Abstract
Conserved from yeast to humans, Elongator is a protein complex implicated in multiple processes including transcription regulation, α-tubulin acetylation, and tRNA modification, and its defects have been shown to cause human diseases such as familial dysautonomia. Elongator consists of two copies of six core subunits (Elp1, Elp2, Elp3, Elp4, Elp5, and Elp6) that are organized into two subcomplexes: Elp1/2/3 and Elp4/5/6 and form a stable assembly of ~ 850 kDa in size. Although the catalytic subunit of Elongator is Elp3, which contains a radical S-adenosyl-l-methionine (SAM) domain and a putative histone acetyltransferase domain, the Elp4/5/6 subcomplex also possesses ATP-modulated tRNA binding activity. How at the molecular level, Elongator performs its multiple functions and how the different subunits regulate Elongator’s activities remains poorly understood. Here, we provide an overview of the proposed functions of Elongator and describe how recent structural studies provide new insights into the mechanism of action of this multifunctional complex.
Keywords: Elongator, Transcription, tRNA modification, X-ray crystallography, Electron microscopy, Familial dysautonomia
Introduction
Discovery of the Elongator complex
The Elongator complex was originally discovered in yeast Saccharomyces cerevisiae from a biochemical purification aimed at determining the composition of the elongating form of RNA polymerase II (RNAPII). This study identified a novel complex containing three proteins of ~ 150, ~ 85, and ~ 60 kDa in mass (renamed Elongator protein 1 (Elp1), Elp2, and Elp3, respectively), that was thought to bind and switch RNAPII from the initiation state to the elongation state [1]. Subsequent improvement in purification procedure led to the finding that holo-Elongator actually contains three additional, but smaller sized subunits (renamed Elp4, Elp5, and Elp6) that form a discrete subcomplex [2–4]. Shortly after, the orthologous six-subunit human Elongator was biochemically isolated from human cells, confirming that the complex is conserved throughout eukaryotes [5]. Furthermore, it was shown in yeast that deletion of any of the six genes encoding Elongator subunits leads to the same growth defects and sensitivity to high salt, caffeine, and 6-azouracil phenotypes, suggesting that all six subunits are essential to the function of this complex [2, 3, 6, 7]. In addition, other research groups working on understanding the inhibition mechanism of zymocin stumbled upon genes encoding Elongator subunits in genetic screens they conducted to search for the target of this anti-fungal toxin from Kluyveromyces lactis [7–10]. However, how Elongator mediates both transcriptional regulation and resistance to zymocin remained obscure.
Sequence analysis showed that Elp3, the putative catalytic subunit of Elongator, contains a domain homologous to members of the GNAT (Gcn5-related N-terminal acetyltransferases) superfamily of lysine acetyltransferases (KAT). This led to the hypothesis that Elongator mediates transcription elongation through acetylating-nucleosomal histones [6]. The fact that Elp3 also possesses a radical SAM (S-adenosyl-l-methionine) domain indicated that Elongator may have other physiological functions. It was initially thought that the Elp3 SAM domain might mediate DNA or histone methylation, because other SAM domain containing proteins seems to possess such activity [11]. However, more recent studies suggested that this SAM domain is more likely to be involved in mediating chemical modification of uridines located in the wobble position of tRNAs instead [12, 13]. To delineate the mechanisms of action of Elongator, a comprehensive understanding of the molecular structure of this multifunctional protein complex is necessary. In this review article, we give a brief overview of the current knowledge and controversy surrounding the physiological functions of Elongator as well as highlight recent advances in understanding the structural properties of Elongator subunits and the full Elongator complex. Readers are also encouraged to check out several excellent recent reviews that provide more in-depth discussion on Elongator’s involvement in translational regulation [14], the functional implications of its structural asymmetry [15], and its engagement with other cofactors [16].
Elongator and transcription control
Soon after the discovery of Elongator, yeast genetic data supporting Elongator’s role in transcription were reported. Notably, deletion of the elp3 gene encoding the catalytic subunit of Elongator in yeast was found to lead to reduction in global histone H3 and H4 acetylation to a similar extent as a Δgcn5 strain [17]. Similarly, deletion mutants of the six elp genes lead to sensitivity to 6-azouracil [2], a phenotype generally caused by defects in transcriptional elongation. The six Elongator genes also showed synthetic genetic lethality with histones and the Gcn5 KAT [18]. Finally, knocking out certain histone deacetylases was found to rescue growth phenotypes of Δelp3 yeast [18]. Despite these genetic data, there was a lack of convincing biochemical evidence supporting the proposed KAT activity of Elongator. For example, the assay method used by the research group that demonstrated the yeast Elp3 has KAT activity required an “in gel” renaturation step of insoluble Elp3 [6]. Along the same line, the group that demonstrated that purified human Elongator can specifically acetylate histone H3 and H4 also showed that human Elongator cannot acetylate-nucleosomal histones [19]. While Elongator subunits were later shown to bind nascent mRNA in yeast cells [20], data from another study indicated that yeast Elongator subunits did not co-purify with known transcription factors or RNA Pol II subunits [21]. Furthermore, two research groups have reported conflicting results when using chromatin immunoprecipitation (ChIP) to assess Elongator binding with promoter and coding regions of different genes [22, 23]. The last piece of data that is inconsistent with Elongator’s involvement in nuclear processes came from localization studies which showed that Elongator subunits are primarily localized to the cytoplasm [5, 19, 23, 24].
Elongator and tRNA modification
For the first few years following the discovery of Elongator, the prominent hypothesis of the field was that this complex mediates transcriptional elongation by acetylating histone tails. A shift in this thought began when it was shown that genetic deletion of the Sin3p, the fission yeast Schizosaccharomyces pombe ortholog of Elp3, leads to a reduction in levels of 5-methoxycarbonylmethyl-2-thiouridine (mcm5s2U), a conserved modified wobble-nucleoside incorporated into the anticodon loop of several tRNAs [12]. Follow-up experiments performed in S. cerevisiae showed that Elp3 as well as an intact 6-subunit Elongator were required for multiple tRNA modifications, including mcm5U (5-methoxycarbonylmethyluridine), mcm5s2U, and ncm5U (5-carbamoylmethyluridine) [12]. In addition, overexpression of two tRNA species (tRNALysUUU and tRNALeuUUG) that are putatively modified by Elongator was sufficient to reverse phenotypes seen in Δelp yeast strains [25]. Perhaps, the most convincing supportive evidence was the finding that the K. lactis γ-toxin, zymocin is actually a RNAse enzyme that targets tRNA anticodons containing modified wobble-nucleoside mcm5s2U [26], which could explain why deletion of Elongator genes led to zymocin resistance.
