Abstract
Peroxiredoxins are antioxidant enzymes that use redox active Cys residues to reduce H2O2 and various organic hydroperoxides to less reactive products, and thereby protect cells against oxidative stress. In yeasts and mammals, the Prx1 proteins are sensitive to hyperoxidation and consequent loss of their peroxidase activity whereas in most bacteria they are not. In this paper we report the characterization of the Prx1 family in the non-parasitic protist Tetrahymena thermophila. In this organism, four genes potentially encoding Prx1 have been identified. In particular, we show that the mitochondrial Prx1 protein (Prx1m) from T. thermophila is relatively robust to hyperoxidation. This is surprising given that T. thermophila is a eukaryote like yeasts and mammals. In addition, the proliferation of the T. thermophila cells was relatively robust to inhibition by H2O2, cumene hydroperoxide and plant natural products that are known to promote the production of H2O2. In the presence of these agents, the abundance of the T. thermophila Prx1m protein was shown to increase. This suggested that the Prx1m protein may be protecting the cells against oxidative stress. There was no evidence for any increase in Prx1m gene expression in the stressed cells. Thus, increasing protein stability rather than increasing gene expression may explain the increasing Prx1m protein abundance we observed.
Keywords: Alveolates, Ciliated protozoa, Antioxidant enzymes, Oxidative stress, Plant natural products
Introduction
The peroxiredoxins (Prxs) are antioxidant enzymes that have been well studied in parasitic protists such as Plasmodium falciparum (super-phylum alveolata, phylum apicomplexa) but not in non-parasitic protists such as Tetrahymena thermophila (super-phylum Alveolata, phylum Ciliophora) [1]. All Prxs have a redox active Cys residue known as the peroxidatic Cys or CysP which they use to reduce H2O2, peroxynitrite and various organic hydroperoxides to less reactive products, and thereby protect cells against oxidative stress [2]. The Prx1 subfamily also has a second Cys residue known as the resolving Cys or CysR. During catalysis, CysP, in its reduced (thiolate) state (CysP-SH), reacts with the peroxide substrate and becomes oxidised to Cys sulfenic acid (CysP-SOH). Subsequently, CysP-SOH forms a disulphide bond with CysR-SH from a separate subunit giving rise to a disulphide-linked dimer. Following this, thioredoxin (Trx), or a similar redox protein, disrupts the disulphide bond and restores CysP to its original (reduced/thiolate) state ready to begin another cycle of catalysis. Occasionally, however, for example when H2O2 concentrations are very high, CysP-SOH becomes further oxidised to Cys sulfinic acid (CysP-SO2H) and the peroxidase activity of the Prx1 proteins is lost [3, 4]. This unusual behaviour led to the “floodgate hypothesis” [4]. According to this hypothesis, inactivation of the Prx1 proteins allows H2O2 to accumulate to act as a cell signalling molecule. The hyperoxidised/inactivated form of the Prx proteins can be restored to the reduced/active form by sulfiredoxin (Srx) enzymes, but this process is relatively slow [5–7]. Thus, there is time for H2O2 to accumulate and exert its effects.
In general, eukaryote Prx1 proteins are “sensitive” to hyperoxidation, whereas prokaryote Prx1 proteins are “robust” and eukaryote Prx1 proteins possess conserved GG(L/I/V)G and Y(F/L) motifs that can explain their sensitivity whereas prokaryote Prx1 proteins do not, but there are exceptions [4, 8]. For example, two different strains of cyanobacteria, Anabaena sp. PCC 7120 and Synechocystis sp. PCC 6803, both of which possess the conserved GG(L/I/V)G and Y(F/L) motifs, display different sensitivities of their Prx1 proteins to hyperoxidation [8]. The Prx1 protein of the Anabaena strain is highly “sensitive” to hyperoxidation (normally associated with eukaryote Prx proteins) whereas that of the Synechocystis strain is relatively “robust”. Additionally, the Anabaena strain has a gene encoding a Srx protein whereas the Synechocystis strain does not. This suggests that the presence/absence of Srx genes rather than the presence/absence of the GG(L/I/V)G and Y(F/L) motifs is a better predictor of the sensitivity of Prx1 proteins to hyperoxidation.
The sensitivity of protozoan Prx1 proteins to hyperoxidation has not been studied, mainly due to the complexity of carrying out such studies in obligate endoparasites such as P. falciparum. Thus, the present study aimed: (1) to survey a selection of parasitic and non-parasitic protozoans from the phylum Alveolata (alveolates) for the presence of genes encoding Prx1 proteins and other antioxidant enzymes; (2) to investigate whether the GG(L/I/V)G and Y(F/L) motifs are present or absent in the Prx1 proteins of these organisms; (3) to determine whether genes encoding Srx proteins are present or absent in these organisms; (4) to test the sensitivity of the mitochondrial Prx1 protein of T. thermophila to hyperoxidation; and (5) to test various plant natural products known to act as pro-oxidants and compare their effects with those of H2O2 in terms of promoting the oxidation/hyperoxidation of the mitochondrial Prx1 protein of T. thermophila.
Materials and methods
Bioinformatic analysis of the number and diversity of antioxidant genes in the genomes of T. thermophila, a selection of other ciliates and a selection of apicomplexans
Four ciliates, T. thermophila, Stylonychia lemnae, Ichthyophthirius multifiliis and Oxytricha trifallax, and six apicomplexans, P. falciparum, Toxoplasma gondii, Cryptosporidium parvum, Eimeria tenella, Theileria annulata and Babesia microti were chosen for this analysis. Searches were done using the Ensembl Protists web browser (http://protists.ensembl.org/index.html) to find the best species to study, i.e., those with the most complete genome sequences. The species chosen are listed in Table 1. Once the species had been chosen, their scientific names, together with the names of the main families of antioxidant enzymes, were used to search the protein database on the National Center for Biotechnology Information (NCBI) web site (https://www.ncbi.nlm.nih.gov/). The names of the enzymes are listed in Table 1. In addition, we also used thioredoxin peroxidase, thioredoxin-dependent peroxide reductase, thiol-specific antioxidant protein, alkyl hydroperoxide reductase and peroxidoxin which are alternative names for the peroxiredoxins. Identical genes were identified either using the information on the NCBI website or by performing multiple sequence alignments and only unique genes were included in the count displayed in Table 1. Following this, the Prx proteins belonging to the Prx1 subfamily were named according to the new naming system proposed by Gretes et al. [1]. This new system seeks to overcome the confusion in the literature arising from the multiple different names used for the same or similar Prx proteins in different organisms. The names and the corresponding accession numbers can be seen in Table 2.
Table 1.
A summary of the number of genes belonging to each of the various antioxidant gene families in a selection of ciliates and apicomplexans
| Species name | Gene name | ||||
|---|---|---|---|---|---|
| SOD | CAT | GPx | PHGPx | Prx | |
| Ciliates | |||||
| Tetrahymena thermophila | 4 | 1 | 3 | 9 | 4 |
| Stylonychia lemnae | 7 | 4 | 11 | 0 | 4 |
| Ichthyophthirius multifiliis | 6 | 1 | 0 | 0 | 7 |
| Oxytricha trifallax | 7 | 4 | 9 | 0 | 6 |
| Apicomplexans | |||||
| Plasmodium falciparum (3D7) | 2 | 0 | 0 | 0 | 5 |
| Toxoplasma gondii (ME49) | 3 | 1 | 0 | 0 | 4 |
| Cryptosporidium parvum | 1 | 0 | 1 | 0 | 1 |
| Eimeria tenella (GCA_000499545) | 3 | 2 | 1 | 0 | 3 |
| Theileria annulata | 2 | 0 | 0 | 0 | 3 |
| Babesia microti | 2 | 0 | 0 | 0 | 2 |
The genes were found by searching the protein database on the National Center for Biotechnology Information (NCBI) web site (https://www.ncbi.nlm.nih.gov/)
The abbreviations indicate SOD superoxide dismutase, CAT catalase, GPx glutathione peroxidase, PHGPx phospholipid hydroperoxide glutathione peroxidase, Prx peroxiredoxin
Table 2.
Predicted subcellular locations and molecular weights (MW) of the Prx1 proteins from a selection of ciliates and apicomplexans
| Species | Protein | Accession number | Score | Location | MW (kD) | ||
|---|---|---|---|---|---|---|---|
| mTP | SP | Other | |||||
| Ciliates | |||||||
| T. thermophila | Prx1a | XP_001029987.1 | 0.10 | 0.32 | 0.60 | – | 23 |
| Prx1b | XP_001031522.1 | 0.42 | 0.05 | 0.52 | – | 26 | |
| Prx1m | EAR92904.2 | 0.73 | 0.24 | 0.10 | M | 28 | |
| Prx1c | EAR84222.2 | 0.50 | 0.31 | 0.30 | – | 21 | |
| S. lemnae | Prx1a | CDW79681.1 | 0.11 | 0.92 | 0.03 | S | 23 |
| Prx1b | CDW72826.1 | 0.11 | 0.08 | 0.90 | – | 29 | |
| Prx1m | CDW79455.1 | 0.90 | 0.05 | 0.10 | M | 25 | |
| I. multifiliis | Prxla | EGR27018.1 | 0.30 | 0.11 | 0.61 | – | 24 |
| Prxlb | EGR27550.1 | 0.20 | 0.11 | 0.70 | – | 23 | |
| Prxlc | EGR30706.1 | 0.01 | 1.00 | 0.14 | S | 24 | |
| Prxld | EGR31994.1 | 0.10 | 0.24 | 0.70 | – | 23 | |
| Prxle | EGR29838.1 | 0.20 | 0.93 | 0.10 | S | 25 | |
| Prx1m | EGR29584.1 | 0.60 | 0.05 | 0.42 | M | 22 | |
| O. trifallax | Prx1a | EJY85734.1 | 0.20 | 0.91 | 0.02 | S | 23 |
| Prx1b | EJY73729.1 | 0.34 | 0.10 | 0.44 | – | 28 | |
| Prx1c | EJY85449.1 | 0.10 | 0.05 | 0.94 | – | 51 | |
| Prx1d | EJY82209.1 | 0.10 | 0.05 | 0.94 | – | 46 | |
| Prx1e | EJY81461.1 | 0.34 | 0.10 | 0.61 | – | 24 | |
| Apicomplexans | |||||||
| P. falciparum | Prx1a | XP_001348542.1 | 0.14 | 0.10 | 0.82 | – | 22 |
| Prx1m | XP_001350554.1 | 0.90 | 0.04 | 0.14 | M | 22 | |
| T. gondii | Prx1a | EPR63775.1 | 0.11 | 0.20 | 0.80 | – | 22 |
| Prx1m | EPR64157.1 | 0.93 | 0.03 | 0.11 | M | 22 | |
| C. parvum | Prx1a | ACV31867.1 | 0.23 | 0.11 | 0.60 | – | 22 |
| E. tenella | Prx1a | XP_013230312.1 | 0.20 | 0.10 | 0.80 | – | 21 |
| Prx1m | CDJ42921 | 0.97 | 0.04 | 0.03 | M | 28 | |
| T. annulata | Prx1a | CAI76104.1 | 0.11 | 0.12 | 0.80 | – | 22 |
| Prx1b | CAI73861.1 | 0.40 | 0.06 | 0.52 | – | 29 | |
| B. microti | Prx1a | XP_012647735.1 | 0.10 | 0.20 | 0.80 | – | 22 |
| Prx1m | XP_012647852.1 | 0.94 | 0.02 | 0.10 | M | 22 | |
The proteins were named according to the new naming system recently proposed by Gretes et al. [1]. According to this system, when there are multiple Prx1 proteins, they are designated ‘a’, ‘b’, ‘c’, etc., except for mitochondrial Prx1 proteins which are designated ‘m’. The subcellular locations were predicted using TargetP version 1.1 (http://www.cbs.dtu.dk/services/TargetP/). The MW values were predicted using the “Compute pI/MW” program accessed via the ExPASy Bioinformatics Resource Portal (https://www.expasy.org/resources). The MW values exclude the mitochondrial targeting peptide when one is present. The abbreviations mTP, SP, M and S indicate mitochondrial targeting peptide, secretory pathway signal peptide, a mitochondrial protein and a secreted protein, respectively. The scores indicate the likelihood of the prediction being correct with 1.0 being the highest possible value
Phylogenetic analysis of the relationships between the Prx1 proteins in our selection of ciliates and apicomplexans
The amino acid sequences of the Prx1 proteins in our selection of ciliates and apicomplexans were aligned using the T-Coffee multiple sequence alignment method [9]. The quality of the alignment was good with a score of 946. ProtTest3 software was used to select the best model to use to analyze the Prx1 protein evolution [10]. One hundred and twenty-two candidate models and three different types of criteria (Akaike information criterion, corrected Akaike information criterion and Bayesian information criterion) were used for this analysis. The software selected the LG + I+G + F model, with a gamma shape value (four rate categories) of 1.495 using all statistical criteria (− lnL = 10,206.67), as the best model. Phylogenetic trees were built using the Bayesian inference (BI) method implemented using MrBayes 3.2 [11]. Four independent runs, each with four simultaneous Markov Chain Monte Carlo (MCMC) chains, were performed for 106 generations sampled every 103 generations. Furthermore, we also used the maximum likelihood (ML) method implemented in PhyML 3.0 [12]. Bootstrap analyses were performed on 105 trees using both kinds of tree topology improvement, i.e., nearest neighbor interchange and subtree pruning and regrafting. FigTree v1.3 software was used to display the final annotated phylogenetic trees.
