Abstract
Metabolic reprogramming has now been accepted as a hallmark of cancer. Compared to normal cells, cancer cells exhibit different metabolic features, including increased glucose uptake, aerobic glycolysis, enhanced glutamine uptake and glutaminolysis, altered lipid metabolism, and so on. Cancer metabolic reprogramming, which supports excessive cell proliferation and growth, has been widely regulated by activation of oncogenes or loss of tumor suppressors. Here, we review that long non-coding RNAs (lncRNAs) can affect cancer metabolism by mutual regulation with oncogenes or tumor suppressors. Additionally, the interaction of lncRNAs with crucial transcription factors, metabolic enzymes or microRNAs can also effectively modulate the processes of cancer metabolism. LncRNAs-derived metabolism reprogramming allows cancer cells to maintain deregulated proliferation and withstand hostile microenvironment such as energy stress. Understanding the functions of lncRNAs in cancer metabolic reprogramming that contributes to carcinogenesis and cancer development may help to develop novel and effective strategies for cancer diagnosis, prognosis and treatment.
Keywords: Long non-coding RNA, Cancer, Metabolism
Introduction
Metabolism reprogramming is regarded as an emerging hallmark of cancer [1]. Cancers share a common trait of uncontrolled and fast cell proliferation, which drives reprogramming of cellular energy metabolism and macromolecules biosynthesis to satisfy the requirement of cancer expansion and dissemination. Reprogramming of cancer metabolism signifies cancer-specific metabolic alterations, including dysregulation of glucose and glutamine metabolism, alterations of lipid synthesis and decomposition, and rewiring of mitochondrial function [2–5]. These phenotypes of cellular metabolism reprogramming are direct and indirect consequences of oncogenic events (activation of oncogenes [6, 7] or loss of tumor suppressors [8, 9]) or constraints imposed by the tumor microenvironment (e.g., hypoxia [10] and nutrient scarcity [11]). As a primary feature during carcinogenesis, metabolic reprogramming contributes to the establishment and maintenance of the cancerogenic state [12, 13].
LncRNAs are defined as a group of transcripts longer than 200 nucleotides, with little or no protein-coding potential. Through interactions with cellular macromolecules including chromatin, protein or RNA, lncRNAs can regulate gene expression by controlling chromatin architecture, promoting the assembly of protein complexes or disrupting protein–protein interactions, sequestering miRNA away from target mRNA as competitive endogenous RNA, and affecting mRNA metabolism including mRNA splicing, stability, and translation, etc [14]. LncRNAs have been determined to be involved in many physiologic and pathologic processes including differentiation, development and disease [15–17]. Multiple evidence verified that lncRNAs are aberrantly expressed in various cancers where they act as oncogenes [18] or tumor suppressors [19], which qualifies their potential utilization for cancer diagnosis, monitoring, prognosis, and prediction for therapeutic responsiveness [14]. Although microRNAs (miRNAs), one class of small non-coding RNAs, have been well documented for their involvement in cancer metabolism [20, 21], the roles of lncRNAs in the regulation of metabolism and energy homeostasis remain largely unknown. Here, we review the recent findings on the functions of lncRNAs in cancer metabolism reprogramming, with particular emphasis on how lncRNAs regulate glucose, glutamine and lipid metabolism, as well as their response to energy stress in cancer cells.
Regulation of central metabolism by lncRNAs
Activation of oncogenes or loss of tumor suppressors promotes metabolism reprogramming in cancer and supports growth and survival of cancer cells [6–9]. The ability of oncogenes (e.g., c-MYC [22, 23] and HIF-1α [24, 25]) and tumor suppressors (e.g., p53 [26] and AMPK [27]) to alter cellular metabolism have been well established. The oncogenic transcription factor c-Myc is a master regulator that not only controls glucose [28, 29], glutamine [30, 31] and lipid metabolism [32, 33] in cancer, but also supports nucleotide [34] and serine biosynthesis [35] and formation of new organelles, particular ribosomes [36] and mitochondria [37]. A amount of metabolic genes are activated by c-Myc, including GLUT1, HK2, LDH-A and MCT in glycolysis, SLC1A5 and GLS1 in glutaminolysis, as well as ACLY, ACACA, FASN and SCD in lipid synthesis [2, 38]. However, tumor suppressor p53 represses glucose transport [39], glycolysis [40] and lipid synthesis [41], and promotes fatty acid oxidation [42], glutaminolysis [43] and mitochondrial respiration [44]. Transcription, protein stability and functional activity of oncogenes or tumor suppressors can be regulated by multiple factors in cancer cells, leading to abnormal metabolism and further tumor progression or inhibition [22, 38, 45].
PCGEM1, an androgen-induced prostate-specific lncRNA, is the first lncRNA that has been identified to function as a master regulator of metabolic reprogramming in cancer [46]. Although PCGEM1 facilitates androgen-induced metabolic gene expression through the activation of androgen receptor, it is c-Myc that predominantly mediates the regulation of metabolic genes by PCGEM1. By directly interacting with c-Myc and functioning as a c-Myc coactivator, PCGEM1 enhances c-Myc transactivation potency and facilitates the recruitment of c-Myc to the chromatin target sites, therefore, controlling expression profiles of multiple key metabolic pathways [46]. On the one hand, PCGEM1 overexpression in prostate cells enhances glucose uptake and lactate production, indicating an increased aerobic glycolysis. On the other hand, elevated cellular levels of citrate, G6PD activity, and NADPH in PCGEM1-overexpressing cells demonstrate increased biosynthesis of fatty acid, nucleotide and redox control by PCGEM1 [46]. The correlation of PCGEM1 with high-risk prostate cancer patients [47] and its specific role in tumor metabolism [46] makes it a promising therapeutic target for prostate cancer. Another lncRNA, colorectal neoplasia differentially expressed transcript (CRNDE), has also been reported to regulate genes involved in central metabolism [48]. Transcripts from the CRNDE gene locus comprise of two categories of lncRNAs: transcripts retaining intronic sequences localized in the nucleus and transcripts lacking intronic sequences enriched in the cytoplasm [48]. Nuclear CRNDE are specifically downregulated by insulin and IGF1/2 in colorectal cancer cells. It appears to regulate genes at both transcriptional and post-transcriptional levels [49], and many of these genes are involved in glucose and lipid metabolism [48].
Regulation of glucose metabolism by lncRNAs
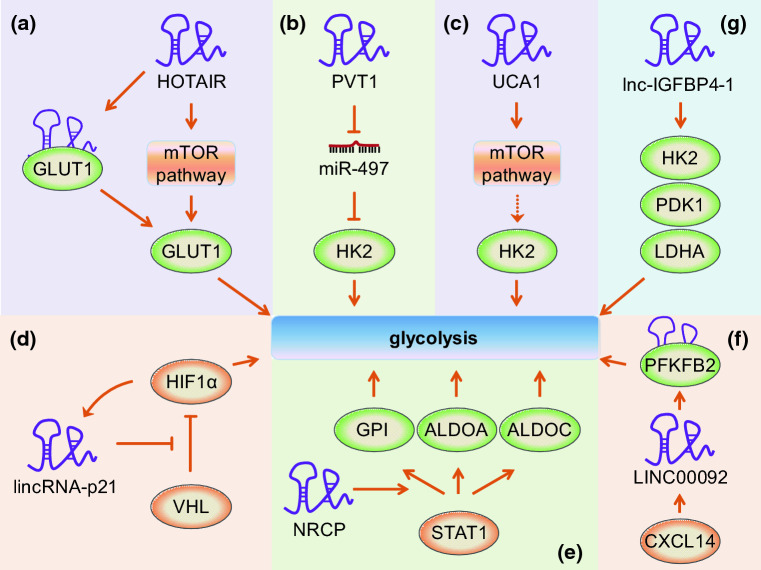
Glucose is a major cellular energy source, providing energy mostly in the form of adenosine triphosphate (ATP). After being imported into cells by transporters, glucose is broken down via glycolysis, generating energy and building blocks for cell growth and proliferation [2, 29]. Owing to an accelerated glucose uptake or undergoing aerobic glycolysis other than oxidative phosphorylation in mitochondria, cancer cells usually exhibit a higher rate of glucose metabolism than normal cells [50]. HOTAIR, a highly expressed lncRNA in hepatocellular carcinoma cells and tissues, promotes cell proliferation by regulating glucose metabolism, partly through inducing glucose transporter 1 (GLUT1) expression via activating the mTOR pathway, or through interacting with GLUT1 directly [51] (Fig. 1a). As a lncRNA overexpressed in osteosarcoma cells and tissues, PVT1 not only promotes uptake of glucose but also enhances production of lactate by acting as a molecular sponge of miR-497, which directly targets and suppresses the expression of the rate-limiting glycolytic enzyme hexokinase 2 (HK2) [52] (Fig. 1b). Another study showed that lncRNA UCA1 promotes glycolysis in bladder cancer cells by activating the mTOR pathway, which mediates the regulation of UCA1 to HK2 through activation of STAT3 and repression of miR-143 [53] (Fig. 1c). Although all above lncRNAs play important roles in regulating glucose metabolism reprogramming in cancer, the studies are limited to in vitro observations.