The potential involvement of Elongator in tRNA modification also appears to be evolutionarily conserved. A subsequent study performed in the nematode Caenorhabditis elegans reconciled the developmental dysfunction of Elongator mutants with the lack of tRNA modification [27]. Arabidopsis thaliana plants lacking Elp3 were found to lack mcm5s5U modifications on tRNAs [28]. In germ cells from mice with a conditional Elp1 deficiency, wobble uridine modification levels were found to be at low level [29]. Finally, brain tissues and fibroblasts isolated from patients with familial dysautonomia (FD), a disease resulting from Elongator malfunction caused by mutations to human Elp1, had lower than normal levels of modified tRNA [30, 31]. With this pool of recent data, the current belief in the field is that Elongator’s primary function is in modifying wobble uridines of tRNA, and the transcription defects observed in Elongator mutants were a consequence of deficiency in tRNA modification, which has been shown to be critical to translation regulation [32].
Other potential functions of Elongator
Knowledge on the molecular function(s) of Elongator was largely derived from studies using the yeast model organism. Unexpectedly, Elongator mutants in multicellular organisms exhibit a plethora of phenotypes in a tissue-specific manner, indicating that this complex might be involved in other cellular processes. In C. elegans, Elongator has been implicated in modulating neuronal cell differentiation and migration through its role in acetylating α-tubulin [33]. Interestingly, it has been shown that yeast Elp2 can bind microtubules [34]. In mice, disruption of Elongator in germ cells impairs spermatogenesis, resulting in infertility [29]. Mouse peripheral neurons lacking Elp1 have impaired target innervation [35]. Furthermore, loss of Elongator in mouse retinal ganglion cells causes cell degeneration eventually leading to blindness [36]. RNAi-mediated knockdown of Elp1, Elp3, or Elp4 in mouse zygotes was found to impair paternal DNA demethylation, and the SAM domain of Elp3 was specifically implicated in this process [37]. Conditional knockout (CKO) of Elp3 in the mouse forebrain was shown to trigger the unfolded protein response (UPR), ultimately resulting in microcephaly [38], while CKO of Elp1 in dorsal root ganglia caused mitochondrial depolarization and fragmentation [39]. Extensive studies in Arabidopsis have revealed that Elongator regulates growth and development, abiotic stress response, and immune response [40–44]. Due to limited amount of molecular investigations of Elongator in these higher eukaryotes, it remains to be confirmed whether these severe phenotypes are caused by the loss of transcriptional/translation control or other undiscovered functions of Elongator.
Elongator and human diseases
Genetic mutations in various subunits of human Elongator have been linked to different human diseases. Most prominently, mutations in the ELP1 gene causes FD, a rare neurodegenerative disease that is associated with growth abnormalities and degradation of sensory functions. The most common disease-causing mutation is a T-to-C polymorphism in the donor splice site of intron 20 in the ELP1 gene, leading to altered splicing causing exon skipping in a tissue-specific manner [45, 46]. In neuronal tissue, this mutation causes a reduction in the total level of Elp1, while splicing and Elp1 expression levels appear to be normal in non-neuronal tissue [47]. While degradation of the mutant transcript by the nonsense-mediated decay pathway has been suggested for this reduction in expression [48, 49], it remains to be experimentally confirmed whether a low level of truncated Elp1 is produced in neuronal tissues [46]. One earlier study showed that knock down of ELP1 in neuronal-derived cells leads to defects in cell motility, suggesting that impaired cell motility could be causing the phenotypes associated with FD [22]. It was also shown that ELP1 depletion resulted in downregulation of genes required for normal cell motility, implicating a role for human Elongator in transcriptional control. As mentioned earlier, two groups recently showed that cells from FD patients exhibit lower level wobble uridine modification in tRNAs [30, 31].
Besides ELP1, genetic studies have revealed that mutations to other ELP genes are associated with different types of neurological disorders, further demonstrating the importance of this complex in the nervous system. Specifically, ELP2 has been implicated in intellectual disability [50], ELP3 has been linked with amyotrophic lateral sclerosis (ALS) [51, 52], and ELP4 was associated with Rolandic epilepsy and intellectual disability [53–55]. In recent years, there have been major advances in characterizing the molecular structures of the core subunits of Elongator as well as the full complex (Table 1). In the next few sections, we will highlight key findings from these structural studies and their implications on the function and mechanism of action of Elongator.
Table 1.
Known structures of Elongator subunits, subcomplexes, holo-Elongator, and known accessory proteins
| Subunit(s) | Organism | Technique | PBD/EMDB | References |
|---|---|---|---|---|
| Elp1 (residues 919–1349) | S. cerevisiae | X-ray crystallography | 5CQS | Xu et al. [59] |
| Elp1 (residues 715–1332) | H. sapiens | X-ray crystallography | 5CQR | Xu et al. [59] |
| Elp2 | S. cerevisiae | X-ray crystallography | 4XFV | Dong et al. [34] |
| Elp2 | S. cerevisiae | X-ray crystallography | 5M2N | Dauden et al. [63] |
| Elp3 | D. mccartyi | X-ray crystallography | 5L7J | Glatt et al. [69] |
| Elp4/5/6 | S. cerevisiae | X-ray crystallography | 4A8J | Glatt et al. [70] |
| Elp4/5/6 | S. cerevisiae | X-ray crystallography | 4EJS | Lin et al. [71] |
| Elp1/2/3 | S. cerevisiae | Electron microscopy | EMD-8291 | Setiaputra et al. [72] |
| Elp1/2/3 | S. cerevisiae | Electron microscopy | EMD-4151 | Dauden et al. [63] |
| Elp1/2/3/4/5/6 | S. cerevisiae | Electron microscopy | EMD-8239 | Setiaputra et al. [72] |
| Elp1/2/3/4/5/6 | S. cerevisiae | Electron microscopy | EMD-4152 | Dauden et al. [63] |
| Kti11 | S. cerevisiae | NMR | 1YOP | Sun et al. [77] |
| Kti11 | S. cerevisiae | X-ray crystallography | 5AX2 | Kumar et al. [Unpublished] |
| Kti13 | S. cerevisiae | X-ray crystallography | 4D4Q | Glatt et al. [68] |
| Kti11/Kti13 | S. cerevisiae | X-ray crystallography | 4D4O | Glatt et al. [68] |
| Kti11/Kti13 | S. cerevisiae | X-ray crystallography | 4D4P | Glatt et al. [68] |
| Kti11/Kti13 | S. cerevisiae | X-ray crystallography | 4X33 | Kolaj-Robin et al. [67] |
Elp1
Originally identified as a regulator of IκB kinase involved in proinflammatory cytokine signaling, Elp1 (also known as IKI3, IKBKAP, and IKAP) is the largest subunit (~ 150 kDa) of Elongator and is highly conserved across eukaryotes. Elp1 is predicted to contain two WD40 repeat domains arranged in tandem followed by a C-terminal tetratricopeptide repeat (TPR) domain (Fig. 1a). The presence of domains involved in protein–protein interactions suggests that Elp1 likely functions as a scaffold for other Elongator subunits and/or mediates interaction with other binding partners of Elongator. Indeed, it was shown shortly after the discovery of Elongator that Elp1 forms a stable subcomplex with Elp2 and Elp3 that is stable under high salt conditions [1]. More recently, a conserved basic region of Elp1 that resembles a nuclear localization signal located at the C-terminal domain of this protein was shown to mediate tRNA binding (Fig. 1a) [56]. Finally, phosphorylation of Elp1 at two sites (Ser-1198 and Ser-1202), regulated by the casein kinase Hrr25 and the phosphatase Sit4 [57], has been shown to be crucial to the tRNA modification activity of the Elongator complex [58]. However, instead of altering interactions of Elp1 with other Elongator’s subunits, this phosphorylation is thought to regulate interactions of Elp1 with accessory proteins Hrr25 and Kti12.
