Abstract
In the past two decades, transmembrane channel-like (TMC) proteins have attracted a significant amount of research interest, because mutations of Tmc1 lead to hereditary deafness. As evolutionarily conserved membrane proteins, TMC proteins are widely involved in diverse sensorimotor functions of many species, such as hearing, chemosensation, egg laying, and food texture detection. Interestingly, recent structural and physiological studies suggest that TMC channels may share a similar membrane topology with the Ca2+-activated Cl− channel TMEM16 and the mechanically activated OSCA1.2/TMEM63 channel. Namely, these channels form dimers and each subunit consists of ten transmembrane segments. Despite this important structural insight, a key question remains: what is the gating mechanism of TMC channels? The major technical hurdle to answer this question is that the reconstitution of TMC proteins as functional ion channels has been challenging in mammalian heterologous systems. Since TMC channels are conserved across taxa, genetic studies of TMC channels in model organisms such as C. elegans, Drosophila, and zebrafish may provide us critical information on the physiological function and regulation of TMCs. Here, we present a comparative overview on the diverse functions of TMC channels in different species.
Keywords: TMC channels, C. elegans, Drosophila, Zebrafish, Membrane excitability, Hearing, Mechanosensation
Introduction
Transmembrane channel-like (TMC) proteins are a novel family of ion channel-like proteins that are conserved from C. elegans to humans [1, 2]. The discovery of TMC1, the founding member of TMC family proteins, benefited tremendously from genetic studies of deafness in mice and humans. In 1958, Deol and Kocher first reported a deafness (dn) mouse strain with a recessive inheritance [3]. More than 20 years later, Steel and Bock examined these deafness mice and found that they lacked cochlear microphonics [4]. Within a few years, human siblings with recessive deafness were identified and its locus was linked to 9qll–q21 (DFNB7) region [5, 6]. Remarkably, this region of the human genome is syntenic to the mouse deafness (dn) locus, implying that mutations of a common gene are likely responsible for the deafness in both humans and mice. In 2002, using positional cloning, Kurima et al. identified eight mutations of a novel gene, Tmc1, in a dominant deafness locus DFNA36 which maps to the human chromosome 9q13–21 in a region overlapping the DFNB7/B11 locus for recessive deafness [7]. They also showed that a 1.6 kb deletion in Tmc1 was causative disruption in the deafness (dn) mouse [7]. Coincidently, an independent study from Vreugde et al. reported that the Beethoven (Bth) mutant mouse, which carried a methionine to lysine substitution of residue 412 in TMC1, is a new mouse model for dominant, progressive hearing loss DFNA36 [8]. Collectively, these exciting studies have paved the way for the discovery of TMC family proteins.
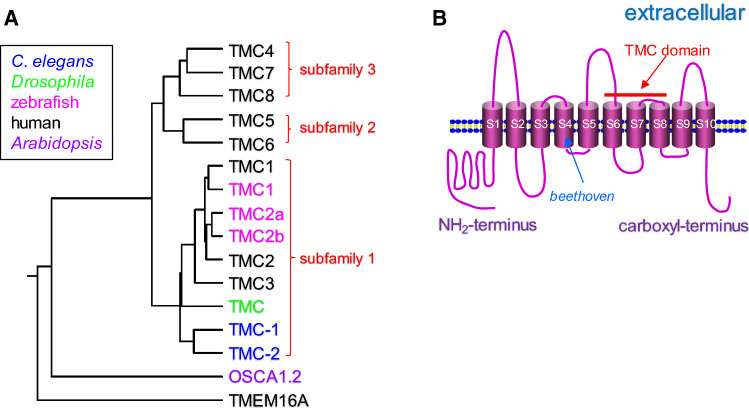
Through sequence homology search, seven additional TMC channels (TMC2–TMC8) were identified in humans and mice (Fig. 1a) [2, 7]. These TMC channels were strongly predicted to encode proteins with 6–10 transmembrane domains and they all contain a novel conserved 120-amino-acid sequence termed TMC domain (Fig. 1b). Based on their amino-acid sequence similarities, TMC channels can be further grouped into three subfamilies that include TMCs 1–3, TMCs 5 and 6, and TMCs 4, 7 and 8 [2]. In addition to mammals, homologues of TMC channels were identified in other species such as C. elegans, Drosophila, and zebrafish, indicating that TMC proteins might have evolutionarily conserved functions. Here, we present an overview of the diverse functions of TMC channels in different species.
Fig. 1.
TMC channels and their predicted membrane topology. a Protein sequences of TMC channels from C. elegans (in blue), Drosophila (in green), zebrafish (in magenta), and human (in black), OSCA1.2 channel from Arabidopsis (in purple), and TMEM16A channel from human (in black) are aligned up using ClustalW (https://www.genome.jp/tools-bin/clustalw). All these ion channels are predicted to share a similar membrane topology. Based on sequence homology, TMC channels form three subfamilies: TMCs1–3 (subfamily 1), TMCs5–6 (subfamily 2), and TMCs4, 7 and 8 (subfamily 3). b Predicted membrane topology of TMC1 channel. In general, TMC1 protein contains ten transmembrane segments (S1–S10) and distinct mutations of TMC1 can cause both dominant and recessive deafness in mammals. Shown is the beethoven missense mutation of TMC1 (M412K) in the fourth transmembrane segment, which causes dominant deafness in mice. The most conserved region among all TMC proteins is the TMC domain which is formed by the S6–S8 segments
TMC channels in C. elegans
Chemosensation
The C. elegans genome encodes two TMC proteins, TMC-1 and TMC-2, which are most similar to mammalian TMC3 [9]. The C. elegans TMC-1 is widely expressed in multiple types of neurons (e.g., ASH, ADF, ASE, ADL, AQR, PQR, URX, and PHA), body wall muscle, and vulval muscle, while TMC-2 is mainly expressed in the body wall muscle and vulval muscle [9–11]. TMC-1 was originally reported as a sodium-sensitive ion channel and it seemed to be required for the ASH neuron-mediated high salt chemosensation [10]. However, Wang et al. later reported that, acting in the ASH neuron, TMC-1 actually mediated the nociceptive response to high pH, but not sodium, allowing worms to avoid strongly alkaline environments, where most animals cannot survive [12].
The C. elegans ASH neuron is a polymodal sensory neuron that is sensitive to multiple nociceptive stimuli such as high osmolarity, alkaline pH, and aversive odorants [13]. Intriguingly, although acid sensation has been well characterized before, we know little about how animals sense alkali in the environment. By screening a collection of ion channel (e.g., TRP, ENaC/DEG, TMC, and CNG channels) mutants for defects in the alkali-induced avoidance behavior, Wang et al. found that both osm-9 (a TRPV channel) and tmc-1 mutants were severely defective in this avoidance behavior [12]. Furthermore, transgenic expression of tmc-1 or osm-9 cDNA in ASH was sufficient to rescue the behavioral phenotype, suggesting that they act in ASH to mediate the alkaline pH-induced avoidance behavior.
Using whole-cell patch clamp recordings, Wang et al. next recorded from the tmc-1;osm-9 double mutant and found that the double mutant abolished the alkali-evoked current in ASH. Moreover, the tmc-1 single mutant has a significantly stronger defect in the alkali-evoked current than does the osm-9 mutant, indicating that TMC-1, but not OSM-9, might be the major contributor of alkali sensing in ASH [12]. Interestingly, while OSM-9 is sensitive to both acidic and basic pH, TMC-1 exhibits a specificity toward alkali, as it is required for the alkali- but not acid-triggered behavioral response and electrical current in ASH [12]. Based on these findings, a working model has been established on how the nociceptive ASH neuron transduces environmental stimuli (Fig. 2a) [14].