Routine maintenance of the Tetrahymena thermophila cells
Tetrahymena thermophila strain CU428.2 cells were obtained from the Tetrahymena Stock Center at Cornell University, USA (https://tetrahymena.vet.cornell.edu/). For routine maintenance, they were kept, at 13 °C without agitation, in modified Neff’s medium [13]. Prior to any experiments, the cells were multiplied up, at 35 °C with gentle agitation, in SSP medium [13]. SSP medium consisted of 2% (w/v) proteose peptone, 0.1% (w/v) yeast extract, 0.2% (w/v) glucose and 33 µM Fe Cl3·6H2O.
Determining the effects of various treatments on the proliferation of the T. thermophila cells
The required number of T. thermophila cells was seeded into the wells of the required number of 24-well plates (4 × 104 cells per well). In addition to the cells, each well contained 1.5 ml of SSP medium plus various concentrations of H2O2 (Thermo Fisher Scientific), cumene hydroperoxide (Sigma-Aldrich), garlic (Allium sativum) oil (Mystic Moments, Fordingbridge, Hampshire, UK), tea tree (Melaleuca alternifolia) oil (Integria Health Care, Australia Pty. Ltd.), diallyl disulphide (DADS; Sigma-Aldrich), diallyl trisulfide (DATS; Cayman Chemical Company, USA) or terpinen-4-ol (VWR International Pty. Ltd., Australia). Stock solutions of the garlic oil, tea tree oil, DADS, DATS and terpinen-4-ol were prepared in dimethyl sulfoxide (DMSO) before they were diluted in the SSP medium. Preliminary experiments determined that the residual DMSO in the culture medium had no effect on the proliferation of the cells. The cells were incubated with gentle agitation in a temperature controlled FLUOstar plate reader (BMG LABTECH) at 35 °C and cell proliferation was measured by monitoring the change in absorbance at 600 nm. The plate reader was programmed to briefly cease agitation during the collection of the absorbance readings.
Analysing the in vivo redox state of the T. thermophila Prx1m protein
The in vivo redox state of the T. thermophila Prx1m protein was analysed using the method of Cox et al. [14], developed for Jurkat T-lymphocytes (human cancer cells). This method uses non-reducing polyacrylamide gel electrophoresis (NR-PAGE), coupled with immunoblotting with anti-Prx antibodies, to distinguish between the ‘reduced’ (CysP in the thiolate state), ‘oxidised’ (CysP forming a disulphide bond with CysR) and ‘hyperoxidised’ (CysP in the sulfinic acid state and unable to form a disulphide bond with CysR) forms of the enzyme. The reduced and hyperoxidised forms run as monomers of ~ 28 kD whereas the oxidised form runs as a dimer of ~ 56 kD. Care must be taken to preserve the in vivo redox state of CysP by including an alkylating agent, such as N-ethylmaleimide (NEM), in the extraction buffer [14]. NEM reacts with the thiolate group of CysP and prevents it from undergoing artefactual oxidation during the cell extraction process. The required number of T. thermophila cells was seeded into the wells of the required number of 6-well plates (4 × 106 cells per well). Each well contained 4 ml of SSP medium plus various concentrations H2O2, cumene hydroperoxide, garlic oil, tea tree oil, DADS, DATS or terpinen-4-ol. The cells were incubated for 10 min at 35 °C and then harvested by centrifugation at 10,000g for 30 s before being resuspended in 100 µl of alkylation buffer. The alkylation buffer contained 40 mM 4-(2-hydroxyethyl)piperazine-1-ethanesulfonic acid (HEPES) (pH 7.4), 50 mM NaCl, 1 mM ethylenediaminetetraacetic acid (EDTA), 1 mM ethylene glycol-bis(2-aminoethylether)-N,N,Nʹ,Nʹ-tetraacetic acid (EGTA), 2 mM phenylmethansulfonyl fluoride (PMSF) and 100 mM N-ethylmaleimide (NEM) [14]. Following the alkylation reaction, the cells were disrupted by the addition of 4 µl of 25% (w/v) 3-((3-cholamidopropyl)dimethylammonium)-1-propanesulfonate (CHAPS) to the extraction buffer and the extracts were then clarified by centrifugation at 15,000g for 5 min at 4 °C. The protein concentration in the clarified extracts was determined using a BCA protein assay kit (Thermo Fisher Scientific) and equal amounts of protein from the various extracts were subjected to NR-PAGE. The NR-PAGE loading buffer contained 62.5 mM Tris–HCl (pH 6.8), 2% (w/v) sodium dodecyl sulfate (SDS), 10% (v/v) glycerol and 0.025% (w/v) bromophenol blue. The cell extracts were mixed 1:1 (v/v) with the loading buffer before being subjected to NR-PAGE. For the NR-PAGE, the stacking gel contained 0.625 M Tris-base (pH 6.8), 4% (w/v) acrylamide:N,N′-methylenebisacrylamide (37.5:1), 0.125% (v/v) N,N,N′,N′-tetramethyl-ethylenediamine (TEMED) and 0.05% (w/v) ammonium persulphate and the resolving gel contained 0.375 M Tris-base (pH 8.8), 15% (w/v) acrylamide:N,N′-methylenebisacrylamide (37.5:1), 0.1% (v/v) TEMED and 0.05% (w/v) ammonium persulphate. The gel was run in a buffer containing 25 mM Tris-base (pH 8.3), 192 mM glycine and 0.1% (w/v) SDS in a Bio-Rad Mini-PROTEAN®II electrophoresis apparatus at a constant voltage of 200 V until the tracking dye (bromophenol blue) had reached the bottom of the gel. At the end of the run, the gel was either stained for protein or the proteins were transferred to a nitrocellulose membrane using a Bio-Rad Mini Trans-Blot® apparatus set to deliver a constant current of 200 mA for 2 h. The transfer buffer contained 50 mM Tris-base, 380 mM glycine, 0.1% (w/v) SDS and 20% (v/v) methanol. Following the transfer, the membrane was blocked for 1 h at room temperature in a blocking buffer containing 50 mM Tris–HCl (pH 7.5), 150 mM NaCl, 0.2% (v/v) Tween-20 and 5% (w/v) skim milk powder and then it was incubated overnight at 4 °C in fresh blocking buffer containing a 1:1000 dilution of anti-(T. thermophila Prx1m) antibodies. The antibodies, which were polyclonal, had been produced by immunizing a New Zealand White rabbit with the peptide CEEYLRLVQAFQYAD conjugated to keyhole limpet hemocyanin. This peptide is found in the T. thermophila Prx1m amino acid sequence and it was chosen using the software tools available on the Immune Epitope Database and Analysis Resource website (http://www.iedb.org). A cysteine residue that was not present in the T. thermophila Prx1m protein was added to the N-terminus of the peptide to facilitate its conjugation to the keyhole limpet hemocyanin carrier protein. Immunization of the rabbit followed standard procedures approved by the South Australian Health and Medical Research Institute Animal Ethics Committee. Following the incubation with the primary antibodies, the membrane was washed 5 × 5 min with washing buffer (blocking buffer minus the milk powder) and then incubated for 1–2 h at room temperature with a secondary antibody preparation diluted 1:1000 in blocking buffer. The secondary antibody preparation consisted of goat anti-(rabbit IgG) conjugated to horseradish peroxidase (Rockland Immunochemicals for Research). Any cross-reacting proteins were detected using the SuperSignal® West Pico Chemiluminescent substrate kit (Thermo SCIENTIFIC) and images of the blots were prepared using a Bio-Rad ChemiDoc™ MP imaging system. When the NR-PAGE gels were stained for protein, the protein staining solution contained 0.1% (w/v) Coomassie Brilliant Blue R-250 stain, 50% (v/v) methanol and 10% (v/v) glacial acetic acid and the destaining solution contained 50% (v/v) methanol and 10% (v/v) glacial acetic acid.
Gene expression analysis
Gene expression was analysed using quantitative polymerase chain reaction (qPCR). The primers required for this analysis were designed based on the nucleotide sequences associated with the accession numbers listed in Table 2. All primers were validated using conventional PCR before being used for qPCR. For both types of PCR, T. thermophila cells were cultured as described above for the analysis of the redox state of the Prx1m protein except that the cells were simply pelleted by centrifugation rather than being resuspended in NEM buffer. RNA was extracted from the cell pellets using an RNeasy® Mini kit (QIAGEN Pty. Ltd.), according to the manufacturer’s instructions. Subsequently, 1 µg of the extracted RNA was used to synthesize first strand cDNA, employing a SuperScript® IV First-Strand Synthesis kit (Invitrogen Pty. Ltd.). Conventional PCR was performed in 50 µl reactions containing 10 µl Promega 5 × Colorless GoTaq® Flexi Buffer, 5 units of New England Biolabs Thermopol DNA Polymerase, 0.2 µM each of the relevant forward and reverse primers (Table 3), 200 µM dNTPs, 500 µM MgCl2 and 5 µl of a 1:5 dilution of the first strand cDNA synthesis reaction obtained for the untreated control T. thermophila cells. The cycling conditions for conventional PCR were a pre-denaturation step at 95 °C for 2 min followed by 50 cycles of denaturation at 95 °C for 30 s, annealing at 55 °C for 30 s and extension at 72 °C for 1 min plus a final extension at 72 °C for 5 min. At the end of the cycling, the products were analysed using agarose gel electrophoresis and nucleotide sequencing. For each primer pair, there was only one product produced and its size and nucleotide sequence were correct (data not shown). The qPCR procedure was performed using a Platinum® SYBR® Green qPCR SuperMix-UDG Kit and a Rotor-Gene® Q thermal cycler (QIAGEN Pty. Ltd.). Each qPCR reaction (total volume 25 µl) contained 12.5 µl Platinum® SYBR® Green qPCR SuperMix-UDG buffer, 1 mM MgCl2, 0.1 µM each of the relevant forward and reverse primers (Table 3) and 5 µl of a 1:5 dilution of the relevant first strand cDNA synthesis reaction. The qPCR cycling conditions, following a uracil-DNA glycosylase (UDG) incubation step at 50 °C for 2 min, were a pre-denaturation step at 95 °C for 2 min followed by either 45 or 50 cycles of denaturation at 95 °C for 15 s, annealing at 55 °C for 30 s and extension at 72 °C for 30 s. The fluorescence curves produced for each sample were used to calculate the threshold cycle (Ct) value. The transcript abundance for the normalisation gene, glyceraldehyde-3-phosphate dehydrogenase (GAPDH), was calculated using the formula 2−∆Ct. The transcript abundance for the gene of interest was normalised to the transcript abundance for GAPDH using the formula 2−∆Ct, where ΔCt = Ct (gene of interest) − Ct [normalization gene (GAPDH)] [15]. Three or four biological replicates and two technical replicates were analysed for each sample. The amplification efficiencies for each set of PCR primers were determined by producing standard curves using a series of tenfold dilutions of the conventional PCR products for each gene. The conventional PCR products had been purified using the Promega Wizard® SV Gel and PCR Clean-Up System before they were used to generate the standard curves. The amplification efficiencies for all primer pairs were ≥ 0.90 indicating that the amplifications were highly efficient. To confirm that only one PCR product was obtained for each of the primers, melt curve analysis was performed at the end of each qPCR run. This involved heating the reaction mixture to either 65 or 72 °C for 45 s and then raising the temperature by 1 °C every 5 s until a maximum temperature of 95 °C was reached while at the same time measuring the change in fluorescence. In all cases, there was only one peak in the fluorescence curve. This confirmed the agarose gel results which had shown that only one product was produced for each pair of PCR primers.
Table 3.
Nucleotide sequence, amplicon size and standard curve parameters for the PCR primers used in this study
| Target gene | Direction | Primer sequence | Amplicon size (bp) | R2 | Slope | Y-intercept | Efficiency (%) |
|---|---|---|---|---|---|---|---|
| GAPDH | Forward | 5′-GTCTTGCTCCCGTTGCTAAG-3′ | 152 | 0.99918 | − 3.485 | 34.726 | 94 |
| Reverse | 5′-GGTTGAGGCAGCTCTACCAG-3′ | ||||||
| Catalase | Forward | 5′-GAACGTGATCCTCGTGGTTT-3′ | 166 | 0.99977 | − 3.484 | 33.765 | 94 |
| Reverse | 5′-TGTTAGCGCACTTGAGGTTG-3′ | ||||||
| Prx1a | Forward | 5′-ACCACTGCTTGGGATGGTAG-3′ | 226 | 0.99965 | − 3.493 | 32.941 | 93 |
| Reverse | 5′-CTTTTCTGGGCTTCTTGCAC-3′ | ||||||
| Prx1b | Forward | 5′-GGAGCAGCGTATAGAGGAACA-3′ | 152 | 0.99736 | − 3.593 | 34.958 | 91 |
| Reverse | 5′-GGACAAACTTCTCCGTGCTC-3′ | ||||||
| Prx1c | Forward | 5′-CCCACTGAATTGGTTGCTTT-3′ | 165 | 0.99640 | − 3.351 | 42.032 | 99 |
| Reverse | 5′-ATCTGCAAGGAGGGGAATCT-3′ | ||||||
| Prx1m | Forward | 5′-CAGAAGGTGGTTTGGGAGAA-3′ | 177 | 0.99849 | − 3.473 | 35.772 | 92 |
| Reverse | 5′-TTCTACCGACTGGGAGATCG-3′ | ||||||
| GPx1 | Forward | 5′-ACAGGCCAAACCTCAAGAGA-3′ | 210 | 0.99972 | − 3.599 | 33.620 | 90 |
| Reverse | 5′-TTCGGTTTCTCAAGCTACTGC-3′ | ||||||
| GPx2 | Forward | 5′-GGAGCTGGCTTTACAGCATT-3′ | 170 | 0.99069 | − 3.559 | 36.136 | 91 |
| Reverse | 5′-CTTTGGTTGCCACCTCCTTA-3′ | ||||||
| GPx3 | Forward | 5′-CCATTTGATGAACCTGCAAT-3′ | 199 | 0.99884 | − 3.374 | 35.077 | 98 |
| Reverse | 5′-AAACTGGTTTTCCCTCAGCA-3′ | ||||||
| PHGPx9 | Forward | 5′-TCATGGGCTAAGAACCTTGG-3′ | 220 | 0.99915 | − 3.602 | 34.828 | 90 |
| Reverse | 5′-CCATCGGGTCCAATTAAAAA-3′ |
Statistical analyses
Statistical analyses were performed using the IBM® SPSS® Statistics 19 software package. The data were analysed using one-way analysis of variance (one-way ANOVA) followed by the Tukey post hoc multiple comparison test. Differences were considered to be statistically significant when P < 0.05.