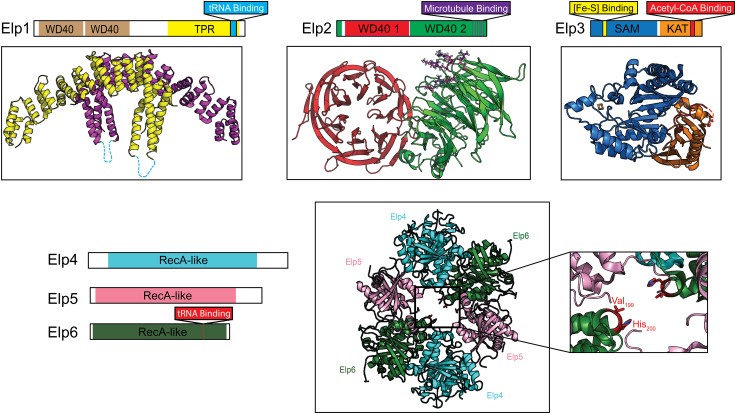
Fig. 1.
Structures of the Elp1, Elp2, and Elp3 subunits and the Elp4/5/6 subcomplex. a Schematic representation of the domain composition of Elp1 and the crystal structure of the Saccharomyces cerevisiae Elp1 dimerization domain (residues 919–1349) (PDB ID: 5CQS). Two copies of the dimerization domain are represented in yellow and purple, respectively. Basic region implicated in tRNA binding is shown in blue. b Domain structure of the Elp2 subunit and the crystal structure of full-length S. cerevisiae Elp2 protein (PDB ID: 5M2N). Residues proposed to mediate microtubule binding are shown in stick representation in purple. c Domain overview of the Elp3 subunit and the structure of Dehalococcoides mccartyi Elp3 (PDB ID: 5L7J). Acetyl-CoA binding loop and the [2Fe–2S] cluster are shown in red and yellow/orange, respectively. d Schematic representation of RecA-like domains found in Elp4 (blue), Elp5 (pink) and Elp6 (green) (left) and the crystal structure of the heterohexameric form of the subcomplex (right) (PDB ID: 4A8J). The inset shows key residues of Elp6 required for the interaction of tRNA with Elp4/5/6
The recent high-resolution structures of the C-terminal domains of human Elp1 (residues 715–1332) and yeast Elp1 (residues 919–1349), determined to 2.7 and 3 Å, respectively by X-ray crystallography, provided critical insights into the function of Elp1 in the context of full Elongator [59]. The human Elp1 C-terminal domain structure showed that this longer fragment is dimeric and adopts a horseshoe-shaped overall architecture, with helices α19 and α21 from one monomer engaging in polar and hydrophobic interactions with α25 and α27 from the other monomer [59] (Fig. 1a). It was estimated that dimerization leads to burial of 1732 Å2 of solvent accessible surface. The yeast Elp1 C-terminal domain structure, which only captured the dimerization motif, but superimposes well with that of the corresponding human Elp1, confirmed the obligate and highly conserved nature of Elp1 dimerization. Indeed, co-precipitation experiments showed that Elp1 dimerization is essential to proper assembly and stability of Elongator in human cells and Elongator function in yeast. Interestingly, self-association of Elp1 was found to be dispensable for tRNA binding. It was proposed, based on the fact that the Elp1 FD disease mutant is devoid of the C-terminal domain, that the impairment of Elongator function in FD patients could be attributed to deficiency in Elp1 dimerization and associated problems in Elongator assembly [59].
The disease importance of Elp1 has led to studies aimed at identifying additional physiological functions of this protein. Tissue-specific depletion of Elp1 in mice was shown to lead to infertility [29], retinal ganglion cell degradation [36], mitochondrial depolarization [39], and cortical neuron development dysfunction [60]. Yeast Elp1 was found to physically interact with the Guanine exchange factor (GEF) Sec2p, and this interaction is required for localization of Sec2p to the plasma membrane [61]. This finding presents a new role for Elp1 in regulating exocytosis of post-Golgi secretory vesicles. While the above studies cannot clearly distinguish whether Elp1 or full Elongator are directly involved in these processes, they did confirm that functional Elp1 is essential for Elongator function.
Elp2
Elp2 is the second largest subunit of the Elongator complex (~ 90 kDa) that was initially identified through its association to Elp1 (and Elp3) forming the “core” Elongator complex. Δelp2 deletion mutants presented similar phenotypes to Δelp1 and Δelp3 mutants in yeast, demonstrating that it is essential to complex function [62]. At the primary sequence level, Elp2 was predicted to contain two tandemly arranged WD40 repeats (Fig. 1b). Two recent crystal structures of yeast Elp2, determined to 2.8 and 3.2 Å resolution, respectively, have revealed that this protein indeed contains two similar sized seven-bladed WD40 β-propellers that are oriented in a “twisted” fashion with respect to one another (Fig. 1b) [34, 63]. These structures further showed that the first propeller was constructed from a continuous string of residues from an N-to-C direction, while the final β strand of blade 14 of the second propeller consists of ten N-terminal residues, generating a “molecular velcro” to maintain the structural integrity of Elp2. Deleting the final β strand of this protein was sufficient to ablate the binding of Elp2-to-Elp1 and Elp3, demonstrating that the stability of the WD40 fold is key to holo-Elongator complex assembly. One of the two structural studies also showed that a set of conserved arginine and lysine residues form a positively charged surface on Elp2 which mediates interaction with binding microtubules, but not histone octamers and nucleosomes in vitro (Fig. 1b). Although Elongator has been previously reported to acetylate neuronal α-tubulin [64], whether Elongator is a true acetyltransferase for α-tubulin remains a highly debated topic in the field. The other structural study showed that mutations in some of these basic residues in yeast yield phenotypes consistent with aberrant tRNA modification, suggesting a role for the region in mediating tRNA modifications [63].
