Abstract
ORP2 is a ubiquitously expressed OSBP-related protein previously implicated in endoplasmic reticulum (ER)—lipid droplet (LD) contacts, triacylglycerol (TG) metabolism, cholesterol transport, adrenocortical steroidogenesis, and actin-dependent cell dynamics. Here, we characterize the role of ORP2 in carbohydrate and lipid metabolism by employing ORP2-knockout (KO) hepatoma cells (HuH7) generated by CRISPR-Cas9 gene editing. The ORP2-KO and control HuH7 cells were subjected to RNA sequencing, analyses of Akt signaling, carbohydrate and TG metabolism, the extracellular acidification rate, and the lipidome, as well as to transmission electron microscopy. The loss of ORP2 resulted in a marked reduction of active phosphorylated Akt(Ser473) and its target Glycogen synthase kinase 3β(Ser9), consistent with defective Akt signaling. ORP2 was found to form a physical complex with the key controllers of Akt activity, Cdc37, and Hsp90, and to co-localize with Cdc37 and active Akt(Ser473) at lamellipodial plasma membrane regions, in addition to the previously reported ER–LD localization. ORP2-KO reduced glucose uptake, glycogen synthesis, glycolysis, mRNA-encoding glycolytic enzymes, and SREBP-1 target gene expression, and led to defective TG synthesis and storage. ORP2-KO did not reduce but rather increased ER–LD contacts under basal culture conditions and interfered with their expansion upon fatty acid loading. Together with our recently published work (Kentala et al. in FASEB J 32:1281–1295, 2018), this study identifies ORP2 as a new regulatory nexus of Akt signaling, cellular energy metabolism, actin cytoskeletal function, cell migration, and proliferation.
Electronic supplementary material
The online version of this article (10.1007/s00018-018-2850-8) contains supplementary material, which is available to authorized users.
Keywords: Akt signaling, CRISPR-Cas9, Glycolysis, OSBPL2, OSBP-related protein, Triacylglycerol
Introduction
Oxysterol-binding protein (OSBP)-related protein 2 (ORP2) belongs to a conserved family of ubiquitous lipid-binding/transfer proteins, characterized by a carboxy-terminal OSBP-related lipid-binding domain (ORD). In addition, most ORPs contain an amino-terminal pleckstrin homology (PH) domain that binds membrane phosphoinositides (PIPs) and a two phenylalanines in an acidic tract (FFAT) motif which targets the ORPs to the ER membranes via interaction with the integral VAMP-associated proteins (VAPs) [28, 37]. The ORD of several ORPs binds cholesterol, oxysterols, or phosphatidylserine (PS), and additionally PI4P or other PIPs [8, 11, 17, 30, 31, 34, 48]. Recent hallmark studies have established that a number of ORPs have the capacity to mediate the counter-current transport of cholesterol or PS in exchange for PIPs, a process in which the synthesis and hydrolysis of PIPs energize the transport of cholesterol or PS against their concentration gradients [8, 31, 32, 34]. For the founder member of the ORP family, OSBP, it was shown that its highest affinity oxysterol ligand, 25-hydroxycholesterol (25OHC), inhibits the function of the protein as a bidirectional cholesterol/PI4P transporter at ER-Golgi contacts [31].
ORP2 is a cytosolic protein which lacks the membrane-targeting PH domain, but is found to transiently localize on the surface of the intracellular lipid droplets and to ER–LD contacts [16, 22]. In cells lacking large lipid droplets or after treatment with the high-affinity oxysterol ligand of ORP2, 22(R)-hydroxycholesterol [22(R)OHC], the protein is often found in the cortical region of the cells [16, 22]. The cellular function of ORP2 was previously linked to cholesterol and triacylglycerol (TG) homeostasis. ORP2 is found to bind cholesterol, 22(R)OHC, 25OHC, and 7-ketocholesterol (7KC) [16, 47]. Furthermore, ORP2 was reported to bind lipid vesicles containing phosphatidylinositol phosphates (PIPs) [15]. In HeLa cells, ORP2 overexpression was shown to increase the efflux of [14C]cholesterol and enhance its intracellular transport [15]. In adrenocortical cells, silencing of ORP2 was shown to increase the levels of cholesterol and to reduce 22(R)OHC and 7KC [12]. Knockdown of ORP2 and its ER anchors, the VAP proteins in HuH7 hepatoma cells were found to enhance the hydrolysis and to decrease the synthesis of [3H]TGs [50].
Our recently published observations revealed a cellular role for ORP2 beyond lipid metabolism. Analysis of ORP2 knockout (ORP2-KO) HuH7 cells indicated a role for ORP2, in addition to lipid metabolism, in actin cytoskeletal regulation as well as in the control of cell adhesion, migration, and proliferation [21]. Moreover, analysis of ORP2 protein interactome in HuH7 cells identified ORP2-binding partners involved in RhoA signaling [21], a central actin regulatory pathway. These findings prompted the hypothesis that ORP2 could mediate a crosstalk between lipid metabolism and actin cytoskeletal functions. In the present study, we demonstrate a defect in Akt signaling and analyze the energy metabolism and lipidome in HuH7 hepatoma cells subjected to ORP2-KO by CRISPR-Cas9 mediated gene editing.
Experimental procedures
Antibodies
The rabbit anti-ORP2 antibody was described in [24]. Anti-β-actin was purchased from Sigma-Aldrich (St. Louis, MO, USA). Rabbit anti-Cdc37, anti-Hsp90, anti-Akt, anti-Akt(Ser473), anti-GSK-3β, and anti-GSK-3β(Ser9) were from Cell Signaling (Danvers, MA, USA). Mouse monoclonal anti-Cdc37 was from Thermo Scientific (Waltham, MA, USA) and mouse monoclonal anti-cortactin from Merck-Millipore (Temecula, CA, USA). The fluorescent secondary fAlexa Fluor-568 goat anti-rabbit and Cy5® goat anti-mouse antibodies were from Molecular Probes/Invitrogen (Carlsbad, CA, USA) and Life Technologies (Eugene, OR, USA), respectively.
Cell culture, ORP2-KO cell lines, and transfections
The HuH7 human hepatoma cell line [35] was cultured as described in [21, 22]. ORP2-KO lines were derived therefrom by CRISPR-Cas9-mediated gene editing by employing vectors described in [42, 44] as reported in [21]. For the generation of ORP2-KO1 guide, RNAs targeting exon 4 of OSBPL2 were used, and for ORP2-KO2 guide RNAs targeting both exon 4 and 5 were used. The Cas9 control HuH7 cells were prepared similarly to the KO cells transfected with the Cas9 expression plasmid without the OSBPL2-specific gRNA constructs. HuH7 cells were transfected with a GFP-ORP2 (human ORP2 cDNA in pEGFP-N1, Clontech/TaKaRa Bio, Mountain View, CA, USA) using Lipofectamine® 2000 (Invitrogen). Human umbilical vein endothelial cells (HUVECs; Lonza, Basel, Switzerland) were cultured in fibronectin–gelatin-coated flasks in endothelial cell growth medium [EGM; 0.1% human epidermal growth factor (EGF), 0.1% hydrocortisone, 0.1% gentamicin–amphotericin-B, 0.4% bovine brain extract, 2% fetal bovine serum; Lonza], and transfected by electroporation using the Nucleofector® Kit for HUVECs (Lonza).
Lentivirus-mediated ORP2 overexpression and knockdown
Generation of recombinant lentiviruses expressing wild-type (wt) ORP2 or its mutants ORP2 mPIP (H178–179A, K423A) and mFFAT (F7–8V, D9V), and a GFP-encoding vector control, as well as cell infections of ORP2-KO HuH7 cells are described in [21]. Knockdown of ORP2 in HUVECs was carried out by transducing the cells for 48 h at a multiplicity of infection 10, with an lentivirus expressing the ORP2-specific shRNA, TRCN0000154381 (Sigma-Aldrich), using non-targeting SHC002 (Sigma-Aldrich) as a control. The viruses were packaged and titrated by the Virus Core of the Biomedicum Functional Genomics Unit (Helsinki Institute of Life Science, HiLIFE).
RNA sequencing and pathway analysis
RNA sequencing was performed as described in [21]. In detail, samples were prepared from four parallel basal cultures each of ORP2-KO1, ORP2-KO2, and Cas9 control HuH7 cells using TruSeq Stranded Total RNA Library Prep Kit with Ribo-Zero (Illumina, San Diego, CA, USA). BWA [26] was used to align output reads to human reference genome (GRCh38). The FeatureCounts program [27] was employed to count mapped fragments against genomic features defined by the Ensembl annotation file (Homo_sapiens.GRCh38.76.gtf). The genes with counts equal or less than 1 in all samples were removed and Deseq 2 was used to analyze the differential gene expression [29]. Benjamini–Hochberg test was used to control the false discovery rate. Genes which differed significantly (adjusted p value < 0.05) between ORP2-KO1 and Cas9 control cells were selected for analysis by Ingenuity Pathways Analysis® tool (IPA, http://www.ingenuity.com). Enrichment of the genes in the pathways was assessed in comparison with a reference set in the whole Ingenuity pathway knowledge base.
Western blotting
Western blot analyses were carried out as described in [50].
