09465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weidmann CA, et al. The RNA binding domain of Pumilio antagonizes poly-adenosine binding protein and accelerates deadenylation. RNA. 2014;20:1298–1319. doi: 10.1261/rna.046029.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ota R, et al. Biochemical characterization of Pumilio1 and Pumilio2 in Xenopus oocytes. J Biol Chem. 2011;286:2853–2863. doi: 10.1074/jbc.M110.155523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mendez R, et al. Differential mRNA translation and meiotic progression require Cdc2-mediated CPEB destruction. EMBO J. 2002;21:1833–1844. doi: 10.1093/emboj/21.7.1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hara M et al (2017) Control of PNG kinase, a key regulator of mRNA translation, is coupled to meiosis completion at egg activation. eLife 6:e22219. 10.7554/eLife.22219 [DOI] [PMC free article] [PubMed]
- 40.Sha QQ, et al. A MAPK cascade couples maternal mRNA translation and degradation to meiotic cell cycle progression in mouse oocytes. Development. 2017;144:452–463. doi: 10.1242/dev.144410. [DOI] [PubMed] [Google Scholar]
- 41.Liu YS, et al. BTG4 is a key regulator for maternal mRNA clearance during mouse early embryogenesis. J Mol Cell Biol. 2016;8:366–368. doi: 10.1093/jmcb/mjw023. [DOI] [PubMed] [Google Scholar]
- 42.Pasternak M et al (2016) The BTG4 and CAF1 complex prevents the spontaneous activation of eggs by deadenylating maternal mRNAs. Open Biol. 10.1098/rsob.160184 [DOI] [PMC free article] [PubMed]
- 43.Yu C, et al. BTG4 is a meiotic cell cycle-coupled maternal-zygotic transition licensing factor in oocytes. Nat Struct Mol Biol. 2016;23:387–394. doi: 10.1038/nsmb.3204. [DOI] [PubMed] [Google Scholar]
- 44.Glisovic T, et al. RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett. 2008;582:1977–1986. doi: 10.1016/j.febslet.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Farley BM, Ryder SP. Regulation of maternal mRNAs in early development. Crit Rev Biochem Mol. 2008;43:135–162. doi: 10.1080/10409230801921338. [DOI] [PubMed] [Google Scholar]
- 46.Schubert CM, et al. MEX-5 and MEX-6 function to establish soma/germline asymmetry in early C-elegans embryos. Mol Cell. 2000;5:671–682. doi: 10.1016/S1097-2765(00)80246-4. [DOI] [PubMed] [Google Scholar]
- 47.Gerber AP, et al. Genome-wide identification of mRNAs associated with the translational regulator PUMILIO in Drosophila melanogaster. Proc Natl Acad Sci USA. 2006;103:4487–4492. doi: 10.1073/pnas.0509260103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Paillard L, et al. EDEN and EDEN-BP, a cis element and an associated factor that mediate sequence-specific mRNA deadenylation in Xenopus embryos. EMBO J. 1998;17:278–287. doi: 10.1093/emboj/17.1.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ramos SBV, et al. The CCCH tandem zinc-finger protein Zfp36I2 is crucial for female fertility and early embryonic development. Development. 2004;131:4883–4893. doi: 10.1242/dev.01336. [DOI] [PubMed] [Google Scholar]
- 50.Yu J, et al. RNA-binding properties and translation repression in vitro by germ cell-specific MSY2 protein. Biol Reprod. 2002;67:1093–1098. doi: 10.1095/biolreprod67.4.1093. [DOI] [PubMed] [Google Scholar]
- 51.Graindorge A, et al. Identification of CUG-BP1/EDEN-BP target mRNAs in Xenopus tropicalis. Nucleic Acids Res. 2008;36:1861–1870. doi: 10.1093/nar/gkn031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Detivaud L, et al. Regulation of EDEN-dependent deadenylation of Aurora A/Eg2-derived mRNA via phosphorylation and dephosphorylation in Xenopus laevis egg extracts. J Cell Sci. 2003;116:2697–2705. doi: 10.1242/jcs.00477. [DOI] [PubMed] [Google Scholar]
- 53.Moraes KCM, et al. CUG-BP binds to RNA substrates and recruits PARN deadenylase. RNA. 2006;12:1084–1091. doi: 10.1261/rna.59606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Roberts R, et al. Altered phosphorylation and intracellular distribution of a (CUG)(n) triplet repeat RNA binding protein in patients with myotonic dystrophy and in myotonin protein kinase knockout mice. Proc Natl Acad Sci USA. 1997;94:13221–13226. doi: 10.1073/pnas.94.24.13221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang JX, et al. The DNA/RNA-binding protein MSY2 marks specific transcripts for cytoplasmic storage in mouse male germ cells. Proc Natl Acad Sci USA. 2005;102:1513–1518. doi: 10.1073/pnas.0404685102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Medvedev S, et al. Absence of MSY2 in mouse oocytes perturbs oocyte growth and maturation, RNA stability, and the transcriptome. Biol Reprod. 2011;85:575–583. doi: 10.1095/biolreprod.111.091710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yu JY, et al. Transgenic RNAi-mediated reduction of MSY2 in mouse oocytes results in reduced fertility. Dev Biol. 2004;268:195–206. doi: 10.1016/j.ydbio.2003.12.020. [DOI] [PubMed] [Google Scholar]