Elp3
Elp3 is the main catalytic subunit of the Elongator complex and the third protein originally identified to form the core Elongator complex [6]. Elp3 contains an N-terminal radical SAM domain and a C-terminal domain that shows sequence homology to members of the Gcn5-related N-acetyltransferase (GNAT) superfamily of lysine acetyltransferases (KAT) (Fig. 1c). Interestingly, Elp3 is the only Elongator subunit that has orthologs present in archaea, bacteria, and viruses [13]. Biochemical characterization of the SAM domain of Elp3 from the thermophilic methanogenic archaeon Methanocaldococcus jannaschii showed that this protein fragment contains an [4Fe–4S] cluster that is coordinated by three conserved cysteines (C96, C101, and C104), and that it can bind and cleave SAM molecules [65]. Mutating these cysteines in yeast Elp3 was subsequently shown to adversely affect interaction of this subunit with other Elongator components and with the accessory proteins/cofactors Kti11 and Kti12 [66].
More recently, a mechanism of wobble uridine modification by Elp3 SAM was proposed based on data from in vitro reconstitution experiments on Methanocaldococcus infernus Elp3 reconstituted with an [4Fe–4S] cluster [13]. In this proposed mechanism, the presence of tRNA causes the formation of a 5′-deoxyadenosyl radical (5′-dA·) from SAM in the radical SAM domain. An acetyl-CoA molecule bound to the KAT domain then loses a hydrogen atom from its methyl group to the 5′-dA·, generating an acetyl radical. The acetyl radical then adds to U34 of the bound tRNA at the C5 position. A carbon–carbon bond then forms between acetyl-CoA and U34 of a tRNA, and a subsequent hydrolysis event results in the formation of a cm5U modification. It has been proposed that the electron required for initial 5′-dA· formation could be donated through Kti11–Kti13, an accessory complex of Elongator that has been shown by recent crystallographic analyses to bind an iron or zinc ion [67, 68]. While the in vitro reconstitution study identified the key steps involved in the tRNA modification reaction, the exact sequence of events and the catalytic residues remain obscure.
A major breakthrough in understanding the structural properties and catalytic mechanism of Elp3 came from the recent 2.15 Å crystal structure of full-length Elp3 from Dehalococcoides mccartyi (DmcElp3) [69], a bacterial Elp3 that shares high sequence identity with both S. cerevisiae Elp3 (41%) and human Elp3 (42%). This high-resolution structure revealed an overall compact architecture of Elp3, with the SAM and KAT domains connected by a zinc-binding domain and forming a large interface that results in the canonical substrate-binding site of the KAT domain being occluded by a blocking loop (Fig. 1c). Replacing this loop with a short linker causes an increase in the binding affinity of DmcElp3 for tRNA, providing an explanation why acetyl-CoA turnover is stimulated upon the addition of tRNA [13]. The crystal structure captured the dimeric form of DmcElp3, which according to gel filtration chromatography analysis, exists both in monomeric and dimeric forms in solution. Interestingly, one [2Fe–2S] cluster was observed at the interface of two DmcElp3 molecules in the dimer. It is believed that this [2Fe–2S] cluster was generated from disintegration of the [4Fe–4S] cluster during either protein purification, crystallization in aerobic conditions, or radiation damage during crystallographic data collection. Residues crucial for DmcElp3 dimerization and coordination of the iron–sulfur cluster are conserved with yeast Elp3. Structure-guided mutagenesis in conjunction with in vivo functional assays showed that these residues are critical to the activity of Elongator. Electrophoretic mobility shift assays further showed that tRNAs bind DmcElp3 at a highly conserved basic cavity located between the SAM and KAT domains. Subsequent RNase footprinting analyses further showed that DmcElp3 binds both the anticodon and the D loop of tRNAs. Although a model of tRNA in complex with Elp3 generated based on the above data predicts that the wobble base of the tRNA would be positioned near the active site of the KAT domain, further structural and biochemical investigations would be needed to define the precise catalytic mechanism.
Elp4, Elp5, and Elp6
The Elp4, Elp5, and Elp6 are the smallest subunits of the Elongator complex that assemble into a discrete subcomplex (Elp4/5/6). Although these three proteins were all identified in the original genetic screen for strains resistant to the K. lactis zymocin toxin, this subcomplex was only recovered as part of Elongator after improvement in purification method [2–4]. It was found that Elp4/5/6 subcomplex can be dissociated from “core” Elongator upon treatment of the complex with high salt concentrations [2]. Crystallographic studies carried out by two different research groups demonstrated that yeast Elp4/5/6 assemble into a heterotrimer that dimerizes to form a heterohexameric ring (Fig. 1d) [70, 71]. The two crystal structures also revealed that all three subunits adopt identical RecA ATPase-like folds that contain a mostly parallel and twisted β-sheet flanked by α-helices. Electrophoretic mobility shift assays showed that the Elp4/5/6 hexamer directly bound to tRNA [70]. However, this interaction was not mediated through positively charged binding loops, as has been observed for other RecA-like proteins. Instead, tRNA was found to bind a central L2 loop of Elp6 but not Elp4 or Elp5. In line with the high degree of observed structural homology to RecA, Elp4/5/6 can bind and hydrolyze ATP [70]. Interestingly, the presence of ATP reduces the affinity of Elp4/5/6 to tRNA in a concentration-dependent manner, suggesting that ATP hydrolysis may promote tRNA dissociation after wobble uridine modification. Finally, the Elp4/5/6 was shown to co-immunoprecipitate with the first 28 residues of yeast histone H3 [71]; however, the relevance of this interaction in the context of histones assembled into nucleosomes remains unclear. Collectively, the two structural investigations showed that Elp4/5/6 adopts a hexameric ring shaped subcomplex that may play a role in substrate binding and/or turnover.
Structure of full Elongator complex
Despite the wealth of high-resolution structural data on individual Elongator subunits, the overall architecture of the Elongator complex remained unknown until very recently. Upon full assembly, the holo-Elongator complex has a molecular mass of ~ 850 kDa, making it a suitable target for structural analysis by single-particle electron microscopy (EM). Indeed, two groups successfully utilized negative stain electron microscopy to determine the 3-dimensional reconstructions of native Elongator isolated from yeast [63, 72]. These structures showed that Elongator adopts an overall “moth-like” bilobal architecture with two lobes connected by a small density (Fig. 1d). Unexpectedly, the overall structure of Elongator is asymmetrical with one of the two lobes containing a hexameric density that corresponds to the Elp4/5/6 subcomplex. The two research groups also managed to isolate and characterize the structure of the Elp1/2/3 subcomplex. Their data showed that Elp1/2/3 subcomplex is also dimeric but symmetric, and it undergoes a conformational change upon binding Elp4/5/6. Since Elongator is inactive in the absence of the Elp4/5/6 subcomplex, the observed conformational change could potentially be a mechanism to activate the complex. However, in vitro reconstitution, experiments showed that upon incubation with an excess of Elp4/5/6, a second copy of this hexameric subcomplex can bind and be incorporated to the other lobe of Elongator [72]. This finding indicated that the observed “single-loaded” nature of native yeast Elongator could potentially be due to lower level of Elp4, Elp5, and Elp6 compared to Elp1, Elp2, and Elp3 within the yeast cell [73].