Fig. 1.
Regulation of glucose metabolism by lncRNAs. a HOTAIR enhances GLUT1 expression through activating the mTOR pathway, or through interacting with GLUT1 directly. b PVT1 acts as a molecular sponge of miR-497, which directly targets and suppresses the expression of HK2. c UCA1 promotes glycolysis by activating the mTOR pathway, which regulates the expression of HK2. d Under conditions of hypoxia, HIF-1α activates the transcription of lincRNA-p21. In return, lincRNA-p21 stabilizes HIF-1α by inhibiting VHL-mediated HIF-1α ubiquitination and degradation. e NRCP promotes glycolysis by enhancing the regulation of STAT1 to its downstream target genes, including GPI, ALDOA, and ALDOC. f LINC00092 is induced by CXCL14 and enhances glycolysis by interaction with PFKFB2. g Lnc-IGFBP4-1 enhances glycolysis by upregulating the expression of metabolic enzyme genes, including HK2, PDK1, and LDHA. Please see more detail in the text. “down arrow” indicates a promotion effect, but “perpendicular sign” indicates an inhibition effect. Solid line indicates a direct effect, but dashed line indicates an indirect effect. HOTAIR HOX transcript antisense intergenic RNA, GLUT1 glucose transporter 1, PVT1 plasmacytoma variant translocation 1, HK2 hexokinase 2, UCA1 urothelial cancer-associated 1, NRCP long non-coding RNA ceruloplasmin, GPI glucose-6-phosphate isomerase, ALDOA aldolase, fructose-bisphosphate A, ALDOC aldolase, fructose-bisphosphate C, PFKFB2 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase, PDK1 pyruvate dehydrogenase kinase 1, LDHA lactate dehydrogenase A
However, several other lncRNAs have been demonstrated to regulate glucose metabolism and affect cancer development both in vitro and in vivo. LincRNA-p21, a class of large intergenic non-coding RNAs, has been reported to be induced by hypoxia and is essential for hypoxia-enhanced glycolysis [54]. Under conditions of hypoxia, HIF-1α is induced and activates lincRNA-p21 at the transcriptional level. LincRNA-p21 in return stabilizes HIF-1α by disrupting the VHL-HIF-1α interaction, which mediates HIF-1α ubiquitination and degradation (Fig. 1d). As a result, HIF-1α is accumulated and promotes glycolysis under hypoxia. Additionally, validation of lincRNA-p21 in promoting tumor growth by mouse xenograft models indicates lincRNA-p21 would be a valuable therapeutic target for cancer [54]. NRCP is a non-coding splice variant of ceruloplasmin-coding gene with deletion of exon 11 and several nucleotide variations in the 3′ end exons [55]. Rupaimoole et al. have observed that NRCP is a top-upregulated lncRNA in ovarian tumor samples compared with normal ovarian tissues [55]. Additionally, in Kaplan–Meier survival analyses, patients with high tumoral NRCP expression have significantly worse overall survival. Furthermore, they revealed that NRCP promotes glycolysis and tumor progression by functioning as an intermediate binding partner between STAT1 and RNA polymerase II, which results in enhanced expression of downstream target genes, including glucose-6-phosphate isomerase (GPI), ALDOA, and ALDOC (Fig. 1e). Together with the finding that NRCP silencing reduced tumor growth and metastasis in vivo in an orthotopic mouse model of ovarian cancer [55], their study highlights the potential for targeting NRCP in the treatment of this kind of cancer. Another lncRNA LINC00092 was also reported to be upregulated and involved in the alteration of glycolysis in ovarian cancer [56]. LINC00092 is induced upon stimulation by CXCL14, which is a premetastatic factor secreted by cancer-associated fibroblasts (CAFs) in ovarian cancer and promotes ovarian cancer metastasis in vitro and in vivo. By directly interacting with 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase (PFKFB2), a glycolytic enzyme, LINC00092 enhances glycolysis, promotes metastasis, and sustains the local supportive function of CAFs within tumor microenvironment [56] (Fig. 1f). Recently, one study reported that lnc-IGFBP4-1 was a significantly up-regulated lncRNA in lung cancer tissues compared to corresponding non-tumor tissues [57]. It increases ATP production and enhances aerobic glycolysis by upregulating the expression of metabolic enzymes, including HK2, PDK1, and LDHA (Fig. 1g). Possibly through reprogramming tumor cell energy metabolism, lnc-IGFBP4-1 plays a positive role in cell proliferation and metastasis. This study suggests lnc-IGFBP4-1 may be a potential biomarker or therapeutic target for lung cancer [57].
Regulation of glutamine metabolism by lncRNAs
As another kind of important cellular energy source, glutamine is also essential for the survival and proliferation of most cancer cells [58]. Through providing nitrogen and carbon, glutamine metabolism is critical for many fundamental cell functions in cancer cells, including energy generation, macromolecular synthesis, activation of cell signaling, and maintenance of redox balance [59]. After being imported into cytoplasm via transporters, glutamine is first converted into glutamate by glutaminase (GLS/GLS2) that is the rate-limiting enzyme of glutaminolysis. Besides regulating glucose metabolism [53], UCA1 has also been reported to promote glutamine metabolism in human bladder cancer [60]. UCA1 and GLS2 were shown to be positively correlated in bladder cancer tissues and cell lines. By binding to miR-16 as a molecular sponge, UCA1 improves GLS2 expression through UCA1–miR-16–GLS2 axis, leading to increased glutaminolysis and repressed ROS formation in bladder cancer cells (Fig. 2a). Another recent study demonstrated that lncRNA HOTTIP was involved in GLS1-mediated glutaminolysis in hepatocellular carcinoma (HCC) [61] (Fig. 2b). MiR-192 and miR-204 are two miRNAs identified to suppress HOTTIP expression via the Argonaute 2-mediated RNA interference pathway. Ectopic expression of miR-192, miR-204 or HOTTIP siRNA significantly represses GLS1 expression, thereby interrupts glutaminolysis and inhibits HCC growth in vitro and in vivo [61]. Therefore, miR-192/-204-HOTTIP axis may be a potential target for prognostic and therapeutic implications in HCC.
Fig. 2.