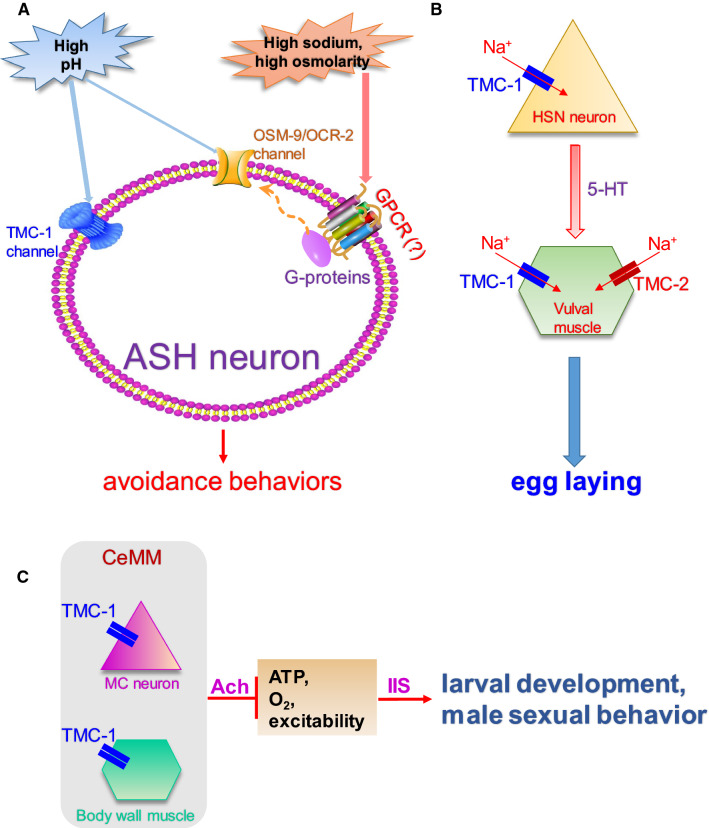
Fig. 2.
Functions of TMC channels in C. elegans. a TMC-1 acts in the nociceptive ASH neuron to mediate the alkaline sensation. Both TMC-1 and TPRV (OSM-9, OCR-2) channels are expressed in the ASH neuron. TMC-1 is a major player in sensing noxious alkaline environment independent of G protein signaling. By contrast, high sodium, high osmolarity seem to activate the C. elegans TRPV channels through G protein signaling and unknown G protein-coupled receptors (GPCRs) [14]. b TMC channels promote C. elegans egg laying by modulating membrane excitability. TMC channels are expressed in the egg-laying circuit of C. elegans (HSN neurons and vulval muscle), where they modulate membrane excitability through a background Na+-leak conductance [9]. The HSN motor neuron secretes serotonin (5HT) to innervate vulval muscle cells. c Synthetic CeMM suppresses larval development and male sexual behaviors in C. elegans, which is mediated by TMC-1 in the pharyngeal MC neurons and body wall muscle cells through cholinergic signaling and insulin/insulin-like growth factor signaling (IIS) [11]
Promoting membrane excitability
Membrane excitability is a fundamental feature for all excitable cells (e.g., neurons, muscle cells, and certain neuroendocrine cells) by determining the responsiveness of these cells. The resting membrane potential is tightly controlled by the counteractions between hyperpolarizing leak conductances (K+ and Cl−) and depolarizing leak conductances (Na+). While the two-pore-domain potassium channels are well established to function as background K+-leak channels and hyperpolarize the plasma membrane [15], much less is known about the depolarizing background leak channels. NCA/NALCN channels mediate a depolarizing Na+-leak conductance in certain types of neurons to modulate many physiological processes including basal locomotion, response to volatile anesthetics, and normal respiratory rhythms [16–20]. However, extra neuronal Na+-leak currents are still present in the absence of NCA/NALCN proteins and the channels mediating muscular background depolarizing leak conductance are unknown [19, 20].
Recently, we identified an unexpected role of TMC channels: both TMC-1 and TMC-2 are required for normal egg laying in C. elegans (Fig. 2b) [9]. The disruption of TMC proteins hyperpolarizes membrane potential and reduces the rhythmic calcium activities of neurons and muscle cells involved in egg laying. Through electrophysiological recordings, we demonstrated that TMC channels can function as another type of background leak channel to modulate membrane excitability in both neurons and muscle cells [9]. Furthermore, we found that TMC channels were non-selective cation channels, though they displayed the highest permeability to Na+. Finally, unlike NCA/NALCN channels that are known to be inhibited by Ca2+, TMC channels appeared to be insensitive to the extracellular Ca2+, supporting the idea that TMC channels and NCA/NALCN channels are distinct types of background leak channels [9].
Since TMC-1 mediates alkaline sensation in the ASH neuron [12], we studied whether the membrane excitability of ASH is affected in the tmc-1 mutant background. Consistent with the result obtained in the C. elegans egg-laying circuit, the resting membrane potential of ASH neurons in the tmc-1 mutant is hyperpolarized and the background Na+-leak current is also significantly reduced [9]. Thus, TMC-1 mediates a background Na+-leak conductance in ASH. However, whether the Na+-leak conductance role of TMC-1 is involved in alkaline sensation remains unclear, as TMC-1 may be directly activated by alkaline pH. On the other hand, since both TMC-1 and TMC-2 are expressed in the body wall muscle [9, 11], it would be interesting to examine whether muscle cells are chemosensitive by responding to alkaline pH or high salt.
A major technical hurdle to study the function and regulation of TMC proteins is that it has been challenging to express them as functional ion channels in mammalian heterologous expression systems. When expressed in mammalian cell lines, most TMC proteins are trapped in the endoplasmic reticulum (ER) component. Intriguingly, ectopic expression of mammalian TMC proteins (TMC1, TMC2, and TMC3) in either neurons or muscle cells can rescue the defective egg laying and resting membrane potential observed in the C. elegans tmc mutants, suggesting that these TMC channels might have an evolutionarily conserved role in modulating membrane excitability.
Regulating development and sexual behavior
In the chemically defined synthetic C. elegans Maintenance Medium (CeMM) which is a suboptimal food source compared to the standard diet E. coli OP50, signaling from the pharyngeal MC neurons and body wall muscles can significantly slow larval development [11]. Interestingly, the tmc-1 mutant exhibited an accelerated development rate in CeMM by promoting the cellular metabolism of MC neurons and body wall muscle (Fig. 2c) [11]. In addition to development, CeMM suppresses male sexual behaviors and tmc-1 mutation attenuated the diet-induced inhibition of male sexual behaviors [11]. Thus, TMC-1 mediates the overall inhibitory effect of CeMM on larval development and male sexual behaviors. Notably, the synthetic CeMM contains no sodium. As TMC-1 promotes membrane excitability by acting as a background Na+-leak channel in the egg-laying circuit [9], could the low extracellular sodium concentration of CeMM trigger the inhibitory effect on development and sexual behaviors? Three lines of evidence argue against this notion. First, adding sodium salt into CeMM had no effect on development for both CeMM-fed wild type and tmc-1 mutant worms [11]. Second, although both TMC-1 and TMC-2 channels mediate the background Na+-leak conductance in egg laying, the tmc-2 mutant worm did not exhibit fast development in CeMM [11], suggesting that TMC-1 and TMC-2 must have different roles in development and male sexual behaviors. Third, TMC proteins enhance membrane excitability in the egg-laying circuit, while TMC-1 suppresses cellular respiration in the CeMM-fed worms [9, 11]. Collectively, TMC-1, but not TMC-2, appears to have Na+-independent functions in suppressing cellular metabolism and delaying development in the nutrient-poor CeMM.
The proper trafficking of TMC channels onto the plasma membrane requires auxiliary subunits as heterologous mammalian TMC proteins are typically retained in intracellular compartments [21]. Notably, although a large portion of C. elegans TMC proteins are expressed on the plasma membrane, intracellular TMCs can also be detected [9]. Since the Na+ conductance function of TMC-1 is not required for CeMM to delay development and suppress sexual behaviors, intracellular TMC-1 or TMC-1-specific auxiliary subunit(s) might mediate TMC-1’s roles in development and male sexual behaviors. Taken together, the C. elegans TMCs appear to be multifunctional depending on which cells they are expressed in. Whether a unifying mechanism could explain the diverse functions of TMCs in C. elegans requires further investigation.