Complementary chemical cross linking coupled with mass spectrometry analysis of native Elongator provided a wealth of information regarding the subunit connectivity [63, 72]. Notably, a network of crosslinks was observed between the Elp1N-terminal domain, Elp2, Elp3, and Elp4, indicating that these regions likely form the interaction surface of the complex. By integrating this data with existing atomic resolution structures of individual subunits, a pseudo-atomic model of Elongator complex was built (Fig. 2). Based on this model, the two copies of the Elp3 subunit are located on each lobe, precluding the possibility of the formation of a dimer, as seen in the D. mccartyi Elp3 structure [69]. In addition, two cavities were identified as potential binding sites for tRNA, one through the center of the Elp4/5/6 hexamer and another formed between Elp1CTD, Elp2, and Elp3. However, due to the low resolution of these structures, it remains difficult to confirm and further define the substrate-binding site. Therefore, further studies are warranted to generate a near-atomic resolution model of Elongator, perhaps in complex with tRNAs, using high-resolution cryo electron microscopy (cryo-EM) approaches.
Fig. 2.
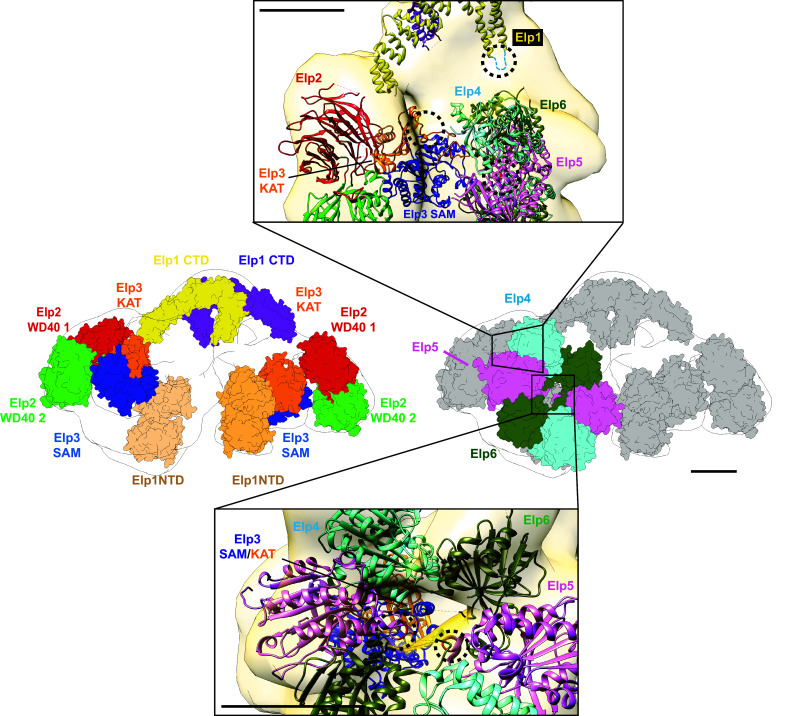
Model of the molecular assembly of the holo-Elongator complex. Three-dimensional reconstruction of S. cerevisiae Elongator obtained from negative stain EM (EMDB accession ID: EMD-8239). The crystal structures of Elp1 C-terminus (PDB ID: 5CQS), Elp2 (PDB ID: 5M2N), D. mccartyi Elp3 (PDB ID: 5L7J), Elp4/5/6 (PDB ID: 4A8J), and homology models of the Elp1 tandem WD40 were fitted into to electron density map to generate a multiscale model of Elongator subunit organization. The proposed tRNA interactions with Elongator are shown within the insets. Approximate locations of proposed tRNA–Elongator interaction sites have been highlighted using dashed circles. All scale bars in this figure represent 50 Å
Future directions
Although recent structural and biochemical investigations have yielded a wealth of novel information on individual Elongator subunits and the full holo-Elongator (Table 1), mechanistically how this complex exerts its diverse physiological functions remains unclear. First, different studies have shown that the Elp1, Elp3, and Elp6 subunits can all bind tRNAs, yet the physiological substrate-binding site remains unknown. Next, the relevance of the Elp2 subunit-binding microtubules [34] and the Elp4/5/6 subcomplex having ATPase activity [70] in the context of overall function of the Elongator complex is yet to be determined. Finally, Elp3 has been shown to interact with S-adenosylmethionine and acetyl-CoA. However, the precise roles of these cofactors in catalysis of wobble uridine modification remains speculative [69]. Although recent negative stain EM studies have enhanced our understanding of the overall architecture and subunit organization of yeast Elongator, addressing the fundamental questions listed above necessitates comprehensive understanding of the structural properties of holo-Elongator and Elongator in complex with tRNA substrates at much higher resolution. With the recent introduction of direct detection devices and new image processing algorithms, it is now routine to obtain near-atomic resolution structural information on diverse proteins and protein complexes using the cryo electron microscopy (cryo-EM) approach [74, 75]. We foresee that cryo-EM will be heavily relied on in the future to investigate the mechanism of action of Elongator.
Another topic that warrants further investigation is the molecular function of Elongator in higher eukaryotes. Although cellular and whole organism-level studies indicated that Elongator could have expanded roles in higher eukaryotes, limited amount of biochemical and structural studies have been conducted on the Elongator subunits and complex in these organisms. The development of new genome editing techniques such as the CRISPR–Cas9 system provides possible means to efficiently isolate native Elongator in higher eukaryotes for biochemical and structural analyses [76]. Investigating the impact of the FD causing mutation on the structural and biochemical properties of human Elongator will provide key information in understanding the molecular etiologies of the FD and other neuropathies. Similarly, the fact that mouse and C. elegans orthologs of Elongator have been implicated in neuronal development suggests that biochemical characterization of Elongator in these organisms is likely to have implications in understanding the molecular basis of neurological disorders.
Acknowledgements
This work was supported by a Natural Sciences and Engineering Research Council of Canada (NSERC) Discovery Grant (418157-2012), a Canadian Institutes of Health Research (CIHR) Foundation Grant (FDN-143228), a Michael Smith Foundation for Health Research Career Investigator Award, a CIHR New Investigator Award to CY, and an NSERC PGS-D fellowship to U.D.