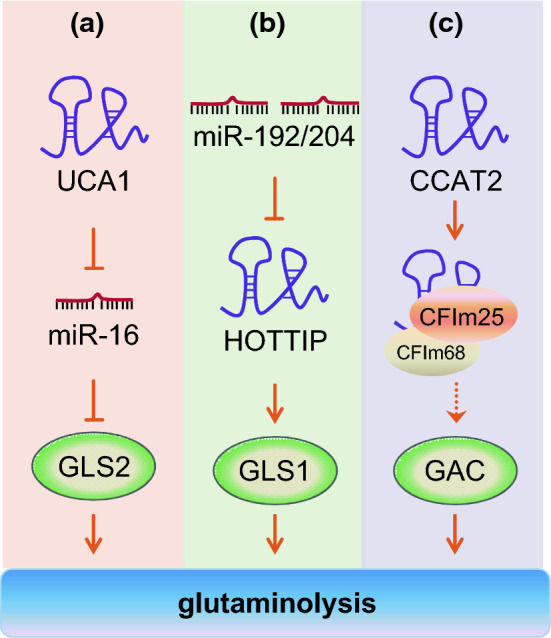
Regulation of glutamine metabolism by lncRNAs. a UCA1 functions as a molecular sponge of miR-16 and releases the expression of its target GLS2, which converts glutamine into glutamate. b HOTTIP, a target of both miR-192 and miR-204, is involved in glutaminolysis through its downstream gene GLS1. c CCAT2 alleles bind the CFIm complex, regulate the alternative splicing of GLS, and induce the production of GAC, which promotes the cancer progression. Please see more detail in the text. GLS glutaminase, HOTTIP HOXA transcript at the distal tip, CCAT2 colon cancer-associated transcript 2, CFIm cleavage factor I, GAC glutaminase isoform C
Another lncRNA Colon Cancer-Associated Transcript 2 (CCAT2), whose coding gene locates at the highly conserved 8q24 region harboring cancer risk-associated rs6983267 SNP, not only boosts glycolysis but also regulates glutamine metabolism in an allele-specific manner in vitro and in vivo [62]. For the two alleles of the rs6983267 SNP, the CCAT2 G allele is related to greater colorectal cancer predisposition than the T allele [63]. Although both CCAT2 G and T allele enhance glucose uptake in colon cancer cells, it is CCAT2 G allele but not T allele that increases oxygen consumption and intracellular glutamate production [62]. Mechanistically, CCAT2 G allele preferentially binds CFIm25, the small 25 kDa subunit of the Cleavage Factor I (CFIm) complex, which binds to UGUA sequences within intron 14 of GLS pre-mRNA. Whereas CCAT2 T allele prefers to bind CFIm68, the larger 68 kDa subunit of the CFIm complex. The interaction between CCAT2 G allele and the CFIm complex regulates alternative splicing of glutaminase (GLS) by selecting the poly(A) site in intron 14 of the GLS pre-mRNA, inducing the production of GAC splice isoform (glutaminase isoform C) other than KGA (glutaminase kidney isoform) (Fig. 2c). For reason of possessing a higher enzymatic activity than KGA, GAC is a more aggressive splicing variant of glutaminase and promotes in vitro cell proliferation and migration and in vivo metastases [62]. This is the first example that a lncRNA serves as a regulator of glutamine metabolism by controlling alternative splicing of glutaminase.
Regulation of lipid metabolism by lncRNAs
In addition to aberrant glucose and glutamine metabolism, dysregulation of lipid metabolism has also been identified as one character of cancer metabolic reprogramming [2]. Lipid metabolic network contains import of exogenous lipids or cholesterol, catabolic pathways of lipids (fatty acid oxidation), de novo synthesis pathways including lipogenesis and cholesterol synthesis, as well as storage in lipid droplets and export of high-density lipoproteins into circulations [64, 65]. Fatty acids need to be transformed to acyl-CoA before they enter into the subsequent anabolism or catabolism. Acyl-CoA synthetases, which catalyze the conversion of fatty acids to fatty acid-CoA, are classified into very long-chain acyl-CoA synthases (ACSVL), long-chain acyl-CoA synthases (ACSL), medium-chain acyl-CoA synthases (ACSM) and short-chain acyl-CoA synthases (ACSS) according to the carbon chain length of the targeted fatty acid [66]. Highly upregulated in liver cancer (HULC) is the first lncRNA identified to be specifically upregulated in hepatocellular carcinoma [67]. Cui et al. revealed that HULC facilitates the deregulation of lipid metabolism through miR-9/PPARA/ACSL1 pathway in hepatoma cells [68] (Fig. 3a). HULC downregulates miR-9 by inducing methylation of CpG islands in its promoter, then releases the inhibition of PPARA by miR-9. Next, the transcriptional factor PPARA activates ACSL1. ACSL1 can catalyze the initial step in cellular long-chain fatty acid metabolism in mammals and participate in the formation of triglycerides and cholesterol in the liver. Furthermore, HULC-modulated abnormal lipid metabolism promotes tumor growth in vivo [68].
Fig. 3.
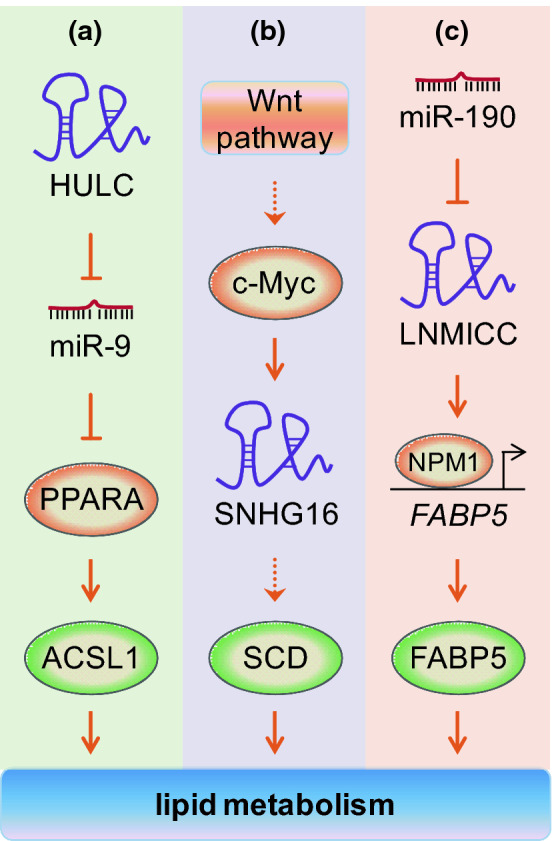
Regulation of lipid metabolism by lncRNAs. a HULC modulates lipid metabolism by miR-9/PPARA/ACSL1 pathway in hepatoma cells. b SNHG16, which is activated by the Wnt pathway/c-Myc axis in colorectal cancer, promotes the expression of genes involved in fatty acid biosynthesis, such as SCD. c LNMICC, a target of miR-190, induces binding of NPM1 to the promoter of FABP5 and activates its expression in cervical cancer. Consequently, FABP5 regulates the reprogramming of fatty acid metabolism. Please see more detail in the text. HULC highly upregulated in liver cancer, PPARA peroxisome proliferator activated receptor alpha, ACSL1 acyl-CoA synthetase long chain family member 1, SNHG16 snoRNA host gene 16, SCD stearoyl-CoA desaturase, LNMICC lncRNA associated with lymph node metastasis in cervical cancer, NPM1 nucleophosmin 1, FABP5 fatty acid binding protein 5
Recently, Andersen’s research group reported lncRNA SNHG16 was involved in lipid metabolism [69]. Their study found that SNHG16 was up-regulated in colorectal adenomas and all stages of adenocarcinomas when compared to adjacent normal colon mucosa samples. The mechanism behind SNHG16 deregulation in colorectal cancer (CRC) may be attributed to the transcriptional activation of the SNHG16 locus by the Wnt signaling pathway-regulated transcription factors, including c-Myc (Fig. 3b). Knockdown of SNHG16 suppresses growth, increases apoptotic death, and decreases migration in vitro. This evidence indicates that SNHG16 plays an oncogenic role in CRC. By binding to AGO and HuR, SNHG16 may function as a microRNA sponge and relieve miRNA-mediated repression of targets, including genes involved in fatty acid biosynthesis, such as fatty acid desaturase family member stearoyl-CoA desaturase (SCD) [69]. Another recent study reported that lncRNA LNMICC, a direct target of miR-190, is upregulated in cervical cancer with lymph node metastasis and associated with poor prognosis [70]. Mechanistically, LNMICC recruits NPM1 to the promoter region of FABP5 and enhance transcription of FABP5, thereby modulates reprogramming of fatty acid metabolism (Fig. 3c). Moreover, through FABP5-mediated fatty acid metabolism, LNMICC facilitate lymph node metastasis of cervical cancer. This study highlight LNMICC as a candidate prognostic biomarker and therapeutic target in cervical cancer.