- 58.Yu JY, et al. Expression of MSY2 in mouse oocytes and preimplantation embryos. Biol Reprod. 2001;65:1260–1270. doi: 10.1095/biolreprod65.4.1260. [DOI] [PubMed] [Google Scholar]
- 59.Medvedev S, et al. CDC2A (CDK1)-mediated phosphorylation of MSY2 triggers maternal mRNA degradation during mouse oocyte maturation. Dev Biol. 2008;321:205–215. doi: 10.1016/j.ydbio.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Svoboda P, et al. Sculpting the transcriptome during the oocyte-to-embryo transition in mouse. Curr Top Dev Biol. 2015;113:305–349. doi: 10.1016/bs.ctdb.2015.06.004. [DOI] [PubMed] [Google Scholar]
- 61.Ma J, et al. Maternally recruited DCP1A and DCP2 contribute to messenger RNA degradation during oocyte maturation and genome activation in mouse. Biol Reprod. 2013;88:11. doi: 10.1095/biolreprod.112.105312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wu E, et al. Pervasive and cooperative deadenylation of 3′UTRs by embryonic microRNA families. Mol Cell. 2010;40:558–570. doi: 10.1016/j.molcel.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lasko P. Translational control during early development. Prog Mol Biol Transl Sci. 2009;90:211–254. doi: 10.1016/S1877-1173(09)90006-0. [DOI] [PubMed] [Google Scholar]
- 65.Bushati N, et al. Temporal reciprocity of miRNAs and their targets during the maternal-to-zygotic transition in Drosophila. Curr Biol. 2008;18:501–506. doi: 10.1016/j.cub.2008.02.081. [DOI] [PubMed] [Google Scholar]
- 66.Bazzini AA, et al. Ribosome profiling shows that miR-430 reduces translation before causing mRNA decay in zebrafish. Science. 2012;336:233–237. doi: 10.1126/science.1215704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tang F, et al. Maternal microRNAs are essential for mouse zygotic development. Genes Dev. 2007;21:644–648. doi: 10.1101/gad.418707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Denli AM, et al. Processing of primary microRNAs by the microprocessor complex. Nature. 2004;432:231–235. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- 69.Ketting RF, et al. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev. 2001;15:2654–2659. doi: 10.1101/gad.927801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Knight SW, Bass BL. A role for the RNase III enzyme DCR-1 in RNA interference and germ line development in Caenorhabditis elegans. Science. 2001;293:2269–2271. doi: 10.1126/science.1062039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Drake M, et al. A requirement for ERK-dependent Dicer phosphorylation in coordinating oocyte-to-embryo transition in C. elegans. Dev Cell. 2014;31:614–628. doi: 10.1016/j.devcel.2014.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Arur S. Context-dependent regulation of Dicer activity and small RNA production: implications to oocyte-to-embryo transition. Worm. 2015;4:e1086062. doi: 10.1080/21624054.2015.1086062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang SF, et al. Proteome of mouse oocytes at different developmental stages. Proc Natl Acad Sci USA. 2010;107:17639–17644. doi: 10.1073/pnas.1013185107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Varshavsky A. The ubiquitin system, autophagy, and regulated protein degradation. Annu Rev Biochem. 2017;86:123–128. doi: 10.1146/annurev-biochem-061516-044859. [DOI] [PubMed] [Google Scholar]
- 75.Dikic I. Proteasomal and autophagic degradation systems. Annu Rev Biochem. 2017;86:193–224. doi: 10.1146/annurev-biochem-061516-044908. [DOI] [PubMed] [Google Scholar]
- 76.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mizushima N, et al. Methods in mammalian autophagy research. Cell. 2010;140:313–326. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li WW, et al. Microautophagy: lesser-known self-eating. Cell Mol Life Sci CMLS. 2012;69:1125–1136. doi: 10.1007/s00018-011-0865-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cuervo AM, Wong E. Chaperone-mediated autophagy: roles in disease and aging. Cell Res. 2014;24:92–104. doi: 10.1038/cr.2013.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mizushima N. Autophagy: process and function. Genes Dev. 2007;21:2861–2873. doi: 10.1101/gad.1599207. [DOI] [PubMed] [Google Scholar]