Results
Bioinformatic analysis of the number and diversity of antioxidant genes in the genomes of a selection of ciliates and apicomplexans
Four species from the phylum Ciliophora (ciliates), including T. thermophila, and six species from the phylum Apicomplexa (apicomplexans), including P. falciparum and T. gondii, were chosen for this analysis (Table 1). The apicomplexans were all obligate endoparasites, whereas the ciliates were all free-living, except for I. multifiliis which can also be ectoparasitic. Interestingly, the ciliates possessed a large number and great diversity of antioxidant genes whereas the apicomplexans were much more limited in this respect. Specifically, the ciliates had from four to seven SOD genes whereas the apicomplexans had only one to three. The ciliates had from one to four CAT genes whereas the apicomplexans had either none or only one or two. Most strikingly, the ciliates, except for the ectoparasitic I. multifiliis, had from nine to twelve GPx or PHGPx genes whereas the apicomplexans had either none or only one. Finally, the ciliates had from four to seven Prx genes whereas the apicomplexans had only one to five. Taking these results all together, it appears as if a reduction in the number and diversity of antioxidant genes is a characteristic of the parasitic way of life.
Predicted subcellular locations and subunit molecular weights of the Prx1 proteins in our selection of ciliates and apicomplexans
The predicted (and in some cases experimentally confirmed) subcellular locations and subunit molecular weights of the Prx1 proteins in our selection of ciliates and apicomplexans can be seen in Table 2. Most of the species had one mitochondrial and at least one other Prx1 protein and in most cases, the subunit molecular weights of these proteins ranged from approximately 20–30 kD. The exceptions to this were the Prx1c and Prx1d proteins of O. trifallax which had subunit molecular weights that were approximately twice this size.
Phylogenetic relationships between the Prx1 proteins in our selection of ciliates and apicomplexans
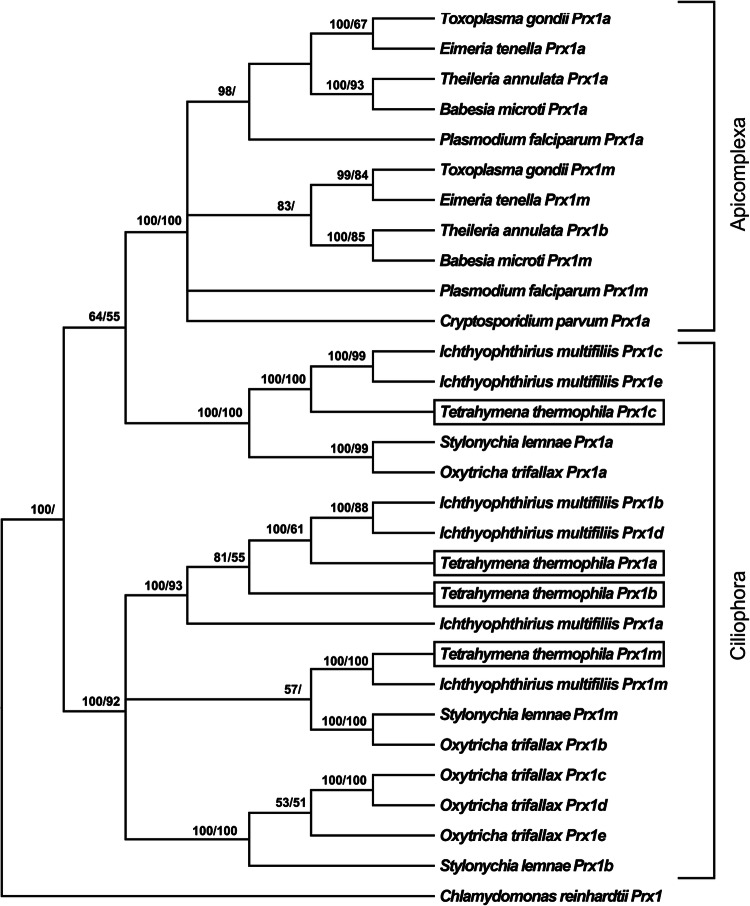
Figure 1 displays the phylogenetic relationships between the Prx1 proteins in our selection of ciliates and apicomplexans. The T. thermophila Prx1 proteins grouped together with orthologues from the other ciliates and clearly separated from the Prx1 proteins of the apicomplexans (posterior probability 100%; bootstrap value 100%). In particular, T. thermophila Prx1a and Prx1b grouped together with Prx1a, Prx1b and Prx1d from I. multifiliis (posterior probability 100%; bootstrap value 93%). Tetrahymena thermophila Prx1m was most closely related to I. multifiliis Prx1m, S. lemnae Prx1m and O. trifallax Prx1b (posterior probability 57). Another cluster, including T. thermophila Prx1c, I. multifiliis Prx1c and Prx1e, S. lemnae Prx1a and O. trifallax Prx1a, was clearly separated from the Prx1 proteins from the other ciliates (posterior probability 100%; bootstrap value 100%).
Fig. 1.
Phylogenetic relationships amongst the Prx1 proteins of various protists reconstructed on the basis of amino acid sequences and using both BI (arithmetic mean = − 10,347.46; harmonic mean = − 10,371.05) and ML (arithmetic mean = − 10,257.5) methods. Bayesian posterior probability (first number) and bootstrap values (second number, if present) higher than 50% are indicated on each node. The T. thermophila Prx1 proteins are boxed
Amino acid sequence comparison of the human, T. thermophila and P. falciparum Prx1 proteins
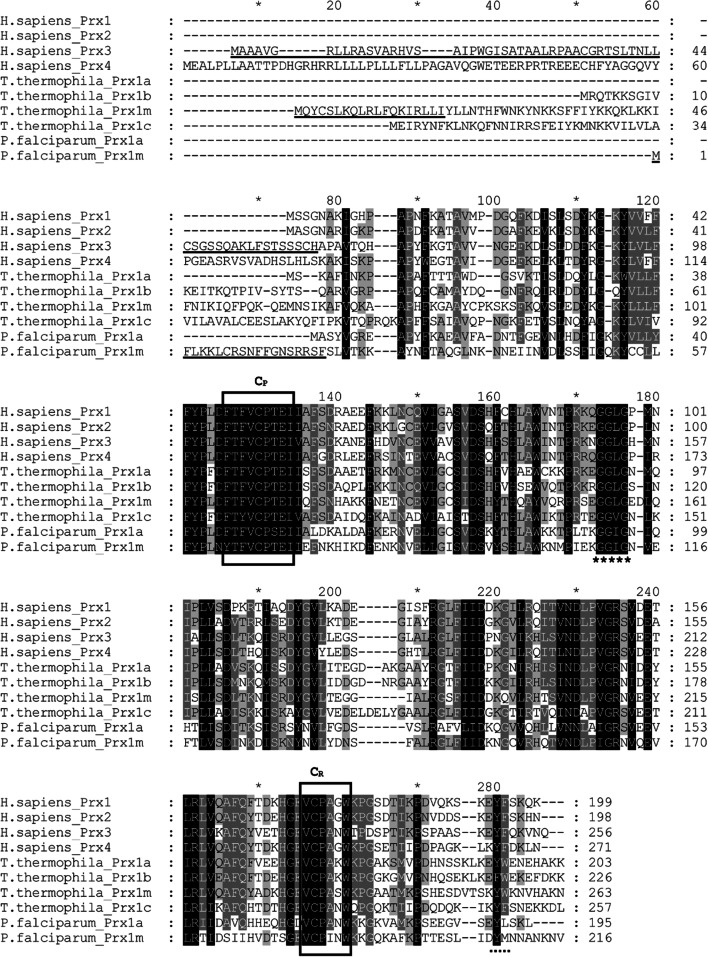
Figure 2 shows an alignment of Prx1 amino acid sequences from Homo sapiens, T. thermophila and P. falciparum. The T. thermophila Prx1 proteins are highly similar to the human and P. falciparum Prx1 proteins, especially in the regions of CP, CR and the GG(L/I/V)G motif. On the other hand, in the region of the Y(F/L) motif there is much more variation. The Y(F/L) motif is conserved only in the human Prx1 proteins, the T. thermophila Prx1c protein and the P. falciparum Prx1a protein. In the T. thermophila Prx1a and Prx1m proteins, the Y(F/L) motif is replaced by a YW motif, and in the T. thermophila Prx1b protein it is replaced by a FW motif. Similarly, in the P. falciparum Prx1m protein, the Y(F/L) motif is replaced by a YM motif.
Fig. 2.
An alignment of Prx1 amino acid sequences from Homo sapiens, T. thermophila and P. falciparum. Note that for the human proteins, the original names have been retained but they are all Prx1 subfamily members, according to the criteria of Gretes et al. [1]. The peroxidatic and resolving Cys residues are indicated by “CP” and “CR”, respectively, and the locations of the conserved GG(L/I/V)G and Y(F/L) motifs are indicated by ***** and ….., respectively. Note that we use the broader definition of these motifs proposed by Pascual et al. [8] rather than the narrower definition originally used by Wood et al. [4]
Correlation of the sensitivity of the Prx1 proteins to hyperoxidation with the presence/absence of the GG(L/I/V)G and Y(F/L) motifs and sulfiredoxin genes
All of the species in our selection of ciliates and apicomplexans possessed the GG(L/I/V)G and Y(F/L) motifs, or variants thereof, in their Prx1 proteins but all of them lacked Srx genes in their genomes (Table 4). The sensitivity of the Prx1 proteins of these species to hyperoxidation has not been studied (except for Prx1m from T. thermophila in the present study) but the presence of the GG(L/I/V)G and Y(F/L) motifs would suggest sensitivity to hyperoxidation whereas the absence of Srx genes would suggest robustness. As explained in the introduction, the presence/absence of Srx genes appears to be a better predictor of the sensitivity/robustness of Prx1 proteins to hyperoxidation than the presence/absence of the GG(L/I/V)G and Y(F/L) motifs. Thus, we predict that all of the Prx1 proteins from all of the species in our selection of cilitates and apicomplexans will be robust to hyperoxidation.
Table 4.
