Abstract
The hindbrain is a key relay hub of the central nervous system (CNS), linking the bilaterally symmetric half-sides of lower and upper CNS centers via an extensive network of neural pathways. Dedicated neural assemblies within the hindbrain control many physiological processes, including respiration, blood pressure, motor coordination and different sensations. During early development, the hindbrain forms metameric segmented units known as rhombomeres along the antero-posterior (AP) axis of the nervous system. These compartmentalized units are highly conserved during vertebrate evolution and act as the template for adult brainstem structure and function. TALE and HOX homeodomain family transcription factors play a key role in the initial induction of the hindbrain and its specification into rhombomeric cell fate identities along the AP axis. Signaling pathways, such as canonical-Wnt, FGF and retinoic acid, play multiple roles to initially induce the hindbrain and regulate Hox gene-family expression to control rhombomeric identity. Additional transcription factors including Krox20, Kreisler and others act both upstream and downstream to Hox genes, modulating their expression and protein activity. In this review, we will examine the earliest embryonic signaling pathways that induce the hindbrain and subsequent rhombomeric segmentation via Hox and other gene expression. We will examine how these signaling pathways and transcription factors interact to activate downstream targets that organize the segmented AP pattern of the embryonic vertebrate hindbrain.
Keywords: Hindbrain; Neural specification and patterning; Hox proteins; Meis and Pbx proteins; FGF, Wnt and retinoic acid signaling; Rhombomere patterning
Introduction
The central nervous system (CNS) has been a classic model system to study pattern formation during early vertebrate development. Vertebrate CNS morphology is strikingly conserved, from fish to mammals. Dorsal ectodermal cells on the outer surface of the embryo are induced to neural fates by neighboring dorsal mesoderm cells. This induced neural plate tissue subsequently thickens, elevating at the embryo’s two lateral edges to form the neural folds. These two arising neural folds merge at the dorsal midline of the embryo, creating the neural tube. The neural tube is asymmetric; the wider, thicker walled anterior region forms the brain, while the narrower, posterior part forms the spinal cord.
The brain subdivides into three regions with distinct antero-posterior (AP) characteristics: the most anterior forebrain (telencephalon and diencephalon), the midbrain (mesencephalon) and most caudally, the hindbrain (rhombencephalon). These regions are the morphological basis of distinct functional units of the brain. In the forebrain, the telencephalon gives rise to the cerebral cortex, basal ganglia and hippocampus. The diencephalon gives rise to the thalamus, hypothalamus and pineal gland. In the lateral diencephalon regions, optic vesicles form the eye structure. The midbrain will generate centers of sensory and motor control, whereas the hindbrain gives rise to the cerebellum, pons and medulla [1]. The hindbrain controls crucial physiological processes such as motor activity, respiration, sleep and blood circulation. It also receives processes and transmits multiple sensory inputs including the auditory, precerebellar and vestibular systems. Different streams of neural crest cells are generated in the hindbrain to give rise to cranial sensory ganglia, Schwann cells, cardiac connective tissue and most of the cranial skeleton [2].
The embryonic hindbrain is divided into seven or eight segmented regions called rhombomeres (r). In the most rostral hindbrain, r1 borders the midbrain in a region called the midbrain–hindbrain boundary, whereas at the most caudal end, r7/r8 borders the most-anterior spinal cord. Each rhombomere has unique gene expression patterns that promote the region-specific fates, differentiation of neurons and production of distinct neural crest streams. Rhombomeric units are highly conserved during vertebrate evolution, acting as templates for the adult hindbrain structure and function, by giving rise to the pons, medulla oblongata and different cerebellar cell layers. The most caudal region of the central nervous system, the spinal cord, forms posterior to the hindbrain, extending to the rear of the body.
This elegant CNS morphogenesis is achieved in the vertebrate embryo through a series of inductive events. In amphibians, Mangold and Spemann in the 1920s found that grafting a dorsal mesodermal lip removed from the blastopore of an early gastrula stage embryo induced a secondary nervous system in the naïve ventral ectoderm of the transplanted host [3]. This region is called “the organizer”, and the phenomenon of this experiment was termed “embryonic induction”. These experiments were confirmed in other vertebrates in analogous fish and bird Spemann–Mangold organizer regions [4, 5]. More recent genetic studies suggest that a neural inducing organizer activity also exists in mammals. The mammalian organizer center may be composed of two regions acting in differing time periods. The earlier anterior visceral endoderm establishes initial embryonic AP polarity, while the later node region induces neural tissue [6].
Seminal experiments of Nieuwkoop, Eyal-Giladi, Toivonen and Saxen suggested that there are two inductive steps involved in neural induction and patterning [7–9]. In the first step, the “organizer” initially induces a general neural tissue having an anterior forebrain fate in a process called “activation”. The second “transformation” step is a caudalizing-posterior induction, which re-specifies the “activated” neuro-ectoderm into more posterior CNS cell fates such as hindbrain and spinal cord [7].
Three different neural inducing proteins (noggin, chordin, follistatin) were initially identified and characterized in amphibians [10–12]. These proteins all induce anterior-pan neural tissue. While structurally different, these proteins share one common activity, the ability to inhibit BMP protein signaling activity [12–14] by blocking BMP ligand–receptor interactions [14–16]. Thus, BMP antagonism serves as the initial “activation” signal inducing ectoderm to rostral neural fates. Since BMP signaling actively drives ectoderm cells to epidermal/non-neural cell fates, BMP signaling inhibition suffices to convert ventral epidermal ectoderm to a more dorsal neural fate [17–19].
Parallel to the discovery of the “activation” signal of BMP antagonism, neural caudalizing “transformation” molecules were also identified. Using Xenopus and chick experimental embryology techniques, in addition to zebrafish and mouse genetics, three signaling pathways were identified that caudalize rostral neural tissue. These include the basic fibroblast growth factor (bFGF), retinoic acid (RA) and canonical Wnt signaling pathways [20–35]. For each pathway, multiple ligands, receptors and antagonists are expressed in different temporal windows and embryonic locales during the neural patterning process. The same pathways seem to have variant spatial and temporal roles in specifying multiple posterior cell fates in the developing vertebrate nervous system. These caudalizing factors induce an initial posterior neural “ground state”, which undergoes fine-tuning into distinct locales such as hindbrain and spinal cord along the neural AP axis.
While the sequence of morphological events that lead to the partition of the CNS into sub-domains is well known, the genetic networks that govern the specification and connectivity of different cell types along the CNS to yield a functional nervous system are only partially resolved (reviewed in [36–39]). This review examines the conserved transcriptional networks and morphogens that orchestrate the intricate regulation of early hindbrain specification and patterning in different vertebrates. It will cover both the earliest stages of hindbrain development/induction, as well as later stages of rhombomere specification. By addressing the complex interactive dynamics between signaling pathways, the earliest activation of Hox and TALE homeodomain proteins, as well as the later expression of non-homeodomain transcription factors, this review provides a unique comprehensive synopsis of the central processes of hindbrain development in relevant vertebrate systems.
The transcription factor blueprint of the hindbrain
The Hox genes are one of the most ancient regulators of body formation in metazoans, being crucial for AP axis formation in the developing vertebrate embryo. Hox proteins partner up with another ancient family of homeodomain proteins, the TALE class proteins, Meis and Pbx. By their joint interactive activities during the earliest stages of neural development, Meis, Pbx and Hox proteins act to induce and specify the hindbrain. Vertebrates typically have thirteen Hox gene paralog groups (PG) expressed along the AP axis. The most anterior extent of Hox expression along the AP axis is the hindbrain, and the PG1–4 group genes are regionally expressed in the hindbrain, and required for its formation. Acting with the Hox genes to induce the hindbrain are the Meis/Pbx proteins. Meis/Pbx expression can precede Hox gene expression. In some systems, Meis/Pbx was shown to be required for the earliest activation of Hox gene expression in the hindbrain. In addition, various dimers or trimer of Meis/Pbx/Hox proteins directly activate target genes in the developing hindbrain. This review will address how these transcription factors interact with signaling pathways to regulate the earliest formation of the hindbrain.
Homeodomain proteins
TALE class homeodomain proteins: Meis, Prep and Pbx proteins
The correct temporal and spatial expression of Hox paralogous group (PG1–4) proteins is crucial for establishing the initial segmentation of the hindbrain. Hindbrain induction is also dependent on Three-Amino acid Loop Extension (TALE) homeodomain proteins, which belong to the MEIS (Meis1–3 proteins), PREP and PBC (Pbx1–4 proteins) groups. TALE proteins are atypical homeodomain-containing transcription factors having three additional amino acids between the first and second helix of the homeodomain [40]. During hindbrain development in zebrafish and Xenopus, meis/pbx genes are activated very early, preceding hox gene expression in the presumptive neural plate. Meis/Pbx and Hox proteins interact at two distinct levels. Early in zebrafish and Xenopus development, Meis/Pbx proteins are required to initially activate PG1–4 hox gene expression in the hindbrain [41–44]. Later, Meis (Prep)/Pbx/Hox protein combinations bind target genes to activate their transcription. Some of these target genes are also hox and meis genes themselves, but non-hox gene targets also lie downstream to Meis/Pbx/Hox (Fig. 1) [45, 46].
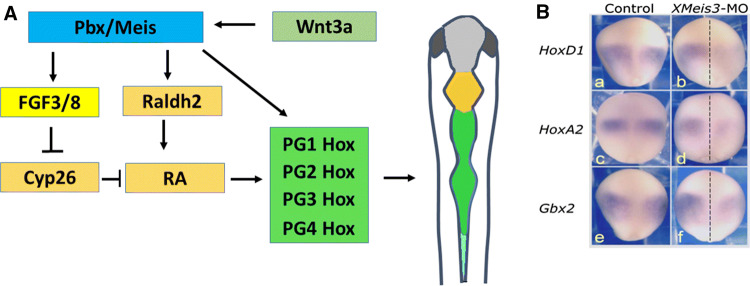
Fig. 1.
Pbx/Meis proteins lie upstream of Hox gene expression in the early hindbrain. A Schematic representation of Pbx and Meis regulatory activity in the early hindbrain. Pbx/Meis proteins are induced in the neural plate by Wnt signaling prior to Hox gene expression. Pbx and Meis activate (individually and additively) the transcription of PG1–4 Hox genes, by binding to their Pbx/Meis-responsive elements. In parallel, Pbx/Meis also activate FGF and RA signaling to promote Hox gene expression. This is mediated by their binding to responsive elements in the Raldh2 gene, the main RA-synthesizing enzyme, as well as by inducing FGF gene expression that in turn inhibits the RA-degrading enzyme Cyp26. Following the initial activation of Hox gene expression, Meis/Pbx synergize with HoxA1 protein to induce expression of other Hox genes (not shown). B Meis3 regulates early homeobox gene expression (from Elkouby et al. [44]). Meis3 knockdown inhibits early HoxD1, HoxA2 and Gbx2 gene expression. Embryos were injected into one blastomere at the two-cell stage with Meis3-MO (10–12.5 ng/blastomere; b, d, f). Gene expression was examined at late gastrula, stage 12.5. All embryos are viewed dorsally, and oriented with anterior at the top, posterior at the bottom. The dashed line in b, d, f indicates the dorsal midline; the XMeis3-MO-injected side is on the right. Early expression of HoxD1, HoxA2 and Gbx2 is inhibited on the Meis3-MO-injected side (100%, n = 16; 88.2%, n = 34; 66.4%, n = 33, respectively). In neurula-stage Meis3-morphant sibling embryos, a typical inhibition of posterior neural cell types was observed
Studies in Xenopus and zebrafish embryos revealed the requirement of Meis1 and Meis3 for proper hindbrain formation [41–44, 47–50]. The expression of meis3 initiates early, at late gastrula stages in the presumptive hindbrain region. At later stages, its expression becomes localized to the r2–r4 region [42, 43, 47, 48]. In contrast, expression of the meis1 and meis2 genes is more general, expanding into more anterior neural regions than meis3 in multiple vertebrate species, such as mouse, chick, Xenopus and zebrafish [43, 51–56]. In Xenopus and zebrafish, loss of Meis/Pbx function, obtained by either dominant-negative proteins, antisense oligonucleotide morpholino (MO) knockdown, or genetic mutation, triggers a loss of the entire hindbrain region, accompanied by a loss in expression of a many hindbrain markers, including hox PG1-4 genes [41–44, 47]. This is concomitant with a posterior expansion and enlargement of more anterior forebrain structures. Meis3 also suffices for hindbrain induction, as its overexpression induces ectopic hindbrain formation while repressing forebrain formation in both Xenopus and zebrafish embryos and explants [42–44, 47, 48, 57]. In mouse and chick, Meis/Prep and Pbx proteins also are required for activation of hindbrain enhancers in the hoxb1 and hoxb2 genes [58–61].
Slightly later in zebrafish development, Meis3 synergizes with HoxB1b (HoxA1 in other vertebrates) protein to induce expression of various other hindbrain markers such as hoxb1a (hoxb1) and krox20 [48]. Similarly, in Xenopus, HoxD1 and Meis3 co-expression enhances hoxb3 and krox20 expression [62]. In mice and zebrafish, the krox20 promoter has a functional r3-specific Hox/Meis/Pbx binding site, although it does not appear to be a direct Meis3-target in Xenopus [44, 63, 64]. Moreover, hoxd1 is a direct-target gene of Meis3 in Xenopus that acts downstream of Meis3 to induce hindbrain cell fates [44, 57, 62]. In Xenopus, the Zic1 and Pax3 transcription factors are required upstream of Meis3. Knockdown of Zic1 or Pax3 prevents early meis3 gene expression, leading to a loss of hindbrain cell fates [57]. Ectopic Meis3 can rescue Zic1, but not Pax3 knockdown phenotypes.
Together with Meis proteins, the PBC family proteins are central for hindbrain induction. Pbx4 directly interacts with Meis1 and Meis3 proteins in zebrafish, and perturbation of either Pbx2 or Pbx4 activities eliminates r2–r6 cell fates in zebrafish [41, 43, 48, 65, 66]. Pbx4 is expressed in the presumptive hindbrain region, where its early expression closely overlaps the meis3 and hoxb1b (hoxa1) genes [48]. In Xenopus, Meis1 and Pbx1 proteins interact to regulate neural cell fate specification, where Pbx1 knockdown also disrupts hindbrain formation [50]. Pbx1 is expressed in the presumptive forebrain, hindbrain and neural crest regions [53]. In Xenopus, Pbx1 and HoxD1 proteins strongly activate a heterologous mouse hoxb1 enhancer, but Meis3 had no additive effect with either protein when tested separately or together [62, 67]. A Pbx protein partner that enhances Meis3 activity in Xenopus has not been identified. In chick and mice, Meis2 and Pbx proteins were found to synergize and to activate a krox20 enhancer element in r3 [63]. Interestingly, in zebrafish, ectopic Meis1 expression rescued hindbrain formation in the absence of zygotic Pbx4 protein. This suggests a potential Pbx-independent mechanism of action, although maternal Pbx protein involvement was not fully ruled out (Fig. 1), [43].
Hox proteins
PG1 Hox proteins are key factors controlling early hindbrain specification. The PG1 Hox proteins are homologs of the Drosophila labial gene and include the HoxA1, HoxB1 and HoxD1 proteins. In all vertebrates, expression of the PG1 hox genes precedes all other hox genes in the presumptive hindbrain region at early gastrula stages and persists through neurula stages [68–77]. PG1 proteins are essential for correct hindbrain induction and segmentation. Combined knockdown of HoxB1a (Mouse Hoxb1) and HoxB1b (Mouse Hoxa1) proteins in zebrafish, or hoxA1 and hoxB1 gene deletion in mouse embryos led to significant hindbrain perturbation [75, 76, 78, 79]. In Xenopus embryos, triple knockdown of all the PG1 genes, hoxa1, hoxb1 and hoxd1 caused a complete loss of r2–6, and the entire hindbrain resembled the Hox non-expressing r1 region [77]. This phenotype is reminiscent of Pbx2 loss-of-function in zebrafish embryos, arguing for cooperative function of Pbx2 and PG1 Hox proteins in the specification of r2–6 regions [66]. Supporting this assumption, HoxA1 hexapeptide mutant proteins that fail to interact with Pbx proteins cause severe hindbrain phenotypes in mice [80]. Moreover, the combined PG1 loss-of-function phenotype is synergistically stronger than that of each of the individual inhibitions, and in Xenopus, PG1 knockdown could be rescued by overexpression of HoxD1 protein alone. This suggests at least partial functional redundancy between PG1 members [77]. To gain insight into the transcriptional network regulated by HoxA1, microarray analysis was performed on the prospective r3–5 region of hoxa1 null and wild-type mouse embryos [81]. Around 300 genes were differentially expressed between the samples. While many of these genes were previously identified to play a role in hindbrain development, new target genes were also found to be downregulated in hoxa1-nulls, such as FGF receptor 3 (fgfr3), zic1, hnf1b, foxd3 and lhx5, suggesting that HoxA1 protein acts in a genetic cascade upstream to many target-genes that control wider aspects of hindbrain development than previously thought. HoxA1 mutations in humans have brainstem function perturbations, and it was suggested that this could be related to improper embryonic hindbrain development [82].