- 81.Mizushima N, et al. The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol. 2011;27:107–132. doi: 10.1146/annurev-cellbio-092910-154005. [DOI] [PubMed] [Google Scholar]
- 82.Mizushima N, Levine B. Autophagy in mammalian development and differentiation. Nat Cell Biol. 2010;12:823–830. doi: 10.1038/ncb0910-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yang Z, Klionsky DJ. An overview of the molecular mechanism of autophagy. Curr Top Microbiol Immunol. 2009;335:1–32. doi: 10.1007/978-3-642-00302-8_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tsukamoto S, et al. Autophagy is essential for preimplantation development of mouse embryos. Science. 2008;321:117–120. doi: 10.1126/science.1154822. [DOI] [PubMed] [Google Scholar]
- 85.Updike D, Strome S. P granule assembly and function in Caenorhabditis elegans germ cells. J Androl. 2010;31:53–60. doi: 10.2164/jandrol.109.008292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhang YX, et al. SEPA-1 mediates the specific recognition and degradation of P granule components by autophagy in C. elegans. Cell. 2009;136:308–321. doi: 10.1016/j.cell.2008.12.022. [DOI] [PubMed] [Google Scholar]
- 87.Xu YN, et al. Autophagy influences maternal mRNA degradation and apoptosis in porcine parthenotes developing in vitro. J Reprod Dev. 2012;58:576–584. doi: 10.1262/jrd.2012-005. [DOI] [PubMed] [Google Scholar]
- 88.Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 89.Pickart CM. Mechanisms underlying ubiquitination. Annu Rev Biochem. 2001;70:503–533. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- 90.Welchman RL, et al. Ubiquitin and ubiquitin-like proteins as multifunctional signals. Nat Rev Mol Cell Biol. 2005;6:599–609. doi: 10.1038/nrm1700. [DOI] [PubMed] [Google Scholar]
- 91.Komander D, Rape M. The ubiquitin code. Annu Rev Biochem. 2012;81(81):203–229. doi: 10.1146/annurev-biochem-060310-170328. [DOI] [PubMed] [Google Scholar]
- 92.Pesin JA, Orr-Weaver TL. Regulation of APC/C activators in mitosis and meiosis. Annu Rev Cell Dev Biol. 2008;24:475–499. doi: 10.1146/annurev.cellbio.041408.115949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Schmidt A, et al. Xenopus polo-like kinase Plx1 regulates XErp1, a novel inhibitor of APC/C activity. Genes Dev. 2005;19:502–513. doi: 10.1101/gad.320705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Shoji S, et al. Mammalian Emi2 mediates cytostatic arrest and transduces the signal for meiotic exit via Cdc20. EMBO J. 2006;25:834–845. doi: 10.1038/sj.emboj.7600953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Nishiyama T, et al. Phosphorylation of Erp1 by p90rsk is required for cytostatic factor arrest in Xenopus laevis eggs. Nature. 2007;446:1096–1099. doi: 10.1038/nature05696. [DOI] [PubMed] [Google Scholar]
- 96.Hansen DV, et al. CaMKII and polo-like kinase 1 sequentially phosphorylate the cytostatic factor Emi2/XErp1 to trigger its destruction and meiotic exit. Proc Natl Acad Sci USA. 2006;103:608–613. doi: 10.1073/pnas.0509549102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Liu JJ, Maller JL. Calcium elevation at fertilization coordinates phosphorylation of XErp1/Emi2 by Plx1 and CaMK II to release metaphase arrest by cytostatic factor. Curr Biol. 2005;15:1458–1468. doi: 10.1016/j.cub.2005.07.030. [DOI] [PubMed] [Google Scholar]
- 98.Lorca T, et al. Calmodulin-dependent protein kinase-II mediates inactivation of Mpf and Csf upon fertilization of Xenopus eggs. Nature. 1993;366:270–273. doi: 10.1038/366270a0. [DOI] [PubMed] [Google Scholar]
- 99.Dupont G. Link between fertilization-induced Ca2+ oscillations and relief from metaphase II arrest in mammalian eggs: a model based on calmodulin-dependent kinase II activation. Biophys Chem. 1998;72:153–167. doi: 10.1016/S0301-4622(98)00131-8. [DOI] [PubMed] [Google Scholar]
- 100.Suzumori N, et al. RFPL4 interacts with oocyte proteins of the ubiquitin–proteasome degradation pathway. Proc Natl Acad Sci USA. 2003;100:550–555. doi: 10.1073/pnas.0234474100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Page AW, OrrWeaver TL. The Drosophila genes grauzone and cortex are necessary for proper female meiosis. J Cell Sci. 1996;109:1707–1715. doi: 10.1242/jcs.109.7.1707. [DOI] [PubMed] [Google Scholar]