Response of lncRNAs to energy stress
Metabolic composition of the extracellular environment around cancer cells is usually altered for their rapid proliferation, which results in tumor microenvironment with characters of hypoxia, downregulated pH, increased redox stress, and nutrient depletion [71]. Tumor cells always ingeniously scheme out various strategies to adapt to the harsh environment, such as energy stress [72–74]. A key signaling pathway involved in energy stress adaptation is the liver kinase B1 (LKB1)–AMP–activated protein kinase (AMPK) pathway [75]. Energy stress strikingly induces phosphorylation of AMPKα by LKB1 and activates AMPK. Then, AMPK phosphorylates a number of downstream targets to inactivate ATP-consuming anabolic processes, such as mammalian target of rapamycin complex 1 (mTORC1)-regulated protein synthesis [76]. AMPK also promotes autophagy and cell survival under energy stress through direct phosphorylation of ULK1, the mammalian autophagy-initiating kinase [77, 78]. Liu et al. reported lincRNA neighbour of BRCA1 gene 2 (NBR2) was involved in metabolic stress response by regulating the function of AMPK [79] (Fig. 4a). Under the condition of energy stress such as glucose starvation, NBR2 expression was induced through the LKB1–AMPK pathway. However, NBR2 depletion attenuated glucose starvation-induced AMPK activation and mTORC1 inactivation. Mechanistically, NBR2 regulates AMPK activity under energy stress by interaction with its kinase domain AMPKα, which is the catalytic subunit of AMPK. Therefore, the functional effects of NBR2 are partially mediated by AMPK, which may serve to explain the phenomena that NBR2 deficiency leads to increased apoptosis and decreased autophagy in response to energy stress [79].
Fig. 4.
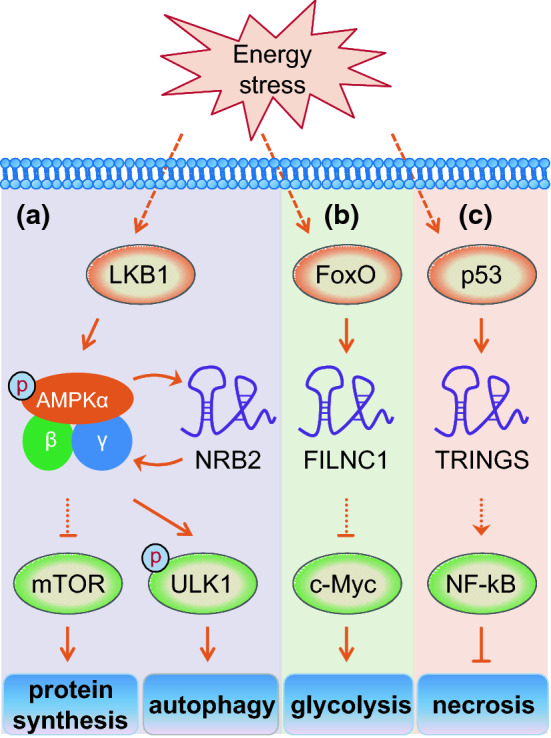
Response of lncRNAs to energy stress. Under the condition of energy stress such as glucose starvation, the expression of lncRNAs NBR2, FILNC1, and TRINGS are activated by LKB1–AMPK pathway, p53, and FoxO, respectively. a NBR2 enhances AMPK activity under energy stress by interaction with its kinase domain AMPKα. On the one hand, AMPK inhibits mTOR signaling that mediates protein synthesis. On the other hand, AMPK directly phosphorylates and activates the autophagy regulator ULK1. b FILNC1 suppresses the protein level of c-Myc, a master regulator of glycolysis. c NF-kB signaling is promoted by TRINGS and protect cancer cells from necrosis. Please see more detail in the text. NBR2 neighbour of BRCA1 gene 2, FILNC1 FoxO-induced long non-coding RNA 1, TRINGS Tp53-regulated inhibitor of necrosis under glucose starvation
Additionally, FoxO-induced long non-coding RNA 1 (FILNC1), another lncRNA, was reported to repress c-Myc-mediated energy metabolism under energy stress in renal cancer [80] (Fig. 4b). In response to energy stress such as glucose starvation, FoxO transcription factors enhanced the expression of FILNC1. Furthermore, glucose starvation also induced the interaction of FILNC1 with AUF1, which can bind to AU-rich elements within 3′ UTR of c-Myc mRNA and promote c-Myc translation without affecting c-Myc mRNA level. As a result, AUF1 was sequestered from binding to c-Myc mRNA, resulting in downregulated protein level of c-Myc, a master transcription factor regulating most of the glycolytic enzymes [2, 38]. Consistently, FILNC1 deficiency increased the expression of various genes involved in glucose uptake, glycolysis, and lactate secretion under glucose starvation condition, thereby led to enhanced glucose uptake and lactate production [80]. Therefore, FILNC1 controls energy metabolism reprogramming under energy stress conditions by repressing c-Myc protein level via a post-transcriptional regulation. Recently, Wu’s research group revealed that lncRNA TRINGS was induced by p53 and protected cancer cells from necrosis induced by glucose starvation [81] (Fig. 4c). p53 activated TRINGS by physically interacting with p53 response elements in the promoter region of TRINGS. As a consequence of p53 up-regulation under glucose starvation, TRINGS was significantly induced in cancer cells and protected cancer cells from necrotic cell death. Mechanistically, glucose starvation-induced TRINGS impairs the interaction between STRAP and GSK3β by competitive binding to STRAP, leading to increased GSK3β phosphorylation at serine 9 that inhibited GSK3β activity [81]. Therefore, inhibition of NF-kB signaling by GSK3β is released to protect cancer cells from necrosis [82]. Taken together, TRINGS promotes cell survival under glucose starvation through the STRAP–GSK3β–NF-kB axis [81].
Conclusions
Carcinogenesis and cancer development are dependent on the reprogramming of cellular metabolism, which is precisely controlled by the activation of oncogenes and inactivation of tumor suppressors. The metabolic reprogramming enables cancer cells acquire increased nutrient and biosynthesis and escape hostile environment, which facilitates cancer development [83–85]. We summarize here that on the one hand, lncRNAs in cancer can be regulated by oncogenes (e.g., c-Myc [69]) or tumor suppressors (e.g., p53 [81]). On the other hand, lncRNAs can also affect the expression or function of oncogenes (e.g., HIF-1α [54] and c-Myc [46, 80]), tumor suppressors (e.g., AMPK [79]), metabolic enzyme genes [48, 57, 61], as well as transcription factors [70] and signaling pathway [51, 53] (Table 1). Additionally, the interaction of lncRNAs with crucial transcription factors [55, 80], metabolic enzymes [56] or miRNAs [52, 60] can effectively regulate their function or activity, modulate the processes of cancer metabolism, and influence cancer progression. Through acting as oncogenes or tumor suppressors, lncRNAs drive metabolic reprogramming that allows cancer cells to maintain deregulated proliferation, withstand metabolic challenges of poor oxygen and nutrient limitation, and sustain the surrounding microenvironment suitable for tumor growth and dissemination.
Table 1.