TMC channels in Drosophila
Detecting food texture
The Drosophila genome encodes a single TMC protein which is required for flies to sense food texture in the tongue. Drosophila can discriminate distinct food based on their hardness and viscosity. Zhang et al. found that the food texture discrimination depends upon a previously unknown multidendritic (md-L) neuron which extends elaborate dendritic arbors innervating the bases of taste hairs [22]. TMC channel is expressed in md-L neurons, where it is required for sensing the hardness and viscosity of food. Specifically, loss of tmc greatly reduced the ability to behaviorally discriminate the preferred softness (1% agarose) and smoothness (sucrose solution only) from harder and stickier food options [22]. Furthermore, mechanical deflection of taste sensilla can induce action potentials in md-L neurons and these mechanically activated action potentials were abolished in tmc mutant flies [22]. Collectively, this nice study revealed the cellular and molecular components that enable external sensory bristles on the fly tongue to communicate textural features to the brain through a pre-ingestive mechanism [22]. Although TMC seems to mediate food texture-triggered mechanical responses in flies, current evidence still cannot provide definitive answer to whether the Drosophila TMC protein is a mechanically gated ion channel as other TMC auxiliary subunits might be the real mechanosensor.
Proprioception
Proprioception, the sense of position, orientation, and movement of body parts, provides important sensory feedback information for animals to maintain proper gestures and coordinate their body movements [23, 24]. It has been known for centuries that proprioception is mediated by mechanosensitive proprioceptors [25]. Similarly, Drosophila larval locomotion, which entails rhythmic body contractions, is controlled by sensory feedback from proprioceptors. However, the molecular mechanism mediating this feedback is little understood. The Drosophila tmc gene is expressed in larval peripheral sensory neurons as well as the larval class I and class II dendritic arborization (da) neurons and bipolar dendrite (bd) neurons [26, 27]. Genetic disruption of tmc led to reduced crawling speed, increased head cast frequency, and enhanced backward locomotion. Importantly, expressing Drosophila TMC or mammalian TMC1 and/or TMC2 in the tmc-positive neurons rescued these mutant phenotypes [26], suggesting the functional conservation of TMC channels in mechanosensation. Interestingly, mechanical stimuli did not trigger any current in the Drosophila tmc-transfected S2 cells, while nompC (encoding a mechanosensitive TRPN channel)-transfected S2 cells exhibited mechanically activated currents [26]. Thus, the Drosophila TMC protein by itself might not be mechanosensitive and other auxiliary subunits may be required for its mechanosensitivity. An alternative explanation is that TMC proteins might not traffic to the plasma membrane in the Drosophila S2 expression system.
TMC channels in zebrafish
Auditory sensation
The zebrafish genome encodes three TMC channels: TMC1, TMC2a, and TMC2b of which the two TMC2 proteins are more closely related to mammalian TMC2. Similar to their mammalian counterparts, both zebrafish TMC1 and TMC2a channels can physically interact with protocadherin 15 (PCDH15), a key component of the mechanotransduction complex in auditory and vestibular hair cells [28]. In zebrafish, TMC channels seem to be restricted to hair cells of the inner ear and lateral line organ, suggesting their roles in hearing and mechanotransduction. In line with this notion, overexpression of a truncated TMC2a channel resulted in mislocalization of PCDH15 within hair bundles and disrupted mechanosensitivity of ear hair cells [28].
Several auxiliary proteins physically interact with zebrafish TMCs and regulate their proper trafficking into hair bundles. For example, TMIE is a two transmembrane domain protein expressed in the stereocilia of hair cells and mutations in TMIE cause deafness in zebrafish, mice, and humans [29–32]. In Tmie mutant zebrafish, TMC proteins fail to target to hair bundles, whereas TMIE overexpression promotes bundle localization of TMC proteins [33]. Mechanistically, the second transmembrane domain and adjacent regions of TMIE seem to be particularly important for targeting TMCs into hair bundles [33]. In addition, the Golgi apparatus-enriched TOMT protein interacts with TMCs within the secretory pathway, which allows TMCs to integrate into the mechanotransduction complex in zebrafish hair cells [34]. Notably, the regulation of TMCs by TOMT is also conserved in mammals as mouse TOMT and TMC1 directly interact with each other through the His183 residue of TOMT [34, 35].
Water motion detection
Fish rely on mechanosensitive hair cells located in neuromasts of the lateral line system on the animal’s surface to detect water motion. In zebrafish, TMC2b is robustly expressed in hair cells of the lateral line and Tmc2b null mutant displayed defective mechanotransduction [36]. Interestingly, neuromasts may contribute differentially to motor behaviors and distinct neuromasts with different orientations appear to express different levels of TMC2b, which may allow fish to detect water flow from different directions [36, 37]. For example, although M2 and MI1 neuromasts display high reliance on TMC2b, IO4 hair cells are largely independent of TMC2b [36]. Therefore, neuromasts may use different molecular mechanisms to encode water motion. Intriguingly, TOMT is highly expressed in both ear hair cells and lateral line neuromasts [34]. If TOMT plays a common role in targeting TMCs into stereocilia tips, it would be interesting to examine whether Tomt mutant fish are also defective in detecting water motion.
TMC channels in mammals
TMC1 and TMC2 in auditory transduction
To date, more than 40 different Tmc1 mutations (both dominant and recessive) have been found to cause deafness in humans and mice [21]. A summary table of mutations in TMCs and their associated diseases in humans and rodents is shown in Table 1. Surprisingly, there are no known spontaneous mutations of Tmc2 that cause a genetic disease, even though deletion mutations of Tmc2 in mice cause deafness if Tmc1 is absent. In rodents and humans, both Tmc1 and Tmc2 are expressed in inner ear hair cells, where they play an essential role in hearing transduction [38]. In addition, Tmc1 can be detected in other organs such as the brain, eye, colon and testis [1]. In the inner ear, the temporal expression pattern differs for the two Tmc genes [38]. Namely, in the cochlea, Tmc2 messenger RNA (mRNA) expression begins to rise at the base around birth and at the apex around postnatal day 2 (P2). After peak expression during the first postnatal week, Tmc2 expression begins to decline to near zero by postnatal day 10. As the expression of Tmc2 declines, Tmc1 expression begins to rise and is maintained into adulthood [38]. In the vestibular organs, although Tmc2 mRNA expression also precedes Tmc1, both Tmc1 and Tmc2 are expressed in mature vestibular hair cells. It should be noted that, in both auditory and vestibular organs, the spatiotemporal expression pattern of Tmc2 is well correlated with the onset of mechanosensitivity of hair cells [38–40]. Interestingly, deletion of either Tmc1 or Tmc2 does not eliminate mechanotransduction in early postnatal mice, while double knockouts display no conventional mechanotransduction in hair cells [38, 41, 42], indicating the potential functional redundancy between TMC1 and TMC2 channels.
Table 1.