- 102.Pesin JA, Orr-Weaver TL. Developmental role and regulation of cortex, a meiosis-specific anaphase-promoting complex/cyclosome activator. PLoS Genet. 2007;3:2208–2220. doi: 10.1371/journal.pgen.0030202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Chu T, et al. Cortex, a Drosophila gene required to complete oocyte meiosis, is a member of the Cdc20/fizzy protein family. Genesis. 2001;29:141–152. doi: 10.1002/gene.1017. [DOI] [PubMed] [Google Scholar]
- 104.Swan A, Schupbach T. The Cdc20 (Fzy)/Cdh1-related protein, Cort, cooperates with Fzy in cyclin destruction and anaphase progression in meiosis I and II in Drosophila. Development. 2007;134:891–899. doi: 10.1242/dev.02784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Whitfield ZJ et al (2013) A meiosis-specific form of the APC/C promotes the oocyte-to-embryo transition by decreasing levels of the polo kinase inhibitor matrimony. PLoS Biol 11:e1001648 [DOI] [PMC free article] [PubMed]
- 106.Mcnally FJ, Vale RD. Identification of katanin, an atpase that severs and disassembles stable microtubules. Cell. 1993;75:419–429. doi: 10.1016/0092-8674(93)90377-3. [DOI] [PubMed] [Google Scholar]
- 107.Srayko M, et al. MEI-1/MEI-2 katanin-like microtubule severing activity is required for Caenorhabditis elegans meiosis. Genes Dev. 2000;14:1072–1084. [PMC free article] [PubMed] [Google Scholar]
- 108.Petroski MD, Deshaies RJ. Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Bio. 2005;6:9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- 109.Pintard L, et al. The BTB protein MEL-26 is a substrate-specific adaptor of the CUL-3 ubiquitin-ligase. Nature. 2003;425:311–316. doi: 10.1038/nature01959. [DOI] [PubMed] [Google Scholar]
- 110.Xu L, et al. BTB proteins are substrate-specific adaptors in an SCF-like modular ubiquitin ligase containing CUL-3. Nature. 2003;425:316–321. doi: 10.1038/nature01985. [DOI] [PubMed] [Google Scholar]
- 111.Furukawa M, et al. Targeting of protein ubiquitination by BTB-Cullin 3-Roc1 ubiquitin ligases. Nat Cell Biol. 2003;5:1001–1007. doi: 10.1038/ncb1056. [DOI] [PubMed] [Google Scholar]
- 112.Bowerman B, Kurz T. Degrade to create: developmental requirements for ubiquitin-mediated proteolysis during early C. elegans embryogenesis. Development. 2006;133:773–784. doi: 10.1242/dev.02276. [DOI] [PubMed] [Google Scholar]
- 113.Enchev RI, et al. Protein neddylation: beyond cullin-RING ligases. Nat Rev Mol Cell Biol. 2015;16:30–44. doi: 10.1038/nrm3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Pintard L, et al. Neddylation and deneddylation of CUL-3 is required to target MEI-1/katanin for degradation at the meiosis-to-mitosis transition in C. elegans. Curr Biol. 2003;13:911–921. doi: 10.1016/S0960-9822(03)00336-1. [DOI] [PubMed] [Google Scholar]
- 115.Quintin S, et al. The mbk-2 kinase is required for inactivation of MEI-1/katanin in the one-cell Caenorhabditis elegans embryo. EMBO Rep. 2003;4:1175–1181. doi: 10.1038/sj.embor.7400029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Pang KM, et al. The minibrain kinase homolog, mbk-2, is required for spindle positioning and asymmetric cell division in early C. elegans embryos. Dev Biol. 2004;265:127–139. doi: 10.1016/j.ydbio.2003.09.024. [DOI] [PubMed] [Google Scholar]
- 117.Stitzel ML, et al. The C. elegans DYRK kinase MBK-2 marks oocyte proteins for degradation in response to meiotic maturation. Curr Biol. 2006;16:56–62. doi: 10.1016/j.cub.2005.11.063. [DOI] [PubMed] [Google Scholar]
- 118.Stitzel ML, et al. Regulation of MBK-2/Dyrk kinase by dynamic cortical anchoring during the oocyte-to-zygote transition. Curr Biol. 2007;17:1545–1554. doi: 10.1016/j.cub.2007.08.049. [DOI] [PubMed] [Google Scholar]
- 119.Cheng KCC, et al. Regulation of MBK-2/DYRK by CDK-1 and the pseudophosphatases EGG-4 and EGG-5 during the oocyte-to-embryo transition. Cell. 2009;139:560–572. doi: 10.1016/j.cell.2009.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Boxem M, et al. The Caenorhabditis elegans gene ncc-1 encodes a cdc2-related kinase required for M phase in meiotic and mitotic cell divisions, but not for S phase. Development. 1999;126:2227–2239. doi: 10.1242/dev.126.10.2227. [DOI] [PubMed] [Google Scholar]