Long non-coding RNAs involved in regulation of cancer metabolic reprogramming
| LncRNAs | Target | Cancer type | References | |
|---|---|---|---|---|
| Central metabolism | PCGEM1 | c-Myc | Prostate cancer | [46] |
| CRNDE | Multiple metabolic genes | Colorectal cancer | [48] | |
| Glucose metabolism | HOTAIR | mTOR pathway, GLUT1 | Hepatocellular carcinoma | [51] |
| PVT1 | miR-497/HK2 axis | Osteosarcoma | [52] | |
| UCA1 | mTOR pathway | Bladder cancer | [53] | |
| lincRNA-p21 | HIF-1α | Ovarian cancer | [54] | |
| NRCP | STAT1 | Ovarian cancer | [55] | |
| LINC00092 | PFKFB2 | Ovarian cancer | [56] | |
| lnc-IGFBP4-1 | HK2, PDK1 and LDHA | Lung cancer | [57] | |
| Glutamine metabolism | UCA1 | miR-16-GLS2 axis | Bladder cancer | [60] |
| HOTTIP | GLS1 | Hepatocellular carcinoma | [61] | |
| CCAT2 | CFIm-GLS axis | Colon cancer | [62] | |
| Lipid metabolism | HULC | miR-9/PPARA/ACSL1 axis | Hepatocellular carcinoma | [68] |
| SNHG16 | Multiple miRNAs | Colorectal cancer | [69] | |
| LNMICC | FABP5 | Cervical cancer | [70] | |
| Energy stress | NBR2 | AMPK | Renal cancer | [79] |
| FILNC1 | AUF1 | Renal cancer | [80] | |
| TRINGS | STRAP | Osteosarcoma | [81] |
Recently, Natalya Pavlova and Craig Thompson have organized known cancer-associated metabolic changes into six hallmarks, including deregulated uptake of glucose and amino acids, increased demand for nitrogen, metabolic interactions with the microenvironment, and so on [71]. In this review, we have highlighted lncRNAs as an important class of regulators in cancer metabolism. Nevertheless, function and species of lncRNAs, which are involved in affecting hallmarks of cancer metabolic reprogramming, remain far from being clarified and need to be further discovered. Comprehensive understanding of which and how lncRNAs mediate regulation of metabolism reprogramming in tumor cells may facilitate the development of lncRNA inhibitors. Consequently, investigation on the functional roles and action mechanisms of lncRNAs in regulating cancer metabolism would help to develop lncRNAs as valuable targets for the diagnosis, prognosis, or treatment of human cancers.
Acknowledgements
This work was financially supported by the National Natural Science Foundation of China (81772552, 81572714, 81372215) and the Fundamental Research Funds for the Central Universities of China (531107051117, 531107051157).
References
- 1.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Li Z, Zhang H. Reprogramming of glucose, fatty acid and amino acid metabolism for cancer progression. Cell Mol Life Sci (CMLS) 2016;73:377–392. doi: 10.1007/s00018-015-2070-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Daye D, Wellen KE. Metabolic reprogramming in cancer: unraveling the role of glutamine in tumorigenesis. Semin Cell Dev Biol. 2012;23:362–369. doi: 10.1016/j.semcdb.2012.02.002. [DOI] [PubMed] [Google Scholar]
- 4.Beloribi-Djefaflia S, Vasseur S, Guillaumond F. Lipid metabolic reprogramming in cancer cells. Oncogenesis. 2016;5:e189. doi: 10.1038/oncsis.2015.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Bari L, Atlante A. Including the mitochondrial metabolism of l-lactate in cancer metabolic reprogramming. Cell Mol Life Sci (CMLS) 2018;75:2763–2776. doi: 10.1007/s00018-018-2831-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mishra P, Tang W, Putluri V, Dorsey TH, Jin F, Wang F, Zhu D, Amable L, Deng T, Zhang S, Killian JK, Wang Y, Minas TZ, Yfantis HG, Lee DH, Sreekumar A, Bustin M, Liu W, Putluri N, Ambs S. ADHFE1 is a breast cancer oncogene and induces metabolic reprogramming. J Clin Investig. 2018;128:323–340. doi: 10.1172/JCI93815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yan W, Wu X, Zhou W, Fong MY, Cao M, Liu J, Liu X, Chen CH, Fadare O, Pizzo DP, Wu J, Liu L, Liu X, Chin AR, Ren X, Chen Y, Locasale JW, Wang SE. Cancer-cell-secreted exosomal miR-105 promotes tumour growth through the MYC-dependent metabolic reprogramming of stromal cells. Nat Cell Biol. 2018;20:597–609. doi: 10.1038/s41556-018-0083-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Faubert B, Vincent EE, Griss T, Samborska B, Izreig S, Svensson RU, Mamer OA, Avizonis D, Shackelford DB, Shaw RJ, Jones RG. Loss of the tumor suppressor LKB1 promotes metabolic reprogramming of cancer cells via HIF-1alpha. Proc Natl Acad Sci USA. 2014;111:2554–2559. doi: 10.1073/pnas.1312570111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou G, Wang J, Zhao M, Xie TX, Tanaka N, Sano D, Patel AA, Ward AM, Sandulache VC, Jasser SA, Skinner HD, Fitzgerald AL, Osman AA, Wei Y, Xia X, Songyang Z, Mills GB, Hung MC, Caulin C, Liang J, Myers JN. Gain-of-function mutant p53 promotes cell growth and cancer cell metabolism via inhibition of AMPK activation. Mol Cell. 2014;54:960–974. doi: 10.1016/j.molcel.2014.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schito L, Rey S. Cell-autonomous metabolic reprogramming in hypoxia. Trends Cell Biol. 2018;28:128–142. doi: 10.1016/j.tcb.2017.10.006. [DOI] [PubMed] [Google Scholar]
- 11.Cassim S, Raymond VA, Dehbidi-Assadzadeh L, Lapierre P, Bilodeau M. Metabolic reprogramming enables hepatocarcinoma cells to efficiently adapt and survive to a nutrient-restricted microenvironment. Cell Cycle. 2018;1:1–14. doi: 10.1080/15384101.2018.1460023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu F, Ma F, Wang Y, Hao L, Zeng H, Jia C, Wang Y, Liu P, Ong IM, Li B, Chen G, Jiang J, Gong S, Li L, Xu W. PKM2 methylation by CARM1 activates aerobic glycolysis to promote tumorigenesis. Nat Cell Biol. 2017;19:1358–1370. doi: 10.1038/ncb3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bu P, Chen KY, Xiang K, Johnson C, Crown SB, Rakhilin N, Ai Y, Wang L, Xi R, Astapova I, Han Y, Li J, Barth BB, Lu M, Gao Z, Mines R, Zhang L, Herman M, Hsu D, Zhang GF, Shen X. Aldolase B-mediated fructose metabolism drives metabolic reprogramming of colon cancer liver metastasis. Cell Metab. 2018;27(1249–1262):e1244. doi: 10.1016/j.cmet.2018.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schmitt AM, Chang HY. Long noncoding RNAs in cancer pathways. Cancer Cell. 2016;29:452–463. doi: 10.1016/j.ccell.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet. 2016;17:47–62. doi: 10.1038/nrg.2015.10. [DOI] [PubMed] [Google Scholar]