Pull-down assay
HuH7 cells were grown in basal culture conditions or alternatively treated with 5 µM Withaferin A for 24 h prior to lysis in 10 mM HEPES pH 7.6, 150 mM NaCl, 0.5 mM MgCl2, 10% glycerol, 0.5% Triton X-100, 0.5% Na-deoxycholate, and protease inhibitor cocktail (Roche Diagnostics, Basel, Switzerland). Lysates were mixed thoroughly by vortexing and incubated on ice for 10 min. Unbroken cells and insoluble material were removed by 10 min centrifugation at 20,000×g. The obtained lysates were incubated with 300 µmol GST-ORP2 or GST [21] and 20 µl Glutathione Sepharose 4B (GE Healthcare, Buckinghamshire, UK) for 2 h at 4 °C. The beads were washed four times with the lysis buffer, resuspended in 30 µl Laemmli sample buffer, boiled for 5 min, and separated by SDS-PAGE. The bound proteins were detected on western blots with anti-Cdc37, anti-Akt, and anti-Hsp90 antibodies.
To study the role of Akt phosphorylation, cell lysates containing 0.1 mg of total protein were treated with 2000 U of Lambda protein Phosphatase (λ-PP, New England BioLabs, Ipswich, MA, USA) in 50 mM HEPES (pH 7.5), 100 mM NaCl, 2 mM DTT, 0.01% Brij 35, 1 mM MnCl2, and protease inhibitor cocktail (Roche Diagnostics) for 1 h at 30 °C, followed by pull-down assay with GST-ORP2 as described above.
Immunofluorescence staining
HuH7 cells were fixed with 4% paraformaldehyde and 0.02% glutaraldehyde in phosphate-buffered saline (PBS) for 30 min. Aldehyde groups were blocked with 50 mM NH4Cl for 15 min and cells were permeabilized with 0.1% Triton X-100 for 5 min. Unspecific binding of antibodies was blocked with 10% FBS for 45 min, followed by incubation with the primary antibodies in 5% FBS for 30 min at 37 °C. The cells were washed with PBS, incubated with fluorescent secondary antibodies in 5% FBS for 45 min at 37 °C, and mounted in a mixture of Mowiol (Calbiochem, La Jolla, CA, USA), 1,4-diazabicyclo-[2.2.2]octane (50 mg/ml, Sigma-Aldrich), and DAPI (5 μg/ml, Molecular Probes).
Fluorescence microscopy
Fluorescence signals were detected with a Zeiss Axio Observer Z1 microscope with a PlnApo 63×/1.40 oil DICII objective and Colibri laser. Images were recorded with the Zen 2 2.0.0 software (Carl Zeiss Imaging Solutions GmbH, Oberkochen, Germany). Image panels were created with Adobe Photoshop7 software.
Assays for glucose and fatty acid uptake
Glucose uptake was measured in ORP2-KO and Cas9 control cells after starving the cells in glucose and serum free Dulbecco’s modified Eagle medium (DMEM, Gibco, Grand Island, NY, USA) for 3 h. The cells were treated with a mixture of 10 µM 2-deoxy-d-glucose (Sigma-Aldrich) and 10 µM [3H]-2-deoxy-d-glucose (100 mCi/mmol) (PerkinElmer, Waltham, MA, USA) for 15 min. The non-specific uptake of glucose was measured by treating the cells 45 min with 40 µM cytochalasin B (Sigma-Aldrich) prior to adding the glucose mixture. The cells were washed three times with PBS and lysed in 0.03% SDS. The radioactivity of the lysed cells was measured by liquid scintillation counting in Optiphase HiSafe 3 scintillation cocktail (PerkinElmer) and normalized to total cellular protein (BCA assay, Thermo Scientific). The non-specific glucose uptake was subtracted from the measurements.
Fatty acid uptake was measured similarly: ORP2-KO and Cas9 control were starved in glucose and serum-free DMEM for 3 h, followed by treatment with a mixture of 20 mM glucose, 5 nM oleic acid-BSA and 1 µCi [3H]-oleic acid (PerkinElmer) for 15 min. The cell lysis and scintillation counting were performed as described above.
Assay for glycogen synthesis
To measure glycogen synthesis, cells were first starved for 2 h in glucose and serum free DMEM (Gibco). The cells were then treated with 0.18 µCi/µmol d-[U-14C]-glucose (PerkinElmer) for 1 h, after which they were washed three times with PBS and lysed in 0.03% SDS. The extracted glycogen was mixed with 1 mg carrier glycogen (Sigma-Aldrich) and boiled for 30 min, followed by precipitation with 98–99% ethanol. The precipitated glycogen was washed with 70% ethanol, centrifuged at 20,000×g for 15 min, and dried from the excess ethanol. The radioactivity of the glycogen was measured with liquid scintillation counting in Optiphase HiSafe 3 scintillation cocktail (PerkinElmer) and normalized to total cellular protein (BCA assay, Thermo Scientific).
Determination of glycolytic activity
To analyze glycolysis, ORP2-KO and Cas9 control cells were seeded on XF96 cell culture microplates in basal growth medium (Agilent Technologies, Santa Clara, CA, USA). After 24 h, the culture media was changed to Seahorse XF base medium (Agilent Tecnologies) with 2 mM l-glutamine (pH 7.35) and incubated for 1 h at 37 °C in a non-CO2 incubator. After incubation, extracellular acidification rate (ECAR) was measured in real time with a Seahorse XFe96 Analyzer (Agilent Technologies) as follows: cells were treated with 10 mM glucose, 1 μM oligomycin and 50 mM 2-deoxy-d-glucose (Sigma-Aldrich) followed by a 20 min measurement between the treatments. Extracellular acidification was normalized to total cellular protein. Glycolysis was calculated by subtracting the non-glycolytic ECAR from the maximum ECAR after glucose injection.
Analysis of cellular TG content and TG synthesis
Thin-layer chromatography used to analyze the TG content of HuH7 ORP2-KO and Cas9 control cells is described in [16, 22] and the radiometric method to analyze TG synthesis in [16].
Lipidome analysis
Methodology of the mass spectrometric lipidome analysis is described in the Supplementary Materials.
Transmission electron microscopy and quantification of LDs and ER–LD contacts
For TEM analysis, cells grown on glass coverslips were fixed, osmicated, dehydrated in a graded ethanol series and acetone, and infiltrated into Epon (TAAB, Aldermaston, UK) as described in [41]. 60-nm thick sections were cut parallel to the cover slip, and post-stained with uranyl acetate and lead citrate. Specimens were observed using Jeol JEM 1400 (Jeol Ltd., Tokyo, Japan) microscope equipped with Orius SC 1000B bottom mounted CCD camera (Gatan Inc., USA) at an acceleration voltage of 80 kV. For systematic random sampling, sixteen cells from each condition were selected at low magnification and four non-overlapping images were acquired from the cell cytoplasmic area at nominal magnification of 6000 x. To analyze LDs and ER–LD contact sites, all complete LDs and the adjacent ER in the TEM micrographs were manually traced using MIB software [4] and the quantity and the length of ER–LD contacts sites were analyzed using a custom made plugin in MIB. The probing distance of 150 nm between the ER and LDs was applied and the profiles closer than 30 nm were assigned as a contact site.
Statistics
In the IPA analysis, Fisher’s exact test was used to determine the significance of each pathway and the Benjamini–Hochberg method was used to control for false discovery rate. Statistical analysis of TLC data and cell biological assays was carried out using Student’s t test (two-tailed, independent).