- 121.Zhang P et al (2009) Proteomic-based identification of maternal proteins in mature mouse oocytes. BMC Genom 10:348 [DOI] [PMC free article] [PubMed]
- 122.Yang Y, et al. The E3 ubiquitin ligase RNF114 and TAB 1 degradation are required for maternal-to-zygotic transition. EMBO Rep. 2017;18:205–216. doi: 10.15252/embr.201642573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Shibuya H, et al. TAB 1: an activator of the TAK1 MAPKKK in TGF-beta signal transduction. Science. 1996;272:1179–1182. doi: 10.1126/science.272.5265.1179. [DOI] [PubMed] [Google Scholar]
- 124.Wang C, et al. TAK1 is a ubiquitin-dependent kinase of MKK and IKK. Nature. 2001;412:346–351. doi: 10.1038/35085597. [DOI] [PubMed] [Google Scholar]
- 125.Moreno-Garcia ME, et al. Kinase-independent feedback of the TAK1/TAB 1 complex on BCL10 turnover and NF-kappaB activation. Mol Cell Biol. 2013;33:1149–1163. doi: 10.1128/MCB.06407-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Nishikimi A, et al. Nuclear translocation of nuclear factor kappa B in early 1-cell mouse embryos. Biol Reprod. 1999;60:1536–1541. doi: 10.1095/biolreprod60.6.1536. [DOI] [PubMed] [Google Scholar]
- 127.Barnard DC, et al. Symplekin and xGLD-2 are required for CPEB-mediated cytoplasmic polyadenylation. Cell. 2004;119:641–651. doi: 10.1016/j.cell.2004.10.029. [DOI] [PubMed] [Google Scholar]
- 128.Mendez R, et al. Phosphorylation of CPEB by Eg2 mediates the recruitment of CPSF into an active cytoplasmic polyadenylation complex. Mol Cell. 2000;6:1253–1259. doi: 10.1016/S1097-2765(00)00121-0. [DOI] [PubMed] [Google Scholar]
- 129.Setoyama D, et al. Mechanism of degradation of CPEB during Xenopus oocyte maturation. Proc Natl Acad Sci USA. 2007;104:18001–18006. doi: 10.1073/pnas.0706952104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Nechama M, et al. An unusual two-step control of CPEB destruction by Pin1. Mol Cell Biol. 2013;33:48–58. doi: 10.1128/MCB.00904-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Li L, et al. A subcortical maternal complex essential for preimplantation mouse embryogenesis. Dev Cell. 2008;15:416–425. doi: 10.1016/j.devcel.2008.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Koyanagi S, et al. Effects of ubiquitin C-terminal hydrolase L1 deficiency on mouse ova. Reproduction. 2012;143:271–279. doi: 10.1530/REP-11-0128. [DOI] [PubMed] [Google Scholar]
- 133.Sato M, Sato K. Dynamic regulation of autophagy and endocytosis for cell remodeling during early development. Traffic. 2013;14:479–486. doi: 10.1111/tra.12050. [DOI] [PubMed] [Google Scholar]
- 134.Sato K, et al. Dynamic regulation of caveolin-1 trafficking in the germ line and embryo of Caenorhabditis elegans. Mol Biol Cell. 2006;17:3085–3094. doi: 10.1091/mbc.e06-03-0211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Balklava Z, et al. Genome-wide analysis identifies a general requirement for polarity proteins in endocytic traffic. Nat Cell Biol. 2007;9:U1035–U1066. doi: 10.1038/ncb1627. [DOI] [PubMed] [Google Scholar]
- 136.Grant B, Hirsh D. Receptor-mediated endocytosis in the Caenorhabditis elegans oocyte. Mol Biol Cell. 1999;10:4311–4326. doi: 10.1091/mbc.10.12.4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Maruyama R, et al. EGG-3 regulates cell-surface and cortex rearrangements during egg activation in Caenorhabditis elegans. Curr Biol. 2007;17:1555–1560. doi: 10.1016/j.cub.2007.08.011. [DOI] [PubMed] [Google Scholar]
- 138.Kadandale P, et al. The egg surface LDL receptor repeat-containing proteins EGG-1 and EGG-2 are required for fertilization in Caenorhabditis elegans. Curr Biol. 2005;15:2222–2229. doi: 10.1016/j.cub.2005.10.043. [DOI] [PubMed] [Google Scholar]
- 139.Hanson PI, Cashikar A. Multivesicular Body Morphogenesis. Annu Rev Cell Dev Biol. 2012;28(28):337–362. doi: 10.1146/annurev-cellbio-092910-154152. [DOI] [PubMed] [Google Scholar]
- 140.Sato M, et al. Rab11 is required for synchronous secretion of chondroitin proteoglycans after fertilization in Caenorhabditis elegans. J Cell Sci. 2008;121:3177–3186. doi: 10.1242/jcs.034678. [DOI] [PubMed] [Google Scholar]
- 141.Traub LM, Lukacs GL. Decoding ubiquitin sorting signals for clathrin-dependent endocytosis by CLASPs. J Cell Sci. 2007;120:543–553. doi: 10.1242/jcs.03385. [DOI] [PubMed] [Google Scholar]