Early PG1 protein expression is a prerequisite for the proper sequential expression of later, more posteriorly expressed PG2–4 Hox genes [62, 77]. In PG2 mutant mice, development of the r3/r4 region is disrupted with poor border formation between r2/r3. Cells in r2 express only hoxa2, the most anterior of all Hox genes, and hoxa2 loss-of-function mutations cause r2 to r1 fate changes [83–87]. Notably, Hox PG1 and PG2 genes are both expressed in r4, but with temporal differences. Single hoxb2 mutant mice had no hindbrain segmentation defects, but in hoxa2/hoxb2 double-mutant mice, the r2/3 and r3/r4 borders were lost, suggesting that the action of both PG2 genes is synergistic [88]. Interestingly, recent sequence analysis of hoxa2 in fugu uncovered the presence of two orthologues, hoxa2a and hoxa2b. Each orthologue contains distinct cis-regulatory elements to drive hoxa2 expression in neural crest or rhombomeres, respectively. This study suggests that these regulatory regions are conserved throughout vertebrate evolution to mediate differential hoxa2 expression and activity during development [89]. A negative regulatory mechanism exists between PG3 and PG2 groups. Studies in mice and chick embryos showed that Hoxb3 protein directly binds to hoxb1 regulatory regions and represses hoxb1 expression posterior to r4 [90]. In multiple PG3 mutants, r5/6 identity was disrupted and r4-specific hoxb1 expression was ectopically activated in r5/6 [91]. In PG4 mutants, in contrast, hindbrain development was normal [92].
A recent study compared the binding targets of HoxA1 proteins in zebrafish and mouse finding that they share many common hindbrain target genes. Many of these targets also shared occupancy with Meis and Pbx proteins [93]. HoxA1 also was found to bind enhancer regions of the meis2 and meis3 genes [94], suggesting that HoxA1 may be regulating the early expression of these meis genes. This coupled to studies in Xenopus showing Meis3 regulation of PG1 Hox gene expression suggests that there is a mutual co-dependence between Meis and Hox PG1 proteins to regulate the earliest stages of vertebrate hindbrain specification.
TALE and Hox proteins cross-talk in the hindbrain
As mentioned previously, during early development, Meis, Pbx and Hox proteins interact at two distinct levels. Initially, Meis–Prep/Pbx and PG1 Hox proteins may reciprocally co-activate each other’s gene expression in the hindbrain. Later, Meis/Pbx/Hox protein combinations bind target genes to further activate hox gene expression in the hindbrain [42]. In zebrafish embryos, ChIP studies showed that Meis/Pbx proteins specifically bind the hoxb1a and hoxb2a promoters in their respective tissues of expression [95]. These promoters were also enriched for histone H4 acetylation. In embryos ectopically expressing dominant negative Pbx proteins, Meis/Pbx activity was inhibited and histone acetylation was highly reduced. Furthermore, Meis/Pbx protein complex formation removes histone deacetylase (HDAC) from Hox-regulated promoters. Additionally, Meis proteins recruit CBP/p300 histone acetylase to hox promoters. Thus, Meis proteins function as direct transcriptional activators of the hoxb1a target gene by controlling the accessibility of HDAC/CBP proteins to its promoter [95]. More recent studies in zebrafish embryos show that TALE protein complexes actively poise hoxb1a promoters for expression by chromatin modification, as early as blastula stages. Later expression of the hoxb1a promoter at gastrula stage is triggered by Hoxb1b protein binding to these TALE protein complexes [96]. Pbx–Hox and Meis–Prep binding sites have also been used to define a shared sequence syntax system for identifying functional hindbrain-specific enhancer elements in zebrafish [97]. In rhombomeric segments, Hox gene expression is positively controlled by auto- and cross-regulatory binding of Meis/Pbx/Hox proteins to enhancer elements. For example, the hoxb1 gene enhancer that drives expression in r4 harbors distinct Meis/Pbx and Hox/Pbx binding sites. In mice, Hoxa1 together with Meis/Pbx proteins initially activates this enhancer, but later, Hoxb1 itself, together with Hoxb2, Meis3 and Pbx2/4 proteins, is required for expression maintenance in r4 [58, 65]. Interestingly, additional Hox proteins of the PG3 group, but not Pbx/Meis proteins, negatively regulate hoxb1 such that it remains restricted to r4 [90, 91].
Many additional examples of such interdependent, cross-regulatory loops are known. For instance, hoxd1, hoxb2 and hoxa2 all require Meis/Pbx for their expression in zebrafish, Xenopus, chick and mouse [43, 44, 49, 59, 66, 98, 99]. In mice, Hoxb2 expression is directly activated by HoxB1 in r4, and HoxB2 protein then drives hoxb1 expression. Thus, HoxB2 indirectly controls its own expression via HoxB1 [59–61, 88, 100]. Hoxa2 expression in r4 is also regulated by conserved vertebrate enhancer elements that bind Meis/Pbx and HoxB1/HoxA2 proteins [98, 99, 101]. In more anterior r2–r3, Meis3/Pbx proteins are required for the early neural expression of hoxa2 in Xenopus [44]. Studies in mouse and chick suggest that r4 activation of hoxa2 requires Meis3/Pbx proteins; however, it is still an open question for the r2 hoxa2 enhancer regions, which contain Sox binding sites and perhaps is not regulated by Meis/Pbx proteins [98, 99]. In mouse and chick, more posteriorly, hoxa3 expression in r5/6 is controlled by an element binding Meis/Pbx and HoxA3/HoxB3 proteins [102]. In r6/7, the expression of hoxb4 and hoxd4 in mice is also regulated by Meis1/Pbx as well as Hoxb4/Hoxd4-responsive elements after initial induction by retinoic acid signaling [103–105]. Finally, adding an additional level of complexity, all rhombomeres fail to develop properly upon Meis3 knockdown in Xenopus, despite the fact that meis3 expression is restricted to r2–4 [42, 44, 47]. The elimination of early r4 fgf3 expression in Meis3-depleted embryos likely triggers these non-autonomous effects [57]. Similarly, r4-specific fgf3 expression is eliminated in zebrafish pbx/hox mutants [66], and this triggers a loss of hindbrain cell fates from r2–r6. In Xenopus or zebrafish, any direct activation of Meis/Pbx/Hox enhancer sequences in more posterior rhombomeres could be mediated by more ubiquitously expressed Meis or Prep proteins (Fig. 1).
Non-homeodomain transcription regulators
Three early expressed transcription factor proteins, variant hepatocyte nuclear factor-1 (Vhnf1), Krox20, and the Mafb/Kreisler/Valentino proteins, are crucial for regulating the earliest expression of hox genes. These transcription factors are thus essential for the establishment of correct rhombomeric AP identity in the early hindbrain. Genetic disruption of these proteins severely perturbs the formation of r3–r6. Vhnf1 is the earliest gene expressed in r5/r6. In zebrafish, the Vhnf1 protein induces both krox20 (r5) and mafb (r5/6) expression, which subsequently determines correct rhombomeric identities. Loss of Vhnf1 reduces r5/6 domains while the hoxb1a-expressing r4 region is expanded [106–108]. In mice, a Vhnf1-binding site was identified in a regulatory element of the kreisler gene. Mutating this site results in loss of kreisler induction, indicating that Vhnf1 is essential for rhombomere-specific kreisler expression in the future r5/r6 domain [109] (Fig. 2).
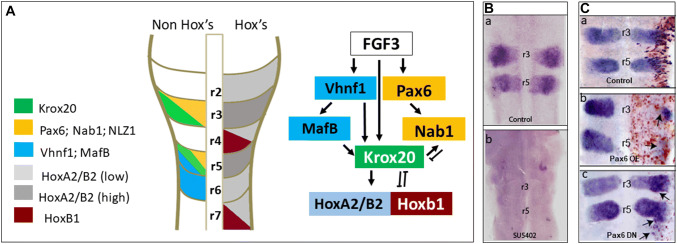
Fig. 2.
Upstream and downstream regulation of Krox20 in the hindbrain. A Induction of Krox20 gene expression in r3/r5 is initiated by FGF signaling. FGF induces Vhnf1 in r5/r6 that upregulates MafB, leading to Krox20 induction in r5. In r3, FGF induces Krox20 expression in a Vhnf1-independent manner. Krox20 expression is restricted to the correct rhombomeres by Pax6, which is also induced by FGF. Pax6, which is expressed in r3/r5 is upregulated, and acts as a negative regulator of Krox20 expression via the induction of Nab1. NLZ is another Krox20-negative regulator that is also co-expressed in r3/r5. In parallel, Hoxb1 is expressed in r4 and represses Krox20 expression in r4. Following the upregulation of Krox20 in r3/r5, Krox20 regulates PG-2 Hox gene expression in r3/r5. It also induces the expression of its negative modulator Nab1. In addition, Krox20 represses Hoxb1 expression in r3/r5, leading to accurate r3/r4/r5 identities. In early stages of development, Krox20 transiently activates Hoxb1 (not shown). B FGF acts upstream of Krox20 in the chick hindbrain. (a) Krox20 is expressed in control embryos, but is missing in (b) hindbrains treated with the FGF receptor inhibitor SU5402 (from Weisinger et al. [142]). C Pax6 is a negative modulator of Krox20 that restricts its expression to r3/r5 in the chick hindbrain. (a) Hindbrains electroporated with control plasmid show normal Krox20 expression. (b) Overexpression (OE) of Pax6 leads to a reduction in Krox20 expression (arrows). (c) Expression of a dominant-negative (DN) Pax6 results in the expansion of Krox20 (arrows) to additional domains (from Kayam et al. [124])
Krox20 is expressed in rhombomeres 3 and 5, and is paramount for r3/r5 regional identity and cell fate, as directly evident by various knockout or knockdown experiments in all vertebrates [107, 110–112]. In addition to Vhnf1, induction of krox20 is triggered by a positive input from Pbx/Meis and HoxB1 [63, 113–115]. In chick, mice and zebrafish, krox20 promoter/enhancer sites have functional r3-specific Hoxb1/Meis/Pbx binding sites, although in Xenopus it does not appear to be a direct Meis3-target [44, 63, 64]. Following the upregulation of krox20 via these multiple regulators, it acts in a positive auto-regulatory loop to maintain its own expression. A recent study revealed that one of the krox20 regulatory regions is required not only to initiate its expression but also to enable the auto-regulatory binding site to function [116]. This region acts with additional cis-regulatory elements, some of which are positioned further away from the krox20 loci. These multiple regulatory modes ensure a proper level of krox20 auto-regulation in r3 and r5. Notably, another member of Egr family, Egr4, was recently identified in Xenopus to be expressed in the posterior hindbrain [117]. Knockdown experiments uncovered a role for Egr4 in krox20 and mafb upregulation in r5 or r5/r6, respectively, indicating that in the frog, Egr4 mediates the effect of Vhnf1 to activate these two genes in the posterior hindbrain (Fig. 2).
Initially, Hoxb1 activates krox20 by binding to its enhancer. Later, when Hoxb1 expression levels increase and are restricted to r4, it represses krox20 expression. Reciprocally, Vhnf1 and Krox20 proteins repress hoxb1 to limit its expression to r4. These dual-negative regulatory interactions lead to the establishment of well-defined rhombomeric identities [107, 108, 112]. Moreover, Krox20 protein directly activates the expression of the ephA4 receptor gene in r3/r5, which in turn sharpens the rhombomeric borders by preventing cell intermixing between segments [118–120]. Interestingly, two negative regulators of krox20 gene expression, Nab and Nlz (also known as Neurl1a), have been identified in zebrafish, mice and chick hindbrains [121–123]. These factors co-localize with krox20, but repress its transcription, restricting its expression to the odd-numbered rhombomeres. Krox20 itself is involved in mediating its negative transcriptional regulation by acting as a SUMO ligase that simulates Nab protein activity [123]. Moreover, Hoxb1 was found to upregulate nlz and thus indirectly repress krox20 expression [115]. The paired-rule gene pax6 is also co-expressed with krox20 in r3/r5 but negatively regulates it, leading to the stabilization of krox20 expression borders in r3/r5 [124]. Pax6 protein negatively modulates krox20 gene expression via its ability to induce nab1 gene expression; nab1 protein subsequently binds to krox20 regulatory elements to repress its expression (Fig. 2).
Mafb/Kreisler/Valentino proteins are expressed in r5/r6, and are obligatory for their specification. In mafb mutants, r5/6 identity is lost and r4 is expanded [125, 126]. Mafb and Vhnf1 mutually regulate gene expression in a positive feedback loop. Mafb protein regulates ephrinB2a expression, which represses r4, and further promotes r5/6 identities [108, 127]. Vhnf1 together with Mafb and Krox20 also activates hoxa3 and hoxb3 expression, which is crucial to specify r5/6 identity [108, 128, 129]. In zebrafish embryos, krox20 transcriptional activation also requires the Vhnf1 protein, which synergizes with FGF activity to induce mafb/kreisler/valentino and krox20 gene expression. Vhnf1 protein in turn represses hoxb1 gene expression, limiting its expression to r4, thus enabling formation of more posterior rhombomeric fates [107, 108]. Subsequently, Vhnf1 together with Mafb and Krox20 activate hoxa3 and hoxb3 expression in r5/6 to specify r5/6 identity [108, 128] (Fig. 2).
Signaling pathways induce the hindbrain
FGF signaling
FGF signaling plays a key role in hindbrain induction and patterning. Pioneer studies in the chick embryo revealed that caudal epiblast cells fated to give rise to hindbrain character are located adjacent and lateral to the anterior primitive streak of stage 4 embryos [130, 131]. As cells in and around the streak express different FGFs [132, 133], a possibility was raised that FGFs participate in the induction of the hindbrain. Exposure of neural plate explants from different stages and axial levels to FGFs confirmed that FGF signaling is required, although not sufficient, to induce cells of hindbrain character [134].
In zebrafish embryos, the FGF3 and FGF8 genes are expressed in the presumptive hindbrain primordia at 80–90% epiboly, before the onset of rhombomere segmentation [135, 136]. At segmentation initiation, FGF3/8 transcripts are expressed in the central r4 region, with r3/r5 overlap. When morphological segments have formed, FGF3 continues to be expressed in r4, but FGF8 expression is extinguished, with newly shifted expression to the more anterior isthmus region. FGF3/8 activities appear to be functionally redundant since a strong hindbrain phenotype is only seen in zebrafish embryos co-injected with morpholino oligonucleotides (MO) to both genes. FGF3/8 knockdown severely inhibited formation of all rhombomeres, except r4. In FGF3/8 morphant embryos, initial hoxB1 expression in r4 was normal but declined with time, suggesting that neighboring rhombomeres are necessary to maintain its expression. However, in FGF3/8 morphants, neurons derived for r1–3 and r5–7 are disrupted, but r4-derived neurons formed fairly typically. The possibility that FGFs are acting redundantly in the hindbrain is also reinforced in mice, where otic-placode induction is severely perturbed in FGF3 null embryos, but hindbrain segmentation remains normal [137].
The expression of FGF3 in hindbrain primordia is conserved in vertebrates [133, 135, 136, 138–141]. Yet, in contrast to the r4-restricted expression in Xenopus and zebrafish, in chick and mice FGF3 is also evident in r2 and r6 [105, 120, 140–142]. It was suggested that early FGF3/8 signaling in r4 forms a primary hindbrain-inducing center since transplanted r4 cells or ectopic expression of FGF3/8 induces expression of r5/6-specific markers in chick and zebrafish embryos [135, 143]. In Xenopus, FGF8 is also involved in hindbrain induction; one of its splice forms, FGF8a, mediates hindbrain specification since its knockdown severely perturbs formation of hindbrain and other posterior neural fates [25]. Noticeably, studies in chick and mice demonstrate that much after rhombomere specification takes place, FGF3 is downregulated from r2/r4/r6 but maintained in all rhombomere boundaries [120, 144]. Rhombomere boundaries display unique cellular and molecular properties that are different from rhombomere bodies [145–149]. In chick, the expression of different boundary markers requires FGF3 [144]. Recent findings suggested that rhombomere boundaries serve as pools of neural-stem-like cells that express Sox2 and nestin and contribute neurons to the hindbrain at stages when rhombomere cells are actively differentiating [150]. Whether FGF signaling is also required for the development and/or maintenance of these neural stem cells in the hindbrain awaits further studies.
A tight cross-talk exists between different transcription factors and FGF signaling in the hindbrain. These interactions play critical early roles in hindbrain induction and segmental patterning. For instance, the activity of Meis proteins is required to induce FGF3 expression as Meis3 knockdown in Xenopus resulted in elimination of early FGF3 expression in r4 [57]. As a consequence, the entire hindbrain failed to form due to the loss in FGF signaling [42, 151], and Meis3 protein cannot induce hindbrain marker expression in the absence of FGF signaling [24, 151]. Moreover, in an attempt to screen for Hoxb1 target genes in zebrafish, a novel gene, ppp1r14al, was identified that is induced by Hoxb1 in r4. Ppp1r14al in turn regulates FGF3 expression in r4, indicating that it is also essential for the establishment of the earliest hindbrain signaling center in r4 [46]. The PG1–3 Hox groups, Vhnf1, Krox20 and Kreisler/MafB/Valentino proteins all act downstream of FGF signaling to induce hindbrain segments. For instance, FGF3 signal indirectly activates hoxa2, hoxb2 and hoxb3 expression via induction of krox20 gene expression [128, 129, 132, 143, 152]. FGF signaling also up-regulates vhnf1 gene expression, which in zebrafish controls caudal hindbrain specification by synergizing with FGF activity to induce mafb/kreisler and krox20 expression. Upon FGF signaling activation, Mafb and Vhnf1 proteins are upregulated and bind to specific enhancer elements in the krox20 promoter. This transcriptional regulation is required for initial krox20 expression in r3/r5, but not for its later maintenance [107, 113]. In addition, Vhnf1 protein also appears to activate fgf3 expression and to repress hoxB1 gene expression, presumably to exclusively limit its expression to r4. Thus, in the hindbrain, the FGF-inducing center acts at a pivotal position, being downstream to Meis/Pbx/Vhnf1/Hox PG1 proteins, but upstream to PG2–3 Hox and Vhnf1/Krox20/Kreisler gene activity.