Results
ORP2-KO interferes with Akt signaling
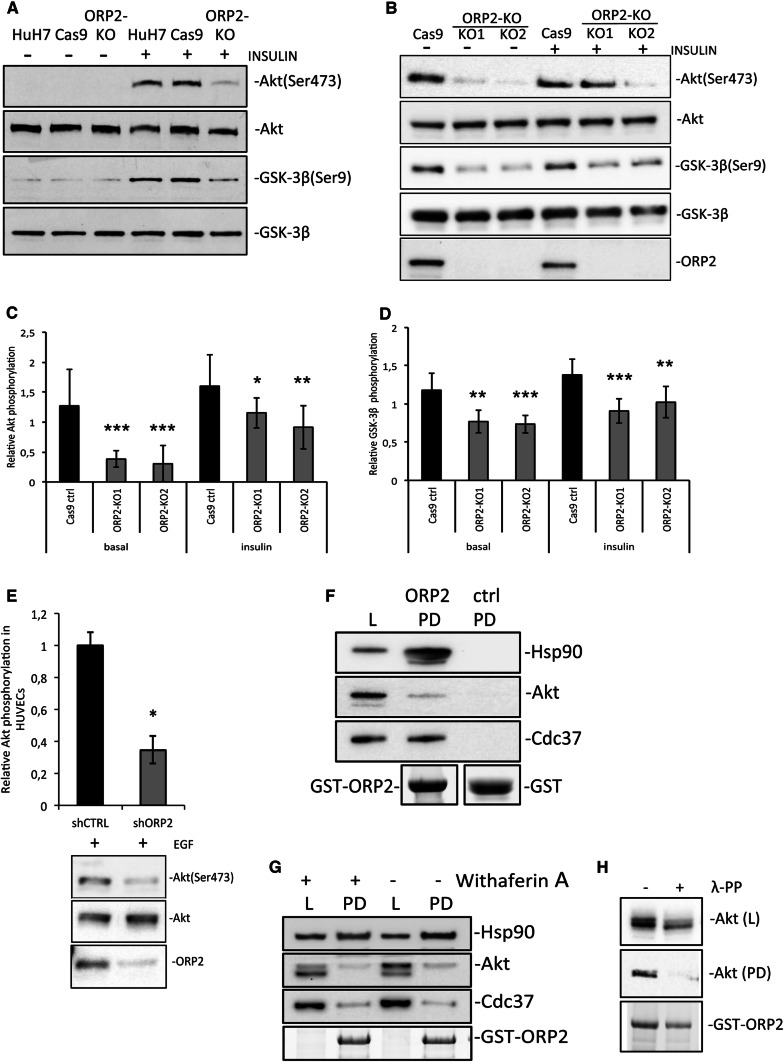
RNA sequencing analysis of HuH7 hepatoma cells subjected to ORP2 knockout by CRISPR-Cas9 gene editing [21] identified 20 components of the PI3K/Akt signaling pathway dysregulated in the ORP2-KO cells (Fig. 1a, b). Nine of the mRNAs were up-regulated (Fig. 1a, shown in green; Fig. 1b) and 11 down-regulated (Fig. 1a, shown in red; Fig. 1b). Ingenuity® pathway analysis of the mRNAs affected by ORP2-KO predicted PI3K/Akt signaling to be significantly inhibited (activation z score − 1.7, p value of overlapping genes 0.03). Prompted by these observations, the activity of the Akt signaling pathway in the two previously characterized ORP2-KO cell lines (ORP2-KO1 and -2; [21]) and Cas9 control cells was studied by western analysis with phosphor-Akt(Ser473) and glycogen synthase kinase 3β [GSK-3β(Ser9)] antibodies. The cells were either serum starved for 2 h (Fig. 2a) or grown in the basal culture medium (Fig. 2b), in the absence or presence of 8 min stimulation with 100 nM insulin, prior to lysis and sample preparation. The levels of Akt(Ser473) or GSK-3β(Ser9) were normalized to the total Akt and GSK-3β signals. The analysis revealed significant inhibition in the phosphorylation of Akt (Fig. 2c) and GSK-3β (Fig. 2d), under both basal and insulin-stimulated conditions (by 35–73 and 30–36%, respectively), indicating a suppression of PI3K/Akt signaling in the ORP2-KO cells.
Fig. 1.
ORP2-KO disturbs Akt signaling. a The Akt signaling components dysregulated in the transcriptome of ORP2-KO cells are shown in green (for up-regulation) or red (for down-regulation). b Relative mRNA expression of Akt signaling components in ORP2-KO versus control cells (values for Cas9 controls were set at 0) analyzed by RNA sequencing of three replicate cultures. *p < 0.05, **p < 0.01, ***p < 0.001 (after BH adjustment)
Fig. 2.
ORP2-KO inhibits Akt phosphorylation and ORP2 interacts with the Cdc37–Hsp90–Akt effector complex. a Western blot analysis of Akt(Ser473) and GSK-3β(Ser9) after 2 h serum starvation followed by an additional 8 min incubation with 100 nM insulin added in the starvation medium. Data are shown for native HuH7 cells, Cas9 control cells, and the ORP2 knockout cell line. b Western blot analysis of Akt(Ser473) and GSK-3β(Ser9) under basal culture conditions or after 8 min incubation with 100 nM insulin added in the basal medium. The ORP2 knockout cell lines are indicated as KO1, KO2, and the Cas9 controls as Cas9. c Quantification of Akt(Ser473) phosphorylation. The data represent four independent western blots of independent cultures similar to that shown in b. d Quantification of the relative GSK-3β(Ser9) phosphorylation from four independent western blots of independent cultures similar to that shown in b. e Western blot quantification of Akt(Ser473) phosphorylation in HUVECs silenced with ORP2-specific shRNA (shORP2) or shRNA control (shCTRL) followed by an epidermal growth factor stimulation [(EGF) 30 ng/ml for 10 min]. The data represent three independent western blots of independent cultures. f HuH7 cells were lysed and subjected to GST-ORP2 pull-down. Cell lysates (L) and pull-downs (PD) were analyzed by western blotting with anti-Cdc37, anti-Hsp90, and anti-Akt antibodies. Plain GST was used as a negative control. g GST-ORP2 pull-down of HuH7 cells treated with 5 µM Withaferin A. Cell lysates (L) and pull-downs (PD) were analyzed by western blotting with anti-Cdc37, anti-Hsp90, and anti-Akt antibodies. h GST-ORP2 pull-down from HuH7 cell lysates treated with lambda protein phosphatase (λ-PP). Cell lysates (L) and pull-downs (PD) were analyzed by western blotting with anti-Akt antibody. *p < 0.05, **p < 0.01, ***p < 0.001 (Student’s t test)
To confirm the effect of ORP2 depletion on Akt(Ser473) phosphorylation also in non-cancerous cells, we subjected primary human umbilical vein endothelial cells (HUVECs) to ORP2 knockdown with a lentivirus expressing ORP2-specific shRNA. In these cells, a similar dampening of Akt activation (− 65%) upon epidermal growth factor (EGF) stimulation as in the HuH7 model was observed (Fig. 2e). Furthermore, to assess whether the Akt phosphorylation defect is directly caused by the lack of ORP2, we overexpressed wild-type (wt) ORP2 or its mutants defective in binding the ER receptors, VAPs (mFFAT), or phosphoinositides (mPIP) in the basally cultured HUVECs with a lentiviral vector, followed by western analysis of Akt(Ser473) phosphorylation. In these experiments, the wt ORP2 but also both of the mutants were capable of enhancing the phosphorylation of Akt. The effect of ORP2(mFFAT) was weaker than of the wt and mPIP proteins, but this may be due to the lower expression level (or reduced stability) of the mFFAT protein (Supplemental Fig. S1A).
ORP2 interacts with the Akt effectors Cdc37 and Hsp90
We previously identified the ORP2 interactome in HuH7 cells by a co-immunoprecipitation approach [21]. Interestingly, the Akt effector heat shock protein 90 (Hsp90) co-chaperone Cdc37 was identified in this analysis as a specific partner of ORP2. We now validated the interaction of ORP2 with endogenous Cdc37 by pull-down assays using recombinant GST-ORP2 and lysates of HuH7 cells. The GST-ORP2 pull-downs contained abundant Cdc37, while the negative controls with plain GST lacked the signal (Fig. 2f), supporting a specific interaction of Cdc37 with ORP2. To clarify whether ORP2 is physically associated with the Akt signaling activating protein complex Hsp90–Cdc37–Akt, the pull-down fractions were probed with anti-Akt and anti-Hsp90 antibodies. Interestingly, the analysis revealed specific signals in the GST-ORP2 pull-downs for both Akt and Hsp90 (Fig. 2f), suggesting that ORP2 may interact with the Hsp90–Cdc37–Akt complex. Of note, the ORP2 interactome [21] contained, in addition to Cdc37, also other established Hsp90-associated proteins: FK506-binding protein 4 (FKBP52) [38] and nuclear autoantigen sperm protein (NASP) [1].
To further analyze the specificity of ORP2 interaction with the Hsp90–Cdc37–Akt complex, cells were treated with 5 μM Withaferin A for 24 h prior to the pull-down assays. Withaferin A is a steroidal lactone which has previously been shown to bind Hsp90, inhibiting its interaction with Cdc37 and hence its chaperone activity. Thereby, this drug reduces the phosphorylation and enhances the degradation of Akt [55]. In our pull-down assays, the Withaferin A treatment did not disrupt the interaction of ORP2 with Hsp90 or Cdc37 (Fig. 2g), bringing up the possibility that ORP2 may interact directly with both of these proteins. The Withaferin A treatment did, however, drastically reduce the phosphorylation of Akt, evident from an Akt mobility shift. We confirmed that this shift truly reflects loss of Akt phosphorylation by dephosphorylation of Akt with lambda protein phosphatase before the SDS-PAGE/western analysis (Fig. 2h). Moreover, GST-ORP2 was only able to pull down the lower mobility, hyperphosphorylated active form of Akt and the interaction was abolished upon dephosphorylation with lambda phosphatase (Fig. 2h), supporting a functional role of ORP2 in the protein complex that promotes Akt activation.