TMC mutations in diseases of humans and rodents
| Gene | Mutations | Predicted effect | Ex/in number | Phenotype | References |
|---|---|---|---|---|---|
| Human | |||||
| TMC1 | c.16 + 1G > T, | Splice disruption | Intron 5 | Hearing impairment | [7, 68–95] |
| c.100C > T, | p.R34X nonsense | Exon 7 | |||
| c.1543T > C, | p.C515R missense | Exon 17 | |||
| c.2004T > G, | p.S668R missense | Exon 21 | |||
| c.64 + 2T > A, | Splice disruption | Intron 6 | |||
| c.1330G > A, | p.G444R missense | Exon 16 | |||
| c.1333C > T, | p.R445C missense | Exon 16 | |||
| c.2030T > C, | p.I677T missense | Exon 21 | |||
| c.1696_2283del, | 431 kb deletion | Exon 19 | |||
| c.195_16del, | 27 kb deletion | Exon 5 | |||
| c.295delA, | p.K99KfsX4 | Exon 8 | |||
| c.536-8T > A, | Splice disruption | Intron 10 | |||
| c.884 + 1G > A, | Splice disruption | Intron 13 | |||
| c.1534C > T, | p.R512X nonsense | Exon 17 | |||
| c.1960A > G, | p.M654V missense | Exon 20 | |||
| c.776 + 1G > A, | Splice disruption | Exon 7 | |||
| c.767delT, | p.F255FfsX14 | Exon 13 | |||
| c.1166G > A, | p.R389Q missense | Exon 15 | |||
| c.1810C > T, | p.R604X nonsense | Exon 20 | |||
| c.1165C > T, | p.R389X nonsense | Exon 15 | |||
| c.1764G > A, | p.W588X nonsense | Exon 20 | |||
| c.237-6T > G, | Splice disruption | Intron 7 | |||
| c.453 + 2T > C, | Splice disruption | Intron 9 | |||
| c.628_630delATC, | p.I210del | Exon 11 | |||
| c.800G > A, | p.G267E missense | Exon 13 | |||
| c.1114G > A, | p.V372M missense | Exon 15 | |||
| c.1566 + 1G > A, | Splice disruption | Intron 17 | |||
| c.596A > T, | p.N199I missense | Exon 11 | |||
| c.1404 + 1G > T, | Splice disruption | Intron 16 | |||
| c.1788C > A, | p.S596R missense | Exon 20 | |||
| c.150delT, | p.N50KfsX25 | Exon 7 | |||
| c.362 + 18A > G, | p.Glu122Tyrfs*10 | Exon 8 | |||
| c.1714G > C, | p.D572N missense | Exon 9 | |||
| c.1714G > A, | p.D572H missense | Exon 9 | |||
| c.236 + 1G > C, | Splice disruption | Intron 7 | |||
| c.458G > A, | p.W153X nonsense | Exon 10 | |||
| c.582G > A, | p.W194X nonsense | Exon 11 | |||
| c.589G > A, | p.G197R missense | Exon 11 | |||
| c.776A > G, | p.Y259C missense | Exon 13 | |||
| c.797T > C, | p.I266T missense | Exon 13 | |||
| c.821C > T, | p.P274Lmissense | Exon 13 | |||
| c.830A > G, | p.Y277C missense | Exon 13 | |||
| c.1083_1087del, | p.R362PfsX6 | Exon 15 | |||
| c.1107C > A, | p.N369K missense | Exon 15 | |||
| c.1171C > T, | p.Q391X nonsense | Exon 15 | |||
| c.1209G > C, | p.W403C missense | Exon 15 | |||
| c.1334G > A, | p.R445H missense | Exon 16 | |||
| c.1396_1398AAC, | p.N466del | Exon 16 | |||
| c.1312G > A, | p.A438T missense | Exon 16 | |||
| c.1253T > A, | p.M418K missense | Exon 16 | |||
| c.1249G > A, | p.G417R missense | Exon 16 | |||
| c.1247T > G, | p.L416R missense | Exon 16 | |||
| c.1589_1590delCT, | p.S530X frameshift | Exon 18 | |||
| c.1718T > A, | p.I573N missense | Exon 19 | |||
| c.1714G > A, | p.D572N missense | Exon 19 | |||
| c.1714G > C, | p.D572H missense | Exon 19 | |||
| c.1763 + 3A > G, | p.W588WfsX81 | Intron 19 | |||
| c.IVS19 + 5G > A, | splice disruption | Intron 19 | |||
| c.1939T > C, | p.S647P missense | Exon 20 | |||
| c.1959C > G, | p.Y653X missense | Exon 20 | |||
| c.1979C > T, | p.P660L missense | Exon 20 | |||
| c.2035G > A, | p.E679K missense | Exon 21 | |||
| c.2050G > A, | p.D684N missense | Exon 21 | |||
| c.2130_1delG, | splice disruption | Exon 22 | |||
| c.2210_2211insCT, | p.E737HfsX2 | Exon 23 | |||
| c.2260 + 2T > A | splice disruption | Intron 23 | |||
| TMC2 | Unknown | Unknown | Unknown | Unknown | |
| TMC3 | Unknown | Unknown | Unknown | Unknown | |
| TMC4 | Unknown | Unknown | Unknown | Unknown | |
| TMC5 | Up-regulated | Unknown | Unknown | Intrahepatic cholangiocarcinoma (ICC) | [96] |
| TMC6 (EVER1) | c.280C → T, | p.R94X nonsense | Exon 5 | Epidermodysplasia verruciformis (EV) | [97, 98] |
| c.1726G → T | p.G576X nonsense | Exon 14 | |||
| TMC7 | Unknown | Unknown | Unknown | Unknown | |
| TMC8 (EVER2) | c.754/755delT, | p.F252fs X283 | Exon 7 | Epidermodysplasia verruciformis (EV) | [97, 98] |
| c.1084G → T | p.E362X nonsense | Exon 9 | |||
| Rodents | |||||
| TMC1 | c.1235T > A (Beethoven mutation), | p.M412K missense | Exon 13 | Deafness | [7, 8, 38, 99, 100] |
| c.1387_1557del (deafness mutation), | p.I463_Q519del | Exon 14 | |||
| c.545A > G (baringo mutation), | p.Y182C missense | Exon 8 | |||
| c.1345T > C (nice mutation), | p.Y449H missense | Exon 13 | |||
| c.1661G > T (stitch mutation), | p.W554L missense | Exon 15 | |||
| Tmc1Δ/Δ | Targeted deletion | ||||
| TMC2 | Unknown | Unknown | Unknown | Unknown | |
| TMC3 | Unknown | Unknown | Unknown | Unknown | |
| TMC4 | Unknown | Unknown | Unknown | Unknown | |
| TMC5 | Unknown | Unknown | Unknown | Unknown | |
| TMC6 | Unknown | Unknown | Unknown | Unknown | |
| TMC7 | Unknown | Unknown | Unknown | Unknown | |
| TMC8 | Unknown | Unknown | Unknown | Unknown | |
All annotations are based on cDNAs (c.) of TMC proteins
p protein, del deletion, fs frame shift, ex exon, in intron
Both TMC1 and TMC2 channels are located at the tips of hair cell stereocilia [43–45], where the mechanotransduction channels are located [46–48]. Very interestingly, hair cells that express Tmc1 but not Tmc2 exhibit smaller single-channel conductance, lower calcium permeability, and faster adaptation. By contrast, hair cells with Tmc2 but not Tmc1 displayed the opposite biophysical features and those with both Tmc1 and Tmc2 expression contain at least four distinct conductances, suggesting the formation of heteromultimers [42]. Furthermore, the Beethoven (Bth) mutation (M412K) of TMC1 altered ionic selectivity and biophysical properties [42, 49, 50], whereas two other TMC1 mutations (M412C, D569C) alter the binding affinity to the pore blocker dihydrostreptomycin [49, 51]. Collectively, these results suggest that TMC1 is a key component of hearing transduction pathway.