Studies in the chick hindbrain demonstrated that FGF signaling activates the MAP kinase signal transduction protein ERK1/2 that in turn induces the expression of the Ets-family transcription factor pea3. This signaling cascade is required for krox20 induction in the early hindbrain [142, 153, 154]. FGF3 was also found to induce the expression of pax6 in r3/r5, that in turn negatively regulates krox20 expression, via the induction of nab1 [124]. Thus, by regulating both krox20 and pax6, which mutually repress each other, FGF3 acts as a guardian to sharply define rhombomere borders. Moreover, Sprouty4 protein, which is also induced throughout the hindbrain by FGF, acts in a negative-feedback loop to define sharp rhombomeric domains. Enhanced FGF signaling by sprouty4 knockdown triggers premature and ectopic krox20 gene expression fusing r3 with r5 that eliminate r4 cell fates [115]. Thus, a multitude of molecular interactions are modulated via the FGF pathway to provide an accurate positional identity of hindbrain rhombomeres.
Wnt signaling
Many Wnt ligands, such as Wnt-1, -3, -3a, -4, -8, -8b and -10, are expressed in the developing CNS [33, 155–161]. Their role in posterior neural development was first shown in Wnt1 and Wnt3a knockout mouse embryos, in which midbrain, hindbrain and spinal cord structures were poorly developed [162–164]. Wnt3 null mouse embryos had posterior truncations, with expanded expression of the otx2 forebrain marker, and a concomitant loss of expression of the hindbrain hoxb1 marker [165]. Similar results were seen in chick embryos implanted with beads soaked with a soluble form of the Frz receptor (mFrz8-CRD) that antagonizes endogenous Wnt ligand signaling. These embryos had an expansion of forebrain markers (otx2 and pax6) and a down-regulation of hindbrain (gbx2) markers [166]. In cultured chick neural explants, mFrz8-CRD also eliminated expression of the gbx2, krox20 and hoxb4 hindbrain markers [166, 167]. Similar results were also confirmed in Xenopus embryos, where expression of dominant-negative Wnt proteins that inhibit canonical Wnt activity anteriorized embryos [34]. Like Wnt3−/− mouse embryos, these embryos also exhibited neural tube closure defects and expression of the xanf1 and otx2 forebrain markers was posteriorly expanded, while expression of the hindbrain krox20 marker was reduced [34]. Specific Wnt3a-MO targeting suggested a role in hindbrain development in both zebrafish and Xenopus embryos [44, 168–170]. Targeted mesodermal Wnt3a knockdown in Xenopus ablated hindbrain formation [44]. Wnt3a morphant embryos exhibited neural convergent and extension defects, having the typical caudal expansion of forebrain markers, with depletion of hindbrain markers. Moreover, during late gastrula stages, expression of homeoproteins that regulate early hindbrain induction such as meis3, hoxd1 and gbx2 was reduced [44]. Wnt3a MO pheno-copied the neural phenotypes of the general canonical Wnt inhibitor Dkk1 [44], further supporting a role for Wnt3a as the primary posterior neural inducer in vertebrates. In contrast, zebrafish mutated for both Wnt1/Wnt10b did not have abnormal hindbrain morphology [171, 172], while knockdown of Wnt3a or Wnt8b did affect hindbrain patterning in zebrafish [172], indicating a potential redundancy in the function of different Wnt ligands for normal hindbrain patterning.
Complementing these loss-of-function studies, Wnt gain-of-function activity induces hindbrain neural cell fates. The Xenopus animal cap (AC) explant system has provided a great tool for examining the role of Wnt signaling in neural patterning. BMP4 antagonism in AC explants induces neural tissue. Such neuralized explants mimic the initial state of the newly induced embryonic neural plate. AC explants express pan-neural and anterior neural markers, and will develop as anterior forebrain/cement gland in the absence of additional caudalizing signals [10]. Neuralized AC explants overexpressing different Wnt ligands or downstream effectors, such as β-catenin or inducible constitutively active Tcf, robustly induced expression of hindbrain markers, while strongly repressing anterior neural marker expression [33, 35, 44, 173–175]. In addition, expression of the earliest hindbrain specifying homeoproteins, meis3, hoxd1, hoxa2 and gbx2, along with caudalizing FGF3 and FGF8 genes, were also induced in gastrula-stage-neuralized AC explants overexpressing either Wnt3a or β-catenin [44, 176]. Chick forebrain explants were also caudally transformed by Wnt3a added to the culture medium (supplemented with FGF), as evident by the induced expression of gbx2 and krox20, instead of otx2 [166]. Canonical Wnt induction of posterior neural cell fates was shown in both frogs and zebrafish to occur specifically during mid-late gastrula stages [35, 44, 169, 170].
Zebrafish embryos overexpressing a heat-shock protein (Hsp)–Wnt8 driver-plasmid induced at gastrula stages had an anterior shift in hindbrain markers, with forebrain markers pushed to the anterior extremity [169]. Zebrafish headless mutants lacked midbrain, eye and forebrain tissues and weakly expressed anf1, six3 and rx3 forebrain markers. Concomitantly, krox20 expression was expanded anteriorly [177]. Headless was identified as a point mutation in the Wnt downstream negative-effector Tcf3 gene, and mutant TCF3 protein was unable to translocate to the nucleus or to bind DNA, thus causing a loss-of-function phenotype [177]. In cells where the canonical Wnt pathway is not activated, Tcf3 represses expression of Wnt-pathway target genes. The loss of Tcf3-mediated repression in the headless mutants reflects an overactive canonical Wnt pathway in the embryos [177]. Xenopus embryos overexpressing an inducible β-catenin protein activated at gastrula stages, or a CMV-promoter driving zygotic Wnt3a expression both induced caudalized embryos with ectopic expansion of hindbrain markers anteriorly, and down-regulation of anterior markers [35, 44]. This anterior transformation in morphology and gene expression pattern to more caudal fates was also evident in Dkk1 null mouse embryos [178] and chick embryos implanted with Wnt3a-soaked beads [166].
The Wnt ligands that induce hindbrain from the paraxial mesoderm may vary between species. In Xenopus, this ligand is Wnt3a, since its specific knockdown inhibits hindbrain formation despite relatively normal wnt8 mesodermal expression levels [44]. In zebrafish, paraxial mesoderm expressed Wnt8a protein is required for hindbrain formation [169]. In mice and chick, Wnt3 and Wnt8 ligands are also expressed during early development. Whether their hindbrain inducing effects are mediated via the mesoderm or directly in the neural plate still needs to be determined.
RA signaling
RA signaling is also seminal for hindbrain patterning. Yet, unlike FGF and Wnt, its availability is largely dependent on diet, as it is produced from vitamin A (reviewed in [179, 180]). The sensitivity of the hindbrain to small perturbations in RA levels has made it an excellent model to study how morphogen gradients govern pattern formation. Studies in frog [32, 74, 181], chick [104, 182], quail [183, 184], mouse [79, 185, 186], rat [187, 188] and zebrafish [189–192] have generated a vast amount of knowledge on the manner by which RA controls hindbrain regional identity. RA signaling is unique among morphogens as it relies not only on its source of production, but on its site of degradation, creating a posteriorhigh–anteriorlow activity gradient.
Initially, the existence of a RA gradient was debatable since depletion of endogenous RA was rescued by applying uniform and high doses of RA throughout the embryo, which resulted in fairly normal hindbrain [180, 193–195]. These findings suggest that a RA gradient is generated not only through diffusion from its posterior source but also from its anterior inactivation. Indeed, there are two main groups of metabolic enzymes that coordinate RA levels. RALDHs convert retinaldehyde into RA, whereas CYP26s inactivate RA via oxidation [196–199]. RA is initially synthesized in the presomitic mesoderm by RALDH, and it diffuses into the adjacent hindbrain in a posteriorhigh–anteriorlow gradient. CYP26 is expressed in the anterior hindbrain, where it leads to RA degradation, further reinforcing the decrease in RA signaling in the rostral embryo [193–195, 200–205]. RA transduces its effects by binding to RA receptors (RARs) and retinoid X receptor (RXRs) [206–210]. Unlike FGF and Wnt, RA uses a nuclear, rather than a membranal, receptor. Upon binding to RA, the receptors act as transcriptional regulators by directly binding specific RA-responsive elements (RAREs) that are positioned at the 3′ or 5′ ends of different hindbrain patterning genes, such as hoxa1, hoxb1, hoxb4 and vhnf1 [79, 104, 108, 211–219]. For instance, in r6/r7, the initial expression of hoxb4 and hoxd4 is activated through RAREs [103–105]. Interestingly, the early transcription of the hoxb1 gene is initiated by a conserved 3′ RARE, but later, when hoxb1 expression is restricted to r4, its repression in r3 and r5 is mediated by a 5′ RARE [220, 221]. Furthermore, the rarβ gene itself contains a HoxB4/HoxD4-responsive element [105], providing a positive feed-forward loop to maintain RA levels and PG4 Hox expression in r6/7.
In addition to controlling the expression of hindbrain segmentation genes in a graded manner, RA regulates its own metabolism in the same manner. RA downregulates the expression of Raldh genes in a negative feedback loop, while upregulating the expression of Cyp26 in a positive feedback loop [191, 194, 195, 202, 222–225]. Yet, RA also induces its own synthesis via a feed-forward mechanism. Experiments in mice and Xenopus embryos revealed that Hoxa1–Pbx1/2–Meis2 directly binds a specific regulatory element that is required for maintaining Raldh2 expression levels. As RA induces the expression of Hoxa1, this study revealed an indirect autoregulation of RA synthesis via Hoxa1 [226]. Moreover, RA positively regulates the expression of the intracellular RA-binding proteins, Crabps, which mediate RA transfer to Cyp26 to trigger its degradation [194]. Intriguingly, Crabps were also found to promote the delivery of RA to its receptors, thus eliciting RA signaling [191, 227, 228]. The duality of RA regulation on its own metabolism requires future research to reveal how its autoregulation is controlled in the hindbrain.
Interestingly, while expression of the RA-degrading enzyme Cyp26a1 gene is tightly regulated by the levels of RA [194], two other members of the cyp26 family, cyp26b1 and cyp26c1, are not directly regulated by RA signaling and display an unexpected segmental expression with lower levels in r3/r5 than in r2/r4/r6 [193, 195, 229]. A recent study in zebrafish found that this segmental expression plays a key role in sharpening r3–r5 segmental gene expression [229]. During hindbrain segmentation, some r3/r5 (Krox20+) cells are initially found in r4, but later switch into r4 (Hoxb1+) identity. This study found that the krox20-intermingled cells are exposed to lower RA levels in r4 due to the elevated activity of cyp26b1 and cyp26c1, which results in downregulation of krox20 and upregulation of hoxb1. The coupling between segmental gene expression and dynamic levels of RA provides the first evidence describing how a signal like RA that is thought to act mainly in a graded caudal–rostral axis orchestrates boundary sharpening via regulating levels of gene expression in alternating rhombomeres [230].
Visualization of the actual RA gradient in live embryos was needed to fully confirm its existence [231]. Initially, this challenge was tackled by generating transgenic mice or zebrafish embryos where GFP/LacZ expression was driven under the control of RAREs through in vivo injection of RA–GFP fused constructs [193, 210, 232, 233]. These strategies supported the shape of a RA gradient, which fitted well with the expression domains of RALDH and CYP26 genes. However, the non-peptidic structure of RA, as well as the failure to observe the GFP signals at early stages, prevented full in vivo validation [194, 210, 234–236]. More recently, imaging tools were developed for this purpose. In one approach, genetically encoded probes for RA (GEPRs) were fused with different GFP variants and introduced into zebrafish. Each GERP displayed a different affinity to RA. Binding of RA to different GERPs led to conformational change that was converted into changes in fluorescence resonance energy transfer (FRET) [237]. This strategy confirmed the concentration gradient of RA in live embryos, where the local source and sink jointly establish the highest RA concentration in the mid-trunk and the lowest in the tail and head. A more recent technology utilized fluorescence lifetime imaging microscopy and phasor analysis to calculate the relative abundances of RA along the hindbrain [238]. This strategy, which is based on the endogenous fluorescence of RA, demonstrated that intracellular free RA forms an anteriorly declining gradient similar to that previously reported with FRET or RARE-GFP/LacZ [193, 237]. This sensitive technology also enabled visualization of random fluctuations in RA levels that can vary rapidly within one hindbrain position. This study suggested that individual cells can actively control the magnitude of random fluctuations of RA levels to preserve the required concentrations for hindbrain segmentation [231].
In general, limiting RA activity results in shortening or loss of the posterior hindbrain and caudal expansion of its anterior part. Yet, the severity of these phenotypes depends on the level in which RA signaling was modified. Complete vitamin A deficiency (VAD) was initially studied in quail embryos, causing a complete loss of r4–r8, and a posterior expansion of r3 [183]. This morphological distortion was combined with loss of posteriorly expressed genes, such as hoxb1, FGF3, and mafB, together with caudal expansion of the r3 stripe of krox20. In rats, VAD was accomplished by a gradual, rather than total reduction of vitamin A [187, 188]. These embryos displayed a correlation between titrated reduced doses of RA and a progressive expansion of anterior rhombomeres at the expense of posterior ones. Reduction of RA-signaling levels was also achieved by modulation of its metabolism. In Xenopus, ectopic Cyp26 protein anteriorized the posterior hindbrain in a dose-dependent manner [239]. Yet, only a partial duplication of anterior rhombomeres was observed in the posterior hindbrain. Similar intermediate effects were found in two zebrafish lines, neckless and no-fin, where the raldh2 gene is mutated [200, 240]. Attenuation of RARs was another means to inhibit RA. This was performed by the generation of RAR null mice or zebrafish, or by expressing dominant negative RARs in mice, chick and frog [32, 74, 181, 183, 203, 241–243]. In most cases, expression of posterior genes was delayed and formation of the posterior rhombomeres was disrupted but not lost. Notably, stronger defects were observed upon combination of different RAR mutations or inhibitors [185–203]. Finally, the less severe hindbrain phenotypes were achieved by manipulating RA-binding sites on target genes. For example, mutations of RAREs in the 3′ ends of hoxa1 and hoxb1 delayed their initial rhombomeric expression, but this was fully restored later in development [78, 79, 213]. These studies demonstrate the complex level of regulation of RA signaling in the hindbrain, which combines different sources for RA synthesis and degradation, the activity of several RA receptor subtypes and the binding to RAREs on different hindbrain genes to deliver the correct patterning outcome.
Cross-talk between hindbrain signaling pathways: cooperative or antagonistic activities?
As mentioned, the role of RA, FGF and Wnt in regulating the expression of key hindbrain genes is well documented. Yet, there are marked differences between these morphogens in terms of their expression/activity patterns. Whereas RA signal spans the entire hindbrain in a gradient manner [180], expression of FGFs and Wnts is more limited: FGF3/FGF8 is expressed in r4 (zebrafish, frog) or r2/r4/r6 (chick and mice) [133, 135, 136, 139, 141]. Wnt8a and Wnt8b are expressed in r4 and r3/r5, respectively [244, 245]. The neural patterning of Wnt3a and Wnt8a ligands, as well as FGFs, is also expressed in the dorsal–lateral mesodermal regions similar to RA, where they are secreted to induce posterior cell fates in adjacent neural tissue (Fig. 3) [44]. How the localized FGF and Wnt signals and the graded RA pathway are integrated to govern the positional identity of the hindbrain is not well understood.
Fig. 3.
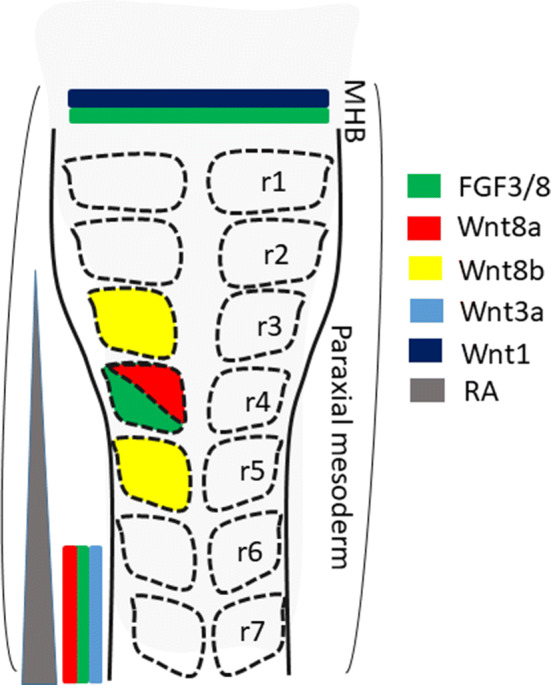
A summary of the expression domains of different signaling factors in the hindbrain. The spatial organization of the multiple signaling factors triggering hindbrain induction and patterning without temporal separation is shown. Before rhombomere specification, FGF3/8 and canonical Wnt 3a/8a are expressed in the paraxial mesoderm adjacent to the caudal hindbrain. RA is also synthesized in the same domain and acts in a caudal–rostral gradient along the hindbrain. Slightly later, FGF3/8 and Wnt8a are expressed in r4, whereas Wnt8b is expressed in r3/r5. FGF3/8 and Wnt1/3a are also secreted from the mid–hindbrain boundary
Signaling cooperativity
Interactions mediating the expression of all these signals have been demonstrated.
For example, in VAD-quail embryos or in mice carrying a null mutation for the raldh2 gene, FGF3 expression is lost [183, 246]. This observation indicated that the induction and/or maintenance of FGFs in hindbrain segments involve RA signaling. Moreover, FGF3-null mice have reduced wnt8a expression in r4, whereas excess FGF8 in hindbrain explants induces ectopic wnt8a expression. These results suggest that the wnt8a expression in the hindbrain is mediated by FGF signaling [247]. Further support for the Wnt–FGF cross-talk was shown by studying sprouty1/2 genes. Sprouty proteins are negative regulators of the FGF signaling pathway and restrict Wnt8a to r4. Mice mutated for Spry1/2 showed expansion of Wnt8a into additional rhombomeres, probably via releasing the negative regulation of FGF signaling [248]. However, no gross hindbrain malformations appeared in the Spry1/2 mutants, thus the interactive roles of FGF and Wnt in regulating hindbrain induction and patterning are still unclear.