ORP2 co-localizes with Cdc37 and phosphorylated Akt at lamellipodia
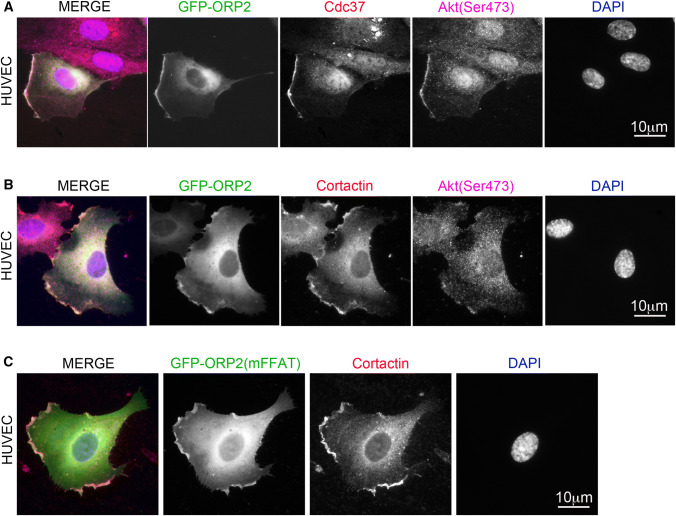
To visualize the distribution of ORP2, Cdc37, and active Akt, cells transfected with ORP2-GFP were after insulin stimulation stained for the endogenous Cdc37 and Akt(Ser473) (Supplemental Fig. S2). Since HuH7 cells are not particularly amenable to morphologic observation, these studies were also carried out in HUVECs (Fig. 3). Interestingly, ORP2 was found to co-localize with both Cdc37 and Akt(Ser473) at specific regions of the cell surface, which were by cortactin staining shown to represent actively expanding lamellipodia (Fig. 3a, b; Supplemental Fig. S2). Of note, we recently showed that a defect in lamellipodia formation is a hallmark phenotype of the ORP2-KO cells, resulting in decreased cell motility [21]. We have previously reported prominent localization of ORP2 at the surface of cytoplasmic lipid droplets or at ER domains that associate with LD surfaces [16, 22, 50]. The lamellipodial localization is easily overlooked in cells that display intense ORP2 staining at the LD. Moreover, in cells treated with the high-affinity oxysterol ligand of ORP2, 22(R)hydroxycholesterol [16, 22], or in ones that do not have large LDs, the emphasis of ORP2 localization is frequently seen at the cell cortex. To analyze whether the lamellipodial localization is dependent of interaction of ORP2 with VAPs, an ORP2 mutant incapable of VAP-binding (mFFAT) was overexpressed in HUVECs and the lamellipodia were visualized with cortactin staining. Similar to wt ORP2, the ORP2(mFFAT) co-localized with cortactin at lamellipodial cell surface protrusions, indicating that the interaction of ORP2 with the ER is not essential for its lamellipodial localization (Fig. 3c).
Fig. 3.
ORP2 co-localizes with Akt(Ser473) and Cdc37 at lamellipodia. a HUVECs transfected with GFP-ORP2, stimulated with 100 nM insulin for 2 min, and stained with Cdc37 [1:100 dilution (Thermo Scientific)] and Akt(Ser473) [1:25 dilution (Cell Signaling)] antibodies and DAPI. b HUVECs transfected with GFP-ORP2, stimulated with 100 nM insulin for 2 min, and stained with Akt(Ser473) and cortactin [10 μg/ml (Merck-Millipore)] antibodies and DAPI. c HUVECs transfected with GFP-ORP2(mFFAT), stimulated with 100 nM insulin for 2 min, and stained with cortactin antibody [10 μg/ml (Merck-Millipore)] and DAPI
ORP2 knockout reduces glucose uptake and glycogen synthesis
The Akt serine/threonine kinase plays a key role in the regulation of glucose homeostasis by elevating glucose uptake and increasing glycogen synthesis via phosphorylation of GSK-3 [9]. The results outlined in “ORP2-KO interferes with Akt signaling” demonstrated that the loss of ORP2 suppresses Akt signaling with possible effects on cellular carbohydrate metabolism. In addition, the ORP2-KO cells’ transcriptome and IPA analyses indicated a close link between ORP2 and carbohydrate metabolism (p value of overlapping genes 0.018). Therefore, the capacity of ORP2-KO cells to take up glucose was measured by incubating glucose- and serum-starved cells with [3H]-2-deoxy-d-glucose for 15 min, followed by measurement of the cellular radioactivity. The analysis demonstrated that the glucose uptake was reduced by 28–33% in the ORP2-KO cells as compared to the controls (Fig. 4a).
Fig. 4.
ORP2-KO inhibits glucose uptake, glycogen synthesis, and glycolysis. a Quantification of [3H]-2-deoxy-d-glucose uptake into ORP2-KO and Cas9 control cells during 15 min. Shown in a relative scale are averages of four independent experiments each performed as four replicates. b Quantification of d-[U-14C]-glucose incorporation into glycogen in ORP2-KO and control cells during 1 h. Shown in a relative scale are averages of three independent experiments each performed as three replicates. c Quantification of d-[U-14C]-glucose incorporation into glycogen during 1 h in ORP2-KO control cells infected with an empty lentiviral vector (ctrl) or a lentivirus overexpressing ORP2 (ORP2 oex). Shown are averages of two independent experiments both performed as five replicates. *p < 0.05, **p < 0.01, ***p < 0.001 (Student’s t test). d Schematic model of the glycolytic pathway and its enzymes down-regulated in the ORP2-KO cells. e Relative expression of glycolytic genes in ORP2-KO versus control cells (Cas9 controls were set at 1) analyzed by RNA sequencing of four replicate cultures. *p < 0.05, **p < 0.01, ***p < 0.001 (after BH adjustment). f Extracellular acidification rate (ECAR) after glucose, oligomycin and 2-deoxy-d-glucose (2-DG) injections of control and ORP2-KO cells measured with a Seahorse XFe96 analyzer in four independent measurements each as 12 replicates. g Calculation of the average glycolysis rate from the measurements shown in f by reducing the non-glycolytic acidification from the maximum ECAR before oligomycin injection; *p < 0.05, **p < 0.01, ***p < 0.001 (Student’s t test). PGK1 phosphoglycerate kinase 1, GAPDH glyceraldehyde-3-phosphate dehydrogenase, ALDOA fructose-bisphosphate aldolase A, ENO enolase, PKM pyruvate kinase, PGAM1 phosphoglycerate mutase 1, TPI1 triose phosphate isomerase 1, PFKFB 6-phosphofructo-2-kinase, GP1 glucose-6-phosphate isomerase
To further characterize the intracellular handling of glucose, the incorporation of d-[U-14C]-glucose into glycogen during 1 h was analyzed in ORP2-KO and control cells. The synthesis of glycogen in ORP2-KO cells was reduced by 50–58% as compared to the control (Fig. 4b), fully consistent with the observed reduction of GSK-3β(Ser9) phosphorylation. We next re-expressed ORP2 in the KO cells using a lentiviral vector to determine if this can rescue the defect in glycogen synthesis. The expression of ORP2 elevated glycogen synthesis in these cells by 37% as compared to the mock-transfected KO control (Fig. 4c), confirming that the synthesis defect is truly due to the loss of ORP2. To study whether the interactions of ORP2 with VAPs or phosphoinositides are necessary for this rescue effect, we also re-expressed the mFFAT and mPIP mutants of ORP2 in the KO cells, followed by analyses of glycogen synthesis. The results revealed that ORP2(mFFAT) rescued the glycogen synthesis, whereas the effect of ORP2(mPIP) varied between experiments, with no statistically significant overall rescue effect detectable (Supplemental Fig. S1B). These observations suggest that the binding of VAPs may be dispensable for the function of ORP2 in glucose metabolism, while the functional significance of PIP binding cannot be reliably inferred from the present data.
ORP2-KO inhibits glycolysis
The RNA sequencing demonstrated that the loss of ORP2 results in down-regulation of 12 major glycolytic genes, glycolysis being indicated as a significantly affected function by the IPA analysis of the ORP2-KO cells (p value of overlapping genes 0.003; Fig. 4d, e). The glycolytic capacity of ORP2-KO and control cells was, therefore, measured with a Seahorse® XFe96 extracellular flux analyser by monitoring their extracellular acidification rate in the presence of glucose substrate (Fig. 4f). Consistent with the suppression of major glycolytic genes, the rate of glycolysis was in the ORP2-KO cells reduced by 22–37% compared to the controls (Fig. 4g).
Knockout of ORP2 reduces the cellular triacylglycerol content
Prompted by our earlier observations on impacts of ORP2 knockdown on cellular TG metabolism [16, 50], we investigated the TG content of ORP2-KO and Cas9 control cells under basal culture conditions by thin-layer chromatography. The analysis revealed a significant, 21–28% reduction of the cellular TGs in the ORP2-KO cells compared to the controls (Fig. 5a). The capacity of ORP2-KO and control cells to synthesize TGs was measured by [3H]-oleic acid labeling during a 3 h period. TG synthesis by the KO cells was reduced by 41–53% as compared to the controls (Fig. 5b). Of note, fatty acid uptake measured by incubating the cells with [3H]-palmitic acid did not differ between the ORP2-KO and the controls (Fig. 5c), consistent with a defect in the activation or incorporation of fatty acids into TGs. We next re-expressed ORP2 in the KO cells with a lentiviral vector and measured the effect on TG synthesis as above. The re-expression of ORP2 increased the TG synthesis by 31% compared to the mock-transfected KO control (Fig. 5d), demonstrating that the TG synthesis defect results from the loss of ORP2. However, no significant difference between the TG synthesis rescue with the wt ORP2 and ORP2 mutant mFFAT and mPIP was detected (Supplemental Fig. S1C), suggesting that the ORP2 TG metabolic function is not directly dependent on ORP2′s ability to bind VAP or PIPs.