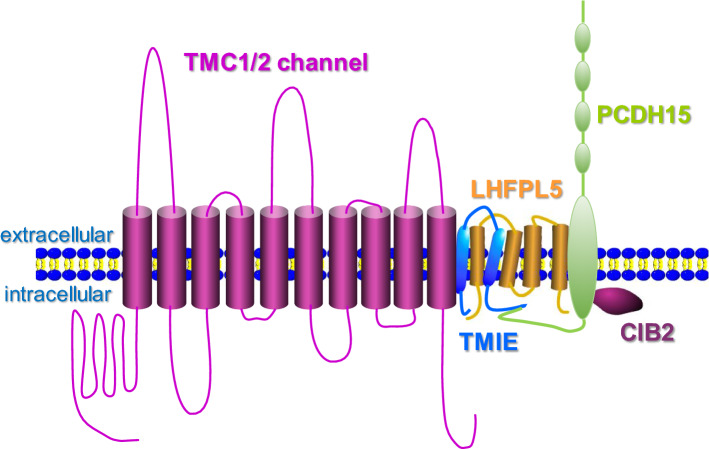
The proper targeting of TMC1 and TMC2 proteins to the tips of hair cell stereocilia and their functional integrity in hearing transduction depend on several accessory binding partners, including PCDH15, TMIE, LHFPL5, and CIB2 (Fig. 3) [52]. For example, the tip link protein PCDH15 may transduce tip link tension into opening of the mechanically activated hearing transduction channel by directly interacting with LHFPL5 and TMC proteins [28, 53, 54]. Meanwhile, TMIE binds to both LHFPL5 and PCDH15 (through the intracellular CD2 domain) in the hair cell mechanotransduction machinery [55]. Intriguingly, mutations of these TMC binding partners also cause deafness and defective mechanotransduction [56]. Therefore, the mechanosensitivity of hair cell transduction channels might involve multiple functional modules.
Fig. 3.
Potential arrangement of proteins within the vertebrate auditory transduction complex. By interacting with multiple auxiliary proteins (TMIE, LHFPL5, PCDH15, and CIB2), TMC1 channel may form the pore of auditory transduction complex (modified from Corey et al. [67]). For simplicity, a TMC monomer is depicted in the model figure (according to recent structural studies, TMC channels form dimers)
Very recently, structural studies of TMC1 suggest that TMC channels may have a similar membrane topology to the Ca2+-activated Cl− channel TMEM16 and the mechanically activated OSCA1.2/TMEM63 channel [51, 57–60]. Namely, TMC1 channels appear to be assembled as dimers and each subunit contains ten transmembrane segments. Importantly, the transmembrane helices S4–S7 which contain the Beethoven (Bth) mutation form a lipid-facing groove that may act as a pore, raising the possibility that TMC1 in hair cells forms the ion conduction pathway for auditory transduction. In support of this view, Pan et al. performed scanning cysteine mutagenesis of TMC1 and found that at least 12 amino-acid residues within the S4–S7 segment influence the mechanically activated current in hair cells [51]. Given the large body of genetic and physiological evidence, TMC1 may function as part of the ion channel pore during hair cell auditory transduction.
Notably, despite the strong structural and physiological evidence on TMC1 channel in auditory transduction, it remains unclear whether the transduction pore is completely bounded by S4–S7 helices or bounded in part by other accessory proteins like TMIE. Alternatively, the pore could be exposed to the lipid membrane. Further studies from cryo-EM or X-ray crystallography may help clarify additional structural details of TMC1 channel, including the pore region, location of the gate, and force-dependent transformation that occurs as TMC1 channels’ transition from closed to open states. In addition, to firmly establish TMC1 as a mechanosensitive hearing transduction ion channel, an important experiment remains unfulfilled: reconstitution of TMC1 channels and mechanosensitivity in a heterologous expression system. As TMC1 does not traffic to the plasma membrane in heterologous cells, if reconstitution of hair cell-like mechanosensitivity in a heterologous system eventually proves successful, it will likely require expression of TMC1 in the correct lipid environment, co-expression with correct binding partners, scaffold proteins, and chaperones. On the other hand, since mammalian TMC1 and TMC2 channels have been ectopically expressed as functional proteins in C. elegans and Drosophila [9, 12, 26], it would be very interesting to examine whether the ectopically expressed mammalian TMC1 channel exhibits mechanosensitivity in worms and flies. If not, could the co-expression of TMC1 with its binding partners (i.e., PCDH15, TMIE, LHFPL5, and CIB2) reconstitute the mechanosensitivity? In this regard, C. elegans and Drosophila might provide useful expression systems to conduct structure–function studies of mammalian TMC channels.
Other mammalian TMC channels
In addition to TMC1 and TMC2, six other TMC channels are present in mammals. However, very little is known about the function and regulation of these TMC proteins. Tmc3 mRNA can be detected in most neuronal organs as well as some non-neuronal organs. The mRNAs of Tmc4, Tmc5, Tmc6, and Tmc7 are expressed in most murine organs tested, while Tmc8 mRNA is detectable in thymus, lung, and spleen [1]. Mutations in Tmc6 (EVER1) and Tmc8 (EVER2) are implicated in epidermodysplasia verruciformis, a recessive disorder comprising susceptibility to cutaneous human papilloma virus infections and associated nonmelanoma skin cancers [2]. Mechanistically, mutations in Tmc8 appears to be linked to up-regulated Zn2+/Ca2+ signaling and anoctamin 1 (TMEM16A) activation [61, 62]. More investigations are clearly required to reveal the physiological roles of these understudied TMC channels.
Discussion
Among eight TMC channels in mammals, TMC1 and TMC2 are best studied because of their essential role in hearing transduction. Similarly, homologues of TMC1 and TMC2 have been implicated in mechanotransduction of Drosophila and zebrafish. This raises an intriguing question: are TMC1 and/or TMC2 channels gated by mechanic force? Mechanosensitive ion channels are ubiquitously present in nearly all organisms [63]. In general, mechanosensitive channels can be divided into two categories depending on whether they require other auxiliary components for the mechanosensitivity [64]. The bacterial Msc channels and mammalian PIEZO channels are inherently mechanosensitive and they can be functionally reconstituted in lipid bilayers [65, 66]. By contrast, TMC channels require multiple auxiliary factors for proper membrane trafficking and mechanosensitivity. In fact, at this stage, we still do not know whether TMC channels are intrinsically mechanosensitive, because many essential binding partners of TMC channels could act as the real mechanosensor. Functional reconstitution of TMC1/2 channels in a heterologous system may help bridge this knowledge gap. In addition, the recent development of cryo-EM microscopy may provide more structural insights into the gating mechanism of TMC channels.
C. elegans TMC channels display the most similarity to Drosophila TMC and vertebrate TMCs1-3 proteins (Fig. 1a) [9]. However, defective mechanosensation has not been reported for the tmc mutant worms. Very interestingly, ectopic expression of mammalian TMC1, TMC2, and TMC3 in the egg-laying circuit of C. elegans can functionally rescue the egg-laying defect and membrane excitability of tmc mutants. This finding has two implications: first, mammalian TMC1–3 channels might also modulate membrane excitability by acting as background Na+-leak channels, and second, we may use C. elegans as a heterologous expression system to study the biophysical and pharmacological features of mammalian TMC channels. Notably, unlike their mammalian counterparts, little is known about the binding partners of TMC channels in C. elegans. As tmc mutant worms display various defects in chemosensation, development, and egg laying, genetic screens using these defective behaviors might reveal novel binding proteins of TMC channels.
In summary, pioneering genetic and physiological studies of TMC proteins have revealed a novel family of ion channels involved in many important cellular processes. Although mammalian TMC1 and TMC2 channels clearly have an essential role in hearing, more studies are needed to define their precise action during auditory transduction. Meanwhile, further investigations are required to understand the physiology of other understudied TMC channels (TMC3–8).
Funding
The work in the Xiao lab is supported by the American Cancer Society Research Scholar Grant (RSG-17-171-01-DMC), UF Center for Smell and Taste Pilot Grant, and the American Federation for Aging Research (AFAR) Research Grant (17146). The work in the Kang lab is supported by the National Foundation of Natural Science of China (31771113, 31900736, 31800878), the Fundamental Research Funds for the Central Universities (2018FZA7004), China Postdoctoral Science Foundation (2018M640551, 2018M642412, 2019T120503, 2019T120505), the 111 project, and the Non-profit Central Research Institute Fund of Chinese Academy of Medical Sciences (2018PT31041).
Compliance with ethical standards
Conflict of interest
The authors declare no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Lijun Kang, Phone: 86-18606504032, Email: kanglijun@zju.edu.cn.