- 16.Uszczynska-Ratajczak B, Lagarde J, Frankish A, Guigo R, Johnson R. Towards a complete map of the human long non-coding RNA transcriptome. Nat Rev Genet. 2018;19:535–548. doi: 10.1038/s41576-018-0017-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ransohoff JD, Wei Y, Khavari PA. The functions and unique features of long intergenic non-coding RNA. Nat Rev Mol Cell Biol. 2018;19:143–157. doi: 10.1038/nrm.2017.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen S, Wu DD, Sang XB, Wang LL, Zong ZH, Sun KX, Liu BL, Zhao Y. The lncRNA HULC functions as an oncogene by targeting ATG7 and ITGB1 in epithelial ovarian carcinoma. Cell Death Dis. 2017;8:e3118. doi: 10.1038/cddis.2017.486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Keckesova Z, Donaher JL, De Cock J, Freinkman E, Lingrell S, Bachovchin DA, Bierie B, Tischler V, Noske A, Okondo MC, Reinhardt F, Thiru P, Golub TR, Vance JE, Weinberg RA. LACTB is a tumour suppressor that modulates lipid metabolism and cell state. Nature. 2017;543:681–686. doi: 10.1038/nature21408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tomasetti M, Amati M, Santarelli L, Neuzil J. MicroRNA in metabolic re-programming and their role in tumorigenesis. Int J Mol Sci. 2016;17:754. doi: 10.3390/ijms17050754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang LF, Jiang S, Liu MF. MicroRNA regulation and analytical methods in cancer cell metabolism. Cell Mol Life Sci. 2017;74:2929–2941. doi: 10.1007/s00018-017-2508-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yue M, Jiang J, Gao P, Liu H, Qing G. Oncogenic MYC activates a feedforward regulatory loop promoting essential amino acid metabolism and tumorigenesis. Cell Rep. 2017;21:3819–3832. doi: 10.1016/j.celrep.2017.12.002. [DOI] [PubMed] [Google Scholar]
- 23.Goetzman ES, Prochownik EV. The role for Myc in coordinating glycolysis, oxidative phosphorylation, glutaminolysis, and fatty acid metabolism in normal and neoplastic tissues. Front Endocrinol. 2018;9:129. doi: 10.3389/fendo.2018.00129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sim J, Cowburn AS, Palazon A, Madhu B, Tyrakis PA, Macias D, Bargiela DM, Pietsch S, Gralla M, Evans CE, Kittipassorn T, Chey YCJ, Branco CM, Rundqvist H, Peet DJ, Johnson RS. The factor inhibiting HIF asparaginyl hydroxylase regulates oxidative metabolism and accelerates metabolic adaptation to hypoxia. Cell Metab. 2018;27(898–913):e897. doi: 10.1016/j.cmet.2018.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang HS, Du GY, Zhang ZG, Zhou Z, Sun HL, Yu XY, Shi YT, Xiong DN, Li H, Huang YH. NRF2 facilitates breast cancer cell growth via HIF1a-mediated metabolic reprogramming. Int J Biochem Cell Biol. 2018;95:85–92. doi: 10.1016/j.biocel.2017.12.016. [DOI] [PubMed] [Google Scholar]
- 26.Gnanapradeepan K, Basu S, Barnoud T, Budina-Kolomets A, Kung CP, Murphy ME. The p53 tumor suppressor in the control of metabolism and ferroptosis. Front Endocrinol. 2018;9:124. doi: 10.3389/fendo.2018.00124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Herzig S, Shaw RJ. AMPK: guardian of metabolism and mitochondrial homeostasis. Nat Rev Mol Cell Biol. 2018;19:121–135. doi: 10.1038/nrm.2017.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim JW, Zeller KI, Wang Y, Jegga AG, Aronow BJ, O’Donnell KA, Dang CV. Evaluation of myc E-box phylogenetic footprints in glycolytic genes by chromatin immunoprecipitation assays. Mol Cell Biol. 2004;24:5923–5936. doi: 10.1128/MCB.24.13.5923-5936.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mikawa T, Lleonart ME, Takaori-Kondo A, Inagaki N, Yokode M, Kondoh H. Dysregulated glycolysis as an oncogenic event. Cell Mol Life Sci (CMLS) 2015;72:1881–189231. doi: 10.1007/s00018-015-1840-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wise DR, DeBerardinis RJ, Mancuso A, Sayed N, Zhang XY, Pfeiffer HK, Nissim I, Daikhin E, Yudkoff M, McMahon SB, Thompson CB. Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc Natl Acad Sci USA. 2008;105:18782–18787. doi: 10.1073/pnas.0810199105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gao P, Tchernyshyov I, Chang TC, Lee YS, Kita K, Ochi T, Zeller KI, De Marzo AM, Van Eyk JE, Mendell JT, Dang CV. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature. 2009;458:762–765. doi: 10.1038/nature07823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eberlin LS, Gabay M, Fan AC, Gouw AM, Tibshirani RJ, Felsher DW, Zare RN. Alteration of the lipid profile in lymphomas induced by MYC overexpression. Proc Natl Acad Sci USA. 2014;111:10450–10455. doi: 10.1073/pnas.1409778111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Edmunds LR, Sharma L, Kang A, Lu J, Vockley J, Basu S, Uppala R, Goetzman ES, Beck ME, Scott D, Prochownik EV. c-Myc programs fatty acid metabolism and dictates acetyl-CoA abundance and fate. J Biol Chem. 2014;289:25382–25392. doi: 10.1074/jbc.M114.580662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mannava S, Grachtchouk V, Wheeler LJ, Im M, Zhuang D, Slavina EG, Mathews CK, Shewach DS, Nikiforov MA. Direct role of nucleotide metabolism in C-MYC-dependent proliferation of melanoma cells. Cell Cycle. 2008;7:2392–2400. doi: 10.4161/cc.6390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sun L, Song L, Wan Q, Wu G, Li X, Wang Y, Wang J, Liu Z, Zhong X, He X, Shen S, Pan X, Li A, Wang Y, Gao P, Tang H, Zhang H. cMyc-mediated activation of serine biosynthesis pathway is critical for cancer progression under nutrient deprivation conditions. Cell Res. 2015;25:429–444. doi: 10.1038/cr.2015.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Greasley PJ, Bonnard C, Amati B. Myc induces the nucleolin and BN51 genes: possible implications in ribosome biogenesis. Nucleic Acids Res. 2000;28:446–453. doi: 10.1093/nar/28.2.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li F, Wang Y, Zeller KI, Potter JJ, Wonsey DR, O’Donnell KA, Kim JW, Yustein JT, Lee LA, Dang CV. Myc stimulates nuclearly encoded mitochondrial genes and mitochondrial biogenesis. Mol Cell Biol. 2005;25:6225–6234. doi: 10.1128/MCB.25.14.6225-6234.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stine ZE, Walton ZE, Altman BJ, Hsieh AL, Dang CV. MYC, metabolism, and cancer. Cancer Discov. 2015;5:1024–1039. doi: 10.1158/2159-8290.CD-15-0507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schwartzenberg-Bar-Yoseph F, Armoni M, Karnieli E. The tumor suppressor p53 down-regulates glucose transporters GLUT1 and GLUT4 gene expression. Can Res. 2004;64:2627–2633. doi: 10.1158/0008-5472.CAN-03-0846. [DOI] [PubMed] [Google Scholar]
- 40.Bensaad K, Tsuruta A, Selak MA, Vidal MN, Nakano K, Bartrons R, Gottlieb E, Vousden KH. TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell. 2006;126:107–120. doi: 10.1016/j.cell.2006.05.036. [DOI] [PubMed] [Google Scholar]
- 41.Berkers CR, Maddocks OD, Cheung EC, Mor I, Vousden KH. Metabolic regulation by p53 family members. Cell Metab. 2013;18:617–633. doi: 10.1016/j.cmet.2013.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Assaily W, Rubinger DA, Wheaton K, Lin Y, Ma W, Xuan W, Brown-Endres L, Tsuchihara K, Mak TW, Benchimol S. ROS-mediated p53 induction of Lpin1 regulates fatty acid oxidation in response to nutritional stress. Mol Cell. 2011;44:491–501. doi: 10.1016/j.molcel.2011.08.038. [DOI] [PubMed] [Google Scholar]