Fig. 5.
ORP2 knockout reduces the TG content and modifies the expression of TG metabolic genes of hepatoma cells. a ORP2-KO and control cells’ TG content was assessed by thin-layer chromatography. Shown is mean of five replicate cultures. b TG synthesis was measured in ORP2-KO and control cells by [3H]-oleic acid labeling for 3 h. Shown is mean of two independent experiments each performed as four replicates. c Quantification of [3H]-palmitic acid uptake into ORP2-KO and control cells during 15 min. Shown in a relative scale are averages of two independent experiments each performed as four replicates. d Quantification of TG synthesis with [3H]-oleic acid labeling for 3 h in ORP2-KO control cells infected with an empty lentiviral vector (ctrl) or a lentivirus overexpressing ORP2 (ORP2 oex). Shown are averages of two independent experiments both performed as five replicates. *p < 0.05, **p < 0.01, ***p < 0.001 (Student’s t test). e Relative mRNA expression of SREBP-1 and its target genes in ORP2-KO versus control cells (values for Cas9 controls were set at 1) analyzed by RNA sequencing of four replicate cultures. f Relative gene expression of TG metabolic genes in ORP2-KO cells versus control cells (Cas9 controls were set at 1) analyzed by RNA sequencing of four replicate cultures. The statistical analysis of the RNA sequencing data is specified in the “Experimental procedures”, section ‘RNA sequencing and pathway analysis’. *p < 0.05, **p < 0.01, ***p < 0.001 (after BH adjustment)
ORP2 knockout modulates the expression of SREBP-1 and TG metabolic genes
The cellular TGs are retained within intracellular lipid droplets (LD), which store energy in the form of neutral lipids. The balance between TG synthesis and hydrolysis is coordinated to respond to the fluctuating energy demands of the cell. One master regulator of TG homeostasis is the transcription factor Sterol regulatory-element-binding protein-1, SREBP-1 [14]. To obtain clues of the mechanisms underlying the phenotypic effects of ORP2-KO, we again analyzed the transcriptome data of these cells [21].
When the mRNAs dysregulated in the ORP2-KO cells were compared to known SREBP-1 target genes [43], 27 established SREBP-1 targets were found to be significantly affected by the ORP2-KO (Fig. 5e). Moreover, we compared the mRNAs affected by ORP2-KO with the list of gene promoters occupied by SREBP-1 in a global chromatin immunoprecipitation analysis [43]. This comparison revealed 168 genes the promoters of which are occupied by SREBP-1 and which are affected by the loss of ORP2 (Supplemental Table S1). IPA analysis of the mRNAs affected by ORP2-KO predicted SREBP-1 transcriptional regulation to be significantly inhibited (activation z score − 2.2, p value of overlapping genes 0.0001). This suggests that the observed reduction in the TG content of ORP2-KO cells is at least in part due to a suppression of SREBP-1 function. In addition to the SREBP-1 targets, we identified among the genes affected by ORP2-KO 11 other mRNAs with established roles in TG metabolism (Fig. 5f). The above results imply that the depletion of cellular TGs by ORP2-KO involves disturbed transcriptional regulation of multiple TG homeostatic genes.
ORP2-KO does not reduce the quantity of ER–LD contacts, but alters their dynamics upon fatty acyl loading
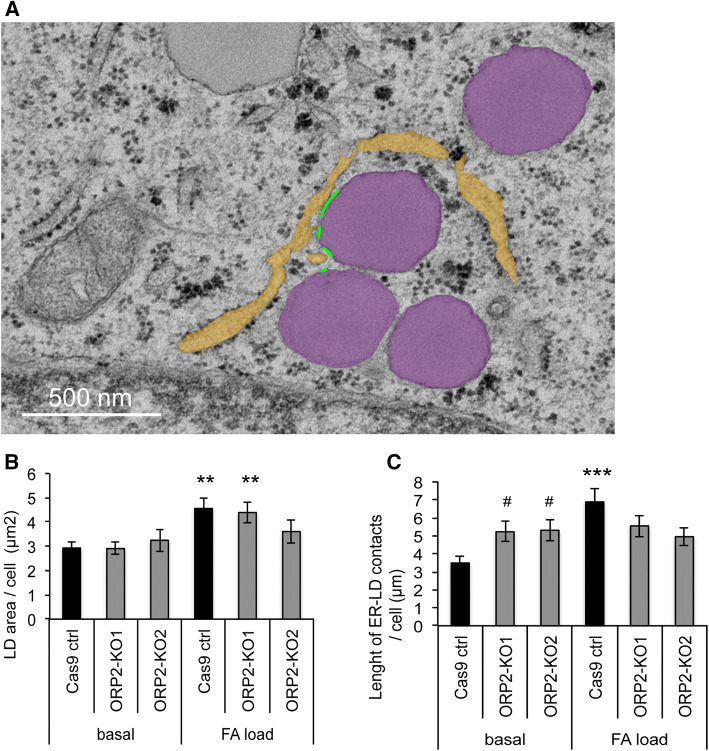
ORP2 was previously found to localize on the surface of intracellular LDs or at ER–LD membrane contact sites [16, 22]. To investigate whether the loss of ORP2 affects the extent of ER–LD contacts, we analyzed by TEM the ORP2-KO cell lines and Cas9 control cells grown under basal conditions or after 3 h fatty acid loading (Fig. 6a). Upon fatty acid loading, the total LD area increased significantly in Cas9 control and ORP2-KO1, the increase being minor and insignificant in ORP2-KO2 (Fig. 6b). However, the total length of ER–LD contacts was only significantly increased in Cas9 control cells upon FA loading, but did not change in ORP2-KO cells. Of note, the length of ER–LD contacts was significantly elevated in ORP2-KO cells under basal conditions compared to Cas9 controls (Fig. 6c). These observations demonstrated that ORP2 is not essential for the formation of ER–LD contacts nor the growth of LD upon FA loading, but may modulate the dynamic expansion of the ER–LD contacts under conditions in which TGs are actively synthesized.
Fig. 6.
ORP2-KO does not reduce ER–LD contact sites, but affects their dynamic expansion after fatty acid (FA) loading. ORP2-KO and control cells were either grown in basal culture medium or loaded for 3 h with FA as described in “Experimental procedures”. a TEM micrograph from KO2 loaded sample showing modeled LDs (depicted in purple) and the ER (depicted in yellow) that is in close vicinity to intact LDs. The ER–LD contact sites that are within 30 nm distance are shown with green lines. Scale bar 500 nm. b Total LD area per cell. c Total length of ER–LD contacts per cell. The data represent mean ± SEM from 16–32 cells, with four images from each analyzed; */#p < 0.05, **/##p < 0.01, ***/###p < 0.001 (Student’s t test); asterisk indicates significant difference between unloaded and loaded cells, and hash symbol indicates significant difference between Cas9 ctrl and ORP2-KO cells
Based on the RNA sequencing analysis, we found no evidence that ORP2-KO would affect the expression of the previously identified LD–ER tethering factors such as insulin-sensitive fatty acid transport protein 1 (FATP1/SLC25A1) [53], diacylglycerol O-acyltransferase 2 (DGAT2) [53], Seipin/BSCL2 [44], or monoacylglycerol acyltransferase 1 (MGAT1) [25]. The RNA sequencing analysis did, however, reveal that ORP2-KO results in the suppression of mRNAs encoding components important for LD formation or growth: perilipin 2 (− 28%; adjusted p = 6.05E−16) [5] and glycerol-3-phosphate acyltransferase 4 (GPAT4; − 13%; adjusted p = 5.8E−5) [51].
Knockout of ORP2 has no significant impact on the quantities of major membrane phospholipids and sterols
Previous research suggests a functional role of ORP2 in cellular neutral lipid and sterol homeostasis [12, 15, 16, 24]. These studies were performed by employing ORP2 overexpression or partial knockdown. Here, we analyzed lipid classes and their molecular species in HuH7 cells with a complete knockout of ORP2, grown in serum-containing basal medium. Of note, the mass spectrometry platform did not cover TGs, which were analyzed by TLC. In addition, the cellular sterol content was manipulated by lipoprotein starvation, mevastatin, or LDL treatments.
We first analyzed ORP2-KO and control hepatocytes’ content of glycerophospholipids [phosphatidylcholines (PCs), lyso-PCs (LPCs), phosphatidylethanolamines (PEs), PE-plasmalogens (PE-Ps), phosphatidylinositols (PIs), -serines (PSs), and -glycerols (PGs)], ceramides, sphingomyelin, free (FC), and esterified cholesterol (CEs), as well as five oxysterols (25OHC, 27OHC, 7αOHC, 7βOHC, and 7KC). The analysis did not reveal significant alterations in any of the analyzed lipid classes nor the oxysterol species in the ORP2-KO cells as compared to the controls (Supplemental Fig. S3A). Neither did the cellular sterol manipulations reveal possible latent effects on the cellular concentrations of FC or CEs, which might have remained undetected under the basal conditions (Supplemental Fig. S3B). Moreover, clustering analysis grouped the molecular species into nine distinct lipid clusters, none of which differed significantly between ORP2-KO and Cas9 cells. However, RNA sequencing of the ORP2-KO cells revealed significant alterations in the mRNA expression of 11 established cholesterol homeostatic genes (Supplemental Table S2), bringing up the possibility that the cells chronically devoid of ORP2 may have adapted to a putative disturbance of cholesterol homeostasis via transcriptional changes. Upon analysis of individual lipid molecular species, very few differences were detectable between the ORP2-KO and control cells, and none of these remained significant after multiple test correction (Supplemental Table S3).