Additional evidence suggests that these morphogens cooperate to induce the hindbrain. In Xenopus embryos and explants, caudalizing canonical Wnt activity regulated early neural FGF3/8 gene expression [35, 57]. Wnt3a protein activates expression of the hindbrain-specific meis3 gene [44]. Meis3 protein then directly activates FGF3/8 gene expression [57, 151]. In Meis3 knockdown embryos, FGF3 expression in r4 is eliminated and hindbrain cell fates are lost [57]. Ectopic FGF ligand expression can partially rescue Meis3 morphant phenotypes in Xenopus embryos [57]. Yet, neither canonical Wnt nor Meis3 protein activities efficiently caudalize the CNS, when downstream FGF signaling is compromised [24, 35, 151]. Moreover, Wnt activation of Meis3 protein modulates expression of RA direct target genes, such as hoxd1 [30, 44, 62]. Hoxd1 is a direct target of RA, Wnt and the Meis3 protein [30, 44, 62, 249, 250]. Meis3/RA act synergistically to optimize hoxd1 gene expression to promote correct hindbrain formation [62].
Combined FGF and RA signaling activity was also found to induce hindbrain cell fate in the chick; exposure of caudal epiblast cells to FGFs led to the induction of krox20 only when caudal paraxial mesoderm was also present [134]. This study suggested that a combination of FGF and paraxial mesoderm-caudalizing activity acts directly on epiblast cells to induce hindbrain character. This activity was later identified as RA [251]. The combined activity of RA and FGFs is also demonstrated in setting up the hindbrain–spinal cord border, as marked by cdx1/4 expression. In zebrafish, loss of cdx1/4 resulted in caudal expansion of hindbrain genes. This phenotype could only be rescued when both FGF and RA were inhibited, suggesting that both signals act together to coordinate the formation of the border between the hindbrain and spinal cord [252]. These observations show that CDX proteins modify the cell competence to respond to both FGFs and RA in the posterior neural tissues, including the hindbrain. Hence, both signals are required to define the precise hindbrain–spinal cord boundary.
At slightly later stages, rhombomere-derived FGFs are required for the specification of r3 and r5, as inhibition of FGF downregulates krox20 expression in both segments [108, 135, 136, 142, 143]. Notably, a recent study in zebrafish elucidated a novel mechanism through which the restriction of krox20 expression to r3/5 is also mediated by different levels of RA in odd versus even segments. This is mediated by the higher expression and activity of Cyp26b in r4, so that Krox20 expressing cells that cross into the r4 territory are exposed to higher levels of RA than in r3/r5, which results in the downregulation of krox20, and the upregulation of hoxb1 gene expression [229]. Yet, although defined RA levels are also necessary for krox20 expression, RA activity is more limited than FGF3; attenuation of RA signaling results in loss of the more posterior r5 stripe of krox20, whereas it remains intact in r3 or even expands posteriorly, depending on the degree of RA modulation [108, 183, 188]. Nevertheless, synergism between RA and FGF is required to induce krox20 through the induction of downstream mediators such as Vhnf1 [108].
From gastrulation stages and onwards, FGF and Wnt are also expressed in the posterior mesoderm, together with RA. All these signals were found to induce posterior and suppress anterior expression of genes involved in rhombomere specification, as detailed in the previous sections. The mesodermal FGFs and Wnts prevent the upregulation of cyp26 in the posterior hindbrain [23, 194] and also maintain raldh2 expression in this domain. RA, on the other hand, was found to induce the expression of wnt1/3A and the downstream FGF target gene, pea3 [253]. In this way, posterior FGF and Wnt are involved in maintaining the hindbrain RA gradient, whereas RA positively regulates the expression/activity of these morphogens in more posterior domains. The tight connection of FGF and Wnt signaling with RA maintains stable and adaptable RA concentration levels along the hindbrain AP axis [180, 192].
Opposing signaling activities
One of the initial indications supporting a negative cross-talk between hindbrain signals came again from VAD-quail embryos, where the posterior hindbrain was abolished, but anterior regionalization occurred normally [183, 184]. The morphogen regulating patterning in the anterior hindbrain is the mid–hindbrain boundary (MHB)-derived FGF8 protein [212, 254–256]. Grafting of MHB or adding FGF8 beads into the posterior hindbrain induced ectopic expression of genes that are normally expressed anteriorly, together with downregulation of posterior genes, like hoxb1. These results were later confirmed by pharmacologic or genetic disruption of RA in other species, where the posterior hindbrain became anteriorized [179, 242, 257, 258]. Conversely, either inhibition of FGF8 or enhancement of RA activity led to the expansion of anterior Hox genes [256, 259]. Knockout of FGF8 also resulted in midbrain Wnt1 expression expanding posteriorly into the isthmus and cerebellum [260]. These and other studies indicate that posterior RA and anterior FGF8 act in opposing manners to specify Hox-positive/negative domains to define the AP borders of the hindbrain.
Intriguingly, these antagonistic FGF8 and RA activities are similar to those seen in the posterior end of the embryo; FGF3/8 are expressed in neuro-mesodermal progenitor cells at the tail and act in a posteriorhigh–anteriorlow gradient to inhibit their differentiation into mesoderm or ectoderm lineages. RA, which is produced in the more rostral paraxial mesoderm, acts in an opposite anteriorhigh–posteriorlow gradient to trigger the differentiation of the progenitors and to induce Hox gene distribution [134, 261–263]. These observations strongly suggest that both signals act in the same direction to pattern the posterior hindbrain, but their interaction is antagonistic in the most anterior or posterior positions along the neural tube. Further support for this conclusion comes from a study where the distinct effects of FGF and RA were examined on different members of the HoxB group [214]. The anterior expression border of hoxb4, which normally lies in r6/7, was expanded anteriorly upon exogenous application of RA or by grafting posterior mesoderm adjacent to anterior rhombomeres. However, the expression pattern of the more caudal gene, hoxb9, was not modified upon manipulating RA signaling. Conversely, application or inhibition of FGF did not change the expression pattern of hoxb4 but led to a dramatic expansion or loss of hoxb9 gene expression, respectively.
The complex cross-talk between RA, FGF and Wnt is also evident in the positioning of the border between the hindbrain and the spinal cord, as marked by the anterior expression of cdx1/4 at the level of somite 3. In zebrafish, loss of cdx1/4 causes a caudal shift of the hindbrain–spinal cord border via posterior expansion of hindbrain genes (such as hoxb4), and caudalization of spinal cord markers, such as hoxb8 [252, 264]. Hence, the spatial regulation of cdx1/4 expression is crucial for positioning the hindbrain–spinal cord border region. Analyzing the regulatory role of RA and FGF revealed that in RA-deficient embryos, cdx4 expression shifts dorsally while in FGF-deficient embryos cdx4 expression shifts caudally. Yet, in embryos lacking both RA and FGF signals, the shift in cdx4 axial expression is rescued and it is aligned at the level of somite 3, indicating that FGF negatively modulated RA activity in regulating cdx4 expression [252, 265, 266]. Thus, for hindbrain patterning, the FGF and RA pathways have additive functions, but with respect to the axial position of the hindbrain/spinal cord border region, they may be antagonistic. In this way, FGF and RA interactions are different in each process: additive in hindbrain patterning, antagonistic in hindbrain size specification and epistatic in neural-mesodermal tissue alignment.
In Wnt-deficient embryos, cdx4 expression shifts caudally, but the simultaneous loss of both Wnt and RA activities results in a more severe caudal shift of cdx4 gene expression, in comparison to solely inhibiting Wnt signaling, indicating that Wnt and RA act together to regulate cdx4 axial positioning in the hindbrain–spinal cord border [170, 215, 267, 268]. These studies clearly demonstrate the multifaceted network of morphogen regulation that is differentially activated or inhibited along the hindbrain AP axis. In the future, it will be important to elucidate in greater detail the spatial and temporal dynamics of these regulatory signaling pathways to fully understand how their joint activities orchestrate rhombomere specification.
Acknowledgements
We wish to thank Dr. Yuval Peretz for his help with the illustrations. DF was supported by grants from the Israel Science Foundation (ISF, 658/15) and the Israel Cancer Research Fund (ICRF). DSD was supported by grants from the ISF (1515/16), The Chief Scientist Office of the Ministry of Health, Israel (3-0000-15441) and United States–Israel Binational Science Foundation (2015087).
Footnotes
Dale Frank and Dalit Sela-Donenfeld have contributed equally to this work.
Contributor Information
Dale Frank, Email: dale@technion.ac.il.
Dalit Sela-Donenfeld, Email: dalit.seladon@mail.huji.ac.il.
References
- 1.Shoja MM, Johal J, Oakes WJ, Tubbs RS. Embryology and pathophysiology of the Chiari I and II malformations: a comprehensive review. Clin Anat. 2018;31:202–215. doi: 10.1002/ca.22939. [DOI] [PubMed] [Google Scholar]
- 2.Shoja MM, Ramdhan R, Jensen CJ, Chern JJ, Oakes WJ, Tubbs RS. Embryology of the craniocervical junction and posterior cranial fossa, part II: embryogenesis of the hindbrain. Clin Anat. 2018;31:488–500. doi: 10.1002/ca.23048. [DOI] [PubMed] [Google Scholar]
- 3.Spemann H, Mangold H. Uber Induktion von Embryonalanlagen durch Implantation artfremder Organisatoren. Arch mikr Anat Entw mech. 1924;100:599–638. [Google Scholar]
- 4.Oppenheimer JM. Transplantation experiments on developing teleosts (Fundulus and Perca) J Exp Biol. 1936;72:409–437. [Google Scholar]
- 5.Waddington CH. Induction by the primitive streak and its derivatives in the chick. J Exp Biol. 1933;10:38–46. [Google Scholar]
- 6.Beddington RS, Robertson EJ. Axis development and early asymmetry in mammals. Cell. 1999;96:195–209. doi: 10.1016/S0092-8674(00)80560-7. [DOI] [PubMed] [Google Scholar]
- 7.Nieuwkoop PD. Activation and organization of the central nervous system in amphibians. III Synthesis of a new working hypothesis. J Exp Zool. 1952;120:83–108. doi: 10.1002/jez.1401200104. [DOI] [Google Scholar]
- 8.Eyal-Giladi H. Dynamic aspects of neural induction. Arc Biol. 1954;65:180–259. [PubMed] [Google Scholar]
- 9.Toivonen S, Saxen L. Morphogenetic interaction of presumptive neural and mesodermal cells mixed in different ratios. Science. 1968;159:539–540. doi: 10.1126/science.159.3814.539. [DOI] [PubMed] [Google Scholar]
- 10.Lamb TM, Knecht AK, Smith WC, Stachel SE, Economides AN, Stahl N, Yancopolous GD, Harland RM. Neural induction by the secreted polypeptide noggin. Science. 1993;262:713–718. doi: 10.1126/science.8235591. [DOI] [PubMed] [Google Scholar]
- 11.Hemmati-Brivanlou A, Kelly OG, Melton DA. Follistatin, an antagonist of activin, is expressed in the Spemann organizer and displays direct neuralizing activity. Cell. 1994;77:283–295. doi: 10.1016/0092-8674(94)90320-4. [DOI] [PubMed] [Google Scholar]
- 12.Sasai Y, Lu B, Steinbeisser H, De Robertis EM. Regulation of neural induction by the Chd and Bmp-4 antagonistic patterning signals in Xenopus. Nature. 1995;376:333–336. doi: 10.1038/376333a0. [DOI] [PubMed] [Google Scholar]
- 13.Re’em-Kalma Y, Lamb T, Frank D. Competition between noggin and bone morphogenetic protein 4 activities may regulate dorsalization during Xenopus development. Proc Natl Acad Sci USA. 1995;92:12141–12145. doi: 10.1073/pnas.92.26.12141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fainsod A, Deissler K, Yelin R, Marom K, Epstein M, Pillemer G, Steinbeisser H, Blum M. The dorsalizing and neural inducing gene follistatin is an antagonist of BMP-4. Mech Dev. 1997;63:39–50. doi: 10.1016/S0925-4773(97)00673-4. [DOI] [PubMed] [Google Scholar]
- 15.Zimmerman LB, De Jesus-Escobar JM, Harland RM. The Spemann organizer signal noggin binds and inactivates bone morphogenetic protein 4. Cell. 1996;86:599–606. doi: 10.1016/S0092-8674(00)80133-6. [DOI] [PubMed] [Google Scholar]
- 16.Piccolo S, Sasai Y, Lu B, De Robertis EM. Dorsoventral patterning in Xenopus: inhibition of ventral signals by direct binding of chordin to BMP-4. Cell. 1996;86:589–598. doi: 10.1016/S0092-8674(00)80132-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hemmati-Brivanlou A, Melton DA. Inhibition of activin receptor signaling promotes neuralization in Xenopus. Cell. 1994;77:273–281. doi: 10.1016/0092-8674(94)90319-0. [DOI] [PubMed] [Google Scholar]
- 18.Wilson PA, Hemmati-Brivanlou A. Induction of epidermis and inhibition of neural fate by Bmp-4. Nature. 1995;376:331–333. doi: 10.1038/376331a0. [DOI] [PubMed] [Google Scholar]
- 19.Wilson SI, Rydstrom A, Trimborn T, Willert K, Nusse R, Jessell TM, Edlund T. The status of Wnt signalling regulates neural and epidermal fates in the chick embryo. Nature. 2001;411:325–330. doi: 10.1038/35077115. [DOI] [PubMed] [Google Scholar]
- 20.Cox WG, Hemmati-Brivanlou A. Caudalization of neural fate by tissue recombination and bFGF. Development. 1995;121:4349–4358. doi: 10.1242/dev.121.12.4349. [DOI] [PubMed] [Google Scholar]
- 21.Lamb TM, Harland RM. Fibroblast growth factor is a direct neural inducer, which combined with noggin generates anterior-posterior neural pattern. Development. 1995;121:3627–3636. doi: 10.1242/dev.121.11.3627. [DOI] [PubMed] [Google Scholar]
- 22.Holowacz T, Sokol S. FGF is required for posterior neural patterning but not for neural induction. Dev Biol. 1999;205:296–308. doi: 10.1006/dbio.1998.9108. [DOI] [PubMed] [Google Scholar]
- 23.Kudoh T, Wilson SW, Dawid IB. Distinct roles for Fgf, Wnt and retinoic acid in posteriorizing the neural ectoderm. Development. 2002;129:4335–4346. doi: 10.1242/dev.129.18.4335. [DOI] [PubMed] [Google Scholar]
- 24.Ribisi S, Mariani FV, Aamar E, Lamb TM, Frank D, Harland RM. Ras-mediated FGF signaling is required for the formation of posterior but not anterior neural tissue in Xenopus laevis. Dev Biol. 2000;227:183–196. doi: 10.1006/dbio.2000.9889. [DOI] [PubMed] [Google Scholar]
- 25.Fletcher RB, Baker JC, Harland RM. FGF8 spliceforms mediate early mesoderm and posterior neural tissue formation in Xenopus. Development. 2006;133:1703–1714. doi: 10.1242/dev.02342. [DOI] [PubMed] [Google Scholar]
- 26.Durston AJ, Timmermans JP, Hage WJ, Hendriks HF, de Vries NJ, Heideveld M, Nieuwkoop PD. Retinoic acid causes an anteroposterior transformation in the developing central nervous system. Nature. 1989;340:140–144. doi: 10.1038/340140a0. [DOI] [PubMed] [Google Scholar]
- 27.Sive HL, Draper BW, Harland RM, Weintraub H. Identification of a retinoic acid-sensitive period during primary axis formation in Xenopus laevis. Genes Dev. 1990;4:932–942. doi: 10.1101/gad.4.6.932. [DOI] [PubMed] [Google Scholar]