Relationship between the presence/absence of the GG(L/I/V)G and Y(F/L) motifs in the Prx1 proteins of various organisms, the sensitivity of the Prx1 proteins to hyperoxidation and the presence/absence of sulfiredoxin genes
| Species name | Protein name | Motif | Sensitive or robust? | Sulfiredoxin | |
|---|---|---|---|---|---|
| Humans | |||||
| Homo sapiens | Prx1 | GGLG | YF | Sensitive | Present |
| Prx2 | GGLG | YF | Sensitive | ||
| Prx3 | GGLG | YF | Sensitive | ||
| Prx4 | GGLG | YF | Sensitive | ||
| Yeasts | |||||
| Schizosaccharomyces pombe | TPx1 | GGLG | YF | Sensitive | Present |
| Saccharomyces cerevisiae | Tsa1 | GGLG | YF | Sensitive | Present |
| Tsa2 | GGLG | YF | Sensitive | ||
| Ciliates | |||||
| Tetrahymena thermophila | Prx1a | GGLG | YW | Not studied | Absent |
| Prx1b | GGLG | FW | Not studied | ||
| Prx1m | GGLG | YW | This study | ||
| Prx1c | GGVG | YF | Not studied | ||
| Stylonychia lemnae | Prx1a | GGLG | YF | Not studied | Absent |
| Prx1b | GGLG | YW | Not studied | ||
| Prx1m | GGLG | YW | Not studied | ||
| Ichthyophthirius multifiliis | Prx1a | GGIA | YW | Not studied | Absent |
| Prx1b | GGLG | YW | Not studied | ||
| Prx1c | GGVG | YF | Not studied | ||
| Prx1d | GGLG | YW | Not studied | ||
| Prx1e | GGVG | YF | Not studied | ||
| Prx1m | GGLG | YW | Not studied | ||
| Oxytricha trifallax | Prx1a | GGLG | YF | Not studied | Absent |
| Prx1b | GGLG | YW | Not studied | ||
| Prx1c | GGLG | YW | Not studied | ||
| Prx1d | GGLG | YW | Not studied | ||
| Prx1e | GGLG | YW | Not studied | ||
| Apicomplexans | |||||
| Plasmodium falciparum | Prx1a | GGIG | YL | Not studied | Absent |
| Prx1m | GGIG | YM | Not studied | ||
| Toxoplasma gondii | Prx1a | GGIG | YL | Not studied | Absent |
| Prx1m | GGIG | YL | Not studied | ||
| Cryptosporidium parvum | Prx1a | GGIG | YL | Not studied | Absent |
| Eimeria tenella | Prx1a | GGLG | YL | Not studied | Absent |
| Prx1m | GGLG | HL | Not studied | ||
| Theileria annulata | Prx1a | AGVG | HL | Not studied | Absent |
| Prx1b | GGVS | YL | Not studied | ||
| Babesia microti | Prx1a | GGIG | HL | Not studied | Absent |
| Prx1m | GGIP | YL | Not studied | ||
| Bacteria | |||||
| Streptococcus mutans | Prx | – | – | Robust | Absent |
| Escherichia coli | Prx | – | – | Robust | Absent |
| Salmonella typhimurium | Prx | – | – | Robust | Absent |
| Cyanobacteria | |||||
| Synechocystis sp. (NP_442066) | Prx | GGIG | YF | Robust | Absent |
| Anabaena sp. (NP_488681) | Prx | GGVG | YF | Sensitive | Present |
The effects of H2O2, cumene hydroperoxide and various plant natural products on the redox status of the Prx1m protein in T. thermophila cells
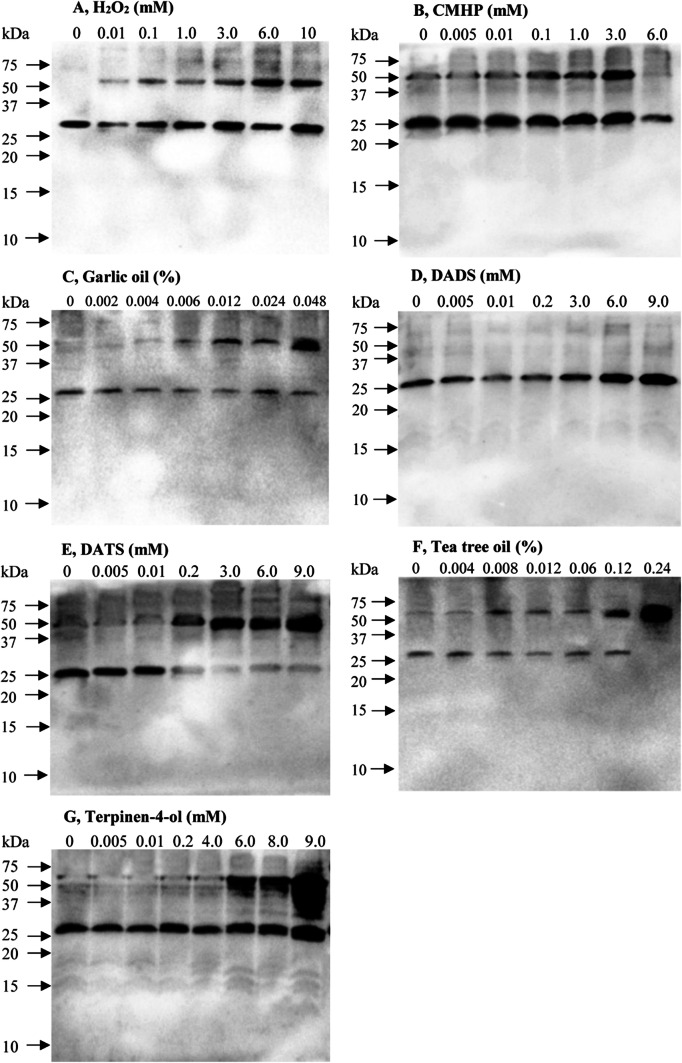
Figure 3 shows the effects of various concentrations of H2O2, cumene hydroperoxide, garlic oil, diallyl disulphide (DADS), diallyl trisulfide (DATS), tea tree oil and terpinen-4-ol on the redox status of the Prx1m protein in T. thermophila. In the absence of any treatment, the Prx1m protein was mostly in the reduced monomer state but when the T. thermophila cells were exposed to increasing concentrations of H2O2, there was increasing conversion of the reduced monomer to the oxidised dimer (Fig. 3a). In addition, there was an increase in the overall amount of both forms of the protein suggesting either that gene expression was increasing or that protein turnover was decreasing. Similar results were obtained when the cells were exposed either to cumene hydroperoxide or garlic oil (Fig. 3b, c). In contrast, when the cells were exposed to DADS, the Prx1m protein remained in its reduced monomer state and there was no evidence of the oxidised dimer state (Fig. 3d). This was quite different to what we observed with DATS. DATS promoted conversion of the reduced monomer state to the oxidised dimer state and, as had been observed for the other treatments, increased the overall amount of the Prx1m protein (Fig. 3e). Tea tree oil had a similar effect to garlic oil but garlic oil was more effective at much lower concentrations than tea tree oil (Fig. 3c, g). The apparent increase in the overall amount of the Prx1m protein was particularly striking in response to increasing concentrations of tea tree oil. Terpinen-4-ol, like tea tree oil, also promoted conversion of the reduced monomer form of the Prx1m protein to the oxidised dimer form and it strongly increased the overall amount of the Prx1m protein. We are confident that the anti-(Prx1m) antibodies were not cross-reacting with either Prx1a or Prx1c because the protein we observed on the immunoblots was too large (> 25 kD) to be either Prx1a or Prx1c (both < 25 kD). The anti-(Prx1m) antibodies might have been cross-reacting with Prx1b, but this is unlikely because the amino acid sequence in the region used to design the peptide to produce the anti-(Prx1m) antibodies was only 70% identical between the Prx1b protein and the Prx1m protein (Fig. 2).
Fig. 3.
The effects of H2O2, cumene hydroperoxide (CMHP) and various plant natural products on the redox status of the Prx1m protein in T. thermophila. The T. thermophila cells were exposed to the indicated concentrations of either the peroxides or the plant natural products for a period of 10 min and then the total protein was extracted from the cells and subjected to NR-PAGE. Equal amounts of protein were loaded into each of the lanes of the gel. Following the NR-PAGE step, the proteins were transferred to nitrocellulose and the Prx1m protein was detected using anti-(T. thermophila Prx1m) antibodies. The monomer and dimers forms of the Prx1m protein are located at ~ 28 and ~ 56 kD, respectively. The molecular size markers were supplied by Bio-Rad
The effects of H2O2, cumene hydroperoxide and various plant natural products on the proliferation of T. thermophila cells
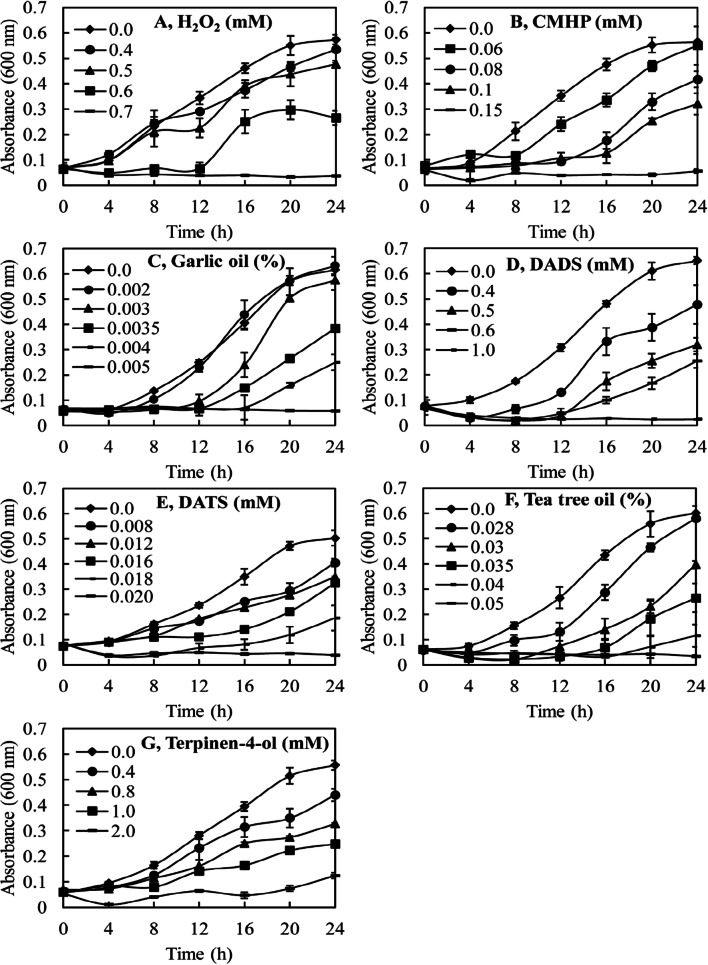
Figure 4 shows the effects of H2O2, cumene hydroperoxide and various plant natural products on the proliferation of the T. thermophila cells. We predicted that the agents that had been the most potent in promoting oxidation of the Prx1m protein would also be the most potent in inhibiting the proliferation of the cells. This prediction was largely borne out by the results. A concentration of 0.7 mM was required for H2O2 to completely inhibit the proliferation of the cells, whereas a concentration of only 0.15 mM was required for cumene hydroperoxide to have the same effect. Garlic oil was an extremely potent inhibitor of the proliferation of the cells with only 0.005% (v/v) required for complete inhibition. DATS was a much stronger inhibitor of cell proliferation than DADS. A concentration of 1.0 mM DADS was required to completely inhibit the proliferation of the cells whereas a concentration of only 0.02 mM of DATS was required. Tea tree oil was a less potent inhibitor of cell proliferation than garlic oil. A concentration of 0.05% (v/v) tea tree oil was required to completely inhibit the proliferation of the cells whereas the concentration of garlic oil required was only 0.005% (v/v), an order of magnitude lower. Terpinen-4-ol was slightly less effective than DADS in inhibiting the proliferation of the cells and much less effective than DATS. The terpinen-4-ol concentration required to completely inhibit the proliferation of the cells was 2.0 mM compared with only 0.02 mM for DATS.
Fig. 4.
The effects of H2O2, cumene hydroperoxide (CMHP) and various plant natural products on the proliferation of T. thermophila cells. The T. thermophila cells were seeded into the wells of 24-well plates at an initial concentration of 4 × 104 cells in 1.5 ml of culture medium containing the various treatments. The proliferation of the cells was monitored at a wavelength of 600 nm with readings being taken automatically every 6 min. Only the readings taken every 4 h are presented
The effects of the various plant natural products on the proliferation of the cells correlated well with their effects on the redox status of the Prx1m protein. Most strikingly, DADS failed to promote conversion of the reduced monomer form of the Prx1m protein to its oxidised dimer form and a relatively high concentration of DADS was required to inhibit the proliferation of the cells. In contrast, DATS was a very potent promoter of the conversion of the reduced monomer form of the Prx1m protein to its oxidised dimer form and also a very potent inhibitor of cell proliferation. Terpinen-4-ol was a weaker promoter of Prx1m oxidation than DATS and also a weaker inhibitor of cell proliferation. Overall these results suggest that the redox state of the Prx1m protein is a good indicator of the extent of oxidative stress being experienced by the cells and, therefore, a good predictor of the extent of inhibition of cell proliferation in response to the different stressors.
Effects of H2O2, cumene hydroperoxide and various plant natural products on the expression of the T. thermophila Prx1 genes
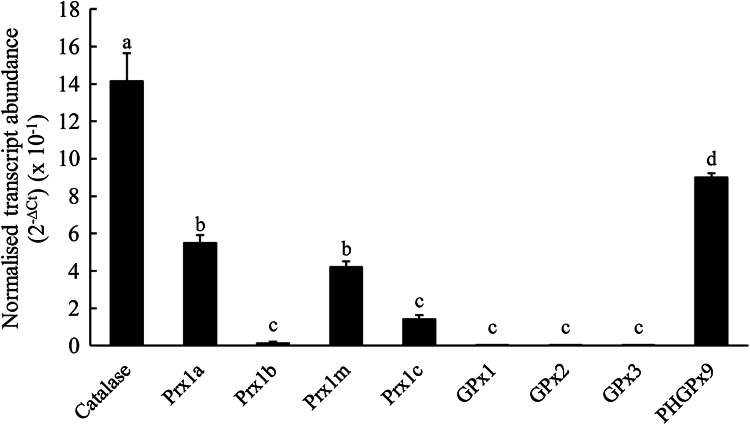
We had observed that exposure of the T. thermophila cells to H2O2, cumene hydroperoxide and various plant natural products increased the abundance of the Prx1m protein, both the reduced monomer form and the oxidised dimer form, but especially the oxidised dimer form (Fig. 3) and we hypothesised that increasing Prx1 protein expression could be due to increasing Prx1m gene expression. To test this, we exposed the cells to the same treatments as for the protein experiments but instead examined gene expression. Prior to this, however, we compared the expression of the four different Prx1 genes with the expression of the single CAT gene, the three GPx genes and one of the nine PHGPx genes that we had identified in T. thermophila (Fig. 5). The Prx1a and Prx1m genes were expressed at similar levels to one another and at much higher levels than either the Prx1b or Prx1c genes. The Prx1a and Prx1m genes were expressed at approximately one-fourth of the level of the CAT gene. Together these data suggest that the Prx1a and Prx1m gene products are important in protecting T. thermophila cells against oxidative stress. The three GPx genes were expressed at extremely low levels, near to the minimum level of detection. In contrast, the one PHGPx gene that we examined was expressed at a relatively high level, somewhat higher than the Prx1a and Prx1m genes and about half as high as the CAT gene.
Fig. 5.