Discussion
We employ in this study a complete CRISPR-Cas9 knockout of ORP2 HuH7 cells to comprehensively analyze the impacts of the loss of ORP2 function. The knockout resulted in reductions of Akt activity, glucose uptake, glycogen synthesis, glycolysis, mRNAs encoding glycolytic enzymes, and SREBP-1 target genes, as well as defective TG synthesis and storage. The defect in Akt activity was observed in both the ORP2-KO HuH7 hepatoma cells and in primary endothelial cells subjected to ORP2 knockdown. ORP2 was found to form a physical complex with key controllers of Akt activity, Cdc37, and Hsp90, and to co-localize with Cdc37 and active Akt(Ser473) at the lamellipodia of both HuH7 cells and the non-cancerous HUVECs. Of note, although ORP2–VAPA complexes are reported to localize at ER–LD contacts [50], our quantitative EM analysis demonstrated that loss of ORP2 did not abolish or reduce the ER–LD membrane contact sites; under the basal culture conditions even a modest increase of the total length and number of such contacts was observed in the KO cells. Interestingly, these parameters increased significantly in the Cas9 controls upon fatty acid loading, while no significant increase occurred in the KO cells, suggesting a functional role of ORP2 or its interaction partners in the dynamic regulation of ER–LD contacts. The knockout did not significantly affect the cellular levels of membrane phospholipids, cholesterol or oxysterols.
In the present study, the discovery of ORP2 function in Akt signaling is of central importance. Our results show that ORP2 is physically associated with an Akt activation complex that contains Cdc37 and Hsp90 [3, 45]. Moreover, we demonstrate that GST-ORP2 selectively pulls down the active, hyperphosphorylated form of Akt, and that the loss of ORP2 inhibits Akt signaling, consistent with the notion that ORP2 plays an important role in Akt activation. In this context, one should also consider our earlier observation that ORP2 interacts in vitro with vesicles containing phosphoinositides, particularly PI(3,4,5)P3 [15], which plays a key role Akt activation, and with IQGAP1 [21], which is suggested to play an important role in PI(3,4,5)P3 formation by scaffolding phosphoinositide kinases as well as PDK1 and Akt into functional proximity [7]. Importantly, our results demonstrate that ORP2-KO inhibits glucose uptake, glycogen synthesis, and glycolysis effects that most likely result from the suppression of Akt signaling in the KO cells. Akt signaling regulates glucose uptake into hepatocytes by increasing the expression and plasma membrane localization of GLUT1 [2, 23], the mRNA of which was significantly suppressed in the ORP2-KO cells (by 30%; adjusted p value 3.3E−18). Akt also regulates glucose uptake indirectly by promoting a concentration gradient: It induces the expression of glucokinase [18], which reduces the intracellular glucose concentration by phosphorylation of glucose. Thus, we envision that the observed impairment of glucose uptake in the ORP2-KO cells is at least in part explained by the suppression of Akt signaling. Moreover, by fold-change, the most up-regulated individual mRNA in ORP2-KO cells was thioredoxin-interacting protein [21], which suppresses the function of GLUT1 [52].
Inhibition of glycogen synthesis observed in the ORP2-KO cells is consistent with the reduced Akt-mediated phosphorylation of GSK-3. Glycogen synthase is regulated by reversible phosphorylation by GSK-3, which itself is phosphorylated and inactivated by Akt [9]. Importantly, activation of Akt promotes the conversion of cellular energy metabolism from oxidative phosphorylation to glycolysis by increasing the expression of glycolytic enzymes [46]. Consistently, the reduction of active Akt in the ORP2-KO cells coincided in the present study with a down-regulation of the mRNAs encoding all major glycolytic enzymes and glycolysis measured as ECAR. Rescue of glycogen synthesis in the ORP2-KO cells by ORP2(mFFAT) suggested that, at least under the overexpression conditions, linking to the ER via VAPs is not required for the function of ORP2 in glucose metabolism. Consistently, the mFFAT mutant defective in VAP-binding localized, similar to the wt ORP2, to the cell cortex where key regulation of Akt activity takes place (see below), and its overexpression enhanced Akt phosphorylation at Ser473. Similar to mFFAT, the PIP binding deficient mutant ORP2(mPIP) did enhance Akt phosphorylation, suggesting that PIP binding by ORP2 is not absolutely necessary for its function in the regulation of Akt, even though we could not obtain conclusive results on the ability of this mutant to rescue glycogen synthesis in the KO cells.
Our latest work suggested an important role of ORP2 in actin-dependent cell adhesion and migration [21], processes tightly connected with glycolysis: glycolytic enzymes such as GAPDH and aldolase associate with F-actin, and the acute energy requirements in motile cell protrusions and filopodia are met by rapid local ATP synthesis via glycolysis [10, 33, 36]. On the other hand, PI(3,4,5)P3 and Akt orchestrate lamellipodia formation and cell migration by inducing actin polymerization at the leading edge of cells. PI(3,4,5)P3 and active Akt are found localizing in lamellipodia, and PI3K/Akt inhibitors disrupt lamellipodia formation and reduce cell motility [6, 20, 49]. Here, we show that ORP2 co-localizes with active Akt(Ser473) and Cdc37 at cortactin-positive lamellipodia, in agreement with a functional interplay of these proteins in cell migration. Consistently, we recently demonstrated that the ORP2-KO cells are defective in lamellipodia formation and migration [21]. Thus, the present observations suggest a new, ORP-mediated regulatory link between cellular energy metabolism and motility.
How does the major lipid phenotype of the ORP2-KO cells, the reduction of TGs, relate to the defective glucose metabolism and Akt signaling? Obviously, reduced glucose uptake and glycolysis are likely to be reflected in a reduced supply of precursors for de novo lipogenesis via the tricarboxylic acid cycle. However, the defective Akt signaling may contribute to the reduction of TG synthesis and storage also via another mechanism. Akt plays an important role in hepatic lipogenesis by enhancing transcription and nuclear localization of the lipogenic transcription factor SREBP-1c [13, 39, 40, 54]. Thus, we envision that the present TG and glucose phenotypes may arise as a combined effect of Akt and SREBP-1 defects, and the Akt defect may be one factor contributing to the disturbance of SREBP-1 target gene expression. A model of ORP2 function in Akt signaling arising from the present results is presented in Fig. 7.
Fig. 7.
A schematic model of ORP2 function in Akt signaling. Akt signaling is activated by extracellular growth factors (e.g., insulin), which binds to plasma membrane (PM) growth factor receptors (GFR) resulting in activation of intracellular PI3-kinase (PI3K). PI3K phosphorylates PIP2 to PIP3, triggering a PM recruitment of phosphoinositide-dependent kinase-1 (PDK1) and Akt. The protein kinases (PI3K/PDK1/Akt) are scaffolded by IQGAP1 which interacts with ORP2. At PM, Akt interacts with its effector complex Hsp90–Cdc37–ORP2 and is phosphorylated by PDK1. Active Akt regulates the activity of several downstream target molecules (e.g., GSK3), resulting in increased uptake of glucose, glycogen synthesis, glycolysis, lipogenesis via SREBP-1, and migration of cells
ORP2 was previously implicated in TG metabolism at ER–LD contacts [50], cholesterol transport [15, 19, 24], adrenocortical steroidogenesis [12], and actin-dependent cell dynamics [21]. The present analysis is consistent with a function of the protein as a regulator TG synthesis and storage. Rescue of the TG synthesis defect by ORP2(mFFAT) and ORP2(mPIP) suggests that, at least under the overexpression conditions, the interactions with VAPs and PIPs are not necessary for the function of ORP2 in TG metabolism. The reported activities of ORP2 in cellular cholesterol homeostasis are not directly supported by the present data. We have previously shown that the loss of ORP2 does not cause significant compensatory changes in the mRNA levels of the ten other ORPs expressed in HuH7 cells or their ER receptors, the VAMP-associated proteins A and B [21]. However, the present RNA sequencing analysis suggests that putative distortions of cholesterol homeostasis caused by the chronic loss of ORP2 may have been compensated for by altered mRNA expression of a number of cholesterol homeostatic genes.
To conclude, the present study, together with our recently published work, identifies ORP2 as a new regulatory nexus of Akt signaling, cellular energy metabolism, actin cytoskeletal function, cell migration, and proliferation.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary material 1 (TIFF 25519 kb)
Supplementary material 2 (TIFF 25523 kb)
Supplementary material 3 (TIFF 25520 kb)
Acknowledgements
We thank Riikka Kosonen and Mervi Lindman for expert technical assistance, and Adj. Prof. Reijo Käkelä (Department of Biosciences, University of Helsinki) for valuable comments on the manuscript. Personnel of the Genome Biology Unit (Biocenter Finland) and the Biomedicum Functional Genomics Unit (Helsinki Institute of Life Science, HiLIFE) are acknowledged for help in generating the recombinant lentiviruses. Prof. Feng Zhang (Broad Institute of Massachusetts Institute of Technology and Harvard, Cambridge, MA, USA) is thanked for kindly providing components of the CRISPR-Cas9 vector system.