- 28.Sharpe CR. Retinoic acid can mimic endogenous signals involved in transformation of the Xenopus nervous system. Neuron. 1991;7:239–247. doi: 10.1016/0896-6273(91)90262-X. [DOI] [PubMed] [Google Scholar]
- 29.Ruiz i Altaba A, Jessell TM. Retinoic acid modifies the pattern of cell differentiation in the central nervous system of neurula stage Xenopus embryos. Development. 1991;112:945–958. doi: 10.1242/dev.112.4.945. [DOI] [PubMed] [Google Scholar]
- 30.Kolm PJ, Sive H. Hindbrain patterning requires retinoid signaling. Dev Biol. 1995;192:1–16. doi: 10.1006/dbio.1997.8754. [DOI] [PubMed] [Google Scholar]
- 31.Papalopulu N, Kintner C. A posteriorising factor, retinoic acid, reveals that anteroposterior patterning controls the timing of neuronal differentiation in Xenopus neuroectoderm. Development. 1996;122:3409–3418. doi: 10.1242/dev.122.11.3409. [DOI] [PubMed] [Google Scholar]
- 32.Blumberg B, Bolado J, Jr, Moreno TA, Kintner C, Evans RM, Papalopulu N. An essential role for retinoid signaling in anteroposterior neural patterning. Development. 1997;124:373–379. doi: 10.1242/dev.124.2.373. [DOI] [PubMed] [Google Scholar]
- 33.McGrew LL, Lai CJ, Moon RT. Specification of the anteroposterior neural axis through synergistic interaction of the Wnt signaling cascade with noggin and follistatin. Dev Biol. 1995;172:337–342. doi: 10.1006/dbio.1995.0027. [DOI] [PubMed] [Google Scholar]
- 34.McGrew LL, Hoppler S, Moon RT. Wnt and FGF pathways cooperatively pattern anteroposterior neural ectoderm in Xenopus. Mech Dev. 1997;69:105–114. doi: 10.1016/S0925-4773(97)00160-3. [DOI] [PubMed] [Google Scholar]
- 35.Domingos PM, Itasaki N, Jones CM, Mercurio S, Sargent MG, Smith JC, Krumlauf R. The Wnt/beta-catenin pathway posteriorizes neural tissue in Xenopus by an indirect mechanism requiring FGF signaling. Dev Biol. 2001;239:148–160. doi: 10.1006/dbio.2001.0431. [DOI] [PubMed] [Google Scholar]
- 36.Schulte D, Frank D. TALE transcription factors during early development of the vertebrate brain and eye. Dev Dyn. 2014;24:99–116. doi: 10.1002/dvdy.24030. [DOI] [PubMed] [Google Scholar]
- 37.Krumlauf R. Hox genes and the hindbrain: a study in segments. Curr Top Dev Biol. 2016;116:581–596. doi: 10.1016/bs.ctdb.2015.12.011. [DOI] [PubMed] [Google Scholar]
- 38.Hernandez-Miranda LR, Müller T, Birchmeier C. The dorsal spinal cord and hindbrain: from developmental mechanisms to functional circuits. Dev Biol. 2017;432:34–42. doi: 10.1016/j.ydbio.2016.10.008. [DOI] [PubMed] [Google Scholar]
- 39.Parker HJ, Krumlauf R. Segmental arithmetic: summing up the Hox gene regulatory network for hindbrain development in chordates. WIRE Dev Biol. 2017 doi: 10.1002/wdev.286. [DOI] [PubMed] [Google Scholar]
- 40.Burglin T. Analysis of TALE superclass homeobox genes (MEIS, PBC, KNOX, Iroquois, TGIF) reveals a novel domain conserved between plants and animals. Nucleic Acids Res. 1997;25:4173–4180. doi: 10.1093/nar/25.21.4173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pöpperl H, Rikhof H, Chang H, Haffter P, Kimmel CB, Moens CB. lazarus is a novel pbx gene that globally mediates hox gene function in zebrafish. Mol Cell. 2000;6:255–267. doi: 10.1016/S1097-2765(00)00027-7. [DOI] [PubMed] [Google Scholar]
- 42.Dibner C, Elias S, Frank D. XMeis3 protein activity is required for proper hindbrain patterning in Xenopus laevis embryos. Development. 2001;128:3415–3426. doi: 10.1242/dev.128.18.3415. [DOI] [PubMed] [Google Scholar]
- 43.Waskiewicz AJ, Rikhof HA, Hernandez RE, Moens CB. Zebrafish Meis functions to stabilize Pbx proteins and regulate hindbrain patterning. Development. 2001;128:4139–4151. doi: 10.1242/dev.128.21.4139. [DOI] [PubMed] [Google Scholar]
- 44.Elkouby YM, Elias S, Casey ES, Blythe SA, Tsabar N, Klein PS, Root H, Liu KJ, Frank D. Mesodermal Wnt signaling organizes the neural plate via Meis3. Development. 2010;137:1531–1541. doi: 10.1242/dev.044750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rohrschneider MR, Elsen GE, Prince VE. Zebrafish Hoxb1a regulates multiple downstream genes including prickle1b. Dev Biol. 2007;309:358–372. doi: 10.1016/j.ydbio.2007.06.012. [DOI] [PubMed] [Google Scholar]
- 46.Choe S, Zhang X, Hirsch N, StraubhaarJ Sagerstrom CG. A screen for hoxb1-regulated genes identifies ppp1r14al as a regulator of the rhombomere 4 Fgf-signaling center. Dev Biol. 2011;358:356–367. doi: 10.1016/j.ydbio.2011.05.676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Salzberg A, Elias S, Nachaliel N, Bonstein L, Henig C, Frank D. A Meis family protein caudalizes neural cell fates in Xenopus. Mech Dev. 1999;80:3–13. doi: 10.1016/S0925-4773(98)00187-7. [DOI] [PubMed] [Google Scholar]
- 48.Vlachakis N, Choe SK, Sagerstrom CG. Meis3 synergizes with Pbx4 and Hoxb1b in promoting hindbrain fates in the zebrafish. Development. 2001;128:1299–1312. doi: 10.1242/dev.128.8.1299. [DOI] [PubMed] [Google Scholar]
- 49.Choe S, Vlachakis N, Sagerstrom CG. Meis family proteins are required for hindbrain development in the zebrafish. Development. 2002;129:585–595. doi: 10.1242/dev.129.3.585. [DOI] [PubMed] [Google Scholar]
- 50.Maeda R, Ishimura A, Mood K, Park EK, Buchberg AM, Daar IO. Xpbx1b and Xmeis1b play a collaborative role in hindbrain and neural crest gene expression in Xenopus embryos. Proc Natl Acad Sci USA. 2002;99:5448–5453. doi: 10.1073/pnas.082654899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Oulad-Abdelghani M, Chazaud C, Bouillet P, Sapin V, Chambon P, Dolle P. Meis2, a novel mouse Pbx-related homeobox gene induced by retinoic acid during differentiation of P19 embryonal carcinoma cells. Dev Dyn. 1997;210:173–183. doi: 10.1002/(SICI)1097-0177(199710)210:2<173::AID-AJA9>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 52.Cecconi F, Proetzel G, Alvarez-Bolado G, Jay D, Gruss P. Expression of Meis2, a Knotted-related murine homeobox gene, indicates a role in the differentiation of the forebrain and the somatic mesoderm. Dev Dyn. 1997;210:184–190. doi: 10.1002/(SICI)1097-0177(199710)210:2<184::AID-AJA10>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 53.Maeda R, Mood K, Jones TL, Aruga J, Buchberg AM, Daar IO. Xmeis1, a protooncogene involved in specifying neural crest cell fate in Xenopus embryos. Oncogene. 2001;20:1329–1342. doi: 10.1038/sj.onc.1204250. [DOI] [PubMed] [Google Scholar]
- 54.Zerucha T, Prince VE. Cloning and developmental expression of a zebrafish meis2 homeobox gene. Mech Dev. 2001;102:247–250. doi: 10.1016/S0925-4773(01)00299-4. [DOI] [PubMed] [Google Scholar]
- 55.Bumsted-O’Brien KM, Hendrickson A, Haverkamp S, Ashery-Padan R, Schulte D. Expression of the homeodomain transcription factor Meis2 in the embryonic and postnatal retina. J Comp Neurol. 2007;505:58–72. doi: 10.1002/cne.21458. [DOI] [PubMed] [Google Scholar]
- 56.Sanchez-Guardado LO, Irimia M, Sanchez-Arrones L, Burguera D, Rodrıguez-Gallardo L, Garcia-Fernandez J, Puelles L, Ferran JL, Hidalgo-Sanchez M. Distinct and redundant expression and transcriptional diversity of MEIS gene paralogs during chicken development. Dev Dyn. 2011;240:1475–1492. doi: 10.1002/dvdy.22621. [DOI] [PubMed] [Google Scholar]
- 57.Gutkovich YE, Ofir R, Elkouby YM, Dibner C, Gefen A, Elias S, Frank D. Xenopus Meis3 protein lies at a nexus downstream to Zic1 and Pax3 proteins, regulating multiple cell-fates during early nervous system development. Dev Biol. 2010;338:50–62. doi: 10.1016/j.ydbio.2009.11.024. [DOI] [PubMed] [Google Scholar]
- 58.Ferretti E, Cambronero F, Tumpel S, Longobardi E, Wiedemann LM, Blasi F, Krumlauf R. Hoxb1 enhancer and control of rhombomere 4 expression: complex interplay between PREP1- PBX1-HOXB1 binding sites. Mol Cell Biol. 2005;25:8541–8552. doi: 10.1128/MCB.25.19.8541-8552.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ferretti E, Marshall H, Popperl H, Maconochie M, Krumlauf R, Blasi F. Segmental expression of Hoxb2 in r4 requires two separate sites that integrate cooperative interactions between Prep1, Pbx and Hox proteins. Development. 2000;127:155–166. doi: 10.1242/dev.127.1.155. [DOI] [PubMed] [Google Scholar]
- 60.Maconochie MK, Nonchev S, Studer M, Chan SK, Popperl H, Sham MH, Mann RS, Krumlauf R. Cross-regulation in the mouse HoxB complex: the expression of Hoxb2 in rhombomere 4 is regulated by Hoxb1. Genes Dev. 1997;11:1885–1895. doi: 10.1101/gad.11.14.1885. [DOI] [PubMed] [Google Scholar]
- 61.Jacobs Y, Schnabel CA, Cleary ML. Trimeric association of Hox and TALE homeodomain proteins mediates Hoxb2 hindbrain enhancer activity. Mol Cell Biol. 1999;19:5134–5142. doi: 10.1128/MCB.19.7.5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dibner C, Elias S, Ofir R, Souopgui J, Kolm PJ, Sive H, Pieler T, Frank D. The Meis3 protein and retinoid signaling interact to pattern the Xenopus hindbrain. Dev Biol. 2004;271:75–86. doi: 10.1016/j.ydbio.2004.02.029. [DOI] [PubMed] [Google Scholar]
- 63.Wassef MA, Chomette D, Pouilhe M, Stedman A, Havis E, Dinh CD, Schneider- Maunoury S, Gilardi-Hebenstreit P, Charnay P, Ghislain J. Rostral hindbrain patterning involves the direct activation of a Krox20 transcriptional enhancer by Hox/Pbx and Meis factors. Development. 2008;135:3369–3378. doi: 10.1242/dev.023614. [DOI] [PubMed] [Google Scholar]
- 64.Stedman A, Lecaudey V, Havis E, Anselme I, Wassef M, Gilardi- Hebenstreit P, Schneider-Maunoury S. A functional interaction between Irx and Meis patterns the anterior hindbrain and activates krox20 expression in rhombomere 3. Dev Biol. 2009;327:566–577. doi: 10.1016/j.ydbio.2008.12.018. [DOI] [PubMed] [Google Scholar]
- 65.Popperl H, Bienz M, Studer M, Chan SK, Aparicio S, Brenner S, Mann RS, Krumlauf R. Segmental expression of Hoxb-1 is controlled by a highly conserved autoregulatory loop dependent upon exd/pbx. Cell. 1995;81:1031–1042. doi: 10.1016/S0092-8674(05)80008-X. [DOI] [PubMed] [Google Scholar]
- 66.Waskiewicz AJ, Rikhof HA, Moens CB. Eliminating zebrafish pbx proteins reveals a hindbrain ground state. Dev Cell. 2002;3:723–733. doi: 10.1016/S1534-5807(02)00319-2. [DOI] [PubMed] [Google Scholar]
- 67.Chan SK, Popperl H, Krumlauf R, Mann RS. An extradenticle-induced conformational change in a HOX protein overcomes an inhibitory function of the conserved hexapeptide motif. EMBO J. 1996;15:2476–2487. doi: 10.1002/j.1460-2075.1996.tb00605.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wilkinson DG, Bhatt S, Cook M, Boncinelli E, Krumlauf R. Segmental expression of Hox-2 homoeobox containing genes in the developing mouse hindbrain. Nature. 1989;341:405–409. doi: 10.1038/341405a0. [DOI] [PubMed] [Google Scholar]
- 69.Sundin OH, Busse HG, Rogers MB, Gudas LJ, Eichele G. Region-specific expression in early chick and mouse embryos of Ghox-lab and Hox 1.6, vertebrate homeobox-containing genes related to Drosophila labial. Development. 1990;108:47–58. doi: 10.1242/dev.108.Supplement.47. [DOI] [PubMed] [Google Scholar]
- 70.Frohman MA, Boyle M, Martin GR. Isolation of the mouse Hox-2.9 gene; analysis of embryonic expression suggests that positional information along the anterior-posterior axis is specified by mesoderm. Development. 1990;110:589–607. doi: 10.1242/dev.110.2.589. [DOI] [PubMed] [Google Scholar]
- 71.Murphy P, Hill RE. Expression of the mouse labial-like homeobox-containing genes, Hox 2.9 and Hox 1.6, during segmentation of the hindbrain. Development. 1991;111:61–74. doi: 10.1242/dev.111.1.61. [DOI] [PubMed] [Google Scholar]
- 72.Frohman MA, Martin GR. Isolation and analysis of embryonic expression of Hox-4.9, a member of the murine labial-like gene family. Mech Dev. 1992;38:55–67. doi: 10.1016/0925-4773(92)90038-L. [DOI] [PubMed] [Google Scholar]
- 73.Godsave S, Dekker EJ, Holling T, Pannese M, Boncinelli E, Durston A. Expression patterns of Hoxb genes in the Xenopus embryo suggest roles in anteroposterior specification of the hindbrain and in dorsoventral patterning of the mesoderm. Dev Biol. 1994;166:465–476. doi: 10.1006/dbio.1994.1330. [DOI] [PubMed] [Google Scholar]
- 74.Kolm PJ, Apekin V, Sive H. Xenopus hindbrain patterning requires retinoid signaling. Dev Biol. 1997;192:1–16. doi: 10.1006/dbio.1997.8754. [DOI] [PubMed] [Google Scholar]
- 75.Rossel M, Capecchi MR. Mice mutant for both Hoxa1 and Hoxb1 show extensive remodeling of the hindbrain and defects in craniofacial development. Development. 1999;126:5027–5040. doi: 10.1242/dev.126.22.5027. [DOI] [PubMed] [Google Scholar]
- 76.McClintock JM, Kheirbek MA, Prince VE. Knockdown of duplicated zebrafish hoxb1 genes reveals distinct roles in hindbrain patterning and a novel mechanism of duplicate gene retention. Development. 2002;129:2339–2354. doi: 10.1242/dev.129.10.2339. [DOI] [PubMed] [Google Scholar]
- 77.McNulty CL, Peres JN, Bardine N, van den Akker WM, Durston AJ. Knockdown of the complete Hox paralogous group 1 leads to dramatic hindbrain and neural crest defects. Development. 2005;132:2861–2871. doi: 10.1242/dev.01872. [DOI] [PubMed] [Google Scholar]
- 78.Gavalas A, Studer M, Lumsden A, Rijli FM, Krumlauf R, Chambon P. Hoxa1 and Hoxb1 synergize in patterning the hindbrain, cranial nerves and second pharyngeal arch. Development. 1998;125:1123–1136. doi: 10.1242/dev.125.6.1123. [DOI] [PubMed] [Google Scholar]
- 79.Studer M, Gavalas A, Marshall H, Ariza-McNaughton L, Rijli FM, Chambon P, Krumlauf R. Genetic interactions between Hoxa1 and Hoxb1 reveal new roles in regulation of early hindbrain patterning. Development. 1998;125:1025–1036. doi: 10.1242/dev.125.6.1025. [DOI] [PubMed] [Google Scholar]
- 80.Remacle S, Abbas L, de Backer O, Pacico N, Gavalas A, Gofflot F, Picard JJ, Rezsohazy R. Loss of function but no gain of function caused by amino acid substitutions in the hexapeptide of Hoxa1 in vivo. Mol Cell Biol. 2004;24:8567–8575. doi: 10.1128/MCB.24.19.8567-8575.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Makki N, Capecchi MR. Identification of novel Hoxa1 downstream targets regulating hindbrain, neural crest and inner ear development. Dev Biol. 2011;357:295–304. doi: 10.1016/j.ydbio.2011.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tischfield MA, Bosley TM, Salih MA, Alorainy IA, Sener EC, Nester MJ, Oystreck DT, Chan WM, Andrews C, Erickson RP, Engle EC. Homozygous HOXA1 mutations disrupt human brainstem, inner ear, cardiovascular and cognitive development. Nat Genet. 2015;37:1035–1037. doi: 10.1038/ng1636. [DOI] [PubMed] [Google Scholar]
- 83.Prince V, Lumsden A. Hoxa-2 expression in normal and transposed rhombomeres: independent regulation in the neural tube and neural crest. Development. 1994;120:911–923. doi: 10.1242/dev.120.4.911. [DOI] [PubMed] [Google Scholar]
- 84.Gavalas A, Davenne M, Lumsden A, Chambon P, Rijli FM. Role of Hoxa-2 in axon pathfinding and rostral hindbrain patterning. Development. 1997;124:3693–3702. doi: 10.1242/dev.124.19.3693. [DOI] [PubMed] [Google Scholar]
- 85.Barrow JR, Stadler HS, Capecchi MR. Roles of Hoxa1 and Hoxa2 in patterning the early hindbrain of the mouse. Development. 2000;127:933–944. doi: 10.1242/dev.127.5.933. [DOI] [PubMed] [Google Scholar]
- 86.Ren S, Angran P, Rijli FM. Targeted insertion results in a rhombomere 2- specific Hoxa2 knockdown and ectopic activation of Hoxa1 expression. Dev Dyn. 2002;225:305–315. doi: 10.1002/dvdy.10171. [DOI] [PubMed] [Google Scholar]
- 87.Oury F, Murakami Y, Renaud J, Pasqualetti M, Charnay P, Ren S, Rijli FM. Hoxa2- and rhombomere-dependent development of the mouse facial somatosensory map. Science. 2006;313:1408–1413. doi: 10.1126/science.1130042. [DOI] [PubMed] [Google Scholar]