The expression of the Prx1 genes in T. thermophila compared with the one catalase gene, the three glutathione peroxidase (GPx) genes and one of the nine phospholipid hydroperoxide glutathione peroxidase (PHGPx) genes. The cells had been cultured under control conditions in SSP medium as described for their routine maintenance. The transcript abundances for the genes of interest have been normalized to the transcript abundance for GAPDH using the formula 2−∆Ct. The columns represent the means and the bars represent the standard errors of the means (n = 3). Different letters indicate statistically significant differences between the means (P < 0.05)
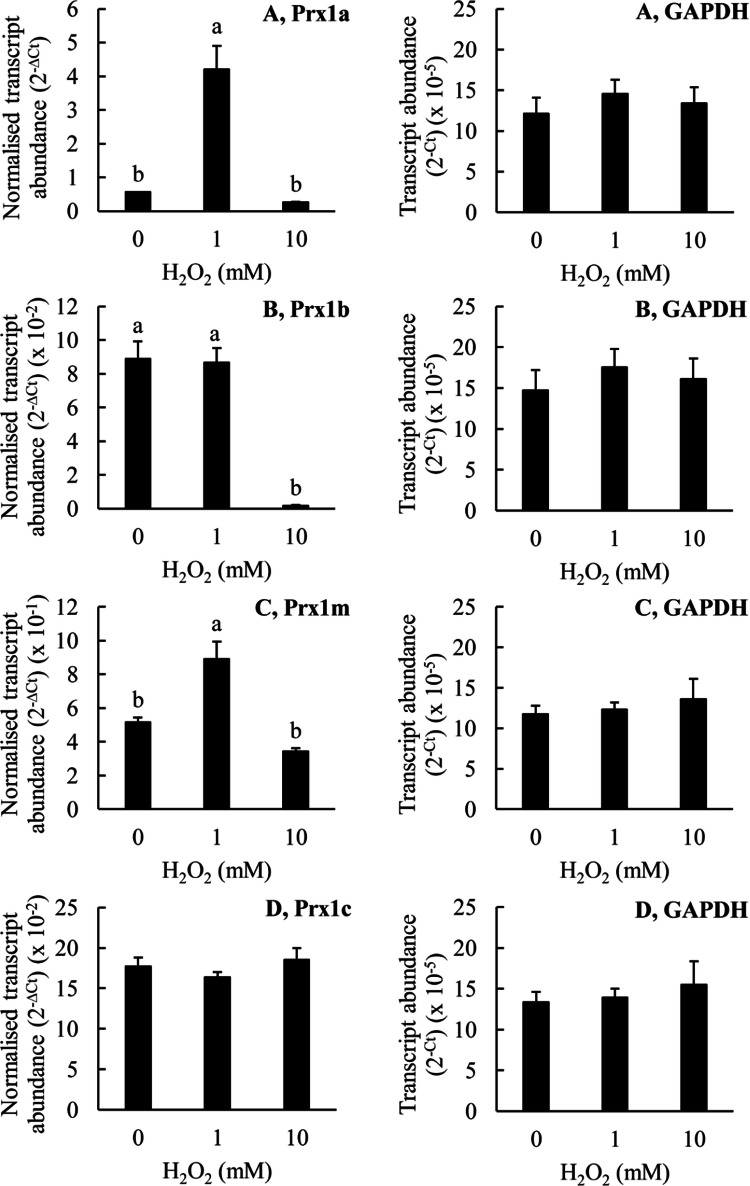
Figures 6, 7, 8, 9 and 10 show the effects of H2O2, garlic oil, DATS, tea tree oil and terpinen-4-ol on the expression of the four T. thermophila Prx1 genes. The Prx1 transcript abundance is expressed relative to the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) transcript abundance. In addition, we also show the ‘raw’ GAPDH transcript abundance data as an indicator of whether the treatments might be causing generalised toxicity to the cells rather than specifically affecting the expression of the Prx1 genes. Treatment of the cells with 1 mM H2O2 greatly increased the transcript abundance for Prx1a and Prx1m, but it did not significantly affect the transcript abundance for either Prx1b or Prx1c (Fig. 6). In contrast, treatment of the cells with 10 mM H2O2 greatly decreased the transcript abundance for Prx1b, but it did not significantly affect the transcript abundances for any of the other Prx1 genes, relative to the control. These results are contrary to the hypothesis that increasing Prx1m gene expression explains the increasing Prx1m protein abundance in T. thermophila in response to H2O2 treatment because the greatest increase in Prx1m protein abundance was seen at a H2O2 concentration of 10 mM whereas no increase in Prx1m transcript abundance was seen at this concentration. It is also important to note that the transcript abundance for GAPDH remained stable indicating that there was no generalized toxicity effect of the H2O2 treatment.
Fig. 6.
The effects of H2O2 on the expression of the Prx genes in T. thermophila. The T. thermophila cells were exposed to the indicated concentrations of the stressor for a period of 10 min before being pelleted by centrifugation, resuspended in 5 ml of SSP medium, counted and then pelleted again. RNA was extracted from 4 × 106 cells as described in the “Materials and methods” section. The transcript abundances for the Prx genes have been normalized to the transcript abundance for GAPDH using the formula 2−∆Ct. The ‘raw’ transcript abundance for GAPDH is also shown, calculated using the formula 2−Ct. The columns represent the means and the bars represent the standard errors of the means (n = 3). Different letters indicate statistically significant differences between the means (P < 0.05)
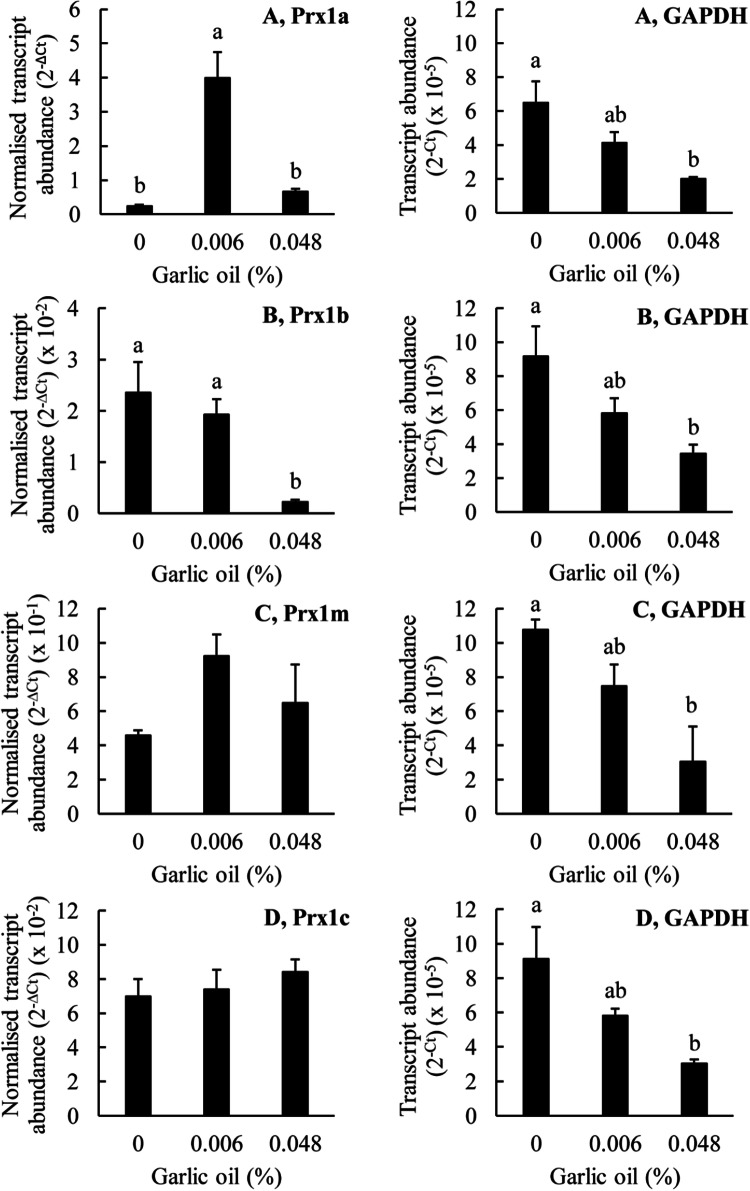
Fig. 7.
The effects of garlic oil on the expression of the Prx genes in T. thermophila. The T. thermophila cells were exposed to the indicated concentrations of the stressor for a period of 10 min before being pelleted by centrifugation, resuspended in 5 ml of SSP medium, counted and then pelleted again. RNA was extracted from 4 × 106 cells as described in the “Materials and methods” section. The transcript abundances for the Prx genes have been normalized to the transcript abundance for GAPDH using the formula 2−∆Ct. The ‘raw’ transcript abundance for GAPDH is also shown, calculated using the formula 2−Ct. The columns represent the means and the bars represent the standard errors of the means (n = 3). Different letters indicate statistically significant differences between the means (P < 0.05)
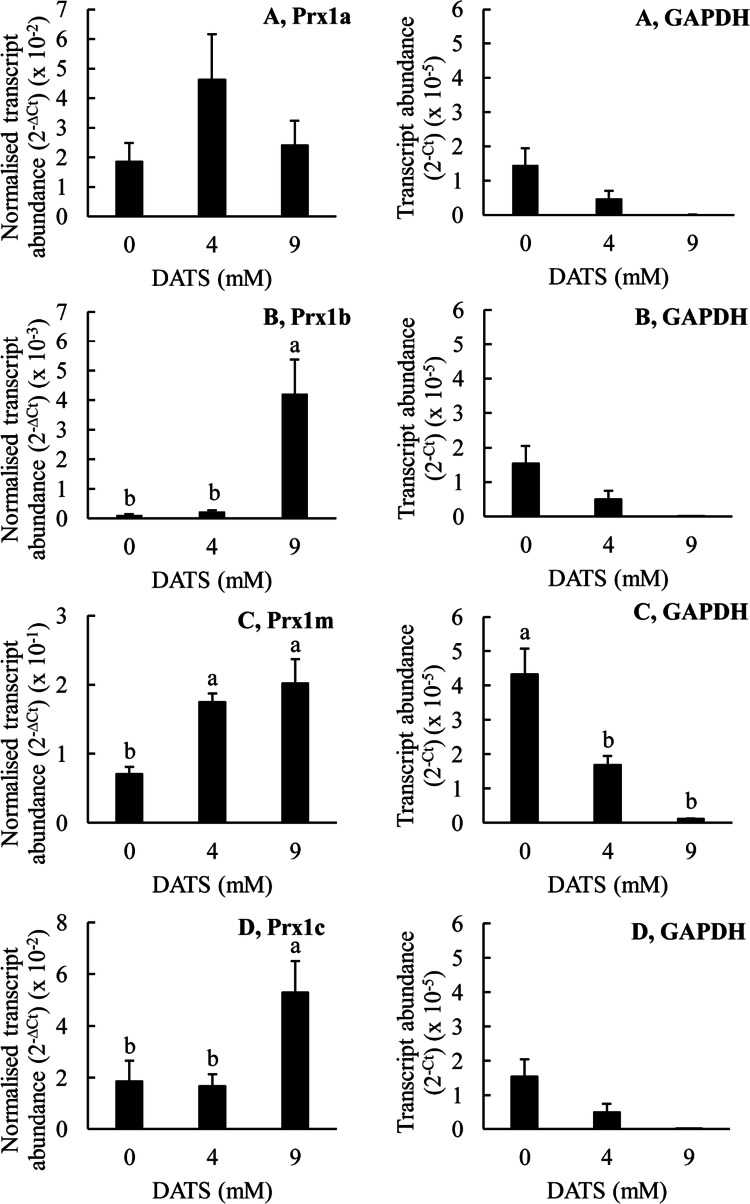
Fig. 8.
The effects DATS on the expression of the Prx genes in T. thermophila. The T. thermophila cells were exposed to the indicated concentrations of the stressor for a period of 10 min before being pelleted by centrifugation, resuspended in 5 ml of SSP medium, counted and then pelleted again. RNA was extracted from 4 × 106 cells as described in the “Materials and methods” section. The transcript abundances for the Prx genes have been normalized to the transcript abundance for GAPDH using the formula 2−∆Ct. The ‘raw’ transcript abundance for GAPDH is also shown, calculated using the formula 2−Ct. The columns represent the means and the bars represent the standard errors of the means (n = 3). Different letters indicate statistically significant differences between the means (P < 0.05)
Fig. 9.
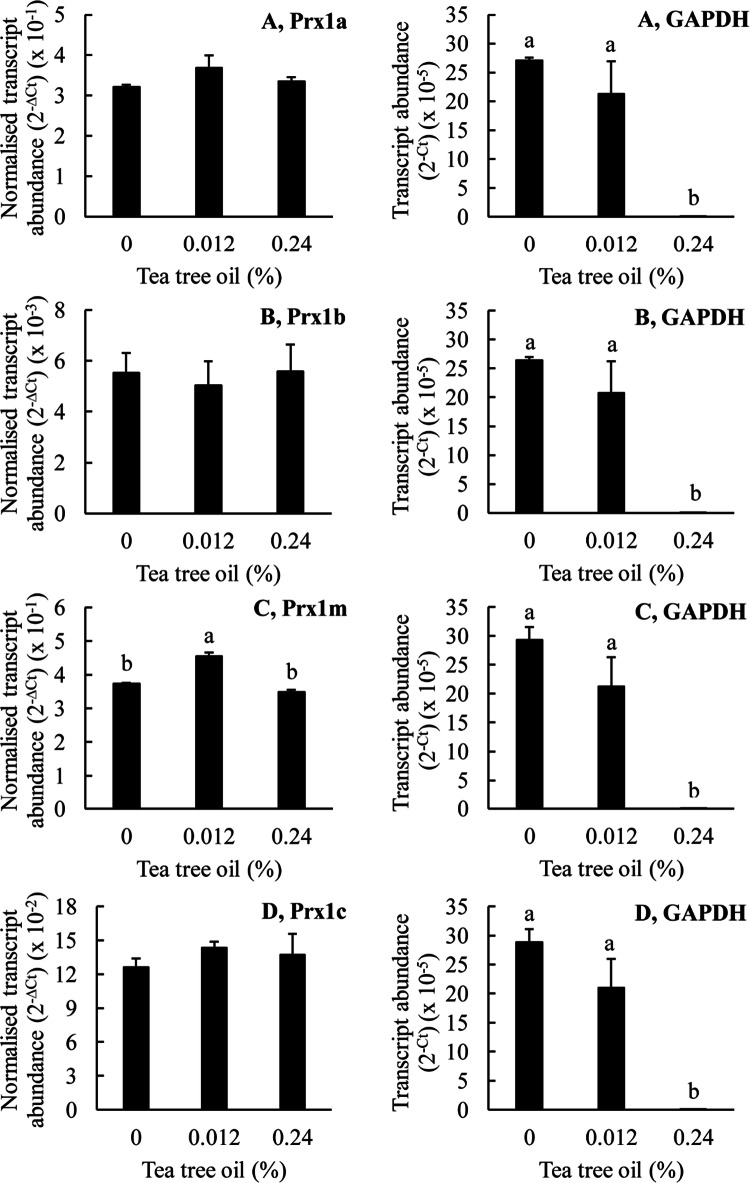
The effects of tea tree oil on the expression of the Prx genes in T. thermophila. The T. thermophila cells were exposed to the indicated concentrations of the stressor for a period of 10 min before being pelleted by centrifugation, resuspended in 5 ml of SSP medium, counted and then pelleted again. RNA was extracted from 4 × 106 cells as described in the “Materials and methods” section. The transcript abundances for the Prx genes have been normalized to the transcript abundance for GAPDH using the formula 2−∆Ct. The ‘raw’ transcript abundance for GAPDH is also shown, calculated using the formula 2−∆Ct. The columns represent the means and the bars represent the standard errors of the means (n = 3). Different letters indicate statistically significant differences between the means (P < 0.05)
Fig. 10.