Abbreviations
- CRISPR
Clustered regularly interspaced short palindromic repeats
- ECAR
Extracellular acidification rate
- EM
Electron microscopy
- ER
Endoplasmic reticulum
- FA
Fatty acid
- GSK
Glycogen synthase kinase
- HUVEC
Human umbilical vein endothelial cell
- IPA
Ingenuity® pathway analysis
- KO
Knockout
- LD
Lipid droplet
- LDL
Low-density lipoprotein
- OSBP
Oxysterol-binding protein
- ORP
OSBP-related protein
- PIP
Phosphatidylinositol phosphate
- TEM
Transmission electron microscopy
- TG
Triacylglycerol
Funding
This study was supported by grants from the Academy of Finland (285223 to V.M.O., 307415 and 312491 to E.I.), the University of Helsinki Doctoral Programme in Biomedicine (H.K.), the Finnish Concordia Fund (H.K.), the Ida Montin Foundation (H.K), the Finnish-Norwegian Medical Foundation (H.K.), the Aarne Koskelo Foundation (H.K.), the Orion Research Foundation sr (H.K.), the Päivikki and Sakari Sohlberg Foundation (H.K.), the Sigrid Juselius Foundation, the Magnus Ehrnrooth Foundation, and the Finnish Foundation for Cardiovascular Research (V.M.O.). Electron Microscopy Unit is supported by Biocenter Finland and Helsinki Institute of Life Science. The funding bodies played no role in the study design, analysis or interpretation of the data, writing of the report or the decision to submit the article for publication.
Compliance with ethical standards
Conflict of interest
The authors declare no conflict of interest.
References
- 1.Alekseev OM, Widgren EE, Richardson RT, O’Rand MG. Association of NASP with HSP90 in mouse spermatogenic cells: stimulation of ATPase activity and transport of linker histones into nuclei. J Biol Chem. 2005;280:2904–2911. doi: 10.1074/jbc.M410397200. [DOI] [PubMed] [Google Scholar]
- 2.Barthel A, Okino ST, Liao J, Nakatani K, Li J, Whitlock JP, Jr, Roth RA. Regulation of GLUT1 gene transcription by the serine/threonine kinase Akt1. J Biol Chem. 1999;274:20281–20286. doi: 10.1074/jbc.274.29.20281. [DOI] [PubMed] [Google Scholar]
- 3.Basso AD, Solit DB, Chiosis G, Giri B, Tsichlis P, Rosen N. Akt forms an intracellular complex with heat shock protein 90 (Hsp90) and Cdc37 and is destabilized by inhibitors of Hsp90 function. J Biol Chem. 2002;277:39858–39866. doi: 10.1074/jbc.M206322200. [DOI] [PubMed] [Google Scholar]
- 4.Belevich I, Joensuu M, Kumar D, Vihinen H, Jokitalo E. Microscopy image browser: a platform for segmentation and analysis of multidimensional datasets. PLoS Biol. 2016;14:e1002340. doi: 10.1371/journal.pbio.1002340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bickel PE, Tansey JT, Welte MA. PAT proteins, an ancient family of lipid droplet proteins that regulate cellular lipid stores. Biochim Biophys Acta. 2009;1791:419–440. doi: 10.1016/j.bbalip.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chanvorachote P, Chunhacha P, Pongrakhananon V. Caveolin-1 induces lamellipodia formation via an Akt-dependent pathway. Cancer Cell Int. 2014;14:52. doi: 10.1186/1475-2867-14-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choi S, Hedman AC, Sayedyahossein S, Thapa N, Sacks DB, Anderson RA. Agonist-stimulated phosphatidylinositol-3,4,5-trisphosphate generation by scaffolded phosphoinositide kinases. Nat Cell Biol. 2016;18:1324–1335. doi: 10.1038/ncb3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chung J, Torta F, Masai K, Lucast L, Czapla H, Tanner LB, Narayanaswamy P, Wenk MR, Nakatsu F, De Camilli P. INTRACELLULAR TRANSPORT. PI4P/phosphatidylserine countertransport at ORP5- and ORP8-mediated ER-plasma membrane contacts. Science. 2015;349:428–432. doi: 10.1126/science.aab1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cross DA, Alessi DR, Cohen P, Andjelkovich M, Hemmings BA. Inhibition of glycogen synthase kinase-3 by insulin mediated by protein kinase B. Nature. 1995;378:785–789. doi: 10.1038/378785a0. [DOI] [PubMed] [Google Scholar]
- 10.De Bock K, Georgiadou M, Carmeliet P. Role of endothelial cell metabolism in vessel sprouting. Cell Metab. 2013;18:634–647. doi: 10.1016/j.cmet.2013.08.001. [DOI] [PubMed] [Google Scholar]
- 11.de Saint-Jean M, Delfosse V, Douguet D, Chicanne G, Payrastre B, Bourguet W, Antonny B, Drin G. Osh4p exchanges sterols for phosphatidylinositol 4-phosphate between lipid bilayers. J Cell Biol. 2011;195:965–978. doi: 10.1083/jcb.201104062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Escajadillo T, Wang H, Li L, Li D, Sewer MB. Oxysterol-related-binding-protein related protein-2 (ORP2) regulates cortisol biosynthesis and cholesterol homeostasis. Mol Cell Endocrinol. 2016;427:73–85. doi: 10.1016/j.mce.2016.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fleischmann M, Iynedjian PB. Regulation of sterol regulatory-element-binding protein 1 gene expression in liver: role of insulin and protein kinase B/cAkt. Biochem J. 2000;349:13–17. doi: 10.1042/bj3490013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Horton JD, Shah NA, Warrington JA, Anderson NN, Park SW, Brown MS, Goldstein JL. Combined analysis of oligonucleotide microarray data from transgenic and knockout mice identifies direct SREBP target genes. Proc Natl Acad Sci USA. 2003;100:12027–12032. doi: 10.1073/pnas.1534923100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hynynen R, Laitinen S, Käkelä R, Tanhuanpää K, Lusa S, Ehnholm C, Somerharju P, Ikonen E, Olkkonen VM. Overexpression of OSBP-related protein 2 (ORP2) induces changes in cellular cholesterol metabolism and enhances endocytosis. Biochem J. 2005;390:273–283. doi: 10.1042/BJ20042082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hynynen R, Suchanek M, Spandl J, Back N, Thiele C, Olkkonen VM. OSBP-related protein 2 is a sterol receptor on lipid droplets that regulates the metabolism of neutral lipids. J Lipid Res. 2009;50:1305–1315. doi: 10.1194/jlr.M800661-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Im YJ, Raychaudhuri S, Prinz WA, Hurley JH. Structural mechanism for sterol sensing and transport by OSBP-related proteins. Nature. 2005;437:154–158. doi: 10.1038/nature03923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iynedjian PB. Molecular physiology of mammalian glucokinase. Cell Mol Life Sci. 2009;66:27–42. doi: 10.1007/s00018-008-8322-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jansen M, Ohsaki Y, Rita Rega L, Bittman R, Olkkonen VM, Ikonen E. Role of ORPs in sterol transport from plasma membrane to ER and lipid droplets in mammalian cells. Traffic. 2011;12:218–231. doi: 10.1111/j.1600-0854.2010.01142.x. [DOI] [PubMed] [Google Scholar]
- 20.Kakinuma N, Roy BC, Zhu Y, Wang Y, Kiyama R. Kank regulates RhoA-dependent formation of actin stress fibers and cell migration via 14-3-3 in PI3K-Akt signaling. J Cell Biol. 2008;181:537–549. doi: 10.1083/jcb.200707022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kentala H, Koponen A, Kivelä AM, Andrews R, Li C, Zhou Y, Olkkonen VM. Analysis of ORP2-knockout hepatocytes uncovers a novel function in actin cytoskeletal regulation. FASEB J. 2018;32:1281–1295. doi: 10.1096/fj.201700604R. [DOI] [PubMed] [Google Scholar]
- 22.Kentala H, Pfisterer SG, Olkkonen VM, Weber-Boyvat M. Sterol liganding of OSBP-related proteins (ORPs) regulates the subcellular distribution of ORP-VAPA complexes and their impacts on organelle structure. Steroids. 2015;99:248–258. doi: 10.1016/j.steroids.2015.01.027. [DOI] [PubMed] [Google Scholar]
- 23.Kohn AD, Summers SA, Birnbaum MJ, Roth RA. Expression of a constitutively active Akt Ser/Thr kinase in 3T3-L1 adipocytes stimulates glucose uptake and glucose transporter 4 translocation. J Biol Chem. 1996;271:31372–31378. doi: 10.1074/jbc.271.49.31372. [DOI] [PubMed] [Google Scholar]
- 24.Laitinen S, Lehto M, Lehtonen S, Hyvärinen K, Heino S, Lehtonen E, Ehnholm C, Ikonen E, Olkkonen VM. ORP2, a homolog of oxysterol binding protein, regulates cellular cholesterol metabolism. J Lipid Res. 2002;43:245–255. [PubMed] [Google Scholar]