- 88.Davenne M, Maconochie MK, Neun R, Pattyn A, Chambon P, Krumlauf R, Rijli FM. Hoxa2 and Hoxb2 control dorsoventral patterns of neuronal development in the rostral hindbrain. Neuron. 1999;22:677–691. doi: 10.1016/S0896-6273(00)80728-X. [DOI] [PubMed] [Google Scholar]
- 89.McEllin JA, Alexander TB, Tümpel S, Wiedemann LM, Krumlauf R. Analyses of fugu hoxa2 genes provide evidence for subfunctionalization of neural crest cell and rhombomere cis-regulatory modules during vertebrate evolution. Dev Biol. 2016;409:530–542. doi: 10.1016/j.ydbio.2015.11.006. [DOI] [PubMed] [Google Scholar]
- 90.Wong EY, Wang XA, Mak SS, Sae-Pang JJ, Ling KW, Fritzsch B, Sham MH. Hoxb3 negatively regulates Hoxb1 expression in mouse hindbrain patterning. Dev Biol. 2011;352:382–392. doi: 10.1016/j.ydbio.2011.02.003. [DOI] [PubMed] [Google Scholar]
- 91.Gaufo GO, Thomas KR, Capecchi MR. Hox3 genes coordinate mechanisms of genetic suppression and activation in the generation of branchial and somatic motoneurons. Development. 2003;130:5191–5201. doi: 10.1242/dev.00730. [DOI] [PubMed] [Google Scholar]
- 92.Horan GS, Ramırez-Solis R, Featherstone MS, Wolgemuth DJ, Bradley A, Behringer RR. Compound mutants for the paralogous hoxa-4, hoxb-4, and hoxd-4 genes show more complete homeotic transformations and a dose-dependent increase in the number of vertebrae transformed. Genes Dev. 1995;9:1667–1677. doi: 10.1101/gad.9.13.1667. [DOI] [PubMed] [Google Scholar]
- 93.De Kumar B, Parker HJ, Paulson A, Parrish ME, Zeitlinger J, Krumlauf R. Hoxa1 targets signaling pathways during neural differentiation of ES cells and mouse embryogenesis. Dev Biol. 2017;432:151–164. doi: 10.1016/j.ydbio.2017.09.033. [DOI] [PubMed] [Google Scholar]
- 94.De Kumar B, Parker HJ, Paulson A, Parrish ME, Pushel I, Singh NP, Zhang Y, Slaughter BD, Unruh JR, Florens L, Zeitlinger J, Krumlauf R. HOXA1 and TALE proteins display cross-regulatory interactions and form a combinatorial binding code on HOXA1 targets. Genome Res. 2017;27:1501–1512. doi: 10.1101/gr.219386.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Choe S, Lu P, Nakamura M, Lee J, Sagerstrom CG. Meis cofactors control HDAC and CBP accessibility at Hox-regulated promoters during zebrafish embryogenesis. Dev Cell. 2009;17:561–567. doi: 10.1016/j.devcel.2009.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Choe SK, Ladam F, Sagerström CG. TALE factors poise promoters for activation by Hox proteins. Dev Cell. 2014;28:203–211. doi: 10.1016/j.devcel.2013.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Grice J, Noyvert B, Doglio L, Elgar G. A simple predictive enhancer syntax for hindbrain patterning is conserved in vertebrate genomes. PLoS One. 2015;10:e0130413. doi: 10.1371/journal.pone.0130413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Tümpel S, Cambronero F, Sims C, Krumlauf R, Wiedemann LM. A regulatory module embedded in the coding region of Hoxa2 controls expression in rhombomere 2. Proc Natl Acad Sci USA. 2008;105:20077–20082. doi: 10.1073/pnas.0806360105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lampe X, Samad OA, Guiguen A, Matis C, Remacle S, Picard JJ, Rijli FM, Rezsohazy R. An ultraconserved Hox-Pbx responsive element resides in the coding sequence of Hoxa2 and is active on rhombomere 4. Nucleic Acids Res. 2008;36:3214–3225. doi: 10.1093/nar/gkn148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gavalas A, Ruhrberg C, Livet J, Henderson CE, Krumlauf R. Neuronal defects in the hindbrain of Hoxa1, Hoxb1 and Hoxb2 mutants reflect regulatory interactions among these Hox genes. Development. 2003;130:5663–5679. doi: 10.1242/dev.00802. [DOI] [PubMed] [Google Scholar]
- 101.Tümpel S, Cambronero F, Ferretti E, Blasi F, Wiedemann LM, Krumlauf R. Expression of Hoxa2 in rhombomere 4 is regulated by a conserved cross-regulatory mechanism dependent upon Hoxb1. Dev Biol. 2007;302:646–660. doi: 10.1016/j.ydbio.2006.10.029. [DOI] [PubMed] [Google Scholar]
- 102.Manzanares M, Bel-Vialar S, Ariza-McNaughton L, Ferretti E, Marshall H, Maconochie MM, Blasi F, Krumlauf R. Independent regulation of initiation and maintenance phases of Hoxa3 expression in the vertebrate hindbrain involve auto- and cross-regulatory mechanisms. Development. 2001;128:3595–3607. doi: 10.1242/dev.128.18.3595. [DOI] [PubMed] [Google Scholar]
- 103.Gould A, Morrison A, Sproat G, White RA, Krumlauf R. Positive crossregulation and enhancer sharing: two mechanisms for specifying overlapping Hox expression patterns. Genes Dev. 1997;11:900–913. doi: 10.1101/gad.11.7.900. [DOI] [PubMed] [Google Scholar]
- 104.Gould A, Itasaki N, Krumlauf R. Initiation of rhombomeric Hoxb4 expression requires induction by somites and a retinoid pathway. Neuron. 1998;21:39–51. doi: 10.1016/S0896-6273(00)80513-9. [DOI] [PubMed] [Google Scholar]
- 105.Serpente P, Tumpel S, Ghyselinck NB, Niederreither K, Wiedemann LM, Dolle P, Chambon P, Krumlauf R, Gould AP. Direct crossregulation between retinoic acid receptor beta and Hox genes during hindbrain segmentation. Development. 2005;132:503–513. doi: 10.1242/dev.01593. [DOI] [PubMed] [Google Scholar]
- 106.Sun Z, Hopkins N. vhnf1, the MODY5 and familial GCKD-associated gene, regulates regional specification of the zebrafish gut, pronephros, and hindbrain. Genes Dev. 2001;15:3217–3229. doi: 10.1101/gad946701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wiellette EL, Sive H. vhnf1 and Fgf signals synergize to specify rhombomere identity in the zebrafish hindbrain. Development. 2003;130:3821–3829. doi: 10.1242/dev.00572. [DOI] [PubMed] [Google Scholar]
- 108.Hernandez RE, Rikhof HA, Bachmann R, Moens CB. vhnf1 integrates global RA patterning and local FGF signals to direct posterior hindbrain development in zebrafish. Development. 2004;131:4511–4520. doi: 10.1242/dev.01297. [DOI] [PubMed] [Google Scholar]
- 109.Kim FA, Sing A, Kaneko T, Bieman M, Stallwood N, Sadl VS, Cordes SP. The vHNF1 homeodomain protein establishes early rhombomere identity by direct regulation of Kreisler expression. Mech Dev. 2005;122:1300–1309. doi: 10.1016/j.mod.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 110.Moens CB, Cordes SP, Giorgianni MW, Barsh GS, Kimmel CB. Equivalence in the genetic control of hindbrain segmentation in fish and mouse. Development. 1998;125:381–391. doi: 10.1242/dev.125.3.381. [DOI] [PubMed] [Google Scholar]
- 111.Voiculescu O, Taillebourg E, Pujades C, Kress C, Buart S, Charnay P, Schneider-Maunoury S. Hindbrain patterning: Krox20 couples segmentation and specification of regional identity. Development. 2001;128:4967–4978. doi: 10.1242/dev.128.24.4967. [DOI] [PubMed] [Google Scholar]
- 112.Aragón F, Vázquez-Echeverría C, Ulloa E, Reber M, Cereghini S, Alsina B, Giraldez F, Pujades C. vHnf1 regulates specification of caudal rhombomere identity in the chick hindbrain. Dev Dyn. 2005;234:567–576. doi: 10.1002/dvdy.20528. [DOI] [PubMed] [Google Scholar]
- 113.Chomette D, Frain M, Cereghini S, Charnay P, Ghislain J. Krox20 hindbrain cis-regulatory landscape: interplay between multiple long-range initiation and autoregulatory elements. Development. 2006;133:1253–1262. doi: 10.1242/dev.02289. [DOI] [PubMed] [Google Scholar]
- 114.Bouchoucha YX, Reingruber J, Labalette C, Wassef MA, Thierion E, Desmarquet-Trin Dinh C, Holcman D, Gilardi-Hebenstreit P, Charnay P. Dissection of a Krox20 positive feedback loop driving cell fate choices in hindbrain patterning. Mol Syst Biol. 2013;9:690. doi: 10.1038/msb.2013.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Labalette C, Wassef MA, Desmarquet-Trin Dinh C, Bouchoucha YX, Le Men J, Charnay P, Gilardi-Hebenstreit P. Molecular dissection of segment formation in the developing hindbrain. Development. 2015;142:185–195. doi: 10.1242/dev.109652. [DOI] [PubMed] [Google Scholar]
- 116.Thierion E, Le Men J, Collombet S, Hernandez C, Coulpier F, Torbey P, Thomas-Chollier M, Noordermeer D, Charnay P, Gilardi-Hebenstreit P. Krox20 hindbrain regulation incorporates multiple modes of cooperation between cis-acting elements. PLoS Genet. 2017;13:e1006903. doi: 10.1371/journal.pgen.1006903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Bae CJ, Jeong J, Saint-Jeannet JP. A novel function for Egr4 in posterior hindbrain development. Sci Rep. 2015;5:7750. doi: 10.1038/srep07750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Theil T, Frain M, Gilardi-Hebenstreit P, Flenniken A, Charnay P, Wilkinson DG. Segmental expression of the EphA4 (Sek-1) receptor tyrosine kinase in the hindbrain is under direct transcriptional control of Krox-20. Development. 1998;125:443–452. doi: 10.1242/dev.125.3.443. [DOI] [PubMed] [Google Scholar]
- 119.Cooke JE, Kemp HA, Moens CB. EphA4 is required for cell adhesion and rhombomere-boundary formation in the zebrafish. Curr Biol. 2005;15:536–542. doi: 10.1016/j.cub.2005.02.019. [DOI] [PubMed] [Google Scholar]
- 120.Sela-Donenfeld D, Kayam G, Wilkinson DG. Boundary cells regulate a switch in the expression of FGF3 in hindbrain rhombomeres. BMC Dev Biol. 2009 doi: 10.1186/1471-213x-9-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Mechta-Grigoriou F, Garel S, Charnay P. Nab proteins mediate a negative feedback loop controlling Krox-20 activity in the developing hindbrain. Development. 2000;127:119–128. doi: 10.1242/dev.127.1.119. [DOI] [PubMed] [Google Scholar]
- 122.Runko AP, Sagerström CG. Nlz belongs to a family of zinc-finger-containing repressors and controls segmental gene expression in the zebrafish hindbrain. Dev Biol. 2003;262:254–267. doi: 10.1016/S0012-1606(03)00388-9. [DOI] [PubMed] [Google Scholar]
- 123.García-Gutiérrez P, Juárez-Vicente F, Gallardo-Chamizo F, Charnay P, García-Domínguez M. The transcription factor Krox20 is an E3 ligase that sumoylates its Nab coregulators. EMBO Rep. 2011;12:1018–1023. doi: 10.1038/embor.2011.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kayam G, Kohl A, Magen Z, Peretz Y, Weisinger K, Bar A, Novikov O, Brodski C, Sela-Donenfeld D. A novel role for Pax6 in the segmental organization of the hindbrain. Development. 2013;140:2190–2202. doi: 10.1242/dev.089136. [DOI] [PubMed] [Google Scholar]
- 125.Moens CB, Yan YL, Appel B, Force AG, Kimmel CB. Valentino: a zebrafish gene required for normal hindbrain segmentation. Development. 1996;122:3981–3990. doi: 10.1242/dev.122.12.3981. [DOI] [PubMed] [Google Scholar]
- 126.Prince VE, Moens CB, Kimmel CB, Ho RK. Zebrafish hox genes: expression in the hindbrain region of wild-type and mutants of the segmentation gene, valentino. Development. 1998;125:393–406. doi: 10.1242/dev.125.3.393. [DOI] [PubMed] [Google Scholar]
- 127.Cooke J, Moens C, Roth L, Durbin L, Shiomi K, Brennan C, Kimmel C, Wilson S, Holder N. Eph signalling functions downstream of Val to regulate cell sorting and boundary formation in the caudal hindbrain. Development. 2001;128:571–580. doi: 10.1242/dev.128.4.571. [DOI] [PubMed] [Google Scholar]
- 128.Manzanares M, Cordes S, Ariza-McNaughton L, Sadl V, Maruthainar K, Barsh G, Krumlauf R. Conserved and distinct roles of kreisler in regulation of the paralogous Hoxa3 and Hoxb3 genes. Development. 1999;126:759–769. doi: 10.1242/dev.126.4.759. [DOI] [PubMed] [Google Scholar]
- 129.Manzanares M, Nardelli J, Gilardi- Hebenstreit P, Marshall H, Giudicelli F, Martinez-Pastor MT, Krumlauf R, Charnay P. Krox20 and Kreisler co-operate in the transcriptional control of segmental expression of Hoxb3 in the developing hindbrain. EMBO J. 2002;21:365–376. doi: 10.1093/emboj/21.3.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Rao BR. The appearance and extension of neural differentiation tendencies in the neurectoderm of the early chick embryo. Wilhelm Roux Arch Entwickl Mech Org. 1968;160:187–236. doi: 10.1007/BF00572650. [DOI] [PubMed] [Google Scholar]
- 131.Garcia-Martinez V, Alvarez IS, Schoenwolf GC. Locations of the ectodermal and nonectodermal subdivisions of the epiblast at stages 3 and 4 of avian gastrulation and neurulation. J Exp Zool. 1993;267:431–446. doi: 10.1002/jez.1402670409. [DOI] [PubMed] [Google Scholar]
- 132.Niswander L, Martin GR. Fgf-4 expression during gastrulation, myogenesis, limb and tooth development in the mouse. Development. 1992;114:755–768. doi: 10.1242/dev.114.3.755. [DOI] [PubMed] [Google Scholar]
- 133.Mahmood R, Kiefer P, Guthrie S, Dickson C, Mason I. Multiple roles for FGF-3 during cranial neural development in the chicken. Development. 1995;121:1399–1410. doi: 10.1242/dev.121.5.1399. [DOI] [PubMed] [Google Scholar]
- 134.Muhr J, Graziano E, Wilson S, Jessell TM, Edlund T. Convergent inductive signals specify midbrain, hindbrain, and spinal cord identity in gastrula stage chick embryos. Neuron. 1999;23:689–702. doi: 10.1016/S0896-6273(01)80028-3. [DOI] [PubMed] [Google Scholar]
- 135.Maves L, Jackman W, Kimmel CB. FGF3 and FGF8 mediate a rhombomere 4 signaling activity in the zebrafish hindbrain. Development. 2002;129:3825–3837. doi: 10.1242/dev.129.16.3825. [DOI] [PubMed] [Google Scholar]
- 136.Walshe J, Maroon H, McGonnell IM, Dickson C, Mason I. Establishment of hindbrain segmental identity requires signaling by FGF3 and FGF8. Curr Biol. 2002;12:1117–1123. doi: 10.1016/S0960-9822(02)00899-0. [DOI] [PubMed] [Google Scholar]
- 137.Wright TJ, Mansour SL. Fgf3 and Fgf10 are required for mouse otic placode induction. Development. 2003;130:3379–3390. doi: 10.1242/dev.00555. [DOI] [PubMed] [Google Scholar]
- 138.Mahmood R, Mason IJ, Morriss-Kay GM. Expression of Fgf-3 in relation to hindbrain segmentation, otic pit position and pharyngeal arch morphology in normal and retinoic acid-exposed mouse embryos. Anat Embryol (Berl) 1996;194:13–22. doi: 10.1007/BF00196311. [DOI] [PubMed] [Google Scholar]
- 139.Lombardo A, Isaacs HV, Slack JM. Expression and functions of FGF-3 in Xenopus development. Int J Dev Biol. 1998;42:1101–1107. [PubMed] [Google Scholar]
- 140.Powles N, Marshall H, Economou A, Chiang C, Murakami A, Dickson C, Krumlauf R, Maconochie M. Regulatory analysis of the mouse Fgf3 gene: control of embryonic expression patterns and dependence upon sonic hedgehog (Shh) signalling. Dev Dyn. 2004;230:44–56. doi: 10.1002/dvdy.20028. [DOI] [PubMed] [Google Scholar]
- 141.Weisinger K, Wilkinson DG, Sela-Donenfeld D. Inhibition of BMPs by follistatin is required for FGF3 expression and segmental patterning of the hindbrain. Dev Biol. 2008;324:213–225. doi: 10.1016/j.ydbio.2008.09.005. [DOI] [PubMed] [Google Scholar]
- 142.Weisinger K, Kayam G, Missulawin-Drillman T, Sela-Donenfeld D. Analysis of expression and function of FGF-MAPK signaling components in the hindbrain reveals a central role for FGF3 in the regulation of Krox20, mediated by Pea3. Dev Biol. 2010;344:881–895. doi: 10.1016/j.ydbio.2010.06.001. [DOI] [PubMed] [Google Scholar]
- 143.Marín F, Charnay P. Hindbrain patterning: FGFs regulate Krox20 and mafB/kr expression in the otic/preotic region. Development. 2000;127:4925–4935. doi: 10.1242/dev.127.22.4925. [DOI] [PubMed] [Google Scholar]
- 144.Weisinger K, Kohl A, Kayam G, Monsonego-Ornan E, Sela-Donenfeld D. Expression of hindbrain boundary markers is regulated by FGF3. Biol Open. 2012;1:67–74. doi: 10.1242/bio.2011032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Guthrie S, Lumsden A. Formation and regeneration of rhombomere boundaries in the developing chick hindbrain. Development. 1991;112:221–229. doi: 10.1242/dev.112.1.221. [DOI] [PubMed] [Google Scholar]