Abbreviations
- ATP
Adenosine triphosphate
- CoA
Coenzyme A
- cm5U
5-carboxymethyluridine
- cryo-EM
Cryo electron microscopy
- Elp
Elongator protein
- EM
Electron microscopy
- FD
Familial dysautonomia
- GEF
Guanine-nucleotide exchange factor
- GNAT
Gcn5-related N-terminal acetyltransferase
- HAT
Histone acetyltransferase
- IKAP
IκB kinase associated protein
- IKBKAP
Inhibitor of kappa light polypeptide gene enhancer in B cells, kinase complex-associated protein
- IKI3
Insensitive to killer toxin-3
- IκB
Inhibitor of kappa-B
- mcm5U
5-Methoxycarbonylmethyluridine
- mcm5s2U
5-methoxycarbonylmethyl-2-thiouridine
- ncm5U
5-carbamoylmethyluridine
- RNAPII
RNA polymerase II
- SAM
S-adenosyl-l-methionine
- TPR
Tetratricopeptide repeat
- WD40
Typtophan-aspartic acid-40
References
- 1.Otero G, et al. Elongator, a multisubunit component of a novel RNA polymerase II holoenzyme for transcriptional elongation. Mol Cell. 1999;3:109–118. doi: 10.1016/S1097-2765(00)80179-3. [DOI] [PubMed] [Google Scholar]
- 2.Krogan NJ, Greenblatt JF. Characterization of a six-subunit holo-elongator complex required for the regulated expression of a group of genes in Saccharomyces cerevisiae. Mol Cell Biol. 2001;21:8203–8212. doi: 10.1128/MCB.21.23.8203-8212.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Winkler GS, Petrakis TG, Ethelberg S, Tokunaga M, Erdjument-Bromage H, Tempst P, Svejstrup JQ. RNA polymerase II elongator holoenzyme is composed of two discrete subcomplexes. J Biol Chem. 2001;276:32743–32749. doi: 10.1074/jbc.M105303200. [DOI] [PubMed] [Google Scholar]
- 4.Li Y, Takagi Y, Jiang Y, Tokunaga M, Erdjument-Bromage H, Tempst P, Kornberg RD. A multiprotein complex that interacts with RNA polymerase II elongator. J Biol Chem. 2001;276:29628–29631. doi: 10.1074/jbc.C100274200. [DOI] [PubMed] [Google Scholar]
- 5.Hawkes NA, et al. Purification and characterization of the human elongator complex. J Biol Chem. 2002;277:3047–3052. doi: 10.1074/jbc.M110445200. [DOI] [PubMed] [Google Scholar]
- 6.Wittschieben BO, et al. A novel histone acetyltransferase is an integral subunit of elongating RNA polymerase II holoenzyme. Mol Cell. 1999;4:123–128. doi: 10.1016/S1097-2765(00)80194-X. [DOI] [PubMed] [Google Scholar]
- 7.Frohloff F, Fichtner L, Jablonowski D, Breunig KD, Schaffrath R. Saccharomyces cerevisiae Elongator mutations confer resistance to the Kluyveromyces lactis zymocin. EMBO J. 2001;20:1993–2003. doi: 10.1093/emboj/20.8.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Butler AR, White JH, Folawiyo Y, Edlin A, Gardiner D, Stark MJ. Two Saccharomyces cerevisiae genes which control sensitivity to G1 arrest induced by Kluyveromyces lactis toxin. Mol Cell Biol. 1994;14:6306–6316. doi: 10.1128/MCB.14.9.6306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kawamoto S, et al. Isolation and characterization of mutants of Saccharomyces cerevisiae resistant to killer toxin of Kluyveromyces lactis. J Ferment Bioeng. 1990;70:222–227. doi: 10.1016/0922-338X(90)90052-X. [DOI] [Google Scholar]
- 10.Kishida M, Tokunaga M, Katayose Y, Yajima H, Kawamura-Watabe A, Hishinuma F. Isolation and genetic characterization of pGKL killer-insensitive mutants (iki) from Saccharomyces cerevisiae. Biosci Biotechnol Biochem. 1996;60:798–801. doi: 10.1271/bbb.60.798. [DOI] [PubMed] [Google Scholar]
- 11.Chinenov Y. A second catalytic domain in the Elp3 histone acetyltransferases: a candidate for histone demethylase activity? Trends Biochem Sci. 2002;27:115–117. doi: 10.1016/S0968-0004(02)02058-3. [DOI] [PubMed] [Google Scholar]
- 12.Huang B, Johansson MJ, Bystrom AS. An early step in wobble uridine tRNA modification requires the Elongator complex. RNA. 2005;11:424–436. doi: 10.1261/rna.7247705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Selvadurai K, Wang P, Seimetz J, Huang RH. Archaeal Elp3 catalyzes tRNA wobble uridine modification at C5 via a radical mechanism. Nat Chem Biol. 2014;10:810–812. doi: 10.1038/nchembio.1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johansson MJO, Xu F, Bystrom AS. Elongator—a tRNA modifying complex that promotes efficient translational decoding. Biochim Biophys Acta. 2017 doi: 10.1016/j.bbagrm.2017.11.006. [DOI] [PubMed] [Google Scholar]
- 15.Dauden MI, Jaciuk M, Muller CW, Glatt S. Structural asymmetry in the eukaryotic Elongator complex. FEBS Lett. 2017 doi: 10.1002/1873-3468.12865. [DOI] [PubMed] [Google Scholar]
- 16.Kolaj-Robin O, Seraphin B. Structures and activities of the Elongator complex and its cofactors. Enzymes. 2017;41:117–149. doi: 10.1016/bs.enz.2017.03.001. [DOI] [PubMed] [Google Scholar]
- 17.Winkler GS, Kristjuhan A, Erdjument-Bromage H, Tempst P, Svejstrup JQ. Elongator is a histone H3 and H4 acetyltransferase important for normal histone acetylation levels in vivo. Proc Natl Acad Sci USA. 2002;99:3517–3522. doi: 10.1073/pnas.022042899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wittschieben BO, Fellows J, Du W, Stillman DJ, Svejstrup JQ. Overlapping roles for the histone acetyltransferase activities of SAGA and elongator in vivo. EMBO J. 2000;19:3060–3068. doi: 10.1093/emboj/19.12.3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim JH, Lane WS, Reinberg D. Human Elongator facilitates RNA polymerase II transcription through chromatin. Proc Natl Acad Sci USA. 2002;99:1241–1246. doi: 10.1073/pnas.251672198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gilbert C, Kristjuhan A, Winkler GS, Svejstrup JQ. Elongator interactions with nascent mRNA revealed by RNA immunoprecipitation. Mol Cell. 2004;14:457–464. doi: 10.1016/S1097-2765(04)00239-4. [DOI] [PubMed] [Google Scholar]
- 21.Krogan NJ, et al. RNA polymerase II elongation factors of Saccharomyces cerevisiae: a targeted proteomics approach. Mol Cell Biol. 2002;22:6979–6992. doi: 10.1128/MCB.22.20.6979-6992.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Close P, et al. Transcription impairment and cell migration defects in elongator-depleted cells: implication for familial dysautonomia. Mol Cell. 2006;22:521–531. doi: 10.1016/j.molcel.2006.04.017. [DOI] [PubMed] [Google Scholar]
- 23.Pokholok DK, Hannett NM, Young RA. Exchange of RNA polymerase II initiation and elongation factors during gene expression in vivo. Mol Cell. 2002;9:799–809. doi: 10.1016/S1097-2765(02)00502-6. [DOI] [PubMed] [Google Scholar]