- 142.Mukhopadhyay D, Riezman H. Proteasome-independent functions of ubiquitin in endocytosis and signaling. Science. 2007;315:201–205. doi: 10.1126/science.1127085. [DOI] [PubMed] [Google Scholar]
- 143.Lauwers E, et al. The ubiquitin code of yeast permease trafficking. Trends Cell Biol. 2010;20:196–204. doi: 10.1016/j.tcb.2010.01.004. [DOI] [PubMed] [Google Scholar]
- 144.Sato M, et al. Fertilization-induced K63-linked ubiquitylation mediates clearance of maternal membrane proteins. Development. 2014;141:U1268–U1324. doi: 10.1242/dev.103044. [DOI] [PubMed] [Google Scholar]
- 145.Jukam D, et al. Zygotic genome activation in vertebrates. Dev Cell. 2017;42:316–332. doi: 10.1016/j.devcel.2017.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Palfy M, et al. The timing of zygotic genome activation. Curr Opin Genet Dev. 2017;43:53–60. doi: 10.1016/j.gde.2016.12.001. [DOI] [PubMed] [Google Scholar]
- 147.Newport J, Kirschner M. A major developmental transition in early Xenopus-embryos. 1. Characterization and timing of cellular-changes at the midblastula stage. Cell. 1982;30:675–686. doi: 10.1016/0092-8674(82)90272-0. [DOI] [PubMed] [Google Scholar]
- 148.Howe JA, et al. Identification of a developmental timer regulating the stability of embryonic cyclin-a and a new somatic a-type cyclin at gastrulation. Genes Dev. 1995;9:1164–1176. doi: 10.1101/gad.9.10.1164. [DOI] [PubMed] [Google Scholar]
- 149.Amodeo AA, et al. Histone titration against the genome sets the DNA-to-cytoplasm threshold for the Xenopus midblastula transition. Proc Natl Acad Sci USA. 2015;112:E1086–E1095. doi: 10.1073/pnas.1413990112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Collart C, et al. Titration of four replication factors is essential for the Xenopus laevis midblastula transition. Science. 2013;341:893–896. doi: 10.1126/science.1241530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Dunican DS, et al. xDnmt1 regulates transcriptional silencing in pre-MBT Xenopus embryos independently of its catalytic function. Development. 2008;135:1295–1302. doi: 10.1242/dev.016402. [DOI] [PubMed] [Google Scholar]
- 152.Brown JL, et al. Repression of the Drosophila–Fushi–Tarazu (Ftz) segmentation gene. EMBO J. 1991;10:665–674. doi: 10.1002/j.1460-2075.1991.tb07995.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Kane DA, Kimmel CB. The zebrafish midblastula transition. Development. 1993;119:447–456. doi: 10.1242/dev.119.2.447. [DOI] [PubMed] [Google Scholar]
- 154.Siefert JC, et al. Cell cycle control in the early embryonic development of aquatic animal species. Comp Biochem Phys C. 2015;178:8–15. doi: 10.1016/j.cbpc.2015.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Rempel RE, et al. Maternal Xenopus Cdk2-cyclin E complexes function during meiotic and early embryonic cell cycles that lack a G1 phase. J Biol Chem. 1995;270:6843–6855. doi: 10.1074/jbc.270.12.6843. [DOI] [PubMed] [Google Scholar]
- 156.Strausfeld UP, et al. Both cyclin A and cyclin E have S-phase promoting (SPF) activity in Xenopus egg extracts. J Cell Sci. 1996;109:1555–1563. doi: 10.1242/jcs.109.6.1555. [DOI] [PubMed] [Google Scholar]
- 157.Zhang M, et al. Cell cycle remodeling and zygotic gene activation at the midblastula transition. Adv Exp Med Biol. 2017;953:441–487. doi: 10.1007/978-3-319-46095-6_9. [DOI] [PubMed] [Google Scholar]
- 158.Kim SH, et al. A maternal form of the phosphatase Cdc25A regulates early embryonic cell cycles in Xenopus laevis. Dev Biol. 1999;212:381–391. doi: 10.1006/dbio.1999.9361. [DOI] [PubMed] [Google Scholar]
- 159.Murakami MS, Vande Woude GF. Analysis of the early embryonic cell cycles of Xenopus; regulation of cell cycle length by Xe-wee1 and Mos. Development. 1998;125:237–248. doi: 10.1242/dev.125.2.237. [DOI] [PubMed] [Google Scholar]
- 160.Sha W, et al. Hysteresis drives cell-cycle transitions in Xenopus laevis egg extracts. Proc Natl Acad Sci USA. 2003;100:975–980. doi: 10.1073/pnas.0235349100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Kang Q, Pomerening JR. Punctuated cyclin synthesis drives early embryonic cell cycle oscillations. Mol Biol Cell. 2012;23:284–296. doi: 10.1091/mbc.e11-09-0768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Guven-Ozkan T, et al. Global transcriptional repression in C. elegans germline precursors by regulated sequestration of TAF-4. Cell. 2008;135:149–160. doi: 10.1016/j.cell.2008.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Veenstra GJ, et al. Translation of maternal TATA-binding protein mRNA potentiates basal but not activated transcription in Xenopus embryos at the midblastula transition. Mol Cell Biol. 1999;19:7972–7982. doi: 10.1128/MCB.19.12.7972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Bartfai R, et al. TBP2, a vertebrate-specific member of the TBP family, is required in embryonic development of zebrafish. Curr Biol. 2004;14:593–598. doi: 10.1016/j.cub.2004.03.034. [DOI] [PubMed] [Google Scholar]