- 146.Martinez S, Geijo E, Sánchez-Vives MV, Puelles L, Gallego R. Reduced junctional permeability at interrhombomeric boundaries. Development. 1992;116:1069–1076. doi: 10.1242/dev.116.4.1069. [DOI] [PubMed] [Google Scholar]
- 147.Heyman I, Faissner A, Lumsden A. Cell and matrix specialisations of rhombomere boundaries. Dev Dyn. 1995;204:301–315. doi: 10.1002/aja.1002040308. [DOI] [PubMed] [Google Scholar]
- 148.Cayuso J, Xu Q, Wilkinson DG. Mechanisms of boundary formation by Eph receptor and ephrin signaling. Dev Biol. 2015;401:122–131. doi: 10.1016/j.ydbio.2014.11.013. [DOI] [PubMed] [Google Scholar]
- 149.Terriente J, Pujades C. Cell segregation in the vertebrate hindbrain: a matter of boundaries. Cell Mol Life Sci. 2015;72:3721–3730. doi: 10.1007/s00018-015-1953-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Peretz Y, Eren N, Kohl A, Hen G, Yaniv K, Weisinger K, Cinnamon Y, Sela-Donenfeld D. A new role of hindbrain boundaries as pools of neural stem/progenitor cells regulated by Sox2. BMC Biol. 2016;14:57. doi: 10.1186/s12915-016-0277-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Aamar E, Frank D. Xenopus Meis3 protein forms a hindbrain inducing center by activating FGF/MAP kinase and PCP pathways. Development. 2004;131:153–163. doi: 10.1242/dev.00905. [DOI] [PubMed] [Google Scholar]
- 152.Maconochie MK, Nonchev S, Manzanares M, Marshall H, Krumlauf R. Differences in Krox20-dependent regulation of Hoxa2 and Hoxb2 during hindbrain development. Dev Biol. 2001;233:468–481. doi: 10.1006/dbio.2001.0197. [DOI] [PubMed] [Google Scholar]
- 153.Lunn JS, Fishwick KJ, Halley PA, Storey KG. A spatial and temporal map of FGF/Erk1/2 activity and response repertoires in the early chick embryo. Dev Biol. 2007;302:536–552. doi: 10.1016/j.ydbio.2006.10.014. [DOI] [PubMed] [Google Scholar]
- 154.Aragon F, Pujades C. FGF signaling controls caudal hindbrain specification through Ras-ERK1/2 pathway. BMC Dev Biol. 2009;9:61. doi: 10.1186/1471-213X-9-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Roelink H, Nusse R. Expression of two members of the Wnt family during mouse development—restricted temporal and spatial patterns in the developing neural tube. Genes Dev. 1991;5:381–388. doi: 10.1101/gad.5.3.381. [DOI] [PubMed] [Google Scholar]
- 156.Molven A, Njolstad PR, Fjose A. Genomic structure and restricted neural expression of the zebrafish wnt-1 (int-1) gene. EMBO J. 1991;10:799–807. doi: 10.1002/j.1460-2075.1991.tb08012.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.McGrew LL, Otte AP, Moon RT. Analysis of Xwnt-4 in embryos of Xenopus laevis: a Wnt family member expressed in the brain and floor plate. Development. 1992;115:463–473. doi: 10.1242/dev.115.2.463. [DOI] [PubMed] [Google Scholar]
- 158.Hume CR, Dodd J. Cwnt-8C: a novel Wnt gene with a potential role in primitive streak formation and hindbrain organization. Development. 1993;119:1147–1160. doi: 10.1242/dev.119.4.1147. [DOI] [PubMed] [Google Scholar]
- 159.Wolda SL, Moody CJ, Moon RT. Overlapping expression of Xwnt-3A and Xwnt-1 in neural tissue of Xenopus laevis embryos. Dev Biol. 1993;155:46–57. doi: 10.1006/dbio.1993.1005. [DOI] [PubMed] [Google Scholar]
- 160.Kelly GM, Lai CJ, Moon RT. Expression of wnt10a in the central nervous system of developing zebrafish. Dev Biol. 1993;158:113–121. doi: 10.1006/dbio.1993.1172. [DOI] [PubMed] [Google Scholar]
- 161.Hollyday M, McMahon JA, McMahon AP. Wnt expression patterns in chick embryo nervous system. Mech Dev. 1995;52:9–25. doi: 10.1016/0925-4773(95)00385-E. [DOI] [PubMed] [Google Scholar]
- 162.Augustine K, Liu ET, Sadler TW. Antisense attenuation of Wnt-1 and Wnt-3a expression in whole embryo culture reveals roles for these genes in craniofacial, spinal cord, and cardiac morphogenesis. Dev Genet. 1993;14:500–520. doi: 10.1002/dvg.1020140611. [DOI] [PubMed] [Google Scholar]
- 163.Augustine KA, Liu ET, Sadler TW. Interactions of Wnt-1 and Wnt-3a are essential for neural tube patterning. Teratology. 1995;51:107–119. doi: 10.1002/tera.1420510209. [DOI] [PubMed] [Google Scholar]
- 164.McMahon AP, Joyner AL, Bradley A, McMahon JA. The midbrain–hindbrain phenotype of Wnt-1-/Wnt-1- mice results from stepwise deletion of engrailed-expressing cells by 9.5 days postcoitum. Cell. 1992;69:581–595. doi: 10.1016/0092-8674(92)90222-X. [DOI] [PubMed] [Google Scholar]
- 165.Liu P, Wakamiya M, Shea MJ, Albrecht U, Behringer RR, Bradley A. Requirement for Wnt3 in vertebrate axis formation. Nat Genet. 1999;22:361–365. doi: 10.1038/11932. [DOI] [PubMed] [Google Scholar]
- 166.Nordstrom U, Jessell TM, Edlund T. Progressive induction of caudal neural character by graded Wnt signaling. Nat Neurosci. 2002;5:525–532. doi: 10.1038/nn0602-854. [DOI] [PubMed] [Google Scholar]
- 167.Nordstrom U, Maier E, Jessell TM, Edlund T. An early role for WNT signaling in specifying neural patterns of Cdx and Hox gene expression and motor neuron subtype identity. PLoS Biol. 2006;4:e252. doi: 10.1371/journal.pbio.0040252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Lekven AC, Thorpe CJ, Waxman JS, Moon RT. Zebrafish wnt8 encodes two wnt8 proteins on a bicistronic transcript and is required for mesoderm and neurectoderm patterning. Dev Cell. 2001;1:103–114. doi: 10.1016/S1534-5807(01)00007-7. [DOI] [PubMed] [Google Scholar]
- 169.Erter CE, Wilm TP, Basler N, Wright CV, Solnica Krezel L. Wnt8 is required in lateral mesendodermal precursors for neural posteriorization in vivo. Development. 2001;128:3571–3583. doi: 10.1242/dev.128.18.3571. [DOI] [PubMed] [Google Scholar]
- 170.Shimizu T, Bae YK, Muraoka O, Hibi M. Interaction of Wnt and caudal-related genes in zebrafish posterior body formation. Dev Biol. 2005;279:125–141. doi: 10.1016/j.ydbio.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 171.Lekven AC, Buckles GR, Kostakis N, Moon RT. Wnt1 and wnt10b function redundantly at the zebrafish midbrain-hindbrain boundary. Dev Biol. 2003;254:172–187. doi: 10.1016/S0012-1606(02)00044-1. [DOI] [PubMed] [Google Scholar]
- 172.Riley BB, Chiang MY, Storch EM, Heck R, Buckles GR, Lekven AC. Rhombomere boundaries are Wnt signaling centers that regulate metameric patterning in the zebrafish hindbrain. Dev Dyn. 2004;231:278–291. doi: 10.1002/dvdy.20133. [DOI] [PubMed] [Google Scholar]
- 173.Kiecker C, Niehrs C. A morphogen gradient of Wnt/beta-catenin signaling regulates anteroposterior neural patterning in Xenopus. Development. 2001;128:4189–4201. doi: 10.1242/dev.128.21.4189. [DOI] [PubMed] [Google Scholar]
- 174.Monsoro-Burq AH, Wang E, Harland RM. Msx1 and Pax3 cooperate to mediate FGF8 and WNT signals during Xenopus neural crest induction. Dev Cell. 2005;8:167–178. doi: 10.1016/j.devcel.2004.12.017. [DOI] [PubMed] [Google Scholar]
- 175.Wu J, Yang J, Klein PS. Neural crest induction by the canonical Wnt pathway can be dissociated from anterior–posterior neural patterning in Xenopus. Dev Biol. 2005;279:220–232. doi: 10.1016/j.ydbio.2004.12.016. [DOI] [PubMed] [Google Scholar]
- 176.Li B, Kuriyama S, Moreno M, Mayor R. The posteriorizing gene Gbx2 is a direct target of Wnt signaling and the earliest factor in neural crest induction. Development. 2009;136:3267–3278. doi: 10.1242/dev.036954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Kim CH, Oda T, Itoh M, Jiang D, Artinger KB, Chandrasekharappa SC, Driever W, Chitnis AB. Repressor activity of Headless/Tcf3 is essential for vertebrate head formation. Nature. 2000;407:913–916. doi: 10.1038/35038097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.MacDonald BT, Adamska M, Meisler MH. Hypomorphic expression of Dkk1 in the doubleridge mouse: dose dependence and compensatory interactions with Lrp6. Development. 2004;131:2543–2552. doi: 10.1242/dev.01126. [DOI] [PubMed] [Google Scholar]
- 179.Gavalas A, Krumlauf R. Retinoid signalling and hindbrain patterning. Curr Opin Genet Dev. 2000;10:380–386. doi: 10.1016/S0959-437X(00)00100-3. [DOI] [PubMed] [Google Scholar]
- 180.Schilling TF, Nie Q, Lander AD. Dynamics and precision in retinoic acid morphogen gradients. Curr Opin Genet Dev. 2012;22:562–569. doi: 10.1016/j.gde.2012.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.van der Wees J, Schilthuis JG, Koster CH, Diesveld-Schipper H, Folkers GE, van der Saag PT, Dawson MI, Shudo K, van der Burg B, Durston AJ. Inhibition of retinoic acid receptor-mediated signalling alters positional identity in the developing hindbrain. Development. 1998;125:545–556. doi: 10.1242/dev.125.3.545. [DOI] [PubMed] [Google Scholar]
- 182.Berggren K, McCaffery P, Dräger U, Forehand CJ. Differential distribution of retinoic acid synthesis in the chicken embryo as determined by immunolocalization of the retinoic acid synthetic enzyme, RALDH-2. Dev Biol. 1999;210:288–304. doi: 10.1006/dbio.1999.9286. [DOI] [PubMed] [Google Scholar]
- 183.Maden M, Gale E, Kostetskii I, Zile M. Vitamin A-deficient quail embryos have half a hindbrain and other neural defects. Curr Biol. 1996;6:417–426. doi: 10.1016/S0960-9822(02)00509-2. [DOI] [PubMed] [Google Scholar]
- 184.Gale E, Zile M, Maden M. Hindbrain respecification in the retinoid-deficient quail. Mech Dev. 1999;89:43–54. doi: 10.1016/S0925-4773(99)00202-6. [DOI] [PubMed] [Google Scholar]
- 185.Dupé V, Ghyselinck NB, Wendling O, Chambon P, Mark M. Key roles of retinoic acid receptors alpha and beta in the patterning of the caudal hindbrain, pharyngeal arches and otocyst in the mouse. Development. 1999;126:5051–5059. doi: 10.1242/dev.126.22.5051. [DOI] [PubMed] [Google Scholar]
- 186.Niederreither K, Vermot J, Schuhbaur B, Chambon P, Dollé P. Retinoic acid synthesis and hindbrain patterning in the mouse embryo. Development. 2000;127:75–85. doi: 10.1242/dev.127.1.75. [DOI] [PubMed] [Google Scholar]
- 187.Dickman ED, Thaller C, Smith SM. Temporally-regulated retinoic acid depletion produces specific neural crest, ocular and nervous system defects. Development. 1997;124:3111–3121. doi: 10.1242/dev.124.16.3111. [DOI] [PubMed] [Google Scholar]
- 188.White JC, Highland M, Kaiser M, Clagett-Dame M. Vitamin A deficiency results in the dose-dependent acquisition of anterior character and shortening of the caudal hindbrain of the rat embryo. Dev Biol. 2000;220:263–284. doi: 10.1006/dbio.2000.9635. [DOI] [PubMed] [Google Scholar]
- 189.Hill J, Clarke JD, Vargesson N, Jowett T, Holder N. Exogenous retinoic acid causes specific alterations in the development of the midbrain and hindbrain of the zebrafish embryo including positional respecification of the Mauthner neuron. Mech Dev. 1995;50:3–16. doi: 10.1016/0925-4773(94)00321-D. [DOI] [PubMed] [Google Scholar]
- 190.Maves L, Kimmel CB. Dynamic and sequential patterning of the zebrafish posterior hindbrain by retinoic acid. Dev Biol. 2005;285:593–605. doi: 10.1016/j.ydbio.2005.07.015. [DOI] [PubMed] [Google Scholar]
- 191.Cai AQ, Radtke K, Linville A, Lander AD, Nie Q, Schilling TF. Cellular retinoic acid-binding proteins are essential for hindbrain patterning and signal robustness in zebrafish. Development. 2012;139:2150–2155. doi: 10.1242/dev.077065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Samarut E, Fraher D, Laudet V, Gibert Y. ZebRA: an overview of retinoic acid signaling during zebrafish development. Biochim Biophys Acta. 2015;1849:73–83. doi: 10.1016/j.bbagrm.2014.05.030. [DOI] [PubMed] [Google Scholar]
- 193.Sirbu IO, Gresh L, Barra J, Duester G. Shifting boundaries of retinoic acid activity control hindbrain segmental gene expression. Development. 2005;132:2611–2622. doi: 10.1242/dev.01845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.White RJ, Nie Q, Lander AD, Schilling TF. Complex regulation of cyp26a1 creates a robust retinoic acid gradient in the zebrafish embryo. PLoS Biol. 2007;5:e304. doi: 10.1371/journal.pbio.0050304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Hernandez RE, Putzke AP, Myers JP, Margaretha L, Moens CB. Cyp26 enzymes generate the retinoic acid response pattern necessary for hindbrain development. Development. 2007;134:177–187. doi: 10.1242/dev.02706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.McCaffery P, Dräger UC. Retinoic acid synthesis in the developing retina. Adv Exp Med Biol. 1993;328:181–190. doi: 10.1007/978-1-4615-2904-0_20. [DOI] [PubMed] [Google Scholar]
- 197.Fujii H, Sato T, Kaneko S, Gotoh O, Fujii-Kuriyama Y, Osawa K, Kato S, Hamada H. Metabolic inactivation of retinoic acid by a novel P450 differentially expressed in developing mouse embryos. EMBO J. 1997;16:4163–4173. doi: 10.1093/emboj/16.14.4163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Moss JB, Xavier-Neto J, Shapiro MD, Nayeem SM, McCaffery P, Dräger UC, Rosenthal N. Dynamic patterns of retinoic acid synthesis and response in the developing mammalian heart. Dev Biol. 1998;199:55–71. doi: 10.1006/dbio.1998.8911. [DOI] [PubMed] [Google Scholar]
- 199.Swindell EC, Thaller C, Sockanathan S, Petkovich M, Jessell TM, Eichele G. Complementary domains of retinoic acid production and degradation in the early chick embryo. Dev Biol. 1999;216:282–296. doi: 10.1006/dbio.1999.9487. [DOI] [PubMed] [Google Scholar]
- 200.Begemann G, Schilling TF, Rauch GJ, Geisler R, Ingham PW. The zebrafish neckless mutation reveals a requirement for raldh2 in mesodermal signals that pattern the hindbrain. Development. 2001;128:3081–3094. doi: 10.1242/dev.128.16.3081. [DOI] [PubMed] [Google Scholar]
- 201.Dobbs-McAuliffe B, Zhao Q, Linney E. Feedback mechanisms regulate retinoic acid production and degradation in the zebrafish embryo. Mech Dev. 2004;121:339–350. doi: 10.1016/j.mod.2004.02.008. [DOI] [PubMed] [Google Scholar]
- 202.Emoto Y, Wada H, Okamoto H, Kudo A, Imai Y. Retinoic acid-metabolizing enzyme Cyp26a1 is essential for determining territories of hindbrain and spinal cord in zebrafish. Dev Biol. 2005;278:415–427. doi: 10.1016/j.ydbio.2004.11.023. [DOI] [PubMed] [Google Scholar]
- 203.Linville A, Radtke K, Waxman JS, Yelon D, Schilling TF. Combinatorial roles for zebrafish retinoic acid receptors in the hindbrain, limbs and pharyngeal arches. Dev Biol. 2009;325:60–70. doi: 10.1016/j.ydbio.2008.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.McCaffery PJ, Adams J, Maden M, Rosa-Molinar E. Too much of a good thing: retinoic acid as an endogenous regulator of neural differentiation and exogenous teratogen. Eur J Neurosci. 2003;18:457–472. doi: 10.1046/j.1460-9568.2003.02765.x. [DOI] [PubMed] [Google Scholar]
- 205.Reijntjes S, Blentic A, Gale E, Maden M. The control of morphogen signalling: regulation of the synthesis and catabolism of retinoic acid in the developing embryo. Dev Biol. 2005;285:224–237. doi: 10.1016/j.ydbio.2005.06.019. [DOI] [PubMed] [Google Scholar]
- 206.Giguere V, Ong ES, Segui P, Evans RM. Identification of a receptor for the morphogen retinoic acid. Nature. 1987;330:624–629. doi: 10.1038/330624a0. [DOI] [PubMed] [Google Scholar]
- 207.Petkovich M, Brand NJ, Krust A, Chambon P. A human retinoic acid receptor which belongs to the family of nuclear receptors. Nature. 1987;330:444–450. doi: 10.1038/330444a0. [DOI] [PubMed] [Google Scholar]
- 208.Mangelsdorf DJ, Evans RM. The RXR heterodimers and orphan receptors. Cell. 1995;83:841–850. doi: 10.1016/0092-8674(95)90200-7. [DOI] [PubMed] [Google Scholar]
- 209.Tang XH, Gudas LJ. Retinoids, retinoic acid receptors, and cancer. Annu Rev Pathol. 2011;6:345–364. doi: 10.1146/annurev-pathol-011110-130303. [DOI] [PubMed] [Google Scholar]