- 25.Lee YJ, Kim JW. Monoacylglycerol O-acyltransferase 1 (MGAT1) localizes to the ER and lipid droplets promoting triacylglycerol synthesis. BMB Rep. 2017;50:367–372. doi: 10.5483/BMBRep.2017.50.7.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 28.Loewen CJ, Roy A, Levine TP. A conserved ER targeting motif in three families of lipid binding proteins and in Opi1p binds VAP. EMBO J. 2003;22:2025–2035. doi: 10.1093/emboj/cdg201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maeda K, Anand K, Chiapparino A, Kumar A, Poletto M, Kaksonen M, Gavin AC. Interactome map uncovers phosphatidylserine transport by oxysterol-binding proteins. Nature. 2013;501:257–261. doi: 10.1038/nature12430. [DOI] [PubMed] [Google Scholar]
- 31.Mesmin B, Bigay J, Moser von Filseck J, Lacas-Gervais S, Drin G, Antonny B. A four-step cycle driven by PI(4)P hydrolysis directs sterol/PI(4)P exchange by the ER-Golgi tether OSBP. Cell. 2013;155:830–843. doi: 10.1016/j.cell.2013.09.056. [DOI] [PubMed] [Google Scholar]
- 32.Mesmin B, Bigay J, Polidori J, Jamecna D, Lacas-Gervais S, Antonny B. Sterol transfer, PI4P consumption, and control of membrane lipid order by endogenous OSBP. EMBO J. 2017 doi: 10.15252/embj.201796687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Minaschek G, Groschel-Stewart U, Blum S, Bereiter-Hahn J. Microcompartmentation of glycolytic enzymes in cultured cells. Eur J Cell Biol. 1992;58:418–428. [PubMed] [Google Scholar]
- 34.Moser von Filseck J, Copic A, Delfosse V, Vanni S, Jackson CL, Bourguet W, Drin G. INTRACELLULAR TRANSPORT. Phosphatidylserine transport by ORP/Osh proteins is driven by phosphatidylinositol 4-phosphate. Science. 2015;349:432–436. doi: 10.1126/science.aab1346. [DOI] [PubMed] [Google Scholar]
- 35.Nakabayashi H, Taketa K, Miyano K, Yamane T, Sato J. Growth of human hepatoma cells lines with differentiated functions in chemically defined medium. Cancer Res. 1982;42:3858–3863. [PubMed] [Google Scholar]
- 36.Nguyen TN, Wang HJ, Zalzal S, Nanci A, Nabi IR. Purification and characterization of beta-actin-rich tumor cell pseudopodia: role of glycolysis. Exp Cell Res. 2000;258:171–183. doi: 10.1006/excr.2000.4929. [DOI] [PubMed] [Google Scholar]
- 37.Olkkonen VM, Li S. Oxysterol-binding proteins: sterol and phosphoinositide sensors coordinating transport, signaling and metabolism. Prog Lipid Res. 2013;52:529–538. doi: 10.1016/j.plipres.2013.06.004. [DOI] [PubMed] [Google Scholar]
- 38.Peattie DA, Harding MW, Fleming MA, DeCenzo MT, Lippke JA, Livingston DJ, Benasutti M. Expression and characterization of human FKBP52, an immunophilin that associates with the 90-kDa heat shock protein and is a component of steroid receptor complexes. Proc Natl Acad Sci USA. 1992;89:10974–10978. doi: 10.1073/pnas.89.22.10974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Porstmann T, Griffiths B, Chung YL, Delpuech O, Griffiths JR, Downward J, Schulze A. PKB/Akt induces transcription of enzymes involved in cholesterol and fatty acid biosynthesis via activation of SREBP. Oncogene. 2005;24:6465–6481. doi: 10.1038/sj.onc.1208802. [DOI] [PubMed] [Google Scholar]
- 40.Porstmann T, Santos CR, Griffiths B, Cully M, Wu M, Leevers S, Griffiths JR, Chung YL, Schulze A. SREBP activity is regulated by mTORC1 and contributes to Akt-dependent cell growth. Cell Metab. 2008;8:224–236. doi: 10.1016/j.cmet.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Puhka M, Vihinen H, Joensuu M, Jokitalo E. Endoplasmic reticulum remains continuous and undergoes sheet-to-tubule transformation during cell division in mammalian cells. J Cell Biol. 2007;179:895–909. doi: 10.1083/jcb.200705112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reed BD, Charos AE, Szekely AM, Weissman SM, Snyder M. Genome-wide occupancy of SREBP1 and its partners NFY and SP1 reveals novel functional roles and combinatorial regulation of distinct classes of genes. PLoS Genet. 2008;4:e1000133. doi: 10.1371/journal.pgen.1000133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Salo VT, Belevich I, Li S, Karhinen L, Vihinen H, Vigouroux C, Magre J, Thiele C, Holtta-Vuori M, Jokitalo E, Ikonen E. Seipin regulates ER-lipid droplet contacts and cargo delivery. EMBO J. 2016;35:2699–2716. doi: 10.15252/embj.201695170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sato S, Fujita N, Tsuruo T. Modulation of Akt kinase activity by binding to Hsp90. Proc Natl Acad Sci USA. 2000;97:10832–10837. doi: 10.1073/pnas.170276797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Semenza GL, Jiang BH, Leung SW, Passantino R, Concordet JP, Maire P, Giallongo A. Hypoxia response elements in the aldolase A, enolase 1, and lactate dehydrogenase A gene promoters contain essential binding sites for hypoxia-inducible factor 1. J Biol Chem. 1996;271:32529–32537. doi: 10.1074/jbc.271.51.32529. [DOI] [PubMed] [Google Scholar]
- 47.Suchanek M, Hynynen R, Wohlfahrt G, Lehto M, Johansson M, Saarinen H, Radzikowska A, Thiele C, Olkkonen VM. The mammalian oxysterol-binding protein-related proteins (ORPs) bind 25-hydroxycholesterol in an evolutionarily conserved pocket. Biochem J. 2007;405:473–480. doi: 10.1042/BJ20070176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tong J, Yang H, Yang H, Eom SH, Im YJ. Structure of Osh3 reveals a conserved mode of phosphoinositide binding in oxysterol-binding proteins. Structure. 2013;21:1203–1213. doi: 10.1016/j.str.2013.05.007. [DOI] [PubMed] [Google Scholar]
- 49.Usatyuk PV, Fu P, Mohan V, Epshtein Y, Jacobson JR, Gomez-Cambronero J, Wary KK, Bindokas V, Dudek SM, Salgia R, Garcia JG, Natarajan V. Role of c-Met/phosphatidylinositol 3-kinase (PI3k)/Akt signaling in hepatocyte growth factor (HGF)-mediated lamellipodia formation, reactive oxygen species (ROS) generation, and motility of lung endothelial cells. J Biol Chem. 2014;289:13476–13491. doi: 10.1074/jbc.M113.527556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weber-Boyvat M, Kentala H, Peränen J, Olkkonen VM. Ligand-dependent localization and function of ORP-VAP complexes at membrane contact sites. Cell Mol Life Sci. 2015;72:1967–1987. doi: 10.1007/s00018-014-1786-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wilfling F, Wang H, Haas JT, Krahmer N, Gould TJ, Uchida A, Cheng JX, Graham M, Christiano R, Frohlich F, Liu X, Buhman KK, Coleman RA, Bewersdorf J, Farese RV, Jr, Walther TC. Triacylglycerol synthesis enzymes mediate lipid droplet growth by relocalizing from the ER to lipid droplets. Dev Cell. 2013;24:384–399. doi: 10.1016/j.devcel.2013.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu N, Zheng B, Shaywitz A, Dagon Y, Tower C, Bellinger G, Shen CH, Wen J, Asara J, McGraw TE, Kahn BB, Cantley LC. AMPK-dependent degradation of TXNIP upon energy stress leads to enhanced glucose uptake via GLUT1. Mol Cell. 2013;49:1167–1175. doi: 10.1016/j.molcel.2013.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xu N, Zhang SO, Cole RA, McKinney SA, Guo F, Haas JT, Bobba S, Farese RV, Jr, Mak HY. The FATP1-DGAT2 complex facilitates lipid droplet expansion at the ER-lipid droplet interface. J Cell Biol. 2012;198:895–911. doi: 10.1083/jcb.201201139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yecies JL, Zhang HH, Menon S, Liu S, Yecies D, Lipovsky AI, Gorgun C, Kwiatkowski DJ, Hotamisligil GS, Lee CH, Manning BD. Akt stimulates hepatic SREBP1c and lipogenesis through parallel mTORC1-dependent and independent pathways. Cell Metab. 2011;14:21–32. doi: 10.1016/j.cmet.2011.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yu Y, Hamza A, Zhang T, Gu M, Zou P, Newman B, Li Y, Gunatilaka AA, Zhan CG, Sun D. Withaferin A targets heat shock protein 90 in pancreatic cancer cells. Biochem Pharmacol. 2010;79:542–551. doi: 10.1016/j.bcp.2009.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material 1 (TIFF 25519 kb)
Supplementary material 2 (TIFF 25523 kb)
Supplementary material 3 (TIFF 25520 kb)