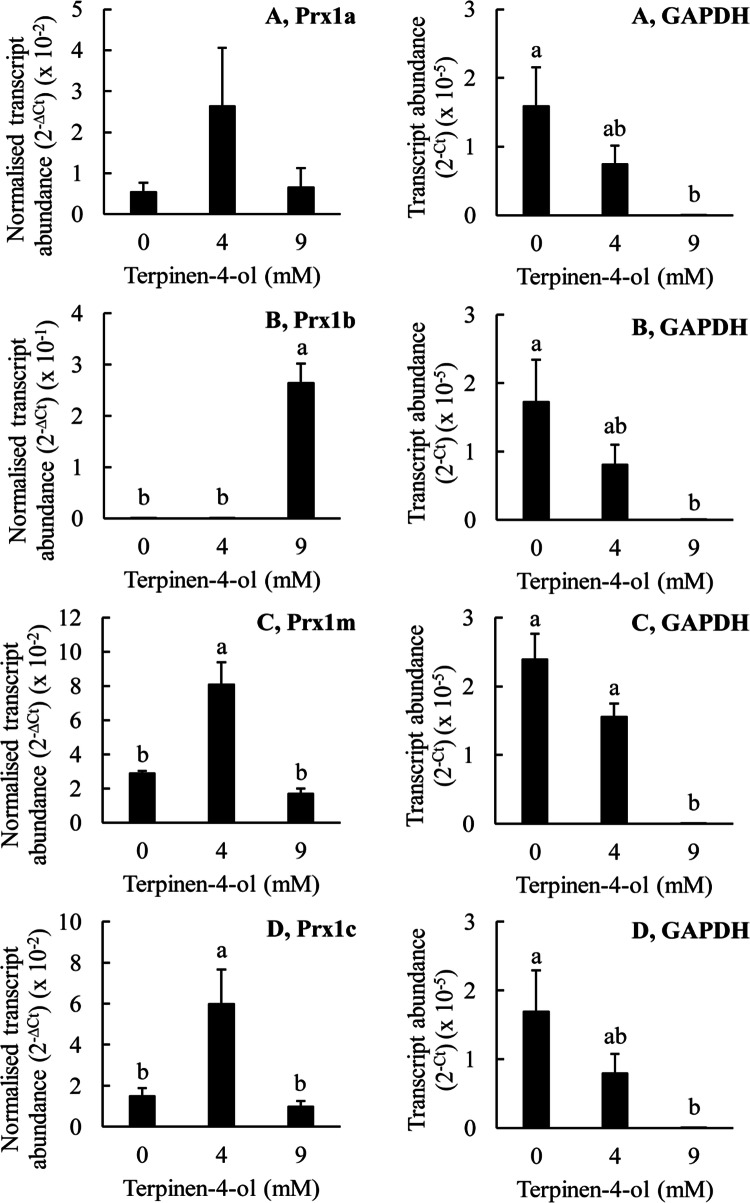
The effects of terpinen-4-ol on the expression of the Prx genes in T. thermophila. The T. thermophila cells were exposed to the indicated concentrations of the stressor for a period of 10 min before being pelleted by centrifugation, resuspended in 5 ml of SSP medium, counted and then pelleted again. RNA was extracted from 4 × 106 cells as described in the “Materials and methods” section. The transcript abundances for the Prx genes have been normalized to the transcript abundance for GAPDH using the formula −∆Ct. The ‘raw’ transcript abundance for GAPDH is also shown, calculated using the formula −∆Ct. The columns represent the means and the bars represent the standard errors of the means (n = 3). Different letters indicate statistically significant differences between the means (P < 0.05)
The results obtained with garlic oil were somewhat different to those obtained with H2O2 (Fig. 7). Treatment of the cells with 0.006% (v/v) garlic oil greatly increased the transcript abundance for Prx1a but it did not significantly affect the transcript abundances for any of the other Prx1 genes, including Prx1m. Thus, 1 mM H2O2 and 0.006% (v/v) garlic oil both increased the transcript abundance for Prx1a but only H2O2 affected the transcript abundance for Prx1m. There was a tendency for the garlic oil treatment to increase the transcript abundance for Prx1m, but it was not statistically significant. At a higher garlic oil concentration of 0.048% (v/v), the transcript abundances for Prx1a, Prx1m and Prx1c were not significantly different from the control values. In contrast, the transcript abundance for Prx1b was greatly decreased. Thus, at the highest concentrations tested, 10 mM for H2O2 and 0.048% (v/v) for garlic oil, there was a dramatic decrease in Prx1b transcript abundance. The transcript abundance for GAPDH, the reference gene, was statistically significantly lower compared with the control at the highest garlic oil concentration. This suggests a generalised toxic effect of this high concentration of garlic oil. Thus, the results obtained with 0.048% (v/v) garlic oil, the higher concentration, should be treated with caution. Overall, these results do not support the conclusion that increasing Prx1m protein abundance in response to garlic oil treatment is due to increasing Prx1m gene expression.
The effects of DATS on the transcript abundances for the various T. thermophila Prx1 genes are shown in Fig. 8. DATS is one of the two major constituents of garlic oil. Thus, it was surprising that the effects of DATS on the Prx1 genes were different to those of garlic oil. DATS at a concentration of 4 mM did not significantly affect the transcript abundance for any of the Prx1 genes whereas at a higher concentration of 9 mM it significantly increased the expression of Prx1b, Prx1m and Prx1c, but did not significantly affect the expression of Prx1a. This was in contrast to the results obtained with garlic oil where there was an increase in Prx1a transcript abundance at 0.006% (v/v) garlic oil, a decrease in Prx1b transcript abundance at 0.048% (v/v) garlic oil and no change in transcript abundance for either Prx1m or Prx1c gene. The differences between the effects of garlic oil and DATS may be due to other constituents in the garlic oil influencing the expression of the genes. The increasing transcript abundance for Prx1m at 9 mM DATS suggests that increasing gene expression may, possibly, be contributing to the increasing Prx1m protein abundance at this DATS concentration. However, Prx1m protein abundance was shown to increase at a DATS concentration of 3 mM and there is no indication of increasing Prx1m transcript abundance until the DATS concentration reaches 9 mM. Thus, we conclude that increasing Prx1m gene expression does not explain the increasing Prx1m protein abundance we observed when the T. thermophila cells were treated with DATS. Additionally, it is important to note that DATS at the higher concentration of 9 mM was apparently toxic to the cells as indicated by the significantly reduced expression of the reference gene GAPDH. Thus, the gene expression results obtained with DATS at this highest concentration must be treated with caution.
Figure 9 shows the effects of tea tree oil treatment on the transcript abundances for the various T. thermophila Prx1 genes. Tea tree oil did not significantly affect the transcript abundance for any of the Prx1 genes except for a small positive effect on the transcript abundance for Prx1m, at a concentration of 0.012% (v/v). In the protein expression experiments, we observed that a tea tree oil concentration of 0.024% (v/v) greatly increased the abundance of the Prx1m protein but in the gene expression experiments we observed no statistically significant effect of this treatment on Prx1m transcript abundance. Thus, we conclude that increasing Prx1m gene expression does not explain the increasing Prx1m protein abundance in response to tea tree oil treatment. As observed for the garlic oil and DATS treatments, the highest concentration of tea tree oil, 0.24% (v/v), was apparently toxic to the cells as indicated by the greatly reduced, in fact barely detectable, expression of the reference gene GAPDH. Thus, again, the data obtained at this highest concentration must be treated with caution.
Lastly, Fig. 10 shows the effects of terpinen-4-ol treatment on the transcript abundances for the Prx1 genes in T. thermophila. Terpinen-4-ol is a major constituent of tea tree oil. Treatment of the T. thermophila cells with 4 mM terpinen-4-ol did not significantly affect the transcript abundance for either Prx1a or Prx1b but it markedly increased the transcript abundance for Prx1m and Prx1c. In contrast, treatment of the cells with 9 mM terpinen-4-ol did not significantly affect the transcript abundances for any of the Prx1 genes except for Prx1b; the transcript abundance for Prx1b was very greatly increased by this treatment. When we consider the protein expression experiments, terpinen-4-ol was by far the most effective of all of the tested substances in increasing the abundance of the Prx1m protein (Fig. 3). However, terpinen-4-ol increased Prx1m gene expression at a concentration of 4 mM, but it did not increase Prx1m protein abundance at this concentration. Conversely, terpinen-4-ol greatly increased Prx1m protein abundance at a concentration of 9 mM but this same concentration had no significant effect on Prx1m transcript abundance. Thus, again, we must conclude that increasing Prx1m gene expression does not explain the increasing Prx1m protein abundance we observed in the T. thermophila cells treated with terpinen-4-ol, or any of the other substances we tested. Taking all of our data together, we conclude that the increasing Prx1m protein abundance we observed in response to the various treatments is not the result of increasing Prx1m gene expression.
Discussion
The alveolates are a clade of protists characterized by the presence of membrane-bound sacs known as alveoli [16]. Two important alveolate phyla are the apicomplexans, which are obligate endoparasites, and the ciliates, which are mostly free-living. We compared the antioxidant enzyme complements of a selection of four ciliates and six apicomplexans and found that the ciliates, except for the facultatively ectoparasitic I. multifiliis, have a plethora of genes encoding antioxidant enzymes whereas the apicomplexans are much more limited in this respect. A recent analysis showed that a large number of genes has been lost in the evolution of apicomplexans from their free-living proto-apicomplexan ancestor [17]. Thus, this presumably explains the more limited repertoire of antioxidant enzymes in the apicomplexans.
It is well established that P. falciparum, the causal organism of the most serious form of human malaria, and T. gondii, the causal organism of toxoplasmosis, have a number of Prx enzymes but a relative dearth, or complete lack, of other peroxide-detoxifying enzymes, i.e., CAT, GPx or PHGPx [1, 18–22]. Because of this, the Prx enzymes of these parasites have been extensively investigated as possible targets for the development of new anti-malarial and anti-toxoplasmosis drug therapies. Interestingly, the other apicomplexans in our survey also had a preponderance of Prx enzymes over either CAT, GPx or PHGPx enzymes. Thus, it seems as if a dependence on Prx enzymes for peroxide detoxification and protection against oxidative stress is characteristic of the parasitic way of life.
Most of the species in our selection of ciliates and apicomplexans had at least one mitochondrial Prx1 protein and at least one other Prx1 protein and most of these Prx1 proteins had predicted molecular weights ranging from approximately 20 to 30 kD. The exceptions to this were two of the five Prx1 proteins of O. trifallax. These were much larger with molecular weights of 46 and 51 kD. An alignment of the O. trifallax Prx1 proteins with the other Prx1 proteins showed that the larger size of the former was due to a long leader sequence, or perhaps another protein, fused at the N-terminus of the Prx1c and Prx1d proteins (data not shown). This leader sequence, or other protein, may explain why the TargetP software we used did not predict a mitochondrial targeting sequence in any of the O. trifallax Prx1 proteins. Perhaps the software is incapable of recognising the mitochondrial targeting sequence employed in O. trifallax.
In T. thermophila four genes potentially encoding Prx1 proteins have been identified. This feature is typical of gene families showing a redundancy due to gene duplication phenomena in their evolution in eukaryotic genomes, and in mammals in particular [23, 24]. In T. thermophila, duplication processes have driven the evolution of several proteins, including metallothioneins [25–28] and superoxide dismutases [29]. Regarding Prx1 enzymes, an interesting result is that the phylum ciliophora enzymes do not group in a single cluster, but are distributed in two main ones: one that includes T. thermophila Prx1c and one from which the other three isoforms emerge. Both clusters contain components of both the Spirotrichea class (to which O. trifallax and S. lemnae belong) and the Oligohymenophorea class (to which I. multifiliis and T. thermophila belong). From our phylogenetic analyses it seems that the first cluster is more closely related to apicomplexa Prx1 enzymes, although posterior probability and bootstrap values are quite low (64% and 55%, respectively). This result could be explained with an evolutionary process characterized by a duplication event that involved the ancestor gene, which probably occurred in distant time, before the speciation event that led to ciliophora and apicomplexa, with a subsequent deletion loss in the latter phylum. An alternative hypothesis could involve an adaptive convergence that could lead to the occurrence of structurally and functionally similar proteins from different ancestor proteins. From our data it is impossible to deduce which of these two hypotheses is the most reliable and it goes without saying that the proposed models are largely speculative, as a finer reconstruction within the Prx1 clade would demand a considerably larger number of available protozoan sequences. Nevertheless, we believe that, even in the present form, our evolutionary reconstruction makes an important contribution to the subject of Prx evolution, describing the very first analysis of multiple Prx sequences from protozoa.