- 165.Dantonel JC, et al. TBP-like factor is required for embryonic RNA polymerase II transcription in C. elegans. Mol Cell. 2000;6:715–722. doi: 10.1016/S1097-2765(00)00069-1. [DOI] [PubMed] [Google Scholar]
- 166.Jallow Z, et al. Specialized and redundant roles of TBP and a vertebrate-specific TBP paralog in embryonic gene regulation in Xenopus. Proc Natl Acad Sci USA. 2004;101:13525–13530. doi: 10.1073/pnas.0405536101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Veenstra GJC, et al. Distinct roles for TBP and TBP-like factor in early embryonic gene transcription in Xenopus. Science. 2000;290:2312. doi: 10.1126/science.290.5500.2312. [DOI] [PubMed] [Google Scholar]
- 168.Liang HL, et al. The zinc-finger protein Zelda is a key activator of the early zygotic genome in Drosophila. Nature. 2008;456:U400–U467. doi: 10.1038/nature07388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Lee MT, et al. Nanog, Pou5f1 and SoxB1 activate zygotic gene expression during the maternal-to-zygotic transition. Nature. 2013;503:360. doi: 10.1038/nature12632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Leichsenring M, et al. Pou5f1 transcription factor controls zygotic gene activation in vertebrates. Science. 2013;341:1005–1009. doi: 10.1126/science.1242527. [DOI] [PubMed] [Google Scholar]
- 171.De Iaco A, et al. DUX-family transcription factors regulate zygotic genome activation in placental mammals. Nat Genet. 2017;49:941. doi: 10.1038/ng.3858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Hendrickson PG, et al. Conserved roles of mouse DUX and human DUX4 in activating cleavage-stage genes and MERVL/HERVL retrotransposons. Nat Genet. 2017;49:925. doi: 10.1038/ng.3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Whiddon JL, et al. Conservation and innovation in the DUX4-family gene network. Nat Genet. 2017;49:935. doi: 10.1038/ng.3846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Walker AK, Blackwell TK. A broad but restricted requirement for TAF-5 (human TAF(II)100) for embryonic transcription in Caenorhabditis elegans. J Biol Chem. 2003;278:6181–6186. doi: 10.1074/jbc.M211056200. [DOI] [PubMed] [Google Scholar]
- 175.Du Z, et al. E3 ubiquitin ligases promote progression of differentiation during C. elegans embryogenesis. Dev Biol. 2015;398:267–279. doi: 10.1016/j.ydbio.2014.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Abbassi L et al (2016) Multiple mechanisms cooperate to constitutively exclude the transcriptional co-activator YAP from the nucleus during murine oogenesis. Biol Reprod 94(5):102 [DOI] [PMC free article] [PubMed]
- 177.Li B, et al. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 178.Newport J, Kirschner M. A major developmental transition in early Xenopus-embryos. 2. Control of the onset of transcription. Cell. 1982;30:687–696. doi: 10.1016/0092-8674(82)90273-2. [DOI] [PubMed] [Google Scholar]
- 179.Wu JY, et al. The landscape of accessible chromatin in mammalian preimplantation embryos. Nature. 2016;534:652. doi: 10.1038/nature18606. [DOI] [PubMed] [Google Scholar]
- 180.Dekker J, Mirny L. The 3D genome as moderator of chromosomal communication. Cell. 2016;164:1110–1121. doi: 10.1016/j.cell.2016.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Ke YW, et al. 3D Chromatin Structures of Mature Gametes and Structural Reprogramming during Mammalian Embryogenesis. Cell. 2017;170:367. doi: 10.1016/j.cell.2017.06.029. [DOI] [PubMed] [Google Scholar]
- 182.Du ZH, et al. Allelic reprogramming of 3D chromatin architecture during early mammalian development. Nature. 2017;547:232. doi: 10.1038/nature23263. [DOI] [PubMed] [Google Scholar]
- 183.Berr A, et al. Chromatin modification and remodelling: a regulatory landscape for the control of Arabidopsis defence responses upon pathogen attack. Cell Microbiol. 2012;14:829–839. doi: 10.1111/j.1462-5822.2012.01785.x. [DOI] [PubMed] [Google Scholar]