Rui Xiao, Phone: 352-273-9389, Email: rxiao@ufl.edu.
References
- 1.Keresztes G, Mutai H, Heller S. TMC and EVER genes belong to a larger novel family, the TMC gene family encoding transmembrane proteins. BMC Genom. 2003;4(1):24. doi: 10.1186/1471-2164-4-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kurima K, et al. Characterization of the transmembrane channel-like (TMC) gene family: functional clues from hearing loss and epidermodysplasia verruciformis. Genomics. 2003;82(3):300–308. doi: 10.1016/s0888-7543(03)00154-x. [DOI] [PubMed] [Google Scholar]
- 3.Deol M, Kocher W. A new gene for deafness in the mouse. Heredity. 1958;12(4):463–466. [Google Scholar]
- 4.Steel KP, Bock GR. The nature of inherited deafness in deafness mice. Nature. 1980;288(5787):159–161. doi: 10.1038/288159a0. [DOI] [PubMed] [Google Scholar]
- 5.Jain PK, et al. A human recessive neurosensory nonsyndromic hearing impairment locus is potential homologue of murine deafness (dn) locus. Hum Mol Genet. 1995;4(12):2391–2394. doi: 10.1093/hmg/4.12.2391. [DOI] [PubMed] [Google Scholar]
- 6.Keats BJ, Nouri N, Huang JM, Money M, Webster DB, Berlin CI. The deafness locus (dn) maps to mouse chromosome 19. Mamm Genome. 1995;6(1):8e10. doi: 10.1007/BF00350886. [DOI] [PubMed] [Google Scholar]
- 7.Kurima K, et al. Dominant and recessive deafness caused by mutations of a novel gene, TMC1, required for cochlear hair-cell function. Nat Genet. 2002;30(3):277–284. doi: 10.1038/ng842. [DOI] [PubMed] [Google Scholar]
- 8.Vreugde S, et al. Beethoven, a mouse model for dominant, progressive hearing loss DFNA36. Nat Genet. 2002;30(3):257–258. doi: 10.1038/ng848. [DOI] [PubMed] [Google Scholar]
- 9.Yue X, et al. TMC proteins modulate egg laying and membrane excitability through a background leak conductance in C. elegans. Neuron. 2018;97(3):571–585.e5. doi: 10.1016/j.neuron.2017.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chatzigeorgiou M, et al. Tmc-1 encodes a sodium-sensitive channel required for salt chemosensation in C. elegans. Nature. 2013;494(7435):95–99. doi: 10.1038/nature11845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang L, et al. TMC-1 attenuates C. elegans development and sexual behaviour in a chemically defined food environment. Nat Commun. 2015;6:6345. doi: 10.1038/ncomms7345. [DOI] [PubMed] [Google Scholar]
- 12.Wang X, et al. TMC-1 mediates alkaline sensation in C. elegans through nociceptive neurons. Neuron. 2016;91(1):146–154. doi: 10.1016/j.neuron.2016.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bargmann CI (2006) Chemosensation in C. elegans. WormBook pp 1–29 [DOI] [PMC free article] [PubMed]
- 14.Spalthoff C, Gopfert MC. Sensing pH with TMCs. Neuron. 2016;91(1):6–8. doi: 10.1016/j.neuron.2016.06.026. [DOI] [PubMed] [Google Scholar]
- 15.Enyedi P, Czirjak G. Molecular background of leak K+ currents: two-pore domain potassium channels. Physiol Rev. 2010;90(2):559–605. doi: 10.1152/physrev.00029.2009. [DOI] [PubMed] [Google Scholar]
- 16.Cochet-Bissuel M, Lory P, Monteil A. The sodium leak channel, NALCN, in health and disease. Front Cell Neurosci. 2014;8:132. doi: 10.3389/fncel.2014.00132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao S, et al. The NCA sodium leak channel is required for persistent motor circuit activity that sustains locomotion. Nat Commun. 2015;6:6323. doi: 10.1038/ncomms7323. [DOI] [PubMed] [Google Scholar]
- 18.Lutas A, et al. The leak channel NALCN controls tonic firing and glycolytic sensitivity of substantia nigra pars reticulata neurons. Elife. 2016 doi: 10.7554/eLife.15271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xie L, et al. NLF-1 delivers a sodium leak channel to regulate neuronal excitability and modulate rhythmic locomotion. Neuron. 2013;77(6):1069–1082. doi: 10.1016/j.neuron.2013.01.018. [DOI] [PubMed] [Google Scholar]
- 20.Lu B, Su Y, Das S, Liu J, Xia J, Ren D. The neuronal channel NALCN contributes resting sodium permeability and is required for normal respiratory rhythm. Cell. 2007;129(2):371–383. doi: 10.1016/j.cell.2007.02.041. [DOI] [PubMed] [Google Scholar]
- 21.Kawashima Y, et al. Transmembrane channel-like (TMC) genes are required for auditory and vestibular mechanosensation. Pflugers Arch. 2015;467(1):85–94. doi: 10.1007/s00424-014-1582-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang YV, et al. The basis of food texture sensation in Drosophila. Neuron. 2016;91(4):863–877. doi: 10.1016/j.neuron.2016.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hasan Z. Role of proprioceptors in neural control. Curr Opin Neurobiol. 1992;2(6):824–829. doi: 10.1016/0959-4388(92)90140-g. [DOI] [PubMed] [Google Scholar]
- 24.Dietz V. Proprioception and locomotor disorders. Nat Rev Neurosci. 2002;3(10):781–790. doi: 10.1038/nrn939. [DOI] [PubMed] [Google Scholar]
- 25.Tuthill JC, Azim E. Proprioception. Curr Biol. 2018;28(5):R194–R203. doi: 10.1016/j.cub.2018.01.064. [DOI] [PubMed] [Google Scholar]
- 26.Guo Y, et al. Transmembrane channel-like (tmc) gene regulates Drosophila larval locomotion. Proc Natl Acad Sci U S A. 2016;113(26):7243–7248. doi: 10.1073/pnas.1606537113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He L, et al. Direction selectivity in Drosophila proprioceptors requires the mechanosensory channel Tmc. Curr Biol. 2019;29(6):945–956.e3. doi: 10.1016/j.cub.2019.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maeda R, et al. Tip-link protein protocadherin 15 interacts with transmembrane channel-like proteins TMC1 and TMC2. Proc Natl Acad Sci U S A. 2014;111(35):12907–12912. doi: 10.1073/pnas.1402152111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mitchem KL, et al. Mutation of the novel gene Tmie results in sensory cell defects in the inner ear of spinner, a mouse model of human hearing loss DFNB6. Hum Mol Genet. 2002;11(16):1887–1898. doi: 10.1093/hmg/11.16.1887. [DOI] [PubMed] [Google Scholar]
- 30.Gleason MR, et al. The transmembrane inner ear (Tmie) protein is essential for normal hearing and balance in the zebrafish. Proc Natl Acad Sci USA. 2009;106(50):21347–21352. doi: 10.1073/pnas.0911632106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cho KI, et al. The circling mouse (C57BL/6J-cir) has a 40-kilobase genomic deletion that includes the transmembrane inner ear (tmie) gene. Comp Med. 2006;56(6):476–481. [PubMed] [Google Scholar]
- 32.Shin MJ, et al. Spatiotemporal expression of tmie in the inner ear of rats during postnatal development. Comp Med. 2010;60(4):288–294. [PMC free article] [PubMed] [Google Scholar]