- 210.Rhinn M. Dollé P Retinoic acid signalling during development. Development. 2012;139:843–858. doi: 10.1242/dev.065938. [DOI] [PubMed] [Google Scholar]
- 211.Popperl H, Featherstone MS. Identification of a retinoic acid response element upstream of the murine Hox-4.2 gene. Mol Cell Biol. 1993;13:257–265. doi: 10.1128/MCB.13.1.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Marin F, Puelles L. Patterning of the embryonic avian midbrain after experimental inversions: a polarizing activity from the isthmus. Dev Biol. 1994;163:19–37. doi: 10.1006/dbio.1994.1120. [DOI] [PubMed] [Google Scholar]
- 213.Dupé V, Davenne M, Brocard J, Dollé P, Mark M, Dierich A, Chambon P, Rijli FM. In vivo functional analysis of the Hoxa-1 3′ retinoic acid response element (3′RARE) Development. 1997;124:399–410. doi: 10.1242/dev.124.2.399. [DOI] [PubMed] [Google Scholar]
- 214.Bel-Vialar S, Itasaki N, Krumlauf R. Initiating Hox gene expression: in the early chick neural tube differential sensitivity to FGF and RA signaling subdivides the HoxB genes in two distinct groups. Development. 2002;129:5103–5115. doi: 10.1242/dev.129.22.5103. [DOI] [PubMed] [Google Scholar]
- 215.Houle M, Prinos P, Iulianella A, Bouchard N, Lohnes D. Retinoic acid regulation of Cdx1: an indirect mechanism for retinoids and vertebral specification. Mol Cell Biol. 2003;20:6579–6586. doi: 10.1128/MCB.20.17.6579-6586.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Nolte C, Amores A, Kovacs EN, Postlethwait J, Featherstone M. The role of a retinoic acid response element in establishing the anterior neural expression border of Hoxd4 transgenes. Mech Dev. 2003;120:325–335. doi: 10.1016/S0925-4773(02)00442-2. [DOI] [PubMed] [Google Scholar]
- 217.Bertrand S, Thisse B, Tavares R, Sachs L, Chaumot A, Bardet PL, Escrivà H, Duffraisse M, Marchand O, Safi R, Thisse C, Laudet V. Unexpected novel relational links uncovered by extensive developmental profiling of nuclear receptor expression. PLoS Genet. 2007;3:e188. doi: 10.1371/journal.pgen.0030188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Pouilhe M, Gilardi-Hebenstreit P, Desmarquet-Trin Dinh C, Charnay P. Direct regulation of vHnf1 by retinoic acid signaling and MAF-related factors in the neural tube. Dev Biol. 2007;309:344–357. doi: 10.1016/j.ydbio.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 219.Ahn Y, Mullan HE, Krumlauf R. Long-range regulation by shared retinoic acid response elements modulates dynamic expression of posterior Hoxb genes in CNS development. Dev Biol. 2014;388:134–144. doi: 10.1016/j.ydbio.2014.01.027. [DOI] [PubMed] [Google Scholar]
- 220.Studer M, Pöpperl H, Marshall H, Kuroiwa A, Krumlauf R. Role of a conserved retinoic acid response element in rhombomere restriction of Hoxb-1. Science. 1994;265:1728–1732. doi: 10.1126/science.7916164. [DOI] [PubMed] [Google Scholar]
- 221.Marshall H, Studer M, Pöpperl H, Aparicio S, Kuroiwa A, Brenner S, Krumlauf R. A conserved retinoic acid response element required for early expression of the homeobox gene Hoxb-1. Nature. 1994;370:567–571. doi: 10.1038/370567a0. [DOI] [PubMed] [Google Scholar]
- 222.Niederreither K, McCaffery P, Dräger UC, Chambon P, Dollé P. Restricted expression and retinoic acid-induced downregulation of the retinaldehyde dehydrogenase type 2 (RALDH-2) gene during mouse development. Mech Dev. 1997;62:67–78. doi: 10.1016/S0925-4773(96)00653-3. [DOI] [PubMed] [Google Scholar]
- 223.Laue K, Jänicke M, Plaster N, Sonntag C, Hammerschmidt M. Restriction of retinoic acid activity by Cyp26b1 is required for proper timing and patterning of osteogenesis during zebrafish development. Development. 2008;135:3775–3787. doi: 10.1242/dev.021238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Strate I, Min TH, Iliev D, Pera EM. Retinol dehydrogenase 10 is a feedback regulator of retinoic acid signalling during axis formation and patterning of the central nervous system. Development. 2009;136:461–472. doi: 10.1242/dev.024901. [DOI] [PubMed] [Google Scholar]
- 225.Feng L, Hernandez RE, Waxman JS, Yelon D, Moens CB. Dhrs3a regulates retinoic acid biosynthesis through a feedback inhibition mechanism. Dev Biol. 2010;338:1–14. doi: 10.1016/j.ydbio.2009.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Vitobello A, Ferretti E, Lampe X, Vilain N, Ducret S, Ori M, Spetz JF, Selleri L, Rijli FM. Hox and Pbx factors control retinoic acid synthesis during hindbrain segmentation. Dev Cell. 2011;20:469–482. doi: 10.1016/j.devcel.2011.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Kim YK, Wassef L, Hamberger L, Piantedosi R, Palczewski K, Blaner WS, Quadro L. Retinyl ester formation by lecithin: retinol acyltransferase is a key regulator of retinoid homeostasis in mouse embryogenesis. J Biol Chem. 2008;283:5611–5621. doi: 10.1074/jbc.M708885200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Zhang YR, Zhao YQ, Huang JF. Retinoid-binding proteins: similar protein architectures bind similar ligands via completely different ways. PLoS One. 2012;7:e36772. doi: 10.1371/journal.pone.0036772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Addison M, Xu Q, Cayuso J, Wilkinson DG. Cell identity switching regulated by retinoic acid signaling maintains homogeneous segments in the hindbrain. Dev Cell. 2018;45:606–620. doi: 10.1016/j.devcel.2018.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Wilkinson DG (2018) Establishing sharp and homogeneous segments in the hindbrain. F1000 Res 7. 10.12688/f1000research.15391.1 [DOI] [PMC free article] [PubMed]
- 231.Schilling TF, Sosnik J, Nie Q. Visualizing retinoic acid morphogen gradients. Methods Cell Biol. 2016;133:139–163. doi: 10.1016/bs.mcb.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Rossant J, Zirngibl R, Cado D, Shago M, Giguère V. Expression of a retinoic acid response element-hsplacZ transgene defines specific domains of transcriptional activity during mouse embryogenesis. Genes Dev. 1991;5:1333–1344. doi: 10.1101/gad.5.8.1333. [DOI] [PubMed] [Google Scholar]
- 233.Perz-Edwards A, Hardison NL, Linney E. Retinoic acid-mediated gene expression in transgenic reporter zebrafish. Dev Biol. 2001;229:89–101. doi: 10.1006/dbio.2000.9979. [DOI] [PubMed] [Google Scholar]
- 234.Waxman JS, Yelon D. Zebrafish retinoic acid receptors function as context-dependent transcriptional activators. Dev Biol. 2011;352:128–140. doi: 10.1016/j.ydbio.2011.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Li J, Hu P, Li K, Zhao Q. Identification and characterization of a novel retinoic acid response element in zebrafish cyp26a1 promoter. Anat Rec. 2012;295:268–277. doi: 10.1002/ar.21520. [DOI] [PubMed] [Google Scholar]
- 236.Mandal A, Rydeen A, Anderson J, Sorrell MR, Zygmunt T, Torres-Vázquez J, Waxman JS. Transgenic retinoic acid sensor lines in zebrafish indicate regions of available embryonic retinoic acid. Dev Dyn. 2013;242:989–1000. doi: 10.1002/dvdy.23987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Shimozono S, Iimura T, Kitaguchi T, Higashijima S, Miyawaki A. Visualization of an endogenous retinoic acid gradient across embryonic development. Nature. 2013;496:363–366. doi: 10.1038/nature12037. [DOI] [PubMed] [Google Scholar]
- 238.Sosnik J, Zheng L, Rackauckas CV, Digman M, Gratton E, Nie Q, Schilling TF. Noise modulation in retinoic acid signaling sharpens segmental boundaries of gene expression in the embryonic zebrafish hindbrain. Elife. 2016;5:e14034. doi: 10.7554/eLife.14034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Hollemann T, Chen Y, Grunz H, Pieler T. Regionalized metabolic activity establishes boundaries of retinoic acid signalling. EMBO J. 1998;17:7361–7372. doi: 10.1093/emboj/17.24.7361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Grandel H, Lun K, Rauch GJ, Rhinn M, Piotrowski T, Houart C, Sordino P, Küchler AM, Schulte-Merker S, Geisler R, Holder N, Wilson SW, Brand M. Retinoic acid signalling in the zebrafish embryo is necessary during pre-segmentation stages to pattern the anterior-posterior axis of the CNS and to induce a pectoral fin bud. Development. 2002;129:2851–2865. doi: 10.1242/dev.129.12.2851. [DOI] [PubMed] [Google Scholar]
- 241.Kastner P, Messaddeq N, Mark M, Wendling O, Grondona JM, Ward S, Ghyselinck N, Chambon P. Vitamin A deficiency and mutations of RXRalpha, RXRbeta and RARalpha lead to early differentiation of embryonic ventricular cardiomyocytes. Development. 1997;124:4749–4758. doi: 10.1242/dev.124.23.4749. [DOI] [PubMed] [Google Scholar]
- 242.Dupé V, Lumsden A. Hindbrain patterning involves graded responses to retinoic acid signalling. Development. 2001;128:2199–2208. doi: 10.1242/dev.128.12.2199. [DOI] [PubMed] [Google Scholar]
- 243.Escriva H, Bertrand S, Germain P, Robinson-Rechavi M, Umbhauer M, Cartry J, Duffraisse M, Holland L, Gronemeyer H, Laudet V. Neofunctionalization in vertebrates: the example of retinoic acid receptors. PLoS Genet. 2006;2:e102. doi: 10.1371/journal.pgen.0020102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Bouillet P, Oulad-Abdelghani M, Ward SJ, Bronner S, Chambon P, Dolle P. A new mouse member of the Wnt gene family, mWnt-8, is expressed during early embryogenesis and is ectopically induced by retinoic acid. Mech Dev. 1996;58:141–152. doi: 10.1016/S0925-4773(96)00569-2. [DOI] [PubMed] [Google Scholar]
- 245.Hans S, Westerfield M. Changes in retinoic acid signaling alter otic patterning. Development. 2007;134:2449–2458. doi: 10.1242/dev.000448. [DOI] [PubMed] [Google Scholar]
- 246.Niederreither K, Subbarayan V, Dollé P, Chambon P. Embryonic retinoic acid synthesis is essential for early mouse post-implantation development. Nat Genet. 1999;21:444–448. doi: 10.1038/7788. [DOI] [PubMed] [Google Scholar]
- 247.Urness LD, Paxton CN, Wang X, Schoenwolf GC, Mansour SL. FGF signaling regulates otic placode induction and refinement by controlling both ectodermal target genes and hindbrain Wnt8a. Dev Biol. 2010;340:595–604. doi: 10.1016/j.ydbio.2010.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Mahoney Rogers AA, Zhang J, Shim K. Sprouty1 and Sprouty2 limit both the size of the otic placode and hindbrain Wnt8a by antagonizing FGF signaling. Dev Biol. 2011;353:94–104. doi: 10.1016/j.ydbio.2011.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.In der Rieden PM, Vilaspasa FL, Durston AJ. Xwnt8 directly initiates expression of labial Hox genes. Dev Dyn. 2010;239:126–139. doi: 10.1002/dvdy.22020. [DOI] [PubMed] [Google Scholar]
- 250.Janssens S, Denayer T, Deroo T, van Roy F, Vleminckx K. Direct control of Hoxd1 and Irx3 expression by Wnt/beta catenin signaling during anteroposterior patterning of the neural axis in Xenopus. Int J Dev Biol. 2010;54:1435–1442. doi: 10.1387/ijdb.092985sj. [DOI] [PubMed] [Google Scholar]
- 251.Diez del Corral R, Storey KG. Opposing FGF and retinoid pathways: a signalling switch that controls differentiation and patterning onset in the extending vertebrate body axis. BioEssays. 2004;26:857–869. doi: 10.1002/bies.20080. [DOI] [PubMed] [Google Scholar]
- 252.Shimizu T, Bae YK, Hibi M. Cdx-Hox code controls competence for responding to Fgfs and retinoic acid in zebrafish neural tissue. Development. 2006;133:4709–4719. doi: 10.1242/dev.02660. [DOI] [PubMed] [Google Scholar]
- 253.Zhao X, Duester G. Effect of retinoic acid signaling on Wnt/beta-catenin and FGF signaling during body axis extension. Gene Expr Patterns. 2009;9:430–435. doi: 10.1016/j.gep.2009.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Martínez S, Marín F, Nieto MA, Puelles L. Induction of ectopic engrailed expression and fate change in avian rhombomeres: intersegmental boundaries as barriers. Mech Dev. 1995;51:289–303. doi: 10.1016/0925-4773(95)00376-2. [DOI] [PubMed] [Google Scholar]
- 255.Crossley PH, Martinez S, Martin GR. Midbrain development induced by FGF8 in the chick embryo. Nature. 1996;380:66–68. doi: 10.1038/380066a0. [DOI] [PubMed] [Google Scholar]
- 256.Irving C, Mason I. Signalling by FGF8 from the isthmus patterns anterior hindbrain and establishes the anterior limit of Hox gene expression. Development. 2000;127:177–186. doi: 10.1242/dev.127.1.177. [DOI] [PubMed] [Google Scholar]
- 257.Wendling O, Ghyselinck NB, Chambon P, Mark M. Roles of retinoic acid receptors in early embryonic morphogenesis and hindbrain patterning. Development. 2001;128:2031–2038. doi: 10.1242/dev.128.11.2031. [DOI] [PubMed] [Google Scholar]
- 258.Linville A, Gumusaneli E, Chandraratna RA, Schilling TF. Independent roles for retinoic acid in segmentation and neuronal differentiation in the zebrafish hindbrain. Dev Biol. 2004;270:186–199. doi: 10.1016/j.ydbio.2004.02.022. [DOI] [PubMed] [Google Scholar]
- 259.Nittenberg R, Patel K, Joshi Y, Krumlauf R, Wilkinson DG, Brickell PM, Tickle C, Clarke JD. Cell movements, neuronal organisation and gene expression in hindbrains lacking morphological boundaries. Development. 1997;124:2297–2306. doi: 10.1242/dev.124.12.2297. [DOI] [PubMed] [Google Scholar]
- 260.Sato T, Joyner AL. The duration of Fgf8 isthmic organizer expression is key to patterning different tectal-isthmo-cerebellum structures. Development. 2009;136:3617–3626. doi: 10.1242/dev.041210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Diez del Corral R, Olivera-Martinez I, Goriely A, Gale E, Maden M, Storey K. Opposing FGF and retinoid pathways control ventral neural pattern, neuronal differentiation, and segmentation during body axis extension. Neuron. 2003;40:65–79. doi: 10.1016/S0896-6273(03)00565-8. [DOI] [PubMed] [Google Scholar]
- 262.Duester G. Retinoid signaling in control of progenitor cell differentiation during mouse development. Semin Cell Dev Biol. 2013;24:694–700. doi: 10.1016/j.semcdb.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Parker HJ, Bronner ME, Krumlauf R. The vertebrate Hox gene regulatory network for hindbrain segmentation: evolution and diversification: Coupling of a Hox gene regulatory network to hindbrain segmentation is an ancient trait originating at the base of vertebrates. BioEssays. 2016;38:526–538. doi: 10.1002/bies.201600010. [DOI] [PubMed] [Google Scholar]
- 264.Skromne I, Thorsen D, Hale M, Prince VE, Ho RK. Repression of the hindbrain developmental program by Cdx factors is required for the specification of the vertebrate spinal cord. Development. 2007;134:2147–2158. doi: 10.1242/dev.002980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Chang J, Skromne I, Ho RK. CDX4 and retinoic acid interact to position the hindbrain-spinal cord transition. Dev Biol. 2016;410:178–189. doi: 10.1016/j.ydbio.2015.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Lee K, Skromne I. Retinoic acid regulates size, pattern and alignment of tissues at the head-trunk transition. Development. 2014;141:4375–4384. doi: 10.1242/dev.109603. [DOI] [PubMed] [Google Scholar]
- 267.Isaacs HV, Pownall ME, Slack JM. Regulation of Hox gene expression and posterior development by the Xenopus caudal homologue Xcad3. EMBO J. 1998;17:3413–3427. doi: 10.1093/emboj/17.12.3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Gaunt SJ, Drage D, Cockley A. Vertebrate caudal gene expression gradients investigated by use of chick cdx-A/lacZ and mouse cdx-1/lacZ reporters in transgenic mouse embryos: evidence for an intron enhancer. Mech Dev. 2003;120:573–586. doi: 10.1016/S0925-4773(03)00023-6. [DOI] [PubMed] [Google Scholar]