- 184.Izzo A, Schneider R. Chatting histone modifications in mammals. Brief Funct Genom. 2010;9:429–443. doi: 10.1093/bfgp/elq024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Freedman BS, Heald R. Functional comparison of H1 histones in Xenopus reveals isoform-specific regulation by Cdk1 and RanGTP. Curr Biol. 2010;20:1048–1052. doi: 10.1016/j.cub.2010.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Chang CC, et al. A maternal store of macroH2A is removed from pronuclei prior to onset of somatic macroH2A expression in preimplantation embryos. Dev Biol. 2005;278:367–380. doi: 10.1016/j.ydbio.2004.11.032. [DOI] [PubMed] [Google Scholar]
- 187.Douet J, et al. MacroH2A histone variants maintain nuclear organization and heterochromatin architecture. J Cell Sci. 2017;130:1570–1582. doi: 10.1242/jcs.199216. [DOI] [PubMed] [Google Scholar]
- 188.Ostrup O, et al. Chromatin-linked determinants of zygotic genome activation. Cell Mol Life Sci. 2013;70:1425–1437. doi: 10.1007/s00018-012-1143-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Puschendorf M, et al. PRC1 and Suv39 h specify parental asymmetry at constitutive heterochromatin in early mouse embryos. Nat Genet. 2008;40:411–420. doi: 10.1038/ng.99. [DOI] [PubMed] [Google Scholar]
- 190.Levine SS, et al. Division of labor in polycomb group repression. Trends Biochem Sci. 2004;29:478–485. doi: 10.1016/j.tibs.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 191.Braun RE. Packaging paternal chromosomes with protamine. Nat Genet. 2001;28:10–12. doi: 10.1038/ng0501-10. [DOI] [PubMed] [Google Scholar]
- 192.Jung YH, et al. Chromatin states in mouse sperm correlate with embryonic and adult regulatory landscapes. Cell Rep. 2017;18:1366–1382. doi: 10.1016/j.celrep.2017.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Lindeman LC, et al. Prepatterning of developmental gene expression by modified histones before zygotic genome activation. Dev Cell. 2011;21:993–1004. doi: 10.1016/j.devcel.2011.10.008. [DOI] [PubMed] [Google Scholar]
- 194.Zenk F, et al. Germ line-inherited H3K27me3 restricts enhancer function during maternal-to-zygotic transition. Science. 2017;357:212–216. doi: 10.1126/science.aam5339. [DOI] [PubMed] [Google Scholar]
- 195.Hontelez S et al (2015) Embryonic transcription is controlled by maternally defined chromatin state. Nat Commun 6:10148 [DOI] [PMC free article] [PubMed]
- 196.Lawrence M, et al. Lateral thinking: how histone modifications regulate gene expression. Trends Genet. 2016;32:42–56. doi: 10.1016/j.tig.2015.10.007. [DOI] [PubMed] [Google Scholar]
- 197.Schuettengruber B, et al. Genome regulation by polycomb and trithorax proteins. Cell. 2007;128:735–745. doi: 10.1016/j.cell.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 198.Vastenhouw NL, et al. Chromatin signature of embryonic pluripotency is established during genome activation. Nature. 2010;464:U143–U922. doi: 10.1038/nature08866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Bernstein BE, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 200.Zhang BJ, et al. Allelic reprogramming of the histone modification H3K4me3 in early mammalian development. Nature. 2016;537:553. doi: 10.1038/nature19361. [DOI] [PubMed] [Google Scholar]
- 201.Dahl JA, et al. Broad histone H3K4me3 domains in mouse oocytes modulate maternal-to-zygotic transition. Nature. 2016;537:548. doi: 10.1038/nature19360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Liu XY, et al. Distinct features of H3K4me3 and H3K27me3 chromatin domains in pre-implantation embryos. Nature. 2016;537:558. doi: 10.1038/nature19362. [DOI] [PubMed] [Google Scholar]
- 203.Nagaraj R, et al. Nuclear localization of mitochondrial TCA cycle enzymes as a critical step in mammalian zygotic genome activation. Cell. 2017;168:210. doi: 10.1016/j.cell.2016.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Du Z, et al. Protein palmitoylation activate zygotic gene expression during the maternal-to-zygotic transition. Biochem Biophys Res Commun. 2016;475:194–201. doi: 10.1016/j.bbrc.2016.05.074. [DOI] [PubMed] [Google Scholar]
- 205.Tung JJ, et al. A role for the anaphase-promoting complex inhibitor Emi2/XErp1, a homolog of early mitotic inhibitor 1, in cytostatic factor arrest of Xenopus eggs. Proc Natl Acad Sci USA. 2005;102:4318–4323. doi: 10.1073/pnas.0501108102. [DOI] [PMC free article] [PubMed] [Google Scholar]