- 33.Pacentine IV, Nicolson T. Subunits of the mechano-electrical transduction channel, Tmc1/2b, require Tmie to localize in zebrafish sensory hair cells. PLoS Genet. 2019;15(2):e1007635. doi: 10.1371/journal.pgen.1007635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Erickson T, et al. Integration of Tmc1/2 into the mechanotransduction complex in zebrafish hair cells is regulated by transmembrane O-methyltransferase (Tomt) Elife. 2017 doi: 10.7554/eLife.28474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cunningham CL, et al. The murine catecholamine methyltransferase mTOMT is essential for mechanotransduction by cochlear hair cells. Elife. 2017 doi: 10.7554/eLife.24318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chou SW, et al. A molecular basis for water motion detection by the mechanosensory lateral line of zebrafish. Nat Commun. 2017;8(1):2234. doi: 10.1038/s41467-017-01604-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Olszewski J, et al. Zebrafish larvae exhibit rheotaxis and can escape a continuous suction source using their lateral line. PLoS One. 2012;7(5):e36661. doi: 10.1371/journal.pone.0036661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kawashima Y, et al. Mechanotransduction in mouse inner ear hair cells requires transmembrane channel-like genes. J Clin Invest. 2011;121(12):4796–4809. doi: 10.1172/JCI60405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Géléoc GS, Holt J. Developmental acquisition of sensory transduction in hair cells of the mouse inner ear. Nat Neurosci. 2003;6:1019–1020. doi: 10.1038/nn1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lelli A, Kazmierczak P, Kawashima Y, Muller U, Holt JR. Development and regeneration of sensory transduction in auditory hair cells requires functional interaction between cadherin-23 and protocadherin-15. J Neurosci. 2010;30:11259–11269. doi: 10.1523/JNEUROSCI.1949-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim KX, et al. The role of transmembrane channel-like proteins in the operation of hair cell mechanotransducer channels. J Gen Physiol. 2013;142(5):493–505. doi: 10.1085/jgp.201311068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pan B, et al. TMC1 and TMC2 are components of the mechanotransduction channel in hair cells of the mammalian inner ear. Neuron. 2013;79(3):504–515. doi: 10.1016/j.neuron.2013.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mahendrasingam S, Furness DN. Ultrastructural localization of the likely mechanoelectrical transduction channel protein, transmembrane-like channel 1 (TMC1) during development of cochlear hair cells. Sci Rep. 2019;9(1):1274. doi: 10.1038/s41598-018-37563-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li X, et al. Localization of TMC1 and LHFPL5 in auditory hair cells in neonatal and adult mice. FASEB J. 2019;33:fj201802155RR. doi: 10.1096/fj.201802155RR. [DOI] [PubMed] [Google Scholar]
- 45.Kurima K, et al. TMC1 and TMC2 localize at the site of mechanotransduction in mammalian inner ear hair cell stereocilia. Cell Rep. 2015;12(10):1606–1617. doi: 10.1016/j.celrep.2015.07.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Beurg M, Fettiplace R, Nam JH, Ricci AJ. Localization of inner hair cell mechanotransducer channels using high-speed calcium imaging. Nat Neurosci. 2009;12(5):553–558. doi: 10.1038/nn.2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jaramillo F, Hudspeth AJ. Localization of the hair cell’s transduction channels at the hair bundle’s top by iontophoretic application of a channel blocker. Neuron. 1991;7(3):409–420. doi: 10.1016/0896-6273(91)90293-9. [DOI] [PubMed] [Google Scholar]
- 48.Hudspeth AJ. Extracellular current flow and the site of transduction by vertebrate hair cells. J Neurosci. 1982;2(1):1–10. doi: 10.1523/JNEUROSCI.02-01-00001.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Corns LF, et al. Tmc1 point mutation affects Ca2+ sensitivity and block by dihydrostreptomycin of the mechanoelectrical transducer current of mouse outer hair cells. J Neurosci. 2016;36(2):336–349. doi: 10.1523/JNEUROSCI.2439-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Beurg M, Goldring AC, Fettiplace R. The effects of Tmc1 Beethoven mutation on mechanotransducer channel function in cochlear hair cells. J Gen Physiol. 2015;146(3):233–243. doi: 10.1085/jgp.201511458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pan B, et al. TMC1 forms the pore of mechanosensory transduction channels in vertebrate inner ear hair cells. Neuron. 2018;99(4):736–753.e6. doi: 10.1016/j.neuron.2018.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cunningham CL, Muller U. Molecular structure of the hair cell mechanoelectrical transduction complex. Cold Spring Harb Perspect Med. 2019 doi: 10.1101/cshperspect.a033167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ge J, et al. Structure of mouse protocadherin 15 of the stereocilia tip link in complex with LHFPL5. Elife. 2018 doi: 10.7554/eLife.38770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xiong W, et al. TMHS is an integral component of the mechanotransduction machinery of cochlear hair cells. Cell. 2012;151(6):1283–1295. doi: 10.1016/j.cell.2012.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhao B, et al. TMIE is an essential component of the mechanotransduction machinery of cochlear hair cells. Neuron. 2014;84(5):954–967. doi: 10.1016/j.neuron.2014.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fettiplace R. Is TMC1 the hair cell mechanotransducer channel? Biophys J. 2016;111(1):3–9. doi: 10.1016/j.bpj.2016.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hahn Y, et al. Anoctamin and transmembrane channel-like proteins are evolutionarily related. Int J Mol Med. 2009;24(1):51–55. doi: 10.3892/ijmm_00000205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Medrano-Soto A, et al. Bioinformatic characterization of the Anoctamin Superfamily of Ca2+ -activated ion channels and lipid scramblases. PLoS One. 2018;13(3):e0192851. doi: 10.1371/journal.pone.0192851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ballesteros A, Fenollar-Ferrer C, Swartz KJ. Structural relationship between the putative hair cell mechanotransduction channel TMC1 and TMEM16 proteins. ELife. 2018 doi: 10.7554/eLife.38433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jojoa-Cruz S, et al. Cryo-EM structure of the mechanically activated ion channel OSCA1.2. Elife. 2018 doi: 10.7554/eLife.41845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lazarczyk M, et al. Regulation of cellular zinc balance as a potential mechanism of EVER-mediated protection against pathogenesis by cutaneous oncogenic human papillomaviruses. J Exp Med. 2008;205(1):35–42. doi: 10.1084/jem.20071311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sirianant L, et al. TMC8 (EVER2) attenuates intracellular signaling by Zn2+ and Ca2+ and suppresses activation of Cl− currents. Cell Signal. 2014;26(12):2826–2833. doi: 10.1016/j.cellsig.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 63.Gillespie PG, Walker RG. Molecular basis of mechanosensory transduction. Nature. 2001;413(6852):194–202. doi: 10.1038/35093011. [DOI] [PubMed] [Google Scholar]
- 64.Xiao R, Xu XZ. Mechanosensitive channels: in touch with Piezo. Curr Biol. 2010;20(21):R936–R938. doi: 10.1016/j.cub.2010.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Syeda R, et al. Piezo1 channels are inherently mechanosensitive. Cell Rep. 2016;17(7):1739–1746. doi: 10.1016/j.celrep.2016.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ranade SS, Syeda R, Patapoutian A. Mechanically activated ion channels. Neuron. 2015;87(6):1162–1179. doi: 10.1016/j.neuron.2015.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Corey DP, Akyuz N, Holt JR. Function and dysfunction of TMC channels in inner ear hair cells. Cold Spring Harb Perspect Med. 2018 doi: 10.1101/cshperspect.a033506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kalay E, et al. Four novel TMC1 (DFNB7/DFNB11) mutations in Turkish patients with congenital autosomal recessive nonsyndromic hearing loss. Hum Mutat. 2005;26(6):591. doi: 10.1002/humu.9384. [DOI] [PubMed] [Google Scholar]
- 69.Meyer CG, et al. Novel TMC1 structural and splice variants associated with congenital nonsyndromic deafness in a Sudanese pedigree. Hum Mutat. 2005;25(1):100. doi: 10.1002/humu.9302. [DOI] [PubMed] [Google Scholar]