- 24.Fichtner L, Frohloff F, Jablonowski D, Stark MJ, Schaffrath R. Protein interactions within Saccharomyces cerevisiae Elongator, a complex essential for Kluyveromyces lactis zymocicity. Mol Microbiol. 2002;45:817–826. doi: 10.1046/j.1365-2958.2002.03055.x. [DOI] [PubMed] [Google Scholar]
- 25.Esberg A, Huang B, Johansson MJ, Bystrom AS. Elevated levels of two tRNA species bypass the requirement for elongator complex in transcription and exocytosis. Mol Cell. 2006;24:139–148. doi: 10.1016/j.molcel.2006.07.031. [DOI] [PubMed] [Google Scholar]
- 26.Lu J, Huang B, Esberg A, Johansson MJ, Bystrom AS. The Kluyveromyces lactis gamma-toxin targets tRNA anticodons. RNA. 2005;11:1648–1654. doi: 10.1261/rna.2172105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen C, Tuck S, Bystrom AS. Defects in tRNA modification associated with neurological and developmental dysfunctions in Caenorhabditis elegans elongator mutants. PLoS Genet. 2009;5:e1000561. doi: 10.1371/journal.pgen.1000561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mehlgarten C, et al. Elongator function in tRNA wobble uridine modification is conserved between yeast and plants. Mol Microbiol. 2010;76:1082–1094. doi: 10.1111/j.1365-2958.2010.07163.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin FJ, Shen L, Jang CW, Falnes PO, Zhang Y. Ikbkap/Elp1 deficiency causes male infertility by disrupting meiotic progression. PLoS Genet. 2013;9:e1003516. doi: 10.1371/journal.pgen.1003516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Karlsborn T, Tukenmez H, Chen C, Bystrom AS. Familial dysautonomia (FD) patients have reduced levels of the modified wobble nucleoside mcm(5)s(2)U in tRNA. Biochem Biophys Res Commun. 2014;454:441–445. doi: 10.1016/j.bbrc.2014.10.116. [DOI] [PubMed] [Google Scholar]
- 31.Yoshida M, et al. Rectifier of aberrant mRNA splicing recovers tRNA modification in familial dysautonomia. Proc Natl Acad Sci USA. 2015;112:2764–2769. doi: 10.1073/pnas.1415525112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johansson MJ, Esberg A, Huang B, Bjork GR, Bystrom AS. Eukaryotic wobble uridine modifications promote a functionally redundant decoding system. Mol Cell Biol. 2008;28:3301–3312. doi: 10.1128/MCB.01542-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Solinger JA, et al. The Caenorhabditis elegans Elongator complex regulates neuronal alpha-tubulin acetylation. PLoS Genet. 2010;6:e1000820. doi: 10.1371/journal.pgen.1000820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dong C, et al. The Elp2 subunit is essential for elongator complex assembly and functional regulation. Structure. 2015;23:1078–1086. doi: 10.1016/j.str.2015.03.018. [DOI] [PubMed] [Google Scholar]
- 35.Jackson MZ, Gruner KA, Qin C, Tourtellotte WG. A neuron autonomous role for the familial dysautonomia gene ELP1 in sympathetic and sensory target tissue innervation. Development. 2014;141:2452–2461. doi: 10.1242/dev.107797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ueki Y, Ramirez G, Salcedo E, Stabio ME, Lefcort F (2016) Loss of Ikbkap causes slow, progressive retinal degeneration in a mouse model of familial dysautonomia. eNeuro. 10.1523/ENEURO.0143-16.2016 [DOI] [PMC free article] [PubMed]
- 37.Okada Y, Yamagata K, Hong K, Wakayama T, Zhang Y. A role for the elongator complex in zygotic paternal genome demethylation. Nature. 2010;463:554–558. doi: 10.1038/nature08732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Laguesse S, et al. A dynamic unfolded protein response contributes to the control of cortical neurogenesis. Dev Cell. 2015;35:553–567. doi: 10.1016/j.devcel.2015.11.005. [DOI] [PubMed] [Google Scholar]
- 39.Ohlen SB, Russell ML, Brownstein MJ, Lefcort F. BGP-15 prevents the death of neurons in a mouse model of familial dysautonomia. Proc Natl Acad Sci USA. 2017;114:5035–5040. doi: 10.1073/pnas.1620212114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nelissen H, et al. DRL1, a homolog of the yeast TOT4/KTI12 protein, has a function in meristem activity and organ growth in plants. Plant Cell. 2003;15:639–654. doi: 10.1105/tpc.007062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nelissen H, et al. The elongata mutants identify a functional Elongator complex in plants with a role in cell proliferation during organ growth. Proc Natl Acad Sci USA. 2005;102:7754–7759. doi: 10.1073/pnas.0502600102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Falcone A, Nelissen H, Fleury D, Van Lijsebettens M, Bitonti MB. Cytological investigations of the Arabidopsis thaliana elo1 mutant give new insights into leaf lateral growth and Elongator function. Ann Bot. 2007;100:261–270. doi: 10.1093/aob/mcm102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou X, Hua D, Chen Z, Zhou Z, Gong Z. Elongator mediates ABA responses, oxidative stress resistance and anthocyanin biosynthesis in Arabidopsis. Plant J. 2009;60:79–90. doi: 10.1111/j.1365-313X.2009.03931.x. [DOI] [PubMed] [Google Scholar]
- 44.DeFraia CT, Zhang X, Mou Z. Elongator subunit 2 is an accelerator of immune responses in Arabidopsis thaliana. Plant J. 2010;64:511–523. doi: 10.1111/j.1365-313X.2010.04345.x. [DOI] [PubMed] [Google Scholar]
- 45.Anderson SL, Coli R, Daly IW, Kichula EA, Rork MJ, Volpi SA, Ekstein J, Rubin BY. Familial dysautonomia is caused by mutations of the IKAP gene. Am J Hum Genet. 2001;68:753–758. doi: 10.1086/318808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Slaugenhaupt SA, et al. Tissue-specific expression of a splicing mutation in the IKBKAP gene causes familial dysautonomia. Am J Hum Genet. 2001;68:598–605. doi: 10.1086/318810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cuajungco MP, et al. Tissue-specific reduction in splicing efficiency of IKBKAP due to the major mutation associated with familial dysautonomia. Am J Hum Genet. 2003;72:749–758. doi: 10.1086/368263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hug N, Longman D, Caceres JF. Mechanism and regulation of the nonsense-mediated decay pathway. Nucleic Acids Res. 2016;44:1483–1495. doi: 10.1093/nar/gkw010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Boone N, Bergon A, Loriod B, Deveze A, Nguyen C, Axelrod FB, Ibrahim EC. Genome-wide analysis of familial dysautonomia and kinetin target genes with patient olfactory ecto-mesenchymal stem cells. Hum Mutat. 2012;33:530–540. doi: 10.1002/humu.22010. [DOI] [PubMed] [Google Scholar]