- 43.Suzuki S, Tanaka T, Poyurovsky MV, Nagano H, Mayama T, Ohkubo S, Lokshin M, Hosokawa H, Nakayama T, Suzuki Y, Sugano S, Sato E, Nagao T, Yokote K, Tatsuno I, Prives C. Phosphate-activated glutaminase (GLS2), a p53-inducible regulator of glutamine metabolism and reactive oxygen species. Proc Natl Acad Sci USA. 2010;107:7461–7466. doi: 10.1073/pnas.1002459107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Matoba S, Kang JG, Patino WD, Wragg A, Boehm M, Gavrilova O, Hurley PJ, Bunz F, Hwang PM. p53 regulates mitochondrial respiration. Science. 2006;312:1650–1653. doi: 10.1126/science.1126863. [DOI] [PubMed] [Google Scholar]
- 45.Lin SC, Hardie DG. AMPK: sensing glucose as well as cellular energy status. Cell Metab. 2018;27:299–313. doi: 10.1016/j.cmet.2017.10.009. [DOI] [PubMed] [Google Scholar]
- 46.Hung CL, Wang LY, Yu YL, Chen HW, Srivastava S, Petrovics G, Kung HJ. A long noncoding RNA connects c-Myc to tumor metabolism. Proc Natl Acad Sci USA. 2014;111:18697–18702. doi: 10.1073/pnas.1415669112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Petrovics G, Zhang W, Makarem M, Street JP, Connelly R, Sun L, Sesterhenn IA, Srikantan V, Moul JW, Srivastava S. Elevated expression of PCGEM1, a prostate-specific gene with cell growth-promoting function, is associated with high-risk prostate cancer patients. Oncogene. 2004;23:605–611. doi: 10.1038/sj.onc.1207069. [DOI] [PubMed] [Google Scholar]
- 48.Ellis BC, Graham LD, Molloy PL. CRNDE, a long non-coding RNA responsive to insulin/IGF signaling, regulates genes involved in central metabolism. Biochem Biophys Acta. 2014;1843:372–386. doi: 10.1016/j.bbamcr.2013.10.016. [DOI] [PubMed] [Google Scholar]
- 49.Guil S, Soler M, Portela A, Carrere J, Fonalleras E, Gomez A, Villanueva A, Esteller M. Intronic RNAs mediate EZH2 regulation of epigenetic targets. Nat Struct Mol Biol. 2012;19:664–670. doi: 10.1038/nsmb.2315. [DOI] [PubMed] [Google Scholar]
- 50.Hay N. Reprogramming glucose metabolism in cancer: can it be exploited for cancer therapy? Nat Rev Cancer. 2016;16:635–649. doi: 10.1038/nrc.2016.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wei S, Fan Q, Yang L, Zhang X, Ma Y, Zong Z, Hua X, Su D, Sun H, Li H, Liu Z. Promotion of glycolysis by HOTAIR through GLUT1 upregulation via mTOR signaling. Oncol Rep. 2017;38:1902–1908. doi: 10.3892/or.2017.5840. [DOI] [PubMed] [Google Scholar]
- 52.Song J, Wu X, Liu F, Li M, Sun Y, Wang Y, Wang C, Zhu K, Jia X, Wang B, Ma X. Long non-coding RNA PVT1 promotes glycolysis and tumor progression by regulating miR-497/HK2 axis in osteosarcoma. Biochem Biophys Res Commun. 2017;490:217–224. doi: 10.1016/j.bbrc.2017.06.024. [DOI] [PubMed] [Google Scholar]
- 53.Li Z, Li X, Wu S, Xue M, Chen W. Long non-coding RNA UCA1 promotes glycolysis by upregulating hexokinase 2 through the mTOR-STAT3/microRNA143 pathway. Cancer Sci. 2014;105:951–955. doi: 10.1111/cas.12461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang F, Zhang H, Mei Y, Wu M. Reciprocal regulation of HIF-1alpha and lincRNA-p21 modulates the Warburg effect. Mol Cell. 2014;53:88–100. doi: 10.1016/j.molcel.2013.11.004. [DOI] [PubMed] [Google Scholar]
- 55.Rupaimoole R, Lee J, Haemmerle M, Ling H, Previs RA, Pradeep S, Wu SY, Ivan C, Ferracin M, Dennison JB, Millward NMZ, Nagaraja AS, Gharpure KM, McGuire M, Sam N, Armaiz-Pena GN, Sadaoui NC, Rodriguez-Aguayo C, Calin GA, Drapkin RI, Kovacs J, Mills GB, Zhang W, Lopez-Berestein G, Bhattacharya PK, Sood AK. Long noncoding RNA ceruloplasmin promotes cancer growth by altering glycolysis. Cell Rep. 2015;13:2395–2402. doi: 10.1016/j.celrep.2015.11.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhao L, Ji G, Le X, Wang C, Xu L, Feng M, Zhang Y, Yang H, Xuan Y, Yang Y, Lei L, Yang Q, Lau WB, Lau B, Chen Y, Deng X, Yao S, Yi T, Zhao X, Wei Y, Zhou S. Long noncoding RNA LINC00092 acts in cancer-associated fibroblasts to drive glycolysis and progression of ovarian cancer. Can Res. 2017;77:1369–1382. doi: 10.1158/0008-5472.CAN-16-1615. [DOI] [PubMed] [Google Scholar]
- 57.Yang B, Zhang L, Cao Y, Chen S, Cao J, Wu D, Chen J, Xiong H, Pan Z, Qiu F, Chen J, Ling X, Yan M, Huang S, Zhou S, Li T, Yang L, Huang Y, Lu J. Overexpression of lncRNA IGFBP4-1 reprograms energy metabolism to promote lung cancer progression. Mol Cancer. 2017;16:154. doi: 10.1186/s12943-017-0722-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Altman BJ, Stine ZE, Dang CV. From Krebs to clinic: glutamine metabolism to cancer therapy. Nat Rev Cancer. 2016;16:619–634. doi: 10.1038/nrc.2016.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yang L, Venneti S, Nagrath D. Glutaminolysis: a hallmark of cancer metabolism. Annu Rev Biomed Eng. 2017;19:163–194. doi: 10.1146/annurev-bioeng-071516-044546. [DOI] [PubMed] [Google Scholar]
- 60.Li HJ, Li X, Pang H, Pan JJ, Xie XJ, Chen W. Long non-coding RNA UCA1 promotes glutamine metabolism by targeting miR-16 in human bladder cancer. Jpn J Clin Oncol. 2015;45:1055–1063. doi: 10.1093/jjco/hyv132. [DOI] [PubMed] [Google Scholar]
- 61.Ge Y, Yan X, Jin Y, Yang X, Yu X, Zhou L, Han S, Yuan Q, Yang M. MiRNA-192 (corrected) and miRNA-204 directly suppress lncRNA HOTTIP and interrupt GLS1-mediated glutaminolysis in hepatocellular carcinoma. PLoS Genet. 2015;11:e1005726. doi: 10.1371/journal.pgen.1005726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Redis RS, Vela LE, Lu W, Ferreira de Oliveira J, Ivan C, Rodriguez-Aguayo C, Adamoski D, Pasculli B, Taguchi A, Chen Y, Fernandez AF, Valledor L, Van Roosbroeck K, Chang S, Shah M, Kinnebrew G, Han L, Atlasi Y, Cheung LH, Huang GY, Monroig P, Ramirez MS, Catela Ivkovic T, Van L, Ling H, Gafa R, Kapitanovic S, Lanza G, Bankson JA, Huang P, Lai SY, Bast RC, Rosenblum MG, Radovich M, Ivan M, Bartholomeusz G, Liang H, Fraga MF, Widger WR, Hanash S, Berindan-Neagoe I, Lopez-Berestein G, Ambrosio ALB, Gomes Dias SM, Calin GA. Allele-specific reprogramming of cancer metabolism by the long non-coding RNA CCAT2. Mol Cell. 2016;61:520–534. doi: 10.1016/j.molcel.2016.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tomlinson I, Webb E, Carvajal-Carmona L, Broderick P, Kemp Z, Spain S, Penegar S, Chandler I, Gorman M, Wood W, Barclay E, Lubbe S, Martin L, Sellick G, Jaeger E, Hubner R, Wild R, Rowan A, Fielding S, Howarth K, Consortium C, Silver A, Atkin W, Muir K, Logan R, Kerr D, Johnstone E, Sieber O, Gray R, Thomas H, Peto J, Cazier JB, Houlston R. A genome-wide association scan of tag SNPs identifies a susceptibility variant for colorectal cancer at 8q24.21. Nature Genet. 2007;39:984–988. doi: 10.1038/ng2085. [DOI] [PubMed] [Google Scholar]