- 70.Santos RL, et al. Novel sequence variants in the TMC1 gene in Pakistani families with autosomal recessive hearing impairment. Hum Mutat. 2005;26(4):396. doi: 10.1002/humu.9374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kitajiri SI, et al. Identities, frequencies and origins of TMC1 mutations causing DFNB7/B11 deafness in Pakistan. Clin Genet. 2007;72(6):546–550. doi: 10.1111/j.1399-0004.2007.00895.x. [DOI] [PubMed] [Google Scholar]
- 72.Hilgert N, et al. Mutation analysis of TMC1 identifies four new mutations and suggests an additional deafness gene at loci DFNA36 and DFNB7/11. Clin Genet. 2008;74(3):223–232. doi: 10.1111/j.1399-0004.2008.01053.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tlili A, et al. TMC1 but not TMC2 is responsible for autosomal recessive nonsyndromic hearing impairment in Tunisian families. Audiol Neurootol. 2008;13(4):213–218. doi: 10.1159/000115430. [DOI] [PubMed] [Google Scholar]
- 74.Sirmaci A, et al. Mutations in TMC1 contribute significantly to nonsyndromic autosomal recessive sensorineural hearing loss: a report of five novel mutations. Int J Pediatr Otorhinolaryngol. 2009;73(5):699–705. doi: 10.1016/j.ijporl.2009.01.005. [DOI] [PubMed] [Google Scholar]
- 75.Hildebrand MS, et al. Mutations in TMC1 are a common cause of DFNB7/11 hearing loss in the Iranian population. Ann Otol Rhinol Laryngol. 2010;119(12):830–835. doi: 10.1177/000348941011901207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yang T, et al. A novel mutation adjacent to the Bth mouse mutation in the TMC1 gene makes this mouse an excellent model of human deafness at the DFNA36 locus. Clin Genet. 2010;77(4):395–398. doi: 10.1111/j.1399-0004.2009.01338.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Brownstein Z, et al. Targeted genomic capture and massively parallel sequencing to identify genes for hereditary hearing loss in Middle Eastern families. Genome Biol. 2011;12(9):R89. doi: 10.1186/gb-2011-12-9-r89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.de Heer AM, et al. Progressive sensorineural hearing loss and normal vestibular function in a Dutch DFNB7/11 family with a novel mutation in TMC1. Audiol Neurootol. 2011;16(2):93–105. doi: 10.1159/000313282. [DOI] [PubMed] [Google Scholar]
- 79.Duman D, et al. Screening of 38 genes identifies mutations in 62% of families with nonsyndromic deafness in Turkey. Genet Test Mol Biomark. 2011;15(1–2):29–33. doi: 10.1089/gtmb.2010.0120. [DOI] [PubMed] [Google Scholar]
- 80.Gao X, et al. Novel compound heterozygous TMC1 mutations associated with autosomal recessive hearing loss in a Chinese family. PLoS One. 2013;8(5):e63026. doi: 10.1371/journal.pone.0063026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Schrauwen I, et al. A sensitive and specific diagnostic test for hearing loss using a microdroplet PCR-based approach and next generation sequencing. Am J Med Genet A. 2013;161A(1):145–152. doi: 10.1002/ajmg.a.35737. [DOI] [PubMed] [Google Scholar]
- 82.Yang T, et al. Genetic etiology study of the non-syndromic deafness in Chinese Hans by targeted next-generation sequencing. Orphanet J Rare Dis. 2013;8:85. doi: 10.1186/1750-1172-8-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ganapathy A, et al. Non-syndromic hearing impairment in India: high allelic heterogeneity among mutations in TMPRSS3, TMC1, USHIC, CDH23 and TMIE. PLoS One. 2014;9(1):e84773. doi: 10.1371/journal.pone.0084773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lin F, et al. Autosomal recessive non-syndromic hearing loss is caused by novel compound heterozygous mutations in TMC1 from a Tibetan Chinese family. Int J Pediatr Otorhinolaryngol. 2014;78(12):2216–2221. doi: 10.1016/j.ijporl.2014.10.016. [DOI] [PubMed] [Google Scholar]
- 85.Nakanishi H, et al. Mutations of TMC1 cause deafness by disrupting mechanoelectrical transduction. Auris Nasus Larynx. 2014;41(5):399–408. doi: 10.1016/j.anl.2014.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Riahi Z, et al. Whole exome sequencing identifies new causative mutations in Tunisian families with non-syndromic deafness. PLoS One. 2014;9(6):e99797. doi: 10.1371/journal.pone.0099797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Shafique S, et al. Genetic spectrum of autosomal recessive non-syndromic hearing loss in Pakistani families. PLoS One. 2014;9(6):e100146. doi: 10.1371/journal.pone.0100146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhao Y, et al. A novel DFNA36 mutation in TMC1 orthologous to the Beethoven (Bth) mouse associated with autosomal dominant hearing loss in a Chinese family. PLoS One. 2014;9(5):e97064. doi: 10.1371/journal.pone.0097064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bakhchane A, et al. A novel mutation in the TMC1 gene causes non-syndromic hearing loss in a Moroccan family. Gene. 2015;574(1):28–33. doi: 10.1016/j.gene.2015.07.075. [DOI] [PubMed] [Google Scholar]
- 90.Chen Y, et al. Targeted next-generation sequencing in Uyghur families with non-syndromic sensorineural hearing loss. PLoS One. 2015;10(5):e0127879. doi: 10.1371/journal.pone.0127879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Davoudi-Dehaghani E, et al. Allelic heterogeneity among Iranian DFNB7/11 families: report of a new Iranian deaf family with TMC1 mutation identified by next-generation sequencing. Acta Otolaryngol. 2015;135(2):125–129. doi: 10.3109/00016489.2014.969383. [DOI] [PubMed] [Google Scholar]
- 92.Gao X, et al. Targeted gene capture and massively parallel sequencing identify TMC1 as the causative gene in a six-generation Chinese family with autosomal dominant hearing loss. Am J Med Genet A. 2015;167A(10):2357–2365. doi: 10.1002/ajmg.a.37206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bademci G, et al. Comprehensive analysis via exome sequencing uncovers genetic etiology in autosomal recessive nonsyndromic deafness in a large multiethnic cohort. Genet Med. 2016;18(4):364–371. doi: 10.1038/gim.2015.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hu J, et al. Exome sequencing identifies a mutation in TMC1 as a novel cause of autosomal recessive nonsyndromic hearing loss. J Transl Med. 2016;14:29. doi: 10.1186/s12967-016-0780-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Imtiaz A, et al. Recessive mutations of TMC1 associated with moderate to severe hearing loss. Neurogenetics. 2016;17(2):115–123. doi: 10.1007/s10048-016-0477-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Subrungruanga I, et al. Gene expression profiling of intrahepatic cholangiocarcinoma. Asian Pac J Cancer Prev. 2013;14(1):557–563. [PubMed] [Google Scholar]
- 97.Ramoz N, et al. A susceptibility locus for epidermodysplasia verruciformis, an abnormal predisposition to infection with the oncogenic human papillomavirus type 5, maps to chromosome 17qter in a region containing a psoriasis locus. J Invest Dermatol. 1999;112(3):259–263. doi: 10.1046/j.1523-1747.1999.00536.x. [DOI] [PubMed] [Google Scholar]
- 98.Ramoz N, et al. Mutations in two adjacent novel genes are associated with epidermodysplasia verruciformis. Nat Genet. 2002;32(4):579–581. doi: 10.1038/ng1044. [DOI] [PubMed] [Google Scholar]
- 99.Manji SS, et al. Identification of three novel hearing loss mouse strains with mutations in the Tmc1 gene. Am J Pathol. 2012;180(4):1560–1569. doi: 10.1016/j.ajpath.2011.12.034. [DOI] [PubMed] [Google Scholar]
- 100.Kim KX, Fettiplace R. Developmental changes in the cochlear hair cell mechanotransducer channel and their regulation by transmembrane channel-like proteins. J Gen Physiol. 2013;141(1):141–148. doi: 10.1085/jgp.201210913. [DOI] [PMC free article] [PubMed] [Google Scholar]