Previous studies have shown that the Prx1 proteins of H. sapiens, the yeasts S. pombe and S. cerevisiae and the cyanobacterium Anabaena sp. are relatively sensitive to hyperoxidation and that these organisms have conserved GG(L/I/V)G and Y(F/L) motifs in their Prx1 proteins and Srx genes in their genomes [4, 8]. In contrast, the Prx1 proteins of the human pathogenic bacteria, S. mutans, E. coli and S. typhimurium are relatively robust to hyperoxidation and these organisms lack the GG(L/I/V)G and Y(F/L) motifs in their Prx1 proteins and Srx genes in their genomes [4]. These data support the hypothesis of Wood et al. that the presence of the GG(L/I/V)G and Y(F/L) motifs is associated with sensitivity to hyperoxidation in Prx1 proteins whereas their absence is associated with robustness [4]. They also support the hypothesis that the presence of Srx genes/proteins is associated with sensitivity to hyperoxidation whereas the absence is not. The difficulty arises when we consider the data for the cyanobacterium Synechocystis sp. The Prx1 protein of this organism possesses the GG(L/I/V)G and Y(F/L) motifs suggesting that it should be sensitive to hyperoxidation when in fact it has been found to be robust [8]. Thus, presence/absence of these motifs may not be a good predictor of the sensitivity of Prx1 proteins to hyperoxidation in all cases. On the other hand, the Synechocystis sp. genome lacks any Srx genes [8]. This is usually associated with robustness to hyperoxidation and indeed the Synechocystis sp. Prx1 protein has been shown to be robust [8]. Thus, there is a consistent pattern that all organisms studied so far that lack Srx genes have Prx1 proteins that are robust to hyperoxidation.
Most of the Prx1 proteins in most of the ciliates we have investigated possessed the conserved GG(L/I/V)G motif but in almost all of them, the Y(F/L) motif was replaced with a YW motif. In other words, the canonical phenylalanine (F) residue was replaced with a tryptophan (W) residue. Tryptophan is relatively polar, whereas phenylalanine is relatively non-polar. In their original article in which they first proposed the “floodgate hypothesis” Wood et al. [4] presented structural modelling data that showed that the “sensitive” type Prx1 proteins from eukaryotes possessed a C-terminal helix containing the conserved YF motif that stabilized the “fully-folded” conformation of these proteins and thereby favoured their hyperoxidation by hindering the formation of the disulphide bond between CysP and CysR. Thus, replacement of phenylalanine with tryptophan in the ciliate Prx1 proteins may fail to hinder the stabilization of their “fully-folded” conformation making them “robust” to hyperoxidation. This is interesting because it suggests a major exception to the general rule that eukaryote Prx1 proteins are generally “sensitive” to hyperoxidation.
The GG(L/I/V)G motifs were mostly well conserved in the Prx1 proteins of the apicomplexans we studied except for several cases where the Y(F/L) motif was replaced with a HL motif. This would have meant replacement of a tyrosine (Y) residue with a histidine (H) residue. Both residues are relatively polar although histidine is more polar then tyrosine. However, since the leucine (L) residue was conserved, we predict that the apicomplexan Prx1 proteins will be “sensitive” to hyperoxidation, like many other eukaryote Prx1 proteins but unlike the ciliate Prx1 proteins.
Mutations in the YF motif, or nearby, can indeed alter the sensitivity of Prx1 proteins to hyperoxidation [30–32]. For example, when eight residues, including the YF motif, were deleted from the C-terminal helix of a Prx1 protein from H. sapiens, the sensitivity of the mutated protein to hyperoxidation was decreased [30]. Similarly, in an earlier study it was discovered that the helminth parasite Schistosoma mansoni has three different Prx1 proteins, one of which is truncated relative to the other two and completely lacks the YF motif [31]. The truncated protein was “robust” to hyperoxidation whereas the other two proteins were “sensitive” to hyperoxidation and when the C-terminal 22 amino acids from one of the “sensitive” proteins were transferred to the “robust” protein, it had the expected effect of converting the “robust” protein to a “sensitive” protein. Conversely, removal of these 22 amino acids from one of the “sensitive” proteins converted it to a “robust” protein. The alterations made in these studies are relatively broad-scale making it difficult to pinpoint exactly which amino acids are important in determining the sensitivity of the Prx proteins to hyperoxidation. In a more targeted study, it was shown that deletion of a lysine (K) residue followed by a histidine (H) residue downstream of the YF motif was sufficient to convert a Schizosaccharomyces pombe Prx1 protein from “sensitive” to “robust” [32]. Thus, the YF motif was not essential in this regard. Subsequently, these authors mutated the downstream histidine and lysine residues to various other residues and found that mutation of the histidine residue to a glutamine residue or mutation of the lysine residue to an arginine residue restored the sensitivity of the S. pombe Prx1 protein to hyperoxidation. Thus, it is clear that a fine-scaled analysis of the amino acids within the C-terminal helix of the Prx1 proteins is required before we can fully understand what exactly determines the sensitivity of these proteins to hyperoxidation. Recently, two new motifs, referred to as motifs A and B, that confer resistance to hyperoxidation in Prx1 proteins, have been discovered and validated using protein crystallography and mutagenesis of human and bacterial Prx1 proteins [33]. Thus, in future it will be important to consider these motifs as well as the GGLG and YF motifs. In addition, modifications of the C-terminus of sensitive Prx1 proteins in the region of the YF motif, such as nitration of the tyrosine (Y) residue in the YF motif or acetylation of a nearby lysine residue, have been shown to increase the resistance of these proteins to hyperoxidation [34, 35].
Using essentially the same methods as we did to treat the cells and to separate and identify the different redox states of the Prx1 proteins within them, i.e., NR-PAGE followed by immunoblotting with anti-(Prx) antibodies, it has been demonstrated that the Prx1 proteins of Jurkat T-lymphocytes (human cancer cells) are relatively “sensitive” to hyperoxidation [36]. Specifically, the oxidised dimer form of the human Prx1 proteins became progressively less and less abundant whereas the hyperoxidised monomer form became progressively more and more abundant as the H2O2 concentration was increased from 5 to 400 µM. This is quite different to what we observed. In our case, the oxidised dimer form of the Prx1 proteins became progressively more and more abundant as the H2O2 concentration was increased (up to as much as 10 mM) and there was never any evidence of the oxidised dimer form being supplanted by the hyperoxidised monomer form as had been observed for the Jurkat T-lymphocytes. Thus, we conclude that the T. thermophila Prx1m protein is relatively “robust” to hyperoxidation, compared with the human Prx1 proteins. This is consistent with the T. thermophila Prx1m protein having a YW motif in its C-terminal helix instead of the YF motif that is found in the human proteins and it adds further credence to our hypothesis that the ciliate Prx1 proteins will be “robust” to hyperoxidation because of the replacement of the relatively non-polar phenylalanine (F) residue with the relatively polar tryptophan (W) residue in their C-terminal helix.
We observed that garlic oil was highly potent at inducing oxidation/dimerization of the Prx1m protein of T. thermophila and that one of its major constituents, DATS, was also highly effective. Surprisingly, DADS, the other major constituent of garlic oil, was completely ineffective. It is known that allyl sulphides, including DADS and DATS, promote redox cycling and that this involves the continuous formation of H2O2 [37]. Thus, formation of the oxidised dimer form of the T. thermophila Prx1m protein is presumably due to this continuous production of H2O2. In rat erythrocytes, DATS was more effective than DADS in promoting the generation of H2O2 [38]. Thus, this may explain why DATS, but not DADS, promoted oxidation of the T. thermophila Prx1m protein. An alternative explanation could be differing effects of these allyl sulphides on thioredoxin reductase (TrxR). In mammals, the reduced monomer form of the Prx1 proteins is regenerated from the oxidised dimer form by thioredoxin (Trx) which in turn is regenerated by TrxR [2]. In the A549 human lung cancer cell line, treatment with DADS (20–200 µM) increased TrxR activity by ~ 50% whereas treatment with DATS (6.25–50 µM) had no effect [39]. Thus, the positive effect of DADS on TrxR activity could antagonise the tendency of H2O2, generated by redox cycling, to promote Prx1m oxidation/dimerization and this could explain the different effects of DADS and DATS on Prx1m.
Interestingly, we found that DATS was more effective than DADS at inhibiting the proliferation of T. thermophila cells. This accorded with our observation that DATS promoted oxidation of the T. thermophila Prx1m protein whereas DADS did not. In other words, oxidation of the Prx1m protein may be causally linked to the inhibition of cell proliferation. From studies with mammalian cells, it is known that both DADS and DATS can induce apoptosis [40] and recently it was shown that the disulphide bond in the oxidised dimer form of a human cytosolic Prx1 protein could be transduced to the apoptosis signalling kinase1 (ASK1) protein [41]. This activated the ASK1 protein and allowed it to initiate apoptosis. It is unknown whether a similar mechanism operates in unicellular eukaryotes such as T. thermophila. Indeed, it appears counterintuitive for a unicellular organism to undergo apoptosis. Nevertheless, there is evidence that T. thermophila, and other unicellular eukaryotes, including P. falciparum, exhibit at least some of the hallmarks of apoptosis, including DNA fragmentation, membrane blebbing and caspase-like activity [42].
A well as garlic oil, we also tested the effects of tea tree oil, and its major constituent terpinen-4-ol, on the redox/oligomerization state of the T. thermophila Prx1m protein. Tea tree oil, from the Australian native plant M. alternifolia, has been used since the 1930s as a topical treatment to disinfect wounds but its mechanism of action is not well understood [43]. In vitro studies with various different bacteria and fungi have shown that tea tree oil compromises the structural and functional integrity of cellular membranes. Similar effects have been observed with the various constituents of tea tree oil, including terpinen-4-ol. In addition, terpinen-4-ol inhibits the proliferation of cancer cells by promoting the production of reactive oxygen species (ROS), including H2O2, and inducing apoptosis [44]. Here we have shown that both tea tree oil and terpinen-4-ol promote the conversion of the reduced monomer form of the T. thermophila Prx1m protein to its oxidised dimer form. We conclude that this is most likely due to increasing ROS production as has been observed in human cancer cells. Thus, the underlying mechanisms of action of garlic oil, tea tree oil and their major constituents may be the same in that they may all act through alterations in the redox state of the Prx1m protein.
Perhaps the most interesting observation of the present study was the increase in the abundance of the Prx1m protein in response to treatment of the T. thermophila cells with high concentrations of either garlic oil, DATS, tea tree oil or terpinen-4-ol. This increase was most pronounced in the tea tree oil and terpinen-4-ol treatments, but was also clearly seen in the garlic oil and DATS treatments, and to some extent in the H2O2 and cumene hydroperoxide treatments. Increasing protein abundance can be due to increasing gene transcription, increasing transcript stability, increasing transcript translation rates, increasing protein stability or a combination of any of these. Our results showed that increasing transcript abundance or transcript stability were not the reasons for the increasing abundance of the Prx1m protein as there were no correlations between the changes in protein abundance and the changes in transcript abundance for any of the treatments we tested. Thus, we conclude that the increasing Prx1m protein abundance is most likely due either to increasing transcript translation rates or to increasing protein stability.
Evidence consistent with increasing protein stability comes from previous studies with the yeast S. cerevisiae. In this yeast, two different Prx1 proteins were shown to undergo a transition from a peroxidase to a chaperone function in response to either oxidative or heat stress [45]. This involved their conversion from a lower molecular weight form to various higher molecular weight forms with the higher molecular weight forms exhibiting chaperone activity. Perhaps something similar is occurring in T. thermophila. Perhaps the T. thermophila Prx1m protein is remaining stable under stressful conditions so that it can act as a chaperone to repair other proteins that have become damaged by the stress. This requires further examination.
In summary, we have shown that the T. thermophila Prx1m protein is “robust” to hyperoxidation. This is the first time that sensitivity to hyperoxidation of Prx1 proteins has been investigated in any ciliated protozoan and it demonstrates, experimentally, for the first time, that not all eukaryote Prx1 proteins are “sensitive” to hyperoxidation. We have also shown that the presence of the canonical GG(L/I/V)G and Y(F/L) motifs is not always a good predictor of the sensitivity of a particular Prx1 protein to hyperoxidation. In addition, we have shown that various plant natural products known to induce the production of ROS also induce the conversion of the reduced monomer form of the T. thermophila Prx1m protein to its oxidised dimer form. Finally, we have shown that increasing gene expression cannot explain the increase in Prx1m protein abundance in response to the treatment of T. thermophila cells with peroxides or peroxide generating plant natural products. Instead we propose that the increasing Prx1m protein abundance in response to these stressful treatments is due to increasing stability of the Prx1m protein.