- 64.Beloribi-Djefaflia S, Vasseur S, Guillaumond F. Lipid metabolic reprogramming in cancer cells. Oncogenesis. 2016;5:e189. doi: 10.1038/oncsis.2015.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Long J, Zhang CJ, Zhu N, Du K, Yin YF, Tan X, Liao DF, Qin L. Lipid metabolism and carcinogenesis, cancer development. Am J Cancer Res. 2018;8:778–791. [PMC free article] [PubMed] [Google Scholar]
- 66.Yan S, Yang XF, Liu HL, Fu N, Ouyang Y, Qing K. Long-chain acyl-CoA synthetase in fatty acid metabolism involved in liver and other diseases: an update. World J Gastroenterol. 2015;21:3492–3498. doi: 10.3748/wjg.v21.i12.3492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Panzitt K, Tschernatsch MM, Guelly C, Moustafa T, Stradner M, Strohmaier HM, Buck CR, Denk H, Schroeder R, Trauner M, Zatloukal K. Characterization of HULC, a novel gene with striking up-regulation in hepatocellular carcinoma, as noncoding RNA. Gastroenterology. 2007;132:330–342. doi: 10.1053/j.gastro.2006.08.026. [DOI] [PubMed] [Google Scholar]
- 68.Cui M, Xiao Z, Wang Y, Zheng M, Song T, Cai X, Sun B, Ye L, Zhang X. Long noncoding RNA HULC modulates abnormal lipid metabolism in hepatoma cells through an miR-9-mediated RXRA signaling pathway. Can Res. 2015;75:846–857. doi: 10.1158/0008-5472.CAN-14-1192. [DOI] [PubMed] [Google Scholar]
- 69.Christensen LL, True K, Hamilton MP, Nielsen MM, Damas ND, Damgaard CK, Ongen H, Dermitzakis E, Bramsen JB, Pedersen JS, Lund AH, Vang S, Stribolt K, Madsen MR, Laurberg S, McGuire SE, Orntoft TF, Andersen CL. SNHG16 is regulated by the Wnt pathway in colorectal cancer and affects genes involved in lipid metabolism. Mol Oncol. 2016;10:1266–1282. doi: 10.1016/j.molonc.2016.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shang C, Wang W, Liao Y, Chen Y, Liu T, Du Q, Huang J, Liang Y, Liu J, Zhao Y, Guo L, Hu Z, Yao S. LNMICC promotes nodal metastasis of cervical cancer by reprogramming fatty acid metabolism. Can Res. 2018;78:877–890. doi: 10.1158/0008-5472.CAN-17-2356. [DOI] [PubMed] [Google Scholar]
- 71.Pavlova NN, Thompson CB. The emerging hallmarks of cancer metabolism. Cell Metab. 2016;23:27–47. doi: 10.1016/j.cmet.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jeon SM, Chandel NS, Hay N. AMPK regulates NADPH homeostasis to promote tumour cell survival during energy stress. Nature. 2012;485:661–665. doi: 10.1038/nature11066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Qian X, Li X, Tan L, Lee JH, Xia Y, Cai Q, Zheng Y, Wang H, Lorenzi PL, Lu Z. Conversion of PRPS hexamer to monomer by AMPK-mediated phosphorylation inhibits nucleotide synthesis in response to energy stress. Cancer Discov. 2018;8:94–107. doi: 10.1158/2159-8290.CD-17-0712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bezawork-Geleta A, Wen H, Dong L, Yan B, Vider J, Boukalova S, Krobova L, Vanova K, Zobalova R, Sobol M, Hozak P, Novais SM, Caisova V, Abaffy P, Naraine R, Pang Y, Zaw T, Zhang P, Sindelka R, Kubista M, Zuryn S, Molloy MP, Berridge MV, Pacak K, Rohlena J, Park S, Neuzil J. Alternative assembly of respiratory complex II connects energy stress to metabolic checkpoints. Nat Commun. 2018;9:2221. doi: 10.1038/s41467-018-04603-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shaw RJ, Kosmatka M, Bardeesy N, Hurley RL, Witters LA, DePinho RA, Cantley LC. The tumor suppressor LKB1 kinase directly activates AMP-activated kinase and regulates apoptosis in response to energy stress. Proc Natl Acad Sci USA. 2004;101:3329–3335. doi: 10.1073/pnas.0308061100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kim LC, Cook RS, Chen J. mTORC1 and mTORC2 in cancer and the tumor microenvironment. Oncogene. 2017;36:2191–2201. doi: 10.1038/onc.2016.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Egan DF, Shackelford DB, Mihaylova MM, Gelino S, Kohnz RA, Mair W, Vasquez DS, Joshi A, Gwinn DM, Taylor R, Asara JM, Fitzpatrick J, Dillin A, Viollet B, Kundu M, Hansen M, Shaw RJ. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science. 2011;331:456–461. doi: 10.1126/science.1196371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13:132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Liu X, Xiao ZD, Han L, Zhang J, Lee SW, Wang W, Lee H, Zhuang L, Chen J, Lin HK, Wang J, Liang H, Gan B. LncRNA NBR2 engages a metabolic checkpoint by regulating AMPK under energy stress. Nat Cell Biol. 2016;18:431–442. doi: 10.1038/ncb3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Xiao ZD, Han L, Lee H, Zhuang L, Zhang Y, Baddour J, Nagrath D, Wood CG, Gu J, Wu X, Liang H, Gan B. Energy stress-induced lncRNA FILNC1 represses c-Myc-mediated energy metabolism and inhibits renal tumor development. Nat Commun. 2017;8:783. doi: 10.1038/s41467-017-00902-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Khan MR, Xiang S, Song Z, Wu M. The p53-inducible long noncoding RNA TRINGS protects cancer cells from necrosis under glucose starvation. EMBO J. 2017;36:3483–3500. doi: 10.15252/embj.201696239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sanchez JF, Sniderhan LF, Williamson AL, Fan S, Chakraborty-Sett S, Maggirwar SB. Glycogen synthase kinase 3beta-mediated apoptosis of primary cortical astrocytes involves inhibition of nuclear factor kappaB signaling. Mol Cell Biol. 2003;23:4649–4662. doi: 10.1128/MCB.23.13.4649-4662.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Liu F, Ma F, Wang Y, Hao L, Zeng H, Jia C, Wang Y, Liu P, Ong IM, Li B, Chen G, Jiang J, Gong S, Li L, Xu W. PKM2 methylation by CARM1 activates aerobic glycolysis to promote tumorigenesis. Nat Cell Biol. 2017;19:1358–1370. doi: 10.1038/ncb3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ji X, Qian J, Rahman SMJ, Siska PJ, Zou Y, Harris BK, Hoeksema MD, Trenary IA, Heidi C, Eisenberg R, Rathmell JC, Young JD, Massion PP. xCT (SLC7A11)-mediated metabolic reprogramming promotes non-small cell lung cancer progression. Oncogene. 2018;37:5007–5019. doi: 10.1038/s41388-018-0307-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Fujiwara N, Nakagawa H, Enooku K, Kudo Y, Hayata Y, Nakatsuka T, Tanaka Y, Tateishi R, Hikiba Y, Misumi K, Tanaka M, Hayashi A, Shibahara J, Fukayama M, Arita J, Hasegawa K, Hirschfield H, Hoshida Y, Hirata Y, Otsuka M, Tateishi K, Koike K. CPT2 downregulation adapts HCC to lipid-rich environment and promotes carcinogenesis via acylcarnitine accumulation in obesity. Gut. 2018;67:1493–1504. doi: 10.1136/gutjnl-2017-315193. [DOI] [PMC free article] [PubMed] [Google Scholar]