- 50.Cohen JS, Srivastava S, Farwell KD, Lu HM, Zeng W, Lu H, Chao EC, Fatemi A. ELP2 is a novel gene implicated in neurodevelopmental disabilities. Am J Med Genet A. 2015;167:1391–1395. doi: 10.1002/ajmg.a.36935. [DOI] [PubMed] [Google Scholar]
- 51.Simpson CL, et al. Variants of the elongator protein 3 (ELP3) gene are associated with motor neuron degeneration. Hum Mol Genet. 2009;18:472–481. doi: 10.1093/hmg/ddn375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kwee LC, et al. A high-density genome-wide association screen of sporadic ALS in US veterans. PLoS One. 2012;7:e32768. doi: 10.1371/journal.pone.0032768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reinthaler EM, et al. Analysis of ELP4, SRPX2, and interacting genes in typical and atypical rolandic epilepsy. Epilepsia. 2014;55:e89–e93. doi: 10.1111/epi.12712. [DOI] [PubMed] [Google Scholar]
- 54.Strug LJ, et al. Centrotemporal sharp wave EEG trait in rolandic epilepsy maps to Elongator Protein Complex 4 (ELP4) Eur J Hum Genet. 2009;17:1171–1181. doi: 10.1038/ejhg.2008.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Addis L, et al. Microdeletions of ELP4 are associated with language impairment, autism spectrum disorder, and mental retardation. Hum Mutat. 2015;36:842–850. doi: 10.1002/humu.22816. [DOI] [PubMed] [Google Scholar]
- 56.Di Santo R, Bandau S, Stark MJ. A conserved and essential basic region mediates tRNA binding to the Elp1 subunit of the Saccharomyces cerevisiae Elongator complex. Mol Microbiol. 2014;92:1227–1242. doi: 10.1111/mmi.12624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mehlgarten C, Jablonowski D, Breunig KD, Stark MJ, Schaffrath R. Elongator function depends on antagonistic regulation by casein kinase Hrr25 and protein phosphatase Sit4. Mol Microbiol. 2009;73:869–881. doi: 10.1111/j.1365-2958.2009.06811.x. [DOI] [PubMed] [Google Scholar]
- 58.Abdel-Fattah W, et al. Phosphorylation of Elp1 by Hrr25 is required for elongator-dependent tRNA modification in yeast. PLoS Genet. 2015;11:e1004931. doi: 10.1371/journal.pgen.1004931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xu H, et al. Dimerization of elongator protein 1 is essential for Elongator complex assembly. Proc Natl Acad Sci USA. 2015;112:10697–10702. doi: 10.1073/pnas.1502597112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chaverra M, et al. The familial dysautonomia disease gene IKBKAP is required in the developing and adult mouse central nervous system. Dis Model Mech. 2017;10:605–618. doi: 10.1242/dmm.028258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rahl PB, Chen CZ, Collins RN. Elp1p, the yeast homolog of the FD disease syndrome protein, negatively regulates exocytosis independently of transcriptional elongation. Mol Cell. 2005;17:841–853. doi: 10.1016/j.molcel.2005.02.018. [DOI] [PubMed] [Google Scholar]
- 62.Fellows J, Erdjument-Bromage H, Tempst P, Svejstrup JQ. The Elp2 subunit of elongator and elongating RNA polymerase II holoenzyme is a WD40 repeat protein. J Biol Chem. 2000;275:12896–12899. doi: 10.1074/jbc.275.17.12896. [DOI] [PubMed] [Google Scholar]
- 63.Dauden MI, et al. Architecture of the yeast Elongator complex. EMBO Rep. 2017;18:264–279. doi: 10.15252/embr.201643353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Creppe C, et al. Elongator controls the migration and differentiation of cortical neurons through acetylation of alpha-tubulin. Cell. 2009;136:551–564. doi: 10.1016/j.cell.2008.11.043. [DOI] [PubMed] [Google Scholar]
- 65.Paraskevopoulou C, Fairhurst SA, Lowe DJ, Brick P, Onesti S. The Elongator subunit Elp3 contains a Fe4S4 cluster and binds S-adenosylmethionine. Mol Microbiol. 2006;59:795–806. doi: 10.1111/j.1365-2958.2005.04989.x. [DOI] [PubMed] [Google Scholar]
- 66.Greenwood C, Selth LA, Dirac-Svejstrup AB, Svejstrup JQ. An iron–sulfur cluster domain in Elp3 important for the structural integrity of elongator. J Biol Chem. 2009;284:141–149. doi: 10.1074/jbc.M805312200. [DOI] [PubMed] [Google Scholar]
- 67.Kolaj-Robin O, McEwen AG, Cavarelli J, Seraphin B. Structure of the Elongator cofactor complex Kti11/Kti13 provides insight into the role of Kti13 in Elongator-dependent tRNA modification. FEBS J. 2015;282:819–833. doi: 10.1111/febs.13199. [DOI] [PubMed] [Google Scholar]
- 68.Glatt S, et al. Structure of the Kti11/Kti13 heterodimer and its double role in modifications of tRNA and eukaryotic elongation factor 2. Structure. 2015;23:149–160. doi: 10.1016/j.str.2014.11.008. [DOI] [PubMed] [Google Scholar]
- 69.Glatt S, et al. Structural basis for tRNA modification by Elp3 from Dehalococcoides mccartyi. Nat Struct Mol Biol. 2016;23:794–802. doi: 10.1038/nsmb.3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Glatt S, Letoquart J, Faux C, Taylor NM, Seraphin B, Muller CW. The Elongator subcomplex Elp456 is a hexameric RecA-like ATPase. Nat Struct Mol Biol. 2012;19:314–320. doi: 10.1038/nsmb.2234. [DOI] [PubMed] [Google Scholar]
- 71.Lin Z, Zhao W, Diao W, Xie X, Wang Z, Zhang J, Shen Y, Long J. Crystal structure of elongator subcomplex Elp4-6. J Biol Chem. 2012;287:21501–21508. doi: 10.1074/jbc.M112.341560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Setiaputra DT, et al. Molecular architecture of the yeast Elongator complex reveals an unexpected asymmetric subunit arrangement. EMBO Rep. 2017;18:280–291. doi: 10.15252/embr.201642548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O’Shea EK, Weissman JS. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 74.Kuhlbrandt W. Biochemistry. The resolution revolution. Science. 2014;343:1443–1444. doi: 10.1126/science.1251652. [DOI] [PubMed] [Google Scholar]
- 75.Smith MT, Rubinstein JL. Structural biology. Beyond blob-ology. Science. 2014;345:617–619. doi: 10.1126/science.1256358. [DOI] [PubMed] [Google Scholar]
- 76.Dalvai M, Loehr J, Jacquet K, Huard CC, Roques C, Herst P, Cote J, Doyon Y. A scalable genome-editing-based approach for mapping multiprotein complexes in human cells. Cell Rep. 2015;13:621–633. doi: 10.1016/j.celrep.2015.09.009. [DOI] [PubMed] [Google Scholar]
- 77.Sun J, et al. Solution structure of Kti11p from Saccharomyces cerevisiae reveals a novel zinc-binding module. Biochemistry. 2005;44:8801–8809. doi: 10.1021/bi0504714. [DOI] [PubMed] [Google Scholar]