Abbreviations
- Prx
Peroxiredoxin
- Prxs
Peroxiredoxins
- Srx
Sulfiredoxin
Funding
Funding was provided by Flinders University and a PhD scholarship awarded to Sarmad Al-Asadi by the Government of Iraq.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Gretes MC, Poole LB, Karplus PA. Peroxiredoxins in parasites. Antioxid Redox Signal. 2012;17(4):608–633. doi: 10.1089/ars.2011.4404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wood ZA, Schröder E, Harris JR, Poole LB. Structure, mechanism and regulation of peroxiredoxins. Trends Biochem Sci. 2003;28(1):32–40. doi: 10.1016/s0968-0004(02)00003-8. [DOI] [PubMed] [Google Scholar]
- 3.Rhee SG, Woo HA. Multiple functions of peroxiredoxins: peroxidases, sensors and regulators of the intracellular messenger H2O2, and protein chaperones. Antioxid Redox Signal. 2011;15(3):781–794. doi: 10.1089/ars.2010.3393. [DOI] [PubMed] [Google Scholar]
- 4.Wood ZA, Poole LB, Karplus PA. Peroxiredoxin evolution and the regulation of hydrogen peroxide signaling. Science. 2003;300(5619):650–653. doi: 10.1126/science.1080405. [DOI] [PubMed] [Google Scholar]
- 5.Biteau B, Labarre J, Toledano MB. ATP-dependent reduction of cysteine-sulphinic acid by S. cerevisiae sulphiredoxin. Nature. 2003;425(6961):980–984. doi: 10.1038/nature02075. [DOI] [PubMed] [Google Scholar]
- 6.Woo HA, Chae HZ, Hwang SC, Yang K-S, Kang SW, Kim K, Rhee SG. Reversing the inactivation of peroxiredoxins caused by cysteine sulfinic acid formation. Science. 2003;300(5619):653–656. doi: 10.1126/science.1080273. [DOI] [PubMed] [Google Scholar]
- 7.Rhee SG, Jeong W, Chang TS, Woo HA. Sulfiredoxin, the cysteine sulfinic acid reductase specific to 2-Cys peroxiredoxin: its discovery, mechanism of action, and biological significance. Kidney Int. 2007;72:S3–S8. doi: 10.1038/sj.ki.5002380. [DOI] [PubMed] [Google Scholar]
- 8.Pascual MB, Mata-Cabana A, Florencio FJ, Lindahl M, Cejudo FJ. Overoxidation of 2-Cys peroxiredoxin in prokaryotes: cyanobacterial 2-Cys peroxiredoxins sensitive to oxidative stress. J Biol Chem. 2010;285(45):34485–34492. doi: 10.1074/jbc.M110.160465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Notredame C, Higgins DG, Heringa J. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J Mol Biol. 2000;302(1):205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 10.Darriba D, Taboada GL, Doallo R, Posada D. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics. 2011;27(8):1164–1165. doi: 10.1093/bioinformatics/btr088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61(3):539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59(3):307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 13.Cassidy-Hanley DM. Tetrahymena in the laboratory: strain resources, methods for culture, maintenance, and storage. In: Collins K, editor. Tetrahymena thermophila. San Diego: Elsevier Academic Press Inc; 2012. pp. 239–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cox AG, Winterbourn CC, Hampton MB. Measuring the redox state of cellular peroxiredoxins by immunoblotting. Methods Enzymol. 2010;474:51–66. doi: 10.1016/S0076-6879(10)74004-0. [DOI] [PubMed] [Google Scholar]
- 15.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 16.Tikhonenkov DV, Janouskovec J, Mylnikov AP, Mikhailov KV, Simdyanov TG, Aleoshin VV, Keeling PJ. Description of Colponema vietnamica sp.n. and Acavomonas peruviana n. gen. n. sp., two new alveolate phyla (Colponemidia nom. nov and Acavomonidia nom. nov.) and their contributions to reconstructing the ancestral state of alveolates and eukaryotes. PLoS One. 2014;9(4):e95467. doi: 10.1371/journal.pone.0095467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Woo YH, Ansari H, Otto TD, Klinger CM, Kolisko M, Michalek J, Saxena A, Shanmugam D, Tayyrov A, Veluchamy A, Ali S, Bernal A, del Campo J, Cihlar J, Flegontov P, Gornik SG, Hajduskova E, Hora A, Janouskovec J, Katris NJ, Mast FD, Miranda-Saavedra D, Mourier T, Naeem R, Nair M, Panigrahi AK, Rawlings ND, Padron-Regalado E, Ramaprasad A, Samad N, Tomcala A, Wilkes J, Neafsey DE, Doerig C, Bowler C, Keeling PJ, Roos DS, Dacks JB, Templeton TJ, Waller RF, Lukes J, Obornik M, Pain A. Chromerid genomes reveal the evolutionary path from photosynthetic algae to obligate intracellular parasites. Elife. 2015;4:e06974. doi: 10.7554/eLife.06974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kwok LY, Schluter D, Clayton C, Soldati D. The antioxidant systems in Toxoplasma gondii and the role of cytosolic catalase in defence against oxidative injury. Mol Microbiol. 2004;51(1):47–61. doi: 10.1046/j.1365-2958.2003.03823.x. [DOI] [PubMed] [Google Scholar]
- 19.Akerman SE, Müller S. Peroxiredoxin-linked detoxification of hydroperoxides in Toxoplasma gondii. J Biol Chem. 2005;280(1):564–570. doi: 10.1074/jbc.M406367200. [DOI] [PubMed] [Google Scholar]
- 20.Clarebout G, Slomianny C, Delcourt P, Leu B, Masset A, Camus D, Dive D. Status of Plasmodium falciparum towards catalase. Br J Haematol. 1998;103(1):52–59. doi: 10.1046/j.1365-2141.1998.00946.x. [DOI] [PubMed] [Google Scholar]
- 21.Sztajer H, Gamain B, Aumann KD, Slomianny C, Becker K, Brigelius-Flohe R, Flohe L. The putative glutathione peroxidase gene of Plasmodium falciparum codes for a thioredoxin peroxidase. J Biol Chem. 2001;276(10):7397–7403. doi: 10.1074/jbc.M008631200. [DOI] [PubMed] [Google Scholar]
- 22.Jortzik E, Becker K. Thioredoxin and glutathione systems in Plasmodium falciparum. Int J Med Microbiol. 2012;302(4–5):187–194. doi: 10.1016/j.ijmm.2012.07.007. [DOI] [PubMed] [Google Scholar]
- 23.Goncalves I, Duret L, Mouchiroud D. Nature and structure of human genes that generate retropseudogenes. Genome. 2000;10(5):672–678. doi: 10.1101/gr.10.5.672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen C, Gentles AJ, Jurka J, Karlin S. Genes, pseudogenes, and Alu sequence organization across human chromosomes 21 and 22. Proc Natl Acad Sci USA. 2002;99(5):2930–2935. doi: 10.1073/pnas.052692099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shang Y, Song X, Bowen J, Corstanje R, Gao Y, Gaertig J, Gorovsky MA. A robust inducible-repressible promoter greatly facilitates gene knockouts, conditional expression, and overexpression of homologous and heterologous genes in Tetrahymena thermophila. Proc Natl Acad Sci USA. 2002;99(6):3734–3739. doi: 10.1073/pnas.052016199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boldrin F, Santovito G, Gaertig J, Wloga D, Cassidy-Hanley D, Clark TG, Piccinni E. Metallothionein gene from Tetrahymena thermophila with a copper-inducible-repressible promoter. Eukaryot Cell. 2006;5(2):422–425. doi: 10.1128/EC.5.2.422-425.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Diaz S, Amaro F, Rico D, Campos V, Benitez L, Martin-Gonzales A, Hamilton EP, Orias E, Gutierrez JC. Tetrahymena metallothioneins fall into two discrete subfamilies. PLoS One. 2007;2(3):e291. doi: 10.1371/journal.pone.0000291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Santovito G, Formigari A, Boldrin F, Piccinni E. Molecular and functional evolution of Tetrahymena metallothioneins: new insights into the gene family of Tetrahymena thermophila. Comp Biochem Physiol C Toxicol Pharmacol. 2007;144(4):391–397. doi: 10.1016/j.cbpc.2006.11.010. [DOI] [PubMed] [Google Scholar]
- 29.Ferro D, Bakiu R, De Pittà C, Boldrin F, Cattalini F, Pucciarelli S, Miceli C, Santovito G. Cu, Zn superoxide dismutases from Tetrahymena thermophila: molecular evolution and gene expression of the first line of antioxidant defenses. Protist. 2015;166(1):131–145. doi: 10.1016/j.protis.2014.12.003. [DOI] [PubMed] [Google Scholar]
- 30.Wang X, Wang LK, Wang X, Sun F, Wang CC. Structural insights into the peroxidase activity and inactivation of human peroxiredoxin 4. Biochem J. 2012;441:113–118. doi: 10.1042/BJ20110380. [DOI] [PubMed] [Google Scholar]
- 31.Sayed AA, Williams DL. Biochemical characterization of 2-Cys peroxiredoxins from Schistosoma mansoni. J Biol Chem. 2004;279(25):26159–26166. doi: 10.1074/jbc.M401748200. [DOI] [PubMed] [Google Scholar]
- 32.Koo KH, Lee S, Jeong SY, Kim ET, Kim HJ, Kim K, Song K, Chae HZ. Regulation of thioredoxin peroxidase activity by C-terminal truncation. Arch Biochem Biophys. 2002;397(2):312–318. doi: 10.1006/abbi.2001.2700. [DOI] [PubMed] [Google Scholar]
- 33.Bolduc JA, Nelson KJ, Haynes AC, Lee J, Reisz JA, Graff AH, Clodfelter JE, Parsonage D, Poole LB, Furdui CM, Lowther WT. Novel hyperoxidation resistance motifs in 2-Cys peroxiredoxins. J Biol Chem. 2018;293(30):11901–11912. doi: 10.1074/jbc.RA117.001690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Randall LM, Manta B, Hugo M, Gil M, Batthyàny C, Trujillo M, Poole LB, Denicola A. Nitration transforms a sensitive peroxiredoxin 2 into a more active and robust peroxidase. J Biol Chem. 2014;289(22):15536–15543. doi: 10.1074/jbc.M113.539213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parmigiani RB, Xu WS, Venta-Perez G, Erdjument-Bromage H, Yaneva M, Tempst P, Marks PA. HDAC6 is a specific deacetylase of peroxiredoxins and is involved in redox regulation. PNAS. 2008;105(28):9633–9638. doi: 10.1073/pnas.0803749105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cox AG, Pearson AG, Pullar JM, Jonsson TJ, Lowther WT, Winterbourn CC, Hampton MB. Mitochondrial peroxiredoxin 3 is more resilient to hyperoxidation than cytoplasmic peroxiredoxins. Biochem J. 2009;421:51–58. doi: 10.1042/BJ20090242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Munday R. Harmful and beneficial effects of organic monosulfides, disulfides, and polysulfides in animals and humans. Chem Res Toxicol. 2012;25(1):47–60. doi: 10.1021/tx200373u. [DOI] [PubMed] [Google Scholar]
- 38.Munday R, Munday JS, Munday CM. Comparative effects of mono-, di-, tri-, and tetrasulfides derived from plants of the allium family: redox cycling in vitro and hemolytic activity and phase 2 enzyme induction in vivo. Free Radic Biol Med. 2003;34(9):1200–1211. doi: 10.1016/s0891-5849(03)00144-8. [DOI] [PubMed] [Google Scholar]
- 39.Hu Y, Urig S, Koncarevic S, Wu XJ, Fischer M, Rahlfs S, Mersch-Sundermann V, Becker K. Glutathione- and thioredoxin-related enzymes are modulated by sulfur-containing chemopreventive agents. Biol Chem. 2007;388(10):1069–1081. doi: 10.1515/BC.2007.135. [DOI] [PubMed] [Google Scholar]
- 40.Wu XJ, Kassie F, Mersch-Sundermann V. Induction of apoptosis in tumor cells by naturally occurring sulfur-containing compounds. Mutat Res Rev Mutat Res. 2005;589(2):81–102. doi: 10.1016/j.mrrev.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 41.Jarvis RM, Hughes SM, Ledgerwood EC. Peroxiredoxin 1 functions as a signal peroxidase to receive, transduce, and transmit peroxide signals in mammalian cells. Free Radic Biol Med. 2012;53(7):1522–1530. doi: 10.1016/j.freeradbiomed.2012.08.001. [DOI] [PubMed] [Google Scholar]
- 42.Deponte M. Programmed cell death in protists. Biochim Biophys Acta Mol Cell Res. 2008;1783(7):1396–1405. doi: 10.1016/j.bbamcr.2008.01.018. [DOI] [PubMed] [Google Scholar]
- 43.Carson CF, Hammer KA, Riley TV. Melaleuca alternifolia (tea tree) oil: a review of antimicrobial and other medicinal properties. Clin Microbiol Rev. 2006;19(1):50–62. doi: 10.1128/CMR.19.1.50-62.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nakayama K, Murata S, Ito H, Iwasaki K, Villareal MO, Zheng YW, Matsui H, Isoda H, Ohkohchi N. Terpinen-4-ol inhibits colorectal cancer growth via reactive oxygen species. Oncol Lett. 2017;14(2):2015–2024. doi: 10.3892/ol.2017.6370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jang HH, Lee KO, Chi YH, Jung BG, Park SK, Park JH, Lee JR, Lee SS, Moon JC, Yun JW, Choi YO, Kim WY, Kang JS, Cheong GW, Yun DJ, Rhee SG, Cho MJ, Lee SY. Two enzymes in one: two yeast peroxiredoxins display oxidative stress-dependent switching from a peroxidase to a molecular chaperone function. Cell. 2004;117(5):625–635. doi: 10.1016/j.cell.2004.05.002. [DOI] [PubMed] [Google Scholar]